- Crop Information Center, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
There is limited advancement on seed number per pod (SNPP) in soybean breeding, resulting in low yield in China. To address this issue, we identified PIN1 and CKX gene families that regulate SNPP in Arabidopsis, analyzed the differences of auxin and cytokinin pathways, and constructed interaction networks on PIN1, CKX, and yield-related genes in soybean and cowpea. First, the relative expression level (REL) of PIN1 and the plasma membrane localization and phosphorylation levels of PIN1 protein were less in soybean than in cowpea, which make auxin transport efficiency lower in soybean, and its two interacted proteins might be involved in serine hydrolysis, so soybean has lower SNPP than cowpea. Then, the CKX gene family, along with its positive regulatory factor ROCK1, had higher REL and less miRNA regulation in soybean flowers than in cowpea ones. These lead to higher cytokinin degradation level, which further reduces the REL of PIN1 and decreases soybean SNPP. We found that VuACX4 had much higher REL than GmACX4, although the two genes essential in embryo development interact with the CKX gene family. Next, a tandem duplication experienced by legumes led to the differentiation of CKX3 into CKX3a and CKX3b, in which CKX3a is a key gene affecting ovule number. Finally, in the yield-related gene networks, three cowpea CBP genes had higher RELs than two soybean CBP genes, low RELs of three soybean-specific IPT genes might lead to a decrease in cytokinin synthesis, and some negative and positive SNPP regulation were found, respectively, in soybean and cowpea. These networks may explain the SNPP difference in the two crops. We deduced that ckx3a or ckx3a ckx6 ckx7 mutants, interfering CYP88A, and over-expressed DELLA increase SNPP in soybean. This study reveals the molecular mechanism for the SNPP difference in the two crops, and provides an important idea for increasing soybean yield.
Introduction
Soybean is a major oil crop in China and an important source of plant protein for human beings. However, the soybean imports of China have increased from 58.38 million tons in 2012 to 100.32 million tons in 2020, which has seriously affected the food security in China. In order to revitalize the soybean industry in China, the key is to increase the yield per unit area. Although seed number per pod (SNPP) in soybean is an important yield component factor, such as the utilization of gene ln in Zhonghuang13 (Zhu et al., 2020), the advance in long-term soybean breeding is limited. Thus, increasing SNPP of soybean is a new direction for increasing its yield per unit area. Although elite genes for important traits in most crops frequently come from their wild or closely related species, and the SNPP of cultivated soybean is almost the same as that of wild soybean, cowpea has much higher SNPP (approximately 12) than soybean (2–4). Therefore, it is necessary to investigate the molecular mechanism of SNPP difference between soybean and cowpea.
In the past decade, efforts have been made to dissect the genetic foundation and molecular mechanism of SNPP. As described by Carlson and Lersten (2004), the SNPP difference is mainly caused by ovule number per pistil. Jeong et al. (2012) cloned a soybean locus ln and proved its pleiotropy for narrow-leaf and higher seed number. Cai et al. (2021) used CRISPR/Cas9 technology to edit gene Ln and its homologous genes in soybean cultivar “Huachun 6” to create a new material carrying gene ln, which is available for the future field breeding. To confirm whether the difference of SNPP between Ln and ln genotypes is caused by ovule number per pistil, Fang et al. (2013) found that ovate leaflet cultivar “Han 2296” with two-seeded pods has two to three ovules per ovary, and narrow leaflet cultivar “Lvbaoshi” with four-seeded pods has three to four ovules per ovary. This indicates that the Ln gene may influence SNPP by regulating ovule number per pistil. In Arabidopsis thaliana, ovules are produced from the placenta as lateral organs, and the ovule number in each ovary is regulated by plant hormones, such as auxin and cytokinin (Bencivenga et al., 2012; Galbiati et al., 2013; Reyes-Olalde et al., 2013; Cucinotta et al., 2014).
Auxin is required for placenta formation and ovule growth, and reduced auxin biosynthesis or transport in plants leads to severe defects in gynoecium development, resulting in loss of placenta tissue and ovule (Nemhauser et al., 2000; Nole-Wilson et al., 2010). The auxin exogenous vector PIN1 is one main element that regulates auxin accumulation at various ovule development stages. The PIN1-dependent auxin efflux mediates primordium development by supplying the apex of the ovule primordium with an auxin maximal zone (Benková et al., 2003; Ceccato et al., 2013). The expression level of PIN1 in Arabidopsis pin1-5 mutants and the number of ovules are reduced, compared with those in wild-type Col-0 (Bencivenga et al., 2012). Yu et al. (2020) showed that ovule primordia initiate asynchronously and new ovule primordia formation still requires the auxin maximal zone. Taken together, it is of great significance to investigate the difference of PIN1-mediated auxin transport in order to dissect the molecular mechanism for the difference of ovule numbers between soybean and cowpea.
Cytokinins (CK) are positive regulators of shoot apical meristem (Werner et al., 2001, 2003; Riefler et al., 2006; Kurakawa et al., 2007) and play important roles in ovule development. The defects in plant cytokinin production or perception affected ovule formation (Higuchi et al., 2004; Nishimura et al., 2004; Bencivenga et al., 2012). After the treatment of synthetic cytokinin 6-benzylaminopurine (BAP) in Arabidopsis thaliana and Brassica napus inflorescence, the expression level of PIN1 in the pistil and the ovule number per pistil increased (Bencivenga et al., 2012; Zuñiga-Mayo et al., 2018). Cytokinin is specifically degraded by Cytokinin dehydrogenase (CKX). As compared to wild types, ckx3 ckx5 mutants in Arabidopsis increased SNPP and seed yield (Bartrina et al., 2011), and bnckx3 bnckx5 sixfold mutant in Brassica napus increased ovule numbers per pistil and pod numbers, resulting in an increase in final seed yield (Schwarz et al., 2020). In conclusion, CKX-mediated cytokinin degradation may be the key to improving crop yield.
To dissect possible molecular mechanisms for the SNPP difference between soybean and cowpea, first, in this study, we identified PIN1 and CKX gene families in soybean and cowpea genomes, and analyzed the differences of auxin and cytokinin pathways between the two crops to mine SNPP-related genes. Then, we constructed interaction networks on PIN1, CKX, SNPP, and yield-related genes in soybean and cowpea to explore possible molecular mechanisms for the SNPP difference in the two legumes. In addition, we discussed how to improve SNPP in soybean.
Materials and Methods
Data Sources
The whole genome protein sequences in Arabidopsis and soybean were downloaded from TAIR database (1 TAIR release 10) and SoyBase database (2 version wm82.a2. V1; Schmutz et al., 2010), respectively, while the whole genome protein sequences in cowpea and kidney bean were download from Legumeinfo database (3 Lonardi et al., 2019; version gnm1.ann1).
Transcriptome datasets were downloaded from Phytozome database4 for soybean flower and embryo, Legumeinfo database (see text footnote 3) for cowpea and kidney bean flower and embryo, NCBI5 for soybean seed, and NCBI6 for cowpea seed.
Seven hundred fifty-six mature miRNA sequences in soybean were downloaded from miRBase database7, while 656 mature miRNA sequences in cowpea were downloaded from Martins et al. (2020).
Identification of Homologous Gene Families and Phylogenetic Tree Construction
The OrthoFinder was used to identify homologous gene families (Emms and Kelly, 2015) when we input the whole genome protein sequence for Arabidopsis, soybean, and cowpea, and adopted the default parameters. Based on the homologous gene families, the number of unique, single-copy, multi-copy, and unclustered gene families and protein numbers in soybean and cowpea were counted.
The phylogenetic tree was constructed by Neighbor–Joining (NJ) approach using the MEGA7 software8 and protein sequence alignment was carried out by the ClustalW method. The number of replicates in Bootstrap Test was set as 1,000, and the other parameters were set as default values. The final result tree file was visualized and beautified using the ITOL online tool9.
Gene Structure and Protein Structure Analyses and Subcellular Localization Prediction
The online tools GSDS2.0 (10 Gene Structure Display Server), pfam11, Wolf PSORT12, and MEME13 were used to graphically display gene structure, search the target protein sequences of conservative domain structure, predict the subcellular localization of target protein, and analyze conservative motif, respectively.
Collinearity and Protein Interaction Analyses
The collinearity among Arabidopsis thaliana, soybean, cowpea, and the collinearity within soybean species, were analyzed using TBtools software (Chen et al., 2020). The sequences of target proteins in Arabidopsis were submitted to the STRING database14 to search for interaction proteins with experimental evidence, active interaction sources in soybean were set up as “all,” and combined score >0.4 was regarded as the cut-off point of significance. In cowpea, there is no interacted protein database available. Thus, interaction protein network in mung bean (Vigna radiata), which is the closest species of cowpea, was used to represent the one in cowpea. Here, comparative genomics analysis between cowpea and mung bean was conducted on Phytozome and OrthoFinder. The protein interaction network was beautified by the Cytoscape software. Gene function annotation was conducted on SoyBase and Phytozome for soybean and TAIR for Arabidopsis.
Expression Pattern Analysis and Relative Expression Level Comparison
The expression patterns of genes in flower, leaf, stem, root, pod, and seed in soybean, cowpea, and kidney bean were analyzed via standardized expression levels, log2(y + 1), where y was real expression level. Relative expression levels were defined as the ratio of the expression level of one gene to average expression level of all the genes in the species (deleting the genes with expression level less than 1.0) (Zhang et al., 2018; Zhou et al., 2019; Cheng et al., 2021) and used to compare the differences of expression levels of genes in the flowers of soybean and cowpea. The TBtools and SigmaPlot software packages were used to draw HeatMap and relative expression levels, respectively.
Prediction of MicroRNAs and Expansion Pattern Analysis of Gene Families
The online tool psRNATarget15, a plant small RNA target analysis server, was used to predict micoRNAs, implemented by Schema V2 (2017 release) where expected value was set as 4 and other parameters were set as their default values. The number of predicted miRNA for each gene was counted and then plotted by SigmaPlot.
In this study, we focused on two types of patterns in gene expansion: tandem and segmental duplications. The above collinearity results were used to determine duplicated gene pairs, and these gene pairs were compared with the gene pairs downloaded from PlantDGD16 to validate the predicted gene pairs. The formula T = Ks/2λ was used to calculate the date of occurrence of repeated events, where λ is equal to 6.1 × 10–9 (Lynch and Conery, 2000).
