
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 September 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.733237
This article is part of the Research TopicMolecular Interactions between Crops and Phytopathogens, Volume III: Vegetables and Other CropsView all 11 articles
 Hongbo Yuan1,2
Hongbo Yuan1,2 Cong Jin1
Cong Jin1 Hongcui Pei1,2
Hongcui Pei1,2 Lifang Zhao1,2
Lifang Zhao1,2 Xue Li1,2
Xue Li1,2 Jiali Li1,2
Jiali Li1,2 Wanting Huang1,3
Wanting Huang1,3 Renchun Fan1,2
Renchun Fan1,2 Wende Liu4
Wende Liu4 Qian-Hua Shen1,2*
Qian-Hua Shen1,2*Powdery mildew is one of the most important fungal pathogen diseases. The genome of barley mildew fungus, Blumeria graminis f. sp. hordei (Bgh), encodes a large number of candidate secreted effector proteins (CSEPs). So far, the function and mechanism of most CSEPs remain largely unknown. Here, we identify a Bgh effector CSEP0027, a member of family 41, triggering cell death in Nicotiana benthamiana. CSEP0027 contains a functional signal peptide (SP), verified by yeast secretion assay. We show that CSEP0027 promotes Bgh virulence in barley infection using transient gene expression and host-induced gene silencing (HIGS). Barley catalase HvCAT1 is identified as a CSEP0027 interactor by yeast two-hybrid (Y2H) screening, and the interaction is verified in yeast, in vitro and in vivo. The coexpression of CSEP0027 and HvCAT1 in barley cells results in altered localization of HvCAT1 from the peroxisome to the nucleus. Barley stripe mosaic virus (BSMV)-silencing and transiently-induced gene silencing (TIGS) assays reveal that HvCAT1 is required for barley immunity against Bgh. We propose that CSEP0027 interacts with barley HvCAT1 to regulate the host immunity and likely reactive oxygen species (ROS) homeostasis to promote fungal virulence during barley infection.
Powdery mildews are widespread fungal diseases that affect more than 10,000 plant species, such as important cereal crops, economic, and ornamental plants (Glawe, 2008; Dean et al., 2012; Takamatsu, 2013). As obligate biotrophic pathogens, powdery mildew fungi totally depend on the living plant cells for survival and reproduction. Mildew conidiospores attach to the epidermal tissue of the host, germinate and produce fungal infection structures, such as the appressorium and penetration peg to penetrate the plant cell wall, subsequently, the haustoria are developed within the lumen of the host cells but separated from the host cell cytoplasm by extrahaustorial membrane (EHM) and extrahaustoral matrix (EHMX) (Panstruga, 2003; Both et al., 2005). The haustorium is believed to be a site for nutrient uptake and signaling exchange (Panstruga and Dodds, 2009; Stergiopoulos and de Wit, 2009), and effector proteins are believed to deliver into the plant cells through haustorium to promote fungal virulence.
Blumeria graminis, the powdery mildew fungus causing disease on the cereal crop species and grasses (Poaceae), has been classified into at least eight formae speciales (f.sp.), each adapted to a host genus (Troch et al., 2014). B. graminis f.sp. hordei (Bgh) and B. graminis f.sp. tritici (Bgt) colonize barley and wheat, respectively. The Bgh and Bgt genomes code for ∼700 and 800 candidate secreted effector proteins (CSEPs), respectively (Godfrey et al., 2010; Spanu et al., 2010; Pedersen et al., 2012; Wicker et al., 2013; Frantzeskakis et al., 2018; Müller et al., 2019). Many Bgh CSEPs are overlapped with the so called Blumeria effector candidate (BEC) proteins identified from the proteomic analyses (Bindschedler et al., 2009, 2016; Godfrey et al., 2009). A majority of these Bgh CSEPs have a predicted amino-terminal signal peptide (SP) and a putative Y/F/WxC motif (Bindschedler et al., 2009; Godfrey et al., 2010; Spanu et al., 2010; Pedersen et al., 2012). A large proportion of Bgh CSEPs (c. 25%) are structurally predicted similar to RNase and/or RNA-binding activity, and these CSEPs are termed as RNase Like Proteins expressed in Haustoria (RALPHs) (Pedersen et al., 2012; Spanu, 2017). Interestingly, most of the so far identified Bgh AVRA effectors, each recognized by a cognate barley MLA receptor, are also RALPHs with fungal RNase folds but lacking the residues required for RNase activity (Lu et al., 2016; Saur et al., 2019; Bauer et al., 2021). So far, several Bgh CSEPs/BECs have been functionally characterized with respect to fungal virulence through transient gene expression and host-induced gene silencing (HIGS) approaches (Bindschedler et al., 2009; Godfrey et al., 2009; Nowara et al., 2010; Spanu et al., 2010; Pedersen et al., 2012; Pliego et al., 2013; Ahmed et al., 2015, 2016; Menardo et al., 2017; Frantzeskakis et al., 2018; Pennington et al., 2019; Li et al., 2021). The host targets have been identified for some CSEPs that are involved in plant immunity and stress responses (Zhang et al., 2012; Schmidt et al., 2014; Ahmed et al., 2015; Pennington et al., 2016, 2019; Saur et al., 2019). Recently, few Bgh CSEPs have been showed or proposed to play a role in regulating the host cell death (Pennington et al., 2019; Li et al., 2021). A CSEP0064/BEC1054, one of the Bgh RALPHs, binds to RNA and may act as a pseudoenzyme to inhibit the action of the host ribosome-inactivating proteins (RIPs) that would otherwise induce cell death (Pennington et al., 2019). The CSEP0139 and CSEP0182 are capable of suppressing programmed cell death (PCD) induced by various cell death inducers in plant cells (Li et al., 2021). Despite these intensive studies, the function and mode of action of many CSEPs remain largely unclear.
Reactive oxygen species (ROS), produced from aerobic metabolism in plants, have been appreciated as major signaling molecules in plant development and in response to the biotic and abiotic stresses (Apel and Hirt, 2004; Nanda et al., 2010; Waszczak et al., 2018). In plant–pathogen interactions, ROS can directly kill the invading pathogens and trigger cell death to stop pathogen invasion, or can serve as signaling molecules to regulate the plant defense responses (Mittler et al., 2011; Mittler, 2017). Hydrogen peroxide (H2O2) and superoxide anion (O2–) are the two major ROS molecules accumulating in the plants in response to the pathogen infections. The plants rely on an intricate network to control the levels of ROS at different subcellular compartments (Hückelhoven and Kogel, 2003; Nanda et al., 2010; Petrov and Van Breusegem, 2012). Catalases are part of “the ROS network,” playing a central role in maintaining the cellular H2O2 balance and in signaling crosstalk (Du et al., 2008; Chaouch et al., 2010; Nanda et al., 2010; Sharma and Ahmad, 2014; Li et al., 2015; Zhang et al., 2015; Murota et al., 2017; Yuan et al., 2017; Chen and Jarosz, 2020; Chen et al., 2020).
In barley/wheat response to B. graminis infection, ROS are involved in immune responses at early and late stages of the pathogen infections (Hückelhoven and Kogel, 2003). In barley under attack by Bgh or Bgt spores, H2O2 is detected to locally accumulate in papillae (cell wall appositions) or in the whole cell, which is generally associated with host cell inaccessibility (Thordal-Christensen et al., 1997; Hückelhoven et al., 1999, 2001, 2003). The ROS are also detected in Bgt-attacked wheat epidermal cells and are involved in both pattern-triggered immunity (PTI) and effector-triggered immunity (ETI; Altpeter et al., 2005; Schweizer, 2008; Chang et al., 2019). On the other hand, superoxide radical anion (O2–) is believed to act in restricting cell death. In barley epidermal cells under attack by Bgh spores, O2– accumulation is strictly associated with a successful penetration and O2– also accumulates in the living cells neighboring the HR cells (Hückelhoven and Kogel, 1998; Hückelhoven et al., 2000). These studies suggest that ROS play a complex role in the plant–biotrophic fungal interactions, not only in early cell wall-associated defense and in late defense signaling but also in the cell-death suppression.
In this study, we screen ∼100 Bgh CSEPs through agroinfiltration in Nicotiana benthamiana and identify CSEP0027 triggering cell death. We show that CSEP0027 promotes fungal virulence in barley infection. We further identify CSEP0027 interactors by yeast two-hybrid (Y2H) screening and barley HvCAT1 is shown to interact with CSEP0027 in yeast, in vitro and in vivo. Coexpression of CSEP0027 and HvCAT1 in barley cells induces the nuclear accumulation of HvCAT1 that is normally localized to the peroxisome. The functional analyses indicate that HvCAT1 is involved in barley immunity against Bgh. We propose CSEP0027 target barley HvCAT1 to regulate host immunity and promote fungal virulence in barley infection.
The Bgh genome encodes several hundreds of potential effectors, and ∼491 effector-like proteins were initially identified to be CSEPs (Spanu et al., 2010; Pedersen et al., 2012). We selected a hundred of these CSEP genes for further characterization based on their expression levels and abundance in haustoria (Godfrey et al., 2010; Pedersen et al., 2012). The cDNA sequences of 101 CSEPs from 34 families were amplified with specific primers using RNA samples derived from barley leaf materials infected with the compatible isolate BghA6 (Supplementary Table 1). All CSEP cDNA sequences excluding the predicted signal peptide (ΔSP) were subcloned into vector pGR107 for Agrobacterium tumefaciens-mediated transient expression in N. benthamiana (Wang et al., 2011). We identified several CSEPs suppressing cell death in plants (Li et al., 2021), but much fewer CSEPs inducing cell death. As shown in Figure 1A, CSEP0027 is one of the CSEPs inducing clear water-soaked-like cell death phenotype in N. benthamiana, as compared to GFP alone, which serves as a negative control. The AVRa13 effector and its cognate receptor MLA13 were also coexpressed and triggered cell death in N. benthamiana (Lu et al., 2016), which severed as a positive and technique control here (Supplementary Figure 1). Trypan blue staining confirmed the localized cell death and immunoblotting verified the expression of the HA-tagged fusion proteins (Figure 1A), and DAB (3, 3′-diaminobenzidine) staining also revealed H2O2 accumulation in the infiltrated area (Supplementary Figure 2).
