
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 August 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.710826
This article is part of the Research TopicInteractions Between Biochemical Pathways Producing Plant Colors and ScentsView all 19 articles
Auxin, an important plant hormone, induces the biosynthesis of various secondary metabolites by modulating the expression of auxin-responsive genes. In the ornamental plant Hedychium coronarium, linalool and methyl benzoate are biosynthesized by the terpene synthase (TPS) HcTPS5 and the benzoic/salicylic acid methyltransferase (BSMT) HcBSMT2, respectively. However, the transcriptional regulation of this process remains unclear. Here, we identified and functionally characterized the R2R3-MYB transcription factors HcMYB1 and HcMYB2 in regulating the biosynthesis of these floral aroma compounds. HcMYB1 and HcMYB2 are specifically expressed in flowers, their expression is correlated with the emission of volatile compounds in flowers, and is induced by auxin. Moreover, HcMYB1 and HcMYB2 interact with the HcBSMT2 promoter region. HcMYB2 activates the expression of the linalool synthase gene HcTPS5. In flowers with HcMYB1 or HcMYB2 silenced, the levels of floral scent compounds were significantly reduced, and HcBSMT2 and HcTPS5 were downregulated compared with the wild type. Moreover, HcMYB1 form protein-protein interaction with key scent-related HcIAA4 protein to regulate floral aroma production. Taken together, these results indicate that HcMYB1 and HcMYB2 play crucial roles in regulating the formation of scent compounds in Hedychium coronarium (H. coronarium) flowers in response to auxin signaling.
Floral scent compounds are among the most important secondary metabolites in plants and comprise three major groups based on their origins: terpenoids, phenylpropanoids/benzenoids, and fatty acid derivatives (Dudareva et al., 2013; Muhlemann et al., 2014; Abbas et al., 2017). These compounds are attractive not only to humans but also to pollinators that facilitate fertilization, and thus they play key roles in plant evolution and the plant lifecycle (Raguso, 2009). The anti-herbivore or antimicrobial activity of volatiles released from flowers protects the vulnerable reproductive parts of the plant against pathogen attack (Dudareva et al., 2006). Floral scent also is an important trait that increases the aesthetic values of ornamental plants to humans (Dudareva et al., 2006; Pichersky and Dudareva, 2007), and scented compounds derived from flowers are widely used as flavorings, in cosmetics and perfumes, and as medicinal substances (Gershenzon and Dudareva, 2007; Muhlemann et al., 2014). However, notwithstanding the importance of floral scents to both plant biology and industry, little is known about the transcriptional regulation of this process.
MYB transcription factors are important regulators of the biosynthesis of plant secondary metabolites, such as phenylpropanoids (Liu L. et al., 2015; Zhou and Memelink, 2016; Ramya et al., 2017). MYB proteins possess two regions: a conserved MYB DNA-binding domain at the N-terminus and a diverse modulator region at the C-terminus that is responsible for their regulatory activity. MYB transcription factors (TFs) are classified into four subunits/groups based on the number of adjacent repeats in the DNA-binding domains: the R2R3-, 1R-, 3R-, and 4R-MYBs (Dubos et al., 2008). Most MYB TFs involved in regulating secondary metabolite biosynthesis in flowers belong to the R2R3-MYB family (Ramya et al., 2017). To date, however, only a few MYB TFs identified from floral scent model species, i.e., snapdragon (Antirrhinum majus) and petunia (Petunia spp.) have been shown to regulate the expression of structural genes related to volatile biosynthetic pathways. The R2R3-MYB TFs ODORANT1 (ODO1) and EMISSION OF BENZENOID II (EOBII) regulate volatile biosynthesis genes in the benzenoid/phenylpropanoid pathway in petunia (Verdonk et al., 2005; Spitzer-Rimon et al., 2012). In snapdragon, the MYBs AmMYB305 and AmMYB340 are involved in regulating the volatile phenylpropanoid/benzenoid metabolic pathway (Uimari and Strommer, 1997; Shin et al., 2002). In addition, the R2R3-MYB TF FaEOBII regulates the production of the volatile eugenol in ripe strawberry (Fragaria × ananassa) receptacles by activating the expression of CINNAMYL ALCOHOL DEHYDROGENASE (FaCAD1) and EUGENOL SYNTHASE (FaEGS2) (Medina-Puche et al., 2015). In the Cymbidium orchid cultivar “Sael Bit,” CsMYB1 is highly expressed in floral organs and is involved in regulating the biosynthesis of floral volatiles such as polyacrylate and 2-methyl butyraldehyde in petals (Ramya et al., 2019).
