- 1State Key Laboratory of Forest Genetics and Tree Breeding, Northeast Forestry University, Harbin, China
- 2Biology Group, Jiamusi No.1 High School, Jiamusi, China
- 3Economic Forest Laboratory, Mudanjiang Branch of Heilongjiang Academy of Forestry, Mudanjiang, China
In eukaryotes, alternative splicing (AS) is a crucial regulatory mechanism that modulates mRNA diversity and stability. The contribution of AS to stress is known in many species related to stress, but the posttranscriptional mechanism in poplar under cold stress is still unclear. Recent studies have utilized the advantages of single molecular real-time (SMRT) sequencing technology from Pacific Bioscience (PacBio) to identify full-length transcripts. We, therefore, used a combination of single-molecule long-read sequencing and Illumina RNA sequencing (RNA-Seq) for a global analysis of AS in two poplar species (Populus trichocarpa and P. ussuriensis) under cold stress. We further identified 1,261 AS events in P. trichocarpa and 2,101 in P. ussuriensis among which intron retention, with a frequency of more than 30%, was the most prominent type under cold stress. RNA-Seq data analysis and annotation revealed the importance of calcium, abscisic acid, and reactive oxygen species signaling in cold stress response. Besides, the low temperature rapidly induced multiple splicing factors, transcription factors, and differentially expressed genes through AS. In P. ussuriensis, there was a rapid occurrence of AS events, which provided a new insight into the complexity and regulation of AS during cold stress response in different poplar species for the first time.
Introduction
In eukaryotes, precursor messenger RNAs (pre-mRNAs) with multiple introns undergo alternative splicing (AS) to generate two or more mature mRNA isoforms, encoding structurally and functionally different proteins (Palusa et al., 2007). AS is a crucial regulatory mechanism that contributes to cellular and functional complexity and has been extensively studied in animals and plants (Wang et al., 2018; Li et al., 2019). Genome-wide investigation of AS has been performed on development or in response to stresses in multiple plants. Studies have revealed that an estimated 60% of Arabidopsis, 50% of soybean, 40% of cotton, and 40% of maize intron-containing genes undergo AS (Marquez et al., 2012; Syed et al., 2012; Li et al., 2019). Five major types of AS events are recognized, namely exon skipping (ES), intron retention (IR), mutually exclusive exon (MXE), alternative 5′ splice site (A5SS), and alternative 3′ splice site (A3SS), of which IR is the most common event in plants (Reddy et al., 2013). Evidence suggests that plants employ AS to achieve phenotypic plasticity in response to different abiotic stresses, such as heat, drought, salinity, and cold (Zhao et al., 2014; Liu et al., 2018; Zhu et al., 2018).
Several studies have identified and reported the complexity of AS in plant species, such as cotton (Wang et al., 2018), cassava (Li et al., 2019), Phyllostachys edulis (Wang et al., 2017), Populus trichocarpa (Bao et al., 2013), Arabidopsis (Marquez et al., 2012), and rice (Zhang et al., 2019). Different from the constitutively spliced isoforms, alternatively spliced ones always show cell-, tissue-, or condition-specific expression patterns, and the extent of AS in plants depends on the complexity of tissues (Kalsotra and Cooper, 2011). Gan et al. (2011) identified multiple varied AS events among the different genotypes of Arabidopsis thaliana. Zhou et al. (2011) discovered a total of 45 AS events in Brassica oleracea in two organs and under two environmental conditions (heat and cold), while the splice sites in the pre-mRNA are recognized by splicing factors (SFs), which recruit the splice osome for intron removal, to generate different alternatively spliced isoforms (Fu and Ares, 2014; Lee and Rio, 2015). Besides, SFs are essential for plant growth and development processes, including control of flowering time, regulation of the circadian rhythms, and response to abiotic stresses (Staiger and Brown, 2013; Schlaen et al., 2015). These studies indicate that the regulated AS of downstream targets is essential for plants. A recent survey by Calixto et al. (2018) has reported a massive and rapid AS response under cold stress that involved changes in hundreds of cold-responsive transcription factors (TFs) and SFRNA-binding proteins in Arabidopsis. Moreover, misexpression of SFs altered cold sensitivity or tolerance in plants (Reddy et al., 2013; Staiger and Brown, 2013). In Arabidopsis, low temperature regulated the expression and splicing patterns of the serine/arginine-rich SFs (Palusa et al., 2007). These studies strongly suggest the central role of SFs and the importance of AS in plant response to cold stress.
Studies have widely used short-read RNA sequencing (RNA-Seq) technology to detect AS events; however, it is challenging to identify full-length splicing isoforms accurately by this method. By contrast, the SMRT developed by Pacific Biosciences (PacBio, CA, USA) offers an improvement in read length over the previous technologies. PacBio technology is valuable for the annotation of newly sequenced genes and the analysis of AS (Eid et al., 2009; Sharon et al., 2013). Researchers have started using PacBio sequencing technology to characterize the complexity of transcriptome in various plant species.
Low temperature is one of the most common stresses that negatively affect plant growth and crop production (Calixto et al., 2018). Numerous studies have demonstrated changes in AS events in response to external cold/chilling stimuli; however, the regulation of AS in poplar species under cold stress is not clear at the whole transcriptomic level. In this study, it was better cold tolerance in P. ussuriensis than in P. trichocarpa. We aim to investigate the reason for this. Our data reveal that cold intensely affects gene expression via alternative splicing regulated by SFs in P. ussuriensis to resistant cold stress and advances our understanding of the high complexity and specificity of species-specific AS regulation in response to cold.
Materials and Methods
Plant Materials and Growth Conditions
Populus trichocarpa “Nisqually-1” and P. ussuriensis Kom. were used in this study. Stem segments (3 cm long) of these poplar species were cultured in ½ MS medium for 1 month. These clonally cultured seedlings were placed at 25 ± 2°C under 16-h/8-h (light/dark) photoperiod. Both 2-week-old plants were transferred to a chamber for cold treatment at 3 and −3°C for 3 h, respectively. The young leaves and shoot apices of the control sample (25°C), 3°C, and −3°C treated samples were collected at the same time and immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Each time point was repeated with three biological replicates. The nine RNAs samples (25, 3, and −3°C with three replicates) from each poplar species were subjected to 150 bp paired-end sequencing using the Illumina HiSeq platform, respectively. The nine RNAs samples from each poplar species were mixed in equal volume and sequenced on the PacBio RS II platform, respectively (Figure 1A).

Figure 1. Phenotypic changes in P. trichocarpa and P. ussuriensis in response to cold stress. P. trichocarpa and P. ussuriensis grown at 25°C for 3 h (A); 3°C for 3h (B); −3°C for 3 h (C); and −3°C for 4 h (D).
RNA Extraction and the SMRT Sequencing Library Construction
Total RNA was extracted from the leaves and shoot apices of P. trichocarpa and P. ussuriensis maintained under different temperatures (25, 3, and −3°C) using the CTAB method (Jaakola et al., 2001). The total RNA was assessed using Agilent Bioanalyzer 2100 (Agilent, https://www.agilent.com). RNA purity was checked using the kaiao K5500® Spectrophotometer (Kaiao, Beijing, China). RNA integrity and concentration were assessed using the Bioanalyzer 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA). Total RNA of the nine samples from each species was pooled in equal amounts, and 1 μg of the pooled RNA was used for cDNA synthesis and SMRTbell library construction. The purified RNA was reversely transcribed into cDNA using the SMARTer PCR cDNA Synthesis Kit (Clontech Laboratories, Inc. Mountain View, CA, USA). The cDNA was amplified using the Kapa HiFi PCR Kit (Kapa Biosystems, Wilmington, MA, USA). Size selection was carried out on a BluePippin (Sage Science, Beverly, MA, USA), and 1–2, 2–3, 3–6, and 5–10 kb fractions were collected. These collected cDNA fractions were treated with a DNA damage repair mix, followed by end repair and ligation of SMRT adapters using the PacBio SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA) to create SMRT sequencing libraries.
PacBio Data Analysis
Raw data generated by the PacBio RSII platform were processed following the Iso-seq method. The PacBio raw reads were processed into error-corrected “reads of inserts” (ROIs) using ToFu (version 2.3.0) (Gordon et al., 2015) with the following parameters: minimum full pass, >1; minimum ROI length, >200 nucleotides; and prediction accuracy, >0.8. The ROIs were classified into circular consensus sequences (CCS) and non-CCS subreads based on the presence or absence of sequencing adapters. CCS subreads were classified into full-length non-chimeric reads (FLNC) or non-FLNC based on the presence of both the primer sequences (the 5′ and 3′ sequences) and the polyA tail signal.
Next, isoform-level clustering algorithm ICE (Iterative Clustering for Error Correction) (Gordon et al., 2015) was applied to all the full-length (FL) transcripts to obtain the consensus transcripts based on the sequence similarity and generate a consensus sequence for each cluster. Quiver was used for error correction to obtain high-quality (HQ) and low-quality (LQ) isoforms (accuracy, ≥99%). Finally, high-quality FL transcripts were combined to obtain all high-quality FL transcripts of each sample.
Library Preparation and Illumina Sequencing
Illumina sequencing was performed to generate data that can be used to validate and quantify the PacBio transcripts. Approximately, 2 μg of RNA per sample was used as input material for the Illumina sequencing. The clustering of the index-coded samples was performed on a cBot cluster generation system (IlluminaHiSeq PE Cluster Kit v4-cBot-HS), following the instructions of the manufacturer. After cluster generation, the libraries were sequenced on an Illumina platform, and 150 bp paired-end reads were generated. In order to guarantee the data quality, the original data (raw data) were filtered. The read counts for each gene in each sample were generated using HTSeq v0.6.0, and FPKM (fragments per kilobase million mapped reads) values were then calculated to estimate the expression level of the genes in each sample. Gene annotation was based on the following databases: Nr (NCBI nonredundant protein sequences), Nt (NCBI nonredundant nucleotide sequences), Swiss-Prot (a manually annotated and reviewed protein sequence database), Pfam (protein family), COG (clusters of orthologous groups of proteins), GO (gene ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes) orthology database (Kanehisa et al., 2008), and GO enrichment analysis of the differentially expressed genes (DEGs) were implemented by the GOseq (Young et al., 2010). Genes with q ≤ 0.05 and |log2_ratio| ≥ 1 are identified as DEGs.
Identification of Differentially Spliced Events
The tool ASprofile (version b-1.0.4) was employed to classify the AS events using the raw.gtffiles assembled from the Illumina RNA-Seq and SMRT-Seq data of P. ussuriensis compared with P. trichocarpa. The AS events, including IR, ES, MXE, A5SS, and A3SS, were extracted and counted. The differential alternative splicing (DAS) events from two poplar comparisons at different temperatures were identified by rMATS.3.2.2 (Shen et al., 2014). The AS events with a false discovery rate (FDR) <0.05 were defined as DAS events. GO enrichment analyses with DAS were conducted using AgriGO (Tian et al., 2017).
Calcium and Physiological Indicators Determination
After cold stress, the leaves of 25, 3, −3°C for P. trichocarpa and P. ussuriensis were collected and dried at 70°C in an oven. Approximately 0.5 mg (dry weight) of samples were weighted and analyzed using an inductively coupled plasma (ICP) emission spectrometer (ICP-OES 5110 VDV, Agilent, USA). These data were averaged from three replicates (n = 3).
For MDA content measurement, about 50 mg of leaves were ground and homogenized in 1 ml of 0.1% (w/v) TCA for 10 min and centrifuged at 10,000 g for 15 min at 4°C. The supernatant was reacted with 20% (w/v) TCA, containing 0.5% (w/v) thiobarbituric acid. After being boiled and cooled, it was centrifuged at 10,000 g for 5 min at 4°C. The detailed method was performed as described by Metwally et al. (2003). These data were averaged from three replicates (n = 3).
