- 1Key Laboratory of Horticultural Plant Biology of Ministry of Education, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, China
- 2Tobacco Research Institute, Chinese Academy of Agricultural Science, Qingdao, China
- 3Hubei Engineering Technology Research Center for Forestry Information, Huazhong Agricultural University, Wuhan, China
As a member of the CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (CLE) family, the dodecapeptide tracheary element differentiation inhibitory factor (TDIF) has a major impact on vascular development in plants. However, the influence of polymorphisms in the TDIF peptide motif on activity remains poorly understood. The model plant, Arabidopsis provides a fast and effective tool for assaying the activity of TDIF homologs. Five TDIF homologs from a group of 93 CLE genes in switchgrass (Panicum virgatum), a perennial biomass crop, named PvTDIF-like (PvTDIFL) genes were studied. The expression levels of PvTDIFL1, PvTDIFL3MR3, and PvTDIFL3MR2 were relatively high and all of them were expressed at the highest levels in the rachis of switchgrass. The precursor proteins for PvTDIFL1, PvTDIFL3MR3, and PvTDIFL3MR2 contained one, three, and two TDIFL motifs, respectively. Treatments with exogenous PvTDIFL peptides increased the number of stele cells in the hypocotyls of Arabidopsis seedlings, with the exception of PvTDIFL_4p. Heterologous expression of PvTDIFL1 in Arabidopsis strongly inhibited plant growth, increased cell division in the vascular tissue of the hypocotyl, and disrupted the cellular organization of the hypocotyl. Although heterologous expression of PvTDIFL3MR3 and PvTDIFL3MR2 also affected plant growth and vascular development, PvTDIFL activity was not enhanced by the multiple TDIFL motifs encoded by PvTDIFL3MR3 and PvTDIFL3MR2. These data indicate that in general, PvTDIFLs are functionally similar to Arabidopsis TDIF but that the processing and activities of the PvTDIFL peptides are more complex.
Introduction
In plants, the CLAVATA3 (CLV3)/EMBRYO SURROUNDING REGION (CLE) gene family plays a vital role in cell division and cell differentiation by mediating intercellular communication (Fletcher et al., 1999; Fiers et al., 2005, 2007). CLE peptides are derived from a pre-propeptide containing an N-terminal signal peptide and a conserved C-terminal motif of 12-13 amino acids. A few CLE genes encode multiple CLE motifs in rice, wheat (Triticum aetivum), Picea, and Amborella, etc. (Goad et al., 2017). CLE genes are ubiquitous in the plant kingdom, and have been comprehensively studied in eudicots (Arabidopsis thaliana, Glycine max, Lycopersicon esculentum, Gossypium hirsutum, Populus trichocarpa, and Vitis vinifera), monocots (Oryza sativa, T. aestivum, and Zea mays), and gymnosperms (Picea abies and P. glauca) (Cock and McCormick, 2001; Sawa et al., 2008; Strabala et al., 2014; Han et al., 2016; Goad et al., 2017; Wang et al., 2019). In A. thaliana, at least 32 CLE genes have been identified (Ito et al., 2006; Kondo et al., 2006; Ohyama et al., 2009; Ogawa-Ohnishi et al., 2013). Based on their biological functions, the Arabidopsis CLE peptides were grouped into two major classes. The A-type CLE peptides which maintain the apical meristems in shoots and roots and include CLV3, CLE1-27, and CLE40. The B-type/H-type CLE peptides, on the other hand, suppress xylem differentiation and promote vascular procambial cell division and include CLE41, CLE42, and CLE44 (Schoof et al., 2000; Ito et al., 2006; Whitford et al., 2008; Stahl et al., 2009; Hirakawa et al., 2010a,b).
One member of the CLE family, a dodecapeptide (H-E-V-P-S-G-P-N-P-I-S-N) named tracheary element differentiation inhibitory factor (TDIF) was first identified in a Zinnia elegans mesophyll cell xylogenesis system. H1 is unique among CLE peptides with TDIF activity, and V3, N8, and N12 are essential for TDIF activity (Ito et al., 2006). In Arabidopsis, TDIF is encoded by two genes, CLE41 and CLE44. CLE42 and CLE46 are classified as TDIF-like (TDIFL) genes, because their CLE motifs (H-G-V-P-S-G-P-N-P-I-S-N and H-K-H-P-S-G-P-N-P-T-G-N, respectively) are highly homologous to the TDIF motif. The TDIF genes are mainly involved in the regulation of vascular development. The TDIF signaling pathway in the vascular meristem has been well-studied (Ito et al., 2006; Ohyama et al., 2008). The TDIF RECEPTOR/PHLOEM INTERCALATED WITH XYLEM (TDR/PXY), a member of the leucine-rich repeat receptor-like kinase (LRR-RLK) family, is the TDIF receptor in procambial cells (Fisher and Turner, 2007; Hirakawa et al., 2008). The TDIF peptide that is produced in the phloem controls vascular procambial or cambial cell proliferation and xylem differentiation by activating TDR/PXY in the procambium or cambium. Two transcription factors, WUSCHEL HOMEOBOX RELATED 4 (WOX4) and WOX14, regulate the proliferation of plant vascular tissue by acting downstream of the TDIF–TDR/PXY signaling pathway (Hirakawa et al., 2010a,b; Etchells et al., 2013; Kucukoglu et al., 2017; Li et al., 2018). In addition, an NAC domain transcription factor, XVP, fine-tunes the TDIF signaling that contributes to vascular development by serving as a negative regulator (Kucukoglu, 2020; Yang et al., 2020). Current reports on TDIF/TDIFL have focused on genes encoding a single motif. Little is known about the function of TDIF/TDIFL genes encoding multiple motifs.
Switchgrass (P. virgatum) is a highly productive herbaceous perennial that thrives in diverse environments and is, therefore, a model perennial biomass crop (PBC) (Sanderson et al., 1996; McLaughlin and Adams Kszos, 2005). It has been a feedstock for the biofuels and specialty chemicals (Parrish and Fike, 2005; Sanderson et al., 2006; Keshwani and Cheng, 2009). Among the four PBCs (poplar, switchgrass, Miscanthus, and Salix), molecular regulation of lignocellulosic formation has been comprehensively studied in poplar trees (Clifton-Brown et al., 2019). Increasing vascular cambial activity results in incremental xylem and phloem, therefore increasing the biomass of wood. The division of vascular cambial cells was stimulated by manipulating CLE41–PXY signaling in hybrid poplar (Etchells and Turner, 2010; Etchells et al., 2015). The WOX4 controls the rate of cambial cell division and hence, the growth of stem girth in a TDIF-dependent manner (Kucukoglu et al., 2017). In a recent report, the CLE gene family in switchgrass, including three PvTDIFL genes, was defined. The statistical analysis demonstrated that no common TDIF/TDIFL motif is shared between monocots and dicots and that the contribution of TDIF/TDIFL to the development of vascular tissue has probably been diverged in monocots and dicots (Zhang et al., 2020). However, the knowledge of TDIF/TDIFL peptide functions in switchgrass remains limited.
Studying the mechanisms that drive rapid growth in switchgrass and other PBCs may help to introduce biomass-related genes, pathways or regulatory modules into dicotyledonous plants. Arabidopsis is widely used in the study of secondary cell wall formation and vascular development because although Arabidopsis is an annual herbaceous plant, it has most of the cell types associated with secondary growth (Zhang et al., 2011; Ursache et al., 2013; Ragni and Hardtke, 2014). The TDIF–TDR/PXY signaling pathway has been well-studied in Arabidopsis (Etchells and Turner, 2010; Hirakawa et al., 2010a,b; Etchells et al., 2013, 2016; Wang et al., 2013; Kondo and Fukuda, 2015; Zhang et al., 2016; Kucukoglu, 2020). Thus, Arabidopsis is suitable for activity screening and functional analysis of TDIF/TDIFL genes from various plants. This study aimed to use bioinformatics tools to identify the CLE gene family, including TDIF/TDIFL genes, at the whole genome level in P. virgatum. Treatment of Arabidopsis with exogenous TDIF/TDIFL peptides and heterologous expression of TDIF/TDIFL genes in Arabidopsis allowed the researchers of this study to test whether particular PvTDIFL peptides influenced plant growth and vascular development. Rapid characterization of PvTDIFL peptides in Arabidopsis can provide a preliminary understanding of the biological activities of PvTDIFL peptides and indicate potential biotechnological applications related to wood formation and biomass improvement. This is not practical to achieve with switchgrass because the genetic transformation of switchgrass is still time-consuming and laborious (Xi et al., 2009; Chen and Song, 2019; Ondzighi-Assoume et al., 2019).
Materials and Methods
Identification of CLE Genes in P. virgatum
The amino acid sequences of Arabidopsis CLE proteins were downloaded from The Arabidopsis Information Resource (TAIR) (https://www.Arabidopsis.org/). The 12-amino acid CLE motifs were used as queries in TBLASTN search of the P. virgatum v1.1 genome with a threshold e-value of 500 in the Phytozome v12.1 database (https://phytozome.jgi.doe.gov/pz/portal.html) (Goodstein et al., 2012). Repeated hits were removed and only one hit at each chromosomal location was kept. Genes at each chromosomal location were identified as potential CLE genes. If there was no annotated gene at the target location, a 5-kb genome fragment with the target location in the center was then retrieved and subjected to an analysis with the online software, FGENESH, for gene predictions on the softberry website (http://linux1.softberry.com/) (Solovyev et al., 2006). Amino acid sequences encoded by each of the potential CLE genes were checked for the C-terminal conserved CLE motifs. All of the newly identified motifs were used as queries in TBLASTN searches, as described in the preceding steps. The analysis was iterated until no more CLE candidate could be identified. All of the CLE candidates were compared with the previously reported CLE genes in P. virgatum (Zhang et al., 2020). The final TDIF/TDIFL gene sequences were obtained using cloning based on the nucleotide sequences from Phytozome (Supplementary Figure 1).
