
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 November 2021
Sec. Plant Symbiotic Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.727741
 Dao-Jun Guo1,2,3†
Dao-Jun Guo1,2,3† Dong-Ping Li4†
Dong-Ping Li4† Rajesh Kumar Singh2,3
Rajesh Kumar Singh2,3 Pratiksha Singh2,3
Pratiksha Singh2,3 Anjney Sharma2,3
Anjney Sharma2,3 Krishan K. Verma2,3
Krishan K. Verma2,3 Ying Qin1
Ying Qin1 Qaisar Khan1
Qaisar Khan1 Zhen Lu1
Zhen Lu1 Mukesh K. Malviya2,3
Mukesh K. Malviya2,3 Xiu-Peng Song2
Xiu-Peng Song2 Yong-Xiu Xing1*
Yong-Xiu Xing1* Yang-Rui Li1,2,3*
Yang-Rui Li1,2,3*Plant endophytic bacteria have many vital roles in plant growth promotion (PGP), such as nitrogen (N) fixation and resistance to biotic and abiotic stresses. In this study, the seedlings of sugarcane varieties B8 (requires a low concentration of nitrogen for growth) and GT11 (requires a high concentration of nitrogen for growth) were inoculated with endophytic diazotroph Enterobacter roggenkampii ED5, which exhibits multiple PGP traits, isolated from sugarcane roots. The results showed that the inoculation with E. roggenkampii ED5 promoted the growth of plant significantly in both sugarcane varieties. 15N detection at 60 days post-inoculation proved that the inoculation with strain ED5 increased the total nitrogen concentration in the leaf and root than control in both sugarcane varieties, which was higher in B8. Biochemical parameters and phytohormones in leaf were analyzed at 30 and 60 days after the inoculation. The results showed that the inoculation with E. roggenkampii ED5 improved the activities of superoxide dismutase (SOD), catalase (CAT), NADH-glutamate dehydrogenase (NADH-GDH), glutamine synthetase (GS), and endo-β-1,4-glucanase, and the contents of proline and indole acetic acid (IAA) in leaf, and it was generally more significant in B8 than in GT11. Tandem Mass Tags (TMT) labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to perform comparative proteomic analysis in the sugarcane leaves at 30 days after inoculation with strain ED5. A total of 27,508 proteins were detected, and 378 differentially expressed proteins (DEPs) were found in the treated sugarcane variety B8 (BE) as compared to control (BC), of which 244 were upregulated and 134 were downregulated. In contrast, a total of 177 DEPs were identified in the treated sugarcane variety GT11 (GE) as compared to control (GC), of which 103 were upregulated and 74 were downregulated. The DEPs were associated with nitrogen metabolism, photosynthesis, starch, sucrose metabolism, response to oxidative stress, hydrolase activity, oxidative phosphorylation, glutathione metabolism, phenylpropanoid metabolic process, and response to stresses in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. To the best of our knowledge, this is the first proteomic approach to investigate the molecular basis of the interaction between N-fixing endophytic strain E. roggenkampii ED5 and sugarcane.
Sugarcane (Scacharum spp. interspecific hybrids) is a major sugar and bioenergy crop worldwide (Li and Yang, 2015; Bordonal et al., 2018; Parsaee et al., 2019). The production of sugarcane requires a large practice of fertilizers, especially nitrogen (N). However, excess N fertilizer contributes to the soil, groundwater, and environmental pollution, as well as high cost for crop production (Basanta et al., 2003; Franco et al., 2011, 2015; Boschiero et al., 2020). Plant growth-promoting bacteria (PGPB) are the microorganisms that can colonize in plant tissues or rhizospheric soil and show beneficial effects on plant development, biotic or abiotic stresses, and nutrient assimilation (Esitken et al., 2010; Piromyou et al., 2011; Orozco-Mosqueda et al., 2020). PGPB promotes the growth of plant through various ways, such as the production of siderophore, solubilization of phosphate, secretion of indole acetic acid (IAA), enhancement of nutrient absorption by plants, and improvement of tolerance to phytopathogens (Lugtenberg and Kamilova, 2009). Some PGPB can convert natural molecular nitrogen into ammonia and provide it to the host plants (Reis and Teixeira, 2015; Igiehon and Babalola, 2018). It is effective to use these N-fixing bacteria to improve nitrogen nutrition in plants which is necessary for crop production, especially in sugarcane (Wei et al., 2014; Singh et al., 2017). At present, some reports are available on the isolation of N-fixing microorganisms with PGP traits from rhizospheric soil or tissues of sugarcane, such as Klebsiella variicola, Kosakonia sacchari, Streptomyces chartreusis, Pseudomonas spp., Herbaspirillum seropedicae, Stenotrophomonas maltophilia, Kosakonia radicincitans, Bacillus spp., Pantoea dispersa, and Enterobacter asburiae (Chen et al., 2014; Wang Z. et al., 2016; Li et al., 2017; Antunes et al., 2019; Singh et al., 2020, 2021b).
At present, some progress has been made in studies of PGPB-plant interaction mechanisms to enhance disease resistance, tolerance to abiotic stresses, and N-fixation in plants (Fernandez et al., 2012; Imran, 2013; Sivasakthi et al., 2014; Kumari et al., 2019). Regarding the advantage of genomics, transcriptomics, proteomics, metabolomics, and other molecular biological technologies can reveal the complete mechanism of the interactions between microorganisms and plants (Shen et al., 2013; Xie et al., 2015; Mhlongo et al., 2018). The mechanism of interaction between microorganisms and plants is widely investigated by using proteomic approaches. Recently, protein labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS) have been widely applied (Chinnasamy, 2006; Kwon et al., 2016; Naher et al., 2018; Bai et al., 2019; Alberton et al., 2020), which is highly efficient in identifying and quantifying differentially expressed proteins (DEPs) in different samples to investigate the mechanisms of salt tolerance (Jiang et al., 2017; Pan et al., 2019; Ma et al., 2020), drought resistance (Xie et al., 2016; Wang Y. et al., 2019; Asghari et al., 2020), and disease resistance (Wang J. et al., 2016; Li et al., 2017; Shi et al., 2019; Singh et al., 2021a) in plants. At present, Tandem Mass Tags (TMT) technology is one of the most powerful analytical approaches for the quantitative analysis of differential proteins in a protein profile and to explain the molecular mechanisms of different experimental treatments (Lü et al., 2016; Zhu et al., 2019; Wu et al., 2020). Earlier, this technique was used to investigate the effect of oxidative stress on the heat tolerance of Pseudomonas fluorescens (Chen et al., 2019). For different wheat varieties infected with Fusarium, a total of 366 DEPs were identified by TMT technology and the enrichment of the DEPs in phenylpropane biosynthesis, secondary metabolite biosynthesis, glutathione metabolism, signal transduction of phytohormones, mitogen-activated protein kinase (MAPK) signal transduction pathway, photosynthesis was observed, revealing the defense mechanism of wheat against Fusarium (Qiao et al., 2021).
According to Guo et al. (2020), Enterobacter roggenkampii ED5 isolated from the sugarcane roots exhibited high nitrogenase activity with multiple PGP traits, as well as enhanced sugarcane growth under greenhouse conditions. Analysis of the complete genome sequence of this isolate also confirmed the presence of multiple PGP genes in its genome, which might be linked to N-fixation and PGP in sugarcane plants (Guo et al., 2020). However, the molecular mechanisms of the interaction between E. roggenkampii ED5 and sugarcane are still unclear.
In this study, the technology of TMT labeling and LC-MS/MS was applied to investigate the comparative protein profiles of the two sugarcane varieties (i.e., B8 and GT11) inoculated with E. roggenkampii ED5 under greenhouse conditions. This study aimed to reveal (i) the response to oxidative kinase activity, (ii) the level of plant hormones, (iii) the pattern in nitrogen use, and (iv) the key proteins involved in sugarcane growth promotion after inoculation with E. roggenkampii ED5, so as to provide a reference for further study of E. roggenkampii ED5 and its application in sugarcane production. To the best of our knowledge, this is the first study to explore the molecular mechanisms of the interaction between E. roggenkampii and sugarcane using a proteomic approach.
The strain E. roggenkampii ED5 was isolated from the sugarcane roots planted at Sugarcane Germplasm Resource Nursery, Guangxi Academy of Agricultural Sciences (GXAAS), Nanning, Guangxi, China. Two sugarcane varieties with different levels of N-fixation, namely, the variety B8 (needs less nitrogen for growth) and the variety GT11 (needs more nitrogen for growth), were used in the experiment. The field soil was sterilized at 121°C, autoclaved for 30 min, and used for pot experiment. The upper diameter of the pots was 20 cm, bottom 16 cm, and a height of 15 cm. The experimental site was located in the greenhouse of Sugarcane Research Institute, GXAAS, Nanning, Guangxi, China. The enzyme analysis kits were obtained from Suzhou Gres Biotech Co., Ltd., China. The ammonium sulfate-15N2 isotope label chemical was obtained from the Bioisotope Engineering Center, Shanghai, China.
The strain E. roggenkampii ED5 was cultured in Luria-Bertani (LB) agar medium and incubated at 32°C and 220 rpm in a shaker (48 h). After incubation, the pellet was collected by centrifugation of bacterial culture at 5,000 × g for 15 min and the pellet was resuspended in sterile water to obtain the desired bacterial concentration (106 CFU/ml) for the inoculation in sugarcane. Sugarcane bud setts were placed in a tray, covered with sterilized soil, and watered appropriately, and grown seedlings with uniform size (∼15 cm) were used for the experiment.
For pot experiment, 60 healthy sugarcane plantlets of each sugarcane variety were divided into two groups (30 plants in each group). The sugarcane seedlings were uprooted from the tray. Before the seedlings were transplanted into pods, their roots were cleaned properly with running tap water and soaked in a culture broth with a concentration of 106 CFU/ml for 1 h. The seedlings root-soaked with sterile water were used as control. The experiment was conducted in a randomized block design (RBD), and each experiment was repeated thrice (n = 3). Sugarcane leaf + 1 (top visible dewlap leaf) samples were collected for analysis at 30 and 60 days after the treatment.
Kjeldahl method of nitrogen determination was used for total nitrogen test (Sáez-Plaza et al., 2013). The 15N isotope dilution method has the potential to measure the BNF in both B8 and GT11 sugarcane varieties. This technique involves inoculating with N-fixing ED5 strain to the sugarcane plants. Soil and sand were blended in a 1:3 (w/w) ratio and sterilized twice for 45 min at 121°C. After cooling at ambient temperature, 10 mg ammonium sulfate-15N (10.12% atom 15N excess) was applied per kilogram of soil and homogenized to ensure consistent dispersion of (15NH4)2SO4. The experiment was conducted according to the method followed by Lin et al. (2012), and nitrogen derived from the air (Ndfa) in the leaf and root of sugarcane tissues was calculated as suggested by Urquiaga et al. (1992).
The quantitative changes in biochemical parameters related to oxidative stress, namely the activities of superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) and the contents of malondialdehyde (MDA) and proline (Pro); those related to nitrogen metabolisms, such as the activities of NADH-glutamate dehydrogenase (NADH-GDH), glutamine synthetase (GS), nitrate reductase (NR); and those related to biocontrol mechanisms, such as the activities of β-1,3-glucanase and endo-β-1,4-glucanase, were observed at 30 and 60 days after the treatment. The analyses were performed using Biochemical Test Kit (Suzhou Grace Biotechnology Co., Ltd., China) following the instructions of the manufacturer. The results of all the analyzed biochemical parameters were presented as the ratio of values of the inoculated treatment and control.
The phytohormones in the sugarcane leaf samples were detected at 30 and 60 days after inoculation with strain ED5. Phytohormones, such as, indole-3-acetic acid (IAA), abscisic acid (ABA), and gibberellins (GA3), were measured by high-performance liquid chromatography (HPLC) (LC-100 PLUS, Shanghai, China). Notably, 0.2 g of sugarcane leaf + 1 samples were grounded with liquid nitrogen and diluted with 1 ml of precooled methanolic acid solution (70–80%) and kept overnight (4°C) for extraction. The extracts were centrifuged at 6,000 × g for 10 min (4°C) and extracted again with 0.5 ml of methanol solution (70–80%) for 2 h (4°C). The supernatants were collected after centrifugation, mixed, and evaporated to one-third volume under reduced pressure at 40°C and then an equal volume of petroleum ether was added for layering. The extraction and decolorization process was repeated 2–3 times. Later, triethylamine was added and the pH was adjusted to 8.0, incubated with shaking at 150 rpm for 20 min after the addition of cross-linked polyvinylpyrrolidone (CTFA). The supernatant was adjusted to pH 3.0 with hydrochloric acid after centrifugation and extraction with ethyl acetate and then evaporated to dry under reduced pressure at 40°C. The mobile-phase solution was added and vortexed to dissolve it, filtering with a syringe filter for measurement. The C18 reversed-phase column (150 mm × 4.6 mm, 5 μm) was used for HPLC at 0.8 ml/min flow rate (25°C). The mobile phase was made up of solvent A and solvent B. Solvent A was 100% methanol and solvent B was 0.1% acetic acid aqueous solution (A:B = 55:45). The injection volume was 20 μl, and the UV wavelength was set at 254 nm.
