
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 January 2022
Sec. Plant Symbiotic Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.748542
The properties of plant rhizosphere are dynamic and heterogeneous, serving as different habitat filters for or against certain microorganisms. Herein, we studied the spatial distribution of bacterial communities in the rhizosphere of pepper plants treated with a disease-suppressive or non-suppressive soil. The bacterial richness was significantly (p < 0.05) higher in plants treated with the disease-suppressive soil than in those treated with the non-suppressive soil. Bacterial richness and evenness greatly differed between root parts, with decrease from the upper taproot to the upper fibrous root, the lower taproot, and the lower fibrous root. As expected, the bacterial community in the rhizosphere differed between suppressive and non-suppressive soil. However, the spatial variation (36%) of the bacterial community in the rhizosphere was much greater than that explained by soils (10%). Taxa such as subgroups of Acidobacteria, Nitrosospira, and Nitrospira were known to be selectively enriched in the upper taproot. In vitro Bacillus antagonists against Phytophthora capsici were also preferentially colonized in the taproot, while the genera such as Clostridium, Rhizobium, Azotobacter, Hydrogenophaga, and Magnetospirillum were enriched in the lower taproot or fibrous root. In conclusion, the spatial distribution of bacterial taxa and antagonists in the rhizosphere of pepper sheds light on our understanding of microbial ecology in the rhizosphere.
The abundance and diversity of microorganisms that inhabit the rhizosphere of plants play key roles in maintaining plant nutrients and health (Berg and Smalla, 2009; Mendes et al., 2011, 2013; Faria et al., 2020; Yu et al., 2020). Rhizospheric microbiomes may increase crop nutrients acquisition and resistance to environmental stresses (Ahkami et al., 2017), thus decreasing the excessive use of chemical fertilizers or pesticides. Mechanisms governing the assembly of microbial communities in the rhizosphere are the foundation for building beneficial rhizospheric microbiomes (Bai et al., 2015; Heijden and Hartmann, 2016; Gu et al., 2020; Trivedi et al., 2020). Progress in high-throughput sequencing and its application in metagenomics have advanced our understanding of microbial ecology in the rhizosphere. Several biotic and abiotic factors, such as plant species and development (Bell et al., 2015; Ma et al., 2019), soil animals (Jiang et al., 2017), viruses (Pratama et al., 2020), inoculation of microbial consortia (Zhang et al., 2019), invasion of pathogens (Yin et al., 2021), land use (Wang C. et al., 2021), fertilization (Ding et al., 2020), and tillage (Li et al., 2021), may cause changes in the composition and functions of microbial communities inhabiting the rhizosphere. Shifts in the microbial community may increase the resistance of plants to diseases, such as the common scab of potato (Shi et al., 2019) and pepper blight (Li et al., 2019; Zhang et al., 2019). In addition, the interactions among rhizospheric microorganisms may also influence the recruitment of beneficial plant microorganisms (Tao et al., 2020).
So far, the microbiome in the rhizosphere of plants has been studied using the whole root system (Barillot et al., 2013). However, the properties of the rhizosphere are very likely dynamic and heterogeneous (Vetterlein et al., 2020). For example, root systems are continuously differentiating and changing during the growth of plants (Hodge, 2006; Gruber et al., 2013). In addition, at finer scales, the soils vary greatly with respect to their redox potential (Smith et al., 2021), pH (Blossfeld et al., 2011), and availability of different nutrients or enzyme activities (Koop-Jakobsen and Wenzhöfer, 2015; Kuzyakov and Razavi, 2019). These factors affect the structures of soil microbial communities (Nunan, 2017). Microbial niches in the rhizosphere may serve as different habitat filters for or against certain other microorganisms. Thus, responses of the microbiome in the rhizosphere to different factors might be a “sum up” of the changes in these microbial niches. The characteristics of microbial niches are likely to be shaped synergistically by plant root exudates, by the physicochemical properties surrounding the root, or by the feedbacks of the microorganisms. Root exudates initiate and mediate the activities of microorganisms and their interactions with plants (Berg and Smalla, 2009). Taproot and fibrous roots differ greatly in terms of the release of root exudates, and most root exudates are released from the fibrous roots or root hairs (Trivedi et al., 2020). Physicochemical and biological gradients along the soil profile also differ in common; therefore, the vertical surroundings of taproots or fibrous roots might differ greatly. Thus, we hypothesize that the composition of the bacterial community recruited in the rhizosphere would depend on the combination of root type and its vertical surroundings and that habitat filter is a major driver of rhizosphere communities at a finer scale.
In this study, the spatial distribution of bacterial communities recruited from the disease-suppressive or non-suppressive soil was explored in the rhizosphere of pepper plants. We focused on two main questions: (1) What is the spatial distribution of the bacterial community and the antagonists in the rhizosphere of peppers plants? (2) To which level such spatial distributions might be influenced by different soil microbiomes?
Soil samples were collected from a long-term greenhouse experiment conducted at the Quzhou Experimental Station (36° 52′ N, 115° 01′ E), Hebei, China. That experiment contains organic and conventional farming systems, which followed the same scheme of crop rotation, tillage, and irrigation. Details of the experiment have been described previously (Han et al., 2017; Ding et al., 2019). Plant diseases such as late blight and powdery mildew on tomato (Solanum lycopersicum L.) or cucumber (Cucumis sativus L.) are less severe in the organic farming system than in the conventional farming systems (Yang et al., 2009a,b). In the climate chamber experiment, an incidence of pepper blight was 41% lower in the soil from the organic farming system than that from the conventional farming system, possibly due to the enhanced Bacillus antagonists in the rhizosphere (Li et al., 2019). Long-term organic farming likely increased the suppressive power of the soil toward plant diseases; henceforth, the soils samples from the organic and conventional farming systems are referred to as disease-suppressive and non-suppressive soils, respectively.
For each system, 75 cores (2 cm) of soil from the top layer (1–20 cm) were collected, mixed thoroughly, and passed through 2-mm mesh and stored at 4°C. The bioassay was performed as follows: after surface sterilization, “Cayenne” pepper (Capsicum annuum) seeds (Zhong liang xin) were germinated at 30°C in the dark, and after germination, the seeds were set at the seedling point suggested on the PhytoTC seed germination pouch (18 cm × 12.5 cm) and soaked in a mixture of 10 g of disease-suppressive or non-suppressive soil and 20 mL sterilized Hoagland solution. The seedlings were grown for 28 days in a growth chamber (Hangzhou Lvbo Instrument Co., Ltd., LB-1000D-LED) at 30°C, 70% relative humidity, and under a 12-h light (15,000 lx) period. Fresh standard Hoagland solution (20 ml) was re-added in the pouch. Each treatment consisted of four replicates, and each replicates contained two plants. The root system adhering to the seed germination paper was dissected into four different parts (e.g., upper taproot, upper fibrous root, lower taproot, and lower fibrous root), and the roots were carefully taken from the pouch. Different parts of root were vortexed vigorously in a 0.85% NaCl solution for 5 min (King et al., 2021). The pellet for DNA extraction was collected by a centrifugation (Eppendorf 5804R) at 6,000g for 5 min. All these samples were stored at −20°C prior to bacterial isolation or DNA extraction.
