
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 September 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.721548
This article is part of the Research Topic Elicitors, Secret Agents at the Service of the Plant Kingdom View all 10 articles
The receptor-like kinase Suppressor of BIR1 (SOBIR1) binds various receptor-like proteins (RLPs) that perceive microbe-associated molecular patterns (MAMPs) at the plasma membrane, which is thought to activate plant pattern-triggered immunity (PTI) against pathogen invasion. Despite its potentially crucial role, how SOBIR1 transmits immune signaling to ultimately elicit PTI remains largely unresolved. Herein, we report that a Nicotiana benthamiana gene NbRLP1, like NbSOBIR1, was highly induced upon Phytophthora parasitica infection. Intriguingly, NbRLP1 is characterized as a receptor-like protein localizing to the endoplasmic reticulum (ER) membrane rather than the plasma membrane. Using bimolecular fluorescence complementation and affinity purification assays, we established that NbRLP1 is likely to associate with NbSOBIR1 through the contact between the ER and plasma membrane. We further found that NbSOBIR1 at the plasma membrane partitions into mobile microdomains that undergo frequent lateral movement and internalization. Remarkably, the dynamics of NbSOBIR1 microdomain is coupled to the remodeling of the cortical ER network. When NbSOBIR1 microdomains were induced by the P. parasitica MAMP ParA1, tobacco cells overexpressing NbRLP1 accelerated NbSOBIR1 internalization. Overexpressing NbRLP1 in tobacco further exaggerated the ParA1-induced necrosis. Together, these findings have prompted us to propose that ER and the ER-localized NbRLP1 may play a role in transmitting plant immune signals by regulating NbSOBIR1 internalization.
As a sessile organism, plants have evolved a series of defense mechanisms to protect themselves against pathogens. Plant cells sense potential pathogens through the function of pattern recognition receptors (PRRs) that recognize a wide range of microbe-associated molecular patterns (MAMPs) to ultimately elicit pattern-triggered immunity (PTI) (Jones and Dangl, 2006; Dodds and Rathjen, 2010). The onset of PTI can rapidly activate a series of downstream plant defense responses, including callose deposition, generation of reactive oxygen species (ROS), activation of mitogen-activated protein kinase cascade, induction of defense genes, and in some cases cell death (Boller and Felix, 2009; Yu et al., 2017).
Plant PRRs are plasma membrane-localized transmembrane protein receptors belonging to the family of receptor-like kinases (RLKs) or receptor-like proteins (RLPs), which differ in the presence or absence of intracellular kinase domains (Zipfel, 2014). The emerging evidence suggests that plant immune response is triggered upon the perception of MAMPs, accompanied by the association of selected PRRs with additional factors, such as BRI1-associated receptor kinase 1 (BAK1), also known as somatic embryogenesis receptor-like kinase 3 (SERK3), and other members of the SERK family to form multimeric kinase complex on the cell surface (Roux et al., 2011; Albert et al., 2020). Moreover, PRRs of the RLP type are known to constitutively form complexes with Suppressor of BIR1 (SOBIR1), an RLK containing a short leucine-rich repeat (LRR) ectodomain (Liebrand et al., 2013).
SOBIR1 was originally identified in Arabidopsis thaliana as a counterplayer of BAK1-interacting receptor-like kinase1 (BIR1) (Gao et al., 2009). When overexpressed, SOBIR1 constitutively activates cell death and plant immune responses. Recent studies further demonstrated that SOBIR1 is indispensable for PTI elicited by various MAMPs, including sclerotinia culture filtrate elicitor 1 (SCFE1) from Sclerotinia sclerotiorum, Botrytis cinerea endopolygalacturonase 1, Phytophthora elicitins, and Phytophthora sojae XEG1, all of which are perceived by PRRs of the LRR-RLP type (Zhang et al., 2013, 2014; Du et al., 2015; Wang et al., 2018). Thus, SOBIR1 appears to function as a common adaptor protein for various LRR-RLP PRRs and thus is thought to play a central role in plant immunity.
Recent studies based on high-resolution microscopy indicate that the immune and growth receptors form distinct nanodomains on the plasma membrane and these organized units potentially provide specificity for the downstream signaling events in plants (Bücherl et al., 2017). Nonetheless, how exactly the upstream perception event communicates with intracellular components during plant immune signaling remains largely unknown. Notably, a variety of organelles are found in close proximity with the plasma membrane in plant cells, such as the endoplasmic reticulum (ER). This organelle occupies a large volume in the cell and exerts multiple functions, including protein and lipid synthesis, calcium storage, vesicular trafficking, as well as biogenesis of other organelles (Phillips and Voeltz, 2016). Moreover, it is known to further compartmentalize into sheets and tubules, along with a variety of microdomains, to enable its versatile functions (English et al., 2009). In Arabidopsis, the ER makes contact with the plasma membrane through the ER–PM contact site (EPCS), with synaptotagmin 1 (SYT1) and vesicle-associated protein 27 (VAP27) as tethers that mediate EPCS formation (Perez-Sancho et al., 2016). The presence of membrane contact site (MCS) defines a unique feature of eukaryotes. More and more pieces of evidence have shown that MCS form transiently between two membrane compartments, creating a bridge for interorganelle communication, such as exchanges of metabolite, cellular stress response, membrane dynamics, and signaling (Prinz et al., 2020). However, to date, not much is known about the association of cellular organelles with the PRRs and their role in plant immune responses.
Previously, we demonstrated that tomato (Solanum lycopersicum) SlSOBIR1 and SlSOBIR1-like are required for the perception of the elicitin ParA1, a MAMP from the oomycete pathogen Phytophthora parasitica, and for plant defense against this pathogen (Peng et al., 2015). We documented that SlSOBIR1 is translocated from the plasma membrane to endosomes in response to ParA1 treatment, which suggests that SlSOBIR1 endocytosis is coupled to the plant immune signaling. To further tackle the function and regulation of SOBIR1, we took the biochemical approach to look for SOBIR1-interacting proteins. In this study, we report a novel RLP from Nicotiana benthamiana, named NbRLP1 that encodes an unconventional RLP protein residing within the ER membrane, but not the plasma membrane. By using bimolecular fluorescence complementation and tandem-affinity purification, we validated the interaction between NbRLP1 and NbSOBIR1. In addition, we observed that NbSOBIR1 formed microdomains on the plasma membrane whose dynamics is coupled to the remodeling of the ER. Further evidence supports that NbRLP1 downregulates the number of NbSOBIR1 microdomains and promotes NbSOBIR1 internalization in the presence of ParA1 that triggers partitioning of NbSOBIR1 into the microdomains. As overexpressing NbRLP1 exaggerated the ParA1-induced necrosis in plants, we propose that ER and unconventional RLPs in the ER, such as NbRLP1, are engaged in SOBIR1-mediated plant immunity through mediating MAMP-induced SOBIR1 internalization.
SlSOBIR1 is involved in PTI response to the P. parasitica MAMP termed ParA1 (Peng et al., 2015). To identify proteins associated with SlSOBIR1 during the process, we treated Nicotiana benthamiana harboring SlSOBIR1-GFP with ParA1, followed by immunoprecipitation with anti-GFP antibody. Among a collection of putative SlSOBIR1-interacting proteins, we selected one of the major candidates, NbS0003586g0006.1 [Niben.0.4.4., Sol genetic network (SGN1)] for further analysis. In contrast to no signal in the control without reverse transcriptase, two major transcripts were detected by reverse transcriptase-PCR reaction (Figure 1A). One is a 2.91 Kb cDNA (GenBank Accession number MW924093), which corresponds to the exact genomic DNA sequence of NbRLP1, and the other shorter cDNA of 1.41 Kb lacks the putative intron sequence (MW924094), reflecting a spliced form of NbRLP1 (Supplementary Table 1). We focused on the 2.91-Kb gene, named NbRLP1 hereafter, that encodes a 969-amino acid protein with a characteristic of RLPs (Figure 1A).
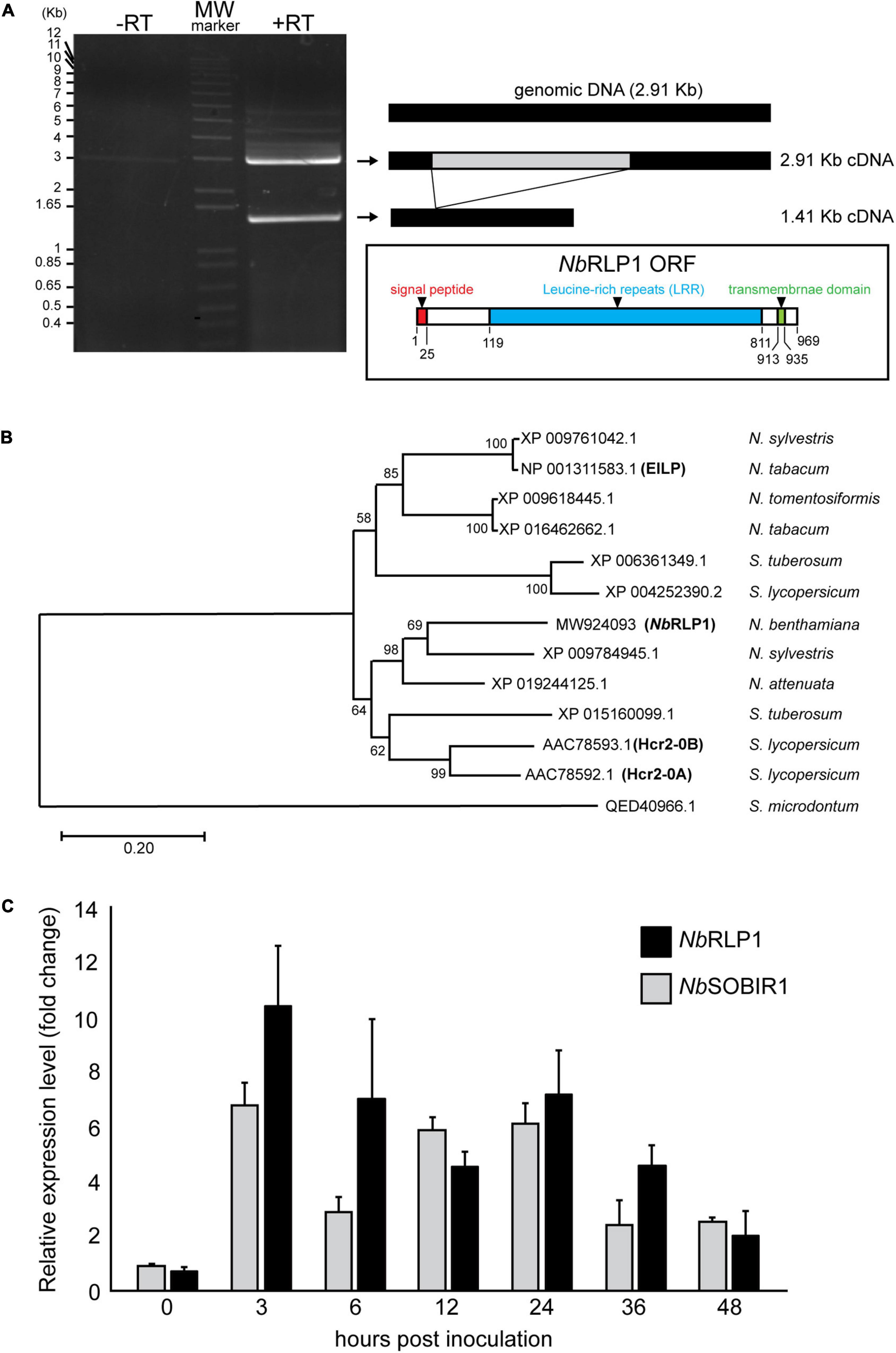
Figure 1. The expression of NbRLP1 is induced in response to infection by Phytophthora parasitica. (A) NbRLP1 is alternatively spliced. Reverse transcriptase-polymerase chain reaction (RT-PCR) gave rise to two amplified products with respective length of 1.41 and 2.91 Kb (+ RT), which were not detected in the absence of reverse transcriptase (-RT). The 2.91-Kb gene, named NbRLP1 (MW924093) encodes a 969-amino acid protein containing a signal peptide (predicted by SignalP), an LRR domain (predicted by InterPro), and a transmembrane domain (predicted by TMHMM) as illustrated in the inset. (B) Phylogenetic analysis of NbRLP1. Multiple sequence alignment of NbRLP1 and its homologs from Nicotiana and Solanum retrieved from the protein database of the National Center for Biological information (NCBI) was performed by using ClustalX. The phylogenetic tree was constructed by employing the maximum likelihood method, followed by bootstrap analysis with 1,000 pseudo-replicates. (C) The expression of the NbRLP1 and NbSOBIR1 is upregulated in response to P. parasitica infection. At the indicated hours post P. parasitica zoospore inoculation of Nicotiana benthamiana, total RNAs were isolated and analyzed by quantitative reverse transcription PCR (RT-PCR). Data are presented as fold-change relative to mock treatment of the same time point. Values are means (±SE) from three independent experiments.
