- 1Key Laboratory of Tea Science of Ministry of Education, Hunan Agricultural University, Changsha, China
- 2National Research Center of Engineering and Technology for Utilization of Botanical Functional Ingredients, Hunan Agricultural University, Changsha, China
- 3Co-Innovation Center of Education Ministry for Utilization of Botanical Functional Ingredients, Hunan Agricultural University, Changsha, China
- 4Tea Research Institution, Chinese Academy of Agricultural Sciences, Hangzhou, China
- 5Institution of Genomics and Bioinformatics, South China Agricultural University, Guangzhou, China
Camellia sinensis cv. ‘Yanlingyinbiancha’ is a leaf-variegated mutant with stable genetic traits. The current study aimed to reveal the differences between its albino and green tissues, and the molecular mechanism underlying the variegation. Anatomic analysis showed the chloroplasts of albino tissues to have no intact lamellar structure. Photosynthetic pigment in albino tissues was significantly lower than that in green tissues, whereas all catechin components were more abundant in the former. Transcriptome analysis revealed most differentially expressed genes involved in the biosynthesis of photosynthetic pigment, photosynthesis, and energy metabolism to be downregulated in albino tissues while most of those participating in flavonoid metabolism were upregulated. In addition, it was found cryptochrome 1 (CRY1) and phytochrome B (PHYB) genes that encode blue and red light photoreceptors to be downregulated. These photoreceptors mediate chloroplast protein gene expression, chloroplast protein import and photosynthetic pigment biosynthesis. Simultaneously, SUS gene, which was upregulated in albino tissues, encodes sucrose synthase considered a biochemical marker for sink strength. Collectively, we arrived to the following conclusions: (1) repression of the biosynthesis of photosynthetic pigment causes albinism; (2) destruction of photoreceptors in albino tissues suppresses photomorphogenesis, leading to abnormal chloroplast development; (3) albino tissues receive sucrose from the green tissues and decompose their own storage substances to obtain the energy needed for survival; and (4) UV-B signal and brassinosteroids promote flavonoid biosynthesis.
Introduction
The tea plant [Camellia sinensis (L.) O. Kuntze] is an important economic crop that is cultivated worldwide. Leaf-color variation in tea plants has been extensively studied. Till date, lots of leaf color phenotypes, such as green, white, yellow, purple, and variegated, have been found in tea plants (Li et al., 2016a; Song et al., 2017; Shen et al., 2018). Leaf color is usually determined by pigments, such as chlorophyll, anthocyanins, and carotenoids. In general, the altered gene expression of leaf-color mutants can directly or indirectly affect the synthesis, degradation, content, and proportion of pigment, thereby blocking photosynthesis and causing abnormal leaf color (Zhao et al., 2020). Furthermore, significant differences exist in the content of main biochemical components, such as catechins, amino acids, flavonoids, and other substances, in tea plants with different leaf colors. Albino tea plants include three types of colors: albino, etiolated, and variegation. Compared to normal green tea plants, albino tea plants are precious, due to the special flavor. Albino tea leaves usually had a higher abundance of free amino acids, along with lower levels of catechins and caffeine, compared with green tea leaves (Feng et al., 2014). Change of these metabolites leads to decreased bitterness and astringency, and enhanced umami taste of the albino tea.
Variegation is a common phenotype in higher plants, with both green and white (or yellow) areas on the same leaf. Variegated mutants are important materials for studying the mechanism of development and maintenance of plant plastids (Sakamoto, 2003). Variegated mutants in Arabidopsis thaliana have been studied in much detail. The immutans (im) mutant of Arabidopsis produces reactive oxygen species due to the deletion of plastid terminal oxidase, thereby resulting in photobleaching to form a variegated phenotype (Carol et al., 1999). VAR1 and VAR2 are the main components of the FtsH complex that repairs photodamaged proteins in the thylakoid membrane. Both var1 and var2 mutants are susceptible to photodamage under intense light, leading to a yellow variegated phenotype (Sakamoto et al., 2003). While variegation may occur due to several reasons, it will eventually lead to the loss or abnormality of chloroplasts. At present, there are fewer variegated mutants in tea plants, such as ‘Yinghongjiuhao’. Ma et al. found that almost all the proteins mapped in the photosynthetic pathway showed decreased expression in ‘Yinghongjiuhao’ variegated mutant compared to green leaves (Ma et al., 2016).
Light, one of the main environmental factors that determine the biogenesis of chloroplasts, controls gene transcription, chlorophyll biosynthesis, and protein degradation through photomorphogenesis (Pogson and Albrecht, 2011). Photomorphogenesis is the process by which plants grow and develop in response to light signals. This process is mediated by a complex network of photoreceptors, including phytochromes (PHYs), cryptochromes (CRYs), and phototropins (PHOTs) (Han et al., 2007). CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), a master negative regulator of light signaling pathway, targets multiple transcription factors of light signaling pathways for repressing photomorphogenesis (Lau and Deng, 2012). Upon irradiation of light, the photoreceptors inactivate COP1 through direct protein–protein interaction and/or nuclear exclusion of COP1, and thereby facilitate photomorphogenesis (Podolec and Ulm, 2018; Xu, 2020). PHYB has also been reported to play an important role in the biosynthesis of chlorophyll and carotenoid, and development of chloroplast membrane (Zhao et al., 2013; Toledo-Ortiz et al., 2014). Photosynthetic pigments are essential for the assembly and protection of photosynthetic complexes, and thereby promoting chloroplast development. CRY has also been confirmed to be involved in inducing the expression of genes related to photosynthesis (Ohgishi et al., 2004). Once the photomorphogenesis is disrupted, photosynthetic pigment biosynthesis and chloroplast development will be blocked, leading to the inhibition of photosynthesis. Subsequently, energy metabolism is also influenced due to insufficient carbon supply. Taken together, photomorphogenesis is likely to play a prominent role in the formation of variegated plants.
A variegated mutant of ‘Yanlingyinbiancha’, with white edge and green main veins, is found in Yanling County, Hunan, China. After multiple generations of asexual reproduction and trial planting, the phenotypic traits have stabilized. In this study, we aimed to conduct transcriptome sequencing of the white and green tissues of ‘Yanlingyinbiancha’ in order to reveal the mechanism underlying variegation.
Materials and Methods
Plant Materials
The tea plant [Camellia sinensis (L.) O. Kuntze cv. ‘Yanlingyinbiancha’] was cultivated in Yanling County, Hunan, China. In 2017, one bud and two leaves were plucked and washed using ultrapure water. The albino and green tissues were then cut separately with scissors, fixed with liquid nitrogen immediately, and stored in a −80°C refrigerator.
Transmission Electron Microscopy Analysis
Fresh leaf was sliced into 1 mm2 sections, then fixed with 2.5% (v/v) glutaraldehyde for 2 h at 20°C and rinse three times with phosphate buffered solution (PBS) pH 7.4 for 15 min each. Leaf cells were post-fixed with 1% (v/v) OsO4 at 20°C for 5 h and then dehydrated in a series ethanol solutions (30, 50, 70, 80, 90, 95, and 100% for 1 h each). Subsequently, the samples were embedded in prepared 100% Epon-812 and polymerized in oven at 60°C for 48 h. The samples were cut into ultrathin sections (60–80 nm) using an ultramicrotome (EM UC7, Leica Microsystems Inc., Mannheim, Germany), and then stained with uranyl acetate and lead citrate for 30 min. Finally, images acquisition and analysis were obtained with a TEM (HT7700, Hitachi Ltd., Tokyo, Japan).
