
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 September 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.741463
Root rot, mainly caused by Fusarium oxysporum, is the most destructive disease affecting lily (Lilium spp.) production. The WRKY transcription factors (TFs) have important roles during plant immune responses. To clarify the effects of WRKY TFs on plant defense responses to pathogens, a WRKY gene (LrWRKY2) was isolated from Lilium regale Wilson, which is a wild lily species highly resistant to F. oxysporum. The expression of LrWRKY2, which encodes a nuclear protein, is induced by various hormones (methyl jasmonate, ethephon, salicylic acid, and hydrogen peroxide) and by F. oxysporum infection. In this study, LrWRKY2-overexpressing transgenic tobacco plants were more resistant to F. oxysporum than the wild-type plants. Moreover, the expression levels of jasmonic acid biosynthetic pathway-related genes (NtAOC, NtAOS, NtKAT, NtPACX, NtJMT, NtOPR, and NtLOX), pathogenesis-related genes (NtCHI, NtGlu2, and NtPR-1), and antioxidant stress-related superoxide dismutase genes (NtSOD, NtCu-ZnSOD, and MnSOD) were significantly up-regulated in LrWRKY2 transgenic tobacco lines. Additionally, the transient expression of a hairpin RNA targeting LrWRKY2 increased the susceptibility of L. regale scales to F. oxysporum. Furthermore, an F. oxysporum resistance gene (LrCHI2) encoding a chitinase was isolated from L. regale. An electrophoretic mobility shift assay demonstrated that LrWRKY2 can bind to the LrCHI2 promoter containing the W-box element. Yeast one-hybrid assay results suggested that LrWRKY2 can activate LrCHI2 transcription. An examination of transgenic tobacco transformed with LrWRKY2 and the LrCHI2 promoter revealed that LrWRKY2 activates the LrCHI2 promoter. Therefore, in L. regale, LrWRKY2 is an important positive regulator that contributes to plant defense responses to F. oxysporum by modulating LrCHI2 expression.
Plants have evolved a series of responsive and adaptive mechanisms to cope with various stresses. The WRKY transcription factors (TFs), which represent one of the largest families of transcriptional regulators in plants, are mainly involved in regulating the plant immune system during responses to stress (Wang P. et al., 2018). These TFs usually contain one or two conserved domains comprising approximately 60 amino acids (Yan et al., 2019a). The domain with a highly conserved WRKYGQK motif at the N-terminal end and a zinc-finger motif (CX4−5CX22−23HXH or CX7CX23HXC) at the C-terminal end was designated as the WRKY domain (Jimmy and Babu, 2019). On the basis of the number of WRKY domains and the structure of the zinc-finger motif, WRKY TFs have been classified into three main groups (I, II, and III) (Dong et al., 2020). The Group I members have two WRKY domains and a C2H2 (CX4−5CX22−23HXH) zinc-finger motif, the Group II members contain one WRKY domain and a C2H2 (CX4−5CX22−23HXH) zinc-finger motif, and the Group III members contain a single WRKY domain and a C2HC (CX7CX23HXC) motif (Nan and Gao, 2019). Moreover, the Group II WRKY TFs have been further divided into Subgroups IIa, IIb, IIc, IId, and IIe according to their primary amino acid sequences (Huang et al., 2015).
Transcription factors bind to cis-acting elements in the promoters of their downstream target genes to regulate expression (Long et al., 2019). The WRKY TFs are a family of plant-specific DNA-binding proteins (Rushton et al., 2010). More specifically, the WRKY domain binds to the W-box (TTGACC/T) in the promoters of target genes to activate, repress, or de-repress transcription. WRKY TFs can also bind to several W-box-like elements (Choi et al., 2015). It is due to WRKY TFs binds to the TGAC core motif of the W-box, whereas the preferred WRKY TF-binding sites are determined by the sequences adjacent to the TGAC core motif (Choi et al., 2015). The WRKYGQK and zinc-finger motifs are required for the high-affinity binding of WRKY proteins to the consensus cis-element W-box present in the promoters of target genes, including those encoding pathogenesis-related (PR) proteins (Karim et al., 2015). The solution structure of the WRKY domain was first reported by Yamasaki et al. (2005). The WRKY domain of the Arabidopsis thaliana WRKY4 TF comprises a four-stranded β-sheet, with zinc-coordinating Cys/His residues forming a zinc-binding pocket (Yamasaki et al., 2005). The WRKYGQK residues form the most N-terminal β-strand, which partly protrudes from the surface of the protein, enabling it to interact with the DNA major groove. The β-strand including the WRKYGQK motif reportedly interacts with an approximately 6-bp region, which is consistent with the length of the W-box (Jiang et al., 2016). The WRKYGQK motif recognizes DNA mainly through apolar associations with the methyl groups of the thymine bases present in the W-box which was reviewed by Viana et al. (2018).
The WRKY TFs have a very complex regulatory role influencing plant disease resistance. They are central components mediating innate immunity, including molecular pattern-triggered immunity, pathogen-associated molecular pattern-triggered immunity, effector-triggered immunity, basal defense, and systemic acquired resistance (Eulgem and Somssich, 2007). The activation of WRKY TFs under stress conditions leads to a thorough transcriptional reprogramming affecting defense-related genes, including genes for secondary metabolites (phytoalexins such as momilactones and phenylamides) and PR proteins (PR3, PR4c, PR10a, PR10b, and root-specific PR10) (Jimmy and Babu, 2019). The constitutive expression of rice (Oryza sativa) WRKY45 in A. thaliana enhances the resistance of the transgenic plants to the pathogen Pseudomonas syringae pv. tomato DC3000 by up-regulating the expression of some PR genes (Qiu and Yu, 2009). In Capsicum annuum, WRKY40b is a negative regulator that directly affects immunity at multiple levels by modulating the production of signal regulatory proteins, TFs, and PR proteins (Ifnan Khan et al., 2018). In a previous study, the overexpression of apple (Malus × domestica) WRKYN1 in the susceptible apple cultivar “Golden Delicious” resulted in increased resistance to the apple leaf spot fungus Alternaria alternata f. sp. mali, which was related to the significantly up-regulated expression of PR genes (Zhang et al., 2017).
Root rot caused by Fusarium oxysporum seriously threatens the growth and development of lily (Lilium spp.). Several wild Lilium species are highly resistant to F. oxysporum, including L. regale Wilson, L. pumilum, and L. dauricum Ker Gawler (He et al., 2014, 2019; Ramekar et al., 2019). Although the germplasm resources of Lilium species resistant to F. oxysporum have been used for breeding disease-resistant lines, little is known about their mechanisms regulating transcription during an infection by F. oxysporum. In a recent study, 35 WRKY genes were identified in L. regale after the transcriptomes of plants inoculated with F. oxysporum were sequenced (Li et al., 2021b). At the transcriptional level, LrWRKY2 is highly responsive to methyl jasmonate (MeJA), salicylic acid (SA), ethephon (ETH), and hydrogen peroxide (H2O2) treatments as well as F. oxysporum infections. Thus, in this study, we focused on unraveling the LrWRKY2 (accession no. MW125547) regulatory mechanism during the L. regale defense response to F. oxysporum. The tobacco is an important cash crop and model plant used for rapid verification of plant gene functions, moreover, the F. oxysporum causes root rot in tobacco (Clemente, 2006; Ding et al., 2021). In addition, the infection rate of F. oxysporum in tobacco seedlings is faster than the lily, and the obvious disease symptoms can appear within a week after inoculation. Therefore, the effects of LrWRKY2 on the resistance to F. oxysporum was evaluated by overexpressing LrWKKY2 in tobacco plants and transiently expressing the hairpin RNA targeting LrWRKY2 in L. regale scales considering the long period of lily genetic transformation and great difficulties to obtain enough transformants for subsequent experiments. Furthermore, LrWRKY2 was characterized regarding its subcellular localization and its ability to activate the promoter of an L. regale PR gene (LrCHI2), which is an F. oxysporum-responsive gene according to transcriptome sequencing data. Additionally, tobacco plants were transformed with the LrCHI2 promoter and LrWRKY2 to analyze the regulatory effects of LrWRKY2 on LrCHI2 expression.
Wild L. regale Wilson plants collected in the Minjiang River Basin of Sichuan province, China, were grown in a greenhouse. An F. oxysporum strain isolated from diseased lily plants exhibiting typical symptoms of Fusarium wilt was characterized and preserved by our research group. Several typical plant fungal pathogens (Colletotrichum gloeosporioides, Fusarium solani, and Alternaria panax) were also selected for the antifungal activity analysis. F.oxysporum, C. gloeosporioides, F. solani, and A. panax stored at 4°C were cultured on potato dextrose agar (PDA) before use. Sterile tobacco seedlings were cultured in a climate-controlled cabinet prior to genetic transformations.
The total RNA was extracted from 1 g wild L. regale roots using the TRIGene kit (Genstar, China). The cDNA was obtained following the reverse transcription of total RNA using the GoScript™ Reverse Transcription System (Promega, USA) and served as the template for isolating LrWRKY2 and LrCHI2. Gene-specific primers (Supplementary Table 1) were designed to amplify the LrWRKY2 and LrCHI2 open reading frames (ORFs) using L. regale cDNA as the template. The PCR products were cloned into the pMD-18T vector (TaKaRa, Japan) and the resulting recombinant vectors pMD-18T-LrWRKY2 and pMD-18T-LrCHI2 were inserted into competent Escherichia coli DH5α cells. The positive clones on a LB plate adding the ampicillin (50 mg/L) were verified by PCR and sequencing in Tsingke Biotechnology Co., Ltd. Bioinformatics analyses were completed as described by Taif et al. (2020).