Total community DNA was extracted using the FastDNA Spin Kit from the soil samples according to the instruction of the manufacturer. Amplification of 16S rRNA fragments was performed using the universal primers 515F (5′-GTGC CAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACVSG GGTATCTAAT-3′) with a 12-bp barcode at the 5′ end of each primer (Lauber et al., 2009). Gene library preparation and the PCR reactions followed a previously reported method (Souza et al., 2021). The amplified PCR product were quantified and mixed at equal molar for gel purification. Sequencing was performed on the Illumina NovaSeq PE250 platform according to the protocol of the manufacturer. All sequences were submitted to the NCBI SRA (PRJNA750233).
Analysis of 16S rRNA sequences was performed as previously described (Ding et al., 2019; Chen et al., 2021). High-quality sequences without a technical region (barcode or primers) were used for the following analysis. Chimera sequences were removed jointly by a local BLASTN analysis using the SILVA database (version 138) and ChimeraSlayer analysis. Operational taxonomic unit (OTU) assignment and classification of representative sequences for each OTU were performed using the vsearch software (VSEARCH: a versatile open-source tool for metagenomics) (Rognes et al., 2016) and RDP Classifier (version 2.13) (Wang et al., 2007), respectively. OTUs affiliated to chloroplasts were removed. Statistical comparison, multiple comparison, and plotting were performed using the R software (version 3.6) with different add-On packages. The alpha-diversity index (Chao1 and Pielous’ evenness) was calculated by resampling, for 100 times, of an equal number of reads from each sample. This method helped reduce the biases caused by different read numbers. The unweighted pair group method with arithmetic mean (UPGMA) cluster, based on Bray–Curtis distance, was used to analyze the bacterial beta-diversity. Variations in the bacterial community explained by different parts of the root or soil were analyzed using the R add-On package “Vegan” (Zhang et al., 2016). Discriminative taxa were identified by multiple comparisons using a negative binomial model (Hothorn et al., 2008). Bacterial genera that were positively correlated with each other (Spearman’s rank coefficient > 0.6 and p < 0.01) were subjected to co-occurrence network analysis using the Gephi software (version 0.91). In vitro antagonists against Phytophthora capsici isolated from a previous study using the same soil (Li et al., 2019) were used to estimate their spatial distribution pattern in the rhizosphere. Subsequences of the 16S rRNA gene between 515F and 806R of each antagonist were extracted, and the unique phylotype was extracted using the software Vsearch. These unique phylotypes were mapped against the 16S rRNA sequence library with a minimum identity of 99% using a standalone BLASTN analysis. All tools mentioned above were implemented in a galaxy instance1.
Bacterial communities in different parts of the root systems were analyzed by Illumina sequencing of the 16S rRNA gene fragments. A total of 13,404,282 sequences were acquired and 146,369 sequences affiliated with chloroplasts were removed for further analysis. The remaining sequences were grouped into 69,622 OTUs. The most abundant phyla were Proteobacteria (44.8%), Firmicutes (15.3%), Verrucomicrobia (6.8%), Acidobacteria (6.2%), Bacteroidetes (5.3%), and Planctomycetes (4.9%) (Figure 1A). The relative abundance of Proteobacteria was lower in the upper taproot than in the other parts of the root for both soil types (Figure 1A and Supplementary Table 1). For both soil types, bacterial richness and evenness in the rhizosphere decreased from the upper taproot to the upper fibrous root, the lower taproot, and the lower fibrous root (Figures 1B,C). A significant difference between the upper taproot and the lower fibrous root was observed for bacterial evenness in both soil treatments and for richness only in the non-suppressive soils (Figures 1B,C). The bacterial richness was significantly (p < 0.05) higher in plants treated with the diseases-suppressive soil than in those treated with the non-suppressive soil (Figure 1B). The bacterial evenness was comparable between the suppressive and non-suppressive soils (Figures 1B,C and Supplementary Table 2). UPGMA cluster analysis indicated that bacterial communities mainly differed between the upper and lower parts of roots (Figure 1D). Variation partition analysis revealed that spatial variation (38.0%) was much greater than that explained by different soils (9.0%) (Figure 1E).
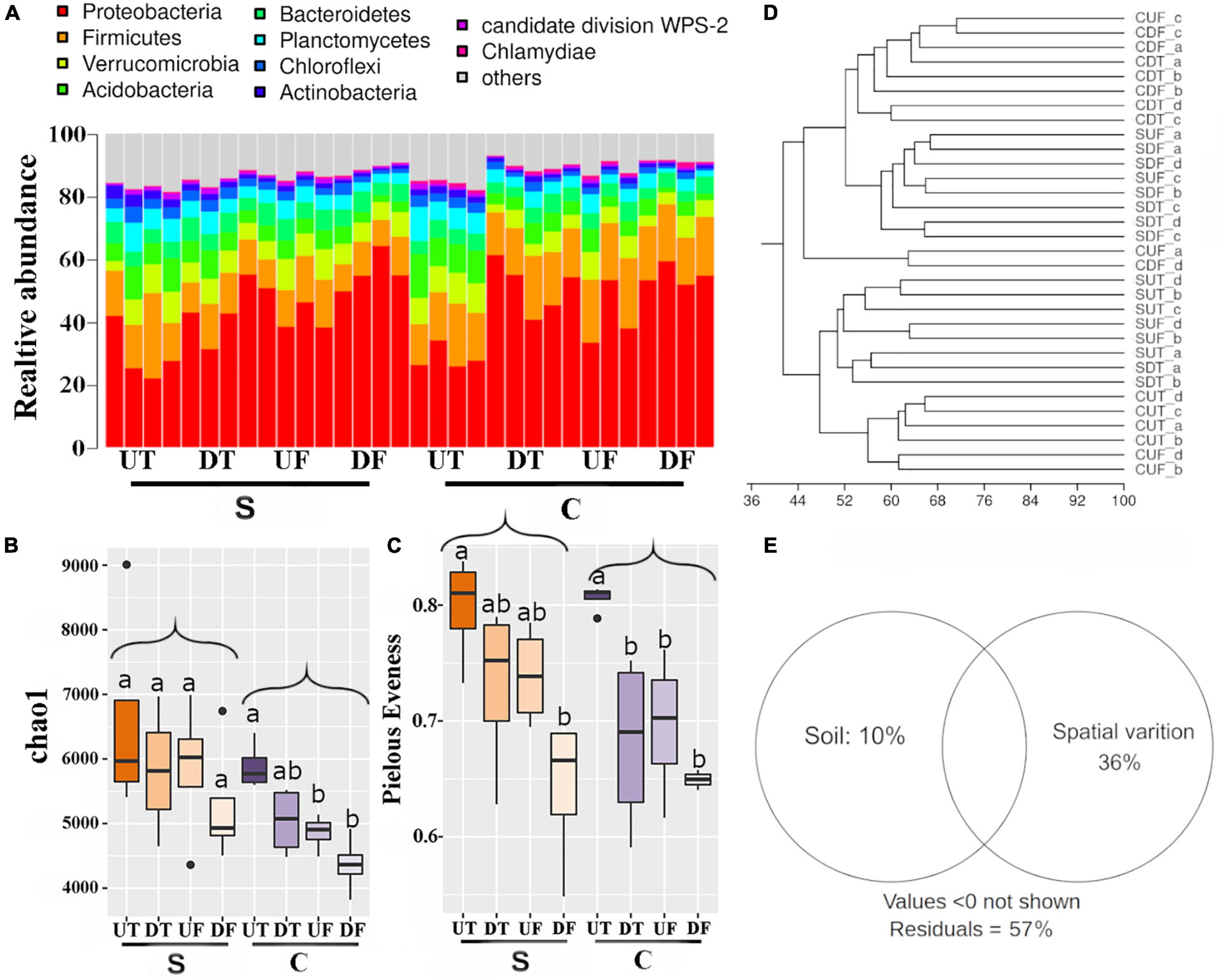
Figure 1. Bacterial diversity in the rhizosphere of the upper (U) or lower (D) part of the taproot (T) or fibrous (F) roots treated by the disease-suppressive (S) or non-suppressive soil (C). Relative abundance of dominant phyla (A), Chao1 richness (B), Pielous’ evenness (C), UPGMA cluster (D), and variation explained by different soils or different parts of roots (E). Significant differences in bacterial alpha-diversity among four parts of root under disease-suppressive or non-suppressive soil are indicated by different letters.
