- Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
Low temperature (non-freezing) is one of the major limiting factors in peanut (Arachis hypogaea L.) growth, yield, and geographic distribution. Due to the complexity of cold-resistance trait in peanut, the molecular mechanism of cold tolerance and related gene networks were largely unknown. In this study, metabolomic analysis of two peanut cultivars subjected to chilling stress obtained a set of cold-responsive metabolites, including several carbohydrates and polyamines. These substances showed a higher accumulation pattern in cold-tolerant variety SLH than cold-susceptible variety ZH12 under cold stress, indicating their importance in protecting peanut from chilling injuries. In addition, 3,620 cold tolerance genes (CTGs) were identified by transcriptome sequencing, and the CTGs were most significantly enriched in the “phenylpropanoid biosynthesis” pathway. Two vital modules and several novel hub genes were obtained by weighted gene co-expression network analysis (WGCNA). Several key genes involved in soluble sugar, polyamine, and G-lignin biosynthetic pathways were substantially higher and/or responded more quickly in SLH (cold tolerant) than ZH12 (cold susceptible) under low temperature, suggesting they might be crucial contributors during the adaptation of peanut to low temperature. These findings will not only provide valuable resources for study of cold resistance in peanut but also lay a foundation for genetic modification of cold regulators to enhance stress tolerance in crops.
Introduction
Peanut (Arachis hypogaea L.) is an important oil and economic crop worldwide, and provides a rich source of nutrients for humans, including fat, protein, sugar, fatty acids, and free amino acids (Zhao et al., 2012; Sorita et al., 2020). Low temperature is one of the major environmental stresses that adversely affect plant development and geographic distribution (Kakani et al., 2002; Wang et al., 2003). The growth of peanut is severely inhibited below 15°C, and chilling stress (non-freezing) is deemed as a limiting factor in peanut cultivation and production (Bell et al., 1994). In northeast China, cold weather often occurs during spring sowing and autumn harvest seasons, leading to a serious decrease of peanut yields per year (Chen et al., 2014). With the increasing demand for peanut in recent years, it is urgent to investigate the mechanism of cold tolerance and cultivate new cold-resistant varieties.
Cold stress causes excessive accumulation of reactive oxygen species (ROS) during respiration and photosynthesis processes in plant, thus increasing cell membrane permeability and the lipid peroxidation level (Zhang et al., 2019). Nevertheless, plants evolve a series of sophisticated mechanisms to defense against cold stress to some extent (Ding et al., 2020). The alterations of gene expression in response to low temperature are a common strategy to cope with cold damage in plants (Suzuki, 2019). During the last two decades, a number of components involved in the cold-responsive-signaling pathway have been isolated and characterized, including messenger molecules (e.g., Ca2+), Ca2+-related protein kinases and several crucial transcription factors (Yuan et al., 2018). One of the best studied examples was the plant ICE-CBF-COR-signaling module (Chinnusamy et al., 2003). C-repeat Binding Factors/Dehydration-Responsive Element-Binding proteins (CBFs/DREBs) were members of APETALA 2/Ethylene Response Factor (AP2/ERF) family and played central roles in cold acclimation (Kim et al., 2015). The transcript levels of CBFs were sharply upregulated by Inducer of CBF Expression protein (ICE), an MYC-type basic helix-loop-helix family transcription factor, and then CBFs activated the expression of downstream cold-responsive (COR) genes via binding to cis-elements in their promoters (Lee et al., 2005; Tang et al., 2020). In addition, a lot of cold-resistant proteins (e.g., late embryogenesis-abundant proteins, IEAs) and protective substances (e.g., soluble sugars and proline) were synthesized in plant cells, functioning as osmolytes to regulate osmotic potential and maintain membrane integrity (Dong et al., 2002; Zuther et al., 2004; Ma et al., 2009).
Transcriptomic analysis has been a highly efficient way to reveal cold-responsive genes in many crops, such as maize (Li et al., 2019), rice (Pan et al., 2020), Brassica napus (Mehmood et al., 2021) and Brassica juncea (Sinha et al., 2015). Microarray analysis of peanut leaves subjected to low temperature got a number of cold-regulated genes and provided some useful insights into cold signal transduction pathways in peanut (Chen et al., 2014). With the rapid development of high-throughput sequencing and mass spectrometer technologies, a massive of multi-omics data (e.g., transcriptomics and metabolomics) have been generated, and it is possible to explore the mechanism of plant cold tolerance by analyzing and mining the big data (Zhao et al., 2019; Cui et al., 2020; Du et al., 2020; Yang et al., 2020; Jiang et al., 2021). Comparative transcriptomic analysis of two peanut varieties with contrasting cold resistance (NH5 and FH18) identified several cold-responsive transcription factors, including bHLH, MYB, and NAC (Jiang et al., 2020). Furthermore, lipidomic analysis of the two varieties revealed that membrane lipid and fatty acid metabolisms contributed substantially to cold resistance in peanut (Zhang H. et al., 2020).
Although there are a few reports about cold tolerance in peanut, the molecular mechanism underlining the signal pathway and related gene networks need to be further investigated due to the complexity of the cold-resistant trait in this important crop (Wang et al., 2017, 2020; Zhang et al., 2019). In the current study, an integrated analysis of transcriptome and metabolome from two peanut cultivars (SLH and ZH12) allowed us to obtain a set of cold-responsive metabolites/genes that possibly protect peanut from chilling damage. These results contributed to a better understanding of the molecular mechanism of cold response in peanut and will be useful for the development of cultivars with enhanced cold-stress tolerance.
Materials and Methods
Plant Materials and Treatments
The cold-resistant peanut cultivar “Silihong” (SLH) and cold-sensitive “Zhonghua 12” (ZH12) were used in this study. Peanut seeds were soaked in distilled water for 4 h, and placed in petri dishes with moistened filter papers, and then germinated in the dark at 28°C for 2 days. The germinated seeds were sown in soil pots and transferred into a growth chamber under a 16-h/8-h (light-dark) cycle at 28°C, with a photosynthetic photon flux density of 700 μmol m–2 s–1. Two-week-old seedlings were used for cold treatment with a 16-h/8-h cycle (light/dark) at 10°C. The third leaves from seedlings were collected at 0, 3, 24, and 48 h, quickly frozen in liquid nitrogen, and stored at −80°C until use. All samples were performed in three independent biological replicates.
Physiological Index Measurements
The two varieties of peanut seeds were immersed in distilled water for 4 h, transferred to petri dishes at 2°C for 72 h, and then returned to normal temperature (28°C) for additional 3 days. The controls were continuously germinated at 28°C for 3 days. The germination rates were measured every day.
Malondialdehyde (MDA) and H2O2 content were determined by Lipid Peroxidation MDA Assay Kit and Hydrogen Peroxide Assay Kit (Beyotime Biotechnology Co., Ltd., Shanghai, China), respectively. The superoxide anion level was analyzed by Micro Superoxide Anion Assay Kit (Beijing Solarbio Science and Technology Co., Ltd., Beijing, China). All the experiments were performed according to the manufacturers’ protocols, and data were derived from three biological replicates. Student’s T-test was performed to calculate the p-values using SPSS 20.0 version.
Metabolite Extraction and Profiling
The frozen leaves were grounded into fine powder in liquid nitrogen. About 100 mg of samples was placed into 2-ml Eppendorf tubes and resuspended with 500-μl extraction liquid (80% methanol and 0.1% formic acid). After vortex for 30 s, the mixtures were incubated on ice for 5 min and then centrifuged at 15,000 rpm, 4°C for 10 min. Some of the supernatant was transferred to a fresh EP tube and diluted to final concentration containing 53% methanol by adding with LC-MS grade water. The samples were centrifuged at 15,000 g, 4°C for 20 min. Finally, the supernatant was injected into the LC-MS/MS system for analysis.
For the positive polarity mode, LC-MS/MS analyses were performed using an ExionLCTM AD system (SCIEX) on a BEH C8 Column (100 mm × 2.1 mm, 1.9 μm), coupled with a QTRAP® 6500 + mass spectrometer (SCIEX). The mobile phase consisted of 0.1% formic acid water (A) and acetonitrile (B) with elution gradient at a flow rate of 0.35 ml/min as follows: 5% B, 1 min; 5–100% B, 24 min; 100% B, 28 min; 100–5% B, 28.1 min, 5% B, 30 min. For the negative polarity mode, samples were injected onto an HSS T3 Column (100 mm × 2.1 mm). The flow rate was set as 0.35 ml/min with solvent gradient as follows: 2% B, 1 min; 2–100% B, 18 min; 100% B, 22 min; 100–5% B, 22.1 min; 5% B, 25 min. The main parameters of QTRAP® 6500 + mass spectrometer were set as follows: Curtain Gas of 35 psi, Collision Gas of Medium, Temperature of 500°C, Ion Spray Voltage of 5,500 V or −4,500 V in a positive or negative mode, respectively.
Metabolite identification was based on the fragmentation patterns of Q1 and Q3, retention time, decluttering potential, and collision energy. Q3 was used to quantify the metabolite according to the in-house database (Novogene Bioinformatics Technology Co., Ltd.).
Transcriptome Sequencing
Total RNA was isolated from the peanut leaves. The concentration and integrity of RNA were checked by the Bioanalyzer 2100 system (Agilent Technologies, CA, United States). About 1 μg of RNA per sample was used as input material for the transcriptome library preparation. The libraries were sequenced on an Illumina NovaSeq platform (Tianjin, China), and 150-bp paired-end reads were generated. The high-quality reads were obtained by removing reads containing adapter, reads containing poly-N, and low-quality reads from raw data. The high-quality paired-end reads were then mapped to the peanut reference genome (Tifrunner.gnm1.ann1.CCJH1) using HISAT2 software (version 2.0.5) with default parameters (Kim et al., 2019). The feature counts program (with a parameter of v1.5.0-p3) (Liao et al., 2014) was used to count the reads numbers mapped to each gene. If a read was mapped to two or more positions in the genome, then it will be filtered out. Only uniquely mapped reads were used for quantification. The gene expression level was calculated by the FPKM values (Fragments per Kilobase of transcript per Million mapped reads) using the Cufflinks program (Trapnell et al., 2010). The Pearson correlation coefficients between different samples were calculated by the cor function in R package and presented as heatmap using pheatmap software2. To identify cold-responsive genes in peanut, the differentially expressed genes (DEGs) were screened using the DESeq2 R package (1.20.0) with the criteria of | log2FC (fold change) | > 1 and an adjusted P-value < 0.01 (Love et al., 2014). Gene ontology (GO) and KEGG enrichment analysis of DEGs were performed by the cluster Profiler R package (Yu et al., 2012).