Results
Identification and Copy Number Analysis of Homologous Gene Families in Arabidopsis, Soybean, and Cowpea
To identify orthologous genes (OGs) in soybean and cowpea, all the genes in Arabidopsis, soybean, and cowpea were clustered using the OrthoFinder software. As a consequence, 113,233 protein-coding genes from the three species were clustered into 21,582 OGs (Supplementary Table 1), with each OG representing a gene family. Among these gene families, 1,541 (7.14%) were identified as soybean-specific gene families, and only 451 (2.09%) were identified as cowpea-specific gene families. The two proportions (9.23%) were very low, indicating the very high similarity in the evolutionary process between soybean and cowpea. Meanwhile, 549 (2.54%) OGs with single-copy soybean and multi-copy cowpea genes was significantly lower than 10,196 (47.24%) OGs with single-copy cowpea and multi-copy soybean genes. This means that soybean underwent a unique allotetraploidy event.
Difference of Auxin Transport Mediated by PIN1 Gene Family in Soybean and Cowpea
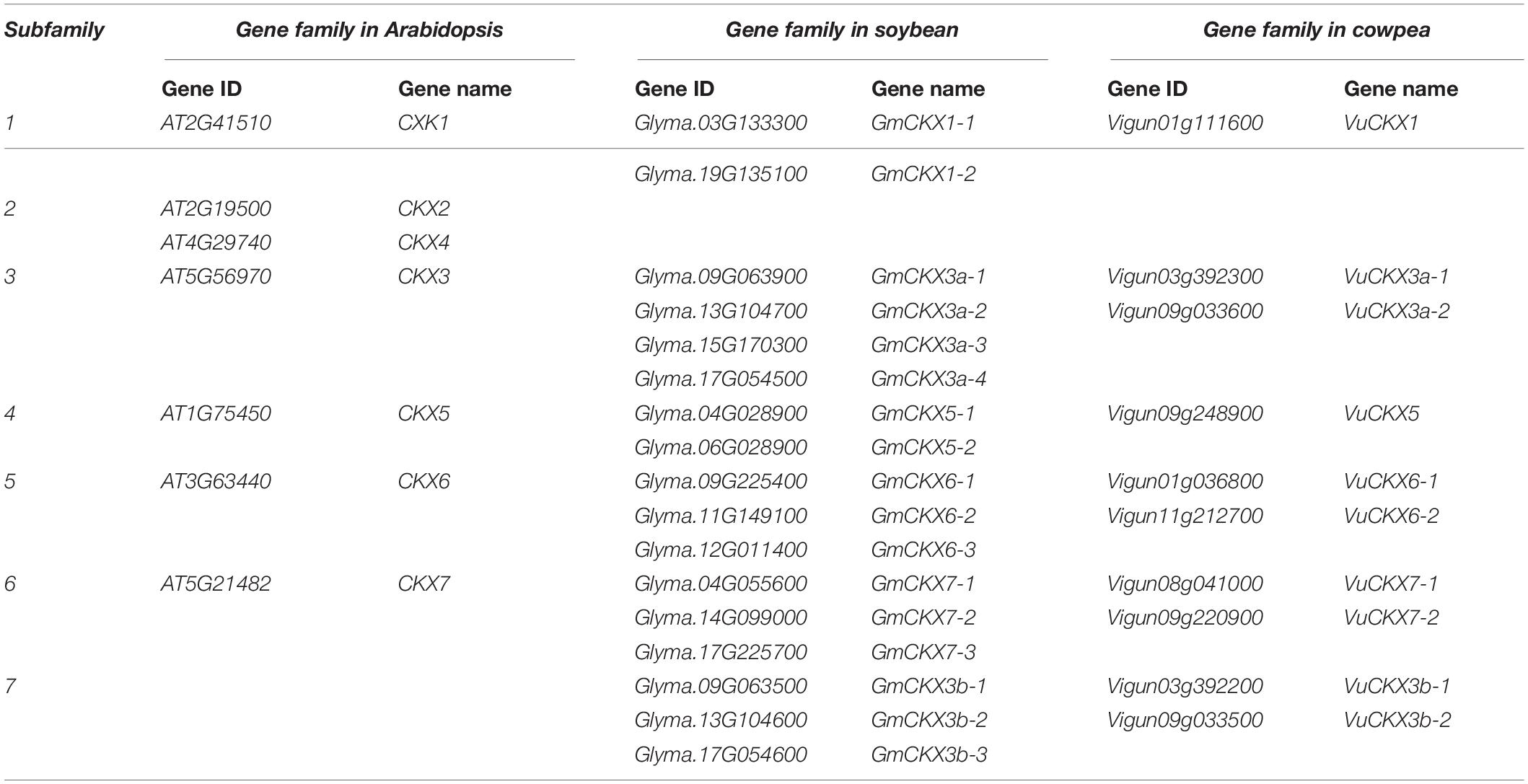
Among all the above OGs, five GmPIN1 genes in soybean and three VuPIN1 genes in cowpea were identified. The genes GmPIN1a, GmPIN1b, GmPIN1c, VuPIN1a, VuPIN1b, and A. thaliana PIN1 were found to be in one gene family, and GmPIN1b and VuPIN1b had the highest relative expression levels in the flowers of soybean and cowpea (Table 1) and the closest genetic distance to A. thaliana PIN1 gene (Figure 1A). All the PIN1 genes were predicted by the Wolf PSORT software to be localized in the plasma membrane.
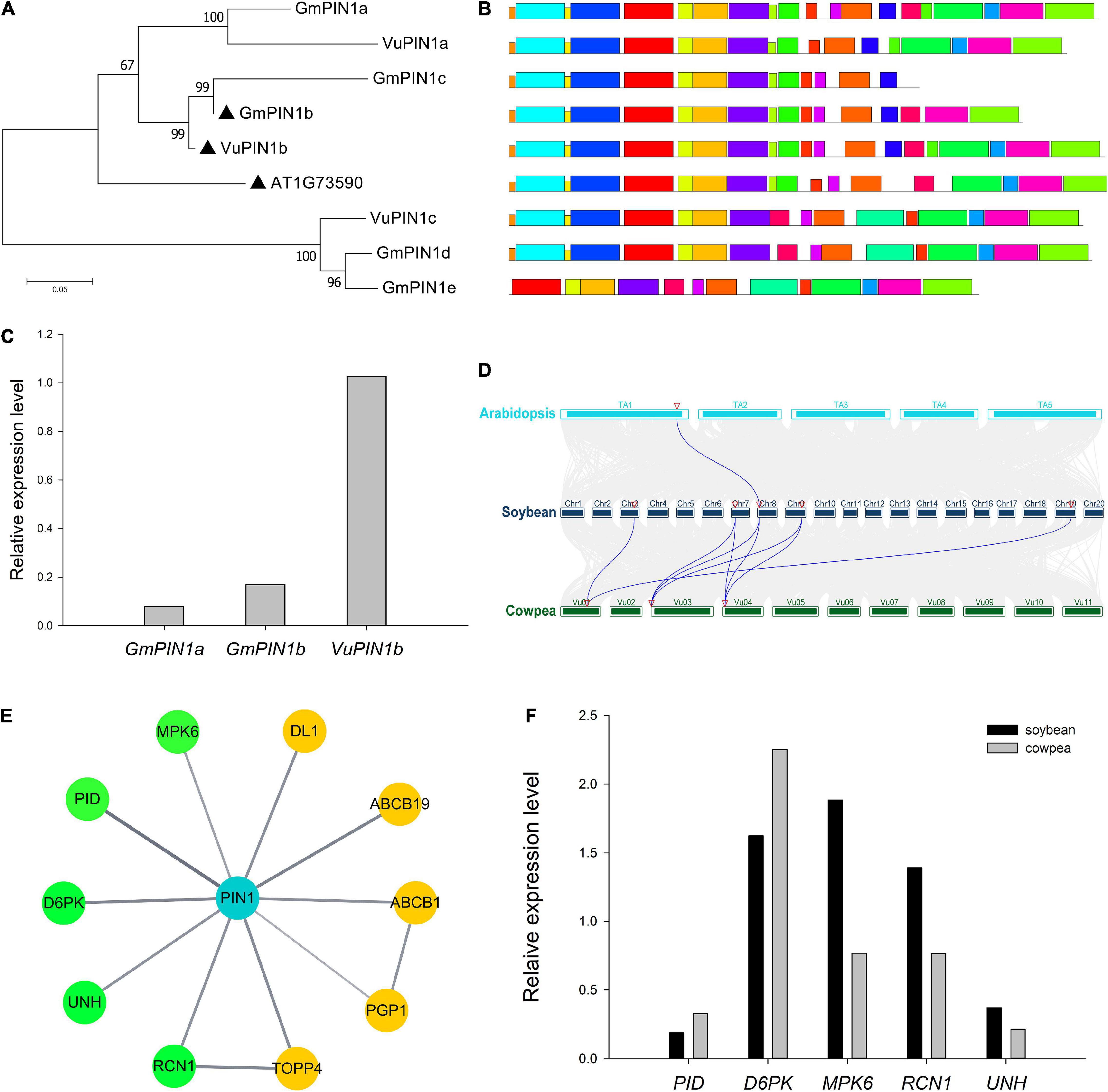
Figure 1. Analysis of PIN1 and its interaction proteins in soybean and cowpea. (A) Phylogenetics tree of PIN1 gene family. (B) Motif analysis of PIN1 proteins. (C) Relative expression levels of targeted PIN1 in soybean and cowpea flowers. (D) Collinearity analysis of PIN1 gene family. (E) The network of proteins interacted with PIN1 in Arabidopsis predicted from the STRING database. The proteins with similar expression levels in soybean and cowpea were marked by yellow color, while those with significantly different expression levels in the two crops were marked by green color. (F) Comparison of the relative expression levels of genes, interacted with PIN1, between soybean and cowpea. As described by Zhang et al. (2018), the relative expression levels was calculated.
In the conserved motif analysis via the MEME software, almost all the PIN1 proteins were structurally relatively conserved (Figure 1B), except for GmPIN1c and GmPIN1e, which may be the repeat proteins with the lack of the sequences at the 3′ end of GmPIN1b and at the 5′ end of GmPIN1d, respectively. In the comparison of relative expression levels, GmPIN1a and GmPIN1b had significantly lower relative expression levels than VuPIN1b (Figure 1C). In the collinearity analysis in Arabidopsis, soybean, and cowpea via the TBtools software (Chen et al., 2020), GmPIN1a was collinear with A. thaliana PIN1 gene; GmPIN1d and GmPIN1e were collinear with VuPIN1c; and GmPIN1a, GmPIN1b, and GmPIN1c were collinear with VuPIN1a and VuPIN1b. The results were consistent with those in the evolutionary tree (Figure 1D).
