- 1Department of Environmental, Biological and Pharmaceutical Sciences and Technologies, University of Campania “Luigi Vanvitelli”, Caserta, Italy
- 2Department of Biology, University of Naples Federico II, Naples, Italy
Several past and recent climatic and geological events have greatly influenced the current distribution of coastal species around the Mediterranean Basin. As a consequence, the reconstruction of the distributional history of these species is challenging. In this study, we used both chloroplast and nuclear SNPs to assess the levels of genetic differentiation, contemporary/historical levels of gene flow, and demographic history for the three only known (one mainland and two insular) populations of Eokochia saxicola, a rare Mediterranean coastal rocky halophyte. Plastid genome analysis revealed very low intraspecific haplotype variation and partial admixture among Capri and Palinuro populations with at least two independent colonization events for the Strombolicchio islet. Nuclear SNPs variation consistently identified three distinct genetic clusters corresponding to our sampling localities. Furthermore, strong genetic isolation was confirmed by both historical and contemporary levels of migration among the three populations. The DIYABC analysis identified two introductions temporally separated from Palinuro to Capri (ca.25 Mya) and subsequently to Strombolicchio (ca.09 Mya) as the most likely hypothesis for the current distribution of E. saxicola. Regardless of their small population sizes, all study sites supported high-genetic diversity maintained by outcrossing and random mating between individuals owing largely to wind pollination, an exclusive trait among Mediterranean narrow endemics. In conclusion, the patterns observed confirm that some Mediterranean endemics are not necessarily “evolutionary dead-ends” but rather represent species that have extensive demographic stability and a strong evolutionary legacy.
Introduction
The Mediterranean Basin is one of the Hotspots of Biodiversity on the Earth (Médail and Quézel, 1999; Myers et al., 2000), largely due to the high number of endemic species (Quezel, 1985; Greuter, 1991). This biodiversity is the result of the current high heterogeneity of environmental matrix, climate, land uses, and geological and climatic events that occurred in the last 15 M years (Thompson, 2005; Blondel et al., 2010).
In particular, the most recent quaternary glacial-interglacial events have greatly influenced the current distribution of many plant species around the Mediterranean Basin (Médail and Diadema, 2009). The role of Glacial Refugia Areas and postglacial migration routes in shaping the current distribution of mainland European plants is well-known (Taberlet et al., 1998; Hewitt, 2001; Heuertz et al., 2004). However, the impact of glaciations on plant species mainly distributed on islands by climatic oscillations remains less explored. The Last Glacial Maximum (LGM) induced the lowering of sea levels, which subsequently led to the creation of several land bridges, facilitating the migration of plant species between the mainland and continental islands (Whittaker and Fernández-Palacios, 2007 quoting Wallace, 1880). Ultimately, the subsequent warming phases led to these temporary land bridges being gradually submerged by rising sea levels (Feliner, 2014). This main isolating barrier favored speciation processes, which resulted in the increase of insular endemics in the Mediterranean Basin (Médail, 2017). In contrast to insular endemics, water level oscillations played a different role in coastal halophytic species. Halophytes are salt-tolerant or salt-resistance species adapted to the very harsh habitat conditions due to high salinity (Ungar, 1991). These species are, therefore, typical of the coastal azonal vegetation [i.e., vegetation occurring where local ecological conditions, such as high salinity, overrule the effect of climate, according to Walter (1985)]. Indeed, for several coastal halophytes, the sea represents the main pathway for gene flow through the dispersal, even at long distances, of ramets or seeds (referred to as hydrochory) (Kadereit and Westberg, 2007; Westberg and Kadereit, 2009). Therefore, the current distribution of halophytic coastal species is the result of both past (main climatic oscillations and related sea-level variations) and recent abiotic factors (i.e., weather conditions, sea storms, and current sea circulations) acting on dispersion processes. Finally, the ecology of both species and plant communities can contribute to the complexity in the interpretation of coastal species distribution (Weising and Freitag, 2007; Feliner, 2014). The reconstruction of the distributional history of coastal halophytes is partly facilitated by their exclusive adaptations to hypersaline habitats that limit their distribution to coastlines, with a linear distribution range and patterns of long-distance dispersal being found to be more influenced by the direction of sea marine currents than by habitat filtering (Kadereit et al., 2005). However, the general pattern largely associated with coastal halophytes inhabiting dune and marshes habitats has not been found in coastal rocky species (Kadereit and Westberg, 2007). Both inland and coastal rocky areas provide suitable habitats for numerous endemic species often with extremely restricted distribution (Davis, 1951; Lavergne et al., 2004; Thompson, 2005) with many of them being considered “narrow endemics” (Kruckeberg and Rabinowitz, 1985; Médail and Baumel, 2018) and threatened by extinction (Montmollin and Strahm, 2005; Orsenigo et al., 2018). Therefore, they play a key role in the biodiversity conservation priority setting (Médail and Baumel, 2018).
Eokochia saxicola (Guss.) Freitag and G. Kadereith (Amaranthaceae) is a perennial evergreen shrub restricted entirely to coastal rocky environments exposed to the salt spray in very few locations around the South Tyrrhenian Sea (Strumia et al., 2020a). Historically, distribution records suggest E. saxicola occurred only on three small islands: Strombolicchio (a recent volcanic islet near Sicily), Capri (a calcareous island block), and Ischia (an old volcanic island, where it is now thought to be extinct). However, a new population has recently been discovered on the mainland coastal area along Campania and represents the only known mainland population of E. saxicola (Strumia et al., 2015). According to the criteria (less than 500 individuals today distributed in five or less populations) proposed by López-Pujol et al. (2013), E. saxicola can be considered an “extremely narrow endemic.” This species is a remnant of old lineages of Camphorosmae and is part of the Chenolea clade, originating in the early Miocene period during the evolution of the Mediterranean Basin (Kadereit and Freitag, 2011). To date, this clade includes only a few highly disjunct halophytic species, which are typical of warmer temperate climates. E. saxicola diverged from two other species [Chenolea diffusa Thunb. and Spirobassia hirsuta (L) Freitag and G. Kadereith] about 10.2 Mya (Kadereit and Freitag, 2011), i.e., largely before the onset of current Mediterranean climate (about 3.2 Mya, Suc, 1984) and is, therefore, considered a palaeoendemism (Favarger and Contandriopoulos, 1961; Stebbins and Major, 1965). Interestingly, E. saxicola distribution sharply contrasts with the general view that coastlines represent linear biogeographic systems connected by sea dispersion as reported for other coastal halophytes (Clausing et al., 2000; Kadereit et al., 2005). Genetic markers are ideal to study seed dispersal and show patterns of populations structuring determined by past and present gene flows (Bermingham and Moritz, 1998). This is particularly true in the case of species without pollen or fossil records. At the same time, the phylogeographical approach may also represent an important tool in conservation (Diniz-Filho et al., 2008) by identifying the populations that possess higher genetic diversity and, therefore, constituting a possible “genetic source” to be considered for future reintroductions (Médail and Baumel, 2018). The main aims of this study were, therefore, to answer the following questions: (1) Do populations of E. saxicola show any phylogeographic structure? (2) What are the primary factors that have shaped the phylogeographic structure? (3) What determines the genetic diversity and genetic structure of extant populations? (4) Can the combination of phylogeography and genetic diversity help define E. saxicola conservation priorities?
Materials and Methods
Study Species
Eokochia saxicola is a multi-branched perennial evergreen shrub found exclusively on maritime rocks (both calcareous and volcanic) close to the sea level in the area occupied by halophilic vegetation (Strumia et al., 2020b). Characteristic features of this species include protogyny, papillose stigma, and recalcitrant pollen, typical of wind pollinated plants (Barone Lumaga et al., 2016). Like other halophytes, the main seed dispersal mechanism is hypothesized to be hydrochory, owing mainly to the presence of diaspores able to float for several days. Moreover, a decrease in the germination rate of aging seeds and the inability for seedlings to anchor at suitable establishment sites have been assessed (Strumia et al., 2020a). According to IUCN, E. saxicola is threatened with extinction as it falls in the category Endangered (EN) (Santangelo et al., 2012; Orsenigo et al., 2018). The main threats are landslides in the few sites of occurrence and supposed reduction of fertility and reproduction success due to the small number of living individuals. We sampled E. saxicola in three locations representing the only known populations of its current distribution: Capri Island near Naples (Campania), Strombolicchio islet (Aeolian Archipelago, Sicily), and Palinuro, along the Cilento and Vallo di Diano and Alburni Nature Park coastline (Campania). Field collections were conducted from 2015 to 2017. Due to the inaccessibility of the cliffs (Strumia et al., 2020a), only the most accessible individuals were sampled. In each sampling site, the maximum estimate number of individuals was recorded through visual census (i.e., by binoculars from a boat) (Table 1).
DNA Isolation, Library Preparation, and Sequencing
Genomic DNAs of 51 Eokochia saxicola individuals were extracted from preserved leaf material using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the protocol of the manufacturer. The quantity and the purity of DNA extractions were checked using the NanoDrop ND 1000 Spectrophotometer (Thermo Fisher Scientific, Delaware), whereas the concentration was assessed with the Qubit fluorometer system (Invitrogen, United States) and the Quant-IT ds-DNA BR Assay kit (Invitrogen). Samples were sequenced using a slight modification to the standard ddRAD protocol (Peterson et al., 2012), with the restriction enzymes EcoRI and TaqI. A single library with barcoded individuals was sequenced in a single Illumina HiSeq 2500 lane for 150-base single-end reads. From a subset of samples (16 E. saxicola + S. hirsuta), a “genome skim” was performed (Straub et al., 2012) to gather sufficient data for complete plastid assembly without prior enrichment or isolation of plastid DNA (Coissac et al., 2016; Supplementary Table SM1). Libraries were constructed according to the Nextera DNA Library Prep Kit (Illumina) protocol and Nextera XT indexes (Kit v2 Set A), which were used to multiplex the individual samples. Libraries were sequenced in a single Illumina HiSeq 2500 lane for 150-base single-end reads.
Preprocessing and Analysis of the Complete Plastome Sequences
The Eokochia saxicola and Spirobassia hirsuta raw sequence reads were quality checked with the FastQC v0.11.91, and low-quality reads and adapters were subsequently trimmed with Trimmomatic v0.39 (Bolger et al., 2014). We implemented a de novo assembly of S. hirsuta plastome by setting kmer = 121 in Velvet v1.2.10 software (Zerbino and Birney, 2008), and trimmed reads were then further assembled using Pilon v1.23 (Walker et al., 2014). We implemented FASTQ Screen v0.14 (Wingett and Andrews, 2018) for filtering the plastid reads from each E. saxicola individual by using a de novo-assembled S. hirsuta plastome (unpublished), and orphan reads were removed through the python script FASTQ Combine Paired End.py2. The E. saxicola filtered reads were mapped to S. hirsuta plastome by using BOWTIE2 v2.3.5.1 (Li and Durbin, 2009). The alignments (BAM files) produced a single-variant call format (vcf) file for all samples, where the SNPs were identified using bcftools v.1.9 with the setting “mpileup -Ou” and called via bcftools using the −mv and –ploidy 1 functions. The vcf file was converted to a Nexus format, and a haplotype network (also including available Amarantaceae plastome sequences) was built by using the TCS method implemented in PopArt v1.7 (Leigh and Bryant, 2015).