Multiple comparisons were performed to identify the dominant (relative abundance of one sample > 0.5%) taxa with contrasting spatial distribution in the rhizosphere of pepper (Figure 2). Most discriminative genera could be assigned to six groups according to their response patterns (Figure 2). The genera in group 1 were commonly enriched in the upper taproot of the plant, and the majority were affiliated with Acidobacteria (e.g., Gp16, Gp3, and Gp6), Planctomycetes (e.g., Pirellula, Gemmata, and Gimesia), Chloroflexi (e.g., Litorilinea and Sphaerobacter), and Proteobacteria (e.g., Steroidobacter and Sphingobium) (Figure 2). In addition, few genera, such as Clostridium sensu stricto, Armatimonadetes gp5, Gemmatimonas, and Parcubacteria_genera_incertae_sedis, which included members were also commonly enriched in the taproots (Figure 2). Genera (e.g., Azotobacter, Hydrogenophaga, Pseudoxanthomonas, Rhizobium, Clostridium IlI, Sporomusa, Clostridium XIVa, and Leptonema) in the group 2 commonly decreased in the taproots (Figure 2). The other four groups consisted of genera specifically enriched (groups 3 and 6) or decreased (groups 4 and 5) in the taproot treated with disease-suppressive or non-suppressive soil (Figure 2). Among them, relatively abundant genera such as Anaeroarcus, Acidovorax, and Magnetospirillum, which are known to contain anaerobes, were also lowest in the taproot of pepper treated with disease-suppressive or non-suppressive soil (Figure 2).
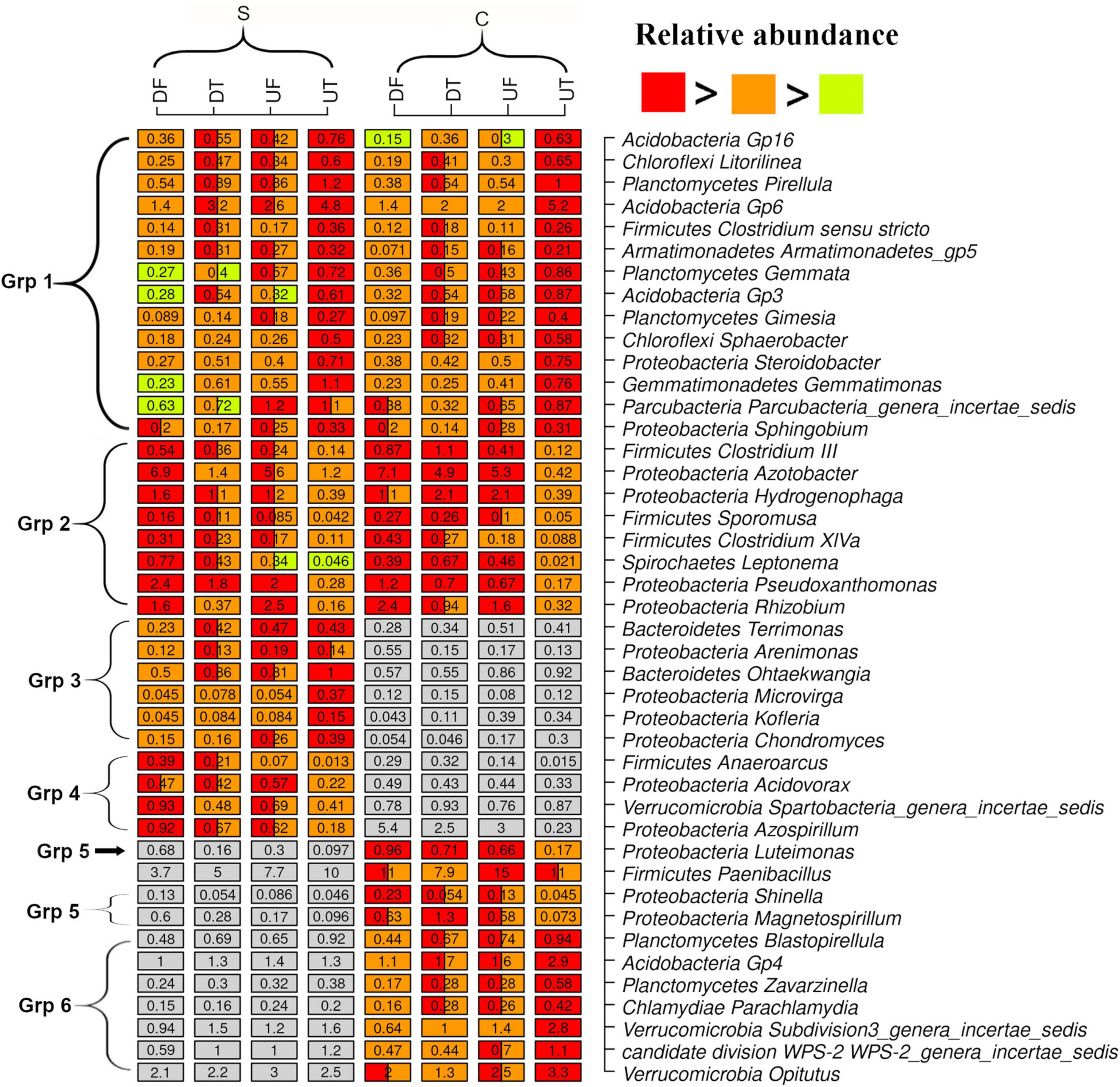
Figure 2. Genera with significantly different relative abundance in the rhizosphere of the upper (U) or lower (L) part of the taproot (T) or fibrous (F) roots treated by the disease-suppressive (S) or non-suppressive soil (C). Significant difference is indicated by a different color. A box with two colors indicates no significant difference from other treatment containing one of the two colors.
