
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 October 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.751965
This article is part of the Research TopicSalinity Tolerance: From Model or Wild Plants to Adapted CropsView all 27 articles
The wall-associated kinase (WAK) multigene family plays critical roles in various cellular processes and stress responses in plants, however, whether WAKs are involved in salt tolerance is obscure. Herein, we report the functional characterization of a rice WAK, WAK112, whose expression is suppressed by salt. Overexpression of OsWAK112 in rice and heterologous expression of OsWAK112 in Arabidopsis significantly decreased plant survival under conditions of salt stress, while knocking down the OsWAK112 in rice increased plant survival under salt stress. OsWAK112 is universally expressed in plant and associated with cell wall. Meanwhile, in vitro kinase assays and salt tolerance analyses showed that OsWAK112 possesses kinase activity and that it plays a negative role in the response of plants to salt stress. In addition, OsWAK112 interacts with S-adenosyl-L-methionine synthetase (SAMS) 1/2/3, which catalyzes SAM synthesis from ATP and L-methionine, and promotes OsSAMS1 degradation under salt stress. Furthermore, in OsWAK112-overexpressing plants, there is a decreased SAMS content and a decreased ethylene content under salt stress. These results indicate that OsWAK112 negatively regulates plant salt responses by inhibiting ethylene production, possibly via direct binding with OsSAMS1/2/3.
As sessile organisms, plants are more susceptible than animals to abiotic stresses, such as salinity, drought, and temperature extremes (Munns and Tester, 2008). Salt stress, an important abiotic stress, can affect all aspects of plant development (Hanin et al., 2016). The main manifestations of salt stress are reduced seed germination, decreased chlorophyll synthesis, premature senescence, and death after long-term exposure to excessive salt (Zhu, 2002). To withstand salt stress, plants have evolved a network of physiological, biochemical, and molecular mechanisms (Hanin et al., 2016). These mechanisms help minimize the harmful effects of ionic stress by sequestering Na+ in the vacuoles or outside of the cell via the classical salt overly sensitive pathway (Deinlein et al., 2014). Osmotic regulators are synthesized that counteract the osmotic stress created by excess salt (Yang and Guo, 2018). Plants may also increase the gene expression and protein activity of reactive oxygen species (ROS)-scavenging enzymes to alleviate oxidative damage (Sreenivasulu et al., 2004; Zhou et al., 2018). In addition, plants can induce the expression of salt stress-related genes to enhance their ability to adapt to high-salt environments (Huang et al., 2012). Still, a comprehensive investigation of the mechanisms that underlie plant cell responses to salt stress is required to understand how plants grow and survive under high-salt conditions.
The phytohormones play a crucial role in plant growth and development, and a sophisticated and efficient role under different environmental conditions. Among the nine well-characterized plant hormones, ethylene, gibberellin, jasmonic acid, and cytokinins are able to be regulated by salt stress (Yu et al., 2020). Numerous studies have shown that ethylene plays a crucial role in the regulation of diverse stress responses, including it to salt stress (Riyazuddin et al., 2020; Zhao et al., 2021a). In plants, ethylene is biosynthesized from S-adenosyl-L-methionine (SAM), which is synthesized from l-methionine to ATP catalyzed by S-adenosyl-L-methionine synthetases (SAMSs). SAM is conversed to 1-aminocyclopropane-1-carboxylicd (ACC) by ACC synthase (ACS). ACC is further oxygenated by ACC oxidase (ACO) to produce ethylene (Pattyn et al., 2021). The roles of ethylene in the regulation of salt tolerance are complex. Several studies have demonstrated that the overproduction of endogenous ethylene increases salinity stress tolerance in plants, while the inhibition of ethylene signaling leads to increased sensitivity to salinity stress (Zhang et al., 2011; Yang et al., 2013; Peng et al., 2014). For example, the loss-of-function mutant eto1, which exhibits increased levels of ethylene, shows enhanced salt tolerance (Jiang et al., 2013). Meanwhile, the Arabidopsis ethylene-insensitive mutants ein2-5 and ein3-1eil1-1 have been shown to exhibit salt-sensitive phenotypes (Lei et al., 2011; Peng et al., 2014). Moreover, ethylene has been found to be an essential positive mediator of salinity stress tolerance in rice. For example, the overexpression of acireductone dioxygenase, a metal-binding metalloenzyme encoded by OsARD1, elevates the endogenous ethylene release rate to reduce the sensitivity of rice plants to drought, salt, and osmotic stresses (Liang et al., 2019). These reports collectively suggest an important role for ethylene in salt stress tolerance in plants.
Receptor-like kinases (RLKs) play important roles in plant growth and development as well as in plant responses to biotic and abiotic stresses (Gao and Xue, 2012). Wall-associated kinases (WAKs) are a unique class of RLKs named for their close association with the plant cell wall (Verica and He, 2002). WAKs contain an extracellular domain that can be linked to molecules in the cell wall; this domain receives exogenous stimuli and then transfers them through the plasma membrane via a cytoplasmic serine/threonine kinase domain to initiate a downstream signaling cascade (Kohorn and Kohorn, 2012). The Arabidopsis genome encodes 5 WAKs and 21 WAK-like (WAKL) proteins. There are 5-fold more WAK members in rice than in Arabidopsis, indicating expansion of the WAK family in monocots (Zhang et al., 2005).
WAKs are involved in many physiological processes in plants (Kohorn and Kohorn, 2012; Kohorn, 2016). For instance, silencing of AtWAK2 results in small rosette leaves (Wagner and Kohorn, 2001), while the suppression of AtWAK4 produces a series of pleiotropic effects, including short primary roots, small rosette leaves, condensed inflorescence stems, unopened miniature flowers, and short siliques (Lally et al., 2001). In rice, silencing of the rice indica OsiWAK1 gene results in dwarf seedlings, impaired root development, and sterility due to anther indehiscence, suggesting an important role in plant development via the regulation of cell expansion (Kanneganti and Gupta, 2011). Further, a loss of OsWAK91/OsDEES1 in rice plants can reduce fertility due to a defect in embryo development (Wang et al., 2012). These reports indicate that WAKs play essential roles in vegetative and reproductive development in plants. WAKs are also suggested to be involved in biotic stress responses. When the pathogen-responsive gene AtWAK1 was knocked down, the expression of several pathogenesis-related genes was downregulated and the plants were more labile than wild type in response to a pathogen attack (He et al., 1998). The WAKL protein AtWAKL22/RFO1 is a novel type of dominant disease-resistance protein that mediates resistance to multiple Fusarium races (Diener and Ausubel, 2005). Another WAKL protein, AtWAKL10, is a positive regulator of basal immune responses and effector-triggered immunity in Arabidopsis (Bot et al., 2019). The first WAK identified in rice, OsWAK1, which is induced by Magnaporthe oryzae, plays a positive role in plant disease resistance (Li et al., 2009). Several other rice WAKs, such as OsWAK14, OsWAK91, OsWAK92, and OsWAK112, act as positive or negative regulators of quantitative resistance to the rice blast fungus (Delteil et al., 2016). In recent decades, the function of the WAK gene family in plant fungal disease resistance has also been characterized in maize, wheat, and tomato plants (Rosli et al., 2013; Hurni et al., 2015; Zuo et al., 2015; Saintenac et al., 2018). WAKs also help regulate plant adaptation to abiotic stresses. For instance, the mRNA and protein levels of AtWAK1 were found to rise quickly in response to aluminum treatment, and overexpression of AtWAK1 retarded the growth inhibition of roots caused by aluminum stress in Arabidopsis (Sivaguru et al., 2003). Meanwhile, impaired expression of AtWAKL4 increased plant hypersensitivity to excess K+, Na+, Cu2+, and Zn2+, while WAKL4 overexpression conferred Ni2+ tolerance (Hou et al., 2005). More recently, AtWAKL10 was found to positively regulate salt stress while negatively regulating drought stress (Bot et al., 2019). In addition, WAK1 in tomato regulates leaf Na+ accumulation and alters the source–sink balance under high-salt conditions (Meco et al., 2020). Although these studies have deepened our understanding of plant WAKs, the molecular mechanisms underlying the functions of WAKs in salt stress responses are undefined, and the direct interacting components of WAKs in salt stress responses are unknown.
Rice, which is a staple food for more than half of the world’s population, faces various biotic and abiotic stresses during its three- to four-month growing season. Although 125 OsWAK genes have been annotated from rice genomes (Zhang et al., 2005), their functions are largely unknown and whether they are involved in salt stress responses are a mystery.
In this study, we investigated the function of OsWAK112 in salt stress responses. Our results show that OsWAK112 negatively regulates salt stress in rice depending on its kinase activity, possibly by inhibiting S-adenosyl-L-methionine synthase (SAMS) accumulation, which in turn decreases ethylene production.
The rice plants used in this study were Oryza sativa japonica (Jap). The Arabidopsis thaliana plants used were wild-type Columbia (Col).
To create 35S promoter-driven Myc-tagged OsWAK112 or OsWAK112K678E or OsWAK112D794A constructs, the OsWAK112 coding sequence was amplified by PCR with the primers OsWAK112-F and OsWAK112-R (Supplementary Table 1) to construct OsWAK112/pENTR using the commercial donor vector pENTR™/SD/D-TOPO™ (K242020; Invitrogen, Carlsbad, CA, United States). Next, OsWAK112K678E and OsWAK112D794A pENTR were generated using the Fast Mutagenesis System (FM111; TransGen Biotech, Beijing, China) with OsWAK112-K678E-F and OsWAK112-K678E-R or OsWAK112-D794A-F and OsWAK112-D794A-R as the overlapping mutagenic primers (Supplementary Table 1) and OsWAK112/pENTR as the template. Sequences encoding both native and mutated versions of OsWAK112 were cloned into the binary vector pG7MH1 (Wang et al., 2013) using the Gateway LR Clonase™ II Enzyme Mix (11,791,020; Invitrogen).
To generate OsWAK112 RNAi transgenic plants, a 581-bp OsWAK112 fragment was amplified with specific primers OsWAK112RNAi-F and OsWAK112RNAi-R (Supplementary Table 1) using the LA Taq with GC Buffer kit (Takara, DRR20AG). The OsWAK112 RNAi fragment was inserted into the RNAi vector pTCK303 (Wang et al., 2004) through its KpnI/SpeI and BamHI/SacI sites.
For the native promoter-driven GUS construct proOsWAK112::GUS, the promoter sequence of OsWAK112 (with 2,108 base pairs upstream of the ATG) was amplified and inserted into the binary vector pCAMBI1300::GUS using HindIII and BamHI.
