- 1Key Laboratory of Tobacco Biology and Processing, Ministry of Agriculture, Tobacco Research Institute, Chinese Academy of Agricultural Sciences, Qingdao, China
- 2Shandong Provincial Key Laboratory of Biochemical Engineering, College of Marine Science and Biological Engineering, Qingdao University of Science and Technology, Qingdao, China
- 3Excellence and Innovation Center, Jiangsu Academy of Agricultural Sciences, Nanjing, China
Secondary wall-associated NAC (SWN) genes are a subgroup of NAC (NAM, ATAF, and CUC) transcription factors (TF) that play a key role in regulating secondary cell wall biosynthesis in plants. However, this gene family has not been systematically characterized, and their potential roles in response to hormones are unknown in Nicotiana tabacum. In this study, a total of 40 SWN genes, of which 12 from Nicotiana tomentosiformis, 13 from Nicotiana sylvestris, and 15 from Nicotiana tabacum, were successfully identified. The 15 SWNs from Nicotiana tabacum were further classified into three groups, namely, vascular-related NAC domain genes (NtVNDs), NAC secondary wall thickening promoting factor genes (NtNSTs), and secondary wall-associated NAC domain genes (NtSNDs). The protein characteristic, gene structure, and chromosomal location of 15 NtSWNs (also named Nt1 to Nt15) were also analyzed. The NtVND and NtNST group genes had five conserved subdomains in their N-terminal regions and a motif (LP[Q/x] L[E/x] S[P/A]) in their diverged C- terminal regions. Some hormones, dark and low-temperature related cis-acting elements, were significantly enriched in the promoters of NtSWN genes. A comprehensive expression profile analysis revealed that Nt4 and Nt12 might play a role in vein development. Others might be important for stem development. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) revealed that in the NtNST group, genes such as Nt7, Nt8, and Nt13 were more sensitive than the genes in NtVND and NtSND groups under abiotic stress conditions. A transactivation assay further suggested that Nt7, Nt8, and Nt13 showed a significant transactivation activity. Overall, SWN genes were finally identified and characterized in diploid and tetraploid tobacco, revealing new insights into their evolution, variation, and homology relationships. Transcriptome, cis-acting element, qRT-PCR, and transactivation assay analysis indicated the roles in hormonal and stress responses, which provided further resources in molecular mechanism and genetic improvement.
Introduction
Secondary cell walls are mainly made up of xylem treachery elements and fibers (Zhong and Ye, 2015a), which are present in the plant cells but absent in the animal cells. They can protect plant cells from potential biotic and abiotic stresses (Brown et al., 2005). They are also the main component of plant materials used for fuel, paper-making, and textiles (Zhong et al., 2010). Secondary wall-associated NAC (SWN) genes are one of the largest transcription factor (TF) families (Su et al., 2013). Among the NAC family, vascular-related NAC domain proteins (VND1 to VND7) and NAC secondary wall thickening-promoting factor proteins (NST1, NST2, and NST3) play a key role in secondary wall biosynthesis (Zhong and Ye, 2015a). Besides, two Arabidopsis SWN domain genes, SND2 and SND3, are also specifically expressed in secondary cell wall development (Hussey et al., 2013; Zhong and Ye, 2014). After a comparative genome analysis across 19 higher plant species, SWN transcriptional factors, including three groups, SND, NST, and VND, were identified as playing a crucial and ruling role in secondary cell wall biosynthesis (Yao et al., 2012).
In Arabidopsis thaliana, AtSWN genes have been proved to be key regulators of secondary cell wall biosynthesis (Wang and Dixon, 2012). AtVND1 to AtVND5 are specifically expressed in xylem vessels, and they can activate the expression of genes involved in secondary wall biosynthesis (Zhou et al., 2014). AtVND6 and AtVND7 can control the formation of metaxylem and protoxylem vessels, and their over-expression will result in the ectopic deposition of the patterned secondary cell walls (Ohashi-Ito et al., 2010). AtNST1 and AtNST2 are redundantly responsible for secondary wall thickening in anther endothecium. AtNST1 and AtNST3 redundantly regulate the secondary wall thickening in the secondary xylem of hypocotyls (Mitsuda et al., 2007). In addition, AtNST1, AtNST2, and AtSND1 regulate secondary cell wall biosynthesis in fibers of stems (Zhong et al., 2006; Zhong and Ye, 2015b). AtSND2 and AtSND3 are shown to promote secondary wall thickening in fibers (Grant et al., 2010). AtVND1–AtVND7 binds directly to the SNBE/TERE-like motif in the AtVND7 promoter region (Nakano et al., 2015). AtVND1–AtVND5 probably triggers xylem tracheary element formation by regulating AtVND7 expression (Fukuda, 2016). AtSND3 is the direct target gene of AtNST1, AtNST1, AtNST2, AtVND6, and AtVND7 (Zhong et al., 2008). AtSND2 is indirectly regulated by AtSND1/AtNST1, but there is no evidence for regulation by AtVND6/AtVND7 (Hussey et al., 2011). Transactivation assays showed that the expression of AtSND1 operates under positive feedback control from itself and that the AtSND1 protein could bind directly to a conserved motif in its promoter (Wang et al., 2011). A two-hybrid assay of yeast revealed the ability of AtVND7 to form homodimers and heterodimers with other AtVND proteins (Yamaguchi et al., 2008). These results indicate that secondary wall biosynthesis is a complex and delicate process; AtSWN genes from the NAC family are key TFs.
Hormones and many TFs also directly regulate or indirectly influence SWN genes. In Populus stems, auxin can repress SND/NST genes associated with fiber and secondary wall formation but induce vessel-specific VND genes; GA (gibberellic acid) induces the expression of SWN genes (Johnsson et al., 2019). In Arabidopsis, auxin inhibits fiber wall thickening by affecting LBD (LATERAL ORGAN BOUNDARIES DOMAIN) genes and repressing the expression of AtNST1- AtNST3 (Lee et al., 2019). LBD15 can directly bind with the upstream sequence of the AtVND7 gene and positively regulate AtVND7 expression (Ohashi-Ito et al., 2018). AtSND1 and AtNST2 are upregulated in the mutants of athb15, which codes a TF that belongs to the HD-ZIP III family and functions as a suppressor of the secondary wall-related regulatory pathway (Du and Wang, 2015). WRKY12 can bind directly to the promoter of AtNST2 to repress its expression, resulting in the suppression of secondary wall biosynthetic genes in pith cells (Wang et al., 2010). E2Fc, a member of the E2F family, can activate the expression of AtVND7 in a dose-dependent manner (Rao and Dixon, 2018). MYB26 is capable of binding to the promoter of AtNST1 and AtNST2 for the regulation of secondary wall thickening (Yang et al., 2007). MYC2 and MYC4 from the bHLH family can also directly bind the promotor region of AtNST1 (Zhang et al., 2018).
In this study, we identified a total of 40 secondary-wall associated NAC transcriptional factors from Nicotiana tabacum (K326), Nicotiana sylvestris, and Nicotiana tomentosiformis. We analyzed 15 NtSWN genes from N. tabacum for further research. After successfully cloning these NtSWN sequences, their gene structures were analyzed, such as motifs, chromosome localization, and expression patterns in different tissues. Additionally, we also analyzed the cis-acting elements of their promoter regions and how they respond to hormones and stress. Finally, functional characterization of the motifs, transactivation activity assay, and subcellular localization were analyzed and studied as well.
