
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 08 October 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.744991
This article is part of the Research TopicStomatal Biology and BeyondView all 16 articles
 Wenxiu Ye1,2†
Wenxiu Ye1,2† Shota Koya3†
Shota Koya3† Yuki Hayashi3
Yuki Hayashi3 Huimin Jiang1
Huimin Jiang1 Takaya Oishi3
Takaya Oishi3 Kyohei Kato3
Kyohei Kato3 Kohei Fukatsu3
Kohei Fukatsu3 Toshinori Kinoshita2,3*
Toshinori Kinoshita2,3*Stomatal guard cells (GCs) are highly specialized cells that respond to various stimuli, such as blue light (BL) and abscisic acid, for the regulation of stomatal aperture. Many signaling components that are involved in the stomatal movement are preferentially expressed in GCs. In this study, we identified four new such genes in addition to an aluminum-activated malate transporter, ALMT6, and GDSL lipase, Occlusion of Stomatal Pore 1 (OSP1), based on the expression analysis using public resources, reverse transcription PCR, and promoter-driven β-glucuronidase assays. Some null mutants of GC-specific genes evidenced altered stomatal movement. We further investigated the role played by ALMT6, a vacuolar malate channel, in stomatal opening. Epidermal strips from an ALMT6-null mutant exhibited defective stomatal opening induced by BL and fusicoccin, a strong plasma membrane H+-ATPase activator. The deficiency was enhanced when the assay buffer [Cl–] was low, suggesting that malate and/or Cl– facilitate efficient opening. The results indicate that the GC-specific genes are frequently involved in stomatal movement. Further detailed analyses of the hitherto uncharacterized GC-specific genes will provide new insights into stomatal regulation.
Stomata that are formed by pairs of guard cells (GCs) in the shoot epidermis of plants are key regulators of gas exchange, such as CO2 uptake for photosynthesis and water loss during transpiration (Shimazaki et al., 2007; Munemasa et al., 2015). GCs respond to internal and external signals, such as light, CO2, phytohormones, and microbial elicitors, where the stomata remain either open or close (Murata et al., 2015; Inoue and Kinoshita, 2017). Many critical signaling components that are involved in GC signaling are preferentially expressed in GCs, such as Open Stomata 1 (OST1) (Mustilli et al., 2002), slow anion channel-associated 1 (SLAC1) (Negi et al., 2008; Vahisalu et al., 2008), high leaf temperature 1 (HT1) (Hashimoto et al., 2006), and aluminum-activated malate transporter 12 (ALMT12) (Meyer et al., 2010; Sasaki et al., 2010), suggesting that GC-specific genes are important candidates in hunting for new GC signaling components.
Blue light (BL) and red light (RL) are major cues for stomatal opening (Shimazaki et al., 2007; Inoue and Kinoshita, 2017). On BL perception, phototropins undergo autophosphorylation, which triggers signaling by BLUS1, BHP1, type 1 protein phosphatase (PP1), and its regulatory subunit PRSL1, in turn leading to phosphorylation of the penultimate threonine (penThr) residues of the plasma membrane (PM) H+-ATPases, and the subsequent binding of 14-3-3 proteins activates the H+-ATPases. More recently, RL was shown to induce the activation of GC PM H+-ATPases by phosphorylation (Ando and Kinoshita, 2018). PM H+-ATPases are important in terms of stomatal movement; the activation induces PM hyperpolarization, triggering a K+ influx through inward-rectifying K+ channels (Shimazaki et al., 2007; Inoue and Kinoshita, 2017). Together with the accumulation of K+, the increase of counter anions, such as malate, biosynthesized in GCs and/or apoplast and Cl– from apoplast, and other osmolytes, such as sucrose, lower the water potential in GCs, leading to an inflow of water, the swelling of GCs, and eventually the stomatal opening (Shimazaki et al., 2007; Santelia and Lawson, 2016). Recently, it has been shown that the activation of PM H+-ATPases occurs upstream of starch degradation associated with BL-induced stomatal opening; this, combined with CO2 fixation in GC chloroplasts, yields the carbon skeletons required for malate synthesis (Horrer et al., 2016). Vacuoles accumulate most of the ions and water that control stomatal movement (Barbier-Brygoo et al., 2011). The electrophysiological experiments revealed that the ALMT6 and ALMT9 vacuole channels facilitated malate and Cl– import (Meyer et al., 2011); ALMT9 played a critical role in the light-induced stomatal opening (De Angeli et al., 2013).
To identify new signaling components involved in the light-induced stomatal opening, we reasoned that GC preferentially expressed genes are good candidates and identified four new such genes by the analyses of public resources, reverse transcription PCR (RT-PCR), and promoter GUS assay in Arabidopsis thaliana. Functional analysis revealed that some of the GC-specific genes in addition to ALMT6 are critical in the light-induced stomatal opening.
All A. thaliana strains were grown in soil under a photon flux density of 50 μmol m–2 s–1 and a 16-h-light/8-h-dark regime. The temperature and the relative humidity were 23 ± 2°C and 55–70%, respectively. All mutants [at5g18430 (SALK_116756), almt6-1 (GABI_259D05; Meyer et al., 2011), at1g33811 (GABI_492D11), osp1-1 (SALK _106116; Tang et al., 2020), and at3g23840 (GABI_180G04)] are in the Columbia ecotype background (Col-0).
Guard cell protoplasts (GCPs) and mesophyll cell protoplasts (MCPs) were isolated from glabra1-1 (gl1) as described previously (Okumura et al., 2016).
