
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 September 2021
Sec. Crop and Product Physiology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.738261
This article is part of the Research TopicMolecular Interactions between Crops and Phytopathogens, Volume I: Wheat and MaizeView all 11 articles
 Chunxia Ge1,2†
Chunxia Ge1,2† Yi-Ge Wang1†
Yi-Ge Wang1† Shouping Lu3
Shouping Lu3 Xiang Yu Zhao4
Xiang Yu Zhao4 Bing-Kai Hou1
Bing-Kai Hou1 Peter J. Balint-Kurti5,6
Peter J. Balint-Kurti5,6 Guan-Feng Wang1*
Guan-Feng Wang1*Maize is one of the major crops in the world; however, diseases caused by various pathogens seriously affect its yield and quality. The maize Rp1-D21 mutant (mt) caused by the intragenic recombination between two nucleotide-binding, leucine-rich repeat (NLR) proteins, exhibits autoactive hypersensitive response (HR). In this study, we integrated transcriptomic and metabolomic analyses to identify differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) in Rp1-D21 mt compared to the wild type (WT). Genes involved in pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) were enriched among the DEGs. The salicylic acid (SA) pathway and the phenylpropanoid biosynthesis pathway were induced at both the transcriptional and metabolic levels. The DAMs identified included lipids, flavones, and phenolic acids, including 2,5-DHBA O-hexoside, the production of which is catalyzed by uridinediphosphate (UDP)-dependent glycosyltransferase (UGT). Four maize UGTs (ZmUGTs) homologous genes were among the DEGs. Functional analysis by transient co-expression in Nicotiana benthamiana showed that ZmUGT9250 and ZmUGT5174, but not ZmUGT9256 and ZmUGT8707, partially suppressed the HR triggered by Rp1-D21 or its N-terminal coiled-coil signaling domain (CCD21). None of the four ZmUGTs interacted physically with CCD21 in yeast two-hybrid or co-immunoprecipitation assays. We discuss the possibility that ZmUGTs might be involved in defense response by regulating SA homeostasis.
To defend against pathogenic microorganisms, plants have evolved a multilayered and sophisticated immune system including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI; McHale et al., 2006; Cui et al., 2015). The PTI is activated by the recognition of PAMPs via the pattern recognition receptors (PRRs) localized at the surface of plant cells, while ETI is triggered when the intracellular receptors termed nucleotide-binding leucine-rich-repeat (NLR) proteins recognize the specific effector proteins secreted from pathogens. Two recent studies (Ngou et al., 2021; Yuan et al., 2021) suggest that PTI and ETI responses are closely connected with ETI potentiating the PTI response. PRR- and NLR-mediated downstream signaling pathways result in some similar immune outputs, such as increased expression of pathogenesis-related (PR) genes and the burst of reactive oxygen species (ROS). A nicotinamide adenine dinucleotide phosphate (NADPH) oxidase called respiratory burst oxidase homolog D (RBOHD) is an indispensable immune component connecting PRR and NLR immune receptors, and the phosphorylation of RBOHD mediates ROS generation and activates disease resistance (Ngou et al., 2021). A distinct feature of NLR-triggered immunity is often accompanied by the hypersensitive response (HR), a form of localized programmed cell death at the pathogen infection sites (Bent and Mackey, 2007; Kourelis and van der Hoorn, 2018; Balint-Kurti, 2019).
Salicylic acid (SA) is a pivotal phytohormone mediating both local and systemic defense responses against biotrophic and semi-biotrophic pathogens (Vlot et al., 2009; Dempsey et al., 2011). The activation of SA biosynthesis, metabolism, and signaling pathways play critical roles for both PTI- and ETI-mediated defense responses (Vlot et al., 2009; Ding et al., 2018). The SA is synthesized by the isochorismate synthase (ICS) and the Phe ammonia-lyase (PAL) pathways which play the major and minor roles in SA biosynthesis, respectively (Dempsey et al., 2011). In Arabidopsis, when the gene EDS16 in the ICS pathway was mutated, the total SA level was reduced to <10% of the wild-type level after Erysiphe orontii infection (Dewdney et al., 2000). Once synthesized, the SA is subject to a number of modifications including glycosylation, hydroxylation, and methylation. Hydroxylated products of SA, including 2,3-dihydroxybenzoic acid (2,3-DHBA) and 2,5-dihydroxybenzoic acid (2,5-DHBA), are the major metabolic forms of SA. The DHBA is cytotoxic and it is generally found in its less toxic glycosylated form in plants (Bartsch et al., 2010). Glycosylation products of DHBA may activate plant defense. DHBA glycoside compounds are increased in Arabidopsis after infection by Pseudomonas syringae pv. tomato (Pst) strain DC3000 or Hyaloperonospora arabidopsidis (Bartsch et al., 2010). When overexpressed, the Arabidopsis uridine diphosphate (UDP)-dependent glycosyltransferase UGT76D1, which catalyzes the formation of DHBA glycosides, leads to the induction of HR, a burst of ROS, the increased expression of PR genes and enhanced resistance to Pst DC3000 (Huang et al., 2018). The UGT76D1 plays important roles in plant immunity by modulating SA homeostasis by glycosylations of DHBA (Huang et al., 2018).
Nucleotide-binding leucine-rich-repeat disease resistance proteins can be divided into two major types depending on their N-terminal domains; the coiled-coil type (CNL) and the Toll/interleukin-1 receptor type (TNL; Monteiro and Nishimura, 2018; Sun et al., 2020). The maize Rp1 locus on the short arm of chromosome 10 carries multiple tandemly repeated CNL genes (Hulbert, 1997). One of these genes, Rp1-D, confers resistance to maize common rust caused by the fungus, Puccini sorghi (Hulbert, 1997). The chimeric gene Rp1-D21 was derived from intragenic recombination between two paralogs, Rp1-D and Rp1-dp2 (Sudupak et al., 1993; Sun et al., 2001; Smith et al., 2010). The Rp1-D21 confers a spontaneous HR phenotype in the absence of pathogen infection (Sun et al., 2001; Smith et al., 2010; Wang et al., 2015a). The severity of this HR is affected by light, temperature, developmental stage, and genetic background (Chintamanani et al., 2010; Negeri et al., 2013). The Rp1-D21-induced HR is entirely suppressed at 30°C and can be activated by reducing the temperature to 22°C (Negeri et al., 2013). The Rp1-D21 had been used as a tool to identify quantitative trait loci (QTL), genes, and pathways associated with modulation of the severity of HR in maize (Chintamanani et al., 2010; Olukolu et al., 2014). Two key enzymes in lignin biosynthesis pathway, caffeoyl-CoA O-methyltransferase (CCoAOMT) and hydroxycinnamoyl transferase (HCT), have been shown to suppress Rp1-D21-induced HR through physical interaction (Wang et al., 2015b; Wang and Balint-Kurti, 2016). The gene encoding CCoAOMT has also been shown to increase resistance to both southern leaf blight and gray leaf spot in maize (Yang et al., 2017).
In recent years, transcriptome and metabolome analyses have provided a powerful comprehensive approach to assess the relationship of genotype, phenotype, and metabolite changes in plants challenged by abiotic and biotic stresses (Etalo et al., 2013; Mo et al., 2019; Ye et al., 2019; Hong et al., 2020). For example, transcriptional profiling combined with targeted metabolite quantification found that the levels of many genes and several metabolites in phenylpropanoid and shikimate pathways are significantly changed by the expression of WtsE, an effector secreted from Pantoea stewartii ssp. Stewartii (Pnss), which can cause Stewart's wilt and leaf blight in maize (Asselin et al., 2015).
Here, using a similar multi-omics approach, we investigated the gene regulatory network modulating Rp1-D21-mediated HR. We identified a number of different pathways associated with resistance response mediated by Rp1-D21, in particular, the SA biosynthesis and metabolism pathway. Four DHBA glucosyl-transferase homologs of ZmUGTs, were highly induced, two of which we show may have important roles in modulating Rp1-D21-mediated HR.
Transcriptional analysis of Rp1-D21-induced hypersensitive response (HR) was undertaken in two different temperature regimes (Supplementary Figure 1). In the first treatment, here called the temperature shift treatment, plants were grown at 30°C, and then were transferred immediately to 22°C to induce a synchronous systematic HR. For the second temperature regime, the constant temperature treatment, the plants were grown at a constant 22°C. Comparisons of two different pairs of near isogenic F1 hybrids were used for these experiments: B73 × H95-Rp1-D21 with B73 × H 95 and Mo17 × H95-Rp1-D21 with Mo17 × H95.
The Rp1-D21 HR phenotype in Mo17 × H95-Rp1-D21 is more severe than in B73 × H95-Rp1-D21 (Chintamanani et al., 2010). In the temperature shift experiment, Rp1-D21-carrying plants displayed HR at 3 days after temperature shift in the Mo17 × H95-Rp1-D21, and at 5 days in the B73 × H95-Rp1-D21 background. For this experiment, samples from near-isogenic wild type (WT) and Rp1-D21 mutant (mt) plants in each genetic background were collected at 3, 6, 24, and 48 h post the temperature shift (hpts). The HR was not observed in either background at these time points. Therefore, in order to confirm that the defense response had indeed been activated by the temperature shift, we used semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) to monitor the transcript levels of PR1 and PR5, the two plant defense response marker genes. The PR1 levels were noticeably induced in mutant plants at 6 hpts in Mo17 × H95 background and at 24 hpts in the B73× H95 background, while the PR5 levels increased in mutant plants at 24 hpts and 48 hpts in B73 × H95 and Mo17 × H95, respectively (Supplementary Figure 2). Thus, we chose samples collected at 6 and 48 hpts for RNA sequencing (RNA-seq) experiments to investigate the early response genes involved in Rp1-D21-induced HR. These samples are referred to below as B73 × H95-WT-6 hpts, B73 × H95-mt-6 hpts, B73 × H95-WT-48 hpts, B73 × H95-mt-48 hpts, Mo17 × H95-WT-6 hpts, Mo17 × H95-mt-6 hpts, Mo17 × H95-WT-48 hpts, and Mo17 × H95-mt-48 hpts. To investigate whether the expression of Rp1-D21 was induced after temperature shift which induced HR, we performed semi-quantitative RT-PCR analysis and found that the transcript levels of Rp1-D21 were increasingly induced from 3 to 48 hpts in both genetic backgrounds (Supplementary Figure 2).
We used the constant temperature experiment to explore the late response genes involved in Rp1-D21-induced HR. In this case, we sampled plants from near isogenic pairs of each genetic background and the sample names were B73 × H95-WT-22°C, B73 × H95-mt-22°C, Mo17 × H95-WT-22°C, and Mo17 × H95-mt-22°C.
Read numbers for each sample ranged from 21.48 to 44.06 million with an average of 32.36 million. In each case, the read number of WT and mt from the same background at the same time point were quite similar (Supplementary Table 1). Hierarchical indexing for spliced alignment of transcripts 2 (HISAT2) was used to map the reads against the B73 reference genome (B73_v4, maizegdb.org). Approximately 87.57–92.92% of the reads were mapped to the reference genome, of which 84.79% were uniquely mapped (Supplementary Table 1). Fragments per kilobase of exon per million mapped reads (FPKM) values were calculated to evaluate the gene expression levels and the reproducibility of biological replicates. Raw counts expression data showed an average Pearson's correlation coefficient of 0.87 ranging from 0.65 to 0.96, indicating high correlation between the two biological replicates in each case (Supplementary Figure 3A). A multidimensional scaling (MDS) plot was generated to assess the sequencing quality (Supplementary Figure 3B). The transcript levels of all the expressed genes were clearly divided into different groups at constant 22°C for WT and Rp1-D21 mt from B73× H95 and Mo17 × H95 backgrounds, but it was difficult to distinguish for samples at 6 hpts, which might be due to the low number of differentially expressed genes (DEGs) detected at this timepoint.
