- 1National Research Center of Engineering and Technology for Utilization of Botanical Functional Ingredients, Hunan Co-innovation Center for Utilization of Botanical Functional Ingredients, Hunan Agricultural University, Changsha, China
- 2Key Laboratory of Tea Science of Ministry of Education, Hunan Agricultural University, Changsha, China
- 3School of Chemistry Biology and Environmental Engineering, Xiangnan University, Chenzhou, China
Polyphenol oxidase (PPO) plays a role in stress response, secondary metabolism, and other physiological processes during plant growth and development, and is also a critical enzyme in black tea production. However, the regulatory mechanisms of PPO genes and their activity in tea plants are still unclear. In this study, we measured PPO activity in two different tea cultivars, Taoyuandaye (TYDY) and Bixiangzao (BXZ), which are commonly used to produce black tea and green tea, respectively. The expression pattern of CsPPO1 was assessed and validated via transcriptomics and quantitative polymerase chain reaction in both tea varieties. In addition, we isolated and identified an R2R3-MYB transcription factor CsMYB59 that may regulate CsPPO1 expression. CsMYB59 was found to be a nuclear protein, and its expression in tea leaves was positively correlated with CsPPO1 expression and PPO activity. Transcriptional activity analysis showed that CsMYB59 was a transcriptional activator, and the dual-luciferase assay indicated that CsMYB59 could activate the expression of CsPPO1 in tobacco leaves. In summary, our study demonstrates that CsMYB59 represents a transcriptional activator in tea plants and may mediate the regulation of PPO activity by activating CsPPO1 expression. These findings provide novel insights into the regulatory mechanism of PPO gene in Camellia sinensis, which might help to breed tea cultivars with high PPO activity.
Introduction
Polyphenol oxidases (PPOs) are a group of terminal copper-containing oxidases commonly found in various tissues and organs of plants, and play an important role in the enzymatic browning reaction. PPOs are mainly divided into two categories in plants: tyrosinase (EC 1.14.18.1) and catechol-oxidases (EC 1.10.3.1). Tyrosinase has monophenolase activity and catalyzes the formation of o-diphenol from monophenols, which is then oxidized to produce the corresponding quinone. Catechol-oxidases show diphenolase activity and catechol oxidase activity, which catalyze the oxidation of diphenolic compounds to generate quinone (Yu et al., 2015; Liu et al., 2019). PPOs are most abundant in young parts and less abundant in mature tissues in plants (Taranto et al., 2017). PPOs exhibit physiological functions in the growth process of plants such as resistance to stress (Mishra and Singh Sangwan, 2019), pests and diseases (Constabel et al., 2000), and regulate the formation of color in flowers (Molitor et al., 2015). In intact plant cells, PPOs are compartmentalized together with their substrates in chloroplast thylakoid membranes and vesicles, respectively (Mayer and Harel, 1979). When the cells are subjected to mechanical damage, the contact of PPO enzymes with the substrates causes rapid oxidation of polyphenols, which is responsible for the negative effects on the appearance, nutritional quality, and commercial value of agricultural products (Hojnik Podrepšek et al., 2020). However, the enzymatic browning reaction associated with PPOs is not entirely unfavorable. PPO is one of the most essential enzymes in black tea processing.
Tea is the most consumed beverage in the world and is popular worldwide due to its unique flavor and numerous health benefits (Sánchez et al., 2020; Zhu et al., 2020a,b). The fundamental difference between black tea and green tea is the occurrence of enzymatic oxidation in the former during the production process (Chen et al., 2020). The activity of PPO determines the efficiency of the withering and oxidation procedures in black tea processing (Chen et al., 2020). PPO affects a variety of tea characteristics, including color and taste, through the inhibition or enhancement of enzyme activity during tea processing. In particular, PPO and peroxidase are crucial enzymes for generating pigments during the fermentation process of black tea (Stodt et al., 2014). PPO catalyzes the oxidization of catechins in tea leaves into the orange-red theaflavins and the reddish-brown thearubigins, as well as catalyze the enzyme-coupled oxidation of fragrance precursors (Solano et al., 1997). Theaflavins and thearubigins are the major polyphenols that determine the quality of black tea. Theaflavins contribute to the color, astringency and the brightness of black tea liquor, and thearubigins are positively correlated aftertaste of astringency in black tea (Owuor et al., 2010; Wang et al., 2014). Therefore, PPO activity plays a key role in the development of black tea characteristics.
Currently, the PPO gene family in Camellia sinensis is thought to contain five to six members (Zhao et al., 2001). Zhang et al. (2020) find that methyl jasmonate enhances PPO activity by activating the expression of CsPPO2 and CsPPO4 gene to improve defense against tea geometrid larvae (Ectropis grisescens) in tea plant. Additionally, mechanical damage or regurgitant of tea geometrid significantly upregulates the transcriptional expression of CsPPO1 and CsPPO2 (Huang et al., 2018). These findings indicate that increasing the expression of PPO genes in tea plants is of interest; however, the molecular mechanism of the regulation of PPO activity in tea plants remains unclear.
Transcription factors (TFs) have been shown to increase gene expression in various systems. TFs represent a specific group of proteins that regulate stress and signaling in plants, and bind to cis-acting elements in the promoters of downstream genes to regulate gene expression (Riechmann et al., 2000; Amoutzias et al., 2007). MYB proteins are a TF superfamily with diverse functions. Based on the number of incomplete repeats within the MYB conserved domain, MYBs are classified into four categories (1R-, R2R3-, R3-, and 4R-) (An et al., 2018). The R2R3-MYB subfamily is the largest class of MYB TFs in plants, regulating a variety of physiological functions at the transcriptional level, including plant growth and development, metabolite synthesis, and resistance to stress (Stracke et al., 2001). Currently, 126, 109, and 140 R2R3-MYB members have been identified in Arabidopsis thaliana (Dubos et al., 2010), Oryza sativa L. (Feller et al., 2011), and C. sinensis (Chen et al., 2021), respectively. In tea plants, R2R3-MYBs are involved in regulating the biosynthesis of flavonoids and theanine (Wang et al., 2018; Wen et al., 2020). In the purple foliage mutant phenotype, the R2R3-MYB TF CsAN1 regulates the expression of anthocyanin biosynthesis-related genes causing ectopic accumulation of pigments in purple tea (Sun et al., 2016). In addition, the Arabidopsis-derived AtMYB96 TF can modulate the expression of abscisic acid and auxin-related genes to regulate drought-related stress response (Seo et al., 2009). Overexpression of MYB49 in tomato significantly enhances the resistance of tomato plants to Phytophthora infestans, and increases the superoxide dismutase activity, peroxidase activity, chlorophyll content, and photosynthetic rate (Cui et al., 2018). Recently, the R1R2R3 TF MnMYB3R1 derived from mulberry plants (Morus notabilis) has been shown to upregulate PPO by binding to MSA in the promoter region of MnPPO1 (Liu et al., 2019). However, no study has reported on whether TFs can regulate PPO activity in tea plants.
In this study, C. sinensis var. sinensis cv. Taoyuandaye (TYDY) with high PPO activity and superior fermentation characteristics & C. sinensis var. sinensis cv. Bixiangzao (BXZ) with low PPO activity and inferior fermentation characteristics were used. Analysis of transcriptomic data was performed between TYDY and BXZ. The expression patterns of CsPPO1 and R2R2-MYB TF CsMYB59 were analyzed via quantitative real time polymerase chain reaction (qRT-PCR). These results improve the current understanding of the regulatory mechanisms of PPO gene in tea leaves, which provide theoretical support for the further innovation of high-quality tea cultivar.
Materials and Methods
Tea Plant Materials
Leaves were obtained from clonal tea plants cultivated in Tea Germplasm Repository of Hunan Agricultural University, Changsha, Hunan, China. The leaves of one bud with two leaves of similar growth and development without mechanical damage were picked, flash frozen using liquid nitrogen, and stored at −80°C.
