
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 October 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.715456
Abiotic stresses, such as salinity, drought, and nutrient deficiency adversely affect nitrogen (N) uptake and assimilation in plants. However, the regulation of N metabolism and N pathway genes in Sophora japonica under abiotic stresses is unclear. Sophora japonica seedlings were subjected to drought (5% polyethylene glycol 6,000), salinity (75mM NaCl), or low N (0.01mM NH4NO3) for 3weeks in a semi-hydroponic phenotyping platform. Salinity and low N negatively affected plant growth, while drought promoted root growth and inhibited aboveground growth. The NH4+/NO3− ratio increased under all three treatments with the exception of a reduction in leaves under salinity. Drought significantly increased leaf NO2− concentrations. Nitrate reductase (NR) activity was unaltered or increased under stresses with the exception of a reduction in leaves under salinity. Drought enhanced ammonium assimilation with increased glutamate synthase (GOGAT) activity, although glutamine synthetase (GS) activity remained unchanged, whereas salinity and low N inhibited ammonium assimilation with decreased GS activity under salt stress and decreased GOGAT activity under low N treatment. Glutamate dehydrogenase (GDH) activity also changed dramatically under different stresses. Additionally, expression changes of genes involved in N reduction and assimilation were generally consistent with related enzyme activities. In roots, ammonium transporters, especially SjAMT1.1 and SjAMT2.1a, showed higher transcription under all three stresses; however, most nitrate transporters (NRTs) were upregulated under salinity but unchanged under drought. SjNRT2.4, SjNRT2.5, and SjNRT3.1 were highly induced by low N. These results indicate that N uptake and metabolism processes respond differently to drought, salinity, and low N conditions in S. japonica seedlings, possibly playing key roles in plant resistance to environmental stress.
Excessive drought, salinity, and nutrient limitations are the main forms of abiotic stresses that significantly affect plant growth and development. Currently, it is estimated that more than 6% of the world’s land and 20% of the total irrigated area are affected by salt with major affliction in the Asian countries (Munns and Tester, 2008; FAO and ITPS, 2015). Forest trees are expected to face more frequent and severe droughts as a result of climate change (Plattner and GianKasper, 2014). Such soil conditions will further reduce nitrogen (N) availability and accelerate N limitation during the vegetative growth periods (Plattner and GianKasper, 2014). In agriculture, N limitation is overcome by the application of N fertilizers. However, trees usually have a high distribution on marginal soils with low N availability and are rarely managed. Plants, particularly long-living trees, have evolved various strategies to cope with different stressful conditions. N is a constituent of many plant cell components, such as chlorophyll, nucleic acids, proteins, amino acids, and some hormones which are involved in the ability of plants to resist abiotic stresses through different mechanisms (Kamel, 2012; Rais et al., 2013). Exploring the N acquisition strategy and improving the N uptake efficiency under stressful conditions are of great significance for maintaining the healthy growth of trees, breeding resistant varieties, and providing feasible management methods.
Nitrogen is one of the essential macronutrients for plant growth and development. Higher plants absorb inorganic NH4+ and NO3− via specific ammonium (AMTs) and nitrate transporters (NRTs; Kaiser et al., 2002; Wang et al., 2018b). Subsequently, nitrate assimilation depends on two enzymes, nitrate reductase (NR) and nitrite reductase (NiR), to convert NO3− to NH4+ (Xu et al., 2012). More than 95% of ammonium assimilation is performed via glutamine synthetase (GS) and glutamate synthase (GOGAT) in plants (Suárez et al., 2002). NH4+ can also be assimilated by the conversion of a-oxoglutarate to glutamate via glutamate dehydrogenase (GDH). N transport and assimilation processes are regulated by internal and external signals, leading to differences in N metabolism of plants in response to different environmental stresses. The variation in nitrogen concentrations, enzyme activities, and transcript levels of genes related to N utilization can reflect the accumulation strategies of nitrogen under different stresses.
Drought and excessive amounts of salt are known to negatively affect plant growth and productivity (Boyer, 1982; Park Williams et al., 2012; McDowell et al., 2018). Previous studies have demonstrated that Sophora japonica showed growth limitation, as indicated by declines in the stem base diameter, height increment, chlorophyll pigment contents, osmotic regulator contents, and altered antioxidative enzyme activities under both drought and salinity (Quy et al., 2017; Lu et al., 2018). Reductions in N uptake and metabolism have been observed in many tree and crop species (GeBler et al., 2005; Zhang et al., 2014; Ahanger et al., 2017; Ashraf et al., 2018). For instance, salinity decreased the net N uptake rate and total N content in Populus canescens (Dluzniewska et al., 2007). Drought stress inhibited ammonium and nitrate uptake in Malus prunifolia and beech (Rennenberg et al., 2006; Huang et al., 2018) and decreased NR, GS, and GOGAT activities in M. prunifolia (Huang et al., 2018). Genes involved in N uptake and metabolic processes also play key roles in plant resistance to extreme conditions. Meng et al. (2015) reported that AMT1.2 may play key roles in NH4+ uptake under both drought and salt stress in Populus. The enzyme-encoding genes GS, GOGAT, and GDH were induced by drought, cold, or heat stress in tomato (Liu et al., 2016). Co-overexpression of GS1.1 and GS2 enhanced tolerance to osmotic and salinity stress in rice (James et al., 2018). The physiological and molecular responses of N uptake and metabolism process to abiotic stress are widely studied in annual plants such as crops or model genera such as Arabidopsis and Populus, whereas information on trees is limited. It is worth noting that although some effects are shared between salinity and drought stress, as both stresses lower the soil water potential, differences in the potential regulatory mechanisms exist, which eventually affect N absorption, reduction, and assimilation differently. Generally, drought reduces N uptake and translocation, as well as NR and GS activities, because of restricted photosynthetic and transpiration rates (Bowen et al., 2018; Iqbal et al., 2020). Apart from the results caused by limited water absorption, salt stress adds additional negative effects. For example, high concentrations of Na+ and Cl− cause ion toxicities and ionic imbalances, which can directly inhibit N transport and assimilation processes (Flores et al., 2000; Khare et al., 2020). Our previous research showed that salinization, 50mM NaCl application for 10days, prominently affected the root system of S. japonica seedlings, causing a less developed root system, but under drought, the roots thrived (Quy et al., 2017). Therefore, it is necessary to verify the underlying regulatory differences between drought and salinity, particularly, in the N utilization process.
Sophora japonica, also known as Chinese scholar tree, is a leguminous tree species native to China and is cultivated in Vietnam, Japan, Korea, and other regions. High stress tolerance contributes to its long lifespan of thousands of years. Sophora japonica has been widely planted recently because of its medicinal and economic value. Most legumes can alleviate N limitation by fixing atmospheric N via nodules in soils, but S. japonica is an exception that lacks this ability, so its growth is inhibited under low N conditions unlike that of other legume species such as Robinia pseudoacacia that can fix N (Yin et al., 2006; Wang et al., 2018a). However, little is known about the mechanisms of N uptake and metabolism of S. japonica under low N conditions. Few have studied the molecular regulation of N utilization due to the lack of genomic resources for S. japonica in the past, but we have discovered related candidate genes through transcriptome sequencing (Tian et al., 2021). This study provides the basis to study the N uptake and assimilation process of S. japonica in response to low N conditions.
In this study, to understand the morphological, physiological, and molecular responses of S. japonica to drought, salinity, and low N stresses, we performed a controlled experiment using a semi-hydroponic phenotyping platform that is quite efficient for studying root architecture (Chen et al., 2011). Morphological (shoot and root traits), physiological (photosynthesis, chlorophyll content, total N concentration, accumulation of NH4+, NO3−, and NO2−, and activities of N metabolism enzymes, including NR, NiR, GS, GOGAT, and GDH), and molecular (transcript levels of genes involved in N uptake and metabolism) indices were assessed. It was hypothesized that (1) the process of N uptake and metabolism would change significantly in response to the three stress treatments and (2) Sophora japonica would exhibit differential root growth under drought and salinity stresses. Studying the effects of different stresses on N uptake and metabolism is of particular importance for the generation of resistant varieties of S. japonica using transgenic approaches to improve N use efficiency and tolerance to abiotic stresses.
Seeds of S. japonica were purchased from Shaanxi Sanyuan Gold Seed Industry Technology Co., Ltd., Yangling Branch. After the seeds germinated, uniform seedlings about 3days old were transferred to a semi-hydroponic phenotyping platform, which was established as Chen et al. (2011) described (Figures 1A,B). Four bins, each accommodating 36 plants, were placed in a greenhouse (natural light, day/night 28/20°C, and 80% relative humidity) at Northwest A&F University, Yangling (34°16’N, 108°4''E). Each bin contained 30L of nutrition solution (Meng et al., 2015) supplemented with 1mM NH4NO3 as the N source. The solution was renewed weekly throughout the experiment.
