
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 December 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.749394
This article is part of the Research TopicRecent Advances in Hazelnut (Corylus spp.)View all 26 articles
 Nadia Valentini†
Nadia Valentini† Ezio Portis†
Ezio Portis† Roberto Botta*
Roberto Botta* Alberto Acquadro
Alberto Acquadro Vera Pavese
Vera Pavese Emile Cavalet Giorsa
Emile Cavalet Giorsa Daniela Torello Marinoni
Daniela Torello Marinoni
An increasing interest in the cultivation of (European) hazelnut (Corylus avellana) is driving a demand to breed cultivars adapted to non-conventional environments, particularly in the context of incipient climate change. Given that plant phenology is so strongly determined by genotype, a rational approach to support these breeding efforts will be to identify quantitative trait loci (QTLs) and the genes underlying the basis for adaptation. The present study was designed to map QTLs for phenology-related traits, such as the timing of both male and female flowering, dichogamy, and the period required for nuts to reach maturity. The analysis took advantage of an existing linkage map developed from a population of F1 progeny bred from the cross “Tonda Gentile delle Langhe” × “Merveille de Bollwiller,” consisting in 11 LG. A total of 42 QTL-harboring regions were identified. Overall, 71 QTLs were detected, 49 on the TGdL map and 22 on the MB map; among these, 21 were classified as major; 13 were detected in at least two of the seasons (stable-major QTL). In detail, 20 QTLs were identified as contributing to the time of male flowering, 15 to time of female flowering, 25 to dichogamy, and 11 to time of nut maturity. LG02 was found to harbor 16 QTLs, while 15 QTLs mapped to LG10 and 14 to LG03. Many of the QTLs were clustered with one another. The major cluster was located on TGdL_02 and consisted of mainly major QTLs governing all the analyzed traits. A search of the key genomic regions revealed 22 candidate genes underlying the set of traits being investigated. Many of them have been described in the literature as involved in processes related to flowering, control of dormancy, budburst, the switch from vegetative to reproductive growth, or the morphogenesis of flowers and seeds.
The European hazelnut (Corylus avellana L., 2n = 2x = 22) is a high-value cash crop, with the two largest producers being Turkey and Italy, and growing industry in several other countries worldwide (Food and Agriculture Organization Corporate Statistical Database [FAOSTAT], 2021). The increased interest in this crop is fueling a demand for cultivars adapted to non-conventional environments, particularly in the context of incipient climate change. The phenological traits are considered key to adaptation (Ghelardini et al., 2014) and include the time of male and female flowering, budburst, and nut maturity.
The flowering behavior of hazelnut is somewhat unusual, as it occurs during the winter. In the northern hemisphere, pollen is shed and the pistils are receptive between mid-December and mid-March, dependent on genotype, year, and location (Mehlenbacher, 1991). Moreover, hazelnut plants are monoecious, wind-pollinated, and most cultivars are dichogamous, that is the male and female reproductive organs mature at different times. Because the species is self-incompatible and even cultivars are cross-incompatible in some parental combinations (Mehlenbacher and Thompson, 1988), it is necessary to include a pollinizer genotype in orchards, since insufficient pollination causes yield reduction. Flowering time in hazelnut, as is similarly the case for most plant species, is both under strong genetic control, and also strongly influenced by the environmental factors (Jung and Müller, 2009) and is a major factor to be considered in climatic adaptation both to warm and frosty areas (Ntladi et al., 2018). The time of budburst is another important limiting factor of the environment in which cultivars can be successfully grown. In pear (Pyrus communis), for example, the failure to satisfy the plant’s chilling requirement compromises vegetative budburst (Gabay et al., 2018).
In hazelnut, budburst dates widely differ in hazelnut cultivars, but in northern Italy, leaf emergence typically happens between mid-March and early April. Otherwise, nuts of the major hazelnut cultivar mature over a period from August to October. Early maturing cultivars are favored by growers, both because harvesting during the dry, warm weather typical of the late summer is easier than in autumn, and the nuts harvested under dry conditions are better able to maintain their high nutritional quality.
Conventional breeding in tree crops is a slow process, but it can be accelerated by the application of marker-assisted selection (MAS). The first genetic linkage map of hazelnut appeared 15 years ago (Mehlenbacher et al., 2006), but since this time others have been added (Gürcan and Mehlenbacher, 2010a,b; Gürcan et al., 2010; Beltramo et al., 2016; Bhattarai and Mehlenbacher, 2017; Colburn et al., 2017; Rowley et al., 2018; Torello Marinoni et al., 2018). The maps have been used to identify several major genes conferring resistance to Eastern Filbert Blight and self-incompatibility (Rowley et al., 2018), as well as to show that traits such as vigor, sucker habit, and timing of budburst are all under polygenic control (Beltramo et al., 2016; Torello Marinoni et al., 2018). Ozturk et al. (2017) exploited a genome-wide association (GWA) mapping approach to identify simple sequence repeat (SSR) markers associated with nut and kernel traits. Some hazelnut genomic resources are also available in the public domain, including its transcriptome (Rowley et al., 2012), a de novo assembled genome of the cultivar “Jefferson” and some resequencing data (Rowley et al., 2018)1. Very recently, a fully assembled and annotated genome sequence of cultivar “Tombul” was published (Lucas et al., 2021)2 as well as that of the cultivar “Tonda Gentile delle Langhe” (TGdL) (Pavese et al., 2021)3.
The present study was designed to determine the genomic regions associated with flowering, dichogamy, and nut maturity. The approach chosen was to apply quantitative trait locus (QTL) mapping to a set of F1 progeny bred from the cross TGdL × “Merveille de Bollwiller” (MB), which has previously been used to identify QTLs associated to the timing of budburst (Beltramo et al., 2016; Torello Marinoni et al., 2018). Based on available annotated genome sequences, these genomic regions were exploited to identify potential candidate genes underlying the phenology of hazelnut.
A total of 275 seedlings of the F1 progeny bred from the cross TGdL × MB, described by Beltramo et al. (2016) and Torello Marinoni et al. (2018), and three individuals obtained from rooted suckers of each of the two parents, were planted at the campus of the University of Torino (Department of Agricultural, Forest and Food Sciences; 45°07′N; 7°58′E; 293 m a.s.l.) in 2009.
The mapping population was a set of 213 individuals, planted in the core of the field. The plants, spaced 4 × 4 m, and trained in an open vase system, were irrigated between mid-June and mid-September using an integral PC drip line (UniRam 20010 AS, Netafim). Local meteorological data (temperature, relative humidity, rainfall) were recorded by Regione Piemonte - Rete Agrometeorologica Regionale using an automatic weather station comprising a set of sensors installed 2 m above the ground, following World Meteorological Organization guidelines.
Across four seasons (2012/13 to 2015/16), records were taken every 5–7 days from the end of December until mid-March with respect to the dates at which 10% of the catkins had released pollen (time of male flowering, tmf) and at which 10% of the female flowers were receptive (time of female flowering, tff), following Germain and Sarraquigne (2004). The data were grouped into nine classes according to International Union for the Protection of New Varieties of Plants [UPOV] (1979), from very early (1) to very late (9). The degree of dichogamy (dc) was scored on a scale of 1 (very protandrous) to 9 (very protogynous), following International Plant Genetic Resources Institute [IPGRI] et al. (2008). Time of nut maturity (tnm) was assessed across three seasons (2014–2016), with measurements being taken every 5–7 days from the end of July until the beginning of October, defined by the date at which 10% of the nuts had dropped from the tree. The data were converted into nine classes, from very early (1) to very late (9), following UPOV guidelines.
Population means, standard deviations, normality (kurtosis and skewness), and trait correlations were calculated using the IBM SPSS Statistics v25.0 package4. Normality, kurtosis, and skewness were tested using the Shapiro Wilks test (α = 0.05). Correlations between the traits, including the time of leaf budburst (tlb), as given by Torello Marinoni et al. (2018), were calculated on the basis of the Spearman coefficient. Segregation was considered as transgressive where the performance of at least one F1 individual either exceeded that of the higher scoring parent or fell short of that of the lower scoring parent by at least two standard deviations.
The QTL analysis was performed using MapQTL v5 software (van Ooijen, 2004), based on the two parental maps (TGdL and MB) developed by Torello Marinoni et al. (2018). The location of putative QTL was based initially on the simple interval mapping procedure (Lander and Botstein, 1989), then confirmed using the multiple QTL mapping procedure (Jansen and Stam, 1994). A mapping step size of 1 cM was used in both analyses. For the multiple QTL mapping, a backward elimination procedure was used to select appropriate co-factors (e.g., significantly associated with each trait at p < 0.02). Genome-wide logarithm of odds (LOD) thresholds (P < 0.05) were determined empirically for each trait, using the PERMUTATION test provided within MapQTL with 1,000 iterations (Churchill and Doerge, 1994). Only QTLs associated with an LOD higher than the genome-wide threshold were considered, and 1-LOD support intervals were determined for each LOD peak (van Ooijen, 1992). The proportion of the overall phenotypic variance (PV) associated with each QTL was estimated from the multiple QTL mapping model. QTL positions were drawn using MapChart (Voorrips, 2002). Each QTL was designated by its trait name (tmf, tff, dc, tnm, and tlb), followed by the relevant linkage group (LG) and the relevant season: thus, for example, tmf_TGdL_02_13 indicates a QTL underlying tmf mapping to LG02 on the TGdL map, as identified from data collected in the 2012/13 season.
Markers for map development and QTL analyses were initially identified using the “Jefferson” genome (Torello Marinoni et al., 2018). Recently, the genome of the cultivar TGdL, containing a wider number of annotated genes, was publicly made available (Pavese et al., 2021). Thus, candidate genes were retrieved by mapping the “Jefferson” scaffolds containing QTLs on the TGdL genome (Pavese et al., 2021) using a BLAST search. The structural/functional annotation of the genes in the QTL was carried out using the gff annotation file provided by Pavese et al. (2021) (see text footnote 3). Genes in the QTL intervals were discussed when showing a clear function/annotation related to flowering-like processes, as inferred from literature.