To facilitate the global identification of genes triggered during Rp1-D21-mediated hypersensitive response (HR), uniquely mapped DEGs were identified by comparing the gene expression levels and the abundance of each transcript in Rp1-D21 mt relative to the WT for each near-isogenic pair at each timepoint. The DEGs were identified with a false discovery rate (FDR) or adjusted p-values (padj) < 0.05 and a |log2 (fold-change) |>1. Since we want to analyze the most robust DEGs, we detected DEGs using both edgeR and DEseq2 software, and only those DEGs identified by both the methods were used for further analysis. The number of DEGs detected by edgeR/DEseq2 methods at each timepoint/condition is shown in Figure 1A and Supplementary Table 2. In every case, the large majority of DEGs were identified by both the methods (Figure 1B). At 6 hpts, seven common DEGs were identified in B73 × H95 and Mo17 × H95 backgrounds using both the methods (Figure 1B). At 48 hpts, 263 and 141 common DEGs were identified in B73 × H95 and Mo17 × H95 backgrounds, respectively, and 116 genes were commonly identified in both the backgrounds (Figures 1B,C). In the constant temperature experiment, 4,463 (2,799 induced, 1,664 repressed) and 5,106 (3,156 induced, 1,950 repressed) DEGs were identified in B73 × H95 and Mo17 × H95 backgrounds, respectively (Figure 1B). Of these, 3,633 common DEGs were detected in both the backgrounds using two methods (Figure 1C).
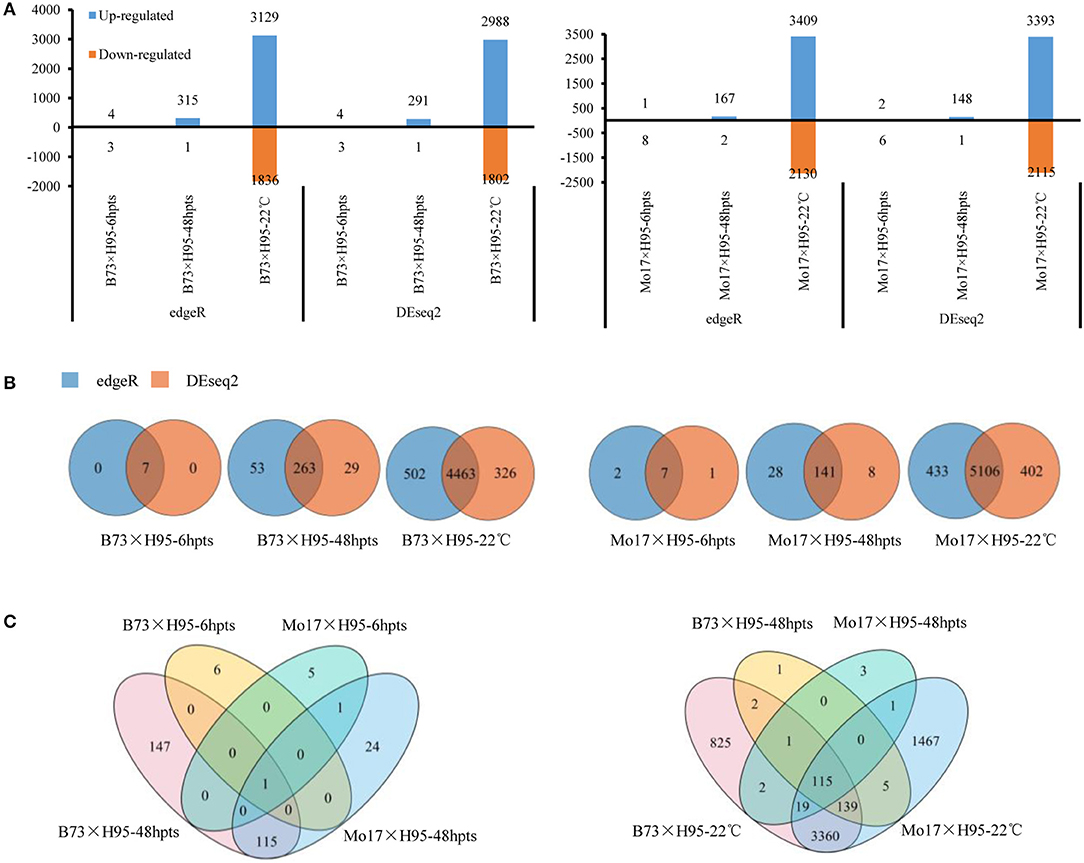
Figure 1. Differentially expressed genes (DEGs) in Rp1-D21 mutant (mt) compared to the wild type (WT) in B73 × H95 and Mo17 × H95 backgrounds. (A) The up and downregulated DEGs in B73 × H95 and Mo17 × H95 backgrounds as calculated by edgeR and DEseq2. (B) Numbers of DEGs from edgeR and DEseq2 methods at different treatments. Venn diagrams of DEGs among different time points and temperature treatments from both edgeR and DEseq2. (C) The common DEGs between both genotypes at 6 and 48 h post temperature shift (hpts) (left), and at constant 22°C (right), respectively.
Many more DEGs were identified in the constant temperature experiment (>5,000) than in the temperature shift experiment (149–316 at 48 hpts) in both the backgrounds. In every case, more DEGs were upregulated than downregulated by the presence or activation of Rp1-D21 (Figure 1A). There were some notable differences between the two genetic backgrounds. For example, at 48 hpts, the number of DEGs in the B73 × H95 background was almost twice that in the Mo17 × H95 background. On the other hand, substantially more DEGs, both up- and downregulated, were detected in the Mo17 × H95 background at constant 22°C.
The maize gene ontology (GO) database was used for the functional annotation of the DEGs identified by both edgeR and DEseq2 in B73 and Mo17 backgrounds. At 6 hpts, 6 out of 7 DEGs were predicted to be involved in adenosine diphosphate (ADP) binding (GO:0043531), and oxidation-reduction process (GO:0055114). At 48 hpts, the DEGs were mainly predicted to be involved in heme binding (GO:0020037), extracellular region (GO:0005576), transmembrane transport (GO:0055085), and defense response (GO:0006952), which included some genes in salicylic acid (SA) biosynthetic process (GO:0080142) (Figure 2A). At constant 22°C, the DEGs in the biological process category mainly included transmembrane transport, protein autophosphorylation, and hormone signaling pathway (Figure 2B). For molecular function category, the enriched GO terms mainly contained genes involved in binding (e.g., cofactor and heme binding) and catalytic activity [e.g., uridine diphosphate (UDP)-glycosyltransferase activity] (Figure 2B). For cellular component (CC) category, the enriched GO terms included plasmodesma, chloroplast thylakoid membrane, and intracellular component of the plasma membrane (Figure 2B).
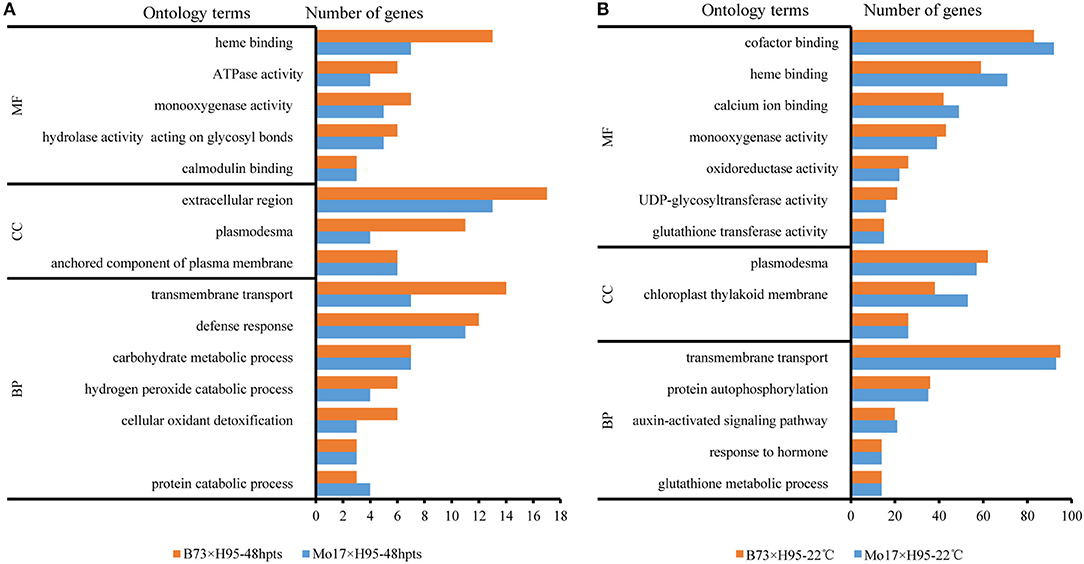
Figure 2. The Gene ontology (GO) enrichment of differentially expressed genes (DEGs) in maize Rp1-D21 mutant (mt) compared to the wild type (WT) for the indicated times. (A) Top 15 GO subcategories of DEGs from Rp1-D21 mt in B73 × H95 and Mo17 × H95 backgrounds at 48 hours post temperature shift (hpts). (B) Top 15 GO subcategories of DEGs from Rp1-D21 mt in B73× H95 and Mo17 × H95 backgrounds at constant 22°C. MF, molecular function; CC, cellular component; BP, biological process.
Upon the initiation of HR, transcriptional activation genes were mainly involved in genes encoding receptor kinases, transcription factors, calcium regulation, and protein degradation (Supplementary Figure 4) as analyzed by MapMan software (Usadel et al., 2009). At constant 22°C conditions, a large number of transcription factors (TFs) were differentially expressed in Rp1-D21 mts from both B73 × H95 and Mo17 × H95 backgrounds, indicating that massive transcription reprogramming occurred at this condition. Most of these genes were related to the establishment of transcriptional reprogramming and the enhancement of immune response for Rp1-D21-mediated HR. Gene regulatory networks (GRNs) represent the maps of potential transcriptional regulation between TFs and their target genes. To investigate the GRNs of Rp1-D21-mediated HR, the GRNs of the 3,633 DEGs detected in both B73 and Mo17 backgrounds at constant 22°C were predicted for the regulatory interactions with their upstream TFs based on the existing TF binding motifs and the conservation of TF binding sites (TFBSs) (Zhou et al., 2020). In total, 73 putative TFs were predicted to bind DEGs at constant 22°C condition under cutoff p-value ≤ 0.05 (Supplementary Table 3). Of those 73 TFs, the majority are WRKY (16, 21.92%), bZIP (14, 19.18%), MYB (9, 12.33%), and NAC (9, 12.33%). Some TFs were predicted to target themselves (Supplementary Table 3). Most of other DEGs which were related to biotic stresses, included genes involved in protein modification, protein degradation, calcium regulation, and hormone biosynthesis. Interestingly, many DEGs encoded receptor kinases, G-proteins, and mitogen activated protein (MAP) kinases, which directly or indirectly regulate the signal perception and activation of immunity, such as SA defense-related pathway.