Bioinformatics Analysis
To classify PvCLE genes, all 12-aa CLE motifs encoded by AtCLE and PvCLE genes were extracted, including the PvCLE genes encoding multiple CLE motifs. Phylogenetic trees containing the 12-aa CLE motif sequences were constructed as previously described (Zhang et al., 2020). Based on the clustering of CLE motifs, PvCLE genes in the same group as AtTDIF/TDIFL genes were predicted as PvTDIFL genes. To further analyze the evolutionary relationship between PvTDIFL and AtTDIF/TDIFL genes, phylogenetic trees containing the full-length amino acid sequences for TDIF/TDIFL from both P. virgatum and Arabidopsis were constructed using the MEGA 5.05 software (Hall, 2013) with the following parameters: alignment, Muscle, phylogeny construct or test, Maximum Likelihood Tree, and number of bootstrap replication = 1000. Signal peptides that target the TDIF/TDIFL proteins to the secretary pathway were predicted with the SMART Server using a normal model (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) (Letunic and Bork, 2018). Gene structure analysis was performed using the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). The Format of Gene Features was set as fast-all (FASTA) Sequence. Other features containing signal peptides and motifs were uploaded in Browser Extensible Data (BED) format. Phylogenetic Tree was uploaded in Newick format based on full-length amino acid sequences (Hu et al., 2015). Multiple alignments were performed using the DNAMAN v6.0 software (Lynnon Biosoft, Quebec, Canada). Weblogo-Create Sequence Logos (http://weblogo.berkeley.edu/logo.cgi) was used for comparative analysis of motif conservation and for conservation analysis of each amino acid site of TDIF/TDIFL motifs in Arabidopsis and switchgrass (Crooks et al., 2004). Isoelectric point (pI) and molecular weight (MW) were calculated using the ExPASy-ProtParam tool (http://web.expasy.org/protparam/) (Bjellqvist et al., 1993).
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) and P. virgatum were used in this study. Arabidopsis seeds were surface sterilized in 75% (v/v) ethanol for 1 min and 5% (v/v) sodium hypochlorite for 15 min, with occasional gentle shaking. The seeds were then washed five times with sterilized distilled H2O (dH2O). After stratification at 4°C for 2 days, the seeds were placed in liquid Murashige and Skoog (MS) medium (pH 5.8) containing 1.5% sucrose (w/v) or on solid MS medium (pH 5.8) containing 1.5% sucrose (w/v) and 0.7% plant agar (w/v). The conical flasks containing liquid medium and the plates containing solid medium were placed in a tissue culture room maintained at 23°C and grown in a photoperiod containing 16 h of light at a fluence rate of 100 μmol photons m−2 s−1 followed by 8 h of dark. The conical flasks were placed on an orbital shaker at a rotational speed of 80 rpm. The plates were positioned vertically. Arabidopsis were grown in soil under the same temperature and light conditions, except that light intensity was set as 150 μmol photons m−2 s−1. The P. virgatum plants used in this study were grown at Huazhong Agricultural University, Wuhan, China.
Exogenous Peptide Treatment
The TDIF/TDIFL peptides derived from Arabidopsis TDIF and PvTDIFL motifs (Supplementary Table 1) were chemically synthesized by GenScript Biotech Corporation (Nanjing, China). Twenty milligrams of each peptide was provided at a purity ≥ 90% (w/w). Peptides were dissolved in double distilled water (ddH2O) at a stock concentration of 1 mg/ml and stored at −80°C for future use. Synthetic TDIF and PvTDIFL peptides were added to liquid medium at a final concentration of 10 μM as previously described (Whitford et al., 2008). Arabidopsis seeds were treated with 10 ml of medium in conical flasks on an orbital shaker at a rotational speed of 80 rpm for 10 days. Each treatment required six flasks, with six seeds per flask.
RNA Extraction and qRT-PCR Analysis of PvTDIFL Genes
Five different tissues of P. virgatum, namely, seed (mature seeds), rachis (bearing mature seeds), leaf (fully expanded leaf), stem (middle internode of seedling), and root were sampled in triplicates and immediately frozen in liquid nitrogen. Total RNA was extracted using the 2 × CTAB method (Li et al., 2008). The pellet was dissolved in 30 μl of RNase free ddH2O. The concentration and quality of RNA were measured with a NanoDrop® 2000 spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA). The RNA samples with A260/A280 ratios that ranged from 1.8 to 2.1 and A260/A230 ratios ≥2 were stored at −80°C for future use.
To quantify the relative expression levels of PvTDIFL genes in different samples, 1 μg of total RNA was used as a template to synthesize cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Dalian, China). The qRT-PCR reactions were then prepared using 2 × HSYBR qPCR Mix without ROX (ZOMANBIO, Beijing, China). A standard 2-step amplification protocol was run in a LightCycler® 96 Real-Time PCR System (Roche, USA). The comparative CT method (ΔΔCt method) was used for relative quantification of real-time PCR (Pfaffl, 2001). PvUBQ6 (Pavir.5NG345900) was used as the reference gene (Gimeno et al., 2014). All gene specific primers (Supplementary Table 2) were designed using the Primer Premier 5.0 software (www.PremierBiosoft.com). All real-time PCR reactions were run in three biological replicates and two technical replicates.
Plasmid Construction and Generation of Transgenic Plants
The coding sequences (CDS) from PvTDIFL3MR3 and PvTDIFL3MR2 were amplified using PCR with gene specific primers (Supplementary Table 2). The CDS of PvTDIFL1 was artificially synthesized (Sunny Technology, Shanghai, China). The products were cloned into a Gateway™ entry vector pDONR201 using the BP recombination reaction (Invitrogen). The clones were analyzed using DNA sequencing. The correct fragments were subsequently recombined into the destination vector, pK2GW7 (Karimi et al., 2002). Transformation of A. thaliana was performed using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998).
Morphological and Histological Analysis of Arabidopsis Seedlings
Arabidopsis seedlings were photographed with a Canon EOS 7D digital camera to obtain high-resolution images. Root length was measured by using the ImageJ software (http://imagej.nih.gov/ij). The images presented in the figures were representative images from images of 24 individual seedlings for each line for the plate-grown plants and 10 individual seedlings for each line for the soil-grown plants, respectively.
Histological analysis of the vasculature was performed by using semi-thin sections. The upper part of the hypocotyl, 2–3 mm in length, was cut with a scalpel. A small amount of tissue from the stem and petiole of the rosette was kept to mark the upper end of the hypocotyl segment. The hypocotyl segments were fixed in formalin-acetic acid-alcohol (FAA) fixative (50% ethanol, 10% glacial acetic acid, 5% formaldehyde, v/v/v) at 4°C for 24 h. Dehydration was carried out by immersing the specimen in an ascending series of ethanol. Lastly, the specimens were infiltrated and embedded with Technovit® 7100 resin (www.kulzer-technik.com). The hypocotyl specimens were transversely sectioned at a thickness of 2.5 μm using a RM2265 Microtome (Leica BIOSYSTEMS, Nussloch, Germany) beginning from the upper end. The rosette tissues were first trimmed and monitored by sectioning and microscopy. The 10th to 20th sections of the hypocotyls were stained with 0.05% (w/v) aqueous toluidine blue and visualized with a BX53 light microscope (OLYMPUS, Tokyo, Japan). The cells in the stele were marked by using the ImageJ software and the number of cells was counted manually.
Statistical analysis was performed with six biological replicates and a t-test was used with double sample variance assumption. The letters a, b, and c and asterisks indicate statistically significant differences relative to the control.
Accession Numbers
Sequence data from this article are from the P. virgatum v1.1 genome in the Phytozome v12.1 database and TAIR10 associated with the following accession numbers: AtEF1-α (At5G60390), PvUBQ6 (Pavir.5NG345900), AtCLE41 (AT3G24770), AtCLE42 (AT2G34925), AtCLE44 (AT4G13195), AtCLE46 (AT5G59305), PvTDIFL1 (Pavir.Ab03264), PvTDIFL2 (Pavir.Aa00134), and PvTDIFL4 (Pavir.J06189). The sequence data for PvTDIFL3MR3 and PvTDIFL3MR2 has been submitted to GenBank and are associated with accession numbers MZ463198 and MZ463199, respectively.
Results
Identification of CLE Genes in P. virgatum
To identify CLE genes in P. virgatum (PvCLE), sequences similar to the Arabidopsis CLE motifs were identified in the P. virgatum v1.1 genome (https://phytozome.jgi.doe.gov/pz/portal.html). The PvCLE genes were predicted and clustered using the recently reported method (Zhang et al., 2020). In total, 93 PvCLE genes were identified, including 91 annotated PvCLEs in the P. virgatum v1.1 genome and two novel PvCLEs identified in this study (Figure 1, Supplementary Table 3). Since there were three PvCLEs that encoded multiple CLE motifs, all PvCLEs encoded a total of 97 CLE motifs. The total number of PvCLEs was nearly three times of the number of AtCLEs. Based on the clustering, PvCLEs were divided into six groups (Figure 1A, Supplementary Figure 2). The Weblogo of CLE motifs showed that Group 1, including eight AtCLEs (AtCLE1-7, and AtCLV3) and 21 PvCLEs, had two conserved amino acids, namely, Asp (D) at the 8th residue and His (H) at 12th residue. These two residues were replaced with Asn (N) in Group 3, which contained 13 AtCLEs (AtCLE8-14, AtCLE16-17, and AtCLE19-22) and 38 PvCLEs. Residue number 12 was Asn (N) in Group 2, which contained five AtCLEs (AtCLE18, AtCLE25-27, and AtCLE45) and 20 PvCLEs (Supplementary Figure 2). The Pavir.Fa00904.1 from Group 2 encoded two repeats of RRVRRGSDPIHN, therefore the Group 2 PvCLEs encoded a total of 21 CLE motifs. Group 4 was comprised of nine TDIF/TDIFL genes, with four members from Arabidopsis (AtCLE41, 42, 44, and 46) and five genes from P. virgatum. In Group 4, there were two PvCLEs encoding multiple TDIFL motifs (see the next paragraph for details), and the total number of TDIFL motifs encoded in P. virgatum was as many as eight. Group 5 had two Arabidopsis members (AtCLE40 and AtCLE43) and three PvCLEs. In general, the motif conservation in Group 5 was lower relative to groups 1 through 4. Six PvCLE genes were found upon encoding CLE motifs that are very different from the AtCLE motifs and thus, fell into “Group others” (Figure 1A, Supplementary Table 3). The CLE motifs are rather conserved between the AtCLEs and PvCLEs (Figures 1B,C).