Phylogenetic analysis of NbRLP1 and its homologs from Solanaceae retrieved from the NCBI databases indicates that XP_009784945 of Nicotiana sylvestris and XP_019244125 of Nicotiana attenuata are close homologs of NbRLP1 (Figure 1B). These genes along with AAC78592 (Hcr2-0A) and AAC78593 (Hcr2-0B), two homologs of tomato disease resistance gene, Cf-5 (Dixon et al., 1998), form a clade distinct from that encompassing elicitor-inducible EILP of Nicotiana tabacum (Takemoto et al., 2000). As well, these two clades are distinguished from ELR of Solanum microdontum (QED40966), an RLP involved in the recognition of Phytophthora elicitins (Du et al., 2015).
To know whether NbRLP1 is induced by pathogen infection, we infected N. benthamiana with P. parasitica zoospores and performed quantitative RT-PCR, with the expression level of elongation factor 1 alpha (NbEF1a) serving as an internal control. The results showed that the expression of NbRLP1, like NbSOBIR1, was largely induced 3 h post-inoculation (hpi) and persists high expression through later infection stage to at least 36 hpi (Figure 1C). Collectively, these data imply a potential role of NbRLP1 for plant defense responses.
To protect against pathogens, plant cells possess a variety of transmembrane RLKs and RLPs on the plasma membranes, many of which contain extracellular LRR domains to enable ligand recognition. Intriguingly, when we analyzed NbRLP1 localization by the use of GFP fused to either amino- or carboxyl-terminus of NbRLP1 for fluorescence microscopy, we observed a distribution pattern of network and/or multiple puncta near the cell cortex (Figure 2A). As it resembles the plant ER network, we next examined cells coexpressing NbRLP1-GFP with the plasma membrane marker AtACA8-mCherry (Figure 2B) or the ER marker mCherry-KDEL (Figure 2C). The results showed that NbRLP1-GFP localization is in large agreement with the luminal ER protein mCherry-KDEL but not with the evenly distributed AtACA8-mCherry signal of the plasma membrane. Thus, we conclude that NbRLP1 is an unconventional RLP localizing to the ER rather than the plasma membrane. Based on sequence prediction results, NbRLP1 is likely to adapt the standard type I transmembrane protein topology in the ER as depicted in Figure 2D. The main moiety of this protein, containing LRR, is in the ER lumen, and the protein also contains a single spanning transmembrane helix and a short cytoplasmic tail.
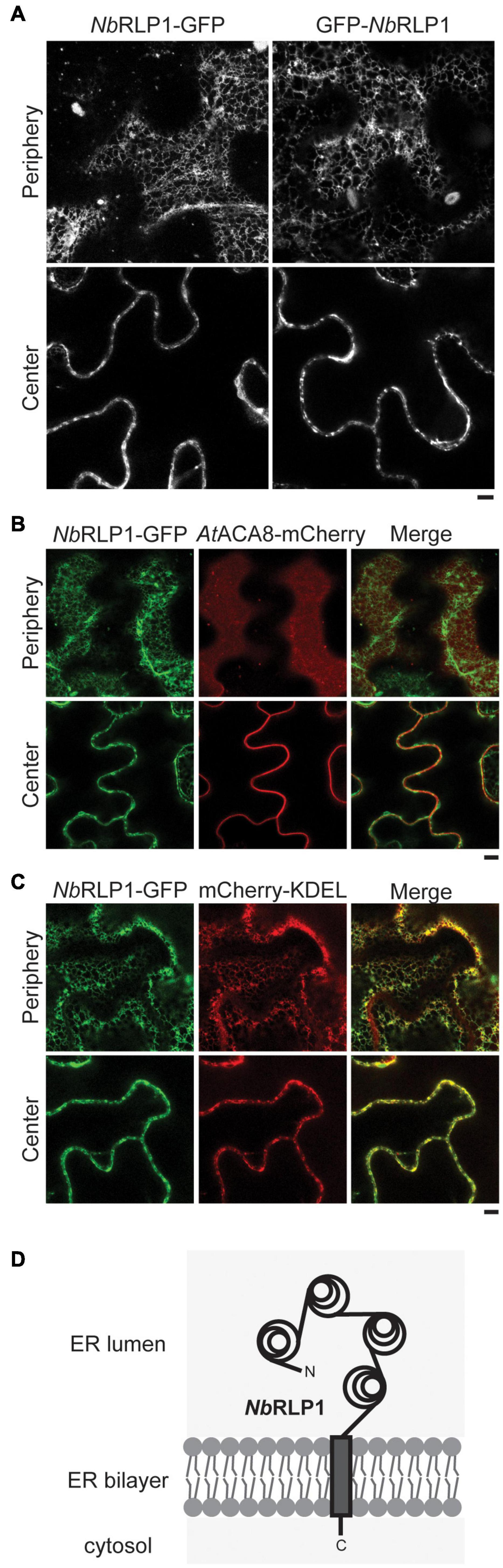
Figure 2. NbRLP1-GFP resides in the endoplasmic reticulum (ER) membrane. (A) Still images of NbRLP1-GFP or GFP-NbRLP1 taken from the periphery or the center of the same Nicotiana benthamiana leaves by Zeiss LSM880 confocal microscope. Scale bar, 5 μm. (B) Same as A, except that NbRLP1-GFP and AtACA8-mCherry are coexpressed in plants and imaged. Also shown are the corresponding merged images. (C) Same as B, except that NbRLP1-GFP and mCherry-KDEL are coexpressed in plants and imaged. (D) A scheme depicting NbRLP1 topology in the ER based on TMHMM.
To consolidate the potential relationship between NbRLP1 and NbSOBIR1, we first employed the bimolecular fluorescence complementation (BiFC) assay, known to provide in situ interacting information in planta. Our results indicate that NbRLP1-Vn, but not Vn-NbRLP1, interacts with NbSOBIR1-Vc in plants, thus showing Venus YFP fluorescence, in contrast to no or dim signals of various negative controls (Figure 3A). The fact that only Vn fused to NbRLP1 at the C-terminus but not to N-terminus gave a positive BiFC signal, which is consistent with the topological prediction of NbRLP1 in the ER (Figure 2D). Since NbRLP1 is an ER protein and NbSOBIR1 is a plasma membrane protein, we hypothesize that their interaction most likely is achieved through close proximity between the ER and plasma membrane. To know whether NbRLP1 may partition into the previously characterized EPCS, NbRLP1-GFP was coexpressed with the EPCS tether AtSYT1-mCherry. Indeed, we observed that signals of the two proteins colocalized at the cell periphery (Supplementary Figure 1). Thus, these results imply that the ER-localized NbRLP1 might interact with the plasma membrane-localized NbSOBIR1 protein in vivo, likely at a position where cortical ER and the plasma membrane join in physical proximity, such as the EPCS tethered by AtSYT1-mCherry.
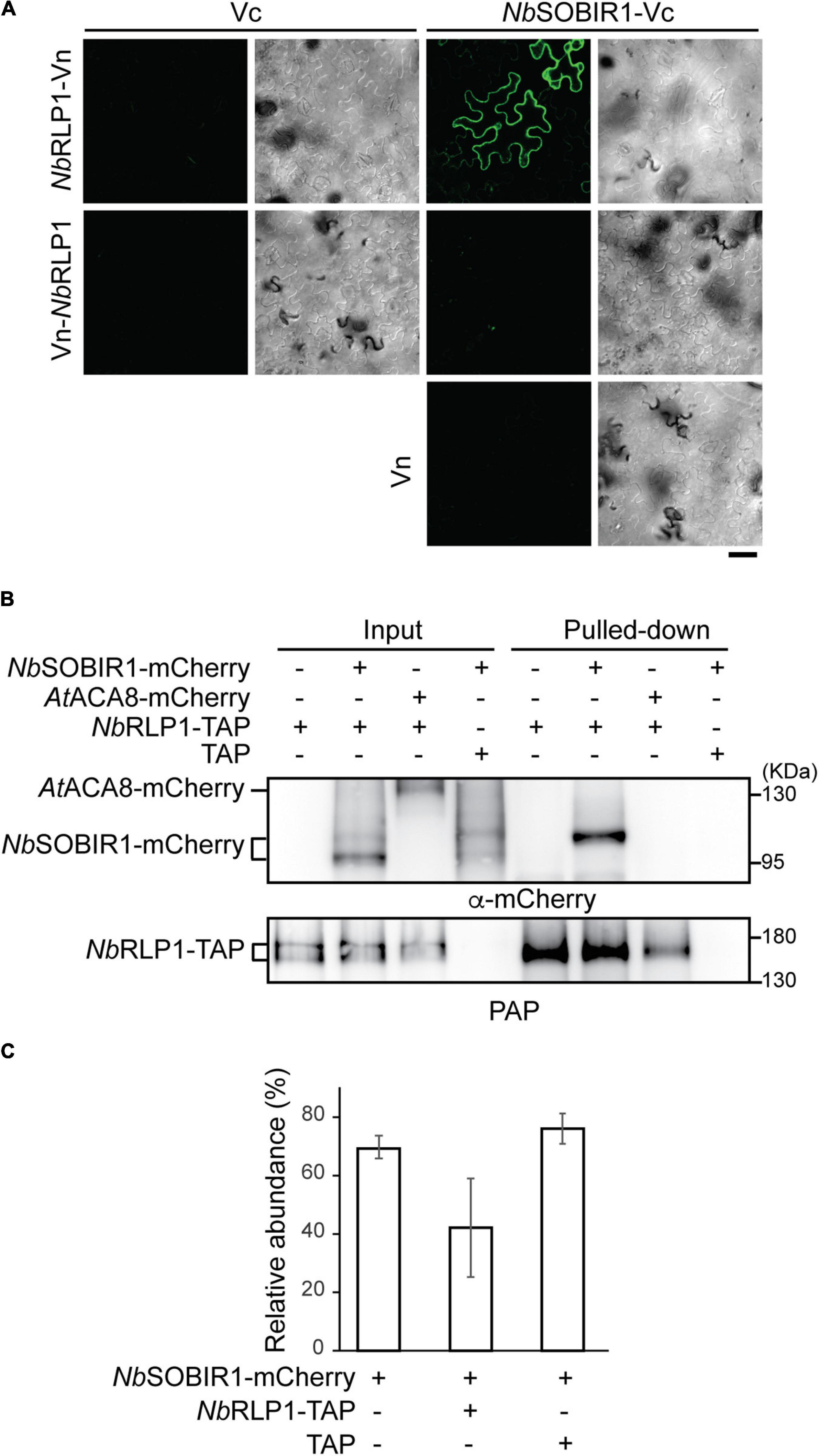
Figure 3. NbRLP1 interacts with NbSOBIR1. (A) The carboxyl half of Venus protein (Vc) alone or fused to NbSOBIR1 was coexpressed with the amino half of Venus (Vn) alone or fused to NbRLP1 as indicated on the Nicotiana benthamiana leaves. Images were taken by Zeiss LSM880 confocal microscope. Complemented Venus fluorescence and differential interference contrast (DIC) images of the same area are shown. Scale bar, 40 μm. (B) N. benthamiana leaves expressing NbSOBIR1-mCherry, AtACA8-mCherry, NbRLP1-TAP, or TAP as indicated were harvested and lyzed. The lysate (input) was subjected to pull-down by IgG Sepharose as described in the Materials and Methods section. The input and the bound (pulled-down) fractions were subjected to SDS-PAGE followed by the Western blot analysis with the use of anti-mCherry and PAP antibodies. (C) N. benthamiana leaves expressing NbSOBIR1-mCherry, NbRLP1-TAP, or TAP as indicated were lyzed and NbSOBIR1-mCherry proteins in the lysate were subjected to SDS-PAGE followed by Western blot analysis with anti-mCherry antibody. The ratio of the upper NbSOBIR1 band (with higher molecular mass) relative to the total NbSOBIR1 (combining both the upper and lower bands of NbSOBIR1) was plotted and compared. Data from three experimental repeats are shown as means (± SD).