Pathogen-triggered immunity (PTI) and effector-triggered immunity (ETI) can function synergistically to protect plants against pathogens. Interestingly, we found that many genes predicted to be involved in maize PTI were differentially expressed in Rp1-D21 mt, including the homologs of BAK1, FLS2, Ca2+-ATPase genes (CAs), and RBOHs (Supplementary Figure 5, Supplementary Table 4). Some key components predicted to act in the plant immunity signaling pathway were also differentially expressed, for instance, the mitogen-activated protein kinases (MAPKs) cascades and receptor-like cytoplasmic kinase VII (RLCK VII) subfamily genes, which act as central players in both PTI and ETI (Liang and Zhou, 2018; Zhou and Zhang, 2020). The SA pathway plays important roles in plant disease resistance and pathogen-induced HR (Zheng et al., 2015; Cui et al., 2017; Zhang et al., 2017). We found that the genes predicted to encode the SA receptors, nonexpressor of pathogenesis-related genes 1 (NPR1) and NPR4, and the SA marker genes, pathogenesis-related 1 (PR1) and PR5 were significantly differentially expressed in Rp1-D21 mt.
To investigate the metabolic changes induced by Rp1-D21, widely targeted metabolome analysis (Chen et al., 2013) was conducted by ultra-high-performance liquid chromatography-tandem mass spectroscopy (UPLC-MS). Due to the lack of seeds in B73 × H95 or Mo17 × H95 backgrounds, we used the isogenic hybrids A632 × H95-Rp1-D21 and A632 × H95 for this experiment. A total of 423 metabolites were detected, including lipid, phenolic acids, flavonoids, and alkaloids. The orthogonal partial least squares discriminant analysis (OPLS-DA) model was performed on the metabolic analysis, and significantly differential accumulated metabolites (DAMs) were selected with |log2 (fold-change)| > 0.6 and variable importance for projection (VIP) > 1. Finally, a total of 103 DAMs (76 upregulated and 27 downregulated in the Rp1-D21 background) were identified (Supplementary Figure 6A, Supplementary Table 5). Pathway enrichment analysis was conducted using Kyoto encyclopedia of genes and genomes (KEGG). These DAMs were mainly divided into “biosynthesis of secondary metabolites,” “biosynthesis of amino acids,” and “phenylpropanoid biosynthesis” (Supplementary Figure 6B).
To study the association between transcriptomic and metabolic analyses in Rp1-D21 mt, the conjoint analysis of DAMs and DEGs at constant temperature were conducted by the KEGG pathway. Seven common pathways were enriched in Rp1-D21 mt (Figure 3, Supplementary Table 5), with phenylpropanoid biosynthesis (Supplementary Figure 7) and α-linolenic acid metabolism as the top two significantly enriched pathways.
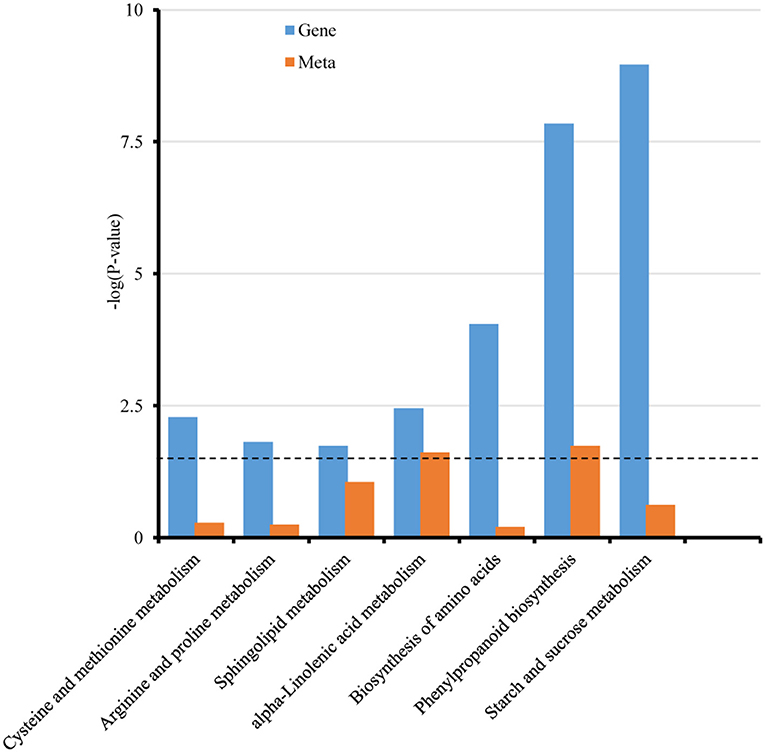
Figure 3. Integrated analyses of transcriptomic and metabonomic results of Rp1-D21 mutant (mt) compared to the wild type (WT). The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment histogram of conjoint analysis of DEGs and differential accumulated metabolites (DAMs). The x-axis represents the metabolic pathways and the y-axis represents the expression as -log (p-value). The blue columns represent the enrichment p-values of DEGs, and the orange columns represent the enrichment p-values of DAMs. The black dotted line represents the threshold for significant enrichment at p < 0.05.
Salicylic acid and its derivatives play important roles in plant disease resistance (Vlot et al., 2009; Ding et al., 2018); therefore, we investigated the DEGs and DAMs in the SA pathway. The SA biosynthesis occurs via two pathways, the isochorismate synthase (ICS) pathway and the phenylalanine ammonia-lyase (PAL) pathway, which play the major and minor roles in SA biosynthesis, respectively (Figure 4A, Dempsey et al., 2011). Interestingly, most genes encoding enzymes in ICS biosynthesis, including ICS1, EDS5, and PBS3, were downregulated in the late stage of HR induction (22°C), while most genes in the PAL pathway had an opposite expression pattern, including PAL and AIM1 homologs (Figure 4B, Supplementary Table 6). The level of phenylalanine, the precursor of the PAL pathway was increased to 1.53 fold in Rp1-D21 mt compared to WT (Supplementary Table 5). In addition to the SA biosynthesis, the regulation of genes predicted to be involved in SA modifications were also investigated (Figure 4B). The genes associated with SA hydroxylation (S5H and S3H) were highly induced, while the genes related to SA methylation (BSMT), amino acid conjugation (GH3.5), and SA sulfonation (SOT) were downregulated in the late stage of HR (Figures 4A,B). In Arabidopsis, AtUGT74F1/2 and AtUGT75B1 modify SA to generate glycosylated SA (Noutoshi et al., 2012; George Thompson et al., 2017). We identified 9 and 4 SA glucosyltransferase (SAGT) homologs when searched the maize genome for sequences homologous to AtUGT74F1/2 and AtUGT75B1, respectively (Supplementary Figure 8). Among these 13 genes encoding SAGTs, 5 and 1 were up- and downregulated in Rp1-D21, respectively (Figure 4B, Supplementary Figure 8).
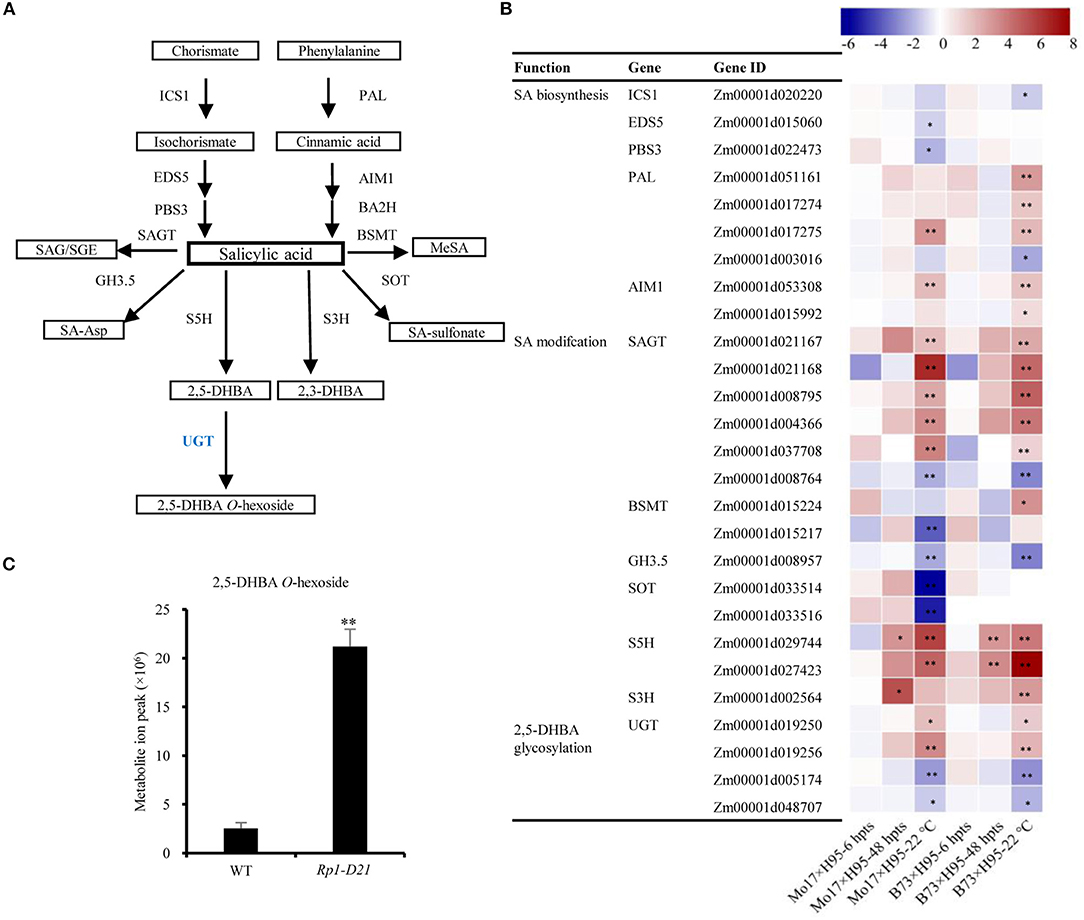
Figure 4. The key enzymes involved in salicylic acid (SA) biosynthesis and modification pathways. (A) The SA biosynthesis and modification pathways. ICS1, isochorismate synthase 1; PAL, phenylalanine ammonia-lyase; EDS5, enhanced disease susceptibility 5; PBS3, AvrPphB susceptible3; AIM1, abnormal inflorescence meristem1; BA2H, benzoic acid 2-hydroxylase; UGT, UDP-glucosyltransferases; SAGT, salicylic acid glucosyltransferase; SAG, SA 2-O-β-Dglucoside; SGE, salicylate glucose ester; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; MeSA, methyl salicyliate; GH3.5, GH3 acyl adenylase family member 3.5; SA-Asp, salicyloyl-L-aspartate; SOT, sulfotransferase; S5H, SA 5-hydroxylase; S3H, SA 3-hydroxylase; 2,5-DHBA, 2,5-dihydroxybenzoic acid; 2,3-DHBA, 2,3-dihydroxybenzoic acid. (B) The log2 fold-change of the genes encoding key enzymes in SA biosynthesis and modification pathways. The fold change was normalized with the FPKM value. (C) The levels of 2,5-DHBA O-hexoside were significantly accumulated in Rp1-D21 mutant (mt) compared to the wild type (WT). Asterisks indicate significant difference between samples, each sample with three biological replicates.