The H2O2 was measured using an Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen, Carlsbad, CA, USA) as described in Xing et al. (2008). Leaves of poplars were frozen in N2 and ground. The phosphate buffer (20 mM K2HPO4, pH6.5) was added to 50 mg of ground frozen tissue. After centrifugation, the supernatant was incubated with 0.2-U ml−1 horseradish peroxidase and 100-μM Amplex Red reagent (10-acetyl-3,7-dihydrophenoxazine) at room temperature for 30 min in darkness. The fluorescence was quantified using FLUOStarOptima (excitation at 560 nm and emission at 590 nm). These data were averaged from three replicates (n = 3).
Validation of Alternative Splicing Transcripts
Total RNA was extracted from the leaves and shoot apices of P. ussuriensis as described above, which was consistent with transcriptome sequencing. The TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, China) was used for simultaneous genomic DNA removal and cDNA synthesis (20 μl) following the instructions of the manufacturer. The AS transcripts were validated by reverse transcription-PCR (RT-PCR) using specific primers listed in Supplementary Table S1.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA extraction and cDNA synthesis were performed as described above. QRT–PCR was performed to validate the expression level of DEGs, DAS isoforms that respond to cold stress, DAS of splicing-associated factors, and TFs. The PuActin gene was used as the internal control. The relative gene expression values in P. ussuriensis were analyzed using the 2−ΔΔCt method compared with P. trichocarpa. All reactions were performed using three biological replicates for each sample. The primers are listed in Supplementary Tables S2–S5.
Statistical Analysis
All statistical analyses were performed using SPSS software version 20 (IBM, Chicago, USA), and significant differences were evaluated using analysis of variance (ANOVA). Statistically significant differences were indicated by *p < 0.05 and **p < 0.01.
Results
Phenotypic Changes in Two Poplar Species in Response to Cold Stress
Low temperature is one of the key environmental stresses, which negatively impairs plant growth and development (Li et al., 2017). To evaluate the tolerance of cold, we compared the phenotypic changes of two poplar species P. trichocarpa and P. ussuriensis, which are tolerant and sensitive to cold stress, respectively. About 5 cm height of seedlings was used for physiological and transcriptome analyses at 3°C for 3 h and −3°C for 3 h. The seedlings cultured at room temperature (Figure 1A) were used as control. Treatment-related changes in the phenotype of P. trichocarpa were observed after 4 h at −3°C (Figure 1D); the leaves were damaged and wilted, while the P. ussuriensis plants were not damaged (Figure 1D). In comparison, no difference was observed after 3 h at 3°C (Figure 1B) and −3°C (Figure 1C) between the two species.
Calcium, MDA, POD, and H2O2 Determination
At room temperature, the content of calcium, MDA, POD, and H2O2 in P. ussuriensis was not different from them in P. trichocarpa (Figure 2). As the temperature fell, the calcium (Figure 2A), MDA (Figure 2B), POD (Figure 2C), and H2O2 (Figure 2D) content in both poplars was increased. But the calcium (Figure 2A), and POD (Figure 2C) content in P. ussuriensis increased more than that in P. trichocarpa, while the MDA (Figure 2B) and H2O2 (Figure 2D) content in P. ussuriensis decreased more than that in P. trichocarpa at 3 and −3°C.
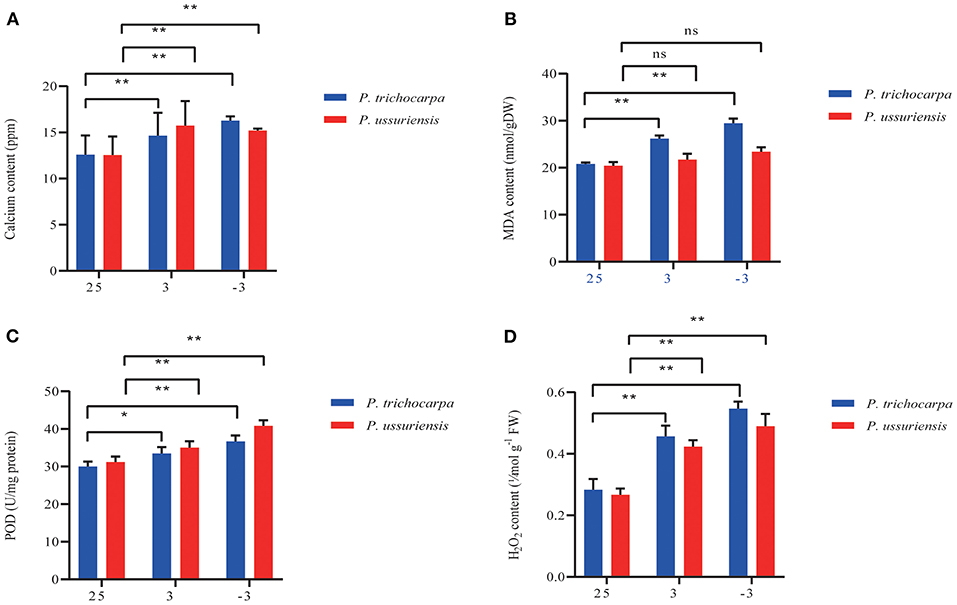
Figure 2. Calcium and physiological indicators measurement in P. trichocarpa and P. ussuriensis. (A) Calcium content measurement using inductively coupled plasma (ICP) emission spectrometer (ICP-OES 5110 VDV, Agilent, USA). (B) MDA, (C) POD, and (D) H2O2 content measurement. *P < 0.05; **P < 0.01. ns denotes no significance.
Overview of PacBio Iso-Seq Data of Two Poplar Species
We used P. ussuriensis, the widely cultivated species in northeast China, to compare with P. trichocarpa for this study. We performed PacBio sequencing and conduce analysis based on P. trichocarpa genome. From the phenotypic point of view, P. ussuriensis was more tolerant to lower temperatures than P. trichocarpa. To better understand the full-length splice variants, novel genes, and alternative polyadenylation (APA) sites of the two species in response to cold stress, the PacBio Iso-Seq method was used to sequence the transcriptomes. A total of two SMRT cells (r64053_20191003_023517_4_H01 for P. trichocarpa; r64053_20191003_023517_4_H01 for P. ussuriensis) mixed with nine samples were constructed to eliminate instrumental bias toward short fragments, respectively. A dataset with more than 20 Gb of clean reads was obtained after filtering using SMRTLink (6.0). Referring to P. trichocarpa genome, a total of 51,153 and 58,082 consensus isoforms were obtained from the two libraries (P. trichocarpa and P. ussuriensis), respectively (Supplementary Table). CCS reads were self-corrected to obtain high-quality (HQ) reads of insert (ROI) when minimum full pass, >1; minimum ROI length, >200 nt; and prediction accuracy, >0.8 for adaptors. Then, a clustering algorithm, isoform-level clustering algorithm (ICE), was applied to all full-length transcripts to generate the consensus transcripts. It grouped them into clusters based on the sequence similarity and generated a consensus isoform for each cluster. Subsequently, the untested complete insert sequence was aligned back to the consensus isoform, and then corrected to obtain HQ isoforms and low-quality (LQ) isoforms. Due to the removal of a lot of redundancy, HQ isoforms were significantly reduced compared to CCS reads. The mean length of consensus isoforms in the two libraries was between 1,683 and 1,910 bp, respectively (Table 1). A total of 50,910 reads of HQ isoforms with a mean length of 1,678 bp were identified from the library of P. trichocarpa (Table 1). A total of 57,872 reads of HQ isoforms with a mean length of 1,905 bp were identified from the library of P. ussuriensis (Table 1). Among which, a total of 228 and 188 low-quality LQ isoforms with a mean length of 1,157 and 1,344 bp were obtained, respectively (Table 1). For SMRT-seq analysis, with a total of 26,153,342 subreads, 330,371 CCS reads, and 35,414 consensus reads appeared after the modification of a site mismatch (Table 2). Mapping to the reference genome of P. trichocarpa RSEM (1.2.31) software (Li and Dewey, 2011), a total of 97.66 and 97.51% of the isoforms detected by PacBio SMRT-Seq were mapped reads in P. trichocarpa (Figure 3A) and P. ussuriensis (Figure 3B), respectively; 1.8 and 1.02% were multi-mapped to the genome in P. trichocarpa (Figure 3A) and P. ussuriensis (Figure 3B), respectively. The mapped isoforms (density) were spread across different chromosomes.
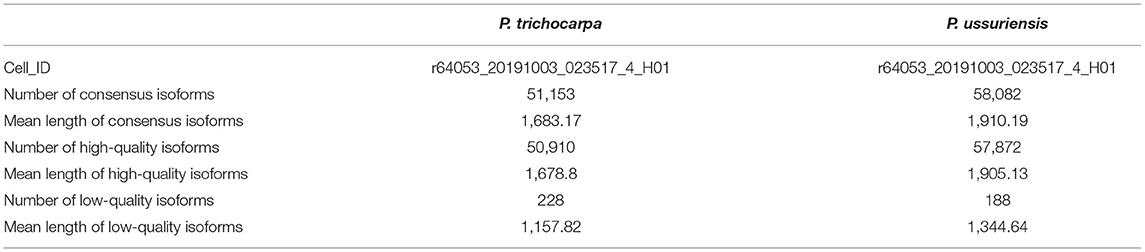
Table 1. Difference in genes as annotated in two poplar species compared with P. trichocarpa genome by the PacBio sequencing data, respectively.
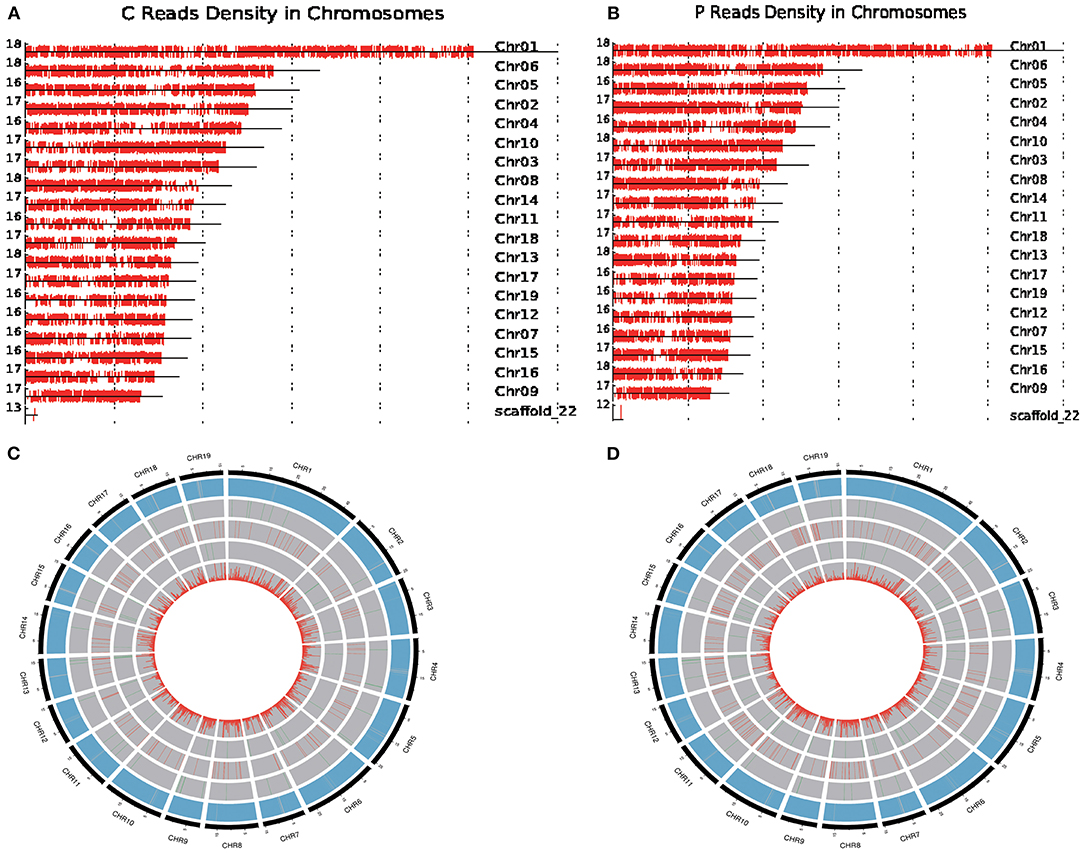
Figure 3. Circos visualization of PacBio Isoseq data in P. trichocarpa and P. ussuriensis compared using SMRT sequencing. (A,B) Distribution of full-length reads density in P. trichocarpa. and P. ussuriensis, respectively, on reference chromosome, respectively. (C,D) The distribution of genetic variants in P. trichocarpa. and P. ussuriensis, respectively.