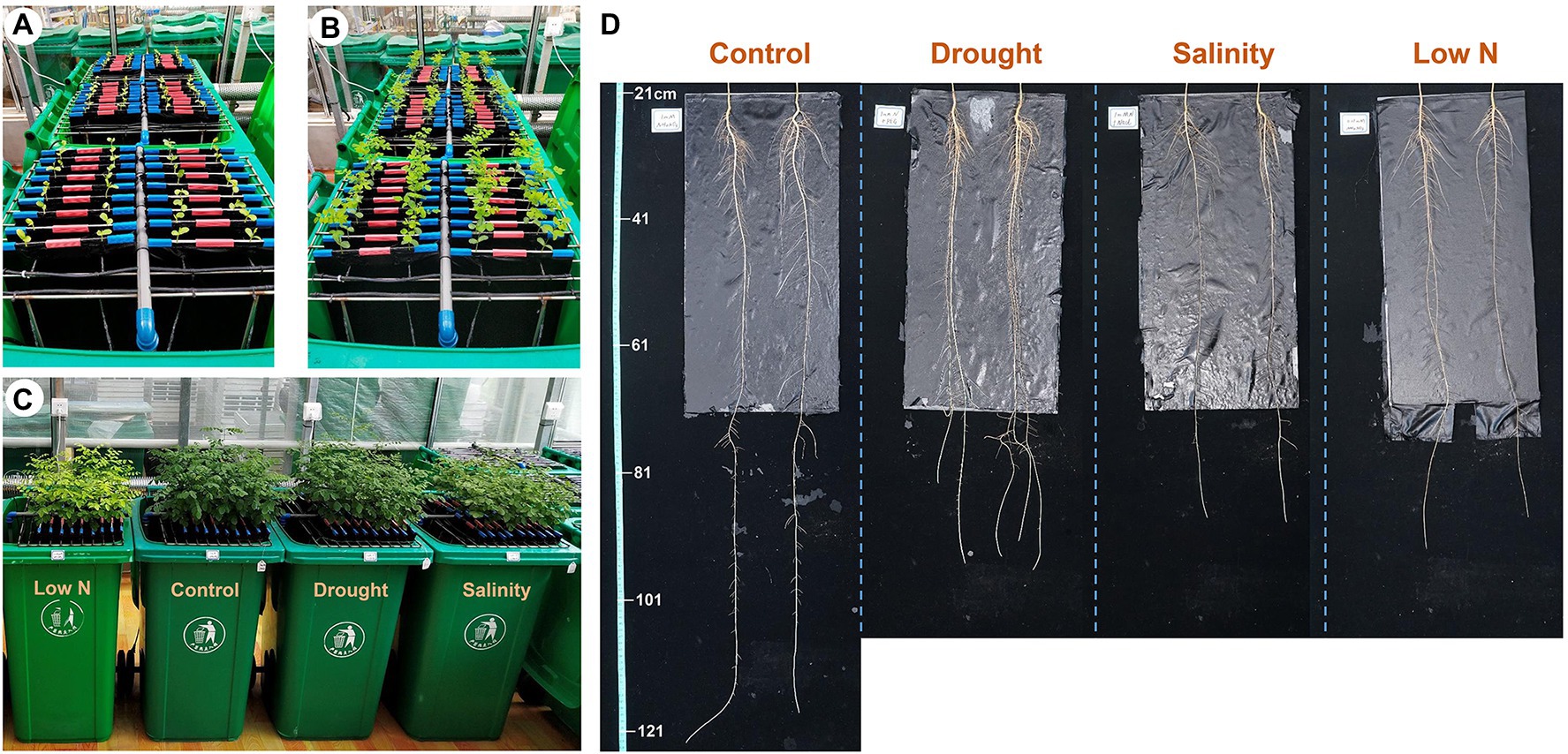
Figure 1. Sophora japonica seedlings grown in a semi-hydroponic phenotyping platform, 1day (A) and 7days (B) after transplanting; aboveground growth (C) and root morphology (D) of plants under control, drought, salinity, and low N treatment at harvest.
We set up three stress treatments (one treatment one bin) in this study. The saplings that grew normally for 7weeks were used as controls. The drought, salinity, and low N treatments commenced 4weeks after transfer and lasted for 3weeks before harvesting. Mild drought stress was applied by adding 5% polyethylene glycol (PEG) 6,000 to the solution. Moderate salt stress was increased gradually by applying 25, 50, and finally 75mM NaCl on the first, fourth, and seventh days of treatment to avoid early lethal damage. Then, NaCl concentration was maintained at 75mM during the remaining 2weeks until harvest. Low N stress was initiated by setting the N concentration at 0.01mM. As Populus is the model tree species used in studies on woody plant molecular biology, the choice of PEG-6000, NaCl, and NH4NO3 concentrations in this study was based on our previous studies on Populus simonii stress resistance in a hydroponic system (Meng et al., 2015, 2016). Samples were harvested when seedlings showed obvious phenotypic differences among treatments at the end of third week after the stresses began.
At harvest, seedlings that were infected with pests or disease or had mechanical damage caused by experimental operations during the stress period were excluded from each treatment. As a result, approximately 80% of the plants were collected for further measurements. Shoots and roots were harvested separately. The morphological indices were determined with eight replicates (one plant one replicate) per treatment. Three biological replicates, each containing a pool of four different plants, were rapidly frozen in liquid nitrogen for all physiological and molecular assays.
The leaf net photosynthetic rate was measured 1day before harvest from 9:00 to 11:00h on the fully expanded leaves of six plants per treatment using a Li-6400 portable photosynthesis system (Li-Cor, Lincoln, NE, United States). The measurement conditions were as follows: attached LED light source 1,000μmol photon m−2s−1, CO2 concentration in the chambers 400μmolmol−1, and air flow 500μmols−1. The chlorophyll content of 12 plants in each treatment was measured with a SPAD 502 plus chlorophyll meter (Minolta SPAD 502 Meter).
The shoot height and stem base diameter were measured using a ruler. The compound leaf number (CLN) was recorded simultaneously. All the lobules of the 4–7th compound leaves from the bottom were flattened on mesh paper (1cm×1cm), photographed with a camera, and processed in Photoshop CS (Adobe Systems, San Jose, United States) to obtain the leaf area (LA). These leaves were also collected to calculate the leaf water content (LWC) as follows:
where FW is the fresh weight of leaves obtained immediately after harvest and DW is the dry weight of leaves obtained after drying at 70°C to constant weight. Root systems were photographed using a portable photo box first. The primary root length was measured manually. Then, subsamples of roots were cut into three sections (0–20, 20–40, and >40cm) according to the root depth along the glass sheet. All root section samples were scanned and analyzed by a WinRHIZO root analyzer system (WinRHIZO version 2007b, Regent Instruments Canada, Montreal, Canada) in the laboratory. Root morphology parameters, such as root length, root surface area (TRSA), and root diameter were generated. After the above determinations, subsamples of roots from one plant were combined into one root sample. Aerial biomass (AB) and total root dry mass were recorded after oven-drying at 70°C for 72h. Leaves and roots were then ground into powder for measurement of the total N concentration by dry combustion using an elemental analyzer (Vario MAX CN Elemental Analyzer, Elementar, Germany).
Concentrations of NH4+ in the leaves and roots were calculated based on the Berthelot reaction (Bräutigam et al., 2007). The concentration of NO3− was analyzed as described by Patterson et al. (2010). The concentration of NO2− was quantified as described by Ogawa et al. (1999). The detailed laboratory experimental processes were reported in our previous study (Meng et al., 2015).
The activities of NR and GOGAT in the roots and leaves were assayed according to the methods previously described (Högberg et al., 1986; Singh and Srivastava, 1986), and they were defined as the catalytic reduction in the amount of nicotinamide adenine dinucleotide (NADH) per gram fresh weight sample per minute. The activity of NiR was defined as the reduction in the amount of NO2− per gram fresh weight sample per hour (Seith et al., 1994). GS activity was analyzed spectrophotometrically (Yu and Zhang, 2012) and defined as the amount of γ-glutamyl isohydroxamic acid produced per gram fresh weight sample per minute. GDH activity was measured using a physiological assay kit (GDH-1-Y, Suzhou Keming Biological Technology Co., Ltd., Suzhou, China) according to the manufacturers’ recommendations.
In our previous study, we conducted RNA-seq of S. japonica roots exposed to 20% PEG-6000 induced drought stress under normal N and N starvation conditions for 3h (Tian et al., 2021). Based on the transcriptomic annotation information, unigenes involved in N transport and metabolism were obtained. These protein sequences were then submitted to the Arabidopsis genome database1 for BLAST homology. When multiple sequences were annotated to the same homologous gene of Arabidopsis thaliana, only the longest was retained. Unigenes with an expression level lower than five fragments per kilobase per million mapped reads (FPKM) and no differential expression were also excluded. A phylogenetic tree of 22 identified NRT protein sequences from S. japonica and 61 NRT family members from Arabidopsis was constructed via the neighbor-joining (1,000 bootstrap replicates) method with MEGA 7.0 software (Center for Evolutionary Medicine and Informatics, Tempe, AZ, United States). According to the clustering results (Supplementary Figure S1), 12 NRTs were clustered together with those in Arabidopsis that have been proven to transport NO3−. Then 12 NRTs, as well as four AMTs and nine genes encoding N assimilation including one NR, one NiR, three GSs, two GOGATs, and two GDHs, were selected for quantitative real-time PCR (qRT-PCR) analysis in this experiment (Table 1). We provisionally named these genes based on the identities in protein BLAST results between S. japonica and A. thaliana sequences. The protein sequences of S. japonica are provided in Supplementary Table S1, and related gene accession numbers in Arabidopsis are listed in Supplementary Table S2.
Total RNA was extracted and purified from leaves and roots with a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, United States). Concentrations of RNA samples were estimated using NanoDrop 20000 (Thermo Scientific, Pittsburgh, PA, United States). Only when OD260/280 ratios were between 1.8 and 2.2 and OD260/230 ratios were higher than 2.0, could the samples be used for further experiments. First-strand cDNA synthesis was performed using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions using an anchored oligo (dT)18 primer and 500ng of total RNA. Reactions were performed on a LightCycler® 96 real-time PCR system (Roche, Mannheim, Germany) using 2×SYBR real-time PCR premixture (Bioteke, Beijing, China) in a 20μl reaction under the following conditions: one cycle of 150s at 95°C, followed by 40cycles at 95°C for 10s, 58°C for 20s, and 72°C for 30s. The primers are listed in Table 1. 18s rRNA was used as the reference gene. For each of the selected genes, three biological replicates with three technical replicates were assayed. The relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Statistical tests were performed with SPSS 23 (SPSS Inc., Chicago, IL, United States) by one-way ANOVA. The normality of all data was tested prior to further analysis. Differences between means were determined by Tukey’s test at the p<0.05 probability level.
