- 1Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 2School of Forestry and Resource Conservation, National Taiwan University, Taipei, Taiwan
The early-diverging eudicot family Berberidaceae is composed of a morphologically diverse assemblage of disjunctly distributed genera long praised for their great horticultural and medicinal values. However, despite century-long studies, generic delimitation of Berberidaceae remains controversial and its tribal classification has never been formally proposed under a rigorous phylogenetic context. Currently, the number of accepted genera in Berberidaceae ranges consecutively from 13 to 19, depending on whether to define Berberis, Jeffersonia, and Podophyllum broadly, or to segregate these three genera further and recognize Alloberberis, Mahonia, and Moranothamnus, Plagiorhegma, and Dysosma, Diphylleia, and Sinopodophyllum, respectively. To resolve Berberidaceae’s taxonomic disputes, we newly assembled 23 plastomes and, together with 85 plastomes from the GenBank, completed the generic sampling of the family. With 4 problematic and 14 redundant plastome sequences excluded, robust phylogenomic relationships were reconstructed based on 93 plastomes representing all 19 genera of Berberidaceae and three outgroups. Maximum likelihood phylogenomic relationships corroborated with divergence time estimation support the recognition of three subfamilies Berberidoideae, Nandinoideae, and Podophylloideae, with tribes Berberideae and Ranzanieae, Leonticeae and Nandineae, and Podophylleae, Achlydeae, Bongardieae tr. nov., Epimedieae, and Jeffersonieae tr. nov. in the former three subfamilies, respectively. By applying specifically stated criteria, our phylogenomic data also support the classification of 19 genera, recognizing Alloberberis, Mahonia, and Moranothamnus, Plagiorhegma, and Diphylleia, Dysosma, and Sinopodophyllum that are morphologically and evolutionarily distinct from Berberis, Jeffersonia, and Podophyllum, respectively. Comparison of plastome structures across Berberidaceae confirms inverted repeat expansion in the tribe Berberideae and reveals substantial length variation in accD gene caused by repeated sequences in Berberidoideae. Comparison of plastome tree with previous studies and nuclear ribosomal DNA (nrDNA) phylogeny also reveals considerable conflicts at different phylogenetic levels, suggesting that incomplete lineage sorting and/or hybridization had occurred throughout the evolutionary history of Berberidaceae and that Alloberberis and Moranothamnus could have resulted from reciprocal hybridization between Berberis and Mahonia in ancient times prior to the radiations of the latter two genera.
Introduction
The early-diverging eudicot family Berberidaceae is composed of a morphological diverse assemblage of genera (Figure 1) long praised for their great horticultural (Ahrendt, 1961; Stearn, 2002) and medicinal values (Peng et al., 2006; Hao, 2018). Although more than 85% of the ca. 700 species of Berberidaceae (Christenhusz and Byng, 2016) are woody shrubs (Yu and Chung, 2017), at the generic level, the family is predominantly represented by mono- and oligotypic temperate herbaceous genera known for several classic examples of biogeographic disjunctions (Liu et al., 2002; Wang et al., 2007; Zhang et al., 2007; Sun et al., 2018).
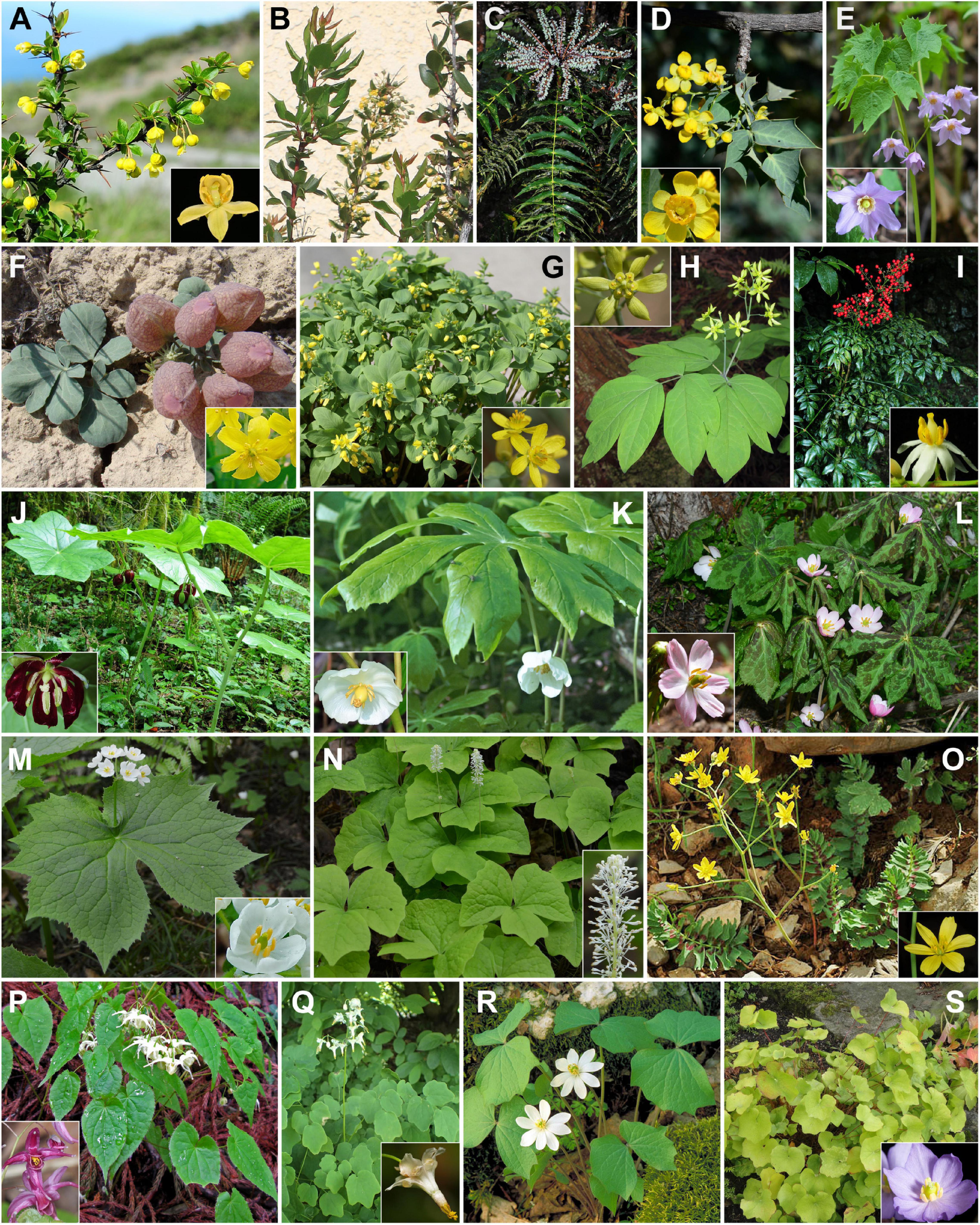
Figure 1. Morphological diversity in Berberidaceae. (A) Berberis morrisonensis and B. mingetsensis (flower). (B) Moranothamnus claireae, courtesy of Bart O’Brien. (C) Mahonia oiwakensis. (D) Alloberberis fremontii and flower photo of A. nevinii by Stan Shebs/CC BY-SA 3.0. (E) Ranzania japonica, courtesy of Takuro Ito, and flower photo by Qwert1234/CC BY-SA 3.0. (F) Leontice incerta, photo by Yuriy Danilevsky/CC BY-SA 3.0 and L. leotopetalum (flowers), photo by Averater/CC BY-SA 3.0. (G) Gymnospermium altaicum, photos by Ettrig/CC BY-SA 4.0. (H) Caulophyllum robustum, photo by Qwert1234/CC BY-SA 3.0, flower photo by Alpsdake/CC BY-SA 4.0. (I) Nandina domestica. (J) Dysosma pleiantha. (K) Podophyllum peltatum, photo by WilderAddict/CC BY-SA 4.0, flower photo by Nicholas A. Tonelli/CC BY 2.0. (L) Sinopodophyllum hexandrum, courtesy of Mu-Tan Hsieh. (M) Diphylleia grayi, courtesy of Takuro Ito, and flower photo by yamatsu/CC0 1.0. (N) Achlys triphylla, courtesy of Takuro Ito. (O) Bongardia chrysogonum, photos by Ori Fragman-Sapir/CC BY 3.0. (P) Epimedium koreanum, photo by Qwert1234/CC BY-SA 3.0 and flower photo of E. grandiflorum var. thunbergianum by Alpsdake/CC BY-SA 3.0. (Q) Vancouveria hexandra, photo by Krzysztof Ziarnek, Kenraiz/CC BY-SA 4.0, and flower photo by Walter Siegmund/CC BY-SA 3.0. (R) Jeffersonia diphylla, photo by Barnes Dr. Thomas G, U.S. Fish and Wildlife Service. (S) Plagiorhegma dubium, photo by Daderot/CC0 1.0 and flower photo by sunoochi/CC BY 2.0.
In the Northern Hemisphere, Berberidaceae exhibits seven intercontinental disjunctions: the East Asian (EA) and western North American (WNA) disjunctions in Achlys (Fukuda, 1967) and Mahonia (Yu and Chung, 2017; Chen et al., 2020), the Eurasian Epimedium and its WNA disjunct sister genus Vancouveria (Stearn, 1938; Zhang et al., 2007), the EA and eastern North American (ENA) disjunctions in Diphylleia (Ying et al., 1984) and Caulophyllum (Loconte and Blackwell, 1985), and the EA monotypic genera Sinopodophyllum (Ying, 1979) and Plagiorhegma (Hutchinson, 1920) and their respective disjunct ENA sister genera Podophyllum and Jeffersonia (Wang et al., 2007). Because of great economic, ecological, and taxonomic interests, Berberidaceae has been studied extensively in seedling morphology (Terabayashi, 1985c), floral morphology (Terabayashi, 1985a; Brückner, 2000), embryology (Sastri, 1969), serology (Jensen, 1973), palynology (Zhang et al., 2017), wood anatomy (Carlquist, 1995), and chromosome cytology (Kuroki, 1970; Adhikari et al., 2014; Huang et al., 2018; Wang et al., 2020).
Historically, however, owing to the heterogeneous composition of the family that is “held together more by a linkage of characteristics than by possession of any set of diagnostic features (Meacham, 1980),” Berberidaceae had been variously segregated into smaller families including Nandinaceae, Leonticaceae, Podophyllaceae, and Ranzaniaceae (e.g., Janchen, 1949; Airy Shaw, 1973; Hutchinson, 1973; Wu et al., 2003; Takhtajan, 2009; Lu and Tang, 2020), and/or classified into different infrafamilial taxa including subfamilies (i.e., Berberidoideae, Epimedioideae, Leonticoideae, Nandinoideae, and Podophylloideae) and tribes (i.e., Achlydeae, Berberideae, Bongardieae, Epimedieae, Leonticeae, Podophylleae, and Ranzanieae) (Table 1). Additionally, as stated in the popular encyclopedia “Flowering Plant Families of the World” that Berberidaceae contains “12 to 16” genera (Heywood et al., 2007), generic delimitation of the family has long been disputed and thus the number of its recognized genera varies greatly (Table 1 and Supplementary Table 1). Indeed, there seems no consensus regarding whether to adopt a broadly defined Berberis (e.g., Sun et al., 2018; Kreuzer et al., 2019), Jeffersonia (e.g., Sun et al., 2016), and Podophyllum (e.g., Shaw, 2002; Christenhusz et al., 2018), or to recognize Alloberberis, Mahonia, and Moranothamnus, Plagiorhegma, and Diphylleia, Dysosma, and Sinopodophyllum as distinct genera separated from the former three genera (Supplementary Table 1). In particular, whether Mahonia (i.e., the compound-leaved Berberis) should be synonymized under a broad sense Berberis (Berberis s.l.) has been debated for more than two centuries (Fedde, 1901; Moran, 1982; Kim et al., 2004b; Adhikari et al., 2015; Yu and Chung, 2017). Please refer to Ahrendt (1961) and Yu and Chung (2017) for more details about the Berberis vs. Mahonia debates.
To resolve Berberidaceae’s taxonomic controversies, early molecular studies using nuclear glyceraldehyde-3-phosphate dehydrogenase gene (Adachi et al., 1995) and chloroplast rbcL gene and restriction site (Kim and Jansen, 1995) both showed that Nandina should be included within the family. Subsequent molecular phylogenetic studies (Kim and Jansen, 1996; Kim et al., 2004a; Wang et al., 2007) revealed three clades within Berberidaceae, resulting in the circumscription of three subfamilies corresponding to three chromosome groups (Wang et al., 2009): Berberidoideae (x = 7), Podophylloideae (x = 6), and Nandinoideae (x = 8 and x = 10). Except for Lu and Tang (2020), Wang et al.’s (2009) subfamilial classification of Berberidaceae has been widely followed (Table 1). Subsequent historical biogeographic analyses based on molecular phylogenetic data also indicate that the Bering Land Bridge had functioned as a crucial pathway for the intercontinental disjunctions (Wen et al., 2010). Based on the internal transcribed spacer (ITS), Kim et al. (2004b) showed that Mahonia is paraphyletic, with Mahonia sect. Horridae sister to the simple-leaved Berberis (i.e., Berberis s.s.). More recently, based on the combined analysis of ITS and chloroplast ndhF gene sequences, Adhikari et al. (2015) further showed that Sect. Horridae is polyphyletic, together with Kim et al. (2004b) arguing for a broadly circumscribed Berberis (i.e., Berberis s.l.) that includes the compound-leaved Mahonia.
However, both Kim et al. (2004b) and Adhikari et al. (2015) suffered from issues including inadequate taxon sampling, problematic outgroup rooting, inclusion of poor-quality DNA sequences from GenBank, and taxon misidentification, undermining their taxonomic conclusion (Yu and Chung, 2017). To resolve the Mahonia vs. Berberis debate that has been lasting for more than two centuries, Yu and Chung (2017) expanded and verified taxon sampling of Mahonia and included Berberis claireae, a unique spineless Baja California endemic species with unifoliolate to 7-foliolate compound leaves (Moran, 1982) that had never been sampled previously. Based on ITS and four cpDNA markers, Yu and Chung’s (2017) phylogenetic analyses of Berberis s.l. revealed four strongly supported clades, Berberis s.s., B. claireae, core Mahonia, and Mahonia sect. Horridae. Because these four clades are ecologically and morphologically distinct and evolutionarily comparable to other genera of Berberidaceae, Yu and Chung (2017) proposed a new classification that recognizes these four clades as genera: Alloberberis (≡ Mahonia sect. Horridae), Berberis (≡ Berberis s.s.), Mahonia (≡ core Mahonia), and, Moranothamnus (≡ B. claireae), “reloading” the two-century long “Mahonia vs. Berberis” debate (see cover of the journal Taxon 66(6); doi:10.1002/tax.666001.