Determination of Polyphenol Oxidase Activity in Tea Leaves
Polyphenol oxidase activity was measured using the spectrophotometric method (Teng et al., 2017) with modifications. In brief, 0.5 g fresh leaves were homogenized with 2 g quartz sands and 0.3 g polyvinylpyrrolidone (Sigma-Aldrich, Shanghai, China) in 4.5 mL, pH 5.6, 100 mM cooled citric acid-phosphate buffer (citric acid and phosphate were purchased from Sinopharm chemical reagent, Shanghai, China), and the volume was set to 10 mL by adding the buffer following extraction at 4°C for 12 h. After filtration, the homogenate was centrifuged at 8000 × g for 15 min at 4°C and the supernatant containing the crude enzyme was collected. Crude enzyme (0.2 mL) was added to a mixture of 100 mM catechol (0.5 mL, Sinopharm chemical reagent, Shanghai, China) and 100 mM citric acid-phosphate buffer (1 mL), and then the absorbance of the resultant mixture was measured at 420 nm for 20 min. One unit (U) of PPO activity was expressed as 0.01 U in Δ420/min, and each sample were measured thrice.
RNA Extraction, Sequencing, and cDNA Synthesis
Total RNA was extracted from tea leaves using an RNA extraction kit (Tiangen, Beijing, China). Total RNA quality and concentration were assessed via agarose gel electrophoresis and RNA Nano 6000 Assay Kit (Agilent Technologies, Santa Clara, CA, United States), respectively. A cDNA library was synthesized using a reverse transcription kit (Takara, Shiga, Japan); duplicates of each sample were measured thrice. The cDNA library was quantified using a Qubit 2.0 fluorometer (Invitrogen, Waltham, MA, United States), and an Agilent 2100 Bioanalyzer (Agilent Technologies) was used for quality assessment. The cDNA library was sequenced using Illumina HiSeqTM 2000 (Illumina, San Diego, CA, United States). Clean reads were filtered from the raw reads by deleting reads containing adaptors and low-quality reads. The C. sinensis reference genome was downloaded from https://pcsb.ahau.edu.cn:8080/CSS/ (Wei et al., 2018). Transcripts were aligned to the reference tea plant genome using Hisat2 v2.0.5 (Kim et al., 2015). Gene expression levels were normalized by calculating fragments per kilobase of transcript per million mapped reads (FPKM).
Gene Cloning and Expression Analysis
The upregulated MYB gene CsMYB59 was selected and identified from the TYDY transcriptome (unpublished) and C. sinensis genome (Wei et al., 2018), and aligned to the reference C. sinensis genome. The full-length sequence of CsMYB59 was amplified with specific primers which were designed according to the reference genome (primers are shown in Supplementary Table 1). CsMYB59 was cloned using 2X PRO Taq Master Mix kit (Accurate Biology, Changsha, China) under the following parameters: pre-denaturation at 94°C for 30 s, subsequent denaturation at 98°C for 10 s, annealing at 58°C for 10 s, and elongation at 72°C for 1 min. Amplification was performed in 35 cycles. Finally, the PCR cloning was completed at 72°C for 5 min. The full-length sequence of CsMYB59 (750 bp) was obtained after cloning and sequencing (Supplementary Table 2). Gene expression analysis was carried out by qRT-PCR with a SYBR Green Premix Pro Taq HS qPCR Kit (Accurate, Hunan, China) on an ABI QuantStudio 3 Real-Time PCR system (Thermo Fisher, United States). The reaction parameters of qRT-PCR were described as our previous published study (Luo et al., 2018).
Gene Identification and Characterization Analysis
Amino acid sequences were translated using the National Centre for Biotechnology Information (NCBI) ORF finder1. The physical and chemical characteristics of the protein were predicted using ProParam tool2. Homology analyses of protein sequences were performed using NCBI basic local alignment search tool (BLAST) database3. The multiple alignment of amino acid sequences was performed using the MUSCLE program in MEGA X (Kumar et al., 2018), and then phylogenetic trees were generated via the Neighbor--Joining method with a bootstrap number 1000. The NCBI Conserved Domain Database (CDD4 was used to identify the domains of the protein sequences that were edited using Genedoc software (Nicholas et al., 1997).
Promoter Isolation and Analysis
DNA was extracted from tea leaves using the cetyl trimethylammonium bromide method. The promoter sequence of the CsPPO1 gene (TEA026892.1) was obtained from the reference tea genome5. Full length CsPPO1 promoter amplification primers (Supplementary Table 1) were designed using Premier 5.0. The CsPPO1 promoter was isolated via PCR. After sequencing, the promoter sequence of CsPPO1 with a length of 1016 bp was obtained. The Plant Care online software6 was used to predict the cis-acting elements of CsPPO1 promoter.
Subcellular Localization of CsMYB59 in Tobacco (Nicotiana benthamiana) Plants
Subcellular localization was predicted using the Plant-Ploc7 online software. To confirm the subcellular localization prediction, the CsMYB59 coding sequence without the stop codon was cloned into a pEAQ-green fluorescence protein (GFP) vector (Sainsbury et al., 2009) to generate the fusion protein CsMYB59-GFP. Subsequently, both the pEAQ-CsMYB59-GFP and pEAQ-GFP vectors (Sainsbury et al., 2009) were transformed into Agrobacterium tumefaciens strain EHA105 (Weidi Biotechnology Co., Ltd, Shanghai, China), and the constructs were injected into tobacco leaves (Yu et al., 2018). After incubation for 2 days, the green fluorescence signals were analyzed using an LSM 710 laser-scanning confocal microscope (Carl Zeiss, Oberkochen, Germany) (Wen et al., 2020).
Transcriptional Activation in Yeast and Tobacco
Transcriptional activity assays were performed using a yeast system. The coding sequence of CsMYB59 was cloned into a pGBKT7 vector (Clontech, Mountain View, CA, United States) to generate a BD-CsMYB59 expression vector. The BD-CsMYB59 vector and a control BD empty vector were transformed into Saccharomyces cerevisiae Y2H Gold (Weidi Biotechnology Co., Ltd, Shanghai, China). The yeast cells were cultivated in synthetic defined (SD) medium (Sangon Biotech Co., Ltd., Shanghai, China) lacking tryptophan (SD/–Trp) at 28°C for 48–96 h. The transformed yeast cells were then cultured in SD medium without tryptophan, histidine, and adenine (SD/-Trp/-His-Ade) at 28°C for 48–72 h. Transcriptional activity of CsMYB59 was determined using X-α-galactosidase (Sangon Biotech Co., Ltd., Shanghai, China). In addition, the full-length sequence of the CsMYB59 TF was cloned into a pBD vector to generate pBD-CsMYB59 as an effector. The double-reporter vector was driven by 35S promoter, and contained GAL4-LUC and an internal control Renilla luciferase (REN). The GAL4-LUC contains 5X GAL4, minimal TATA region, and the Luciferase (LUC) (Vo et al., 2017). Mixtures of the effector and reporter were injected into tobacco leaves. After incubation for 2 days, The LUC/REN ratio in tobacco leaves was determined using a Dual Luciferase Reporter Assay Kit (Vazyme Biotech, Nanjing, China) as described by Wen et al. (2020) to confirm the transcriptional activity of CsMYB59 (Luo et al., 2018).
Dual-Luciferase Reporter Assay
To investigate the transcriptional effect of the CsMYB59 TF on the promoter of CsPPO1, the CsMYB59-pEAQ vector was used as an effector, whereas a pGreen II 0800-LUC vector containing the promoter fragments of CsPPO1 was used as a reporter. The mixtures of effector and reporter were injected into tobacco leaves. After incubation for 2 days, The LUC/REN ratio in tobacco leaves was determined to evaluate the transcriptional effect of CsMYB59 on the CsPPO1 promoter gene as described by Luo et al. (2018).
Statistical Analysis
All the experiments were performed in triplicate and all data are presented as the mean ± standard deviation. Statistical analysis was performed using Student’s t-tests. A value of P < 0.05 or P < 0.01 was considered significant.
Results
Analysis of Polyphenol Oxidase Enzyme Activity in Leaves of Different Tea Plant Cultivars
To investigate the difference in PPO activity between TYDY and BXZ, spectrophotometric analysis was performed to determine the PPO activity. The PPO activity in TYDY was significantly higher than that in BXZ (Figure 1A; P < 0.01).
