- 1The Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
- 2Key Laboratory of Plant Resources/Beijing Botanical Garden, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 3Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
Kudzu, Pueraria lobata, is a traditional Chinese food and medicinal herb that has been commonly used since ancient times. Kudzu roots are rich sources of isoflavonoids, e.g., puerarin, with beneficial effects on human health. To gain global information on the isoflavonoid biosynthetic regulation network in kudzu, de novo transcriptome sequencings were performed using two genotypes of kudzu with and without puerarin accumulation in roots. RNAseq data showed that the genes of the isoflavonoid biosynthetic pathway were significantly represented in the upregulated genes in the kudzu with puerarin. To discover regulatory genes, 105, 112, and 143 genes encoding MYB, bHLH, and WD40 transcription regulators were identified and classified, respectively. Among them, three MYB, four bHLHs, and one WD40 gene were found to be highly identical to their orthologs involved in flavonoid biosynthesis in other plants. Notably, the expression profiles of PlMYB1, PlHLH3-4, and PlWD40-1 genes were closely correlated with isoflavonoid accumulation profiles in different tissues and cell cultures of kudzu. Over-expression of PlMYB1 in Arabidopsis thaliana significantly increased the accumulation of anthocyanins in leaves and proanthocyanidins in seeds, by activating AtDFR, AtANR, and AtANS genes. Our study provided valuable comparative transcriptome information for further identification of regulatory or structural genes involved in the isoflavonoid pathway in P. lobata, as well as for bioengineering of bioactive isoflavonoid compounds.
Introduction
Pueraria lobata, commonly known as kudzu, belongs to the Leguminosae family. The roots of kudzu are enriched in starch, which has traditionally been used as a source of food consumption and beverage production in East Asia. Additionally, kudzu has been used for centuries in Chinese traditional medicine as an antipyretic, antidiarrheic, and antiemetic agent (Keung and Vallee, 1998; Wong et al., 2011). Kudzu roots are rich resources of natural product isoflavonoids, including daidzein, genistein, formononetin, and puerarin (also called daidzein 8-C-glycoside) (Prasain et al., 2003; Li et al., 2014). Among these isoflavonoids, puerarin is the major effective ingredient with antioxidative, antidiabetic, and antithrombotic effects (Hien et al., 2010; Liu et al., 2013), and it could also help to cure non-alcoholic fatty liver diseases, alcohol-induced adipogenesis, and osteonecrosis (Xia et al., 2013). The beneficial therapeutic effects of isoflavonoids, in particular puerarin, have made P. lobata an interesting plant species in investigating isoflavonoid biosynthesis and regulation.
Isoflavonoids are almost exclusively limited to the family of Leguminosae, the biosynthesis of which share the common upstream pathway with flavonoids (Supplementary Figure 1). Three molecules of malony-CoA were condensed with one molecule of 4-coumaroyl-CoA to form naringenin-chalcone or isoliquiritigenin, under the catalysis of chalcone synthase (CHS) or chalcone synthase/chalcone reductase (CHR). Chalcone isomerase (CHI) then catalyzes the following reaction to form naringenin and liquiritigenin, respectively. Subsequently, these two products were converted to daizein and genistein by the sequential actions of isoflavone synthase (IFS) and hydroxyflavanone dehydratase (HID). Finally, UDP-glycosyltransferase (UGT) could add glucose moiety to these two aglycone intermediates to form different O-glucosides at C7 or C5 position (He et al., 2011). Puerarin has been revealed to be synthesized via daidzein (He et al., 2011; Li et al., 2014; Wang et al., 2017). Although several structural genes, such as 4-Coumarate: Coenzyme A ligases, and UGTs involved in the isoflavonoid pathway have been identified in kudzu (He et al., 2011; Li et al., 2014; Wang et al., 2017), transcription factors were less identified for the regulation of isoflavonoid biosynthesis in kudzu.
Transcriptional regulation of flavonoid pathway has been extensively studied in many plant species, such as Zea mays (Grotewold et al., 1998; Carey et al., 2004), Arabidopsis thaliana (Nesi et al., 2000, 2001; Zhu et al., 2009; Wada et al., 2014; Shin et al., 2015), Malus domestica (Espley et al., 2007), and Vitis vinifera (Terrier et al., 2009; Hichri et al., 2010; Ali et al., 2011; Huang et al., 2014). These studies have established the key roles of transcription factors in the regulation of most structural genes. Up to now, at least six distinct types of transcription regulators, which are MYB, bHLH, WD40, WRKY, Zinc finger, and MADS-box proteins, have been proven to be involved in the regulation of secondary metabolite biosynthetic pathway (Sun et al., 2019; Deng et al., 2020a, b; Hao et al., 2020, 2021; Anguraj Vadivel et al., 2021; Fan et al., 2021; Liu et al., 2021; Mao et al., 2021; Zhou et al., 2021a, b). Among them, transcription regulators of MYB, bHLH, and WD40 function individually or collaborate with each other as MBW complex to control multiple enzymatic steps in the flavonoid pathway (Ramsay and Glover, 2005; Gonzalez et al., 2008; Xu et al., 2014).
MYB proteins, R2R3-MYBs in particular, are major players as the positive or negative regulators toward key biosynthetic genes required for the production of flavonoids (Jiu et al., 2021; Li et al., 2021; Shi et al., 2021). In Arabidopsis, flavonoid biosynthesis is mainly regulated by a set of R2R3-MYB transcription factors. Transparent Testa 2 (AtTT2), Production of Anthocyanin Pigments 1 (AtPAP1), and AtMYB11 could activate flavonoid biosynthesis, whereas AtMYBL2 and others repressed the biosynthesis of flavonoids (Xu et al., 2014).
The first bHLH (the basic helix-loop-helix) protein Lc was reported from maize, which cooperates with the MYB transcription factors C1 and PL1 to regulate the anthocyanin biosynthetic pathway in maize (Ludwig et al., 1989). Several corresponding orthologs of the Lc gene have been identified in other plant species (Nesi et al., 2000; Ramsay et al., 2003; Park et al., 2007; Montefiori et al., 2015). The bHLH type protein AtTT8 from Arabidopsis is required for normal expression of late flavonoid biosynthetic genes in the siliques of Arabidopsis (Nesi et al., 2000), which interacts directly with MYB transcription factors for flavonoid biosynthesis.
WD40 repeat domain proteins are not considered to be real transcription factors in regulating flavonoid pathway, in contrast, they act more likely as a docking platform or receptor for recruiting other proteins (van Nocker and Ludwig, 2003; Zhao et al., 2013). In Arabidopsis, the WD40 protein Transparent Testa Glabra 1 (AtTTG1) acts primarily via interplaying with AtTT8 partner in the TT2-TT8-TTG1 complex regulating the expression of AtANR gene (Nesi et al., 2001; Baudry et al., 2004). Several genes encoding WD40 proteins involved in the regulation of flavonoid pathway have been identified from several plant species, including AN11 from Petunia × hybrid (de Vetten et al., 1997), PFWD from Perilla frutescens (Sompornpailin et al., 2002), ZmPAC1 from maize (Carey et al., 2004), MtWD40-1 from Medicago truncatula (Pang et al., 2009), and WDR1 and WDR2 from grapevine (Matus et al., 2010).
The biosynthesis of flavonoids/isoflavonoids has been extensively studied in model plants, but it was left behind in non-model plants due to the lack of genomic and genetic information. The current fast-growing RNA-Seq sequencing technique makes it possible to take the advantage of global gene expression data in some species without available genomic information. In this study, we integrated comparative transcriptome information from two kudzu genotypes with contrasting isoflavonoid concentrations in roots and identified a set of putative genes that are possibly involved in isoflavonoid production in kudzu roots, with an emphasis on transcription factors (MYB, bHLH, and WD40). Moreover, our investigation demonstrated that PlMYB1 was a critical transcription factor involved in the isoflavonoid pathway in P. lobata. These findings, thus, provided insights into the regulation network of isoflavonoid biosynthesis in kudzu and offered an important target for bioengineering of bioactive isoflavonoids with beneficial effects on human health.
Results
Transcriptome Sequencing and Gene Annotation
In a previous report, it was revealed that a kudzu genotype (No. 1, Figure 1A) accumulates a massive amount of puerarin in the roots, whereas the other genotype did not (No. 2, Figure 1B; He et al., 2011). In the present study, we further carried out transcriptome sequencing using the roots of these two types of kudzu plants, attempting to obtain global transcriptome information and to discover new genes involved in the regulation of isoflavonoid biosynthesis by transcriptome comparison.
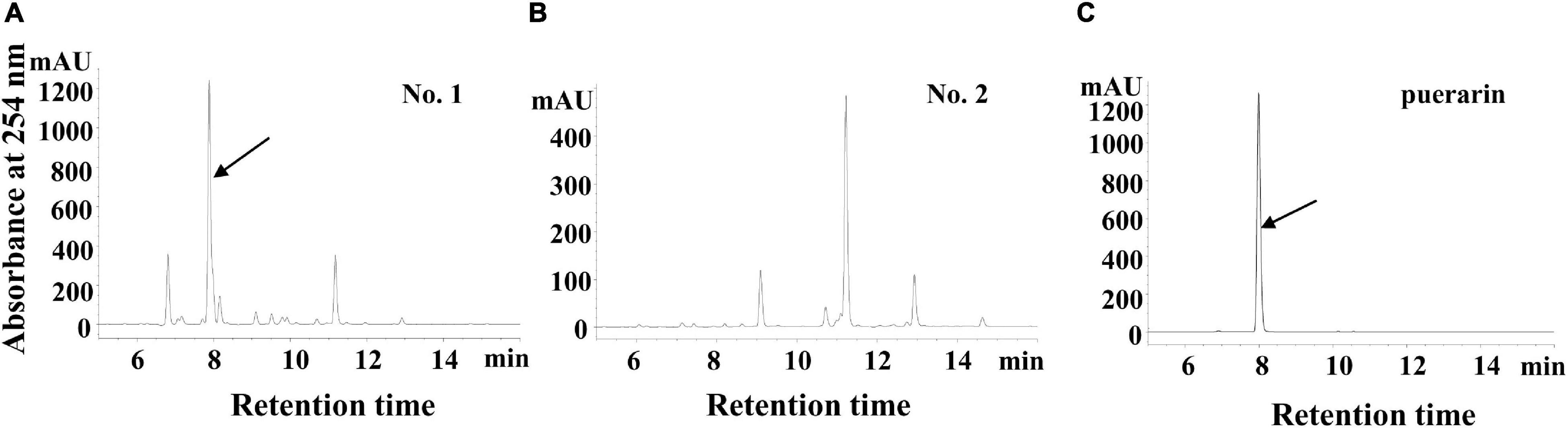
Figure 1. The high-performance liquid chromatography (HPLC) of flavonoid profiles in two types of kudzu plants. (A) HPLC chromatogram of the flavonoids in No.1 kudzu plant, the arrowhead indicates puerarin. (B) HPLC chromatogram of the flavonoids in No.2 kudzu plant. (C) HPLC chromatogram of the puerarin authentic standard.
