- 1College of Science, China Agricultural University, Beijing, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
Patatin-like proteins (PLPs) have non-specific lipid acyl hydrolysis (LAH) activity, which can hydrolyze membrane lipids into fatty acids and lysophospholipids. The vital role of PLPs in plant growth and abiotic stress has been well documented. However, the function of PLPs in plant defense responses against pathogens is still poorly understood. Here, we isolated and identified a novel cotton (Gossypium hirsutum) PLP gene GhPLP2. The expression of GhPLP2 was induced upon treatment with Verticillium dahliae, the signaling molecules jasmonic acid (JA) and ethylene (ETH) in cotton plants. Subcellular localization revealed that GhPLP2 was localized to the plasma membrane. GhPLP2-silenced cotton plants were more susceptible to infection by V. dahliae, while the overexpression of GhPLP2 in Arabidopsis enhanced its resistance to V. dahliae, which was apparent as mild symptoms, and a decrease in the disease index and fungal biomass. The hypersensitive response, deposition of callose, and H2O2 accumulation triggered by V. dahliae elicitor were reduced in GhPLP2-silenced cotton plants. The overexpression of GhPLP2 in Arabidopsis resulted in the accumulation of linoleic acid (LA, 18:2) and α-linolenic acid (ALA, 18:3) and facilitated the biosynthesis of JA and JA-mediated defensive responses. GhPLP2 silencing in cotton plants consistently reduced the accumulation of linoleic acid (LA, 18:2) and α-linolenic acid (ALA, 18:3) and suppressed the biosynthesis of JA and the defensive responses mediated by JA. These results indicate that GhPLP2 is involved in the resistance of cotton to V. dahliae by maintaining fatty acid metabolism pools for JA biosynthesis and activating the JA signaling pathway.
Introduction
Lipids in plants play a critical role in cell membrane components, the storage of sustainable energy, and signaling transduction in response to biotic and abiotic stress (Wang, 2004; Li et al., 2016; Lim et al., 2017). The accumulation of products of hydrolysis owing to the activity of lipid acyl hydrolase (LAH) in lipid metabolism affects pathogenesis and the mechanisms of resistance in plant-microbe interactions (Shah, 2005). As the key hydrolytic products of membrane lipids, fatty acids (FAs) participate in modulating the accumulation of endogenous nitric oxide and the biosynthesis of azelaic acid (AzA) which contribute to the development of systemic acquired resistance (SAR), production of reactive oxygen species (ROS), and subsequent defense responses in plants (Mandal et al., 2012; Yu et al., 2013; Wang et al., 2014). Oxidation and further conversions of polyunsaturated FAs by α-dioxygenase (α-DOX) and lipoxygenases (LOXs) generate various products that are collectively termed oxylipins (Savchenko et al., 2014; Cook et al., 2021). Jasmonates (JAs) constitute a crucial member of oxylipins, many of which are involved in plant defenses against pathogens, nematodes, and herbivores (Campos et al., 2014; Wasternack and Strnad, 2016, 2017). Moreover, oxylipins are related to the initiation of hypersensitive response (HR) in the interactions between plants and incompatible bacterial or fungal elicitors (Coll et al., 2011; García-Marcos et al., 2013; Sun et al., 2014).
Patatin proteins constitute a significant family of LAHs that possess non-specific LAH activity toward several membrane lipids; they were first identified and isolated as vacuolar storage proteins in potato tubers (Hendriks et al., 1991; Scherer et al., 2010). They generate free fatty acids and lysophospholipids by catalyzing the non-specific hydrolysis of membrane lipids, including phospholipids, glycolipids, sulfolipids, and mono- and diacylglycerols (Li and Wang, 2014). Various functions of patatin proteins have been demonstrated in responses to auxin, the production of cellulose, salt tolerance, drought response, and plant growth (Li et al., 2011; Yang et al., 2012; Dong et al., 2014; Li J. et al., 2020). More importantly, patatin and patatin-like proteins (PLPs) play an essential role in defense response to various pathogens. In Arabidopsis, patatin-related phospholipase I (AtPLAI) with the activity to hydrolyze acyls plays a role in maintaining the homeostatic pool of basal free fatty acids and basal jasmonic acid production and conferring resistance to infection with Botrytis cinerea (Yang et al., 2007). The Arabidopsis patatin-like protein 2 (AtPLP2), a pathogen-induced patatin-like LAH, provides fatty acids precursors for the biosynthesis of oxylipins and differentially affects the resistance to pathogens with distinct lifestyles (La Camera et al., 2009). Overexpression of Arabidopsis patatin-related phospholipase IIIα (AtpPLAIIIα) enhanced resistance to turnip crinkle virus (TCV), which was accompanied by a higher ratio of salicylic acid to jasmonic acid, and increased the level of expression of defense gene PR1 (Jang et al., 2020). The silencing of pepper (Capsicum annuum) CaPLP1 compromised the defense responses to avirulent Xanthomonas campestris pv. vesicatoria (Xcv) (Kim et al., 2014). Several PLPs are involved in releasing FAs from lipid membranes and serving as biosynthetic precursors of jasmonic acid synthesis in the grapevine Vitis vinifera cv. Regent (resistant cultivars) after infection with Plasmopara viticola (Laureano et al., 2018). Furthermore, three lipid acyl hydrolases (NtPATs) were rapidly induced in tobacco during its HR response to the Tobacco mosaic virus (TMV) (Dhondt et al., 2000).
Cotton (Gossypium spp.) is widely cultivated around the world because of its significant economic value to the textile industry (Gao et al., 2013). However, the yield and quality of cotton can be seriously affected by Verticillium wilt, caused by the soil-borne pathogen Verticillium dahliae Kleb (Gong et al., 2017; Dhar et al., 2020). V. dahliae is a hemibiotrophic fungus that undergoes biotrophic and necrotrophic phases during the process of infecting the host (Pantelides et al., 2010; Lo Presti et al., 2015; Zhang L. et al., 2017). It colonizes the plants through young roots, the sites of lateral root formation or puncture wounds to the xylem and leads to the browning of vasculature, defoliation, and wilting (Chen et al., 2016; Shaban et al., 2018). This pathogen can infect more than 200 species of dicotyledonous plants and is particularly difficult to control by fungicides because the fungus resides in the woody vascular tissues (Fradin and Thomma, 2006; Mo et al., 2016). Moreover, the resting structures microsclerotia of V. dahliae can survive in the soil for up to 14 years even without a host (Short et al., 2015). The cultivation of resistant cotton varieties has proven to be an effective strategy to control Verticillium wilt (Gayoso et al., 2010). The identification and isolation of candidate gene involved in defense against Verticillium wilt are essential to study the mechanism of resistance to this disease. Therefore, studying the function and mechanism of patatin proteins in the resistance of cotton to infection by V. dahliae may provide a new strategy for controlling Verticillium wilt.
Jasmonic acid (JA) is a vital lipid-derived phytohormone that confers the host resistance to biotrophic and hemibiotrophic pathogens (Ghorbel et al., 2021). Previous genetic evidence indicates that the JA signaling pathway is integral to the resistance of cotton to V. dahliae (Li et al., 2014; He et al., 2016; Dhar et al., 2020). The homeodomain transcription factor HDTF1 identified from cotton plants infected with V. dahliae is a negative regulator of the JA signaling pathway. HDTF1 silencing resulted in the activation of the JA signaling pathway and enhanced the resistance to V. dahliae and B. cinerea (Gao et al., 2016). The GhJAZ2 protein can interact with GhbHLH171 and inhibit its transcriptional activity, resulting in an impaired JA-mediated defense response against V. dahliae (He et al., 2018). The calcium-dependent protein kinase GhCPK33 from upland cotton (Gossypium hirsutum) phosphorylates GhOPR3, which leads to the enhanced instability of GhOPR3, which consequently limits the biosynthesis of JA and the subsequent resistance to V. dahliae (Hu et al., 2018). Silencing of the BEL1-Like transcription factor GhBLH7-D06 enhanced the resistance of cotton to Verticillium wilt by activating the biosynthesis of lignin and the JA signaling pathway (Ma et al., 2020). The constitutive expression of the GSK3-like kinase BIN2 (Brassinosteroid insensitive 2) attenuated the resistance of plants to V. dahliae by regulating the endogenous content of JA and the expression of JA-responsive marker genes (Song et al., 2021).
In this study, we report the identification and characterization of PLP gene GhPLP2 in cotton (G. hirsutum) that is induced by V. dahliae. The patterns of transcriptional expression of GhPLP2 were studied in response to biotic and abiotic stress. Sequence analyses showed that GhPLP2 had conserved catalytic dyad residues, which are critical for the LAH activity of patatins. The recombinant GhPLP2 protein further confirmed its in vitro LAH activity. Reduced HR response during the V. dahliae elicitor infection was observed in GhPLP2-silenced plants, suggesting the role of GhPLP2 in HR-like cell death signaling. The positive role of GhPLP2 in regulating the resistance of cotton to V. dahliae was examined by virus-induced gene silencing (VIGS) technology in cotton plants and overexpression in Arabidopsis. Further experiments demonstrated that GhPLP2 positively regulates the resistance of cotton to V. dahliae by mediating the metabolism of fatty acids and activation of the JA signaling pathway. Taken together, this research reveals the molecular mechanism of GhPLP2 in the defense of cotton against V. dahliae and provides new insights to improve the resistance of cotton to Verticillium wilt.
Materials and Methods
Plant Growth and Culturing of Verticillium dahliae
Seeds of the upland cotton (G. hirsutum) resistant cultivar Zhongzhimian 2 (original strain no. GK44) were provided by the Cotton Research Institute, Chinese Academy of Agricultural Sciences (Anyang, Henan). The germinated seeds were cultured in nutrient soil and vermiculite (2:1, w/w) under a photoperiod of 16 h/8 h. After vernalization for 3 days at 4°C in Murashige-Skoog (MS) culture medium, both the wild-type (Columbia-0) and transgenic Arabidopsis plants were grown in a mixture of nutrient soil and vermiculite (1:1, w/w) under a controlled environment with 16 h light (25°C)/8-h dark (22°C). The aggressive defoliating isolate Vd991 of V. dahliae was cultured on potato dextrose agar (PDA) at 25°C for 7 days and then inoculated in Czapek liquid media. The experiments were performed using conidial suspensions of 1 × 107 conidia/mL. Briefly, the cotton plants (2 weeks after VIGS) and Arabidopsis plants (4-week-old) were gently uprooted and the roots were dipped in conidial suspensions (1 × 107 conidia/mL) for 5 min. Then they were replanted in a mixture of nutrient soil and vermiculite.
Isolation of GhPLP2 cDNA
Total RNA was extracted using an RNA extraction kit (Biomed Gene Technology Co., Ltd., Wuhan, China), and the cDNA was synthesized using the manufacturer’s instructions with a FASTQuant cDNA RT Kit (Tiangen Biotech Co., Ltd., Beijing, China). The forward primer 5′-ACGCGTCGACATGGAAAAAAGTACTGGAAAC-3′ (SalI, underlined restriction site) and the reverse primer 5′-ATAGACTAGTTGGGCCAAGCTTGTGCAA-3′ (SpeI, underlined restriction site) were designed according to the GhPLP2 sequence (Gene ID: Gh_D02G0923) from cotton protein databases (G. hirsutum, NAU assembly) in the Cotton Functional Genomics Database1 to amplify the open reading frame (ORF) using the cDNA template. The PCR product was transferred into the pMD18-T vector according to the manufacturer’s instructions (TaKaRa, Dalian, China), and the positive clone was sequenced.
Bioinformatic Analyses
The theoretical isoelectric point (pI) and molecular mass were calculated with ProtParam.2 All the sequences were retrieved from the website of the National Center for Biotechnology Information database,3 and the accession numbers of the patatin proteins are listed in Supplementary Table 1. Multiple amino acid sequence alignments were performed using Clustal Omega,4 and the multiple alignment file was shaded with BoxShade.5 A phylogenetic tree was constructed with the neighbor-joining method using MEGA 7 with bootstrap values from 1,000 replicates indicated at the nodes, and the motifs were annotated using MEME6 (Bailey et al., 2009; Kumar et al., 2016). The homology model of GhPLP2 was generated using SWISS-MODEL, and three-dimensional models were analyzed and visualized using EzMol, Version 1.20 (Bordoli et al., 2009; Reynolds et al., 2018). The crystal structure of SeMet Patatin (1oxw.1.B) was selected as a template to predict the theoretical model.
Treatments With Methyl Jasmonate, Ethylene, PEG 6000, and Verticillium dahliae
For abiotic stresses and hormone treatments, 2-week-old cotton seedlings were gently uprooted and the roots were immersed in Hoagland solution that contained 100 μM methyl jasmonate (Sigma-Aldrich, St. Louis, MO, United States), 2 mM ethylene (Sigma-Aldrich), and 2.5% (w/v) PEG 6000, respectively (Champion et al., 2009; Pei et al., 2019). For the treatments with methyl jasmonate and ethylene, the whole plant was collected at 0, 0.5, 3, 6, 9, 12, 24, and 30 h for RNA extraction. For PEG 6000 treatment, the whole plant was collected at 0, 1, 3, 6, 12, 24, and 48 h for RNA extraction. For V. dahliae treatment, seedling roots were inoculated with conidial suspensions for 5 min and then transplanted into sterile soil. Control samples were treated with sterile water. The whole plant was collected at 0, 0.5, 12 h, 1, 3, 5, and 7 days for RNA extraction.
Expression and Purification of the Recombinant GhPLP2 Protein
GhPLP2 was cloned into the 6 × His-Tagged protein expression vector pET-22b (+) (Novagen, Madison, WI, United States). The primers are listed in Supplementary Table 2. The recombinant vector was expressed in Escherichia coli BL21 (DE3). Single colonies were cultured at 37°C in LB broth that contained 100 μg/mL ampicillin. When the OD600 reached 0.6, the culture was induced with 0.4 mM IPTG for another 12 h at 22°C at 200 rpm. The protein was purified using a 6 × His-Tagged Protein Purification Kit (CWbio, Beijing, China) following the manufacturer’s instructions and detected using SDS-PAGE. The concentration of purified protein was determined using a Bradford Protein Assay Kit (TaKaRa, Dalian, China).