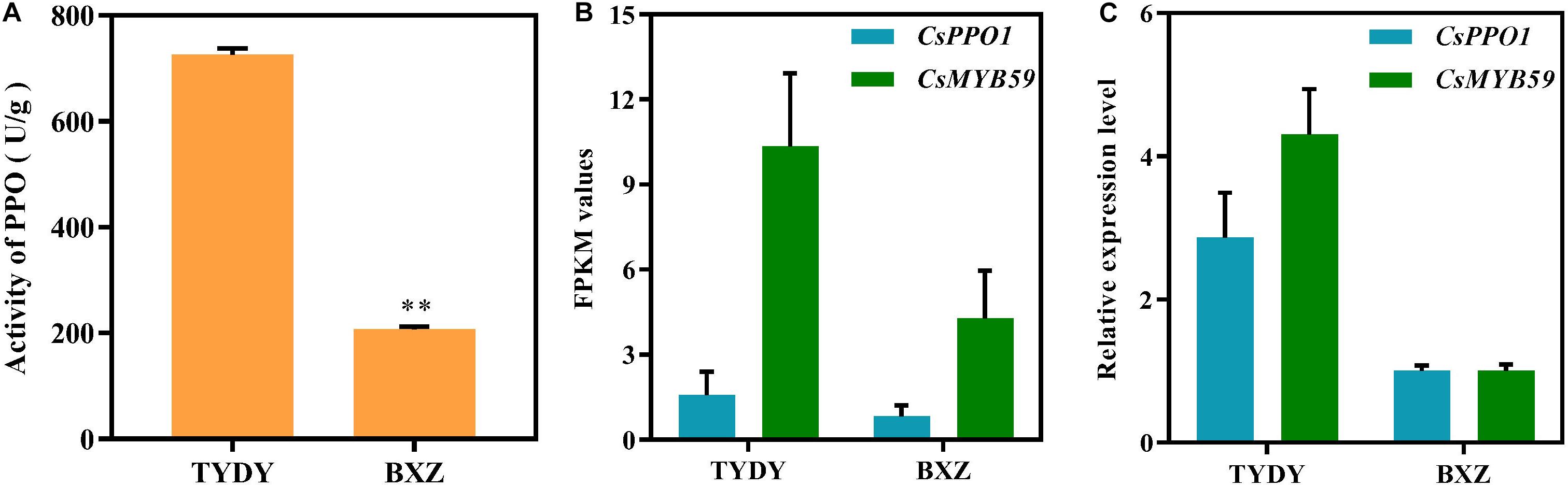
Figure 1. Polyphenol oxidase activity and gene expression of CsPPO1 and CsMYB59 in different tea cultivar leaves. (A) PPO activity in TYDY and BXZ. (B) The FPKM values of CsPPO1 and CsMYB59 in TYDY and BXZ. (C) The relative expression level of CsPPO1 and CsMYB59 in TYDY and BXZ. PPO, polyphenol oxidase; TYDY, Taoyuandaye; BXZ, Bixiangzao; FPKM, fragments per kilobase of transcript per million mapped reads. **represents significant difference at P < 0.01.
Sequencing and Transcriptome Mapping
Six cDNA libraries of TYDY and BXZ (three biological replicates for each cultivar) were generated for transcriptome sequencing. Based on the reference C. sinensis transcriptome assembly (Wei et al., 2018), a total of 339 million reads were identified after raw-read quality filtering, with an average of 56.6 million clean reads in each library. The average Q30 and GC content were 91.96 and 44%, respectively. Reference-based mapping of quality reads yielded 89.18% (TYDY) and 89.01% (BXZ). In addition, the uniquely mapped reads were 83.79–84.43% (Supplementary Table 3).
Analysis of the Expression Pattern of CsMYB59 and CsPPO1
The PPO activity in tea leaves is regulated by the expression of CsPPO genes. To investigate the regulatory mechanism of CsMYB59 TF in the effect on PPO activity in tea leaves, we measured the FPKM values of CsMYB59 and CsPPO1 genes via RNA sequencing (RNA-seq). By analyzing the differences in PPO activity between the two tea cultivars, we found that the expression level of CsPPO1 was positively correlated with PPO activity (Figures 1A,B). In addition, the FPKM value in TYDY was higher than that in BXZ which was consistent with the trend of CsPPO1 expression.
The gene expression levels of CsPPO1 and CsMYB59 were analyzed via qRT-PCR. The expression level trends of CsPPO1 and CsMYB59 genes were consistent with the transcriptomic data (Figures 1B,C). These results indicated that the CsMYB59 TF may have a positive correlation with CsPPO1 expression and PPO activity.
Identification and Bioinformatics Analysis of CsMYB59
The CsMYB59 sequence cloned from TYDY was analyzed using the ExPASy software for physicochemical properties (Supplementary Table 4). The open reading frame of CsMYB59 was 750 bp and encoded a protein containing 249 amino acids with a predicted molecular mass of 28.93 kDa. The theoretical isoelectric point was 6.55, the instability index was 76.57, and the average hydrophilic coefficient was −0.859. These data indicated that CsMYB59 may be an unstable hydrophilic protein. Analysis using the NCBI CDD database revealed that CsMYB59 was similar to other homologous MYB TFs and contained a conserved R2R3-MYB DNA-binding domain at the N-terminus (Figure 2A), which may perform analogous functions.
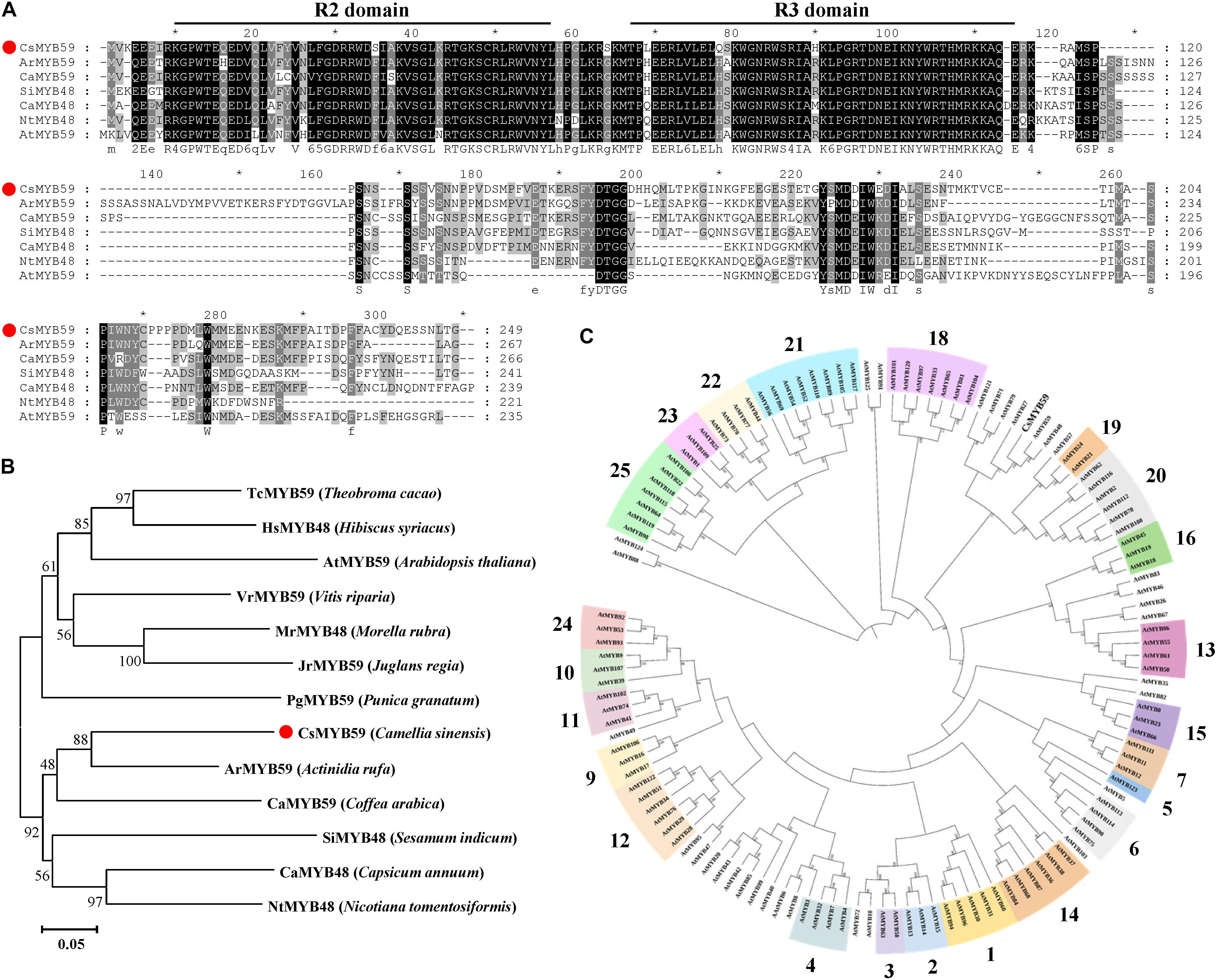
Figure 2. Bioinformatics analysis of the CsMYB59 protein sequence. (A) Protein sequence alignment between CsMYB59 and ArMYB59, CaMYB59, SiMYB48, CaMYB48, NtMYB48, and AtMYB59. (B) Phylogenetic tree generated using sequences of CsMYB59 and homologous MYB transcription factors of 12 other plant species; CsMYB59 is labeled with a red dot. The plants are specified in the brackets. The accession numbers of the protein sequences are as follows: TcMYB59 (EOY34142.1), HsMYB48 (XP_038991988.1), AtMYB59 (Q4JL84.2), VrMYB59 (XP_034687869.1), MrMYB48 (KAB1226039.1), JrMYB59 (XP_035545886.1), PgMYB59 (XP_031387608.1), ArMYB59 (GFS42876.1), CaMYB59 (XP_027108891.1), SiMYB48 (XP_011089426.1), CaMYB48 (KAF3662000.1), and NtMYB48 (XP_009590049.1). (C) Phylogenetic analysis of CsMYB59 and Arabidopsis-derived R2R3-MYB sequence. The bold numerals represent subgroups of R2R3-MYB TFs in A. thaliana. *means positions that have a single, fully conserved residue. CsMYB59 is marked with a red dot.