Floral and fruit volatiles are also regulated by the essential plant hormone auxin, which induces the biosynthesis of numerous secondary metabolites by regulating the expression of auxin-responsive genes (Zhou and Memelink, 2016). Treatment with exogenous auxin increases the emission of the volatile compound linalool in apple (Malus domestica) scions (Kim et al., 2011) and modifies the quantity of fruit flavor compounds. In strawberry, auxin treatment enhances the aggregation of phenolic volatiles such as 2-phenylethanol, phenylacetaldehyde, and methyl benzoate, and inhibits the production of benzyl cyanide, 2-isobutylthiazole, 1-hexanol, and 1-nitro-2-phenylethane (Wu et al., 2018). Exogenous auxin treatment also modifies the expression of several key genes associated with the biosynthetic pathways of scent volatiles, including PHENYL ALDEHYDE REDUCTASE 1 (SlPAR1), SlPAR2, and SlSAMT1, in tomato (Solanum lycopersicum) (Wu et al., 2018). In grapefruit (Citrus × paradisi), auxin treatment influences sugar accumulation in various ways, as well as the accumulation of volatile compounds and the expression of aroma-related genes (Jia et al., 2017). Many MYB TF genes respond to auxin signalings, such as the Arabidopsis thaliana genes AtMYB44, AtMYB77, and AtMYB108 (Shin et al., 2007; Dubos et al., 2010) and ten R2R3-MYB genes in cassava (Manihot esculenta) (Liao et al., 2016). Nevertheless, how auxin is involved in regulating the phenylpropanoid and terpenoid biosynthetic pathway via MYB TFs was not known.
Hedychium coronarium is a perennial herb of the Zingiberaceae family that is cultivated as a cut flower, garden plant, and medicinal plant and for aromatic oil production. At blooming, H. coronarium flowers emit large amounts of volatile compounds, including the monoterpenes linalool and (E,Z)-β-ocimene, and benzenoids such as methyl benzoate (Fan et al., 2003, 2007; Li and Fan, 2011; Lan et al., 2013; Yue et al., 2015). We previously identified several structural genes in the H. coronarium volatile biosynthetic pathway, including genes encoding terpene synthases (TPSs) and benzoic/salicylic acid methyltransferase (BSMT). A total of 12 HcBSMT and 62 HcTPS genes were found in H. coronarium. HcBSMT2 specifically expressed in flowers, its expression level was enormously high among all HcBSMT genes and correlated with flower development (Supplementary Figure 6). Likewise, the expression values of HcTPS3 and HcTPS5 were tremendously high among all the HcTPSs and specifically expressed in H. coronarium flowers (Supplementary Figure 7). Moreover, their expression pattern positively correlated with flower development as well as with the emission of monoterpenes, and their encoded enzymes localize to plastids (Yue et al., 2015). Our functional characterization indicated that HcTPS3 functions in (E)-β-ocimene production, HcTPS5 functions in linalool production, and HcBSMT2 functions in methyl benzoate production (Yue et al., 2015, 2021). Analysis of previously generated RNA-seq data showed that six HcMYB family members were clustered in a group involved in regulating secondary metabolism. The expression levels of these MYB family members were analyzed in different tissues (flowers, bracts, leaves, and rhizomes). The RNA-sequence data showed that among six HcMYBs, HcMYB1, and HcMYB2 were highly flower-specific and the abundance of their transcripts correspond with the flower development as well as with the emission of floral volatile contents. The relative transcript abundance of six HcMYB family members has been provided in Supplementary Figure 3. However, the hormone-responsive transcriptional regulation of these genes has not been elucidated. Furthermore, we comprehensively analyzed Aux/IAA genes in H. coronarium genome. The genome−wide analysis and characterization of Aux/IAA genes reveal the potential role of HcIAA2 and HcIAA4 in floral aroma production in H. coronarium (Ke et al., 2019).
In the current study, we functionally characterized two R2R3-MYB TF genes (HcMYB1 and HcMYB2) that are expressed in an auxin-responsive manner specifically in flowers. These MYB TFs regulate phenylpropanoid/benzenoid and terpenoid biosynthesis specifically in H. coronarium flowers by activating HcBSMT2 and HcTPS5 expression. Furthermore, the interaction of HcMYB1 with key scent-related auxin protein (HcIAA4) was revealed via yeast two-hybrid (Y2H) assay and bimolecular fluorescence complementation (BiFC) assays. These findings shed light on the mechanism underlying the emission of floral scent compounds in H. coronarium.
In a previous transcriptomic analysis, we identified a clade of genes whose expression rose throughout flower development and with increasing floral scent emissions (Yue et al., 2015). Among these genes, HcMYB1 and HcMYB2 are specifically expressed in flowers. The full-length complementary DNA (cDNA) sequences of HcMYB1 and HcMYB2 contain open reading frames (ORFs) of 618 and 693 bp, encoding polypeptides of 205 and 230 amino acid residues with molecular masses of 23.26 and 24.68 kDa, respectively. Analysis of the predicted protein sequences of HcMYB1 and HcMYB2 revealed the presence of 2R and 3R repeat signatures at the N-termini: these features of R2R3 DNA-binding MYB proteins (Figure 1A) are essential for their interactions with regulatory sequences in the promoters of their target genes (Kranz et al., 1998; Dubos et al., 2010; Medina-Puche et al., 2014).
