- 1Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 2Plant Germplasm Division, Taiwan Agricultural Research Institute, Taichung, Taiwan
- 3Taiwan Banana Research Institute, Pingtung, Taiwan
Bananas (Musa spp.) are some of the most important fruit crops in the world, contributing up to US$10 billion in export values annually. In this study, we use high-throughput sequencing to obtain genomic resources of high-copy DNA molecules in bananas. We sampled 13 wild species and eight cultivars that represent the three genera (Ensete, Musa, and Musella) of the banana family (Musaceae). Their plastomic, 45S rDNA, and mitochondrial scaffolds were recovered from genome skimming data. Two major clades (Clades I & II) within Musa are strongly supported by the three genomic compartment data. We document, for the first time, that the plastomes of Musaceae have expanded inverted repeats (IR) after they diverged from their two close relatives, Heliconiaceae (the lobster-claws) and Strelitziaceae (the traveler's bananas). The presence/absence of rps19 within IR regions reinforces the two intra-generic clades within Musa. Our comparisons of the bananas' plastomic and mitochondrial DNA sequence trees aid in identifying hybrid bananas' parentage. As the mitochondrial genes of Musa have elevated substitution rates, paternal inheritance likely plays an influential role on the Musa mitogenome evolution. We propose genome skimming as a useful method for reliable genealogy tracing and phylogenetics in bananas.
Introduction
Musaceae, the banana family, comprises ca. 91 species in three genera: Ensete, Musella, and Musa (Christenhusz and Byng, 2016). Bananas are one of the major fruit crops in many developing countries (Padam et al., 2014). It is estimated that >100 million tons of bananas are produced annually since 2009, with the export value of US$13.5 billion in 2019 (Food Agriculture Organization of the United Nations, 2021). Despite their enormous socio-economic value, cultivated bananas are threatened by various diseases and pests, such as Fusarium wilt (Panama disease), black leaf streak, and nematode infection, and by frequent drought due to climate change (Bakry et al., 2009; Brown et al., 2020). Genetic improvements through hybridization have thus been the preferred way to obtain resistant cultivars that ensure sustainable and safe banana production (Ortiz, 2013).
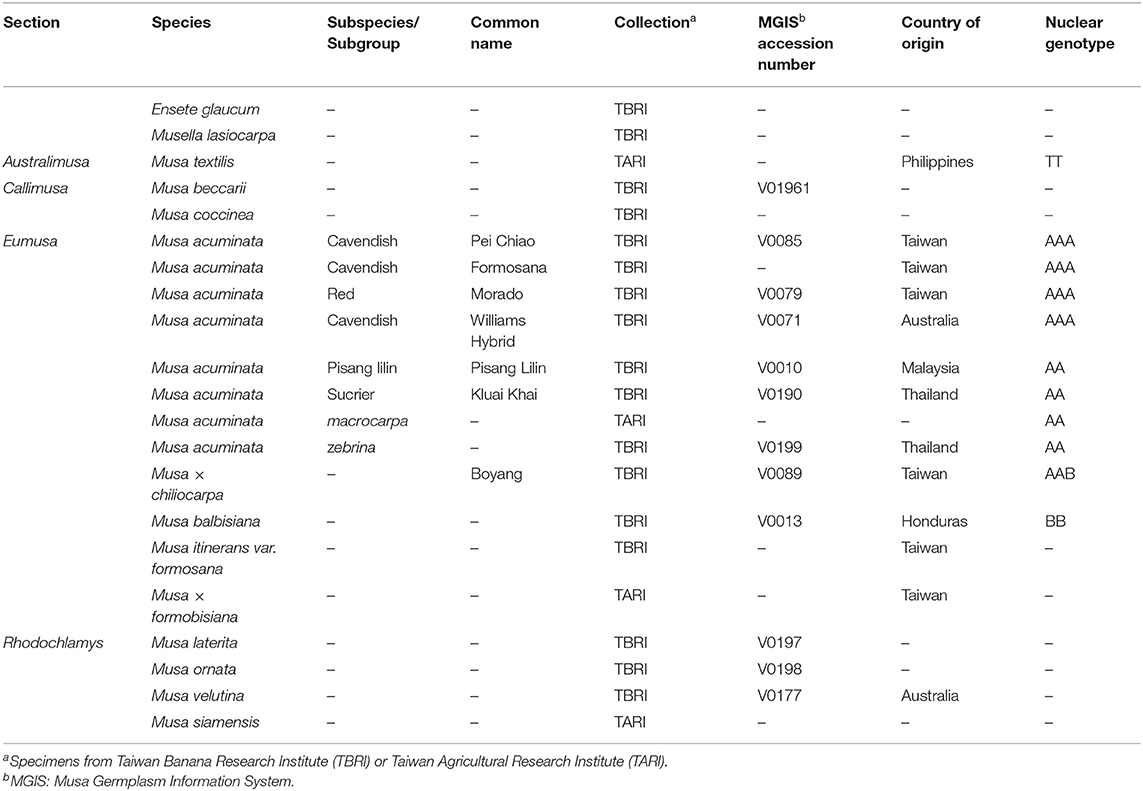
Musa L., the largest banana genus, contains four sections: Australimusa, 2n = 2x = 20; Callimusa, 2x = 18 or 20; Eumusa, 2x = 22; Rhodochlamys, 2x = 22 (Cheesman, 1947). However, this traditional classification has been revised with Eumusa and Rhodochlamys merged into the sect. Musa (or Clade I), and Australimusa and Callimusa into the sect. Callimusa (Clade II) based on numerous molecular studies (e.g., Li et al., 2010; Christelová et al., 2011; Janssens et al., 2016; Lamare et al., 2017). Many edible bananas arose from intra- or inter-specific hybridizations between Musa acuminata (Eumusa; A genome) and M. balbisiana (Eumusa; B genome), resulting in various cultivars with AA, BB, AB, AAA, AAB, ABB, AAAA, AAAB, AABB, and ABBB genomes (Simmonds and Shepherd, 1995). In addition, M. schizocarpa (Eumusa; S genome) and M. textilis (Australimusa; T genome) have contributed to the genetic pool in banana breeding programs (Pereira and Maraschin, 2015; Christelová et al., 2017).
