
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 October 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.746517
As a subfamily of basic helix-loop-helix (bHLH) transcription factors, phytochrome-interacting factors (PIFs) participate in regulating light-dependent growth and development of plants. However, limited information is available about PIFs in pepper. In the present study, we identified six pepper PIF genes using bioinformatics-based methods. Phylogenetic analysis revealed that the PIFs from pepper and some other plants could be divided into three distinct groups. Motif analysis revealed the presence of many conserved motifs, which is consistent with the classification of PIF proteins. Gene structure analysis suggested that the CaPIF genes have five to seven introns, exhibiting a relatively more stable intron number than other plants such as rice, maize, and tomato. Expression analysis showed that CaPIF8 was up-regulated by cold and salt treatments. CaPIF8-silenced pepper plants obtained by virus-induced gene silencing (VIGS) exhibited higher sensitivity to cold and salt stress, with an obvious increase in relative electrolyte leakage (REL) and variations in the expression of stress-related genes. Further stress tolerance assays revealed that CaPIF8 plays different regulatory roles in cold and salt stress response by promoting the expression of the CBF1 gene and ABA biosynthesis genes, respectively. Our results reveal the key roles of CaPIF8 in cold and salt tolerance of pepper, and lay a solid foundation for clarifying the biological roles of PIFs in pepper and other plants.
Plants have developed multiple signaling transduction processes for regulating growth and development, and modulating complex molecular mechanisms by driving transcriptional or translational changes in transcription factors under diverse environmental conditions (Haak et al., 2017; Yang et al., 2019). Light provides photosynthate and energy through photosynthesis as well as acts as an essential environmental signal that participates in plant development and stress response. Plants perceive light via photoreceptors and rapidly respond to light intensity, quality, and photoperiod (Galvão and Fankhauser, 2015; Li and Mathews, 2016; Roeber et al., 2021). Phytochromes (phys) are red (R)/far-red (FR) light photoreceptors synthesized in cytoplasm and can be translocated to the nucleus for mediating light-regulated modulation of plant growth and development by interacting with various downstream signal transduction elements (Klose et al., 2015; Pham et al., 2018; Paik and Huq, 2019). There are five phytochromes from phytochrome A (phyA) to phyE in Arabidopsis, and their regulatory mechanisms exhibit distinct characteristics (Wang and Wang, 2015; Pham et al., 2018). Specially, phytochrome-interacting factors (PIFs) are a subfamily of basic helix-loop-helix (bHLH) transcription factors acting as the downstream targets of phytochromes. Under light exposure, phytochromes undergo physical interactions with PIFs and further inhibit their functions by promoting the phosphorylation and ubiquitin proteasome-dependent proteolysis and/or sequestration of PIFs (Shen et al., 2005; Al-Sady et al., 2006; Park et al., 2018).
PIF proteins possess two conserved domains: bHLH and APB (active phyB-binding), and some PIFs have an additional APA (active phyA-binding) domain (Pham et al., 2018). In Arabidopsis, the APA and APB domains are responsible for the physical interaction of PIFs with phyA and phyB, respectively. PIF proteins are widely distributed from bryophytes to angiosperms, with only one member in Marchantia polymorpha (Inoue et al., 2016), six in Physcomitrella patens (Possart et al., 2017), six in Oryza sativa (Nakamura et al., 2007), seven in Zea mays (Zhang et al., 2018; Wu et al., 2019), seven in Malus domestica (Zheng et al., 2020), eight in Arabidopsis thaliana (Lee and Choi, 2017), eight in Solanum lycopersicum (Rosado et al., 2016), 11 in Populus trichocarpa (Ding et al., 2021), and 12, 16, and 30 in Brassica rapa, B. oleracea, and B. napus, respectively (Li W. et al., 2021).
PIFs have been shown to be a negative regulator in the light signal transduction pathway that can promote skotomorphogenic development and suppress photomorphogenesis under dark conditions. In Arabidopsis, most PIFs redundantly promote skotomorphogenesis partly by promoting cell elongation, and mutation of PIF1, PIF3, PIF4, and PIF5 (usually named as pifq) would result in a striking constitutive photomorphogenesis phenotype in the dark (Leivar et al., 2008; Shin et al., 2009; Leivar and Monte, 2014). Unlike that of these PIFs, mutation of PIF2 would lead to elongated hypocotyls and small cotyledons under continuous R and FR light, indicating that PIF2 acts as a positive regulator in photomorphogenesis (Luo et al., 2014). In addition, AtPIF1, AtPIF3, AtPIF4, AtPIF5, and AtPIF7 promote the elongation of hypocotyl in response to red light, while AtPIF8 promotes the elongation of hypocotyl in response to blue and/or far-red light but not to red light (Pham et al., 2018; Oh et al., 2020). AtPIF4 and AtPIF5 not only act downstream of cryptochromes to mediate hypocotyl elongation in response to low blue light (Pedmale et al., 2016), but also participate in the UV-B photoreceptor UVR8 mediated hypocotyl elongation regulation (Tavridou et al., 2020).
PIFs can bind to a core G-box DNA-sequence motif (CACGTG) or PIF binding E-box (CANNTG) in the promoters of a great variety of target genes to activate or repress their expression (Zhang et al., 2013; Leivar and Monte, 2014; Luo et al., 2014; Sakuraba et al., 2017; Xi et al., 2021). Besides the vital role in photomorphogenesis, PIFs are also involved in the regulation of many other cellular processes, including fruit ripening, flowering time, and hormone response (Paik et al., 2017). For example, Arabidopsis PIFs have important functions in shade avoidance response (SAS) mainly through auxin biosynthesis and auxin signaling (Hornitschek et al., 2012; Hersch et al., 2014; Pacín et al., 2016; Jia et al., 2020), as well as via other phytohormones including gibberellin (GA) and brassinosteroid (BR) (Feng et al., 2008; Sorin et al., 2009; Xie et al., 2017; Hayes et al., 2019; Wang X. et al., 2020). Additionally, PIF1, PIF3, PIF4, PIF5, and PIF7 are necessary for the phy-mediated seasonal regulation of hypocotyl elongation in Arabidopsis (Leivar et al., 2020). In Populus, both PtPIF4 and PtPIF8 have conserved and redundant functions in SAS, but only PtPIF8 participates in regulating seasonal growth (Ding et al., 2021). Rosa chinensis RcPIFs function as a suppressor of flowering in response to light intensity by interacting with RcCO and thus impair the binding of RcCO to the RcFT promoter (Sun et al., 2021). AtPIF4 and AtPIF5 can activate dark-, age-, and heat-induced leaf senescence via regulation of ORESARA 1 (ORE1) and other known senescence-associated genes (SAGs) (Sakuraba et al., 2014; Song et al., 2014; Kim et al., 2020; Li N. et al., 2021). PIFs are also involved in the regulation of biotic and abiotic stress responses in plants. Arabidopsis PIFs were found to have redundant control over plant defense response to Botrytis cinerea (Xiang et al., 2020). Both PIF4 and PIF7 have central functions in thermomorphogenesis downstream of phyB, and PIF4 plays a more crucial role than PIF7 in promoting auxin response during thermomorphogenesis in Arabidopsis (Koini et al., 2009; Zhu et al., 2016; Chung et al., 2020; Fiorucci et al., 2020). Maize ZmPIF3.1 can confer drought resistance by regulating stomatal closure under the mediation of abscisic acid (ABA) in rice (Gao et al., 2018). In addition, overexpression of apple MdPIF3 would decrease the cold tolerance but promote drought resistance of both Arabidopsis plants and apple callus (Zheng et al., 2021).
Pepper (Capsicum annuum L.) is an economically important vegetable crop worldwide for its high contents of many essential nutrients. However, its growth and development are usually negatively influenced by various stresses, causing a serious loss of its yield and quality (Guo et al., 2015; Zhang R.X. et al., 2020). Identification of resistance genes in pepper will provide valuable guidance and resources for breeding to enhance its yield and quality under stress conditions. Although previous studies have revealed the functions of PIFs in regulating the development and stress response of plants, the functions of PIFs in pepper remain poorly understood. In this study, six PIF family members were identified from the pepper genome, and their phylogenetic relationship, gene structure, conserved motif, and expression profile were analyzed. Besides, virus-induced gene silencing (VIGS) of CaPIF8 was found to reduce the tolerance of pepper to cold and salt stress. Our results provide important information for further revealing the role of CaPIFs in pepper.
To screen the PIF proteins in pepper, all capsicum proteins were acquired from three public databases: PepperHub server,1 Solanaceae Genome Database,2 and NCBI.3 Then, the amino acid sequences of AtPIFs were obtained from TAIR,4 and used as queries to search for the homologous sequences from the capsicum proteome. The candidate pepper PIFs were submitted to SMART5 and InterPro6 for checking the presence of conserved bHLH and APB domains.
The molecular weight (MW) and isoelectric point (pI) of the candidate PIFs were calculated with ExPasy.7 In order to study the evolutionary relationships of the PIFs between pepper and other plant species, the protein sequences of PIFs from pepper, Arabidopsis, tomato, rice, and maize were aligned by Clustal Omega, and the alignment results were used to create an NJ (neighbor-joining) phylogenetic tree with the MEGA7.0 software using the pairwise deletion option, Poisson correction and 1,000 bootstrap replicates. The online MEME tool was used to search the conserved motifs of the pepper, Arabidopsis, tomato, rice, and maize, and the results were visualized by TBtools (Chen et al., 2020). The online tool Gene Structure Display Server v2.08 was employed to analyze the gene structures of PIFs from different plant species.
The full-length coding region without the stop codon (1,416 bp) of CaPIF8 (XM_016689836.1) was cloned into the Super1300-GFP vector between the Xba I and Kpn I sites. The pSuper1300:CaPIF8:GFP vector was transformed into Agrobacterium tumefaciens strain GV3101. The pSuper1300:CaPIF8:GFP and pSuper1300:GFP (as control) were grown overnight, re-suspended in the induction medium (10 mM MgCl2, 10 mM MES, and 200 μmol L–1 acetosyringone), and injected into Nicotiana benthamiana. GFP fluorescence was photographed using a confocal laser-scanning microscope (Nikon C2-ER, Tokyo, Japan).