Analysis of Gene Expression by Real-Time Quantitative RT-PCR
Total RNA was extracted from the peanut leaves of three replicates using an EASYspin plant RNA extraction kit (Aidlab Biotechlogies, Co., Ltd., China) according to the manufacturer’s instructions. First-strand cDNA was synthesized from DNase I-treated total RNA (about 1 μg) using an MMLV reverse transcriptase kit (Thermofisher Scientific, United States). Quantitative RT-PCR (qRT-PCR) assays were performed on the Bio-Rad CFX96 RT-PCR Detection system (Bio-Rad, Hercules, CA, United States) using Hieff qPCR SYBR Green Master Mix (YEASEN, Shanghai, China). The relative transcript levels among different samples were quantified by the 2–ΔΔCt method (Livak and Schmittgen, 2001), using A. hypogaea Actin gene (accession number: Aradu.W2Y55) as a reference gene for normalization. The expression level of 0 h in SLH was used as a control, whose value was set to 1. The relative expression levels of other samples (including a 0-h time point in ZH12) were determined relative to the control. In this way, we can compare all samples across the two varieties within the same gene. The information of genes for qRT-PCR analysis is listed in Supplementary Table 1.
Weighted Gene Co-expression Network Analysis
Weighted gene co-expression network analysis (WGCNA) can be used for identifying genes with similar expression patterns that may participate in specific biological functions (Langfelder and Horvath, 2008). A total of 3,620 cold tolerance core genes were used as an input in the R package WGCNA (version 1.47) to construct weighted co-expression modules with following parameters: weighted network = unsigned, power = 10, minimum module size = 30, minimum height for merging modules = 0.25. Then, the calculated module eigen genes and Pearson’s correlation coefficient values were used to determine the association of modules with the metabolite contents (soluble sugars, polyamines, amino acids, and phenols) for the 24 samples. The number of edges in a node represented the hubness of the gene. Hub genes were ranked based on the module eigengene values and edge numbers, and they often exhibit the most connections with other genes within a module. The gene networks and top 20 hub genes within a module were visualized by Cytoscape software (Shannon et al., 2003).
Results
Physiological Differences Between SLH and ZH12 in Response to Cold Stress
Two peanut cultivars with differential cold tolerance (SLH and ZH12) were selected for investigating their physiological traits in response to chilling stress (Zhang et al., 2021). SLH has been characterized as a cold-tolerant germplasm, and it is popularly planted in northeast China (a relative high latitude region, 48°N–55°N) where cold weather often happens in the early sowing and harvest seasons. ZH12 (cold susceptible) is a representative variety released by Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, and it is commonly planted in Hubei province, central China (a low latitude region, 29°N–33°N), which has a warm climate in the sowing and harvest season. It was found that the two cultivars showed similar germination rates under normal condition but were severely affected for ZH12 in comparison to SLH when they were exposed to low temperature (Figures 1A,B). Additionally, the levels of hydrogen peroxide (H2O2) and superoxide anion (O2–) were significantly higher in the seedlings of ZH12 than those of SLH within 24 h of cold treatment (10°C) (Figures 1C,D). It is known that ROS overproduction brings about lipid peroxidation, and the malondialdehyde (MDA) level is an important indicator for membrane damage (Jambunathan, 2010). In the current study, the leaf MDA contents in cold-treated ZH12 increased about 2-fold compared with control (0 h) and displayed higher levels than those of SLH after 48-h cold treatment (Figure 1E). The pronounced enhancement of MDA content in susceptible peanut cultivar ZH12 was presumably caused by excessive accumulation of O2– and H2O2 under chilling stress. Thus, the difference of physiological responses to low temperature confirmed that SLH was relatively stable, while ZH12 was more susceptible to cold damage.
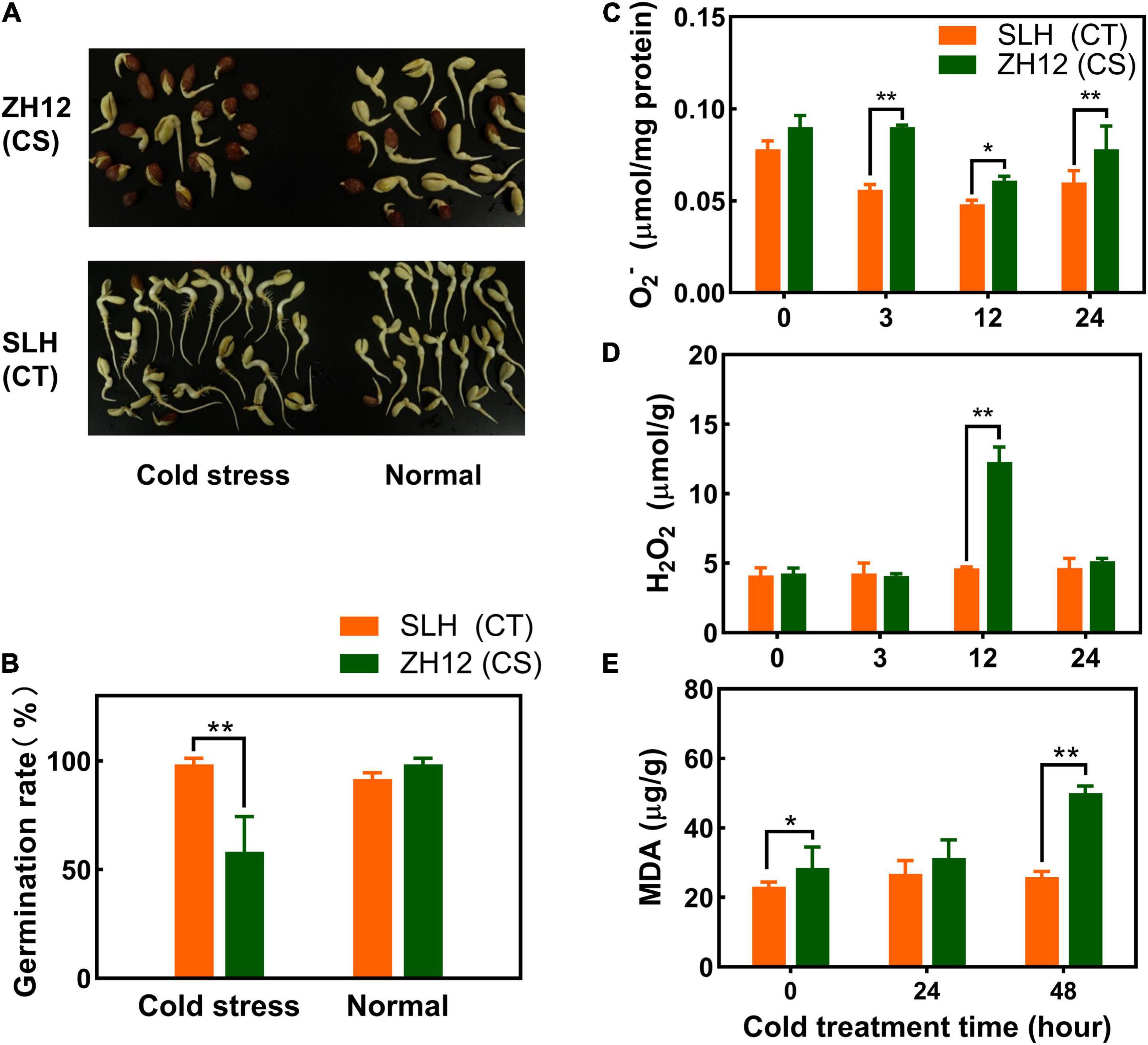
Figure 1. Differential physiological responses of SLH and ZH12 peanut cultivars to cold stress. (A,B) Effects of low temperature on germination rates of the two peanut cultivars. For cold treatment, seeds were immersed in distilled water for 4 h, and then transferred to petri dishes at 2°C for 72 h in the dark, and, finally, returned to normal temperature (28°C) for additional 3 days for germination. The controls were continuously germinated at 28°C for 3 days. SLH (CT), the cold-tolerance cultivar; ZH12 (CS), the cold-susceptible cultivar. The level of O2– (C), H2O2 (D), and malondialdehyde [MDA, (E)] of peanut seedlings in response to chilling stress. Error bars represent the SD of the means of three biological replications. *An asterisk indicates significant difference between the two cultivars as determined by the Student’s T-test (*p < 0.05; **p < 0.01).
Metabolome Profiling of Peanut in Response to Cold Stress
To investigate metabolome profiling of the two peanut genotypes exposed to low temperature, leaves were collected at the four different treatment time points (with three biological replicates per point), and further subjected to UPLC-MS for analyzing their metabolic changes. As a result, a total of 563 metabolites were successfully detected, and these could be mainly classified into different categories, including amino acids, carbohydrates, nucleosides, lipids, alkaloids, flavonoids, terpenoids, etc., (Supplementary Table 2). Principle component analysis (PCA) of metabolites profiling showed that the two cultivars were separated by PC1 (21.70%), and samples collected at different time points were separated by PC2 (15.97%) (Figure 2A). Moreover, the control and cold-treated samples were separated, indicating that low temperature had profound impacts on the compound accumulation patterns in peanut.
