
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 January 2022
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.751429
This article is part of the Research TopicAccelerating Genetic Gains in PulsesView all 15 articles
 Kuldeep Tripathi1*
Kuldeep Tripathi1* Jyoti Kumari1
Jyoti Kumari1 Padmavati G. Gore1
Padmavati G. Gore1 Dwijesh C. Mishra2
Dwijesh C. Mishra2 Amit Kumar Singh1
Amit Kumar Singh1 Gyan P. Mishra3
Gyan P. Mishra3 Gayacharan C.1
Gayacharan C.1 H. K. Dikshit3
H. K. Dikshit3 Neeta Singh1
Neeta Singh1 D. P. Semwal1
D. P. Semwal1 Reena Mehra4
Reena Mehra4 Rakesh Bhardwaj1
Rakesh Bhardwaj1 Ruchi Bansal1
Ruchi Bansal1 J. C. Rana5
J. C. Rana5 Ashok Kumar1*
Ashok Kumar1* Veena Gupta1
Veena Gupta1 Kuldeep Singh1
Kuldeep Singh1 Ashutosh Sarker4*
Ashutosh Sarker4*Lentil (Lens culinaris Medik.) is one of the major cool-season pulse crops worldwide. Its increasing demand as a staple pulse has led to the unlocking of diverse germplasm collections conserved in the genebanks to develop its superior varieties. The Indian National Genebank, housed at the Indian Council of Agricultural Research (ICAR)-National Bureau of Plant Genetic Resources, New Delhi, India, currently has 2,324 accessions comprising 1,796 indigenous and 528 exotic collections. This study was conducted to unveil the potential of lentil germplasm by assessing its agro-morphological characteristics and diversity, identifying trait-specific germplasm, and developing a core set. The complete germplasm set was characterized for two years, i.e., 2017–2018 and 2018–2019, and data were recorded on 26 agro-morphological traits. High phenotypic variability was observed for nine quantitative and 17 qualitative traits. A core set comprising 170 accessions (137 Indian and 33 exotic) was derived based on the characterization data as well as geographical origin using a heuristic method and PowerCore software. This core set was found to be sufficiently diverse and representative of the entire collection based on the comparison made using Shannon–Weaver diversity indices and χ2 test. These results were further validated by summary statistics. The core set displayed high genetic diversity as evident from a higher coefficient of variance in comparison to the entire set for individual traits and overall Shannon–Weaver diversity indices (entire: 1.054; core: 1.361). In addition, the total variation explained by the first three principal components was higher in the core set (70.69%) than in the entire collection (68.03%). Further, the conservation of pairwise correlation values among descriptors in the entire and core set reflected the maintenance of the structure of the whole set. Based on the results, this core set is believed to represent the entire collection, completely. Therefore, it constitutes a potential set of germplasm that can be used in the genetic enhancement of lentils.
Lentil (Lens culinaris Medik.; 2n = 2 × = 14) is a diploid, self- pollinating, and cool-season pulse that is grown worldwide (Kumar and Gupta, 2020). It is regarded as one of the founder crops of neolithic agriculture (Zohary and Hopf, 1973). In addition, the global productivity of lentils has increased from an average yield of 806 kg ha–1 to 1194.6 kg ha–1 during the last two decades (FAOSTAT, 2021). In India, lentil ranks as the second-most important winter pulse crop after chickpea. In 2019, 1.23 million tons of lentil was produced in India, with a mean productivity of average yield of 901 kg ha–1 (FAOSTAT, 2021). However, India’s current yield of lentils is considerably lower than that of several other countries because of the poor yield of cultivars.
Poor seedling vigor, low biomass, delicate stem, low harvest index, lodging, less conversion of flower to the pod, and climate-induced stresses are the primary yield-reducing factors in lentils (Erskine et al., 2009). In addition, the narrow genetic base or limited parentage of modern varieties has emerged as a major concern for lentil improvement. Consequently, the potential genetic gains in lentil productivity could not be achieved. Genebanks are the source of genes exhibiting valuable traits that can be utilized not only to develop superior varieties but also those with tolerance to stresses induced due to changing climate (Díez et al., 2018). Therefore, it is essential to search genebanks and identify novel germplasm for its use in lentil-breeding programs to enhance cultivar productivity and resilience to climate change. The efficient use of genetic diversity in the varietal development program is an effective method to enhance yield gains and cope with the emerging climate-induced stresses.
To identify novel accessions corresponding to the genes with desired traits, it is prudent to characterize the germplasm collections conserved in the genebanks. However, the characterization of extensive collections by plant breeders is time- and resource-consuming, genebank curators have developed a concept of a core set from entire collections that are characterized at once to identify the targeted germplasm and use it efficiently (Frankel and Brown, 1984; Brown, 1989; van Hintum et al., 2000). The core set refers to a minimum set of germplasm that captures the entire range of genetic variability of any crop, with minimum repetitiveness. Being smaller in size and diverse in nature, the core set can be efficiently used as a kickoff point to enhance genetic gains, including the use of phenomics and genomics tools in less time. Globally, legume germplasm curators have developed core sets in soybean (Oliveira et al., 2010), cowpea (Mahalakshmi et al., 2007), pigeonpea (Reddy et al., 2005), groundnut (Upadhyaya et al., 2003), chickpea (Upadhyaya et al., 2001), lentil (Simon and Hannan, 1995; Tullu et al., 2001), and lablab bean (Vaijayanthi et al., 2015) to enhance the germplasm use. The Indian Council of Agricultural Research-National Bureau of Plant Genetic Resources (ICAR-NBPGR) has developed core sets for Indian mustard (Nanjundan et al., 2021), wheat (Phogat et al., 2020), chickpea (Archak et al., 2016), wild lens (Singh et al., 2014), brinjal (Gangopadhyay et al., 2010), and mungbean (Bisht et al., 1998). Therefore, the aim of the present study was to develop a core set of cultivated lentil germplasm conserved in Indian national genebank based on agro-morphological characterization and diversity indices for accelerating utilization of germplasm in lentil breeding programs.
The study material consisted of 2,324 cultivated lentil germplasm accessions comprising 1,796 indigenous collections (ICs) and 528 exotic collections (ECs). The indigenous accessions were collected from different lentil-growing areas in India after exploring several crops from 1976 to 2017. Geo-coordinates of each collection site were mapped using geographic information system (GIS) tools (Figure 1). Geo-referenced maps were prepared with WGS84 datum and geographic projection system using GIS tools (Semwal et al., 2014). The germplasm was characterized during the winter season of 2017–2018 and 2018–2019 at Research Farm of ICAR-NBPGR located at a latitude of 28°38′N and longitude of 77°10′ E and an altitude of 228.61 m above the mean sea level. The soil of the location was sandy loam type having an optimal pH range. Lentil germplasm accessions were sown under natural field conditions. The recommended package of practices for growing lentil was followed. To meet the crop’s balanced nutrient demand, 20 kg ha–1 nitrogen and 40 kg ha–1 phosphorus was applied as a basal dose before sowing. One presowing irrigation was given to ensure proper germination in the field, while irrigation was also given at the pod formation stage. The seeds were treated with a mixure of thiram (2g) and carbendazim (1 g) per kg of seed. Manual weeding was completed at 25–30 and 45–50 days after sowing. The experimental design consisted of an augmented block (Federer and Raghavarao, 1975) with four popular checks, namely, DPL62, IPL316, IPL526, and L4717. The experiment was replicated in 24 blocks. A hundred accessions were sown per block uniformly in 23 blocks, whereas, the 24th block was incomplete with 24 accessions. The accessions were sown in paired rows of two m length, with a row-to-row distance of 30 cm. The detailed timeline of the experiment is shown in Figure 2.
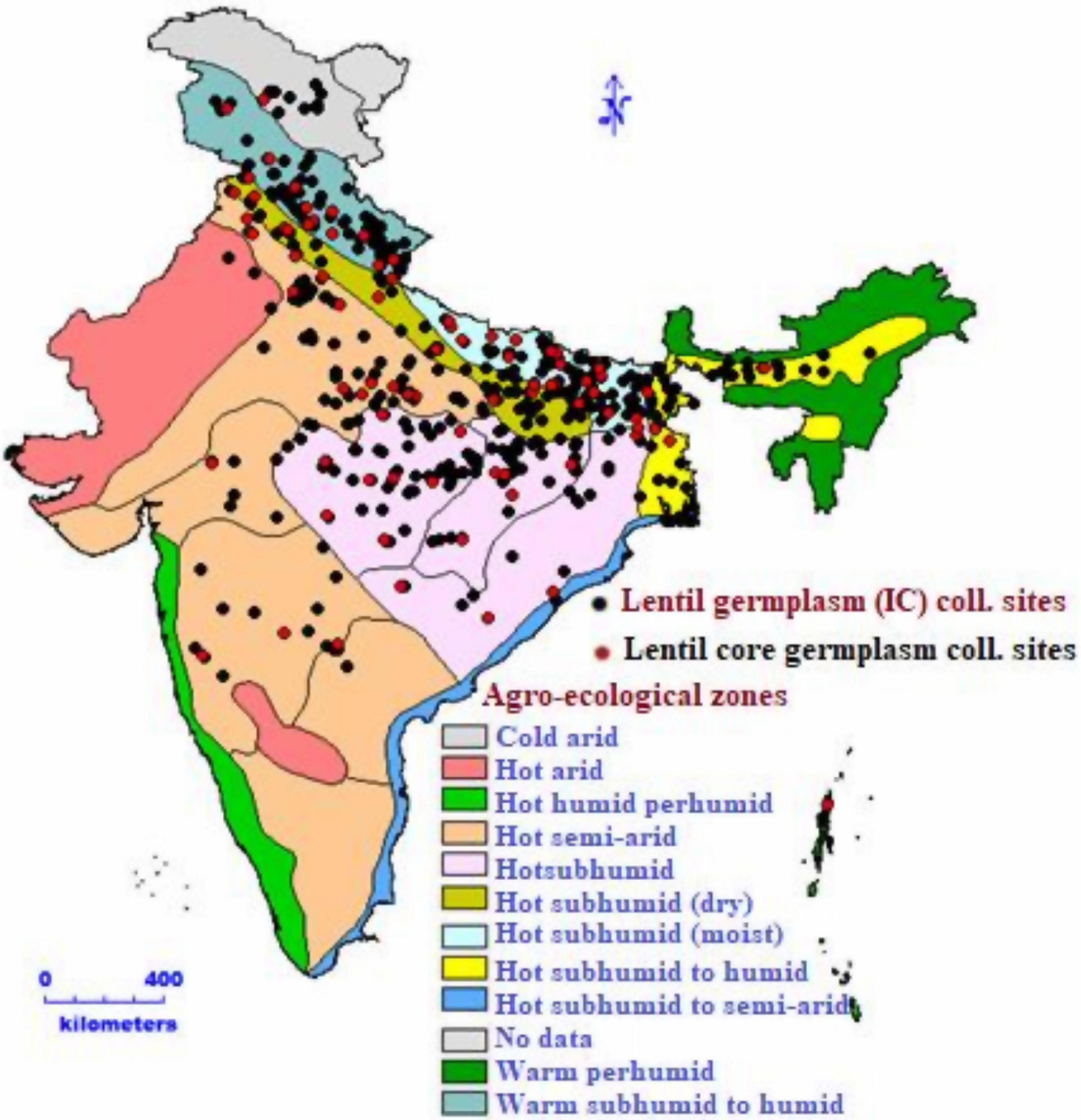
Figure 1. Indigenous lentil collection sites of national genebank from different agro-ecological zones.