The set of phenological data for both the parents and the mapping population (Table 1) showed a significant degree of season-to-season variation (P < 0.05) between TGdL and MB for tmf, tff, and tnm, but not for dc. For TGdL, tmf was either “very early” (class 1) or “very early to early” (class 2), while tff ranged from “very early” (class 1) to “early” (class 3); for MB, tmf and tff were scored as, respectively, “medium-late” (class 6) to “late” (class 7). MB plants were consistently scored as “homogamous” (dc, class 5), while TGdL plants were “slightly protandrous” (class 4) in three of the 4 years, and “slightly protogynous” (class 6) in 2013. With respect to tnm, TGdL was scored as “very early to early” (class 2) in 2 of the 3 years and “early” (class 3) in 1 year, while MB was scored as either “medium” or “medium-late” (classes 5 to 6).
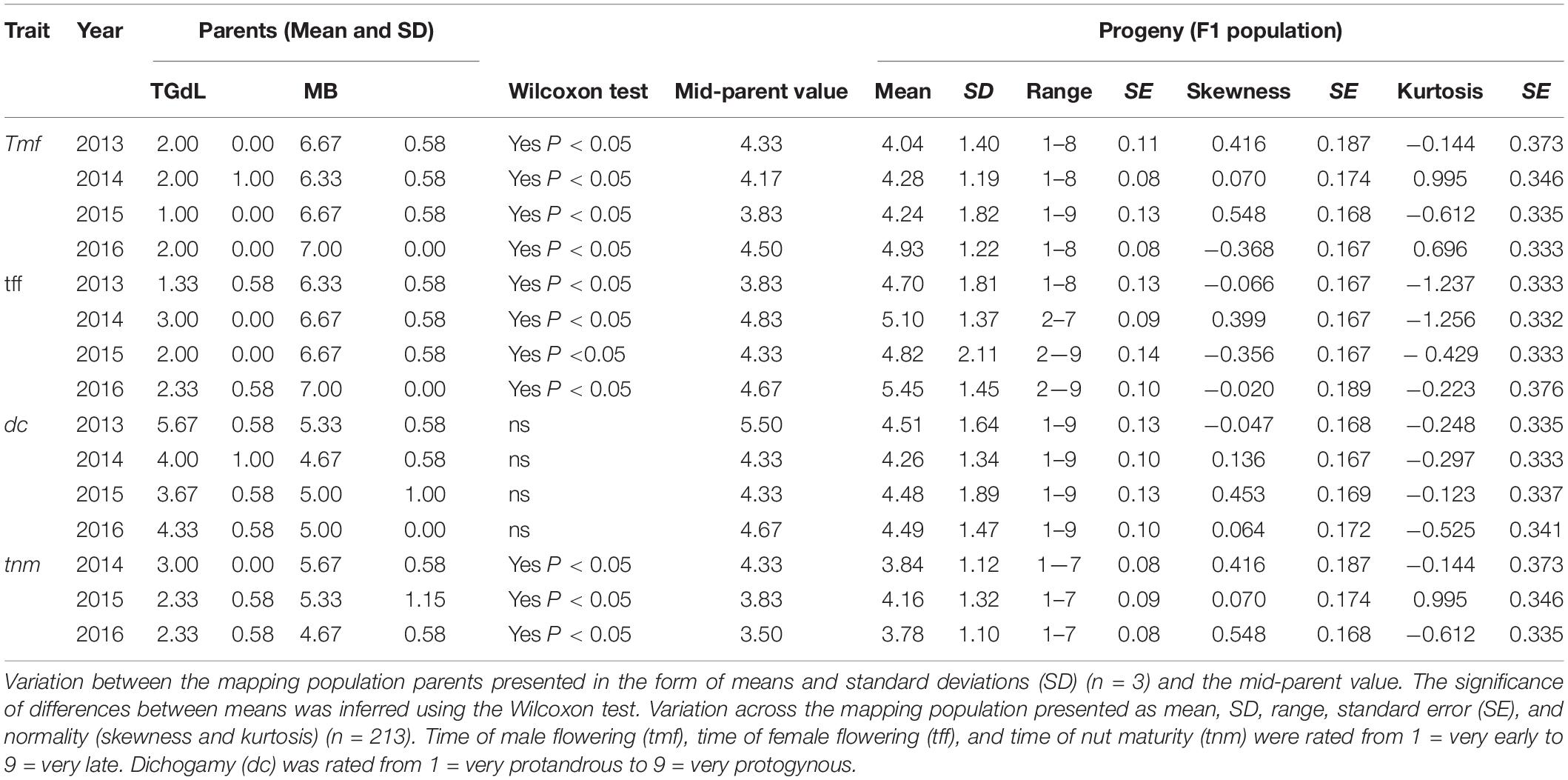
Table 1. Variation for time of male flowering (tmf), time of female flowering (tff), dichogamy (dc), and time of nut maturity (tnm).
Across the 213 individuals of the mapping population, flowering was initiated between late December/early January and continued to late February/early March. Nut fall began in late July and continued until mid-September (Supplementary Table 1). The tmf scores largely fell between those of the parents, with only a few examples of transgressive segregation (negative for two plants and positive for one, in 2013; positive in 2014 and 2015, respectively, for two and nine plants; positive for one plant and negative for five plants in 2016, Figure 1). For tff, the population mean consistently laid above the mid-parent value. Positive transgressive segregation was associated with ten individuals in both 2013 and 2016, and with 39 in 2015; however, in 2014, only one negative transgression was detected. The mapping population mean dc was intermediate between the parental values, and the distribution was normal. There was a substantial level of transgressive segregation in each year, affecting from 25.8% of the population in 2014 to 52.1% in 2013 (Figure 1). On the contrary, there was little evidence of transgressive segregation for tnm across the mapping population.
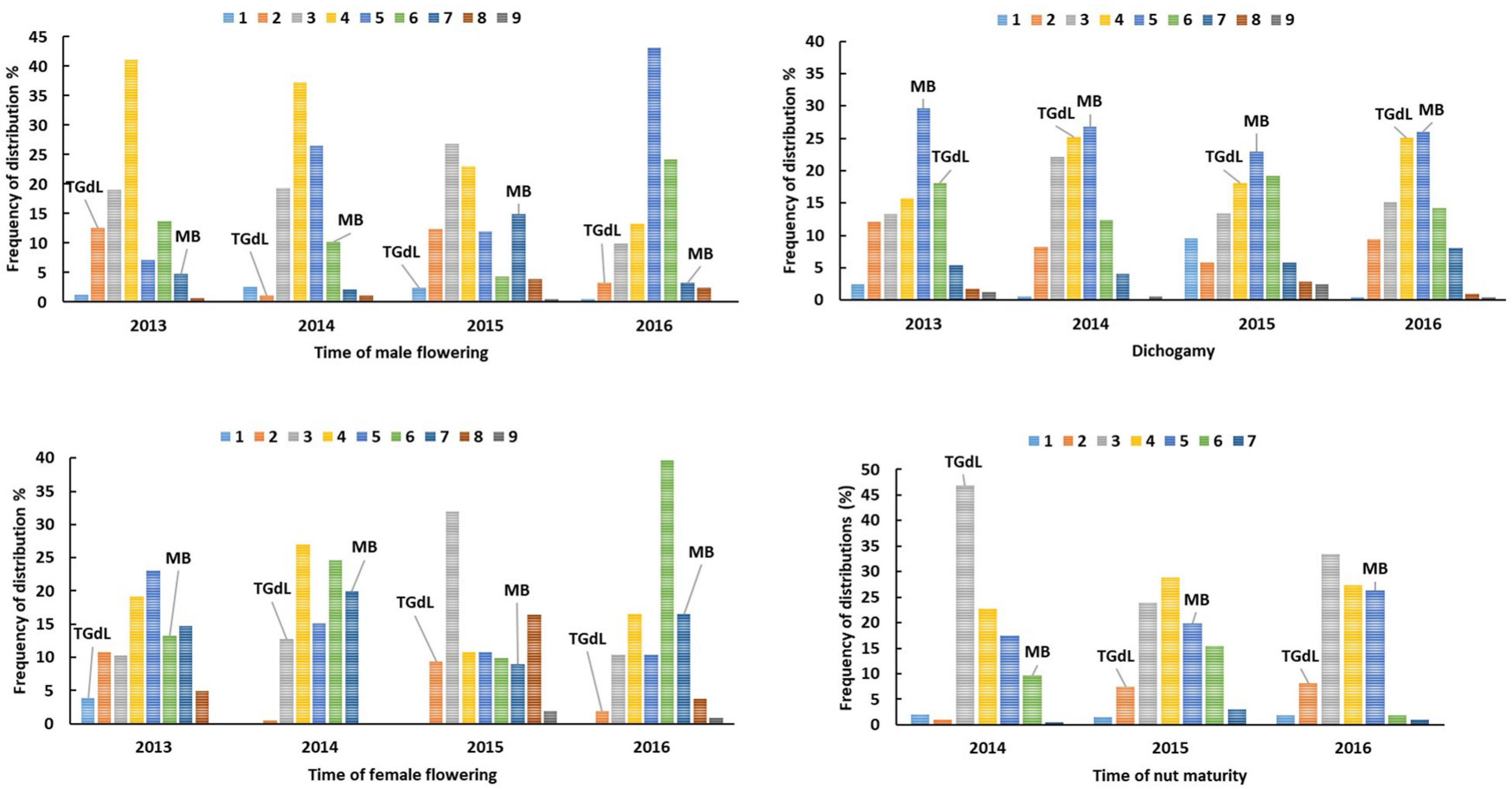
Figure 1. Frequency distribution plot for the mapping population with respect to time of male flowering (tmf), time of female flowering (tff), dichogamy (dc) and time of nut maturity (tnm) over four seasons. Data are grouped in classes from 1 = very early to 9 = very late. For dichogamy (dc), data are grouped in classes from 1 = very protandrous to 9 = very protogynous (5 = homogamous). The positions (means of three individuals) of the parents TGdL and MB are shown in each histogram.
