
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 October 2021
Sec. Plant Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.743568
Polyamines have been implicated in ameliorating the detrimental effects of drought and saline conditions on plant growth and development. The independent impact of these two abiotic stresses on polyamine (PA) biosynthesis, catabolism, and homeostasis, as well as on their transcript abundance in tomato leaves, is presented here. We show that the total levels of putrescine (PUT), spermidine (SPD), and spermine (SPM) increase up to 72 h during drought and up to 48 h during salinity stress before their precipitable drop thereafter. Thus, tomato plants maintain survivability to drought as well as salinity stress for up to 3 and 2 days, respectively. Independent multivariant analyses of drought and salinity stress kinetic data separately showed a closer association with levels of free, conjugated, and bound forms of SPD and SPM, but not with free or bound PUT. However, combined multivariant analyses showed a closer association of free SPD, conjugated SPD, and bound SPD with both stresses; SPD-bound and SPM conjugated with drought; and free SPM and conjugated PUT with salinity stress, respectively. PA biosynthesis genes, ARG1, SPDS1, and SAMDc3, segregated with drought and SPDS2 with salinity stress. PA catabolic genes CuAO4-like and PAO4 were associated with drought and salinity stresses, respectively, suggesting differential involvement of PA biosynthesis and catabolic genes in drought and salinity stresses. Pearson correlation indicated mostly positive correlations between the levels of free, conjugated, and bound forms of PUT, SPD, and SPM under drought and salinity stress. However, negative correlations were mostly seen between the levels of various forms of the PAs and their biosynthesis/catabolic genes. Levels of different PA forms had a twofold higher negative correlation during drought as compared to salinity stress (66 vs. 32) and with transcript levels of PA biosynthesis and catabolic genes. Transcripts of light-harvesting chlorophyll a/b-binding genes were generally positively associated with different forms of PAs but negatively to carbon flow genes. Most of the PA biosynthesis genes were coordinately regulated under both stresses. Collectively, these results indicate that PAs are distinctly regulated under drought and salinity stress with different but specific homologs of PA biosynthesis and catabolic genes contributing to the accumulation of free, conjugated, and bound forms of PAs.
Abiotic environmental factors, such as drought and salinity, are significant plant stressors with a major impact on plant development and productivity, thus, causing serious agricultural yield losses (Flowers, 2004; Godfray et al., 2010; Tester and Langridge, 2010; Golldack et al., 2014). The complex regulatory processes of plant adaptation to drought and salt involve control of water flux and cellular osmotic adjustment via biosynthesis of osmoprotectants (Hasegawa et al., 2000; Flowers, 2004; Munns, 2005; Ashraf and Akram, 2009). Additionally, drought and salinity have major detrimental impacts on the cellular energy supply and redox homeostasis that are balanced by global reprogramming of plant primary metabolism and altering cellular architecture (Chen et al., 2005; Baena-González et al., 2007; Jaspers and Kangasjärvi, 2010; Miller et al., 2010; Zhu et al., 2010). One important class of cellular metabolites acting as osmoreceptors is polyamines (PAs). PAs as osmo-protectants have been shown to protect plants against adverse environmental conditions (Alcázar et al., 2010; Mattoo et al., 2015b; Gong et al., 2016; Handa et al., 2018), notably against drought and salinity in various crops and crop models (Capell et al., 2004; Cona et al., 2006; Kusano et al., 2008; Alcázar et al., 2010; Minocha et al., 2014; Mattoo et al., 2015b).
Polyamines are found either as free, conjugated, bound, or non-covalently conjugated (NCC) forms in nature [reviewed by Chen et al. (2019), and references there in]. Free PAs covalently bound with biomacromolecules, such as proteins, nucleic acids, uronic acids, or lignin by ionic and hydrogen bonds, are known as bound PAs (Chen et al., 2019). In the physiological pH range, free-PAs are fully protonated and positively charged, and can electrostatically combine with negatively charged biomacromolecules (acidic proteins, membrane phospholipids, and nucleic acids) in the organism and known as conjugated PAs (Igarashi and Kashiwagi, 2015; Chen et al., 2019). The conjugated PAs have a wide range of biological functions in plant growth and development. They have been associated with the regulation of enzyme activity, DNA replication, gene transcription, cell division, and membrane stability (Chen et al., 2019). However, little is known about the association of different polyamine forms with abiotic stresses, particularly in tomatoes.
Polyamines (SPM, SPD, and PUT) were shown to regulate the stomata pore opening and closing to control water loss from plants during drought conditions (Liu et al., 2000). In alfalfa, PUT treatment was shown to improve seed germination and increase the growth indexes under polyethylene glycol (PEG 4000)-mediated drought stress both in vitro and in a pot experiment (Zeid and Shedeed, 2006). Arabidopsis mutant acl5/Spm, which lacks SPM, is hypersensitive to high salt and drought and was rescued by SPM pretreatment but not by PUT or SPD, suggesting that the drought hypersensitivity of the mutant is due to SPM deficiency (Yamaguchi et al., 2007). Arabidopsis ADC2 deletion mutant was found extremely sensitive to salt stress, which was alleviated by applying exogenous PUT (Naka et al., 2010). Furthermore, SPM was found strongly associated with drought resistance in apple (Liu et al., 2010) and cherry tomatoes (Montesinos-Pereira et al., 2014). Moreover, SPD and SPM relieved the inhibitory effects of drought stress and promoted grain filling and drought resistance in wheat, while PUT had the opposite effect (Liu et al., 2016). The application of different types and varying concentrations of exogenous PAs was shown to alleviate the effects of NaCl stress in Brassica juncea and strawberry seedlings (Verma and Mishra, 2005; Li et al., 2010). Similarly, the application of SPM and SPD were found to result in increased reactive oxygen metabolism and photosynthesis, which, in turn, improved plant growth and reduced the inhibitory effects of salt stress (Meng et al., 2015; Baniasadi et al., 2018). Interestingly, recent information has indicated that SPD and thermospermine (TSPM) share molecular functions related to quality control pathways for tightly regulated mRNAs at the level of translation [Poidevin et al. (2019) and references therein].
The plant polyamine metabolic pathway is mediated by several enzymes in the sequential synthesis of PUT, SPD, and SPM (Mattoo et al., 2015a). Arginine (Arg) decarboxylase (ADC; EC 4.1.1.19) converts Arg to PUT while, alternatively, Ornithine (Orn) decarboxylase (ODC; EC 4.1.1.7) converts Orn to PUT (Mattoo et al., 2015a; Tsaniklidis et al., 2016). Arginase (ARG; EC 3.5.3.1) balances both pathways by converting Arg to Orn. Higher PAs, SPD, and SPM are synthesized from PUT by spermidine synthase (SPDS; EC 2.5.1.16) and spermine synthase (SPMS; EC 2.5.1.22), respectively. Decarboxylation of SAM by SAM-decarboxylase (SAMDc; EC 4.1.1.50) adds amino propyl groups in a sequential manner to PUT and SPD for the synthesis of SPD and SPM, respectively. A sequentially back conversion of PAs from SPM to SPD to PUT under specific conditions/processes is achieved by PA oxidase (PAO; EC 1.5.3.11) and copper-containing amine oxidase (CuAO; EC 1.4.3.6). Such interconversion of PAs is thought to contribute to the tight regulation of PAs homeostasis (Møller and McPherson, 1998; Moschou et al., 2012; Tavladoraki et al., 2016), which results in the production of the signaling molecule H2O2 as a byproduct (Sebela et al., 2001; Cona et al., 2003, 2006; Angelini et al., 2008; Moschou et al., 2008). Differential activation of PA metabolic pathway genes in tomato leaves under heat and cold stress conditions has also been determined (Upadhyay et al., 2020).
Novel approaches that utilized genetic engineering to accumulate PAs in agronomic and model plants, including tobacco, rice, tomato, Arabidopsis, pear, and potato, and overexpressed SAM decarboxylase were found to be tolerant against given stresses, such as salt, osmotic, and heat (Wi et al., 2006; Alcázar et al., 2010), while overexpression of spermidine synthase led to tolerance against drought, salt, and oxidative stresses (Kasukabe et al., 2004, 2006; He et al., 2008; Wen et al., 2008; Alcázar et al., 2010; Neily et al., 2011). Similarly, transgenic Lotus tenuis plants expressing ADC adjusted better to osmotic adjustment (5.8-fold) under salinity stress (Espasandin et al., 2018).
A plant leaf is a photosynthetically active aerial tissue mainly responsible for carbon assimilation activities and is highly prone to environmental changes, which in turn, affect the yield and productivity of the whole plant. How PA biosynthesis/catabolism pathways are modulated in tomato leaf tissue in response to drought or salinity stress and how they coordinate with specific changes in PA levels in their various forms (free, conjugated, and bound) are yet to be fully determined. Here, we present genetic data involving the quantification of 18 tomato gene transcripts involved in PA biosynthesis and catabolism in relation to drought and salt stress of leaves, and expression dynamics of other 22 genes that include nuclear-encoded photosynthetic and plastid-encoded protein genes, carbon fixation-encoding genes, together with stress-specific marker genes to establish correlative indices with PA abundance. The results obtained outline specific gene flow responses to conditions of drought and salt stress and influence differential PA abundance.
Wild-type tomato (Solanum lycopersicum cv. Ailsa Craig) plants were grown in a temperature-controlled (22°C ± 3) greenhouse under natural light conditions in metro-mix 360 soil (sun-grow). After transplanting, plants were grown for 4 weeks (28 days) before initiating stress treatments. All the harvested samples were immediately frozen in liquid nitrogen and stored at −80°C until used.
The transplanted tomato plants were grown in soil for 28 days and used for stress experiments. Salt and drought treatments were given in a temperature-controlled (22°C ± 3) greenhouse as described previously (Upadhyay et al., 2019). Briefly, drought treatment involved withholding watering of plants for 7 days (168 h). Leaf samples were collected at 0, 24 (day 1), 48 (day 2), 72 (day 3), and 168 h (day 7) after initiating the drought treatment. Control samples were also collected at indicated time points. Salt (200-mM NaCl) was used for salinity stress. The plants were daily irrigated with 200 ml of NaCl (200 mM) solution. Leaf samples of salt-treated plants were collected at 0, 2, 6, 48, and 96 h, along with control samples.
Gene expression of tomato PA metabolic pathway enzymes was carried out (listed in Supplementary Table S1). Briefly, for PUT accumulation, two arginase-encoding genes (SlARG1, 2); two arginine decarboxylase-encoding genes (SlADC1, 2); agmatine iminohydrolase/deiminase (SlAIH); N-carbamoyl putrescine amidase (SlCPA); and two ornithine decarboxylase-encoding genes (SlODC1, 2) were selected; for SPD and SPM accumulation, three S-adenosylmethionine decarboxylase-encoding genes (SlSAMDc1, 2, and 3); two spermidine synthase-encoding genes (SlSPDS1, 2); and one spermine synthase-encoding (SPMS) gene were chosen for transcript analysis. Concurrently, four catabolic genes, including two flavin-dependent polyamine oxidases (SlPAO4-like and SlPAO2), and two copper-dependent amine oxidases (SlCuAO4 and SlCuAO-like) were chosen to study PA catabolism along with biosynthesis (Supplementary Table S1). To study the flow of ornithine toward the proline metabolic pathway, a single copy ornithine aminotransferase-encoding gene (SlOAT1) was chosen to address the PA pathway shift toward the proline pathway in drought and salinity conditions (Upadhyay et al., 2020).
For drought stress responses, eight genes, namely, SlDREB1 (Solyc06g050520) and SlDREB2 (Solyc05g052410); SlNCED1 (Solyc07g056570) and SlNCED2 (Solyc08g016720); SlRD29A (Solyc03g025810) and SlDELLA (Solyc11g011260) genes; SlWIRKY57 (Solyc05g012500) and SlJUB1 (Solyc05g021090) served as drought marker genes. Three salt responsive genes, SlSOS1 (Solyc01g005020), SlNHX3 (Solyc01g067710), and SlNHX4 (Solyc01g098190) were used as salt-stress-marker genes. Sequences were extracted from the tomato genome database [International Tomato Genome Sequencing Consortium (SGN1) as Arabidopsis homologs].
The effect of stress on the carbon assimilation of the plant was tested using known genes that encode proteins involved in photosynthesis, such as the nuclear-encoded light-harvesting chlorophyll a/b-binding protein genes [SlLhcb1 (Solyc03g005770.1), SlLhcb2 (Solyc12g006140.1), SlLhcb3 (Solyc07g063600.2), SlLhcb4 (Solyc09g014520.2), SlLhcb5 (Solyc06g063370.2), and SlLhcb6 (Solyc01g105050.2)], as well as plastid-encoded photosystem II (PSII) protein genes SlpsbA and SlaccD. Tomato SlpsbA gene [encoding the D1 protein of PSII (NC_007898.3)] and accD gene (encoding the beta-carboxyl transferase subunit of acetyl-CoA carboxylase) information were extracted from tomato chloroplast genome (NC_007898.3). SlLhcb1-6 genes, phosphoenolpyruvate carboxylase (SlPEPC) (AJ243417.1/Solyc07g062530.2) and isocitrate dehydrogenase (SlICDH) (XM_010314428.2/Solyc11g011930.1.1) genes were used as markers for the carbon flow during drought/salt stress (Supplementary Table S2).
