
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 November 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.748531
This article is part of the Research Topic Harnessing Genebanks: High-Throughput Phenotyping and Genotyping of Crop Wild Relatives and Landraces View all 14 articles
 Noritoshi Inagaki1*†
Noritoshi Inagaki1*† Hidenori Asami2,3†
Hidenori Asami2,3† Hideyuki Hirabayashi4
Hideyuki Hirabayashi4 Akira Uchino5
Akira Uchino5 Toshiyuki Imaizumi3
Toshiyuki Imaizumi3 Ken Ishimaru4
Ken Ishimaru4To maximize crop growth, crops need to capture sunlight efficiently. This property is primarily influenced by the shape of the crops such as the angle, area, and arrangement of leaves. We constructed a rice (Oryza sativa L.) inbred line that displayed an ideal transition of plant shapes in terms of sunlight receiving efficiency. During vegetative growth, this line exhibited tiller spreading with increased tiller number, which formed a parabolic antenna-like structure. The architecture probably improved light reception efficiency of individuals compared with the recurrent parent. The line achieved not only acceleration of the vegetative growth, but also significant suppression of weed growth under the canopy. The increased light reception efficiency of the line has consequently reduced the amount of incident light to the ground and supplied significant competitiveness against weeds. The spread tillers became erect from the entry of the reproductive growth phase, adaptively sustaining light reception efficiency in thicker stands. The line carries a small chromosomal segment from Oryza rufipogon Griff., a putative progenitor of Asian cultivated rice. The introduced chromosome segment had little effect on grain yield and quality. Our results shed light on potentials hidden in the wild rice chromosome segment to achieve the valuable traits.
Asian cultivated rice, Oryza sativa (O. sativa), is one of the world’s most important crops, sustaining several billion people as a staple food (Global Rice Science Partnership, 2013). To ensure permanence of humanity, improvement of rice cultivation systems should be required not only from economic perspectives, but also from viewpoints based on sustainability.
O. sativa was domesticated from a wild rice species, Oryza rufipogon (O. rufipogon), which is distributed throughout Asia and Oceania (Chen et al., 2019). Human ancestors empirically selected beneficial lines along their cropping systems from the progenitor. Such domestication processes substantially narrowed the genetic diversity at genetic bottlenecks formed during these winnowing steps (Doebley et al., 2006). Comprehensive genome sequencing and analyses of O. rufipogon and O. sativa quantitatively ascertained the vast reduction in genetic diversity that occurred during domestication (Zhu et al., 2007; Huang et al., 2012; Chen et al., 2019). On the other hand, modern breeding has certainly attained some success in constructing elite cultivars using the limited amount of genetic variation still retained in cultivated rice plants and acquired mutations in key genes involved in critical agricultural traits; however, this strategy has been sometimes confronted with limits attributed to a low level of genetic variation. Therefore, the positive resurrection of genetic variations lost during domestication will be a powerful tool to solve deadlocks in genetic improvement (Kamboj et al., 2020).
During the rice domestication, one of the most drastic events was exclusion of prostrate plants, mostly exhibited by wild rice, which triggered to form majority of the erect plants found in almost all the modern rice cultivars. This selection probably preceded at the early phase of the domestication, which conferred an increase in yield per area by supplying the ability for dense planting. This characteristic was extremely important in the ancient times when farmland expanding ability was low and/or farmland with appropriate conditions was restricted.
In 2008, the domestication locus responsible for the erect tiller trait was identified and named the Prostrate growth 1 (Prog1), a gene located on the short arm of chromosome 7 (Jin et al., 2008; Tan et al., 2008). The Prog1 gene encodes a C2H2-type zinc finger transcription factor (Agarwal et al., 2007). Recently, Wu et al. (2018) reported that O. rufipogon (accession DXCWR) possesses approximately 110 kbp of an additional chromosomal segment named the RICE PLANT ARCHITECTURE DOMESTICATION (RPAD) in the near vicinity of the Prog1 gene. The RPAD segment contains seven tandem Prog1-like genes; at least three of the Prog1-like genes were confirmed to modify plant architecture. The erect habit of cultivated rice plants was presumed to be guided by the synergistic effects of sequence changes in the Prog1 and deletion of the RPAD segment from the O. rufipogon genome (Huang et al., 2020).
Growth of crops, including rice plants, certainly requires the adequate sunlight as a basal energy source for photosynthesis and is greatly affected by the efficiency of sunlight reception. In fact, Monteith (1977) experimentally found that primary crop production is proportional to the amount of intercepted light energy. Therefore, improving the light-intercepting characteristic of plant shape is expected to promote crop growth. The exact plant shape, which is most effective in perceiving solar radiation, is a point of debate. Theoretical studies examining the relationship between leaf angles and dry matter production have suggested that erect leaves are preferable for increasing the capture of sunlight and enhancing photosynthetic production during the late vegetative growth phase when leaves grow thicker and become mutually shaded (Monteith, 1965; Duncan et al., 1967). In terms of growth promotion during the late vegetative phase, rice lines possessing erect leaves and/or panicles have been constructed that can achieve some increase in yield (Sakamoto et al., 2006; San et al., 2018; Fei et al., 2019). In contrast, Monteith (1965) and Duncan et al. (1967) also reported that inclined leaves are more efficient in perceiving light when there is no mutual shading as typically found in plants with a low leaf area index (LAI), implying that a spreading phenotype is preferable for plants during the early vegetative growth phase. These interpretations indicate that the ideal plant shape for efficient light capture changes depending on the growth phase of the crop.
Among biotic factors interfering with crop yield, weeds constantly invade and spread in fields and are a major cause of reduced yield in all the regions of the globe (Chauhan, 2020). Conventional weed control by manually removing weeds from the field is burdensome and the frequent application of herbicides is expensive and comes with ecological costs such as environmental destruction risk and/or the appearance of herbicide-resistant weeds. Fewer herbicide applications relieve not only the economic burden, but also the ecological issues; however, these applications alone will reduce yields of crops due to insufficient weed control. More ecological approaches, which can substantially reduce herbicide applications, i.e., construction of cultivars with an appropriate plant shape that preserves strong competitiveness against weeds, have been considered (Johnson et al., 1998; Fischer et al., 2001; Dass et al., 2017).
The ideal plant shape for rice to improve weed competitiveness is a plant with many tillers and sloping leaves (Gibson et al., 2003; Koarai and Morita, 2003). These traits would form a deep canopy, limit light penetration onto the ground, and then suppress the growth of weeds under the canopy. From these perspectives, researchers have sought and examined cultivars displaying such ideal plant shapes for strong weed competitiveness. However, it was feared that such traits would lead to a severe decrease in yield due to energy loss by mutual shading of the crop leaves, i.e., the features conflict with the preferable plant shape for high yields in the late vegetative phase. Therefore, cultivars with compatible plant shape are earnestly demanded, especially in developing countries with little economic capacity, which would reduce the weeding costs. Even in developed countries, the decrease of herbicide application has great merits to relieve not only environmental loads, but also appearance of herbicide-resistant weeds (Annett et al., 2014; Heap and Duke, 2018; Islam et al., 2018).
From the perspective of agricultural management, sparse planting of rice seedlings is demanded to save labor and the production cost. However, the sparse planting brings to a new weakness in weed control because the canopy closure of rice plants, which represses weed growth, is delayed. We have to examine ideal plant shapes for the sparse planting that maintain yield and are compatible with weed control.
In this study, we established the rice near-isogenic line (NIL) of Koshihikari holding ideal plant shapes that are compatible with yield and weed control even under sparse planting condition. Koshihikari is the most widely accepted japonica cultivar for staple food in Japan due to its good taste and is recognized to be a monumental existence that has made great contributions to rice breeding in Japan as a mating parent. Koshihikari has erect tillers, which characteristic is generally found in common cultivated rice. The Koshihikari-background NIL contains a genomic segment of chromosome 7 from an accession of O. rufipogon collected in Thailand, which segment carries the novel Prog1 sequence with the RPAD. The NIL displayed the spreading tiller phenotype in the vegetative growth phase; however, the tillers began to erect from the entry of the reproductive growth phase. This transition between the plant shapes complies with maintenance of optimal light receiving efficiency throughout plant development; therefore, the NIL balances promoted growth and significant weed suppression. These results suggest that the ancestral genetic resources used in this study have a great ability to upgrade current rice farming to innovative systems fitting with the United Nations Sustainable Development Goals (SDGs)1.
In this study, Koshihikari, a cultivar of japonica rice (O. sativa), was used as the control. We used GP9-7, the NIL for Koshihikari, containing a segment of chromosome 7 from an accession of O. rufipogon collected in Thailand (IRGC Acc. No. 104814). This line was selected by marker-assisted breeding from lines obtained in the BC4F2 generation of KRIL31 backcrossed to Koshihikari. KRIL31 is a line in the chromosome segment substitution lines (CSSLs) containing chromosomal segments from wild relatives in the background of japonica cultivars, which has been constructed in our former work (Hirabayashi et al., 2010). This study was conducted at the Tsukuba-Kannondai test fields for NARO (N-36.0°, E-140.1°, 22 m above sea level) and at the Western Region Agricultural Research Center, Fukuyama test fields for NARO (N-34.5°, E-133.4°, 1 m above sea level). Details of the field conditions in our experiments were described in the text or the legends of the figures. A summary of climate conditions in the years of the field experiments indicates in Supplementary Table 1.