GhPLP2 Enzyme Activity Assay Using the Substrates p-Nitrophenyl Palmitate and Phospholipids
The LAH activity of the GhPLP2 protein was assayed using the artificial substrate p-nitrophenyl palmitate (p-NPP) and phospholipids (Hong et al., 2008). Briefly, the reaction mixture was composed of 200 μL of Tris-HCl (pH 8.0), 10 mM CaCl2, 0.05% (v/v) Triton X-100, and 2 mM p-nitrophenyl palmitate to which 25 μg of purified protein was added and incubated at 37°C for 30 min. A volume of 700 μL of absolute ethanol was added and centrifuged for 2 min at 7,000 rpm. The absorbance of p-nitrophenol (NP) liberated from p-NPP was measured at 405 nm.
The substrates for phospholipase assay included 1,2-distearoyl-sn-glycerol-3-phosphoethanolamine (PE), 1,2-distearoyl-sn-glycerol-3-phosphocholine (PC), or L-α-phosphatidylglycerol (PG) (Aladdin, Shanghai, China). The reaction mixture was composed of 50 mM Tris-HCl (pH 8.0), 10 mM CaCl2, 0.05% (v/v) Triton X-100, 500 μg substrate, and 10 μg purified protein in a final volume of 600 μL. The reactions were performed at 37°C for 1 h and terminated by the addition of 500 μL chloroform: methanol (2:1, v/v). The fatty acids products released from the substrates were separated and analyzed as previously described (La Camera et al., 2005). Individual fatty acid methyl esters (FAME) were quantified using gas chromatography (GC) equipped with an Agilent column (Agilent Technologies, Santa Clara, CA, United States) (30 m by 0.25 mm, 0.25 μm film) and a flame ionization detector (FID).
Subcellular Localization and Generation of Transgenic Plants
The PCR product with a SalI/SpeI restriction site was inserted into a modified pCAMBIA 1300 vector that harbored hygromycin phosphor-transferase (hptII) and green fluorescent protein (GFP) under the control of the CaMV 35S promoter (Supplementary Figure 1; Wang et al., 2011). By particle bombardment (PDS-1000; Bio-Rad, United States), the CaMV 35S:GhPLP2-GFP and 35S:GFP empty vector were transformed into onion epidermal cells. The transformed cells were incubated on the MS medium (with 20 g/L sucrose) in the dark for 18–24 h at 22°C.
For the generation of GhPLP2-transgenic plants, the construct was introduced into Agrobacterium tumefaciens strain GV3101 by the freeze-thaw method (Jyothishwaran et al., 2007), and Arabidopsis was transformed by the floral dip method (Clough and Bent, 1998). Transgenic Arabidopsis seeds were screened on MS plates (with 20 g/L sucrose) that contained 25 mg/mL hygromycin B and T3 homozygous lines that displayed 100% hygromycin resistance were used for additional experiments.
Construction of Virus-Induced Gene Silencing Vectors and Agrobacterium-Mediated Virus-Induced Gene Silencing
The virus-induced gene silencing (VIGS) transient expression methods were previously described by Gao et al. (2011). The silenced fragments of GhCLA1 (Cloroplastos alterados 1) and GhPLP2 were amplified from cotton cDNA and inserted into the TRV:00 vector to generate the TRV:GhCLA1 and TRV:GhPLP2 vectors. Then plasmids of TRV:GhCLA1 and TRV:GhPLP2 were transformed into A. tumefaciens strain GV3101 by heat shock (Dong et al., 2007). TRV:GhCLA1 plants were used as positive controls. Two weeks after infiltration, when the GhCLA1-silenced plants showed clear signs of albinism in the leaves, the efficiency of GhPLP2 was evaluated by quantitative real-time reverse transcriptase–PCR (qRT-PCR). GhUBQ7 (DQ116441) was amplified as an internal control with the primers qUBQ7-F/R. All the primers are listed in Supplementary Table 2.
Verticillium dahliae Inoculation and Disease Investigation
Two weeks after VIGS, when the true leaves of GhCLA1-silenced cotton plants showed clear signs of albinism, the plants were inoculated with V. dahliae (1 × 107 conidia/mL) as previously described (Liu et al., 2018). The plant disease index (DI) was monitored using the following formula as described by Wang et al. (2020): DI = [Σ (n × the number of seedlings at level n)/(4 × the number of total seedlings)] × 100, n denotes disease level, cotton seedlings were divided into five levels based on their disease severity after inoculation with V. dahliae (level 0, 1, 2, 3, 4). For Arabidopsis plants, 4-week-old GhPLP2-transgenic and wild-type (WT) Arabidopsis plants were inoculated with V. dahliae spores as previously described (Gao et al., 2013). The controls were dipped in sterilized water. The disease index and classification of symptoms were determined as previously reported (Pei et al., 2019). Data were collected from three independent technical replicates (n ≥ 30).
The V. dahliae recovery assay was conducted and modified as described by Fradin et al. (2009), and the antibiotic in the PDA is kanamycin in this study. The stem section above the cotyledons was taken from cotton plants 3 weeks after inoculation with V. dahliae. Six slices were sterilized with 75% ethanol and 10% sodium hypochlorite solution and transferred onto PDA supplemented with kanamycin (50 mg/L) after the slices were washed three times with sterile water. The V. dahliae biomass was quantified as previously described (Ellendorff et al., 2009). Inoculated cotton (first internode) and Arabidopsis plants (whole plants) were harvested at 21 days post-inoculation (dpi) and 14 dpi, respectively. DNA was extracted from 100 mg of the fine powder. V. dahliae biomass was determined by qRT-PCR using fungus-specific ITS1-F (Liguori et al., 2007) and V. dahliae-specific ST-Ve1-R primers (Fradin et al., 2011). GhUBQ7-F/R (DQ116441) was used as a reference gene in the cotton plants, and AtEF1α-F/R (AT5G60390) was used as a reference gene in the Arabidopsis plants. All the primers are listed in Supplementary Table 2.
Analysis of Hypersensitive Response in Cotton Induced by the PevD1 Elicitor From Verticillium dahliae
The primers were designed to obtain the PevD1 fragment from V. dahliae (Supplementary Table 2), which was then cloned into the pET-28a (+) vector. The recombinant vector was expressed in E. coli BL21 (DE3), and the protein was induced with 0.1 mM IPTG for 10 h at 22°C while shaken at 200 rpm. The protein was purified using a 6 × His-Tagged Protein Purification Kit (CWbio, Beijing, China) following the manufacturer’s instructions and detected using SDS-PAGE. The concentration of purified protein was determined using a Bradford Protein Assay Kit (TaKaRa, Dalian, China). The treatment with PevD1 elicitor was performed as described by Li et al. (2019). The expression of HR marker genes GhHSR203 and GhHIN1 triggered by PevD1were detected in cotton leaves at 24 h after infection. Callose deposition was stained using 0.1% aniline blue as previously described (Wang et al., 2018). Images were observed using FLUOVIEW FV1000 on a confocal laser scanning microscope (Olympus, Tokyo, Japan). The level of H2O2 was measured by freshly made FOX reagent (Fe[NH4]2∙[SO4]2) (250 mM), xylenol orange (100 μM), sorbitol (100 μM), and H2SO4 (25 mM), and 1% ethanol as described by Pei et al. (2019).
Fatty Acids Analysis and the Quantification of Hormones
Four-week-old Arabidopsis and 2-week GhPLP2-silenced cotton plants after VIGS were used to analyze the composition of fatty acids and quantify the hormones. The hormones were quantified as previously described (Li D. et al., 2020). Fresh samples were ground into fine powder in liquid nitrogen. A total of 100 mg powder was extracted with 1 mL of 80% aqueous methanol that contained 1% formic acid using an ultrasonic treatment at 4°C for 10 min and centrifuged at 12,000 rpm at 4°C for 10 min. The supernatant was transferred into a 2 mL centrifuge tube that contained 50 mg of primary secondary amine (CNW Technologies GmbH, Dusseldorf, Germany), dried with nitrogen, and added to 100 μL of 80% aqueous methanol that contained 1% formic acid. The samples were analyzed by an Agilent 6410B Triple Quadrupole HPLC-MS/MS (Agilent Technologies), which is equipped with an HPLC reverse phase C18 column (Athena C18-WP 2.1 × 50 mm, 3 μm). The flow rate was 0.3 mL/min. Methanol and ultrapure water were used as the mobile phases A and B, respectively. Isocratic elution was conducted for 5 min at a 65(A):35(B) ratio. MS was performed using the multiple reaction monitoring modes and negative electrospray ionization. All the parameters were performed as described by Li D. et al. (2020).
The fatty acid composition was analyzed by gas chromatography (GC) on Agilent 7890A (Agilent Technologies, Santa Clara, CA, United States) with a flame ionization detector (FID). Fatty acid methyl esterification was performed as described by Liu et al. (2019) with slight modification. The plant samples were dried at 90°C to a constant weight, and 50 mg of dry powder was weighed. Free fatty acids were methylated in 1 mL 0.5 mol/L KOH-methanol at 60°C for 2 h. Next, 1 mL of hexane that contained 0.01% butylated hydroxytoluene (BHT) and 0.1 mg methyl non-adecanoate was added. After shaking and standing for stratification, the supernatant that contained the FAMEs was separated. The GC parameters were as follows: 180°C for 10 min followed by a ramp to 190°C at 1°C/min, a hold at 190°C for 3 min, and then heating up to 220°C with a gradient at 4°C/min. The final temperature was maintained for 3 min.
Plant Drought Treatment
Arabidopsis drought treatment was carried out as previously described (Wang et al., 2018). Briefly, water was withheld from 3-week-old Arabidopsis plants for 16 days, followed by watering for 2 days from the bottom. All the Arabidopsis plants were grown in a controlled environment with 16 h light (25°C)/8-h dark (22°C) and a relative humidity at 80%.
Gene Expression Analysis by Quantitative Real-Time PCR
Total RNA was obtained from the plant samples with an RNA extraction kit with DNase I (Biomed Gene Technology, Co., Ltd., Beijing, China). The cDNA was synthesized using a PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa Bio, Dalian, China). qRT-PCR was performed using SYBR Premix Ex Taq (Tli RNaseH Plus) (TaKaRa, Dalian, China) on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, United States). AtEF1a (AT5G60390) and GhUBQ7 (DQ116441) were utilized as internal standard genes in Arabidopsis and cotton, respectively. The relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The melting curves for each gene studied (with a no template control) are provided in Supplementary Figures 2, 3. The primer sequences are listed in Supplementary Table 2.
Data Analysis
The data were obtained from three independent technical or biological replicates per treatment and presented as the mean ± standard error. The statistical analyses were performed by Student’s t-test. Asterisks indicates a significant difference between the groups (∗P < 0.05, ∗∗P < 0.01).
Results
Identification and Sequence Analysis of GhPLP2
From the transcriptome analysis of the cotton cultivar Zhongzhimian 2 that is resistant to infection by V. dahliae Vd991, the PLP gene GhPLP2 was highly induced when the plants were infected with V. dahliae. We isolated the full-length clone of GhPLP2 that consists of a 68 bp 5′untranslated region (5′ UTR), 138 bp 3′ UTR, and 1,218 bp open reading frame (ORF) that encoded a 405 amino acid protein with a theoretical pI of 8.21 and a molecular weight of 44.44 kDa. The GhPLP2 protein contains the conserved serine (S) hydrolase motif GXSXG at residues 63–67 and a conserved aspartic acid (D) residue at 214 within the patatin domain (residues 21–221) (Supplementary Figure 4). The catalytic dyad of S and D residues is critical for the LAH activity of patatin (Rydel et al., 2003; Rietz et al., 2010). Using SeMet patatin, an isozyme of patatin 17 from Solanum cardiophyllum as the template, the homology modeling results indicate that the Ser-Asp catalytic dyad is essential for the LAH activity of GhPLP2 (Supplementary Figure 5). GhPLP2 lacks the transmembrane domain or signal peptide. To study the possible role of GhPLP2 protein in cotton plants, a phylogenetic analysis was generated with homologous patatin proteins with precise functions that had been previously reported from other species. The phylogenetic analysis showed that plant patatins were divided into three groups, and GhPLP2 is located in group II that contains NtPat1 (Cacas et al., 2005), GhPat1 (Cacas et al., 2009), CaPLP1 (Kim et al., 2014), and AtPLP2 (La Camera et al., 2009; Figure 1A). The patatin homologs were divided into three groups based on the conserved motif alignments, which are consistent with the phylogenetic analysis result (Figure 1B). Group II members have the canonical S-D dyad esterase motif constituted by GxSxG and DGG/A in patatin catalytic centers, a conserved anion binding element DGGGxxG and the proline motif APP. GhPLP2 is a patatin group II member with the conserved catalytic dyad residues.
