- 1Institution of Genomics and Bioinformatics, South China Agricultural University, Guangzhou, China
- 2Crop Biotech Institute, Graduate School of Biotechnology, Kyung Hee University, Yongin, South Korea
- 3Department of Plant Bioscience, Pusan National University, Miryang, South Korea
Photoperiod sensitivity is a dominant determinant for the phase transition in cereal crops. CCT (CONSTANS, CO-like, and TOC1) transcription factors (TFs) are involved in many physiological functions including the regulation of the photoperiodic flowering. However, the functional roles of CCT TFs have not been elucidated in the wild progenitors of crops. In this study, we identified 41 CCT TFs, including 19 CMF, 17 COL, and five PRR TFs in Oryza rufipogon, the presumed wild ancestor of Asian cultivated rice. There are thirty-eight orthologous CCT genes in Oryza sativa, of which ten pairs of duplicated CCT TFs are shared with O. rufipogon. We investigated daily expression patterns, showing that 36 OrCCT genes exhibited circadian rhythmic expression. A total of thirteen OrCCT genes were identified as putative flowering suppressors in O. rufipogon based on rhythmic and developmental expression patterns and transgenic phenotypes. We propose that OrCCT08, OrCCT24, and OrCCT26 are the strong functional alleles of rice DTH2, Ghd7, and OsPRR37, respectively. The SD treatment at 80 DAG stimulated flowering of the LD-grown O. rufipogon plants. Our results further showed that the nine OrCCT genes were significantly downregulated under the treatment. Our findings would provide valuable information for the construction of photoperiodic flowering regulatory network and functional characterization of the CCT TFs in both O. rufipogon and O. sativa.
Introduction
Oryza rufipogon Griff. is widely considered as the perennial progenitor of Asian cultivated rice (Oryza sativa L.) and serves as promising sources of elite alleles for rice improvement (Khush, 1997; Stein et al., 2018; Zhao et al., 2018). Modern rice varieties have expanded from their primitive domesticated regions to a wide range of latitudes from 53°N to 40°S as a result of the photoperiodic diversification during rice domestication (Koo et al., 2013). In cereal crops, photoperiod sensitivity, the dominant determinant for the phase transition from vegetative growth to reproductive growth, is regulated by the interaction between endogenous circadian clocks and exogenous day lengths which varies based on the difference in geographical latitudes (Koo et al., 2013). As a result of adaptation, flowering plants have a suitable flowering time to propagate offspring by sensing the seasonal cues. When the external solar rhythm agrees with the circadian rhythm, the time signal promotes the synthesis of CO/Hd1 that activates the expression of florigens which move from leaves to the shoot apical meristem (SAM) to trigger flowering (Song et al., 2015). In rice, the two flowering pathways, OsGI-Hd1-Hd3a/RFT1 under short-day (SD) and OsGI-(Hd1/Ghd7/DTH8)-Ehd1-H3da/RFT1 under long-day (LD), have been well elucidated (Hori et al., 2016; Zong et al., 2021). In addition, some flowering regulators are not involved in the two main flowering pathways, for example, DTH2 activates flowering by directly upregulating Hd3a and RFT1 (Wu et al., 2013).
CCT (CONSTANS, CO-like, and TOC1) transcription factors (TFs) that possess a conserved CCT domain are commonly present in flowering plants (Strayer et al., 2000). The CCT TFs can be divided into the three subfamilies depending on their domains (Li and Xu, 2017). The CCT motif (CMF) family proteins, like Ghd7, possess a CCT domain. The CONSTANS-like (COL) subfamily proteins, such as CO and Hd1, are characterized by one or two zinc finger B-box (BBOX) and a CCT domain. The members of pseudo-response regulator (PRR) subfamily encode a response-regulator (REC) domain at the N-terminus and the CCT domain at the C-terminus (Cockram et al., 2012). CCT genes regulate photoperiodic flowering, circadian rhythms, vernalization as well as defense against abiotic stresses (Zhang J. et al., 2015; Omolade et al., 2016; Li and Xu, 2017; Liu et al., 2020). It was reported that eighteen rice OsCCT genes are involved in flowering regulation (Zhang et al., 2020). Hd1, Ghd2, Ghd7, OsCCT1, OsCOL4, OsCOL10, and DTH7 inhibited the expression of Ehd1 under LD. Under SD, the expression of Ehd1 is suppressed by OsCO3, OsCOL4, and OsCOL10, while Hd1 and DTH2 induce Ehd1 (Li and Xu, 2017). In addition, OsCCT3, OsCCT22, OsCCT38, and OsCCT41 were found as flowering regulators (Zhang et al., 2020).
As the wild progenitor of Asian cultivated rice, O. rufipogon has attracted great attention to investigating population genetics, adaptation, speciation, and gene flow (Morishima et al., 1961; Gao and Hong, 2000; Gao et al., 2001; Gao, 2002, 2004; Zheng and Ge, 2010; Huang et al., 2012; Stein et al., 2018; Li et al., 2020a; Xie et al., 2020; Xu et al., 2020). Our previous investigation suggested that natural populations of O. rufipogon exhibited clinal variation in flowering time from north to south within its range in China (Gao et al., 2000). The CCT TFs have been identified and functionally elucidated in several crop species, such as rice (Zhang et al., 2017), maize (Huang et al., 2018), wheat (Yan et al., 2004), barley (Turner et al., 2005), and Medicago truncatula (Ma et al., 2020). However, functional roles of CCT TFs have not been elucidated in their wild progenitors, such as O. rufipogon in this study. It is widely recognized that O. rufipogon has very strong photoperiod sensitivity for flowering, which inhibits flowering under LD and induces flowering only under SD (Zong et al., 2021). But its response to photoperiod remains to be investigated in O. rufipogon.
In this study, we performed a genome-wide identification of the OrCCT TFs in O. rufipogon. Our results showed that, under LD, most OrCCT genes displayed rhythmic expression and regulated flowering time as suppressors. We also found that, compared with O. sativa, O. rufipogon plants took nearly double time for vegetative growth to reach the point when the plants can respond to the SD-induction to induce flowering. Our findings presented here would provide valuable information for the construction of photoperiod response, flowering regulatory network, and functional characterization of the CCT gene family in both O. rufipogon and O. sativa.
Materials and Methods
Materials and Growth Conditions
Oryza sativa ssp. japonica cv. Nipponbare and O. rufipogon (named CWR1) which were collected from Yuanjiang County, Yunnan Province, China, were studied in this study (Li et al., 2020a). They both display photoperiod sensitivity, in which flowering is delayed under LD conditions and induced under SD conditions. Seeds were germinated on ½ Murashige and Skoog medium for 10 days. Seedlings were transplanted to plastic pots and grown in the controlled growth room under either LD (14/10 light/dark cycle, 28/22°C) or SD (10/14 light/dark cycle, 28/22°C) conditions. Light intensity was approximately 1,000 μmol m–2 s–1 with humidity of approximately 50%.
Identification of CCT Transcription Factors
The two genome assemblies of O. rufipogon (Li et al., 2020b) and Nipponbare (Ouyang et al., 2007) were retrieved to identify CCT TFs. The Nipponbare reference genome (RGAP_7) was downloaded from RGAP database1. The longest isoforms were extracted using the Fast Get Representative program of TBtools2. Unless otherwise stated, the longest isoform was used throughout the study. HMMER 3.0 was employed to screen the protein sets with the Hidden Markov Model (HMM)3 file of CCT (PF06203), BBOX (PF00643), and REC (PF00072) as queries (cutoff = 0.01, other parameters of default). The putative CCT proteins in which the length of the aligned domain is smaller than 50% of what HMM file annotated were filtered out. The redundant sequences were discarded after BLASTP searches (E-value < 10––10). Proteins containing CCT domain and lacking BBOX and REC domains were classified as CMF genes. Proteins with CCT domain and additional BBOX or REC domain toward their amino-terminus were defined as COL or PRR genes, respectively. The deduced CCT TFs were further checked for the existence of the corresponding domain by using the Conserved Domain Database4. We named the CCT TFs with initials of genus and species and numerical symbols based on their chromosomal locations.
The molecular weight (D) and isoelectric point (Pi) of OrCCT TFs were calculated by ExPASy5. The web-server BUSCA was used to predict the subcellular localization of OrCCT proteins6. The information of position on chromosomes, exons, introns, and UTR regions of OrCCT genes was extracted from the gene finding format (GFF3) file. MEME software7 was used to identify the conserved motifs with the width of each motif = 10–100 amino acid residues, maximum number of motifs = 10, and other parameters of default values (Bailey et al., 2009). The visualization of gene structure and conserved domain (including classification) were conducted using the Gene Structure View tool of TBtools (Chen et al., 2020).
Identification of Orthologous CCT Genes Between O. rufipogon and O. sativa
Multiple Collinearity Scan toolkit (MCScanX) is often used to scan multiple genomes to detect putative homologous chromosomal regions using genes as anchors (Wang et al., 2012). To identify the putative orthologous CCT genes between O. rufipogon and O. sativa, the inter-species collinear relationship was identified using MCScanX with the parameters recommended by MCScanX’s manual (Wang et al., 2012). The collinear and syntenic gene pairs of CCT genes were extracted from the MCScanX output files. In this step, the data sets include both paralogs and orthologs. To remove the possible paralogs, the genes that showed the same order on chromosomes were selected as orthologous CCT genes between O. rufipogon and O. sativa.
Gene duplication events within CCT TFs were detected by MCScanX (Wang et al., 2012), and then visualized by Advanced Circos software (see text footnote 2). Non-synonymous (ka) and synonymous (ks) substitution of the paired CCT genes were calculated using KaKs_Calculator 2.0 (Wang et al., 2010). Gene duplication events were approximately dated according to the eq. T = Ks/2λ (λ = 6.5 × 10–9) (Yu et al., 2005). The comparative synteny relationships of CCT TFs between O. rufipogon and O. sativa were constructed by Multiple Synteny Plotter software (see text footnote 2).
Phylogenetic Analysis
All identified CCT TFs were divided into the three subfamilies according to their domains. The sequence of CCT TFs from Brachypodium distachyon and O. sativa ssp. indica was downloaded from the Phytozome database v138. The sequence of OnCCT TFs was downloaded from the Gramene database9. Multiple sequence alignment of CCT full proteins from the four species was performed by using MAFFT 7.243 with E-INS-i algorithm (Katoh and Standley, 2013). The Neighbor-Joining (NJ) phylogenetic tree was inferred by MEGA6 (Kumar et al., 1994) with bootstraps = 1,000.