We next performed biochemical studies to further strengthen the notion that NbRLP1 interacts with NbSOBIR1. To achieve this, we generated an NbRLP1 harboring a TAP tag (an S-tag followed by a protein A tag) at its C-terminus and asked whether the fusion protein (termed NbRLP1-TAP) binds NbSOBIR1-mCherry in detergent-solubilized plant extracts. To unambiguously judge the interaction, we also tested whether NbRLP1-TAP binds another plasma membrane protein AtACA8-mCherry. Our results showed that NbSOBIR1-mCherry, but not AtACA8-mCherry, was pulled down by NbRLP1-TAP, examined by Western blotting using an antibody against mCherry (Figure 3B) as well as mass spectrometry (not shown). In contrast, NbSOBIR1 signal has never been detected in the pulled-down fractions derived from TAP-alone expressing samples, indicative of specificity. Intriguingly, Western blotting data revealed two major forms of NbSOBIR1-mCherry with a size difference of ∼8.5 kD in the inputs; however, only NbSOBIR1-mCherry with the higher molecular weight was pulled down by NbRLP1-TAP (Figure 3B). Notably, the expression of NbRLP1-TAP but not TAP alone in plants reduced the proportion of the larger molecular weight form of NbSOBIR1 in the steady state (Figure 3C), implying that NbRLP1-TAP not only binds to this unique form of NbSOBIR1 but may further reduce its expression or accelerate its turnover.
The BiFC data supports that the short tail of NbRLP1 fused with Vn interacts with the cytoplasmic domain of NbSOBIR1 fused with Vc. To know the contribution of the N-terminal LRR domain and the short C-terminal cytoplasmic tail of NbRLP1 to its interaction with NbSOBIR1, we also prepared different truncated versions of NbRLP1. Analysis by using the biochemical pulled-down assay indicates NbRLP1-ΔC-TAP with the short cytoplasmic tail removed still bound with NbSOBIR1 (Supplementary Figure 2A). In contrast, NbRLP1-ΔN-TAP, having signal peptide but lacking the large LRR domain, is bound to NbSOBIR1 with reduced affinity (Supplementary Figure 2A). Although the NbRLP1 with cytoplasmic tail and a transmembrane anchor seems to be sufficient for binding with NbSOBIR1, the result implies that other proteins in the ER or the plasma membrane may bridge or stabilize the interaction between NbRLP1 and NbSOBIR1 (Supplementary Figure 2B). Collectively, both in vivo and in vitro data support the notion that the ER-localized NbRLP1 interacts with the plasma membrane-localized NbSOBIR1.
Having established that NbRLP1 interacts with NbSOBIR1, we next explored the potential regulatory mechanism underlying their interplay in planta. The observation of two major forms of NbSOBIR1-mCherry, while only one form is capable of binding with and being regulated by NbRLP1, prompted us to investigate whether NbSOBIR1 may associate with distinct structures on the plasma membrane. We thus performed confocal microscopy with Airyscan to gain insights into NbSOBIR1-mCherry localization on the plasma membrane. Interestingly, in addition to the dispersed signal along the plasma membrane, NbSOBIR1-mCherry was identified in several punctate structures, which was not observed when another plasma membrane protein, AtACA8-mCherry, was subjected to imaging by the Airyscan microscopy (Figure 4A). Thereby, we suggest that a portion of NbSOBIR1 is likely partitioned into microdomains on the plasma membrane.
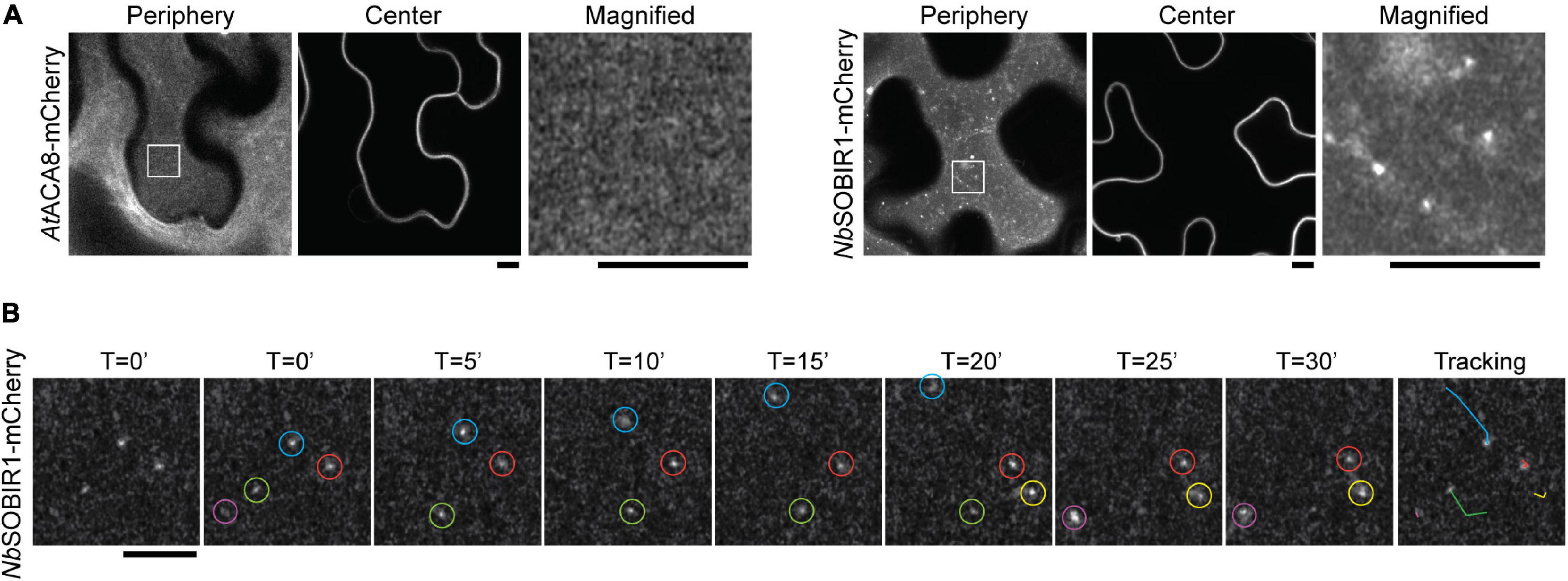
Figure 4. NbSOBIR1-mCherry forms microdomains in the plant plasma membrane. (A) Nicotiana benthamiana leaves expressing AtACA8-mCherry or NbSOBIR1-mCherry were imaged at the periphery or center sections using Zeiss LSM880 confocal microscope with Airyscan. The box area in the image of the periphery section was enlarged and displayed to show the microdomains of NbSOBIR1 that were not detectable on AtACA8-mCherry images. Scale bar, 5 μm. (B) N. benthamiana leaves expressing NbSOBIR1-mCherry was subjected to time-lapse microscopy by Zeiss LSM880 confocal microscope with the Airyscan. The total image acquisition length was 16 min with a time interval of 5 s (Supplementary Movie 1). Representative area was selected for particle tracking analysis using the ImageJ, and the image of each time point is shown. Five microdomains, each circled by different colors were identified in the displayed image and their tracks are shown in the right-most image.
To further understand the nature of NbSOBIR1-mCherry microdomains on the plasma membrane, we imaged these fine structures using time-lapse microscopy with a time interval of 5 s. The results clearly showed that these plasma membrane microdomains of NbSOBIR1 are mobile structures, although a small portion of them appeared relatively more static (Figure 4B and Supplementary Movie 1). Particle tracking analysis data indicated that the NbSOBIR1-mCherry microdomains can move laterally, showing an irregular pattern (Figure 4B). During the course of our analysis, we also observed the disappearance (Figure 4B, green circle) and emergence (Figure 4B, yellow circle) of these microdomains on the plasma membrane. In either case, it appears to involve a quick biogenesis and turnover process, detected within the time interval of 5 s. Considering SOBIR1 functions as an adaptor protein that assembled together with various LRR-RLPs into a receptor complex on the plasma membrane awaiting ligand binding, partitioning of SOBIR1 into microdomains may serve as a platform to facilitate such a process. Overall, these observations have led us to hypothesize that the dynamics of these microdomains of NbSOBIR1 is a regulated process and that the microdomain structure may represent one of the functional forms of NbSOBIR1.
Given that the aforementioned NbSOBIR1-NbRLP1 interaction depends on close proximity between the ER and the plasma membrane, we further asked whether these NbSOBIR1 microdomains may have an association with the ER. The eukaryotic ER is organized into a complicated network containing various degrees of sheets and tubules (Shibata et al., 2006; English et al., 2009). The highly dynamic ER disperses throughout the entire cytoplasm and is thought to fine-tune cell physiology to cope with environmental cues. When the localization of NbRLP1-GFP was compared with NbSOBIR1-mCherry, we noticed that NbSOBIR1-mCherry was either associated with the edge of the ER sheets or surrounded by the three-way junction of the ER tubules labeled with NbRLP1 at the cell periphery (Figure 5A).