Materials and Methods
Plant Materials and Hormone Treatments
Nicotiana tabacum (K326) was used in all the experiments. The seedlings were grown in a culturing room at 25°C under a 16 h:8 h light/dark cycle. To clone the sequences of NtSWNs (Supplementary Table 4), a mixture of complementary DNA (cDNA) extracted from the root, stem, and leaf of 4-week-old seedlings were used as an amplification template. Forward/reverse primers (Supplementary Table 3) were designed in the nearest upstream/downstream regions close to the start/stop codon of the coding sequences (CDS). To obtain correct cDNA sequences, the 2 × Taq Plus Master Mix (Dye Plus) (P213-01; Vazyme Biotech Co. Ltd., Nanjing, China) was used to amplify the NtSWNs, and sequenced with Sanger technology. To investigate the response of NtSWN genes to hormones, 3-week-old seedlings, under normal conditions, were separately given 10 μM GA (GA3, G8040; Solarbio, Beijing, China), 200 μM abscisic acid (ABA, A8060; Solarbio, Beijing, China), 2 mM methyl salicylate (MeSA, SB8540; Solarbio, Beijing, China) in ultrapure water, 100 μM methyl jasmonate (MeJA, M8640; Solarbio, Beijing, China) in.01% (V/V) ethanol, 0.5 μM α-naphthaleneacetic acid (NAA, N8010; Solarbio, Beijing, China), 0.5 μM 2,4-epi-brassionlade (epiBL, B8780; Solarbio, Beijing, China), dark, and 4°C treatments. The seedlings were collected at 0, 1, 3, 6 h after treatment initiation, and non-treated seedlings were used as a control. The samples were collected and immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Three biological replicates were employed per sample.
Transcriptome Sequencing and Expression Analysis
Total RNA from the root, stem, and leaf of 4-week-old seedlings was extracted with the RNAprep Pure Plant Kit (432; TIANGEN, Sichuan, China) and submitted into ribonucleic acid sequencing using the Illumina HiSeq × Ten system with the paired-end method by Annoroad Gene Technology (Beijing) Co. Ltd. For transcriptome analysis, reference genome and annotation files with a gene model were downloaded from the SOL genomics network (SGN, http://www.sgn.cornell.edu) database directly. Raw reads were trimmed for low quality and length by Fastp v0.20.0 (Chen et al., 2018). Clean reads were aligned to the reference genome using Hisat v2.0.5 (Pertea et al., 2016) with the splice-aware method. The gene expression levels from RNA-seq were quantified by feature counts (Liao et al., 2014) and normalized by fragments per kb per million reads (FPKM) values. Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (1.16.1) (Love et al., 2014). The p-values were adjusted using Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted p < 0.05 found by DESeq2 were assigned as differentially expressed. The raw data were deposited in the NCBI SRA database (Accession: PRJNA605912).
Identification and Phylogenetic Analysis of the NAC Genes in Nicotiana spp
The sequences of N. tabacum (K326), N. sylvestris, and N. tomentosiformis were downloaded from the SGN database (Solanaceae Genomics Network, https://solgenomics.net/organism/Nicotiana_tabacum/genome) (Edwards et al., 2017). Previously reported AtSWN protein sequences (AtSWNs) (Zhou et al., 2014) were retrieved from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org) database, and then used as seed sequences to blast against the SGN database with the parameter E-value of 10–5. After manually removing the redundant sequences, candidate SWN protein sequences with a conserved domain (PF02365) were analyzed with the NCBICDD online program (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Multiple sequence alignments of the SWN protein sequences at the amino acid level were performed using the ClustalX program. A neighbor-joining (NJ) phylogenetic tree was then generated based on the alignment result using MEGA 7.0 with the following parameters: Poisson model, pairwise deletion, and bootstrap values (1,000 replicates) (Kumar et al., 2016). Chromosomal locations were determined based on the chromosomal information derived from the SGN database. The positions of the NtSWNs were physically mapped to each chromosome according to their coordinates on the tobacco genome. Finally, SWN genes in N. tabacum (NtSWNs) were identified and named based on their physical locations across the chromosomes and scaffolds.
Gene Structure, Conserved Motif Analysis, and cis-Acting Regulatory Element Analysis
The number of amino acids, molecular weight (MW), and isoelectric point (pI) for each SWN protein were calculated with the DNAStar software (Version 7.0) (Burland, 2000). The subcellular localizations were predicted with ProtComp 9.0 (http://linux1.softberry.com). Exon-intron structures were analyzed by comparing the CDSs and genomic sequences obtained from the SNG database. Conserved motifs were identified with Multiple Em for Motiv Elicitation (MEME) 5.0.5 (Bailey et al., 2015). Parameters were set as follows: distribution of motif occurrences, zero or one per sequence; maximum number of motifs, 25; optimum motif width, ≥10 and ≤24; sites per motif, ≥2 and ≤14. All the results, together with the NJ tree, were visualized with TBtools V0.665 (Chen et al., 2020). The upstream region sequences (3,000 bp) for each gene were also extracted from genome sequences and used for cis-acting element analysis using the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
First-strand cDNA synthesis was performed using 1 μg total RNA with the HiScript®IIIRT SuperMix for qPCR [+ genomic DNA (gDNA) wiper] (R323-01; Vazyme Biotech Co. Ltd., Nanjing, China). The qRT-PCR was performed on a QuantStudio™ 3 Real-Time PCR System machine in a 20 μl reaction with ChamQTM Universal SYBR® qPCR Master Mix (Q711-02/03, Vazyme Biotech Co. Ltd., Nanjing, China). The qRT-PCR assay results were obtained from three independent replicates. Expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001) and normalized by comparison with the NtActin gene (Accession:XM_016618658.1).
Transactivation Activity Assay
The CDS of Nt7, Nt8, Nt10, and Nt13 were amplified using cDNA from K326 leaves with gene-specific primers (Supplementary Table 6). The PCR products were combined into the pBD-GAL4 vector via EcoRI/SalI sites with specific primers (639636; Clontech, CA, United States) (Xiao et al., 2018), and with the pBD-GAL4 vector as the negative control. The yeast strain AH109 (YC1010; Weidi, Shanghai, China), containing the HIS3, ADE2, and MEL1 reporter genes, was used to test transcriptional activation activity. These constructs, together with pGADT7, were transfected into AH109 following the instructions of the yeast strain manufacturer. The transformed yeast cells were incubated on a synthetic dropout (SD) medium lacking leucine and tryptophan (DDO, SD/-Leu-Trp) plates at 30°C for 4 days. Then, the positive colonies were transferred to the SD medium lacking leucine, tryptophan, histidine, and adenine (QDO, SD/-Leu-Trp-His-Ade) supplemented with X-α-Gal plates at 30°C for 4 days.
Subcellular Localization
The full length of Nt7 and the NAC domain of Nt8 and Nt13, excluding the terminator codon, was amplified and ligated into the subcellular localization vector pYG57, which contains the CaMV35S promoter and a C-terminal green fluorescent protein (GFP) domain. Positive clones were confirmed by DNA sequencing, and the recombinant constructs and control were transformed into Agrobacterium tumefaciens strain GV3101. Forty-eight hours after Agrobacterium-mediated transient expression in the 4-week-old N. benthamiana seedlings, fluorescence signals were captured with a confocal microscope (TCS-SP8; Leica, Wetzlar, Germany). DNA dye 4,6-diamidino-2-phenylindole (DAPI) staining was performed to show the position of the nucleus.