RNA-Sequencing and Data Analyses
Total RNA was extracted from the leaves of 90-day-old plants using the QIAGEN plant RNA kit (Hilden, Germany). The concentration and quality of RNA were evaluated using NanoDrop 2000 UV-VIS spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). Paired-end reads were generated on a HiSeq 2000 platform following the manufacturer’s instructions (Illumina, United States). RNA-sequencing (RNA-seq) data were mapped on the reference genome with HISAT2 2.1.0 (Kim et al., 2019). FeatureCounts 1.6.2 was used to count the number of reads mapped on exons (Liao et al., 2014). Differentially expressed genes (DEGs) were evaluated by edgeR 3.32.0 (Robinson et al., 2010). Genes with p < 0.05 and log2 fold-changes >1 were considered as DEGs. Further screening among the initial DEGs was performed based on fragments per kilo-base per million fragments mapped (FPKM) values.
RNA Isolation and Quantitative Real-Time PCR
Total RNAs were extracted from the leaves using RNAiso Plus (TaKaRa, Shiga, Japan). The first cDNA strand was synthesized with 2 μg total RNA, using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI, United States) with 10 ng of the oligo(dT) 18 primer and 2.5 mM deoxyribonucleotide triphosphate. Synthesized cDNAs were used as templates for quantitative real-time PCR (qRT-PCR) with SYBR Premix Ex Taq II (TaKaRa) and the Rotor-Gene 6000 instrument system (Corbett Research, Sydney, NSW, Australia). The primers used for qRT-PCR were designed according to O. rufipogon reference sequences. The specificity of primers in both O. rufipogon and Nipponbare was checked by melting curve. The relative expression levels were calculated with rice Ubi1 as an internal control. Each dataset was collected from five independent biological repeats. The primers used are listed in Supplementary Table 1.
Vector Construction and Transformation
The 2,427-bp full-length genome DNA sequence of OrCCT24 was amplified from CWR1 using PCR with specific primers (CATAAGCTTTATCCGTTCATGTCGATGGGA and CC GGTACCCTATCTGAACCATTGTCCAAGC, where underlined sequences indicate HindIII and KpnI enzyme sites, respectively). The PCR fragments were cloned into the pGEM-T Easy vector for blue-white screening. After checking the insert by DNA sequencing, the cloned fragment from the positive clone was moved into the overexpression binary vector pGA3426 under the control of the maize ubiquitin 1 promoter (Kim et al., 2009). After checking its quality by DNA-sequencing, the recombinant vector was transformed into Nipponbare via Agrobacterium-mediated co-cultivation (An et al., 1989). Transgenic rice plants were generated through the stable transformation method as previously reported (An et al., 1989). The putative positive calli were transferred to shoot induction medium that contains 40 mg L–1 hygromycin.
Results
Identification, Classification, and Structure of CCT Transcription Factors
We identified 41 candidate OrCCT TFs in O. rufipogon (PRJCA002637)10. The proteins were named as OrCCT01 to OrCCT41 according to their chromosomal locations (Figure 1 and Supplementary Table 2). In addition, 41 OsCCT TFs were identified in the Nipponbare reference genome, as previously reported (Zhang et al., 2017). The molecular weight of OrCCT proteins ranged from 9,689.86 D (OrCCT40) to 171,328.16 D (OrCCT13). Their isoelectric points varied from 4.09 (OrCCT20) to 11.44 (OrCCT40) (Supplementary Table 2). Our results suggest that OrCCT proteins varied greatly among molecular features. Our prediction using BUSCA (Savojardo et al., 2018) suggested that 32 OrCCTs were located in the nucleus, while others were in chloroplast (5), extracellular space (3), and mitochondrion (1) (Supplementary Table 2).

Figure 1. Phylogenetic relationship, gene structure, and conserved motifs of OrCCT TFs in O. rufipogon. (A) Phylograms of OrCCT TFs were constructed based on the full-length protein sequences. Different subfamilies are highlighted with different colors. PRR in yellow, COL in green, and CMF in red. (B) Exon-intron structure and conserved domains of OrCCT TFs. (C) The motif patterns of OrCCT proteins. The sequence information for each motif is given in Supplementary Table 3.
The phylogram of CCT genes in O. rufipogon showed that OrCCT TFs were grouped into the three clusters based on their conserved domains (Figure 1A). The first cluster was the CMF subfamily with 19 members, the second was the COL subfamily with 17 members, and the third was the PRR subfamily with five members. The number of the possessed exons ranged from 1 (OrCCT36) to 33 (OrCCT28) (Figure 1B). The motif number of OrCCT genes alternated from 1 to 6. All CCT members possessed motif 1. The CMF members, OrCCT38 and OrCCT39, had the most motif, which possessed additional motif 5, 6, 7, 8, and 9. COL members had additional motif 2, while motif 10 was specifically presented in OrCCT08 and OrCCT21. PRR members possessed motif 1 and motif 3 (Figure 1C). The results suggest that the classification of OrCCT genes is coincident with their conserved motifs. The sequence information for each motif was present in Supplementary Table 3.
Chromosomal Distribution, Synteny, and Evolutionary Analysis of CCT Genes in O. rufipogon and O. sativa
Our results showed that the OrCCT genes were unevenly distributed on the 12 chromosomes of O. rufipogon. Chromosome 1 contained the largest number of OrCCT TFs (8), and chromosomes 1 and 4 had only one OrCCT TF (Figure 2A). The distribution of OsCCT genes on chromosomes is similar to that in O. rufipogon (Figure 2B). Our results showed that there were 11 duplicated OrCCT gene pairs in O. rufipogon (Figure 2A). OrCCT37, OrCCT38, OrCCT39, and OrCCT40 were present as tandem duplicated genes on Chromosome 12 (Figure 2A). Thirty-eight OrCCT genes had the orthologous genes in O. sativa (Figure 2 and Supplementary Table 4). However, the orthologs of OrCCT27, OrCCT36, and OrCCT40 were absent in O. sativa (Figure 2C). In addition, we failed to identify orthologs of OsCCT19, OsCCT25, and OsCCT37 in O. rufipogon, indicating that they are likely O. sativa-specific (Figure 2 and Supplementary Table 4).
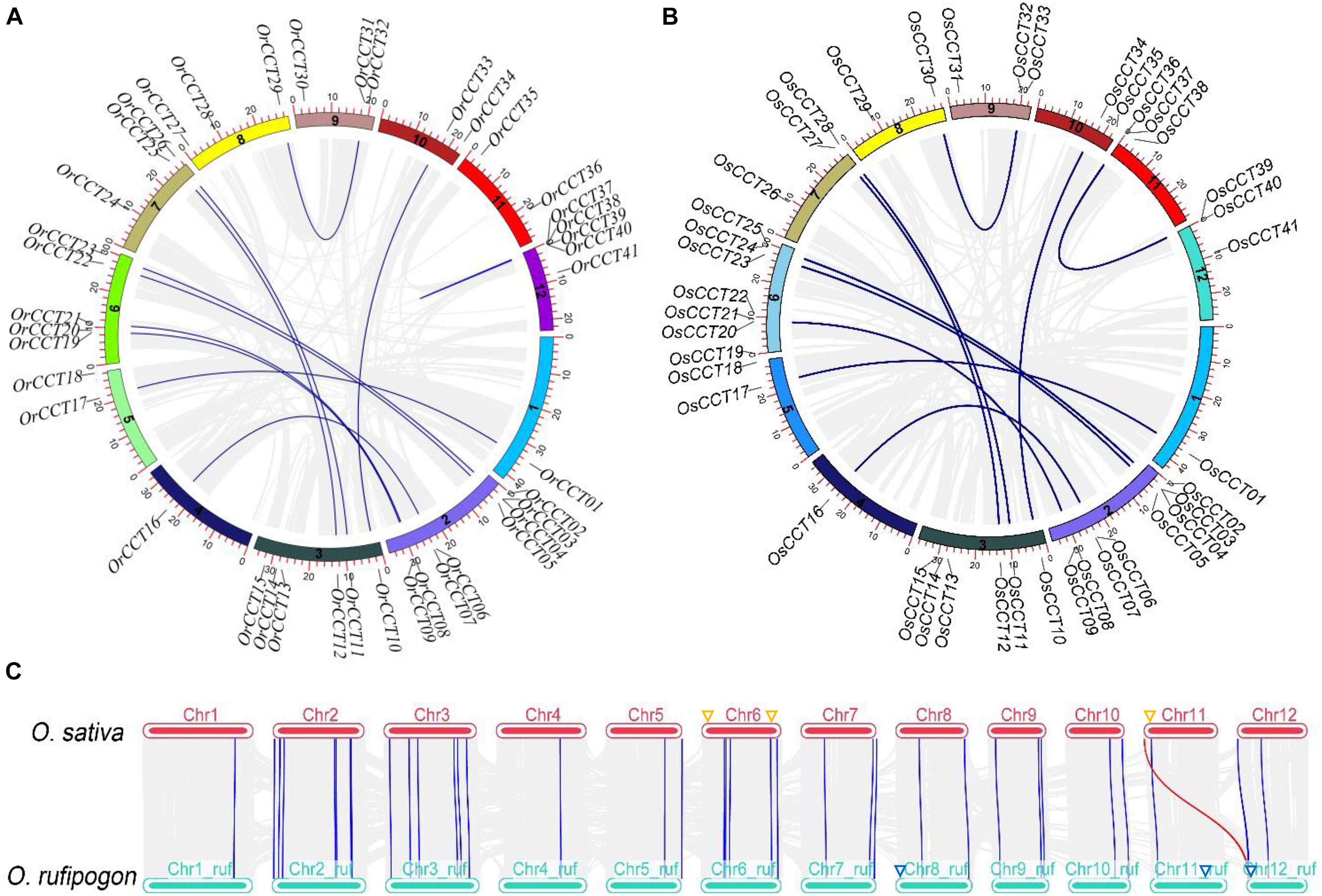
Figure 2. The inter-chromosomal relationship among CCT genes. (A,B) The chromosome distribution and gene duplication events in O. rufipogon (A) and O. sativa (B). The approximate location of each CCT gene is marked on corresponding chromosomes. The blue lines indicate the duplicated OrCCT genes, and gray lines in the background represent all duplication blocks within genomes. (C) The collinear relationship of CCT genes between O. rufipogon and O. sativa. The blue lines indicate orthologous gene pairs, while the red line shows that the CCT gene was likely generated after the domestication of O. sativa. The specific CCT genes of O. sativa and O. rufipogon are marked with orange and blue triangles on the corresponding positions of chromosomes, respectively.