After sequence cleaning and assembling by using Trinity (trinityrnaseq_r2013-02-25) program (Grabherr et al., 2011), a total of 88,398 unigenes were obtained from these two kudzu plants, and the number of unigenes decreased with the length ranging from short (201 bp) to long (more than 3 kb) (Supplementary Figure 2 and Supplementary Table 1). Among them, 27,515 and 25,175 unigenes were detected solely in Nos. 1 and 2 kudzu, respectively. For the genes expressed in both samples, the reads per kilobases per million reads (RPKM) value of 22, 389 unigenes changed less than twofold between the samples, whereas 5,938 and 7,373 unigenes increased or decreased significantly more than twofold in roots of No. 1 kudzu than in No. 2 kudzu (Supplementary Table 1).
The function of 14,896 significantly upregulated unigenes in roots of the No. 1 sample was further annotated and classed into 25 groups based on the Cluster of Orthologous Groups of proteins (COG) database. Among them, the highest amounts (2,289) are those having signal transduction mechanisms (Figure 2A). Overall, 863 unigenes were predicted to be involved in the biosynthesis, transport, and catabolism of secondary metabolites (Figure 2A). Among them, 20 unigenes encoding enzymes were related to the isoflavonoid pathway, including 1 IFS, 14 CHS, 2 CHR, and 3 CHI genes (Supplementary Table 2).
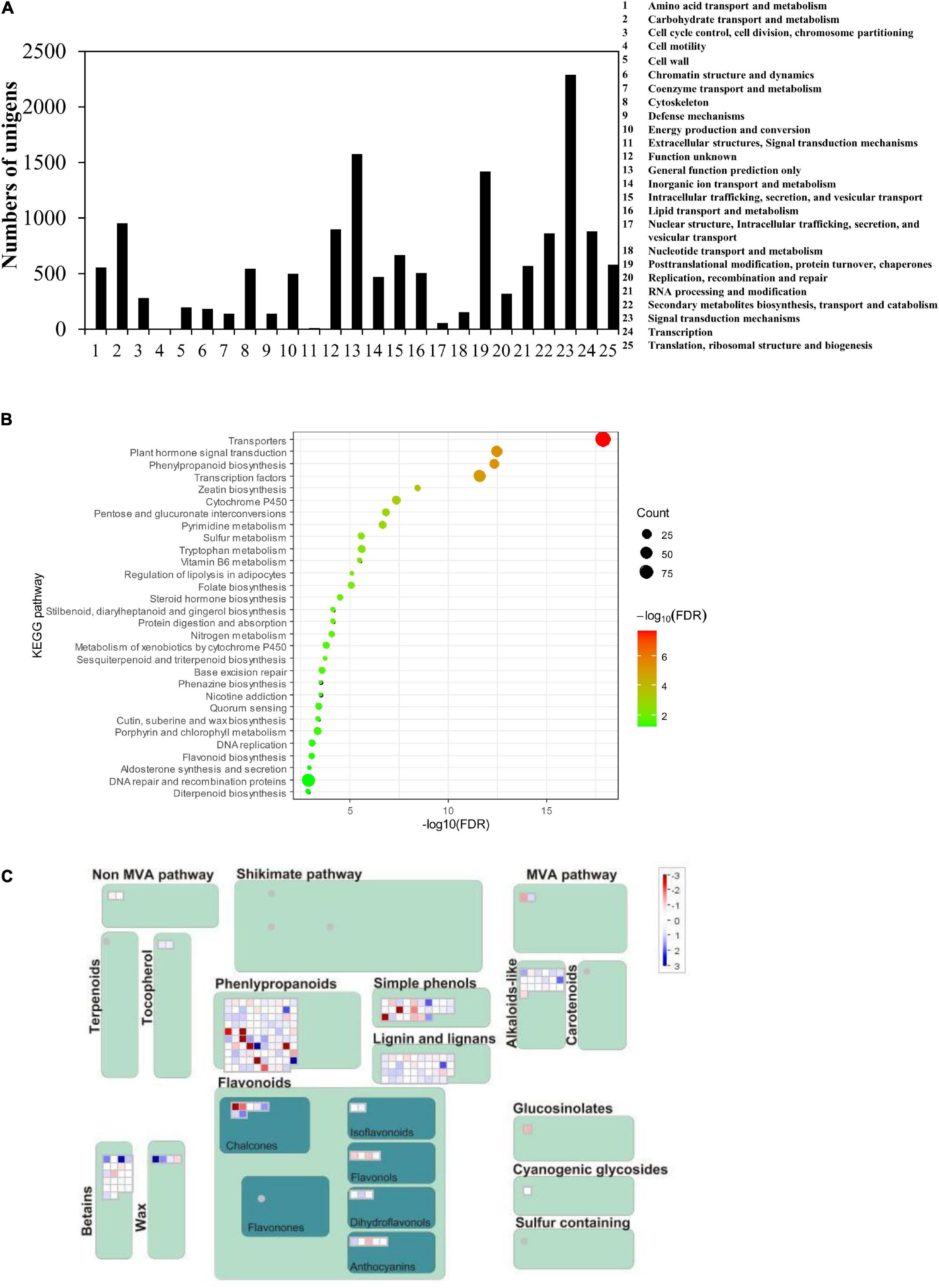
Figure 2. Statistical analysis of the kudzu root transcriptome data. (A) Cluster of Orthologous Groups (COG) classification of unigenes of P. lobata. A total of 14,896 significantly upregulated unigenes were assigned to 25 classifications. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched pathway highlighting the significantly upregulated genes in the secondary metabolic pathway in the two kudzu transcriptome. (C) MapMan overview maps related to flavonoid and phenylpropanoid pathways show evident differences in transcript levels between two kudzu plants. Red indicated upregulated genes and blue indicated downregulated genes.
In particular, KEGG pathway analyses showed that these upregulated genes were significantly enriched in the phenylpropanoid pathway and flavonoid pathway (Figures 2B,C). It was speculated that the accumulation of a higher amount of isoflavonoids resulted from the higher expression of entire pathway genes, like CHS, CHI, IFS, and so on, which should be coordinately regulated by some unknown transcription factors. Therefore, this study mainly focused on the discovery of transcription factor genes, specially MYB, bHLH, and WD40 genes.
Identification and Classification of MYBs in the Isoflavonoid Biosynthetic Pathway
In this study, 105 MYB genes encoding transcription factors from kudzu roots were identified and confirmed by their conserved domains (Supplementary Table 3). These putative MYB transcription factors were classified into seven different super-families, including DNA_pol_phi superfamily, GAT_SF superfamily, H15 superfamily, Myb_CC_LHEQLE superfamily, SANT superfamily, SKIP_SNW superfamily, and VHS_ENTH_ANTH superfamily (Supplementary Table 3). Among them, the SANT superfamily is the largest superfamily with 65 unigenes (Supplementary Table 3).
R2R3-MYB transcription factors of the SANT superfamily in Arabidopsis were divided into 25 different subgroups, and the members from Sg4, Sg5, Sg6, Sg7, and Sg15 subgroups were reported to be involved in the regulation of anthocyanin and proanthocyanidin biosynthesis (Stracke et al., 2001; Hichri et al., 2011). In kudzu, two MYB genes PlMYB4-1 (comp739_c0_seq1) and PlMYB4-2 (comp41186_c0_seq1) were classed in the Sg4 subgroup, and they shared 34% identity with AtMYB32 and 88% identity with AtMYB4, respectively. The available transcript of PlMYB4-1 (204 bp) and PlMYB4-2 (1,110 bp) were predicted to encode a truncated peptide and a full-length protein of 370 amino acid residues, respectively.
Additionally, among MYB unigenes predicted in kudzu, PlMYB1 (comp36832_c0_seq1_3, Genbank accession No. KR698796) showed the highest identity with GmMYB176 (68%) and AtTT2 (63%) that were key regulators of isoflavonoid and proanthocyanidin biosynthesis in soybean and Arabidopsis, respectively (Nesi et al., 2001; Yi et al., 2010). Moreover, PlMYB1 (657 bp) was predicted to be full-length and it encodes a deduced protein comprising 219 amino acid residues.
Sequence alignments of the deduced PlMYB1, PlMYB4-1, and PlMYB4-2 proteins showed that the R3 regions were highly conserved compared to other known MYB proteins involved in flavonoid pathway in other species, in particular in the bHLH binding motif (Figure 3A). It is obvious that PlMYB4-2 belonged to the R2R3 group, while PlMYB1 belonged to the R3 group. Phylogenetic analysis showed that PlMYB4-2 was clustered into a group with AtMYB4, FaMYB1, and AmMYB308 that were involved in the general flavonoid pathway, while PlMYB1 was grouped with DkMYB1 and VvMYBPA1 that were involved in the proanthocyanidin biosynthesis (Figure 3B). However, the R2R3 region was missing in PlMYB4-1, which was not pursued further in the current investigation.