However, debates on generic concepts of Berberidaceae are not restricted to Mahonia vs. Berberis. Neither do controversies end with phylogenetic and phylogenomic data. In both Kim et al. (2004a) and Wang et al. (2007), Berberidaceae were regarded as having 17 genera; however, in the first molecular-based formal infrafamilial classification of Berberidaceae, Wang et al. (2009) sampled “all 16 genera of Berberidaceae,” neglecting Leontice L. that had never previously been synonymized (Table 1). In a recent phylogenomic study using plastome sequences, Sun et al. (2018) recognized 18 genera in Berberidaceae, accepting Yu and Chung’s (2017) new genera Alloberberis and Moranothamnus and yet subsuming Plagiorhegma under Jeffersonia (Table 2). However, in a subsequent study aiming to develop clade-specific DNA barcodes of Berberis using plastome sequences, sampled Alloberberis nevinii, Mahonia nervosa, and M. polyodonta were all treated as Berberis s.l. (Kreuzer et al., 2019). The flux of Berberidaceae’s generic delimitation is also manifested across major biodiversity databases and online resources (Supplementary Table 1). Nevertheless, Yu and Chung’s (2017) classification has been taken by taxonomic (Colin et al., 2021), floristic (Galasso et al., 2018), paleobotanical (Doweld, 2018), and biogeographic (Chen et al., 2020) studies.
The disparity on generic concepts across different studies and online resources (Supplementary Table 1) illustrates the lack of consensus on precise and objective criteria for generic delimitation (Humphreys and Linder, 2009) in Berberidaceae. Indeed, most of the abovementioned studies and online resources did not specify references or explicitly state reasons for their adoption of a particular generic treatment. To achieve an objective generic delimitation of Berberis s.l., Yu and Chung (2017) followed strictly five criteria advocated by Backlund and Bremer (1998), Linder et al. (2010), and Heenan and Smissen (2013) to delimit Berberis s.l.: (1) prioritizing primary (i.e., family, genus, and species) over secondary ranks (i.e., subgenus, section, etc.), (2) maximizing phylogenetic information and reducing redundancy in a classification, (3) recognizing evolutionarily equivalent (i.e., clade age, phylogenetic distance, and morphology) groups as the same rank, (4) delimiting genus that is morphologically, ecologically, and geographically homogenous, and (5) taking into account the full taxonomic history of the group and minimizing name changes to maintain nomenclatural stability. However, such objective generic delimitation has not been applied to other genera of Berberidaceae.
In recent years, rapid advances in high-throughput sequencing technology have made plastome sequences accessible for resolving recalcitrant phylogenetic relationships not attainable previously using Sanger sequences (Wicke and Schneeweiss, 2015; Tonti-Filippini et al., 2017; Gitzendanner et al., 2018). Several phylogenomic studies of Berberidaceae have been conducted using whole plastome sequences (Zhang et al., 2016; Sun et al., 2018; Ye et al., 2018; Kreuzer et al., 2019); however, no phylogenomic studies have yet sampled all 19 genera and covered adequate infrageneric diversity needed to resolve the taxonomic controversies. In this study, we report 23 newly assembled plastome sequences that complete the generic sampling of Berberidaceae. By implementing explicit criteria of generic delimitation, an infrafamilial classification representing monophyletic subdivisions of Berberidaceae is proposed, aiming to settle the taxonomic controversies and debates that have been fraught for centuries.
Materials and Methods
Classification Adopted
For clarity, the classification of 19 genera in Berberidaceae (Table 1) that recognizes Alloberberis, Berberis, Mahonia, and Moranothamnus (Yu and Chung, 2017), Jeffersonia and Plagiorhegma (Hutchinson, 1920), and Diphylleia, Dysosma, Podophyllum, and Sinopodophyllum (Wang et al., 2009) as opposed to the broadly defined Berberis s.l., Jeffersonia s.l., and Podophyllum s.l., respectively, is followed in all subsequent discussion unless otherwise stated.
Taxon Sampling
A total of 85 plastomes representing 60 species and two additional varieties in 17 genera of Berberidaceae available (accessed 25 March 2021) on GenBank were downloaded (Supplementary Table 2). To complete generic (N = 19) and infrageneric sampling of Berberidaceae, 23 species of Berberideae, including 3 species of Alloberberis, the monotypic Moranothamnus, 8 species of Berberis (7 species of Group Septentrionales and 1 species of Group Australes), and 11 species of Mahonia (5 species of Group Orientales and 6 of Group Occidentales), were sampled (Supplementary Table 2) for plastome assembly. Although we only sampled 11 species and two additional varieties of the ca. 500 species of Berberis and 11 of the ca. 100 species of Mahonia, our sampling is geographically and phylogenetically sufficient (Yu and Chung, 2017; Yu, 2018) to address issues of generic circumscription in Berberidaceae. Based on recent studies (e.g., Lane et al., 2018), plastomes of Ranunculus macrantha (Ranunculaceae), Stephania japonica (Menispermaceae), and Akebia quinata (Lardizabalaceae) were also downloaded from GenBank as outgroups (Supplementary Table 2).
DNA Extraction and Next-Generation Sequencing
CTAB method (Doyle and Doyle, 1987) was used to extract total genomic DNA from silica-dried and herbarium leaf materials. The DNA concentration was quantified by Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States). The DNAs were sent to the Genomic Core Lab of Institute of Molecular Biology, Academia Sinica for library preparation using KAPA LTP Library Preparation Kits (KAPA Biosystems, Wilmington, MA, United States), and for whole genome shotgun (WGS) sequencing using Illumina NextSeq 500 (Illumina Inc., San Diego, CA, United States) with pair-end mode, read length = 150 bp, and insert size = ca. 300 bp.
Plastome Assembly and Annotation
The quality of raw reads was assessed by FastQC v.0.11.9 (Andrews, 2010). Reads were trimmed using Trimmomatic v.0.39 (Bolger et al., 2014) with the setting “LEADING:25 TRAILING:25 SLIDINGWINDOW:4:20 CROP:149 MINLEN:100.” The de novo assembly of the plastome was performed by GetOrganelle v.1.7.5 (Jin et al., 2020) with the setting of “-R 10 -t 3 -w 0.8 -k 37,55,65,85,105,127,131 -F embplant_pt –reduce-reads-for-coverage inf,” using Berberis amurensis (GenBank accession: KM057374) as a reference for assembly. The resulting sequences generated by GetOrganelle were imported into Geneious Prime (Biomatters Ltd., Auckland, New Zealand) (Kearse et al., 2012) for validation and/or final assembly completion. For samples not assembled into a complete plastome using GetOrganelle, the “Map to Reference” function with “High Sensitivity” and default setting of Geneious was implemented to generate the draft genome, using the consensus of the mapping file to temporarily fill the “unassembled regions.” The “unassembled regions” were corrected by mapping the trimmed reads to the draft genome using the “Map to Reference” function with “Medium-Low Sensitivity” and default setting in order to complete the assembly. All complete plastome sequences were further verified by read mapping.
Newly assembled plastomes were annotated by transferring the annotations of published Berberidaceae plastomes to the newly sequenced ones under the alignment generated by MAFFT v.7.388 (Katoh and Standley, 2013) launched in Geneious. The presence of start and stop codons of each protein-coding gene was checked and adjusted manually. Genes with any premature stop codon that might interrupt translations from half of the original reading frame were annotated as pseudogenes. The correct length and identity of tRNA genes were further confirmed using the web server tRNAscan-SE 2.0 (Lowe and Chan, 2016). The boundaries of IRs were annotated by GeSeq (Tillich et al., 2017) and manually checked with self-dot plots under Geneious. Plastome maps were drawn using OGDRAW (Greiner et al., 2019).
Plastome Phylogenetic Analyses
Our initial matrix comprised 111 plastomes, including 108 of Berberidaceae (81 species and 2 additional varieties in 19 genera) and 3 outgroups (Supplementary Table 2). The sequence MG593045 (Dysosma delavayi) was excluded because high sequence variation was detected between its two inverted repeats (IRs). Of the remaining 80 species of Berberidaceae, 18 species were represented by multiple sequences. To lessen computational loading, we conducted a preliminary maximum likelihood (ML) analysis of the 110 sequences using IQ-TREE v.1.6.12 (Nguyen et al., 2015). Based on the preliminary ML tree (Supplementary Figure 1), 14 redundant and 3 problematic sequences were further excluded (see section “Results”), leaving a total of 93 plastomes representing 80 species and 2 additional varieties in all 19 genera of Berberidaceae and 3 outgroups for subsequent analyses.
Prior to phylogenetic analyses, IRB was removed. To accommodate substitution rate heterogeneity across plastomes, sequences were partitioned by the four gene categories [i.e., coding sequences (CDSs) of protein-coding genes, introns, RNA (tRNA and rRNA) genes, and intergenic spacers (IGSs)] as well as codon position of CDS. Each category was extracted, concatenated, and aligned individually by MAFFT using Geneious. For CDS, after excluding pseudogenes and partially duplicated genes, the remaining 76 genes (Supplementary Table 3) were concatenated and aligned using the “Translation Align” function based on bacterial genetic codes implemented by MAFFT under Geneious, with manual adjustments. The final concatenated alignment contains six partitions (i.e., plastid partition scheme): CDS1, CDS2, CDS3, introns, RNA genes, and IGS. Sites with more than 97% gaps were excluded using “Mask Alignment” function in Geneious. The number and proportion of parsimony informative sites of the concatenated plastome alignment were calculated by AMAS (Borowiec, 2016).
IQ-TREE was used with the “-m MFP+MERGE -bb 5000” option to conduct the following analyses: (1) searching for the best-fit partition scheme, (2) determining the best-fit nucleotide model for each partition by ModelFinder (Kalyaanamoorthy et al., 2017), and (3) reconstructing phylogenies based on ML method with 5000 replicates using ultrafast bootstrap approximation approach (Minh et al., 2013). The final tree with ultrafast bootstrap support (UFBS) values was visualized using FigTree v.1.4.2.1
Nuclear Ribosomal DNA Assembly and Analysis
Nuclear ribosomal DNA (nrDNA) sequences, spanning across partial external transcribed spacer (ETS), 18S rRNA gene, ITS 1, 5.8S rRNA gene, ITS2, 26S rRNA gene, and partial non-transcribed spacer (NTS) were assembled from raw reads of the 23 newly generated WGS sequencing using GetOrganelle with the setting of “-R 15 -t 10 -w 0.7 -k 37,69,85,115,127,131,135,139 -F embplant_nr –reduce-reads-for-coverage inf.” Additionally, nrDNA were also assembled for B. amurensis, B. koreana, B. weiningensis, Bongardia chrysogonum, and Podophyllum peltatum from WGS sequencing reads downloaded from NCBI Sequence Read Archive (SRA) using NCBI SRA Toolkit v.2.1.11. The nrDNA of these samples were assembled by executing GetOrganelle with customized settings (Supplementary Table 7). All nrDNA were verified by read mapping with the same procedure as verifying plastome sequences.
The 28 nrDNA sequences were aligned and partitioned (i.e., partial ETS, 18S, ITS1, 5.8S, ITS2, 26S, and partial NTS) by MAFFT implemented in Geneious. We employed IQ-TREE with “-m MFP+MERGE -bb 5000” options to conduct the same analyses as the plastome dataset. Concurrently, a plastome tree including 28 species sampled for the nrDNA was generated by IQ-TREE using the same partition scheme and analytical settings of the 93-plastome dataset.
Divergence Times Estimation
For divergence times estimation, we kept only one sequence for each species to further reduce the computational time. As a result, the matrix including 83 plastome sequences of 80 species in 19 genera of Berberidaceae and 3 outgroups (Supplementary Table 2) was analyzed using BEAST v.2.6.0 (Bouckaert et al., 2019) on CIPRES Science Gateway v.3 (Miller et al., 2011). Parameters and priors of the input xml file were set via BEAUti launched in the software package of BEAST v.2.6.0. With IRB excluded, the analysis was performed with the plastid partition scheme, and the prior of site models were set according to the best-fit nucleotide models and partition scheme determined by ModelFinder in IQ-TREE with the options “-m TESTMERGEONLY -mset mrbayes.” To accommodate rate heterogeneity across different Berberidaceae lineages (Yu and Chung, 2017; Sun et al., 2018), we used relaxed clock log normal as the prior of the clock model. The tree prior was set as a Yule model, and the remaining parameters followed default settings except for specifying three fossil calibration points to constrain the ages of three nodes. In Yu and Chung (2017), the age of the fossil Leefructus mirus (Sun et al., 2011; Wang et al., 2016) at 124.4 million years ago (Ma) was taken as the crown age of the Berberidaceae + Ranunculaceae clade. However, because of concern over the authenticity of the fossil of L. mirus (Zhou, 2014), three alternative fossils were used instead. First, the fossil of Prototinomiscium vangerowii dated back to the Turonian at ca. 91 Ma was assigned as the stem age of Menispermaceae (Anderson et al., 2005; Wang et al., 2012) with a lognormal distribution (mean = 92 in real space, SD = 0.06). Second, the fossil of Mahonia simplex from the Oligocene dated back to ca. 28.45 Ma (Huang et al., 2016) was designated as the crown age of Mahonia with the lognormal distribution (mean = 28.45 in real space, sigma = 0.1). Third, the fossil of Alloberberis obliqua from the Oligocene at ca. 35.55 Ma (MacGinitie, 1953; Doweld, 2018) was chosen as the crown age of Alloberberis with a lognormal distribution (mean = 35.55 in real space, SD = 0.05). We conducted two independent runs of Markov Chain Monte Carlo (MCMC), one with 400 million generations of MCMC and the other with 200 million generations. Both runs were sampled every 1000 steps for log files and every 50,000 steps for tree files. To evaluate the convergence of each parameter, the log file of each run was summarized and visualized by Tracer v.1.7.1 (Rambaut et al., 2018). The tree files were then combined by LogCombiner v.2.6.2 (launched in BEAST v.2.6.0) with the first 100 million trees discarded as burn-in for each run. Finally, we used TreeAnnotator v.2.6.0 (launched in the software package of BEAST v.2.6.0) to summarize the combined tree file into a maximum clade credibility tree with 95% highest posterior density (HPD) interval of age of each node calculated by mean heights, and visualized the tree using FigTree.