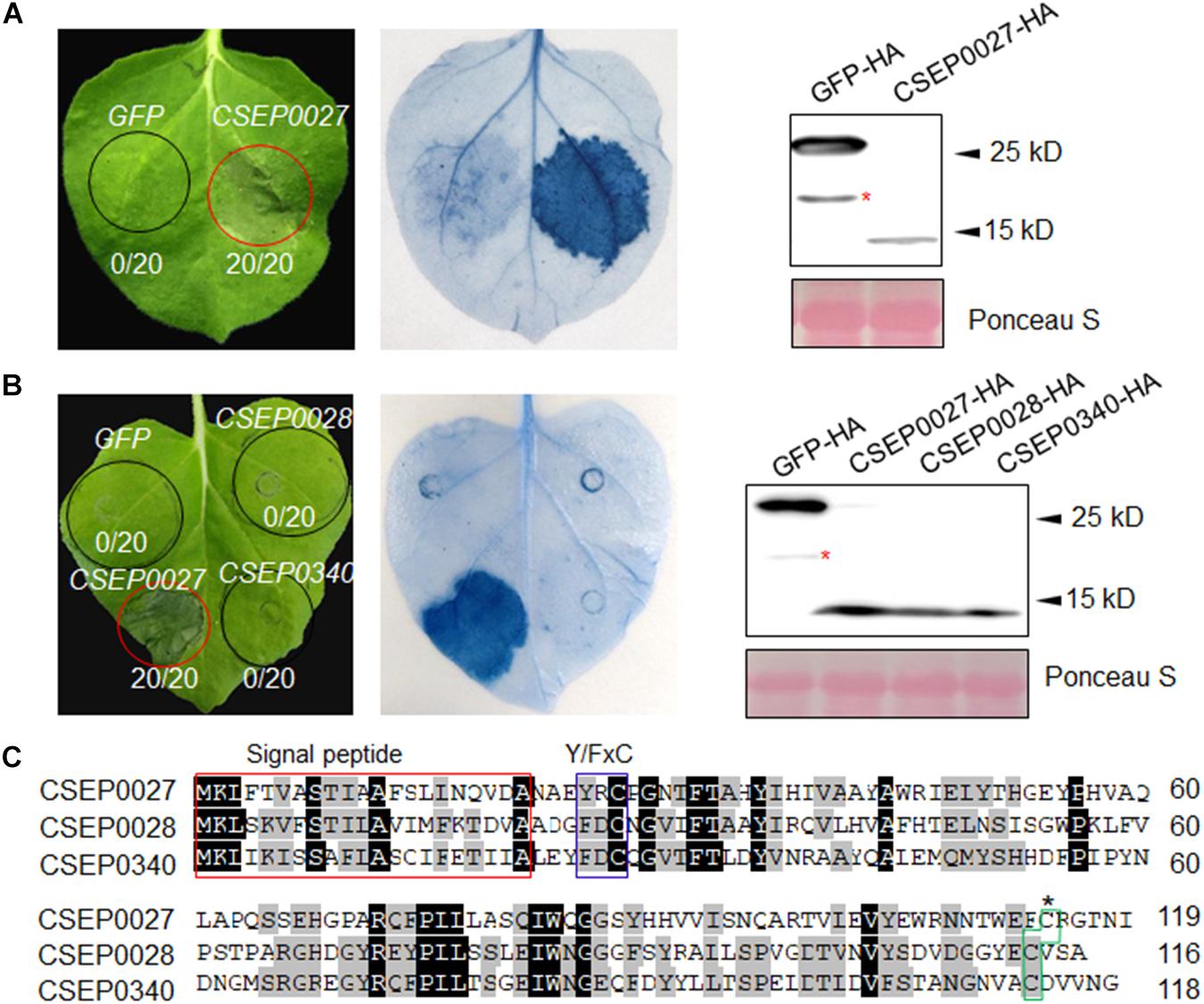
Figure 1. CSEP0027 triggers cell death in Nicotiana benthamiana. (A) Expression of CSEP0027 triggers cell death in N. benthamiana. Agrobacterium tumefaciens was used to transiently express CSEP0027 or GFP in N. benthamiana leaves. The picture was taken at 5 dpi, and cell death was visualized by trypan blue staining. The numbers in each circled area indicate numbers of cell death in total number of leaf areas infiltrated with the construct. Total protein extract was obtained from N. benthamiana leaves at 60 hpi and protein expressions were confirmed by immunoblotting using anti-HA antibody. Ponceau staining was used to show equal loading. (B) CSEP0028 and CSEP0340 do not trigger cell death in N. benthamiana. The experimental procedures are the same as in (A). The stars in the Western blots in panels (A,B) indicate non-specific signals. (C) Sequence alignment of CSEP0027, CSEP0028, and CSEP0340, performed using the DNAMAN software. The signal peptides are highlighted in red box, Y/FxC motif in blue box, and C-terminal conserved cysteine in green box.
The CSEP0027, CSEP0028, and CSEP0340 are the three members from the same Bgh CSEP family 41 (Pedersen et al., 2012), in addition, BgtE-10117 and BgtE-20000 are the two potential Bgt homologs being identified as highly related sequences to CSEP0027 (Supplementary Figure 3A) (Praz et al., 2017). All these five CSEPs harbor a predicted SP, a Y/FxC motif, and a conserved C-terminal cysteine, with some conserved residues in the middle (Figure 1C and Supplementary Figure 3A). We tested if any of the other four CSEPs trigger cell death, unexpectedly none of them induced cell death in N. benthamiana (Figure 1B and Supplementary Figure 3B). CSEP0027, thus represents a unique Bgh effector protein to induce cell death in N. benthamiana.
To validate the secretory function of the CSEP0027 signal peptide, we used a yeast secretion assay based on invertase secretion and yeast growth on sucrose or raffinose media (Lee et al., 2006; Oh et al., 2009). The predicted SPs were fused in frame to the mature sequence of yeast invertase in the vector pSUC2 and expressed in the invertase mutant yeast strain YTK12 that otherwise cannot grow on YPRAA medium (Gu et al., 2011). CSEP0027-SP derived construct enabled transformed yeast cells to grow on YPRAA plate (with raffinose instead of sucrose as the carbon source), and so did the PsAvr1b-SP from the oomycete Avr1b effector as a positive control (Figure 2, middle panel). The first 25 amino acids of Mg87, a Magnaporthe grisea cytoplasmic protein as a negative control, did not enable yeast to grow (Figure 2). In addition, the secretion of the invertase was confirmed by the conversion of 2, 3, 5-triphenyltetrazolium chloride (TTC) to the insoluble red-colored triphenylformazan (Figure 2, bottom panel). These results suggest that CSEP0027 is a secreted protein carrying a functional SP.
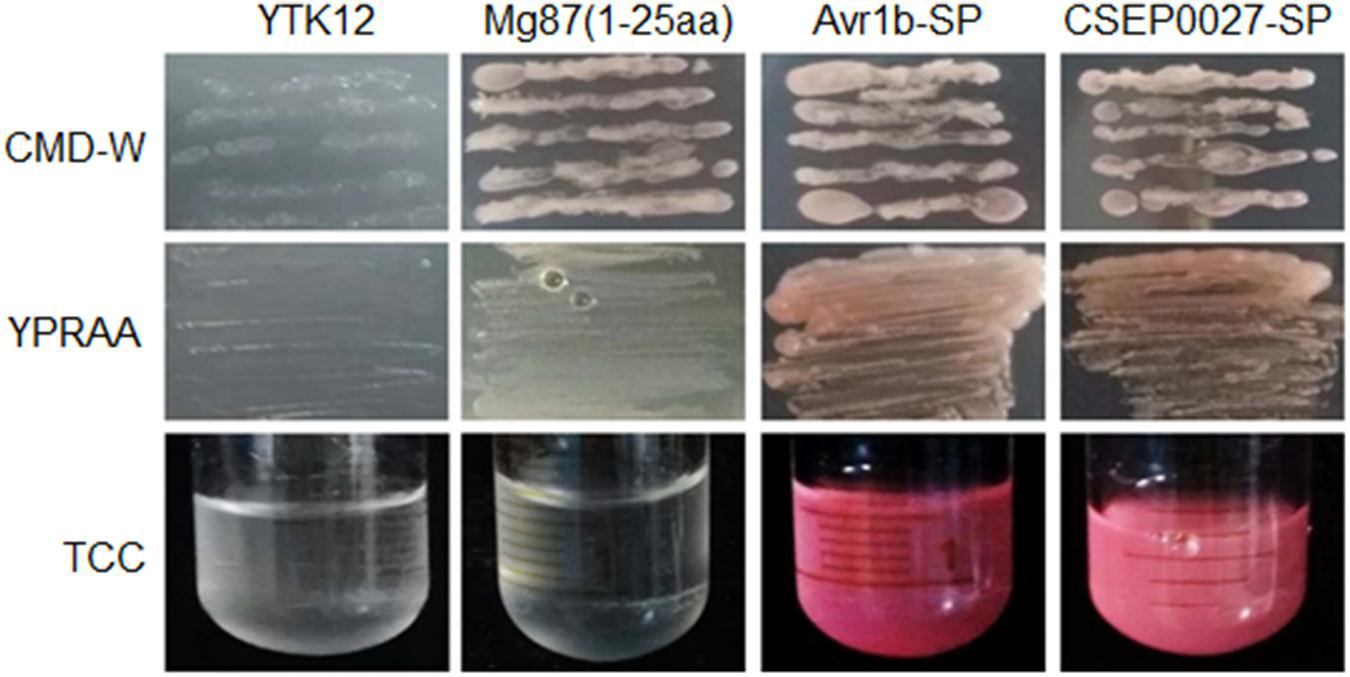
Figure 2. CSEP0027 is a secreted protein. Yeast invertase secretion assay was used to confirm the function of the predicted SP of CSEP0027. A construct expressing a fusion of SP sequence of CSEP0027 and a yeast invertase was transformed into the yeast strain YTK12 and tested in the assay, with the N-terminal sequence of Magnaporthe oryzae Mg87 protein and SP sequence of Phytophthora sojae PsAvr1b used as negative and positive controls, respectively. CMD-W plates were used to select yeast strain YTK12 carrying the pSUC2 vector. YPRAA media were used to indicate invertase secretion. An enzymatic activity test based on the reduction of 2, 3, 5-triphenyltetrazolium chloride (TTC) to red-colored formazan was also used to confirm invertase secretion.
To investigate the function of CSEP0027 in fungal virulence, we first overexpressed CSEP0027 in barley epidermal cells through single-cell transient gene expression followed by BghA6 infection in a compatible interaction (Bai et al., 2012). The expression of mature CSEP0027 (CSEP0027ΔSP) in barley cells led to markedly increased haustorial formation rate (i.e., haustorium index) to ∼68%, as compared to ∼52% in the empty vector control (EV) (Figure 3A). By contrast, silencing CSEP0027 through HIGS significantly decreased haustorium formation rate by ∼40%, relative to the EV control (Figure 3B). Similarly, the silencing of CSEP0105, an effector gene used as a positive control (Nowara et al., 2010; Ahmed et al., 2015), led to a stronger effect on the reduction of haustorium index by ∼60%, also relative to the EV (Figure 3B). These data indicate that CSEP0027 contributes to Bgh virulence.