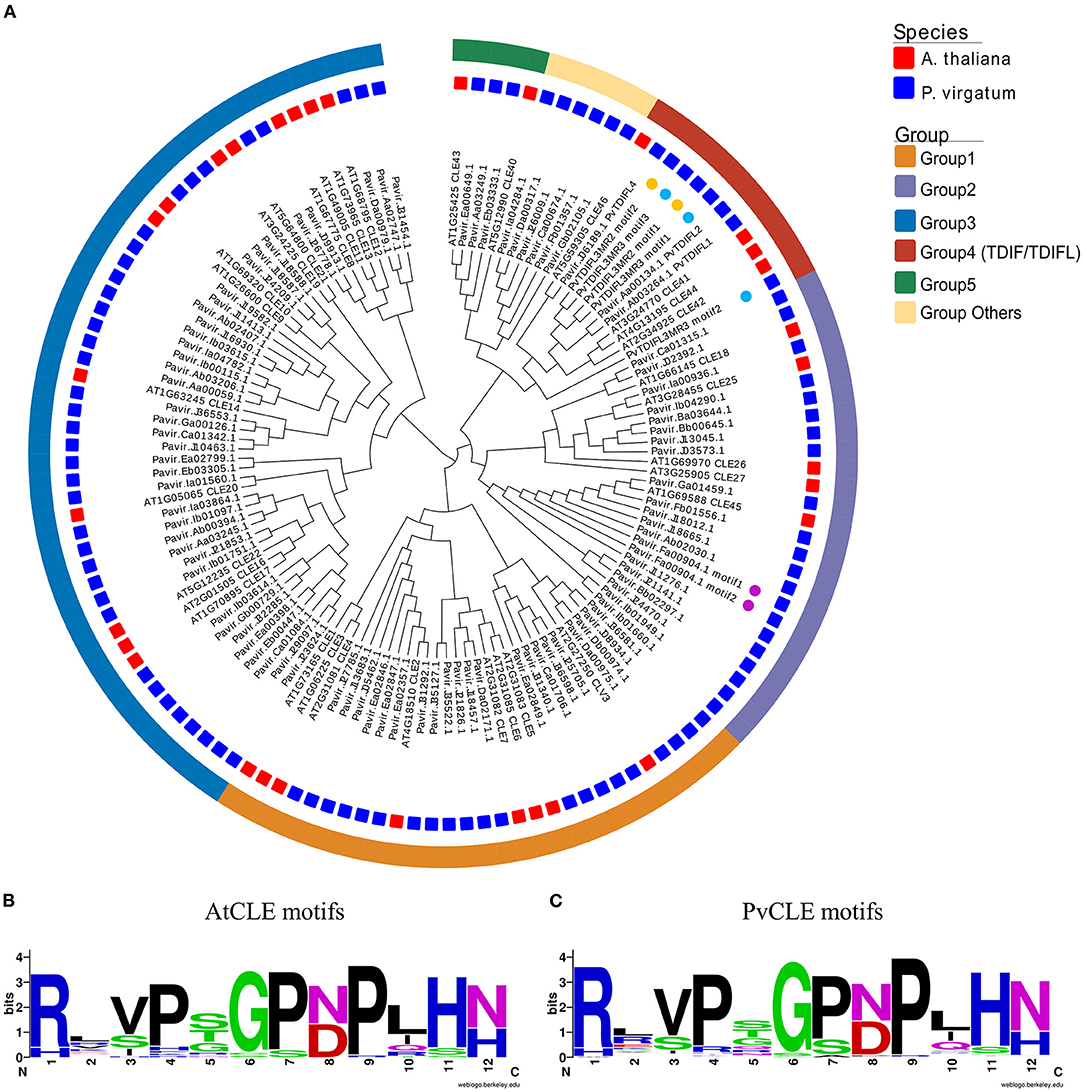
Figure 1. Phylogenetics and conservation analysis of CLE genes in P. virgatum and A. thaliana. (A) Phylogeny of 12-aa motif sequences from PvCLEs and AtCLEs. The phylogenetic tree was constructed as previously described (Zhang et al., 2020). Red and blue quadrates in the middle layer indicate AtCLEs and PvCLEs, respectively. Colored boxes in the outermost layer indicate different groups; colored dots in inner layer indicate genes with multiple motifs. (B,C) Weblogo images of CLE motifs from AtCLEs (B) and PvCLEs (C), created by using the Weblogo online tool with the amino acid sequences from CLE peptides.
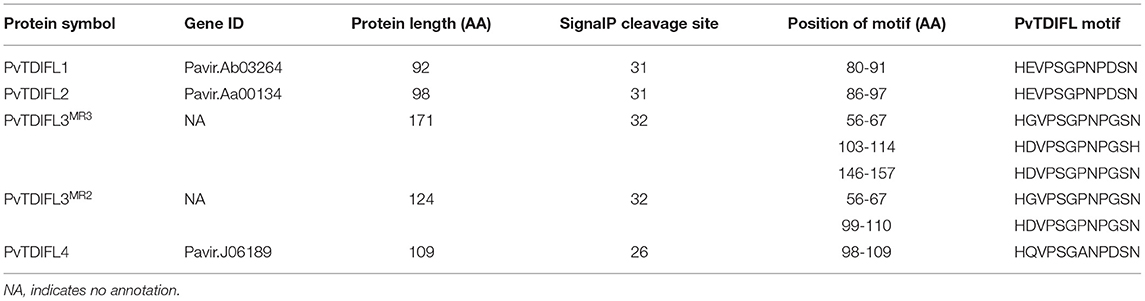
In total, five TDIF homologous genes were identified in the P. virgatum genome (Figure 2, Supplementary Figure 1, Table 1). The three TDIF homologs that were annotated are Pavir.Ab03264, Pavir.Aa00134, and Pavir.J06189, namely PvTDIF-like1 (PvTDIFL1), PvTDIFL2, and PvTDIFL4 in this study, respectively. A novel TDIFL gene, PvTDIFL3, was identified from TBLASTN searches of the P. virgatum v1.1 genome with a threshold e-value of 500. The predicted PvTDIFL3 protein product contained three potential TDIFL motifs and hence named PvTDIFL3MR3. The MR3 is referring to that it has three motif repeats (Figure 2, Table 1). When gene-specific primers were used to amplify the CDS of PvTDIFLs, a shorter fragment derived from PvTDIFL3MR3 was also cloned that was named PvTDIFL3MR2 (Figures 2A–C). The amino acid sequence of PvTDIFL3MR2 is identical to that of PvTDIFL3MR3, except that 47 amino acid residues are missing from the middle. The missing 47 amino acid residues constitute the second TDIFL motif and its flanking sequences of PvTDIFL3MR3 (Figure 2C). The PvTDIFL3MR2 matched to the same chromosomal location as PvTDIFL3MR3 in TBLASTN search of the P. virgatum v1.1 genome. In a phylogenetic analysis, amino acid sequences from the five PvTDIFL genes clustered in the same clade as the amino acid sequences from Arabidopsis TDIF/TDIFL genes (Figures 1, 2A). The nucleotide sequences were subjected to the online gene prediction program FGENESH on the Softberry website (http://linux1.softberry.com/), to evaluate the genomic sequence and gene structure of these five genes.
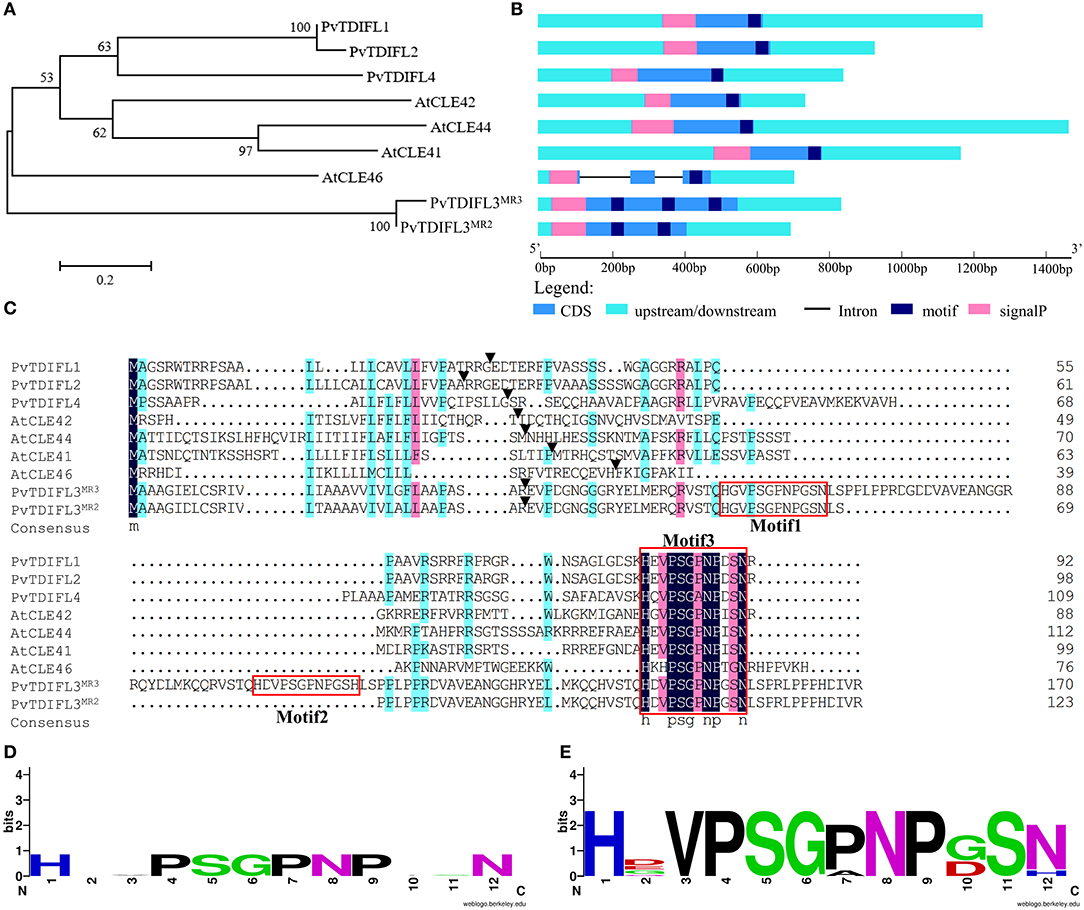
Figure 2. Phylogenetic analysis and genomic organization of tracheary element differentiation inhibitory factor (TDIF) in P. virgatum and A. thaliana. (A) Phylogeny of full-length protein sequences from PvTDIFLs and AtTDIF. The alignment was generated by using Muscle with the Maximum Likelihood method and 1,000 bootstrap iterations. Numbers on the nodes indicate clade credibility values. (B) Gene structures of PvTDIFLs and AtTDIF from nucleotide sequences. Gene structure analysis was performed using the gene structure display server 2.0. Cyan box, 3′ UTR and 5′ UTR; blue box, CDS; pink box, signal peptide; navy-blue box, TDIF peptide; black line, intron. (C) Multiple amino acid sequence alignment of PvTDIFL and AtTDIF proteins. Multiple alignments were performed using the DNAMAN v6.0 software. TDIF peptides are boxed in red. Black triangles indicate the SignalP cleavage site. (D,E) Weblogo images of AtTDIF (D) and PvTDIFL (E). The images were created by using the Weblogo online tool with the amino acid sequences from AtTDIF and PvTDIFL peptides, respectively.