Results
Plastome Features of Berberidoideae
All newly generated plastomes of Berberideae were assembled into circular molecules with sizes ranging from 164,553 (Alloberberis trifoliolata) to 168,208 bp (Berberis hayatana). The average coverages of the newly assembled plastomes ranged from 64× (B. dictyophylla) to 1127.8× (Mahonia harrisoniana) (Table 2). The GC contents vary only slightly (38.0–38.2%), and the genome structures are found to represent the typical quadripartite configuration (Figure 2 and Supplementary Figures 2–5), consisting of a large single copy (LSC) ranging from 72,349 (A. trifoliolata) to 73,669 bp (Mahonia fortunei), a small single copy (SSC) ranging from 16,194 (B. kawakamii) to 18,932 bp (Moranothamnus claireae), and two IRs ranging from 36,445 (M. tikushiensis) to 39,343 bp (B. hayatana) (Table 2). Referring to early-diverging eudicots (Sun et al., 2016), both gene orders (Figure 2 and Supplementary Figures 2–5) and gene contents (Supplementary Table 3) of the 23 newly assembled plastomes are consistent with the published plastome of Mahonia bealei (Ma et al., 2013), which has experienced significant IR expansions at IRB/LSC boundary from rps19 into the spacer between clpP and psbB (Supplementary Table 2 and Supplementary Figure 6). In addition to Berberideae, IR expansion was also detected in MG234280 of Ranzania (Wang et al., 2018) and Epimedium ecalcaratum (MN939634). On the other hand, IR contraction was found in MN371716 of Epimedium brevicornu (Zheng et al., 2019). However, both IR expansion and contraction are not previously known in Epimedium. Comparison of IR/SC boundaries across Berberidaceae is shown in Supplementary Figure 6. Additionally, as noted in Ma et al. (2013), rpoA gene is lost in all our newly sequenced plastomes of Berberideae. However, while Ma et al. (2013) reported that ndhK had degenerated into a pseudogene in M. bealei, ndhK gene does not contain any internal stop codon in all our newly assembled plastomes.
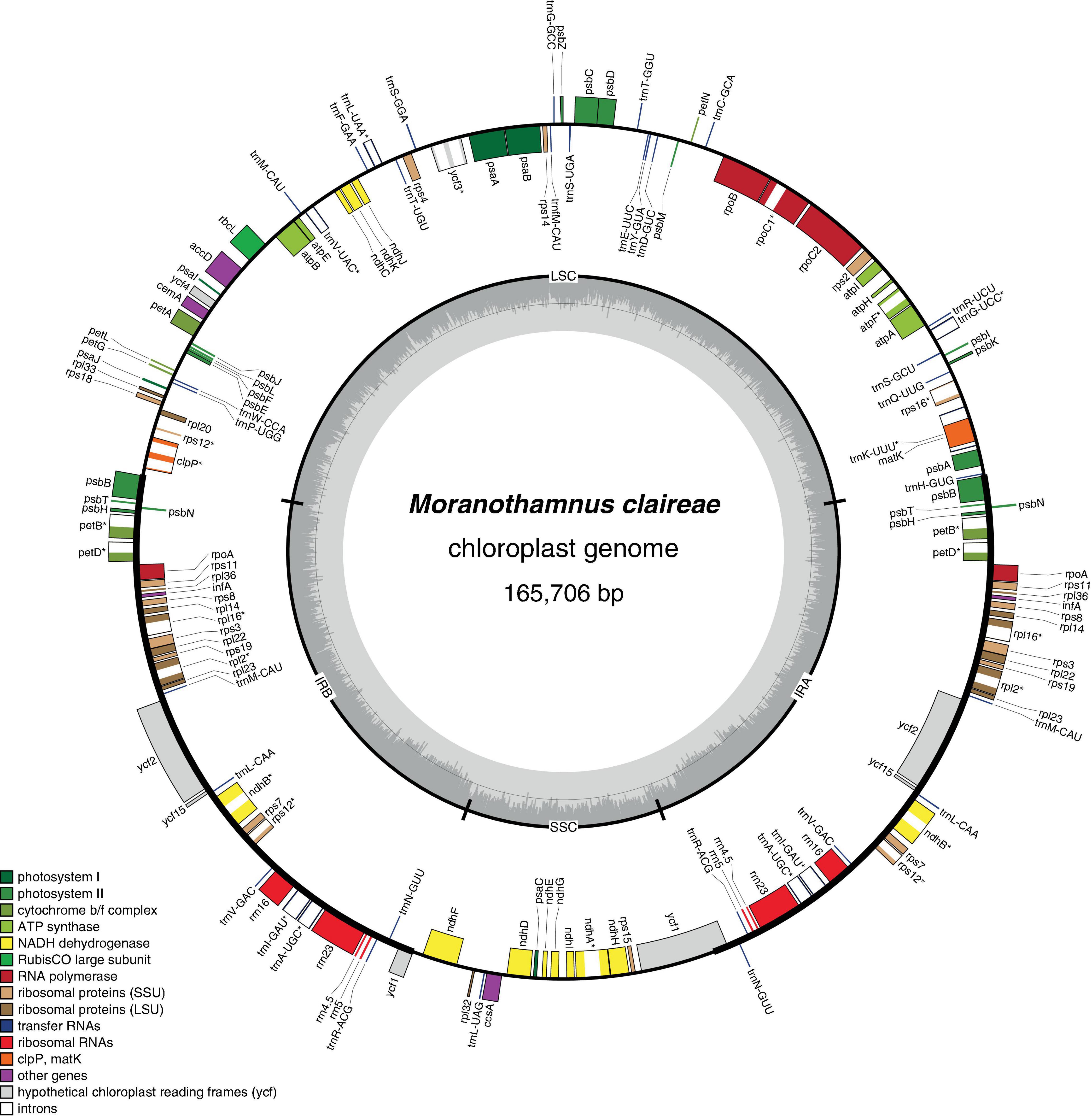
Figure 2. Chloroplast genome map of Moranothamnus claireae. The genes drawn on the inner side of the outer circle are transcribed clockwise, and those on the outer side are transcribed counterclockwise. IRs are shown in bold lines in the outer circle. The darker gray areas of the inner circle indicate GC contents across the genome with lighter gray areas indicating AT contents. Genes belonging to different functional groups were shown in different color as in the legend.
Together with all newly assembled plastomes, we also noticed a substantial length variation in accD genes in Berberidaceae (Supplementary Figure 7), especially in Berberidoideae (Supplementary Figure 8). All sampled plastomes of Alloberberis and Mahonia share a 216-bp deletion close to the 3′ end of the reading frame, with two additional deletions of 120 and 30 bp unique to the former genus (Supplementary Figure 8). However, the greatest sequence variation of accD locates in the central part of the gene. Visualizing the translation alignment revealed that the length variation in accD is featured by repeats composed of five amino acid sequences. In Berberidoideae, a total of 33 types of the amino acid repeats translated from 37 types of 15-bp DNA sequences were identified (Supplementary Table 4). The total number of these repeats in each species varies from 6 in B. dictyophylla to 27 in Berberis aristata (MN746308) and B. saxicola. Of the 33 amino acid repeats, R19 (120 copies) and R22 (87 copies) are the two most numerous copies, found in almost all plastomes of Berberideae (Supplementary Table 5 and Supplementary Figure 8). Some repeats were detected in certain groups and thus appear to be clade specific. For example, R21 and R31 occur exclusively in Asian Mahonia clade (Group Orientales) except for M. nervosa, R10 is unique to Mahonia, and R8, R20, and R23 were found only in Alloberberis (Supplementary Table 5 and Supplementary Figure 8).
Plastid Phylogenomic Analyses
Our preliminary ML analyses of the 110 plastome dataset indicated that, of the 18 species represented by multiple sequences, 10 species (B. amurensis, M. fortunei, Ranzania japonica, Dysosma pleiantha, Diphylleia sinensis, Sinopodophyllum hexandrum, E. brevicornu, E. tianmenshanensis, E. wushanense, and Plagiorhegma dubium) were recovered as monophyletic groups and two species (Achlys triphylla and E. pseudowushanese) were paraphyletic (Supplementary Figure 1). Of the two plastome sequences of A. triphylla, MG461315 was deleted for its poor sequence quality (Ye et al., 2018). For the two plastome sequences of R. japonica, MH423072 (Sun et al., 2018) was selected because MG234280 (Wang et al., 2018) contains expanded IRs (Supplementary Figure 6) that was not reported by early chloroplast restriction site mapping study (Kim and Jansen, 1994). For the remaining eight monophyletic and one paraphyletic species, one plastome sequence was randomly selected for each species for subsequent analyses (Supplementary Figure 1). Seven species (B. aristata, Mahonia oiwakensis, M. bealei, Dysosma versipellis, P. peltatum, Epimedium mikinorii, and E. sagittatum) were shown to be polyphyletic (Supplementary Figure 1) and all their sequences were retained except for MG593052 (P. peltatum) that shares 99.19% of “% Identity” with S. hexandrum (KT445939) and yet only 87.2% with its conspecific sequence.
With IRB excluded and partitions concatenated, alignment of the remaining 93 plastomes is 153,461 bp in length, and contains 36,726 parsimony informative sites (23.93% of the alignment). The substitution models for each partition determined by ModelFinder are listed in Supplementary Table 6. Rooted by A. quinata, ML phylogeny shows that Ranunculaceae (Ranunculus macranthus) is sister to Berberidaceae (Figure 3A) with a strong support (UFBS: 100). Within Berberidaceae, three clades corresponding to the three subfamilies Berberidoideae, Nandinoideae, and Podophylloideae were recovered with full support (UFBS: 100).
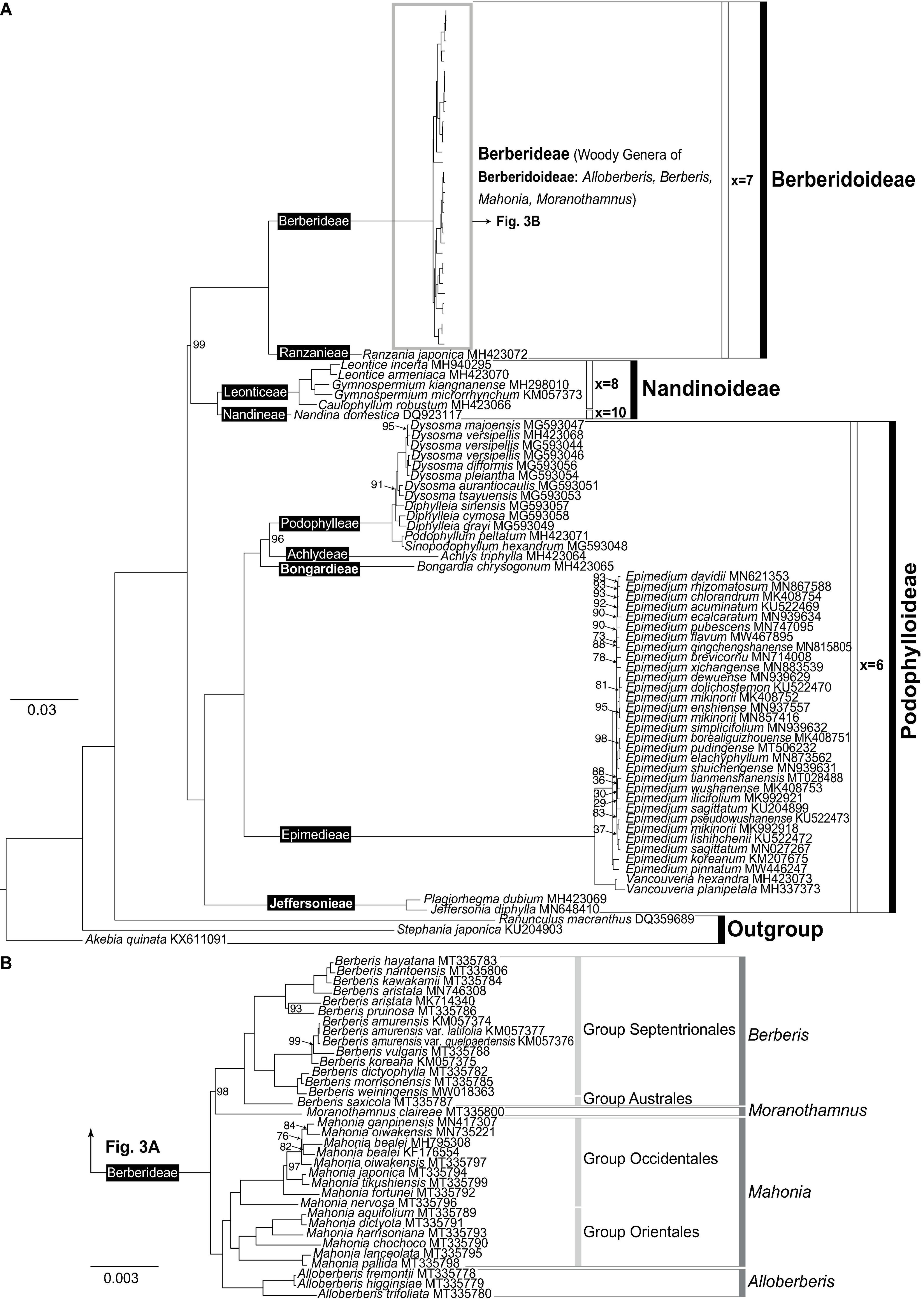
Figure 3. Best-scoring maximum likelihood tree based on the whole plastome sequences reconstructed under IQ-TREE. All branches are fully supported (UFBS: 100) except for those marked by exact numbers (UFBS <100). (A) ML phylogram of Berberidaceae labeled with infrafamilial classification proposed in current study; (B) phylogenetic relationships of Berberideae with labels following Yu and Chung (2017).
Within Nandinoideae, Nandina (i.e., Nandineae; x = 10) is sister to the clade of Leontice + Gymnospermium + Caulophyllum (i.e., Leonticeae; x = 8). Within Podophylloideae, five long-branched clades (Figure 3A) corresponding to clade Jeffersonia + Plagiorhegma (i.e., Jeffersonieae tri. nov.), clade Epimedium + Vancouveria (i.e., Epimedieae), Bongardia (i.e., Bongardieae tr. nov.), Achlys (i.e., Achlydeae), and clade Dysosma + Diphylleia + Podophyllum + Sinopodophyllum (i.e., Podophylleae) were recovered, with each successive sister to the remaining clades within the subfamily (Figure 3A). Within Epimedieae, the monophyletic WNA Vancouveria is sister to the monophyletic Eurasian Epimedium. Within Epimedium, interspecific relationships in general are poorly supported and different relationships have been recovered between the 110-plastome (Supplementary Figure 1) and 93-plastome datasets (Figure 3); however, in both datasets, E. pinnatum (Subgenus Rhizophyllum) and E. koreanum (Sect. Macroceras) form a strongly supported clade sister to the clade of Sect. Diphyllon (UFBS: 100). Within Sect. Diphyllon, two moderately to strongly supported clades A and B each characterized by slightly different IRB/LSC boundaries were recovered (Supplementary Figure 9). Within Podophylleae, Podophyllum and Sinopodophyllum form a clade sister to Dysosma + Diphylleia, though Diphylleia is paraphyletic with D. sinensis sister to Dysosma (Figure 3A).
Within Berberidoideae (Figure 3A), our ML analysis also reveals that R. japonica (i.e., Ranzanieae) is sister to Berberideae that is composed of four clades corresponding to Alloberberis, Berberis, Mahonia, and Moranothamnus with full supports, confirming Yu and Chung’s (2017) classification. However, while Alloberberis was resolved as the sister clade of Berberis + Mahonia + Moranothamnus in Yu and Chung (2017), the genus was placed as the sister group of Mahonia with full support in current analysis. Although our sampling of Berberis is too limited to test the infrageneric classification of Berberis and Mahonia (Ahrendt, 1961), the monophyly of Group Septentrionales sister to Group Australes is strongly supported (Figure 3B). Within Mahonia, the monophyly of the New World Group Occidentales and the predominant Old World Group Orientales are also both fully supported (Figure 3B).