Ten duplicated gene pairs were present in both O. rufipogon and O. sativa (Figure 2 and Supplementary Table 5). We used the formula T = Ks/2λ to evaluate approximate dates of duplicated genes (DEs). The dates of shared DEs of CCT genes varied from 23.05 to 89.31 million years ago (Mya) in O. rufipogon and O. sativa (Supplementary Table 5). The DE OsCCT37-OsCCT40, which was estimated to generate about 0.7 Mya, was absent in O. rufipogon, indicating that it probably occurred after the domestication of O. sativa. ω (dN/dS) is a good indicator of selective pressure at both nucleotide and protein levels. It is often expected that ω > 1, ω = 1, and ω < 1 imply positive selection, neutral selection, and purifying selection, respectively (Zhang et al., 2014). Our results suggest that nearly all duplicated CCT gene pairs underwent negative selection in both O. rufipogon and O. sativa (Supplementary Table 5).
We further investigated the diversification of CCT TFs in Brachypodium distachyon, Oryza nivara, and O. sativa ssp. indica (Supplementary Table 6). All investigated species possessed five PRR genes. Our results showed that O. rufipogon, O. nivara, and O. sativa ssp. japonica possessed the same composition of CCT subfamilies (19 CMF, 17 COL, and five PRR) (Figure 3A and Supplementary Table 6). A phylogenetic tree of CCT TFs was constructed using the complete protein sequences from the four species, including B. distachyon, O. rufipogon, O. nivara, O. sativa ssp. japonica, and O. sativa ssp. indica. As shown in Figure 3B, the CCT proteins could be divided into three clusters with nine clades (A to I). Clade A contained all PRR proteins. Clade B, C, F, and I consisted of CMF sub-family proteins. Clade D possessed COL proteins. The clade E, G, and H were composed of COL and few CMF proteins. Interestingly, the COL proteins were closely related to CMF proteins, suggesting that the COL proteins might originate from CMF proteins by gaining the BBOX domain. Alternatively, the CMF proteins were derived from COL proteins due to the loss of the BBOX domain.
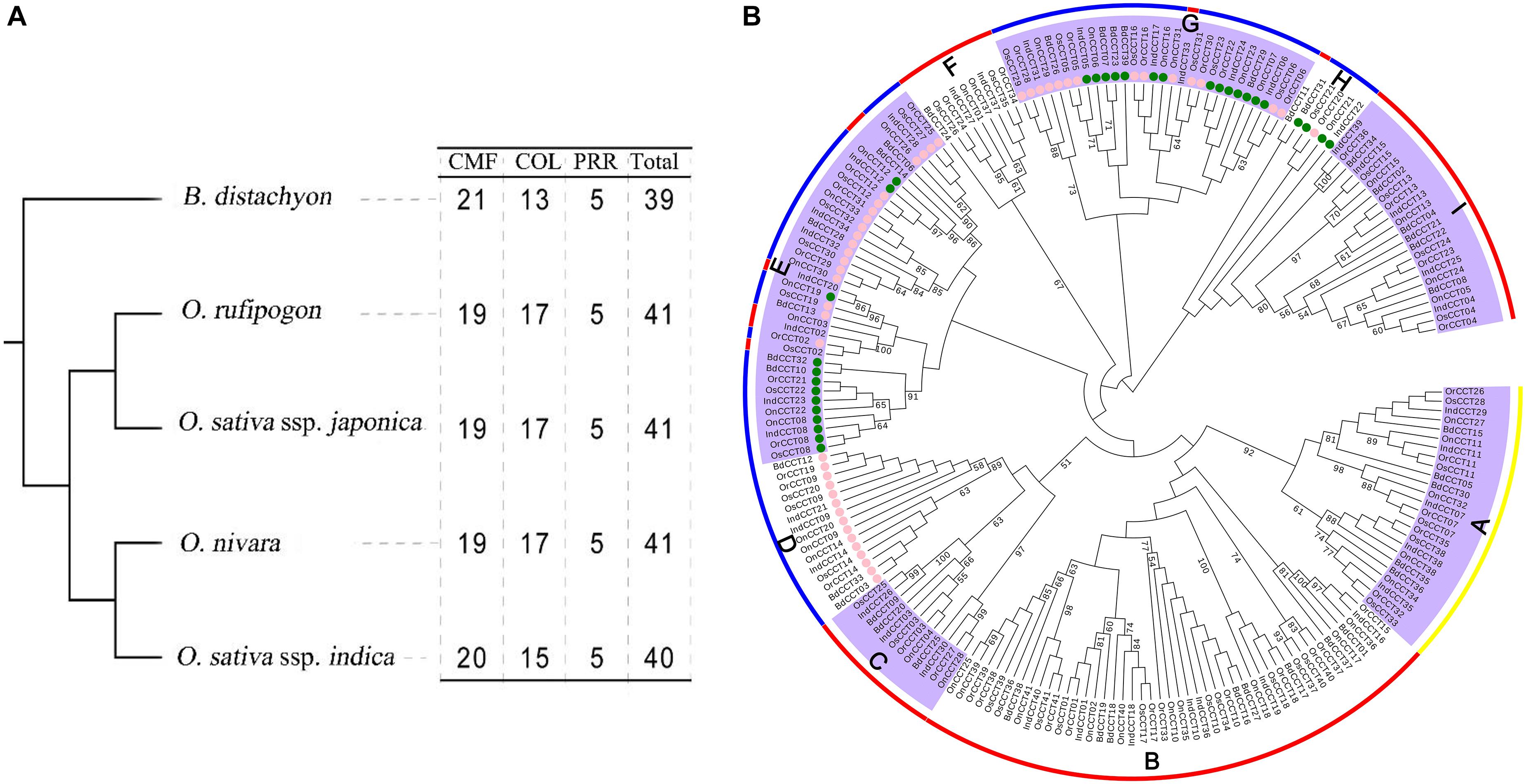
Figure 3. Phylogenetic relationships of CCT TFs among B. distachyon, O. sativa ssp. japonica, O. sativa ssp. indica, O. nivara, and O. rufipogon. (A) The species tree following the number of CCT TFs among the investigated species; (B) Phylogenetic tree representing relationships among CCT TFs from the four plant species. The pink and green circles of the terminal node indicate COL gene with 1 and 2 BBOX, respectively. The prefixes of tree labels are Bd, B. distachyon; Os, O. sativa ssp. japonica; Ind, O. sativa ssp. indica; On, O. nivara, and Or, O. rufipogon. The subfamilies are marked with red line: CMF; blue: COL; yellow: PRR. The locus of CCT TFs presenting here is listed in Supplementary Table 6.
Daily Expression Profiling of OrCCTs in Long-Day and Short-Day Conditions
Rice senses the day length by endogenous genetic factors to onset reproductive growth (Cho et al., 2017). Previous results suggested that 18 OsCCT genes are flowering regulators in O. sativa (Zhang et al., 2020). To investigate whether OrCCT can respond to photoperiod, the O. rufipogon plants were grown under LD and SD, and the second leaves from the top of main stems were collected at Zeitgeber time (ZT)-2 h, 8 h, and 15 h at 90 days after germination (DAG), respectively. RNA-seq experiments generated temporal expression profiles of all 41 OrCCT genes with three independent replicates.
Our results showed that thirty OrCCT genes showed significantly different expression levels among ZT-2 h, ZT-8 h, and ZT-15 h under LD (Figure 4A). Seven genes (OrCCT06, OrCCT14, OrCCT16, OrCCT22, OrCCT24, OrCCT30, and OrCCT34) were highly expressed at ZT-2 h and weakly expressed at ZT-15 h, suggesting that they are morning-peak genes. Ghd7, the rice ortholog of OrCCT24, was highly expressed in the morning (Xue et al., 2008). Twelve genes (OrCCT04, OrCCT08, OrCCT09, OrCCT12, OrCCT13, OrCCT19, OrCCT20, OrCCT21, OrCCT025, OrCCT027, OrCCT33, and OrCCT36) were highly expressed at ZT-15 h and weakly at ZT-2 h and ZT-8 h, suggesting that they are evening peak genes (Figure 4A). OrCCT20 is the ortholog of rice Hd1 that is highly expressed in the evening (Cho et al., 2018). Eleven genes (OrCCT01, OrCCT03, OrCCT05, OrCCT07, OrCCT11, OrCCT15, OrCCT17, OrCCT26, OrCCT31, OrCCT32, and OrCCT35) exhibited a high expression level at ZT-8 h compared to ZT-2 h and ZT-15 h (Figure 4A).