All the TDIF/TDIFL proteins from Arabidopsis and switchgrass contained a signal peptide for the secretory pathway at the N-terminus (Figures 2B,C). Similar to the Arabidopsis TDIF proteins, PvTDIFL1, 2, and 4 had a single TDIFL motif at the C-terminus. However, PvTDIFL3MR3 and PvTDIFL3MR2 had three and two TDIFL motifs, respectively (Figures 2B,C). The PvTDIFL proteins ranged from 92 to 175 amino acid residues in length. Their theoretical molecular weights (MW) and isoelectric points (pI) ranged from 10.01 to 18.50 kDa and from 5.81 to 11.94, respectively (Table 1).
A multiple sequence alignment analysis revealed that PvTDIFL1 and 2 proteins shared the same motif, HEVPSGPNPDSN, which is different from the Arabidopsis TDIF motif at the 10th residue in that an Ile (I) residue is changed to an Asp (D) residue. Motif 1 and motif 3 of PvTDIFL3MR3/PvTDIFL3MR2 were different at the 2nd and 10th residues relative to the TDIF motif. The 12th residue of Motif 2 from PvTDIFL3MR3 had an Asn (N) to His (H) substitution. The 2nd, 7th, and 10th residues of the motif from PvTDIFL4 were different from the TDIF motif (Figure 2C). The conserved TDIF/TDIFL dodecapeptides in Arabidopsis and switchgrass were compared by using a WebLogo analysis. Similar to the Arabidopsis TDIF, the 4th and 7th residues of the TDIFL motifs in PvTDIFL proteins were conserved Pro (P) residues. However, their 2nd, 10th and 12th residues were less conserved (Figures 2D,E).
Tissue-Specific Expression Analysis of PvTDIFL Genes in P. virgatum
In Arabidopsis, CLE41 is expressed in the phloem and the neighboring pericycle cells in the root and hypocotyl. CLE44 is expressed in the phloem, pericycle and endodermal cells (Hirakawa et al., 2008). To analyze the expression pattern of PvTDIFL genes in the different tissues of switchgrass, in silico expression analysis and qRT-PCR were both performed.
The in silico expression data for PvTDIFL1, 2, and 4 were downloaded from Phytozome. PvTDIFL1 and 2 were both expressed at the highest levels in the inflorescence panicle, rachis and shoot. The PvTDIFL3MR3 and PvTDIFL3MR2 were not annotated in Phytozome and thus, no in silico expression data was available. The expression of PvTDIFL4 was barely detectable in the tissues studied (Figure 3A).
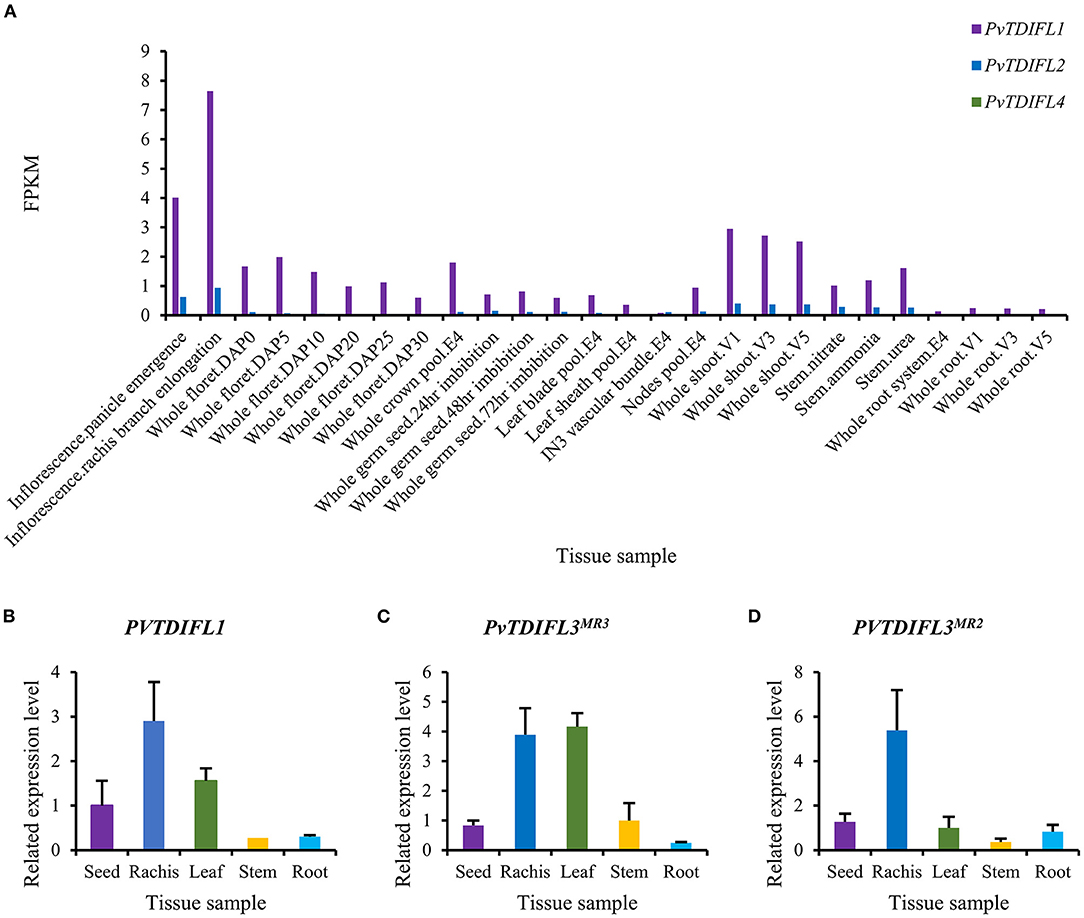
Figure 3. Expression analysis of PvTDIFL genes in P. virgatum. (A) Expression of PvTDIFL1, 2 and 4 in different samples of P. virgatum. Fragments Per Kilobase per Million (FPKM) values of PvTDIFL1, 2, and 4 were obtained from in silico expression data in Phytozome. Samples listed are as follows: Inflorescence.Panicle emergence, Inflorescence.rachis branch elongation, Whole floret.DAP0, 5, 10, 20, 25, 30, Whole crown pool.E4, Whole germ seed 24 h imbibition, 48 h imbibition, 72 h imbibition, Leaf blade pool.E4, Leaf sheath pool.E4, IN3 vascular bundle.E4, Nodes pool.E4, Whole shoot.V1, 3, 5, Stem.nitrate, Stem.ammonia, Stem.urea, Whole root system.E4, Whole root.V1, 3, 5. (B–D) Relative expression levels of PvTDIFL1 (B), PvTDIFL3MR3 (C), and PvTDIFL3MR2 (D) quantified using qRT-PCR. Seed (mature), rachis (bearing mature seeds), leaf (fully expanded leaf), stem (middle internode), and root of switchgrass grown on campus were collected in triplicate in July. PvUBQ6 was used as the reference gene. The comparative CT method (ΔΔCt method) was used for relative quantification of real-time PCR.
For qRT-PCR, five switchgrass tissue samples were collected in triplicates. These tissues included seed, rachis, leaf, stem, and root. The PveEF-1α, PvACT12, and PvUBQ6 were evaluated as internal reference genes (Gimeno et al., 2014). The PvUBQ6 was selected because of its uniform expression levels in different tissues. The qRT-PCR results showed that the expression levels of PvTDIFL1 were in the rachis, which is consistent with the in silico data (Figure 3B). The expression of PvTDIFL3MR3 was relatively high in the rachis and leaf. In contrast, the expression of PvTDIFL3MR2 was more specific to the rachis (Figures 3C,D). However, the expression of PvTDIFL2 and PvTDIFL4 was undetectable, probably due to low levels of expression. The relatively high levels of expression in the rachis provides evidence for PvTDIFL genes contributing to vascular development, similar to the Arabidopsis TDIF genes.
In vitro PvTDIFL Peptide Treatment Increased Hypocotyl Stele Cell Numbers in Arabidopsis
Exogenous TDIF peptide promotes procambial cell divisions, which leads to a remarkable increase in the vascular development of the hypocotyl in Arabidopsis (Hirakawa et al., 2010a). To study the activity and functional conservation of PvTDIFL peptides, four types of PvTDIFL peptides corresponding to the TDIFL motifs from PvTDIFL1, PvTDIFL3MR3 and PvTDIFL3MR2 were chemically synthesized and exogenously applied to Arabidopsis seedlings, at a concentration of 10 μM. The Arabidopsis TDIF peptide was used as the positive control (Whitford et al., 2008).
