- 1State Key Laboratory of Crop Stress Biology in Arid Areas, College of Agronomy and Yangling Branch of China Wheat Improvement Center, Northwest A&F University, Yangling, China
- 2Australia-China Joint Research Centre for Abiotic and Biotic Stress Management in Agriculture, Horticulture and Forestry, Yangling, China
The lateral organ boundaries domain (LBD) genes, as the plant-specific transcription factor family, play a crucial role in controlling plant architecture and stress tolerance. Although it has been thoroughly characterized in many species, the LBD family was not well studied in wheat. Here, the wheat LBD family was systematically investigated through an in silico genome-wide search method. A total of 90 wheat LBD genes (TaLBDs) were identified, which were classified into class I containing seven subfamilies, and class II containing two subfamilies. Exon–intron structure, conserved protein motif, and cis-regulatory elements analysis showed that the members in the same subfamily shared similar gene structure organizations, supporting the classification. Furthermore, the expression patterns of these TaLBDs in different types of tissues and under diverse stresses were identified through public RNA-seq data analysis, and the regulation networks of TaLBDs involved were predicted. Finally, the expression levels of 12 TaLBDs were validated by quantitative PCR (qPCR) analysis and the homoeologous genes showed differential expression. Additionally, the genetic diversity of TaLBDs in the landrace population showed slightly higher than that of the genetically improved germplasm population while obvious asymmetry at the subgenome level. This study not only provided the potential targets for further functional analysis but also contributed to better understand the roles of LBD genes in regulating development and stress tolerance in wheat and beyond.
Introduction
The lateral organ boundaries domain (LBD), also called the AS2/LOB gene family, is one class of plant-specific transcription factors, which plays an important role in regulating lateral organ development, morphogenesis, and metabolism in plants (Majer and Hochholdinger, 2011). LBD genes are expressed in a band of cells at the adaxial base of all lateral organs formed from the shoot apical meristem and at the base of lateral roots. LBD transcription factors are typically defined by an N-terminal LBD domain, which generally comprises a C-domain containing four highly conserved cysteine (C) residues in a CX2CX6CX3C zinc finger-like motif. Besides, some LBD proteins also possessed the glycine residue and leucine zipper-like motif LX6LX3LX6L (Shuai et al., 2002; Matsumura et al., 2009). Generally, the LBD proteins can be divided into class I and class II subfamily according to their conserved domains. In detail, class I members usually contain four highly conserved cys residues in a CX2CX6CX3C motif, together with a leucine zipper-like motif (LX6LX3LX6L) that can form the coiled-coiled structure for protein interactions, while class II members have the conserved cys residues and leucine zipper-like domain that cannot form the coiled-coil structure (Landschaslz, 1998). The majority of LBD proteins belong to class I and widely participate in regulating plant development and signal transduction (Majer and Hochholdinger, 2011; Yu et al., 2020).
Up to now, extensive studies have been conducted to investigate the LBD gene family in many plants, such as 34 putative LBD genes found in barley, 44 in maize, and 35 in rice, respectively (Yang et al., 2006; Zhang et al., 2014; Guo et al., 2016). Meanwhile, the biological function of some LBD genes has been well studied. In Arabidopsis, AS1 and AS2 genes were found to be involved in the establishment of a prominent midvein and other veins and in the formation of the symmetric leaf lamina, which might be related to the repression of knotted 1 homeobox genes in leaves (Semiarti et al., 2001). AtLBD16 and AtLBD18 functioned in the initiation and emergence of lateral root formation via different pathways, and they could interact with downstream ARF7 and ARF19 (Lee et al., 2009). AtASL9 was found to be exclusively regulated by the plant hormone cytokinin (Naito et al., 2007). In rice, OsARL1, encoded the protein with an LBD domain, was an auxin-responsive factor involved in the auxin-mediated cell dedifferentiation (Liu et al., 2005). OsAS2 was required for shoot differentiation and leaf development (Ma et al., 2009). OsLBD3-7 might act as the upstream regulatory gene of bulliform cell development to regulate leaf rolling (Li et al., 2016). In soybean, the expression of GmLBD12 could be induced by drought, salt, cold, and hormones (Yang et al., 2017).
Wheat is one of the most important crops all over the world, occupying approximately 17% of global cultivated lands (Shewry, 2009). Wheat provides plenty of proteins, starches, and vitamins for humans, acting as an essential plant protein resource. Genetically, wheat is an allohexaploid, derived from three diploid donor species through two naturally interspecific hybridization events (Berkman et al., 2013; Thomas et al., 2014). As a result, wheat has a huge and complicated genome with three subgenomes (A, B, and D), making it an ideal model for polyploidization and homoeologous gene interaction studies in plants (Wicker et al., 2011). The completion of its reference sequence provides the opportunity to investigate the genomic organization and evolution dynamics of the wheat gene family at the genomic level (IWGSC, 2018). At present, the systematical investigation of the LBD gene family has not yet been performed in wheat. Here, we conducted an in silico genome-wide search of the LBD gene family in wheat. Then, the phylogenetic relationship, chromosome localization, gene structure, conserved protein domain, cis-elements, expression profiles, and regulatory network and genetic variations of the putative wheat LBD genes (TaLBDs) were systematically analyzed. These results provided global information on the composition, structure, and evolution of the TaLBD family, which will facilitate the further functional study of this important transcript factor family in wheat and beyond.
Materials and Methods
Databases Search and Sequences Analysis
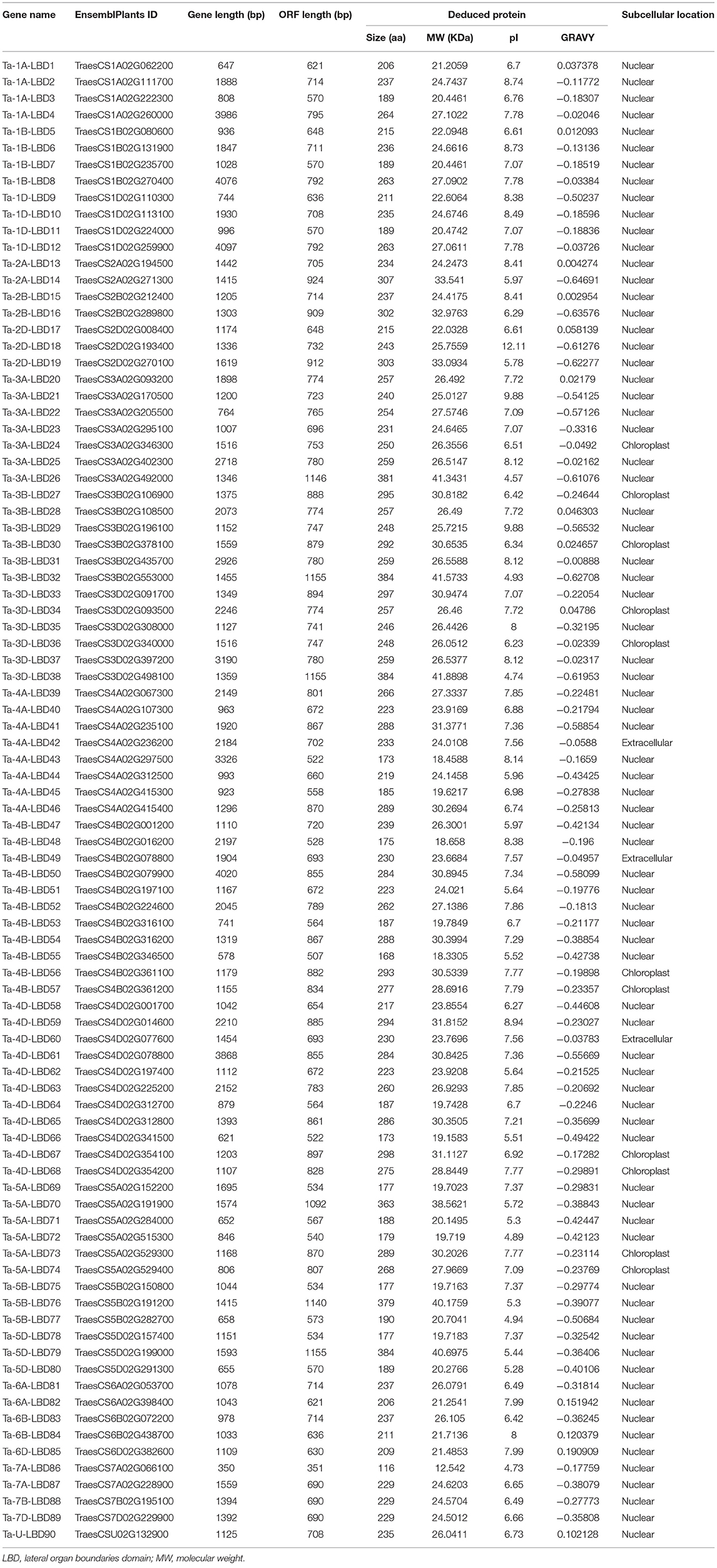
The reference sequence (release-49) of the Chinese Spring genome and its annotated proteins were downloaded from the EnsemblPlants database (ftp://ftp.ensemblgenomes.org/pub/plants/release_49/fasta/triticum_aestivum/) (IWGSC, 2018). Then, two methods were adopted to identify putative TaLBDs. Firstly, the LBD proteins of rice and Arabidopsis were retrieved and downloaded from RGAP (http://rice.plantbiology.msu.edu/index.shtml) and TAIR (https://www.arabidopsis.org/) databases, respectively, and then used as the queries to perform a BLASTP search against the local protein database of wheat (IWGSC_v1.1) with an e-value of 1e-5 and identity of 50% as the threshold. Furthermore, the LBD domain (PF03195) obtained from the PFAM database (http://pfam.xfam.org/) was used as the query for the hidden Markov model (HMM) search using HMMER 3.0 program (http://hmmer.org/). The protein sequences identified by both aforementioned methods were integrated and parsed by manual editing to remove the redundancy. The remaining proteins were considered as the putative TaLBDs. The chromosome locations of these LBD genes were obtained by referring the gff annotation file and then were visualized using the Circos tool (v0.67) (Krzywinski et al., 2009). Finally, the putative TaLBDs were submitted to the NCBI-CDD server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and the SMART database (http://smart.embl.de/) to confirm the existence of the LBD domain. The theoretical isoelectric point (PI) and molecular weight (MW) of the identified LBD proteins were calculated by the ExPASy server (http://www.expasy.org/). The subcellular localization prediction of each protein was predicted using the cello software (Yu et al., 2006).
