- 1State Key Laboratory of Natural Medicines, Department of Pharmacognosy, Institute of Pharmaceutical Science, China Pharmaceutical University, Nanjing, China
- 2School of Traditional Chinese Pharmacy, China Pharmaceutical University, Nanjing, China
Salvianolic acids, a group of secondary metabolites produced by Salvia miltiorrhiza, are widely used for treating cerebrovascular diseases. Copper is recognized as a necessary microelement and plays an essential role in plant growth. At present, the effect of copper on the biosynthesis of SalAs is unknown. Here, an integrated metabolomic and transcriptomic approach, coupled with biochemical analyses, was employed to dissect the mechanisms by which copper ions induced the biosynthesis of SalAs. In this study, we identified that a low concentration (5 μM) of copper ions could promote growth of S. miltiorrhiza and the biosynthesis of SalAs. Results of the metabolomics analysis showed that 160 metabolites (90 increased and 70 decreased) were significantly changed in S. miltiorrhiza treated with low concentration of copper ions. The differential metabolites were mainly involved in amino acid metabolism, the pentose phosphate pathway, and carbon fixation in photosynthetic organisms. The contents of chlorophyll a, chlorophyll b, and total chlorophyll were significantly increased in leaves of low concentration of copper-treated S. miltiorrhiza plants. Importantly, core SalA biosynthetic genes (laccases and rosmarinic acid synthase), SalA biosynthesis-related transcription factors (MYBs and zinc finger CCCH domain-containing protein 33), and chloroplast proteins-encoding genes (blue copper protein and chlorophyll-binding protein) were upregulated in the treated samples as indicated by a comprehensive transcriptomic analysis. Bioinformatics and enzyme activity analyses showed that laccase 20 contained copper-binding motifs, and its activity in low concentration of copper ions-treated S. miltiorrhiza was much higher than that in the control. Our results demonstrate that enhancement of copper ions of the accumulation of SalAs might be through regulating laccase 20, MYBs, and zinc finger transcription factors, and photosynthetic genes.
Introduction
Copper, a necessary microelement in plants, plays an essential role in various physiological activities during plant growth and development, such as cell wall metabolism, hormone signaling, photosynthesis, and redox reactions (Choudhary et al., 2012; De Caroli et al., 2020; Ding et al., 2020; Saleem et al., 2020). Copper is a redox-active transition metal, and two valence states (Cu2+ and Cu+) exist under physiological conditions. Copper can work as a metal cofactor in metalloproteins involved in electron transport and oxidative stress response (Quist et al., 2017). In chloroplasts, copper is a constituent of plastocyanin (Pc), the most abundant copper protein in plant chloroplasts, which acts as an electron carrier in primary photosynthetic reactions (Droppa et al., 1984). Copper also works as a constituent of Cu/Zn-superoxide dismutase (Cu/Zn-SOD), localized in the stroma that protects against reactive oxygen species (Yruela, 2009). Due to these characters, copper ion plays an important role in regulating plant physiology.
Although copper is indispensable in plant development, most studies have reported that excessive copper causes toxicity to plants via the production of reactive oxygen species (ROS) that can oxidize biological macromolecules, such as lipids and nucleic acids, and cause enzyme inactivation (Andre et al., 2010). It has been reported that excessive copper causes rice cell death by increasing the ROS level in rice radicles (Zhang et al., 2017). However, a low dose of copper can enhance enzyme activity to increase the production of plant secondary metabolites (Ibrahim et al., 2017). Treatment of Arabidopsis seedlings with different concentrations of copper showed that the right amount of copper increased the meristem size of the seedlings (Song et al., 2017). Although pieces of research on copper have reported its role in several biological functions involving cytochrome c oxidase, plastocyanin, and ethylene receptors (Burkhead et al., 2009; Song et al., 2017), the majority of previous studies have not focused on whether copper ions are involved in plant secondary metabolism. Therefore, studies are needed to improve our understanding of the role of copper in plant secondary metabolism.
Salvia miltiorrhiza, a traditional Chinese herb, is widely used in the treatment of cardiovascular diseases. Water-soluble phenolic acids, such as salvianolic acid A, salvianolic acid B (Sal B), and rosmarinic acid (RA), are the essential active compounds in the roots of S. miltiorrhiza (Huang et al., 2019). There are two biosynthetic routes for phenolic acid in plants: the tyrosine-derived and phenylpropanoid pathways. In the tyrosine-derived pathway, tyrosine is converted to 4-hydroxyphenyllactate under the action of various enzymes, including tyrosine aminotransferase and 4-hydroxyphenylpyruvate reductase. In the phenylpropanoid pathway, phenylalanine is converted to 4-coumaroyl-CoA under the action of various enzymes, including phenylalanine ammonia-lyase, cinnamic acid 4-hydroxylase, and 4-coumarate: CoA ligase. Rosmarinic acid synthase catalyzes the transformation of 4-coumaroyl-CoA and 4-hydroxyphenyllactate to rosmarinic acid (Ma et al., 2013; Shi et al., 2019). The molecular mechanism by which rosmarinic acid converts to salvianolic acid B is still not completely understood. Recently, laccase has been found to catalyze the conversion of rosmarinic acid to salvianolic acid E, which is then transformed into salvianolic acid B and other compounds (Li et al., 2019a; Wang et al., 2019). Furthermore, many transcription factors also regulate the biosynthesis of phenolic acids (Xing et al., 2018; Sun et al., 2019; Deng et al., 2020a; Yin et al., 2020).
As ceruploplasmin oxidases, laccases are widely distributed in all kinds of organisms, including bacteria, fungi, insects, and plants. Laccases belong to the multicopper oxidases family, and four copper ions existed in laccase (Hoegger et al., 2006). Laccases contain three types of catalytic sites (T1, T2, and T3), and T1 site is capable of oxidating substrates, while T2/3 sites are responsible for the consumption of oxygen and the generation of water (Solomon et al., 1996; Riva, 2006). T1 site containing one blue copper ion is mainly responsible for taking the electrons from the substrate after the laccase protein binds to the substrate. The remained three copper ions existed in T2 and T3 sites; they, together, form into the T2/T3 trinuclear center (copper cluster) (Patel et al., 2019). After the T1-copper ion obtains electrons, the electrons will be transferred to the T2/T3 trinuclear center via the amino acid bridge, and oxygen molecules obtained from the environment are reduced to produce water (Jones et al., 2015). In a word, the process of laccase catalyzes the oxidation by taking electrons from the substrate and transferring them to the corresponding domains and reacts with oxygen in the environment to form water. Meanwhile, the substrate loses electrons and becomes free radicals. To date, a large number of laccases have been identified and reported to exert their function in plant lignin biosynthesis, cell wall biosynthesis, and flavonoid biosynthesis (Turlapati et al., 2011). In Arabidopsis, 17 laccases have been annotated, which are clustered into six distinct clades (McCaig et al., 2005; Turlapati et al., 2011). In S. miltiorrhiza, laccases were also systemically investigated and demonstrated to play a great role in plant growth, development, and secondary metabolites biosynthesis (Zhou et al., 2018; Li et al., 2019a).
When a plant encounters adverse stress, the plant's own stress resistance mechanism will be activated and related genes will control the process of protein synthesis to protect themselves against adversity. Here, metabolomics and transcriptomics approaches were applied to reveal how copper affects the biosynthesis of phenolic acids in S. miltiorrhiza. The genes that respond to copper and the genes in the biosynthetic pathway of salvanoic acids (SalAs) were analyzed. Our findings are of great significance in understanding the underlying mechanism of the induction of the biosynthesis of SalAs under copper treatment and provide potential approach to improve SalA content in S. miltiorrhiza.
Methods
Plant Materials and Treatment
Salvia miltiorrhiza was cultivated at 25°C with a 16-h-light/8-h-dark cycle in MS basal medium containing 3% sugar and 8% agar. CuSO4·5H2O (Sinopharm Chemical Reagent Company., Ltd, China) was dissolved in distilled water to make concentrations of 5, 25, and 100 mM. All the prepared CuSO4·5H2O were sterilized through 0.22-μm filters and then added to MS basal medium to a final concentration of 5, 25, or 100 μM. S. miltiorrhiza was treated with different concentrations of CuSO4·5H2O for 5 days, and then plant matter was collected for the following experiments.
Phenotype and Fresh Weight
After 5 days of treatment with different concentrations of CuSO4·5H2O, a photo of S. miltiorrhiza planted in a culture flask was taken. To measure fresh weight, the whole plant of S. miltiorrhiza was taken out from the culture flask, washing away culture medicum, and measured with electronic balance. To accurately reflect fresh weight changes, at least four plants were used each time, and average weight was calculated.