Catechin, Caffeine, Gallic Acid, and Theobromine Content
Catechin, CAF, GA, and TB contents were determined by high-performance liquid chromatography. Catechin and CAF contents were determined based on protocols from the National Standards of the People's Republic of China (GB/T 8313-2018 and GB/T 8314-2013). The HPLC system (Waters 590; Waters Corp.) was equipped with a Hypersil ODS 2 C18 column (5 ml, 4.6 mm 250 mm, 35 c) and the detection wavelength was set at 280 nm. Solvents A (2% acetic acid) and B (acetonitrile) were run in a linear gradient for 20 min, from 93 to 55%, and which the flow rate was maintained for 5 min at 1.4 mL min−1. Caffeine and catechin were quantitatively determined by comparing the peak areas of samples with known standards.
Photosynthetic Pigments
The methods used to determine the photosynthetic pigments (chlorophyll a, chlorophyll b, and carotenoids) contents of leaves were as follows: 20 mg of fresh tea leaves were cut into several filaments of ≥20 mm length and ≤1 mm width along the central vein. The strips were placed in 5 mL of 95% ethanol and incubated in the dark for 12 h. The extracts were filtered and analyzed with a UV-2550 spectrophotometer (Shimadzu, Japan), and their absorbance was recorded at the following wavelengths: 665 nm for chlorophyll a, 649 nm for chlorophyll b, and 470 nm for total carotenoids. Calculation formula: Chla (mg/L) = 13.95A665 – 6.88A649; Chlb (mg/L) = 24.96A649 – 7.32A665; Car (mg/L) = (1000A470 – 2.05Chla – 114.8Chlb)/245.
Transcriptome Analysis
RNA Preparation, Library Construction, and Sequencing
Total RNA of albino and green tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA integrity was assessed using the RNA 6000 Nano Assay Kit in the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. The library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. At last, the library preparations were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated.
Data Analysis
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts, including removing reads containing adapter, reads containing ploy-N and low quality reads from raw data, and calculating Q20, Q30, and GC content the clean data. The remaining high-quality clean reads were aligned to C.sinensis reference genome using Hisat2 (v2.0.5) (Wei et al., 2018). For gene annotation, all UniGenes were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases by BLAST (E value < 1.0E−6).
Differential Expression Analysis
featureCounts (v1.5.0-p3) was used to count the reads numbers mapped to each gene. Then, fragments per kilobase per million (FPKM) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis was performed using the DESeq2 R package (v1.20.0). Genes with an adjusted P-value < 0.05 found by DESeq2 were assigned as differentially expressed.
GO and KEGG Enrichment Analysis
GO and KEGG enrichment analysis of differentially expressed genes (DEGs) was implemented by the clusterProfiler R package, in which gene length bias was corrected. GO terms/KEGG pathways with corrected P-value < 0.05 were considered significantly enriched by differential expressed genes.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted using an RNA extraction kit (Tiangen, Beijing, China), and complementary DNA (cDNA) was synthesized using a PrimeScript™ RT Reagent Kit (Takara, Dalian, China). Supplementary Table 1 lists the primer pairs used for qRT-PCR; β-actin was used as the reference gene. The thermal cycling protocol followed the manufacturer's instructions. Three independent biological replicates of each reaction were conducted, and the relative transcript levels of target genes were calculated against those of β-actin using the formula (Livak and Schmittgen, 2001).
Statistical Analysis
Analysis of variance was performed with SPSS software (v22.0, SPSS Inc., Chicago, IL) to determine the least significant differences between different treatments (p < 0.05), and Duncan's multiple range test was used to compare the averages. Mean and standard deviation were calculated based on six independent biological replicates per tissue. GraphPad Prism (v8.0.1) was used to process the data and generate the figures.
Results
Changes in Ultrastructures and Photosynthetic Pigments
‘Yanlingyinbiancha’ is a variegated tea plant with stable phenotypic traits, which is not affected by light or temperature. As shown in Figures 1A,B, the leaves of ‘Yanlingyinbiancha’ displayed a variegated phenotype, with green and albino tissues. TEM analysis showed the chloroplasts of albino tissues to have no intact lamellar structure, whereas those of green tissues were fully developed (Figures 1C,D). As shown in Figure 1E, chlorophyll a, chlorophyll b, and carotenoid contents in green tissues were 3.66, 3.48, and 4.05 times higher than in albino tissues (p < 0.05). These indicate that chloroplast developmental disorders and a deficiency in chlorophylls and carotenoids are important factors leading to the variegated phenotype of ‘Yanlingyinbiancha’.
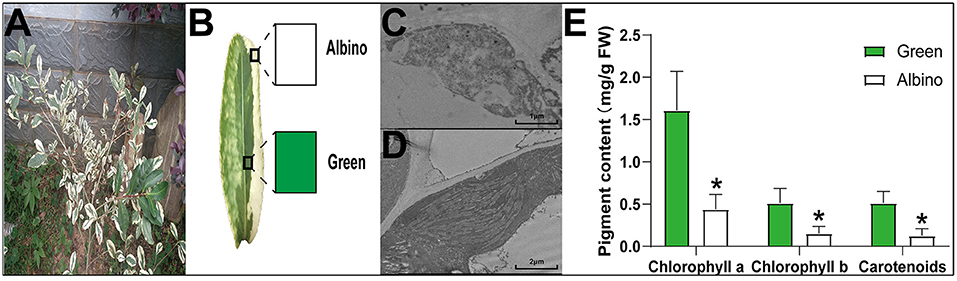
Figure 1. The anatomical characteristics and photosynthetic pigment concentration of ‘Yanlingyinbiancha’. (A,B) Phenotypic characteristics of ‘Yanlingyinbiancha’; (C) Chloroplast ultrastructure of albino tissues; (D) Chloroplast ultrastructure of green tissues; (E) Photosynthetic pigments of albino and green tissues. Data were assessed by one-way ANOVA followed by Duncan's multiple range test. * indicates p < 0.05.
Changes in Catechins, Caffeine, Gallic Acid, and Theobromine Contents
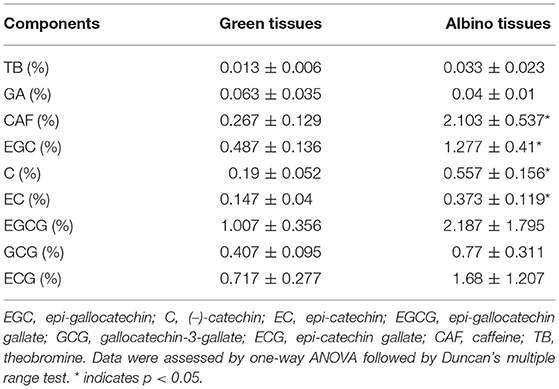
To explore the differences in the biochemical components of albino and green tissues, the contents of catechins (EGC, C, EC, EGCG, GCG, and ECG), caffeine (CAF), gallic acid (GA), and theobromine (TB) in the tested tea leaf samples were determined. Table 1 shows the concentration of nine biochemical components in both the tissues to range from 0.013 ± 0.006% to 2.187 ± 1.795%. TB, CAF and catachins contents in albino tissues were higher than in green tissues, while GA content in albino tissues was lower than in green tissues. Among them, CAF and non-galloylated catechins (EGC, C, and EC) contents in albino tissues were 7.88-fold, 2.62-fold, 2.93-fold, and 2.53-fold higher than in green tissues, respectively (p < 0.05, each). Catechins, together with CAF, are mainly responsible for the bitter and astringent taste, and overall quality of the tea (Chaturvedula and Prakash, 2011). Compared with green leaves, albino tea leaves usually contain lower levels of catechins and CAF (Feng et al., 2014). This is contrary to our results and needs to be further explored.