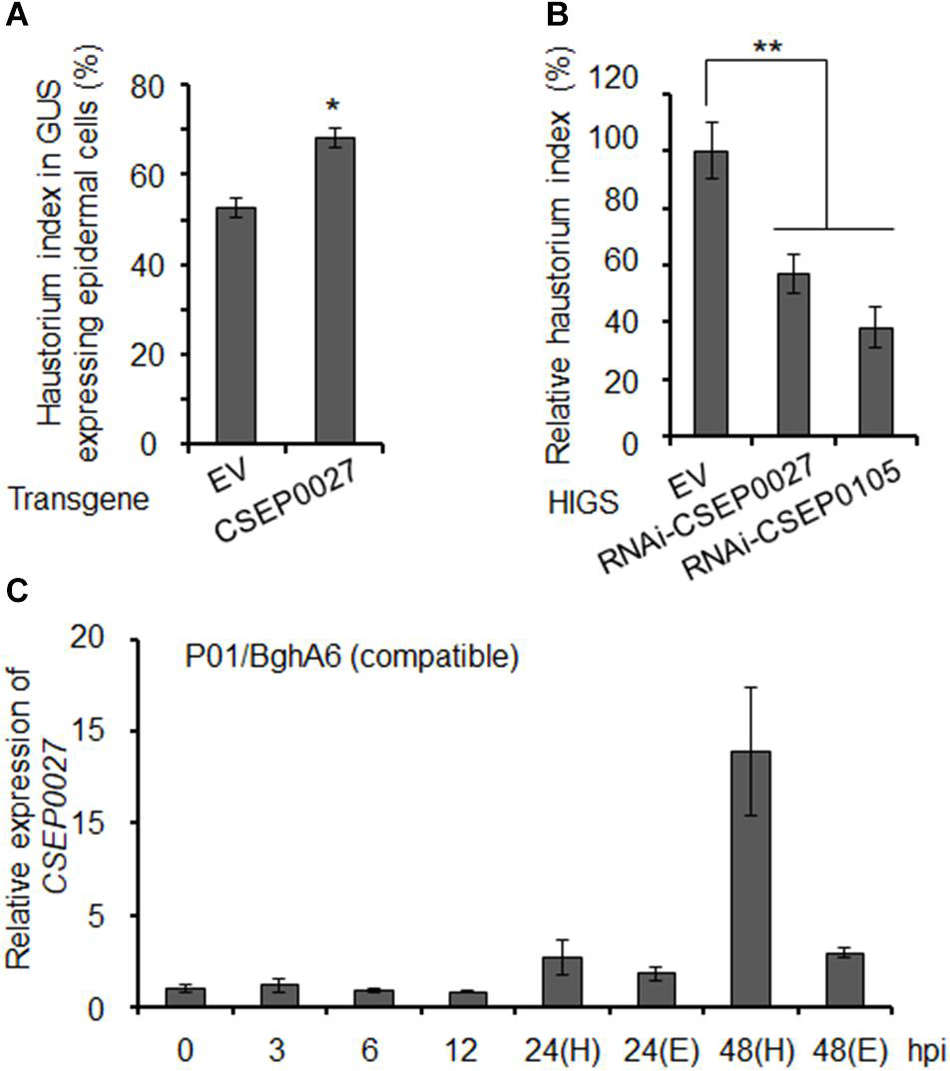
Figure 3. CSEP0027 contributes to Bgh virulence. (A) Overexpression of CSEP0027 promotes Bgh haustorial formation rate. One-week-old barley leaves (P01) were bombarded with EV or CSEP0027 construct plus GUS reporter construct, and inoculated with compatible isolate BghA6. Bgh haustorium was microscopically scored, and haustorium index (HI%) was calculated as the number of cells containing haustorium in glucuronidase (GUS) expression cells divided by the total number of GUS expression cells with germinated Bgh spores and an attached appressorium. Data show the average values and SD are from three representative experiments. (B) Silencing of CSEP0027 by host induced gene silencing (HIGS) reduces Bgh haustorial formation rate. One-week-old barley leaves (P01) were bombarded with indicated construct plus GUS reporter construct. The bombarded leaves were inoculated with the virulent isolate BghA6 at 48 h after bombardment. Bgh haustorium index was microscopically scored at 48 hpi, and the relative Bgh haustorium index in silencing experiment was standardized to EV (pIKP007) control, which was arbitrarily set to 100%. Data show the average values and SD from three representative replicates. RNAi-CSEP0105 is used as a positive control. *p < 0.05 and **p < 0.01; show significant difference by Student’s t test. (C) Expression pattern of CSEP0027 at early stages of Bgh infection. The barley P01 was inoculated with the virulent isolate BghA6. Total RNA was isolated from Bgh-infected barley leaves at 0, 3, 6, 12, 24, and 48 hpi for quantitative real-time PCR (qRT-PCR) analysis. H denotes leaf samples containing haustorium, and E denotes epiphytic Bgh tissues removed from the leaf surface. Relative expression was determined by comparing with time point 0 hpi, arbitrarily set to 1. Bgh glyceraldehyde 3-phosphate dehydrogenase was used as the reference gene. Error bars indicate SD of three replicates. The experiments were repeated two times with similar results.
The expression of many predicted or functionally confirmed CSEP genes is induced during barley infection (Godfrey et al., 2009; Spanu et al., 2010; Pedersen et al., 2012; Hacquard et al., 2013; Schmidt et al., 2014). To further analyze the expression pattern of CSEP0027, we conducted a time course experiment (Figure 3C). The transcript level of CSEP0027 remained low from 0 to12 hpi and was markedly induced at 24 and 48 hpi in both the haustorial containing samples (H) and epiphytic structures (E), with highly enriched transcripts in H sample but not in E sample at 48 hpi (Figure 3C). This expression pattern supports CSEP0027 functioning during barley infection and likely at the post-penetration stages.
To identify host targets of CSEP0027, we performed a Y2H screening of a cDNA prey library derived from Bgh infected barley leaves. Using a bait of CSEP0027 without the SP, we identified two independent clones harboring the fragments of a barley catalase gene, HvCAT1. The targeted Y2H analysis showed that CSEP0027 interacted with full-length HvCAT1 but not with HvCAT2 (Figure 4A), another reported barley catalase that shares more than 70% amino acid identity with HvCAT1 (Supplementary Figure 4; Skadsen et al., 1995). Further interaction analysis indicated that HvCAT1 interacts with CSEP0027 likely through the N-terminal catalase domain but not the C-terminal domain (Supplementary Figure 5).
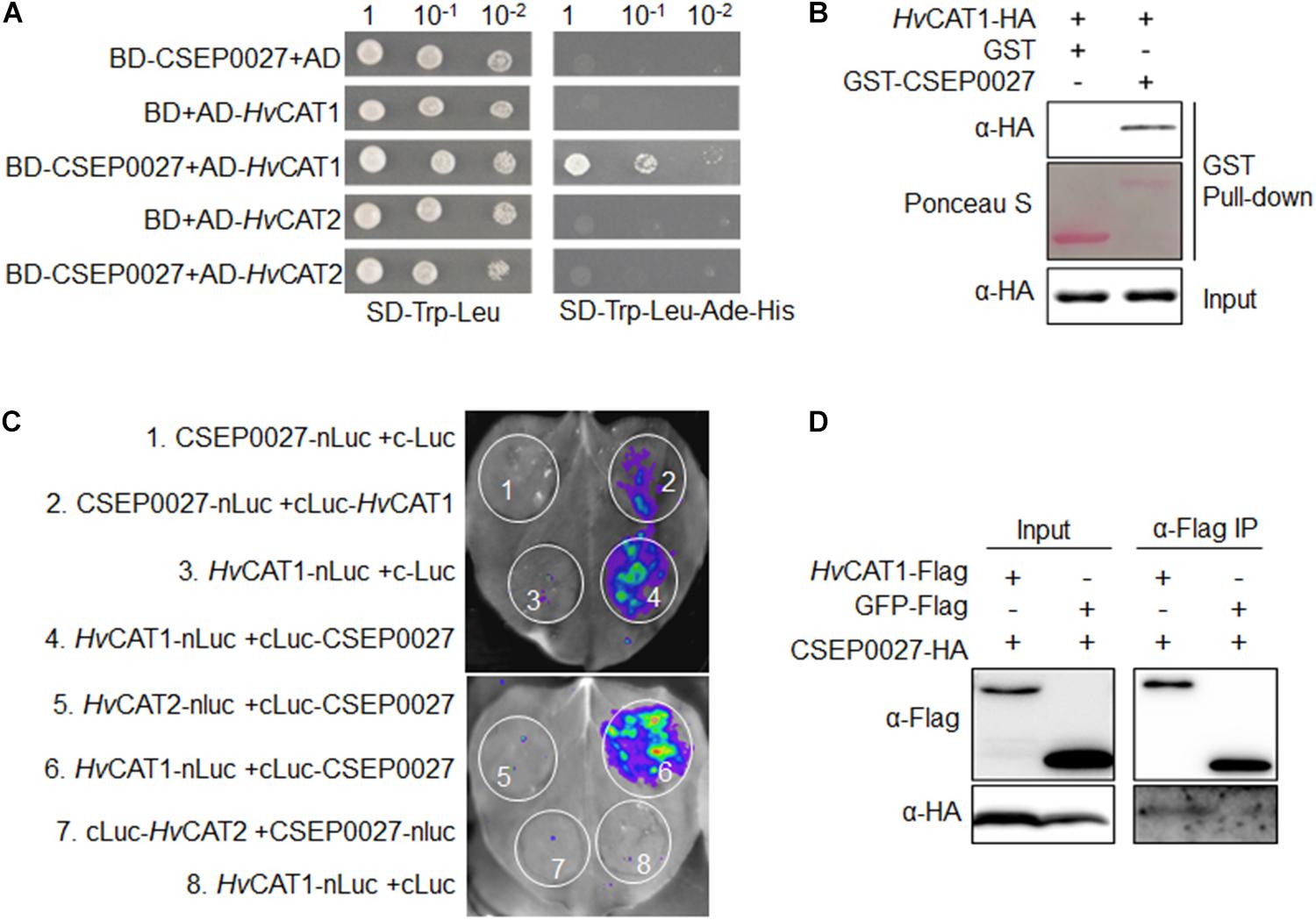
Figure 4. CSEP0027 specifically interacts with barley catalase HvCAT1. (A) Yeast two-hybrid (Y2H) assay shows CSEP0027-HvCAT1 interaction. Yeast was transformed with indicated bait and prey constructs. Serial dilutions from cell suspension of yeast expressing bait and prey constructs are shown. Growth on SD-Trp-Leu plates indicates yeast clones carrying the bait and prey constructs. The interactions were detected as yeast growth on SD-Trp-Leu-Ade-His plates. (B) Glutathione S-transferase (GST) pull-down assay confirms CSEP0027-HvCAT1 interaction. HvCAT1-HA was extracted from N. benthamiana leaves at 2 dpi, while GST-CSEP0027 and GST alone were purified from E. coli. GST pull-down fractions were detected by immunoblotting using anti-HA antibody and by Ponceau staining. (C) LCI assay confirms CSEP0027-HvCAT1 interaction. The N- or C- terminal fragment of LUC (nLuc or cLuc) was fused with indicated proteins. Indicated fusion pairs were coexpressed in N. benthamiana by agroinfiltration. The luminescent signal was collected at 48 hpi with a charge-coupled device (CCD) imaging apparatus. (D) Co-immunoprecipiation (Co-IP) analysis validates CSEP0027 and HvCAT1 interaction. HvCAT1-Flag or GFP-Flag was transiently coexpressed with CSEP0027-HA in N. benthamiana. The crude proteins were extracted at 48 hpi and subjected to Co-IP analysis.