RNAs from gl1 GCPs, MCPs, rosette leaves, roots, petioles, stems, flowers, and etiolated seedlings were extracted using the RNeasy Plant Mini Kit (QIAGEN) according to the protocol of the manufacturer. Complementary DNA was synthesized using the PrimeScript II 1st strand cDNA Synthesis Kit (Takara). The PCR primers are listed in Supplementary Table 1.
The promoter regions (3-kb upstream of the start codons) of AT5G18430, ALMT6, AT1G33811, OSP1, AT3G23840, and AT3G17070 were amplified in two PCR steps using the primers listed in Supplementary Table 2 and cloned into pCR8/GW/TOPO followed by subcloning into pGWB433 binary vector. The vectors were transformed into Agrobacterium GV3101, which were then used to transform Col-0 by floral dip. Transformants were selected using kanamycin and carbenicillin and subjected to GUS staining at various developmental stages.
The stomatal aperture measurement was performed as described previously (Tomiyama et al., 2014; Toh et al., 2018). Epidermal tissues and leaf disks were prepared from dark-adapted plants and subjected to light illumination and fusicoccin (FC) treatment. The apertures were measured under a microscope (Olympus).
The immunohistochemical staining was performed as described previously (Hayashi et al., 2011). Epidermal tissues were prepared from dark-adapted plants and subjected to light illumination and FC treatment. RL (50 μmol m–2 s–1) was illuminated for 20 min (Red), and BL (10 μmol m–2 s–1) was illuminated with superimposed on RL for 2.5 min (Red + Blue). FC at 10 μM was applied to the epidermal tissue for 5 min in the dark (FC). PM H+-ATPases and the phosphorylation level of the penThr were detected using specific antibodies against the catalytic domain of AHA2 (anti-PM H+-ATPase antibody) and phosphorylated Thr-947 in AHA2 (anti-pThr) (Hayashi et al., 2010).