Comparison of Proteins Interacted With PIN1, CKX3, and CKX5 Between Soybean and Cowpea
Ten proteins in A. thaliana were found in STRING database to be interacted with A. thaliana PIN1 based on experimental evidence. These proteins were used to mine homologous genes in soybean and cowpea. If one protein had multiple homologous genes in soybean or cowpea, the gene with the highest relative expression level was selected. In the large gene family of TOPP4; meanwhile, further phylogenetic analysis was conducted to determine the closest homologous relationship (Supplementary Figure 1A). The relative expression levels of homologous genes selected in soybean and cowpea were compared to find the differences of these homologous genes in soybean and cowpea. As a result, MPK6, PID, D6PK, UNH, and RCN1 had significant differences of relative expression levels in soybean and cowpea (Figure 1F), while ABCB1, ABCB19, DL1, PGP1, and TOPP4 had similar relative expression levels in soybean and cowpea (Figure 1E and Supplementary Table 1). Here we focused on the first five proteins.
First, we discussed their functions. PID and D6PK have been confirmed to positively regulate auxin transport through phosphorylation at the PIN1 serine S1 (S231), S2 (S252), S4 (S271), and S3 (S290), which plays an important role in ovular formation (Benjamins et al., 2001; Lee and Cho, 2006; Zourelidou et al., 2014). The S1 to S4 phosphate sites in A. thaliana were also found in the five soybean and cowpea PIN1 proteins, which are in the same OG as A. thaliana PIN1 (Supplementary Figure 2). RCN1 is found to inhibit the phosphorylation of PIN1 protein to antagonize the polar transport of auxin (Rashotte et al., 2001; Zhou et al., 2004; Michniewicz et al., 2007; Dai et al., 2012), while MPK6-mediated phosphorylation of PIN1 leads to the loss of the plasma membrane localization of PIN1, affecting auxin polar transport (Dory et al., 2018). Up to now, only one phosphate site of MPK6, S337, has been confirmed (Jia et al., 2016), and this site was found to be conserved in soybean and cowpea PIN1 proteins (Supplementary Figure 2). UNH is important in reducing PIN1 expression level in marginal cells, possibly through the localization of PIN1 into vacuoles for degradation (Pahari et al., 2014).
Then, we compared their relative expression levels. As a result, PID and D6PK, which are conducive to the phosphorylation of PIN1, had lower relative expression levels in soybean than in cowpea, while RCN1 and UNH, which affect plasma membrane localization, and MPK6, which affects the phosphorylation of PIN1, had higher relative expression levels in soybean than in cowpea. We speculated that an important reason for the SNPP difference between soybean and cowpea lies in the difference of relative expression level of PIN1. Owing to the difference of relative expression levels of genes encoding the interaction proteins of PIN1, PIN1 protein in soybean was less located on plasma membrane, and there was lower phosphorylation level in soybean. Thus, lower auxin transport efficiency results in less auxin maximum zone and fewer ovules and SNPP.
More importantly, the above method was used to predict the proteins that interact with PIN1, CKX3, and CKX5 homologies in soybean and cowpea. The results are shown in Figure 2A and Supplementary Table 7. In the proteins interacted with PIN1 homologies, two soybean proteins, GmABCB19 (Glyma.13G063700) and GmPID (Glyma.13G220100), were consistent with those in Arabidopsis, while six cowpea proteins, VuABCB1 (Vigun01g162000), VuABCB2 (Vigun07g072700), VuABCB19 (Vigun04g051400), VuMPK6 (Vigun03g181200), VuD6PK (Vigun06g148700), and VuPID (Vigun06g179800), were consistent with those in Arabidopsis. Based on the functions and annotations of these interacted genes, they commonly focus on auxin transport and serine phosphorylation. Interestingly, Glyma.02G186700 and Glyma.10G106900 with high expression levels in soybean ovary may be involved in the hydrolysis of serine, which is the active site of PIN1, because their homology AT2G41530 (AtSFGH) had been proven to encode a serine hydrolase in Arabidopsis (Cummins et al., 2006). CKX3 and CKX5 homologies in the two legumes commonly focus on embryo development and substance metabolism (Figure 2B). GmSLD5 (Glyma.08g194400, and Glyma.07g011200) and VuSLD5 (Vigun10g174500) are the homologies of AT5G49010 (SLD5), whose mutant can cause defective embryo development in Arabidopsis (Meinke, 2020). Another pair of genes, GmACX4 (Glyma.18g202800) and VuACX4 (Vigun10g081200), are the homologies of ACX4 (AT3G51840), which encodes a short-chain acyl-CoA oxidase. ACX4 is essential at early embryo development stages, and the acx3acx4 double mutants abort during the first embryo development phase (Rylott et al., 2003). VuACX4 (1.98) had much higher relative expression level than GmACX4 (0.51). This may lead to better embryo development and more seeds in cowpea than in soybean. Thus, we infer that the hindrance of ovular formation via serine hydrolysis in soybean and the promotion of embryo development via VuACX4 in cowpea may be responsible for the SNPP difference in the two legumes.
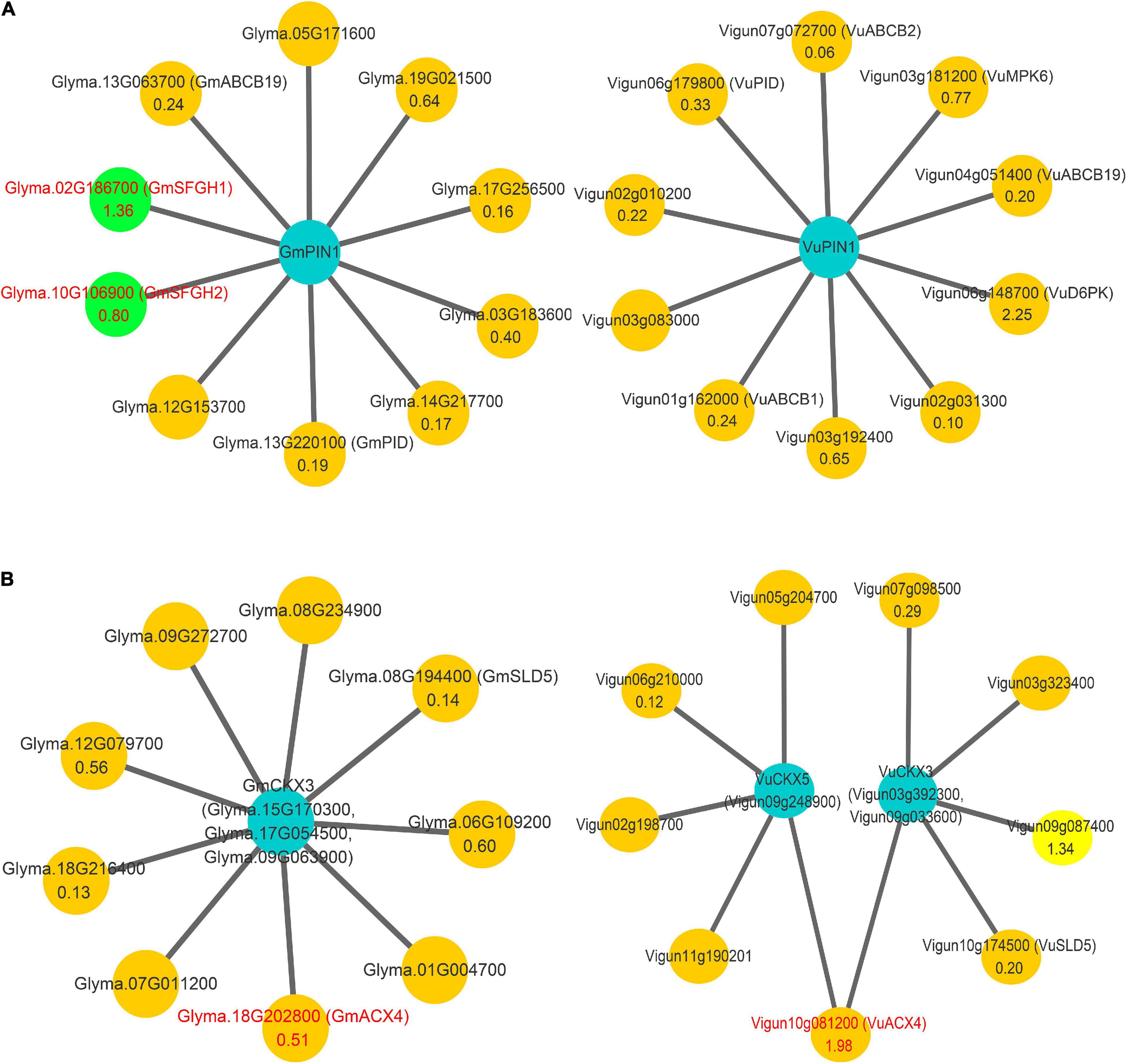
Figure 2. The interaction networks on PIN1, CKX3, and CKX5 in soybean and cowpea. (A) The interaction networks on GmPIN1 and VuPIN1. (B) The interaction networks on GmCKX3, VuCKX3, and VuCKX5. All the genes that encode serine hydrolase in soybean and short-chain acyl-CoA oxidase in cowpea were marked by red color and green background.
Difference of Cytokinin Degradation Mediated by CKX Gene Family Between Soybean and Cowpea
Phylogenetic and Structural Analyses of CKX Gene Family
Based on the protein sequence homology, the collinearity of gene sequences, and seven A. thaliana CKX genes, it was determined that a total of 17 soybean CKX genes and 10 cowpea CKX genes were in the same CKX gene family (Figures 3A,B). In the gene family, two kinds of soybean genes were homologous to the CKX3 Arabidopsis gene, higher homologous genes were named CKX3a, and other ones generated by tandem duplication were named CKX3b (Table 2). This tandem duplication was also found in acacia bean, Medicago truncatula (Medtr4G126160) and chickpea rather than in oilseed rape and rice. We deduced that the tandem duplications of CKX3 are a specific event experienced in the evolutionary process of leguminosae.
Figure 3. Bioinformatics analysis of CKX gene family in soybean. (A) Collinearity analysis of CKX gene family in soybean. (B) Phylogenetics tree of CKX gene family in Arabidopsis, Glycine max, and Vigna unguiculata. (C) The conserved motifs of GmCKX gene family.
