- 1State Key Laboratory for Plant Gene and Protein Research, School of Life Sciences, School of Advanced Agriculture Sciences, Peking University, Beijing, China
- 2PKU Core Facility of Mass Spectrometry, School of Chemistry and Molecular Engineering, Peking University, Beijing, China
SPINDLY is involved in some aspects of plant development. However, the nature of this protein as an O-fucosyltransferase was recently discovered. In this study, we show that SPINDLY (SPY) interacts with CPN20 in yeast two-hybrid and split-luc assays, and the interaction is promoted by ABA. CPN20 is a chloroplast-localized co-chaperonin that negatively regulates ABAR-mediated ABA signaling. By using Electron Transfer Dissociation-MS/MS analysis, two O-fucosylation sites, e.g., 116th and 119th threonines, were detected in ectopically expressed CPN20 in mammalian cells and in Arabidopsis. The O-fucosylation at both threonine residues was confirmed by in vitro peptide O-fucosylation assay. We further show that CPN20 accumulates in the chloroplast of spy mutants, suggesting that SPY negatively regulates CPN20 localization in the chloroplast. In vivo protein degradation assay along with CPN20 localization behavior suggest that import of CPN20 into the chloroplast is negatively regulated by SPY. Genetic analysis shows that ABA insensitive phenotypes of spy-3 in terms of seed germination and early seedling development are partially suppressed by the cpn20 mutation, suggesting that CPN20 acts downstream of SPY in this ABA signaling pathway and that there may exist other pathways in parallel with CPN20. Collectively, the above data support the notion that the O-fucosylation of CPN20 by SPY fine-tunes ABA signaling in Arabidopsis.
Introduction
Glycosylation is a highly elaborate protein post-translational modification that occurs in eukaryotes and procaryotes (Wacker et al., 2002; Young et al., 2002). Other than the more studied N-linked and mucin-type O-linked modifications, O-linked monosaccharide glycosylations such as O-GlcNAc and O-fucosylation have been studied for the past 20 years (Hofsteenge et al., 2001; Okajima and Irvine, 2002; Panin et al., 2002; Luther and Haltiwanger, 2009; Lira-Navarrete et al., 2011; Bond and Hanover, 2015; Zentella et al., 2017). O-GlcNAc transferases (OGTs) catalyze the modification of hydroxyl groups of threonine and serine residues of a protein. Protein O-fucosyltransferases modify target proteins by transferring O-fucose from GDP-L-fucose to the hydroxy oxygen of serine and threonine residues. O-fucosylation occurs in the endoplasmic reticulum (ER), and proteins with EGF-like repeats (EGFs), such as Notch protein or proteins with Thrombospondin Repeats (TSRs), are also O-fucose modified (Hofsteenge et al., 2001; Okajima and Irvine, 2002; Luther and Haltiwanger, 2009; Lira-Navarrete et al., 2011; Chen et al., 2012). The role of O-fucosylation on the regulation of Notch signaling has been extensively studied (Reviewed by Rana and Haltiwanger, 2011, and references therein). O-fucosylation was found to facilitate the transmission of malaria (Lopaticki et al., 2017). However, how O-fucosylation functions in planta are less well-known.
Mutations affecting SPINDLY (SPY) suppress the phenotypes of gai and gibberellin acid (GA) deficient mutants in Arabidopsis. In contrast to wild-type, germination of spy mutants is not inhibited by the GA-synthesis inhibitor paclobutrazol, and spy mutants display GA overdosed phenotypes (Jacobsen and Olszewski, 1993; Wilson and Somerville, 1995). Although SPY was discovered from the investigation on GA signaling, a detailed phenotypic examination of spy mutants suggested multiple roles for SPY (Swain et al., 2001). SPY interacts with GIGANTEA to regulate the rhythms of cotyledon movement and it is involved in light responses (Tseng et al., 2004). SPY also plays a positive role in cytokinin (CK) signaling (Greenboim-Wainberg et al., 2005) and interacts with TCP14/15 in CK responses (Steiner et al., 2012), and it is responsible for TCP14 stability (Steiner et al., 2016). Spy mutants exhibit salt and drought tolerance, and some stress-responsive genes including DREB1E, LEA, RD20, AREB1-like are up-regulated in spy mutants (Qin et al., 2011). In addition, SPY was also shown to be involved in redox-mediated cortex proliferation in the root (Cui and Benfey, 2009; Cui et al., 2014; Cui, 2015). The SPY gene (Jacobsen et al., 1996) encodes a protein that is similar to mammalian OGT (Robertson et al., 1998). SPY and mammalian OGTs have similar overall structures: TPR domains at N-terminus and a catalytic domain at C-terminus of the protein (Thornton et al., 1999; Roos and Hanover, 2000). SPY had long been considered as a putative OGT but its OGT activity was never biochemically confirmed. In contrast, SPY was recently demonstrated to be an O-fucosyltransferase that O-fucosylates the DELLA protein, resistance gene analog (RGA) (Zentella et al., 2017). SPY, as a O-fucosyltransferase, plays an opposite role in regulating DELLA (Zentella et al., 2016, 2017; Camut et al., 2017) to SECRET AGENT (SEC), the sole putative Arabidopsis OGT, with sequence and structure similarities to SPY (Hartweck et al., 2002, 2006). Very recently, nuclear SPY was shown to facilitate PSEUDO-RESPONSE REGULATOR 5 (PRR5) proteolysis via its O-fucosylation to modulate Arabidopsis circadian clock (Wang et al., 2020).
The PYR/PYL/RCAR ABA receptor-mediated abscisic acid (ABA) signaling pathway has been widely accepted as the core ABA signaling pathway (Reviewed by Park et al., 2009; Santiago et al., 2009; Hsu et al., 2021). Except for the PYR/PYL/RCAR family of ABA receptors, a type of G-protein coupled receptor (GPCR) GCR1 (Liu et al., 2007), GPCR-type G proteins (GTG1 and GTG2) (Pandey et al., 2009), and Mg-chelatase H subunit (CHLH) (Shen et al., 2006) were also reported as putative ABA receptors. Mg-chelatase H subunit (CHLH), also called ABAR, resides across double membranes of chloroplasts (Shen et al., 2006; Wu et al., 2009; Shang et al., 2010). ABA-bound ABAR initiates ABA signaling by inhibiting the expression and nuclear localization of WRKY40, which is a transcription repressor that suppresses the expression of ABA-responsive transcription factors such as ABI5/ABI4 (Shang et al., 2010; Liu et al., 2012). CPN20, a co-chaperonin localized in chloroplast, an interaction partner of ABAR, is a negative regulator of ABA signaling at the same node with ABAR upstream of the WRKY40 transcription factor (Zhang et al., 2013). ABA inhibits the expression of CPN20 and suppresses interaction between CPN20 and ABAR, leading to enhanced interaction between WRKY40 and ABAR (Zhang et al., 2014).
Given that SPY plays important roles in multiple biological processes including stress responses, we were interested in further exploring the mechanisms of how SPY executes its functions. Genetic analyses showed that SPY has a complex relationship with ABI5, ABI3, and ABI4, in which SPY is generally upstream of ABI5, ABI3, and ABI4. However, SPY bypasses the core PYR-mediated ABA signaling pathway, as demonstrated by genetic analyses. Alternatively, SPY likely acts in the ABAR-mediated ABA signaling pathway (Liang et al., 2018).
When performing a yeast-two-hybrid screen for putative SPY interactors, we identified the co-chaperonin CPN20. In the present study, we showed that SPY-facilitated O-fucosylation of CPN20 may modulate ABA responses during seed germination and seedling development.
Materials and Methods
Plant Materials and Growth Conditions
All the Arabidopsis materials used in the study were in Col-0 background. Seeds were sown on Murashige and Skoog (MS) media (Sigma-Aldrich), stratified at 4°C in the dark, and then transferred to a chamber, germinated and grown for about 10 days at 22°C with a 16-h-light/8-h-dark cycle (light intensity of 120 μmol m−2 s−1). Then seedlings were potted in soil and transferred to a growth room at 22°C with a 16-h-light/8-h-dark cycle (light intensity of 120 μmol m−2 s−1).
Generation of Double Mutants
The gene cpn20-1 is a T-DNA insertion mutant (Zhang et al., 2013), while spy-3 is a point mutation (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). ♀spy-3♂cpn20-1 was generated by making a cross between spy-3 and cpn20-1. F1 progeny was identified by sequencing using primers SPY-3F and SPY-3R in Supplementary Table 1. F2 progeny germinating under 35 mg/L paclobutrazol were transferred to soil and checked for homozygosity of spy-3 by sequencing, and of cpn20-1 by PCR. Homozygous ♀spy-3♂cpn20-1 were confirmed by sequencing and PCR-based identification (Supplementary Table 1).
Seed Germination and Early Seedling Development Assays
For seed germination assays, different genotypes were grown in the same conditions and harvested almost at the same time. For each comparison, seeds were sown on MS medium supplemented with 1% sucrose and 0.8% agar with pH 5.8 with or without different concentrations of ABA. After 4-day stratification at 4°C in the dark, plants were transferred to a chamber at 22°C with a 16-h-light/8-h-dark cycle (light intensity of 120 μmol m−2 s−1). Rates of seed germination were scored and calculated at different days after germination. Germinated seeds were defined as the apparent emergence of the radicle out of the seed coat. The rates of green cotyledons were also calculated. Green cotyledons were defined as green and wide open cotyledons. The germinating seeds and seedlings were photographed at different times after germination.
Construction of Vectors
All the constructs used in this study are described below.
Constructs for Yeast Two-Hybrid Screening and Yeast Two-Hybrid Assays
pGBKT7-SPY
The SPY coding sequence was PCR amplified from cDNA and cloned to pGBKT7 vector (Clontech) between NcoI and SmaI using primers, SPY-Y2H-CDS-F1 and SPY-Y2H-CDS-R1 (Supplementary Table 1).
pGBKT7-11TPR
The 11-TPR domain was PCR amplified from cDNA and cloned to pGBKT7 vector in NcoI using SPY-Y2H-CDS-F1 and SPY-11TPR-R (Supplementary Table 1).
pGADT7-CPN20
The CPN20 coding sequence was PCR amplified from cDNA and cloned to pGADT7 between EcoRI and BamHI using CPN20-EcoR1-F and CPN20-BamH1-R (Supplementary Table 1).
Constructs for Bimolecular Fluorescence Complementation (BiFC) Assays
SPY-YFPN and CPN20-YFPC
Protein coding sequences (CDSs) of SPY and CPN20 were amplified by PCR using primer pairs 736-SPY-Sac1-V-F/736-SPY-Spe1-V-R and 735-CPN20-Sac1-V-F/735-CPN20-Spe1-V-R (Supplementary Table 1), respectively, and were inserted into pSY736 and pSY735 (Bracha-Drori et al., 2004), respectively, using One Step Cloning Kit (Vazyme) according to the user manual.
Constructs for Split-Luciferase Complementation (Split-Luc) Assays
Nluc-SPY, Cluc-CPN20, and Cluc-HFR1
CDSs of SPY, CPN20, and HFR1 were amplified by PCR and inserted into Nluc, Cluc, or Cluc vectors, respectively, between KpnI and SalI using primers SPY-CDS-Kpn1-F/SPY-CDS-Sal1-R and CPN20-Kpn1-F/CPN20-Sal1-R and HFR1-Kpn1-F/HFR1-Sal1-R (Supplementary Table 1).