Results
Genome-Wide Identification of SWN Family Genes in Tobacco
Twelve AtSWN proteins were employed to search for potential SWNs against the tobacco database by blast and hmmsearch programs. Redundant sequences were then removed through manual reconstruction. Based on conserved domains, the protein sequences shared the same clades, and the AtVND, AtNST, and AtSND groups were used for phylogenetic analysis. A total of 40 NtSWN proteins were successfully identified, including 12 from N. tomentosiformis (Nto), 13 from N. Sylvestris (Nsy), and 15 from N. tabacum (Nt), which were further renamed as Nto1 to Nto12, Nsy1 to Nsy13, and Nt1 to Nt15 (Supplementary Table 1). The full-length amino acid sequences of NtSWNs varied from 279 to 399, and the average amino acid length was 339. The MWs of NtSWN proteins ranged from 31.6 to 45.5 kDa, with the average being 38.9 kDa. The pI values ranged from 5.11 to 8.89, with an average of 6.6. The predicted sub-cellular localization revealed that all the NtSWN proteins were localized to the nucleus (Supplementary Table 2). Furthermore, the NtSWN genes were aligned to the reference genome by the blast program and only 7 of the 15 NtSWN genes were successfully mapped on six chromosomes. Nt4 and Nt5 genes were localized on chromosome 17, and five other NtSWNs were mapped on Chr4, Chr6, Chr12, Chr22, and 23 (Figure 3C).
Phylogenetic Analysis
To investigate the evolutionary relationships among the NtSWN proteins, 52 SWN proteins from A. thaliana, N. tomentosiformis, N. sylvestris, and N. tabacum were aligned by the ClustalX program with multiple alignment method. A neighbor-joint tree was then generated by the MEGA7 program (Figure 1). Based on the phylogenetic tree, the SWN proteins could be clustered into three groups, corresponding to VND, NST, and SND. Every group was highlighted with different colors. The NtSWN proteins from the three tobacco species could be further classed into eight subfamilies (I–VIII) (Figure 1). Among them, the VND and NST groups were categorized separately into three subgroups, while the SND group was divided into two subgroups.
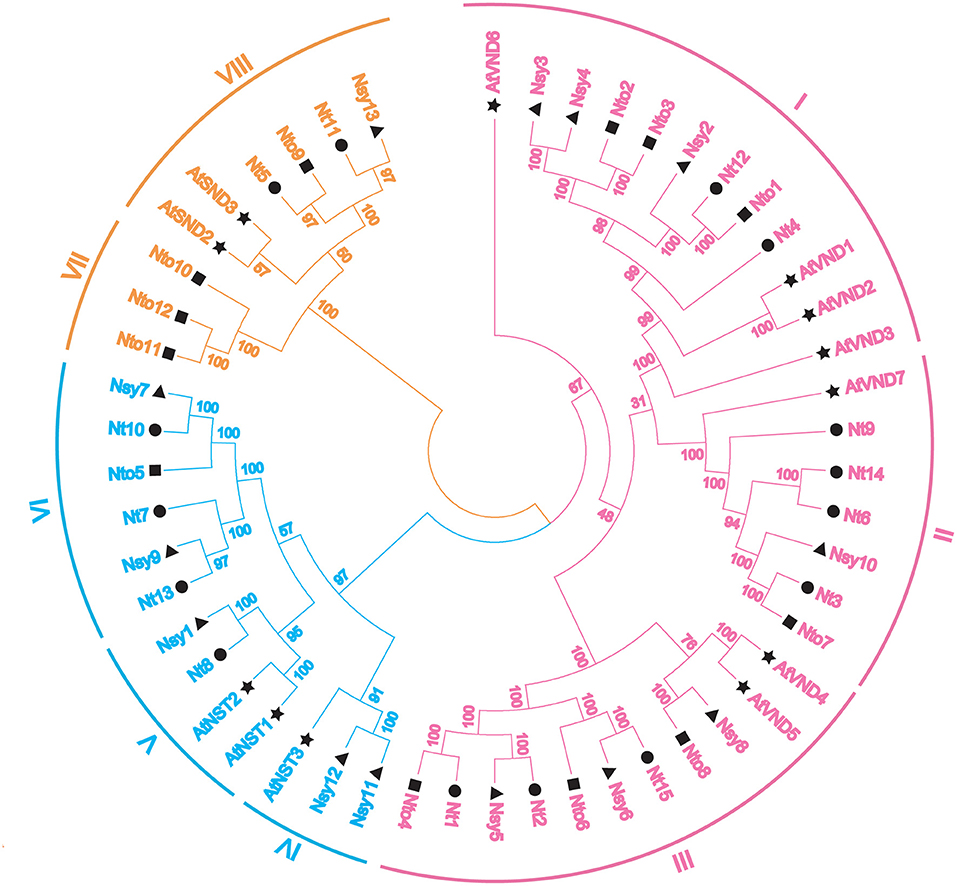
Figure 1. Phylogenetic tree of secondary wall-associated NAC genes (SWNs) with Arabidopsis thaliana, Nicotiana tomentosiformis, Nicotiana sylvestris, and Nicotiana tabacum. Fifty-two SWN proteins were classed into three groups, which are highlighted in specific colors: vascular-related NAC domain genes (VNDs) in pink, NAC secondary wall thickening promoting factor genes (NSTs) in blue, and secondary wall-associated NAC domain genes (SNDs) in orange. Black graphic symbols represent different species: stars indicate A. thaliana, triangles indicate N. Sylvestris, squares indicate N. tomentosiformis, and circles indicate N. tabacum. Peripheral arcs indicated the subgroups of each group, and Roman numerals on the arc represent the subgroups of each group.
Exon–Intron Organization of SWN Genes
To gain further insight into the phylogenetic relationship among the 15 SWN proteins from N. tabacum, a NJ tree was generated individually (Figure 2A). The phylogenetic tree was also classed into three groups, homologous to AtVND, AtNST, and AtSND groups. Patterns of exon–intron architecture variation of the NtSWN genes are analyzed, as shown in Figure 2B. Most of NtSWN genes had three exons and two introns, but Nt4 and Nt9 genes had two exons, and Nt13 had four exons. The length of the second intron of genes in the NtSND subgroup was ~2 kb, which was longer than what was found in the other genes (Figures 2A,B). The genes in the same subgroup generally shared similar exon/intron structures. These results showed that NtSND and NtNST genes were more conserved in the exon–intron structure than NtVND, and showed the gene structure divergence of different subfamilies during evolution (Figure 2C).
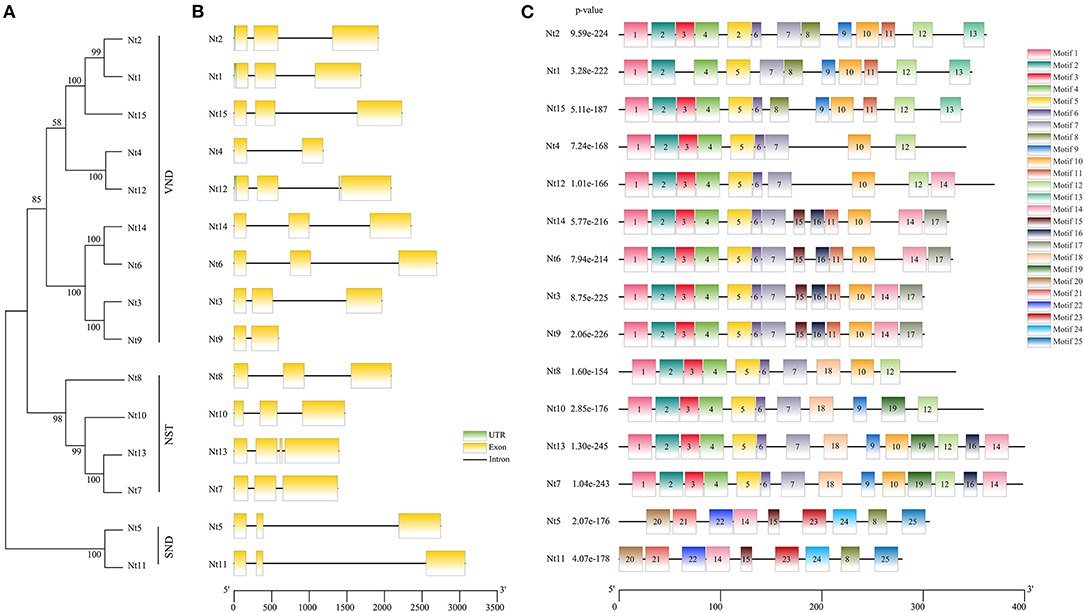
Figure 2. Neighbor-joining (NJ) phylogenetic relationships, gene structure, and motif analysis of NtSWNs. (A) In the phylogenetic tree, the NtVND, NtNST, and NtSND groups were marked in a straight line on the right. (B) In gene structures, untranslated region (UTRs) were indicated by green boxes, exons were represented by yellow boxes, and introns were shown in black lines. (C) Motif compositions were detected with Multiple Em for Motiv Elicitation (MEME); in motif analysis, the motifs were represented by different colored boxes, and each gene contained 9 to 14 motifs.