Salicylic acid can be hydroxylated by S5H to form 2,5-dihydroxy benzoic acid (2,5-DHBA), which can be further glycosylated by UGT76D1 to generate 2,5-DHBA glucosides in Arabidopsis (Zhang et al., 2017; Huang et al., 2018). We further measured the total and free SA levels of H95-Rp1-D21 crossed into different backgrounds when they were grown at 22°C, and the result showed that Rp1-D21 mt in different backgrounds had more SA accumulation than their corresponding WT (Figure 5A). Interestingly, the SA levels were largely positively related with the HR strength in different backgrounds (Figure 5B). To investigate whether SA affect Rp1-D21-mediated HR, we grew Mo17x H95-Rp1-D21 at 30°C for 8 days, and then treated the seedlings with H2O and 1.2 mM benzothiadiazole S-methyl ester (BTH), an analog of SA, and put the plant at 22°C to induce HR. We found that BTH significantly reduced the severity of Rp1-D21-mediated HR compared to H2O (Figure 6). Among the DAMs, 2,5-DHBA O-hexoside was identified as one of the top DAMs, which accumulated at 8.34-fold in Rp1-D21 mt than the WT (Figure 4C, Supplementary Table 5).
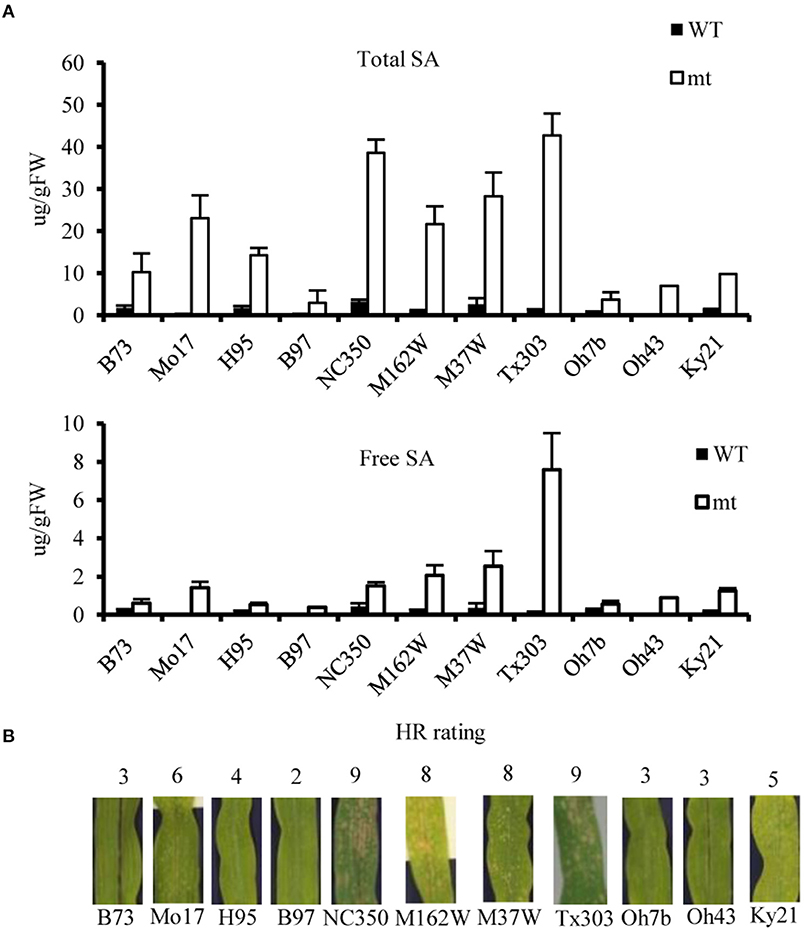
Figure 5. The salicylic acid (SA) levels and hypersensitive response (HR) severity of Rp1-D21 in different genetic backgrounds. (A) The levels of total SA and free SA were significantly accumulated in Rp1-D21 mutant (mt) compared to the wild type (WT) in different backgrounds crossed with H95-Rp1-D21 when they were grown at 22°C. (B) The HR rating of Rp1-D21 mt in different backgrounds crossed with H95-Rp1-D21grown at 22°C condition.
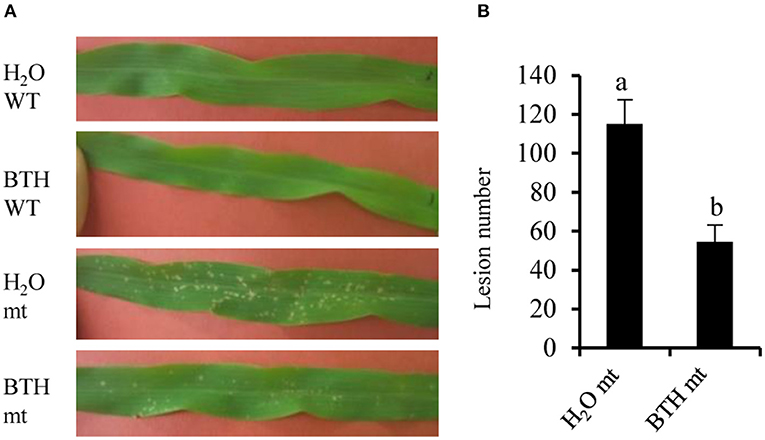
Figure 6. Benzothiadiazole (BTH) treatment reduced Rp1-D21-mediated HR. (A) Plants in Mo17 × H95 background were grown at 30°C for 8 days, then the plants were treated by H2O or 1.2 mM BTH. After treatment, the temperature was dropped to 22°C to induce hypersensitive response (HR), and the pictures of the third leaves were taken at 4 days after temperature shift. (B) The lesion number in mutant (mt was calculated after H2O and BTH treatment. Significant differences (p < 0.05) between samples are indicated by different letters (a,b). Wild type (WT) and Rp1-D21 mutant (mt).
In Arabidopsis, AtUGT76D1 (AT2G26480) is responsible for the conversion from 2,5-DHBA to 2,5-DHBA glucoside (Huang et al., 2018). We identified UDP-glucuronosyltransferase (UGT) homologs from maize (ZmUGTs) according to AtUGT76D1 and performed a phylogenetic analysis (Figure 7A). Of the nine ZmUGT homologs identified, Zm00001d019250 (ZmUGT9250) and Zm00001d019256 (ZmUGT9256) were upregulated at the late stage of HR induction in the Rp1-D21 mt compared to the corresponding WT in both B73 and Mo17 backgrounds (Figure 7A), while Zm00001d005174 (ZmUGT5174) and Zm00001d048707 (ZmUGT8707) had the opposite expression pattern (Supplementary Table 7). Other ZmUGT homologs were not differentially expressed under these conditions (Supplementary Table 7).
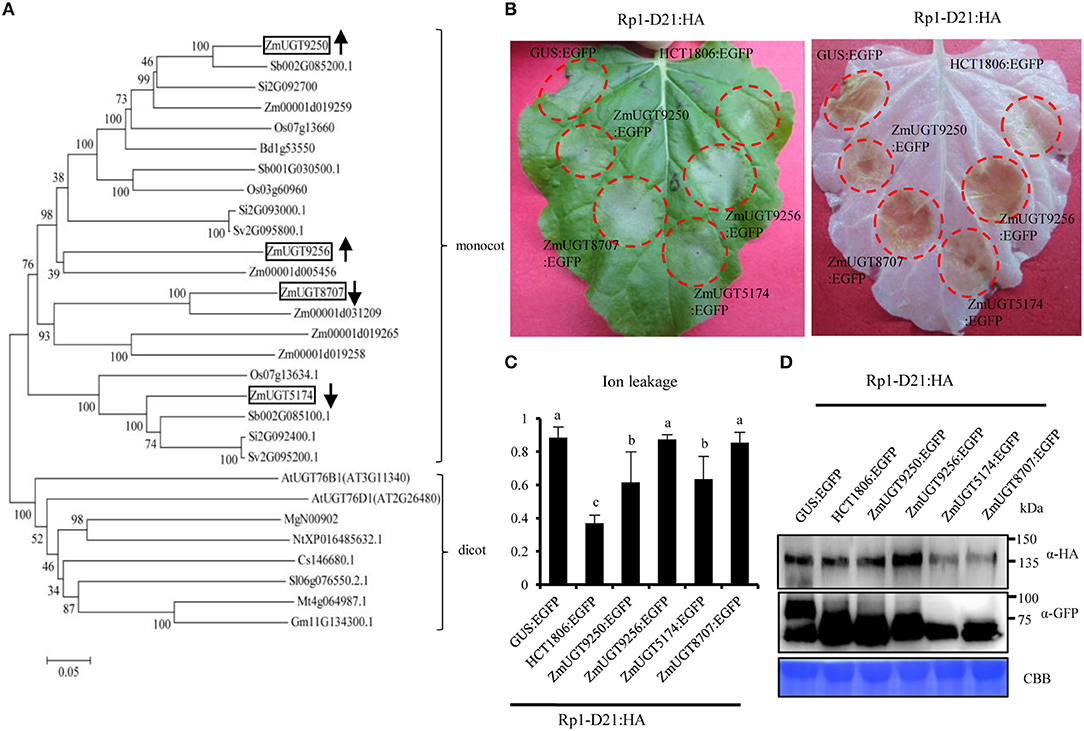
Figure 7. Phylogenetic analysis of UDP-glucuronosyltransferase (UGT) proteins and the function of ZmUDP-glucuronosyltransferases (UGTs) in Rp1-D21-mediated hypersensitive response (HR). (A) The phylogenetic tree was constructed using MEGA 6.0 software. At, Arabidopsis thaliana; Bd, Brachypodiumdistachyon; Cs, Cucumissativus; Gm, Glycine max; Mg, Mimulusguttatus; Mt, Medicagotruncatula; Nt, Nicotianatabacum; Os, Oryza sativa; Sb, Sorghum bicolor; Si, Setaria italica; Sl, Solanum lycopersicum; Sv, Setaria viridis; Zm, Zea mays. The boxes indicate the UGT homologs of maize which were differentially expressed in Rp1-D21. (B) ZmUGTs were transiently co-expressed with Rp1-D21 into Nicotiana benthamiana. The representative leaf was photographed at 3 days after inoculation (left), and the same leaf was cleared by ethanol (right). (C) Ion leakage conductivity [average ± standard error (SE), n > 5] was measured at 60 h after co-expression of GUS, HCT1806, or ZmUGTs with Rp1-D21. Significant differences (p < 0.05) between samples are indicated by different letters (a–c). (D) Total protein was extracted from agro-infiltrated leaves at 40 hour post inoculation (hpi). Anti-HA was used to detect the expression of Rp1-D21, and anti-GFP was used to detect the expression of GUS, HCT1806, and ZmUGTs. Equal loading of protein samples was shown by Coomassie brilliant blue (CBB) staining. The experiments were performed three times with similar results.
To investigate the possible roles of the four differentially regulated ZmUGTs in Rp1-D21-mediated HR, we used the agrobacteria-mediated transient expression in Nicotiana benthamiana. The four ZmUGTs were fused with a C-terminal enhanced green fluorescent protein (EGFP) tag. GUS:EGFP and HCT1806:EGFP were used as negative and positive controls, respectively. The HCT1806 had previously been shown to be a strong suppressor of Rp1-D21-mediated HR (Wang et al., 2015b; Wang and Balint-Kurti, 2016; Murphree et al., 2020). When transiently co-expressed with Rp1-D21 fused with C-terminal 3 × hemagglutinin (HA) tag in N. benthamiana, ZmUGT9250 and ZmUGT5174 partially suppressed Rp1-D21-induced HR compared to HCT1806, while ZmUGT9256 and ZmUGT8707 did not suppress Rp1-D21-induced HR (Figure 7B). Ion leakage conductivity data further verified our visual observations (Figure 7C). Co-expression of ZmUGTs did not significantly change the protein accumulation of Rp1-D21 which was expressed at substantial and broadly comparable levels (Figure 7D).