After 3weeks of exposure to drought, salinity, and low N stresses, the shoot traits of S. japonica seedlings were indicative of growth inhibition under all three stresses, mainly showing decreases in the shoot height (SH) and leaf water content (LWC; Figure 1C; Table 2). The stem base diameter (SBD) was only reduced under salinity stress, while the CLN was only reduced under the low N treatment. LA and AB were negatively affected by both salinity and low N but had no change under drought (Table 2).
For global root traits, there was no significant difference in the average root diameter (ARD) and total root volume (TRV), but the PRL was markedly decreased under all three stresses (Figure 1D; Table 2). Drought increased the total length (TRL), surface area (TRSA), and biomass of the roots (TRDW), whereas salinity and low N had the opposite effect. The root-to-shoot dry mass ratio (RSM) had a markedly higher increase under drought, followed by low N, but did not change under salinity (Table 2).
Root traits at different depths showed different responses to stress treatments (Table 2). Root length (RL-upper) and surface area (RA-upper) in the upper layer and root surface area in the middle layer (RA-middle) were promoted by drought; salinity mainly reduced root growth in the lower section; root traits in both the upper and lower layers were inhibited by low N with the exception of root volume in the upper layer (RV-upper; Table 2).
These results suggested that the growth inhibition of S. japonica under low N was stronger than that under salt stress. Drought had a minimal negative effect on the aboveground parts but promoted root growth instead, especially in the upper root system.
In addition to the plant morphology, photosynthesis and nutrient accumulation also distinctly responded to the three abiotic stresses. Both the net photosynthetic rate and the chlorophyll content in the leaves declined under all three treatments (Figures 2A,B). Neither the total N concentrations in leaves nor roots were affected by drought. Nitrogen accumulation decreased only in leaves under salinity, and was greatly reduced in both leaves and roots under low N treatment (Figures 2C,D).
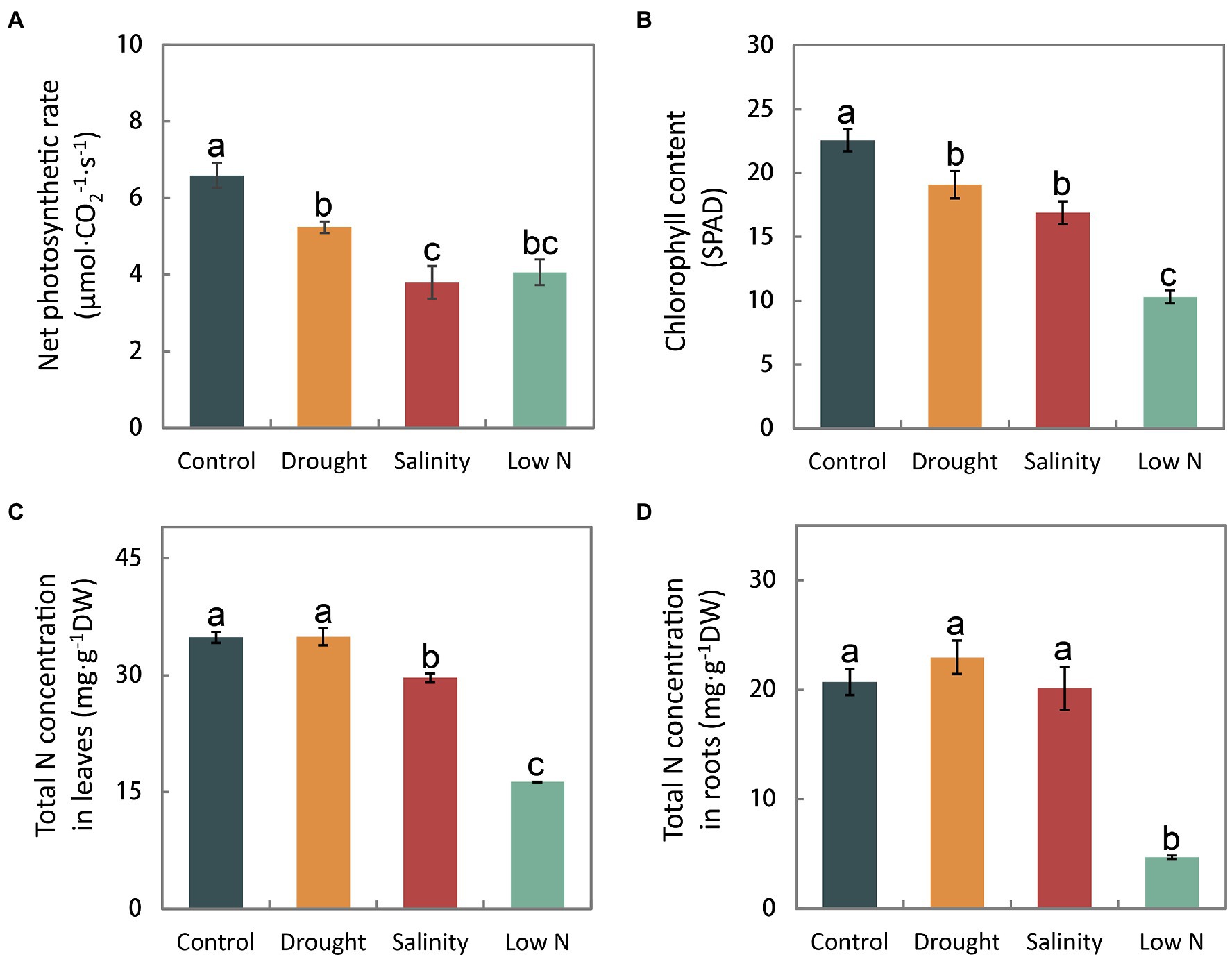
Figure 2. Net photosynthetic rate (A), chlorophyll content (B), and total N concentrations in leaves (C) and roots (D) of S. japonica plants in response to drought, salinity, and low N treatment. Data indicate means±SE of three replicates (12 plants). Bars labeled with different letters indicate significant differences between treatments (p<0.05).
We measured the NH4+, NO3−, and NO2 − concentrations in the roots and leaves of plants. Stresses caused the NH4+ concentration to increase in the roots (Figure 3A). Only low N treatment resulted in NH4+ increases in leaves (Figure 3B). The level of NO3− in leaves and roots was reduced in response to drought and low N, but under salt stress, it was not altered in roots and increased in leaves (Figures 3C,D). Moreover, the NO2− content was not significantly affected by stresses, with the exception of the increase in leaves under drought (Figures 3E,F). Furthermore, changes in the accumulation of NH4+ and NO3− led to a notable increase in NH4+/NO3− in both leaves and roots under drought and low N treatments, but the increase was higher under low N. However, different from its increase in roots under salinity, the ratio of NH4+/NO3− decreased in leaves when compared with the control level (Figures 3G,H).
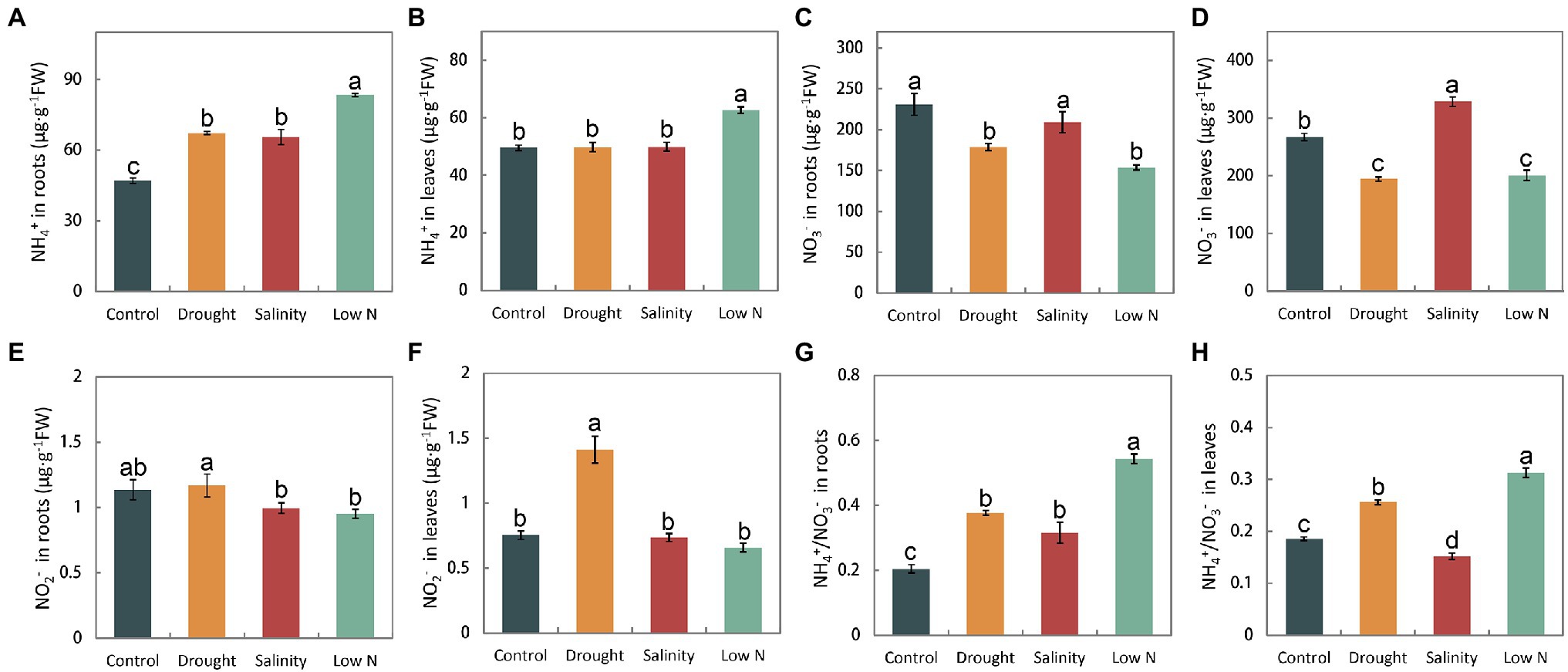
Figure 3. NH4+, NO3−, and NO2− contents, as well as NH4+/NO3− in roots (A,C,E,G) and leaves (B,D,F,H) of S. japonica as affected by drought, salinity, and low N treatment. Data indicate means±SE of three replicates (12 plants). Bars labeled with different letters indicate significant differences between treatments (p<0.05).