Phylogenetic, Gene Structure, and Conserved Motif Analysis
All of the identified TaLBD proteins together with LBD proteins of rice and Arabidopsis were used to perform a multiple sequence alignment using the DNAMAN tools (Woffelman, 2004) with the default parameters. An unrooted neighbor-joining tree with 1,000 bootstrap replications was constructed using MEGA 8.0 (Kumar et al., 2018) based on the full-length protein alignment. The exon–intron organizations and splicing phase of these predicted LBD genes were retrieved from the annotation file of the wheat genome and then graphically displayed by the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Conserved motifs or domains were predicted using the MEME tool (http://meme-suite.org/), with the following parameters: the maximum number of motifs set at 15 and the optimum width of each motif falls between 5 and 200 residues.
Promoter Analysis and Identification of miRNAs Targets
The upstream 1.5 kilobases (kb) genomic DNA sequences of each predicted TaLBDs were extracted from the wheat genome and then submitted to the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/) to identify the putative cis-regulatory elements in the promoter regions. Furthermore, all the identified LBD transcripts were searched against the published wheat microRNAs (miRNAs) in the miRBase using the psRNATarget tool (Dai et al., 2018) to predict the TaLBDs targeted by miRNA.
Gene Expression and Regulatory Network Analysis
To study the expression profiles of TaLBDs, a total of 49 public available RNA-seq samples from five tissues (root, stem, leaf, spike, and grain) and four stressed conditions (cold, salt, heat, and drought) were downloaded from WHEAT URGI (https://urgi.versailles.inra.fr/files/RNASeqWheat/) and NCBI Sequence Read Archive (SRA) database. The accession numbers and sample information were listed in Supplementary Table 1. The FPKM value (fragments per kilobase of transcript per million fragments mapped) was calculated for each LBD gene by Hisat2 and Stringtie software (Kim et al., 2015) and the spatial-temporal expression patterns of them were obtained. The heatmap was drawn based on the log 10-transformed (FPKM + 1) values through pheatmap package in R software.
To obtain the regulation relationship between TaLBDs and other wheat genes, the interaction networks, where these putative TaLBDs were involved in, were investigated based on the orthologous genes between wheat and Arabidopsis using the STRING tool (http://string-db.org/cgi) and the AraNet V2 tool (http://www.inetbio.org/aranet/). The predicted interaction network was displayed through the Cytoscape software (Otasek et al., 2019).
Genetic Variations of TaLBDs
The genetic diversity of the LBD family in wheat landrace and genetically improved germplasm populations was investigated based on the resequencing data (Zhou et al., 2020). VCF file was downloaded from the Genome Variation Map (https://bigd.big.ac.cn/gvm) and the single-nucleotide polymorphisms (SNPs) in TaLBDs were extracted from the VCF file. Then, the nucleotide diversity (Pi value) in landrace and genetically improved germplasm populations was calculated using the vcftools (v 0.1.16).
qRT-PCR Validation of TaLBDs Under Salt Stress
Quantitative PCR analysis was performed using the previously described method with some modifications (Lei et al., 2021). In brief, the seeds of cv. Chinese Spring were germinated in Petri dishes and grown in water at a growth chamber with the controlled condition (23°C ± 1°C, 16-h light/8-h dark cycle). The three-leaf seedlings were subjected to salt stress treatment. The healthy plants were incubated in 150 mM NaCl solution for 6, 12, and 24 h with the plants under normal conditions used as the control. Leaves of all these samples were collected with three biological replications. Total RNA was isolated by Plant RNA Kit reagent (Omega Bio-Tek, GA, USA) according to the instructions of the manufacturer. A total of 12 TaLBDs belonging to four homoeologous groups were selected to investigate their expression levels under salt stress conditions by QPCR analysis using the primers as listed in Supplementary Table 2. QPCR reactions were performed using the QuantStudioTM 7 Flex platform (Thermo Fisher Scientific, USA) with SYBR® Premix Ex Taq™ II reagent following the manufacturer's protocol (TaKaRa, Dalian, China). The thermal cycling condition of QPC analysis was as follow: 95°C for 30 s followed by 40 cycles of 95°C for 3 s, 60°C for 30 s.
Results and Discussion
Identification of the LBD Family in Wheat
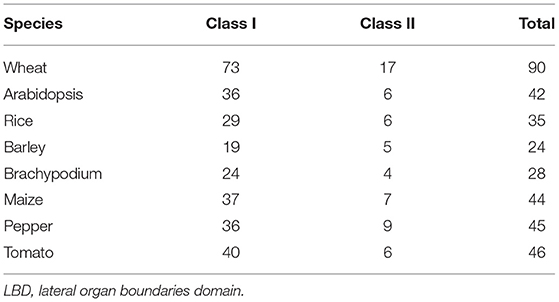
Using the aforementioned method, a total of 90 putative LBD proteins were identified in the wheat genome, which represented the most abundant LBD family among plants, indicating that the LBD gene family expanded significantly in wheat (Tables 1, 2). Chromosome location analysis showed that 89 TaLBDs were unevenly distributed on all of the 21 wheat chromosomes and only one (Ta-U-LBD90) was not mapped on any chromosome, of which 4B and 4D had the most abundant LBD genes with each containing 11, followed by 4A with 8, 3A with 7, and 3B, 3D, and 5A with 6, respectively, while 6B, 7B, and 7D only possessed one LBD gene (Figure 1). A total of 31, 29, and 29 LBD genes are non-randomly distributed in the wheat A, B, and D subgenomes, respectively, indicating that there was no significant difference in the LBD abundance at the subgenome level. Since there is no standard nomenclature, the predicted TaLBDs were then named based on their chromosomal distribution and physical location.