To obtain the expression profiles of CaPIF genes in various tissues including bud, flower, leaf, root and stem, transcriptomic data of pepper cultivar “Zunla” were obtained from publicly available GEO database under the accession numbers of GSE45037 and GSE45154 (Qin et al., 2014). The genome-wide transcriptome data of an elite breeding pepper (Capsicum annuum) line 6,421 treated with 200 mM NaCl and 10°C were obtained under PepperHub server (see text footnote 1) (Liu et al., 2017). The gene expression levels were measured following the TopHat/Cufflinks pipeline based on the FPKM (fragments per kilobase of exon per million fragments mapped) values according to the previous reports (Haq et al., 2019; Wu et al., 2020). The heatmaps of the gene log2 transformed values were generated with TBtools software.
Hangjiao12 (Capsicum annuum L.) seedlings were planted in a greenhouse under 25/18°C (day/night) temperature with a 12 h photoperiod. For cold stress, TRV2:CaPIF8 and TRV2:00 seedlings at the four-leaf stage were placed in 4°C incubator. RNA samples were collected from leaves after 6 h of cold treatment. For salt stress, 300 mM NaCl (90 mL per plant) was used to irrigate the seedlings. RNA samples were collected from the leaves after 12 h of salt treatment. Harvested samples were rapidly frozen by using liquid nitrogen and stored at –80°C.
Tobacco rattle virus (TRV) based VIGS was used to generate CaPIF8-silenced pepper plants. Partial cDNA fragment of CaPIF8 (300 bp) was amplified with the VIGS tool.9 The PCR-amplified fragment was cloned into the pTRV2 vector by using the restriction sites of EcoR I and Xho I. The empty vector of TRV2:00 served as the control, and the TRV2:CaPDS (phytoene desaturase gene) vector was set as positive control due to the photobleaching phenotype, and above TRV2 and TRV1 were transformed into Agrobacterium tumefaciens strain GV3101. Two-week-old pepper seedlings were used for silencing the PIF8 gene according to the previously described method (Yang et al., 2015).
Total RNA extraction was conducted with the total RNA Miniprep Kit (Axygen Biosciences, Union City, CA, United States). Approximately 5 μg of RNA was reverse transcribed with the ReverTra Ace qPCR-RT Kit (Toyobo, Japan) according to the manufacturer’s protocol. qRT-PCR was carried out in triplicate using the iCycler iQTM Real-time PCR Detection System (Bio-Rad, Hercules, CA, United States) with the procedures as previously described (Yang et al., 2019). The pepper actin gene was used as an internal control. The primers of actin and CaPIF genes are listed in Supplementary Table 1.
To assess membrane permeability, electrolyte leakage assays were performed as described previously by using a conductivity meter (ST3100C, Changzhou, China). Relative electrolyte leakage (REL) was calculated based on the previously described method (Haq et al., 2019; Zhang H. et al., 2020). To determine the ABA content, 0.25 g of frozen leaves was ground into powder in liquid nitrogen. ABA was extracted in 1.5 mL of 80% aqueous methanol overnight at 4°C. After centrifugation, the supernatant was removed and extracted twice with 2 mL petroleum ether, the pH was adjusted to 2–3 with citric acid, and 2 mL ethyl acetate was used to extract the supernatant twice. The upper organic phase was transferred to a new EP tube, blown dry under the flow of N2 (gas). The dried samples were dissolved and mixed with 0.2 mL of mobile phase, and filtered through a needle filter. ABA content was analyzed using a high-performance liquid chromatography (Waters 2695, Milford, MA, United States).
Significant differences (P < 0.05) were tested by Duncan’s test in the statistical product and service solution (SPSS) software between the data from the control and treatments, and different letters were used to indicate differences.
In total, six PIF genes were identified from the pepper genome, which were designated as CaPIF1, CaPIF3, CaPIF4, CaPIF7a, CaPIF7b, and CaPIF8. Table 1 presents the physiological and biochemical properties of these six CaPIF genes. The gene lengths of CaPIFs varied from 1,950 to 11,887 bp, and the amino acid sequence lengths were from 342 to 633 aa. The predicted pI of various proteins ranged from 5.55 to 7.31 (Table 1).
To analyze the residue conservation features in these CaPIFs, the full-length CaPIFs and AtPIFs were aligned by Clustal Omega. As a result, the bHLH and APB domains were found in all six CaPIF proteins, with the exception of CaPIF7b, which contained an incomplete Helix2 in the bHLH domain (Table 1 and Figure 1). In addition, CaPIF1 and CaPIF3 contained an additional APA domain, which was also observed in PIF proteins from other plants, such as tomato SlPIF1a, SlPIF1b, and SlPIF3 (Rosado et al., 2016), Arabidopsis AtPIF1 and AtPIF3 (Lee and Choi, 2017), and apple MdPIF1–MdPIF5 (Zheng et al., 2020).
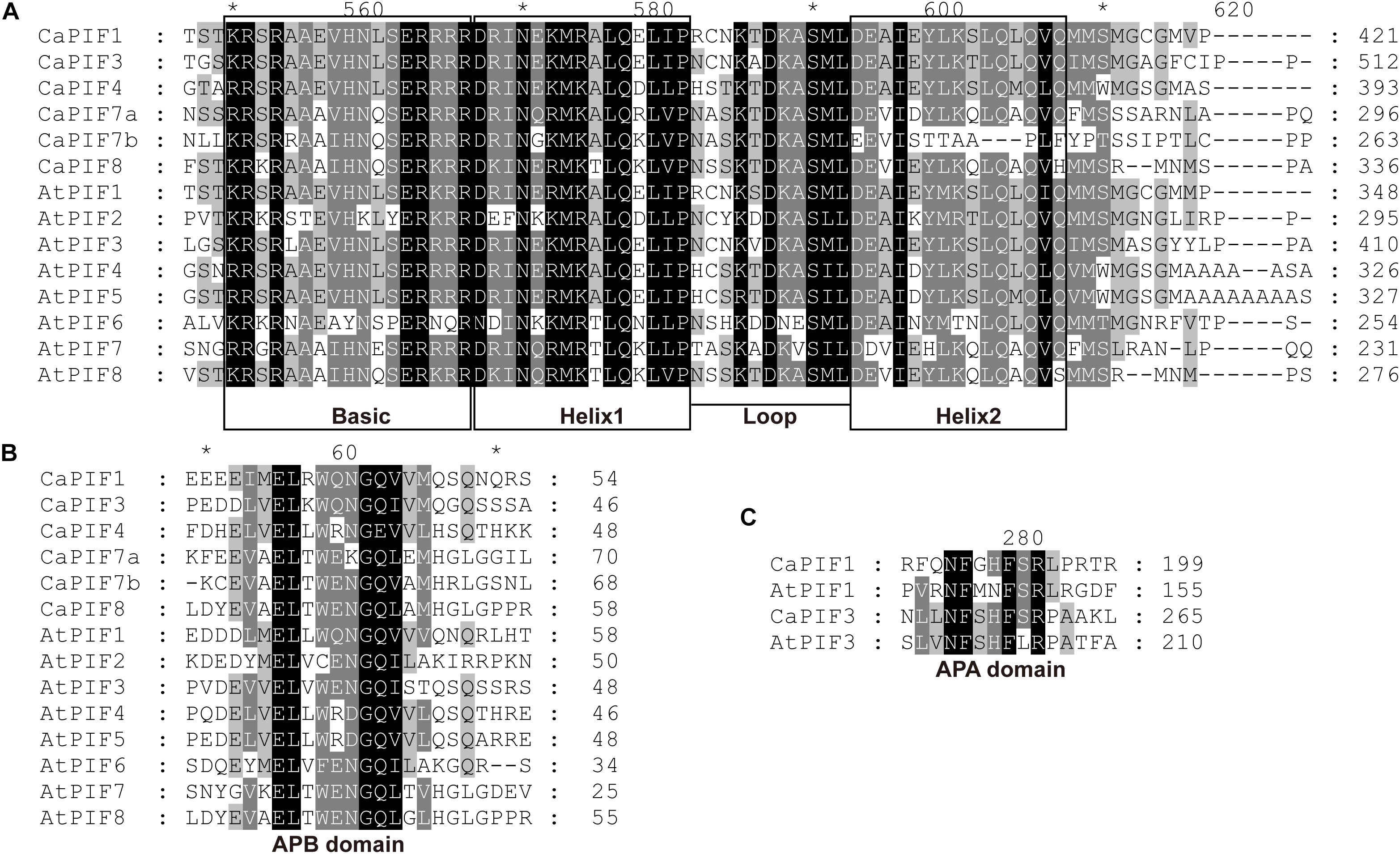
Figure 1. Multiple sequence alignment of the conserved bHLH (A), APB (B), and APA (C) domains of PIFs from pepper and Arabidopsis. Multiple sequence alignment was performed with Clustal Omega and displayed with the GeneDoc software. Asterisks were automatically generated by the GeneDoc software.
To elucidate the evolution of PIFs in pepper and other plant species, a NJ phylogenetic tree was created on the basis of multiple sequence alignment of full-length PIF protein sequences from pepper, Arabidopsis (Lee and Choi, 2017), tomato (Rosado et al., 2016), rice (Nakamura et al., 2007), and maize (Wu et al., 2019). In the phylogenetic tree, these PIF proteins could be divided into three distinct groups, namely PIF1/4/5, PIF2/3/6, and PIF7/8, and CaPIFs displayed a closer relationship with tomato SlPIFs and Arabidopsis AtPIFs than with rice OsPIFs and maize ZmPIFs (Figure 2A).