High-Performance Liquid Chromatography (HPLC)
To extract phenolic acids, fresh roots of copper-treated or non-treated S. miltiorrhiza were collected from a culture flask, dried at 55°C until the weight remained constant, and then grounded into powder. Each sample of 0.1 g was added to 1-ml methanol: water (70: 30, v v-1), followed by 1-h ultrasound and centrifugation at 12,000 rpm for 20 min. The supernatant was then removed and filtered through a 0.22-μm filter. HPLC was performed on an Agilent 1290 with a DAD detector (Agilent Technologies, USA), equipped with an Agilent ODS-SP 5-μm 4.6 mm × 250 mm column. The mobile phase was H2O (0.1% formic acid, A) and acetonitrile (B). The HPLC program was set to the following line gradient: 10–25% of B for 0–10 min, 25–27% of B for 10–15 min, 27–55% of B for 15–20 min, 55–95% of B for 20–21 min, held 5 min, 95–10% of B for 26–27 min, and 10% of B for 27–30 min. The flow rate was 1 ml/min, the column temperature was set to 35°C, sample injection volume was 10 μl, and the wavelength was 280 nm. Sal B and RA standards (Shanghai Yuanye Bio-Technology Co., Ltd, China) were configured as a mixed standard solution with a concentration of 0.25 mg/ml. The standard curve method was used to calculate the content of Sal B and RA. The experiment was performed with four repetitions, and data were expressed as mean ± SD.
Ultra-Performance Liquid Chromatography-Quadrupole-Time of Flight-Mass Spectrometry (UPLC-QTOF MS)
For metabolomics analysis, the same samples were analyzed by LC-MS using an Agilent 6545A Q/TOF mass spectrometer equipped with an electrospray ion (ESI) source. In the negative ion mode, the following conditions were used: a drying N2 gas flow rate, 8 L/min; temperature, 320°C; nebulizer, 35 psig; capillary, 3,000 V; skimmer, 65 V; OCT RFV, 750 V. Mass spectra were recorded in a full-scan mode from 100 to 1,200 m/z range. Agilent 1290 Infinity Ultra-performance liquid chromatography (UPLC, Agilent technologies, USA), equipped with an Agilent ODS-SP 5 μm 4.6 mm × 250 mm column, was used to affect the separations. The mobile phase consisted of acetonitrile and 0.03% acetic acid (mobile phase A) and H2O with 0.03% acetic acid (mobile phase B), which were eluted as follows: 10–13% of B at 0–8 min, 13–23% of B at 8–13 min, 23% of B at 25 min, 23–27% of B at 25–45 min, 27–36% of B at 45–54 min, 36–70% of B at 54–65 min, 70–73% of B at 65–75 min, 73–93% of B at 75–85 min, 93–100% of B at 85–87 min, and then held for 5 min. The flow rate was 0.5 ml/min; the column temperature was set at 35°C, sample injection volume was 5 μl, and post-run time was 12 min.
For data analysis, raw LC-MS data were converted to mzData format using DA reprocessor software (Agilent), and the XCMS package in R was used to perform peak finding, filtering, and alignment. Metabioananlyst 4.0 (https://www.metaboanalyst.ca) was used to obtain normalized data. Metabolites with P < 0.05 and fold change (copper treatment/blank) >1.5 or < 0.5 were regarded as differential metabolites. Metlin database, Human Metabolome database, and our in-house laboratory database were used to identify metabolites. The experiment was performed with four repetitions, and data were expressed as mean ± SD.
Global Transcriptomic Analysis
Total RNA was isolated from fresh root of copper-treated or non-treated S. miltiorrhiza using RNeasy Plus Kit with the genomic DNA removal step. The concentration and the quality of extracted RNA were evaluated. cDNA library construction and sequencing were performed by the Biomarker Technologies Corporation (Beijing, China). The raw sequence data were filtered to obtain clean data, which were then compared with the S. miltiorrhiza reference genome using HISAT2 software (https://ccb.jhu.edu/software/hisat2/manual.shtml). Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) was used to calculate gene expression. Differential gene expression analysis used edgeR (Robinson et al., 2010), and genes with p < 0.05 and fold change >1.5 or <0.5 are regarded as differentially expressed genes (DEGs). The DEGs were classified into functional categories by blasting against the clusters of orthologous groups against the eukaryotic complete genomes (KOG) database (http://www.ncbi.nlm.gov/KOG) and the gene ontology (GO) database (http://geneontology.org/). The experiment was performed with three repetitions, and data were expressed as mean ± SD.
Total RNA Extraction and Reverse Transcription-Quantitative PCR (RT-qPCR)
Total RNA extraction was achieved using a plant total RNA extraction kit (Tiangene Biotech Co., Ltd., China). Briefly, a 0.5-g fresh S. miltiorrhiza root was grounded into powder with liquid nitrogen, and extraction was carried out according to the instructions. RNA reverse transcription (Hieff™ First Strand cDNA Synthesis Super Mix, Yeasen, China) and gene quantification (Hieff™ qPCR SYBR Green Master Mix kit, Yeasen, China) were performed following the protocols of the manufacturer. The qPCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 70°C for 90 s. mRNA expression levels were calculated using the 2−ΔΔCt method and presented as a ratio to β-actin. Primer sequences are shown in Supplementary Table 4.
Quantification of Laccase Enzyme Activity
A Laccase Activity Detection kit (Solarbio Life Sciences, Beijing, China) was used to detect laccase activity. To detect laccase activity, a 0.1-g fresh S. miltiorrhiza root was added to a 1-ml extraction buffer, the mixture was homogenized on ice, and then centrifuged at 12,000 rpm for 20 min at 4°C. The supernatant was then moved to a new centrifuge tube, and the absorbance was measured at 562 nm to determine protein concentration of supernatant using a BCA kit (Good Laboratory Practice Bioscience, Montclair, CA, USA). Subsequently, 150 μL of supernatant was mixed with 850 μL of a working buffer and heated in an oven at 45°C for 3 min. As negative control, 150 μL of an extraction buffer was mixed with 850 μL of the working buffer and heated in an oven at 45°C for 3 min. Then, the produced ABTS radical was determined through measuring mixture absorbance at 420 nm. Enzyme activity was calculated through measuring ABTS radical content change according to the instructions of the manufacturer.
Phylogenetic Construction of Laccase
The whole genome sequence and protein data from S. miltiorrhiza (BioProject: PRJNA287594) were downloaded from the Genome Warehouse in the BIG Data Center (http://gigadb.org/dataset/100164). The whole genome data and protein data of Arabidopsis thaliana were downloaded from the Arabidopsis thaliana information resource website (TAIR, https://www.arabidopsis.org/). In order to select all the laccases in the genomes of Salvia miltiorrhiza and Arabidopsis thaliana, we downloaded the unique HMM model (the hidden Markov model) of laccase from the Pfam database. HMMER 3.0 software with hmmsearch was used to search for laccase-conserved domain models, including PF00394, PF07731, and PF07732, in the protein database of S. miltiorrhiza and Arabidopsis thaliana. A phylogenetic tree was constructed using the maximum likelihood (ML) method with bootstraps in MEGA X software.
Correlation Analysis
FPKM values of genes in copper ion-treated and non-treated groups were used to calculate correlation coefficient. Gene co-expression analysis between the candidate transcription factors and the key genes was performed by the mean of the Pearson correlation test using IBM SPSS Statistics 20. Data are shown as the mean ± SD (n = 3) (*p < 0.05 and **p < 0.01). The correlation coefficient >0.8 was considered to be co-expressed, and the coexpression network was visualized using Cytoscape.
Copper-Response Element Predication in Laccases and Transcription Factors
The promoter sequences of analyzed genes (from 2,000 bp, relative to a translation start site) were extracted using gff file of S. miltiorrhiza genome. Core motif of copper-response element (GTAC) (Quinn et al., 2000; Kropat et al., 2005) and metal response-element (TGCxCxC) (Murphy et al., 1999) were predicted in extracted 2,000 bp promoter sequences of laccases and transcription factors using the Plantcare database (Lescot et al., 2002) and visualized by TB tools (Chen et al., 2020). In addition, transcription factors-binding motifs were also predicated in extracted 2,000 bp promoter sequences of laccases.