Sequence data can be found in the Arabidopsis genome database TAIR10 under the following accession numbers: ALMT6 (AT2G17470), OSP1 (AT2G04570), ALMT9 (AT3G18440), AT1G02980, AT1G12030, AT1G33811, AT2G32830, AT3G17070, AT3G23840, and AT5G18430.
We analyzed publicly available microarray data on GCPs and MCPs (Yang et al., 2008), and those of the Arabidopsis eFP browsers1. The inclusion criteria were as follows: (1) a microarray GCP signal unique to GCPs or at least fourfold higher than the MCP signal and (2) the “Tissue-Specific” criteria of the Arabidopsis eFP browsers indicated GC-specific expression. We retrieved 124 candidate genes and checked their expression levels by RT-PCR in various cells, tissues, and organs of A. thaliana (Figure 1A). A total of 10 genes were strongly expressed in GCPs but not in MCPs and roots; these included SLAC1, the cation/H+ exchanger-encoding AtCHX20, and genes encoding the GDSL lipases OSP1 and ALMT6 (which are preferentially expressed in GCs; Padmanaban et al., 2007; Negi et al., 2008; Vahisalu et al., 2008; Tang et al., 2020) and six functionally uncharacterized genes. The RT-PCR data for OSP1, ALMT6, and the uncharacterized genes are shown in Figure 1B.
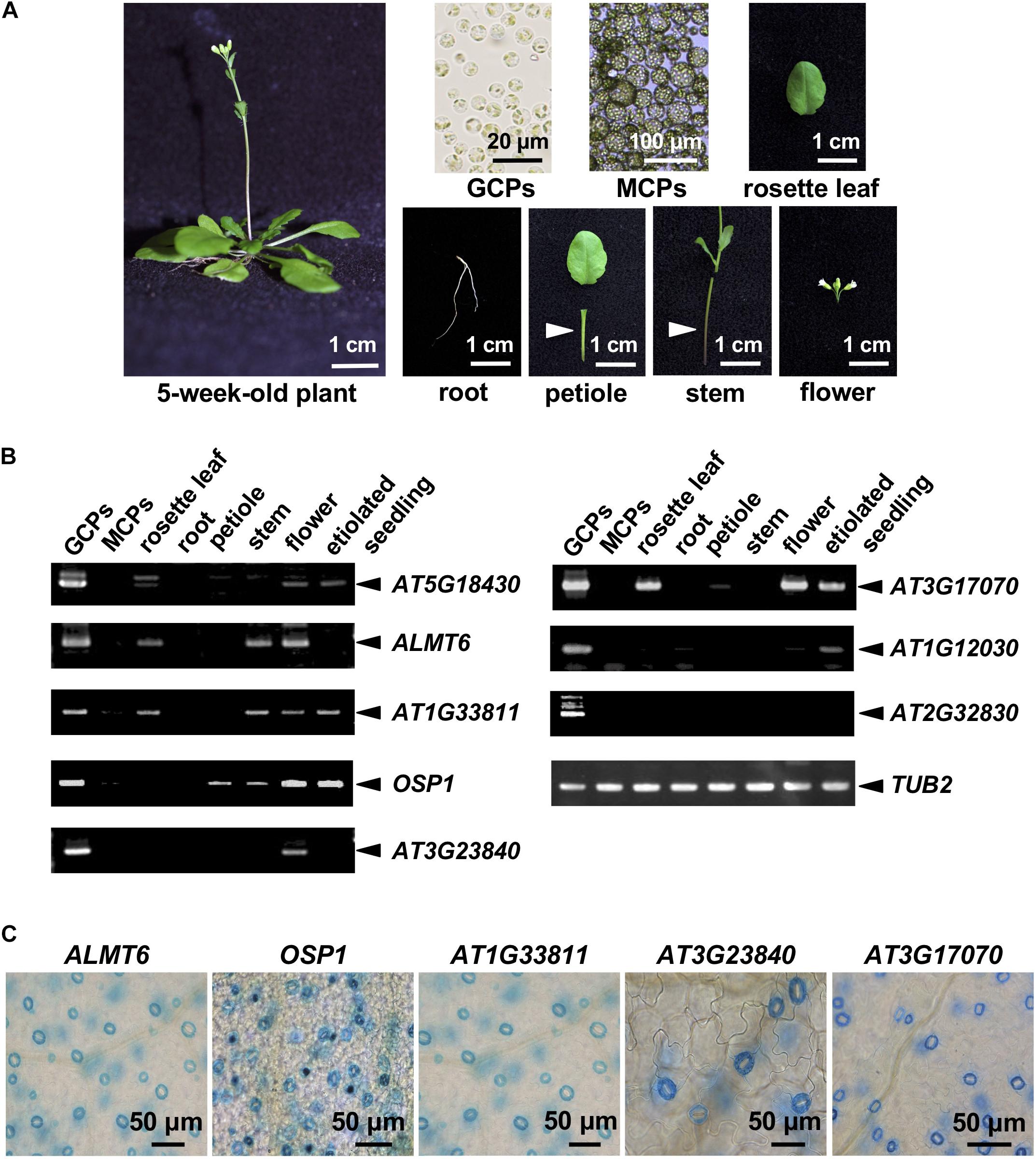
Figure 1. Guard cell (GC)-preferentially expressed genes. (A) The plant organs, tissues, and cells used in the analysis. (B) GC-preferentially expressed genes as revealed by RT-PCR using the materials shown in panel (A). (C) GC-preferentially expressed genes in GUS-reporter-bearing plants.
To further confirm preferential GC expression in intact plants, we constructed transgenic plants expressing the reporter β-GUS-encoding gene driven by promoter regions ranging to about 3-kb upstream of the start codons. pALMT6:GUS and pOSP1:GUS exhibited the high-level GUS activity in GCs (in particular) (Figure 1C), consistent with previous findings (Meyer et al., 2011; Tang et al., 2020). Also, the AT1G33811, AT3G23840, AT3G17070, and AT5G18430 promoters drove GC-preferential GUS expression (Figures 1C, 2A). The