The LrCHI2 expression pattern in L. regale was analyzed in a qRT-PCR assay. The roots, leaves, stems, flowers, and scales of healthy L. regale plants were used to analyze tissue-specific LrCHI2 expression. Regarding the fungal inoculation, the root tips were wounded using surgical scissors and inoculated with a fresh F. oxysporum spore suspension (5 × 106 spores/mL) for 30 min. The control plants were treated with sterile water instead of the spore suspension. The roots were sampled at 12, 24, 48, and 72 h post-inoculation. Finally, total RNA extracted from each sample was reverse transcribed into cDNA for a qRT-PCR analysis of LrCHI2 expression levels using the GoScript™ Reverse Transcription System (Promega, USA). The SYBR green I-based qRT-PCR analysis was conducted using the ABI Prism 7500 Sequence Detection System (Applied Biosystems, USA) and Eastep® qPCR Master Mix (Promega, USA) according to an established method (Zhao et al., 2020). The L. regale glyceraldehyde-3-phosphate dehydrogenase gene (LrGAPDH, GenBank No. KJ543468.1) was used as an internal reference gene to calculate the relative expression values of LrCHI2. The LrCHI2 expression levels were calculated according to the 2−ΔΔCt method. The qRT-PCR analysis was completed using three biological replicates, and there were three technical repetitions included in each biological replicate.
The subcellular localization of LrWRKY2 and LrCHI2 was predicted using the PSORT online program (https://www.genscript.com/psort.html) and then confirmed by transiently expressing green fluorescent protein (GFP)-tagged fusion proteins in onion (Allium cepa) epidermal cells. More specifically, the LrWRKY2 and LrCHI2 ORFs lacking the stop codon were amplified from pMD-18T-LrWRKY2 and pMD-18T-LrCHI2, respectively. The PCR products were then inserted into the pBIN m0-gfp5-ER vector to generate the LrWRKY2-GFP and LrCHI2-GFP constructs. The two recombinant vectors were transferred into Agrobacterium tumefaciens EHA105 cells using a CaCl2 freeze–thaw method (Holsters et al., 1978). An empty pBIN m-gfp5-ER vector served as a control. The transformed A. tumefaciens cells were used to insert the recombinant or control vector into onion epidermal cells, which were then cultured in darkness for 48 h. To analyze transient gene expression, GFP fluorescence in the onion epidermal cells was monitored using a confocal microscope (Nikon, JPN) as described by Liu et al. (2018).
The plasmids pMD-18T-LrWRKY2 and pCAMBIA2300s were digested with BamHI and XbaI, while the pMD-18T-LrCHI2 and pCAMBIA2300s plasmids were digested with BamHI and EcoRI for constructing the overexpression vectors of LrWRKY2 and LrCHI2, respectively. After which the pCAMBIA2300s-LrWRKY2 and pCAMBIA2300s-LrCHI2 recombinant plasmids were generated using the T4 DNA ligase. The recombinant plasmids were inserted into separate competent A. tumefaciens LBA4404 cells. The positive clones identified by PCR were used to transform tobacco leaf disks as described by Horsch et al. (1985). Genomic DNA was extracted from T0 transgenic tobacco plants using cetyltrimethylammonium bromide (CTAB) as described by Allen et al. (2006), and then the genomic DNA was used as the template for a PCR conducted to confirm the transgenic lines carried the correct transgene, with wild-type (WT) plants serving as the negative control. The transgenic tobacco plants were grown in a greenhouse to produce T2 generation lines.
Total RNA was extracted from T2 generation LrWRKY2 transgenic tobacco lines and reverse transcribed into cDNA with the forementioned method. The LrWRKY2 transcription levels in the transgenic tobacco lines were determined by qRT-PCR with the tobacco actin gene (NtACT, GenBank No. AB158612.1) as an internal reference gene. Moreover, the roots and leaves of several LrWRKY2 transgenic tobacco lines and WT plants were inoculated with F. oxysporum to evaluate the disease resistance of the transgenic tobacco lines. After the wounded tobacco roots were immersed in an F. oxysporum spore suspension (5 × 106 spores/mL) for 30 min, the inoculated tobacco plants were grown under hydroponic conditions in an illumination incubator for 1 week. Regarding the leaf inoculation, leaves were wounded, inoculated with 20 μL F. oxysporum spore suspension (5 × 106 spores/mL), and then placed in a humid illumination incubator for 1 week. The leaves of T2 generation LrCHI2 transgenic tobacco lines were similarly inoculated. The disease symptoms caused by the F. oxysporum infections were examined.
The expression levels of the following defense-related genes in the LrWRKY2 transgenic tobacco lines were analyzed by qRT-PCR with the forementioned method: jasmonic acid (JA) biosynthetic pathway-related genes (NtAOC, NtAOS, NtKAT, NtPACX, NtJMT, NtOPR, and NtLOX), PR genes (NtCHI, NtGlu2, and NtPR-1), and antioxidant stress-related superoxide dismutase (SOD) genes (NtSOD, NtCu-ZnSOD, and MnSOD). Sequence details regarding the defense-related genes were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/) and used to design gene-specific primers (Supplementary Table 2). Three LrWRKY2 transgenic tobacco lines were randomly selected for an analysis of the expression of these defense-related genes.
Primers with the attB linker were designed to amplify the LrWRKY2 RNA interference (RNAi) fragment (Supplementary Table 1). The PCR product was incorporated into the RNAi vector pHellsgate2 via a BP recombination reaction using the Gateway® BP Clonase™ II Enzyme Mix kit (Invitrogen, USA). The pHellsgate2-LrWRKY2 recombinant plasmid was inserted into competent E. coli DH10B cells. Positive clones were selected on agar-solidified Luria-Bertani medium containing spectinomycin (90 mg/L). The plasmids of the positive clones were digested with XbaI and XhoI to confirm the LrWRKY2 fragment was correctly recombined. The pHellsgate2-LrWRKY2 recombinant plasmid and the empty pHellsgate2 vector were inserted into separate A. tumefaciens EHA105 cells, which were then cultured in MGL medium at 28°C for 5 h in a constant-temperature shaker (150 rpm). Fresh L. regale scales were washed with sterile water and rubbed with sandpaper to form uniformly sized wounds. The wounded L. regale scales were transformed with A. tumefaciens liquid containing the pHellsgate2-LrWRKY2 recombinant plasmid or the empty pHellsgate2 vector. The L. regale scales were placed on filter paper moistened with sterile water and incubated in a climate-controlled cabinet set at 25°C for 24 h, and then inoculated with 20 μL F. oxysporum spore suspension (5 × 106 spores/mL). The inoculated scales were collected at 72 h for an analysis of LrWRKY2 expression and disease symptoms. The L. regale scales in which the empty pHellsgate2 vector was transiently expressed were used as the control.
The SignalP 5.0 Server (https://www.cbs.dtu.dk/services/SignalP/) was used for predicting the presence of a signal peptide. The LrWRKY2 ORF amplified by PCR from pMD-18T-LrWRKY2 was digested with HindIII and BamHI and then subcloned into the pET-32a vector. Gene-specific primers (Supplementary Table 1) were designed to amplify the LrCHI2 ORF without the signal peptide-encoding sequence. The PCR product was inserted into pMD-18T. The resulting recombinant plasmid was digested with EcoRI and EcoRV and then the LrCHI2 fragment was incorporated into the pET-32a vector. The pET-32a-LrWRKY2 and pET-32a-LrCHI2 plasmids were transferred into competent E. coli BL21 cells. The empty pET-32a vector was used as a control. The production of the LrWRKY2 and LrCHI2 recombinant proteins was induced and the inclusion body proteins were denatured and renatured as described by Zhao et al. (2020). The recombinant proteins were purified using the Ni-NTA Sepharose Column (Sangon Biotech, China).
The in vitro antifungal effects of the LrCHI2 recombinant protein were examined using F. oxysporum, C. gloeosporioides, F. solani, and A. panax. Agar plugs (1 cm diameter) removed from the outer edge of actively growing fungus on agar-solidified PDA medium in plates were used to inoculate fresh agar-solidified PDA medium. When the diameter of the fungal colonies reached 2 cm, LrCHI2 (5, 10, and 20 μg) was added to the plates. Sterile water and phosphate buffer (pH 8.0) were used as controls. After a 3-day incubation at 28°C, the fungal colonies were photographed and the average growth inhibition zones (mm2) were calculated using Photoshop 7.0.
The LrCHI2 promoter region was isolated from L. regale genomic DNA by genome walking using the Universal GenomeWalker™ 2.0 kit (TaKaRa, Japan). Two nested primers were designed specifically for the LrCHI2 unigene sequence (Supplementary Table 1). The sequence upstream of LrCHI2 was amplified by two rounds of PCR. The resulting PCR product was cloned into pMD-18T. The promoter fragment was confirmed by sequencing and an alignment with the LrCHI2 unigene sequence, and a 491-bp LrCHI2 promoter fragment was obtained. Additionally, the promoter cis-elements were predicted using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
A 50-bp LrCHI2 promoter fragment with one W-box was used as the probe. The mutant probe had one mutation in the W-box core sequence (Supplementary Table 1). These two probe sequences were synthesized and labeled with biotin (Sangon Biotech, China). The unlabeled probe served as the competitor. The EMSA reaction mixtures contained 0.5 μg purified LrWRKY2 protein and 7 μL 1 × gel shift binding buffer [100 mM Tris, 500 mM KCl, 10 mM DTT, 50% glycerol, 100 mM MgCl2, 1% NP-40, 1 M KCl, 200 mM EDTA, and 1 μg/μL poly (dI•dC)] in a total volume of 20 μL. After a 20-min incubation at room temperature, 2 μL biotin-labeled probes were added and the incubation was continued for 20 min. For the competitor probe, the binding reaction components were incubated for 40 min. The DNA–protein complexes were electrophoresed on 6.5% non-denaturing polyacrylamide gels in an ice-cold water bath and then transferred to a nylon membrane. Finally, the membrane was dried and analyzed using the LightShift Chemiluminescent EMSA Kit (Pierce, USA).