The N-terminal coiled-coil (CC) domain of Rp1-D21 protein (CCD21) conferred an obvious autoactive HR when it was fused with EGFP and transiently expressed in N. benthamiana (Wang et al., 2015b). To determine whether ZmUGTs can suppress CCD21-mediated HR, we co-expressed them and CCD21:EGFP in N. benthamiana. The results showed that ZmUGT9250 suppressed CCD21-mediated HR which was similar to HCT1806, ZmUGT5174 partially suppressed CCD21-mediated HR, and ZmUGT9256 and ZmUGT8707 had no obvious effect (Supplementary Figure 9).
Several Rp1-D21 regulators we identified previously interact with CCD21 and suppress CCD21-mediated HR (Wang et al., 2015b; Wang and Balint-Kurti, 2016; Zhu et al., 2020; Liu et al., 2021; Luan et al., 2021). To investigate whether ZmUGTs can interact with CCD21, we performed yeast two-hybrid assays. We did not detect interaction between any of the four ZmUGTs investigated and CCD21 (Supplementary Figures 10A,B). A co-immunoprecipitation assay between CCD21 and ZmUGT9250, which had the strongest suppression effect on CCD21-mediated HR, also did not detect any interaction (Supplementary Figure 10C).
To investigate whether ZmUGTs could suppress HR induced by other nucleotide-binding leucine-rich-repeats (NLRs), we co-expressed them with Arabidopsis RPM1(D505V) and barley MLA10(D502V), which confer an autoactive HR when transiently expressed in N. benthamiana (Gao et al., 2011; Bai et al., 2012). The results showed that all the four ZmUGTs had no obvious suppression effect on either MLA10(D502V)- or RPM1(D505V)-induced HR (Supplementary Figure 11). We further co-expressed ZmUGTs with BAX or INF, the death-promoting protein or cell death elicitor (Kamoun et al., 1998; Lacomme and Santa Cruz, 1999), and found that neither of them substantially suppressed either BAX- or INF-induced HR (Supplementary Figure 11).
In this study, we used transcriptome and metabolome analyses to investigate the molecular responses of Rp1-D21-mediated hypersensitive response (HR). The conditions used for the transcriptome analysis and metabolome analysis differed substantially in terms of the growth conditions and the genotypes used. Despite this, the salicylic acid (SA) pathway and the phenylpropanoid biosynthesis pathway were induced at both the transcriptional and metabolic levels. Especially, the genes predicted to encode 2,5-DHBA UGTs were differentially expressed in Rp1-D21 mutant (mt) and the 2,5-DHBA O-hexoside catalyzed by UGTs were high accumulated in Rp1-D21 mt. We further examined that two Zm UDP-glucuronosyltransferases (UGTs) partially suppress Rp1-D21-mediated HR.
Autoimmune mutants caused by inappropriate activation of nucleotide-binding leucine-rich-repeats (NLRs) are widely used for mechanistic investigation of immune signaling components in different plants, e.g., Arabidopsis snc1 and mkk1mkk2 (Kong et al., 2012; Dong et al., 2016; Chakraborty et al., 2018). Rp1-D21 is an NLR protein which confers autoimmunity in maize (Sun et al., 2001; Smith et al., 2010). The transcript levels of Rp1-D21 were greatly increased over time after temperature dropping from 30 to 22°C, especially at 48 h post the temperature shift (hpts) (Supplementary Figure 2). At constant 22°C, the transcript levels of Rp1-D21 were even much higher (Supplementary Figure 2). These data suggested that the onset of HR might be related with the increased expression of Rp1-D21. Consistent with the trends of the transcript levels of Rp1-D21, more differentially expressed genes (DEGs) were identified at constant 22°C than at 48 and 6 hpts, indicating that these DEGs were induced by the increased levels of Rp1-D21. Several transcription factors (TF)s, including WRKY and bZIP which are known to act in plant immunity (Jakoby et al., 2002; Kaminaka et al., 2006; Pandey and Somssich, 2009), were predicted as the major TFs to bind the promoters of DEGs (Supplementary Table 3), indicating that Rp1-D21-mediated HR is under tight transcriptional regulation. We showed that many genes predicted to act in pathogen-triggered immunity (PTI) were differentially expressed in the defense response triggered by the NLR Rp1-D21, including gene homologs encoding FLS2, BAK1, RBOHs, RLCKs, and MAPK cascades (Supplementary Figure 5). Consistent with our results, it was recently reported that effector-triggered immunity (ETI) can increase the defense strength through transcriptional induction of PTI signaling components and PTI is required for disease resistance mediated by several NLRs (Ngou et al., 2021; Yuan et al., 2021).
Several studies have used the Rp1-D21 mt to identify genes or loci associated with modulation of the severity of HR (Chintamanani et al., 2010; Chaikam et al., 2011; Olukolu et al., 2014). We have verified that two homolog genes encoding enzymes of lignin biosynthesis pathway ZmHCT1806 and ZmCCoAOMT2 (Wang et al., 2015b; Wang and Balint-Kurti, 2016), two flavone synthase homologs of ZmFNSIs (Zhu et al., 2020), the nicotinate N-methyl transferase ZmNANMT(Liu et al., 2021), and the metacaspase homologs, ZmMC1 and ZmMC2 (Luan et al., 2021), can suppress Rp1-D21-mediated HR likely through physical interaction with the cellular component (CC) signaling domain of Rp1-D21 when transiently expressed in Nicotiana benthamiana. Interestingly, ZmCCoAOMT2 was also proved to confer quantitative resistance to both southern leaf blight and gray leaf spot in maize (Yang et al., 2017). The homologs of these genes in different plant species also act in plant immunity (Coll et al., 2010; Senthil-Kumar et al., 2010; Gallego-Giraldo et al., 2011; Zeilmaker et al., 2015), suggesting the functional association between disease resistance and HR regulation. Recently, it was reported that the transcriptional responses triggered by Rp1-D21 are broadly similar to those triggered by its wild type (WT) counterpart Rp1-D, which is activated by Puccinia sorghi infection (Kim et al., 2021). The DEGs identified in Rp1-D21 are highly correlated with disease resistance; therefore, Rp1-D21 can be used as a quick and effective tool for exploring new HR regulation and disease resistance genes.
Except for the SA pathway, we found that the α-linolenic acid pathways and the phenylpropanoid biosynthesis pathway were induced in Rp1-D21 at both the transcriptional and metabolic levels according to the integrated transcriptome and metabolome analyses (Figure 3). The α-linolenic acid is produced by two metabolic pathways, 9-lipoxygenase (9-LOX) pathway, and 13-lipoxygenase (13-LOX) pathway (Wasternack, 2007). LOX-derived oxylipins have been implicated in plant growth and development, senescence, and resistance to pathogens (Blée, 2002; Mosblech et al., 2009). When infected by different pathogens, the transcript levels and the related metabolites in 9-LOX pathway were increased in tobacco, potato, and maize with the occurrence of HR (Gobel et al., 2001, 2003; Davoine et al., 2006; Gao et al., 2009). The 9-hydroxyoctadecatrienoic acid (9-HOT) and 9,10-epoxyoctadecenoic acid (9,10-EOT) in the 9-LOX pathways were among the most active oxylipins (Prost et al., 2005; Vicente et al., 2012). The mutant insensitive to 9-HOT displays enhanced susceptibility to Pseudomonas infection (Vellosillo et al., 2007). In our metabolites analysis, we found that the transcript levels of genes and metabolite products in the α-linolenic acid metabolism pathway were increased in the Rp1-D21 mutant (Figure 3, Supplementary Table 5). In particular, we observed a massive accumulation of 9-HOT and 9,10-EOToxylipins derived from the 9-LOX pathway suggested that they play pivotal roles in Rp1-D21-mediated HR and plant disease resistance.
The phenylpropaniod biosynthesis pathway, mainly including lignin intermediates, anthocyanins, isoflavonoidphytoalexins, and phenolic compounds, has been implicated as one of the major pathways for defense against pathogens (Ranjan et al., 2019). As displayed in Figure 3, transcriptome and metabolome conjoint analysis indicated that differentially expressed genes (DEGs) and differential accumulated metabolites (DAMs) associated with the phenylpropaniod pathway were significantly enriched (Supplementary Table 5). Many DEGs in the lignin biosynthesis pathway, including PAL, cinnamoyl-CoA reductase (CCR), CCoAOMT, and HCT play important roles in plant defense response (Kawasaki et al., 2006; Gallego-Giraldo et al., 2011; Wang and Balint-Kurti, 2016; He et al., 2020). Many genes involved in phenylpropanoid pathway are differentially expressed after inoculation with the pathogen Fusarium graminearum (Liu et al., 2016). These results suggest that the phenylpropaniod biosynthesis pathway constituted a key class of enzymes and metabolites associated with modulating the HR induced by Rp1-D21 and disease resistance in maize defense response.
We have previously shown that several regulators interact with CCD21 and modulate Rp1-D21-mediated HR (Wang et al., 2015b; Wang and Balint-Kurti, 2016; Zhu et al., 2020; Liu et al., 2021; Luan et al., 2021). Here we found that ZmUGTs suppressed Rp1-D21- and CCD21-mediated HR, but they did not interact with CCD21 (Supplementary Figure 10). These data suggested that the regulation of Rp1-D21-mediated HR by ZmUGTs might not be through the formation of a protein complex with Rp1-D21.
The plant hormone, SA has profound importance in both local resistance against biotrophic and hemi-biotrophic pathogens and systemic acquired resistance (Vlot et al., 2009). In local pathogen infection, high levels of SA induce defense gene expression by activating the transcriptional activator NPR1 and lead to cell death (Fu and Dong, 2013). In the maize lesion mimic mutant, Les4, the free and total SA levels were significantly increased at the lesion-developed stage compared to the prelesion stage (Morris et al., 1998). In this study, many gene homologs in SA biosynthesis and metabolism pathways were differentially expressed in Rp1-D21 (Figure 4B). Interestingly, the genes in the isochorismate synthase 1 (ICS1) and phenylalanine ammonia-lyase (PAL) pathways were mostly down- and upregulated in Rp1-D21 mt compared to WT at the late stage, respectively (Figure 4B). The level of phenylalanine was also increased in Rp1-D21 mt compared to WT (Supplementary Table 5). However, cinnamic acid, chorismate, and isochorismate were not identified under our conditions. The levels of total SA and free SA were significantly elevated in Rp1-D21 mts compared to the corresponding WT and there was a positive correlation between SA levels and the HR strength in different backgrounds (Figure 5). These data suggested that the activation of the SA pathway might be associated with the formation of HR lesions in plants carrying Rp1-D21. The upregulation of genes in the PAL pathway might act to keep the SA level at an appropriate level to trigger HR while the downregulation of genes in the ICS1 pathway might act to prevent the over-accumulation of SA at the late stage. On the other hand, the exogenous application of SA or SA analogs suppresses ETI-mediated HR in Arabidopsis (Zavaliev et al., 2020). Similarly, we found that exogenous application of benzothiadiazole (BTH), an analog of SA, also suppressed Rp1-D21-mediated HR in maize (Figure 6). These studies suggest that SA plays important roles in suppressing NLR-mediated HR. Therefore, SA plays dual roles in HR regulation, likely through the induction of positive defense regulators to activate HR or induction of negative regulators to suppress HR (Radojicic et al., 2018; Peng et al., 2021).