Transcriptome Analysis
De-novo Assembly
In total, 41.14 Gb clean data with a Q30 value of 92.27% and GC content of 44.26% were obtained after rigorous quality control and data cleansing, indicating high-quality sequencing results for subsequent analysis (Supplementary Table 2). On an average, 82.43% of clean reads were aligned to C. sinensis reference genome (Wei et al., 2018); the mapping statistics are presented in Supplementary Table 3. Uniformity of the mapping result for each sample suggested that they were comparable. A Pearson's correlation coefficient heatmap and principal component analysis between each pair of biological replicates were created (Supplementary Figure 1). Results showed the sequencing data used in the present study to be highly reliable. This dataset has been deposited in NCBI with BioProject number PRJNA733288.
GO and KEGG Analysis of Differentially Expressed Genes
In total, 7,995 DEGs, including 3,407 upregulated and 4,588 downregulated in albino tissues, were identified (Supplementary Figure 2). As shown in Figure 2, “photosynthesis”, “cellular carbohydrate biosynthetic process”, and “cellular carbohydrate metabolic process” were the most highly enriched GO terms in the biological process (BP) category, whereas “thylakoid”, “thylakoid part”, and “photosystem” were highly enriched in the cellular component (CC) category. GO molecular function (MF) analysis showed “carbohydrate phosphatase activity”, “sugar-phosphatase activity”, “fructose 1,6-bisphosphate 1-phosphatase activity” to be the most enriched terms. In addition, the largest number of DEGs were annotated to tetrapyrrole binding, iron ion binding, and oxidoreductase activity. KEGG enrichment analysis revealed most of the DEGs to be enriched in “carbon metabolism” (55), and “starch and sucrose metabolism” (36). “Photosynthesis”, “photosynthesis-antenna proteins”, “glycine, serine, and threonine metabolism”, “phenylpropanoid biosynthesis”, and “starch and sucrose metabolism” were the most highly enriched terms in KEGG pathway (Figure 2). GO and KEGG enrichment analysis indicate that photosynthesis, energy metabolism, chloroplast development and chlorophyll metabolism in albino tissue were significantly affected compared to green tissues. Therefore, we conducted a further analysis of the transcriptome around the above pathways.
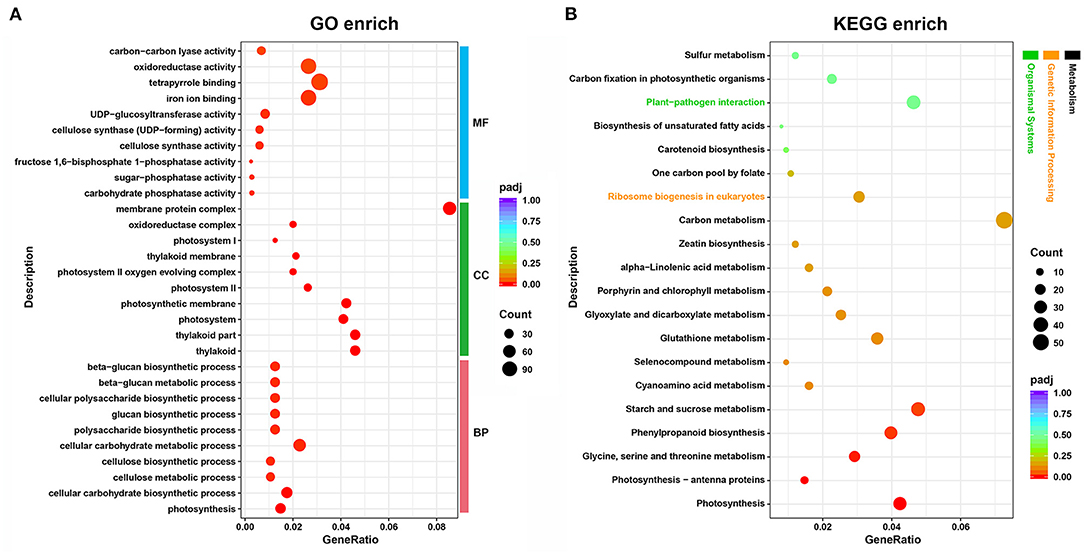
Figure 2. Functional annotation and classification of DEGs identified in the tea plant transcriptome, as determined using the GO and KEGG databases. (A) GO enrichment analysis; (B) KEGG enrichment analysis. GeneRatio, the ratio between DEGs annotated to a particular KEGG pathway/GO term and all DEGs annotated to KEGG pathway/GO category; padj, p-values corrected for multiple hypothesis testing, a lower padj indicates more significant enrichment of the DEGs; Count, the number of DEGs annotated to KEGG pathway/GO term; MF, molecular function; CC, cellular component; BP, biological process.
Biosynthesis of Photosynthetic Pigments
We conducted pathway analysis of DEGs involved in chlorophyll and carotenoid metabolism. As shown in Figure 3, most DEGs related to chlorophyll metabolism were downregulated in albino tissues, including six DEGs (HEMD, PPOX, GUN5, CRD1, PCB2, and CH1) associated with chlorophyll biosynthesis and four DEGs (NYC1, HCAR, CLH1, and ACD2) mediated chlorophyll degradation. Correlation analysis between pigments and DEGs showed that the expression of HEMD, CH1, and ACD2 gene was significantly correlated with chlorophyll content. All carotenoid synthesis-related DEGs in albino tissues were downregulated, including zeta-carotene isomerase (Z-ISO), zeta-carotene desaturase (ZDS), lycopene epsilon-cyclase (LUT2), beta-hydroxylase 1 (BETA-OHASE_1), zeaxanthin epoxidase (ZEP), and nine-cis-epoxycarotenoid dioxygenase 4 (NCED4) genes. Correlation analysis showed that the expression of ZDS and ZEP genes was significantly related to carotenoid content. These results showed the decrease of photosynthetic pigments in albino tissues to possibly be due to the deficit in chlorophyll and carotenoid biosynthesis.
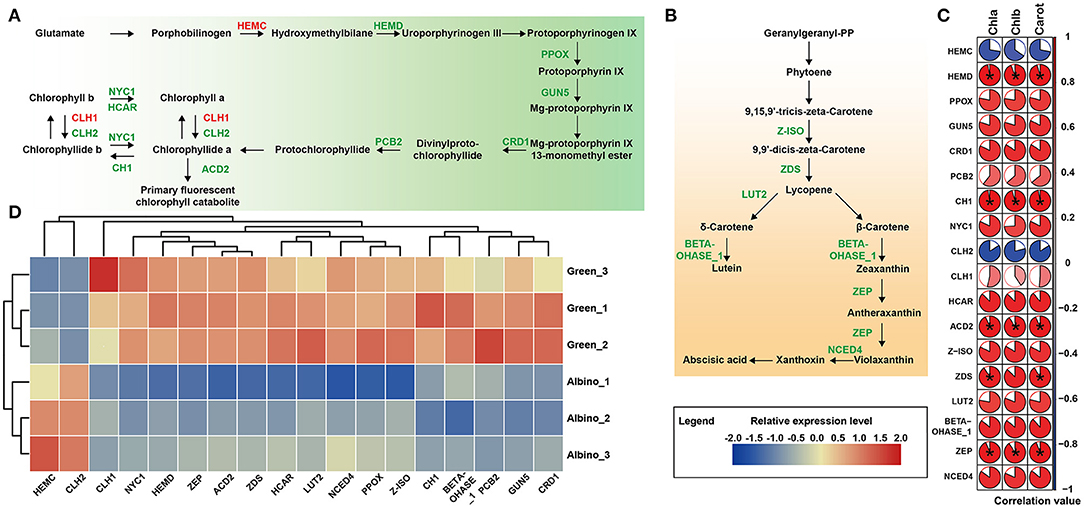
Figure 3. DEGs involved in photosynthetic pigment pathways. (A) Chlorophyll biosynthesis; (B) Carotenoid biosynthesis (Enzymes in red indicate that the gene encoding the enzyme was upregulated in albino tissue while those in green indicate that the gene encoding the enzyme was downregulated in albino tissue compared to that in green tissues); (C) Pearson's correlation analysis between pigments and DEGs (Data were assessed by two-tailed t-test. *indicate p < 0.05. The area of the white sector indicates the p value of the statistical test of the correlation value); (D) Relative expression level of DEGs in both metabolisms. HEMC, hydroxymethylbilane synthase; HEMD, uroporphyrinogen III synthase; PPOX, protoporphyrinogen oxidase; GUN5, magnesium-chelatase subunit chlH; CRD1, magnesium-protoporphyrin IX monomethyl ester cyclase; PCB2, divinyl protochlorophyllide 8-vinyl reductase; NYC1, chlorophyll(ide) b reductase; CH1, chlorophyll a oxygenase; CLH1, chlorophyllase 1; CLH2, chlorophyllase 2; HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase; ACD2, red chlorophyll catabolite reductase; Z-ISO, zeta-carotene isomerase; ZDS, zeta-carotene desaturase; LUT2, lycopene epsilon-cyclase; BETA-OHASE_1, beta-hydroxylase 1; ZEP, zeaxanthin epoxidase; NCED4, nine-cis-epoxycarotenoid dioxygenase 4.