Conservation and Divergence of C-Terminal Domain Structure in NtSWN Proteins
A typical NAC protein comprises a conserved N-terminal NAC domain (~150 amino acids) and a diversified C-terminal transcription regulatory (TR) region (Puranik et al., 2012). The NAC domain contains five subdomains, namely, A, B, C, D, and E (Satheesh et al., 2014), and the NtVND and NtNST groups have conserved motif patterns, which apparently constitute a conserved NAC domain. In the N-terminal region of NAC, seven motifs were identified in the NAC domain corresponding to subdomains A, B, C, D, and E, and the amino acids in these motifs were conserved (Figure 3A). Among them, Nt1 was without motif 3 in the C subdomain and motif 6 in the D subdomain and Nt15 was without motif 7 in the E subdomain. Compared with the N-terminal, the C-terminal of genes in the NtVND and NtNST groups were highly diverged. The type, quantity, and distribution of the motifs were relatively conserved only in the same subgroup. Motif 18 existed in all NtNSTs and was unique to this group. Except for Nt10, NtVNDs and NtNSTs had motif 10 (Figure 3B). Interestingly, although the C-terminal was highly divergent, motif 10 (LP[Q/x] L[E/x] S[P/A]) was the highly conserved motif across many species (Shen et al., 2009), which indicated that it might be a functional element. The NtSND group was clustered into an independent branch, which is highly divergent in the NAC domain. NtSNDs had nine motifs, such as motifs 14, 8, and 15, which were common with those motifs in the TR region of the NtVND and NtNST groups, while six other motifs were NtSND-specific. These suggested that potential functional differences in NtSNDs existed.

Figure 3. Sequence-specific MEME motifs for NtVNDs and NtNSTs. (A) A–E subdomains were marked in the top left corner, and their corresponding motif numbers are shown in Figure 2C were indicated in the top right corner. (B) The LP [Q/x] L[E/x] S [P/A] motif, namely motif 10, is also shown.
cis-Acting Element Analysis of NtSWN Genes
Promoter regions that contain many cis-acting elements play important roles in the regulation of gene expression response to stresses (Ding et al., 2016). The 3 kb upstream regions of NtSWNs were submitted into the PlantCARE database for the detection of useful information on the regulatory mechanism (Figure 4). All the 15 NtSWN genes contained light-responsive, abiotic stress-related, and hormone-responsive elements. Except for Nt6, Nt8, Nt9, Nt11, and Nt14, the other 10 gene promoters had development-related elements. Furthermore, only Nt3, Nt9, Nt13, and Nt14 had circadian control elements. These results implied that NtSWNs were related to plants responding to light, stress, hormone, and development, and that NtSWNs could be regulated by TFs. The circadian control elements in Nt3, Nt9, Nt13, and Nt14 gene promoters suggest that they might have a distinct diurnal expression pattern. The most abundant cis-acting elements were MYB-related (Myb, MYB, MYB recognition site, Myb-binding site, MYB-like sequence) and MYC-related (Myc, MYC). The highest diverged cis-acting elements were light-responsive. Auxin (AuxRR-core, TGA-box, TGA-element), gibberellin (GARE-motif, P-box, TATC-box), ABA (ABRE), MeJA (CGTCA-motif, TGACG-motif), and salicylic acid (TCA-element) responsiveness elements were also found in the promoters of NtSWNs, which indicated that auxin, gibberellin, jasmonic acid, and salicylic acid participated in the expression regulation of NtSWN genes. The elements involved in ABA and MeJA were the most abundant, followed by ABA and gibberellin elements.
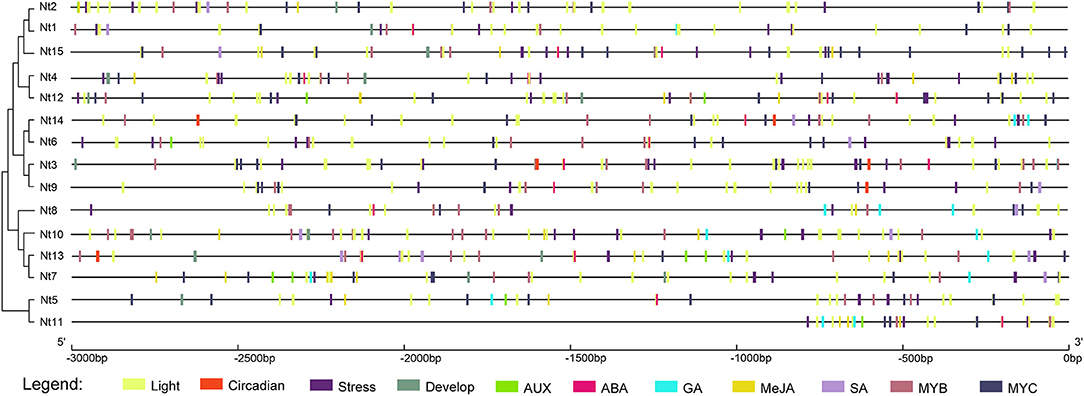
Figure 4. Distribution of main cis-acting elements in the 3 kb upstream promoter regions of NtSWN genes. The different types of cis-acting elements were represented using rectangles with different colors. The scale above was used to measure nucleoside length.
NtSWN Gene Isolation and Expression Pattern in Different Tissues
Expression patterns of these genes were then analyzed using the RNA-seq data. Genes in the same subgroup tended to have similar expression patterns (Figure 5). The expression level of NtSWN genes was highest in the stem, followed by leaf and vein and in root it was minimum. Interestingly, the expression patterns in the stem and vein of Nt4 and Nt12 in the VND group were opposite to the other genes. The gene expression profiles in different tissues provided preliminary clues for their function. These results showed that most NtSWN genes are involved in stem development, and that Nt4 and Nt12 might participate in vein development. Most of the NtSWN genes have a similar expression pattern, which indicated that the secondary cell wall thickening mainly in stem at this development stage, and these transcriptional factors were co-regulated in this process.
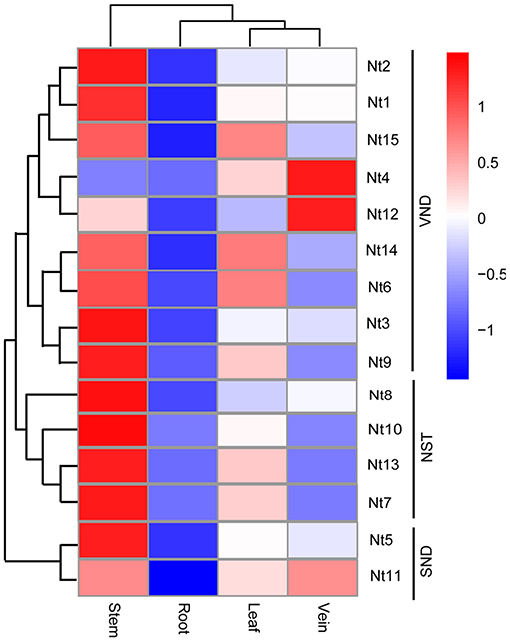
Figure 5. Expression profile of NtSWN genes in tissues. Expression patterns of the NtSWNs in the root, stem, leaf, and vein of 3-week-old seedlings.