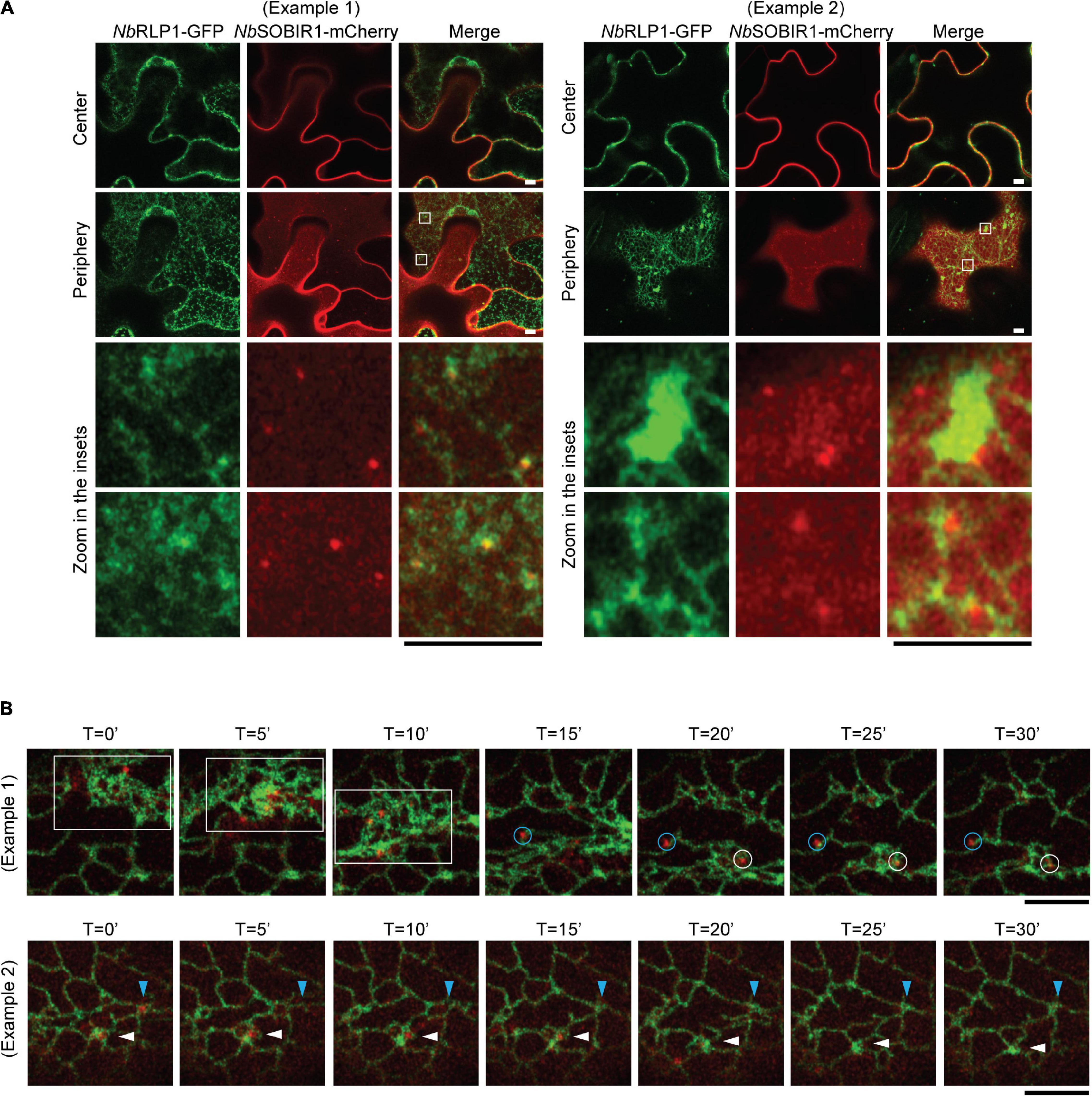
Figure 5. The NbSOBIR1 microdomains in the plasma membrane move as the cortical ER changes the morphology. (A) Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP were subjected to confocal microscopy by Zeiss LSM880 with use of Airyscan. The periphery and center sections of the epidermal cell of the same leaf were imaged, and the insets were enlarged to reveal the association between NbSOBIR1-mCherry and NbRLP1-GFP. Examples 1 and 2 show different degrees of ER tubules and sheets marked by NbRLP1-GFP to reveal how NbSOBIR1 microdomains are associated with the ER. (B) N. benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP were subjected to time-lapse microscopy by Zeiss LSM880 confocal microscope with use of Airyscan. The total image acquisition length was 16 min with a time interval of 5 s (Supplementary Movie 2). Representative area was selected for display by time course. (Example 1) White box area, containing several NbSOBIR1 microdomains, represents movement of these structures after a quick remodeling of the ER harboring high degree of sheets. Blue and white circles denote two NbSOBIR1 microdomains associated with ER tubules. (Example 2) Blue and white arrowheads denote two NbSOBIR1 microdomains that are internalized from the cell surface as the ER transformed from tubules to sheets. Scale bar, 5 μm.
Employing time-lapse microscopy, we observed that ER labeled by NbRLP1-GFP constantly undergoes morphological remodeling (Figure 5B and Supplementary Movie 2). Most importantly, the lateral movement of NbSOBIR1 microdomains was correlated with the dynamic changes of the ER, as the ER labeled with NbRLP1 always associated with the movement of the NbSOBIR1 microdomains (Figure 5B, blue and white circles, and Supplementary Movie 2). The correlation of the NbSOBIR1 microdomains with the cortical ER suggests that these microdomain structures are likely where NbSOBIR1 makes contact with NbRLP1 at the first place in their native state. Intriguingly, we observed that NbSOBIR1-mCherry microdomains diminished in the condition when a wave of ER sheets transit through the cell cortex during ER remodeling (Figure 5B, white box). In addition, we observed NbSOBIR1 microdomains originally surrounded by ER tubules disappeared in the next time point with the same area being replaced with ER sheets (Figure 5B, white and blue arrowheads). Collectively, the evidence in which NbSOBIR1 microdomain dynamics is correlated with ER remodeling raises an interesting possibility that ER may contribute to NbSOBIR1-mediated endocytosis and/or plant immunity.
In our previous paper, we have established that SlSOBIR1 endocytosis is triggered by the perception of an oomycete MAMP termed, ParA1. If the NbSOBIR1 microdomain defines a functional unit for the protein, we suspect that this structure is likely responsive to ParA1. Accordingly, we treated the NbSOBIR1-overexpressing plants with ParA1 to compare with the treatment with MES buffer, followed by Airyscan confocal microscopy to monitor the extent of NbSOBIR1 microdomain formation on the plasma membrane. Our results clearly showed that the number of NbSOBIR1 microdomain formed on the plasma membrane increased significantly in the ParA1-treated condition compared to the MES control, and the quantification results based on a number of cells examined further support the notion (Figure 6A). Remarkably, ParA1 treatment resulted in the formation of more NbSOBIR1-mCherry microdomains of a larger size which appeared more static (Figure 6A and Supplementary Movie 3). Time-lapse microscopy experiments performed with N. benthamiana plants coexpressing NbRLP1 identified that these larger NbSOBIR1-mCherry puncta though relatively more static can become diminished and/or disappear quickly (Figure 6B, blue and yellow circles, and Supplementary Movie 4), representing the occurrence of either endocytosis or diffusion upon ParA1 perception. While focusing on the focal plane containing the ParA1-induced static NbSOBIR1 microdomains, time-lapse microscopy revealed that these structures move inward from the cell cortex, which supports that endocytosis of NbSOBIR1 occurs directly through the microdomain structures upon ParA1 perception (Supplementary Figure 3).
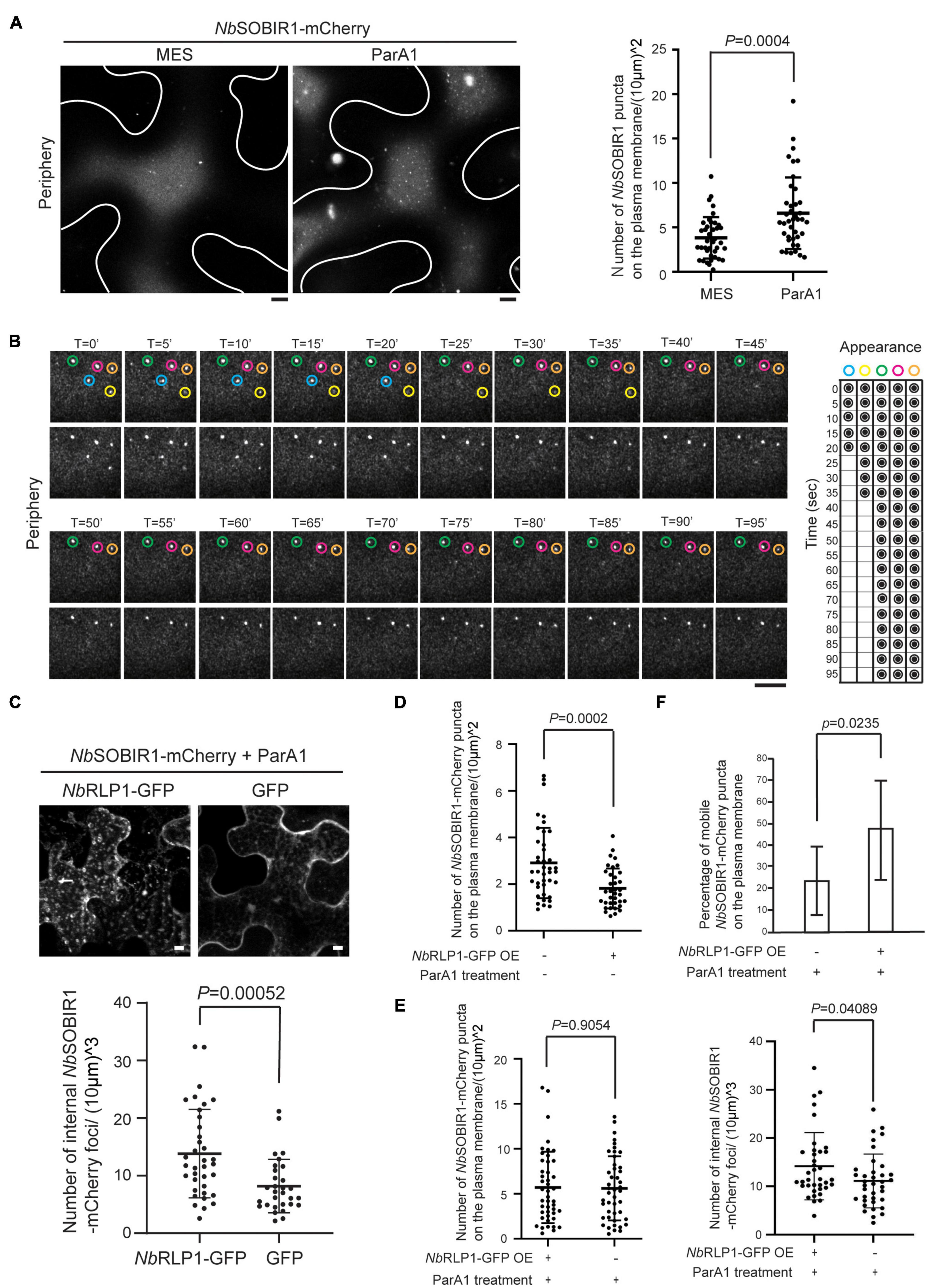
Figure 6. NbRLP1-GFP overexpression promotes NbSOBIR1-mCherry endocytosis upon ParA1 elicitin treatment. (A) Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry were infiltrated with MES buffer alone (control) or 0.3 μMParA1 in MES buffer. After 30 min, the leaves were subjected to microscopy, focusing only on the periphery of the leaf epidermal cell, using the Zeiss LSM880 confocal microscope with an Airyscan. Representative images are shown on the left panel with an outline of the cell, and the quantification data using the scatter plot is shown on the right. Statistics is carried out by the two-tailed student’s t-test and the p-value is displayed. (B) N. benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP were infiltrated with 0.3 μM ParA1 in MES buffer. After 30 min, leaves were subjected to time-lapse microscopy using Zeiss LSM880 confocal microscope with Airyscan. The total image acquisition length was 16 min with a time interval of 5 s (Supplementary Movie 4). Representative area was selected for display by time course, and the appearance and absence of each microdomain at each displayed time point was summarized on the right. Blue and yellow circles mark two NbSOBIR1 microdomains with a larger size undergoing endocytosis during the imaging time frame. Green, pink, and orange circles mark NbSOBIR1 microdomains with a larger size that remain static during the imaging time frame. (C) N. benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP or GFP alone were infiltrated with 0.3 μM ParA1 in MES buffer. After 30 min, leaves were subjected to microscopy with five Z stack for a total of 8.20 mm stack size using Zeiss LSM 510 confocal microscope. The maximal projection images are shown. The number of internal NbSOBIR1-mCherry puncta was quantified as described in the Materials and Methods section and displayed using a scatter plot. Statistical analysis was carried out with the two-tailed student’s t-test and the p-value is shown. (D) N. benthamiana leaves expressing NbSOBIR1-mCherry alone or coexpressed with NbRLP1-GFP were subjected to microscopy using the Zeiss LSM880 confocal microscope with an Airyscan. Microdomain number was quantified as described in the Materials and Methods section and shown as scatter plots. Statistics is carried out with the two-tailed student’s t-test and the p-value is displayed. (E) N. benthamiana leaves expressing NbSOBIR1-mCherry alone or coexpressing with NbRLP1-GFP were treated with 0.3 μM ParA1. After 30 min, leaves were subjected to microscopy by Zeiss LSM880 confocal microscope with the use of Airyscan. Microdomain number (left) was quantified as in (D). The same leaves were subjected to Zeiss LSM880 confocal imaging and quantified for internal NbSOBIR1-mCherry foci number (right) as in (C). Statistics is carried out with the two-tailed student’s t-test and the p-values are displayed. (F) N. benthamiana leaves expressing NbSOBIR1-mCherry alone or coexpressing with NbRLP1-GFP were infiltrated with 0.3 μM ParA1. After 30 min, leaves were subjected to time-lapse microscopy by Zeiss LSM880 confocal microscope with the use of Airyscan. The ParA1-induced microdomains were quantified if their size was bigger than 0.4 mm in diameter. A total of 10 cells for each treatment was quantified within an imaging timeframe of 60 s and data are shown as percentage (number of mobile puncta/number of total puncta). Statistics is carried out with the two-tailed student’s t-test and the p-value is displayed.