Numerous molecular techniques have been developed to differentiate banana cultivars, including restriction fragment length polymorphism (RFLP; Gawel et al., 1992), cytogenetics (Osuji et al., 1997; CíŽková et al., 2015), microsatellites (SSR; Mattos et al., 2010; Christelová et al., 2017), diversity arrays technology markers (DArT; Sardos et al., 2016), and DNA barcodes (Hribová et al., 2011; Dhivya et al., 2020). However, the complex breeding history of bananas may further complicate efforts to identify accessions and trace their origins. For example, extensive studies showed frequent occurrences of chromosomal recombination (e.g., reciprocal translocation or chromosomal inversion) in bananas (Baurens et al., 2018; Martin et al., 2020a,b; Cenci et al., 2021). These genome structural diversities have been applied to study the origins of banana cultivars and Musa evolution (Martin et al., 2020b).
Fauré et al. (1994) firstly reported the differential inheritance modes of organellar genomes in bananas; the plastid genomes (plastomes) are maternally inherited, while the mitochondria (mitogenomes) are passed down from fathers. This unusual cytoplasmic inheritance mode facilitates the tracing of hybrid banana origins (Carreel et al., 2002; Boonruangrod et al., 2008). However, previous studies relied on complex molecular marker patterns to estimate genealogies. We therefore propose an alternative method, genome skimming, to retrieve high-copy DNA sequences (e.g., nuclear ribosomal DNA (rDNA), plastomes, and mitogenomes; Straub et al., 2012; Dodsworth, 2015) to determine parentage in bananas. Genome skimming enables generation of sequence assemblies from high-copy DNA molecules without the need for primer design, PCR amplification, electrophoretic assays, cloning, and Sanger sequencing. In the era of next generation sequencing (NGS), this approach is more reliable and less tedious than genotyping or RFLP analyses but has not yet been applied in estimating banana genealogies or phylogenies.
To verify if our proposed method can be used in lineage and hybrid identification, we used an artificial interspecific hybrid, M × formobisiana, as one of our study materials. This hybrid cultivar originated from a controlled cross between a maternal parent M. itinerans var. formosana and a paternal parent M. balbisiana (Chiu et al., 2017). We assembled the plastomes, 45S rDNAs, and mitochondrial gene sequences of 21 banana accessions using the genome skimming approach. We further show that these sequences are useful to study the banana phylogeny.
Moreover, the distinct inheritance modes between plastomes and mitogenomes of bananas provide an excellent opportunity to compare mutational loads in paternal and maternal gametes, as proposed by Greiner et al. (2015). Mitogenome mutation rates of land plants are known to be the slowest among the three genetic compartments (Wolfe et al., 1987; Drouin et al., 2008). In this study, we examine whether this still holds in bananas.
Materials and Methods
Material Collection, DNA Extraction, Sequencing, and Assembly
We sampled 21 accessions from live collections of the Taiwan Banana Research Institute and the Taiwan Agricultural Research Institute. These taxa were identified by experts and represent all three genera of the banana family (Table 1). For each sample, approximately 2 g of young leaves were harvested for DNA extraction using a modified CTAB method that includes 0.1% of polyvinylpyrrolidone (PVP-40, Sigma) in the extraction buffer (Stewart and Via, 1993). DNA libraries were constructed using Ovation Rapid Library Preparation kits (NuGEN) and sequenced on an Illumina HiSeq 4000 platform in Tri-I Biotech Company (New Taipei City, Taiwan) to generate pair-end reads of 2 × 150 bp. After quality trimming using Trimmomatic 0.38 (Bolger et al., 2014) with the parameters of leading = 3, trailing = 3, slidingwindow = 4:15, and minlen = 50, these reads were de-novo assembled using SPAdes 3.14.0 (Bankevich et al., 2012) with kmer lengths = 21, 33, 55, 77, and 91. We used NOVOPlasty 4.3.1 (Dierckxsens et al., 2017) and GetOrganelle 1.7.4 (Jin et al., 2020) to obtain complete plastome sequences with the reference from the plastid scaffolds generated by SPAdes. Base-scale corrections were conducted using Pilon 1.24 (Walker et al., 2014).
Sequence Identification and Annotation
Using BLAST+ 2.10.0 (Camacho et al., 2009) with a maximum E-value of 1E−10, 45S rDNA, plastid, and mitochondrial scaffolds were searched against an in-house database that includes publicly available 45S rDNA, plastid, and mitochondrial sequences of monocots. Protein-coding genes on the scaffolds were annotated using the “Transfer Annotation” function in Geneious 11.1.5 (https://www.geneious.com), followed by manual adjustments.
Sequence Alignment and Phylogenetic Tree Construction
Protein-coding and rDNA sequences were aligned using MUSCLE (Edgar, 2004) implemented in MEGA 7.0.26 (Kumar et al., 2016) with “Codons” and “DNA” options, respectively. These alignments were concatenated to generate specific data matrices for plastid, 45S rDNA, and mitochondrial sequences. We used IQ-TREE 1.5.12 (Nguyen et al., 2015) for partition analysis and model testing and then inferred maximum-likelihood (ML) trees with the standard non-parametric bootstrap of 1,000 replicates. The resulting trees were condensed using a 50% majority-rule consensus and visualized in MEGA.
Nucleotide Substitution Rates and Statistic Test
Pairwise nucleotide substitution rates between Musa accessions were estimated in MEGA with the substitution model = Kimura 2-parameter model, rates among sites = uniform rates, and data treatment = pairwise deletion. Paired sample t-tests were performed with the Bonferroni correction in R 3.6.2 (https://www.R-project.org/).
Results
Copy-Number Differentiation Among Plastid, 45S, and Mitochondrial DNAs
We generated a total of 1.9–2.3 Gb of sequence consisting of 12.7–15.7 million reads for the 21 sampled taxa (Supplementary Table 1). After de novo assembly, we removed scaffolds that significantly deviated from the mean read coverage because they were likely DNA fragments of intracellular horizontal transfers, such as nuclear organelle DNA (norgs) and mitochondrial plastid DNA (mtpts). Among the retained scaffolds, the 45S rDNA scaffold has the highest read coverage, followed by plastid and mitochondrial scaffolds (Figure 1). The read coverage differences are due to copy number variation among scaffolds in the three subgenomes.
