- 1College of Agricultural and Environmental Sciences, Makerere University, Kampala, Uganda
- 2National Crops Resources Research Institute, Kampala, Uganda
- 3National Root Crops Research Institute, Umudike, Nigeria
- 4Makerere University Regional Center for Crop Improvement, Kampala, Uganda
- 5International Institute of Tropical Agriculture, Ibadan, Nigeria
- 6Cornell University Root Crops Research Institute, Ithaca, NY, United States
Cassava mosaic geminiviruses (CMGs) and cassava brown streak viruses (CBSVs) cause the highest yield losses in cassava production in Africa. In particular, cassava brown streak disease (CBSD) is and continues to be a significant constraint to optimal cassava production in Eastern and Southern Africa. While CBSD has not been reported in West Africa, its recent rapid spread and damage to cassava productivity in Eastern, and Southern Africa is alarming. The aim of this study was to evaluate Nigerian cassava genotypes in order to determine their responses to CBSD, in the event that it invades Nigeria, the world’s largest cassava producer. The study gathered information on whether useful CBSD resistance alleles are present in the elite Nigerian cassava accessions. A total of 1,980 full-sib cassava seedlings from 106 families were assessed in the field at the seedling stage for a year. A subset of 569 clones were selected and assessed for another year at the clonal stage in Namulonge, central Uganda, a known hotspot for CBSD screening. Results indicated that foliar and root incidences and severities varied significantly (p ≤ 0.01, p ≤ 0.001) except for CBSD foliar incidence at 6 months (CBSD6i). Highest and lowest plot-based heritability estimates for CBSD were registered for CBSD root severity (CBSDrs) (0.71) and CBSD6i (0.5). Positive and highly significant correlations were noted between CBSD root incidence (CBSDri) and CBSDrs (r = 0.90***). Significant positive correlations were also noted between CBSD foliar severity at 3 months (CBSD3s) and CBSD foliar incidence at 6 months (CBSD6i) (r = 0.77***), CBSD3s and CBSDrs (r = 0.35***). Fresh root weight (FreshRW) negatively correlated with CBSDri and CBSDrs, respectively (r = −0.21*** and r = −0.22***). Similarly, CBSD3s correlated negatively with cassava mosaic disease severity at 3 (CMD3s) and 6 months (CMD6s), respectively (r = −0.25*** and r = −0.21***). Fifteen clones were selected using a non-weighted summation selection index for further screening. In conclusion, results revealed that the elite Nigerian accessions exhibited significant susceptibility to CBSD within 2 years of evaluation period. It is expected that this information will aid future breeding decisions for the improvement of CBSD resistance among the Nigerian cassava varieties.
Introduction
Cassava (Manihot esculenta Crantz) is a perennial plant cultivated as an annual crop. It originated in Latin America and is widely grown across the subtropical and tropical regions of the world. It is an essential source of carbohydrate and a major staple food for over 800 million people globally (Hammond et al., 2013). Cassava is popular because of its hardy nature and ability to adapt to drought and low nutrient availability in the soil. The crop is also amenable to piecemeal harvesting for a period between 8 and 24 months after planting, an attribute that makes it popular among smallholder farmers. These inherent characteristics make cassava one of the most resilient food security crops of the 21st century. Approximately 70% of cassava is currently grown in Africa and Asia, with an estimated cultivated area of more than 22 million hectares (Food and Agricultural Organization of the United Nations [FAOSTAT], 2020). Africa accounts for 61% of 277 million tons of cassava production worldwide, most of which comes from Nigeria with an estimated 59 million tons (Food and Agricultural Organization of the United Nations [FAOSTAT], 2020).
Cassava is a basic staple food for more than 65% of the population in Nigeria who consume it in different forms at least once a day (Eke-okoro and Njoku, 2012). It is processed into over 50 food forms, such as garri, lafun, bread, flakes, and flour (Eke-okoro and Njoku, 2012). With the increasing demand for cassava for both food and non-food uses in Nigeria, efforts must be devoted to sustain and/or increase cassava production and productivity.
Significant cassava yield gaps are commonplace in most tropical countries. For example, in Nigeria, where more than 40 different varieties are grown by farmers (Bankole, 2019), the national average yield of 13.6 metric tons per hectare reflects a shortfall of 65.9% when compared with potential yield estimates of 40 metric tons per hectare (Food and Agricultural Organization of the United Nations [FAOSTAT], 2020). This yield gap is due to an array of biotic and abiotic constraints, notably poor agronomic practices, susceptibility to prevalent pests and diseases, drought, a deficit of clean planting materials, and drudgery associated with most farm operations. Among these constraints, diseases are by far the main impediments to optimal cassava production (Ntawuruhunga et al., 2013).
In Africa, cassava production is mostly limited by two viral diseases: cassava mosaic disease (CMD) and cassava brown streak disease (CBSD). CMD causes yield losses of up to 70% in susceptible varieties and can kill or stunt plants to varying degrees (Tomlinson et al., 2018). CBSD causes yield losses of up to 100% in susceptible varieties and destroys the edible roots even when the rest of the plant looks healthy (Hillocks et al., 2008). CMD is distributed across the major African cassava growing belt and is considered one of the most severe and widespread diseases of cassava in Nigeria. In contrast, CBSD is restricted to Eastern, Central, and some parts of Southern Africa and is currently considered the world’s most dangerous threat to cassava production (Legg et al., 2011; Patil et al., 2014; Eni et al., 2018; Tomlinson et al., 2018).
Based on complete genome sequences, CBSD is caused by two distinct virus species: cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV; Mbanzibwa et al., 2009; Winter et al., 2010). Both species belong to the genus Ipomovirus and family Potyviridae (Alicai et al., 2016) and can be found in low- and mid-altitude areas (Munga, 2008; Mrema, 2016). Symptomatically, CBSD constricts and necrotizes cassava roots, rendering them unpalatable and unmarketable. On most instances, blotchy yellow chlorosis or feathery chlorosis appears on the minor veins of the leaves of infected plants. Brown, round, or elongated streak-like lesions can also occur on the young green portion of the infected stems. These lesions develop in severe infections to cause dieback and possibly kill the whole plant. The most prevalent viruses that are causal agents of CMD in Africa are single-stranded DNA bipartite cassava mosaic begomoviruses (CMBs), African cassava mosaic virus (ACMV), and East African cassava mosaic virus (EACMV). In Nigeria, CMD reached epidemic status in the 1950s and breeding for its resistance began in 1970 (Hahn et al., 1980). Since then, CMD resistance has been a backbone for all cassava varieties being developed and released by the national breeding program.
Both CBSD and CMD are transmitted over short distances by the whitefly vector, Bemisia tabaci (Gennadius) (Maruthi et al., 2005), whereas they are transmitted over long distances through the transport of infected planting materials (Patil et al., 2015). Management practices for both diseases include planting of clean (symptomless) cassava cuttings, uprooting and destroying cassava plants showing disease symptoms, sterilization of farm implements, especially when cutting cassava stems for multiplication, and using tolerant/resistant varieties. Cultivation of tolerant/resistant varieties remains the most viable management practice (Munga, 2008).
Previously regarded as a low-altitude disease, CBSD has recently been confirmed to exist in both lowland and highland areas, with the recent reports indicating a wide spread of CBSD in Uganda, Kenya, Malawi, Burundi, Rwanda, and some parts of the Democratic Republic of Congo (DRC; Hillocks et al., 2002; Bigirimana et al., 2011; Hillocks and Maruthi, 2015; Chipeta et al., 2016; Koima et al., 2018; Munganyinka et al., 2018). With the existence of CBSD in DRC (Mulimbi et al., 2012; Casinga et al., 2021), there is a possibility of the disease spreading towards West Africa. The potential reach of CBSD into West Africa, a region with no previous reports of the disease, is considered disturbing.
Unlike the East African cassava breeding programs (Munga, 2008; Abaca et al., 2012; Kawuki et al., 2016; Ferguson et al., 2017), West African cassava germplasm has not been undergoing selection for CBSD resistance because of the absence of the disease in the region and is, thus, likely to exhibit susceptibility. A westward drift of CBSD (Bigirimana et al., 2011; Mulimbi et al., 2012; Casinga et al., 2021), as well as projected studies indicating the presence of CBSD in West Africa by 2030 (Jarvis et al., 2012), is a cause for great concern. The recent surge in B. tabaci populations in Eastern and Central Africa, which might have played a role in CBSD outbreaks in mid-altitude regions (Legg et al., 2011, 2014), equally troubling. Preemptive breeding strategies to avert future impact of CBSD in West Africa is, therefore, critical since cassava is a major staple and industry crop in the region. Thus, the objectives of this study were to (i) determine the reaction of elite Nigerian cassava genotypes to CBSD in Uganda and (ii) determine the relationship between CBSD field response and other important agronomic attributes in the Nigerian cassava genotypes assessed in Uganda.
Materials and Methods
Study Site
This study was conducted at the National Crops Resources Research Institute (NaCRRI), Namulonge (0.5232°N, 32.6158°E) which is located at an altitude of 1,200 m above sea level and characterized by the bimodal rainfall distribution and an average annual temperature ranging between 24 and 30°C (Sebuwufu et al., 2015). The soil is characterized by acidic ferralsols, with pH of below 6.5–7.0 (Sebuwufu et al., 2015). NaCRRI is located 27 km north of Kampala, the capital city of Uganda, and has a tropical wet and mild dry climate with annual rainfall ranging between 1,000 and 1,450 mm with slight humid condition (average: 65%) (Sebuwufu et al., 2015). Namulonge, geographically located in central Uganda, is considered a hotspot for CBSD screening (Alicai et al., 2007; Sebuwufu et al., 2015). The site is also associated with high B. tabaci populations (Kawuki et al., 2016). These characteristics qualify NaCRRI as an optimal site for CBSD and CMD resistance screening (Abaca et al., 2012). In this study, we, therefore, assessed the response of introduced Nigerian germplasm to CBSD infestation at NaCRRI, a known hotspot in Uganda.
Test Materials
In response to the CBSD spread across Africa, National Root Crops Research Institute (NRCRI), Umudike, Nigeria, took the initiative to screen some crosses of elite-by-elite cassava genotypes in Uganda, a hotspot for CBSD. A total of 5,000 botanical seeds were generated from various biparental crosses involving 48 elite progenitors. The progenitors were part of Cycle 1 of the NRCRI cyclic population and were selected based on their yielding ability and resistance to CMD. Accordingly, these seeds were then shipped and planted in a seedling nursery at Namulonge. The progenies were screened in Uganda in order to guide the NRCRI breeding program, Nigeria, in developing breeding strategies for CBSD resistance. A total of 1,980 botanical seeds successfully emerged into seedlings. These 1,980 seedlings represented 106 families (Supplementary Table 1).
Seedling Evaluation Trial
In a field trial, the 1,980 seedlings were planted during the second rainfall of 2018 (September/October) for an evaluation period of one year and was harvested during the second rainfall of 2019 (September/October). The field trial was established in a completely randomized design, comprising six blocks, with each block consisting of 33 ranges. Each row comprised of 10 individuals (unique genotypes). Since replication of seedlings at a seedling trial is not possible (Barandica et al., 2016), we stratified the families across blocks such that sibs were planted in at least three separate blocks. Seedlings were planted at a spacing of 1 m × 1 m. The clone TME 204, which is highly susceptible to CBSD, was planted along the experimental borders as spreaders to augment CBSD pressure in the evaluation plots. The experiment was carried out under rainfed conditions, without the application of pesticides and fertilizers, and was kept weed-free by regular hand weeding.