Preprocessing ddRAD Data
Raw reads were demultiplexed in individual paired-end libraries and filtered for low-quality reads with the process_radtags pipeline in STACKS v2.4 (Rochette et al., 2019). Quality control of demultiplexed reads was performed with FastQC v0.11.9. The catalog RAD-loci was built with de novo assembly using the de novo_map.pl wrapper in STACKS v2.4 by using parameters m = 1, M = 8, and n = 1, following the optimization procedure described in Paris et al. (2017). The population program in a Denovo_map.pl wrapper was used for collecting the polymorphic SNPs in a vcf file. The vcf files (with 48 individuals) were analyzed with the –missing function of PLINK v1.07 (Purcell et al., 2007) for calculating the percentage of missing data (approximately ∼81% of missing data). Lastly, to avoid the risk of calling false SNPs, we reduced the dataset to include all sites with a minor allele frequency (MAF) greater than or equal to 0.05 and with a (–max-obs-het) setting of 0.70 (as also suggested in Gargiulo et al., 2021). Finally, to account for the effects of missing data per individual, VCFTOOLS v0.1.14 (Danecek et al., 2011) was implemented to filter the minimum number of ddRAD loci shared by at least 70% of individuals (i.e., the full dataset). To minimize the potential effect of missing data in altering the results, some population demographic analyses were also or only performed on a subsample of individuals by maximizing the number of reads per sample and minimizing the amount of missing data. Therefore, a reduced dataset was constructed with 25 individuals (i.e., only those ranging between 2,000,000 and 500,000 reads, namely 10 individuals from Palinuro, eight from Capri, seven from Strombolicchio, see Supplementary Text 1.1), which were filtered so that missing data were present in two or less individuals for each locus (i.e., ddRAD loci present in 95% of the individuals).
Analysis of ddRAD Data
Analyses on the Full (48 Individuals) and Reduced Dataset (25 Individuals)
Maximum Likelihood Tree (ML) trees were generated from both datasets. GTRCAT was used as the substitution model for nucleotide sequences, as implemented in RAxML v8.2.12 (Stamatakis, 2014). The resulting trees were drawn to scale with Figtree v1.4.43.
The R package AWclust (Gao and Starmer, 2008) was implemented as a non-parametric distance-based clustering model (allele-shared distance) for the analysis of populations structure with a HapMap format file as input. We estimated the best K using 100 simulations with an interval of K from 1 to 6. To visualize the genetic relationships among individuals, we performed a multidimensional scaling (MDS) based on the optimal number of clusters (best K).
The vcf file (48 individuals) converted into a genind object with the function vcfR2genlight in the R package adegenet v2.02. (Jombart, 2008) was subsequently used for the Discriminant Analysis of Principal Components (DAPC) (Supplementary Text 1.2 and Supplementary Figures 5–7). Lastly, the optimal number of clusters (K) was then determined by the lowest Bayesian Information Criterion (BIC), and the DAPC plot was constructed with the function scatter.dapc.
Pairwise Fst among individuals and populations was calculated with the function stamppFst (in the R package adegenet), and the result was visualized in a heatmap plot with function heatmap 0.2. The fineRAD structure was implemented to infer population structure via shared ancestry in the ddRAD dataset, focusing on the most recent coalescence (common ancestry) among the sampled individuals (Malinsky et al., 2018). Samples were assigned to populations using 100,000 iterations as burn-in before sampling 100,000 iterations. The trees were built using 10,000 iterations and the output was visualized using fineStructure GUI.
Analyses on the Reduced Dataset (25 Individuals)
The following analyses, more sensitive to the excess of missing data, were conducted with the reduced dataset containing 25 individuals and filtered for a minimum number of ddRAD loci shared by 95% of individuals. The function basic.stats in R package hierfstat (Goudet, 2005) was implemented for the estimation of observed heterozygosity (Ho), expected heterozygosity (He), and the within-population inbreeding coefficient (Fis).
We focused on the timing of the connectivity change based on the estimates of historical and contemporary migration by using a coalescent-based method (MIGRATE) and a disequilibrium-based method (BayesAss), respectively. The historical migration rate (Supplementary Text 1.4 and Supplementary Table SM3) was estimated using Bayesian inference in MIGRATE (Beerli, 2006), while the contemporary migration rates (Supplementary Text 1.3; Supplementary Table SM2; Supplementary Figures 8, 9) were estimated from the current generation and two past generations using Bayesian inference in BayesAss v3 (Rannala, 2007).
Alternative scenarios of origin, the direction of migration, and relationships among E. saxicola populations were tested on the ddRAD dataset under the coalescent-based approximate Bayesian computation in DIYABC Random Forest v1.0 (Collin et al., 2021). This enables the comparison of different population demographic models and determines the estimation of their parameters without calculating complex likelihood functions (Beaumont, 2010). Different historical demographic scenarios were tested with the following Ne-prior parameters set for each population: Npalinuro (100–500); Ncapri (10–500) and NStrombolicchio (10–50). This low value of Ne priors is based on the rarity of E. saxicola among the different locations (Table 1).
For Eokochia saxicola, a generation time of 10 years was considered (ex situ cultivated 5-year-old plants have not bloomed yet). Then, in accordance with the geological age of the sites-hosting current populations (Vezzoli, 1988; Gillot and Keller, 1993; Santangelo et al., 2011), we set the split time (t) as a number of generations from 10 to 50,000. Conditions were set as t1 > t2, according to the timing of events. Bottleneck priors (db time) were set from 10 to 100 generations and the number of founders (Nb) from 10 to 50 individuals. Since previous analyses based on genetic structure did not show admixture events, we did not include admixture events into scenarios.
Four scenarios (Supplementary Figure 10) were designed to explore alternative hypotheses regarding the origin and mainland-island migrations (Scenario 1). The colonization of Capri occurred through a long-distance dispersal event of a small fraction of the Palinuro population at time t1. Then, the bottleneck event in the newly founded population (i.e., Capri) was modeled through a limited number of founders (Nb) during a short period (t1-db). At time t2, the Strombolicchio population was born through a long-distance dispersal event of a small fraction of the Capri population. Then, the bottleneck event in the newly founded population (i.e., Strombolicchio) was modeled through a limited number of founders (Nb) during a short period (t2-db) (Scenario 2). The colonization of Capri occurred through a long-distance dispersal event of a small fraction of the Palinuro population at time t1. Then, the bottleneck event in the newly founded population (i.e., Capri) was modeled through a limited number of founders (Nb) during a short period (t1-db). At time t2, the Strombolicchio population was born through a long-distance dispersal event of a small fraction of the Palinuro population. Then, the bottleneck event in the newly founded population (i.e., Strombolicchio) was modeled through a limited number of founders (Nb) during a short period (t2-db) (Scenario 3). An ancestral population split between Capri and Palinuro populations occurred at time t1. At time t2, Strombolicchio colonization occurred through a long-distance dispersal event of a small fraction of the Capri population. Then, the bottleneck event in the newly founded population (i.e., Strombolicchio) was modeled through a limited number of founders (Nb) during a short period (t2-db) (Scenario 4). An ancestral population split between Capri and Palinuro populations occurred at time t1. At time t2, Strombolicchio colonization occurred through a long-distance dispersal event of a small fraction of the Palinuro population. Then, the bottleneck event in the newly founded population (i.e., Strombolicchio) was modeled through a limited number of founders (Nb) during a short period (t2-db) (Supplementary Figure 10 and figure caption).
We processed 1,000,000 simulations datasets, ∼250,000 simulations datasets per scenario. For scenario choice, a linear discriminant analysis (LDA) processed on the summary statistics was used before processing Random Forests (RF) predictions (Supplementary Figure 11). The 250,000 simulation datasets (i.e., higher than the suggested 20,000 simulation datasets) of the best supported scenario were used to evaluate the model choice and parameter estimation in RF analysis. The number of trees in the constructed random forests were fixed to n = 1,500, the minimum number to ensure a stable accuracy measure of the best scenario and to estimate the mean value and the lower and upper 95% quartiles of the posterior distributions (Chapuis et al., 2020; Supplementary Figure 12).
Results
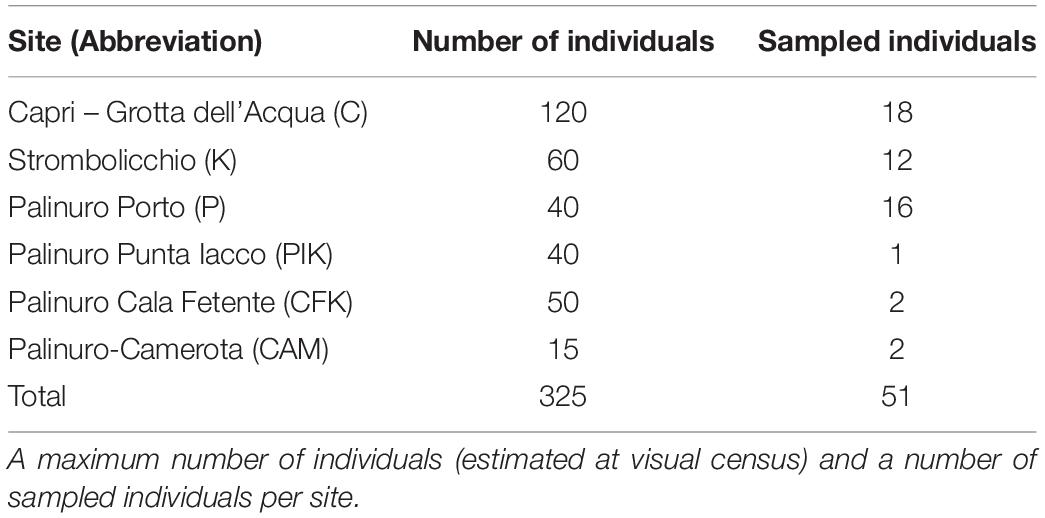
For the Illumina sequencing for the analysis of SNPs variation in the complete plastome sequences of 16 E. saxicola + S. hirsuta, 41,187,562 raw reads (150 bp for average read length) were generated; of which, 34,528,403 reads remained after performing the preprocessing procedure (Supplementary Text 1.1 and Supplementary Table SM1). Variant calling revealed 333 SNPs among both S. hirsuta and E. saxicola individuals as opposed to only three SNPs detected among E. saxicola individuals. The location, nucleotide change, annotation, and mutation type of these three SNPs in E. saxicola are provided in Supplementary Table 1. These SNPs identified three haplotypes in the E. saxicola populations, with haplotype H2 present in all three locations and haplotype H1 shared between the mainland and Capri Island, whereas the haplotype H3 was exclusive to Strombolicchio islet (Figure 1; Supplementary Table 1). All other Amarantaceae plastomes, including the newly assembled S. hirsuta, complete plastome (data not shown) differed largely from E. saxicola plastome both in size and number of SNPs.