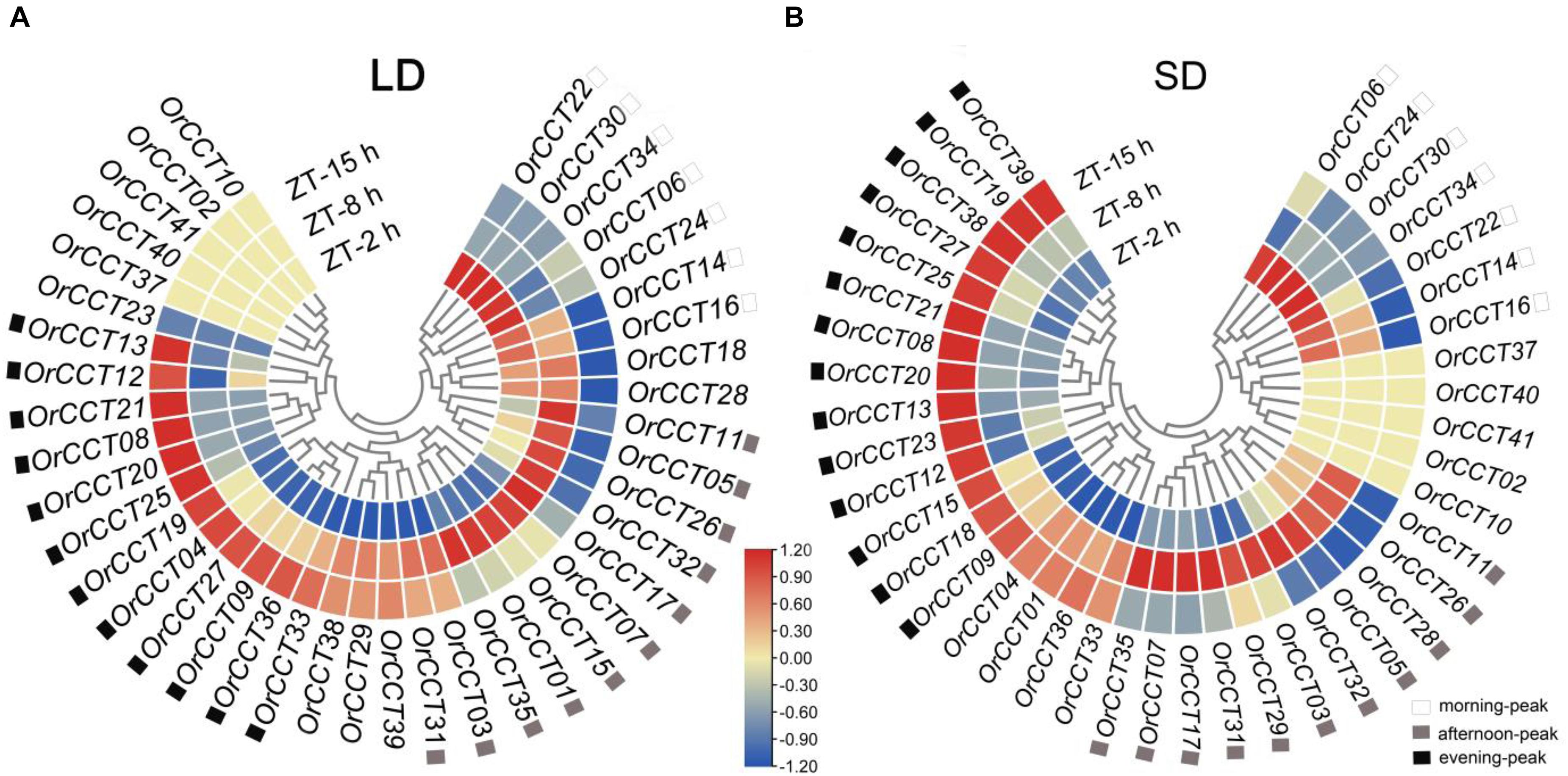
Figure 4. The daily expression profiles of OrCCT genes under LD (A) and SD conditions (B) at 90 DAG in O. rufipogon (CWR1). The heatmaps were drawn by FPKM values with row scale normalization (n = 3). The prefix ZT-2 h, ZT-8 h, and ZT-15 h indicate 2 h, 8 h, 15 h ZT, respectively.
Under SD, daily expression patterns of OrCCT genes were similar to those observed from LD (Figure 4B). All seven genes that were expressed most highly at ZT-2 h also showed a similar morning-peak expression under SD. Among 12 evening-peak genes, nine exhibited similar daily expression patterns between LD and SD. However, three genes (OrCCT04, OrCCT33, and OrCCT36) were similarly expressed at ZT-8 h and ZT-15 h under SD conditions. Instead, five genes (OrCCT15, OrCCT18, OrCCT23, OrCCT38, and OrCCT39) that failed to show evening-peak under LD displayed a high expression at ZT-15 h under SD. It is well known that ZT-15 h is at the beginning of the dark period under LD whereas the time is at near midnight under SD. Thus, the difference in some CCT genes might be due to the day-length.
To validate the veracity of our RNA-seq results, we tested twenty OrCCT TFs that showed rhythmic expression by using qRT-PCR experiments. The relative expression patterns of the selected genes were almost consistent with those of RNA-seq analysis (Supplementary Figure 1).
Developmental Expression Profiling of OrCCT Genes Under Long-Day Condition
The expression patterns of 16 OrCCT and 14 OsCCT genes were measured by qRT-PCR at different developmental stages under LD. Four flowering regulators, Ehd1, Hd3a, RFT1, and OsGI, were included to monitor the developmental stages of plants. The penultimate leaves of the main stems were sampled from O. rufipogon (CWR1) and Nipponbare plants at ZT-2 h, ZT-8 h, and ZT-15 h at 4 days intervals. The time points for qRT-PCR corresponded to the expression peak as shown in Figure 4. In Nipponbare, the transcript level of Ehd1 rapidly started to increase at 46 DAG, peaking at 75 DAG (Figure 5A). Hd3a and RFT1 also exhibited similar expression patterns with Ehd1 in Nipponbare plants (Figures 5B,C). However, all three genes did not express at a detectable level during the experimental period in CWR1 (Figures 5A–C). OsGI kept a high expression level until 54 DAG and then rapidly declined in Nipponbare, while it remained at a high level in CWR1 (Figure 5D). The phenotypic observation showed that Nipponbare flowered at 86–90 DAG, while CWR1 showed a non-flowering phenotype when grown for >213 DAG. Our results indicate that Nipponbare can complete the floral transition with the promotion of Ehd1, Hd3a, and RFT1 under LD. However, O. rufipogon remained at the vegetative growth phase during the investigated period.

Figure 5. The transcript levels of Ehd1 (A), Hd3a (B), RFT1 (C), OsGI (D), and 16 CCT genes (E–T) in leaf blades of Nipponbare and O. rufipogon (CWR1) at different developmental stages. Leaf blade samples were isolated at ZT-2 h, ZT-8 h, and ZT-15 h at 4 days intervals starting from 38 DAG. Transcript levels are relative to OsUbi1. Error bars indicate standard deviation for five biological replicates. ∗, ∗∗ significant differences by Student’s t-test at P ≤ 0.05 and P ≤ 0.01, respectively.
The transcript level of Hd1 that is a major photoperiod-sensitive floral regulator stayed at a relatively constant level in Nipponbare (Figure 5L). A similar expression pattern was observed for OrCCT20 that is an ortholog of Hd1 in O. rufipogon. The expression level of Ghd7 decreased to a low level at 61 DAG after floral transition in Nipponbare, but the transcript amount of OrCCT24 remained at a much high level and did not decline during the experimental period in CWR1 (Figure 5N). Similarly, the transcript levels of Ghd2 and OsCCT22 decreased after floral transition in Nipponbare, while their orthologs in O. rufipogon, OrCCT09 and OrCCT21, respectively, remained at relatively high levels during the investigated stages (Figures 5H,M). The content of OsPRR59 was high before the floral transition and the level declined after floral transition in Nipponbare, but its ortholog, OrCCT35, was lowly expressed at all stages in CWR1 (Figure 5R).
Three CCT genes interestingly exhibited opposite expression patterns between CWR1 and Nipponbare plants. The transcript levels of OsCCT01, OsCCT04, and NRR decreased as the Nipponbare plants grew up, whereas gene expression levels of their orthologs, OrCCT01, OrCCT04, and OrCCT18, increased during the experimental period in CWR1 (Figures 5E,F,K). The expression level of OsPRR37 was relatively low and slightly increased after 75 DAG in Nipponbare, but the level of its ortholog OrCCT26 increased rapidly after 61 DAG in O. rufipogon (Figure 5O), suggesting that OrCCT26 is a strongly functional allele of OsPRR37. Transcript level of DTH2 that is a rice flowering activator gradually increased after floral transition in Nipponbare, while OrCCT08 remained at a relatively high level during the experiment in CWR1 (Figure 5G).
Several genes showed similar expression patterns between Nipponbare and O. rufipogon. The transcript levels of both OsCO3 and OrCCT30 were high at 42 DAG and declined to low levels at 68 DAG in Nipponbare and O. rufipogon (Figure 5Q). Expression levels of OsPRR73 (OrCCT11) and OsCOL15 (OrCCT29) did not vary significantly during the experimental period in both Nipponbare and CWR1 (Figures 5I,P). The developmental expression pattern of OsCOL10, a floral repressor downstream of Ghd7 (Tan et al., 2016), was similar to its ortholog OrCCT14 (Figure 5J), indicating that they may function similarly.
The transcript levels of O. rufipogon-specific CCT genes, OrCCT36 and OrCCT40, were at a relatively low level and did not change significantly during the experimental period, indicating that they may not involve in controlling flowering (Figures 5S,T). Sequence similarity and developmental expression patterns suggest that OrCCT08, OrCCT24, and OrCCT26 are the functional alleles of DTH2, Ghd7, and OsPRR37, respectively. Expression levels of OrCCT01, OrCCT04, OrCCT09, OrCCT18, OrCCT21, OrCCT24, and OrCCT26 were high during the vegetative phase, suggesting that they may function as the flowering suppressor in O. rufipogon under LD.
Effects of Short-Day Treatment on Flowering Time in Rice and Its Wild Progenitor
Flowering is induced by 1 week SD treatment in O. sativa (Doi et al., 2004). To examine whether SD treatment induces flowering in O. rufipogon, we applied SD treatment to LD-grown CWR1 with Nipponbare as a control. At 40 DAG, rice plants were transferred to the SD growth room. After 10 days treatment, these plants were transplanted back to the LD growth room until flowering (Figure 6A). All SD-treated Nipponbare plants flowered evenly 13.5 days earlier than the mock-control (continuously grown under LD) plants, suggesting that, as expected, 10 days SD treatment induced flowering in Nipponbare (Figures 6A,E,F). However, O. rufipogon plants treated in the same way did not induce flowering even at 180 days after the treatment (Figures 6A,E,G). For the Nipponbare plants, Ehd1 and Hd3a were induced after 3 days of SD treatment, and the transcript level of RFT1 increased after 7 days of treatment (Figures 6B–D). However, these three genes were expressed at low levels in the SD-treated O. rufipogon plants (Figures 6B–D).