Furthermore, we compared the genome-wide genetic variants, including chromosome distribution, gene density, novel gene, novel long noncoding RNA (lncRNA), APA site, and single-nucleotide polymorphism (SNP), between P. trichocarpa and P. ussuriensis (Figure 3) using the Circos program. A total of 852 (Figure 3C) and 1,093 (Figure 3D) novel genes and 25 (Figure 3C) and 42 (Figure 3D) APA sites were detected in P. trichocarpa and P. ussuriensis, respectively. A total of 235,900 (Figure 3C) and 49,781 (Figure 3D) SNPs were detected in P. trichocarpa and P. ussuriensis, respectively.
Identification of Known and Novel Alternative Splicing (AS) Events by SMRT-Seq
To investigate the role of AS in response to cold stress, we surveyed the transcript isoforms in the two poplar species. We examined the five major types of AS events (IR, ES, MXE, A5SS, and A3SS) in the isoforms using SMRT-Seq (Figure 4A). We compared the frequency of AS types from the two poplar species at 25, −3, and 3°C, respectively. Based on the reference genome (P. trichocarpa), 35.7% of the AS events were IR, 30.3% were A5SS, 24.7% were A3SS, 9.% were ES, and 0.3% were MXE in P. trichocarpa (Figure 4B) while 34.7% of the AS events were IR, 32.7% were A5SS, 24.5% were A3SS, 7.9% were ES, and 0.2% were MXE in P. ussuriensis (Figure 4C). The number of AS events in P. ussuriensis under cold stress was more than that in P. trichocarpa (Figure 4D). Among the AS types, IR, with a frequency of more than 30%, was the most abundant type under cold stress (Figure 4E). Interestingly, the percentage of different AS types changed with a decrease in temperature in both poplar species, especially IR dramatically increased and ES decreased at −3°C compared with that at 3°C both in P. trichocarpa and P. ussuriensis (Figures 4E,F).
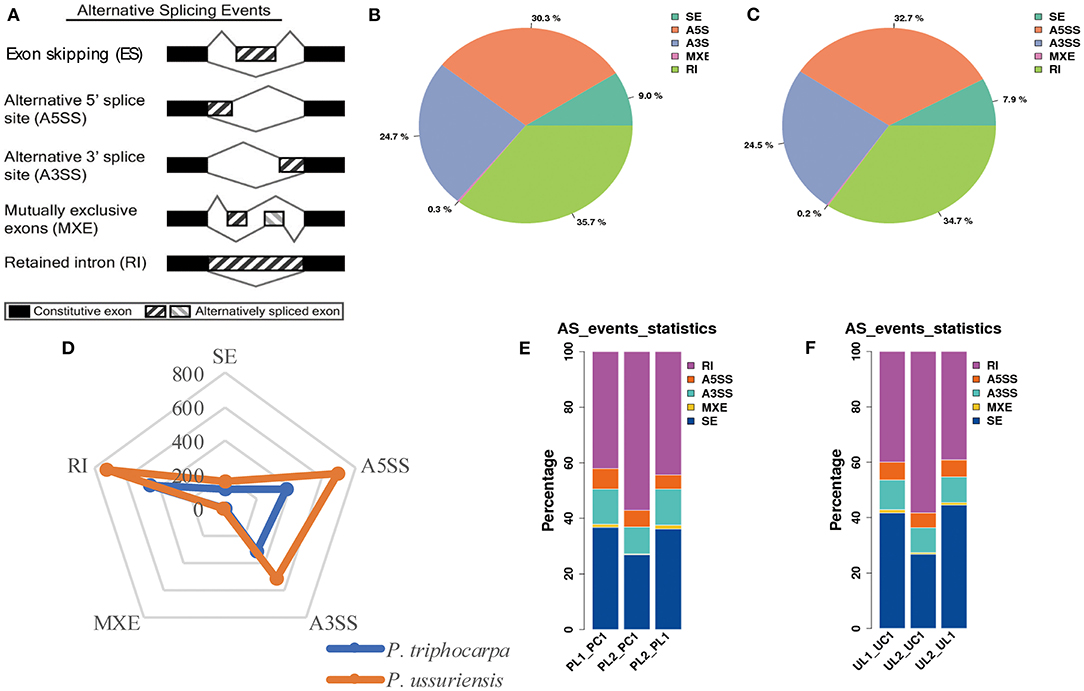
Figure 4. Comparison of alternative splicing (AS) events in response to cold stress in P. trichocarpa and P. ussuriensis identified by PacBio sequencing. (A) AS types. (B) The proportion of different AS types in response to cold stress in P. trichocarpa. (C) The proportion of different AS types in response to cold stress in P. ussuriensis. (D) A radar plot showing the percentage of AS types in response to cold stress in P. trichocarpa and P. ussuriensis. (E) Comparison of the AS types at different cold temperatures in P. trichocarpa. PL1_PC1: AS percentage statistical analysis at 3°C compared with 25°C in P. trichocarpa; PL2_PC1: AS percentage statistical analysis at −3°C compared with 25°C in P. trichocarpa; PL2_PL1: AS percentage statistical analysis at −3°C compared with 3°C in P. trichocarpa. (F) Comparison of the AS types at different cold temperatures in P. ussuriensis. UL1_UC1: AS percentage statistical analysis at 3°C compared with 25°C in P. ussuriensis; UL2_UC1: AS percentage statistical analysis at −3°C compared with 25°C in P. ussuriensis; UL2_UL1: AS percentage statistical analysis at −3°C compared with 3°C in P. ussuriensis.
Analysis of DEGs, Differential Alternative Splicing (DAS) Events, and Corresponding Cold-Responsive Genes by RNA-Seq
RNA-Seq helps understand the network via AS influence gene expression and transcriptome reprogramming. To improve the accuracy of AS analysis, we investigated the transcriptional changes and compared the two poplar species in response to cold stress by RNA-seq. Compared to P. trichocarpa, 6,060 (Supplementary Data S1), 5,208 (Supplementary Data S2), and 5,867 (Supplementary Data S3) DEGs (p < 0.05;|log2 (FC)|>1) were identified at 25, 3, and −3°C, respectively, in P. ussuriensis. We further analyzed the most significantly enriched GO terms for DEGs (Figures 5A–I) and DAS (Figures 6A–I) genes. The most significantly enriched GO terms for DEGs were “response to stimulus and stress” in biological process (BP, Figures 5A–C), an “intrinsic component of membrane” in a cellular component (CC, Figures 5D–F), and “catalytic activity” in molecular function (MF, Figures 5G–I). The most significantly enriched GO terms for DAS genes are shown in Figures 6A–I. As the temperature decreased, the “response to abiotic stimulus” item of BP increased significantly, which is consistent with the GO terms of DEGs.
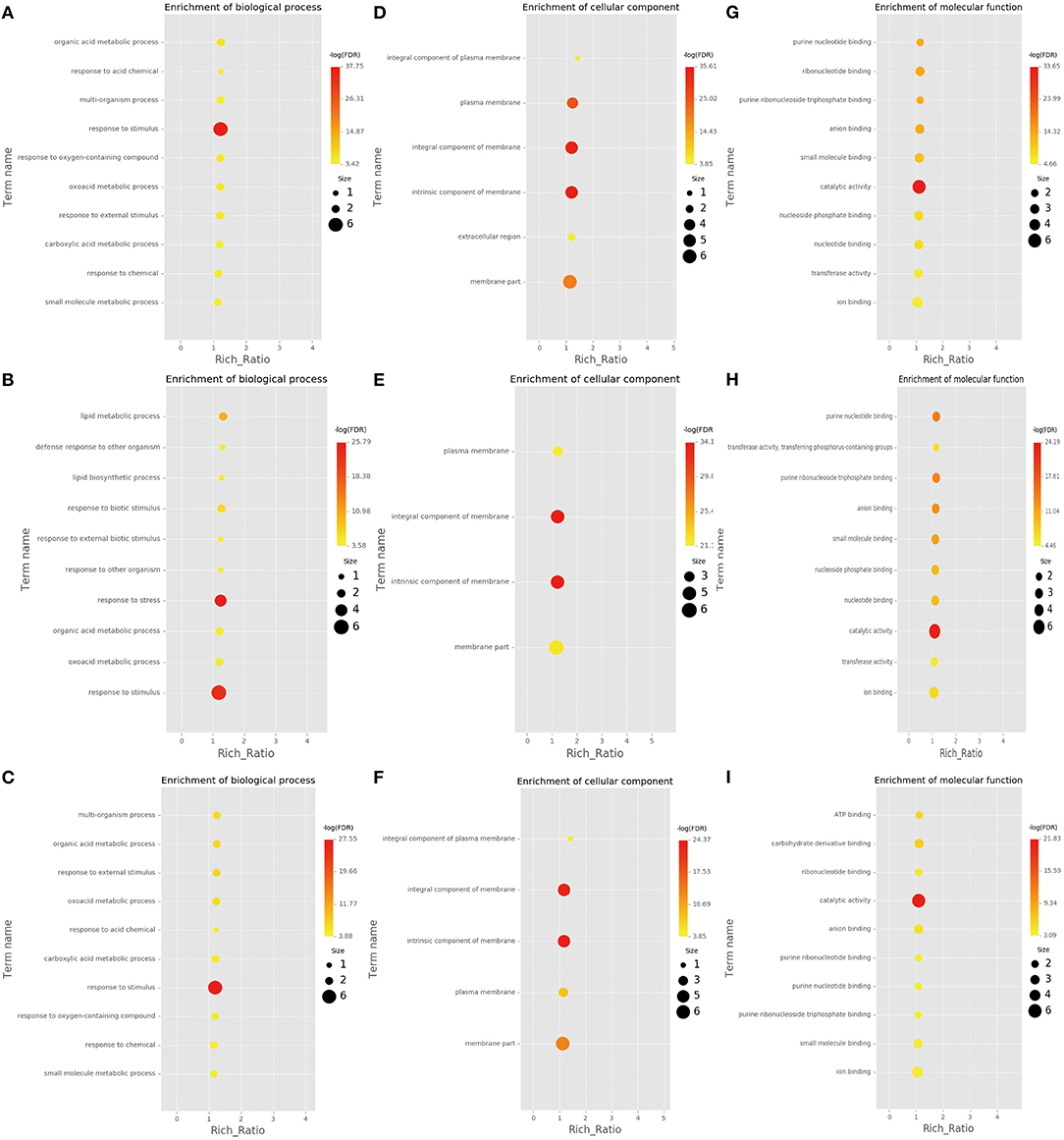
Figure 5. Most significantly enriched gene ontology (GO) terms for differentially expressed genes (DEGs) identified by RNA sequencing analysis of two poplar species under cold conditions. (A–C) GO terms for DEGs between P. ussuriensis and P. trichocarpa at 25°C. (D–F) GO terms for DEGs between P. ussuriensis and P. trichocarpa at 3°C. (G–I) GO terms for DEGs between P. ussuriensis and P. trichocarpa at −3°C.
