- 1Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Northeast Region), Ministry of Agriculture and Rural Affairs, Harbin, China
- 2College of Horticulture and Landscape Architecture, Northeast Agricultural University, Harbin, China
Heterosis is a common biological phenomenon in nature. It substantially contributes to the biomass yield and grain yield of plants. Moreover, this phenomenon results in high economic returns in agricultural production. However, the utilization of heterosis far exceeds the level of theoretical research on this phenomenon. In this review, the recent progress in research on heterosis in plants was reviewed from the aspects of classical genetics, parental genetic distance, quantitative trait loci, transcriptomes, proteomes, epigenetics (DNA methylation, histone modification, and small RNA), and hormone regulation. A regulatory network of various heterosis-related genes under the action of different regulatory factors was summarized. This review lays a foundation for the in-depth study of the molecular and physiological aspects of this phenomenon to promote its effects on increasing the yield of agricultural production.
Introduction
Heterosis is a common biological phenomenon in nature. It refers to the heterozygote produced by hybridization between two or more parents with different genetic bases. Hybrids are superior to parents in terms of yield, growth rate, viability, and disease resistance (Hochholdinger and Hoecker, 2007). In agricultural production, heterosis is widely utilized in animals, such as silkworms, and plants, such as rice, maize, vegetables, and some perennials (Vaillancourt et al., 1995; Arcade et al., 1996; Kopp et al., 2002; Marcelo et al., 2007; Costa et al., 2014; Chen et al., 2020; Wang et al., 2021; Yu et al., 2021; Zhang et al., 2021). The use of heterosis has gradually improved the yield, quality, and disease resistance of animals and plants, thereby enhancing the social and economic benefits of agricultural production. However, even after over 100 years of research, the mechanism by which heterosis forms remains obscure. Therefore, although heterosis is widely utilized, our theoretical understanding of this phenomenon is incomplete. Moreover, limited research on the genetic basis of heterosis restricts its further applications in agricultural production. After Darwin (1876) was the first to propose the idea of heterosis after he noted that the yield of hybrid generations produced by inbred maize hybrids was 25% higher than that of their parents. Since then, several hypotheses have been offered to explain its genetic basis. Heterosis is a complex physiological and biochemical genetic phenomenon that scientists have been trying to understand from different aspects, and have obtained some relevant progress, such as classical genetics, molecular genetics, epigenetics, and physiology. Here, the recent researches on plant heterosis were summarized from different aspects including classical genetics, parental genetic distance (GD), quantitative trait loci (QTL) transcriptomes, proteomes, epigenetics, hormone regulation and gene regulation, and can provide theoretical reference for improving plant yield in breeding in the future.
Plant Heterosis According to Classical Genetics
Prior to the advent of modern molecular biology and molecular genetics, the mechanism of heterosis was explained by three primary hypotheses based on classical genetics. The first is the dominant hypothesis, which emphasizes the dominant complementarity of favorable alleles in hybrid lines; this hypothesis states that the harmful genes of one parent can be covered by the favorable dominant genes of the other parent. This hypothesis was first proposed by Bruce (1910). Jones (1917) further improved this hypothesis and proposed the dominance of linked genes hypothesis. Analysis of the heterozygosity and detection of the QTL of 141 restriction fragment length polymorphisms revealed that the heterozygosity of rice heterosis is not related to the heterozygosity of its whole genome. Xiao et al. (1995) reported that the comprehensive performance of recombinant lines screened in F8 generations is better than that of inbred lines, further proving that the dominant effect of parental genes substantially affects rice heterosis. This observation supports the dominance hypothesis. Heterozygous fragments containing qSS7 and qHD8 exerted dominant effects that contributed to the heterosis of the hybrid rice variety “Liangyoupei9” (LYP9) (Lin T. et al., 2020). Large-scale genetic analysis of the offspring of three excellent maize hybrids and examination of the heterosis patterns of 628 related loci revealed that most loci have complete–incomplete dominant (main) or overdominant (secondary) effects on hybrid genotypes; moreover, the proportion of dominant alleles from two parental lines was almost equal, which was the main reason for the strong heterosis (Liu H. et al., 2019). The next hypotheses based on classical genetics is the superdominance hypothesis, which emphasizes that the interaction of heterozygous alleles in hybrid lines has a stronger growth heterosis than the parental homozygous alleles (Shull, 1908). A 100 years later, Krieger et al. (2010) and Guo et al. (2013) found the superdominant QTLs in tomato and cotton. Chen L. et al. (2018) analyzed the transcriptome of the development of young panicles of the rice variety WFYT025, and they argued that the superdominant effect may be the main reason for the heterosis its grain number. Differentially expressed genes at the same position as grain number QTL is considered a candidate gene that can provide valuable targets for cloning and functional analysis of these grain number QTLs. Through genome-wide comparative transcriptome analysis, Shahzad et al. (2020) found that the circadian rhythm pathway genes related to LATE ELONGATED HYPOCOTYL (LHY) and many root genes involved in peroxidase activity also show overdominant expression in hybrids. They added that the overdominance of gene expression levels plays a key role in the early biomass vigor of cotton hybrids. Tian M. et al. (2018) performed transcriptome analysis of F1 hybrids and found that the genes involved in nicotine synthesis and metabolism (ADC, PMT, MPO, QPT, AO, QS, QPT, A622, and BBLs) and nicotine-transport genes (JAT2, MATE1, MATE2, NUP1, and NUP2) are upregulated, indicating that the overdominant expression of nicotine-metabolism genes plays a key role in F1 heterosis. These results strongly proved that superdominance is an important genetic basis for heterosis. Another hypotheses based on classical genetics is the epistatic hypothesis. Powers (1944) argued that the interaction of non-alleles in hybrids at different loci is the core of heterosis; this assumption had been confirmed in the analysis of heterosis in maize and rice (Yu et al., 1997; Tang et al., 2010), especially in terms of grains per panicle and grain weight per panicle of rice (Li et al., 1997; Zhang et al., 2021). Furthermore, harmful alleles can repress other favorable QTLs by epistasis during floral transition in maize (Xiao et al., 2021). The aforementioned theories explain the genetic basis of heterosis from different angles. These hypotheses highlight the most essential genetic basis of heterosis that genetic heterogeneity is required among hybrid parents. However, the three models of dominance, superdominance, and epistasis are not mutually exclusive, and they are all related to plant heterosis. Furthermore, dividing heterosis into three independent proportions (i.e., the contribution of dominance, superdominance, or epistasis) is difficult because heterosis is a non-linear effect of multiple heterozygous gene combinations in agricultural production (Liu J. et al., 2020).
Parental Genetic Distance and Plant Heterosis
The genetic basis of phenotypic differences between hybrids and their parents originates from differences in genomic composition. With the rapid development of the fields of molecular genetics and functional genomics, genetic differences between hybrid parents and their relationship with heterosis can now be evaluated via molecular analysis to determine the molecular mechanism of heterosis. Genetic differences between hybrid parents can be expressed by GD, and the degree of heterosis is strongly correlated with the GD of both parents. In a broad sense, the degree of heterosis increases with the increase of GD (Usatov et al., 2014; Boeven et al., 2020), and the farther the GD between the parent inbred lines is, the more scattered the gene expression in the hybrids will be (Ma et al., 2006; Stupar and Springer, 2006; Swanson-Wagner et al., 2006; Zhang and Borevitz, 2009; Birchler and Veitia, 2010; He et al., 2010; Paschold et al., 2012). Studies on a variety of plants have shown that GD has a considerable correlation with heterosis. In rice, Jaikishan et al. (2010) found that compared with rice genome simple sequence repetition (SSR), expressed sequence tag (EST) derived SSR (EST-SSRs) can lead to a better correlation between GD and grain yield heterosis. In addition to the rice, Sudi et al. (2010) conducted molecular, morphological, and genealogical analyses, finding that the genetic diversity of wheat is moderately to substantially correlated with plant height. Furthermore, they noted that the genetic diversity of wheat according to morphological markers is moderately to considerably correlated with grain number per spike. Hao et al. (2015) used single nucleotide polymorphism (SNP) to evaluate the genetic diversity, indicating that the subpopulation structure of waxy corn and common maize populations is related to heterosis. On the basis of SSR markers related to yield, GD, mid-parent heterosis (MPH), and best-parent heterosis (BPH) have a consistent and substantial correlation with the yields of seed cotton and lint in the Alar area (Li X. et al., 2019).
However, the experimental results of many scholars are opposite to the above views. They believed that GD estimated by molecular markers is not significant correlated with heterosis, and could not be used to predict heterosis. For example, Arcade et al. (1996) found that there is no correlation between some quantitative traits of larch and GD based on random amplified polymorphic DNA (RAPD). Rajendran et al. (2014) noted that the GD in rice is no correlated with heterosis through SSR marker. Kawamura et al. (2015) found that the GD estimated by SSR and cleaved amplified polymorphic sequence (CAPS) markers is not correlated with the heterosis of Chinese cabbage. Chen et al. (2020) found that the correlation between GD and heterosis in eucalyptus is poor by EST-SSR markers. Similarly, some studies have shown that GD also does not predict heterosis levels in maize, wheat and melon (Dreisigacker et al., 2005; Maruthi et al., 2019; Asaf et al., 2021). But Tian et al. (2017) argued that the GD estimated using random SSR and sequence related amplified polymorphism (SRAP) primers is not correlated with the heterosis of Brassica napus, whereas the GD estimated by the F1 heterozygous marker is highly correlated with the number of pods, thousand-seed weight, and single-species yield of this plant.
In addition, it has been reported that GD is highly positive correlation with geographical distance in Linum austriacum (Sheidai et al., 2014), Trachypogon plumosus (Baruch et al., 2004), Sinocalycanthus chinensis (Li et al., 2012), and Erodium ciconium (Xi et al., 2021). And in rough fescue, GD estimated by RAPD markers is related to its geographical distance. Genetic diversity seems to increase from west to east in Alberta, and the populations farther east have greater adaptive adaptability (Zhao et al., 2008). In addition, it has highly positive correlation between GD and geographical distance when using SRAP markers to determine the heterosis of pepper. Further studies showed that when the GD is less than 0.4051, the yield heterosis increases with the increase of GD, and when the GD is greater than 0.4051, the yield heterosis decreases with the increase of GD (Wu et al., 2012). However, Upadhyay et al. (2011) found that geographical distance do not contribute to genetic divergence in dolichos beans. This suggests that predicting levels of heterosis needs to be done within a valid geographical range. Overall, when species diversity is more abundant, they are more able to extend their geographical range and adapt better to new environments.
In summary, although the calculation of GD based on molecular markers is widely used, it cannot be applied to predict heterosis strictly and accurately. However, with the development of high-throughput sequencing technology, the application of molecular markers will be more helpful to predict heterosis. In addition, it can be found that the potential of molecular markers to predict crossing performance may largely depend on plant materials, phenotype, prediction methods and planting environment. Unless the DNA markers used in the analysis are associated with the genes affecting relative traits, hybrid performance cannot be accurately predicted by GD. Restriction-fragment length polymorphism (RFLP), SSR and other random markers may not be associated with relative traits, while other molecular markers whose primers are designed according to target genes have relatively high correlation to relative traits. So the correlation between molecular markers and heterosis cannot be accurately described.