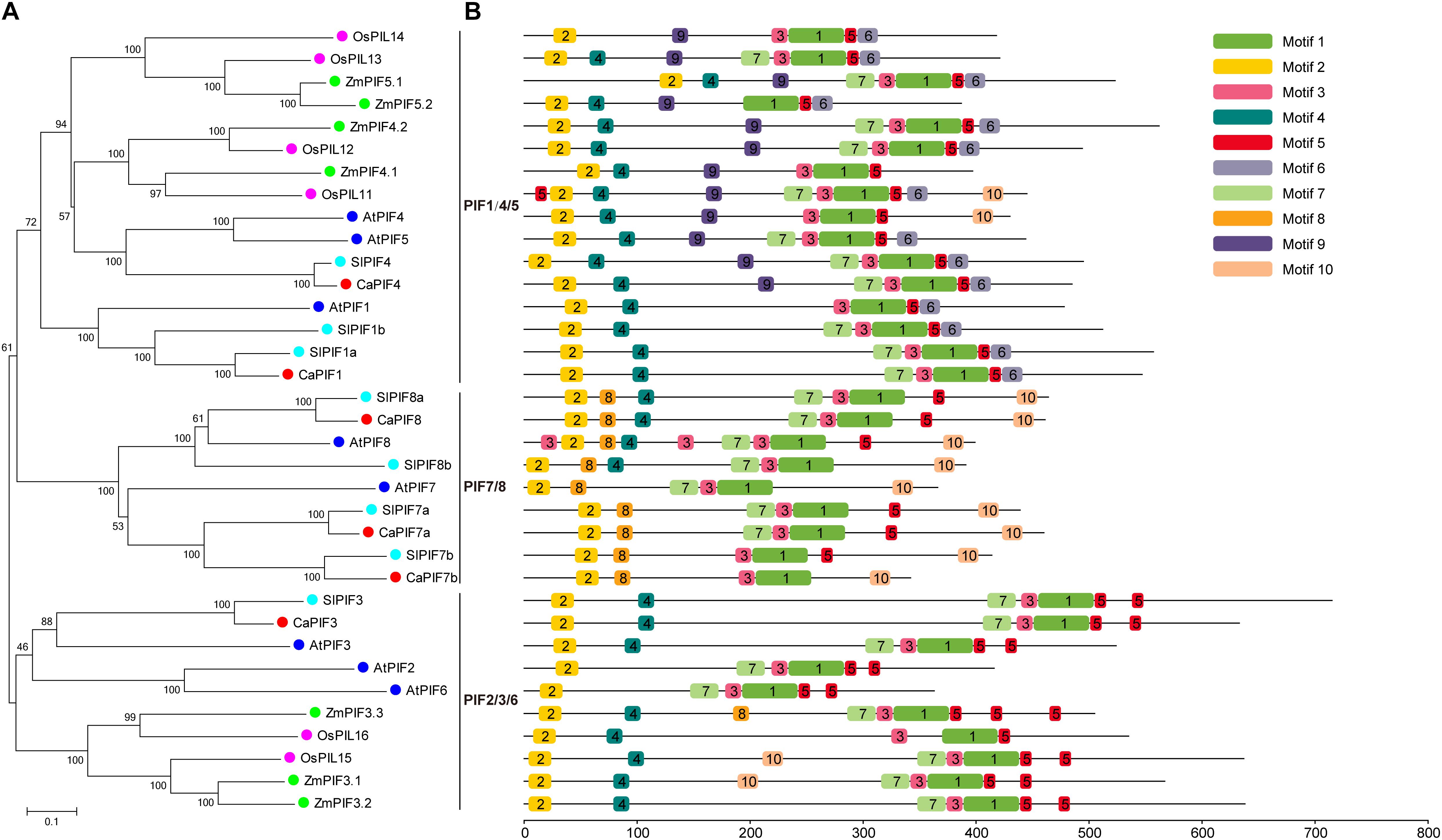
Figure 2. Phylogenetic analysis and conserved motif analysis of the PIF proteins of pepper, Arabidopsis, tomato, rice, and maize. (A) Evolutionary relationships of PIF proteins in pepper, Arabidopsis, tomato, rice, and maize. The phylogenetic tree was created with the neighbor-joining method and 1,000 bootstrap replications conducted in MEGA 7.0. The accession numbers of PIFs used to construct the phylogenetic tree are listed in Supplementary Table 2. (B) Distribution of the 10 conserved motifs in the PIF proteins of pepper, Arabidopsis, tomato, rice, and maize indicated by the analysis with MEME. Motifs are colored by different boxes and their sequences are listed in Supplementary Table 3.
Conserved motif analysis was also conducted with the MEME tool, resulting in the identification of 10 conserved motifs (Figure 2B). Among these motifs, motif 1, and motif 2 were annotated as the bHLH and APB domains, which are present in all PIF proteins. In addition, motifs 3, 5, and 7 were also widely distributed in these PIF proteins. However, several motifs were unique to specific groups. For example, motif 9 was identified specifically in PIF4/5 members. Motif 8 was only observed in ZmPIF3.3 and all PIF7/8 members, and ZmPIF3.3 was lack of motif 10, which was widely found in PIF7/8 group (Figure 2B). In addition, PIF7 proteins were lack of motif 4, which was widely present in PIF proteins of other groups, except for OsPIL14, AtPIF2, and AtPIF6 (Figure 2B).
We also analyzed the structure of the PIF genes from pepper and other plant species. As a result, most of the PIF genes possessed 4–6 introns, especially those from dicots. The number of introns in CaPIF genes ranged from five to seven, while that of PIF genes in other plants varied more greatly, ranging from one (SlPIF1a) to nine (ZmPIF5.1) (Figure 3). In addition, some closely related PIF genes exhibited similar gene structures in intron number and CDS length, such as ZmPIF4.2 and OsPIL12, SlPIF8a and CaPIF8, SlPIF7a and CaPIF7a (Figure 3).
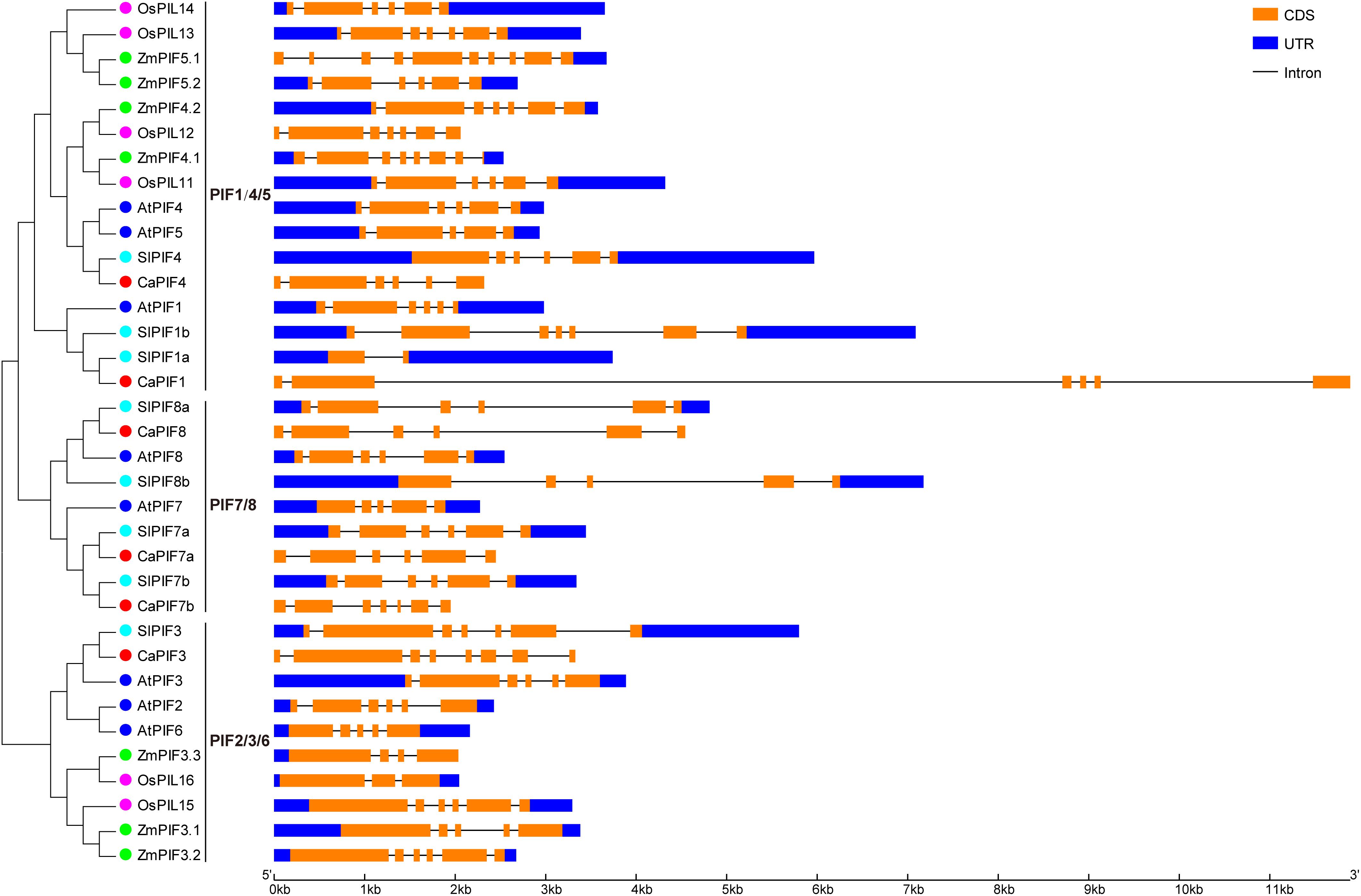
Figure 3. Structural analysis of PIF genes from pepper and other plant species according to the phylogenetic relationship. The CDS and untranslated region (UTR) are presented by orange and blue boxes, respectively.