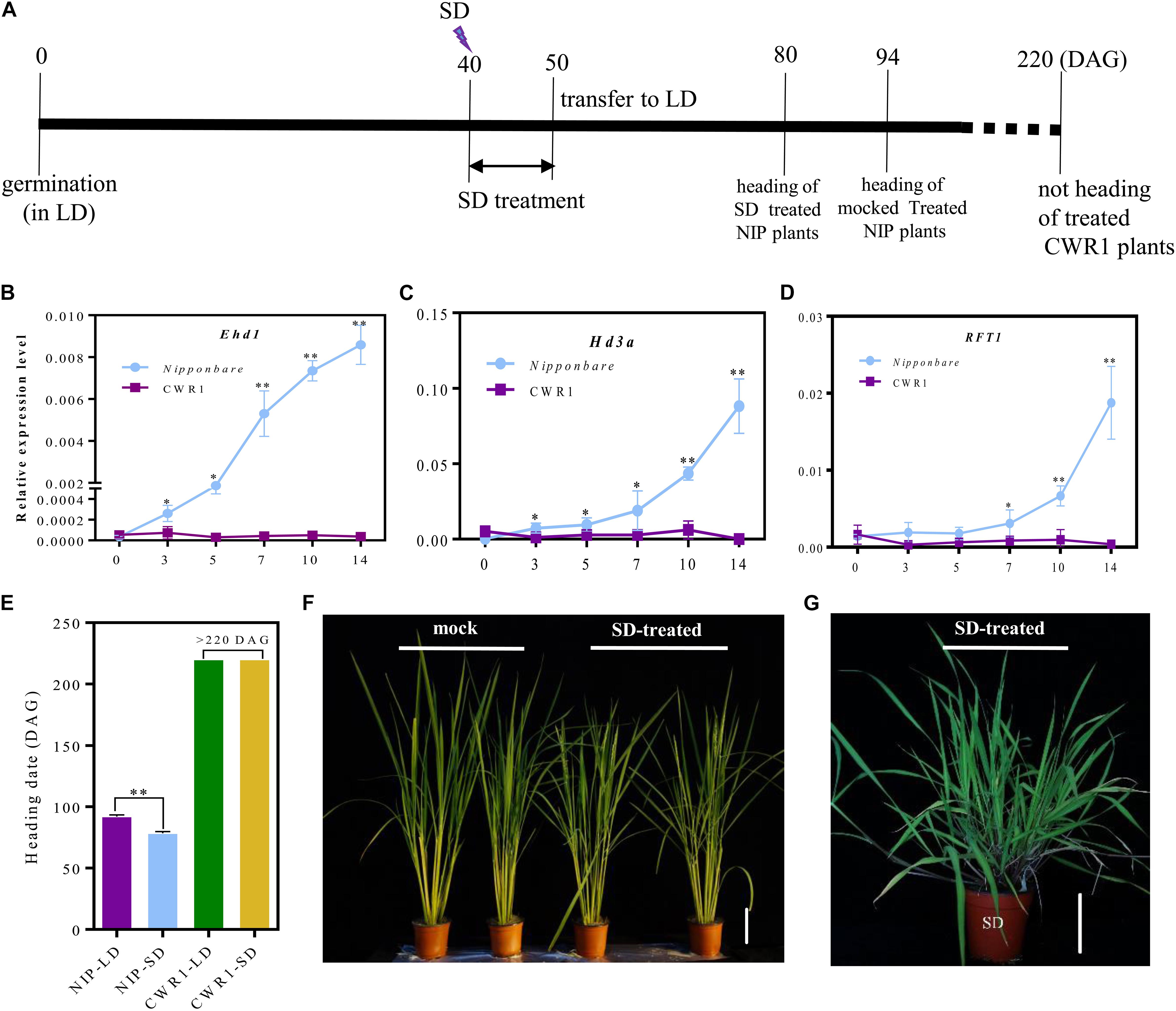
Figure 6. The photoperiod response to 10 days SD-treatment from 40 DAG to 50 DAG in Nipponbare and O. rufipogon (CWR1) plants. (A) Scheme for SD treatment. (B–D) The expression pattern of Ehd1, Hd3a, and RFT1 in the plants of Nipponbare and O. rufipogon (CWR1) with 10 days SD-treatment. The y-axis shows the relative expression levels of genes with rice OsUbi1 as an internal control; the x-axis presents the day of SD treatment. Values are means ± SD (n = 5). *, ** significant differences by student’s t-test at P ≤ 0.05 and P ≤ 0.01, respectively. (E) The heading date for LD-grown plants and SD-treated plants. (F) Phenotypes of mock (left) and SD-treated Nipponbare plants (right). (G) The phenotype of CWR1 with 10 days SD-treatment at 80 DAG. Bar = 10 cm in (F,G).
Because SD treatment at 40 DAG did not induce the expression of flowering regulatory genes, we assumed that O. rufipogon requires a longer vegetative growth period than Nipponbare before the onset of phase transition. Therefore, SD treatment was imposed on the 80-DAG CWR1 plants that were first grown under LD (Figure 7A). The SD-treated O. rufipogon plants flowered at 132–140 DAG, while mock plants did not flower even after growing for >223 DAG (Figures 7A,F). Compared with the mock plants, the expression of OrEhd1 was induced after 5 days of SD treatment (Figure 7B). Similarly, transcript levels of OrRFT1 and OrHd3a were increased with the treatment (Figures 7C,D).
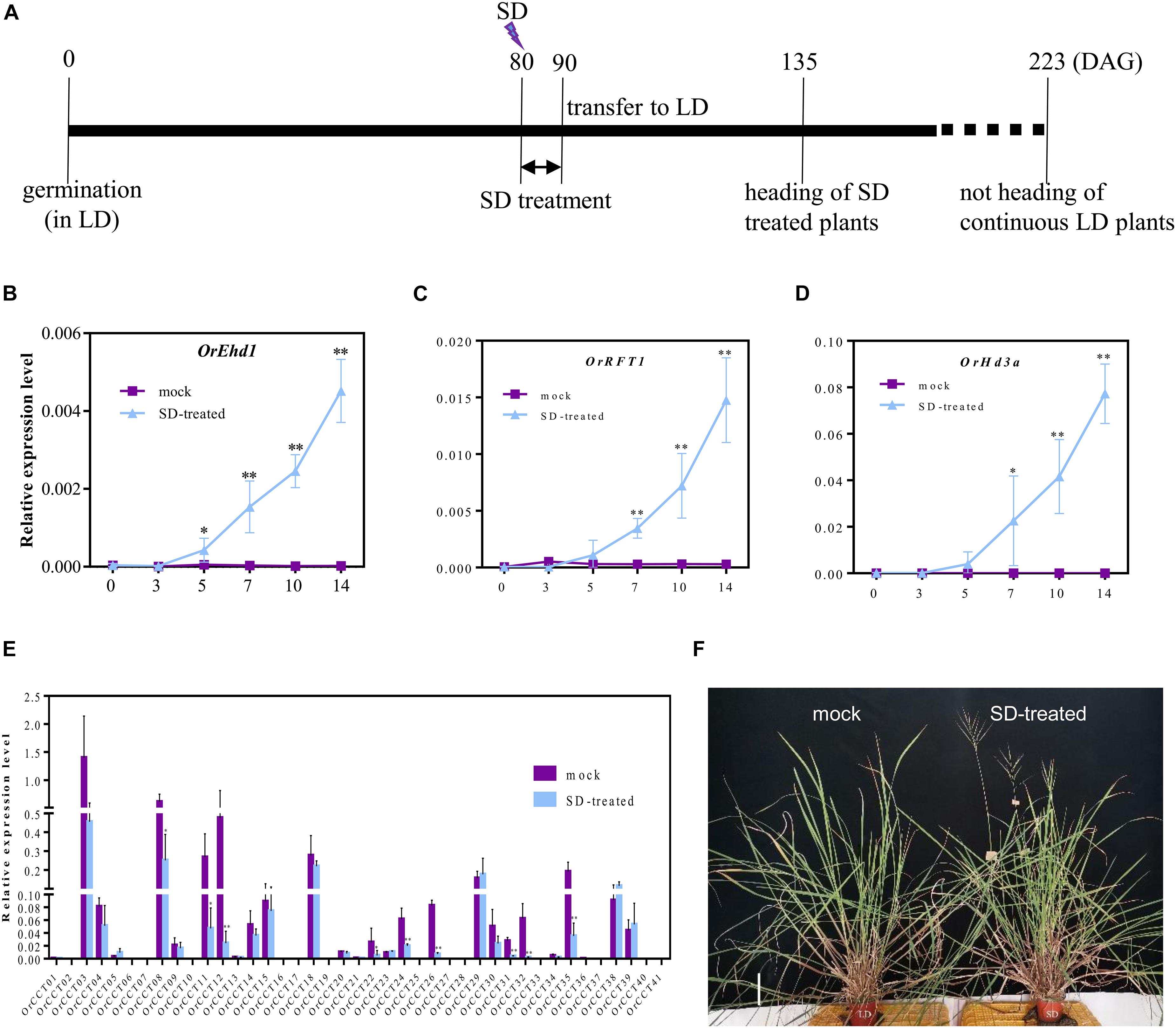
Figure 7. The response to SD-induction starting from 80 DAG in O. rufipogon (CWR1). (A) Scheme of SD treatment; (B–D) The expression level of OrEhd1 (B), OrRFT1 (C), and OrHd3a (D) in the leaf blades of CWR1 plants with mock and SD-treatment. The x-axis presents the days of SD treatment. (E) The transcript level of 41 OrCCT genes in the leaf blades of O. rufipogon (CWR1) after 10 days of SD-treatment. The transcript levels were relative to OsUbi1. Values are means ± SD (n = 5). *, ** significant differences by Student’s t-test at P ≤ 0.05 and P ≤ 0.01, respectively. (F) The phenotypes of O. rufipogon (CWR1) plants with mock and SD-treatment (right) at 140 DAG. Bar = 10 cm in (F).
The expression levels of 41 OrCCT genes further showed that, compared with mock plant, the expression of OrCCT08, OrCCT11, OrCCT12, OrCCT22, OrCCT24, OrCCT26, OrCCT31, OrCCT32, and OrCCT35 were significantly downregulated in the SD-treated plants, indicating that these CCT genes function as flowering suppressors in O. rufipogon (Figure 7E). It was reported that DTH2, the orthologous gene of OrCCT08, may induce flowering under LD. The expression level of DTH2 peaked at the beginning of the dark period and gradually reduced after that time (Wu et al., 2013). ZT-15 h is at the beginning of dark in LD, whereas the time is 5 h after dark in SD treatment. Therefore, the significantly decreased expression of OrCCT08 in SD-treated plants might be due to the change of day-length.
