- 1Institute of Crop Sciences, National Wheat Improvement Center, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Hebei Laboratory of Crop Genetics and Breeding, Institute of Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang, China
- 3Shandong Provincial Key Laboratory of Dryland Farming Technology, College of Agronomy, Qingdao Agricultural University, Qingdao, China
- 4International Maize and Wheat Improvement Center (CIMMYT) China Office, Beijing, China
Preharvest sprouting (PHS) significantly reduces grain yield and quality. Identification of genetic loci for PHS resistance will facilitate breeding sprouting-resistant wheat cultivars. In this study, we constructed a genetic map comprising 1,702 non-redundant markers in a recombinant inbred line (RIL) population derived from cross Yangxiaomai/Zhongyou9507 using the wheat 15K single-nucleotide polymorphism (SNP) assay. Four quantitative trait loci (QTL) for germination index (GI), a major indicator of PHS, were identified, explaining 4.6–18.5% of the phenotypic variances. Resistance alleles of Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-7BL were from Yangxiaomai, and Zhongyou9507 contributed a resistance allele in Qphs.caas-4AL. No epistatic effects were detected among the QTL, and combined resistance alleles significantly increased PHS resistance. Sequencing and linkage mapping showed that Qphs.caas-3AL and Qphs.caas-3DL corresponded to grain color genes Tamyb10-A and Tamyb10-D, respectively, whereas Qphs.caas-4AL and Qphs.caas-7BL were probably new QTL for PHS. We further developed cost-effective, high-throughput kompetitive allele-specific PCR (KASP) markers tightly linked to Qphs.caas-4AL and Qphs.caas-7BL and validated their association with GI in a test panel of cultivars. The resistance alleles at the Qphs.caas-4AL and Qphs.caas-7BL loci were present in 72.2 and 16.5% cultivars, respectively, suggesting that the former might be subjected to positive selection in wheat breeding. The findings provide not only genetic resources for PHS resistance but also breeding tools for marker-assisted selection.
Introduction
Preharvest sprouting (PHS) refers to the germination of physiologically mature grains in spikes before harvest under rainy weather or humid environment (Groos et al., 2002). PHS is a major problem in cereal production and causes losses in seed vitality, yield, and quality (Xu et al., 2019). Wheat (Triticum aestivum L.) is one of the most important staple crops. The average annual loss of wheat caused by PHS exceeds $1 billion worldwide (Shao et al., 2018). Identification of genetic loci for PHS should be helpful for breeding resistant wheat cultivars.
Preharvest sprouting is a complex trait influenced by genetic and environmental factors (Barrero et al., 2015; Wang et al., 2019). Seed dormancy, an adaptive trait that prevents seeds from germinating, even under favorable conditions, is a major genetic factor for PHS (Née et al., 2017). Germination index (GI) is a common parameter to quantify genetic mechanisms underlying seed dormancy and PHS (Barrero et al., 2015). Some non-dormancy factors, such as spike erectness, spike and awn structure, and openness of florets, are also associated with PHS (Zhu et al., 2019).
Molecular markers have an important role in determining the genetic basis of agronomic traits in wheat (Collard and Mackill, 2008). Markers tightly linked with genes for PHS resistance can be used in marker-assisted selection (MAS). Using diverse mapping populations, many quantitative trait loci (QTL) for PHS resistance or seed dormancy on all 21 wheat chromosomes have been reported (Cao et al., 2016; Lin et al., 2016; Yang et al., 2019; Zhu et al., 2019; Tai et al., 2021). Among them, QTL on group 3 chromosomes and chromosome 4AL have major effects on PHS (Mori et al., 2005; Chen et al., 2008; Ogbonnaya et al., 2008; Shao et al., 2018; Vetch et al., 2019). A few genes for PHS in wheat were also isolated by map-based cloning. For example, TaPHS1, an MFT homolog, is the causal gene in Qphs.pseru-3AS (Liu et al., 2013; Jiang et al., 2018; Wang et al., 2020); Tamyb10 genes at the R loci on chromosomes 3A, 3B, and 3D control grain coat color by regulating the accumulation of anthocyanins (Himi and Noda, 2005; Wang et al., 2016; Mares and Himi, 2021); Mitogen-activated protein kinase kinase 3 (MKK3) is the causal gene of Phs1-4AL for seed dormancy in wheat (Torada et al., 2016; Martinez et al., 2020); and tandem duplicated plasma membrane protein genes (PM19) have been validated as candidates for a major dormancy QTL on chromosome 4AL through transcriptome analysis (Barrero et al., 2015; Shorinola et al., 2016). Homology-based cloning approaches were also used to identify PHS-related genes, such as TaSdr (Zhang et al., 2014, 2017), Vp-1 (McCarty et al., 1991; Yang et al., 2007, 2013; Feng et al., 2017; Zhou et al., 2017), Qsd1 (Sato et al., 2016; Onishi et al., 2017), and DOG1 (Ashikawa et al., 2010; Nakabayashi et al., 2012; Rikiishi and Maekawa, 2014; Nishimura et al., 2018).
Yangxiaomai, a red-seeded Chinese landrace, has a high level of PHS resistance, whereas white-seeded Zhongyou9507 with good processing quality is susceptible to PHS. The objectives of this study are to mine QTL for PHS resistance in a recombinant inbred line (RIL) population derived from a Yangxiaomai/Zhongyou9507 cross and to develop breeding-friendly markers for selection of PHS-resistant varieties.
Materials and Methods
Plant Materials and Field Trials
The parents Yangxiaomai and Zhongyou9507 and 194 F6 RILs were planted at Beijing and Shijiazhuang (Hebei Province) in the 2011–2012 cropping season and at Gaoyi (Hebei Province) and Xinxiang (Henan Province) in the 2019–2020 cropping season. Field experiments were arranged in randomized complete blocks with three replications. Each plot was 1 m single row in which 30 seeds were sown. A panel of 101 wheat cultivars (Zhang et al., 2017) was used to determine the genetic effects of the QTL of interest.
