- 1State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai'an, China
- 2Agronomy College of Shandong Agricultural University, Tai'an, China
- 3State Key Laboratory of Crop Stress Biology for Arid Areas and College of Agronomy, Northwest A&F University, Yangling, China
Thinopyrum ponticum (2n = 10x = 70) is a wild relative of wheat with high tolerance to both biotic and abiotic stresses; it has been wildly used in wheat genetic improvement. A disomic substitution line named SN19647 was derived from a cross between Triticum aestivum and the wheat–Th. ponticum partial amphiploid SNTE20 (2n = 8x = 56). It was evaluated for disease resistance and characterized via sequential fluorescence in situ hybridization (FISH)-genomic in situ hybridization (GISH) and molecular markers. The results showed that SN19647 carried resistance to both powdery mildew and leaf rust. It contained 42 chromosomes with a pair of wheat chromosome 1B replaced by a pair of JS chromosomes from Th. ponticum. In addition to chromosomal substitution events, structural variation also occurred on wheat chromosomes 2A, 5A, 6B, and 7B. Based on marker analysis, 19 markers specific to the JS chromosome were obtained, of which seventeen markers belonged to homoeologous group one. These results indicated that SN19647 was a 1JS (1B) substitution line. Compared with the known 1JS (1D) substitution line CH10A5, it was found that 17 markers generated different specific bands to Th. ponticum, confirming the novelty of the 1JS chromosome in SN19647. Therefore, SN19647, resistant to powdery mildew and leaf rust, was a novel 1JS (1B) substitution line that can be used in wheat genetic improvement.
Introduction
Wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is cultivated in diverse geographical regions, environments, and production systems, occupying approximately 220 million ha worldwide (Singh et al., 2016). With the global population estimated to exceed 9 billion by 2050, a 1.6% annual increase in wheat production has been expected to satisfy the increasing population, with the projection of an enhanced yield from 3 to 5 tons/ha (Tilman et al., 2002; Singh et al., 2016). However, climate change and disease occurrence are threatening wheat productivity (Price et al., 2013; Curtis and Halford, 2014; Lobell, 2019).
Powdery mildew is a devastating foliar disease of wheat especially in regions with a cool, maritime climate. It is caused by the fungus Blumeria graminis f. sp. tritici (Bgt). The epidemic that powdery mildew causes will destroy leaf and sheath tissues and significantly reduces tiller number, grain number and kernel weight (Conner et al., 2003), resulting in yield losses. Leaf rust, caused by the fungus Puccinia triticina (Pt), is another major wheat disease that frequently occurs (Bolton et al., 2008). It causes significant yield losses by affecting biomass, kernel weight and kernel number per acre (Herrera-Foessel et al., 2006). Although fungicides can be used to control these two kinds of diseases, breeding resistant cultivars are considered a cost-effective and eco-friendly approach (Wang et al., 2005; Liu et al., 2017). This breeding process is based on exploration and utilization of resistance genes.
So far, there are 68 (Pm1 to Pm68) powdery mildew resistance genes and 79 (Lr1 to Lr79) leaf rust resistance genes which have been cataloged in wheat and its relatives (Qureshi et al., 2018; He et al., 2021). However, many of them are overcome by new virulent pathogens. Previous studies showed that resistance genes from wheat-related species generally had a wider resistance spectrum and better durability than those from common wheat (Klymiuk et al., 2018; Wang et al., 2018). Therefore, it has become increasingly important to identify new sources of disease resistance from wild relatives via distant hybridization (Wang et al., 2016; Mo et al., 2017).
Thinopyrum ponticum (2n = 10x = 70, EeEeEbEbExExStStStSt or JJJJJJJSJSJSJS), a perennial wild relative of wheat, has proven to be a valuable source for resistance to various wheat diseases (Li and Wang, 2009). Up to now, there have been 11 genes formally designated from Th. ponticum, including powdery mildew resistance gene Pm51 and leaf rust resistance genes Lr19, Lr24 and Lr29 (Li and Wang, 2009; Zhan et al., 2014). However, none of them originated from the homoeologous group one of Th. ponticum. Recently, the 1JS chromosome of Th. ponticum in the 1JS (1D) disomic substitution line CH10A5 was reported to be probably responsible for resistance to both powdery mildew and stripe rust (Wang et al., 2020).
In this study, a novel wheat–Th. ponticum 1JS (1B) disomic substitution line with resistance to powdery mildew and leaf rust was developed from a hybrid of the common wheat cultivar Jimai22 and wheat-Th. ponticum octoploid SNTE20, and named SN19647. Methods of disease evaluation, genomic in situ hybridization (GISH), fluorescence in situ hybridization (FISH) and molecular markers were employed to identify its disease resistance, chromosomal composition and variation. The alien chromosomes in SN19647 were also compared with those in CH10A5.