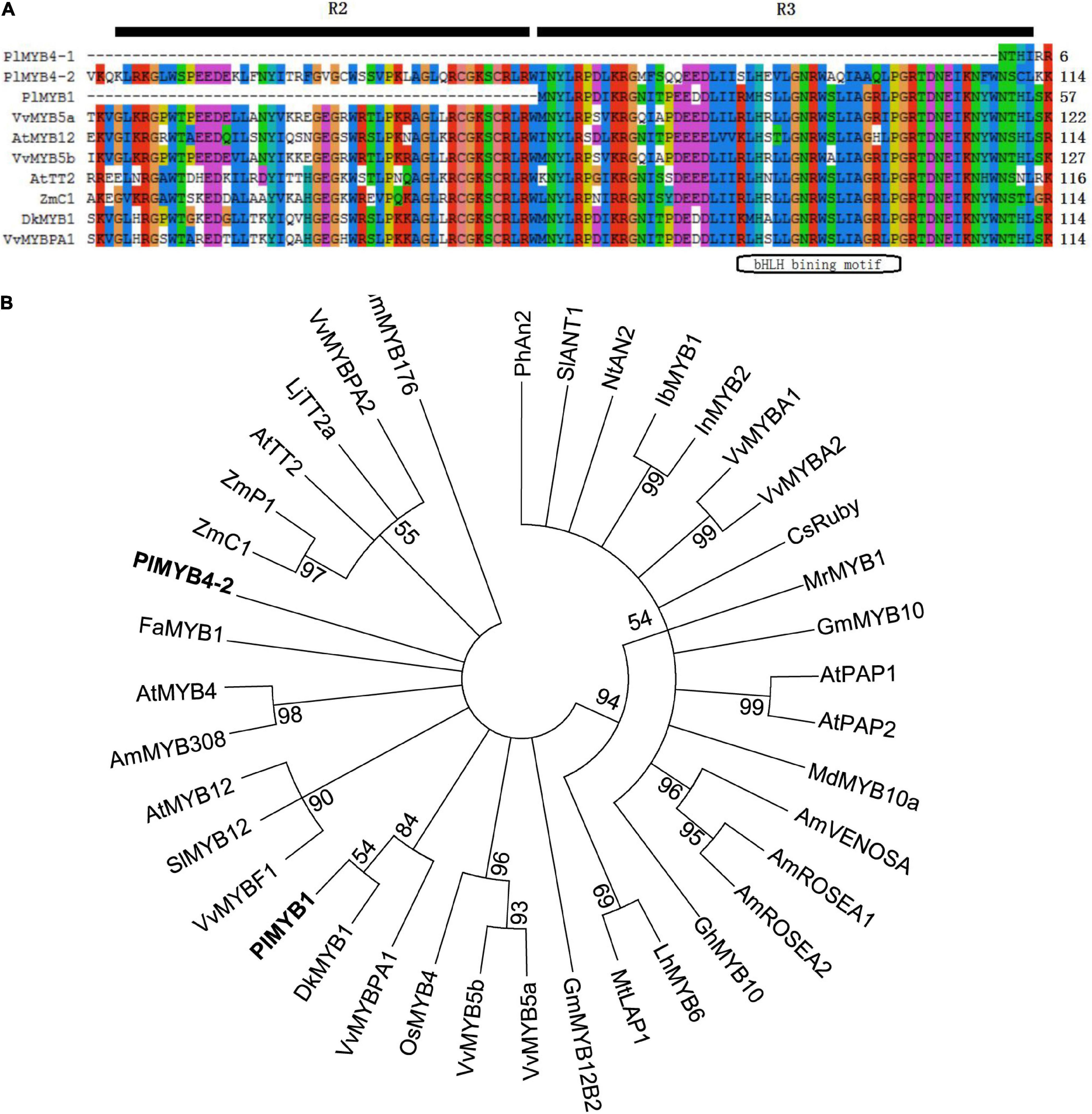
Figure 3. Sequence analysis of MYBs regulating flavonoid biosynthesis at amino acid level. (A) Alignment of several MYBs regulating flavonoid biosynthesis. (B) Phylogenetic analysis of MYB transcription factors regulating flavonoid metabolism. All the sequences used in the figure are retrieved from the GenBank database, the accession number are as followed (shown in parenthesis): Antirrhinum majus AmROSEA1 (ABB83826); AmROSEA2 (ABB83827); AmVENOSA (ABB83828); AmMYB308 (P81393); Arabidopsis thaliana AtPAP1 (AAG42001); AtPAP2 (AAG42002); AtTT2 (NP_198405); AtMYB12 (ABB03913); AtMYB4 (NP_195574); Citrus sinensis CsRuby (AFB73913); Diospyros kaki DkMYB4 (BAI49721); Fragaria × ananassa FaMYB1 (AAK84064); Garcinia mangostana GmMYB10 (ACM62751); Gerbera hybrid GhMYB10 (CAD87010); Ipomoea batatas IbMYB1 (BAF45114); Ipomoea nil InMYB2 (BAE94709); Solanum lycopersicum SlANT1 (AAQ55181); Solanum lycopersicum SlMYB12 (ACB46530); Lilium hybrid LhMYB6 (BAJ05399); Lotus japonicus TT2a (BAG12893); Malus × domestica MdMYB10a (ABB84753); Medicago truncatula MtLAP1(ACN79541); Morella rubra MrMYB1 (ADG21957); Nicotiana tabacum NtAN2 (ACO52470); Oryza sativa OsMYB4 (BAA23340); Petunia × hybrida PhAn2 (AAF66727); Vitis vinifera VvMYBA1 (BAD18977); VvMYBA2 (BAD18978); VvMYBPA1 (CAJ90831); VvMYBPA2 (ACK56131); VvMYBF1 (ACV81697); VvMYB5a (AAS68190); VvMYB5b (AAX51291); Zea mays ZmC1 (AAA33482); ZmPl (AAA19819); Glycine max GmMYB176 (NP_001236048); GmMYB12B2 (AEC13303); P. lobata PlMYB1 (AKR04122).
Identification and Classification of bHLH Genes in the Isoflavonoid Biosynthetic Pathway in Kudzu
A total of 111 bHLH unigenes were predicted in the transcriptome of kudzu roots, and they were classified into corresponding subgroups by sequence identity comparison with orthologs in Arabidopsis (Supplementary Table 4). In particular, four kudzu bHLH proteins were grouped into Subgroup III, the ortholog of which was involved in flavonoid biosynthesis in Arabidopsis (Heim et al., 2003). PlbHLH3-1 (Comp36398_c2_seq1, 201 bp) encoding protein shared the highest identity at amino acid level with AtbHLH93 (62%) that belongs to Subgroup IIIb. The other three were assigned to Subgroup IIId. Among them, both PlbHLH3-2 (comp37021_c0_seq1, 474 bp) and PlbHLH3-3 (comp37021_c1_seq1, 1,332 bp) shared 63% and 45% identity with AtbHLH13, respectively. PlbHLH3-4 (comp45038_c0_seq1, 1,539 bp, Genbank Accession No. KT236099) shared the highest identity (48%) with AtbHLH3.
Sequence alignments of the deduced PlbHLH3-1, PlbHLH3-2, PlbHLH3-3, and PlbHLH3-4 proteins with other representative bHLH proteins involved in the flavonoid pathway showed that only PlbHLH3-4 had the intact bHLH-MYC_N region (Figure 4A), which is usually present in the N-terminal of bHLH transcription factors regulating phenylpropanoid biosynthesis (Marchler-Bauer et al., 2015). The bHLH-MYC_N domain commonly has the specific DNA-binding ability attributed to the amphipathic affinity of its N-terminus. This was especially apparent in the critical His-Glu-Arg (H-E-R) residues located at positions 5, 9, and 13 in the basic region for PlbHLH3-3 and PlbHLH3-4 (Figure 4A), which could bind to DNA target as reported previously (Atchley and Fitch, 1997; Massari and Murre, 2000; Toledo-Ortiz et al., 2003). The bHLH-MYC_N region was composed of two hydrophobic α-helices linked by a divergent loop, but only PlbHLH3-3 and PlbHLH3-4 contained an intact domain in this region (Figure 4A).
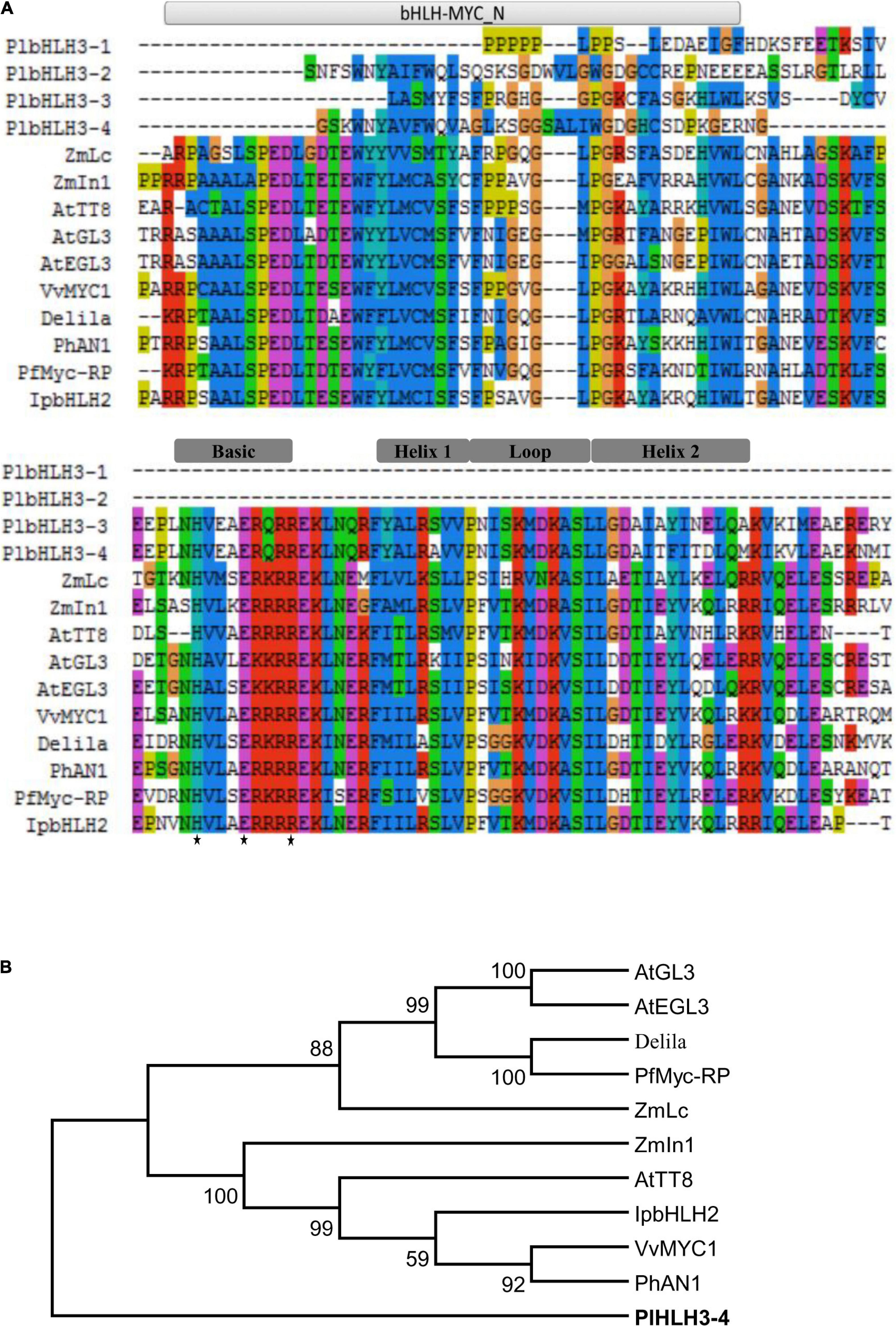
Figure 4. Analysis of bHLHs regulating flavonoid biosynthesis at the amino acid level. (A) Alignment of bHLHs regulating flavonoid biosynthesis. (B) Phylogenetic analysis of bHLH transcription factors regulating flavonoid metabolism. All the sequences used in the figure are derived from the GenBank database, the accession number are as followed (shown in parenthesis): Zea mays ZmLc (NP_001105339); ZmIn1 (AAB03841); Arabidopsis thaliana AtTT8 (Q9FT81); AtGL3 (AED94664); AtEGL3 (Q9CAD0); Delila (AAA32663); Vitis vinifera VvMYC1 (ACC68685); Petunia × hybrid PhAN1 (AAG25928), PhMyc-rp (BAA75513); Ipomoea purpurea IpbHLH2 (ABW69688), and P. lobata PlbHLH3-4 (AKR04123). The His5-Glu9-Arg13 (H-E-R) motif is indicated with a black star.
The phylogenic relationship showed that PlbHLH3-3 and PlbHLH3-4 were separated from the other bHLH transcription factors regulating the anthocyanin pathway, such as AtTT8 from Arabidopsis and Lc from maize (Figure 4B), suggesting that they are likely involved in other flavonoid branch pathways, e.g., isoflavonoid pathway.