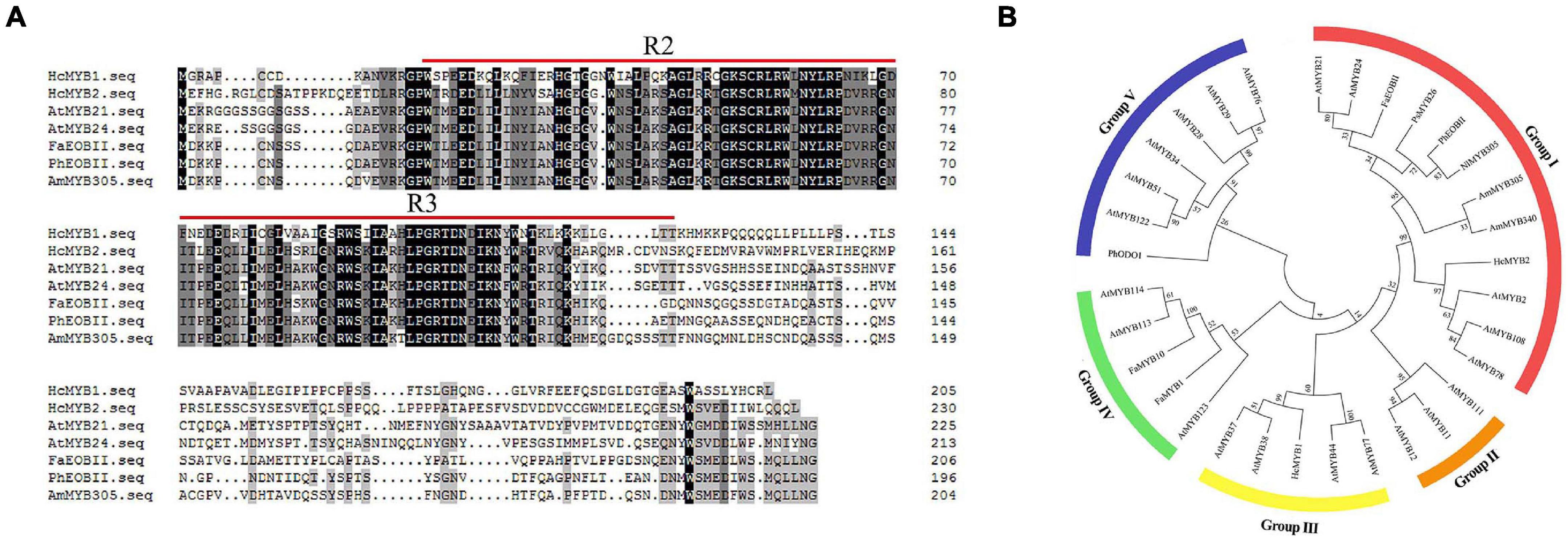
Figure 1. Multiple sequence alignment and phylogenetic analysis of R2R3-MYB proteins. (A) Multiple sequence alignment of HcMYB1 and HcMYB2 with other related MYBs from different plants. The conserved R2 and R3 motifs are underlined. Multiple sequence alignment was performed using Clustal Ω and edited with GeneDoc. (B) Phylogenetic analysis of HcMYB1 and HcMYB2 together with selected R2R3-MYB proteins from different plants. Amino acid sequence alignment was performed using Clustal Ω, and the tree was constructed using the NJ method in MEGA X. The bootstrap values were set to 1000. The accession numbers for the protein sequences are listed in Supplementary Table 3. The sequence info of HcMYB1 and HcMYB2 is given in Supplementary Material.
We performed a phylogenetic analysis of HcMYB1 and HcMYB2 compared to R2R3-MYBs involved in secondary metabolism in other plant species. HcMYB1 and HcMYB2 clustered into different groups (Figure 1B). HcMYB1 belongs to Group III and shares high amino acid homology with AtMYB77 and AtMYB44 (Aharoni et al., 2001; Shin et al., 2007; Jaradat et al., 2013). In contrast, HcMYB2 belongs to Group I, whose members include AmMYB305, AmMYB340 (A. majus), AtMYB24 (A. thaliana), AtMYB21 (A. thaliana), PhEOBII (Petunia × hybrida), FaEOBII (Fragaria × ananassa) NlMYB305 (Nicotiana langsdorffii), and PsMYB26 (Pisum sativum) (Uimari and Strommer, 1997; Shin et al., 2002; Li et al., 2006b; Liu et al., 2009; Spitzer-Rimon et al., 2010; Medina-Puche et al., 2015). The two proteins clustered into different groups and may have different functions and/or operate through different pathways to take part in floral volatile production. Furthermore, to interrogate the evolutionary relationship of six HcMYB family members with Arabidopsis MYBs, a phylogenetic tree was built. The phylogenetic analysis revealed that all MYB proteins can be clustered into five different groups (G A–G F). HcMYB1 was clustered into group G B, HcMYB2/6/5 belongs to group G D, while HcMYB3/4 was grouped into G A (Supplementary Figure 1).
The accumulation of floral volatiles increases as flower development proceeds (Yue et al., 2015; Abbas et al., 2019). To study this process, we divided the flower development process into six stages (Figure 2A). The emission of floral volatiles was low during the bud period (F1 and F2), substantially increased beginning at the initial flowering stage (F3), peaked during the full-bloom stage (F4 and F5), and declined at the senescence stage (F6) (Figure 2B).