Inter-trait correlations both within and between years are shown in Table 2. The correlation across years within a trait was consistently positive and highly significant (P < 0.01): for tmf, the levels ranged from 0.81 to 0.87, for tff from 0.77 to 0.89, for dc from 0.71 to 0.76, for tnm from 0.48 to 0.67, and for tlb from 0.78 to 0.86 (tlb data from Torello Marinoni et al., 2018).
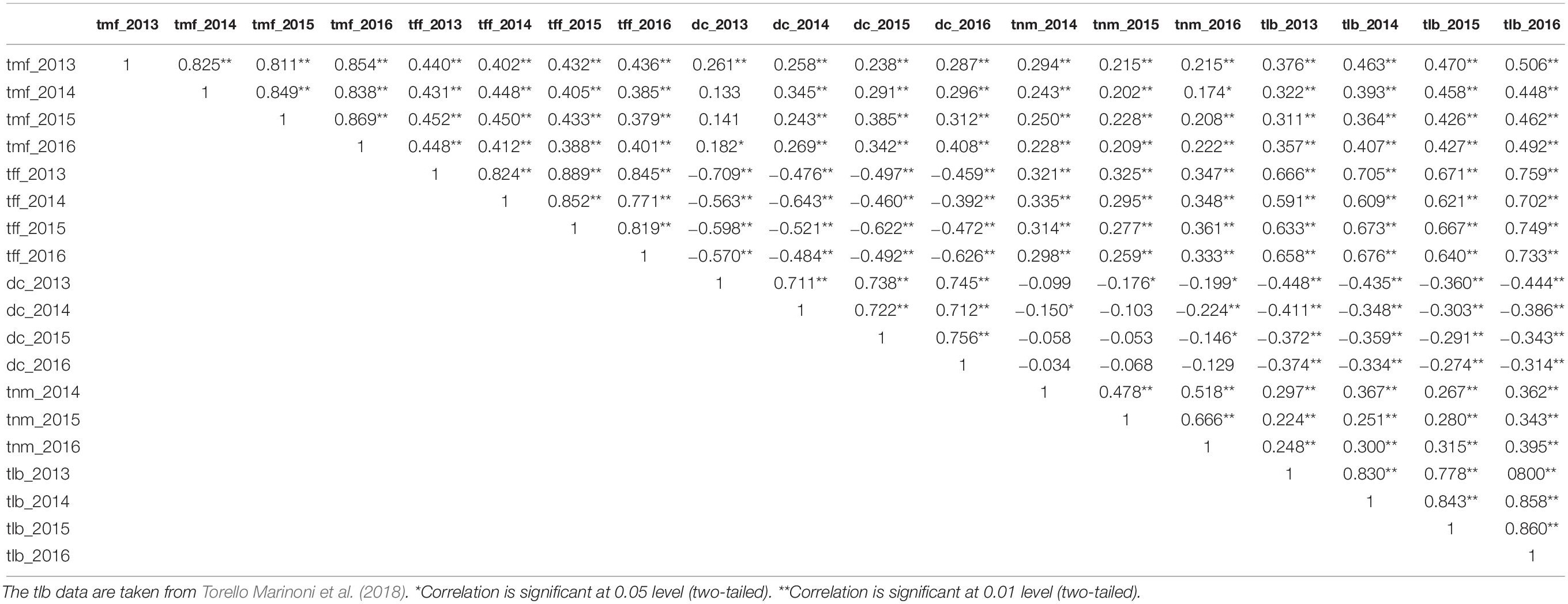
Table 2. Spearman correlation coefficients across years between each of the five traits: time of male flowering (tmf), time of female flowering (tff), dichogamy (dc), time of nut maturity (tnm), time of leaf budburst (tlb) and year of detection (n = 213).
The correlations between different traits were significant: tmf was positively correlated with each of the remaining traits, with values ranging from 0.38 to 0.45 for tff, from 0.22 to 0.45 for dc, from 0.29 to 0.51 for tlb, and from 0.15 to 0.28 for tnm. The tff was also positively correlated with both tlb (coefficients ranging from 0.59 to 0.73) and tnm (0.26–0.36), but was negatively correlated with dc (from −0.46 to −0.71). The tlb was positively correlated with tnm (from 0.22 to 0.39) and negatively with dc (from −0.27 to −0.45). The only trait combination showing any evidence of any correlation within a year was tnm vs dc, in both 2015 and 2016.
In all, 71 QTLs (49 on the TGdL map and 22 on the MB map) were identified. The loci were distributed across ten of the eleven LGs (only LG06 lacked at least one QTL). In all, 21 of the QTLs accounted for at least 10% of the phenotypic variance (PV) and are here indicated as “major” QTLs. The QTL associated with both the highest LOD (41.1) and the highest proportion of the PV (55.2%) was tff_TGdL_02 (Table 3). Overall, among the major QTLs, 13 were detected in at least two of the seasons (stable-major QTLs), while three were specific to 2013/14, two were specific to 2014/15, and three were specific to 2015/16. Four QTLs were classified as minor in one of the seasons but as major in the other seasons (stable QTLs). The map locations of these QTLs are given in Figure 2. LG02 was found to harbor 16 QTLs (15 on TGdL, one on MB), while 15 QTLs mapped to LG10 (seven on TGdL map, eight on MB map) and 14 to LG03 (11 on TGdL map, and three on MB map).
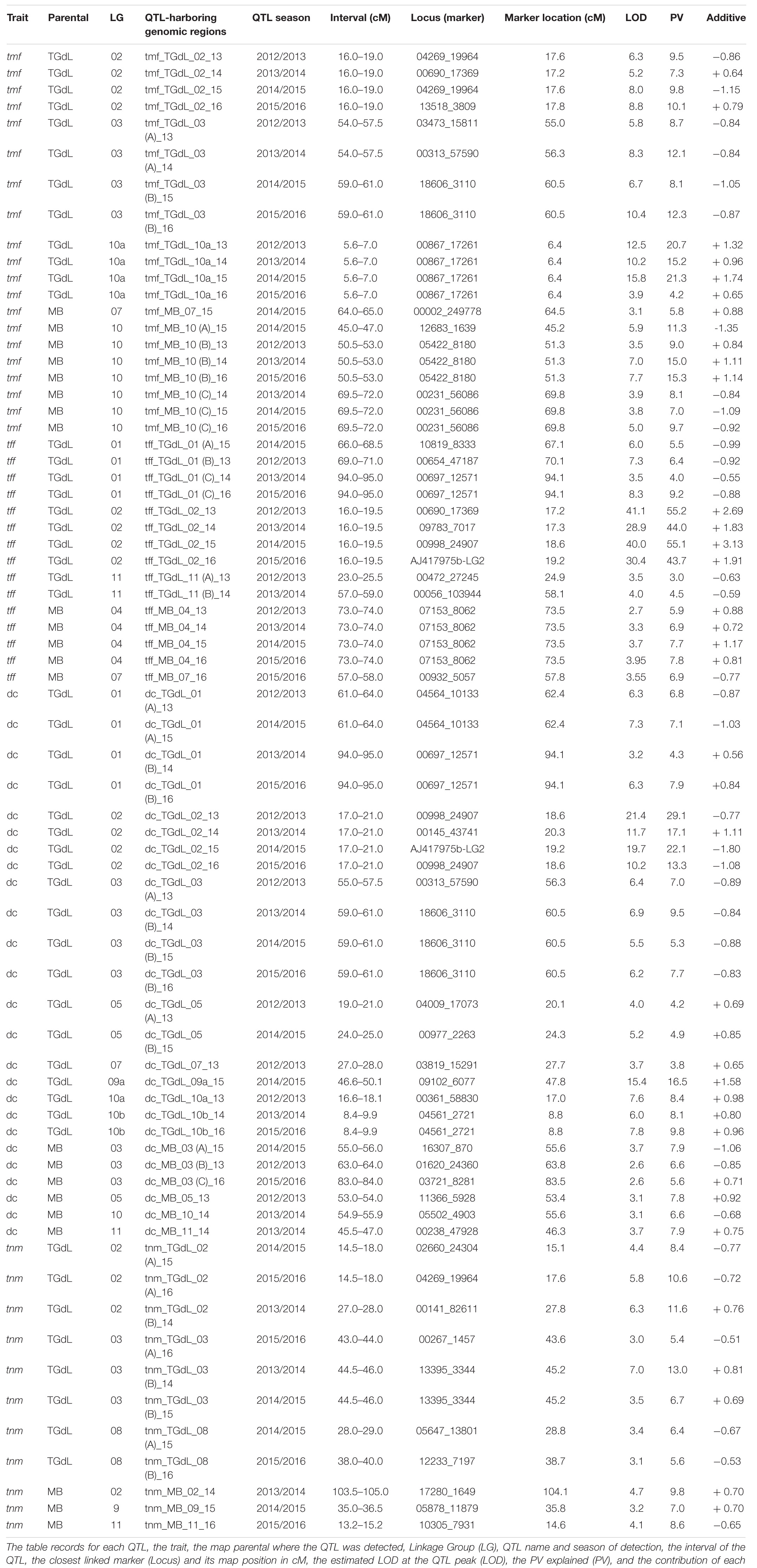
Table 3. Time of male flowering (tmf), time of female flowering (tff), dichogamy (dc), and time of nut maturity (tnm) QTL identified using either the TGdL or the MB linkage map.