Identification and Classification of Unigenes Encoding WD40 Repeat Domain Proteins in Kudzu
The repeat proteins WD40 are key components in the MBW complex, therefore, we searched and identified a total of 143 unigenes encoding WD40 repeat domain proteins in the transcriptome of kudzu roots (Supplementary Table 5). They were grouped into 16 different super-families, and the WD40 super-family was the largest super-family (96 members, Supplementary Table 5). Among all these predicted WD40 repeat domain proteins, the deduced amino acid sequence of PlWD40-1 (comp42449_c1_seq1_24, Genbank Accession No. AKR04124.1) showed the highest identity (64%) to Transparent testa glabra 1 (AtTTG1) from Arabidopsis, 62% to MtWD40-1 from M. truncatula, and 61% to PhAN1 from P. hybrid. The WD40 repeat domains are highly conserved among these WD40 proteins (Figure 5A). Phylogenetic analysis showed that PlWD40-1 is most closely related to ZmPAC1 that regulates anthocyanins production in Zea mays (Figure 5B). As these WD40 orthologs are involved in the flavonoid pathway, the high identity and close phylogenetic relationship of PlWD40-1 with them suggested that PlWD40-1 may have a similar function in the isoflavonoid pathway in kudzu.

Figure 5. Analysis of PlWD40-1 proteins with WD40 proteins of other plant species at the amino acid level. (A) Alignment of WD40s regulating flavonoid biosynthesis. The typical WD domains were indicated in solid lines, and the last two amino acids of each repeat domain are marked with red stars. (B) Phylogenetic analysis of WD40 transcription factors regulating flavonoid metabolism. All the sequences used in the figure are derived from the GenBank database; the accession number is as followed (shown in parenthesis): Arabidopsis thaliana AtTTG1 (CAB45372); Malus domestica MdTTG1 (ADI58760); Petunia × hybrid PhAN11 (AAC18914); Ipomoea nil InWDR (BAE94407); Perilla frutescens PFWD (BAB58883); Zea mays ZmPAC1 (AAM76742) and Medicago truncatula MtWD40-1 (ABW08112), and P. lobata PlWD40-1 (AKR04124.1).
Expression of Key Transcription Factor Genes Was Closely Correlated With Total Flavonoid Accumulation in Kudzu
To further screen candidate transcription factors that play key roles in the isoflavonoid pathway, the association between flavonoid accumulation level and the expression level of candidate genes was investigated. We found that the total flavonoid level was relatively higher in leaves than in roots or stems in both two kudzu genotypes (Figure 6A). Furthermore, total flavonoid content was higher in the roots of No. 1 than in No. 2, which was essentially contributed by puerarin as confirmed on high performance liquid chromatography (HPLC) in the present study (Figure 1). By contrast, total flavonoids were the lowest in stems in both kudzu genotypes (Figure 6A). Accordingly, the transcript level of PlMYB1 determined by qPCR was relatively higher in leaves than in other tissues of both kudzu plants (Figure 6C). Notably, the transcript level of PlMYB1 is exactly consistent with the accumulation levels of total flavonoids in various tissues. This result implied that PlMYB1 was likely involved in the regulation of isoflavonoid biosynthesis. By contrast, the PlMYB4-2 gene was highly expressed in the roots of both kudzu genotypes, with a very low level in stems and leaves (Figure 6E), implying a weaker association with total flavonoid accumulation.
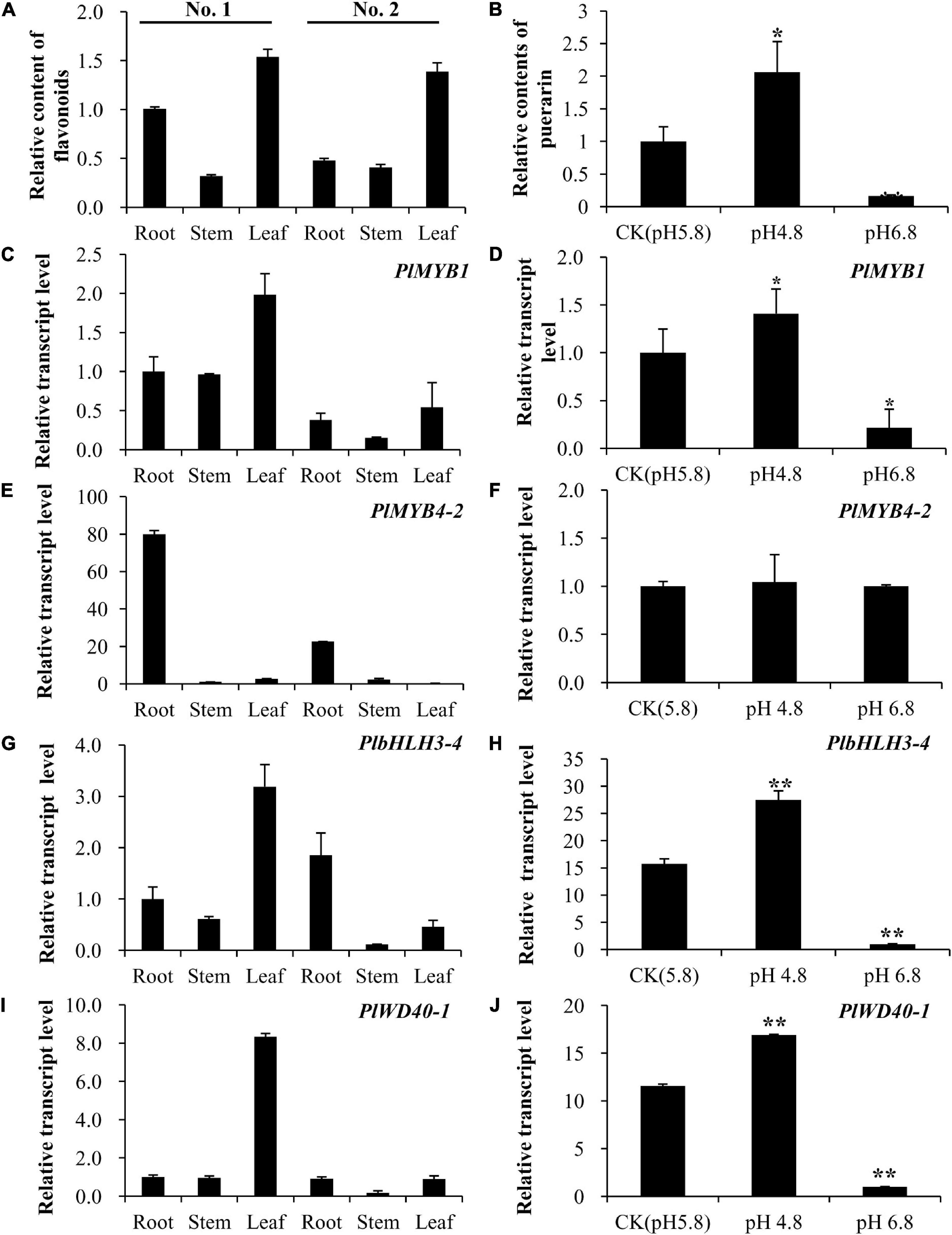
Figure 6. Association analyses of flavonoid accumulation and expression levels of key transcription factor genes in various tissues and cell culture under different pH conditions. (A) Accumulation of flavonoids in roots, stems, and leaves of two types of kudzu plants. (B) Accumulation of puerarin in cell cultures under different pH conditions. (C,E,G,I) The relative transcript levels of PlMYB1 (C), PlMYB4-2 (E), PlbHLH3-4 (G), and PlWD40-1 (I) in roots, stems, and leaves of two types of kudzu plants. (D,F,H,J) The relative transcript levels of kudzu PlMYB1 (D), PlMYB4-2 (F), PlbHLH3-4 (H), and PlWD40-1 (J) in cell cultures under pH 4.8, pH 5.8, and pH 6.8.
It was revealed that PlbHLH3-4 was expressed at the highest level in leaves of No. 1 kudzu genotype plants, but it accumulated at a low level in the leaves of the No. 2 kudzu genotype (Figure 6G), which is far more different from the total flavonoid accumulation pattern. Same as PlbHLH3-4, the transcript level of PlWD40-1 was relatively high (more than 8-fold than in other tissues) in leaves of the No. 1 kudzu plant, but lower in other tissues of the No. 1 kudzu plant (Figure 6I, left), implying less correlation with total flavonoid accumulation.
Key Transcription Factor Genes Were Consistently Associated With Puerarin Content in Cell Cultures
Puerarin is the predominant isoflavonoid compound in kudzu. It was revealed that the cell culture produced from the No. 1 kudzu genotype plant accumulated an evident amount of puerarin, and its content was not significantly affected by sugar or naphthyl acetic acid (NAA) concentration, or the presence of SA, MeJA, or light, but by the pH value of medium (Supplementary Figure 3). In comparison with the control pH value of 5.8, the puerarin content doubled when the pH value dropped to 4.8, whereas the puerarin content reduced about 6 folds when the pH value was increased to 6.8 (Figure 6B, right).
Quantitative PCR analyses showed that the transcript level of PlMYB1 was relatively higher at a pH value of 4.8 than at 5.8 or 6.8 (Figure 6D). The transcript profile of PlMYB1 is consistent with the accumulation levels of puerarin in cell cultures grown under different pH conditions. This result implied that PlMYB1 is likely involved in the regulation of isoflavonoids, in particular puerarin biosynthesis. However, the expression level of PlMYB4-2 was not affected by the pH value of the cell culture medium (Figure 6F, right), suggesting a weaker association with puerarin biosynthesis.
Similar to PlMYB1, the expression level of PlbHLH3-4 in cell culture was increased at a pH value of 4.8 and decreased at a pH value of 6.8 as compared to the control at a pH value of 5.8 (Figure 6H). In addition, the transcript level of PlWD40-1 was also higher at a pH value of 4.8 but lower at a pH value of 6.8 when compared with that of control at a pH value of 5.8 (Figure 6J, right).
Taken together, PlMYB1, PlbHLH3-4, and PlWD40-1 were highly expressed in cell culture under a pH value of 4.8, and their expression patterns were highly correlated with levels of puerarin in cell cultures under various pH treatments, suggesting they might cooperate as an MBW complex to regulate the isoflavonoids, e.g., puerarin biosynthesis, under different pH treatments. Especially, the expression pattern of PlMYB1 matched very well with the accumulation of puerarin, implying it is likely a key player in the MBW complex.
Subcellular Localization of PlMYB1
To validate the function of the putative transcription factor in the regulation of the isoflavonoid pathway, the PlMYB1 gene was successfully cloned for further characterization. The open reading frame (ORF) of PlMYB1 was fused with green fluorescent protein (GFP) at the C-terminus and transferred into Arabidopsis leaf protoplasts. Green fluorescence signals for PlMYB1:GFP were detected in the nucleus (Figure 7A), which was evidently distinct from that of the control GFP in the cytosol (Figure 7B). This result indicated that PlMYB1 is localized in the nucleus as a transcription factor to exert its function.