Materials and Methods
Plant Materials
The wild relatives used in this study included Th. ponticum (2n = 10x = 70, EeEeEbEbExExStStStSt or JJJJJJJSJSJSJS) and Pseudoroegneria spicata (2n = 2x = 14, StSt). Th. ponticum (accession No. R431) was provided by Prof. Zhensheng Li, formerly of the Northwest Institute of Botany, the Chinese Academy of Sciences, Yangling, China. P. spicata was provided by Prof. Lihui Li, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China. Other materials included SNTE20, Yannong15 (YN15), Shannongfu63 (SNF63), Jimai22 (JM22), Huixianhong (HXH), and CH10A5. Wheat-Th. ponticum octoploid SNTE20, developed from the cross of Th. ponticum/YN15//SNF63, was used as the female parent. A predominant wheat cultivar in Shandong Province which is JM22 was used as the male parent to cross with SNTE20 and backcrossed with the resulting F1 progeny, and then SN19647 was selected in the generation BC1F4. For disease resistance evaluation, HXH served as the susceptible control. The 1JS (1D) substitution line CH10A5, provided by Pro. Changyou Wang, Agronomy College of Northwest A&F University, Yangling, China, was used to compare the 1JS chromosomes in the two substitution lines.
Evaluation of Powdery Mildew and Leaf Rust Resistance
Seedling powdery mildew resistance was performed in a growth chamber using the Bgt isolate E09, following the method stated in the study by Zhao et al. (2013). Seedlings were grown in rectangular plastic trays (5 × 5 cm, 10 plants per tray) and inoculated with fresh Bgt conidiospores obtained from the susceptible cultivar HXH at the one-leaf stage. After ~2 weeks, when symptoms were severe on the susceptible HXH, infection types (ITs) on the plants were described using a 0–4 infection scale: 0–2 scores indicate resistance while 3–4 indicate susceptibility. At the adult stage, resistance to powdery mildew and leaf rust was evaluated after natural infection in field-grown plants at the Experimental Station of Shandong Agricultural University over three growing seasons (2018–2020). The most severe reaction type in a given year was considered to be the final resistance results. Fifteen plants were grown in each 1.5-m long row, with 25 cm spacing between the rows. HXH was planted perpendicular and adjacent to the test rows to serve as an inoculum spreader and a susceptible control. The disease symptoms were recorded three times at weekly intervals after flowering, and the most severe infection score was used as the final response. The infection types of powdery mildew and leaf rust at the adult stage were scored using a 0–9 scale (Li et al., 2011) and a 0–4 scale (Roelfs et al., 1992), respectively.
In situ Hybridization
The chromosomes were prepared following the method described in the study by Kato et al. (2004). The purified total genomic DNA extracted from Th. ponticum and Ps. spicata were labeled with fluorescein-12-dUTP and used as probes, with the sheared genomic DNA from YN15 (AABBDD) as a blocker. Genomic in situ hybridization analysis was performed as described in the study by Fu et al. (2012). For FISH analysis, oligonucleotide probes, including TAMRA (6-carboxytetramethylrhodamine)-labeled oligonucleotides pAs1-1, pAs1-3, pAs1-4, pAs1-6, AFA-3 and AFA-4, and FAM (6-carboxyfluorescein)-labeled oligonucleotides pSc119.2-1 and (GAA)10, were used. All probes were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Fluorescence in situ hybridization analysis was performed as described in the study by Huang et al. (2018). The chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI), and the images were captured with a fluorescence microscope (Olympus BX60, Tokyo, Japan) equipped with a charge-coupled device (CCD) camera.
Molecular Marker Analysis
A total of 914 markers (Table 1), including simple sequence repeat (SSR)/expressed sequence tag (EST), which belonged to the first homologous group (https://wheat.pw.usda.gov/GG3/), intron targeting (IT) markers (Zhang et al., 2017), and PCR-based landmark unique gene (PLUG) markers (Wang et al., 2020), were synthesized by Sangon Biotech Co., Ltd. and used to identify the homeologous group of the alien chromosomes in SN19647 and compare their identity with those in CH10A5. PCR amplification system was a 10-μL reaction mixture, containing 40 ng genomic DNA, 2 μM each of forward and reverse primers, 2.5 mM each of the dNTPs, 2.5 mM MgCl2, 1× PCR buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl), and 0.5 U Taq DNA polymerase. It was carried out on a PTC-200 thermal cycler (Bio-Rad, Hercules, CA). The PCR products were resolved on 8% non-denaturing polyacrylamide gels, and the band patterns were visualized by silver staining.
Results
Assessment of Powdery Mildew and Leaf Rust Resistance
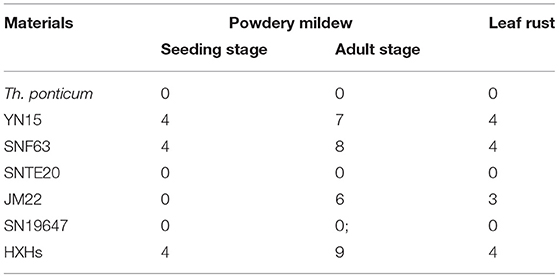
At the seedling stage, SN19647 and its parents were inoculated with the Bgt isolate E09, then the disease reaction was assessed once the susceptible control HXH was thoroughly infected. The results showed that Th. ponticum, SNTE20, JM22 and SN19647 were all resistant with an IT score of 0. Whereas the common wheat parents YN15 and SNF63 with an IT score of 4 appeared susceptible. These results suggested that the resistance to powdery mildew at the seeding stage in SN19647 originated from either Th. ponticum or JM22 (Figure 1A, Table 2).