We further investigated the potential role of NbRLP1 for NbSOBIR1 endocytosis under ParA1 treatment based on a condition we reported previously (Peng et al., 2015). First, we asked whether coexpressing NbRLP1 and NbSOBIR1 affect NbSOBIR1 endocytosis in response to ParA1 treatment. Compared to the control coexpressing GFP and NbSOBIR1-mCherry, more intracellular vesicles harboring NbSOBIR1-mCherry were observed on tobacco leaves coexpressing NbRLP1-GFP and NbSOBIR1-mCherry in response to ParA1 treatment (Figure 6C). Given that ParA1 treatment induced the formation of NbSOBIR1 microdomains and that NbRLP1 overexpression promoted ParA1-induced NbSOBIR1 endocytosis, it seems plausible to predict that NbRLP1 overexpression may facilitate NbSOBIR1 endocytosis, thereby reducing the number of NbSOBIR1 microdomains on the plasma membrane. As shown in Figure 6D, in the absence of ParA1, a condition when fewer NbSOBIR1 microdomains were visualized, NbRLP1 overexpression caused a significant reduction in the number of NbSOBIR1 microdomains present on the plasma membrane. In the presence of ParA1, although overexpressing NbRLP1 did not significantly reduce the number of NbSOBIR1 microdomains on the plasma membrane (Figure 6E, left panel), it appears to accelerate the mobility of NbSOBIR1 microdomains as reflected by the higher percentage of mobile NbSOBIR1 puncta detected on the plasma membrane (Figure 6F). Moreover, overexpressing NbRLP1 led to the detection of more NbSOBIR1-mCherry-labeled structures inside the plant cells, indicative of more active NbSOBIR1 endocytosis (Figure 6E, right panel). Collectively, these data have led us to propose that the ParA1 elicitin-inducible NbSOBIR1 microdomain formed on the plasma membrane likely represents an active unit of NbSOBIR1 for subsequent receptor-mediated endocytosis and that NbRLP1 might act as a positive regulator for NbSOBIR1 internalization.
Our data have supported that the unconventional NbRLP1 in ER underlies part of the regulatory network for NbSOBIR1, the essential adaptor for various LRR-RLPs, and thus it seems likely that NbRLP1 might contribute to the NbSOBIR1-associated PTI. To explore this possibility, we analyzed the effect of NbRLP1 overexpression on ParA1-induced necrosis, a downstream output of plant immune response. Consistent with our previous findings, ParA1 induced the formation of necrotic lesions on N. benthamiana leaves at 24 h post-treatment (Figure 7). NbRLP1 silencing under either NbRLP1 endogenous or overexpressing conditions did not cause a significant change for the ParA1-induced necrosis on N. benthamiana leaves when compared to the control group. However, when ParA1 treatment was carried out in the N. benthamiana plants overexpressing NbRLP1-GFP, a condition when more NbSOBIR1 endocytic vesicles were observed (Figure 6C), we observed that necrotic lesions on N. benthamiana leaves were more exaggerated as evaluated by ANOVA. Altogether, these results support that NbRLP1 acting as a regulator lies within the repertoire of NbSOBIR1-mediated PTI.
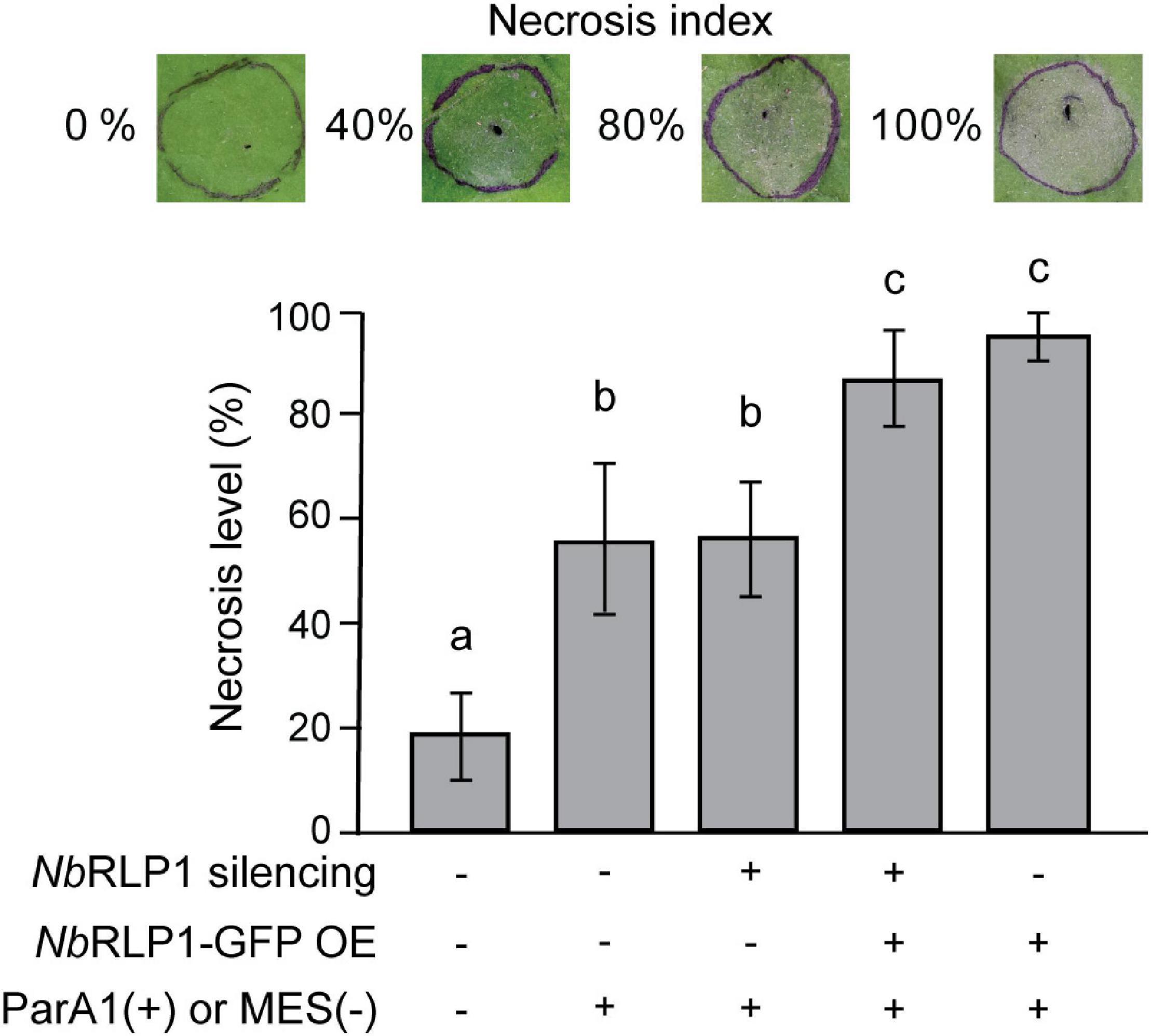
Figure 7. NbRLP1 overexpression exaggerated the ParA1-induced necrosis on Nicotiana benthamiana leaves. N. benthamiana leaves that are either silenced for NbRLP1 expression or overexpressing NbRLP1 are infiltrated with 0.3 μM ParA1 or 10 mM MES (control) as indicated. After 24 h, the development of cell death was analyzed based on the representative necrosis index, defined by the percentage of necrosis area over the infiltrated area. The experiment has been repeated four times with similar results, and only one set of the experimental result is shown. Values are means (± SD) of 18 biological repeats. Different letters indicate significant differences based on LSD (p < 0.01).
To combat pathogen infection, plants have evolved PRRs, which detect MAMPs at the cell surface to elicit PTI. Studies in various plant systems demonstrated that SOBIR1 plays a central role in PTI involving various PRRs of the LRR-RLP type (Tang et al., 2017; Domazakis et al., 2018). SOBIR1 moves from the plasma membrane to endocytic vesicles via endocytosis in response to MAMP treatment (Liebrand et al., 2013; Peng et al., 2015). However, how exactly SOBIR1 functions in plant immune response remains largely elusive.
Located at the outermost boundary of the cell as to communicate with the environment, the plasma membrane is known to be compartmentalized into subdomains, where immune complexes may be formed even in the absence of matching ligand, enabling rapid transmission and activation of diverse immune responses (Jarsch et al., 2014; Somssich et al., 2015; Bücherl et al., 2017). In this study, we report the identification of dynamic microdomains of NbSOBIR1 on the plasma membrane, which reinforces the important concept of nano- and micro-domains in plant immune response.
We document in this study that the ParA1 elicitin treatment induced both in the number and size of NbSOBIR1-labeled microdomaind, formed on the plasma membrane (Figure 6A), concomitant with the induction of NbSOBIR1 internalization, reflecting that the protein forms clusters in response to elicitin perception in N. benthamiana. The importance of nano- and micro-domains for plant innate immunity has been demonstrated in a number of recent studies. In the response of rice (Oryza sativa) to chitin elicitor, microdomains are required for the dynamics of the plasma membrane-anchored Rac/ROP small GTPase Rac1 (a molecular switch in defense signaling) and NADPH oxidase-encoding respiratory burst oxidase homologs (RBOHs) (Nagano et al., 2016). As well, the immune receptor FLS2 required for the perception of flg22 also localizes to the plasma membrane nanodomains of Arabidopsis (Bücherl et al., 2017). Consistently, NbSOBIR1 microdomain we discovered here plays a crucial role in the PTI regulated through PRRs of the RLP type.