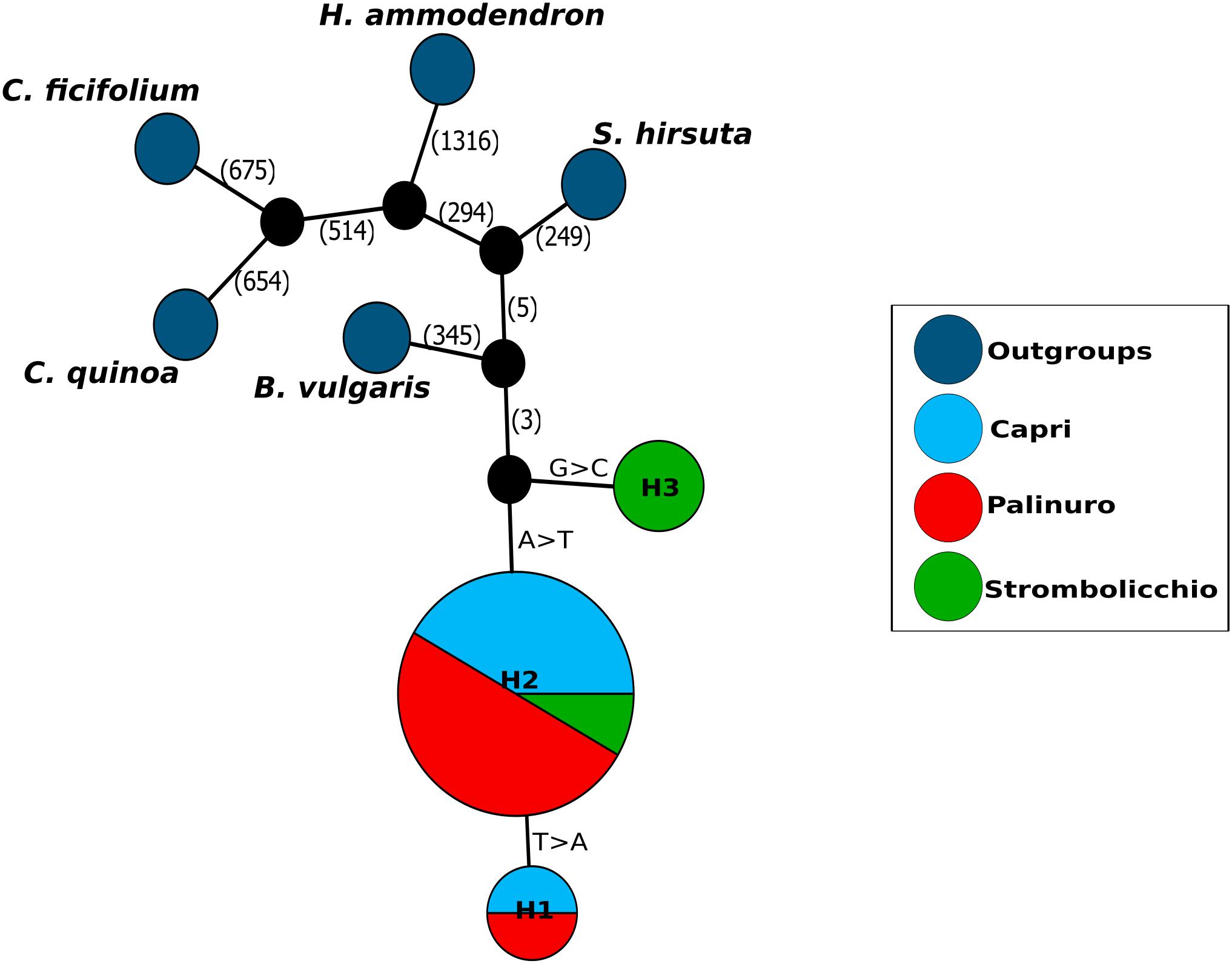
Figure 1. Plastid haplotype network. Each circle represents a different haplotype, where circle size is proportional to the number of individuals that carry the same haplotype. Black circles indicate missing intermediate haplotypes. Mutational steps are reported on the lines of the haplotype networks. Colors show the genotypes belonging to different geographic origins.
From ddRAD sequencing, a total of 30,022,310 raw reads were obtained. After which, 29,276,067 reads were identified through de-multiplexing and filtering using STACKS pipeline to remove any low-quality read, ambiguous barcodes, and sequences without cut sites. We obtained, on average, 609,918 raw reads per sample and 224,637 mapped reads per sample (Supplementary Table SM1).
The pipeline de novo_map.pl generated 6,476,170 loci and assembled ∼ 99.9% of loci into contigs. The total length of pair-end contigs (the generated de novo assembly) was approximately 175 Mbp, as calculated with the software Bandage (Wick et al., 2015). The average mapping efficiency of our samples to this de novo assembly was two reads per locus, as calculated with module gstack of the Stack pipeline. In total, 10,782,592 reads were aligned with the generated genome, i.e., 70% of raw reads did not align (i.e., they were unmapped unique reads). Due to this low coverage, SNPs were called by adopting a variant calling strategy of 3×-minimum coverage. With our filtering approach, in the 48 individuals/3,962 SNPs datasets, we never observed any case of multiple variants in the same locus (suggestive of a diploid status for E. saxicola) or of variants called in less than five individuals. With this conservative approach, we may have lost some rare variants, but we have reduced the risk of calling false SNPs due to low coverage.
An ML tree was built with 3,962 SNPs present in at least 70% of samples. The 48 individuals clustered distinctly in three separate clades. The separation of three distinct clades was further emphasized when using the reduced dataset of 25 individuals with 120 SNPs shared by 95% of individuals (Supplementary Figure 1).
Genetic Clustering with Awclust software identified K = 3 as the optimal number, with each cluster corresponding to a population (Figure 2A). The 48 individuals were split into three distinct clusters or geographical regions in the MDS analysis too (Figure 2B). Moreover, the AWclust dendrogram plot displays a clade with two clusters (the island populations) and a clade with the mainland Palinuro population (Figure 2C).
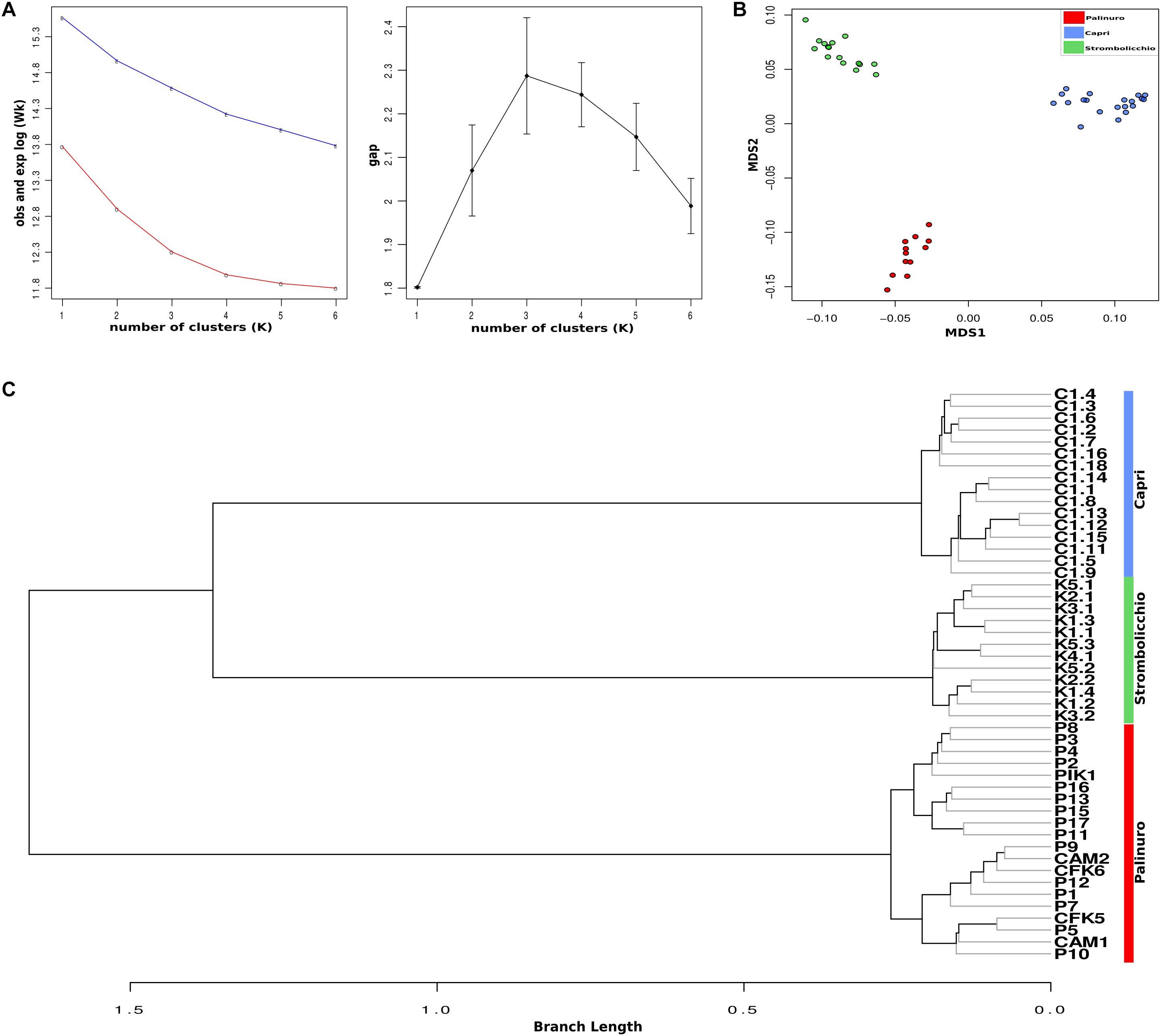
Figure 2. Genetic clustering of sampled individuals. (A) Left: The red line represents the log of the pooled within-cluster sum of squares (Wk) from the observed data. The blue line represents the expected log of Wk from a uniform distribution. Right: the actual gap statistic. (B) A clustering by size reduction (an MDS plot) is represented. Individuals are distributed as points in a two-dimensional vector space. (C) The ancestry-inferred tree with AWclust.
Visualization of population structure using a DAPC (Discriminant Analysis of Principal Components) concordantly revealed three distinct genetic clusters, which included the two island populations (Strombolicchio and Capri) and the mainland Palinuro population (Figure 3; Supplementary Figure 7).
Figure 3. Discriminate analysis of principal components (DAPC). The analysis was drawn using 3,962 SNPs across 48 individuals (SNPs present in at least 70% individuals) and was constructed using 20 principal components (PCs) and two discriminate functions. Dots represent individuals, with colors denoting sampling origin.