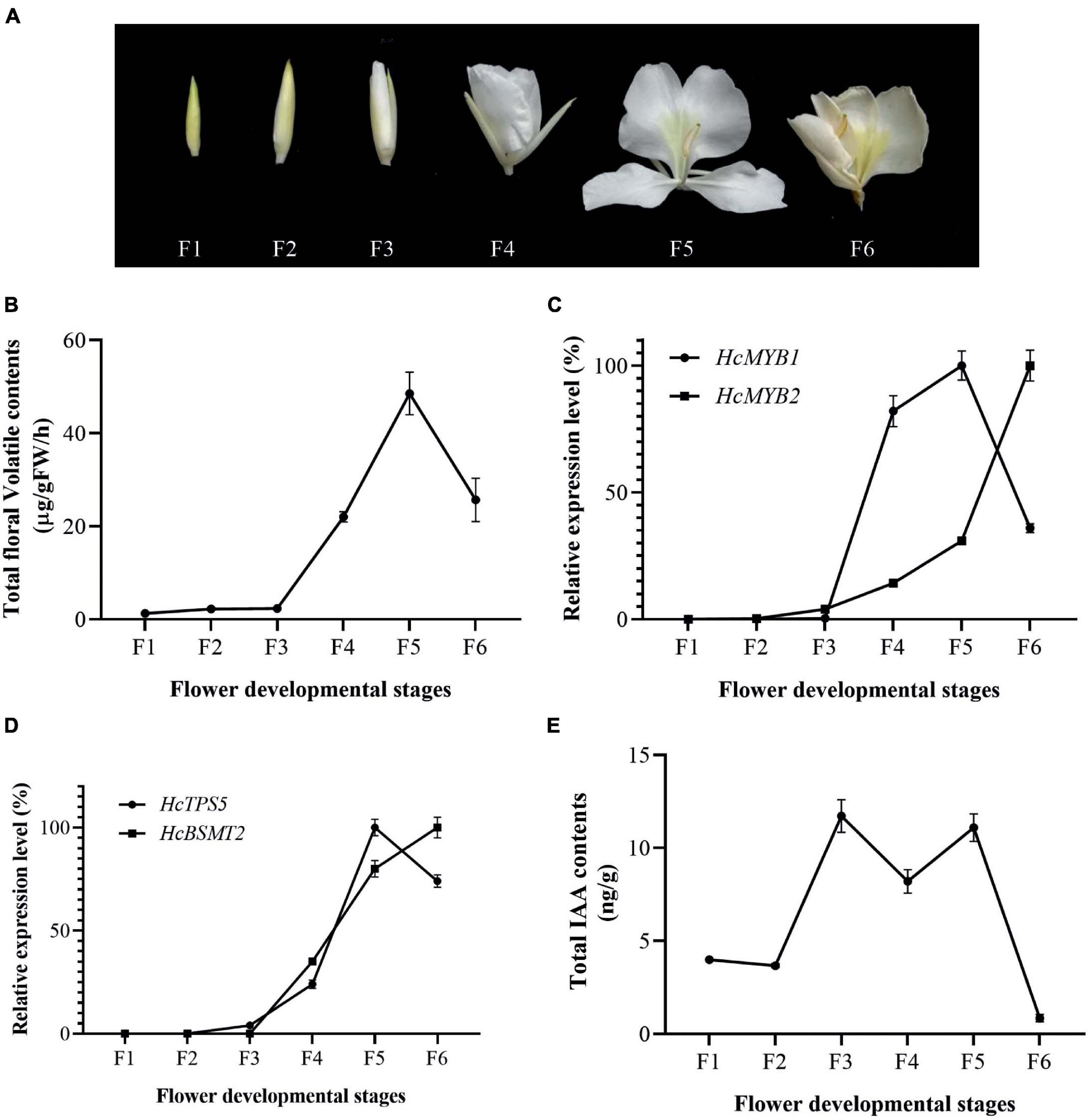
Figure 2. Changes in gene expression, IAA, and volatiles over the course of flower development (A) Pictorial view of different stages in floral development in H. coronarium. (B) Total floral volatile contents during flower development (F1–F6). (C) Expression analysis of HcMYB1 and HcMYB2 during different stages of flower development. (D) Relative expression levels of HcTPS5 and HcBSMT2. (E) Endogenous auxin contents during flower development. Bars represent the means ± SD (n = 3–5).
To examine the relationship between HcMYB1 and HcMYB2 and key volatile biosynthesis genes (HcTPS5 and HcBSMT2) involved in floral volatile contents, we measured the expression levels of these genes. HcMYB1 and HcMYB2 transcript levels were low during early flower development and substantially increased thereafter. HcMYB1 expression peaked at the full-bloom stage (F4–F5) and decreased at the senescence stage (F6), whereas HcMYB2 was most strongly expressed at F6 (Figure 2C). We detected similar expression patterns for HcTPS5 and HcBSMT2 during flower development (Figure 2D). Moreover, the expression levels of HcMYB1 and HcMYB2 were positively correlated with the emissions of floral volatiles; this correlation was highly significant for HcMYB1 (Supplementary Figure 2). Moreover, HcMYB1 showed a highly significant correlation with the emission of linalool contents (Supplementary Figure 3). These results suggest that these genes play important roles in floral scent formation in H. coronarium.