Our findings uncovered NbRLP1 as a novel player in underlying the network of NbSOBIR1-mediated PTI response, which led to a yet unexplored area regarding the engagement of ER during NbSOBIR1 endocytosis and/or plant immunity. Using the in vivo BiFC assay and the in vitro pulled-down assay, we document in this study that the plasma membrane-localized NbSOBIR1 interacts with the ER-localized transmembrane protein NbRLP1, whose LRR domain resides within the ER lumen (Figure 3). This result also predicts that NbRLP1 and NbSOBIR1 might interact via a close association between the ER and the plasma membrane, such as EPCS or the ER-endosomes interface (Dong et al., 2016). However, BiFC fluorescence may emit artificially due to close proximity of target proteins, and the assay results in the irreversible formation of fluorescent proteins, thus limiting its applications to further explore the dynamics and transient interactions between NbRLP1 and NbSOBIR1 (Shyu and Hu, 2008). In addition, as reported by Tao et al. (2019), the BiFC assay in plants has the tendency to artificially induce membrane contact especially when involving the overexpression of two proteins which might interact with ER and plasma membrane interfaces. Thus, where exactly does the interaction between NbRLP1 and NbSOBIR1 occur awaits further investigation.
To compensate for this limitation, we studied the localization and dynamics of NbRLP1 and NbSOBIR1 in plants by transient overexpression followed by confocal microscopy. Remarkably, our findings clearly uncovered that the dynamics of NbSOBIR1 microdomains on the plasma membrane, including its lateral movement and downregulation via endocytosis, is regulated through the remodeling of cortical ER underneath the plasma membrane (Figures 4, 5). We suspect that the NbRLP1 and NbSOBIR1 interaction may contribute to binding the two compartments together to enable the coupled motility. However, when we performed biochemical studies to learn about the contribution of NbRLP1 domains to the NbRLP1-NbSOBIR1 complex formation, the data suggested that the interaction between NbRLP1 and NbSOBIR1 might be bridged by other yet undiscovered factors (Supplementary Figure 2). Further evidence indicated that NbRLP1, devoid of its cytoplasmic tail, showed slightly compromised activity to downregulate the number of NbSOBIR1 microdomains on the plasma membrane (Supplementary Figure 2C), despite its binding with NbSOBIR1. These observations have prompted us to hypothesize that the interaction between NbRLP1 and NbSOBIR1 is achieved transiently and/or involves remodeling of a complicated protein complex in order to regulate NbSOBIR1 endocytosis. Along this line, understanding the exact composition of the NbRLP1 and NbSOBIR1-associated complex would be necessary to know how the ER contributes to NbSOBIR1-mediated PTI response and NbSOBIR1 endocytosis.
The NbSOBIR1 microdomains in our studies showed an interesting pattern of either abutting or surrounded by the ER (Figure 5). In contrast to the flat ER sheets, the ER tubules are highly curved structures, shaped by the evolutionarily conserved reticulon family proteins that adopt a wedge-like topology in the ER. The reticulons localized to the border of the ER sheets, the ER tubules, and the three-way junction, all of which define the areas of the ER membrane that are highly curved (Nziengui et al., 2007). Intriguingly, these areas appear to be preferred by the NbSOBIR1 microdomains on the plasma membrane to associate with the ER. Recently, EPCSs marked by SYT1-mCherry are identified at static ER tubules in plant (Ishikawa et al., 2018). In Arabidopsis thaliana, AtSYT1 plays an essential role in maintaining cell integrity and virus movement (Uchiyama et al., 2014; Perez-Sancho et al., 2015). Notably, another plant EPCS tether AtVAP27-1 has recently been shown to bind clathrin and phosphoinositides in Arabidopsis and vap27-1/-3 mutant showed endocytosis defects, thus raising the possibility that endocytosis may occur at EPCS sites in plants (Stefano et al., 2018). In non-plant systems, EPCSs have been reported to get involved in diverse functions including lipid homeostasis, calcium influx, signaling, and endocytosis (Wakana et al., 2015; van der Burgh et al., 2019). Although the relationship between EPCSs and plant PRRs has not been established, the changes of EPCS in space and time in response to pathogen infection are definitely of great interest to be explored as a missing link in plant immunity.
Given that NbSOBIR1 moves from the plasma membrane to endocytic vesicles in response to the ParA1 elicitin (Peng et al., 2015), it seems most likely that NbRLP1 might regulate NbSOBIR1 endocytosis. Indeed, overexpressing of NbRLP1 in the absence of ParA1 treatment significantly reduced the number of NbSOBIR1 microdomains on the plasma membrane (Figure 6D). In the presence of ParA1, NbRLP1 overexpression though showed no effect on the total number of NbSOBIR1 microdomains on the plasma membrane (Figure 6E, left) significantly accelerated the mobility of the ParA1-induced microdomains (Figure 6F) and ParA1-triggered NbSOBIR1 endocytosis (Figure 6E, right). The SOBIR1 is involved in PTI elicited by various proteinaceous MAMPs. Whether NbRLP1 similarly regulates NbSOBIR1 endocytosis that is triggered by other elicitors await further investigation. It would also be interesting to know whether endocytosis of the corresponding RLP-PRRs is regulated by NbRLP1 and exactly how endocytosis regulation is accomplished in this scenario.
In this study, we detected on the immune blot, two forms of NbSOBIR1 which differ in molecular weight of around ∼8.5 kD (Figure 3B), implying post-translational protein modification on NbSOBIR1. Interestingly, overexpression of NbRLP1 resulted in the reduction of the higher molecular weight form of NbSOBIR1 that is also the form that binds NbRLP1 (Figure 3C). Our hunch is that the NbSOBIR1 of greater molecular weight may reside in the microdomains to mediate NbRLP1 interaction. Since NbRLP1 overexpression reduced the NbSOBIR1 microdomain on the plasma membrane (Figure 6D), through facilitating its endocytosis, it seems plausible to predict that the reduction in protein abundance is due to its endocytic turnover. It has been reported that SOBIR1 when overexpressed constitutively activates immune responses and is highly phosphorylated in A. thaliana, likely through the kinase activity of SOBIR1 itself and BAK1 (van der Burgh et al., 2019). However, using an antibody specifically against phosphoserine failed to recognize the NbSOBIR1 pulled down by NbRLP1-TAP (our unpublished data). In addition, the larger NbSOBIR1 that binds NbRLP1 also showed resistance to phosphatase treatment (our unpublished data). Thus, this unique form of NbSOBIR1 does not seem to represent a phosphorylated and activated form of this protein kinase at least in N. benthamiana. It would be of great interest to further identify the PTM and know whether it plays an important regulatory role for MAMP perception and PTI response in plants.
As shown by phylogenetic analysis, NbRLP1 clustered with two genes from N. sylvestris and N. attenuata, respectively. This clade is distinct from that encompassing EILP from N. tabacum (Takemoto et al., 2000) but closer to that containing two Cf-5 homologs (Hcr2-0A and Hcr2-0B) from S. lycopersicum (Dixon et al., 1998). Therefore, NbRLP1 appears not as an ortholog of EILP. Regarding the plant defense response, we document that NbRLP1 overexpression enhanced ParA1-induced necrosis (Figure 7). Although its silencing did not show a profound effect on ParA1-induced necrosis, SOBIR1 endocytosis, and plant resistance against P. parasitica (Supplementary Figure 3), a previous report showed overexpressing a fragment of NbRLP1 (termed NbEILP previously) in plants enhanced the accumulation of Bamboo mosaic virus, whereas gene silencing reduced its accumulation (Chen et al., 2017). Collectively, it seems likely that this ER RLP actively participates in a variety of host–pathogen interactions. It would be necessary to further investigate the exact molecular function of NbRLP1 in the ER in planta.
Overall, our studies on NbRLP1 and NbSOBIR1 have provided many new insights into the tale of NbSOBIR1-mediated PTI. The discovery of the NbSOBIR1 microdomain at the plasma membrane and the control of its dynamics through the contact with the ER not only advance our current knowledge on this important adaptor protein but also reinforce the concept of microdomain formation as an important platform during MAMP perception. Though more needs to be learned from how SOBIR1 works in concert with various RLPs to achieve plant immunity, it seems equally important to think beyond the plasma membrane toward a potential role of ER either in transmitting plant immunity signals or even in the organization of immune receptor complex upon MAMP perception.
Nicotiana benthamiana was grown in a mixture of peat moss, perlite, and vermiculite (4:1:1) at 28°C under 12-h light/dark. Phytophthora parasitica (isolate 94069) was cultured on 10% V8 juice agar [10% V8 juice (Campbell, NJ, United States), 0.02% CaCO3, and 1.5% select agar (Thermo Fisher Scientific, Waltham, MA, United States)] at 25°C.
Total RNAs were isolated from N. benthamiana leaves using the Plant Total RNA extraction kit (Viogene, New Taipei City, Taiwan) followed by using Turbo-DNA-free kit (Thermo Fisher Scientific, Waltham, MA, United States) to remove the residual DNA. N. benthamiana complementary DNA (cDNA) was prepared by using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, United States). The NbRLP1 was amplified by using NbRLP1_F1-3 and NbRLP1_R1-2 primers (Supplementary Table 1), followed by cloning into the pENTRTM/D-TOPOTM vector (Thermo Fisher Scientific, Waltham, MA, United States) for sequencing and subsequent subcloning. The signal peptide of NbRLP1 was predicted by using Signal P-5.02. Positions of leucine-rich repeat and transmembrane domain of NbRLP1 were predicted by InterPro3 and TMHMM Server v.2.04, respectively. Multiple sequence alignment involved the use of Clustal X. Phylogenetic tree was generated by the maximum likelihood algorithm implemented in MEGA (v.10.0.5) with the default parameters. Nodal support of the tree was estimated by bootstrapping with 1,000 pseudoreplicate data sets.
At 0, 3, 6, 12, 24, 36, and 48 h after inoculation of P. parasitica zoospores, total RNAs from the 7th and 8th leaves of N. benthamiana were isolated and cDNA was synthesized as described in the previous section. Quantitative PCR (qPCR) was performed using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, United States) with the primers listed in Supplementary Table 1 by using the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, United States). Raw data were normalized to the level of NbEF1α (as an internal control) and displayed as a fold-change relative to the transcript level of mock-treated plants of the same time points.
Virus-induced gene silencing (VIGS) experiments were performed as described by Peng et al. (2015). Briefly, a fragment of NbRLP1 amplified by PCR with primers listed in Supplementary Table 1 was cloned into pTRV2 (pYL279) by Gateway cloning (Thermo Fisher Scientific, Waltham, MA, United States) to generate TRV2:NbRLP1, which was transformed into Agrobacterium tumefaciens GV3101 strain. After growth in Luria-Bertani (LB) broth amended with rifampicin and kanamycin at 28°C for 18 h, the bacteria were diluted with induction medium (10 mM MES, pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone) to OD600 of 0.6, which was then mixed with an equal volume of GV3101 bacteria harboring TRV1 (pYL192). The mixture was infiltrated onto leaves of 18-day-old N. benthamiana seedlings by the use of 1-mL syringes. Downregulation of gene expression was verified by qRT-PCR at 21 days post-agroinfection.