Evaluation of PHS Resistance
The GI was used as an indicator of PHS. Five spikes were harvested from each plot at physiological maturity characterized by loss of green color from the spike (Liu et al., 2013). The harvested spikes were air-dried for 2 days at room temperature, hand-threshed to avoid damage to embryos, and then stored in a refrigerator at −20°C to maintain dormancy until phenotyping (Zhang et al., 2017). Seeds were sterilized with 1% (V/V) of NaClO for 20 min, followed by three rinses with sterile water. Notably, 100 healthy seeds of each line were incubated in a 90 mm Petri dish containing a filter paper and 8 ml of distilled water at 20°C for 7 days. Germinated seeds were counted every day and removed. GI was calculated according to the following formula (Walker-Simmons, 1988): , where n1, n2, …, n7 are the number of seeds germinated on the first, second, and subsequent days until the seventh day.
Statistical Analyses
Phenotypic correlation coefficients among environments, the best linear unbiased prediction (BLUP) values, ANOVA, and t-tests were carried out using SAS 9.4 software (SAS Institute Inc., Cary, NC, USA). Broad-sense heritability (H2) for PHS was calculated using the following formula: H2 /(/e+ /re), where , , and are the variances of genotype, genotype-environment interaction, and residual error, respectively, r is the number of replicates, and e is the number of environments (Nyquist and Baker, 1991).
Genotyping and Linkage Map Construction
The 194 RILs and parents were genotyped with the wheat 15K single-nucleotide polymorphism (SNP) chips containing 13,947 SNP markers at China Golden Marker (Beijing) Biotech Co., Ltd. (http://www.cgmb.com.cn/). To reduce the impact of low-quality SNPs on mapping results, SNP data were processed as follows: (1) Heterozygous loci were treated as missing data, and (2) SNPs with low minor allele frequencies (<0.3) and missing values (>0.2) were excluded using Tassel version 5.0 (Bradbury et al., 2007). Redundant markers were eliminated by the BIN function in QTL IciMapping version 4.2 (Meng et al., 2015). Joinmap version 4.0 was used for linkage map construction (Stam, 1993), and genetic distances between markers were calculated according to the Kosambi mapping function (Kosambi, 1943).
QTL Analysis
Composite interval mapping (CIM) was used to search QTL of phenotypic traits from each environment and BLUP value by Windows QTL Cartographer version 2.5 (Zeng, 1994; Wang et al., 2012). Significant QTLs were identified if the logarithm of odds (LOD) values were more than the threshold of 2.5 (Yan et al., 2006). According to International Wheat Genome Sequencing Consortium (IWGSC) RefSeq 1.0 [(International Wheat Genome Sequencing Consortium (IWGSC), 2018) http://plants.ensembl.org/index.html], the physical positions of QTL were figured out by the closely linked flanking markers. The genetic maps of QTL were drawn using MapChart version 2.3 (Voorrips, 2002). The analysis of epistatic effects among the QTL was performed using IciMapping version 4.2.
KASP Marker Development and Validation
Kompetitive allele-specific PCR primers (Supplementary Table 5) were designed using PolyMarker (Ramirez-Gonzalez et al., 2015). Primer mixture was prepared with 46 μl of H2O, 30 μl of common primer (100 μM), and 12 μl of each tailed primer (100 μM). PCR was performed in a 384-well plate, and each reaction of ~3 μl comprising 20–30 ng of genomic DNA, 1.5 μl of 2× KASP master mix (V4.0, LGC Genomics, Hoddesdon, UK), 0.0336 μl of primer mixture, and 1.5 μl of H2O. Thermal cycling profile of PCR consisted of hot start at 95°C for 15 min, 10 touchdown cycles (95°C for 20 s and touchdown at 65 and −1°C per cycle for 25 s), and followed by 35 additional cycles (95°C for 20 s and 57°C for 60 s). The 384-well optically clear plates were read on PHERAstarplus SNP (BMG Labtech GmbH, Ortenberg, Germany), and data analysis was carried out using KlusterCaller (LGC, Hoddesdon, UK).
Results
Phenotypic Evaluation
The parents Yangxiaomai and Zhongyou9507 and RILs were evaluated for PHS resistance in four environments. The phenotypes of seed germination in parents Yangxiaomai and Zhongyou9507 were depicted in Supplementary Figure 1. Yangxiaomai had a significantly lower GI (4.3%) than Zhongyou9507 (72.3%) across environments (Supplementary Figure 2). GI for the RIL population showed continuous variation, indicating polygenic inheritance (Supplementary Figure 2). The GI frequencies were skewed toward resistance, suggesting the presence of major genetic loci. GI was significantly correlated among environments with correlation coefficients of 0.53–0.73 (Supplementary Table 1). ANOVA indicated that genotypes and environments, as well as their interactions, had significant effects on GI (Supplementary Table 2). The broad-sense heritability of GI was high (0.88) across environments, denoting that GI variation was mainly determined by genotypes.
Linkage Map Construction and QTL Analysis
The RIL population was genotyped by 15K SNP chips, and 4,515 polymorphic markers were used to construct a genetic map with 1,702 bin markers, spanning 2,630.9 cM on 21 wheat chromosomes (Supplementary Table 3 and Supplementary Figure 3). The average linkage group was 125.3 cM with an average marker interval of 1.6 cM. Overall, 1,743 (38.6%), 1,750 (38.8%), and 1,022 (22.6%) markers were mapped to the A, B, and D sub-genomes with average marker densities of 1.5, 1.2, and 2.2 cM, respectively (Supplementary Table 3 and Supplementary Figure 3).
Four QTLs for PHS were detected by CIM on the linkage groups 3AL (Qphs.caas-3AL), 3DL (Qphs.caas-3DL), 4AL (Qphs.caas-4AL), and 7BL (Qphs.caas-7BL) (Table 1 and Figure 1). Alleles for resistance to PHS on chromosome arms 3AL, 3DL, and 7BL loci were from Yangxiaomai, whereas the resistance allele on 4AL was contributed by Zhongyou9507. Qphs.caas-3DL was identified across all four environments and explained 8.9–18.5% of the phenotypic variances; Qphs.caas-3AL and Qphs.caas-4AL were detected in three of four environments, explaining 10.5–13.5 and 4.6–10.6% of the phenotypic variances, respectively; and Qphs.caas-7BL accounting for 5.0–6.7% of the phenotypic variances was detected in two environments.