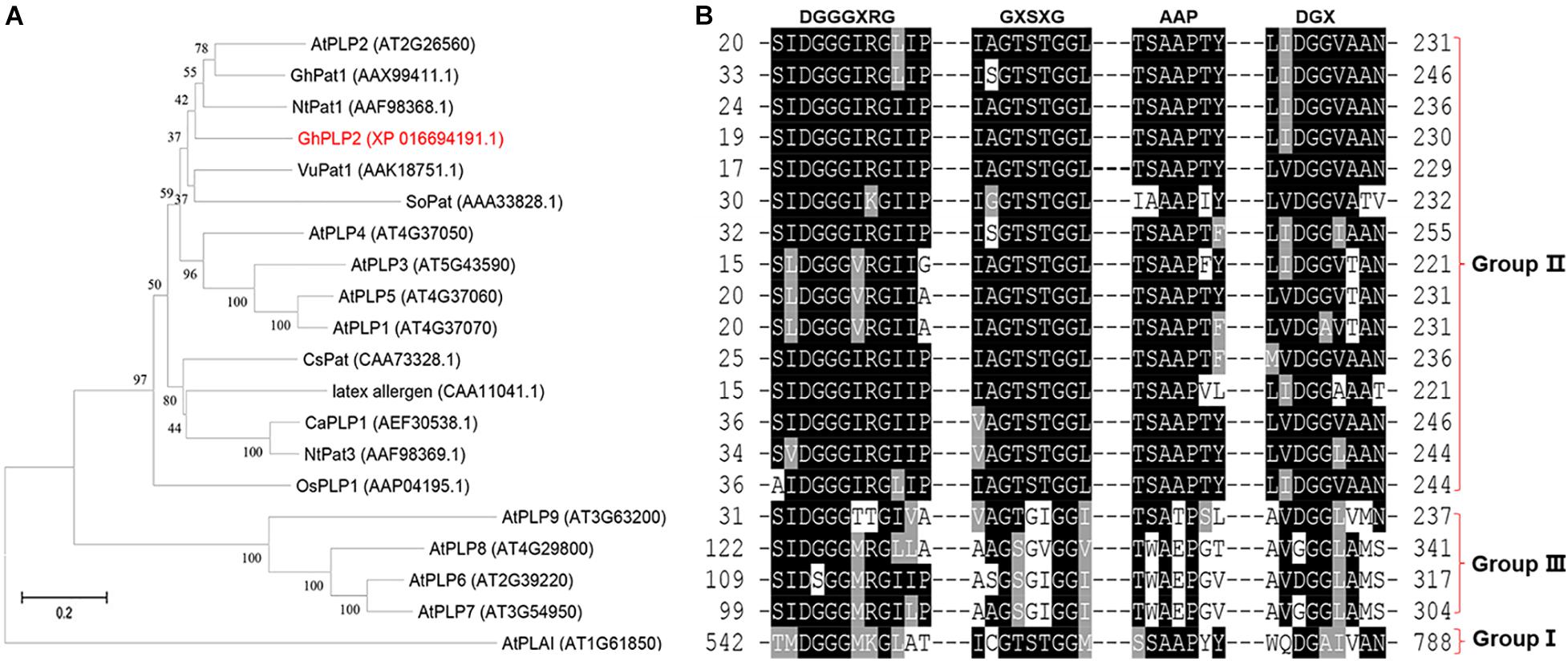
Figure 1. Sequence analyses of GhPLP2 and other patatin-like proteins. (A) Phylogenetic analysis of GhPLP2 and other plant patatin-like proteins. At1g61850 (AtPLAI) was used as the outgroup. The numbers on the tree represent bootstrap scores. (B) Alignment of the conserved motifs of patatin-like proteins. Esterase box GxSxG and residues DGX constitute the S-D catalytic dyad. Anion binding element: DGGGxxG and proline motif: APP.
Lipid Acyl Hydrolase Activity Assay of GhPLP2
The LAH activity of patatin proteins can hydrolyze membrane lipids into free fatty acids and lysophosphatidic acid, which lead to a series of signaling pathways involved in growth development and defense responses (Ryu, 2004). To understand the role of GhPLP2 in more detail, the LAH activity of the recombinant GhPLP2 protein was studied. The recombinant protein GhPLP2 was produced and purified from E. coli BL21 (DE3). The GhPLP2 protein was induced with 0.4 mM IPTG at 22°C and as early as 1 h after induction (Figure 2A). The GhPLP2 fusion protein was purified using Ni columns, and the elution fractions were confirmed by SDS-PAGE (Figure 2B). When p-NPP was used as the substrate, the His-GhPLP2 protein exhibited high LAH activity (97 nmol min–1 mg–1), and the vector control was totally deficient in enzyme activity (Figure 2C). In addition, when the phospholipids PC, PE, and PG were used as substrates, the release of free fatty acids was quantified. The purified GhPLP2 protein released fatty acids from these phospholipids and exhibited variant enzyme activities in different phospholipid substrates (Figure 2D). The results indicate that GhPLP2 may function as a LAH to release free fatty acids in plants.
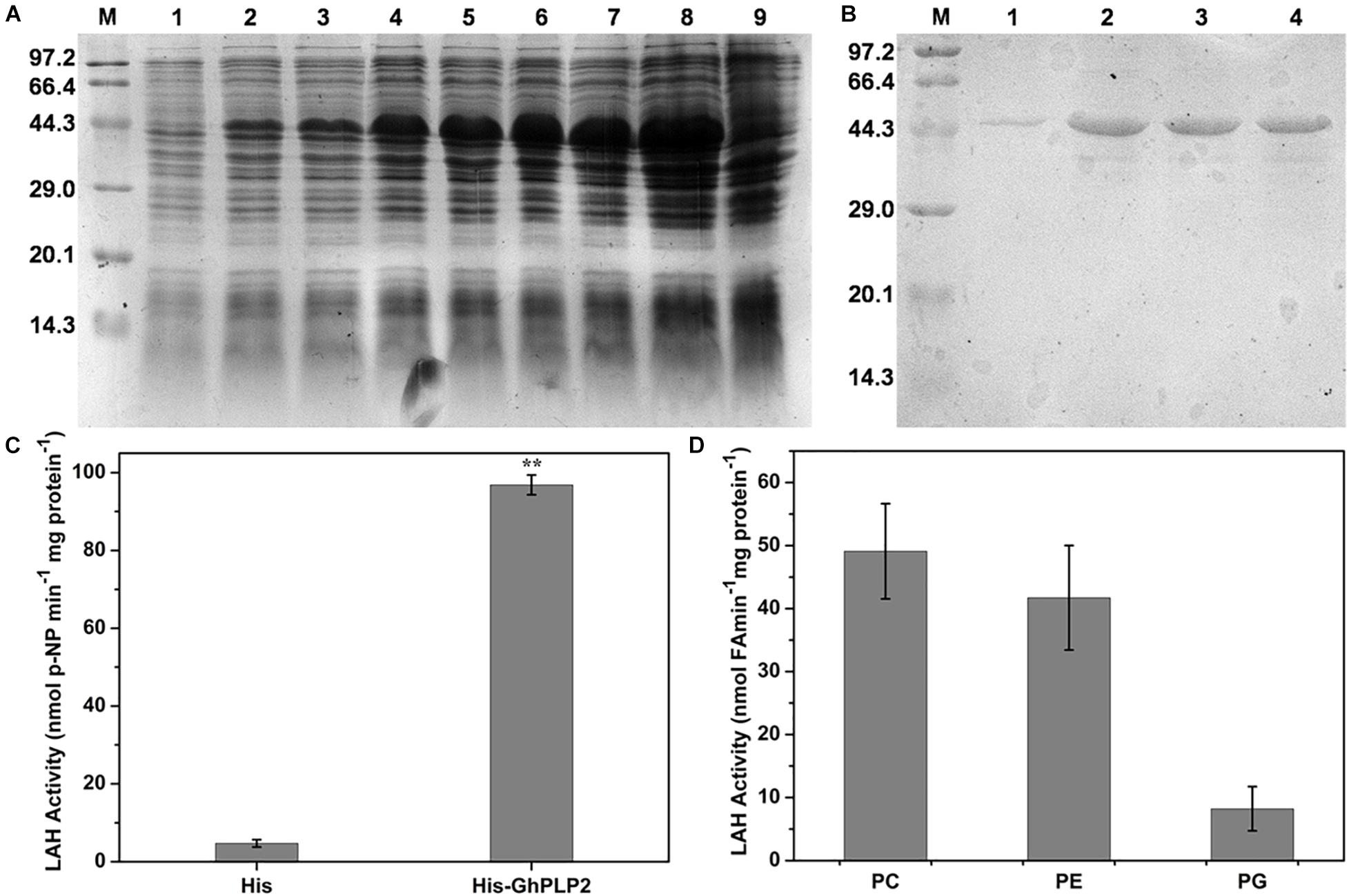
Figure 2. Induction and purification of the His-GhPLP2 protein and the LAH activity assay. (A) Proteins from induced bacteria were analyzed by SDS-PAGE. M: molecular mass of markers (kDa); 1: uninduced crude bacterial proteins; 2–8: IPTG-induced crude bacterial proteins at 22°C with different time points (1, 2, 4, 6, 12, 16, and 24 h); 9: pET-22b empty vector was induced at 24 h. (B) GhPLP2 fusion proteins purified on a Ni-column. 1–4: a series of eluted proteins. (C) p-nitrophenol (NP) was liberated from p-nitrophenyl palmitate by the LAH activity of GhPLP2 protein. Asterisks indicate a significant difference compared with the empty vector control (**P < 0.01, Student’s t-test). (D) Fatty acids were released by the activity of LAH from the GhPLP2 protein when PC, PE, and PG were used as substrates. Data were obtained from three independent biological replicates and presented as the mean ± standard error.
Subcellular Localization of the GhPLP2 Protein
To determine the subcellular localization of GhPLP2 protein, the GFP signals of GhPLP2-GFP fusion protein and 35S:GFP empty vector were monitored by Nikon A1R SI Confocal (Nikon, Japan). The GFP signal of 35S:GFP empty vector was diffused throughout the cell (Figures 3A–C), whereas the GhPLP2-GFP fusion protein was found to accumulate exclusively in the cell wall or plasma membrane (Figures 3D–F). To further clarify the location of GhPLP2, the onion epidermal cells were treated with 0.8 M mannitol for 10 min. After plasmolysis, the GFP signals of GhPLP2-GFP fusion protein was found in the plasma membrane (Figures 3G–I). These results revealed that GhPLP2 was localized to the plasma membrane.
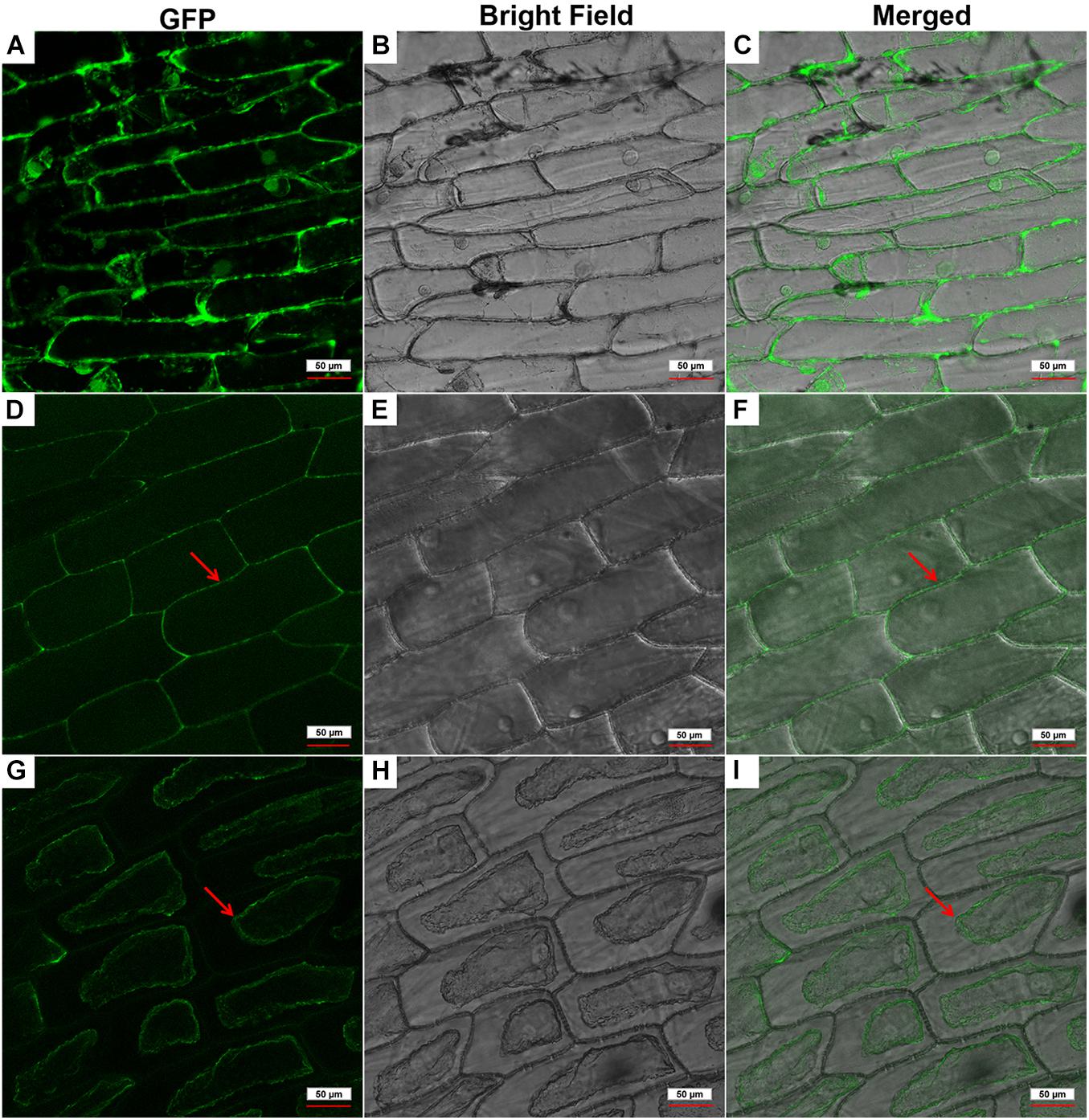
Figure 3. Subcellular localization of the GhPLP2-GFP fusion protein in onion epidermal cells. (A–C) 35S:GFP empty vector was used as a control. (D–F) Red arrows indicated the localization of the GhPLP2-GFP fusion protein. (G–I) Red arrows indicated the localization of the GhPLP2-GFP fusion protein after plasmolysis. Scale bar represents 50 μm.
Induction of GhPLP2 by Various Stresses
Expression of the GhPLP2 gene in cotton was examined by qRT-PCR. GhPLP2 was the most abundant in cotton roots (Figure 4A), which are the first physical barrier in cotton plants against infection by V. dahliae. The change in expression of GhPLP2 in response to various stresses was investigated. The expression of GhPLP2 was significantly increased by 9.4- and 7.1-fold at 0.5 h and 7 days after inoculation with V. dahliae, respectively (Figure 4B). We investigated the expression of GhPLP2 after treatment with the defense-related signaling molecules methyl jasmonate and ethylene in more detail. The expression of GhPLP2 was significantly higher at 3 h by 6.2-fold, and another 6.0-fold peak appeared at 24 h after treatment with JA (Figure 4C). In contrast, ethylene increased the expression of GhPLP2 at 0.5 h, with a maximum level of 4.7-fold observed at 3 h (Figure 4C). The treatment with 2.5% PEG 6000 (w/v) markedly increased the gene expression at 1 and 3 h with 5.6- and 3.7-fold, respectively (Figure 4D).