Clonal Evaluation Trial
The clonal evaluation trial (CET) was established at NaCRRI, Namulonge, during the second rainfall of 2019 (September/October) for a one year evaluation period and was harvested during the second rainfall of 2020 (September/October). The clonal trial constituted 569 clones advanced from the seedling stage. Advancement from seedling evaluation trial (SET) to CET was based mainly on the ability of a clone to produce ample planting materials, i.e., at least 10 stem cuttings. This is because 10 stem cuttings were required to constitute a row, which would represent a single clone. Briefly, the trial was established in an augmented design, comprising 24 blocks; each clone was planted in a single row of 10 stem cuttings, at a spacing of 1 m × 1 m within rows. Within each block, the selected test clones were planted along with three randomized checks, namely, TME 204, NAROCASS 1, and Mkumba. TME 204 was the most susceptible check to CBSD, whereas NAROCASS 1 and Mkumba were tolerant checks, officially released in Uganda and Tanzania, respectively. TME 204 also acted as a spreader to augment CBSD pressure in the field trial. Similar to the seedling trial, the experiment was carried out under rainfed conditions, without the application of pesticides and fertilizers, and was kept weed-free by regular hand weeding.
Data Collection
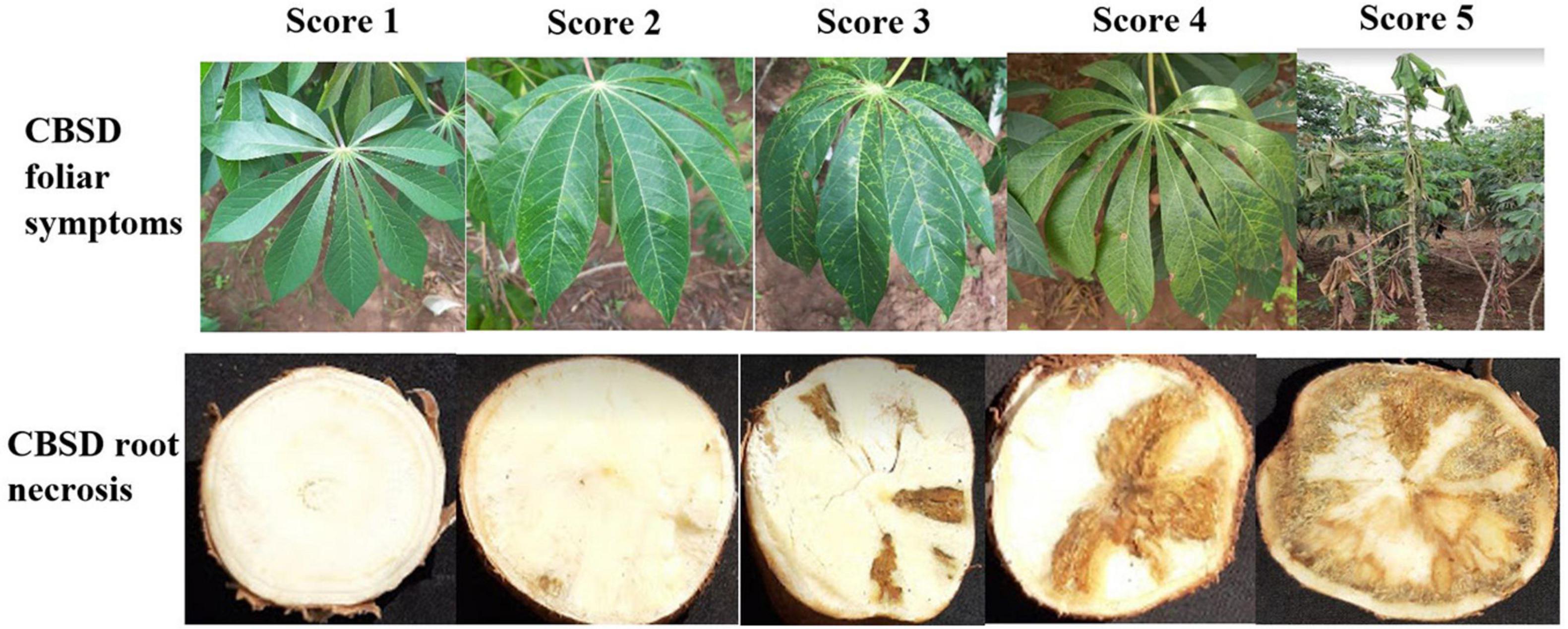
For both SET and CET trials, data were collected on CBSD and CMD. Foliar disease symptoms were assessed on each plant at three and six months after planting (MAP). For assessment of CBSD, plants were assigned disease severity scores based on the standard five-point scoring scale (Gondwe et al., 2003), where 1 = no apparent symptoms; 2 = slight foliar feathery chlorosis and no stem lesions; 3 = prominent foliar feathery chlorosis, mild-stem lesions, and no dieback; 4 = severe foliar feathery chlorosis, severe stem lesions, and no dieback; and 5 = defoliation, severe stem lesions, and dieback (Figure 1).
Similarly, CMD was assessed using the 1–5 scoring scale, where 1 = cassava plant showing no leaf symptom; 2 = mild distortion and mild chlorosis on the leaves; 3 = significant distortion and chlorosis on one-third of most leaves; 4 = extreme distortion and presence of mosaic patterns on two-third of most leaves and general reduction of leaf size; and 5 = very severe mosaic symptoms on all leaves, appearance of distortion, twisting, misshapen, and severe leaf reduction of most leaves accompanied by severe stunting of plants (Thresh and Cooter, 2005).
At 12MAP (during harvest), individual plants (in SET) and plots (in CET) were uprooted, and roots were sliced for CBSD root necrosis assessment. The CBSD root severity was assessed on all harvested roots, from a plant, using the 1–5 scoring scale (Gondwe et al., 2003), where 1 = no apparent necrosis, 2 ≤5% of root necrosis, 3 = 6–10% of root necrosis, 4 = 11–25% of root necrosis and mild root constrictions, and 5 ≥25% of root necrosis with severe root constriction (Figure 1).
Finally, for both SET and CET, data were collected on total carotenoid content and fresh root weight (FRW). The total carotenoid content was assessed visually using a qualitative 1–6 standard root color scoring scale developed by the International Center for Tropical Agriculture (CIAT), where 1 = white, 2 = light cream, 3 = cream, 4 = light yellow, 5 = yellow, and 6 = deep yellow. To estimate fresh root yield (FRYD) (tons ha–1) from FRW, we used the formula: FRYD = [(FRW/Number of plants harvested per plot) × 10,000]/1,000 (Tumuhimbise et al., 2014).
Data Analyses
Datasets generated in the seedling trial were described using summary statistics in R (R Core Team, 2020). Relationships between CBSD severity and other evaluated key agronomic traits were visualized using the ggscatter function in R (R Core Team, 2020). The Pearson’s correlation test was conducted, and the coefficient of correlation was extracted using the cor.test. All the functions mentioned above are available in ggpubr package in R.
For clonal trial datasets, linear mixed model effects using lme4 package in R was used to estimate variance components and, thus, to enable the estimation of best linear unbiased predictors (BLUPs). The BLUPs were computed to enable genotype comparison for the unbalanced dataset (Bernardo, 2010). The linear model used to generate the analysis of variance for single-site analysis was as follows:
where Yijk is the observed response of the i-th clone in the j-th block for the k-th test genotype, μ is the general mean of the genotypes, Ci is the fixed effect of the checks, Bj is the random effect of the block, Gk is the random effect of the test genotypes, and εijk is the random error associated with the ijk-th observation. The broad-sense heritability estimates for each trait were calculated using the following formula:
where H2 is the broad-sense heritability, σ2G is the genotype variance, and σ2e is the error variance. To extract the variance components for computing broad-sense heritability, the effects of the block and test genotypes were considered random, whereas the check effects were considered fixed.
A non-weighted rank summation selection index (SI) was used to compare the performance of the test genotypes. Briefly, genotype BLUP values generated from CBSD foliar and root scores were ranked and subsequently summed up to compute the SI (Mulamba and Mock, 1978).
Results
Field Reaction of Cassava Genotypes at the Seedling Stage
A total of 1,980 genotypes were planted and evaluated in the seedling trial. However, by the third month, 38% of the genotypes had succumbed to CBSD pressure (Figure 2). About 0.01% of genotypes were lost by the 6th month, and by the 12th month, an additional 15% of genotypes had succumbed to CBSD pressure. Hence, only 47% of the genotypes survived, and the root necrosis assessment were undertaken on them at 12MAP. For the genotypes that survived up to 3 months after planting (3MAP), the CBSD severity ranged from 1 to 3 at 3MAP, while at 6MAP and 12MAP, it ranged from 1 to 5 (Figure 3). Percentage CBSD root incidence ranged from 0 to 100%, with a mean score of 56.8. Mean severity scores for CBSD at 3MAP, 6MAP, and 12MAP were 1.03, 1.21, and 2.30, respectively.
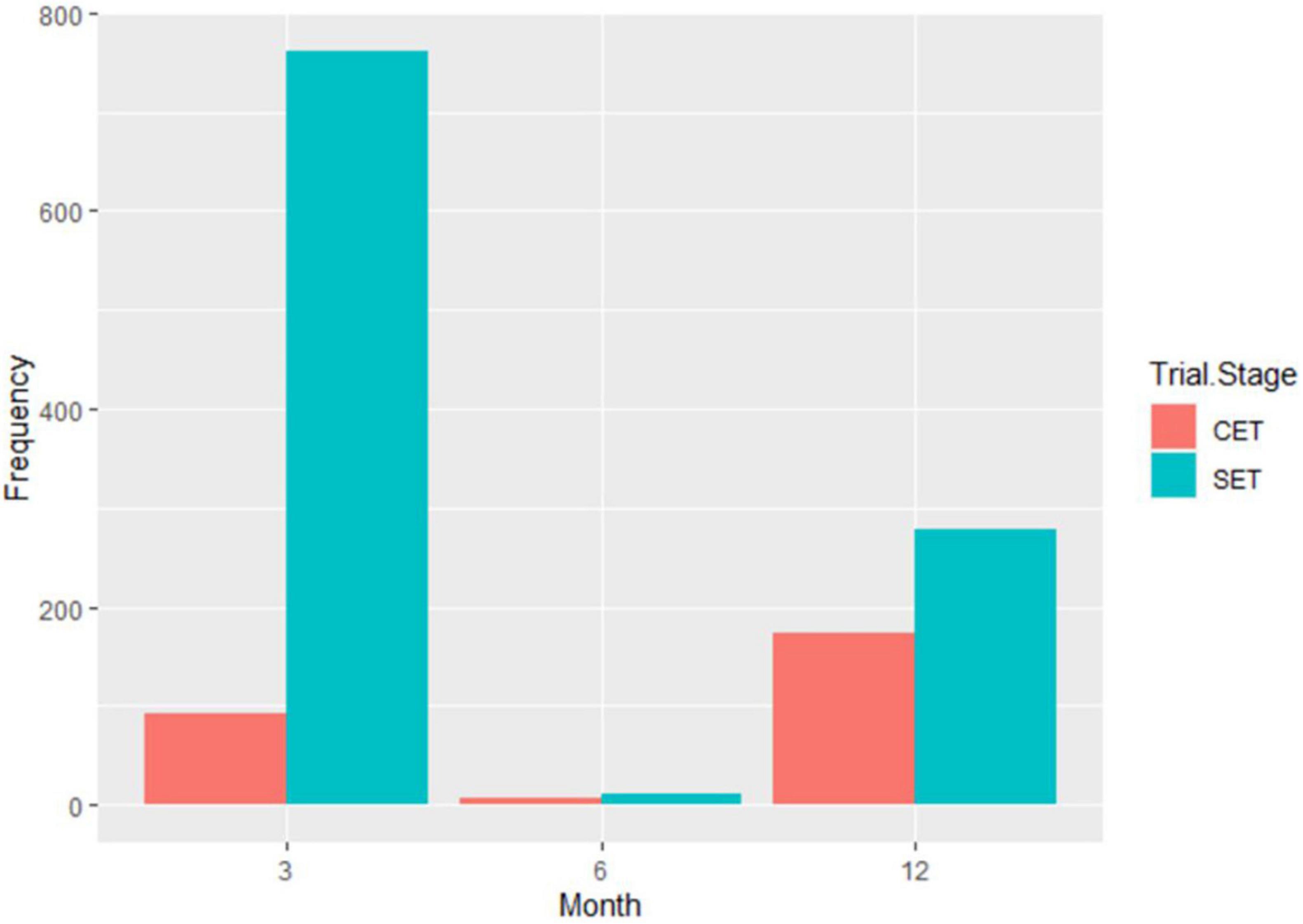
Figure 2. Dieback and death inflicted by CBSD on the Nigerian germplasm evaluated in Uganda at SET and CET.