For our bimolecular fluorescence complementation (BiFC) assays, the coding sequences of OsSAMS1/2/3 were amplified by PCR using the primers OsSAMS1/2/3-F and OsSAMS1/2/3-R (Supplementary Table 1) to construct OsSAMS1/2/3/pENTR. Then, the coding sequences of OsWAK112 and OsSAMS1/2/3 were individually introduced into 35S::X-NYFP and 35S::X-CCFP (Wang et al., 2013) using OsWAK112/pENTR and OsSAMS1/2/3/pENTR as templates.
For the co-immunoprecipitation (Co-IP) assays, the coding sequences of OsSAMS1, OsSAMS2, OsSAMS3, and the OsWAK112 kinase domain (OsWAK112KD) were cloned into pMDC83 and pG7MH1 to produce ectopic expression of OsSAMS1/2/3-GFP and OsWAK112KD-Myc, respectively.
The above-mentioned binary expression constructs were introduced into Agrobacterium tumefaciens strain EHA105 and then transformed into Arabidopsis or rice for stable transformation or co-infiltrated into 4-week-old Nicotiana benthamiana leaves for transient assays.
For prokaryotic protein expression, the coding sequence of the OsWAK112KD was amplified by PCR using the primers OsWAK112-KD-F and OsWAK112-KD-R (Supplementary Table 1) and inserted into pGEX-4T-1 to produce glutathione S-transferase (GST)-OsWAK112KD. Next, GST-OsWAK112K678E, GST-OsWAK112K791R, and GST-OsWAK112D794A were generated using the Fast Mutagenesis System (FM111; TransGen Biotech) with OsWAK112-K678E-F/R, OsWAK112-K791R-F/R, or OsWAK112-D794A-F/R as the overlapping mutagenic primers (Supplementary Table 1) and GST-OsWAK112KD as the template. For myelin basic protein (MBP)-OsSAMS1/2/3, the coding sequences of OsSAMS1/2/3 were cloned into the destination vector gc-PML2C (Xin et al., 2021) using OsSAMS1/2/3/pENTR as the donor vector. The above-mentioned constructs were transformed into Escherichia coli strain BL21 for protein purification.
All rice plants were grown in a greenhouse with a 12-h/12-h light-dark cycle at 28°C (100μmolm−2 s−1 intensity) or in a paddy field under natural conditions (May to October in Hebei, China) for general growth and seed production. The Arabidopsis seedlings were grown either in a growth chamber under continuous light (100μmolm−2 s−1 intensity) or in a culture room under 16h of light/8h of darkness (90μmolm−2 s−1 intensity) at 22°C for seed production.
For salt sensitivity analyses of the rice in hydroponic culture, seeds were germinated in water for 2days, grown in a hydroponic culture solution (Ren et al., 2005) for 10days, and treated with a 200mM NaCl-containing culture solution for another 4days. After 8days of recovery, the survive rate was determined. To assess the salt response in solid medium, seeds were germinated on 0.5×Murashige and Skoog (MS) solid medium for 2days then transplanted to 0.5×MS solid medium without (0mM) or with 150mM or 200mM NaCl for 20days at 28°C in a light chamber under a 14-h/10-h light-dark cycle.
Arabidopsis seeds were sown on 0.5×MS medium with or without 125mM NaCl, kept for 2days in the dark at 4°C, grown under 16h of light/8h of darkness at 22°C for 10days, and then observed to determine the phenotype.
Seedlings that were still green and continuing to produce new leaves were registered as survivors. All experiments were repeated at least three times.
Total RNA from 7-day-old rice seedlings was isolated using TRIzol reagent (15596–026; Invitrogen) following the manufacturer’s instructions. cDNA was synthesized from 0.5 to 1μg of total RNA with a RevertAid First Strand cDNA Synthesis Kit (K1622; Thermo Fisher Scientific, Waltham, MA, United States) using oligo (dT)16 primer. To analyze OsWAK112, quantitative semi-quantitative reverse transcription (RT-qPCR) was performed using the SYBR Premix Ex Taq Reagent (RR420A; Takara, Otsu, Japan); the Os18S rRNA gene was used as an internal control. The primers used are listed in Supplementary Table 1.
The roots of 7-day-old rice plants treated with or without 150mM NaCl for 30min were stained with 5-(and-6)-chloromethyl-2ʹ,7ʹ-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Thermo Fisher Scientific). The fluorescence intensity was determined with an Axio Imager (M2; Carl Zeiss, Jena, Germany); all pictures were analyzed using ImageJ. The fluorescent intensity of Jap at 0mM NaCl is set to 1.
To investigate the subcellular localization of OsWAK112, the coding sequence of OsWAK112 from OsWAK112/pENTR was introduced into binary vector pMDC83 to construct the vector 35S:: OsWAK112-GFP, and 35S::GFP was used as a negative control vector. Both constructs were transferred to A. tumefaciens strain GV3101 and then transiently transformed into tobacco (N. benthamiana) leaves or transformed into onion (Allium cepa) epidermal cells by particle bombardment using the Bio-Rad PDS-1000/He system according to the manufacturer’s protocol. After 40 to 48h infiltration, localization of the protein was examined using a Zeiss LSM 510 confocal microscopy system with a 488nm laser for excitation from 500 to 515nm for GFP emission. 0.9M mannitol was used to induce plasmolysis.
Recombinant GST-OsWAK112KD, GST-WAK112KDK678E, GST-WAK112KDK791R, and GST-WAK112KDD794A were affinity purified using glutathione agarose beads (GE Healthcare, Chicago, IL, United States) and subjected to an in vitro kinase assay, which included incubation with 1μL of cold ATP (1mM) in kinase reaction buffer (50mM HEPES, 10mM MgCl2, 5mM MnCl2, and 1mM ATP, 10μCi of [γ-32P]ATP) at 30°C for 3h. The mixture was separated by 10% SDS-PAGE and detected with a Typhoon 9,410 imager (GE Healthcare).
For our BiFC assays, OsWAK112-NYFP and OsSAMSs-CCFP or YCE (control) were co-expressed in N. benthamiana leaves. Fluorescence from the BiFC signals was observed under a confocal microscope (Meta710-LCM; Carl Zeiss).
For the overlay assays, gel blots (polyvinylidene fluoride [PVDF] filters) containing GST-WAK112KD and GST were incubated with 2μg of MBP-OsSAMS1, MBP-OsSAMS2, and MBP-OsSAMS3, followed by anti-MBP-HRP (E8038S; New England BioLabs, Ipswich, MA, United States).
In our Co-IP assays, N. benthamiana leaves transiently co-expressing OsSAMSs-GFP and OsWAK112KD-Myc, or expressing OsSAMSs-GFP or OsWAK112KD-Myc alone, were harvested and extracted with IP buffer (50mM Tris-HCl, pH 7.5, 150mM NaCl, 2mM EDTA, and 0.1% NP-40) and then incubated with anti-GFP agarose at 4°C for 2h. Next, the proteins were washed with IP buffer five times and then boiled at 100°C in water for 5min with 2×loading buffer. The proteins were then separated by SDS-PAGE. Anti-Myc (019M4760V; Sigma-Aldrich, St. Louis, MO, United States) and anti-GFP antibodies (HT801-01; TransGen Biotech) were used to detect OsWAK112KD and OsSAMS1/2/3, respectively.
For ethylene measurement, 7-day-old wild-type (Jap) and OsWAK112-overexpressing (OsWAK112OE) rice seedlings treated with or without 200mM NaCl for 24h were placed into 15-mL vials containing 3ml of 0.5×MS medium and closed with a rubber cap on the top; the vials were then allowed to sit for 24h (16-h light/8-h dark) at 28°C. A total of 1mL of the air from the headspace of each vial was taken to inject into a gas chromatograph to measure the ethylene content.
For SAMS measurement, total protein from 7-day-old Jap and OsWAK112OE seedlings treated with or without 200mM NaCl for 24h was extracted and used for SAMS content measurement with a SAMS ELISA Kit (SY-P09712; Shanghai Win-Win Biotechnology Co., Ltd., Shanghai, China). The assay was performed according the manufacturer’s protocol.
To test whether OsWAK112 affects OsSAMSs protein stability, OsSAMS1-GFP alone or OsSAMS1 together with OsWAK112-Myc was filtrated in tobacco leave. After 40 to 48h, tobacco leave was separated into two parts, treated in liquid 1/2 MS medium with or without 200mM NaCl for 2h, and then, the total protein was extracted for Western blotting.
In this study, we examined the involvement of OsWAK112 in plant salt responses. OsWAK112 bound directly to and inhibited OsSAMSs to suppress the induction of ethylene production so as to negatively regulate salt tolerance.
WAKs are protein kinases with important roles not only in plant growth and development but also in plant responses to exogenous stresses (Hou et al., 2005; Bot et al., 2019; Meco et al., 2020); however, their role in salt stress responses is poorly understood. Published microarray data revealed that the expression levels of several WAKs are regulated by salt (Mishra et al., 2018). Herein, the expression pattern of one of them, OsWAK112 (Os10g10130; Mishra et al., 2018), was examined under high-salt conditions. Its transcript level decreased gradually as the treatment time increased; at 24h, it was reduced to nearly half of the control level (Supplementary Figure S1), indicating that OsWAK112 is a potential salt-related regulator in rice.
To determine the biological function of OsWAK112 in salt responses, ectopic expression lines using a 35S promoter-driven OsWAK112 coding sequence with a Myc tag were generated in a wild-type Jap background. Two independent OsWAK112OE lines named OE7 and OE8 were used in our subsequent experiments because they exhibited high transcript levels (approximately 2.6- or 3.2-fold that in Jap; Figure 1A). Under normal growth conditions, the OsWAK112OE plants exhibited no clear difference from Jap plants (Figure 1B, before treatment). However, after being treated with 200mM NaCl for 4days and subsequently allowed to recover in regular hydroponic culture for another 8days, the OsWAK112OE plants were drier and yellower than the Jap plants (Figure 1B), in accordance with their lower survival rates (50% for OE7 and 43% for OE8) compared to Jap (63%) (Figure 1C), indicating that OsWAK112 induces poor salt resistance in rice.