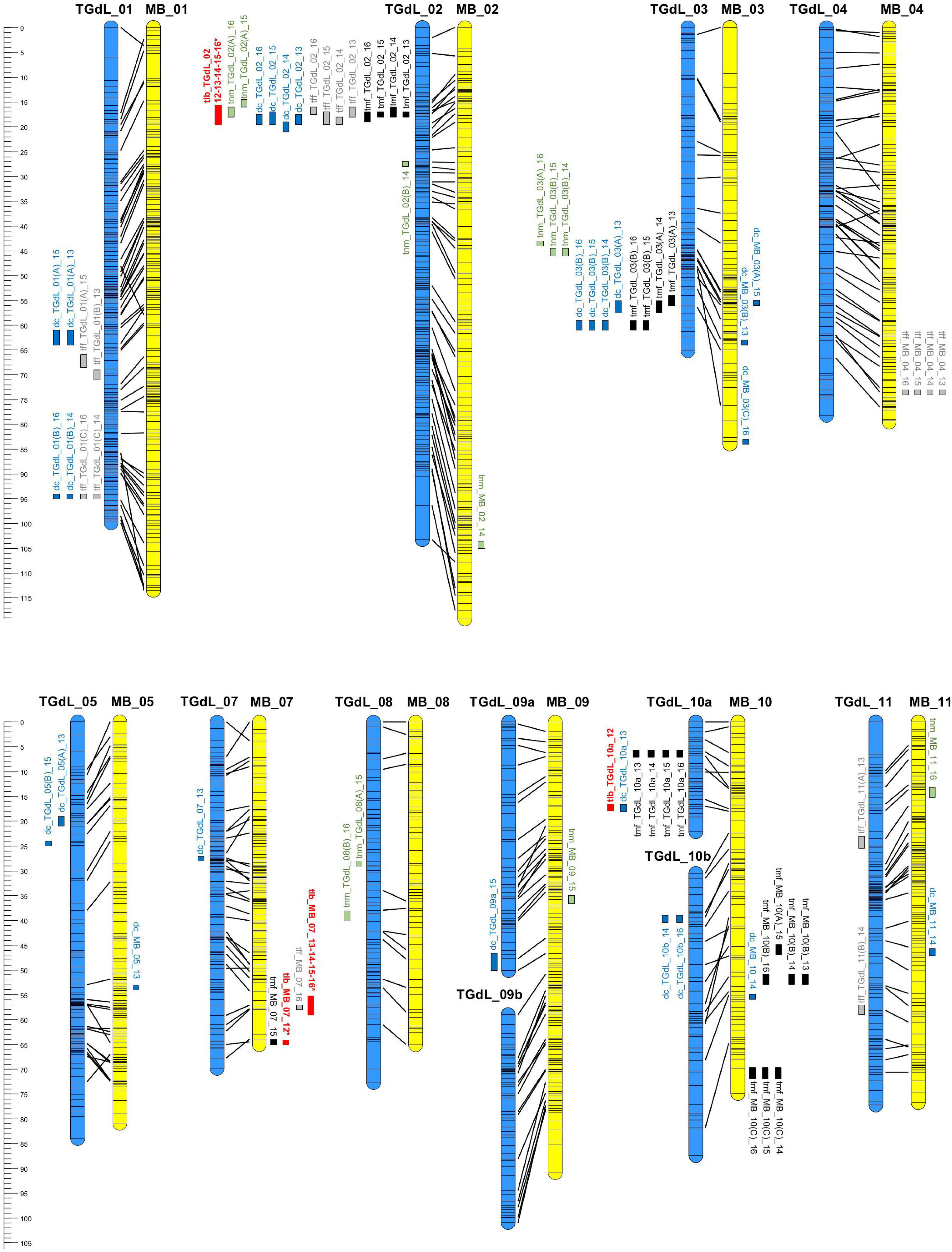
Figure 2. Linkage maps derived from TGdL (female parent of the mapping population) and MB (male parent). TGdL LGs are shown in blue on the left and MB ones in yellow on the right. The maps have been aligned based on shared scaffolds. The locations of the tlb QTL are shown in red. LGs not harboring any QTL have not been included. The ruler on the left represents the physical length of the LGs, while QTL locations are indicated in cM.
Many of the QTLs were clustered with one another, as expected given the extensive degree of inter-trait correlation (Table 2). The major cluster on TGdL_02 consisted of mainly major QTL governing all of the traits. Two year-specific clusters were identified on TGdL_03 associated with tmf and dc, one comprising QTLs detected in both 2012/13 and 2013/14, and the other QTLs expressed in both 2014/15 and 2015/16. A cluster on TGdL_01 harbored minor QTLs for dc and tff, while three smaller clusters involving tlb (Torello Marinoni et al., 2018) included minor QTLs underlying dc (on TGdL_10a), tmf, and tff (in two distinct clusters at the bottom of MB_07).
Of the 20 QTLs identified as contributing to tmf, 12 were mapped on the TGdL map and eight on the MB map (Table 3), altogether 477 genes. With respect to the former set of QTLs, four genomic regions were identified, one on each of TGdL_02 and 10a, and two on TGdL_03. The LG10a QTL, linked to marker AJ_00867_17261, was responsible for 15–21% of the PV in the first three seasons (with LOD peak values of 10–16 and associated with an additive effect of 0.96–1.74 classes out of the nine detected), but only 4% in the fourth season (maximum LOD 3.90 and 0.65 additive value). The LG02 QTLs were detected in all four seasons and explained 7–10% of the PV, while the two regions on LG03 together accounted for 8–12% of the PV. The four regions together harbored 383 genes, of which six are implicated in control over aspects of flowering (Supplementary Table 2). In detail, tmf_TGdL_02 carried 194 genes (3.484 – 5.116 Mb), tmf_TGdL_10a carried 155 genes (22.364–23.894 Mb), and tmf_TGdL_10b carried 34 genes (7.310 – 7.611 Mb). Of the four genomic regions identified in the MB map, three mapped to LG10; two of these harbored a QTL expressed in three of the four seasons, accounting for, respectively, 9–15% and 7–10% of the PV (with LOD peak values of 3.5–7.7 and 3.8–5.0, respectively, and both associated with additive effects of 0.8–1.1). A further QTL, responsible for 11% of the PV, was detected only in the 2014/15 season. The four regions together housed 94 genes, of which two are implicated in control over aspects of flowering (Supplementary Table 2). In detail, tmf_MB_07 carried 42 genes (0.360–0.768 Mb) and tmf_MB_10 carried 52 genes (3.353–3.774 Mb; 5.952–6.258 Mb).
Of the 15 tff QTLs detected, ten were located on the TGdL map and five on the MB map (Table 3), carrying 813 genes. The former set mapped within six genomic regions, distributed across LG01 (three regions), LG02, and LG11 (two regions). The largest effect QTL mapped between 17.2 and 19.2 cM along LG02, was detected in every season and explained 44–55% of the PV (with LOD peak values of 29–41 and associated with an additive effect of 1.8–3.1). The six genomic regions together harbored 731 genes, 19 of which are implicated in control over aspects of flowering (Supplementary Table 2). In detail, tff_TGdL_01 carried 205 genes (33.032–34.473 Mb; 33.840–50.216 Mb), tff_TGdL_02 carried 187 genes (2.743–5.038 Mb), tff_TGdL_11 (A) carried 257 genes (23.371–26.613 Mb), and tff_TGdL_11 (B) carried 82 genes (2.940 – 3.648 Mb). The loci located using the MB map were placed on two LGs. The one on LG04 was expressed in each year, but only explained 6–8% of the PV (maximum LOD 3.0–3.2 and 0.7–1.2 additive effect). The two regions housed 82 genes, but only one of these is implicated in control over aspects of flowering (Supplementary Table 2). In detail, tff_MB_04 carried 45 genes (36.031 – 36.381 Mb) and tff_MB_07 carried 37 genes (1.553 - 1.877 Mb).
Of the 25 dc QTLs, 19 were located using the TGdL map and six the MB map (Table 3), carrying 1,688 genes. With respect to the former set, eleven genomic regions, distributed over eight LGs, were identified, including two regions each on LG01, LG03, LG05, and one region each on LG02, LG07, LG09a, LG10a, and LG10b. The largest effect QTL, which was detected in each of the four seasons, laid between 18.6 and 20.3 cM of LG02 and explained 13–29% of the PV (10–21 LOD peak values, 0.8–1.8 days of additive effect). A major QTL on LG09a (responsible for 16.5% of the PV) was detected only in the 2014/15 season. A minor QTL, explaining 5–9% in three of the 4 years was identified on LG03, as were loci expressed in 2 of the years on both LG01 and LG10b. The eleven regions housed 1,345 genes, of which 18 are implicated in potential control over dichogamy (Supplementary Table 2). In detail, dc_TGdL_01 (A) carried 108 genes (31.054 – 32.516 Mb), dc_TGdL_01 (B) carried 26 genes (49.781–50.216 Mb), dc_TGdL_02 carried 233 genes (3.884–6.049 Mb), dc_TGdL_05 (A) carried 70 genes (31.646 – 32.384 Mb), dc_TGdL_05 (B) carried 18 genes (30.843 – 30.987 Mb), dc_TGdL_07 carried 262 genes (13.173 – 20.404 Mb), dc_TGdL_09a carried 481 genes (22.804 – 29.178 Mb), dc_TGdL_10a carried 14 genes (19.215 – 19.364 Mb), and dc_TGdL_10b carried 133 genes (9.400 – 12.937 Mb). The six regions located using the MB map harbored only minor QTLs, each of which was only detected in a single season; these regions mapped to LG03 (three QTLs), LG05, LG10, and LG11, and housed 343 genes, of which four were associated with potential control over dichogamy (Supplementary Table 2). In detail, dc_MB_05 carried 193 genes (15.072 – 20.183 Mb) and dc_MB_10 carried 150 genes (2.692 – 3.873 Mb).