The Tris-balanced phenol extraction method (BPP method) was used to extract protein from the sugarcane leaves on 30-day inoculated strain ED5. Notably, 0.2 g of sugarcane leaf + 1 samples stored at −80°C in a freezer were transferred into a shaking tube and mixed with the appropriate amount of borax/polyvinyl-polypyrrolidone/phenol (BPP) solution and grounded for six times with a grinder (180 s each time). After centrifugation at 12,000 × g for 20 min at 4°C, the supernatant was mixed with an equal volume of Tris-saturated phenol, stored at 4°C for 30 min, and then vortexed for 10 min. Again, the supernatant was centrifuged at 12,000 × g for 20 min under 4°C, and the phenol phase was collected and added with an equal volume of BPP solution and then vortexed for 10 min at 4°C. Then supernatant was centrifuged for 20 min at 12,000 × g under 4°C. The phenol phase was collected and mixed with five volumes of precooled ammonium acetate methanol solution, and protein was precipitated overnight at −20°C. After incubation in a water bath at 25°C, the extracts were centrifuged at 12,000 × g for 20 min under 4°C. The supernatant was discarded and 1 ml 90% precooled acetone was added, mixed well, centrifuged, and again the supernatant was discarded. This step was repeated twice. The precipitates were dissolved in 1 ml protein lysate (8 M urea + 1% sodium dodecyl sulfate, a cocktail containing protease inhibitor) for precipitation and sonicated for 2 min on ice. The mixtures were centrifuged at 12,000 × g for 20 min under 4°C to obtain the protein supernatant. Notably, 100 μg of the protein extracts were added with appropriate amount of lysate to 1 ml kept for 60 min in 37°C water bath, and finally 1 ml of 10 mM Tris(2-carboxyethyl)phosphine (protein-reducing agent) was added. Then, 1 ml of 40 mM iodoacetamide was added and reacted in dark at room temperature (RT) for 40 min. Later, 1 ml of precooled acetone was added to each tube, precipitated at 4°C for 4 h, and centrifuged at 10,000 × g for 20 min. The precipitates were collected and fully dissolved in 1 ml of 100 nM TEAB, and trypsin was added according to the mass ratio of 1:50 (enzyme:protein), hydrolyzed overnight at 37°C, and then stored in a dry place. Notably, 100 μg of the peptide obtained by enzymatic digestion were added with 1 ml TMT reagent (Thermo Fisher Catalog No. 90111) and incubated at RT for 2 h. Again, 1 ml hydroxylamine was added and kept at RT for 15 min. Equal amount of the TMT-labeled product was mixed in a tube and vacuum-concentrated. The detailed information of all proteins obtained from the three biological replicates of sugarcane leaf + 1 samples is presented in Supplementary Tables 1, 2.
The peptide samples were reconstituted with the one-dimensional ultraperformance liquid chromatography loading buffer, and the reverse phase C18 column (ACQUITY UPLC BEH C18 Column 1.7 μm, 2.1 mm × 150 mm, Waters, United States) was used for high pH liquid-phase separation. Phase A (2% acetonitrile) and phase B (80% acetonitrile) were used for liquid chromatography. A total of 20 fractions were collected according to the peak type and time and combined into 10 fractions. After concentration by vacuum centrifugation, they were dissolved in the mass spectrometry loading buffer (2% acetonitrile and 0.1% formic acid) for the two-dimensional analysis, and the second dimension adopts nanoliter volume flow HPLC liquid system. Easy-nLC1200 was used for separation. The peptides were diluted in mass spectrometry loading buffer and separated by chromatography for 120 min after loading with a volume flow rate of 300 μl/min. EASY-nLC liquid phase underwent gradient elution with phase A 2% acetonitrile (ammonia water, pH 10) and phase B 80% acetonitrile (ammonia water, pH 10). The elution gradients were as follows: 0–1 min, 5% solvent B; 1–63 min, 5–23% solvent B; 63–77 min, 23–29% solvent B; 77–86 min, 29–38% solvent B; 86–88 min, 38–48% solvent B; 88–89 min, 48–100% solvent B; 89–95 min, 100% solvent B; and 95–96 min, 100–0% solvent B.
Proteome Discoverer, TM Software version 2.1 was used to analyze the original files from the mass spectrometer. The significance p-value of the difference between samples was calculated using the t-test function in the R language. The screening criterion for significantly DEPs was p < 0.05, and the differences > 0.83 and > 1.2 between the two samples indicated downregulated and upregulated proteins, respectively. Molecular functions of the identified DEPs were annotated by Cluster of Orthologous Groups of Proteins (COG), Pfam, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Non-Redundant Protein Sequence (NR) databases. The software BLAST2GO version 2.5.0 was used for the enrichment of GO function and KOBAS version 2.1.1 was used for the enrichment of KEGG GO function.
Plant biochemical data were analyzed using Excel 2010, and SPSS version 20.0 software was used to perform the analysis of variance (ANOVA). The difference in significance at p ≤ 0.05 was used to assess the comparison between the means. The statistical analysis for DEPs was carried out using Majorbio I-Sanger Cloud Platform, a free online platform1.
The results of biochemical analyses are shown in Figure 1. The SOD activity in sugarcane variety GT11 was enhanced at 30 days and decreased at 60 days after inoculation with ED5 as compared to control; however, it was higher in sugarcane variety B8 after the inoculation of strain ED5 at both time points (30 and 60 days) (Figure 1A). The CAT activity was found higher in both varieties inoculated with ED5 as relative to control, except for GT11 at 30 days (Figure 1B). The POD activity was found higher in GT11 and did not change in B8 at 30 days, whereas it was reduced in GT11 and increased in B8 at 60 days (Figure 1C). The MDA content was lower in GT11 and higher in B8 at specific time points as compared to control (Figure 1D). The proline content was initially decreased (30 days) and then increased (60 days) in GT11, whereas it initially did not change significantly and then it was higher at 60 days in B8 as compared to control (Figure 1E).
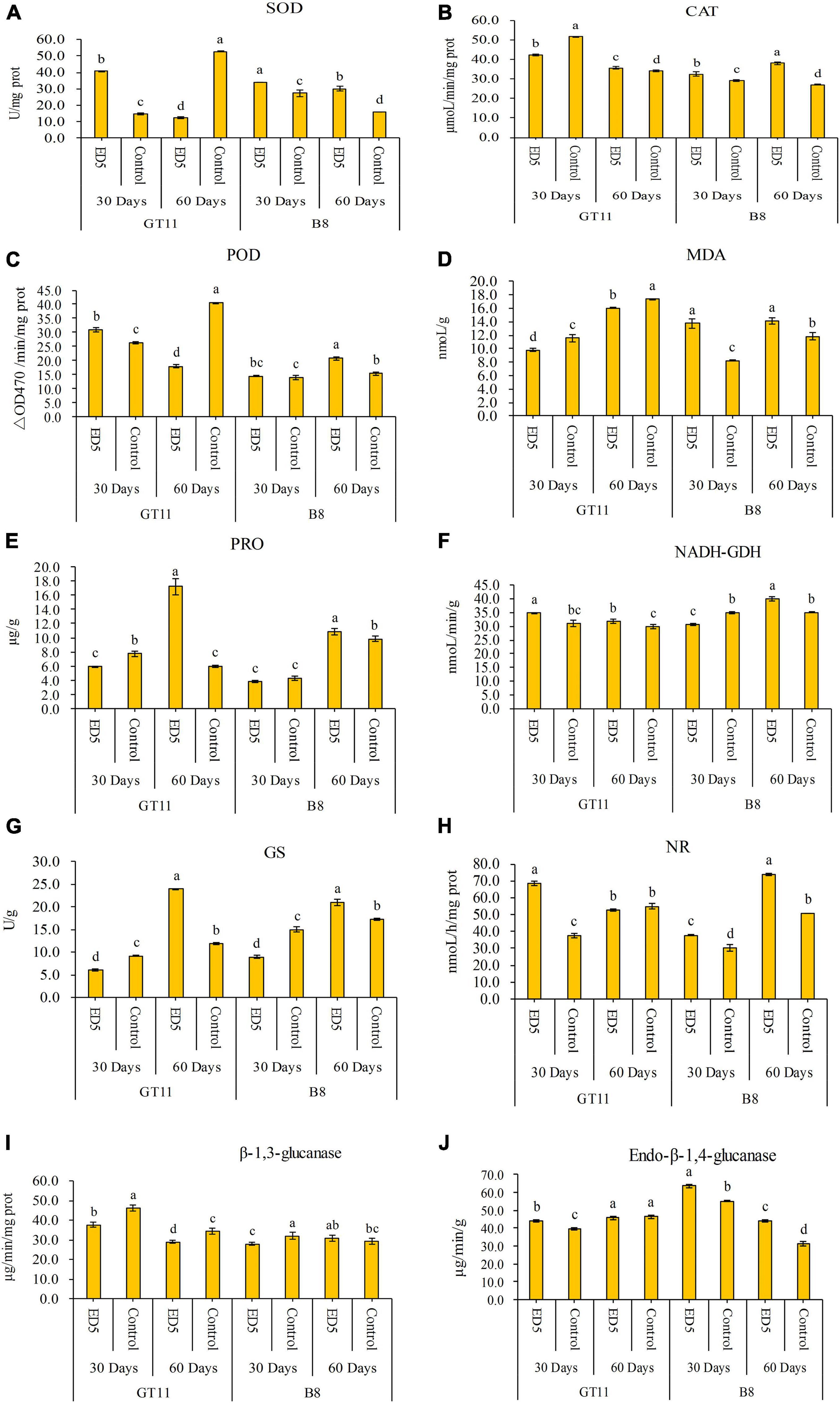
Figure 1. Variations in biochemical parameters in the leaf of sugarcane varieties GT11 and B8 inoculated with E. roggenkampii ED5 as compared to control. (A) Superoxide dismutase (SOD) activity, (B) catalase (CAT) activity, (C) peroxidase (POD) activity, (D) malondialdehyde (MDA) content, (E) proline (Pro) content, (F) NADH-glutamate dehydrogenase (NADH-GDH) activity, (G) glutamine synthetase (GS) activity, (H) nitrate reductase (NR) activity, (I) β-1,3-glucanase (β-1,3-GA) activity, and (J) endo-β-1,4-glucanase activity. Plant samples were collected at 30 and 60 days after the inoculation. At the level of p < 0.05, different letters indicate significant changes between treatments.
The NADH-GDH activity was higher at both time points in GT11 and lower at 30 days and higher at 60 days in B8 (Figure 1F). The GS activity was observed lower and then higher in both varieties at 30 and 60 days after inoculation with strain ED5 (Figure 1G). The NR activity showed different patterns with different sugarcane varieties, which was initially increased and then did not change significantly in GT11, but higher in B8 at both periods (Figure 1H).
The β-1,3-glucanase activity was lower in GT11 at each time point and did not change in B8 as compared to control (Figure 1I). The endo-β-1,4-glucanase activity in B8 was higher at both time points, and that in GT11 it was found higher at 30 days and did not change 60 days after inoculation with ED5 (Figure 1J).
The contents of phytohormones, such as IAA, GA3, and ABA, were measured in both sugarcane varieties GT11 and B8 at 30 and 60 days after inoculation with E. roggenkampii ED5. The IAA content was higher in both varieties at 30 and 60 days after the inoculation as compared to control (Figure 2A). The ABA content was increased in both varieties at 30 days, but did not change in GT11 at 60 days and was reduced in B8 at 60 days (Figure 2B). For GA3, the content was increased in GT11 and decreased in B8 at 30 days after the inoculation, but did not change significantly in GT11 and was decreased in B8 at 60 days (Figure 2C).