Results
A Low Concentration of Copper Ions Promoted Plant Growth
To explore the effect of copper on the growth of S. miltiorrhiza plants, we exposed the plants to predetermined copper concentrations (0, 5, 25, and 100 μM) and determined the phenotypic changes thereafter. After 5 days of cultivation, the S. miltiorrhiza plants wilted under treatments with a greater copper concentration. Compared with the 0 and 5 μM treatments, S. miltiorrhiza plants grew slowly and the leaves withered and yellowed in the 25- and 100-μM treatments (Figure 1A). It is noteworthy that low concentration of copper (5 μM) significantly promoted plant growth compared with the blank (0 μM) group (Figure 1A). Thus, 5 μM of copper was optimum for the growth of the S. miltiorrhiza plants.
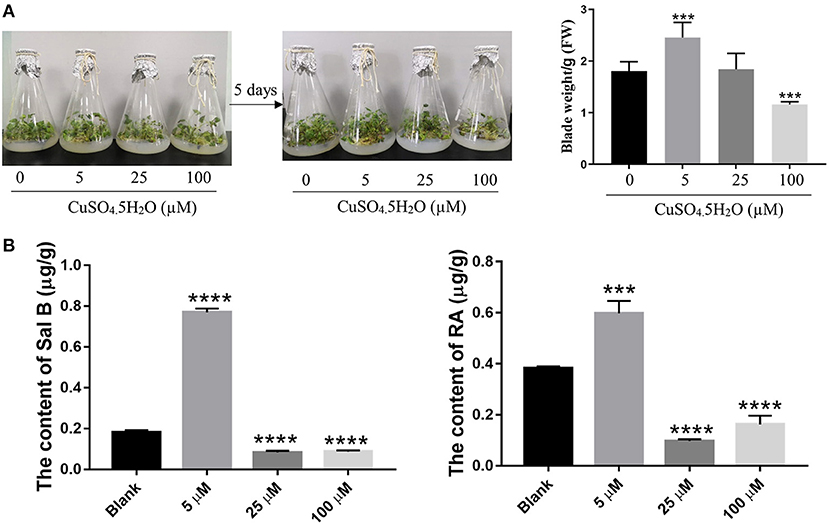
Figure 1. Effect of copper ion treatment on the growth of Salvia miltiorrhiza and biosynthesis of salvinolic acids (SalAs). Two-month-old S. miltiorrhiza was treated with different concentrations of CuSO4.5H2O (0, 5, 25, and 100 μM) for 5 days. (A) Phenotype change and weight change of S. miltiorrhiza were recorded before and after the treatment. (B) The content of salvianolic acid B and rosemarinic acid in untreated and treated S. miltiorrhiza using high-performance liquid chromatography (HPLC) with standards. Data represent the mean ± SD of three independent biological replicates. Significant differences between the treatment and control samples were determined using a Student's T-test (***P < 0.001 and ****P < 0.0001).
A Low Concentration of Copper Ions Promoted the Accumulation of Sal B and RA
As the major SalAs, Sal B, and RA are important active secondary metabolites in S. miltiorrhiza. To ascertain the most suitable concentration of copper that promotes SalAs biosyntheses, we used HPLC to determine the contents of Sal B and RA in the copper-stressed groups and the blank group. The low concentration of copper treatment increased Sal B by an average of 4.27-fold compared with the control, but the medium and high concentrations of copper treatments decreased Sal B by 2.16- and 2.06-fold, respectively, compared with the control (Figure 1B and Supplementary Figure 1). Compared with the blank group, low concentration of copper increased RA content 1.55-fold while medium and high concentrations of copper decreased its amount 4.22- and 2.37-fold, respectively (Figure 1B and Supplementary Figure 1). Based on these findings, it is evident that low concentration of copper promotes the biosyntheses of Sal B and RA.
A Low Concentration of Copper Increases the Content of SalAs
To further confirm the holistic effect of low copper on the accumulation of SalAs in S. miltiorrhiza, the plant samples exposed to low concentration of copper and the blank group were analyzed using UPLC-QTOF-MS. In the negative iron model, the total ion chromatogram was different between the control and the low copper treatment (Supplementary Figure 2). A total of 2,041 ions were detected (Figure 2A). Using the selection criteria of change ratio >1.5 (significant increase; P < 0.05) or < 0.5 (significant decrease; P < 0.05), 90 increased and 70 decreased compounds were detected. The principal component analysis (PCA) analysis showed that the copper stress treatment and control clearly clustered into two categories (Figure 2B). Among the different metabolites, 13 SalAs, including Sal B, RA, lithospermic acid, salvianolic acid D, salvianolic acid A, and danshensu, were significantly increased in the low concentration of copper treatment compared with the control (Figure 3A), indicating that low copper stress can positively regulate the synthesis of SalAs. The pathway enrichment analysis of differential metabolites showed that copper ions had significant effects on amino acid synthesis and metabolism, the pentose phosphate pathway, and carbon fixation in photosynthesis in S. miltiorrhiza (Figure 3B).
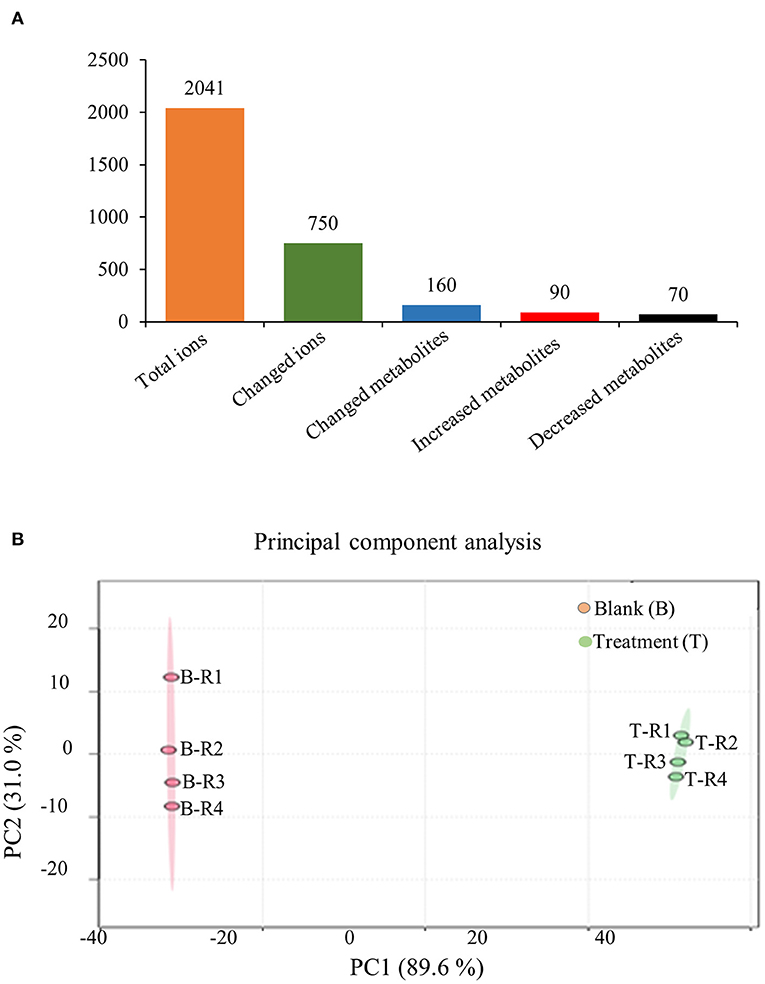
Figure 2. Metabolomic analysis of low concentration of copper ion (LCCI) on the biosynthesis of salvinolic acids (SalAs) in Salvia miltiorrhiza. Two-month-old S. miltiorrhiza was treated with CuSO4.5H2O (5 μM) for 5 days. Metabolites were extracted from roots and subjected to metabolomics analysis by untargeted ultra-performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q/TOF-MS). The total ions were counted within 90 min. (A) The statistical number of identified ions, metabolites, and significantly changed metabolites is listed. (B) Principal component analysis (PCA) and a volcano plot based on identified ions in control and treated samples. To characterize their ion profiles, the identified ions were estimated using a principal component analysis. The significantly changed ions are depicted with a volcano plot. Red dots indicate significantly changed ions.