The activity of enzymes, such as NR, NiR, GS, GOGAT, and GDH reflects the process of N assimilation, which occurs after NH4+ and NO3− are taken into roots. In this study, we found that NR activity in the roots under drought and low N was on par with that in the control but increased significantly under salt stress (Figure 4A). NR activity in the leaves was enhanced by drought and inhibited by salinity but was not altered under the low N treatment (Figure 4B). Moreover, all three stress treatments had no effect on NiR activity in either roots or leaves (Figures 4C,D).
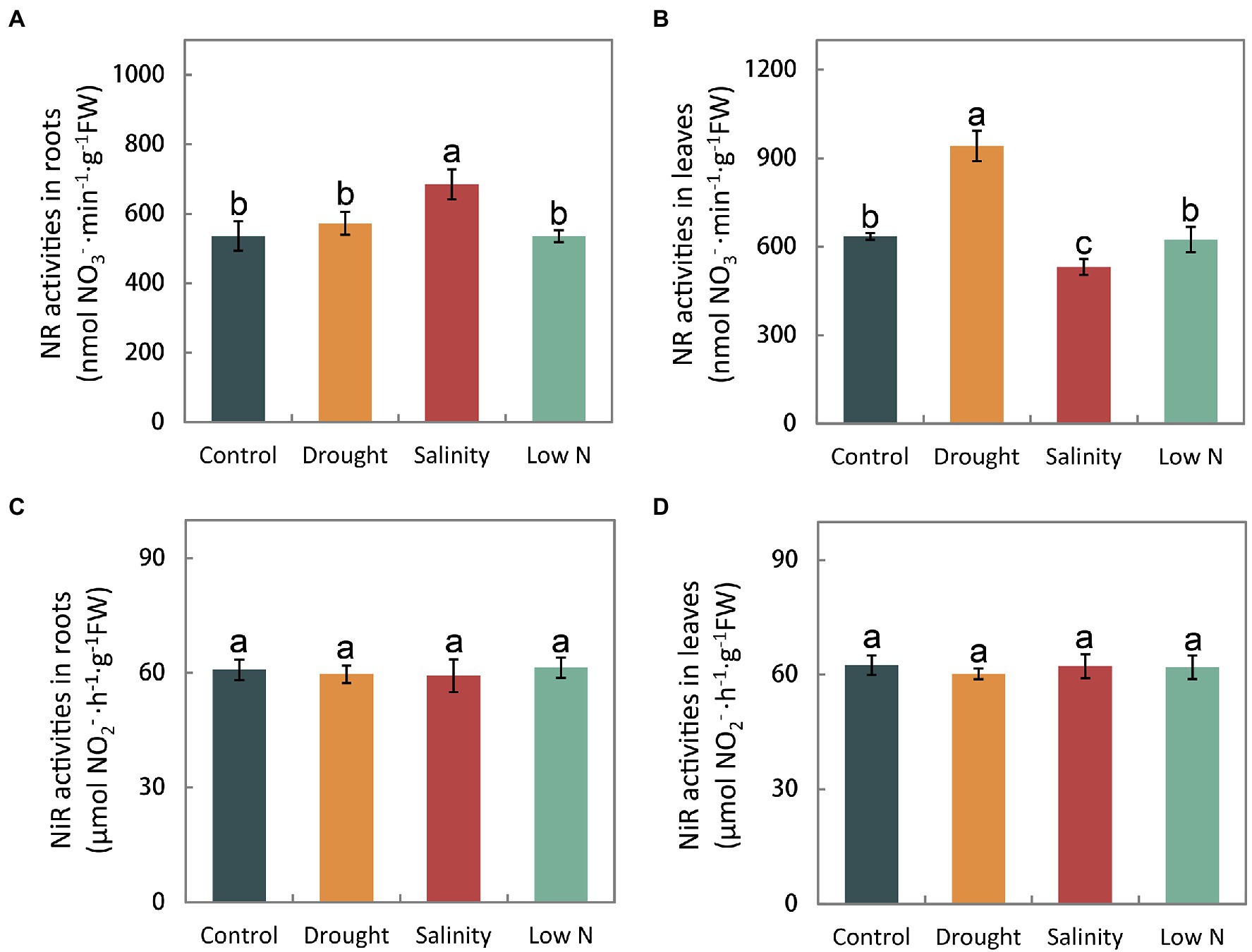
Figure 4. Activities of nitrate reductase and nitrite reductase in roots (A,C) and leaves (B,D) of S. japonica as affected by drought, salinity, and low N treatment. Data indicate means±SE of three replicates (12 plants). Bars labeled with different letters indicate significant differences between treatments (p<0.05).
There was no significant difference in the GS activities of roots and leaves under drought, but a reduction was observed under salinity. Low N also decreased GS activity in roots (Figures 5A,B). GOGAT activities increased significantly in both leaves and roots under drought treatment, greatly decreased in both leaves and roots under low N treatment, and highly increased in roots but reduced in leaves under salinity (Figures 5C,D). Additionally, GDH activity in leaves and roots increased significantly under both drought and salinity treatments but more so under salt stress. However, GDH activity was markedly reduced in both roots and leaves in response to low N treatment (Figures 5E,F).
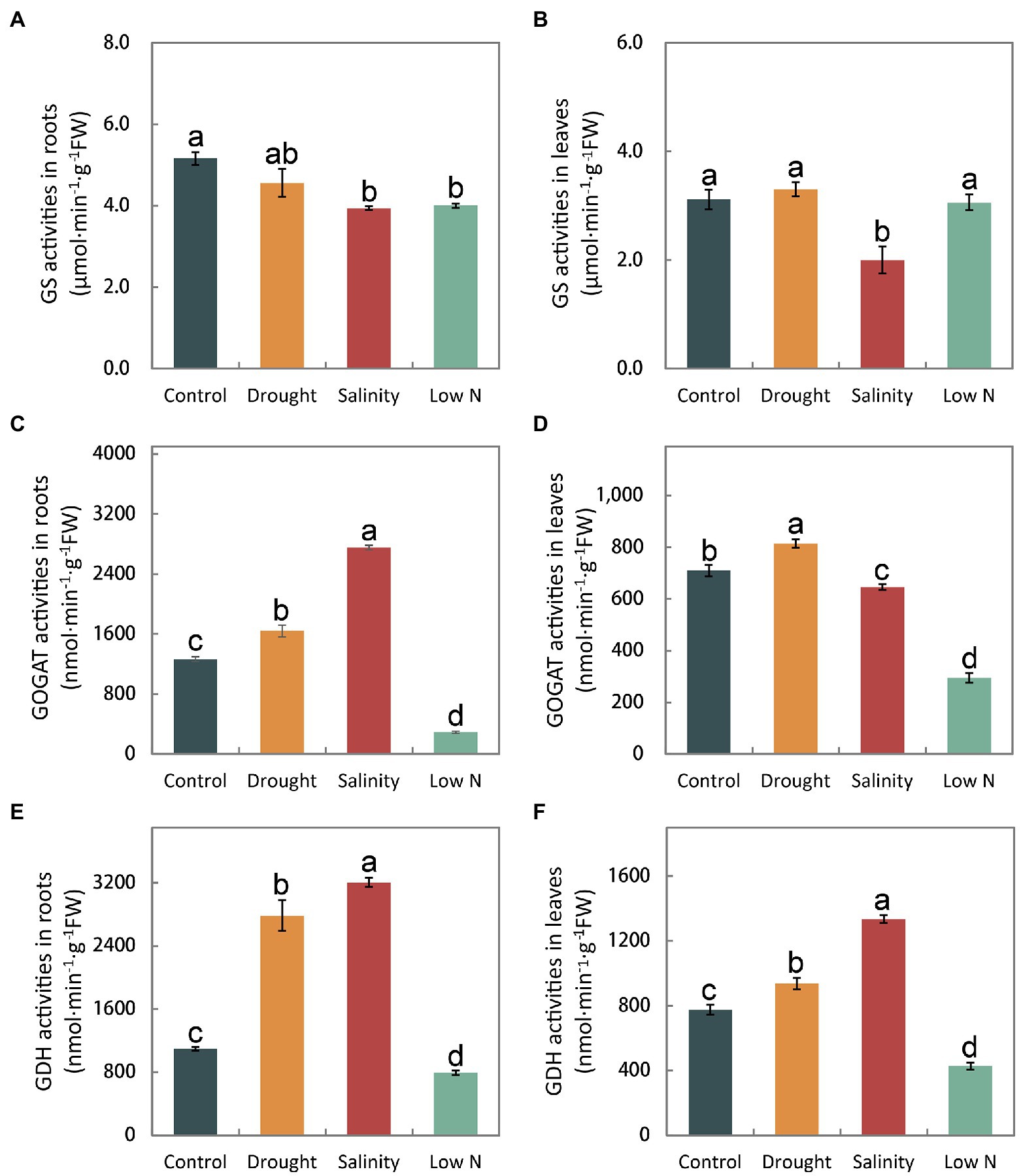
Figure 5. Activities of glutamine synthetase, glutamate synthase, and glutamate dehydrogenase in roots (A,C,E) and leaves (B,D,F) of S. japonica as affected by drought, salinity, and low N treatment. Data indicate means±SE of three replicates (12 plants). Bars labeled with different letters indicate significant differences between treatments (p<0.05).