After emasculation of pollen on female plants by soaking panicles in hot water at 43°C for 7 min, mating was accomplished by sprinkling the pollen of male plants onto emasculated female stigmata. DNA was extracted from a small piece of a leaf tip using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) following the instruction of the manufacturer. Genotyping was carried out by PCR using the single sequence repeat (SSR) markers (Temnykh et al., 2000; McCouch et al., 2002; International Rice Genome Sequencing Project, 2005) listed in Supplementary Table 2. The PCR mixture (10 μl) consisted of 0.5 μl of template DNA, 5 μl of GoTaq Green Master Mix (Promega, Madison, WI, United States), and 0.6 μl of 10 μM primers. Amplification was performed for 35 cycles of 94°C (30 s), 55°C (30 s), and 72°C (30 s). Amplified DNA products were electrophoresed in 3.0% (w/v) NuSieve 3:1 Agarose Gels (Lonza, Basel, Switzerland, United Kingdom).
At designed intervals, above ground parts of randomly selected seven plants grown in a paddy field (500 m2; interplant space = 18 cm × 25 cm; 22.2 plants m–2) were harvested. Leaf area was measured with a leaf area meter (AAM-9, Hayashi Denko Corporation Ltd., Tokyo, Japan) and LAI was calculated. All the samples were oven dried for 2 days prior to measuring dry weight. The crop growth rate (CGR) (g m–2 day–1) and net assimilation rate (NAR) (g m–2 day–1) were calculated using the following equations:
where, K indicates the plant density (plants m–2), W1 and W2 represent the above ground dry weight (g) at times t1 and t2, respectively, and A1 and A2 represent the leaf area (m2) at t1 and t2, respectively.
Tiller inclination angles were measured as the angle from the horizontal to a tiller located on the outermost circumference. The vegetation cover rates were calculated from binarized images of plants taken from directly above as the ratio of pixels corresponding to the plant body to the total number of pixels. The binarized images were generated from the a∗ signals in the CIELAB (Commission internationale de l’éclairage L∗ a∗ b∗) color space of the original images converted with the ImageJ software version 1.52 k2 with a color space converter plugin (LPX color; LPIXEL, Tokyo, Japan).
In this study, Echinochloa crus-galli (L.) Beauv. var. formosensis Ohwi (E. crus-galli var. formosensis), a well-known grass weed in Japan, was used. At 10 days after planting (DAP) of rice seedlings on paddy fields (interplant space = 30 cm × 30 cm; 11.1 plants m–2), E. crus-galli var. formosensis at the first leaf stage was transplanted to the middle of rice rows. The number of tillers of E. crus-galli var. formosensis was measured every other week. A total of 10 weed plants per plot (1.8 m × 3.3 m = 5.94 m2) were examined in three repetitions. In addition, all the E. crus-galli var. formosensis naturally emerging in the three plots of the field were sampled 10 days before rice harvest and the number of emerging weeds, their tiller number, and their dry matter weight were measured.
Vertical transitions of the RPPFD in the rice stand were measured on a cloudy day at noon using a quantum sensor (LI-190SB, LI-COR, Lincoln, NE, United States). Simultaneous measurements were carried out for locations every 10 cm from the ground surface in the rice stand and above the canopy (1 m from the ground surface) using the measurements taken above the stand as 100%. The measurements were recorded three times and the mean value was used in the analysis.
Transitions of the RPPFD in the rice stand during the growth were measured using a line quantum sensor (366813M; Ollie Corporation Ltd., Osaka, Japan). Simultaneous measurements were carried out in the rice stand (ground surface) and outside the stand (1 m from the ground surface) using measurements taken outside the stand as 100%. Measurements were recorded once a week from transplanting to harvest. The measurements were recorded three times and the mean value was used in the analysis. At the same time, the number of rice tillers was measured for three plots that consist of 10 plants.
A total of 12 individual plants randomly selected from 100 plants grown in a field (4 m × 3.3 m = 13.2 m2; interplant space = 36 cm × 36 cm; 7.7 plants m–2) were harvested as one plot. The panicles in each plot (three plot replicates) were mechanically threshed, the obtained grains were dried, and the weight of the paddy rice was measured to determine grain yield. Samples for the taste test were obtained from plants growing in the same field as those used for grain yield measurements. Taste tests were conducted at the AiHO Rice Cooking Research Institute (Toyokawa, Aichi, Japan). Protein and amylose contents were measured using a Rice Composition Analyzer (Shizuoka Seiki Corporation Ltd., Fukuroi, Japan). Mido Meter (Toyo Rice, Wakayama, Japan) and Rice Taste Analyzer (Satake Corporation, Higashi-Hiroshima, Japan) were used for objective comparisons of taste-related factors. Values for texture of cooked rice were examined by a Tensipresser (Takemoto Electric Incorporation, Tokyo, Japan).
Statistical analyses were conducted using the statistical computing software R, version 3.6.33.
For the first trial, we examined the agricultural traits of KRILs (Hirabayashi et al., 2010) in field conditions in which seedlings were planted in sparse (interplant space = 36 cm; 7.7 plants m–2) or dense (interplant space = 18 cm; 30.9 plants m–2) plant densities. The KRILs consist of 40 lines, each of which possess one or few chromosomal segments of O. rufipogon (IRGC accession number 104814) in Koshihikari background. This CSSL (KRILs) were constructed to examine whether the wild rice genetic resources can be used to overcome weaknesses and/or add new features to Koshihikari. Among the KRILs, KRIL31 displayed wide spreading with an increased number of tillers, especially in the sparsely planted condition (Supplementary Figure 1). Therefore, we focused on these traits that altered vegetative growth presumably caused by a change in their light-receiving posture, which could also provide strong weed competitiveness. Interestingly, KRIL31 did not have a clear dwarf phenotype, even though its tiller number had more than doubled compared with that of Koshihikari. The BC1F1 plants derived from a cross between KRIL31 and Koshihikari showed both the traits, indicating these traits were inherited dominantly (data not shown). KRIL31 possesses almost all of the long arm of chromosome 9 and three small segments in chromosomes 1, 3, and 7 from O. rufipogon (Hirabayashi et al., 2010; Supplementary Figure 2A). KRIL31 tillers maintained a strong spreading trait throughout plant development. When the chromosome 9 segment from O. rufipogon replaced that of Koshihikari, the spreading trait in the reproductive phase disappeared (data not shown). Yu et al. (2007) indicated that the Tiller Angle Control 1 (TAC1) gene, which mediates tiller angle during the reproductive stage (late compact stage), is localized in the long arm of chromosome 9. This study aligns with our observation that when this segment was replaced with Koshihikari, resultant progenies lost tiller inclination during the reproductive growth phase. In contrast, the segment of chromosome 7 from O. rufipogon was linked to leaning tillers only during the vegetative growth phase. To define the segment responsible for the spreading of tillers, we conducted phenotypic and genotypic analyses of 430 lines of the BC2F2 generation, which indicated that the loci for spreading and increasing tiller number were detected between two SSR markers, RM20967 and RM20999 (Supplementary Figure 2B). We selected the NIL, GP9-7 from 300 lines of the BC3F4 generation by genotypic analyses based on the above information. The chromosomes of GP9-7 had almost reverted to Koshihikari form except for the region between two SSR markers, RM20973 and RM21002 (Figure 1A). We used this line in all the subsequent studies.
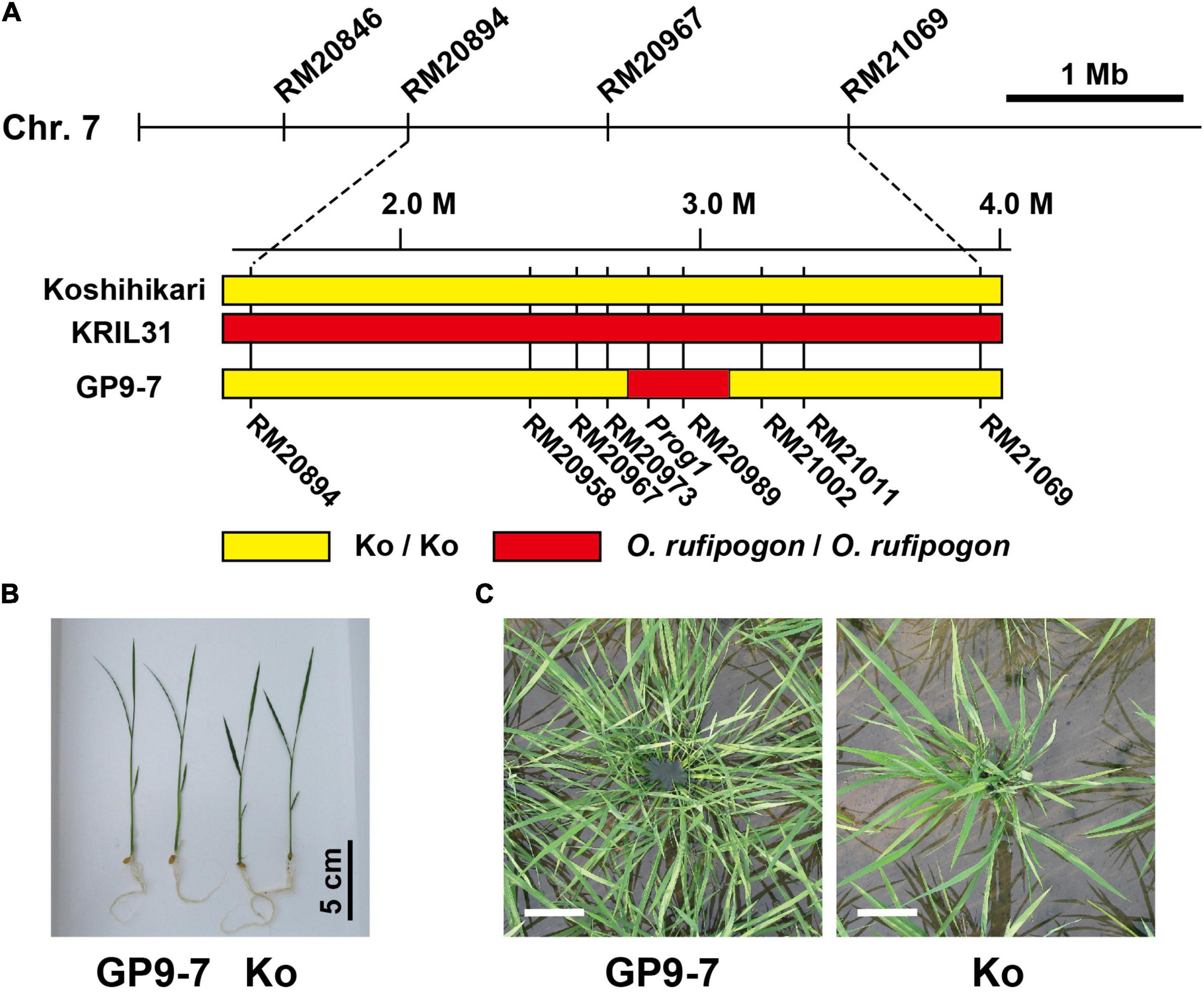
Figure 1. Basic properties of GP9-7. (A) A comparative diagram showing the genotyping results of GP9-7, KRIL31, and Koshihikari. The yellow zones indicate the segments derived from Koshihikari and the red zones represent the segments derived from Oryza rufipogon (O. rufipogon). (B,C) Typical plant shapes of the rice plants are used in this study. The photograph (B) shows typical nursling seedlings of Koshihikari and GP9-7 at 10 days after sowing. The photographs (C) indicate typical plant shapes in paddy fields taken from directly above the plants at 33 days after planting (DAP). Scale bars: 10 cm.