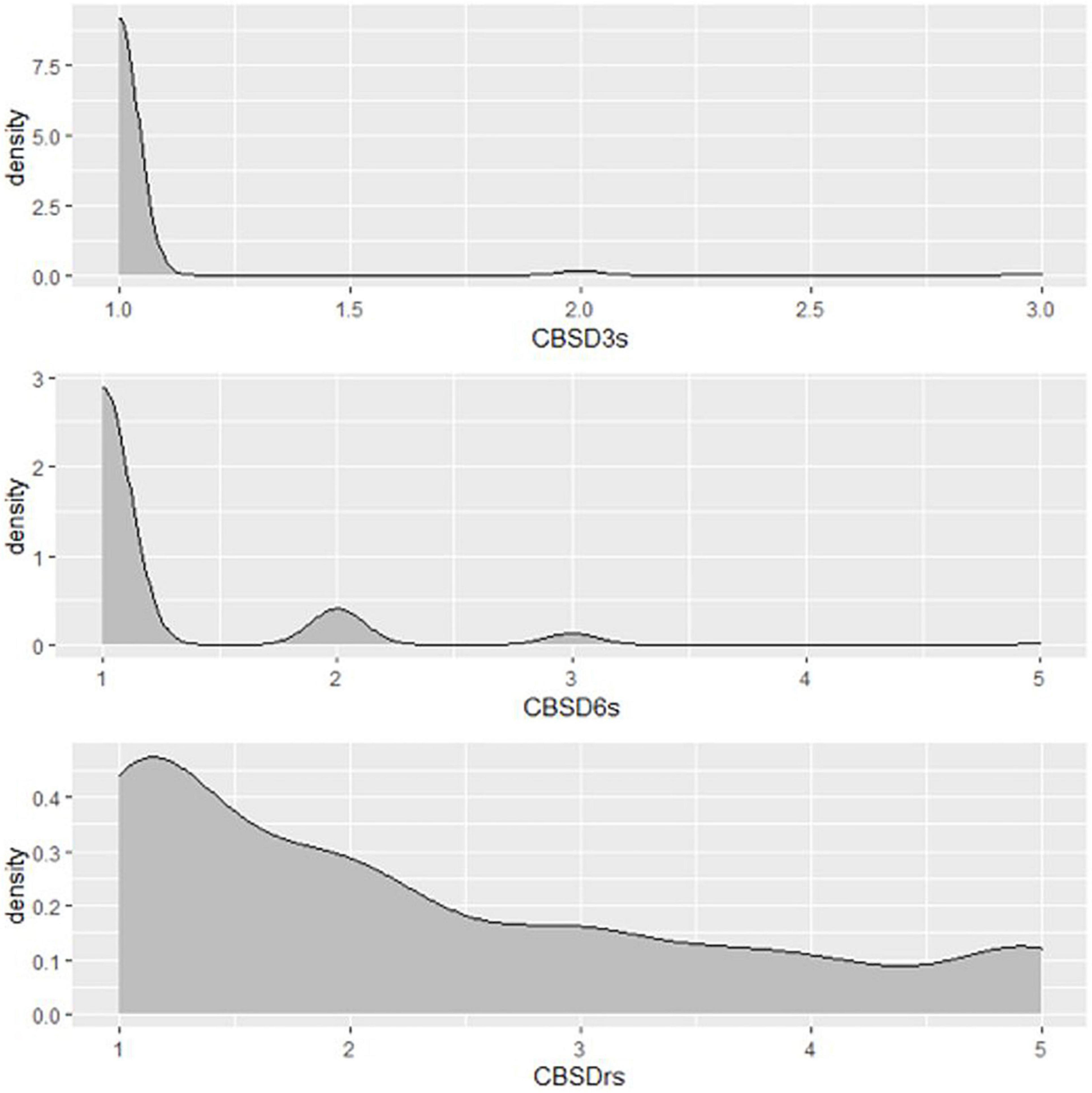
Figure 3. Distribution of CBSD foliar and root severity scores of Nigerian germplasm evaluated at SET in Namulonge, Uganda in 2018. CBSD3s, CBSD severity assessed at three months after planting, CBSD severity assessed at six months after planting, CBSDrs, CBSD root severity at 12MAP.
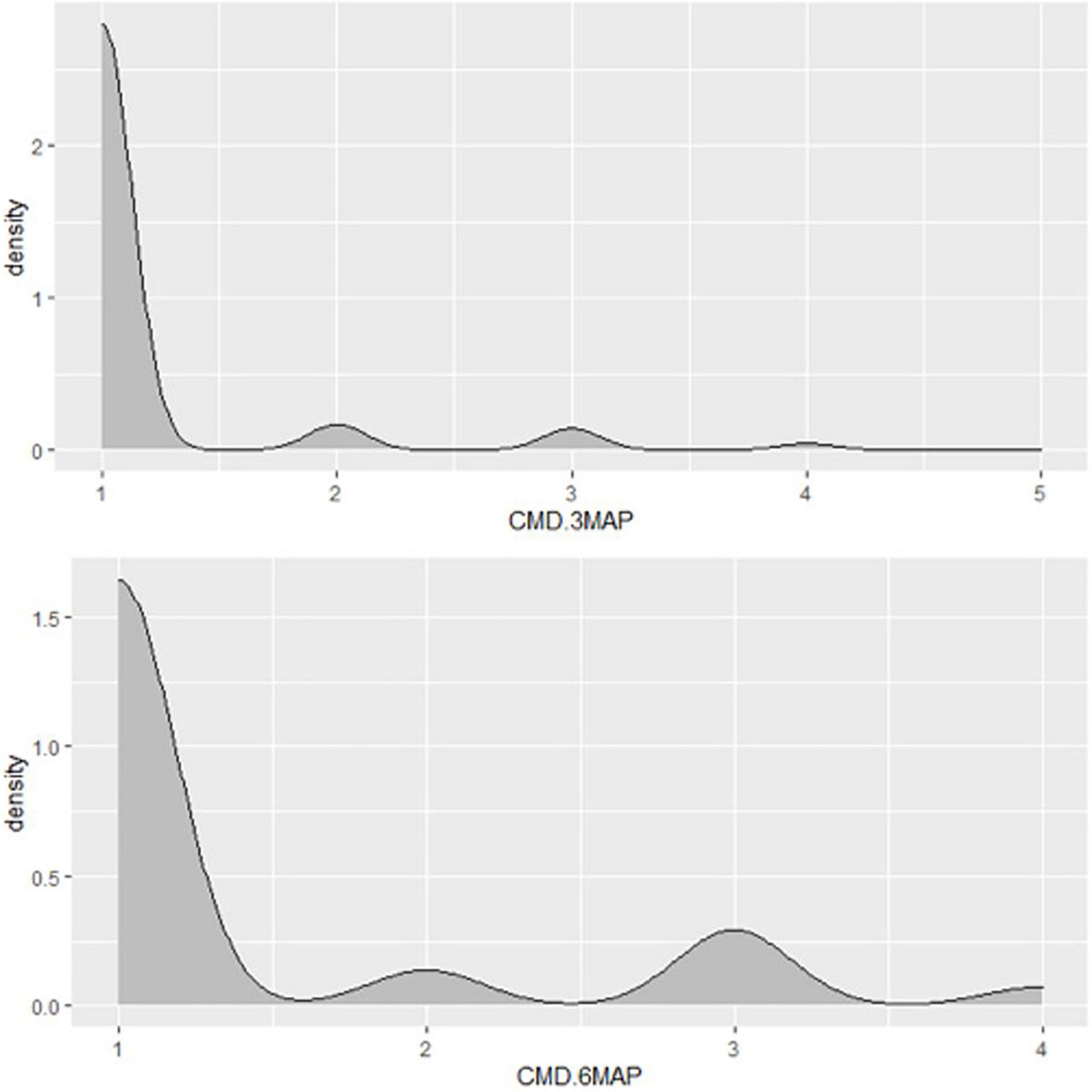
Figure 4. Distribution of CMD severity scores of Nigerian germplasm evaluated at SET in Namulonge, Uganda in 2018. CMD.3MAP, CMD severity assessed at three months after planting, CBMD severity assessed at six months after planting.
A total of 1,014 (out of 1,206) genotypes had low CBSD foliar severity score (≤1.5) at 6 months. In contrast, 330 out of 927 genotypes assessed during harvest exhibited low root necrosis severity score (≤1.5). Overall, 51% of the 927 accessions evaluated had yellow roots. Notably, there was a gradual increase in severity from 3MAP to 6MAP, with the highest average score at 12MAP (Supplementary Table 2).
Response of Cassava Genotypes to Cassava Brown Streak Disease at the Clonal Stage
A total of 569 clones were selected from the seedling trial for further evaluation at the clonal stage during the 2019–2020 season. Overall, 272 accessions succumbed to disease pressure during the evaluation period: 92 accessions at 3 months, 6 accessions at 6 months, and 174 accessions at 12 months (Figure 2). Hence, 297 accessions survived until root necrosis assessment at 12MAP. CBSD severity scores ranged from 1 to 4 at 3MAP and 6MAP and 1 to 5 at 12MAP (Figure 5). The percentage CBSD incidence ranged from 0 to 100 at 3MAP, 6MAP, and 12MAP. Mean scores for CBSD severity at 3MAP, 6MAP, and 12MAP were 2.3, 2.5, and 2.5, respectively. Mean scores of 79.7 and 84.4 were also recorded for CBSD foliar incidence and 59.3 for root incidence (Table 1). Approximately 80% of the genotypes had CBSD scores between 2 and 3 (Figure 5). In contrast, approximately 30% of the accessions had CBSD root necrosis scores ranging from 1 to 2. For CBSD incidence, more than 90% of the genotypes exhibited incidence scores between 75 and 100% at 3 and 6 months, while 25% of the genotypes registered root incidences between 75 and 100% for CBSDri. For CMD, more than 90% of the genotypes exhibited severity scores between 1 and 2, and incidences between 0 and 25% at 6MAP (Figure 6).