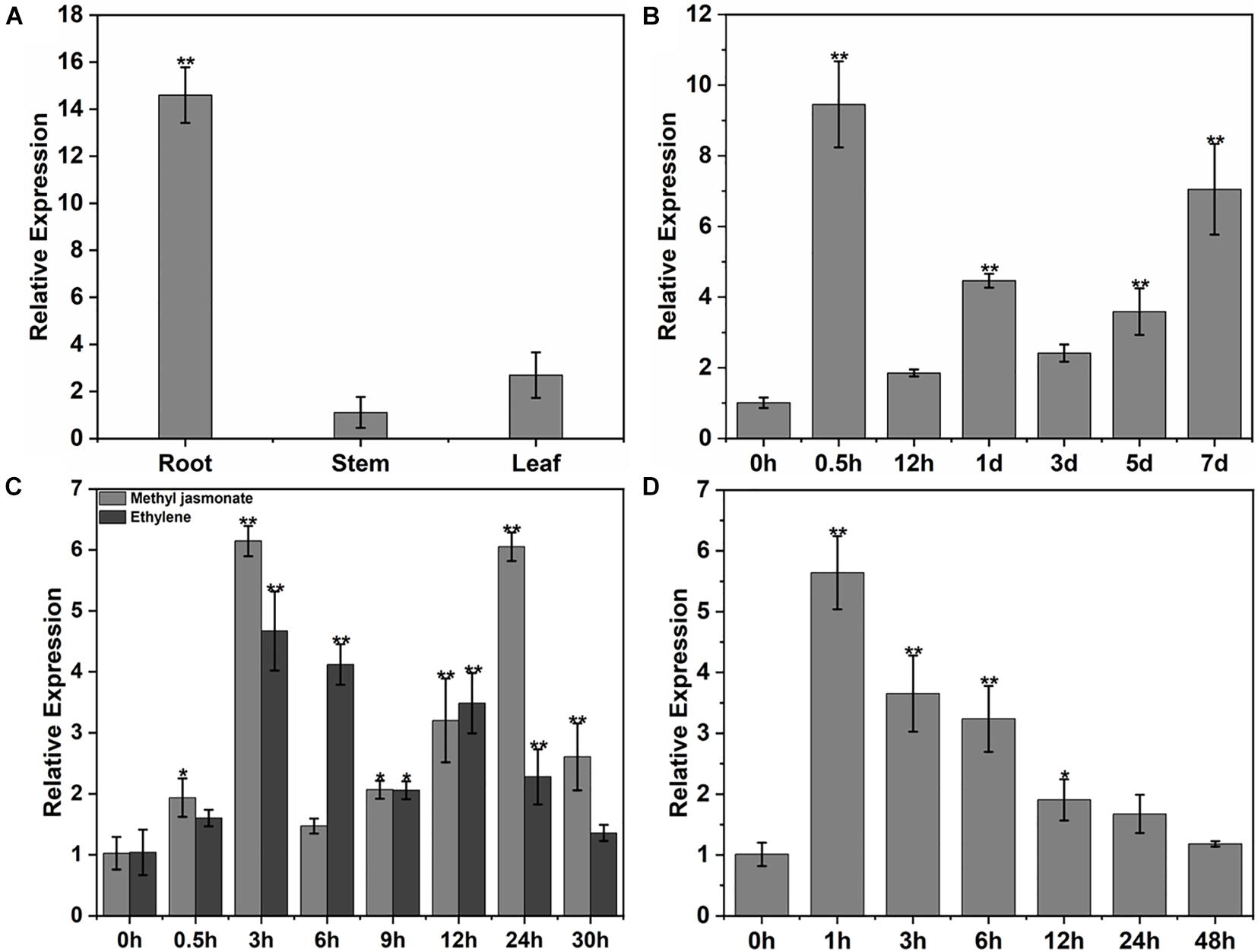
Figure 4. Patterns of expression of GhPLP2 in cotton subjected to different types of stress. (A) The patterns of expression of GhPLP2 in different tissues. (B) Expression of GhPLP2 after inoculation with Verticillium dahliae. (C) Expression of GhPLP2 after treatment with 100 μM methyl jasmonate and 2 mM ethylene. (D) Expression of GhPLP2 after treatment with 2.5% PEG 6000 (w/v). Data were obtained from three independent biological replicates and presented as the mean ± standard error. Asterisks indicate a significant difference compared with the control. (*P < 0.05, **P < 0.01, Student’s t-test).
Enhanced Disease Susceptibility of GhPLP2-Silenced Cotton Plants to Verticillium dahliae
The GhPLP2 gene was silenced by VIGS in cotton plants to clarify its function by Gao et al. (2013). The cotton gene GhCLA1, which is involved in chloroplast development, was used as a positive control (Gao et al., 2011). To assess its gene silencing efficiency, the expression of GhPLP2 was monitored by semi-quantitative PCR and qRT-PCR in TRV:00 and TRV:GhPLP2 cotton true leaves after 2 weeks of VIGS (Figure 5G and Supplementary Figure 6). The results indicated that the expression of GhPLP2 was effectively reduced in GhPLP2-silenced cotton plants.
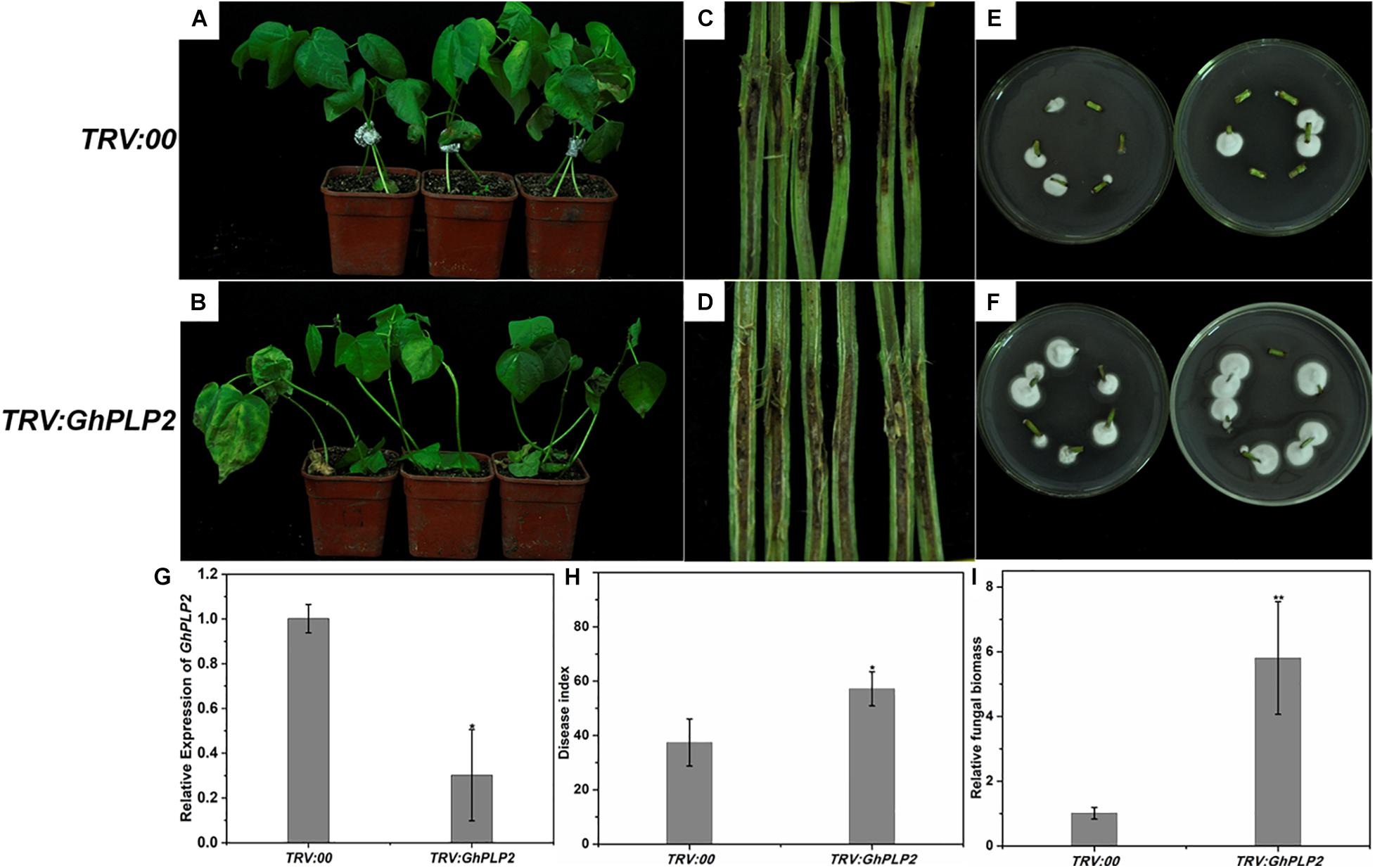
Figure 5. Attenuated resistance of GhPLP2-silenced cotton plants to Verticillium dahliae. Disease phenotypes of the control (A) and GhPLP2-silenced cotton plants (B) inoculated with V. dahliae at 14 days. Vascular browning in the stems of control (C) and GhPLP2-silenced cotton plants (D) infected by V. dahliae at 21 days. The fungal renewal cultivation of control (E) and GhPLP2-silenced cotton plants (F) infected by V. dahliae at 21 days. (G) The VIGS efficiency of GhPLP2 was evaluated by qRT-PCR after 2 weeks. Data were obtained from three independent biological replicates. Asterisks indicate a significant difference compared with the TRV:00. (H) The disease index of control and GhPLP2-silenced cotton plants inoculated with V. dahliae at 14 days. Data were obtained from three independent technical replicates. Asterisks indicate a significant difference compared with TRV:00. (I) Relative fungal biomass of the control and GhPLP2-silenced cotton plants infected by V. dahliae at 21 days. Data represent the means ± standard error of three independent biological replicates. Asterisks indicate a significant difference compared with TRV:00. (*P < 0.05, **P < 0.01, Student’s t-test).
To analyze the involvement of GhPLP2 in the interaction between cotton and V. dahliae, cotton plants were inoculated with this pathogen. After inoculation, typical disease symptoms appeared at 10 days in GhPLP2-silenced cotton. At 14 days, leaf chlorosis and necrosis were more severe in the GhPLP2-silenced plants (Figures 5A,B). Dark and necrotic vascular bundles of the dissected stems were more apparent in GhPLP2-silenced plants at 21 days (Figures 5C,D). The fungal renewal cultivation showed that more fungi were recovered from GhPLP2-silenced plants compared with the control (Figures 5E,F). The plant disease index of the silenced plants was higher than that of the control plants (Figure 5H). In addition, the fungal biomass of the stems from the GhPLP2-silenced plants were higher than that in the control plants determined by qRT-PCR analysis (Figure 5I). These results suggest that the silencing of GhPLP2 attenuates the resistance of cotton to V. dahliae.
Silencing of GhPLP2 Compromises the Hypersensitive Response Symptoms Triggered by Verticillium dahliae Elicitor PevD1
The HR is an efficient and immediate resistance reaction during the attack of pathogens, which rapidly causes cell death around the attempted infection site that restricts the spread of pathogens (Vailleau et al., 2002). During the HR, plant immune responses, including the accumulation of H2O2, deposition of callose, and the expression of HR-related marker genes were rapidly induced (Hamada et al., 2005; Simon et al., 2010; Rossi et al., 2011). PLPs have been shown to participate in HR in response to infection by the Gram-negative bacterial pathogen Xanthomonas campestris pv. vesicatoria in pepper plants and the response to Tobacco mosaic virus (TMV) in tobacco plants (Dhondt et al., 2000; Choi and Hwang, 2011; Kim et al., 2014). Therefore, GhPLP2 may regulate the resistance to V. dahliae by mediating an HR induced by V. dahliae elicitors. To test our hypothesis, we expressed and purified PevD1 from V. dahliae (Supplementary Figure 7), which triggered HR and resistance responses in cotton, Arabidopsis, and tobacco (Wang et al., 2012; Bu et al., 2014; Liu et al., 2016). The HR-like cell death was initiated at 24 h after injection with PevD1, while it was significantly reduced in GhPLP2-silenced leaves (Figure 6A). Moreover, PevD1 triggered the deposition of callose, and the expression of HR marker genes GhHIN1 and GhHSR203 were compromised in the GhPLP2-silenced leaves at 24 h after the infiltration (Figures 6B–D). In addition, the silencing of GhPLP2 significantly compromised the production of H2O2 induced by PevD1 (Figure 6E).
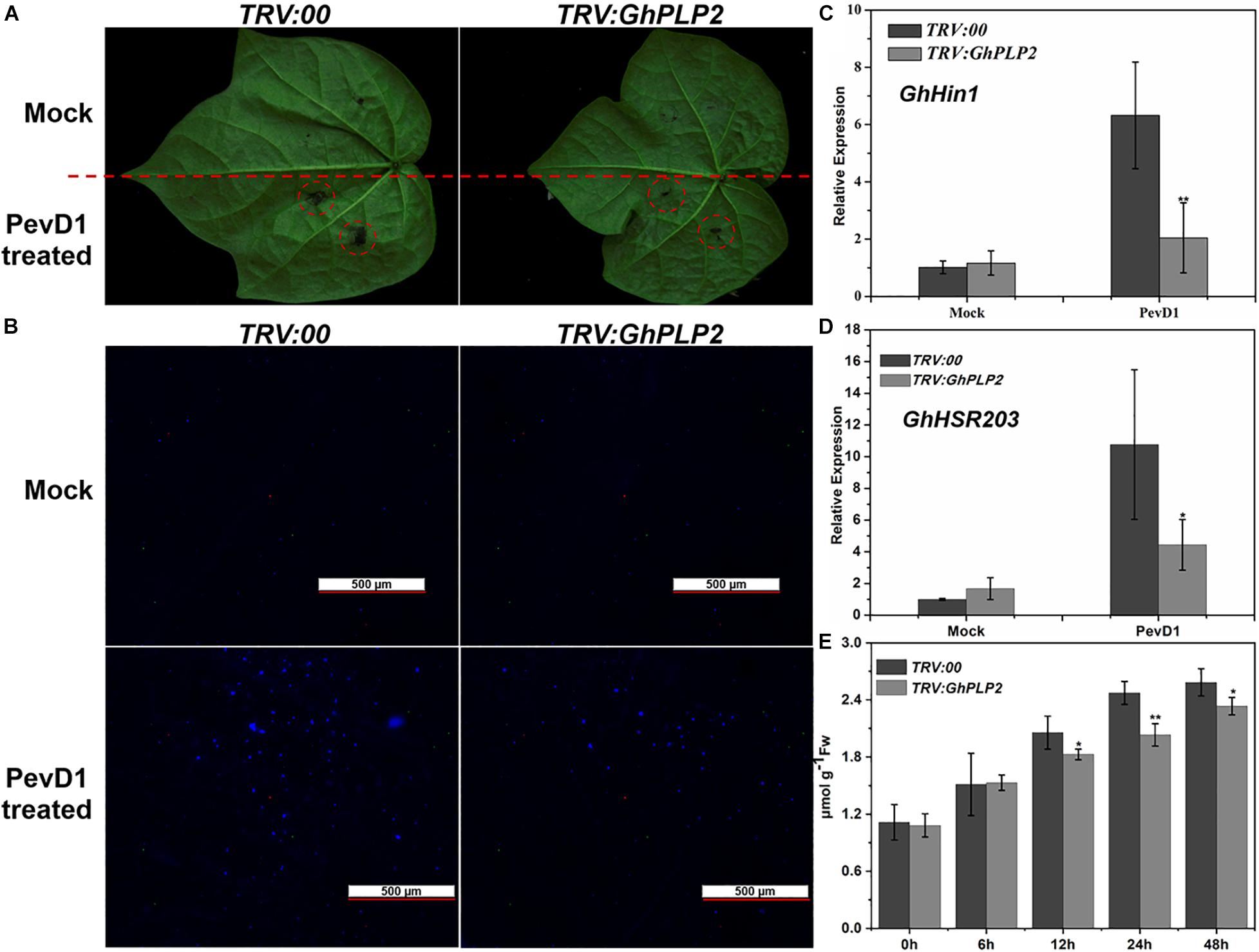
Figure 6. GhPLP2-mediated HR triggered by the Verticillium dahliae elicitor PevD1. (A) HR phenotypes developed on the leaves after injection with the PevD1 elicitor. (B) Callose deposition on the infected leaves at 24 h after injection. Scale bar represents 500 μm. (C,D) The expression of HR marker genes GhHIN1 and GhHSR203. (E) Detection of H2O2 in the leaves during injection with PevD1. Data were obtained from three independent biological replicates. Asterisks indicate a significant difference, as determined by Student’s t-test. (*P < 0.05, **P < 0.01).