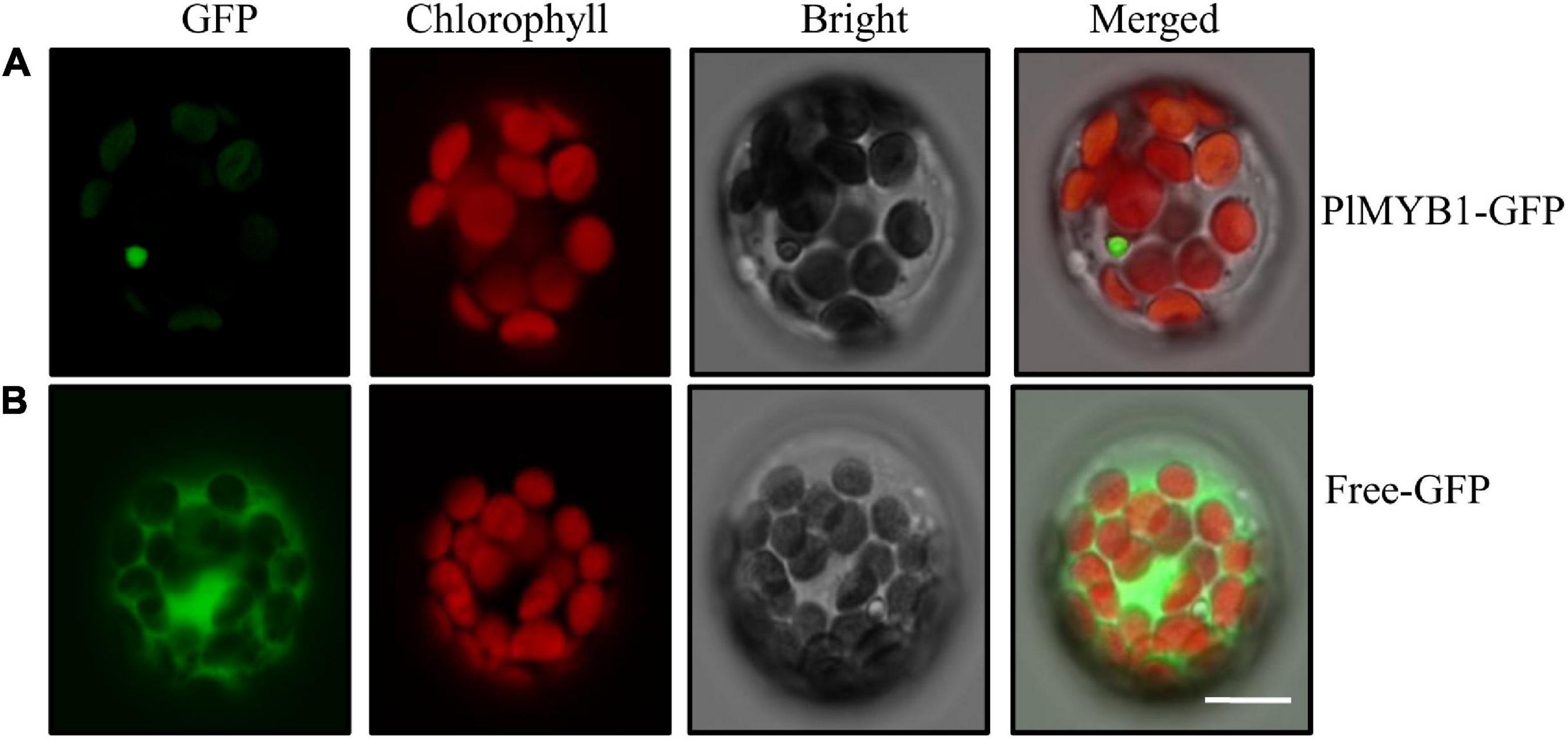
Figure 7. Subcellular localization of PlMYB1 protein. Subcellular localization assays of PlMYB1 fused with GFP in Arabidopsis protoplast. Fluorescence signals were visualized using confocal laser scanning microscopy. From left to right: green fluorescence, autofluorescence of chloroplast, bright field, and merged images of PlMYB1-GFP fusion protein (A), and GFP (B). Bar = 10μm.
In vivo Functional Characterization of PlMYB1 in Arabidopsis
To further determine the regulatory function of PlMYB1, it was also over-expressed in the wild-type Arabidopsis. The expression levels of PlMYB1 were confirmed by qPCR analysis in different lines (Figure 8A). Quantitative analysis revealed that anthocyanin levels increased by more than 0.4, 1.4, and 0.9-fold in rosette leaves of three transgenic lines as compared to the wild-type control (Figure 8B and Supplementary Figure 4A). In addition, soluble and insoluble proanthocyanidins in the mature seeds of these transgenic lines increased from 0.6- to 1.5-fold and 0.7- to 1-fold than the wild-type control, respectively (Figure 8C and Supplementary Figure 4B).
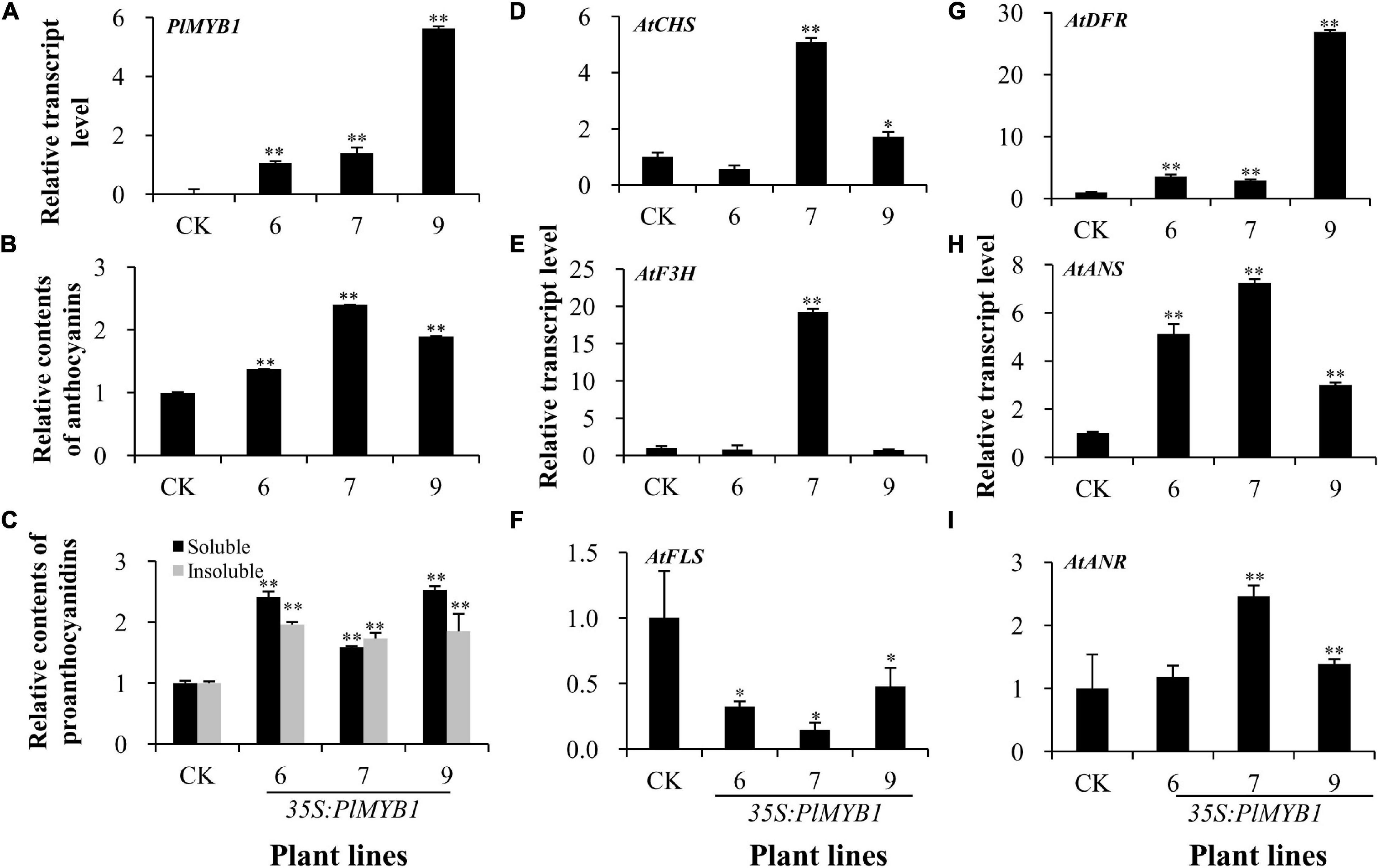
Figure 8. Over-expression of PlMYB1 in Arabidopsis. (A) The relative transcript level of PlMYB1 in the wild type of Arabidopsis. (B,C) The relative content of anthocyanins in rosette leaves (B) and proanthocyanidins in mature seeds (C) in the PlMYB1 over-expressing homozygous Arabidopsis. Soluble proanthocyanidins were measured by using the DMACA-based method and insoluble proanthocyanidins were measured by using the butanol/HCl hydrolysis method. (D–I). Relative transcript levels of AtCHS, AtF3H, AtFLS, AtDFR, AtANS, and AtANR in the rosette leaves of the transgenic and wild type Arabidopsis determined by qPCR analysis.
To further investigate the effects of PlMYB1 on the expression of anthocyanin/proanthocyanidin pathway genes, the expression levels of several key pathway genes were determined in rosette leaves by qPCR analysis (Figures 8D–I). For the early pathway genes of AtCHS and AtF3H, they were both increased more significantly in the No. 7 transgenic line (Figures 8D,E). Especially, it showed that the expression levels of three later pathway genes AtDFR, AtANS, and AtANR genes were highly increased in these transgenic lines (Figures 8G–I). Notably, the expression level of AtFLS was significantly decreased in the transgenic lines than that in the wild-type control (Figure 8F). Taken together, these data indicated that the over-expression of PlMYB1 activated the expression of anthocyanin/proanthocyanidin pathway genes, and consequently increased the accumulation of these compounds.
Discussion
Pueraria lobata is a legume plant endemic to China, which is well known for its special accumulation of health-beneficial isoflavonoid compound of puerarin in the roots. In a previous study, we found that a kudzu genotype produces high puerarin content, while another genotype has low puerarin content in the roots (He et al., 2011; Figure 1). In the present study, we explored the transcriptomes of roots from two previously established kudzu plants. Transcriptome comparative analysis revealed several transcription factor genes were differentially expressed in two kudzu genotypes. Among them, the expression of PlMYB1 showed a close correlation with the biosynthesis of puerarin, and it was able to significantly increase the accumulation of anthocyanins/proanthocyanidins in transgenic Arabidopsis through activating AtDFR, AtANR, and AtANS (Figure 8).
PlMYB1 Regulates Flavonoid Pathway in Kudzu and Arabidopsis
It is well known that flavonoid and isoflavonoid pathway genes were mainly regulated by the MBW complex comprising of MYB, bHLH, and WD40, and the regulatory function of MBW is conserved in most plants (Goodrich et al., 1992). In the MBW complex, MYB transcription factors are the major player, and they were classified into distinct groups. The MYBs in the Sg4 subgroup share the ERF-associated amphiphilic repression (EAR) motif core (Goodrich et al., 1992). The Sg4 subgroup members were involved in stress responses and plant evolution (Bedon et al., 2010), and also acted as a repressor factor of phenolic acid metabolism and lignin biosynthesis (Zhao et al., 2013). The MYBs members in Arabidopsis (AtMYB3, AtMYB4, AtMYB7, and AtMYB32) of the Sg4 subgroup are able to repress the biosynthesis of polyphenols by interacting with bHLH proteins (Zhao et al., 2013). AtMYB4 has been shown to be a transcriptional repressor involved in the inhibition of genes in the polyphenol biosynthetic pathway, such as the Cinnamate 4-Hydroxylase gene (C4H) (Jin et al., 2000).