Photosynthesis
As shown in Figure 4, all DEGs that mediated the light reactions in albino tissues were downregulated, including twelve genes involved in the photosystem II (PSII) subunit, six in the photosystem I (PSI) subunit, one related to cytochrome b6/f complex, four mediating the photosynthetic electron transport, nine encoding F-type ATPase, and eleven involved in the light-harvesting chlorophyll protein complex (LHC). Except for triosephosphate isomerase gene (TIM), all DEGs involved in the Calvin cycle were downregulated. The results indicated that development of chloroplast organization in albino tissues was blocked, which affected photosynthesis remarkably in albino tissues.
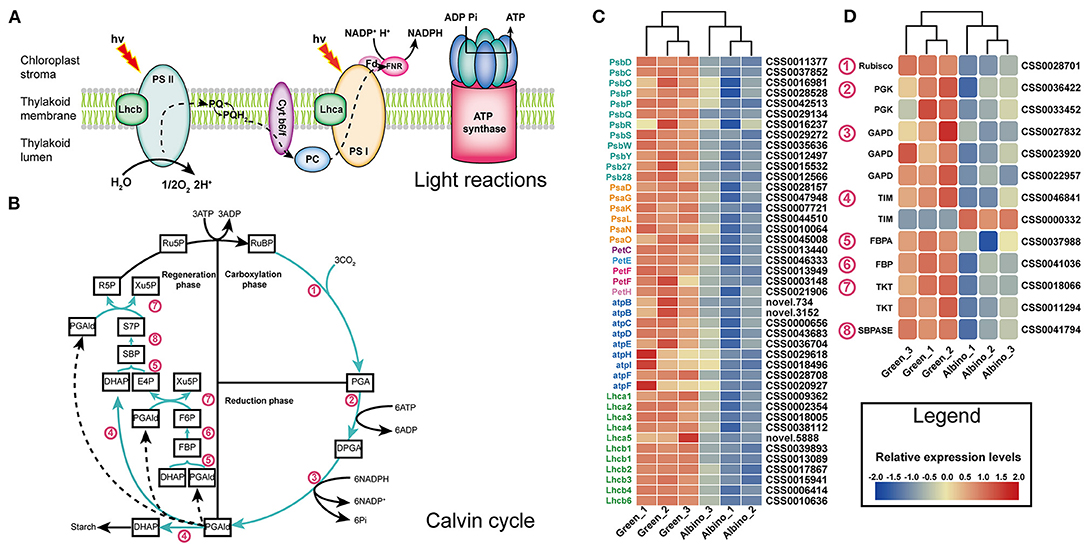
Figure 4. DEGs associated with light reactions (A) and Calvin cycle (B). (C) Relative expression level of DEGs in light reactions; (D) relative expression level of DEGs in Calvin cycle. Lhcb, light harvesting complex of photosystem II; PS II, photosystem II; PQ, plastoquinone; Cyt b6/f, cytochrome b6/f; PC, plastocyanin; Lhca, light harvesting complex of photosystem I; PS I, photosystem I; Fd, ferredoxin; FNR, ferredoxin-NADP+reductase; RuBP, D-Ribulose 1:5-bisphosphate; PGA, 3-Phosphoglycerate; DPGA, 1:3-Bisphospho-D-glycerate; PGAld, Glyceraldehyde 3-phosphate; DHAP, Dihydroxyacetone phosphate; FBP, D-Fructose 1:6-bisphosphate; F6P, D-Fructose 6-phosphate; E4P, D-Erythrose 4-phosphate; Xu5P, D-Xylulose 5-phosphate; SBP, Sedoheptulose 1:7-bisphosphate; S7P, Sedoheptulose 7-phosphate; R5P, Ribose 5-phosphate; Ru5P, Ribulose 5-phosphate; Rubisco, ribulose bisphosphate carboxylase; PGK, phosphoglycerate kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TIM, triosephosphate isomerase; FBA, fructose-bisphosphate aldolase; FBP, fructose 1,6-bisphosphate 1-phosphatase; TKL, transketolase; SBPASE, sedoheptulose-bisphosphatase.
Energy Metabolism
As shown in Figure 5, most DEGs involved in energy metabolism in albino tissues, including phosphofructokinase gene (PFK), hexokinase gene (HXK), glucose-6-phosphate dehydrogenase gene (G6PD), and isocitrate dehydrogenase gene (ICDH), were downregulated. They encoded key enzymes involved in glycolysis, pentose phosphate pathway, and citrate cycle. Although the genes encoding pyruvate kinase (PKP) and citrate synthase (CSY), which are key enzymes in glycolysis and citrate cycle, were upregulated, energy metabolism in albino tissues was severely disturbed, and green tissues might supply energy to albino tissues.