In the domain analysis of the CKX proteins, these proteins contained cytokinin-bind and FAD binding 4 domains, which affect the binding of CKX proteins to cytokinins and FAD cofactors, respectively. Note that the lack of the first half of FAD binding domain sequences in GmCKX5-1, GmCKX5-2, and GmCKX7-1 may affect the binding of these proteins with FAD cofactors, and thus affect their functions and expression levels (Supplementary Figure 3).
In the phylogenetics tree, all the above 34 CKX genes were divided into seven classes, including CKX1, CKX2 and CKX4, CKX3a, CKX5, CKX6, CKX7, and CKX3b classes (Figure 3B). Among these classes, CKX3b is close to CKX3a, and CKX1 is close to CKX6 in evolutionary relationship (Figure 3B). For the structural analysis of 17 GmCKX genes, all the structural information is shown in Supplementary Figure 4. We found that GmCKX1 and GmCKX3a gene families had the highest similarity in the number, distribution, and length of CDS. In the motif analysis of the 17 CKX genes in soybean using MEME online tool, 23 conserved motifs were identified. Among these motifs, GmCKX6 and GmCKX1 had the same motif due to having similar function, GmCKX5 had no specific motifs, and the other GmCKX families had some specific motifs, i.e., motif15 is specific to GmCKX3a proteins, and motif19 is specific to GmCKX3b proteins. These specific motifs may be related to their specific functions (Figure 3C).
Expression Pattern and Expression Level Analyses of CKX Gene Family
We downloaded and analyzed the expression levels of these CKX genes in different tissues of soybean and cowpea. In soybean, GmCKX5 with incomplete domain and GmCKX3b-1 were hardly expressed in each tissue, while GmCKX3a, GmCKX7-2, and GmCKX6-1 were expressed mainly in flowers, and GmCKX3b-2 and GmCKX3b-3 were expressed in roots. Meanwhile, VuCKX5 with a complete domain in cowpea was expressed in flowers, roots, and pods, VuCKX6 and VuCKX7 genes were highly expressed in roots, VuCKX3a gene was expressed in flowers, and VuCKX3b gene was not expressed in any tissues (Figure 4A). In addition, we identified 10 CKX genes in kidney bean, which correspond to 10 VuCKX genes in the evolutionary tree (Supplementary Figure 1B and Supplementary Table 4). Their expression patterns in various tissues were the same as those in cowpea. For example, PvCKX3b were hardly expressed in all tissues, PvCKX7-1 and PvCKX6-2 were highly expressed in roots, and PvCKX3a was expressed in flowers (Figure 4A).
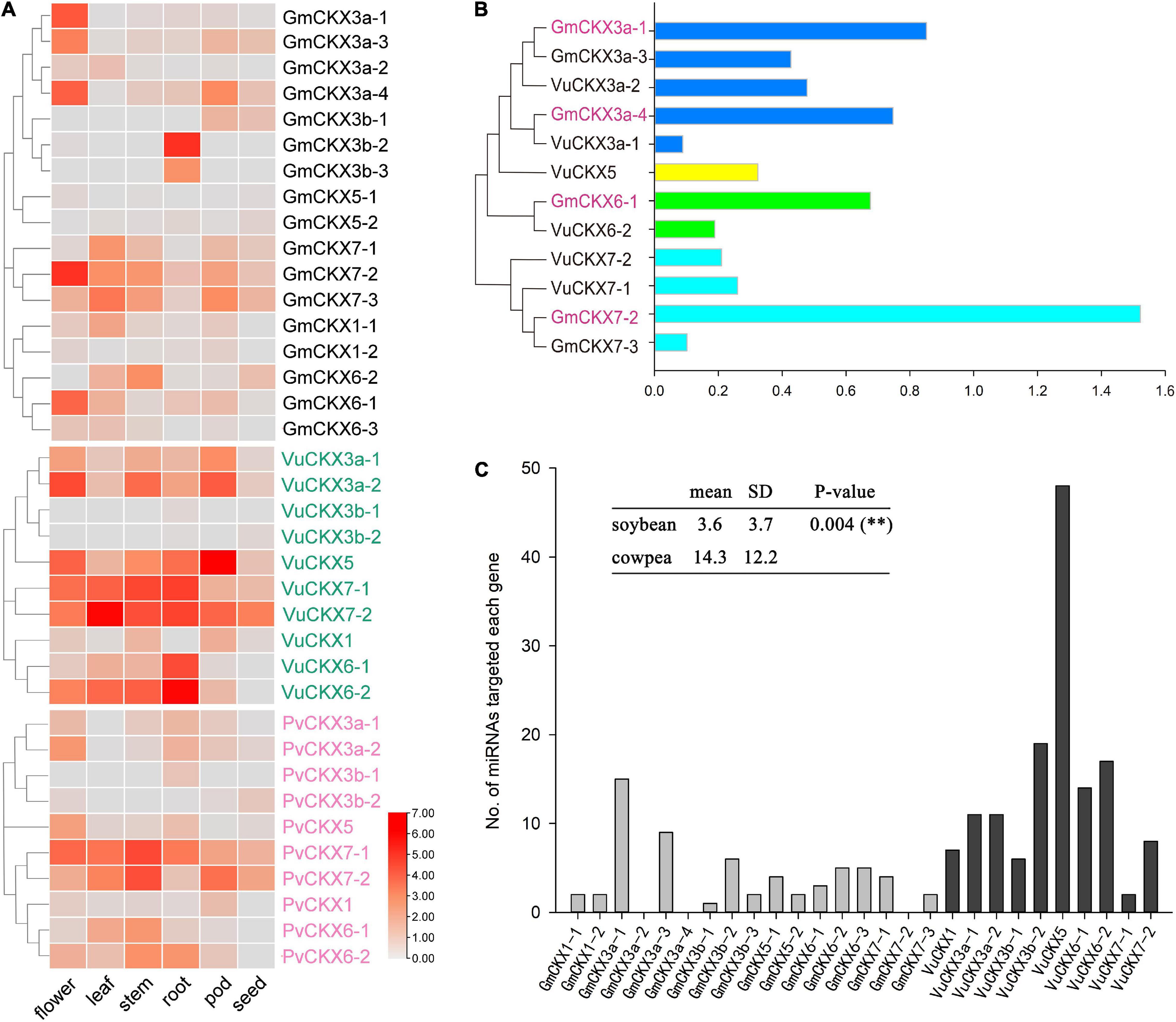
Figure 4. Expression patterns and regulation of CKX gene family. (A) Expression levels of CKX gene family in flower, leaf, stem, root, pod, and seed of soybean, cowpea and kidney bean. (B) Relative expression levels of CKX gene family in soybean and cowpea flowers. (C) The numbers of miRNAs targeted each CKX gene.
Recently, Wang et al. (2021) identified the function of Medtr4G126160 in Medicago truncatula, which is the highest homology with GmCKX3b-2 and GmCKX3b-3 (Supplementary Table 3). In detail, the Medtr4G126160 mutant significantly reduced main root length and increased the number of lateral roots, indicating the important role of Medtr4G126160 in root development. In this study, CKX3b genes were found to have two expression patterns in different leguminosae crops. In soybean and Medicago truncatula, the CKX3b genes were highly expressed in roots, which may play an important role in root development, while, in cowpea and kidney bean, the CKX3b genes were hardly expressed in various tissues, indicating the functional differentiation of CKX3b tandem duplication in plant evolution.
In addition, we compared the relative expression levels of the CKX gene family in soybean and cowpea flowers (Figure 4B). As a result, although GmCKX5 with incomplete structure domains had significantly lower relative expression level in soybean than in cowpea, CKX3, CKX6, and CKX7 had much higher relative expression levels in soybean than in cowpea. These highly expressed GmCKX genes, especially the GmCKX3a genes, reduce cytokinins content in soybean in the process of ovule formation. Less cytokinins result in low expression of auxin efflux carrier PIN1. Thus, there are lower number of ovules and SNPP (Bartrina et al., 2011; Bencivenga et al., 2012; Zuñiga-Mayo et al., 2018).
Prediction of miRNA and ROCK1 Gene Regulation in CKX Gene Family
To predict all the miRNAs for targeted soybean and cowpea CKX genes via the online software PsRNATarget, we downloaded 756 soybean miRNA sequences from the miRBase database and 656 cowpea miRNA sequences from Martins et al. (2020). As a result, 62 miRNAs were predicted to be targeted with soybean CKX genes, while 144 miRNAs were predicted to be targeted with cowpea CKX genes (Figure 4C). In the cowpea CKX gene family, VuCKX5 was predicted to be regulated by 48 miRNAs (maximum), while VuCKX7-1 was predicted to be regulated by 2 miRNAs (minimum). In the soybean CKX gene family, GmCKX3-1 was predicted to be regulated by 15 miRNAs (maximum), while no miRNAs were predicted to regulate GmCKX3a-2, GmCKX3a-4, and GmCKX7-2 (Figure 4C). We speculated that less miRNA regulation in soybean may be one reason for relatively high expression levels of soybean CKX genes.
In Niemann et al. (2015), ROCK1 was a positive regulator of CKX protein activity in Arabidopsis thaliana, while in this study, soybean ROCK1 genes (GmROCK1 and Glyma.08G135800) had much higher relative expression level than cowpea ROCK1 genes (VuROCK1 and Vigun03G076200). This may result in higher activity of soybean CKX protein than cowpea CKX protein, which would enhance soybean cytokinin degradation.
Expansion Patterns of the CKX Gene Family in Soybean
Using the database PlantDGD (Qiao et al., 2019), we obtained 13 pairs of duplicate genes in soybean, which were consistent with our collinearity analysis results. Among these duplicate genes, nine pairs were normal and four pairs were abnormal, namely, three pairs of GmCKX3a and GmCKX3b on chromosomes 9, 13, and 17 owing to the mismatch in collinearity analysis, and one pair of GmCKX6-2 and GmCKX1-2 (Supplementary Table 6). Here, we replaced GmCKX3b with its adjacent GmCKX3a and calculated the Ks values of these gene pairs to estimate their replication times (Supplementary Table 5). As a result, the duplications for six, six, and one pairs of GmCKX genes occurred, respectively, between 8 and 18 mya, between 70 and 74 mya, and between 133 and 169 mya. This indicates that most GmCKX duplications occurred at 10–15 mya [a soybean-specific whole genome duplication (WGD) event] and 59 mya (a legume-specific WGD event), and individual GmCKX duplications occurred at approximately 150 mya.