Quantitative Trait loci-Associated Heterosis in Plant
Various QTLs have been identified for general or specific combination ability in hybrids. In the research of rice, heterosis can be affected by genes that regulate spikelets, such as SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (SPL14) and Gnarley1 (Ashikari et al., 2005; Jiao et al., 2010; Miura et al., 2010). And Huang et al. (2016) detected different dominant loci in different types of hybrid rice, such as Heading date 3a (Hd3a) and TILLER ANGLE CONTRO 1 (TAC1) in the hybrid rice system of cytoplasmic male sterility (CMS) for female parental lines; LAX PANICLE 1 (LAX1) and Grain number, plant height and heading date 8 (Ghd8) in the two-line system of environmentally sensitive male sterility; and DENSE AND ERECT PANICLE 1 (DEP1), IDEAL PLANT ARCHITECTURE 1 (IPA1), and NARROW LEAF 1 (NAL1) in the system of the two populations from rice indica–japonica crosses. Li D. et al. (2016) re-sequenced the recombinant inbred lines population of the super hybrid rice LYP9. Combined with transcriptome analysis of early panicle development, they found the main heterosis locus RH8 and photoperiod-sensitive genes DAYS TO HEADING 8 (DTH8)/Ghd8/LATE HEADING DATE 1 (LHD1). Ghd8 is the primary gene for heterosis in most Indica–Indica hybrid systems (Huang et al., 2016) and LYP9 (Li D. et al., 2016). Another study showed that the heterosis loci on chromosomes 11 and 12 of rice have strong heterosis effects on biomass, panicle weight, grain yield, and other traits (Lin Z. et al., 2020). In addition to the study of heterosis in rice, a study that adopted different methods in different combinations and even different species showed that flowering-related genes are related to heterosis, such as the tomato flowering gene SINGLE FLOWER TRUSS contributes to the heterosis of tomato yield (Krieger et al., 2010). Li et al. (2011) found that the histone deacetylase genes in hybrid rice cause early flowering by regulating the expression of some non-additive genes and the key genes during flowering, such as Ghd7 and Hd3a. Wang et al. (2018a) examined the heterosis of four maize core-related traits in two experimental populations of chromosome segment substitution lines derived from Reid × TSPT. They identified 63 and 57 different QTL loci from the two populations, which provided the basis for fine mapping of heterosis loci in grain size. Sarfraz et al. (2018) focused on 32 heterosis QTLs related to cotton fiber quality traits. They found 96 unique favorable alleles that are chiefly related to fiber quality. Liu C. et al. (2019) reported that triploid loquat has a more remarkable heterosis than diploid and tetraploid loquat, and they found that the heterosis of triploid loquat is related to the clock genes TIMING OF CAB EXPRESSION 1 (TOC1), LHY, and GIGANTEA (GI). Liu Y. et al. (2020) detected six QTLs of maize kernel-related traits and the MPH of these traits. They found that the heterosis of grain size and the genetic mechanism of kernel length, kernel width, kernel thickness, and hundred-kernel weight are not completely independent. In summary, QTLs related to heterosis have a complex regulatory network, and environment, genetic background and ploidy can impact on this regulatory network. This is also one of the difficulties in heterosis research.
Transcriptomics and Proteomics Reveal Plant Heterosis
Different genes are involved in the formation of heterosis in different tissues and developmental stages. In turn, these heterosis-related genes are involved in transcription, translation, cell division, transportation, signal transduction, defense and stress response, biological regulation, development, energy metabolism, protein metabolism, amino acid metabolism, biosynthesis of secondary metabolites, photosynthesis, carbon fixation, chlorophyll synthesis, carbohydrate metabolism, photorespiration, nitrogen absorption, and cofactor and vitamin metabolism (Bao et al., 2005; Lai et al., 2006; Meyer et al., 2007; Song et al., 2007, 2010; Zhang et al., 2008, 2012; Li et al., 2009; Fujimoto et al., 2012). Liu et al. (2021) determined that the growth heterosis of hybrids is determined by the combined ability of cell division and photosynthesis, and the early development of hybrid leaves might enhance the growth heterosis of hybrids. In the past century, some scholars proposed a multiple gene model with complementary alleles and gene expression variations, all of which may be an important factor that leads to heterosis. With the emergence of DNA microarray, QTL analysis, allele expression analysis, real-time quantitative single nucleotide site typing detection of allele fluorescence, and high-throughput sequencing technology, differences in allele expression can now be detected. Differential gene expression may be a reflection of differential gene expression in parents, differential expression of primary and secondary metabolic genes, and changes in metabolic profiles. The research of Yi et al. (2020) indicated that heterosis may related to changes in primary and secondary metabolic balance.
Preliminary studies on hybrid rice have shown that the polymorphism of differentially expressed transcription factors and promoter elements are two important factors for rice heterosis (Zhang et al., 2008). Shao et al. (2019) observed that allele-specific expression (ASE) genes were significantly enriched in the genomic regions of differential selection in the process of rice breeding, indicating that ASE is strongly affected by the expression level of the parent genes. In addition, the study on the relationship between transcriptomics and heterosis suggested that the additive and non-additive expression of genes are the main patterns that constitute the difference in gene expression between hybrids and parents. In maize hybrids, Paschold et al. (2012) found that 10% of the genes are non-additively expressed, and 14% of the genes are ASE. Li et al. (2017) examined the heterosis of traits related to panicle weight by using excellent maize inbred lines. They proved that combinations of heterosis loci depend on the genotype. To identify heterosis-related genes in Brassica species, Yi et al. (2017) used the Br300K microarray to perform transcriptome analysis at three developmental stages in non-heading Chinese cabbage. They observed that numerous genes were differentially expressed in F1 hybrids, with prominent non-additive expression. The genes specifically expressed in the three stages of F1 hybrids were some unidentified genes specific to Brassica and several genes related to defense. Zhang et al. (2016) found that the chromatin remodeler DECREASE IN DNA METHYLATION 1 (DDM1) affects the heterosis of Arabidopsis by regulating salicylic acid metabolism, and this gene can promote the non-additive expression of related genes. However, transcriptome analysis of hybrid broccoli revealed that the entire gene expression profile of the hybrids and their parents are similar, and only a few genes showed significant differential expression levels in the hybrids and their parents (Li et al., 2018). Guo et al. (2010) found no significant correlation between maize heterosis and the frequency of non-additive expression but observed a positive correlation between maize heterosis and the proportion of additive expression. These findings were supported by the results of Meyer et al. (2012) on Arabidopsis Different selections among different subgroups form different heterosis alleles. The male and female parents of hybrid rice have genetic variations. When the female genome is introduced from other subspecies and shows a high level, allele differences between the male and the female parents at the heterosis loci will result in the formation of heterosis loci in hybrid rice (Lin Z. et al., 2020).
Gene expression in hybrids is affected by cis-acting elements, trans-acting factors, and their interaction (Birchler and Veitia, 2010). Cis-regulatory elements are short DNA sequences containing trans-factor-specific binding sites that are used to control the expression of their associated genes (Bao et al., 2019; Luo et al., 2021). It can lead to changes in gene expression, and in hybrids, different regulatory factors and transcriptional networks recombine (Birchler et al., 2003; Riddle and Birchler, 2003; Zhang et al., 2008; Bougas et al., 2010). ASE is closely related to the parental origin effect and regulated by the complex interaction of cis- and trans-acting factors, a feature considered a key factor to differences between hybrids and their parents (Botet and Keurentjes, 2020). The genome study of Zhang and Borevitz (2009) revealed that over 40% of DEGs in parents showed ASE in hybrids, indicating that cis-regulatory variations have an effect on hybrid loci. Liu et al. (2018b) concluded that the differences in gene expression between parental alleles in triploid loquat are largely due to cis-regulatory variations. Li et al. (2021) demonstrated that trans-regulatory factors have a greater influence on differences in parental expression than cis-regulatory factors. In hybrids, genes can appear as homozygous or heterozygous pairs. Homozygous genes may be differentially expressed in different genetic backgrounds, indicating that transregulated variation has an additive effect on phenotypes. By comparison, different alleles of heterozygous genes can be differentially expressed in the same genetic background, indicating that cis-regulation or ASE mutation occurs (Metzger et al., 2016).
At the gene expression level, many recent studies have shown that heterosis can be revealed not only at the transcriptional level but also at the proteome level. Proteomics analysis indicated that indole-3-acetic acid (IAA) content was positively correlated with the length of the eighth internode of maize, but negatively correlated with the extent of the heterosis of the length of the eighth internode (Chen Y. et al., 2018). Analysis of the proteomics of popcorn hybrid combinations revealed that 22 kinds of biological processes are related to non-additive proteins. The heterosis of the popcorn hybrid at the early stage of plant development is related to the upregulation of protein synthesis and energy metabolism (Rockenbach et al., 2018). Marcon et al. (2013) scrutinized the proteome of maize seminal roots to identify the molecular basis of development vitality of hybrid seedlings. They detected 85 proteins that non-additively accumulate in at least one hybrid, and the result indicated that the increase of hybrid protein synthesis rate might be related to the early performance of hybrid vigor in seminal roots. Wang et al. (2021) identified over 2000 proteomes from maize hybrids and their parents’ seedling leaves via label-free quantification. Moreover, they verified four stress-related proteins and eight photosynthetic-related proteins via parallel reaction monitoring. Among these proteins, 10 were substantially different from the mid-parent’s proteins. Consistent with changes in the gene expression of hybrids and allopolyploids, both additive and non-additive proteomic patterns have been found in embryos (Marcon et al., 2010), roots (Hoecker et al., 2008), and spikes (Dahal et al., 2012) of maize hybrids; mature embryos of rice hybrids (Wang et al., 2008), and leaves of Arabidopsis autopolyploids and allopolyploids (Ng et al., 2012). Similar non-additive proteomes have also been found in the embryos and roots of maize hybrids and embryos of rice hybrids (Hoecker et al., 2008; Marcon et al., 2010), indicating that the hybrids of different plants have common regulatory changes.
In summary, the molecular mechanism of plant heterosis is mainly related to the gene expression in the growth and development-related pathways, involving photosynthesis, transportation, nutrition, resistance and epigenetic network. A number of studies at the transcriptome level have shown that the differential expression of genes, gene additive and non-additive expression between hybrid progeny and parents are the most critical factors to explain the heterosis mechanism. In the proteome comparison between F1 and its parents, most of the differentially expressed proteins are non-additive, and the proteins are the performers and executors of gene functions. Therefore, proteomics is a necessary supplement and effective proof of transcriptomics. However, neither transcriptomics nor proteomics can fully explain the mechanism of heterosis. It is necessary to combine multi-omics research and make a comprehensive analyze from multiple levels to reveal the molecular genetic mechanism of plant heterosis.
Epigenetics and Plant Heterosis
Given that gene expression is affected by DNA methylation, histone modification and non-coding RNA, breeders have adopted epigenetic breeding. Epigenetics refers to a phenotypic variation that is not caused by changes in gene functions due to changes in DNA sequences. Epigenetic regulation can regulate plant gene expression from three levels, namely, DNA methylation, histone modification, and non-coding RNA, all of which are closely related to the formation of heterosis. Epigenetic factors are among the key factors that determine hybrid performance. The heterosis of Arabidopsis hybrids can be directly or indirectly triggered by epigenetic differences between parental lines without being affected by genetic changes (Lauss et al., 2018; Rockenbach et al., 2018). Kawanabe et al. (2016) found that the heterosis of Arabidopsis is reduced by the knockout of METHYLTRANSFERASE 1, and the reduction of heterosis is highly correlated with the decrease in methylation levels of some important genes.