The heatmap plot confirmed the existence of the three populations (Figure 4). Overall, all populations revealed a moderate level of intrapopulation co-ancestry (Figure 5). Comparable results were gathered with the reduced dataset (by only using 120 SNPs present in at least 95% individuals across 25 individuals) but with a higher intrapopulation co-ancestry value as it was found inversely correlated with a number of SNPs and missing data (Supplementary Text 1.1; Supplementary Figure 4).
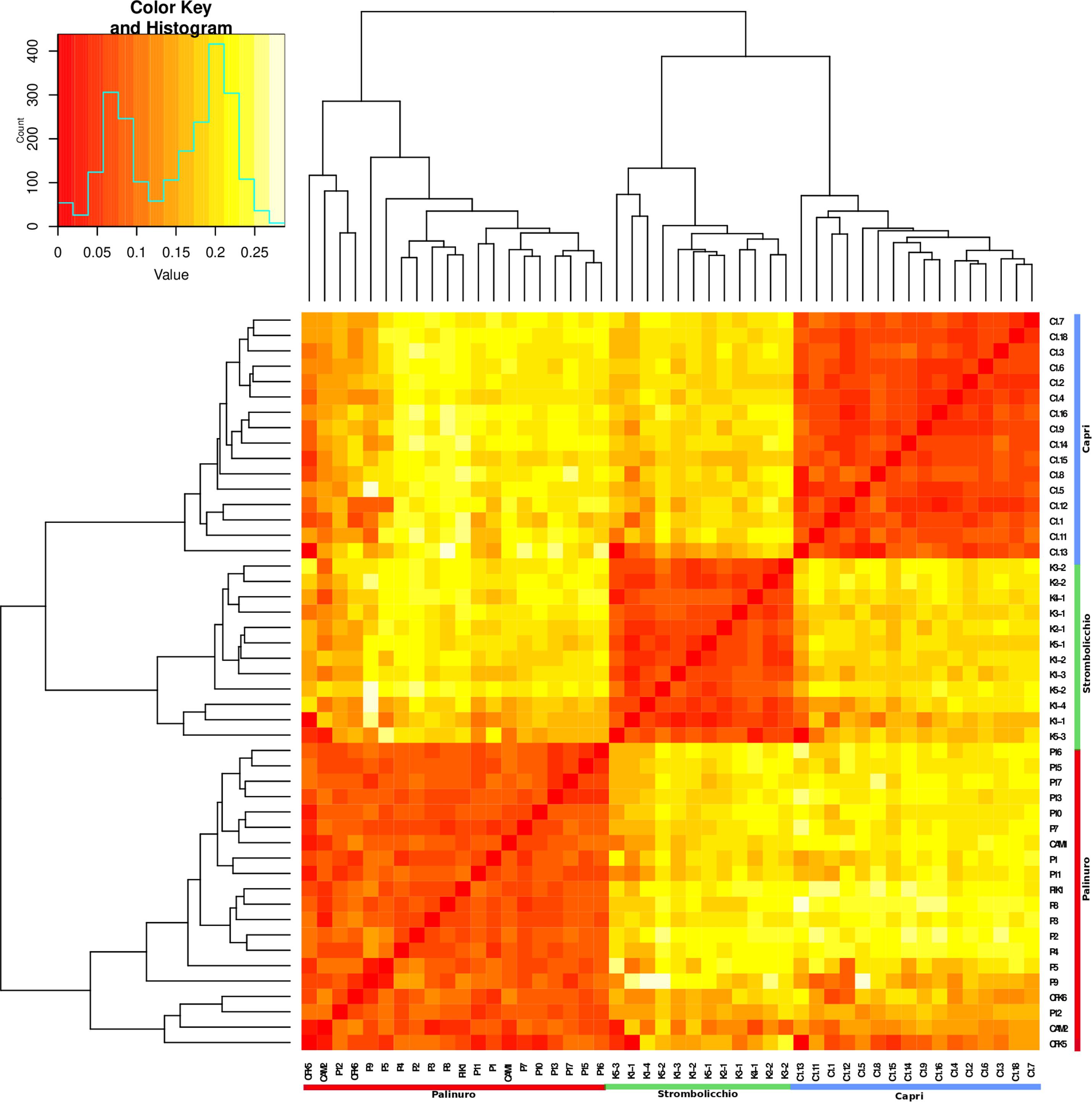
Figure 4. A heatmap graph of Fst was built by using 3,962 SNPs across 48 individuals (SNPs present in at least 70% of individuals). The heatmap illustrates the Fst pairwise matrix based on a color code: orange/red color (low Fst) to white (high Fst). Along the axes of the matrix, phylogenetic trees are constructed using the Neighbor Joining method. The legend (top left) depicts the color-coded Fst values.
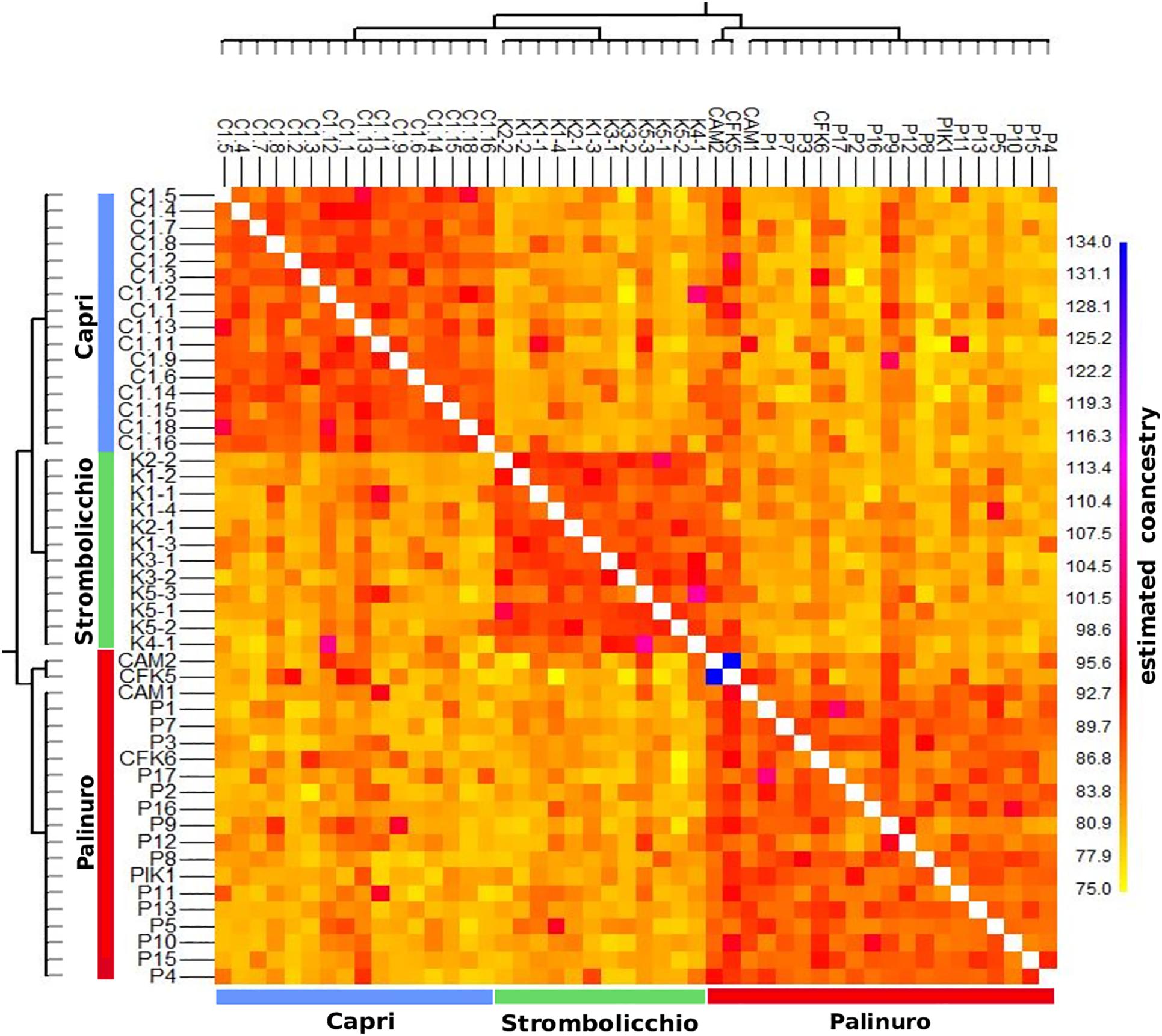
Figure 5. The co-ancestry matrix was shared among 48 individuals (SNPs present in at least 70% individuals). The co-ancestry is calculated as the number of different sequences (i.e., SNPs) between pairs of individuals. The color of each cell in the matrix indicates the expected number of chunks imported from a donor genome (horizontal axis) into a recipient genome (vertical axis). Therefore, highly different cells are indicated in yellow, while indistinguishable cells are represented by blue/purple. Lastly, two phylogenetic trees with the Neighbor-Joining method are represented along the x and y axes.
The mean genetic diversity parameters among populations were 0.0941 for expected Heterozygosity (He), 0.0841 for observed Heterozygosity (Ho), and 0.0781 for the Fixation index (Fis), respectively. Interestingly, the Palinuro population had the highest inbreeding value (Fis = 0.2343) (Table 2).
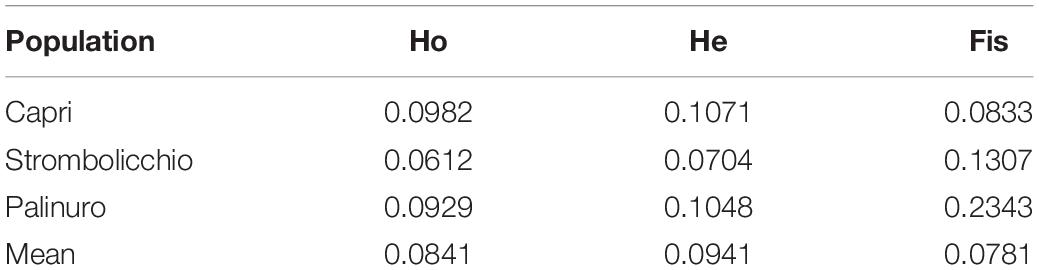
Table 2. Estimates of genetic diversity parameters over 25 individuals (SNPs present in at least 95% individuals) implemented with function basic.stats in the hierfstat R package.
Multiple runs of BAYESASS yielded low levels of contemporary gene flow (mc, fraction of individuals that are immigrants) among the three populations. Concordantly, also, the historical rates of migration as calculated in MIGRATE were generally very low (Figure 6; Supplementary Table 2). Low mc and mh values (mh lower 95% CI includes zero) suggest that populations have become demographically independent in the past. Despite the reduced geographic distance, historical rates of migration between Palinuro and Strombolicchio have the lowest values. Some hidden environmental barriers (likely North heading sea currents) may have impeded any past migration between these two populations.