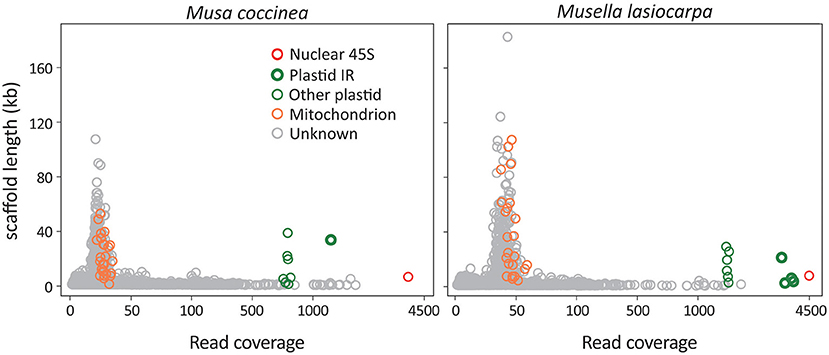
Figure 1. Scatter plots of scaffold lengths vs. read coverage. Musa coccinea and Musella lasiocarpa are used as the representatives. Red, green, and orange circles indicate 45S rDNA, plastid, and mitochondrial scaffolds, while gray are “unknown” scaffolds that could not be identified using BLAST searches against our in-house database. Some plastid scaffolds are identified as IR regions because their read coverage is approximately double of the other plastid scaffolds.
Plastome scaffolds fall into two separate groups in our analyses (Figure 1). One of the scaffolds has approximately twice as many reads as the other, indicating that the former is the inverted repeat (IR) sequence, while the latter represents the single-copy (SC) region. These plastid scaffolds were subsequently used as the initial seed for successive rounds of read extension, eventually yielding circular plastomes ranging from 167,700 to 171,982 bp in length (Supplementary Figure 1). In summary, we obtained a single scaffold for both plastomes and 45S rDNAs and 17–32 scaffolds for mitogenomes of each sequenced taxon (Supplementary Table 1). All sequences utilized in this study and their GenBank accession numbers are listed in Supplementary Table 2.
Plastid Phylogenomics of Musaceae
We inferred an ML tree for the three banana genera based on the concatenated plastid protein-coding genes (Figure 2). Figure 2 shows that Ensete and Musella form a monophyletic group sister to Musa. Within Musa, two clades (Clades I and II), are strongly supported, each containing taxa from several sections. Clade I includes taxa from three sections: Australimusa, Eumusa, and Rhodoclamys, while Clade II includes those from two sections: Australimusa and Callimusa. None of the four sections appears to be monophyletic in this ML tree. Moreover, we found that M. textilis, M. ornata, and M. acuminata are paraphyletic. M. ornata (NC_042874), M. siamensis, and M. laterita are nested within the accessions of M. acuminata (Figure 2). The publicly available M. textilis plastome sequence (NC_022926) and our sequenced M. textilis are placed separately in Clade I and II. Similarly, our newly sequenced M. ornata is sister to M. velutina rather than to its conspecific accession NC_042874. To clarify if this species-level paraphyly was due to taxonomic misidentification, we constructed a matK-based tree with additional sampling (Supplementary Figure 2). In the matK tree, our M. textilis and M. ornata accessions are each clustered together with at least two conspecific bananas, while no accession is sister to either NC_022926 or NC_042874 (Supplementary Figure 2). These data strongly suggest that NC_022926 and NC_042874 might have been mistakenly labeled in previous studies. By removing both problematic sequences, Clade I contains only Eumusa and Rhodoclamys, in good agreement with several previous molecular studies (Li et al., 2010; Christelová et al., 2011; Janssens et al., 2016; Lamare et al., 2017).
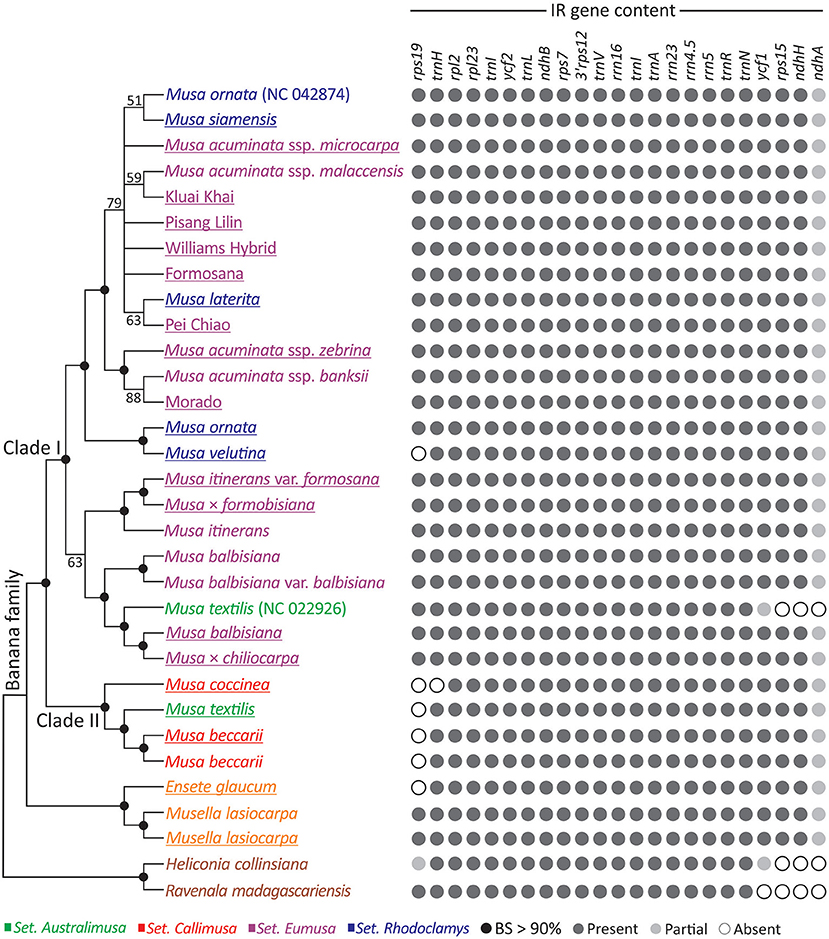
Figure 2. Plastid phylogeny and IR gene content of banana species. An ML tree inferred from the concatenation of 83 plastid genes is shown in the left panel, while the IR gene content of the associated taxa is illustrated in the right panel. The underlined taxa are sequenced in this study. BS, bootstrap support.
The Presence of rps19 in the IR Distinguishes the Two Musa Clades
To explore the IR dynamics and its implication across the banana family, patterns of IR gene content were plotted next to the ML tree (Figure 2). We found that the IR of M. textilis (NC_022926) lacks ycf1, rps15, and ndhA, but these genes are retained in the IRs of other bananas, including our sequenced M. textilis. We excluded NC_022926 from our IR comparison because of the uncertainty in its identity discussed above. Martin et al. (2013) reported that an IR expansion has resulted in duplication of ycf1, rps15, ndhH, and partial ndhA in Musa. Using both Ensete and Musella plastomes as the outgroup, we found that the IR expansion seems to be a synapomorphic trait that distinguishes the banana family from its two close relatives, Heliconiaceae (represented by Heliconia) and Strelitziaceae (represented by Ravenala). Within Musa, Clade I is separated from Clade II by the presence of rps19 in IRs, except for M. velutina, which has a species-specific IR reduction (Figure 2). These structural differences reinforce distinctive intra-generic clades within Musa.