Frozen tomato leaf tissue was ground to a fine powder (100 mg) to extract total RNA using Plant RNeasy kit (Qiagen, Hilden, Germany). DNase (Qiagen) was used to remove genomic DNA, followed by a cleanup with a RNeasy mini kit (Qiagen). RNA quality was checked by spectrophotometer, and the RNAs with an A260/280 with ratios of 1.8–2 were subjected to agarose gel electrophoresis to ensure the presence of intact rRNA band before their selection for cDNA preparation (Upadhyay and Mattoo, 2018). RNA (2 μg) was used for cDNA synthesis using an iScriptadvanced cDNA synthesis kit (Bio-Rad, Hercules, CA, United States), followed by a 10-fold dilution for further use. Quantitative real-time PCR (qRT-PCR) was performed using Sso Advanced Universal SYBR Green Supermix (Bio-Rad) in a Bio-Rad cycler (CFX96 Bio-Rad Real-Time PCR machine). PCR conditions were sequentially 95°C for 5 min, 95°C for 15 s, and 60°C for 60 s (40 cycles), followed by melt curve analysis (Shukla et al., 2017). Gene expression was quantified using the ΔΔCT method (Livak and Schmittgen, 2001). Two reference genes (SlTIP41 and SlUBI3) were used for normalizing the expression of the target genes (Expósito-Rodríguez et al., 2008; Mascia et al., 2010). MIQE (Minimum Information for Publication of Quantitative real-time PCR Experiments) guidelines were followed for quantification of genes (Bustin et al., 2009). Relative fold changes were calculated as previously described (Shukla et al., 2017). Primer sequences for 42 genes used for qRT-PCR are listed in Supplementary Table S2. Relative qRT-PCR data represent average ± SD from a minimum of three independent biological replicates.
Freeze-dried tomato leaf material was extracted and dansylated as described previously (Torrigiani et al., 2012; Anwar et al., 2019) with some modifications. About 50 mg of each finely ground leaf sample was suspended in 800 μl of 5% ice-cold perchloric acid (PCA) and homogenized in a handheld homogenizer. After centrifugation at 20,000 g for 30 min at 4°C, the supernatant (free and conjugated PAs) and residue pellet (bound PAs) were collected separately. The 20,000-g pellet was washed two times with 5% PCA and resuspended in 800 μl of 5% cold PCA. An aliquot (0.5 ml) of supernatant and suspended pellet was hydrolyzed in an equal volume of 6-N HCl for 18 h at 110°C to release conjugated and bound insoluble PAs, respectively. About 100-μl aliquots of soluble supernatant before and after hydrolysis, and of the suspended pellet were then quantified (soluble, conjugated, and bound PAs, respectively). To each aliquot, saturated sodium carbonate (200 μl) and 1,7-heptanediamine (400 μl, as an internal standard) were added and then dansylated with dansyl chloride for 60 min at 60°C in the dark. Dansylation was terminated by adding 100-μl proline and incubating the reaction mixture for 30 min at 60°C. Other details were the same as described earlier (Anwar et al., 2019). For PAs recovery and calibration curves, authentic PA standards (Sigma-Aldrich, St. Louis, MO, United States) were used as control. PAs were integrated and quantified using Millennium 4.0 Chromatography Manager software from Waters Corporation. Samples of PCA-soluble, PCA-soluble hydrolyzed with HCl, and PCA-insoluble after hydrolysis were quantified, and are designated as free, free + conjugated, and bound forms of each PA, respectively (Anwar et al., 2019).
Data were examined with Graph-Pad Prism (version 8.2.1) statistical software package, and two-way ANOVA (a mixed model) with recommended Geisser–Greenhouse Correction was performed for deriving statistical significance. Multiple comparisons against “oh” within data points were performed using Tukey/Dunnett test. Differences among treatments were considered significant at p < 0.05. PCA, correlation, and Cytoscape analyses were performed using XLSTAT, Excel, and Cytoscape (Shannon et al., 2003) programs, respectively. The correlation coefficients were determined using the Microsoft EXCEL program and analyzed using the Expression Correlation App of Cytoscape program (Shannon et al., 2003). All values were normalized as% of the initial value (0-time point) before determining the correlation coefficient r. Gene abbreviations are the same as in Supplementary Tables S1, S2. Only correlations > 0.8 were plotted. All plotted correlations were significant at a p-value of 0.001.
Figure 1A shows the changes in total (Free + Conjugate + Bound) cellular content of PUT, SPD, and SPM and PAs during the increasing period of drought stress. The total cellular levels of PUT continued to decline with an increasing drought period, whereas the total cellular contents of SPD and SPM increased until 72 h of drought before showing precipitable declines after 168-h drought (Figure 1A). Collectively, these results indicate that the cellular metabolic activities of the PAs pathway continued up to at least 72 h after initiation of drought stress.
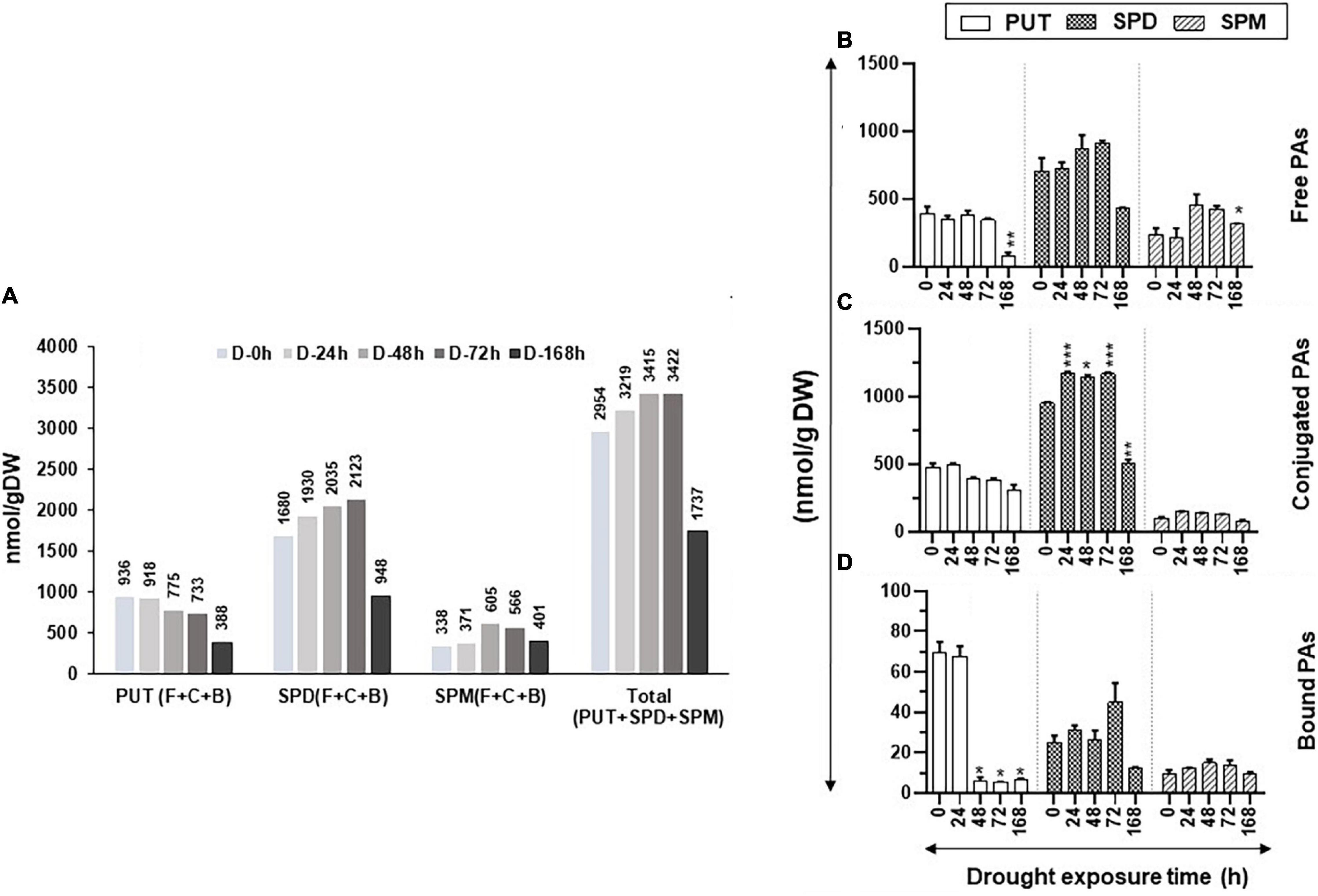
Figure 1. Changes in the levels of free, conjugated, and bound forms of putrescine (PUT), spermidine (SPD), and spermine (SPM) during drought stress in tomato leaves. The drought stress was imposed on tomato plants by withholding water. Leaf samples from a minimum of three plants were harvested at the indicated time periods and immediately frozen in liquid N2 and then stored at –80°C until analyzed. High-pressure liquid chromatography (HPLC) was used to determine the levels of free, bound, and conjugated forms of polyamines (PAs) as described in the Materials and Methods section. (A) Changes in the total amount of free, bound, and conjugated PUT, SPD, and SPM under drought stress; (B) free PUT, SPD, and SPM; (C) conjugated PUT, SPD, and SPM; (D) bound PUT, SPD, and SPM, respectively, during the increasing period of drought stress. Shown are mean + STE from three independent biological replicates. Statistical significance (p-value) was derived using the Graph prism pad program via Tukey test/t-test. *, **, *** indicate significance at p-value < 0.05, <0.005, and <0.0005, respectively.
The increasing period of drought stress did not significantly change the levels of the free PUT (PUT-F), except after prolonged drought stress, when its levels precipitously dropped at 168 h of drought (Figure 1B). Similar trends were seen for the SPD-F as its level declined after 168 h of drought (Figure 1B). The levels of SPM-F, however, showed a significant increase after 48 and 72 h of drought before its decline at 168 h of drought (Figure 1B). The conjugated PUT (PUT-C) levels steadily declined throughout the experimental period, but these changes were not significantly different at all sampling periods (Figure 1C). The SPD-C levels, however, significantly increased after 24, 48, and 72 h of drought and declined after 168 h of withholding water (Figure 1C). The levels of SPM-C did not show noticeable changes during the experiment (Figure 1C). The levels of bound PUT (PUT-B) steeply declined after 24 h of drought (Figure 1D) and, by 48, 72, and 168 h, the levels were insignificant (Figure 1D). The small contribution of PUT-B levels to total PUT did not appreciably affect total PUT (Figure 1A). We interpret the sharp decline in PUT-B to be possibly due to the onset of drought stress, leading to the breakdown of PUT-binding macromolecules. The levels of SPD-B and SPM-B did not appreciably change during the increasing period of drought. Taken together, these results suggest that only prolonged drought stress affects the levels of PAs, especially PUT-F and PUT-B. A significant increase in SPD-C but not in SPD-F after 24–72 h of drought stress indicates that plants maintain homeostasis for SPD-F during the drought stress by conjugating SPD. The higher levels of SPM-F but not SPM-C suggest a limited role for SPM conjugation during drought stress. Statistical differences for polyamine abundances during drought stress kinetics are shown in Supplementary Table S3.
Prolonged drought stress affected the accumulation of transcript levels of several PAs biosynthesis pathway genes. The steady state levels of SlARG1/2, SlAIH, SlSPDS1, and SlOAT1 transcripts increased significantly only after 168 h of drought stress (Figures 2A,D,F), whereas the steady state levels of SlODC2 and SlSPMS transcripts significantly increased after 24 h, continuing until 72 h before declining at 168 h of drought (Figures 2B,D). Levels of SlADC1, SlSAMdDc1/2, and SlODC1 steadily declined during the increasing period of drought stress (Figures 2B,C). Transcript levels of SlOAT1 SlARG1, SlAIH, SlSAMdc2, SlSPDS1, SlCuAO4, and SlCuAO4-like genes in PA metabolism had significant increases only after 168 h of drought. The molecular basis of their increase after 168 h is not clear. It is possible that these genes respond only upon prolonged drought, but this remains to be tested.
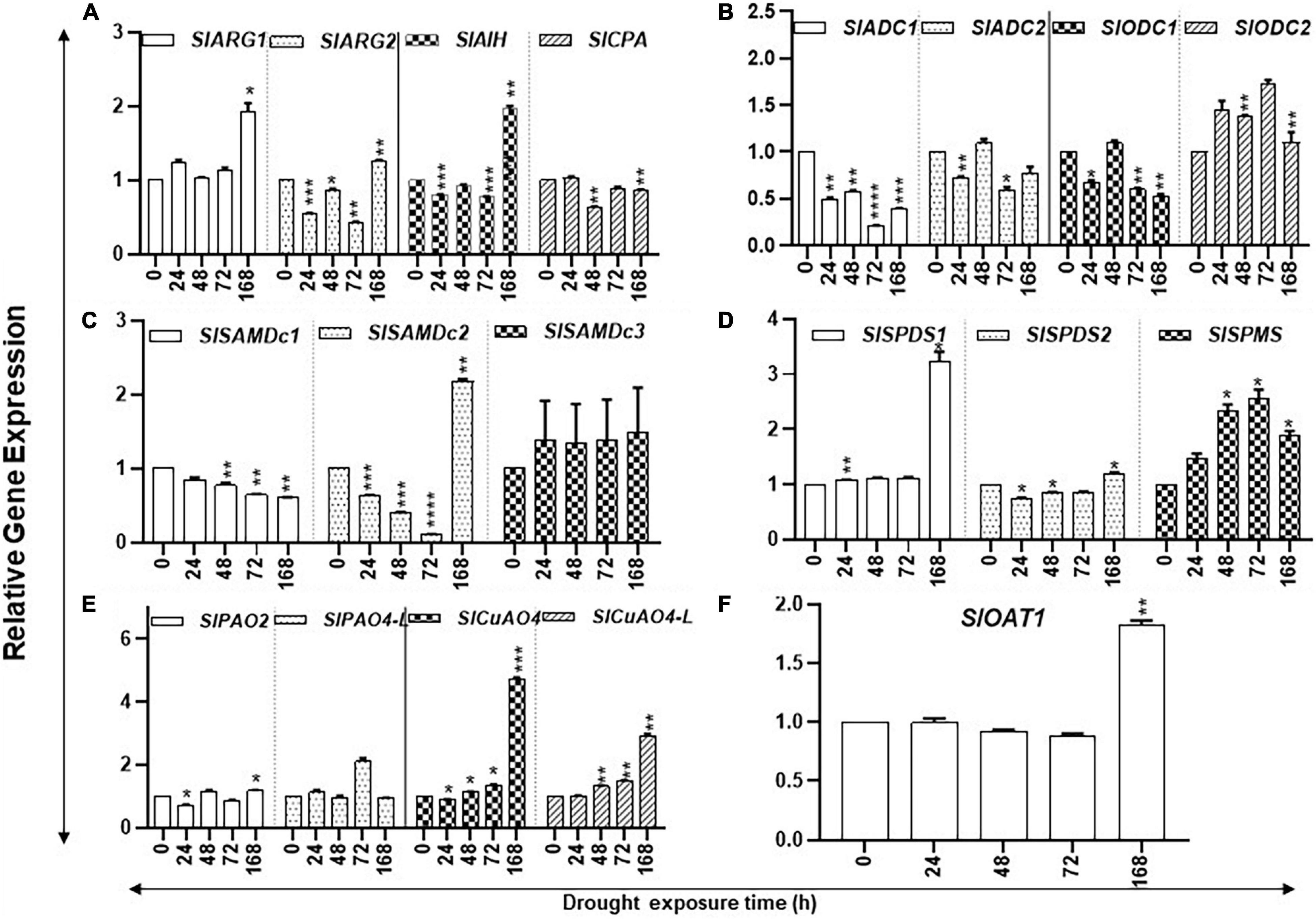
Figure 2. Kinetics of drought-induced expression of PA metabolic pathway genes in tomato leaves. Total RNAs were extracted from tomato leaves harvested after withholding water for 0, 24, 48, 72, and 168 h, respectively. The steady state levels of transcripts of various PAs biosynthesis and catabolism genes were determined by qRT-PCR as described in section “Materials and Methods.” Genes included were of PUT biosynthesis (SlARG1, 2, SlADC1, 2 SlODC1, 2, SlAIH, and SlCPA), SPD, and SPM biosynthesis (SlSAMDc1, 2, 3, SlSPDS1, 2, and SlSPMS); and catabolism (SlPAO2 and SlPAO4-Like; SlCuAO and SlCuAO4-like) pathways, respectively. SlUBI3 and SlTIP41 were used as reference genes. Other details were the same as in Figure 1. **** indicates significance at p-value of <0.00005.