Of the 11 tnm QTLs identified, eight were located using the TGdL map and three the MB map (Table 3), carrying 816 genes. The former fell within LG02, LG03, and LG08, with each LG housing two regions. Major QTLs mapped to the two regions on LG02, each accounting for 11–12% of the PV (maximum LOD 5.8–6.3; additive value 0.7–0.8). A QTL mapping to 45.2 cM along LG03 explained 7–13% of the PV in two of the four seasons. Together, the six regions harbored 507 genes, of which five were implicated in control over seed development (Supplementary Table 2). In detail, tnm_TGdL_02 (A) carried 237 genes (2.825–4.915 Mb), tnm_TGdL_02 (B) carried 67 genes (8.660–9.716 Mb), tnm_TGdL_08 (A) carried 91 genes (6.777–8.646 Mb), and tnm_TGdL_08 (B) carried 112 genes (13.405– 15.761 Mb). Using the MB map, a minor QTL was mapped on each of the LG02, LG09, and LG11; all of them were only expressed in a single season. The three regions together housed 309 genes, of which just two genes were associated with control over seed development (Supplementary Table 2). In detail, tnm_MB_02 carried 88 genes (44.867–46.099 Mb), tnm_MB_09 carried 64 genes (26.813–27.570 Mb), and tnm_MB_11 carried 157 genes (26.925–28.570 Mb).
The present analysis was based on patterns of segregation for key phenological traits among progeny bred from a cross between two disparate parents: one of these (TGdL) is adapted to the climatic conditions prevalent in NW Italy, where it flowers early and its nuts mature early, whereas the other (MB), which was adapted to a rather cooler environment (Germany–East France), flowers later and its nuts are harvested later (Črepinšek et al., 2012). Exploiting a linkage map built from several hundred molecular markers (SNP and SSR markers) led to the recognition of a number of genomic regions harboring genes influencing the timing of both male and female flowering, dichogamy, and nut maturity. In several of these regions, the location of two or more QTLs responsible either for different traits (reflecting a set of tightly linked loci or, more likely, a single pleiotropic locus) and/or for the same trait across seasons overlapped. As an example, the major cluster on TGdL_02 consisted of mainly major QTLs governing all of the traits; this is the same region where Torello Marinoni et al. (2018) mapped a major QTL controlling leaf budburst trait.
The variation for the various traits was typically not normally distributed, and furthermore, was not constant from 1 year to the next; the latter behavior reflects the major influence of the climate (particularly temperature) on these traits. The dates of tmf and tff in the progeny were similar over the years but the influence of temperature on the number of individuals in each class of distributions is evident. The effect of temperature on phenophase timing has been reported for numerous trees (Howe et al., 2003; Cooke et al., 2012; Ghelardini et al., 2014), including fruit tree species such as hazelnut (Črepinšek et al., 2012). As also noted by Črepinšek et al. (2012), the length of the period over which female flowering in hazelnut takes place can range from under one to over 10 weeks, depending on mean air temperature; early maturing cultivars such as TGdL are particularly labile in this respect. These authors also suggest that male and female flowers respond more strongly to increased air temperature than leafing.
As a consequence, the extent of dichogamy too is influenced jointly by genotype and climatic conditions (Bastias and Grau, 2005; Črepinšek et al., 2012); the dichogamy type of certain hazelnut cultivars, including MB, can be affected by the climate (Turcu et al., 2001; Črepinšek et al., 2012). Transgressive segregation in both directions was observed for each of the traits (Figure 1). This even included dc, even though the two parental lines did not differ significantly from one another for this trait. Trait transgression typically arises as a result of the inheritance of novel combinations of distinct alleles present in each parent (de Vicente and Tanksley, 1993). For each of the traits, there was a high positive correlation (P < 0.01) between the performance of given progeny in the various seasons, while there were also extensive, mostly positive correlations between pairs of traits (the exception was the negative correlation between dc and both tff and tlb). The trait which was least well correlated with others was tnm.
The genetic basis of phenology is complex as a result of the numerous physiological pathways that are involved. Until now, the only phenological trait of hazelnut to be genetically analyzed has been tlb (Torello Marinoni et al., 2018). The present research has extended the knowledge based on phenological traits by revealing that several QTLs underlie variation for each of the traits investigated. While most of these QTLs individually explained less than 10% of the PV, about a quarter of them proved to be stable over years and some were responsible for quite a high proportion of the PV; these latter loci in principle could be targeted for marker-assisted selection. The most substantial tff QTL, which accounted for as much as 55% of the PV, mapped between 17 and 19 cM on TGdL_02. Meanwhile, the largest effect tmf QTL mapped near one end (6.4 cM) of TGdL_10a was detected; the observation that this QTL was rather poorly expressed in one of the four seasons suggests that it is regulated by gene(s) which are responsive to environmental conditions.
Of particular note is the coincident map location of a number of the QTLs, which implies that a degree of pleiotropy underlies variation for these traits. The most striking example relates to the region of TGdL_02 lying between 17.2 and 27.8 cM, within which QTLs controlling tff, tmf, dc, and tnm were consistently mapped across years (Figure 2). The same region also harbors a QTL for tlb (Torello Marinoni et al., 2018). A useful marker for this cluster of QTLs is the microsatellite locus AJ417975b. A second important genomic region along TGdL_03 harbored QTsL involved in the determination of tmf, dc, and tnm; while dc and tmf QTLs both mapped in the segment between 55 and 60 cM, the tnm QTL was located in a somewhat less distal segment (43–45 cM). There was also a coincident location for dc and tff on TGdL_01; the association of these two traits is understandable, since dichogamy, is strongly influenced by climatic conditions (Črepinšek et al., 2012).
The genetic basis of flowering time in hazelnut remains poorly researched, in contrast to the situation in pear (Pyrus communis) (Gabay et al., 2018; Ntladi et al., 2018), apple (Malus domestica) (Allard et al., 2016), apricot (Prunus armeniaca) (Kitamura et al., 2018), and willow (Salix spp.) (Ghelardini et al., 2014). In both pear (Gabay et al., 2018) and apple (Allard et al., 2016), the location of some QTLs underlying variation in flowering time coincides with those associated with vegetative budburst, just as was the case with hazelnut. Similar pleiotropic effects observed in willow have been taken to imply that the determination of these phenological traits shares common components (Ghelardini et al., 2014), a conclusion which is not unexpected given that they are all strongly influenced by temperature and/or photoperiod.
The number of genes present in the regions harboring tmf, tff, dc, and tnm QTLs were, respectively: 477 (383 in the TGdL and 94 in the MB maps), 813 (731 in the TGdL and 82 in the MB), 1,688 (1,345 in the TGdL and 193 in the MB), and 816 (507 in the TGdL and 309 in the MB). Some of these were expected to include genes involved in the control of dormancy, budburst, the switch from vegetative to reproductive growth, or the morphogenesis of flowers and seeds. Accordingly, many of them have been described in the literature as involved in processes related to flowering, at the molecular level (Supplementary Figure 1) and to specific plant ontology (PO) terms: pollen-tube-cell (PO:0025195), plant sperm cell (PO:0000084), pollen (PO:0025281), inflorescence-meristem (PO:0000230), carpel (PO:0009030) stamen (PO:0009029), and flower (PO:0009046). Genes in the QTL intervals were thus discussed when showing a clear function/annotation related to flowering-like processes in a range of plant species, as inferred from literature (Supplementary Table 2).
Five genes appeared strongly related to flowering phenotype (Supplementary Table 2). Haze_17445, in tmf_MB_07, is a homolog of TEM1, which in Arabidopsis thaliana encodes a protein acting to delay flowering by inhibiting the production of both FT and the hormone gibberellic acid, until such time as the plant has matured beyond a particular growth stage (Castillejo and Pelaz, 2008; Matías-Hernández et al., 2014). Haze_04134, in tmf_TGdL_02, is a homolog of the A. thaliana gene RFI2. It encodes an E3 ubiquitin-protein ligase (Chen and Ni, 2006) known to negatively regulate the CONSTANS/FLOWERING LOCUS T (CO/FT) module, which is key to the promotion of flowering in response to photoperiod (Chen and Ni, 2006; Turck et al., 2008). The other three genes are homologs of genes involved in determining the timing of anthesis; Haze_21037, in tmf_TGdL_10a, a homolog of MSP1 which encodes a leucine-rich repeat receptor protein kinase; Haze_17456, in tmf_MB_07, a homolog of PP2AA2 (serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A), and Haze_20983, in tmf_TGdL_10a, a homolog of CPL2 (RNA polymerase II C-terminal domain phosphatase-like 2) (Supplementary Table 2). In A. thaliana, these latter three genes are all involved in sporogenesis and the regulation of floral development through phosphorylation/dephosphorylation circuits (Nonomura et al., 2003; Ueda et al., 2008; Kataya et al., 2015). Other three genes are potential homologs of DPD1 (Tang et al., 2012), MBD9 (Peng et al., 2006) and APD1 (Luo et al., 2012).