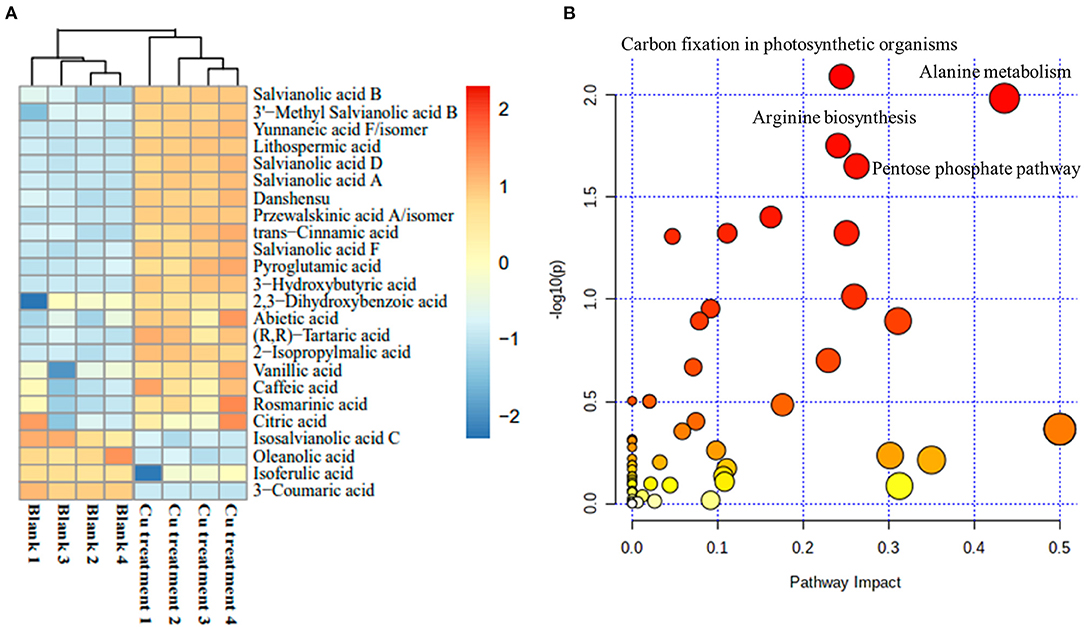
Figure 3. Change of identified salvinolic acids (SalAs). Two-month-old S. miltiorrhiza was treated with CuSO4.5H2O (5 μM) for 5 days. (A) The changes of identified SalAs in control and treated samples were analyzed using a Cluster analysis. (B) Pathway enrichment analysis of significantly changed metabolites was performed using the KEGG database.
Transcriptomics Analysis to Identify Genes That Responded to Copper Treatment in S. miltiorrhiza
To explore how the low concentration of copper treatment regulated the biosynthesis of SalAs in S. miltiorrhiza, a transcriptome analysis was performed between the control and low copper treatments. Using the selection criteria of the change ratio P < 0.05 and fold change >1.5 and <0.5, a total of 418 differential genes were obtained, of which 207 were upregulated and 211 were downregulated in the low copper-treated group compared with the blank group (Supplementary Table 2). Gene Ontology (GO) analysis showed that these significantly changed genes were mainly involved in copper ion binding, oxidoreductase, and ROS metabolism (Figure 4A). Pathway enrichment analysis showed that the differentially expressed genes are related to tyrosine and phenylalanine metabolism, chlorophyll biosynthesis, plant pathogene interaction, photosynthesis, and plant hormone signal transduction (Figure 4B).
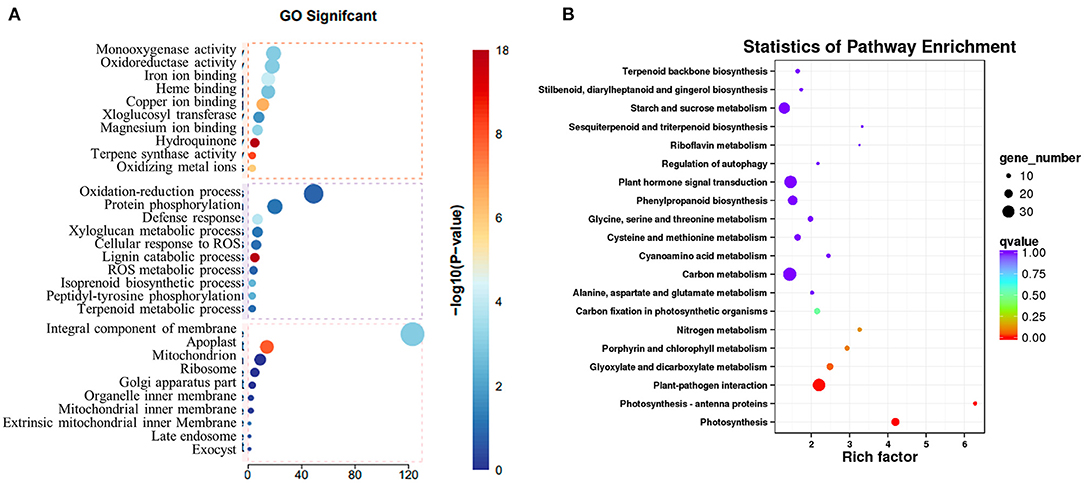
Figure 4. Transcriptomic analysis of Salvia miltiorrhiza with low concentration copper treatment. (A) Two-month-old S. miltiorrhiza was treated with CuSO4.5H2O (5 μM) for 5 days, following which mRNA was extracted from roots and subjected to transcriptomic analysis. (A) Differentially expressed genes were identified by comparing gene expression in copper treated vs. untreated samples. Function of identified significantly changed genes was analyzed using the gene ontology (GO) database. (B) Pathway mapping of significantly changed genes was performed using the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/).
A further gene expression pattern indicated that SalAs biosynthesis-related genes (laccase 20, laccase 13, laccase 7, and rosmarinate synthase), photosynthesis-related genes (blue copper protein, basic blue protein, and chlorophyll A-B-binding protein), and zinc transporter 5 were upregulated in the copper-treated group compared with the blank group (Figure 5A and Table 1). In contrast, copper transport-related genes, such as copper transporter 6, copper transporter 5, copper transport protein CCH, and copper-transporting ATPase RAN1, were downregulated in the copper-treated group (Figure 5A and Table 1). In addition, transcription factors, such as ethylene-responsive transcription factor 5, ERF012, ethylene-responsive transcription factor RAP2-7, ethylene-responsive transcription factor TINY, transcription factor MYB14, MYB33, MYB73, basic helix-loop-helix transcription factor, and transcription factor bHLH69, were responsive to low copper treatment (Figure 5A and Table 1).
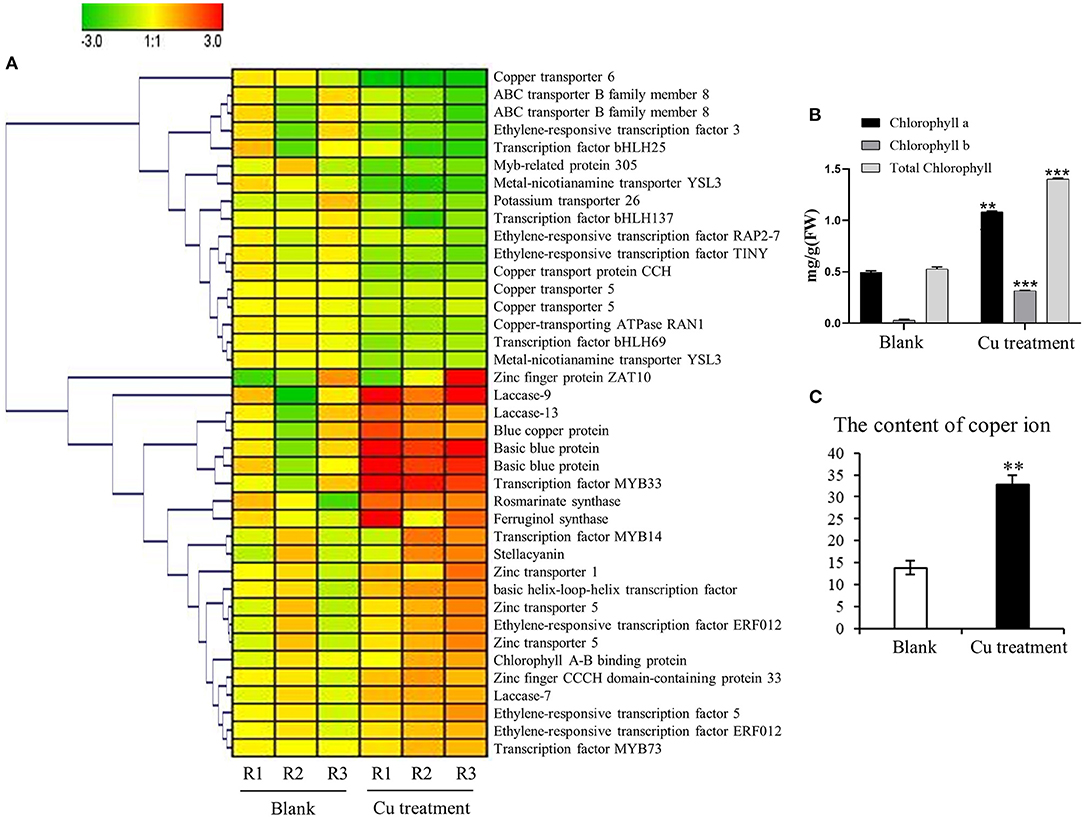
Figure 5. Impact of the copper treatment on biosynthesis of salvinolic acids (SalAs), chlorophyll metabolism, and ion transport. (A) The change tendency of differentially expressed genes involved in the biosynthesis of SalAs, chlorophyll metabolism, and ion transport was analyzed based on their expression levels in untreated and treated samples. (B) The content of chlorophyll a, chlorophyll b, and total chlorophyll in untreated and treated samples was determined. (C) The content of copper ions in untreated and treated samples was measured. Data represent the mean ± SD of three independent biological replicates. Significant differences between treatment and control samples were determined using a Student's T-test (**P < 0.01, ***P < 0.001).