Arabidopsis seedlings were grown in liquid MS media either with or without TDIF/TDIFL peptides for 10 d. To test whether these peptide treatments affected the vasculature of the hypocotyl, semi-thin transverse sections were prepared. The results showed that the application of Arabidopsis TDIF peptides induced increases in the size of the stele with significant increases in cell numbers, as previously reported (Figures 4A,B,G) (Whitford et al., 2008). Similarly, the number of cells in the stele increased significantly in the seedlings treated with PvTDIFL_1p, 2p, and 3p, but not as much as in the TDIF-treated seedlings (Figures 4A–E,G). In contrast, the PvTDIFL_4p treatment caused an unexpected decrease in the number of cells in the stele, due to the His (H) substitution for Asn (N) at the 12th residue of PvTDIFL_4p (Figures 4A,F,G). These data indicate that the activities of the three peptides (PvTDIFL_2p, 3p, and 4p) encoded by PvTDIFL3 diverged.
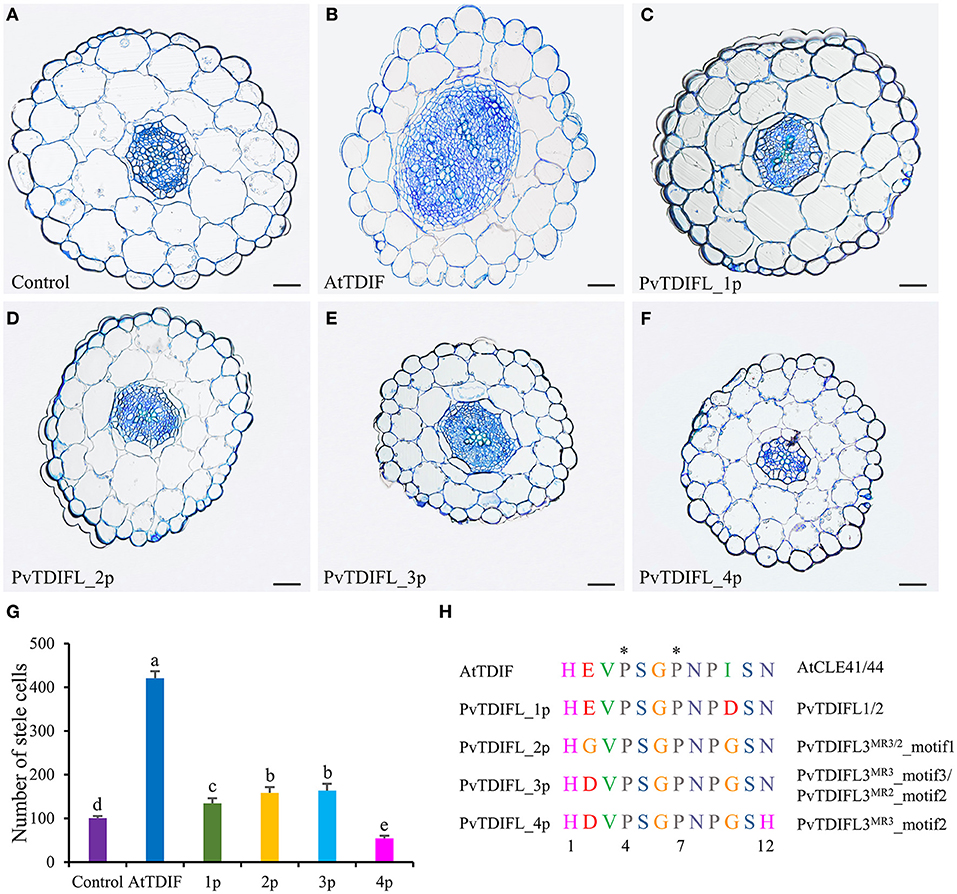
Figure 4. Effects of AtTDIF and PvTDIFL peptides on stele development of 10-day-old Arabidopsis hypocotyls grown in liquid media. (A) Transverse section of hypocotyl from Arabidopsis plants without peptide-treatment. (B–F) Transverse sections of hypocotyls from Arabidopsis plants treated with the AtTDIF (B), PvTDIFL_1p (C), PvTDIFL_2p (D), PvTDIFL_3p (E), and PvTDIFL_4p (F) synthetic peptides. Sections were 2.5 μm thick and stained with 0.05% (w/v) aqueous toluidine blue. Bar = 50 μm. (G) Numbers of stele cells in the hypocotyls of non-treated and synthetic peptide-treated plants. Biological replicates were sections from 6 different hypocotyls. Different letters indicate statistically significant differences (p < 0.05), n = 6. (H) Sequences and modifications of AtTDIF and PvTDIFL peptides. Peptide names are indicated at the left. Proteins containing the indicated peptides are indicated at the right. The asterisk indicates hydroxyproline.
Influence of Heterologous Expression of PvTDIFL Genes on Plant Morphology and Vascular Development in Arabidopsis
As described above, PvTDIFL1, 3MR3 and 3MR2 differ in their motif numbers, motif sequences and peptide activities. To further investigate their functions in plant development, the 35S promoter was used to drive the expression of PvTDIFL1, 3MR3 and 3MR2 in Arabidopsis. The relative expression levels of the PvTDIFL transgenes in Arabidopsis were analyzed by RT-PCR (Figure 5A). The transgene expression levels and morphologies of two representative lines for each transgene were compared to Col-0. Four-week-old soil-grown plants were photographed (Figures 5B–H). In comparison with the wild-type plants (Figure 5B). Both lines harboring the 35S:PvTDIFL1 transgene developed smaller rosettes with small, round, and bushy leaves relative to Col-0 (Figures 5B–D). Similar results were obtained when AtCLE42 and 44 were overexpressed in Arabidopsis (Strabala et al., 2006). In contrast, the morphological changes of 35S:PvTDIFL3MR3 and 35S:PvTDIFL3MR2 plants were less severe and less consistent (Figures 5E–H).
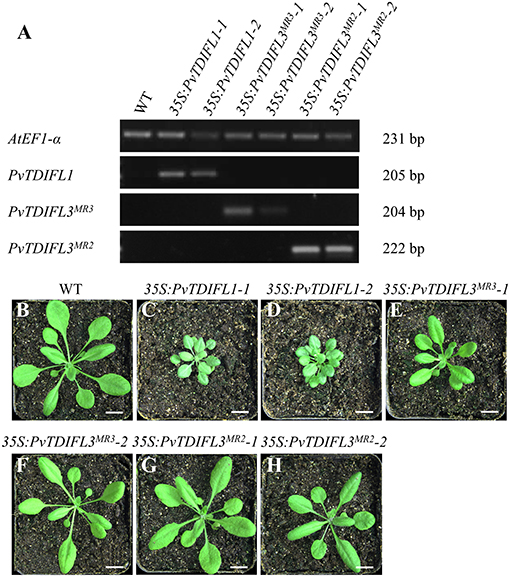
Figure 5. Reverse Transcription PCR (RT-PCR) analysis and morphology of 4-week-old Arabidopsis seedlings expressing PvTDIFLs. (A) RT-PCR analysis of 35S:PvTDIFL lines used in this study. AtEF1-α was used as the reference gene. (B) 4-week-old wild type plant. (C–H) 4-week-old transgenic plants harboring 35S:PvTDIFL1-1 (C), 35S:PvTDIFL1-2 (D), 35S:PvTDIFL3MR3-1 (E), 35S:PvTDIFL3MR3-2 (F), 35S:PvTDIFL3MR2-1 (G), and 35S:PvTDIFL3MR2-2 (H) transgenes grown in soil. Bar = 1 cm in (B–H).
To further quantify the phenotypic changes of the 35S:PvTDIFL plants, the above-mentioned lines were grown on vertical plates for 2 weeks to observe the morphological changes in roots. A significant decrease in the length of the primary root was observed in all heterologous expression lines, except for line 35S:PvTDIFL3MR2-1 (Figures 6A–G,O). Similar short-root phenotypes were observed in rice and pine after treatments with synthetic TDIF peptides (Kinoshita et al., 2007; Strabala et al., 2014). The heights of the inflorescence of 35S:PvTDIFL plants were measured after 6 weeks of growth in soil, 2 weeks after the initiation of flowering. Extreme dwarfism was observed in the lines harboring the 35S:PvTDIFL1 transgene. The heights of their inflorescences were 40% of the wild-type plants (p ≤ 0.001, n = 10). The height of the inflorescence was also decreased in the lines harboring the 35S:PvTDIFL3MR3 transgene. The heights of the inflorescence from the line 35S:PvTDIFL3MR3-1 and -2 were 54% (p ≤ 0.001, n = 10) and 34% (p ≤ 0.001, n = 10) of the wild type plants, respectively. In contrast, no significant decrease in the height of the inflorescence was observed in the lines harboring the 35S:PvTDIFL3MR2 transgene (Figures 6H–N,P).