DNA Methylation and Heterosis
The methylation level of DNA in hybrids is remarkably different from that of their parents, especially when the methylation level of the epiallele region of the parents is considerably different, as it often causes substantial changes in the methylation level of the hybrids. Homozygosity or heterozygosity of methylated DNA may be involved in regulating inbreeding depression or heterosis (Nakamura and Hosaka, 2010). On the basis of the sequences of different ecotypes of Arabidopsis hybrids and their parents, Shen et al. (2012) found that the methylation levels of hybrids generally increase, and most of them occur in regions with different parental methylation levels. They proposed that the decrease in methylation levels in hybrids may affect growth vigor. Identically, differential DNA methylation affects gene regulation and heterosis phenotypes (Liu et al., 2018a). Analysis of the heterosis of broccoli bulb yield revealed that the DNA methylation rate of hybrids is higher than that of their parents, and the loci with different methylation levels are dominant in intergenic regions (Li et al., 2018). Moreover, the degree of DNA methylation of hybrids is proportional to the degree of genetic relationship between parents. The farther the genetic relationship between parents is, the greater the change in DNA methylation degree of F1 will be (Kawanabe et al., 2016). To study epigenetic modification and its relationship with gene expression (Chodavarapu et al., 2012), sequenced the Japonica rice Nipponbare and the Indica rice 9311 and their hybrid F1. They found that 7.48% of cytosine methylation levels between Nipponbare and 9311 were different, whereas the difference between parents and F1 was only 0.79%, suggesting that the heterosis of F1 is related to its DNA methylation level. Xiong et al. (1999) found that the methylation level of rice hybrid F1 (18%) is higher than the average methylation level of its parents (16.3%). Some methylation variations change the transcription level, which play a role in the increase in biomass heterosis (Greaves et al., 2012). Wang et al. (2018b) analyzed soybean hybrids by using MSAP and found that the increase in node number is promoted by hypomethylation, and the stem diameter of hybrids can be increased by hypermethylation. More interestingly, the methylation level of alleles from both parents will change when the DNA methylation level greatly varies. For example, the hybridization of Col-0 ecotype Arabidopsis with low tRNA ADENOSINE DEAMINASE 3 (TAD3) methylation level and Nok-1 ecotype Arabidopsis with high TAD3 methylation level showed that the methylation level of TAD3 from Col-0 increased and that of TAD3 from Nok-1 decreased in F1 (Astrid et al., 2017).
Histone Modification and Plant Heterosis
Histone modifications include acetylation, methylation, ubiquitination, phosphorylation, glycosylation, and carbonylation. Studies on acetylation and methylation are very thorough because they are closely related to gene expression regulation. The acetylation of histone lysine is established by histone acetyltransferases and eliminated by histone deacetylases, which is usually related to gene activation. Methylation is established by histone lysine methyltransferase and removed by histone demethylase, and it is associated with transcriptional activation or inhibition (Berger, 2007; Kouzarides, 2007; Shahbazian and Grunstein, 2007; Liu et al., 2010; He et al., 2011). Histone lysine methylation is different in terms of the number of methyl groups, such as monomethyl, dimethyl, or trimethyl lysine. The heterosis of Arabidopsis is related to the degree of histone modification enriched in the promoter regions of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LHY. CCA1 and LHY are two genes that can regulate the circadian clock process. CCA1 and LHY, as well as their regulators TOC1 and GI, are positively correlated with the levels of histone H3-Lys 9 acetylation (H3K9ac) and histone H3-Lys 4 dimethylation (H3K4me2). Changes in the degree of H3K9ac and H3K4me2 modification can directly lead to the decrease in CCA and LHY expression, promoting the expression of downstream genes that control photosynthesis and starch metabolism, thereby increasing the photosynthetic efficiency, starch accumulation, and growth advantage of F1 (Ni et al., 2009). CCA1 is also a key gene in plant disease resistance heterosis. When pathogens attack, the expression of CCA1 in F1 hybrids is accurately regulated by its rhythmic histone modification at different time points during the day (Yang et al., 2021). The expression level of TOC1 is related to the time regulation of histone H3 acetylation. The expression peak of TOC1-luciferase report can be induced by the inhibition of histone deacetylases by trichostatin A (Perales and Mas, 2007). Li et al. (2011) established that histone modification plays a role in the pattern of changes in non-additive expression in hybrid rice by overexpression and inactivation of histone deacetylase coding genes in hybrid rice. Histone modification is allele-specific in hybrids. For example, in rice hybrid, ASE is primarily regulated by allele-specific histone modifications–histone H3 lysine 36 rather than histone H3 lysine 27 (H3K27me3) (Guo et al., 2015). Parent specificity and tissue specificity are also observed in hybrids. For example, parent-specific differences in H3K27me3 have been detected in Arabidopsis endosperm, especially in transposons (Moreno-Romero et al., 2016). However, the activity of panicle primordia meristem in rice is directly related to panicle development and grain yield. In Arabidopsis allotetraploids, most non-additively expressed genes (Wang et al., 2006), including circadian clock genes (Ni et al., 2009), are related to high levels of H3K9ac and H3K4me3 (Ha et al., 2011).
Small RNA and Plant Heterosis
Aside from DNA methylation and histone modification, small RNA, including small interfering RNAs (siRNAs), microRNAs (miRNAs), and trans-acting ta-siRNAs, is also an important part of epigenetics. HUA ENHANCER 1 (HEN1) is a gene-encoding RNA methyltransferase and a key factor in the biological occurrence of small RNAs in plants. The production of functional small RNA can be inhibited by the mutations of HEN1, and it will inhibit the growth vigor of Arabidopsis hybird (Shen et al., 2012). Shen et al. (2017) found that the non-additive effect of miRNA in hybrid offspring can affect the expression of target genes and regulate heterosis. And this view is supported in the research of maize and rice (Zhang et al., 2014; Zhao et al., 2015). Chen et al. (2010) conducted miRNA microarray to compare the expression levels of miRNA and the high abundance small RNAs in rice hybrid combinations. They confirmed that miRNAs are involved in regulating gene differential expression in hybrids. He et al. (2010) further found that the differential expression of miRNAs is negatively correlated with the differential expression of its target genes between the hybrid rice and its parents. While Crisp et al. (2020) suggested that hybridization affect the expression of small RNAs at specific sites, and small RNAs show more variations between inbred lines compared with gene expression variations. In addition, Hu et al. (2021) stated that 32.5% of differential alternative splicing contains or lacks at least one annotated binding site of maize miRNA in the maize hybrid and parents. They suggested that it may be involved in miRNA-mediated post-transcriptional regulation. The upregulated expression of miR156, miR159, and miR319 in hybrid offspring can regulate the target genes SPL, MYB, and TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP), suggesting that they can change the plant morphology of B. napus to adapt to the environment. The accumulation of MYB mRNA can be reduced by the overexpression of miR159 and may result in male sterility, whereas the overexpression of miR159-resistant MYB33 results in leaf curling up, dwarfing, and petiole shortening (Alonso-Peral et al., 2010). The developmental processes of leaf size, leaf shape, and flower symmetry are guided by TCP transcription factors (Danisman et al., 2012). Overexpression of miR319 results in inconsistent leaf shape and delayed flowering, whereas TCP mRNA can be specifically downregulated by miR319 (Shen et al., 2017). Overexpression of miR408a improves leaf area, petiole length, plant height, flower size and silique length in Arabidopsis, as well as increases photosynthetic rate in hybrid maize, resulting in high biomass and seed yield (Hou et al., 2020). miR164, miR166, miR167, and miR390 have been shown to target ARF genes and are involved in the IAA response pathway (Mallory et al., 2005). These miRNAs are highly conserved in plant species and their targets in B. napus (Shen et al., 2015). The sequencing results of a small RNA library of 21 maize inbred lines showed that variations in parental small RNA expression are negatively correlated with the heterosis of grain yield (Seifert et al., 2018). The expression of Pol IV-dependent siRNA is also negatively correlated with the expression of a group of AGMADS-like (AGL) genes, which encode type I MADS-box transcription factors and are expressed in endosperm and involved in regulating seed size (Lu et al., 2012; Chen, 2013).
In conclusion, these epigenetic modifications are of great significance to explain heterosis, especially DNA methylation, small RNA and histone modification. Generally speaking, the methylation level of hybrids with heterosis is higher than the average methylation level of parents, but not all high levels of methylation have a role in heterosis. Heterosis has a strong correlation with the change of methylation level and pattern of specific sites. The differential expression of small RNAs in hybrids and their regulatory effects on target genes may affect the biomass and grain yield of hybrids to varying degrees. Some histones found in hybrids may be related to heterosis, such as H3K9ac, H3K4me2, and H3K4me3. And different types of histone modification can regulate genes related to photosynthesis, circadian rhythm and metabolic level, leading to the heterosis in hybrids. However, due to the large number of histones and the variety of their modification methods, the regulatory mechanism between histones and heterosis needs further research.
Hormone Regulation and Plant Heterosis
DEG-targeted hormone-mediated signaling pathways mainly involve abscisic acid, jasmonic acid, salicylic acid, brassinosteroids, and auxin. These hormones have been shown to play an important role in heterosis formation (Li Y. et al., 2016; Zhang et al., 2016; Hu et al., 2017). Pathways involved in various biological and abiotic stresses have also been confirmed to participate in heterosis (Groszmann et al., 2015; Miller et al., 2015; Wang et al., 2015). Ethylene is a plant hormone that promotes fruit maturation but inhibits hypocotyl elongation. The application of exogenous ethylene can eliminate the biomass activity of Arabidopsis F1 hybrids (Song et al., 2018), and it plays a negative role in heterosis and salicylic acid can inhibit ETHYLENE RESPONSE FACTORS (Shen et al., 2017). The monosaccharide transporter gene is downregulated by the overexpression of the ethylene receptor gene ETR2 in rice, thereby preventing the sugar from transferring from stem to grains, resulting in the reduction of grain weight (Wuriyanghan et al., 2009). Consistent with these studies, Katara et al. (2020) also observed the downregulation of the ethylene receptor gene ETR in two hybrid sterile lines. In WFYT025 hybrid rice, regulatory changes in gibberellin and abscisic acid biosynthetic genes can lead to heterosis, and it might have a stronger gibberellin biosynthesis potential than its parents, thereby promoting the increase in spikelet primordium number of the hybrid WFYT025. Moreover, the phosphorylation of SNF1-related protein kinase 2 is necessary for the kinase activity of downstream targets, which is related to abscisic acid biosynthesis (Chen L. et al., 2018). Smith (2019) noted that a decrease in the salicylic acid level in hybrids with abnormally high salicylic acid level would promote the hybrid’s growth. By contrast, the salicylic acid-regulated defense genes of hybrids that show heterosis are downregulated. Both Arabidopsis C24/Ler F1 hybrids and C24 bacterial degradative enzyme salicylate 1 hydroxylase may promote changes in the main regulatory factor TL1 BINDING TRANSCRIPTION FACTOR 1 (TBF1) for defense and growth by regulating the level of salicylic acid, thereby promoting plant growth (Gonzalez-Bayon et al., 2019). Hydrogen peroxide (H2O2) is also an important signal molecule that can interact with different plant hormone signaling pathways, such as abscisic acid, salicylic acid, jasmonic acid, auxin, and brassinosteroids; regulate plant development and stress response (Saxena et al., 2016; Yuan et al., 2017); and induce the oxidation of BRASSINAZOLE-RESISTANT 1 (BZR1) transcription factor, a major regulator of the brassinosteroid signaling pathway. H2O2 oxidation of BZR1 enhances its interaction with PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and AUXIN RESPONSE FACTOR 6 (ARF6) (Tian Y. et al., 2018). The upregulation of the auxin biosynthesis gene YUCCA (YUC) can increase the level of IAA, which in turn can be activated by the transcription factor PIF4 (Sun et al., 2012, 2013; Wang et al., 2017). The yield heterosis for improving the plant structure of B. napus can be enhanced by the degradation mutation of the potential gene BnaA3.IAA7 that encodes auxin/indole acetic acid proteins (Li H. et al., 2019). TCP4 directly activate YUC5 transcription, all of which integrate organ morphogenesis with auxin and brassinosteroid reactions that promote hypocotyl cell elongation. In addition, TCP4 requires brassinosteroid response to promote hypocotyl growth (Reddy et al., 2016). These studies confirmed the contribution of epigenetic regulation of hormone signals and gene expression to heterosis. However, hormone level is only a part of plant growth and heterosis.