The interaction between CSEP0027 and HvCAT1 was further verified by in vitro and in vivo assays (Figures 4B–D). For glutathione S-transferase (GST) pull-down assay, GST-CSEP0027 fusion or GST alone derived from E. coli was incubated with HvCAT1-HA containing crude lysate of N. benthamiana. An immunoblotting analysis indicated that GST-CSEP0027 pulled down HvCAT1-HA whereas GST did not (Figure 4B). In luciferase complementation imaging (LCI) assays, CSEP0027-nLuc interacted with cLuc-HvCAT1, thus generated luminescence signal, the reciprocal pair HvCAT1-nLuc and cLuc-CSEP0027 also generated strong luminescence signal in N. benthamiana (Figures 2, 4), while two pairs of negative control did not produce any detectable signal (Figures 1, 3, 4C). In addition, the HvCAT2-nLuc and cLuc-CSEP0027 did not generate detectable signal (Figures 4C, 5). In co-immunoprecipitation (co-IP) analysis, the HvCAT1-Flag fusion did immuno-precipitate with CSEP0027-HA in N. benthamiana, whereas GFP-Flag did not (Figure 4D).
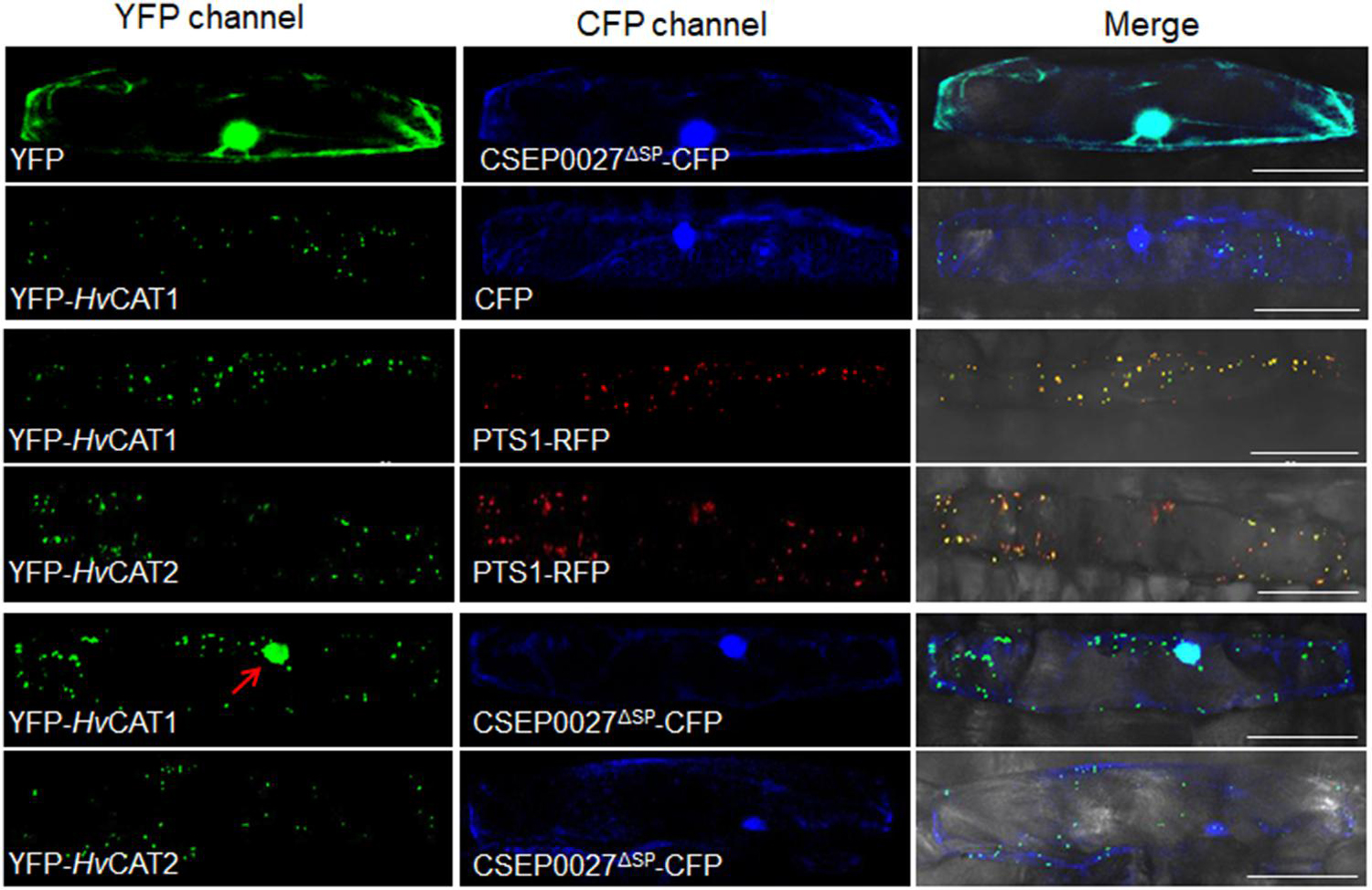
Figure 5. CSEP0027 affects the subcellular localization of HvCAT1. One-week-old barley leaves (P01) were bombarded with combination of indicated constructs coexpressing YFP-HvCAT1/PTS1-RFP, YFP-HvCAT2/PTS1-RFP, YFP-HvCAT1/CSEP0027-CFP, or YFP-HvCAT2/CSEP0027-CFP, respectively. Photographs were taken at 2 days after bombardment using a Nikon confocal laser scanning microscope. Bar = 50 μm.
Together, these results indicate that CSEP0027 specifically interacts with barley HvCAT1.
Since CSEP0027 interacts with HvCAT1, we examined the subcellular localization of CSEP0027 and catalases in barley cells. The plasmids expressing CSEP0027ΔSP-CFP (Cyan Fluorescent Protein), YFP (Yellow Fluorescent Protein)-HvCAT1, and YFP-HvCAT2 fusions were constructed and delivered into barley cells by particle bombardment. Confocal imaging indicated that CSEP0027ΔSP-CFP was localized in both cytosol and nucleus, similar to YFP alone (Figure 5, the top panels), while YFP-HvCAT1 was localized in many small dots in the cytoplasm, totally different from that of CFP alone (Figure 5, 2nd panels). Since many plant catalases are localized to peroxisomes, we tested the localization of YFP-HvCAT1 in peroxisomes by coexpression of YFP-HvCAT1 with a peroxisomal marker, PST1-RFP (Red Fluorescent Protein). As expected, YFP-HvCAT1 was almost fully co-localized with PST1-RFP in many cytoplasmic foci in the same cells (Figure 5, 3rd panels). Interestingly, YFP-HvCAT2 was also co-localized with PST1-RFP in most of the cytoplasmic dots (Figure 5, 4th panels). Next, we tested the localization of CSEP0027ΔSP-CFP and YFP-HvCAT1 in barley cells by coexpression analysis. Remarkably, confocal imaging indicated that YFP-HvCAT1 was detected not only in the peroxisomal dots but also in the nucleus, and CSEP0027ΔSP-CFP appeared to co-localize with YFP-HvCAT1 in some of the cytoplasmic dots but fully overlapped with YFP-HvCAT1 in the nucleus (Figure 5, 5th panels). Interestingly, when YFP-HvCAT2 was coexpressed with CSEP0027ΔSP-CFP in barley cells, YFP-HvCAT2 remained to localize in the peroxisomal dots and some dots appeared to overlap with CSEP0027ΔSP-CFP in the cytoplasm (Figure 5, the bottom panels). These localization analyses suggest that HvCAT1 and CSEP0027 have overlapped subcellular localization in the cytosol and CSEP0027 specifically induces the nuclear localization of HvCAT1.
The plant catalases play an important role in biotic stress responses by regulating ROS signaling and homeostasis (Du et al., 2008; Chaouch et al., 2010; Sharma and Ahmad, 2014). To evaluate the function of HvCAT1 in barley immunity, we knocked down the HvCAT1 expression through barley stripe mosaic virus vector (BSMV)-mediated virus-induced gene silencing (VIGS) approach followed by the inoculation of a compatible Bgh isolate. An antisense fragment of HvCAT1 used efficiently silenced HvCAT1 but not HvCAT2 (Figure 6A and Supplementary Figure 3). Scoring of Bgh microcolony formation rate (i.e., microcolony index, MI%) in barley leaf cells at 60–72 hpi indicated that the relative MI% increased by ∼30% in the HvCAT1-silenced leaves as compared to the EV control (Figure 6B). Staining of the Bgh infected barley leaves showed more microcolonies and better hyphae growth on the leaf surface of HvCAT1-silenced barley, as compared with the EV control (Figure 6C). Furthermore, transiently-induced gene silencing (TIGS) technique was used to silence HvCAT1 in barley leaf epidermal cells (Himmelbach et al., 2007; Bai et al., 2012). The RNAi-HvCAT1 construct was delivered into the barley cells by particle bombardment followed by Bgh spores inoculation. Relative haustorium formation rate (i.e., relative haustorium index, HI%) scored at 48 hpi also significantly increased by ∼50% as compared with the EV control (Figure 6D). By contrast, TIGS-silencing of the barley Mlo, a gene required for full susceptibility to Bgh (Kusch and Panstruga, 2017), drastically reduced Bgh HI% in barley cells by ∼80% (Figure 6D). Together, these data indicate that HvCAT1 is involved in barley immunity against Bgh.