Constructs for Stable Transformation Into Arabidopsis
CDS of CPN20 was amplified by PCR and inserted into pJim19-GFP (Sun et al., 2016) between XbaI and XhoI using primer pairs CPN20-Xba1-F/CPN20-Xho1-R.
Constructs for Protein Expression in Mammalian Cells
pEGFP-C1-CPN20 and -CPN10α1 The coding sequences of CPN20 and CPN10α1 were cloned into plasmid pEGFP-C1 between EcoRIand BamH using primer pair CPN20-EcoR1-F2 (CGGAATTCTATGGCGGCGACTCAACTTA)/CPN20-BamH1-R2(CGGGATCCCTAAGAAAGTATAGCCATCACATC), CPN10α1-EcoR1-F1(CG GAATTCTATGATGAAGCGTCTGATCCCAAC), and CPN10α1-BamH1-R1(CGGGATCCATCCTCGTGCAAAGTTCCCAA).
Transient Expression in Arabidopsis Mesophyll Cell Protoplasts and Stable Expression in Arabidopsis Plants for Subcellular Localization Assays
Transient expression in Arabidopsis mesophyll cell protoplasts was performed essentially according to Walter et al. (2004). Protoplasts were isolated from the leaves of 4-week-old plants of wild-type Col-0 or spy-3 mutant and transiently transformed with the constructs described above essentially according to the protocol of Sheen laboratory (http://genetics.mgh.harvard.edu/sheenweb/).
For stable expression of the CPN20-GFP protein in Arabidopsis Col-0 and spy-3 plants, the cDNA encoding CPN20 was cloned using the same primers as described above for transient expression in mesophyll cell protoplasts. The cDNA was cloned into the binary vector pJim19 and fused with the C-terminal GFP flag under the control of the CaMV 35S promoter. The resultant pJim19-CPN20-GFP constructs were introduced into the Agrobacterium tumefaciens GV3101 strain and transformed into Arabidopsis Col-0 and spy-3 plants by floral dip transformation method to generate transgenic CPN20-GFP/Col-0 and CPN20-GFP/spy-3. Isolation of homozygous transgenic lines that contained a single insertion site was done according to Hu et al., 2008.
Construction of cDNA Library for Yeast Two-Hybrid (Y2H) Screening
Total RNAs were extracted from 5-day-old Arabidopsis seedlings pretreated with 50 μM ABA (QIAGEN). The cDNA library was constructed into pGADT7 vector (Clontech) according to Matchmaker Gold Yeast Two-Hybrid System (Clontech, Cat. no. 630489). The cDNA library in pGADT7 and pGBKT7-SPY were co-transformed into Yeast strain Y2H Gold (Clontech) to ensure no autoactivation before the screening, and library screening was performed according to “Yeastmaker Yeast Transformation System 2 User Manual” (Clontech, PT1172-1).
Yeast Two-Hybrid Assays
To verify specific interaction between SPY and CPN20, the CPN20 coding sequence was PCR amplified from cDNA and cloned into pGADT7 vector (Clontech) using primer pair CGGAATTCGCGGCGACTCAACTTACA and CGGGATCCCTAAGAAAGTATAGCCATCACATC. 11-TPR-domain of SPY was amplified and cloned into pGBKT7 vector using primer pair CATGCCATGG AGAATATTCTTCGGGCAAGAAACA, AAAACTGCAGAAGCTTCTGCATATGTGGGATT, and ACGCGTCGACTTAGAAGTCTGGTCGTAGAAGCA. No autoactivation was detected before the validation of one-to-one interaction. The interaction was determined by growth on SD/-Trp/-Leu/-His/AbA/X-α-Gal or SD/-Trp/-Leu/-His/-Ade/AbA/X-α-Gal (Clontech, PT1172-1).
Bimolecular Fluorescence Complementation (BiFC) Assays
Protoplasts were isolated from leaves of 4–5-week-old plants of Arabidopsis (Col-0). Arabidopsis mesophyll cell protoplast isolation and transient expression assay were essentially performed as described previously (Yoo et al., 2007).
YFP (yellow fluorescent protein) was observed under a confocal microscope (Zeiss LSM 710) after incubation at 23°C for 16 h.
Split-Luciferase Complementation (Split-Luc) Assays
The CDSs of SPY, CPN20, and HFR1 were amplified by PCR and inserted into Nluc, Cluc, or Cluc vectors, respectively, between KpnI and SalI using primers SPY-CDS-Kpn1-F/SPY-CDS-Sal1-R, CPN20-Kpn1-F/CPN20-Sal1-R, and HFR1-Kpn1-F/HFR1-Sal1-R (Supplementary Table 1).
Split-Luciferase Complementation assay was performed as described previously (Chen et al., 2008; Li et al., 2014). The plasmids constructed as described above were transformed into Agrobacterium tumefaciens strain GV3101 which was injected into leaves of Nicotiana benthamiana. After 48 h infection at room temperature, LUC activity was measured after 1 mM luciferin was sprayed onto the leaves. Six to eight hours before observation, 10 μM ABA or H2O were injected into corresponding leaves. Luciferase (LUC) images were captured by cooled CCD imaging apparatus. Relative LUC activities were measured as the average of eight leaf disks punching from leaf area injected with GV3101.
Expression in 293T Cells and Purification of Proteins for Western Blot and MS/MS Assays
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS), 100 μg/ml penicillin, and streptomycin. Lipofectamine 3000 Transformation Kit (Invitrogen, L3000-015) was used for cell transformation. 293T cells transformed with pEGFP-C1-CPN20 were cultured for another 24 h. After three-time washes in PBS, cells were dissolved in cell lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1% TritonX-100, cocktail, 10 μM PUGNAc, 20 μM MG132) at 4°C for 15 min. After centrifugation at 4°C at 14,000 rpm for 30 min, the supernatant was incubated with GFP-Trap beads (Chromo Tek, gtma-20) and mildly rotated for 2 h at 4°C. Precipitated immunocomplexes were washed with cell lysis buffer and PBS three times, respectively, and were mixed with 1 × SDS loading buffer at 80°C for 10 min. The precipitates were subjected to SDS-PAGE and stained by coomassie brilliant blue. The desired band was cut down for ETD-MS/MS analysis.
Purification of GFP-Tagged CPN20 From Arabidopsis for MS/MS Assays
CPN20-GFP was purified from germinating seeds of CPN20-GFP/Col-0, CPN20-GFP/spy-3, and CPN20-GFP/HA-SPY grown under MS medium with or without ABA. Plant materials were ground in liquid N2 and homogenized in buffer A [50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 2.5 mM 2-mercaptoethanol, 1x Plant Protease Inhibitor Cocktail (Roche), 20 μM MG132 (Merck) and 10 μM PuGNAc (Sigma-Aldrich)]. After centrifugation at 14,000 rpm for 20 min, supernatants were filtered with a.45 μm syringe filter, and 20 μl GFP-Trap beads (ChromTek) were added. After 1 h rotation at 4°C, beads were recovered and washed with buffer A for three times. Proteins on beads were separated by SDS PAGE and used for MS/MS analysis.
Identification of O-fucosylation Sites by Liquid Chromatography (LC)-ETD Tandem MS (MS/MS) Assays and Data Processing
Each gel band of target protein was excised and digested in-gel with 10 ng/μl sequencing grade trypsin in 50 mM ammonium bicarbonate overnight at 37°C. Prior to the addition of the enzyme, gel pieces were dehydrated in acetonitrile, incubated in 10 mM DTT in 50 mM ammonium bicarbonate at 56°C for 40 min, and incubated in 55 mM iodoacetamide in 50 mM ammonium bicarbonate at ambient temperature for 1 h in the dark, then dehydrated again. The resulting peptides were extracted twice with 5% formic acid/50% acetonitrile, then vacuum-centrifuged to dryness.
For LC-MS/MS analysis, the samples were reconstituted in.2% formic acid, loaded onto a 100 μm × 2 cm pre-column, and separated on a 75 μm × 15 cm capillary column with a laser-pulled sprayer. Both columns were packed in-house with 4 μm C18 bulk material (InnosepBio, China). An Easy nLC 1000 system (Thermo Scientific, USA) was used to deliver the following HPLC gradient: 5–35% B in 60 min, 35–75% B in 4 min, then held at 75% B for 10 min (A = 0.1% formic acid in the water, B = 0.1% formic acid in acetonitrile) at a flow rate of 300 nl/min. The eluted peptides were sprayed into a Velos Pro Orbitrap Elite mass spectrometer (Thermo Scientific, USA) equipped with a nano-ESI source. The mass spectrometer was operated in data-dependent mode with a full MS scan (375–1,600 M/z) in FT mode at a resolution of 120,000 followed by ETD (Electron Transfer Dissociation) MS/MS scans on the 10 most abundant ions in the initial MS scan. Automatic gain control (AGC) targets were 1e6 ions for orbitrap scans and 5e4 for MS/MS scans, and the AGC for the fluoranthene ions used for ETD was 5e5. ETD activation time was 100 ms. Supplemental activation of the charge-reduced species was used in the ETD analysis to improve fragmentation. For dynamic exclusion, the following parameters were used: isolation window, 2 m/z; repeat count, 1; repeat duration, 25 s; and exclusion duration, 25 s.
The raw data files were converted to mascot generic format (“.mgf”) using MSConvert before being submitted for database search. Mascot (version 2.3.02) carried out all database search with the following parameters: Carbamidomethyl (Cys) as fixed modification, Oxidation (Met), HexNAc for O-GlcNAc (Ser/Thr), dHex for fucose (Ser/Thr), and Phosphorylation (Ser/Thr) as a variable modification; +/– 10 ppm for peptide pass tolerance and +/– 0.6 Da for fragment mass tolerance; max missed cleavages 2. Assignments of all modified peptides were checked manually.
In vitro Assays of O-Fucosyltransferase Activity of SPY by ETD-MS/MS
Based on the identification of fucosylation sites in mammalian cells and in planta assays, CPN20 peptide was synthesized (GenScript) for the in vitro enzyme assays, KIDITVPTGAQIIYSK (amino acid residues 112-127). Each 20 μl of reaction included 150 μM CPN20 peptide, 200 μM GDP-fucose, and 10 μg 3TPR-SPY. The reaction buffer contained 50 mM Tris-HCl, pH 8.2, 50 mM NaCl, and 5 mM MgCl2. After incubation for 8 h at 25°C, the sample was loaded onto a pre-column connected to the analytical column. The peptide was analyzed by tandem MS with an Orbitrap Elite mass spectrometer equipped with ETD.
Fluorescence Microscopy of CPN20-GFP in Chloroplasts
CPN20-GFP signal and chloroplast signal from rosette leaves and cotyledons of CPN20-GFP/Col-0 and CPN20-GFP/spy-3 were observed and photographed under a confocal microscope (Zeiss LSM 710). GFP fluorescence emission spectra were collected at 508 nm wavelength after being excited at 488 nm wavelength with argon laser. Identical parameters were set for the comparison of a particular signal between different genotypes.
Extraction of Total Proteins and Chloroplast Proteins
Total proteins were isolated from rosette leaves of 4-week-old Arabidopsis plants using buffer A [50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 2.5 mM 2-mercaptoethanol, 1x Plant Protease Inhibitor Cocktail (Roche), 20 μM MG132 (Merck), and 10 μM PuGNAc (Sigma-Aldrich)] after grounded into powder in liquid N2.