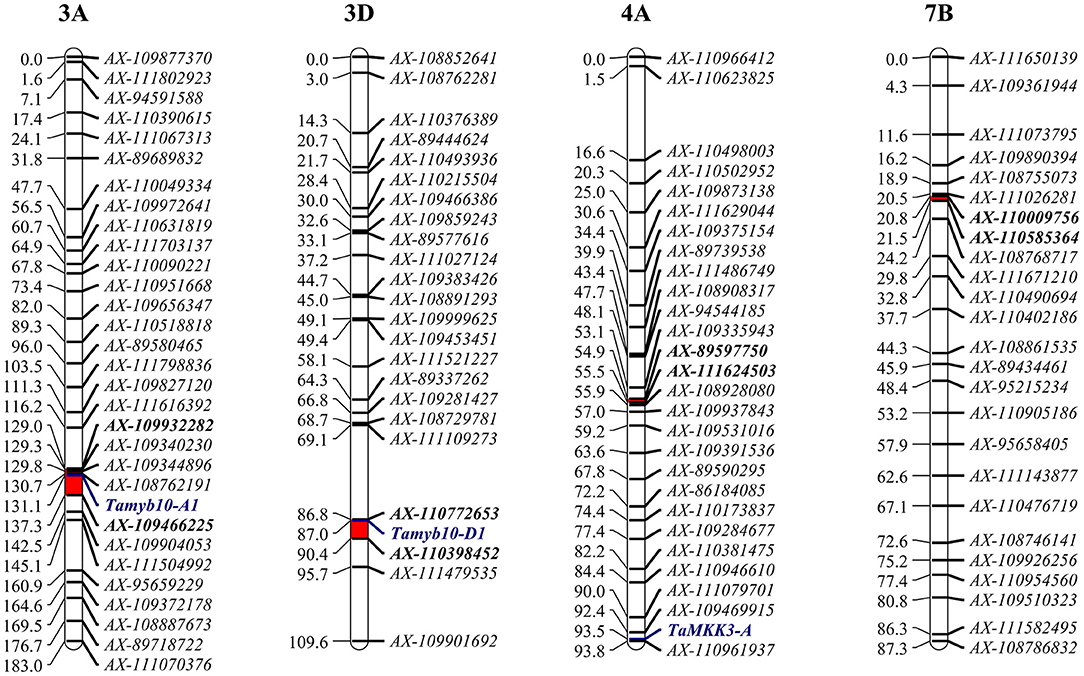
Figure 1. Genetic mapping of Qphs.caas-3AL, Qphs.caas-3DL, Qphs.caas-4AL, and Qphs.caas-7BL in the Yangxiaomai/Zhongyou9507 recombinant inbred line (RIL) population. Target regions of the quantitative trait loci (QTL) are indicated as red bars; gene-specific markers are shown in blue script; and flanking markers are shown in bold.
Combinational Effects of the Stable QTL for PHS Resistance
Quantitative trait loci for a given trait detected in more than one-half of tested environments can be considered stable genetic loci (Cao et al., 2020). The QTL Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-4AL fulfilled that criterion. To confirm their genetic effects on PHS, the population was classified into eight groups based on the closest flanking SNPs for each QTL (Supplementary Table 4). Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-4AL were temporarily designated as the loci 1, 2, and 3, respectively, and R and S represented resistance and susceptible alleles, respectively. The GI values of eight groups (i.e., 1R2R3R, 1R2R3S, 1R2S3R, 1S2R3R, 1R2S3S, 1S2R3S, 1S2S3R, and 1S2S3S) were compared across four environments (Figure 2 and Supplementary Table 4).
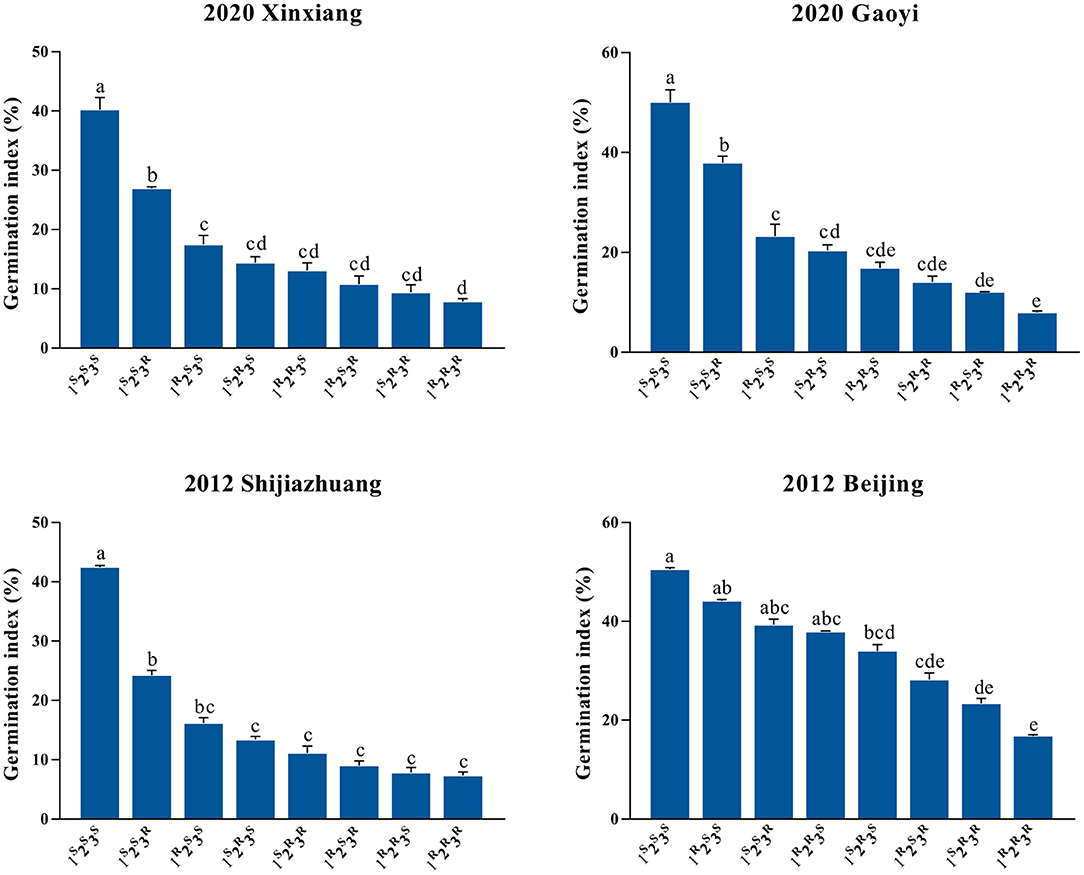
Figure 2. Distributions of germination index (GI) among eight genotypic combinations of Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-4AL grown in four environments. The x-axis shows the genotypic groups, and the y-axis indicates GI (%). The numbers 1, 2, and 3 represent Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-4AL, respectively; the superscript letters R and S represent resistance and susceptible alleles, respectively. Genotypes with different letters indicate significant differences (P < 0.05) in GI, and those with the same letters show no significant differences (P > 0.05). Error bar, standard error of each group mean.