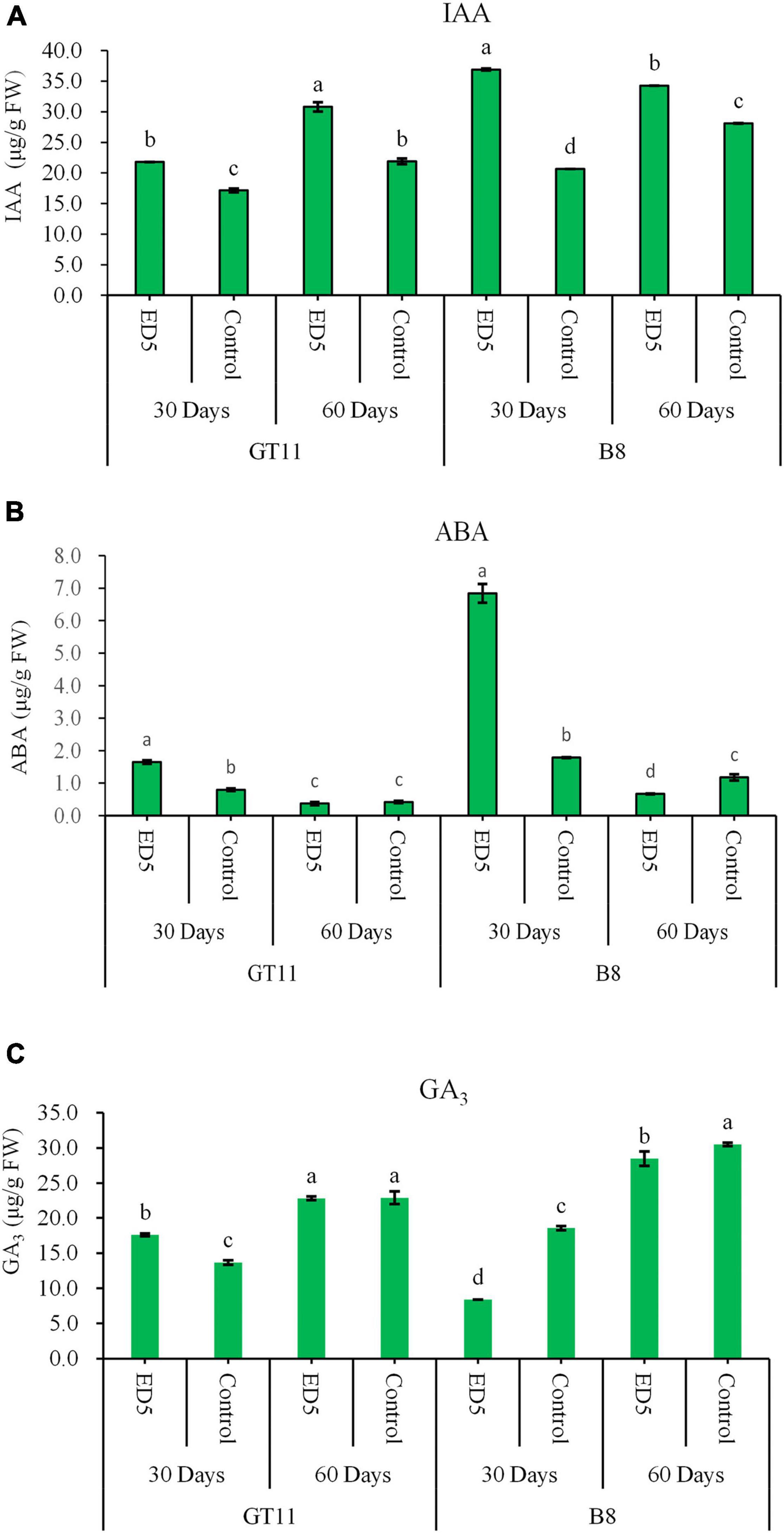
Figure 2. Variations in contents of phytohormones in the leaf of sugarcane varieties GT11 and B8 inoculated with E. roggenkampii ED5 as compared to control at 30 and 60 days after the treatment. (A) Indole acetic acid (IAA), (B) abscisic acid (ABA), and (C) gibberellin (GA3). At the level of p < 0.05, different letters indicate significant changes between treatments.
The percentage of 15N and total nitrogen was also estimated in both sugarcane varieties at 60 days post-inoculation of ED5 (Table 1). The percentage of 15N in the root tissues of both sugarcane varieties was significantly higher in the treatment inoculated with ED5 than control. In leaf tissues, it was higher in B8 and did not change significantly in GT11 (p < 0.05; Table 1). However, both sugarcane varieties GT11 and B8 had significantly higher total nitrogen concentration in leaf and root of sugarcane inoculated with ED5 than control (p < 0.05; Table 1). The growth of both sugarcane varieties was increased after the inoculation of strain E. roggenkampii ED5 as compared to control (Figure 3).
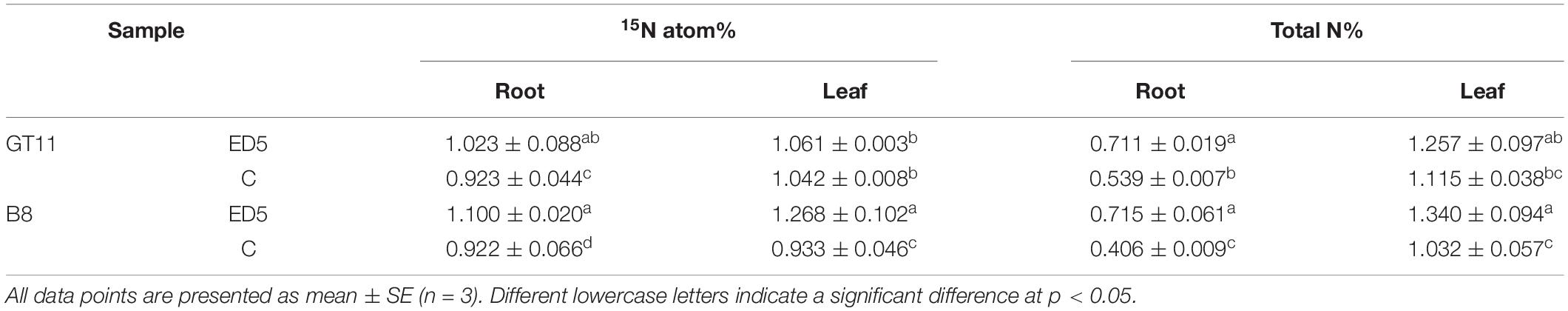
Table 1. Effect of strain ED5 on percentage of 15N and total nitrogen content in two sugarcane varieties.

Figure 3. Sugarcane growth promotion by inoculation of E. roggenkampii ED5 as compared to control. (A) GT11 (30 days), (B) B8 (30 days), (C) GT11 (60 days), and (D) B8 (60 days).
Tandem Mass Tags labeling was used to identify the DEPs in GT11 and B8 varieties inoculated with E. roggenkampii ED5 as relative to control. A sum of 27,508 proteins from 73,823 peptides, matching 301,280 spectrograms, was observed (Supplementary Table 1). The distribution of peptide matching error, peptide number, peptide length, and protein molecular is shown in Supplementary Figures 1A–D, respectively. The percentage of protein sequences with coverage of (0, 1%) was 2.83% and the coverage of (1, 5%) and (40, 60%) was 15.4%. The percentage of protein sequences with the coverage of (5, 10%), (10, 20%), (20, 40%), and (60, 80%) was 14.17, 20.04, 26.23, and 5.41%, respectively, whereas the protein sequences with coverage more than 80% accounted for 0.52% (Supplementary Figure 2). The heat map was used for correlation analysis of different duplicate samples and the data showed that the duplicate samples were clustered together (Supplementary Figure 3). The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium2 via the iProX partner repository with the data set identifier PXD026390 (Ma et al., 2019).
When compared to control, the DEPs were identified by using a fold-change threshold of > 1.20 or < 0.83 (p < 0.05). We analyzed the DEPs in both sugarcane varieties GT11 and B8 between the treatments inoculated with E. roggenkampii ED5 (GE, BE) and controls (GC, BC). A total of 177 DEPs, 103 upregulated and 74 downregulated (74), were marked in GT11 between GC and GE (Figure 4A). In contrast, a total of 378 DEPs, 244 upregulated and 134 downregulated, were identified in B8 between BC and BE (Figure 4B). A Venn diagram was constructed to show the common and different DEP numbers between GT11 and B8 varieties (Figure 5). Annotation of the DEPs in the GO, COG, and KEGG biological databases obtained the relevant annotation information (Supplementary Figures 4–6). The key DEPs were related to the growth of plant and mainly involved in the biological functions, such as nitrogen metabolism, photosynthesis, response to oxidative stress, starch and sucrose metabolism, hydrolase activity, oxidative phosphorylation, glutathione metabolism, phenylpropanoid metabolic process, response to stress, and proline catabolic process (Table 2).
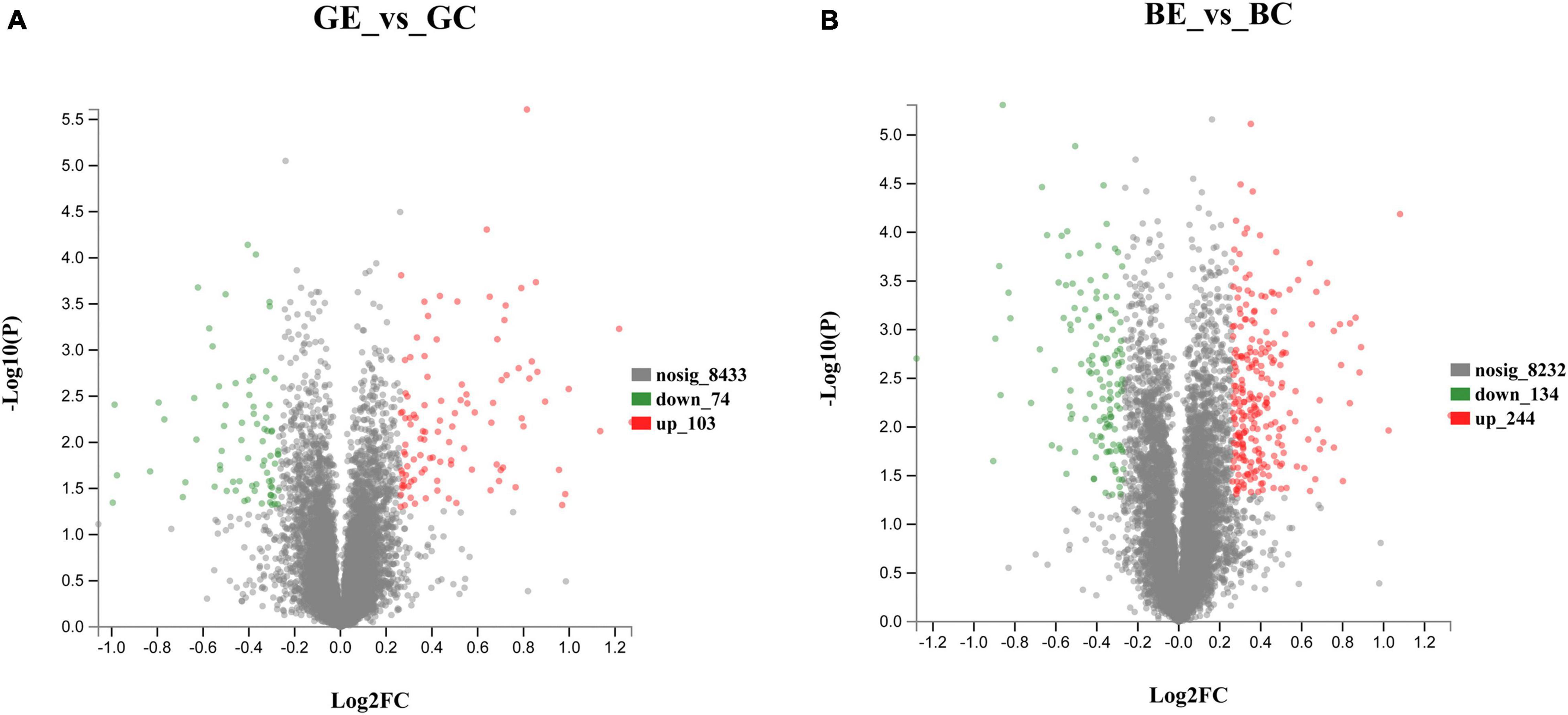
Figure 4. Volcano plots showing the differentially expressed protein allocation in the sugarcane varieties GT11 (A) and B8 (B) after the inoculation of E. roggenkampii ED5 as compared to control. Each data point represented the differential expression of the protein in the volcano plot. Downregulated differentially expressed proteins were shown with green dots, upregulated differentially expressed proteins were marked with red dots, and non-differentially expressed proteins were labeled by gray dots. Protein ratios with p-value < 0.05, fold change > 1.20 (upregulated), or < 0.83 (downregulated) were significantly differently expressed.