Twenty-six agro-morphological descriptors (nine quantitative and 17 qualitative) were selected for phenotypic characterization using descriptor lists of lentil (IBPGR, 1985; Mahajan et al., 2000) (Table 1). The qualitative characters were (1) early plant vigor (EPV), (2) seedling stem pigmentation (SSP), (3) growth habit (GH), (4) leaf color (LC), (5) leaf pubescence (LP), (6) leaflet size (LS), (7) flower ground color (FGC), (8) tendril length (TL), (9) biomass score (BS), (10) lodging score (LoD), (11) pod pigmentation (PP), (12) pod dehiscence (PD), (13) pod shedding (PS), (14) seed shape (SS), (15) seed coat color (SCC), (16) pattern of seed testa (PST), and (17) cotyledon color (CC). The quantitative traits included (1) days to 50% flowering (DF), (2) days to 80% maturity (DM), (3) plant height (PH), (4) number secondary branches per plant (SBP), (5) seed diameter (SD), (6) seed thickness (ST), (7) pods per plant (PPP), (8) seeds per pod (SPP), and (9) 100-seed weight (SW).
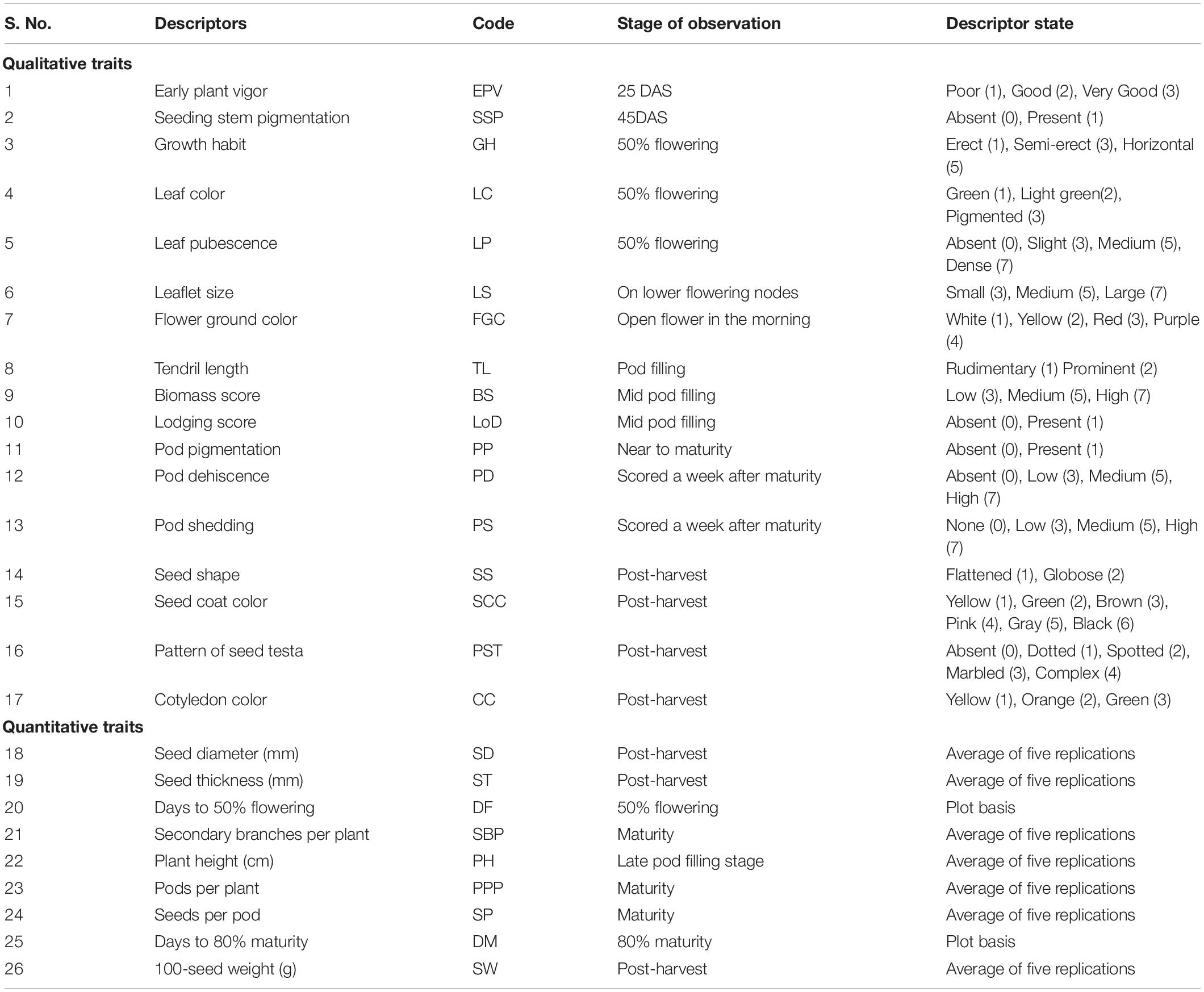
Table 1. Descriptors used for agro-morphological characterization of lentil germplasm of Indian national genebank.
A statistical software, PowerCore, was used to constitute a core collection based on the characterization data generated using 26 different descriptor traits (Kim et al., 2007). Continuous variables were classified into different categories based on Sturges’ rule (Sturges, 1926) (K = 1 + log2n), where n and K mean the number of observed accessions and number of classes, respectively. The number of classes can be modified. The modified heuristic algorithm was implemented to identify and select accessions of core collections using the strategy of advanced maximization or allelic richness. Subsets created using PowerCore represented all observation classes with the least allelic redundancy, ultimately ensuring a highly reproducible entry list. Core sets were extracted with different combinations of variables to assess their efficacy using PowerCore and using qualitative traits, quantitative traits, a combination of quantitative and qualitative traits, and a combination of quantitative and qualitative traits and passport data. Another method, known as principal component score strategy (PCSS), was used to extract the core (Noirot et al., 1996). This method uses principal component analysis (PCA) to eliminate collinearity between variables and sample individuals based on their cumulative relative contribution. The Newman–Keuls procedure was followed to compare the means of the entire collection and developed core sets (Newman, 1939; Keuls, 1952) for descriptor traits. The similarity of distribution frequencies in the core and entire sets was evaluated using the χ2 test. The Shannon–Weaver diversity index (SDI) was used to compare the representativeness of the whole set and the germplasm selected as core entries.
The data recorded for nine quantitative traits were first analyzed separately for each year, after which a combined analysis was performed using the general linear model (PROC GLM) method of SAS 9.3 for augmented design. The accessions were considered random and year as a fixed effect. Levene’s test was performed to assess the homogeneity of error variances before performing the combined analysis (Levene, 1960). Descriptive statistics were estimated for the complete set and the core collections separately. Frequency distribution for different classes of qualitative traits was obtained using Excel, and histograms for quantitative traits were drawn using IBM statistical package for the social sciences (SPSS) statistics (version 20.0). Variability in descriptor traits between ICs and ECs was compared using boxplots developed using Statistical Analysis Software-JMP 14 software. Hierarchical cluster analysis was performed using the Euclidean distance matrix following Ward’s minimum variance method for accession number grouping and estimating genetic relationships. Core sets were extracted using PowerCore and the PCS strategy. The principles laid by Oliveira et al. (2010) were used to calculate the range retention percentage. In addition, the distribution of homogeneity for each of the descriptor traits was analyzed using the χ2 test. For the quantitative traits, classes were formed based on Sturges’ formula. The observed number of accessions in the core set in each class was determined and was tested against the expected number of accessions using the χ2 test. Further, the SDI (H’) was computed as a measure of phenotypic variability using the phenotypic frequencies of quantitative and qualitative traits (Shannon and Weaver, 1949) in the entire and different core sets extracted using the following formula.
where pi is the proportion of germplasm accessions in the ith class of an n-class characteristic and n is the number of phenotypic categories for a character. Phenotypic correlation coefficients (r) among descriptors in the core collection (Cored) and entire set were estimated. Moreover, the contribution of different descriptor traits to multivariate polymorphism and conservation of contribution in the core collection were assessed based on PCA. The SAS 9.3 program was used to estimate the correlation and PCA (SAS Institute, 2012).
The actual germplasm collection sites of lentils in India (Figure 1) were presented using geo-referenced map. These maps revealed that most of the lentil germplasm were collected from Uttar Pradesh, Bihar, Uttarakhand, and Madhya Pradesh. Geo-referenced map showed that lentils were collected from Western Himalayas (Kashmir, Himachal Pradesh, and Uttarakhand) to Eastern Himalayas (Assam and Arunachal Pradesh). It was collected from high-altitude areas in north (Leh) to low-lying areas of eastern part of India (West Bengal). Collections from dry areas of Rajasthan to wet areas of North-east Himalayas indicated that India has sufficient diversity in its collections. Indian lentil collections showed the unique distribution pattern having representation from four global biodiversity hotspots, the Himalayas, the Western Ghats, the Indo-Burma region, and the Sundaland (Includes Nicobar group of Islands). All 2,324 accessions and four checks were characterized under the agro-climatic region in the north-western Indian conditions for two consecutive years (2017–2018 and 2018–2019). The experimental field view and seed coat color diversity are presented in Figure 3.

Figure 3. (A) Field view of agro-morphological characterization of lentil germplasm and (B) seed coat diversity.
The frequency distribution of the entire set was not normal and varied among traits. In certain cases, a particular trait was predominant, whereas it was dispersed in other cases (Figure 4). The majority of the accessions (2,061) showed purple pigmentation on the stem at the seedling stage, whereas leaf color was recorded as green. A slight to medium leaf pubescence was prominent, whereas 48 accessions were glabrous. EPV of most of the accessions (2,044) was good, whereas 275 accessions were highly vigorous. Prominent tendril was recorded in 1,360 accessions, whereas 954 accessions showed rudimentary tendril. High biomass was observed at the mid pod filling stage in 1,438 accessions, whereas 72 accessions showed low biomass. Semi-erect plant growth was more prominent (1,648 accessions), and horizontal growth was recorded in 525 accessions, whereas the growth in 151 accessions was erect. Around 67% of the accessions showed lodging at the time of pod filling. The maximum number of accessions (2,159) were of purple flower ground color. After a week of maturity, medium pod shedding was recorded in 180 accessions, whereas most of the accessions (1,671) showed no pod shedding. Pod dehiscence was absent in 1,765 accessions, whereas low to medium pod dehiscence was noted in 549 accessions. Pod pigmentation was absent in the majority of the accessions, whereas brown seed coat color and dotted testa pattern were prominent. A flattened seed shape was observed in 2,060 accessions, whereas 254 accessions showed the globose seed shape. There was substantial variation in the cotyledon color with a maximum of orange type.
The results revealed significant variations as evident from the mean, range, and coefficient of variations (Supplementary Table 1 and Figure 5). The days to 50% flowering and maturity ranged from 51 to 123 and 93 to 140 days, respectively, whereas 100-seed weight varied from 0.75 to 7.6 g. The average seed thickness and seed diameters were 2.42 and 4.18 cm, respectively. The mean plant height and the number of secondary branches were 35.34 and 28.64 cm, respectively. The coefficient of variation ranged from low (<10.17%; days to flowering, seed thickness, days to maturity, and number of seeds per pod) to high (>17.34%; the number of secondary branches, 100-seed weight, number of pods per plant, and seed diameter) for key descriptor traits. In addition, substantial variation in the range for the number of pods per plant, number of secondary branches per plant, 100-seed weight, and earliness was observed. The least variation was observed in the number of seeds per pod, with the majority of the accessions with two seeds per pod in studied accessions. ICs showed earliness, small seeds, and low biomass as compared to the ECs. The days to 50% flowering, days to 80% maturity, and seeds per pod were negatively skewed, indicating that a majority of the accessions had higher mean values for these traits and days to 80% maturity and seeds per pod showed high kurtosis values with a relatively higher peak. Boxplot analysis revealed a comparison of trait distribution between exotic and indigenous accessions. The average performance for plant height, days to 50% flowering, pods per plant, and number of secondary branches was lower in ECs than in ICs. However, the mean value of seed weight was higher in ECs than in ICs (Figure 6).