Key genes implicated in N uptake and metabolism were obtained from the transcriptome data in our previous study (Tian et al., 2021). We performed transcriptional expression analysis of these genes to explore the internal molecular changes of N assimilation in response to different abiotic stresses (Figure 6).
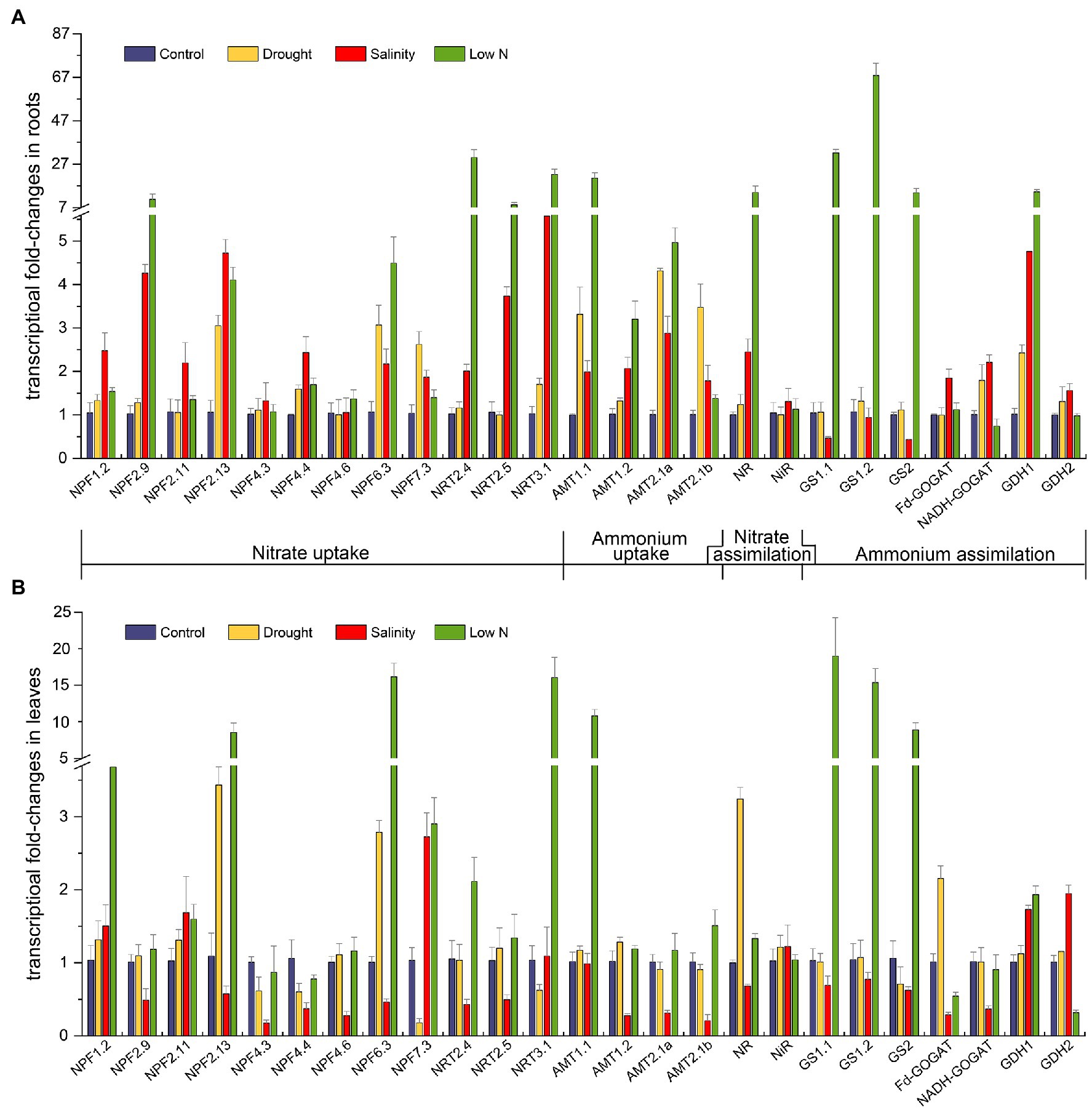
Figure 6. Transcriptional fold-changes of genes involved in N uptake and assimilation in roots (A) and leaves (B) of S. japonica as affected by drought, salinity, and low N treatment. Data indicate means±SE of three replicates (12 plants). Bars labeled with different letters indicate significant differences between treatments (p<0.05). NPF, nitrate transporter 1(NRT1); NRT2, nitrate transporter 2; NRT3, nitrate transporter 3; AMT1, ammonium transporter 1; AMT2, ammonium transporter 2; NR, nitrate reductase; NiR, nitrite reductase; GS, glutamine synthetase; Fd-GOGAT, ferredoxin-dependent glutamate synthase; NADH-GOGAT, NADH-dependent glutamate synthase; and GDH, glutamate dehydrogenase.
The transcript abundance of NRTs, which play key roles in NO3− uptake, was diverse. Under drought, the transcript levels of NRTs remained unaltered except that NPF2.13 and NPF6.3 showed higher expression, and the transcript level of NPF7.3 increased in the roots but decreased in the leaves (Figures 6A,B). Under salinity treatment, 10 of the 12 NRTs in the roots were upregulated, especially NRT3.1, but eight of them were downregulated in the leaves (Figures 6A,B). In contrast, the expression of most NRTs increased under the low N condition, especially NPF2.9, NPF2.13, NPF6.3, NRT2.4, NRT2.5, and NRT3.1 in the roots (Figure 6A) and NPF1.2, NPF2.13, NPF6.3, NPF7.3, and NRT3.1 in the leaves (Figure 6B).
The uptake of NH4+ is largely determined by specific transporters known as AMTs. In the roots, four AMTs in our study exhibited higher transcript levels under all three kinds of stresses (Figure 6A). The transcription of AMT1.1 and AMT2.1a ranked as follows: low N>drought>salinity. AMT1.2 had the highest expression under the low N treatment, and AMT2.1b showed the highest expression under drought (Figure 6A). In the leaves, four AMTs were not altered by drought; salinity suppressed the mRNA levels of AMT1.2, AMT2.1a, and AMT2.1b, whereas low N strongly induced the expression of AMT1.1 (Figure 6B).
The imposition of stresses also affected the transcript levels of genes encoding enzymes involved in N metabolism. Drought upregulated the expression of NADH-GOGAT and GDH1 in the roots as well as NR and Fd-GOGAT in the leaves (Figures 6A,B). Salinity inhibited the transcription of GS1.1, GS1.2, and GS2 but enhanced the transcription of GDH1 and GDH2 in both roots and leaves. The mRNA levels of NR, Fd-GOGAT, and NADH-GOGAT under salt stress were higher in roots but decreased in leaves (Figures 6A,B). When treated with low N, three genes encoding GS and GDH1 were all intensively induced in roots and leaves, and NR also showed extremely high expression in the roots. Finally, NiR expression was not affected by any of the three stresses in both roots and leaves (Figure 6).
In this study, we observed that the shoot height, LWC, net photosynthetic rate, and chlorophyll content of S. japonica were inhibited under all three treatments (Table 2; Figures 2A,B). Carbohydrates generated through photosynthesis are the main components of plant biomass production. Slowing down plant growth as a result of slower physiological activities such as reduced photosynthetic and transpiration rates may help S. japonica seedlings redistribute limited resources, which is an adaptation response in order to enhance survival under stress (Skirycz et al., 2011).
The effects of the three stresses on the root growth of S. japonica seedlings were different. As expected, there was better growth of both aboveground parts and roots under drought than under salinity (Table 2). Even when compared with the control, the root systems under drought were more thriving with greater total root length, total root surface area, and root biomass, which were mainly contributed by upper roots (Table 2). Roots are the main organs to absorb water, which can first perceive environmental changes and send chemical signals to the aboveground parts. The extension of the root system depends not only on the species but also on the degree of drought (Jin et al., 2009; Ustun et al., 2018; Alam et al., 2020). The findings showed that S. japonica had high adaptation ability to mild drought, which greatly stimulated lateral fine root growth to acquire more water and nutrients. In contrast, sapling growth was most restricted under a low N concentration as indicated by decreases in both shoot and root growth as well as extremely low total N accumulation of the whole plants (Table 2; Figures 2C,D). These data suggest that a low nitrogen concentration of 0.01mM NH4NO3 cannot meet the requirements of the normal growth of S. japonica.