Auxin plays a crucial role throughout flower development (Krizek, 2011; Ke et al., 2018). We, therefore measured total auxin levels in H. coronarium during flower development (F1–F6). The total auxin contents were low during F1 and F2, peaked at F3–F5, dropped slightly at F4, and declined further at F6 (Figure 2E). The emission of total floral volatiles was correlated with indole-3-acetic acid (IAA) contents, suggesting that auxin might play a crucial role in the biosynthesis of these compounds. Under IAA treatment, the contents of the major floral volatiles ocimene, linalool, and methyl benzoate increased by 16, 17, and 20%, respectively, compared to those in control flowers not treated with IAA (CK) (Figure 3A). Moreover, the expression of key structural volatile biosynthesis genes (HcTPS1, HcTPS3, HcTPS5, HcTPS8, HcPAL, and HcBSMT2) was upregulated by this treatment (Figure 3B).
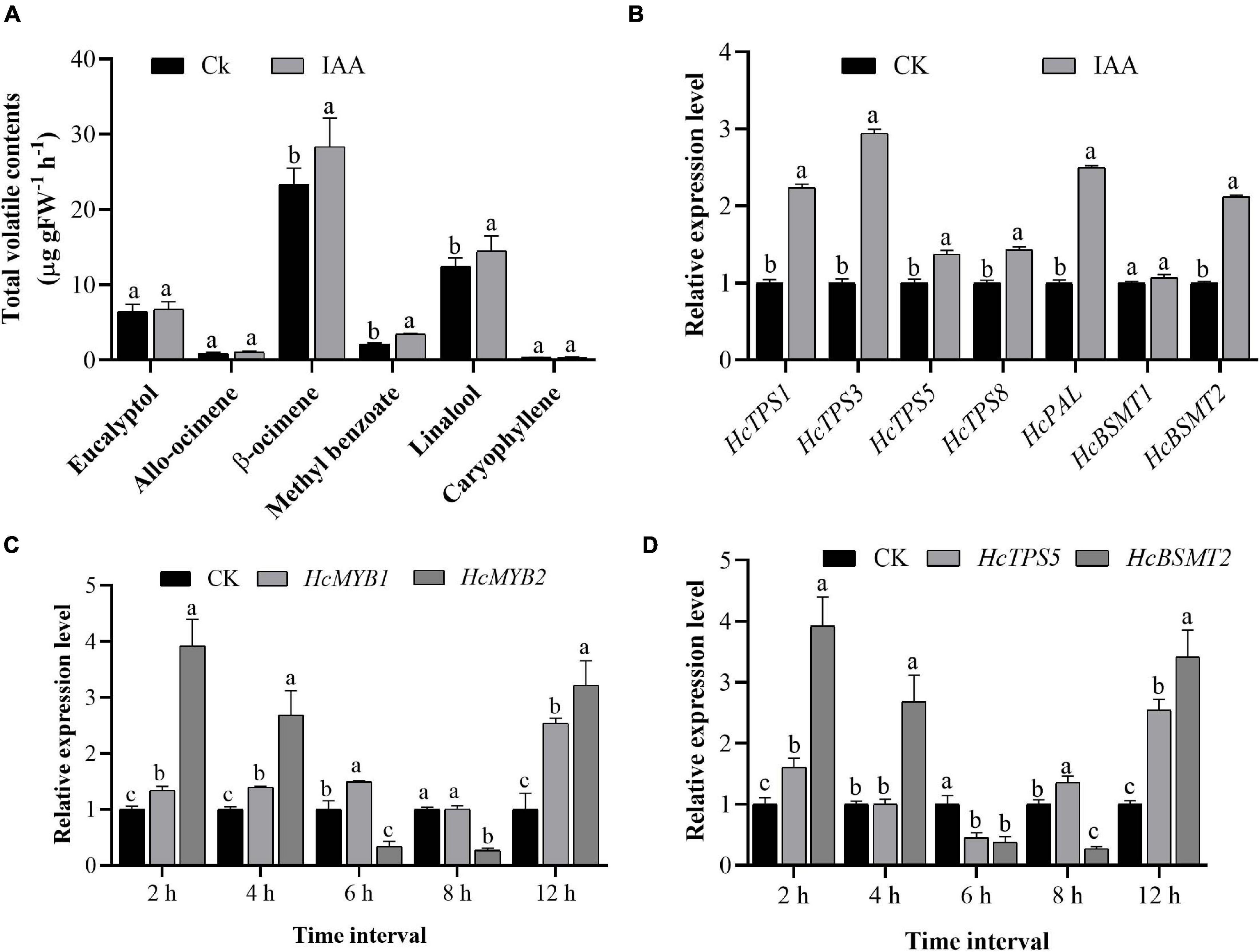
Figure 3. Exogenous auxin treatment influences the expression of HcMYB1, HcMYB2, and key volatile biosynthesis genes. (A) The emission of major floral volatile compounds in H. coronarium after auxin treatment. (B) Relative expression levels of key volatile biosynthesis genes after auxin treatment. (C,D) Expression patterns of HcMYB1/2 and key structural genes (HcTPS5 and HcBSMT2) in response to auxin treatment. Bars represent the means ± SD (n = 3–5). Different letters on bars indicate statistically significant differences (P < 0.05).