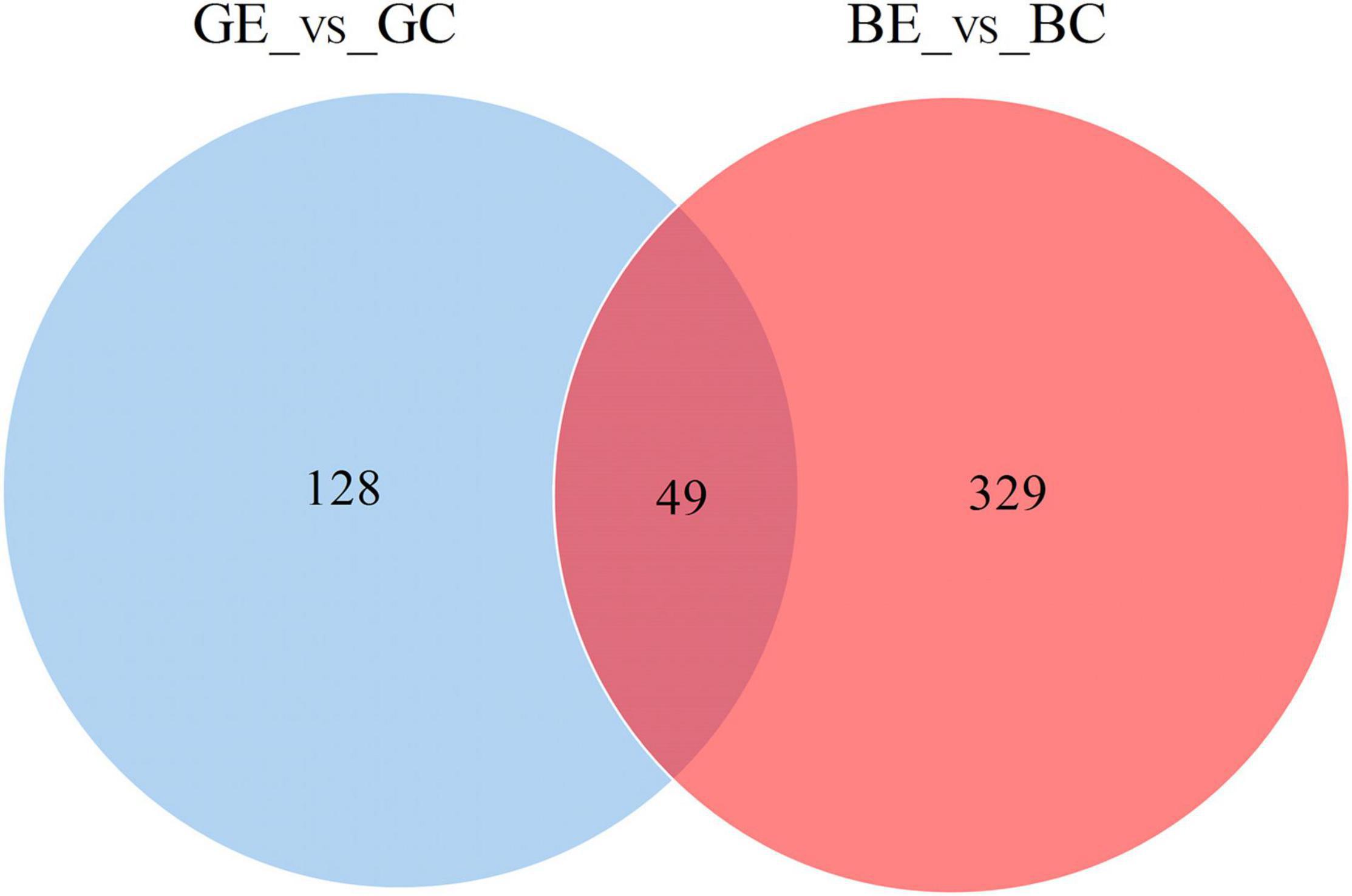
Figure 5. Venn diagrams showing the differentially expressed protein number allocation among the high-efficiency nitrogen-fixing variety B8 and low-efficiency nitrogen-fixing variety GT11 of sugarcane after the inoculation of E. roggenkampii ED5 as compared to control.
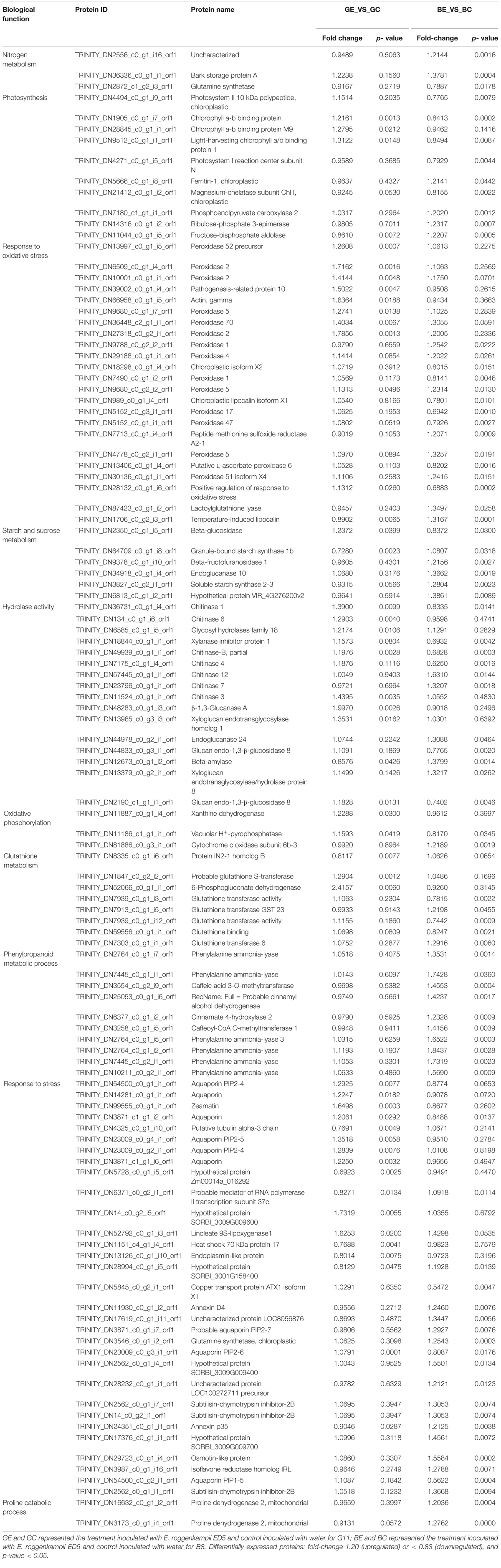
Table 2. List of differentially expressed proteins in two sugarcane varieties inoculated with the diazotroph E. roggenkampii ED5 as compared to control.
The putative functions of DEPs were investigated in the two sugarcane varieties GT11 and B8 inoculated with E. roggenkampii ED5 using GO enrichment analysis and classified into the groups related to biological, cellular, and molecular processes (Figure 6). The results of the GO enrichment analysis showed that response to stimulus (GO: 0050896) was the most significantly enriched term under the biological process for GT11. Additionally, the terms response to stress (GO: 0006950), hydrogen peroxide catabolic process (GO: 0042744), photosynthesis (GO: 0009765), response to oxidative stress (GO: 0006979), response to water (GO: 0009415), and response to acid chemical (GO: 0001101) were enriched in GT11 (Figure 6A). The GO term enrichment for B8 is mainly focused on phenylpropanoid metabolic process (GO: 0009698), secondary metabolite biosynthetic process (GO: 0044550), phenylpropanoid biosynthetic process (GO: 0009699), serine-type endopeptidase inhibitor activity (GO: 0004867), peptidase regulator activity (GO: 0061134), and peptidase inhibitor activity (GO: 0030414) (Figure 6B).
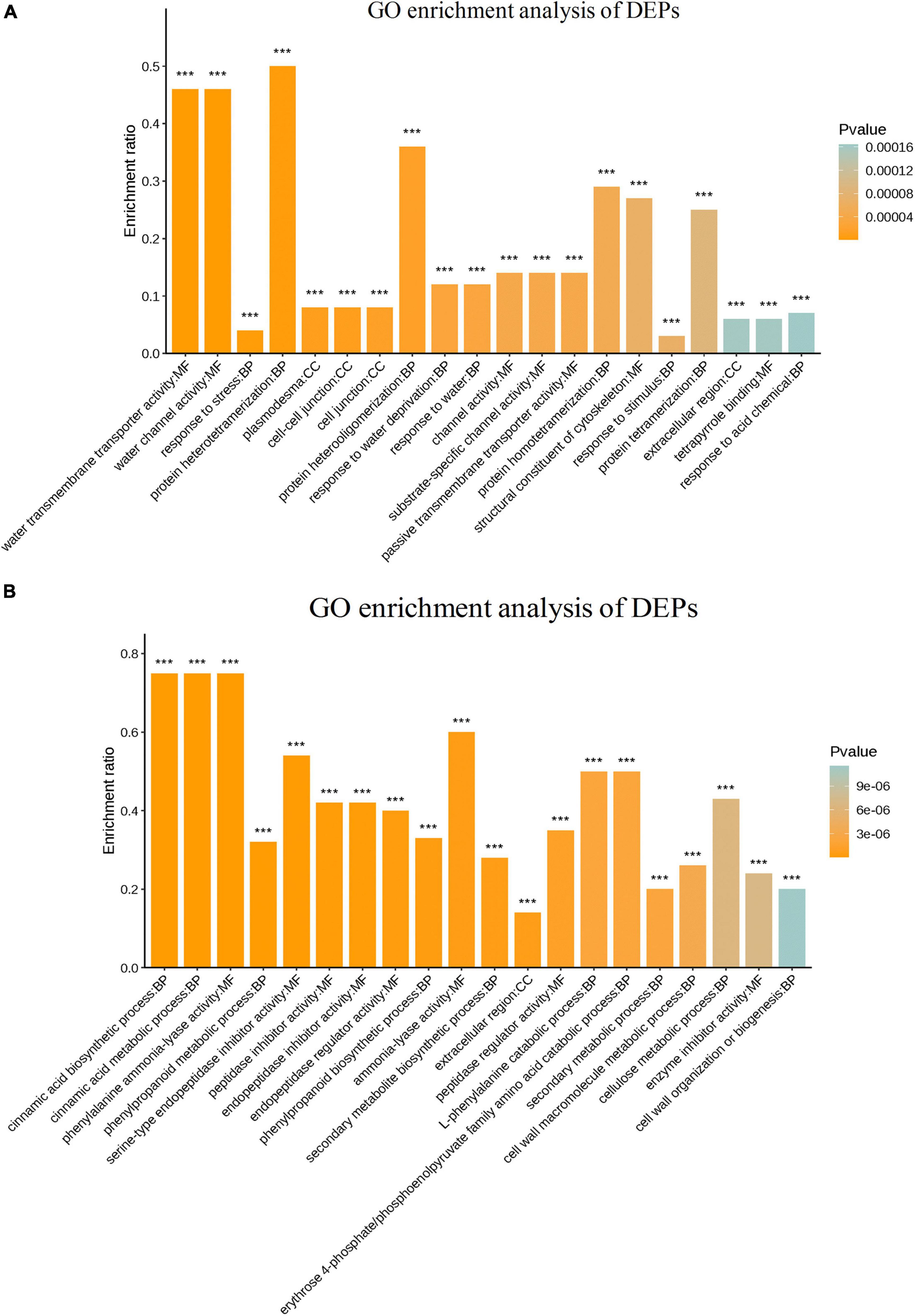
Figure 6. GO enrichment of differentially expressed proteins in (A) GT11 and (B) B8 varieties of sugarcane inoculated with E. roggenkampii ED5 as compared to control. The abscissa represents the GO term and the ordinate represents the enrichment rate. The color gradient of the column represents the significance of enrichment, where p or FDR < 0.001 was marked as ***, p or FDR < 0.01 was marked as **, and p or FDR < 0.05 was marked as *.