Intact chloroplasts were isolated from rosette leaves of 4-week-old Arabidopsis plants as described previously (Shang et al., 2010). Briefly, leaves were homogenized in homogenization buffer containing 330 mM sorbitol, 5 mM MgCl2, 2 mM EDTA, 1 mM MnCl2, and 50 mM HEPES/ KOH, pH 7.8. The homogenate was filtered and centrifuged, and the pellet was re-suspended in 100 ml suspending buffer containing 330 mM sorbitol, 5 mM MgCl2, 50 mM HEPES/KOH, pH 7.8, and a complete protease inhibitor cocktail. The re-suspended chloroplasts were loaded onto a two-step Percoll gradient and were centrifuged in a swinging-bucket rotor at 1,500 g for 10 min. The band that appeared between the two phases contained intact chloroplasts and was recovered. The intact chloroplasts were washed with the suspending buffer at a rate of chloroplast to buffer 1/10 (v/v) by inverting the tubes carefully. The chloroplasts were centrifuged in a swinging-bucket rotor at 1,000 g for 3 min, and the pellet was recovered and re-suspended in the suspending buffer. The intactness and fluorescence of the chloroplasts were checked under a confocal microscope (Zeiss LSM 710) before extraction of proteins with freezing and thawing with liquid nitrogen.
In vivo Protein Stability Assays
In vivo protein stability assays were performed according to Jung et al. (2015). Two-week-old seedlings of 35S:CPN20-GFP in Col-0 and spy-3 backgrounds were incubated in liquid MS medium with 40 μM MG132 (Merck) for 16 h to enrich the CPN20-GFP protein for stability assay. A total of 40 μM MG132 treated seedlings were washed five times before being transferred to a liquid MS medium supplemented with 100 μM cycloheximide (Sigma-Aldrich) to block de novo protein synthesis. The treated seedlings were collected at the indicated time points for protein preparation and immunoblot analysis.
Protein Immunoblot Analyses
For immunoblot analyses, seedlings of tested Arabidopsis genotypes were harvested in protein extraction buffer composed of 50 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10 mM NaF, 2 mM Na3VO4, 25 mM β-glycerol phosphate, 10% (vol/vol) glycerol, 0.1% (vol/vol) Nonidet P-40, 1 mM PMSF and 1 × complete Protease Inhibitor Mixture, and pH 7.5. Briefly, protein samples were separated by SDS-PAGE and transferred to a polyvinylidene fluoride film. After being blocked in 5% milk-containing 1 × PBST buffer, the film was incubated with the selected primary antibody at desired dilution overnight at 4°C, washed three times with 1 × PBST (5 min each), and incubated with the selected secondary antibody at desired dilution for 1 h at room temperature. After three washes with 1 × PBST (5 min each), the film was illuminated and photographed under a Bio-Rad illumination detection device.
Co-localization Analyses
Co-localization of fluorescent signals was analyzed by Pearson's correlation coefficient equation by using the Coloc2 plugin in the ImageJ program.
Statistical Analysis
Most of the experiments had been repeated three times. Representative results were shown. A significant difference between treatments and corresponding control was analyzed using the Student's t-test.
Results
SPY Interacts With CPN20 and the Interaction Is Promoted by ABA
The N-terminal TPR domain of SPY is responsible for protein-protein interaction (Jacobsen et al., 1996). Spy mutants display manifold phenotypes not constrained in GA signaling (Swain et al., 2001), suggesting that SPY participates in multiple biological processes. In order to identify substrates of SPY and to elucidate the mechanisms by which SPY takes part in developmental and hormone signaling processes, we carried out a yeast two-hybrid screen, using BD-SPY as bait to search for its interacting proteins on the premise that there was no auto-activation of BD-SPY and AD (Supplementary Figure 1). Three prey clones expressing CPN20 were recovered from this screen. CPN20 is a co-chaperonin identified from chloroplasts of pea (Bertsch et al., 1992) and of Arabidopsis (Hirohashi et al., 1999). In order to confirm the interaction between SPY and CPN20, we cloned full-length CPN20 into prey vectors. In yeast, SPY interacted with CPN20 through the N-terminal TPR domain (Figure 1A). To test this interaction in vivo, we performed two types of bimolecular fluorescence complementation (BiFC) assays: Split-YFP and Split-Luc. In Split-YFP assay, SPY interacted with CPN20 likely in the cytosol around the chloroplast of Arabidopsis mesophyll cell protoplasts (Figure 1B). In Split-Luc assay, SPY interacted with CPN20 in tobacco leaves and, surprisingly, ABA at 10 μM promoted this interaction (Figures 1C,D). From the experiments above, we conclude that SPY interacts with CPN20 and ABA promotes this interaction.
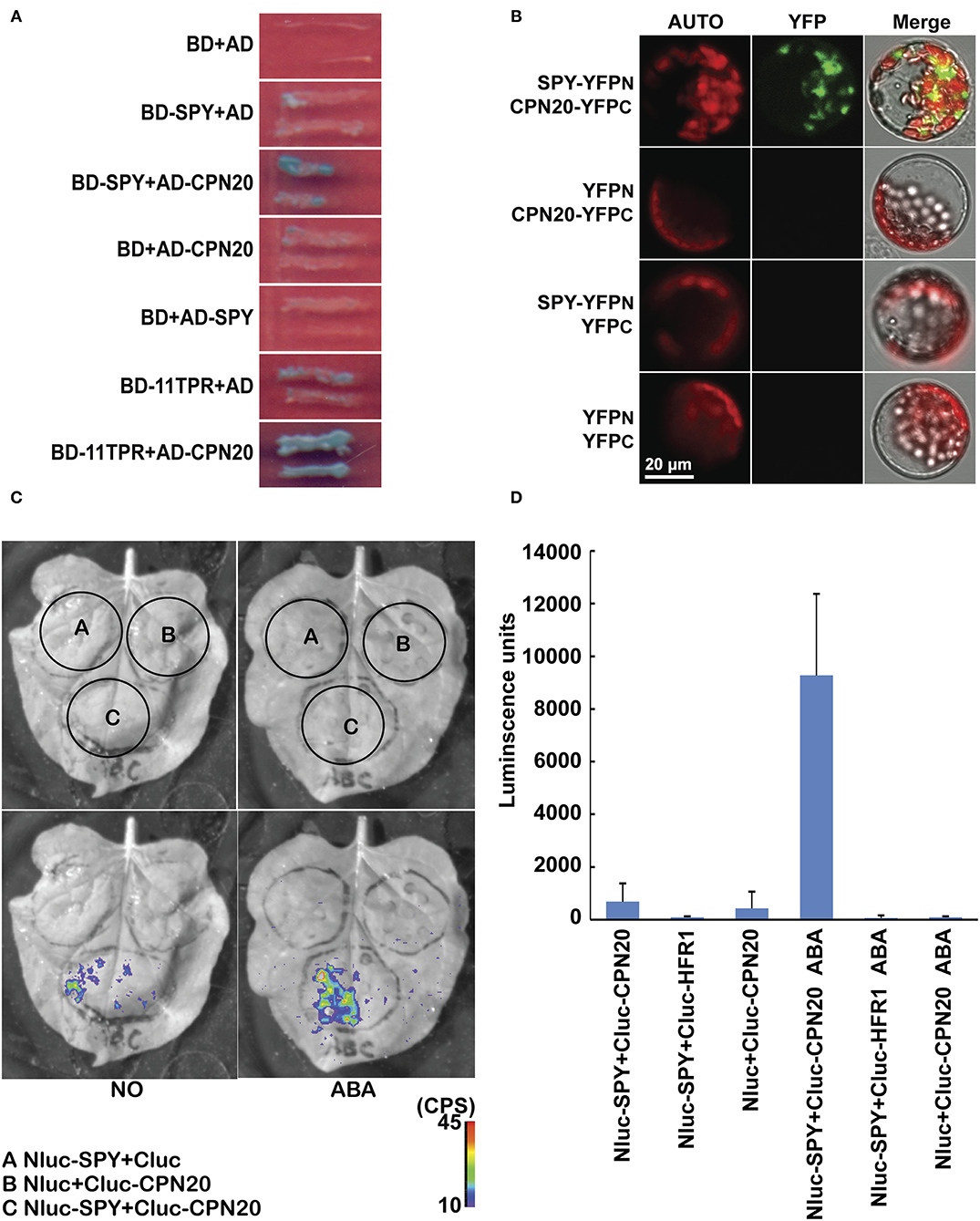
Figure 1. SPY interacts with CPN20. (A) SPY interacts with CPN20 through TPR domains in yeast two-hybrid assay. Pictures were taken after yeast cells were grown on SD-3/X/A for 4 days. BD, DNA-binding domain; AD, activation domain; SD-3/X/A, SD medium lacking Leu, Trp, and His and adding X-α-Gal and Aureobasidin A; (B), SPY interacts with CPN20 in BiFC assay. Protoplasts were transformed with pairs of indicated vectors and observed under a confocal microscope using identical parameters. AUTO, chloroplast autofluorescence; YFP, the fluorescence of the YFP protein; Merged, merged image of YFP, AUTO, and the bright-field image. (C,D) ABA promotes interaction between SPY and CPN20 in Split-Luc assay. Leaves of tobacco were transformed by infiltration of GV3101 with indicated construct pairs. LUC images were captured by cooled CCD imaging apparatus (C). Relative LUC activities were measured by GLOMAX 96 microplate luminometer. Data are shown as means ± SE measured from 8 leaf disks punched from leaf area injected with GV3101 (D). The concentration was 10 μM when ABA was applied.
CPN20 Is O-fucosylated in Mammalian Cells and in planta
A novel finding that SPY O-fucosylates the growth repressor DELLA and affects functions of DELLA's interactors (Zentella et al., 2017) prompted us to test the possible O-fucosylation of CPN20.
The GFP-tagged CPN20 and CPN10α1 (TAIR ID: AT1g14980) were successfully expressed in mammalian cells, immune-purified, and detected by anti-GFP and Coomassie blue (Supplementary Figure 2). GFP-tagged CPN20 and CPN10α1 were subjected to ETD-MS/MS analysis. Two O-fucosylation sites, e.g., 116th and 119th threonines, were detected only in CPN20-GFP (Figures 2A,C), but not in the control protein CPN10α1-GFP, suggesting that the O-fucosyltransferase in mammalian cells specifically O-fucosylate ectopically expressed CPN20 instead of CPN10α1. In this assay, we intended not to introduce exogenous SPY together with CPN20-GFP or its control CPN10α-GFP into mammalian cells considering that mammalian cells have endogenous O-fucosyltransferases. The pertinent conclusion would be that CPN20 can be O-fucosylated by either SPY or mammalian cell endogenous O-fucosyltransferases even if SPY was co-transformed. To identify O-fucosylation sites of CPN20 in vivo, GFP-tagged CPN20 was isolated from germinating seeds of CPN20-overexpressing plants (Supplementary Figure 3). After ETD-MS/MS, O-fucosylation sites at 116th and 119th threonines were found in GFP-tagged CPN20 under certain circumstances (Figures 2A,B,D), which were identical to O-fucose modified sites identified in CPN20-GFP expressed in mammalian cells (Figures 2A,C).