A total of 16 genes strongly related to flowering were present in the genomic regions harboring tff QTLs (Supplementary Table 2). The first of these is Haze_16689, in tff_TGdL_11 (A), a homolog of rice HDR1 and an ortholog of CO, genes that regulate the photoperiod-dependent flowering pathway (Sun et al., 2016). The second is Haze_16712, in tff_TGdL_11 (A), and a homolog of SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1) which encodes a transcription activator of LEAFY (Gregis et al., 2009). The third is Haze_15371, in tff_TGdL_11 (B), a potential homolog of A. thaliana AP2 which encodes a transcription factor activating the floral meristem (Krogan et al., 2012). Haze_15391, in tff_TGdL_11 (B), is a strong candidate for tff given that the product of its homolog is probably involved in the auxin-mediated control of gynoecium patterning (Gremski et al., 2007). The five genes Haze_02311, in tff_TGdL_01 (B), Haze_16714 and Haze_16827, in tff_TGdL_11 (A), Haze_15445, in tff_TGdL_11 (B), and Haze_17592, in tff_MB_07, are also homologs of relevant transcription factors in A. thaliana, namely SRS3 (Kuusk et al., 2006), MADS3 (Fernandez et al., 2013), AHL18 (Xiao et al., 2009), HHO5 (Moreau et al., 2016), and KAN4 (Gao et al., 2010), respectively. The tenth potential candidate gene is Haze_04134, in tff_TGdL_02, which as noted above, is a homolog of A. thaliana RFI2 (Chen and Ni, 2006). A group of six genes: Haze_02317, Haze_02318, in tff_TGdL_01 (B), and Haze_02223, in tff_TGdL_01 (A), Haze_04095, in tff_TGdL_02, Haze_16862, in tff_TGdL_11 (A), Haze_15376, in tff_TGdL_11 (B), likely encoding proteins responsible for histone methylation/demethylation/acetylation, are also plausible as candidates, because it is known that the transcript level of the key genes FT and FC is regulated epigenetically (Jeong et al., 2015). These six genes are potential homologs of, respectively, WDR5A (Jiang et al., 2009), SHL (Lopez-Gonzalez et al., 2014), TAF14B (Bieluszewski et al., 2015), MBD9 (Yaish et al., 2009), ELF6 (Jeong et al., 2009) and CLF (Saleh et al., 2007). Other four genes are homologs of MEE40 (Pagnussat et al., 2005), QKY (Fulton et al., 2010), FLD (Liu et al., 2007) and NFD4 (Portereiko et al., 2006).
Many species of plants have developed mechanisms that prevent self-pollination. Although the hazelnut typically exhibits sporophytic self-incompatibility, many cultivars are also dichogamous. Fourteen genes were related to the dichogamy trait were identified (Supplementary Table 2). The genes Haze_02127, in dc_TGdL_01 (A), Haze_04221, in dc_TGdL_02, Haze_11214, Haze_11325, Haze_11453, in dc_TGdL_09a, Haze_20664, in dc_TGdL_10a, Haze_13480, in dc_MB_05, and Haze_19677, in dc_MB_10, encode eight transcription factors involved in the determination of cell fate and organ development; these genes were homologs of, respectively, AMS (Lou et al., 2014), NFYC9 (Hou et al., 2014), TCX2 (Sijacic et al., 2011), SUP (Hiratsu et al., 2002), CO3 (Kim et al., 2008), AP2-3 (Lee et al., 2007), MADS1 (Poupin et al., 2007), and OFP13 (Wang et al., 2011). A further set of candidate genes: Haze_04202, in dc_TGdL_02, Haze_18674, in dc_TGdL_07, Haze_11374, Haze_11526, and Haze_11258, in dc_TGdL_09a, potentially encode five regulators of flowering. Haze_11374 is a homolog of CRY2, the product of which is a major photoreceptor regulating FT and FLC (Endo et al., 2007). Haze_11526 is a homolog of BLI, the product of which controls cotyledon and leaf patterning by inhibiting premature differentiation (Schatlowski et al., 2010); in particular, this gene is required for the activation of FLC and is involved in the response to chilling. Haze_18674 is a homolog of MED8, the product of which is involved in the regulation of flowering time (Lalanne et al., 2004; Kidd et al., 2009). Haze_04202 is a homolog of MIRO1, the product of which is a mitochondrial GTPase required during gametogenesis (Sormo et al., 2011). Haze_11258 is a homolog of LFR, the product of which is a nuclear protein required for the formation of anthers and may be a key component of the genetic network regulating anther development (Wang et al., 2012). The final potential candidate is Haze_14427, in dc_TGdL_05 (B), a homolog of RPK2, the product of which is an LRR receptor-like serine/threonine-protein kinase, involved in the regulation of anther development, including tapetum degradation during pollen maturation (Mizuno et al., 2007; Nodine et al., 2007). Other genes are potential homologs of NFD4 (Portereiko et al., 2006), CLPS3 (Xing et al., 2008), HUA1 (Cheng et al., 2003), EDA40 (Pagnussat et al., 2005), CLO (Liu et al., 2009), RIE1 (Xu and Li, 2003), JASON (De Storme and Geelen, 2011), LIS (Voelz et al., 2012).
In hazelnut, the ovary has yet to form at the time when the pistil is mature. The differentiation of several layers of ovary primordial cells occurs only after fertilization has been achieved. The identity of the genes that regulate ovary and ovule development in hazelnut is largely unknown.
Five genes potentially influencing seed development were placed within a genomic region harboring the tnm QTLs. These genes were Haze_03973, Haze_11440, Haze_04470, Haze_25121 and Haze_16946. Haze_03973, in tnm_TGdL_02 (A), and Haze_11440, in tnm_MB_09 are homologs of ACS3 and ACS1, respectively. The product of ACS3 is 1-aminocyclopropane-1-carboxylate synthase 3, while that of ACS1 is 1-aminocyclopropane-1-carboxylate synthase; both of these enzymes catalyze 1-aminocyclopropane-1-carboxylic acid, a direct precursor of ethylene (Peng et al., 2005). In A. thaliana, the absence of this compound results in abnormal embryo morphogenesis and embryo lethality (Baud et al., 2003), while Cheng et al. (2019) have suggested that since the mature hazelnut kernel is rich in unsaturated fatty acids, its absence impairs the conversion of citrate into long-chain fatty acids, thereby compromising the development of the ovule. This is a very interesting finding since the occurrence of blank nuts in hazelnut is still poorly understood, but represents a serious problem to solve to avoid important losses of yields in orchards. Haze_04470, in tnm_TGdL_02 (B) is a homolog of EIN4, the product of which is a negative regulator of ethylene signaling (Hua et al., 1998). Haze_25121, in tnm_TGdL_08 (B) encodes a protein thought to be involved in the abscission of the mature nut (Liljegren et al., 2004); the gene is a homolog of IND, the product of which is a transcription factor involved in the differentiation of various cell types required for fruit dehiscence (Liljegren et al., 2004). Finally, Haze_16946, in tnm_MB_11, is a homolog of AP2-2, the product of which is a member of AP2/ERF family of transcription factors involved in the control of inflorescence architecture and floral meristem establishment (Dai et al., 2016). Other two genes are potential homologs of GPA3 (Ren et al., 2014) and IDL4 (Stenvik et al., 2008).
The present study explores the genetic architecture of phenology traits in a progeny of Corylus avellana. The QTL mapping described here has identified for the first time (except for tlb trait) several major QTLs underlying phenological-related traits in hazelnut (time of male and female flowering, dichogamy, and nut maturity). Several regions were identified where many QTLs co-localized for different traits or for the same trait across years. The 26% of identified QTLs were very robust, stable, being therefore promising for the use in marker-assisted selection.
The search of genes along the scaffolds linked to the QTL of interest has shown some interesting match with orthologous genes involved in flowering and nut growth processes in other plant species. The presence of homologs of genes known to be involved in the determination of flowering time and seed development in the relevant genomic regions has provided some leads toward gaining an understanding of the genetic and molecular basis of these important traits.
The regions surrounding the inferred locations of the major QTLs harbor hundreds of genes, some of which will likely represent fruitful targets for future investigations. Resequencing of strong candidate genes could be used to reveal the extent of allelic variation present in phenotypically contrasting cultivars. The availability of a mapping population will be useful in narrowing the search for genuine candidate genes.
The need to develop genomic tools able to accelerate hazelnut breeding is increasing. The data reported here will make a contribution toward the formulation of a biotechnology-based strategy designed to efficiently select trees more likely to be able to adapt to a changing environment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
RB conceived and designed the experiments. NV and DTM performed field investigations and analyzed field data. DTM and EP performed QTL analysis. AA, VP, and ECG performed candidate genes and functional annotation analysis. DTM, NV, and EP wrote the manuscript. All authors contributed to the discussion, revised, and approved the final manuscript.
This research was carried out in collaboration with the Ferrero Hazelnut Company and we thank them for their financial support. This research was also supported by the Fondazione CRT (Cassa di Risparmio di Torino) and University of Torino-Local Research, 2018.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.749394/full#supplementary-material
Supplementary Figure 1 | Candidate genes underlying variation in phenology in hazelnut. (A) A four-way Venn diagram illustrating overlap in the genes mapping within the regions harboring tmf, tff, dc, and tnm QTL. (B) Functional categorization of the 121 genes potentially underlying variation in tmf, tff, dc, and tnm. (C) Functional categorization of candidate genes mapping within regions harboring either tmf, tff, dc, or tnm QTL.
Supplementary Table 1 | Dates of male (tmf) and female flowering (tff) and nut maturity (tnm): the data refer to the earliest and latest flowering and maturing segregants recorded over four seasons.
Supplementary Table 2 | The gene content of the regions harboring time of male flowering (tmf), time of female flowering (tff), dichogamy (dc), and time of nut maturity (tnm) QTL. Hazelnut genes (Haze_Xx) were identified using the TGdL genome.gff annotation file (https://zenodo.org/record/4454484).