Analysis of the KEGG pathways was done to understand the effective functional information of identified DEPs in sugarcane inoculated with ED5. The 177 DEPs in GT11 were linked to 61 pathways, and 378 DEPs in B8 were mapped to 77 pathways (Supplementary Figure 6). Besides, the function enrichment of DEPs in KEGG pathways was analyzed using Fisher’s exact test. When the corrected p-value is < 0.05, the DEP was considered to be significantly enriched. The top 50 target proteins with a large number of annotation pathways and the pathways that contained these target proteins in the enrichment results and the pathways with the significance p-value of the top 15 are displayed in the string diagram (Figure 7). The pathways for GT11 were mainly enriched with phenylpropanoid biosynthesis (map00940), photosynthesis-antenna proteins (map00196), beta-alanine metabolism (map00410), tryptophan metabolism (map00380), cutin, suberin, and wax biosynthesis (map00073), fatty acid degradation (map00071), and circadian rhythm plant (map04712) (Figure 7A). The pathways for B8 were mainly enriched with phenylpropanoid biosynthesis (map00940), phenylalanine metabolism (map00360), glutathione metabolism (map00480), galactose metabolism (map00052), amino sugar and nucleotide sugar metabolism (map00520), nitrogen metabolism (map00910), arginine and proline metabolism (map00330), tryptophan metabolism (map00380), and ascorbate and aldarate metabolism (map00053) (Figure 7B).
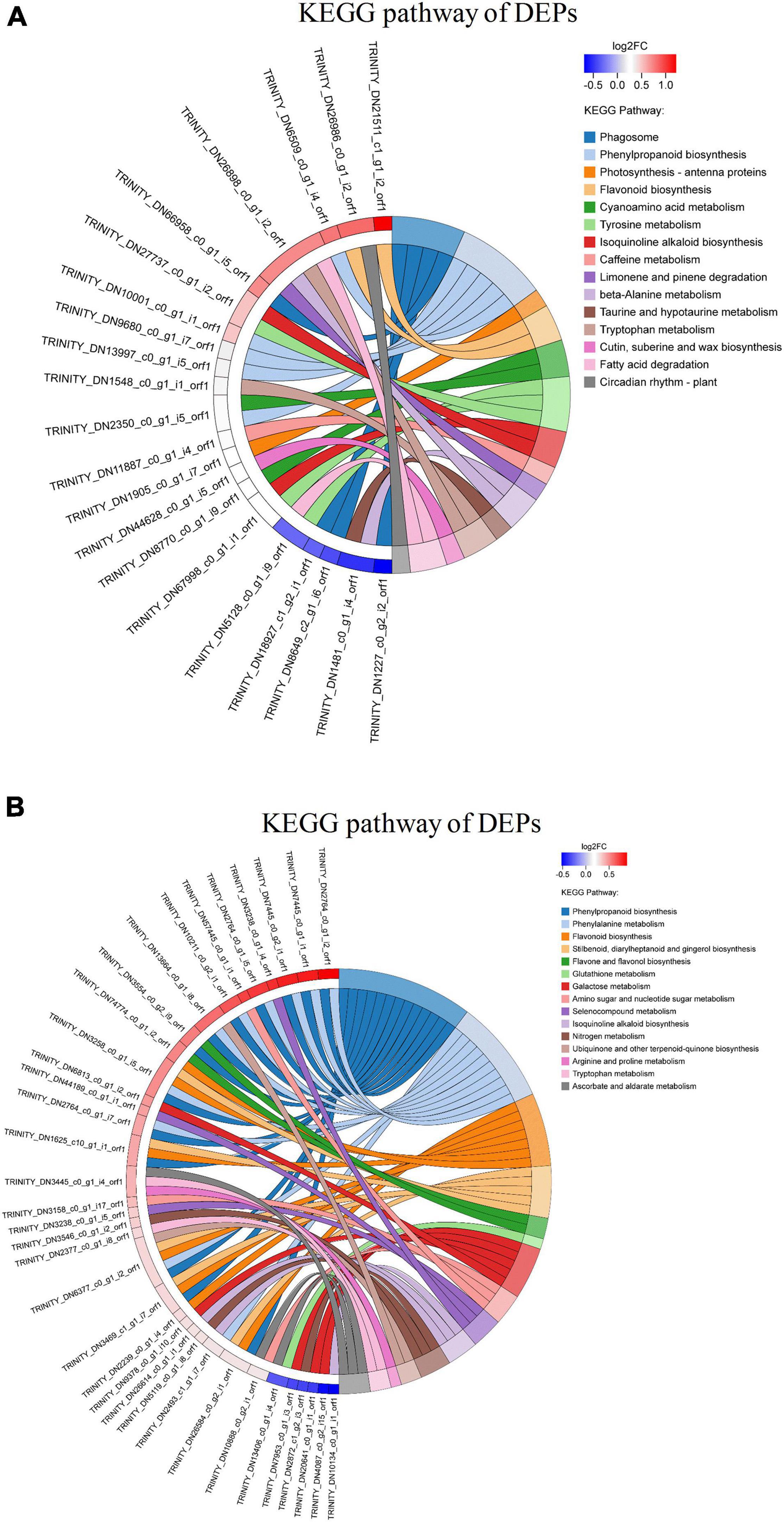
Figure 7. KEGG enrichment of differentially expressed proteins in (A) GT11 and (B) B8 varieties of sugarcane inoculated with E. roggenkampii ED5 as compared to control. The KEGG-enriched string diagram depicted the relationship between the target protein set and the KEGG pathway annotation and enrichment. The left side indicated the proteins and the log2FC was displayed in order from top to bottom. The larger the log2FC, the greater the difference in the expression of the upregulated proteins. The smaller the log2FC, the greater the difference in the expression of the downregulated proteins. The closer the log2FC was to 0, the smaller the difference in the fold of differential expression of the proteins. The right gave the name of the KEGG pathway that enriched the target protein and the Z score. The count represented the number of proteins associated with this pathway. For the total number of target proteins, up represented the number of upregulated proteins linked in this pathway, and down represented the number of downregulated proteins involved in this pathway. Z score > 0 meant that there were more upregulated proteins than downregulated proteins involved in this pathway, and this pathway was more likely to be activated. Z score < 0 indicated that there were fewer upregulated proteins than downregulated proteins involved in this pathway, and this pathway was more likely to be inhibited. In this enrichment chart, the top 50 target proteins with a large number of annotation pathways were selected. The pathways contained these target proteins in the enrichment and the pathways with the enrichment significance p-value of these target proteins were ranked in the top 15 for display.
Plant growth-promoting bacteria are important for the growth of plant, and their use in agricultural production has a lot of advantages (Bhattacharyya and Jha, 2012; Kumari et al., 2019). The mechanism of PGPB in promoting the growth of plant is multifaceted, including direct or indirect mechanisms (Goswami et al., 2016). In this study, the inoculation of PGP N-fixing E. roggenkampii ED5 strain isolated from the sugarcane roots enhanced the growth and nitrogen fixation in the two sugarcane varieties GT11 and B8 under greenhouse conditions as compared to control (Figure 3). In addition, the results of biochemical analyses, phytohormone detection, and comparative proteome analysis revealed the physiological, biochemical, and molecular mechanisms of the interaction between E. roggenkampii ED5 and sugarcane.
Plant growth-promoting bacteria has positive effects on the enhancement of plant antioxidant enzyme activities and growth enhancement (Islam et al., 2014; Benidire et al., 2021). This study also found that after the inoculation of ED5, stress-related enzyme activities (SOD and CAT) and osmotic regulator Pro content in both the sugarcane cultivars were dramatically increased. In contrast, the results of protein expression analysis showed differential expression of Pro and POD in ED5-inoculated sugarcane varieties. Enhanced expression of proline dehydrogenase 2 was observed in B18, but remained unchanged in GT11. Also, increased expression of peroxidase 2 in GT11 and decreased expression of peroxidase 17 in B8 variety were observed.
In addition, some PGPB can produce endogenous hormones required for the growth of plant, such as auxin, cytokinin, gibberellin, ABA, ethylene, salicylic acid, and jasmonic acid, thereby directly regulating the performance of plant (Tsukanova et al., 2017). In this study, the sugarcane endogenous hormone IAA was found higher in both varieties after the inoculation of strain ED5. It implied that the E. roggenkampii ED5 has the potential to promote the growth of sugarcane and was consistent with the plant growth of the two sugarcane varieties in the greenhouse. To stimulate the growth of plant, the ABA plays a crucial role in the adaptive response to abiotic environmental challenges, but it is usually linked to growth inhibition of plants or organs (Leung and Giraudat, 1998; Kuromori et al., 2010). In this study, the ABA content of the two sugarcane varieties was higher at 30 days, but did not change in GT11 and lower in B8 at 60 days, suggesting that the inoculation of E. roggenkampii ED5 could improve the environment adaptive ability of sugarcane at early stage of growth by increasing the ABA level but this effect is not necessary after the plants have adopted the environment and begun to grow fast. Based on the results of this study, it seems that the effect of inoculation of E. roggenkampii ED5 on gibberellin content in sugarcane is variety-dependent. However, no significant changes were observed in the expression of phytohormone-related proteins in both sugarcane varieties inoculated with ED5.
Nitrogen fixation is one of the growth-promoting factors of some PGPB (Jha and Saraf, 2015). Therefore, enhanced activities of nitrogen metabolism-related enzymes are directly associated with nitrogen fixation by PGPB. In this study, the activities of nitrogen metabolism-related enzymes GS and NADH-GDH in the sugarcane leaves were found to be higher in the treatment inoculated with E. roggenkampii ED5 than control, indicating the improvement of the nitrogen metabolism by the treatment inoculated with ED5. The endo-β-1,4-glucanase activity was also found higher in the treatment inoculated with E. roggenkampii ED5 than control, which could be connected to the faster growth of sugarcane plants in the treatment inoculated with ED5. The activities of NR and β-1,3-glucanase were sugarcane variety- and growth stage-dependent in this study. We also found increased expression of some proteins was related to hydrolase activity and nitrogen metabolism in both sugarcane varieties after the inoculation of strain ED5.
Taking advantage of the nitrogen fixation function of PGPB to improve the efficiency of plant nitrogen utilization is an important way to achieve a sustainable development of crop productivity (Figueiredo et al., 2008; Baset et al., 2010; Matse et al., 2020; Basu et al., 2021). As seen earlier, the strain E. roggenkampii ED5 showed high nitrogenase activity (29.60 nmoL C2H4 mg protein/h) through acetylene reduction assay, indicating its N-fixation potential (Guo et al., 2020). To confirm this hypothesis, the ammonium sulfate-15N2 isotope label dilution method was applied to analyze the utilization rate of nitrogen in sugarcane plant organs such as root and leaf, and the results showed that the concentration of N content in the sugarcane root and leaf was significantly enhanced after the inoculation of E. roggenkampii ED5 in both varieties GT11 and B8 as relative to normal plants. Therefore, the E. roggenkampii ED5 can effectively fix nitrogen and supply to host sugarcane plants.
Previously, the PGPB in sugarcane rhizosphere is widely investigated and some potent isolates were obtained (Lamizadeh et al., 2016; Wang Z. et al., 2019; Singh et al., 2020). However, the information using proteomics to analyze the interaction mechanism between PGPB and sugarcane is limited. In this study, we used the TMT labeling method to identify the DEPs of two different N-fixing sugarcane varieties GT11 (needs more nitrogen for growth) and B8 (needs less nitrogen for growth) after the inoculation of the diazotroph strain ED5. The results of KEGG enrichment of DEPs indicated that there were three proteins enriched in the N metabolism pathway (map00910) in B8, but not in GT11. Earlier, some researchers have shown that different sugarcane varieties have different N-fixation abilities (Boddey et al., 1991; de Matos Nogueira et al., 2005). Obviously, the inconsistent enrichment of KEGG N-fixing metabolic pathways in the sugarcane varieties with different N-fixation abilities was caused by the interactions of sugarcane varieties with the diazotroph strain ED5.