As mentioned above, GP9-7 contains a small segment of chromosome 7 from O. rufipogon (IRGC accession number 104814) between two SSR markers, RM20973 and RM21002 (Figure 1A). The segment between the two markers corresponds to 358 kbp based on the Nipponbare genome sequence (International Rice Genome Sequencing Project, 2005). This segment includes the Prog1 gene that is involved in key processes required for domestication and has been reported to determine tiller inclination angle and number of tillers (Jin et al., 2008; Tan et al., 2008). The Prog1 gene encodes a single C2H2 zinc-finger transcription factor (Agarwal et al., 2007; Supplementary Figure 3). The Prog1 sequences of O. rufipogon collected in China have been sequenced comprehensively, but the Prog1 sequence of GP9-7, which is from an accession of O. rufipogon (IRGC accession number 104814) collected in Thailand, is novel. This sequence is similar to that of cultivated rice with a deletion of 9 bp in the open reading frame that causes a deletion of three residues near an EAR (ethylene-responsive element binding factor-associated amphiphilic repression)-like motif (Kagale et al., 2010; Supplementary Figure 3). Wu et al. (2018) proposed that the RPAD region is also involved in determining plant shape, located on the vicinity of the Prog1 gene in the O. rufipogon genome. To verify the insertion of the RPAD segment in GP9-7, a PCR assay was performed using a primer set to detect the RPAD insertion. The assay revealed that GP9-7 probably possesses the RPAD segment (Supplementary Figure 4) that is likely to confer the ability to alter plant shape together with the Prog1 gene.
Nursling seedlings of GP9-7 were slightly thinner and more elongated compared with Koshihikari (Figure 1B). After planting on paddy fields in sparse condition (interplant space = 36 cm × 36 cm; 7.7 plants m–2), the number of GP9-7 tillers increased and began leaning more each day (Supplementary Figure 5; Supplementary Movie 1), comparable with previously reported inbred lines containing similar chromosomal segments of O. rufipogon (Jin et al., 2008; Tan et al., 2008). The number of GP9-7 tillers was roughly equivalent to those of Koshihikari until about 15 DAP; however, afterward, the number of GP9-7 tillers increased significantly, reaching more than three times those of Koshihikari by 43 DAP (Supplementary Figure 5). The plastochron of GP9-7 was almost equivalent to that of Koshihikari (data not shown). This observation and the biased increase in tiller number toward the later developmental stages suggested that higher order tillers that normally do not appear in Koshihikari did emerge from GP9-7. Tillers that elongate too much can lead to mutual shading, but the emerging GP9-7 tillers spread in all the directions with very few overlaps and then assumed a parabolic antenna-like structure (Figure 1C). Therefore, leaves on the tillers of GP9-7 did not compete with each other for light.
The erect tiller trait in cultivated rice, including Koshihikari, is caused by the upward curving of laterally emerging non-elongated internodes (Supplementary Figure 6A). Tan et al. (2008) reported that the curved internodes are due to their asymmetric development in which the near-ground border, the outermost cell layer of the tiller base closest to the ground, is longer than the border further away from the ground. Cell sizes for both the borders were almost equivalent suggesting that the curved internodes are due to an increase in cell number on the near-ground border. In contrast, the laterally emerging non-elongated internodes of GP9-7 were straight (Supplementary Figure 6A), probably due to symmetric development between the near-ground and the far-ground borders. This observation does not mean that GP9-7 is not gravitropic. Coleoptiles of dark-germinated Koshihikari and GP9-7 seedlings were clearly gravitropic in an experiment in which seedlings were rotated horizontally at the midpoint of an incubation period (Supplementary Figure 6B). Furthermore, tillers of GP9-7 began to rise during the transition from the vegetative to the reproductive growth phase (Figure 2A; Supplementary Movie 2). This trait was derived from a clear gravitropic bend in GP9-7 nodes, especially the node between internodes III and IV (Supplementary Figure 6C), resulting in erect panicles (Figure 2B). This trait favorably increasing the light reception efficiency in the thicker stand as is often the case in the reproductive growth phase (Sakamoto et al., 2006; San et al., 2018; Fei et al., 2019). In addition, this trait is preferable for modern agricultural harvesting methods using combine harvesters. As mentioned above, GP9-7 exhibited a clear transition in plant shape from the vegetative growth phase during which plants had inclined tillers to the reproductive growth phase when the tillers were erect.
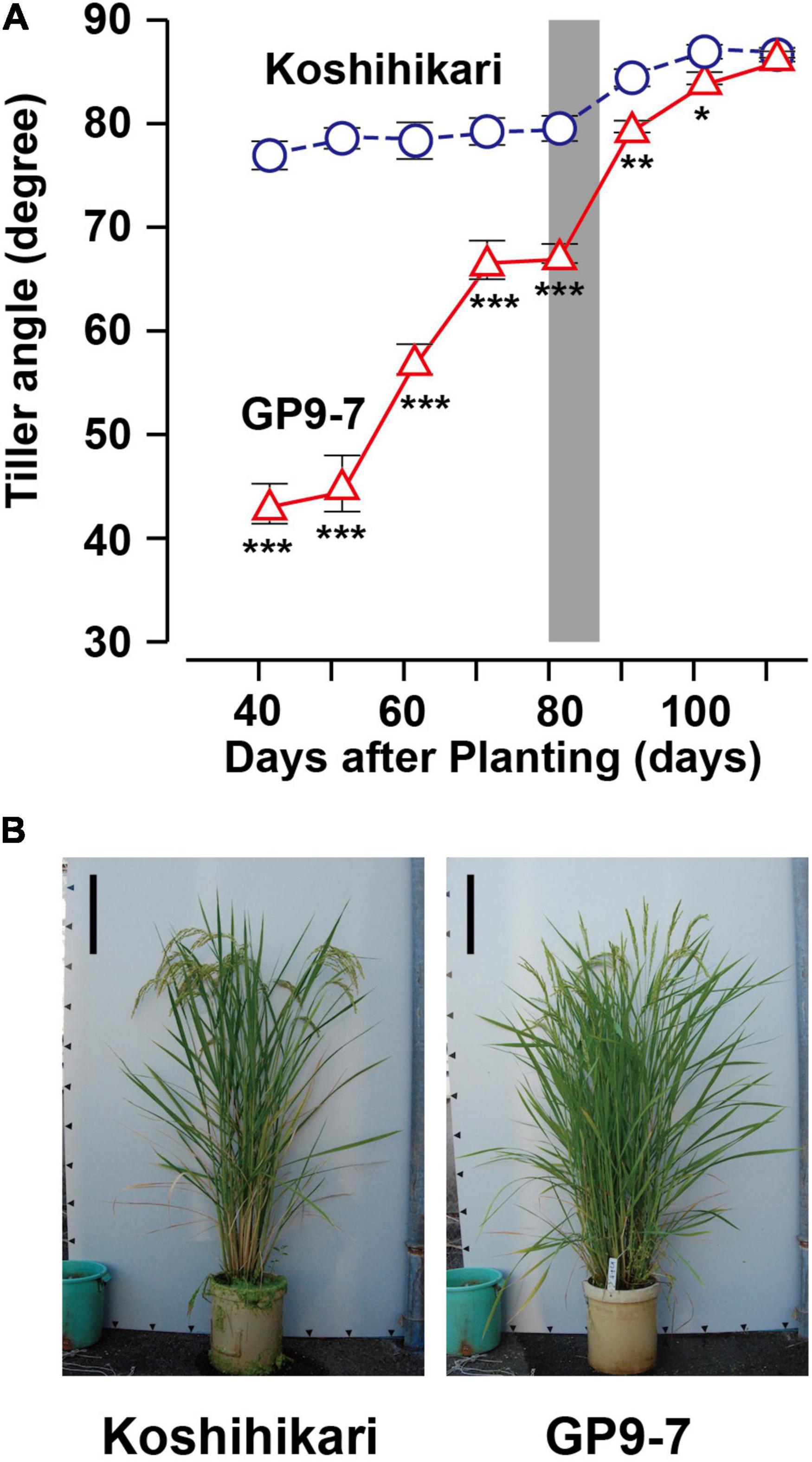
Figure 2. Basic properties of GP9-7 toward the reproductive growth phase. (A) Developmental transitions of tiller angles from horizontal in Koshihikari (blue circles with broken lines) and GP9-7 (red triangles with solid lines) grown in a test field at Tsukuba, Japan in the summer of 2020 (planting date: May 21, 2020). The tiller angles of the same six plants were continuously measured and the mean values with SE were shown in the graph. The period for heading is indicated by a vertical gray bar. Symbols, ***, **, and * indicate statistically significant differences compared to Koshihikari at P < 0.001, P < 0.01, and P < 0.05, respectively, calculated by the Welch’s t-test. The photographs (B) display typical plant shapes of Koshihikari and GP9-7 during the ripening phase (25 days after heading). Scale bars: 20 cm.