A total of 91 genera that were positively correlated with each other were subjected to co-occurrence network analysis (Figure 3). The majority of them were affiliated with Proteobacteria (32 genera), Firmicutes (14 genera), Bacteroidetes (10 genera), Acidobacteria (8 genera), Planctomycetes (7 genera), and Verrucomicrobia (5 genera). These co-occurring genera formed six hubs, including two enriched hubs (modules 3 and 5 in green and cyan, respectively) and one decreased hub (module 1 in purple), in the taproot of pepper (Figure 3A). Correlation analysis further revealed that modules 3 and 5 were positively correlated with each other (Figure 3H), and both were negatively correlated with module 1 (Figures 3I,J). Interestingly, all co-occurring genera affiliated with Acidobacteria or Planctomycetes were among module 3 or 5 (Figure 3A), suggesting that these taxa might prefer the microbial niche in the taproot of pepper. In addition, module 3 also contained nitrifying bacterial taxa, such as Nitrosospira and Nitrospira (Figure 3A). Module 1 consisted of several bacterial genera adapted to anaerobic and anoxic environments, such as Clostridium III or Clostridium XIVa, Magnetospirillum, and Anaeroarcus (Figure 3A). No clear spatial distribution pattern was observed for modules 0 and 4 (Figure 3A). Module 4 was significantly enriched in the treatment with disease-suppressive soil in contrast to module 0 (Figures 3F,G).
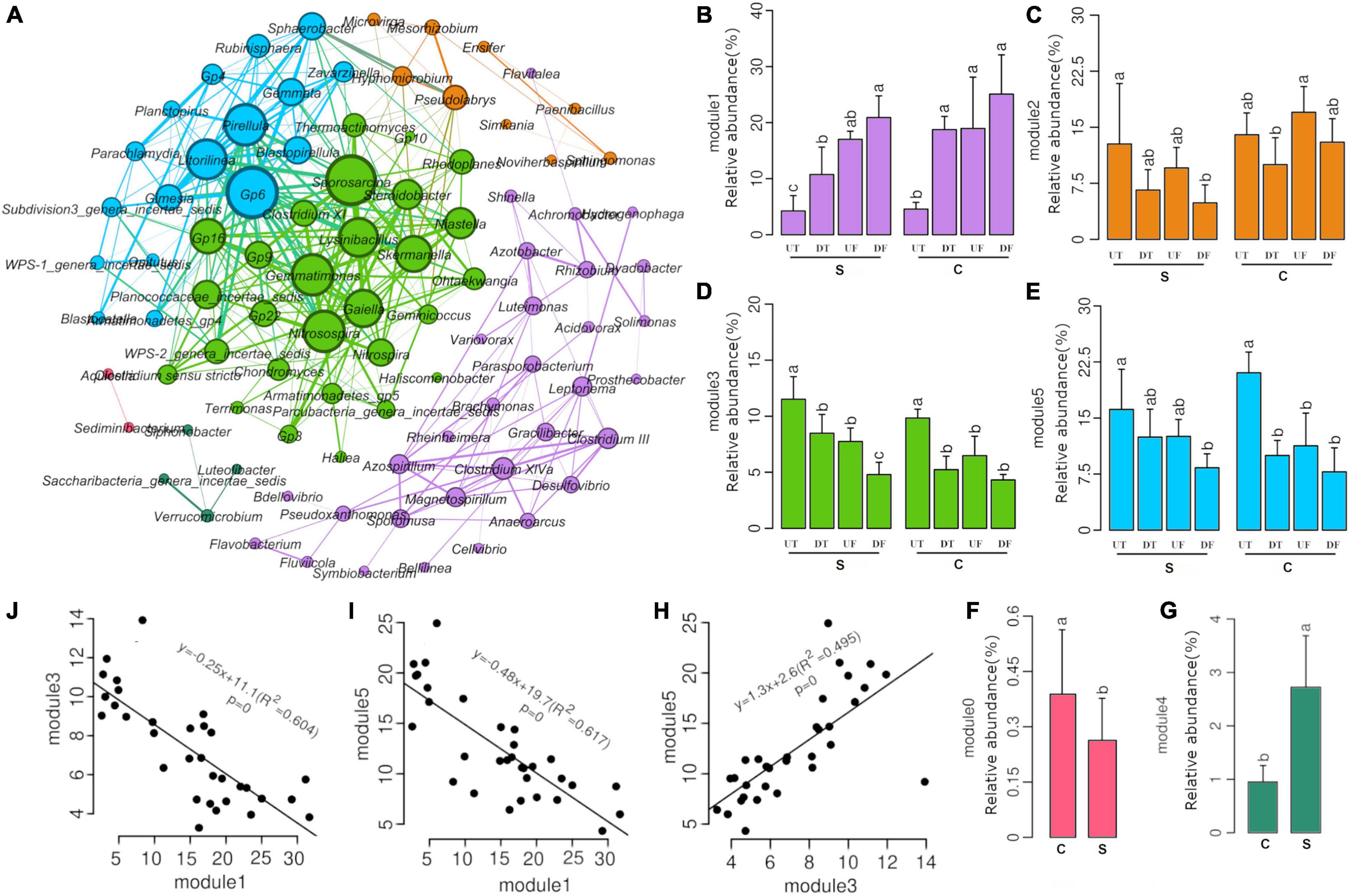
Figure 3. Co-occurrence network analysis of dominant bacteria (A), relative abundance of modules (B–G), and significantly correlated modules (H–J).
To estimate the spatial distribution of in vitro antagonists against P. capsici along the rhizosphere of pepper, 24 unique phylotypes acquired from a previous study using the same soil microbiome were mapped against the 16S rRNA gene fragment. A total of 19 phylotypes could be mapped to 132,713 sequences, accounting for 1.0% of the 16S rRNA gene fragments acquired. Interestingly, the percentages of mapped sequences varied between the different parts of the roots (Figure 4). Only 0.37 and 0.43% of sequences could be mapped for the upper taproot of pepper treated by the disease-suppressive or non-suppressive soil, respectively, in contrast to 1.7% for the fibrous roots (Supplementary Figure 1). Among them, the phylotypes similar to Beijerinckia fluminensis were most abundant and its relative abundance tended to be higher in the taproot than in the fibrous roots (Figure 4). Seven phylotypes were distributed distinctly along the rhizosphere of pepper treated by the disease-suppressive soil, in contrast to the two phylotypes in the non-suppressive soil (Figure 4). Four out of these seven phylotypes were similar to Beijerinckia tquilensis, Beijerinckia aerophilus, Beijerinckia cereus, and Beijerinckia firmus, and their relative abundance was highest in the taproot of pepper (Figure 4). In the treatment with the non-suppressive soil, the phylotype similar to Beijerinckia licheniformis was also more abundant in the taproot of pepper (Figure 4).