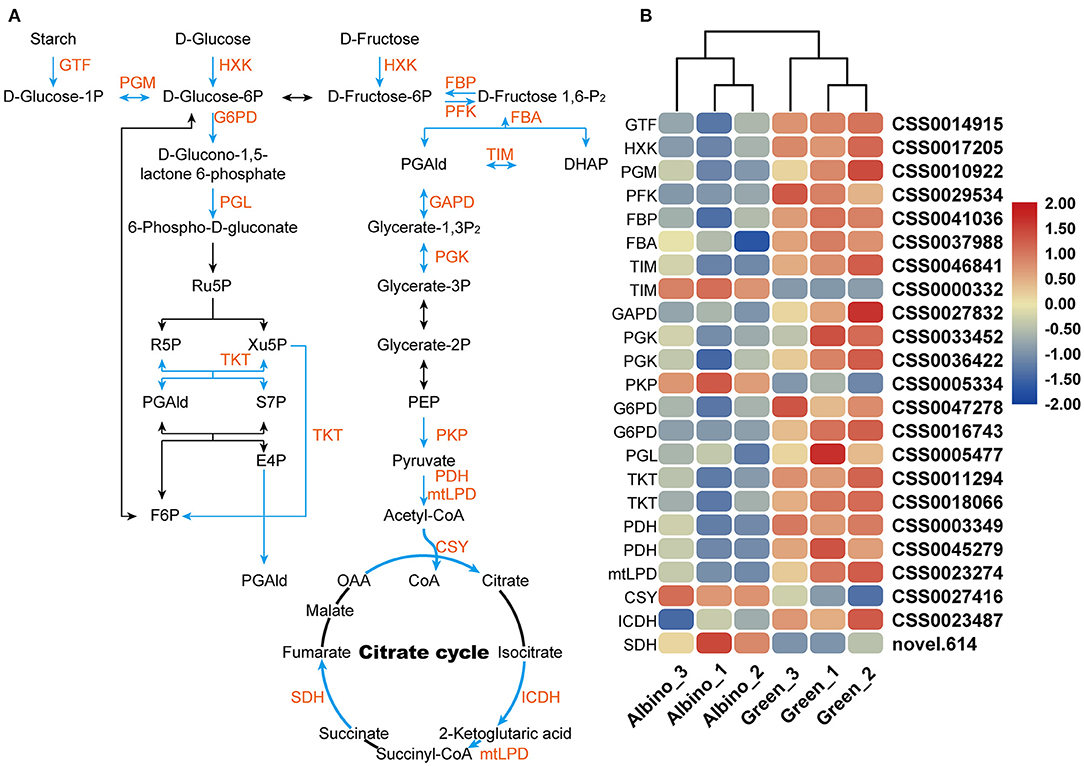
Figure 5. DEGs involved in glycolysis, pentose phosphate pathway, and citrate cycle. (A) Energy metabolism; (B) relative expression level of DEGs in energy metabolism. GTF, 4-alpha-glucanotransferase; HXK, hexokinase; PGM, phosphoglucomutase; PFK, phosphofructokinase; FBP, fructose 1,6-bisphosphate 1-phosphatase; FBA, fructose-bisphosphate aldolase; TIM, triosephosphate isomerase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PKP, pyruvate kinase;G6PD, glucose-6-phosphate dehydrogenase; PGL, 6-phosphogluconolactonase; TKL, transketolase; PDH, pyruvate dehydrogenase; mtLPD, mitochondrial lipoamide dehydrogenase; CSY, citrate synthase; ICDH, isocitrate dehydrogenase; SDH, succinate dehydrogenase.
Flavonoid Metabolism
As presented in Figure 6, more than half of the DEGs involved in flavonoid metabolism in albino tissues were upregulated, including four 4-coumarate: CoA ligase (4CL) genes, chitinase (CHI) gene, flavonol synthase (FLS) gene, and anthocyanidin reductase (ANR) gene, consistent with the increase in catechin levels in albino tissue. Pearson's correlation analysis showed that 4CL (CSS0003013) gene and FLS gene were positively and significantly correlated with (–)-catechin content. Anthocyanidin reductase converts anthocyanidins to epi-flavan-3-ols (EGC, EC), indicating that the significant increase in EC and EGC contents in albino tissues was mainly attributed to the upregulation of ANR gene. In addition to catechins, the contents of other flavonoids in albino tissues was also likely to increase due to the upregulation of FLS gene, such as quercetin, kaempferol and myricetin.
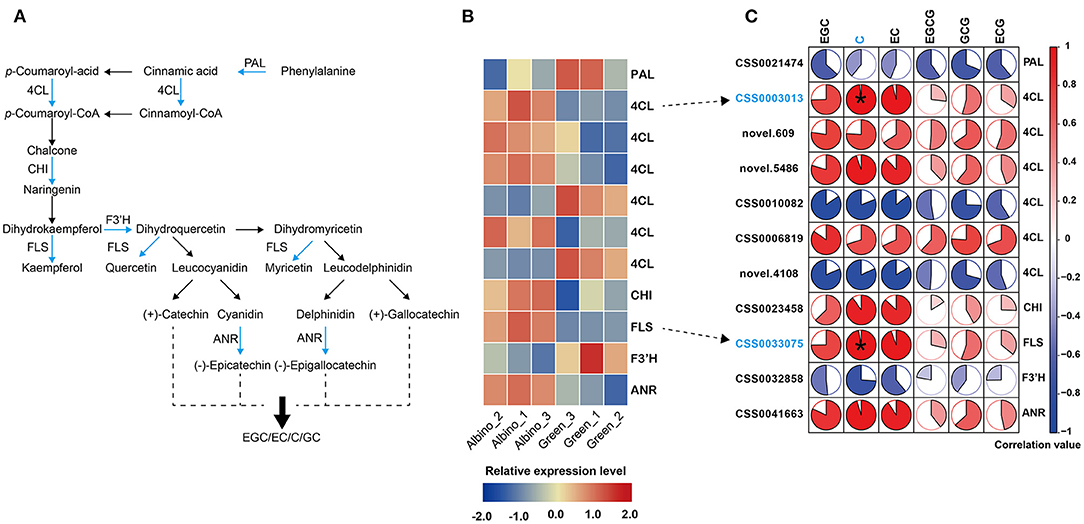
Figure 6. Analysis of flavonoid metabolism. (A): flavonoid biosynthesis; (B): differential expressed genes (DEGs) in different tissues; (C): correlation analysis between catechins and DEGs (Data were assessed by two-tailed t-test. *indicate p < 0.05. The area of the white sector indicates the p value of the correlation coefficient.). PAL, phenyl alanine ammonia-lyase; 4CL, 4-coumarate: CoA ligase; CHI, chitinase; FLS, flavonol synthase; F3'H, flavanone 3-hydroxylase; ANR, anthocyanidin reductase.
Photomorphogenesis
As presented in Figure 7, DEGs encoding positive regulators of photomorphogenesis involved in chloroplast development were all downregulated in albino tissues, including cryptochrome 1 (CRY1), phytochrome B (PHYB), and chloroplast heat shock protein 70-2 (cpHsc70-2) genes. As shown in Figures 7A–C, CRY1 and PHYB genes encode blue and red light photoreceptors, respectively, and mediate chloroplast nuclear gene expression, chloroplast protein import and photosynthetic pigment biosynthesis gene expression. DEGs participating in protein ubiquitination and degradation were all upregulated, including RING-box 1 (RBX1), damaged DNA binding protein 1A (DDB1A), and ubiquitin 4 (UBQ4) genes. These genes can participate in the degradation of positive regulators of photomorphogenesis. In addition, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) gene that mediates UV-B signaling pathway was upregulated. UV-B signaling pathway is closely related to flavonoid metabolism, and UV-B signal can promote flavonoid biosynthesis through transcriptional regulation.
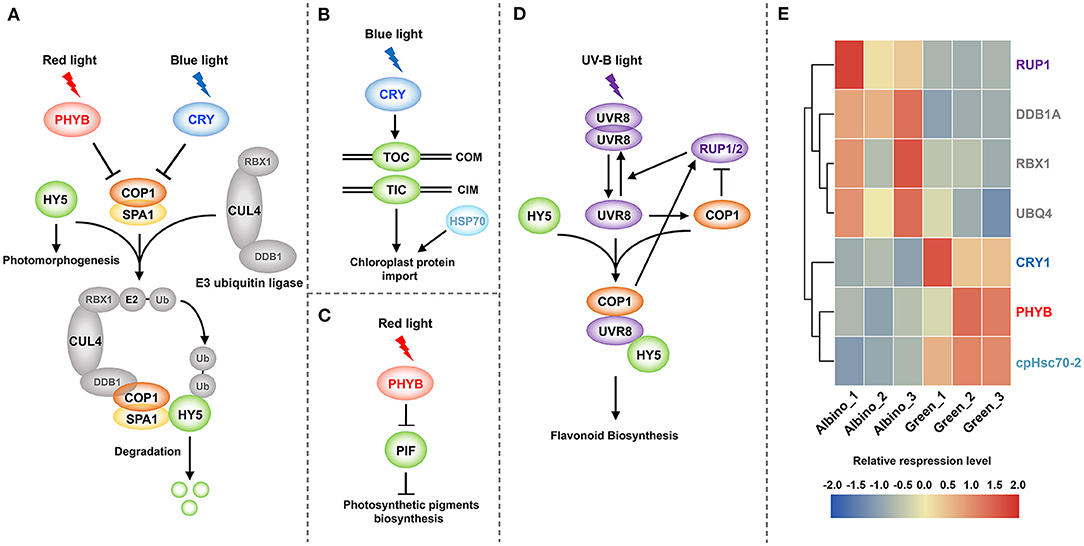
Figure 7. Photomorphogenesis mediates chloroplast development and flavonoid biosynthesis. (A) Chloroplast protein-related nuclear gene expression; (B) Chloroplast protein import; (C): Photosynthetic pigment biosynthesis; (D) UV-B signal mediated flavonoid biosynthesis; (E) DEGs involved in photomorphogenesis. PHYB, phytochrome B; CRY, cryptochrome; HY5, ELONGATED HYPOCOTYL 5; COP1, CONSTITUTIVE PHOTOMORPHOGENIC 1; SPA1, SUPPRESSOR OF PHYA-105 1; RBX1, RING-box 1; CUL4, cullin4; DDB1, damaged DNA binding protein 1; Ub, ubiquitin; TOC, translocon at the outer envelope membrane of chloroplasts; TIC, translocon at the inner envelope membrane of chloroplasts; COM, chloroplast outer membrane; CIM, chloroplast inner membrane; HSP70, heat shock protein 70; PIF, phytochrome interacting factor; UVR8, UV RESISTANCE LOCUS 8; RUP1/2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1/2.