Photosynthesis is the major physiological process related to the growth and development of plant, and the relative growth rate (RGR) was positively correlated with the total biomass, leaf mass, and leaf area expansion in plants (Kruger and Volin, 2006). In this study, photosynthesis-related DEPs, such as photosystem II 10 kDa polypeptide, chlorophyll a-b binding protein, photosystem I reaction center subunit N, phosphoenolpyruvate carboxylase 2, and fructose-bisphosphate aldolase, were detected in the treatment inoculated with E. roggenkampii ED5 as compared to control. Chlorophyll a and b and Chl a/b involve in the light absorption and energy transfer to the photosystem and the reaction center of charge separation in green plants (Gobets et al., 2001; Horn et al., 2007; Kjaer et al., 2020). The results in this study showed that there was a significant upregulation of photosystem-II-related proteins after inoculation with E. roggenkampii ED5, which is beneficial to photosynthesis and plant growth. Meanwhile, other photosynthesis-related enzymes, such as photosystem II 10 kDa polypeptide, light-harvesting chlorophyll a/b, and magnesium-chelatase subunit Chl I, were also upregulated, which further confirmed our previous findings that E. roggenkampii ED5 is an efficient diazotroph PGPB strain (Guo et al., 2020). According to relevant research studies, PGPB was able to affect plant photosynthetic capacity, thereby promoting the growth of plant (Su et al., 2015; Samaniego-Gámez et al., 2016; Mathivanan et al., 2017).
Sugarcane is a well-known sugar and bioenergy crop worldwide (Li and Yang, 2015). Breeding high sucrose sugarcane varieties is the main research purpose of breeders (Jackson, 2005). In this experiment, differential expression of proteins related to sucrose metabolism, such as beta-glucosidase, granule-bound starch synthase 1b, beta-fructofuranosidase 1, endoglucanase 10, and soluble starch synthase 2-3, was identified after the inoculation of ED5 in sugarcane. Among them, the protein soluble starch synthase 2-3 (ID: TRINITY_DN3827_c0_g2_i1_orf1) was upregulated with fold-change of 1.237 (p-value = 0.002) in B8 (Table 2). Soluble starch synthase (SSs) is mainly associated with the synthesis of amylopectin (Cao et al., 1999; Myers et al., 2000), including SSI, SSII, SSIII, and SSIV subtypes (Kossmann et al., 1999). SSII and SSIII usually participate in the extension of short chains (Edwards et al., 1999; Delvallé et al., 2005). Moreover, SSs can transfer the glucose residues of adenosine diphosphate glucose (ADPG) to the non-reducing end of the glucan chain through α-1,4-glycosidic bonds and play a major role in starch and sucrose metabolisms (Komor, 2000; Xue et al., 2019). These results implied that E. roggenkampii ED5 might contribute to enhance the activity of sucrose synthesis in sugarcane.
Plant growth-promoting bacteria enhances the resistance of plants to stress, thereby indirectly promoting the growth of plant. Some studies have shown that PGPB can effectively enhance the response of plants to stresses, such as water-deficit conditions (Ngumbi and Kloepper, 2016), salinity (Raklami et al., 2019), pathogens (Siddiqui, 2005), and heat and heavy metals (Raklami et al., 2019). Our previous research also predicted that the E. roggenkampii ED5 genome had some vital abiotic stress-related genes, such as those encoding cold and heat shock proteins (HSPs), heavy metal, and drought resistance proteins, and we speculated that the strain ED5 has the potential to resist abiotic stress (Guo et al., 2020). In this study, the GO enrichment of DEPs was focused on several abiotic stress-related proteins, including aquaporin PIP2-4 (protein ID: TRINITY_DN54500_c0_g1_i1_orf1), heat shock 70 kDa protein 17 (protein ID: TRINITY_ DN1151 _c4_g1_i4_orf1), copper transport protein ATX1 isoform X1 (protein ID: TRINITY_DN5845 _c0 _g2 _i1_orf1), GS and chloroplastic (protein ID: TRINITY_DN3546_c0_g1_i2_orf1), and osmotin-like protein (protein ID: TRINITY_DN29723_c0_g1_i4_orf1). These results further confirmed that the tolerance of sugarcane to abiotic stress may be improved after the inoculation of E. roggenkampii ED5. Recently, proteomic approaches have been widely used in revealing the molecular mechanism of resistance of plant to abiotic stress, especially in salt and drought tolerance in crops under field conditions (Gautam et al., 2020). In this study, some DEPs related to abiotic stress were observed with TMT proteomics, which provides a reference for further exploring the molecular mechanisms of the interaction between the strain ED5 and sugarcane to improve resistance of sugarcane to abiotic stress.
In addition, GO enrichment analysis showed some DEPs involving in glutathione metabolism pathway, such as glutathione S-transferase (protein ID: TRINITY_DN1847_c0_g2_i2_orf1), 6-phosphogluconate dehydrogenase (protein ID: TRINITY_DN52066_c0_g1_i1_orf1, glutathione transferase (TRINITY_DN7939_c0_g1_i3_orf1), glutathione transferase GST 23 (protein ID: TRINI TY_DN7913_c0_g1_i5_orf1), glutathione binding (protein ID: TRINITY_DN59556_c0_g1_i1_orf1), and glutathione transferase 6 (protein ID: TRINITY_DN7303_c0_g1_i1_orf1), were detected after the inoculation of ED5. The glutathione plays a significant role in the maintenance of antioxidant ability of plant tissues and regulation of redox-sensitive signal transduction (Sen, 1999; Cnubben et al., 2001; Hérouart et al., 2002). The level of glutathione was closely related to the tolerance of plants to various biological heterologous substances and environmental stresses (May et al., 1998; Nianiou-Obeidat et al., 2017). In this study, some glutathione metabolism-related DEPs were identified, which were related to the improvement of tolerance of plant to various stresses, the enhancement of plant defense system, and the enhanced plant growth in sugarcane after the inoculation of ED5. Peroxidase is a key enzyme responsible for the synthesis of resistance inducers (Perkins-Veazie et al., 1996). In this study, the significant enrichment of peroxidase was observed in sugarcane after inoculation with strain ED5.
Moreover, in this study, the phenylpropanoid metabolic process of DEPs was also enriched by the KEGG and GO databases. Recent studies have shown that plants respond to environmental stresses during growth and several factors participate in the metabolism of propanol and regulation of the homeostasis of phenylpropanol (Dong and Lin, 2021). The phenylpropanoid metabolic process also contributes to abiotic and biotic stresses (Nakabayashi and Saito, 2015; Sharma et al., 2019). Some phenylalanine ammonia-lyases were enriched in GO analysis. The most fold-change of GO term in phenylpropanoid metabolic was phenylalanine ammonia-lyase (protein ID: TRINITY_DN27 64_c0 _g1_i2_orf1), 1.844 (p-value = 0.003). Our previous study also found the presence of key genes related to abiotic stress resistance in the ED5 genome (Guo et al., 2020). Similarly, proteomics analysis revealed several vital stress-related DEPs, including enrichment of phenylpropanoid metabolic proteins in the GO and KEGG databases. All these results implied that sugarcane may enhance resistance and strengthen self-defense ability through various metabolic pathways after inoculation with E. roggenkampii ED5.
Some key hydrolases, such as chitinase, glycosyl hydrolases, and glucan endo-1,3-β-glucosidase, play a major role in resistance of plant to biotic or abiotic stresses (Flach et al., 1992; Beffa et al., 1993; Gaudioso-Pedraza and Benitez-Alfonso, 2014). Previously, we found that the E. roggenkampii ED5 secreted hydrolases such as cellulase, chitinase, endoglucanase, and protease, and the related genes were also predicted in its genome (Guo et al., 2020). In this study, the GO enrichment of DEPs displayed some hydrolases, with the chitinases being the most enriched hydrolases, including chitinase 1, chitinase 3, chitinase 4, chitinase 6, and chitinase 12. Chitinase plays a crucial role in plant defense, particularly against diseases (Sharma et al., 2011; Kumar et al., 2018; Liu et al., 2020). The strain ED5 has a high inhibitory effect on five kinds of pathogenic fungi in vitro plate confrontation test, and several biocontrol-related coding genes were predicted in its genome (Guo et al., 2020). In this study, higher enrichment of the proteins related to hydrolases confirmed our previous hypothesis that the TMT proteomics study can further verify the biocontrol potential of the strain ED5. This isolate may have value in the application of sugarcane biocontrol in future.
In this study, TMT labeling and LC-MS/MS analyses were carried out to study the comparative protein profiles of two different sugarcane varieties GT11 and B8 inoculated with E. roggenkampii ED5. Some key proteins, such as those involved in nitrogen metabolism, photosynthesis, starch and sucrose metabolism, response to oxidative stress, hydrolysis, oxidative phosphorylation, glutathione metabolism, phenylpropanoid metabolic process, and response to stresses, were enriched in the GO and KEGG databases. Furthermore, the research revealed that E. roggenkampii ED5 has positive effects on improving nitrogen utilization in sugarcane and promoting plant growth in sugarcane. Increases in the contents of IAA and proline and activities of oxidation defensive enzymes and nitrogen metabolism enzymes were found after inoculated with strain ED5. Therefore, this strain may have the potential to improve sugarcane productivity and eco-environmental sustainability.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
D-JG, RS, PS, and Y-RL designed the experiments. D-JG, D-PL, RS, and PS accomplished the experiments. AS, KV, YQ, QK, ZL, X-PS, and MM analyzed the data. D-JG, RS, and PS drafted the manuscript. Y-XX and Y-RL critically revised and finalized the article. All authors reviewed the article and approved it for publication.