To further evaluate the molecular properties of CsMYB59, phylogenetic trees were generated. Homology analysis was performed using the amino acid sequences of CsMYB59 and MYB proteins from 12 other plant species (Figure 2B). Moreover, a neighbor-joining analysis was performed using the CsMYB59 and A. thaliana R2R3-MYB amino acid sequences (Figure 2C). Results showed that CsMYB59 was homologous to Actinidia rufa-derived ArMYB59 (64.23%). MYB59 has been reported to play a central role in biotic and abiotic stress responses in plants (Hickman et al., 2017; Fasani et al., 2019). AtMYB48 and AtMYB59 are homologous genes involved in the response to nutrient deprivation in Arabidopsis (Nishida et al., 2017). In addition, AtMYB59 participates in modulating calcium homeostasis and signal transduction induced by excessive cadmium levels (Fasani et al., 2021). These findings indicate that CsMYB59 may be involved in the response to stress and secondary metabolite biosynthesis in tea plants.
Analysis of Subcellular Localization of CsMYB59
Subcellular localization analysis predicted that CsMYB59 was localized in the nucleus and may be a nuclear protein. Subcellular localization assays using tobacco plants verified this result (Figure 3). The control GFP fluorescence signal was observed in tobacco leaf epidermal cells, whereas the CsMYB59-GFP signal was detected only in the nuclear region. This result indicated that CsMYB59 may be a nuclear protein showing transcriptional activity.
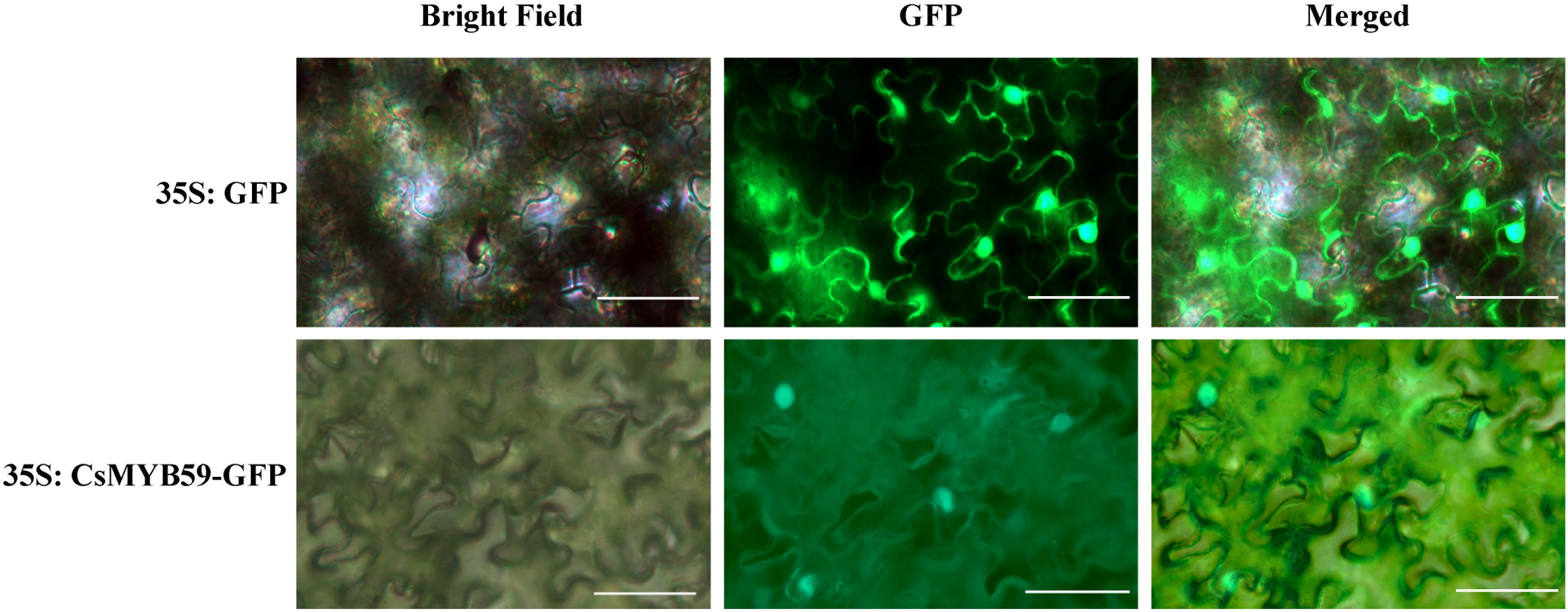
Figure 3. Subcellular localization analysis of CsMYB59 protein. The GFP- and CsMYB59-GFP-fused proteins were transfected into the epidermal cells in Nicotiana benthamiana leaves. The white bars represent 50 μm. GFP, green fluorescent protein.
Transcriptional Activity Analysis of CsMYB59
To investigate the transcriptional activity of CsMYB59, we used a GAL4-responsive reporter system in yeast. The transformed yeast carrying CsMYB59-BD showed growth when cultivated in SD/-Trp/-His/-Ade selective medium and showed α-galactosidase activity which indicated that CsMYB59 activates transcription as an activator (Figure 4A). In addition, the transcriptional activity of CsMYB59 was evaluated via dual-luciferase assays in N. benthamiana leaves. Compared to the pBD-empty vector, the ratio of LUC/REN in the pBD-CsMYB59 was significantly higher than that in the control (Figures 4B,C). Therefore, the role of CsMYB59 as a transcriptional activator was verified.