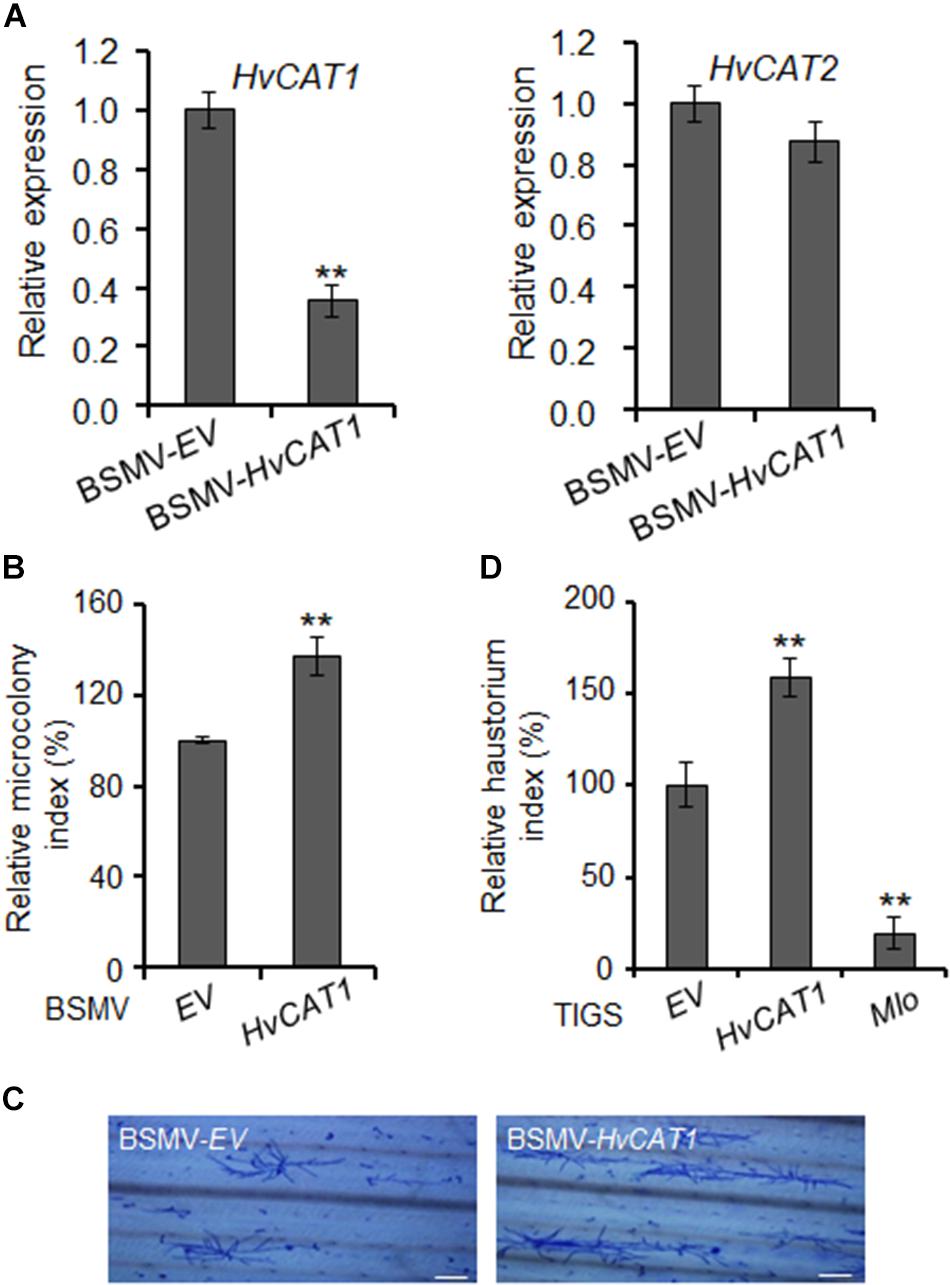
Figure 6. HvCAT1 is involved in barley immunity. (A) Barley stripe mosaic virus (BSMV)-HvCAT1 specifically silenced the expression of HvCAT1 but not HvCAT2. HvCAT1 (left) and HvCAT2 (right) gene expression levels were determined by qRT-PCR. (B) BSMV-VIGS of HvCAT1 affected barley immunity to Bgh. Bgh microcolony index was scored upon silencing of HvCAT1 in barley leaves by using BSMV-HvCAT1 after inoculation with a compatible isolate BghA6, and standardized to the BSMV-EV control that was reset to 100%. At least 1,000 interacting sites were microscopically evaluated in one experiment. (C) Representative pictures of Bgh microcolony and hyphae growth on barley leaf surface in BMSV-VIGS assays. Bar = 200 μm. (D) Transiently-induced gene silencing (TIGS) silencing of HvCAT1 affected barley immunity to Bgh. Indicated RNAi construct was bombarded into barley epidermal cells, and Bgh haustorium index was scored and standardized to that of EV control. RNAi-Mlo construct was used as a control for silencing of Mlo that caused dramatic reduction of HI%. The data were presented as average ± SD from three representative replicates. *p < 0.05; **p < 0.01, show significant difference by Student’s t test.
The genomes of many filamentous plant pathogens interacting biotrophically with plants encode hundreds of predicted effectors, and yet loss of function of some individual effectors can have measurable effect on fungal virulence. B. graminis fungi also encode several hundreds of CSEPs, and it is expected that many of the CSEPs contribute to the obligate biotrophy life style of the B. graminis fungi, for example, co-survival with the host cells or tissues. It is thus of particular interests to understand the functions and mechanisms of CSEPs in regulating host immune responses and cell-death related processes. Here, we identify Bgh CSEP0027 that triggers cell death when heterologously expressed in N. benthamiana. Importantly, CSEP0027 promotes fungal virulence in barley and interacts with HvCAT1 that is involved in host immunity, most likely, in the maintenance of ROS homeostasis in host cells. In this study, the primary aim in ectopically expressing the Bgh CSEPs in N. benthamiana is to identify those who may have cell-death related functions, either suppressing or inducing cell death, hoping to better understand the biotrophic lifestyle of the Bgh fungus. Indeed, we have predominately identified CSEPs suppressing cell death in N. benthamiana (Li et al., 2021), but unexpectedly, CSEP0027 triggering cell death as shown in the present study. We speculate that this cell death activity of CSEP0027 and related pathway may not be fully conserved in dicots and monocots. For example, the co-receptors BAK1 and SOBIR1 are important immune signaling components required for PTI and cell death in dicots (Liu et al., 2016; van der Burgh et al., 2019), while whether the co-receptors are also required for CSEP0027-induced cell death in N. benthamiana is not yet resolved here, and importantly, whether the signaling pathway for CSEP0027 induced cell death is shared between N. benthamiana and barley awaits for further investigation. Nevertheless, this study findings suggest that B. graminis fungi may utilize the CSEPs to target host catalase, a likely component of host ROS networks, presumably to manipulate the ROS homeostasis and signaling for the benefit of the pathogens.
The well-established HIGS technique has been used for identifying Bgh CSEPs with functions in promoting fungal virulence (Nowara et al., 2010). So far, a few dozens of Bgh CSEPs have been shown to contribute to Bgh pathogenicity (Nowara et al., 2010; Zhang et al., 2012, 2019; Pliego et al., 2013; Aguilar et al., 2015; Ahmed et al., 2015, 2016; Pennington et al., 2019; Li et al., 2021). In the present study, HIGS of CSEP0027 led to the reduction of HI% by ∼37% in the infected barley cells. Together with the transient overexpression results, our data support the role of CSEP0027 in promoting fungal virulence during barley infection. Our data also suggest that CSEP0027 is most likely a cytoplasmic effector and HvCAT1 is one of its virulence targets. By affecting the subcellular localization of HvCAT1, CSEP0027 may facilitate Bgh infection of host barley.
Bgh CSEP genes are usually induced and/or differentially expressed during the infection of barley. Some CSEP genes are induced at early stages of barley infection, for example, from ∼6 to 12 hpi, whereas others are induced at later stages from 24 to 48 hpi (Godfrey et al., 2009; Zhang et al., 2012; Hackenberg et al., 2013; Schmidt et al., 2014; Aguilar et al., 2015; Ahmed et al., 2015, 2016). CSEP0027 is induced from 24 to 48 hpi and is enriched in fungal haustoria (Figure 3B). We thus believe CSEP0027 functions at later stages of infection, most likely during and after haustorial formation.
Reactive oxygen species, as major regulatory and signaling molecules, can be generated in different subcellular compartments of plant cells and are regulated by an array of antioxidant systems (Waszczak et al., 2018). During plant–fungus interaction, one of the early events in plant response to fungal penetration is an oxidative burst in the apoplastic space, generated mainly by the phagocyte respiratory burst oxidase homologous nicotinamide adenine dinucleotide phosphate (NADPH) oxidases, cell wall peroxidases, and oxalate oxidases (Hückelhoven, 2007; Lehmann et al., 2015). In barley/wheat and B. graminis interactions, H2O2 and some other ROS molecules are generated in plant cells during the early stages of fungal penetration, participating in the cell wall lignification and apposition (Zhang et al., 1995; Thordal-Christensen et al., 1997; Hückelhoven et al., 1999; Hückelhoven et al., 2001, 2003; Hückelhoven, 2007; Schweizer, 2008; Li et al., 2015). Interestingly, Bgh fungus also secrets an extracellular catalase that may function in H2O2 scavenging in the apoplastic space of host cells (Zhang et al., 2004). The catalases have been known as a class of ROS scavenging enzymes catalyzing the conversion of H2O2 into H2O and O2, thereby regulating the homeostasis of the intracellular ROS level (Mhamdi et al., 2010; Sharma and Ahmad, 2014). ROS homeostasis is maintained in a very complex manner, involving different ROS-scavenging enzymes, such as catalases, ascorbate peroxidases, glutathione, superoxide dismutases (Mittler et al., 2004; Torres et al., 2006). The peroxisomal ROS levels are closely regulated by CAT activity, and in Arabidopsis, the primary peroxisomal H2O2 scavenger is CAT2 (Mhamdi et al., 2012). Here, we show that barley HvCAT1 and HvCAT2 are also peroxisomal catalases. It is unclear whether and when these two HvCAT1 and HvCAT2 are involved in the H2O2 decomposition and signaling in peroxisomes during barley interaction with Bgh fungus. Since CSEP0027 expression is induced and most likely functions post haustorium formation, we speculate that HvCAT1 may play a role in regulating ROS homeostasis at later stages, e.g., during and post haustorium formation. Our preliminary data suggest that CSEP0027 triggered-cell death involves H2O2 accumulation in N. benthamiana, however, it is not clear if the expression CSEP0027 induced disturbance of ROS homeostasis thus cell death, or vice versa. On the other hand, it is also not yet clear if CSEP0027 has activity in cell death during barley interaction with Bgh fungus. Undoubtedly, more data are needed for fully understanding the role of CSEP0027 in interacting with barley catalases and in regulating ROS homeostasis during barley interaction with Bgh fungus, particularly, the cell death signaling pathway that might be a primary target of the biotrophic fungal pathogen.