The eight groups were ranked according to the GI in four environments and BLUP values. In general, more resistance alleles conferred lower GI, demonstrating cumulative effects of resistance alleles at the three loci (Figure 2). RILs with genotype 1R2R3R had the lowest GI, and those with 1S2S3S exhibited the highest GI across all environments. However, the GI of RILs with 1R2R3S were higher than those with 1S2R3S in Beijing 2012, suggesting that the genetic effect of locus 1, Qphs.caas-3AL, was significantly affected by the environment in some cases. No epistatic effects were detected among the QTL. Regression analysis also showed that the lines carrying more resistance alleles had higher PHS resistance in individual environments and the BLUP data (Supplementary Figure 4). Thus, the pyramiding of resistance alleles was effective in improving PHS resistance.
Relationship of Qphs.caas-3AL and Qphs.caas-3DL With Tamyb10
According to IWGSC RefSeq 1.0, Qphs.caas-3AL and Qphs.caas-3DL were delimited in the intervals of 700.4–709.2 Mb and 570.2–575.1 Mb, respectively, based on their flanking markers. This placed PHS-related genes Tamyb10-A1 (~703.9 Mb) and Tamyb10-D1 (~570.8 Mb), in the regions of Qphs.caas-3AL and Qphs.caas-3DL, respectively (Himi et al., 2011). To confirm the genetic relationship of the two genes with Qphs.caas-3AL and Qphs.caas-3DL, we re-genotyped the Yangxiaomai/Zhongyou9507 RIL population using a KASP marker for Tamyb10-A1 and an STS marker for Tamyb10-D1 (Supplementary Table 5) (Himi et al., 2011; Wang et al., 2016). In these analyses, Tamyb10-A1 and Tamyb10-D1 were mapped to the genetic regions of Qphs.caas-3AL and Qphs.caas-3DL, respectively (Figure 1). Thus, Tamyb10-A1 and Tamyb10-D1 were likely causal genes in Qphs.caas-3AL and Qphs.caas-3DL, respectively.
Relationship Between Qphs.caas-4AL and Reported PHS Resistance Genes on Chromosome 4AL
TaMKK3-A was reported as a major gene controlling seed dormancy on chromosome 4AL (Torada et al., 2016). Based on the IWGSC RefSeq 1.0, TaMKK3-A is located at the site of ~609.1 Mb (GenBank accession number: LC091368.1) (Liton et al., 2021). Qphs.caas-4AL spans the interval of 489.1–532.2 Mb according to flanking markers AX-89597750 and AX-111624503, suggesting that TaMKK3-A is not in the target region of Qphs.caas-4AL. We sequenced the TaMKK3-A gene in Yangxiaomai and Zhongyou9507 to confirm the relationship between TaMKK3-A and Qphs.caas-4AL. Sequence analysis showed that TaMKK3-A had no polymorphic sites between two parents in all exons, but three SNPs were detected in the introns (Supplementary Figure 5). A KASP marker KASP-6464 was developed for TaMKK3-A. Linkage mapping showed that KASP-6464 was out of the target region of Qphs.caas-4AL (Figure 1). Association analysis also indicated that TaMKK3-A had no significant effect on GI in three environments (Supplementary Table 6). Therefore, TaMKK3-A was not a candidate gene in Qphs.caas-4AL.
PM19-A1 is a second PHS-related gene in chromosome 4AL. However, it is positioned at ~604.1 Mb, which is far from Qphs.caas-4AL (489.1–532.2 Mb) according to the IWGSC RefSeq 1.0. We also compared the sequences of PM19-A1 between Yangxiaomai and Zhongyou9507 and found no polymorphic sites in the open reading frame and the previously reported 18 bp indel in the promoter (Barrero et al., 2015; Shorinola et al., 2016, 2017). Therefore, PM19-A1 was not the causal gene in Qphs.caas-4AL.
No genes related to PHS were isolated on 7B so far, so no candidate genes could be used to perform sequencing and genetic mapping analyses for Qphs.caas-7BL. Qphs.caas-7BL is defined in the interval of 522.6–529.7 Mb based on its flanking markers according to IWGSC RefSeq 1.0. QTL related to PHS, which have been reported on 7B, were summarized in Supplementary Table 7. The location of Qphs.caas-7BL is different from those of the previously reported QTL based on their flanking markers.