The PAs catabolism pathway involves catabolism of PUT by copper amine oxidases (CuAO4 and CuAO4-like) and of SPD/SPM by flavin adenine dinucleotide (FAD)-dependent PA oxidases (PAO2 and PAO4-like) (Cona et al., 2006; Angelini et al., 2008; Tavladoraki et al., 2016). Expression of SlPAO2 remained low during the drought stress and significantly declined at 24 h, followed by an increase in 168-h samples (Figure 2E). The transcript levels of SlPAO4-like remained similar at most time points of kinetics, except for an increase after 72 h of drought. The steady state levels of SlCuAO4 and SlCuAO4-like gene transcripts significantly increased during the experiment and continued to increase up to 168 h of water withholding (Figure 2E). We interpret these results to suggest that tomato leaves exhibit noticeable changes in PA biosynthesis and catabolism until 72 h but undergo dramatic changes after prolonged drought stress. This has ramifications in the recovery of plants from drought stress as it is likely that plants have the potential to recover within 72 h of drought while prolonged drought stress becomes irreversible.
The study state levels of carbon assimilation genes during the increasing period of drought stress are shown in Figure 3. The transcript levels of six nuclear-encoded light-harvesting chlorophyll a/b-binding genes (SlLhcb1, SlLhcb2, SlLhcb3, SlLhcb4, SlLhcb5, and SlLhcb6) steadily declined during the 168-h course of water withholding (Figure 3A). The levels of these genes significantly declined after 24 h of drought and continued to decline during the prolonged 168-h drought. The two plastid-encoded photosystem II (PSII) genes, SlpsbA and SlaccD, significantly declined at 168 h of drought compared to control of the 0-h time point but transiently increased in the 48-h samples (Figure 3B). Patterns of transcript levels of SlPEPC (phosphoenolpyruvate carboxylase) and SlICDH (isocitrate dehydrogenase) genes were analyzed as markers for the carbon flow in response to drought stress (Figure 3C). The transcript levels of SlPEPC were significantly higher at 72 h and greatly increased by 168 h while SlICDH transcript levels were significantly higher at 24 and 168 h with a dip in their levels at 24 and 72 h.
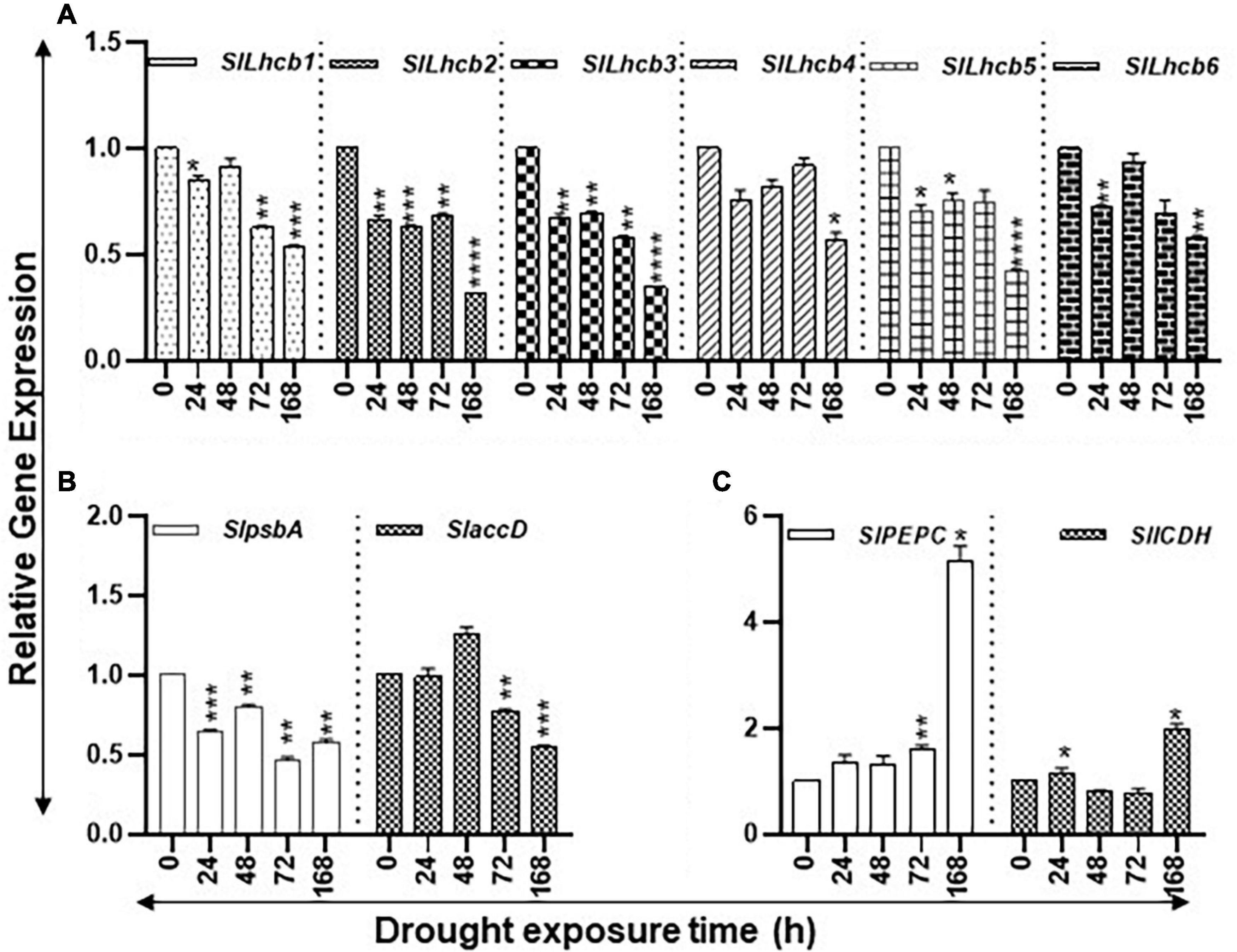
Figure 3. Effect of drought on the steady state abundance of chlorophyll a/b-binding, plastid-encoded photosystem II (PSII), and carbon flow gene transcripts in tomato leaf tissue. Quantitative PCR was used to determine the steady state levels of transcripts of (A) nuclear-encoded light-harvesting chlorophyll a/b-binding protein genes (Lhcb1-6); (B) Plastid-encoded photosystem II (PSII) genes (psbA and accD); and (C) phosphoenolpyruvate carboxylase (PEPC) and isocitrate dehydrogenase (ICDH) genes. Other details are the same as in Figure 1. **** indicates significance at p-value of <0.00005.
Expression of desiccation-responsive SlRD29A, SlDELLA, SlWIRKY57, SlJUB1, SlDREB1, and SlDREB2 genes, together with ABA biosynthesis pathway genes SlNCED1 and SlNCED2, was determined since their induction in response to drought stress has been reported in the literature. After an initial decrease in the transcript levels of SlDREB1/2, SlRD29, SlNCED1, and SlJUB1, their steady state transcript levels were found to significantly increase after 168 h of drought (Figure 4). Transcript levels of SlDELLA and SlWRKY57 declined during the increasing drought period, whereas mixed patterns of accumulation of SlNCED2 transcripts were apparent during the stress period with their decline seen at 168 h (Figure 4). Taken together, these results indicate that seedlings perceive drought stress within 24 h that becomes severe upon prolonged drought.
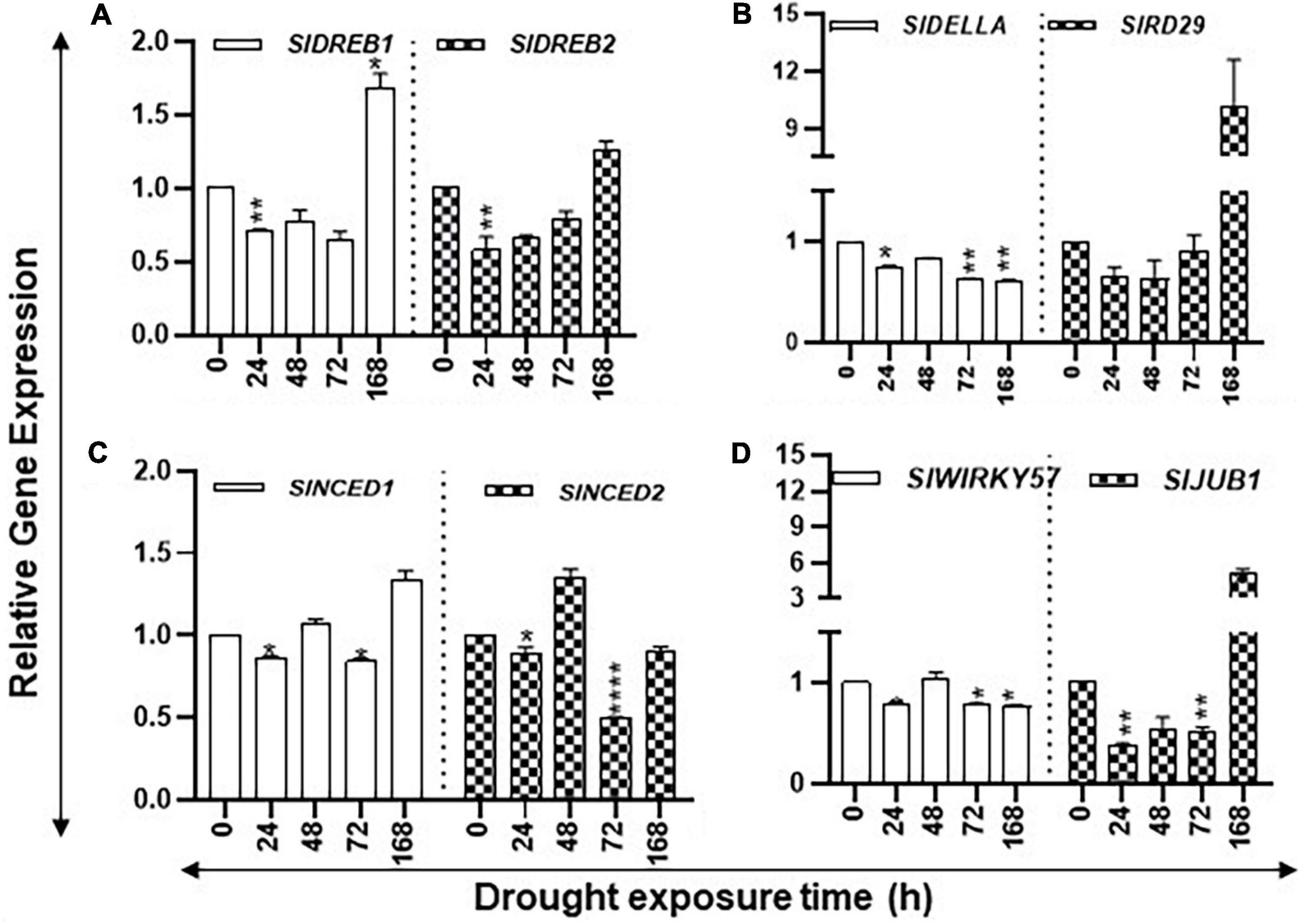
Figure 4. Changes in the expression levels of drought stress-related marker genes during the increasing period of drought in tomato leaves. Quantitative PCR was used to determine the steady state levels of transcripts of SlDREB1/2, SlNCED1/2, SlRD29A, SlWIRKY57, and SlJUB1 genes. Other details are the same as in Figure 1.