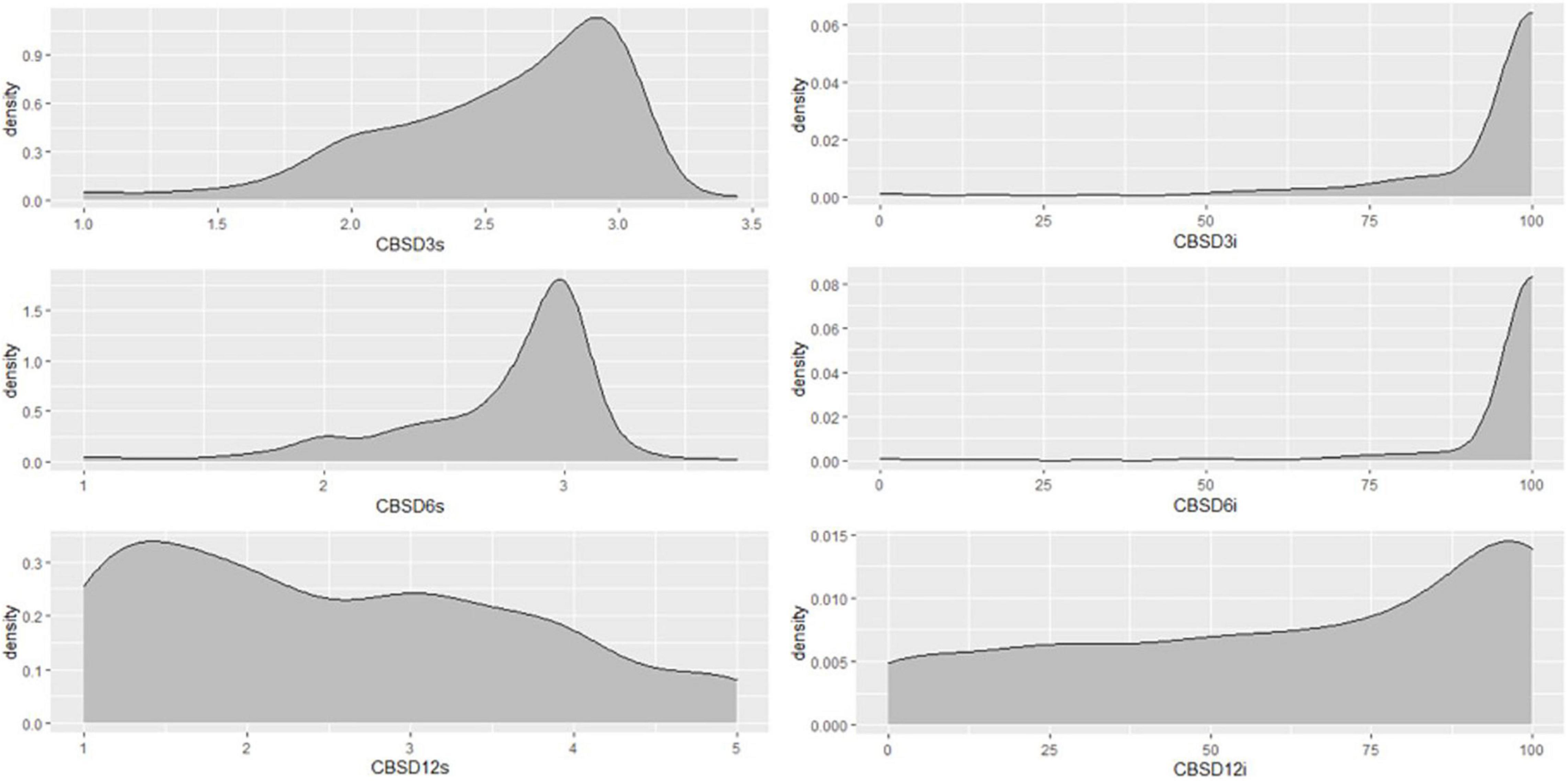
Figure 5. Distribution of CBSD foliar and root severities and incidences in Nigerian germplasm evaluated in clonal trial Uganda in 2019. CBSD3s, CBSD severity 3MAP; CBSD6s, CBSD severity 6MAP; CBSD12s, CBSD root severity at 12MAP; CBSD3i, CBSD incidence 3MAP; CBSD6i, CBSD incidence 6MAP; CBSD12i, CBSD root incidence at 12MAP. Analysis based on 297 genotypes.
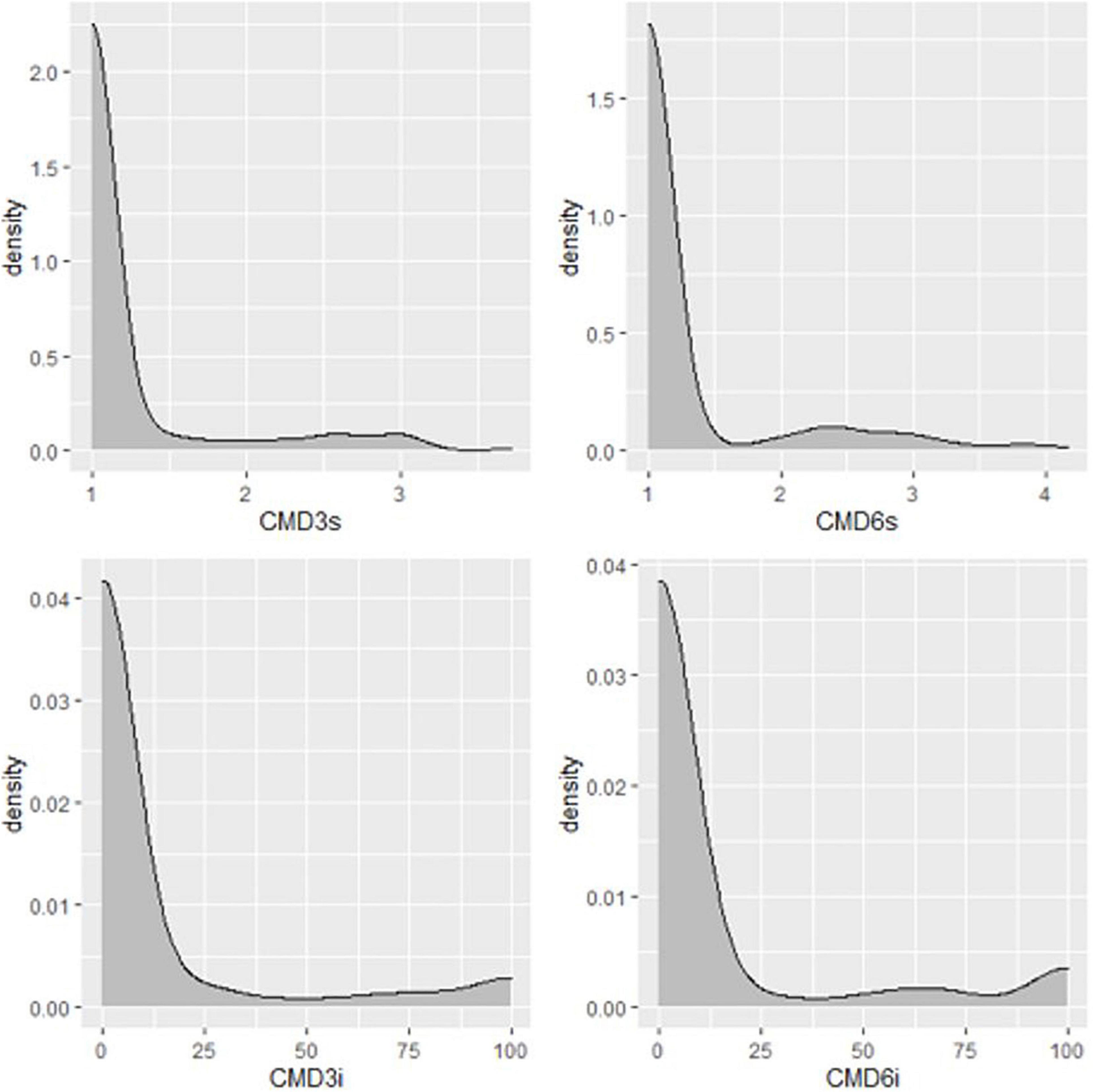
Figure 6. Distribution of CMD severities in Nigerian germplasm evaluated in clonal trial in Uganda in 2019. CMD3s, CMD severity assessed at three months after planting, CBMD severity assessed at six months after planting, CMD incidence assessed at three months after planting, CMD incidence assessed at six months after planting.
Correlations Between Cassava Brown Streak Disease and Other Related Traits in the Seedling Trial
Foliar severity at six months had a significant but low positive correlation with CBSD root severity (r = 0.1376**) and CBSD root incidence (0.1223**) (Table 2). However, CBSD root severity positively correlated with CBSD root incidence (r = 0.8296***). There were no correlations between CBSD and CMD. However, CMD severity at six months had a low negative correlation with root weight (r = −0.1306**). Furthermore, root weight was negatively and marginally correlated with CBSD root necrosis incidence and root necrosis severity (−0.0946* and −0.1782***, respectively). There were marginal correlations between total carotenoid content and root weight (0.1894***). There were also marginal negative correlations between total carotenoids and CBSD root severity (−0.0612).

Table 2. Relationship between CBSD and other related traits in Nigerian genotypes evaluated in seedling trial at Namulonge in 2018.
Correlations Between Cassava Brown Streak Disease and Other Related Traits at the Clonal Evaluation
Correlations between CBSD and other agronomic traits evaluated at the clonal stage are presented in Table 3. Analyses between traits showed a significant positive relationship between CBSD root incidence and CBSD root severity (r = 0.90***). CBSD foliar severity at 3 months and CBSD foliar incidence at three months (CBSD3i) also had highly significant positive correlations of r = 0.89***. The same pattern was noted for CBSD foliar incidence at six months and CBSD foliar severity at six months (r = 0.89***). CBSD foliar incidence at three months also correlated positively and significantly with CBSD foliar incidence at six months (r = 0.86***) and CBSD foliar severity at six months (r = 0.78***). However, correlations were lower between CBSD root incidence (r = 0.40***) and CBSD root severity (r = 0.34***). Similarly, CBSD foliar severity at 3 months correlated positively with CBSD foliar incidence at six months (r = 0.77***) and CBSD foliar severity at six months (r = 0.78***), but lower with CBSD root incidence (r = 0.40***) and CBSD root severity (r = 0.35***). In contrast, CBSD foliar severity at three months correlated negatively with CMD incidence at three months (CMD3i) (r = −0.25***), CMD severity at 3 months (r = −0.25***), CMD incidence at six months (CMD6i) (r = −0.20***), and CMD severity at six months (r = −0.21***).