Excessive levels of SA can be toxic to plants; therefore SA can be modified by different conjugations to less toxic derivatives (Dempsey et al., 2011). In Arabidopsis, two SAGTs, UGT74F1, and UGT76B1 can glycosylate SA to produce SAG (Noutoshi et al., 2012). Loss of UGT74F1 and UGT76B1 confers resistance to both Pst DC3000 and a strain which can be recognized by the NLR protein RPM1 (Noutoshi et al., 2012). Here we found that several maize SAGT homologs were upregulated in Rp1-D21 and the total SA levels were increased in Rp1-D21 (Figures 4, 5), suggesting that these SAGT homologs might play a role in Rp1-D21-mediated HR.
2,5-dihydroxybenzoic acid (2,5-DHBA) and 2,3-DHBA are two major catabolic products of SA and they are catalyzed by hydroxylating SA via S5H and S3H, respectively (Figure 4B, Zhang et al., 2017). 2,5-DHBA is widely distributed in planta and is induced by the pathogen, PstDC3000 in Arabidopsis (Zhang et al., 2017). In a previous study (Zhu et al., 2020), we found that the transcript levels of ZmFNSIs (also known as ZmS5Hs) and ZmS3H were upregulated in Rp1-D21 mt (Figure 4B), and ZmS5Hs but ZmS3H does not suppress Rp1-D21- and CCD21-mediated HR (Zhu et al., 2020), indicating that ZmS5Hs play important roles in maize defense response. The 2,5-DHBA and 2,3-DHBA can be further glycosylated by a unique UDP-glycosyltransferase UGT76D1 in Arabidopsis (Huang et al., 2018). Expression of the AtUGT76D1 is induced by Pst DC3000. Overexpression of AtUGT76D1 leads to high SA accumulation, upregulation of defense genes and the autoactive HR phenotype, while the immune responses were compromised when AtUGT76D1 is knocked out implying that AtUGT76D1 is a positive regulator in plant innate immunity (Huang et al., 2018). Interestingly, AtUGT76B1, a close homolog which had 38.8% similarity of amino acid with AtUGT76D1, was recently found to use N-hydroxy-pipecolic acid (NHP) as substrate and act in systemic-acquired resistance (Bauer et al., 2021; Holmes et al., 2021; Mohnike et al., 2021). Loss-of-function mts of AtUGT76B1 confer a dwarf phenotype and constitutive defense response, with high NHP and SA accumulation and enhanced disease resistance to PstDC3000 (Bauer et al., 2021; Mohnike et al., 2021). The 2,5-DHBA glucosides were also accumulated in tomato and cucumber after infection with the pathogen, citrus exocortis viroid (CEVd) and prunus necrotic ringspot virus (PNRSV) (isolate NCM1), respectively (Fayos et al., 2006). In maize, 147 UGTs belonging to 17 groups were identified (Li et al., 2014), and none of them have been functionally investigated in disease resistance. Here we also found that the transcript levels of four DHBA-glycosyltransferase ZmUGT homologs of AtUGT76D1 were significantly differentially expressed and higher levels of SA and 2,5-DHBA O-hexoside were accumulated in Rp1-D21 mt compared to WT (Figures 4, 5, 7). We further showed that the two ZmUGTs, ZmUGT9250 and ZmUGT5174, suppressed Rp1-D21-mediated HR. These results indicated that ZmUGTs play negative roles in NLR protein Rp1-D21-mediated defense response, which is different from the positive role of AtUGT76D1 in plant immunity. Interestingly, ZmUGT9250 and ZmUGT5174 have opposite expression patterns in the lines harboring Rp1-D21 (Figures 4, 7). The transcript level of ZmUGT9250 was increased in Rp1-D21, suggesting that it might mainly act to inhibit the further spreading of HR when Rp1-D21 is activated, while the transcript level of ZmUGT5174 was decreased in Rp1-D21, suggesting that it might mainly act to keep NLR protein in the inhibited state in WT. The two ZmUGTs had no obvious effects on RPM1 (D505V), MLA (D502V), INF1, or Bax-mediated cell death (Supplementary Figure 11), indicating that they are not general cell death suppressors and they might have different mechanisms with Rp1-D21 for triggering cell death. It was reported that DHBA glycosyltransferases play important roles in the innate immune response through regulating the SA homeostasis (Huang et al., 2018). In this way, we hypothesized that the accumulation of DHBA glycosides which might act as an endogenous modulator of SA levels to switch the activity of enzymes in SA-mediated signaling pathways, thus regulating the HR phenotype in Rp1-D21. Since the enzyme activity of the four ZmUGThomologs has not been investigated yet, it is not clear which member has the DHBA glycosyltransferase activity. Since these ZmUGTs were homologous to both AtUGT76D1 and AtUGT76B1 (Figure 7A), it will be interesting to further investigate whether these ZmUGTs are also functionally similar to AtUGT76B1.
In summary, we used transcriptomic and metabolomic analyses to identify DEGs and DAMs involved in Rp1-D21-meidiated HR, which provides a useful resource for exploring maize disease resistance genes. The SA metabolic pathway and the phenylpropanoid biosynthesis pathway were induced at both the transcriptional and metabolic levels. We further demonstrated that two of four ZmUGTs partially suppressed the HR triggered by Rp1-D21 or its N-terminal CCD21 domain, which forms the basis for further investigating their roles in plant immunity.
Maize (Zea mays) line B73 was used for isolating ZmUGTs. The Rp1-D21-H95 line was generated by repeatedly backcrossing a line carrying Rp1-D21 as a female to the H95 inbred. As previously reported (Chaikam et al., 2011), Rp1-D21 mutant is maintained in the heterozygous state in H95 background (H95-Rp1-D21) due to its sterile nature in the homozygous state. The H95-Rp1-D21 line was then crossed as female to B73 and Mo17 to produce the isogenic hybrid pairs, B73 × H95 and B73 × H95-Rp1-D21 and the isogenic pair, Mo17 × H95 and Mo17 × H95-Rp1-D21, respectively. The only substantial genetic difference between the components of each isogenic pair was at the Rp1-D21 locus (Chaikam et al., 2011). A632 × H95-Rp1-D21 used for the metabolic analysis and other lines used for SA assays were generated similarly. For the RNA sequencing (RNA-seq) analysis, plants were grown in the growth chamber in the North Carolina State University (NCSU) Phytotron at temperatures of 22 or 30°C in a 12 h light/12 h dark cycle. The experiments have two temperature treatments: (1) Plants were grown at constant 22°C for 18 days, and the fourth leaves were collected for RNA-seq analysis. (2) Plants were grown at 30°C for 14 days; then the temperature was dropped to 22°C to induce HR, and the fourth leaves were collected at 3, 6, 24, and 48 h post the temperature shift (hpts). Each sample was composed of pooled fourth leaves from five randomly chosen plants with two biological replicates. For the metabolic analysis, the isogenic hybrid pairs, A632 × H95 and A632 × H 95-Rp1-D21 were grown in the field (Qingdao, China) and the fifth leaves at the V7 stage were collected with three biological replicates. For SA assays, the isogenic hybrid pairs in different genetic backgrounds were grown in the growth chamber for 18 days at a constant 22°C, and the fourth leaves were collected for SA measurement with three biological replicates.
Wild type Nicotiana benthamiana was grown at 24°C with a cycle of 16 h light and 8 h dark.
All primers used in this study are listed in Supplementary Table 8. Semi-quantitative RT-PCR was used to measure the gene expression as reported in our previous study (Negeri et al., 2013). Total RNA was extracted from the maize leaf tissue using Trizol (Life Technologies, Carlsbad, CA, USA) according to the instructions of the manufacturer. For complementary DNA (cDNA) synthesis, 1 μg of total RNA was reverse-transcribed using M-MLV (Life Technologies Corporation, Carlsbad, USA) following standard protocols. The amplification conditions for PCR consisted of 32 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, using 250 μM of each primer and 1–2 μL of the 5 × diluted cDNA per reaction.
Total RNA was extracted from the fourth leaves pooled from five individuals collected from Rp1-D21 mt and corresponding WT in B73 and Mo17 backgrounds at two treatments. The procedures for RNA sequencing (RNA-seq) analysis were performed according to our previous studies (Olukolu et al., 2014; Wang et al., 2015b). The quality and quantity of RNA were monitored by a NanoDrop 2000c spectrophotometer (Thermo Scientifc, Wilmington, DE, USA) and agarose gel electrophoresis. The messenger RNA (mRNA) was isolated from the total RNA by Dynabeadsoligo (dT25) (Invitrogen Life Technologies, MA, USA). The RNA-seq libraries were constructed according to the TruSeq RNA Sample Prep v2 LS as per the instructions of the manufacturer (Illumina Inc., CA, USA). The normalized libraries with individual index were loaded onto IlluminaHiSeq 2000 platform (Illumina Inc., San Diego, CA, USA) for cluster generation and sequencing. The data from two biological replicates were obtained by single end and paired-end reads (100-bp), respectively.
The sequencing reads were aligned to the maize B73 reference genome (ZmB73_RefGen_v38, ftp://ftp.ensemblgenomes.org) using hierarchical indexing for spliced alignment of transcripts 2 (HISAT2) (Kim et al., 2015) by the default parameter settings. Fragments per kilobase of exon model per million mapped reads (FPKM) and Pearson's correlation test were used for the estimation of gene transcription levels and the correlation between different biological replicates, respectively. To avoid taking the log of a number <1, all FPKM values were increased by 1. Differentially expressed genes (DEGs) were identified using the software package, edgeR, and DESeq2 from the Bioconductorsuite (Robinson et al., 2010; Love et al., 2014). The common DEGs were identified by both the methods with a false discovery rate (FDR) or adjusted p-values (padj) < 0.05 and a | Log2 (fold-change) | ≥1. Functional enrichment analysis of DEGs was performed based on Gene ontology (GO) Consortium database (http://www.geneontology.org). Network of DEGs and differentially accumulated metabolites (DAMs) were constructed based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Kanehisa et al., 2017). A regulation overview of DEGs was accomplished using the MapMantool (Usadel et al., 2009). The common DEGs from 48 hpts and at 22°C grown condition treatments were selected to infer the gene regulatory networks (GRNs) using PlantTFDB v5.0 (http://plantregmap.cbi.pku.edu.cn; Zhou et al., 2020). The functional assignment of predicted transcription factors was done based on the functional classification information from the MapMan Toolkit.
The fifth leaves of maize seedlings at V7 stage were collected with three replications for widely targeted metabolites analysis by ultra-performance liquid chromatography-mass spectrometry/Mass spectrometry (UPLC-MS/MS) (Chen et al., 2013). As described previously (Li et al., 2018; Mo et al., 2019), ~100 mg of freeze-dried leaf powder was dissolved in 0.6 mL of 70% methanol at 4°C overnight. Supernatant after centrifugation at 10, 000 g for 10 min was filtrated with a 0.22 μm pore size membrane for UPLC-MS/MS analysis using an LC-ESI-MS/MS system (HPLC, Shim-pack UFLC SHIMADZU CBM30A system, https://www.shimadzu.com.cn/; MS, Applied Biosystems 4500 Q TRAP, http://www.appliedbiosystems.com.cn/). Metabolite quantification is acquired with triple quadrupole scans using multi-reaction monitoring (MRM) model. The DAMs were identified by orthogonal partial least squares-discriminant analysis (OPLS-DA) according to the criteria of |fold-change| ≥ 1.5 and the variable importance in project (VIP) ≥1.