Overexpression of GhPLP2 in Arabidopsis Confers Enhanced Resistance to Verticillium dahliae
To further evaluate the role of GhPLP2 in response to V. dahliae, we examined the resistance of wild-type (WT) and transgenic Arabidopsis seedlings to V. dahliae infection. Three homozygous transgenic (T3 generation) lines with the highest levels of expression of GhPLP2 (Lines 1, 4, and 8) were selected for additional experiments (Supplementary Figure 8).
Four-week-old transgenic and WT plants were infected with V. dahliae spores by the root dipping method (Pei et al., 2019). After inoculation, the WT plants showed more serious wilting, yellowish coloring and necrosis compared with the transgenic plants at 14 dpi (Figure 7A); this was consistent with the disease index investigation (Figure 7B). The fungal biomass of the WT plants was remarkably high compared with that on the transgenic plants as determined by qRT-PCR analysis (Figure 7C). These results indicate that the overexpression of GhPLP2 in Arabidopsis plants confers enhanced resistance to V. dahliae.
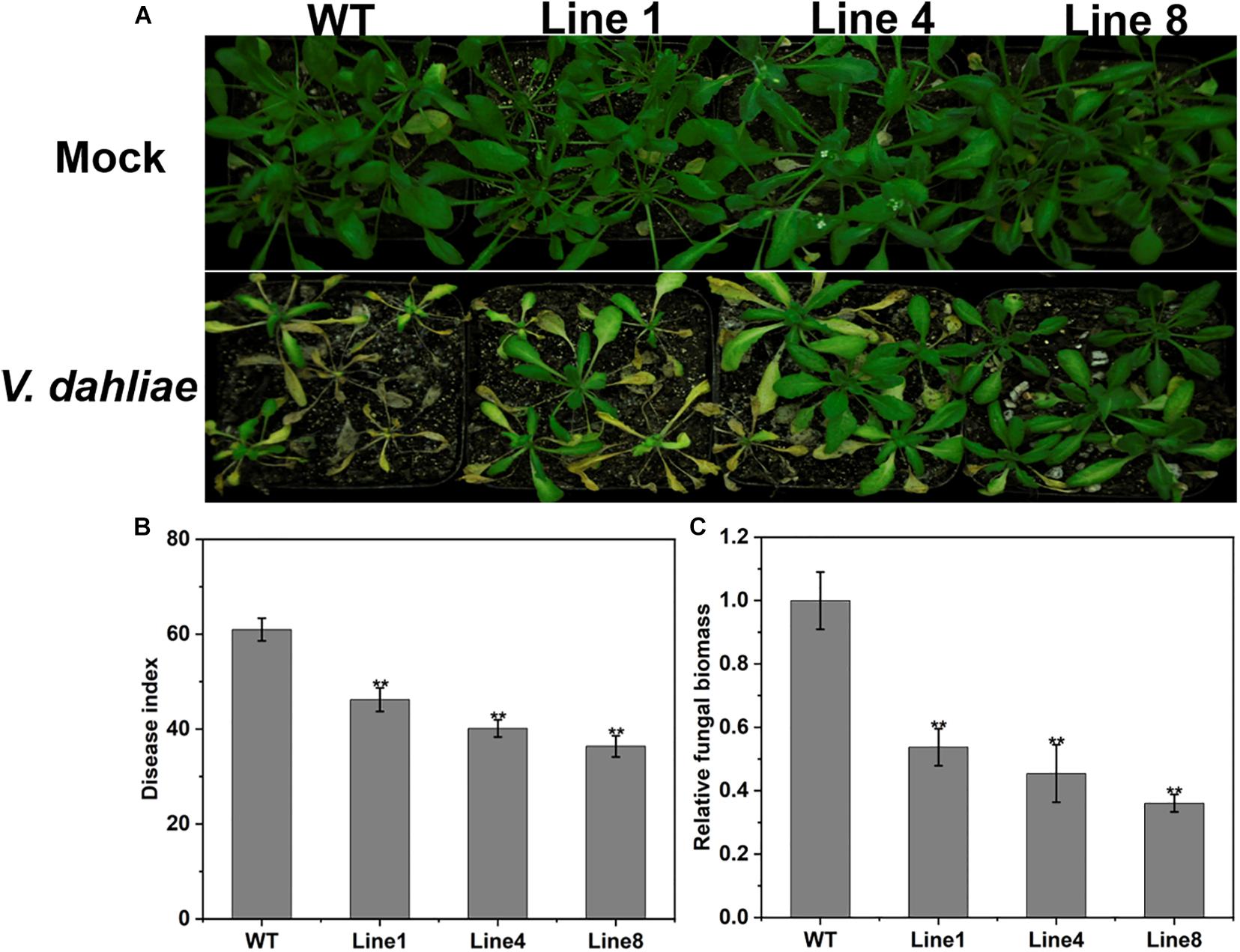
Figure 7. Increased resistance to Verticillium dahliae in GhPLP2-overexpressed Arabidopsis. (A) Arabidopsis plants were inoculated with sterile water and V. dahliae by root dipping, respectively. Images were taken at 14 dpi. (B) Disease index at 14 dpi. Data represent the means ± standard error of three independent technical replicates. Asterisks indicate a significant difference compared with the WT. (**P < 0.01, Student’s t-test). (C) The fungal biomass of Arabidopsis plants at 14 dpi. Data represent the means ± standard error of three independent biological replicates. Asterisks indicate a significant difference compared with the WT. (**P < 0.01, Student’s t-test).
GhPLP2 Involved in the Accumulation of Fatty Acids and Jasmonic Acid Signaling Pathway
Free fatty acids (FFAs) are among the hydrolyzates of membrane lipids owing to PLPs with LAH activity (Rivas and Heitz, 2014). The endogenous LAH activity of crude proteins from different genotypes plants was assayed using p-NPP as the substrate (Lin et al., 2011). The LAH activity of GhPLP2-overexpressed Arabidopsis lines was higher than that of the WT plants, whereas the LAH activity decreased in TRV:GhPLP2 cotton plants compared with the TRV:00 plants (Supplementary Figure 9). To further assess the metabolic differences between different genotypes of plants, the composition of fatty acids was examined in Arabidopsis and cotton plants 3 days after the inoculation of V. dahliae and water (mock) (Figure 8). As shown in Figure 8A, the analyses of the composition of fatty acids indicated that the levels of linoleic acid (LA, 18:2) and α-linolenic acid (ALA, 18:3) were higher in GhPLP2-overexpressed Arabidopsis plants than in the WT plants under treatment with water. After inoculation with V. dahliae, the content of different types of fatty acids increased in both genotypes of Arabidopsis plants, and the increase in GhPLP2-overexpressed Arabidopsis plants was more apparent compared with the WT (Figure 8A). Moreover, GhPLP2-silenced cotton plants exhibited a significant reduction in their contents of LA and ALA regardless of the treatment with V. dahliae (Figure 8B). In addition, the content of palmitic acid (16:0) and stearic acid (18:0) also increased significantly after the inoculation of cotton plants with V. dahliae.
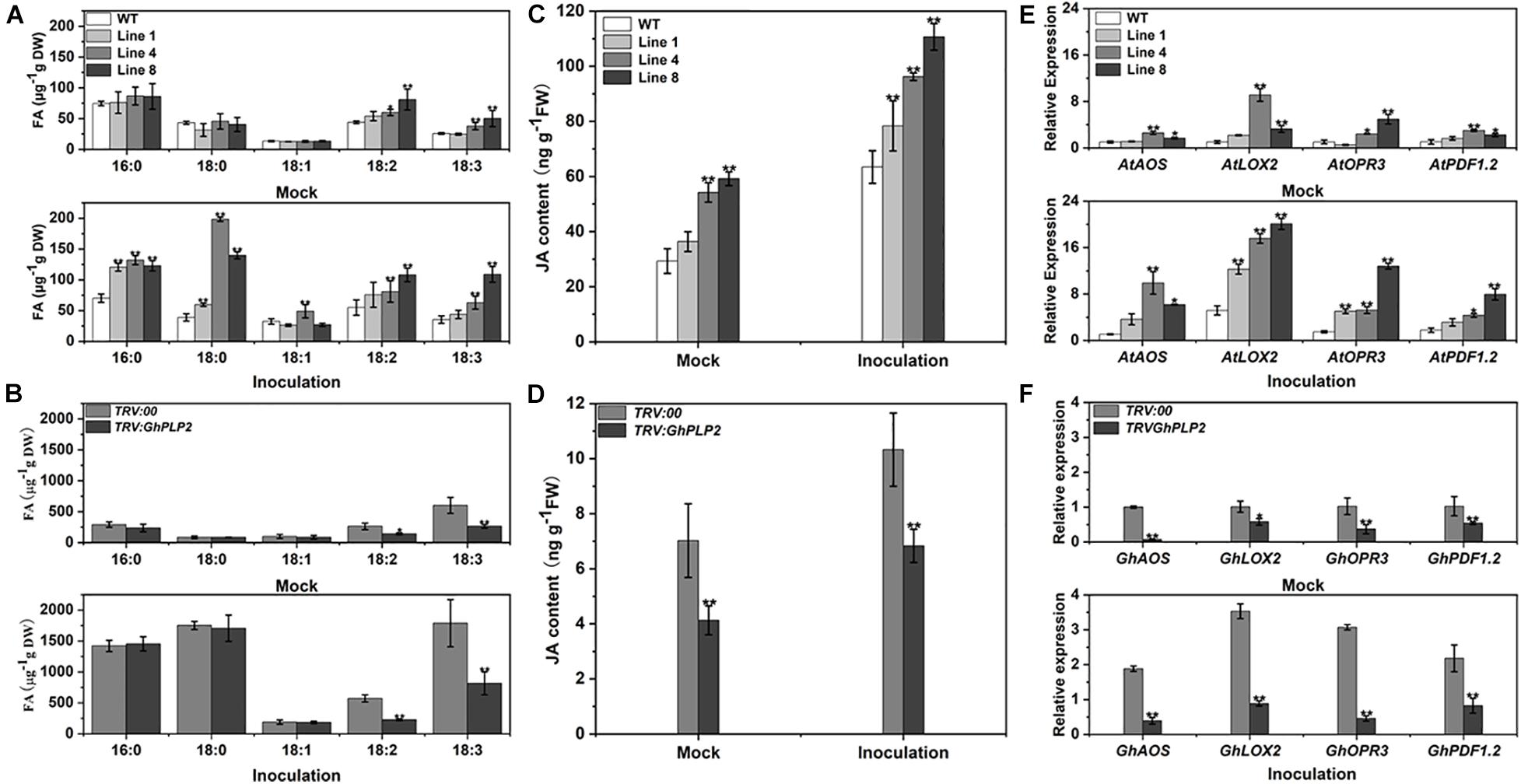
Figure 8. The composition of fatty acids, accumulation with JA, and gene expression were determined in Arabidopsis and cotton plants 3 days after inoculation with water (mock) and Verticillium dahliae. (A) Analysis of the fatty acids composition in Arabidopsis plants 3 days after inoculation with water (mock) and V. dahliae. (B) Analysis of the fatty acids composition in cotton plants 3 days after inoculation with water (mock) and V. dahliae. (C) Accumulation of JA in Arabidopsis plants 3 days after inoculation with water (mock) and V. dahliae. (D) Accumulation of JA in cotton plants 3 days after inoculation with water (mock) and V. dahliae. (E) The relative level of expression of the genes associated with the JA signaling pathway in WT and GhPLP2-overexpressed Arabidopsis plants 3 days after inoculation with water (mock) and V. dahliae. AtEF1α (AT5G60390) was used as the standard internal gene. (F) The relative expression of genes associated with the JA signaling pathway in TRV:00 and GhPLP2-silenced cotton plants 3 days after inoculation with water (mock) and V. dahliae. GhUBQ7 (DQ116441) was used as the standard internal gene. Data represent the means ± standard error of three independent biological repeats. Asterisks indicate a significant difference compared with WT or TRV:00, respectively. (*P < 0.05, **P < 0.01, Student’s t-test).