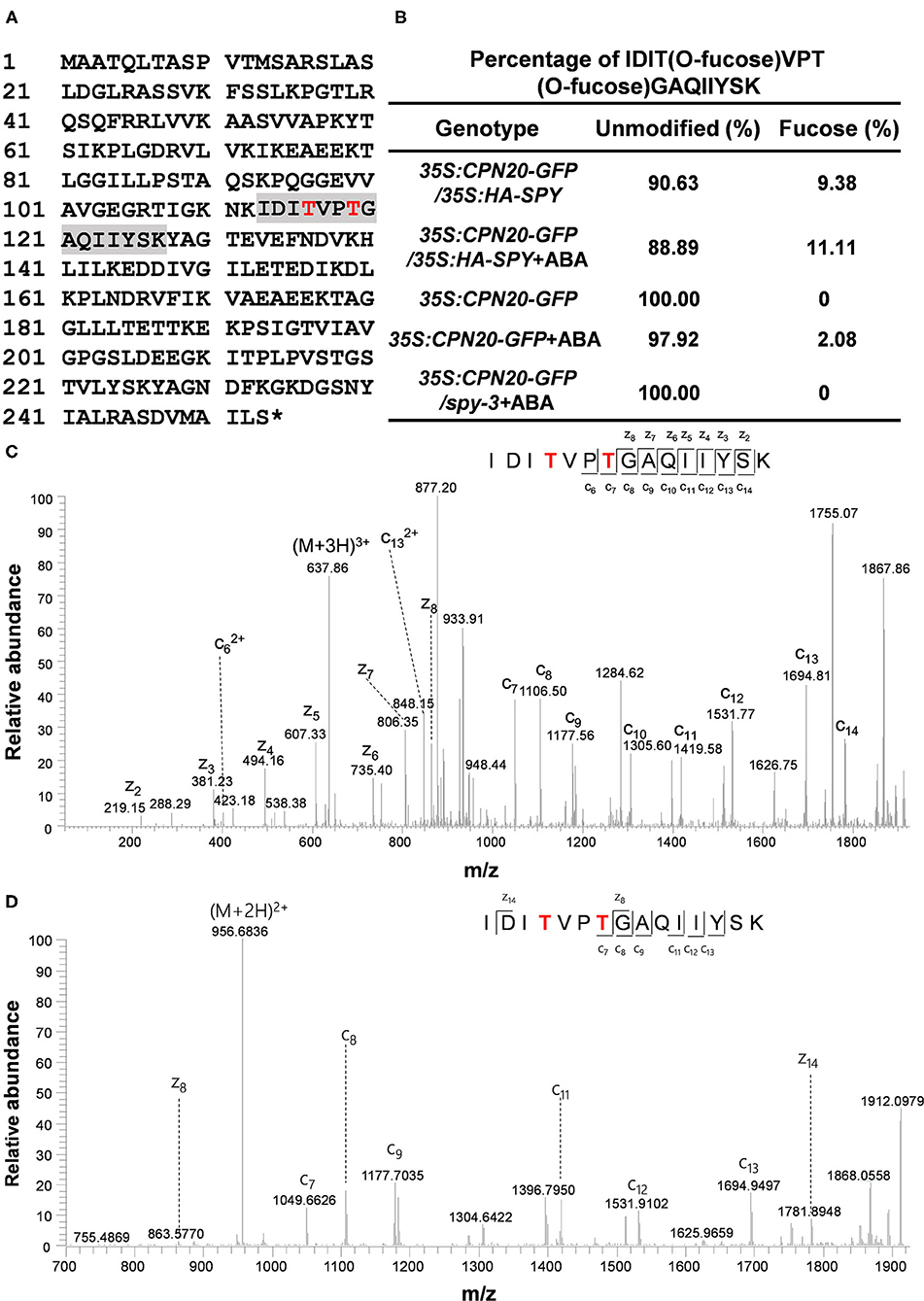
Figure 2. CPN20 is O-fucosylated in mammalian cells and in planta. (A) O-fucosylation sites in CPN20 determined by ETD-MS/MS. A sequence in gray is an O-fucose modified peptide within CPN20 isolated from germinating seeds of CPN20-GFP/35S:HA-SPY. Red letters indicate confirmed O-fucosylating sites. (B) Percentage of IDITO−fucoseVPTO−fucoseGAQIIYSK vs. IDITVPTGAQIIYSK identified by ETD-MS/MS from CPN20-GFP purified from germinating seeds of indicated materials under indicated conditions. The percentage was calculated by spectra counting. The concentration was 10 μM when ABA was applied. (C) ETD MS2 spectrum of the tryptic peptide IDITO−fucoseVPTO−fucoseGAQIIYSK from CPN20-GFP immune-purified from 293T cells ectopically expressing this fusion protein. (D) ETD MS2 spectrum of the tryptic peptide IDITO−fucoseVPTO−fucoseGAQIIYSK from CPN20-GFP prepared from germinating seeds of CPN20-GFP/35S: HA-SPY.
The MS/MS identification was repeated three times with plant materials, and we calculated the percentage of O-fucosylation that was successfully detected at the 116th and 119th threonines. O-fucosylation at the 116th and 119th threonines was not detected in plants that express either CPN20-GFP alone or CPN20-GFP/spy-3. The percentage of O-fucosylation at the 116th and 119th threonines was somehow increased in plants that overexpress SPY (Figure 2B). In addition, ABA treatment at 10 μM appeared to mildly increase the percentage of success in detecting O-fucosylation at the 116th and 119th threonines within CPN20 although an accurate quantitative comparison cannot be made due to technical difficulties for MS/MS (Figure 2B).
SPY O-fucosylates the Peptide of CPN20 in in vitro Assay by ETD-MS/MS
Based on the identified fucosylation sites of CPN20 in vivo, a CPN20 peptide KIDITVPTGAQIIYSK (amino acid residues 112–127) was synthesized for further confirmation of O-fucosylation sites in the in vitro assays of O-fucosyltransferase activity of SPY by ETD-MS/MS. After a 20 μl reaction mixture was subjected to tandem MS with an Orbitrap Elite mass spectrometer equipped with ETD, O-fucosylation of the 116th and 119th threonines, and 126th serine was identified (Figure 3; Supplementary Figure 4). Thus, this assay further confirmed the O-fucosylation at the 116th and 119th threonines but recognized a new site, e.g., 126th serine (Figure 3; Supplementary Figure 4), that was not detected in mammalian cells and plant materials.
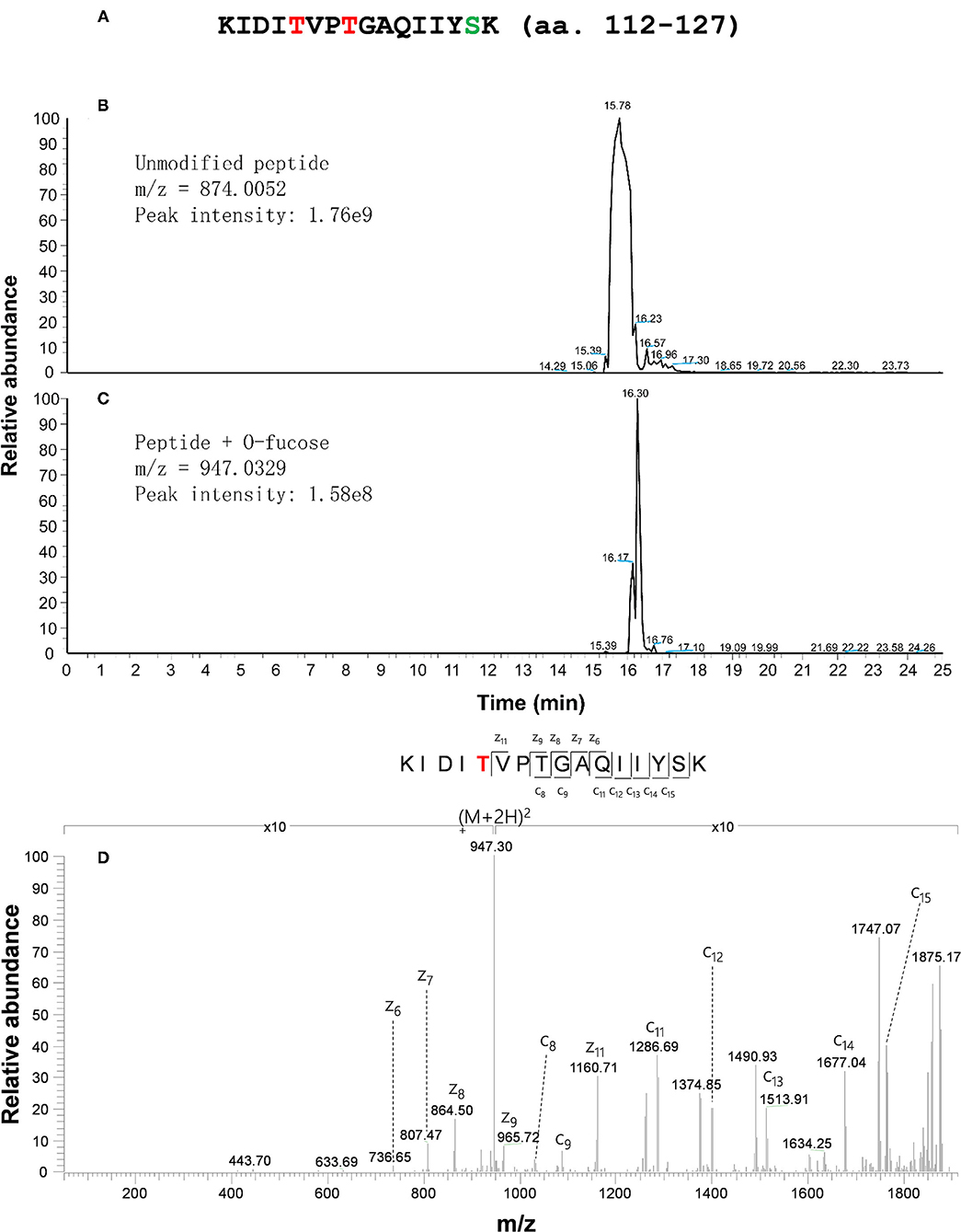
Figure 3. CPN20 peptide is O-fucosylated in an in vitro SPY activity assay. (A) Amino acid sequence of synthesized CPN20 peptide (amino acid residues 112–127). The letters T in red and S in light blue indicate the residues mono-O-fucosylated. (B) EIC (Extracted ion chromatogram) of un-O-fucosylated CPN20 peptide. (C) EIC (Extracted ion chromatogram) of mono-O-fucosylated CPN20 peptide. (D) ETD MS2 spectrum of synthesized CPN20 peptide KIDITO−fucoseVPTGAQIIYSK. The CPN20 peptide reacted with GDP-fucose and 3TPR-SPY in the reaction buffer for 8 h at 25°C before being analyzed by ETD-MS/MS.
SPY Regulates Accumulation of CPN20 in Chloroplasts via O-fucosylation of CPN20
In order to achieve a functional chloroplast, proteins are transported into and accumulated in it, which should be precisely controlled. Considering that CPN20 is a chloroplast-localized protein (Bertsch et al., 1992; Koumoto et al., 2001) and that CPN20 is O-fucosylated, we were curious about whether SPY affects chloroplast-localization of CPN20 via O-fucosylation. We generated transgenic lines with GFP-tagged CPN20 in Col-0 and spy-3 backgrounds. The alleles with similar mRNA levels (Figure 4A) were chosen for further experiments. Immunoblot analyses were employed to detect the localization of the CPN20 protein. Total and chloroplast fractions were extracted from mesophyll cells of rosette leaves of 40-day-old CPN20-GFP transgenic plants in Col-0 and spy-3 backgrounds. It was shown that no HSP70 (as a cytosol marker) was detected in the chloroplast fraction as compared to clear detection of HSP70 from the total fraction (Figure 4B), confirming that the chloroplast fraction was not contaminated by cytosolic proteins such as HSP70. In order to test the content of CPN20 in two different genetic backgrounds, anti-GFP was used. Interestingly, chloroplast-localized CPN20 was increased in spy-3 compared with Col-0 (Figure 4C). To avoid the bias caused by possible different levels of CPN20 in total proteins in Col-0 and spy-3, we detected the levels of CPN20 in the total fraction. The level of CPN20 in spy-3 was comparable to that in Col-0 (Figure 4D). We also examined the distribution of the CFP20-GFP fluorescence signal by confocal microscopy. The intensity of GFP signal in the chloroplast of either mesophyll cell or guard cells was remarkably increased in the spy-3 background (Figure 4E; Supplementary Figures 5A–D). Following a careful observation, we noted that the localization of GFP signal in mesophyll cells was different between Col-0 and spy-3 (Supplementary Figures 5C,D). In Col-0, a large portion of the GFP signal was associated with the mesophyll cell chloroplasts, but quite a large portion of the GFP signal remained outside, appearing as discrete green granules (Supplementary Figure 5C). In spy-3, the majority of GFP signal located inside or associated with the mesophyll cell chloroplasts, while only a small portion of granular GFP signal was outside the mesophyll chloroplasts (Supplementary Figure 5D). Quantitative analysis by Pearson's correlation coefficient analysis supported these observations. Pearson's correlation coefficient R-value was 0.24 and 0.79 in Col-0 and spy-3 backgrounds, respectively. This indicates that the GFP signal is more correlated with the chloroplast in spy-3 than that in Col-0.