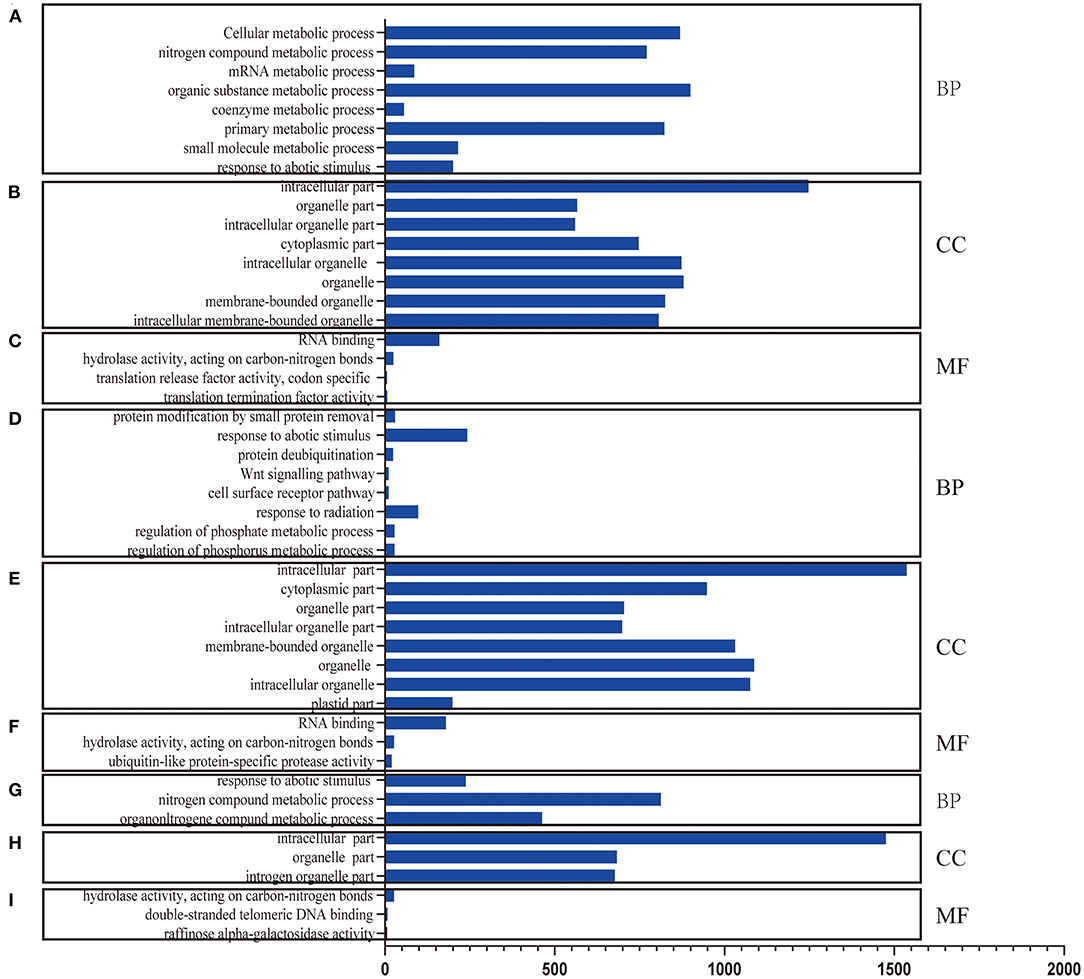
Figure 6. Differential alternative splicing (DAS) genes identified by RNA sequencing analysis of two poplar species under cold conditions. (A–C) GO terms for DAS genes between P. ussuriensis and P. trichocarpa at 25°C. (D–F) GO terms for DAS genes between P. ussuriensis and P. trichocarpa at 3°C. (G–I) GO terms for DAS genes between P. ussuriensis and P. trichocarpa at −3°C.
From the RNA-Seq data, U1–U9 samples were classified into six subclasses of genes that were upregulated or downregulated in P. ussuriensis compared with P. trichocarpa (P1–P9 samples) in response to cold stress with different patterns of expression (Supplementary Figure S1). For example, genes of subclass 3 in P. ussuriensis were upregulated compared with that in P. trichocarpa (Figure 7A). These upregulated genes included genes involved in calcium signaling, such as calmodulin (CAM), calcium-dependent protein kinase (CPK), and CBL-interacting protein kinase (CIPK) (Table 3), and many mitogen-activated protein kinase (MAPK) cascades (Supplementary Data S4). Additionally, a 9-cis-epoxycarotenoid dioxygenase (NCED) gene encoding the key enzyme of abscisic acid (ABA) biosynthesis was upregulated under cold stress (Table 3). Many peroxidase (POD) genes were also upregulated in P. ussuriensis (Table 3). We also used the RNA-Seq data to identify genes that were DAS between contrast poplar species. In total, 2,785 (IR, 50.4%; A5SS, 10.9%; A3SS, 18.2%; MXE, 1.7%; ES, 18.8%), 3,613 (IR, 37.9%; A5SS, 8.7%; A3SS, 15.%; MXE, 3.5%; ES, 34.8%), and 3,408 (IR, 43.1%; A5SS, 10.9%; A3SS, 16.5%; MXE, 4.%; ES, 26.3%) DAS genes were identified at 25, 3, and −3°C, respectively, in P. ussuriensis compared with that in P. trichocarpa by RNA-seq (Figure 7B). MATS identified the DAS events in the two species under different temperature conditions. A model pattern example from IGV view of Potri.001G016200 alternative splicing (ES) type is shown in Figure 7C. Venn analysis revealed a substantial overlap among the DEGs, DAS genes, and the cold-responsive DEGs identified in P. ussuriensis compared with P. trichocarpa (Figures 7D–F). About half of the DEGs were DAS genes (Figures 7D–F), of which 18.1, 28.2, and 35.1% DEGs at 25, 3, and −3°C, respectively, in response to cold stress were DAS genes (Figures 7D–F); these genes with significant expression (p < 0.05;|log2 (FC)|>1) are shown in Table 4 based on GO. We found that several DEGs related to cold stress, such as cold-regulated inner membrane protein 1 (CORIMP1, Potri.001G353500) at 25°C, low-temperature- and salt-responsive proteins (LTI6A, Potri.013G001600 and CORIMP1Potri.001G353500) at 3°C, and late elongated hypocotyl (LHY, Potri.014G106800), low-temperature-induced integral membrane protein (LTI6A, Potri.013G001600), and calcium-binding protein (CML, Potri.002G182500, Potri.014G108200) at −3°C in P. ussuriensis (Table 4) had more isoforms based on P. trichocarpa genome. Additionally, we investigated the dynamics of different AS types under cold stress; the result showed that the number of up and downregulated DEGs showing an upward trend as the temperature goes down in almost all AS types. In contrast, more genes were downregulated than upregulated in P. ussuriensis compared with P. trichocarpa (Figure 8). RT-PCR validation revealed more than one isoform for these genes (Figure 9). Meanwhile, the qRT-PCR result indicated that some genes that generated more transcripts were responsive to cold stress (Figure 10). We also randomly selected some DEGs using qRT-PCR to validate the reliability of the RNA-seq data. The result showed that the qRT-PCR was consistent with RNA-seq data (Supplementary Figure S2).
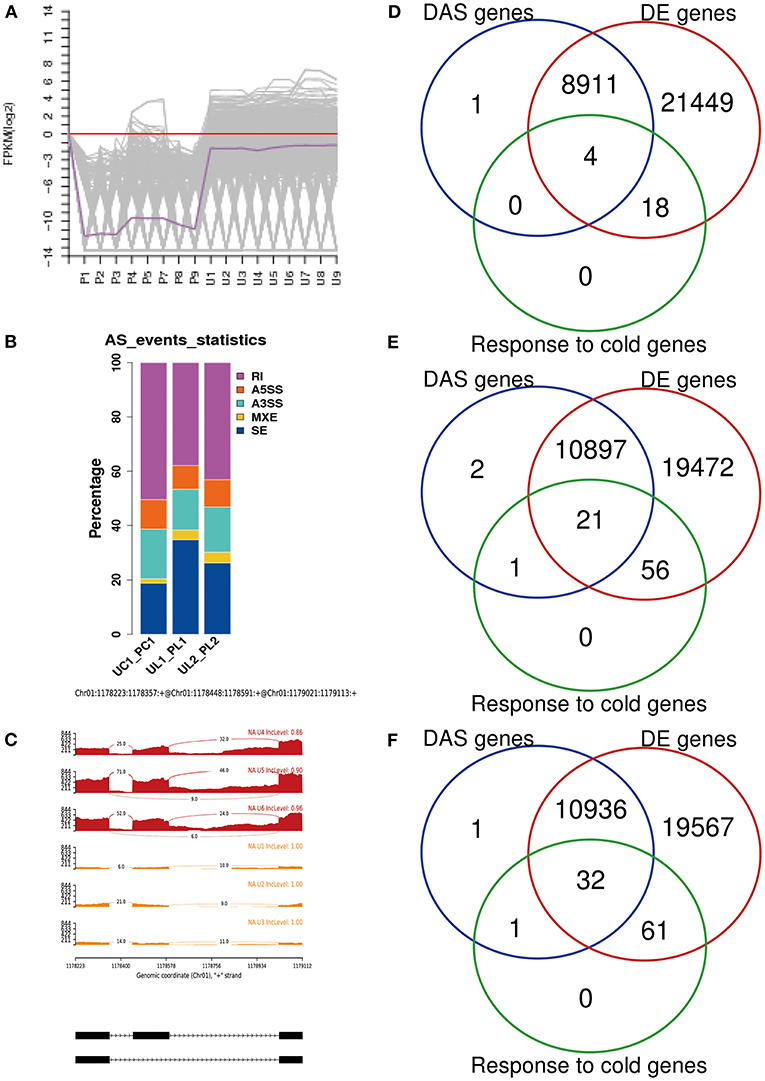
Figure 7. Changes in DEGs and DAS genes in response to cold stress. (A) One subclass of differentially expressed genes (DEGs) clustering in P. ussuriensis compared with P. trichocarpa in response to cold stress identified by RNA sequencing analysis. (B) AS events analysis between P. trichocarpa and P. ussuriensis UC1_PC1: AS percentage statistical analysis at 25°C in P. trichocarpa compared with P. ussuriensis; UL1_PL1: AS percentage statistical analysis at 3°C in P. trichocarpa compared with P. ussuriensis; UL2_PL2: AS percentage statistical analysis at −3°C in P. trichocarpa compared with P. ussuriensis. (C) A model pattern example from the IGV view of Potri.001G016200 alternative splicing (ES) type. (D) The Venn view of DAS genes, DE genes, and genes response to cold at 25°C. (E) The Venn view of DAS genes, DE genes, and genes response to cold at 3°C. (F) The Venn view of DAS genes, DE genes, and genes response to cold at −3°C.