Using the database PlantDGD (Qiao et al., 2019), we checked the fragment duplications of CKX gene family in kidney bean and Arabidopsis. As a result, a pair of PvCKX1 and PvCKX6 was obtained, while three pairs of possible fragment replicators CKX2 and CKX3, CKX2 and CKX4, and CKX3 and CKX4 in Arabidopsis were observed. Thus, we speculated that CKX2 and CKX4 were a segmental duplication event that occurred in other species after legume differentiation, while one copy of CKX6 gene was a segmental duplication of CKX1 in legumes. The CKX6 copy from CKX1 is different from the other CKX6 copies in expression pattern. The former was not expressed in flowers, while the latter was expressed in flowers.
Comparison of Relative Expression Levels for PIN1 and CKX Gene Families in Soybean and Kidney Bean
To confirm whether the above SNPP relationship and the difference of expression levels of PIN1 and CKX gene families exists in kidney bean, we compared their relative expression levels in soybean and kidney bean. As a result, Phvul.004G150600 had much higher relative expression level in kidney bean flowers than GmPIN1a and GmPIN1b in soybean flowers, while GmCKX gene family had higher relative expression levels in flowers than PvCKX gene family (Supplementary Figure 5). The results were consistent with those in soybean and cowpea. The differences of relative expression levels of PIN1 and CKX gene families in soybean and cowpea, along with the differences in soybean and kidney bean, may be an important reason for the SNPP difference.
Comparison of Seed Number per Pod-Related Interaction Networks in Soybean and Cowpea
Candidate SNPP genes in this and previous (Schwarz et al., 2020; Fang et al., 2021) studies and known gene Ln were used to construct interaction networks in soybean and cowpea. All the results are shown in Figure 5 and Supplementary Table 7. In soybean network, low SNPP may be due to two reasons. First, four-seed-pod-related gene Glyma.10G002200, in Fang et al. (2021), was interacted with GmCBP-1 and GmCBP-2, while its homology Vigun07g002900 in cowpea was interacted with VuCBP-1, VuCP1, and VuCAM4. The five interacted genes in soybean and cowpea belong to the calmodulin and calcium-binding protein gene (CBP) family. In previous studies, calmodulin and calcium-binding proteins in plants not only directly affected SNPP (Midhat et al., 2018), but were also critical for the biosynthesis of brassinosteroid (Du and Poovaiah, 2005), which plays an important role in determining the number of ovules and seeds via positive regulator BZR1 (Huang et al., 2013). However, VuCBP-1 (5.67), VuCP1 (4.30), and VuCAM4 (4.94) had much higher relative expression levels than GmCBP-1 (0.38) and GmCBP-2 (0.90), which are inhibited by gma-miR4405. Although Vigun03g412600 was inhibited by 016048_minus, its binding degree was relatively low (Supplementary Table 8). Thus, we speculated that the high expression may increase SNPP in cowpea.
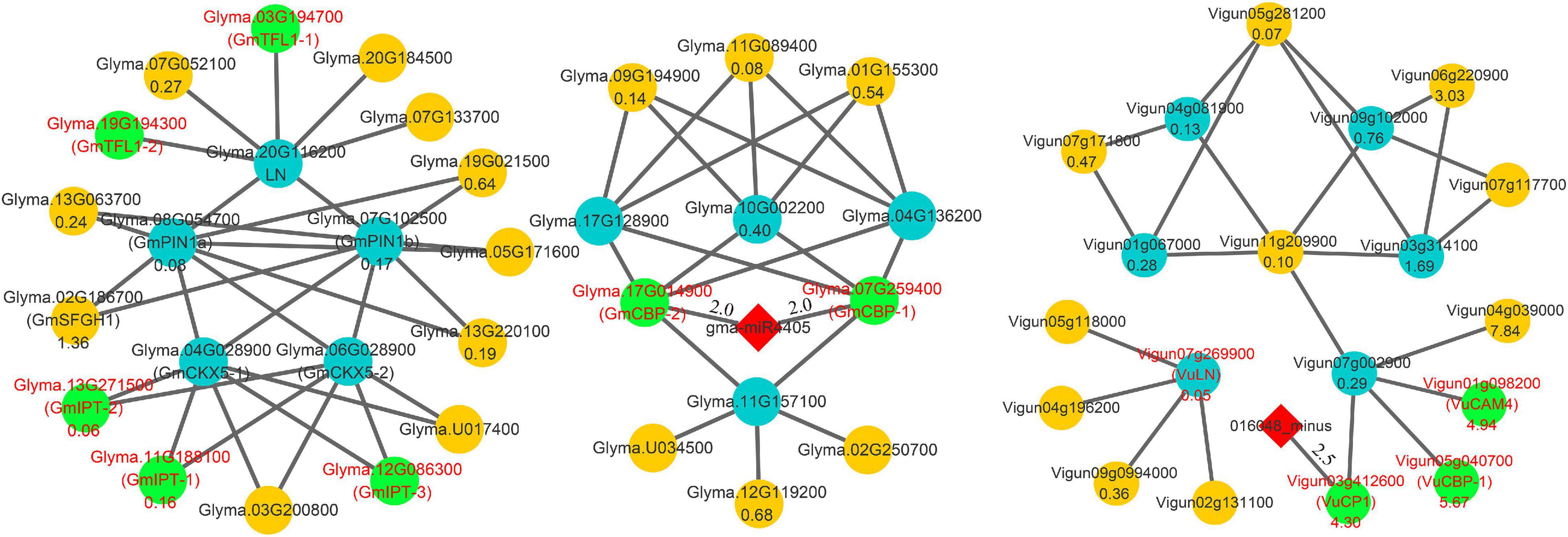
Figure 5. Main interaction networks on SNPP-related genes in soybean and cowpea. All the genes that encode Isopentenyltransferase, TERMINAL FLOWER 1, and calmodulin and calcium-binding proteins were marked by red color characters and green background.
Then, SNPP-related genes, GmCKX5-1 and GmCKX5-2, were interacted with soybean genes GmIPT-1 (Glyma.11G188100), GmIPT-2 (Glyma.13G271500), and GmIPT-3 (Glyma.12G086300). Their homologies in Arabidopsis all belong to Isopentenyltransferase (IPT) gene family, which plays an important role in cytokinin biosynthesis (Miyawaki et al., 2004, 2006), and overexpressing IPT can increase the levels of endogenous cytokinins (Décima Oneto et al., 2016). Their low expression levels in soybean may lead to a decrease in cytokinin synthesis, which further affects ovule numbers in soybean. The negative regulation was not found in cowpea.
In addition, a known SNPP gene Ln was interacted with Glyma.03G194700 (GmTFL1-1) and Glyma.19G194300 (GmTFL1-2), for which their Arabidopsis homology AT5G03840 (TFL1) determines seed size, and loss-of-function mutants exhibit a large seed phenotype (Zhang et al., 2020). Low expression levels of GmTFL1-1 (0.10) and GmTFL1-2 (0.01) may lead to large seed in soybean. The interacted network was not found in cowpea.
Comparison of Yield-Related Gene Network in Soybean and Cowpea
In this study, all the known yield-related soybean genes in Zhang et al. (2021), along with the above known and candidate SNPP genes, were used to construct a comprehensive network for the two legumes. The results are shown in Figure 6 and Supplementary Table 9.
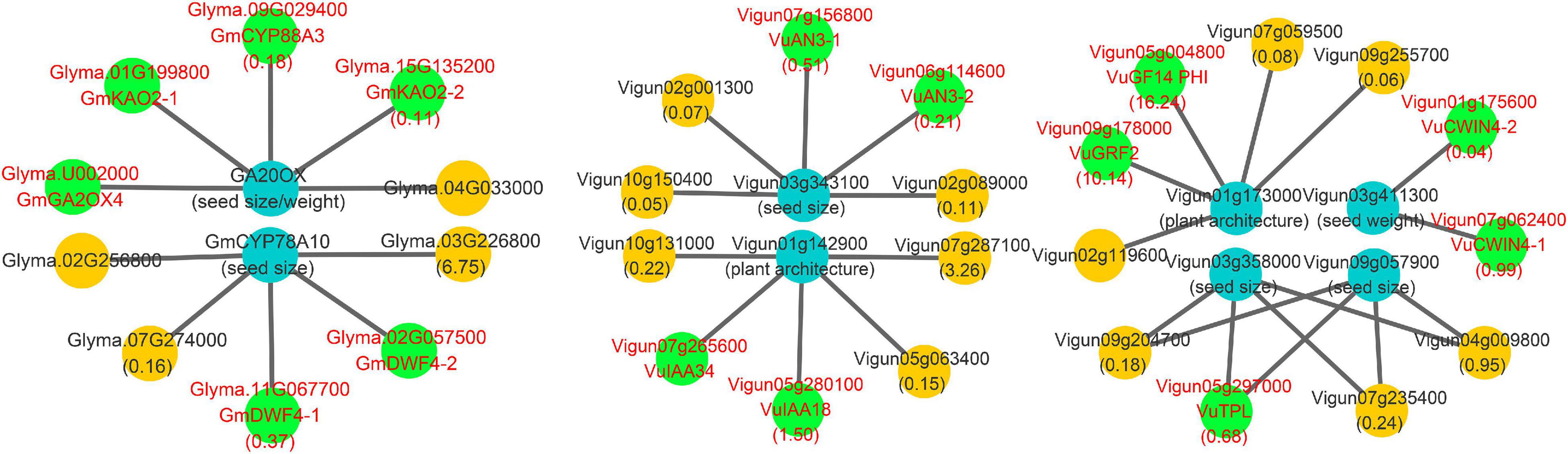
Figure 6. Main interaction networks on yield-related genes in soybean and cowpea. All the genes that encode CYP88A gene family, gibberellin oxidase, 22α hydroxylase, 14-3-3 gene family, CWIN4, and AN3 were marked by red color characters and green background.
In the network, some negative regulation was found in soybean. First, known seed size/weight gene Glyma.07G081700 was found to interact with Glyma.09G029400, Glyma.01G199800, and Glyma.15G135200 of cytochrome P450 subfamily 88A (CYP88A) gene family, which catalyzes the conversion of KA (ent-kaurenoic acid) to GA12 (the precursor of all gibberellins) and catalyzes three steps of gibberellin biosynthesis pathway in Arabidopsis (Helliwell et al., 2001; Regnault et al., 2014). It should be noted that GA was found to negatively regulate the number of ovules in Fuentes et al. (2012) and Carrera et al. (2012). Then, Glyma.07G081700 was found to interact with Glyma.U002000 that encodes gibberellin oxidase, whose expression decreases cytokinin activity (Jasinski et al., 2005). Finally, known seed-size-gene Glyma.05g019200 was found to interact with Glyma.11G067700 and Glyma.02G057500, which encode 22α hydroxylase that is an inhibitor of Brassinosteroid (BR) biosynthesis in Arabidopsis (Fujiyama et al., 2019). The negative regulation may result in lower SNPP in soybean than in cowpea.