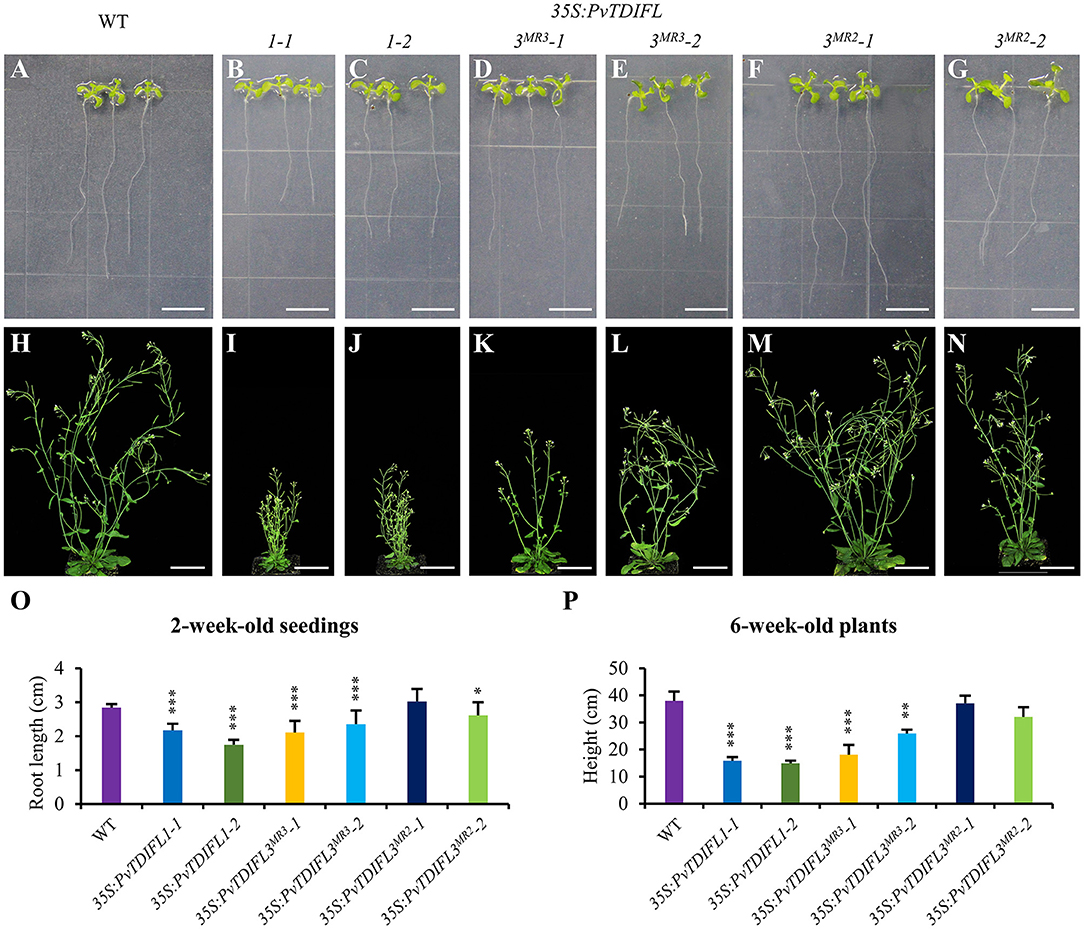
Figure 6. Morphology of 2 and 6-week-old Arabidopsis expressing PvTDIFL transgenes. (A–G) Morphological observations of 2-week-old seedlings from WT (A), and transgenic plants harboring 35S:PvTDIFL1-1 (B), 35S:PvTDIFL1-2 (C), 35S:PvTDIFL3MR3-1 (D), 35S:PvTDIFL3MR3-2 (E), 35S:PvTDIFL3MR2-1 (F), 35S:PvTDIFL3MR2-2 (G) transgenes grown on vertical plates. (H–N) Morphological observation of 6-week-old seedlings from WT (H) and transgenic plants harboring 35S:PvTDIFL1-1 (I), 35S:PvTDIFL1-2 (J), 35S:PvTDIFL3MR3-1 (K), 35S:PvTDIFL3MR3-2 (L), 35S:PvTDIFL3MR2-1 (M), 35S:PvTDIFL3MR2-2 (N) transgenes grown in soil. (O) Quantitation analysis of root length of 2-week-old seedlings. (P) Heights of 6-week-old plants. *, **, and *** indicates statistically significant differences from WT at p < 0.05, p < 0.01, and p < 0.001, respectively. Sample size n = 24 (biological replicates) for the vertical plates and n = 10 (biological replicates) for plants grown in soil. Bar = 1 and 5 cm in (A–G) and (H–N), respectively.
To investigate the functions of PvTDIFL genes in vascular development, the hypocotyls of 6-week-old Arabidopsis were sectioned and analyzed using light microscopy. In comparison with Col-0 (Figures 7A,E), heterologous expression of PvTDIFL1 induced a drastic increase in the number of cells in the stele, suppressed the differentiation of xylem and disrupted the organization of the vascular tissue (Figures 7B,F). Heterologous expression of PvTDIFL3MR3 induced a reduction in the size of xylem and an increase in the size of phloem without disrupting the organization of the vascular tissue and did not influence the diameter of the hypocotyl (Figures 7C,G). Heterologous expression of PvTDIFL3MR2 led to a similar albeit somewhat attenuated phenotype relative to the heterologous expression of PvTDIFL1. For instance, the size of treachery elements was not reduced in the plants expressing PvTDIFL3MR2 (Figures 7D,H). The above results are consistent with the previous reports on the TDIF genes in Arabidopsis and Populus (Hirakawa et al., 2008; Whitford et al., 2008; Etchells and Turner, 2010; Etchells et al., 2015; Li et al., 2018). Although PvTDIFL1 contains only one single TDIFL motif, it had the greatest influence on vascular development. On the contrary, multiple TDIFL motifs did not increase the activity of either PvTDIFL3MR3 or PvTDIFL3MR2 relative to PvTDIFL1.
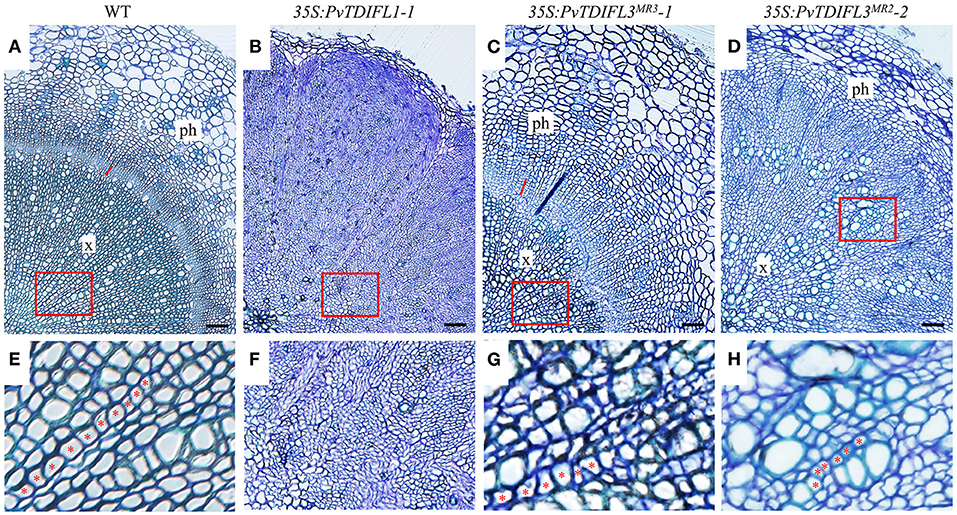
Figure 7. Histology of 6-week-old Arabidopsis lines expressing PvTDIFL transgenes. (A–D) Histological observation of hypocotyls from WT (A) and transgenic seedlings harboring 35S:PvTDIFL1-1 (B), 35S:PvTDIFL3MR3-1 (C), and 35S:PvTDIFL3MR2-2 (D) transgenes. Transverse sections that were 2.5 μm thick were stained with 0.05% (w/v) aqueous toluidine blue. Bar = 50 μm. × and ph indicate xylem and phloem, respectively. The red line indicates cambium in (A) and (C). (E–H) High magnification images from the areas in red boxes from (A–D), respectively. Red asterisks show examples of organized files of cells.
Discussion
Identification of TDIF/TDIFL Genes in P. virgatum
In this study, 93 putative CLE genes were identified in the genome of P. virgatum, by using a novel method that was recently developed (Zhang et al., 2020) followed by gene cloning. In Arabidopsis, all of the 32 CLE genes encode proteins with a single CLE motif, except for CLE18, which contains a second CLEL motif (Meng et al., 2012). Surprisingly, five CLE proteins containing multiple CLE motifs were identified in O. sativa, T. aestivum, and Medicago truncatula (Oelkers et al., 2008). A total of 59 CLE proteins in 27 plant species contain multiple CLE motifs (Goad et al., 2017). Genes encoding proteins containing multiple CLE motifs have been recently identified in various plants, despite that the CLE motifs from the same protein might or might not be identical to each other (Goad et al., 2017). Most functional studies on CLE genes have been conducted with genes encoding a single CLE motif. The knowledge of the CLE genes encoding multiple motifs remains limited. In this study of CLE genes in P. virgatum, it was found that three PvCLE genes—Pavir.Fa00904.1, PvTDIFL3MR3, and PvTDIFL3MR2 encode proteins containing multiple CLE motifs, which allowed the functions of the CLE motif-containing proteins encoded by these genes to be studied.
Among the three PvCLEs that encode multiple CLE motifs, PvTDIFL3MR3 and 3MR2 appear to encode CLE peptides that are homologous to the TDIF peptide. The function of the TDIF peptide in vascular development has been well-studied in several species, such as Arabidopsis, Populus, Marchantia polymorpha (Ito et al., 2006; Hirakawa et al., 2008, 2010a,b; Etchells and Turner, 2010; Etchells et al., 2015; Kondo and Fukuda, 2015). However, these previously studied TDIF/TIDFL genes encode a single TDIF/TDIFL motif. Therefore, functional analysis of the TDIFL peptides from the PvTDIFL3MR3 and 3MR2 proteins could provide a better understanding of the regulatory activities of TDIF/TDIFL peptides.
In total, five PvTDIFL-encoding genes were identified in P. virgatum. The motif sequences of PvTDIFL1/2 and TDIF are very similar. Indeed, only the 10th amino acid residue of the motif is different between PvTDIFL1/2 and TDIF. The TDIFL peptide from PvTDIFL1/2 represents the largest group of TDIF/TIDFL peptides in monocots (Zhang et al., 2020). The number of different residues in the TDIFL motifs of PvTDIFL3MR3, 3MR2, and 4 ranges from two to three (Figures 2B,C). The PvTDIFL3MR3 and 3MR2 contain three and two TDIFL motifs, respectively. Sequence alignments of PvTDIFL proteins show that PvTDIFL3MR3 had one more repeat of the TDIFL motif within its flanking sequence relative to PvTDIFL3MR2. The PvTDIFL3MR2 was a “byproduct” of cloning PvTDIFL3MR3 by using gene specific primers for PvTDIFL3MR3. Meanwhile, PvTDIFL3MR3 and 3MR2 were matched to the same chromosomal locus, which provides evidence that these two genes could be alleles. An alternative explanation is that PvTDIFL3MR2 is simply missing from the v1.1 genome sequence of P. virgatum.
Activities of PvTDIFL Peptides in Arabidopsis
Alanine scanning mutagenesis indicated that the 2nd, 5th, 7th, 10th, and 11th residues of the TDIF motif do not contribute to its activity. However, the H1, V3, G6, N8, P9, and N12 substitutions caused severe losses of TDIF activity (Ito et al., 2006). In order to investigate the activities of PvTDIFL peptides based on the predicted motifs, four PvTDIFL peptides were synthesized and applied to Arabidopsis seedlings. The peptide activities were evaluated based on their ability to influence cell numbers and cell arrangement in the vasculature of the hypocotyl.