A Y1H assay was performed to analyze the LrCHI2 promoter-binding capability of LrWRKY2. The recombinant prey (pGADT7-LrWRKY2) and bait (pLrCHI2-pAbAi) vectors were constructed. The reporter strains were generated by integrating linearized pLrCHI2-pAbAi or empty pAbAi plasmids into the genome of yeast strain Y1HGold. Positive clones identified by PCR were used to inoculate synthetic defined (SD) medium containing different aureobasidin A (AbA) concentrations to determine the appropriate AbA inhibitory concentration. The recombinant prey plasmid (pGADT7-LrWRKY2) was integrated into yeast cells containing pLrCHI2-pAbAi or empty pAbAi, whereas the recombinant prey plasmid (pGADT7-p53) was inserted into Y1HGold yeast cells carrying the p53-pAbAi plasmid (positive control). Positive clones were confirmed by PCR. The protein–DNA interaction was identified on the basis of the activation of the AbA resistance gene when a prey protein from the library bound to the bait sequence. Thus, the three types of Y1HGold yeast cells were used to inoculate SD/–Leu medium supplemented with the appropriate AbA inhibitory concentration. The Y1H assay was performed using the Matchmaker™ Gold Yeast One-Hybrid Library Screening System (Takara, USA).
The LrCHI2 promoter was cloned to study whether LrWRKY2 regulates LrCHI2 expression. The pMD-18T-pLrCHI2 plasmid was digested with BamHI and HindIII and then the LrCHI2 promoter fragment was subcloned into the pBI121-GUS vector digested with the same restriction enzymes. The pBI121-pLrCHI2-GUS plasmid was integrated into WT and LrWRKY2 overexpressing tobacco leaf disks via A. tumefaciens-mediated transformation. As controls, WT and LrWRKY2 transgenic tobacco leaf disks were transformed with the empty pBI121-GUS vector. The genomic DNA of the putative transgenic tobacco lines served as the template for a PCR amplification using β-glucuronidase (GUS) gene-specific primers to verify the transgenic plants were transformed correctly (Supplementary Table 1). The GUS enzyme activity (pM 4-MU min−1 μg−1 protein) of the confirmed transgenic tobacco lines was analyzed using a fluorescence spectrophotometer (Hitachi F-4600, Japan) as described by Chen et al. (2017).
Relative gene expression levels, fungal growth inhibition zone sizes, tobacco leaf and lily scale lesion sizes, and GUS enzyme activities are herein presented as the mean ± standard deviation. The data were analyzed using the SPSS software (version 17.0) and the significance of any differences was determined according to Student's t-test and Duncan's multiple range test. All experiments were performed independently at least three times under the same conditions.
Researchers previously identified 35 WRKY genes by sequencing the wild L. regale transcriptome during an F. oxysporum infection; they also revealed that LrWRKY2 expression is induced by F. oxysporum and is substantially affected by MeJA, SA, ETH, and H2O2 treatments (Li et al., 2021a). Thus, LrWRKY2 was functionally characterized. The L. regale LrWRKY2 cDNA was amplified by RT-PCR using gene-specific primers. The full-length LrWRKY2 cDNA comprised 1,302 bp, including a 1,032-bp ORF encoding a Subgroup IIe WRKY protein consisting of 343 amino acid residues. A sequence analysis confirmed the putative LrWRKY2 protein contained a typical WRKY domain with the highly conserved WRKYGQK sequence and a C2H2-type (CX4−5CX22−23HXH) zinc-finger motif, which is a typical feature of Group II members (Figure 1A). The deduced LrWRKY2 amino acid sequence was highly similar to the sequences of several WRKY proteins from monocots, including Phoenix dactylifera WRKY14 (accession no. XP_008788702.2), Elaeis guineensis WRKY14 (accession no. XP_010920047.1), and Cocos nucifera WRKY14 (accession no. KAG1354178.1) (Figure 1B).
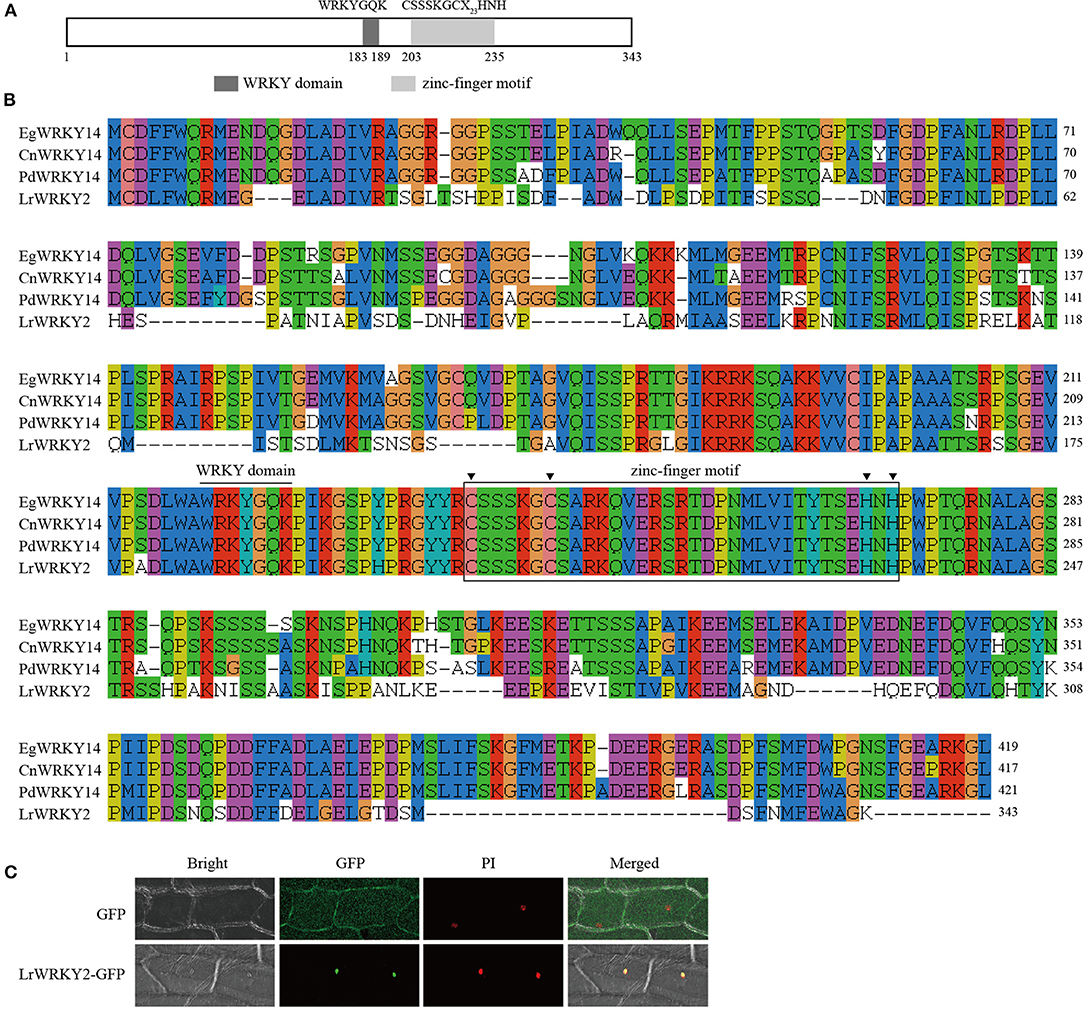
Figure 1. The sequence and subcellular localization analyses of LrWRKY2. (A) The conserved domains predicted in the LrWRKY2. (B) The multiple alignment of the amino acid sequence of LrWRKY2 and three homologous WRKYs were performed with the ClustalW. (C) Subcellular localization analysis of LrWRKY2-GFP fusion protein. The LrWRKY2-GFP fusion protein was transiently expressed in nucleus after genetic transform mediated by Agrobacterium tumefaciens. GFP, fluorescent light; Bright, white light; PI, the nucleus was displayed by propidium iodide (PI) staining; Merged, overlaid of fluorescent and the nucleus displayed by PI staining.
The PSORT program predicted that LrWRKY2 contains a putative NLS “KRPR” sequence, implying LrWRKY2 may be localized in the nucleus. To experimentally determine the subcellular localization of LrWRKY2, the LrWRKY2 coding sequence was fused to a GFP reporter gene, and the resulting fusion construct was inserted into onion epidermal cells. Green fluorescence was detected throughout the onion cells infected with A. tumefaciens carrying the empty pBIN m-gfp5-ER vector, whereas it was limited to the nucleus in cells infected with A. tumefaciens carrying the LrWRKY2-GFP construct. Moreover, the LrWRKY2-GFP fusion protein colocalized with the nuclear localization marker PI (Figure 1C). These observations indicate LrWRKY2 is a nuclear protein.
The integration of LrWRKY2 into transgenic tobacco plants was verified by PCR. There were no differences in the phenotype and growth status of the WT and LrWRKY2 transgenic tobacco plants. Twelve confirmed transgenic lines were selected for the subsequent investigation. The analysis of gene expression by qRT-PCR indicated that LrWRKY2 was stably expressed in the transgenic lines (Figure 2A). Among the transgenic lines, LrWRKY2 expression levels were highest in W2-32, W2-38, W2-39, and W2-44. Thus, these four lines were selected for further analyses.