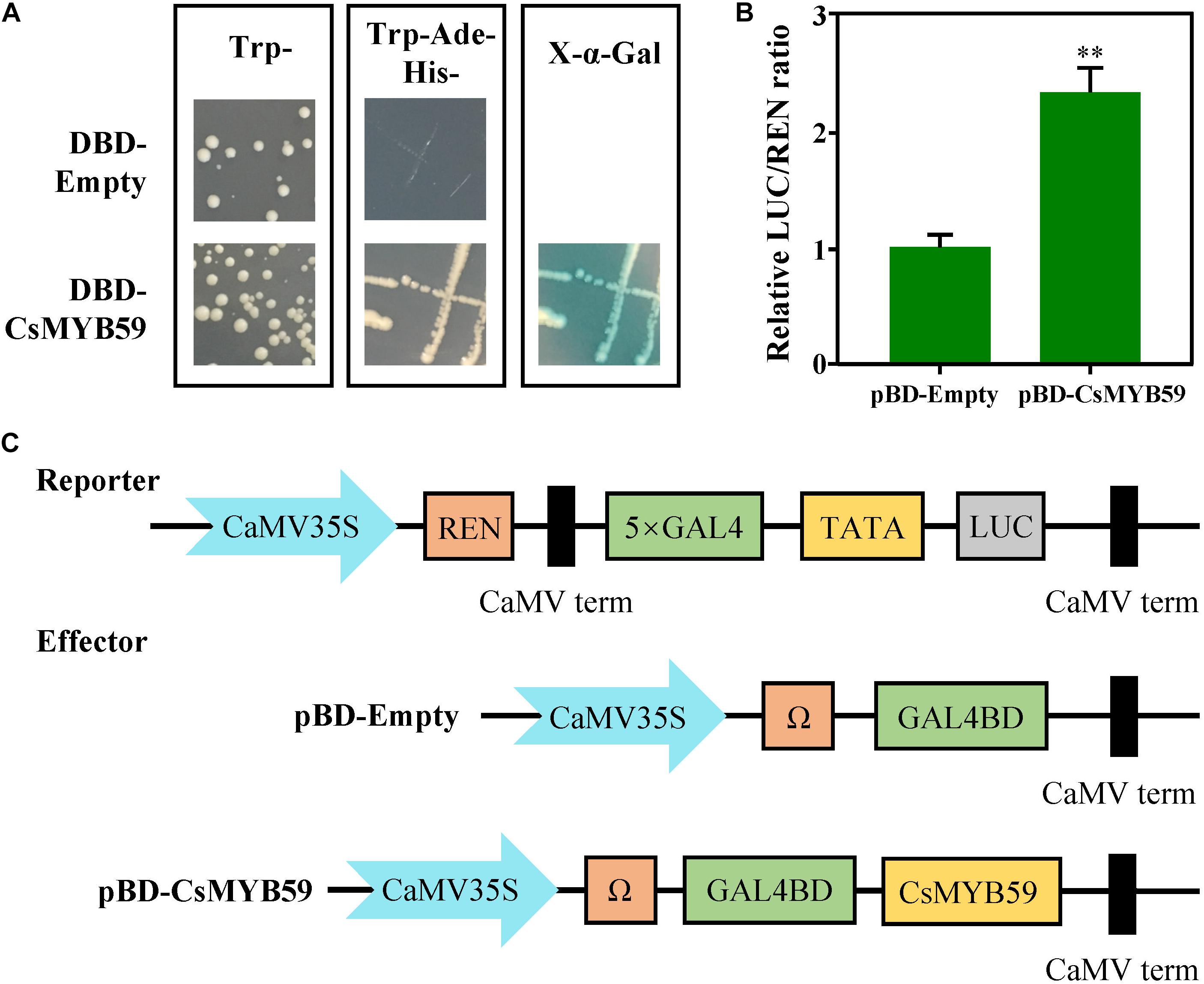
Figure 4. Transcriptional activity analysis of CsMYB59. (A) Transcriptional activation of CsMYB59 in Saccharomyces cerevisiae. CsMYB59 was subcloned into a pGBKT7 vector and then transformed into yeast cells. The transformed yeast cells were grown on selective SD mediums (SD/-Trp/-His/-Ade), and further selected via X-Gal assays. (B,C) The relative REN/LUC ratio of CsMYB59 and pBD-empty was calculated to evaluate the transcriptional activity of CsMYB59. Transcriptional activation of CsMYB59 in tobacco. The transcriptional activation of CsMYB59 was verified via dual-luciferase reporter assays using Nicotiana benthamiana leaves. ∗∗ represents significant difference at P < 0.01. SD, synthetic defined; -Trp/-His/-Ade, media lacking tryptophan, histidine, and adenine; LUC, luciferase; REN, Renilla luciferase; X-Gal, X-α-galactosidase.
CsMYB59 TF Regulated the Expression of CsPPO1 Promoter
Analysis of the cis-acting elements (Figure 5A) showed that the promoter region of CsPPO1 contained several specific MYB-binding cis-elements such as MYB (5′-CAACCA-3′), MBSI (5′-TTTTTACGGTTA-3′), and MER (5′-AACCTAA-3′) (Smita et al., 2015). To further investigate the transcriptional regulatory effects of CsMYB59 on CsPPO1, dual-luciferase assays were performed using N. benthamiana leaves. The constructed CsMYB59-pEAQ vector was used as the effector, and CsPPO1 pro-LUC was used as the reporter. Compared to the control which was transformed with an empty vector, the co-transformation of CsMYB59 and CsPPO1 pro-LUC significantly increased the ratio of LUC/REN (Figure 5B). This result suggested that CsMYB59 may activate the expression of CsPPO1, and may be positively correlated with PPO activity.
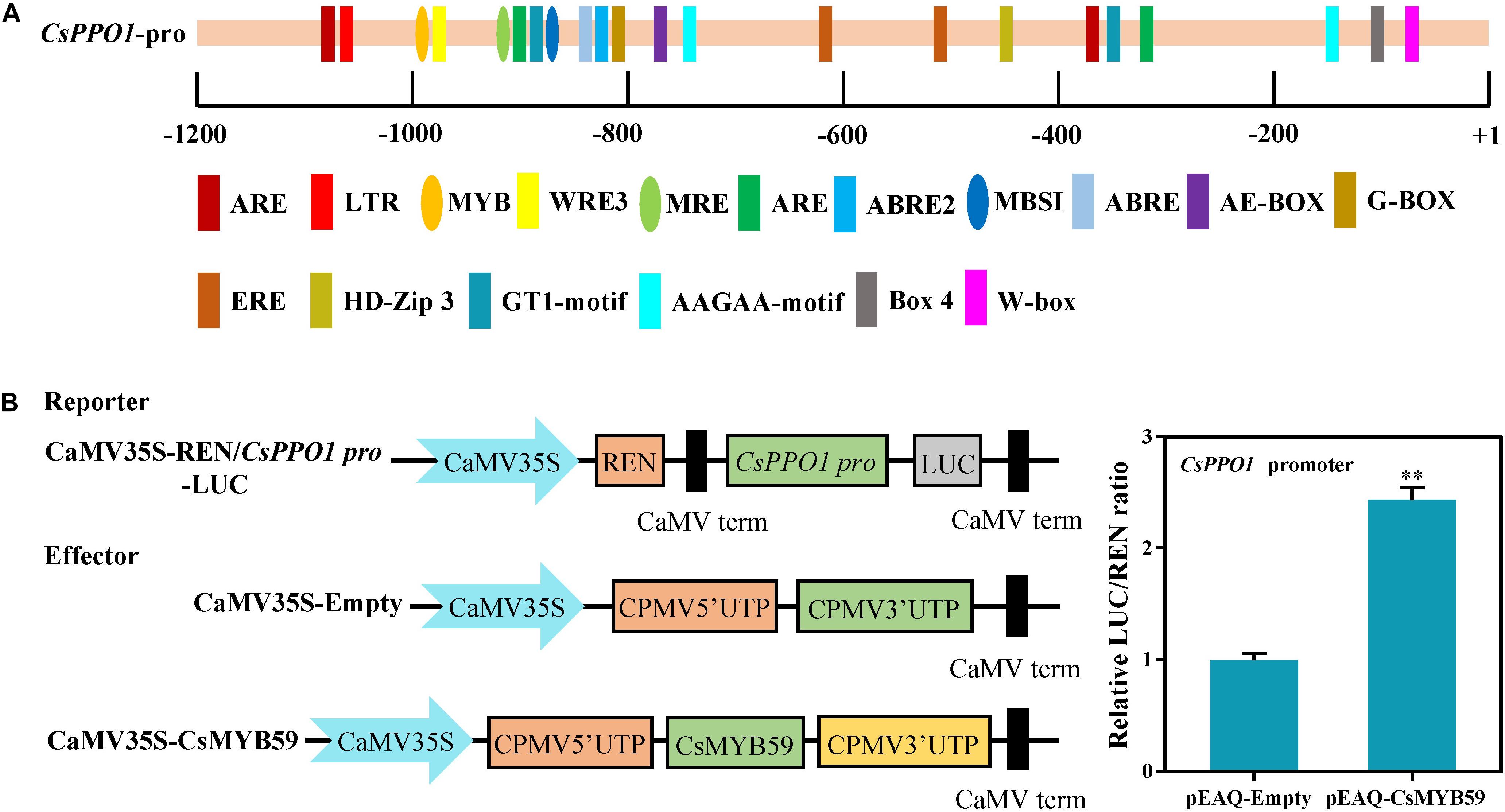
Figure 5. (A) Cis element analysis of CsPPO1 promoter gene. The axis represents the cloned CsPPO1 promoter sequence. Ellipses represent elements that can bind to MYB transcription factors. Rectangles represent other elements. (B) Ability of CsMYB59 to activate transcription of CsPPO1 promoter in Nicotiana benthamiana. The relative REN/LUC ratio of pEAQ-CsMYB59 and pEAQ-empty was calculated to evaluate the transcriptional regulatory effects of CsMYB59 on CsPPO1 promoter. ∗∗ represents significant difference at P < 0.01. LUC, luciferase; REN, Renilla luciferase.