Effect of Overexpressed OrCCT24 on Flowering Time Under Long-Day Condition
OrCCT24, the ortholog of rice Ghd7, was highly expressed in O. rufipogon compared to Ghd7 in Nipponbare (Figure 5N). Sequence analysis showed that three single nucleotide polymorphisms (SNPs) were found in the coding region of OrCCT24 in CWR1 compared with Nipponbare. Among them, two SNPs caused amino acid substitutions, and one SNP was synonymous mutation (Figure 8A). To examine whether the high expression of OrCCT24 caused late flowering in O. rufipogon, we constructed the overexpressed OrCCT24 vector, and then transformed it into Nipponbare (Figure 8A). From 15 independently transformed plants, two lines with high levels of expression of OrCCT24 were selected (Figure 8B). The developmental expression patterns showed that the expression levels of Ehd1, Hd3a, and RFT1 rapidly increased from 50 DAG to 75 DAG in Nipponbare. Hence, the transcript levels of the three genes were measured at ZT-2 h from the transgenic plants at 60 DAG under LD. qRT-PCR experiments showed that the expression of Ehd1, Hd3a, and RFT1 was induced in the wild type (WT), whereas their expression was strongly suppressed in the overexpressed OrCCT24 plants (Figures 8C–E). The transgenic plants did not flower up to 220 DAG, while their WT controls flowered at 85–90 DAG (Figures 8F,G). Our results indicated that OrCCT24 is a strong inhibitor of flowering by suppressing the expression of Ehd1, Hd3a, and RFT1 in O. rufipogon.
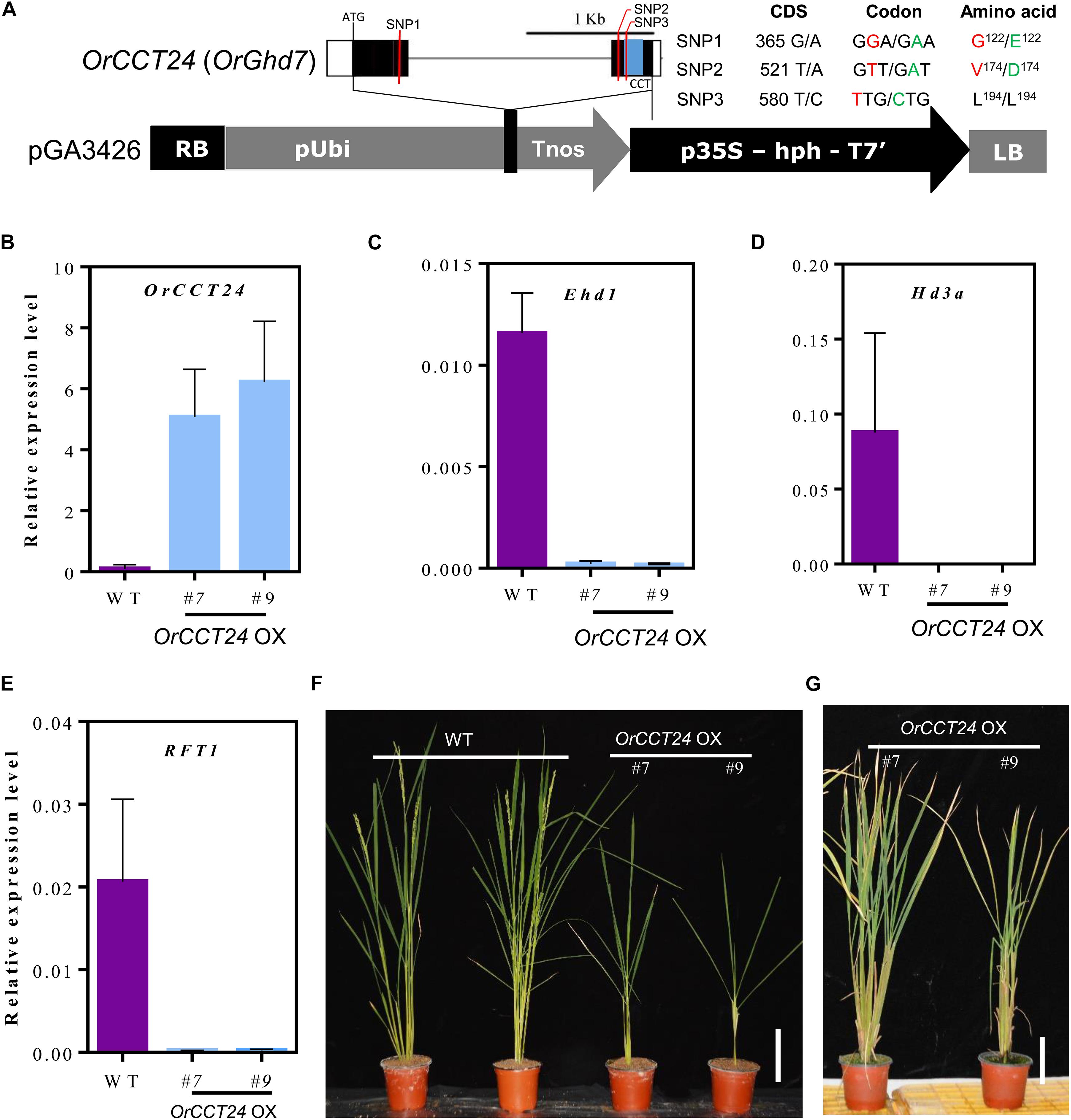
Figure 8. Expression and phenotype analyses of OrCCT24-overexpression plants under LD condition. (A) Scheme of OrCCT24 overexpressed vector. (B–E) The expression level of OrCCT24 (B), Ehd1 (C), Hd3a (D), and RFT1 (E) in WT and OrCCT24-overexpressed plants. The transcript levels were relative to OsUbi1. Error bars indicate standard deviations (n = 5). Leaf blades were harvested at ZT-2 h at 60 DAG. (F) Phenotypes of OrCCT24-overexpressed plants (T0 generation) compared with WT at 90 DAG. (G) Phenotypes of OrCCT24 overexpressed plants at 220 DAG. Scale bar = 10 cm.
Discussion
Flowering Regulation of OrCCT Genes in O. rufipogon
Crop wild relatives play an extremely important role in crops’ adaptation to farming practices, market demands, and climatic conditions (Dempewolf et al., 2017). Over the past decade, the reference genomes of approximately 15 Oryza species have been deciphered, which have greatly facilitated comprehensive allele mining in these Oryza species (Zhang et al., 2014; Stein et al., 2018; Li et al., 2020b; Shi et al., 2020). There have been great successes in introducing desired traits from wild rice into cultivated rice, such as cytoplasmic male sterile source (Lin and Yuan, 1980). In this study, we obtained OrCCT genes with the strategy of the reference genome-based gene family identification, which is time-efficient compared with traditional methods of genetic mapping (Peng et al., 2019).
In rice and Arabidopsis, the PRR subfamily is a crucial component of feedback loops of the core oscillator for the circadian clock (Mizuno and Nakamichi, 2005). In the present study, the expression levels of PRR TFs (OrCCT07, OrCCT11, OrCCT26, OrCCT32, and OrCCT35) was significantly divergent among ZT-2 h, 8 h, and 15 h both under LD and SD in O. rufipogon plants, suggesting that OrPRR genes are relevant to the circadian clock. In rice, two florigens, Hd3a and RFT1, activate floral transition by inducing the expression of MADS14 and MADS15 (Shrestha et al., 2014). The OsGI-Ghd7-Ehd1-RFT1/Hd3a pathway regulates rice flowering under LD. In this pathway, Ghd7 represses the cereal-specific flowering inducer gene Ehd1, thereby delays flowering by decreasing expression of Hd3a and RFT1 (Doi et al., 2004; Xue et al., 2008; Cho et al., 2016). OsPRR37, OsCCT01, Ghd2 negatively regulate flowering by downregulating Ehd1 (Koo et al., 2013; Zhang L. et al., 2015; Liu et al., 2016). DTH2 activates flowering by directly upregulating Hd3a and RFT1 (Wu et al., 2013). Overexpressed NRR decreases the expression of Hd3a and RFT1, which consequently delays flowering (Zhang et al., 2013). In addition, Hd1 suppresses flowering under LD when functional Ghd7 is present (Fujino et al., 2019; Zhang et al., 2019).
In the present study, a typical O. rufipogon accession (CWR1) did not flower up to 213 DAG under LD condition. The developmental expression profiles revealed that the orthologs of Ehd1 and florigens were not expressed in CWR1 under LD condition. Several CCT genes, including OrCCT01, OrCCT04, OrCCT09, OrCCT18, OrCCT21, OrCCT24, and OrCCT26, were highly expressed in CWR1 compared to Nipponbare, suggesting that they are repressors of flowering in O. rufipogon. Among these genes, orthologs of six OrCCTs except for OrCCT04 are flowering suppressors in rice (Xue et al., 2008; Koo et al., 2013; Zhang et al., 2013; Zhang L. et al., 2015; Zhang et al., 2020; Liu et al., 2016). In addition, the expression of OrCCT08, OrCCT11, OrCCT12, OrCCT22, OrCCT24, OrCCT26, OrCCT31, OrCCT32, and OrCCT35 were significantly downregulated in the SD-treated plants. With the combination of previous findings and our obtained results in this study, we propose a model for flowering regulation of OrCCT TFs in O. rufipogon under LD (Figure 9). In the model, the florigen genes OrHd3a and OrRFT1 are induced by OrEhd1 that are repressed by OrCCT01, OrCCT09, OrCCT24, and OrCCT26. Among the repressors, OrCCT24 and OrCCT26 are the strongest suppressors. OrGI positively controls OrCCT20 and OrCCT24 expression. In addition, OrCCT11, OrCCT12, OrCCT22, OrCCT31, OrCCT32, and OrCCT35 may negatively regulate flowering (Figure 9). However, their up- and down-stream genes are still unknown. Further efforts are thus needed to elucidate the roles of OrCCT TFs under SD.