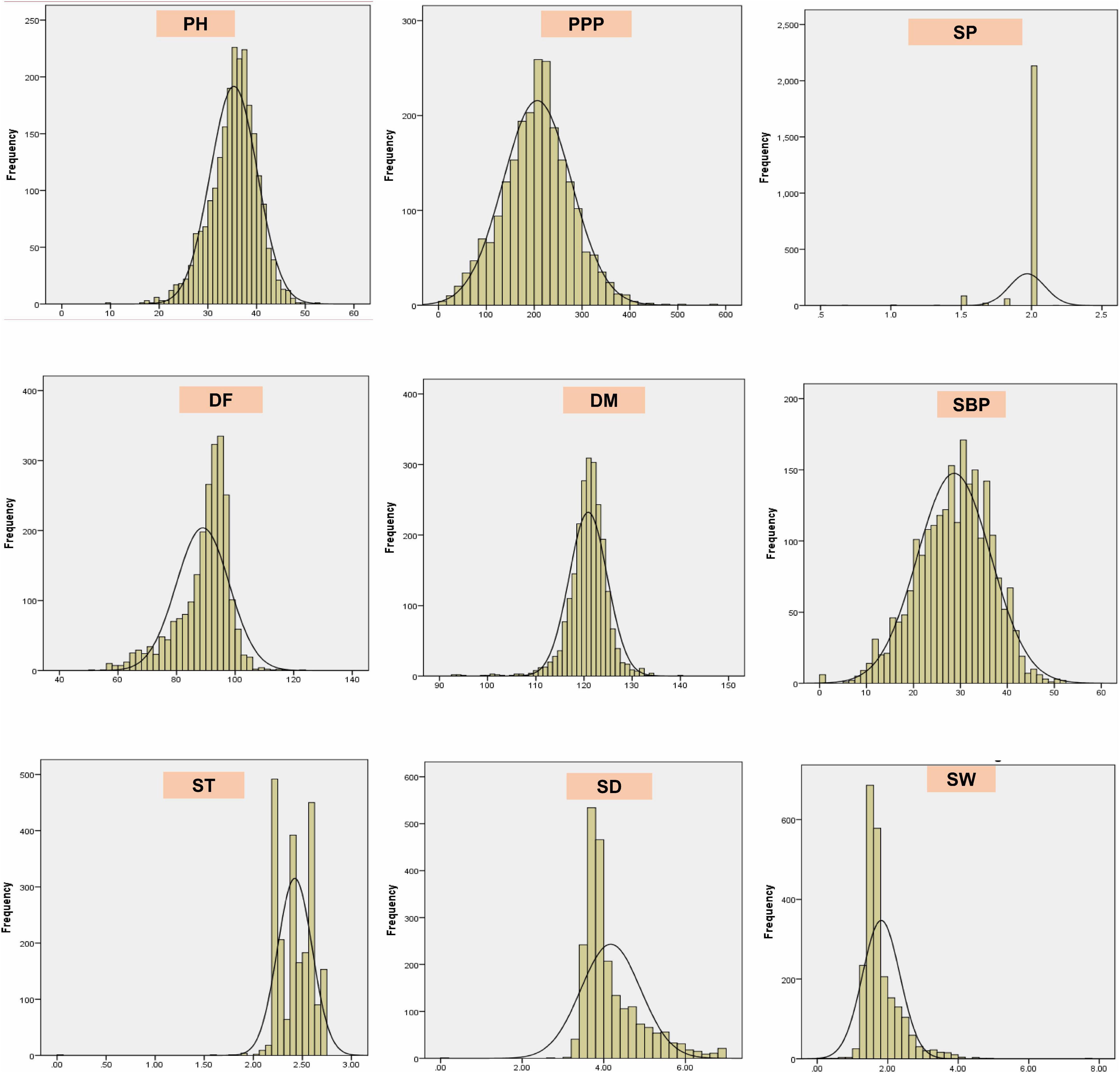
Figure 5. Frequency distribution plots showing the variability of nine quantitative traits in the entire lentil germplasm conserved in Indian national genebank.
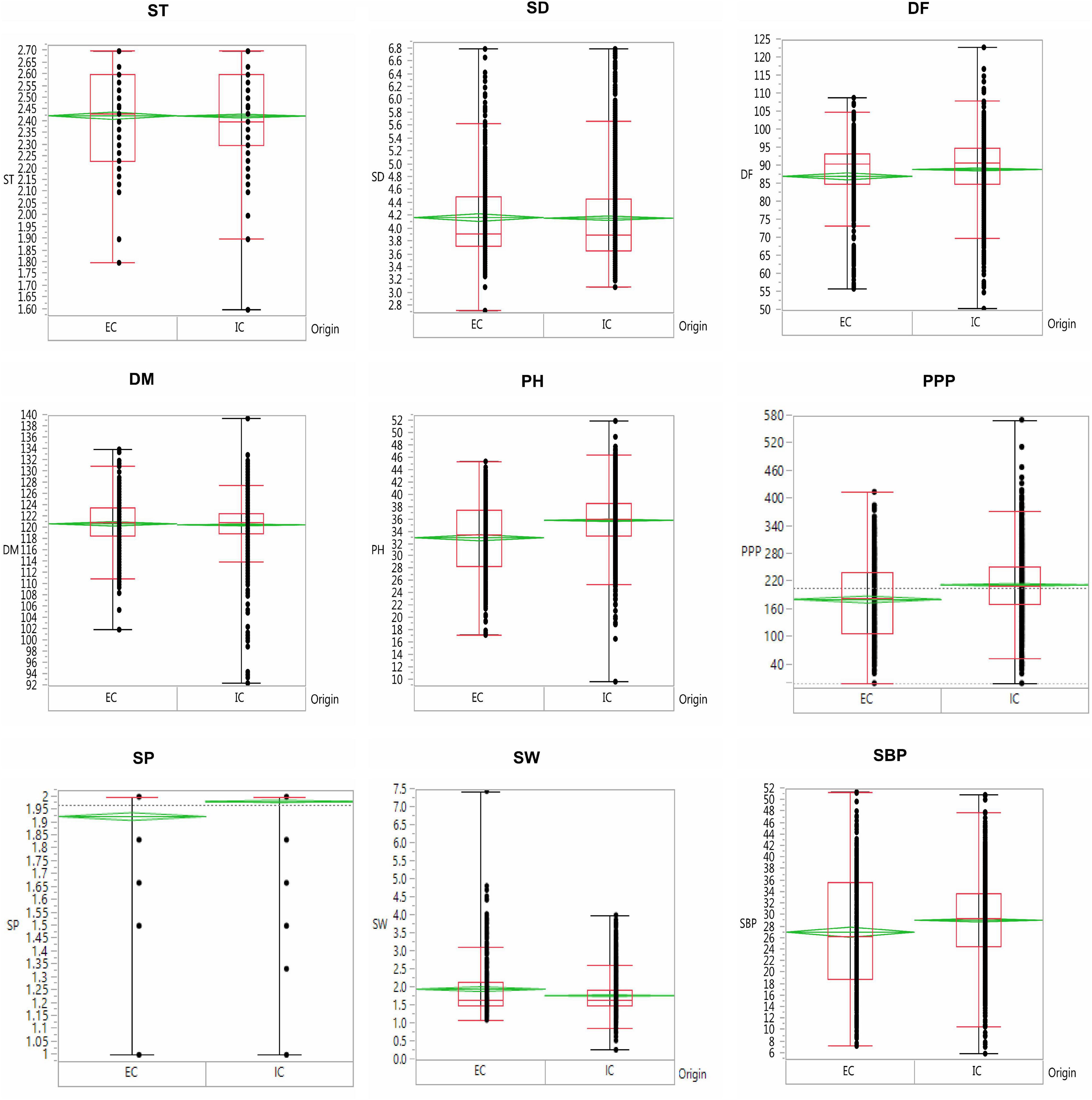
Figure 6. Boxplot depicting the variability of traits in the indigenous collection (IC) and exotic collection (EC) of the entire lentil germplasm conserved in Indian national genebank.
Grouping several accessions into certain homogenous clusters assists in selecting diverse parents. This allowed the accurate comparison among all probable pairs of individuals and brought together gene constellations, yielding desired progenies through crossing between different parents. Hierarchical cluster analysis with Ward’s method of minimum variance was applied to assess genetic diversity. Based on nine quantitative traits, phenotypic relationships among lentil accessions were determined using Euclidean distances. The Indian national gene bank (INGB) lentil accessions were grouped into 12 clusters with a varied number of accessions in each cluster (Figure 7 and Supplementary Table 2). A list of core set accessions with cluster number is given in Supplementary Table 3. The cluster also showed heatmap distribution of accessions, showing trait variability in different clusters represented by different colors. Cluster II was the largest with 393 accessions followed by Clusters III and IV with 382 and 253 accessions, respectively. Cluster II grouped 320 ICs and 73 ECs accessions; whereas Cluster XI was the smallest with only 38 accessions (24 ECs and 14 ICs). The cluster mean values of studied traits showed that Clusters XI and XII included extra-early accessions, such as IC241529, IC241531, and IC241532, whereas early to medium maturing accessions were grouped into Clusters VI and VII. Late flowering accessions such as IC73690, IC73692, IC381127, and IC398044 grouped in cluster I. Proportionately, exotic accessions were relatively dominant in Clusters X, XI, and XII, and indigenous were dominant in Clusters II to IX. Cluster XII comprised all exotic accessions from Syria. Cluster V contained accessions having very tall plant types (IC329110, IC398793, and IC59038, a high number of pods per plant (IC78387 and IC78398), and a high number of secondary branches (IC148333, IC16453). The accessions such as EC223214 and IC241543 grouped in cluster XII had highest mean value for seed weight (3.239 g) whereas cluster III had accessions with mean value of 1.523 g.