ALMT6, AT1G33811, OSP1, AT3G17070, and AT3G23840 promoters drove GUS expression in stipules, lateral roots, and trichomes (Figure 3A); however, the AT5G18430 promoter was active in GCs only (Figure 2A). The AT1G12030 and AT2G32830 promoters never drove GUS expression, rendering the analyses difficult. This is probably due to the lack of or weak activity of the 3-kb upstream promoter regions of these two genes.
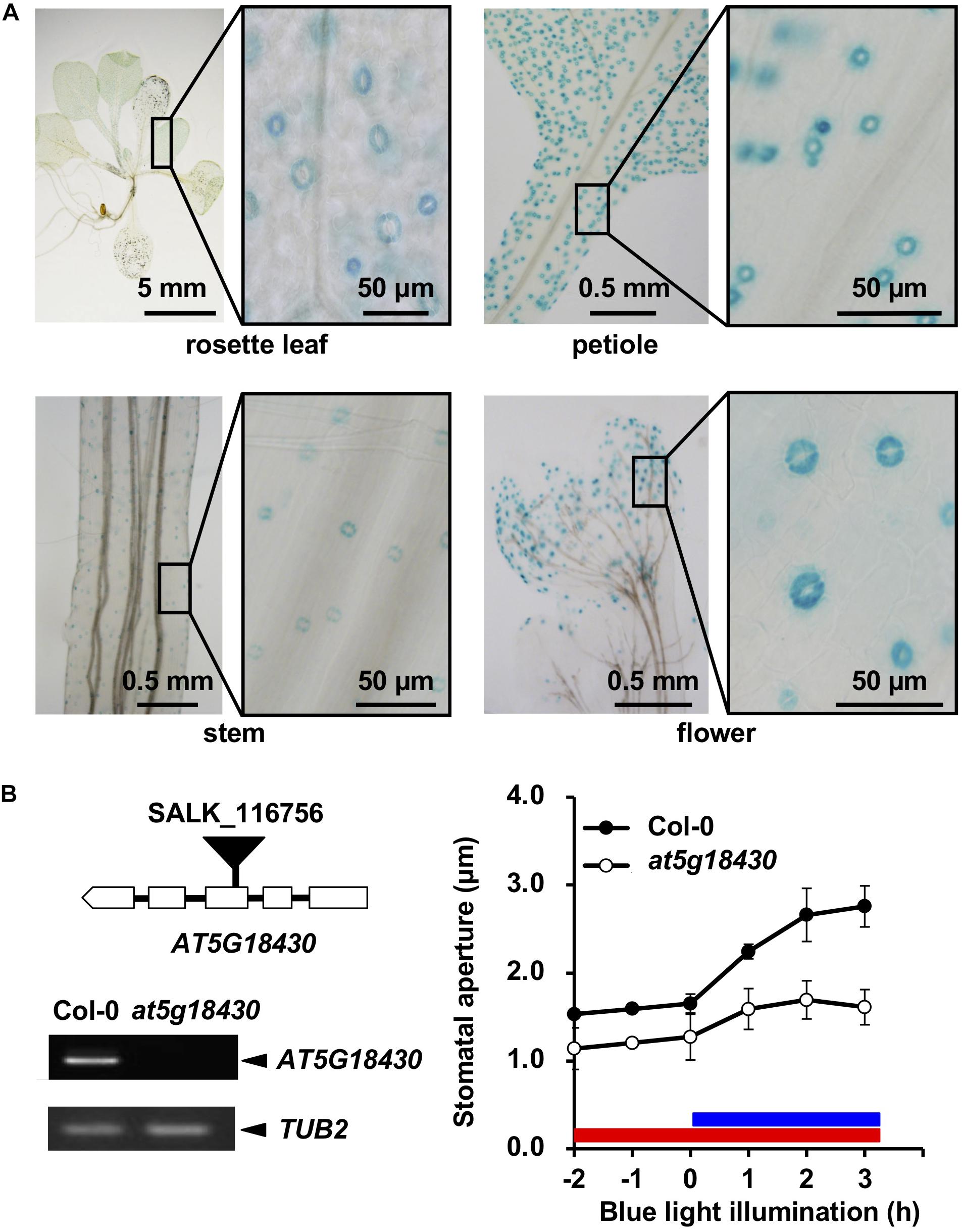
Figure 2. Expression of At5g18430 and the stomatal phenotype of the at5g18430 mutant. (A) Expression pattern of At5g18430 in GUS-reporter-bearing plants. (B) The stomatal phenotype of the at5g18430 mutant. Left: A schematic of the T-DNA insertion site in at5g18430 and the transcript levels in leaves. Right: blue light (BL)-induced stomatal opening in Col-0 and at5g18430 epidermal tissues. The red bar indicates the red light (RL) illumination period (50 μmol m–2 s–1). The blue bar indicates the BL illumination period (10 μmol m–2 s–1). Epidermal tissues were incubated in 50 mM KCl, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5). Averages from three independent experiments are shown. Error bars: SDs (n = 3).
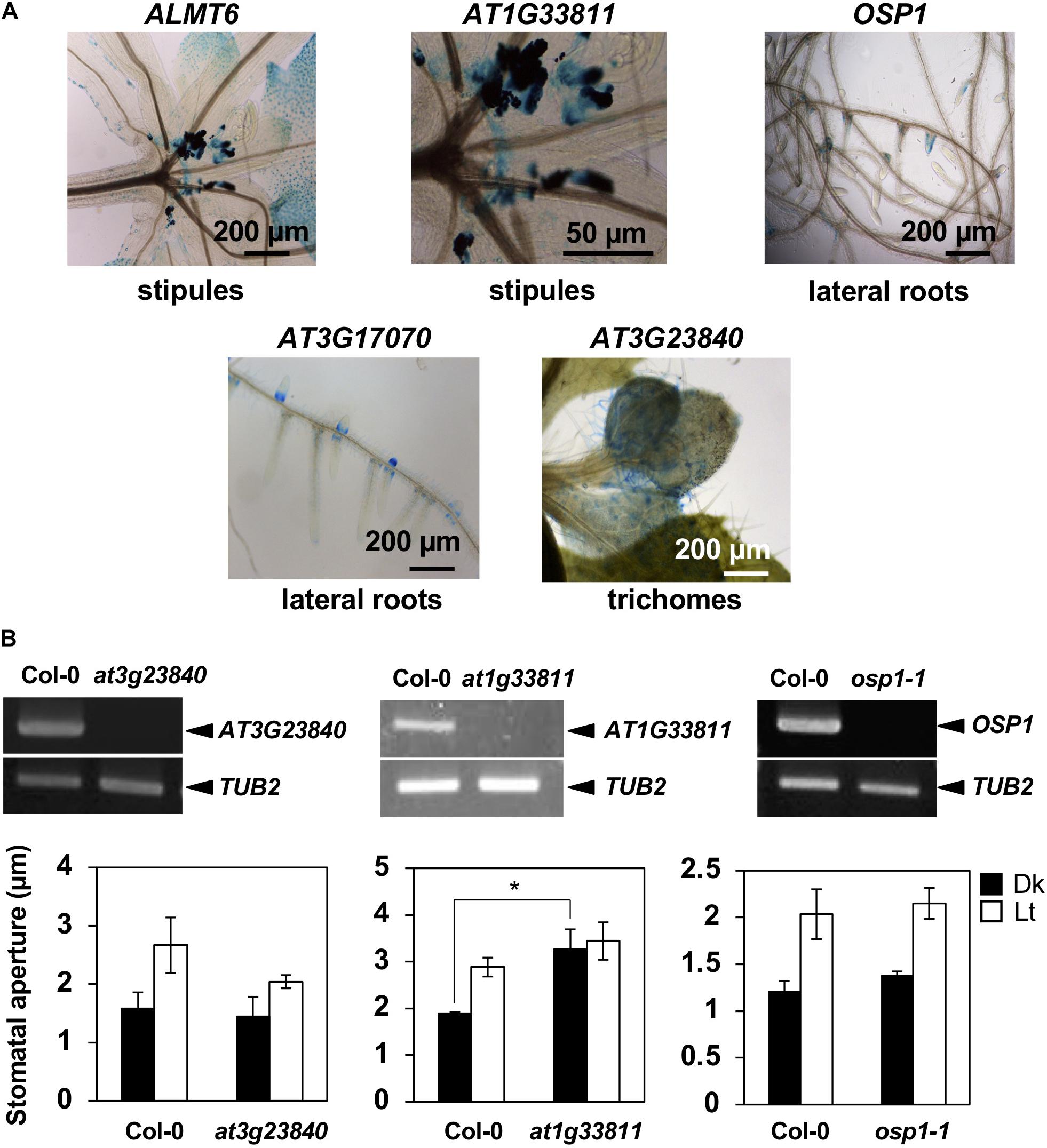
Figure 3. Expression patterns of GC-preferentially expressed genes and the stomatal phenotypes of knockout mutants. (A) Gene expression in tissues other than stomata as revealed by the GUS-reporter assay. (B) Upper panels show the RT-PCR of target genes. Lower graphs show BL-induced stomatal opening in at3g23840, at1g33811, and osp1-1 null mutants. Epidermal tissues (at3g23840 and at1g33811 mutants) or leaf disks (osp1-1 mutant) were incubated in 50 mM KCl, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5). Dark bars, dark treatment for 3 h (Dk); white bars, light treatment for 3 h (RL, 50 μmol m–2 s–1; BL, 10 μmol m–2 s–1) (Lt). Average values from three independent experiments are shown. Error bars: SDs (n = 3).