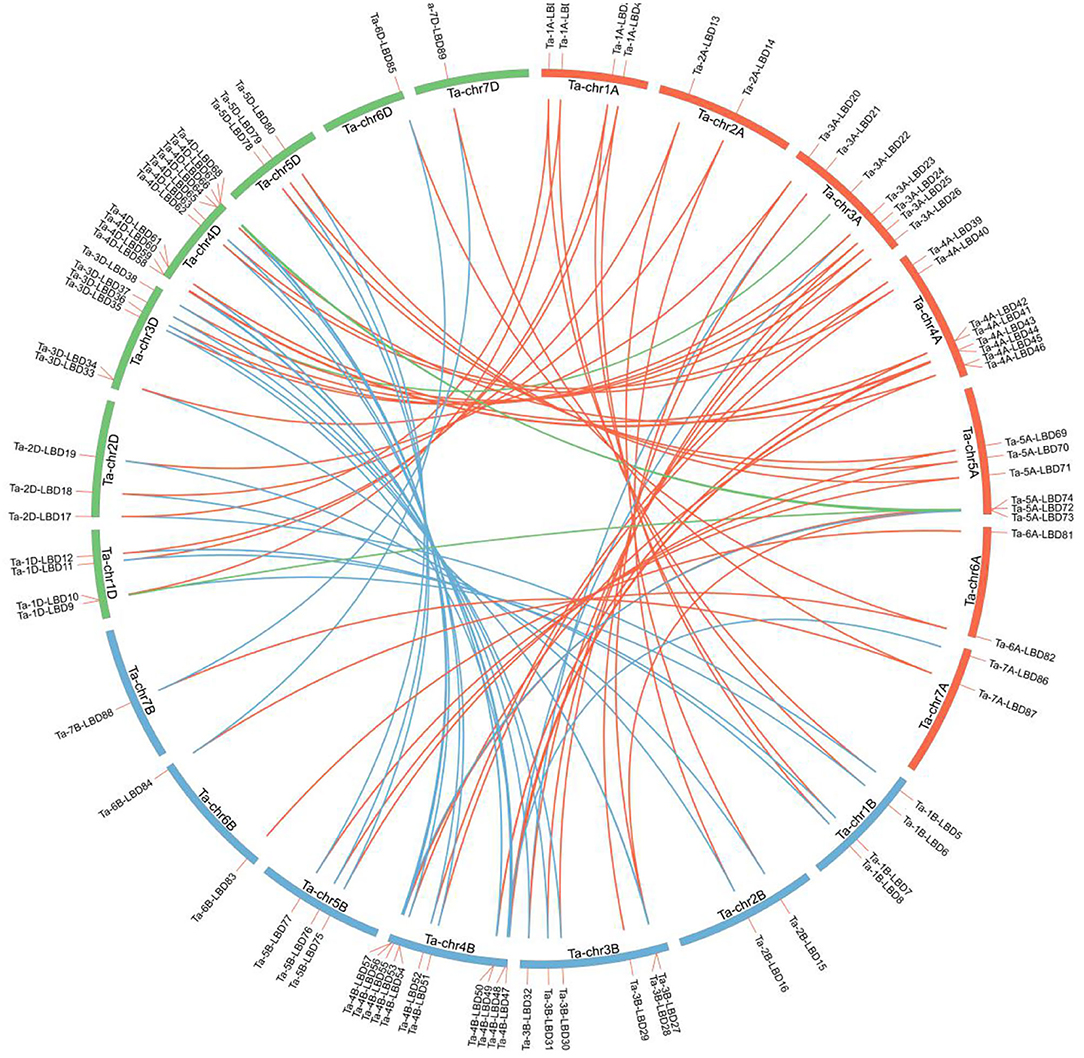
Figure 1. Distribution of the identified 90 TaLBDs across the seven chromosome groups and A (red), B (blue), and D (green) subgenomes of wheat. All wheat chromosomes are drawn to scale based on their actual physical lengths. TaLBDs, wheat lateral organ boundaries domain genes.
As reported in rice and Arabidopsis, the LBD gene family could be divided into two major groups, namely, class I and class II, according to the conserved domain organization (Yang et al., 2006; Matsumura et al., 2009). We further investigated the conserved motif in these TaLBDs. Results showed that all the putative wheat LBDs possessed the conserved cysteine-rich C-motif (CX2CX6CX3C) signature motif (Supplementary Figure 1). Among them, 73 TaLBDs shared the complete cysteine-rich C-motif, GAS-block, and leucine zipper-like structure, which could be categorized into class I, while the remaining 17 TaLBDs had an incomplete leucine zipper motif, belonging to class II. Furthermore, the length of putative TaLBD proteins ranged from 116 to 384 amino acids, with the putative MW ranging from 12.5 to 41.9 KDa and theoretical PI ranging from 4.57 to 12.11, respectively. Meanwhile, the subcellular localization prediction found that most of the TaLBDs (76) were localized in the nuclear region, except for 11 in chloroplast and 3 in the extracellular region (Table 2).
Phylogenetic Relationship, Conserved Motif, and Gene Structure Analysis
To evaluate the evolutionary relationships of TaLBDs, phylogenetic analysis was further conducted based on multiple protein sequence alignment of all of the TaLBDs together with rice and Arabidopsis LBD proteins. Phylogenetic tree clustered these LBD genes into two major clades (class I and class II), which was consistent with the categorization depending on their domain composition as found in previous studies (Yang et al., 2006; Matsumura et al., 2009) (Figure 2). Class I could be further divided into eight groups (IA–IH), and class II was divided into two groups according to the phylogenetic relationship. It is obvious that the phylogenetic tree was monophyletic and the TaLBDs clustered together with their orthologous counterpart in rice and Arabidopsis in each subgroup, respectively (Figure 3A). Furthermore, a clear paralogous expansion by gene tandem duplication was found in the TaLBD family and each gene had two or three homoeologous copies, indicating that the allohexaploidization together with tandem duplication contributed to the expansion of TaLBDs (Qiao et al., 2015; Gombos et al., 2017). However, compared to rice and Arabidopsis, no significant paralogous expansion happened on the wheat LBD family. Interestingly, the IE subgroup only contained AtLBDs without rice and wheat orthologs, suggesting an absence of this subgroup in rice and wheat (Figure 2).
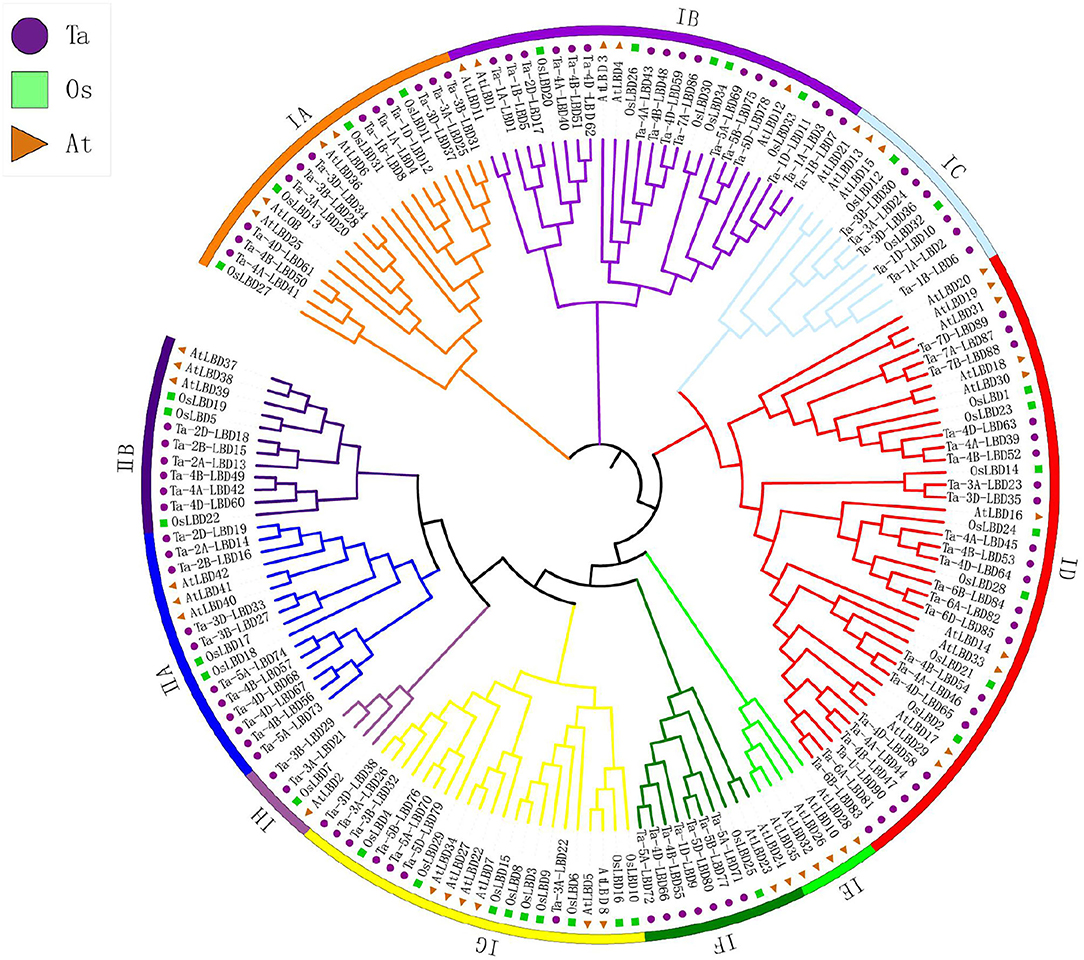
Figure 2. Phylogenetic analysis of LBD gene family in wheat. Tree was constructed by MEGA8.0 using the neighbor-joining method with 1,000 bootstraps. Purple, green, and orange colors represent LBD protein sequences from Triticum aestivum (Ta), Oryza sativa (Os), and Arabidopsis thaliana (At), respectively. The branch length represents the magnitude of genetic change. Different clade colors represent subfamilies, respectively. LBD, lateral organ boundaries domain.