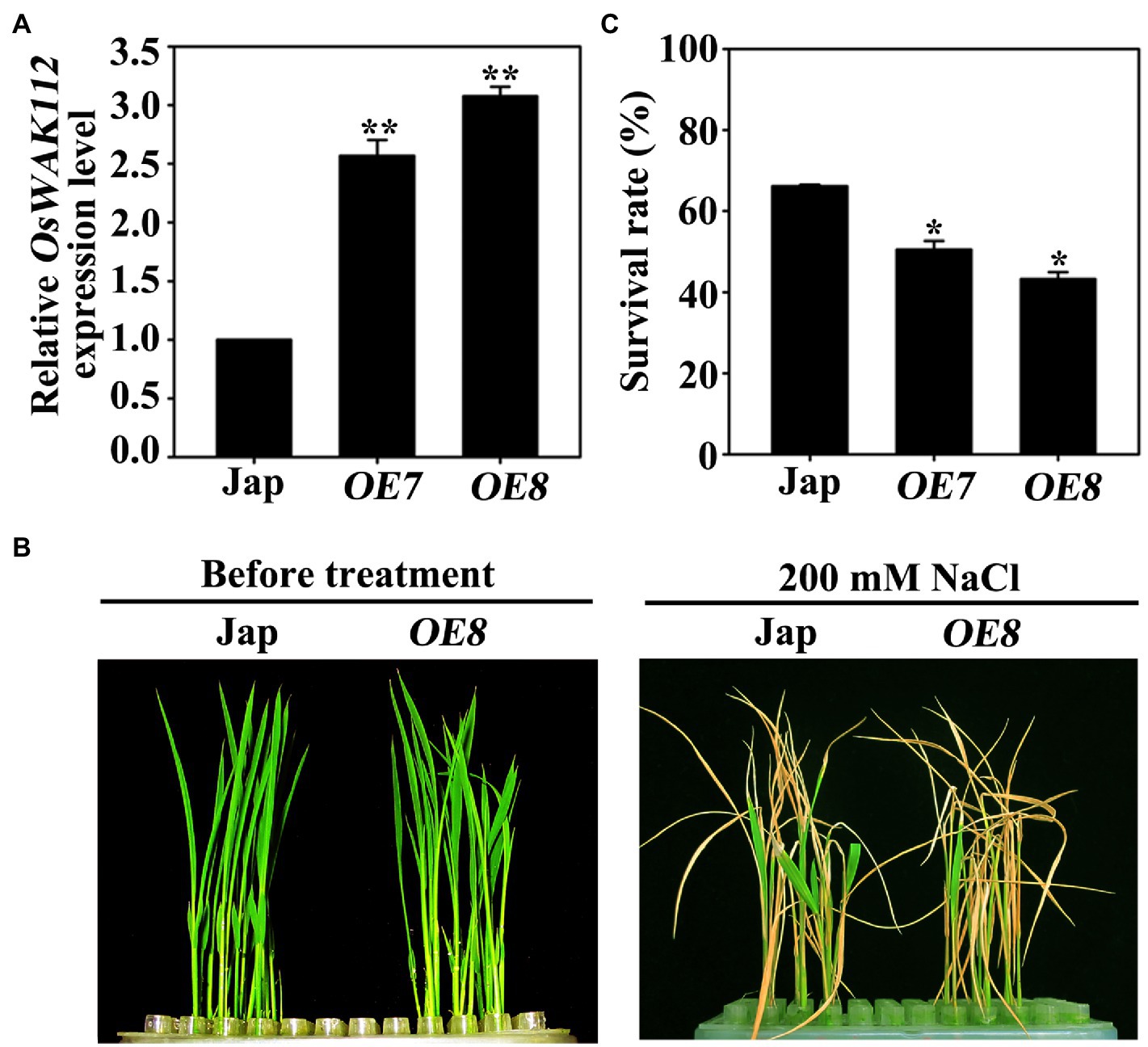
Figure 1. Phenotypes of the OsWAK112OE lines under salt stress. (A) The OsWAK112 transcript levels in 7-day-old Jap and OsWAK112OE lines as determined by RT-qPCR. (B) A comparison of the phenotypes of Jap plants and a representative OsWAK112OE line (OE8) before and after salt treatment. Ten-day-old seedlings were used as described in the Materials and Methods. (C) Survival rates of Jap and two individual OsWAK112OE lines following salt exposure. In (A,C), each data point represents the mean ± standard error (SE) from three biological repeats (n≥50). Asterisks indicate a difference relative to Jap (Student’s t-test, *p<0.05 and **p<0.01).
To confirm the function of OsWAK112 in plant salt tolerance, the salt responses of OE7 and OE8 plants were examined on solid medium. After 2 days of germination on regular medium, seeds were transplanted into medium containing 0, 150, or 200mM NaCl. The OE7 and OE8 seedlings did not show any difference from Jap seedlings when grown on regular medium (Figure 2A, 0mM NaCl); however, they showed greater growth inhibition by salt than Jap plants depending on the NaCl concentration (Figure 2A). The shoot length and shoot fresh weight of OE7 and OE8 were both lower than those of Jap (Figures 2B,C).
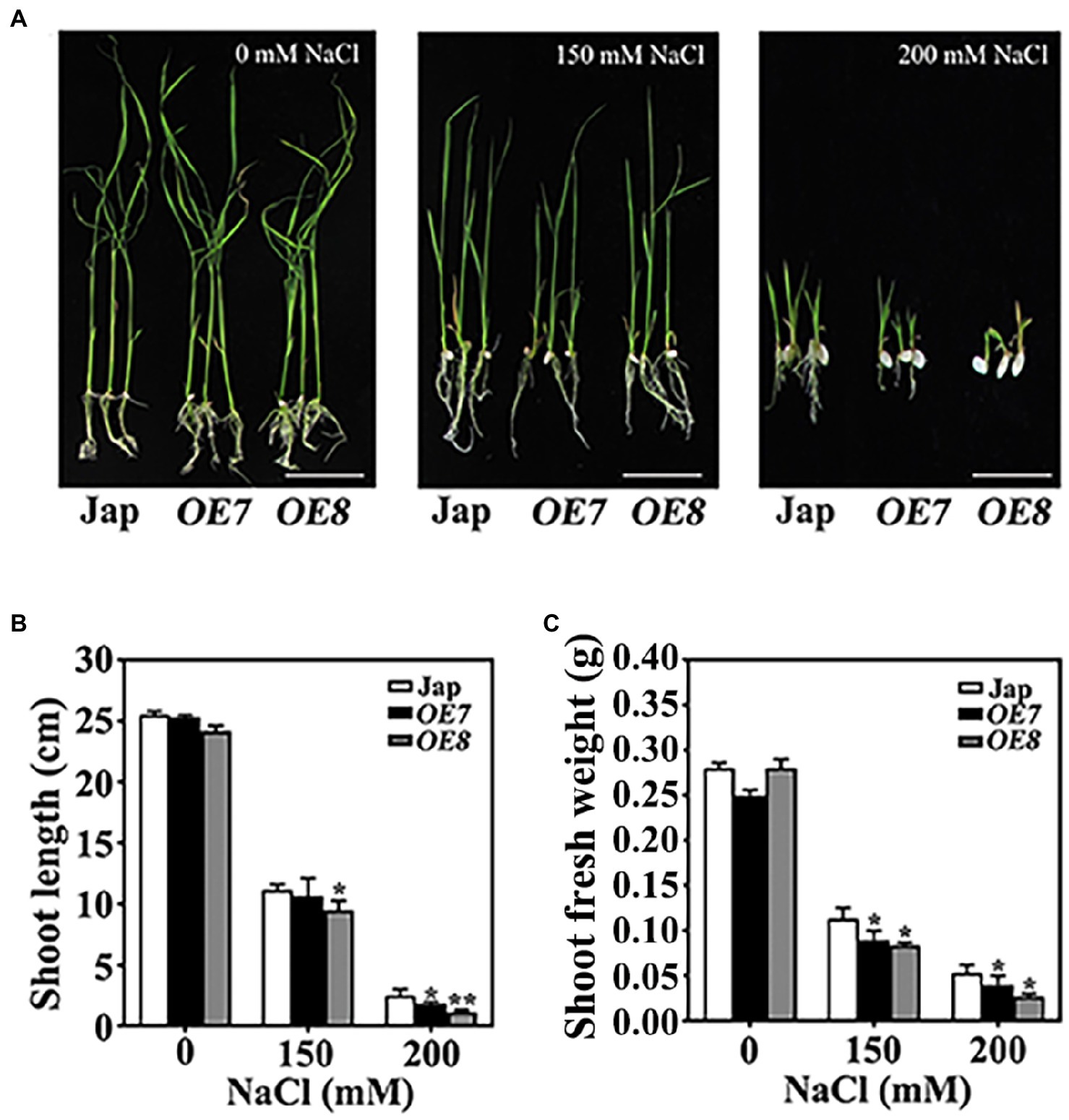
Figure 2. OsWAK112 overexpression confers salt hypersensitivity in rice. (A) The performance of OsWAK112OE seedlings compared with Jap on media containing the indicated concentrations of NaCl. Scale bar=2cm. The shoot lengths (B) and fresh shoot weights (C) of 10-day-old seedlings were analyzed. The experiments were repeated three times. Each data point represents the mean±SE (n≥10). Asterisks indicate a difference relative to Jap (Student’s t-test, *p<0.05 and **p<0.01) in (B,C).
To completely investigate the function of OsWAK112, the RNAi of OsWAK112 transgenic rice plants was also generated by introducing the vector pTCK303. Two independent OsWAK112 RNAi lines (RNAi1 and RNAi3, which showed around 4 and 7% of OsWAK112 mRNA level) were assessed under salt stress conditions. Without NaCl treatment, the OsWAK112 RNAi plants were indistinguishable from wild-type japonica (Jap; Supplementary Figure S2). After treatment with 200mM NaCl for 4 days and subsequent growth by regular hydroponic culture for 8 days, the RNAi plants showed significantly higher survival rates compared with Jap (Supplementary Figure S2).
Taken together, these findings (Figures 1, 2 and Supplementary Figure S2) demonstrate that OsWAK112 plays a negative role in salt tolerance.