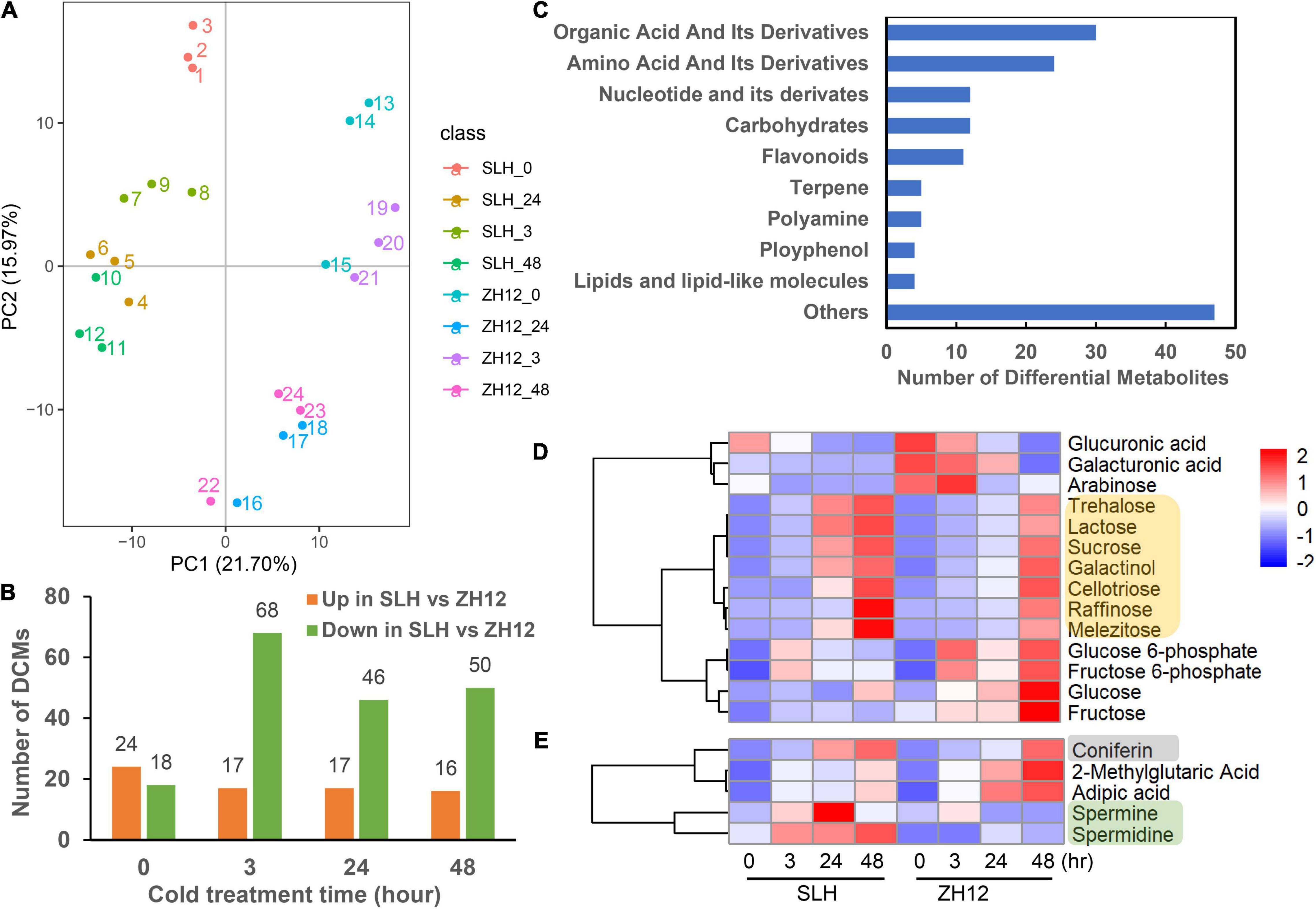
Figure 2. Metabolite profiling of two peanut varieties exposed to cold stress. (A) Principle component analysis (PCA) of metabolome data derived from SLH and ZH12 during four time points under cold stress (0, 3, 24, 48 h, with three biological replicates per point). (B) The differentially changed metabolites (DCMs) between the SLH and ZH12 under cold stress. The DCMs were screened by the criteria of | log2 (fold change) | > 1 and P-value < 0.05. (C) Classification of 145 DCMs between the two cultivars. (D,E) The accumulation pattern of carbohydrates (indicated by yellow), polyamines (marked by green), and phenols (gray) under cold stress in SLH and ZH12. The relative contents of metabolites (represented by the LC-MS/MS peak area) in different samples were used for z-scale normalization in the heatmap. The red color indicated relative high content of substances, while blue was low.
To identify the metabolites that contribute to cold tolerance in peanut, we performed comparative analysis of the metabolomic changes between SLH and ZH12. Using thresholds for | log2 (fold change) | > 1 and P-value < 0.05, a total of 42, 85, 63, and 56 differentially changed metabolites (DCMs) were found between the two cultivars in the four time points of cold treatment, respectively (Figure 2B and Supplementary Table 3). Altogether, we identified 145 DCMs between the two cultivars, which were mainly involved in organic acid, amino acids, nucleotides and carbohydrates (Figure 2C and Supplementary Table 4). Among the changed carbohydrates, the contents of seven sugars (e.g., raffinose, trehalose, lactose, galactinol, cellotriose, melezitose, and sucrose) were found to be significantly increased under cold stress. Moreover, it appeared that SLH exhibited at an earlier induction time point (at 24 h) and displayed more enhanced levels of the sugars compared to ZH12 in response to low temperature (Figure 2D and Supplementary Table 5). Similarly, other metabolites, such as polyamines (spermine, spermidine) and phenols (coniferin), also exhibited higher accumulation patterns in SLH than ZH12 (Figure 2E). As some soluble sugars, polyamines and phenols were reported to be involved in plant cold response (Fuertauer et al., 2019; Alcazar et al., 2020), the differential accumulation patterns of these substances probably provided stronger cold tolerance capability in SLH.
Transcriptomic Analysis of Peanut Exposed to Cold Stress
General Description of Transcriptome Data
Total mRNA extracted from peanut leaves was subjected to construct sequencing libraries using the Illumina paired-end platform. With three biological replicates per time point, the 24 samples yielded more than 185.44 Gb high-quality data with an average Q30 score of 93.09% (Supplementary Table 6). A total of 48,374 and 46,844 unigenes were obtained from SLH and ZH12 with FPKM values > 0.1 in at least one time point, respectively (Supplementary Figures 2A,B). There were 38,741 and 37,705 transcripts expressed at all-time points in SLH and ZH12, respectively. Over 90% (45,302) of the detected transcripts were shared by the two genotypes (Supplementary Figure 2C). Pearson correlation analysis of RNA-seq data displayed high reproducibility between biological replicates (Supplementary Figure 3). PCA analysis indicated that the samples from the different time points could be separated by the PC1 (41.93%), and the genotypes were separated by the PC2 (14.72%) (Figure 3A). Additionally, the number of cold-responsive genes in 24 or 48 h is greater than that in 3 h (Supplementary Figure 4). This suggested that prolonged duration of cold stress resulted in more profound changes of a transcriptome profile in peanut.
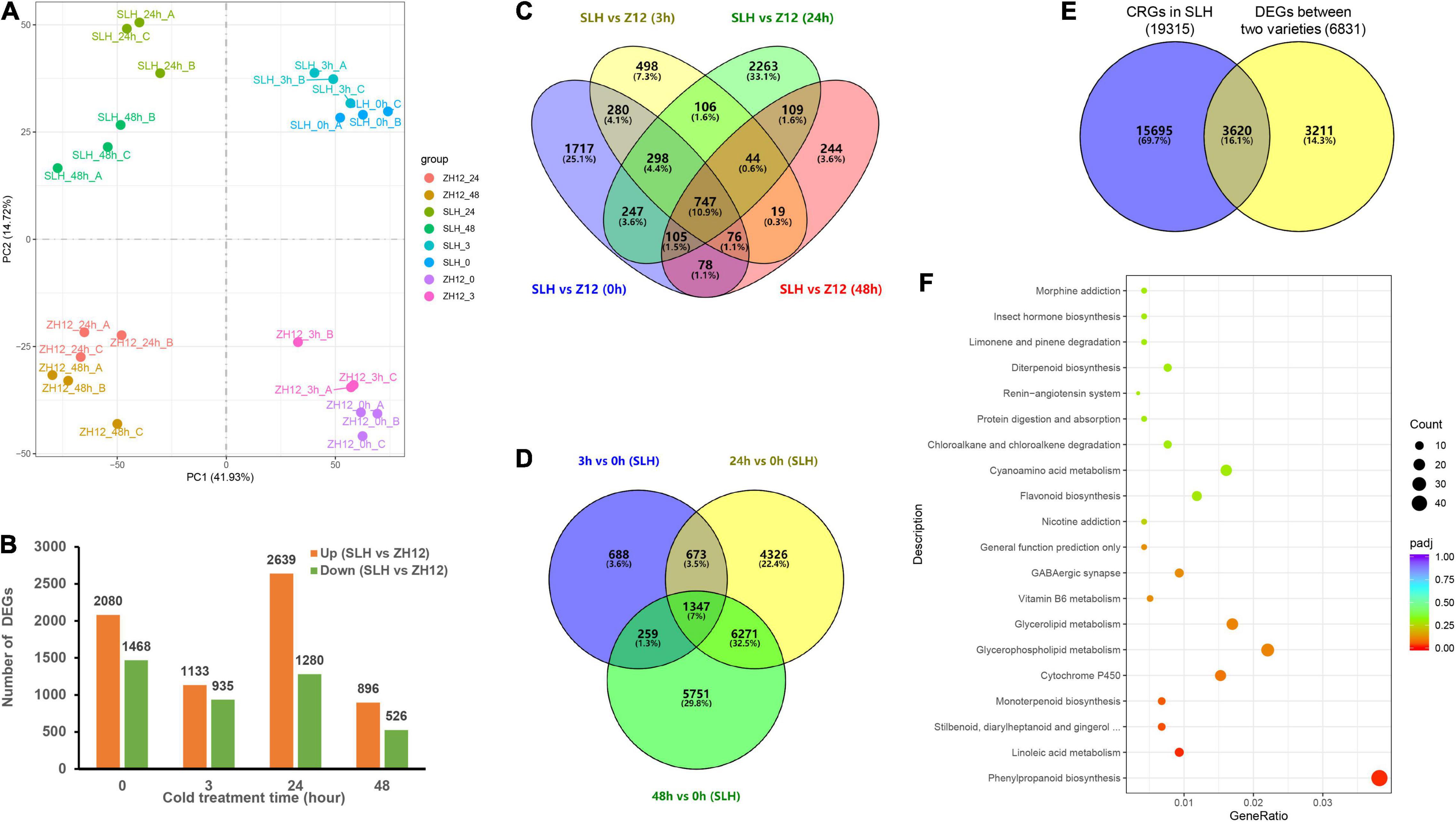
Figure 3. Transcriptome analysis of two peanut cultivars in response to cold stress. (A) Principle component analysis (PCA) of transcriptome data derived from SLH and ZH12 during four time points under cold stress (0, 3, 24, 48 h, with three biological replicates per point). (B) The number of differentially expressed genes (DEGs) between SLH and ZH12 exposed to cold treatment. DEGs were filed out using the DESeq2 R package with the parameter of | log2 (fold change)| > 1 and an adjusted P-value < 0.01. (C) The Venn diagram of DEGs between the two cultivars at four different cold treatment time points. (D) The Venn diagram of Cold Responsive Genes (CRGs) in SLH in the three cold exposure times (3, 24, and 48 h) compared to control (0 h). (E) The Venn diagram showing 3,620 putative Cold Tolerance Genes (CTG) in peanut. The yellow circle refers to the total 6,831 DEGs during the cold treatments between the two cultivars as shown in panel (C), and the blue circle represents the total 19,315 cold-responsive genes (CRGs) in SLH as indicated in panel (D). (F) KEGG enrichment analysis of the 3,620 Cold Tolerance Genes (CTG) in peanut.