Expression Dynamic of NtSWN Gene Response to Hormones and Stresses by qRT-PCR
To identify the NtSWN genes responsive to hormones and abiotic stresses, the expression patterns of NtSWNs in plants subjected to GA, ABA, MeSA, MeJA, NAA, epiBL, dark, and low temperature treatments were investigated by qRT-PCR (Figure 6). The PCR primers used in this study are shown in Supplementary Table 5, and the 0 h seedlings were used as control. In statistics, differential expression patterns of NtSWNs were analyzed by t-test (Supplementary Table 7). The expression level of most of the NtSWNs were upregulated with the ABA, MeJA, NAA, epiBL, dark, and low temperature treatments, but those genes were downregulated with the GA3 and MeSA treatments. For the GA3 and MeJA treatments, the expression level of most of the NtSWNs reached peaks at 1 h and then kept decreasing. In the ABA, epiBL, and low temperature treatments, the expression level of most of the NtSWNs reached peaks at 3 h. For the MeSA, NAA, and dark treatments, the expression level of most of the NtSWNs reached peaks at 6 h. Nt9 was remarkably induced by ABA, MeSA, and NAA, when its transcription level increased more than 14-fold at 3 h, 7-fold at 6 h, and 53-fold at 3 h, respectively. In the GA3 and dark treatments, the expression level of Nt13 was upregulated by 2-fold and 28-fold at 1 h, respectively. Nt1was upregulated with MeJA by 12-fold at 1 h; Nt7 upregulated with epiBL by 17-fold at 3 h; Nt6 was upregulated with low temperature by 7-fold at 3 h. Genes in the NtNST group were used for further research, because they were more sensitive to hormone and abiotic stresses than those in NtVND and NtSND.
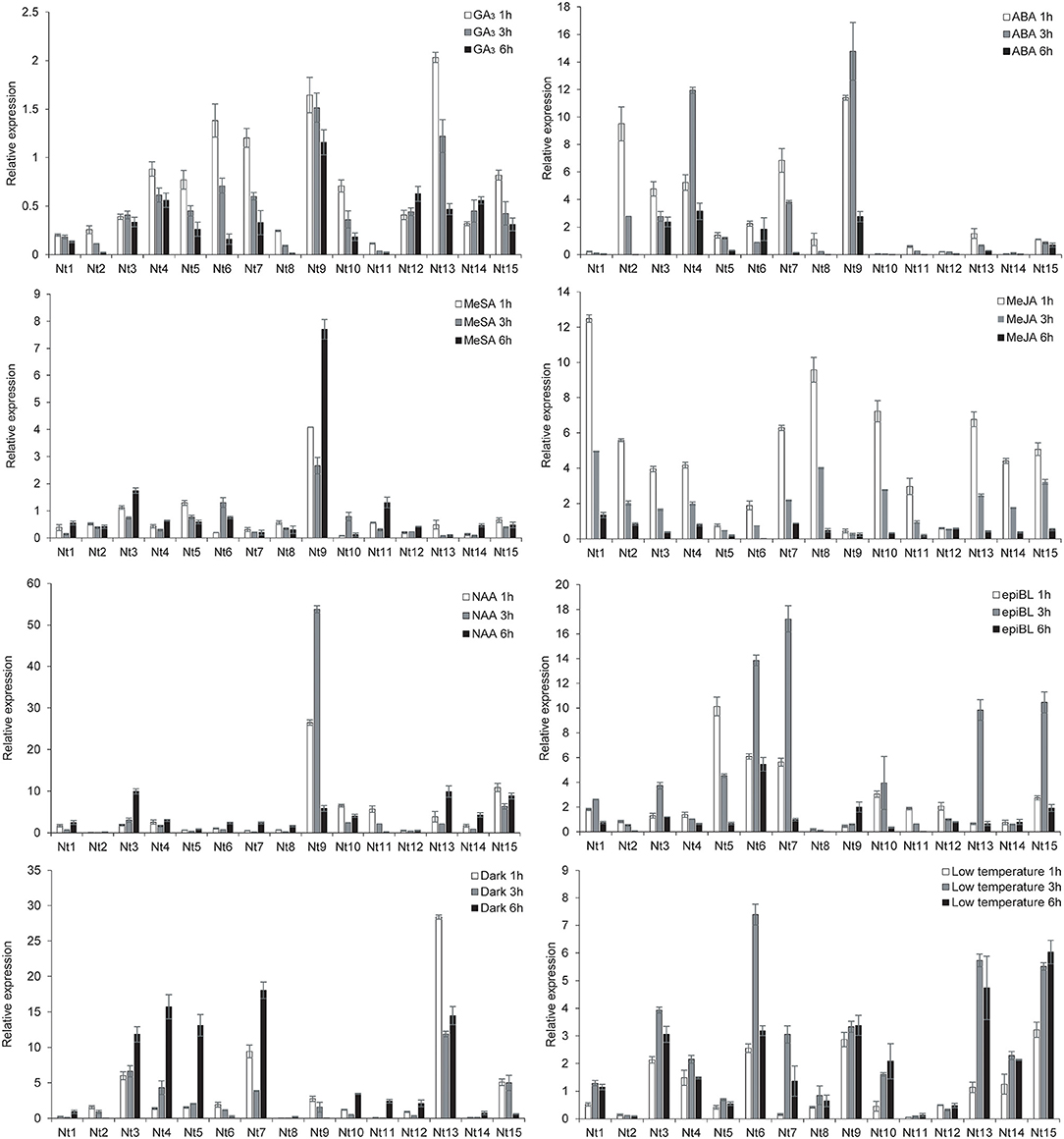
Figure 6. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of NtSWN expression in response to hormone treatments and abiotic stresses. Tree-week-old seedlings were treated with 10 μM gibberellic acid (GA3), 200 μM abscisic acid (ABA), 2 mM methyl salicylate (MeSA), 100 μM methyl jasmonate (MeJA), 0.5 μM α-naphthaleneacetic acid (NAA), 0.5 μM 2,4-epi-brassionlade (epiBL), dark and 4°C. Error bars indicate SE.
Transactivation Activity Assay
The full-length CDS of the NtNST group (Nt7, Nt8, Nt10, and Nt13) were used to investigate the transactivation activity with the reporter genes of AH109, such as lacZ, HIS3, ADE2, and MEL1. The transgenic yeasts were selected on the SD medium (SD/-Leu-Trp), followed by X-α-Gal activity monitoring on the SD medium (SD/-Leu-Trp-His-Ade). The yeast transformant constructed with pBD-GAL4-t7, pBD-GAL4-Nt8, and pBD-GAL4-Nt13 turned blue in the presence of X-α-Gal and grown normally on the SD medium (SD/-Leu-Trp-His-Ade), whereas the pBD-GAL4-Nt10 and negative control did not (Figure 7). These results suggested that Nt7, Nt8, and Nt13 could regulate the expression of downstream target genes, and that Nt10 perhaps could not.