Salt stress induces the rapid accumulation of ROS, and an imbalance in ROS levels often triggers serious membrane damage in plants (You and Chan, 2015; Yang and Guo, 2018). To examine whether the increased salt sensitivity of OsWAK112OE was related to the ROS level, the quantity of H2O2, a major and stable ROS, was evaluated using CM-H2DCFDA with 7-day-old roots from Jap and OsWAK112OE lines with or without 200mM NaCl treatment. This probe can be transported into cells, where its acetate groups are passively cleaved by intracellular esterases, producing the fluorescent compound dichlorodihydrofluorescein (DCF; Wang et al., 2021). As shown in Supplementary Figure S3A, without salt treatment, the H2O2 levels in the OsWAK112OE lines were slightly lower than that in Jap. After salt treatment, the H2O2 levels in Jap and in the two OsWAK112OE lines were dramatically increased, indicating the induction of H2O2 production by salt as described previously (Li et al., 2014). However, the H2O2 levels in the OsWAK112OE lines were significantly higher (110 and 103% higher than in OE7 and OE8 [individual controls], respectively) than in Jap (46% higher than its control; Supplementary Figures S3A,B), revealing that the OsWAK112OE plants accumulated more ROS during salt treatment.
To investigate the tissue-specific expression pattern of OsWAK112, the 2,108-base pair promoter sequence was inserted into a pCAMBIA1300 vector harboring the gene encoding GUS. This proOsWAK112::GUS construct was introduced into rice plants by A. tumefaciens-mediated callus transformation. Histochemical staining revealed OsWAK112 expression in coleoptiles (Supplementary Figure S4A), ligules (Supplementary Figure S4B), leaf blades (Supplementary Figure S4C), stems (Supplementary Figures S4D,E), roots (Supplementary Figure S4F), and the radicles of germinated seeds (Supplementary Figure S4G) during vegetative growth. During reproductive growth, OsWAK112 was observed in paleae (Supplementary Figure S4H), stamens, and stigmas (Supplementary Figure S4I). These results show that OsWAK112 is expressed in most plant tissues.
Online sequence analyses have shown that OsWAK112 possesses a typical receptor kinase structure, including an extracellular domain, a transmembrane domain, and an intracellular kinase domain.1 Thus, it is speculated to be a plasma membrane protein (Figure 3A); however, the sub-location of OsWAK112 is unclear. Thus, the full-length coding sequence of OsWAK112 fused with the sequence encoding green fluorescence protein (GFP) was generated and expressed transiently in tobacco (N. benthamiana) and onion (A. cepa) epidermal cells. As shown in Figure 3, GFP alone was expressed ubiquitously in the intracellular region of the tobacco epidermal cells (Figure 3B, GFP) and onion epidermal cells (Figure 3C, GFP), whereas OsWAK112-GFP was detected only in the region of the plasma membrane (Figures 3B,C, OsWAK112-GFP). To verify whether OsWAK112 is associated with the cell wall, onion epidermal cells were treated with 0.9 M mannitol to induce plasmolysis. After plasmolysis, OsWAK112-GFP was detected in the plasma membrane region and in the cell wall region (Figure 3C, OsWAK112-GFP plasmolyzed). In contrast, the control protein (GFP) was detected only in the intracellular region (Figure 3C, GFP plasmolyzed). These data indicate that OsWAK112 is localized to the plasma membrane and that it may connect tightly to the cell wall. Moreover, these data suggest that OsWAK112 is a wall-associated protein in plant cells.
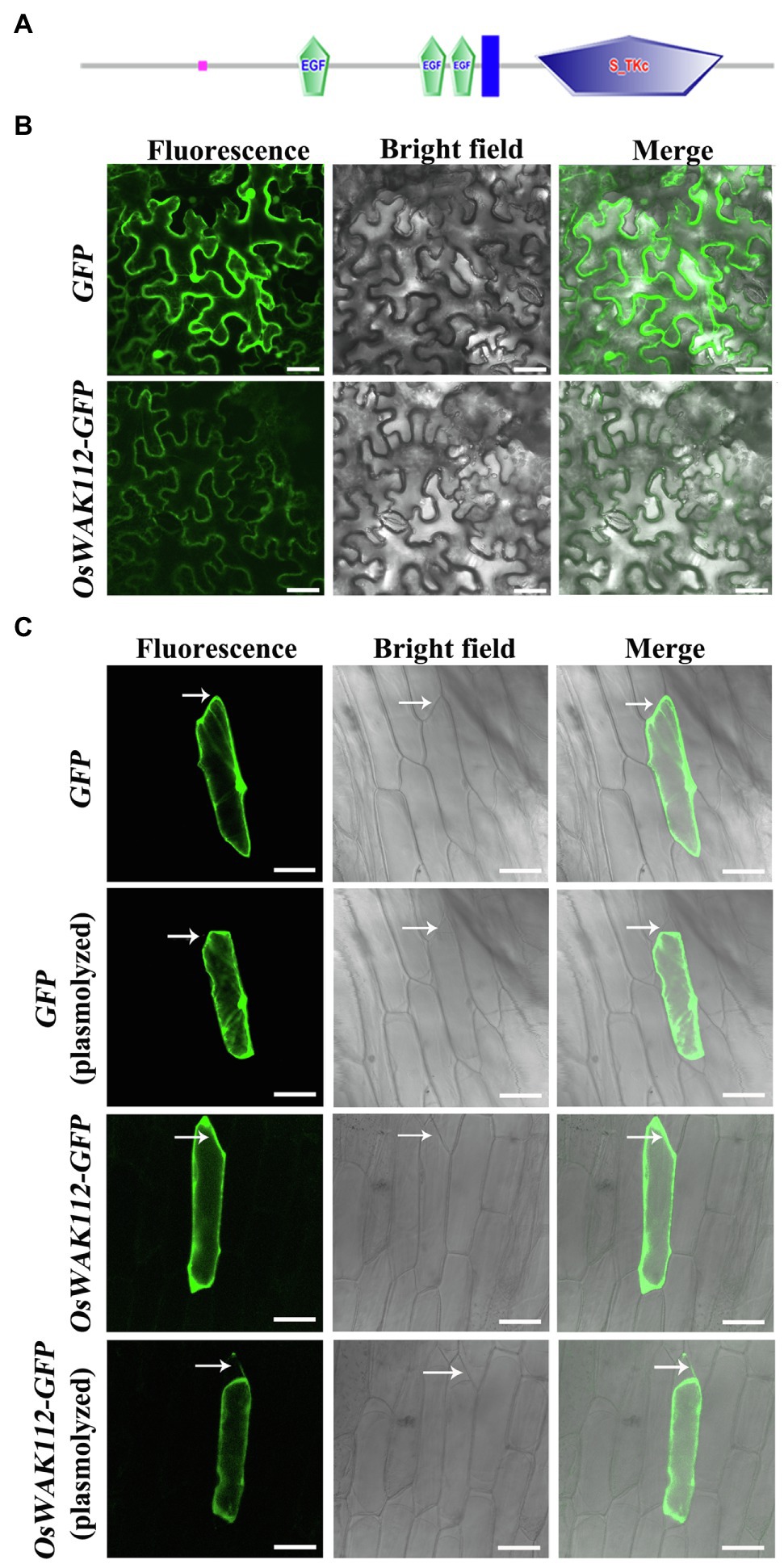
Figure 3. The subcellular localization of OsWAK112. (A) The protein structure of OsWAK112 as predicted by SMART. Pink box: low complexity region. EGF: epidermal growth factor-like domain. Blue box: transmembrane region. S-TKc: serine/threonine protein kinase, catalytic domain. (B,C) Subcellular localization of OsWAK112-GFP in tobacco and onion epidermal cells. Arrows in (C) indicate the cell wall region where plasmolysis happened. Scale bar=50μm in (B) and 100μm in (C).
To investigate the function of OsWAK112 further, we performed a sequence alignment and homology analysis between OsWAK112 and multiple RLKs in Arabidopsis using GenBank from the National Center for Biotechnology Information.2 The sequence of OsWAK112 showed 40–60% similarity with those of WAK and cytosolic receptor kinase family members in Arabidopsis, and most of the conserved sequences were in the kinase domain (Supplementary Figure S5).
The sequence alignment of the kinase domain from OsWAK112 with those from several kinases in Arabidopsis is shown in Figure 4A. A typical ATP-binding motif and kinase active site were located in the intracellular domain of OsWAK112 (Figure 4A), indicating that it is an active kinase. Although it is predicted that OsWAK112 is linked to the cell wall via its cytosolic kinase domain (Figure 4A), whether WAK112 has kinase activity is unknown. To test this, the lysine (Lys-678) in the ATP-binding region and lysine (Lys-791) and aspartic acid (Asp794) in the activation loop of the kinase domain (KD) were mutated to generate WAK112KDK678E, WAK112KDK791R, and WAK112KDD794A, respectively (Figure 4B). The wild-type and mutated forms of the WAK112 KD were produced as fusions with GST. An in vitro kinase assay showed that the OsWAK112KD produced strong autophosphorylation and phosphorylation of MBP (Figure 4B, lanes 1 and 6). WAK112KDK791R also showed strong autophosphorylation (Figure 4B, lane 3), but the mutated forms (GST-OsWAK112K678E and GST-OsWAK112D794A; Figure 4B, lanes 2 and 4) and GST alone (Figure 4B, lane 5) did not exhibit autophosphorylation activity, indicating that OsWAK112 is an active RLK and that both Lys-678 and Asp-794 are necessary for its activity.