We prepared null mutants of OSP1, AT1G33811, AT3G23840, AT3G17070, and AT5G18430. Although we failed to obtain the knockout mutants of at3g17070 (SALK_121694), the null mutants of at5g18430 (SALK_116756) and at3g23840 (GABI_180G04) were impaired in BL-induced stomatal opening (Figure 2B) and light-induced stomatal opening (Figure 3B), respectively. Interestingly, the stomata of at1g33811 (GABI_492D11) null mutant were open even in the dark (Figure 3B). The null mutant of OSP1, i.e., osp1-1, exhibited a normal stable-status stomatal opening in the light (Figure 3B and Supplementary Figure 1), which is consistent with a previous report (Tang et al., 2020). Thus, the previously uncharacterized GC-specific genes AT1G33811, AT3G23840, and AT5G18430 may be involved in stomatal movement.
The ALMT6 is a vacuolar malate channel (Meyer et al., 2011). We explored the stomatal movements of the almt6-1 mutant in detail. The expression of ALMT9 in GCs is not altered in almt6-1 (Supplementary Figure 2). As shown in Figure 4A, the stomata of almt6-1 were slightly narrower than wild type under the dark and RL condition, as well as opened but less efficiently to a similar size of those of wild type on BL illumination. Less efficiency of almt6-1 stomatal opening in response to 10 μM FC, an activator of PM H+-ATPase, was more prominent compared with the case of BL-induced stomatal opening (Figure 4B). Stomatal apertures in the almt6-1 mutant were comparable to those in wild type when epidermal peels were treated with light or FC for more than 3 or 4 h, respectively (Figures 4A,B). Thus, ALMT6 may be required for stomatal opening induced by BL and FC. It is worthy of note that usually BL-insensitive mutants, such as phot1 phot2 double mutant, show completely insensitive phenotype to BL but open normally in response to FC (Kinoshita et al., 2001). The less efficient phenotype of stomatal opening in almt6-1 is very similar to a kincless mutant (Lebaudy et al., 2008) and aks1 aks2 mutant (Takahashi et al., 2013), which shows the low activity of PM inward K+ channels in GCs, suggesting that the deficient of ion transport for stomatal opening leads to less efficiency of stomatal opening.
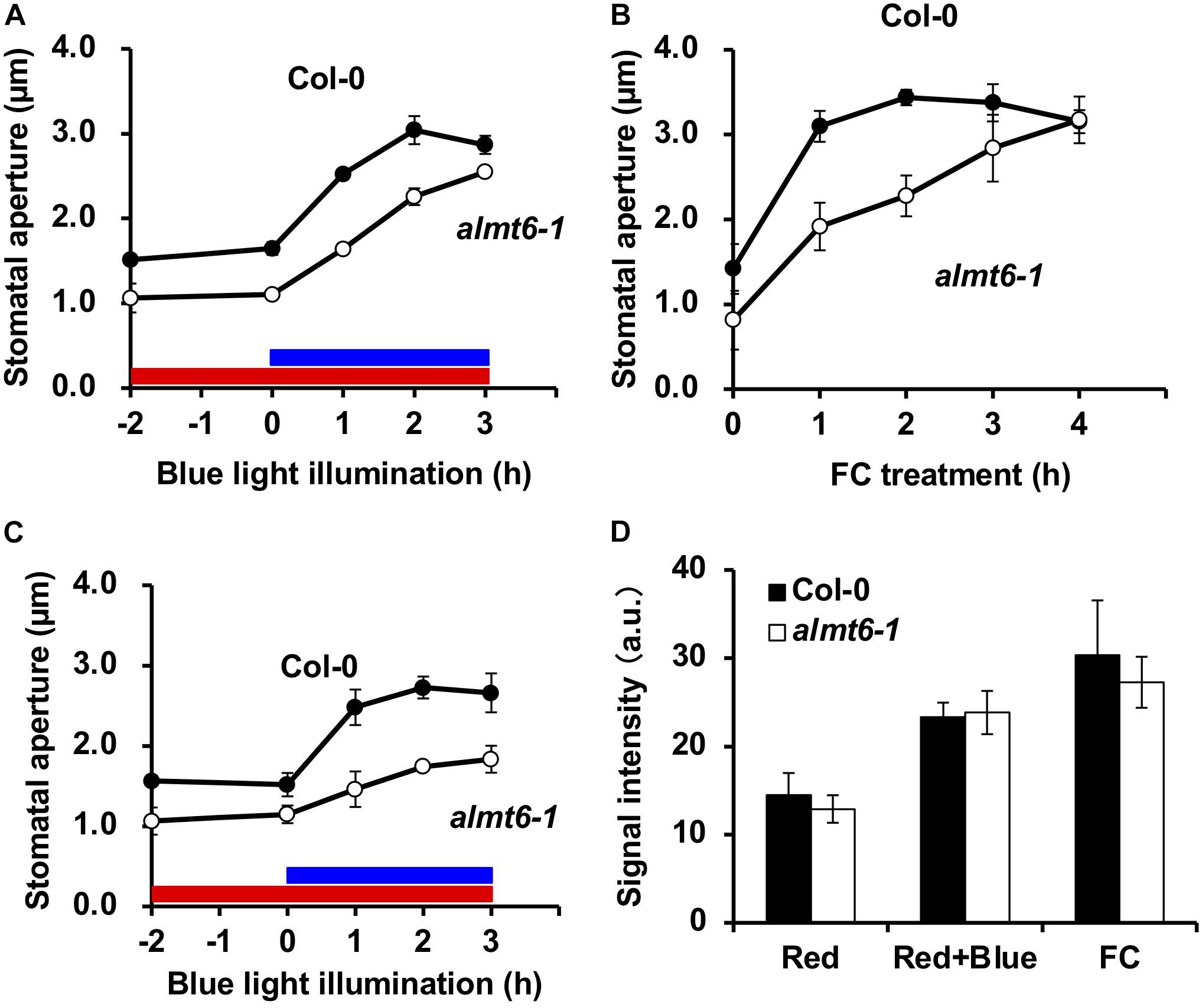
Figure 4. Stomatal phenotypes in wild-type Col-0 and almt6-1 mutant. (A) BL-induced stomatal opening in wild-type Col-0 and almt6-1 mutant epidermal tissues. Red bar: RL illumination period (50 μmol m–2 s–1). Blue bar: BL illumination period (10 μmol m–2 s–1). (B) Fusicoccin (FC)-induced stomatal opening in Col-0 and almt6-1 epidermal tissues. In panels (A,B), epidermal tissues were incubated in 50 mM KCl, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5) and then subjected to light and 10 μM FC treatments. (C) BL-induced stomatal opening in low Cl– (0.2 mM) buffer of Col-0 and almt6-1 epidermal tissues. Epidermal tissues were incubated in 50 mM potassium gluconate, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5). Averages from three independent experiments are shown. Error bars: SDs (n = 3). The other conditions are those in panel (A). (D) Immunohistochemical detection of BL- and 10 μM FC-induced phosphorylation of PM H+-ATPase in Col-0 and almt6-1 GCs. GC fluorescence was quantified using an anti-pThr antibody and Alexa Fluor 488-conjugated secondary antibody as described in section “Materials and Methods.” Bars: Averages from three independent experiments. Error bars: SDs (n = 3). a.u., arbitrary units.