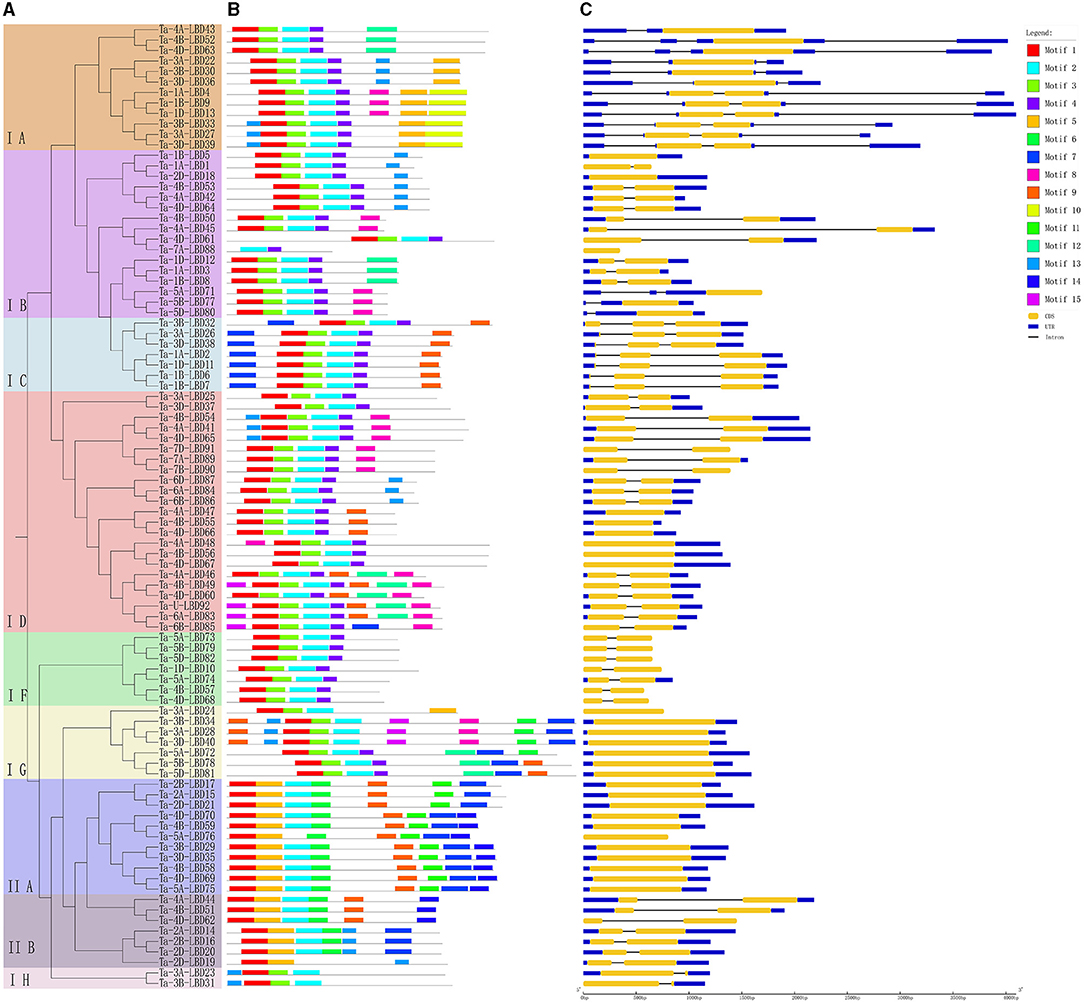
Figure 3. Phylogenetic relationship (A), conserved motifs (B), and gene structure (C) of LBD genes in wheat. The conserved motifs analysis of the LBDs based on their phylogenetic relationship were identified using MEME software. The different colors represent the 15 identified motifs. The yellow and blue rectangles represent the coding sequences (CDSs) and untranslated regions (UTRs), respectively, and the black lines represent introns. The lengths of the CDSs, UTRs, and introns for each LBD gene are shown proportionally. LBD, lateral organ boundaries domain.
Furthermore, the deduced TaLBD proteins were submitted to the MEME web server and 15 conserved motifs were identified. Results showed that motif 1 and motif 3 were the most conserved motifs among them, which were located in the LBD domain and shared by all of the wheat LBD proteins (Figure 3B). Further analysis found that the class I and class II LBD proteins possessed a specific motif composition. All the proteins belonging to class I harbored motif 1, motif 2, and motif 3, while all the members of class II contained motif 1, motif 3, and motif 5 (Figure 3B). Interestingly, motif 5 was uniquely found in class II, which might be class II specific. It is noteworthy that the LBD proteins within the same subgroup were usually found to share a similar motifs composition. For example, motif 9 and motif 10 were shared by the three members belonging to the IE subfamily, while motif 13 was specifically shared by the nine members within the class II subfamily. These results confirmed our phylogeny-based groupings.
Gene structure can provide important clues for analyzing evolutionary features and phylogenetic relationships of a gene family (Zhu et al., 2020). Therefore, we analyzed the intron–exon structure of these TaLBDs. Results showed that most of the TaLBDs were intron-less and the number of exons ranged from 1 to 4 (Figure 3C). A total of 25 TaLBDs had no exon, 42 had one exon, and 23 genes had multiple exons. The subfamily IA had a more sophisticated structure than other subfamilies because of the various number of intron. Furthermore, the members within the same subfamily shared a similar intron–exon structure and gene size, which supported their close evolutionary relationship and the classification of subfamilies. Through comparing the LBD gene structure among wheat, Arabidopsis, and barley (Matsumura et al., 2009; Guo et al., 2016), we found that they shared similar intron–exon structures, proving that the LBD gene family was rather conserved. Based on the phylogenetic tree and gene structure, the homoeologous groups of these 90 TaLBDs were identified. A total of 38 homoeologous groups were found, of which 22 groups with each containing A, B, and D homoeologous copies, 8 groups with each containing 2 of the 3 homoeologous copies, and the remaining 8 genes had only homologous copy, suggesting that homoeologous-copy-loss event might occur in the LBD family during wheat polyploidization. The specific retention and dispersion patterns of TaLBDs in homoeologous chromosomes provided important insight into the mechanism of wheat chromosome evolution and interaction (Wang et al., 2016). Interestingly, although the chromosome location of Ta-U-LBD90 was unknown, its location could be deduced to anchor on 6D based on the homoeologous grouping.
Cis-Elements and miRNA Targets Analysis
The cis-elements are the important regulatory factors that are involved in the transcriptional regulation of genes during plant growth and development, and stress response (Le et al., 2012). The 1.5 kb genomic sequences upstream from 5'-UTR of these 90 TaLBDs were extracted from wheat genome sequences and then used to predict cis-elements. A total of 36 cis-elements were identified belonging to different functional categories (Supplementary Tables 3 and 4). Results showed that a lot of phytohormone-responsive elements were widely found in the promoters of the TaLBDs. In detail, 78 TaLBDs contained ABA-responsive elements in their promoters, suggesting that these TaLBDs were probably induced by ABA, also 62 TaLBDs contained MeJA-responsive elements (TGACG-motif and CGTCA-motif) (Supplementary Figure 2). In addition, three gibberellin-responsive elements, namely, GARE-motif, P-box motif, and TATC-box were also found, which was consistent with the previous study that LBD genes may be involved in signaling transduction such as gibberellin pathway (Lee et al., 2009). Furthermore, light-responsive elements were also found in the promoter region of TaLBDs, for example, sp1 element and G-box element. A total of 63 out of 90 TaLBDs had the G-box element. According to the previous study, the light-responsive elements could combine with other cis-elements, such as ABA-related, to mediate signaling transduction in plant defense processes (Kumar et al., 2009), suggesting that these LBD genes might be involved in plant defense responses.