To elucidate the possible functions of CaPIF genes, their tissue expression profiles were analyzed with publicly available transcriptomic data. As a result, CaPIF1, CaPIF3, CaPIF4, CaPIF7a, and CaPIF8 were constitutively expressed in all the tissues tested, while CaPIF7b was only expressed in leaves and stems (Figure 4). Notably, all CaPIF genes had high expression in the leaves and stems, but there were some differences among CaPIF genes. For example, the expression of CaPIF8 was much higher than that of other PIF genes in leaves and stems. Only one gene, CaPIF7a, showed a higher expression level in the leaves than in the stems, and other genes exhibited no obvious difference in expression between the two tissues (Figure 4).
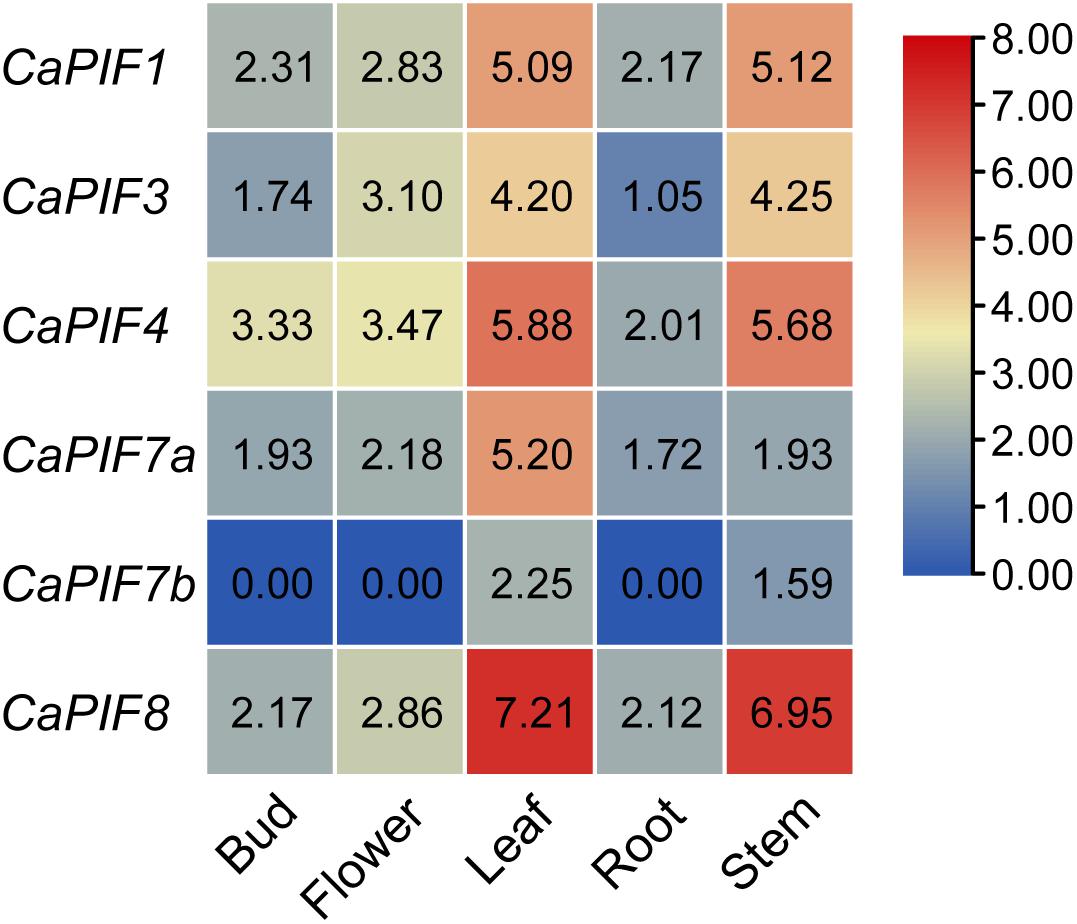
Figure 4. Relative expression levels of CaPIF genes in different tissues based on RNA-seq data. RNA-seq data of five tissues in pepper cultivar “Zunla” were used to analyze the expression pattern. Roots, stems, and leaves were collected from plants at the full-bloom stage, and the unopened flower buds (buds) and fully open flowers (flowers) were collected from mature plants. The heat map was drawn in log2 transformed expression values.
To investigate the possible functions of CaPIFs under abiotic stresses, we examined their expression profiles under cold and salt stress based on public RNA-seq data. Under cold stress treatment, CaPIF4 and CaPIF8 displayed up-regulated expression at certain time points (3 h and/or 6 h), but the expression decreased at 24 h (Figure 5A). The expression of CaPIF7a and CaPIF7b was down-regulated at the later time points (24 h) (Figure 5A). Under salt stress treatment, CaPIF4, CaPIF7a, and CaPIF7b were down-regulated at 12 h, while CaPIF8 was up-regulated at 3 h (Figure 5B). Since CaPIF8 showed noticeably high expression under cold and salt stress, and its homolog in citrus (CsPIF8) was found to enhance the tolerance of citrus to cold stress (He et al., 2020), we then chose CaPIF8 for further research in cold and salt stress.
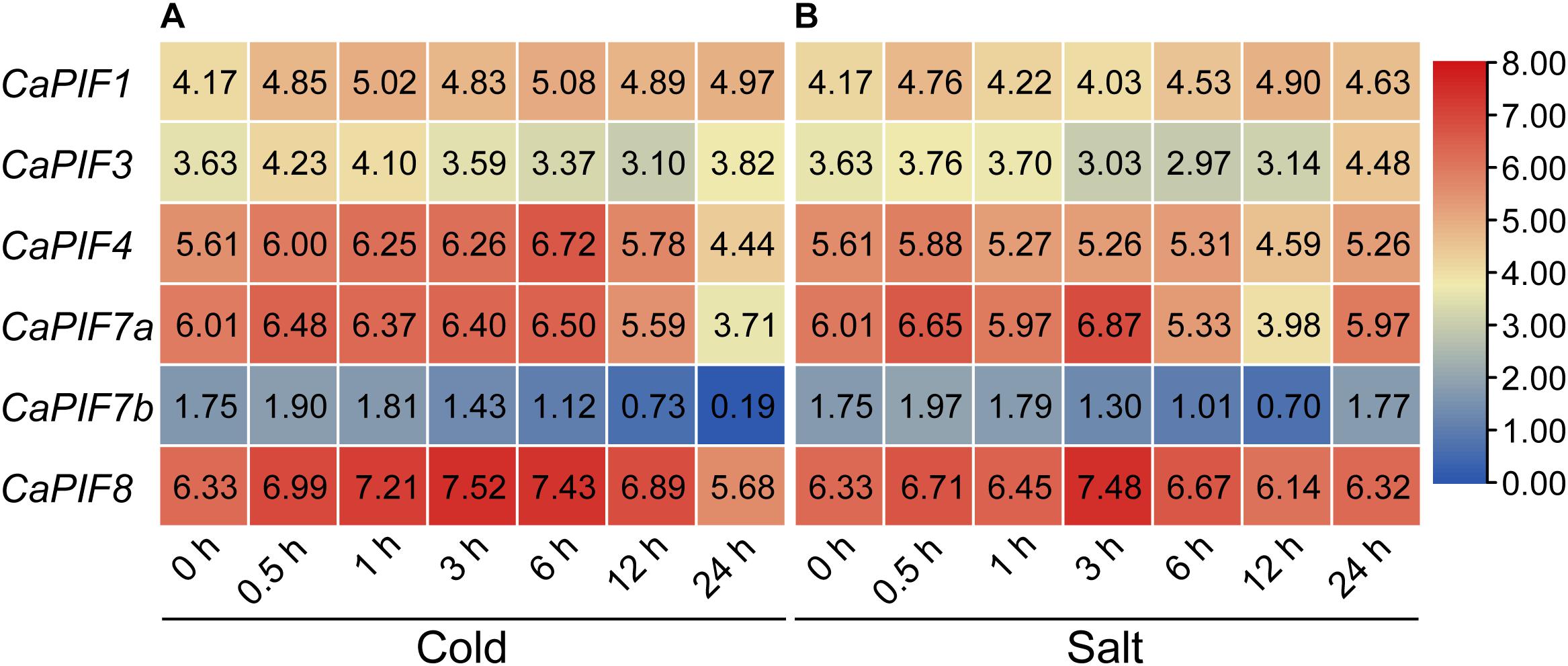
Figure 5. Expression profiles of CaPIF genes under cold (A) and salt (B) stress based on RNA-seq data. Cold and salt stress treatments were applied on 40-day old seedlings of an elite breeding pepper line 6,421 treated with 10°C and 200 mM NaCl, respectively, and leaf samples were collected at 0, 0.5, 1, 3, 6, 12, and 24 h post treatment. Numbers in the boxes show the expression based on log2 (FPKM +1) values.
For subcellular localization analysis of CaPIF8, the pSuper1300:CaPIF8:GFP and pSuper1300:GFP (as control) vectors were infiltrated into Nicotiana benthamiana epidermal cells by transient Agrobacterium-mediated transformation. Confocal laser scanning microscopy revealed that the GFP fluorescence of CaPIF8-GFP fusion protein was exclusively located in the nucleus, while that of 35S::GFP control was ubiquitously distributed in the whole cells (Figure 6), indicating that CaPIF8 is localized to the nucleus.