The ALMT6 transports (principally) malate and fumarate but Cl– to a lesser extent (Meyer et al., 2011); we thus explored how Cl– affected BL-induced stomatal opening. The usual stomatal assay buffer contains 50 mM KCl, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5). To exclude exogenous Cl–, we evaluated stomatal opening in a buffer with 50 mM potassium gluconate, 0.1 mM CaCl2, and 10 mM Mes-BTP (pH 6.5). Gluconate does not readily cross the PM. Figure 4C shows that the 3-h BL-induced stomatal opening at a low [Cl–] was impaired in the almt6-1 mutant in terms of both speed and amplitude.
Both BL and FC induce the phosphorylation of the penThr of PM H+-ATPases, in GCs, which provides a driving force for stomatal opening (Inoue and Kinoshita, 2017). Thus, we immunohistochemically investigated the effects of BL and FC on the phosphorylation status of GC PM H+-ATPase; such phosphorylation was not impaired in the almt6-1 mutant (Figure 4D). The amount of PM H+-ATPase in almt6-1 under BL and FC was comparative to that in wild type (Supplementary Figure 3). The almt6 mutation did not affect PM H+-ATPase phosphorylation and amount in response to BL and FC.
In this study, we identified 10 genes including SLAC1, Cation/H+ Exchanger AtCHX20, GDSL lipases OSP1 and ALMT6, preferentially expressed in GCs based on the analyses of public resources, RT-PCR, and promoter GUS assay (Figures 1–3). Of these, AT1G33811, AT3G17070, AT3G23840, and AT5G18430 have not been functionally characterized in stomata. Among these four genes, three genes, namely, AT1G33811, AT3G23840, and AT5G18430, were found involved in the stomatal movement (Figures 2, 3). Remarkably, two of them, AT1G33811 and AT5G18430, are members of the GDSL family of serine esterases/lipases (Akoh et al., 2004), indicating the importance of this family in regulating stomatal movement. Tang et al. (2020) found that a GDSL lipase, i.e., OSP1, is preferentially expressed in GCs, and osp1 mutants showed low stomatal conductance and high leaf temperature due to a high percentage of occluded stomata. Detail biological and/or biochemical analyses revealed that OSP1 is required for wax biosynthesis and proper formation of the stomatal outer cuticular ledge (Tang et al., 2020). Interestingly, osp1 mutants were also impaired in abscisic acid (ABA)-induced stomatal closure, indicating a potential role of OSP1 in stomatal movement (Tang et al., 2020). It would be very interesting to investigate whether AT1G33811 and AT5G18430 have similar functions as OSP1. AT3G23840, previously named as CER26-like, is probably related to very long-chain fatty acid metabolism (Pascal et al., 2013), and a knockout mutant, at3g23840, showed reduced light-induced stomatal opening (Figure 3B). Future study is needed to provide a detailed mechanism mediated by those genes in stomatal movement.