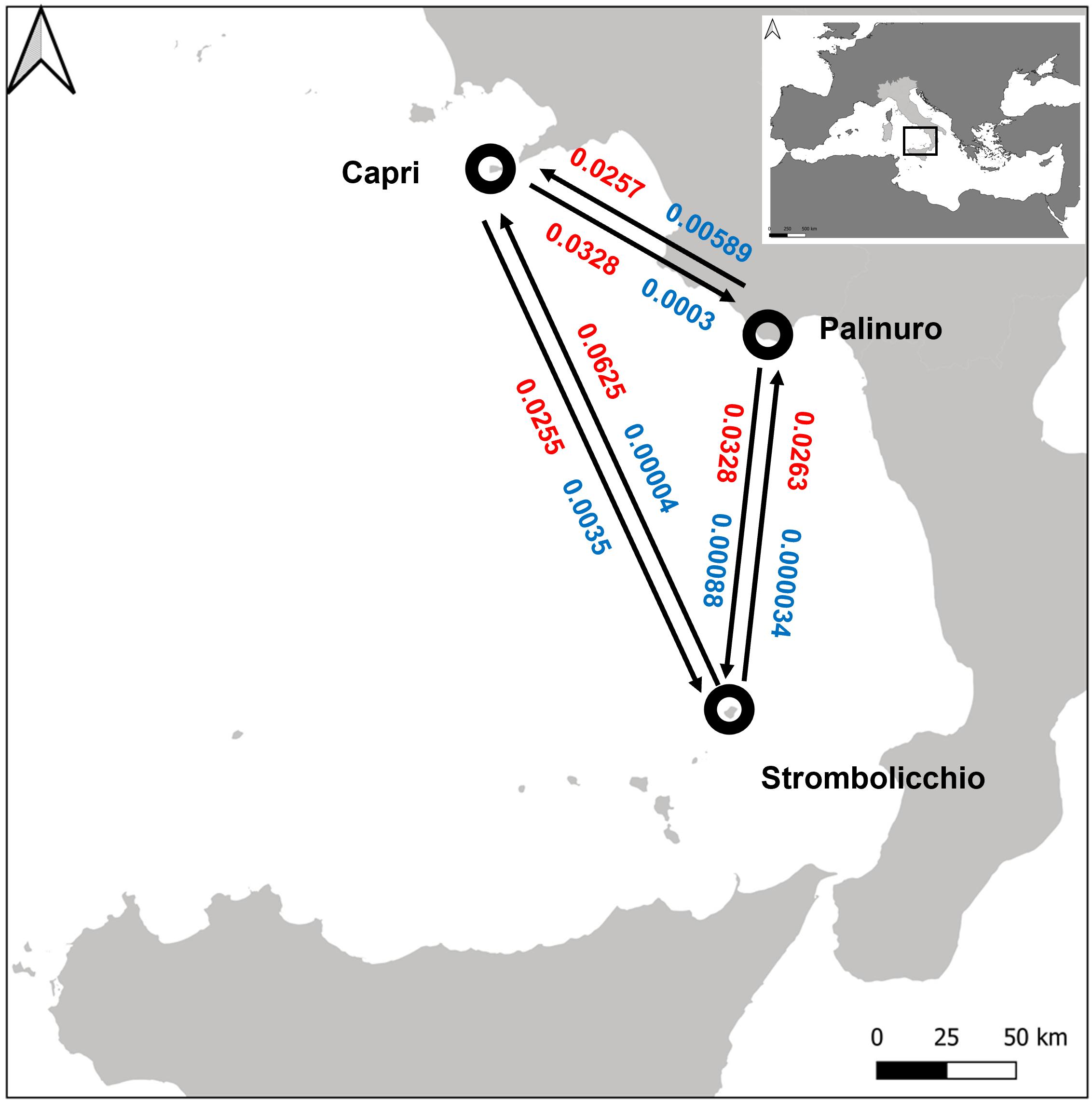
Figure 6. Estimates of the mean posterior recent (blue) and historical (red) migration rate among 25 individuals (SNPs present in at least 95% individuals). The circles represent populations and arrows indicate the direction of migrant individuals between different populations. The inset plot at the top right of the figure reports the position of the map in the Mediterranean Basin.
The E. saxicola dispersion was inferred to have occurred by temporally separated introductions from the Palinuro to Capri and by the latter, to Strombolicchio as revealed by DIYABC model selection, scenario 1 (Figure 7). This scenario was supported by the model (votes = 781; posterior probability p = 0.501). The alternative scenario (2) where Strombolicchio originated from Palinuro received 231 votes. Instead, the scenarios (3 and 4) suggesting the split of an ancestral population between Capri and Palinuro and the subsequent colonization of Strombolicchio from Capri or Palinuro received 308 and 180 of the votes, respectively.
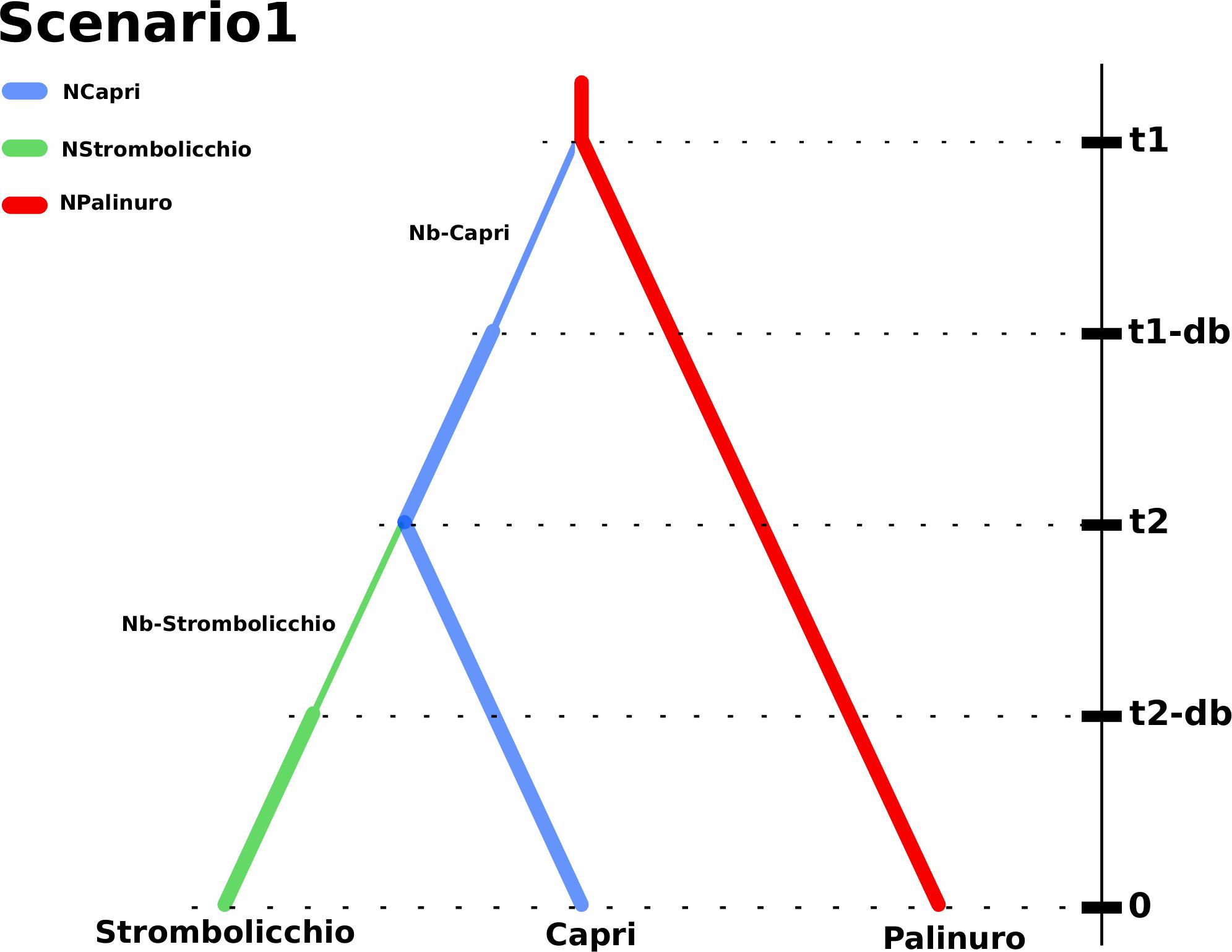
Figure 7. Demographic history of Eokochia saxicola populations implemented by DIY ABC. Scenario 1 represents the most likely hypotheses of the demographic history: the Strombolicchio population was derived from the Capri population, which originated from the Palinuro population. The branch colors indicate discrete population size parameters in the model. t1 represents the split time between the Palinuro population and Capri population, while t2 represents the split time between the Capri population and Strombolicchio population. The thin branch width indicates bottlenecks of duration db (t1-db and t2-db) with effective population sizes of Nb (Nb-Capri and Nb-Strombolicchio). Note: Time is not to scale.
The colonization of Capri from the mainland population was predicted to have occurred around 24,615 generations ago while the following colonization of Strombolicchio only occurred 9,003 generations ago. Interestingly, the relatively narrow posterior parameter distribution of the population introduction from the mainland Palinuro to the Capri Island suggests this single event was followed by a bottleneck period (db) of at least 43 generations with a relatively small founding population of 10 individuals. The bottleneck size of the Strombolicchio population (Nb-Strombolicchio) was around nine individuals. However, at time 0, the effective population size of Strombolicchio is 32 individuals, while the effective population size of Capri is 369 (249–484), i.e., comparable with 379 (150–483) of Palinuro (Table 3).
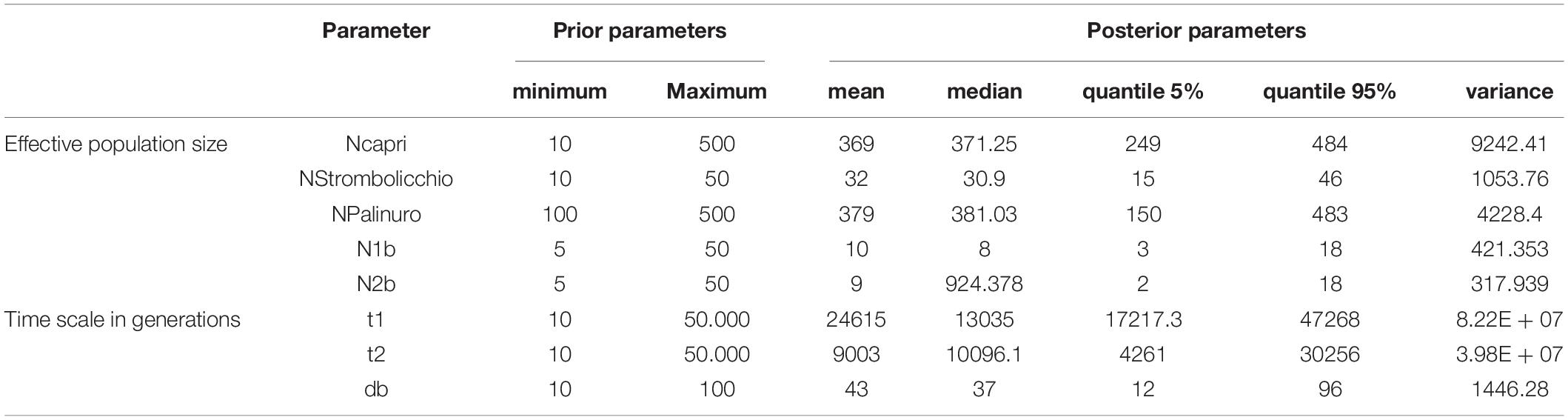
Table 3. Prior values (minimum and maximum, with uniform distribution) for the parameters used for the demographic scenarios and posterior values (mean; median; quantile 5%, and 95% and variance) were estimated from Scenario 1 in the DIYABC approach.