Meanwhile, some positive regulation was found in cowpea. First, the homology Vigun01g142900 of known plant-architecture gene Glyma.19G164600 was found to be interacted with Vigun07g265600 and Vigun05g280100, while the homology Vigun03g358000 of known seed size gene Glyma.17G112800 was found to be interacted with Vigun05g297000. Vigun07g265600 (VuIAA34), Vigun05g297000 (VuTPL), and Vigun05g280100 (VuIAA18) focused on auxin synthesis and transport (Liscum and Reed, 2002). Then, the homology Vigun01g173000 of known plant-architecture gene Glyma.19G194300 was found to be interacted with Vigun05g004800 and Vigun09g178000 in 14-3-3 gene family, which is involved in PIN auxin carrier, auxin transport-related development, and brassinosteroid signal transduction (Gampala et al., 2007; Keicher et al., 2017). In this study, Vigun05g004800 and Vigun09g178000 were found to have high expression levels at ovule developmental stages. Third, the homology Vigun03g411300 of known seed-weight gene Glyma.17g036300 was found to be interacted with cell wall invertase genes Vigun01g175600 and Vigun07g062400, in which their homology AT2G36190 (CWIN4) may regulate ovule formation by modulating downstream auxin signaling and MADS-box transcription factors in Arabidopsis (Liao et al., 2020). Finally, the homology Vigun06g114600 of known seed size gene Glyma.17G112800 was found to be interacted with Vigun06g114600 and Vigun07g156800, in which their homology AT5G28640 (AN3) can regulate seed embryo development together with AT1G55600 (MINI3), and its mutant line had lower seed/silique number, silique length, and seed/silique weight than wild-type plants (Meng et al., 2016). The positive regulation results in higher SNPP in cowpea than in soybean.
Known seed size or weight genes Glyma.01G061100 (GmCYP78A70), Glyma.02G119600 (GmCYP78A57), Glyma.19G240800 (GmCYP78A72), and Glyma.05G019200 (GmCYP78A10) belong to the CYP78A gene family, which is found to be associated with seed size in Arabidopsis (Adamski et al., 2009; Fang et al., 2012). In cowpea, there were only two homologous copies Vigun02g047800 and Vigun03g343100 of the four soybean genes. We found that the first three soybean genes had high expression levels at middle and later seed development stages, while all the two cowpea copies had low expression levels (Supplementary Table 10). This may explain why soybean seed is larger than cowpea seed.
In summary, species-specific traits in crops may be derived from species-specific gene networks.
Discussion
Molecular Mechanisms for Seed Number per Pod Difference in Soybean and Cowpea
In this study, we observed four interesting results. The results are showed in Figure 7. First, PIN1 had lower expression level in soybean flowers than in cowpea flowers. Then, among the proteins, PID, D6PK, RCN1, UNH, and MPK6 in Arabidopsis, that interacted with PIN1 and their homologies were differentially expressed between soybean and cowpea, lower PID and D6PK and higher RCN1 expression levels in soybean resulted in lower phosphorylation level in soybean flowers, while high UNH and MPK6 expression levels decreased plasma membrane localization level in soybean flowers, as compared with those in cowpea flowers. Thus, we speculate that lower PIN1 expression level and lower phosphorylation and plasma membrane localization levels derived from the above five differential expression proteins interacted with PIN1 make auxin transport efficiency lower in soybean flowers than in cowpea flowers, which forms a lower auxin maximum zone (Benková et al., 2003; Ceccato et al., 2013). Next, some differential genes in the interaction networks on PIN1 and CKX gene families were found. SFGH, which hydrolyzes serine, was found in soybean rather than in cowpea, and ACX4 had much higher relative expression level in cowpea than in soybean. These may lead to better embryo development and more seeds in cowpea. Finally, in the interaction networks on yield-related genes, higher expression levels of the CBP genes in cowpea, as compared with those in soybean, increase SNPP; low expression levels of three soybean-specific IPT genes may inhibit CK synthesis. The above results may lead to lower number of ovules and ultimately lower SNPP in soybean than in cowpea.
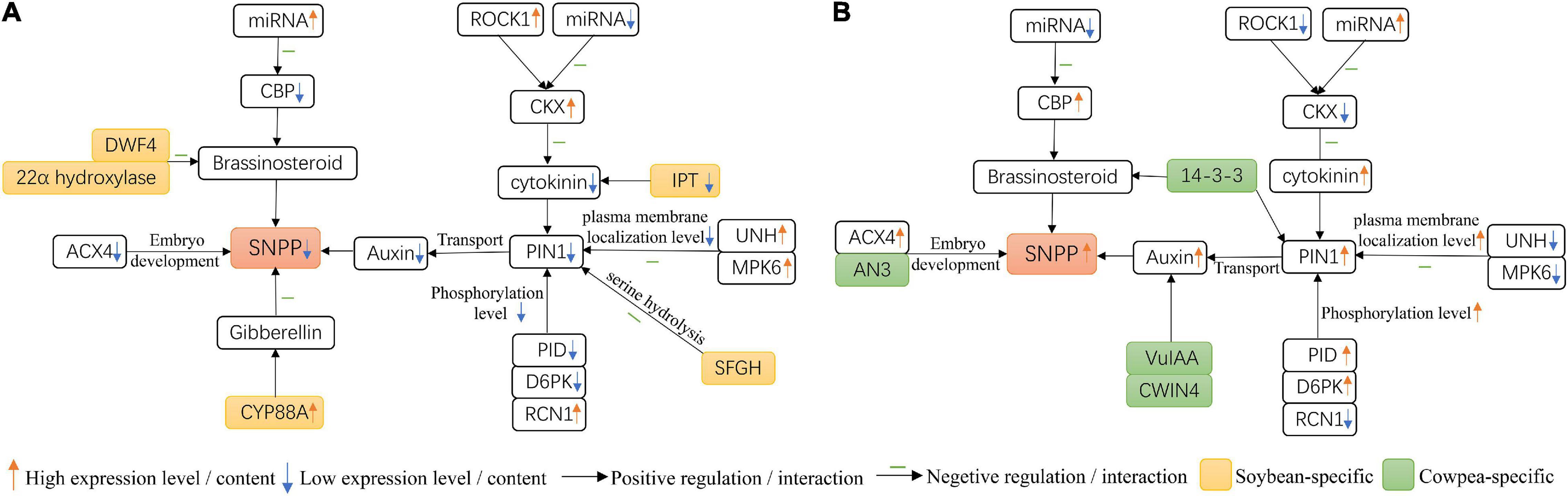
Figure 7. Molecular mechanisms for the difference of seed number per pod (SNPP) in (A) soybean and (B) cowpea.
In a previous study, one locus for soybean four-seed pod was located by Yang et al. (2013) on chromosome 13. Around this locus, there was one candidate gene GmMIF2 (Glyma.13G063100). Here we found that soybean gene GmMIF2 had much higher relative expression level in flower than cowpea gene VuMIF2 (Vigun04g052300). More importantly, their homologous gene MIF2 (AT3G28917) in Arabidopsis was found to hinder its ovule development (Bollier et al., 2018). Thus, MIF2 may be a potential gene for SNPP difference between soybean and cowpea.
In addition, we explained the reasons for low expression level of PIN1 gene in soybean. In this study we observed another two interesting phenomena. First, CKX genes and their positive regulator ROCK1 had higher relative expression levels in soybean flower than in cowpea flower. Then, fewer miRNAs were predicted to be targeted with soybean CKX genes than with cowpea CKX genes. These two results lead to higher cytokinin degradation level in soybean than in cowpea, which decreases cytokinin level in soybean (Supplementary Table 11) and affecting the expression level of PIN1 in soybean (Bencivenga et al., 2012; Zuñiga-Mayo et al., 2018).
Auxin Transport Efficiency Difference May Cause Seed Number per Pod Difference in Soybean and Cowpea
In previous studies, three aspects of low auxin transport efficiency in Arabidopsis have been observed. First, PIN1 expression level affects auxin transport efficiency. Okada et al. (1991) compared the polar transport efficiency of auxin in the inflorescence tissue between pin1 mutants and their wild type, indicating the significant decreases of polar transport efficiency between pin1-1 (↓ 86%) and pin1-2 (↓ 93%) mutants and their wild type. Then, plasma membrane localization level of PIN1 affects auxin transport efficiency. Wisniewska et al. (2006) modified PIN1 polarity and examined auxin translocation direction in Arabidopsis thaliana. As a result, PIN1 polarity determines a primary direction in auxin transport of meristematic tissues. Finally, some kinases have been found to positively regulate auxin transport through PIN1 phosphorylation in Arabidopsis, such as PID and D6PK (Benjamins et al., 2001; Lee and Cho, 2006; Zourelidou et al., 2014). In addition, GmPIN1 is expressed and polarly localized in nodule primordium cells, and controls nodule formation by transporting auxin to form an auxin maximal zone in soybean (Gao et al., 2021). As we know, nodule primordium and ovule primordium are meristematic tissues. Thus, these findings provide evidence for the role of GmPIN1 in the formation of soybean ovule primordium. In this study, we found that the relative expression level of PIN1 gene was much lower in soybean flowers than in cowpea flowers, and the differences of relative expression levels of the above five proteins interacting with PIN1 might cause lower phosphorylation and plasma membrane localization levels in soybean than in cowpea. These results may cause lower auxin transport efficiency in soybean than in cowpea.
During ovule primordium formation, Benková et al. (2003) found that auxin accumulated in large amounts at the apex of ovule primordium, and this auxin maximum zone is a necessary condition for ovule formation. Bencivenga et al. (2012) showed that lower auxin transport efficiency significantly reduced ovule number per pistil from wild-type Col-0 (48 ovules) to weak mutant pin1-5 (9 ovules). Carlson and Lersten (2004) and Yang et al. (2017) observed that ovule number difference could cause SNPP difference. In summary, auxin transport efficiency difference in soybean and cowpea may cause lower formation efficiency of auxin maximum zone in soybean, resulting in lower ovule number and SNPP in soybean.