Nuclear 45S rDNAs as Promising Super-Barcodes
A single 45S rDNA scaffold containing an array of 5'ETS, 18S, ITS1, 5.8S, ITS2, 26S, and 3'ETS (Figure 3A) was recovered from each sampled individual. These nrDNA sequences range from 6,532 to 6,816 bp. They exhibit high sequence divergence at four loci, (3'ETS, 5'ETS, ITS1, and ITS2), where identical sites are nearly absent.
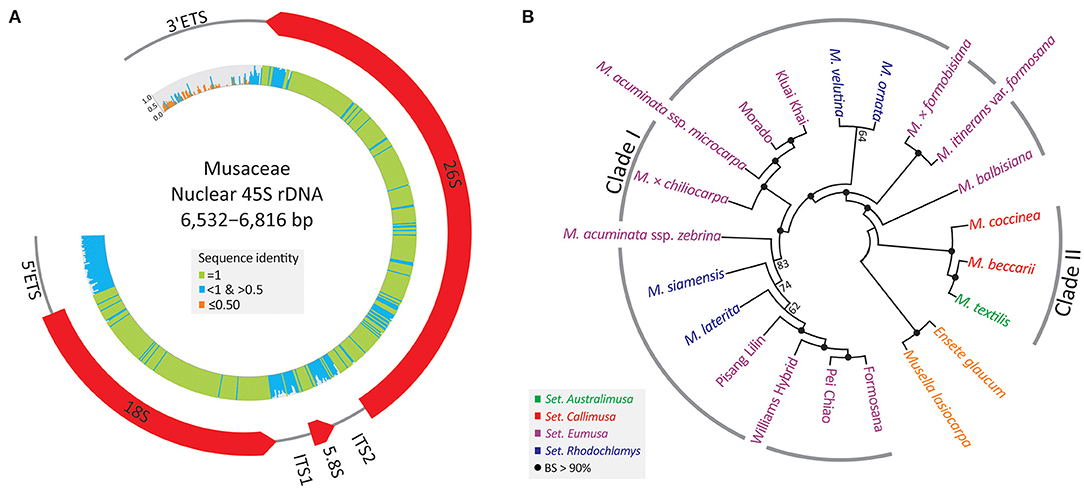
Figure 3. Assemblies of 45S rDNAs in the banana family. (A) A map depicting the banana 45S rDNA, including 5'ETS, 18S, ITS1, 5.8S, ITS2, 26S, and 3'ETS. Sequence identity across the 45S rDNA is shown in the inner histograms. (B) An unrooted ML tree inferred from the entire 45S rDNA sequence. BS, bootstrap support.
Figure 3B depicts an unrooted ML tree inferred from all 45S rDNA sequences. The major discrepancy between 45S rDNA and plastid trees is the relative position of a triploid (AAB) banana, M. × chiliocarpa. This banana is clustered with M. balbisiana in the plastid tree (Figure 2), but it is nested within the accessions of M. acuminata in the 45S rDNA tree (Figure 3B). Nonetheless, the two trees are congruent in support of the two intra-generic clades (i.e., Clades I and II) within Musa. Moreover, all taxa, including cultivars, are fully resolved using a 50% majority consensus rule in the 45S rDNA tree (Figure 3B), suggesting that 45S rDNA is an effective super-barcode for discriminating banana species and cultivars.
Mitochondrial Gene Repertoire of Musaceae
We obtained 42 mitochondrial gene sequences, including 39 protein-coding and three rRNA genes (Figure 4A), from all sampled taxa. Three genes (i.e. sdh4, rpl10, and rps8) were not detected in our mitochondrial scaffolds though they are often present in angiosperm mitochondria (Chen et al., 2017). Although sdh3 was previously detected in Musa based on Southern blot hybridizations (Adams et al., 2002), it was reported as either truncated at the 5'-region or premature due to early stop codons. Therefore, we treated sdh3 as a pseudogene in all sampled Musa taxa, except for M. acuminata, M. × chiliocarpa, M. laterita, and M. siamensis whose sdh3 remains functional (Supplementary Figure 3; Figure 4A). We did not detect sdh3 in Ensete and Musella. They also lack the intron in ccmFc (Figure 4B). In addition, there are four nad7 introns in Musa, but only one (intron 4) and three (introns 1, 2, and 4) are retained in Ensete and Musella, respectively (Figure 4C), suggesting that Ensete likely lost nad7i1 and nad7i2 after it diverged from Musella.
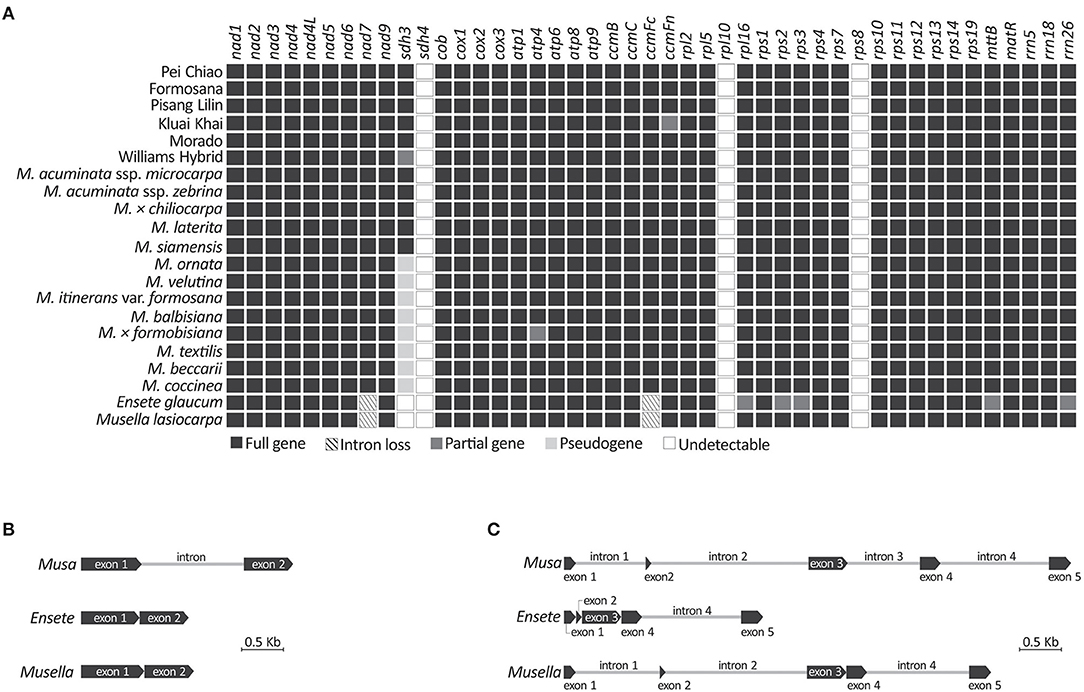
Figure 4. Comparisons of mitochondrial genes recovered from genome skimming. (A) Presence or absence of mitochondrial genes in the sampled banana genome assemblies. (B) Intron loss from ccmFc. (C) Intron loss from nad7.