The results showed that PvTDIFL_1p, 2p, and 3p had similar and significant activities in promoting cell division, disrupting vascular cell patterns and inhibiting xylem development, although their activities were weaker than the Arabidopsis TDIF peptide (Figures 4A–E,G). On the contrary, PvTDIFL_4p (corresponding to the 2nd motif of PvTDIFL3MR3) had no TDIF activity. Instead, PvTDIFL_4p induced a 50% reduction in the number of cells in the hypocotyl, which is consistent with PvTDIFL_4p serving as an antagonist of peptides that possess TDIF activity (Figures 4F,G). The sequence alignment of the PvTDIFL peptides that included the TDIF peptide showed that PvTDIFL_1p, 2p, and 3p have different residues at the 2nd and/or 10th residues, which are not critical positions for TDIF activity (Ito et al., 2006). The PvTDIFL_4p has an extra substitution, with a His (H) instead of an Asn (N) at the 12th residue (Figure 4H), which is one of the critical positions (Ito et al., 2006). This H-to-N substitution is consistent with its inhibitory activity in the exogenous peptide treatment experiments.
Functional Analysis of PvTDIFL Genes in Arabidopsis
It has been hypothesized that a full CLE protein precursor carrying multiple motifs can release several active peptides after processing, which could play an amplification effect (Oelkers et al., 2008). To better understand the function of PvTDIFL genes, PvTDIFL1 (one motif), PvTDIFL3MR3 (three motifs), and PvTDIFL3MR2 (two motifs) were expressed in Arabidopsis. Heterologous expression of PvTDIFL1 mimicked the phenotypes of the Arabidopsis plants that overexpressed the endogenous TDIF genes, such as CLE41 and CLE44 (Strabala et al., 2006; Etchells and Turner, 2010). However, heterologous expression of PvTDIFL3MR3 and 3MR2 produced more subtle phenotypes than the heterologous expression of PvTDIFL1 (Figures 5–7). An amplification of TDIFL activity in Arabidopsis plants expressing PvTDIFL3MR3 or 3MR2 was not observed although these genes encode peptides with multiple motifs. The lack of the amplification effect has several possible explanations. Firstly, PvTDIFL3MR3 and 3MR2 were heterologously expressed in Arabidopsis. Because Arabidopsis does not have any CLE proteins with multiple CLE motifs, it might not be able to process PvTDIFL3MR3 or 3MR2 protein precursors efficiently. Secondly, the motif sequences of PvTDIFL peptides are not the same as the endogenous TDIF/TDIFL peptides of Arabidopsis. Therefore, the affinity of the PXY/TDR receptor in Arabidopsis for the PvTDIFL peptides is difficult to predict and requires more experimentation to understand. Thirdly, although the typical sequence of the TDIF peptide is HEVPSGPNPISN (Ito et al., 2006), the conserved sequences flanking the TDIFL motifs from PvTDIFL3MR3 and 3MR2 provide evidence that possible variants of the mature motifs are encoded by these two genes. Finally, the experiments in this study have multiple variables, therefore it is hard to know whether all the TDIFL motifs from PvTDIFL3MR3 and 3MR2 have been successfully processed. The proteomics analysis of small proteins based on liquid chromatography with tandem mass spectrometry (LC-MS/MS) technique (Wang et al., 2020) can be applied to the analysis of transgenic lines harboring the transgenes that encode multiple PvTDIFL motifs with various alanine substitutions. This may help to analyze the processing of multiple PvTDIFL motifs.
The PvTDIFL3MR3 contains a 47-amino acids insertion between the amino acid residues 77 and 123. This insertion introduces an extra TDIFL motif in PvTDIFL3MR3 relative to PvTDIFL3MR2 (Figure 2C). The extra TDIFL motif is corresponding to PvTDIFL_4p (Figure 4H). A moderate TDIF-overexpression phenotype was apparent in the hypocotyl sections of the 6-week-old 35S:PvTDIFL3MR2 plants, which provides evidence that at least one of the PvTDIFL peptides (motif 1 and motif 3) was successfully processed. On the contrary, although vascular development in 35S:PvTDIFL3MR3 plants was reduced, the vascular organization of the hypocotyl was well maintained. These data suggest a processing of motif 2 from PvTDIFL3MR3 and indicate that PvTDIFL_4P may serve as an antagonist of PvTDIFL_2P (PvTDIFL3MR3_motif 1), PvTDIFL_3P (PvTDIFL3MR3_motif 3), and the endogenous TDIF peptide. The inhibitory effect derived from motif 2 of PvTDIFL3MR3 in the PvTDIFL3MR3 heterologous expression experiments is consistent with the inhibition of stele development by PvTDIFL_4p that was observed during the exogenous peptide treatments in 10-day-old Arabidopsis seedlings (Figures 4F,G).
Previous reports showed that although TDIF has no inhibitory effect on root elongation in Arabidopsis (Ito et al., 2006; Whitford et al., 2008), TDIF mildly inhibits root elongation in rice and pine (Kinoshita et al., 2007; Strabala et al., 2014). In this study, heterologous expression of PvTDIFLs significantly shortened the roots of Arabidopsis, especially in the 35S:PvTDIFL1 lines (Figures 6A–G). These data are consistent with the diversification of TDIF/TDIFL gene function in different species. However, little is known about the mechanism responsible for these differences in peptide activity.
Tissue-Specific Expression of TDIF to Increase Biomass in Plant
As a PBC, P. virgatum is a commonly used material to study the synthesis of biomass. In this study, heterologous expression of PvTDIFLs in Arabidopsis caused dwarf seedlings and a disordered vasculature, indicating that constitutive heterologous expression of PvTDIFLs reduced biomass. It is consistent with the previous studies on overexpression of the endogenous TDIF genes in Arabidopsis (Hirakawa et al., 2008; Whitford et al., 2008; Etchells and Turner, 2010). However, the phloem-specific expression of PttCLE41 leads to increased woody biomass in Populus and thus, demonstrates that it is possible to increase the biomass by manipulating the TDIF–PXY/TDR signaling module in plants (Etchells et al., 2015). In hybrid poplar, the PttWOX4 genes act downstream of PXY/TDR to control cell division activity in the vascular cambium and hence, to increase stem girth (Kucukoglu et al., 2017). Overexpression of the WOX gene STF (STENOFOLIA) improves biomass yields in grasses (Wang et al., 2017). In this study, five TDIF/TDIFL genes in P. virgatum were identified. Three of these genes in Arabidopsis were cloned and heterologous expressed. Overexpression technologies and exogenous peptide treatment experiments gave a better understanding of the functions and activities of various TDIFL peptides, including PvTDIFL peptides, and also provide clues that will drive future applied research on biomass improvement by manipulating the TDIF–PXY/TDR–WOX4 signaling pathway in plants.
Conclusion
In this study, 93 genes that are homologous to CLE were identified in P. virgatum and were divided into 6 groups based on a phylogenetic analysis. A total of five PvCLE members were assigned to the fourth group that consisted of the H-type CLEs. The five genes that were most similar to Arabidopsis TDIF were named PvTDIFL1, PvTDIFL2, PvTDIFL3MR3, PvTDIFL3MR2, and PvTDIFL4. PvTDIFL3MR3 and 3MR2 contain three and two TDIFL peptide motifs, respectively. Expression analysis showed that PvTDIFL genes were highly expressed in the rachis, which is rich in vascular tissue. Experiments with exogenous polypeptides demonstrated that, apart from PvTDIFL_4p, the ability to influence the development of the stele in the hypocotyl is conserved in both PvTDIFL and Arabidopsis TDIF peptides. The heterologous expression of PvTDIFL1, 3MR3, and 3MR2 all affected the growth of Arabidopsis plants and the development of the vascular tissue in the hypocotyl to varying degrees. The Arabidopsis plants that stably expressed PvTDIFL3MR3 and 3MR2 did not develop elevated levels of CLE peptide activity that might be expected from the multiple CLE motifs contained in the proteins encoded by these genes. At present, there are few studies on the processing and activity of plant CLEs containing multiple motifs. This study will shape future work that aims to increase biomass by modifying the TDIF signaling pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
BZ and XS conceived and designed the experiments. XS, ZZ, and LL identified the PvCLE genes. LL, KD, JT, and DT performed the experiments and analyzed the corresponding results. DT drafted the manuscript. RL, BZ, and XS revised this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (2662020YLPY026, 2662018PY071), and the National Natural Science Foundation of China (31770639, 31370673).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.737219/full#supplementary-material
Supplementary Figure 1. Nucleotide sequences of PvTDIFL3MR3 and PvTDIFL3MR2. The nucleotide sequences were obtained by clones. Green, blue, and pink font represent 5′-UTR, CDS, and 3′-UTR, respectively. The sequences corresponding to the TDIFL motifs are underlined.
Supplementary Figure 2. Weblogo images of CLE motifs from six groups in Arabidopsis and switchgrass. Weblogo images were created by using the Weblogo online tool with the amino acid sequences from the predicted CLE motifs.
Supplementary Table 1. Amino acid sequences from TDIF/TDIFL used for chemical synthesis.
Supplementary Table 2. Primers used in this study.
Supplementary Table 3. Information of PvCLE proteins identified in this study.