MicroRNAs are a class of small non-coding regulatory RNAs that are involved in controlling gene expression through guiding target mRNA cleavage or translation inhibition (Sunkar and Zhu, 2004). Recently, some miRNAs were found to regulate abiotic stress tolerance in diverse plants through targeting transcription factors (Akdogan et al., 2016; Yan et al., 2016; Zhou and Tang, 2019). To investigate the potential regulatory association between LBD transcription factor and miRNA, the putative miRNA-TaLBDs relationship was predicted using the available wheat miRNAs to search against these identified 90 TaLBDs through the psRNAtarget tool. Results showed that a total of 30 TaLBDs were predicted to be targeted by 21 miRNAs (Figure 4), of which tae-miR5384-3p could target 11 TaLBDs, tae-miR444, tae-miR9677b, and tae-miR9659-3P could target on three TaLBDs. Most of the TaLBDs were targeted by one miRNA, but Ta-4A-LBD39, Ta-3A-LBD25, Ta-3B-LBD31, and Ta-3D-LBD37 were targeted by three miRNAs. What is more, Ta-3A-LBD25, Ta-3B-LBD31, and Ta-3D-LBD37 belonged to the same homoeologous group but were targeted by three different miRNAs. The specific miRNA-LBD pairs provided the crucial information for precisely manipulating the function of these LBD genes through miRNA-based method. Although most of the TaLBDs were silenced by miRNAs through transcript cleavage, some genes were silenced by translation inhibition, such as Ta-4B-LBD55. In addition, we found that the homoeologous group genes Ta-6A-LBD82, Ta-6B-LBD84, and Ta-6D-LBD85 were regulated by tae-miR444a and tae-miR444b. According to the previous study, the miR444 family played important roles in root development, tiller formation, and stress response (Yan et al., 2014), indicating that they might be involved in wheat development and stress tolerance through miRNA regulation. The miRNAs-LBD complex identified in this study would be useful in interpreting the posttranscriptional control of gene expression during various stress-induced physiological and cellular processes in wheat and other cereal crops.
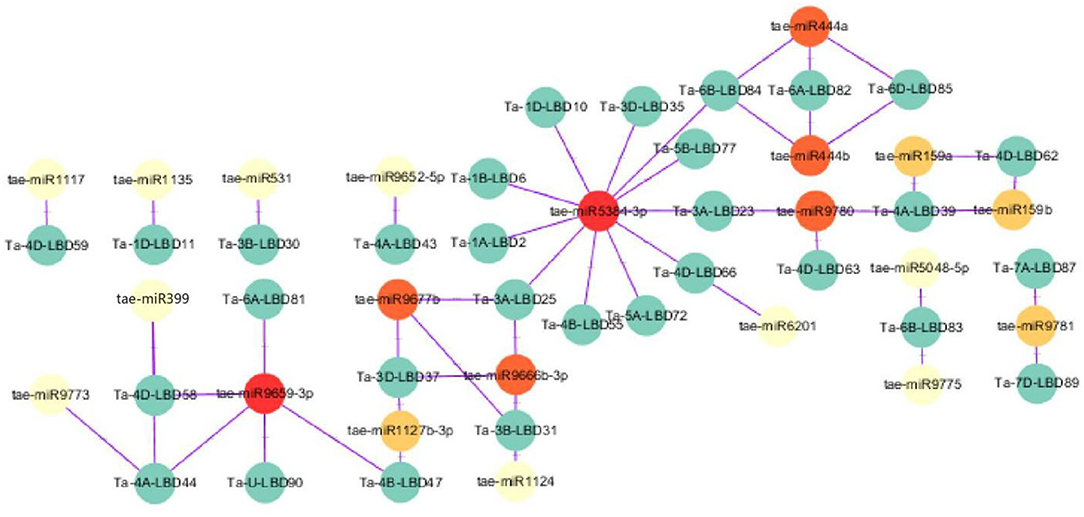
Figure 4. A schematic representation of the regulatory network relationships between the putative miRNAs and their targeted TaLBDs. The miRNAs that regulate one TaLBD are shown in yellow circles, miRNAs that regulate two TaLBDs are shown in orange circles, and miRNAs that regulate more than two TaLBDs are shown in red circles. The miRNAs and TaLBDs linked by the purple line indicate a putative regulatory relationship. TaLBDs, wheat lateral organ boundaries domain genes.
Regulatory Network Between LBDs and Other Genes in Wheat
LBDs transcription factor superfamily plays a crucial role in regulating the complex processes of plant growth, development, and stresses response through interacting with other functional genes (Naito et al., 2007; Ma et al., 2009). To get the preliminary information about the interaction relationship between LBD and other genes in wheat, we constructed the interaction network they were involved in using the orthology-based method (Figure 5). The results revealed that these TaLBDs widely interacted with the functional genes associated with organ development and morphogenesis (KNAT, EXPA17, and PRR1) and also interacted with other transcription factors to form signal transduction cascades (ARF19, NAC075, and WOX). Among them, Ta-4A-LBD40 was found to interact with KAN and KAN2 genes, which were reported to be involved in the molecular mechanism of lateral axis-dependent development of lateral organs in seed plants (Bowman et al., 2002), suggesting that Ta-4A-LBD40 might act as a regulator in wheat lateral axis-dependent development. Previous studies found that LBD18/ASL20 could upregulate EXPA17 to promote lateral root formation through auxin signaling in Arabidopsis (Lee et al., 2009; Lee and Kim, 2013). Here, we found that Ta-3A-LBD21, the orthology of AtLBD18, could interact with EXPA17, indicating that it might regulate root development in wheat. In conclusion, the co-expression network analysis of LBD genes provided vital information for a better understanding of LBD transduction pathways in wheat.
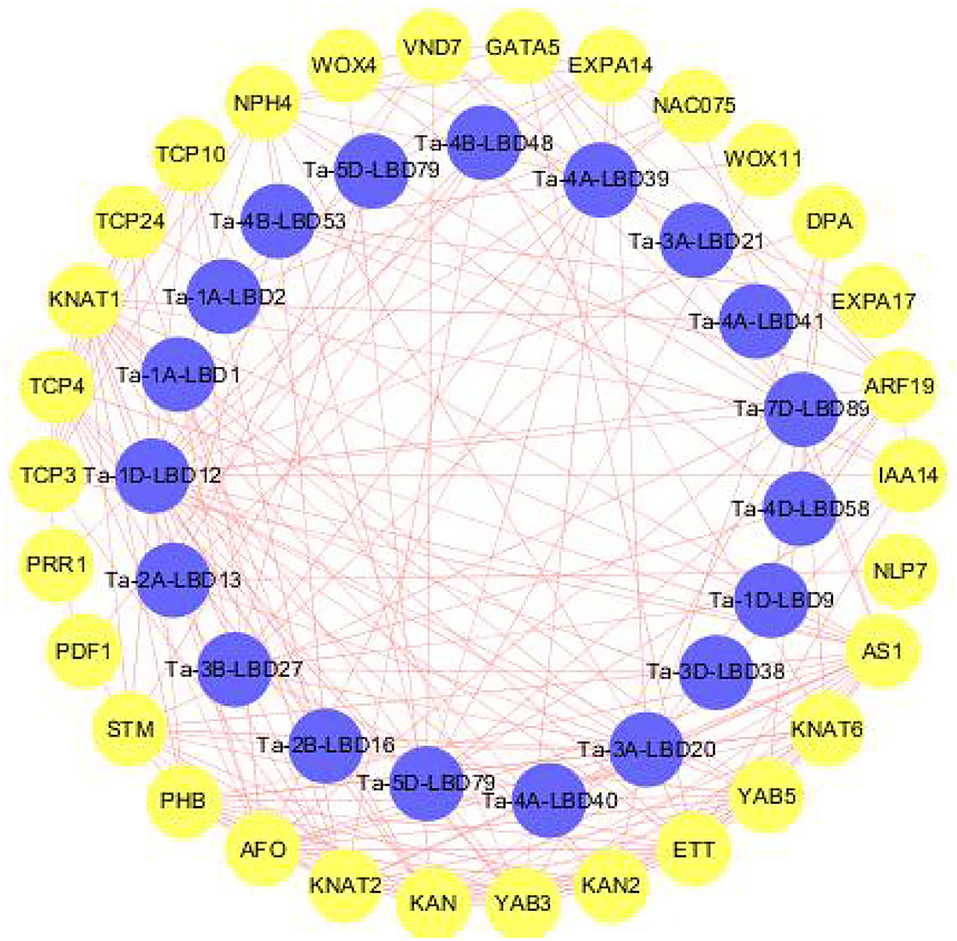
Figure 5. Predicted protein-protein interaction networks of TaLBD proteins with other wheat proteins using STRING tool. The blue circles represent wheat LBD proteins, and the circles on the outside represent proteins that interact with TaLBDs. The two circles connected by the red line represent the interaction between the proteins. LBD: lateral organ boundaries domain, TaLBDs, wheat lateral organ boundaries domain genes.