Identification of Putative Cold Tolerance Genes in Peanut
To get a deep insight into the mechanism of cold tolerance in peanut, we performed comparative analysis of the transcriptional difference between SLH and ZH12 at the four time points of cold treatment (Figures 3B,C). In total, 6,831 differentially expressed genes (DEGs) were found between the two contrasting cultivars (Supplementary Table 7). Since genes involved in cold tolerance were probably induced and/or depressed by low temperature, we identified a total of 19,315 cold-responsive genes (CRGs) in SLH by comparing the transcriptome profile in 3, 24, and 48 h with that of control (0 h) (Figure 3D and Supplementary Table 8). As a result, there were 3,620 common genes shared by DEGs between the two cultivars and cold-responsive genes in SLH (Figure 3E). These common genes were believed to be a putative “Cold Tolerance Gene set” (CTGs) in peanut (Supplementary Table 9). Several transcription factors that were known to be involved in the regulating cold-signaling pathway were included in CTGs, such as two DREBs (Arahy.73DHZN and Arahy.SE8WTS) and two phytochrome interacting factors (Arahy.SDDU5A and Arahy.F2RES5) (Figure 4). Also, some structure genes in the carbohydrate metabolism were found in CTGs, including two raffinose synthases (Arahy.UR837P and Arahy.DBZB80) and a galactinol synthase (Arahy.N6N04K). The gene expression changes of nine CTGs were analyzed by qRT-PCR. Their transcript levels were higher and/or induce a data earlier time point by cold stress in SLH (cold tolerant) than ZH12 (cold susceptible), which had a good correlation with RNA-seq results (Figure 4 and Supplementary Figure 5).
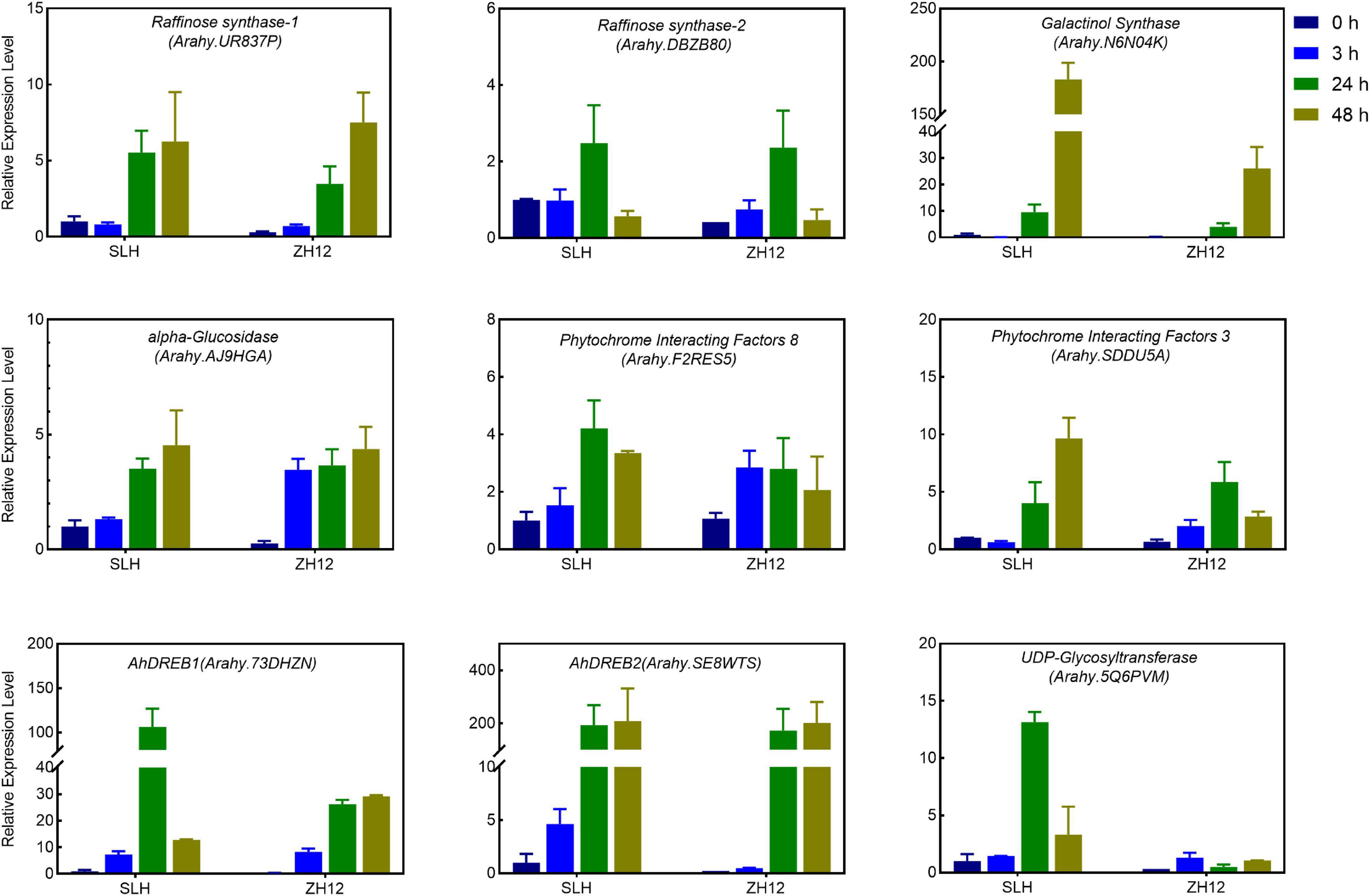
Figure 4. Quantitative RT-PCR (qRT-PCR) analysis of the expression levels of selected Core Cold Tolerance Genes in the peanut seedlings subjected to cold stress. The relative transcript levels among different samples were quantified by the 2− ΔΔCt method, with A. hypogaea Actin gene as the reference gene for normalization. Error bars indicate the S.D. of the means of three biological replicates.
Functional Enrichment Analysis of Cold Tolerance Genes in Peanut
Based on the GO enrichment analysis, the 3,620 CTGs were distributed into different known GO terms (Supplementary Table 10 and Supplementary Figure 6). Among the “biological process” category, the most significantly enriched terms are “response to biotic stimulus” and “defense response.” In addition, the “metal ion transport” term included 14 genes encoding for heavy metal-associated isoprenylated plant proteins (HIPPs), which were reported to play important roles in response to biotic/abiotic stresses (Zhang et al., 2015). The top enriched GO terms in “molecular functions” were related to “transferring acyl groups” and “glucosyltransferase activity.” Of these terms, 12 genes were annotated as putative cellulose synthases involved in the biosynthesis of the hemicellulose backbone, which might be helpful to confer abiotic stress tolerance to plants (Endler et al., 2015; Huang et al., 2021; Yuan et al., 2021).
KEGG pathway enrichment analysis of CTGs showed that the top significant enriched pathways were “phenylpropanoid biosynthesis” and “linoleic acid metabolism,” and “stilbenoid, diarylheptanoid, and gingerol biosynthesis” (Figure 3F and Supplementary Table 11). As one of the major branches of the phenylpropanoid pathway, lignin biosynthetic genes were upregulated by cold stress. In addition, the “linoleic acid metabolism” pathway included 11 lipoxygenase genes that were known to be related cold acclimation in plants (Upadhyay et al., 2019). These results indicate that both lignin and lipids take a great part in response to cold stress in peanut.
Identification of Key Genes and Modules in Response to Cold Stress by Weighted Gene Co-expression Network Analysis
Weighted gene co-expression network analysis is a popular systems biology method used to not only construct gene networks but also detect gene modules and identify the central players (i.e., hub genes) within modules. To obtain a comprehensive understanding of the molecular mechanism of cold tolerance in peanut, the 3,620 CTGs were put into the WGCNA software R package to build a gene co-expression network (Langfelder and Horvath, 2008). Based on pairwise correlations analysis of gene expression, 13 merged co-expression modules marked with different colors are shown in Figure 5A and could be further clustered into two main branches (Figure 5A and Supplementary Table 12). Analysis of the module-trait relationships for the 24 samples revealed that amino acids (Arginine and Alanine), spermidine, and two sugar compounds (raffinose and melezitose) were tightly associated with a red module (R > 0.5 and p < 0.01), while coniferin and the other three soluble sugars (trehalose, glucose, and sucrose) were more related to the tan module (Figure 5B). Based on the values of WGCNA edge weight and node scores, the top 20 hub genes were identified in the red module (Supplementary Table 13). These genes encoded some important proteins involved in abiotic stress processes, including dehydrin (arahy.4JU6QM), 6-phosphogluconate dehydrogenase (6PGDH, arahy.RH103U), pentatricopeptide repeat family protein (PPR, arahy.97P92R), nucleoporin (arahy.9L03M9), and S-adenosylmethionine decarboxylase (SAMDC, arahy.0034KJ) and GTP-binding nuclear protein (Ran, arahy.G34P0I) (Figure 5C). For the tan module, the top 20 genes encoded transcription factors (such as heat shock transcription factor and NAC-domain protein) and several stress-related proteins/enzymes, including cytochrome P450 (P450, arahy.MG688V), serine hydroxy-methyltransferase 6 (SHMT6, arahy. HK9TK1), F-box protein (arahy.C9YQVF), and SRO5 (arahy.7WJT5G) (Figure 5D and Supplementary Table 13). Notably, the transcript levels of hub genes in both of the modules were remarkably upregulated in SLH but were hardly expressed or only slightly enhanced in ZH12 at a late treatment point (48 h) (Figures 5E,F).