It has been reported that the oxidation and further conversion of C18 polyunsaturated FAs with lipoxygenases (LOXs) may generate the production of JA (Walley et al., 2013; Sun et al., 2014; Fu et al., 2015). We then investigated the JA contents in GhPLP2-overexpressed Arabidopsis and GhPLP2-silenced cotton plants. The contents of JA in GhPLP2-overexpressed Arabidopsis lines 4 and 8 were 84.97 and 101.99% higher than that of the WT, respectively, when they had not been inoculated with V. dahliae. After inoculation with V. dahliae, the JA contents of GhPLP2-overexpressed Arabidopsis lines 4 and 8 were 51.63 and 74.42% higher than that of the WT, respectively (Figure 8C). The levels of JA in the TRV:GhPLP2 cotton plants were 41.21 and 33.83% lower than that of the TRV:00 cotton plants under mock and V. dahliae inoculations, respectively (Figure 8D).
We also conducted qRT-PCR to detect the expression of a series of the JA-synthesis and -signaling genes from Arabidopsis and cotton plants, namely AOS, LOX2, OPR3, and PDF1.2. All of these genes were highly induced by infection with V. dahliae in both Arabidopsis and cotton at 3 dpi (Figures 8E,F). Consistent with the contents of JA, we found that the expression of these genes was significantly upregulated in GhPLP2-overexpressed Arabidopsis plants compared with the WT and downregulated in TRV:GhPLP2 cotton plants relative to the TRV:00 plants in both treatments (Figures 8E,F). Taken together, the results showed that GhPLP2 contributes to the accumulation of fatty acids and is involved in the JA signaling pathway in both Arabidopsis and cotton plants.
GhPLP2 Overexpression Has No Effect on Arabidopsis Tolerance to Drought
Based on our observation that PEG 6000 induced the expression of GhPLP2, we investigated the drought tolerance of GhPLP2-overexpressed Arabidopsis plants. However, the GhPLP2-overexpressed Arabidopsis did not show the distinct phenotype of drought sensitivity compared with the WT plants (Supplementary Figure 10).
Discussion
Patatin-like proteins (PLPs) are a significant family of lipases with LAH activity and play an essential role in lipid metabolism during the immunity that arises from plant defenses (Rivas and Heitz, 2014). In this study, we identified the novel PLP gene GhPLP2 from cotton and demonstrated its role in fatty acid metabolism and the JA signaling pathway that confers resistance to V. dahliae in cotton.
PLPs have a conserved S-D catalytic dyad that is composed of serine of esterase motifs (GxSxG) and a central aspartic acid, which is essential for the LAH activity (Rydel et al., 2003). These residues were also identified in human Ca2+-independent phospholipase A2 (iPLA2s) and many microbial proteins (Burke and Dennis, 2009; Mansfeld, 2009; Heller et al., 2018). Similar to other patatin proteins in plants, GhPLP2 has the canonical esterase motif and the conserved S-D catalytic dyad for its LAH activity (Figure 1B). GhPLP2 is localized in the plasma membrane (Figure 3), which was consistent with the localization of several PLPs (Holk et al., 2002; Kim et al., 2014). The localization on the plasma membrane suggests that GhPLP2 may be involved in plant physiology and defense responses through the activation of lipid signaling by the hydrolysis of membrane lipids. The in vitro enzyme activity assay confirmed that GhPLP2 has LAH activity (Figures 2C,D) as it has been reported for other plant PLPs (Holk et al., 2002; Li and Wang, 2014). The GhPLP2-overexpressed Arabidopsis plants and GhPLP2-silenced cotton plants were utilized to investigate the in vivo function of GhPLP2 on plant fatty acid metabolism. The overexpression of GhPLP2 in Arabidopsis enhanced the levels of linoleic acid (LA, 18:2) and α-linolenic acid (ALA, 18:3) relative to the WT (Figure 8A) not only after inoculation with V. dahliae but also after the water treatment. In contrast, the knockdown of GhPLP2 in cotton plants reduced the contents of both types of FAs (Figure 8B). The functional LAH activity of GhPLP2 indicates its ability to release fatty acids from membrane lipids.
Many studies have suggested that PLPs with soluble LAH activity are necessary for the response of plants to various environmental stresses, such as drought, wounding, or pathogen infections (Matos and Pham-Thi, 2009; Rivas and Heitz, 2014). PLP proteins in tobacco, Arabidopsis, pepper, and grape show distinct resistance mechanisms in different host-pathogen interactions (Cacas et al., 2005; Yang et al., 2007; La Camera et al., 2009; Kim et al., 2014; Laureano et al., 2018). The strong upregulation of the GhPLP2 gene upon infection by V. dahliae (Figure 4B) suggests its crucial role in the interaction of cotton plants with V. dahliae. The silencing of GhPLP2 in cotton plants enhanced their susceptibility to infection by V. dahliae (Figure 5), and the overexpression of GhPLP2 in Arabidopsis plants visibly enhanced their resistance to disease through the appearance of merely mild symptoms of disease (Figure 7), demonstrating that GhPLP2 played a positive role in the plant defense to infection with V. dahliae.
Previous studies have shown that the JA biosynthetic and signaling pathway are important for plant resistance against V. dahliae (Li et al., 2014; He et al., 2016; Dhar et al., 2020). GhPLP2 was highly induced after JA treatment (Figure 4C), suggesting that GhPLP2 is likely to involve JA-dependent defense pathway. Several studies have shown that PLPs mediate plant immune responses through the regulation of FA metabolism and the JA signaling pathway. In tobacco, PLP was rapidly induced before the accumulation of JA in response to infection by the Tobacco mosaic virus (TMV) (Dhondt et al., 2002). AtPLAI targets the chloroplasts and plays a critical role in maintaining the homeostatic pool of free FA and basal JA, thus, increasing resistance to B. cinerea (Yang et al., 2007). The branch of the JA family (OPDA, JA, and dinor-OPDA) are fed by the primary cytoplasmic localization protein AtPLP2 (La Camera et al., 2009). The overexpression of the PLP gene OSAG78 in Arabidopsis increased the amounts of linoleic acid and linolenic acid and induced the expression levels of the JA-related defense genes PDF 1.2 and PR4 (Lin et al., 2011). In this study, the content of JA and the levels of expression of the JA biosynthetic-related genes (LOX2, AOS, OPR3) were increased in GhPLP2-overexpressed plants and decreased when GhPLP2 was knocked down (Figures 8C–F). As expected, the JA-responsive marker gene PDF1.2 exhibited a similar trend, which is upregulated upon infection with V. dahliae and positively related to the content of JA and the level of expression of GhPLP2 in plants. Therefore, GhPLP2 may be involved in the biosynthesis of JA in Arabidopsis and cotton, which contributed to the plant defenses against V. dahliae.
JA is known to take part in various physiological processes including salinity, drought, and temperature (low/high) stress (Ghorbel et al., 2021). The application of exogenous JA not only increased the total carbohydrate, soluble sugar, free amino acid, total proline, and protein contents, but also the activities of catalase (CAT), POD, and SOD in maize plants (Zea mays) (Ali and Baek, 2020). In pearl millet, the exogenous application JA weakens drought stress effects on pearl millet plantlets by increasing the fresh weight of seedlings, root length and shoot length (Awan et al., 2021). In this study, the phenotype of drought sensitivity has no significant difference between GhPLP2-overexpressed and WT Arabidopsis plants (Supplementary Figure 10). It seems that GhPLP2 altered JA level is not related to the plant drought response. Whether it plays a role in other abiotic stresses sharing a similar pathway with PEG 6000 treatment needs to be further determined.
Moreover, the alteration in JA-mediated pathways can also affect the vegetative and reproductive growth of plants (Qi et al., 2016; Ghorbel et al., 2021). The Arabidopsis mutant defective in anther dehiscence1 (dad1) displays the reduced accumulation of JA and defects in anther dehiscence, pollen maturation, and flower opening. The exogenous application of JA or linolenic acid could rescue the defects (Ishiguro et al., 2001). In rice (Oryza sativa), a plastid-targeted lipase EXTRA GLUME 1 (EG1) participates in the biosynthesis of JA during the development of rice spikelets. The eg1 mutant exhibits altered spikelet morphology with changes in floral organ identity and number, as well as defective floral meristem determinacy, resulting in a reduction in reproductive success and the yield of grains (Cai et al., 2014). In this study, GhPLP2-silenced cotton plants and GhPLP2-overexpressed Arabidopsis plants have altered JA content under mock treatment but no changes in the growing phenotype (Supplementary Figures 11A,B). This suggests that an alteration in the JA pathway through the modification of GhPLP2 does not affect the normal growth of plants. Additionally, the overexpression of GhPLP2 in Arabidopsis does not affect the weight of the grains (Supplementary Figure 11C). Whether the change in the JA-mediated pathway related to GhPLP2 affects fertility and reproductive output in cotton plants is yet to be determined.
Fatty acids are not only major membrane components of the cell, but they also directly or indirectly participate in a series of immune responses in plants (Walley et al., 2013; Lim et al., 2017; He and Ding, 2020). In fact, C16:0 and 18:0 FAs serve as precursors for the synthesis of cutin and wax polymers, which provide a physical barrier to defend against insects and pathogens (Raffaele et al., 2009; Yu et al., 2013). Moreover, C18:0 fatty acid can be converted to unsaturated fatty acids including oleic acid (18:1), linoleic acid (18:2), and α-linolenic acid (18:3) after a series of desaturation reactions (Kachroo and Kachroo, 2009). Previous studies confirmed that linoleic acid (18:2) and α-linolenic acid (18:3) are precursors for JA biosynthesis (Hung and Kao, 1998; Li et al., 2003; Banilas et al., 2007). The Arabidopsis defective in anther dehiscence1 (DAD1) protein is a chloroplastic phospholipase A1 that catalyzes the initial step of JA biosynthesis by the release of JA precursor α-linolenic acid (Ishiguro et al., 2001). In cotton plants, cytochrome P450 CYP82D (SSN) competes for C18 fatty acids substrates with LOXs. The inhibition of SSN leads to free fatty acids that enter the metabolism of LOXs to synthesize JA and enhance the levels of JA and disease resistance to V. dahliae (Sun et al., 2014). The long non-coding RNA (lncRNA) GhlncLOX3 positively regulates the resistance to V. dahliae through the modulation of the expression of GhLOX3 implicated in α-linolenic acid (18:3) metabolism associated with JA biosynthesis (Wang et al., 2021). In this study, the contents of palmitic acid (16:0) and stearic acid (18:0) significantly increased after inoculation with V. dahliae in cotton and increased slightly in Arabidopsis plants. However, the difference in the contents between GhPLP2-silenced and control cotton plants was not significant. It is not clear whether GhPLP2 enhances the resistance of cotton against V. dahliae via the regulation of accumulation of palmitic acid (16:0) and stearic acid (18:0). However, the contents of linoleic acid (18:2) and α-linolenic acid (18:3) in both Arabidopsis and cotton plants were not only closely related to the expression of GhPLP2 under mock treatment but were also increased following infection with V. dahliae (Figures 8A,B). These findings suggest that GhPLP2 mediates the production of linoleic acid (18:2) and α-linolenic acid (18:3), which are involved in plant defense against V. dahliae. Free C18:2 and C18:3 was reduced to stable hydroxy FA in the cytosol or plastids and then converted to oxylipins which including JAs (Feussner and Wasternack, 2002; La Camera et al., 2009). Therefore, we hypothesize that the production of linoleic acid and α-linolenic acid by GhPLP2 may promote the biosynthesis and signaling of JA.
HR is one of the defense mechanisms that confers the plant with the ability to inhibit pathogen infection and further induce a rapid defense response (Kombrink and Schmelzer, 2001; Zhang Y. et al., 2017). Interestingly, several oxylipins that are derived from fatty acids in the 9- and 13-LOX pathways were found to be sufficient to initiate HR in different pathosystems (García-Marcos et al., 2013; Sun et al., 2014). Increasing amounts of evidence showed that the PLPs play an essential role in the execution of cell death in HR in plants. In Arabidopsis, AtPLP2 contributes to the resistance response to Cucumber mosaic virus (CMV) upon HR-dependent process (La Camera et al., 2009). CaPLP1 regulates the production of phospholipid-derived molecules, which leads to the HR-like cell death during the incompatible interaction of pepper plants with avirulent Xcv (Kim et al., 2014). The elicitor protein PevD1 from V. dahliae can induce typical HR-like necrosis in tobacco and trigger innate immunity in cotton plants. GhPLP2 silencing has reduced the degree of HR and expression of the HR marker gene GhHIN1 and GhHSR203 during the induction of V. dahliae elicitor PevD1 in cotton leaves (Figure 6). This suggests that GhPLP2 is required for resistance (R) gene-mediated disease resistance in cotton plants. However, further research is merited to reveal the relationship between GhPLP2 and PevD1-triggered HR during the interaction between cotton and V. dahliae.
Taken together, we propose a potential model of the involvement of GhPLP2 in the resistance of plants against V. dahliae (Figure 9). GhPLP2 contributes to the build-up of free fatty acids pools, including linoleic acid and α-linolenic acid. The accumulation of linoleic acid and α-linolenic acid may directly or indirectly lead to the activation of the JA-synthesis and -signaling pathway in plants; GhPLP2 is also involved in PevD1-triggered HR. Both in concert enhance the resistance of cotton plants to disease caused by infection with V. dahliae.