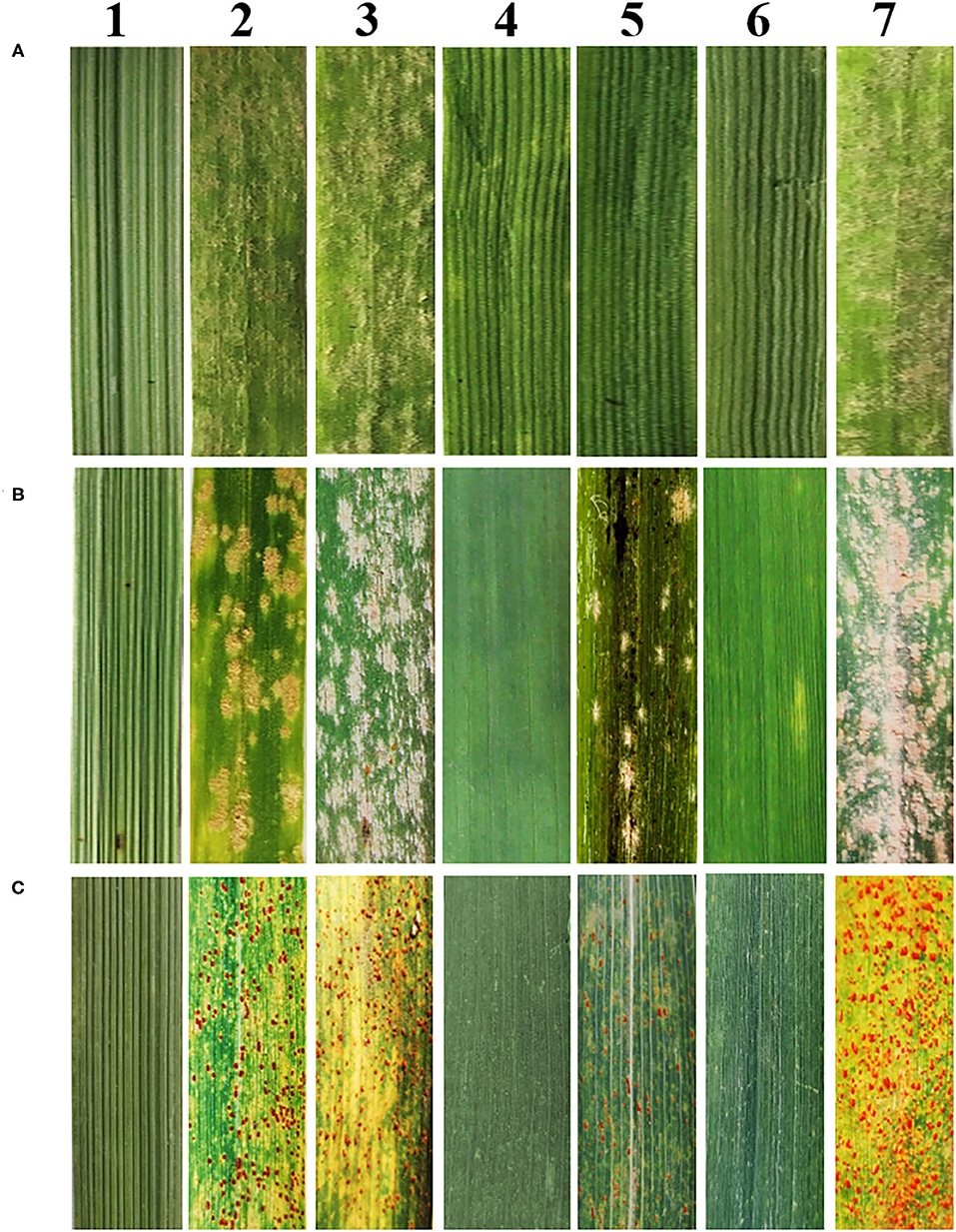
Figure 1. Reactions to powdery mildew and leaf rust. (A) Reactions to powdery mildew at the seedling stage. (B) Reactions to powdery mildew at the adult stage. (C) Reactions to leaf rust at the adult stage. 1–7 refers to Th. ponticum, YN15, SNF63, SNTE20, JM22, SN19647 and HXH, respectively.
At the adult stage, resistance to powdery mildew and leaf rust was tested in the field over three growing seasons (2018–2020), and the most severe reaction type observed in a given year was considered to be the final result. It was found that SN19647, Th. ponticum, and SNTE20 were resistant to powdery mildew, while YN15, SNF63 and JM22 showed susceptibility (Figure 1B, Table 2). Similar results were observed for leaf rust resistance at the adult stage in all the examined materials (Figure 1C, Table 2). Thus, these results collectively indicated that the resistance to two types of disease of SN19647 was inherited from Th. ponticum at the adult stage.
Cytological Characterization
Genomic in situ hybridization and FISH were performed to determine the chromosomal composition of SN19647. When probed with total genomic DNA of Th. ponticum and blocked with DNA of YN15, GISH showed that SN19647 contained two Th. ponticum chromosomes with distinct and uniform hybridization signals in addition to 40 wheat chromosomes counterstained by DAPI (Figure 2A). When the St-genomic DNA of Ps. spicata was used as a probe, the two alien chromosomes showed red signals in the centromeric regions and occasionally in the telomeric regions, which suggested that they belonged to the JS genome (Figure 2B).