Our study showed that two MYB members of the Sg4 subgroup were present in kudzu, namely PlMYB4-1, and PlMYB4-2. Among them, the expression pattern of PlMYB4-2 was not consistent with the accumulation profiles of puerarin (Figures 6E,F), therefore, PlMYB4-2 is unlikely related to puerarin accumulation in kudzu. By contrast, PlMYB1 showed a high expression level in roots and leaves in both kudzu plants, which is completely consistent with the accumulation pattern of total flavonoid in various tissues of kudzu.
Moreover, a previous study demonstrated that pH condition is a key regulatory factor in flavonoid biosynthesis. It was found that treatment with a high medium pH value induced a dramatic decrease in the concentration of cyanidin in crabapple leaves (Zhang et al., 2014), whereas high medium pH values increased the content of flavones and flavonols (Zhang et al., 2014). Several MYB TFs have been suggested to be involved in the regulation of pH responses (Zhang et al., 2014). In our study, we found that low pH treatment increased puerarin content in kudzu cell cultures, and the transcript level of the PlMYB1 gene was also accelerated (Figure 6). Particularly, PlMYB1 was evidently increased under low pH value treatment and significantly decreased under high pH value treatment (Figure 6), which is consistent with the accumulation levels of puerarin under various pH value conditions. Therefore, PlMYB1 possibly affected puerarin contents via regulating the transcript level of downstream key structural genes.
Furthermore, the ectopic over-expression of PlMYB1 led to significant increases in the expression level of several pathway structural genes, e.g., AtDFR, AtANS, AtANR, Figure 8), and accordingly increased the content of anthocyanins and proanthocyanidins in Arabidopsis. These results demonstrated that PlMYB1 functioned as an activator for the anthocyanins/proanthocyanidins branch. Notably, the transcript level of FLS was reduced significantly (Figure 9), which might block the flavonol branch and switch the flux to the anthocyanidins/proanthocyanidins branch as illustrated in Figure 9, indicating PlMYB functioned as a repressor for the flavonol branch. Therefore, PlMYB1 acted dual role as activator and repressor in the flavonoid biosynthetic pathway in Arabidopsis, which is similar to VviMYB86 which oppositely regulates different flavonoid subpathways in grape berries (Cheng et al., 2020).
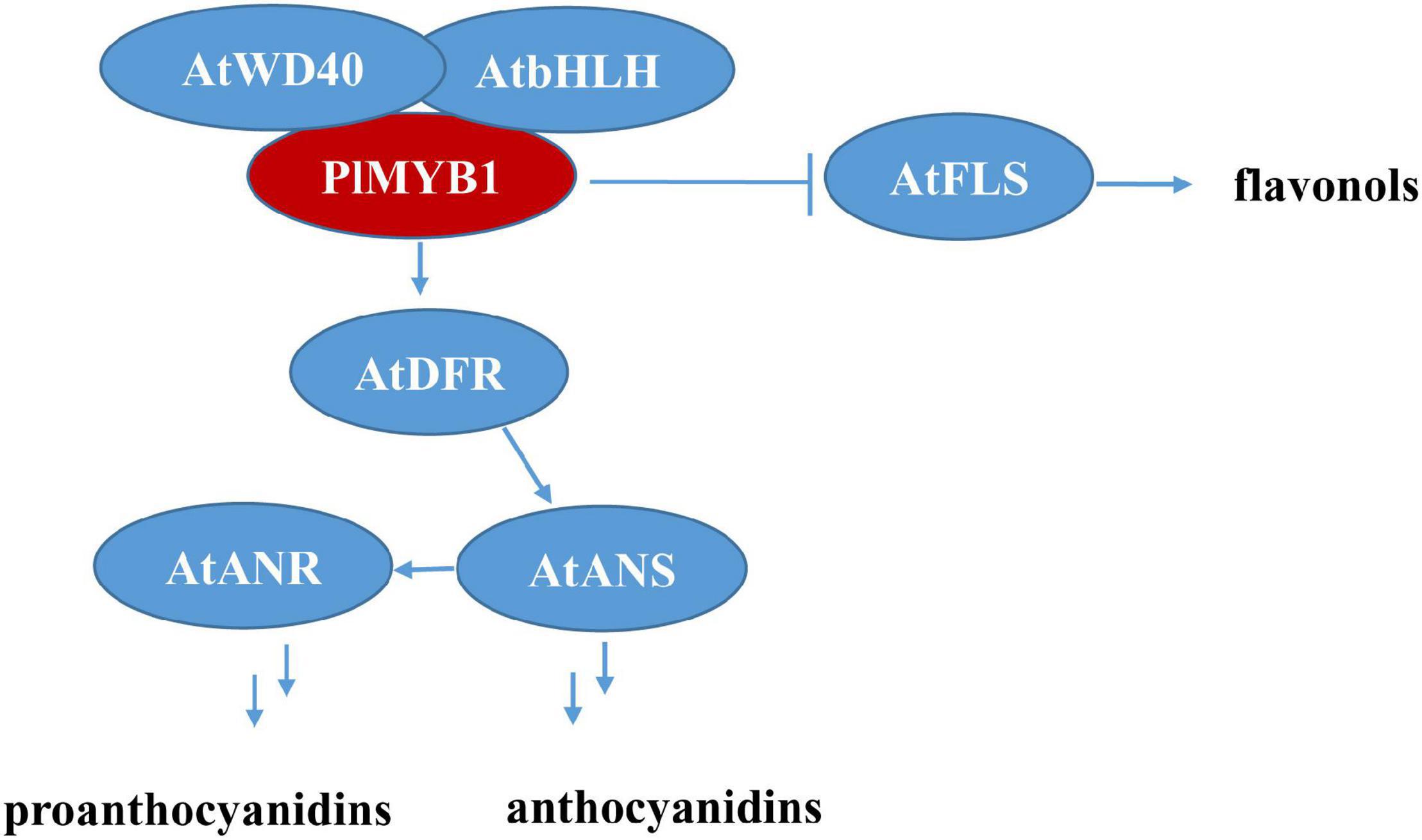
Figure 9. Predicted regulatory mechanism of PlMYB1 in Arabidopsis. FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase.
PlbHLH3-4 and PlWD40-1 Co-expressed With PlMYB1 Under Various pH Treatments
Most bHLH proteins can interact with R3 repeat domains of MYB proteins at the N-terminal acidic region to form the MYB–bHLH complex which frequently occurred in flavonoid biosynthetic pathways (Zhao et al., 2013). The bHLH members in subgroup IIIf were involved in anthocyanin biosynthesis (Spelt et al., 2000), seed coat differentiation, trichome formation, and root hair formation (Nesi et al., 2001). In addition, bHLH genes in subfamily 2 were found to generally respond to wounds, insects, drought, oxidative stress, jasmonic acid, abscisic acid, and chitin, but they also are able to regulate anthocyanin metabolism (Carretero-Paulet et al., 2010). AtbHLH13 is a member of subfamily 2 (Heim et al., 2003) and subgroup IIId (Song et al., 2013) as well.
In this study, three bHLH genes in kudzu, namely PlbHLH3-2, PlbHLH3-3, PlbHLH3-4, belong to the subfamily 2. They showed high similarities to AtbHLH13, with 63%, 65%, and 48% identity at amino acid level, respectively. The expression patterns of the PlbHLH3-4 gene were well consistent with total flavonoid content in the roots and leaves of high puerarin kudzu, but were inconsistent with that in low puerarin kudzu, implying PlbHLH3-4 might be involved in isoflavonoid biosynthesis but is not the determinant factor in kudzu. Interestingly, the expression of the PlbHLH3-4 gene was evidently increased under low pH medium and significantly decreased under high pH conditions, which is completely consistent with the accumulation of puerarin, suggesting PlbHLH3-4 was possibly responsible for the biosynthesis regulation of puerarin under various pH conditions. Moreover, PlbHLH3-4 was found to be localized in the nucleus as PlMYB1 at the subcellular level (Supplementary Figure 5), implying that it is likely to interact with PlMYB1 in the nucleus as a major regulator for the flavonoid pathway.
Except for bHLH, Arabidopsis WD40 like AtTTG1 also plays a key role in the regulation of flavonoid biosynthesis. AtTTG1 plays an important part in the regulation of AtDFR, AtANS, and AtANR by interacting with bHLH transcription factors (AtGL3, AtEGL3, or AtTT8) and MYB transcription factors (AtPAP1, AtPAP2, AtMYB113, or AtMYB114) in the MBW complex. This ternary MBW complex was known for controlling proanthocyanidin accumulation in seeds and anthocyanin accumulation in leaves (Walker et al., 1999; Payne et al., 2000; Baudry et al., 2004; Hichri et al., 2011). MdTTG1 identified from M. domestica was capable of fully replacing AtTTG1 to activate AtBAN promoter in cooperation with AtTT2 and AtTT8 in a co-transfection system (An et al., 2012). In M. truncatula, the deficiency of MtWD40-1 expression strongly suppressed the expression of flavonoid structural genes and thus blocked the accumulation of a range of flavonoid compounds (Pang et al., 2009).
In the present study, kudzu PlWD40-1 showed high sequence similarity with MdTTG1 and MtWD40-1 (Figure 5). The expression profiles of the PlWD40-1 gene were less correlated with the accumulation of total flavonoids in either high or low puerarin kudzu plants. However, the expression of the PlWD40-1 gene was highly consistent with the puerarin accumulation under various pH treatments, suggesting PlWD40-1 possibly involved in the biosynthesis regulation of puerarin under various pH conditions.
As PlbHLH3-4 and PlWD40-1 showed very high similarity with AtTT8 and AtTTG1 of Arabidopsis, and they displayed similar transcript profiles to PlMYB1 under various pH treatments (Figure 6), thus PlMYB1, PlbHLH3-4, and PlWD40-1 might form an MBW complex to regulate the accumulation of isoflavonoids in kudzu. In particular, PlbHLH3-4 and PlWD40-1 possibly play key roles in the isoflavonoid biosynthesis under various stresses like pH stimuli.
In summary, PlMYB1 acted as a potent transcript factor to regulate the production of various flavonoids/isoflavonoids. The expression profile of the PlMYB1 gene was significantly consistent with the total flavonoid level in kudzu. Furthermore, expression levels of PlMYB1 together with PlbHLH3-4 and PlWD40-1 were consistent with the puerarin content under various pH treatments. Therefore, it is reasonable to speculate that PlMYB1, PlbHLH3-4, and PlWD40-1 should cooperate together to finely tune the production of various isoflavonoids in kudzu. Overexpression of PlMYB1 induced a significant increase of anthocyanins/proanthocyanidins as well as related biosynthesis pathway genes. Our investigation could shed some light on the regulation network of isoflavonoid biosynthesis in kudzu and provide a potential gene target for the bioengineering of particular flavonoids in plants.