Discussion
The integration of different nuclear and plastid genetic markers provided novel insight into the temporal and spatial distribution, a colonization pathway, and dispersal patterns of E. saxicola. In particular, both markers contributed to disclosing the roles of contemporary versus historical processes in shaping the current genetic variation of E. saxicola populations. Despite its paleoendemic status, the present distribution and partition of genetic diversity in E. saxicola revealed a rather recent dispersion/fragmentation, most likely occurring during the last quaternary climatic oscillations, which resulted in the almost complete interruption of ongoing gene flow among living populations.
It is well-known that different filtering strategies influence both the estimation of genetic diversity and differentiation (Cozzolino et al., 2020; Gargiulo et al., 2021). Therefore, for testing the robustness and reliability of our results using the ML tree, pairwise Fst and co-ancestry among individuals and populations were calculated both with the full dataset (3,962 SNPs across 48 individuals, SNPs present in at least 70% individuals) and with the reduced dataset (25 individuals by selecting 120 SNPs shared by at least 95% of individuals). We found largely overlapping results between the two datasets (Supplementary Figures 1–3). Still, as expected, when using different stringency criteria in loci selection, when comparing datasets with SNPs present in 70% vs. 95% of the samples, we found a decrease of between-lineage differentiation of the employed loci as measured by global FST and an increase in the average overall estimated co-ancestry (Supplementary Text 1.1. and Supplementary Figure 4). Thus, our conservative approach may have not precisely estimated the values of some population genetic parameters (e.g., heterozygosity, FST, Ne, and migration rates).
Do Populations Show Any Phylogeographic Structure?
Analysis of full plastid genome variation in E. saxicola revealed a surprisingly low level of intraspecific variation (only three haplotypes differing for a maximum of two mutation steps) among the three isolated populations with a considerable number of differences (in total plastome length too) with the related S. hirsuta. Since large interspecific differences can be expected between paleoendemic species separated approximately 10 Mya (cfr. Kadereit and Freitag, 2011), both haplotype admixture in populations and very low haplotype variation at the intraspecific level support a recent separation of current E. saxicola populations. The Palinuro and Capri populations share two haplotypes (H1 and H2), differing by a single-base mutation (Figure 1). The most common haplotype (H2) was found in the Strombolicchio population, but, here, the “distant” haplotype H3 (i.e., differing by two mutational steps) was also detected (Figure 1). Even if the scenario of dispersal from Palinuro is the best supported, actual co-occurrence of two related haplotypes in both insular Capri and mainland Palinuro does not rule out an alternative scenario where these populations were likely part of a larger, ancestral population that later split into today isolated populations without a bottleneck phase. Interestingly, this scenario is the second-best supported scenario (i.e., Scenario 3) in the DYABC analysis based on nuclear markers (discussed below).
What Are the Primary Factors That Have Shaped the Phylogeographic Structure?
According to the morpho-bathymetric reconstruction of the Southern Tyrrhenian Sea (Aiello et al., 2015) and considering that, during the last glacial phases, the sea level decreased up to 100 meters, Capri was once connected to the mainland cliffs of Southern Italy. Moreover, the coastline shape between Capri and Palinuro drastically changed during the Quaternary due to the combination of tectonics, sedimentary inputs, volcanism, and sea-level oscillations (Santangelo et al., 2017). In this scenario, which occurred in the last 1.8 Mya, both frequency and surface extension of unsuitable (defined as sandy coasts related to alluvial plains) and suitable (defined as rocky coasts related to pre-quaternary geological substrates) habitats for E. saxicola were transformed from the sea-level oscillations. Moreover, these oscillations most likely reduced the “jumping” distance from the Palinuro cliffs to Capri, favoring a linear “hopping” migration along the coastline, as also reported for several other Mediterranean halophytes (Clausing et al., 2000; Kadereit et al., 2005; Weising and Freitag, 2007). The current distribution of E. saxicola along the Palinuro cliffs (ca. < 15 km) further supports the evidence of a “hopping” dispersal ability along with these coastal rocky habitats.
In contrast, the relatively young volcanic islet of Strombolicchio (0.2 Mya) has been completed isolated from any mainland, even during the Quaternary seal-level oscillations. Accordingly, long distance dispersal or “jumping” is the only plausible scenario for the colonization of E. saxicola on Strombolicchio. Surprisingly, despite the rarity of E. saxicola populations and the small size of the islet, at least two occasional events (two different plastid haplotypes) of colonization likely occurred as founder events for the Strombolicchio population (Figure 1). Hydrochory, as for many other halophytes of the Mediterranean Sea (Kadereit et al., 2005; Arafeh and Kadereit, 2006; Weising and Freitag, 2007; De Castro et al., 2020), may have contributed to the long-distance dispersal of E. saxicola seeds. Indeed, sea-drifted seeds depend entirely on currents for their dispersal, and there is some evidence from the surface circulation observed in the southern Tyrrhenian Sea (Vetrano et al., 2004; Rinaldi et al., 2010) that is consistent with the known distribution of E. saxicola. Changes in sea currents due to the presence of a vortex north of Capri renders the north and south Tyrrhenian Sea “isolated” from each other (Procaccini et al., 2001) and impedes seeds dispersal toward the north.
According to the average reported speed of south Tyrrhenian surface circulation (Rinaldi et al., 2010), the floating time ability of the E. saxicola (Barone Lumaga et al., 2016) would have been sufficient to reach Strombolicchio islet from the Palinuro/Capri coastlines. Haplotype admixture between Capri and Palinuro does not allow detecting the source population, but nuclear data point to the Capri population as more proximate to the Strombolicchio one. Nevertheless, the presence of a second haplotype in the young islet, assuming that this haplotype may not have locally evolved or that it was present in Palinuro/Capri and gone undetected/extinct, points to a second colonization event from an unknown source population. This latter hypothesis is supported by the distinctiveness of exclusive Strombolicchio haplotype compared with those found in Palinuro/Capri (i.e., two mutation steps). The presumed source population, likely distributed along the south Tyrrhenian coastline, could be extinct today or even still existing. Despite the long history of floristic investigations in the Mediterranean Basin, the existence of unknown populations of E. saxicola could not be excluded due to the difficulty in recognizing the species (Strumia et al., 2015). Implementation of botanical surveys along the South Tyrrhenian coastlines, considering the Sea surface circulation and suitable rocky habitats, could fill this gap. Furthermore, a previous study on the genus Limonium showed that rising sea levels during the warm phases of the glaciation had led to the steady decline of coastal populations (Koutroumpa et al., 2021). We, therefore, cannot exclude that this flooding may have played a similar role in the extinction of some E. saxicola populations along the coastline.
What Determines the Genetic Diversity and Genetic Structure of Extant Populations?
The complete analysis of plastid genome variation revealed partial admixture and a very low level of between population differentiation (only 1–2 mutation steps). The different analyses of nuclear markers, performed with different settings, concordantly shows three main genetic clusters matching the three sampling localities of Palinuro, Capri, and Strombolicchio (Figure 2C). In contrast to plastidial admixture, the nuclear markers revealed a present story of strong genetic isolation for the three living populations (Figures 2–4; Table 3). Indeed, all sampled individuals have been correctly assigned to their source population in all analyses, thus almost indicating a complete absence of genetic admixture and ongoing gene flow (with corresponding high Fst values).
MIGRATE and BayesAss estimates revealed that historical (mh) and contemporary (mc) levels of migration between the three genetically distinct populations are very small and similar in magnitude (Supplementary Table 2), as also supported by the large generation times estimated by the simulation performed in DIYABC. The low levels of contemporary migration are not surprising because geographic distance and cliff habitat make dispersal between populations highly unlikely. However, as historical migration rates are also very low, concordant patterns strongly imply that the high levels of genetic structure currently observed among extant populations stem from a low colonization ability, which thus seems to be a long-standing life-history trait of E. saxicola. One evolutionary implication of similar low contemporary and historical migration rates over time is that E. saxicola has a long history of living in relatively small, isolated populations (still preserving enough genetic variation). In contrast to other island narrow endemics (Molins et al., 2009; Mayol et al., 2012; López-Pujol et al., 2013), for which the sea level oscillations represented a switch-off-switch-on the barrier to gene flow, sea levels theoretically do not represent a major barrier for seed dispersal of E. saxicola, as well as for other halophytes of both sandy and rocky coasts. Nonetheless, phases of sea lowering could have led to both the increase in available habitat through land emersion and the promotion of hopping colonization by the decrease in distance between isolated populations. However, phases of the current sea rising do not totally halt the potential dispersion of E. saxicola but decrease the probability of its occurrence due to the increased distance between a few suitable sites.
The DIYABC analysis identified two introductions temporally separated from the Palinuro to Capri, and from there to Strombolicchio (DIYABC model selection, Scenario 1) as the most likely hypothesis for E. saxicola presents distribution. By assuming a generation time of 10 years, we can consider that the introduction to Capri, approximately 0.25 Mya, and the colonization of Strombolicchio (0.09 Mya) took place around the late Pleistocene. The presence of inaccessible known plant spots (on Capri) and the possible occurrence of other unknown plant spots along the Palinuro cliffs suggest we probably have underestimated the census population sizes (N) for the three populations. Still, Ne estimations are near and even larger than N (Table 3), i.e., most of the individuals are breeding individuals in the population. Overall, the effective population size of E. saxicola is historically low because climatic oscillations during the quaternary glacial-interglacial cycles periodically forced populations to track their optimum into smaller cliff areas of suitable habitat. The almost exclusive preference of E. saxicola for north-facing cliffs suggests an adaptation to cooler conditions, which further highlights the possibility of different phases of population expansion/contraction during these oscillation cycles.
Can the Combination of Phylogeography and Genetic Diversity Help Define E. saxicola Conservation Priorities?