GP9-7 and Koshihikari were grown in a field in Tsukuba, a region in eastern Japan, using local agricultural practices (interplant space = 18 cm × 25 cm; 30.9 plants m–2). Leaf area and shoot dry matter weight were measured at intervals; LAI, CGR, and NAR were calculated from these values (Figure 3). Early vegetative growth of GP9-7 up to 50 DAP was indistinguishable from Koshihikari in terms of dry matter weight per plant; however, beginning at 57 DAP, the increase in dry matter weight of GP9-7 was significantly accelerated (Figure 3A). Therefore, the CGR values for GP9-7 during these periods were higher than those of Koshihikari (Figure 3B). Since the CGR is recognized as the product of LAI and NAR, it is easy to distinguish which factor contributes more to the increase in the CGR. The factor that promoted the CGR of GP9-7 from 50 to 57 DAP was attributed to a temporary increase in the NAR during the same period (Figures 3B,D). In contrast, the higher CGR in the subsequent period (57 to 85 DAP) was due to an increase in LAI (Figures 3B,C). The temporary increase in the NAR may be due to improved light-intercepting characteristics in the stand rather than from an increase in the photosynthetic activity of leaves, as discussed in more detail in the next section.
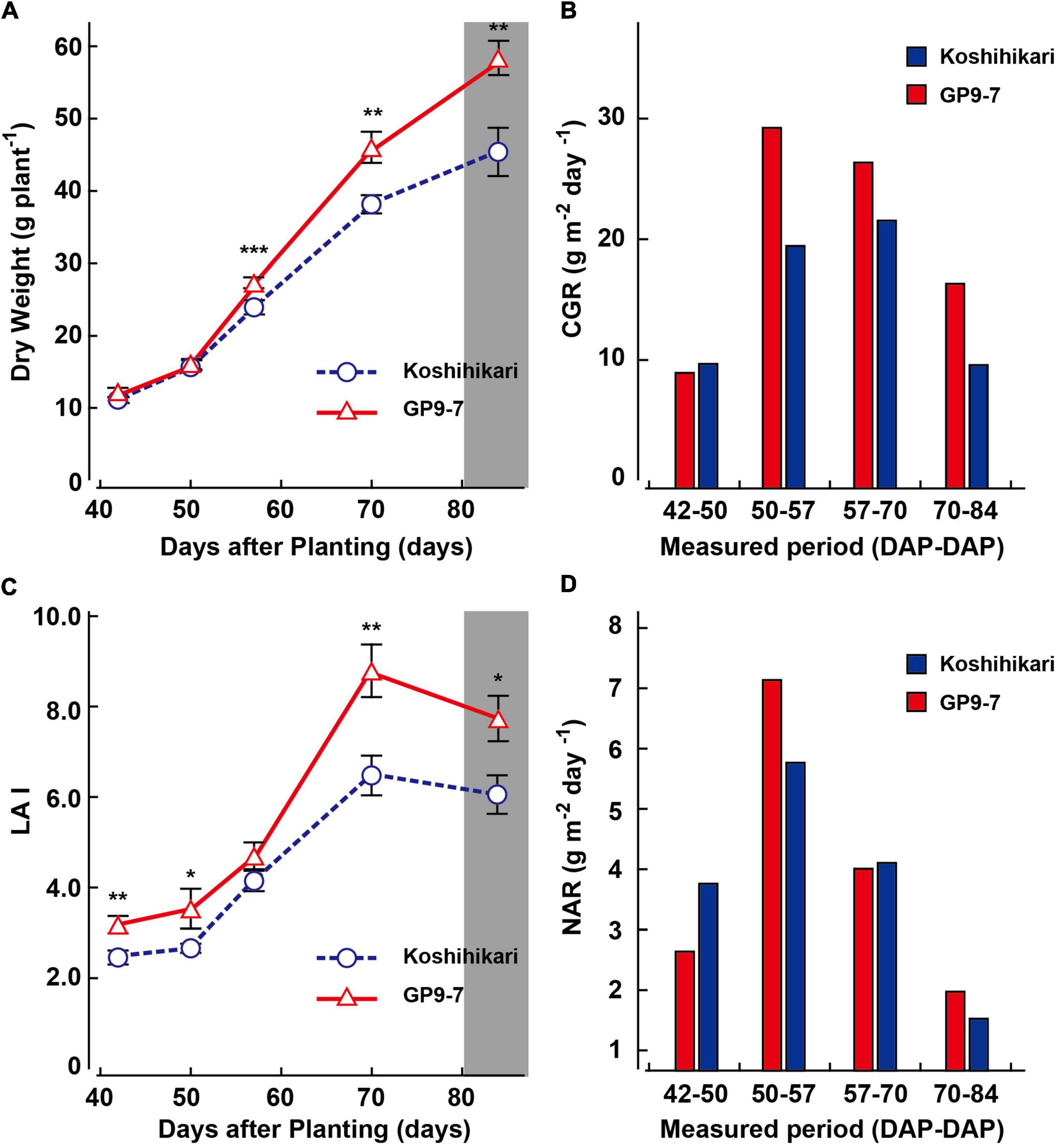
Figure 3. Developmental transitions of growth traits of Koshihikari (blue circles with broken lines or blue bars) and GP9-7 (red triangles with solid lines or red bars) from the vegetative to the reproductive growth phase. Transitions of dry weight per plant (A), crop growth rate (CGR) (B), leaf area index (LAI) (C), and net assimilation rate (NAR) (D) of Koshihikari and GP9-7 grown in a test field at Tsukuba, Japan in the summer of 2018 (planting date: May 9, 2018). Mean values of the dry weights and leaf area indexes obtained from seven plants are plotted. The SE values are shown as error bars in the graphs. The period for heading is indicated by a vertical gray bar. Symbols, ***, **, and * indicate statistically significant differences compared to Koshihikari at P < 0.001, P < 0.01, and P < 0.05, respectively, calculated by the Welch’s t-test.
The increased growth performance of GP9-7 is probably due to the enhanced amount of light energy received by the individuals attributed to their distinctive shape. Tiller emergence of GP9-7 plants was greater than that of Koshihikari and the tillers were arranged radially causing less mutual shielding (Figure 1C; Supplementary Figure 5). Therefore, the vegetation cover ratios of GP9-7 in the midvegetative phase were significantly higher than those of Koshihikari (Figure 4A). This result suggests that the light-intercepting efficiency of GP9-7 was increased by the plant shape. The RPPFD of GP9-7 in the stand at 53 DAP had a characteristic vertical profile (Figure 4B) when the NAR of GP9-7 was at its peak (Figure 3D). The vertical RPPFD profile in cultivated rice plants with erect tillers usually indicates a gradual decrease in light intensity from the canopy to the ground surface. In alignment with this expectation, the vertical RPPFD of Koshihikari gradually decreased below 60 cm (Figure 4B). In contrast, an extreme decrease in the RPPFD was detected below 40 cm in GP9-7 (Figure 4B). This characteristic was attributed to the parabolic antenna-like morphology of GP9-7 that had many uniformly spread tillers with a constant slope, forming wide shadows without mutual shielding (Figure 1C). Under the condition, slightly excessive LAI should be connected to effective light capture. In addition, tilted leaf sheaths of GP9-7 were fully exposed to the sky (Figure 1C), which implies that photosynthetic assimilation in the leaf sheaths contributes growth promotion of GP9-7. Actually, Guo et al. (2011) reported that rice leaf sheaths had active photosynthetic apparatus such as leaf blades. Moreover, the leaf sheath photosynthesis was accounted for 10 to 20% of the final yield. Our observation that leaf sheaths of GP9-7 were dark green, such as active photosynthetic organs, is consistent with these interpretations. Thus, efficient light receiving of GP9-7 attributed to the distinctive plant shape during the period from 50 to 57 DAP that could be associated with the temporary improvement of the NAR during this period (Figure 3D). In the subsequent period (57 to 85 DAP), GP9-7 had the extremely higher LAI, which raises the risk of mutual shielding as an inevitable consequence. However, the plant shape transition of GP9-7 from 57 DAP, when tillers began to erect (Figure 2A), could adaptively sustain a preferred light receiving character in the thicker stand.