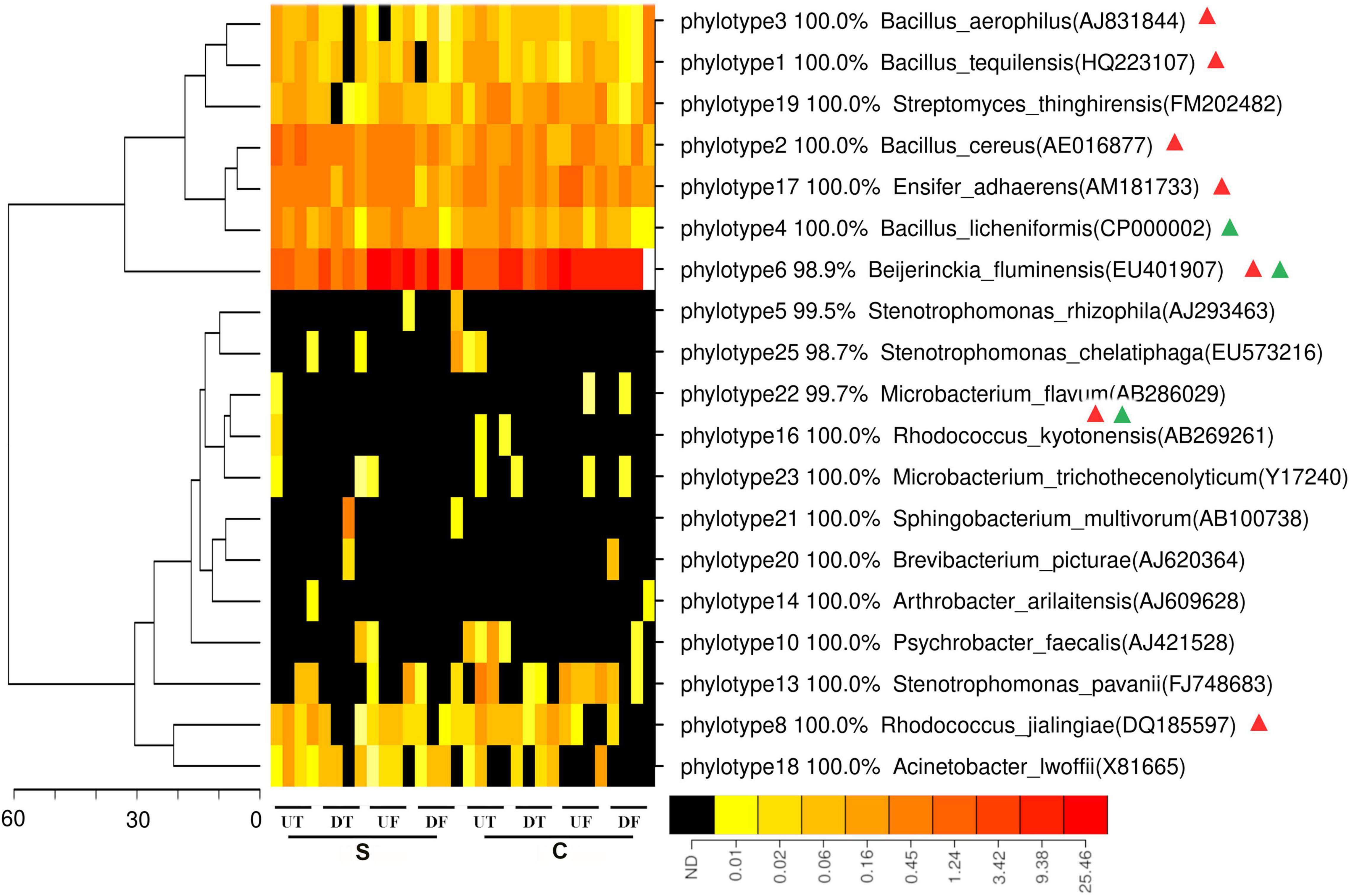
Figure 4. Heatmap analysis of in vitro antagonist belonging to each phylotype in the rhizosphere of the upper (U) or lower (D) part of the taproot (T) or fibrous (F) roots treated by the disease-suppressive (S) or non-suppressive soil (C). Those phylotypes with significantly different relative abundance at four parts of roots are indicated by green or red triangle for the disease-suppressive or non-suppressive soil, respectively. The minus symbol indicates decreased relative abundance in the suppressive soil. Phylotype that significantly differed at relative abundance in the rhizosphere of different root parts under suppressive or non-suppressive soil is indicated by red or green triangle, respectively.
Previously, the rhizospheric microbiome has been frequently studied by using the whole system, in which the heterogeneity in the rhizosphere may be neglected. Herein, a distinct spatial distribution of the bacterial community along the rhizosphere of pepper was detected, indicating that niche differentiation within the rhizosphere might also be considerable. Bacterial alpha-diversity was frequently lower in the rhizosphere than in the corresponding bulk soil, possibly because the plant root only recruits a fraction of soil microorganisms into its rhizosphere (Fierer, 2017; Zhou et al., 2020). Here, the lowest bacterial richness and evenness were detected at the lower fibrous roots in both treatments, suggesting that the selective pressure exerted by plant roots might be strongest at the lower fibrous root of pepper. Root exudates are the major driving forces of microbial communities in the rhizosphere (Berg and Smalla, 2009; Turner et al., 2013). Root hair or fibrous roots often release large quantities of metabolites into the soil (Pandey et al., 2021). Interestingly, the bacterial alpha-diversity in the upper part of fibrous root tends to be higher than in the lower part. Compared with the lower parts, oxygen might be more easily dissolved in the upper parts, which may favor the decomposition of metabolites released by fibrous roots. A slight increase in bacterial richness in the rhizosphere of pepper treated by the disease-suppressive soil agrees with the findings of a previous study (Li et al., 2019). However, it is still premature to state that high bacterial diversity in the rhizosphere may lead to increased resistance to plant diseases, as several other physicochemical properties may influence the assemblage of microbiome in the rhizosphere (Schreiter et al., 2014; Wahdan et al., 2021).