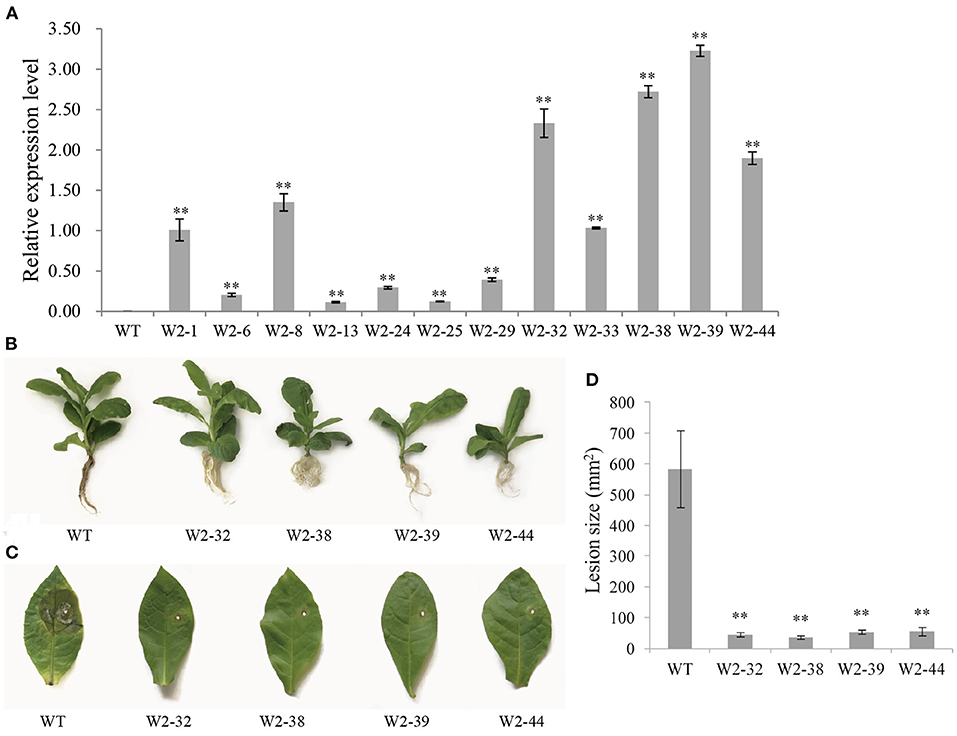
Figure 2. Gene expression and resistance analyses of T2 generation LrWRKY2 transgenic tobacco lines. (A) The LrWRKY2 was stably expressed in the 12 T2 generation LrWRKY2 transgenic tobacco lines (W2-1/6/8/13/24/25/29/32/33/38/39/44). (B) The root inoculation assay revealed the enhanced resistance of four T2 LrWRKY2 transgenic tobacco lines (W2-32/38/39/44) to F. oxysporum infection. (C) The leaf inoculation assay revealed the increased resistance of four T2 LrWRKY2 transgenic tobacco lines (W2-32/38/39/44) to F. oxysporum infection. (D) The lesion areas in the inoculated leaves of the four T2 LrWRKY2 transgenic tobacco lines were significantly smaller than that in the WT (**p < 0.01). Bars represent the standard errors of three biological replicates, and the statistical analysis was performed with the t test (**p < 0.01).
The roots of WT and LrWRKY2 transgenic tobacco plants were inoculated with an F. oxysporum spore suspension (5 × 106 spores/mL). After a 7-day incubation, the inoculated WT plants had curled, yellow, and withered leaves as well as blackened or rotted roots. In contrast, the LrWRKY2 transgenic tobacco leaves and roots were healthy and growing well (Figure 2B). The leaf inoculation assay produced similar results. More specifically, obvious disease symptoms were detected surrounding the inoculation site of the WT leaves (Figure 2C). The WT leaves were yellow and rotted, which was in sharp contrast to the healthy LrWRKY2 transgenic leaves. The average leaf lesion size for the transgenic lines and WT control was <100 mm2 and ~580 mm2, respectively (Figure 2D). These results imply that LrWRKY2 overexpression can substantially increase the resistance of transgenic tobacco to F. oxysporum.
Several defense response-related genes were selected to examine the transcriptional changes in the LrWRKY2 transgenic tobacco lines. The JA biosynthetic pathway-related genes (NtAOC, NtAOS, NtKAT, NtPACX, NtJMT, NtOPR, and NtLOX), PR genes (NtCHI, NtGlu2, and NtPR-1), and SOD genes (NtSOD, NtCu-ZnSOD, and MnSOD) showed higher expression levels in the LrWRKY2 transgenic lines than in the WT plants (Figure 3). The JA biosynthetic pathway-related genes NtAOC, NtKAT, NtJMT, NtOPR, and NtLOX as well as the PR gene NtGlu2 were most highly expressed in line W2-1, with the NtGlu2 expression level 15-fold higher than that in the WT plants. The JA biosynthetic pathway-related genes NtAOS and NtPACX, the PR genes NtCHI and NtPR-1, and the SOD genes NtSOD, NtCu-ZnSOD, and MnSOD displayed the highest expression levels in line W2-33, with the NtPR-1 expression level 17-fold higher than that in the WT plants. The expression levels of all analyzed genes were higher in line W2-8 than in the WT plants. These observations suggest that LrWRKY2 overexpression in tobacco up-regulates the expression of JA signaling pathway genes and induces the expression of some PR and SOD genes.
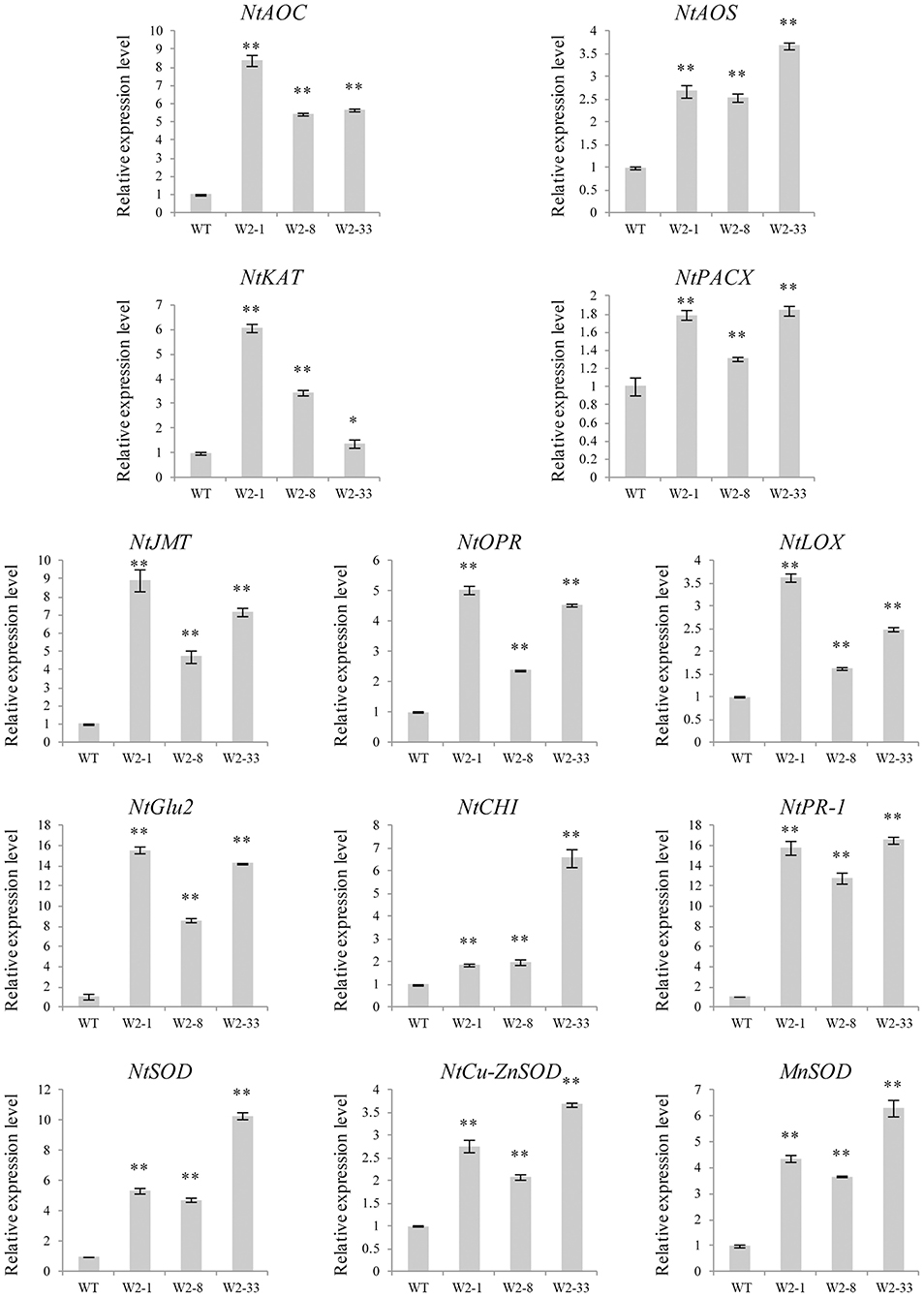
Figure 3. The expression levels of some JA biosynthesis and defense-related genes including the NtAOC, NtAOS, NtKAT, NtPACX, NtJMT, NtOPR, NtLOX, NtCHI, NtGlu2, NtPR-1, NtSOD, NtCu-ZnSOD, and MnSOD in the three T2 LrWRKY2 transgenic tobacco lines (W2-1/8/33) were evaluated by qRT-PCR. Bars represent the standard errors of three biological replicates. The results was calculated by the 2−ΔΔCt method and analyzed by the t test (*p < 0.05, **p < 0.01).
To clarify the effect of silencing LrWRKY2 expression, the LrWRKY2-RNAi construct was transiently expressed in L. regale scales, which were then inoculated with F. oxysporum. After a 3-day incubation, the scales expressing the LrWRKY2-RNAi fragment had dark brown wounds and were rotted, whereas the scales transformed with the empty RNAi vector had light brown wounds and were only slightly rotted (Figure 4A). Moreover, the average lesion size was greater for the scales expressing the LrWRKY2-RNAi fragment (~100 mm2) than for the scales transformed with the empty RNAi vector (~28 mm2) (Figure 4B). At 3 days after the inoculation with F. oxysporum, the LrWRKY2 expression level was significantly lower in the L. regale scales expressing the LrWRKY2-RNAi fragment than in the L. regale scales with the empty RNAi vector (Figure 4C). The lily scales expressing the LrWRKY2-RNAi fragment were more susceptible to F. oxysporum than the control scales, which was consistent with the relatively low LrWRKY2 expression level due to the RNAi fragment. These findings imply that LrWRKY2 functions as a positive regulator of L. regale responses to an F. oxysporum infection.