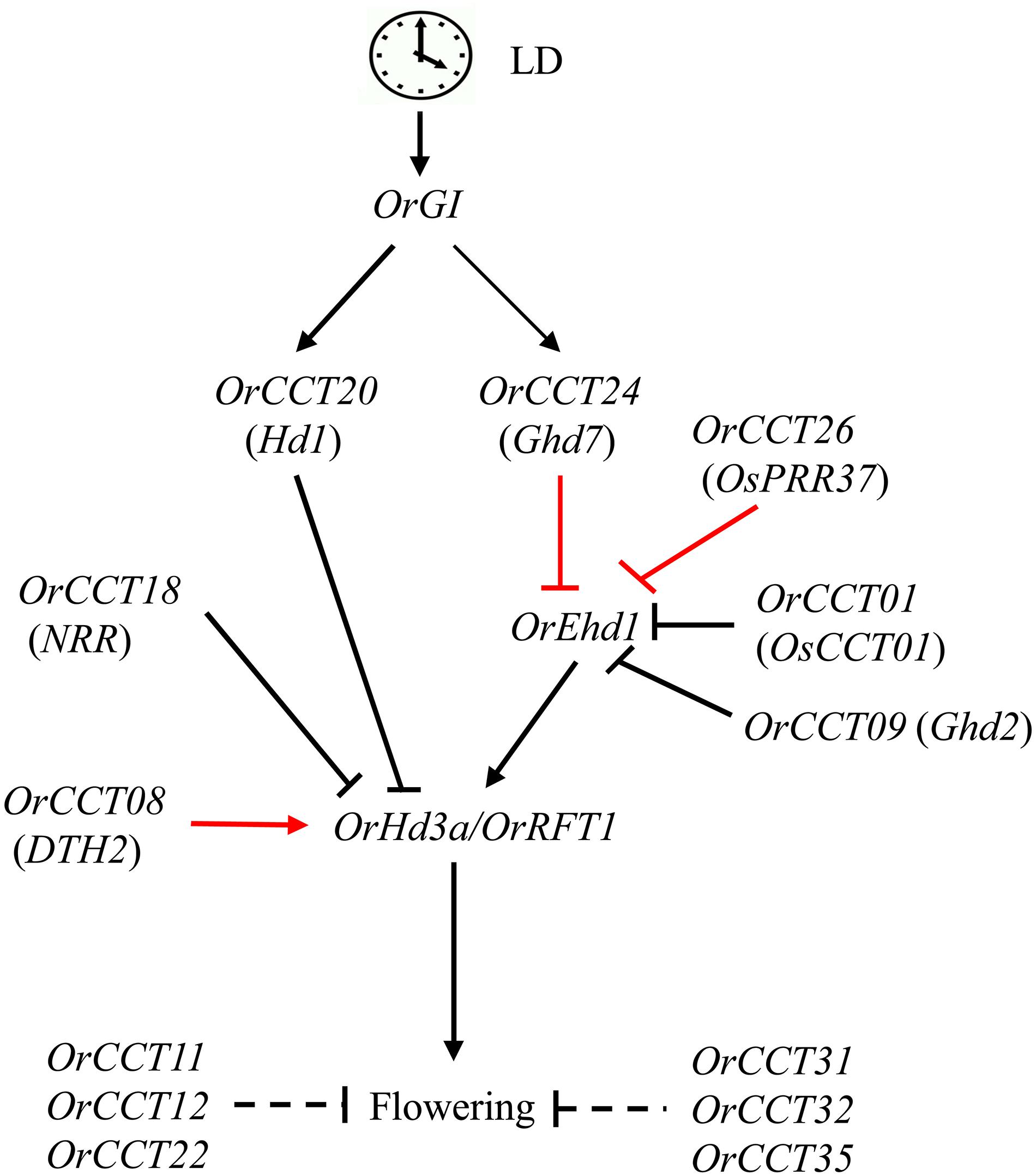
Figure 9. CCT TFs involved regulatory network for flowering time of O. rufipogon under LD. The clock at the top designates the circadian clock. Black arrows represent induction, and black bars indicate suppression. Red arrows show strong induction, and red bars denote strong suppression. The virtual line shows indirect effect.
OrCCT24, the ortholog of Ghd7, was highly expressed during all examined developmental stages in CWR1 under LD compared to Nipponbare, suggesting that OrCCT24 is a strong repressor of flowering in O. rufipogon. We observed that OrCCT24 was highly expressed in the OrCCT24 over-expressed plants compared with WT. However, the transgenic plants did not flower up to 220 DAG. RFT1 is a major florigen that functions to induce reproductive development in the SAM. It is documented that overexpression of RFT1 resulted in the direct formation of spikelets from most of the transgenic calli (Pasriga et al., 2019). In all, we confirm that 13 OrCCT TFs have played important roles in controlling flowering time, but functional roles of other OrCCT genes remain largely unknown.
Photoperiod Sensitivity of O. rufipogon
Crops are distinguished from their wild progenitors by some typical alterations, such as the loss of seed dormancy and shattering mechanisms, reduced branching, increased fruit or seed size, and changes in photoperiod sensitivity (Olsen and Wendel, 2013). The growth of O. rufipogon is limited to tropical regions (Gao, 2004; Zhao et al., 2013). In this study, O. rufipogon plants did not flower under LD conditions, which is indicative of its high photoperiod sensitivity. In most well-known examples, the members of CCT TFs are involved in the adaptation for photoperiod and flowering, including Tof11 and Tof12 in soybean (Lu et al., 2020), as well as Hd1, Ghd7, and OsPRR37 in rice (Koo et al., 2013; Zong et al., 2021). Our findings suggest that the daily expression patterns of 36 OrCCT genes (of a total of 41 members) changed with the circadian rhythm, indicating that they can respond to the light signal. We also found that 13 OrCCT genes are likely the flowering suppressors based on their expression patterns, and OrCCT08, OrCCT24, and OrCCT26 serve as the strong functional alleles of rice DTH2, Ghd7, and OsPRR37, respectively. Ghd7 and OsPRR37 are the pivotal determinants for strong photoperiod sensitivity in rice (Koo et al., 2013; Zhang J. et al., 2015; Zong et al., 2021). As discussed above, we conclude that the 13 OrCCT TFs have likely contributed to the strong photoperiod sensitivity in O. rufipogon, resulting in extremely delayed flowering under LD.
Rice cultivation has been expanded from its primitive domesticated regions to wide regions due to the long-term natural and artificial selection during rice domestication and subsequent modern improvement (Fujino et al., 2013). In this study, 10 days SD-treatment at the early developmental stage (40 DAG) did not promote flowering in O. rufipogon. Such a result indicates that the wild rice plants may require a long vegetative growth stage before responding to SD induction. When 80 days LD-grown plants were treated with 10 days of SD-induction, the treated O. rufipogon plants flowered at 52–60 days after the treatment while untreated control plants did not flower. During the SD treatment, nine OrCCT genes were significantly downregulated, indicating that they can respond to SD to regulate flowering in O. rufipogon. The photoperiod-responding OrCCT genes may be applied to breeding new rice varieties that possess higher biomass and increased grain yields.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Genomics Data Center, accession no: PRJCA006767.
Author Contributions
L-ZG and GA conceived and designed the study. XP, WT, S-FD, J-YL, Q-JZ, G-YY, JY, and L-HC conducted the experiments. XP and WT analyzed the data. XP wrote the draft of the manuscript. L-ZG, XP, and GA revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by a grant from Startup Grant from South China Agricultural University, National Research Foundation of Korea (Grant No. NRF-2020R1A2C2006826), Doctor Overseas Study Program from South China Agricultural University (Grant No. 2019LHPY014), National Natural Science Foundation of China (Grant No. 31801326), and Guangdong Special Support Program (2017TQ04N847).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Kyungsook An for generating the transgenic lines and handling the seed stock. We appreciate the reviewers for their comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.736419/full#supplementary-material
Footnotes
- ^ http://rice.uga.edu/
- ^ https://github.com/CJ-Chen/TBtools
- ^ http://pfam.xfam.org/
- ^ https://www.ncbi.nlm.nih.gov/cdd/
- ^ https://web.expasy.org/protparam/
- ^ http://busca.biocomp.unibo.it/
- ^ http://meme-suite.org/tools/meme
- ^ https://phytozome.jgi.doe.gov/
- ^ http://archive.gramene.org/
- ^ https://bigd.big.ac.cn/gsa/
References
An, G., Ebert, P. R., Mitra, A., and Ha, S. B. (1989). “Binary vectors,” in Plant Molecular Biology Manual, ed. Kluwer Academic Publisher (Dordrecht: Springer), 29–47.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cho, L., Yoon, J., and An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Cho, L., Yoon, J., Pasriga, R., and An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Cho, L. H., Yoon, J., Wai, A. H., and An, G. (2018). Histone deacetylase 701 (HDT701) induces flowering in rice by modulating expression of OsIDS1. Mol. Cells 41, 665–675. doi: 10.14348/molcells.2018.0148
Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., Bailey, P. C., et al. (2012). Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7:e45307. doi: 10.1371/journal.pone.0045307
Dempewolf, H., Baute, G., Anderson, J., Kilian, B., Smith, C., and Guarino, L. (2017). Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. doi: 10.2135/cropsci2016.10.0885
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Gene. Dev. 18, 926–936. doi: 10.1101/gad.1189604
Fujino, K., Yamanouchi, U., Nonoue, Y., Obara, M., and Yano, M. (2019). Switching genetic effects of the flowering time gene Hd1 in LD conditions by Ghd7 and OsPRR37 in rice. Breed. Sci. 69, 127–132. doi: 10.1270/jsbbs.18060
Fujino, K., Yamanouchi, U., and Yano, M. (2013). Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor. Appl. Genet. 126, 611–618. doi: 10.1007/s00122-012-2005-5
Gao, L. (2004). Population structure and conservation genetics of wild rice Oryza rufipogon (Poaceae): a region-wide perspective from microsatellite variation. Mol. Ecol. 13, 1009–1024. doi: 10.1111/j.1365-294X.2004.02108.x
Gao, L., Song, G., and Hong, D. (2000). A preliminary study on ecological differentiation within the common wild rice Oryza rufipogon Griff. Acta Agron. Sin. 26, 210–213.