As we all know, plant growth is inseparable from hormones. In recent years, great progress has been made in the study of plant hormones involved in plant growth and development, including gibberellin promoting plant height, abscisic acid promoting seed germination, and the interaction between hormones leading to the changes of reproductive organ number and grain weight. At the same time, these traits show relatively obvious heterosis, but hormone regulation is effective in a certain range. If the hormone level is too high, it will have a negative effect. Therefore, the differences in metabolism and regulation of plant hormones between parents and hybrid progenies may become a research hotspot in the future, which will be conducive to further analysis of the mechanism of plant heterosis.
Regulation Network of Biomass Heterosis and Grain Heterosis
Yield is the top priority of plant production. Breeders pursue different yields for different plants. In some plants, they focus more on biomass yields, such as leafy vegetables and graminaceous crops. Some pursue root and tuber yield, such as potato and sweet potato. However, people may pursue different yields within the same crop, such as grain yield for conventional maize and biomass yield for forage silage maize.
Cell division and photosynthesis of hybrid plants are important components of growth vigor (Liu et al., 2021). Leaf growth in early development is the key to biomass heterosis in Arabidopsis. However, heterosis is not caused by the increase in photosynthetic efficiency per leaf area but by the increase in total leaf area and total photosynthesis per plant (Liu P. et al., 2020). In addition to photosynthesis and sucrose and starch pathways, oxidative phosphorylation and the tricarboxylic acid cycle may also play a role in heterosis (Yao et al., 2005). The yield and biomass heterosis of hybrids may be due to altered expression patterns of genes that control biological functions (such as carbon fixation, sugar metabolism and circadian rhythm) (Chen, 2013). Dry matter yield, which represents the biomass of barley, is closely related to grain yield. Grain yield is mainly determined by additive genetic effect (Zhang et al., 2015), while dry matter yield is affected by both additive and non-additive genetic components (Madić et al., 2014). Singh et al. (2013) found that dry matter yield could also be affected by general combining ability and special combining ability. And Mühleisen et al. (2013) found that special combining ability has a significant effect on grain yield. Similarly, in cultivated tomato, biomass and yield are correlated in terms of heterosis (Semel et al., 2006). In addition, ploidy effect is also related to biomass and grain yield heterosis. Studies have shown that the effect of ploidy on plant size seems to have a dosage compensation mechanism (Miller et al., 2012). In maize, plant size increases with increasing ploidy (from haploid to triploid), but decreased in tetraploid (Riddle et al., 2006). In the study of Arabidopsis, the effect of ploidy on biomass is not obvious, but the increase of ploidy level is positively correlated with seed size and weight (Miller et al., 2012).
Rice, which represents graminaceous plants, show substantial heterosis in economic yield, especially in grain yield. In plant breeding, the most effective study of heterosis has been conducted in rice. Rice yield and quality have been greatly improved. In 2017, the yield of the super hybrid rice reached the peak of 1149 kg/666.67 m2 (Li et al., 2020). The grain yield heterosis of rice is controlled by Hd3a; TAC1 of CMS lines; LAX1 and Ghd8 of the environment-sensitive Indica hybrid; and Sd-1, GW6a, DEP1, IPA1, NAL1, and N11q25 of the Indica × Japonica rice. Ghd8 is the main heterosis gene in most Indica–Indica hybrid rice systems and also the primary heterosis gene in the yield of LYP9 (Huang et al., 2016; Li D. et al., 2016). Early flowering of hybrid rice can be caused by the key genes Ghd7 and Hd3a (Li et al., 2011), whereas Ghd7 encode a CCT (CO, CO-like, and TOC1) domain protein, and they play a key QTL role in controlling rice yield, plant height, and heading date, Ghd7 is also expressed under long-day conditions, inhibiting the expression of Early heading date 1 (Ehd1), thereby inhibiting long-day flowering (Xue et al., 2008). Overall, rice grain yield is most affected by the flowering time genes SOC1, Ghd8, and Ghd7 (Tadege et al., 2003; Xue et al., 2008; Yan et al., 2011). Heading date 1 (Hd1), CONSTANS (CO) homolog of Arabidopsis in rice, activates Hd3a [a homolog of FLOWERING LOCUS T (FT)] under short-day conditions and inhibits Hd3a under long-day conditions (Yano et al., 2000; Hayama and Coupland, 2004). Ehd1 can up-regulate Hd3a expression and promote short-day-dependent flowering in rice, which is inhibited by GI. Furthermore, Ehd1 can activate RICE FLOWERING LOCUS T1. In rice, overexpression of the histone deacetylase gene HDT1 can repress the non-additive expression of Hd1 and GI in the hybrid (Komiya et al., 2009; Li et al., 2011; Doi et al., 2016). A highly conserved R2R3 MYB domain transcription factor family is encoded by the gibberellin- and abscisic acid-regulated MYB (GAMYB) or GAMYB-like genes, in which the expression of GAMYB is induced by gibberellic acid (Woodger et al., 2003; Aya et al., 2009; Murray et al., 2010). Moreover, GAMYB is negatively regulated by the miRNA family miR159 (Tsuji et al., 2006; Alonso-Peral et al., 2010).
Heterosis in biomass yield is represented by the model plant Arabidopsis. Previous studies on Arabidopsis heterosis revealed potential heterosis phenotype candidate genes in circadian clock, flavonoid biosynthesis, auxin transport, salicylic acid metabolism, and response pathways (Ni et al., 2009; Shen et al., 2012; Groszmann et al., 2015; Zhang et al., 2016; Lauss et al., 2018). Early seedling biomass heterosis is related to the earlier expression in photosynthetic pathway relative to the parents, and high IAA levels may be attributed to the early photosynthesis of hybrid seedlings (Zhu et al., 2020). The transcription factor PIF4 also plays an important role in heterosis (Wang et al., 2017). DDM1 affects Arabidopsis heterosis in seedlings by regulating salicylic acid metabolism (Zhang et al., 2016). The metabolic level of salicylic acid can promote changes in the main regulatory factor TBF1 of defense and growth, thereby promoting plant growth (Gonzalez-Bayon et al., 2019). The changes in the expression of time gene CCA1 can enhance carbon and starch accumulation and promote the formation of heterosis in biomass (Ko et al., 2016). Epigenetic activation of CCA1 can also promote salicylic acid accumulation in hybrids; thus, heterosis works in defense (Yang et al., 2021). CCA1 and LHY regulate the biomass clock process, and their regulators TOC1 and GI are positively correlated with the levels of H3K9ac and H3K4me2 (Ni et al., 2009). In the morning-phased loop, LHY and CCA1 activate the expression of Pseudo-response Regulator 7 (PRR7) and PRR9 genes, which maintain period length and amplitude in plants (Shahzad et al., 2020). Downregulation of CCA1 during the daytime indirectly inhibits the expression of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) in hybrids to reduce ethylene production. PIF4 and PIF5 usually activate the expression of ACS at night (Song et al., 2018). The transcription factors LEAFY (LFY) is upregulated by FT through SOC1 in Arabidopsis (Das et al., 2019). LFY and FUL are directly activated by the miRNA-targeted transcription factor SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (Yamaguchi et al., 2009). GAMYB transcription induces the expression of LFY (Achard et al., 2004). In addition, in the study of miRNA, MiR160 is complementary to AUXIN RESPONSE FACTOR 10 (ARF10), and miR167 is complementary to ARF6 and ARF8 (Rhoades et al., 2002).
Heterosis is a complex biomass phenomenon. Numerous studies have shown that the strength and formation mechanism of heterosis of different biological species, different varieties of the same crop, and different traits may be different. The key to heterosis formation lies in the genetic differences between parents. The genomic sequence information of hybrids from parents does not change, and the phenotype and protein and gene expression substantially change. The orderly expression of many genes under the action of various regulatory factors forms a network system that affects the heterosis of grain yield and biomass yield by regulating different ways, as shown in Figure 1.
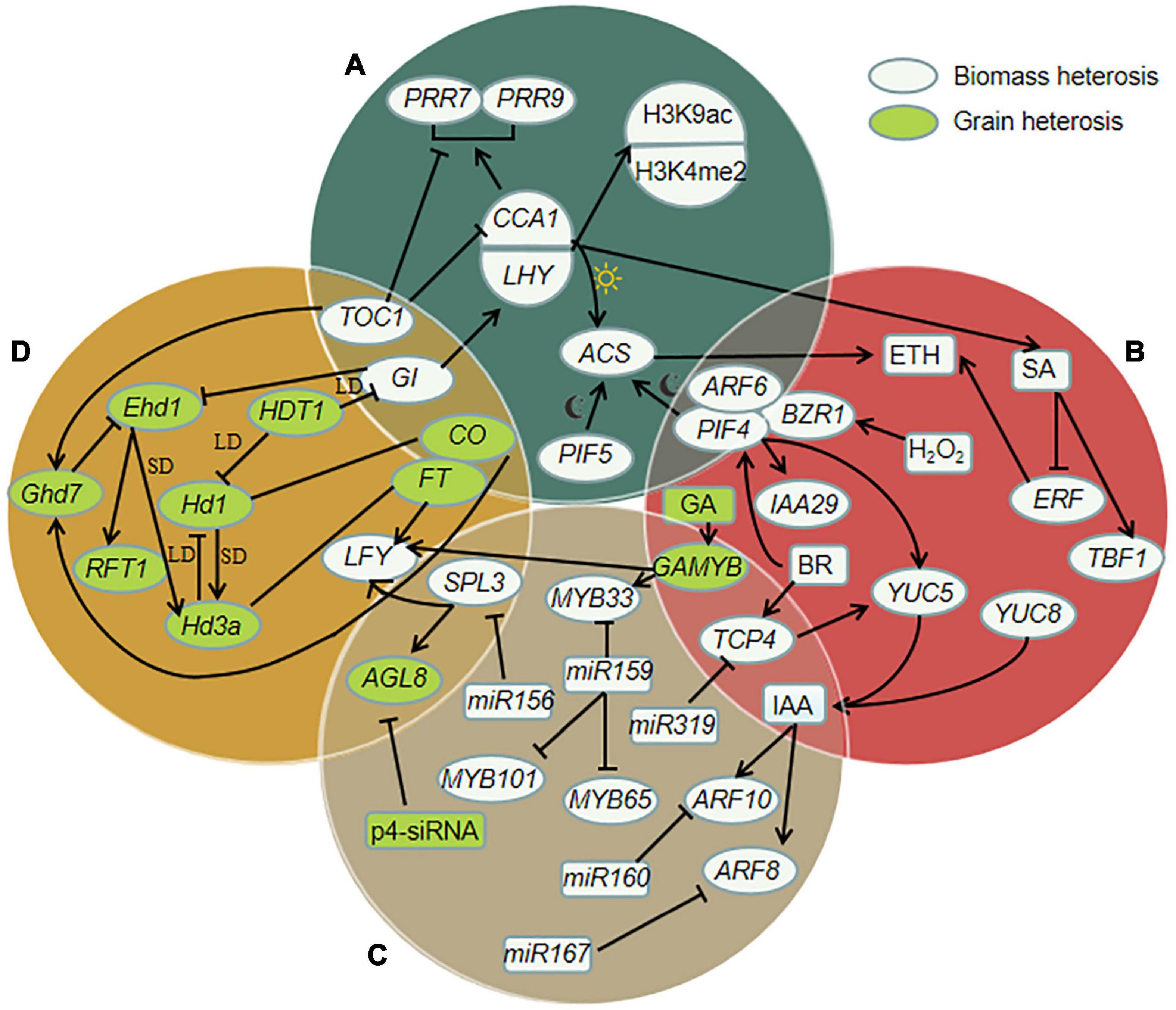
Figure 1. Schematic diagram of molecular regulation in biomass and grain heterosis. Solid arrows represent positive regulation, inhibitory arrows represent negative regulation; straight lines represent homology. (A) Heterosis regulation by photoperiod rhythm related genes. CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATEDD HYPOCOTYL (LHY) and their regulators TIMING OF CAB EXPRESSION 1 (TOC1) and GIGANTEA (GI) are positively correlated with histone H3-Lys 9 acetylation (H3K9ac) and histone H3-Lys 4 dimethylation (H3K4me2). LHY and CCA1 can activate the expression of PSEUDO-RESPONSE REGULATOR 7 (PRR7) and PRR9, and TOC1 can inhibit the expression of PRR7 and PRR9. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5 can often activate expression of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) (ai night), down-regulation of CCA1 inhibites the expression of ACS (at daytime), and ACS can promote the increase of ethylene. (B) Regulation of heterosis involving hormones and related genes. CCA1 can promote the increase of salicylic acid (SA) in hybrids. SA promotes the expression of TL1 BINDING TRANSCRIPTION FACTOR 1 (TBF1) and inhibites the expression of ETHYLENE RESPONSE FACTORS (ERF). Hydrogen peroxide (H2O2) and brassinosteroid (BR) enhance the interaction of BRASSINAZOLE-RESISTANT 1 (BZR1) with PIF4 and AUXIN RESPONSE FACTOR 6 (ARF6). PIF4 can activate the expression of YUCCA8 (YUC8) and IAA29, and the up-regulation of YUC genes leads to the increase of IAA level. TEOSINTE BRANCHED 1, CYCLODEA, PROLIFERATING CELL FACTORS 4 (TCP4) directly activates the expression of YUC5, and the reaction of BR is helpful for the expression of TCP4. The expression of gibberellin- and abscisic acid-regulated MYB (GAMYB) is induced by GA and negatively regulated by miR159, and GAMYB can promote the expression of LEAFY (LFY) and MYB33. (C) Regulation of heterosis involving small RNA and related genes. Overexpression of microRNA159 (miR159) can inhibit MYB33, MYB65, MYB101. MiR319 can inhibit TCP4. MiR160 targets ARF10, miR167 targets ARF8 genes, and they are regulated by IAA. The expression of Pol IV-dependent siRNAs (p4-siRNA) is correlated with the expression of FRUITFULL (AGL8, also known as FUL). And the expression of AGL8 is inhibited by SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3), SPL3 can be inhibited by miR156. (D) Heterosis regulation by flowering related genes. Grain number, plant height and heading date 7 (Ghd7) can be promoted by CONSTANS (CO) and TOC1. Heading date 1 (Hd1, CO homologous genes) and Early heading date 1 (Ehd1) up-regulate the expression of Heading date 3a (Hd3a, FLOWERING LOCUS T, FT homologous genes) under short-day (SD) conditions. Under long-day (LD) conditions, the expression of Ehd1 is inhibited by Ghd7, while Hd1 is the inhibitor of Hd3a, histone deacetylase gene HDT1 inhibites Hd1 and GI. Inhibition of GI can promote the expression of Ehd1. And the increase of Ehd1 induces the expression of RICE FLOWERING LOCUS T1 (RFT1). SPL3 and FT can promote the expression of LFY.