To characterize the expression levels of HcMYB1 and HcMYB2 in response to IAA treatment, we performed qRT-PCR analysis. HcMYB1 and HcMYB2 transcript levels strongly increased after IAA treatment, reaching their highest levels at 12 h after treatment. In contrast, a rapid increase in HcMYB2 and HcTPS5 expression was observed at 2 h after treatment (Figure 3C). Volatile biosynthesis genes (HcTPS5 and HcBSMT2) were also upregulated at 12 h after IAA treatment (Figure 3D). The results suggest that the biosynthesis of floral volatiles is spatially and temporally regulated by HcMYB1 and HcMYB2, which are strongly associated with auxin-induced volatile emissions in H. coronarium.
We also examined the effect of p-chlorophenoxyisobutyric acid (PCIB) (inhibit auxin action) on the floral volatile compounds (Figure 4). In contrast to auxin, the emission of floral volatile compounds decreases. Under PCIB treatment, the contents of eucalyptol, allo-ocimene, β-ocimene, methyl benzoate, and linalool were decreased by 57, 81, 89, 100, and 42%, respectively, compared to those in control flowers not treated with PCIB (Figure 4A). As expected, similar to IAA, the volatile contents of caryophyllene do not change significantly. We perform qRT-PCR analysis to characterize the expression level of key genes under PCIB treatment. The expression level of key volatile biosynthesis genes HcTPS1, HcTPS3, HcTPS5, HcTPS8, HcPAL, HcBSMT1, HcBSMT2, HcMYB1, and HcMYB2 were downregulated by 79, 93, 64, 89, 33, 97, 96, 75, and 72%, respectively, relative to control (Figure 4B). The data endorse the aforementioned findings that auxin plays a crucial role in the biosynthesis of floral volatile compounds.
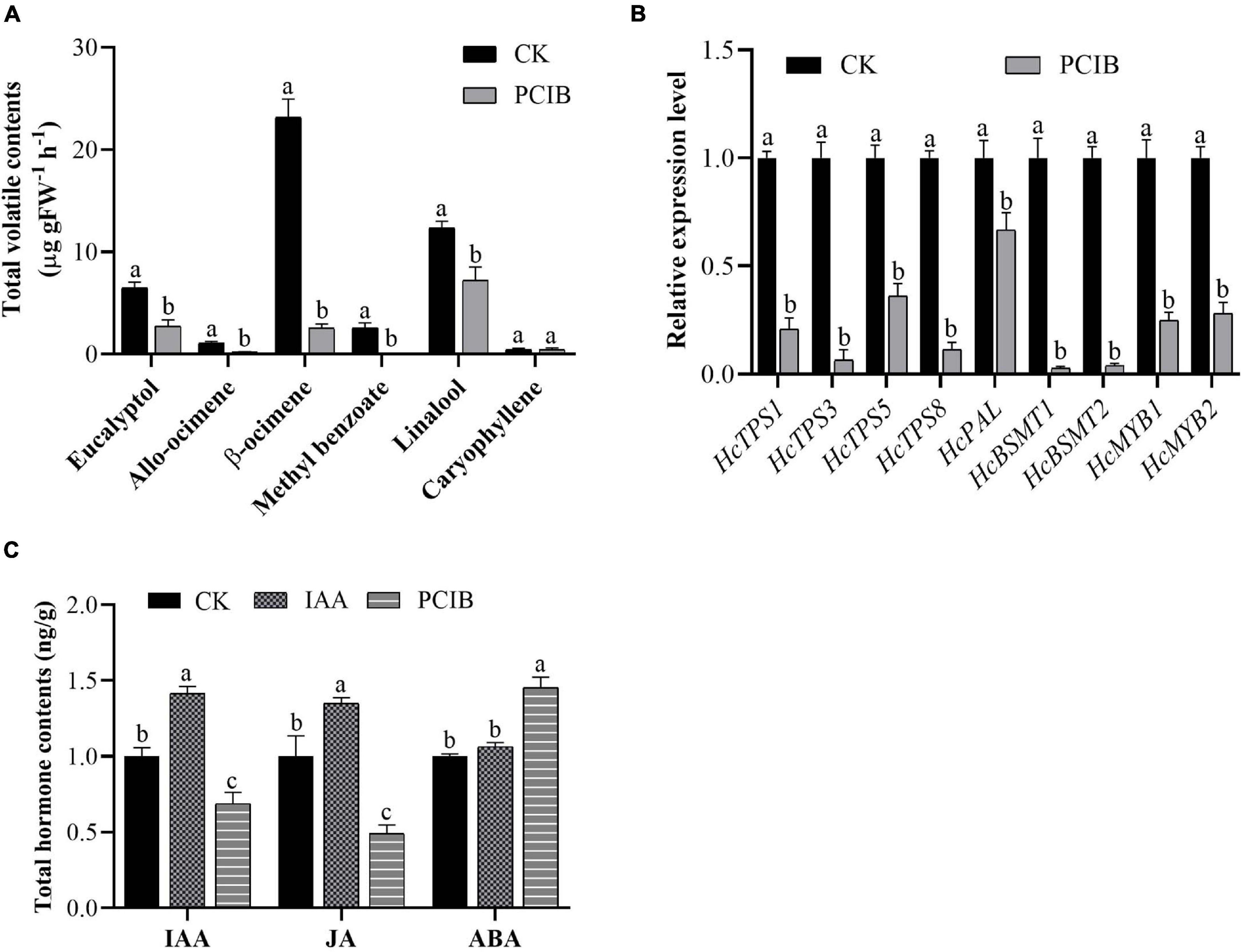
Figure 4. Marked decrease in scent emission, decreased expression level of key structural volatile synthesis genes, and total IAA, JA, and ABA contents under PCIB treatment. (A) Headspace analyses of individual scent compounds under PCIB treatment using GC–MS. (B) Relative expression level of key structural genes under PCIB treatment. GAPDH was used as an endogenous control. Bars represent the means ± SD (n = 3–5). (C) Total pool contents of IAA, JA, and ABA in response to auxin and PCIB treatment. Each time point represents the average of three to four independent experiments, with SD indicated by vertical lines. Graphs represent the average of three independent experiments. Different letters on bars indicate statistically significant differences (P < 0.05).