Organellar Genome Phylogenies as a Useful Method to Study Banana Genealogy
A concatenated mitochondrial gene matrix was generated from the 21 newly sequenced accessions and two (M. acuminata ssp. malaccensis and M. acuminata ssp. banksii) from the Banana Genome Hub database (Supplementary Table 2). Subsequently, this matrix was used to construct an unrooted ML tree (Figure 5: left panel). To compare the mitochondrial to the plastid phylogeny, we reconstructed a plastid phylogenomic tree of a reduced set of taxa (Figure 5: right panel). Both trees support Clades I and II within Musa but differ in the placements of M. × formobisiana and M. × chiliocarpa. The former is placed as sister to M. balbisiana with a bootstrap value >90% in the mitochondrial tree, but as a sister of M. itinerans var. formosana in the plastid trees (Figures 2, 5). Since M. × formobisiana was a hybrid between M. itinerans var. formosana (maternal) and M. balbisiana (paternal; Chiu et al., 2017), this discrepancy implies heterogeneous transmission of cytoplasmic DNA, in line with the different modes of inheritance of these organelles (Fauré et al., 1994). In addition, M. × chiliocarpa is nested within the accessions of M. acuminata in the mitochondrial tree rather than a sister to M. balbisiana in the plastid gene trees (Figures 2, 5). Thus, we infer that M. × chiliocarpa's AAB genotype is likely derived from a maternal parent M. balbisiana and a paternal parent M. acuminata. Collectively, our results demonstrate that comparisons of the two organellar phylogenies facilitate identification of bananas' parentage.
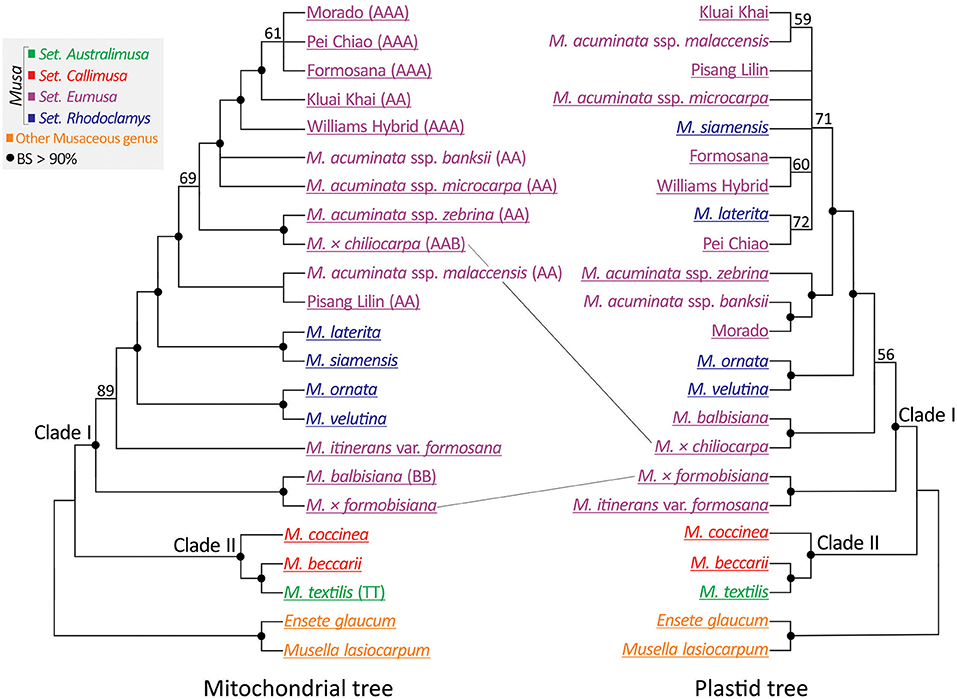
Figure 5. Comparing mitochondrial (left panel) and plastid (right panel) trees reveals different placement of two interspecific hybrid bananas. Genome types (if known) are indicated within parentheses. Taxa underlined are sequenced in this study. BS, bootstrap support.
Paternally Inherited Banana Mitogenomes Show Significantly Elevated Nucleotide Substitution Rates
Musa mitogenomes are paternally inherited, unlike most seed plants where they are passed down from maternal parents (Greiner et al., 2015). This raises a question: did the paternal inheritance mode affect mitogenome evolution in Musa? To address this question, we estimated and compared synonymous (dS) and non-synonymous (dN) substitution rates between plastid and mitochondrial genes in Musa. We used pairwise rather than tree-based methods in estimating the substitution rates because of the differences in topology between the two organellar trees (Figure 5). Paired t-tests indicate that mitochondrial genes exhibit significantly higher nucleotide substitution rates than plastid loci at both dN (P = 1.19 × 10−42) and dS (P = 2.77 × 10−42) sites after the Bonferroni correction (Figure 6). Similarly, significantly elevated rates of nucleotide substitutions are also observed in mitochondrial rRNA genes compared to plastids (Supplementary Figure 4). These results contradict the previous suggestions that mitochondrial genes evolve slower than plastid loci (Wolfe et al., 1987; Drouin et al., 2008; Smith, 2015). Therefore, paternal inheritance likely plays an influential role on the evolution of Musa mitogenomes.