The detailed analyses of expression pattern by GUS-reporter assay of ALMT6, OSP1, AT1G33811, AT3G17070, AT3G23840, and AT5G18430 revealed that promoter regions, i.e., 3-kb upstream of the start codon, drive GC-preferential gene expression with AT5G18430 promoter showing the most GC-selective property (Figures 1–3). The 1-kb upstream region of the AT3G23840 start codon was promoter-active in flowers (Pascal et al., 2013). The 3-kb promoter region of AT3G23840 studied here showed strong GC signals in addition to flower (Figure 1B). The 3-kb promoter region of ALMT6 studied here was more GC-specific than a 1.8-kb region used previously, which exhibited strong activities in floral tissues, such as sepals, petals, and anthers (i.e., not only GCs; Meyer et al., 2011) (Figures 1C, 3A). These results suggest that specific elements and their patterns determine the GC-specific activity of promoters. So far, several GC-preferentially expressed genes, such as GC1 (Yang et al., 2008) and MYB60 (Cominelli et al., 2005), were reported. However, these genes were out of our strict criteria, indicating that the genes reported in this study are more specific in GCs (Supplementary Figure 4). Cell type-specific promoters are important tools to study gene function and of great application potential (Imlau et al., 1999; Yang et al., 2008; Wang et al., 2014). The promoters identified in this study thus add the options for promoter engineering for GC-specific gene regulation.
Both ALMT6 and ALMT9 were initially identified as channels mediating malate accumulation in vacuoles (Kovermann et al., 2007; Meyer et al., 2011). Later, ALMT9 was shown to be a Cl– channel regulated by cytosol malate and to be required for the light-induced stomatal opening (De Angeli et al., 2013). As shown in Figures 4A,B, almt6-1 mutant opened stomata with less efficiency in response to BL illumination and FC treatment, suggesting that ALMT6 is also required for BL- and FC-induced stomatal opening. Notably, almt6-1 mutant showed significant impairment in BL-induced stomatal opening under low Cl– condition (Figure 4C), suggesting that ALMT6, such as ALMT9, also contributes Cl– influx to vacuole during stomatal opening. Since both almt6 and almt9 single mutants are impaired in the light-induced stomatal opening and there is no compensation of ALMT9 expression in almt6 (Supplementary Figure 2), it is possible that ALMT6 and ALMT9 function in an additive and/or cooperative manner. In the cooperative mode, a bold hypothesis is that ALMT6 and ALMT9 form heteromeric channels mediating anion accumulation in GC vacuoles as they were shown to form tetramer channels in heterosystems (Zhang et al., 2013). Future electrophysiological and genetic studies such as phenotyping using almt6 almt9 double mutant are needed to clarify the contribution of ALMT6 and ALMT9 in stomatal opening.
In this study, we identified preferentially expressed genes in GCs and found that some uncharacterized genes are involved in stomatal movement. Especially, to our knowledge, AT5G18430 shows the most specific expression in GCs. In addition, we showed evidence that ALMT6 is important for BL- and FC-induced stomatal opening. Further detailed analyses of the uncharacterized GC-specific genes will provide novel understandings for stomatal movement.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
WY, SK, and TK designed the experiments. WY, SK, YH, HJ, TO, KK, and TK performed the experiments. WY, SK, YH, and TK wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Grants-in-Aid for Scientific Research from MEXT (Nos. 15H05956, 20H05687, and 20H05910 to TK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Koji Takahashi and Shin-ichiro Inoue for providing technical advice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.744991/full#supplementary-material
Akoh, C. C., Lee, G., Liaw, Y., Huang, T., and Shaw, J. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552. doi: 10.1016/j.plipres.2004.09.002
Ando, E., and Kinoshita, T. (2018). Red light-induced phosphorylation of plasma membrane H+-ATPase in stomatal guard cells. Plant Physiol. 178, 838–849. doi: 10.1104/pp.18.00544
Barbier-Brygoo, H., De Angeli, A., Filleur, S., Frachisse, J. M., Gambale, F., Thomine, S., et al. (2011). Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 62, 25–51. doi: 10.1146/annurev-arplant-042110-103741
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200.
De Angeli, A., Zhang, J., Meyer, S., and Martinoia, E. (2013). AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 4:1804.