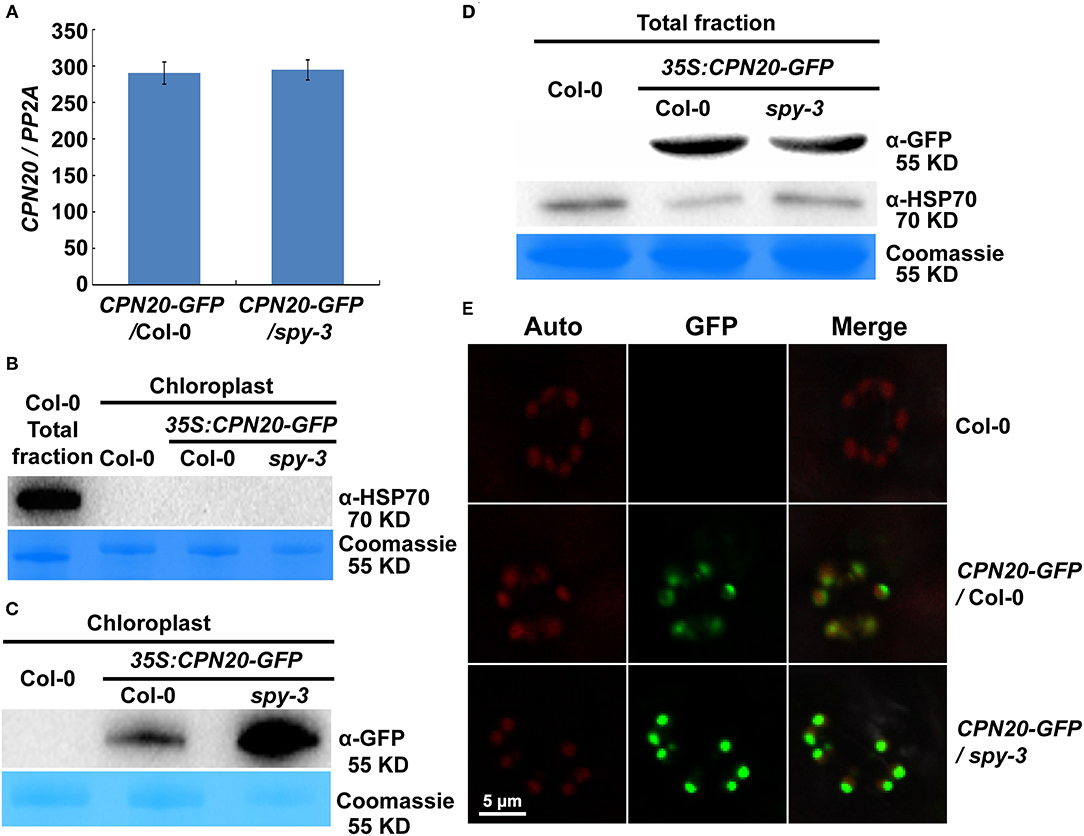
Figure 4. SPY regulates the accumulation of CPN20 in guard cells and mesophyll cell chloroplasts. (A) mRNA level of CPN20 in different transgenic materials. CPN20-GFP/Col-0 or CPN20-GFP/spy-3 represents the CPN20-overexpressing line in the background of Col-0 or and spy-3. (B) Purity of chloroplasts confirmed by cytosolic marker protein HSP70 on western blot. Total proteins and chloroplast proteins were isolated from leaves of indicated materials before being subjected to SDS PAGE and western blotting using α-HSP70. Coomassie blue staining of RbcL was used as a loading control. (C) CPN20 is more abundant in chloroplasts from spy-3. GFP-tagged CPN20 was detected by α-GFP while coomassie blue staining of RbcL was used as a loading control. GFP fluorescence of CPN20-GFP in chloroplasts of stomatal guard cells in CPN20-GFP/Col-0 and CPN20-GFP/spy-3. Leaves of 40-day-old plants were observed under a confocal microscope using identical parameters. Auto, GFP, and Merge indicate chlorophyll autofluorescence, the fluorescence of CPN20-GFP, and merged image of Auto and GFP. (D) Total CPN20 abundance is largely unchanged in CPN20-GFP/spy-3 compared with CPN20-GFP/Col-0. CPN20-GFP fusion proteins in extracts from indicated genotypes were detected by western blotting using α-GFP or α-HSP70. Coomassie blue staining of RbcL was used as a loading control. (E) GFP fluorescence of CPN20-GFP in chloroplasts of stomatal guard cells in CPN20-GFP/Col-0 and CPN20-GFP/spy-3. Leaves of 40-day-old plants were observed under a confocal microscope using identical parameters. Auto, GFP, and Merge indicate chlorophyll autofluorescence, the fluorescence of CPN20-GFP, and merged image of Auto and GFP.
The Mutation of O-fucosylation Sites Increases Chloroplast-Accumulation of CPN20
Based on the data that CPN20 is O-fucosylated at the 116th and 119th threonines and 126th serine, and that the spy mutation increases chloroplast-localization of CPN20, we speculated that the mutation of some or all of these sites in CPN20 may impact the CPN20 localization in the chloroplast. We made the construct carrying a mutated form of CPN20 in which 116th and 119th threonines were replaced with alanine and transformed Arabidopsis mesophyll cell protoplasts. As shown in Figure 5A, the substitution of the 116th and 119th threonines for alanine apparently strengthened the fluorescence signal of CPN20-GFP in the chloroplast. This implicates that O-fucosylation of the 116th and 119th threonines may reduce chloroplast-localization of CPN20. Thus, we tentatively conclude that SPY-catalyzed O-fucosylation of CPN20 may inhibit CPN20 accumulation in the chloroplast.
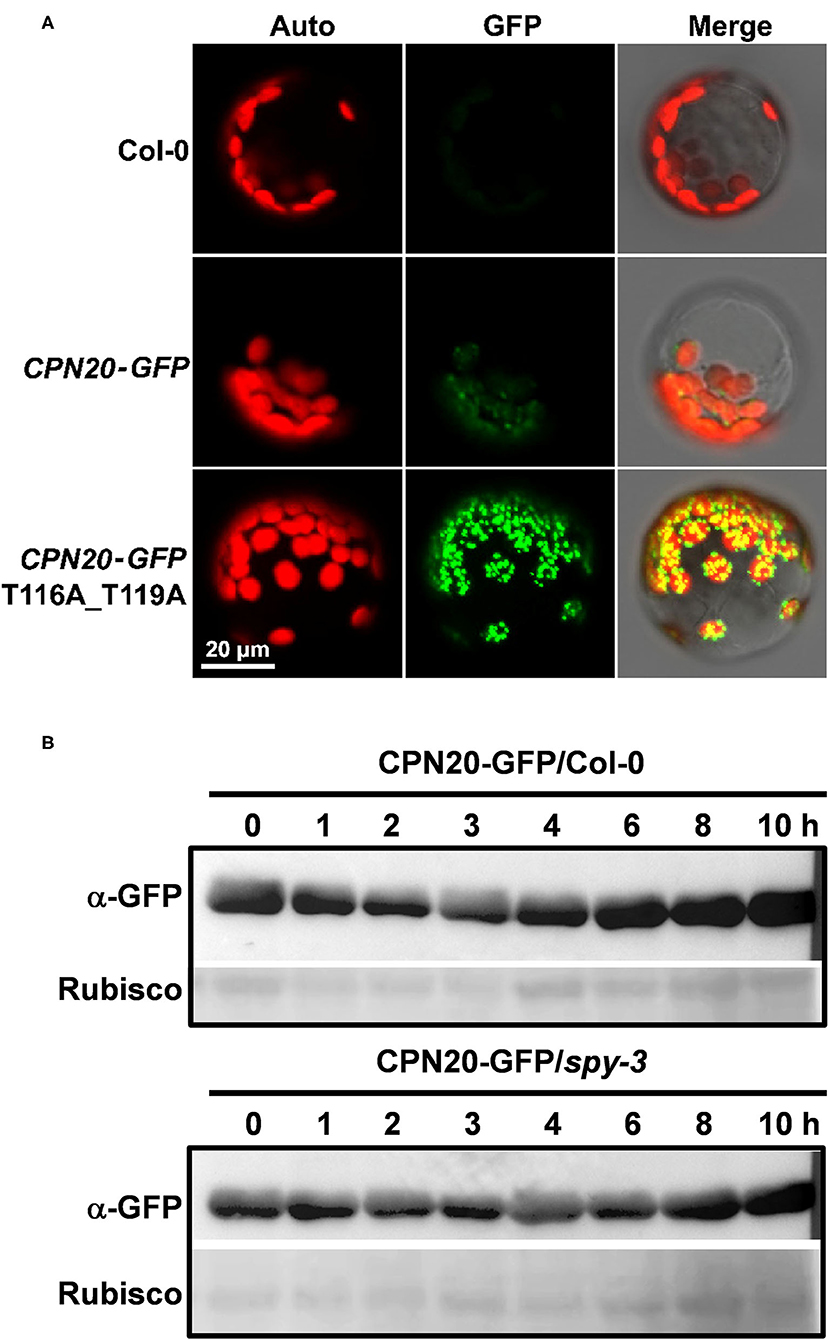
Figure 5. Effect of CPN20 mutation at the O-fucosylation sites or SPY mutation on chloroplast localization or stability of CPN20. (A) CPN20 mutation at the O-fucosylation sites affects the localization of CPN20 in the chloroplast. Plasmids of GFP fused downstream of wild-type full-length ORF of CPN20 and mutated CPN20 with T116a and T119A were transformed into mesophyll protoplasts prepared from leaves of 30-day-old Col-0 plants. Auto, GFP, and Merge indicate chlorophyll autofluorescence, the fluorescence of CPN20-GFP, and merged image of Auto and GFP. (B) the stability of CPN20 is unaltered by SPY mutation. Two-week-old seedlings of 35S:CPN20-GFP in Col-0 and spy-3 backgrounds were incubated in liquid MS medium supplemented with 40 μM MG132 for 16 h and washed five times before being transferred to liquid MS supplemented with 100 μM cycloheximide for further incubation. Proteins were extracted at the indicated time points and detected by an anti-GFP antibody. Rubisco levels as detected by ponceau staining were used as loading controls.