Validation of DEG Transcript Levels
qRT-PCR was used for verification of the RNA-seq results (Supplementary Figure 3). Sixteen DEGs were randomly selected for qRT-PCR verification. Of them, four were involved in chlorophyll biosynthesis (HEMD, CH1, NYC1, and ACD2), two were involved in carotenoid biosynthesis (ZDS and ZEP), four mediated the light reaction (Lhca1, Lhcb1, PsbS, and PsbY), two were related to Calvin cycle (Rubisco and PGK), three were involved in energy metabolism (GTF, FBA, and PGK), and two were involved in flavonoid metabolism (PAL and FLS). Expression pattern of these genes was consistent with RNA-seq data, indicating that the RNA-seq data are reliable.
Discussion
Repression of Photosynthetic Pigment Biosynthesis Leads to Albinism
Chlorophyll metabolism, including its biosynthesis, cycling, and degradation, is a complex biological process in plants. In our study, six (HEMD, PPOX, GUN5, CRD1, PCB2, and CH1) and four (NYC1, HCAR, CLH1, and ACD2) DEGs associated with chlorophyll biosynthesis and chlorophyll degradation, respectively, were found to be downregulated. Previous work had shown that mutation in the GUN5 genecan cause the formation of albinos (Mochizuki et al., 2001). Plants with reduced CHL27/CRD1 display chlorosis and excessive MgProtoME accumulation (Tottey et al., 2003; Bang et al., 2008; Peter et al., 2010). Moreover, chlorophyll content of PCB2 mutant decreased significantly compared to that in wild-type (Nakanishi et al., 2005). Therefore, we considered variegated phenotype to possibly arise from impaired chlorophyll biosynthesis in albino tissues, and the DEGs involved in chlorophyll degradation to possibly be downregulated due to the substrate inhibition effect.
In plants, carotenoids serve as accessory pigments in photosynthesis and protect against photooxidative stress (Britton et al., 2004). Previous studies had shown that plants deficient in genes encoding carotenoid biosynthesis pathway enzymes exhibit an albino phenotype. For example, Chen et al. had reported that Arabidopsis Z-ISO mutant plants contained lower levels of carotenoids and chlorophylls than the wild type (Chen et al., 2010). Furthermore, in cases of ZDS deficiency, maize showed defective chloroplast development and reduced photosynthetic pigments, leading to an albino phenotype (Wang et al., 2020). In this study, five DEGs involved in carotenoid biosynthesis were downregulated in albino tissues, indicating the decrease in carotenoid to be due to the reduction in carotenoid biosynthesis. Carotenoids contribute to the maintenance of chlorophyll levels (Wang et al., 2014; Shen et al., 2018); hence, inhibition of carotenoid biosynthesis in albino tissues might be one of the reasons for reduction of chlorophyll and variegated phenotype.
Photomorphogenesis Affects Chloroplast Development
Chloroplast is essential for photosynthesis as well as the production of hormones and metabolites (Pogson and Albrecht, 2011). Our study found that chloroplasts have no intact lamellar structure in albino tissues, and that photosynthesis was suppressed. Similar results were also observed in other albino or variegated plants (Sun et al., 2017; Shih et al., 2019; Li et al., 2020). As shown in Figure 4A, four major photosynthetic complexes in plants, including PSII, PSI, the cytochrome b6/f (cytb6/f) complex, and plastid ATP synthase, are located in the chloroplast thylakoid membrane (Rochaix, 2004). These components convert water into oxygen and produce NADPH and ATP. Subsequently, NADPH and ATP are used for CO2 assimilation by the Calvin cycle (Fromme et al., 2001; Rochaix, 2011; Allahverdiyeva et al., 2013). In this study, fifty-five DEGs involved in the above processes were downregulated in albino tissues, indicating the light reaction to be severely suppressed and Calvin cycle to be inhibited by the shortage of ATP and NAPDH.
One major environmental factor determining the biogenesis of chloroplast is light, which acts via photomorphogenic pathways to control the transcription of genes encoding chloroplast-targeted protein (Pogson and Albrecht, 2011). The perception of light has been shown to require the activation of photoreceptors, such as phytochromes (PHYs) and cryptochromes (CRYs). As shown in Figure 7A, phytochromes and cryptochromes inhibit the function of COP1 protein upon irradiation of red and blue light. In the dark, COP1-SPA1 complex can combine with E3 ubiquitin ligase to degrade ELONGATED HYPOCOTYL 5 (HY5) transcription factor, and thereby inhibits photomorphogenesis. In our study, PHYB and CRY1 genes were dramatically downregulated in albino tissues. Moreover, based on the observation that RBX1, DDB1A, and UBQ4 genes that encode E3 ubiquitin ligase were upregulated in albino tissues, we speculated that the destruction of photoreceptors in albino tissues causes COP1 to accumulate in the nucleus. Subsequently, COP1 mediates the degradation and ubiquitination of positive transcription factors of photomorphogenesis and prevents light-induced gene expression. These results show that downregulation of PHYB and CRY1 genes in albino tissues might lead to downregulation of the gene encoding chloroplast protein that is involved in the light reaction.
In addition to control the transcription of genes encoding chloroplast protein, light can also affect the process of protein import into chloroplast stroma. Previous study had shown that induction of translocon at the outer envelope membrane of chloroplasts (TOC) and translocon at the inner envelope membrane of chloroplasts (TIC) genes, which encode components of the chloroplast protein import apparatus, is mediated by CRY1 (Fukazawa et al., 2020). This suggested that downregulation of CRY1 in albino tissues might inhibit the expression of TOC and TIC genes, leading to a blockage in chloroplast protein import. Moreover, we also observed the downregulation of cpHsc70-2 gene. Previous study had shown that cpHsc70-2 gene is important for protein import into chloroplasts (Su and Li, 2010). Hence, we considered the disruption of chloroplast protein import pathway in albino tissues to be one of the reasons for abnormal chloroplast development.
Chlorophylls and carotenoids are essential for the assembly and protection of photosynthetic apparatus. PHYB, as the predominant member of PHY family, promotes the ubiquitination and degradation of phytochrome interacting factors (PIFs) (Leivar et al., 2012). PIFs are negative regulators of tetrapyrrole biosynthesis genes and carotenoid biosynthesis genes (Gommers and Monte, 2018; Xu, 2020). As shown in Figure 7C, downregulation of PHYB might lead to the accumulation of PIFs in albino tissues, and thus inhibited the biosynthesis of photosynthetic pigment. Deficit in chlorophylls and carotenoids affected the assembly and maintenance of photosynthetic apparatus, which is highly likely another reason for abnormal chloroplast development.