Hashimoto, M., Negi, J., Young, J., Israelsson, M., Schroeder, J. I., and Iba, K. (2006). Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 8, 391–397. doi: 10.1038/ncb1387
Hayashi, M., Inoue, S., Takahashi, K., and Kinoshita, T. (2011). Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in Stomatal Guard Cells. Plant Cell Physiol. 52, 1238–1248. doi: 10.1093/pcp/pcr072
Hayashi, Y., Nakamura, S., Takemiya, A., Takahashi, Y., Shimazaki, K., and Kinoshita, T. (2010). Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol. 51, 1186–1196. doi: 10.1093/pcp/pcq078
Horrer, D., Flütsch, S., Pazmino, D., Matthews, J. S. A., Thalmann, M., Nigro, A., et al. (2016). Blue light induces a distinct starch degradation pathway in guard cells for stomatal opening. Curr. Biol. 26, 362–370. doi: 10.1016/j.cub.2015.12.036
Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322. doi: 10.2307/3870862
Inoue, S., and Kinoshita, T. (2017). Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiol. 174, 531–538. doi: 10.1104/pp.17.00166
Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660. doi: 10.1038/414656a
Kovermann, P., Meyer, S., Hörtensteiner, S., Picco, C., Scholz-Starke, J., Ravera, S., et al. (2007). The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 52, 1169–1180. doi: 10.1111/j.1365-313x.2007.03367.x
Lebaudy, A., Vavasseur, A., Hosy, E., Dreyer, I., Leonhardt, N., Thibaud, J. B., et al. (2008). Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 5271–5276. doi: 10.1073/pnas.0709732105
Meyer, S., Mumm, P., Imes, D., Endler, A., Weder, B., Al-Rasheid, K. A. S., et al. (2010). AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63, 1054–1062. doi: 10.1111/j.1365-313x.2010.04302.x
Meyer, S., Scholz-Starke, J., De Angeli, A., Kovermann, P., Burla, B., Gambale, F., et al. (2011). Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67, 247–257. doi: 10.1111/j.1365-313x.2011.04587.x
Munemasa, S., Hauser, F., Park, J., Waadt, R., Brandt, B., and Schroeder, J. I. (2015). Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28, 154–162. doi: 10.1016/j.pbi.2015.10.010
Murata, Y., Mori, I. C., and Munemasa, S. (2015). Diverse stomatal signaling and the signal integration mechanism. Annu. Rev. Plant Biol. 66, 369–392. doi: 10.1146/annurev-arplant-043014-114707
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Negi, J., Matsuda, O., Nagasawa, T., Oba, Y., Takahashi, H., Kawai-Yamada, M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486. doi: 10.1038/nature06720
Okumura, M., Inoue, S., Kuwata, K., and Kinoshita, T. (2016). Photosynthesis activates plasma membrane H+-ATPase via sugar accumulation. Plant Physiol. 171, 580–589. doi: 10.1104/pp.16.00355
Padmanaban, S., Chanroj, S., Kwak, J. M., Li, X., Ward, J. M., and Sze, H. (2007). Participation of endomembrane cation/H+ exchanger AtCHX20 in osmoregulation of guard cells. Plant Physiol. 144, 82–93. doi: 10.1104/pp.106.092155
Pascal, S., Bernard, A., Sorel, M., Pervent, M., Vile, D., Haslam, R. P., et al. (2013). The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 73, 733–746. doi: 10.1111/tpj.12060
Santelia, D., and Lawson, T. (2016). Rethinking guard cell metabolism. Plant Physiol. 172, 1371–1392. doi: 10.1104/pp.16.00767
Sasaki, T., Mori, I. C., Furuichi, T., Munemasa, S., Toyooka, K., Matsuoka, K., et al. (2010). Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 51, 354–365. doi: 10.1093/pcp/pcq016
Shimazaki, K., Doi, M., Assmann, S. M., and Kinoshita, T. (2007). Light regulation of stomatal movement. Annu. Rev. Plant Biol. 58, 219–247. doi: 10.1146/annurev.arplant.57.032905.105434
Takahashi, Y., Ebisu, Y., Kinoshita, T., Doi, M., Okuma, E., Murata, Y., et al. (2013). bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 6:ra48. doi: 10.1126/scisignal.2003760
Tang, J., Yang, X., Xiao, C., Li, J., Chen, Y., Li, R., et al. (2020). GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 228, 1880–1896. doi: 10.1111/nph.16741
Toh, S., Inoue, S., Toda, Y., Yuki, T., Suzuki, K., Hamamoto, S., et al. (2018). Identification and characterization of compounds that affect stomatal movements. Plant Cell Physiol. 59, 1568–1580. doi: 10.1093/pcp/pcy061
Tomiyama, M., Inoue, S., Tsuzuki, T., Soda, M., Morimoto, S., Okigaki, Y., et al. (2014). Mg-chelatase I subunit 1 and Mg-protoporphyrin IX methyltransferase affect the stomatal aperture in Arabidopsis thaliana. J. Plant Res. 127, 553–563. doi: 10.1007/s10265-014-0636-0
Vahisalu, T., Kollist, H., Wang, Y. F., Nishimura, N., Chan, W. Y., Valerio, G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491. doi: 10.1038/nature06608
Wang, Y., Noguchi, K., Ono, N., Inoue, S., Terashima, I., and Kinoshita, T. (2014). Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc. Natl. Acad. Sci. U.S.A. 111, 533–538. doi: 10.1073/pnas.1305438111
Yang, Y., Costa, A., Leonhardt, N., Siegel, R. S., and Schroeder, J. I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4:6.
Keywords: ALMT6, Arabidopsis, blue light, malate, proton pump, stomatal opening
Citation: Ye W, Koya S, Hayashi Y, Jiang H, Oishi T, Kato K, Fukatsu K and Kinoshita T (2021) Identification of Genes Preferentially Expressed in Stomatal Guard Cells of Arabidopsis thaliana and Involvement of the Aluminum-Activated Malate Transporter 6 Vacuolar Malate Channel in Stomatal Opening. Front. Plant Sci. 12:744991. doi: 10.3389/fpls.2021.744991
Received: 21 July 2021; Accepted: 13 September 2021;
Published: 08 October 2021.
Edited by:
Tatsuru Masuda, The University of Tokyo, JapanReviewed by:
June M. Kwak, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South KoreaCopyright © 2021 Ye, Koya, Hayashi, Jiang, Oishi, Kato, Fukatsu and Kinoshita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshinori Kinoshita, a2lub3NoaXRhQGJpby5uYWdveWEtdS5hYy5qcA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.