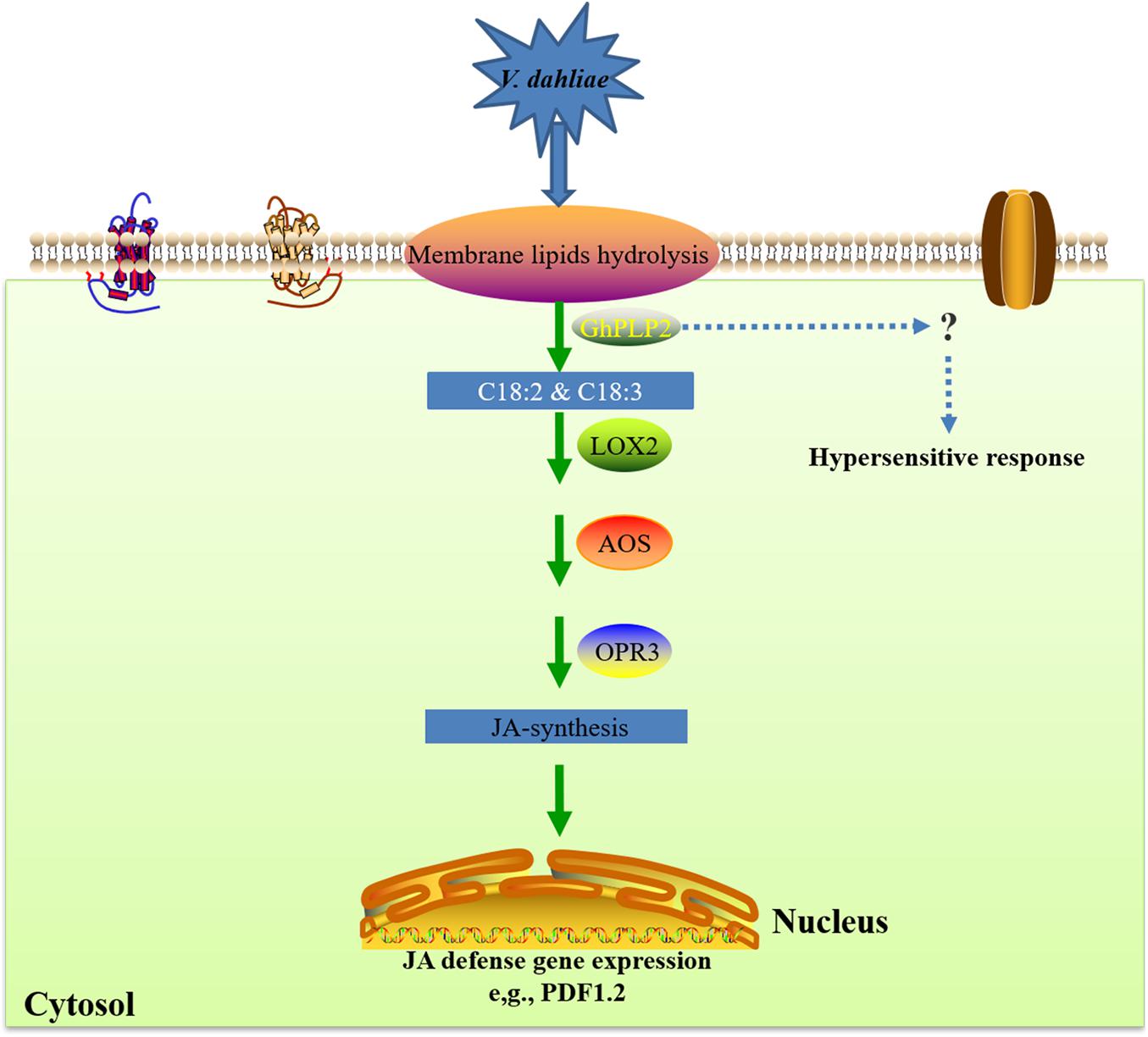
Figure 9. A proposed model for the involvement of GhPLP2 in plant resistance against Verticillium dahliae. LOX, lipoxygenase; AOS, allene oxide synthase; OPR3, cis-(+)-12-oxo-phytodienoic acid reductase 3.
Conclusion
Our study provides evidence that the patatin-like protein GhPLP2 with LAH activity plays a positive role in the enhancement of plant disease resistance. The silencing of GhPLP2 in cotton plants attenuated resistance against V. dahliae, and the overexpression of GhPLP2 in Arabidopsis enhanced resistance against infection with V. dahliae. The resistance mechanism of GhPLP2 may be owing to its release of the synthetic precursors to jasmonic acid, including linoleic acid and α-linolenic acid, via its LAH activity, followed by the accumulation of JA and activation of the JA signaling pathway. In addition, GhPLP2 is involved in triggering a hypersensitive response for disease resistance by a V. dahliae elicitor. These results stressed some insights for the mechanism of GhPLP2 in the response of plant defenses and highlighted the potential application of GhPLP2 in genetic engineering to increase the disease resistance of cotton germplasm.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YH, FL, and YZ conceived and designed the study. YZ conducted most of the experiments and wrote the manuscript. XH, LG, and ZG provided technical assistance to YZ. YP and PW provided analysis tools. XG contributed reagents and materials. All authors reviewed and approved the final manuscript.
Funding
This work was sponsored by the National Key R&D Program of China “Chemical Fertilizers and Pesticide Reduction Efficiency Integrated Technology Research and Development” (Grant No. 2017YFD0201900), the “Seven Crop Breeding” National Major Project (Grant No. 2016YFD0101006), the State Key Laboratory of Cotton Biology (Grant No. CB2021B03), and the Chinese Universities Scientific Fund (Grant No. 2021TC108).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Yule Liu from Tsinghua University for the VIGS constructs. We also thank Dong Li (College of Science, China Agricultural University) and Chunran Zhou (College of Science, China Agricultural University) for hormone quantitation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.749630/full#supplementary-material
Footnotes
- ^ https://cottonfgd.org/
- ^ http://web.expasy.org/protparam/
- ^ http://www.ncbi.nlm.nih.gov
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://embnet.vital-it.ch/software/BOX_form.html
- ^ https://meme-suite.org/meme/tools/meme
References
Ali, M. S., and Baek, K. H. (2020). Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 21, 621–640. doi: 10.3390/ijms21020621
Awan, S. A., Khan, I., Rizwan, M., Zhang, X., Brestic, M., Khan, A., et al. (2021). Exogenous abscisic acid and jasmonic acid restrain polyethylene glycol-induced drought by improving the growth and antioxidative enzyme activities in pearl millet. Physiol. Plant 172, 809–819. doi: 10.1111/ppl.13247
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Banilas, G., Nikiforiadis, A., Makariti, I., Moressis, A., and Hatzopoulos, P. (2007). Discrete roles of a microsomal linoleate desaturase gene in olive identified by spatiotemporal transcriptional analysis. Tree Physiol. 27, 481–490. doi: 10.1093/treephys/27.4.481
Bordoli, L., Kiefer, F., Arnold, K., Benkert, P., Battey, J., and Schwede, T. (2009). Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13. doi: 10.1038/nprot.2008.197
Bu, B., Qiu, D., Zeng, H., Guo, L., Yuan, J., and Yang, X. (2014). A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 33, 461–470. doi: 10.1007/s00299-013-1546-7
Burke, J. E., and Dennis, E. A. (2009). Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50, S237–S242. doi: 10.1194/jlr.R800033-JLR200
Cacas, J. L., Marmey, P., Montillet, J. L., Sayegh-Alhamdia, M., Jalloul, A., Rojas-Mendoza, A., et al. (2009). A novel patatin-like protein from cotton plant, GhPat1, is co-expressed with GhLox1 during Xanthomonas campestris-mediated hypersensitive cell death. Plant Cell Rep. 28, 155–164. doi: 10.1007/s00299-008-0622-x
Cacas, J.-L., Vailleau, F., Davoine, C., Ennar, N., Agnel, J.-P., Tronchet, M., et al. (2005). The combined action of 9 lipoxygenase and galactolipase is sufficient to bring about programmed cell death during tobacco hypersensitive response. Plant Cell Environ. 28, 1367–1378. doi: 10.1111/j.1365-3040.2005.01369.x
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476–3489. doi: 10.1038/ncomms4476
Campos, M. L., Kang, J. H., and Howe, G. A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675. doi: 10.1007/s10886-014-0468-3
Champion, A., Hebrard, E., Parra, B., Bournaud, C., Marmey, P., Tranchant, C., et al. (2009). Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and Xanthomonas. Mol. Plant Pathol. 10, 471–485. doi: 10.1111/j.1364-3703.2009.00549.x
Chen, J. Y., Xiao, H. L., Gui, Y. J., Zhang, D. D., Li, L., Bao, Y. M., et al. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 7:1709. doi: 10.3389/fmicb.2016.01709
Choi, D. S., and Hwang, B. K. (2011). Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–842. doi: 10.1105/tpc.110.082081
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Cook, R., Lupette, J., and Benning, C. (2021). The role of chloroplast membrane lipid metabolism in plant environmental responses. Cells 10, 706–726. doi: 10.3390/cells10030706
Dhar, N., Chen, J. Y., Subbarao, K. V., and Klosterman, S. J. (2020). Hormone signaling and its interplay with development and defense responses in Verticillium-plant interactions. Front. Plant Sci. 11:584997. doi: 10.3389/fpls.2020.584997
Dhondt, S., Geoffroy, P., Stelmach, B. A., Legrand, M., and Heitz, T. (2000). Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J. 23, 431–440. doi: 10.1046/j.1365-313x.2000.00802.x
Dhondt, S., Gouzerh, G., Müller, A., Legrand, M., and Heitz, T. (2002). Spatio-temporal expression of patatin-like lipid acyl hydrolases and accumulation of jasmonates in elicitor-treated tobacco leaves are not affected by endogenous levels of salicylic acid. Plant J. 32, 749–762. doi: 10.1046/j.1365-313x.2002.01465.x
Dong, Y., Burch-Smith, T. M., Liu, Y., Mamillapalli, P., and Dinesh-Kumar, S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and –2 in floral development. Plant Physiol. 145, 1161–1170. doi: 10.1104/pp.107.107391
Dong, Y., Li, M., Zhang, P., Wang, X., Fan, C., and Zhou, Y. (2014). Patatin-related phospholipase pPLAIIIδ influences auxin-responsive cell morphology and organ size in Arabidopsis and Brassica napus. BMC Plant Biol. 14:332. doi: 10.1186/s12870-014-0332-1
Ellendorff, U., Fradin, E. F., de Jonge, R., and Thomma, B. P. (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. doi: 10.1093/jxb/ern306
Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi: 10.1146/annurev.arplant.53.100301.135248
Fradin, E. F., Abd-El-Haliem, A., Masini, L., van den Berg, G. C., Joosten, M. H., and Thomma, B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Fradin, E. F., and Thomma, B. P. H. J. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. doi: 10.1111/j.1364-3703.2006.00323.x
Fradin, E. F., Zhang, Z., Juarez Ayala, J. C., Castroverde, C. D., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Fu, W., Shen, Y., Hao, J., Wu, J., Ke, L., Wu, C., et al. (2015). Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 5:11790. doi: 10.1038/srep11790
Gao, W., Long, L., Xu, L., Lindsey, K., Zhang, X., and Zhu, L. (2016). Suppression of the homeobox gene HDTF1 enhances resistance to Verticillium dahliae and Botrytis cinerea in cotton. J. Integr. Plant Biol. 58, 503–513. doi: 10.1111/jipb.12432
Gao, X., Li, F., Li, M., Kianinejad, A. S., Dever, J. K., Wheeler, T. A., et al. (2013). Cotton GhBAK1 mediates Verticillium wilt resistance and cell death. J. Integr. Plant Biol. 55, 586–596. doi: 10.1111/jipb.12064
Gao, X., Wheeler, T., Li, Z., Kenerley, C. M., He, P., and Shan, L. (2011). Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. doi: 10.1111/j.1365-313X.2011.04491.x
García-Marcos, A., Pacheco, R., Manzano, A., Aguilar, E., and Tenllado, F. (2013). Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair potato virus X-potato virus Y and tomato spotted wilt virus. J. Virol. 87, 5769–5783. doi: 10.1128/jvi.03573-12
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., and de Ilárduya, O. M. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232. doi: 10.1186/1471-2229-10-232
Ghorbel, M., Brini, F., Sharma, A., and Landi, M. (2021). Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 40, 1471–1494. doi: 10.1007/s00299-021-02687-4
Gong, Q., Yang, Z., Wang, X., Butt, H. I., Chen, E., He, S., et al. (2017). Salicylic acid-related cotton (Gossypium arboreum) ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae. BMC Plant Biol. 17:59. doi: 10.1186/s12870-017-1007-5
Hamada, H., Takeuchi, S., Kiba, A., Tsuda, S., Suzuki, K., Hikichi, Y., et al. (2005). Timing and extent of hypersensitive response are critical to restrict local and systemic spread of pepper mild mottle virus in pepper containing the L3 gene. J. Gen. Plant Pathol. 71, 90–94. doi: 10.1007/s10327-004-0164-1
He, M., and Ding, N. Z. (2020). Plant unsaturated fatty acids: multiple roles in stress response. Front. Plant Sci. 11:562785. doi: 10.3389/fpls.2020.562785
He, X., Zhu, L., Wassan, G. M., Wang, Y., Miao, Y., Shaban, M., et al. (2018). GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 19, 896–908. doi: 10.1111/mpp.12575
He, X., Zhu, L., Xu, L., Guo, W., and Zhang, X. (2016). GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 35, 2167–2179. doi: 10.1007/s00299-016-2027-6
Heller, J., Clavé, C., Gladieux, P., Saupe, S. J., and Glass, N. L. (2018). NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. U. S. A. 115, E2292–E2301. doi: 10.1073/pnas.1719705115
Hendriks, T., Vreugdenhil, D., and Stiekema, W. J. (1991). Patatin and four serine proteinase inhibitor genes are differentially expressed during potato tuber development. Plant Mol. Biol. 17, 385–394. doi: 10.1007/bf00040633
Holk, A., Rietz, S., Zahn, M., Quader, H., and Scherer, G. F. (2002). Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130, 90–101. doi: 10.1104/pp.006288
Hong, J. K., Choi, H. W., Hwang, I. S., Kim, D. S., Kim, N. H., Choi, D. S., et al. (2008). Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227, 539–558. doi: 10.1007/s00425-007-0637-5
Hu, Q., Zhu, L., Zhang, X., Guan, Q., Xiao, S., Min, L., et al. (2018). GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 178, 876–889. doi: 10.1104/pp.18.00737
Hung, K. T., and Kao, C. H. (1998). Involvement of lipid peroxidation in methyl jasmonate-promoted senescence in detached rice leaves. Plant Growth Regul. 24, 17–21. doi: 10.1023/A:1005988727235
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. doi: 10.1105/tpc.010192
Jang, J. H., Nguyen, N. Q., Légeret, B., Beisson, F., Kim, Y.-J., Sim, H.-J., et al. (2020). Phospholipase pPLAIIIα increases germination rate and resistance to turnip crinkle virus when overexpressed. Plant Physiol. 184, 1482–1498. doi: 10.1104/pp.20.00630
Jyothishwaran, G., Kotresha, D., Selvaraj, T., Srideshikan, S. M., Rajvanshi, P. K., Jayabaskaran, C., et al. (2007). A modified freeze-thaw method for efficient transformation of Agrobacterium tumefaciens. Curr. Sci. 93, 770–772. doi: 10.3891/cs/v93/i6/770-772
Kachroo, A., and Kachroo, P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Kim, D. S., Jeun, Y., and Hwang, B. K. (2014). The pepper patatin-like phospholipase CaPLP1 functions in plant cell death and defense signaling. Plant Mol. Biol. 84, 329–344. doi: 10.1007/s11103-013-0137-x
Kombrink, E., and Schmelzer, E. (2001). The hypersensitive response and its role in local and systemic disease resistance. Eur. J. Plant Pathol. 107, 69–78. doi: 10.1023/A:1008736629717
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
La Camera, S., Balagué, C., Göbel, C., Geoffroy, P., Legrand, M., Feussner, I., et al. (2009). The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol. Plant Microbe Interact. 22, 469–481. doi: 10.1094/mpmi-22-4-0469