Discussion
Polyphenol oxidase exhibits multiple important physiological effects such as resistance to stress, pests and diseases during growth and development (Huang et al., 2018). During the manufacturing processes of black tea, the flavonoids present in the leaves are oxidized to o-quinones by PPO that are associated with the formation of pigments such as thearubigins, theaflavins, and theabrownin. The oxidized catechins can promote the accumulation of aroma compounds in tea by oxidizing amino acids, carotenoids, fatty acids, and other substances (Shi et al., 2014). Therefore, PPO activity is of great importance with regard to the quality improvement of black tea. The PPO gene family members are typically encoded by small gene families. Seven members of the PPO family have been identified in tomato plants (Kowalski et al., 1992), six members have been identified in the family of potato plants (Thipyapong and Steffens, 1997), and five to six members have been identified in C. sinensis (Zhao et al., 2001). PPOs of different gene family members contain highly conserved copper-binding domains; however, the gene expression of PPOs is specific and functionally diverse. The PPO1 gene in barley is a major determinant in regulation of the phenol reaction in awns (Taketa et al., 2010). Cis-acting elements that interact with MYB TFs have been detected in the SmePPO1 gene in eggplants, which regulates various metabolic reactions (Shetty et al., 2012). Domain analysis of CsPPO1 and CsPPO2 in C. sinensis showed that CsPPO1 and CsPPO2 both contain Tyrosinase, PPO1_DWL, and PPO1_KFDV domains. Among them, CsPPO1 contain two tyrosinase domains, Tyrosinase_1 and Tyrosinase_2, whereas CsPPO2 contains only Tyrosinase_2 (Supplementary Table 5; Cary et al., 1992). CsPPO1 may play various roles due to the presence of two Tyrosinase domains. In this study, the PPO activity of two different tea cultivars was analyzed, and we found that the PPO activity in TYDY was significantly higher than that in BXZ. Furthermore, the CsPPO1 gene expression was significantly higher in TYDY than in BXZ based on the RNA-seq data, and this result was consistent with the trend of PPO activity; qRT-PCR analysis verified this result. Therefore, increasing the expression of CsPPO1 may positively regulate PPO activity in tea plants.
TF CsMYB59, whose expression follows the same pattern of variation in PPO activity among different tea cultivars, was isolated based on the transcriptomic data. Bioinformatics analysis indicated that CsMYB59 belonged to the R2R3-MYB TF family (Figure 2A). In C. sinensis, 122 R2R3-CsMYB genes have been identified and classified into 25 subgroups; CsMYB59 belongs to the S24 subgroup (Chen et al., 2021). Phylogenetic analysis showed that CsMYB59 was highly homologous to ArMYB59 (64.23%, A. rufa) compared to MYB TF sequences from other plants, suggesting that these proteins may have similar functions. It is well known that the R2R3-MYB TF families are involved in plant growth and development (Stracke et al., 2007), stress response (Wong et al., 2016), and primary and secondary metabolism (Blanco et al., 2018). Nishida et al. (2017) found that two homologous genes, AtMYB48 and AtMYB59, show altered DNA-binding sites created by alternative splicing in response to environmental stresses of potassium deficiency. AtMYB59 is involved in the response to Heterodera schachtii infestation in A. thaliana (Wisniewska et al., 2021). In addition, an R2R3 TF ZmMYB31 isolated from Zea mays can enhance superoxide dismutase and ascorbate peroxidase activities to protect plants against low temperature stress (Li et al., 2019). These findings suggest that R2R3-MYB TFs play an important role in plant metabolite synthesis and resistance to stress. Due to the fact that one of the main functions of PPO is involved in resistance to stress in tea plant, the CsMYB59 may has a regulatory role on PPO activity.
Cis-acting element analysis revealed that CsPPO1 contained multiple MYB-binding sites such as G-BOX and MRE which are involved in the light response (Hartmann et al., 1998, 2005), MBSI which is involved in the regulation of flavonoid biosynthetic genes (Zhang et al., 2016), and ERE and MYB of unknown functions. These binding sites may control downstream gene expression by binding MYB TFs. Recently, an R1R2R3 TF MnMYB3R1 is isolated from Morus notabilis which upregulates PPO by binding to MSA in the promoter region of MnPPO1 (Liu et al., 2019). Therefore, it is important to increase the PPO activity by regulating TFs so as to cultivate high stress resistant tea cultivars and improve the quality of black tea. However, there are no studies on tea plant-derived TFs regulating PPO gene expression. Our results suggest that an R2R3-MYB TF CsMYB59 expression may be positively correlated with the gene expression of CsPPO1. In addition, qRT-PCR verified the expression pattern of CsMYB59 in association with CsPPO1; the transcriptomic data were consistent with the qRT-PCR data. Based on these results, we speculate that CsMYB59 may regulate the CsPPO1 promoter.
In total, 98.36% of R2R3-CsMYB TFs are localized in the nucleus (Chen et al., 2021). Similarly, our study showed that CsMYB59 may represent a nuclear protein. MYB TFs have been reported to operate as transcriptional activators as well as transcriptional repressors. NtMYB3 inhibits flavonol biosynthesis in Narcissus by suppressing its FLS promoter activity (Anwar et al., 2019). CsMYB73 in C. sinensis binds to the promoter regions of CsGS1 and CsGS2 via MYB recognition sequences and represses the transcriptional activity of CsGS1 and CsGS2, thereby participating in the biosynthesis ofL-theanine in tea plants (Wen et al., 2020). AtMYB2 activates miR399f expression and regulates the Pi-starvation responses in Arabidopsis by operating as a transcriptional activator that binds directly to the MYB-binding site in the miR399f precursor promoter (Baek et al., 2013). In the present study, we found that CsMYB59 showed transcriptional activity in yeast cells, and the results were verified via transcriptional activity analysis in tobacco leaves which confirmed that CsMYB59 may function as a transcriptional activator. Furthermore, dual-luciferase assays indicated that CsMYB59 activated the transcription of the promoter of CsPPO1, and may regulate PPO activity in tea plants by activating CsPPO1.
Currently, several studies suggest that MYB interacts with transcription factors such as bHLH and WD40 to regulate the expression of promoters (Grotewold et al., 2000; Zimmermann et al., 2004). For example, Liang et al. (2014) showed that AtMYB82 can interact with GL3, and is integrated into the WD40-bHLH-MYB complex to participate in Arabidopsis trichome development. Tea plant-derived CsWD40 interacts with two bHLH TFs (CsGL3 and CsTT8) and two MYB TFs (CsAN2 and CsMYB5e) to form a ternary WBM complex that regulates anthocyanin and pro-anthocyanidin biosynthesis, and trichome development (Liu et al., 2018). In this study, we found that the CsPPO1 promoter contains a large number of elements that can bind to other transcription factors, such as W-Box. Therefore, further studies on the regulatory relationships between CsMYB59 and other CsPPOs, and the interactions of other transcription factors may help us further understand the regulatory mechanisms of PPO in tea plants, and will provide a theoretical basis for the screening and breeding of premium tea plant germplasm resources.