Expression Profiles of TaLBDs
The spatiotemporal expression specificity of genes will provide helpful information to understand their function in growth and development (Tong et al., 2013). In this study, the tissue-specific expression profiles of the 90 TaLBDs in different developmental organs (grain, leaf, root, shoot, and spike) were investigated using RNA-Seq data (Figure 6). Based on the log 10-transformed (FPKM + 1) values, we found that the expression levels of TaLBDs varied significantly in different types of tissues and showed obvious tissue specificity. In detail, a total of 72 TaLBDs were detected to express in at least one of the tested tissues, while 18 showed no expression in all of these tissues. Most of the TaLBDs were much more highly expressed in the root and grain, while fewer genes showed specific expression in stem and leaves. Ta-1A-LBD2, Ta-6B-LBD84, and Ta-4B-LBD47 displayed especially high expression in root. Ta-5A-LBD73, Ta-4B-LBD56, and Ta-4D-LBD67 showed high expression in spikes, while low expression in other tissues, suggesting that they might be involved in seed development and mature. Ta-3B-LBD28, Ta-4A-LBD46, and Ta-4B-LBD54 were specially expressed in stem with relatively low levels, while Ta-2B-LBD18 displayed high expression in leaves but low expression in other tissues. Furthermore, the majority of the homoeologous genes shared similar expression patterns among different types of tissues, nevertheless, we also found that some homoeologous genes showed different expression patterns. For example, Ta-4D-LBD61 was highly expressed in spike, while its homoeologous genes Ta-4A-LBD41 and Ta-4B-LBD50 showed no expression in spike.
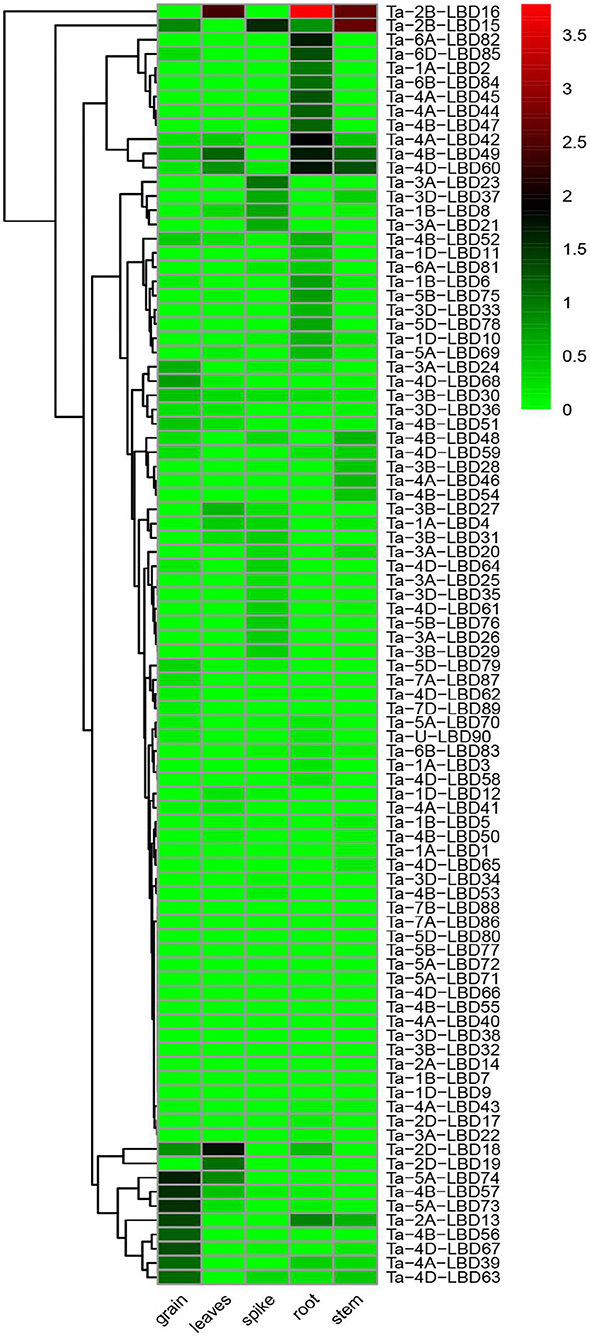
Figure 6. The expression profiles of 90 TaLBDs in different types of tissues, namely, grain, leaves, spike, root, and stem. The X axis represents the samples of different types of tissues. The Y axis represents the 90 different TaLBDs. The color scale represents log 2 expression values. The expression level is equal to the mean values and transforms log 2 values. TaLBDs, wheat lateral organ boundaries domain genes.
Previous studies have demonstrated that the LBD genes, especially class II members, were involved in regulating stress tolerance (Kong et al., 2017; Zhang et al., 2019). To gain some insights into the expression profiles and putative functions of LBD genes in response to stresses, the expression patterns of TaLBDs in four abiotic stresses (cold, heat, drought, and salt) were investigated using RNA-seq data (Figure 7). Results revealed that most of the TaLBDs showed differential expression patterns under these abiotic stresses, particularly under salt stress. Under salt stress, 54 TaLBDs showed differential expression at different time points. Ta-6B-LBD81, Ta-4B-LBD51, and Ta-U-LBD90 displayed high expression at all of the four time points, suggesting their important role in salt stress response. Ta-2B-LBD16, Ta-2D-LBD19, and Ta-1B-LBD5 showed specifically upregulated expression at 6 h, while Ta-5A-LBD69, Ta-4B-LBD54, and Ta-4D-LBD65 expressed specifically at 24 h after salt stress. Furthermore, the expression levels of Ta-1A-LBD1 and Ta-4D-LBD62 increased continually from 6 to 24 h, while Ta-lA-LBD2, Ta-1D-LBD10, and Ta-4B-LBD47 displayed high expression at 6, 12, and 24 h and then showed downregulated expression at 48 h. These results suggested that LBD genes were widely involved in salt stress induction in wheat and the different members played differential roles in regulating the salt stress response. Under cold stress, Ta-4A-LBD40 and Ta-4D-LBD62 showed enhanced expression at 4 h while Ta-4B-LBD49 showed special expression at 23 h. In addition, Ta-2A-LBD13, Ta-2B-LBD15, and Ta-2D-LBD18 showed upregulated expression under drought stress. Ta-5A-LBD72, Ta-5D-LBD79, Ta-5B-LBD76, and Ta-3D-LBD37 showed upregulated expression at 1 h after heat stress treatment and then downregulated expression at 6 h. The spatiotemporal expression profiles of these TaLBDs provided useful information to better understand the roles of TaLBDs in the growth, development, and stress tolerance.
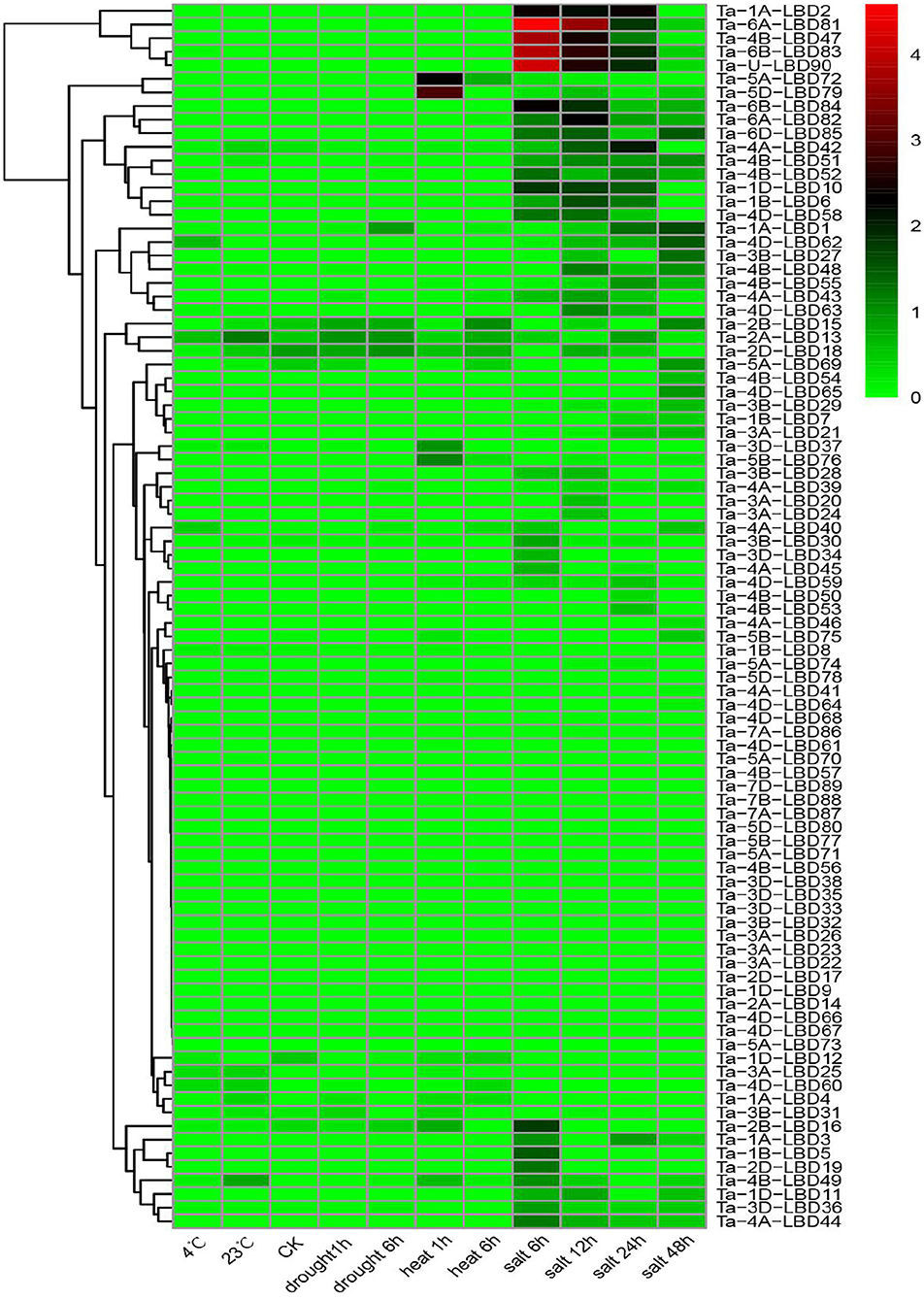
Figure 7. The expression profiles of 90 TaLBDs under different stresses, namely, drought, salinity, cold, and heat stresses. The X axis represents the samples of diverse stresses. The Y axis represents the 90 different TaLBDs. The color scale represents log 2 expression values. The expression level is equal to the mean values and transforms log 2 values. TaLBDs, wheat lateral organ boundaries domain genes.