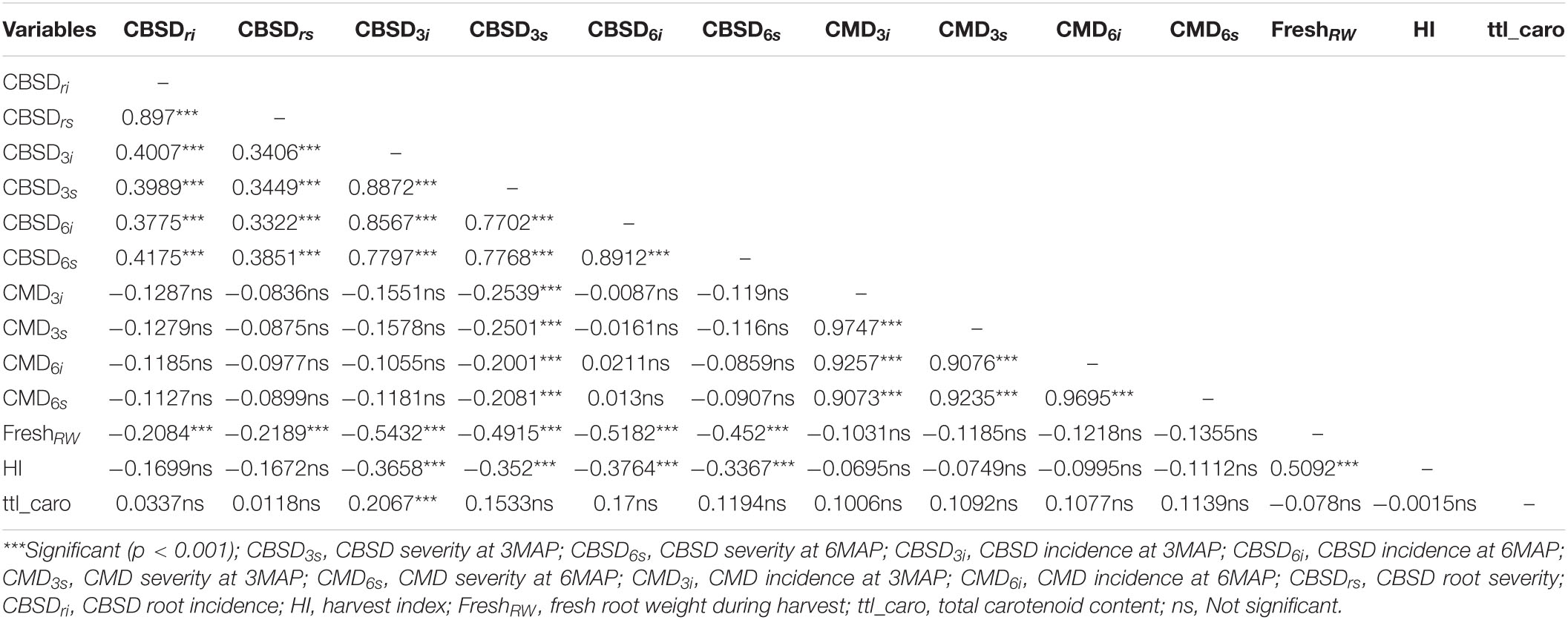
Table 3. Relationship between CBSD and other related traits in Nigerian genotypes evaluated in the clonal trial at Namulonge in 2019.
Fresh root weight during harvest had a significant negative correlation with CBSD foliar incidence at 3 months (r = −0.54***), CBSD foliar severity at 3 months (r = −0.49***), CBSD foliar incidence at 6 months (r = −0.52***), CBSD foliar severity at 6 months (r = −0.45***), CBSD root incidence (r = −0.21***), and CBSD root severity (r = −0.22***). However, there was a significant positive correlation between FRW and harvest index (r = 0.51***). There was a slight non-significant negative correlation between harvest index and CBSD root incidence (r = −0.17). CBSD root severity also had a slight negative non-significant correlation with harvest index (r = −0.17). Negative correlations were consistently observed between harvest index and CBSD foliar incidence at 3 months (r = −0.37***), CBSD foliar severity at 3 months (r = −0.36), CBSD foliar incidence at six months (r = −0.38), and CBSD foliar severity at six months (r = −0.34).
Correlations Between Cassava Brown Streak Disease and Cassava Mosaic Disease at Both Seedling Evaluation Trial and Clonal Evaluation Trial
Correlations between seedling and clonal evaluations are presented in Table 4. There were significant correlations for CBSDri at both SET and CET (r = 0.48***) (Table 4). Correlations between CBSDrs at SET and CET were also significant (r = 0.53***). CMD6s score at the seedling stage was positively correlated with CMD3s at the clonal stage (r = 0.68***) and CMD6s at the clonal stage (r = 0.67***). Negative correlations were consistently observed between CMD6s at the seedling stage and CBSD3s at the clonal stage (r = −0.36***), and also with CMD6s at the seedling stage and CBSD6s at the clonal stage (r = 0.28***). There were slightly non-significant negative correlations between S_CMD3s and C_CBSD3s (r = −0.07), C_CBSD6s (r = −0.05), and C_CBSDrs (r = −0.03) (Table 4). There were also slightly positive non-significant correlations between S_CBSD3s and C_CBSD3s (r = 0.05), S_CBSD3s and C_CBSDrs (r = 0.01), and S_CBSD3s and C_CBSDri (r = 0.03).
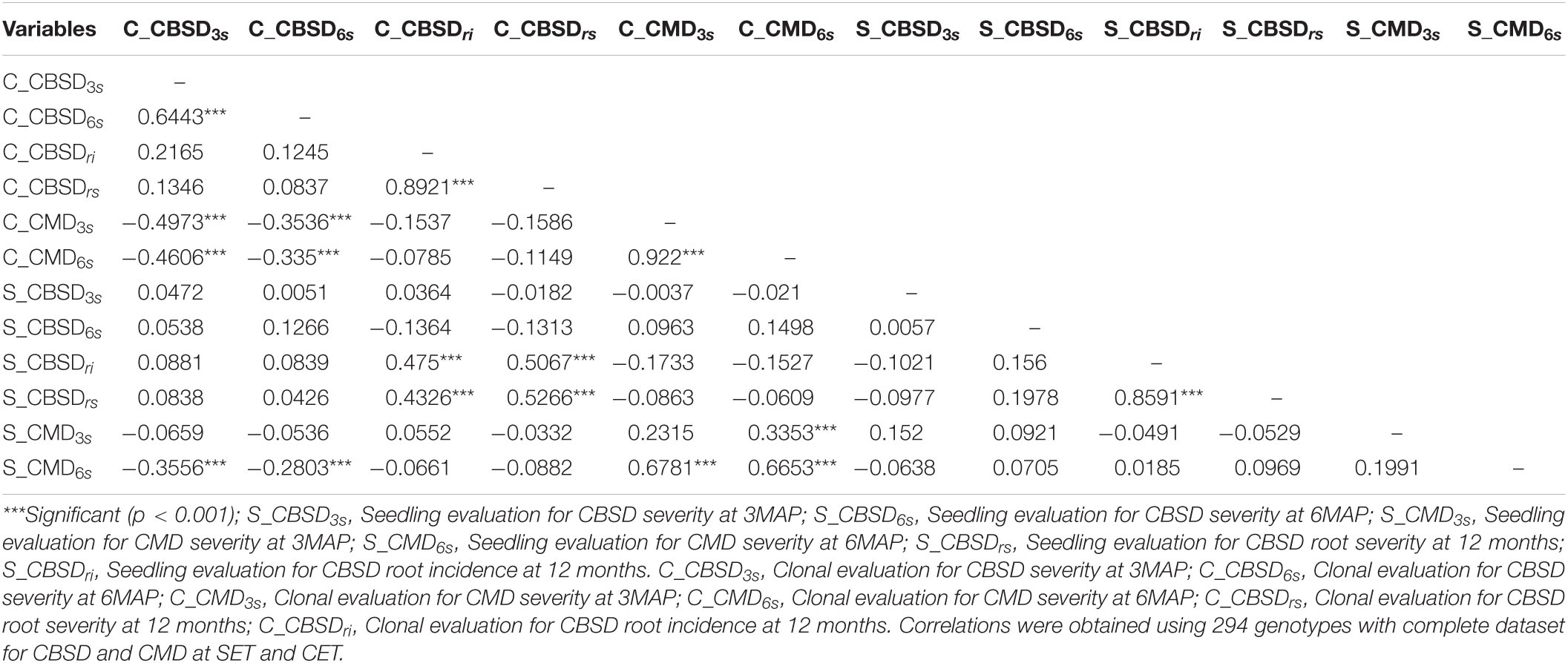
Table 4. Relationships between CBSD and CMD for the genotypes between the seedling and clonal trial.
Variation, Heritabilities for Cassava Brown Streak Disease and Other Important Traits at the Clonal Evaluation
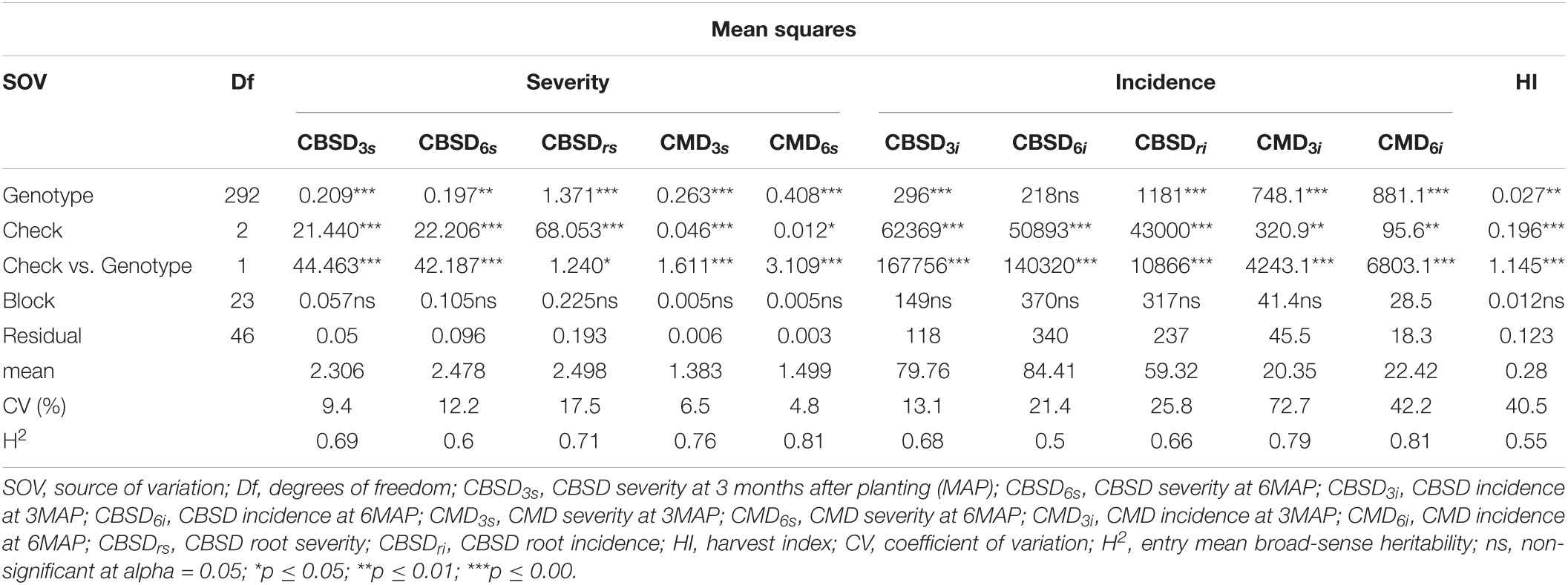
Analysis of variance (ANOVA) results showed significant differences among test genotypes for CBSD severity at 3 (p ≤ 0.001), 6 (p ≤ 0.01), and 12 months (p ≤ 0.001) (Table 1). There were also significant differences among test genotypes for CBSD incidence at 3 and 12 months (p ≤ 0.001) but not at 6 months. Comparisons between test genotype means and means of the checks also showed significant differences (p ≤ 0.05 and p ≤ 0.001) for both CBSD incidence and severity, except CBSDrs (p ≤ 0.05), which exhibited significant marginal differences.
Entry mean-based broad-sense heritability estimates ranged from moderate to high (Table 1). Overall, disease traits had higher heritability estimates than agronomic traits. For disease traits, CBSD6i and CBSD6s had the lowest heritability estimates with moderate H2 of 0.50 and 0.60, respectively. CMD severity and incidence at 6 months had the highest estimates of heritability with H2 = 0.81 and 0.81, respectively. Heritability estimates for CBSD (incidence and severity) at 3, 6, and 12 months varied from moderate to high (CBSD3i H2 = 0.68, CBSD6i H2 = 0.50, CBSDri H2 = 0.66, and CBSD3s H2 = 0.79, CBSD6s H2 = 0.60, CBSDrs H2 = 0.71).