Expression of His-tagged ParA1 in Escherichia coli and the subsequent protein purification were performed as described by Peng et al. (2015) and Hofzumahaus and Schallmey (2013) with some modifications. Escherichia coli strain C43 (DE) harboring pET-20b(+):ParA1 was grown in terrific broth (TB) [1.2% tryptone, 2.4% yeast extract, 0.5% glycerol, and 1 M TB salts (0.17 M KH2PO4 and 0.72 M K2HPO4)] at 37°C. When OD600 reached 1.0, 0.4 mM isopropyl β- d-1-thiogalactopyranoside (IPTG) was added and the bacteria culture was grown at 30°C with constant shaking at 150 rpm. After 24 h, bacteria were harvested and disrupted in a solution [50 mM potassium phosphate buffer (pH 6.5), 500 mM NaCl, and 10 mM imidazole] by the use of a high-pressure homogenizer (Avestin EF-C3, Ottawa, ON, Canada) and His-tagged ParA1 recombinant protein was purified with the use of Ni-NTA agarose according to the protocol of the manufacturer (Qiagen, Hilden, Düsseldorf, Germany). For the ParA1-induced necrosis assay, 0.3 μM recombinant protein in 10 mM MES (pH 5.6) was infiltrated onto leaves of 4.5-week-old N. benthamiana. After 24 h, the respective areas of infiltration and necrotic lesion for each leaf were measured by using the ImageJ software, and necrosis index was calculated accordingly by dividing the necrosis area with the infiltrated area.
All plasmid constructs used in this study were made by using the Gateway cloning system (Thermo Fisher Scientific, Waltham, MA, United States). To generate the GFP-NbRLP1 construct, the nucleotide sequence of NbRLP1ORF in the pENTR plasmid was first changed from T79C80C81A84 to A79G80T81T84, which introduced a ScaI site, whereas maintaining the same amino acid sequence. Then, a GFP-encoding DNA sequence was inserted into the ScaI site, followed by amplification and subcloning of the GFP-NbRLP1 fragment into the pK7WG2 vector (Karimi et al., 2002) by Gateway cloning to get pK7WG2:GFP-NbRLP1. To generate the NbRLP1-ΔN construct, a DNA fragment encompassing nucleotides 2,737–2,910 of NbRLP1 ORF was amplified by PCR, which was then used to replace NbRLP1 devoid of the signal peptide in pENTR:NbRLP1 and then to get pK7FWG2:NbRLP1-ΔN by Gateway cloning. To generate the NbRLP1-GFP and NbRLP1-ΔC constructs, DNA fragments corresponding to the full-length NbRLP1 ORF or NbRLP1 lacking its cytoplasmic tail (with nucleotides 1–2805) were amplified by PCR and cloned into pK7FWG2 to get pK7FWG2:NbRLP1 and pK7FWG2:NbRLP1-ΔC, respectively. To prepare the constructs for the BiFC experiments, fragments of Venus N-terminal half (Vn) and Venus C-terminal half (Vc) were amplified (Sung and Huh, 2007) using primers listed in Supplementary Table 1 and cloned into pENTR-NbRLP1 and pENTR-NbSOBIR1 to get pK7WG2:NbRLP1-Vn and pK7WG2:NbSOBIR1-Vc, respectively. To get pK7WG2:Vn-NbRLP1, Vn fragment was amplified to replace GFP in pENTR:GFP-NbRLP1 followed by Gateway cloning. To get TAP-tagged expression constructs, TAP sequence (Puig et al., 2001) synthesized by Genomics (Xizhi, New Taipei City, Taiwan) was first cloned to pENTR:NbRLP1, pENTR:NbRLP1-ΔN, and pENTR:NbRLP1-ΔC, respectively, followed by Gateway cloning. To prepare the AtSYT1-mCherry construct, the AtSYT1 amplified from the cDNA of A. thaliana was cloned into p35S-C-mCherry (Wu et al., 2011), followed by PCR amplification of the AtSYT1-mCherry fragment and subcloned into pK7WG2. The sequences for all primers are listed in Supplementary Table 1.
For transient gene expression on N. benthamiana, the plasmid constructs described in the section above were transformed into Agrobacterium tumefaciens C58C1 for agroinfiltration as described by Peng et al. (2015). In brief, A. tumefaciens strains carrying constructs were cultured in LB broth amended with corresponding antibiotic at 28°C. After 18 h, the bacteria cells were harvested by centrifugation, resuspended with MMA (10 mM MES, pH 5.6, 10 mM MgCl2, 200 μM acetosyringone) to OD600 of 0.2 for NbRLP1-GFP, 0.05 for NbSOBIR1-mCherry, and 0.2 for AtSYT1-mCherry, respectively. Bacteria were infiltrated onto the newly expanding leaves of 4-week-old N. benthamiana seedlings by the use of 1-mL needleless syringes. To suppress the silencing response, all treatments were coinfiltrated with A. tumefaciens C58C1 harboring P19-expressing construct (OD600of 0.1). For the BiFC experiments, A. tumefaciens C58C1 carrying pK7WG2:NbSOBIR1-Vc or pK7WG2:Vc was adjusted to OD600 of 0.05 and those carrying pK7WG2:Vn-NbRLP1, pK7WG2:NbRLP1-Vn, or pK7WG2:Vn were adjusted to OD600 of 0.2. Fluorescence signals were visualized using the Zeiss LSM 510 Meta confocal microscope, Zeiss LSM 880 confocal microscope, or Leica Stellaris 8 confocal microscopy. The Zeiss LSM 880 confocal microscope with the Airyscan super resolution mode was used for imaging microdomains on the plasma membrane. GFP: excitation of 488 nm and emission from 500 to 550 nm; mCherry: excitation of 543 nm and emission from 565 to 615 nm; YFP: excitation of 514 nm and emission from 510 to 560 nm.
To quantify NbSOBIR1 microdomains, we took still images specifically focused on the plasma membrane section using the Zeiss LSM880 with Airyscan mode. We used the ImageJ freehand selection tool followed by the ROI manager tool to define and calculate the area of the focused plasma membrane region. We manually counted the number of microdomains (usually ∼0.4–0.8 μm) using the multi-points tool of ImageJ. The number of NbSOBIR1 puncta on plasma membrane was calculated using the number of microdomains divided by the area of the plasma membrane. To analyze the dynamics of NbSOBIR1 microdomains, we performed particle tracking analysis using the ImageJ plugin Trackmate. We selected LoG detector tool to analyze particle diameter and intensity with estimated blob diameter of 0.8 μm and a threshold of 6.0. For analyses, we selected HyperStack displayer as a viewer and the simple lack tracker tool with the setting of linking max distance of 8 μm, gap-closing max distance of 8 μm, and gap-closing frame gap 2. To quantify endocytic structures, five z-stack images beneath the plasma membrane, each with 2.05 μm overlapped, were maximally projected to a final stack size of 8.20 μm. We used ImageJ to define and calculate the selected ROI area, followed by manually counting the number of endocytic structures. The number of NbSOBIR1 endocytic vesicles was calculated using the number of endocytic structures divided by the volume of the stack.
Agrobacterium tumefaciens C58C1 carrying pK7WG2:NbRLP1-TAP, pK7WG2:NbRLP1-ΔN-TAP, or pK7WG2:NbRLP1-ΔC-TAP (at a final OD600 of 0.2) was mixed with C58C1 harboring pK7WG2:NbSOBIR1-mCherry (at a final OD600 of 0.05) and infiltrated onto young expanding leaves of 4-week-old N. benthamiana. After 46 h, 1.5 g leaves were ground into fine powder in the presence of liquid nitrogen, followed by resuspension in 3 mL GTEN buffer (10% glycerol, 25 mM Tris–HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10 mM DTT, and 1.5% triton-X100) (Sacco et al., 2007) supplemented with protease inhibitors cocktail (Roche Molecular Systems, NJ, United States). After thawed, debris present in the mixture was removed by a quick spin of 1,000 rpm for 1 min, followed by another spin of 12,000 g for 10 min, both at 4°C. The resulting supernatant was collected and incubated with 100 μL IgG SepharoseTM 6 FF at 4°C with gentle rotation for 3 h. After washing the beads four times with 10 ml GTEN buffer containing 1.25 mM PMSF, the bound proteins were eluted with 80 μL MURB (50 mM sodium phosphate, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, and 5% β-mercaptoethanol) and boiled for 5 min. Samples were subjected to SDS-PAGE followed by the Western blot analysis with the use of PAP antibody (Jackson immune research, PA, United States) or our homemade anti-mCherry antibody (Wu et al., 2011). Signals on the blots were detected by using the UVP ChemiDoc-It imager (Upland, CA, United States).
For inoculation, zoospore suspension (2∗104 spores/mL) prepared as described by Yan and Liou (2006), or water as controls, was sprayed evenly on 5.5-week-old N. benthamiana till run-off, and the inoculated plants were maintained at 28°C in the dark in a moisture chamber for the indicated periods of time. Disease severity was scored, as follows, on a scale from 0 to 4, according to symptoms developed on the plants: 0, healthy, no water-soaking lesions; 1, slight water-soaking, with less than 50% wilting leaves; 2, obvious water-soaking lesions, with more than 50% wilting leaves; 3, severe water-soaking and wilting; 4, complete wilting along with the appearance of mycelia. The disease severity index was calculated as [sum (number of plants × disease index)])/[(total number of plants) × (maximal disease index)] × 100.
All experiments were repeated at least three times. The two-tail student’s t-test was used for two-paired comparison, and the p-value was displayed. Multiple comparison analysis testing was carried out with ANOVA and LSD (p < 0.05).
The original contributions presented in the study are publicly available. This data can be found here: The 2.91- and 1.41-kb NbRLP1 sequences can be found in the GenBank under the following accession numbers: MW924093 and MW924094, respectively.
R-FL and C-WW planned, designed the research, and wrote the manuscript. Y-HL, W-CS, and T-YK executed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
This study was supported by the Academia Sinica Intramural Funds and Career Development Award (AS-CDA-104-L11) (to C-WW) and by a grant (MOST 102-2628-B-002-021-MY3) from the Ministry of Science and Technology, Taiwan (to R-FL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Ji-Ying Huang and Mei-Jane Fang at the Cell Biology Core Lab (IPMB, Academia Sinica) for the microscopy advice. We thank Rey-Huei Chen (IMB, Academia Sinica) for sharing lab resources and providing suggestions for our investigations. We also thank Tsung-Luo Jinn (Institute of Plant Biology, NTU) for providing the plasmid construct mCherry-KDEL.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.721548/full#supplementary-material
Supplementary Figure 1 | Colocalization of NbRLP1-GFP and AtSYT1-mCherry. Nicotiana benthamiana leaves expressing NbRLP1-GFP and AtSYT1-mCherry were imaged at the periphery sections using Leica Stellaris 8 confocal microscope. Scale bar, 10 μm. The box area in the image of the periphery section is enlarged to show the colocalization of NbRLP1-GFP and AtSYT1-mCherry signals.
Supplementary Figure 2 | C-terminus of NbRLP1 is sufficient for binding with NbSOBIR1. (A) Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-TAP, NbRLP1-ΔN-TAP, NbRLP1-ΔC-TAP, or TAP as indicated were harvested and lyzed. The lysate (input) was subjected to pull-down by IgG Sepharose as described in the Materials and Methods section. The input and the bound (pulled-down) fractions were subjected to SDS-PAGE followed by the Western blot analysis with use of anti-mCherry or PAP antibodies. (B) A scheme depicting the current model of how NbRLP1-Vn in the ER might interact with NbSOBIR1-Vc on the plasma membrane. X, Y, and Z indicate putative proteins that might bridge the NbSOBIR1–NbRLP1 interaction. (C) Deletion of the C-terminus but not N-terminus of NbRLP1 showed slightly compromised activity to downregulate the number of NbSOBIR1 microdomains on the plasma membrane. N. benthamiana leaves expressing NbSOBIR1-mCherry alone or coexpressing with NbRLP1-GFP, NbRLP1-ΔN-GFP or NbRLP1-ΔC-GFP were subjected to microscopy by using the Zeiss LSM880 confocal microscope with use of Airyscan. Microdomain number was quantified as described in the Materials and Methods section and shown as scatter plots. Different letters indicate significant differences based on LSD (p < 0.01).