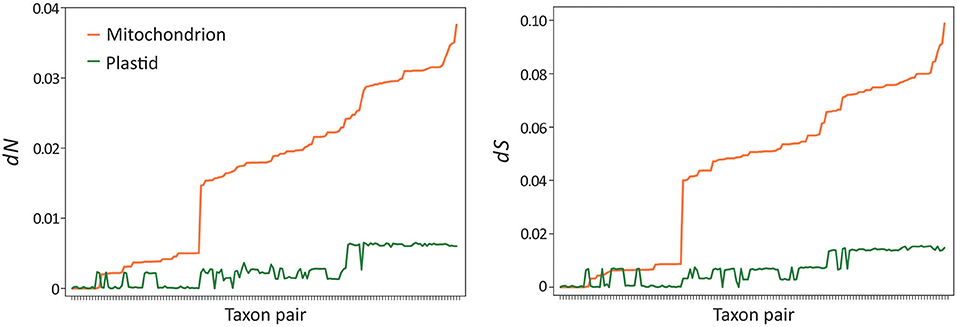
Figure 6. Elevated nucleotide substitution rates in banana mitogenomes. Paired t-tests indicate that both non-synonymous (dN, P = 1.19 × 10−42) and synonymous (dS, P = 2.77 × 10−42) rates are significantly elevated in mitochondria compared to plastids after the Bonferroni correction. Taxon pairs were sorted based on mitochondrial rates from the slowest to the fastest.
Discussion
In this study, we employed genome skim data and recovered plastomes, 45S rDNAs, and mitochondrial gene sequences to re-assess the banana phylogeny and organelle inheritance modes. Numerous molecular studies have concluded that the three banana genera are monophyletic and Musa can be subdivided into two clades rather than four sections (see review in Häkkinen, 2013). Our results also support the two intrageneric clades in Musa, across trees inferred from plastid, 45S rDNA, or mitochondrial sequences. The plastid tree shown in Figure 2 reveals species-level paraphyly in M. textilis, M. ornata, and the accessions of M. acuminata. The former two are likely due to specimen misidentification as shown in our matK tree (Supplementary Figure 2). Taxonomic misidentification often leads to conflicting results because of difficulty in determining banana species, particularly when the specimen is incomplete (Li et al., 2010) or dried. However, taxonomic misidentification cannot account for the paraphyly of M. acuminata in the plastid tree as we found that this species is monophyletic in our mitochondrial tree. Previously, the paraphyly of M. acuminata and its cultivars was noted in several molecular studies based on the combination of a few plastid loci and ITS (Li et al., 2010; Janssens et al., 2016; Lamare et al., 2017) or low-copy nuclear genes (Christelová et al., 2011). Here we propose mitochondrial genes as reliable markers for banana phylogenetics because they eliminated species-level paraphyly from incomplete lineage sorting or genetic introgression (Funk and Omland, 2003; McKay and Zink, 2010). Particularly, rapid radiation was proposed to take place after the divergence of M. acuminata from M. balbisiana (Rouard et al., 2018).
Tracing the origins of hybrid bananas has previously heavily relied on restriction fragment length polymorphisms from several loci, including ITS, plastid, and mitochondrial sequences (Carreel et al., 2002; Nwakanma et al., 2003; Swangpol et al., 2007; Boonruangrod et al., 2008). We demonstrate that we can confidently infer two cultivated bananas' parentages from plastid and mitochondrial genes retrieved using the genome skimming method (Figure 5). Distinct transmission of these two organellar genomes allows identification of both paternal and maternal parents. M. × chiliocarpa is the second banana cultivar with the AAB genotype from a paternal M. acuminata and a maternal M. balbisiana after Pisang Rajah (Boonruangrod et al., 2008; De Langhe et al., 2010). M × formosibiana was also correctly inferred as the hybrid of a paternal parent M. balbisiana and a maternal parent M. itinerans var. formosana (Chiu et al., 2017).
Paternal inheritance of mitochondrial genomes is rare in seed plants. It has only been reported in some conifers (see review in Mogensen, 1996), bananas (Fauré et al., 1994; this study), cucumbers (Havey, 1997; Park et al., 2021), and melons (Zhao et al., 2014). These exceptional cases contradict the view that female-derived organelles are preferred as an evolutionary consequence of avoiding higher mutational loads in male gametes (Greiner et al., 2015). Indeed, the paternal organelles have significantly elevated rates of nucleotide substitutions in both conifers (Whittle and Johnston, 2002) and bananas (this study). Organellar genomes of sperm cells have low copy numbers and may exert stronger genetic bottleneck compared to egg cells, eventually leading to genetic drifts and elevated rates of nucleotide substitutions (Greiner et al., 2015). Additionally, low genome copy numbers also disrupt DNA repairs through recombination, resulting in accumulations of mismatches, as proposed in the Silene conica mitogenomes (Broz et al., 2021).
Yet, how these paternally inherited organelles deal with high mutational load and avoid mutational meltdown remain unknown. When and how did paternally-inherited mitochondria emerge in banana also requires further scrutiny. Shen et al. (2015) demonstrated that mitogenomes in the male generative cells of cucumbers are protected from DNA degradation by DNA-binding proteins, thus enabling paternally inherited mitochondria in cucumbers. Whether a similar mechanism also applies to male generative cells in banana species is worthy of investigation in future studies.
In conclusion, we show that 45S rDNA sequences and organellar genome sequences are informative for banana genealogy and phylogeny. As the cost of NGS continues to decrease, recovery of reliable genomic data from genome skimming will be affordable and routinely applicable for banana phylogenetic and genealogical studies. Additionally, our study supports the male-driven evolution in bananas: paternally inherited mitochondrial genes have higher substitution rates than maternally inherited plastid loci. This case further supports the idea that maternal inheritance evolved as the predominant organellar genome inheritance mode to escape higher mutational load in male gametes.
Data Availability Statement
The original contributions presented in the study are publicly available. The DNA data generated in this study are deposited in GenBank with the accession numbers: LC603789–LC612134 and the SRA bioproject: PRJNA744736.
Author Contributions
S-MC conceived and designed the study. C-SW performed the analyses. H-LC and C-PC prepared the banana specimens. C-SW, ES, and S-MC wrote the manuscript. All the authors checked and approved the final version.