La Camera, S., Geoffroy, P., Samaha, H., Ndiaye, A., Rahim, G., Legrand, M., et al. (2005). A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J. 44, 810–825. doi: 10.1111/j.1365-313X.2005.02578.x
Laureano, G., Figueiredo, J., Cavaco, A. R., Duarte, B., Caçador, I., Malhó, R., et al. (2018). The interplay between membrane lipids and phospholipase A family members in grapevine resistance against Plasmopara viticola. Sci. Rep. 8:14538. doi: 10.1038/s41598-018-32559-z
Li, C., He, X., Luo, X., Xu, L., Liu, L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, C., Liu, G., Xu, C., Lee, G. I., Bauer, P., Ling, H. Q., et al. (2003). The tomato suppressor of prosystemin-mediated responses 2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15, 1646–1661. doi: 10.1105/tpc.012237
Li, D., An, Q., Wu, Y., Li, J.-Q., and Pan, C. (2020). Foliar application of selenium nanoparticles on celery stimulates several nutrient component levels by regulating the α-Linolenic acid pathway. ACS Sustain. Chem. Eng. 8, 10502–10510. doi: 10.1021/acssuschemeng.0c02819
Li, J., Li, M., Yao, S., Cai, G., and Wang, X. (2020). Patatin-related phospholipase pPLAIIIγ involved in osmotic and salt tolerance in Arabidopsis. Plants 9, 650–664. doi: 10.3390/plants9050650
Li, M., Bahn, S. C., Guo, L., Musgrave, W., Berg, H., Welti, R., et al. (2011). Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23, 1107–1123. doi: 10.1105/tpc.110.081240
Li, M., and Wang, X. (2014). “pPLA: Patatin-related phospholipase as with multiple biological functions,” in Phospholipases in Plant Signaling, ed. X. Wang (Berlin: Springer), 93–108. doi: 10.1007/978-3-642-42011-5_5
Li, N., Xu, C., Li-Beisson, Y., and Philippar, K. (2016). Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. doi: 10.1016/j.tplants.2015.10.011
Li, X., Sun, Y., Liu, N., Wang, P., Pei, Y., Liu, D., et al. (2019). Enhanced resistance to Verticillium dahliae mediated by an F-box protein GhACIF1 from Gossypium hirsutum. Plant Sci. 284, 127–134. doi: 10.1016/j.plantsci.2019.04.013
Liguori, G., Lucariello, A., Colella, G., De Luca, A., and Marinelli, P. (2007). Rapid identification of candida species in oral rinse solutions by PCR. J. Clin. Pathol. 60, 1035–1039. doi: 10.1136/jcp.2006.040782
Lim, G. H., Singhal, R., Kachroo, A., and Kachroo, P. (2017). Fatty acid- and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 55, 505–536. doi: 10.1146/annurev-phyto-080516-035406
Lin, C. C., Chu, C. F., Liu, P. H., Lin, H. H., Liang, S. C., Hsu, W. E., et al. (2011). Expression of an oncidium gene encoding a patatin-like protein delays flowering in Arabidopsis by reducing gibberellin synthesis. Plant Cell Physiol. 52, 421–435. doi: 10.1093/pcp/pcq206
Liu, F., Ma, L., Wang, Y., Li, Y., Zhang, X., Xue, F., et al. (2019). GhFAD2-3 is required for anther development in Gossypium hirsutum. BMC Plant Biol. 19:393. doi: 10.1186/s12870-019-2010-9
Liu, M., Khan, N. U., Wang, N., Yang, X., and Qiu, D. (2016). The protein elicitor PevD1 enhances resistance to pathogens and promotes growth in Arabidopsis. Int. J. Biol. Sci. 12, 931–943. doi: 10.7150/ijbs.15447
Liu, N., Sun, Y., Wang, P., Duan, H., Ge, X., Li, X., et al. (2018). Mutation of key amino acids in the polygalacturonase-inhibiting proteins CkPGIP1 and GhPGIP1 improves resistance to Verticillium wilt in cotton. Plant J. 96, 546–561. doi: 10.1111/tpj.14048
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Ma, Q., Wang, N., Ma, L., Lu, J., Wang, H., Wang, C., et al. (2020). The cotton BEL1-like transcription factor GhBLH7-D06 negatively regulates the defense response against Verticillium dahliae. Int. J. Mol. Sci. 21, 7126–7143. doi: 10.3390/ijms21197126
Mandal, M. K., Chandra-Shekara, A. C., Jeong, R. D., Yu, K., Zhu, S., Chanda, B., et al. (2012). Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24, 1654–1674. doi: 10.1105/tpc.112.096768
Mansfeld, J. (2009). Plant phospholipases A2: perspectives on biotechnological applications. Biotechnol. Lett. 31, 1373–1380. doi: 10.1007/s10529-009-0034-1
Matos, A. R., and Pham-Thi, A. T. (2009). Lipid deacylating enzymes in plants: old activities, new genes. Plant Physiol. Biochem. 47, 491–503. doi: 10.1016/j.plaphy.2009.02.011
Mo, H. J., Sun, Y. X., Zhu, X. L., Wang, X. F., Zhang, Y., Yang, J., et al. (2016). Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 243, 1023–1039. doi: 10.1007/s00425-015-2463-5
Pantelides, I. S., Tjamos, S. E., and Paplomatas, E. J. (2010). Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol. Plant Pathol. 11, 191–202. doi: 10.1111/j.1364-3703.2009.00592.x
Pei, Y., Li, X., Zhu, Y., Ge, X., Sun, Y., Liu, N., et al. (2019). GhABP19, a novel Germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium Wilt Pathogens. Front. Plant Sci. 10:583. doi: 10.3389/fpls.2019.00583
Qi, J., Li, J., Han, X., Li, R., Wu, J., Yu, H., et al. (2016). Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 58, 564–576. doi: 10.1111/jipb.12436
Raffaele, S., Leger, A., and Roby, D. (2009). Very long chain fatty acid and lipid signaling in the response of plants to pathogens. Plant Signal. Behav. 4, 94–99. doi: 10.4161/psb.4.2.7580
Reynolds, C. R., Islam, S. A., and Sternberg, M. J. E. (2018). EzMol: a web server wizard for the rapid visualization and image production of protein and nucleic acid structures. J. Mol. Biol. 430, 2244–2248. doi: 10.1016/j.jmb.2018.01.013
Rietz, S., Dermendjiev, G., Oppermann, E., Tafesse, F. G., Effendi, Y., Holk, A., et al. (2010). Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Mol. Plant 3, 524–538. doi: 10.1093/mp/ssp109
Rivas, S., and Heitz, T. (2014). “Phospholipase A in plant immunity,” in Phospholipases In Plant Signaling, ed. X. Wang (Berlin: Springer), 183–205. doi: 10.1007/978-3-642-42011-5_10
Rossi, F. R., Gárriz, A., Marina, M., Romero, F. M., Gonzalez, M. E., Collado, I. G., et al. (2011). The sesquiterpene botrydial produced by Botrytis cinerea induces the hypersensitive response on plant tissues and its action is modulated by salicylic acid and jasmonic acid signaling. Mol. Plant Microbe Interact. 24, 888–896. doi: 10.1094/mpmi-10-10-0248
Rydel, T. J., Williams, J. M., Krieger, E., Moshiri, F., Stallings, W. C., Brown, S. M., et al. (2003). The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42, 6696–6708. doi: 10.1021/bi027156r
Ryu, S. B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9, 229–235. doi: 10.1016/j.tplants.2004.03.004
Savchenko, T. V., Zastrijnaja, O. M., and Klimov, V. V. (2014). Oxylipins and plant abiotic stress resistance. Biochemistry 79, 362–375. doi: 10.1134/s0006297914040051
Scherer, G. F., Ryu, S. B., Wang, X., Matos, A. R., and Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700. doi: 10.1016/j.tplants.2010.09.005
Shaban, M., Miao, Y., Ullah, A., Khan, A. Q., Menghwar, H., Khan, A. H., et al. (2018). Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 125, 193–204. doi: 10.1016/j.plaphy.2018.02.011
Shah, J. (2005). Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 43, 229–260. doi: 10.1146/annurev.phyto.43.040204.135951
Short, D. P., Sandoya, G., Vallad, G. E., Koike, S. T., Xiao, C. L., Wu, B. M., et al. (2015). Dynamics of Verticillium species microsclerotia in field soils in response to fumigation, cropping patterns, and flooding. Phytopathology 105, 638–645. doi: 10.1094/phyto-09-14-0259-r
Simon, C., Langlois-Meurinne, M., Bellvert, F., Garmier, M., Didierlaurent, L., Massoud, K., et al. (2010). The differential spatial distribution of secondary metabolites in Arabidopsis leaves reacting hypersensitively to Pseudomonas syringae pv. tomato is dependent on the oxidative burst. J. Exp. Bot. 61, 3355–3370. doi: 10.1093/jxb/erq157
Song, Y., Zhai, Y., Li, L., Yang, Z., Ge, X., Yang, Z., et al. (2021). BIN2 negatively regulates plant defense against Verticillium dahliae in Arabidopsis and cotton. Plant Biotechnol. J. 19, 2097–2112. doi: 10.1111/pbi.13640
Sun, L., Zhu, L., Xu, L., Yuan, D., Min, L., and Zhang, X. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372–5384. doi: 10.1038/ncomms6372
Vailleau, F., Daniel, X., Tronchet, M., Montillet, J. L., Triantaphylidès, C., and Roby, D. (2002). A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. U. S. A. 99, 10179–10184. doi: 10.1073/pnas.152047199
Walley, J. W., Kliebenstein, D. J., Bostock, R. M., and Dehesh, K. (2013). Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 16, 520–526. doi: 10.1016/j.pbi.2013.06.011
Wang, B., Yang, X., Zeng, H., Liu, H., Zhou, T., Tan, B., et al. (2012). The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl. Microbiol. Biotechnol. 93, 191–201. doi: 10.1007/s00253-011-3405-1
Wang, C., El-Shetehy, M., Shine, M. B., Yu, K., Navarre, D., Wendehenne, D., et al. (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. doi: 10.1016/j.celrep.2014.03.032
Wang, G., Wang, X., Zhang, Y., Yang, J., Li, Z., Wu, L., et al. (2021). Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum. BMC Plant Biol. 21:68. doi: 10.1186/s12870-021-02835-8
Wang, P., Sun, Y., Pei, Y., Li, X., Zhang, X., Li, F., et al. (2018). GhSNAP33, a t-SNARE protein from Gossypium hirsutum, mediates resistance to Verticillium dahliae infection and tolerance to drought stress. Front. Plant Sci. 9:896. doi: 10.3389/fpls.2018.00896
Wang, P., Zhou, L., Jamieson, P., Zhang, L., Zhao, Z., Babilonia, K., et al. (2020). The Cotton Wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell 32, 3978–4001. doi: 10.1105/tpc.19.00950
Wang, Q., Zhang, X., Li, F., Hou, Y., Liu, X., and Zhang, X. (2011). Identification of a UDP-glucose pyrophosphorylase from cotton (Gossypium hirsutum L.) involved in cellulose biosynthesis in Arabidopsis thaliana. Plant Cell Rep. 30, 1303–1312. doi: 10.1007/s00299-011-1042-x
Wang, X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7, 329–336. doi: 10.1016/j.pbi.2004.03.012
Wasternack, C., and Strnad, M. (2016). Jasmonate signaling in plant stress responses and development-active and inactive compounds. N. Biotechnol. 33, 604–613. doi: 10.1016/j.nbt.2015.11.001
Wasternack, C., and Strnad, M. (2017). Jasmonates are signals in the biosynthesis of secondary metabolites-Pathways, transcription factors and applied aspects. N. Biotechnol. 48, 1–11. doi: 10.1016/j.nbt.2017.09.007
Yang, W., Devaiah, S. P., Pan, X., Isaac, G., Welti, R., and Wang, X. (2007). AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J. Biol. Chem. 282, 18116–18128. doi: 10.1074/jbc.M700405200
Yang, W. Y., Zheng, Y., Bahn, S. C., Pan, X. Q., Li, M. Y., Vu, H. S., et al. (2012). The patatin-containing phospholipase A pPLAIIα modulates oxylipin formation and water loss in Arabidopsis thaliana. Mol. Plant 5, 452–460. doi: 10.1093/mp/ssr118
Yu, K., Soares, J. M., Mandal, M. K., Wang, C., Chanda, B., Gifford, A. N., et al. (2013). A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278. doi: 10.1016/j.celrep.2013.03.030
Zhang, L., Ni, H., Du, X., Wang, S., Ma, X. W., Nürnberger, T., et al. (2017). The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 215, 368–381. doi: 10.1111/nph.14537
Keywords: Gossypium hirsutum, patatin-like protein, Verticillium dahliae, fatty acid metabolism, jasmonic acid, disease resistance
Citation: Zhu Y, Hu X, Wang P, Gao L, Pei Y, Ge Z, Ge X, Li F and Hou Y (2021) GhPLP2 Positively Regulates Cotton Resistance to Verticillium Wilt by Modulating Fatty Acid Accumulation and Jasmonic Acid Signaling Pathway. Front. Plant Sci. 12:749630. doi: 10.3389/fpls.2021.749630
Received: 29 July 2021; Accepted: 08 October 2021;
Published: 02 November 2021.
Edited by:
Sotiris Tjamos, Agricultural University of Athens, GreeceReviewed by:
Nikhilesh Dhar, University of California, Davis, United StatesKrzysztof Zienkiewicz, Nicolaus Copernicus University in Toruń, Poland
Copyright © 2021 Zhu, Hu, Wang, Gao, Pei, Ge, Ge, Li and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuguang Li, YXlsaWZ1Z0AxMjYuY29t; Yuxia Hou, eXV4aWFjYXVAMTYzLmNvbQ==
 Yutao Zhu
Yutao Zhu Xiaoqian Hu1
Xiaoqian Hu1 Xiaoyang Ge
Xiaoyang Ge Fuguang Li
Fuguang Li Yuxia Hou
Yuxia Hou