While geographic isolation seems the obvious explanation for the current population genetic structure, it partly contrasts with the levels of genetic diversity still detected within each population. Such genetic diversity within populations is unlikely to have resulted from recent or contemporary gene flow but rather points at relative demographic stability (as suggested by Ne estimations) both in recent and ancient timescales. All sampled individuals, despite a clumped distribution due to the accessibility of the cliff faces, were genetically different, indicating that clonality in this species is rare. Moreover, at least at the intrapopulation level, the long lifespan of adult plants, together with their high resprouting ability and wind pollination, may contribute to maintaining a high level of outcrossing and multiple random mating among the few individuals (i.e., observed heterozygosity was comparable to the expected one). Interestingly, wind pollination is an almost unique trait among Mediterranean narrow endemics. Indeed, anemophily is a plesiomorphic trait in E. saxicola and Amarantaceae, whereas, in the few other known wind-pollinated endemics, this mechanism is rather a secondary adaptation to the low availability of biotic pollination (Traveset and Navarro, 2017).
Regardless of the extremely small number of individuals in each population, the moderate degree of intrapopulation co-ancestry (Figure 5) suggests that a consistent level of crosses among unrelated individuals occurs. Even if the reproductive success of individual plants is unknown, the pattern of genetic relatedness among individuals of these small populations suggests that most of the adult plants contribute to the next generation, i.e., that random mating occurs. This reproductive behavior is consistent with the observation of Ho not different from He and a moderate excess of homozygotes in all populations compared with Hardy–Weinberg equilibrium expectations (Fis average, 0.10). The relative excess of homozygotes (higher Fis) observed in Palinuro denotes some heterogeneity in this mainland population compared to the islets, consisting of populations composed of several subgroups (the so-called Wahlund effect). Indeed, in contrast to the other two insular populations sampled on single cliffs, the Palinuro population was sampled along different cliffs. This subpopulation structure is also partially evident in the heatmap graph of Fst (Figure 4).
As reported for other extremely narrow endemics (see Médail and Baumel, 2018), E. saxicola still maintains some degree of genetic diversity even at the intrapopulation level (despite its small population sizes); this genetic trait may have represented life insurance for the long-term survivorship of this rare species. This survivorship was further aided by the low level of anthropogenic disturbance and high stability typical of cliff plant communities (Davis, 1951). Indeed, maintaining genetic diversity can reduce the probability of extinction of small populations by providing the standing genetic variation for local adaptation (Leimu and Fischer, 2008). Therefore, the present Palinuro population still holds enough genetic diversity to represent a valuable “donor population” for future reintroduction programs of this species. The high heterozygosity can be relevant in terms of conservation guidelines as genetic diversity of E. saxicola can be preserved in the long term even in its current isolated populations, confirming that narrow endemics are not necessarily “evolutionary dead-ends,” but rather may represent species that have a strong evolutionary legacy (Médail and Baumel, 2018). From a conservation perspective, we suggest that, at least in the short term, genetic factors may have little impact on the persistence of small populations of this rare paleoendemic species, but, rather, ecological factors (as persistence of suitable habitat) may play a larger role in determining whether populations survive in the long term. In this respect, both the IUCN category and the potential threats of extinction of this species (Santangelo et al., 2012; Orsenigo et al., 2018) should be reconsidered.
Conclusion
Even if short and long seed dispersion is still possible, why E. saxicola is so rare compared with other littoral halophytes species (Davis, 1951)? Indeed, the strong genetic differentiation detected among living populations suggests that the long-distance colonization typical of halophytes is merely occasional. We recognize that, independently of its colonization potential (through hydrochory), the rarity of this species must be considered also in terms of its specific requirements for habitat type as well as its biological features. Currently, E. saxicola has distributed exclusively on north facing (± 45° deviation from North) rocky shores (Strumia et al., 2015), but we do not have experimental evidence to support this restricted distribution. Since long-distance dispersal and colonization are different processes [see Feliner (2014) and references therein], we hypothesize that other factors determine the successful long-distance colonization in E. saxicola: namely, landing in a suitable microhabitat, seed germination, and, finally, seedling anchorage (Strumia et al., 2020b). Thus, the rarity of E. saxicola must be attributed not only in terms of the longevity and the ability of seeds to float but also, if not predominantly, of the habitat suitability of reached cliffs. These findings confirm the distinctiveness of E. saxicola from other halophytes and narrow endemics of the Mediterranean Basin. In conclusion, E. saxicola shows habitat specificity with individuals found predominately in small and sporadic populations. However, anemophilous pollination maintains a high-genetic diversity in this species, thus preventing the rapid decline, which is usually expected in small populations, thus ruling out the possibility of genetic impoverishment and inbreeding depression as immediate causes of threat.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject PRJNA756897.
Author Contributions
SS, AS, and DC: conceptualization. SS and AS: plant material collection. SC and DC: data curation. SC, DC, and TG: formal analysis. DC and SS: funding acquisition. DC and TG: methodology. SS, SC, DC, and AS: writing – original draft preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC were partially funded by a grant provided by Regione Campania – Direzione Generale per l’Ambiente e l’Ecosistema – Dip. 50 DG 06 U.O.D. 07.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the Cilento and Vallo di Diano and Alburni National Park and the Sicily Region – Regional Department of Rural and Land Development – Messina Office, Service 15 that authorized the activities in the field. The authors are extremely grateful to Mariacristina Villani for collecting S. hirsuta and to Antonio Federico for his kind support on the Capri Island and Giovanni Cammarano, Biagio Fedullo, and Diego Errico for their help in collecting plant materials in Palinuro. The authors are grateful to Marco Balducci for language revision, Simone Fior, and Nunzio D’Agostino for their comments on the manuscript. The authors are extremely grateful to two reviewers for their constructive criticism and helpful comments that significantly improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.737111/full#supplementary-material
Footnotes
- ^ www.bioinformatics.babraham.ac.uk/projects/fastqc
- ^ https://github.com/enormandeau/Scripts/blob/master/fastqCombinePairedEnd.py
- ^ http://tree.bio.ed.ac.uk/software/figtree
References
Aiello, G., Marsella, E., and D’Insanto, C. (2015). New morpho-bathymetric and tectono-stratigraphic data on Naples and Salerno Gulfs (Southern Tyrrhenian Sea, Italy) derived from bathymetric and sismic data analysis and integrated geological interpretation. Curr. Dev. Oceanogr. 8, 1–42. doi: 10.3390/geosciences10080319
Arafeh, R., and Kadereit, J. W. (2006). Long-distance seed dispersal, clone longevity and lack of phylogeographical structure in the European distributional range of the coastal Calystegia soldanella (L.) R. Br. (Convolvulaceae). J. Biogeogr. 33, 1461–1469. doi: 10.1111/j.1365-2699.2006.01512.x
Barone Lumaga, M. R., Santangelo, A., and Strumia, S. (2016). Morpho-functional traits influencing the fitness of highly endangered Eokochia saxicola (Guss.) Freitag & G.Kadereit (Amaranthaceae). Flora 218, 11–17. doi: 10.1016/j.flora.2015.11.005
Beaumont, M. A. (2010). Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. 41, 379–406. doi: 10.1146/annurev-ecolsys-102209-144621
Beerli, P. (2006). Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22, 341–345. doi: 10.1093/bioinformatics/bti803
Bermingham, E., and Moritz, C. (1998). Comparative phylogeography: concepts and applications. Mol. Ecol. 7, 367–369. doi: 10.1046/j.1365-294x.1998.00424.x
Blondel, J., Aronson, J., Bodiou, J., and Boeuf, G. (2010). The Mediterranean Region: Biological Diversity in Space and Time, 2nd Edn. New York, NY: Oxford University Press.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chapuis, M. P., Raynal, L., Plantamp, C., Meynard, C. N., Blondin, L., Marin, J. M., et al. (2020). A young age of subspecific divergence in the desert locust inferred by ABC random forest. Mol. Ecol. 29, 4542–4558. doi: 10.1111/mec.15663
Clausing, G., Vickers, K., and Kadereit, J. W. (2000). Historical biogeography in a linear system: genetic variation of Sea Rocket (Cakile maritima) and Sea Holly (Eryngium maritimum) along European coasts. Mol. Ecol. 9, 1823–1833. doi: 10.1046/j.1365-294x.2000.01083.x
Coissac, E., Hollingsworth, P. M., Lavergne, S., and Taberlet, P. (2016). From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 25, 1423–1428. doi: 10.1111/mec.13549
Collin, F. D., Durif, G., Raynal, L., Lombaert, E., Gautier, M., Vitalis, R., et al. (2021). Extending approximate Bayesian computation with supervised machine learning to infer demographic history from genetic polymorphisms using DIYABC Random Forest. Mol. Ecol. Resour. doi: 10.1111/1755-0998.13413 [Epub ahead of print].
Cozzolino, S., Scopece, G., Roma, L., and Schlüter, P. M. (2020). Different filtering strategies of genotyping-by-sequencing data provide complementary resolutions of species boundaries and relationships in a clade of sexually deceptive orchids. J. Syst. Evol. 58, 133–144. doi: 10.1111/jse.12493
Danecek, P., Auton, A., Abecasis, G., Cornelis, G., Albers, C. A., Banks, E., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Davis, P. H. (1951). Cliff vegetation in the Eastern mediterranean. J Ecol 39, 63–93. doi: 10.2307/2256628
De Castro, O., Innangi, M., and Menale, B. (2020). Message in a bottle: the Mediterranean Sea currents acted as protagonists in shaping the distribution of the sea daffodil (Pancratium maritimum, Amaryllidaceae). Bot. J. Linn. Soc. 194, 207–220. doi: 10.1093/botlinnean/boaa037
Diniz-Filho, J. A. F., Campos Telles, M., Bonatto, S., Eizirik, E., De Freitas, T. R., De Marco, P. Jr., et al. (2008). Mapping the evolutionary twilight zone:molecular markers, populations and geography. J. Biogeogr. 35, 753–763. doi: 10.1111/j.1365-2699.2008.01912.x
Favarger, C., and Contandriopoulos, J. (1961). Essai sur l‘endémisme. Biol. Soc. Bot. Suisse 71, 384–408.
Feliner, G. N. (2014). Patterns and processes in plant phylogeography in the Mediterranean Basin. A review. Perspect. Plant Ecol. 16, 265–278. doi: 10.1016/j.ppees.2014.07.002
Gao, X., and Starmer, J. D. (2008). AWclust: point-and-click software for non-parametric population structure analysis. BMC Bioinformatics 9:77. doi: 10.1186/1471-2105-9-77
Gargiulo, R., Kull, T., and Fay, M. F. (2021). Effective double-digest RAD sequencing and genotyping despite large genome size. Mol. Ecol. Resour. 21, 1037–1055. doi: 10.1111/1755-0998.13314
Goudet, J. (2005). Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186.
Greuter, W. (1991). Botanical diversity, endemism, rarity, and extinction in the Mediterranean area: an analysis based on the published volumes of Med-Checklist. Bot. Chron. 10, 63–79.