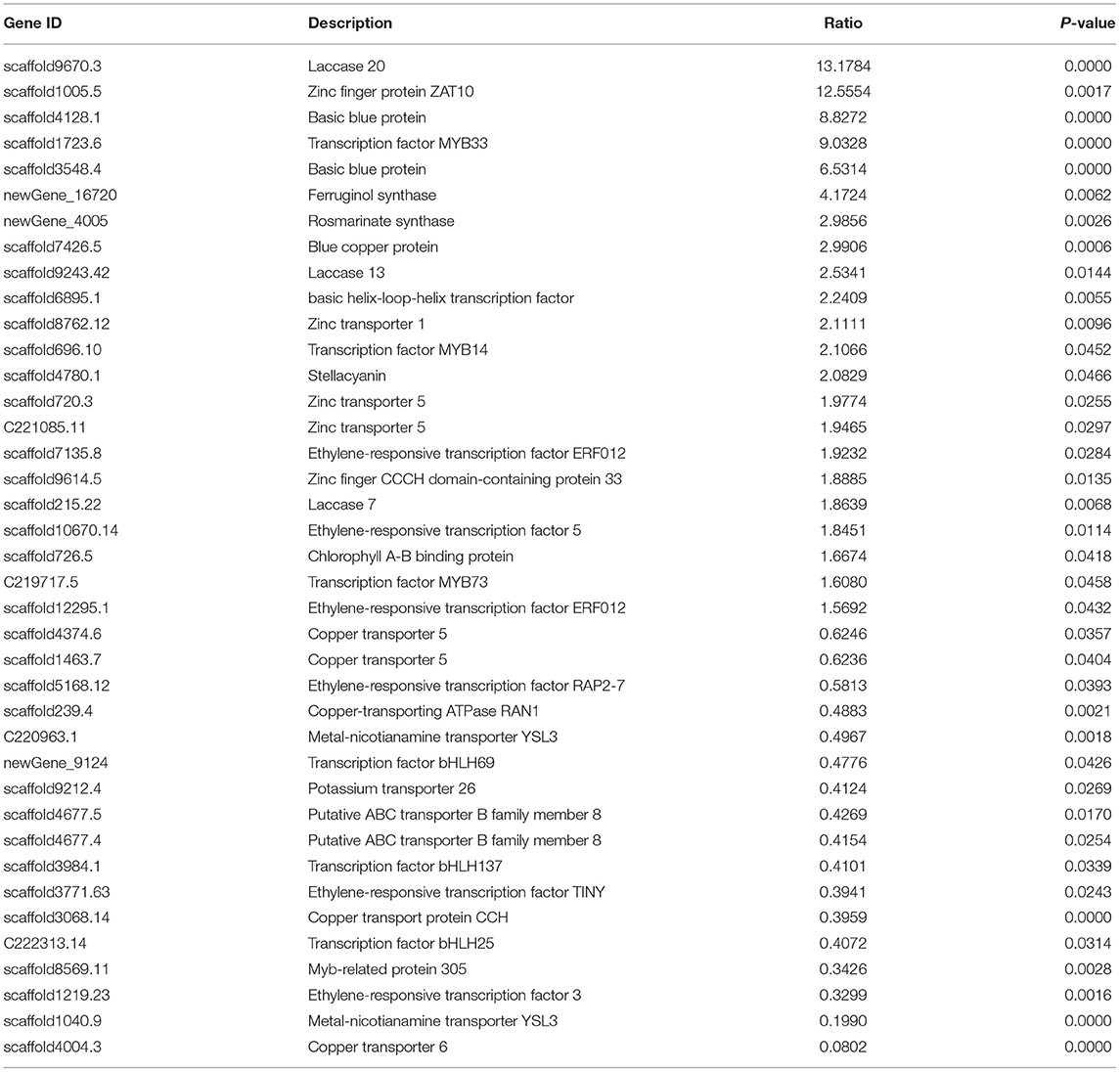
Table 1. Key genes involved in salvianolic acid biosynthesis that showed a significantly change in abundance in leaves and roots of Salvia miltiorrhiza in response to treatment with a low concentration of copper ions.
Effect of Copper Ions on the Chlorophyll Content in the Leaves of S. miltiorrhiza
To determine whether the gene expression changes corresponded to physiological changes, chlorophyll content and copper content in S. miltiorrhiza were measured. Chlorophyll, which plays an extremely important role in photosynthesis, is the main component of chloroplasts in leaves. After the copper ion treatment, we detected the difference in chlorophyll content between the 5-μM copper treatment and the control (Figure 5B). The chlorophyll a, chlorophyll b, and total chlorophyll of S. miltiorrhiza in the 5-μM copper treatment were 1.08, 0.31, and 1.39 mg/g, respectively. The chlorophyll a, chlorophyll b, and total chlorophyll of S. miltiorrhiza in the control were 0.49, 0.03, and 0.52 mg/g, respectively. Compared with the control, the 5-μM copper treatment increased chlorophyll a, chlorophyll b, and total chlorophyll by 2.19, 9.98, and 2.66 times, respectively. Copper content in the treatment group was also significantly increased compared with the control (Figure 5C). The greater chlorophyll content increased the photosynthetic ability of the copper treatment group, the growth state of the plant was healthier, the ability of the plant to resist adverse environments was increased, and the treatment was conducive to the accumulation of active products in S. miltiorrhiza.
Bioinformatics and Enzyme Activity Analysis of Copper Ion-Induced Laccase
Based on the transcriptomics result, we verified the mRNA expression of laccase 20, laccase 13, and laccase 7 using RT-qPCR. The results obtained showed 2.3 and 1.4-fold increases in laccase 20 and laccase 13 expressions, respectively, in the low-copper group relative to the control group. Consequently, laccase enzyme activity increased 3.19-fold in the low-copper group compared with the control group (Figure 6A). These findings suggest the pivotal role of laccase in the synthesis of salvianolic acids in the presence of low copper. To explore the evolutionary relationships among laccases, the sequences of laccases were extracted from S. miltiorrhiza and A. thaliana, and a phylogenetic tree was constructed using the ML method with bootstraps in MEGA X software (Figure 6B). Laccases in S. miltiorrhiza and Arabidopsis thaliana were grouped into seven clusters (I, II, III, IV, V, VI, and VII) based on similarity of amino acid sequence. The copper-induced laccases (marked with a red pentagram) were positioned in Cluster II and Cluster V (Figure 6B). Functional domain analysis found that the three key laccases had the same three motifs and conserved domains, which implies that they have functional similarity (Figure 6C).