Validation of the Expression of Wheat LBD Genes Using QPCR Analysis
Based on the expression patterns, 12 TaLBDs belonging to four homoeologous groups with significantly differential expression based on RNA-seq analysis were selected to investigate their expression levels under salt stress by QPCR analysis (Figure 8). The overall expression trend of these genes obtained by QPCR analysis was basically consistent with that of RNA-seq analysis. In detail, group A (comprising of Ta-1A-LBD1, Ta-1B-LBD5, and Ta-2D-LBD17) and group B (comprising Ta-2A-LBD14, Ta-2B-LBD16, and Ta-2D-LBD19) showed upregulated expression under salt stress, while group A showed the highest expression at 6 h and then decreased expression continually but group B showed the highest expression at 24 h after salt stress. Comparing to their A and D homoeologous genes, Ta-1B-LBD5 and Ta-2B-LBD16 showed higher expression levels at all of the four tested time points (Figures 8A,B). At the same time, the genes in group C (Ta-4A-LBD44, Ta-4B-LBD47, and Ta-4D-LBD58) and group D (Ta-6A-LBD81, Ta-6B-LBD83, and Ta-U-LBD90) showed downregulated expression under salt stress except for Ta-4A-LBD44 at 6 h (Figures 8C,D). The expression divergence among homoeologous genes was also analyzed. For group C, Ta-4A-LBD44 had the highest expression level at 6 and 12 h while its B homoeologous copy Ta-4B-LBD47 showed the highest expression level at 24 and 48 h, respectively (Figure 8C). In group D, Ta-6B-LBD83 had higher expression compared to its homoeologous genes Ta-6A-LBD81 and Ta-U-LBD90 at all of the four time points (Figure 8D). Furthermore, the expression levels of the homoeologous genes showed significant divergence, suggesting that subfunctionalization has occurred in these homoeologous genes when responding to salt stress. Further studies on the underlying effect of subfunctionalization will facilitate a better understanding of the roles of LBD genes in salt tolerance in wheat.
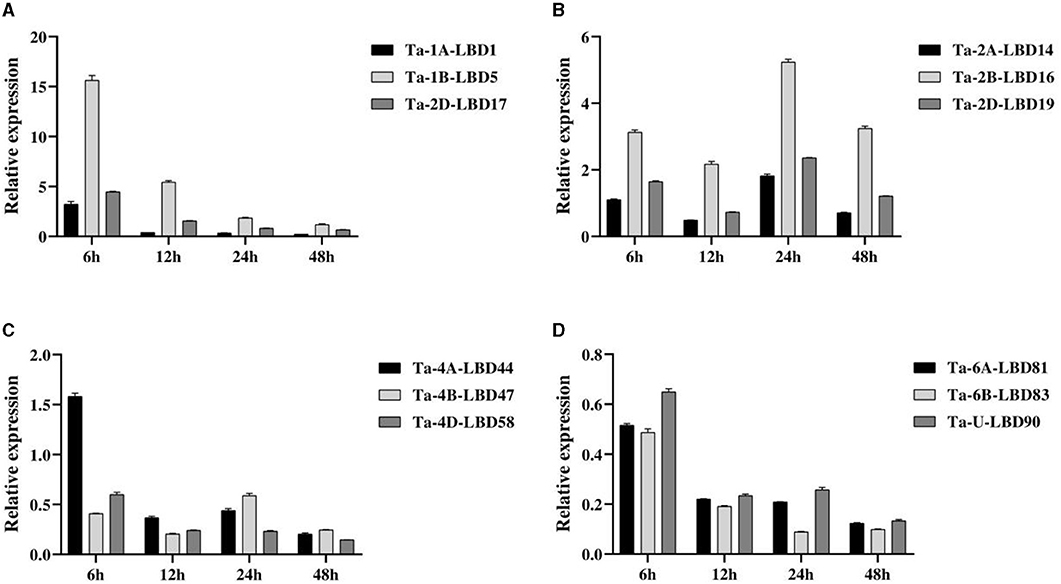
Figure 8. The expressions of the 12 TaLBDs belonging to four homologous groups under salt stress were investigated using QPCR method. The relative expression levels were calculated and compared. Error bars represent standard errors of three independent replicates. TaLBDs, wheat lateral organ boundaries domain genes.
Genetic Diversity of Wheat LBD Genes
To obtain information on genetic variations of TaLBDs, we further investigated their genetic diversities using the available wheat resequencing data (Zhou et al., 2020). A total of 404 (A: 183, B: 172, and D: 49) and 550 (A: 231, B: 218, and D: 101) SNPs were found in LBD genes of wheat landrace and genetically improved germplasm populations, respectively (Supplementary Table 5). The average genetic diversity (Pi value) of the landrace population was 0.889E−04, compared to that of the genetically improved germplasm population with 0.886E−04 (Supplementary Figure 3). No significant genetic diversity variation was observed between them, suggesting no severe bottleneck occurred on the LBD gene family during wheat improvement. Furthermore, at the subgenome level, the average genetic diversities of A, B, and D subgenomes in the landrace population were 1.31E−04, 1.15E−04, and 2.08E−05, respectively, while those in the genetically improved germplasm population were 1.06 E−04, 1.31E−04, and 2.87E−05 (Supplementary Table 6). The D subgenome of the genetically improved germplasm population showed higher diversity than that of the landraces, which was consistent with the previous studies based on the whole genome resequencing data (Cheng et al., 2019; Zhou et al., 2020). It demonstrated that during modern wheat breeding, genetically improved germplasm processes, introgressive hybridization with its wild D relatives had the genetic effect on the LBD gene family to enrich its diversity. At the same time, the B subgenome of the population also had higher diversity than that of landraces, which was not consistent with the general finding that landrace population owed higher diversity on B subgenome (Ormoli et al., 2015; Cheng et al., 2019; Zhou et al., 2020). These results suggested that some specific alien introgression events might occur in the TaLBDs on B subgenome. Further association of the variations with the agronomic traits will contribute to revealing the function and the evolutionary mechanism of the LBD family in wheat.
Conclusion
This study systematically identified and characterized the LBD family in wheat. A total of 90 putative TaLBDs were obtained, which were classified into two classes based on the conserved motif signatures and phylogenetic relationship. Furthermore, the intron–exon structure and conserved motif composition of them supported the classification and the members belonging to the same subfamily shared similar gene structures. The interaction network and miRNA-TaLBDs pairs provided useful clues for revealing the LBD-mediated regulation pathway. The tissue-specific or stress-responsive TaLBDs were identified based on RNA-seq data, of which 12 were validated by QPCR analysis, proving subfunctionalization of homoeologous genes has occurred in wheat. Finally, the genetic diversity of TaLBDs showed obvious asymmetry at the subgenome level. This study not only provides the candidates for further functional analysis of the TaLBDs but also contributes to a better understanding of the regulatory mechanism of LBD genes in regulating growth, development, and stress tolerance in wheat.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XN and WZ conceived and designed the study. ZW and RZ performed the analysis and also drafted the manuscript. YC and PL contributed to plant material collection and QPCR analysis. WS revised the manuscript. All authors have read and approved the final manuscript.