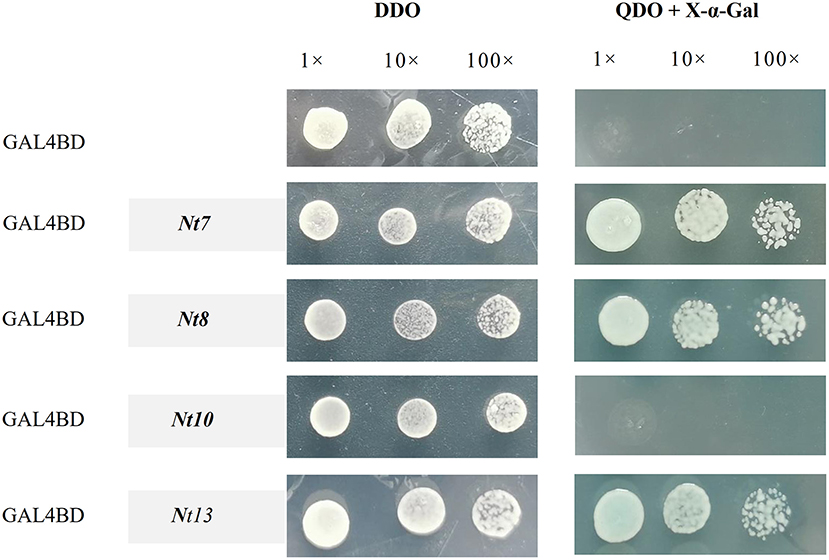
Figure 7. Transactivation analysis of Nt7, Nt8, Nt10, and Nt13. Genes were combined into the GAL4 (BD) DNA binding domain in pBD-GAL4, with the empty pBD-GAL4 vector as the negative control. The transformed yeast cells were incubated on DDO (SD/-Leu-Trp) and QDO (SD/-Leu-Trp-His-Ade) with X-α-Gal. The yeast was diluted 10 and 100 times.
Subcellular Localization
We chose the genes with transactivation activity to further determine their subcellular localization (Figure 8). The open reading frame (ORF) of Nt7, Nt8, and Nt13, excluding the stop codon, was cloned to be in a frame with a GFP reporter gene, which was under the control of the CaMV-35S promoter. Fusion protein, including Nt7-GFP, Nt8-GFP, and Nt13-GFP with full-length CDS, were constructed. But no GFP signal were observed in Nt8-GFP and Nt13-GFP. NAC-domain of Nt8 and Nt13 were further cloned and inserted into pYG57 vector for subcellular localization. The GFP signals from full-length CDS of Nt7 and the NAC domains of Nt8 and Nt13 were all localized in the cell nucleus and merged with DAPI. These results suggested that fusion proteins were specifically localized in the nucleus.
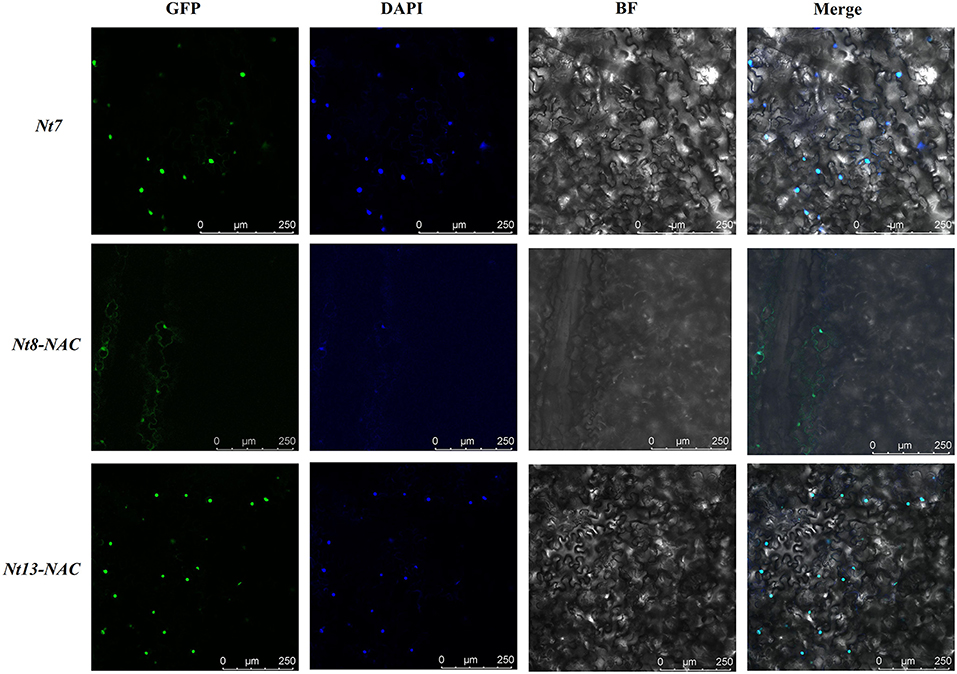
Figure 8. Subcellular localization of Nt7 and the NAC domain of Nt8 and Nt13. The fusion constructs and green fluorescent protein (GFP) driven by the 35S promoter were transiently expressed in tobacco. DNA dye 4,6-diamidino-2-phenylindole (DAPI) was used as a nucleus marker, and the green fluorescent signals of these genes can be merged with DAPI.
Discussion
The major constituent tissue of wood is the xylem, which includes tracheary elements, fibers, and rays (Zhou et al., 2011). Compared with Arabidopsis, tobacco has typical xylem (Chaffey et al., 2002) and is easier to do gene transformation. Therefore, tobacco is an ideal model plant in xylem development research (Edwards et al., 2017). Secondary wall formation is an important process in xylem differentiation (Zhong and Ye, 2015a). SWNs are master TFs in the process of secondary wall deposition (Ohashi-Ito and Fukuda, 2010). To date, SWNs have been identified and characterized in many plants; however, few studies on SWNs in tobacco have been reported. As a result, it is of great importance to isolate SWNs from the tobacco genome and explore their characteristics.
Nicotiana tabacum is an allotetraploid, and it is generated via natural crossing between N. sylvestris and N. tomentosiformis. In this study, we successfully isolated 15 NtSWNs, and the N. tabacum SWNs were fewer probably because of low-sequence conservation or incomplete genome information of N. tabacum (Figure 1) (Li et al., 2018). These 15 NtSWNs, together with AtSWNs, divided into VND, NST, SND, suggesting that the ancestors of SWNs differentiated before the divergence between tobacco and Arabidopsis (Ding et al., 2016).
The exon-intron organization in the same subgroup was similar, indicating a close evolutionary relationship (Figure 2B). In general, among the NAC proteins of different plant species, NAC domains are conserved when TR regions are diverse. However, in this study, NtSNDs did not have a conserved NAC domain at their N-terminal, while NtVNDs and NtNSTs had conserved patterns at their C-terminal (Figure 2C). The motifs in the N-terminal of NtSNDs were different from the A to E subdomains (Shen et al., 2009). Moreover,motifs A, D, and E were present in all the NAC genes, whereas motifs B and C were absent in several NAC genes, which were grouped together in the phylogeny (Satheesh et al., 2014).
The analysis of cis-acting elements helped us to predict the genes responsive to hormones, environment, and stress. Seven kinds of cis-acting elements exist in the promoters of NtSWNs, such as light-responsive elements, stress-related elements, hormone-responsive elements, development-related elements, circadian control elements, and MYB and MYC elements (Figure 5). The ubiquitous MYB and MYC elements in the promoters of NtSWNs suggested that these TFs might be important upstream regulators. MYB46 and MYB83 are direct targets of SWNs in the regulation of secondary cell wall biosynthesis (Zhong and Ye, 2012), but up to now, only MYB26 was reported as the upstream regulatory factor of SWNs (Yang et al., 2007). The ubiquitous MYB elements suggest that there might exist many other MYBs that can directly bind the promoters of NtSWNs. Light-responsive elements are the most variable cis-acting elements in the promoters of NtSWNs, which indicated that different colors and intensities of light might regulate the expression of NtSWNs through different pathways.