References
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino-acid-sequences. Electrophoresis 14, 1023–1031. doi: 10.1002/elps.11501401163
Chen, Q., and Song, G. Q. (2019). Protocol for agrobacterium-mediated transformation and transgenic plant production of switchgrass. Methods Mol. Biol. 1864, 105–115. doi: 10.1007/978-1-4939-8778-8_8
Clifton-Brown, J., Harfouche, A., Casler, M. D., Jones, H. D., Macalpine, W. J., Murphy-Bokern, D., et al. (2019). Breeding progress and preparedness for mass-scale deployment of perennial lignocellulosic biomass crops switchgrass, miscanthus, willow and poplar. GCB Bioenergy 11, 118–151. doi: 10.1111/gcbb.12566
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cock, J. M., and McCormick, S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126, 939–942. doi: 10.1104/pp.126.3.939
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Etchells, J. P., Mishra, L. S., Kumar, M., Campbell, L., and Turner, S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055. doi: 10.1016/j.cub.2015.02.023
Etchells, J. P., Provost, C. M., Mishra, L., and Turner, S. R. (2013). WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 140, 2224–2234. doi: 10.1242/dev.091314
Etchells, J. P., Smit, M. E., Gaudinier, A., Williams, C. J., and Brady, S. M. (2016). A brief history of the TDIF-PXY signalling module: balancing meristem identity and differentiation during vascular development. New Phytol. 209, 474–484. doi: 10.1111/nph.13642
Etchells, J. P., and Turner, S. R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774. doi: 10.1242/dev.044941
Fiers, M., Golemiec, E., Xu, J., van der Geest, L., Heidstra, R., Stiekema, W., et al. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553. doi: 10.1105/tpc.105.034009
Fiers, M., Ku, K. L., and Liu, C. M. (2007). CLE peptide ligands and their roles in establishing meristems. Curr. Opin. Plant Biol. 10, 39–43. doi: 10.1016/j.pbi.2006.11.003
Fisher, K., and Turner, S. (2007). PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 17, 1061–1066. doi: 10.1016/j.cub.2007.05.049
Fletcher, L. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Gimeno, J., Eattock, N., Van Deynze, A., and Blumwald, E. (2014). Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS ONE 9:e91474. doi: 10.1371/journal.pone.0091474
Goad, D. M., Zhu, C., and Kellogg, E. A. (2017). Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 216, 605–616. doi: 10.1111/nph.14348
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–1186. doi: 10.1093/nar/gkr944
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Han, H., Zhang, G., Wu, M., and Wang, G. (2016). Identification and characterization of the Populus trichocarpa CLE family. BMC Genomics 17:174. doi: 10.1186/s12864-016-2504-x
Hirakawa, Y., Kondo, Y., and Fukuda, H. (2010a). Regulation of vascular development by CLE peptide-receptor systems. J. Integr. Plant Biol. 52, 8–16. doi: 10.1111/j.1744-7909.2010.00904.x
Hirakawa, Y., Kondo, Y., and Fukuda, H. (2010b). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629. doi: 10.1105/tpc.110.076083
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. doi: 10.1073/pnas.0808444105
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., et al. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. doi: 10.1126/science.1128436
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Keshwani, D. R., and Cheng, J. J. (2009). Switchgrass for bioethanol and other value-added applications: a review. Bioresource Technol. 100, 1515–1523. doi: 10.1016/j.biortech.2008.09.035
Kinoshita, A., Nakamura, Y., Sasaki, E., Kyozuka, J., Fukuda, H., and Sawa, S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825. doi: 10.1093/pcp/pcm154
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848. doi: 10.1126/science.1128439
Kondo, Y., and Fukuda, H. (2015). The TDIF signaling network. Curr. Opin. Plant Biol. 28, 106–110. doi: 10.1016/j.pbi.2015.10.002
Kucukoglu, M. (2020). A novel NAC domain transcription factor XVP controls the balance of xylem formation and cambial cell divisions. New Phytol. 226, 5–7. doi: 10.1111/nph.16400
Kucukoglu, M., Nilsson, J., Zheng, B., Chaabouni, S., and Nilsson, O. (2017). WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 215, 642–657. doi: 10.1111/nph.14631
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, X., Yang, H., Wang, C., Yang, S., and Wang, J. (2018). Distinct transgenic effects of poplar TDIF genes on vascular development in Arabidopsis. Plant Cell Rep. 37, 799–808. doi: 10.1007/s00299-018-2268-7
Li, Z., Liu, G., Zhang, J., Zhang, J., and Bao, M. (2008). Extraction of high-quality tissue-specific RNA from London plane trees (Platanus acerifolia), permitting the construction of a female inflorescence cDNA library. Funct. Plant Biol. 35, 159–165. doi: 10.1071/FP07212
McLaughlin, S. B., and Adams Kszos, L. (2005). Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 28, 515–535. doi: 10.1016/j.biombioe.2004.05.006
Meng, L., Buchanan, B. B., Feldman, L. J., and Luan, S. (2012). CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 1760–1765. doi: 10.1073/pnas.1119864109
Oelkers, K., Goffard, N., Weiller, G. F., Gresshoff, P. M., Mathesius, U., and Frickey, T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8:1. doi: 10.1186/1471-2229-8-1
Ogawa-Ohnishi, M., Matsushita, W., and Matsubayashi, Y. (2013). Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9, 726–730. doi: 10.1038/nchembio.1351
Ohyama, K., Ogawa, M., and Matsubayashi, Y. (2008). Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160. doi: 10.1111/j.1365-313X.2008.03464.x
Ohyama, K., Shinohara, H., Ogawa-Ohnishi, M., and Matsubayashi, Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580. doi: 10.1038/nchembio.182
Ondzighi-Assoume, C. A., Willis, J. D., Ouma, W. K., Allen, S. M., King, Z., Parrott, W. A., et al. (2019). Embryogenic cell suspensions for high-capacity genetic transformation and regeneration of switchgrass (Panicum virgatum L.). Biotechnol Biofuels 12:290. doi: 10.1186/s13068-019-1632-3
Parrish, D. J., and Fike, J. H. (2005). The biology and agronomy of switchgrass for biofuels. Crit. Rev. Plant Sci. 24, 423–459. doi: 10.1080/07352680500316433
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Ragni, L., and Hardtke, C. S. (2014). Small but thick enough–the Arabidopsis hypocotyl as a model to study secondary growth. Physiol. Plantarum 151, 164–171. doi: 10.1111/ppl.12118
Sanderson, M. A., Adler, P. R., Boateng, A. A., Casler, M. D., and Sarath, G. (2006). Switchgrass as a biofuels feedstock in the USA. Can. J. Plant Sci. 86, 1315–1325. doi: 10.4141/P06-136
Sanderson, M. A., Reed, R. L., McLaughlin, S. B., Wullschleger, S. D., Conger, B. V., Parrish, D. J., et al. (1996). Switchgrass as a sustainable bioenergy crop. Bioresource Technol. 56, 83–93. doi: 10.1016/0960-8524(95)00176-X
Sawa, S., Kinoshita, A., Betsuyaku, S., and Fukuda, H. (2008). A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signal. Behav. 3, 337–339. doi: 10.4161/psb.3.5.5344
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7:(Suppl. 1):S10.1–12. doi: 10.1186/gb-2006-7-s1-s10
Stahl, Y., Wink, R. H., Ingram, G. C., and Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909–914. doi: 10.1016/j.cub.2009.03.060
Strabala, T. J., O'Donnell, P., Smit, A. M., Ampomah-Dwamena, C., Martin, E. J., et al. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331–1344. doi: 10.1104/pp.105.075515
Strabala, T. J., Phillips, L., West, M., and Stanbra, L. (2014). Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 14:47. doi: 10.1186/1471-2229-14-47
Ursache, R., Nieminen, K., and Helariutta, Y. (2013). Genetic and hormonal regulation of cambial development. Physiol. Plantarum 147, 36–45. doi: 10.1111/j.1399-3054.2012.01627.x
Wang, H., Niu, L., Fu, C., Meng, Y., Sang, D., Yin, P., et al. (2017). Overexpression of the WOX gene STENOFOLIA improves biomass yield and sugar release in transgenic grasses and display altered cytokinin homeostasis. PLoS Genet. 13:e1006649. doi: 10.1371/journal.pgen.1006649
Wang, J., Kucukoglu, M., Zhang, L., Chen, P., Decker, D., Nilsson, O., et al. (2013). The Arabidopsis LRR-RLK, PXC1, is a regulator of secondary wall formation correlated with the TDIF-PXY/TDR-WOX4 signaling pathway. BMC Plant Biol. 13:94. doi: 10.1186/1471-2229-13-94
Wang, P., Wang, Y., and Ren, F. (2019). Genome-wide identification of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) family in grape (Vitis vinifera L.). BMC Genomics 20:553. doi: 10.1186/s12864-019-5944-2
Wang, S., Tian, L., Liu, H., Li, X., Zhang, J., Chen, X., et al. (2020). Large-scale discovery of non-conventional peptides in Maize and Arabidopsis through an integrated peptidogenomic pipeline. Mol. Plant 13, 1078–1093. doi: 10.1016/j.molp.2020.05.012
Whitford, R., Fernandez, A., De Groodt, R., Ortega, E., and Hilson, P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. U.S.A. 105, 18625–18630. doi: 10.1073/pnas.0809395105
Xi, Y., Ge, Y., and Wang, Z. Y. (2009). Genetic transformation of switchgrass. Methods Mol. Biol. 581, 53–59. doi: 10.1007/978-1-60761-214-8_4
Yang, J. H., Lee, K.-H., Du, Q., Yang, S., Yuan, B., Qi, L., et al. (2020). A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 226, 59–74. doi: 10.1111/nph.16289
Zhang, H., Lin, X., Han, Z., Wang, J., Qu, L. J., and Chai, J. (2016). SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol. Plant 9, 1406–1414. doi: 10.1016/j.molp.2016.07.004
Zhang, J., Elo, A., and Helariutta, Y. (2011). Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22, 293–299. doi: 10.1016/j.copbio.2010.11.008
Keywords: switchgrass, CLE, TDIF, vascular development, biomass
Citation: Tian D, Tang J, Luo L, Zhang Z, Du K, Larkin RM, Shi X and Zheng B (2021) Influence of Switchgrass TDIF-like Genes on Arabidopsis Vascular Development. Front. Plant Sci. 12:737219. doi: 10.3389/fpls.2021.737219
Received: 06 July 2021; Accepted: 17 August 2021;
Published: 23 September 2021.
Edited by:
Qingyu Wu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Alison W. Roberts, University of Rhode Island, United StatesGuodong Wang, Shaanxi Normal University, China
Copyright © 2021 Tian, Tang, Luo, Zhang, Du, Larkin, Shi and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueping Shi, eHBzaGlAbWFpbC5oemF1LmVkdS5jbg==; Bo Zheng, Ym8uemhlbmdAbWFpbC5oemF1LmVkdS5jbg==
 Dongdong Tian1,2
Dongdong Tian1,2 Kebing Du
Kebing Du Xueping Shi
Xueping Shi Bo Zheng
Bo Zheng