Funding
This study was mainly supported by Tang Scholar in NWSUAF and the National Science Foundation of China (Grant No. 31561143005) and partially supported by the Shaanxi Province Key Research and Development Program, China (Grant No. 2019NY-014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for the insightful and constructive comments of two reviewers on our manuscript. We also appreciated all people whose data were used in this study and thank the High-Performance Computing Center of Northwest A&F University for providing computational resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.721253/full#supplementary-material
References
Akdogan, G., Tufekci, E. D., Uranbey, S., and Unver, T. (2016). miRNA-based drought regulation in wheat. Funct. Integrat. Genom. 16, 221–233. doi: 10.1007/s10142-015-0452-1
Berkman, P. J., Visendi, P., Lee, H. C., Stiller, J., Manoli, S., Lorenc, M. T., et al. (2013). Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol. J. 11, 564–571. doi: 10.1111/pbi.12044
Bowman, J. L., Eshed,. Y., and Baum, S. F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141. doi: 10.1016/S0168-9525(01)02601-4
Cheng, H., Liu, J., Wen, J., Nie,. X. J., Xu,. L. H., Chen, N., et al. (2019). Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 20, 136. doi: 10.1186/s13059-019-1744-x
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Gombos, M., Zombori, Z., Szecsenyi, M., Sandor, G., Kovacs, H., and Gyorgyey, J. (2017). Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep. 36, 61–79. doi: 10.1007/s00299-016-2057-0
Guo, B. J., Wang, J., Lin, S., Tian, Z., Zhou, K., Luan, H. Y., et al. (2016). A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 17, 763–774. doi: 10.1631/jzus.B1500277
IWGSC (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:7191. 10.1126/science.aar7191
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kong, Y., Xu, P., Jing, X., Chen, L., Li, L., and Li, X. (2017). Decipher the ancestry of the plant-specific LBD gene family. BMC Genom. 18:951. doi: 10.1186/s12864-016-3264-3
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, G. M., Mamidala, P., and Podile, A. R. (2009). Regulation of Polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics 2, 238–249. doi: 10.3316/INFORMIT.091004413239069
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Landschaslz, W. (1998). The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. doi: 10.1126/science.3289117
Le, D. T., Nishiyama, R., Watanabe, Y., Vankova, R., Tanaka, M., Seki, M., et al. (2012). Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 7:e42411. doi: 10.1371/journal.pone.0042411
Lee, H. W., and Kim, J. M. (2013). EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 54, 1600–1611. doi: 10.1093/pcp/pct105
Lee, H. W., Kim, N. Y., Lee, D. J., and Kim., J. M. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151, 1377–1389. doi: 10.1104/pp.109.143685
Lei, P. Z., Wei, X. L., Gao, R. T., Huo, F. L., Nie, X. J., Tong, W., et al. (2021). Genome-wide identification of PYL gene family in wheat: evolution, expression and 3D structure analysis. Genomics 113, 854–866. doi: 10.1016/j.ygeno.2020.12.017
Li, C., Zou, X., Zhang, C., Shao, Q., Liu, J., Liu, B., et al. (2016). OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS ONE 11:e0156413. doi: 10.1371/journal.pone.0156413
Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., et al. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43, 47–56. doi: 10.1111/j.1365-313X.2005.02434.x
Ma, Y., Wang, F., Guo, J., and Zhang, X. S. (2009). Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 52, 374–381. doi: 10.1007/s12374-009-9048-4
Majer, C., and Hochholdinger, F. (2011). Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. doi: 10.1016/j.tplants.2010.09.009
Matsumura, Y., Iwakawa, H., Machida, Y., and Machida, C. (2009). Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58, 525–537. doi: 10.1111/j.1365-313X.2009.03797.x
Naito, T., Yamashino, T., Koizumi, N., Kojima, M., Sakakibara, H., and Mizuno, T. (2007). A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1269–1278. doi: 10.1271/bbb.60681
Ormoli, L., Costa, C., Negri, S., Perenzin, M., and Vaccino, P. (2015). Diversity trends in bread wheat in Italy during the 20th century assessed by traditional and multivariate approaches. Sci. Rep. 5:8574. doi: 10.1038/srep08574
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., and Demchak, B. (2019). Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 20, 1–15 doi: 10.1186/s13059-019-1758-4
Qiao, L., Zhang, X., Han, X., Zhang, L., Li, X., Zhan, H., et al. (2015). A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 6:770. doi: 10.3389/fpls.2015.00770
Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. doi: 10.1242/dev.128.10.1771
Shuai, B., Reynaga-Pena, C. G., and Springer, P. S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Sunkar, R., and Zhu, J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
Thomas, M., Sandve, S. R., Heier, L., Spannagl, M., Pfeiferet, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092. doi: 10.1126/science.1250092
Tong, C., Wang, X., Yu, J., Wu, J., Li, W., Huang, J., et al. (2013). Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 14:689. doi: 10.1186/1471-2164-14-689
Wang, M., Yue, H., Feng, K. W., Deng, P. C., Weining, S., and Nie, X. J. (2016). Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 17:668. doi: 10.1186/s12864-016-2993-7
Wicker, T., Mayer, K. F., Gundlach, H., Martis, M., Steuernagel, B., Scholz, U., et al. (2011). Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23, 1706–1718. doi: 10.1105/tpc.111.086629
Woffelman, C. (2004). DNAMAN for Windows, Version 5.2.10. Leidens: Lynon Biosoft, Institute of Molecular Plant Sciences, Leiden University.
Yan, J., Zhao, C., Zhou, J., Yang, Y., Wang, P., Zhu, X., Tang, G., Bressan, R. A., and Zhu, J. K. (2016). The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 12:e1006416. 10.1371/journal.pgen.1006416
Yan, Y. S., Wang, H. C., Hamera, S., Chen, X. Y., and Fang, R. X. (2014). miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 78, 44–55. doi: 10.1111/tpj.12446
Yang, H., Shi, G., Du, H., Wang, H., Zhang, Z., Hu, D., et al. (2017). Genome-wide analysis of soybean-containing genes: a functional investigation of GmLBD12. Plant Genome 10:plantgenome2016-2017. doi: 10.3835/plantgenome2016.07.0058
Yang, Y., Yu, X., and Wu, P. (2006). Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 39, 248–262. doi: 10.1016/j.ympev.2005.09.016
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins Struct. Function Bioinform. 64, 643–651. doi: 10.1002/prot.21018
Yu, J. W., Xie, Q. W., Li, C., Dong, Y. T., Zhu, S. J., and Chen, J. H. (2020). Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 251:81. doi: 10.1007/s00425-020-03364-8
Zhang, X., He, Y., He, W., Su, H., Wang, Y., Hong, G., et al. (2019). Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci. Rep. 9:15651. doi: 10.1038/s41598-019-52027-6
Zhang, Y. M., Zhang, S. Z., and Zheng, C. C. (2014). Genomewide analysis of LATERAL ORGAN BOUNDARIES domain gene family in Zea mays. J. Genet. 93, 79–91. doi: 10.1007/s12041-014-0342-7
Zhou, M., and Tang, W. (2019). MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol. Genet. Genom. 294, 379–393. doi: 10.1007/s00438-018-1516-4
Zhou, Y., Zhao, X., Li, Y., Xu, J., Bi, A., Kang, L., et al. (2020). Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. doi: 10.1038/s41588-020-00722-w
Keywords: abiotic stress, expression profile, genetic variation, LBD gene family, wheat
Citation: Wang Z, Zhang R, Cheng Y, Lei P, Song W, Zheng W and Nie X (2021) Genome-Wide Identification, Evolution, and Expression Analysis of LBD Transcription Factor Family in Bread Wheat (Triticum aestivum L.). Front. Plant Sci. 12:721253. doi: 10.3389/fpls.2021.721253
Received: 06 June 2021; Accepted: 09 August 2021;
Published: 03 September 2021.
Edited by:
Xiujin Lan, Triticeae Research Institute, Sichuan Agricultural University, ChinaCopyright © 2021 Wang, Zhang, Cheng, Lei, Song, Zheng and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijun Zheng, emhlbmd3ZWlqdW5AbndhZnUuZWR1LmNu; Xiaojun Nie, c21hbGxAbndzdWFmLmVkdS5jbg==
†These authors have contributed equally to this work
 Zhenyu Wang1†
Zhenyu Wang1† Weijun Zheng
Weijun Zheng Xiaojun Nie
Xiaojun Nie