The spy Mutation Does Not Impact the Stability of CPN20
As the CPN20 accumulation in the chloroplast is altered by the mutation of SPY, it remained unclear how this phenomenon occurred. The data shown in Figure 4D hints that the spy mutation appears not to affect the stability of CPN20. To confirm this speculation, we carried out an in vivo protein degradation experiment to check whether the content of CPN20 is altered by protein degradation and whether the spy mutation affects the possible degradation of CPN20. As shown in Figure 5B, the content of CPN20 appeared largely unchanged in the seedlings of both in Col-0 and spy3 backgrounds during the incubation in cycloheximide-supplemented media. This result suggests that CPN20 is stable in vivo, and the mutation of SPY does not alter the stability of CPN20. This result supports the notion that greater accumulation of CPN20 in the chloroplast of spy-3 background plants is likely ascribed to the enhanced import caused by the mutation of SPY. In other words, the non-O- fucosylated CPN20 is more readily imported into the chloroplast.
The cpn20 Mutation Suppresses the Insensitivity of spy-3 to ABA
CPN20 is a negative regulator involved in the ABAR-mediated ABA signaling pathway. It acts downstream of or at the same node of CHLH/ABAR, a chloroplast-localized ABA receptor, antagonizing WRKY-domain containing transcription repressors to turn on ABA signaling (Shen et al., 2006; Wu et al., 2009; Shang et al., 2010; Du et al., 2012; Liu et al., 2012; Yang et al., 2012) and upstream of the WRKY40 transcription factor (Zhang et al., 2013). If CPN20 is a substrate of SPY in the plant, spy mutants may likely have ABA-related phenotypes. As reported previously by Steber et al. (1998) and by Liang et al. (2018), different alleles of spy displayed reduced sensitivity to ABA treatment during seed germination and early seedling development. ABA-promoted SPY-CPN20 interaction further led us to propose that SPY may be involved in the ABAR-mediated ABA signaling pathway. In order to dissect the genetic relationship between SPY and CPN20, we generated double mutants ♀spy-3♂cpn20-1. The development of seedlings of ♀spy-3♂cpn20-1 displayed less insensitive phenotypes than those of spy-3 under ABA treatments (Figure 6A), showing that the cpn20 mutation partially suppresses the ABA-insensitive phenotypes of spy-3, e.g., the ABA-insensitive phenotypes of spy-3 is in part dependent on functional CPN20. In germination and cotyledon greening assays, ♀spy-3♂cpn20-1 germinated and turned green a little later than spy-3 and earlier than Col-0 (Figures 6B,C; Supplementary Figure 6). This also supports that the cpn20 mutation partially suppresses the ABA-insensitive phenotypes of spy-3. These experiments suggest the interdependent roles of SPY and CPN20 in ABA signaling.
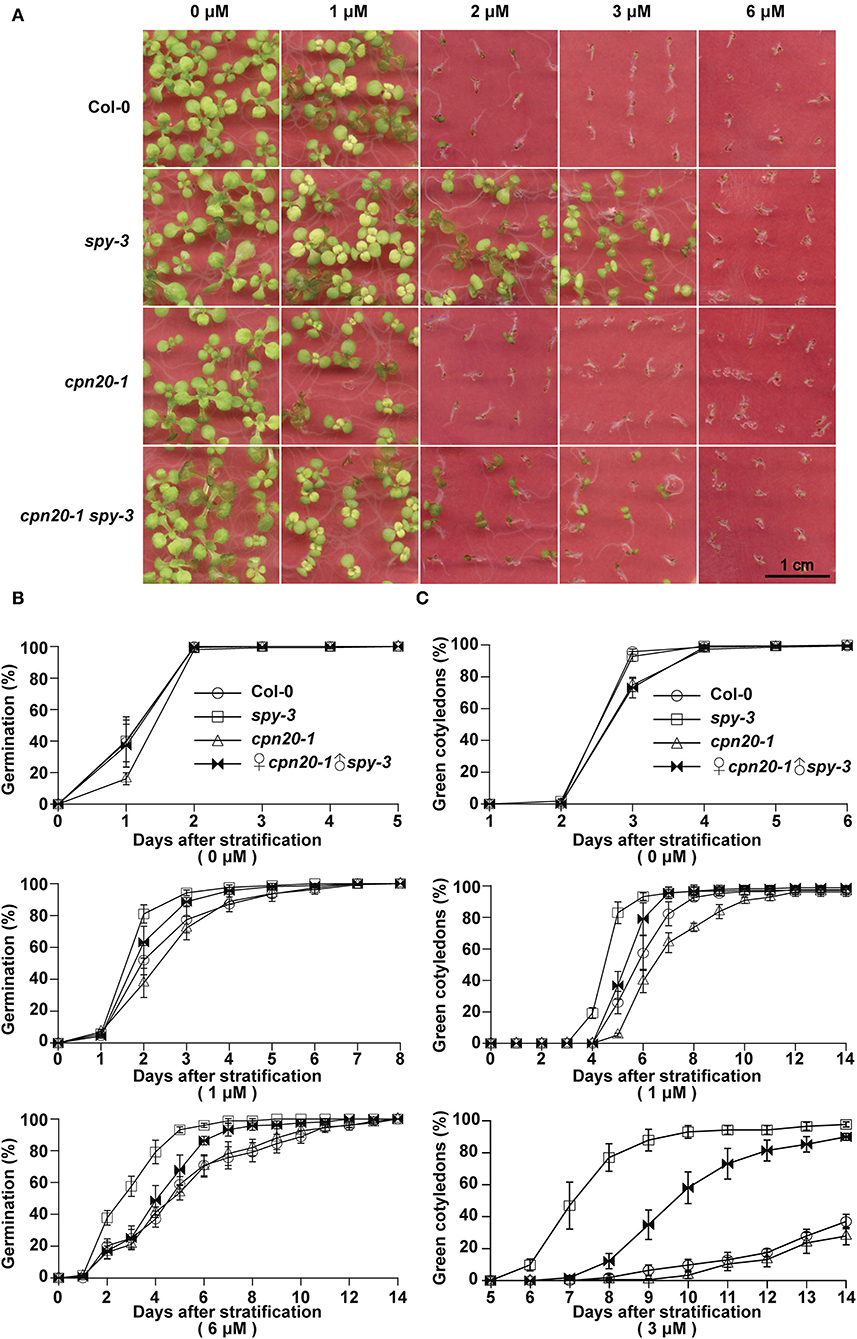
Figure 6. cpn20-1 suppresses the insensitivity of spy-3 to ABA in seed germination and early seedling development. Germination rates (B) and cotyledon greening rates (C) of ♀cpn20-1♂spy-3, cpn20-1, spy-3, and Col-0 under indicated concentrations of ABA were scored and calculated at indicated time after 4-day stratification at 4°C. Data are shown as means ± SE of four replicates. Images of 2-week seedlings were taken (A).
Discussion
Apart from the core PYR/PYL/RCAR-mediated ABA signaling pathway, the ABAR-mediated pathway is a parallel mechanism for ABA signaling (Shen et al., 2006; Wu et al., 2009; Du et al., 2012). In this mechanism, CPN20 plays a negative role interacting with ABAR, the putative chloroplast-localized ABA receptor. This interaction antagonizes ABAR suppression of a group of WRKY-domain transcription repressors and thereby inhibits ABA-responsive genes (Shang et al., 2010; Liu et al., 2012; Yang et al., 2012). In the present study, we demonstrate that putative plant O-fucosyltransferase SPY interacts with CPN20 and that the interaction between SPY and CPN20 is promoted by ABA, CPN20 is modified by O-fucosylation, and that SPY-mediated O-fucosylation may alter CPN20 localization in the chloroplast and influence the function of CPN20 in ABA signaling. These findings gain new insights into ABA signaling mechanisms, in particular the ABAR-mediated ABA signaling pathway.
The SPY had long been considered as a putative OGT although no O-GlcNAc activity was ever detected for it. SPY was demonstrated to be a novel O-fucosyltransferase, mono-O-fucosylating RGA, and RGL1 (Zentella et al., 2017). SPY, as an O-fucosyltransferase, plays an opposite role in regulating DELLA compared to SEC, an OGT in Arabidopsis (Zentella et al., 2016, 2017). Intriguingly, SPY is localized not only in the nucleus but also in the cytosol (Swain et al., 2002). In the present study, we discovered that plant O-fucosyltransferase SPY O-fucosylates a chloroplast-localized protein, CPN20, based on several lines of evidence. First, ectopically expressed CPN20 in mammalian cells is O-fucosylated (Figure 2C). Although no O-fucosyltransferase has been demonstrated before, our repeated measurements by MS demonstrated the O-fucosylation of ectopically expressed CPN20. Second, CPN20 expressed in Arabidopsis is modified by O-fucosylation, which is promoted by the co-expression of SPY to some extent (Figure 2B). In addition, in vitro assays of O-fucosyltransferase activity of SPY with CPN20 peptide confirmed that CPN20 is O-fucosylated by SPY (Figure 3). Thus, we show that the SPY-catalyzed O-fucosylation of a chloroplast protein.
Repressor of GA1-3 (RGA), the first reported plant substrate for SPY-catalyzed O-fucosylation, is a transcriptional regulator, which is located in the nucleus. Its O-fucosylation is responsible for modulating the functions of RGA-interacting proteins PIF3, PIF4, and BZR1 (Zentella et al., 2017). This suggests that SPY-catalyzed O-fucosylation of RGA may occur in the nucleus. However, the possibility that SPY-catalyzed O-fucosylation of RGA occurs in the cytosol cannot be excluded as solid evidence is still lacking (reviewed by Olszewski et al., 2011). It was previously reported that SPY may also function in the cytosol in that ectopically expressed cytosol-localized SPY regulates cytokinin responses via a DELLA-independent pathway (Maymon et al., 2009). In this study, we reported that CPN20, a putative plant substrate of O-fucosylation, is a protein that is transcribed in the nucleus and translated on ribosomes, and eventually imported into and localized in the chloroplast. Yet, whether CPN20 is O-fucosylated in the cytosol or in the chloroplast remains an open question. As reported by Swain et al. (2002), SPY is mostly localized in the nucleus and less in the cytosol. Our immunoblot analysis also supports that SPY is localized in the cytosol and not in the chloroplast (Supplementary Figure 7). Our BiFC assay also showed that SPY interacts with CPN20 mostly outside the chloroplast, likely in the cytosol or the compartments in close proximity of the chloroplast (Figure 1B). Thus, we propose that CPN20 is likely to interact with and be O-fucosylated by SPY in the cytosol.
Most of the substrates reported for O-fucosylation in animals are involved in protein localization or secretion. Our immunoblot, fluorescence assays, and protein stability assay suggest that SPY-catalyzed O-fucosylation inhibits the import of CPN20 into the chloroplast. Yet, the underlying molecular mechanism remains unclear. During the import of soluble proteins into the chloroplast, newly synthesized preproteins are chaperoned by the guidance complex or by chaperone Hsp90 alone. The guidance complex is composed of chaperone Hsp70 and regulatory 14-3-3 proteins. Hsp70-chaperoned preproteins are recognized by Toc159 and Toc34, followed by delivery to the import channel Toc75 (reviewed by Li and Chiu, 2010, and references therein; Sjuts et al., 2017). In our Mass-Spectrometry identification of SPY-interacting proteins, Hsp70-2 is amongst the candidates. Thus, it is plausible that the SPY-facilitated O-fucosylation of CPN20 may modulate its accessibility by HSP70-2 and thereby prompt subsequent import into the chloroplast. Yet, whether SPY, Hsp70-2, and CPN20 form a complex remains to be resolved.