Changes in total (free + conjugated + bound) cellular content of PUT, SPD, and SPM during salinity stress are shown in Figure 5A. The total cellular levels of PUT continued to decline with the increasing saline stress period, whereas the total cellular contents of SPD and SPM increased until 48 h of saline stress before their precipitable decline after 168 h of stress (Figure 5A). The trend of total cellular PA content (all forms of PUT + SPD + SPM) followed a pattern similar to total SPD and SPM, likely since SPD was found to be the dominating cellular PA in tomato leaves under saline stress (Figure 5A). These results are similar to those obtained under drought stress, indicating that, during drought and salinity stresses, the cellular metabolic activities of the PAs pathway continue up to at least 72 h for drought and 48 h for salinity stress (Figures 1A, 5A).
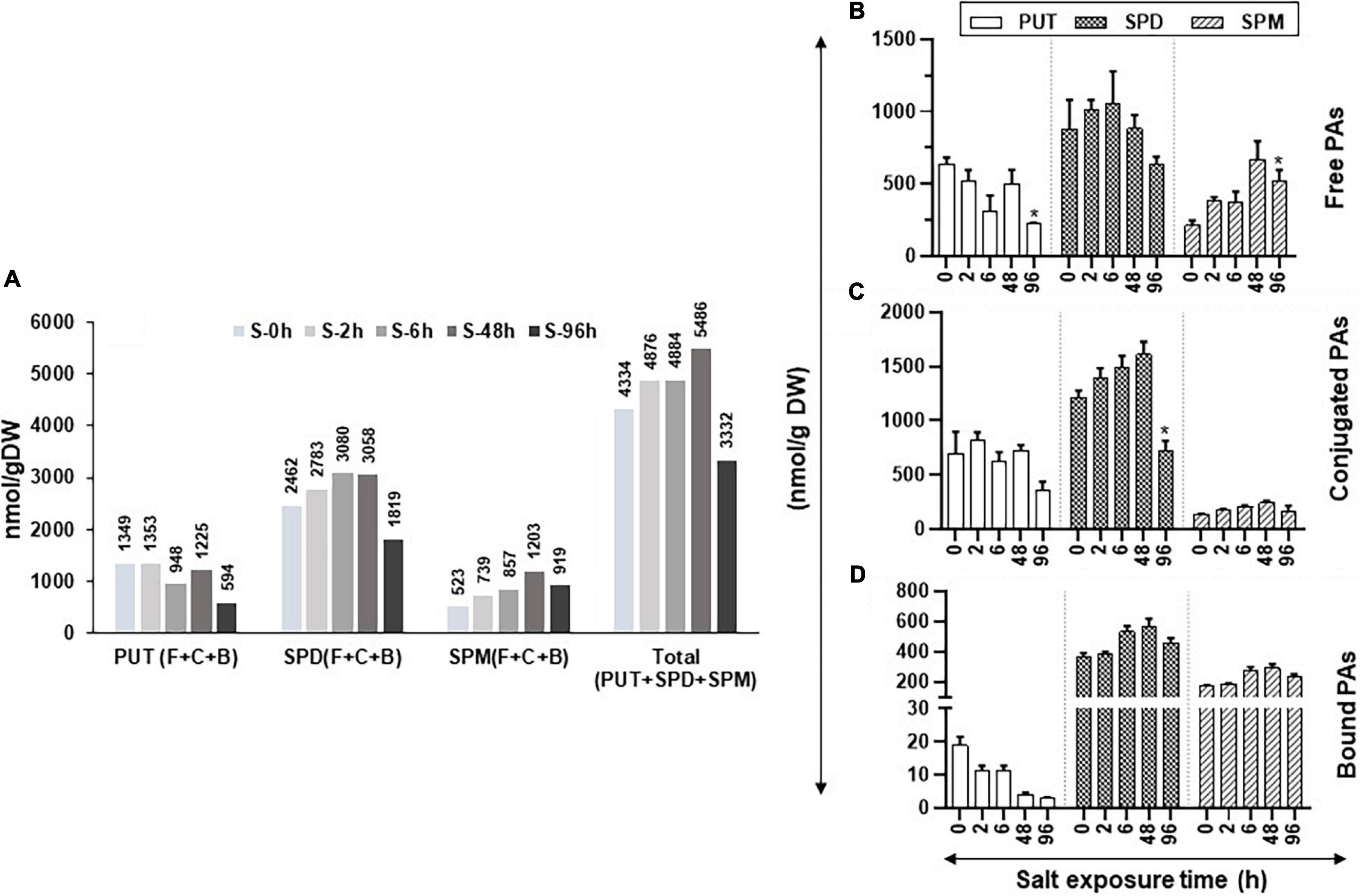
Figure 5. Changes in the levels of free, conjugated, and bound forms of PUT, SPD, and SPM in tomato leaves during salt stress. Salt stress was imposed on tomato plants by irrigating plants daily with 200-mM NaCl (salt stress). Leaf samples from a minimum of three plants were harvested at 0, 2, 6, 48, and 96 h and immediately frozen in liquid N2 and then stored at –80°C until analyzed. HPLC was used to determine the levels of free, bound, and conjugated forms of PAs. (A) Changes in the total amount of free, bound, and conjugated forms of PUT, SPD, and SPM; (B) free PUT, SPD, and SPM; (C) conjugated PUT, SPD, and SPM; (D) bound PUT, SPD, and SPM, respectively, during salt stress, respectively. Other details were the same as in Figure 1.
During the increasing period of salt stress, the levels of free PUT (PUT-F) gradually decreased, except at 48-h treatment, while a significant decline occurred after 96 h of salt treatment (Figure 5B). Levels of SPD-F steadily increased until 6 h before declining thereafter until 96 h of salt treatment while SPM-F levels steadily increased until 48 h before their decline at 96 h of treatment (Figure 5B). The PUT-C levels showed mixed patterns with a precipitable decline observed after 96 h of salt treatment (Figure 5C). Levels of SPD-C increased up to 48-h salt-stress before their steep decline at 96 h (Figure 5C). In contrast, the SPM-C levels were low and remained similar throughout the salt-treatment period (Figure 5C).
The bound form of PUT (PUT-B) steadily declined throughout the salt treatment while both SPD-B and SPM-B levels increased after 6 h of salt treatment and remained higher than that at 0-h control up to 96 h of salt exposure (Figure 5D). These results indicate that exposure to salinity alters cellular levels of PAs, SPD, and SPM in particular. Not only did the salt treatment increase the cellular levels of SPD-F and SPM-F but also converted some of the SPD-F to SPD-C and SPD-B, likely to maintain the homeostasis of cellular SPD (Figures 5B–D). Taken together, these results suggest that both SPD and SPM play key roles in the salinity response of tomato leaves. Quantified levels of free, conjugated and bound PAs and their statistical analyses are shown in Supplementary Table S4.
The steady state levels of transcripts of SlARG1/2, SlAIH, SlSAMDc3, and SlOAT1 declined within the first 2 h of salt treatment; after which, their levels increased (Figure 6). Transcripts of SlCPA, SlADC1/2, SlODC1, SlSAMDc1,2, and SlSPDS1 also showed an initial decline, but their patterns thereafter showed mixed patterns (Figure 6). However, SlSPMS did not show any initial decline after salt treatment and its transcript levels increased after 6 h and continued up to 96 h of treatment (Figure 6).
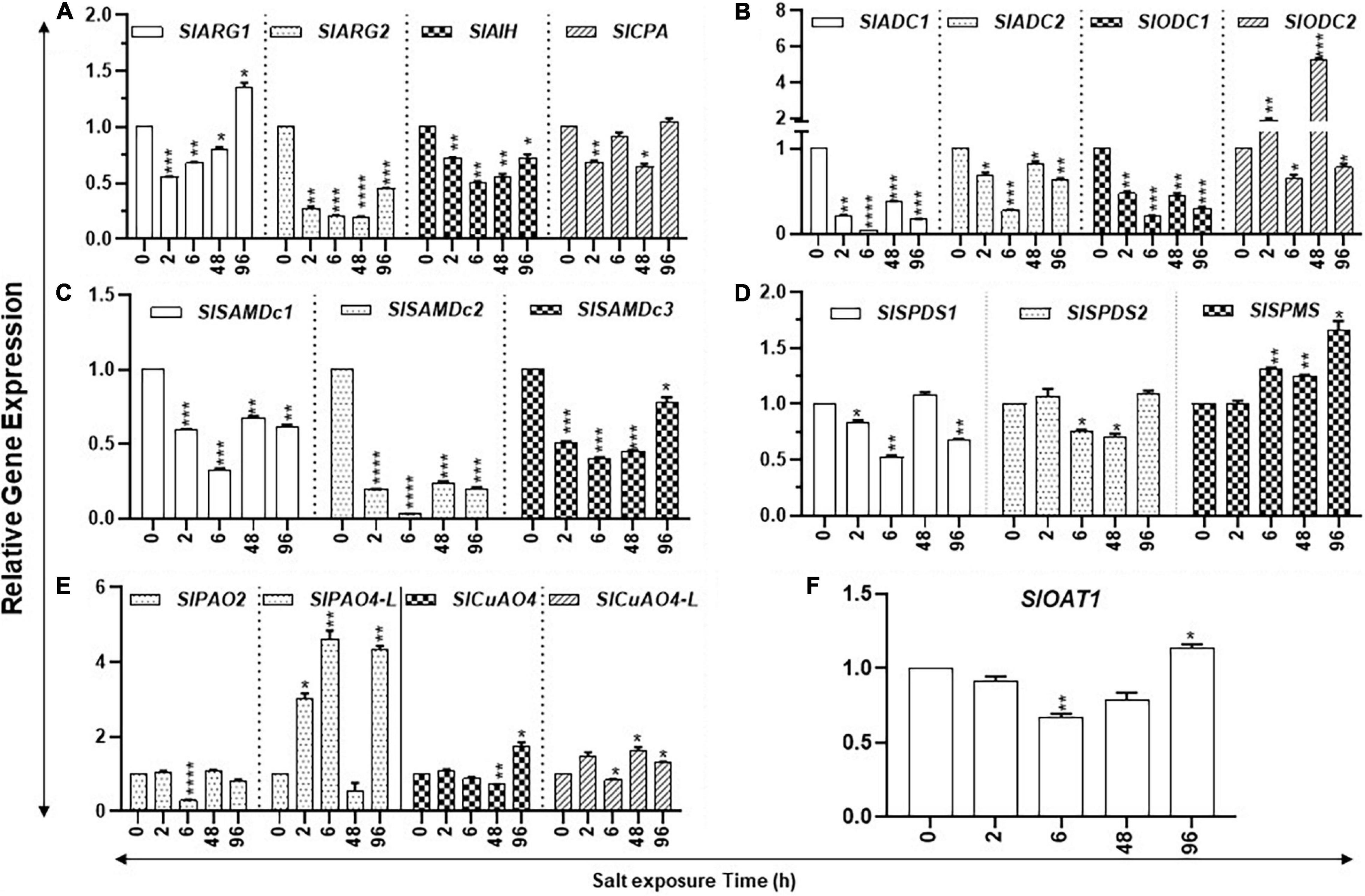
Figure 6. Kinetics of salt stress-induced expression of PA metabolic pathway genes in tomato leaves. Total RNAs were extracted from tomato leaves harvested after salinity stress for 0, 24, 48, and 96 h. The steady state levels of transcripts of various PAs biosynthesis and catabolism genes were determined by qRT-PCR as described in Materials and Methods. Genes included were those of PUT biosynthesis (SlARG1, 2, SlADC1, 2 SlODC1, 2, SlAIH, and SlCPA), SPD, and SPM biosynthesis (SlSAMDc1, 2, 3, SlSPDS1, 2, and SlSPMS); and catabolism (SlPAO2 and SlPAO4-Like; SlCuAO4 and SlCuAO4-like) pathways, respectively. SlOAT1 expression was assessed for the flow of ornithine toward the proline pathway. Mean data points and SE were derived from a minimum of three biological replicates. SlUB13 and SlTIP41 were used as reference genes. Other details were the same as in Figure 1. **** indicates significance at p-value of <0.00005.
Polyamine catabolism pathway genes had specific patterns (Figure 6). For instance, transcript levels of SlPAO4-like increased several-fold within 2 h of salt treatment and remained elevated at 6 h but declined at 48 h. By 96 h of the salt treatment, SlPAO4-like increased substantially (Figure 6). On the other hand, different patterns of SlPAO2 under salinity stress were apparent with a sharp decline at 6 h and with only a slight variation at the remaining time points (Figure 6). In regard to catabolic genes, SlCuAO4 and SlCuAO4-like increased at 2 h, declined at 6 h, but then increased significantly at 48 and 96 h of salt treatment.
The study state levels of carbon assimilation genes during the increasing period of salt stress are shown in Figure 6. The transcript levels of six nuclear-encoded light-harvesting chlorophyll a/b-binding genes (SlLhcb1, SlLhcb2, SlLhcb3, SlLhcb4, SlLhcb5, and SlLhcb6) had variable patterns during the increasing period of salt stress (Figure 7A). SlLhcb1 gene had a steady decline while SlLhcb2-5 declined after 6 h, followed by significant increases thereafter, while Slhcb6 transcript levels increased at 48 h (Figure 7A). Among the two plastid-encoded PSII genes evaluated, SlpsbA expression first declined and then increased, whereas SlaccD had variable patterns with an increase at 48 h (Figure 7B). Transcript levels of SlPEPC increased over twofold after 48- and 96-h salt treatment, while transcript levels of SlICDH significantly increased after 96 h (Figure 7C). Statistical differences in salt stress data points are enlisted separately inSupplementary Table S4.