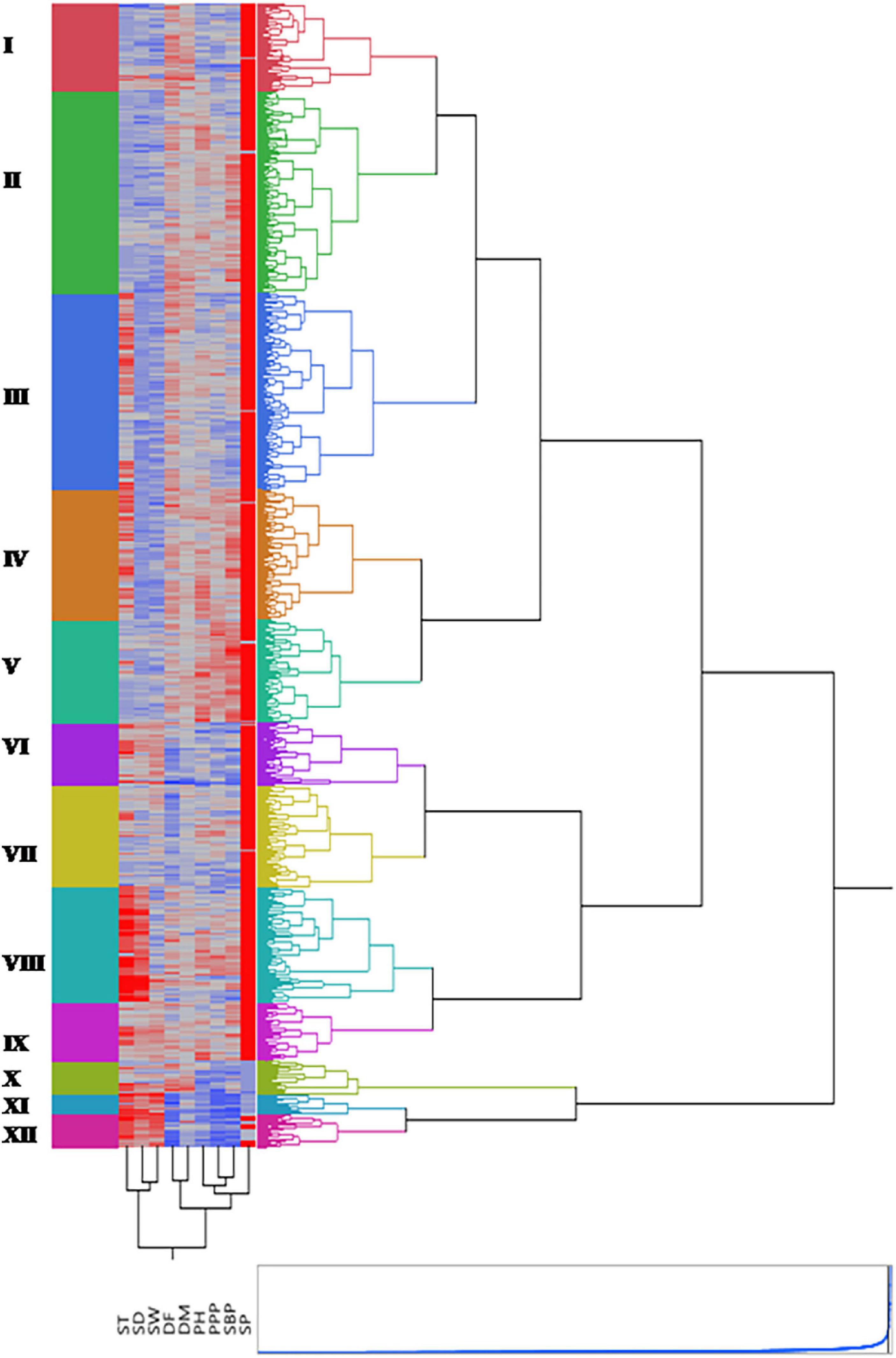
Figure 7. Dendrogram along with heatmap of entire germplasm generated by performing hierarchical cluster analysis. Twelve clusters are represented by different colors. The two-dimensional heatmap is represented by columns and rows. Each column represents different quantitative trait and each row an accession. The higher the trait value, the brighter is the red and similarly, lower the trait value, brighter is the blue color, respectively.
Potential accession numbers for unique traits were identified from characterization datasets and validated over the years and locations at NBPGR, New Delhi, The International Center for Agricultural Research in the Dry Areas (ICARDA), Amlaha, and NBPGR, Regional Station, Ranchi. Some of them are depicted in Figure 8. The accession IC317520 was identified with a unique seed morphotype having extended funiculus. A total of 50% of plants with accessions such as IC241532 and IC241529 exhibited very early flowering (51 days) relative to all checks used in the study. The accession EC267615 displayed a novel type of erect plant architecture, indicating its suitability for mechanical harvesting. Pod setting in this genotype was observed at more than 15 cm above the ground. The accessions IC199461 and IC201716 had a high number of secondary branches (>50) and pods per plant (>400) relative to all checks used in the study. The accession EC499760 was of bold seeded type (100-seed weight, 7.1–7.83 g) and was imported from the United States of America (USA). A novel and unique multiflower (MF) germplasm accession, IC241473, was identified the first time that formed up to 16 flowers per peduncle (FPP) at multiple flowering nodes. In addition, this accession showed fasciation of the main stem.

Figure 8. Trait-specific lentil germplasm, (A) IC317520: unique seed morphotype with an intact funiculus; (B) IC241532: early maturing accession (circled in red) relative to surrounding germplasm; (C) EC267615: suitable to mechanical harvesting; (D) IC199461: high number of secondary branches; (E) EC499760: bold seeded type with high 100-seed weight; (F) IC241473: multiflowering germplasm.
The core sets were extracted using combinations of different datasets using the PowerCore software. Additionally, the PCSS was used to develop the core sets. These core sets were designated as Corea, Coreb, Corec, Cored, and Coree when generated using qualitative traits, quantitative traits, a combination of both traits, a combination of both traits and passport data, and PCSS. A total of 170 accessions were selected based on the trio of qualitative and quantitative traits and passport data using PowerCore (Table 2 and Supplementary Table 3). A total of 67 accessions were selected when the data of only qualitative variables were used, whereas 71 accessions and 79 accessions were selected, respectively, when data of only quantitative characteristics and both variables were used for the analysis. Accessions with the maximum relative contribution corresponding to 10% representation were used as the criteria to select the core; thus, 221 accessions were selected based on the PCS strategy. Relative SDI was estimated for both quantitative and qualitative traits in entire lentil accessions and in core sets developed using a different strategy that revealed adequate diversity. The lentil germplasm was more diverse (H’ > 0.5) for all descriptors, except for EPV, SSP, FGC, PP, SS, CC, and SP. The average SDI for quantitative traits was higher than qualitative traits. The SDIs were consistently higher in the core sets derived from different datasets than in the entire set, signifying an enhanced representation of the available diversity in each core set. PowerCore yielded a better representation of diversity than PCSS. The average SDIs for all traits revealed that Cored (1.361, 170 accessions) was more diverse, followed by Coreb (1.340, 71 accessions) and Corea (1.302, 67 accessions) (Table 2).
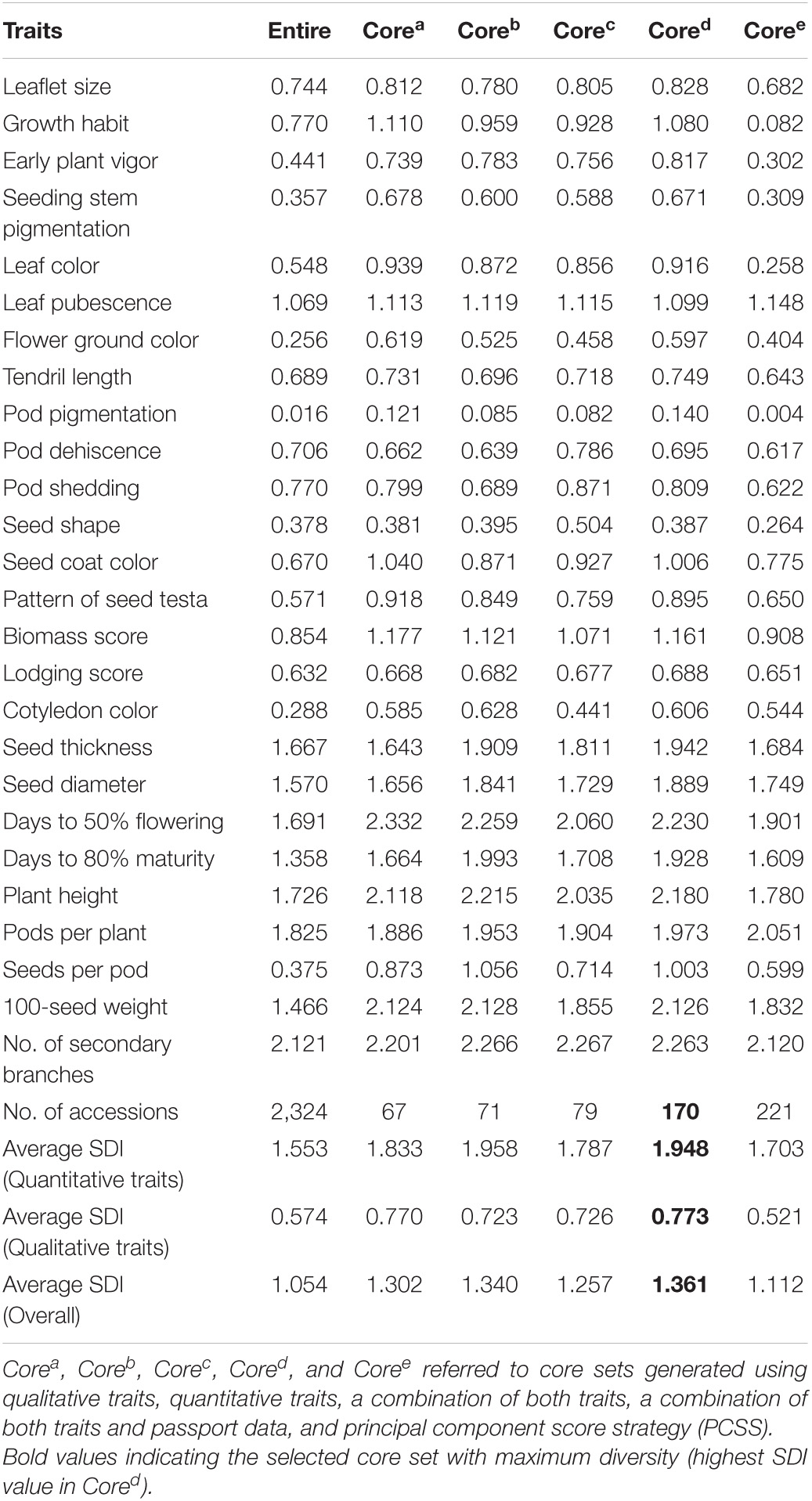
Table 2. Comparison of the Shannon–Weaver diversity index (SDI) of the entire lentil collections to lentil core sets derived from different datasets and strategies.
Qualitative traits in the INGB collections were represented in the core sets developed by all strategies, except the core developed by quantitative method and PCSS strategy, indicating that the Corea, Corec, and Cored captured the allelic richness of the entire INGB collections. The range of quantitative descriptors was classified into 12 groups for frequency distribution and χ2 analysis based on PowerCore. Frequency distribution analysis indicated the homogeneity of distribution of several traits by Corea(10), Coreb(13), Corec(11), Cored (18), and Coree(9) (Table 3). Thus, the core set developed by combining qualitative and quantitative traits and passport data well-represented the structure of the entire lentil germplasm. The results showed that the variability existing in the entire set was well-symbolized in the core collection. Differences in the means of the entire collection and Cored were insignificant for quantitative descriptors, except days to flowering and seed weight. In addition, the range retention percentage was 100, capturing a complete range of diversity, including all extremes (Table 4).