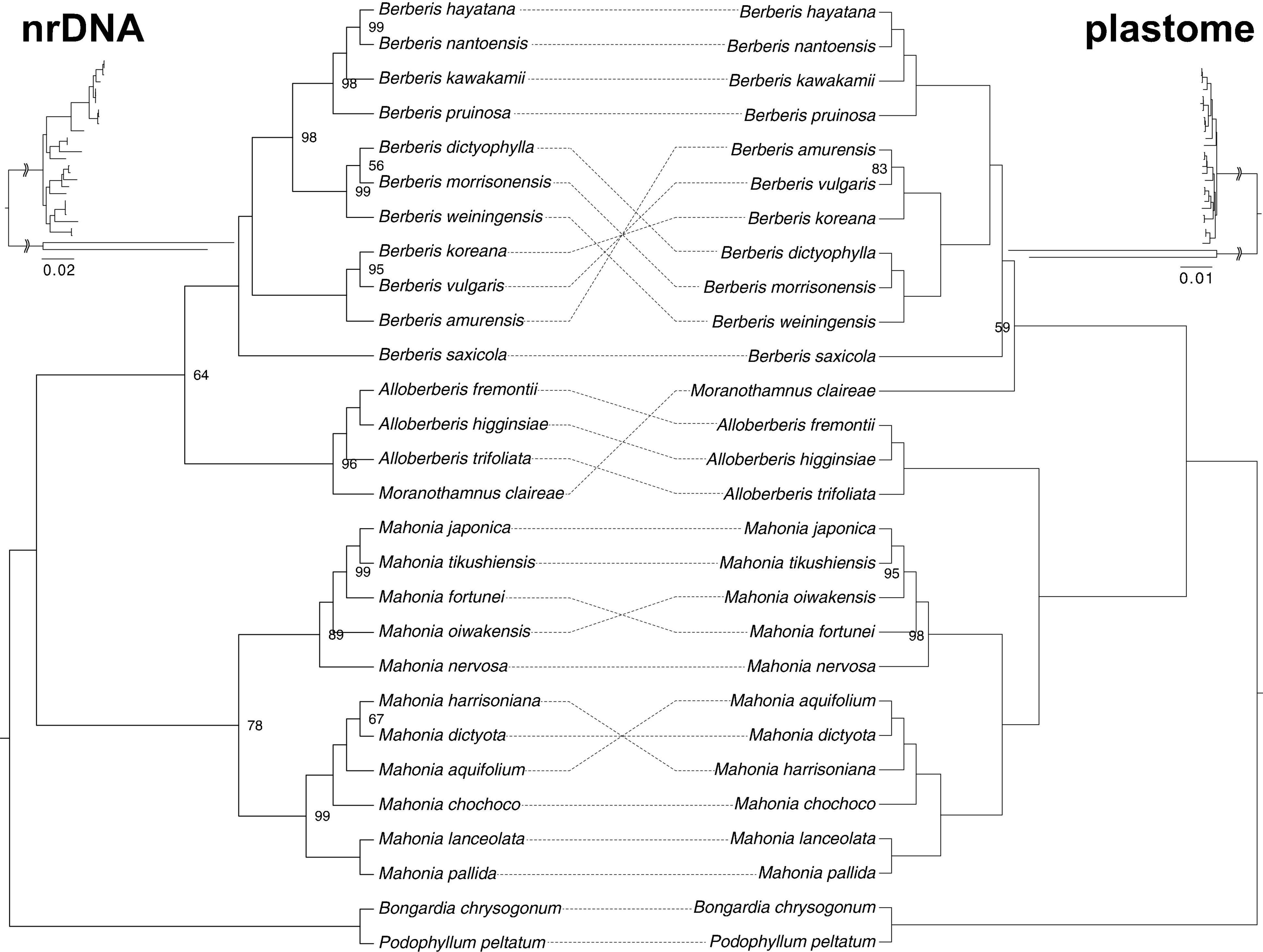
Nuclear Ribosomal DNA and Analyses
Table 2 and Supplementary Table 7 summarizes details of the nrDNA assembly. The average coverage of each species, which was calculated by read mapping, ranges from 876.4× in M. chochoco to 3325.9× in M. japonica (Table 2). The final matrix consists of 7530 aligned base pairs with 855 parsimony informative sites (11.35% of the alignment). The best-fit substitution model for each partition under the best-fit partition scheme was determined by ModelFinder (Supplementary Table 7).
Rooted by Bongardia and Podophyllum, ML analysis of nrDNA using IQ-TREE supports the monophyly of Alloberberis, Berberis, and Mahonia (Figure 4), though the support for the monophyly of Mahonia is low (UFBS: 78). Within Berberideae, Mahonia is sister to the clade composed of Berberis + Alloberberis + Moranothamnus, with the clade Alloberberis + Moranothamnus sister to Berberis with low support (UFBS: 64). As shown in Figure 4, relationships in nrDNA tree among the four genera of Berberideae are in conflict with the plastome tree in which Alloberberis and Moranothamnus are placed sister to Mahonia and Berberis, respectively, though the support for the latter sister relationship is low (UFBS: 59).
Divergence Time Estimation
The best-fit substitution model and partition scheme as the site model prior for BEAST2 analyses evaluated by ModelFinder were summarized in Supplementary Table 7. Using BEAST2, the stem and crown ages of Berberidaceae were estimated to be 91.69 Ma (95% HPD: 103.18–79.93 Ma) and 81.57 Ma (95% HPD: 93.08–70.71 Ma), respectively, falling within the Late Cretaceous (Table 3 and Figure 5). Within Podophylloideae, tribe Jeffersonieae tr. nov. diversified from the rest of Podophylloideae at ca. 74.62 Ma (95% HPD: 86.50–62.97 Ma), with the split between Jeffersonia and Plagiorhegma at ca. 23.15 Ma (95% HPD: 52.72–3.65 Ma). Within the clade of the remaining Podophylloideae, Epimedieae separated from Bongardieae + Achlydeae + Podophylleae at ca. 65.41 Ma (95% HPD: 78.04–53.40 Ma), while Bongardieae split from Achlydeae + Podophylleae at ca. 52.09 Ma (95% HPD: 66.53–36.22 Ma). The split between Achlydeae and Podophylleae was estimated at ca. 43.68 Ma (95% HPD: 57.10–28.36 Ma). The split of Berberidoideae from Nandinoideae was estimated to have occurred at ca. 76.54 Ma (95% HPD: 88.47–64.26 Ma). Within Nandinoideae, Nandineae diverged from the Leonticeae at ca. 48.70 Ma (95% HPD: 73.76–24.75 Ma). Within Berberidoideae, the crown age of Berberidoideae was estimated at ca. 62.24 Ma (95% HPD: 72.09–52.20 Ma). The crown ages of the clades Alloberberis + Mahonia and Berberis + Moranothamnus were estimated at ca. 36.23 Ma (95% HPD: 41.36–31.41 Ma) and ca. 32.89 Ma (95% HPD: 43.13–20.59 Ma), respectively. The crown ages of Alloberberis, Berberis, and Mahonia were estimated to be ca. 33.42 Ma (95% HPD: 36.77–31.32 Ma), 20.47 Ma (95% HPD: 33.09–10.08 Ma), and 28.30 Ma (95% HPD: 33.67–23.14 Ma), respectively.
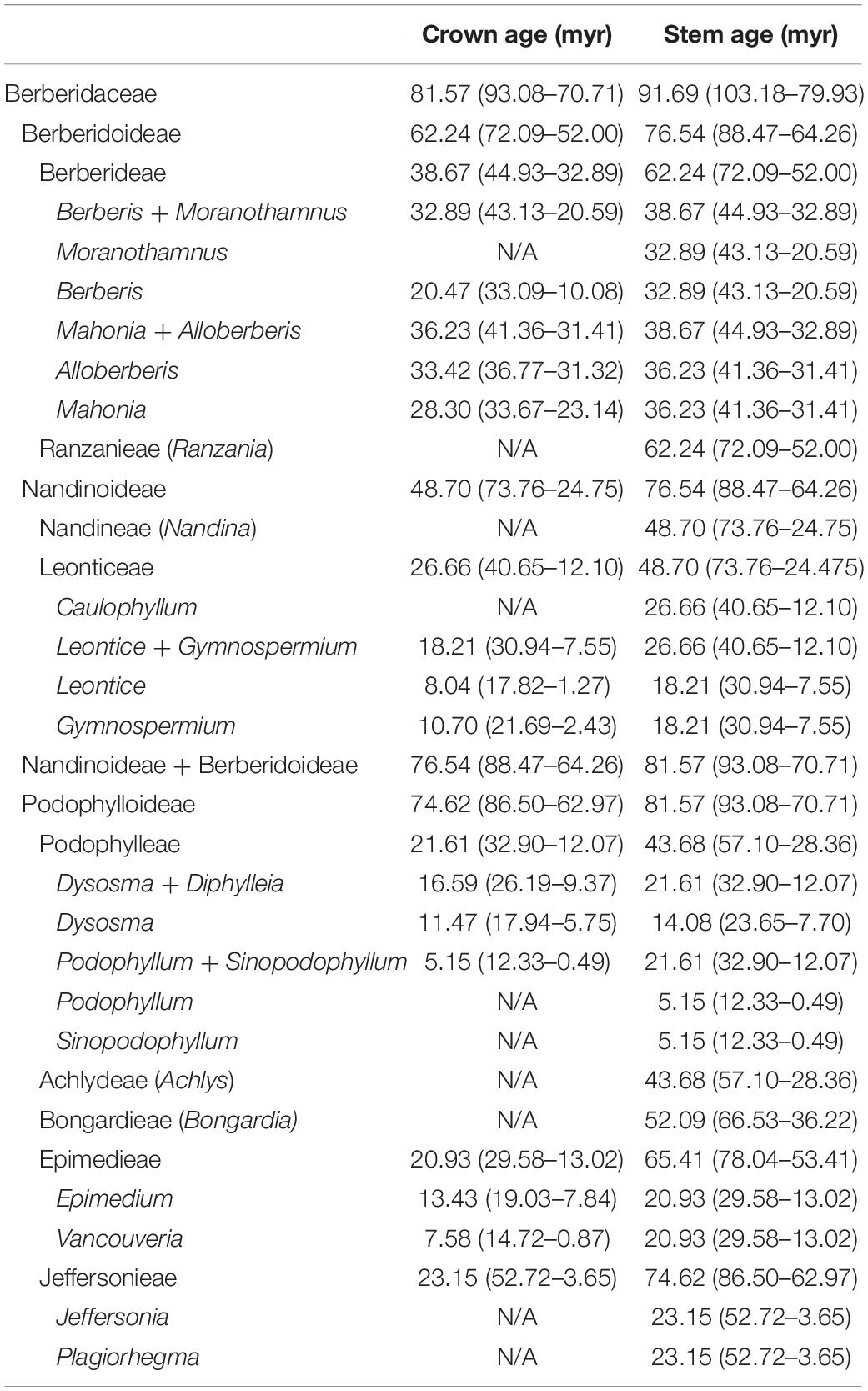
Table 3. Summary of divergence times estimated for genera, tribes, and subfamilies of Berberidaceae by BEAST2.
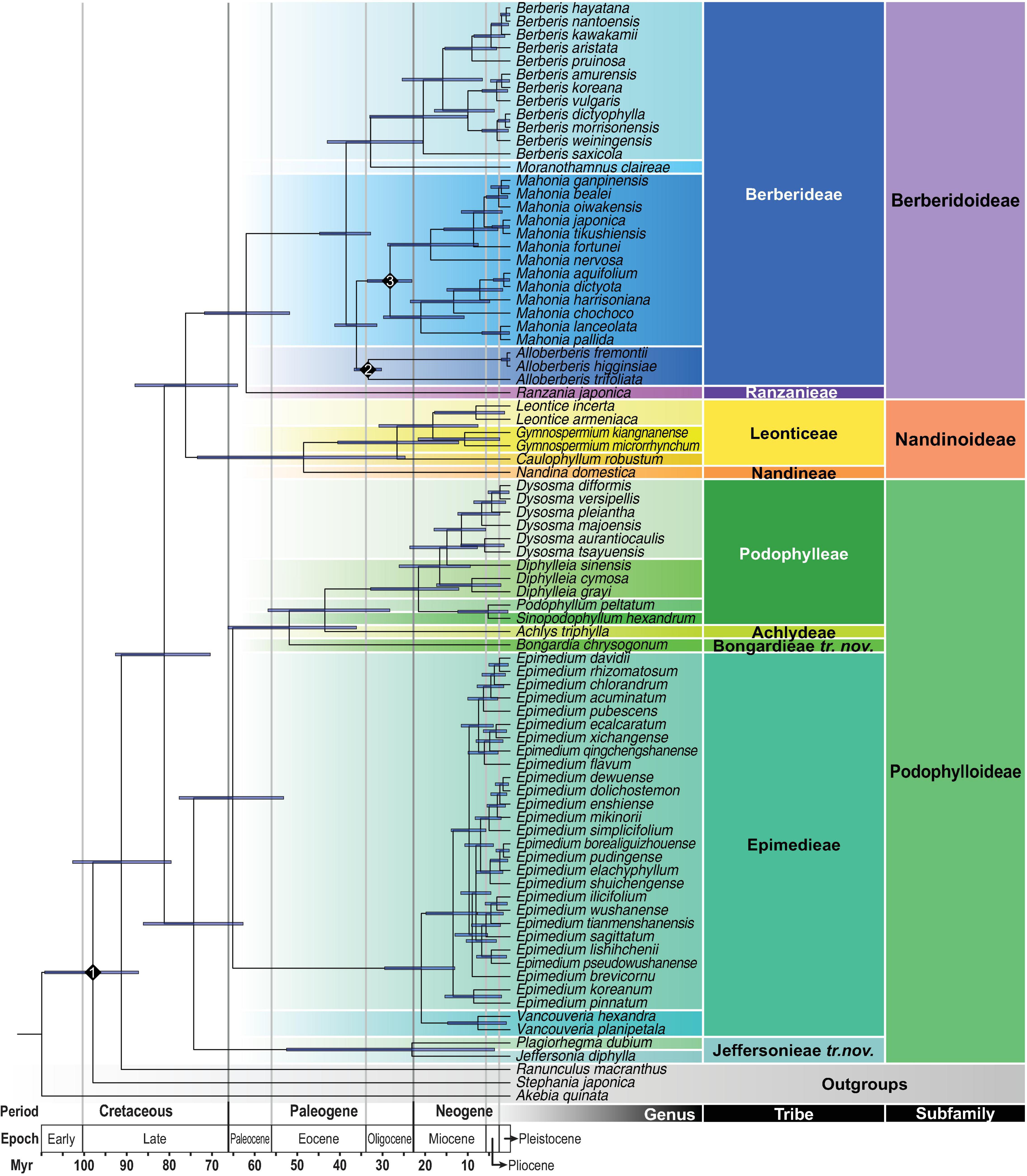
Figure 5. Maximum clade credibility chronogram estimated by BEAST2. The 95% highest posterior density (HPD) date ranges are shown by the node bars. Numbered diamonds are three calibration points.