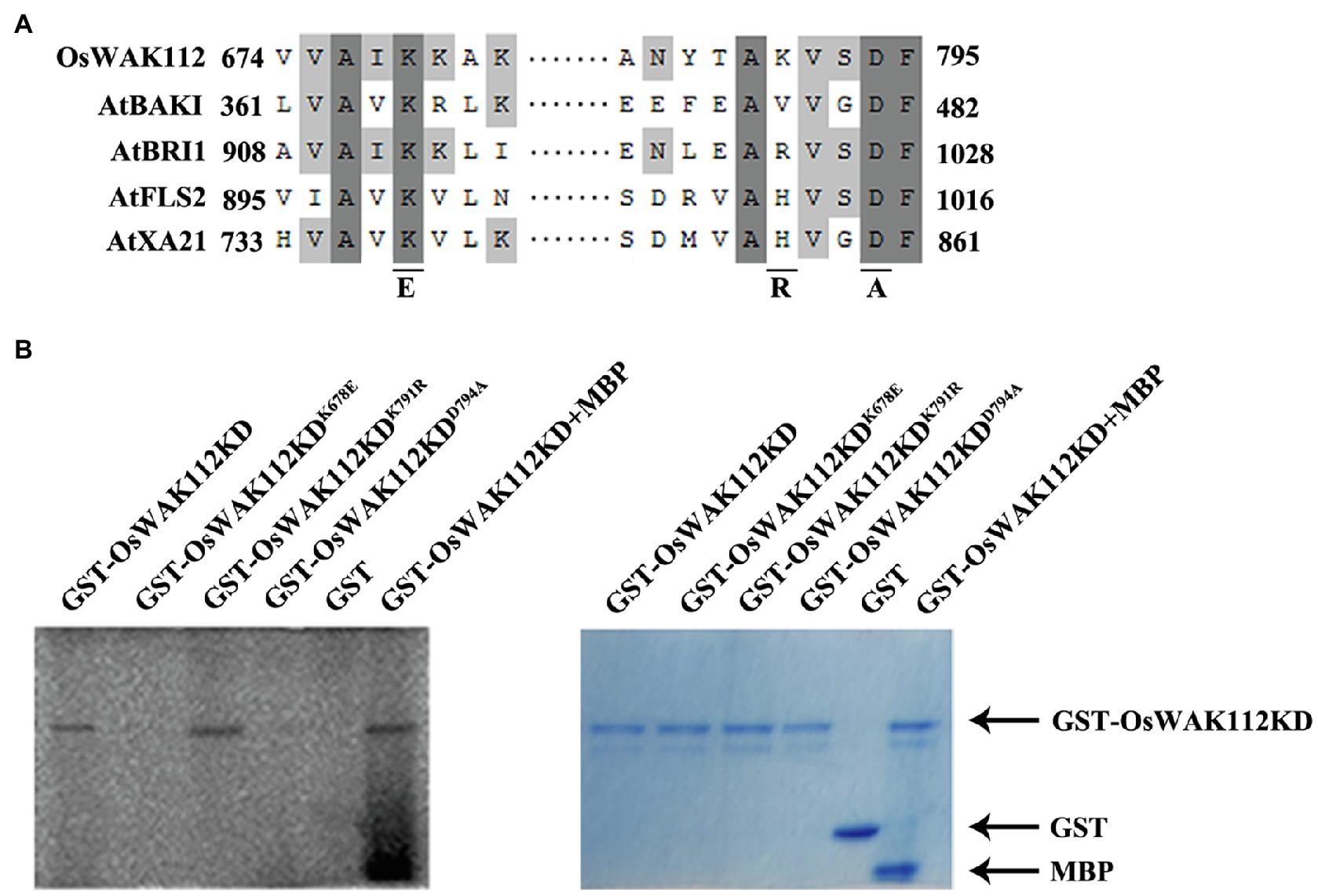
Figure 4. OsWAK112 possesses kinase activity. (A) Alignment of a conserved motif from the OsWAK112KD with that from other known receptor-like kinases (RLKs). Underlines indicate conserved residues in the active kinase that were mutated to the indicated amino acid in the following experiment. (B) GST-OsWAK112KD showed autophosphorylation and substrate (MBP) phosphorylation. The mutated form GST-WAK112KDK791R also showed autophosphorylation, whereas the mutated forms GST-OsWAK112KDK678E and GST-OsWAK112KDD794A did not. The left panel shows autoradiography and the right panel Coomassie blue (CBB) staining of the gel.
To assess the contribution of the kinase activity of OsWAK112 to plant salt sensitivity, we generated transgenic Arabidopsis expressing OsWAK112, OsWAK112K678E, and OsWAK112D794A as Myc fusions under the control of the 35S promoter. Western blotting showed that wild-type and mutated forms of OsWAK112 were well expressed in Arabidopsis (Supplementary Figure S6). On regular medium, little difference was noted among the seedlings (Figures 5A,C, 0mM NaCl). On 0.5×MS medium containing 125mM NaCl, the WAK112-overexpressing seedlings (OE10 and OE11) exhibited more strongly bleached cotyledons (Figure 5A) and lower survival rates than wild-type Col seedlings (57% for OE10 and 50% for OE11, compared with 63% for Col; Figure 5B). This is consistent with the effects of overexpressed OsWAK112 on salt hypersensitivity in rice (Figures 1, 2). However, the Arabidopsis 35S::OsWAK112K678E (named OE12 and OE13) and 35S::OsWAK112D794A (named OE14 and OE15) seedlings showed less damage in response to salt exposure, with around 70–86% survival (higher than that of Col; Figures 5C,D). These results suggest that the kinase activity of OsWAK112 affects salt resistance as a negative regulator.
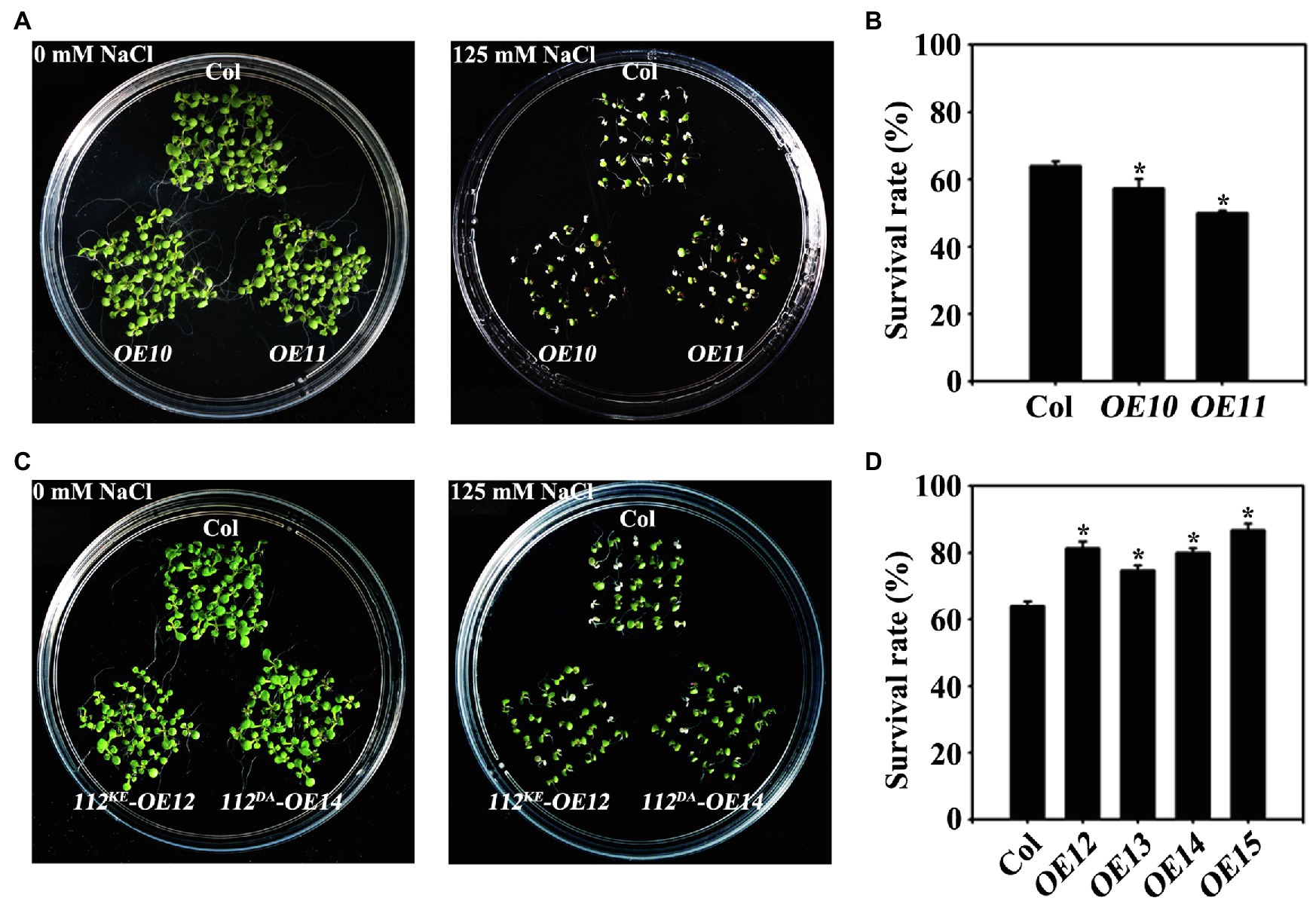
Figure 5. OsWAK112 mediates salt sensitivity depending on its kinase activity. Arabidopsis plants overexpressing OsWAK112 showed hypersensitivity to salt compared with Col (A,B), while Arabidopsis plants overexpressing OsWAK112K678E and OsWAK112D794A showed hyposensitivity to salt compared with Col (C,D). The indicated seedlings were grown on 0.5×MS medium supplemented with or without 125mM NaCl for 10days, and the salt survival rate was calculated. OE10 and OE11 represent two independent 35S:OsWAK112-Myc/Col transgenic lines. OE12 and OE13 represent two independent 35S:OsWAK112K678E-Myc/Col transgenic lines. OE14 and OE15 represent two independent 35S:OsWAK112D794A-Myc/Col transgenic lines. The experiments were repeated three times. Each data point represents the mean ± SE (n≥25). Asterisks indicate a difference relative to Col (Student’s t-test, *p<0.05) in (B-D).
Our work indicated that OsWAK112 plays a negative role in salt tolerance. As a wall-associated protein, it is likely that OsWAK112 confers salt response by directly binding to specific target proteins. Then, we sought to identify its interacting proteins to gain insight into the roles of OsWAK112 in salt response.