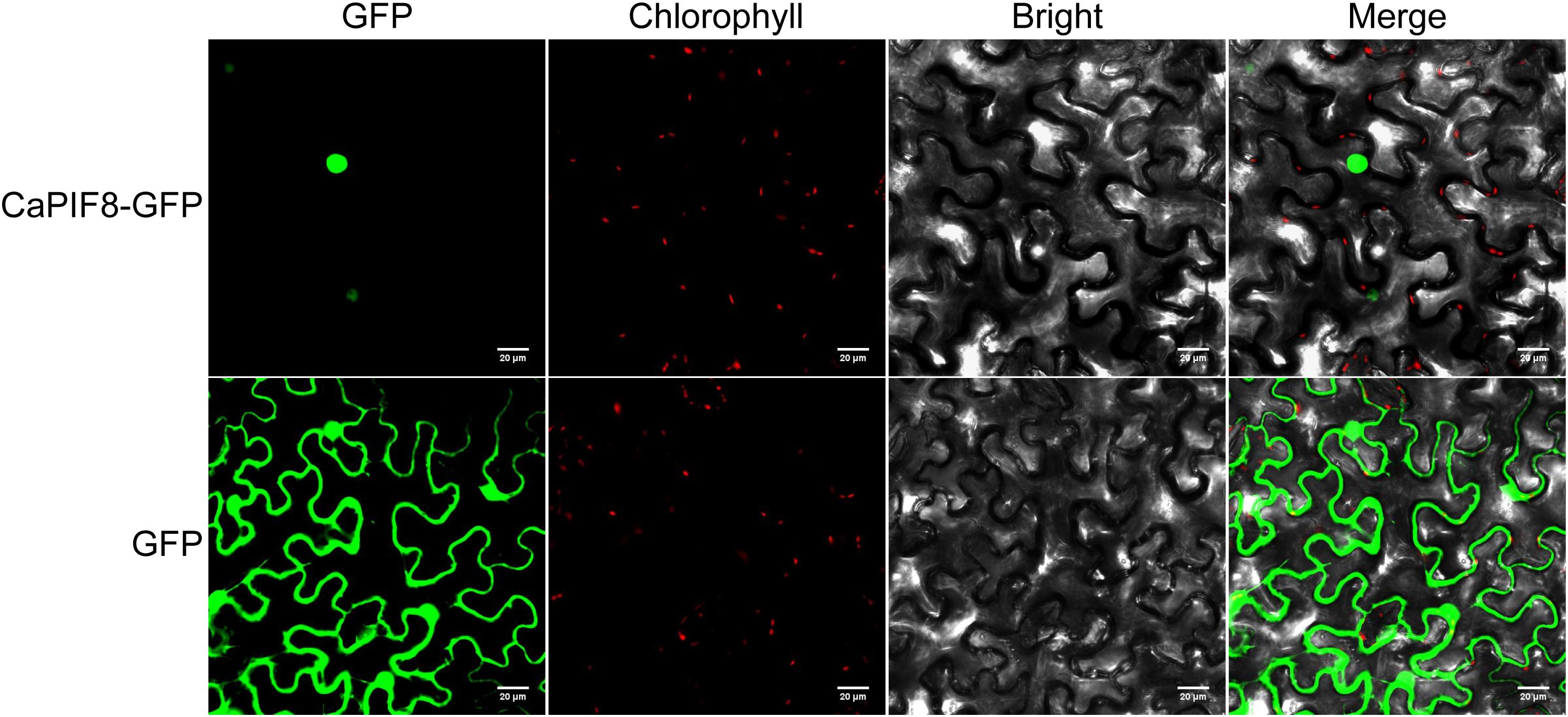
Figure 6. Subcellular localization of CaPIF8 in Nicotiana benthamiana epidermal cells. The subcellular localization of CaPIF8-GFP in N. benthamiana epidermal cells was visualized using confocal laser scanning microscopy.
Since the expression of CaPIF8 markedly increased at specific time points under cold and salt treatments, it can be speculated that CaPIF8 plays certain roles in stress response. Hence, we silenced CaPIF8 in pepper plants by VIGS to study its possible role in cold and salt stress tolerance. After approximately 1 month of virus injection, the leaves of TRV2:CaPDS plants were photo-bleached, while those of plants infected with TRV2:00 and TRV2:CaPIF8 showed no obvious symptoms (Figure 7A). However, the TRV2:CaPIF8 plants had significantly lower height than the TRV2:00 plants (Figure 7B). The qRT-PCR results showed that the CaPIF8 transcription in TRV2:CaPIF8 plants was reduced by 39% compared with that in TRV2:00 plants (Figure 7C), suggesting the successful silencing of CaPIF8.
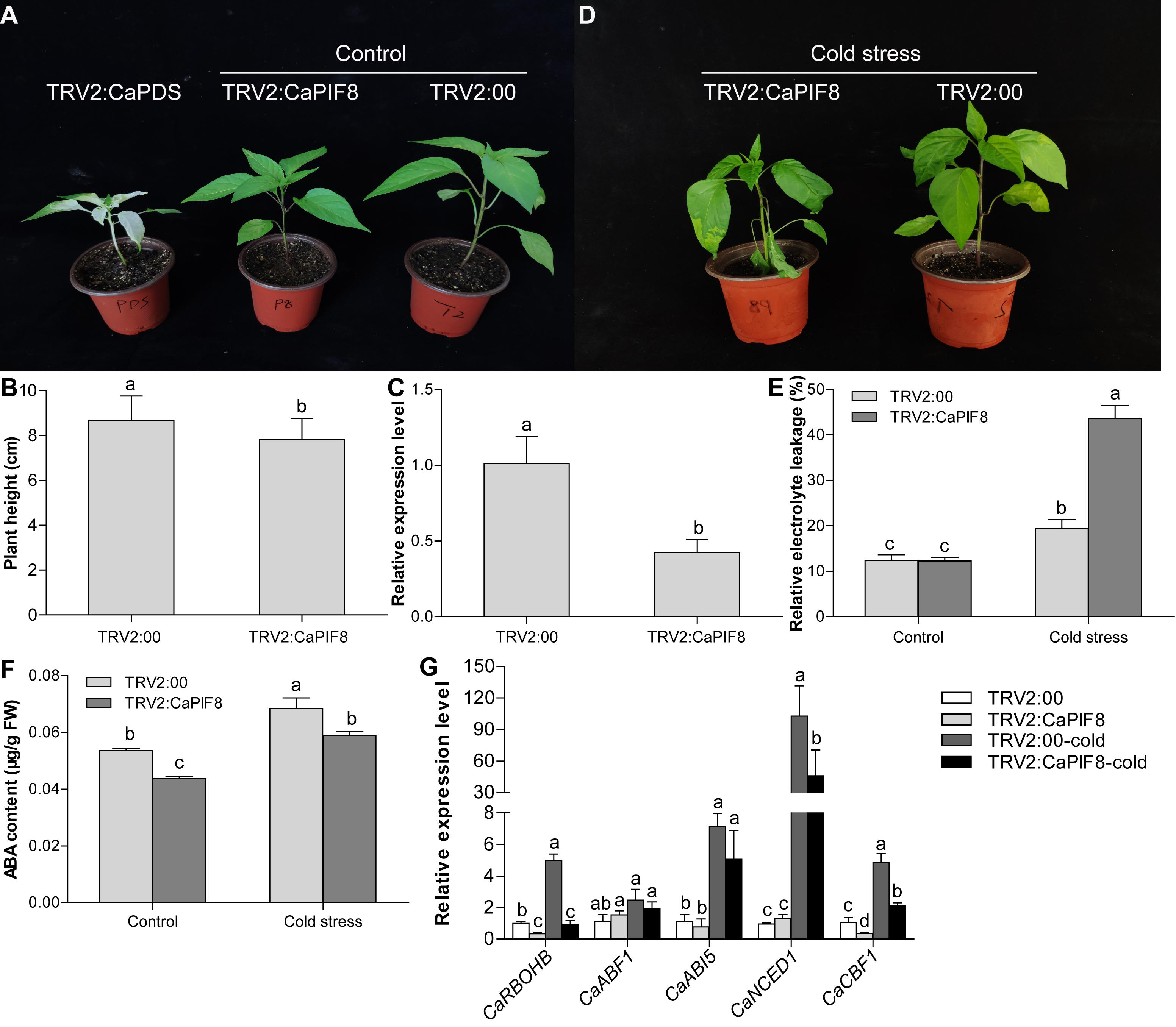
Figure 7. Effect of CaPIF8 silencing on the cold stress tolerance of pepper. (A) Phenotypes of pepper plants after approximately 1 month of virus injection. TRV2:CaPDS, plants with the TRV2:CaPDS vector; TRV2:CaPIF8, CaPIF8-silenced plants; TRV2:00, control plants with the empty TRV2 vector. (B) Height of TRV2:00 and TRV2:CaPIF8 pepper plants before cold stress. (C) Relative expression of CaPIF8 in leaves from CaPIF8-scilenced and control pepper plants. Leaf samples from TRV2:00 and TRV2:CaPIF8 plants were collected before cold stress. (D) Phenotypes of TRV2:00 and TRV2:CaPIF8 plants after 4°C cold stress for 3 days. (E) REL of TRV2:00 and TRV2:CaPIF8 plants. Leaf samples from TRV2:00 and TRV2:CaPIF8 plants were collected without and with cold stress for 3 days. (F) ABA content of TRV2:00 and TRV2:CaPIF8 plants. Leaf samples from TRV2:00 and TRV2:CaPIF8 plants were collected without and with cold stress for 12 h. (G) Relative expression levels of stress-related genes examined by qRT-PCR in leaves from TRV2:00 and TRV2:CaPIF8 plants without and with cold stress for 6 h. Each value is shown as mean ± standard error of three biological replicates, and the multiple comparison test for each gene was presented. Different letters stand for significant differences between TRV2:00 and TRV2:CaPIF8 plants without and with cold stress (Duncan’s test, P < 0.05).
To investigate the response of TRV2:CaPIF8 plants to cold stress, the TRV2:00 and TRV2:CaPIF8 plants were subjected to 4°C cold treatment for 3 days. After the treatment, the TRV2:CaPIF8 plants displayed severe growth retardation compared with the TRV2:00 plants, including serious wilting of leaves (Figure 7D). We also measured the REL and ABA level in leaves of the TRV2:00 and TRV2:CaPIF8 plants. Before cold stress treatment, no difference in REL was found between TRV2:00 and TRV2:CaPIF8 plants, while the TRV2:CaPIF8 plants exhibited markedly higher REL than the TRV2:00 plants after cold stress (Figure 7E). The ABA content in the TRV2:CaPIF8 plants was significantly lower than that in the TRV2:00 plants before and after cold stress treatment, though cold stress treatment led to increases in the ABA content of both plants (Figure 7F).