CKX Genes Play an Important Role in Ovule Formation
Cytokinin dehydrogenase (CKX) can specifically degrade cytokinin, although cytokinin can increase ovules number by promoting PIN1 expression (Bencivenga et al., 2012). Bartrina et al. (2011) compared single and double CKX gene mutations with their wild types in Arabidopsis thaliana. As a result, no significant change in the overall plant morphologies of single CKX gene mutants was observed, indicating the redundant role of CKX gene family. Multiple double mutations with CKX3-1 allele could form more flowers, especially for ckx3 ckx5 double mutant, which formed more ovules. This increased SNPP and led to 55% higher seed yield. Schwarz et al. (2020) obtained similar results in Brassica napus. In detail, compared with the wild type, the bnckx3 bnckx5 sixfold mutant increased the number of flowers, ovule number per pistil, and pod numbers on main stem, increasing seed yield by 20–32%. These results suggest that CKX gene family plays an important role in ovule formation. In this study, we identified 17 soybean and 10 cowpea CKX genes. Among these genes, most were not expressed in flowers, and multiple copy genes CKX3, CKX6, and CKX7 were much higher in soybean flowers than in cowpea flowers. These high expression CKX genes in soybean may lead to more cytokinin degradation and lower cytokinin content, decreasing PIN1 expression level, ovule number per pistil, and SNPP.
At present, there have been limited studies on CKX gene regulator, and only one regulator was reported by Niemann et al. (2015). In detail, the enhanced CKX activity in 35S:CKX1, 35S:CKX2, and 35S:CKX3 plants was reduced through rock1 introgression by 90.5, 64, and 100%, respectively. Meanwhile, rock1 mutant enhanced the activity of apical meristem and organ formation rate in Arabidopsis thaliana. Cytokinin content in inflorescence was increased and the numbers of flowers and pods on main stem were 50% higher in rock1 mutant than in its wild type, which was very similar to the phenotypic changes in the ckx3 ckx5 mutant. These results indicate that ROCK1 acts as a positive regulator of CKX protein. In this study, we found one ROCK1 homologous gene in soybean or cowpea, and ROCK1 had higher relative expression level in soybean flowers than in cowpea flowers. The higher ROCK1 gene expression level may increase the activity of soybean CKX protein. In addition, fewer miRNAs were predicted to target GmCKX genes. In other words, less miRNA regulation may be an important reason for higher GmCKX expression level in soybean. Thus, higher ROCK1 expression and fewer miRNAs enhanced cytokinin degradation by regulating GmCKX, so ovule number and SNPP in soybean decreased.
Breeding by Design for Seed Number per Pod in Soybean
Molecular design breeding has been widely used in soybean with some success (Haun et al., 2014; Bao et al., 2019; Han et al., 2019; Le et al., 2020). As we know, cowpea has more SNPP and smaller seeds, and soybean has fewer SNPP and larger seeds. However, cowpea has significantly higher yield than soybean. We also notice that relatively few SNPP in wild and cultivated soybeans may hinder the increase of soybean yield. Therefore, it is possible and necessary to mine yield-related novel genes in cowpea to conduct molecular design breeding in soybean, especially in the current situation of very sharp contradiction between soybean supply and demand in China.
First, Gmckx3a quadruple mutant may be used to increase SNPP and yield in soybean. In previous studies, ckx3 ckx5 mutant in Arabidopsis and bnckx3 bnckx5 sixfold mutant in Brassica napus can increase their yields (Bartrina et al., 2011; Schwarz et al., 2020). In soybean, we found high expression of four CKX3a and no expression of two CKX5 and three CKX3 tandem-duplication-derived genes (CKX3b) in flowers (Figure 4). Thus, ckx3a quadruple mutant may be used to increase SNPP and soybean yield. In addition, GmCKX6-1 and GmCKX7-2 were found to be highly expressed in flowers in this study. Another possible way is to obtain gmckx3a gmckx6 (or gmckx7) mutant.
Second, we can identify elite alleles of GmCKX3a, GmCKX6 and GmCKX7 from existing four-seed pod cultivars, and transfer these elite alleles into current excellent cultivars via cross and backcross approaches. This method has been confirmed to be effective in Zhu et al. (2020).
Third, we may interfere with the expression of CYP88A genes (Glyma.09G029400, Glyma.01G199800, and Glyma.15G135200) and increase DELLA protein to decrease GA content in soybean. Carrera et al. (2012) reported that inhibiting the synthesis and function of GA via DELLA protein in tomato can increase ovule number.
Finally, we may over-express the CBP genes in soybean (Glyma.17G019400 and Glyma.07G259400) and transfer an excellent mutant gene GmBZL2(P216L) (GmBZL2*) into soybean (Zhang et al., 2016) to increase SNPP via enhancing the synthesis and signal transduction of BR.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
Y-MZ conceived and supervised the study. L-ML, H-QZ, and KC carried out the experimental works and analyzed the data. L-ML, H-QZ, and Y-MZ wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32070557 and 31871242) and Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (Grant No. 2014RC020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Hanwen Zhang (Hence Education Ltd., Vancouver, BC, Canada; aHl3ZW56aGFuZ0BoZW5jZWVkdS5jb20=) for improving the language of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.749902/full#supplementary-material
Footnotes
- ^ https://www.arabidopsis.org/index.jsp
- ^ http://www.soybase.org/
- ^ https://legumeinfo.org/data/public
- ^ https://phytozome.jgi.doe.Gov/phytomine/template.do?Name=Gene_Expression
- ^ https://www.ncbi.nlm.nih.gov/bioproject/?term=GSE42871
- ^ https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna389300
- ^ http://www.mirbase.org/
- ^ http://www.megasoftware.net/
- ^ http://itol.embl.de/
- ^ http://gsds.gao-lab.org/index.php
- ^ https://pfam.xfam.org/search
- ^ https://wolfpsort.hgc.jp/
- ^ http://meme-suite.org/tools/meme
- ^ https://string-db.org/
- ^ http://plantgrn.noble.org/psRNATarget/analysis
- ^ http://pdgd.njau.edu.cn:8080/
References
Adamski, N. M., Anastasiou, E., Eriksson, S., O’Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. doi: 10.1073/pnas.0907024106
Bao, A., Chen, H., Chen, L., Chen, S., Hao, Q., Guo, W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19:131. doi: 10.1186/s12870-019-1746-6
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80. doi: 10.1105/tpc.110.079079
Bencivenga, S., Simonini, S., Benková, E., and Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067.
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/s0092-8674(03)00924-3
Bollier, N., Sicard, A., Leblond, J., Latrasse, D., Gonzalez, N., Gévaudant, F., et al. (2018). At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30, 83–100. doi: 10.1105/tpc.17.00653
Cai, Z., Xian, P., Cheng, Y., Ma, Q., Lian, T., Nian, H., et al. (2021). CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low latitude soybean variety Huachun 6. Plant Biotechnol. J. 19, 1898–1900. doi: 10.1111/pbi.13673
Carlson, J. B., and Lersten, N. R. (2004). “Reproductive morphology,” in Soybeans: Improvement, Production, and Uses, 3rd Edn, eds H. R. Boerma and J. E. Specht (Madison, WI: ASA), 59–95.
Carrera, E., Ruiz-Rivero, O., Peres, L. E., Atares, A., and Garcia-Martinez, J. L. (2012). Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 160, 1581–1596. doi: 10.1104/pp.112.204552
Ceccato, L., Masiero, S., Sinha Roy, D., Bencivenga, S., Roig-Villanova, I., Ditengou, F. A., et al. (2013). Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One 8:e66148. doi: 10.1371/journal.pone.0066148
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, K., Pan, Y.-F., Liu, L.-M., Zhang, H.-Q., and Zhang, Y.-M. (2021). Integrated transcriptomic and bioinformatics analyses reveal the molecular mechanisms for the differences in seed oil and starch content between Glycine max and Cicer arietinum. Front. Plant Sci. 12:743680. doi: 10.3389/fpls.2021.743680
Cucinotta, M., Colombo, L., and Roig-Villanova, I. (2014). Ovule development, a new model for lateral organ formation. Front. Plant Sci. 5:117. doi: 10.3389/fpls.2014.00117
Cummins, I., McAuley, K., Fordham-Skelton, A., Schwoerer, R., Steel, P. G., Davis, B. G., et al. (2006). Unique regulation of the active site of the serine esterase S-formylglutathione hydrolase. J. Mol. Biol. 359, 422–432. doi: 10.1016/j.jmb.2006.03.048
Dai, M., Zhang, C., Kania, U., Chen, F., Xue, Q., McCray, T., et al. (2012). A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24, 2497–2514. doi: 10.1105/tpc.112.098905
Décima Oneto, C., Otegui, M. E., Baroli, I., Beznec, A., Faccio, P., Bossio, E., et al. (2016). Water deficit stress tolerance in maize conferred by expression of an isopentenyltransferase (IPT) gene driven by a stress- and maturation-induced promoter. J. Biotechnol. 220, 66–77. doi: 10.1016/j.jbiotec.2016.01.014
Dory, M., Hatzimasoura, E., Kállai, B. M., Nagy, S. K., Jäger, K., Darula, Z., et al. (2018). Coevolving MAPK and PID phosphosites indicate an ancient environmental control of PIN auxin transporters in land plants. FEBS Lett. 592, 89–102. doi: 10.1002/1873-3468.12929
Du, L., and Poovaiah, B. W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437, 741–745. doi: 10.1038/nature03973
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Fang, C., Li, W., Li, G., Wang, Z., Zhou, Z., Ma, Y., et al. (2013). Cloning of Ln gene through combined approach of map-based cloning and association study in soybean. J. Genet. Genomics 40, 93–96. doi: 10.1016/j.jgg.2013.01.002
Fang, T., Bai, Y. W., Huang, W. X., Wu, Y. Y., Yuan, Z. H., Luan, X. Y., et al. (2021). Identification of potential gene regulatory pathways affecting the ratio of four-seed pod in soybean. Front. Genet. 12:1583. doi: 10.3389/fgene.2021.717770
Fang, W., Wang, Z., Cui, R., Li, J., and Li, Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70, 929–939. doi: 10.1111/j.1365-313X.2012.04907.x
Fuentes, S., Ljung, K., Sorefan, K., Alvey, E., Harberd, N. P., and Østergaard, L. (2012). Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24, 3982–3996. doi: 10.1105/tpc.112.103192
Fujiyama, K., Hino, T., Kanadani, M., Watanabe, B., Jae Lee, H., Mizutani, M., et al. (2019). Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants 5, 589–594. doi: 10.1038/s41477-019-0436-6
Galbiati, F., Sinha Roy, D., Simonini, S., Cucinotta, M., Ceccato, L., Cuesta, C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 76, 446–455. doi: 10.1111/tpj.12309
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189. doi: 10.1016/j.devcel.2007.06.009
Gao, Z., Chen, Z., Cui, Y., Ke, M., Xu, H., Xu, Q., et al. (2021). GmPIN-dependent polar auxin transport is involved in soybean nodule development. Plant Cell 33, 2981–3003. doi: 10.1093/plcell/koab183
Han, J., Guo, B., Guo, Y., Zhang, B., Wang, X., and Qiu, L. J. (2019). Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front. Plant Sci. 10:1446. doi: 10.3389/fpls.2019.01446
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201
Helliwell, C. A., Chandler, P. M., Poole, A., Dennis, E. S., and Peacock, W. J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 98, 2065–2070. doi: 10.1073/pnas.041588998
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. U.S.A. 101, 8821–8826.