Genetic Effects of Qphs.caas-4AL and Qphs.caas-7BL on GI in a Panel of Cultivars
The causal genes of Qphs.caas-4AL and Qphs.caas-7BL remained unknown although they could explain 4.6–10.6% of the phenotypic variances. To further decipher the importance of Qphs.caas-4AL and Qphs.caas-7BL, we attempted to identify their genetic effects in a panel of cultivars. An SNP AX-89597750 closely linked with Qphs.caas-4AL was converted to a KASP marker. The KASP marker was mapped at the position of AX-89597750, indicating that it could act as a closely linked marker of Qphs.caas-4AL. We detected the genetic effect of Qphs.caas-4AL on GI using the KASP marker in the cultivar panel (Supplementary Table 8). Genotypes with the resistance allele had significantly lower GI than those with the susceptible allele (Table 2). Moreover, a majority of genotypes (72.2%) possessed the resistance allele of Qphs.caas-4AL, suggesting that it had been subjected to selection in wheat breeding (Table 2). Another KASP marker was developed based on SNP AX-9496498 closely linked to Qphs.caas-7BL. The KASP marker was mapped to the target region of Qphs.caas-7BL in the mapping population. Like Qphs.caas-4AL, Qphs.caas-7BL was also significantly associated with PHS resistance (Table 2), but only 16.5% of cultivars carried the resistance allele.
Discussion
Grain Color Is a Major Factor Modulating PHS
The red-grain Yangxiaomai and white-grain Zhongyou9507 have a relatively low and higher GI, respectively. In this study, we confirmed that Qphs.caas-3AL and Qphs.caas-3DL co-localized with Tamyb10-A1 and Tamyb10-D1, respectively, at the R loci for grain color (Himi et al., 2011; Lang et al., 2021; Mares and Himi, 2021). Wang et al. (2016) also observed that Tamyb10-D1 was significantly (P < 0.001) associated with GI in a natural population. Thus, grain color is probably regulated by Tamyb10 alleles in Qphs.caas-3AL and Qphs.caas-3DL, which also cause significant differences in GI between Yangxiaomai and Zhongyou9507. Pleiotropic QTL for grain color and PHS resistance on chromosomes 3AL and 3DL were identified in a genome-wide association study conducted by Lin et al. (2016). These findings also confirm that grain color has a great effect on PHS resistance in wheat breeding.
Qphs.caas-4AL Has Potential Value for Wheat Breeding
Qphs.caas-4AL, as a stable QTL, accounted for 4.6–10.6% of the phenotypic variances. Although quite a few QTL for wheat PHS resistance on chromosome 4A have also been reported (Kato et al., 2001; Mares et al., 2005; Torada et al., 2005; Imtiaz et al., 2008; Rasul et al., 2009; Singh et al., 2010; Kulwal et al., 2012; Cabral et al., 2014; Jiang et al., 2015; Cao et al., 2016; Zhou et al., 2017; Martinez et al., 2018; Zuo et al., 2019; Liton et al., 2021), Qphs.caas-4AL appears to be unique based on genetic mapping and physical locations of the flanking SNPs according to IWGSC RefSeq 1.0 (Supplementary Table 9). QTL pyramiding plays an important role in breeding, and resistance allele combinations of QTL for PHS have been reported previously (Imtiaz et al., 2008; Shao et al., 2018; Liton et al., 2021). In this study, we identified that the RILs combining resistance alleles in Qphs.caas-3AL, Qphs.caas-3DL, and Qphs.caas-4AL had the lowest GI (Figure 2). Qphs.caas-4AL also improved resistance to PHS in the absence of the alleles for red-grain color (Figure 2 and Supplementary Table 4). This indicated that Qphs.caas-4AL could function independently of grain color. We converted an SNP tightly linked to Qphs.caas-4AL into a KASP marker. Genotyping by the KASP marker showed that most of the cultivars (72.2%) carried the resistance allele in Qphs.caas-4AL in the test panel (Table 2), indicating that the resistance allele of Qphs.caas-4AL might undergo positive selection in breeding programs. Qphs.caas-4AL is also significantly associated with GI (Table 2). Thus, the KASP marker will be useful for MAS to improve PHS tolerance in wheat. Grain color is an important parameter for wheat appearance quality. Red-grain wheat usually has high resistance to PHS but is adverse to make Chinese traditional food, such as steamed bread and noodles (Fakthongphan et al., 2016; Shao et al., 2018). Thus, Qphs.caas-4AL is a better choice for improvement of PHS than Qphs.caas-3AL and Qphs.caas-3DL at least in Chinese wheat breeding. Overall, these findings indicate that Qphs.caas-4AL is a valuable genetic locus for PHS in wheat breeding.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author Contributions
LL and SC wrote the manuscript. LL, YiZ, ML, DX, XT, JS, and XL performed the experiments. SC and LL analyzed the data. YoZ, LX, and DW participated in the field trials. SC and YaZ designed the experiments. XX and ZH assisted in writing the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 91935304 and 31571663), the National Key Research and Development Programs of China (Grant No. 2016YFD0100502), and the CAAS Science and Technology Innovation Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Prof. R. A. McIntosh, Plant Breeding Institute, University of Sydney, for critical review of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.749206/full#supplementary-material
Supplementary Figure 1. Seed germination of Yangxiaomai and Zhongyou9507. YXM, Yangxiaomai; ZY9507, Zhongyou9507.
Supplementary Figure 2. Distributions of GI in the Yangxiaomai/Zhongyou9507 RIL population in four environments. Black arrows point to the GI of the parents. GI, germination index; RIL, recombinant inbred line; YXM, Yangxiaomai; ZY9507, Zhongyou9507.
Supplementary Figure 3. Genetic map of the Yangxiaomai/Zhongyou9507 RIL population determined from 15K SNP arrays. RIL, recombinant inbred line; SNP, single-nucleotide polymorphism.
Supplementary Figure 4. Linear regression between the resistance allele number of the QTL and GI in four environments and BLUP values in the Yangxiaomai/Zhongyou9507 population. The QTL include Qphs.caas-3AL, Qphs.caas-3DL, Qphs.caas-4AL, and Qphs.caas-7BL. QTL, quantitative trait loci; GI, germination index; BLUP, best linear unbiased prediction.