Sophora japonica seedlings have distinct internal NH4+, NO3−, and NO2−concentrations in response to drought, salinity, and low N treatments. In our study, all three stresses increased the ratios of NH4+ and NO3− with the exception of leaves under salinity (Figures 3G,H), which may have been because NH4+ assimilation requires less energy than NO3− (Guerrero et al., 1981). Higher NH4+ concentrations could promote root growth and enhance stress resistance in plants (Fernández-Crespo et al., 2014; Zhang et al., 2014; Ding et al., 2015; Meng et al., 2016; Miranda et al., 2016; Hessini et al., 2020). The reasons for the increase in the ratio of NH4+ and NO3− in plants may be increases in NO3− assimilation or NH4+ uptake and reductions in NH4+ assimilation or NO3− uptake. Based on the changes in NH4+ and NO3− concentrations as well as NR and GS activities in this study (Figures 3A–D, 4A,B, 5A,B), we conclude that the higher ratio of NH4+ and NO3− in roots of S. japonica may be caused mainly by (1) reductions in NO3− uptake+increases in NH4+ uptake under drought; (2) increases in NO3− assimilation+reductions in NH4+ assimilation under salinity; and (3) reductions in NO3− uptake+reductions in NH4+ assimilation under low N stress. Additionally, we noted that drought and low N treatment reduced the NO3− concentration in both roots and leaves, but unexpectedly, salinity increased the NO3− content in leaves and did not affect it in roots (Figures 3C,D). Generally, drought could impair the acropetal translocation of some nutrients, and salinity could suppress NO3− absorption because of the competition between Cl- and NO3− (Abdelgadir et al., 2005). The results of NO3− accumulation under salinity of S. japonica may be explained by (i) increases in the NO3− concentration compensated osmotically for reductions in soluble carbohydrates when photosynthesis was reduced (McIntyre, 1997); (ii) less absorption of Cl− and promotion of NO3− uptake by Na+. Similar results were also observed in some halophyte plants (Yuan et al., 2010; Kaburagi et al., 2015). Additionally, (iii) NR activity was inhibited. Obviously, this is not the reason for the unchanged NO3− content in roots with a higher increased NR activity. Therefore, we speculated that S. japonica may maintain the NO3− uptake by reducing Cl− absorption in the roots and store more NO3− in vacuoles as an osmotic regulator to promote water absorption by inhibiting NO3− assimilation in the leaves to resist salt stress. However, as the levels of Cl− were not measured in the current study, further experiments should be carried out to verify the speculation. In nitrate assimilation process, NO2− is produced by converting NO3− via NR. Our results showed that the changes of nitrite content were inconsistent with that of NR activities (Figures 3E,F, 4A,B). It may be related to non-enzymatic, chemical reduction of nitrite in plants.
Different enzymes involved in N metabolism are dependent on substrates and the flux of inorganic N into organic compounds, which is crucial for N assimilation. The N assimilating enzyme response differs depending not only on the stress type, but also on the plant species, cultivar and analyzed tissue (Meng et al., 2015, 2016; Wang et al., 2019). Nitrate reductase (NR) is the key enzyme of NO3− assimilation. Our results showed that NR activities were enhanced in leaves or not affected in roots under drought (Figures 4A,B), which is opposite to the results obtained for P. simonii and M. prunifolia (Meng et al., 2015; Huang et al., 2018). In contrast, the NR activity remained unchanged under low N in S. japonica (Figures 4A,B), similar to previous studies (Meng et al., 2015; Huang et al., 2018). It is thought that higher plants have developed a complex regulatory system controlling NR against environmental factors. Various factors, including NO3− itself, carbon and N metabolites, light, phytohormones, CO2 concentration, synthesis of the NR protein, and the energy required for the assimilation process including NADH and adenosine triphosphate (ATP), are involved in the regulation of NR activity (Bungard et al., 2002). In S. japonica, in addition to the primary signal NO3−, the induction of NR activity may be affected in other ways, but the underlying mechanism should be further studied.
Glutamine synthetase (GS) and GOGAT are key enzymes of NH4+ assimilation. GS activities remained unchanged and GOGAT activities increased in both roots and leaves under drought (Figures 5A–D), indicating that S. japonica seedlings accelerated the NH4+ assimilation process to resist drought stress. However, shoot growth was inhibited, which may have been caused by the toxicity of excessive NO2− to cells (Figures 1C, 3F). The decreased GS activities in both roots and leaves under salt stress as well as reduced GOGAT activities in both roots and leaves under low N treatment may suggest that the NH4+ assimilation process was slowed down, eventually resulting in the growth inhibition of both roots and shoots (Figures 5A–D). GDH is considered an alternative enzyme for the GS/GOGAT cycle under abiotic stresses (El-Shora and Abo-Kaseem, 2001). GDH activity increased significantly in the drought and salinity treatments but markedly diminished in response to the low N treatment (Figures 5E,F). Both the aminating and deaminating properties of GDH were considered to be helpful under stress conditions. Amination plays key roles in NH4+ detoxification and in the replenishment of the glutamate pool, which is largely required to produce protective metabolites such as proline and phytochelatins (Cammerts and Jacobs, 1985; Ashraf et al., 2018). Deamination provides an intermediate to the tricarboxylic acid (TCA) cycle and thus sustained carbohydrate metabolism (Kumar et al., 2000). Abiotic stress severely affected the process of ammonia assimilation in S. japonica.
Exposure to stresses also led to changes in gene expression at the molecular level. Prior to NO3− assimilation, nitrate is taken up mainly with the help of two types of NRTs, low affinity nitrate transporters (NRT1/PTR) and high affinity nitrate transporters (NRT2). In Arabidopsis, nitrate transport activity has been proven for 16 out of 53 NRT1/PTR proteins (Léran et al., 2014). Thus, we identified 12 putative nitrate transporters in S. japonica through phylogenetic tree and BLAST methods (Supplementary Figure S1), as well as four AMTs, one NR, one NiR, three GSs, two GOGATs, and two GDHs for qRT-PCR. The regulation of these genes might reflect their importance in plant adaptation to various stresses. In our study, the expression of NPF6.3 was found to be upregulated in roots of S. japonica under all the stress treatments (Figure 6A). In Arabidopsis, AtNPF6.3 (NRT1.1) not only displays dual affinity for nitrate but also participates in auxin transport and nitrate sensing (Krapp et al., 2014). The upregulation of SjNPF6.3 may help plants withstand stress by altering root morphology. AtNPF2.13 (NRT1.7) is responsible for transporting nitrate out of older leaves and into younger leaves (Fan et al., 2009). The increases in SjNPF2.13 transcripts in leaves under drought and low N treatment (Figure 6B) may imply the active process of nitrate source-to-sink remobilization. NRT1.5 is related to the long-distance transport of nitrate from roots to shoots (Lin et al., 2008). Downregulation of AtNPF7.3 (NRT1.5) has been found to be involved in stress tolerance mechanisms in Arabidopsis (Li et al., 2010; Chen et al., 2012). In leaves, the transcript level of SjNPF7.3 increased under salinity and low N conditions but decreased under drought (Figure 6B). Reduced upward transport of nitrate might be beneficial for root development under drought. It is well known that members of the NRT2 family play a role in nitrate uptake when the external nitrate concentration is less than 250μM (Crawford and Glass, 1998). It was reported that overexpressing MdNRT2.4 from apple enhanced low N and osmotic stress tolerance in Arabidopsis (Wang et al., 2019); the nrt2.5 mutation reduced nitrate levels in N-starved Arabidopsis (Lezhneva et al., 2014); AtNRT3;1a is a key player in a two-component system, which includes NRT2s, for nitrate transport (Yong et al., 2010; Kotur et al., 2012). SjNRT2.4, SjNRT2.5, and SjNRT3.1 were highly induced in roots by low N treatment in the present study (Figure 6A), indicating their great importance in nitrate uptake and stress resistance in S. japonica. Upregulation of most NRTs under salt stress (Figure 6A) demonstrated that S. japonica had high salinity resistance that could maintain high NO3− uptake ability in the roots. Additionally, all three stresses positively regulated the expression of AMT genes in the roots of S. japonica (Figure 6A). This further proved that S. japonica enhanced NH4+ uptake with less energy consumption than NO3− to resist stressful environments, which is consistent with the changes in NH4+ and NO3− contents (Figures 3A,C). In Puccinellia tenuiflora, PutAMT1.1 could mediate toxic MeA, alleviate environmental pH stress, and improve salt tolerance during the early root growth stage after seed germination (Bu et al., 2019). SjAMT1.1 may perform the same function in leaves under low N treatment. In both the leaves and roots of S. japonica, GS transcription was more than 10-fold higher than that in the control under low N (Figure 6). As Díaz et al. (2010) stated, proline metabolism is dependent on GS. Overexpression of pine GS1a resulted in enhanced tolerance to drought (Molina-Rueda et al., 2013). Strong activation of GS would aid S. japonica in adapting to a low N environment. The trends of other genes involved in N metabolism were generally consistent with the related enzyme activities.
Collectively, our research is the first to study how the N uptake and metabolism process of S. japonica responded to different abiotic stresses at the morphological, physiological, and transcriptional levels. This research greatly advances our understanding of the underlying mechanisms of N uptake and metabolism strategies in response to different abiotic stresses, which could provide insights for stress resistance studies of leguminous tree species. The different responses under the three abiotic stresses provide physiological and molecular basis to evaluate the nitrogen absoption ability of S. japonica, which is also the first step to further explore the longivity mechanism of ancient trees. Additionally, this study is also of great significance for maintaining the healthy growth of S. japonica, the breeding of resistant varieties, as well as the formulation of feasible management methods.