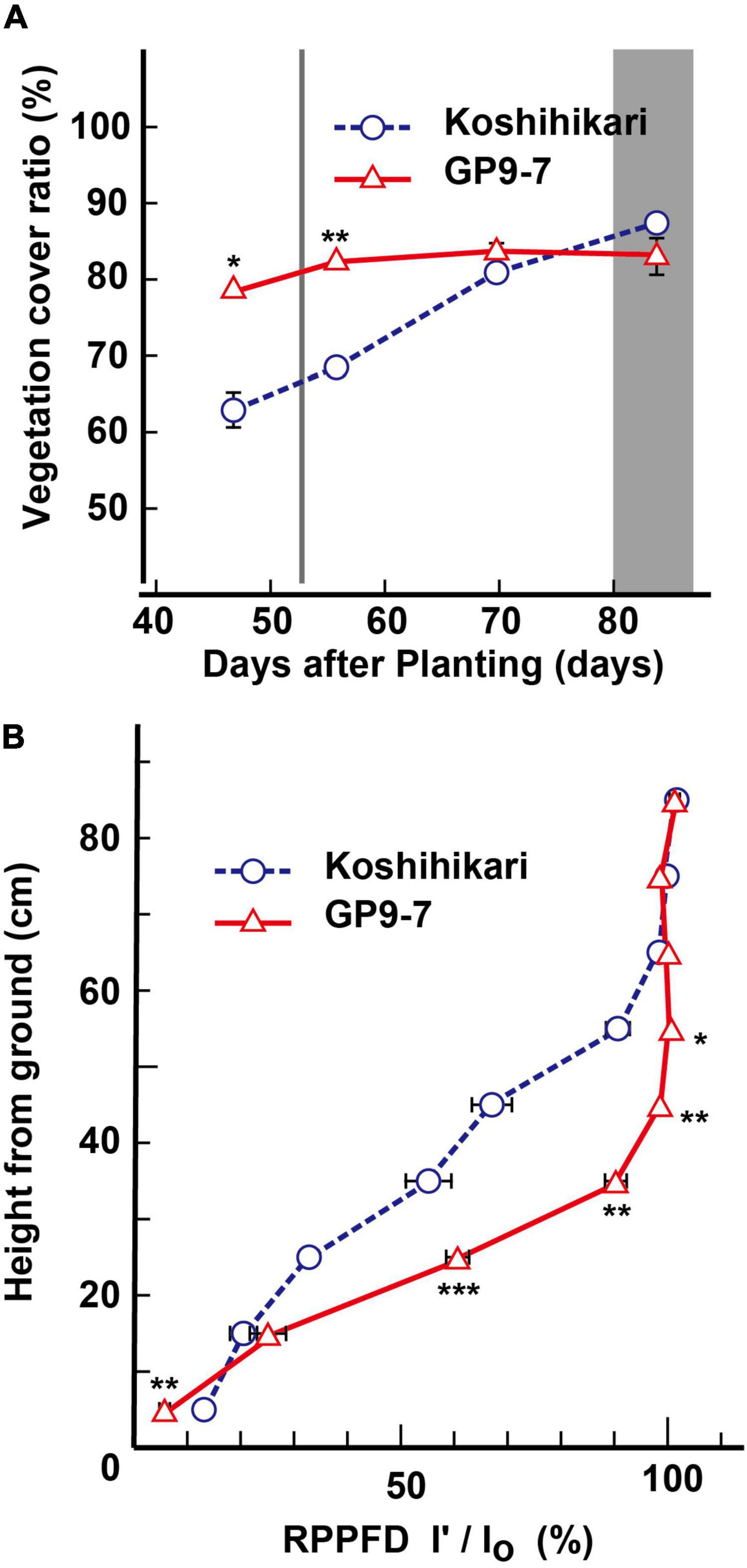
Figure 4. Light reception-related properties of GP9-7. (A) Developmental transitions in vegetative coverage of Koshihikari and GP9-7 in the test field at Tsukuba, Japan in the summer of 2018 (planting date: May 28, 2018). The period for heading is indicated by a vertical gray bar. This analysis was performed according to Materials and Methods, using images continuously taken at the same three points in the field, and the mean values with SE were indicated in the graph. (B) Vertical transitions of the relative photosynthetic photon flux density (RPPFD) of Koshihikari and GP9-7 measured on the 52nd DAP that is shown by a vertical gray line in panel (A). Symbols, ***, **, and * indicate statistically significant differences compared to Koshihikari at P < 0.001, P < 0.01, and P < 0.05, respectively, calculated by the Welch’s t-test. Values for Koshihikari are indicated with blue circles with broken lines and values for GP9-7 are represented with red triangles with solid lines.
Ground coverage proceeded more rapidly in GP9-7 than in Koshihikari (Figure 4A), a factor that restricted light penetration onto the ground surface. Furthermore, the RPPFD just above the ground surface under the GP9-7 canopy at 53 DAP was 5% of that above the canopy and significantly lower than that of Koshihikari (Figure 4B). Since the lower ground level RPPFD is preferable for optimal weed control (Gibson et al., 2003; Koarai and Morita, 2003), the RPPFD of the ground level in GP9-7 stands was examined in detail in Fukuyama, a region in western Japan. In this study, the planting date was mid-June (June 15, 2019), about a month later than that at Tsukuba (mid-May). Although the heading date relative to the DAP shown in Figure 5 was significantly different from experiments conducted in Tsukuba (Figures 2–4), the progression of vegetative growth was essentially similar to that of Tsukuba-grown plants. After planting, the number of GP9-7 tillers increased (Figure 5A) and spread uniformly, significantly decreasing RPPFD of the ground level at approximately 30 DAP (Figure 5A). After 35 DAP, Koshihikari and GP9-7 canopies covered the soil as shown by their similar and continued low RPPFD values. Tiller emergence of the transplanted weed, E. crus-galli var. formosensis, one of the most serious weeds in Japan, was significantly suppressed in GP9-7 stand (Figure 5B). The preceding attenuation of incident light under the GP9-7 canopy (Figure 5A) acted to suppress initial weed growth to a level insufficient to increase weed spread. Also, GP9-7 exhibited remarkable weed suppressive activity against the naturally occurring weed, E. crus-galli var. formosensis (Figure 6). Especially, tiller numbers and dry matter weights of naturally emerging weeds were significantly repressed under GP9-7 canopies (Figures 6B,C). Although GP9-7 did not completely eradicate the weeds in our experiment, the significant repression of weed growth under GP9-7 canopy may reduce nutrients interception by weeds and inhibit weed seed production, two factors that should help relieve farmers of weeding costs. Reduced numbers of herbicide applications will also decrease the environmental load and the risk arising herbicide-resistant weeds (Annett et al., 2014; Heap and Duke, 2018; Islam et al., 2018).
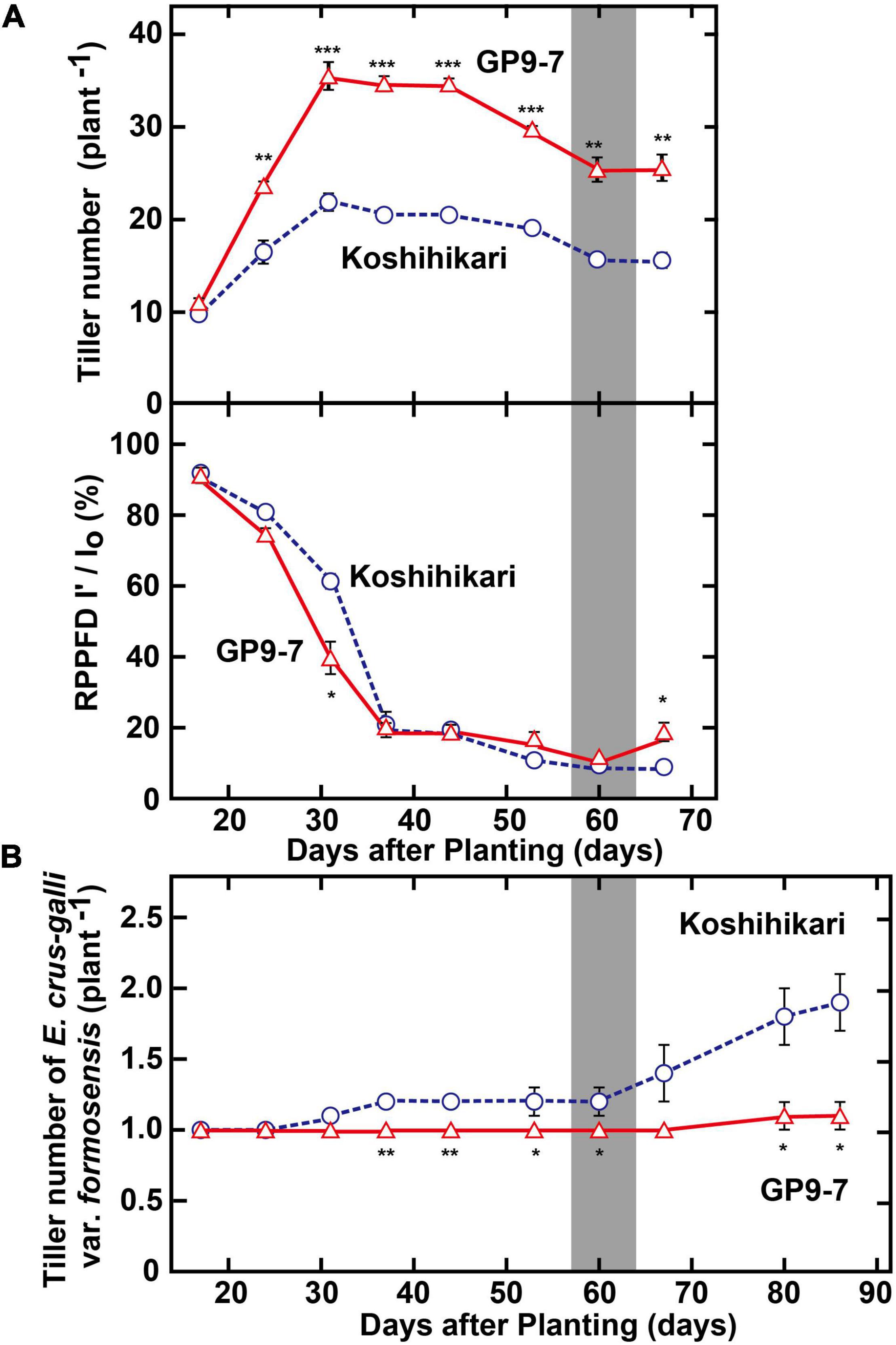
Figure 5. Weed suppression-related properties of GP9-7. (A) Developmental transitions in tiller numbers and the RPPFD at ground level of Koshihikari and the GP9-7 in the test field in Fukuyama, Japan in the summer of 2019 (planting date: June 15, 2019). The periods for heading are indicated by vertical gray bars. (B) Developmental transitions in tiller numbers of the transplanted weed, E. crus-galli var. formosensis under the canopy formed by rice stands. Symbols, ***, **, and * indicate statistically significant differences compared to Koshihikari at P < 0.001, P < 0.01, and P < 0.05, respectively, calculated by the Student’s t-test. Values for Koshihikari are indicated with blue circles with broken lines and values for GP9-7 are represented with red triangles with solid lines.