Discussion
Variation in Berberidoideae Plastome Structure
Despite the functional importance of chloroplasts in photosynthesis and ostensibly the conserved nature of plastid genomes in both structures and contents (Mower and Vickrey, 2018), IR expansions/contractions have been reported across land plants (Goulding et al., 1996; Zhu et al., 2016). In Berberidaceae, early chloroplast restriction site mapping study (Kim and Jansen, 1994) had revealed IR expansion in Berberis and Mahonia (including Alloberberis). Kim and Jansen’s (1994) observation was attested first by the whole plastome sequence of M. bealei (Ma et al., 2013) and subsequent phylogenomic analyses (Sun et al., 2018). In current study, all 23 newly assembled plastomes of Alloberberis, Berberis, and Mahonia, as well as the genus Moranothamnus that has never been sampled previously, are featured by significant IR expansions (Supplementary Figure 6), further corroborating previous studies. However, IR expansion was also reported in R. japonica (MG234280) by Wang et al. (2018), contradicting to its conspecific plastome sequence MH423072 (Sun et al., 2018) and early chloroplast restriction site mapping study (Kim and Jansen, 1994). Although the inclusion of MG234280 did not affect phylogenetic relationships of R. japonica with the rest of Berberidaceae (Supplementary Figure 1), further investigation (e.g., PCR validation) is urgently needed to clarify the SC/IRs junctions in its plastome sequence. Additionally, our analyses also revealed IR expansion and contraction in E. ecalcaratum (MN939634) and E. brevicornu (MN381716; Zheng et al., 2019), respectively, that have never been reported previously in Epimedium. However, in MN803415 (Yao et al., 2020) and MN714008 (Zhang et al., 2020) that are conspecific with MN381716 (Zheng et al., 2019), IR contraction is not detected (Supplementary Figures 6, 9). Further study will be needed to clarify the plastome structure in E. brevicornu specifically and Epimedium in general.
In addition to IR expansion, substantial length variation in accD gene featured by insertions and deletions of repeat sequences was revealed in all sampled Berberidoideae plastomes (Supplementary Figures 7, 8). AccD encodes the β-carboxyl transferase subunit of acetyl-CoA carboxylase (ACCase), which is a functionally essential multi-subunit enzyme in charge of the biosynthesis of fatty acids in plants (Kode et al., 2005) including non-photosynthetic parasitic plants (e.g., Su et al., 2019). However, pseudogenized accD has been reported in Primula sinensis (Liu T.J. et al., 2016) and Vaccinium macrocarpon (Fajardo et al., 2013). Additionally, accD has been lost independently from chloroplast genomes and relocated to the nucleus in gymnosperms, i.e., gnetophytes (Sudianto and Chaw, 2019) and Sciadopitys verticillata (Li et al., 2016), and multiple angiosperm species of Acoraceae (Goremykin et al., 2005), Campanulaceae (Hong et al., 2017), Fabaceae (Magee et al., 2010), Geraniaceae (Guisinger et al., 2008), Oleaceae (Lee et al., 2007), and Poales (Harris et al., 2013). Despite the extensive length variation, accD genes in Berberidoideae appear to be functional as their reading frames are intact without frameshift and the residual sequences at the 3′ end are highly conserved (Supplementary Figure 7). Such length variation characterized by repeat sequences in accD has also been reported in the legume species Medicago truncatula (Gurdon and Maliga, 2014) and the cupressophytes (Li et al., 2018). Gurdon and Maliga (2014) attributed the intragenic expansion and contraction of accD in M. truncatula to the presence of repeat sequences that could have triggered replication slippage. Li et al. (2018) also hypothesized that the presence of accD repeat sequences could have promoted the acceleration of substitution rate and mediated the rearrangement of plastomes in cupressophytes. Further analyses will be conducted to understand the intriguing accD length variation in Berberidoideae.
Ancient Origins of Berberidaceae Genera
Calibrated by fossils of Menispermaceae from the Turonian and Alloberberis and Mahonia from the Oligocene, the stem age of Berberidaceae was estimated to be 91.69 Ma (95% HPD: 103.18–79.93 Ma), largely congruent with that estimated by Magallón et al. (2015) at 80.28 Ma (95% HPD: 95.84–68.17 Ma), Li et al. (2019) at 87.4 Ma (95% HPD: 98.9–72.9 Ma), and Ramírez-Barahona et al. (2020) at 101.21 Ma (95% HPD: 117.74–87.28 Ma; constrained calibration of a complete set 238 fossils). However, our estimated crown ages of the three subfamilies are much older than those estimated by Sun et al. (2018) [Berberidoideae: 62.24 (95% HPD: 72.09–52 Ma) vs. ca. 16 Ma (95% HPD: 28–6 Ma); Nandinoideae: 48.70 (95% HPD: 73.76–24.75) vs. ca. 24 Ma (95% HPD: 33–13 Ma); Podophylloideae: 74.62 (95% HPD: 86.50–62.97) vs. ca. 32.5 Ma (95% HPD: 36–27 Ma)], in which the divergence times were estimated by constraining the minimum age of the crown group of Berberidaceae at 33.9 Ma. The disparity of age estimates between Sun et al. (2018) and a majority of studies including current one reflects the dubious application of the Mahonia fossil to calibrate a deeper node in the former study, resulting in underestimates of ages within the family (Donoghue and Benton, 2007). Indeed, Sun et al. (2018) adopted Magallón et al.’s (2015) calibration strategy that applied the upper Eocene (33.9 Ma) fossil of Mahonia as the minimum crown age of Berberidaceae, apparently underestimating the age for the family. Given this, our results (Table 3 and Figure 5) present a more reliable divergence time estimation of the infrafamilial taxa of Berberidaceae than those of Sun et al. (2018).
Within Berberidaceae, our estimated stem ages of genera range from 5.12 Ma (95% HPD: 13.43–0.34 Ma) in Podophyllum and Sinopodophyllum to 59.74 Ma (95% HPD: 68.56–51.41 Ma) in Ranzania (Table 3 and Figure 5). Except for the former two genera that splitted in the early Pliocene, all genera of Berberidaceae were estimated to have originated prior to the early Miocene. While early Pliocene origins of Podophyllum and Sinopodophyllum are consistent with Liu et al. (2002; 6.52 ± 1.98 Ma) and Wang et al. (2007; 5.8 ± 0.6 Ma), Yu and Chung (2017) has estimated 20.46 Ma (95% HPD: 34.56–2.66 Ma) and 13.78 Ma (95% HPD: 24.47–1.7 Ma) for the stem ages of Podophyllum and Sinopodophyllum, respectively.
The late Cretaceous origins of the three subfamilies of Berberidaceae estimated in present study are consistent with recent studies of temperate eudicots (e.g., Hypericaceae, Juglandaceae, and Ranunculaceae) in which major lineage diversification had occurred during the Late Cretaceous and Paleocene (Nürk et al., 2015; He et al., 2021; Zhang et al., 2021). Considering the paleoclimate of the Cretaceous, our dating estimation also suggests that the early lineages of Berberidaceae should have adapted to warmer environments, implying niche shifts experienced by extant species (Folk et al., 2020). Notably, the unusually long branch between stem and crown ages of Epimedieae within Podophylloideae also suggests the occurrence of extinction and/or rapid diversification if the sampling bias is ignored (Antonelli and Sanmartín, 2011). Additionally, while stem ages of most genera within each subfamily were estimated during the Oligocene and Early Miocene, our result suggests the association between the rise of these genera and global climatic deterioration (i.e., temperature cooling and enhanced seasonality) since the Neogene (Smith and Donoghue, 2010). In contrast, the later divergence between Podophyllum and the montane Sinopodophyllum may be more likely related to the uplift history of the Pan-Himalayan region (Xing and Ree, 2017).
Conflicts Between Plastome and Nuclear Phylogenies
With the inclusion of Alloberberis and Moranothamnus and expanded sampling of Berberis and Mahonia, our plastome phylogenomic analyses support the monophyly of Berberideae, its sister relationship with Ranzania, and the monophyly of Berberidoideae, Nandinoideae, and Podophylloideae (Figure 3), corroborating infrafamilial classification in Berberidaceae (Wang et al., 2009; Yu and Chung, 2017; Sun et al., 2018). However, while previous (Sun et al., 2018) and our current plastome trees both place Berberidoideae sister to Nandinoideae (Figure 3), Nandinoideae was resolved as the sister group of Podophylloideae in the combined ML tree of Yu and Chung (2017) and the recently released Kew Tree of Life (KToL) reconstructed using the Hyb-Seq Angiosperms 353 bait set (Johnson et al., 2019; Baker et al., 2021). The conflicting subfamilial relationships of Berberidaceae could have resulted from multiple causes including sampling issues, incomplete lineage sorting (ILS), and hybridization/introgression (Wendel and Doyle, 1998). Given the congruent results between the nuclear trees, i.e., ITS (Yu and Chung, 2017) and the Angiosperms 353 bait set (Baker et al., 2021), the conflicting relationships between plastomes and nuclear datasets observed at the deep level in Berberidaceae seems more likely due to hybridization in the ancient time (Stull et al., 2020).
Within the tribe Berberideae, our phylogenomic analyses (Figure 3) reveal four clades corresponding to Alloberberis, Berberis, Mahonia, and Moranothamnus, supporting Yu and Chung’s (2017) classification. However, relationships of the four genera differ between the plastome and the nrDNA phylogenies (Figure 4), as well as Yu and Chung (2017). Specifically, while Mahonia and Alloberberis are placed in one clade sister to clade Berberis + Moranothamnus in the plastome tree, in the nrDNA tree Alloberberis and Moranothamnus formed a strongly supported clade (UFBS: 96) sister to Berberis, with Mahonia further sister to the clade Berberis + Alloberberis + Moranothamnus (Figure 4). Although ILS could have led to this phylogenetic incongruence (Wendel and Doyle, 1998), the conflicting relationships between plastome and nrDNA tree can also be explained by hybridization between Berberis and Mahonia (García et al., 2017). Under this scenario, Berberis and Mahonia should be the maternal parents for Moranothamnus and Alloberberis (Figure 3), respectively, given cytoplasmic DNA is known to be maternally inherited in Berberidaceae (Zhang et al., 2003). Coupled with the ancient splits of the four genera (Figure 5), Alloberberis and Moranothamnus could have resulted from ancient reciprocal hybridization (Popelka et al., 2019) between Berberis and Mahonia preceding subsequent radiations of the two parental genera (García et al., 2017). Additionally, the hybrid origins of Alloberberis and Moranothamnus could also explain their combined morphology and more restricted geographic distributions relative to Berberis and Mahonia (Yu and Chung, 2017). Given that Alloberberis and Moranothamnus are both distributed in western North America, the ancestral ranges of Berberis and Mahonia are likely also in the New World, as suggested in recent biogeographic study (Chen et al., 2020). Because contemporary intergeneric hybrids between Berberis and Mahonia (×Mahoberberis) rarely occur naturally (Ahrendt, 1961; Rounsaville and Ranney, 2010), the proposition on the hybrid origins of Alloberberis and Moranothamnus implies a weaker reproductive isolation between the two parental genera in the ancient time.
Within Podophylloideae, although relationships among the five major clades (Figure 3A) are largely congruent with previous studies (Wang et al., 2007; Sun et al., 2018), substantial conflicts exist within tribe Podophylleae between current and previous studies. First, in current and four previous studies (Wang et al., 2007; Sun et al., 2018; He et al., 2019; Li and Dong, 2020), Sinopodophyllum is placed sister to Podophyllum; however, Sinopodophyllum was resolved as sister to Diphylleia + Dysosma + Podophyllum in Ye et al. (2018) and Dysosma + Diphylleia in Yu and Chung (2017). Second, while our plastome tree resolves Diphylleia as a paraphyletic grade sister to Dysosma, Dysosma was resolved sister to Diphylleia + Podophyllum in Ye et al. (2018), paraphyletic grade sister to Diphylleia + Podophyllum + Sinopodophyllum in He et al. (2019) and Li and Dong (2020), and polyphyletic in Mao et al. (2016). Third, while all three samples species of Diphylleia form a clade in Ye et al. (2018), He et al. (2019), and Li and Dong (2020), the genus is paraphyletic in Mao et al. (2016) and current study (Figure 3A). One important issue that could have contributed to the conflicting results is the very different strategies utilized to analyze the plastome sequences. In Sun et al. (2018) and Ye et al. (2018), only protein-coding genes (CDS) were analyzed, while He et al. (2019) and Li and Dong (2020) used whole plastomes for phylogenetic reconstruction. Because rates of molecular evolution are in general slower in woody species than the herbaceous members of the same taxonomic group (Smith and Donoghue, 2008; Smith and Beaulieu, 2009), we used the full plastome sequences specifically to increase phylogenetic resolution within Berberidoideae. Another factor that might lead to conflicting relationships is partitioning (Kainer and Lanfear, 2015). While no information regarding partitioning were reported in Ye et al. (2018), He et al. (2019), and Li and Dong (2020), we partitioned the plastome sequences into CDS, RNA regions, introns, and IGS, with CDS further partitioned into three parts by codon positions, to take into account rate variation. Additionally, while the monophyly of Diphylleia was supported by a combined tree of cpDNA (matK and rbcL) and ITS2 (Wang et al., 2007) and ITS (Mao et al., 2014), matK and rbcL alone did not provide enough phylogenetic information for the monophyly of Diphylleia in Wang et al. (2007). Interestingly, Bayesian phylogenetic analysis of CYP719A, a podophyllotoxin biosynthesis gene that could have experienced relaxed purifying selection, showed that both Diphylleia and Dysosma are not monophyletic (Mao et al., 2016). These conflicting relationships within Podophylleae again could have resulted from ILS and/or hybridization.
To examine whether ILS or hybridization has contributed to conflicting phylogenetic relationships between plastome and nuclear phylogenies, a robust species tree reconstructed from multi-locus genome data (Morales-Briones et al., 2018) such as Angiosperm 353 bait set (Johnson et al., 2019) could provide a promising solution to resolve conflict phylogenetic relationships between plastome and nuclear genes (Shee et al., 2020).