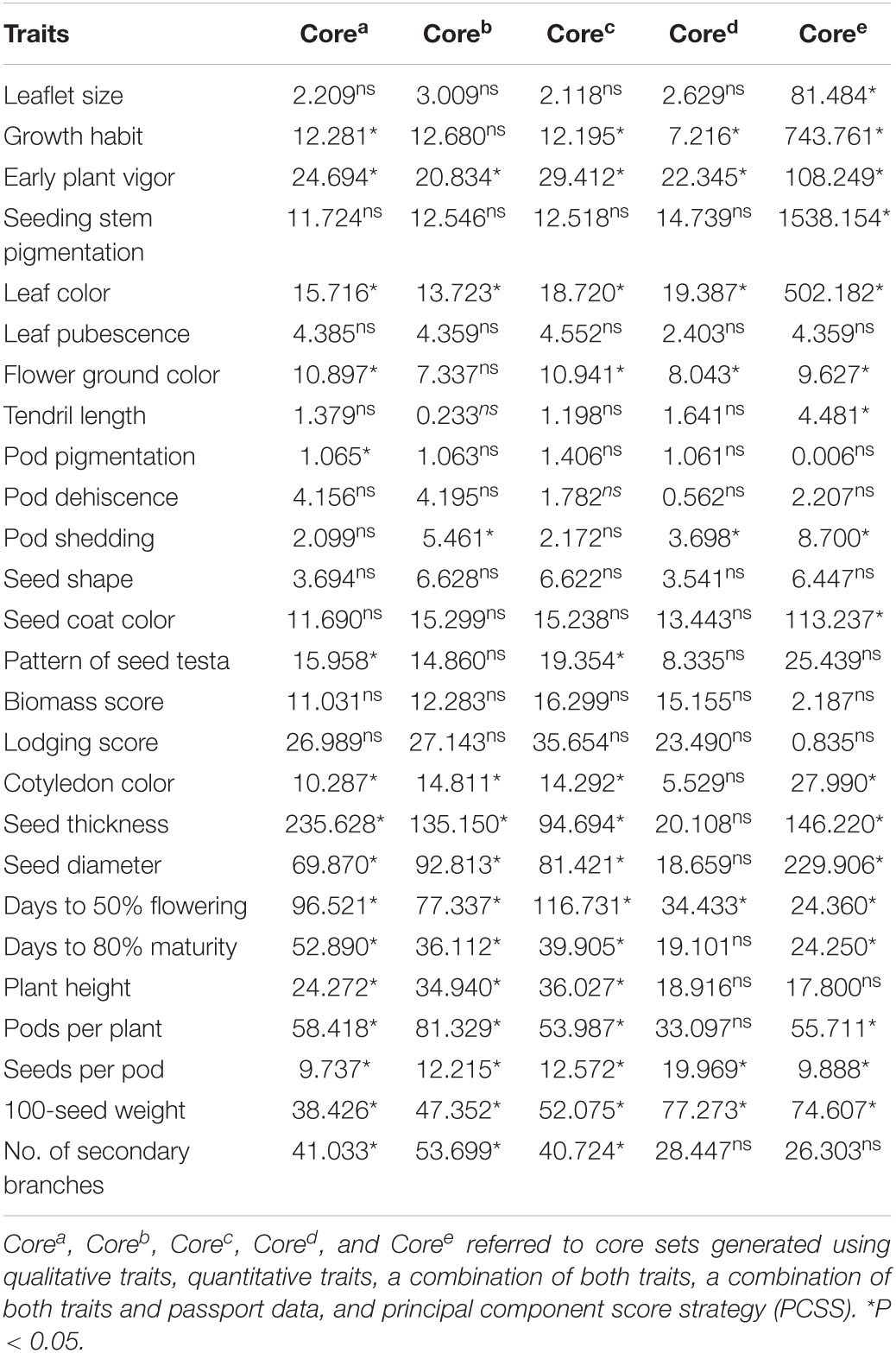
Table 3. Chi-square test for comparison of frequency distribution for descriptor traits studied in lentil core sets derived from different datasets and strategies.
The lentil germplasm accessions recorded significant and positive correlations between seed thickness and seed diameter, equally in both entire set and core set (r = 0.539) and between days to 50% flowering and days to 80% maturity (r = 0.582 in the entire set, r = 0.731 in the core set). Plant height was positively correlated with days to 50% flowering (i = 0.378), days to 80% maturity (r = 0.287), pods per plant (r = 0.496), seeds per pod (r = 0.521), and number of secondary branches (r = 0.542) and negatively with seed thickness (r = –0.034) and seed diameter (r = –0.234) in the core set. Similar trends were recorded in the entire lentil collection. The number of secondary branches was positively correlated with days to 50% flowering, days to 80% maturity, plant height, pods per plant, and seeds per pod negatively with 100-seed weight, both in the core collection and entire set (Figures 9, 10).
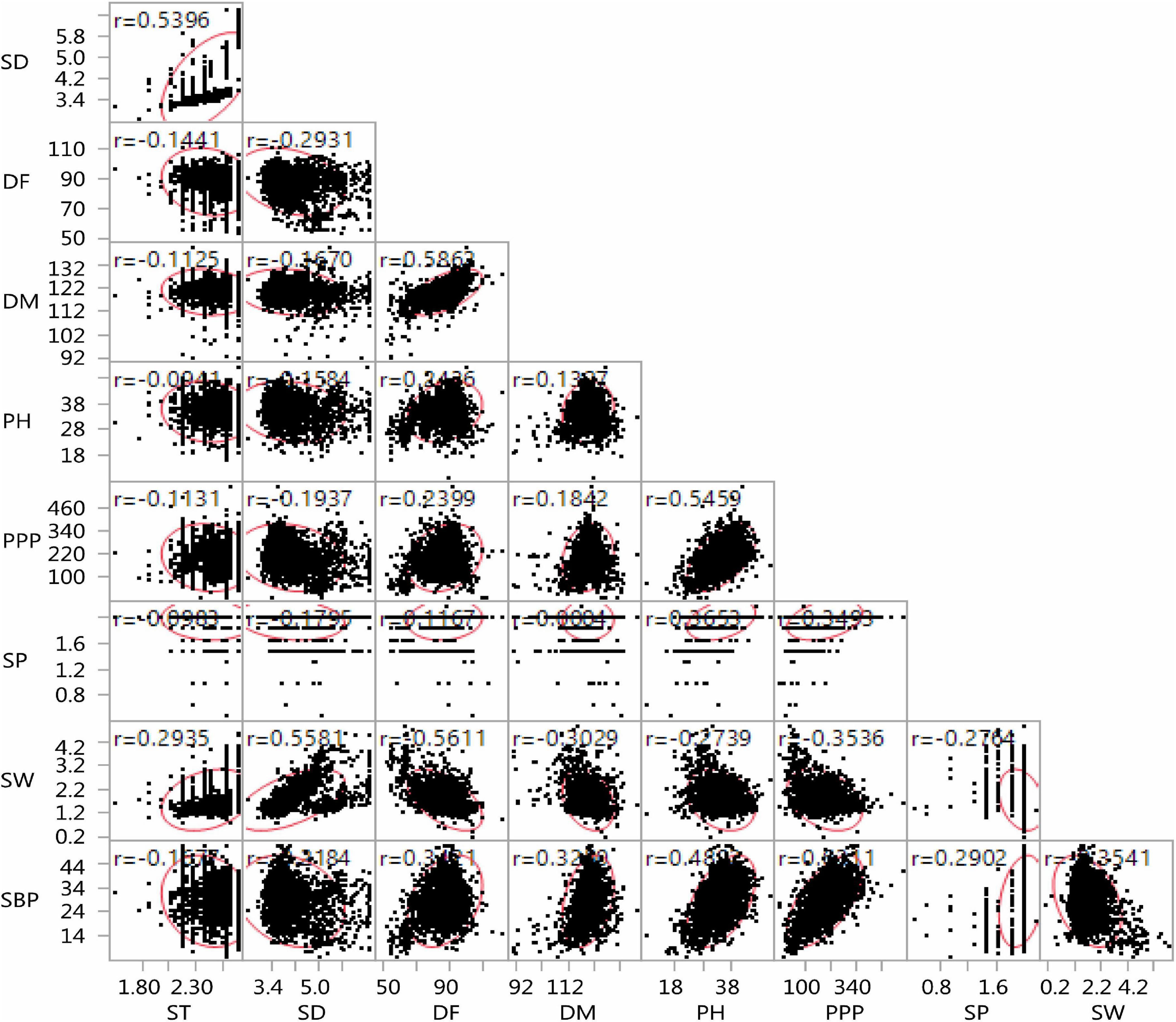
Figure 9. Scatter plot matrix showing the correlation between nine agro-morphological traits of the entire lentil germplasm conserved in the Indian national genebank.
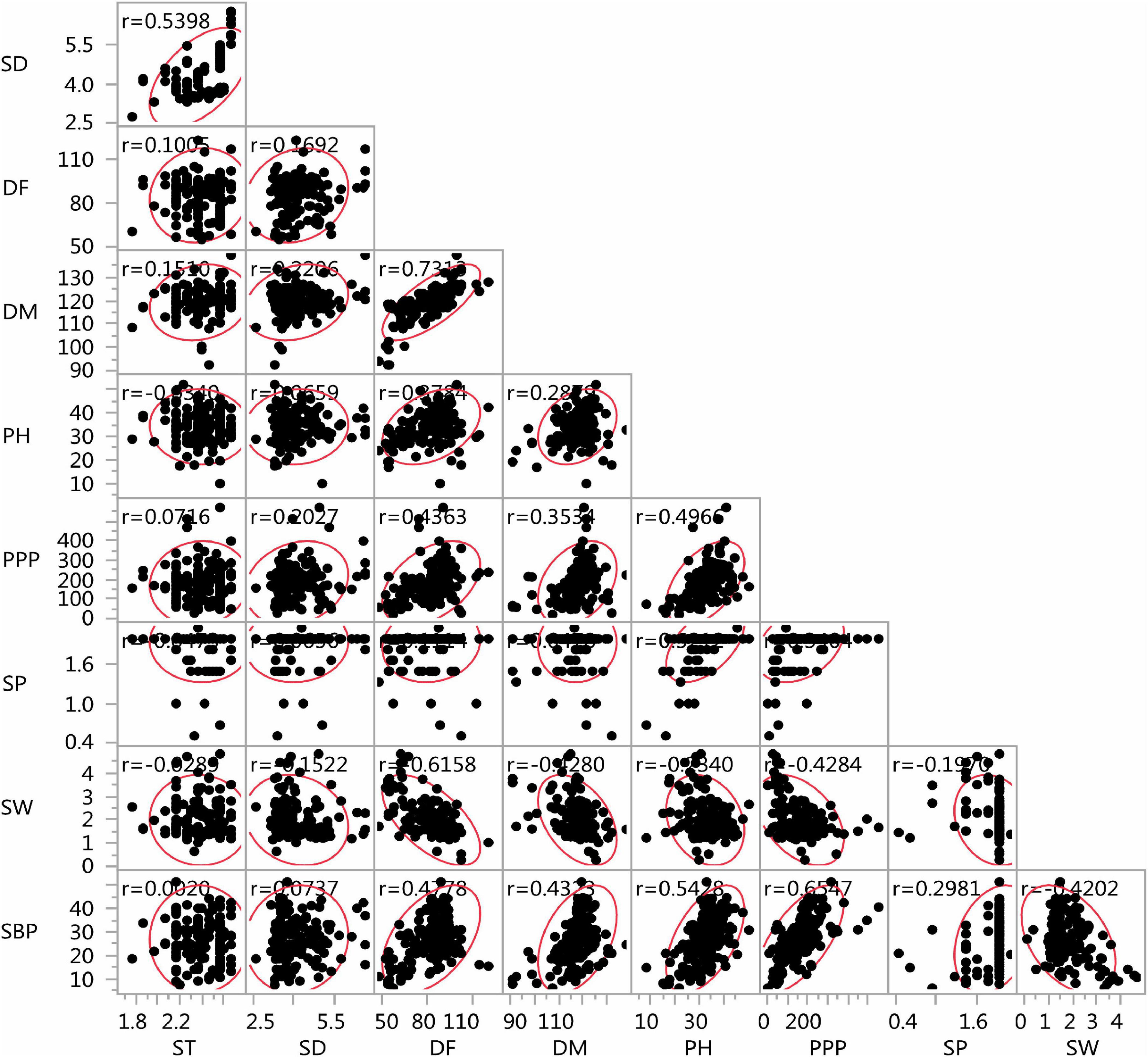
Figure 10. Scatter plot matrix showing the correlation between nine agro-morphological traits of the Indian national genebank lentil core set.