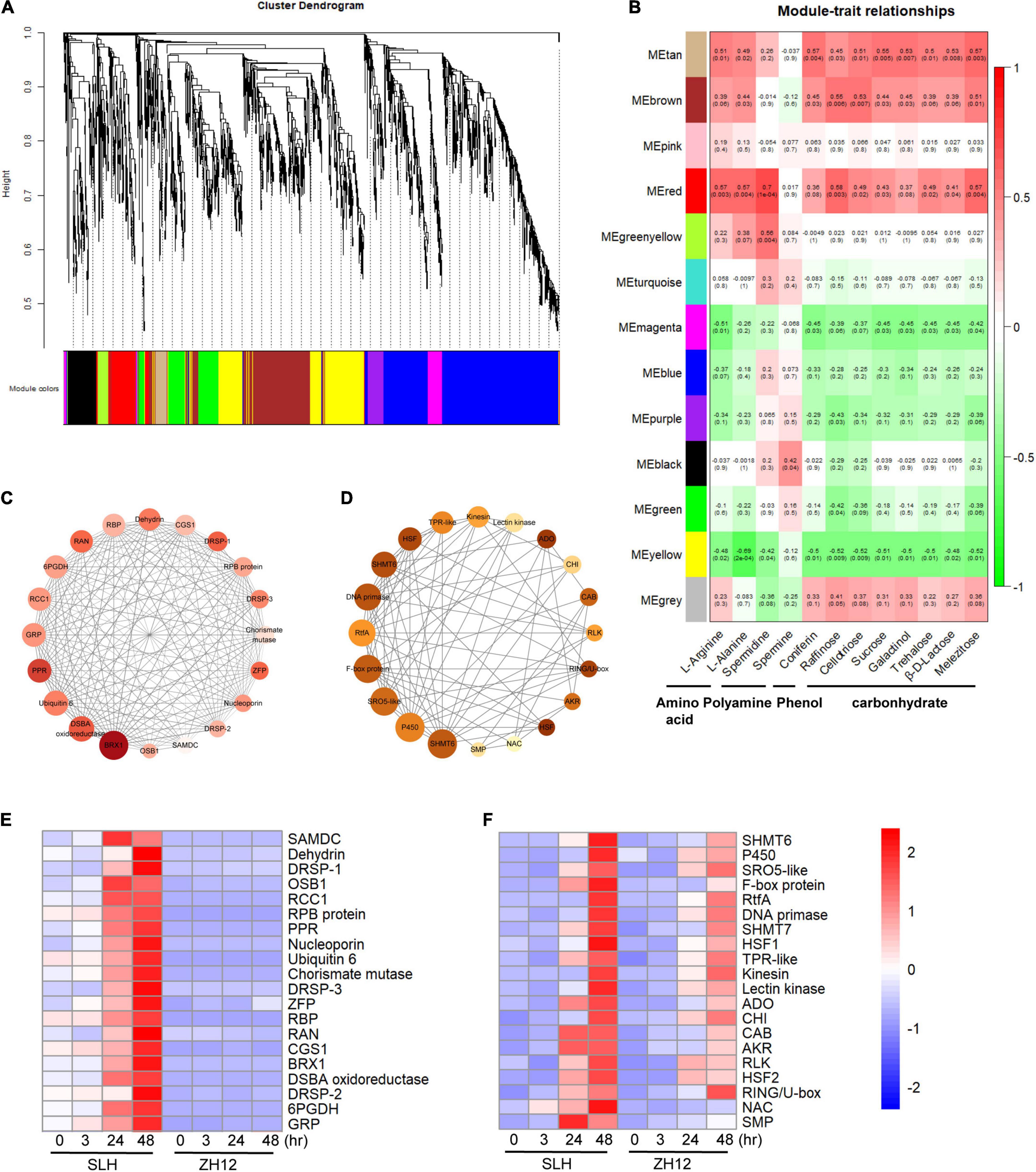
Figure 5. Identification of hub genes and key modules responsive to cold stress in peanut by WGCNA. (A) The hierarchical cluster tree shows 13 modules of co-expressed genes. Different modules are marked with different colors. (B) Correlations of metabolite contents with WGCNA modules. Each row corresponds to a specific module eigengene and a column to a trait. The values in each cell represent the correlation coefficients (r), and the p-values (in parentheses) of the module-trait association. The deeper the red or green in a cell, the higher the positive or negative correlation between the module eigengene and a trait feature. Visualization of the key co-expression network red module (C) and the tan (D) module by Cytoscape. The top 20 hub genes with the highest degree of connection to other genes are shown in the nodes. The color of node corresponds to degree value, and size means module membership value calculated by WGCNA. The darker and larger of the nodes indicates the greater hubness of the gene. Hierarchical cluster analysis of the top 20 hub genes in the red module (E) and the tan (F) module. The values in the heatmap (E,F) represent the z-score of the FPKM (transcript levels) in different samples. Red color indicates a high expression level, while blue is low.
Integrated Metabolome and Transcriptome Analysis to Reveal Crucial Pathways Responsive to Cold Stress
An integrated analysis of transcriptome and metabolome data revealed some common enriched pathways, including “carbohydrates metabolism” and “polyamines metabolism.” For carbohydrates metabolism, most of raffinose family oligosaccharides (RFOs) and sucrose-related metabolites, such as galactinol, raffinose, melibiose, and trehalose, were markedly increased in the two genotypes under cold stress. It should be noted that the fold changes of sugar compounds were consistently higher in cold-tolerant (SLH) than in cold-susceptible genotypes (ZH12), suggesting their vital roles in protection of peanut against low temperature stress (Figures 2C, 6A). Accordingly, the transcriptional profile of most genes involved in sugar biosynthetic pathways correlated well with the accumulation pattern in the two cultivars exposed to cold stress (Figures 2D, 6B). For instance, four genes-encoding galactinol synthases (EC 2.4.1.123, which catalyze the key steps for galactinol synthesis (Sengupta et al., 2015; Shimosaka and Ozawa, 2015), were greatly upregulated in SLH, but their transcripts were not changed or only slightly upregulated under cold stress in ZH12. Raffinose synthase, also called as galactinol-sucrose galactosyltransferase (EC 2.4.1.82), was a rate-limiting enzyme in biosynthesis of raffinose (ElSayed et al., 2014). Interestingly, expression levels of all the six raffinose synthase were strong cold induced in SLH, while moderately enhanced in ZH12. Besides, most of the genes involved in the biosynthesis pathways of sucrose, melibiose, and trehalose were upregulated in both genotypes in response to low temperature, but their expression levels increased more significantly in SLH than in ZH12. These results suggested that the cold tolerance of SLH might be associated with its stronger ability to regulate carbohydrates metabolism, thus leading to a higher accumulation of sucrose, raffinose, and trehalose in SLH than in cold-susceptible genotype (ZH12).
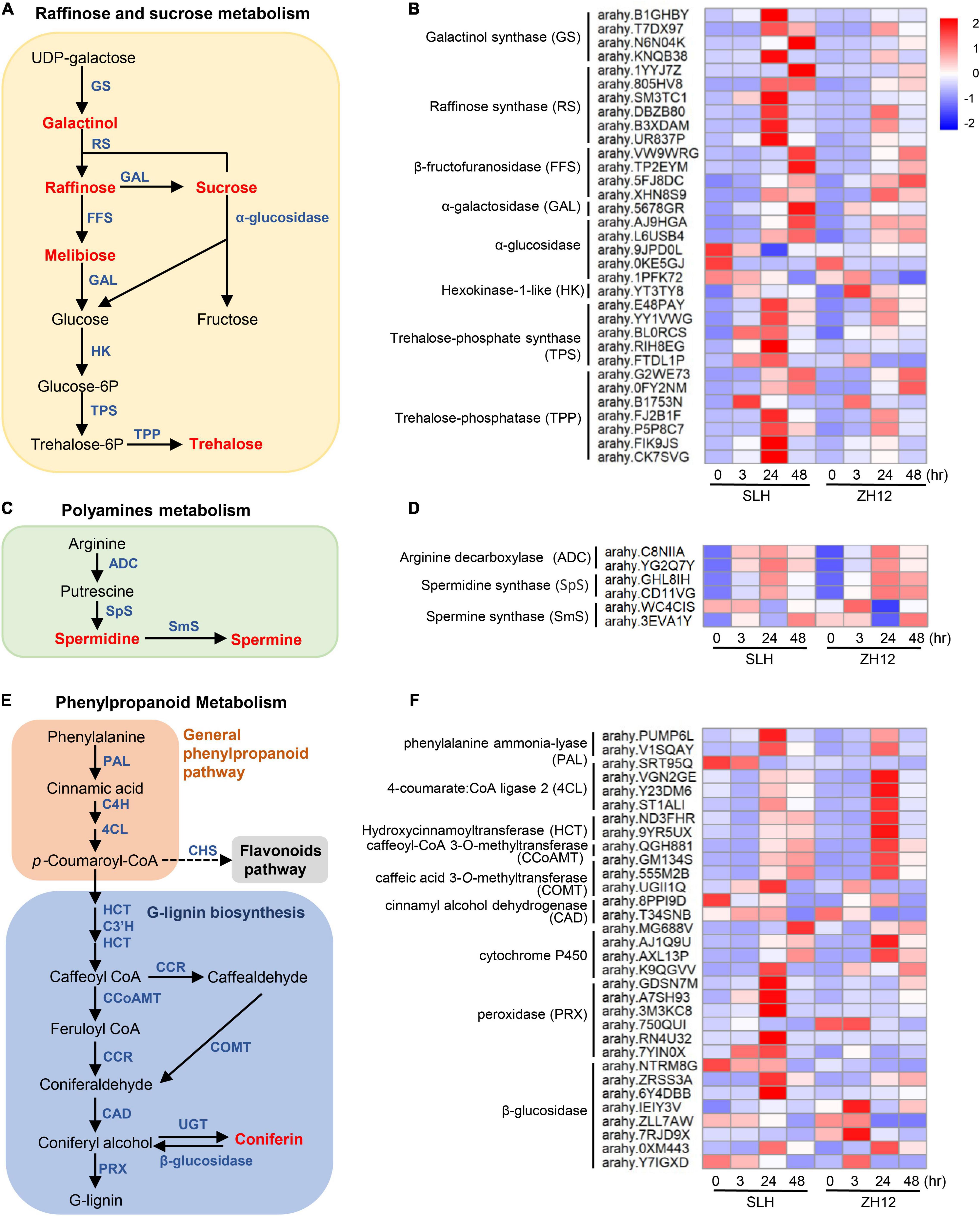
Figure 6. Integrated analysis of metabolomic and transcriptomic profiles of peanut under cold stress. The schematic diagrams of raffinose and sucrose (A), polyamines (C), and phenylpropanoid (E) metabolism pathways. In the diagrams, the metabolites are marked by red, and structural enzymes are indicated by blue. The transcriptional patterns of several structural genes involved in the raffinose and sucrose (B), polyamines (D), and phenylpropanoid (F) metabolism pathway under cold stress. The heatmap values represent the Z-score of the FPKM (transcript levels) in different samples. Red color indicates a high expression level, while blue is low.