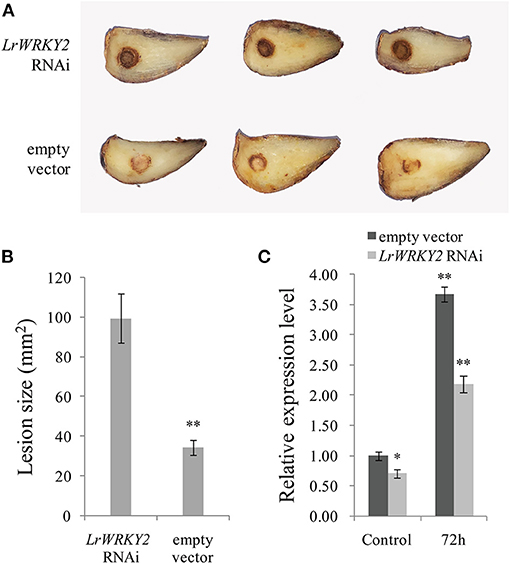
Figure 4. Analysis of the hairpin RNA targeting LrWRKY2 transiently expressed in L. regale scales. (A) The symptoms of L. regale scales after F. oxysporum inoculation, in which the LrWRKY2 RNAi vector and the empty RNAi vector were expressed, respectively. (B) The lesion areas in LrWRKY2-RNAi fragment expressed scales were significantly bigger than that in the empty RNAi vector expressed scales (**p < 0.01). (C) The expression levels of LrWRKY2 in LrWRKY2-RNAi fragment expressed scales and the empty RNAi vector expressed scales were evaluated by qRT-PCR. Bars represent the standard errors of three biological replicates. The results was calculated by the 2−ΔΔCt method and analyzed by the t test (*p < 0.05, **p < 0.01).
To explore the regulatory effects of the LrWRKY2 TF on PR gene expression, an L. regale chitinase gene (LrCHI2) responsive to F. oxysporum was cloned on the basis of the L. regale transcriptome sequencing data (unpublished). Gene expression level of LrCHI2 was estimated as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). The FPKM of LrCHI2 in L. regale roots without inoculation with F. oxysporum was 5.29, while its FPKM in L. regale roots inoculated with F. oxysporum for 96 h was 1015.59. Therefore, the log2FC (log2 fold changes) was 6.97, indicating a significant upregulation of LrCHI2 gene in L. regale roots during F. oxysporum infection. The obtained full-length LrCHI2 cDNA (accession no. MZ272344) consisted of 1,313-bp, with a 933-bp ORF, a 19-bp 5′ untranslated region, and a 361-bp 3′ untranslated region. The encoded protein comprising 310 amino acid residues was ~32.46 kDa and had an isoelectric point of about 5.65. A sequence analysis indicated the deduced LrCHI2 protein possesses a chitin-binding domain (ChtBD) and a Glyco_hydro_19 (GH19) catalytic domain. Additionally, a 491-bp LrCHI2 promoter fragment (accession no. MZ272345) was obtained by TAIL-PCR. A number of cis-elements were identified in the LrCHI2 promoter region, including ABRELATERD1 (abscisic acid-responsive element), GT1CONSENSUS (gibberellic acid-responsive element), and the W-box (Supplementary Table 3).
The qRT-PCR analysis indicated LrCHI2 was expressed at relatively low levels in the stems, leaves, flowers, and scales (Figure 5A), but it was highly expressed in the roots. Moreover, the F. oxysporum infection significantly up-regulated LrCHI2 expression in the L. regale roots (Figure 5B). Specifically, its expression level was about 3-fold and 5.5-fold higher than that in the control (not inoculated) at 24 and 48 h post-inoculation, respectively. Accordingly, LrCHI2 appears to be predominantly expressed in the roots and its expression is induced by F. oxysporum.
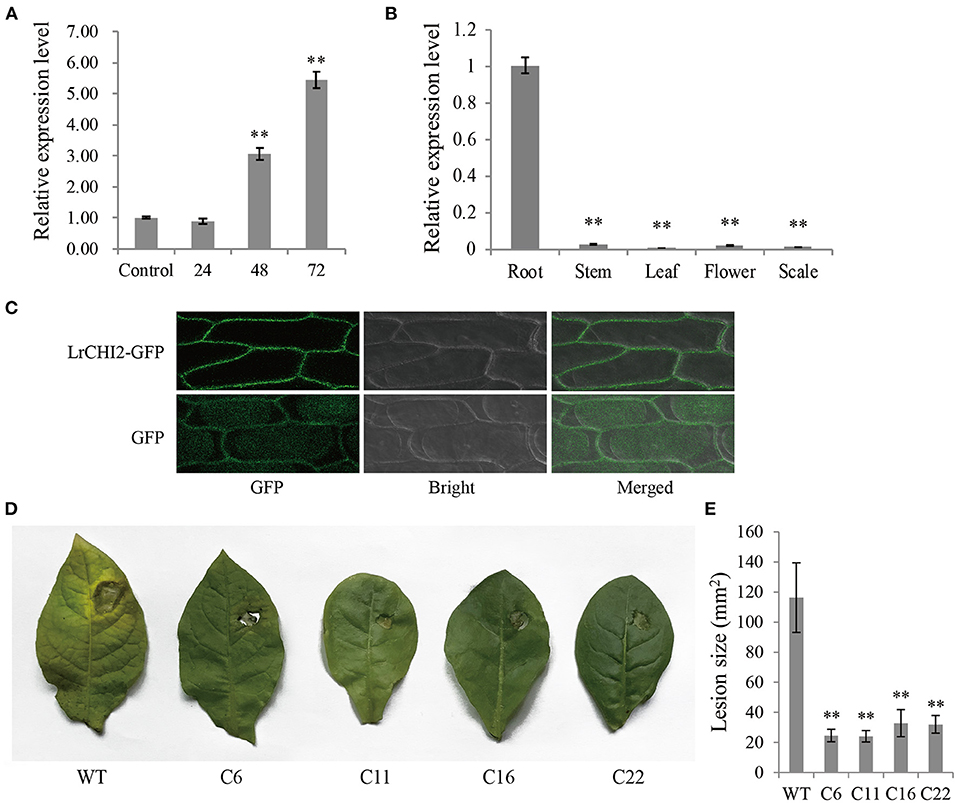
Figure 5. The functional analysis of LrCHI2. (A) The expression levels of LrCHI2 in L. regale roots after inoculation with F. oxysporum were analyzed by qRT-PCR. The L. regale roots were inoculated with F. oxysporum, and then were collected at 4, 24, 48, and 72 hpi. The roots inoculated with sterile water were used as control sample. (B) The expression levels of LrCHI2 in various tissues of L. regale under normal conditions were analyzed by qRT-PCR. (C) Subcellular localization analysis of LrCHI2-GFP fusion protein. The LrCHI2-GFP fusion protein was transiently expressed in cell wall after genetic transform mediated by A. tumefaciens. GFP, fluorescent light; Bright, white light; Merged, overlaid of fluorescent and white light. (D) The leaf inoculation assay revealed the four T2 LrWRKY2 transgenic tobacco lines (C6, C11, C16, and C22) showed enhanced resistance to F. oxysporum infection. (E) The lesion areas in the inoculated leaves of the four T2 generation LrCHI2 transgenic tobacco lines were significantly smaller than that in the WT (**p < 0.01). Bars represent the standard errors of three biological replicates. The statistical analysis was performed with the t test (**p < 0.01).
An N-terminal signal peptide was detected in LrCHI2, indicating this protein is secreted. The subcellular localization of LrCHI2 was determined by expressing a GFP-tagged fusion protein in onion epidermal cells. The fluorescence of LrCHI2-GFP was exclusively detected in the cell wall, suggesting LrCHI2 is a cell wall protein (Figure 5C).
Four T2 transgenic tobacco lines overexpressing LrCHI2 (C6, C11, C16, and C22) were randomly selected and examined regarding their resistance to F. oxysporum. The WT and LrCHI2 transgenic tobacco leaves were inoculated with an F. oxysporum spore suspension (5 × 106 spores/mL). After a 7-day incubation, transgenic tobacco leaves infected with F. oxysporum had relatively small wounds with slightly yellow edges, whereas the infected WT leaves had large wounds and were more obviously decayed (Figure 5D). Hence, LrCHI2 overexpression considerably enhanced the resistance of the transgenic tobacco lines to F. oxysporum. The lesions on the leaves of the four transgenic tobacco lines (C6, C11, C16, and C22) were smaller than 40 mm2, whereas the WT leaf lesions were almost 120 mm2 (Figure 5E).
The LrCHI2 recombinant protein lacking a signal peptide was expressed in E. coli cells. The SDS-PAGE analysis indicated that the His-tagged LrCHI2 protein produced in cells induced by isopropyl-β-D-1-thiogalactopyranoside (IPTG) was ~47 kDa, which was consistent with the predicted size (Figure 6A). The LrCHI2 recombinant protein in inclusion bodies was solubilized and then purified by nickel-affinity chromatography, with most of the protein eluted by the 100 mM imidazole elution buffer. The lack of extra bands during the SDS-PAGE analysis reflected the purity of the obtained LrCHI2 recombinant protein (Figure 6B). The in vitro antifungal activity of the purified LrCHI2 recombinant protein was examined. Compared with the blank controls, the LrCHI2 recombinant protein significantly inhibited the growth of F. oxysporum, C. gloeosporioides, F. solani, and A. panax (Figures 6C–F). Moreover, the inhibitory effect increased as the concentration of the recombinant protein solution used to treat the tested fungi increased (Figure 6G).