Principal component analysis revealed the association among different descriptors and their contribution toward variability. The first three PCA components provided a realistic summary of the data and explained 70.69% of the total variation in the core set. The first principal component (PC1) accounted for 39.36% of the total variation, whereas PC2 and PC3 accounted for 17.73% and 13.60% variance, respectively. These values were comparable with those of the entire lentil collection, explaining 68.03% of the variation (PC1:37.49%; PC2:16.62%, and PC3:13.91%). The negative variation in PC1 was mainly contributed (>50%) by days to 50% flowering, days to 80% maturity, plant height, number of secondary branches per plant, and pods per plant and 100-seed weight in the entire and lentil core set (Supplementary Table 4 and Supplementary Figure 1).
Genebanks are reservoirs of germplasm variability containing vital genes. These play a crucial role in crop-breeding programs (Qiu et al., 2013; Sandhu and Singh, 2021). Efficient and effective use of genebank collections in crop improvement depends on a thorough understanding of the existing genetic variability, knowledge of the genes present in individual accessions, and their value for use (Ortiz, 2002). Characterization and evaluation are the most crucial activities of plant genetic resources (PGR) management (Jones, 1984). Its scope has increased due to the increasing emphasis on conservation. The use of genebank accessions through characterization and evaluation has resulted in the development of genetic stocks, breeding lines, and commercial varieties (Figure 11; Carvalho et al., 2013).
Figure 11. Flowchart depicting the stepwise utilization of Indian national genebank lentil collections.
Geo-referenced maps of India revealed that most of the Indian accessions were from the Gangetic plains and eastern part of India belonging to Uttar Pradesh (254), Bihar (203), Uttarakhand (181), and Madhya Pradesh (96). These are the most populated regions of India, with the massive scope of pulse production in their rice fallow areas (Chowdhury et al., 2020). The majority of the germplasm was collected from hot subhumid (dry and moist) to hot subhumid agro-ecological zones of India. In contrast, the ECs, conserved in INGB, represented all six continents, primarily Asia.
The major steps in crop improvement are assessing the variability for desired traits and their utilization in breeding programs (Kumar et al., 2015). Previous researchers adopted the piecemeal approach to characterize the agro-morphological traits in lentil germplasm collections (Gautam et al., 2013; Jha et al., 2014; Kumar and Solanki, 2014). However, in this study, the entire genebank collections representing all agro-ecological zones were characterized in one go. Significant differences among lentil accessions based on agro-morphological characterization indicated their potential for use in current and future lentil breeding programs. The results showed significant variations in certain traits such as days to flowering, seed thickness, days to maturity, number of secondary branches, 100-seed weight, number of pods per plant, and seed diameter. The wide range of variability obtained was attributable to the diverse collection assessed, involving native and introduced germplasm originating from diverse geographical origins with different genetic makeup (Phogat et al., 2020).
Lentil is a less vigorous grain legume that faces competition from weeds during the early growth stage. Lentil germplasm with increased vigor can dominate the weeds, ultimately enhancing the yield (Sharma et al., 2018). The previous researchers reported that EPV and rapid canopy development as key traits for escaping drought at the terminal stage because these traits lead to the early onset of maturity (Sarker et al., 2005; Kumar et al., 2012). The majority of exotic accessions had light-green leaves, whereas indigenous varieties were dominated by green to dark green canopy. In addition, the purple pigmentation was observed to be disappeared with the increase in the ambient temperature. Similarly, in subterranean clover, Nichols et al. (1996) reported prominence of purple pigmentation during winter, which is appeared with the arrival of spring. Wide variations were recorded in leaf pubescence, ranging from the absence of pubescence to dense pubescence. Low or moderate leaf pubescence has promoted tolerance to aphids (Kumari et al., 2009). However, Tripathi et al. (2020) indicated that pubescence on the pod is a unique feature of cowpea, a marker-trait for insect resistance. Variation was also reported in leaflet size, tendril length, flower ground color, pod shedding, pod dehiscence, testa color, and seed shape.
Elias et al. (1979) suggested that the protein quality is affected by seed coat color in beans. Diversity in seed color among accessions may provide valuable genetic resources for biofortified lentils. Orange cotyledons are preferred by Indians, which is also reflected in Indian genebank collections (Singh et al., 2014). These agro-morphological descriptors can facilitate the differentiation of distinct phenotypic classes and are used as diagnostic keys for taxonomic delineation (Pundir et al., 1985; Gore et al., 2019). The mode of inheritance of these traits can be investigated using the principles of classical genetics. This mega characterization program showed that most of the Mediterranean germplasm were found with low biomass and poor yield. The direct use of these accessions appeared difficult in Indian conditions, leading to the ubiquitous selection of Indian germplasm with high biomass and better yield traits.
The entire lentil germplasm was more diverse for quantitative traits such as number of secondary branches per plant, pods per plant, plant height, days to 50% flowering, seed thickness and qualitative traits, leaf pubescence, biomass score, pod shedding, growth habit, leaflet size, and pod dehiscence. Plant height (cm) in the INGB lentil core set ranged from 9.70 to 52.13 with mean value of 34.04, while it was ranged from 15 to 40 cm with mean value of 23 cm in the USDA core set sown at Pullman, Washington, United States, in 1996 (Tullu et al., 2001). An average value of days to flowering and maturity is lower in the USDA lentil core set than the INGB lentil core set (Tullu et al., 2001). The grouping of germplasm in different clusters allowed us to study their genetic diversity, whereas the heatmap depicted the trait diversity. Germplasm accessions belonging to diverse clusters can be used in cross-breeding programs and recombination studies; for example, Clusters XI comprised early maturing accessions such as IC241529, IC241531, and IC241532 (less than 100 days), and cluster X comprised late maturing accessions such as EC267554, EC267615, IC258265, and EC440747 > 125 days. Similarly, Clusters XI had accessions with low plant height IC241488, IC241501, IC241529, and IC241531 (<20 cm) and cluster V had accessions with larger plant height, IC329110, IC398793, and IC59038 (>50 cm). The accessions such as EC223214, IC241543 (with high 100-seed weight) grouped in cluster XII and may be crossed with accessions with low seed weight IC361467, IC521438 (<1.4 g) from cluster III for genetics and mapping purposes.
The ultimate aim of germplasm characterization is to support crop-breeding programs (He and Li, 2020; Guerra-García et al., 2021). Similarly, the present work unfolded the treasure of lentil collections conserved in the NGB. The accession IC317520 collected from Rajasthan was identified as a unique seed morphotype with intact funiculus at maturity (Tripathi et al., 2019). This was reported for the first time and is registered as a genetic stock (INGR19072) for its utilization in lentil breeding programs. An Intact funiculus may provide resilience to drought stress, resulting in high yields. This trait may help to gain area in rice-fallow areas of eastern India, where the crop is sown at conditions of conserved moisture (Lamichaney et al., 2021; Tripathi et al., 2021). Rice–wheat and rice–rice systems in the Indo-Gangetic plains dominate South Asia’s cereal systems (Kumar et al., 2013). A substantial reduction in the acreage under pulses is attributed to the expansion of high-yielding wheat and boro rice in northern India and eastern to north-eastern India, respectively. This trend of pushing out pulses is likely to continue unless extra-early varieties that fit into these cropping systems are developed (Erskine et al., 1993; Shrestha et al., 2006). Similarly, in the rice-fallow areas of South Asia, the top soil layer generally dries at the harvesting stage, making it impossible to sow the next crop. Under such conditions, extra-early lentil germplasm can be used as donors to breed early maturing varieties to fit into the cropping system and convert these mono-cropped areas into double-cropped areas, thereby increasing the production of lentils under rice-based systems. In a previous study, a lentil accession with 59 days to flower was reported (Kumar and Solanki, 2014). However, the present experiment discovered accessions, IC241529 and IC241532, flowered in 51 days. The number of pods per plant and the number of secondary branches are the primary quantitative descriptor traits that can be used as kickstarting points for selecting high-yielding germplasm. Further, the seed size and shape of lentils are important traits because they affect the market class, cooking time, and the quality and yield of milled lentils (Fedoruk, 2013). Indigenous and exotic accessions are broadly classified into small- and large-seeded types, respectively. Descriptor traits, such as tall, erect, top pod-bearing habits, thick stems, synchronous maturity, lodging tolerance, and non-shattering reported contributing to the development of varieties with machine harvestability (Kumar et al., 2013). Genetic variability for these traits was recorded in the INGB lentil germplasm. In an earlier study, Idleb-2 variety was released for machine harvesting (El-Ashkar et al., 2004). Identified germplasm, EC267615, may provide avenues for breeding machine harvestable varieties in India. Multiflowering accessions can be identified with the expression of three or more flowers at one or multiple flowering nodes (Gaur and Gour, 2002; Benlloch et al., 2015; Sanwal et al., 2016; Devi et al., 2018). In several legumes, previous researchers have documented multiflowering clusters for a few flowering nodes in Cicer (up to nine FPP) (Gaur and Gour, 2002), Pisum (up to five FPP) (Devi et al., 2018), and Lens (up to seven FPP) (Mishra et al., 2020). However, this study found a unique multiflowering germplasm accession, IC241473, in cultivated lentils, formed up to 16 FPP at multiple flowering nodes. This trait can fulfill the aim of genebanks to work in the direction of conservation and utilization. Trait specific germplasm (TSG) developed in this study were shared with the National Agricultural Research System (NARS) partners under the material transfer agreement for utilization in the varietal development programs.
Globally, the first and foremost reason behind the limited use of conserved germplasm by plant breeders is huge and uncharacterized germplasm collections in certain genebanks (Ortiz et al., 2008). Characterized germplasm and smaller subsets can enhance germplasm use in trait discovery (Dutta et al., 2015). Core sets are subsets of large and diffused germplasm collections, containing selected accessions that represent the genetic variability of the entire set and acts as the entry point to the entire collection for trait-specific evaluation (Ortiz et al., 1998, 1999; Huamán et al., 1999; Upadhyaya et al., 2003). PowerCore is a fast and reliable tool for extracting a core set with a higher percentage of diversity than other methods used (Kim et al., 2007; Zhang et al., 2011; Dutta et al., 2015).
A subsampling procedure based on 26 agro-morphological descriptors and geographical origin data resulted in the selection of 170 germplasm accessions as the lentil core set, whereas only 79 accessions were sampled without passport information. Therefore, the inclusion of stratification into the geographical origin and places should be a choice for the sampling procedure. The INGB lentil core set amounted to 7.31% of the entire collection (2,324 accessions) conserved in the INGB, New Delhi. Similarly, Archak et al. (2016) selected 1,103 accessions (7.5%) as the chickpea core set from 14,651 accessions conserved in the INGB. As expected, the INGB lentil core set comprised 80.58% accessions belonging to Indian origin. However, only 12.89% (37 accessions) of the Indian accessions contributed to a lentil core set developed for agro-morphological descriptor traits by United States Department of Agriculture (USDA) (Tullu et al., 2001). The USDA lentil core consists of 12% of the 2,390 lentils (Simon and Hannan, 1995; Tullu et al., 2001). The maximum representation of germplasm accessions in INGB lentil core collections was from India (137), followed by Syria (22), indicating the rich diversity of cultivated lentils in India. A total of 95.88% of the core set belonged to Asian countries, including 15.29% from other Asian countries (Syria [mainly from ICARDA], Bangladesh, Israel, and Pakistan). As lentil is the main crop of Asia (Laskar et al., 2019), the INGB core set can be used as a diversity hub to meet the present and future challenges of the Asian lentil breeding program. Simon and Hannan (1995) also reported the utility of the legume cores in chickpea, lentil, and pea developed by USDA for directing users toward desirable germplasm from targeted geographic regions, and facilitating users at the preliminary stages of germplasm evaluation.