Supplementary Figure 5. Schematic of the open reading frame of TaMKK3-A and its polymorphic sites between Yangxiaomai (YXM) and Zhongyou9507 (ZY9507). Bold bars and thin lines indicate exons and introns, respectively.
Supplementary Table 1. Correlation coefficients of GI from the Yangxiaomai/Zhongyou9507 RIL population among four environments.
Supplementary Table 2. Variance analysis for GI of the Yangxiaomai/Zhongyou9507 RIL population.
Supplementary Table 3. Information of genetic map construction for Yangxiaomai/Zhongyou9507 RIL population using 15K SNP chips.
Supplementary Table 4. GI of eight genotypic groups defined by three stable QTL in the Yangxiaomai/Zhongyou9507 RIL population.
Supplementary Table 5. Primers used in this study.
Supplementary Table 6. Association analysis of TaMKK3-A and GI in the Yangxiaomai/Zhongyou9507 RIL population.
Supplementary Table 7. Information of reported QTL for PHS on the chromosome 7B.
Supplementary Table 8. Genotypes of Qphs.caas-4AL and Qphs.caas-7BL and GI of 101 wheat cultivars.
Supplementary Table 9. Information of reported QTL for PHS on the chromosome 4A.
References
Ashikawa, I., Abe, F., and Nakamura, S. (2010). Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 179, 536–542. doi: 10.1016/j.plantsci.2010.08.002
Barrero, J. M., Cavanagh, C., Verbyla, K. L., Tibbits, J. F., Verbyla, A. P., Huang, B. E., et al. (2015). Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 16:93. doi: 10.1186/s13059-015-0665-6
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cabral, A. L., Jordan, M. C., McCartney, C. A., You, F. M., Humphreys, D. G., MacLachlan, R., et al. (2014). Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 14:340. doi: 10.1186/s12870-014-0340-1
Cao, L., Hayashi, K., Tokui, M., Mori, M., Miura, H., and Onishi, K. (2016). Detection of QTLs for traits associated with pre-harvest sprouting resistance in bread wheat (Triticum aestivum L.). Breed. Sci. 66, 260–270. doi: 10.1270/jsbbs.66.260
Cao, S., Xu, D., Hanif, M., Xia, X., and He, Z. (2020). Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 133, 1811–1823. doi: 10.1007/s00122-020-03562-8
Chen, C., Cai, S., and Bai, G. (2008). A major QTL controlling seed dormancy and pre-harvest sprouting resistance on chromosome 4A in a Chinese wheat landrace. Mol. Breed. 21, 351–358. doi: 10.1007/s11032-007-9135-5
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Phil. Trans. R. Soc. B. 363, 557–572. doi: 10.1098/rstb.2007.2170
Fakthongphan, J., Bai, G., St Amand, P., Graybosch, R. A., and Baenziger, P. S. (2016). Identification of markers linked to genes for sprouting tolerance (independent of grain color) in hard white winter wheat (HWWW). Theor. Appl. Genet. 129, 419–430. doi: 10.1007/s00122-015-2636-4
Feng, Y., Qu, R., Liu, S., and Yang, Y. (2017). Rich haplotypes of Viviparous-1 in Triticum aestivum subsp. spelta with different abscisic acid sensitivities. J. Sci. Food Agric. 97, 497–504. doi: 10.1002/jsfa.7751
Groos, C., Gay, G., Perretant, M. R., Gervais, L., Bernard, M., Dedryver, F., et al. (2002). Study of the relationship between pre-harvest sprouting and grain color by quantitative trait loci analysis in a white × red grain bread-wheat cross. Theor. Appl. Genet. 104, 39–47. doi: 10.1007/s001220200004
Himi, E., Maekawa, M., Miura, H., and Noda, K. (2011). Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theor. Appl. Genet. 122, 1561–1576. doi: 10.1007/s00122-011-1555-2
Himi, E., and Noda, K. (2005). Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica 143, 239–242. doi: 10.1007/s10681-005-7854-4
Imtiaz, M., Ogbonnaya, F. C., Oman, J., and van Ginkel, M. (2008). Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178, 1725–1736. doi: 10.1534/genetics.107.084939
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Jiang, H., Zhao, L., Chen, X., Cao, J., Wu, Z., Liu, K., et al. (2018). A novel 33-bp insertion in the promoter of TaMFT-3A is associated with pre-harvest sprouting resistance in common wheat. Mol. Breed. 38:69. doi: 10.1007/s11032-018-0830-1
Jiang, Y., Wang, J., Luo, W., Wei, Y., Qi, P., Liu, Y., et al. (2015). Quantitative trait locus mapping for seed dormancy in different post-ripening stages in a Tibetan semi-wild wheat (Triticum aestivum ssp. tibetanum Shao). Euphytica 203, 557–567. doi: 10.1007/s10681-014-1266-2
Kato, K., Nakamura, W., Tabiki, T., Miura, H., and Sawada, S. (2001). Detection of loci controlling seed dormancy on group 4 chromosomes of wheat and comparative mapping with rice and barley genomes. Theor. Appl. Genet. 102, 980–985. doi: 10.1007/s001220000494
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugenic. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Kulwal, P., Ishikawa, G., Benscher, D., Feng, Z., Yu, L. X., Jadhav, A., et al. (2012). Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor. Appl. Genet. 125, 793–805. doi: 10.1007/s00122-012-1872-0
Lang, J., Fu, Y., Zhou, Y., Cheng, M., Deng, M., Li, M., et al. (2021). Myb10-D confers PHS-3D resistance to pre-harvest sprouting by regulating NCED in ABA biosynthesis pathway of wheat. New Phytol. 230, 1940–1952. doi: 10.1111/nph.17312