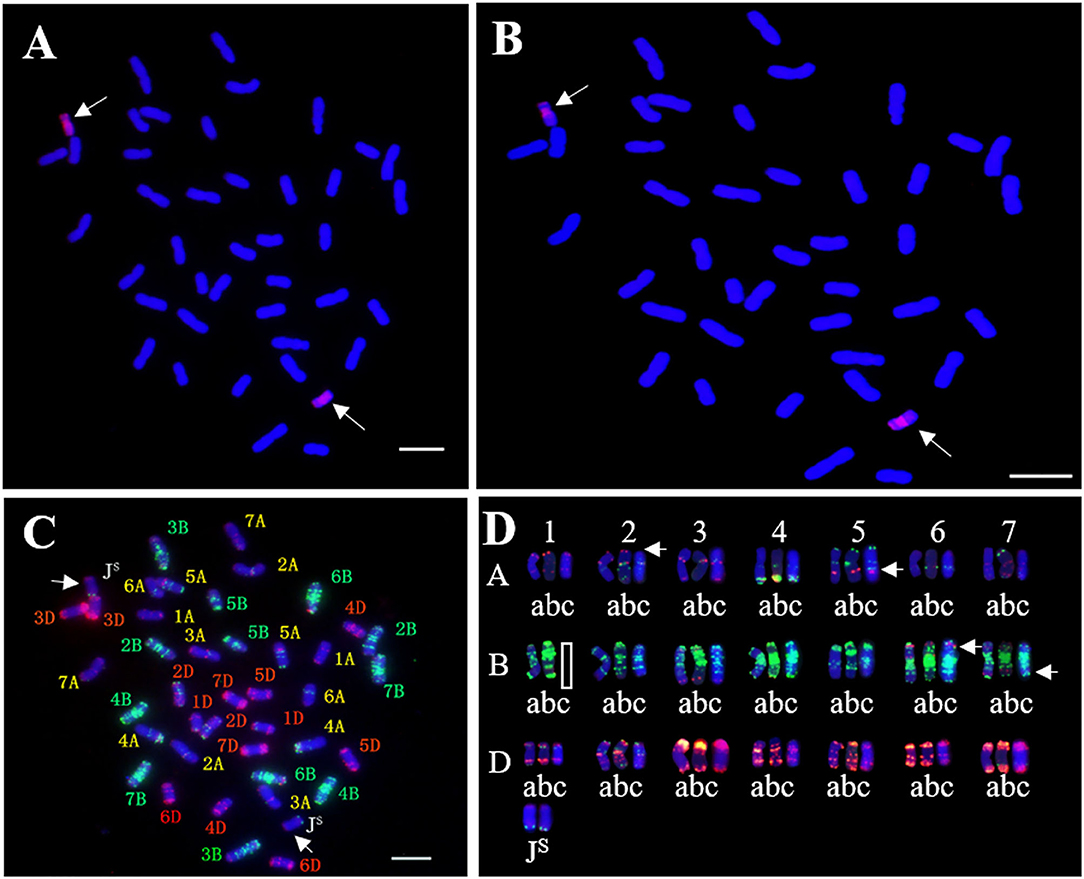
Figure 2. In situ hybridization analysis of SN19647. (A) GISH pattern of SN19647 probed with Th. ponticum genomic DNA. (B) GISH pattern of SN19647 probed with Ps. spicata genomic DNA. (C) FISH pattern of SN19647. (D) Comparison of wheat chromosomes in SNTE20 (a), JM22 (b) and SN19647 (c). Arrows indicate the chromosomes JS or locations with different FISH bands. Scale bars = 10 μm.
After removing the GISH signals, the slide was subjected to FISH analysis using eight probes. The JS chromosome pair produced red and green signals in the terminal regions of the short and long arms, respectively (Figure 2C). The wheat chromosome pair 1B was eliminated and 14 A-(1A-7A), 12 B-(2B-7B), and 14 D-(1D-7D) chromosomes were detected, indicating SN19647 as a disomic substitution line with a Th. ponticum JS chromosome pair replacing a wheat 1B chromosome pair. Additionally, FISH patterns of the chromosomes in SN19647 were compared with those in its parents, SNTE20 and JM22. Differences were found in the terminal region of 2AS, the middle of 5AL and 6BS, and the region close to the terminal in 7BL (Figure 2D), which suggested that these chromosomes underwent structural variations with the formation of SN19647.
Molecular Marker Analysis
To determine the homoeologous group of the JS chromosome introduced to SN19647, 841 IT and 65 EST/SSR primer pairs were screened among Th. ponticum, SN19647 and its parents, SNTE20 and JM22. In total, 12 IT markers, 3 EST markers and one SSR marker showed amplification with prominent bands specific to Th. ponticum in SN19647, which were absent in the common wheat parent JM22 (Figure 3). Among them, 14 markers, including one SSR, three EST and 10 IT markers belonged to the first homoeologous group (Table 3), indicating 1JS as the introduced Th. ponticum chromosome in SN19647. Thus, SN19647 was confirmed as a wheat-Th. ponticum 1JS (1B) disomic substitution line.