Gao, L., Wei, C., Yang, Q., Hong, D., and Song, G. (2001). Intra-population genetic structure of Oryza rufipogon Griff. in Yunnan, China. J. Plant Res. 114, 107–113. doi: 10.1007/PL00013973
Gao, L. Z. (2002). Assessment of population genetic structure of common wild rice Oryza rufipogon Griff. detected by microsatellite DNA and allozyme loci. Theor. Appl. Genet. 106, 173–180. doi: 10.1007/s00122-002-1027-9
Gao, L. Z., and Hong, D. Y. (2000). Allozymic diversity and genetic structure of common wild rice Oryza rufipogon Griff., China. Theor. Appl. Genet. 101, 494–502. doi: 10.1007/s001220051508
Hori, K., Matsubara, K., and Yano, M. (2016). Genetic control of flowering time in rice: integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129, 2241–2252. doi: 10.1007/s00122-016-2773-4
Huang, C., Sun, H., Xu, D., Chen, Q., Liang, Y., Wang, X., et al. (2018). ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. U.S.A. 115, E334–E341. doi: 10.1073/pnas.1718058115
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khush, G. S. (1997). Origin, dispersal cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. doi: 10.1023/A:1005810616885
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, S., Lee, D., Yang, J., Moon, S., and An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73–78. doi: 10.1007/s12374-008-9008-4
Koo, B., Yoo, S., Park, J., Kwon, C., Lee, B., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Kumar, S., Tamura, K., and Nei, M. (1994). MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 10, 189–191.
Li, W., Li, K., Huang, Y., Shi, C., Hu, W., Zhang, Y., et al. (2020a). SMRT sequencing of the Oryza rufipogon genome reveals the genomic basis of rice adaptation. Commun. Biol. 3, 153–167. doi: 10.1038/s42003-020-0890-8
Li, W., Zhang, Q. J., Zhu, T., Tong, Y., Li, K., Shi, C., et al. (2020b). Draft genomes of two outcrossing wild rice, Oryza rufipogon and O. longistaminata, reveal genomic features associated with mating-system evolution. Plant Direct 4:e00232. doi: 10.1002/pld3.232
Li, Y., and Xu, M. (2017). CCT family genes in cereal crops: a current overview. Crop J. 5, 449–458. doi: 10.1016/j.cj.2017.07.001
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general-purpose read summarization program. Bioinformatics 30, 923–930.
Lin, S. C., and Yuan, L. P. (1980). “Hybrid rice breeding in China,” in Proceedings of the Innovative Approaches to Rice Breeding: Selected Papers From the 1979 International Rice Research Conference, (Los Banos: International Rice Research Institute).
Liu, H., Zhou, X., Li, Q., Wang, L., and Xing, Y. (2020). CCT domain-containing genes in cereal crops: flowering time and beyond. Theor. Appl. Genet. 133, 1385–1396. doi: 10.1007/s00122-020-03554-8
Liu, J., Shen, J., Xu, Y., Li, X., Xiao, J., and Xiong, L. (2016). Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 67, 5785–5798. doi: 10.1093/jxb/erw344
Lu, S., Dong, L., Fang, C., Liu, S., Kong, L., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436. doi: 10.1038/s41588-020-0604-7
Ma, L., Yi, D., Yang, J., Liu, X., and Pang, Y. (2020). Genome-wide identification, expression analysis and functional study of CCT gene family in Medicago truncatula. Plants 9, 513. doi: 10.3390/plants9040513
Mizuno, T., and Nakamichi, N. (2005). Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 46, 677–685. doi: 10.1093/pcp/pci087
Morishima, H., Oka, H., and Chang, W. (1961). Directions of differentiation in populations of wild rice, Oryza perennis and O. sativa f. spontanea. Evolution 15, 326–339.
Olsen, K. M., and Wendel, J. F. (2013). A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64, 47–70. doi: 10.1146/annurev-arplant-050312-120048
Omolade, O., Müller, A. E., Jung, C., and Melzer, S. (2016). BvPRR7 is a cold responsive gene with a clock function in beet. Biol. Plant. 60, 95–104. doi: 10.1007/s10535-015-0568-0
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl976
Pasriga, R., Yoon, J., Cho, L., and An, G. (2019). Overexpression of RICE FLOWERING LOCUS T 1 (RFT1) induces extremely early flowering in rice. Mol. Cells 42, 406–417. doi: 10.14348/molcells.2019.0009
Peng, X., Luo, L., Cui, H., Wang, H., Guo, T., Liu, Y., et al. (2019). Characterization and fine mapping of a leaf wilt mutant, m3, induced by heavy ion irradiation of rice. Crop Sci. 59, 2679–2688. doi: 10.2135/cropsci2019.03.0167
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Savojardo, C., Martelli, P. L., Fariselli, P., Profiti, G., and Casadio, R. (2018). BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 46, W459–W466. doi: 10.1093/nar/gky320
Shi, C., Li, W., Zhang, Q., Zhang, Y., Tong, Y., Li, K., et al. (2020). The draft genome sequence of an upland wild rice species, Oryza granulata. Sci. Data 7:131. doi: 10.1038/s41597-020-0470-2
Shrestha, R., Gómez-Ariza, J., Brambilla, V., and Fornara, F. (2014). Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Ann. Bot. 114, 1445–1458. doi: 10.1093/aob/mcu032
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Stein, J. C., Yu, Y., Copetti, D., Zwickl, D. J., Zhang, L., Zhang, C., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296. doi: 10.1038/s41588-018-0040-0
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Más, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Tan, J., Jin, M., Wang, J., Wu, F., Sheng, P., Cheng, Z., et al. (2016). OsCOL10, a CONSTANS-like gene, functions as a flowering time repressor downstream of Ghd7 in Rice. Plant Cell Physiol. 57, 798–812. doi: 10.1093/pcp/pcw025
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wu, W., Zheng, X. M., Lu, G., Zhong, Z., Gao, H., Chen, L., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. U.S.A. 110, 2775–2780. doi: 10.1073/pnas.1213962110
Xie, X., Du, H., Tang, H., Tang, J., Tan, X., Liu, W., et al. (2020). A chromosome-level genome assembly of the wild rice Oryza rufipogon facilitates tracing the origins of Asian cultivated rice. Sci. China Life Sci. 64, 282–293. doi: 10.1007/s11427-020-1738-x
Xu, X., Meng, Q. L., Geng, M. F., Ren, N. N., Zhou, L., Du, Y. S., et al. (2020). Divergence in flowering time is a major component contributing to reproductive isolation between two wild rice species (Oryza rufipogon and O. nivara). Sci China Life Sci. Sci 63, 1714–1724. doi: 10.1007/s11427-019-1678-6
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644.
Yu, J., Wang, U., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The Genomes of Oryza sativa: a history of duplications. Plos Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Zhang, J., Fan, X., Hu, Y., Zhou, X., He, Q., Liang, L., et al. (2020). Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J. Integr. Plant Biol. 63, 913–923.
Zhang, J., Hu, Y., Xu, L., He, Q., Fan, X., and Xing, Y. (2017). The CCT domain-containing gene family has large impacts on heading date, regional adaptation, and grain yield in rice. J. Integr. Agric. 16, 2686–2697. doi: 10.1016/S2095-3119(17)61724-6
Zhang, J., Zhou, X., Yan, W., Zhang, Z., Lu, L., Han, Z., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066. doi: 10.1111/nph.13538
Zhang, L., Li, Q., Dong, H., He, Q., Liang, L., Tan, C., et al. (2015). Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 5:7663. doi: 10.1038/srep07663
Zhang, Q., Zhu, T., Xia, E., Shi, C., Liu, Y., Zhang, Y., et al. (2014). Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, E4954–E4962. doi: 10.1073/pnas.1418307111
Zhang, Y., Zhang, G., Xiao, N., Wang, L., Fu, Y., Sun, Z., et al. (2013). The rice ‘nutrition response and root growth’ (NRR) gene regulates heading date. Mol. Plant 6, 585–588. doi: 10.1093/mp/sss157
Zhang, Z., Zhang, B., Qi, F., Wu, H., Li, Z., and Xing, Y. (2019). Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long-day conditions in rice. Mol. Breed. 39:92. doi: 10.1007/s11032-019-1001-8
Zhao, Q., Feng, Q., Lu, H., Li, Y., Wang, A., Tian, Q., et al. (2018). Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284. doi: 10.1038/s41588-018-0041-z
Zhao, Y., Vrieling, K., Liao, H., Xiao, M., Zhu, Y., Rong, J., et al. (2013). Are habitat fragmentation, local adaptation and isolation-by-distance driving population divergence in wild rice Oryza rufipogon? Mol. Ecol. 22, 5531–5547. doi: 10.1111/mec.12517
Zheng, X. M., and Ge, S. (2010). Ecological divergence in the presence of gene flow in two closely related Oryza species (Oryza rufipogon and O. nivara). Mol. Ecol. 19, 2439–2454.
Keywords: Oryza rufipogon, rice, CCT genes, genomic synteny, expression profiles, photoperiodic flowering regulation
Citation: Peng X, Tun W, Dai S-f, Li J-y, Zhang Q-j, Yin G-y, Yoon J, Cho L-h, An G and Gao L-z (2021) Genome-Wide Analysis of CCT Transcript Factors to Identify Genes Contributing to Photoperiodic Flowering in Oryza rufipogon. Front. Plant Sci. 12:736419. doi: 10.3389/fpls.2021.736419
Received: 05 July 2021; Accepted: 08 October 2021;
Published: 08 November 2021.
Edited by:
Siegbert Melzer, University of Kiel, GermanyReviewed by:
Nazgol Emrani, University of Kiel, GermanyShowkat Ganie, Royal Holloway, University of London, United Kingdom
Copyright © 2021 Peng, Tun, Dai, Li, Zhang, Yin, Yoon, Cho, An and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gynheung An, Z2VuZWFuQGtodS5hYy5rcg==; Li-zhi Gao, TGdhb2dlbm9taWNzQDE2My5jb20=
 Xin Peng
Xin Peng Win Tun
Win Tun Shuang-feng Dai1
Shuang-feng Dai1 Jia-yue Li
Jia-yue Li Guo-ying Yin
Guo-ying Yin Jinmi Yoon
Jinmi Yoon Gynheung An
Gynheung An Li-zhi Gao
Li-zhi Gao