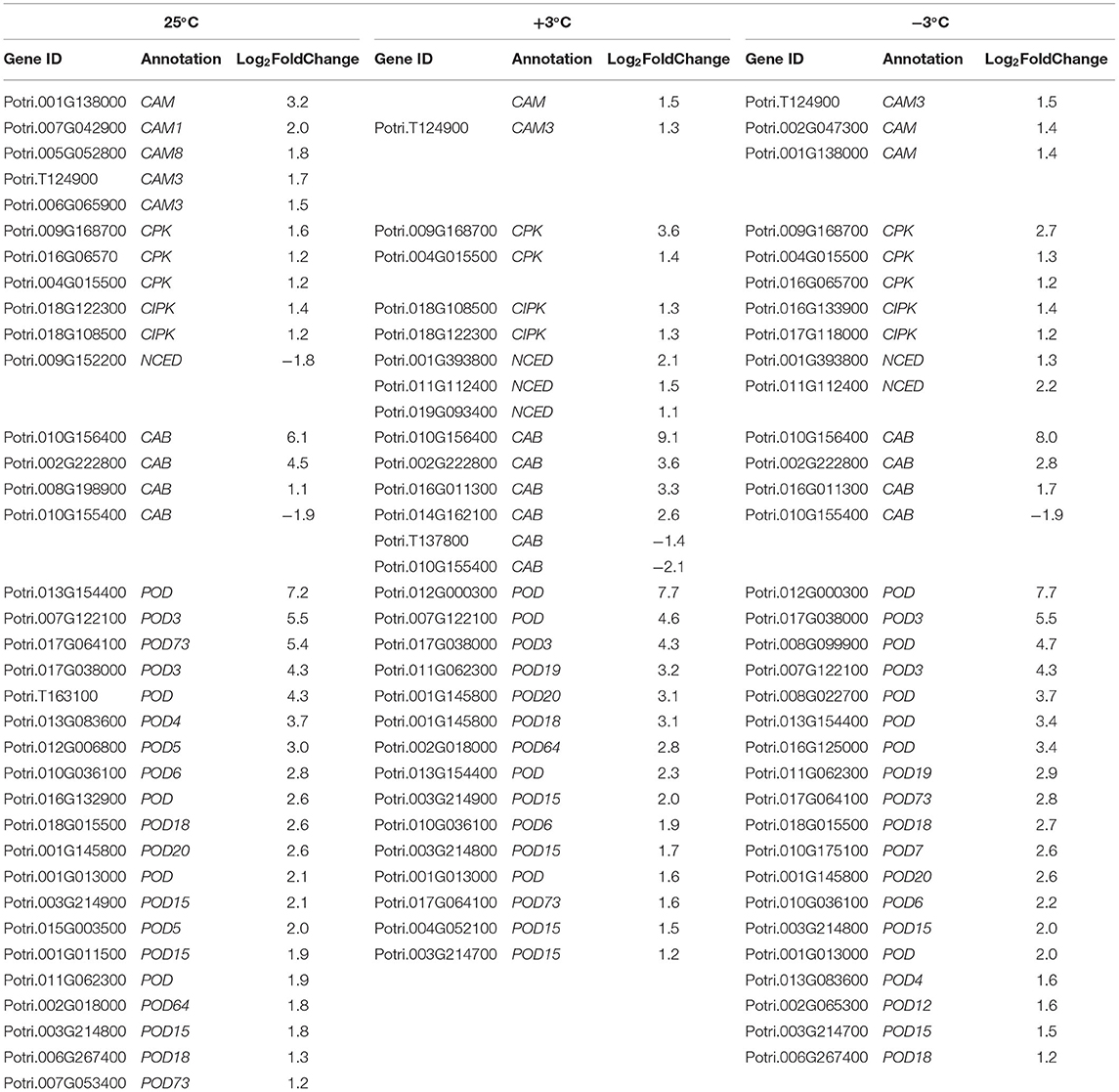
Table 3. Partial differentially expressed genes (DEGs) response to cold in P. ussuriensis compared with P. trichocarpa.
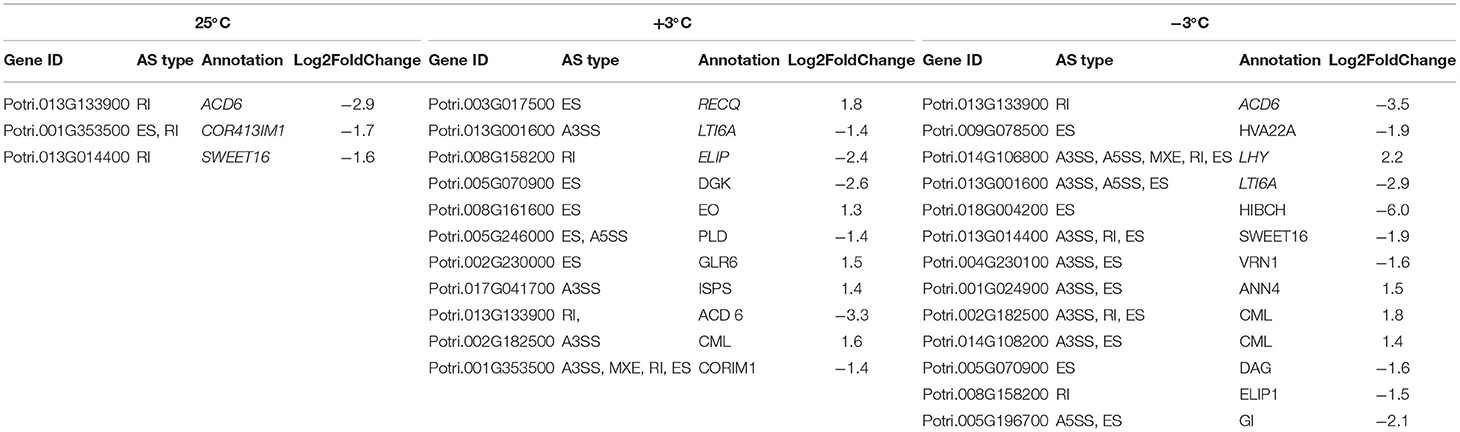
Table 4. Differential alternative splicing events in response to cold in P. ussuriensis compared with P. trichocarpa.
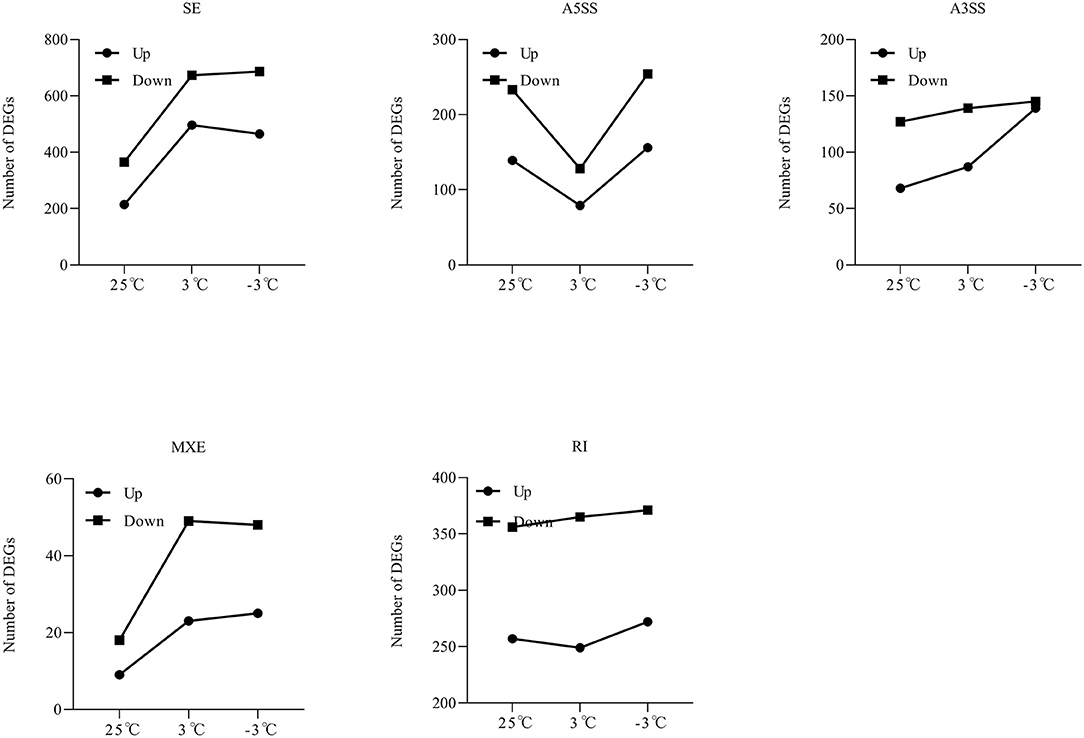
Figure 8. The dynamics of AS types under cold stress in P. ussuriensis compared with P. trichocarpa.
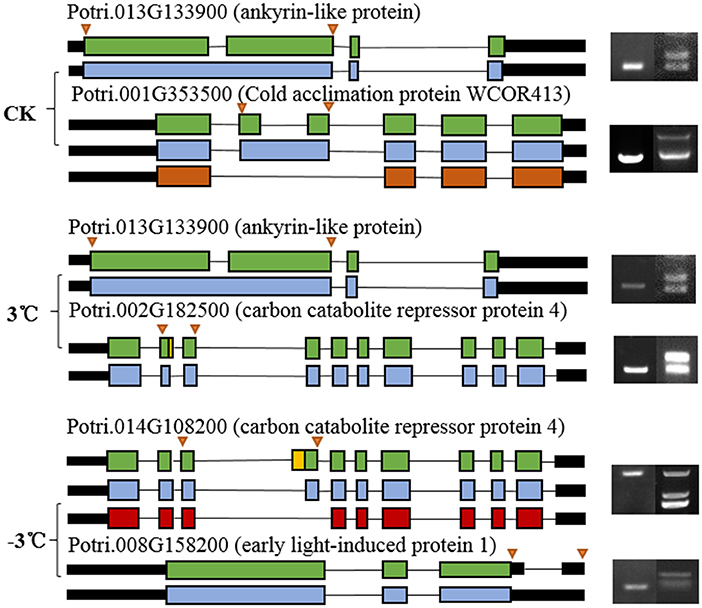
Figure 9. Characteristic of randomly selected AS events and validation of randomly selected alternative splicing events detected by RNA sequencing using reverse transcription-PCR (RT-PCR). The arrow on the left side indicates the position of PCR primers used for RT-PCR. The electropherogram on the right side shows the alternatively spliced product bands in P. trichocarpa and P. ussuriensis.