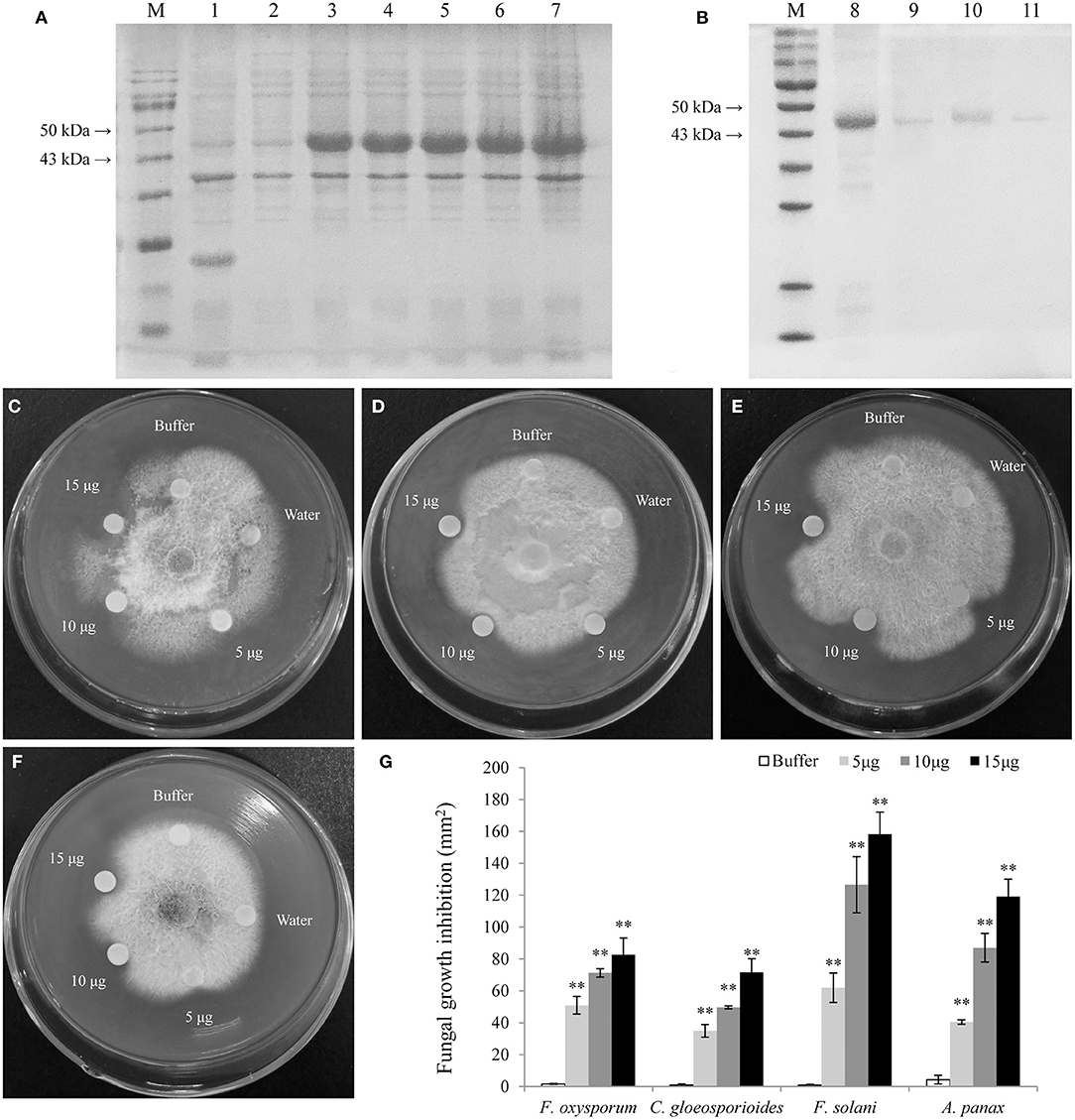
Figure 6. Expression, purification, and antifungal assay of LrCHI2 recombinant protein. (A) The induced expression of LrCHI2 recombinant protein under 1 mM IPTG condition. M, protein marker; Line 1, the protein of E. coli with empty vector pET-32a was detected after induction; Line 2, the expression of LrCHI2 protein was detected without induction; Line 3-7, the expression of LrCHI2 protein was detected at 6, 8, 10, 12, and 24 h after induction, respectively. (B) The purification of LrCHI2 recombinant protein. M, protein marker; Line 8, the supernatant after treatment with the inclusion body solubilizing solution; Line 9-11, the purified LrCHI2 recombinant protein with 50, 100, 150 mM imidazole washing buffer, respectively. (C–F) The LrCHI2 recombinant protein has evident antifungal activity to F. oxysporum (C), C. gloeosporioides (D), F. solani (E) and A. panax (F). (G) The fungal growth inhibition areas (mm2). Bars represent the standard errors of three biological replicates. The statistical analysis was performed with the t test (**p < 0.01).
The WRKY TFs often target the W-box cis-element to activate or suppress target gene expression. The EMSA results confirmed the LrWRKY2 recombinant protein purified from E. coli cells was able to bind directly to the W-box sequence in the LrCHI2 promoter. The biotin-labeled probes alone revealed a lack of band shifts in the gel (lane 1, Figure 7A). However, LrWRKY2 was able to bind to the biotin-labeled probes at the LrCHI2 promoter fragment, resulting in a mobility shift (lane 2, Figure 7A). The inclusion of both biotin-labeled and unlabeled probes resulted in the detection of a band shift in the gel (lane 3, Figure 7A), reflecting the binding of the two probes to LrWRKY2. Because the unlabeled probes were 50-fold more abundant than the biotin-labeled probes, LrWRKY2 mainly bound to the competitor (unlabeled probe). Thus, the band shift was less intense in lane 3 than in lane 2 (Figure 7A). A band shift was undetectable in lane 4 (Figure 7A), which corresponded to the analysis involving the mutant probe and LrWRKY2, implying LrWRKY2 binds specifically to the W-box in the LrCHI2 promoter.
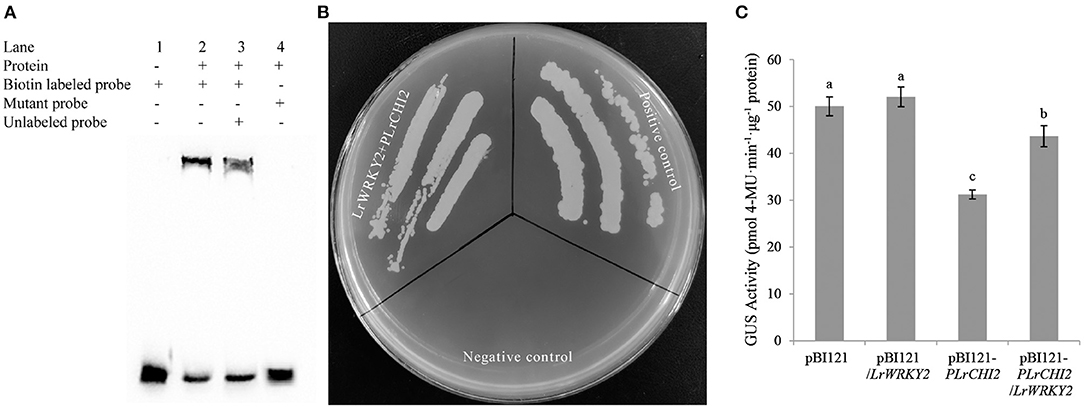
Figure 7. Analysis of the interaction between LrWRKY2 and LrCHI2 promoter. (A) The specific binding of LrWRKY2 with the LrCHI2 promoter fragment was verified by EMSA. Line 1, the biotin labeled probes containing the W-box sequence. Line 2, LrWRKY2 can interact with the biotin labeled probes containing the W-box sequence 3, LrWRKY2 can interact with both the biotin labeled and unlabeled probes containing the W-box sequence. 4, LrWRKY1 can't interact with mutant probes. (B) Analysis of the transactivation effect of LrWRKY2 recombinant protein on LrCHI2 promoter. LrWRKY2 + pLrCHI2, the interaction of pGADT7-LrWRKY2 and LrCHI2 promoter; Positive control, the interaction of pGADT7-p53 and pAbAi-p53; Negative control, the interaction of pAbAi-p53 and pGADT7-LrWRKY2. (C) GUS activity was higher in transgenic tobacco lines co-expressing of LrWRKY2 and LrCHI2 promoter than in the transgenic tobacco lines expressing of LrCHI2 promoter. Bars represent the standard errors of three biological replicates. The statistical differences were analyzed by Duncan's multiple range test (Different letters indicate a significant difference).
The ability of LrWRKY2 to activate transcription was evaluated in a Y1H assay, in which Y1HGold yeast cells were transformed with pGADT7-LrWRKY2 and pLrCHI2-pAbAi. The assay results indicated that the yeast cells co-transformed with pGADT7-LrWRKY2 and pLrCHI2-pAbAi grew well, whereas the yeast cells transformed with pGADT7-LrWRKY2 and the empty pAbAi vector failed to survive on the SD/–Leu selective medium containing 50 ng/mL AbA (Figure 7B). These observations indicate LrWRKY2 can bind to the LrCHI2 promoter in vivo and activate transcription in yeast cells.
The transcriptional activation of LrCHI2 by LrWRKY2 was verified by inserting an expression construct comprising a GUS-encoding gene under the control of the LrCHI2 promoter into LrWRKY2 transgenic tobacco. A total of 23 transgenic tobacco plants carrying both LrWRKY2 and pLrCHI2-GUS and 27 transgenic tobacco plants transformed with pLrCHI2-GUS alone were identified by PCR. The GUS activity in the pLrCHI2-GUS transgenic tobacco plants was significantly lower than that in the transgenic tobacco carrying both LrWRKY2 and pLrCHI2-GUS (Figure 7C). The activity of the GUS expressed under the control of the LrCHI2 promoter increased by approximately 1.4-fold in the presence of LrWRKY2. There were no significant differences in the GUS activities of the two kinds of transgenic tobacco plants transformed with the empty pBI121-GUS vector. This implies that LrWRKY2 does not affect the CaMV 35S promoter. Therefore, LrWRKY2 positively regulates LrCHI2 expression.