The current data are in line with the notion that ROS, in particular cellular H2O2, may play an important role in the barley interactions with the B. graminis fungi. It is not unexpected that peroxisomal ROS signaling/homeostasis and ROS signaling cross-talk among the organelles are integral and important parts of barley defense responses to the biotrophic Bgh fungal pathogen.
Apart from being regulated at transcriptional level, plant catalases are also regulated at post-translational level (Mhamdi et al., 2010). A variety of plant proteins have been reported to affect the activity and stability of plant catalases (Yang and Poovaiah, 2002; Fukamatsu et al., 2003; Verslues et al., 2007; Li et al., 2013, 2015; Zou et al., 2015; Kneeshaw et al., 2017). In addition, some pathogen secreted proteins are also identified to interact with the plant catalases and affect their activity, stability, and subcellular localization (Inaba et al., 2011; Mathioudakis et al., 2013; Zhang et al., 2015; Murota et al., 2017; Sun et al., 2017). In line with these examples, the current study data provide new evidence that biotrophic fungal pathogen also secretes an effector to target and affect host catalase subcellular localization in plants.
The plant catalases are mostly peroxisomal proteins and imported into the peroxisome matrix via the peroxisomal targeting signal 1 (PTS1) pathway, i.e., relying on the C-terminal tripeptide PTS1 signal to interact with a peroxisomal receptor and translocate into the peroxisome (Gatto et al., 2000; Lanyon-Hogg et al., 2010). Barley HvCAT1 and HvCAT2, each contains a typical PTS1 signal, PNM or PSM, respectively (Supplementary Figure 4; Mhamdi et al., 2012), and both are localized to the peroxisomes of barley cells in a transient expression analysis (Figure 5). Although different mechanisms may account for the specific re-localization of HvCAT1 upon co-expression with CSEP0027, one scenario can be that CSEP0027 interacts with HvCAT1 but not HvCAT2 in the cytoplasm thus interferes with the interaction of PTS1 signal of HvCAT1 with the peroxisomal receptor. However, how HvCAT1 is specifically regulated by CSEP0027 is not yet clear. Further investigation of the subcellular localization, trafficking, and post-translational modification of HvCAT1 will help to better understand the functions of the catalase and the virulence strategies of the biotrophic fungus.
Barley (Hordeum vulgare L.) cultivars (cv) in this study include Golden Promise and “P01” (isogenic line from cv Pallas containing Mla1). Barley seedlings were grown in a growth chamber at 20°C with 16 h light and 8 h dark cycles. N. benthamiana plants were grown in greenhouse at 24 ± 1°C with a long-day cycle (16 h light/8 h dark).
The barley powdery mildew (B. graminis f.sp. hordei [Bgh]) isolates A6 (AvrMla6, AvrMla10, and virMla1) and K1 (AvrMla1, virMla6, and virMla10) used in this study were maintained on Golden Promise.
Total RNA was extracted from P01 barley leaves inoculated with Bgh isolate A6 using Trizol solution (Invitrogen; 15596-026) and the cDNA was synthesized using reverse transcriptase M-MLV (Invitrogen; C28025). Candidate CSEP sequences excluding the signal peptide (ΔSP) were amplified using the specific primer pairs (Supplementary Table 2) and subcloned into pGR107 vector through restriction enzyme digestion and ligation for agroinfiltration in N. benthamiana (Wang et al., 2011), all candidates confirmed by sequencing.
The analysis of CSEP0027 expression profile was performed as previously described (Ahmed et al., 2015). In brief, total RNA was isolated from P01 leaves at 0, 3, 6, 12, 24, and 48 hpi inoculated with virulent isolate A6. The epiphytic Bgh tissues and the remaining leaf tissues containing Bgh haustoria were separately collected at 24 and 48 hpi. The epiphytic tissues were collected from leaf surfaces by dipping the Bgh-infected leaves into 10% cellulose acetate according to previously described (Ahmed et al., 2015). A quantitative real-time PCR (qRT-PCR) was performed on Applied Biosystems step-one real time PCR system with indicated primers (Supplementary Table 2). Bgh glyceraldehyde 3-phosphate dehydrogenase was used as the reference gene. The statistical significance was evaluated by Student’s t test. The assays were repeated two times with three replicates each time.
The yeast invertase secretion was previously described (Gu et al., 2011). Briefly, the predicted SP sequence of CSEP0027 and Avr1b, or the first 25 amino acids of Magnaporthe oryzae Mg87 were fused in frame with the yeast invertase lacking its own SP in the vector pSUC2. The pSUC2-derived constructs were transformed into the invertase secretion-deficient yeast strain YTK12, and yeasts were then placed on CMD-W medium (0.67% yeast N base without amino acids, 0.075% tryptophan dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar). The positive yeast clones were transferred onto YPRAA medium (1% yeast extract, 2% peptone, 2% raffinose, 2 μg L–1 antimycin, and 2% agar) for growth testing. Invertase activity was also detected by monitoring conversion of TTC to the insoluble red-colored triphenylformazan.
Single-cell transient gene expression assay was carried out as previously described (Bai et al., 2012). Briefly, a construct expressing a gene-of-interest was bombarded in barley leaf epidermal cells together with a vector expressing ß-glucuronidase (GUS) reporter. The leaves were inoculated with a compatible Bgh isolate at 4 h after the bombardment and then stained with GUS staining solution at 48 hpi. The fungal haustorium index was scored as previously described in the barley leaves after inoculated with Bgh spores. The statistical significance was evaluated by Student’s t test with data from three replicate experiments that have been repeated for three times.
For transient gene silencing assay, the specific gene fragments were cloned into pIPK007 to form a hairpin structure and expression driven by 35S promoter as previously described (Himmelbach et al., 2007). The remaining steps were the same as the transient gene expression assay, except that leaves were inoculated with Bgh isolates at 48 h after bombardment.
Yeast two-hybrid screening was performed according to the protocols of the manufacturer (Clontech; PT4048-1). In total, 5 × 107 transformants were screened. In brief, yeast strain Y2HGold expressing pGBKT7-CSEP0027 (ΔSP) was used for mating with yeast strain Y187 harboring a cDNA prey library derived from Bgh-infected barley leaves and placed onto SD-Leu-Trp-His-Ade plates at 30°C. After 35 days, the resistant clones were selected for further verification.
For Y2H assay, the corresponding bait and prey vectors were co-transformed into yeast strain Y2HGold and plated onto SD-Leu-Trp plates. The positive interactions were detected by placing the strains onto SD-Leu-Trp-His-Ade plates at 30°C.
Luciferase complementation imaging assays were performed according to previously described by Chen et al. (2008). Briefly, the coding region of CSEP0027 (ΔSP) and HvCAT1 were subcloned into vectors pCAMBIA-Cluc or pCAMBIA-Nluc, respectively, to generate constructs for expressing Cluc-CSEP0027 and Cluc-HvCAT1, or CSEP0027-Nluc and HvCAT1-Nluc. The NLuc-/CLuc-derivative constructs were transformed into the A. tumefaciens strain GV3101. The overnight agrobacteria cultures were resuspended with infiltration buffer (2% sucrose, 0.5% MS, 100 μM acetosyringone, and 10 mM MES) into OD600 = 1.0. Equal volume of agrobacteria resuspension carrying the nLUC and cLUC derivative constructs were mixed and co-infiltrated into the N. benthamiana leaves. The infiltrated area was examined for the luciferase activity 40–50 h post agroinfiltration with a cooled charge-coupled device (CCD) imaging apparatus. For each pair of constructs, at least 10 leaves were co-infiltrated in one experiment, and three independent replicates were conducted.
Pull-down assays were performed according to previously described with some modifications (Chang et al., 2013). Briefly, 500 ng of GST-CSEP0027 and GST proteins purified from Escherichia coli were incubated with 150 μl of Glutathione Sepharose 4B beads for 1 h at 4°C, then, beads were sealed with 100 μg BSA for 1 h and incubated with 1.0 g crude protein extracted from N. benthamiana leaves expressing HvCAT1-HA. After incubation for 2 h, the beads were washed five times with RB buffer, then resuspended with 30 μl of 2 × Laemmli buffer, and loaded for sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-HA antibody. GST-CSEP0027 and GST proteins were detected by Ponceau staining.
For Co-IP assay, the total proteins extracted from N. benthamiana coexpressing GFP-Flag/CSEP0027-HA or HvCAT1-Flag/CSEP0027-HA were incubated with anti-FLAG antibody-coupled beads for 2 h, then washed five times with extraction buffer, proteins were further eluted from the beads using 0.5 mg ml–1 3 × Flag peptide and used for immunoblotting with anti-HA antibody, or anti-Flag antibody.
For subcellular localization analysis, the coding sequences of CSEP0027ΔSP, HvCAT1 and HvCAT2 were subcloned into vectors pUBI-mYFP-GW and pUBI-GW-CFP to generate pUBI-CSEP0027 ΔSP-CFP, pUBI-mYFP-HvCAT1, and pUBI-mYFP-HvCAT2 constructs. A pair of constructs was delivered into barley leaf epidermal cells by the particle bombardment for coexpression of the indicated fusion proteins, and confocal imaging was conducted at 48 h post-particle delivery. Laser illumination was set at 405 nm for CFP, 488 nm for YFP, and 561 nm for RFP using a Nikon confocal microscope. This assay was repeated three independent times and at least 20 cells were examined for each coexpression.