Figure 3. Molecular marker analysis of SN19647. (A) CINAU851. (B) CINAU857. (C) CINAU859. (D) CINAU862. (E) CINAU865. (F) CINAU866. (G) CINAU870. (H) CINAU880. (I) CINAU889. (J) CINAU890. (K) CINAU971. (L) CINAU1616. (M) Xgwm268. (N) SWES650. (O) Xmag3030. (P) BE405232. Lanes: M, DL2000 marker; 1-4 refer to Th. ponticum, SNTE20, JM22, and SN19647, respectively. Arrows indicate the specific bands of Th. ponticum.
Recently, CH10A5 was reported as a wheat–Th. ponticum 1JS (1D) disomic substitution line (Wang et al., 2020). To further compare the 1JS chromosomes in SN19647 and CH10A5, eight markers specific to the 1JS chromosome of CH10A5 were used to screen the two substitution lines. The results showed that only Xcfd63 produced the same Th. ponticum band in SN19647 and CH10A5, while others exhibited differences (Figure 4). Four markers, including two PLUG markers (TNAC1044-HaeIII and TNAC1088- HaeIII) and two SSR markers (Xwmc432 and Xwmc93), worked only in CH10A5. Two PLUG markers (TNAC1021-TaqI and TNAC1026-HaeIII) and one EST marker (BG313767) showed amplification in both substitution lines but generated different bands specific to Th. ponticum.
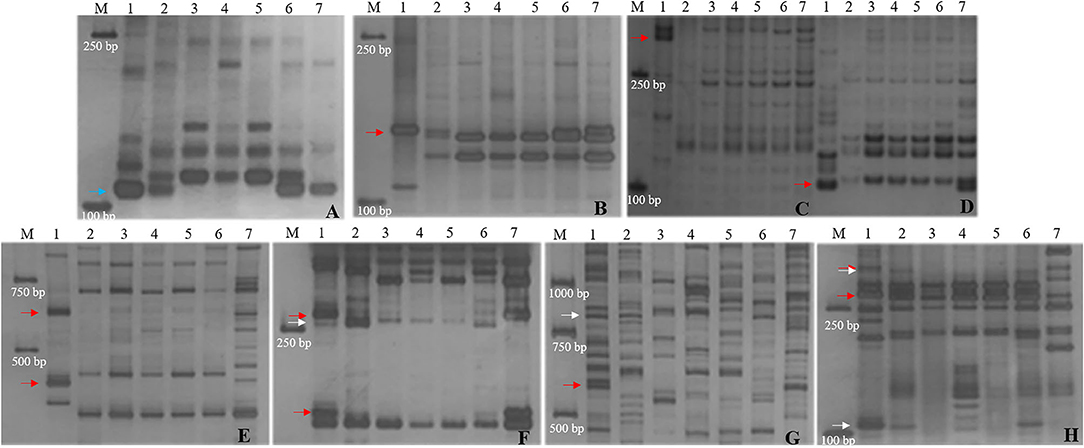
Figure 4. Molecular marker analysis of SN19747 and CH10A5 (specific markers in CH10A5). (A) Xcfd63. (B) TNAC1044-HaeIII. (C) Xwmc432. (D) Xwmc93. (E) HaeIII-TNAC1088. (F) TNAC1021-TaqI. (G) BG313767. (H) TNAC1026-HaeIII. Lanes: M, DL2000; 1-7 refer to Th. ponticum, SNTE20, SNF63, YN15, JM22, SN19647 and CH10A5, respectively. The blue, red and white arrows indicate the bands shared between SN19647 and CH10A5, specific to CH10A5 and SN19647, respectively.
Furthermore, the 14 markers specific to the 1JS chromosome of SN19647 were used to screen the two substitution lines. Four markers produced the same Th. ponticum band in SN19647 and CH10A5, and others exhibited differences (Figure 5). Ten markers, including five IT markers (CINAU851, CINAU859, CINAU865, CINAU870 and CINAU890), three EST markers (SWES650, BE405232 and Xmag3030) and one SSR marker (Xgwm268), worked only in SN19647. One IT marker (CINAU880) worked in both substitution lines but generated different bands specific to Th. ponticum. These observations indicated that SN19647 carried a novel pair of 1JS chromosomes different from that of CH10A5. Besides, three new Th. ponticum-specific markers were detected in SN19647 (TNAC1021-TaqI, BG313767, and TNAC1026-HaeIII) compared with CH10A5, among a total of nineteen Th. ponticum-specific markers. The results above indicated that the alien chromosomes in SN19647 were different from those in CH10A5.