Lin, M., Zhang, D., Liu, S., Zhang, G., Yu, J., Fritz, A. K., et al. (2016). Genome-wide association analysis on pre-harvest sprouting resistance and grain color in U.S. winter wheat. BMC Genom. 17:794. doi: 10.1186/s12864-016-3148-6
Liton, M., McCartney, C. A., Hiebert, C. W., Kumar, S., Jordan, M. C., and Ayele, B. T. (2021). Identification of loci for pre-harvest sprouting resistance in the highly dormant spring wheat RL4137. Theor. Appl. Genet. 134, 113–124. doi: 10.1007/s00122-020-03685-y
Liu, S., Sehgal, S. K., Li, J., Lin, M., Trick, H. N., Yu, J., et al. (2013). Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195, 263–273. doi: 10.1534/genetics.113.152330
Mares, D., and Himi, E. (2021). The role of TaMYB10-A1 of wheat (Triticum aestivum L.) in determining grain coat colour and dormancy phenotype. Euphytica 217:89. doi: 10.1007/s10681-021-02826-8
Mares, D., Mrva, K., Cheong, J., Williams, K., Watson, B., Storlie, E., et al. (2005). A QTL located on chromosome 4A associated with dormancy in white-and red-grained wheats of diverse origin. Theor. Appl. Genet. 111, 1357–1364. doi: 10.1007/s00122-005-0065-5
Martinez, S. A., Godoy, J., Huang, M., Zhang, Z., Carter, A. H., Campbell, K. A., et al. (2018). Genome-wide association mapping for tolerance to preharvest sprouting and low falling numbers in wheat. Front. Plant Sci. 9:141. doi: 10.3389/fpls.2018.00141
Martinez, S. A., Shorinola, O., Conselman, S., See, D., Skinner, D. Z., Uauy, C., et al. (2020). Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype. Theor. Appl. Genet. 133, 719–736. doi: 10.1007/s00122-019-03503-0
McCarty, D. R., Hattori, T., Carson, C. B., Vasil, V., Lazar, M., and Vasil, I. K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. doi: 10.1016/0092-8674(91)90436-3
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Mori, M., Uchino, N., Chono, M., Kato, K., and Miura, H. (2005). Mapping QTLs for grain dormancy on wheat chromosome 3A and the group 4 chromosomes, and their combined effect. Theor. Appl. Genet. 110, 1315–1323. doi: 10.1007/s00122-005-1972-1
Nakabayashi, K., Bartsch, M., Xiang, Y., Miatton, E., Pellengahr, S., Yano, R., et al. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. doi: 10.1105/tpc.112.100214
Née, G., Xiang, Y., and Soppe, W. J. (2017). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. doi: 10.1016/j.pbi.2016.09.002
Nishimura, N., Tsuchiya, W., Moresco, J. J., Hayashi, Y., Satoh, K., Kaiwa, N., et al. (2018). Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 9:2132. doi: 10.1038/s41467-018-04437-9
Nyquist, W. E., and Baker, R. J. (1991). Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 10, 235–322. doi: 10.1080/07352689109382313
Ogbonnaya, F. C., Imtiaz, M., Ye, G., Hearnden, P. R., Hernandez, E., Eastwood, R. F., et al. (2008). Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 116, 891–902. doi: 10.1007/s00122-008-0712-8
Onishi, K., Yamane, M., Yamaji, N., Tokui, M., Kanamori, H., Wu, J., et al. (2017). Sequence differences in the seed dormancy gene Qsd1 among various wheat genomes. BMC Genom. 18:497. doi: 10.1186/s12864-017-4046-2
Ramirez-Gonzalez, R. H., Uauy, C., and Caccamo, M. (2015). Poly-Marker: a fast polyploid primer design pipeline. Bioinformatics 31:2038–2039. doi: 10.1093/bioinformatics/btv069
Rasul, G., Humphreys, D. G., Brûlé-Babel, A., McCartney, C. A., Knox, R. M., and Somers, D. J. (2009). Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross ‘RL4452/AC domain'. Euphytica 168, 363–378. doi: 10.1007/s10681-009-9934-3
Rikiishi, K., and Maekawa, M. (2014). Seed maturation regulators are related to the control of seed dormancy in wheat (Triticum aestivum L.). PLoS ONE 9:e107618. doi: 10.1371/journal.pone.0107618
Sato, K., Yamane, M., Yamaji, N., Kanamori, H., Tagiri, A., Schwerdt, J. G., et al. (2016). Alanine aminotransferase controls seed dormancy in barley. Nat. Commun. 7:11625. doi: 10.1038/ncomms11625
Shao, M., Bai, G., Rife, T. W., Poland, J., Lin, M., Liu, S., et al. (2018). QTL mapping of pre-harvest sprouting resistance in a white wheat cultivar Danby. Theor. Appl. Genet. 131, 1683–1697. doi: 10.1007/s00122-018-3107-5
Shorinola, O., Balcárková, B., Hyles, J., Tibbits, J., Hayden, M. J., Holušova, K., et al. (2017). Haplotype analysis of the pre-harvest sprouting resistance locus Phs-A1 reveals a causal role of TaMKK3-A in global germplasm. Front. Plant Sci. 8:1555. doi: 10.3389/fpls.2017.01555
Shorinola, O., Bird, N., Simmonds, J., Berry, S., Henriksson, T., Jack, P., et al. (2016). The wheat Phs-A1 pre-harvest sprouting resistance locus delays the rate of seed dormancy loss and maps 0.3 cM distal to the PM19 genes in UK germplasm. J. Exp. Bot. 67, 4169–4178. doi: 10.1093/jxb/erw194
Singh, R., Matus-Cádiz, M., Båga, M., Hucl, P., and Chibbar, R. N. (2010). Identification of genomic regions associated with seed dormancy in white-grained wheat. Euphytica 174, 391–408. doi: 10.1007/s10681-010-0137-8
Stam, P. (1993). Construction of integrated genetic linkage maps by means of a new computer package: JOINMAP. Plant J. 3, 739–744. doi: 10.1111/j.1365-313X.1993.00739.x