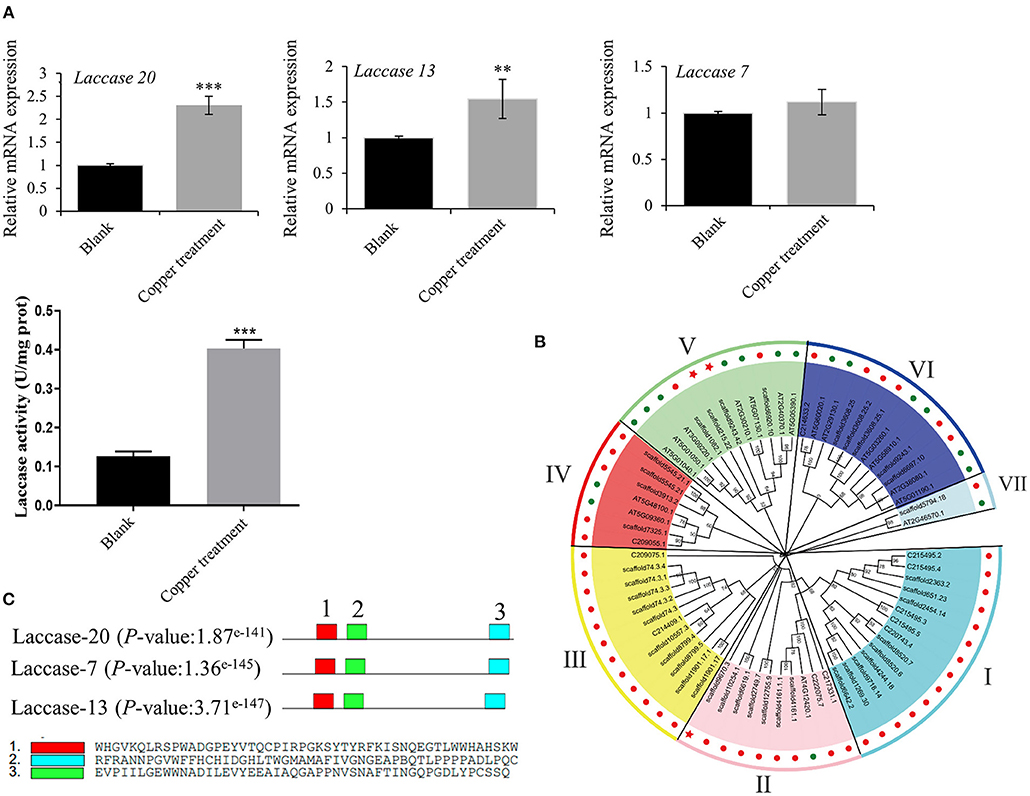
Figure 6. Bioinformatics and enzyme activity analyses of copper ion-induced laccase. (A) mRNA expression levels of laccase 20, laccase 13, and laccase 7 in untreated and treated samples were checked by reverse transcription-quantitative PCR (RT-qPCR). Crude proteins were extracted from root of Salvia miltiorrhiza, and laccase activity was measured in untreated and treated samples using an ELISA kit. (B) A phylogenetic tree of laccases in Salvia miltiorrhiza (red dots) and Arabidopsis thaliana (green dots) was constructed using the maximum likelihood (ML) method with bootstraps in MEGA X software. Our target laccases (laccase 20, laccase 13, and laccase 7) were marked with a red pentagram. (C) A motif analysis of three key laccases was performed using MEME Suite. **P-value < 0.01, ***P-value < 0.001.
To identify the upstream transcription factor involved in the regulation of copper-induced biosynthetic enzymes of SalAs, a coexpression analysis of biosynthetic genes related to SalAs (laccases and rosmarinate synthase) and 12 key transcription factors was carried out. As shown in Figure 7, transcription factors were positively correlated with key candidate genes in the biosynthetic pathways of SalAs. Among them, zinc finger protein ZAT10, Myb family transcription factor PHL5, ethylene-responsive transcription factor 5, and bZIP transcription factor 18 were significantly coexpressed with laccase 20 (gene 3); Zinc finger CCCH domain-containing protein 33/29, zinc finger A20 and AN1 domain-containing stress-associated protein 5, transcription factor MYB73, and ethylene-responsive transcription factor ERF012 were significantly coexpressed with laccase 7 (gene 1); Transcription factor MYB33 was significantly coexpressed with laccase 13 (gene 3) (Supplementary Table 3). To further explore the underlying mechanism by which copper induces expression of laccases and transcription factors, a copper-response element and metal-response elements were searched against 2,000 bp promoter sequences of laccases and transcription factors. As shown in Supplementary Figure 3, a core motif of a copper-response element (GTAC) (Quinn et al., 2000; Kropat et al., 2005) and a metal-response element (TGCxCxC) (Murphy et al., 1999) were identified in extracted 2,000 bp promoter sequences of laccases (laccase 20 and laccase 13) and transcription factors (zinc finger protein ZAT10, Zinc finger CCCH domain-containing protein 33, transcription factor MYB73, and transcription factor MYB33). In addition, MYB transcription factor-binding motifs were also identified in promoter sequences of laccase 20 and laccase 13 (Supplementary Figure 3).
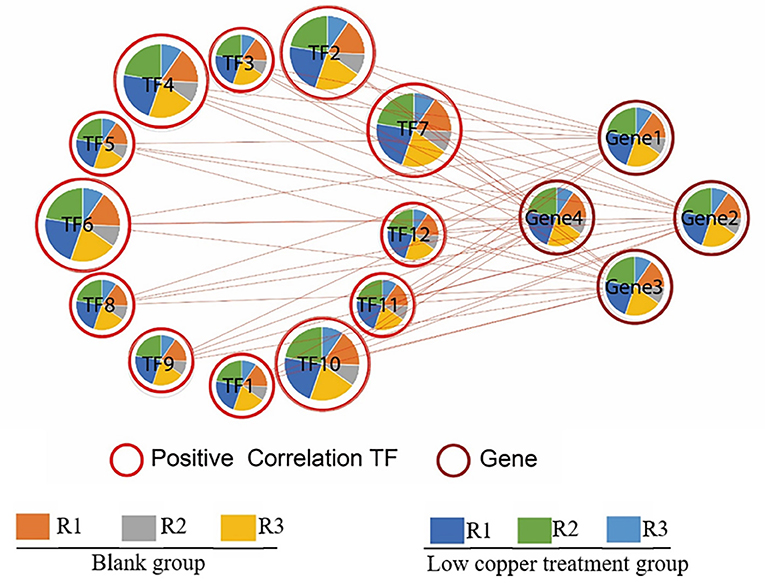
Figure 7. Correlation analysis between significantly changed transcription factors and enzymes related to SalA biosynthesis. The correlation between significantly changed SalA biosynthetic enzymes and the relative expression of candidate transcription factors was analyzed using R software. A Spearman correlation analysis was performed to calculate the correlation coefficient.
Discussion
SalAs are important water-soluble compounds in S. miltiorrhiza, which contribute to the treatment of cerebrovascular and cardiovascular diseases (Chen et al., 2016; Zhou et al., 2016). Recently, increasing research has been conducted to increase the content of SalA in S. miltiorrhiza (Xiao et al., 2009; Zhang et al., 2010). In this study, we reported that a low concentration of copper (5 μM) could promote the biosynthesis of SalA in S. miltiorrhiza. Then, we conducted comprehensive, integrated metabolomic and transcriptomic analyses to provide insights into the mechanisms underlying low copper-induced SalA biosynthesis. Genes-encoding SalAs biosynthetic enzymes (laccases and rosmarinate synthase) and transcription factors were identified as potential key factors involved in low copper-induced SalA biosynthesis.
A Low Copper Concentration Promotes SalA Biosynthesis in S. miltiorrhiza Through Regulating Plant Growth and Photosynthesis
Our results revealed that the growth status and fresh weight of S. miltiorrhiza were significantly improved in the low copper concentration treatment compared with the control. The metabolites involved in carbon fixation in photosynthesis were significantly increased by the low-copper concentration treatment. Importantly, chlorophyll a, chlorophyll b, and total chlorophyll, which play an important role in photosynthesis (Baker, 2008), showed an increase in accumulation in low copper-treated plants. Photosynthesis contributes greatly to the biosynthesis of secondary metabolites in plants (Landi et al., 2020). A previous study has shown that copper played a role in photosynthesis through strengthening the photosynthetic capacity of plants (Douglas and Jaqueline, 2019). In our study, blue copper protein and basic blue protein, which are involved in promoting photosynthesis through acting as electron acceptors (Skyes, 1991), were upregulated in low copper-treated plants. Sal B and RA, the two most representative secondary metabolites of phenolic acids in S. miltiorrhiza (Zhao et al., 2015; Shi et al., 2016, 2019; Zhou et al., 2017; Huang et al., 2019), were significantly increased in S. miltiorrhiza treated with a low concentration of copper. These results suggest that low concentrations of copper induce the accumulation of SalAs might be through regulating photosynthesis related genes such as blue copper proteins in S. miltiorrhiza.