While sampling for a core set, the phenotypic associations in the core set need to be preserved for maintaining coadapted genetic complexes (Ortiz et al., 1998) and efficient utilization of germplasm (Upadhyaya et al., 2001). Nine traits were studied and 36 comparisons were performed between the pairs of characteristics. All trait combinations maintained similarity in correlation coefficient value between the entire INGB collection and the INGB core set. Earlier studies on the development of core in various crops such as chickpea (Upadhyaya et al., 2001; Archak et al., 2016), eggplant (Gangopadhyay et al., 2010), and wheat (Phogat et al., 2020) also reported the preservation of trait association in the core set. The strong correlation among some of the traits, such as days to 50% flowering and days to maturity (r = 0.52 in the entire collection, r = 0.731 in core set), and secondary branches per plant and pods per plant (r = 0.63 in the entire collection, r = 0.65 in core set), showed that future germplasm characterization might use days to 50% flowering and secondary branches per plant during preliminary evaluation. This trait is easier to measure than other traits, such as days to maturity and pods per plant. Similar observations were reported by Upadhyaya et al. (2003). Examination of the entries’ spatial distribution and explaining the variance through PCA were also reported as an exploratory criterion for evaluating the core set in the study of Bisht et al. (1998) and Wang et al. (2008). In the present study, the first three PCs collectively explained 70.39% of the total variation in the core sets and 68.03% in the entire set. Germplasm accessions were majorly distributed in one large group and one small group in both the complete set and the core collection of INGB lentil germplasm.
The present study was based on the characterization of 2,324 accessions of cultivated lentils that resulted in the development of a core set of 170 accessions. The core set possessed a high diversity and allelic richness for major descriptor traits as revealed by the summary statistics, chi-square test, and SDI. The lentil core set described in this study represents the variability existing in the germplasm conserved in the INGB and provides insights into the traits for earliness, seed size, growth habit, and other vital agronomic traits. Identified and validated TSGs could serve as a potential source of novel alleles and genes. Characterization data of INGB lentil germplasm and developed multipurpose core set may serve as a valuable resource for lentil workers and open the avenues for germplasm utilization in the selection, mapping, genomics, and trait discovery to attain sustainable lentil production under climate changing regime.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
KT and AK planned and designed the research. KT, PG, AKS, GC, RBh, RBa, and RM assisted in data recording. JK, KT, and DM analyzed the data. NS, VG, and PG contributed to the materials. DS and PG performed the passport data analysis and geo-referenced map. KT and PG prepared the manuscript. AS, GM, JR, and HD edited the manuscript. KS provided the necessary support for inter-institutional collaboration. All the authors contributed to the article and approved the submitted version.
This research was funded by the NBPGR-ICARDA collaborative research program on lentils (Project code: 1012548).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the support from the ICAR, NBPGR, and ICARDA. The authors would also like to thank Mr. Baburam and Mr. B. L. Meena for their technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.751429/full#supplementary-material
Archak, S., Tyagi, R. K., Harer, P. N., Mahase, L. B., Singh, N., Dahiya, O. P., et al. (2016). Characterization of chickpea germplasm conserved in the Indian National Genebank and development of a core set using qualitative and quantitative trait data. Crop J. 5, 417–424. doi: 10.1016/j.cj.2016.06.013
Benlloch, R., Berbel, A., Ali, L., Gohari, G., Millan, T., and Madueno, F. (2015). Genetic control of inflorescence architecture in legumes. Front. Plant Sci. 6:543. doi: 10.3389/fpls.2015.00543
Bisht, I. S., Mahajan, R. K., and Patel, D. P. (1998). The use of characterisation data to establish the Indian mungbean core collection and assessment of genetic diversity. Genet. Resour. Crop Evol. 45, 127–133. doi: 10.1023/A:1008670332570
Brown, A. H. D. (1989). Core collections: a practical approach to genetic resourcesmanagement. Genome 3, 818–824. doi: 10.1139/g89-144
Carvalho, M., Bebeli, P., Bettencourt, E., Costa, G., Dias, S., Santos, T., et al. (2013). Cereal landraces genetic resources in worldwide genebanks: a review. Agron. Sustain. Dev. 33, 177–203. doi: 10.1007/s13593-012-0090-0
Chowdhury, M., Dash, S., Sar, K., and Gulati, J. M. L. (2020). Pulses in rice fallow: a way towards achieving nutritional security: a review. Agric. Rev. 41, 264–271. doi: 10.18805/ag.R-2013
Devi, J., Mishra, G. P., Sanwal, S. K., Dubey, R. K., Singh, P. M., and Singh, B. (2018). Development and characterization of penta-flowering and triple-flowering genotypes in garden pea (Pisum sativum L. var. hortense). PLoS One 13:e0201235. doi: 10.1371/journal.pone.0201235
Díez, M. J., Rosa, D. l. L, Martín, I., Guasch, L., Cartea, M. E., Mallor, C., et al. (2018). Plant genebanks: present situation and proposals for their improvement. the case of the Spanish network. Front. Plant Sci. 9:1794. doi: 10.3389/fpls.2018.01794
Dutta, M., Phogat, B. S., Kumar, S., Kumar, N., Kumari, J., Pandey, A. C., et al. (2015). Development of Core Set of Wheat (Triticum spp.) Germplasm Conserved in the National Genebank in India. Tokyo: Springer, doi: 10.1007/978-4-431-55675-6_4
El-Ashkar, F., Sarker, A., Erskine, W., Bayaa, B., El-Hassan, H., Kadah, N., et al. (2004). Registration of ‘Idlib-4’ lentil. Crop Sci. 44, 2261–2263. doi: 10.2135/cropsci2004.2261a
Elias, L. G., Fernandez, D. G. D. E., and Bressani, R. (1979). Possible effects of seed coat polyphenolics on the nutritional quality of bean protein. J. Food Sci. 44, 524–527. doi: 10.1111/j.1365-2621.1979.tb03827
Erskine, W., Muehlbauer, F. J., Sarker, A., and Sharma, B. (2009). The Lentil: Botany, Production and Uses. Wallingford: CABI publishing, doi: 10.1079/9781845934873.0000
Erskine, W., Tufail, M., Russell, A., Tyagi, M. C., Rahman, M. M., and Saxena, M. C. (1993). Current and future strategies in breeding lentil for resistance to biotic and abiotic stresses. Euphytica 73, 127–135. doi: 10.1007/BF00027189
FAOSTAT (2021). data/QC/Visualize. Available online at: http://www.fao.org/faostat/en/ (accessed July 10, 2021)
Federer, W. T., and Raghavarao, D. (1975). On augmented designs. Biometrics 31, 29–35. doi: 10.2307/2529707
Fedoruk, M. (2013). Linkage and Association Mapping of Seed Size and Shape in Lentil. Ph.D. dissertation. Saskatoon, SK: University of Saskatchewan.
Frankel, O. H., and Brown, A. H. D. (1984). “Plant genetic resources today: a critical appraisal,” in The Crop Genetic Resources: Conservation and Evaluation, eds J. H. W. Holden and J. T. Williams (London: George Allen & Unwin), 249–257.
Gangopadhyay, K. K., Mahajan, R. K., Kumar, G., Yadav, S. K., Meena, B. L., Pandey, C., et al. (2010). Development of a core set in brinjal (Solanum melongena L.). Crop Sci. 50, 755–762. doi: 10.2135/cropsci2009.03.0151
Gaur, P. M., and Gour, V. K. (2002). A gene producing one to nine flowers per flowering node in chickpea. Euphytica 128, 231–235. doi: 10.1023/A:1020845815319
Gautam, N. K., Singh, M., Khan, Z., Roy, A., Akhtar, J., and Ram, B. (2013). Assessment of lentil germplasm with respect to agromonic performance and major biotic stress. Legum Res. 36, 214–219.
Gore, P. G., Tripathi, K., Pratap, A., Bhat, K. V., Umdale, S. D., Gupta, V., et al. (2019). Delineating taxonomic identity of two closely related Vigna species of section Aconitifoliae: V. trilobata (L.) Verdc. and V. stipulacea (Lam.) Kuntz in India. Genet. Resour. Crop Evol. 5, 1155–1165. doi: 10.1007/s10722-019-00767-9
Guerra-García, A., Gioia, T., Wettberg, V. E., Logozzo, G., Papa, R., Bitocchi, E., et al. (2021). Intelligent characterization of lentil genetic resources: evolutionary history, genetic diversity of germplasm, and the need for well-represented collections. Curr. Protoc. 1:e134. doi: 10.1002/cpz1.134
He, T., and Li, C. (2020). Harness the power of genomic selection and the potential of germplasm in crop breeding for global food security in the era with rapid climate change. Crop J. 8, 688–700. doi: 10.1016/j.cj.2020.04.005
Huamán, Z., Aguilar, C., and Ortiz, R. (1999). Selecting a Peruvian sweetpotato core collection on the basis of morphological, eco-geographical, and disease and pest reaction data. Theor. Appl. Genet. 98, 840–844. doi: 10.1007/s001220051142
Jha, G. K., Mazumder, C., Kumari, J., and Singh, G. (2014). Nonlinear principal component based fuzzy clustering: a case study of lentil genotypes. Indian J. Genet. Plant Breed. 74, 189–196.
Jones, Q. (1984). “A National plant germplasm system,” in The Conservation of Crop Germplasm-An International Perspective, eds W. L. Brown Chm, T. T. Chang, M. M. Goodman, and Q. Jones (Madison, WI: Crop Science Society of America), 27–33. doi: 10.2135/cssaspecpub8.c3
Keuls, M. (1952). The use of the studentized range in connection with an analysis of variance. Euphytica 1, 112–122. doi: 10.1007/BF01908269
Kim, K. W., Chung, H. K., Cho, G. T., Ma, K. H., Chandrabalan, D., Gwag, J. G., et al. (2007). PowerCore: a program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 16, 2155–2162. doi: 10.1093/bioinformatics/btm313
Kumar, J., and Gupta, S. D. (2020). Prospects of next generation sequencing in lentil breeding. Mol. Biol. Rep. 47, 9043–9053. doi: 10.1007/s11033-020-05891-9
Kumar, J., and Solanki, R. K. (2014). Evaluation of lentil germplasm for agro-morphological traits. J. Food Legumes 4, 302–306. doi: 10.1371/journal.pone.0229554
Kumar, J., Basu, P. S., Srivastava, E., Chaturvedi, S. K., Nadarajan, N., and Kumar, S. (2012). Phenotyping of traits imparting drought tolerance in lentil. Crop Pasture Sci. 6, 547–554. doi: 10.1071/CP12168
Kumar, S., Barpete, S., Kumar, J., Gupta, P., and Sarker, A. (2013). Global lentil production: constraints and strategies. SATSA Mukhapatra Annu. Tech. 17, 1–13. doi: 10.1098/rsif.2014.1092
Kumar, S., Rajendran, K., Kumar, J., Hamwieh, A., and Baum, M. (2015). Current knowledge in lentil genomics and its application for crop improvement. Front. Plant Sci. 6:78. doi: 10.3389/fpls.2015.00078
Kumari, J., Ahmad, R., Chandra, S., and Jha, G. K. (2009). Determination of morpholological attributes imparting resistance against aphids (Aphis craccivora Koch) in lentil (Lens culinaris Medik). Arch. Phytopathol. Pflanzenschutz 1, 52–57. doi: 10.1080/03235400600940806
Lamichaney, A., Parihar, A. K., Dixit, G. P., Singh, A. K., Kumar, N., and Singh, N. P. (2021). Intact funiculus in mature harvested seeds of field pea (Pisum sativum L.): preliminary investigation and possible implications. Crop Sci. 61, 2863–2871. doi: 10.1002/csc2.20582
Laskar, R. A., Khan, S., Deb, C. R., Tomlekova, N., Wani, M. R., Raina, A., et al. (2019). “Lentil (Lens culinaris Medik.) diversity, cytogenetics and breeding,” in Advances in Plant Breeding Strategies: Legumes, eds J. Al-Khayri, S. Jain, and D. Johnson (Cham: Springer), 319–369. doi: 10.1007/978-3-030-23400-3_9
Levene, H. (1960). “Robust tests for equality of variances,” in The Contributions to Probability and Statistics: Essays in Honour of Harold Hotelling, ed. I. Olkin (Stanford, CA: Stanford University Press), 278–292.