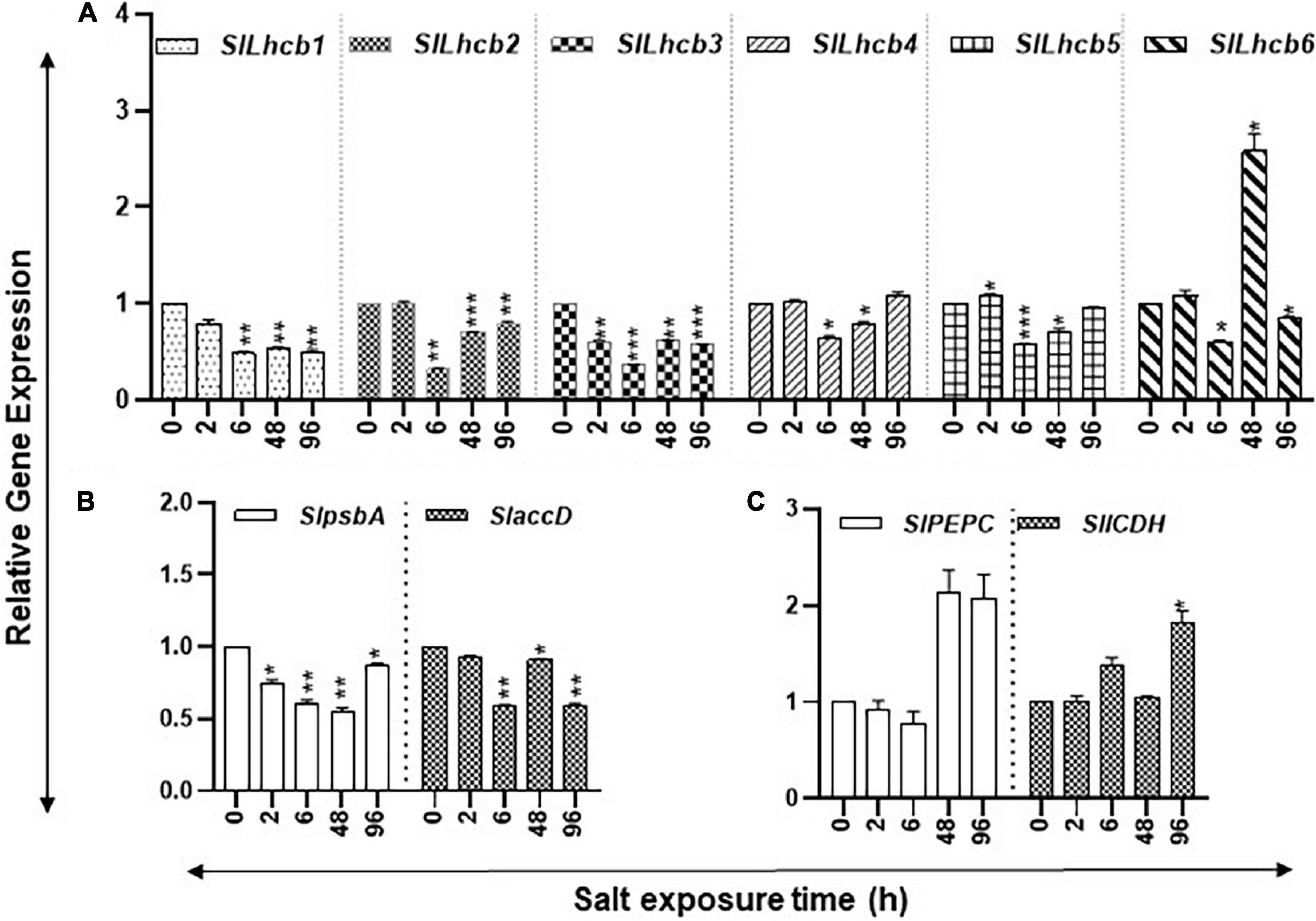
Figure 7. Salt stress and abundance of chlorophyll a/b-binding, photosystem II (PSII), and carbon flow gene transcripts of tomato leaves. Quantitative PCR was used to determine the steady state levels of transcripts of (A) nuclear-encoded light-harvesting chlorophyll a/b-binding protein genes (Lhcb1-6); (B) Plastid-encoded photosystem II (PSII) genes (psbA and accD); and (C) phosphoenolpyruvate carboxylase (PEPC) and isocitrate dehydrogenase (ICDH) genes. Other details were the same as in Figure 1.
Salt Overly Sensitive SlSOS1 and the Na+/H+ antiporter (NHX) gene family are known to be upregulated during salt stress (Rodríguez-Rosales et al., 2008; Ji et al., 2013; Yang et al., 2017). We quantified the expression of SlSOS1, SlNHX3, and SlNHX4 as indicators for salt stress. Transcript levels of SlSOS1 increased significantly only after 96 h of salt treatment (Figure 8). On the other hand, the transcript levels of SlNHX3 were significantly higher at 6 h and 96 h but had significant dips at 2 and 48 h of salt treatment (Figure 8). The SlNHX4 gene transcripts significantly increased at 2, 6, and 96 h after salt treatment, but their significant decline was apparent after 48 h of salt treatment (Figure 8). These patterns suggest that plants were acclimating during early hours of stress and showed signs of seedling revival after 96 h of salt treatment (Figure 8). Statistical differences in salt stress data points are separately listed in Supplementary Table S4.
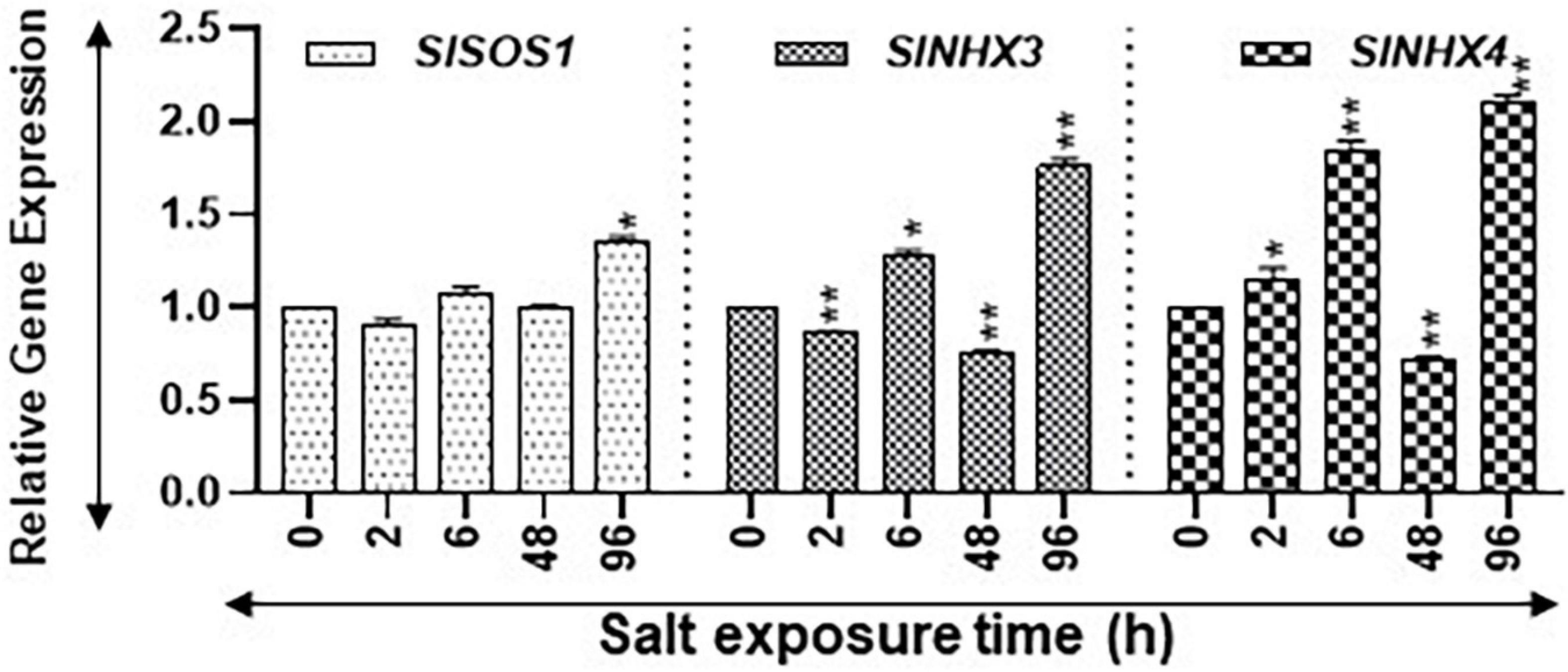
Figure 8. Expression levels of salt stress-related marker genes. Quantitative PCR was used to determine the steady state levels of transcripts of salt-induced marker genes SlSOS1, SlNHX3, and SlNHX4. SlUBI3 and SlTIP41 were used as reference genes. Other details are the same as in Figure 1.
Pearson correlation coefficient among free, conjugated, and bound forms of PAs was determined during increasing periods of drought and salinity stresses (Supplementary Table S5). Collectively, out of 81 possible combinations among the free, conjugated, and bound forms of PUT, SPD, and SPM for each of the two stresses, 59 and 47 combinations were significant at positive + r (>0.4) for both drought and salinity stresses, respectively (Supplementary Table S5). Only four combinations for both drought and salinity were significant with −r (< −0.4) (Supplementary Table S5). Strong (negative) −r (<−0.8) was observed only for SPM-F, while PUT-B showed −r under both drought and salinity stresses (Supplementary Table S5). Under the drought stress, PUT-F had strong + r (>0.8) with PUT-C, SPD-F/C/B, and SPD-C/B but not with SPM-F and PUT-B (Supplementary Table S5). A very different pattern was observed under the salinity stress; PUT-F showed strong + r only with PUT-C (Supplementary Table S5). SPD-F exhibited + r > 0.8 with SPD-C/B and SPM-C/B under drought conditions and with PUT-C/B, SPD-C under the salinity stress, respectively (Supplementary Table S5). Under drought stress, SPD-C and SPM-C had similar patterns since they exhibited + r > 0.8 with each other and with PUT-F/C and SPM-B, while SPM-C did not show any correlation (Supplementary Table S5). Under the saline conditions, SPD-C exhibited + r > 0.8 with SPD-F/C, PUT-C, and SPM-C, whereas SPM-C had strong + r > 0.8 with SPD-C/B and SPM-B (Supplementary Table S5). Taken together, these results reveal coordinately positive regulation of free, conjugated, and bound PAs levels in drought and salinity stresses, most likely to maintain homeostasis.
Although PA biosynthesis genes had differential levels of transcripts under the drought and saline conditions, their correlations with different forms of PAs were generally negative (Supplementary Table S6). Out of 135 correlation coefficients among the 15 potential PAs biosynthesis genes and 9 forms of PAs (free, conjugated, and bound forms of PUT, SPD, and SPM), correlations for 51 and 40 sets were significantly negative with 33 and 29 combinations with significant + r for drought and salinity stress, respectively (Supplementary Table S6). The genes that showed + r values > 0.8 with any form of PAs included ODC2, SAMDc1, and SPMS during the drought stress and ADC1 and ODC1/2 under the salinity stress, respectively. The PAs biosynthesis genes SlARG1/2, SlAIH1, SlCPA, SlSAMDc2/3, SlSPDS1/2, SlSPMS, and SlOAT1 showed −r under drought. Under salinity stress, SlARG1/2, SlADC1, SlAIH1, SlCPA, SlSAMDc3, SlSPDS2, and SlSPMS showed −r with several forms of PAs. Collectively, these data indicate that the PA biosynthesis pathway was negatively influenced under both drought and salinity stresses but to a higher extent under drought conditions. Data also indicated that, in some instances, different homologs of genes were affected under the two stresses (Supplementary Table S6). PA catabolism genes were negatively influenced more under drought than salt stress (Supplementary Table S6). Transcript levels of SlPAO2 and SlCuAO4-like were affected by both drought and salt stresses (Supplementary Table S6). This analysis strongly indicates that both drought and salt stresses negatively affect PA metabolism, while drought stress (86 combinations) affects PA metabolism to a higher degree than salinity stress (65 combinations) (Supplementary Table S6).
To evaluate the effects of drought and salinity stresses on the nuclear-encoded light-harvesting chlorophyll a/b-binding, plastid-encoded PSII, carbon flow genes, drought, and salinity stress-associated genes, the Pearson correlation between the transcript levels of selected genes with the levels of free, conjugated, and bound PUT, SPD, and SPM were determined (Supplementary Table S7). Both drought and salinity stress negatively affected these genes in general, but more genes had significantly −r under drought (64 combinations) than salt (53 combinations) stress. SlPEPC and SlICDH had −r with most forms of PAs except SPM-F and PUT-B under both drought and salinity stresses. During the drought stress, the Lhcb1-6 genes had + r with several forms of PAs, whereas they showed limited associations with PA forms under salinity stress (Supplementary Table S7). Drought-associated markers SlRD29A, SlNCED1/2, and SlWIRKY57 showed strong −r with most forms of PAs but weak correlation with SPM-F and PUT-B. Among the salinity-associated genes, SlSOS1 showed −r with SPD-F, PUT-C, and SlNHX4 with SPD/SPM-C and SPD/SPM-B. SlNHX3 did not have a strong association with any form of PAs (Supplementary Table S7).
Coordinate regulation of both drought and salinity stresses with the PAs pathway was examined as well. SlRD29A, SlNCED1/2, SlWIRKY57 exhibited + r with SlARG1/2, SlAIH1, SlODC2, SlSAMDc2/3, SlSPDS1/2, SlOAT1, SlPAO2, SlPAO4-like, and SlCuAO4-like genes and −r with SAMDC1 genes during the drought stress. DREB1/2 and Della exhibited + r with different sets of the PAs pathway that included ADC1/2, ODC1, and SAMDc1 (Supplementary Table S8). SlNHX3 and SlNHX4 generally exhibited + r with some of the PAs pathway genes. These results suggest that transcription of all drought and salinity stress maker genes tested in this investigation is not coordinately regulated with PAs biosynthesis or catabolism pathways.