To elucidate the possible function of CaPIF8 in cold stress response, the expression profiles of five stress-related genes were determined by qRT-PCR. As a result, cold treatment did not significantly change the expression of ABA responsive genes (CaABF1 and CaABI5) in both TRV2:00 and TRV2:CaPIF8 plants (Figure 7G). However, the TRV2:CaPIF8 plants showed observably lower expression levels of CaRBOHB, CaCBF1, and ABA biosynthesis gene (CaNCED1) than the TRV2:00 plants with or without cold treatment (Figure 7G).
Additionally, we tested the response of TRV2:CaPIF8 plants to salt stress. After salt stress treatment for 7 days, the TRV2:CaPIF8 plants exhibited more serious symptoms than TRV2:00 plants, including leaf shrinkage, yellowing, and even wilting (Figure 8A). Also, no significant difference was observed in REL and ABA content between TRV2:00 and TRV2:CaPIF8 plants without salt stress. Upon salt stress treatment, the TRV2:CaPIF8 plants exhibited remarkably higher REL (Figure 8B) and a nearly 50% decrease in ABA content (Figure 8C) relative to the TRV2:00 plants. As for the stress-related genes, CaABI5 and CaNHX2 showed no significant changes in expression with and without salt treatment in both TRV2:00 and TRV2:CaPIF8 plants (Figure 8D). Salt treatment increased the expression of CaRBOHA, CaCAT2, and ABA biosynthesis genes (CaNCED1, CaNCED3, and CaNCED6) in both TRV2:CaPIF8 and TRV2:00 plants, and the expression was much lower in TRV2:CaPIF8 plants than in TRV2:00 plants (Figure 8D).
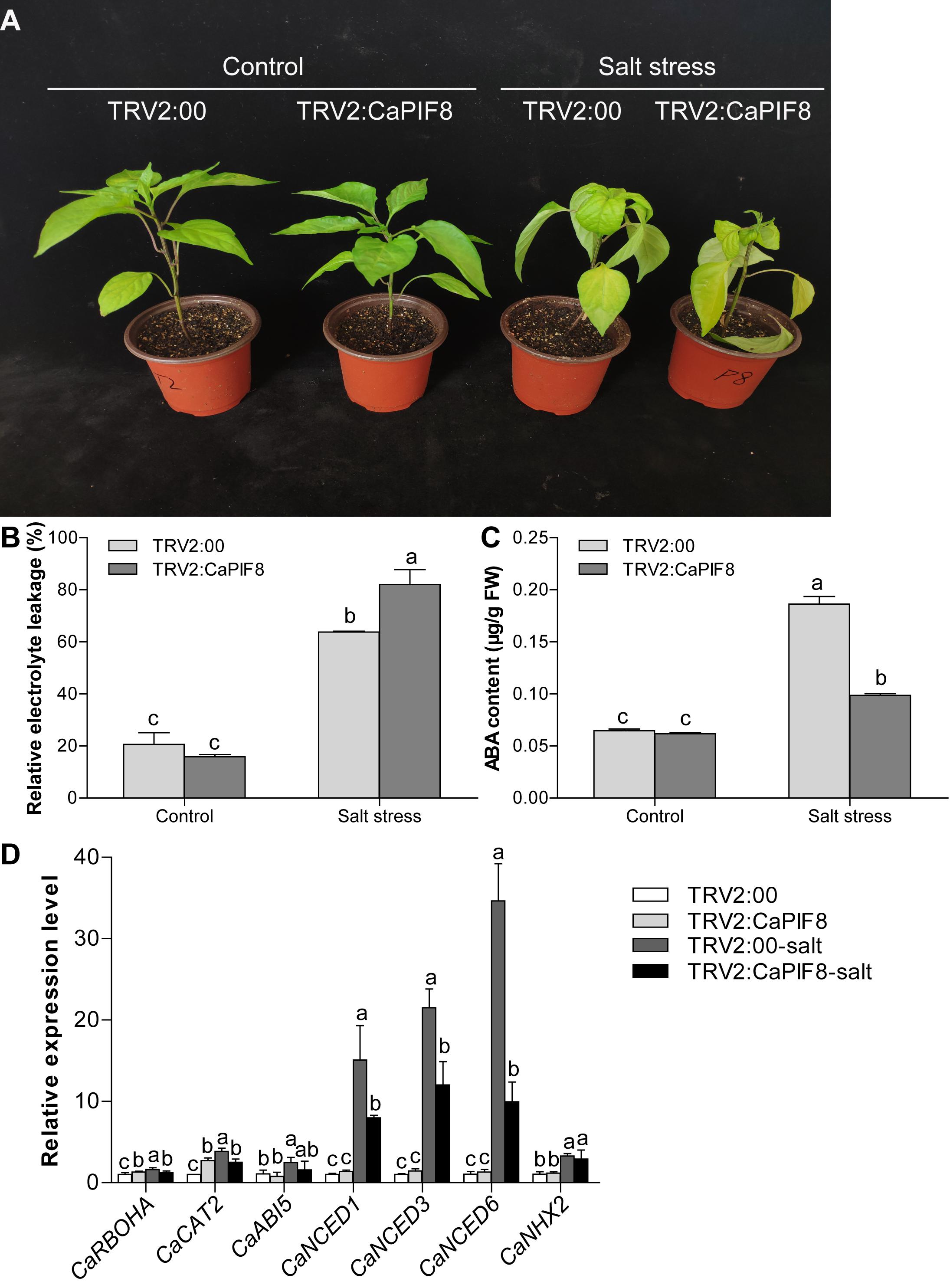
Figure 8. Effect of CaPIF8 silencing on the salt stress tolerance of pepper. (A) Phenotypes of TRV2:00 and TRV2:CaPIF8 pepper plants without and with salt stress treatment. The TRV2:00 and TRV2:CaPIF8 plants were subjected to salt stress by irrigation of 300 mM NaCl solution for 7 days. (B) REL of TRV2:00 and TRV2:CaPIF8 plants. Leaf samples from TRV2:00 and TRV2:CaPIF8 plants were collected without and with salt stress for 7 days. (C) ABA content of TRV2:00 and TRV2:CaPIF8 plants. Leaf samples from TRV2:00 and TRV2:CaPIF8 plants were collected without and with salt stress for 3 days. (D) Relative expression levels of stress-related genes examined by qRT-PCR in leaves from TRV2:00 and TRV2:CaPIF8 plants without and with salt stress for 12 h. Each value is shown as mean ± standard error of three biological replicates, and the multiple comparison test for each gene was presented. Different letters stand for significant differences between TRV2:00 and TRV2:CaPIF8 plants without and with salt stress (Duncan’s test, P < 0.05).
In this work, six PIF genes were identified from the pepper genome, and the number was comparable to that in many other plants, such as P. patens (six genes) (Possart et al., 2017), O. sativa (six genes) (Nakamura et al., 2007), Z. mays (seven genes) (Zhang et al., 2018; Wu et al., 2019), M. domestica (seven genes) (Zheng et al., 2020), A. thaliana (eight genes) (Lee and Choi, 2017), and S. lycopersicum (eight genes) (Rosado et al., 2016). Amino acid sequence alignment revealed that all the six CaPIF proteins harbor a conserved bHLH and an APB domain (Figures 1A,B), which is a typical feature of PIF proteins. However, CaPIF7b has an incomplete Helix2 in its bHLH domain, which may affect its function to interact with other proteins. Notably, an amino acid substitution (Q to E) in the APB domain was observed in CaPIF4 (Figure 1B), which was also identified in MdPIF8 (Zheng et al., 2020), suggesting that they possibly have similar functions. Similar to tomato SlPIF1s and SlPIF3 (Rosado et al., 2016), maize ZmPIF3s (Wu et al., 2019), Arabidopsis AtPIF1 and AtPIF3 (Leivar and Quail, 2011), CaPIF1 and CaPIF3 also contain the conserved functional APA domain (Figure 1C), which was found to play a vital role in modulating the interaction with phyA.
In the phylogenetic tree, PIF proteins from different plants (pepper, Arabidopsis, tomato, rice, and maize) could be divided as three distinct groups, and members of the same group have similar conserved proteins motif arrangements (Figure 2). PIF proteins did not cluster in a species-specific manner; however, CaPIFs had a much closer phylogenetic relationship with PIFs from dicots than with those from monocots (Figure 2A). Additionally, the PIF1/4/5 and PIF2/3/6 groups comprise PIF proteins from both dicots and monocots, while the PIF7/8 group exclusively includes PIF proteins from dicots (Figure 2A), indicating the evolution of the PIF genes after the monocotyledon–dicotyledon differentiation, and the PIF genes in the PIF7/8 group might have been derived from those in the PIF1/4/5 and PIF2/3/6 groups. Compared with Arabidopsis, pepper is lack of PIF2, PIF5, and PIF6 homologs (Table 1 and Figure 2A), and the intron numbers of CaPIF genes are relatively stable compared with those of PIFs in other plants (Figure 3). It can be speculated that pepper PIF genes might have undergone relatively small variations and gene loss during evolution.