Mahajan, R. K., Sapra, R. L., Srivastava, U., Singh, M., and Sharma, G. D. (2000). Minimal Descriptors for Agri-Horticultural Crops. Part I. New Delhi: National Bureau of Plant Genetic Resources, 22–25.
Mahalakshmi, V., Ng, Q., Lawson, M., and Ortiz, R. (2007). Cowpea [Vigna unguiculata (L.)Walp.] core collection defined by geographical, agronomical and botanical descriptors. Plant Genet. 5, 113–119. doi: 10.1017/S1479262107837166
Mishra, G. P., Dikshit, H. K., Kumari, J., Tripathi, K., Devi, J., Aski, M., et al. (2020). Identification and characterization of novel penta-podded genotypes in the cultivated lentil. Crop Sci. 4, 1974–1985. doi: 10.1002/csc2.20156
Nanjundan, J., Aravind, J., Radhamani, J., Singh, K. H., Kumar, A., Thakur, A. K., et al. (2021). Development of Indian mustard [Brassica juncea (L.) Czern.] core collection based on agro-morphological traits. Genet. Resour. Crop Evol. 69, 145–162. doi: 10.1007/s10722-021-01211-7
Newman, D. (1939). The distribution of range in samples from a normal population expressed in terms of an independent estimate of standard deviation. Biometrika 31, 20–30. doi: 10.1093/biomet/31.1-2.20
Nichols, P. G. H., Collins, W. J., and Barbetti, M. J. (1996). Registered Cultivars of Subterranean Clover-their Characteristics, Origin and Identification, Bulletin No. 4327. South Perth, WA: Agriculture Western Australia.
Noirot, M., Hamon, S., and Anthony, F. (1996). The principal component scoring: a new method of constituting a core collection using quantitative data. Genet. Resour. Crop Evol. 43, 1–6. doi: 10.1007/BF00126934
Oliveira, M. F., Nelson, R. L., Geraldi, O. I., Cru, C. D., José, C., and de Toledoa, J. F. F. (2010). Establishing a soybean germplasm core collection. Field Crops Res. 119, 277–289. doi: 10.1016/j.fcr.2010.07.021
Ortiz, R. (2002). No Just Seed Repositories: A More Proactive role for Gene Banks. Gene Conserve. Wallingford: CAB International.
Ortiz, R., Braun, H. J., Crossa, J., Crouch, J. H., Davenport, G., Dixon, J., et al. (2008). Wheat genetic resources enhancement by the International Maize and Wheat Improvement Center (CIMMYT).Genet. Resour. Crop Evol. 7, 1095–1140. doi: 10.1007/s10722-008-9372-4
Ortiz, R., Madsen, S., Ruiz-Tapia, E. N., Jacobsen, S. E., Mujica-Sánchez, A., Christiansen, J. L., et al. (1999). Validating a core collection of Peruvian quinoa germplasm.Genet. Resour. Crop Evol. 3, 285–290. doi: 10.1023/A:1008636232584
Ortiz, R., Ruiz-Tapia, E. N., and Mujica-Sanchez, A. (1998). Sampling strategy for a core collection of Peruvian quinoa germplasm. Theor. Appl. Genet. 96, 475–483. doi: 10.1007/s001220050764
Phogat, B. S., Kumar, S., Kumari, J., Kumar, N., Pandey, A. C., Singh, T. P., et al. (2020). Characterization of wheat germplasm conserved in the Indian National Genebank and establishment of a composite core collection. Crop Sci. 61, 604–620. doi: 10.1002/csc2.20285
Pundir, R. P. S., Rao, N. K., and Van den Maesen, L. J. G. (1985). Distribution of qualitative traits in the world germplasm of chickpea (Cicer arietinum L.). Euphytica 34, 697–703. doi: 10.1007/BF00035406
Qiu, L. J., Xing, L. L., Guo, Y., Wang, J., Jackson, S. A., and Chang, R. Z. (2013). A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol. Biol. 83, 41–50. doi: 10.1007/s11103-013-0076-6
Reddy, L. J., Upadhyaya, H. D., Gowda, C. L. L., and Singh, S. (2005). Development of core collection in pigeonpea [Cajanus cajan (L.) Millspaugh] using geographic and qualitative morphological descriptors.Genet. Resour. Crop Evol. 52, 1049–1056. doi: 10.1007/s10722-004-6152-7
Sandhu, K., and Singh, A. (2021). Strategies for the utilization of the USDA mung bean germplasm collection for breeding outcomes. Crop Sci. 61, 422–442. doi: 10.1002/csc2.20322
Sanwal, S. K., Kumar, R., and Singh, B. (2016). VRP-500 (IC610501; INGR15009), a garden pea (Pisum sativum) germplasm with triple pods at every node. Indian J. Plant Genet. Resour. 29:90.
Sarker, A., Erskine, W., and Singh, M. (2005). Variation in shoot and root characteristics and their association with drought tolerance in lentil landraces. Genet. Resour. Crop Evol. 52, 89–97. doi: 10.1007/s10722-005-0289-x
Semwal, D. P., Pandey, A., Bhandari, D. C., Dhariwal, O. P., and Sharma, S. K. (2014). Variability study in seed morphology and uses of indigenous rice (Oryza sativa L.) landraces collected from West Bengal, India. Aust. J. Crop Sci. 8, 860–867.
Shannon, C. E., and Weaver, W. (1949). The Mathematical Theory of Communication. Champaign, IL: The University of Illinois Press.
Sharma, U., Singh, S. R., Aggarwal, S., Kaur, N., Gill, J., Kushwah, R. K., et al. (2018). Genetic variation for tolerance to post-emergence herbicide, imazethapyr in lentil (Lens culinaris Medik.) Arch. Agron. Soil Sci. 64, 1818–1830. doi: 10.1080/03650340.2018.1463519
Shrestha, R., Turner, N. C., Siddique, K. H. M., Turner, D. W., and Speijers, J. (2006). A water deficit during pod development in lentils reduces flower and pod numbers but not seed size.Aust. J. Agric. Res. 57, 427–438. doi: 10.1071/Ar05225
Simon, C. J., and Hannan, R. (1995). Development and use of core subsets of cool season food legume germplasm collections. Hort. Sci. 304:907.
Singh, M., Bisht, I. S., Kumar, S., Dutta, M., Bansal, K. C., Karale, M., et al. (2014). Global wild annual lens collection: a potential resource for lentil genetic base broadening and yield enhancement. PLoS One 9:e107781. doi: 10.1371/journal.pone.0107781
Sturges, H. A. (1926). The choice of a class-interval. J. Am. Stat. Assoc. 21, 65–66. doi: 10.1080/01621459.1926.10502161
Tripathi, K., Gore, P. G., Bansal, R., Gayacharan, C., Kumari, S., Kumar, V., et al. (2021). Identification and revealing the potential traits of the unique germplasm with extended funiculus in pea (Pisum sativum L.). Genet. Resour. Crop Evol. 68, 3125–3132. doi: 10.1007/s10722-021-01275-5
Tripathi, K., Gore, P. G., Pandey, A., Bhardwaj, R., Singh, N., Chawla, G., et al. (2019). Seed morphology, quality traits and imbibition behaviour study of atypical lentil (Lens culinaris Medik.) from Rajasthan. India. Genet. Resour. Crop Evol. 66, 697–706. doi: 10.1007/s10722-019-00745-1
Tripathi, K., Parihar, A. K., Murthy, N., Wankhede, D. P., Singh, N., Deshpande, S. K., et al. (2020). Identification of a unique accession in cowpea with dense pubescence. J. Food Legumes 33, 278–279.
Tullu, A., Kusmenoglu, I., McPhee, K. E., and Muehlbauer, F. J. (2001). Characterization of core collection of lentil germplasm for phenology, morphology, seed and straw yields. Genet. Resour. Crop Evol. 48, 143–152. doi: 10.1023/A:1011254629628
Upadhyaya, H. D., Bramel, P. J., and Singh, S. (2001). Development of a chickpea core subset using geographic distribution and quantitative traits. Crop Sci. 41, 206–210.
Upadhyaya, H. D., Ortiz, R., and Bramel, P. J. (2003). Development of a groundnut corecollection using taxonomical, geographical and morphological descriptors. Genet. Resour. Crop Evol. 50, 139–148. doi: 10.1023/A:1022945715628
Vaijayanthi, P. V., Ramesh, S., Gowda, M. B., Rao, A. M., and Keerthi, C. M. (2015). Development of core sets of Dolichos bean (Lablab purpureus L. Sweet) germplasm. J. Crop Improv. 29, 405–419. doi: 10.1080/15427528.2015.1036955
van Hintum, T. J. L., Brown, A. H. D., Spillane, C., and Hodgkin, T. (2000). Core Collections of Plant Genetic Resources. IPGRI Technical Bulletin No. 3. Rome: International Plant Genetic Resources Institute, 48.
Wang, J. C., Hu, J., Huang, X., and Xu, S. C. (2008). Assessment of different genetic distances in constructing cotton core subset by genotypic values. J. Zhejiang. Univ. Sci. 9, 356–362. doi: 10.1631/jzus.B0710615
Zhang, H., Zhang, D., Wang, M., Sun, J., Qi, Y., Li, J., et al. (2011). A core collection and mini core collection of Oryza sativa L. in China. Theor. Appl. Genet. 122, 49–61. doi: 10.1007/s00122-010-1421-7
Keywords: Lens culinaris, characterization, core-set, genetic diversity, trait-specific germplasm
Citation: Tripathi K, Kumari J, Gore PG, Mishra DC, Singh AK, Mishra GP, C G, Dikshit HK, Singh N, Semwal DP, Mehra R, Bhardwaj R, Bansal R, Rana JC, Kumar A, Gupta V, Singh K and Sarker A (2022) Agro-Morphological Characterization of Lentil Germplasm of Indian National Genebank and Development of a Core Set for Efficient Utilization in Lentil Improvement Programs. Front. Plant Sci. 12:751429. doi: 10.3389/fpls.2021.751429
Received: 01 August 2021; Accepted: 21 December 2021;
Published: 27 January 2022.
Edited by:
Patricia L. Polowick, National Research Council Canada (NRC-CNRC), CanadaCopyright © 2022 Tripathi, Kumari, Gore, Mishra, Singh, Mishra, C, Dikshit, Singh, Semwal, Mehra, Bhardwaj, Bansal, Rana, Kumar, Gupta, Singh and Sarker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuldeep Tripathi, a2R0cmlwYXRoaTg5QGdtYWlsLmNvbQ==; Ashok Kumar, YXNob2sua3VtYXIyOEBpY2FyLmdvdi5pbg==; Ashutosh Sarker, YS5zYXJrZXJAY2dpYXIub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.