Funding
This study was supported by research grants from Biodiversity Research Center, Academia Sinica (AS), Taiwan and partially from AS-23-23 to S-MC, and partially by the 2019 AS postdoctoral fellowship to ES.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Taiwan Banana Research Institute, Taiwan Agricultural Research Institute, and Taipei Botanical Garden for providing plant materials. We also thank the two reviewers for critical comments that improved this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.713216/full#supplementary-material
References
Adams, K. L., Qiu, Y. L., Stoutemyer, M., and Palmer, J. D. (2002). Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 99, 9905–9912. doi: 10.1073/pnas.042694899
Bakry, F., Carreel, F., Jenny, C., and Horry, J. P. (2009). “Genetic improvement of banana,” in Breeding Plantation Tree Crops: Tropical Species, eds. S. M. Jain and P. M. Priyadarshan (New York, NY: Springer-Verlag), 3–50.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baurens, F. -C., Martin, G., Hervouet, C., Salmon, F., Yohome, D., Ricci, S., et al. (2018). Recombination and large structural variations shape interspecific edible bananas genomes. Mol. Biol. Evol. 36, 97–111. doi: 10.1093/molbev/msy199
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boonruangrod, R., Desai, D., Fluch, S., Berenyi, M., and Burg, K. (2008). Identification of cytoplasmic ancestor gene-pools of Musa acuminata Colla and Musa balbisiana Colla and their hybrids by chloroplast and mitochondrial haplotyping. Theor. Appl. Genet. 118, 43–55. doi: 10.1007/s00122-008-0875-3
Brown, A., Carpentier, S. C., and Swennen, R. (2020). “Breeding climate-resilient bananas,” in Genomic Designing of Climate-Smart Fruit Crops, ed C. Kole (Cham: Springer International Publishing), 91–115.
Broz, A. K., Waneka, G., Wu, Z., Gyorfy, M. F., and Sloan, D. B. (2021). Detecting de novo mitochondrial mutations in angiosperms with highly divergent evolutionary rates. Genetics 218:iyab039. doi: 10.1093/genetics/iyab039
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carreel, F., Gonzalez, de Leon, D., Lagoda, P., Lanaud, C., Jenny, C., et al. (2002). Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45, 679–692. doi: 10.1139/g02-033
Cenci, A., Sardos, J., Hueber, Y., Martin, G., Breton, C., Roux, N., et al. (2021). Unravelling the complex story of intergenomic recombination in ABB allotriploid bananas. Ann. Bot. 127, 7–20. doi: 10.1093/aob/mcaa032
Cheesman, E. E. (1947). Classification of the bananas II. The genus Musa L. Kew Bull. 2, 106–117. doi: 10.2307/4109207
Chen, Z., Zhao, N., Li, S., Grover, C. E., Nie, H., Wendel, J. F., et al. (2017). Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit. Rev. Plant Sci. 36, 55–69. doi: 10.1080/07352689.2017.1327762
Chiu, H. L., Shii, C. T., and Aleck Yang, T. Y. (2017). Musa × formobisiana (Musaceae), a new interspecific hybrid banana. Taiwania 62, 147–150. doi: 10.6165/tai.2017.62.147
Christelová, P., Langhe, E. D., Hribová, E., CíŽková, J., Sardos, J., Hušáková, M., et al. (2017). Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 26, 801–824. doi: 10.1007/s10531-016-1273-9
Christelová, P., Valárik, M., Hribová, E., De Langhe, E., and DoleŽel, J. (2011). A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evol. Biol. 11:103. doi: 10.1186/1471-2148-11-103
Christenhusz, M. J. M., and Byng, J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261, 201–217. doi: 10.11646/phytotaxa.261.3.1
CíŽková, J., Hribová, E., Christelová, P., Van den Houwe, I., and Häkkinen, M., Roux, N., et al. (2015). Molecular and cytogenetic characterization of wild Musa species. PLoS ONE 10:e0134096. doi: 10.1371/journal.pone.0134096
De Langhe, E., Hribová, E., Carpentier, S., DoleŽel, J., and Swennen, R. (2010). Did backcrossing contribute to the origin of hybrid edible bananas? Ann. Bot. 106, 849–857. doi: 10.1093/aob/mcq187
Dhivya, S., Ashutosh, S., Gowtham, I., Baskar, V., Baala Harini, A., Mukunthakumar, S., et al. (2020). Molecular identification and evolutionary relationships between the subspecies of Musa by DNA barcodes. BMC Genomics 21:659. doi: 10.1186/s12864-020-07036-5
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. doi: 10.1093/nar/gkw955
Dodsworth, S. (2015). Genome skimming for next-generation biodiversity analysis. Trends Plant Sci 20, 525–527. doi: 10.1016/j.tplants.2015.06.012
Drouin, G., Daoud, H., and Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenetics Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fauré, S., Noyer, J. L., Carreel, F., Horry, J. P., Bakry, F., and Lanaud, C. (1994). Maternal inheritance of chloroplast genome and paternal inheritance of mitochondrial genome in bananas (Musa acuminata). Curr. Genet. 25, 265–269. doi: 10.1007/BF00357172
Food Agriculture Organization of the United Nations (2021). FAOSTAT Statistical Database. Available online at: http://www.fao.org/faostat/en/ (accessed June 18, 2021).
Funk, D., J., and Omland, K. E. (2003). Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 34, 397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421
Gawel, N. J., Jarret, R. L., and Whittemore, A. P. (1992). Restriction fragment length polymorphisms (RFLP)-based phylogenetic analysis of Musa. Theor. Appl. Genet. 84, 286–290. doi: 10.1007/BF00229484
Greiner, S., Sobanski, J., and Bock, R. (2015). Why are most organelle genomes transmitted maternally? Bioessays 37, 80–94. doi: 10.1002/bies.201400110
Häkkinen, M. (2013). Reappraisal of sectional taxonomy in Musa (Musaceae). Taxon 62, 809–813. doi: 10.12705/624.3
Havey, M. (1997). Predominant paternal transmission of the mitochondrial genome in cucumber. J. Hered. 88, 232–235. doi: 10.1093/oxfordjournals.jhered.a023094
Hribová, E., CíŽková, J., Christelová, P., Taudien, S., de Langhe, E., and DoleŽel, J. (2011). The ITS1-5.8S-ITS2 sequence region in the Musaceae: structure, diversity and use in molecular phylogeny. PLoS ONE 6:e17863. doi: 10.1371/journal.pone.0017863
Janssens, S. B., Vandelook, F., De Langhe, E., Verstraete, B., Smets, E., Vandenhouwe, I., et al. (2016). Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 210, 1453–1465. doi: 10.1111/nph.13856