The present study provided unique and valuable observations on how the N uptake and metabolism process of S. japonica responded to different abiotic stresses at the morphological, physiological, and transcriptional levels. Under drought, root growth was promoted, which may be a result of increases in NH4+ uptake and NH4+ assimilation ability. This can be confirmed by the upregulated AMTs (SjAMT1.1, SjAMT2.1a, and SjAMT2.1b) and increased GOGAT activity. However, the root growth under stress is influenced by many other factors apart from N uptake and assimilation. Salinity and low N negatively affected plant growth, probably because NH4+ assimilation was slowed down due to the reduced GS activity. It was found that most NRTs were upregulated and the NO3− concentration was unaltered even if NR activity increased in roots under salinity, indicating high salinity resistance and that S. japonica could retain high NO3− uptake ability. Low N treatment most restricted plant growth, which suggested that 0.01mM NH4NO3 cannot meet the requirements of S. japonica for its normal growth. However, plants could strongly activate the expression of genes in the N pathway, especially NRT2s and AMTs, as well as three GS genes to adapt to N limitation. Moreover, SjNPF2.13, SjNPF6.3, SjNPF7.3, and GDH1 were highly induced by the three treatments. They may play key roles in plant root resistance under environmental stress. All the findings suggested that the resistance of S. japonica seedlings to different stresses in our study could be ranked as follows: drought>salinity>low N treatment. The distinct changes in plant growth, N accumulation, enzyme activity, and gene expression indicate that S. japonica could plastically regulate N metabolism strategies with the changes of environmental conditions. It provides insights for stress resistance studies of leguminous tree species and long longivity studies of ancient trees. However, further functional identification of the key genes involved in N utilization in response to stress is essential and necessary for maintaining the healthy growth of S. japonica, the breeding of resistant varieties, as well as the formulation of feasible management methods.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JT and ZZ conceived the study and its design, drafted the manuscript, and revised the paper. JT and YP performed the experiments and contributed to collect and analyze the data. All authors contributed to the article and approved the submitted version.
This research was supported by the National Forestry Industry Research Special Funds for Public Welfare Projects (China; 201404302). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We sincerely thank Yinglong Chen from Institute of Soil and Water Conservation, Northwest A&F University, for providing the semi-hydroponic phenotyping platform. We also thank Tingting An and Baoxing Liu for their great help in the sample collection. We also thank Zhu Hailan for her assistance in laboratory.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.715456/full#supplementary-material
Abdelgadir, E. M., Oka, M., and Fujiyama, H. (2005). Characteristics of nitrate uptake by plants under salinity. J. Plant Nutr. 28, 33–46. doi: 10.1081/PLN-200042156
Ahanger, M. A., Tittal, M., Mir, R. A., and Agarwal, R. M. (2017). Alleviation of water and osmotic stress-induced changes in nitrogen metabolizing enzymes in Triticum aestivum L. cultivars by potassium. Protoplasma 254, 1953–1963. doi: 10.1007/s00709-017-1086-z
Alam, H., Khattak, J., Saleem, M. H., Fahad, S., Sohail, H., Alkahtani, J., et al. (2020). Negative impact of long-term exposure of salinity and drought stress on native Tetraena mandavillei L. Physiol. Plant. 172, 1336–1351. doi: 10.1111/ppl.13273
Ashraf, M., Shahzad, S. M., Imtiaz, M., and Rizwan, M. S. (2018). Salinity effects on nitrogen metabolism in plants focusing on the activities of nitrogen metabolizing enzymes: a review. J. Plant Nutr. 41, 1065–1081. doi: 10.1080/01904167.2018.1431670
Bowen, L., Changqing, M., Zhijun, Z., Zhiwei, W., Tengteng, G., Qi, Z., et al. (2018). Long-term exogenous application of melatonin improves nutrient uptake fluxes in apple plants under moderate drought stress. Environ. Exp. Bot. 155, 650–661. doi: 10.1016/j.envexpbot.2018.08.016
Boyer, J. S. (1982). Plant productivity and environment. Science 218, 443–448. doi: 10.1126/science.218.4571.443
Bräutigam, A., Gagneul, D., and Weber, A. P. (2007). High-throughput colorimetric method for the parallel assay of glyoxylic acid and ammonium in a single extract. Anal. Biochem. 362, 151–153. doi: 10.1016/j.ab.2006.12.033
Bu, Y., Takano, T., and Liu, S. (2019). The role of ammonium transporter (AMT) against salt stress in plants. Plant Signal. Behav. 14:1625696. doi: 10.1080/15592324.2019.1625696
Bungard, R. A., Wingler, A., Morton, J. D., Andrews, M., and Scholes, J. D. (2002). Ammonium can stimulate nitrate and nitrite absence of nitrate in Clematis vitalba. Plant Cell Environ. 22, 859–866. doi: 10.1046/j.1365-3040.1999.00456.x
Cammerts, D., and Jacobs, M. (1985). A study of the role of glutamate dehydrogenase in the nitrogen metabolism of Arabidopsis thaliana. Planta 163, 517–526. doi: 10.1007/bf00392709
Chen, Y. L., Dunbabin, V. M., Diggle, A. J., Siddique, K. H. M., and Rengel, Z. (2011). Development of a novel semi-hydroponic phenotyping system for studying root architecture. Funct. Plant Biol. 38, 355–363. doi: 10.1071/fp10241
Chen, C.-J., Lv, X.-F., Li, Y.-J., Yi, H.-Y., and Gong, J.-M. (2012). Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590. doi: 10.1104/pp.112.199257
Crawford, N. M., and Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Cell 3, 389–395. doi: 10.1016/s1360-1385(98)01311-9
Díaz, P., Betti, M., Sánchez, D. H., Udvardi, M. K., Monza, J., and Márquez, A. J. (2010). Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188, 1001–1013. doi: 10.1111/j.1469-8137.2010.03440.x
Ding, L., Gao, C., Li, Y., Li, Y., Zhu, Y., Xu, G., et al. (2015). The enhanced drought tolerance of rice plants under ammonium is related to aquaporin (AQP). Plant Sci. 234, 14–21. doi: 10.1016/j.plantsci.2015.01.016
Dluzniewska, P., Gessler, A., Dietrich, H., Schnitzler, J. P., Teuber, M., and Rennenberg, H. (2007). Nitrogen uptake and metabolism in Populus×canescens as affected by salinity. New Phytol. 173, 279–293. doi: 10.1111/j.1469-8137.2006.01908.x
El-Shora, H. M., and Abo-Kaseem, E. M. (2001). Kinetic characterization of glutamate dehydrogenase of marrow cotyledons. Plant Sci. 161, 1047–1053. doi: 10.1016/s0168-9452(01)00463-0
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H., and Tsay, Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, ls responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
FAO and ITPS (2015). Status of the World’s Soil Resources (SWSR) – Main Report. Rome: Food and Agriculture Organization of the United Nations.
Fernández-Crespo, E., Gómez-Pastor, R., Scalschi, L., Llorens, E., Camañes, G., and García-Agustín, P. (2014). NH4+ induces antioxidant cellular machinery and provides resistance to salt stress in citrus plants. Trees 28, 1693–1704. doi: 10.1007/s00468-014-1078-y
Flores, P., Botella, M. A., Martínez, V., and Cerdá, A. (2000). Ionic and osmotic effects on nitrate reductase activity in tomato seedlings. J. Plant Physiol. 156, 552–557. doi: 10.1016/S0176-1617(00)80172-8
GeBler, A., Jung, K., Gasche, R., Papen, H., Heidenfelder, A., Börner, E., et al. (2005). Climate and forest management influence nitrogen balance of European beech forests: microbial N transformations and inorganic N net uptake capacity of mycorrhizal roots. Eur. J. For. Res. 124, 95–111. doi: 10.1007/s10342-005-0055-9
Guerrero, M. G., Vega, J. M., and Losada, M. (1981). The assimilatory nitrate-reducing system and its regulation. Annu. Rev. Plant Physiol. 32, 169–204. doi: 10.1146/annurev.pp.32.060181.001125
Hessini, K., Jeddi, K., Siddique, K. H. M., and Cruz, C. (2020). Drought and salinity: a comparison of their effects on the ammonium-preferring species Spartina alterniflora. Physiol. Plant. 172:13241. doi: 10.1111/ppl.13241
Högberg, P., Granström, A., Johansson, T., Lundmark-Thelin, A., and Näsholm, T. (1986). Plant nitrate reductase activity as an indicator of availability of nitrate in forest soils. Can. J. For. Res. 16, 1165–1169. doi: 10.1139/x86-207
Huang, L., Li, M., Zhou, K., Sun, T., Hu, L., Li, C., et al. (2018). Uptake and metabolism of ammonium and nitrate in response to drought stress in Malus prunifolia. Plant Physiol. Biochem. 127, 185–193. doi: 10.1016/j.plaphy.2018.03.031
Iqbal, A., Dong, Q., Wang, X., Gui, H., Zhang, H., Zhang, X., et al. (2020). High nitrogen enhance drought tolerance in cotton through antioxidant enzymatic activities, nitrogen metabolism and osmotic adjustment. Plan. Theory 9:178. doi: 10.3390/plants9020178
James, D., Borphukan, B., Fartyal, D., Ram, B., Singh, J., Manna, M., et al. (2018). Concurrent overexpression of OsGS1;1 and OsGS2 genes in transgenic rice (Oryza sativa L.): impact on tolerance to abiotic stresses. Front. Plant Sci. 9:786. doi: 10.3389/fpls.2018.00786
Jin, B. H., Chen, Y. J., Wu, Y. H., Ying, A. N., and Qi, X. H. (2009). Response of root distribution and biomass allocation of different Poa L.varieties to drought stress. Acta Agrestia Sinica. 17, 813–816.