Low Concentration of Copper Induces the Accumulation of SalAs by Activating Key Enzymes Involved in SalA Biosynthesis
Both tyrosine- and phenylalanine-derived pathways are the starting point for the biosynthesis of SalAs s (Hao et al., 2020). The data obtained in the transcriptomic analysis showed that rosmarinate synthase and laccase were upregulated in plants treated with a low copper concentration compared with the control. Rosmarinate synthase is the key enzyme in the pathway of SalAs biosynthesis in S. miltiorrhiza (Zhou et al., 2018). A previous study has reported that phenolic acids, such as rosmarinic acid and lithospermic acid B, were decreased in rosmarinate synthase-knockdown hairy root lines of S. miltiorrhiza (Zhou et al., 2018). However, overexpression of rosmarinate synthase resulted in a higher content of phenolic acids (up to over 3-fold) in transgenic lines compared with the control (Fu et al., 2020). Recently, scientists have demonstrated that laccase plays a critical role in the biosynthesis of Sal B. The laccase gene family in S. bowleyana has undergone expansion, and it might catalyze the oxidative reaction from RA to Sal B (Zheng et al., 2021). At present, 29 laccase candidates have been found in S. miltiorrhiza, and they contained three signature Cu-oxidase domains (Li et al., 2019b). It is known that laccases are copper-containing enzymes, and their catalytic activity requires the presence of copper ions (Arregui et al., 2019). Here, a core motif of a copper-response element (GTAC) and a metal response-element (TGCxCxC) were identified in the promoter region of laccase 20 and laccase 13. Overexpression and silencing assays demonstrated SmLAC7 and SmLAC20 played an important role in the biosynthesis of Sal B (Li et al., 2019b). In our study, laccase 20, laccase 13, laccase 7, and rosmarinate synthase were significantly upregulated in S. miltiorrhiza in the low-copper treatment compared with the control, suggesting that treatment with a low concentration of copper may promote the accumulation of SalAs via the regulation of laccase 20/13 and rosmarinate synthase.
In addition to the upregulation of laccase 20/13 and rosmarinate synthase, transcription factors, such as ethylene-responsive transcription factor (ERF), transcription factor MYBs, and basic helix-loop-helix transcription factor (bHLH), were also responsive to the low-copper treatment. The content of SalAs was increased in SmERF115-overexpressing hairy roots and was decreased in silencing lines (Sun et al., 2019). Previous studies have reported that SmMYB1 was a positive activator that improved the accumulation of phenolic acids in S. miltiorrhiza (Deng et al., 2020b; Zhou et al., 2021). However, SmMYB36 was reported to inhibit the accumulation of phenolic acids (Ding et al., 2017). For the bHLH transcription factor, the overexpression of SmbHLH37 substantially decreased yields of Sal B (Du et al., 2018). Here, zinc finger protein ZAT10, Myb family transcription factor PHL5, ethylene-responsive transcription factor 5, and bZIP transcription factor 18 were significantly upregulated and coexpressed with laccase 20 under low concentration of copper treatment in S. miltiorrhiza. In our study, a core motif of a copper-response element (GTAC) and a metal-response element (TGCxCxC) were identified in the promoter region of transcription factors (zinc finger protein ZAT10, Zinc finger CCCH domain-containing protein 33, transcription factor MYB73, and transcription factor MYB33). In addition, MYB transcription factor-binding motifs were identified in promoter sequences of laccase 20 and laccase 13. In Arabidopsis, copper has been shown to induce expression changes of transcription factors, including bHLH, MYB, and ERF (Jakubowicz et al., 2010; Perea-García et al., 2010; Cai et al., 2021). Combined, these results suggest that transcription factors, such as ERF, MYB, and bHLH, play a role in copper-induced SalA biosynthesis in S. miltiorrhiza.
We systematically studied the effects of copper on SalA biosynthesis in S. miltiorrhiza. We identified a suitable copper ion concentration to promote the biosynthesis of SalA and provide a potential regulatory mechanism by identifying a series of copper-responsive genes. Our work provides an effective approach to induce SalAs and demonstrates a promising future for the metabolic regulation of S. miltiorrhiza.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/, PRJNA645746.
Author Contributions
XY conceived and designed the research and revised the manuscript. YX, WS, XW, and JD analyzed the data and drafted the manuscript. WS and XW performed the experiments. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81803654).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.746117/full#supplementary-material
References
Andre, C. M., Larondelle, Y., and Evers, D. (2010). Dietary antioxidants and oxidative stress from a human and plant perspective: a review. Curr. Nutr. Food Sci. 6, 2–12. doi: 10.2174/157340110790909563
Arregui, L., Ayala, M., Gómez-Gil, X., Gutiérrez-Soto, G., Hernández-Luna, C. E., Herrera de Los Santos, M., et al. (2019). Laccases: structure, function, and potential application in water bioremediation. Microb. Cell Fact. 18:200. doi: 10.1186/s12934-019-1248-0
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Burkhead, J. L., Reynolds, K. A., Abdel-Ghany, S. E., Cohu, C. M., and Pilon, M. (2009). Copper homeostasis. N. Phytol. 182, 799–816. doi: 10.1111/j.1469-8137.2009.02846.x
Cai, Y., Li, Y., and Liang, G. (2021). FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis. Plant Cell Environ. 44, 1679–1691. doi: 10.1111/pce.14000
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Cheng, J., Chen, L., Zhang, G., Huang, H., Zhang, Y., Xu, L., et al. (2016). Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 170, 2136–45. doi: 10.1104/pp.15.01733
Choudhary, S. P., Oral, H. V., Bhardwaj, R., Yu, J. Q., and Tran, L. S. (2012). Interaction of brassinosteroids and polyamines enhances copper stress tolerance in Raphanus sativus. J. Exp. Bot. 63, 5659–5675. doi: 10.1093/jxb/ers219
De Caroli, M., Furini, A., DalCorso, G., Rojas, M., and DiSansebastiano, G. P. (2020). Endomembrane reorganization induced by heavy metals. Plants 9:482. doi: 10.3390/plants9040482
Deng, C., Shi, M., Fu, R., Zhang, Y., Wang, Q., Zhou, Y., et al. (2020a). ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71, 5948–5962. doi: 10.1093/jxb/eraa295
Deng, C., Wang, Y., Huang, F., Lu, S., Zhao, L., Ma, X., et al. (2020b). SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 62, 1688–1702. doi: 10.1111/jipb.12943
Ding, K., Pei, T., Bai, Z., Jia, Y., Ma, P., and Liang, Z. (2017). SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci. Rep. 7:5104. doi: 10.1038/s41598-017-04909-w
Ding, Y., Ding, L., Xia, Y., Wang, F., and Zhu, C. (2020). Emerging roles of micrornas in plant heavy metal tolerance and homeostasis. J. Agric. Food Chem. 68, 1958–1965. doi: 10.1021/acs.jafc.9b07468
Douglas, H. B., and Jaqueline, C. S. (2019). The effects of copper on photosynthesis and biomolecules yield in Chlorolobion braunii. J. Phycol. 55, 1335–1347. doi: 10.1111/jpy.12914
Droppa, M., Terry, N., and Horvath, G. (1984). Effects of Cu deficiency on photosynthetic electron transport. Proc. Natl. Acad. Sci. U.S.A. 81, 2369–2373. doi: 10.1073/pnas.81.8.2369
Du, T., Niu, J., Su, J., Li, S., Guo, X., Li, L., et al. (2018). SmbHLH37 functions antagonistically with SmMYC2 in regulating jasmonate-mediated biosynthesis of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 9:1720. doi: 10.3389/fpls.2018.01720
Fu, R., Shi, M., Deng, C., Zhang, Y., Zhang, X., Wang, Y., et al. (2020). Improved phenolic acid content and bioactivities of Salvia miltiorrhiza hairy roots by genetic manipulation of RAS and CYP98A14. Food Chem. 331:127365. doi: 10.1016/j.foodchem.2020.127365
Hao, X., Pu, Z., Cao, G., You, D., Zhou, Y., Deng, C., et al. (2020). Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J Adv Res. 23, 1–12. doi: 10.1016/j.jare.2020.01.012
Hoegger, P. J., Kilaru, S., James, T. Y., Thacker, J. R., and Kues, U. (2006). Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 273, 2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x
Huang, Q., Sun, M., Yuan, T., Wang, Y., Shi, M., Lu, S., et al. (2019). The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 274, 368–375. doi: 10.1016/j.foodchem.2018.08.119
Ibrahim, M. H., Kong, Y., and Zain, N. A. (2017). Effect of cadmium and copper exposure on growth, secondary metabolites and antioxidant activity in the medicinal plant Sambung Nyawa (Gynura procumbens (Lour.) Merr). Molecules 22:1623. doi: 10.3390/molecules22101623
Jakubowicz, M., Gałgańska, H., Nowak, W., and Sadowski, J. (2010). Exogenously induced expression of ethylene biosynthesis, ethylene perception, phospholipase D, and Rboh-oxidase genes in broccoli seedlings. J. Exp. Bot. 61, 3475–3491. doi: 10.1093/jxb/erq177
Jones S. M Solomon E. I. (2015). Electron transfer and reaction mechanism of laccases. Cell Mol Life Sci. 72, 869–883.