Infrafamilial Classification of Berberidaceae
Based on the robust (Figure 3) and dated phylogenomic relationships (Figure 5) reconstructed using completed generic sampling of plastome sequences of Berberidaceae, we evaluate different generic concepts outlined in Supplementary Table 1 using criteria advocated by Backlund and Bremer (1998), Linder et al. (2010), and Heenan and Smissen (2013). Accordingly, our current plastome phylogenomic study (Figures 3, 5) corroborates the classification of four genera within Berberideae (Yu and Chung, 2017). Within Podophylleae, although Diphylleia is paraphyletic in our plastome tree (Figure 3), the apparent morphological (Figure 1M), ecological, phytochemical, anatomical, cytological, and palynological coherence of the genus (Ying et al., 1984; Stearn, 2002) and monophyly as revealed by ITS trees (Wang et al., 2007; Mao et al., 2014) also favor the generic status of this long-recognized genus, though hybridization probably also had occurred in the past. We also support the generic status of the EA Sinopodophyllum given its morphological (Figure 1L), geographic, and evolutionary distinctness (Figure 5) from the ENA Podophyllum (Ying, 1979). As a member of the earliest diversified clade (i.e., Jeffersonieae) sister to the rest of Podophylloideae, the generic status of Plagiorhegma should also be maintained given its early Miocene split from Jeffersonia (Figure 5) and morphological (Figure 1S) and geographic uniqueness (Hutchinson, 1920).
The maintenance of the generic status of Alloberberis, Mahonia, Moranothamnus, Plagiorhegma, and Sinopodophyllum that are often synonymized (Supplementary Table 1) not only acknowledges their morphological, ecological, and evolutionary distinctness, but also underscores the critical conservation status of these genera. Since the 17th century, Berberis has been a major target for eradication around the world because barberry species (and a few species of Mahonia) are alternative hosts of rust fungi (Peterson, 2018; Barnes et al., 2020). However, Alloberberis (Breckenridge, 1983; Harms, 2007), Moranothamnus (Moran, 1982), and a majority of Asian Mahonia are highly endangered threatened by habitat destruction and overexploitation (Boufford, 2013) for traditional Chinese medicines (He and Mu, 2015). Subsuming Alloberberis, Mahonia, and Moranothamnus under a broadly defined Berberis s.l. would likely further exacerbate their critical conservation status given the stereotypical impression of Berberis as agricultural weeds. Additionally, because both P. dubium (Lee et al., 2018) and S. hexandrum (Liu W. et al., 2016) are also rare and exploited for traditional medicines, recognizing and elevating these two distinct species to the generic rank also confers an effective conservation strategy.
Throughout the taxonomic history of Berberidaceae, several tribes (Janchen, 1949; Terabayashi, 1985b; Loconte, 1993; Takhtajan, 1997; Wu et al., 2003) had been proposed; however, tribal classification has not been implemented under a molecular phylogenetic context. Based on our phylogenomic analyses (Figures 3, 5), we propose to recognize nine clades as tribes within Berberidaceae. We consulted Reveal’s (1955–onward) “Indices Nominum Supragenericorum Plantarum Vascularium” for priority of the tribal names. Within Berberidoideae, we follow Terabayashi (1985b) and Wu et al. (2003), recognizing tribes Berberideae (including Alloberberis, Berberis, Mahonia, and Moranothamnus) and Ranzanieae (including Ranzania). Within Nandinoideae, tribes Leonticeae (including Caulophyllum, Gymnospermium, and Leontice) and Nandineae (including Nandina) have long been recognized (Supplementary Table 1) and thus are followed here. These two tribes are also characterized by chromosome numbers x = 8 and x = 10, respectively. Within Podophylloideae, we propose to recognize the five distinct and long-branched clades as tribes (Figures 3, 5). However, while the names Achlydeae, Epimedieae, and Podophylleae are available, the designation Bongardieae (Takhtajan, 1997) was not validly published according to the Code (Turland et al., 2018) and the clade Jeffersonia + Plagiorhegma has never been named. We provide a description for the valid publication of Bongardieae and propose the tribe Jeffersonieae for the latter clade.
Key to Subfamilies, Tribes, and Genera of Berberidaceae
1. Stamens sensitive; pollen exine psilate and imperforate……………………………………………….2 (Berberidoideae)
1. Stamens not sensitive; pollen exine sculptured and perforate………………………………………………………………………………6
2. Herbaceous……………………………………….Ranzanieae (Ranzania)
2. Woody…………………………………………………………. 3 (Berberideae)
3. Stem dimorphic………………………………………………………………….. 4
3. Stem monomorphic……………………………………………………………. 5
4. Stem spineless; leaves 3–9-foliolate…………………….. Alloberberis
4. Stem almost always spiny; leaves unifoliolate…………….Berberis
5. Leaves imparipinnate, 5–40-foliolate………………………. Mahonia
5. Leaves uni- to 7-foliolate………………………………Moranothamnus
6. Chromosome base number x = 8 or 10……… 7 (Nandinoideae)
6. Chromosome base number x = 6………… 10 (Podophylloideae)
7. Woody……………………………………………….. Nandineae (Nandina)
7. Herbaceous…………………………………………………….. 8 (Leonticeae)
8. Rhizomatous; inflorescence cymose, bracts subulate; flowers calyculate………………………………………………………….Caulophyllum
8. Tuberous; inflorescence a raceme or panicle; bracts foliaceous; flowers excalyculate…………………………………………………………….. 9
9. Leaf solitary, stipulate; seeds exposed by papery pericarp…………………………………………………….. Gymnospermium
9. Leaves 2–4, sheathing, seeds enclosed in an inflated bladder………………………………………………………………………Leontice
10. Perianth abscent ………………………………………Achlydeae (Achlys)
10. Perianth present……………………………………………………………….. 11
11. Leaves pinnate, with more than six pinnae ……………………………………………………….Bongardieae (Bongardia)
11. Leaves simple, lobed, or ternately compound……………………. 12
12. Nectaries absent; aril present…………………………………………….. 13
12. Nectaries present; aril absent …………………….16 (Podophylleae)
13. Evergreen, petiolules presence, multicellular leaf pubescence present; more than one flowers in an inflorescence………………………………………………..14 (Epimedieae)
13. Deciduous petiolules absence, multicellular leaf pubescence abscent; one flower in an inflorescence……………………………………………….15 (Jeffersonieae)
14. Leaves cauline and basal, margins spinose; flowers 2-merous, stamens 4……………………………………………………………..Epimedium
14. Leaves basal, margins not spinose; flowers 3-merous, stamens 6……………………………………………………………………….. Vancouveria
15. Leaves compound; stamens 8……………………………….. Jeffersonia
15. Leaves simple; stamens 6…………………………………. Plagiorhegma
16. All leaves with petiole attached to the leaf base…………………………………………………………… Sinopodophyllum
16. All leaves peltate……………………………………………………………….. 17
17. Anther dehiscence valvate; ovule anatropous……….. Diphylleia
17. Anther dehiscence longitudinal; ovule hemitropous…………. 18
18. Flowers several in fascicle; stamens 6………………………. Dysosma
18. Flowers solitary; stamens more than 8………………. Podophyllum
Conspectus of the Infrafamilial Classification of Berberidaceae
Subfamily Berberidoideae Eaton (1836)
Tribe Berberideae Rchb. (1832)
Alloberberis C.C.Yu & K.F.Chung, Berberis L., Mahonia Nutt., Moranothamnus C.C.Yu & K.F.Chung
Tribe Ranzanieae Kumaz. ex Terab. (1985)
Ranzania T.Ito
Subfamily Nandinoideae Heintze (1927)
Tribe Leonticeae (Spach) Kosenko (1980)
Caulophyllum Michx., Gymnospermium Spach, Leontice L.
Tribe Nandineae Bernh. (1833)
Nandina Thunb.
Subfamily Podophylloideae Eaton (1836)
Tribe Achlydeae Bernh. (1833)
Achlys DC.
Tribe Bongardieae Takht. ex C.L.Hsieh, C.C.Yu & K.F.Chung, tr. nov.
Bongardia C.A.Mey.
Tribe Epimedieae Dumort. (1829)
Epimedium L., Vancouveria C.Morren & Decne.
Tribe Jeffersonieae C.L.Hsieh, C.C.Yu & K.F.Chung, tr. nov.
Jeffersonia Barton, Plagiorhegma Maxim.
Tribe Podophylleae DC. (1817)
Diphylleia Michx., Dysosma Woodson, Podophyllum L., Sinopodophyllum T.S.Ying
Tribe Bongardieae Takht. ex C.L.Hsieh, C.C.Yu & K.F.Chung, tr. nov. – Type: Bongardia C.A.Mey.
Bongardieae Takht., Diversity and Classification of Flowering Plants 91. 1997, num. nud.
Diagnosis. – Perennial herbs, tuberous. Tuber subglobose. Leaves glabrous, somewhat fleshy, petiolate, imparipinnate with 7–17 leaflets; leaflets sometimes in whorls of 3 or 4, sessile, obovate to oblong, glaucous-green, usually coarsely toothed from the tip. Inflorescence a loose panicle with long scape, 20–60 cm tall. Flowers long-stalked; sepals 6, concave, suborbicular or ovate, caducous; petals 6, yellow, oblong–ovate, lanceolate or elliptic-oblong, tips sometimes irregularly crenate. Stamens 6. Ovary with 5–6 basal ovules, ovoid. Fruit a capsule, ovoid, papery, opening from the top by short, acute valves; seeds 1–4, black, pruinose.
Accepted genus. – This tribe contains one genus Bongardia C.A.Mey., which is distributed from southern Greece, northern Africa, Middle East to as far east as Pakistan.
Note. – As far as we can track, Bongardieae was first seen in Takhtajan (1997), reiterated in Takhtajan (2009), and adopted by Wu et al. (2003) and Lu and Tang (2020). However, when Takhtajan (1997) published Bongardieae, he did not provide a clear indication of the rank (Code Article 37.1), a description/diagnosis (Code Article 38.1) in Latin (Code Article 39.1), nor a type designation (Code Article 40.1). Consequently, Bongardieae Takht. (1997) was not validly published and thus the designation is a nomen nudum (Turland et al., 2018).
Tribe Jeffersonieae C.L.Hsieh, C.C.Yu & K.F.Chung, tr. nov. – Type: Jeffersonia Barton
Diagnosis. – Perennial herbs, rhizomatous, deciduous. Rhizome short, slender; aerial stems absent. Leaves basal; petiole long, slender; leaf blade suborbicular or reniform-orbicular in overall outline, simple or divided into 2 sessile leaflets, palmately veined, margin entire or shallowly lobed. Flowers scapose, solitary. Sepals 3 or 4, caducous. Petals 6 or 8, obovate, pale-purple or white. Stamens 6 or 8, antipetalous. Ovary with many ovules, placentation marginal. Fruit a capsule, dehiscing transversely or longitudinally; seeds numerous.
Accepted genera. – This tribe contains two monotypic genera, Jeffersonia Barton and Plagiorhegma Maxim., which are disjunctly distributed in eastern North America and East Asia (northeastern China, South Korea, and Russia along Amur River), respectively.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Supplementary Table 2.
Author Contributions
K-FC designed the research and provided the research resources. C-CY, Y-LH, and K-FC collected plant materials. Y-LH collected the genomic data. C-LH and Y-LH assembled the plastomes. C-LH analyzed the data and prepared the figures and tables. C-LH, C-CY, and K-FC wrote the manuscript. All authors read and confirmed the manuscript.
Funding
This project was supported by the Minister of Science and Technology, Taiwan (MOST 106-2621-B-001-003-MY3).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Julian F. Harber, Missouri Botanical Garden Herbarium, Peckerwood Garden, and Rancho Santa Ana Botanic Garden for providing plant materials, the Genomics Core Lab of the Institute of Molecular Biology, Academia Sinica and the High Throughput Genomic Core Lab of Biodiversity Research Center for sequencing, and Bart O’Brien, Takuro Ito, and Mu-Tan Hsieh for permission to use their photographs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.720171/full#supplementary-material
Footnote
References
Adachi, J., Kosuge, K., Denda, T., and Watanabe, K. (1995). Phylogenetic relationships of the Berberidaceae based on partial sequences of the gapA gene. Plant Syst. Evol. 9, 351–353. doi: 10.1007/978-3-7091-6612-3_37
Adhikari, B., Milne, R., Pennington, R. T., Särkinen, T., and Pendry, C. A. (2015). Systematics and biogeography of Berberis s.l. inferred from nuclear ITS and chloroplast ndhF gene sequences. Taxon 64, 39–48. doi: 10.12705/641.21
Adhikari, B., Pendry, C. A., and Möller, M. (2014). New chromosome counts of Berberis L. (Berberidaceae) suggest that polyploid does not play a significant role in the diversification of the genus in the Nepal Himalaya. Edinburgh J. Bot. 71, 297–308. doi: 10.1017/S0960428614000158
Ahrendt, L. W. A. (1961). Berberis and Mahonia. A taxonomic revision. Bot. J. Linn. Soc. 57, 1–410. doi: 10.1111/j.1095-8339.1961.tb00889.x
Airy Shaw, H. K. (1973). J. C. Willis’ A Dictionary of the Flowering Plants and Ferns, 8th Edn. Cambridge, England: Cambridge University Press.
Anderson, C. L., Bremer, K., and Friis, E. M. (2005). Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am. J. Bot. 92, 1737–1748. doi: 10.3732/ajb.92.10.1737
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Available Online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Antonelli, A., and Sanmartín, I. (2011). Mass extinction, gradual cooling, or rapid radiation? Reconstructing the spatiotemporal evolution of the ancient angiosperm genus Hedyosmum (Chloranthaceae) using empirical and simulated approaches. Syst. Biol. 60, 596–615. doi: 10.1093/sysbio/syr062
Backlund, A., and Bremer, K. (1998). To be or not to be—principles of classification and monotypic plant families. Taxon 47, 391–400. doi: 10.2307/1223768
Baker, W. J., Bailey, P., Barber, V., Barker, A., Bellot, S., Bishop, D., et al. (2021). A comprehensive phylogenomic platform for exploring the angiosperm tree of life. Syst. Biol. 2021:syab035. doi: 10.1093/sysbio/syab035
Barnes, G., Saunders, D. G. O., and Williamson, T. (2020). Banishing barberry: The history of Berberis vulgaris prevalence and wheat stem rust incidence across Britain. Plant Pathol. 69, 1193–1202. doi: 10.1111/ppa.13231
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borowiec, M. L. (2016). AMAS: a fast tool for alignment manipulation and computing of summary statistics. Peer J. 4:e1660. doi: 10.7717/peerj.1660
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchene, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Boufford, D. E. (2013). Mahonia (Berberidaceae) in Asia: typification, synonymy, and notes. Mem. N.Y. Bot. Gard. 108, 251–283.
Breckenridge, F. G. I. (1983). Berberis swaseyi—Facing natural extinction? Texas Native Pl. Soc. News 1:1.
Brückner, C. (2000). Clarification of the carpel number in Papaverales, Capparales, and Berberidaceae. Bot. Rev. 66, 155–307. doi: 10.1007/Bf02858151
Carlquist, S. (1995). Wood anatomy of Berberidaceae: Ecological and phylogenetic considerations. Aliso 14, 85–103.
Chen, X.-H., Xiang, K.-L., Lian, L., Peng, H.-W., Erst, A. S., Xiang, X.-G., et al. (2020). Biogeographic diversification of Mahonia (Berberidaceae): Implications for the origin and evolution of East Asian subtropical evergreen broadleaved forests. Mol. Phylogenet. Evol. 151:e106910. doi: 10.1016/j.ympev.2020.106910
Christenhusz, M. J. M., and Byng, J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217. doi: 10.11646/phytotaxa.261.3.1
Christenhusz, M. J. M., Fay, M. F., and Byng, J. W. (2018). The Global Flora: In GLOVAP Nomenclature Part 1, Special Edn, Vol. 4. Bradford: Plant Gateway Ltd.