Taken together, as presented in Figures 7A–C, we conjectured that the destruction of photoreceptors suppressed photomorphogenesis in albino tissues, leading to the inhibition of chloroplast protein gene transcription, blockage of chloroplast protein import, and damage to the assembly and protection of photosynthetic apparatus. These factors together prevented the development of chloroplasts in albino tissues, resulting in a weakened capacity for photosynthesis.
Survival Strategy of Albino Tissues Under Carbon Starvation Condition
Considering the inhibition of photosynthesis due to defects in chloroplast development, carbon supply for the survival of albino tissues is insufficient. Therefore, we speculated that the albino tissues implemented three strategies to compensate for this deficit. First, albino tissues may require sucrose delivery from green tissues. Sucrose synthase (SUS) has long been considered a biochemical marker for sink strength (Xu et al., 2012). It catalyzes the hydrolysis of sucrose to UDP-glucose and fructose, thus forming a gradient that promotes sucrose transport. Upregulation of SUS gene was observed in albino tissues, so we speculated that albino tissues may receive sucrose from green tissues, as a sink tissue. The same conclusion has been obtained in the study of variegated mutant in Arabidopsis (Aluru et al., 2009). Second, fatty acid respiration is probably a backup mechanism under energy shortage conditions (Kunz et al., 2009). The DEGs involved in β-oxidation of fatty acid in albino tissues, including long-chain acyl-CoA synthetase (LACS), acyl-CoA oxidase (ACX) and peroxisomal 3-ketoacyl-CoA thiolase (PKT) genes, were upregulated. Our results suggested that the albino tissues treat fatty acid oxidation as energy supplement under carbon starvation conditions induced by the inhibition of photosynthesis. Third, anaerobic respiration in albino tissues might increase to compensate for the repression of mitochondrial respiration. Aldehyde dehydrogenase 2 (ALDH2) participates in the detoxification process of alcohol fermentation (Kirch et al., 2004). We found ALDH2B4 and ALDH2B7 genes to be significantly upregulated in albino tissues, in agreement with our speculation.
UV-B Signal and Brassinosteroids Mediate Flavonoid Biosynthesis
The higher content of catechins in albino tissues may result from two reasons. Previous study had shown that the expression of flavonoid pathway genes, such as FLS, 4CL, and ANR, could be induced in response to UV-B signal, leading to the accumulation of catechins in Camellia sinensis (Fan et al., 2018; Liu et al., 2018). While COP1 is a negative regulator of photomorphogenesis, it acts as a positive regulator in the UV-B signaling pathway (Jin and Zhu, 2019). The accumulation of COP1 in albino tissues may promote UV-B signaling and thus increase the level of catechins. As presented in Figure 7D, RUP1/2 were transcriptionally activated by UV-B in a COP1, UVR8, and HY5-dependent manner, and negatively regulated the UV-B pathway to prevent overstimulation by UV-B (Gruber et al., 2010). The upregulation of RUP1 indirectly proved the UV-B signaling pathway to possibly be activated by COP1. Moreover, upregulation of DEGs involved in brassinosteroid (BR) biosynthesis and signal transduction, such as brassinosteroid-6-oxidase 2 (BR6OX2), CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD), and BR-signaling kinase 2 (BSK2) genes, was observed. BR can upregulate the expression of genes involved in catechin synthesis by increasing the accumulation of nitric oxide (NO) (Li et al., 2016b, 2017). Our results suggested that the increase in endogenous BR levels might lead to the higher content of catechins in albino tissues.
Conclusion
‘Yanlingyinbiancha’ has a variegated phenotype owing to the lack of pigment on the edge of the leaves. Analysis of the transcriptome, in this study, showed that chloroplast development in albino tissues might be repressed by three factors: inhibition of chloroplast protein gene transcription, blockage of chloroplast protein import, and damage of the assembly and protection of photosynthetic apparatus. Abnormal chloroplast development inevitably leads to the destruction of photosynthesis and insufficient energy. Therefore, the albino tissues might receive sucrose from the green tissues and decompose their own storage substances to obtain the energy needed for survival. Further, catechins, as a characteristic secondary metabolite of tea plants, accumulated in albino tissues through UV-B signals and hormone signals. Variegated mutants are powerful tools for understanding mechanisms of chloroplast biogenesis. This study preliminarily reveals the difference between albino and green tissues of ‘Yanlingyinbiancha’ and the molecular mechanism underlying variegation.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI repository, accession number: PRJNA733288.
Author Contributions
XG: conceptualization, writing—original draft, and mapping. CZ: writing—review and editing and mapping. CL: investigation, resources, and supervision. MW and NX: validation. JianC: formal analysis. YL: data curation. JiaC: visualization. CS: project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Joint Funds of National Natural Science Foundation of China (grant number U19A2030); the National Natural Science Foundation of China (grant number 31271789); Central Committee Guides Local Science and Technology Development Program (grant number 2019XF5041); and the Special Project for the Construction of Modern Agricultural Industrial Technology Systems in Hunan Province (Xiangcai Nongzhi) (grant number 2020112).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Fuquan Luo for supplying experimental materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.720800/full#supplementary-material
References
Allahverdiyeva, Y., Suorsa, M., Rossi, F., Pavesi, A., Kater, M. M., Antonacci, A., et al. (2013). Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 75, 671–684. doi: 10.1111/tpj.12230
Aluru, M. R., Zola, J., Foudree, A., and Rodermel, S. R. (2009). Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiol. 150, 904–923. doi: 10.1104/pp.109.135780
Bang, W. Y., Jeong, I. S., Kim, D. W., Im, C. H., Ji, C., Hwang, S. M., et al. (2008). Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol. 49, 1350–1363. doi: 10.1093/pcp/pcn111
Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Mache, R., et al. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. doi: 10.1105/tpc.11.1.57
Chaturvedula, V. S. P., and Prakash, I. (2011). The aroma, taste, color and bioactive constituents of tea. J. Med. Plants Res. 5, 2110–2124. doi: 10.5897/JMPR.9001187
Chen, Y., Li, F., and Wurtzel, E.T. (2010). Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 153, 66–79. doi: 10.1104/pp.110.153916
Fan, F., Li, C., Luo, Z., and Tang, G. (2018). “UV-B radiation-induced changes in tea metabolites and related gene expression,” in Stress Physiology of Tea in the Face of Climate Change, ed. W. Y. Han, X. Li, G. J. Ahammed (Singapore: Springer), 111–133.