Materials and Methods
Transcriptome Sequencing and de novo Assembly
The roots of the two previously reported kudzu plants were propagated and collected separately at 7, 14, and 21 days after rooting. Kudzu plant of No. 1 accumulates puerarin, but No. 2 does not (He et al., 2011). Root materials were immediately frozen in liquid nitrogen (LN) and stored at –80°C prior to further analysis. Total RNAs were extracted with Tri-reagent according to the protocol of the manufacturer (Invitrogen, Waltham, MA, United States), followed by cleaning and purification with the DNase I. Equal amount of RNA from 7-, 14-, and 21-day-old root samples from Nos. 1 and 2 kudzu plant were pooled together, respectively, for sequencing with a biological triplicate. Poly(A) mRNA was purified from total RNA with polyoligo d(T) attached magnetic beads and then broken into short fragments, which were used as templates for double-stranded cDNA synthesis using random hexamer primers. The double-stranded cDNAs were purified, connected with sequencing adapters, and were separated by gel electrophoresis (Liuyi, Beijing, China). The purified double-stranded cDNAs with an average insert size of 400 bp were sequenced by the Illumina sequencing platform (San Diego, CA, United States). Reads were then assembled into contigs using Trinity software.
Functional Annotation of Unigenes
Functional annotations of the unigenes were performed by alignment of the assembly with unigenes against the NCBI Nr, SwissProt (UniProt Consortium, Switzerland), KOG1, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases2 using BLASTX (E-value < 10–5). The proteins from the Nr database with the highest hits to the unigenes were used to assign functional annotations. GoPipe software was used to analyze the GO annotations and GO functional classifications (Chen et al., 2005) (IGDB-CAS, Beijing, China).
The expression levels of unigenes were calculated using the reads per kb per million reads (RPKM) method, which eliminates the influence of different gene lengths and sequencing discrepancies (Mortazavi et al., 2008). Thus, this method can be used directly for comparing the differences in gene expression levels between two types of P. lobata roots. The fold-change in each gene expression in the two samples was calculated from the ratio of the RPKMs. In this study, differentially expressed genes (DEGs) were screened with an absolute value of log2 ratio > 2 and a threshold of false discovery rate (FDR) value lower than 0.005 (Wang et al., 2010). The identified DEGs were mapped to each term of the GO database3 and we calculated the gene numbers in each GO term. In addition, DEGs were also used in pathway enrichment analysis by calculating the gene numbers which mapped to KEGG in each pathway (Kanehisa et al., 2010).
Sequence Analysis
Alignments were performed using Clustal W algorithm-based AlignX module (UCD, Dublin, Ireland). The rooted trees were constructed using the ML method with MEGA X Software (Kumar et al., 2018). Tree nodes were evaluated by bootstrap analysis for 1,000 replicates (pairwise deletion, uniform rates, and Poisson correction options). Evolutionary distances were computed using the p-distance method and expressed in units of amino acid differences per site. All positions containing gaps and missing data were eliminated prior to the construction of phylogenetic trees.
Cloning and Expression of Candidate Genes in P. lobata
Putative transcription factor genes were cloned from the roots of P. lobata No. 1. First-strand cDNA was synthesized from total RNAs of roots using FastQuant RT Kit (TIANGEN, China). The primary PCR was performed using cDNA from the roots and the PCR conditions were as following: 5 min of initial denaturation at 94°C, followed by 94°C for 30 s, 54°C for 30 s, and 72°C for 1 min in a 35-cycle reaction, and a final elongation step of 72°C for 7 min. Primers sequences were listed in Supplementary Table 6.
Quantitative PCRs were performed on a Bio-Rad CFX96TM Real-time PCR System using SYBR Real Master Mix (Kangwei, China). The Arabidopsis pp2A gene (accession No. U39568) was used as an internal reference gene for the calculation of relative transcript levels. Primers used for genes from kudzu were listed in Supplementary Table 6, and primers for genes from Arabidopsis were the same as in our previous study (Su et al., 2020). Each reaction (20 μl) contained 1 μM each primer, 1 μl cDNA (1:10 diluted), 7 μl RNase-free H2O, and 10 μl PCR buffer of SYBR Real Master Mix. Thermal cycling conditions were as follows: pre-incubation at 95°C for 10 min, followed by 95°C for 20 s, 60°C for 30 s, and 72°C for 20 s for 40 cycles. Data were calculated from three biological replicates, and each biological replicate was examined in triplicate.
Subcellular Localization Analysis of Candidate genes
The coding region of transcription factor genes was amplified with prime pairs containing Sal I and Bam HI restriction sites, respectively (Supplementary Table 6). The resulting amplification product was digested and ligated to the same enzyme digested destination vector PJIT163hGFP. After confirmation by sequencing, the recombinant constructs were used in Arabidopsis protoplast transformation as described previously (Sheen, 2002). GFP fluorescence in Arabidopsis protoplast cells was detected by laser scanning confocal microscopy using Leica TCS SP5 (Wetzlar, Germany). The emission was collected for GFP from 500 to 560 nm, and for the chlorophyll from 605 to 700 nm.
Establishment and Treatment of P. lobata Cell Suspension Culture
Stems of kudzu plant No. 1 were surface sterilized in 75% (v/v) ethanol for 1–2 min, followed by three washes in sterile distilled water, 15 min in 10% hydrogen peroxide, and another three washes in distilled water. Then the axenic stems were cut into pieces in 1 cm length and planted on MS basal medium (pH = 5.8) with 3% sucrose, 0.8% agarose, 1 mg/L L-NAA, and 2 mg/L 6-benzylaminopurine (BA). Two weeks later, the emerged calli were transformed to B5 liquid medium (pH = 5.8), supplemented with 1 mg/L 2, 4-dichlorophenoxyacetic acid (2, 4-D), 1 mg/L NAA, 0.5 mg/L kinetin (KT), and 1% casein hydrolysate. Calli were then sub-cultured every week and incubated at 25 ± 2°C with a 16/8 h photoperiod. Cell culture was incubated in a flask on a rotary shaker (110–130 rpm) under the same photoperiod at 25°C. After about 1 month, soft, loose, and pale green calli were obtained. Once established, cultures were periodically sub-cultured into 100 ml flasks by transferring 15 ml 7-day-old cells into 40 ml fresh B5 liquid medium for treatment. For the treatment with salicylic acid (SA, 0.1 mg/L) and methyl jasmonate (MeJA, 1 mg/L), the cell cultures were collected at time points of 0, 2, 4, 8, 16, 24, 48, and 72 h. The fresh samples were collected and used for flavonoid analysis on HPLC with a triplicate.
Generation of Transgenic Arabidopsis Plants
To produce PlMYB1-overexpressing Arabidopsis plants, the 651-bp CDS (coding sequence) fragment was amplified by PCR, and then cloned into the binary vector pCXSN at the Xcm I site for gene over-expression in the wild-type A. thaliana. The resulting sequenced pCXSN-PlMYB1 construct was transformed into Agrobacterium strain GV3101 and used to generate transgenic A. thaliana plants by using the inflorescence dip method (Clough and Bent, 1998). The transgenic A. thaliana plants were screened on mass spectrometry (MS) medium supplied with hygromycin (30 mg/L). The 30-day-old rosette leaves and seeds of T3 generation homozygous plants were collected for further analyses.
Analyses of Flavonoid Compounds
For the extraction and quantification of total flavonoids, plant materials were all ground into powder and freeze-dried. Ten milligram dry powders were extracted by sonication for 30 min with 500 μl of 80% methanol in biological triplicates. After an additional overnight extraction at 4°C, the extracts were centrifuged at 12,000 rpm for 10 min. Deionized water amounting to 400 μl and 30 μl 5% NaNO2 was added to every 100 μl supernatant, followed by the addition of 30 μl 10% AlCl3 after 5 min. Later, 200 μl of 1 M NaOH were added 10 min, followed by the addition of 240 μl deionized water to make the final volume to 1 ml. The absorbance at 510 nm was measured using a spectrophotometer with quercetin as standard (BIO-RAD, CA, United States).
The above methanolic extracts were also applied for the identification and quantification of isoflavonoids on HPLC. The analyses were carried out using an Agilent 1260 chromatographic system (Santa Clara, CA, United States) equipped with a quaternary pump, an autosampler, a photodiode array detector, and Eclipse XDB-C18 reverse-phase column (4.6 mm × 250 mm, 5 μm). Flavonoids were separated with a linear eluting gradient (5–70% solvent B over 30 min) with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) at a flow rate of 1 ml/min and detected at 254 nm. The synthetic standard used in this study were all purchased from Sigma-Aldrich (Darmstadt, Germany).
Anthocyanins were extracted in acidified MeOH (0.1% HCl) overnight in the dark at 4°C, followed by sonication for 30 min. After centrifugation at 12,000 rpm for 10 min, the supernatant was mixed with the same volume of water and extracted with chloroform. The supernatant was then measured at 530 nm using a spectrophotometer. Three independent replicates were collected for each infiltration.
Proanthocyanidins were extracted three times with 70% acetone (0.5% acetic acid). Pooled supernatants were then extracted with chloroform and hexane. The soluble and insoluble proanthocyanidins were determined by dimethylaminocinnamaldehyde (DMACA) staining and butanol-hydrochloric acid (HCl) hydrolysis, respectively, as previously reported (Pang et al., 2007).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The RNA-Seq data presented in the study are deposited in the NCBI SRA repository with accession number PRJNA747842. The sequences of PlMYB1, PlbHLH3-4, and PlWD40-1 genes are deposited at Genbank with accession Nos. KR698796, KT236099, and AKR04124.1, respectively.
Author Contributions
GS and YP: conceptualization and funding acquisition. GS, RW, and YP: methodology. GS: software and data analysis. RW and YX: investigation. YP: resources, writing-review and editing, and supervision. GS and RW: writing-original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation of China (No. 31570306), the Key Program of the National Natural Science Foundation of China (No. U20A2004), the CAMS Innovation Fund for Medical Sciences (No. CIFMS, 2017-I2M-3-013), and the Agricultural Science and Technology Innovation Program (No. ASTIP-IAS10).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This project was initiated in Dixon lab at the Noble Foundation (Ardmore, United States) and we thank Prof. Richard A. Dixon, Drs. Xianzhi He, and Yuhong Tang for the initial supervision, sampling, and data presses of this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.743518/full#supplementary-material
Footnotes
References
Ali, M. B., Howard, S., Chen, S., Wang, Y., Yu, O., Kovacs, L. G., et al. (2011). Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol. 11:7. doi: 10.1186/1471-2229-11-17
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., and Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Anguraj Vadivel, A. K., McDowell, T., Renaud, J. B., and Dhaubhadel, S. (2021). A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 4:356. doi: 10.1038/s42003-021-01889-1886
Atchley, W. R., and Fitch, W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U S A. 94, 5172–5176. doi: 10.1073/pnas.94.10.5172
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bedon, F., Bomal, C., Caron, S., Levasseur, C., Boyle, B., Mansfield, S. D., et al. (2010). Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J. Exp. Bot. 61, 3847–3864. doi: 10.1093/jxb/erq196
Carey, C. C., Strahle, J. T., Selinger, D. A., and Chandler, V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16, 450–464. doi: 10.1105/tpc.018796
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martinez-Garcia, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, Z. Z., Xue, C. H., Zhu, S., Zhou, F. F., and Chen, L. B. (2005). GoPipe: streamlined gene ontology annotation for batch anonymous sequences with statistics. Prog. Biochem. Biophys. 32, 187–190. doi: 10.1360/crad20050909
Cheng, J., Yu, K., Shi, Y., Wang, J., and Duan, C. (2020). Transcription factor VviMYB86 oppositely regulates proanthocyanidin and snthocyanin biosynthesis in grape berries. Front. Plant Sci. 11, 613677. doi: 10.3389/fpls.2020.613677
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434. doi: 10.1101/gad.11.11.1422
Deng, C., Shi, M., Fu, R., Zhang, Y., Wang, Q., Zhou, Y., et al. (2020a). ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71, 5948–5962. doi: 10.1093/jxb/eraa295
Deng, C., Wang, Y., Huang, F., Lu, S., Zhao, L., Ma, X., et al. (2020b). SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 62, 1688–1702. doi: 10.1111/jipb.1
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor. MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Fan, Y., Peng, J., Wu, J., Zhou, P., He, R., Allan, A. C., et al. (2021). NtbHLH1, a JAF13-like bHLH, interacts with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus. BMC Plant Biol. 21:275. doi: 10.1186/s12870-021-03050-3051
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Goodrich, J., Carpenter, R., and Coen, E. S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68, 955–964. doi: 10.1016/0092-8674(92)90038-e
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., et al. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10, 721–740.