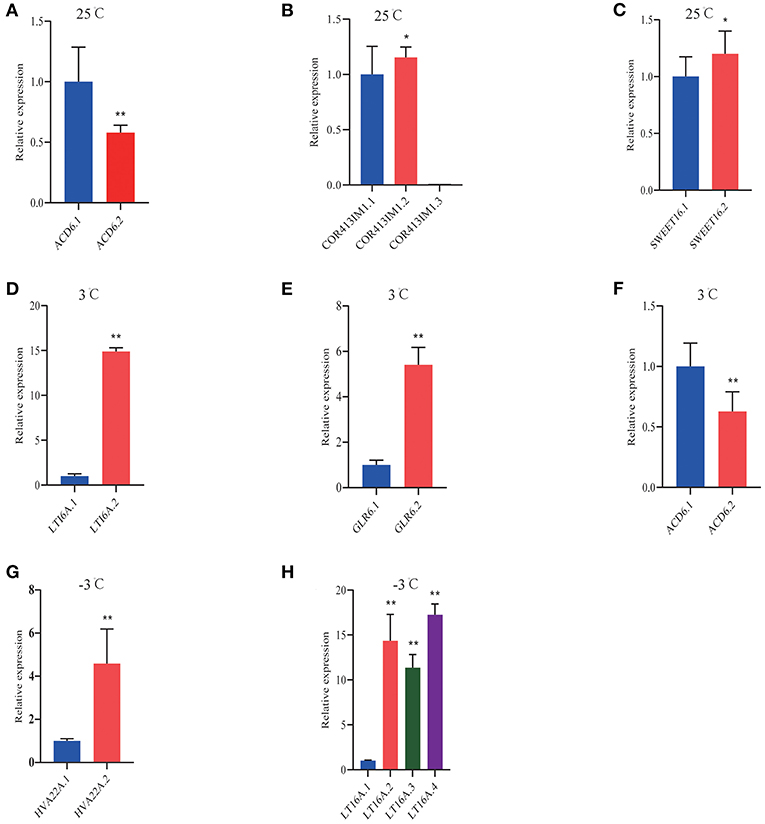
Figure 10. QRT-PCR analysis of DAS isoforms response to cold stress in P. ussuriensis compared with P. trichocarpa. (A–C) DAS genes at 25°C. (D–F) DAS genes at 3°C. (G,H) DAS genes at −3°C.
Cold-induced as in Splicing Factors (SFs) and Transcription Factors (TFs)
The splice site of pre-mRNA was recognized by SFs that recruit the spliceosome to remove the introns away (Chen and Moore, 2015). A number of SFs reported the functions in diverse environmental conditions (Laloum et al., 2018). We focused on the SFs that probably regulated the AS of downstream genes by different spliceosome complexes under a cold condition. The AS events occurred in multiple SFs, including small nuclear ribonucleo proteins (snRNPs), such as spliceosome-associated proteins (DEAH box helicase 1, YT521-B, LSM, AAR2, SART-1, and Prp series; Table 4, p < 0.05;|log2 (FC)|> 1). At 25°C, only two SFs were differentially expressed; five SFs, including the two SFs at 25°C, were differentially expressed at 3°C. Many SFs, including SFs of the two temperatures (25 and 3°C), were also responsive at −3°C (Table 4). Most importantly, all the SFs were upregulated. As shown in Figure 11, the induced SFs, such as CEF1, SR45a, Prp18, SLU7A, DEAH9, and SFRS1, have more AS isoforms in P. ussuriensis compared with P. trichocarpa. In short, more pre-mRNAS Fs responded in P. ussuriensis than that in P. trichocarpa as the temperature decreased.
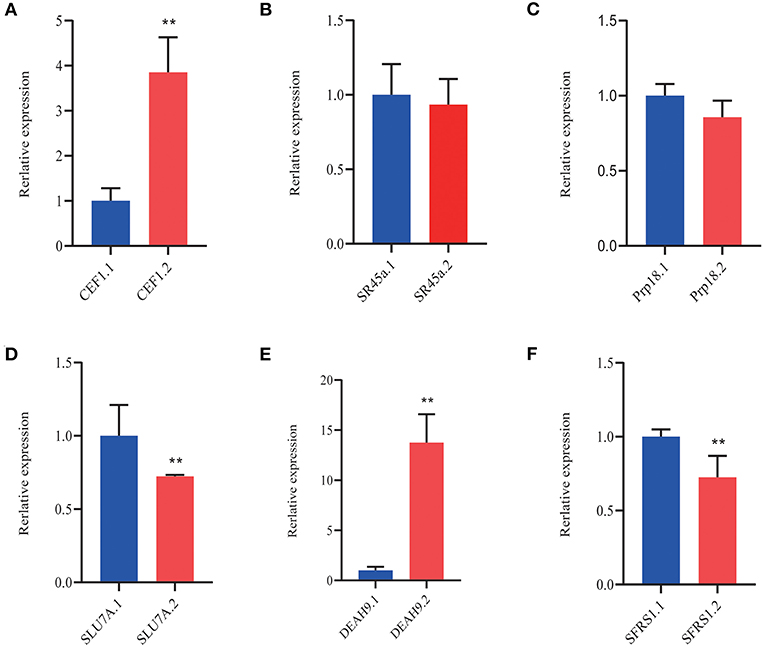
Figure 11. QRT-PCR analysis of DAS isoforms of splicing-associated factors in P. ussuriensis compared with P. trichocarpa. (A) CEF1. (B) SR45a. (C) Prp18. (D) SLU7A. (E) DEAH9. (F) SFRS1.
Analysis based on the reference genome revealed AS of numerous TFs. Table 3 shows significantly induced differentially expressed TFs in P. ussuriensis compared with P. trichocarpa. Under normal conditions, AS events of these TFs were different between the two poplar species and identified, such as WRKY, MYB, B3, and CAMTA. More and more AS events happened as the temperature lowered (Table 5). We selected some differentially expressed TFs that likely related to cold stress to conduct qRT-PCR analysis; the result showed that the isoforms of bHLH, HD-ZIP, CAMTA, WRKY, bZIP, MYB, NAC, and B3 TFs were differentially expressed in P. ussuriensis compared with P. trichocarpa (Figure 12).
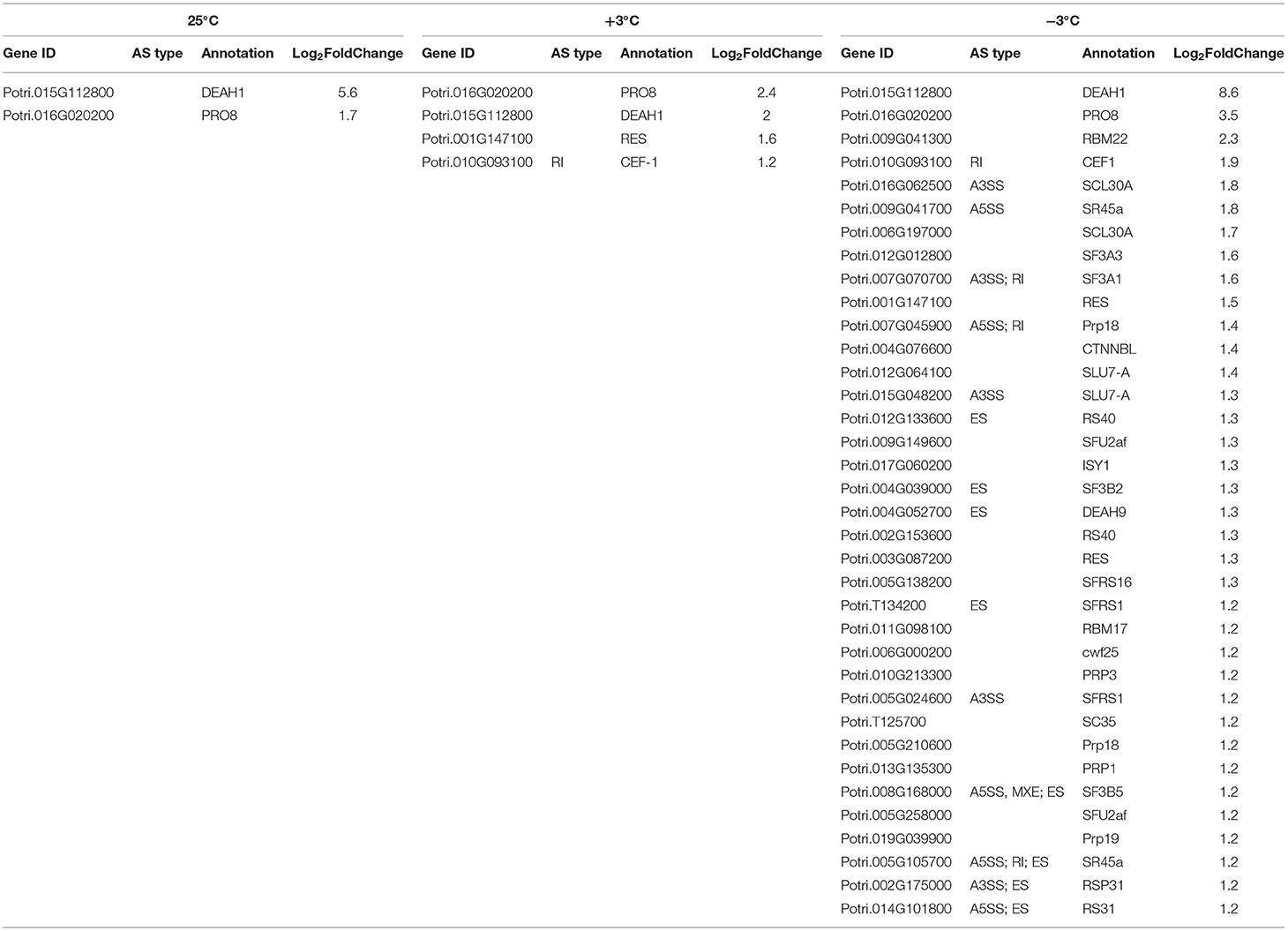
Table 5. Expression change of splicing-associated factors in P. ussuriensis compared with P. trichocarpa at different temperatures.
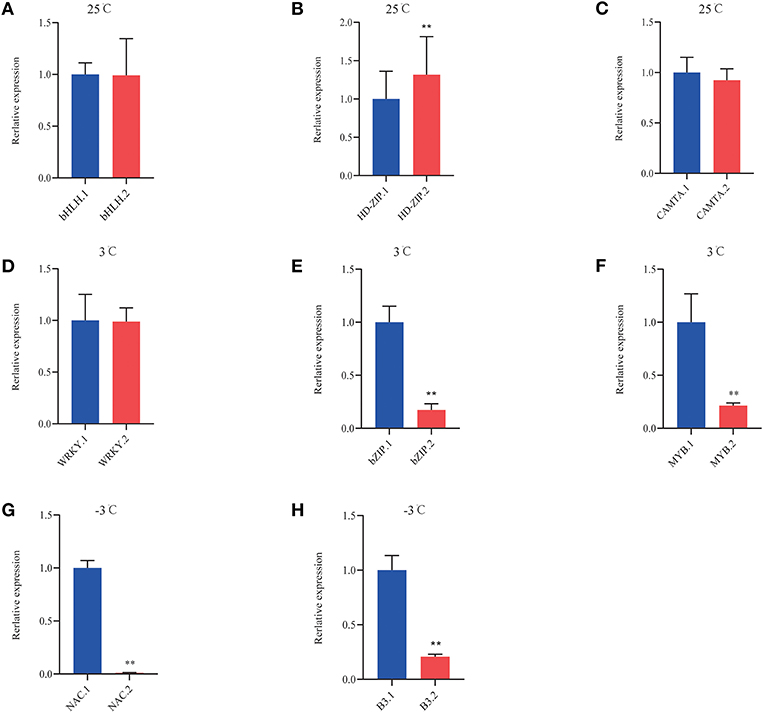
Figure 12. QRT-PCR analysis of DAS isoforms of TFs in P. ussuriensis compared with P. trichocarpa. (A–C) DAS genes of TFs at 25°C. (D–F) DAS genes of TFs at 3°C. (G,H) DAS genes of TFs at −3°C.
Discussion
Plants cope with adverse environmental conditions by reprogramming the gene expression and metabolism in a strict manner (Li et al., 2019). Although the expression of genes in some plant species under cold stress has been studied extensively, pre-mRNA splicing during the transcriptional changes in response to cold is still not clear in poplar species. In the current study, the result showed that the P. ussuriensis had more cold tolerance than P. trichocarpa, probably two poplar species have different abilities to deal with a cold condition. We used SMRT sequencing and RNA-Seq (Illumina) for the first time for a global survey of AS in two poplar species under cold stress. The PacBio Iso-Seq data revealed that 97.66 and 97.51% of the isoforms detected were mapped reads based on the reference genome of P. trichocarpa, and 1.8 and 1.02% were multi-mapped to the genome (Table 6; Figure 13).