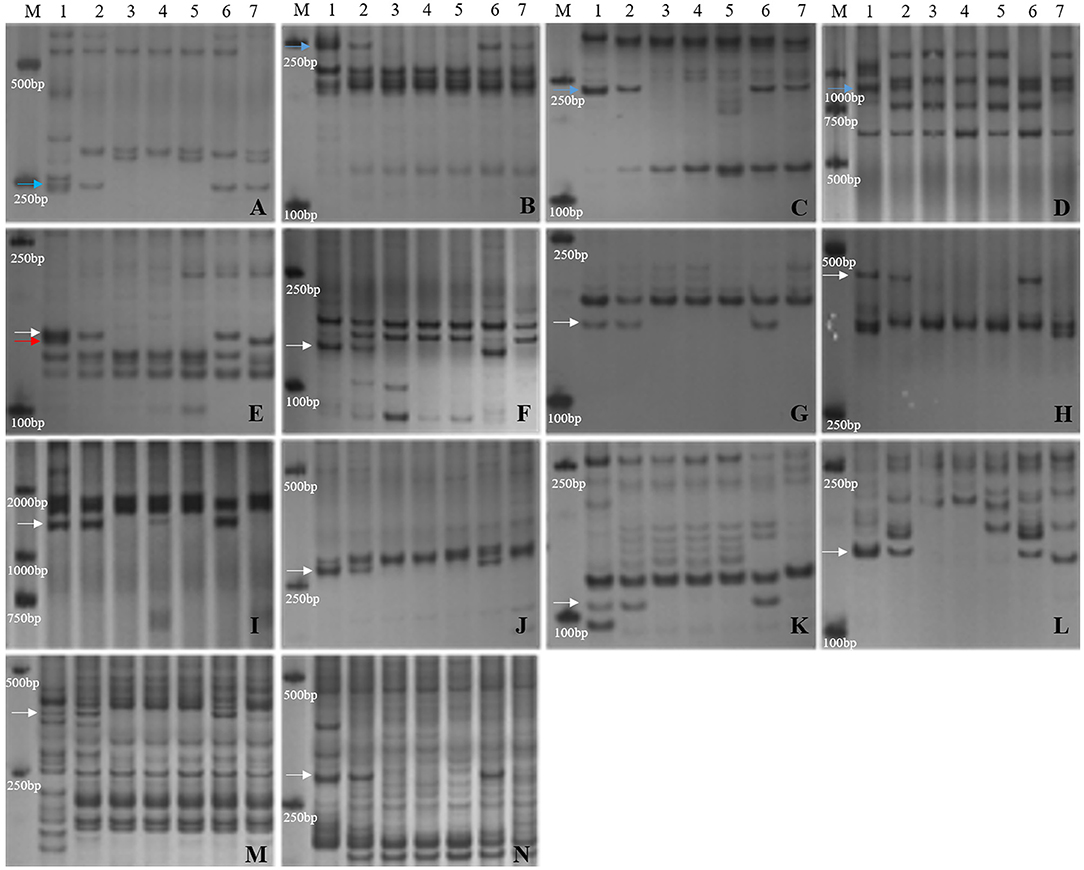
Figure 5. Molecular marker analysis of SN19647 and CH10A5 (specific markers in SN19647). (A) CINAU857. (B) CINAU889. (C) CINAU886. (D) CINAU862. (E) CINAU880. (F) CINAU851. (G) CINAU859. (H) CINAU865. (I) CINAU870. (J) CINAU890. (K) SWES650. (L) Xgwm268. (M) BE405232. (N) Xmag3030. Lanes: M, DL2000; 1-7 refer to Th. ponticum, SNTE20, SNF63, YN15, JM22, SN19647 and CH10A5, respectively. The blue, red, and white arrows indicate the band shared between SN19647 and CH10A5, specific to CH10A5 and SN19647, respectively.
Discussion
Gene transfer from wild relatives to common wheat has proven to be an effective approach for improving resistance. As Th. ponticum carried resistance to various diseases, many wheats—Th. ponticum chromosome lines, including partial amphiploid, addition, substitution and translocation, were developed. Among 11 genes cataloged from Th. ponticum, there were 1 and 3 resistant to powdery mildew and leaf rust, respectively. Furthermore, Pm51 was located on homoeologous 2 (Zhan et al., 2014); Lr19 and Lr29 were located on homoeologous 7, and Lr24 was derived from homoeologous 3 (Li and Wang, 2009). There has been no resistance originating from homoeologous 1 of Th. ponticum until the 1JS (1D) substitution line CH10A5 was reported to be resistant to powdery mildew and stripe rust in 2020 (Wang et al., 2020). In this study, the 1JS (1B) substitution line SN19647 exhibited resistance to powdery mildew and leaf rust. This is the first report of leaf rust resistance probably associated with the chromosome 1JS of Th. ponticum. As for powdery mildew, SN19647, Th. ponticum, SNTE20 and JM22 showed seedling immunity while other parents showed susceptibility. Hence, the origin of powdery mildew resistance is uncertain since JM22 carried PmJM22 resistant to the isolate E09 at the seedling stage (Yin et al., 2009; Fu et al., 2012). At the adult stage, PmJM22 seemed to be ineffective, resulting in susceptibility to powdery mildew. Compared to the resistance response with that of the parents, it was deductive that the 1JS chromosomes of Th. ponticum in SN19647 was responsible for powdery mildew and leaf rust resistance at the adult stage.
Among 22 markers, involving 8 markers specific to the 1JS chromosomes in CH10A5 and 14 markers specific to the 1JS chromosomes in SN19647, 17 were found to generate different specific bands of Th. ponticum between SN19647 and CH10A5. It demonstrated that the 1JS chromosomes in SN19647 were different from those in CH10A5. As known to us, the allodecaploid nature of Th. ponticum results in its large and complex genome. Chromosomes belonging to the same genome and homoeologous group may be different ones. According to the study of Chen et al. (1998), Th. ponticum possessed 14 pairs of JS-genomic chromosomes. Of them, there seemed to be more than one chromosome belonging to homoeologous group one in the JS genome, as shown here.