Although our data suggest that CPN20 may be downstream of SPY in ABAR-mediated ABA signaling, our genetic analyses of the relationship between CPN20 and SPY showed that the cpn20 mutation partially suppresses the ABA-insensitive phenotypes of spy-3 in terms of seed germination and seedling development (Figure 6). SPY catalyzes O-fucosylation (Figures 2, 3) and thereby regulates CPN20 localization in the chloroplast (Figure 4; Supplementary Figure 5), which explains the ability of cpn20 to suppress the ABA-insensitivity associated with the spy-3 mutation. The partial suppression of the ABA-insensitive phenotypes of spy-3 by cpn20-1 indicates that other pathways in parallel with CPN20 to mediate SPY in ABAR-involved ABA signaling may exist. Other signaling substrates for SPY remain to be determined.
Given that GA is an antagonist of ABA in seed germination and that SPY is a negative regulator of GA signaling (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996), it is of importance to elucidate whether SPY is involved in ABA-inhibited seed germination via regulating GA signaling. GA promotes Arabidopsis seed germination mainly via derepressing RGL2 (GAI/RGA-like2) and RGL3 DELLA repressors (Lee et al., 2002; Piskurewicz and Lopez-Molina, 2009). RGL2 has been shown to stimulate ABA biosynthesis and the activity of ABI5, the major positive regulator in ABA signaling (Piskurewicz et al., 2008). Considering that SPY O-fucosylates RGA and RGL1 (Zentella et al., 2017) which are homologous to RGL2 and RGL3, it is possible that RGL2 and RGL3 may also be substrates for SPY. Genetic analyses have shown previously that SPY and ABI5 have a complex relationship. Low concentrations of ABA, SPY, and ABI5 appear to be in the same pathway during seed germination, but at high concentrations of ABA, they tend to diverge into parallel pathways at the cotyledon greening stage (Liang et al., 2018). The relationships of ABI5 with both SPY and RGL2 and RGL3 implicate that SPY may regulate ABI5 possibly through RGL2 and RGL3. Based on the data in the present and previous studies, we cannot exclude the possibility that SPY may also regulate ABA signaling via O-fucosylation of RGL2 and RGL3 despite knowing that SPY regulates ABA signaling via O-fucosylation of CPN20. This possibility remains to be elucidated in future studies.
Based on our experiments and known information on ABAR-mediated ABA signaling in the chloroplast (Shen et al., 2006; Wu et al., 2009; Shang et al., 2010; Du et al., 2012; Liu et al., 2012; Yang et al., 2012), we proposed a working model for O-fucosylation of CPN20 and its functional consequences (Figure 7). In the absence of ABA, un-activated ABA receptor ABAR, which spans the chloroplast envelopes, associates with CPN20 (Shang et al., 2010) and dissociates from WRKY40, which upregulates the expression and nuclear localization of WRKY40 and thereby suppresses expression of ABA-responsive transcription factors such as ABI5/ABI4 (Shang et al., 2010; Liu et al., 2012). In the presence of ABA, ABA-bound ABAR dissociates from CPN20 and associates with WRKY40, leading to the initiation of ABA signaling by inhibiting the expression and nuclear localization of WRKY40. This thereby de-represses expression of ABA-responsive transcription factors such as ABI5/ABI4 (Shang et al., 2010; Liu et al., 2012; Zhang et al., 2014). Meanwhile, ABA inhibits the expression of CPN20 and causes CPN20 dissociation from ABAR while promoting CPN20 association with SPY which thereby enhances CPN20 O-fucosylation and inhibits its accumulation in the chloroplast. This process aids in the interaction between ABAR and WRYK40, and in relieving repression of ABA-responsive transcription factors by WRKY40.
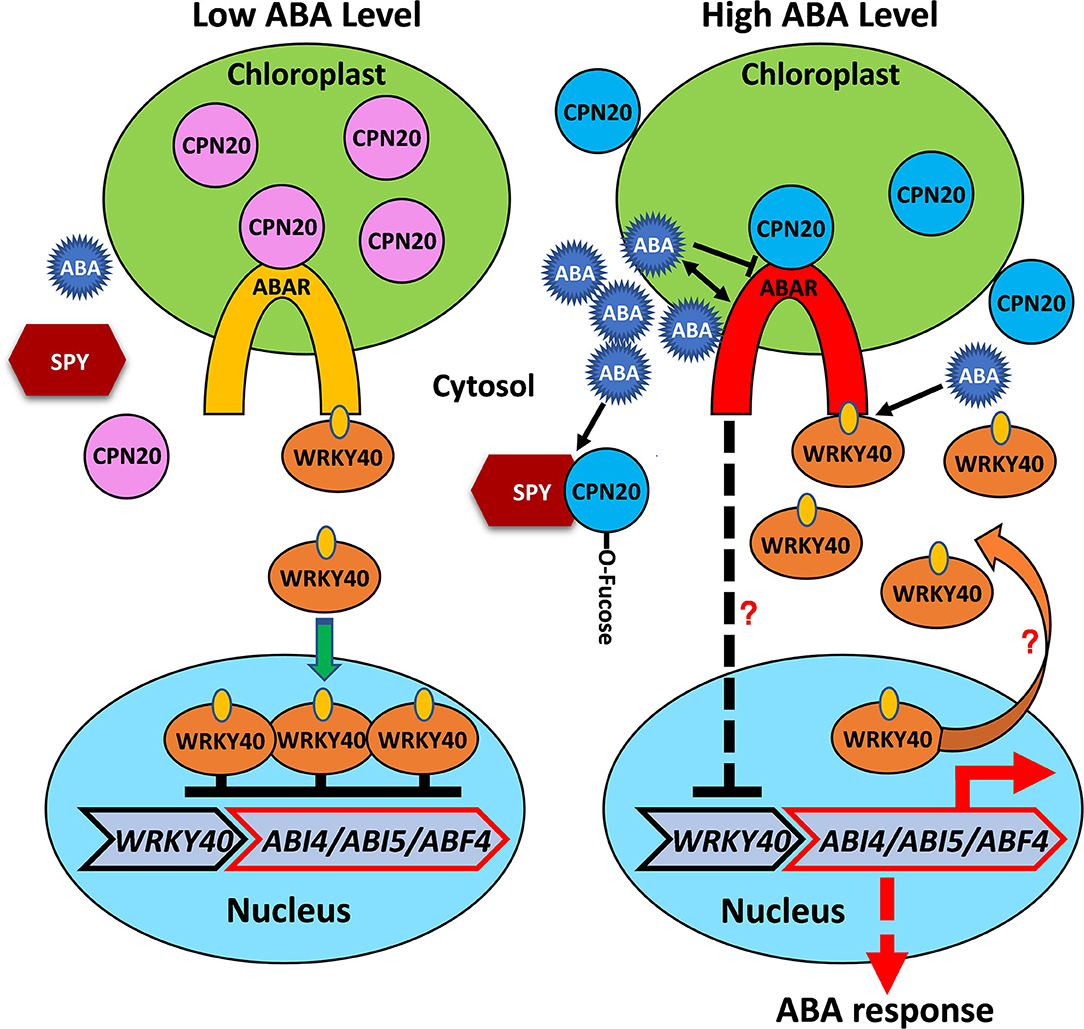
Figure 7. Proposed working model for O-fucosylatation of CPN20 and its functional consequences. The chloroplast envelop-localized ABA receptor ABAR interacts with the WRKY40 transcription factor, a central negative regulator, which inhibits expression of ABA-responsive genes, such as ABI5, ABI4, and ABF4. A high level of ABA signal promotes ABAR-WRKY40 interaction and translocation of WRKY40 from the nucleus to the cytosol, represses WRKY40 expression with unknown mechanisms. These processes lead to relief of the inhibition of ABI5 expression by nucleus-localized WRKY40. In the presence of a high level of ABA, enhanced SPY-CPN20 interaction facilitates O-fucosylation of CPN20 and reduces its localization in the chloroplast. This de-represses CPN20 inhibition of ABAR-initiated ABA signaling along with ABA-inhibited ABAR-CPN20 interaction. Note that dash line indicates possibly with an indirect process, question mark represents with unknown mechanism.
Author's Note
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD027272 with a link at https://www.ebi.ac.uk/pride/archive/projects/PXD027272/private.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ebi.ac.uk/pride/archive/, PXD027272.
Author Contributions
L-MF and LL designed the experiments. LL and QW constructed the plant materials and plasmids, performed protein-protein interaction, imaging, immunoblot, and seed germination assays. LL conducted gene expression analysis while WZ and QW conducted Mass Spectrometry analyses. QW and YW conducted in vitro peptide O-fucosylation. QW conducted in vivo protein degradation assay. ZS and YW conducted part of immunoblot analyses, microscopic observation, and seed germination assays. QL constructed part of plasmids. JY performed yeast screen. YB conducted part of seed germination assays. L-MF, LL, QW, and ZS analyzed the data and wrote the article. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (# 31570248, # 31770276, and # 31370288).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Drs. Da-Peng Zhang, Tai-ping Sun, and Neil Olszewski for providing some of the plant materials and valuable comments on the study and manuscript. We thank Dr. Xing Chen for valuable discussions and for providing the equipment for some of the experiments. We thank Drs. Jianmin Zhou, Hongwei Guo, and Xing Wang Deng for providing plasmids. We thank Drs. Jianmin Zhou and Xiaofeng Zheng for providing the equipment for luciferase assay. We thank Dr. Chuanmao Zhang for providing some of the plasmids. We thank Dr. Yiqun Liu for the image analysis. We thank an linguistic expert from service for the correction of the writing. Part of the imaging analyses was assisted by Yiqun Liu at the core facility of the School of Life Sciences, Peking University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.724144/full#supplementary-material
Supplementary Figure 1. BK-SPY does not have autoactivation.
Supplementary Figure 2. Purification of GFP-tagged CPN20 and CPN10α1 by immuno-precipitation from 293T cells.
Supplementary Figure 3. GFP-tagged CPN20 purified from germinating seeds of CPN20-GFP-overexpressing plant.
Supplementary Figure 4. CPN20 peptide is O-fucosylated in an in vitro SPY activity assay.
Supplementary Figure 5. CPN20-GFP localization in chloroplasts of cotyledon mesophyll and guard cells of 30-day-old seedlings.
Supplementary Figure 6. cpn20-1 suppresses insensitivity of spy-3 to ABA.
Supplementary Figure 7. SPY is absent in the chloroplast.
Supplementary Table 1. Primers used in this article.