Barley stripe mosaic virus-mediated gene silencing in barley was performed as previously described (Yuan et al., 2011). Briefly, an antisense fragment of HvCAT1 was cloned into the pCaBS-γbLIC vector to create pCaBS-γb-HvCAT1 construct with indicated primers (Supplementary Table 2). pCaBS-α, pCaBS-β, and pCaBS-γb-HvCAT1 constructs were transformed into A. tumefaciens strain EHA105, respectively. The agrobacteria were resuspended in infiltration buffer to OD600 = 1.0 and mixed at 1:1:1 ratio to infiltrate N. benthamiana. After 12 days, N. benthamiana leaf sap was extracted to inoculate 10-day-old barley leaves. After 2–3 weeks of inoculation, the newly emerged leaves with virus caused symptoms were used for Bgh infection, and microcolony scoring was done at ∼60–72 hpi. For each treatment, at least four barley leaves were chosen for analysis, and three independent replicates were conducted. The statistical significance was evaluated by Student’s t test.
Agrobacterium tumefaciens-mediated transient gene expression in N. benthamiana assays were performed as described previously (Wang et al., 2011). A. tumefaciens strain GV3101 was transformed with indicated constructs. Agrobacteria were cultured overnight at 28°C, at 200 rpm, then resuspended in 10 mM MgCl2 to a final OD600 = 0.5 and infiltrated into 4-week-old N. benthamiana leaves. The cell death symptoms were photographed at 5 days post-infiltration. For trypan blue staining, the leaves were boiled in a 1:1 mixture of ethanol and staining solution for 5 min as described before (Bai et al., 2012). The leaves were de-stained with chloral hydrate solution (2.5 g ml–1) for 2 days.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
HY and Q-HS designed the research. HY, CJ, and HP performed the experiments with helps from XL, JL, and WH. HY, CJ, LZ, XL, RF, and Q-HS, analyzed the data. WL and RF provided the reagents. Q-HS, HY, and LZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2016YFD0100602 and 2018YFD1000703), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB16), the Ministry of Agriculture and Rural Affairs of China (2016ZX08009-003-001), and the National Natural Science Foundation of China (31530061).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Yuanchao Wang, Daolong Dou from the Nanjing Agricultural University, and Wenxian Sun from the Chinese Agricultural University for providing vectors. We thank Hans Thordal-Christensen from the University of Copenhagen for critical reading of an initial version of the manuscript and suggestions, and constructive comments from various reviewers.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.733237/full#supplementary-material
Supplementary Figure 1 | The MLA13/AVRa13 triggers cell death in N. benthamiana. Expression of MLA13/AVRa13 and GFP in N. benthamiana. The experimental procedure used here was the same as that in Figure 1A.
Supplementary Figure 2 | CSEP0027 induces H2O2 production in N. benthamiana. DAB staining was performed at 2 days after infiltration to detect H2O2 accumulation in the areas infiltrated with agrobacteria transformed with a corresponding construct.
Supplementary Figure 3 | Bgt homologs of CSEP0027 do not trigger cell death in N. benthamiana. (A) Protein sequence alignment of CSEP0027 and its Bgt homologs. Alignment was performed using the DNAMAN software. The red box indicates signal peptides, blue box indicates the Y/FxC motif, and green box indicates a C-terminal conserved cysteine. (B) Expression of CSEP0027 and its Bgt homologs in N. benthamiana. The experimental procedure used here was the same as that in Figure 1A.
Supplementary Figure 4 | Sequence alignment of amino acids of HvCAT1 and HvCAT2.
Supplementary Figure 5 | CSEP0027 N-terminus interacts with HvCAT1 in yeast. (A) Schematic diagram of HvCAT1 constructs, and HvCAT1-NT (1–401) and HvCAT1-CT (402–492) used in the Y2H assay. Catalase core domain and catalase-related immune responsive domain (catalase-rel) are indicated. (B) Y2H analysis of the interaction between NT- or CT-fragments of HvCAT1 and CSEP0027. Yeast was transformed with the indicated bait and prey constructs. Serial dilutions from cell suspension of yeast expressing bait and prey constructs are shown. Growth on SD-Leu-Trp plates indicates yeast clone carrying bait and prey constructs. Interactions were detected on SD-Leu-Trp-His-Ade plates.
Aguilar, G. B., Pedersen, C., and Thordal-Christensen, H. (2015). Identification of eight effector candidate genes involved in early aggressiveness of the barley powdery mildew fungus. Plant Pathol. 65, 953–958. doi: 10.1111/ppa.12476
Ahmed, A. A., Pedersen, C., Schultz-Larsen, T., Kwaaitaal, M., Jorgensen, H. J., and Thordal-Christensen, H. (2015). The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 168, 321–333. doi: 10.1104/pp.15.00278
Ahmed, A. A., Pedersen, C., and Thordal-Christensen, H. (2016). The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS One 11:e0157586. doi: 10.1371/journal.pone.0157586
Altpeter, F., Varshney, A., Abderhalden, O., Douchkov, D., Sautter, C., Kumlehn, J., et al. (2005). Stable expression of a defense-related gene in wheat epidermis under transcriptional control of a novel promoter confers pathogen resistance. Plant Mol. Biol. 57, 271–283. doi: 10.1007/s11103-004-7564-7
Apel, K., and Hirt, H. (2004). REACTIVE OXYGEN SPECIES: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Bauer, S., Yu, D., Lawson, A. W., Saur, I. M. L., Frantzeskakis, L., Kracher, B., et al. (2021). The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold. PLoS Pathog. 17:e1009223. doi: 10.1371/journal.ppat.1009223
Bindschedler, L. V., Burgis, T. A., Mills, D. J., Ho, J. T., Cramer, R., and Spanu, P. D. (2009). In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol. Cell Proteomics 8, 2368–2381. doi: 10.1074/mcp.M900188-MCP200
Bindschedler, L. V., Panstruga, R., and Spanu, P. D. (2016). Mildew-omics: how global analyses aid the understanding of life and evolution of powdery mildews. Front. Plant Sci. 7:123. doi: 10.3389/fpls.2016.00123
Both, M., Csukai, M., Stumpf, M. P., and Spanu, P. D. (2005). Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell. 17, 2107–2122. doi: 10.1105/tpc.105.032631
Chang, C., Yu, D., Jiao, J., Jing, S., Schulze-Lefert, P., and Shen, Q. H. (2013). Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25, 1158–1173. doi: 10.1105/tpc.113.109942
Chang, X. L., Luo, L. Y., Liang, Y. P., Hu, Y. T., Luo, P. G., Gong, G. S., et al. (2019). Papilla formation, defense gene expression and HR contribute to the powdery mildew resistance of the novel wheat line L699 carrying Pm40 gene. Physiol. Mol. Plant Pathol. 106, 208–216. doi: 10.1016/j.pmpp.2019.02.009
Chaouch, S., Queval, G., Vanderauwera, S., Mhamdi, A., Vandorpe, M., Langlois-Meurinne, M., et al. (2010). Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153, 1692–1705. doi: 10.1104/pp.110.153957
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Chen, L., Wu, R., Feng, J., Feng, T., Wang, C., Hu, J., et al. (2020). Transnitrosylation mediated by the non-canonical catalase ROG1 regulates nitric oxide signaling in plants. Dev. Cell 53, e445. doi: 10.1016/j.devcel.2020.03.020
Chen, Y. R., and Jarosz, D. F. (2020). Both ROSy and grim: the landscape of protein redox during aging. Cell Metab. 31, 662–663. doi: 10.1016/j.cmet.2020.03.016
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Du, Y.-Y., Wang, P.-C., Chen, J., and Song, C.-P. (2008). Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 50, 1318–1326. doi: 10.1111/j.1744-7909.2008.00741.x
Frantzeskakis, L., Kracher, B., Kusch, S., Yoshikawa-Maekawa, M., Bauer, S., Pedersen, C., et al. (2018). Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics 19:381. doi: 10.1186/s12864-018-4750-6
Fukamatsu, Y., Yabe, N., and Hasunuma, K. (2003). Arabidopsis NDK1 is a component of ROS signaling by interacting with three catalases. Plant Cell Physiol. 44, 982–989. doi: 10.1093/pcp/pcg140
Gatto, G. J. Jr., Geisbrecht, B. V., Gould, S. J., and Berg, J. M. (2000). Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095. doi: 10.1038/81930
Glawe, D. A. (2008). The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 46, 27–51. doi: 10.1146/annurev.phyto.46.081407.104740
Godfrey, D., Bohlenius, H., Pedersen, C., Zhang, Z., Emmersen, J., and Thordal-Christensen, H. (2010). Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11:317. doi: 10.1186/1471-2164-11-317
Godfrey, D., Zhang, Z., Saalbach, G., and Thordal-Christensen, H. (2009). A proteomics study of barley powdery mildew haustoria. Proteomics 9, 3222–3232. doi: 10.1002/pmic.200800645
Gu, B., Kale, S. D., Wang, Q., Wang, D., Pan, Q., Cao, H., et al. (2011). Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLoS One. 6:e27217. doi: 10.1371/journal.pone.0027217
Hackenberg, T., Juul, T., Auzina, A., Gwizdz, S., Malolepszy, A., Van Der Kelen, K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25, 4616–4626. doi: 10.1105/tpc.113.117192
Hacquard, S., Kracher, B., Maekawa, T., Vernaldi, S., Schulze-Lefert, P., and Ver Loren van Themaat, E. (2013). Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl. Acad. Sci. U.S.A. 110, E2219–E2228. doi: 10.1073/pnas.1306807110
Himmelbach, A., Zierold, U., Hensel, G., Riechen, J., Douchkov, D., Schweizer, P., et al. (2007). A set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192–1200. doi: 10.1104/pp.107.111575
Hückelhoven, R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325
Hückelhoven, R., Dechert, C., and Kogel, K.-H. (2001). Non-host resistance of barley is associated with a hydrogen peroxide burst at sites of attempted penetration by wheat powdery mildew fungus. Mol. Plant Pathol. 2, 199–205. doi: 10.1046/j.1464-6722.2001.00067.x
Hückelhoven, R., Dechert, C., and Kogel, K.-H. (2003). Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. Proc. Natl. Acad. Sci. U.S.A. 100, 5555–5560. doi: 10.1073/pnas.0931464100
Hückelhoven, R., Fodor, J., Preis, C., and Kogel, K.-H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119:1251. doi: 10.1104/pp.119.4.1251
Hückelhoven, R., Fodor, J., Trujillo, M., and Kogel, K. H. (2000). Barley Mla and Rar mutants compromised in the hypersensitive cell death response against Blumeria graminis f.sp. hordei are modified in their ability to accumulate reactive oxygen intermediates at sites of fungal invasion. Planta 212, 16–24. doi: 10.1007/s004250000385