Concluding Remarks and Future Perspectives
In recent years, changes in gene expression levels and expression patterns between hybrids and parents have been compared via molecular biology and molecular genetics techniques. The DEGs obtained by these analyses are mainly focused on photosynthesis, carbohydrate metabolism, and energy metabolism. However, the molecular mechanism of heterosis formation cannot be explained well by these processes. Heterosis formation is a complex process that is affected by various factors, such as genes, environment, and the expression regulation of several genes related to physiological metabolism. Gene expression is characterized by spatiotemporal expression, but it is also affected by the surrounding environment. The material basis of heterosis is the hybridization of genotypes and not the simple embodiment of the overall heterozygosity between two parents. In terms of epigenetics, the issue of whether circular RNAs and long non-coding RNAs participate in heterosis remains unresolved. With the development of biological technology, an increasing number of multi-omics methods have been adopted to analyze problems in heterosis. Heterosis is a complex trait that is regulated by multiple genes. Therefore, genomics, transcriptomics, proteomics, metabolomics, phenomics, and ionomics can be possibly combined to analyze effective genes. Comprehensive analysis from multiple levels helps to better understand metabolic networks, gene functions, biochemical pathways and their correlations, and establish the interdependence between different cell components to better describe molecular phenomena. These results will provide support for revealing the molecular genetic mechanism of plant heterosis. In addition, the use of multi-omics combined with high-throughput tools can bring revolutionary changes to plant biology, because it provides real-time readings of hundreds of genes, proteins, metabolites and ions at different developmental stages and different environmental conditions, which is bound to promote the breakthrough in molecular theory of heterosis breeding. The agronomic traits of hybrids are better than those of parents. Hence, the selection of parental combinations is also a challenge, which may have general heterosis, MPH, and BPH. In the study of heterosis, finding the main QTLs or genes that regulate metabolism will be a hot topic. In addition, the rise of research on bacterial defense heterosis (Yang et al., 2021), stock heterosis (Asaf et al., 2021), sterile line gene editing systems, MiMe (Cas9) systems, and even new biotechnology approaches (Yu et al., 2021) has aroused the interest of researchers involved in heterosis research.
Author Contributions
XW wrote the manuscript and YL assisted the work. RG and YZ designed the review. RG was responsible for the revision of this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Foundation for the National Key Research and Development Program of China (2016YFD0101701), by Ministry of Science and Technology of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achard, P., Herr, A., Baulcombe, D. C., and Harberd, N. P. (2004). Modulation of floral development by a gibberellin-regulatedmicroRNA. Development 131, 3357–3365. doi: 10.1242/dev.01206
Alonso-Peral, M. M., Li, J., Li, Y., Allen, R. S., Schnippenkoetter, W., Ohms, S., et al. (2010). The MicroRNA159-Regulated GAMYB-like genes inhibit growth and promote programmed Cell Death in Arabidopsis. Plant Physiol. 154, 757–771. doi: 10.1104/PP.110.160630
Arcade, A., Faivre-Rampant, P., Le Guerrou, B., Pques, L. E., and Prat, D. (1996). Heterozygosity and hybrid performance in larch. Theor. Appl. Genet. 93, 1274–1281. doi: 10.1007/BF00223460
Asaf, D., Ilan, H., Elad, O., Tal, I., Galil, T., Ayala, M., et al. (2021). Underground heterosis for melons yield. J. Exp. Bot. doi: 10.1093/jxb/erab219 [Epub Online ahead of print].
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Astrid, A., Stéphanie, D., Elisa, F., Cécile, B., Isabelle, G., Matthieu, S., et al. (2017). An Arabidopsis Natural Epiallele Maintained by a Feed-Forward Silencing Loop between Histone and DNA. PLoS Genet. 13:e1006551. doi: 10.1371/journal.pgen.1006551
Aya, K., Ueguchi-Tanaka, M., Kondo, M., Hamada, K., Yano, K., Nishimura, M., et al. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21, 1453–1472. doi: 10.1105/TPC.108.062935
Bao, J., Lee, S., Chen, C., Zhang, X., Zhang, Y., Liu, S., et al. (2005). Serial Analysis of Gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol. 138, 1216–1231. doi: 10.1104/PP.105.060988
Bao, Y., Hu, G., Grover, C. E., Conover, J., Yuan, D., and Wendel, J. F. (2019). Unraveling cis and trans regulatory evolution during cotton domestication. Nat. Commun. 10:5399. doi: 10.1038/s41467-019-13386-w
Baruch, Z., Nassar, J. M., and Bubis, J. (2004). Quantitative trait, genetic, environmental, and geographical distances among populations of the C4 grass Trachypogon plumosus in Neotropical savannas. Divers. Distrib. 10, 283–292. doi: 10.1111/j.1366-9516.2004.00102.x
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Birchler, J. A., and Veitia, R. A. (2010). The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186, 54–62. doi: 10.1111/j.1469-8137.2009.03087.x
Birchler, J. A., Auger, D. L., and Riddle, N. C. (2003). In Search of the molecular basis of heterosis. Plant Cell 15, 2236–2239. doi: 10.1105/TPC.151030
Boeven, P. H. G., Zhao, Y., Thorwarth, P., Liu, F., Maurer, H. P., Gils, M., et al. (2020). Negative dominance and dominance-by-dominance epistatic effects reduce grain-yield heterosis in wide crosses in wheat. Sci. Adv. 6:eaay4897. doi: 10.1126/sciadv.aay4897
Botet, R., and Keurentjes, J. J. B. (2020). The Role of transcriptional regulation in hybrid vigor. Front. Plant Sci. 11:410. doi: 10.3389/fpls.2020.00410
Bougas, B., Granier, S., Audet, C., and Bernatchez, L. (2010). The Transcriptional Landscape of Cross-Specific Hybrids and Its Possible Link With Growth in Brook Charr (Salvelinus fontinalis Mitchill). Genetics 186, 97–107. doi: 10.1534/genetics.110.118158
Bruce, A. (1910). The mendelian theory of heredity and the augmentation of vigor. Science 32, 627–628. doi: 10.1126/science.32.827.627-a
Chen, F., He, G., He, H., Chen, W., Zhu, X., Liang, M., et al. (2010). Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J. Integr. Plant Biol. 52, 971–980. doi: 10.1111/j.1744-7909.2010.00985.x
Chen, L., Bian, J., Shi, S., Yu, J., Khanzada, H., Wassan, G. M., et al. (2018). Genetic analysis for the grain number heterosis of a super-hybrid rice WFYT025 combination using RNA-Seq. Rice 11, 1–13. doi: 10.1186/s12284-018-0229-y
Chen, S., Zhou, C., He, X., Weng, Q., Li, F., Li, M., et al. (2020). Enhanced correlations of EST-SSR-based genetic distance with hybrid performance, specific hybridizing ability, and heterosis using effect-increasing and effect-decreasing alleles: a case study in Eucalyptus L’Hér. Tree Genet. Genomes 16, 1274–1281. doi: 10.1007/s11295-019-1410-1
Chen, Y., Zhou, Q., Tian, R., Ma, Z., Zhao, X., Tang, J., et al. (2018). Proteomic analysis reveals that auxin homeostasis influences the eighth internode length heterosis in maize (Zea mays). Sci. Rep. 8, 60–66. doi: 10.1038/s41598-018-23874-6
Chen, Z. J. (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14, 471–482. doi: 10.1038/nrg3503
Chodavarapu, R. K., Feng, S., Ding, B., Simon, S. A., Lopez, D., Jia, Y., et al. (2012). Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. U. S. A. 109, 12040–12045. doi: 10.1073/pnas.1209297109
Costa, E. S. J., Potts, B. M., and Lopez, G. A. (2014). Heterosis may result in selection favouring the products of long-distance pollen dispersal in Eucalyptus. PLoS One 9:e93811. doi: 10.1371/journal.pone.0093811
Crisp, P. A., Hammond, R., Zhou, P., Vaillancourt, B., Lipzen, A., Daum, C., et al. (2020). Variation and Inheritance of Small RNAs in Maize Inbreds and F1 Hybrids. Plant Physiol. 182, 318–331. doi: 10.1104/pp.19.00817
Dahal, D., Mooney, B. P., and Newton, K. J. (2012). Specific changes in total and mitochondrial proteomes are associated with higher levels of heterosis in maize hybrids. Plant J. 72, 70–83. doi: 10.1111/j.1365-313X.2012.05056.x
Danisman, S., Van, D. W. F., Dhondt, S., Waites, R., De, F. S., Bimbo, A., et al. (2012). Arabidopsis Class I and Class II TCP Transcription Factors Regulate Jasmonic Acid Metabolism and Leaf Development Antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/PP.112.200303
Darwin, C. R. (1876). The Effects of Cross- and Self-fertilisation in the Vegetable Kingdom. London: John Murry.