This study was supported by Fund for Guangxi Innovation Teams of Modern Agriculture Technology (nycytxgxcxtd-2021-03-01), Fund of Guangxi Academy of Agricultural Sciences (2021YT11, GNKB2017028, GNKB2018034, and GuiNongKe2021JM87), Guangxi Special Fund for Scientific Base and Talent (GKAD17195100), and the National Natural Science Foundation of China (31471449, 31171504, and 31101122).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.727741/full#supplementary-material
Alberton, D., Valdameri, G., Moure, V. R., Monteiro, R. A., Pedrosa, F. D. O., Müller-Santos, M., et al. (2020). What did we learn from plant growth-promoting rhizobacteria (PGPR)-grass associations studies through proteomic and metabolomic approaches? Front. Sustain. Food Syst. 4:607343. doi: 10.3389/fsufs.2020.607343
Antunes, J. E. L., Freitas, A. D. S., Oliveira, L., Delyra, M., Fonseca, M. A., et al. (2019). Sugarcane inoculated with endophytic diazotrophic bacteria: effects on yield, biological nitrogen fixation and industrial characteristics. Anais Acad. Brasil. Ciências 91:3765201920180990. doi: 10.1590/0001-3765201920180990
Asghari, B., Khademian, R., and Sedaghati, B. (2020). Plant growth promoting rhizobacteria (PGPR) confer drought resistance and stimulate biosynthesis of secondary metabolites in pennyroyal (Mentha pulegium L.) under water shortage condition. Sci. Horticul. 263:109132. doi: 10.1016/j.scienta.2019.109132
Bai, L., Sun, H. B., Liang, R. T., and Cai, B. Y. (2019). iTRAQ proteomic analysis of continuously cropped soybean root inoculated with Funneliformis mosseae. Front. Microbiol. 10:61. doi: 10.3389/fmicb.2019.00061
Basanta, M. V., Dourado-Neto, D., Reichardt, K., Bacchi, O. O. S., Oliveira, J. C. M., Trivelin, P. C. O., et al. (2003). Management effects on nitrogen recovery in a sugarcane crop grown in Brazil. Geoderma 116, 235–248. doi: 10.1016/s0016-7061(03)00103-4
Baset, M. M., Shamsuddin, Z., Wahab, Z., and Marziah, M. (2010). Effect of plant growth promoting rhizobacterial (PGPR) inoculation on growth and nitrogen incorporation of tissue-cultured’musa’plantlets under nitrogen-free hydroponics condition. Austral. J. Crop Sci. 4, 85–90. doi: 10.3390/su13031140
Basu, A., Prasad, P., Das, S. N., Kalam, S., Sayyed, R., Reddy, M., et al. (2021). Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: recent developments, constraints, and prospects. Sustainability 13:1140. doi: 10.1073/pnas.90.19.8792
Beffa, R. S., Neuhaus, J.-M., and Meins, F. (1993). Physiological compensation in antisense transformants: specific induction of an“ ersatz” glucan endo-1, 3-beta-glucosidase in plants infected with necrotizing viruses. Proc. Natl. Acad. Sci. 90, 8792–8796. doi: 10.1073/pnas.90.19.8792
Benidire, L., Madline, A., Pereira, S., Castro, P., and Boularbah, A. (2021). Synergistic effect of organo-mineral amendments and plant growth-promoting rhizobacteria (PGPR) on the establishment of vegetation cover and amelioration of mine tailings. Chemosphere 262:127803. doi: 10.1016/j.chemosphere.2020.127803
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Boddey, R., Urquiaga, S., Reis, V., and Döbereiner, J. (1991). Biological nitrogen fixation associated with sugarcane. Nitrogen fixation 1991, 105–111. doi: 10.1007/978-94-011-3486-6_22
Bordonal, R. D. O., Carvalho, J. L. N., Lal, R., de Figueiredo, E. B., de Oliveira, B. G., and La Scala, N. (2018). Sustainability of sugarcane production in Brazil. A review. Agronomy Sustain. Dev. 38:0490–x. doi: 10.1007/s13593-018-0490-x
Boschiero, B. N., Mariano, E., Torres-Dorante, L. O., Sattolo, T. M. S., Otto, R., Garcia, P. L., et al. (2020). Nitrogen fertilizer effects on sugarcane growth, nutritional status, and productivity in tropical acid soils. Nutrient Cycling Agroecosyst. 117, 367–382. doi: 10.1007/s10705-020-10074-w
Cao, H., Imparl-Radosevich, J., Guan, H., Keeling, P. L., James, M. G., and Myers, A. M. (1999). Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol. 120, 205–216. doi: 10.1104/pp.120.1.205
Chen, J., Wang, X., Tang, D., and Wang, W. (2019). Oxidative stress adaptation improves the heat tolerance of Pseudomonas fluorescens SN15-2. Biol. Control 138:104070. doi: 10.1016/j.biocontrol.2019.104070
Chen, M., Zhu, B., Lin, L., Yang, L., Li, Y., and An, Q. (2014). Complete genome sequence of Kosakonia sacchari type strain SP1(T.). Stand Genomic Sci. 9, 1311–1318. doi: 10.4056/sigs.5779977
Chinnasamy, G. (2006). A Proteomics Perspective on Biocontrol and Plant Defense Mechanism. Biocontr. Biofertiliz. 2006, 233–255. doi: 10.1007/1-4020-4152-7_9
Cnubben, N. H., Rietjens, I. M., Wortelboer, H., van Zanden, J., van Bladeren, P. J., and pharmacology. (2001). The interplay of glutathione-related processes in antioxidant defense. Environ. Toxicol. 10, 141–152. doi: 10.1016/s1382-6689(01)00077-1
de Matos Nogueira, E., Olivares, F. L., Japiassu, J. C., Vilar, C., Vinagre, F., Baldani, J. I., et al. (2005). Characterization of glutamine synthetase genes in sugarcane genotypes with different rates of biological nitrogen fixation. Plant Sci. 169, 819–832. doi: 10.1016/j.plantsci.2005.05.031
Delvallé, D., Dumez, S., Wattebled, F., Roldán, I., Planchot, V., Berbezy, P., et al. (2005). Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J. 43, 398–412. doi: 10.1111/j.1365-313X.2005.02462.x
Dong, N. Q., and Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integrat. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Edwards, A., Fulton, D. C., Hylton, C. M., Jobling, S. A., Gidley, M., Rössner, U., et al. (1999). A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J. 17, 251–261. doi: 10.1046/j.1365-313x.1999.00371.x
Esitken, A., Yildiz, H. E., Ercisli, S., Figen Donmez, M., Turan, M., and Gunes, A. (2010). Effects of plant growth promoting bacteria (PGPB) on yield, growth and nutrient contents of organically grown strawberry. Sci. Horticult. 124, 62–66. doi: 10.1016/j.scienta.2009.12.012
Fernandez, O., Theocharis, A., Bordiec, S., Feil, R., Jacquens, L., Clément, C., et al. (2012). Burkholderia phytofirmans PsJN acclimates grapevine to cold by modulating carbohydrate metabolism. Mol. Plant Microbe Interact. 25:496. doi: 10.1094/mpmi-09-11-0245
Figueiredo, M., Martinez, C., Burity, H., and Chanway, C. (2008). Plant growth-promoting rhizobacteria for improving nodulation and nitrogen fixation in the common bean (Phaseolus vulgaris L.). World J. Microbiol. Biotechnol. 24, 1187–1193. doi: 10.1007/s11274-007-9591-4
Flach, J., Pilet, P.-E., and Jolles, P. (1992). What’s new in chitinase research? Experientia 48, 701–716. doi: 10.1007/bf02124285
Franco, H. C. J., Otto, R., Faroni, C. E., Vitti, A. C., Almeida, de Oliveira, E. C., et al. (2011). Nitrogen in sugarcane derived from fertilizer under Brazilian field conditions. Field Crops Res. 121, 29–41. doi: 10.1016/j.fcr.2010.11.011
Franco, H. C. J., Otto, R., Vitti, A. C., Faroni, C. E., Oliveira, E. C. D. A., Fortes, C., et al. (2015). Residual recovery and yield performance of nitrogen fertilizer applied at sugarcane planting. Sci. Agricola 72, 528–534. doi: 10.1590/0103-9016-2015-0170
Gaudioso-Pedraza, R., and Benitez-Alfonso, Y. (2014). A phylogenetic approach to study the origin and evolution of plasmodesmata-localized glycosyl hydrolases family 17. Front. Plant Sci. 5:212. doi: 10.3389/fpls.2014.00212
Gautam, A., Pandey, P., and Pandey, A. K. (2020). Proteomics in relation to abiotic stress tolerance in plants. Plant Life Under Changing Environ. 2020, 513–541. doi: 10.1016/b978-0-12-818204-8.00023-0
Gobets, B., Kennis, J. T., Ihalainen, J. A., Brazzoli, M., Croce, R., van Stokkum, I. H., et al. (2001). Excitation energy transfer in dimeric light harvesting complex I: a combined streak-camera/fluorescence upconversion study. J. Phys. Chem. B 105, 10132–10139. doi: 10.1021/jp011901c
Goswami, D., Thakker, J. N., Dhandhukia, P. C., and Agriculture. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agricult. 2:1127500. doi: 10.1080/23311932.2015.1127500
Guo, D. J., Singh, R. K., Singh, P., Li, D. P., Sharma, A., Xing, Y. X., et al. (2020). Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 11:580081. doi: 10.3389/fmicb.2020.580081
Hérouart, D., Baudouin, E., Frendo, P., Harrison, J., Santos, R., Jamet, A., et al. (2002). Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume–Rhizobium symbiosis? Plant Physiol. 40, 619–624.
Horn, R., Grundmann, G., and Paulsen, H. (2007). Consecutive binding of chlorophylls a and b during the assembly in vitro of light-harvesting chlorophyll-a/b protein (LHCIIb). J. Mol. Biol. 366, 1045–1054. doi: 10.1016/j.jmb.2006.11.069
Igiehon, N. O., and Babalola, O. O. (2018). Rhizosphere Microbiome Modulators: Contributions of Nitrogen Fixing Bacteria towards Sustainable Agriculture. Int. J. Environ. Res. Public Health 15:ijerh15040574. doi: 10.3390/ijerph15040574
Imran, A. (2013). Salt-tolerant PGPR strain Planococcus rifietoensis promotes the growth and yield of wheat (Triticum aestivum L.) cultivated in saline soil. Pak. J. Bot. 45, 1955–1962.
Islam, F., Yasmeen, T., Ali, Q., Ali, S., Arif, M. S., Hussain, S., et al. (2014). Influence of Pseudomonas aeruginosa as PGPR on oxidative stress tolerance in wheat under Zn stress. Ecotoxicol. Environ. Safety 104, 285–293. doi: 10.1016/j.ecoenv.2014.03.008
Jackson, P. A. (2005). Breeding for improved sugar content in sugarcane. Field Crops Res. 92, 277–290. doi: 10.1016/j.fcr.2005.01.024
Jha, C. K., and Saraf, M. D. (2015). Plant growth promoting rhizobacteria (PGPR): a review. J. Agricult. Res. Dev. 5, 108–119.
Jiang, Q., Li, X., Niu, F., Sun, X., Hu, Z., and Zhang, H. (2017). iTRAQ based quantitative proteomic analysis of wheat roots in response to salt stress. Proteomics 17:1600265. doi: 10.1002/pmic.201600265
Kjaer, C., Gruber, E., Nielsen, S. B., and Andersen, L. H. (2020). Color tuning of chlorophyll a and b pigments revealed from gas-phase spectroscopy. Phys. Chem. Chemical Phys. 22, 20331–20336. doi: 10.1039/d0cp03210g
Komor, E. (2000). Source physiology and assimilate transport: the interaction of sucrose metabolism, starch storage and phloem export in source leaves and the effects on sugar status in phloem. Funct. Plant Biol. 27, 497–505. doi: 10.1071/pp99127
Kossmann, J., Abel, G. J., Springer, F., Lloyd, J. R., and Willmitzer, L. (1999). Cloning and functional analysis of a cDNA encoding a starch synthase from potato (Solanum tuberosum L.) that is predominantly expressed in leaf tissue. Planta 208, 503–511. doi: 10.1007/s004250050587
Kruger, E. L., and Volin, J. C. (2006). Reexamining the empirical relation between plant growth and leaf photosynthesis. Funct. Plant Biol. 33, 421–429. doi: 10.1071/FP05310
Kumar, M., Brar, A., Yadav, M., Chawade, A., Vivekanand, V., and Pareek, N. (2018). Chitinases—potential candidates for enhanced plant resistance towards fungal pathogens. Agriculture 8:88. doi: 10.3390/agriculture8070088
Kumari, B., Mallick, M., Solanki, M. K., Solanki, A. C., Hora, A., and Guo, W. (2019). Plant growth promoting rhizobacteria (PGPR): modern prospects for sustainable agriculture. Plant Health Under Biotic Stress 2019, 109–127. doi: 10.1007/978-981-13-6040-4_6
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. PNAS 107, 2361–2366. doi: 10.1073/pnas.0912516107
Kwon, Y. S., Lee, D. Y., Rakwal, R., Baek, S. B., Lee, J. H., Kwak, Y. S., et al. (2016). Proteomic analyses of the interaction between the plant-growth promoting rhizobacterium Paenibacillus polymyxa E681 and Arabidopsis thaliana. Proteomics 16, 122–135. doi: 10.1002/pmic.201500196
Lamizadeh, E., Enayatizamir, N., Motamedi, H., and Sciences, A. (2016). Isolation and identification of plant growth-promoting rhizobacteria (PGPR) from the rhizosphere of sugarcane in saline and non-saline soil. Int. J. Curr. Microbiol. 5, 1072–1083. doi: 10.20546/ijcmas.2016.510.113
Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Biol. 49, 199–222.
Li, M., Yang, Y., Feng, F., Zhang, B., Chen, S., Yang, C., et al. (2017). Differential proteomic analysis of replanted Rehmannia glutinosa roots by iTRAQ reveals molecular mechanisms for formation of replant disease. BMC Plant Biol. 17:116. doi: 10.1186/s12870-017-1060-0
Li, Y.-R., and Yang, L.-T. (2015). Sugarcane agriculture and sugar industry in China. Sugar Tech 17, 1–8. doi: 10.1007/s12355-014-0342-1
Lin, L., Guo, W., Xing, Y.-X., Zhang, X.-C., Li, Z.-Y., Hu, C.-J., et al. (2012). The actinobacterium Microbacterium sp. 16SH accepts pBBR1-based pPROBE vectors, forms biofilms, invades roots, and fixes N 2 associated with micropropagated sugarcane plants. Appl. Microbiol. Biotechnol. 93, 1185–1195. doi: 10.1007/s00253-011-3618-3
Liu, M., Gong, Y., Sun, H., Zhang, J., Zhang, L., Sun, J., et al. (2020). Characterization of a novel chitinase from sweet potato and its fungicidal effect against ceratocystis fimbriata. J. Agricult. Food Chem. 68, 7591–7600. doi: 10.1021/acs.jafc.0c01813
Lü, Y., Zhang, S., Wang, J., and Hu, Y. (2016). Quantitative proteomic analysis of wheat seeds during artificial ageing and priming using the isobaric tandem mass tag labeling. PLoS One 11:e0162851. doi: 10.1371/journal.pone.0162851
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 541–556.
Ma, J., Chen, T., Wu, S., Yang, C., Bai, M., Shu, K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217.