Polyamines, including putrescine, spermidine, and spermine, are aliphatic nitrogen substances with low molecular weight and polycation characteristics, which have important roles in improving abiotic stress tolerance, including low-temperature tolerance (Alcazar et al., 2011). Several key enzyme genes involved in the “spermidine biosynthetic process,” including spermidine synthase (E.C. 2.5.1.16) and spermine synthase (E.C. 2.5.1.22), were found to be upregulated in response to cold stress (Figures 6C,D). This was in accordance with the accumulation pattern of polyamines in both peanut cultivars. Moreover, as compared with susceptible peanut (ZH12), the cold-tolerant one (SLH) had higher cellular levels of spermidine and spermine, suggesting it has a larger capacity to enhance polyamine biosynthesis under low temperature.
It has been reported that lignin serves as one of the major contributors to abiotic stress resistance (Cesarino, 2019). Coniferin, the glucoside of coniferyl alcohol, was considered as the most possible storage form of monolignol for G-lignin biosynthesis, which helps plants to fight against several abiotic stresses (Tsuji et al., 2005; Sharma et al., 2019). In this study, cold stress greatly increased coniferin contents in the peanut seedlings (Figures 2E, 6E). Accordingly, several genes in the general phenylpropanoid pathway and downstream G- lignin biosynthesis were significantly upregulated, including two phenylalanine ammonia lyases (PALs, the entry enzyme for phenylpropanoids), four 4-coumarate-CoA ligases (4CLs), two caffeic acid 3-O-methyltransferases (COMTs), and one caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) (Figure 6F and Supplementary Table 12). However, the transcript levels of these genes showed a differential expression pattern between the two cultivars. For instance, two PALs and one COMT gene had significant higher transcript levels in SLH than in ZH12, while the expression levels of four 4CLs, two hydroxycinnamoyltransferases (HCTs), and aCCoAOMT gene were higher in ZH12 compared to SLH after 24-h cold treatment (Figure 6F).
Discussion
Cold weather accounts for serious reductions in crop yield every year, since many agriculturally important crops, including rice (Oryza sativa) and peanut, are chilling sensitive and can only be cultivated in tropical or subtropical regions (Wu et al., 2020). Despite the molecular mechanisms of cold-induced reprogramming of gene expression and the metabolite accumulation pattern were extensively studied in model plants (Hoermiller et al., 2016; Jin et al., 2017; Pagter et al., 2017; Dasgupta et al., 2020), only a handful of reports were related to peanut (Jiang et al., 2020; Wu et al., 2020; Zhang H. et al., 2020). In this study, we attempted to decipher the regulatory mechanisms of peanut in response to cold stress through combining transcriptome with metabolome analysis.
Analysis of Signal Transduction and Protein Kinases in Response to Cold Stress
Ca2+ is an important signaling messenger in response to cold stress (Yuan et al., 2018). The calmodulins (CAMs) and calcium-dependent protein kinases (CDPKs) are vital calcium sensors and could transduce cold signals to activate the expression of downstream cold-response gene (CORs), including DREBs/CBFs and other components (Yang et al., 2010; Zeng et al., 2015). Overexpression of a salt-induced peanut CDPK gene in tobacco could alleviate PSII photoinhibition under abiotic stress, suggesting it played an important role in stress tolerance in peanut (Li et al., 2015). In the current study, the transcription of three calmodulin-binding proteins (CBPs, Arahy.1Y8NUF, Arahy.R2NPVK, and Arahy.GM51F1) and four CDPKs (Arahy.706LQI, Arahy.D2QNFF, Arahy.D2U3E6, and Arahy.P2RW1V) were significantly induced by low temperature (Supplementary Figures 7A,B and Supplementary Table 14) and have a relative higher transcript abundance in SLH (cold tolerant) than ZH12. Also, a total of 17 CBF transcripts were identified in our RNA-seq database (Supplementary Table 15). Among them, seven CBFs were not expressed at any time points in both of the varieties, while most of the other transcripts were increased in response to cold treatment (Supplementary Figure 7). It should be noted that some of the upregulated CBFs (arahy.73DHZN, arahy.WP3ECB, and arahy.5KI8FH) were induced at an earlier time point and/or have relative higher expression levels in cold tolerance variety (SLH) than that in cold susceptible (ZH12) (Figure 4 and Supplementary Figure 7), implying their significance in cold-signal perception and transduction processes in peanut.
In the present study, the transcript expression of two Heat Shock transcription Factors (HSFs) (Arahy.8U41WB and Arahy.W19Z82) was induced by cold stress in both cultivars (Figure 5F), and their transcript levels exhibited much higher in the cold-tolerant (SLH) than susceptible cultivar (ZH12) at 24- or 48-h treatment. Plant HSFs are encoded by large gene families with variable structure, expression, and function. They control responses not only to high temperatures but also to a number of abiotic stresses, such as cold, drought, and salt stress (Andrási et al., 2021). Some reports have shown that transcript abundances of HSF (HSFA4A, HSFA6B, HSFA8, and HSFC1) were enhanced by cold stress in Arabidopsis, while the induction effect was reduced in the ice1 mutant, implicating HSFs were involved in the cold-acclimation pathway (Swindell et al., 2007; Olate et al., 2018). In addition, overexpression of HSFs can stimulate the synthesis of protective metabolites, such as galactinol and raffinose, to improve abiotic stress tolerance in plants (Nishizawa et al., 2006; Lang et al., 2017). Therefore, it was proposed that the relative higher transcript levels of HSFs in SLH could be more beneficial for it to cope with cold stress.
Carbohydrate Metabolism Contributes Greatly to Cold Tolerance in Peanut
Carbohydrates are the primary products of photosynthesis. They act as nutrients as well important regulators that are required for plant growth, energy metabolism, and stress responses (Fuertauer et al., 2019). It was known that soluble sugars can not only function as osmoprotectants (Zuther et al., 2004; Lunn et al., 2014; Ma et al., 2021) but also to be ROS scavengers to provide cold tolerance in plants (Smirnoff and Cumbes, 1989; Van den Ende, 2013; Matros et al., 2015). In our study, the concentrations of soluble sugars (e.g., sucrose, trehalose, and raffinose) greatly raised in the two peanut cultivars exposed to cold stress. Notably, their contents were higher and/or induced at an earlier time point in cold-resistant peanut (SLH) than those in susceptible cultivar (ZH12) (Figure 2D). It has been reported that chilling stress could lead to a significant increase in H2O2 concentration in peanut (Wu et al., 2020). Similarly, we found that low temperature caused a sudden increase in the H2O2 in ZH12 rather than in SLH after 12-h treatment. Thus, it was proposed that the higher accumulation of soluble sugars probably led to maintain a low level of ROS in the cold-tolerant cultivar (SLH), which facilitated to its relative stronger cold-tolerance ability than the susceptible one (ZH12). On the other hand, ROS (including O2– and H2O2) can be detoxified by enzymatic and non-enzymatic antioxidant systems to maintain them at non-toxic levels in a delicate balancing state. In our transcriptome data, the transcript levels of several antioxidant enzymes (including superoxide dismutase and peroxidase) were enhanced in ZH12 (and also in SLH) during the early cold treatment period (3 and 24 h) (Supplementary Figures 7D,E). Thereby, we hypothesize that the upregulation of these antioxidant enzymes might efficiently scavenge the excess H2O2, rendering it recovered to the normal level (0 h) in 24 h.
In addition, the accumulation patterns of raffinose and sucrose between the two cultivars correlated well with expression profiles of their structural enzyme genes in the pathway (Figures 2D,E, 6A,B). Several studies have highlighted the role of these genes in cold stress (Lunn et al., 2014; Sengupta et al., 2015). For instance, overexpression of cold-inducible wheat galactinol synthase increased levels of galactinol and raffinose, and conferred higher tolerance to chilling stress in transgenic rice (Shimosaka and Ozawa, 2015). Simultaneously, exogenous application of sucrose improved chilling tolerance in cucumber seedlings by increasing antioxidant enzyme activity, and enhanced proline and soluble sugar contents. These results indicated that genes and metabolites involved in carbohydrate metabolism contributed much to cold tolerance in peanut.
Polyamine Metabolism Plays Central Roles in Cold Resistance in Peanut
Several studies have shown that the levels of polyamine were substantially elevated in plants exposed to stressful conditions, including drought, salinity, chilling, and heat (Chen et al., 2019). The improvement of cold-stress tolerance could be achieved by exogenous application of polyamines or genetic manipulation of endogenous polyamine levels (Kou et al., 2018; Alcazar et al., 2020). In the present study, spermine and spermidine were specifically accumulated in SLH (cold resistant) under chilling stress. Meanwhile, transcriptomic and WGCNA analysis showed that several transcripts and/or hub genes in polyamines biosynthesis have a relatively higher level in SLH than ZH12. For instance, S-adenosylmethionine decarboxylase (SAMDC, arahy.0034KJ), a key enzyme in the polyamine biosynthetic process, was identified as one of the hub genes in the red module (Figures 5C,E). It has been reported that overexpression of a bermudagrass SAMDC (CdSAMDC1) gene in tobacco enhanced plant cold tolerance through involvement of H2O2 and NO signaling (Luo et al., 2017). Like the transcriptional patterns of other hub genes in this module, the expression level of SAMDC was increased upon prolonged cold stress in SLH, but was not induced in ZH12 samples. These results suggested polyamine metabolism played crucial roles in cold resistance in peanut.