Conclusion
In this study, we evaluated the PPO activity in two different tea cultivars, TYDY and BXZ, and isolated and identified a TF, CsMYB59, that may exert regulatory effects on PPO activity in tea plants. CsMYB59 is a transcriptional activator that may specifically activate the expression of CsPPO1, thereby positively regulating PPO activity. These findings expand the knowledge of the transcriptional regulation mechanism of R2R3-MYB TFs involved in PPO metabolism in tea plants and provide potential theoretical supports for the molecular breeding and innovation of tea cultivar resources with high PPO activity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KW and YL designed the research. SO carried out experiments. XH processed the data and wrote the manuscript. MZ, HL, QL, and JL revised the manuscript. All authors approved the manuscript.
Funding
This work was supported by National Key Research and Development Program of China (2018YFC1604403), the National Natural Science Foundation of China (U19A2030 and 32072629), Project of Central Government for Local Science and Technology Development of Hunan Province (2019XF5041) and Hunan Natural Science Foundation of China (2020JJ5525).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.739951/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/orffinder/
- ^ https://web.expasy.org/protparam/
- ^ http://www.ncbi.nih.gov/BLAST/
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ https://pcsb.ahau.edu.cn:8080/CSS/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/plant/
References
Amoutzias, G. D., Veron, A. S., Weiner, J., Robinsonrechavi, M., Bornbergbauer, E., Oliver, S. G., et al. (2007). One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-Binding site specificity. Mol. Biol. Evol. 24, 827–835. doi: 10.1093/molbev/msl211
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
Anwar, M., Yu, W., Yao, H., Zhou, P., Allan, A. C., and Zeng, L. (2019). NtMYB3, an R2R3-MYB from narcissus, regulates flavonoid biosynthesis. Int. J. Mol. Sci. 20:5456.
Baek, D., Park, H. C., Kim, M. C., and Yun, D. J. (2013). The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal. Behav. 8:e23488. doi: 10.4161/psb.23488
Blanco, E., Sabetta, W., Danzi, D., Negro, D., Passeri, V., Lisi, A., et al. (2018). Isolation and characterization of the flavonol regulator CcMYB12 from the globe artichoke [Cynara cardunculus var. scolymus (L.) Fiori]. Front. Plant Sci. 9:941. doi: 10.3389/fpls.2018.00941
Cary, J. W., Lax, A. R., and Flurkey, W. H. (1992). Cloning and characterization of cDNAs coding for Vicia faba polyphenol oxidase. Plant Mol. Biol. 20, 245–253. doi: 10.1007/BF00014492
Chen, X., Wang, P., Gu, M., Lin, X., Hou, B., Zheng, Y., et al. (2021). R2R3-MYB transcription factor family in tea plant (Camellia sinensis): genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 113, 1565–1578. doi: 10.1016/j.ygeno.2021.03.033
Chen, Y., Zeng, L., Liao, Y., Li, J., Zhou, B., Yang, Z., et al. (2020). Enzymatic reaction-related protein degradation and proteinaceous amino ccid metabolism during the black tea (Camellia sinensis) manufacturing process. Foods 9:66. doi: 10.3390/foods9010066
Constabel, C. P., Yip, L., Patton, J. J., and Christopher, M. E. (2000). Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory. Plant Physiol. 124, 285–295. doi: 10.1104/pp.124.1.285
Cui, J., Jiang, N., Zhou, X., Hou, X., Yang, G., Meng, J., et al. (2018). Tomato MYB49 enhances resistance to Phytophthora infestans and tolerance to water deficit and salt stress. Planta 248, 1487–1503. doi: 10.1007/s00425-018-2987-6
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fasani, E., DalCorso, G., Costa, A., Zenoni, S., and Furini, A. (2019). The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol. Biol. 99, 517–534. doi: 10.1007/s11103-019-00833-x
Fasani, E., DalCorso, G., and Furini, A. (2021). MYB59 transcription factor behaves differently in metallicolous and non-metallicolous populations of Arabidopsis halleri. Funct. Plant Biol. doi: 10.1071/FP20356 (in press)
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Grotewold, E., Sainz, M. B., Tagliani, L., Hernandez, J. M., Bowen, B., and Chandler, V. L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. U. S. A. 97, 13579–13584. doi: 10.1073/pnas.250379897
Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., and Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171. doi: 10.1007/s11103-004-6910-0
Hartmann, U., Valentine, W. J., Christie, J. M., Hays, J., Jenkins, G. I., and Weisshaar, B. (1998). Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol. Biol. 36, 741–754. doi: 10.1023/a:1005921914384
Hickman, R., Van Verk, M. C., Van Dijken, A. J. H., Mendes, M. P., Vroegop-Vos, I. A., Caarls, L., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 29, 2086–2105. doi: 10.1105/tpc.16.00958
Hojnik Podrepšek, G., Knez, Ž, and Leitgeb, M. (2020). The influence of supercritical carbon dioxide on graham flour enzyme polyphenol oxidase activity. Molecules 25:5981. doi: 10.3390/molecules25245981
Huang, C., Zhang, J., Zhang, X., Yu, Y., Bian, W., Zeng, Z., et al. (2018). Two new polyphenol oxidase genes of tea plant (Camellia sinensis) respond differentially to the regurgitant of tea geometrid, Ectropis obliqua. Int. J. Mol. Sci. 19:2414. doi: 10.3390/ijms19082414
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kowalski, S. P., Eannetta, N. T., Hirzel, A. T., and Steffens, J. C. (1992). Purification and characterization of polyphenol oxidase from glandular trichomes of Solanum berthaultii. Plant Physiol. 100, 677–684. doi: 10.1104/pp.100.2.677
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, M., Lin, L., Zhang, Y., and Sui, N. (2019). ZmMYB31, a R2R3-MYB transcription factor in maize, positively regulates the expression of CBF genes and enhances resistance to chilling and oxidative stress. Mol. Biol. Rep. 46, 3937–3944. doi: 10.1007/s11033-019-04840-5
Liang, G., He, H., Li, Y., Ai, Q., and Yu, D. (2014). MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 65, 3215–3223. doi: 10.1093/jxb/eru179
Liu, D., Meng, S., Xiang, Z., Yang, G., and He, N. (2019). An R1R2R3 MYB transcription factor, MnMYB3R1, regulates the polyphenol oxidase gene in Mulberry (Morus notabilis). Int. J. Mol. Sci. 20:2602. doi: 10.3390/ijms20102602
Liu, Y., Hou, H., Jiang, X., Wang, P., Dai, X., Chen, W., et al. (2018). A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin cccumulation through the formation of MYB-bHLH-WD40 ternary complexes. Int. J. Mol. Sci. 19:1686. doi: 10.3390/ijms19061686
Luo, Y., Yu, S., Li, J., Li, Q., Wang, K., Huang, J., et al. (2018). Molecular characterization of WRKY transcription factors that act as negative regulators of O-Methylated catechin biosynthesis in tea plants (Camellia sinensis L.). J. Agric. Food Chem. 66, 11234–11243. doi: 10.1021/acs.jafc.8b02175
Mayer, A. M., and Harel, E. (1979). Polyphenol oxidase in plants. Phytochemistry 18, 193–215. doi: 10.1016/0031-9422(79)80057-6
Mishra, B., and Singh Sangwan, N. (2019). Amelioration of cadmium stress in Withania somnifera by ROS management: active participation of primary and secondary metabolism. Plant Growth Regul. 87, 403–412. doi: 10.1007/s10725-019-00480-8
Molitor, C., Mauracher, S. G., Pargan, S., Mayer, R. L., Halbwirth, H., and Rompel, A. (2015). Latent and active aurone synthase from petals of C. grandiflora: a polyphenol oxidase with unique characteristics. Planta 242, 519–537. doi: 10.1007/s00425-015-2261-0
Nicholas, K. B., Nicholas, H. B., and Deerfield, D. W. (1997). GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14.