The WRKY proteins are critical plant TFs mediating diverse biological processes. However, little is known about the functions of L. regale WRKY genes. Protein functions are closely related to the distribution of the proteins in cells. The localization of LrWRKY2 in the nucleus is in accordance with its putative function as a TF that must exist in the nucleus to regulate the transcription of target genes (Liu et al., 2013; Wang et al., 2013; Sun et al., 2015). Similarly, maize (Zea mays) WRKY106 (Wang C. T. et al., 2018) and cotton (Gossypium hirsutum) WRKY33 are also nuclear proteins (Wang et al., 2019). In fact, the vast majority of the WRKY TFs identified to date are localized in the nucleus. Accordingly, LrWRKY2 is a nuclear protein that regulates cellular processes. In contrast, LrCHI2 was localized to the cell wall in this study. The plant cell wall is the first line of defense against a pathogen invasion. It is possible that LrCHI2 participates in chitinase–pathogen interactions. Some Cucumis sativus chitinases are also localized in the cell wall, wherein they interact directly or indirectly with pathogen elicitors to trigger downstream defense pathways (Bartholomew et al., 2019).
The LrWRKY2 TF belongs to Subgroup IIe of the WRKY family (Li et al., 2021a). Previous research proved that many Subgroup IIe WRKY TFs positively regulate plant responses to biotic and abiotic stresses. For example, the overexpression of the Subgroup IIe CmWRKY10 chrysanthemum (Chrysanthemum morifolium) gene reportedly leads to increased drought tolerance (Jaffar et al., 2016). Transgenic tobacco plants overexpressing the C. annuum WRKY27 gene enhances the resistance to Ralstonia solanacearum (i.e., milder disease symptoms and inhibited pathogen growth) (Dang et al., 2014). Herbaceous peony (Paeonia lactiflora) plants in which the Subgroup IIe gene WRKY65 is silenced exhibit increased susceptibility to Alternaria tenuissima, suggesting PlWRKY65 is a positive regulator of peony defense responses to A. tenuissima (Wang et al., 2020). It is well-known that the lily is difficult to perform the genetic transform, and only a few lily transformations have been achieved thus far (Yan et al., 2019b). The currently established lily transformation system still has some problems, such as the strong genotype dependence, low efficiency of stable transformation, poor genetic stability, and difficult regeneration after transformation (Yan et al., 2019b; Song et al., 2020). It is not so bad that the Agrobacterium-mediated transformation of tobacco using leaf disks has provided a valuable tool for rapid evaluation of function of the transgenes in higher plants (Clemente, 2006). Therefore, the stable genetic transformation of LrWRKY2 was completed in the model plant tobacco in order to further understand the function of LrWRKY2 in the present study. The LrWRKY2-overexpressing transgenic tobacco plants were more resistant to F. oxysporum than the WT control plants. Additionally, L. regale scales transiently expressing LrWRKY2-RNAi were more susceptible to F. oxysporum than the control scales. Our results suggest that LrWRKY2 is an important TF influencing L. regale disease resistance.
The ectopic expression of TF genes facilitated by transgenic technology and the subsequent analysis of the transcriptional changes in the generated transgenic plants may provide new insights regarding transcriptional regulatory networks. As crucial components of plant defense systems, WRKY proteins regulate the expression of some defense-related genes. The overexpression of rice Subgroup IIa WRKY genes (OsWRKY62, OsWRKY28, OsWRKY71, and OsWRKY76) up-regulates the PR10 expression level (Peng et al., 2010). The constitutive overexpression of PtrWRKY18 and PtrWRKY35 in poplar (Populus trichocarpa) enhances the resistance to the biotrophic pathogen Melampsora by inducing PR gene expression (Jiang et al., 2017). The ectopic overexpression of the cotton (G. hirsutum) WRKY44 gene increases the resistance of Nicotiana benthamiana plants to fungal infections and activates the expression of PR genes, including PR1a, PR4, PR5, and NPR1 (Li et al., 2015). The ectopic expression of rice OsWRKY11 results in the constitutive expression of defense-associated genes (CHIT2, PR10, and Betv1), whereas down-regulating OsWRKY11 expression adversely affects the expression of defense-related genes during pathogen invasions, suggesting that OsWRKY11 activates defense responses (Lee et al., 2018). The heterologous expression of rice OsWRKY6 in A. thaliana can dramatically induce the expression of defense-related genes, including PR1, PDF1, NPR4, and a glucanase gene (Hwang et al., 2011). In the present study, the expression levels of several PR genes (e.g., NtCHI, NtGlu2, and NtPR-1) were clearly up-regulated in LrWRKY2 transgenic tobacco lines. Additionally, the expression levels of JA biosynthetic pathway-related genes (NtAOC, NtAOS, NtKAT, NtPACX, NtJMT, NtOPR, and NtLOX) were higher in the LrWRKY2-overexpressing transgenic tobacco lines than in the WT tobacco plants, implying that the JA signaling pathway was activated in response to LrWRKY2 overexpression. Consistent with our findings, a previous study revealed that in transgenic A. thaliana lines overexpressing an L. regale WRKY gene, the expression levels of some JA-responsive genes, including AtLOX, AtMYC2, and AtPDF1.2, are up-regulated (Cui et al., 2018). Moreover, overexpressing PtrWRKY40 in A. thaliana activates the expression of JA-related defense genes, ultimately leading to the resistance to the necrotrophic fungus Botrytis cinerea (Karim et al., 2015). Furthermore, the expression levels of SOD genes (NtSOD, NtCu-ZnSOD, and MnSOD) increased significantly in the LrWRKY2-overexpressing tobacco lines, suggesting that LrWRKY2 confers enhanced disease resistance by activating the JA signaling pathway and inducing the expression of defense-related genes.
The PR proteins have important functions related to plant defense responses to pathogens. They accumulate after pathogen invasions, and may act as antimicrobial agents mediating cell wall hydrolysis, contact toxicity, and perhaps defense signaling (Zhang et al., 2017). A recent study demonstrated that Panax notoginseng PR-like proteins can inhibit F. solani and C. gloeosporioides mycelial growth (Li et al., 2021b). Another study proved that overexpressing the moss (Physcomitrella patens) PR10 gene in A. thaliana enhances plant resistance to Pythium irregulare (Castro et al., 2016). In the current study, LrCHI2 isolated from L. regale was characterized as a new PR gene. This gene was mainly expressed in the roots, and its expression was induced by an F. oxysporum infection. Additionally, LrCHI2 exhibited in vitro antifungal activity that inhibited F. oxysporum, C. gloeosporioides, F. solani, and A. panax mycelial growth. The overexpression of LrCHI2 in tobacco enhanced the resistance of the transgenic plants to F. oxysporum. Thus, LrCHI2 expression contributes to the resistance of L. regale to F. oxysporum.
Previous research proved that various PR genes are targeted by WRKY TFs, which bind to the canonical W-box sequence (TTGACC/T) in the gene promoters. A recent Y1H assay confirmed that rice OsWRKY6 can bind directly to the W-box derived from the rice PR1 promoter in yeasts (Hwang et al., 2011). An in vivo chromatin immunoprecipitation assay and in vitro EMSA experiments revealed that C. annuum WRKY40 binds directly to a C. annuum defensin gene promoter and the WRKY33 promoter, both of which contain a W-box; on the contrary, with mutations to the W-box sequence preventing the binding (Chakraborty et al., 2019). The LrWRKY1 TF induces the transcription of LrPR10-5, which has a promoter containing three W-boxs (Li et al., 2021a). The co-expression of LrWRKY1 and LrPR10-5 in tobacco indicated LrWRKY1 can activate the LrPR10-5 promoter. In this study, the regulatory effect of LrWRKY2 on the LrCHI2 promoter was examined. This promoter contains hormone-responsive, biotic and abiotic stress-related elements as well as one W-box. The EMSA results proved that LrWRKY2 has a high in vitro affinity for the LrCHI2 promoter containing a W-box. Additionally, the Y1H assay results verified the interaction between LrWRKY2 and the LrCHI2 promoter fragment in yeast. These findings suggest that LrWRKY2 binds to the W-box in the LrCHI2 promoter to activate expression. This likely contributes to the LrWRKY2-regulated L. regale defense response to F. oxysporum. Because the transcript levels of several PR genes, including NtCHI, NtGlu2, and NtPR-1, increased in LrWRKY2 transgenic tobacco lines, future studies should investigate whether LrWRKY2 regulates the expression of other PR genes in L. regale.
The nuclear protein LrWRKY2 belongs to Subgroup IIe of the WRKY family. Transgenic tobacco lines overexpressing LrWRKY2 are highly resistant to F. oxysporum, which may be related to the significantly up-regulated expression of many defense-related genes, including JA biosynthetic pathway-related genes, PR genes, and SOD genes. Additionally, the transient expression of the LrWRKY2-RNAi fragment in L. regale scales leads to increased sensitivity to F. oxysporum. Moreover, LrWRKY2 is a positive regulator that mediates L. regale defense responses to F. oxysporum infection by regulating the chitinase gene expression, which is a broad-spectrum resistance gene showing antifungal activity to some important phytopathogens including F. oxysporum. The results of this study may be relevant for future research aimed at elucidating the transcriptional regulatory mechanisms underlying the interaction between L. regale and F. oxysporum. In addition, the lily genetic transformation mediated by the Agrobacterium has been carried out in our research group. It is believed that a more in-depth understanding of the defense response regulation mechanism in L. regale against the Fusarium wilt will be obtained in the near future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SL: methodology, data curation, experiment execution, and writing—original draft preparation. JH: methodology, material collection, and supervision. ZW: investigation and software. JD: validation. TL: supervision. LS: conceptualization. DL: methodology, supervision, writing—reviewing, and editing. All authors contributed to the article and approved the submitted version.