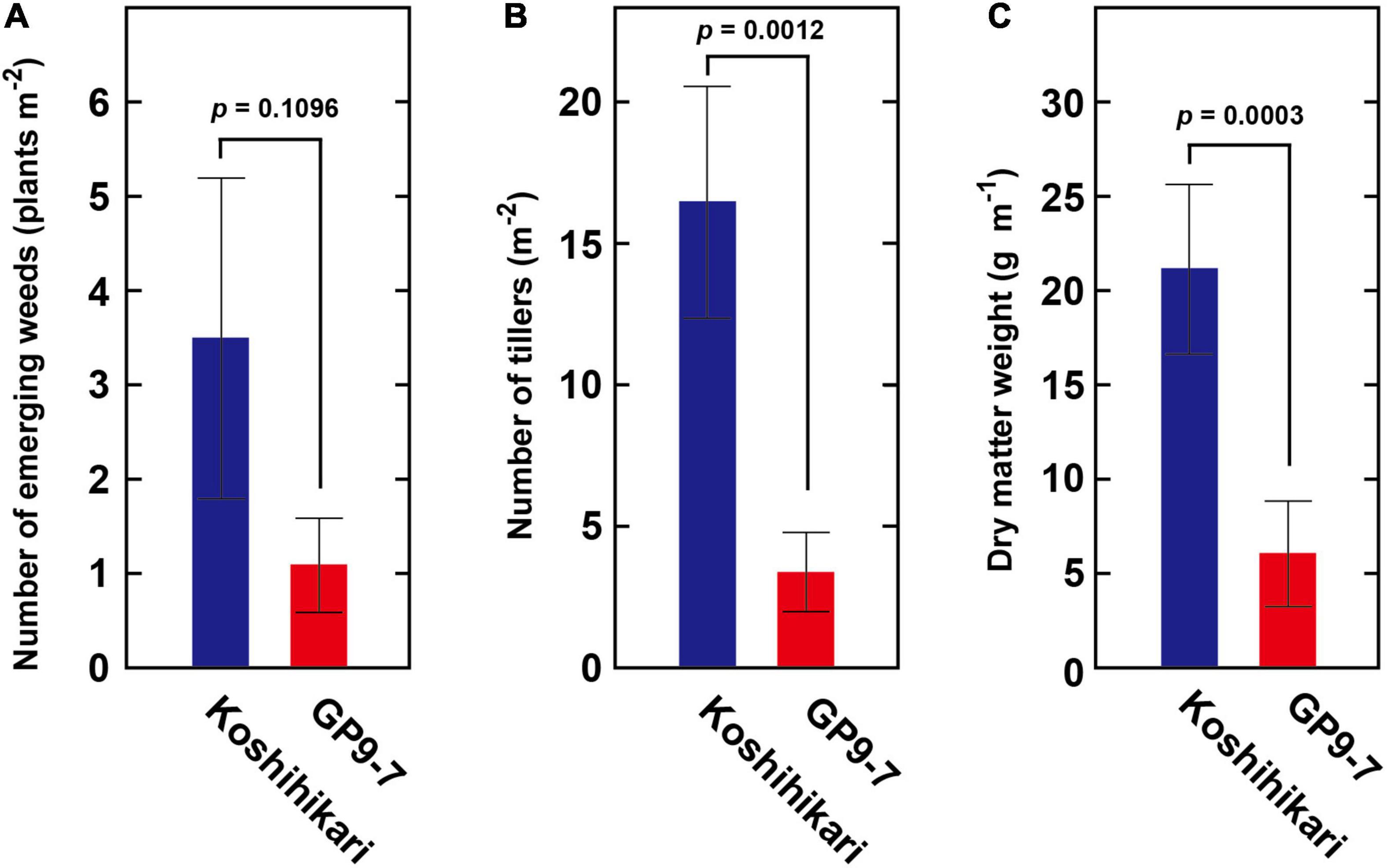
Figure 6. The weed competitiveness of GP9-7 was evaluated by measuring the growth of naturally occurring weeds in the summer of 2019 (planting date: June 15, 2019). The number of emerging weeds (A), their tiller number (B), and the dry matter weight (C) of naturally occurring E. crus-galli var. formosensis under the canopies of Koshihikari or GP9-7 are indicated. Mean values obtained from three plots for Koshihikari and GP9-7 with the SE values are indicated with blue and red bars, respectively. The p-values were calculated by the Student’s t-test and are reported above the bar graphs.
GP9-7 was constructed using the genetic background of Koshihikari, the most widely accepted japonica cultivar for staple food in Japan due to its good taste. Since GP9-7 contains a small segment of chromosome 7 from O. rufipogon (Figure 1A), GP9-7 displayed slightly delayed heading (Figure 7A), almost twice the number of panicles per m2 (Figure 7B), almost half the number of spikelets per panicles (Figure 7C), and a nearly equivalent amount of ripened grain (Figure 7D). The thousand grain weight of GP9-7 was also equivalent to that of Koshihikari (Figure 7E). Consequently, the final yield of GP9-7 was about the same as Koshihikari (Figure 7F). Significant increase of panicle number per plant (Figure 7B) should compensate the panicle number per area, which increase is restricted in existing cultivars under the sparse planting condition. On the other hand, significant promotion of GP9-7 vegetative growth (Figure 3A) did not result in an increase in yield (Figure 7F). We conducted yield tests on the Tsukuba fields from 2016 to 2020 and in the Fukuyama field in 2018 and 2019 (Supplementary Table 3). Despite the cultivation trials over the several summers, no significant increase in yield was detected. Possibly optimal cultivation conditions that bring out the best performance from GP9-7 have not been identified yet. Although the source ability of rice was improved in this study, the sink ability was not manipulated. This possibility may be an alternative reason for why the yield of the NIL did not increase in our experiment. Nevertheless, the introduction of a chromosomal segment that promoted vegetative growth and comparable grain yields is a significant advance since the yields of inbred lines to which a similar chromosomal segment was introduced were reported to be halved (Tan et al., 2008; Hua et al., 2016; Wu et al., 2018). Furthermore, in the yield tests conducted on the Fukuyama fields, no significant reduction of the grain yields of GP9-7 compared with those of Koshihikari was detected even in the semi-dense planting density (Supplementary Table 3), which cultivation condition is close to the previous reports (Tan et al., 2008; Hua et al., 2016; Wu et al., 2018).