Allard, A., Bink, M. C. A. M., Martinez, S., Kelner, J. J., Legave, J. M., di Guardo, M., et al. (2016). Detecting Qtls and putative candidate genes involved in budbreak and flowering time in an apple multiparental population. J. Exp. Bot. 67, 2875–2888. doi: 10.1093/jxb/erw130
Bastias, R., and Grau, P. (2005). Floral phenology of commercial cultivars and chilean pollinizers of hazelnut (corylus avellana l.) in Chile. Acta Hortic. 686, 151–156. doi: 10.17660/ActaHortic.2005.686.19
Baud, S., Guyon, V., Kronenberger, J., Wuillème, S., Miquel, M., Caboche, M., et al. (2003). Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J. 33, 75–86. doi: 10.1046/j.1365-313x.2003.016010.x
Beltramo, C., Valentini, N., Portis, E., Torello Marinoni, D., Boccacci, P., Sandoval Prando, M. A., et al. (2016). Genetic mapping and QTL analysis in European hazelnut (Corylus avellana L.). Mol. Breed. 36, 27. doi: 10.1007/s11032-016-0450-6
Bhattarai, G., and Mehlenbacher, S. A. (2017). In silico development and characterization of tri-nucleotide simple sequence repeat markers in hazelnut (Corylus avellana L.). PLoS One 12:e0178061. doi: 10.1371/journal.pone.0178061
Bieluszewski, T., Galganski, L., Sura, W., Bieluszewska, A., Abram, M., Ludwikow, A., et al. (2015). AtEAF1 is a potential platform protein for Arabidopsis NuA4 acetyltransferase complex. BMC Plant Biol. 15:75. doi: 10.1186/s12870-015-0461-1
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Chen, M., and Ni, M. (2006). RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J. 46, 823–833. doi: 10.1111/j.1365-313X.2006.02740.x
Cheng, Y., Jiang, S., Zhang, X., He, H., and Liu, J. (2019). Whole-Genome Re-Sequencing of Corylus heterophylla Blank-Nut Mutants Reveals Sequence Variations in Genes Associated with Embryo Abortion. Front. Plant Sci. 10:1465. doi: 10.3389/fpls.2019.01465
Cheng, Y., Kato, N., Wang, W., Li, J., and Chen, X. (2003). Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev. Cell 4, 53–66. doi: 10.1016/S1534-5807(02)00399-4
Churchill, G. A., and Doerge, R. W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. doi: 10.1093/genetics/138.3.963
Colburn, B. C., Mehlenbacher, S. A., and Sathuvalli, V. R. (2017). Development and mapping of microsatellite markers from transcriptome sequences of European hazelnut (Corylus avellana L.) and use for germplasm characterization. Mol. Breed. 37:16. doi: 10.1007/s11032-016-0616-2
Cooke, J. E. K., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Črepinšek, Z., Štampar, F., Kajfcž-Bogataj, L., and Solar, A. (2012). The response of Corylus avellana L. phenology to rising temperature in north-eastern Slovenia. Int. J. Biometeorol. 56, 681–694. doi: 10.1007/s00484-011-0469-7
Dai, Z., Wang, J., Zhu, M., Miao, X., and Shi, Z. (2016). OsMADS1 represses microRNA172 in elongation of palea/lemma development in rice. Front. Plant Sci. 7:1891. doi: 10.3389/fpls.2016.01891
De Storme, N., and Geelen, D. (2011). The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol. 155, 1403–1415. doi: 10.1104/pp.110.170415
de Vicente, M. C., and Tanksley, S. D. (1993). QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134, 585–596. doi: 10.1093/genetics/134.2.585
Endo, M., Mochizuki, N., Suzuki, T., and Nagatani, A. (2007). CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19, 84–93. doi: 10.1105/tpc.106.048157
Food and Agriculture Organization Corporate Statistical Database [FAOSTAT] (2021). Food and Agriculture Organization of the United Nations [FAO]. Available online at: http://www.fao.org/faostat/en/#data/QC (Accessed January 21, 2021).
Fernandez, L., Chaib, J., Martinez-Zapater, J. M., Thomas, M. R., and Torregrosa, L. (2013). Mis-expression of a PISTILLATA-like MADS box gene prevents fruit development in grapevine. Plant J. 73, 918–928. doi: 10.1111/tpj.12083
Fulton, L., Vaddepalli, P., Yadav, R. K., Batoux, M., and Schneitz, K. (2010). Inter-cell-layer signalling during Arabidopsis ovule development mediated by the receptor-like kinase STRUBBELIG. Biochem. Soc. Trans. 38, 583–587. doi: 10.1042/BST0380583
Gabay, G., Dahan, Y., Izhaki, Y., Faigenboim, A., Ben-Ari, G., Elkind, Y., et al. (2018). High-resolution genetic linkage map of European pear (Pyrus communis) and QTL fine-mapping of vegetative budbreak time. BCM Plant Biol. 18:175. doi: 10.1186/s12870-018-1386-2
Gao, P., Li, X., Cui, D., Wu, L., Parkin, I., and Gruber, M. Y. (2010). A new dominant Arabidopsis transparent testa mutant, sk21-D, and modulation of seed flavonoid biosynthesis by KAN4. Plant Biotechnol. J. 8, 979–993. doi: 10.1111/j.1467-7652.2010.00525.x
Ghelardini, L., Berlin, S., Weih, M., Lagercrantz, U., and Gyllenstrand, N. (2014). Genetic architecture of spring and autumn phenology in Salix. BMC Plant Biol. 14:31. doi: 10.1186/1471-2229-14-31
Gregis, V., Sessa, A., Dorca-Fornell, C., and Kater, M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626–637. doi: 10.1111/j.1365-313X.2009.03985.x
Gremski, K., Ditta, G., and Yanofsky, M. F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593–3601. doi: 10.1242/dev.011510
Gürcan, K., and Mehlenbacher, S. A. (2010a). Development of microsatellite loci for European hazelnut (Corylus avellana L.) from ISSR fragments. Mol. Breed. 26, 551–559. doi: 10.1007/s11032-010-9464-7
Gürcan, K., and Mehlenbacher, S. A. (2010b). Transferability of microsatellite markers in the Betulaceae. J. Am. Soc. Hortic. Sci. 135, 159–173. doi: 10.21273/JASHS.135.2.159
Gürcan, K., Mehlenbacher, S. A., Botta, R., and Boccacci, P. (2010). Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet. Genomes 6, 513–531. doi: 10.1007/s11295-010-0269-y
Hiratsu, K., Ohta, M., Matsui, K., and Ohme-Takagi, M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514, 351–354. doi: 10.1016/S0014-5793(02)02435-3
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5, 4601–4601. doi: 10.1038/ncomms5601
Howe, G. T., Aitken, S. N., Neale, D. B., Jermstad, K. D., Wheeler, N. C., and Chen, T. H. (2003). From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can. J. Bot. 81, 1247–1266. doi: 10.1139/b03-141
Hua, J., Sakai, H., Nourizadeh, S., Chen, Q. G., Bleecker, A. B., Ecker, J. R., et al. (1998). EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10, 1321–1332. doi: 10.1105/tpc.10.8.1321
International Plant Genetic Resources Institute [IPGRI], Food and Agriculture Organization [FAO], and International Centre for advanced Mediterranean Agronomics Studies [CIHEAM] (2008). Descriptors for hazelnut (Corylus avellana L.). Rome: Bioversity International.
International Union for the Protection of New Varieties of Plants [UPOV] (1979). Hazelnut (Corylus avellana L. & Corylus maxima Mill.): guidelines for the conduct of tests for distinctness, uniformity and stability. Hazelnut/Noisetier/Haselnuss, 79-03-28. Doc. no. TG/71/3. Geneva: UPOV.
Jansen, R. C., and Stam, P. (1994). High-resolution of quantitative traits into multiple loci via interval mapping. Genetics 136, 1447–1455. doi: 10.1093/genetics/136.4.1447
Jeong, H. J., Yang, J., Yi, J., and An, G. (2015). Controlling flowering time by histone methylation and acetylation in arabidopsis and rice. J. Plant Biol. 58, 203–210. doi: 10.1007/s12374-015-0219-1
Jeong, J. H., Song, H. R., Ko, J. H., Jeong, Y. M., Kwon, Y. E., Seol, J. H., et al. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 4:e8033. doi: 10.1371/journal.pone.0008033
Jiang, D., Gu, X., and He, Y. (2009). Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733–1746. doi: 10.1105/tpc.109.067967
Jung, C., and Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Kataya, A. R., Heidari, B., Hagen, L., Kommedal, R., Slupphaug, G., and Lillo, C. (2015). Protein phosphatase 2A holoenzyme is targeted to peroxisomes by piggybacking and positively affects peroxisomal beta-oxidation. Plant Physiol. 167, 493–506. doi: 10.1104/pp.114.254409
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The Mediator Complex Subunit PFT1 Is a Key Regulator of Jasmonate-Dependent Defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Kim, S. K., Yun, C. H., Lee, J. H., Jang, Y. H., Park, H. Y., and Kim, J. K. (2008). OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228, 355–365. doi: 10.1007/s00425-008-0742-0
Kitamura, Y., Habu, T., Yamane, H., Nishiyama, S., Kajita, K., Sobue, T., et al. (2018). Identification of QTLs controlling chilling and heat requirements for dormancy release and bud break in Japanese apricot (Prunus mume). Tree Genet. Genome 14:33. doi: 10.1007/s11295-018-1243-3
Krogan, N. T., Hogan, K., and Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. doi: 10.1242/dev.085407
Kuusk, S., Sohlberg, J. J., Magnus Eklund, D., and Sundberg, E. (2006). Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 47, 99–111. doi: 10.1111/j.1365-313X.2006.02774.x
Lalanne, E., Michaelidis, C., Moore, J. M., Gagliano, W., Johnson, A., Patel, R., et al. (2004). Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167, 1975–1986. doi: 10.1534/genetics.104.030270
Lander, E. S., and Botstein, D. (1989). Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199. doi: 10.1093/genetics/121.1.185
Lee, D. Y., Lee, J., Moon, S., Park, S. Y., and An, G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49, 64–78. doi: 10.1111/j.1365-313X.2006.02941.x
Liljegren, S. J., Roeder, A. H. K., Kempin, S. A., Gremski, K., Ostergaard, L., Guimil, S., et al. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843–853. doi: 10.1016/S0092-8674(04)00217-X
Liu, F., Quesada, V., Crevillen, P., Baeurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407. doi: 10.1016/j.molcel.2007.10.018
Liu, M., Yuan, L., Liu, N. Y., Shi, D. Q., Liu, J., and Yang, W. C. (2009). GAMETOPHYTIC FACTOR 1, involved in pre-mRNA splicing, is essential for megagametogenesis and embryogenesis in Arabidopsis. J. Integr. Plant Biol. 51, 261–271. doi: 10.1111/j.1744-7909.2008.00783.x
Lopez-Gonzalez, L., Mouriz, A., Narro-Diego, L., Bustos, R., Martinez-Zapater, J. M., Jarillo, J. A., et al. (2014). Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 26, 3922–3938. doi: 10.1105/tpc.114.130781
Lou, Y., Xu, X. F., Zhu, J., Gu, J. N., Blackmore, S., and Yang, Z. N. (2014). The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 5, 3855–3855. doi: 10.1038/ncomms4855
Lucas, S. J., Kahraman, K., Avşar, B., Buggs, R. J., and Bilge, I. (2021). A chromosome-scale genome assembly of European hazel (Corylus avellana L.) reveals targets for crop improvement. Plant J. 105, 1413–1430. doi: 10.1111/tpj.15099
Luo, G., Gu, H., Liu, J., and Qu, L. J. (2012). Four closely-related RING-type E3 ligases, APD1-4, are involved in pollen mitosis II regulation in Arabidopsis. J. Integr. Plant Biol. 54, 814–827. doi: 10.1111/j.1744-7909.2012.01152.x
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marín-González, E., Suárez-López, P., and Pelaz, S. (2014). RAV genes: regulation of floral induction and beyond. Ann. Bot. 114, 1459–1470. doi: 10.1093/aob/mcu069
Mehlenbacher, S. A. (1991). Genetic Resources of Temperate Fruit and Nut Crops. Technical Communications of ISHS. Number 290-XVII. Hazelnuts (Corylus). Acta Hortic. 17, 791–836. doi: 10.17660/ActaHortic.1991.290.18
Mehlenbacher, S. A., Brown, R. N., Nouhra, E. R., Gökirmak, T., Bassil, N. V., and Kubisiak, T. L. (2006). A genetic linkage map for hazelnut (Corylus avellana L.) based on RAPD and SSR markers. Genome 49, 122–133. doi: 10.1139/g05-091
Mehlenbacher, S. A., and Thompson, M. M. (1988). Dominance relationships among S-alleles in Corylus avellana L. Theor. Appl. Genet. 76, 669–672. doi: 10.1007/BF00303511
Mizuno, S., Osakabe, Y., Maruyama, K., Ito, T., Osakabe, K., Sato, T., et al. (2007). Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 50, 751–766. doi: 10.1111/j.1365-313X.2007.03083.x
Moreau, F., Thevenon, E., Blanvillain, R., Lopez-Vidriero, I., Franco-Zorrilla, J. M., Dumas, R., et al. (2016). The Myb-domain protein ULTRAPETALA1 INTERACTING FACTOR 1 controls floral meristem activities in Arabidopsis. Development 143, 1108–1119. doi: 10.1242/dev.127365
Nodine, M. D., Yadegari, R., and Tax, F. E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev. Cell 12, 943–956. doi: 10.1016/j.devcel.2007.04.003
Nonomura, K., Miyoshi, K., Eiguchi, M., Suzuki, T., Miyao, A., Hirochika, H., et al. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15, 1728–1739.