Kropat, J., Tottey, S., Birkenbihl, R. P., Depège, N., Huijser, P., and Merchant, S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735. doi: 10.1073/pnas.0507693102
Landi, M., Zivcak, M., Sytar, O., Brestic, M., and Allakhverdiev, S. I. (2020). Plasticity of photosynthetic processes and the accumulation of secondary metabolites in plants in response to monochromatic light environments: a review. Biochim. Biophys. Acta Bioenerg. 1861:148131. doi: 10.1016/j.bbabio.2019.148131
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, C., Li, D., Zhou, H., Li, J., and Lu, S. (2019a). Analysis of the laccase gene family and miR397-/miR408-mediated posttranscriptional regulation in Salvia miltiorrhiza. Peer J. 7:e7605. doi: 10.7717/peerj.7605
Li, Q., Feng, J., Chen, L., Xu, Z., Zhu, Y., Wang, Y., et al. (2019b). Genome-wide identification and characterization of Salvia miltiorrhiza laccases reveal potential targets for salvianolic acid B biosynthesis. Front. Plant Sci. 10:435. doi: 10.3389/fpls.2019.00435
Ma, P., Liu, J., Zhang, C., and Liang, Z. (2013). Regulation of water-soluble phenolic acid biosynthesis in Salvia miltiorrhiza Bunge. Appl. Biochem. Biotechnol. 170, 1253–1262. doi: 10.1007/s12010-013-0265-4
McCaig, B. C., Meagher, R. B., and Dean, J. F. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221, 619–636. doi: 10.1007/s00425-004-1472-6
Murphy, B. J., Andrews, G. K., Bittel, D., Discher, D. J., McCue, J., Green, C. J., et al. (1999). Activation of metallothionein gene expression by hypoxia involves metal response elements and metal transcription factor-1. Cancer Res. 59, 1315–1322
Patel, N., Shahane, S., Shivam, M. R., and Mishra, U. (2019). Mode of action, properties, production, and application of laccase: a review. Recent Pat. Biotechnol. 13, 19–32. doi: 10.2174/1872208312666180821161015
Perea-García, A., Andrés-Colás, N., and Peñarrubia, L. (2010). Copper homeostasis influences the circadian clock in Arabidopsis. Plant Signal. Behav. 5, 1237–1240. doi: 10.4161/psb.5.10.12920
Quinn, J. M., Barraco, P., Eriksson, M., and Merchant, S. (2000). Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275, 6080–6089. doi: 10.1074/jbc.275.9.6080
Quist, D. A., Diaz, D. E., Liu, J. J., and Karlin, K. D. (2017). Activation of dioxygen by copper metalloproteins and insights from model complexes. J. Biol. Inorg. Chem. 22, 253–288. doi: 10.1007/s00775-016-1415-2
Riva, S. (2006). Laccases: blue enzymes for green chemistry. Trends Biotechnol. 24, 219–226. doi: 10.1016/j.tibtech.2006.03.006
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saleem, M. H., Ali, S., Rehman, M., Hasanuzzaman, M., Rizwan, M., Irshad, S., et al. (2020). Jute: a potential candidate for phytoremediation of metals-a review. Plants 9:258. doi: 10.3390/plants9020258
Shi, M., Huang, F., Deng, C., Wang, Y., and Kai, G. (2019). Bioactivities, biosynthesis and biotechnological production of phenolic acids in Salvia miltiorrhiza. Crit. Rev. Food Sci. Nutr. 59, 953–964. doi: 10.1080/10408398.2018.1474170
Shi, M., Luo, X., Ju, G., Li, L., Huang, S., and Zhang, T. (2016). Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agric. Food Chem. 64, 2523–2530. doi: 10.1021/acs.jafc.5b04697
Skyes, A. G. (1991). Active site properties of blue copper proteins. Adv. Inorg. Chem. 36:377. doi: 10.1016/S0898-8838(08)60044-6
Solomon, E. I., Sundaram, U. M., and Machonkin, T. E. (1996). Multicopper oxidases and oxygenases. Chem. Rev. 96, 2563–2605. doi: 10.1021/cr950046o
Song, Y. J., Zhou, L. C., Yang, S. H., Wang, C. L., Zhang, T. J., and Wang, J. H. (2017). Dose-dependent sensitivity of Arabidopsis thaliana seedling root to copper is regulated by auxin homeostasis. Environ. Exp. Bot. 139, 23–30. doi: 10.1016/j.envexpbot.2017.04.003
Sun, M., Shi, M., Wang, Y., Huang, Q., Yuan, T., Wang, Q., et al. (2019). The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 70, 243–245. doi: 10.1093/jxb/ery349
Turlapati, P. V., Kim, K. W., Davin, L. B., and Lewis, N. G. (2011). The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233, 439–470. doi: 10.1007/s00425-010-1298-3
Wang, J., Xu, J., Gong, X., Yang, M., Zhang, C., and Li, M. (2019). Biosynthesis, chemistry, and pharmacology of polyphenols from Chinese salvia species: a review. Molecules 24:155. doi: 10.3390/molecules24010155
Xiao, Y., Gao, S. H., Di, P., Chen, J., Chen, W., and Zhang, L. (2009). Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol. Plantarum 137, 1–9. doi: 10.1111/j.1399-3054.2009.01257.x
Xing, B., Liang, L., Liu, L., Hou, Z., Yang, D., Yan, K., et al. (2018). Overexpression of SmbHLH148 induced biosynthesis of tanshinones as well as phenolic acids in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 37, 1681–1692. doi: 10.1007/s00299-018-2339-9
Yin, X., Fan, H., Chen, Y., Li, L. Z., Song, W., Fan, Y., et al. (2020). Integrative omic and transgenic analyses reveal the positive effect of ultraviolet-B irradiation on salvianolic acid biosynthesis through upregulation of SmNAC1. Plant J. 104, 781–799. doi: 10.1111/tpj.14952
Yruela, I. (2009). Copper in plants: acquisition, transport and interactions. Funct. Plant Biol. 36, 409–430. doi: 10.1071/FP08288
Zhang, H., Lv, S., Xu, H., Hou, D., Li, Y., and Wang, F. (2017). H2O2 is involved in the metallothionein-mediated rice tolerance to copper and cadmium toxicity. Int. J. Mol. Sci. 18:2083. doi: 10.3390/ijms18102083
Zhang, Y. A., Yan, Y. P., and Wang, Z. Z. (2010). The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza. J. Agric. Food Chem. 58, 12168–12175. doi: 10.1021/jf103203e
Zhao, S., Zhang, J., Tan, R., Yang, L., and Zheng, X. (2015). Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J. Exp. Bot. 66, 7211–7226. doi: 10.1093/jxb/erv418
Zheng, X., Chen, D., Chen, B., Liang, L., Huang, Z., Fan, W., et al. (2021). Insights into salvianolic acid B biosynthesis from chromosome-scale assembly of the Salvia bowleyana genome. J. Integr. Plant Biol. 63, 1309–1323. doi: 10.1111/jipb.13085
Zhou, W., Huang, Q., Wu, X., Zhou, Z., Ding, M., and Shi, M. (2017). Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep.7:10554. doi: 10.1038/s41598-017-10215-2
Zhou, W., Shi, M., Deng, C., Lu, S., Huang, F., Wang, Y., et al. (2021). The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic Res. 8:10. doi: 10.1038/s41438-020-00443-5
Zhou, Y., Sun, W., Chen, J., Tan, H., Xiao, Y., Li, Q., et al. (2016). SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6:22852. doi: 10.1038/srep22852
Keywords: copper ion treatment, metabolomics, transcriptomic, Salvia miltiorrhiza, salvianolic acids
Citation: Xiang Y, Wang X, Song W, Du J and Yin X (2021) Integrative Omics Analyses Reveal the Effects of Copper Ions on Salvianolic Acid Biosynthesis. Front. Plant Sci. 12:746117. doi: 10.3389/fpls.2021.746117
Received: 23 July 2021; Accepted: 21 September 2021;
Published: 21 October 2021.
Edited by:
Lei Zhang, Second Military Medical University, ChinaReviewed by:
Shujuan Zhao, Shanghai University of Traditional Chinese Medicine, ChinaZhichao Xu, Northeast Forestry University, China
Copyright © 2021 Xiang, Wang, Song, Du and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojian Yin, YWppYW4uNTE3QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Yaping Xiang
Yaping Xiang Xiaoxiao Wang
Xiaoxiao Wang Wei Song1†
Wei Song1† Xiaojian Yin
Xiaojian Yin