Supplementary Figure 3 | Endocytosis occurs directly via the translocation of NbSOBIR1 microdomains on the plasma membrane. Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry were infiltrated with 0.3 μM ParA1 in MES buffer. After 30 min, leaves were subjected to time-lapse microscopy by focusing on a few microdomains at the center of the cell using the Zeiss LSM880 confocal microscope with an Airyscan. The total image acquisition length was 16 min with a time interval of 5 s. Representative area was selected for display by time course.
Supplementary Figure 4 | NbRLP1 silencing did not show a profound effect on SlSOBIR1 endocytosis or plant resistance against Phytophthora parasitica. (A) SlSOBIR1-GFP was expressed on Nicotiana benthamiana leaves pre-silenced for NbRLP1 expression (TRV-NbRLP1) or infected with TRV-GFP as the control. Around 44 h post-agroinfiltration, the leaves were treated with 0.3 μM ParA1 in MES buffer for 30 min, followed by microscopy with five Z stack for a total of 8.20 stack size using Zeiss LSM 510 confocal microscope. The maximal projection images are shown. The number of internal NbSOBIR1-mCherry puncta was quantified as described in the Materials and Methods section and displayed using a scatter plot. Statistical analysis was carried out with the two-tailed student’s t-test and the p-value is shown. (B) N. benthamiana silenced for NbRLP1 expression (TRV-NbRLP1) or treated as the control carrying TRV-GFP were inoculated with zoospores of P. parasitica. At indicated hours post-inoculation (hpi), the development of disease symptoms was examined and the disease severity index was calculated as detailed in the Materials and methods section. The experiment has been repeated three times with similar results, and only one set of the experimental data is shown.
Supplementary Table 1 | A list of primers used in this study and the sequence of 1.41- and 2.91-Kb NbRLP1.
Supplementary Movie 1 | Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry were subjected to time-lapse microscopy, focusing only on the periphery of the leaf epidermal cell, using Zeiss LSM880 confocal microscope with Airyscan. The total image acquisition length was 16 min with a time interval of 5 s.
Supplementary Movie 2 | Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP were subjected to time-lapse microscopy, focusing only on the periphery of the leaf epidermal cell, using Zeiss LSM880 confocal microscope with Airyscan. The total image acquisition length was 16 min with a time interval of 5 s.
Supplementary Movie 3 | Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry were treated with 0.3 μM ParA1 in MES buffer. After 30 min, the leaves were subjected to microscopy, focusing only on the periphery of the leaf epidermal cell, by Zeiss LSM880 confocal microscope with use of Airyscan. The total image acquisition length was 16 min with a time interval of 5 s.
Supplementary Movie 4 | Nicotiana benthamiana leaves expressing NbSOBIR1-mCherry and NbRLP1-GFP were treated with 0.3 μM ParA1 in MES buffer. After 30 min, the leaves were subjected to microscopy, focusing only on the periphery of the leaf epidermal cell, by using Zeiss LSM880 confocal microscope with Airyscan. The total image acquisition length was 16 min with a time interval of 5 s.
Albert, I., Hua, C., Nurnberger, T., Pruitt, R. N., and Zhang, L. (2020). Surface sensor systems in plant immunity. Plant Physiol. 182, 1582–1596. doi: 10.1104/pp.19.01299
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bücherl, C. A., Jarsch, I. K., Schudoma, C., Segonzac, C., Mbengue, M., Robatzek, S., et al. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6:e25114.
Chen, I. H., Huang, Y. P., Tseng, C. H., Ni, J. T., Tsai, C. H., Hsu, Y. H., et al. (2017). Nicotiana benthamiana elicitor-inducible leucine-rich repeat receptor-like protein assists bamboo mosaic virus cell-to-cell movement. Front. Plant Sci. 8:1736. doi: 10.3389/fpls.2017.01736
Dixon, M. S., Hatzixanthis, K., Jones, D. A., Harrison, K., and Jones, J. D. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10, 1915–1925. doi: 10.2307/3870913
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548.
Domazakis, E., Wouters, D., Visser, R. G. F., Kamoun, S., Joosten, M., and Vleeshouwers, V. (2018). The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol. Plant Microbe Interact. 31, 795–802. doi: 10.1094/mpmi-09-17-0217-r
Dong, R., Saheki, Y., Swarup, S., Lucast, L., Harper, J. W., and De Camilli, P. (2016). Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408–423. doi: 10.1016/j.cell.2016.06.037
Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L. C., Zhou, J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034.
English, A. R., Zurek, N., and Voeltz, G. K. (2009). Peripheral ER structure and function. Curr. Opin. Cell Biol. 21, 596–602. doi: 10.1016/j.ceb.2009.04.004
Gao, M., Wang, X., Wang, D., Xu, F., Ding, X., Zhang, Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. doi: 10.1016/j.chom.2009.05.019
Hofzumahaus, S., and Schallmey, A. (2013). Escherichia coli-based expression system for the heterologous expression and purification of the elicitin beta-cinnamomin from Phytophthora cinnamomi. Protein Expr. Purif. 90, 117–123. doi: 10.1016/j.pep.2013.05.010
Ishikawa, K., Tamura, K., Ueda, H., Ito, Y., Nakano, A., Hara-Nishimura, I., et al. (2018). Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER Tubules. Plant Physiol. 178, 641–653. doi: 10.1104/pp.18.00498
Jarsch, I. K., Konrad, S. S., Stratil, T. F., Urbanus, S. L., Szymanski, W., Braun, P., et al. (2014). Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26, 1698–1711.
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/s1360-1385(02)02251-3
Liebrand, T. W., van den Berg, G. C., Zhang, Z., Smit, P., Cordewener, J. H., America, A. H., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Nagano, M., Ishikawa, T., Fujiwara, M., Fukao, Y., Kawano, Y., Kawai-Yamada, M., et al. (2016). Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 28, 1966–1983. doi: 10.1105/tpc.16.00201
Nziengui, H., Bouhidel, K., Pillon, D., Der, C., Marty, F., and Schoefs, B. (2007). Reticulon-like proteins in Arabidopsis thaliana: structural organization and ER localization. FEBS Lett. 581, 3356–3362. doi: 10.1016/j.febslet.2007.06.032
Peng, K. C., Wang, C. W., Wu, C. H., Huang, C. T., and Liou, R. F. (2015). Tomato SOBIR1/EVR Homologs are involved in Elicitin perception and plant defense against the oomycete pathogen Phytophthora parasitica. Mol. Plant Microbe Interact. 28, 913–926. doi: 10.1094/mpmi-12-14-0405-r
Perez-Sancho, J., Tilsner, J., Samuels, A. L., Botella, M. A., Bayer, E. M., and Rosado, A. (2016). Stitching Organelles: organization and function of specialized membrane contact sites in plants. Trends Cell Biol. 26, 705–717. doi: 10.1016/j.tcb.2016.05.007
Perez-Sancho, J., Vanneste, S., Lee, E., McFarlane, H. E., Esteban Del Valle, A., Valpuesta, V., et al. (2015). The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol. 168, 132–143. doi: 10.1104/pp.15.00260
Phillips, M. J., and Voeltz, G. K. (2016). Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17, 69–82. doi: 10.1038/nrm.2015.8
Prinz, W. A., Toulmay, A., and Balla, T. (2020). The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7–24. doi: 10.1038/s41580-019-0180-9
Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., et al. (2001). The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229. doi: 10.1006/meth.2001.1183
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Sacco, M. A., Mansoor, S., and Moffett, P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 52, 82–93. doi: 10.1111/j.1365-313x.2007.03213.x
Shibata, Y., Voeltz, G. K., and Rapoport, T. A. (2006). Rough sheets and smooth tubules. Cell 126, 435–439. doi: 10.1016/j.cell.2006.07.019
Shyu, Y. J., and Hu, C. D. (2008). Fluorescence complementation: an emerging tool for biological research. Trends Biotechnol. 26, 622–630. doi: 10.1016/j.tibtech.2008.07.006
Somssich, M., Ma, Q., Weidtkamp-Peters, S., Stahl, Y., Felekyan, S., Bleckmann, A., et al. (2015). Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci. Signal 8:ra76.
Stefano, G., Renna, L., Wormsbaecher, C., Gamble, J., Zienkiewicz, K., and Brandizzi, F. (2018). Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep. 23, 2299–2307. doi: 10.1016/j.celrep.2018.04.091
Sung, M. K., and Huh, W. K. (2007). Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24, 767–775. doi: 10.1002/yea.1504
Takemoto, D., Hayashi, M., Doke, N., Mishimura, M., and Kawakita, K. (2000). Isolation of the gene for EILP, an elicitor-inducible LRR receptor-like protein, from tobacco by differential display. Plant Cell Physiol. 41, 458–464. doi: 10.1093/pcp/41.4.458
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor Kinases in Plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tao, K., Waletich, J. R., Arredondo, F., and Tyler, B. M. (2019). Manipulating endoplasmic reticulum-plasma membrane tethering in plants through fluorescent protein complementation. Front. Plant Sci. 10:635. doi: 10.3389/fpls.2019.00635
Uchiyama, A., Shimada-Beltran, H., Levy, A., Zheng, J. Y., Javia, P. A., and Lazarowitz, S. G. (2014). The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 5:584. doi: 10.3389/fpls.2014.00584
van der Burgh, A. M., Postma, J., Robatzek, S., and Joosten, M. (2019). Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20, 410–422. doi: 10.1111/mpp.12767
Wakana, Y., Kotake, R., Oyama, N., Murate, M., Kobayashi, T., Arasaki, K., et al. (2015). CARTS biogenesis requires VAP-lipid transfer protein complexes functioning at the endoplasmic reticulum-Golgi interface. Mol. Biol. Cell 26, 4686–4699. doi: 10.1091/mbc.e15-08-0599
Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 9:594.
Wu, C. H., Lee, S. C., and Wang, C. W. (2011). Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 193, 521–535. doi: 10.1083/jcb.201006023
Yan, H. Z., and Liou, R. F. (2006). Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet. Biol. 43, 430–438.
Yu, X., Feng, B., He, P., and Shan, L. (2017). From Chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. doi: 10.1146/annurev-phyto-080516-035649
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Zhang, W., Fraiture, M., Kolb, D., Loffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Keywords: ER, plasma membrane, microdomain, SOBIR1, pattern-triggered immunity, receptor-like protein, microbe-associated molecular pattern, Phytophthora parasitica
Citation: Li Y-H, Ke T-Y, Shih W-C, Liou R-F and Wang C-W (2021) NbSOBIR1 Partitions Into Plasma Membrane Microdomains and Binds ER-Localized NbRLP1. Front. Plant Sci. 12:721548. doi: 10.3389/fpls.2021.721548
Received: 07 June 2021; Accepted: 02 August 2021;
Published: 01 September 2021.
Edited by:
Rachid Lahlali, Ecole Nationale d’Agriculture de Meknès, MoroccoReviewed by:
Pengwei Wang, Huazhong Agricultural University, ChinaCopyright © 2021 Li, Ke, Shih, Liou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruey-Fen Liou, cmZsaW91QG50dS5lZHUudHc=; Chao-Wen Wang, Y3d3YW5nMDJAZ2F0ZS5zaW5pY2EuZWR1LnR3
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.