Ma, Q., Shi, C., Su, C., and Liu, Y. (2020). Complementary analyses of the transcriptome and iTRAQ proteome revealed mechanism of ethylene dependent salt response in bread wheat (Triticum aestivum L.). Food Chem. 325:126866. doi: 10.1016/j.foodchem.2020.126866
Mathivanan, S., Chidambaram, A., Robert, G. A., and Kalaikandhan, R. (2017). Impact of PGPR inoculation on photosynthetic pigment and protein. J. Sci. Agricult. 1, 29–36. doi: 10.25081/jsa.2017.v1i0.24
Matse, D. T., Huang, C.-H., Huang, Y.-M., and Yen, M.-Y. (2020). Effects of coinoculation of Rhizobium with plant growth promoting rhizobacteria on the nitrogen fixation and nutrient uptake of Trifolium repens in low phosphorus soil. J. Plant Nutrit. 43, 739–752. doi: 10.1080/01904167.2019.1702205
May, M. J., Vernoux, T., Leaver, C., Montagu, M. V., and Inzé, D. (1998). Glutathione homeostasis in plants: implications for environmental sensing and plant development. J. Exp. Bot. 49, 649–667. doi: 10.1093/jexbot/49.321.649
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., and Dubery, I. A. (2018). The Chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9:112. doi: 10.3389/fpls.2018.00112
Myers, A. M., Morell, M. K., James, M. G., and Ball, S. G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–998. doi: 10.1104/pp.122.4.989
Naher, U. A., Panhwar, Q. A., Othman, R., Shamshuddin, J., and Zhou, E. J. P. J. O. B. (2018). Proteomic study on growth promotion of PGPR inoculated aerobic rice (Oryza sativa L.) cultivar MR219-9. Pak. J. Bot. 50, 1843–1852.
Nakabayashi, R., and Saito, K. (2015). Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 24, 10–16. doi: 10.1016/j.pbi.2015.01.003
Ngumbi, E., and Kloepper, J. (2016). Bacterial-mediated drought tolerance: current and future prospects. Appl. Soil Ecol. 105, 109–125. doi: 10.1016/j.apsoil.2016.04.009
Nianiou-Obeidat, I., Madesis, P., Kissoudis, C., Voulgari, G., Chronopoulou, E., Tsaftaris, A., et al. (2017). Plant glutathione transferase-mediated stress tolerance: functions and biotechnological applications. Plant Cell Rep. 36, 791–805. doi: 10.1007/s00299-017-2139-7
Orozco-Mosqueda, M. D. C., Glick, B. R., and Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 235:126439. doi: 10.1016/j.micres.2020.126439
Pan, J., Peng, F., Xue, X., You, Q., Zhang, W., Wang, T., et al. (2019). The growth promotion of two salt-tolerant plant groups with PGPR inoculation: a meta-analysis. Sustainability 11:378. doi: 10.3390/su11020378
Parsaee, M., Kiani Deh, Kiani, M., and Karimi, K. (2019). A review of biogas production from sugarcane vinasse. Biomass Bioener. 122, 117–125. doi: 10.1016/j.biombioe.2019.01.034
Perkins-Veazie, P., Collins, J., Russo, V., and Cartwright, B. (1996). Aphids Stimulate Peroxidase Activity but Not ACC Oxidase Activity in Watermelon Plants Inoculated with Anthracnose. HortScience 31, 677d–677d.
Piromyou, P., Buranabanyat, B., Tantasawat, P., Tittabutr, P., Boonkerd, N., and Teaumroong, N. (2011). Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur. J. Soil Biol. 47, 44–54. doi: 10.1016/j.ejsobi.2010.11.004
Qiao, F., Yang, X., Xu, F., Huang, Y., Zhang, J., Song, M., et al. (2021). TMT-based quantitative proteomic analysis reveals defense mechanism of wheat against the crown rot pathogen Fusarium pseudograminearum. BMC Plant Biol. 21:82. doi: 10.1186/s12870-021-02853-6
Raklami, A., Oufdou, K., Tahiri, A.-I., Mateos-Naranjo, E., Navarro-Torre, S., Rodríguez-Llorente, I. D., et al. (2019). Safe cultivation of Medicago sativa in metal-polluted soils from semi-arid regions assisted by heat-and metallo-resistant PGPR. Microorganisms 7:212. doi: 10.3390/microorganisms7070212
Reis, V. M., and Teixeira, K. R. (2015). Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. J. Basic Microbiol. 55, 931–949. doi: 10.1002/jobm.201400898
Sáez-Plaza, P., Michałowski, T., Navas, M. J., Asuero, A. G., and Wybraniec, S. (2013). An overview of the kjeldahl method of nitrogen determination. part i. early history, chemistry of the procedure, and titrimetric finish. Crit. Rev. Analyt. Chem. 43, 178–223. doi: 10.1080/10408347.2012.751786
Samaniego-Gámez, B. Y., Garruña, R., Tun-Suárez, J. M., Kantun-Can, J., Reyes-Ramírez, A., and Cervantes-Díaz, L. (2016). Bacillus spp. inoculation improves photosystem II efficiency and enhances photosynthesis in pepper plants. Chilean J. Agricult. Res. 76, 409–416. doi: 10.4067/s0718-58392016000400003
Sen, C. K. (1999). Glutathione homeostasis in response to exercise training and nutritional supplements. Stress Adaptat. Prophylaxis Treat. 1999, 31–42. doi: 10.1007/978-1-4615-5097-6_4
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., and Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24:2452. doi: 10.3390/molecules24132452
Sharma, N., Sharma, K., Gaur, R., and Gupta, V. (2011). Role of chitinase in plant defense. Asian J. Biochem. 6, 29–37. doi: 10.3923/ajb.2011.29.37
Shen, X., Hu, H., Peng, H., Wei, W., and Zhang, X. (2013). Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14, 271–271. doi: 10.1186/1471-2164-14-271
Shi, L., Ge, B., Wang, J., Liu, B., Ma, J., Wei, Q., et al. (2019). iTRAQ-based proteomic analysis reveals the mechanisms of Botrytis cinerea controlled with Wuyiencin. BMC Microbiol. 19:280. doi: 10.1186/s12866-019-1675-4
Siddiqui, Z. A. (2005). PGPR: prospective biocontrol agents of plant pathogens. PGPR Biocontr. Biofertiliz. 2005, 111–142. doi: 10.1007/1-4020-4152-7_4
Singh, P., Singh, R. K., Li, H. B., Guo, D. J., Sharma, A., Lakshmanan, P., et al. (2021b). Diazotrophic bacteria Pantoea dispersa and Enterobacter asburiae promote sugarcane growth by inducing nitrogen uptake and defense-related gene expression. Front. Microbiol. 11:600417. doi: 10.3389/fmicb.2020.600417
Singh, P., Singh, R. K., Song, Q.-Q., Li, H.-B., Guo, D.-J., Malviya, M. K., et al. (2021a). Comparative analysis of protein and differential responses of defense-related gene and enzyme activity reveals the long-term molecular responses of sugarcane inoculated with Sporisorium scitamineum. J. Plant Interact. 16, 12–29. doi: 10.1080/17429145.2020.1867770
Singh, R. K., Singh, P., Li, H.-B., Guo, D.-J., Song, Q.-Q., Yang, L.-T., et al. (2020). Plant-PGPR interaction study of plant growth-promoting diazotrophs Kosakonia radicincitans BA1 and Stenotrophomonas maltophilia COA2 to enhance growth and stress-related gene expression in Saccharum spp. J. Plant Interact. 15, 427–445. doi: 10.1080/17429145.2020.1857857
Singh, R. K., Singh, P., Li, H.-B., Yang, L.-T., and Li, Y.-R. (2017). Soil–plant–microbe interactions: use of nitrogen-fixing bacteria for plant growth and development in sugarcane. Plant Microbe Interact. Agro Ecol. Perspect. 2017, 35–59. doi: 10.1007/978-981-10-5813-4_3
Sivasakthi, S., Usharani, G., and Saranraj, P. (2014). Biocontrol potentiality of plant growth promoting bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agricult. Res. 9, 1265–1277.
Su, F., Jacquard, C., Villaume, S., Michel, J., Rabenoelina, F., Clément, C., et al. (2015). Burkholderia phytofirmans PsJN reduces impact of freezing temperatures on photosynthesis in Arabidopsis thaliana. Front. Plant Sci. 6:00810. doi: 10.3389/fpls.2015.00810
Tsukanova, K., Meyer, J., and Bibikova, T. (2017). Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 113, 91–102. doi: 10.1016/j.sajb.2017.07.007
Urquiaga, S., Cruz, K., and Boddey, R. M. (1992). Contribution of nitrogen fixation to sugar cane: nitrogen-15 and nitrogen-balance estimates. Soil Sci. Soc. Am. J. 56, 105–114. doi: 10.2136/sssaj1992.03615995005600010017x
Wang, Z., Solanki, M. K., Pang, F., Singh, R. K., Yang, L.-T., Li, Y.-R., et al. (2016). Identification and efficiency of a nitrogen-fixing endophytic actinobacterial strain from sugarcane. Sugar Tech 19, 492–500. doi: 10.1007/s12355-016-0498-y
Wang, J., Wang, X. R., Zhou, Q., Yang, J. M., Guo, H. X., Yang, L. J., et al. (2016). iTRAQ protein profile analysis provides integrated insight into mechanisms of tolerance to TMV in tobacco (Nicotiana tabacum). J. Proteomics 132, 21–30. doi: 10.1016/j.jprot.2015.11.009
Wang, Y., Zhang, X., Huang, G., Feng, F., Liu, X., Guo, R., et al. (2019). iTRAQ-based quantitative analysis of responsive proteins under PEG-induced drought stress in wheat leaves. Int. J. Mol. Sci. 20:ijms20112621. doi: 10.3390/ijms20112621
Wang, Z., Solanki, M. K., Yu, Z.-X., Yang, L.-T., An, Q.-L., Dong, D.-F., et al. (2019). Draft genome analysis offers insights into the mechanism by which Streptomyces chartreusis WZS021 increases drought tolerance in sugarcane. Front. Microbiol. 9:3262. doi: 10.3389/fmicb.2018.03262
Wei, C.-Y., Lin, L., Luo, L.-J., Xing, Y.-X., Hu, C.-J., Yang, L.-T., et al. (2014). Endophytic nitrogen-fixing Klebsiella variicola strain DX120E promotes sugarcane growth. Biol. Fertil. Soils 50, 657–666. doi: 10.1007/s00374-013-0878-3
Wu, S., Cao, G., Adil, M. F., Tu, Y., Wang, W., Cai, B., et al. (2020). Changes in water loss and cell wall metabolism during postharvest withering of tobacco (Nicotiana tabacum L.) leaves using tandem mass tag-based quantitative proteomics approach. Plant Physiol. Biochem. 150, 121–132. doi: 10.1016/j.plaphy.2020.02.040
Xie, H., Yang, D.-H., Yao, H., Bai, G., Zhang, Y.-H., and Xiao, B.-G. (2016). iTRAQ-based quantitative proteomic analysis reveals proteomic changes in leaves of cultivated tobacco (Nicotiana tabacum) in response to drought stress. Biochem. Biophys. Res. Commun. 469, 768–775. doi: 10.1016/j.bbrc.2015.11.133
Xie, S., Wu, H., Chen, L., Zang, H., Xie, Y., and Gao, X. (2015). Transcriptome profiling of Bacillus subtilis OKB105 in response to rice seedlings. BMC Microbiol. 15:21. doi: 10.1186/s12866-015-0353-4
Xue, J., Tang, Y., Wang, S., Xue, Y., Liu, X., Zhang, X., et al. (2019). Evaluation of dry and wet storage on vase quality of cut peony based on the regulation of starch and sucrose metabolism. Postharv. Biol. 155, 11–19. doi: 10.1016/j.postharvbio.2019.05.007
Keywords: Enterobacter roggenkampii ED5, endophyte, N-fixation, PGP, proteome, sugarcane, TMT
Citation: Guo D-J, Li D-P, Singh RK, Singh P, Sharma A, Verma KK, Qin Y, Khan Q, Lu Z, Malviya MK, Song X-P, Xing Y-X and Li Y-R (2021) Differential Protein Expression Analysis of Two Sugarcane Varieties in Response to Diazotrophic Plant Growth-Promoting Endophyte Enterobacter roggenkampii ED5. Front. Plant Sci. 12:727741. doi: 10.3389/fpls.2021.727741
Received: 19 July 2021; Accepted: 18 October 2021;
Published: 23 November 2021.
Edited by:
Ying Ma, University of Coimbra, PortugalReviewed by:
Prabhat Nath Jha, Birla Institute of Technology and Science, IndiaCopyright © 2021 Guo, Li, Singh, Singh, Sharma, Verma, Qin, Khan, Lu, Malviya, Song, Xing and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Xiu Xing, RG9jdW1lbnQxMjZAMTI2LmNvbQ==; Yang-Rui Li, bGl5ckBneGFhcy5uZXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.