Salicylic acid was extracted as described previously (Wang et al., 2011). Briefly, 200 mg of each sample with three replications was ground into powder and extracted with 1.5 mL of 90% methanol followed by extraction with 1.5 mL of 100% methanol. 500 ng of o-anisic acid (Sigma, 169978) was added in each sample as the internal control. For each sample, 40 units of β-glucosidase (Sigma, G-0395) were added to digest glucosyl-conjugated SA (total SA) for 1.5 h at 37°C, and then were treated with an equal volume of 10% trichloroacetic acid (TCA) and centrifuged at 10,000 g for 10 min. A Dionex AS50 HPLC instrument with an Acclaim 120 C18 reverse column (4.6 3 250 mm) was used to detect the SA contents.
For phylogenetic analysis, the protein sequences from UDP-glucuronosyltransferase (UGT) family were aligned using Clustal X v2.1. Based on this alignment, a neighbor-joining tree was constructed using MEGA 6.0 software with 1,000 bootstrap replicates (Tamura et al., 2007). The neighbor-joining and p-distance methods were used with the pairwise deletion option to deal with gaps in the amino acid sequences.
Rp1-D21:HA, GUS:EGFP, and CCD21:EGFP were generated previously (Wang et al., 2015b). The cDNA sequences of ZmUGTs were isolated from B73 line and cloned into pENTR directional TOPO cloning vector (D-TOPO, Invitrogen, MA, USA). After sequencing, they were constructed into pSITEII-N1-EGFP vector (Martin et al., 2009) by LR reactions.
The Agrobacterium tumefaciens strain, GV3101 (pMP90) transformed with binary vector constructs was grown at 28°C overnight in 10 mL of L-broth medium supplemented with appropriate antibiotics. The detailed procedures were performed according to our previous study (Wang et al., 2015b). Unless otherwise indicated, all the experiments were repeated three times with similar results.
Ion leakage conductivity was measured with a conductivity meter (METTLER TOLEDO, Zurich, Switzerland) according to our previous studies (Wang and Balint-Kurti, 2016; Zhu et al., 2020).
ZmUGTs and CCD21 were respectively cloned into the pGADT7 (AD) and pGBKT7 (BD) vectors (Clontech, Mountain View, CA, USA) through LR reactions. The different combinations of AD- and BD-derived constructs were co-transformed into the yeast strain, Y2HGold. The Y2H assay was performed according to the protocol provided by the protocol of the manufacturer (Clontech, Mountain View, CA, USA).
For protein analysis as described in our previous study, Myc-, EGFP- and HA-tagged constructs were transiently co-expressed in Nicotiana benthamiana and three leaf disk (~1.2 cm in diameter) from different N. benthamiana plants were collected at 30 h post inoculation (hpi). Total input protein was extracted in 150 μl extraction buffer (Wang et al., 2015b). Proteins for Co-IP assay were extracted from 0.6 g of leaf tissue collected at 40 hpi in 2.4 ml extraction buffer (Wang et al., 2015a; Wang and Balint-Kurti, 2016). The Western blot assays were performed according to our previous studies (Wang and Balint-Kurti, 2016; Zhu et al., 2020).
The original contributions presented in the study are publicly available. This data can be found at: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA288794 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA288794).
G-FW conceived the original research plans and supervised and designed the experiments. CG, Y-GW, and G-FW performed the experiments. CG, Y-GW, SL, XZ, B-KH, PB-K, and G-FW analyzed the data. CG and G-FW wrote the manuscript. PB-K revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by grants from the National Natural Science Foundation of China (31871944, 31571263, and 32072405), Qilu Scholarship from Shandong University of China (11200086963061), Agricultural Variety Improvement Project of Shandong Province (2017LZN034), Natural Science Foundation of the USA Plant Genome grants (0822495 and 1444503), and the China Postdoctoral Science Foundation (2019M652391).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.738261/full#supplementary-material
Asselin, J. E., Lin, J., Perez-Quintero, A. L., Gentzel, I., Majerczak, D., Opiyo, S. O., et al. (2015). Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoeastewartii. Plant Physiol. 167, 1117–1135. doi: 10.1104/pp.114.253120
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Balint-Kurti, P. (2019). The plant hypersensitive response, concepts, control and consequences. Mol. Plant Pathol. 20, 1163–1178. doi: 10.1111/mpp.12821
Bartsch, M., Bednarek, P., Vivancos, P. D., Schneider, B., von RoepenackLahaye, E., Foyer, C. H., et al. (2010). Accumulation of isochorismate-derived 2, 3-dihydroxybenzoic 3-O-b-D-xyloside in Arabidopsis resistance to pathogens and ageing of leaves. J. Biol. Chem. 285, 25654–25665. doi: 10.1074/jbc.M109.092569
Bauer, S., Mekonnen, D. W., Hartmann, M., Yildiz, I., Janowski, R., Lange, B., et al. (2021). UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid and controls basal pathogen defense. Plant Cell 33, 714–734. doi: 10.1093/plcell/koaa044
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes, the new paradigm and a lifetime supply of questions. Ann. Rev. Phytopathol. 45, 399–436. doi: 10.1146/annurev.phyto.45.062806.094427
Blée, E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7, 315–322. doi: 10.1016/S1360-1385(02)02290-2
Chaikam, V., Negeri, A., Dhawan, R., Puchaka, B., Ji, J., Chintamanani, S., et al. (2011). Use of mutant-assisted gene identification and characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theoret. Appl. Genet. 123, 985–997. doi: 10.1007/s00122-011-1641-5
Chakraborty, J., Ghosh, P., and Das, S. (2018). Autoimmunity in plants. Planta 248, 751–767. doi: 10.1007/s00425-018-2956-0
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites, application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Chintamanani, S., Hulbert, S. H., Johal, G. S., and Balint-Kurti, P. J. (2010). Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization. Genetics 184, 813–825. doi: 10.1534/genetics.109.111880
Coll, N. S., Vercammen, D., Smidler, A., Clover, C., Van Breusegem, F., Dangl, J. L., et al. (2010). Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. doi: 10.1126/science.1194980
Cui, H., Gobbato, E., Kracher, B., Qiu, J., Bautor, J., and Parker, J. E. (2017). A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 213, 1802–1817. doi: 10.1111/nph.14302
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity, from pathogen perception to robust defense. Ann. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Davoine, C., Falletti, O., Douki, T., Iacazio, G., Ennar, N., Montillet, J. L., et al. (2006). Adducts of oxylipin electrophiles to glutathione reflect a 13 specificity of the downstream lipoxygenase pathway in the tobacco hypersensitive response. Plant Physiol. 140, 1484–1493. doi: 10.1104/pp.105.074690
Dempsey, D. M. A., Vlot, A. C., Wildermuth, M. C., and Klessig, D. F. (2011). Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. doi: 10.1199/tab.0156
Dewdney, J., Reuber, T. L., Wildermuth, M. C., Devoto, A., Cui, J., Stutius, L. M., et al. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. doi: 10.1046/j.1365-313x.2000.00870.x
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Oppositeroles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Dong, O. X., Meteignier, L. V., Plourde, M. B., Ahmed, B., Wang, M., Jensen, C., et al. (2016). Arabidopsis TAF15b localizes to RNA processing bodies and contributes to snc1-mediated autoimmunity. Mol. Plant Microbe Interact. 29, 247–257. doi: 10.1094/MPMI-11-15-0246-R
Etalo, D. W., Stulemeijer, I. J., van Esse, H. P., de Vos, R. C., Bouwmeester, H. J., and Joosten, M. H. (2013). System-wide hypersensitive response-associated transcriptome and metabolome reprogramming in tomato. Plant Physiol. 162, 1599–1617. doi: 10.1104/pp.113.217471
Fayos, J., Bellés, J. M., López-Gresa, M. P., Primo, J., and Conejero, V. (2006). Induction of gentisic acid 5-O-beta-D-xylopyranoside in tomato and cucumber plants infected by different pathogens. Phytochemistry 67, 142–148. doi: 10.1016/j.phytochem.2005.10.014
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance, turning local infection into global defense. Ann. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gallego-Giraldo, L., Escamilla-Trevino, L., Jackson, L. A., and Dixon, R. A. (2011). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. U. S. A. 108, 20814–20819. doi: 10.1073/pnas.1117873108
Gao, X., Brodhagen, M., Isakeit, T., Brown, S. H., Göbel, C., Betran, J., et al. (2009). Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 22, 222–231. doi: 10.1094/MPMI-22-2-0222
Gao, Z., Chung, E. H., Eitas, T. K., and Dangl, J. L. (2011). Plant intracellular innate immune receptor resistance to Pseudomonas syringaepv. maculicola1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 108, 7619–7624. doi: 10.1073/pnas.1104410108
George Thompson, A. M., Iancu, C. V., Neet, K. E., Dean, J. V., and Choe, J. Y. (2017). Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 7:46629. doi: 10.1038/srep46629
Gobel, C., Feussner, I., and Rosahl, S. (2003). Lipid peroxidation during the hypersensitive response in potato in the absence of 9-lipoxygenases. J. Biol. Chem. 278, 52834–52840. doi: 10.1074/jbc.M310833200
Gobel, C., Feussner, I., Schmidt, A., Scheel, D., Sanchez-Serrano, J., Hamberg, M., et al. (2001). Oxylipin profiling reveals the preferential stimulation of the 9-Lipoxygenase pathway in elicitor-treated potato cells. J. Biol. Chem. 276, 6267–6273. doi: 10.1074/jbc.M008606200
He, J., Liu, Y., Yuan, D., Duan, M., Liu, Y., Shen, Z., et al. (2020). An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. U. S. A. 117, 271–277. doi: 10.1073/pnas.1902771116
Holmes, E. C., Chen, Y. C., Mudgett, M. B., and Sattely, E. S. (2021). Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid andinactivates systemic acquired resistance in tomato. Plant Cell 33, 75–765. doi: 10.1093/plcell/koaa052
Hong, Y., Ni, S. J., and Zhang, G. P. (2020). Transcriptome and metabolome analysis reveals regulatory networks and key genes controlling barley malting quality in responses to drought stress. J. Biol. Chem. 152, 1–11. doi: 10.1016/j.plaphy.2020.04.029
Huang, X. X., Zhu, G. Q., Liu, Q., Chen, L., Li, Y. J., and Hou, B. K. (2018). Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoicacids. Plant Physiol. 176, 3103–3119. doi: 10.1104/pp.17.01530
Hulbert, S. H. (1997). Structure and evolution of the rp1 complex conferring rust resistance in maize. Ann. Rev. Phytopathol. 35, 293–310. doi: 10.1146/annurev.phyto.35.1.293
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Kaminaka, H., Nake, C., Epple, P., Dittgen, J., Schutze, K., Chaban, C., et al. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25, 4400–4411. doi: 10.1038/sj.emboj.7601312
Kamoun, S., van West, P., Vleeshouwers, V. G., de Groot, K. E., and Govers, F. (1998). Resistance of nicotianabenthamiana to phytophthorainfestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1426. doi: 10.1105/tpc.10.9.1413
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG, new perspectives on genomes, pathways, diseases and drugs. Nucl. Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kawasaki, T., Koita, H., Nakatsubo, T., Hasegawa, K., Wakabayashi, K., Takahashi, H., et al. (2006). Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPaseRac in defense signaling in rice. Proc. Natl. Acad. Sci. U. S. A. 103, 230–235. doi: 10.1073/pnas.0509875103