G-Lignin Biosynthesis Is Involved in Cold-Stress Response
Phenylpropanoid biosynthesis is one of the most important metabolisms in plants, generating an enormous array of secondary metabolites, such as lignin and flavonoid (Dong and Lin, 2021). Recent studies have revealed that cold stress induced the expression of structural genes in the phenylpropanoid pathway, including chalcone synthase (CHS) and 4-coumarate-CoA ligase (4CL), and, as a consequence, flavonoids and lignin accumulated to facilitate the adaptation to low-temperature environments in A. thaliana (Catalá et al., 2011), loquat fruit (Eriobotrya japonica) (Zhang J. et al., 2020), and apple (M. domestica) (An et al., 2020). In the current study, we found that some cold-responsive genes involved in the common phenylpropanoid metabolism and downstream G-lignin biosynthesis showed higher expression levels in cold-tolerant cultivar (SLH) than those in ZH12 (Figure 6F). For instance, PAL is the rate-limiting enzyme that catalyzes the deamination of L-phenylalanine to trans-cinnamate and forms the entry point into the synthesis of all phenylpropanoids. The higher transcript level of two PALs in SLH compared with those in ZH12 could drive more carbon flux into the phenylpropanoid pathway, probably leading to more coniferin accumulation and better cold tolerance in SLH.
Conclusion
In the present study, combining metabolomic and transcriptomic analysis of peanut seedlings exposed to chilling stress identified a set of important cold-responsive genes and metabolites, some of which showed differential expressions and/or accumulation patterns between the two cultivars. The signaling molecule Ca2+ and its related components first sense the cold stress, and then activate downstream pathways. During the adaptation of peanut to low temperature, soluble sugars, polyamines, and G-lignin might be the significant contributors. These results facilitate a better understanding of the molecular mechanism of cold response in peanut and could be helpful to genetic improvement of cold tolerance of crops. So far, we have heterogeneously overexpressed several peanut cold-tolerance-related genes in Arabidopsis, and obtained some transgenic lines. We are now analyzing their phenotypes under normal and stressful conditions. However, this part has not been finished yet. We hope to take a deep insight into the phenotypic characterization of transgenic Arabidopsis lines. It will be more profound if we get the relevant positive phenotypic results in the future.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA751249.
Author Contributions
XW, YuL, ZH, YC, DH, YK, and ZW conducted the experiments. YoL and BL organized and supervised the overall project. XW, LY, and HJ performed the data analysis and wrote the manuscript. BL edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Key Research and Development Project of China (No. 2018YFD1000900).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.752474/full#supplementary-material
Supplementary Table 1 | qRT-PCR primers in this study.
Supplementary Table 2 | A list of metabolites detected in the samples.
Supplementary Table 3 | The list of differential changed metabolites between the two peanut varieties under cold stress.
Supplementary Table 4 | Classification of differential changed metabolites between two peanut cultivars under cold stress.
Supplementary Table 5 | The accumulation pattern of metabolites during cold stress in peanut.
Supplementary Table 6 | Statistical analyses and mapping results of RNA sequencing reads.
Supplementary Table 7 | Differentially expressed genes between cold-tolerant and cold-susceptible cultivars.
Supplementary Table 8 | A list of cold-responsive genes in SLH and ZH12 peanut.
Supplementary Table 9 | The detail information of 3,620 cold-tolerance genes.
Supplementary Table 10 | GO enrichment analysis cold-tolerance genes.
Supplementary Table 11 | KEGG pathway enrichment analysis of cold tolerance genes.
Supplementary Table 12 | A gene list of WGCNA modules.
Supplementary Table 13 | The top 20 hub genes in red and tan modules.
Supplementary Table 14 | The transcription levels of antioxidative genes and calcium signal genes in the two cultivars under cold stress.
Supplementary Table 15 | The transcription levels of CBF genes in the two cultivars under cold stress.
Footnotes
References
Alcazar, R., Bueno, M., and Tiburcio, A. F. (2020). Polyamines: small amines with large effects on plant abiotic stress tolerance. Cells 9:2373. doi: 10.3390/cells9112373
Alcazar, R., Cuevas, J. C., Planas, J., Zarza, X., Bortolotti, C., Carrasco, P., et al. (2011). Integration of polyamines in the cold acclimation response. Plant Sci. 180, 31–38. doi: 10.1016/j.plantsci.2010.07.022
An, J. P., Wang, X. F., Zhang, X. W., Xu, H. F., and Hao, Y. J. (2020). An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 18, 337–353.
Andrási, N., Pettkó-Szandtner, A., and Szabados, L. (2021). Diversity of plant heat shock factors: regulation, interactions and functions. J. Exp. Bot. 72, 1558–1575. doi: 10.1093/jxb/eraa576
Bell, M. J., Gillespie, T. J., Roy, R. C., Micheals, T. E., and Tollenaar, M. (1994). Peanut leaf photosynthetic activity in cool field environments. Crop Sci. 34, 1023–1029. doi: 10.2135/cropsci1994.0011183X003400040035x
Catalá, R., Medina, J., and Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012
Chen, D., Shao, Q., Yin, L., Younis, A., and Zheng, B. (2019). Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9:1945. doi: 10.3389/fpls.2018.01945
Chen, N., Yang, Q., Hu, D., Pan, L., Chi, X., Chen, M., et al. (2014). Gene expression profiling and identification of resistance genes to low temperature in leaves of peanut (Arachis hypogaea L.). Sci. Hortic. 169, 214–225. doi: 10.1016/j.scienta.2014.01.043
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X. H., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Cui, P., Li, Y., Cui, C., Huo, Y., Lu, G., and Yang, H. (2020). Proteomic and metabolic profile analysis of low-temperature storage responses inIpomoea batataLam. Tuberous roots. BMC Plant Biol. 20:435. doi: 10.1186/s12870-020-02642-7
Dasgupta, P., Das, A., Datta, S., Banerjee, I., Tripathy, S., and Chaudhuri, S. (2020). Understanding the early cold response mechanism in IR64 indica rice variety through comparative transcriptome analysis. BMC Genomics 21:425. doi: 10.1186/s12864-020-06841-2
Ding, Y. L., Shi, Y. T., and Yang, S. H. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564. doi: 10.1016/j.molp.2020.02.004
Dong, C. N., Danyluk, J., Wilson, K. E., Pocock, T., Huner, N. P. A., and Sarhan, F. (2002). Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol. 129, 1368–1381. doi: 10.1104/pp.001925
Dong, N. Q., and Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Du, S., Cui, M., Cai, Y., Xue, A., Hao, Y., Huang, X., et al. (2020). Metabolomic analysis of chilling response in rice (Oryza sativa L.) seedlings by extractive electrospray ionization mass spectrometry. Environ. Exp. Bot. 180:104231. doi: 10.1016/j.envexpbot.2020.104231
ElSayed, A. I., Rafudeen, M. S., and Golldack, D. (2014). Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol. 16, 1–8. doi: 10.1111/plb.12053
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
Fuertauer, L., Weiszmann, J., Weckwerth, W., and Naegele, T. (2019). Dynamics of plant metabolism during cold acclimation. Int. J. Mol. Sci. 20:5411. doi: 10.3390/ijms20215411
Hoermiller, I., Naegele, T., Augustin, H., Stutz, S., Weckwerth, W., and Heyer, A. (2016). Subcellular reprogramming of metabolism during cold acclimation in Arabidopsis thaliana. Plant Cell Environ. 40, 602–610. doi: 10.1111/pce.12836
Huang, T., Luo, X., Fan, Z., Yang, Y., and Wan, W. (2021). Genome-wide identification and analysis of the sucrose synthase gene family in cassava (Manihot esculenta Crantz). Gene 769:145191. doi: 10.1016/j.gene.2020.145191
Jambunathan, N. (2010). Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol. Biol. 639, 292–298. doi: 10.1007/978-1-60761-702-0_18
Jiang, C. J., Zhang, H., Ren, J. Y., Dong, J. L., Zhao, X. H., Wang, X. G., et al. (2020). Comparative transcriptome-based mining and expression profiling of transcription factors related to cold tolerance in peanut. Int. J. Mol. Sci. 21:1921. doi: 10.3390/ijms21061921
Jiang, J., Hou, R., Yang, N., Li, L., Deng, J., Qin, G., et al. (2021). Physiological and TMT-labeled proteomic analyses reveal important roles of sugar and secondary metabolism in Citrus junos under cold stress. J. Proteomics 237:104145. doi: 10.1016/j.jprot.2021.104145
Jin, J. J., Zhang, H., Zhang, J. F., Liu, P. P., Chen, X., Li, Z. F., et al. (2017). Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics 18:496. doi: 10.1186/s12864-017-3871-7
Kakani, V. G., Prasad, P., Craufurd, P. Q., and Wheeler, T. R. (2002). Response of in vitro pollen germination and pollen tube growth of groundnut (Arachis hypogaea L.) genotypes to temperature. Plant Cell Environ. 25, 1651–1661. doi: 10.1046/j.1365-3040.2002.00943.x
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, Y. S., Lee, M., Lee, J.-H., Lee, H.-J., and Park, C.-M. (2015). The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 89, 187–201. doi: 10.1007/s11103-015-0365-3
Kou, S., Chen, L., Tu, W., Scossa, F., Wang, Y., Liu, J., et al. (2018). The arginine decarboxylase gene ADC1, associated to the putrescine pathway, plays an important role in potato cold-acclimated freezing tolerance as revealed by transcriptome and metabolome analyses. Plant J. 96, 1283–1298. doi: 10.1111/tpj.14126
Lang, S., Liu, X., Xue, H., Li, X., and Wang, X. (2017). Functional characterization of BnHSFA4a as a heat shock transcription factor in controlling the re-establishment of desiccation tolerance in seeds. J. Exp. Bot. 68, 2361–2375. doi: 10.1093/jxb/erx097
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lee, B. H., Henderson, D. A., and Zhu, J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Li, M., Sui, N., Lin, L., Yang, Z., and Zhang, Y. H. (2019). Transcriptomic profiling revealed genes involved in response to cold stress in maize. Funct. Plant Biol. 46, 830–844. doi: 10.1071/fp19065
Li, Y., Fang, F., Guo, F., Meng, J.-J., Li, X.-G., Xia, G.-M., et al. (2015). Isolation and functional characterisation of CDPKs gene from Arachis hypogaea under salt stress. Funct. Plant Biol. 42, 274–283. doi: 10.1071/fp14190
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–Δ Δ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lunn, J. E., Delorge, I., Figueroa, C. M., Van Dijck, P., and Stitt, M. (2014). Trehalose metabolism in plants. Plant J. 79, 544–567. doi: 10.1111/tpj.12509
Luo, J., Liu, M., Zhang, C., Zhang, P., Chen, J., Guo, Z., et al. (2017). Transgenic centipedegrass (Eremochloa ophiuroides [Munro] Hack.) overexpressing S-adenosylmethionine decarboxylase (SAMDC) gene for improved cold tolerance through involvement of H2O2 and no signaling. Front. Plant Sci. 8:1655. doi: 10.3389/fpls.2017.01655
Ma, S., Lv, J., Li, X., Ji, T., Zhang, Z., and Gao, L. (2021). Galactinol synthase gene 4 (CsGolS4) increases cold and drought tolerance in Cucumis sativus L. by inducing RFO accumulation and ROS scavenging. Environ. Exp. Bot. 185:104406. doi: 10.1016/j.envexpbot.2021.104406
Ma, Y., Zhang, Y., Lu, J., and Shao, H. (2009). Roles of plant soluble sugars and their responses to plant cold stress. Afr. J. Biotechnol. 8, 2004–2010.