Heuertz, M., Fineschi, S., Anzidei, M., Pastorelli, R., Salvini, D., Paule, L., et al. (2004). Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol. Ecol. 13, 3437–3452. doi: 10.1111/j.1365-294X.2004.02333.x
Hewitt, G. M. (2001). Speciation, hybrid zones and phylogeography: or seeing genes in space and time. Mol. Ecol. 10, 537–549. doi: 10.1046/j.1365-294x.2001.01202.x
Jombart, T. (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Kadereit, G., and Freitag, H. (2011). Molecular phylogeny of Camphorosmeae (Camphorosmoideae, Chenopodiaceae): implications for biogeography, evolution of C4 photosynthesis and taxonomy. Taxon 60, 51–78. doi: 10.1002/tax.601006
Kadereit, J. W., Arafeh, R., Somogyi, G., and Westberg, E. (2005). Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale. Taxon 54, 861–876. doi: 10.2307/25065567
Kadereit, J. W., and Westberg, E. (2007). Determinants of phylogeographic structure: a comparative study of seven coastal flowering plant species across their European range. Watsonia 26, 229–238.
Koutroumpa, K., Warren, B. H., Theodoridis, S., Coiro, M., Romeiras, M. M., Jiménez, A., et al. (2021). Geo-Climatic changes and apomixis as major drivers of diversification in the Mediterranean Sea Lavenders (Limonium Mill.). Front. Plant Sci. 11:612258. doi: 10.3389/fpls.2020.612258
Kruckeberg, A. R., and Rabinowitz, D. (1985). Biological aspects of endemism in higher plants. Annu. Rev. Ecol. Syst. 16, 447–479. doi: 10.1146/annurev.es.16.110185.002311
Lavergne, S., Thompson, J. D., Garnier, E., and Debussche, M. (2004). The biology and ecology of narrow endemic and widespread plants: a comparative study of trait variation in 20 congeneric pairs. Oikos 107, 505–518. doi: 10.1111/j.0030-1299.2004.13423.x
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Leimu, R., and Fischer, M. (2008). A meta-analysis of local adaptation in plants. PLoS One 3:e4010. doi: 10.1371/journal.pone.0004010
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
López-Pujol, J., Martinell, M. C., Massò, S., Blanché, C., and Sáez, L. (2013). The ‘paradigm of extremes’: extremely low genetic diversity in an extremely narrow endemic species, Coristospermum huteri (Umbelliferae). Plant Syst. Evol. 299, 439–446. doi: 10.1007/s00606-012-0732-3
Malinsky, M., Trucchi, E., Lawson, D. J., and Falush, D. (2018). RADpainter and fineRADstructure: population inference from RADseq data. Mol. Biol. Evol. 35, 1284–1290. doi: 10.1093/molbev/msy023
Mayol, M., Palau, C., Rosselló, J. A., González-Martínez, S. C., Molins, A., and Riba, M. (2012). Patterns of genetic variability and habitat occupancy in Crepis triasii (Asteraceae) at different spatial scales: insights on evolutionary processes leading to diversification in continental islands. Ann. Bot. London 109, 429–441. doi: 10.1093/aob/mcr298
Médail, F. (2017). The specific vulnerability of plant biodiversity and vegetation on Mediterranean islands in the face of global change. Reg. Environ. Change 17, 1775–1790. doi: 10.1007/s10113-017-1123-7
Médail, F., and Baumel, A. (2018). Using phylogeography to define conservation priorities: the case of narrow endemic plants in the Mediterranean Basin hotspot. Biol. Conserv. 224, 258–266. doi: 10.1016/j.biocon.2018.05.028
Médail, F., and Diadema, K. (2009). Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr. 36, 1333–1345. doi: 10.1111/j.1365-2699.2008.02051.x
Médail, F., and Quézel, P. (1999). Hot-spots analysis for conservation of plant biodiversity in the Mediterranean Basin. Cons. Biol. 13, 1510–1513. doi: 10.1016/j.biocon.2018.05.028
Molins, A., Mayol, M., and Rosselló, J. A. (2009). Phylogeographical structure in the coastal species Senecio rodriguezii (Asteraceae), a narrowly distributed endemic Mediterranean plant. J. Biogeogr. 36, 1372–1383. doi: 10.1111/j.1365-2699.2009.02108.x
Montmollin, B. D., and Strahm, W. (eds) (2005). “The Top 50 Mediterranean Island plants: wild plants at the brink of extinction, and what is needed to save them,” in IUCN/SSC Mediterranean Islands Plant Specialist Group, (Gland: IUCN).
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Orsenigo, S., Montagnani, C., Fenu, G., Gargano, D., Peruzzi, L., Abeli, T., et al. (2018). Red Listing plants under full national responsibility: extinction risk and threats in the vascular flora endemic to Italy. Biol. Cons. 224, 213–222. doi: 10.1016/j.biocon.2018.05.030
Paris, J. R., Stevens, J. R., and Catchen, J. M. (2017). Lost in parameter space: a road map for stacks. Methods Ecol. Evol. 8, 1360–1373. doi: 10.1111/2041-210X.12775
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S., and Hoekstra, H. E. (2012). Double digest RADseq: an inexpensive method for De Novo SNP discovery and genotyping in model and non-model species. PLoS One 7:e37135. doi: 10.1371/journal.pone.0037135
Procaccini, G., Orsini, L., Ruggiero, M. V., and Scardi, M. (2001). Spatialpatterns of genetic diversity in Posidonia oceanica, anendemic Mediterranean seagrass. Mol. Ecol. 10, 1413–1421. doi: 10.1046/j.1365-294x.2001.01290.x
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Quezel, P. (1985). “Definition of the Mediterranean region and the origin of its flora,” in Plant Conservation in the Mediterranean Area, ed. C. Gomez Campo (Dordrecht: W. Junk), 9–24.
Rinaldi, E., Buongiorno Nardelli, B., Zambianchi, E., Santoleri, R., and Poulain, P.-M. (2010). Lagrangian and Eulerian observations of the surface circulation in the Tyrrhenian Sea. J. Geophys. Res. 115:C04024. doi: 10.1029/2009JC005535
Rochette, N. C., Rivera-Colón, A. G., and Catchen, J. M. (2019). Stacks 2: analytical methods for paired-end sequencing improve radseq-based population genomics. Mol. Ecol. 28, 4737–4754. doi: 10.1111/mec.15253
Santangelo, A., Croce, A., Lo Cascio, P., Pasta, S., Strumia, S., and Troìa, A. (2012). Eokochia saxicola (Guss.) Freitag and G. Kadereit. Inf. Bot. Ital. 44, 428–431.
Santangelo, N., Di Donato, V., Lebreton, V., Romano, P., and Russo Ermolli, E. (2011). Palaeolandscapes of Southern Apennines during the late early and the Middle Pleistocene. Quat. Int. 267, 20–29. doi: 10.1016/j.quaint.2011.02.036
Santangelo, N., Romano, P., Ascione, A., and Russo Ermolli, E. (2017). Quaternary evolution of the Southern Apennines coastal plains. A review. Geol. Carpath. 68, 43–56. doi: 10.1515/geoca-2017-0004
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stebbins, G. L., and Major, J. (1965). Endemism and speciation in the California flora. Ecol. Monogr. 35, 1–35. doi: 10.2307/1942216
Straub, S. C., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., and Liston, A. (2012). Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 99, 349–364.
Strumia, S., Buonanno, M., Aronne, G., Santo, A., and Santangelo, A. (2020a). Monitoring of plant species and communities on coastal cliffs: is the use of unmanned aerial vehicles suitable? Diversity 12:149. doi: 10.3390/d12040149
Strumia, S., Santangelo, A., and Barone Lumaga, M. R. (2020b). Seed germination and seedling roots traits of four species living on Mediterranean coastal cliffs. Plant Biosyst. 154, 990–999. doi: 10.1080/11263504.2020.1837284
Strumia, S., Croce, A., and Santangelo, A. (2015). New distributional data of the rare endemic species Eokochia saxicola (Guss.) Freitag and G.Kadereit (Chenopodiaceae): effects on biogeography and conservation. Plant Biosyst. 149, 559–564. doi: 10.1080/11263504.2013.870246
Suc, J. P. (1984). Origin and evolution of the Mediterranean vegetation and climate in Europe. Nature 307, 429–432. doi: 10.1038/307429a0
Taberlet, P., Fumagalli, L., Wust-Saucy, A. G., and Cosson, J. F. (1998). Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 7, 453–464. doi: 10.1046/j.1365-294x.1998.00289.x
Traveset, A., and Navarro, L. (2017). State of the art of the plant reproductive ecology and evolution in the Mediterranean Islands. Plant Biol. 20(Suppl. 1), 63–77. doi: 10.1111/plb.12636
Vetrano, A., Gasparini, G. P., Molcard, R., and Astraldi, M. (2004). Water flux estimates in the central Mediterranean Sea from an inverse box model. J. Geophys. Res. 109:C0101903. doi: 10.1029/2003JC001903
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wallace, A. R. (1880). Island Life: or The Phenomena and Causes of Insular Faunas and Floras, Including a Revision and Attempted Solution of the Problem of Ecological Climates. London: MacMillan and Co.
Walter, H. (1985). Vegetation of the Earth and Ecological Systems of the Geobiosphere, 3rd Edn. Berlin: Springer-Verlag.
Weising, K., and Freitag, H. (2007). Phylogeography of halophytes from European coastal and inland habitats. Zool. Anz. 246, 279–292. doi: 10.1016/j.jcz.2007.07.005
Westberg, E., and Kadereit, J. W. (2009). The influence of sea currents, past disruption of gene flow and species biology on the phylogeographical structure of coastal flowering plants. J. Biogogr. 36, 1398–1410. doi: 10.1111/j.1365-2699.2008.01973.x
Whittaker, R. J., and Fernández-Palacios, J. M. (2007). Island Biogeography, Ecology, Evolutionand Conservation, 2nd Edn. Oxford: Oxford University Press.
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352.
Wingett, S. W., and Andrews, S. (2018). FastQ screen: a tool for multi-genome mapping and quality control. F1000Research 7, 1338. doi: 10.12688/f1000research.15931.2
Keywords: contemporary and historical gene flow, ddRAD, DIYABC, halophytes, IUCN, long distance dispersal, plant conservation, quaternary sea level oscillations
Citation: Strumia S, Santangelo A, Galise TR, Cozzolino S and Cafasso D (2021) Hopping or Jumping on the Cliffs: The Unusual Phylogeographical and Demographic Structure of an Extremely Narrow Endemic Mediterranean Plant. Front. Plant Sci. 12:737111. doi: 10.3389/fpls.2021.737111
Received: 06 July 2021; Accepted: 07 October 2021;
Published: 10 November 2021.
Edited by:
Gonzalo Nieto Feliner, Real Jardín Botánico (RJB), Spanish National Research Council (CSIC), SpainReviewed by:
Giovanni Zecca, Independent Researcher, Milan, ItalyWaldir M. Berbel-Filho, Swansea University, United Kingdom
Copyright © 2021 Strumia, Santangelo, Galise, Cozzolino and Cafasso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salvatore Cozzolino, Y296em9saW5AdW5pbmEuaXQ=
 Sandro Strumia
Sandro Strumia Annalisa Santangelo
Annalisa Santangelo Teresa Rosa Galise
Teresa Rosa Galise Salvatore Cozzolino2*
Salvatore Cozzolino2* Donata Cafasso
Donata Cafasso