Differential regulation for the PA biosynthesis and catabolism gene transcript levels were observed during drought and salinity stresses. The steady state levels of transcripts for SlARG1/2, SlSAMDc2, and SlCuAO4-like showed coordinate regulation under both drought and salinity stresses (Supplementary Table S8). The genes that showed + r under the drought conditions were SlSAMDc3, SlSPDS2, SlOAT1, and SlPAO2 genes; those under the salinity stress were SlCPA, SlADC1/2, SlODC1, SlSAMDc1, and SlPAO4-like genes (Supplementary Table S8). SlODC2 had generally −r under drought and +r under salinity with some of the genes. In general, among the 284 correlations obtained within the PAs biosynthesis and catabolism genes, 239 exhibited + r > 0.8 and 45 exhibited –r < −0.8, indicating that coordinate regulation of PAs underlies perception and acclimation during both drought and salinity stresses.
Since photosynthesis plays an essential role in both fresh and dry weight gains leading to total crop yield, we examined coordination among the PAs pathway with a selected set of photosynthesis genes (Figures 3, 7 and Supplementary Table S8). Among the 170 correlations obtained within the PAs metabolic pathway and photosynthesis pathway genes under drought and salinity stresses, 137 showed + r > 0.8 and 33 –r < −0.8, respectively. However, differential correlation patterns were also seen for several genes during the drought and salinity stresses (Supplementary Table S8). These data indicate that the PA pathway generally contributes positively to the overall expression of selected photosynthesis genes.
Under both drought and salinity stresses, both SlPEPC and SlICDH showed + r > 0.8 among themselves, but SlpsbA and SlaccD exhibited + r > 0.8 only under the drought conditions (Supplementary Table S9). SlLhcb1-5 showed mostly + r > 0.8 among themselves under both drought and salinity stresses. These genes displayed -r < −0.8 with SlPEPC and SlICDH under drought but no correlation under the saline conditions (Supplementary Table S9). Drought-associated marker genes had differential behavior. SlDREB1/2 and SlDELLA were generally + r > 0.8 with some photosynthesis genes. SlRD29A, SlNCED1/2, and SlWIRKY57 were coordinately expressed with respect to one another and with + r > 0.8 but showed –r < −0.8 for SlLhcb2-5. Among the salinity-associated marker genes, SlSOS1 and SlNHX4 had self-coordination but not with SlNHX3 transcript levels. SlNHX3 had + r > 0.8 with SlLhcb3-6, SlpsbA, and SlaccD (Supplementary Table S9). These results suggest that factors other than PAs also play roles in the regulation of photosynthesis and marker genes.
Multivariant analyses of all variables evaluated under the drought and salt stresses are shown in Figures 9A,B. During drought stress, all active variables representing drought samples were present in +PC1 along with SPD-F, SPM-F, SPD-C, SPM-C, SPD-B, and SPM-B but away from the three forms of PUT, suggesting that SPD and SPM but not PUT play roles during perception/acclimation to drought stress. The genes that co-segregated with all the time points of drought included SlARG1, SlSAMDc3, SlODC2, SlSPDS1, SlSPMS, SlPAO4-like, and SlCuAO4 in +PC1 (Figure 9A). Co-segregation of many SPD/SPM biosynthesis and catabolism genes with drought marker genes SlDREB2 and SlPEPC during drought support the hypothesis that SPD and SPM are significant players in drought stress. Like, under drought stress, all active variables representing salt-treated samples were present in +PC1 (Figure 9B). However, in addition to SPD-F, SPM-F, SPD-C, SPM-C, SPD-B, and SPM-B, PUT-C that segregated with drought stress also co-segregated with salt stress samples, suggesting role(s) of these PAs forms, including PUT-C but not PUT-F and PUT-B in salinity stress (Figure 9B). The genes that showed association with various saline stress samples included SlODC2, SlOAT1, SlSPDS2, SlSPMS, SlPAO4, SlPAO4-Like, SlCuAO4, SlCuAO4-like, and salinity marker gene SlNHX4 (Figure 9B). Among the photosynthesis-related genes SlLhcb 1-6, only SlLhcb6 shared + PC1 with salinity samples. Since the first two components, PC1 and PC2 accounted for >86% and >90% variability for drought and saline stresses, respectively, we interpret these results to suggest that the PA biosynthesis and catabolic genes, namely, ODC2, SPMS, PAO4-like, and CuAO4, participate in both drought and salinity stresses. However, some of the other genes of biosynthesis and catabolism genes were more associated with drought (ARG1, SAMDc3, and SPDS1) and salinity (OAT1, SPDS2, PAO4, and CuAO4-like, respectively).
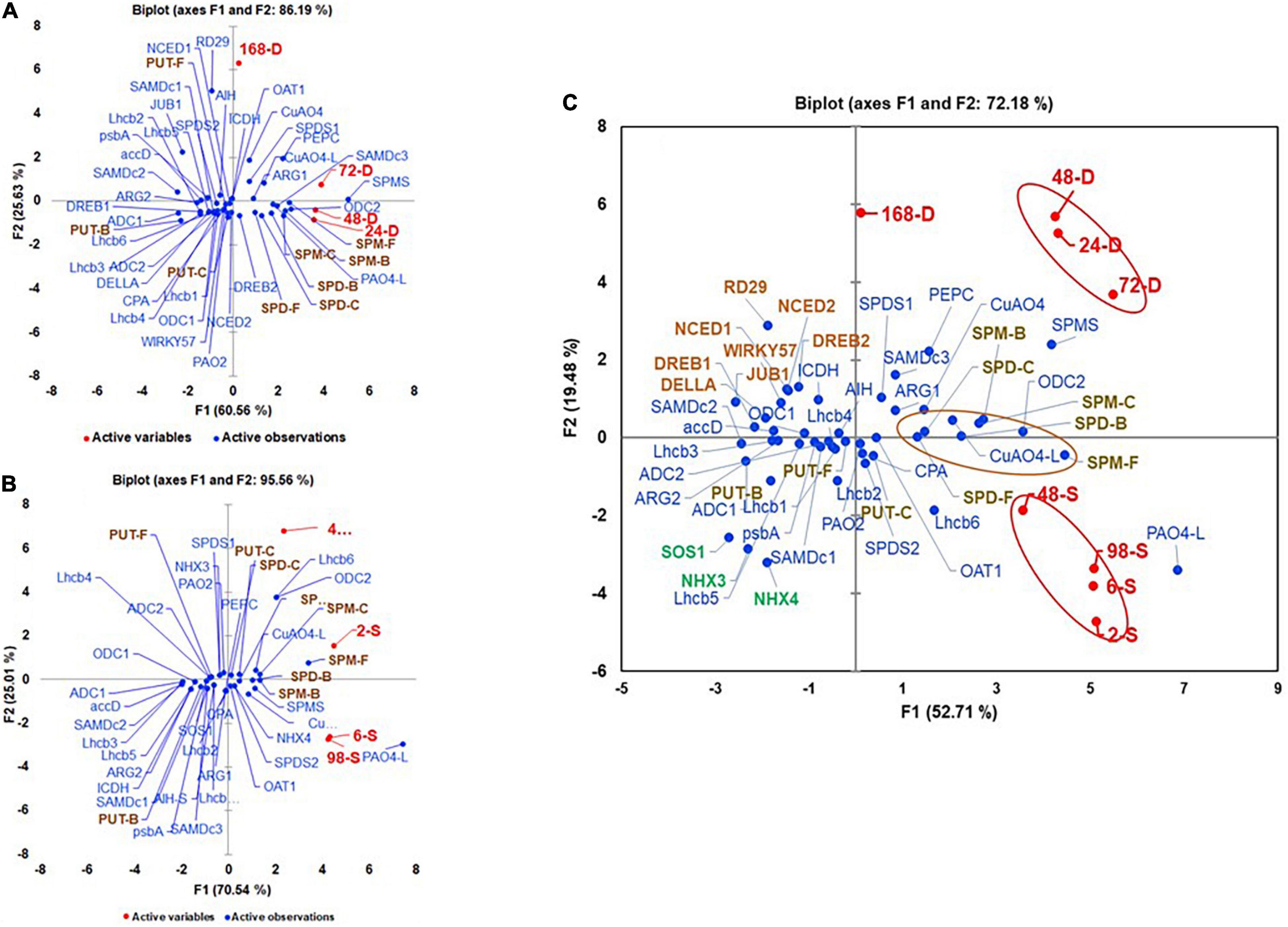
Figure 9. Perchloric Acid analyses of active and variable parameters during (A) drought, (B) salt stress, and (C) their pooled data. The active parameters included leaves exposed to 24, 48, 72, and 168 h of water withholding (A) and 2, 6, 48, and 98 h of 200-mM NaCl treatment (B) and data from two stresses pooled for each parameter before analysis (C). The variable parameters included levels of free (F), conjugated (C), and bound (B) PUT, SPD, and SPM (PUT-F, SPD-F, SPM-F, PUT-C, SPD-C, SPM-C, PUT-B, SPD-B, and SPM-B). Shown are the transcript levels of PAs biosynthesis and catabolizing enzymes-encoding genes, drought, and salt stress maker genes and photosynthetic machinery-related genes. Other details as stated in the material and method section. Gene abbreviations were the same as in Supplementary Tables S1, S2.
We also performed combined multivariant analyses of drought and salinity stress datasets to evaluate similarities of responses under the two stresses (Figure 9C). The first two components, PC1 (53.71) and PC2 (19.48) accounted for over 73% of the total variability (Figure 9C). The drought and salinity stress active variables are separated into different quadrants of PCA. All drought active variables (24-D, 48-D, 72-D, and 168-D) were present in +PC1+/+PC2 and that of salt (2-S, 6-S, 48-S, and 96-S) in +PC1/-PC2, indicating that, in spite of the similarities, the two stresses differentially affect PA metabolism in tomato leaves. Although all forms of SPD, SPM, and PUT-C were present close to the X-axis of +PC1, SPD-F, SPD-C, and SPD-B had a close association with both the stresses, while SPD-B and SPM-C were closely associated with the drought stress and SPM-F and PUT-C co-segregated with the salinity stress (Figure 9C). These results suggest that, during drought, SPD and SPM homeostasis is tilted toward their bound and conjugated forms, while free SPM and conjugated PUT are tilted more toward salinity stress. As observed in independent drought and salinity analyses, SlSAMDc3, SlODC2, SlCuAO4, and SlCuAO4-like were present in +PC1/+PC2, likely indicating that these genes regulate levels of various forms of SPD and SPM during drought stress. Similarly, the association of the salinity stressed samples with SlSPDS2 and SlPAO4-like genes suggests their roles in adaption to salinity stress (Figure 9C). A large number of the PAs pathway and selected photosynthesis genes in −PC1 segregated away from drought and salinity stress samples indicate that their expression is impaired under both stresses and likely responsible for the growth inhibition obtained under these stresses.
Cytoscape analyses of the Pearson correlations among observed parameters under the drought and salinity stress were performed to visualize any possible network among these parameters (Figure 10). Free, conjugated, and bound PAs had highly positive Pearson correlations with one another under both stresses. Under drought stress, PUT-F, SPM-C, and SPD-F showed strong negative (−ve) association with transcript levels of CuAO4-like, SlRD29A, SlNCED1/2, SlSAMDc1/2, SlpsbA, SlICDH, and strong positive (+ve) association with SlSPDS1/2, SlNCED2, SlCuAO4, SlCuAO4-like, SlWIRKY57, SlPEPC, and SlaacD. Other observable components had weak positive and negative correlations (Figure 10A). Cytoscape analyses of the Pearson correlations among the observed parameters under the salt stress showed a negative association with SlSPDS1/2, SlCuOA4, SlPEPC, SlpsbA, SlLhcb1,5, and SlaccD (Figure 10B). Multiple genes were coordinatedly associated among themselves, such as SlODC2, SlSPMS, SlADC2, SlPAO4-like, SlCuAO4-like, SlNHX3/4, SlpsbA, SlLhcb3,4,6, SlPEPC, SlICDH, and SlSOS1. These results were consistent with the PCA analyses of drought and salinity stress samples (Figures 9A,B). Pooling the data from drought and salinity stresses showed strong negative associations for PUT-F, SPD-C, and SPM-C with SlRD29A, SlSOS1, SlLhcb2, SlNCED1, SlJUB1, and SlNHX4. However, a very strong positive association was seen among certain pairs of genes that included SlARG2, SlSAMDc2/3, SlSPDS1/2, SlNHX3/4, SlLhcb3, SlPEPC, SlpsbA, SlICDH, and SlSOS1. The correlation coefficients are shown in Supplementary Tables S5–S9.