Fluorescence in situ hybridization is a technique that has been widely used to identify chromosome composition and detect specific loci. It has been recently modified to describe and track alien chromosomes introduced into the common wheat background based on the high-resolution karyotype patterns (Du et al., 2017; Kong et al., 2018). In the present study, the FISH pattern of the chromosome 1JS of Th. ponticum established 1JS by using the probe combination of AFA-3 (red), AFA-4 (red), pAs1-1 (red), pAs1-3 (red), pAs1-4 (red), pAs1-6 (red), pSc119.2-1 (green), and (GAA)10 (green). Additionally, structural variations were detected on wheat chromosomes 2A, 5A, 6B, and 7B. Studies have reported chromosomal recombination, chromosomal modifications, or genomic changes during interspecific hybridization (Liu et al., 2009; Li et al., 2015; Zheng et al., 2015). Moreover, selective breeding leads to structural rearrangements in the chromosomes (Huang et al., 2018). Therefore, the genomic variations detected in SN19647 may have occurred due to the introduction of Th. ponticum chromosomes and the subsequent artificial selection for increased adaptability.
Powdery mildew and leaf rust resistance genes transferred from alien genomes of wild Triticeae species are helpful to attenuate the yield losses due to powdery mildew and leaf rust. The development of new resistant materials through distant hybridization will lay the foundation for wheat genetic improvement. Thus, the wheat-Th. ponticum 1JS (1B) disomic substitution line SN19647, with resistance to powdery mildew and leaf rust, is a potential resource for exporating positive genes from Th. ponticum and breeding disease-resistant wheat lines. In order to improve the usefulness of SN19647, translocations with positive traits are being developed via pollen irradiation.
Conclusions
A disomic substitution line referred to as SN19647 has a novel pair of 1JS chromosomes from Th. ponticum. Its chromosome composition is 14A+12B+14D+2(1JS). In the formation process of SN19647, wheat chromosomes 2A, 5A, 6B, and 7B underwent structural variations. Additionally, 19 markers were identified as specific markers to the 1JS chromosome of Th. ponticum. Moreover, SN19647 possesses resistance to both powdery mildew and leaf rust. It can serve as positive germplasm for exploring resistance genes from Th. ponticum and breeding disease-resistant wheat lines.
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.
Author Contributions
YB designed the research. ML and YW performed the research. XiaL contributed to the disease assessment. XinL and HW contributed to the development of the materials. ML, YW, and YB wrote the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2016YFD0102004-02) and the Major Basic Research Program of Shandong Natural Science Foundation (ZR2019ZD15).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript.
References
Bolton, M. D., Kolmer, J. A., and Garvin, D. F. (2008). Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575. doi: 10.1111/j.1364-3703.2008.00487.x
Chen, Q., Conner, R. L., Laroche, A., and Thomas, J. B. (1998). Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 41, 580–586. doi: 10.1139/g98-055
Conner, R. L., Kuzyk, A. D., and Su, H. (2003). Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can. J. Plant Sci. 83, 725–728. doi: 10.4141/P03-043
Curtis, T., and Halford, N. G. (2014). Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 164, 354–372. doi: 10.1111/aab.12108
Du, P., Zhuang, L., Wang, Y., Yuan, L., Wang, Q., Wang, D., et al. (2017). Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60, 93–103. doi: 10.1139/gen-2016-0095
Fu, S., Lv, Z., Qi, B., Guo, X., Li, J., Liu, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium Head Blight. J. Genet. Genomics 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Crossa, J., Yuen, J., and Djurle, A. (2006). Effect of leaf rust on grain yield and yield traits of durum wheats with race-specific and slow-rusting resistance to leaf rust. Plant Dis. 90, 1065–1072. doi: 10.1094/PD-90-1065
Huang, X., Zhu, M., Zhuang, L., Zhang, S., Wang, J., Chen, X., et al. (2018). Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 131, 1967–1986. doi: 10.1007/s00122-018-3126-2
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Klymiuk, V., Yaniv, E., Huang, L., Raats, D., Fatiukha, A., Chen, S., et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9:3735. doi: 10.1038/s41467-018-06138-9
Kong, L., Song, X., Xiao, J., Sun, H., Dai, K., Lan, C., et al. (2018). Development and characterization of a complete set of Triticum aestivum–Roegneria ciliaris disomic addition lines. Theor. Appl. Genet. 131, 1793–1806. doi: 10.1007/s00122-018-3114-6