Matros, A., Peshev, D., Peukert, M., Mock, H. P., and Van den Ende, W. (2015). Sugars as hydroxyl radical scavengers: proof-of-concept by studying the fate of sucralose in Arabidopsis. Plant J. 82, 822–839. doi: 10.1111/tpj.12853
Mehmood, S. S., Lu, G., Luo, D., Hussain, M. A., Raza, A., Zafar, Z., et al. (2021). Integrated analysis of transcriptomics and proteomics provides insights into the molecular regulation of cold response in Brassica napus. Environ. Exp. Bot. 187:104480. doi: 10.1016/j.envexpbot.2021.104480
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Olate, E., Jiménez-Gómez, J. M., Holuigue, L., and Salinas, J. (2018). NPR1 mediates a novel regulatory pathway in cold acclimation by interacting with HSFA1 factors. Nat. Plants 4, 811–823. doi: 10.1038/s41477-018-0254-2
Pagter, M., Alpers, J., Erban, A., Kopka, J., Zuther, E., and Hincha, D. (2017). Rapid transcriptional and metabolic regulation of the deacclimation process in cold acclimated Arabidopsis thaliana. BMC Genomics 18:731. doi: 10.1186/s12864-017-4126-3
Pan, Y., Liang, H., Gao, L., Dai, G., Chen, W., Yang, X., et al. (2020). Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice. BMC Plant Biol. 20:371. doi: 10.1186/s12870-020-02569-z
Sengupta, S., Mukherjee, S., Basak, P., and Majumder, A. L. (2015). Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6:656. doi: 10.3389/fpls.2015.00656
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., and Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24:2452. doi: 10.3390/molecules24132452
Shimosaka, E., and Ozawa, K. (2015). Overexpression of cold-inducible wheat galactinol synthase confers tolerance to chilling stress in transgenic rice. Breed. Sci. 65, 363–371.
Sinha, S., Raxwal, V. K., Joshi, B., Jagannath, A., Katiyar-Agarwal, S., Goel, S., et al. (2015). De novo transcriptome profiling of cold-stressed siliques during pod filling stages in Indian mustard (Brassica juncea L.). Front. Plant Sci. 6:932. doi: 10.3389/fpls.2015.00932
Smirnoff, N., and Cumbes, Q. J. (1989). Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 28, 1057–1060. doi: 10.1016/0031-9422(89)80182-7
Sorita, G. D., Leimann, F. V., and Ferreira, S. R. S. (2020). Biorefinery approach: is it an upgrade opportunity for peanut by-products? Trends Food Sci. Technol. 105, 56–69. doi: 10.1016/j.tifs.2020.08.011
Swindell, W. R., Huebner, M., and Weber, A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. doi: 10.1186/1471-2164-8-125
Tang, K., Zhao, L., Ren, Y., Yang, S., Zhu, J.-K., and Zhao, C. (2020). The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tsuji, Y., Fang, C., Yasuda, S., and Fukushima, K. (2005). Unexpected behavior of coniferin in lignin biosynthesis of Ginkgo biloba L. Planta 222, 58–69.
Upadhyay, R. K., Handa, A. K., and Mattoo, A. K. (2019). Transcript abundance patterns of 9-and 13-lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to each Stress. Genes 10:683. doi: 10.3390/genes10090683
Van den Ende, W. (2013). Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 4:247. doi: 10.3389/fpls.2013.00247
Wang, C., Cheng, B., Zheng, Y., Sha, J., Li, A., and Sun, X. (2003). Effects of temperature to seed emergence, seedling growth and anthesis of peanut. J. Peanut Sci. 32, 7–11. doi: 10.14001/j.issn.1002-4093.2003.04.002
Wang, X. J., Shen, Y., Sun, D. L., Bian, N. F., Shi, P. X., Zhang, Z. M., et al. (2020). iTRAQ-based proteomic reveals cell cycle and translation regulation involving in peanut buds cold stress. Russ. J. Plant Physiol. 67, 103–110. doi: 10.1134/s1021443720010239
Wang, X. J., Sun, D. L., Bian, N. F., Shen, Y., Zhang, Z. M., Wang, X., et al. (2017). Metabolic changes of peanut (Arachis hypogaea L.) buds in response to low temperature (LT). S. Afr. J. Bot. 111, 341–345. doi: 10.1016/j.sajb.2017.04.009
Wu, D., Liu, Y. F., Pang, J. Y., Yong, J. W. H., Chen, Y. L., Bai, C. M., et al. (2020). Exogenous Calcium alleviates nocturnal chilling-induced feedback inhibition of photosynthesis by improving sink demand in peanut (Arachis hypogaea). Front. Plant Sci. 11:607029. doi: 10.3389/fpls.2020.607029
Yang, C., Yang, H., Qijun, X., Wang, Y., Sang, Z., and Yuan, H. (2020). Comparative metabolomics analysis of the response to cold stress of resistant and susceptible Tibetan hulless barley (Hordeum distichon). Phytochemistry 174:112346. doi: 10.1016/j.phytochem.2020.112346
Yang, T., Chaudhuri, S., Yang, L., Du, L., and Poovaiah, B. W. (2010). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126. doi: 10.1074/jbc.M109.035659
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi: 10.1089/omi.2011.0118
Yuan, P., Yang, T., and Poovaiah, B. W. (2018). Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 19:3896.
Yuan, W., Liu, J., Takac, T., Chen, H., Li, X., Meng, J., et al. (2021). Genome-wide identification of banana csl gene family and their different responses to low temperature between chilling-sensitive and tolerant cultivars. Plants 10:122. doi: 10.3390/plants10010122
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zhang, H., Dong, J. L., Zhao, X. H., Zhang, Y. M., Ren, J. Y., Xing, L. T., et al. (2019). Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front. Plant Sci. 10:838. doi: 10.3389/fpls.2019.00838
Zhang, H., Jiang, C. J., Ren, J. Y., Dong, J. L., Shi, X. L., Zhao, X. H., et al. (2020). An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Front. Plant Sci. 11:1110. doi: 10.3389/fpls.2020.01110
Zhang, H., Jiang, C. J., Yin, D. M., Dong, J. L., Ren, J. Y., Zhao, X. H., et al. (2021). Establishment of comprehensive evaluation system for cold tolerance and screening of cold-tolerance germplasm in peanut. Acta Agron. Sin. 47, 1753–1767. doi: 10.3724/SP.J.1006.2021.04182
Zhang, J., Yin, X.-R., Li, H., Xu, M., Zhang, M.-X., Li, S.-J., et al. (2020). Ethylene response factor EjERF39- EjMYB8 complex activates cold-induced lignification of loquat fruit, via the biosynthetic gene Ej4CL1. J. Exp. Bot. 71, 3172–3184. doi: 10.1093/jxb/eraa085
Zhang, X., Feng, H., Feng, C., Xu, H., Huang, X., Wang, Q., et al. (2015). Isolation and characterisation of cDNA encoding a wheat heavy metal-associated isoprenylated protein involved in stress responses. Plant Biol. 17, 1176–1186. doi: 10.1111/plb.12344
Zhao, X., Chen, J., and Du, F. (2012). Potential use of peanut by-products in food processing: a review. J. Food Sci. Technol. 49, 521–529. doi: 10.1007/s13197-011-0449-2
Zhao, Y., Zhou, M., Xu, K., Li, J. H., Li, S. S., Zhang, S. H., et al. (2019). Integrated transcriptomics and metabolomics analyses provide insights into cold stress response in wheat. Crop J. 7, 857–866. doi: 10.1016/j.cj.2019.09.002
Keywords: Arachis hypogaea, RNA-Seq, metabolome, carbohydrate, polyamine, lignin, phenylpropanoid
Citation: Wang X, Liu Y, Han Z, Chen Y, Huai D, Kang Y, Wang Z, Yan L, Jiang H, Lei Y and Liao B (2021) Integrated Transcriptomics and Metabolomics Analysis Reveal Key Metabolism Pathways Contributing to Cold Tolerance in Peanut. Front. Plant Sci. 12:752474. doi: 10.3389/fpls.2021.752474
Received: 03 August 2021; Accepted: 21 October 2021;
Published: 24 November 2021.
Edited by:
Dhruv Lavania, University of Alberta, CanadaReviewed by:
Sibaji Kumar Sanyal, Jawaharlal Nehru University, IndiaVivek Kumar Raxwal, Central European Institute of Technology (CEITEC), Czechia
Copyright © 2021 Wang, Liu, Han, Chen, Huai, Kang, Wang, Yan, Jiang, Lei and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Lei, bGVpeW9uZ0BjYWFzLmNu; Boshou Liao, bGlhb2Jvc2hvdUBjYWFzLmNu
†These authors have contributed equally to this work
 Xin Wang
Xin Wang Yue Liu†
Yue Liu† Yuning Chen
Yuning Chen Dongxin Huai
Dongxin Huai Zhihui Wang
Zhihui Wang Huifang Jiang
Huifang Jiang Yong Lei
Yong Lei Boshou Liao
Boshou Liao