Different genes had the same cis-acting elements in their promoter-region, which might provide clues for further exploration of different molecular functions of NtSWNs. For example, three NAC transcription factors, ANAC019, ANAC055, and ANAC072, were proved to bind MYC-like sequence (CATGTG) in Arabidopsis (Tran, 2004). Besides, BES1(bri1-EMS-suppressor 1), a positive regulator in brassinosteroid (BR) signaling pathway, interacts with BIM1 (BES1-interacting MYC-like 1) to synergistically bind to E box (CANNTG) sequences present in many BR-induced promoters in Arabidopsis (Yin et al., 2005). It was suggested that both NAC and BES1 might regulate the expression of NtSWNs by binding to an MYC element. The widespread G-box, mentioned as a light-responsive element, was also the target of BZR1 (an important gene in the BR signal pathway.) (Oh et al., 2012; Zhang et al., 2014). It was suggested that both light and BR could regulate the expression of NtSWNs by binding to G-box. Due to BR also playing an important role in photomorphogenesis (Kang et al., 2001, Paulisic et al., 2017), the light might regulate the expression of NtSWNs through the BR signal pathway. Moreover, the same factor might regulate the NtSWNs genes through different cis-acting elements. For instance, jasmonate (JA) and MYC TFs, which emerged as both activators and repressors of the JA signaling pathway, can bind to G-box (Yang et al., 2018). It was suggested that JA might regulate NtSWNs through G-box. Besides G-box, JA could regulate NtSWNs through the GARE-motif, P-box, and TATC-box as well. Auxin regulates cambial activity; GA can regulate xylem differentiation and xylem fiber elongation (Elo et al., 2009); BR can promote xylem differentiation (Zhang et al., 2011); JA can stimulate secondary growth (Sehr et al., 2010). Cold, light, and salicylic acid can stimulate lignin biosynthesis; BR can regulate cellulose biosynthesis (Didi et al., 2015; Le Gall et al., 2015). VND2, VND4, VND5, and NST3 are upregulated by ABA, and VND6 is downregulated by auxin and inducted by BR (Didi et al., 2015). According to previous studies, we explored the expression profiles of NtSWNs comprehensively, and it is the first time to report the expression profiles of NtSWNs with different hormones and stresses (Figure 7). Statistical results show that most of NtSWNs could be upregulated by ABA, MeJA, NAA, epiBL, dark, and low temperature, and downregulated by GA3 and MeSA. With the ABA, MeJA, NAA, and low temperature treatments, the expression pattern of Nt12 was different from that of the other genes, indicating that the function of Nt12 in hormonal and stress responses might be opposite to that of the other genes. In comparison with the other genes, Nt9 was more sensitive to ABA, NAA, and MeSA, indicating that Nt9 might play an important role in the response to these hormones. The gene expression profiles in different tissues provided preliminary clues for their function. These results showed that most of the NtSWN genes are involved in stem development, and that Nt4 and Nt12 might participate in vein development. Most of the NtSWN genes have a similar expression pattern, which indicated that the secondary cell wall thickening, mainly in the stem at this development stage and transcriptional factors, were co-regulated in this process.
A transactivation activity assay is always performed to investigate whether a gene has transactivation activity and performs the function as a TF in regulating the expression of downstream target genes. Information on the subcellular localization of proteins can help to clarify their functions and be used to predict the subcellular localization of homologous genes in other species. In this study, Nt7, Nt8, and Nt13 localized in the cell nucleus and had a transactivation activity, which indicated that Nt7, Nt8, and Nt13 are functional genes. This analysis provides preliminary results of the putative functions of NtSWNs for further research on the regulation of secondary cell wall biosynthesis and xylem development.
Conclusions
In this study, a total of 40 SWN genes in tobacco were systematically identified and characterized. Fifteen NtSWN genes from the NtVND, NtNST, and NtSND groups were further investigated for their expression patterns, dynamic change, and transactivation with hormones and stresses. Transcriptome and qRT-PCR analysis revealed that the NtNST group genes, such as Nt7, Nt8, and Nt13, were more sensitive to abiotic stresses. The transactivation assay further suggested that Nt7, Nt8, and Nt13 showed a significant transactivation activity. These results will provide useful insights on secondary cell wall-associated NAC genes for further molecular mechanism and genetic improvement in tobacco.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The RNA-Seq data were deposited in the NCBI SRA database (Accession: PRJNA605912). The accession numbers of NtNAC genes can be found in the article/Supplementary Material.
Author Contributions
WS and YL conceived and designed the experiments. NX and LM performed the experiments. LS, XL, SD, and FH participated in data collection and analysis. NX wrote the manuscript. WS and YL revised the manuscript. All the authors have read and approved the final manuscript.
Funding
This study was financially supported by the Agricultural Science and Technology Innovation Program (ASTIP-TRIC03), the Science Foundation for Young Scholars of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (2018B03), and the National Nature Science Foundation of Shandong Province (ZR2019BC066).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Jingjuan Yu (State Key Laboratory of Agrobiotechnology) for the kind technical guidance and for providing the transactivation activity experimental system. We also thank Dr. Wei Li (Tobacco Research Institute, Chinese Academy of Agricultural Sciences) for the critical discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.712254/full#supplementary-material
Supplementary Table 1. Protein sequences of the secondary wall-associated NAC genes (SWN) gene family members in Arabidopsis, Nicotiana sylvestris, Nicotiana tomentosiformis, and Nicotiana tabacum (K326).
Supplementary Table 2. Supplementary Table 2. Characteristics of NtSWNs identified in Nicotiana tabacum.
Supplementary Table 3. Specific primers for the amplification of NtSWNs.
Supplementary Table 4. Complementary deoxyribonucleic acid (cDNA) sequences of NtSWN genes.
Supplementary Table 5. Specific primers for quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Supplementary Table 6. Specific primers for transactivation activity assay.
Supplementary Table 7. Statistical differential expression analysis of 15 NtSWNs between treatments and control.