Das, A., Geetha, G. A., Ravishankar, K. V., Shivashankara, K. S., Roy, T. K., and Dinesh, M. R. (2019). Interrelations of growth regulators, carbohydrates and expression of flowering genes (FT, LFY, AP1) in leaf and shoot apex of regular and alternate bearing mango (Mangifera indica L.) cultivars during flowering. Sci. Hortic. 253, 263–269. doi: 10.1016/j.scienta.2019.04.027
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2016). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Gene Dev. 18, 926–936. doi: 10.1101/gad.1189604
Dreisigacker, S., Melchinger, A. E., Zhang, P., Ammar, K., Flachenecker, C., Hoisington, D., et al. (2005). Hybrid performance and heterosis in spring bread wheat, and their relations to SSR-based genetic distances and coefficients of parentage. Euphytica 144, 51–59. doi: 10.1007/s10681-005-4053-2
Fujimoto, R., Taylor, J. M., Shirasawa, S., Peacock, W. J., and Dennis, E. S. (2012). Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. U. S. A. 109, 7109–7114. doi: 10.1073/pnas.1204464109
Gonzalez-Bayon, R., Shen, Y., Groszmann, M., Zhu, A., Wang, A., Allu, A. D., et al. (2019). Senescence and defense pathways contribute to heterosis. Plant Physiol. 180, 240–252. doi: 10.1104/pp.18.01205
Greaves, I. K., Groszmann, M., Ying, H., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2012). Trans Chromosomal Methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. U. S. A. 109, 3570–3575. doi: 10.1073/pnas.1201043109
Groszmann, M., Gonzalez-Bayon, R., Lyons, R. L., Greaves, I. K., Kazan, K., Peacock, W. J., et al. (2015). Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. U. S. A. 112, 6397–6406. doi: 10.1073/pnas.1519926112
Guo, M., Rupe, M. A., Danilevskaya, O. N., Yang, X., and Hu, Z. (2010). Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. Cell Mol. Biol. 36, 30–44. doi: 10.1046/j.1365-313X.2003.01852.x
Guo, X., Guo, Y., Ma, J., Wang, F., Sun, M., Gui, L., et al. (2013). Mapping Heterotic Loci for Yield and Agronomic Traits Using Chromosome Segment Introgression Lines in Cotton. J. Integr. Plant Biol. 55, 759–774. doi: 10.1111/jipb.12054
Guo, Z., Song, G., Liu, Z., Qu, X., Chen, R., Jiang, D., et al. (2015). Global epigenomic analysis indicates that epialleles contribute to Allele-specific expression via Allele-specific histone modifications in hybrid rice. BMC Genomics 16:232. doi: 10.1186/s12864-015-1454-z
Ha, M., Ng, W. K., Li, W. H., and Chen, Z. J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21, 590–598. doi: 10.1101/gr.116467.110
Hao, D., Zhang, Z., Cheng, Y., Chen, G., Lu, H., Mao, Y., et al. (2015). Identification of Genetic Differentiation between Waxy and Common Maize by SNP Genotyping. PLoS One 10:e142585. doi: 10.1371/journal.pone.0142585
Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135, 677–684. doi: 10.1104/PP.104.042614
He, G., Elling, A. A., and Deng, X. W. (2011). The Epigenome and Plant Development. Annu. Rev. Plant Biol. 62, 411–435. doi: 10.1146/annurev-arplant-042110-103806
He, G., Zhu, X., Elling, A. A., Chen, L., Wang, X., Guo, L., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33. doi: 10.1105/tpc.109.072041
Hochholdinger, F., and Hoecker, N. (2007). Towards the molecular basis of heterosis. Trends Plant Sci. 12, 427–432. doi: 10.1016/j.tplants.2007.08.005
Hoecker, N., Keller, B., Muthreich, N., Chollet, D., Descombes, P., Piepho, H. P., et al. (2008). Comparison of maize (Zea mays L.) F1-hybrid and parental inbred line primary root transcriptomes suggests organ-specific patterns of nonadditive gene expression and conserved expression trends. Genetics 179, 1275–1283. doi: 10.1534/genetics.108.088278
Hou, G., Dong, Y., Zhu, F., Zhao, Q., Li, T., Dou, D., et al. (2020). MicroRNA transcriptomic analysis of the sixth leaf of maize (Zea mays L.) revealed a regulatory mechanism of jointing stage heterosis. BMC Plant Biol. 20:541. doi: 10.1186/s12870-020-02751-3
Hu, S., Wang, C., Sanchez, D. L., Lipka, A. E., Liu, P., Yin, Y., et al. (2017). Gibberellins Promote Brassinosteroids Action and Both Increase Heterosis for Plant Height in Maize (Zea mays L.). Front. Plant Sci. 8:1039. doi: 10.3389/fpls.2017.01039
Hu, X., Wang, H., Li, K., Liu, X., Liu, Z., Wu, Y., et al. (2021). Genome-wide alternative splicing variation and its potential contribution to maize immature-ear heterosis. Crop J. 9, 476–486. doi: 10.1016/J.CJ.2020.09.003
Huang, X., Yang, S., Gong, J., Zhao, Q., Feng, Q., Zhan, Q., et al. (2016). Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633. doi: 10.1038/nature19760
Jaikishan, I., Rajendrakumar, P., Ramesha, M., Viraktamath, B., Balachandran, S., Neeraja, C., et al. (2010). Prediction of heterosis for grain yield in rice using ‘key’ informative EST-SSR markers. Plant Breed. 129, 108–111. doi: 10.1111/j.1439-0523.2009.01633.x
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Jones, D. F. (1917). Dominance of Linked Factors as a Means of Accounting for Heterosis. Proc. Natl. Acad. Sci. U. S. A. 3, 310–312.
Katara, J. L., Verma, R. L., Parida, M., Ngangkham, U., Molla, K. A., Barbadikar, K. M., et al. (2020). Differential expression of genes at panicle initiation and grain filling stages implied in heterosis of rice hybrids. Int. J. Mol. Sci. 21, 1–17. doi: 10.3390/ijms21031080
Kawamura, K., Kawanabe, T., Shimizu, M., Nagano, A. J., Saeki, N., Okazaki, K., et al. (2015). Genetic distance of inbred lines of Chinese cabbage and its relationship to heterosis. Plant Gene 5, 1–7. doi: 10.1016/j.plgene.2015.10.003
Kawanabe, T., Ishikura, S., Miyaji, N., Sasaki, T., Wu, L. M., Itabashi, E., et al. (2016). Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 113, E6704–E6711. doi: 10.1073/pnas.1613372113
Ko, D. K., Rohozinski, D., Song, Q., Taylor, S. H., Juenger, T. E., Harmon, F. G., et al. (2016). Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS Genet. 12:e1006197. doi: 10.1371/journal.pgen.1006197
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding amobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Kopp, F., Smart, B., Maynard, A., Tuskan, A., and Abrahamson, P. (2002). Predicting within-family variability in juvenile height growth of Salix based upon similarity among parental AFLP fingerprints. Theor. Appl. Genet. 105, 106–112. doi: 10.1007/s00122-001-0855-3
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Krieger, U., Lippman, Z. B., and Zamir, D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–63. doi: 10.1038/ng.550
Lai, Z., Gross, B. L., Zou, Y., Andrews, J., and Rieseberg, L. H. (2006). Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 15, 1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x
Lauss, K., Wardenaar, R., Oka, R., Van, H. M. H. A., Guryev, V., Keurentjes, J. J. B., et al. (2018). Parental DNA Methylation States Are Associated with Heterosis in Epigenetic Hybrids. Plant Physiol. 176, 1627–1645. doi: 10.1104/PP.17.01054
Li, B., Zhang, D. F., Jia, G. Q., Dai, J. R., and Wang, S. C. (2009). Genome-wide Comparisons of Gene Expression for Yield Heterosis in Maize. Plant Mol. Biol. Rep. 27, 162–176. doi: 10.1007/s11105-008-0068-x
Li, C., Huang, L., Xu, C., Zhao, Y., and Zhou, D. (2011). Altered levels of histone deacetylase oshdt1 affect differential gene expression patterns in hybrid rice. PLoS One 6:e21789. doi: 10.1371/journal.pone.0021789
Li, D., Huang, Z., Song, S., Xin, Y., Mao, D., Lv, Q., et al. (2016). Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proc. Natl. Acad. Sci. U. S. A. 113, E6026–E6035. doi: 10.1073/pnas.1610115113
Li, H., Jiang, S., Li, C., Liu, L., Lin, Z., He, H., et al. (2020). The hybrid protein interactome contributes to rice heterosis as epistatic effects. Plant J. 102, 116–128. doi: 10.1111/tpj.14616
Li, H., Li, J., Song, J., Zhao, B., Guo, C., Wang, B., et al. (2019). An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol. 222, 837–851. doi: 10.1111/nph.15632
Li, H., Yang, Q., Gao, L., Zhang, M., Ni, Z., and Zhang, Y. (2017). Identification of Heterosis-Associated Stable QTLs for Ear-Weight-Related Traits in an Elite Maize Hybrid Zhengdan 958 by Design III. Front. Plant Sci. 8:561. doi: 10.3389/fpls.2017.00561
Li, H., Yuan, J., Wu, M., Han, Z., Li, L., Jiang, H., et al. (2018). Transcriptome and DNA methylome reveal insights into yield heterosis in the curds of broccoli (Brassica oleracea L var. italic). BMC Plant Biol. 18:168. doi: 10.1186/s12870-018-1384-4
Li, J., Jin, Z., and Gu, J. (2012). Genetic isolation by distance in the endangered plant sinocalycanthus Chinensis endrmic to China. Pak. J. Bot. 44, 1275–1280. doi: 10.1105/tpc.112.240861
Li, S., Jayasinghege, C. P. A., Guo, J., Zhang, E., Wang, X., and Xu, Z. (2021). Comparative Transcriptomic Analysis of Gene Expression Inheritance Patterns Associated with Cabbage Head Heterosis. Plants 10:275. doi: 10.3390/PLANTS10020275
Li, X., Shahzad, K., Guo, L., Qi, T., Zhang, X., Wang, H., et al. (2019). Using yield quantitative trait locus targeted SSR markers to study the relationship between genetic distance and yield heterosis in upland cotton (Gossypium hirsutum). Plant Breed. 138, 105–113. doi: 10.1111/pbr.12668
Li, Y., Wang, C., Liu, X., Li, J. S., Sui, Z., Zhang, M., et al. (2016). Up-regulating the abscisic acid inactivation gene ZmABA8ox1b contributes to seed germination heterosis by promoting cell expansion. J. Exp. Bot. 67, 2889–2900.