The spatial variation of bacterial community composition along the rhizosphere of pepper was much greater than that explained by different soils, suggesting that the properties of microbial niches in the rhizosphere might be largely determined by the type of root (fibrous root or taproot) and the surroundings at the microscale. This finding agrees with the fact that microbial scale heterogeneity in the soil/rhizosphere, which was largely disregarded, is important for understanding the microbial ecology in soil (Nunan, 2017). Additionally, several biotic/abiotic factors, such as nutrients and bulk density, which could shift the microbial community in the rhizosphere, were also able to change the topology of plant roots (Glimskär, 2000; Wang et al., 2015; Tang et al., 2020). Thus, it is possible that some changes in microbial communities in the rhizosphere were attributed to alterations in the root system in the soil. Additionally, bacterial communities in the rhizosphere of pepper were significantly different between the disease-suppressive and non-suppressive soils, which is in agreement with previous studies. Our results suggest that such differences might be also vary spatially along the rhizosphere and that the upper taproot might be most influenced by the soil. Taken together, all these findings indicate that different roots and soil microbiomes might synergistically shape the spatial distribution of microbiomes in the rhizosphere of pepper.
Acidobacteria are abundant in soils and represent a significant fraction of the soil bacterial community. Previously, Acidobacteria were largely regarded as oligotrophy (Fierer et al., 2007; Kielak et al., 2016), which may not outcompete in the rhizosphere with excessive nutrients. Here, acidobacterial subgroups 3, 4, 6, and 16 were more abundant in the rhizosphere of the upper taproot. The prevalence of acidobacterial subgroups was also detected in the rhizosphere of several other crops, such as potatoes and leek (Rocha et al., 2013), soybean (Navarrete et al., 2013), and tea (Wang M. et al., 2021), suggesting that some Acidobacteria might also be competent in utilizing root depositions. So far, only dozens of Acidobacteria have been cultivated, which limits our understanding of their physiology (Kalam et al., 2020). However, physiological and genomic studies on cultivable acidobacterial have indicated that some Acidobacteria can degrade xylan (Kielak et al., 2016), which is a major polysaccharide in primary cell walls. However, it remains to be elucidated whether the enrichment of acidobacterial subgroups in the rhizosphere at the upper taproot is associated with their ability to degrade polysaccharides. Interestingly, the module enriched at the taproot also contains Nitrosospira and Nitrospira, members that have the ability to live an aerobic chemoautotrophic lifestyle by ammonia oxidation (Norton et al., 2008; Lagostina et al., 2015; Kuypers et al., 2018). The relative abundance of anaerobic genera such as Clostridium IlI, Sporomusa, and Clostridium XIVa was enriched in the lower taproot or fibrous roots, and these taxa were often prevalent in anaerobic environments with abundant organic materials (Nevin et al., 2011; Edwards et al., 2013; Ðapa et al., 2013). Genera such as Rhizobium, Azotobacter, and Azospirillum were enriched at the lower fibrous roots, which is in agreement with the fact that nitrogen fixation occurs only under anaerobic conditions (Hayat et al., 2010; Meena et al., 2017; Huang et al., 2021). Other genera such as Hydrogenophaga and Magnetospirillum are also known to adapt to anaerobic conditions by nitrate respiration (Golby et al., 2012). In summary, these findings indicate that the availability of oxygen may also play an important role in the spatial distribution of different taxa in the rhizosphere of pepper, in addition to root exudates.
Competition for microbial niches has been proposed as a mechanism employed by beneficial microorganisms to fight against phytopathogens (Götz et al., 2006). Previously, Bacillus antagonists in the disease-suppressive soil used in this study may contribute to the suppression of pepper light caused by P. capsici (Li et al., 2019). In silicon analysis revealed that four isolated Bacillus antagonists preferentially colonized the taproot of pepper treated with disease-suppressive soils. This result indicated that the upper taproot might be a hot spot where the Bacillus antagonists interacted with the phytopathogen P. capsici. This finding agrees with several other studies based on green fluorescence protein-labeled antagonists, in which many antagonists, including Bacillus, were preferentially colonized at the lateral root junctions (Liu et al., 2006).
It is also worth noting that the spatial distribution of the bacterial community was studied by dividing the roots systems into four rough compartments, and the microbial communities at finer scale have not been resolved. In addition, the physicochemical conditions of the root pouch differed from those in the soil. Thus, further analysis of the microbial community at a finer scale may provide more details on the spatial distribution of the bacterial community in the rhizosphere. In conclusion, distinct spatial distribution of bacterial community in the rhizosphere of pepper with largely aerobic heterotrophic (including Bacillus antagonists against P. capsici) and chemotrophic taxa at the upper root and anaerobic taxa (including heterotrophic or diazotrophic, nitrate respiration) at the lower taproot or fibrous roots was found, thus highlighting the importance of root exudates and the availability of oxygen for the reassembling of the rhizosphere microbiome.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA750233.
G-CD, HL, NW, JL, and YW designed the experiments, wrote the manuscript, and analyzed the data. HL, JD, YL, and XD performed the experiments. All authors participated in the survey, sample collection of organic greenhouse agriculture in different regions, and reviewed the manuscript.
This work was supported by the National Key Research and Development Program of China (grant number 2019YFD1002000) and the National Natural Science Foundation of China (grant number 32071552).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.748542/full#supplementary-material
Supplementary Figure 1 | Average relative abundance of reads mapped against all in vitro antagonists. Significant differences (p < 0.05, multiple comparisons under negative binomial model) are indicated by different letters.