Comparisons and Ranking of Cassava Clones Based on the Selection Index
Genotype ranking based on the non-weighted rank summation SI revealed the overall best performer as UGIN181367, a yellow-fleshed clone (Table 5). Other top nine performing genotypes included UGIN180528, UGIN181535, UGIN180247, UGIN181791, UGIN181326, UGIN181154, UGIN181371, UGIN180602, and UGIN181346. Notably, 8 out of the best 15 clones were yellow clones. Families of the best and worst performers in terms of CBSD resistance are presented in Table 6. Only family F036 had 2 progenies in the best 15 categories, and other families, such as F001, F004, F014, and F018, had one progeny each in the best performing category.

Table 5. Rankings of the best-15 and worst-15 genotypes following CBSD evaluations at Namulonge in 2019.
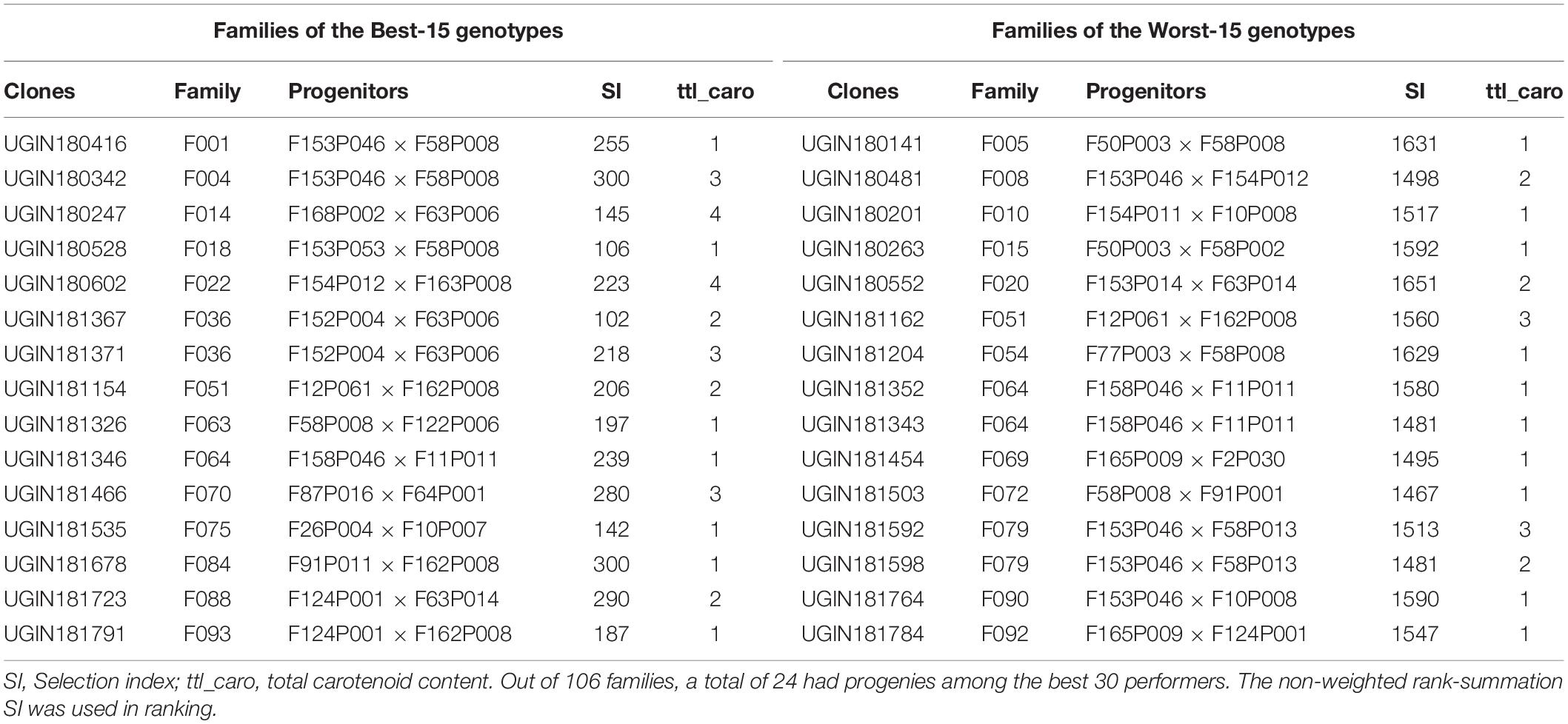
Table 6. Family based responses to CBSD of the best-15 and worst-15 Nigerian genotypes evaluated at Namulonge in 2019.
Comparison of checks and test genotypes is shown in Figure 7. CBSD-resistant checks (Mkumba and NAROCASS 1) performed markedly better than all other test genotypes for CBSD resistance. However, 65% of test genotypes performed better than the susceptible check (TMEB204) in terms of CBSD resistance (Figure 7).
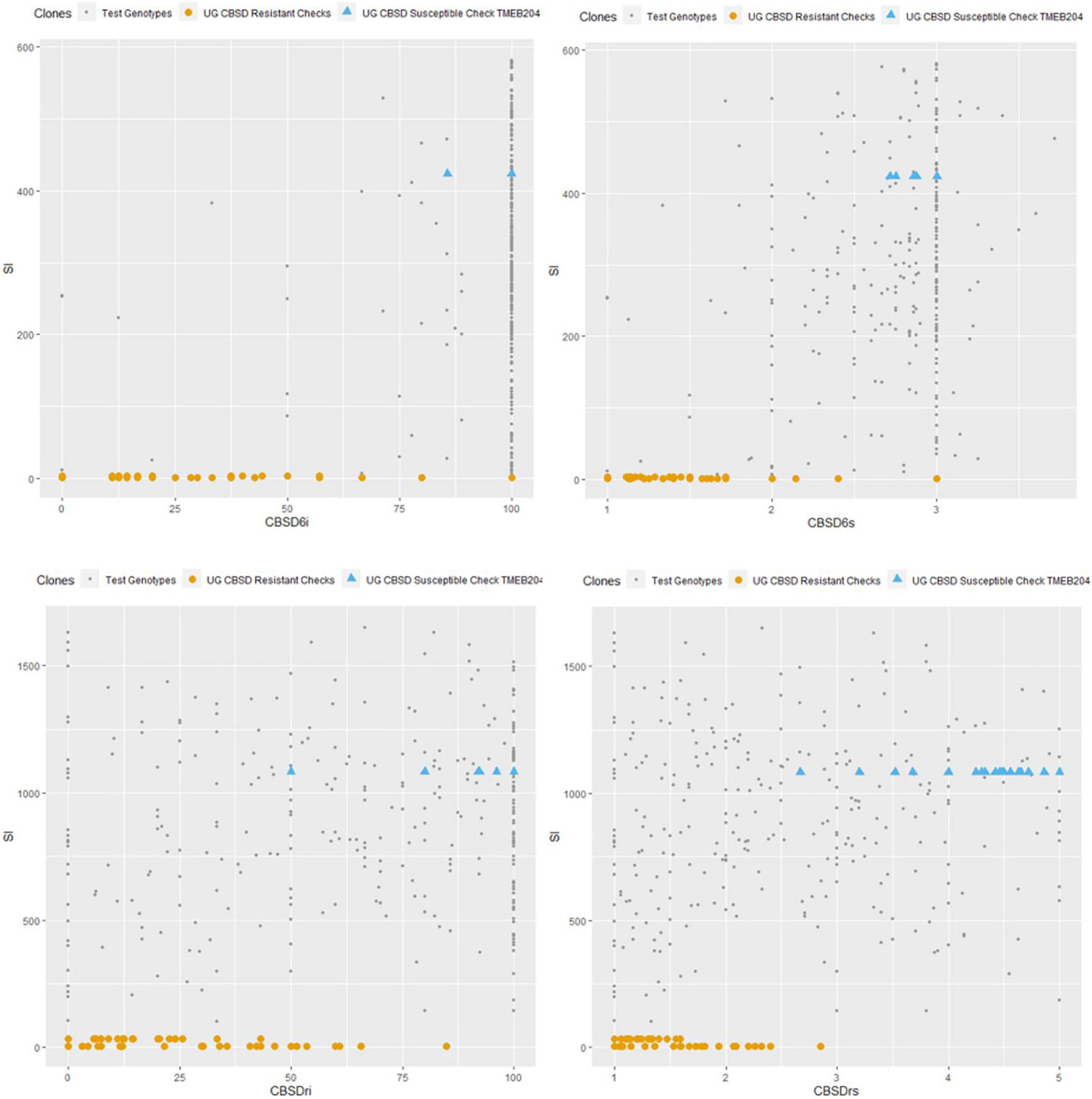
Figure 7. Comparison of CBSD susceptible and resistant checks to test genotypes at 6 and 12 months. CBSD6i, CBSD incidence at 6MAP; CBSD6s, CBSD severity at 6MAP; CBSDri, CBSD root incidence; CBSDrs, CBSD root severity; SI, Selection Index.
From the seedling to clonal evaluation stage, family progression with regard to CBSD is presented in Supplementary Figure 1. Mean CBSD severities at 3, 6, and 12 months in the seedling stage were 1.03, 1.18, and 2.28, respectively. However, at the clonal stage, mean CBSD severities at 3, 6, and 12 months were 2.50, 2.70, and 2.57, respectively. Evidently, there was a general increase in CBSD from seedling to clonal stage. The survival rate of progenies per family from seedling to clonal evaluation ranged from 5 to 80% (Supplementary Figure 2). The family F059 had the highest survival rate, with 80% of its progenies surviving until harvest at 12MAP. Only two families had higher CBSD root severity means than that of the susceptible check TME 204 (4.33) at 12 months, with F040 being the most susceptible with a mean score of 5.0. Progenies per family for the seedling trial ranged from 10 to 130. In contrast, progenies advanced to CET after selection ranged from 1 to 18, averaging four progenies per family. Overall, 30 families succumbed to CBSD pressure during the seedling evaluation.
Discussion
Cassava brown streak disease is one of the topmost threats to food security (Patil et al., 2015), undermining any investments made to maximize the benefits of cassava value chains. For this reason, concerted efforts are devoted toward finding sustainable disease management options. Based on the lessons learned in East Africa, where less attention was given to CBSD until it reached epidemic levels after several decades of existence at low altitudes of coastal areas in Tanzania (Storey, 1936; Legg et al., 2011, 2015), it has become necessary to take preemptive measures to prevent the spread of CBSD in West Africa, a major cassava-producing zone in Africa, and limit its impact. Accordingly, this study aimed at contributing toward development of improved cassava varieties with enhanced CBSD resistance in Nigeria.
Consequently, 1,980 seedlings were evaluated for their responses to CBSD. Of these, 569 clones were selected for further evaluation at the clonal level. By the first year of field evaluation, 38% of the 1,980 seedlings succumbed to CBSD pressure during the first three months after planting, and by the 12th month, only 46% of the genotypes survived for root necrosis assessment. This magnitude of CBSD severity can be attributed to a lack of CBSD resistance alleles in the Nigerian germplasm. A similar outcome was noted by Elegba et al. (2020), who reported less than 50% survival rate for Ghanaian cassava genotypes tested against CBSV and UCBSV under screen-house conditions. Likewise, Winter et al. (2019) reported moderate to severe foliar symptoms when 238 South American cassava genotypes were assessed for resistance to CBSVs. Viral inoculum buildup occurs when diseased stem cuttings are recycled (Kaweesi et al., 2014). Indeed, significant correlations between seedling and clonal CBSD root incidences and severities (S_CBSDri and C_CBSDri r = 0.48***; S_CBSDrs and C_CBSDrs r = 0.53***) suggest viral inoculum buildup in the course of the evaluations. For these reasons, conclusions cannot be deducted on resistance status at seedling trials.