Colin, O., Hinsinger, D. D., and Strijk, J. S. (2021). Mahonia lancasteri (Berberidaceae), a new species originating from Sichuan (China) described from cultivation. Phytotaxa 482, 45–54. doi: 10.11646/phytotaxa.482.1.5
Donoghue, P. C. J., and Benton, M. J. (2007). Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 22, 424–431. doi: 10.1016/j.tree.2007.05.005
Doweld, A. B. (2018). New names of fossil Berberidaceae. Phytotaxa 351, 72–80. doi: 10.11646/phytotaxa.351.1.6
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fajardo, D., Senalik, D., Ames, M., Zhu, H. Y., Steffan, S. A., Harbut, R., et al. (2013). Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet. Genomes 9, 489–498. doi: 10.1007/s11295-012-0573-9
Folk, R. A., Siniscalchi, C. M., and Soltis, D. E. (2020). Angiosperms at the edge: Extremity, diversity, and phylogeny. Plant Cell Environ. 43, 2871–2893. doi: 10.1111/pce.13887
Galasso, G., Conti, F., Peruzzi, L., Ardenghi, N. M. G., Banfi, E., Celesti-Grapow, L., et al. (2018). An updated checklist of the vascular flora alien to Italy. Plant Biosyst. 152, 556–592. doi: 10.1080/11263504.2018.1441197
García, N., Folk, R. A., Meerow, A. W., Chamala, S., Gitzendanner, M. A., de Oliveira, R. S., et al. (2017). Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae). Mol. Phylogenet. Evol. 111, 231–247. doi: 10.1016/j.ympev.2017.04.003
Gitzendanner, M. A., Soltis, P. S., Yi, T.-S., Li, D.-Z., and Soltis, D. E. (2018). “Plastome phylogenetics: 30 Years of inferences into plant evolution,” in Advances in Botanical Research, Vol. 85, eds S.-M. Chaw and R. K. Jansen (Cambridge: Academic Press), 293–313. doi: 10.1016/bs.abr.2017.11.016
Goremykin, V. V., Holland, B., Hirsch-Ernst, K. I., and Hellwig, F. H. (2005). Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol. Biol. Evol. 22, 1813–1822. doi: 10.1093/molbev/msi173
Goulding, S. E., Olmstead, R. G., Morden, C. W., and Wolfe, K. H. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 252, 195–206. doi: 10.1007/Bf02173220
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guisinger, M. M., Kuehl, J. N. V., Boore, J. L., and Jansen, R. K. (2008). Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. U.S.A. 105, 18424–18429. doi: 10.1073/pnas.0806759105
Gurdon, C., and Maliga, P. (2014). Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula. DNA Res. 21, 417–427. doi: 10.1093/dnares/dsu007
Hao, D.-C. (2018). Ranunculales Medicinal Plants: Biodiversity, Chemodiversity and Pharmacotherapy. London: Academic Press, doi: 10.1016/C2017-0-01185-0
Harms, R. T. (2007). A field study of hybridization between Berberis swaseyi and B. trifoliolata (Berberidaceae) in Hays County, Texas. Lundellia 10, 18–31.
Harris, M. E., Meyer, G., Vandergon, T., and Vandergon, V. O. (2013). Loss of the acetyl-CoA carboxylase (accD) gene in Poales. Plant Mol. Biol. Rep. 31, 21–31. doi: 10.1007/s11105-012-0461-3
He, J., Lyu, R., Luo, Y., Xiao, J., Xie, L., Wen, J., et al. (2021). A phylotranscriptome study using silica gel-dried leaf tissues produces an updated robust phylogeny of Ranunculaceae. bioRxiv 2021:454256. doi: 10.1101/2021.07.29.454256
He, J. M., and Mu, Q. (2015). The medicinal uses of the genus Mahonia in traditional Chinese medicine: An ethnopharmacological, phytochemical and pharmacological review. J. Ethnopharmacol. 175, 668–683. doi: 10.1016/j.jep.2015.09.013
He, P., Ma, Q., Dong, M., Yang, Z., and Liu, L. (2019). The complete chloroplast genome of Leontice incerta and phylogeny of Berberidaceae. Mitochondrial DNA Part B 4, 101–102. doi: 10.1080/23802359.2018.1536489
Heenan, P. B., and Smissen, R. D. (2013). Revised circumscription of Nothofagus and recognition of the segregate genera Fuscospora, Lophozonia, and Trisyngyne (Nothofagaceae). Phytotaxa 146, 1–31. doi: 10.11646/phytotaxa.146.1.1
Heywood, V. H., Brummitt, R. K., Culham, A., and Selders, D. (2007). Flowering Plant Families of the World. Ontario: Firefly Books.
Hong, C. P., Park, J., Lee, Y., Lee, M., Park, S. G., Uhm, Y., et al. (2017). accD nuclear transfer of Platycodon grandiflorum and the plastid of early Campanulaceae. BMC Genomics 18:e607. doi: 10.1186/s12864-017-4014-x
Huang, J., Su, T., Lebereton-Anberree, J., Zhang, S. T., and Zhou, Z. K. (2016). The oldest Mahonia (Berberidaceae) fossil from East Asia and its biogeographic implications. J. Plant. Res. 129, 209–223. doi: 10.1007/s10265-015-0775-y
Huang, Y.-L., Tseng, Y.-H., Chung, K.-F., and Yu, C.-C. (2018). Chromosome numbers of Berberis Sect. Wallichianae from Taiwan: a new basis for taxonomic and evolutionary implications. Taiwania 63, 111–118. doi: 10.6165/tai.2018.63.111
Humphreys, A. M., and Linder, H. P. (2009). Concept versus data in delimitation of plant genera. Taxon 58, 1054–1074. doi: 10.1002/tax.584002
Hutchinson, J. (1973). The Families of Flowering Plants: Arranged According to a New System Based on Their Probable Phylogeny, 3rd Edn. Oxford: Oxford University Press.
Janchen, E. (1949). Die systematische Gliederung der Ranunculaceen und Berberidaceen. Denkschr. Akad. Wiss. Wien, Math.-Naturwiss. Kl. 108, 1–82.
Jensen, U. (1973). “The interpretation of comparative serological results,” in Chemistry in Botanical Classification: Proceedings of the Twenty Fifth Nobel Symposium, eds G. Bendz and J. Santesson (New York and London: Academic Press), 217–227.
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome. Biol. 21:e241. doi: 10.1186/s13059-020-02154-5
Johnson, M. G., Pokorny, L., Dodsworth, S., Botigue, L. R., Cowan, R. S., Devault, A., et al. (2019). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 68, 594–606. doi: 10.1093/sysbio/syy086
Kainer, D., and Lanfear, R. (2015). The effects of partitioning on phylogenetic inference. Mol. Biol. Evol. 32, 1611–1627. doi: 10.1093/molbev/msv026
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/Nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, Y. D., and Jansen, R. K. (1994). Characterization and phylogenetic distribution of a chloroplast DNA rearrangement in the Berberidaceae. Plant Syst. Evol. 193, 107–114. doi: 10.1007/Bf00983544
Kim, Y.-D., and Jansen, R. K. (1995). Phylogenetic implications of chloroplast DNA variation in the Berberidaceae. Plant Syst. Evol. 9, 341–349. doi: 10.1007/978-3-7091-6612-3_36
Kim, Y. D., and Jansen, R. K. (1996). Phylogenetic implications of rbcL and ITS sequence variation in the Berberidaceae. Syst. Bot. 21, 381–396. doi: 10.2307/2419666
Kim, Y.-D., Kim, S.-H., Kim, C.-H., and Jansen, R. K. (2004a). Phylogeny of Berberidaceae based on sequences of the chloroplast gene ndhF. Biochem. Syst. Ecol. 32, 291–301. doi: 10.1016/j.bse.2003.08.002
Kim, Y. D., Kim, S. H., and Landrum, L. R. (2004b). Taxonomic and phytogeographic implications from ITS phylogeny in Berberis (Berberidaceae). J. Plant Res. 117, 175–182. doi: 10.1007/s10265-004-0145-7
Kode, V., Mudd, E. A., Iamtham, S., and Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Kreuzer, M., Howard, C., Adhikari, B., Pendry, C. A., and Hawkins, J. A. (2019). Phylogenomic approaches to DNA Barcoding of herbal medicines: Developing clade-specific diagnostic characters for Berberis. Front. Plant Sci. 10:586. doi: 10.3389/fpls.2019.00586
Kuroki, Y. (1970). Chromosome study in four species of Berberidaceae. Mem. Ehime Univ., Sect. 2 Nat. Sci., Ser. B. 6, 215–221.
Lane, A. K., Augustin, M. M., Ayyampalayam, S., Plant, A., Gleissberg, S., Di Stilio, V. S., et al. (2018). Phylogenomic analysis of Ranunculales resolves branching events across the order. Bot. J. Linn. Soc. 187, 157–166. doi: 10.1093/botlinnean/boy015
Lee, H.-L., Jansen, R. K., Chumley, T. W., and Kim, K.-J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24, 1161–1180. doi: 10.1093/molbev/msm036
Lee, S.-R., Kim, B.-Y., and Kim, Y.-D. (2018). Genetic diagnosis of a rare myrmecochorous species, Plagiorhegma dubium (Berberidaceae): Historical genetic bottlenecks and strong spatial structures among populations. Ecol. Evol. 8, 8791–8802. doi: 10.1002/ece3.4362
Li, H.-T., Yi, T.-S., Gao, L.-M., Ma, P.-F., Zhang, T., Yang, J.-B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. doi: 10.1038/s41477-019-0421-0
Li, J., Gao, L., Chen, S. S., Tao, K., Su, Y. J., and Wang, T. (2016). Evolution of short inverted repeat in cupressophytes, transfer of accD to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci. Rep. 6:e20934. doi: 10.1038/srep20934
Li, J., Su, Y. J., and Wang, T. (2018). The repeat sequences and elevated substitution rates of the chloroplast accD gene in Cupressophytes. Front. Plant Sci. 9:533. doi: 10.3389/fpls.2018.00533
Li, R., and Dong, M. (2020). The complete plastid genome of Jeffersonia diphylla and its phylogenetic position inference. Mitochondrial DNA Part B 5, 77–78. doi: 10.1080/23802359.2019.1696250
Linder, H. P., Baeza, M., Barker, N. P., Galley, C., Humphreys, A. M., Lloyd, K. M., et al. (2010). A generic classification of the Danthonioideae (Poaceae). Ann. MO Bot. Gard. 97, 306–364. doi: 10.3417/2009006
Liu, J.-Q., Chen, Z.-D., and Lu, A.-M. (2002). Molecular evidence for the sister relationship of the eastern Asia-North American intercontinental species pair in the Podophyllum group (Berberidaceae). Bot. Bull. Acad. Sinica 43, 147–154.
Liu, T.-J., Zhang, C.-Y., Yan, H.-F., Zhang, L., Ge, X.-J., and Hao, G. (2016). Complete plastid genome sequence of Primula sinensis (Primulaceae): structure comparison, sequence variation and evidence for accD transfer to nucleus. Peer J. 4:e2101. doi: 10.7717/peerj.2101
Liu, W., Wang, J., Yin, D. X., Yang, M., Wang, P., Han, Q. S., et al. (2016). Genetic diversity and structure of the threatened species Sinopodophyllum hexandrum (Royle) Ying. Genet. Mol. Res. 15:15028130. doi: 10.4238/gmr.15028130
Loconte, H. (1993). “Berberidaceae,” in The Families and Genera of Vascular Plants, vol. 2, Flowering plants, Dicotyledons, Magnoliid, Hamamelid and Caryophylliid Families, eds K. Kubitzki, J. G. Rohwer, and V. Bittrich (Berlin: Springer-Verlag), 147–152. doi: 10.1007/978-3-662-02899-5_14
Loconte, H., and Blackwell, W. H. (1985). Intrageneric taxonomy of Caulophyllum (Berberidaceae). Rhodora 87, 463–469.
Loconte, H., Campbell, L. M., and Stevenson, D. W. (1995). Ordinal and familial relationships of Ranunculid genera. Plant Syst. Evol. 9, 99–118. doi: 10.1007/978-3-7091-6612-3_10
Loconte, H., and Estes, J. R. (1989). Phylogenetic systematics of Berberidaceae and Ranunculales (Magnoliidae). Syst. Bot. 14, 565–579.
Lowe, T. M., and Chan, P. P. (2016). tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54–W57. doi: 10.1093/nar/gkw413
Lu, A., and Tang, Y. (2020). The Origin and Evolution of Primitive Angiosperms. Beijing: Science Press.
Ma, J., Yang, B. X., Zhu, W., Sun, L. L., Tian, J. K., and Wang, X. M. (2013). The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 528, 120–131. doi: 10.1016/j.gene.2013.07.037
MacGinitie, H. D. (1953). Fossil plants of Florissant beds, Colorado. Publ. Carnegie Inst. Washington 599, 1–198.
Magallón, S., Gomez-Acevedo, S., Sanchez-Reyes, L. L., and Hernandez-Hernandez, T. (2015). A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453. doi: 10.1111/nph.13264
Magee, A. M., Aspinall, S., Rice, D. W., Cusack, B. P., Semon, M., Perry, A. S., et al. (2010). Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20, 1700–1710. doi: 10.1101/gr.111955.110
Mao, Y.-R., Zhang, Y. H., Nakamura, K., Guan, B.-C., and Qiu, Y.-X. (2014). Developing DNA barcodes for species identification in Podophylloideae (Berberidaceae). J. Syst. Evol. 52, 487–499. doi: 10.1111/jse.12076
Mao, Y. R., Zhang, Y. H., Xu, C., and Qiu, Y. X. (2016). Comparative transcriptome resources of two Dysosma species (Berberidaceae) and molecular evolution of the CYP719A gene in Podophylloideae. Mol. Ecol. Resour. 16, 228–241. doi: 10.1111/1755-0998.12415
Meacham, C. A. (1980). Phylogeny of the Berberidaceae with an evaluation of classifications. Syst. Bot. 5, 149–172.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2011). The CIPRES science gateway: a community resource for phylogenetic analyses,” in Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Vol. 41. New York, NY: ACM, doi: 10.1145/2016741.2016785
Minh, B. Q., Nguyen, M. A. T., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Morales-Briones, D. F., Liston, A., and Tank, D. C. (2018). Phylogenomic analyses reveal a deep history of hybridization and polyploidy in the Neotropical genus Lachemilla (Rosaceae). New Phytol. 218, 1668–1684. doi: 10.1111/nph.15099
Moran, R. (1982). Berberis claireae, a new species from Baja California; and why not Mahonia. Phytologia 52, 221–226.