Li, H., Guo, X., Wang, C., and Ji, W. (2015). Spontaneous and divergent hexaploid triticales derived from common wheat x rye by complete elimination of D-genome chromosomes. PLoS ONE 10:e0120421. doi: 10.1371/journal.pone.0120421
Li, H., and Wang, X. (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. J. Genet. Genomics 36, 557–565. doi: 10.1016/S1673-8527(08)60147-2
Li, H. J., Wang, X. M., Song, F. J., Wu, C. P., Wu, X. F., Zhang, N., et al. (2011). Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron. Sin. 37, 943–954. doi: 10.3724/sp.j.1006.2011.00943
Liu, B., Xu, C., Zhao, N., Qi, B., Kimatu, J. N., Pang, J., et al. (2009). Rapid genomic changes in polyploid wheat and related species: implications for genome evolution and genetic improvement. J. Genet. Genomics 36, 519–528. doi: 10.1016/S1673-8527(08)60143-5
Liu, W., Koo, D. H., Xia, Q., Li, C., Bai, F., Song, Y., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
Lobell, D.B. (2019). Climate trends and global crop production since 1980. Meditat. Self. 333, 616–621. doi: 10.7551/mitpress/8876.003.0036
Mo, Q., Wang, C. Y., Chen, C. H., Wang, Y. J., Zhang, H., Liu, X. L., et al. (2017). Molecular cytogenetic identification of a wheat-Thinopyrum ponticum substitution line with stripe rust resistance. Cereal Res. Commun. 45, 564–573. doi: 10.1556/0806.45.2017.037
Price, J. A., Simmons, A. R., Rashed, A., Workneh, F., and Rush, C. M. (2013). Winter wheat cultivars with temperature-sensitive resistance to wheat streak mosaic virus do not recover from early-season infections. Plant Dis. 98, 525–531. doi: 10.1094/PDIS-04-13-0455-RE
Qureshi, N., Bariana, H., Kumran, V. V., Muruga, S., Forrest, K. L., Hayden, M. J., et al. (2018). A new leaf rust resistance gene Lr79 mapped in chromosome 3BL from the durum wheat landrace Aus26582. Theor. Appl. Genet. 131, 1091–1098. doi: 10.1007/s00122-018-3060-3
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Diseases of Wheat: Concepts and Methods of Disease Management. Mexico, DF: CIMMYT.
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustaiability and intensive production practices. Nature 418, 671–677. doi: 10.1038/nature01014
Wang, Y., Cao, Q., Zhang, J., Wang, S., Chen, C., Wang, C., et al. (2020). Cytogenetic analysis and molecular marker development for a new wheat–Thinopyrum ponticum 1JS (1D) disomic substitution line with resistance to stripe rust and powdery mildew. Front. Plant Sci. 11:1282. doi: 10.3389/fpls.2020.01282
Wang, Y., Quan, W., Peng, N., Wang, C., Yang, X., Liu, X., et al. (2016). Molecular cytogenetic identification of a wheat–Aegilops geniculata Roth 7Mg disomic addition line with powdery mildew resistance. Mol. Breed. 36, 1–10. doi: 10.1007/s11032-016-0463-1
Wang, Y., Zhang, H., Xie, J., Guo, B., Chen, Y., Zhang, H., et al. (2018). Mapping stripe rust resistance genes by BSR-Seq: YrMM58 and YrHY1 on chromosome 2AS in Chinese wheat lines Mengmai 58 and Huaiyang 1 are Yr17. Crop J. 6, 91–98. doi: 10.1016/j.cj.2017.03.002
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/PD-89-0457
Yin, G., Li, G., He, Z., Liu, J., Wang, H., and Xia, X. (2009). Molecular mapping of powdery mildew resistance gene in Wheat cultivar Jimai 22. Acta Agron. Sin. 35, 1425–1431. doi: 10.3724/sp.j.1006.2009.01425
Zhan, H., Li, G., Zhang, X., Li, X., Guo, H., Gong, W., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE 9:e113455. doi: 10.1371/journal.pone.0113455
Zhang, X., Wei, X., Xiao, J., Yuan, C., Wu, Y., Cao, A., et al. (2017). Whole genome development of intron targeting (IT) markers specific for Dasypyrum villosum chromosomes based on next-generation sequencing technology. Mol. Breed. 37, 1–11. doi: 10.1007/s11032-017-0710-0
Zhao, Z., Sun, H., Song, W., Lu, M., Huang, J., Wu, L., et al. (2013). Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor. Appl. Genet. 126, 3081–3089. doi: 10.1007/s00122-013-2194-6
Keywords: Thinopyrum ponticum, substitution line, GISH, FISH, powdery mildew, leaf rust
Citation: Li M, Wang Y, Liu X, Li X, Wang H and Bao Y (2021) Molecular Cytogenetic Identification of a Novel Wheat–Thinopyrum ponticum 1JS (1B) Substitution Line Resistant to Powdery Mildew and Leaf Rust. Front. Plant Sci. 12:727734. doi: 10.3389/fpls.2021.727734
Received: 19 June 2021; Accepted: 25 August 2021;
Published: 01 October 2021.
Edited by:
Sergio J. Ochatt, INRA UMR1347 Agroécologie, FranceReviewed by:
Peter Michael Dracatos, The University of Sydney, AustraliaMasahiro Kishii, International Maize and Wheat Improvement Center, Mexico
Copyright © 2021 Li, Wang, Liu, Li, Wang and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinguang Bao, YmFveWluZ3VhbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Mingzhu Li
Mingzhu Li Yanzhen Wang
Yanzhen Wang Xiaojuan Liu
Xiaojuan Liu Xingfeng Li
Xingfeng Li Honggang Wang
Honggang Wang Yinguang Bao
Yinguang Bao