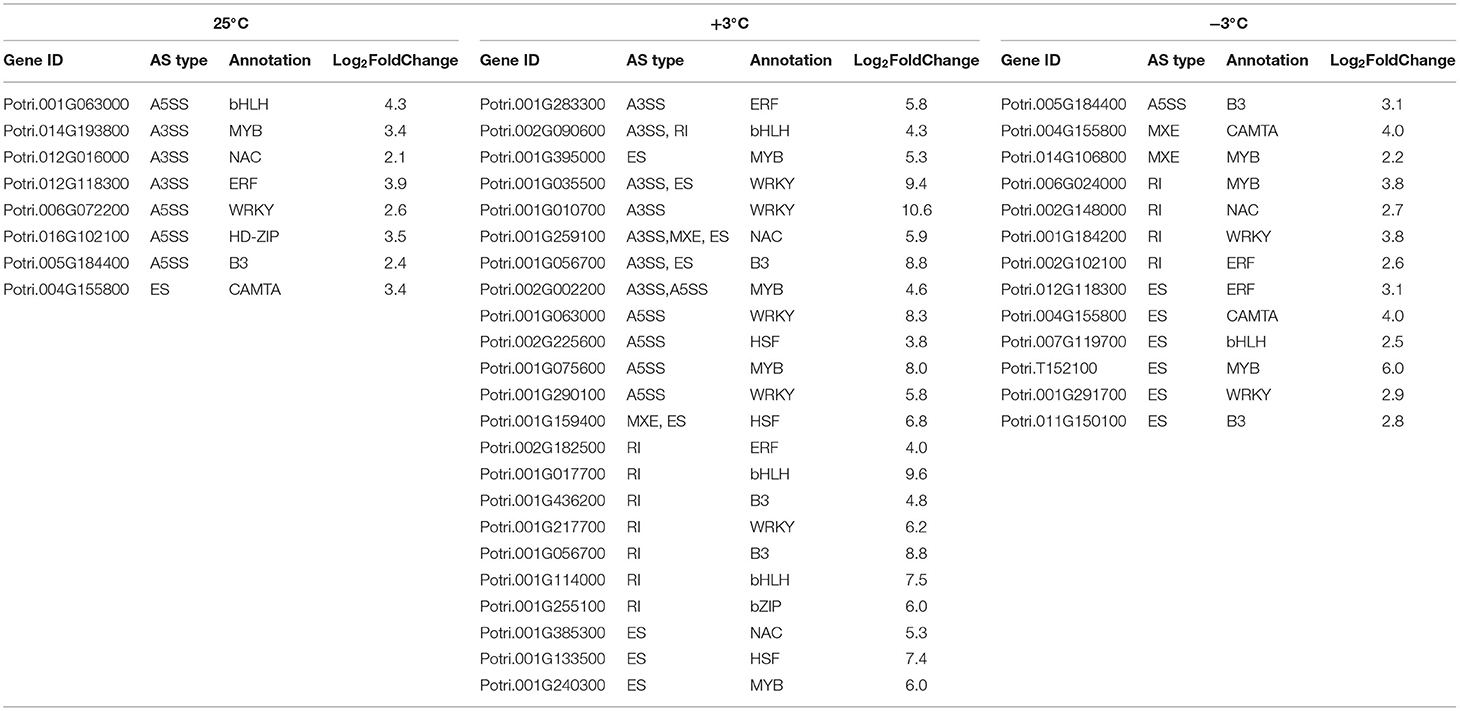
Table 6. Comparison of alternative splicing (AS) events in differentially expressed TFs of two poplar species by the PacBio sequencing data, respectively.
Plant cells sense cold stress through membrane rigidification (Chinnusamy et al., 2007). The rigidification subsequently activates mechanosensitive or ligand-activated calcium channels that lead to transient accumulation of Ca2+ in the cytosol. Studies have reported higher levels of calcium, which lead to signal amplification and act as the initial signals for cold stress response in plants (Vergnolle et al., 2005; Chinnusamy et al., 2007). Ca2+ and MAPK-signaling cascades play significant roles in the cold response pathway of Arabidopsis (Li et al., 2017; Zhao et al., 2017). In our study, P. ussuriensis exhibited more cold tolerance than P. trichocarpa. We found that calcium signaling-related genes, such as CAM, CPK, and CIPK, and many MAPK cascades were upregulated in P. ussuriensis at 25, 3, and −3°C compared with P. trichocarpa. Meanwhile, the Ca2+ content in P. ussuriensis increased more than that in P. trichocarpa under cold stress in a short time (3 h) at different temperatures (Figure 5A), which indicates that the cold response pathway is calcium dependent in poplar species. Subsequently, calcium signal amplification and a range of signals might be involved in cold stress signaling as reported (Chinnusamy et al., 2007).
First, the abovementioned cytoplasmic Ca2+ is sensed by calcium sensors, such as calcium-dependent protein kinases (CPKs), which interact with downstream-signaling components, including hormones, such as ABA and reactive oxygen species (ROS) (Lv et al., 2018). ROS also activate signal transduction pathways in response to biotic and abiotic stresses (Miller et al., 2010). As the temperature lowed, the H2O2 content was increased, and the increased level was greater in P. trichocarpa compared with that in P. ussuriensis. In our study, low temperature upregulated many POD genes in P. ussuriensis, which indicate the influence of ROS on cold stress regulation of gene expression. Besides, ROS can simulate Ca2+ accumulation that affects cold tolerance in plants (Chinnusamy et al., 2007). In response to cold stress, plants usually accumulate an increased amount of ABA. ABA accumulates in plants to regulate stress-responsive genes (Lv et al., 2018). ABA can also act as a secondary signal to change Ca2+ levels that finally influence cold signaling (Boudsocq and Sheen, 2013). In this study, the NCED gene encoding the key enzyme involved in ABA biosynthesis at low temperature was upregulated. Meanwhile, ABA-enhanced stress tolerance is associated with ROS (Lv et al., 2018), suggesting an intensive cross-talk among ABA, ROS, and Ca2+. Together, we conclude that Ca2+ signaling, ROS, and ABA might have led to cold tolerance in a short time. Secondly, researchers have identified the cold-responsive ICE-CBF pathway, which was critical to the regulation of cold-responsive transcriptome in plants. One such cold-responsive pathway involved the binding of ICE1 (Inducer of CBF expression 1) to the promoter of CBFs (C-repeat binding factors)-inducing expression; ICE1 enhances the expression of COR (cold-regulated) genes also during cold acclimation (Ritonga and Chen, 2020). However, the expression of these cold-responsive genes did not change in the two poplar species. Therefore, we speculated this pathway is not the main reason that P. ussuriensis had more cold tolerance than P. trichocarpa.
As reported, Ca2+-dependent kinases activate or repress TFs or SFs. These, in turn, regulate the transcription or AS of downstream genes, thereby driving cascades of cold-induced gene expression (Calixto et al., 2018). Differential expression of SFs is a significant factor that determines the changes in stress-induced AS (Punzo et al., 2020). In the present study, all the SFs identified via GO enrichment analysis were upregulated in P. ussuriensis compared with P. trichocarpa. Among these upregulated SFs, SC35, an arginine-serine-rich protein, is known to regulate plant development by modulating splicing; and transcription of a subset of genes was identified (Yan et al., 2017). Scarecrow-like (SCL) genes are involved in plant information transmission via signaling networks (Liu et al., 2017), and in the regulation of plant abiotic stresses, such as drought or salt stress (Golldack et al., 2013) were identified. The SF RS40, also an arginine-serine-rich protein, participates in miRNA biogenesis (Chen and Moore, 2015). Therefore, the role of SFs in cold-induced AS changes needs further investigation. Additionally, studies have reported numerous TFs that are subjected to AS and subsequently contribute to the regulation of gene expression. In this study, a lot of TFs were differentially expressed in P. ussuriensis compared with P. trichocarpa at different temperatures. Among these, a few TFs had isoforms in P. ussuriensis compared with P. trichocarpa even at room temperature. As the temperature decreased, the amount of AS in TFs increased in P. ussuriensis. It will be interesting to investigate the functions of the novel and already known cold-responsive TFs regulated by AS under cold stress.
Abiotic stress triggers AS events in plants (Pajoro et al., 2017; Laloum et al., 2018). These events regulate proteome diversity and gene expression to adopt an adverse environment (Thatcher et al., 2016). Therefore, the DAS events identified in our study under cold stress were not accidental. The extensive AS information identified here demonstrates the complexity of cold stress response. Studies based on the analysis of DEGs alone have significantly underestimated this regulatory mechanism. We also compared the AS events in two poplar species in response to cold stress. The comparison of the different AS types identified IR events in a large proportion, followed by A3SS in both types of poplars. Studies have reported IR as the most common splicing event in plant species, such as Arabidopsis (Marquez et al., 2012; Calixto et al., 2018) and cassava (Li et al., 2019). Calixto et al. (2018) reported differential splicing events very early under cold stress in Arabidopsis. The AS exhibited significant changes within only 40–60 min under cold stress (Calixto et al., 2018). To identify DAS, we investigated the transcriptional changes in two poplar species in response to cold stress after 3 h by MATS. We identified few DAS events in P. ussuriensis when compared with P. trichocarpa. Many DAS genes regulated by both transcription and AS were differentially expressed in P. ussuriensis compared with P. trichocarpa. Accelerated cell death 6 (ACD6, Potri.013G133900) was downregulated in P. ussuriensis and subjected to IR under different low-temperature conditions. ACD6 encodes a transmembrane protein with intracellular ankyrin repeats, which positively controls cell death and defense (Lu et al., 2003; Yao and Greenberg, 2006). Yao and Greenberg reported that ACD can prevent plant chlorophyll breakdown that induces programmed cell death (PCD) (Yao and Greenberg, 2006). In this study, ACD6 was downregulated. This might have happened because PCD was not activated in P. ussuriensis at any temperature conditions, and thus, P. ussuriensis exhibited higher cold tolerance than P. trichocarpa. LHY is a protein that functions in floral growth (Yon et al., 2016) and stress response (Adams et al., 2018). Studies have reported the influence of temperature changes on LHY transcript in Arabidopsis; moreover, AS in LHY was temperature-dependent (James et al., 2018a,b). In the present study, LHY (Potri.014G106800) was upregulated, and five types isoforms (A3SS, A5SS, MXE, RI, and ES) were identified at −3°C, which indicates the role of LHY in cold tolerance in poplar. Together, our results suggest that AS and DEGs play critical roles in the cold response. We have proposed a network (Figure 9) that reflects the rapid changes in DEGs and DAS genes during the cold response.
Conclusions
In summary, SMRT-Seq and Illumina RNA-Seq revealed that poplar trees rapidly responded at the pre-mRNA alternative splicing (AS) stage, and AS regulated the transcript abundance to adjust to cold stress. Isoform abundance and rapid response in P. ussuriensis, indicate that the changes in AS transcripts might be the most element that resistant to cold stress compared with P. trichocarpa species. Splicing factors and transcription factors are likely important for the regulation of DEGs and DAS. Our findings will support further research on the functions of AS and the coordination between transcriptional and AS responses to confer cold tolerance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JY and CL conceived and planned the experiments. JY, WL, and LS performed the experiments. HL, YF, and ZW collected the materials. CY and AC analyzed the data. JY wrote the manuscript with support from all the authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31971671), the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (2021A02), and the Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge Top Edit LLC for the linguistic editing and proofreading during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.737004/full#supplementary-material
Supplementary Data S1. Differentially expressed genes (DEGs) between Populus ussuriensis and Populus trichocarpa identified by RNA sequencing analysis.