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT, a fast-spliced aligner with low memory requirements. Nat. Method. 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S. B., Van den, B. L., Karre, S., Choi, H., Christensen, S. A., Wang, G.-F., et al. (2021). Analysis of the transcriptomic, metabolomic and gene-regulatoryresponses to Pucciniasorghi in maize. Mol. Plant Pathol. 22, 465–479. doi: 10.1111/mpp.13040
Kong, Q., Qu, N., Gao, M., Zhang, Z., Ding, X., Yang, F., et al. (2012). TheMEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinasekinasekinase in Arabidopsis. Plant Cell 24, 2225–2236. doi: 10.1105/tpc.112.097253
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines, 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U. S. A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Li, S., Dong, X., Fan, G., Yang, Q., Shi, J., Wei, W., et al. (2018). Comprehensive profiling and inheritance patterns of metabolites in foxtail millet. Front. Plant Sci. 9:1716. doi: 10.3389/fpls.2018.01716
Li, Y., Li, P., Wang, Y., Dong, R., Yu, H., and Hou, B. (2014). Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239, 1265–1279. doi: 10.1007/s00425-014-2050-1
Liang, X., and Zhou, J. M. (2018). Receptor-like cytoplasmic kinases,central players in plant receptor kinase-mediated signaling. Ann. Rev. Plant Biol. 69, 267–299. doi: 10.1146/annurev-arplant-042817-040540
Liu, M., Li, Y.-J., Zhu, Y.-X., Sun, Y., and Wang, G.-F. (2021). Maize nicotinate N-methyltransferase interacts with the NLR protein Rp1-D21 and modulates the hypersensitive response. Mol. Plant Pathol. 22,564–579. doi: 10.1111/mpp.13044
Liu, Y., Guo, Y., Ma, C., Zhang, D., Wang, C., and Yang, Q. (2016). Transcriptome analysis of maize resistance to Fusariumgraminearum. BMC Genom. 17:477. doi: 10.1186/s12864-016-2780-5
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Luan, Q.-L., Zhu, Y.-X., Ma, S., Sun, Y., Liu, X. Y., Liu, M., et al. (2021). Maize metacaspases modulate the defense response mediated by the NLR protein Rp1-D21 likely by affecting its subcellular localization. Plant J. 105, 151–166. doi: 10.1111/tpj.15047
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., and Goodin, M. M. (2009). Transient expression in Nicotianabenthamiana fluorescent marker lines provides enhanced definition of protein localization, movement andinteractions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
McHale, L., Tan, X., Koehl, P., and Michelmore, R. W. (2006). Plant NBS-LRR proteins, adaptable guards. Genome Biol. 7:212. doi: 10.1186/gb-2006-7-4-212
Mo, X., Zhang, M., Liang, C., Cai, L., and Tian, J. (2019). Integration of metabolome and transcriptome analyses highlights soybean roots responding to phosphorus deficiency by modulating phosphorylated metabolite processes. J. Biol. Chem. 139, 697–706. doi: 10.1016/j.plaphy.2019.04.033
Mohnike, L., Rekhter, D., Huang, W., Feussner, K., Tian, H., Herrfurth, C., et al. (2021). Theglycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis andplant immunity. Plant Cell 33, 735–749. doi: 10.1093/plcell/koaa045
Monteiro, F., and Nishimura, M. T. (2018). Structural, functional, and genomic diversity of plant NLR proteins,An evolved resource for rational engineering of plant immunity. Ann. Rev. Phytopathol. 56, 243–267. doi: 10.1146/annurev-phyto-080417-045817
Morris, S. W., Vernooij, B., Titatarn, S., Starrett, M., Thomas, S., Wiltse, C. C., et al. (1998). Induced resistance responses in maize. Mol. Plant Microbe Interact. 11, 643–658. doi: 10.1094/MPMI.1998.11.7.643
Mosblech, A., Feussner, I., and Heilmann, I. (2009). Oxylipins, structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47, 511–517. doi: 10.1016/j.plaphy.2008.12.011
Murphree, C., Kim, S. B., Karre, S., Samira, R., and Balint-Kurti, P. (2020). Use of virus-induced gene silencing to characterize genes involved in modulating hypersensitive cell death in maize. Mol. Plant Pathol. 21, 1662–1676. doi: 10.1111/mpp.12999
Negeri, A., Wang, G. F., Benavente, L., Kibiti, C. M., Chaikam, V., Johal, G., et al. (2013). Characterization of temperature and light effects on the defense response phenotypes associated with the maize Rp1-D21 autoactive resistance gene. BMC Plant Biol. 13:106. doi: 10.1186/1471-2229-13-106
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Noutoshi, Y., Okazaki, M., Kida, T., Nishina, Y., Morishita, Y., Ogawa, T., et al. (2012). Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24, 3795–3804. doi: 10.1105/tpc.112.098343
Olukolu, B. A., Wang, G.-F., Vontimitta, V., Venkata, B. P., Marla, S., Ji, J., et al. (2014). A genome-wide association study of the maize hypersensitive defense response identifies genes that cluster in related pathways. PLoS Genet. 10:e1004562. doi: 10.1371/journal.pgen.1004562
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi: 10.1146/annurev-arplant-081320-092855
Prost, I., Dhondt, S., Rothe, G., Vicente, J., Rodriguez, M. J., Kift, N., et al. (2005). Evaluation of the antimicrobial activities of plant oxylipins supports theirinvolvement in defense against pathogens. Plant Physiol. 139, 1902–1913. doi: 10.1104/pp.105.066274
Radojicic, A., Li, X., and Zhang, Y. (2018). Salicylic acid: a double-edged sword for programed cell death in plants. Front. Plant Sci. 9:1133. doi: 10.3389/fpls.2018.01133
Ranjan, A., Westrick, N. M., Jain, S., Piotrowski, J. S., Ranjan, M., Kessens, R., et al. (2019). Resistance against Sclerotiniasclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 17, 1567–1581. doi: 10.1111/pbi.13082
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR, a Bioconductor package for differential expression analysis of digital gene expression data. Biogeosciences 26, 139–140. doi: 10.1093/bioinformatics/btp616
Senthil-Kumar, M., Hema, R., Suryachandra, T. R., Ramegowda, H. V., Gopalakrishna, R., Rama, N., et al. (2010). Functional characterization of three water deficit stress-induced genes in tobacco and Arabidopsis, an approach based on gene down regulation. J. Biol. Chem. 48, 35–44. doi: 10.1016/j.plaphy.2009.09.005
Smith, S., Steinau, M., Trick, H., and Hulbert, S. (2010). Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol. Genet. Genom. 283, 591–602. doi: 10.1007/s00438-010-0536-5
Sudupak, M. A., Bennetzen, J. L., and Hulbert, S. H. (1993). Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics 133, 119–125. doi: 10.1093/genetics/133.1.119
Sun, Q., Collins, N., Ayliffe, M., Smith, S. M., Drake, J., Pryor, T., et al. (2001). Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158, 423–438. doi: 10.1093/genetics/158.1.423
Sun, Y., Zhu, Y. X., Balint-Kurti, P. J., and Wang, G.-F. (2020). Fine-tuning immunity, players and regulators for plant NLRs. Trends Plant Sci. 25, 695–713. doi: 10.1016/j.tplants.2020.02.008
Tamura, K., Stecher, G., Paterson, D., Filipski, A., and Kumar, S. (2007). Mega6, molecular evolutionary genetics analysis software version 6.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Usadel, B., Poree, F., Nagel, A., Lohse, M., Czedik-Eysenberg, A., and Stitt, M. (2009). A guide to using MapMan to visualize and compare omics data in plants, a case study in the crop species, Maize. Plant Cell Environ. 32, 1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x
Vellosillo, T., Martinez, M., Lopez, M., Vicente, J., Cascon, T., Dolan, L., et al. (2007). Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19, 831–846. doi: 10.1105/tpc.106.046052
Vicente, J., Cascón, T., Vicedo, B., García-Agustín, P., Hamberg, M., and Castresana, C. (2012). Role of 9-Lipoxygenase and α-Dioxygenaseoxylipin pathways as modulators of local and systemic defense. Mol. Plant 5, 914–928. doi: 10.1093/mp/ssr105
Vlot, A. C., Dempsey, D. M. A., and Klessig, D. F. (2009). Salicylic acid, a multifacetedhormone to combat disease. Ann. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, G.-F., He, Y., Strauch, R., Olukolu, B. A., Nielsen, D., Li, X., et al. (2015b). Maize homologs of hydroxycinnamoyltransferase, a key enzyme in lignin biosynthesis, bind the nucleotide binding leucine-rich repeat Rp1 proteins to modulate the defense response. Plant Physiol. 169, 2230–2243. doi: 10.1104/pp.15.00703
Wang, G.-F., Ji, J., Ei-Kasmi, F., Dangl, J. L., Johal, G., and Balint-Kurti, P. J. (2015a). Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 11:e1004674. doi: 10.1371/journal.ppat.1004674
Wang, G. F., and Balint-Kurti, P. J. (2016). Maize homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR Rp1 protein to modulate the defense response. Plant Physiol. 171, 2166–2177. doi: 10.1104/pp.16.00224
Wang, G. F., Seabolt, S., Hamdoun, S., Ng, G., Park, J., and Lu, H. (2011). Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156, 1508–1519. doi: 10.1104/pp.111.176776
Wasternack, C. (2007). Jasmonates, an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 4, 681–697. doi: 10.1093/aob/mcm079
Yang, Q., He, Y., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., et al. (2017). A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–1372. doi: 10.1038/ng.3919
Ye, W., Liu, T., Zhang, W., Li, S., Zhu, M., Li, H., et al. (2019). Disclosure of the molecular mechanism of wheat leaf spot disease caused by bipolarissorokiniana through comparative transcriptome and metabolomics analysis. Int. J. Mol. Sci. 20:6090. doi: 10.3390/ijms20236090
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediatedplant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zavaliev, R., Mohan, R., Chen, T., and Dong, X. (2020). Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell 182, 1093–1108 e1018. doi: 10.1016/j.cell.2020.07.016
Zeilmaker, T., Ludwig, N. R., Elberse, J., Seidl, M. F., Berke, L., Van Doorn, A., et al. (2015). Downy Mildew Resistant 6 and DMR6-Like Oxygenase 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222. doi: 10.1111/tpj.12719
Zhang, Y., Zhao, L., Zhao, J., Li, Y., Wang, J., Guo, R., et al. (2017). S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 175, 1082–1093. doi: 10.1104/pp.17.00695
Zheng, X., Zhou, M., Yoo, H., Pruneda-Paz, J., Spivey, N., Kay, S., et al. (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. U. S. A. 112, 9166–9173. doi: 10.1073/pnas.1511182112
Zhou, J. M., and Zhang, Y. (2020). Plant immunity, danger perception and signaling. Cell 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Zhou, P., Li, Z., Magnusson, E., Gomez Cano, F., Crisp, P. A., Noshay, J. M., et al. (2020). Meta gene regulatory networks in maize highlight functionally relevant regulatory interactions. Plant Cell 32, 1377–1396. doi: 10.1105/tpc.20.00080
Keywords: disease resistance, ETI, hypersensitive response, maize, NLR, salicylic acid, UGT
Citation: Ge C, Wang Y-G, Lu S, Zhao XY, Hou B-K, Balint-Kurti PJ and Wang G-F (2021) Multi-Omics Analyses Reveal the Regulatory Network and the Function of ZmUGTs in Maize Defense Response. Front. Plant Sci. 12:738261. doi: 10.3389/fpls.2021.738261
Received: 08 July 2021; Accepted: 26 August 2021;
Published: 24 September 2021.
Edited by:
Meixiang Zhang, Nanjing Agricultural University, ChinaReviewed by:
Qinhu Wang, Northwest A and F University, ChinaCopyright © 2021 Ge, Wang, Lu, Zhao, Hou, Balint-Kurti and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guan-Feng Wang, Z2Z3YW5nQHNkdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.