Li, Z., Pinson, S. R. M., Park, W. D., Paterson, A. H., and Stansel, J. W. (1997). Epistasis for three grain yield components in rice (Oryza sativa L.). Genetics 145, 453–465. doi: 10.1093/GENETICS/145.2.453
Lin, T., Zhou, C., Chen, G., Yu, J., Wu, W., Ge, Y., et al. (2020). Heterosis-associated genes confer high yield in super hybrid rice. Theor. Appl. Genet. 133, 3287–3297. doi: 10.1007/s00122-020-03669-y
Lin, Z., Qin, P., Zhang, X., Fu, C., Deng, H., Fu, X., et al. (2020). Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. Proc. Natl. Acad. Sci. U. S. A. 117, 4623–4631. doi: 10.1073/pnas.1919086117
Liu, C., Liu, T., Ohlson, E. W., Wang, L., Wu, D., Guo, Q., et al. (2019). Loquat (Eriobotrya japonica (Thunb.) circadian clock gene cloning and heterosis studies of artificial triploid loquat. Sci. Hortic. 246, 328–337. doi: 10.1016/j.scienta.2018.10.068
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, C., Wu, D., Wang, L., Dang, J., He, Q., Guo, Q., et al. (2018b). Cis-regulated additively expressed genes play a fundamental role in the formation of triploid loquat (Eriobotrya japonica (Thunb.) Lindl.) Heterosis. Mol. Genet. Genomics 293, 967–981. doi: 10.1007/s00438-018-1433-6
Liu, C., Wang, M., Wang, L., Guo, Q., and Liang, G. (2018a). Extensive genetic and DNA methylation variation contribute to heterosis in triploid loquat hybrids. Genome 61, 437–447. doi: 10.1139/gen-2017-0232
Liu, H., Wang, Q., Chen, M., Ding, Y., Yang, X., Liu, J., et al. (2019). Genomewide identification and analysis of heterotic loci in three maize hybrids. Plant Biotechnol. J. 18, 185–194. doi: 10.1111/pbi.13186
Liu, J., Li, M., Zhang, Q., Wei, X., and Huang, X. (2020). Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 62, 287–298. doi: 10.1111/jipb.12804
Liu, P., Peacock, W. J., Wang, L., Furbank, R., Larkum, A., and Dennis, E. S. (2020). Leaf growth in early development is key to biomass heterosis in Arabidopsis. J. Exp. Bot. 71, 2439–2450. doi: 10.1093/jxb/eraa006
Liu, W., He, G., and Deng, X. W. (2021). Biological pathway expression complementation contributes to biomass heterosis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118:e2023278118. doi: 10.1073/PNAS.2023278118
Liu, Y., Yi, Q., Hou, X., Hu, Y., Li, Y., and Yu, G. (2020). Identification of quantitative trait loci for kernel-related traits and the heterosis for these traits in maize (Zea mays L.). Mol. Genet. Genomics 295, 1–13. doi: 10.1007/s00438-019-01608-1
Lu, J., Zhang, C., Baulcombe, D. C., and Chen, Z. J. (2012). Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. U. S. A. 109, 5529–5534. doi: 10.1073/pnas.1203094109
Luo, J., Wang, M., Jia, G., and He, Y. (2021). Transcriptome-wide analysis of epitranscriptome and translational efficiency associated with heterosis in maize. J. Exp. Bot. 72, 2933–2946. doi: 10.1093/JXB/ERAB074
Ma, J., Morrow, D. J., Fernandes, J., and Walbot, V. (2006). Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Biol. 7, 334–337. doi: 10.1186/gb-2006-7-3-r22
Madić, M., Djurović, D., Knezevic, D., Paunović, A., and Tanasković, S. (2014). Combining abilities for spike traits in a diallel cross of barley. J. Central Eur. Agric. 15, 108–116. doi: 10.5513/JCEA01/15.1.1419
Mallory, A. C., Bartel, D. P., and Bartel, B. (2005). MicroRNA-Directed Regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17, 1360–1375. doi: 10.1105/TPC.105.031716
Marcelo, S. D. A., Daniel, F. F., Aurélio, M. A., Odair, B., Gabriel, D. S. P. R., and Dario, G. (2007). Potential of hybrids among elite clones of eucalypt by microsatellite markers. Pesqui. Agropecu. Bras. 42, 1007–1012. doi: 10.1590/S0100-204X2007000700013
Marcon, C., Lamkemeyer, T., Malik, W. A., Ungrue, D., Piepho, H. P., and Hochholdinger, F. (2013). Heterosis-associated proteome analyses of maize (Zea mays L.) seminal roots by quantitative label-free LC-MS. J. Proteomics 93, 295–302. doi: 10.1016/j.jprot.2013.04.015
Marcon, C., Schützenmeister, A., Schütz, W., Madlung, J., Piepho, H., and Hochholdinger, F. (2010). Nonadditive Protein Accumulation Patterns in Maize (Zea mays L.) Hybrids during Embryo Development. J. Proteome Res. 9, 6511–6522. doi: 10.1021/pr100718d
Maruthi, R. T., Gadge, R. N., Bhat, J. S., and Karjagi, C. G. (2019). Heterotic grouping of late-maturing maize inbred lines based on combining ability and molecular marker studies. J. Environ. Biol. 40, 705–710. doi: 10.22438/jeb/40/4/MRN-921
Metzger, B. P. H., Fabien, D., Yuan, D. C., Stephen, T., Bing, Y., and Wittkopp, P. J. (2016). Contrasting Frequencies and Effects of cis- and trans-Regulatory Mutations Affecting Gene Expression. Mol. Biol. Evol. 33, 1131–1146. doi: 10.1093/molbev/msw011
Meyer, R. C., Witucka-Wall, H., Becher, M., Blacha, A., Boudichevskaia, A., Dörmann, P., et al. (2012). Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 71, 669–683. doi: 10.1111/j.1365-313X.2012.05021.x
Meyer, S., Pospisil, H., and Scholten, S. (2007). Heterosis associated gene expression in maize embryos 6days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol. Biol. 63, 381–391. doi: 10.1007/s11103-006-9095-x
Miller, M., Song, Q., Shi, X., Juenger, T. E., and Chen, Z. J. (2015). Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Commun. 6, 69–74. doi: 10.1038/ncomms8453
Miller, M., Zhang, C., and Chen, Z. J. (2012). Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 2, 505–513. doi: 10.1534/g3.112.002162
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., and Asano, K. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Moreno-Romero, J., Jiang, H., Santos-González, J., and Khler, C. (2016). Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J. 35, 1298–1311. doi: 10.15252/embj.201593534
Mühleisen, J., Maurer, H. P., Stiewe, G., Bury, P., and Reif, J. C. (2013). Hybrid breeding in barley. Crop Sci. 53, 819–824. doi: 10.2135/cropsci2012.07.0411
Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2010). A role for HvGAMYB in anther development. Plant J. 33, 481–491. doi: 10.1046/j.1365-313X.2003.01641.x
Nakamura, S., and Hosaka, K. (2010). DNA methylation in diploid inbred lines of potatoes and its possible role in the regulation of heterosis. Theor. Appl. Genet. 120, 205–214. doi: 10.1007/s00122-009-1058-6
Ng, W. K., Zhang, C., Miller, M., Shen, Z., Briggs, S. P., and Chen, Z. J. (2012). Proteomic divergence in Arabidopsis autopolyploids and allopolyploids and their progenitors. Heredity 108, 419–430. doi: 10.1038/hdy.2011.92
Ni, Z., Kim, E. D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Paschold, A., Jia, Y., Marcon, C., Lund, S., Larson, N. B., Yeh, C., et al. (2012). Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Res. 22, 2445–2454. doi: 10.1101/gr.138461.112
Perales, M., and Mas, P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19, 2111–2123. doi: 10.1105/TPC.107.050807
Powers, L. (1944). An expansion of Jones’s theory for the explanation of heterosis. Am. Nat. 78, 275–280. doi: 10.1086/281199
Rajendran, A., Muthian, A., Joel, J., Shanmugasundaram, P., and Raju, D. (2014). Heterotic grouping and patterning of quality protein maize inbreds based on genetic and molecular marker studies. Proc. Natl. Acad. Sci. U. S. A. 38, 10–20. doi: 10.3906/biy-1303-6
Reddy, C. K., Pooja, A., and Utpal, N. (2016). Activation of YUCCA5 by the Transcription Factor TCP4 Integrates Developmental and Environmental Signals to Promote Hypocotyl Elongation in Arabidopsis. Plant Cell 28, 2117–2130. doi: 10.1105/TPC.16.00360
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of Plant MicroRNA Targets. Cell 110, 513–520.
Riddle, N. C., and Birchler, J. A. (2003). Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19, 597–600. doi: 10.1016/j.tig.2003.09.005
Riddle, N. C., Kato, A., and Birchler, J. A. (2006). Genetic variation for the response to ploidy change in Zea mays L. Theor. Appl. Genet. 114, 101–111. doi: 10.1007/s00122-006-0414-z
Rockenbach, M. F., Corrêa, C. C. G., Heringer, A. S., Freitas, I. L. J., Santa-Catarina, C., Do, A. A. T., et al. (2018). Differentially abundant proteins associated with heterosis in the primary roots of popcorn. PLoS One 13:e197114. doi: 10.1371/journal.pone.0197114
Sarfraz, Z., Iqbal, M. S., Pan, Z., Jia, Y., He, S., Wang, Q., et al. (2018). Integration of conventional and advanced molecular tools to track footprints of heterosis in cotton. BMC Genomics 19:776. doi: 10.1186/s12864-018-5129-4
Saxena, I., Srikanth, S., and Chen, Z. (2016). Cross Talk between H2O2 and Interacting Signal Molecules under Plant Stress Response. Front. Plant Sci. 7:570. doi: 10.3389/fpls.2016.00570
Seifert, F., Thiemann, A., Grant-Downton, R., Edelmann, S., Rybka, D., Schrag, T. A., et al. (2018). Parental Expression Variation of Small RNAs Is Negatively Correlated with Grain Yield Heterosis in a Maize Breeding Population. Front. Plant Sci. 9:13. doi: 10.3389/fpls.2018.00013
Semel, Y., Nissenbaum, J., Menda, N., Zinder, M., Krieger, U., Issman, N., et al. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. U. S. A. 103, 12981–12986. doi: 10.1073/pnas.0604635103
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Shahzad, K., Zhang, X., Guo, L., Qi, T., Tang, H., Zhang, M., et al. (2020). Comparative transcriptome analysis of inbred lines and contrasting hybrids reveals overdominance mediate early biomass vigor in hybrid cotton. BMC Genomics 21:140. doi: 10.1186/s12864-020-6561-9
Shao, L., Xing, F., Xu, C., Zhang, Q., Che, J., Wang, X., et al. (2019). Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. U. S. A. 116, 5653–5658.
Sheidai, M., Afshar, F., Keshavarzi, M., Talebi, S., Noormohammadi, Z., and Shafaf, T. (2014). Genetic diversity and genome size variability in Linum austriacum (Lineaceae) populations. Biochem. Syst. Ecol. 57, 20–26. doi: 10.1016/j.bse.2014.07.014
Shen, E., Zou, J., Hubertus, B. F., Chen, L., Ye, C., Dai, S., et al. (2015). Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 66, 7241–7253. doi: 10.1093/jxb/erv420
Shen, H., He, H., Li, J., Chen, W., Wang, X., Guo, L., et al. (2012). Genome-Wide analysis of dna methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24, 875–892. doi: 10.1105/TPC.111.094870
Shen, Y., Sun, S., Hua, S., Shen, E., Ye, C. Y., Cai, D., et al. (2017). Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 91, 874–893. doi: 10.1111/tpj.13605
Shull, G. H. (1908). The Composition of a Field of Maize. J. Hered. 4, 296–301. doi: 10.1093/jhered/os-4.1.296
Singh, B., Sharma, A., Joshi, N., Mittal, P., and Singh, S. (2013). Combining ability for grain yield and its components in barley (Hordeum vulgare). Indian J. Agric. Sci. 83, 96–98.
Smith, L. M. (2019). Salicylic acid, senescence, and heterosis. Plant Physiol. 180, 3–4. doi: 10.1104/pp.19.00260
Song, G., Zhai, H., Peng, Y., Zhang, L., Wei, G., Chen, X., et al. (2010). Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant 3, 1012–1025. doi: 10.1093/mp/ssq046
Song, Q., Ando, A., Xu, D., Fang, L., Zhang, T., Huq, E., et al. (2018). Diurnal down-regulation of ethylene biosynthesis mediates biomass heterosis. Proc. Natl. Acad. Sci. U. S. A. 115, 5606–5611. doi: 10.1073/pnas.1722068115
Song, S., Qu, H., Chen, C., Hu, S., and Yu, J. (2007). Differential gene expression in an elite hybrid rice cultivar (Oryza sativa, L) and its parental lines based on SAGE data. BMC Plant Biol. 7:49. doi: 10.1186/1471-2229-7-49
Stupar, R. M., and Springer, N. M. (2006). Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173, 2199–2210. doi: 10.1534/GENETICS.106.060699
Sudi, S., Bains, N. S., Nanda, G. S., Singh, K., and Arya, L. (2010). Genetic diversity and heterosis in wheat. SABRAO J. Breed. Genet. 42, 96–105.