Ahkami, A. H., WhiteIII, R., Handakumbura, P. P., and Christer, J. (2017). Rhizosphere engineering: enhancing sustainable plant ecosystem productivity. Rhizosphere 3, 233–243. doi: 10.1016/j.rhisph.2017.04.012
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. doi: 10.1038/nature16192
Barillot, C. D., Sarde, C. O., Bert, V., Tarnaud, E., and Cochet, N. (2013). A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 63, 471–476. doi: 10.1007/s13213-012-0491-y
Bell, C. W., Asao, S., Calderon, F., Wolk, B., and Wallenstein, M. (2015). Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem 85, 170–182. doi: 10.1016/j.soilbio.2015.03.006
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Blossfeld, S., Gansert, D., Thiele, B., Kuhn, A. J., and Lösch, R. (2011). The dynamics of oxygen concentration, pH value, and organic acids in the rhizosphere of Juncus spp. Soil Biol. Biochem. 43, 1186–1197. doi: 10.1016/j.soilbio.2011.02.007
Chen, C., Han, H., Meng, Y., Gong, H., Jia, R., Xu, T., et al. (2021). Total and denitrifying bacterial communities associated with the interception of nitrate leaching by carbon amendment in the subsoil. Appl. Microbiol. Biotechnol. 105, 2559–2572. doi: 10.1007/s00253-021-11189-y
Ðapa, T., Leuzzi, R., Ng, Y. K., Baban, S. T., Adamo, R., Kuehne, S. A., et al. (2013). Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555. doi: 10.1128/JB.01980-12
Ding, G. C., Bai, M. H., Han, H., Li, H. X., Ding, X. Y., Yang, H. Y., et al. (2019). Microbial taxonomic, nitrogen cycling and phosphorus recycling community composition during long-term organic greenhouse farming. FEMS Microbiol. Ecol. 95:fiz042. doi: 10.1093/femsec/fiz042
Ding, Y. Q., Jin, Y. L., He, K. Z., Yi, Z. L., Tan, L., Liu, L. S., et al. (2020). Low nitrogen fertilization alter rhizosphere microorganism community and improve sweetpotato yield in a nitrogen-deficient rocky soil. Front. Microbiol 11:678. doi: 10.3389/fmicb.2020.00678
Edwards, A. N., Suárez, J. M., and McBride, S. M. (2013). Culturing and maintaining Clostridium difficile in an anaerobic environment. J. Vis. Exp. 2013:50787. doi: 10.3791/50787
Faria, M. R. D., Costa, L., Chiaramonte, J. B., Bettiol, W., and Mendes, R. (2020). The rhizosphere microbiome: functions, dynamics, and role in plant protection. Trop Plant Pathol. 46, 13–25. doi: 10.1007/s40858-020-00390-5
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Glimskär, A. (2000). Estimates of root system topology of five plant species grown at steady-state nutrition. Plant Soil 227, 249–256. doi: 10.1023/A:1026531200864
Golby, S., Ceri, H., Gieg, L. M., Chatterjee, I., Marques, L. L. R., and Turner, R. J. (2012). Evaluation of microbial biofilm communities from an Alberta oil sands tailings pond. FEMS Microbiol. Ecol. 79, 240–250. doi: 10.1111/j.1574-6941.2011.01212.x
Götz, M., Gomes, N. C. M., Dratwinski, A., Costa, R., Berg, G., Peixoto, R., et al. (2006). Survival of gfp-tagged antagonistic bacteria in the rhizosphere of tomato plants and their effects on the indigenous bacterial community. FEMS Microbiol. Ecol. 56, 207–218. doi: 10.1111/j.1574-6941.2006.00093.x
Gruber, B. D., Giehl, R. F. H., Friedel, S., and Wirén, N. V. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Gu, S., Wei, Z., Shao, Z., Friman, V.-P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Han, H., Teng, Y. M., Yang, H. F., and Li, J. (2017). Effects of long-term use of compost on N2O and CO2 fluxes in greenhouse vegetable systems. Compost. Sci. Util. 25, 61–69. doi: 10.1080/1065657X.2016.1238786
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/S13213-010-0117-1
Heijden, M. G. A. V. D., and Hartmann, M. (2016). Networking in the plant microbiome. PLoS Biol. 14:e1002378. doi: 10.1371/journal.pbio.1002378
Hodge, A. (2006). Plastic plants and patchy soils. J. Exp. Bot. 57, 401–411. doi: 10.1093/jxb/eri280
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biochem. J. 50, 346–363. doi: 10.1002/bimj.200810425
Huang, W. S., Wang, L. T., Chen, J. S., Chen, Y. T., Wei, S. T.-S., Chiang, Y. R., et al. (2021). Vibrio nitrifigilis sp. nov., a marine nitrogen-fixing bacterium isolated from the lagoon sediment of an islet inside an atoll. Antonie Van Leeuwenhoek 114, 933–945. doi: 10.1007/s10482-021-01567-x
Jiang, Y. J., Liu, M. Q., Zhang, J. B., Chen, Y., Chen, X. Y., Chen, L. J., et al. (2017). Nematode grazing promotes bacterial community dynamics in soil at the aggregate level. ISME J. 11, 2705–2717. doi: 10.1038/ismej.2017.120
Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., Veen, J. A. V., and Kuramae, E. E. (2016). The ecology of acidobacteria: moving beyond genes and genomes. Front Microbiol 7:744. doi: 10.3389/fmicb.2016.00744
King, W. L., Yates, C. F., Guo, J., Fleishman, S. M., Trexler, R. V., Centinari, M., et al. (2021). The hierarchy of root branching order determines bacterial composition, microbial carrying capacity and microbial filtering. Commun. Biol. 4:483. doi: 10.1038/s42003-021-01988-4
Koop-Jakobsen, K., and Wenzhöfer, F. (2015). The dynamics of plant-mediated sediment oxygenation in Spartina anglica rhizospheres—a planar optode study. Estuar. Coast. 38, 951–963. doi: 10.1007/S12237-014-9861-Y
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Kuzyakov, Y., and Razavi, B. S. (2019). Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol. Biochem. 135, 343–360. doi: 10.1016/j.soilbio.2019.05.011
Lagostina, L., Goldhammer, T., Røy, H., Evans, T. W., Lever, M. A., Jørgensen, B. B., et al. (2015). Ammonia-oxidizing Bacteria of the Nitrosospira cluster 1 dominate over ammonia-oxidizing Archaea in oligotrophic surface sediments near the South Atlantic Gyre. Environ. Microbiol. Rep. 7, 404–413. doi: 10.1111/1758-2229.12264
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Li, H. X., Cai, X. X., Gong, J. Y., Xu, T., Ding, G. C., and Li, J. (2019). Long-term organic farming manipulated rhizospheric microbiome and Bacillus antagonism against pepper blight (Phytophthora capsici). Front. Microbiol. 10:483. doi: 10.3389/fmicb.2019.00342
Li, T., Li, Y., Gao, X. C., Wang, S. N., Wang, Z. T., Liu, Y., et al. (2021). Rhizobacterial communities and crop development in response to long-term tillage practices in maize and soybean fields on the Loess Plateau of China. Catena 202:105319. doi: 10.1016/J.CATENA.2021.105319
Liu, X., Zhao, H., and Chen, S. (2006). Colonization of maize and rice plants by strain Bacillus megaterium C4. Curr. Microbiol. 52, 186–190. doi: 10.1007/s00284-005-0162-3
Ma, S., Frenne, P. D., Boon, N., Brunet, J., Cousins, S. A. O., Decocq, G., et al. (2019). Plant species identity and soil characteristics determine rhizosphere soil bacteria community composition in European temperate forests. FEMS Microbiol. Ecol. 95:fiz063. doi: 10.1093/femsec/fiz063