Ntladi, S. M., Human, J. P., Bester, C., Vervalle, J., Roodt-Wilding, R., and Tobutt, K. R. (2018). Quantitative trait loci (QTL) mapping of blush skin and flowering time in a European pear (Pyrus communis) progeny of ‘Flamingo x Abate Fetel’. Tree Genet. Genomes 14:70. doi: 10.1007/s11295-018-1280-y
Ozturk, S. C., Ozturk, S. E., Celik, I., Stampar, F., Veberic, R., Doganlar, S., et al. (2017). Molecular genetic diversity and association mapping of nut and kernel traits in Slovenian hazelnut (Corylus avellana) germplasm. Tree Genet. Genomes 13:16.
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614.
Pavese, V., Cavalet Giorsa, E., Barchi, L., Acquadro, A., Torello Marinoni, D., Portis, E., et al. (2021). Whole-genome assembly of cv ‘Tonda Gentile delle Langhe’ using linked-reads (10X Genomics). G3 11:jkab152. doi: 10.1093/g3journal/jkab152
Peng, H. P., Lin, T. Y., Wang, N. N., and Shih, M. C. (2005). Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol. Biol. 58, 15–25. doi: 10.1007/s11103-005-3573-4
Peng, M., Cui, Y., Bi, Y. M., and Rothstein, S. J. (2006). AtMBD9: a protein with a methyl-CpG-binding domain regulates flowering time and shoot branching in Arabidopsis. Plant J. 46, 282–296.
Portereiko, M. F., Sandaklie-Nikolova, L., Lloyd, A., Dever, C. A., Otsuga, D., and Drews, G. N. (2006). NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 141, 957–965.
Poupin, M. J., Federici, F., Medina, C., Matus, J. T., Timmermann, T., and Arce-Johnson, P. (2007). Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404, 10–24.
Ren, Y., Wang, Y., Liu, F., Zhou, K., Ding, Y., Zhou, F., et al. (2014). GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 26, 410–425.
Rowley, E. R., Fox, S. E., Bryant, D. W., Sullivan, C. M., Givan, S. A., Mehlenbacher, S. A., et al. (2012). Assembly and characterization of the European hazelnut (Corylus avellana L.) ‘Jefferson’ transcriptome. Crop Sci. 52, 2679–2686. doi: 10.2135/cropsci2012.02.0065
Rowley, E. R., Van Buren, R., Bryant, D. W., Priest, D. W., Mehlenbacher, S. A., and Mockler, T. C. (2018). A Draft Genome and High-Density Genetic Map of European Hazelnut (Corylus avellana L.). bioArxiv [Preprint]. doi: 10.1101/469015
Saleh, A., Al-Abdallat, A., Ndamukong, I., Alvarez-Venegas, R., and Avramova, Z. (2007). The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 35, 6290–6296.
Schatlowski, N., Stahl, Y., Hohenstatt, M. L., Goodrich, J., and Schubert, D. (2010). The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. Plant Cell 22, 2291–2305.
Sijacic, P., Wang, W., and Liu, Z. (2011). Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 7:e1002352. doi: 10.1371/journal.pgen.1002352
Sormo, C. G., Brembu, T., Winge, P., and Bones, A. M. (2011). Arabidopsis thaliana MIRO1 and MIRO2 GTPases are unequally redundant in pollen tube growth and fusion of polar nuclei during female gametogenesis. PLoS One 6:e18530. doi: 10.1371/journal.pone.0018530
Stenvik, G.-E., Tandstad, N. M., Guo, Y., Shi, C.-L., Kristiansen, W., Holmgren, A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817.
Sun, X., Zhang, Z., Wu, J., Cui, X., Feng, D., Wang, K., et al. (2016). The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLoS Genet. 12:e1005927. doi: 10.1371/journal.pgen.1005927
Tang, L. Y., Matsushima, R., and Sakamoto, W. (2012). Mutations defective in ribonucleotide reductase activity interfere with pollen plastid DNA degradation mediated by DPD1 exonuclease. Plant J. 70, 637–649.
Torello Marinoni, D., Valentini, N., Portis, E., Acquadro, A., Beltramo, C., Mehlenbacher, S. A., et al. (2018). High density SNP mapping and QTL analysis for time of leaf budburst in Corylus avellana L. PLoS One 13:e0195408. doi: 10.1371/journal.pone.0195408
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Turcu, E., Turcu, I., and Botu, M. (2001). Flowering of hazelnut cultivars in Oltenia Romania. Acta Hortic. 556, 365–370. doi: 10.17660/ActaHortic.2001.556.54
Ueda, A., Li, P., Feng, Y., Vikram, M., Kim, S., Kang, C. H., et al. (2008). The Arabidopsis thaliana carboxyl-terminal domain phosphatase-like 2 regulates plant growth, stress and auxin responses. Plant Mol. Biol. 67, 683–697.
van Ooijen, J. W. (1992). Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 84, 803–811.
van Ooijen, J. W. (2004). MapQTL 5. Software for the mapping of quantitative trait loci in experimental populations of diploid species. Wageningen: Kyazma BV.
Voelz, R., von Lyncker, L., Baumann, N., Dresselhaus, T., Sprunck, S., and Gross-Hardt, R. (2012). LACHESIS-dependent egg-cell signaling regulates the development of female gametophytic cells. Development 139, 498–502.
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78.
Wang, S., Chang, Y., Guo, J., Zeng, Q., Ellis, B. E., and Chen, J. G. (2011). Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 6:e23896. doi: 10.1371/journal.pone.0023896
Wang, X. T., Yuan, C., Yuan, T. T., and Cui, S. J. (2012). The Arabidopsis LFR gene is required for the formation of anther cell layers and normal expression of key regulatory genes. Mol. Plant 5, 993–1000.
Xiao, C., Chen, F., Yu, X., Lin, C., and Fu, Y. F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50.
Xing, D., Zhao, H., Xu, R., and Li, Q. Q. (2008). Arabidopsis PCFS4, a homologue of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J. 54, 899–910.
Xu, R., and Li, Q. Q. (2003). A RING-H2 zinc-finger protein gene RIE1 is essential for seed development in Arabidopsis. Plant Mol. Biol. 53, 37–50.
Keywords: quantitative trait loci, flowering time, dichogamy, nut maturity time, Corylus avellana
Citation: Valentini N, Portis E, Botta R, Acquadro A, Pavese V, Cavalet Giorsa E and Torello Marinoni D (2021) Mapping the Genetic Regions Responsible for Key Phenology-Related Traits in the European Hazelnut. Front. Plant Sci. 12:749394. doi: 10.3389/fpls.2021.749394
Received: 29 July 2021; Accepted: 24 November 2021;
Published: 23 December 2021.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Nahla Victor Bassil, National Clonal Germplasm Repository, Agricultural Research Service, United States Department of Agriculture, United StatesCopyright © 2021 Valentini, Portis, Botta, Acquadro, Pavese, Cavalet Giorsa and Torello Marinoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Botta, cm9iZXJ0by5ib3R0YUB1bml0by5pdA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.