Supplementary Data S2. DEGs between Populus ussuriensis and Populus trichocarpa at 3°C identified by RNA sequencing analysis.
Supplementary Data S3. DEGs between Populus ussuriensis and Populus trichocarpa at −3°C identified by RNA sequencing analysis.
Supplementary Data S4. DEGs of cluster 3 in Populus ussuriensis compared with Populus trichocarpa identified by RNA sequencing analysis.
Supplementary Figure S1. Clustering of differentially expressed genes (DEGs) in Populus ussuriensis compared with Populus trichocarpa in response to cold stress identified by RNA sequencing analysis.
Supplementary Figure S2. Validation of randomly selected DEGs using quantitative real-time PCR (qRT-PCR).
Supplementary Table S1. Primers used for reverse transcription-PCR (RT-PCR).
Supplementary Table S2. Primers used for quantitative real-time PCR (qRT-PCR).
Supplementary Table S3. Primers used for qRT-PCR analysis of DAS isoforms response to cold stress in P. ussuriensis compared to P. trichocarpa.
Supplementary Table S4. Primers used for qRT-PCR analysis of DAS isoforms of splicing-associated factors in P. ussuriensis compared to P. trichocarpa.
Supplementary Table S5. Primers used for qRT-PCR analysis of DAS isoforms of TFs in P. ussuriensis compared to P. trichocarpa.
References
Adams, S., Grundy, J., Veflingstad, S. R., Dyer, N. P., Hannah, M. A., Ott, S., et al. (2018). Circadian control of abscisic acid biosynthesis and signaling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 220, 893–907. doi: 10.1111/nph.15415
Bao, H., Li, E., Mansfield, S. D., Cronk, Q. C. B., Kassaby, Y. A., and Douglas, C. J. (2013). The developing xylem transcriptome and genome-wide analysis of alternative splicing in Populustrichocarpa (black cottonwood) populations. BMC Genomics 14:359. doi: 10.1186/1471-2164-14-359
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Calixto, C. P. G., Guo, W., James, A. B., Tzioutziou, N. A., Entizne, J. C., Panter, P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30, 1424–1444. doi: 10.1105/tpc.18.00177
Chen, W., and Moore, M. J. (2015). Spliceosomes. Curr. Biol. 25, 181–183. doi: 10.1016/j.cub.2014.11.059
Chinnusamy, V., Zhu, J., and Zhu, J. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Fu, X. D., and Ares, M. Jr. (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701. doi: 10.1038/nrg3778
Gan, X., Stegle, O., Behr, J., Steffen, J. G., Drewe, P., Hildebrand, K. L., et al. (2011). Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477, 419–423. doi: 10.1038/nature10414
Golldack, D., Li, C., Mohan, H., and Probst, N. (2013). Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions. Plant Cell Rep. 32, 1007–1016. doi: 10.1007/s00299-013-1409-2
Gordon, S. P., Tseng, E., Salamov, A., Zhang, J., Meng, X., Zhao, Z., et al. (2015). Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS ONE 10:e0132628. doi: 10.1371/journal.pone.0132628
Jaakola, L., Pirttila, A. M., Halonen, M., and Hohtola, A. (2001). Isolation of highquality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19, 201–203. doi: 10.1385/MB:19:2:201
James, A. B., Calixto, C. P. G., Tzioutziou, N. A., Guo, W., Zhang, R., Simpson, C. G., et al. (2018a). How does temperature affect splicing events? Isoform switching of splicing factors regulates splicing of LATE ELONGATED HYPOCOTYL (LHY). Plant Cell Environ. 41, 1539–1550. doi: 10.1111/pce.13193
James, A. B., Sullivan, S., and Nimmo, H. G. (2018b). Global spatial analysis of Arabidopsis natural variants implicates 5′UTR splicing of LATE ELONGATED HYPOCOTYL in responses to temperature. Plant Cell Environ. 41, 1524–1538. doi: 10.1111/pce.13188
Kalsotra, A., and Cooper, T. A. (2011). Functional consequences of developmentallyregulated alternative splicing. Nat. Rev. Genet. 12, 715–729. doi: 10.1038/nrg3052
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, 480–484. doi: 10.1093/nar/gkm882
Laloum, T., Martín, G., and Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci. 23, 140–150. doi: 10.1016/j.tplants.2017.09.019
Lee, Y., and Rio, D. C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323. doi: 10.1146/annurev-biochem-060614-034316
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Ding, Y., Shi, Y., Zhang, X., Zhang, S., Gong, Z., et al. (2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43, 630–642.e4. doi: 10.1016/j.devcel.2017.09.025
Li, P., Li, Y. J., Zhang, F. J., Zhang, G. Z., Jiang, X. Y., Yu, H. M., et al. (2017). The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103. doi: 10.1111/tpj.13324
Li, S., Yu, X., Cheng, Z., Zeng, C., Li, W., Zhang, L., et al. (2019). Large-scale analysis of the cassava transcriptome reveals the cold stress on alternative splicing. J. Exp. Bot. 71, 422–434. doi: 10.1093/jxb/erz444
Liu, H., Qin, J., Fan, H., Cheng, J., Li, L., and Liu, Z. (2017). Genome-wide identification, phylogeny and expression analyses of SCARECROW-LIKE(SCL) genes in millet (Setariaitalica). Physiol. Mol. Biol. Plant 23, 629–640. doi: 10.1007/s12298-017-0455-6
Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H., et al. (2018). Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 16, 714–726. doi: 10.1111/pbi.12822
Lu, H., Rate, D. N., Song, J. T., and Greenberg, J. T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420. doi: 10.1105/tpc.015412
Lv, X., Li, H., Chen, X., Xiang, X., Guo, Z., Yu, J., et al. (2018). The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 69, 4127–4139. doi: 10.1093/jxb/ery212
Marquez, Y., Brown, J. W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. doi: 10.1101/gr.134106.111
Metwally, A., Finkemeier, I., Georgi, M., and Dietz, K. J. (2003). Salicylic acid alleviates the cadmium toxicity in barley seedlings. Plant Physiol. 132, 272–281. doi: 10.1104/pp.102.018457
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Pajoro, A., Severing, E., Angenent, G. C., and Immink, R. G. H. (2017). Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 18:102. doi: 10.1186/s13059-017-1235-x
Palusa, S. G., Ali, G. S., and Reddy, A. S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49, 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Punzo, P., Ruggiero, A., Possenti, M., Perrella, G., Nurcato, R., Costa, A., et al. (2020). DRT111/SFPS splicing factor controls abscisic acid sensitivity during seed development and germination. Plant Physiol. 183, 793–807. doi: 10.1104/pp.20.00037
Reddy, A. S., Marquez, Y., Kalyna, M., and Barta, A. (2013). Complexity of the alternativesplicing landscape in plants. Plant Cell 25, 3657–3683. doi: 10.1105/tpc.113.117523
Ritonga, F. N., and Chen, S. (2020). Physiological and Molecular Mechanism Involved in Cold Stress Tolerance in Plants. Plants Basel. 9:560. doi: 10.3390/plants9050560
Schlaen, R. G., Mancini, E., Sanchez, S. E., Perez-Santángelo, S., Rugnone, M. L., Simpson, C. G., et al. (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. USA. 112, 9382–9387. doi: 10.1073/pnas.1504541112
Sharon, D., Tilgner, H., Grubert, F., and Snyder, M. (2013). A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014. doi: 10.1038/nbt.2705
Shen, S., Park, J. W., Lu, Z., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS:robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 111, 5593–5601. doi: 10.1073/pnas.1419161111
Staiger, D., and Brown, J. W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656. doi: 10.1105/tpc.113.113803
Syed, N. H., Kalyna, M., Marquez, Y., Barta, A., and Brown, J. W. (2012). Alternative splicing in plants: coming of age. Trends Plant Sci. 17, 616–623. doi: 10.1016/j.tplants.2012.06.001
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170, 586–599. doi: 10.1104/pp.15.01267
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 45, 122–129. doi: 10.1093/nar/gkx382
Vergnolle, C., Vaultier, M. N., Taconnat, L., Renou, J. P., Kader, J. C., Zachowski, A., et al. (2005). The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol. 139, 1217–1233. doi: 10.1104/pp.105.068171
Wang, M., Wang, P., Liang, F., Ye, Z., Li, J., Shen, C., et al. (2018). A global survey of alternative splicing in allpolyploid cotton: landscape, complexity and regulation. New Phytol. 217, 163–178. doi: 10.1111/nph.14762
Wang, T., Wang, H., Cai, D., Gao, Y., Zhang, H., Wang, Y., et al. (2017). Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J. 91, 684–699. doi: 10.1111/tpj.13597
Xing, Y., Jia, W. S., and Zhang, J. H. (2008). AtMKK1 mediates ABA-inducedCAT1 expression and H2O2 produced via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x
Yan, Q., Xia, X., Sun, Z., and Fang, Y. (2017). Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genetic 13:e1006663. doi: 10.1371/journal.pgen.1006663
Yao, N., and Greenberg, J. T. (2006). Arabidopsis accelerated cell death2 modulates programmed cell death. Plant Cell 18, 397–411. doi: 10.1105/tpc.105.036251
Yon, F., Joo, Y., Llorca, L. C., Rothe, E., Baldwin, I. T., and Kim, S. G. (2016). Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol. 209, 1058–1066. doi: 10.1111/nph.13681
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, G., Sun, M., Wang, J., Lei, M., Li, C., Zhao, D., et al. (2019). PacBio full-length cDNA sequencing integrated with RNA-seq reads drastically improves the discovery of splicing transcripts in rice. Plant J. 97, 296–305. doi: 10.1111/tpj.14120
Zhao, C., Wang, P., Si, T., Hsu, C. C., Wang, L., Zayed, O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 618–629. doi: 10.1016/j.devcel.2017.09.024
Zhao, Y., Sun, J., Xu, P., Zhang, R., and Li, L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulate cell wall thickening during fiber development in Populus Species. Plant Physiol. 164, 765–776. doi: 10.1104/pp.113.231134
Zhou, R., Moshgabadi, N., and Adams, K. L. (2011). Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc. Natl. Acad. Sci. USA. 108, 16122–16127. doi: 10.1073/pnas.1109551108
Keywords: Populus trichocarpa, Populus ussuriensis, alternative splicing, cold stress, PacBio, RNA-Seq
Citation: Yang J, Lv W, Shao L, Fu Y, Liu H, Yang C, Chen A, Xie X, Wang Z and Li C (2021) PacBio and Illumina RNA Sequencing Identify Alternative Splicing Events in Response to Cold Stress in Two Poplar Species. Front. Plant Sci. 12:737004. doi: 10.3389/fpls.2021.737004
Received: 06 July 2021; Accepted: 02 September 2021;
Published: 07 October 2021.
Edited by:
László Szabados, Hungarian Academy of Sciences (MTA), HungaryReviewed by:
Liuyin Ma, Fujian Agriculture and Forestry University, ChinaQingzhang Du, Beijing Forestry University, China
Copyright © 2021 Yang, Lv, Shao, Fu, Liu, Yang, Chen, Xie, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenghao Li, Y2hsaTBAMTYzLmNvbQ==
 Jingli Yang
Jingli Yang Wanqiu Lv
Wanqiu Lv Liying Shao
Liying Shao Yanrui Fu
Yanrui Fu Haimei Liu2
Haimei Liu2 Chengjun Yang
Chengjun Yang Aihua Chen
Aihua Chen Zhiwei Wang
Zhiwei Wang Chenghao Li
Chenghao Li