References
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–49. doi: 10.1093/nar/gkv416
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. doi: 10.1105/tpc.105.031542
Burland, T. G. (2000). DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91. doi: 10.1385/1-59259-192-2:71
Chaffey, N., Cholewa, E., Regan, S., and Sundberg, B. (2002). Secondary xylem development in Arabidopsis: a model for wood formation. Physiol. Plant. 114, 594–600. doi: 10.1034/j.1399-3054.2002.1140413.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Didi, V., Jackson, P., and Hejatko, J. (2015). Hormonal regulation of secondary cell wall formation. J. Exp. Bot. 66, 5015–5027. doi: 10.1093/jxb/erv222
Ding, A. M., Marowa, P., and Kong, Y. Z. (2016). Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol. Gene. Genom. 291, 1891–1907. doi: 10.1007/s00438-016-1226-8
Du, Q., and Wang, H. Z. (2015). The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 10:e1078955. doi: 10.1080/15592324.2015.1078955
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18:448. doi: 10.1186/s12864-017-3791-6
Elo, A., Immanen, J., Nieminen, K., and Helariutta, Y. (2009). Stem cell function during plant vascular development. Semin. Cell Developm. Biol. 20, 1097–1106. doi: 10.1016/j.semcdb.2009.09.009
Fukuda, H. (2016). Signaling, transcriptional regulation, and asynchronous pattern formation governing plant xylem development. Proceed. Japan Acad. Series B-Physic. Biol. Sci. 92, 98–107. doi: 10.2183/pjab.92.98
Grant, E. H., Fujino, T., Beers, E. P., and Brunner, A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337–352. doi: 10.1007/s00425-010-1181-2
Hussey, S. G., Mizrachi, E., Creux, N. M., and Myburg, A. A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4:325. doi: 10.3389/fpls.2013.00325
Hussey, S. G., Mizrachi, E., Spokevicius, A. V., Bossinger, G., Berger, D. K., and Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11:173. doi: 10.1186/1471-2229-11-173
Johnsson, C., Jin, X., Xue, W. Y., Dubreuil, C., Lezhneva, L., and Fischer, U. (2019). The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol. Plant. 165, 673–689. doi: 10.1111/ppl.12766
Kang, J. G., Yun, J., Kim, D. H., Chung, K. S., Fujioka, S., Kim, J. I., et al. (2001). Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell 105, 625–636. doi: 10.1016/S0092-8674(01)00370-1
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Lee, K. H., Du, Q., Zhuo, C. L., Qi, L. Y., and Wang, H. Z. (2019). LBD29-involved auxin signaling represses NAC master regulators and fiber wall biosynthesis. Plant Physiol. 181, 595–608. doi: 10.1104/pp.19.00148
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, X., Hamyat, M., Liu, C., Ahmad, S., Gao, X., Guo, C., et al. (2018). Identification and characterization of the WOX family genes in five Solanaceae species reveal their conserved roles in peptide signaling. Genes 9:260. doi: 10.3390/genes9050260
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Nakano, Y., Yamaguchiz, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Oh, E., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–U864. doi: 10.1038/ncb2545
Ohashi-Ito, K., and Fukuda, H. (2010). Transcriptional regulation of vascular cell fates. Curr. Opin. Plant Biol. 13, 670–676. doi: 10.1016/j.pbi.2010.08.011
Ohashi-Ito, K., Iwamoto, K., and Fukuda, H. (2018). LOB DOMAIN-CONTAINING PROTEIN 15 positively regulates expression of VND7, a Master regulator of tracheary elements. Plant Cell Physiol. 59, 989–996. doi: 10.1093/pcp/pcy036
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Paulisic, S., Molina-Contreras, M. J., Roig-Villanova, I., and Martinez-Garcia, J. F. (2017). Approaches to study light effects on brassinosteroid sensitivity. Methods Mol. Biol. 1564, 39–47. doi: 10.1007/978-1-4939-6813-8_5
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, String tie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Rao, X. L., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Satheesh, V., Jagannadham, P. T., Chidambaranathan, P., Jain, P. K., and Srinivasan, R. (2014). NAC transcription factor genes: genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.). Mol. Biol. Rep. 41, 7763–7773. doi: 10.1007/s11033-014-3669-5
Sehr, E. M., Agusti, J., Lehner, R., Farmer, E. E., Schwarz, M., and Greb, T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63, 811–822. doi: 10.1111/j.1365-313X.2010.04283.x
Shen, H., Yin, Y. B., Chen, F., Xu, Y., and Dixon, R. A. (2009). A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res. 2, 217–232. doi: 10.1007/s12155-009-9047-9
Su, H. Y., Zhang, S. Z., Yuan, X. W., Chen, C. T., Wang, X. F., and Hao, Y. J. (2013). Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1,2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 71, 11–21. doi: 10.1016/j.plaphy.2013.06.022
Tran, L.-S. P. (2004). Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Wang, H. Z., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Wang, H. Z., and Dixon, R. A. (2012). On-Off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, H. Z., Zhao, Q., Chen, F., Wang, M. Y., and Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x
Xiao, W., Yang, Y., and Yu, J. (2018). ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.). Plant Sci. 266, 83–94. doi: 10.1016/j.plantsci.2017.03.012
Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. doi: 10.1111/j.1365-313X.2008.03533.x
Yang, C. Y., Xu, Z. Y., Song, J., Conner, K., Barrena, G. V., and Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548. doi: 10.1105/tpc.106.046391
Yang, Y. F., Zhang, K. K., Yang, L. Y., Lv, X., Wu, Y., Liu, H. W., et al. (2018). Identification and characterization of MYC transcription factors in Taxus sp. Gene 675, 1–8. doi: 10.1016/j.gene.2018.06.065
Yao, D. X., Wei, Q., Xu, W. Y., Syrenne, R. D., Yuan, J. S., and Su, Z. (2012). Comparative genomic analysis of NAC transcriptional factors to dissect the regulatory mechanisms for cell wall biosynthesis. BMC Bioinform. 13:S10. doi: 10.1186/1471-2105-13-S15-S10
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Zhang, D., Ye, H., Guo, H., Johnson, A., Lin, H., and Yin, Y. (2014). Transcription factors involved in brassinosteroid repressed gene expression and their regulation by BIN2 kinase. Plant Signal. Behav. 9:e27849. doi: 10.4161/psb.27849
Zhang, J., Elo, A., and Helariutta, Y. (2011). Arabidopsis as a model for wood formation. Curr. Opin. Biotechnol. 22, 293–299. doi: 10.1016/j.copbio.2010.11.008
Zhang, Q., Xie, Z., Zhang, R., Xu, P., Liu, H. T., Yang, H. Q., et al. (2018). Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-directed transcriptional network in Arabidopsis. Plant Cell 30, 2512–2528. doi: 10.1105/tpc.18.00315
Zhong, R. Q., Demura, T., and Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R. Q., Lee, C. H., and Ye, Z. H. (2010). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15, 625–632. doi: 10.1016/j.tplants.2010.08.007
Zhong, R. Q., Lee, C. H., Zhou, J. L., McCarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R. Q., and Ye, Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. doi: 10.1093/pcp/pcr185
Zhong, R. Q., and Ye, Z. H. (2014). Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 229, 193–207. doi: 10.1016/j.plantsci.2014.09.009
Zhong, R. Q., and Ye, Z. H. (2015a). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56, 195–214. doi: 10.1093/pcp/pcu140
Zhong, R. Q., and Ye, Z. H. (2015b). The Arabidopsis NAC transcription factor NST2 functions together with SND1 and NST1 to regulate secondary wall biosynthesis in fibers of inflorescence stems. Plant Signal. Behav. 10:e989746. doi: 10.4161/15592324.2014.989746
Zhou, J., Sebastian, J., and Lee, J. Y. (2011). Signaling and gene regulatory programs in plant vascular stem cells. Genesis 49, 885–904. doi: 10.1002/dvg.20795
Keywords: NAC transcription factors, secondary cell wall, hormonal and stress responses, transcriptional activation, subcellular localization, Nicotiana tabacum
Citation: Xu N, Meng L, Song L, Li X, Du S, Hu F, Lv Y and Song W (2021) Identification and Characterization of Secondary Wall-Associated NAC Genes and Their Involvement in Hormonal Responses in Tobacco (Nicotiana tabacum). Front. Plant Sci. 12:712254. doi: 10.3389/fpls.2021.712254
Received: 20 May 2021; Accepted: 12 August 2021;
Published: 14 September 2021.
Edited by:
Mario A. Pagnotta, University of Tuscia, ItalyReviewed by:
Aqeel Ahmad, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaChristell Van Der Vyver, Stellenbosch University, South Africa
Copyright © 2021 Xu, Meng, Song, Li, Du, Hu, Lv and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanda Lv, THlkMDUyN0AxMjYuY29t; Wenjing Song, c29uZ3dlbmppbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Na Xu
Na Xu Lin Meng
Lin Meng Lin Song
Lin Song Xiaoxu Li
Xiaoxu Li Shasha Du
Shasha Du Fengqin Hu
Fengqin Hu Yuanda Lv
Yuanda Lv Wenjing Song
Wenjing Song