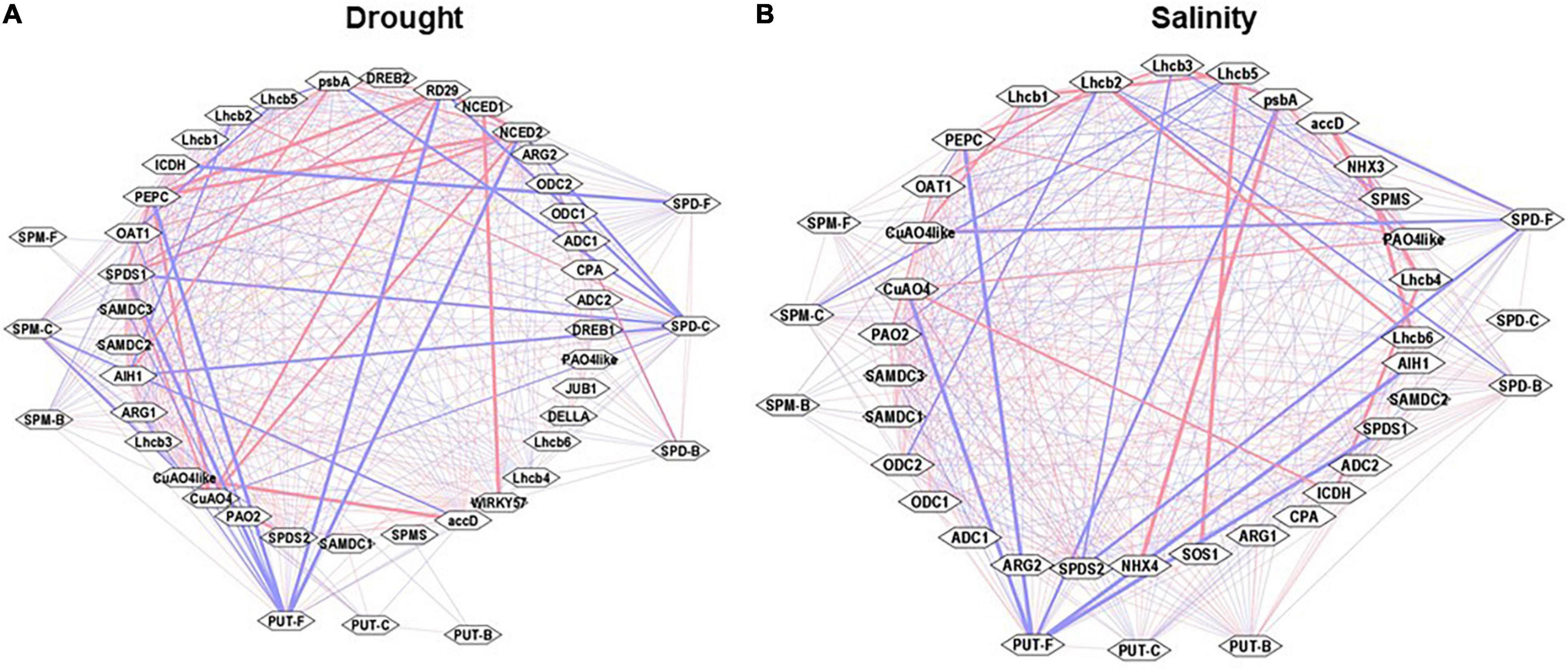
Figure 10. Cytoscape analyses (interactive networks) of different PAs form with the transcript abundances of genes analyzed during drought and salinity stress. Positive (red) and negative (blue) correlations of cellular levels of PUT-F, SPD-F, SPM-F, PUT-C, SPD-C, SPM-C, PUT-B, SPD-B, and SPM-B, and transcript abundance of PAs biosynthesis and catabolizing enzymes genes are shown. Also included in the Cytoscape analysis are the Pearson correlations of selected drought (A) and salinity (B) stress maker genes and photosynthesis processes and carbon fixation pathway genes. Other details are given in the section “Materials and Methods.”
Polyamines, as plant growth regulators, are known to regulate a myriad of developmental and physiological processes, including longevity in plants under both normal and stress conditions (Gill and Tuteja, 2010; Nambeesan et al., 2010, 2012; Minocha et al., 2014; Mattoo et al., 2015b; Handa et al., 2018; Upadhyay et al., 2020). They have been implicated in developing tolerance or survival under harsh environmental conditions, such as heat, cold, drought, and salinity stress (Kusano et al., 2008; Alcázar et al., 2010; Berberich et al., 2015; Handa et al., 2018; Upadhyay et al., 2020). However, only in limited investigations, the response of plants at different development stages has been analyzed for metabolic changes during time kinetics after induction of drought or salt stress. In this study, we report the complexity as well as unique changes in polyamine metabolic events in tomato leaves during independent drought or salt stress. We particularly demonstrate the effects on the PA biosynthesis and catabolic genes and their impact on the expression of photosynthesis protein-encoding genes in tomato leaves.
Given the fact that drought and salt stresses are far apart in their nature, we found similar changes, by and large, in PA abundance during both stress situations. Even though the levels were not exactly similar, they seemed to mirror each other. The patterns of total PAs and that of free, conjugated, and bound PUT, SPD, and SPM were similar under both types of stresses. However, the cellular levels of each type of PAs were much higher under the salt than drought stress (Figures 1A, 5A). Furthermore, levels of PUT-F decreased similarly in drought and salt stress during later stages, and a similar pattern of increase was apparent in the levels of SPD-F and SPM-F under both the stresses. The data presented suggest that pools of free, conjugated, and bound PAs may be important for the maintenance of PA homeostasis under the two stresses investigated. Moreover, among the three forms of PAs (PUT, SPD, and SPM), both stresses impacted PUT levels negatively, while SPD and SPM were upregulated, suggesting positive roles of SPD and SPM in comparison to the negative role of PUT as previously shown (Handa and Mattoo, 2010; Mattoo et al., 2010).
Interestingly, the levels of total PAs increased with the increasing period of drought (up to 72 h) and salinity (up to 48 h) before declining steeply during prolonged drought (168 h) and salt (96 h) stress (Figures 1, 5). This is in contrast with other reports in which progressive loss of free SPD was observed under water stress in wheat (Marciñska et al., 2013; Hura et al., 2015). The increase in PAs under both stresses was largely due to higher accumulation of SPD with measurable contribution from SPM, while PUT levels gradually declined during the drought and salt stress periods. Moreover, a higher increase in SPD (2,783–3,058 nmol g–1 DW) and SPM (739–1,203 nmol g–1 DW) was apparent under salt stress than during drought (1,930–21,238 nmol g–1 DW SPD and 371- to 605-nmol g–1 DW SPM). It is known that higher levels of SPD/SPM contribute to overcoming reactive oxygen species (ROS) by enhancing antioxidative enzymes in plants to overcome salt toxicity (Rider et al., 2007; Chen et al., 2019). The decrease in SPD and SPM levels after a prolonged period of stress likely indicates a loss of cellular viability that results in decreased metabolic activity or oxidation consumption by polyamine oxidase (Cvikrová et al., 2013). However, it remains to be determined if the decreased levels of SPD and SPM are the cause or the effect of loss of cellular vitality after prolonged abiotic stress.
Several investigations have implicated PAs to play significant roles in drought and salinity stress (Kasukabe et al., 2006; Wi et al., 2006; Yamaguchi et al., 2007; He et al., 2008; Alcázar et al., 2010; Liu et al., 2010; Naka et al., 2010; Neily et al., 2011; Chen et al., 2019). Since PAs have been implicated in numerous biological processes, there has been great interest in learning how endogenous PA homeostasis is achieved. Myriad processes are involved in maintaining the optimum cellular levels of various forms of PUT, SPD, and SPM, including their biosynthesis, catabolism, conjugation, and binding to cellular components (Moschou et al., 2008). Interestingly, during heat or cold stress, the change in the levels of free and conjugated PUT, SPD, and SPM is less significant, suggesting that PAs may also utilize different routes to protect plants from cold and high-temperature stress (Upadhyay et al., 2020).
Long-term drought stress has been found to induce structural and functional reorganization of photosystem II of chloroplasts (Giardi et al., 1996). In this context, it is noteworthy that exogenous PAs were found to improve photosynthetic efficiency (Farooq et al., 2009). Similarly, there are some reports that describe the effects of polyamines on the functionality of photosynthetic membrane in vivo and in vitro in isolated chloroplast from tobacco plants (Ioannidis and Kotzabasis, 2007). Furthermore, it has been shown that saline alkalinity repressed gene expression of psbA gene and protein levels of the D1 protein, and exogenous application of spermidine alleviates expression of psbA gene and D1 protein in salinity-alkalinity stress in tomato seedlings (Hu et al., 2015). As studied here, we found that nuclear light-harvesting chlorophyll a/b-binding protein genes (SlLhcb1-6) were downregulated during drought stress and SlLhcb1-6 were downregulated during salt stress, while during early response to salt, SlLhcb4-6 genes were downregulated (Figures 3A, 7A). The plastid-encoded photosystem II (PSII) protein genes SlpsbA (encodes for D1 protein) and SlaccD are similarly regulated in both stresses (Figures 3B, 7B). It is also known that Lhcb proteins are downregulated during drought conditions in Arabidopsis, but PSII protein downregulation occurs late in response to drought (Chen et al., 2016), which may relate to the fact that PSII is reorganized during drought stress (Giardi et al., 1996). Thus, these data reveal that gene transcription is inhibited earlier during drought stress as compared to the plastid-encoded gene expression for photosynthesis-related genes. Expression analysis of tomato phosphoenolpyruvate carboxylase (SlPEPC) and isocitrate dehydrogenase (SlICDH) genes indicated their late activation under both stresses (Figures 3C, 7C). This is indicative of the assertion that markers for the carbon flow in response to drought or salt stress are regulated by a common genetic module. This also suggests that, during acute stress conditions, plants increase carbon fixation and utilization likely by activating the expression of PEPC and ICDH genes.
Based on the PCA analyses of combined data, SPD-F, SPD-C, and SPD-B cohabit with both drought and salinity stresses. Also, it was clear that SPD-C, SPM-B, and SPM-C segregated with drought stress, while PUT-C and SPM-F segregated with salinity stress. We interpret these results to indicate that differential conjugation of PUT, SPD, and SPM underlies the maintenance of overall PA homeostasis under the two stresses evaluated, and that different conjugated moieties of these PAs play differential roles in stress responses (Figure 9C). Among the PA biosynthesis genes, SlARG1, SlSPDS1, and SlSAMDc3 segregated with drought, while different homologs of SlSAMDc and SlSPDS2 segregated with salt stress, suggesting that specific enzymes, including different isozymes, may regulate drought and salt stress responses. A similar conclusion emerged from the response of catabolic enzymes since SlCuAO4-like was associated with drought, while SlPAO4 was associated with salt response. Moreover, such responses also emerged from the Cytoscape network analyses of various parameters (Figure 10).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
RU and AM: conceived and designed the study. RU: bioinformatics and performed the experiments. TF: HPLC/polyamine analysis. RU, AM, AH, and TF: analyzed the final data. AH and AM: facilitated the research, funding grant, and finalized the study. RU, AH, and AM: wrote the manuscript drafts. All authors: contributed to the article and approved the submitted version.
This research was funded by the Agricultural Research Service, United States Department of Agriculture, Project No. 8042-21000-143-00D, AM. Research of AH was supported by USDA/NIFA Hatch project INDO11872H.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
RU was employed at Beltsville Agriculture Research Center through a cooperative agreement with Purdue University. The mention of trade names or commercial products in this publication is solely for providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.743568/full#supplementary-material
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. doi: 10.1007/s00425-010-1130-0
Angelini, R., Tisi, A., Rea, G., Chen, M. M., Botta, M., Federico, R., et al. (2008). Involvement of polyamine oxidase in wound healing. Plant Physiol. 146, 162–177. doi: 10.1104/pp.107.108902
Anwar, R., Fatima, S., Mattoo, A. K., and Handa, A. K. (2019). Fruit architecture in polyamine-rich tomato germplasm is determined via a medley of cell cycle, cell expansion, and fruit shape genes. Plants 8:387. doi: 10.3390/plants8100387
Ashraf, M., and Akram, N. A. (2009). Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnol. Adv. 27, 744–752. doi: 10.1016/j.biotechadv.2009.05.026
Baena-González, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Baniasadi, F., Saffari, V. R., and Maghsoudi Moud, A. A. (2018). Physiological and growth responses of Calendula officinalis L. plants to the interaction effects of polyamines and salt stress. Sci. Hortic. 234, 312–31769. doi: 10.1016/j.scienta.2018.02.069
Berberich, T., Sagor, G. H. M., and Kusano, T. (2015). “Polyamines in plant stress response,” in Polyamines eds T. Kusano and H. Suzuki (Tokyo: Springer). Available online at: https://doi.org/10.1007/978-4-431-55212-3_13
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Capell, T., Bassie, L., and Christou, P. (2004). Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc. Natl. Acad. Sci. U.S.A. 101, 9909–9914. doi: 10.1073/pnas.0306974101
Chen, D., Shao, Q., Yin, L., Younis, A., and Zheng, B. (2019). Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9:1945.
Chen, Y.-E., Liu, W.-J., Su, Y.-Q., Cui, J.-M., Zhang, Z.-W., Yuan, M., et al. (2016). Different response of photosystem II to short and long-term drought stress in Arabidopsis thaliana. Physiol. Plant. 158, 225–235. doi: 10.1111/ppl.12438
Chen, Z., Hong, X., Zhang, H., Wang, Y., Li, X., Zhu, J.-K., et al. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283. doi: 10.1111/j.1365-313x.2005.02452.x
Cona, A., Cenci, F., Cervelli, M., Federico, R., Mariottini, P., Moreno, S., et al. (2003). Polyamine oxidase, a hydrogen peroxide-producing enzyme, is up-regulated by light and down-regulated by auxin in the outer tissues of the maize mesocotyl. Plant Physiol. 131, 803–813. doi: 10.1104/pp.011379
Cona, A., Rea, G., Angelini, R., Federico, R., and Tavladoraki, P. (2006). Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88. doi: 10.1016/j.tplants.2005.12.009
Cvikrová, M., Gemperlová, L., Martincová, O., and Vanková, R. (2013). Effect of drought and combined drought and heat stress on polyamine metabolism in proline-over-producing tobacco plants. Plant Physiol. Biochem. 73, 7–15. doi: 10.1016/j.plaphy.2013.08.005
Espasandin, F. D., Calzadilla, P. I., Maiale, S. J., Ruiz, O. A., and Sansberro, P. A. (2018). Overexpression of the arginine decarboxylase gene improves tolerance to salt stress in Lotus tenuis plants. J. Plant Growth Regulation 37, 156–165. doi: 10.1007/s00344-017-9713-7
Expósito-Rodríguez, M., Borges, A. A., Borges-Pérez, A., and Pérez, J. A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8:131. doi: 10.1186/1471-2229-8-131
Farooq, M., Wahid, A., and Lee, D.-J. (2009). Exogenously applied polyamines increase drought tolerance of rice by improving leaf water status, photosynthesis and membrane properties. Acta Physiol. Plant. 31, 937–945. doi: 10.1007/s11738-009-0307-2
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55, 307–319. doi: 10.1093/jxb/erh003
Giardi, M. T., Cona, A., Geiken, B., Kučera, T., Masojídek, J., and Mattoo, A. K. (1996). Long-term drought stress induces structural and functional reorganization of photosystem II. Planta 199, 118–125. doi: 10.1007/bf00196888
Gill, S. S., and Tuteja, N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5, 26–33. doi: 10.4161/psb.5.1.10291
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818.