According to a structural analysis conducted using WebLab ViewerLite (Accelrys, San Diego, CA), several salt-associated proteins harbor important binding elements for OsWAK112, including GRP1, GRP2, GRP3, OsRP-1, PSBP1, and SAMSs (without OsACS and OsACO) as WAK112 candidate interacting proteins. Firstly, we performed BiFC assay to test whether OsWAK112 interacted with these proteins. The sequences encoding GRP1, GRP2, GRP3, OsRP-1, OsPSBP1, SAMS1/2/3, and OsWAK112 were fused with those sequences encoding the N-terminal half and C-terminal half of YFP to generate GRPs-CCFP, OsRP-1-CCFP, OsPSBP1-CCFP, OsSAMSs-CCFP, and OsWAK112-NYFP, respectively. The constructs were then co-infiltrated into N. benthamiana leaves. When the OsGRP1, OsGRP2, OsGRP3, OsRP-1, and OsPSB1 fusion with CCFP co-filtrated in OsWAK112 fusion with NYFP into N. benthamiana leaves, no fluorescence signal was observed (data not shown). However, co-expression of OsSAMS1/2/3-CCFP and OsWAK112-NYFP resulted in a strong YFP signal, while the negative control (OsWAK112-NYFP+YCE) produced almost no signal (Figure 6A), demonstrating the specific interaction of OsWAK112 with OsSAMSs in vivo. The direct interaction between OsWAK112 and OsSAMS1/2/3 was then examined in overlay assays. Purified MBP-OsSAMS1, MBP-OsSAMS2, and MBP-OsSAMS3 were used as primary antibodies and incubated with PVDF membranes containing GST and GST-OsWAK112KD; MBP-HRP antibodies were used as secondary antibodies. MBP-OsSAMS1, MBP-OsSAMS2, and MBP-SAMS3 were detected at positions corresponding to the size of GST-OsWAK112KD, but no band was detected at the position of the GST tag, suggesting a direct interaction between OsWAK112 and OsSAMS1/2/3 in vitro (Figure 6B). In addition, the overlay assay showed that D794A mutation in OsWAK112 did not affect its interaction with OsSAMS1 (Supplementary Figure S7), indicating the conserved aspartic acid or kinase activity was not required for this interaction. Furthermore, their interaction was further tested using Co-IP assays. The sequences encoding OsSAMS1/2/3 and OsWAK112KD were fused with GFP and Myc to generate OsSAMSs-GFP and OsWAK112KD-Myc, respectively. The constructs were then co-infiltrated into N. benthamiana leaves. Co-IP was achieved using N. benthamiana leaves transiently expressing OsSAMSs-GFP and OsWAK112KD-Myc. OsWAK112KD-Myc could be co-precipitated only when OsSAMSs-GFP was present (Figure 6C), confirming the interaction between OsWAK112 and OsSAMS1, OsSAMS2, or OsSAMS3 in vivo.
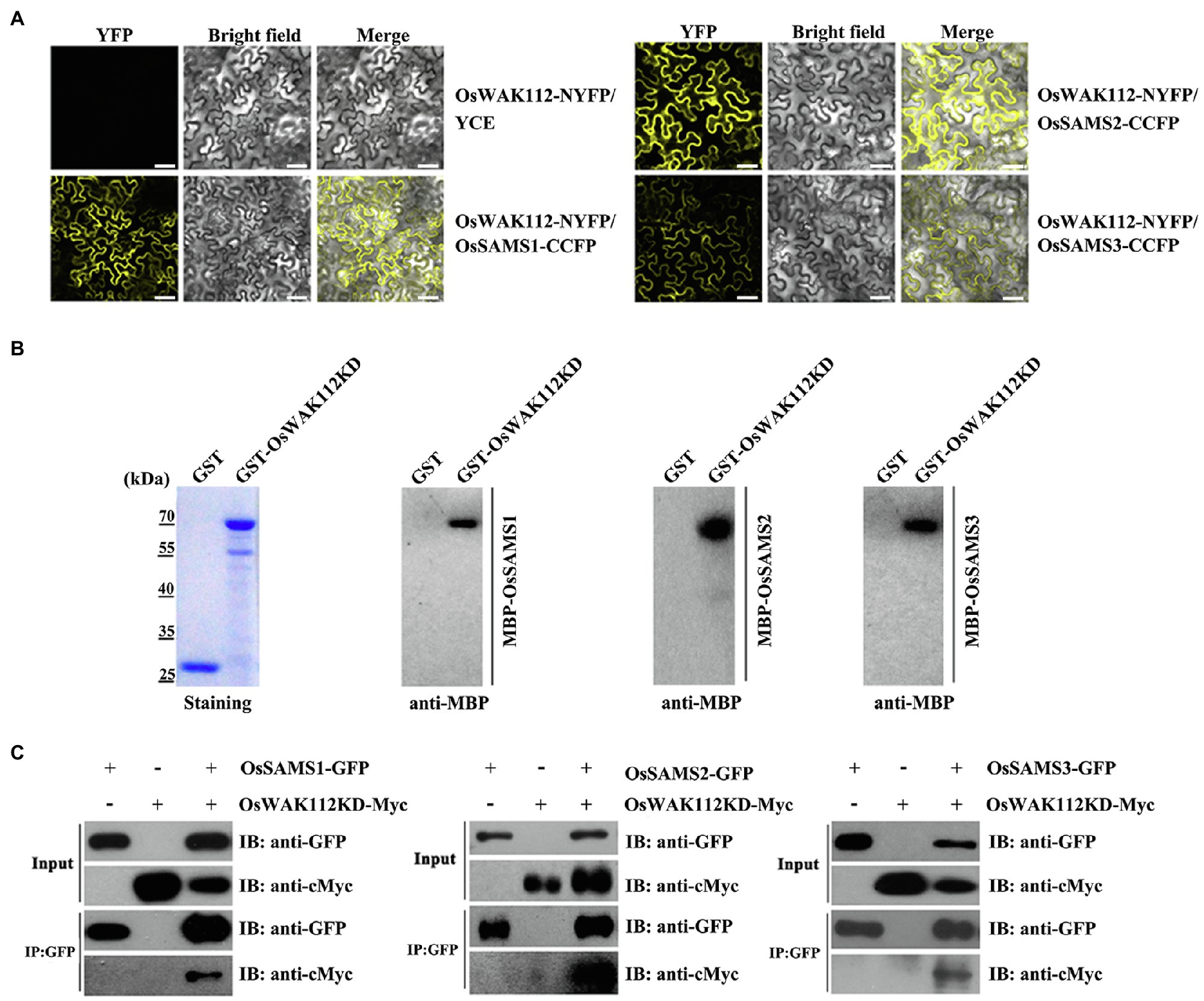
Figure 6. OsWAK112 interacts with OsSAMS1, OsSAMS2, and OsSAMS3 in vitro and in vivo. (A) MBP-OsSAMSs bound to the GST-WAK112KD fusion but not to GST in a gel blot overlay assay. Coomassie blue staining in the left panel indicates the loading of GST-WAK112KD and GST. Bound proteins were probed with anti-MBP-HRP antibodies. OsWAK112 interacted with OsSAMSs in BiFC assays (B) and Co-IP assays (C) in transiently transformed N. benthamiana leaves. Scale bar=50μm in (B).
Together, these results suggest that OsWAK112 interacts physically with OsSAMS1/2/3 in vitro and in vivo. Next, the biological function of the OsWAK112–OsSAMS1 interaction was investigated in tobacco plants. A co-infiltration assay showed that OsSAMS1-GFP was well expressed in tobacco. Compared with mock treatment, 2h of treatment with 200mM NaCl decreased the levels of OsSAMS1 when co-expressed with OsWAK112, indicating that OsWAK112 affects OsSAMS1 stability under saline conditions (Supplementary Figure S8). Together, these data suggest that OsWAK112 interacts with OsSAMS1/2/3 and promotes OsSAMS1 degradation in the presence of salt.
To investigate the influence of OsWAK112 on OsSAMS contents in transgenic plants, the SAMS contents were measured in OE7, OE8, and Jap plants. The data showed that SAMS contents were not significantly different between Jap and OE7/8 under normal conditions, but the SAMS levels were lower in OE7 and OE8 than in Jap following salt treatment (Figure 7A).
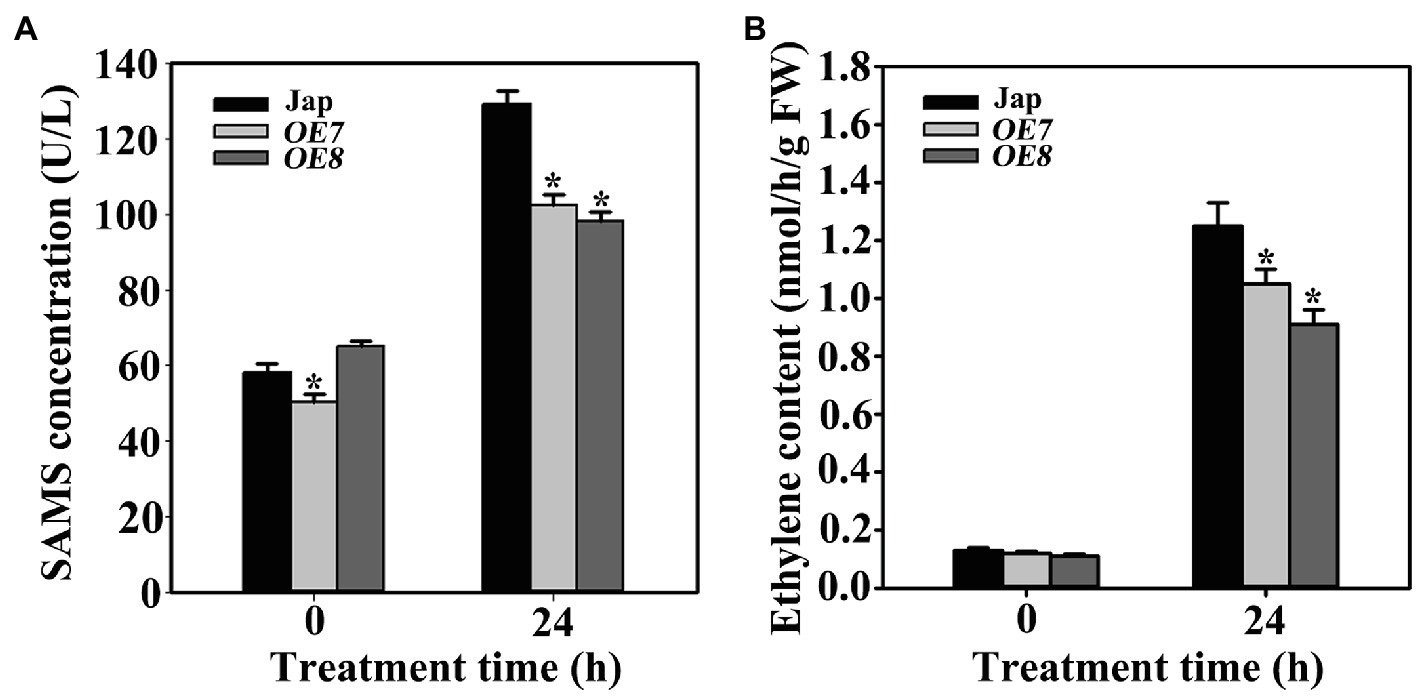
Figure 7. OsWAK112 affects ethylene production under salt stress. (A) The amount of ethylene per gram fresh weight in 7-day-old Jap, OE7, and OE8 seedlings grown under normal conditions or treated with 200mM NaCl for 24h. (B) The SAMS contents of 7-day-old Jap, OE7, and OE8 seedlings grown under normal conditions or treated with 200mM NaCl for 24h. In (A,B), Student’s t-tests were conducted using data from transgenic seedlings compared to those from Jap. Asterisks indicate a significant difference at p<0.05.