Sun, J., Qi, L., Li, Y., Chu, J., Li, C., and Qu, L. J. (2012). PIF4–Mediated Activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8:e1002594. doi: 10.1371/journal.pgen.1002594
Sun, J., Qi, L., Li, Y., Zhai, Q., and Li, C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25, 2102–2114. doi: 10.1105/TPC.113.112417
Swanson-Wagner, R. A., Jia, Y., Decook, R., Borsuk, L. A., Nettleton, D., and Schnable, P. S. (2006). All possible modes of gene action observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. U. S. A. 103, 6805–6810. doi: 10.1073/pnas.0510430103
Tadege, M., Sheldon, C. C., Helliwell, C. A., Upadhyaya, N. M., Dennis, E. S., and Peacock, W. J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1, 361–369. doi: 10.1046/j.1467-7652.2003.00034.x
Tang, J., Yan, J., Ma, X., Teng, W., Wu, W., Dai, J., et al. (2010). Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor. Appl. Genet. 120, 333–340. doi: 10.1007/s00122-009-1213-0
Tian, H. Y., Channa, S. A., and Hu, S. W. (2017). Relationships between genetic distance, combining ability and heterosis in rapeseed (Brassica napus L.). Euphytica 213:1. doi: 10.1007/s10681-016-1788-x
Tian, M., Nie, Q., Li, Z., Zhang, J., Liu, Y., Long, Y., et al. (2018). Transcriptomic analysis reveals overdominance playing a critical role in nicotine heterosis in Nicotiana tabacum L. BMC Plant Biol. 18:48. doi: 10.1186/s12870-018-1257-x
Tian, Y., Fan, M., Qin, Z., Lv, H., Wang, M., Zhang, Z., et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9:1063. doi: 10.1038/s41467-018-03463-x
Tsuji, H., Aya, K., Tanaka, M. U., Shimada, Y., Nakazono, M., Watanabe, R., et al. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47, 427–444. doi: 10.1111/j.1365-313X.2006.02795.x
Upadhyay, D., Mehta, N., Singh, J., and Sahu, M. (2011). Genetic divergence in dolicho bean (Dolichos Lablab L.). Electron. J. Plant Breed. 2, 146–148. doi: 10.1007/s11104-010-0526-8
Usatov, A. V., Klimenko, A. I., Azarin, K. V., Gorbachenko, O. F., Markin, N. V., Tikhobaeva, V. E., et al. (2014). The relationship between heterios and genetic distances based on SSR markers in helianthus annuus. Am. J. Agric. Biol. Sci. 9, 270–276. doi: 10.3844/ajabssp.2014.270.276
Vaillancourt, R., Potts, B., Watson, M., Reid, J., and West, A. (1995). Detection and prediction of heterosis in Eucalyptus globulus. For. Genet. 2, 11–19.
Wang, D., Mu, Y., Hu, X., Ma, B., Wang, Z., Zhu, L., et al. (2021). Comparative proteomic analysis reveals that the Heterosis of two maize hybrids is related to enhancement of stress response and photosynthesis respectively. BMC Plant Biol. 21:34. doi: 10.1186/S12870-020-02806-5
Wang, J., Tian, L., Lee, H., Wei, N. E., Jiang, H., Watson, B., et al. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172, 507–517. doi: 10.1534/GENETICS.105.047894
Wang, L., Greaves, I. K., Groszmann, M., Wu, L. M., Dennis, E. S., and Peacock, W. J. (2015). Hybrid mimics and hybrid vigor in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 112, 4959–4967. doi: 10.1073/pnas.1514190112
Wang, L., Wu, L. M., Greaves, I. K., Zhu, A., Dennis, E. S., and Peacock, W. J. (2017). PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 114, E3555–E3562. doi: 10.1073/pnas.1703179114
Wang, W., Meng, B., Ge, X., Song, S., Yang, Y., Yu, X., et al. (2008). Proteomic profiling of rice embryos from a hybrid rice cultivar and its parental lines. Proteomics 8, 4808–4821. doi: 10.1002/pmic.200701164
Wang, Y., Zhang, X., Shi, X., Sun, C., Jin, J., Tian, R., et al. (2018a). Heterotic loci identified for maize kernel traits in two chromosome segment substitution line test populations. Sci. Rep. 8, 296–301. doi: 10.1038/s41598-018-29338-1
Wang, Y., Zhang, K., Sun, L., Han, X., Fan, S., Li, X., et al. (2018b). Study on the relationship between genetic variation of DNA methylation and heterosis in soybean leaves. Euphytica 214:1. doi: 10.1007/s10681-018-2161-z
Woodger, F. J., Millar, A., Murray, F., Jacobsen, J. V., and Gubler, F. (2003). The Role of GAMYB Transcription Factors in GA-Regulated Gene Expression. J. Plant Growth Regul. 22, 176–184. doi: 10.1007/s00344-003-0025-8
Wu, L., Lin, Y., Luo, Y., Xu, X., Zeng, S., and Zhu, B. (2012). Analysis on correlation between genetic distance of pepper parents and heterosis of yield with SRAP molecular marker. J. Fujian Agric. For. Univ. 41, 595–600.
Wuriyanghan, H., Zhang, B., Cao, W., Ma, B., Lei, G., Liu, Y., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21, 1473–1494. doi: 10.1105/TPC.108.065391
Xi, F., Ruan, X., and Minarifar, A. (2021). ISSR markers and population differentiations in Erodium ciconium (L.) L’her Ex Aiton. Bangl. J. Plant Taxon. 28, 61–73. doi: 10.3329/bjpt.v28i1.54208
Xiao, J., Li, J., Yuan, L., and Tanksley, S. D. (1995). Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140, 745–754. doi: 10.1093/GENETICS/140.2.745
Xiao, Y., Jiang, S., Cheng, Q., Wang, X., Yan, J., Zhang, R., et al. (2021). The genetic mechanism of heterosis utilization in maize improvement. Genome Biol. 22:148. doi: 10.1186/S13059-021-02370-7
Xiong, L. Z., Xu, C. G., Maroof, M. A. S., and Zhang, Q. (1999). Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261, 439–446. doi: 10.1007/s004380050986
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yamaguchi, A., Wu, M., Yang, L., Wu, G., Poethig, S., and Wagner, A. D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Yan, W., Wang, P., Chen, H., Zhou, H., Li, Q., Wang, C., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yang, L., Liu, P., Wang, X., Jia, A., Ren, D., Tang, Y., et al. (2021). A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs. Nat. Commun. 12:2317. doi: 10.1038/S41467-021-22268-Z
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant cell 12, 2473–2484. doi: 10.1105/TPC.12.12.2473
Yao, Y., Ni, Z., Zhang, Y., Chen, Y., Ding, Y., Han, Z., et al. (2005). Identification of differentially expressed genes in leaf and root between wheat hybrid and its parental inbreds using PCR-based cDNA subtraction. Plant Mol. Biol. 58, 367–384. doi: 10.1007/s11103-005-5102-x
Yi, G., Shin, H., Park, H. R., Park, J. E., Ahn, J. H., Lim, S., et al. (2020). Revealing biomass heterosis in the allodiploid xBrassicoraphanus, a hybrid between Brassica rapa and Raphanus sativus, through integrated transcriptome and metabolites analysis. BMC Plant Biol. 20:252. doi: 10.1186/s12870-020-02470-9
Yi, H., Lee, J., Song, H., Dong, X., and Hur, Y. (2017). Genome-wide analysis of heterosis-related genes in non-heading Chinese cabbage. J. Plant Biotechnol. 44, 208–219. doi: 10.5010/JPB.2017.44.3.208
Yu, D., Gu, X., Zhang, S., Dong, S., Miao, H., Gebretsadik, K., et al. (2021). Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 8:120. doi: 10.1038/S41438-021-00552-9
Yu, S. B., Li, J. X., Xu, C. G., Tan, Y. F., Gao, Y. J., Li, X. H., et al. (1997). Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. U. S. A. 94, 9226–9231. doi: 10.1073/pnas.94.17.9226
Yuan, H., Liu, W., and Lu, Y. (2017). CATALASE2 Coordinates SA-Mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21, 143–155. doi: 10.1016/j.chom.2017.01.007
Zhang, H., He, H., Chen, L., Li, L., Liang, M., Wang, X., et al. (2008). A Genome-Wide Transcription Analysis Reveals a Close Correlation of Promoter INDEL Polymorphism and Heterotic Gene Expression in Rice Hybrids. Mol. Plant 1, 720–731. doi: 10.1093/mp/ssn022
Zhang, L., Peng, Y., Wei, X., Dai, Y., Yuan, D., Lu, Y., et al. (2014). Small RNAs as important regulators for the hybrid vigour of super-hybrid rice. J. Exp. Bot. 65, 5989–6002. doi: 10.1093/jxb/eru337
Zhang, Q., Li, Y., Xu, T., Srivastava, A. K., Wang, D., Zeng, L., et al. (2016). The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2, 71–88. doi: 10.1038/celldisc.2016.27
Zhang, S., Huang, X., and Han, B. (2021). Understanding the genetic basis of rice heterosis: advances and prospects. Crop J. 9, 688–692. doi: 10.1016/j.cj.2021.03.011
Zhang, T., Li, B., Zhang, D., Jia, G., Li, Z., and Wang, S. (2012). Genome-Wide Transcriptional Analysis of Yield and Heterosis-Associated Genes in Maize (Zea mays L.). J. Integr. Agr. 11, 1245–1256. doi: 10.1016/S2095-3119(12)60121-X
Zhang, X., and Borevitz, J. O. (2009). Global Analysis of Allele-Specific Expression in Arabidopsis thaliana. Genetics 182, 943–954. doi: 10.1534/genetics.109.103499
Zhang, X., Lv, L., Lv, C., Guo, B., and Xu, R. (2015). Combining Ability of Different Agronomic Traits and Yield Components in Hybrid Barley. PLoS One 10:e0126828. doi: 10.1371/journal.pone.0126828
Zhao, M., Han, B., and Walter, D. W. (2008). Detection of genetic diversity in rough fescue (Festuca campestris Rydb) populations of southern Alberta and British Columbia, Canada, using RAPD markers. Can J. Plant Sci. 88, 307–312. doi: 10.4141/CJPS07071
Zhao, P., Ding, D., Zhang, F., Zhao, X., Xue, Y., Li, W., et al. (2015). Investigating the molecular genetic basis of heterosis for internode expansion in maize by microRNA transcriptomic deep sequencing. Funct. Integr. Genomic 15, 261–270. doi: 10.1007/s10142-014-0411-2
Keywords: heterosis, plant, biomass yield, grain yield, gene
Citation: Wu X, Liu Y, Zhang Y and Gu R (2021) Advances in Research on the Mechanism of Heterosis in Plants. Front. Plant Sci. 12:745726. doi: 10.3389/fpls.2021.745726
Received: 22 July 2021; Accepted: 06 September 2021;
Published: 27 September 2021.
Edited by:
Yongzhong Xing, Huazhong Agricultural University, ChinaReviewed by:
Yuepeng Song, Beijing Forestry University, ChinaWeibo Xie, Huazhong Agricultural University, China
Copyright © 2021 Wu, Liu, Zhang and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaowei Zhang, emhhbmd5YW93ZWlAbmVhdS5lZHUuY24=; RanGu, Z3VyYW5fY2F1QG5lYXUuZWR1LmNu
 Xilin Wu1,2
Xilin Wu1,2 Yan Liu
Yan Liu Yaowei Zhang
Yaowei Zhang