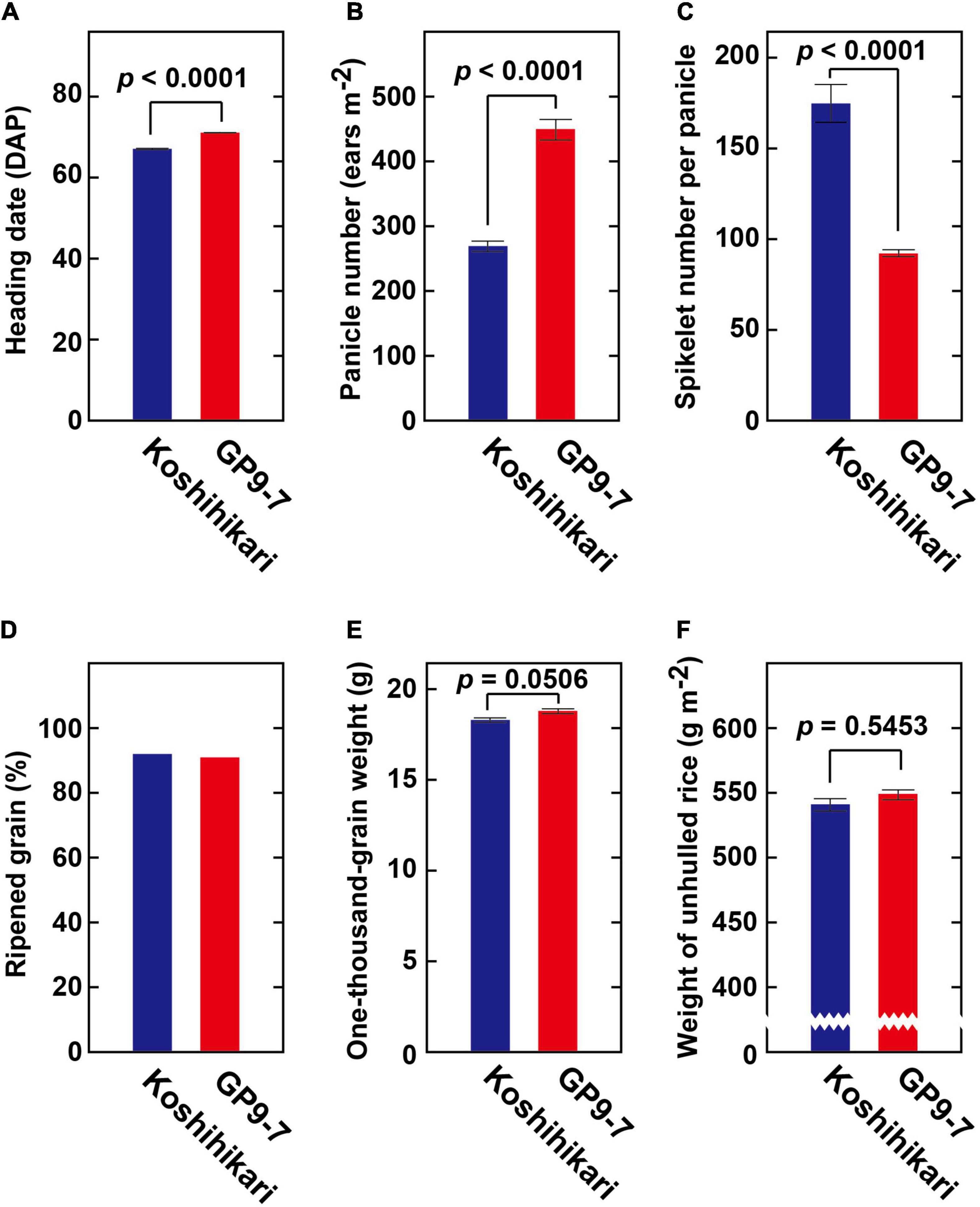
Figure 7. Yield-related traits of GP9-7. (A) Heading date (n = 12), (B) panicle number per m2 (n = 12), (C) spikelet number per panicle (n = 12), (D) percent ripened grain (n = 5), (E) one-thousand grain weight (n = 3), and (F) weight of unhulled rice (n = 3) of Koshihikari and GP9-7 cultivated in the test field at Tsukuba, Japan in the summer of 2018 (planting date: May 28, 2018) under sparse conditions (interplant space = 36 cm; 7.7 plants m–2). P-values were calculated by the Welch’s t-test and are reported above the bar graphs. Mean values for Koshihikari and GP9-7 with SE values are indicated with blue and red bars, respectively.
A similar inbred line, YIL18, was derived from a cross between the indica cultivar, Tequing, as the recipient parent and O. rufipogon (accession: YJCWR), as the donor parent (Tan et al., 2008). Another similar inbred line, DIL29, was derived from a cross between the indica cultivar, Guichao 2, as the recipient parent and O. rufipogon (accession: DXCWR), as the donor parent (Wu et al., 2018). Both the donor accessions, unlike ours, were collected in China. The grain yields of YIL18 and DIL29 were about 57% of Tequing and about 63% of Guichao 2, significantly lead to inferior yields (Tan et al., 2008; Hua et al., 2016; Wu et al., 2018). Hua et al. (2016) examined the canopy structure of YIL18 using three-dimensional digitizing analysis and found a significant decrease in the LAI of YIL18 compared with those of the recurrent parent, Tequing. This report also mentioned that YIL18 had greater mutual shading due to its prostrate plant shape.
We attribute this major discrepancy to three reasons: (1) Inbred lines YIL18 and DIL29 contain several wild rice chromosome segments in addition to the chromosome 7 segment. These additional chromosome segments may be responsible for the yield reduction; (2) In these cases, the recurrent parents were the indica cultivars, whereas we used Koshihikari, a japonica cultivar; and (3) The chromosomal donor in this study was collected from Thailand, whereas the donors in previous studies were collected from China. It is possible that differences in the sequence of this chromosomal segment may impart different traits. In fact, the Prog1 sequence of GP9-7 was different from those of accessions YJCWR and DXCWR (Supplementary Figure 3B). Progress in whole-genome sequence analysis of these accessions may shed light on the reasons for these discrepancies. Identifying which of the three types of O. rufipogon (Or-I, -II, and -III) (Huang et al., 2012) by genome-wide association analyses that accurately describe these accessions may sort out these confusing results.
The surface of GP9-7 milled grains was the white-like Koshihikari (Figure 8). The taste-related factors of GP9-7 were nearly identical except for the amylose content that was very slightly higher in GP9-7 compared to Koshihikari (Figure 8B). Cooked rice from GP9-7 had relatively softer texture factors (Figure 8B), suggesting that the introduced chromosomal segment of wild rice in this study did not negatively influence grain quality, including its taste.
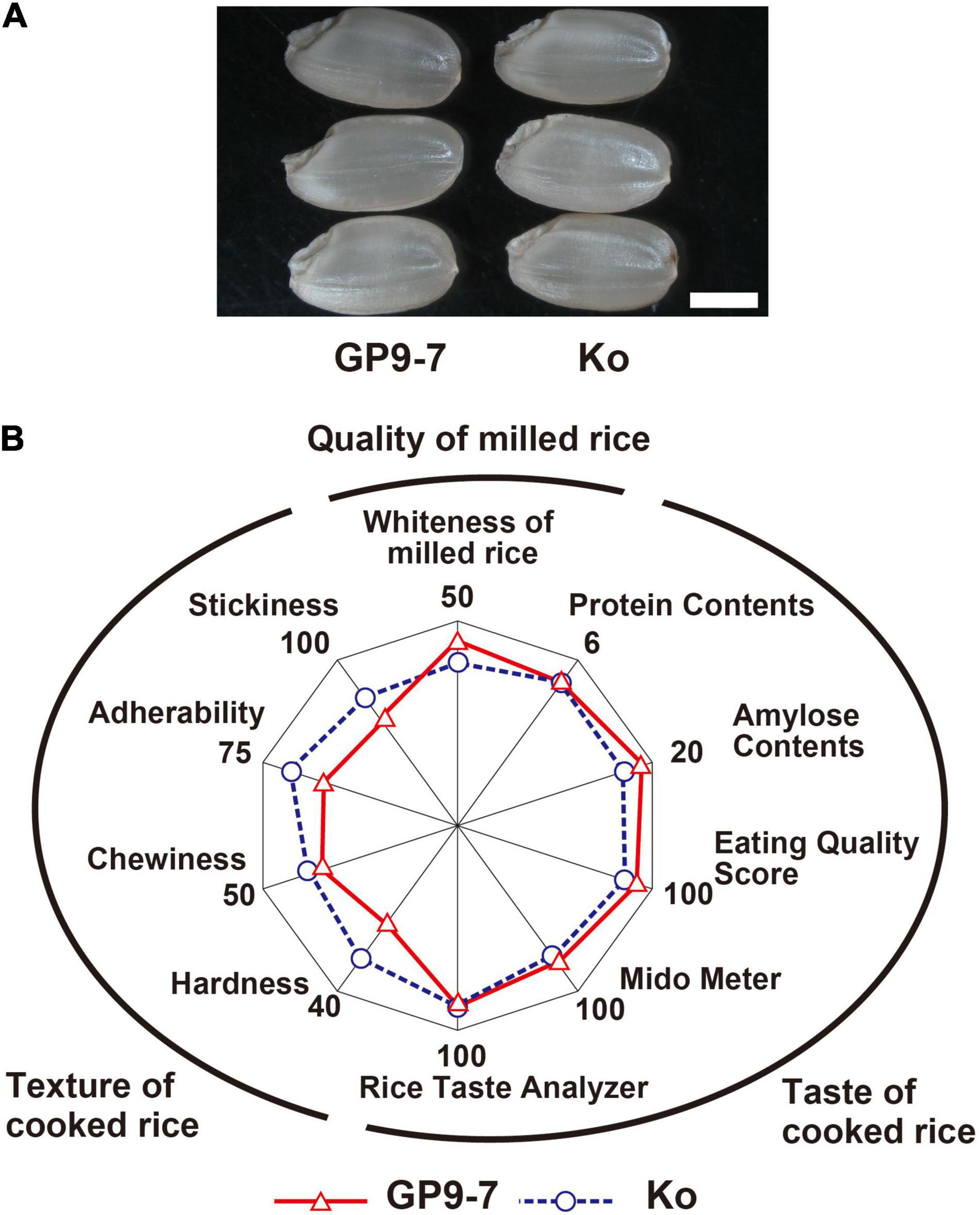
Figure 8. Grain quality-related traits of GP9-7 harvested in the Tsukuba field in the fall of 2018. (A) A morphological comparison of typical hulled rice grains of Koshihikari and GP9-7. Scale bar: 2 mm. (B) Summary of rice texture and taste quality tests. Values for Koshihikari are indicated with blue circles with broken lines and values for GP9-7 are represented with red triangles with solid lines.