Mower, J. P., and Vickrey, T. L. (2018). “Structural diversity among plastid genomes of land plants,” in Advances in Botanical Research, Vol. 85, eds S.-M. Chaw and R. K. Jansen (Amsterdam: Elsevier), 263–292. doi: 10.1016/bs.abr.2017.11.013
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nürk, N. M., Uribe-Convers, S., Gehrke, B., Tank, D. C., and Blattner, F. R. (2015). Oligocene niche shift, Miocene diversification—cold tolerance and accelerated speciation rates in the St. John’s Worts (Hypericum, Hypericaceae). BMC Evol. Biol. 15:80. doi: 10.1186/s12862-015-0359-4
Peng, Y., CHen, S.-B., Liu, Y., Chen, S.-L., and Xiao, P.-G. (2006). A pharmacophylogenetic study of the Berberidaceae (s.l.). Acta Phytotax. Sin. 44, 241–257. doi: 10.1360/aps040149
Peterson, P. D. (2018). The barberry eradication program in Minnesota for stem rust control: A case study. Annu. Rev. Phytopathol. 56, 203–223. doi: 10.1146/annurev-phyto-080417-050133
Popelka, O., Sochor, M., and Duchoslav, M. (2019). Reciprocal hybridization between diploid Ficaria calthifolia and tetraploid Ficaria verna subsp. verna: evidence from experimental crossing, genome size and molecular markers. Bot. J. Linn. Soc. 189, 293–310. doi: 10.1093/botlinnean/boy085
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ramírez-Barahona, S., Sauquet, H., and Magallón, S. (2020). The delayed and geographically heterogeneous diversification of flowering plant families. Nat. Ecol. Evol. 4, 1232–1238. doi: 10.1038/s41559-020-1241-3
Reveal, J. L. (1955–onward). Indices Nominum Supragenericorum Plantarum Vascularium. Available Online at: http://www.plantsystematics.org/reveal/pbio/fam/allspgnames.html
Rounsaville, T. J., and Ranney, T. G. (2010). Ploidy levels and genome sizes of Berberis L. and Mahonia Nutt. species, hybrids, and cultivars. HortScience 45, 1029–1033. doi: 10.21273/HORTSCI.45.7.1029
Sastri, R. L. N. (1969). Floral morphology, embryology, and relationships of the Berberidaceae. Austral. J. Bot. 17, 69–79. doi: 10.1071/BT9690069
Shaw, J. M. H. (2002). “The genus Podophyllum,” in The Genus Epimedium and other Herbaceous Berberidaceae including the Genus Podophyllum, ed. W. T. Stearn (Kew: The Royal Botanic Gardens), 239–314.
Shee, Z. Q., Frodin, D. G., Cámara-Leret, R., and Pokorny, L. (2020). Reconstructing the complex evolutionary history of the Papuasian Schefflera radiation through herbariomics. Front. Plant Sci. 11:258. doi: 10.3389/fpls.2020.00258
Smith, S. A., and Beaulieu, J. M. (2009). Life history influences rates of climatic niche evolution in flowering plants. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 276, 4345–4352. doi: 10.1098/rspb.2009.1176
Smith, S. A., and Donoghue, M. J. (2008). Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89. doi: 10.1126/science.1163197
Smith, S. A., and Donoghue, M. J. (2010). Combining historical biogeography with niche modeling in the Caprifolium clade of Lonicera (Caprifoliaceae, Dipsacales). Syst. Biol. 59, 322–341. doi: 10.1093/sysbio/syq011
Stearn, W. T. (1938). Epimedium and Vancouveria (Berberidaceae), a monograph. J. Linn. Soc. Bot. 51, 409–535. doi: 10.1111/j.1095-8339.1937.tb01914.x
Stearn, W. T. (2002). The Genus Epimedium and other Herbaceous Berberidaceae including the Genus Podophyllum. Kew: The Royal Botanic Gardens.
Stull, G. W., Soltis, P. S., Soltis, D. E., Gitzendanner, M. A., and Smith, S. A. (2020). Nuclear phylogenomic analyses of asterids conflict with plastome trees and support novel relationships among major lineages. Am. J. Bot. 107, 790–805. doi: 10.1002/ajb2.1468
Su, H.-J., Barkman, T. J., Hao, W. L., Jones, S. S., Naumann, J., Skippington, E., et al. (2019). Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc. Natl. Acad. Sci. U.S.A. 116, 934–943. doi: 10.1073/pnas.1816822116
Sudianto, E., and Chaw, S. M. (2019). Two independent plastid accD transfers to the nuclear genome of Gnetum and other insights on acetyl-CoA carboxylase evolution in Gymnosperms. Genome Biol. Evol. 11, 1691–1705. doi: 10.1093/gbe/evz059
Sun, G., Dilcher, D. L., Wang, H.-S., and Chen, Z.-D. (2011). A eudicot from the early cretaceous of China. Nature 471, 625–628. doi: 10.1038/nature09811
Sun, Y., Moore, M. J., Landis, J. B., Lin, N., Chen, L., Deng, T., et al. (2018). Plastome phylogenomics of the early-diverging eudicot family Berberidaceae. Mol. Phylogenet. Evol. 128, 203–211. doi: 10.1016/j.ympev.2018.07.021
Sun, Y. X., Moore, M. J., Zhang, S. J., Soltis, P. S., Soltis, D. E., Zhao, T. T., et al. (2016). Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol. Phylogenet. Evol. 96, 93–101. doi: 10.1016/j.ympev.2015.12.006
Takhtajan, A. (1997). Diversity and Classification of Flowering Plants. New York, NY: Columbia University Press.
Takhtajan, A. (2009). Flowering Plants, 2nd Edn. New York, NY: Springer, doi: 10.1007/978-1-4020-9609-9
Terabayashi, S. (1985c). Seedling morphology of the Berberidaceae. Acta Phytotax. Geobot. 38, 63–74. doi: 10.18942/bunruichiri.KJ00002992235
Terabayashi, S. (1985a). The comparative floral anatomy and systematics of the Berberidaceae I. Morphol. Mem. Fac. Sci. Kyoto Univ. Ser. Biol. 10, 73–90.
Terabayashi, S. (1985b). The comparative floral anatomy and systematics of the Berberidaceae II. Systematic consideration. Acta Phytotax. Geobot. 36, 1–13. doi: 10.18942/bunruichiri.KJ00001078521
Thorne, R. F. (1992). Classification and geography of the flowering plants. Bot. Rev. 58, 225–327. doi: 10.1007/Bf02858611
Thorne, R. F. (2000). The classification and geography of the flowering plants: Dicotyledons of the Class Angiospermae (Subclasses Magnoliidae, Ranunculidae, Caryophyllidae, Dilleniidae, Rosidae, Asteridae, and Lamiidae). Bot. Rev. 66, 441–647. doi: 10.1007/BF02869011
Thorne, R. F., and Reveal, J. L. (2007). An updated classification of the class Magnoliopsida (”Angiospermae”). Bot. Rev. 73, 67–181.
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Tonti-Filippini, J., Nevill, P. G., Dixon, K., and Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. 90, 808–818. doi: 10.1111/tpj.13491
Turland, N. J., Wiersema, J. H., Barrie, F. R., Greuter, W., Hawksworth, D. L., Herendeen, P. S., et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Glashütten: Koeltz Botanical Books, doi: 10.12705/Code.2018
Wang, L.-J., Gao, M.-D., Sheng, M.-Y., and Yin, J. (2020). Cluster analysis of karyotype similarity coefficients in Epimedium (Berberidaceae): insights in the systematics and evolution. Phytokeys 161, 11–26. doi: 10.3897/phytokeys.161.51046
Wang, M. L., Chen, Y., Hina, F., Ohi-Toma, T., and Li, P. (2018). The complete chloroplast genome of Ranzania japonica, an endangered species native to Japan. Conserv. Genet. Resour. 10, 671–674. doi: 10.1007/s12686-017-0898-7
Wang, W., Chen, Z.-D., Liu, Y., Li, R.-Q., and Li, J.-H. (2007). Phylogenetic and biogeographic diversification of Berberidaceae in the northern hemisphere. Syst. Bot. 32, 731–742. doi: 10.1043/06-16.1
Wang, W., Dilcher, D. L., Sun, G., Wang, H.-S., and Chen, Z.-D. (2016). Accelerated evolution of early angiosperms: Evidence from ranunculalean phylogeny by integrating living and fossil data. J. Syst. Evol. 54, 336–341. doi: 10.1111/jse.12090
Wang, W., Lu, A.-M., Ren, Y., Endress, M. E., and Chen, Z.-D. (2009). Phylogeny and classification of Ranunculales: Evidence from four molecular loci and morphological data. Perspect. Plant Ecol. Evol. Syst. 11, 81–110. doi: 10.1016/j.ppees.2009.01.001
Wang, W., Ortiz, R. D., Jacques, F. M. B., Xiang, X.-G., Li, H.-L., Lin, L., et al. (2012). Menispermaceae and the diversification of tropical rainforests near the Cretaceous-Paleogene boundary. New Phytol. 195, 470–478. doi: 10.1111/j.1469-8137.2012.04158.x
Wen, J., Ickert-Bond, S., Nie, Z.-L., and Li, R. (2010). “Timing and modes of evolution of Eastern Asian-North American biogeographic disjunctions in seed plants,” in Darwin’s Heritage Today: Proceedings of the Darwin 200 Beijing International Conference, eds M. Long, H. Gu, and Z. Zhou (Beijing: Higher Education Press), 252–269.
Wendel, J. F., and Doyle, J. J. (1998). “Phylogenetic incongruence: Window into genome history and molecular evolution,” in Molecular Systematics of Plants II: DNA Sequencing, eds D. E. Soltis, P. S. Soltis, and J. J. Doyle (Dordrecht: Kluwer Academic), 265–296. doi: 10.1007/978-1-4615-5419-6_10
Wicke, S., and Schneeweiss, G. M. (2015). “Next-generation organellar genomics: Potentials and pitfalls of high-throughput technologies for molecular evolutionary studies and plant systematics,” in Next-Generation Sequencing in Plant Systematics, eds E. Hörandl and M. Appelhans (Oberreifenberg: Koeltz Botanical Books), 1–42. doi: 10.14630/000002
Wu, Z., Lu, A., Tang, Y., Chen, Z., and Li, D. (2003). The Families and Genera of Angiosperms in China: A Comprehensive Analysis. Beijing: Science Press.
Xing, Y.-W., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yao, Y., Liu, X., Yang, Q., Luo, Y., Zhang, C., Xu, C., et al. (2020). The complete chloroplast genome of Epimedium brevicornu (Berberidaceae), a traditional Chinese medicinal herb. Mitochondrial DNA Part B 5, 887–888. doi: 10.1080/23802359.2020.1718027
Ye, W.-Q., Yap, Z. Y., Li, P., Comes, H. P., and Qiu, Y.-X. (2018). Plastome organization, genome-based phylogeny and evolution of plastid genes in Podophylloideae (Berberidaceae). Mol. Phylogenet. Evol. 127, 978–987. doi: 10.1016/j.ympev.2018.07.001
Ying, T.-S. (1979). On Dysosma Woodson and Sinopodophyllum Ying, gen. nov. of the Berberidaceae. Acta Phytotax. Sin. 17, 15–23.
Ying, T.-S., Terabayashi, S., and Boufford, D. E. (1984). A monograph of Diphylleia (Berberidaceae). J. Arnold Arbor. 65, 57–94.
Yu, C.-C. (2018). Molecular Phylogenetics and Historical Biogeography of Berberis L. (Berberidaceae). Ph.D. Dissertation. Taipei: National Taiwan University.
Yu, C.-C., and Chung, K.-F. (2017). Why Mahonia? Molecular recircumscription of Berberis s.l., with the description of two genera, Alloberberis and Moranothamnus. Taxon 66, 1371–1392. doi: 10.12705/666.6
Zhang, M. L., Uhink, C. H., and Kadereit, J. W. (2007). Phylogeny and biogeography of Epimedium/Vancouveria (Berberidaceae): Western North American-East Asian disjunctions, the origin of European mountain plant taxa, and East Asian species diversity. Syst. Bot. 32, 81–92. doi: 10.1600/036364407780360265
Zhang, M. Y., Lu, L., Wortley, A. H., Wang, H., Li, D. Z., and Blackmore, S. (2017). Evolution of angiosperm pollen: 4. Basal eudicots. Ann. MO. Bot. Gard. 102, 141–182. doi: 10.3417/2015035
Zhang, Q., Liu, Y., and Sodmergen. (2003). Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 44, 941–951. doi: 10.1093/pcp/pcg121
Zhang, Q., Ree, R. H., Salamin, N., Xing, Y., and Silvestro, D. (2021). Fossil-informed models reveal a boreotropical origin and divergent evolutionary trajectories in the Walnut Family (Juglandaceae). Syst. Biol. 2021:syab065. doi: 10.1093/sysbio/syab030
Zhang, Y., Huang, R., Wu, L., Wang, Y., Jin, T., and Liang, Q. (2020). The complete chloroplast genome of Epimedium brevicornu Maxim (Berberidaceae), a traditional Chinese medicine herb. Mitochondrial DNA Part B 5, 588–590. doi: 10.1080/23802359.2019.1710593
Zhang, Y. J., Du, L. W., Liu, A., Chen, J. J., Wu, L., Hu, W. M., et al. (2016). The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 7:306. doi: 10.3389/fpls.2016.00306
Zheng, G., Zhang, C., Yang, J., and Xu, X. (2019). Characterization of the complete chloroplast genome of Epimedium brevicornu (Berberidaceae). Mitochondrial DNA Part B 4, 3681–3682. doi: 10.1080/23802359.2019.1678429
Zhou, Z.-H. (2014). The jehol biota, an early cretaceous terrestrial lagerstätte: new discoveries and implications. Natl. Sci. Rev. 1, 543–559. doi: 10.1093/nsr/nwu055
Keywords: accD length variation, cytonuclear discordance, IR expansion, molecular dating, tribal classification
Citation: Hsieh C-L, Yu C-C, Huang Y-L and Chung K-F (2022) Mahonia vs. Berberis Unloaded: Generic Delimitation and Infrafamilial Classification of Berberidaceae Based on Plastid Phylogenomics. Front. Plant Sci. 12:720171. doi: 10.3389/fpls.2021.720171
Received: 03 June 2021; Accepted: 15 October 2021;
Published: 06 January 2022.
Edited by:
Susann Wicke, Humboldt University of Berlin, GermanyReviewed by:
Ana Otero, Field Museum of Natural History, United StatesRoss McCauley, Fort Lewis College, United States
Diego F. Morales-Briones, Ludwig Maximilian University of Munich, Germany
Copyright © 2022 Hsieh, Yu, Huang and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuo-Fang Chung, Ym9jaHVuZ0BnYXRlLnNpbmljYS5lZHUudHc=
 Chia-Lun Hsieh
Chia-Lun Hsieh Chih-Chieh Yu
Chih-Chieh Yu Yu-Lan Huang1
Yu-Lan Huang1 Kuo-Fang Chung
Kuo-Fang Chung