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151.
Gong, B., Wang, X., Wei, M., Yang, F., Li, Y., and Shi, Q. (2016). Overexpression of S-adenosylmethionine synthetase 1 enhances tomato callus tolerance to alkali stress through polyamine and hydrogen peroxide cross-linked networks. Plant Cell Tiss Organ Cult. 124, 377–391. doi: 10.1007/s11240-015-0901-5
Handa, A. K., and Mattoo, A. K. (2010). Differential and functional interactions emphasize the multiple roles of polyamines in plants. Plant Physiol. Biochem. 48, 540–546. doi: 10.1016/j.plaphy.2010.02.009
Handa, A. K., Fatima, T., and Mattoo, A. K. (2018). Polyamines: bio-Molecules with diverse functions in plant and human health and disease. Front. Chem. 6:10.
Hasegawa, P. M., Bressan, R. A., Zhu, J.-K., and Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499.
He, L., Ban, Y., Inoue, H., Matsuda, N., Liu, J., and Moriguchi, T. (2008). Enhancement of spermidine content and antioxidant capacity in transgenic pear shoots overexpressing apple spermidine synthase in response to salinity and hyperosmosis. Phytochemistry 69, 2133–2141. doi: 10.1016/j.phytochem.2008.05.015
Hu, L., Xiang, L., Li, S., Zou, Z., and Hu, X. H. (2015). Beneficial role of spermidine in chlorophyll metabolism and D1 protein content in tomato seedlings under salinity-alkalinity stress. Physiol. Plant. 156, 468–477. doi: 10.1111/ppl.12398
Hura, T., Dziurka, M., Hura, K., Ostrowska, A., and Dziurka, K. (2015). Free and cell wall-bound polyamines under long-term water stress applied at different growth stages of ×Triticosecale Wittm. PLoS One 10:e0135002. doi: 10.1371/journal.pone.0135002
Igarashi, K., and Kashiwagi, K. (2015). Modulation of protein synthesis by polyamines. IUBMB Life 67, 160–169. doi: 10.1002/iub.1363
Ioannidis, N. E., and Kotzabasis, K. (2007). Effects of polyamines on the functionality of photosynthetic membrane in vivo and in vitro. Biochim. Biophys. Acta 1767, 1372–1382. doi: 10.1016/j.bbabio.2007.10.002
Jaspers, P., and Kangasjärvi, J. (2010). Reactive oxygen species in abiotic stress signaling. Physiol. Plant 138, 405–413. doi: 10.1111/j.1399-3054.2009.01321.x
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Kasukabe, Y., He, L., Nada, K., Misawa, S., Ihara, I., and Tachibana, S. (2004). Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 45, 712–722. doi: 10.1093/pcp/pch083
Kasukabe, Y., He, L., Watakabe, Y., Otani, M., Shimada, T., and Tachibana, S. (2006). Improvement of environmental stress tolerance of sweet potato by introduction of genes for spermidine synthase. Plant Biotechnol. 23, 75–83. doi: 10.5511/plantbiotechnology.23.75
Kusano, T., Berberich, T., Tateda, C., and Takahashi, Y. (2008). Polyamines: essential factors for growth and survival. Planta 228, 367–381. doi: 10.1007/s00425-008-0772-7
Li, Q. Y., Ge, H. B., and Hu, S. M. (2010). Effects of exogenous putrescine and calcium on growth and ions assimilation of strawberry seedlings under NaCl stress. Acta Hortic. 856, 187–192. doi: 10.17660/ActaHortic.2010.856.25
Liu, K., Fu, H., Bei, Q., and Luan, S. (2000). Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol. 124, 1315–1326. doi: 10.1104/pp.124.3.1315
Liu, Y., Liang, H., Lv, X., Liu, D., Wen, X., and Liao, Y. (2016). Effect of polyamines on the grain filling of wheat under drought stress. Plant Physiol. Biochem. 100, 113–129. doi: 10.1016/j.plaphy.2016.01.003
Liu, Y., Zuo, Z., and Hu, J. (2010). Effects of exogenous polyamines on growth and drought resistance of apple seedlings. J. Northwest For. Univ. 25, 000039–000042.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marciñska, I., Czyczyło-Mysza, I., Skrzypek, E., Grzesiak, M. T., Janowiak, F., Filek, M., et al. (2013). Alleviation of osmotic stress effects by exogenous application of salicylic or abscisic acid on wheat seedlings. Int. J. Mol. Sci. 14, 13171–13193. doi: 10.3390/ijms140713171
Mascia, T., Santovito, E., Gallitelli, D., and Cillo, F. (2010). Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 11, 805–816. doi: 10.1111/j.1364-3703.2010.00646.x
Mattoo, A. K., Minocha, S. C., Minocha, R., and Handa, A. K. (2010). Polyamines and cellular metabolism in plants: transgenic approaches reveal different responses to diamine putrescine versus higher polyamines spermidine and spermine. Amino Acids 38, 405–413. doi: 10.1007/s00726-009-0399-4
Mattoo, A. K., Upadhyay, R. K., and Rudrabhatla, S. (2015b). “Abiotic stress in crops: candidate genes, osmolytes, polyamines, and biotechnological intervention,” in Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives, Vol 2, ed. G. K. Pandey (New York, NY: Springer), 415–437. doi: 10.1007/978-1-4939-2540-7_15
Mattoo, A. K., Fatima, T., Upadhyay, R. K., and Handa, A. K. (2015a). “Polyamines in plants: biosynthesis from arginine, and metabolic, physiological and stress-response roles,” in Amino Acids in Higher Plants, ed. J. P. F. D’Mello (Wallingford: CABI).
Meng, D.-Y., Hou, L.-L., Yang, S., Meng, J.-J., Guo, F., Li, X.-G., et al. (2015). Exogenous polyamines alleviating salt stress on peanuts (Arachis hypogaea) grown in pots. Chinese J. Plant Ecol. 39:1209. doi: 10.17521/cjpe.2015.0117
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Minocha, R., Majumdar, R., and Minocha, S. C. (2014). Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 5:175.
Møller, S. G., and McPherson, M. J. (1998). Developmental expression and biochemical analysis of the Arabidopsis atao1 gene encoding an H2O2-generating diamine oxidase. Plant J. 13, 781–791. doi: 10.1046/j.1365-313x.1998.00080.x
Montesinos-Pereira, D., Barrameda-Medina, Y., Romero, L., Ruiz, J. M., and Sánchez-Rodríguez, E. (2014). Genotype differences in the metabolism of proline and polyamines under moderate drought in tomato plants. Plant Biol. 16, 1050–1057.
Moschou, P. N., Paschalidis, K. A., Delis, I. D., Andriopoulou, A. H., Lagiotis, G. D., Yakoumakis, D. I., et al. (2008). Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20, 1708–1724. doi: 10.1105/tpc.108.059733
Moschou, P. N., Wu, J., Cona, A., Tavladoraki, P., Angelini, R., and Roubelakis-Angelakis, K. A. (2012). The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J. Exp. Bot. 63, 5003–5015. doi: 10.1093/jxb/ers202
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Naka, Y., Watanabe, K., Sagor, G. H. M., Niitsu, M., Pillai, M. A., Kusano, T., et al. (2010). Quantitative analysis of plant polyamines including thermospermine during growth and salinity stress. Plant Physiol. Biochem. 48, 527–533. doi: 10.1016/j.plaphy.2010.01.013
Nambeesan, S., AbuQamar, S., Laluk, K., Mattoo, A. K., Mickelbart, M. V., Ferruzzi, M. G., et al. (2012). Polyamines attenuate ethylene-mediated defense responses to abrogate resistance toBotrytis cinerea in tomato. Plant Physiol. 158, 1034–1045. doi: 10.1104/pp.111.188698
Nambeesan, S., Datsenka, T., Ferruzzi, M. G., Malladi, A., Mattoo, A. K., and Handa, A. K. (2010). Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J. 63, 836–847. doi: 10.1111/j.1365-313x.2010.04286.x
Neily, M. H., Baldet, P., Arfaoui, I., Saito, T., Li, Q., Asamizu, E., et al. (2011). Overexpression of apple spermidine synthase 1 (MdSPDS1) leads to significant salt tolerance in tomato plants. Plant Biotechnol. 28, 33–42. doi: 10.5511/plantbiotechnology.10.1013a
Poidevin, L., Unal, D., Belda-Palazón, B., and Ferrando, A. (2019). Polyamines as quality control metabolites operating at the post-transcriptional level. Plants 8:109. doi: 10.3390/plants8040109
Rider, J. E., Hacker, A., Mackintosh, C. A., Pegg, A. E., Woster, P. M., and Casero, R. A. (2007). Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 33, 231–240. doi: 10.1007/s00726-007-0513-4
Rodríguez-Rosales, M. P., Jiang, X., Gálvez, F. J., Aranda, M. N., Cubero, B., and Venema, K. (2008). Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 179, 366–377. doi: 10.1111/j.1469-8137.2008.02461.x
Sebela, M., Radová, A., Angelini, R., Tavladoraki, P., Frébort, I., and Pec, P. (2001). FAD-containing polyamine oxidases: a timely challenge for researchers in biochemistry and physiology of plants. Plant Sci. 160, 197–207. doi: 10.1016/s0168-9452(00)00380-0
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shukla, V., Upadhyay, R. K., Tucker, M. L., Giovannoni, J. J., Rudrabhatla, S. V., and Mattoo, A. K. (2017). Transient regulation of three clustered tomato class-I small heat-shock chaperone genes by ethylene is mediated by SlMADS-RIN transcription factor. Sci. Rep. 7:6474.
Tavladoraki, P., Cona, A., and Angelini, R. (2016). Copper-Containing Amine oxidases and FAD-dependent polyamine oxidases are key players in plant tissue differentiation and organ development. Front. Plant Sci. 7:824.
Tester, M., and Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi: 10.1126/science.1183700
Torrigiani, P., Bressanin, D., Ruiz, K. B., Tadiello, A., Trainotti, L., Bonghi, C., et al. (2012). Spermidine application to young developing peach fruits leads to a slowing down of ripening by impairing ripening-related ethylene and auxin metabolism and signaling. Physiol. Plant. 146, 86–98. doi: 10.1111/j.1399-3054.2012.01612.x
Tsaniklidis, G., Kotsiras, A., Tsafouros, A., Roussos, P. A., Aivalakis, G., Katinakis, P., et al. (2016). Spatial and temporal distribution of genes involved in polyamine metabolism during tomato fruit development. Plant Physiol. Biochem. 100, 27–36. doi: 10.1016/j.plaphy.2016.01.001
Upadhyay, R. K., and Mattoo, A. K. (2018). Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 231, 318–328. doi: 10.1016/j.jplph.2018.10.001
Upadhyay, R. K., Fatima, T., Handa, A. K., and Mattoo, A. K. (2020). Polyamines and their biosynthesis/catabolism genes are differentially modulated in response to heat versus cold stress in tomato leaves (Solanum lycopersicum L.). Cells 9:1749. doi: 10.3390/cells9081749
Upadhyay, R. K., Handa, A. K., and Mattoo, A. K. (2019). Transcript abundance patterns of 9- and 13-lipoxygenase subfamily gene members in response to abiotic stresses (heat, cold, drought or salt) in tomato (Solanum lycopersicum L.) highlights member-specific dynamics relevant to each stress. Genes 10:683. doi: 10.3390/genes10090683
Verma, S., and Mishra, S. N. (2005). Putrescine alleviation of growth in salt stressed Brassica juncea by inducing antioxidative defense system. J. Plant Physiol. 162, 669–677. doi: 10.1016/j.jplph.2004.08.008
Wen, X.-P., Pang, X.-M., Matsuda, N., Kita, M., Inoue, H., Hao, Y.-J., et al. (2008). Over-expression of the apple spermidine synthase gene in pear confers multiple abiotic stress tolerance by altering polyamine titers. Transgenic Res. 17, 251–263. doi: 10.1007/s11248-007-9098-7
Wi, S. J., Kim, W. T., and Park, K. Y. (2006). Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. 25, 1111–1121. doi: 10.1007/s00299-006-0160-3
Yamaguchi, K., Takahashi, Y., Berberich, T., Imai, A., Takahashi, T., Michael, A. J., et al. (2007). A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 352, 486–490. doi: 10.1016/j.bbrc.2006.11.041
Yang, L., Liu, H., Fu, S. M., Ge, H. M., Tang, R. J., Yang, Y., et al. (2017). Na+/H+ and K+/H+ antiporters AtNHX1 and AtNHX3 from Arabidopsis improve salt and drought tolerance in transgenic poplar. Biol. Plant. 61, 641–650. doi: 10.1007/s10535-017-0724-9
Zeid, I. M., and Shedeed, Z. A. (2006). Response of alfalfa to putrescine treatment under drought stress. Biol. Plant. 50:635. doi: 10.1007/s10535-006-0099-9
Keywords: polyamine, putrescine (PUT), spermidine (SPD), spermine (SPM), tomato, drought, salt
Citation: Upadhyay RK, Fatima T, Handa AK and Mattoo AK (2021) Differential Association of Free, Conjugated, and Bound Forms of Polyamines and Transcript Abundance of Their Biosynthetic and Catabolic Genes During Drought/Salinity Stress in Tomato (Solanum lycopersicum L.) Leaves. Front. Plant Sci. 12:743568. doi: 10.3389/fpls.2021.743568
Received: 18 July 2021; Accepted: 13 September 2021;
Published: 15 October 2021.
Edited by:
Alejandro Ferrando, Universitat Politècnica de València, SpainReviewed by:
Kamala Gupta, Government General Degree College, Singur, IndiaCopyright © 2021 Upadhyay, Fatima, Handa and Mattoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Autar K. Mattoo, YXV0YXIubWF0dG9vQHVzZGEuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.