The remaining flaw of GP9-7 was lowered lodging resistance attributed to the smaller stem diameters that are a trade-off for the increased number of tillers. We believe that improvements such as the introduction of alleles conferring lodging resistance are possible. Lodging resistance loci have been well studied and exploitation of them will overcome this flaw (Shah et al., 2019).
This study attained that construction of the NIL capable of accelerating vegetative growth and suppressing weeds using wild rice genetic resources without undesirable consequences and yield reduction. The practical use of the NIL or the chromosomal segment that causes these traits may reduce weed control costs and environmental consequences associated with herbicide application, which open a way to solve some obstacles to the United Nations SDGs. This study also indicates that the genetic diversity of wild rice deserted in the domestication process is a valuable resource for breeding new cultivars with desired traits. The Japanese concept of MOTTAINAI has to be pushed to the front in this case; we have an obligation to excavate and apply these precious resources hidden in wild relatives in order to realize a bright future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank/EMBL/DDBJ Acc. Nos. LC573903 and LC573904.
NI, HA, and KI designed the research and performed the experiments. HH contributed to the construction of the NIL. AU and TI provided critical supports for assaying weed control. NI and HA wrote most of the manuscript with help from all the other authors. All authors read and approved the manuscript.
This study was supported by grants from commissioned project studies on the “Development of Labor-Saving Management of Serious Weeds to Expand Cultivation of Direct-Seeded Rice (19190995)” and “Genomics-Based Technology for Agricultural Improvement (RBS-2010),” Ministry of Agriculture, Forestry and Fisheries, Japan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Tachibana (Western Region Agricultural Research Center/NARO), Koarai (Institute for Plant Protection/NARO), and Sentoku (Institute of Agrobiological Sciences/NARO) for supplying their informative insights and knowledge.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.748531/full#supplementary-material
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485. doi: 10.1007/s11103-007-9199-y
Annett, R., Habibi, H. R., and Hontela, A. (2014). Impact of glyphosate and glyphosate-based herbicides on the freshwater environment. J. Appl. Toxicol. 34, 458–479. doi: 10.1002/jat.2997
Chauhan, B. S. (2020). Grand challenges in weed management. Front. Agron. 1:3. doi: 10.3389/fagro.2019.00003
Chen, E., Huang, X., Tian, Z., Wing, R. A., and Han, B. (2019). The genomics of Oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 70, 639–665. doi: 10.1146/annurev-arplant-050718-100320
Dass, A., Shekhawat, K., Choudhary, A. K., Sepat, S., Rathore, S. S., Mahajan, G., et al. (2017). Weed management in rice using crop competition-a review. Crop Prot. 95, 45–52. doi: 10.1016/j.cropro.2016.08.005
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi: 10.1016/j.cell.2006.12.006
Duncan, W. G., Loomis, R. S., Williams, W. A., and Hanau, R. (1967). A model for simulating photosynthesis in plant communities. Hilgardia 38, 181–205. doi: 10.3733/hilg.v38n04p181
Fei, C., Yu, J., Xu, Z., and Xu, Q. (2019). Erect panicle architecture contributes to increased rice production through the improvement of canopy structure. Mol. Breed. 39:128. doi: 10.1007/s11032-019-1037-9
Fischer, A. J., Ramírez, H. V., Gibson, K. D., and Da Silveira, and Pinheiro, B. (2001). Competitiveness of semidwarf upland rice cultivars against palisadegrass (Brachiaria brizantha) and signalgrass (B. decumbens). Agron. J. 93, 967–973. doi: 10.2134/agronj2001.935967x
Gibson, K. D., Fischer, A. J., Foin, T. C., and Hill, J. E. (2003). Crop traits related to weed suppression in water-seeded rice (Oryza sativa L). Weed Sci. 51, 87–93.
Global Rice Science Partnership (2013). Rice Almanac, 4th Edn. Los Banϸos: International Rice Research Institute.
Guo, Z.-W., Deng, H.-F., Li, S.-Y., Xiao, L.-T., Huang, Z.-Y., et al. (2011). Characteristics of the mesophyllous cells in the sheaths of rice (Oryza sativa L.). Agri. Sci. China 10, 1354–1364. doi: 10.1016/S1671-2927(11)60128-4
Heap, I., and Duke, S. O. (2018). Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 74, 1040–1049. doi: 10.1002/ps.4760
Hirabayashi, H., Sato, H., Nonoue, Y., Kuno-Takemoto, Y., Takeuchi, Y., Kato, H., et al. (2010). Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breed. Sci. 60, 604–612. doi: 10.1270/jsbbs.60.604
Hua, S., Cao, B., Zheng, B., Li, B., and Sun, C. (2016). Quantitative evaluation of influence of PROSTRATE GROWTH 1 gene on rice canopy structure based on three-dimensional structure model. Field Crops Res. 194, 65–74. doi: 10.1016/j.fcr.2016.05.004
Huang, L., Liu, H., Wu, J., Zhao, R., Li, Y., Melaku, G., et al. (2020). Evolution of plant architecture in Oryza driven by the PROG1 locus. Front. Plant Sci. 11:876. doi: 10.3389/fpls.2020.00876
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Islam, F., Wang, J., Farooq, M. A., Khan, M. S. S., Xu, L., Zhu, J., et al. (2018). Potential impact of the herbicide 2,4-dichlorophenoxyacetic acid on human and ecosystems. Environ. Int. 111, 332–351. doi: 10.1016/j.envint.2017.10.020
Jin, J., Huang, W., Gao, J. P., Yang, J., Shi, M., Zhu, M. Z., et al. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. doi: 10.1038/ng.247
Johnson, D. E., Dingkuhn, M., Jones, M. P., and Mahamane, M. C. (1998). The influence of rice plant type on the erect of weed competition on Oryza sativa and Oryza glaberrima. Weed Res. 38, 207–216. doi: 10.1046/j.1365-3180.1998.00092.x
Kagale, S., Links, M. G., and Rozwadowski, K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109–1134. doi: 10.1104/pp.109.151704
Kamboj, R., Singh, B., Mondal, T. K., and Bisht, D. S. (2020). Current status of genomic resources on wild relatives of rice. Breed. Sci. 70, 135–144. doi: 10.1270/jsbbs.19064
Koarai, A., and Morita, H. (2003). Evaluation of the suppression ability of rice (Oryza sativa) on Monochoria vaginalis by measuring photosynthetic photon flux density below rice canopy. Weed Biol. Manag. 3, 172–178. doi: 10.1046/j.1445-6664.2003.00104.x
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Monteith, J. L. (1965). Light distribution and photosynthesis in field crops. Annal. Bot. 29, 17–37. doi: 10.1093/oxfordjournals.aob.a083934
Monteith, J. L. (1977). Climate and the efficiency of crop production in Britain. Philos. Trans. Royal Soc. B 281, 277–294. doi: 10.1098/rstb.1977.0140
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
San, N. S., Ootsuki, Y., Adachi, S., Yamamoto, T., Ueda, T., Tanabata, T., et al. (2018). A near-isogenic rice line carrying a QTL for larger leaf inclination angle yields heavier biomass and grain. Field Crops Res. 219, 131–138. doi: 10.1016/j.fcr.2018.01.025
Shah, L., Yahya, M., Shah, S. M. A., Nadeem, M., Ali, A., Ali, A., et al. (2019). Improving lodging resistance: using wheat and rice as classical examples. Int. J. Mol. Sci. 20:17. doi: 10.3390/ijms20174211
Tan, L., Li, X., Liu, F., Sun, X., Li, C., Zhu, Z., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197
Temnykh, S., Park, W. D., Ayres, N., Cartinhour, S., Hauck, N., Lipovich, L., et al. (2000). Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100, 697–712. doi: 10.1007/s001220051342
Wu, Y., Zhao, S., Li, X., Zhang, B., Jiang, L., Tang, Y., et al. (2018). Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat. Commun. 9:4157. doi: 10.1038/s41467-018-06509-2
Yu, B., Lin, Z., Li, H., Li, X., Li, J., Wang, Y., et al. (2007). Tac1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Keywords: ancestral genetic resource, Asian cultivated rice (Oryza sativa L), light reception efficiency, plant shape, tiller angle, vegetative growth, weed control, wild rice (Oryza rufipogon Griff.)
Citation: Inagaki N, Asami H, Hirabayashi H, Uchino A, Imaizumi T and Ishimaru K (2021) A Rice Ancestral Genetic Resource Conferring Ideal Plant Shapes for Vegetative Growth and Weed Suppression. Front. Plant Sci. 12:748531. doi: 10.3389/fpls.2021.748531
Received: 28 July 2021; Accepted: 28 October 2021;
Published: 26 November 2021.
Edited by:
Monica Fernandez-Aparicio, Institute for Sustainable Agriculture, Spanish National Research Council (CSIC), SpainReviewed by:
Stanley Omar Samonte, Texas A&M AgriLife Research and Extension Center at Beaumont, United StatesCopyright © 2021 Inagaki, Asami, Hirabayashi, Uchino, Imaizumi and Ishimaru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noritoshi Inagaki, bmluYWdha2lAYWZmcmMuZ28uanA=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.