This work was financially supported by two grants received from the National Natural Sciences Foundation of China (grant number 31760586) and Yunnan Ten Thousand Talents Plan Young & Elite Talents Project, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.741463/full#supplementary-material
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Bartholomew, E. S., Black, K., Feng, Z., Liu, W., Shan, N., Zhang, X., et al. (2019). Comprehensive analysis of the chitinase gene family in cucumber (Cucumis sativus L.): from gene identification and evolution to expression in response to Fusarium oxysporum. Int. J. Mol. Sci. 20:5309. doi: 10.3390/ijms20215309
Castro, A., Vidal, S., and Ponce de León, I. (2016). Moss pathogenesis-related-10 protein enhances resistance to Pythium irregulare in Physcomitrella patens and Arabidopsis thaliana. Front. Plant Sci. 7:580. doi: 10.3389/fpls.2016.00580
Chakraborty, J., Ghosh, P., Sen, S., Nandi, A., and Das, S. (2019). CaMPK9 increases the stability of CaWRKY40 transcription factor which triggers defense response in chickpea upon Fusarium oxysporum f. sp. ciceri Race1 infection. Plant Mol. Biol. 100, 411–431. doi: 10.1007/s11103-019-00868-0
Chen, R., He, H., Yang, Y., Qu, Y., Ge, F., and Liu, D. (2017). Functional characterization of a pathogenesis-related protein family 10 gene, LrPR10-5, from Lilium regale Wilson. Australas. Plant Pathol. 46, 251–259. doi: 10.1007/s13313-017-0485-0
Choi, C., Hwang, S. H., Fang, I., Kwon, S. I., Park, S., Ahn, I., et al. (2015). Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. N. Phytol. 208, 846–859. doi: 10.1111/nph.13516
Clemente, T. (2006). Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). Methods Mol. Biol. 343, 143–154. doi: 10.1385/1-59745-130-4:143
Cui, Q., Yan, X., Gao, X., Zhang, D., He, H., and Jia, G. (2018). Analysis of WRKY transcription factors and characterization of two Botrytis cinerea-responsive LrWRKY genes from Lilium regale. Plant Physiol. Biochem. 127, 525–536. doi: 10.1016/j.plaphy.2018.04.027
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. doi: 10.1111/ppl.12093
Ding, Y., Chen, Y., Lin, Z., Tuo, Y., Li, H., and Wang, Y. (2021). Differences in soil microbial community composition between suppressive and root rot-conducive in tobacco fields. Curr. Microbiol. 78, 624–633. doi: 10.1007/s00284-020-02318-3
Dong, Q., Zheng, W., Duan, D., Huang, D., Wang, Q., Liu, C., et al. (2020). MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 299:110611. doi: 10.1016/j.plantsci.2020.110611
Eulgem, T., and Somssich, I. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
He, H., Liu, D., Zhang, N., Zheng, W., Han, Q., Ji, B., et al. (2014). The PR10 gene family is highly expressed in Lilium regale Wilson during Fusarium oxysporum f. sp. lilii infection. Genes Genom. 36, 497–507. doi: 10.1007/s13258-014-0185-x
He, X., Li, W., Zhang, W., Jin, X., Shenkute, A. G., Aynalem, T., et al. (2019). Transcriptome sequencing analysis provides insights into the response to Fusarium oxysporum in Lilium pumilum. Evol. Bioinform. Online. 15:1176934319838818. doi: 10.1177/1176934319838818
Holsters, M., Waele, D., Depicker, A., Messens, E., Van Montagu, M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/BF00267408
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science. 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Huang, X., Li, K., Xu, X., Yao, Z., Jin, C., and Zhang, S. (2015). Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics 16:1104. doi: 10.1186/s12864-015-2233-6
Hwang, S., Yie, S. W., and Hwang, D. (2011). Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 181, 316–323. doi: 10.1016/j.plantsci.2011.06.007
Ifnan Khan, M., Zhang, Y., Liu, Z., Hu, J., Liu, C., Yang, S., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19:1403. doi: 10.3390/ijms19051403
Jaffar, M. A., Song, A., Faheem, M., Chen, S., Jiang, J., Liu, C., et al. (2016). Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 17:693. doi: 10.3390/ijms17050693
Jiang, J., Ma, S., ye, N., Jiang, M., Cao, J., and Zhang, J. (2016). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59. 86–101. doi: 10.1111/jipb.12513
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Jimmy, J. L., and Babu, S. (2019). Variations in the structure and evolution of rice WRKY genes in Indica and Japonica genotypes and their co-expression network in mediating disease resistance. Evol. Bioinform. Online. 15:1176934319857720. doi: 10.1177/1176934319857720
Karim, A., Jiang, Y., Guo, L., Ling, Z., Ye, S., Duan, Y., et al. (2015). Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 35, 1129–1139. doi: 10.1093/treephys/tpv084
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11:5. doi: 10.1186/s12284-018-0199-0
Li, J., Wang, J., Wang, N., Guo, X., and Gao, Z. (2015). GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tiss. Org. Cult. 121, 127–140. doi: 10.1007/s11240-014-0688-9
Li, S., Liu, G., Pu, L., Liu, X., Wang, Z., Zhao, Q., et al. (2021a). WRKY transcription factors actively respond to Fusarium oxysporum in Lilium regale Wilson. Phytopathology doi: 10.1094/PHYTO-10-20-0480-R. [Epub ahead of print].
Li, S., Wang, Z., Tang, B., Zheng, L., Chen, H., Cui, X., et al. (2021b). A pathogenesis-related protein-like gene is involved in the Panax notoginseng defense response to the root rot pathogen. Front. Plant Sci. 11:610176. doi: 10.3389/fpls.2020.610176
Liu, D. Q., Han, Q., Shah, T., Chen, C. Y., Wang, Q., Tang, B. F., et al. (2018). A hybrid proline-rich cell-wall protein gene JsPRP1 from Juglans sigillata Dode confers both biotic and abiotic stresses in transgenic tobacco plants. Trees 32, 1199–1209. doi: 10.1007/s00468-018-1702-3
Liu, Q. L., Zhong, M., Li, S., Pan, Y. Z., Jiang, B. B., Jia, Y., et al. (2013). Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol. Biochem. 69, 27–33. doi: 10.1016/j.plaphy.2013.04.016
Long, L., Yang, W. W., Liao, P., Guo, Y. W., Kumar, A., and Gao, W. (2019). Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci. 281, 72–81. doi: 10.1016/j.plantsci.2019.01.012
Nan, H., and Gao, L. Z. (2019). Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Genet. 10:363. doi: 10.3389/fgene.2019.00363
Peng, Y., Bartley, L. E., Canlas, P., and Ronald, P. C. (2010). OsWRKY IIa transcription factors modulate rice innate immunity. Rice 3, 36–42. doi: 10.1007/s12284-010-9039-6
Qiu, Y., and Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Ramekar, R. V., Park, K. C., Kwak, M., and Choi, I. Y. (2019). The complete chloroplast genome of a rare species in Korea, Lilium Dauricum Ker Gawl. Mitochondrial DNA B Resour. 4, 3591–3592. doi: 10.1080/23802359.2019.1676174
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Song, S., Yan, R., Wang, C., Wang, J., and Sun, H. (2020). Improvement of a genetic transformation system and preliminary study on the function of LpABCB21 and LpPILS7 based on somatic embryogenesis in Lilium pumilum DC. Fisch. Int. J. Mol. Sci. 21:6784. doi: 10.3390/ijms21186784
Sun, J., Hu, W., Zhou, R., Wang, L., Wang, X., Wang, Q., et al. (2015). The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34, 23–35. doi: 10.1007/s00299-014-1684-6
Taif, S., Zhao, Q., Pu, L., Li, X., Liu, D., and Cui, X. (2020). A β-1,3-glucanase gene from Panax notoginseng confers resistance in tobacco to Fusarium solani. Ind. Crops Prod. 143:111947. doi: 10.1016/j.indcrop.2019.111947
Viana, V., Busanello, C., da Maia, L., Pegoraro, C., and Costa de Oliveira, A. (2018). Activation of rice WRKY transcription factors: an army of stress fighting soldiers? Curr. Opin. Plant Biol. 45, 268–275. doi: 10.1016/j.pbi.2018.07.007
Wang, C., Deng, P., Chen, L., Wang, X., Ma, H., Hu, W., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 8:e65120. doi: 10.1371/journal.pone.0065120
Wang, C. T., Ru, J. N., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., et al. (2018). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19:3046. doi: 10.3390/ijms19103046
Wang, N. N., Xu, S. W., Sun, Y. L., Liu, D., Zhou, L., Li, Y., et al. (2019). The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep. 9, 724–724. doi: 10.1038/s41598-018-37035-2
Wang, P., Xu, X., Tang, Z., Zhang, W., Huang, X. Y., and Zhao, F. J. (2018). OsWRKY28 regulates phosphate and arsenate accumulation, root system architecture and fertility in rice. Front. Plant Sci. 9:1330. doi: 10.3389/fpls.2018.01330
Wang, X., Li, J., Guo, J., Qiao, Q., Guo, X., and Ma, Y. (2020). The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 7, 57–57. doi: 10.1038/s41438-020-0267-7
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17, 944–956. doi: 10.1105/tpc.104.026435
Yan, H., Li, M., Xiong, Y., Wu, J., Teixeira da Silva, J. A., and Ma, G. (2019a). Genome-wide characterization, expression profile analysis of WRKY family genes in Santalum album and functional identification of their role in abiotic stress. Int. J. Mol. Sci. 20:5676. doi: 10.3390/ijms20225676
Yan, R., Wang, Z., Ren, Y., Li, H., Liu, N., and Sun, H. (2019b). Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int. J. Mol. Sci. 20:2920. doi: 10.3390/ijms20122920
Zhang, Q., Li, Y., Zhang, Y., Wu, C., Wang, S., Hao, L., et al. (2017). Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front. Plant Sci. 8:526. doi: 10.3389/fpls.2017.00526
Keywords: Lilium regale, Fusarium oxysporum, WRKY transcription factors, transcriptional regulation, W-box, chitinase
Citation: Li S, Hai J, Wang Z, Deng J, Liang T, Su L and Liu D (2021) Lilium regale Wilson WRKY2 Regulates Chitinase Gene Expression During the Response to the Root Rot Pathogen Fusarium oxysporum. Front. Plant Sci. 12:741463. doi: 10.3389/fpls.2021.741463
Received: 14 July 2021; Accepted: 30 August 2021;
Published: 27 September 2021.
Edited by:
David W. M. Leung, University of Canterbury, New ZealandReviewed by:
Vívian Ebeling Viana, Federal University of Pelotas, BrazilCopyright © 2021 Li, Hai, Wang, Deng, Liang, Su and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diqiu Liu, RGlxaXVsaXVAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.