Progenies per family ranged from 10 to 130 in the SET, with an average of 18 progenies per family. In CET, this varied between 1 and 18, with an average of four progenies per family (Supplementary Figure 1). This variation in the number of progenies from each progenitor is related to the highly erratic flowering behavior of different cassava genotypes (Ceballos et al., 2012). Overall, family averages at 6 and 12 months in the CET indicated that most families had considerably low resistance to CBSD owing to high susceptibility scores (Supplementary Figure 1). Some families had seedlings that exhibited marginal CBSD symptoms. A good example is family F036, which had two of its progenies (UGIN181367 and UGIN181371) among the best performers for CBSD at CET. This situation presents an opportunity to select superior genotypes for further evaluation of traits of interests. Similar reports suggested by Mrema (2016) also highlighted on cassava families with varying number of seedlings that exhibited CBSD symptoms and seedlings that were symptomless.
A similar phenomenon was reported by Shirima et al. (2019), who noted the degeneration of cassava genotypes due to increased viral inoculum resulting from stem recycling. Escape variants underscore why the selection for CBSD resistance among previously untested genotypes should not focus on seedling evaluations alone. This is because, at SET, the viral load in stem cuttings would be low compared to that in the clonal level. Also, seedling evaluations and data collection are based on a single plant, as opposed to 10 plants at the clonal stage. Thus, the evaluation at the clonal stage allows for better phenotypic data computation and exposes escape variants, such as the case of UGIN180447 and other accessions that exhibited marginal CBSD symptoms in the seedling trial.
Significant differences were recorded among test genotypes for all traits in the CET, except CBSD incidence at six months (Table 1). A mean score of 84.4 for CBSD6i indicates that most genotypes exhibited high levels of CBSD at six months, explaining why there were no significant variations among the test genotypes. This reinforces the lack of resistance alleles in the Nigerian germplasm. Significant differences in CBSD root severity at three, six, and 12 months, however, indicate differential responses of the genotypes. Similar findings were reported by Abaca et al. (2012), Kaweesi et al. (2014), and Pariyo et al. (2015), who identified varied resistance/susceptibility categorizations of foliar and root severities. This observation is in accordance with earlier studies by Jennings (1960) and Hillocks and Thresh (2000), who reported that different cultivars respond differently to CBSD.
For the clonal trial, the significant positive correlations between CBSD shoot severity and incidence, with CBSD root severity and incidence (Table 3), agree with the reports suggested by Abaca et al. (2012) and Mrema (2016). Results also indicated negative correlations between CBSD and CMD, which were largely non-significant except for CBSD3s (Table 3). The negative correlation suggests that the traits were under the control of different genetics. The negative correlations between harvest index and CBSD illustrate the damage inflicted by CBSD, as observed in previous studies by Nuwamanya et al. (2015) and Mrema (2016). The non-significant negative correlation between harvest index and CMD is likely due to high CMD resistance among the evaluated germplasm. CMD resistance breeding has been ongoing in Nigeria since 1932 (Hahn et al., 1980). Thus, most of the Nigerian germplasm was resistant as evidenced by mean severities and incidences of about 1.4 and 20% respectively (Table 1).
A non-weighted rank summation SI, which comprised the summation of genotype rank for CBSD incidence and severity data at three, six, and 12 months, was used to rank the performance of the test genotypes (Table 5). UGIN181367 (SI = 102) was the overall best genotype in its response to CBSD. This genotype exhibited marginal symptoms to CBSD foliar and root severity and incidence at both SET and CET. However, UGIN181367 was ranked third after NAROCASS 1 and Mkumba, the two resistant checks. NAROCASS 1 and Mkumba had SI values of 6 and 35, respectively. They are varieties that exhibit high tolerance to CBSD (Mukiibi et al., 2019). Interestingly, a few other genotypes, including UGIN180528, UGIN181535, UGIN180247, also had a high SI ranking and exhibited marginal symptoms for CBSD foliar and root severity and incidence (Table 5). These identified genotypes merit for further evaluations to confirm their CBSD responses.
Broad-sense heritability estimates ranged from moderate to high for all traits in the clonal trial (Table 1). Except for CBSDrs and CBSDri, heritability estimates of CBSD incidence and severity at six and 12 months were comparable to those presented by Kayondo et al. (2018) and Ozimati et al. (2018) (CBSDrs H2 = 0.25 and 0.26, respectively). The heritability estimates for CBSDrs in the clonal trial were also similar to the reports suggested by Okul Valentor et al. (2018) (H2 = 0.69). Estimates of broad-sense heritability for CBSD incidence were generally lower than those of CBSD severity in the clonal trial (Table 1). This contrasts with the findings suggested by Okul Valentor et al. (2018), who reported higher broad-sense heritability estimates for CBSD incidence. Accordingly, heritability estimates for CBSDs and CBSDi in the clonal trials suggest that effective selection can be made at the clonal stage.
Conclusion
This study was initiated on the premise of determining the field reaction of Nigerian cassava genotypes to CBSD. Therefore, this could inform the extent of CBSD resistance/susceptible allele distribution in the elite Nigerian germplasm used for cultivation and/or genetic improvement. Based on the generated datasets, it can be concluded that most of the genotypes that constituted the Nigerian cassava population exhibited significant susceptibility to CBSD within the 2-year evaluation period. Fortunately, 15 clones have been identified to have either limited or no CBSD symptoms. These genotypes can be re-evaluated at higher plot capacities and in diverse sites to confirm their resistance/tolerance to CBSD. In addition, crossing the 15 identified clones with CBSD-resistant Ugandan cassava varieties would ensure the introduction of more resistance alleles into the genome of the Nigerian population. The introgression of CBSD resistance from East African clones is, therefore, critical as a preemptive breeding strategy for CBSD resistance in Nigeria. This will help the Nigerian cassava breeding program to develop varieties with more durable resistance to the disease. Furthermore, continued screening and characterization of Nigerian cassava germplasm for responses to CBSD infection is important to identify more genotypes with CBSD tolerance or resistance traits. Deploying such tolerant or resistant cultivars can help in the effective control of the disease spread and reduction in losses associated with it.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
CA was involved in conceptualization, data collection, data analysis, and manuscript writing. MO-S was involved in conceptualization, manuscript writing, manuscript editing, and supervision. AI and DN were involved in manuscript editing and supervision. AO involved in the data collection, data analysis, and manuscript editing. PG was involved in conceptualization, supervision, and manuscript editing. RK was involved in conceptualization, data collection, data analysis, supervision, manuscript writing, and manuscript editing. JO was involved in conceptualization, manuscript editing, fund acquisition, and supervision. CE was involved in conceptualization, manuscript writing, manuscript editing, fund acquisition, and supervision. All authors contributed to the article and approved the submitted version.
Funding
The authors thank the UK’s Foreign, Commonwealth and Development Office (FCDO) and Bill and Melinda Gates Foundation (Grant INV-007637, http://www.gatesfoundation. org) for their financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank all technicians and support staff of Root Crops Programme, NaCRRI, who participated in CBSD data collection for experiments conducted during the 2018/2019 and 2019/2020 planting period. The authors appreciate the collaborative support with the National Root Crops Research Institute (NRCRI), Nigeria, where the breeding materials originated. The authors acknowledge the contribution of Makerere University Regional Center for Crop Improvement (MaRCCI). This study is a part of graduation study of CA at Makerere University, Uganda.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.720532/full#supplementary-material
References
Abaca, A., Kawuki, R., Tukamuhabwa, P., Baguma, Y., Pariyo, A., Alicai, T., et al. (2012). Evaluation of local and elite Cassava genotypes for resistance to cassava brown streak disease in Uganda. J. Agronomy 11, 65–72. doi: 10.3923/ja.2012.65.72
Alicai, T., Ndunguru, J., Sseruwagi, P., Tairo, F., Okao-Okuja, G., Nanvubya, R., et al. (2016). Cassava brown streak virus has a rapidly evolving genome: implications for virus speciation, variability, diagnosis and host resistance. Sci. Rep. 6:36164. doi: 10.1038/srep36164
Alicai, T., Omongo, C. A., Maruthi, M. N., Hillocks, R. J., Baguma, Y., Kawuki, R., et al. (2007). Re-emergence of Cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. doi: 10.1094/PD-91-0024
Bankole, E. (2019). Cassava Production in Nigeria by State. Available online at: https://www.legit.ng/1208794-cassava-production-nigeria-by-state.html (accessed February 7, 2021)
Barandica, O. J., Pérez, J. C., Lenis, J. I., Calle, F., Morante, N., Pino, L., et al. (2016). Cassava breeding II: phenotypic correlations through the different stages of selection. Front. Plant Sci. 7:1649. doi: 10.3389/fpls.2016.01649
Bernardo, R. (2010). Breeding for Quantitative Traits in Plants, 2nd Edn. Saint Paul, MN: Stemma Press.
Bigirimana, S., Barumbanze, P., Ndayihanzamaso, P., Shirima, R., and Legg, J. P. (2011). First report of cassava brown streak disease and associated Ugandan cassava brown streak virus in Burundi. New Dis. Rep. 24:5197.
Casinga, C. M., Shirima, R. R., Mahungu, N. M., Tata-Hangy, W., Bashizi, K. B., Munyerenkana, C. M., et al. (2021). Expansion of the cassava brown streak disease epidemic in eastern democratic republic of Congo. Plant Dis [Online ahead of print] DIS–05–20–1135. doi: 10.1094/PDIS-05-20-1135-RE
Ceballos, H., Hershey, C., and Becerra-López-Lavalle, L. A. (2012). New approaches to cassava breeding. Plant Breed. Rev. 36, 427–504. doi: 10.1002/9781118358566.ch6
Chipeta, M. M., Shanahan, P., Melis, R., Sibiya, J., and Benesi, I. R. M. (2016). Farmers’ knowledge of cassava brown streak disease and its management in Malawi. Int. J. Pest Manag. 62, 175–184. doi: 10.1080/09670874.2016.1167268
Eke-okoro, O. N., and Njoku, D. N. (2012). A review of cassava development in nigeria from 1940-2010. APRN J. Agric. Biol. Sci. 7, 59–65.
Elegba, W., Gruissem, W., and Vanderschuren, H. (2020). Screening for resistance in farmer-preferred cassava cultivars from Ghana to a mixed infection of CBSV and UCBSV. Plants 9, 1–16. doi: 10.3390/PLANTS9081026
Eni, A. O., Efekemo, O. P., Soluade, M. G., Popoola, S. I., and Atayero, A. A. (2018). Incidence of cassava mosaic disease and associated whitefly vectors in South West and North Central Nigeria: Data exploration. Data Brief 19, 370–392. doi: 10.1016/j.dib.2018.05.016
Ferguson, M., Masumba, E. A., Kapinga, F., Mkamilo, G., Salum, K., Kulembeka, H., et al. (2017). QTL associated with resistance to cassava brown streak and cassava mosaic diseases in a bi-parental cross of two Tanzanian farmer varieties, Namikonga and Albert. Theoretical Appl. Genet. 130, 2069–2090. doi: 10.1007/s00122-017-2943-z
Food and Agricultural Organization of the United Nations [FAOSTAT] (2020). Production Quantities of Cassava in the World. Rome: Food and Agricultural Organization of the United Nations.
Gondwe, F. M. T., Mahungu, N. M., Hillocks, R. J., Raya, M. D., Moyo, C. C., Soko, M. M., et al. (2003). “Economic losses experienced by small-scale farmers in Malawi due to cassava brown streak virus disease,” in Cassava Brown Streak Virus Disease: Past, Present and Future: Proceedings of an International Workshop, Mombasa, 27-30 October 2002, (Natural Resources International Ltd), 28–38.