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lamare, A., Otaghvari, A. M., and Rao, S. R. (2017). Phylogenetic implications of the internal transcribed spacers of nrDNA and chloroplast DNA fragments of Musa in deciphering the ambiguities related to the sectional classification of the genus. Genet. Resour. Crop Evol. 64, 1241–1251. doi: 10.1007/s10722-016-0433-9
Li, L. F., Häkkinen, M., Yuan, Y. M., Hao, G., and Ge, X. J. (2010). Molecular phylogeny and systematics of the banana family (Musaceae) inferred from multiple nuclear and chloroplast DNA fragments, with a special reference to the genus Musa. Mol. Phylogenet. Evol. 57, 1–10. doi: 10.1016/j.ympev.2010.06.021
Martin, G., Baurens, F. C., Cardi, C., Aury, J. M., and D'Hont, A. (2013). The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution. PLoS ONE 8:e67350. doi: 10.1371/journal.pone.0067350
Martin, G., Baurens, F. -C., Hervouet, C., Salmon, F., Delos, J. -M., Labadie, K., et al. (2020a). Chromosome reciprocal translocations have accompanied subspecies evolution in bananas. Plant J. 204, 1698–1711. doi: 10.1111/tpj.15031
Martin, G., Cardi, C., Sarah, G., Ricci, S., Jenny, C., Fondi, E., et al. (2020b). Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana. Plant J. 102, 1008–1025. doi: 10.1111/tpj.14683
Mattos, L. A., Amorim, E. P., Amorim, V. B. D. O., Cohen, K. D. O., and Ledo, C. A. D. S. (2010). Agronomical and molecular characterization of banana germplasm. Pesqui. Agropecu. Bras. 45, 146–154. doi: 10.1590/S0100-204X2010000200005
McKay, B. D., and Zink, R. M. (2010). The causes of mitochondrial DNA gene tree paraphyly in birds. Mol. Phylogenet. Evol. 54, 647–650. doi: 10.1016/j.ympev.2009.08.024
Mogensen, H. L. (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 83, 383–404. doi: 10.1002/j.1537-2197.1996.tb12718.x
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nwakanma, D. C., Pillay, M., Okoli, B. E., and Tenkouano, A. (2003). PCR-RFLP of the ribosomal DNA internal transcribed spacers (ITS) provides markers for the A and B genomes in Musa L. Theor. Appl. Genet. 108, 154–159. doi: 10.1007/s00122-003-1402-1
Ortiz, R. (2013). Conventional banana and plantain breeding. Acta Hortic. 986, 117–194. doi: 10.17660/ActaHortic.2013.986.19
Osuji, J. O., Harrison, G., Crouch, J., and Heslop-Harrison, J. S. (1997). Identification of the genomic constitution of Musa L. lines (bananas, plantains and hybrids) using molecular cytogenetics. Ann. Bot. 80, 787–793. doi: 10.1006/anbo.1997.0516
Padam, B. S., Tin, H. S., Chye, F. Y., and Abdullah, M. I. (2014). Banana by-products: an under-utilized renewable food biomass with great potential. J. Food Sci. Technol. 51, 3527–3545. doi: 10.1007/s13197-012-0861-2
Park, H. S., Lee, W. K., Lee, S. C., Lee, H. O., Joh, H. J., Park, J. Y., et al. (2021). Inheritance of chloroplast and mitochondrial genomes in cucumber revealed by four reciprocal F (1) hybrid combinations. Sci. Rep. 11:2506. doi: 10.1038/s41598-021-81988-w
Pereira, A., and Maraschin, M. (2015). Banana (Musa spp) from peel to pulp: ethnopharmacology, source of bioactive compounds and its relevance for human health. J. Ethnopharmacol. 160, 149–163. doi: 10.1016/j.jep.2014.11.008
Rouard, M., Droc, G., Martin, G., Sardos, J., Hueber, Y., Guignon, V., et al. (2018). Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 10, 3129–3140. doi: 10.1093/gbe/evy227
Sardos, J., Perrier, X., DoleŽel, J., Hribová, E., Christelová, P., Van den houwe, I., et al. (2016). DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Ann. Bot. 118, 1269–1278. doi: 10.1093/aob/mcw170
Shen, J., Zhao, J., Bartoszewski, G., Malepszy, S., Havey, M., and Chen, J. (2015). Persistence and protection of mitochondrial DNA in the generative cell of cucumber is consistent with its paternal transmission. Plant Cell Physiol. 56, 2271–2282. doi: 10.1093/pcp/pcv140
Simmonds, N. W., and Shepherd, K. (1995). The taxonomy and origins of the cultivated bananas. J. Linn. Soc., Bot. 55, 302–312. doi: 10.1111/j.1095-8339.1955.tb00015.x
Smith, D. R. (2015). Mutation rates in plastid genomes: they are lower than you might think. Genome Biol. Evol. 7, 1227–1234. doi: 10.1093/gbe/evv069
Stewart, C. N. Jr., and Via, L. E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–750.
Straub, S. C., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., and Liston, A. (2012). Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 99, 349–364. doi: 10.3732/ajb.1100335
Swangpol, S., Volkaert, H., Sotto, R. C., and Seelanan, T. (2007). Utility of selected non-coding chloroplast DNA sequences for lineage assessment of Musa interspecific hybrids. BMB Rep. 40, 577–587. doi: 10.5483/BMBRep.2007.40.4.577
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Whittle, C. A., and Johnston, M. O. (2002). Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol. Biol. Evol. 19, 938–949. doi: 10.1093/oxfordjournals.molbev.a004151
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Keywords: banana, genome skimming, cytoplasmic inheritance, nrDNA, mitogenome, plastome
Citation: Wu C-S, Sudianto E, Chiu H-L, Chao C-P and Chaw S-M (2021) Reassessing Banana Phylogeny and Organelle Inheritance Modes Using Genome Skimming Data. Front. Plant Sci. 12:713216. doi: 10.3389/fpls.2021.713216
Received: 22 May 2021; Accepted: 16 July 2021;
Published: 11 August 2021.
Edited by:
Boas Pucker, University of Cambridge, United KingdomReviewed by:
Mathieu Rouard, Alliance Bioversity International and CIAT, FranceFernando Alexander Garcia-Bastidas, KeyGene, Netherlands
Copyright © 2021 Wu, Sudianto, Chiu, Chao and Chaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu-Miaw Chaw, c21jaGF3QHNpbmljYS5lZHUudHc=
†ORCID: Chung-Shien Wu orcid.org/0000-0003-0000-6253
Edi Sudianto orcid.org/0000-0002-0771-0385
Shu-Miaw Chaw orcid.org/0000-0003-2499-7071
Hui-Lung Chiu orcid.org/0000-0002-3104-807X
Chih-Ping Chao orcid.org/0000-0002-5282-0858
‡These authors have contributed equally to this work
 Chung-Shien Wu
Chung-Shien Wu Edi Sudianto
Edi Sudianto Hui-Lung Chiu
Hui-Lung Chiu Chih-Ping Chao3†
Chih-Ping Chao3† Shu-Miaw Chaw
Shu-Miaw Chaw