Hao, X., Pu, Z., Cao, G., You, D., Zhou, Y., Deng, C., et al. (2020). Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 23, 1–12. doi: 10.1016/j.jare.2020.01.012
Hao, X., Xie, C., Ruan, Q., Zhang, X., Wu, C., Han, B., et al. (2021). The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Hortic. Res. 8:7. doi: 10.1038/s41438-020-00437-433
He, X., Blount, J. W., Ge, S., Tang, Y., and Dixon, R. A. (2011). A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata). Planta 233, 843–855. doi: 10.1007/s00425-010-1344-1341
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hichri, I., Heppel, S. C., Pillet, J., Leon, C., Czemmel, S., Delrot, S., et al. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3, 509–523. doi: 10.1093/mp/ssp118
Hien, T. T., Kim, H. G., Han, E. H., Kang, K. W., and Jeong, H. G. (2010). Molecular mechanism of suppression of MDR1 by puerarin from Pueraria lobata via NF-kappaB pathway and cAMP-responsive element transcriptional activity-dependent up-regulation of AMP-activated protein kinase in breast cancer MCF-7/adr cells. Mol. Nutr. Food. Res. 54, 918–928. doi: 10.1002/mnfr.200900146
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New. Phytol. 201, 795–809. doi: 10.1111/nph.12557
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jiu, S., Guan, L., Leng, X., Zhang, K., Haider, M. S., Yu, X., et al. (2021). The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 19, 1216–1239. doi: 10.1111/pbi.13543
Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M., and Hirakawa, M. (2010). KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360. doi: 10.1093/nar/gkp896
Keung, W. M., and Vallee, B. L. (1998). Kudzu root: an ancient Chinese source of modern antidipsotropic agents. Phytochemistry 47, 499–506. doi: 10.1016/s0031-9422(97)00723-721
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Li, L., He, Y., Ge, H., Liu, Y., and Chen, H. (2021). Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 302:110696. doi: 10.1016/j.plantsci.2020.110696
Li, Z. B., Li, C. F., Li, J., and Zhang, Y. S. (2014). Molecular cloning and functional characterization of two divergent 4-coumarate: coenzyme a ligases from kudzu (Pueraria lobata). Biol. Pharm. Bull. 37, 113–122. doi: 10.1248/bpb.b13-00633
Liu, C. M., Zheng, G. H., Ming, Q. L., Sun, J. M., and Cheng, C. (2013). Protective effect of puerarin on lead-induced mouse cognitive impairment via altering activities of acetyl cholinesterase, monoamine oxidase and nitric oxide synthase. Environ. Toxicol. Pharmacol. 35, 502–510. doi: 10.1016/j.etap.2013.02.009
Liu, Q., Li, L., Cheng, H., Yao, L., Wu, J., Huang, H., et al. (2021). The basic helix-loop-helix transcription factor TabHLH1 increases chlorogenic acid and luteolin biosynthesis in Taraxacum antungense Kitag. Hortic. Res. 8:195. doi: 10.1038/s41438-021-00630-y
Ludwig, S. R., Habera, L. F., Dellaporta, S. L., and Wessler, S. R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. U S A. 86, 7092–7096. doi: 10.1073/pnas.86.18.7092
Mao, Z., Jiang, H., Wang, S., Wang, Y., Yu, L., Zou, Q., et al. (2021). The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 306:110848. doi: 10.1016/j.plantsci.2021.110848
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Massari, M. E., and Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440. doi: 10.1128/MCB.20.2.429-440.2000
Matus, J. T., Poupin, M. J., Canon, P., Bordeu, E., Alcalde, J. A., and Arce-Johnson, P. (2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol. Biol. 72, 607–620. doi: 10.1007/s11103-010-9597-9594
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. doi: 10.1105/tpc.010098
Pang, Y., Peel, G. J., Wright, E., Wang, Z., and Dixon, R. A. (2007). Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 145, 601–615. doi: 10.1104/pp.107.107326
Pang, Y., Wenger, J. P., Saathoff, K., Peel, G. J., Wen, J., Huhman, D., et al. (2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 151, 1114–1129. doi: 10.1104/pp.109.144022
Park, K. I., Ishikawa, N., Morita, Y., Choi, J. D., Hoshino, A., and Iida, S. (2007). A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J. 49, 641–654. doi: 10.1111/j.1365-313X.2006.02988.x
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362.
Prasain, J. K., Jones, K., Kirk, M., Wilson, L., Smith-Johnson, M., Weaver, C., et al. (2003). Profiling and quantification of isoflavonoids in kudzu dietary supplements by high-performance liquid chromatography and electrospray ionization tandem mass spectrometry. J. Agric. Food Chem. 51, 4213–4218. doi: 10.1021/jf030174a
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Ramsay, N. A., Walker, A. R., Mooney, M., and Gray, J. C. (2003). Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol. Biol. 52, 679–688. doi: 10.1023/a:1024852021124
Sheen, J. (2002). A Transient Expression Assay Using Arabidopsis Mesophyll Protoplasts. http://genetics.mgh.harvard.edu/sheenweb/ (accessed March 5, 2015).
Shi, L., Chen, X., Wang, K., Yang, M., Chen, W., Yang, Z., et al. (2021). MrMYB6 from Chinese bayberry (Myrica rubra) negatively regulates anthocyanin and proanthocyanidin accumulation. Front. Plant Sci. 12:685654. doi: 10.3389/fpls.2021.685654
Shin, D. H., Cho, M., Choi, M. G., Das, P. K., Lee, S. K., Choi, S. B., et al. (2015). Identification of genes that may regulate the expression of the transcription factor production of anthocyanin pigment 1 (PAP1)/MYB75 involved in Arabidopsis anthocyanin biosynthesis. Plant Cell Rep. 34, 805–815. doi: 10.1007/s00299-015-1743-1747
Sompornpailin, K., Makita, Y., Yamazaki, M., and Saito, K. (2002). A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol. Biol. 50, 485–495. doi: 10.1023/a:1019850921627
Song, S., Qi, T., Fan, M., Zhang, X., Gao, H., Huang, H., et al. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet 9:e1003653. doi: 10.1371/journal.pgen.1003653
Spelt, C., Quattrocchio, F., Mol, J. N., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1632. doi: 10.1105/tpc.12.9.1619
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-190
Su, X., Xia, Y., Jiang, W., Shen, G., and Pang, Y. (2020). GbMYBR1 from Ginkgo biloba represses phenylpropanoid biosynthesis and trichome development in Arabidopsis. Planta 252:68. doi: 10.1007/s00425-020-03476-1
Sun, M., Shi, M., Wang, Y., Huang, Q., Yuan, T., Wang, Q., et al. (2019). The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 70, 243–254. doi: 10.1093/jxb/ery349
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verries, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
van Nocker, S., and Ludwig, P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4:50. doi: 10.1186/1471-2164-4-50
Wada, T., Kunihiro, A., and Tominaga-Wada, R. (2014). Arabidopsis CAPRICE (MYB) and GLABRA3 (bHLH) control tomato (Solanum lycopersicum) anthocyanin biosynthesis. PLoS One 9:e109093. doi: 10.1371/journal.pone.0109093
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. doi: 10.1105/tpc.11.7.1337
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, X., Li, C., Zhou, C., Li, J., and Zhang, Y. (2017). Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. 90, 535–546. doi: 10.1111/tpj.13510
Wong, K. H., Li, G. Q., Li, K. M., Razmovski-Naumovski, V., and Chan, K. (2011). Kudzu root: traditional uses and potential medicinal benefits in diabetes and cardiovascular diseases. J. Ethnopharmacol. 134, 584–607. doi: 10.1016/j.jep.2011.02.001
Xia, D. Z., Zhang, P. H., Fu, Y., Yu, W. F., and Ju, M. T. (2013). Hepatoprotective activity of puerarin against carbon tetrachloride-induced injuries in rats: a randomized controlled trial. Food Chem. Toxicol. 59, 90–95. doi: 10.1016/j.fct.2013.05.055
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thevenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Yi, J., Derynck, M. R., Li, X., Telmer, P., Marsolais, F., and Dhaubhadel, S. (2010). A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62, 1019–1034. doi: 10.1111/j.1365-313X.2010.04214.x
Zhang, Y., Zhang, J., Song, T., Li, J., Tian, J., Jin, K., et al. (2014). Low medium pH value enhances anthocyanin accumulation in Malus crabapple leaves. PLoS One 9:e97904. doi: 10.1371/journal.pone.0097904
Zhao, L., Gao, L., Wang, H., Chen, X., Wang, Y., Yang, H., et al. (2013). The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genomics 13, 75–98. doi: 10.1007/s10142-012-0301-304
Zhou, W., Li, S., Maoz, I., Wang, Q., Xu, M., Feng, Y., et al. (2021a). SmJRB1 positively regulates the accumulation of phenolic acid in Salvia miltiorrhiza. Ind. Crops Products 164:113417. doi: 10.1016/j.indcrop.2021.113417
Zhou, W., Shi, M., Deng, C., Lu, S., Huang, F., Wang, Y., et al. (2021b). The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic. Res. 8:10. doi: 10.1038/s41438-020-00443-445
Keywords: Pueraria lobata, transcriptome sequencing, transcription factors, MYB, isoflavonoid biosynthesis
Citation: Shen G, Wu R, Xia Y and Pang Y (2021) Identification of Transcription Factor Genes and Functional Characterization of PlMYB1 From Pueraria lobata. Front. Plant Sci. 12:743518. doi: 10.3389/fpls.2021.743518
Received: 18 July 2021; Accepted: 13 September 2021;
Published: 08 October 2021.
Edited by:
Wansheng Chen, Second Military Medical University, ChinaReviewed by:
Zhihua Liao, Southwest University, ChinaGuoyin Kai, Zhejiang Chinese Medical University, China
Copyright © 2021 Shen, Wu, Xia and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhen Pang, cGFuZ3lvbmd6aGVuQGNhYXMuY24=
 Guoan Shen1
Guoan Shen1 Yaying Xia
Yaying Xia Yongzhen Pang
Yongzhen Pang