Besides their crucial functions in photomorphogenesis, PIFs were also found to either positively or negatively regulate the response to different environmental stresses. For instance, both AtPIF1 and AtPIF4 were upregulated under heat and cold stress conditions; AtPIF6 was upregulated by ABA and salt stress; while AtPIF7 was suppressed by cold stress. AtPIF1, AtPIF4, and AtPIF5 were destabilized by cold stress depending on phytochromes, and they negatively regulated freezing tolerance in Arabidopsis (Jiang et al., 2020). AtPIF4 functions as negative regulator of salt tolerance by directly regulating the expression of genes associated with salt stress response, including ORE1, SAG29, and JUNGBRUNNEN1 (JUB1)/ANAC042 (Sakuraba et al., 2017). ZmPIF3.1 was significantly induced by drought and salt stress, and rice overexpressing maize ZmPIF3.1 showed enhanced tolerance to drought and salt stress (Gao et al., 2015). OsPIL14 overexpression or loss-of-function of the DELLA protein SLENDER RICE1 (SLR1) could also enhance the salt tolerance of rice seedlings by integrating light and GA signals (Mo et al., 2020). In this study, CaPIF4, CaPIF7a, CaPIF7b, and CaPIF8 were differentially regulated by cold or salt stress, indicating their critical and different roles in regulating the response to the two stresses (Figure 5). Among these four genes, CaPIF8 was induced under both cold and salt stress, suggesting that this gene may positively regulate plant resistance to cold or salt stress. To verify this inference, we generated CaPIF8-silenced pepper plants by using the VIGS technique. As a result, the TRV2:CaPIF8 plants exhibited more serious symptoms under cold and salt stress with increased REL than the TRV2:00 plants (Figures 7, 8). Compared with the TRV2:00 plants, the TRV2:CaPIF8 plants exhibited observable decreases in the expression of CaRBOHB under cold treatment, and CaRBOHA and CaCAT2 under salt treatment, respectively (Figures 7G, 8D), suggesting that reactive oxygen species (ROS) homeostasis was affected in TRV2:CaPIF8 plants under cold or salt stress. As the citrus homolog of CaPIF8 and AtPIF8, CsPIF8 can enhance the tolerance of citrus to cold stress through the regulation of SOD activity and the expression of CsSOD by binding to the E-box in its promoter (He et al., 2020). Therefore, CaPIF8 may participate in the regulation of the ROS homeostasis under cold and salt stress.
Various adverse stresses can result in ABA accumulation in plants, which can regulate stress-related genes involved in plant adaptation to environmental stimuli (Vishwakarma et al., 2017; Zhou et al., 2018). Some PIF genes were reported to participate in plant response to multiple stresses by activating or inhibiting downstream genes in the ABA-dependent signaling pathway. For example, overexpression of either ZmPIF3.1 or ZmPIF3.2 in rice conferred ABA-dependent drought tolerance (Gao et al., 2015, 2018). Myrothamnus flabellifolia MfPIF1 also positively regulates the response to drought and salt stress by activating the expression of both ABA biosynthesis and ABA-responsive genes (Qiu et al., 2020). In this study, the TRV2:CaPIF8 plants showed lower tolerance to salt stress with a significant decrease in ABA content (Figure 8). qRT-PCR analysis revealed that compared with those in the TRV2:00 plants, the ABA biosynthesis genes (CaNCED1, CaNCED3, and CaNCED6) were down-regulated but the ABA responsive gene showed no obvious change in TRV2:CaPIF8 plants (Figure 8D). Hence, we speculate that CaPIF8 has a crucial function in salt stress tolerance by activating ABA biosynthesis genes and then enhancing the ABA level under salt stress.
The TRV2:CaPIF8 plants showed higher sensitivity to cold stress, but the expression of ABA responsive genes (CaABF1 and CaABI5) had no obvious change when compared with those in the control. However, CaCBF1 was observably down-regulated in TRV2:CaPIF8 plants with and without cold stress (Figure 7G). The C-repeat binding factor (CBF) genes were found to enhance low temperature tolerance of plants. A previous study has suggested that AtPIF4 and AtPIF7 can reduce freezing tolerance through direct repression of the CBF genes under long day conditions (Lee and Thomashow, 2012). Arabidopsis CBF1 can interact with PIF3 but not with PIF1, PIF4, and PIF5, and stabilize PIF3 and phyB protein under cold stress (Jiang et al., 2020). In tomato, SlPIF4 can positively regulate low R/FR light-induced cold tolerance by inducing the CBF genes (Wang F. et al., 2020). Therefore, the decrease in cold stress tolerance of the TRV2:CaPIF8 plants may be mainly attributed to the significant decrease in the expression of the CBF genes.
In this study, six PIF members were identified in pepper, which were then classified into three distinct groups together with PIFs from other plants. Subsequently, their conserved domains, gene structures and expression profiles were analyzed. Amongst the identified CaPIF genes, CaPIF8 was upregulated under cold and salt stress, and silencing of CaPIF8 reduced the tolerance of pepper to cold and salt stress, suggesting that CaPIF8 can positively regulate tolerance to cold and salt stress. CaPIF8 enhances cold stress tolerance probably by inducing the CBF genes, while mediates salt stress tolerance by activating the ABA biosynthesis genes. Therefore, it can be inferred that CaPIF8 plays differential roles in mediating cold and salt tolerance of pepper. Overall, these findings provide important information for further revealing the mechanisms through which CaPIF8 controls the cold and salt stress response of pepper.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YY, FW, and YZ conceived and designed the experiments. YY, YG, FW, YC, WY, XX, SL, and YZ performed the experiments. YY, YG, and YZ analyzed the data. YY and YZ wrote the manuscript. All authors have read and approved the manuscript.
This work was funded by the Training Plan for Academic and Technical Leaders of Major Disciplines in Jiangxi Province, China-Young Talents Project (No. 20204BCJL23044), the National Natural Science Foundation of China (Nos. 32002030, 32160739, and 31801904), and the Natural Science Foundation of Jiangxi Province, China (No. 20202BABL205002), the Foundation of Jiangxi Educational Committee (No. GJJ190219), the Liao Ning Revitalization Talents Program (No. XLYC1807020), the Shenyang Young and Middle-aged Science and Technology Innovation Talent Support Program (No. RC200449), and the Liaoning BaiQianWan Talents Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.746517/full#supplementary-material
Al-Sady, B., Ni, W., Kircher, S., Schäfer, E., and Quail, P. H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446. doi: 10.1016/j.molcel.2006.06.011
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chung, B. Y. W., Balcerowicz, M., Di Antonio, M., Jaeger, K. E., Geng, F., Franaszek, K., et al. (2020). An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522–532. doi: 10.1038/s41477-020-0633-3
Ding, J., Zhang, B., Li, Y., André, D., and Nilsson, O. (2021). Phytochrome B and phytochrome interacting factor8 modulate seasonal growth in trees. New Phytol. doi: 10.1111/nph.17350
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Fiorucci, A. S., Galvão, V. C., Ince, Y., Boccaccini, A., Goyal, A., Allenbach Petrolati, L., et al. (2020). Phytochrome interacting factor 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 226, 50–58. doi: 10.1111/nph.16316
Galvão, V. C., and Fankhauser, C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34, 46–53. doi: 10.1016/j.conb.2015.01.013
Gao, Y., Jiang, W., Dai, Y., Xiao, N., Zhang, C., Li, H., et al. (2015). A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol. Biol. 87, 413–428. doi: 10.1007/s11103-015-0288-z
Gao, Y., Wu, M., Zhang, M., Jiang, W., Ren, X., Liang, E., et al. (2018). A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 16, 1375–1387. doi: 10.1111/pbi.12878
Guo, M., Liu, J. H., Lu, J. P., Zhai, Y. F., Wang, H., Gong, Z. H., et al. (2015). Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 6:806. doi: 10.3389/fpls.2015.00806
Haak, D. C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G., et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8:1564. doi: 10.3389/fpls.2017.01564
Haq, S. U., Khan, A., Ali, M., Gai, W. X., Zhang, H. X., Yu, Q. H., et al. (2019). Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta 250, 2127–2145. doi: 10.1007/s00425-019-03290-4
Hayes, S., Pantazopoulou, C. K., van Gelderen, K., Reinen, E., Tween, A. L., Sharma, A., et al. (2019). Soil salinity limits plant shade avoidance. Curr. Biol. 29, 1669–1676.e1664.