As a major abiotic stress, salt can trigger the production of stress-related hormones and initiate endogenous hormone signaling, resulting in plant resistance reactions (Zhao et al., 2021b). Ethylene is a stress hormone closely related with salt stress (Riyazuddin et al., 2020). In plants, ethylene is biosynthesized from S-adenosyl-L-methionine (SAM; Zhao et al., 2021a), which is synthesized from l-methionine and ATP catalyzed by SAMSs. To explore the mechanism underlying the function of OsWAK112 in plant salt responses, the ethylene content was measured in Jap and OsWAK112OE plants. Under normal growth conditions, the ethylene contents did not differ much between Jap and the OsWAK112OE lines. After salt treatment, the ethylene contents increased in all of the plants; however, they were lower in OE7 and OE8 than in Jap (Figure 7B), while they were higher in RNAi1 and RNAi3 than in Jap (Supplementary Figure S9). Together, these data suggest that OsWAK112 interacts with OsSAMS1/2/3 and promotes OsSAMS1 degradation, resulting in decreased ethylene production in the presence of salt.
Soil salinity is a significant threat to the growth, productivity, and quality of crop plants. During plant salt stress responses, multiple signaling cascade components are activated. RLKs have previously been shown to be involved in salt responses (de Lorenzo et al., 2009; Ouyang et al., 2010; Lin et al., 2020; Passricha et al., 2020; Yuchun et al., 2021); however, the function of a unique group of RLKs, known as WAKs, in salt tolerance remains unclear. WAKs likely associate with pectin in the cell wall and are necessary for both cell expansion during development and mediation of the response to stresses (Kanneganti and Gupta, 2011; Wang et al., 2012; Delteil et al., 2016). In the present study, we found that OsWAK112 functions as a kinase to negatively mediate plant salt responses by reducing the ethylene content via a direct interaction with OsSAMS1.
To our knowledge, OsWAK112 has only been shown to play a negative role in biotic stress tolerance in rice (Delteil et al., 2016); there are no reports of its function in response to abiotic stresses. The time-dependent suppression of OsWAK112 by NaCl treatment (Supplementary Figure S1 ) suggests that OsWAK112 is closely related to plant responses to high salinity. When OsWAK112 was overexpressed in rice, two independent lines (OE7 and OE8) showed hypersensitivity to salt stress. Further, the higher OsWAK112 expression level and the lower the survival rate in OE8 and OE7 (Figures 1, 2), and the less OsWAK112 expression level and the higher survival rate in RNAi lines compared with Jap (Supplementary Figure S2), indicate that the OsWAK112 expression level correlates negatively with plant salt resistance. These findings from morphological and physiological measurements indicate that OsWAK112 is a negative regulator of salt resistance reactions in rice.
Salinity stress induces ROS accumulation. Low levels of ROS usually act as signal molecules in salt signaling networks, but excessive accumulation of ROS is highly toxic to proteins, lipids, and nucleic acids, resulting in cell death (You and Chan, 2015). H2O2 is an especially stable ROS in plant cells. We found that more H2O2 accumulated in OsWAK112OE (OE7/8) plants under salt stress and that the variation in their H2O2 levels was consistent with that of their OsWAK112 expression levels (Supplementary Figure S3). Excessive accumulation of H2O2 accompanied by strong OsWAK112 expression under conditions of salt stress may damage plant cells, resulting in reduced survival.
As a kinase, how does OsWAK112 coordinately regulate plant responses to salt stress? It would be a worthwhile question to investigate.
Each WAK protein contains an extracellular calcium-binding domain, a transmembrane domain, and a cytoplasmic serine/threonine kinase domain (Kohorn and Kohorn, 2012). The extracellular domain senses the signal, while the cytosolic protein kinase domain translates the signal to downstream components via phosphorylation. OsWAK112 contains an arginine residue in its kinase catalytic motif (RDxxxxN) and is classified as a WAK-RD kinase (de Oliveira et al., 2014). Several RLKs have been shown to be involved in salt responses depending on their kinase activity. For example, SIT1, a lectin RLK, negatively regulates salt responses in rice, and it kinase activity plays an important role in its salt response (Li et al., 2014). Another receptor-like cytoplasmic kinase, STRK1, phosphorylates and stimulates the activity of catalase C to adjust H2O2 concentrations and improve salt tolerance (Zhou et al., 2018). Herein, we used an in vitro kinase assay to show that OsWAK112 has autophosphorylation activity and strong substrate activity (Figure 4). We found that Lys-791 near the DFG motif of kinase domain VII produced the same autophosphorylation signal as wild-type OsWAK112 in vitro (Figure 4); therefore, it appears that Lys-791 is not essential for the kinase activity of the enzyme in vitro. That may be because Lys-791 is not conserved in OsWAK112 compared with other known kinases. Our results show that OsWAK112 exhibited kinase activity depending on its conserved residues Lys-678 and Asp-794 (Figure 4). Lys-678 is in the AXK motif of kinase domain II for interaction with the phosphates of ATP. Asp-794 in the DFG motif of kinase domain VII chelates a Mg2+ ion to orient the γ-phosphate of ATP for transfer (Hanks and Hunter, 1995). When Lys-678 or Asp-794 was mutated, OsWAK112 lost its kinase activity (Figure 4). Exogenous expression of OsWAK112 in Arabidopsis made the transgenic plants hypersensitive to salt (Figure 5), indicating that OsWAK112 functions as a negative regulator of salt responses in both Arabidopsis and rice. However, exogenous overexpression of OsWAK112K678E or OsWAK112D794A did not increase the sensitivity of Arabidopsis to salt (Figure 5). A plausible explanation is that kinase-dead forms of OsWAK112 could still sense the salt stimulus and bind to downstream substrates, but the signal could not be transduced due to the loss of kinase activity (Figure 4), resulting in a dominant-negative phenotype. For example, in plants, when ERECTA lack the cytoplasmic kinase domain it acts as dominant-negative receptors by blocking the normal activity of the endogenous counterparts (Shpak et al., 2003). And in plant hormone brassinosteroids signaling pathway, the receptor kinase BRI1 phosphorylates the substrate BSK3 on S215. When S215 mutated into A, S215A had a dominant-negative effect: Plants overexpressing the S215A version of OsBSK3 were smaller and dwarfed than the nontransgenic controls when grown in the light under the same conditions (Zhang et al., 2016).
Ethylene homeostasis is tightly controlled to maintain its dual functions in growth inhibition and growth stimulation. Under normal conditions, the ethylene concentration is low and only increases dramatically at defined developmental stages, such as fruit ripening (Zhao et al., 2021a). A variety of environmental stimuli, including salt stress, can also induce ethylene production (Nascimento et al., 2018; Riyazuddin et al., 2020), in which the ACC synthase-induced conversion of SAM to ACC is a key step. SAM is synthesized by SAMSs, which play important roles in regulating plant–environment interactions. Studies show that SAMSs can be induced by multiple stress treatments and that they play important roles in regulating plant tolerance to environmental changes, especially soil salinity and drought. For example, in sugar beet, BvM14-SAMS2 is induced by salt, and its ectopic expression in Arabidopsis plants increased salt and oxidation resistance (Ma et al., 2017). In tobacco, overexpression of SlSAMS1 affects ethylene emission and improves drought and salt stress tolerance (Zhang et al., 2020). We found that OsWAK112 interacted directly with OsSAMS1/2/3 in overlay, BiFC, and Co-IP assays (Figure 6). Among these three SAMSs in rice, OsSAMS1 plays a predominant role and is expressed abundantly in every tissue (Li et al., 2011). We found that OsWAK112 promoted OsSAMS1 degradation under high-salt conditions (Supplementary Figure S8). There was a lower OsSAMS content and a reduced ethylene content in our OsWAK112OE plants, following salt treatment (Figure 7), while a higher ethylene existed in OsWAK112 RNAi plants content under salt stress. Consistent with our study, OsARD1, a metal-binding metalloenzyme involved in the methionine salvage pathway, enhances ethylene biosynthesis and reduces the salt sensitivity of rice (Liang et al., 2019). These data suggest that OsWAK112 is an active kinase that interacts with OsSAMS1/2/3, promoting OsSAMS1 degradation, and reducing ethylene production under saline conditions.
Soil salinity is a serious threat to crop growth and agricultural productivity. Our findings demonstrate that salt sensitivity in rice is mediated by OsWAK112 depending on its kinase activity. These findings provide important new information for engineering salt-tolerant crops.
Sequence data from this article can be found in GenBank/EMBL under the following accession numbers: OsWAK112 (Os10g10130), OsSAMS1 (Os05g04510), OsSAMS2 (Os01g22010), and OsSAMS3 (Os01g18860).
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.
LZ conceived the project and designed the research. LL and YW carried out most of the experiments, phenotypic observations, RT-qPCR analysis, the rice and Arabidopsis transgenic experiments, and Western blot analysis and data analysis. XL participated in the data analysis. LZ and J-XS wrote the article with contributions from all authors and revised and proofread the manuscript. All authors participated in revising the manuscript and approved the final version.