Hückelhoven, R., and Kogel, K. H. (1998). Tissue-specific superoxide generation at interaction sites in resistant and susceptible near-isogenic barley lines attacked by the powdery mildew fungus (Erysiphe graminis f. sp. hordei). Mol. Plant Microbe Interact. 11, 292–300. doi: 10.1094/MPMI.1998.11.4.292
Hückelhoven, R., and Kogel, K. H. (2003). Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance? Planta 216, 891–902. doi: 10.1007/s00425-003-0973-z
Inaba, J., Kim, B. M., Shimura, H., and Masuta, C. (2011). Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 156, 2026–2036. doi: 10.1104/pp.111.180042
Kneeshaw, S., Keyani, R., Delorme-Hinoux, V., Imrie, L., Loake, G. J., Le Bihan, T., et al. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc. Natl. Acad. Sci. U.S.A. 114:8414. doi: 10.1073/pnas.1703344114
Kusch, S., and Panstruga, R. (2017). mlo-based resistance: an apparently universal “Weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Lanyon-Hogg, T., Warriner, S. L., and Baker, A. (2010). Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol. Cell 102, 245–263. doi: 10.1042/bc20090159
Lee, S. J., Kim, B. D., and Rose, J. K. (2006). Identification of eukaryotic secreted and cell surface proteins using the yeast secretion trap screen. Nat. Protoc. 1, 2439–2447. doi: 10.1038/nprot.2006.373
Lehmann, S., Serrano, M., L’Haridon, F., Tjamos, S. E., and Metraux, J. P. (2015). Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112, 54–62. doi: 10.1016/j.phytochem.2014.08.027
Li, J., Liu, J., Wang, G., Cha, J. Y., Li, G., Chen, S., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27, 908–925. doi: 10.1105/tpc.114.135095
Li, X., Cong, J., Yuan, H., Huang, W. T., Liu, F., Fan, R. C., et al. (2021). The barley powdery mildew effectors CSEP0139 and CSEP0182 suppress cell death and promote B. graminis fungal virulence in plants. Phytopathol. Res. 3:7. doi: 10.1186/s42483-021-00084-z
Li, Y., Chen, L., Mu, J., and Zuo, J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. doi: 10.1104/pp.113.225805
Liu, Y., Huang, X., Li, M., He, P., and Zhang, Y. (2016). Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol. 212, 637–645. doi: 10.1111/nph.14072
Lu, X., Kracher, B., Saur, I. M., Bauer, S., Ellwood, S. R., Wise, R., et al. (2016). Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E6486–E6495. doi: 10.1073/pnas.1612947113
Mathioudakis, M. M., Veiga, R. S., Canto, T., Medina, V., Mossialos, D., Makris, A. M., et al. (2013). Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 14, 589–601. doi: 10.1111/mpp.12034
Menardo, F., Praz, C. R., Wicker, T., and Keller, B. (2017). Rapid turnover of effectors in grass powdery mildew (Blumeria graminis). BMC Evol. Biol. 17:223. doi: 10.1186/s12862-017-1064-2
Mhamdi, A., Noctor, G., and Baker, A. (2012). Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 525, 181–194. doi: 10.1016/j.abb.2012.04.015
Mhamdi, A., Queval, G., Chaouch, S., Vanderauwera, S., Van Breusegem, F., and Noctor, G. (2010). Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. doi: 10.1093/jxb/erq282
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Müller, M. C., Praz, C. R., Sotiropoulos, A. G., Menardo, F., Kunz, L., Schudel, S., et al. (2019). A chromosome-scale genome assembly reveals a highly dynamic effector repertoire of wheat powdery mildew. New phytol. 221, 2176–2189. doi: 10.1111/nph.15529
Murota, K., Shimura, H., Takeshita, M., and Masuta, C. (2017). Interaction between Cucumber mosaic virus 2b protein and plant catalase induces a specific necrosis in association with proteasome activity. Plant Cell Rep. 36, 37–47. doi: 10.1007/s00299-016-2055-2
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., and Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. doi: 10.1111/j.1744-7909.2010.00933.x
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Oh, S. K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21, 2928–2947. doi: 10.1105/tpc.109.068247
Panstruga, R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6, 320–326. doi: 10.1016/s1369-5266(03)00043-8
Panstruga, R., and Dodds, P. N. (2009). Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science (New York, N.Y.) 324, 748–750. doi: 10.1126/science.1171652
Pedersen, C., Ver Loren van Themaat, E., McGuffin, L. J., Abbott, J. C., Burgis, T. A., Barton, G., et al. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics. 13:694. doi: 10.1186/1471-2164-13-694
Pennington, H. G., Gheorghe, D. M., Damerum, A., Pliego, C., Spanu, P. D., Cramer, R., et al. (2016). Interactions between the powdery mildew effector BEC1054 and barley proteins identify candidate host targets. J. Proteome Res. 15, 826–839. doi: 10.1021/acs.jproteome.5b00732
Pennington, H. G., Jones, R., Kwon, S., Bonciani, G., Thieron, H., Chandler, T., et al. (2019). The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 15:e1007620. doi: 10.1371/journal.ppat.1007620
Petrov, V. D., and Van Breusegem, F. (2012). Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012:ls014. doi: 10.1093/aobpla/pls014
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Praz, C. R., Bourras, S., Zeng, F., Sanchez-Martin, J., Menardo, F., Xue, M., et al. (2017). AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 213, 1301–1314. doi: 10.1111/nph.14372
Saur, I. M., Bauer, S., Kracher, B., Lu, X., Franzeskakis, L., Muller, M. C., et al. (2019). Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. Elife 8:e44471. doi: 10.7554/eLife.44471
Schmidt, S. M., Kuhn, H., Micali, C., Liller, C., Kwaaitaal, M., and Panstruga, R. (2014). Interaction of a Blumeria graminis f. sp. hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target. Mol. Plant Pathol. 15, 535–549. doi: 10.1111/mpp.12110
Schweizer, P. (2008). Tissue-specific expression of a defence-related peroxidase in transgenic wheat potentiates cell death in pathogen-attacked leaf epidermis. Mol. Plant Pathol. 9, 45–57. doi: 10.1111/j.1364-3703.2007.00446.x
Sharma, I., and Ahmad, P. (2014). Catalase: a versatile antioxidant in plants. Oxidative Damage Plants 2014, 131–148. doi: 10.1016/b978-0-12-799963-0.00004-6
Skadsen, R. W., Schulze-Lefert, P., and Herbst, J. M. (1995). Molecular cloning, characterization and expression analysis of two catalase isozyme genes in barley. Plant Mol. Biol. 29, 1005–1014.
Spanu, P. D. (2017). Cereal immunity against powdery mildews targets RNase-Like Proteins associated with Haustoria (RALPH) effectors evolved from a common ancestral gene. New Phytol. 213, 969–971. doi: 10.1111/nph.14386
Spanu, P. D., Abbott, J. C., Amselem, J., Burgis, T. A., Soanes, D. M., Stuber, K., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science (New York, N.Y.) 330, 1543–1546. doi: 10.1126/science.1194573
Stergiopoulos, I., and de Wit, P. J. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Sun, Y., Li, P., Deng, M., Shen, D., Dai, G., Yao, N., et al. (2017). The Ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell Microbiol. 19:e12736. doi: 10.1111/cmi.12736
Takamatsu, S. (2013). Molecular phylogeny reveals phenotypic evolution of powdery mildews (Erysiphales, Ascomycota). J. General Plant Pathol. 79, 218–226. doi: 10.1007/s10327-013-0447-5
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Troch, V., Audenaert, K., Wyand, R. A., Haesaert, G., Höfte, M., and Brown, J. K. (2014). Formae speciales of cereal powdery mildew: close or distant relatives? Mol. Plant Pathol. 15, 304–314. doi: 10.1111/mpp.12093
van der Burgh, A. M., Postma, J., Robatzek, S., and Joosten, M. (2019). Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20, 410–422. doi: 10.1111/mpp.12767
Verslues, P. E., Batelli, G., Grillo, S., Agius, F., Kim, Y. S., Zhu, J., et al. (2007). Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Mol. Cell Biol. 27, 7771–7780. doi: 10.1128/mcb.00429-07
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082
Waszczak, C., Carmody, M., and Kangasjärvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69, 209–236. doi: 10.1146/annurev-arplant-042817-040322
Wicker, T., Oberhaensli, S., Parlange, F., Buchmann, J. P., Shatalina, M., Roffler, S., et al. (2013). The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. doi: 10.1038/ng.2704
Yang, T., and Poovaiah, B. W. (2002). Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc. Natl. Acad. Sci. U.S.A. 99, 4097–4102. doi: 10.1073/pnas.052564899
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6:e26468. doi: 10.1371/journal.pone.0026468
Yuan, H.-M., Liu, W.-C., and Lu, Y.-T. (2017). CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe 21, 143–155. doi: 10.1016/j.chom.2017.01.007
Zhang, M., Li, Q., Liu, T., Liu, L., Shen, D., Zhu, Y., et al. (2015). Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. doi: 10.1104/pp.114.252437
Zhang, W. J., Pedersen, C., Kwaaitaal, M., Gregersen, P. L., Morch, S. M., Hanisch, S., et al. (2012). Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x
Zhang, Y., Xu, K., Yu, D., Liu, Z., Peng, C., Li, X., et al. (2019). The highly conserved barley powdery mildew effector BEC1019 confers susceptibility to biotrophic and necrotrophic pathogens in wheat. Int. J. Mol. Sci. 20, 4376.
Zhang, Z., Collinge, D. B., and Thordal-Christensen, H. (1995). Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J. 8, 139–145. doi: 10.1046/j.1365-313X.1995.08010139.x
Zhang, Z., Henderson, C., and Gurr, S. J. (2004). Blumeria graminis secretes an extracellular catalase during infection of barley: potential role in suppression of host defence. Mol. Plant Pathol. 5, 537–547. doi: 10.1111/j.1364-3703.2004.00251.x
Zou, J.-J., Li, X.-D., Ratnasekera, D., Wang, C., Liu, W.-X., Song, L.-F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Keywords: powdery mildew, Blumeria graminis, effector, CSEP, virulence, barley catalase
Citation: Yuan H, Jin C, Pei H, Zhao L, Li X, Li J, Huang W, Fan R, Liu W and Shen Q-H (2021) The Powdery Mildew Effector CSEP0027 Interacts With Barley Catalase to Regulate Host Immunity. Front. Plant Sci. 12:733237. doi: 10.3389/fpls.2021.733237
Received: 30 June 2021; Accepted: 19 August 2021;
Published: 09 September 2021.
Edited by:
Meixiang Zhang, Nanjing Agricultural University, ChinaReviewed by:
Xiangxiu Liang, China Agricultural University, ChinaCopyright © 2021 Yuan, Jin, Pei, Zhao, Li, Li, Huang, Fan, Liu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian-Hua Shen, cWhzaGVuQGdlbmV0aWNzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.