Tai, L., Wang, H., Xu, X., Sun, W., Chen, K., Ju, L., et al. (2021). Cereal pre-harvest sprouting: a global agricultural disaster regulated by complex genetic and biochemical mechanisms. J. Exp. Bot. 72, 2857–2876. doi: 10.1093/jxb/erab024
Torada, A., Ikeguchi, S., and Koike, M. (2005). Mapping and validation of PCR-based markers associated with a major QTL for seed dormancy in wheat. Euphytica 143, 251–255. doi: 10.1007/s10681-005-7872-2
Torada, A., Koike, M., Ogawa, T., Takenouchi, Y., Tadamura, K., Wu, J., et al. (2016). A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr. Biol. 26, 782–787. doi: 10.1016/j.cub.2016.01.063
Vetch, J. M., Stougaard, R. N., Martin, J. M., and Giroux, M. J. (2019). Revealing the genetic mechanisms of pre-harvest sprouting in hexaploidy wheat (Triticum aestivum L.). Plant Sci. 281, 180–185. doi: 10.1016/j.plantsci.2019.01.004
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Walker-Simmons, M. (1988). Enhancement of ABA responsiveness in wheat embryos by high temperature. Plant Cell Environ. 11, 769–775. doi: 10.1111/j.1365-3040.1988.tb01161.x
Wang, D., Pang, Y., Dong, L., Li, A., Kong, L., and Liu, S. (2020). Allelic impacts on pre-harvest sprouting resistance and favorable haplotypes in TaPHS1 of Chinese wheat accessions. Crop J. 8, 515–521. doi: 10.1016/j.cj.2019.12.003
Wang, S., Basten, C., and Zeng, Z. (2012). Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. Available online at: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wang, X., Liu, H., Liu, G., Mia, M. S., Siddique, K., and Yan, G. (2019). Phenotypic and genotypic characterization of near-isogenic lines targeting a major 4BL QTL responsible for pre-harvest sprouting in wheat. BMC Plant Biol. 19:348. doi: 10.1186/s12870-019-1961-1
Wang, Y., Wang, X., Meng, J., Zhang, Y., He, Z., and Yang, Y. (2016). Characterization of Tamyb10 allelic variants and development of STS marker for pre-harvest sprouting resistance in Chinese bread wheat. Mol. Breed. 36:148. doi: 10.1007/s11032-016-0573-9
Xu, F., Tang, J., Gao, S., Cheng, X., Du, L., and Chu, C. (2019). Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 100, 1036–1051. doi: 10.1111/tpj.14501
Yan, J., Tang, H., Huang, Y., Zheng, Y., and Li, J. (2006). Quantitative trait loci mapping and epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica 149, 121–131. doi: 10.1007/s10681-005-9060-9
Yang, J., Tan, C., Lang, J., Tang, H., Hao, M., Tan, Z., et al. (2019). Identification of qPHS.sicau-1B and qPHS.sicau-3D from synthetic wheat for pre-harvest sprouting resistance wheat improvement. Mol. Breed. 39:132. doi: 10.1007/s11032-019-1029-9
Yang, Y., Ma, Y., Xu, Z., Chen, X., He, Z., Yu, Z., et al. (2007). Isolation and characterization of Viviparous-1 genes in wheat cultivars with distinct ABA sensitivity and pre-harvest sprouting tolerance. J. Exp. Bot. 58, 2863–2871. doi: 10.1093/jxb/erm073
Yang, Y., Zhang, C., Liu, S., Sun, Y., Meng, J., and Xia, L. (2013). Characterization of the rich haplotypes of Viviparous-1A in Chinese wheats and development of a novel sequence-tagged site marker for pre-harvest sprouting resistance. Mol. Breed. 33, 75–88. doi: 10.1007/s11032-013-9935-8
Zeng, Z. B. (1994). Precision mapping of quantitative trait loci. Genetics 136, 1457–1468. doi: 10.1093/genetics/136.4.1457
Zhang, Y., Miao, X., Xia, X., and He, Z. (2014). Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor. Appl. Genet. 127, 855–866. doi: 10.1007/s00122-014-2262-6
Zhang, Y., Xia, X., and He, Z. (2017). The seed dormancy allele TaSdr-A1a associated with pre-harvest sprouting tolerance is mainly present in Chinese wheat landraces. Theor. Appl. Genet. 130, 81–89. doi: 10.1007/s00122-016-2793-0
Zhou, Y., Tang, H., Cheng, M. P., Dankwa, K. O., Chen, Z. X., Li, Z. Y., et al. (2017). Genome-wide association study for pre-harvest sprouting resistance in a large germplasm collection of Chinese wheat landraces. Front. Plant Sci. 8:401. doi: 10.3389/fpls.2017.00401
Zhu, Y., Wang, S., Wei, W., Xie, H., Liu, K., Zhang, C., et al. (2019). Genome-wide association study of pre-harvest sprouting tolerance using a 90K SNP array in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 132, 2947–2963. doi: 10.1007/s00122-019-03398-x
Keywords: dormancy, KASP marker, QTL mapping, SNP chip, Triticum aestivum
Citation: Li L, Zhang Y, Zhang Y, Li M, Xu D, Tian X, Song J, Luo X, Xie L, Wang D, He Z, Xia X, Zhang Y and Cao S (2021) Genome-Wide Linkage Mapping for Preharvest Sprouting Resistance in Wheat Using 15K Single-Nucleotide Polymorphism Arrays. Front. Plant Sci. 12:749206. doi: 10.3389/fpls.2021.749206
Received: 29 July 2021; Accepted: 13 September 2021;
Published: 14 October 2021.
Edited by:
Lee Hickey, The University of Queensland, AustraliaReviewed by:
Jirui Wang, Sichuan Agricultural University, ChinaCheng Chang, Anhui Agricultural University, China
Copyright © 2021 Li, Zhang, Zhang, Li, Xu, Tian, Song, Luo, Xie, Wang, He, Xia, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuanghe Cao, Y2Fvc2h1YW5naGVAY2Fhcy5jbg==; Yan Zhang, emhhbmd5YW4wN0BjYWFzLmNu
†These authors have contributed equally to this work
 Lingli Li1†
Lingli Li1†