This work was supported by grants from the National Natural Science Foundation of China (31770297 to LZ and 31872830 to J-XS) and the Department of Education of Hebei Province (BJ2019025 to J-XS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.751965/full#supplementary-material
Bot, P., Mun, B. G., Imran, Q. M., Hussain, A., Lee, S. U., Loake, G., et al. (2019). Differential expression of AtWAKL10 in response to nitric oxide suggests a putative role in biotic and abiotic stress responses. Peer J. 7:e7383. doi: 10.7717/peerj.7383
de Lorenzo, L., Merchan, F., Laporte, P., Thompson, R., Clarke, J., Sousa, C., et al. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21, 668–680. doi: 10.1105/tpc.108.059576
de Oliveira, L. F. V., Christoff, A. P., de Lima, J. C., de Ross, B. C. F., Sachetto-Martins, G., Margis-Pinheiro, M., et al. (2014). The wall-associated kinase gene family in rice genomes. Plant Sci. 229, 181–192. doi: 10.1016/j.plantsci.2014.09.007
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Delteil, A., Gobbato, E., Cayrol, B., Estevan, J., Michel-Romiti, C., Dievart, A., et al. (2016). Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16, 1–10. doi: 10.1186/s12870-016-0711-x
Diener, A. C., and Ausubel, F. M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. doi: 10.1534/genetics.105.042218
Gao, L. L., and Xue, H. W. (2012). Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 5, 143–153. doi: 10.1093/mp/ssr062
Hanin, M., Ebel, C., Ngom, M., Laplaze, L., and Masmoudi, K. (2016). New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 7:1787. doi: 10.3389/fpls.2016.01787
Hanks, S. K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596. doi: 10.1096/fasebj.9.8.7768349
He, Z. H., He, D., and Kohorn, B. D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63. doi: 10.1046/j.1365-313x.1998.00092.x
Hou, X., Tong, H., Selby, J., Dewitt, J., Peng, X., and He, Z. H. (2005). Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 139, 1704–1716. doi: 10.1104/pp.105.066910
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci 112, 8780–8785. doi: 10.1073/pnas.1502522112
Jiang, C., Belfield, E. J., Cao, Y., Smith, J. A., and Harberd, N. P. (2013). An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25, 3535–3552. doi: 10.1105/tpc.113.115659
Kanneganti, V., and Gupta, A. K. (2011). RNAi mediated silencing of a wall associated kinase, OsiWAK1 in Oryza sativa results in impaired root development and sterility due to anther indehiscence: wall associated kinases from Oryza sativa. Physiol. Mol. Biol. Plants 17, 65–77. doi: 10.1007/s12298-011-0050-1
Kohorn, B. D. (2016). Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67, 489–494. doi: 10.1093/jxb/erv467
Kohorn, B. D., and Kohorn, S. L. (2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3:88. doi: 10.3389/fpls.2012.00088
Lally, D., Ingmire, P., Tong, H. Y., and He, Z. H. (2001). Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13, 1317–1331. doi: 10.1105/tpc.13.6.1317
Lei, G., Shen, M., Li, Z. G., Zhang, B., Duan, K. X., Wang, N., et al. (2011). EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 34, 1678–1692. doi: 10.1111/j.1365-3040.2011.02363.x
Li, W., Han, Y., Tao, F., and Chong, K. (2011). Knockdown of SAMS genes encoding S-adenosyl-l-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J. Plant Physiol. 168, 1837–1843. doi: 10.1016/j.jplph.2011.05.020
Li, C. H., Wang, G., Zhao, J. L., Zhang, L. Q., Ai, L. F., Han, Y. F., et al. (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. doi: 10.1105/tpc.114.125187
Li, H., Zhou, S. Y., Zhao, W. S., Su, S. C., and Peng, Y. L. (2009). A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69, 337–346. doi: 10.1007/s11103-008-9430-5
Liang, S., Xiong, W., Yin, C., Xie, X., Jin, Y. J., Zhang, S., et al. (2019). Overexpression of OsARD1 improves submergence, drought, and salt tolerances of seedling through the enhancement of ethylene synthesis in rice. Front. Plant Sci. 10:1088. doi: 10.3389/fpls.2019.01088
Lin, F., Li, S., Wang, K., Tian, H., Gao, J., Zhao, Q., et al. (2020). A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 296:110465. doi: 10.1016/j.plantsci.2020.110465
Ma, C., Wang, Y., Gu, D., Nan, J., Chen, S., and Li, H. (2017). Overexpression of S-adenosyl-l-methionine synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. Int. J. Mol. Sci. 18:847. doi: 10.3390/ijms18040847
Meco, V., Egea, I., Ortiz-Atienza, A., Drevensek, S., Esch, E., Yuste-Lisbona, F. J., et al. (2020). The salt sensitivity induced by disruption of cell wall-associated kinase 1 (SlWAK1) tomato gene is linked to altered osmotic and metabolic homeostasis. Int. J. Mol. Sci. 21:6308. doi: 10.3390/ijms21176308
Mishra, P., Singh, N., Jain, A., Jain, N., Mishra, V., G, P., et al. (2018). Identification of cis-regulatory elements associated with salinity and drought stress tolerance in rice from co-expressed gene interaction networks. Bioinformation 14, 123–131. doi: 10.6026/97320630014123
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nascimento, F. X., Rossi, M. J., and Glick, B. R. (2018). Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant-bacterial interactions. Front. Plant Sci. 9:114. doi: 10.3389/fpls.2018.00114
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Passricha, N., Saifi, S. K., Kharb, P., and Tuteja, N. (2020). Rice lectin receptor-like kinase provides salinity tolerance by ion homeostasis. Biotechnol. Bioeng. 117, 498–510. doi: 10.1002/bit.27216
Pattyn, J., Vaughan-Hirsch, J., and Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229, 770–782. doi: 10.1111/nph.16873
Peng, J., Li, Z., Wen, X., Li, W., Shi, H., Yang, L., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10:e1004664. doi: 10.1371/journal.pgen.1004664
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. doi: 10.1038/ng1643
Riyazuddin, R., Verma, R., Singh, K., Nisha, N., Keisham, M., Bhati, K. K., et al. (2020). Ethylene: a master regulator of salinity stress tolerance in plants. Biomol. Ther. 10:959. doi: 10.3390/biom10060959
Rosli, H. G., Zheng, Y., Pombo, M. A., Zhong, S., Bombarely, A., Fei, Z., et al. (2013). Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 14, 1–15. doi: 10.1186/gb-2013-14-12-r139
Saintenac, C., Lee, W. S., Cambon, F., Rudd, J. J., King, R. C., Marande, W., et al. (2018). Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50, 368–374. doi: 10.1038/s41588-018-0051-x
Shpak, E. D., Lakeman, M. B., and Torii, K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. doi: 10.1105/tpc.010413
Sivaguru, M., Ezaki, B., He, Z. H., Tong, H., Osawa, H., Baluska, F., et al. (2003). Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 132, 2256–2266. doi: 10.1104/pp.103.022129
Sreenivasulu, N., Miranda, M., Prakash, H. S., Wobus, U., and Weschke, W. (2004). Transcriptome changes in foxtail millet genotypes at high salinity: identification and characterization of a PHGPX gene specifically upregulated by NaCl in a salt-tolerant line. J. Plant Physiol. 161, 467–477. doi: 10.1078/0176-1617-01112
Verica, J. A., and He, Z. H. (2002). The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129, 455–459. doi: 10.1104/pp.011028
Wagner, T. A., and Kohorn, B. D. (2001). Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13, 303–318. doi: 10.1105/tpc.13.2.303
Wang, Z., Chen, C., Xu, Y., Jiang, R., Han, Y., Xu, Z., et al. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant. Mol. Biol. Rep. 22, 409–417. doi: 10.1007/BF02772683
Wang, N., Huang, H. J., Ren, S. T., Li, J. J., Sun, Y., Sun, D. Y., et al. (2012). The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 160, 696–707. doi: 10.1104/pp.112.203943
Wang, C., Shang, J. X., Chen, Q. X., Oses-Prieto, J. A., Bai, M. Y., Yang, Y., et al. (2013). Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics. 12, 3653–3665. doi: 10.1074/mcp.M113.029256
Wang, W., Zhang, J., Ai, L., Wu, D., Li, B., Zhang, L., et al. (2021). Cyclic nucleotide-gated ion channel 6 mediates thermotolerance in Arabidopsis seedlings by regulating hydrogen peroxide production via cytosolic calcium ions. Front. Plant Sci. 12:708672. doi: 10.3389/fpls.2021.700928
Xin, J., Li, C., Ning, K., Qin, Y., Shang, J. X., and Sun, Y. (2021). AtPFA-DSP3, an atypical dual-specificity protein tyrosine phosphatase, affects salt stress response by modulating MPK3 and MPK6 activity. Plant Cell Environ. 44, 1534–1548. doi: 10.1111/pce.14002
Yang, Y., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yang, L., Zu, Y. G., and Tang, Z. H. (2013). Ethylene improves Arabidopsis salt tolerance mainly via retaining K+ in shoots and roots rather than decreasing tissue Na+ content. Environ. Exp. Bot. 86, 60–69. doi: 10.1016/j.envexpbot.2010.08.006
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yuchun, R. A. O., Ran, J., Sheng, W., Xianmei, W. U., Hanfei, Y. E., Chenyang, P. A. N., et al. (2021). SPL36 encodes a receptor-like protein kinase that regulates programmed cell death and defense responses in rice. Rice 14, 1–14. doi: 10.1186/s12284-021-00475-y
Zhang, X., Bao, Z. L., Gong, B., and Shi, Q. H. (2020). S-adenosylmethionine synthetase 1 confers drought and salt tolerance in transgenic tomato. Environ. Exp. Bot. 179:104226. doi: 10.1016/j.envexpbot.2020.104226
Zhang, S., Chen, C., Li, L., Meng, L., Singh, J., Jiang, N., et al. (2005). Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139, 1107–1124. doi: 10.1104/pp.105.069005
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, B., Wang, X., Zhao, Z., Wang, R., Huang, X., Zhu, Y., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170, 1149–1161. doi: 10.1104/pp.15.01668
Zhao, H., Yin, C. C., Ma, B., Chen, S. Y., and Zhang, J. S. (2021a). Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J. Integr. Plant Biol. 63, 102–125. doi: 10.1111/jipb.13028
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., and Wang, P. (2021b). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22:4609. doi: 10.3390/ijms22094609
Zhou, Y. B., Liu, C., Tang, D. Y., Yan, L., Wang, D., Yang, Y. Z., et al. (2018). The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell 30, 1100–1118. doi: 10.1105/tpc.17.01000
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: Oryza sativa, salt, wall-associated kinase, ethylene, S-adenosyl-L-methionine synthetases
Citation: Lin W, Wang Y, Liu X, Shang J-X and Zhao L (2021) OsWAK112, A Wall-Associated Kinase, Negatively Regulates Salt Stress Responses by Inhibiting Ethylene Production. Front. Plant Sci. 12:751965. doi: 10.3389/fpls.2021.751965
Received: 02 August 2021; Accepted: 06 September 2021;
Published: 05 October 2021.
Edited by:
Quan-Sheng Qiu, Lanzhou University, ChinaReviewed by:
Changxi Yin, Huazhong Agricultural University, ChinaCopyright © 2021 Lin, Wang, Liu, Shang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqun Zhao, emhhb2xxNzBAMTYzLmNvbQ==, orcid.org/0000-0001-6718-8130; Jian-Xiu Shang, c2hhbmdqaWFueGl1QG1haWwuaGVidHUuZWR1LmNu, orcid.org/0000-0002-9007-4294
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.