Nishida, S., Kakei, Y., Shimada, Y., and Fujiwara, T. (2017). Genome-wide analysis of specific alterations in transcript structure and accumulation caused by nutrient deficiencies in Arabidopsis thaliana. 91, 741–753. doi: 10.1111/tpj.13606
Owuor, P. O., Wachira, F. N., and Ng’etich, W. K. (2010). Influence of region of production on relative clonal plain tea quality parameters in Kenya. Food Chem. 119, 1168–1174. doi: 10.1016/j.foodchem.2009.08.032
Riechmann, J., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sánchez, M., González-Burgos, E., Iglesias, I., Lozano, R., and Gómez-Serranillos, M. P. (2020). The pharmacological activity of Camellia sinensis (ıL.ı) ıKuntzeı on metabolic and endocrine disorders: a systematic review. Biomolecules 10:603. doi: 10.3390/biom10040603
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shetty, S. M., Chandrashekar, A., and Venkatesh, Y. P. (2012). Promoter analyses and transcriptional profiling of eggplant polyphenol oxidase 1 gene (SmePPO1) reveal differential response to exogenous methyl jasmonate and salicylic acid. 169, 718–730. doi: 10.1016/j.jplph.2012.01.007
Shi, J., Wang, L., Ma, C. Y., Lv, H. P., Chen, Z. M., and Lin, Z. (2014). Aroma changes of black tea prepared from methyl jasmonate treated tea plants. J. Zhejiang Univ. Sci. B 15, 313–321. doi: 10.1631/jzus.B1300238
Smita, S., Katiyar, A., Chinnusamy, V., Pandey, D. M., and Bansal, K. C. (2015). Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 6:1157. doi: 10.3389/fpls.2015.01157
Solano, F., Garcia, E., Perez, D., and Sanchez-Amat, A. (1997). Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium. Appl. Environ. Microbiol. 63, 3499–3506. doi: 10.1128/aem.63.9.3499-3506
Stodt, U. W., Blauth, N., Niemann, S., Stark, J., Pawar, V., Jayaraman, S., et al. (2014). Investigation of processes in black tea manufacture through model fermentation (oxidation) experiments. J. Agri. Food Chem. 62, 7854–7861. doi: 10.1021/jf501591j
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, B., Zhu, Z., Cao, P., Chen, H., Chen, C., Zhou, X., et al. (2016). Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 6:32534. doi: 10.1038/srep32534
Taketa, S., Matsuki, K., Amano, S., Saisho, D., Himi, E., Shitsukawa, N., et al. (2010). Duplicate polyphenol oxidase genes on barley chromosome 2H and their functional differentiation in the phenol reaction of spikes and grains. J. Exp. Bot. 61, 3983–3993. doi: 10.1093/jxb/erq211
Taranto, F., Pasqualone, A., Mangini, G., Tripodi, P., Miazzi, M. M., Pavan, S., et al. (2017). Polyphenol oxidases in crops: biochemical, physiological and genetic aspects. Int. J. Mol. Sci. 18:377. doi: 10.3390/ijms18020377
Teng, J., Gong, Z., Deng, Y., Chen, L., Li, Q., Shao, Y., et al. (2017). Purification, characterization and enzymatic synthesis of theaflavins of polyphenol oxidase isozymes from tea leaf (Camellia sinensis). LWT 84, 263–270. doi: 10.1016/j.lwt.2017.05.065
Thipyapong, P., and Steffens, J. C. (1997). Tomato polyphenol oxidase (differential response of the polyphenol oxidase F promoter to injuries and wound signals). Plant Physiol. 115, 409–418. doi: 10.1104/pp.115.2.409
Vo, K. T. X., Kim, C. Y., Hoang, T. V., Lee, S. K., Shirsekar, G., Seo, Y. S., et al. (2017). OsWRKY67 plays a positive role in basal and XA21-mediated resistance in rice. Front. Plant Sci. 8:2220. doi: 10.3389/fpls.2017.02220
Wang, K., Chen, Q., Lin, Y., Li, S., Lin, H., Huang, J., et al. (2014). Comparison of phenolic compounds and taste of Chinese black tea. Food Sci. Technol. Res. 20, 639–646. doi: 10.3136/fstr.20.639
Wang, W. L., Wang, Y. X., Li, H., Liu, Z. W., Cui, X., and Zhuang, J. (2018). Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) O. Kuntze]. BMC Plant Biol. 18:288. doi: 10.1186/s12870-018-1502-3
Wei, C., Hua, Y., Wang, S., Jian, Z., and Wan, X. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U. S. A. 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Wen, B., Luo, Y., Liu, D., Zhang, X., Peng, Z., Wang, K., et al. (2020). The R2R3-MYB transcription factor CsMYB73 negatively regulates l-Theanine biosynthesis in tea plants (Camellia sinensis L.). Plant Sci. 298:110546. doi: 10.1016/j.plantsci.2020.110546
Wisniewska, A., Wojszko, K., Rozanska, E., Lenarczyk, K., Kuczerski, K., and Sobczak, M. (2021). Arabidopsis thaliana myb59 gene is involved in the response to heterodera schachtii infestation, and its overexpression disturbs regular development of nematode-induced syncytia. Int. J. Mol. Sci. 22:6450. doi: 10.3390/ijms22126450
Wong, D. C. J., Schlechter, R., Vannozzi, A., Holl, J., Hmmam, I., Bogs, J., et al. (2016). A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 23, 451–466. doi: 10.1093/dnares/dsw028
Yu, B., Yan, S., Zhou, H., Dong, R., Lei, J., Chen, C., et al. (2018). Overexpression of CsCaM3 improves high temperature tolerance in cucumber. Front. Plant Sci. 9:797. doi: 10.3389/fpls.2018.00797
Yu, Z. H., Han, Y. N., and Xiao, X. G. (2015). A PPO promoter from betalain-producing red swiss chard, directs petiole- and root-preferential expression of foreign gene in anthocyanins-producing plants. Int. J. Mol. Sci. 16, 27032–27043. doi: 10.3390/ijms161126011
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2016). Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 6:30050. doi: 10.1038/srep30050
Zhang, J., Zhang, X., Ye, M., Li, X. W., Lin, S. B., and Sun, X. L. (2020). The jasmonic acid pathway positively regulates the polyphenol oxidase-based defense against tea geometrid caterpillars in the tea plant (Camellia sinensis). J. Chem. Ecol. 46, 308–316. doi: 10.1007/s10886-020-01158-6
Zhao, D., Liu, Z. S., and Biao, X. (2001). Cloning and alignment of polyphenol oxidase cDNA of tea plant. J. Tea Sci. 2, 94–98.
Zhu, M. Z., Li, N., Zhou, F., Ouyang, J., Lu, D. M., Xu, W., et al. (2020a). Microbial bioconversion of the chemical components in dark tea. Food Chem. 312:126043. doi: 10.1016/j.foodchem.2019.126043
Zhu, M. Z., Lu, D. M., Ouyang, J., Zhou, F., Huang, P. F., Gu, B. Z., et al. (2020b). Tea consumption and colorectal cancer risk: a meta-analysis of prospective cohort studies. Eur. J. Nutr. 59, 3603–3615. doi: 10.1007/s00394-020-02195-3
Keywords: Camellia sinensis, polyphenol oxidase activity, transcription factor, CsMYB59, CsPPO1 expression
Citation: Huang X-x, Ou S-q, Li Q, Luo Y, Lin H-y, Li-J, Zhu M-z and Wang K-b (2021) The R2R3 Transcription Factor CsMYB59 Regulates Polyphenol Oxidase Gene CsPPO1 in Tea Plants (Camellia sinensis). Front. Plant Sci. 12:739951. doi: 10.3389/fpls.2021.739951
Received: 12 July 2021; Accepted: 14 October 2021;
Published: 04 November 2021.
Edited by:
Wenyan Han, Tea Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Xinghui Li, Nanjing Agricultural University, ChinaAshutosh Pandey, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2021 Huang, Ou, Li, Luo, Lin, Li, Zhu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Li, eGl4aV9sakAxMjYuY29t; Mingzhi Zhu, bXp6aHVjbkBob3RtYWlsLmNvbQ==; Kunbo Wang, d2tib28xNjNAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xiangxiang Huang
Xiangxiang Huang Shuqiong Ou1,2†
Shuqiong Ou1,2† Mingzhi Zhu
Mingzhi Zhu Kunbo Wang
Kunbo Wang