Feng, L., Gao, M. J., Hou, R. Y., Hu, X. Y., Zhang, L., Wan, X. C., et al. (2014). Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 155, 98–104. doi: 10.1016/j.foodchem.2014.01.044
Fromme, P., Jordan, P., and Krauss, N. (2001). Structure of photosystem I. Biochim. Biophys. Acta 1507, 5–31. doi: 10.1016/S0005-2728(01)00195-5
Fukazawa, H., Tada, A., Richardson, L., Kakizaki, T., Uehara, S., Ito-Inaba, Y., et al. (2020). Induction of TOC and TIC genes during photomorphogenesis is mediated primarily by cryptochrome 1 in Arabidopsis. Sci. Rep. 10:20255. doi: 10.1038/s41598-020-76939-w
Gommers, C., and Monte, E. (2018). Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. doi: 10.1104/pp.17.01460
Gruber, H., Heijde, M., Heller, W., Albert, A., Seidlitz, H. K., and Ulm, R. (2010). Negative feedback regulation of UV-B–induced photomorphogenesis and stress acclimation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107, 20132–20137. doi: 10.1073/pnas.0914532107
Han, Y. J., Song, P. S., and Kim, J. l. (2007). Phytochrome-mediated photomorphogenesis in plants. J. Plant Biol. 50, 230–240. doi: 10.1007/BF03030650
Jin, H., and Zhu, Z. (2019). Dis-RUP for COP1 role-switch under UV-B light. Trends Plant Sci. 24, 569–571. doi: 10.1016/j.tplants.2019.04.006
Kirch, H. H., Bartels, D., Wei, Y., Schnable, P. S., and Wood, A. J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9, 371–377. doi: 10.1016/j.tplants.2004.06.004
Kunz, H. H., Scharnewski, M., Feussner, K., Feussner, I., Flügge, U. I., Fulda, M., et al. (2009). The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749. doi: 10.1105/tpc.108.064857
Lau, O. S., and Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Leivar, P., Monte, E., Cohn, M. M., and Quail, P. H. (2012). Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB–PIF feedback loop. Mol. Plant 5, 734–749. doi: 10.1093/mp/sss031
Li, C. F., Xu, Y. X., Ma, J. Q., Jin, J. Q., Huang, D. J., Yao, M. Z., et al. (2016a). Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’(Camellia sinensis). BMC Plant Biol. 16:195. doi: 10.1186/s12870-016-0885-2
Li, X., Ahammed, G. J., Li, Z. X., Zhang, L., Wei, J. P., Shen, C., et al. (2016b). Brassinosteroids improve quality of summer tea (Camellia sinensis L.) by balancing biosynthesis of polyphenols and amino acids. Front. Plant Sci. 7:1304. doi: 10.3389/fpls.2016.01304
Li, X., Zhang, L., Ahammed, G. J., Li, Z. X., Wei, J. P., Shen, C., et al. (2017). Nitric oxide mediates brassinosteroid-induced flavonoid biosynthesis in Camellia sinensis L. J. Plant Physiol. 214, 145–151. doi: 10.1016/j.jplph.2017.04.005
Li, Y., Li, W., Hu, D., Shen, P., Zhang, G., and Zhu, Y. (2020). Comparative analysis of the metabolome and transcriptome between green and albino zones of variegated leaves from Hydrangea macrophylla ‘Maculata’ infected by hydrangea ringspot virus. Plant Physiol. Biochem. 157, 195–210. doi: 10.1016/j.plaphy.2020.10.012
Liu, L., Li, Y., She, G., Zhang, X., Jordan, B., Chen, Q., et al. (2018). Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 18:233. doi: 10.1186/s12870-018-1440-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, C. Y., Cao, J.X., Li, J. K., Zhou, B., Tang, T. C., and Miao, A. Q. (2016). Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 6:33369. doi: 10.1038/srep33369
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit. Proc. Natl. Acad. Sci. USA 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Nakanishi, H., Nozue, H., Suzuki, K., Kaneko, Y., Taguchi, G., and Hayashida, N. (2005). Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol. 46, 467–473. doi: 10.1093/pcp/pci053
Ohgishi, M., Saji, K., Okada, K., and Sakai, T. (2004). Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 2223–2228. doi: 10.1073/pnas.0305984101
Peter, E., Rothbart, M., Oelze, M. L., Shalygo, N., Dietz, K. J., and Grimm, B. (2010). Mg protoporphyrin monomethylester cyclase deficiency and effects on tetrapyrrole metabolism in different light conditions. Plant Cell Physiol. 51, 1229–1241. doi: 10.1093/pcp/pcq071
Podolec, R., and Ulm, R. (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25. doi: 10.1016/j.pbi.2018.04.018
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Rochaix, J. D. (2004). Genetics of the biogenesis and dynamics of the photosynthetic machinery in eukaryotes. Plant Cell 16, 1650–1660. doi: 10.1105/tpc.160770
Rochaix, J. D. (2011). Assembly of the photosynthetic apparatus. Plant Physiol. 155, 1493–1500. doi: 10.1104/pp.110.169839
Sakamoto, W. (2003). Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 78, 1–9. doi: 10.1266/ggs.78.1
Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15, 2843–2855. doi: 10.1105/tpc.017319
Shen, J., Zou, Z., Zhang, X., Zhou, L., Wang, Y., Fang, W., et al. (2018). Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 5:7. doi: 10.1038/s41438-017-0010-1
Shih, T. H., Lin, S. H., Huang, M. Y., Huang, W. D., and Yang, C. M. (2019). Transcriptome profile of the variegated Ficus microcarpa c.v. milky stripe fig leaf. Int. J. Mol. Sci. 20, 1338. doi: 10.3390/ijms20061338
Song, L., Ma, Q., Zou, Z., Sun, K., Yao, Y., Tao, J., et al. (2017). Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in Camellia sinensis cultivar ‘Huangjinya’. Front. Plant Sci. 8:803. doi: 10.3389/fpls.2017.00803
Su, P. H., and Li, H. M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531. doi: 10.1105/tpc.109.071415
Sun, Y. H., Hung, C. Y., Qiu, J., Chen, J., Kittur, F. S., Oldham, C. E., et al. (2017). Accumulation of high OPDA level correlates with reduced ROS and elevated GSH benefiting white cell survival in variegated leaves. Sci. Rep. 7:44158. doi: 10.1038/srep44158
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10:e1004416. doi: 10.1371/journal.pgen.1004416
Tottey, S., Block, M. A., Allen, M., Westergren, T., Albrieux, C., Scheller, H. V., et al. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. U. S. A. 100, 16119–16124. doi: 10.1073/pnas.2136793100
Wang, L., Yue, C., Cao, H., Zhou, Y., Zeng, J., Yang, Y, et al. (2014). Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 14:352. doi: 10.1186/s12870-014-0352-x
Wang, M., Zhu, X., Li, Y., and Xia, Z. (2020). Transcriptome analysis of a new maize albino mutant reveals that zeta-carotene desaturase is involved in chloroplast development and retrograde signaling. Plant Physiol. Biochem. 156, 407–419. doi: 10.1016/j.plaphy.2020.09.025
Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C., Gao, L., et al. (2018). Draft genome sequence of Camellia sinensis var. Sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U. S. A. 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Xu, D. (2020). COP1- and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Xu, S. M., Brill, E., Llewellyn, D. J., Furbanka, R. T., and Ruanb, Y. L. (2012). Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 5, 430–441. doi: 10.1093/mp/ssr090
Zhao, J., Zhou, J. J., Wang, Y. Y., Gu, J. W., and Xie, X. Z. (2013). Positive regulation of phytochrome B on chlorophyll biosynthesis and chloroplast development in rice. Rice Sci. 20, 243–248. doi: 10.1016/S1672-6308(13)60133-X
Keywords: Camellia sinensis, variegated, transcriptome sequencing, chloroplast development, photomorphogenesis
Citation: Gao X, Zhang C, Lu C, Wang M, Xie N, Chen J, Li Y, Chen J and Shen C (2021) Disruption of Photomorphogenesis Leads to Abnormal Chloroplast Development and Leaf Variegation in Camellia sinensis. Front. Plant Sci. 12:720800. doi: 10.3389/fpls.2021.720800
Received: 05 June 2021; Accepted: 28 July 2021;
Published: 09 September 2021.
Edited by:
Santiago Signorelli, Universidad de la República, UruguayReviewed by:
Pablo Ignacio Calzadilla, The University of Manchester, United KingdomDiep R. Ganguly, Australian National University, Australia
Copyright © 2021 Gao, Zhang, Lu, Wang, Xie, Chen, Li, Chen and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengwen Shen, c2N3NjlAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xizhi Gao
Xizhi Gao Chenyu Zhang1,2,3,4†
Chenyu Zhang1,2,3,4† Cui Lu
Cui Lu Chengwen Shen
Chengwen Shen