Meena, V. S., Meena, S. K., Verma, J. P., Kumar, A., Aeron, A., Mishra, P. K., et al. (2017). Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: a review. Ecol. Eng. 107, 8–32. doi: 10.1016/J.ECOLENG.2017.06.058
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mendes, R., Kruijt, M., Bruijn, I. D., Dekkers, E., Voort, M. V. D., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Navarrete, A. A., Kuramae, E. E., Hollander, M. D., Pijl, A. S., Veen, J. A. V., and Tsai, S. M. (2013). Acidobacterial community responses to agricultural management of soybean in Amazon forest soils. FEMS Microbiol. Ecol. 83, 607–621. doi: 10.1111/1574-6941.12018
Nevin, K. P., Hensley, S. A., Franks, A. E., Summers, Z. M., Ou, J., Woodard, T. L., et al. (2011). Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl. Environ. Microbiol. 77, 2882–2886. doi: 10.1128/AEM.02642-10
Norton, J. M., Klotz, M. G., Stein, L. Y., Arp, D. J., Bottomley, P. J., Chain, P. S. G., et al. (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl. Environ. Microbiol. 74, 3559–3572. doi: 10.1128/AEM.02722-07
Nunan, N. (2017). The microbial habitat in soil: scale, heterogeneity and functional consequences. J. Plant. Nutr. Soil Sci 180, 425-429. doi: 10.1002/jpln.201700184
Pandey, K. D., Patel, A. K., Singh, M., Kumari, V. A., and Jalaluddin, M. (2021). Secondary metabolites from bacteria and viruses. Nat. Bioact. Comp. 2021, 19–40. doi: 10.1016/b978-0-12-820655-3.00002-1
Pratama, A. A., Terpstra, J., Oliveria, A. L. M., and FalcãoSalles, J. (2020). The role of rhizosphere bacteriophages in plant health. Trends Microbiol. 28, 709–718. doi: 10.1016/j.tim.2020.04.005
Rocha, U. N. D., Plugge, C. M., George, I., Elsas, J. D. V., and Overbeek, L. S. V. (2013). The rhizosphere selects for particular groups of acidobacteria and verrucomicrobia. PLoS One 8:e82443. doi: 10.1371/journal.pone.0082443
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Schreiter, S., Ding, G., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Shi, W., Li, M., Wei, G., Tian, R., Li, C., Wang, B., et al. (2019). The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 7:14. doi: 10.1186/s40168-019-0629-2
Smith, G. J., Mcdowell, R. W., Daly, K., Huallacháin, D. Ó, Condron, L. M., and Fenton, O. (2021). Estimating and modelling the risk of redox-sensitive phosphorus loss from saturated soils using different soil tests. Geoderma 398:115094. doi: 10.1016/J.GEODERMA.2021.115094
Souza, L. F. D., Obregón, D., Domeignoz-Horta, L. A., and Gomes, F. V. (2021). Maintaining grass coverage increases methane uptake in Amazonian pasture soils. bioRxiv [Preprint] doi: 10.1101/2021.04.26.441496
Tang, M., Li, L., Wang, X. L., You, J., Li, J. N., and Chen, X. (2020). Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 10:12442. doi: 10.1038/s41598-020-69441-w
Tao, C. Y., Li, R., Xiong, W., Shen, Z. Z., Liu, S. S., Wang, B. B., et al. (2020). Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 8:137. doi: 10.1186/s40168-020-00892-z
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Turner, T. R., James, E. K., and Poole, P. S. (2013). The plant microbiome. Genome Biol. 14:209. doi: 10.1186/gb-2013-14-6-209
Vetterlein, D., Carminati, A., Kögel-Knabner, I., Bienert, G. P., Smalla, K., Oburger, E., et al. (2020). Rhizosphere spatiotemporal organization–a key to rhizosphere functions. Front. Agron. 2:8. doi: 10.3389/fagro.2020.00008
Wahdan, S. F. M., Heintz-Buschart, A., Sansupa, C., Tanunchai, B., Wu, Y. T., Schädler, M., et al. (2021). Targeting the active rhizosphere microbiome of Trifolium pratense in grassland evidences a stronger-than-expected belowground biodiversity-ecosystem functioning link. Front. Microbiol. 12:629169. doi: 10.3389/fmicb.2021.629169
Wang, C., Zheng, M. M., Chen, J., and Shen, R. F. (2021). Land-use change has a greater effect on soil diazotrophic community structure than the plant rhizosphere in acidic ferralsols in southern China. Plant Soil 462, 445–458. doi: 10.1007/s11104-021-04883-3
Wang, M., Sun, H., Xu, L., and Xu, Z. (2021). Bacterial diversity in tea plant (Camellia sinensis) rhizosphere soil from Qinling Mountains and its relationship with environmental elements. Plant Soil 460, 403–415. doi: 10.1007/s11104-020-04822-8
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequethisnces into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Y., Dong, X., Wang, H., Wang, Z., and Gu, J. (2015). Root tip morphology, anatomy, chemistry and potential hydraulic conductivity vary with soil depth in three temperate hardwood species. Tree Physiol. 36, 99–108. doi: 10.1093/treephys/tpv094
Yang, H. F., Fan, J. F., Ge, Z. Q., Shen, G. C., Lu, R. H., and Li, J. (2009a). Main diseases and control effects of organic, integrated and conventional cultivation patterns of greenhouse tomato. Chin. J. Econ. Agric. 17, 933–937. doi: 10.3724/SP.J.1011.2009.00933
Yang, H. F., Fan, J. F., Liang, L. N., Meng, Y. L., Zhang, S. K., and Ji, L. I. (2009b). Studies on the main diseases and control effects under organic, integrated and conventional cultivation patterns of cucumber in greenhouse. Acia Agric. Boreali Sjn. 24, 240–245. doi: 10.7668/hbnxb.2009.S1.056
Yin, C. T., Vargas, J. M. C., Schlatter, D. C., Hagerty, C. H., Hulbert, S. H., and Paulitz, T. C. (2021). Rhizosphere community selection reveals bacteria associated with reduced root disease. Microbiome 9:86. doi: 10.1186/s40168-020-00997-5
Yu, Y. Y., Xu, J. D., Huang, T. X., Zhong, J., Yu, H., Qiu, J. P., et al. (2020). Combination of beneficial bacteria improves blueberry production and soil quality. Food Sci. Nutr. 8, 5776–5784. doi: 10.1002/fsn3.1772
Zhang, L. N., Wang, D. C., Hu, Q., Dai, X. Q., Xie, Y. S., Li, Q., et al. (2019). Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol 10:1668. doi: 10.3389/fmicb.2019.01668
Zhang, X., Gu, Q., Long, X.-E., Li, Z.-L., Liu, D.-X., Ye, D.-H., et al. (2016). Anthropogenic activities drive the microbial community and its function in urban river sediment. J. Soils Sed. 16, 716–725. doi: 10.1007/s11368-015-1246-8
Keywords: spatial distribution, microbial community, rhizosphere, pepper blight, disease- suppressive soil
Citation: Li H, Wang N, Ding J, Liu Y, Ding X, Wei Y, Li J and Ding G-c (2022) Spatial Distribution of the Pepper Blight (Phytophthora capsici) Suppressive Microbiome in the Rhizosphere. Front. Plant Sci. 12:748542. doi: 10.3389/fpls.2021.748542
Received: 28 July 2021; Accepted: 23 December 2021;
Published: 21 January 2022.
Edited by:
Camille Eichelberger Granada, Universidade do Vale do Taquari (Univates), BrazilReviewed by:
Xiaogang Li, Nanjing Forestry University, ChinaCopyright © 2022 Li, Wang, Ding, Liu, Ding, Wei, Li and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-chun Ding, Z2NfZGluZ0BjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.