Furthermore, we quantify the total hormone contents under IAA and PCIB treatment using ultra-performance liquid chromatography-tandem mass spectrometer (UPLC-MS/MS). The data showed that under IAA treatment, the total IAA and jasmonic acid (JA) contents were increased by 42 and 35% compared to control. Likewise, the total IAA and JA contents were decreased by 31 and 51%, respectively, while abscisic acid (ABA) contents were increased by 45% under PCIB treatment (Figure 4C). However, the ABA contents do not change significantly under IAA treatment, suggesting that auxin might play a key role in the biosynthesis of floral volatile compounds via crosstalk with the abovementioned hormones.
Most MYB TFs specifically localize to the nucleus (Zou et al., 2008; Zhu et al., 2015; Zhou et al., 2017). However, some MYB TFs localize to both the nucleus and cytoplasm (Li et al., 2006a). The nuclear localization prediction server WoLF PSORT1 predicted that HcMYB1 and HcMYB2 localize to the nucleus. To assess this prediction, we generated HcMYB1-GFP and HcMYB2-GFP constructs in which these genes were driven by the CaMV 35S promoter and used them to transform Arabidopsis protoplasts. In HcMYB1-GFP- and HcMYB2-GFP-transformed protoplasts, observed green fluorescent protein (GFP) signals specifically in the nuclei, whereas control (GFP) protoplasts showed a ubiquitous distribution of GFP throughout the protoplasts (Figure 5A). We included nuclear localization signal (NLS)-mCherry in each transformation as a marker for nuclear localization. These results demonstrate that HcMYB1 and HcMYB2 are nucleus-localized proteins, which is in keeping with their expected roles as transcription factors.
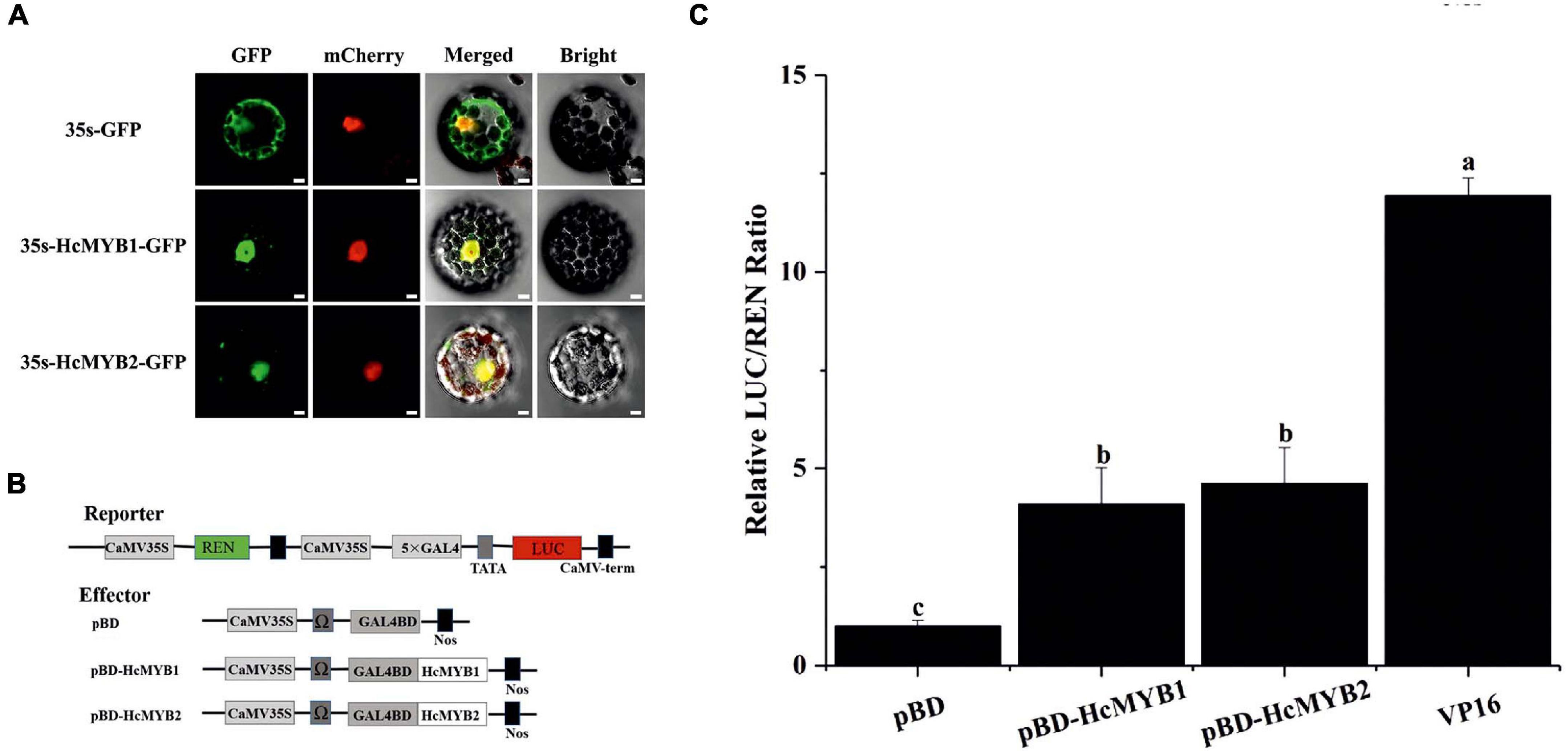
Figure 5. HcMYB1 and HcMYB2 show nuclear localization in Arabidopsis protoplasts. (A) The images were obtained using a confocal laser scanning microscope and processed in adobe Photoshop. GFP: green fluorescent protein. mCherry: nuclear marker. Merged: combined GFP and mCherry signals. Bright: bright field. Bars: 10 μm. (B) Diagram of the reporter and effector plasmids. (C) Transcriptional activity of HcMYB1 and HcMYB2 together with DNA via GAL4 BD (pBD-HcMYB1 and pBD-HcMYB2) transiently co-expressed in N. benthamiana cells. The activity is presented as the ratio of LUC to REN luminescence. The VP16 and pBD represent the positive and negative control, respectively. The ratio of LUC/REN of the empty pBD vector is used as the calibrator (set as 1). Data are represented as means ± standard deviation of three replicates. Compared with the pBD, significant differences at the level of P < 0.05 analyzed by the student’s t-test, are indicated by different letters above the bar.