Hahn, S. K., Terry, E. R., and Leuschner, K. (1980). Breeding cassava for resistance to cassava mosaic disease. Euphytica 29, 673–683. doi: 10.1007/BF00023215
Hammond, W., Lawan, J., Hoogeveen, J., Josef, K., Lava, K., Chikelu, M., et al. (2013). Save and Grow: Cassava, a Guide to Sustainable Production Intensification. Rome: FAO.
Hillocks, R. J., and Maruthi, M. N. (2015). Post-harvest impact of cassava brown streak disease in four countries in eastern Africa. Food Chain 5, 116–122. doi: 10.3362/2046-1887.2015.008
Hillocks, R. J., Raya, M. D., Mtunda, K., and Kiozia, H. (2008). Effects of brown streak virus disease on yield and quality of cassava in Tanzania. J. Phytopathol. 149, 389–394. doi: 10.1111/j.1439-0434.2001.tb03868.x
Hillocks, R. J., and Thresh, J. M. (2000). Cassava mosaic and cassava brown streak virus diseases in Africa: a comparative guide to symptoms and aetiologies. Roots 7, 1–8.
Hillocks, R. J., Thresh, J. M., Tomas, J., Botao, M., Macia, R., and Zavier, R. (2002). Cassava brown streak disease in northern Mozambique. Int. J. Pest Manag. 48, 178–181. doi: 10.1080/09670870110087376
Jarvis, A., Ramirez-Villegas, J., Campo, B. V. H., and Navarro-Racines, C. (2012). Is cassava the answer to african climate change adaptation? Tropical Plant Biol. 5, 9–29. doi: 10.1007/s12042-012-9096-7
Jennings, D. L. (1960). Observations on virus diseases of cassava in resistant and susceptible varieties. II. Brown streak disease. Empire J. Exp. Agric. 28, 261–270.
Kaweesi, T., Kawuki, R., Kyaligonza, V., Baguma, Y., Tusiime, G., and Ferguson, M. E. (2014). Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virol. J. 11:216. doi: 10.1186/s12985-014-0216-x
Kawuki, R. S., Kaweesi, T., Esuma, W., Pariyo, A., Kayondo, I. S., Ozimati, A., et al. (2016). Eleven years of breeding efforts to combat cassava brown streak disease. Breed. Sci. 66, 560–571. doi: 10.1270/jsbbs.16005
Kayondo, S. I., Del Carpio, D. P., Lozano, R., Ozimati, A., Wolfe, M., Baguma, Y., et al. (2018). Genome-wide association mapping and genomic prediction for CBSD resistance in Manihot esculenta. Sci. Rep. 8:1549. doi: 10.1038/s41598-018-19696-1
Koima, I., Orek, C., and Nguluu, S. (2018). Distribution of cassava mosaic and cassava brown streak diseases in agro-ecological zones of lower eastern Kenya. Int. J. Innovative Sci. Res. Technol. 3, 391–399.
Legg, J. P., Jeremiah, S. C., Obiero, H. M., Maruthi, M. N., Ndyetabula, I., Okao-okuja, G., et al. (2011). Comparing the regional epidemiology of the cassava mosaic and cassava brown streak virus pandemics in Africa. Virus Res. 159, 161–170. doi: 10.1016/j.virusres.2011.04.018
Legg, J. P., Lava Kumar, P., Makeshkumar, T., Tripathi, L., Ferguson, M., Kanju, E., et al. (2015). Cassava virus diseases: Biology, epidemiology, and management. Adv. Virus Res. 91, 85–142. doi: 10.1016/bs.aivir.2014.10.001
Legg, J. P., Sseruwagi, P., Boniface, S., Okao-Okuja, G., Shirima, R., Bigirimana, S., et al. (2014). Spatio-temporal patterns of genetic change amongst populations of cassava Bemisia tabaci whiteflies driving virus pandemics in East and Central Africa. Virus Res. 186, 61–75. doi: 10.1016/j.virusres.2013.11.018
Maruthi, M. N., Hillocks, R. J., Mtunda, K., Raya, M. D., Muhanna, M., Kiozia, H., et al. (2005). Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–321. doi: 10.1111/j.1439-0434.2005.00974.x
Mbanzibwa, D. R., Tian, Y. P., Tugume, A. K., Mukasa, S. B., Tairo, F., Kyamanywa, S., et al. (2009). Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154, 353–359. doi: 10.1007/s00705-008-0301-9
Mrema, E. F. (2016). Value of Within and Between Cluster Crosses for Cassava Brown Streak Disease Resistance. Kampala: Makerere University.
Mukiibi, D. R., Alicai, T., Kawuki, R., Okao-Okuja, G., Tairo, F., Sseruwagi, P., et al. (2019). Resistance of advanced cassava breeding clones to infection by major viruses in Uganda. Crop. Protect. 115, 104–112. doi: 10.1016/j.cropro.2018.09.015
Mulimbi, W., Phemba, X., Assumani, B., Kasereka, P., Muyisa, S., Ugentho, H., et al. (2012). First report of Ugandan cassava brown streak virus on cassava in Democratic Republic of Congo. New Disease Rep. 26:11. doi: 10.5197/j.2044-0588.2012.026.011
Mulamba, N. N., and Mock, J. J. (1978) Improvement of yield potential of the eto blanco maize (Zea mays L.) population by breeding for plant traits. Egypt. J. Genet. Cytol. 1, 40–51.
Munga, T. L. (2008). Breeding for Cassava Brown Streak Resistance in Coastal Kenya. Durban: Univerisity of KwaZulu-Natal.
Munganyinka, E., Ateka, E. M., Kihurani, A. W., Kanyange, M. C., Tairo, F., Sseruwagi, P., et al. (2018). Cassava brown streak disease in Rwanda, the associated viruses and disease phenotypes. Plant Pathol. 67, 377–387. doi: 10.1111/ppa.12789
Ntawuruhunga, P., Kanju, E., Ssemakula, G., Okechukwu, R., Whyte, J., and Schofield, J. (2013). Successful innovations and lessons learnt in cassava improvement and deployment by IITA in Eastern African Region. AJRTC 10, 41–51.
Nuwamanya, E., Baguma, Y., and Alicai, T. (2015). Effect of cassava brown streak disease (CBSD) on cassava (Manihot esculenta Crantz) root storage components, starch quantities and starch quality properties. Int. J. Plant Physiol. Biochem. 7, 12–22.
Okul Valentor, A., Ochwo-Ssemakula, M., Kaweesi, T., Ozimati, A., Mrema, E., Mwale, E. S., et al. (2018). Plot based heritability estimates and categorization of cassava genotype response to cassava brown streak disease. Crop Protect. 108, 39–46. doi: 10.1016/j.cropro.2018.02.008
Ozimati, A., Kawuki, R., Esuma, W., Kayondo, I. S., Wolfe, M., Lozano, R., et al. (2018). Training population optimization for prediction of cassava brown streak disease resistance in West African Clones. G3: Genes Genom. Genet. 8, 3903–3913. doi: 10.1534/g3.118.200710
Pariyo, A., Yona, B., Titus, A., Robert, K., Edward, K., Anton, B., et al. (2015). Stability of resistance to cassava brown streak disease in major agro-ecologies of Uganda. J. Plant Breed. Crop Sci. 7, 67–78. doi: 10.5897/JPBCS2013.0490
Patil, B. L., Delhi, N., Delhi, N., Patil, B. L., Patil, B. L., Legg, J. P., et al. (2014). Cassava brown streak disease : a threat to food security in Africa. J. Gen. Virol. 96, 956–968. doi: 10.1099/jgv.0.000014
Patil, B. L., Legg, J. P., Kanju, E., and Fauquet, C. M. (2015). Cassava brown streak disease: a threat to food security in Africa. J. Gen. Virol. 96(Pt 5), 956–968. doi: 10.1099/vir.0.000014
R Core Team. (2020). R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna: R Core Team.
Sebuwufu, G., Mazur, R., Ugen, M., and Westgate, M. (2015). Using improved varieties and fertility enhancements for increasing yield of common beans (Phaseolus vulgaris L.) grown by small-landholder farmers in Uganda. Afr. J. Agric. Res. 10, 4795–4805. doi: 10.5897/AJAR2015.9638
Shirima, R. R., Maeda, D. G., Kanju, E. E., Tumwegamire, S., Ceasar, G., Mushi, E., et al. (2019). Assessing the degeneration of cassava under high-virus inoculum conditions in Coastal Tanzania. Plant Dis. 103, 2652–2664. doi: 10.1094/PDIS-05-18-0750-RE
Storey, H. H. (1936). Virus diseases of east African plants. East Afr. Agric. J. 1, 333–337. doi: 10.1080/03670074.1936.11663679
Thresh, J. M., and Cooter, R. J. (2005). Strategies for controlling cassava mosaic virus disease in Africa. Plant Pathol. 54, 587–614. doi: 10.1111/j.1365-3059.2005.01282.x
Tomlinson, K. R., Bailey, A. M., Alicai, T., Seal, S., and Foster, G. D. (2018). Cassava brown streak disease: historical timeline, current knowledge and future prospects. Mol. Plant Pathol. 19, 1282–1294. doi: 10.1111/mpp.12613
Tumuhimbise, R., Melis, R., Shanahan, P., and Kawuki, R. (2014). Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava. Crop J. 2, 329–337. doi: 10.1016/j.cj.2014.04.008
Winter, S., Koerbler, M., Stein, B., Pietruszka, A., Paape, M., and Butgereitt, A. (2010). Analysis of cassava brown streak viruses reveals the presence of distinct virus species causing cassava brown streak disease in East Africa. J. Gen. Virol. 91, 1365–1372. doi: 10.1099/vir.0.014688-0
Keywords: breeding, cassava brown streak disease (CBSD), resistance screening, preemptive strategies, cassava germplasm, elite genotypes, cassava brown streak virus (CBSV), Ugandan cassava brown streak virus (UCBSV)
Citation: Ano CU, Ochwo-Ssemakula M, Ibanda A, Ozimati A, Gibson P, Onyeka J, Njoku D, Egesi C and Kawuki RS (2021) Cassava Brown Streak Disease Response and Association With Agronomic Traits in Elite Nigerian Cassava Cultivars. Front. Plant Sci. 12:720532. doi: 10.3389/fpls.2021.720532
Received: 04 June 2021; Accepted: 18 October 2021;
Published: 22 November 2021.
Edited by:
Nicolas Rispail, Institute for Sustainable Agriculture, Spanish National Research Council (CSIC), SpainReviewed by:
Robooni Tumuhimbise, National Agricultural Research Organisation, UgandaPrabhakaran Sambasivam, Griffith University, Australia
Clerisse Casinga, International Institute of Tropical Agriculture (IITA), Democratic Republic of the Congo
Andy Bailey, University of Bristol, United Kingdom
Copyright © 2021 Ano, Ochwo-Ssemakula, Ibanda, Ozimati, Gibson, Onyeka, Njoku, Egesi and Kawuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert S. Kawuki, a2F3dWtpc2V6aXJvYmVydEBnbWFpbC5jb20=
 Chukwuka Ugochukwu Ano
Chukwuka Ugochukwu Ano Mildred Ochwo-Ssemakula1
Mildred Ochwo-Ssemakula1 Angele Ibanda
Angele Ibanda Alfred Ozimati
Alfred Ozimati Paul Gibson
Paul Gibson Chiedozie Egesi
Chiedozie Egesi Robert S. Kawuki
Robert S. Kawuki