Huang, H. Y., Jiang, W. B., Hu, Y. W., Wu, P., Zhu, J. Y., Liang, W. Q., et al. (2013). BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 6, 456–469. doi: 10.1093/mp/sss070
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jeong, N., Suh, S. J., Kim, M. H., Lee, S., Moon, J. K., Kim, H. S., et al. (2012). Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell 24, 4807–4818. doi: 10.1105/tpc.112.104968
Jia, W., Li, B., Li, S., Liang, Y., Wu, X., Ma, M., et al. (2016). Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 14:e1002550. doi: 10.1371/journal.pbio.1002550
Keicher, J., Jaspert, N., Weckermann, K., Möller, C., Throm, C., Kintzi, A., et al. (2017). Arabidopsis 14-3-3 epsilon members contribute to polarity of PIN auxin carrier and auxin transport-related development. eLife 6:e24336. doi: 10.7554/eLife.24336
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Le, H., Nguyen, N. H., Ta, D. T., Le, T., Bui, T. P., Le, N. T., et al. (2020). CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front. Plant Sci. 11:612942. doi: 10.3389/fpls.2020.612942
Lee, S. H., and Cho, H. T. (2006). PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18, 1604–1616. doi: 10.1105/tpc.105.035972
Liao, S., Wang, L., Li, J., and Ruan, Y. L. (2020). Cell wall invertase is essential for ovule development through sugar signaling rather than provision of carbon nutrients. Plant Physiol. 183, 1126–1144. doi: 10.1104/pp.20.00400
Liscum, E., and Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400.
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 98, 767–782. doi: 10.1111/tpj.14349
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Martins, T. F., Souza, P., Alves, M. S., Silva, F., Arantes, M. R., Vasconcelos, I. M., et al. (2020). Identification, characterization, and expression analysis of cowpea (Vigna unguiculata [L.] Walp.) miRNAs in response to cowpea severe mosaic virus (CPSMV) challenge. Plant Cell Rep. 39, 1061–1078.
Meinke, D. W. (2020). Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 226, 306–325. doi: 10.1111/nph.16071
Meng, L. S., Wang, Y. B., Loake, G. J., and Jiang, J. H. (2016). Seed embryo development is regulated via an AN3-MINI3 gene cascade. Front. Plant Sci. 7:1645. doi: 10.3389/fpls.2016.01645
Michniewicz, M., Zago, M. K., Abas, L., Weijers, D., Schweighofer, A., Meskiene, I., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056. doi: 10.1016/j.cell.2007.07.033
Midhat, U., Ting, M. K. Y., Teresinski, H. J., and Snedden, W. A. (2018). The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol. Biol. 96, 375–392. doi: 10.1007/s11103-018-0703-3
Miyawaki, K., Matsumoto-Kitano, M., and Kakimoto, T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37, 128–138. doi: 10.1046/j.1365-313x.2003.01945.x
Miyawaki, K., Tarkowski, P., Matsumoto-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16598–16603. doi: 10.1073/pnas.0603522103
Nemhauser, J. L., Feldman, L. J., and Zambryski, P. C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888.
Niemann, M. C., Bartrina, I., Ashikov, A., Weber, H., Novák, O., Spíchal, L., et al. (2015). Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proc. Natl. Acad. Sci. U.S.A. 112, 291–296. doi: 10.1073/pnas.1419050112
Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365–1377. doi: 10.1105/tpc.021477
Nole-Wilson, S., Azhakanandam, S., and Franks, R. G. (2010). Polar auxin transport together with aintegumenta and revoluta coordinate early Arabidopsis gynoecium development. Dev. Biol. 346, 181–195. doi: 10.1016/j.ydbio.2010.07.016
Okada, K., Ueda, J., Komaki, M. K., Bell, C. J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684. doi: 10.1105/tpc.3.7.677
Pahari, S., Cormark, R. D., Blackshaw, M. T., Liu, C., Erickson, J. L., and Schultz, E. A. (2014). Arabidopsis UNHINGED encodes a VPS51 homolog and reveals a role for the GARP complex in leaf shape and vein patterning. Development 141, 1894–1905. doi: 10.1242/dev.099333
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20:38. doi: 10.1186/s13059-019-1650-2
Rashotte, A. M., DeLong, A., and Muday, G. K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13, 1683–1697. doi: 10.1105/tpc.010158
Regnault, T., Davière, J. M., Heintz, D., Lange, T., and Achard, P. (2014). The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 80, 462–474. doi: 10.1111/tpj.12648
Reyes-Olalde, J. I., Zuñiga-Mayo, V. M., Chávez Montes, R. A., Marsch-Martínez, N., and de Folter, S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18, 644–655. doi: 10.1016/j.tplants.2013.08.002
Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Rylott, E. L., Rogers, C. A., Gilday, A. D., Edgell, T., Larson, T. R., and Graham, I. A. (2003). Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J. Biol. Chem. 278, 21370–21377. doi: 10.1074/jbc.M300826200
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Schwarz, I., Scheirlinck, M. T., Otto, E., Bartrina, I., Schmidt, R. C., and Schmülling, T. (2020). Cytokinin regulates the activity of the inflorescence meristem and components of seed yield in oilseed rape. J. Exp. Bot. 71, 7146–7159. doi: 10.1093/jxb/eraa419
Wang, C., Wang, H., Zhu, H., Ji, W., Hou, Y., Meng, Y., et al. (2021). Genome-wide identification and characterization of cytokinin oxidase/dehydrogenase family genes in Medicago truncatula. J. Plant Physiol. 256:153308. doi: 10.1016/j.jplph.2020.153308
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550. doi: 10.1105/tpc.014928
Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. U.S.A. 98, 10487–10492. doi: 10.1073/pnas.171304098
Wisniewska, J., Xu, J., Seifertová, D., Brewer, P. B., Ruzicka, K., Blilou, I., et al. (2006). Polar PIN localization directs auxin flow in plants. Science 312:883. doi: 10.1126/science.1121356
Yang, Y., Wang, Y., Zhan, J., Shi, J., Wang, X., Liu, G., et al. (2017). Genetic and cytological analyses of the natural variation of seed number per pod in rapeseed (Brassica napus L.). Front. Plant Sci. 8:1890. doi: 10.3389/fpls.2017.01890
Yang, Z., Xin, D., Liu, C., Jiang, H., Han, X., Sun, Y., et al. (2013). Identification of QTLs for seed and pod traits in soybean and analysis for additive effects and epistatic effects of QTLs among multiple environments. Mol. Genet. Genomics 288, 651–667. doi: 10.1007/s00438-013-0779-z
Yu, S. X., Zhou, L. W., Hu, L. Q., Jiang, Y. T., Zhang, Y. J., Feng, S. L., et al. (2020). Asynchrony of ovule primordia initiation in Arabidopsis. Development 147:dev196618. doi: 10.1242/dev.196618
Zhang, B., Li, C., Li, Y., and Yu, H. (2020). Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants 6, 1146–1157. doi: 10.1038/s41477-020-0749-5
Zhang, M., Liu, S., Wang, Z., Yuan, Y., Zhang, Z., Liang, Q., et al. (2021). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 2021:10.1111/bi.13682. doi: 10.1111/pbi.13682
Zhang, Y., Zhang, Y. J., Yang, B. J., Yu, X. X., Wang, D., Zu, S. H., et al. (2016). Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci. Rep. 6:31134. doi: 10.1038/srep31134
Zhang, Z., Dunwell, J. M., and Zhang, Y. M. (2018). An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol. 18:328. doi: 10.1186/s12870-018-1542-8
Zhou, H. W., Nussbaumer, C., Chao, Y., and DeLong, A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16, 709–722. doi: 10.1105/tpc.018994
Zhou, Y., Zhu, J., Tong, T., Wang, J., Lin, B., and Zhang, J. (2019). A statistical normalization method and differential expression analysis for RNA-seq data between different species. BMC Bioinform. 20:163. doi: 10.1186/s12859-019-2745-1
Zhu, B., Tian, Z., Zhou, G., Pan, Y., Chen, X., Wang, Z., et al. (2020). A Molecular Breeding Method for a New Soybean Line with High Yield. China Patent CN108260522B. Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. Beijing.
Zourelidou, M., Absmanner, B., Weller, B., Barbosa, I. C., Willige, B. C., Fastner, A., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3:e02860. doi: 10.7554/eLife.02860
Keywords: seed number per pod, ovule number, PIN1, CKX, soybean, cowpea, interaction network, yield
Citation: Liu L-M, Zhang H-Q, Cheng K and Zhang Y-M (2021) Integrated Bioinformatics Analyses of PIN1, CKX, and Yield-Related Genes Reveals the Molecular Mechanisms for the Difference of Seed Number Per Pod Between Soybean and Cowpea. Front. Plant Sci. 12:749902. doi: 10.3389/fpls.2021.749902
Received: 30 July 2021; Accepted: 29 October 2021;
Published: 29 November 2021.
Edited by:
Sean Mayes, University of Nottingham, United KingdomReviewed by:
Changwei Zhang, Nanjing Agricultural University, ChinaWeibing Yang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), China
Copyright © 2021 Liu, Zhang, Cheng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan-Ming Zhang, c295emhhbmdAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Lü-Meng Liu
Lü-Meng Liu Han-Qing Zhang†
Han-Qing Zhang† Kun Cheng
Kun Cheng Yuan-Ming Zhang
Yuan-Ming Zhang