Kaburagi, E., Yamada, M., and Fujiyama, H. (2015). Sodium, but not potassium, enhances root to leaf nitrate translocation in Swiss chard (Beta vulgaris var. cicla L.). Environ. Exp. Bot. 112, 27–32. doi: 10.1016/j.envexpbot.2014.11.007
Kaiser, B. N., Rawat, S. R., Siddiqi, M. Y., Masle, J., and Glass, A. D. (2002). Functional analysis of an Arabidopsis T-DNA ‘knockout’ of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol. 130, 1263–1275. doi: 10.1104/pp.102.010843
Kamel, M. (2012). Improving the tolerance of Vicia faba against environmental salinity resulted from the irrigation with sea water by using KNO3 and (NH4)2SO4 as chemical osmo-regulators. Acta Biol. Colomb. 17, 295–308. doi: 10.1109/STAST.2011.6059254
Khare, T., Srivastava, A. K., Suprasanna, P., and Kumar, V. (2020). Individual and additive stress impacts of Na+ and cl- on proline metabolism and nitrosative responses in rice. Plant Physiol. Biochem. 152, 44–52. doi: 10.1016/j.plaphy.2020.04.028
Kotur, Z., Mackenzie, N., Ramesh, S., Tyerman, S. D., Kaiser, B. N., and Glass, A. D. (2012). Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 194, 724–731. doi: 10.1111/j.1469-8137.2012.04094.x
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A.-S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Kumar, R. G., Shah, K., and Dubey, R. S. (2000). Salinity induced behavioural changes in malate dehydrogenase and glutamate dehydrogenase in rice seedlings of differing salt tolerance. Plant Sci. 156, 23–34. doi: 10.1016/S0168-9452(00)00224-7
Léran, S., Varala, K., Boyer, J.-C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Lezhneva, L., Kiba, T., Feria-Bourrellier, A.-B., Lafouge, F., Boutet-Mercey, S., Zoufan, P., et al. (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 80, 230–241. doi: 10.1111/tpj.12626
Li, J. Y., Fu, Y. L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Lin, S. H., Kuo, H. F., Canivenc, G., Lin, C. S., Lepetit, M., Hsu, P. K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Liu, L., Wang, J., Han, Z., Sun, X., Li, H., Zhang, J., et al. (2016). Molecular analyses of tomato GS,GOGAT and GDH gene families and their response to abiotic stresses. Acta Physiol. Plant. 38, 229–242. doi: 10.1007/s11738-016-2251-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, X., Xue, X., and Zhou, X. (2018). Response of growth and other physiological characteristics of Sophora Japonica L. saplings to drought stress. IOP Conf. Ser. Earth Environ. Sci. 170:052029. doi: 10.1088/1755-1315/170/5/052029.7
McDowell, N., Allen, C. D., Anderson-Teixeira, K., Brando, P., Brienen, R., Chambers, J., et al. (2018). Drivers and mechanisms of tree mortality in moist tropical forests. New Phytol. 219, 851–869. doi: 10.1111/nph.15027
McIntyre, G. I. (1997). The role of nitrate in the osmotic and nutritional control of plant development. Aust. J. Plant Physiol. 24, 103–118. doi: 10.1071/PP96064
Meng, S., Su, L., Li, Y., Zhang, C., and Zhao, Z. (2016). Nitrate and ammonium contribute to the distinct nitrogen metabolism of Populus simonii during moderate salt stress. PLoS One 11:e0150354. doi: 10.1371/journal.pone.0150354
Meng, S., Zhang, C., Su, L., Li, Y., and Zhao, Z. (2015). Nitrogen uptake and metabolism of Populus simonii in response to PEG-induced drought stress. Environ. Exp. Bot. 123, 78–87. doi: 10.1016/j.envexpbot.2015.11.005
Miranda, R. D. S., Gomes-Filho, E., Prisco, J. T., and Alvarez-Pizarro, J. C. (2016). Ammonium improves tolerance to salinity stress in Sorghum bicolor plants. Plant Growth Regul. 78, 121–131. doi: 10.1007/s10725-015-0079-1
Molina-Rueda, J. J., Tsai, C. J., and Kirby, E. G. (2013). The Populus superoxide dismutase gene family and its responses to drought stress in transgenic poplar overexpressing a pine cytosolic glutamine synthetase (GS1a). PLoS One 8:e56421. doi: 10.1371/journal.pone.0056421
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Ogawa, T., Fukuoka, H., Yano, H., and Ohkawa, Y. (1999). Relationships between nitrite reductase activity and genotype-dependent callus growth in rice cell cultures. Plant Cell Rep. 18, 576–581. doi: 10.1007/s002990050625
Park Williams, A., Allen, C. D., Macalady, A. K., Griffin, D., Woodhouse, C. A., Meko, D. M., et al. (2012). Temperature as a potent driver of regional forest drought stress and tree mortality. Nat. Clim. Chang. 3, 292–297. doi: 10.1038/nclimate1693
Patterson, K., Cakmak, T., Cooper, A., Lager, I., Rasmusson, A. G., and Escobar, M. A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 33, 1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x
Plattner,, and GianKasper, (2014). IPCC, 2014: climate change 2014: synthesis report. Contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change. J. Roman. Stud. 4, 85–88. doi: 10.3167/147335304782369122
Quy, N. V., Yi, Z. Q., and Zhong, Z. (2017). Cytokinin ameliorates the abiotic stress severity in Chinese Scholartree (Sophora Japonica L.) through regulation of chlorophyll fluorescence, antioxidative response and proline metabolism. Res. J. Biotechnol. 12, 11–18.
Rais, L., Masood, A., Inam, A., and Khan, N. (2013). Sulfur and nitrogen coordinately improve photosynthetic efficiency, growth and proline accumulation in two cultivars of mustard under salt stress. J. Plant Biochem. Physiol. 1:101. doi: 10.4172/2329-9029.1000101
Rennenberg, H., Loreto, F., Polle, A., Brilli, F., Fares, S., Beniwal, R., et al. (2006). Physiological responses of forest trees to heat and drought. Plant Biol. 8, 556–571. doi: 10.1055/s-2006-924084
Seith, B., Setzer, B., Flaig, H., and Mohr, H. (1994). Appearance of nitrate reductase, nitrite reductase and glutamine synthetase in different organs of the scots pine (Pinus sylvestris) seedling as affected by light, nitrate and ammonium. Physiol. Plant. 91, 419–426. doi: 10.1111/j.1399-3054.1994.tb02969.x
Singh, R. P., and Srivastava, H. S. (1986). Increase in glutamate synthase (NADH) activity in maize seedlings in response to nitrate and ammonium nitrogen. Physiol. Plant. 66, 413–416. doi: 10.1111/j.1399-3054.1986.tb05944.x
Skirycz, A., Vandenbroucke, K., Clauw, P., Maleux, K., De Meyer, B., Dhondt, S., et al. (2011). Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29, 212–214. doi: 10.1038/nbt.1800
Suárez, M. F., Avila, C., Gallardo, F., Cantón, F. R., García-Gutiérrez, A., Gonzalo Claros, M., et al. (2002). Molecular and enzymatic analysis of ammonium assimilation in woody plants. J. Exp. Bot. 53, 891–904. doi: 10.1093/jexbot/53.370.891
Tian, J., Pang, Y., and Zhao, Z. (2021). Comparative transcriptome analysis of Sophora japonica (L.) roots reveals key pathways and genes in response to PEG-induced drought stress under different nitrogen conditions. Forests 12:650. doi: 10.3390/f12050650
Ustun, S., Melek, E., Selda, O., Metin, T., Suzan, Y., and Ertan, Y. (2018). Effects of individual and combined effects of salinity and drought on physiological, nutritional and biochemical properties of cabbage (brassica oleracea var. capitata). Sci. Hortic. 240, 196–204. doi: 10.1016/j.scienta.2018.06.016
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018b). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, X., Guo, X., Yu, Y., Cui, H., Wang, R., and Guo, W. (2018a). Increased nitrogen supply promoted the growth of non-N-fixing woody legume species but not the growth of N-fixing Robinia pseudoacacia. Sci. Rep. 8:17896. doi: 10.1038/s41598-018-35972-6
Wang, Q., Liu, C., Huang, D., Dong, Q., Li, P., and Ma, F. (2019). High-efficient utilization and uptake of N contribute to higher NUE of ‘Qinguan’ apple under drought and N-deficient conditions compared with ‘Honeycrisp’. Tree Physiol. 39, 1880–1895. doi: 10.1093/treephys/tpz093
Xu, G. H., Fan, X. R., and Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yin, A., Jin, H., Han, Z., and Han, S. (2006). Study on the relationship between chromosome numbers and nodulation of 18 species of leguminous trees. Scientia Silvae Scnicae 42, 26–28. doi: 10.11707/j.1001-7488.20060105
Yong, Z., Kotur, Z., and Glass, A. D. (2010). Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 63, 739–748. doi: 10.1111/j.1365-313x.2010.04278.x
Yu, X., and Zhang, F. (2012). Activities of nitrate reductase and glutamine synthetase in rice seedlings during cyanide metabolism. J. Hazard. Mater. 225-226, 190–194. doi: 10.1016/j.jhazmat.2012.05.027
Yuan, J. F., Tian, C. Y., and Feng, G. (2010). Effects of sodium on nitrate uptake and osmotic adjustment of Suaeda physophora. J. Arid. Land. 2, 190–196. doi: 10.3724/SP.J.1227.2010.00190
Keywords: Sophora japonica, drought, salinity, low nitrogen, AMTs and NRTs , nitrogen uptake and metabolism
Citation: Tian J, Pang Y and Zhao Z (2021) Drought, Salinity, and Low Nitrogen Differentially Affect the Growth and Nitrogen Metabolism of Sophora japonica (L.) in a Semi-Hydroponic Phenotyping Platform. Front. Plant Sci. 12:715456. doi: 10.3389/fpls.2021.715456
Received: 27 May 2021; Accepted: 10 September 2021;
Published: 01 October 2021.
Edited by:
Mario A. Pagnotta, University of Tuscia, ItalyReviewed by:
Ratna Karan, University of Florida, United StatesCopyright © 2021 Tian, Pang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Zhao, emhhb3pobHVud2VuMjAxMEAxMjYuY29t; emhhb3poQG53YWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.