He, Z., Zhao, T., Yin, Z., Liu, J., Cheng, Y., and Xu, J. (2020). The phytochrome-interacting transcription factor CsPIF8 contributes to cold tolerance in citrus by regulating superoxide dismutase expression. Plant Sci. 298:110584. doi: 10.1016/j.plantsci.2020.110584
Hersch, M., Lorrain, S., de Wit, M., Trevisan, M., Ljung, K., Bergmann, S., et al. (2014). Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 6515–6520. doi: 10.1073/pnas.1320355111
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., López-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Inoue, K., Nishihama, R., Kataoka, H., Hosaka, M., Manabe, R., Nomoto, M., et al. (2016). Phytochrome signaling is mediated by phytochrome interacting factor in the liverwort Marchantia polymorpha. Plant Cell 28, 1406–1421. doi: 10.1105/tpc.15.01063
Jia, Y., Kong, X., Hu, K., Cao, M., Liu, J., Ma, C., et al. (2020). PIFs coordinate shade avoidance by inhibiting auxin repressor ARF18 and metabolic regulator QQS. New Phytol. 228, 609–621. doi: 10.1111/nph.16732
Jiang, B., Shi, Y., Peng, Y., Jia, Y., Yan, Y., Dong, X., et al. (2020). Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 13, 894–906. doi: 10.1016/j.molp.2020.04.006
Kim, C., Kim, S. J., Jeong, J., Park, E., Oh, E., Park, Y. I., et al. (2020). High ambient temperature accelerates leaf senescence via phytochrome-interacting factor 4 and 5 in Arabidopsis. Mol. Cells 43, 645–661. doi: 10.14348/molcells.2020.0117
Klose, C., Viczián, A., Kircher, S., Schäfer, E., and Nagy, F. (2015). Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 206, 965–971. doi: 10.1111/nph.13207
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. doi: 10.1016/j.cub.2009.01.046
Lee, C. M., and Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Lee, N., and Choi, G. (2017). Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 35, 54–60. doi: 10.1016/j.pbi.2016.11.004
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56–78. doi: 10.1105/tpc.113.120857
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Leivar, P., Martín, G., Soy, J., Dalton-Roesler, J., Quail, P. H., and Monte, E. (2020). Phytochrome-imposed inhibition of PIF7 activity shapes photoperiodic growth in Arabidopsis together with PIF1, 3, 4 and 5. Physiol. Plant 169, 452–466. doi: 10.1111/ppl.13123
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Li, F. W., and Mathews, S. (2016). Evolutionary aspects of plant photoreceptors. J. Plant Res. 129, 115–122. doi: 10.1007/s10265-016-0785-4
Li, N., Bo, C., Zhang, Y., and Wang, L. (2021). PIF4 and PIF5 proteins promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot 72:erab158. doi: 10.1093/jxb/erab158
Li, W., Liu, Y., Wang, W., Liu, J., Yao, M., Guan, M., et al. (2021). Phytochrome-interacting factor (PIF) in rapeseed (Brassica napus L.): genome-wide identification, evolution and expression analyses during abiotic stress, light quality and vernalization. Int. J. Biol. Macromol. 180, 14–27. doi: 10.1016/j.ijbiomac.2021.03.055
Liu, F., Yu, H., Deng, Y., Zheng, J., Liu, M., Ou, L., et al. (2017). PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132. doi: 10.1016/j.molp.2017.03.005
Luo, Q., Lian, H. L., He, S. B., Li, L., Jia, K. P., and Yang, H. Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. doi: 10.1105/tpc.113.121657
Mo, W., Tang, W., Du, Y., Jing, Y., Bu, Q., and Lin, R. (2020). Phytochrome-interacting factor-like14 and slender rice1 interaction controls seedling growth under salt stress. Plant Physiol. 184, 506–517. doi: 10.1104/pp.20.00024
Nakamura, Y., Kato, T., Yamashino, T., Murakami, M., and Mizuno, T. (2007). Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 71, 1183–1191. doi: 10.1271/bbb.60643
Oh, J., Park, E., Song, K., Bae, G., and Choi, G. (2020). Phytochrome interacting factor8 inhibits phytochrome a-mediated far-red light responses in Arabidopsis. Plant Cell 32, 186–205. doi: 10.1105/tpc.19.00515
Pacín, M., Semmoloni, M., Legris, M., Finlayson, S. A., and Casal, J. J. (2016). Convergence of constitutive photomorphogenesis 1 and phytochrome interacting factor signalling during shade avoidance. New Phytol. 211, 967–979. doi: 10.1111/nph.13965
Paik, I., and Huq, E. (2019). Plant photoreceptors: multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 92, 114–121. doi: 10.1016/j.semcdb.2019.03.007
Paik, I., Kathare, P. K., Kim, J. I., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046. doi: 10.1016/j.molp.2017.07.002
Park, E., Kim, Y., and Choi, G. (2018). Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30, 1277–1292. doi: 10.1105/tpc.17.00913
Pedmale, U. V., Huang, S. C., Zander, M., Cole, B. J., Hetzel, J., Ljung, K., et al. (2016). Cryptochromes interact directly with pifs to control plant growth in limiting blue light. Cell 164, 233–245. doi: 10.1016/j.cell.2015.12.018
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038. doi: 10.1104/pp.17.01384
Possart, A., Xu, T., Paik, I., Hanke, S., Keim, S., Hermann, H. M., et al. (2017). Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 29, 310–330. doi: 10.1105/tpc.16.00388
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Qiu, J. R., Xiang, X. Y., Wang, J. T., Xu, W. X., Chen, J., Xiao, Y., et al. (2020). MfPIF1 of resurrection plant Myrothamnus flabellifolia plays a positive regulatory role in responding to drought and salinity stresses in Arabidopsis. Int. J. Mol. Sci. 21:3011. doi: 10.3390/ijms21083011
Roeber, V. M., Bajaj, I., Rohde, M., Schmülling, T., and Cortleven, A. (2021). Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 44, 645–664. doi: 10.1111/pce.13948
Rosado, D., Gramegna, G., Cruz, A., Lira, B. S., Freschi, L., de Setta, N., et al. (2016). Phytochrome interacting factors (PIFs) in Solanum lycopersicum: diversity, evolutionary history and expression profiling during different developmental processes. PLoS One 11:e0165929. doi: 10.1371/journal.pone.0165929
Sakuraba, Y., Bülbül, S., Piao, W., Choi, G., and Paek, N. C. (2017). Arabidopsis early flowering3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92, 1106–1120. doi: 10.1111/tpj.13747
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Shen, H., Moon, J., and Huq, E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44, 1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., and Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Sorin, C., Salla-Martret, M., Bou-Torrent, J., Roig-Villanova, I., and Martínez-García, J. F. (2009). ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 59, 266–277. doi: 10.1111/j.1365-313X.2009.03866.x
Sun, J., Lu, J., Bai, M., Chen, Y., Wang, W., Fan, C., et al. (2021). Phytochrome-interacting factors interact with transcription factor constans to suppress flowering in rose. Plant Physiol. 186, 1186–1201. doi: 10.1093/plphys/kiab109
Tavridou, E., Pireyre, M., and Ulm, R. (2020). Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J. 101, 507–517. doi: 10.1111/tpj.14556
Vishwakarma, K., Upadhyay, N., Kumar, N., Yadav, G., Singh, J., Mishra, R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. doi: 10.3389/fpls.2017.00161
Wang, F., Chen, X., Dong, S., Jiang, X., Wang, L., Yu, J., et al. (2020). Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 18, 1041–1055. doi: 10.1111/pbi.13272
Wang, H., and Wang, H. (2015). Phytochrome signaling: time to tighten up the loose ends. Mol. Plant 8, 540–551. doi: 10.1016/j.molp.2014.11.021
Wang, X., Gao, X., Liu, Y., Fan, S., and Ma, Q. (2020). Progress of research on the regulatory pathway of the plant shade-avoidance syndrome. Front. Plant Sci. 11:439. doi: 10.3389/fpls.2020.00439
Wu, G., Zhao, Y., Shen, R., Wang, B., Xie, Y., Ma, X., et al. (2019). Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. 181, 789–803. doi: 10.1104/pp.19.00239
Wu, Z., Cheng, J., Hu, F., Qin, C., Xu, X., and Hu, K. (2020). The SnRK2 family in pepper (Capsicum annuum L.): genome-wide identification and expression analyses during fruit development and under abiotic stress. Genes Genom. 42, 1117–1130. doi: 10.1007/s13258-020-00968-y
Xi, Y., Yang, Y., Yang, J., Zhang, X., Pan, Y., and Guo, H. (2021). IAA3-mediated repression of PIF proteins coordinates light and auxin signaling in Arabidopsis. PLoS Genet. 17:e1009384. doi: 10.1371/journal.pgen.1009384
Xiang, S., Wu, S., Zhang, H., Mou, M., Chen, Y., Li, D., et al. (2020). The PIFs redundantly control plant defense response against Botrytis cinerea in Arabidopsis. Plants (Basel) 9:1246. doi: 10.3390/plants9091246
Xie, Y., Liu, Y., Wang, H., Ma, X., Wang, B., Wu, G., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8:348. doi: 10.1038/s41467-017-00404-y
Yang, Y. X., Wang, M. M., Yin, Y. L., Onac, E., Zhou, G. F., Peng, S., et al. (2015). RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16:120. doi: 10.1186/s12864-015-1228-7
Yang, Y., Li, J., Li, H., Yang, Y., Guang, Y., and Zhou, Y. (2019). The bZIP gene family in watermelon: genome-wide identification and expression analysis under cold stress and root-knot nematode infection. PeerJ 7:e7878. doi: 10.7717/peerj.7878
Zhang, H., Ma, F., Wang, X., Liu, S., Saeed, U. H., Hou, X., et al. (2020). Molecular and functional characterization of CaNAC035, an NAC transcription factor from pepper (Capsicum annuum L.). Front. Plant Sci. 11:14. doi: 10.3389/fpls.2020.00014
Zhang, R. X., Zhu, W. C., Cheng, G. X., Yu, Y. N., Li, Q. H., Haq, S. U., et al. (2020). A novel gene, CaATHB-12, negatively regulates fruit carotenoid content under cold stress in Capsicum annuum. Food Nutr. Res. 64:3729. doi: 10.29219/fnr.v64.3729
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18:235. doi: 10.1186/s12870-018-1441-z
Zhang, Y., Mayba, O., Pfeiffer, A., Shi, H., Tepperman, J. M., Speed, T. P., et al. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9:e1003244. doi: 10.1371/journal.pgen.1003244
Zheng, P. F., Wang, X., Yang, Y. Y., You, C. X., Zhang, Z. L., and Hao, Y. J. (2020). Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica. Int. J. Mol. Sci. 21:7350. doi: 10.3390/ijms21197350
Zheng, P. F., Yang, Y. Y., Zhang, S., You, C. X., Zhang, Z. L., and Hao, Y. J. (2021). Identification and functional characterization of MdPIF3 in response to cold and drought stress in Malus domestica. Plant Cell Tissue Organ Cult. 144, 435–447. doi: 10.1007/s11240-020-01968-2
Zhou, Y., Hu, L., Ye, S., Jiang, L., and Liu, S. (2018). Molecular cloning and functional characterization of a Cu/Zn superoxide dismutase gene (CsCSD1) from Cucumis sativus. Plant Cell Tissue Organ Cult. 135, 309–319. doi: 10.1007/s11240-018-1465-y
Keywords: pepper, phytochrome-interacting factor (PIF), expression profile, salt stress, cold stress
Citation: Yang Y, Guang Y, Wang F, Chen Y, Yang W, Xiao X, Luo S and Zhou Y (2021) Characterization of Phytochrome-Interacting Factor Genes in Pepper and Functional Analysis of CaPIF8 in Cold and Salt Stress. Front. Plant Sci. 12:746517. doi: 10.3389/fpls.2021.746517
Received: 24 July 2021; Accepted: 04 October 2021;
Published: 25 October 2021.
Edited by:
Xiaojuan Li, Beijing Forestry University, ChinaReviewed by:
Yasuhito Sakuraba, The University of Tokyo, JapanCopyright © 2021 Yang, Guang, Wang, Chen, Yang, Xiao, Luo and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhou, eW9uZ3pob3VAanhhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.