References
Bertsch, U., Soll, J., Seetharam, R., and Viitanen, P.V. (1992). Identification, characterization, and DNA sequence of a functional “double” groES-like chaperonin from chloroplasts of higher plants. Proc. Natl. Acad. Sci. U.S.A. 89, 8696–8700. doi: 10.1073/pnas.89.18.8696
Bond, M. R., and Hanover, J. A. (2015). A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880. doi: 10.1083/jcb.201501101
Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., et al. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427. doi: 10.1111/j.1365-313X.2004.02206.x
Camut, L., Daviere, J. M., and Achard, P. (2017). Dynamic regulation of DELLA protein activity: SPINDLY and SECRET AGENT unmasked! Mol. Plant 10, 785–787. doi: 10.1016/j.molp.2017.05.003
Chen, C. I., Keusch, J. J., Klein, D., Hess, D., Hofsteenge, J., and Gut, H. (2012). Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 31, 3183–3197. doi: 10.1038/emboj.2012.143
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Cui, H. (2015). Cortex proliferation in the root is a protective mechanism against abiotic stress. Plant Signal. Behav. 10:e1011949. doi: 10.1080/15592324.2015.1011949
Cui, H., and Benfey, P. N. (2009). Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 58, 1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x
Cui, H., Kong, D., Wei, P., Hao, Y., Torii, K. U., Lee, J. S., et al. (2014). SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the Arabidopsis root. Mol. Plant 7, 1727–1739. doi: 10.1093/mp/ssu106
Du, S. Y., Zhang, X. F., Lu, Z., Xin, Q., Wu, Z., Jiang, T., et al. (2012). Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol. Biol. 80, 519–537. doi: 10.1007/s11103-012-9965-3
Greenboim-Wainberg, Y., Maymon, I., Borochov, R., Alvarez, J., Olszewski, N., Ori, N., et al. (2005). Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17, 92–102. doi: 10.1105/tpc.104.028472
Hartweck, L. M., Genger, R. K., Grey, W. M., and Olszewski, N. E. (2006). SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J. Exp. Bot. 57, 865–875. doi: 10.1093/jxb/erj071
Hartweck, L. M., Scott, C. L., and Olszewski, N. E. (2002). Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161, 1279–1291. doi: 10.1093/genetics/161.3.1279
Hirohashi, T., Nishio, K., and Nakai, M. (1999). cDNA sequence and overexpression of chloroplast chaperonin 21 from Arabidopsis thaliana. Biochim. Biophys. Acta 1429, 512–515. doi: 10.1016/S0167-4838(98)00268-4
Hofsteenge, J., Huwiler, K. G., Macek, B., Hess, D., Lawler, J., Mosher, D. F., et al. (2001). C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 276, 6485–6498. doi: 10.1074/jbc.M008073200
Hsu, P. K., Dubeaux, G., Takahashi, Y., and Schroeder, J. I. (2021). Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 105, 307–321.
Hu, J., Mitchum, M. G., Barnaby, N., Ayele, B. T., Ogawa, M., Nam, E., et al. (2008). Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20, 320–336. doi: 10.1105/tpc.107.057752
Jacobsen, S. E., Binkowski, K. A., and Olszewski, N. E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 9292–9296. doi: 10.1073/pnas.93.17.9292
Jacobsen, S. E., and Olszewski, N. E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. doi: 10.1105/tpc.5.8.887
Jung, C., Zhao, P., Zhao, Seo, J. S., Mitsuda, N., Deng, S., and Chua, N. H. (2015). PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27, 2016–2031. doi: 10.1105/tpc.15.00385
Koumoto, Y., Shimada, T., Kondo, M., Hara-Nishimura, I., and Nishimura, M. (2001). Chloroplasts have a novel Cpn10 in addition to Cpn20 as co-chaperonins in Arabidopsis thaliana. J. Biol. Chem. 276, 29688–29694. doi: 10.1074/jbc.M102330200
Lee, S., Cheng, H., King, K. E., Wang, W., He, Y., Hussain, A., et al. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. doi: 10.1101/gad.969002
Li, H. M., and Chiu, C. C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61, 157–180. doi: 10.1146/annurev-arplant-042809-112222
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Liang, L., Yang, J., Gao, Z., Wang, Q., Liang, Q., Song, Z., et al. (2018). SPINDLY is involved in ABA signaling bypassing the PYR/PYLs/RCARs-mediated pathway and partly through functional ABAR. Environ. Exp. Bot. 151, 43–54. doi: 10.1016/j.envexpbot.2018.04.004
Lira-Navarrete, E., Valero-Gonzalez, J., Villanueva, R., Martinez-Julvez, M., Tejero, T., Merino, P., et al. (2011). Structural insights into the mechanism of protein O-fucosylation. PLoS ONE 6:e25365. doi: 10.1371/journal.pone.0025365
Liu, X., Yue, Y., Li, B., Nie, Y., Li, W., Wu, W. H., et al. (2007). A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315, 1712–1716. doi: 10.1126/science.1135882
Liu, Z. Q., Yan, L., Wu, Z., Mei, C., Lu, K., Yu, Y. T., et al. (2012). Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 63, 6371–6392. doi: 10.1093/jxb/ers293
Lopaticki, S., Yang, A. S. P., John, A., Scott, N. E., Lingford, J. P., O'Neill, M. T., et al. (2017). Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat. Commun. 8:561. doi: 10.1038/s41467-017-00571-y
Luther, K. B., and Haltiwanger, R. S. (2009). Role of unusual O-glycans in intercellular signaling. Int. J. Biochem. Cell Biol. 41, 1011–1024. doi: 10.1016/j.biocel.2008.10.001
Maymon, I., Greenboim-Wainberg, Y., Sagiv, S., Kieber, J. J., Moshelion, M., Olszewski, N., et al. (2009). Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J. 58, 979–988. doi: 10.1111/j.1365-313X.2009.03840.x
Okajima, T., and Irvine, K. D. (2002). Regulation of notch signaling by O-linked fucose. Cell 111, 893–904. doi: 10.1016/S0092-8674(02)01114-5
Olszewski, N. E., West, C. M., Sassi, S. O., and Hartweck, L. M. (2011). O-GlcNAc protein modification in plants: evolution and function. Biochim. Biophys. Acta. 1800, 49–68. doi: 10.1016/j.bbagen.2009.11.016
Pandey, S., Nelson, D. C., and Assmann, S. M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136–148. doi: 10.1016/j.cell.2008.12.026
Panin, V. M., Shao, L., Lei, L., Moloney, D.J., Irvine, K. D., and Haltiwanger, R. S. (2002). Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 277, 29945–29952 doi: 10.1074/jbc.M204445200
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2c protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Piskurewicz, U., and Lopez-Molina, L. (2009). The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal. Behav. 4, 63–65. doi: 10.4161/psb.4.1.7331
Qin, F., Kodaira, K. S., Maruyama, K., Mizoi, J., Tran, L. S., Fujita, Y., et al. (2011). SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. doi: 10.1104/pp.111.187302
Rana, N. A., and Haltiwanger, R. S. (2011). Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 21, 583–589. doi: 10.1016/j.sbi.2011.08.008
Robertson, M., Swain, S. M., Chandler, P. M., and Olszewski, N. E. (1998). Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10, 995–1007. doi: 10.1105/tpc.10.6.995
Roos, M. D., and Hanover, J. A. (2000). Structure of O-linked GlcNAc transferase: mediator of glycan-dependent signaling. Biochem. Biophys. Res. Commun. 271, 275–280. doi: 10.1006/bbrc.2000.2600
Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S. Y., Jamin, M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. doi: 10.1038/nature08591
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Y. Y., Wang, X. F., Wu, F. Q., Du, S. Y., Cao, Z., Shang, Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826. doi: 10.1038/nature05176
Sjuts, I., Soll, J., and Bolter, B. (2017). Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front. Plant Sci. 8:168. doi: 10.3389/fpls.2017.00168
Steber, C. M., Cooney, S. E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. doi: 10.1093/genetics/149.2.509
Steiner, E., Efroni, I., Gopalraj, M., Saathoff, K., Tseng, T. S., Kieffer, M., et al. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24, 96–108. doi: 10.1105/tpc.111.093518
Steiner, E., Livne, S., Kobinson-Katz, T., Tal, L., Pri-Tal, O., Mosquna, A., et al. (2016). The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits Class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171, 1485–1494. doi: 10.1104/pp.16.00343
Sun, N., Wang, J., Gao, Z., Dong, J., He, H., Terzaghi, W., et al. (2016). Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. U.S.A. 113, 6071–6076. doi: 10.1073/pnas.1604782113
Swain, S. M., Tseng, T. S., and Olszewski, N. E. (2001). Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 126, 1174–1185. doi: 10.1104/pp.126.3.1174
Swain, S. M., Tseng, T. S., Thornton, T. M., Gopalraj, M., and Olszewski, N. E. (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 129, 605–615. doi: 10.1104/pp.020002
Thornton, T. M., Swain, S. M., and Olszewski, N. E. (1999). Gibberellin signal transduction presents the SPY who O-GlcNAc'd me. Trends Plant Sci. 4, 424–428. doi: 10.1016/S1360-1385(99)01485-5
Tseng, T. S., Salome, P. A., McClung, C. R., and Olszewski, N. E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16, 1550–1563. doi: 10.1105/tpc.019224
Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S.J., et al. (2002). N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793. doi: 10.1126/science.298.5599.1790
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wang, Y., He, Y., Su, C., Zentella, R., Sun, T. P., and Wang, L. (2020). Nuclear localized O-fucosyltransferase SPY facilitates PRR5 proteolysis to fine-tune the pace of Arabidopsis circadian clock. Mol. Plant 13, 446–458. doi: 10.1016/j.molp.2019.12.013
Wilson, R. N., and Somerville, C. R. (1995). Phenotypic suppression of the Gibberellin-Insensitive Mutant (gai) of Arabidopsis. Plant Physiol. 108, 495–502. doi: 10.1104/pp.108.2.495
Wu, F. Q., Xin, Q., Cao, Z., Liu, Z. Q., Du, S. Y., Mei, C., et al. (2009). The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol. 150, 1940–1954. doi: 10.1104/pp.109.140731
Yang, S., Zeng, X., Li, T., Liu, M., Zhang, S., Gao, S., et al. (2012). AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis. J. Exp. Bot. 63, 3959–3973. doi: 10.1093/jxb/ers072
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Young, N. M., Brisson, J. R., Kelly, J., Watson, D. C., Tessier, L., Lanthier, P. H., et al. (2002). Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539. doi: 10.1074/jbc.M206114200
Zentella, R., Hu, J., Hsieh, W. P., Matsumoto, P. A., Dawdy, A., Barnhill, B., et al. (2016). O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 30, 164–176. doi: 10.1101/gad.270587.115
Zentella, R., Sui, N., Barnhill, B., Hsieh, W. P., Hu, J., Shabanowitz, J., et al. (2017). The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 13, 479–485. doi: 10.1038/nchembio.2320
Zhang, X., Jiang, T., Yu, Y., Wu, Z., Jiang, S., Lu, K., et al. (2014). Arabidopsis co-chaperonin CPN20 antagonizes Mg-chelatase H subunit to derepress ABA-responsive WRYK40 transcription repressor. Sci. China Life Sci. 57, 11–21. doi: 10.1007/s11427-013-4587-9
Keywords: Arabidopsis thaliana, SPINDLY, CPN20, O-fucosylation, abscisic acid (ABA), seed germination, seedling development, chloroplast
Citation: Liang L, Wang Q, Song Z, Wu Y, Liang Q, Wang Q, Yang J, Bi Y, Zhou W and Fan L-M (2021) O-fucosylation of CPN20 by SPINDLY Derepresses Abscisic Acid Signaling During Seed Germination and Seedling Development. Front. Plant Sci. 12:724144. doi: 10.3389/fpls.2021.724144
Received: 12 June 2021; Accepted: 10 September 2021;
Published: 12 October 2021.
Edited by:
Randy D. Allen, Oklahoma State University, United StatesReviewed by:
Jing Bo Jin, Chinese Academy of Sciences (CAS), ChinaPingfang Yang, Hubei University, China
Copyright © 2021 Liang, Wang, Song, Wu, Liang, Wang, Yang, Bi, Zhou and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Zhou, d2VuLnpob3VAcGt1LmVkdS5jbg==; Liu-Min Fan, bG1mYW5AcGt1LmVkdS5jbg==
†These authors have contributed equally to this work
‡These authors have contributed equally to this work and share last authorship
 Lin Liang
Lin Liang Qi Wang1†
Qi Wang1† Zihao Song
Zihao Song Wen Zhou
Wen Zhou Liu-Min Fan
Liu-Min Fan