Transcription factors regulate their target genes via transactivation activity. To investigate the transactivation activities of HcMYB1 and HcMYB2, we performed transient expression analysis in Nicotiana benthamiana leaves. We fused five copies of the GAL4 DNA-binding element (GAL4BD) and the minimal TATA region (5′-TATAAA-3′) of the 35S promoter to the firefly luciferase (LUC) reporter; the Renilla luciferase (REN) reporter gene driven by the 35S promoter as the reporter vector. The LUC/REN ratio from the reporter vector was used as an internal control. We constructed effector plasmids harboring the ORFs of HcMYB1 and HcMYB2 (Figure 5B). Unlike the GAL4BD negative control (empty vector, pBD), HcMYB1 and HcMYB2 activated the LUC reporter gene. The LUC/REN ratios of HcMYB1, HcMYB2, and GAL4BD-VP16 were 3. 1-, 3. 7-, and 11.1-fold higher, respectively, compared to the negative control (Figure 5C). These results indicate that HcMYB1 and HcMYB2 function as transcriptional activators.
To investigate the potential involvement of HcMYB1 and HcMYB2 in floral scent formation, we suppressed their expression through virus-induced gene silencing (VIGS) in flowers (Renner et al., 2009; Yuan et al., 2011). We confirmed that this led to significant decreases in HcMYB1 and HcMYB2 transcript levels compared to those in unsilenced control flowers (Figures 6A,B). The contents of the volatiles methyl benzoate and linalool in flowers decreased by approximately 57 and 21%, respectively, in response to HcMYB1 silencing, whereas the eucalyptol and ocimene contents did not change significantly (Figure 6C). In HcMYB2-silenced flowers, the contents of methyl benzoate, linalool, ocimene, and eucalyptol decreased by 68, 37, 18, and 17%, respectively, compared to the control (Figure 6C). We also analyzed the expression levels of key volatile biosynthesis genes (HcTPS3, HcTPS5, and HcBSMT2) in H. coronarium. In HcMYB1-silenced flowers, HcTPS5 and HcBSMT2 were significantly downregulated, whereas HcTPS3 did not exhibit any significant changes in expression, compared to the control. Furthermore, in HcMYB2-silenced flowers, HcTPS3, HcTPS5, and HcBSMT2 were all significantly downregulated compared to the control (Figure 6D). These results indicate that HcMYB1 and HcMYB2 play important and overlapping roles in the formation of floral volatiles in H. coronarium.
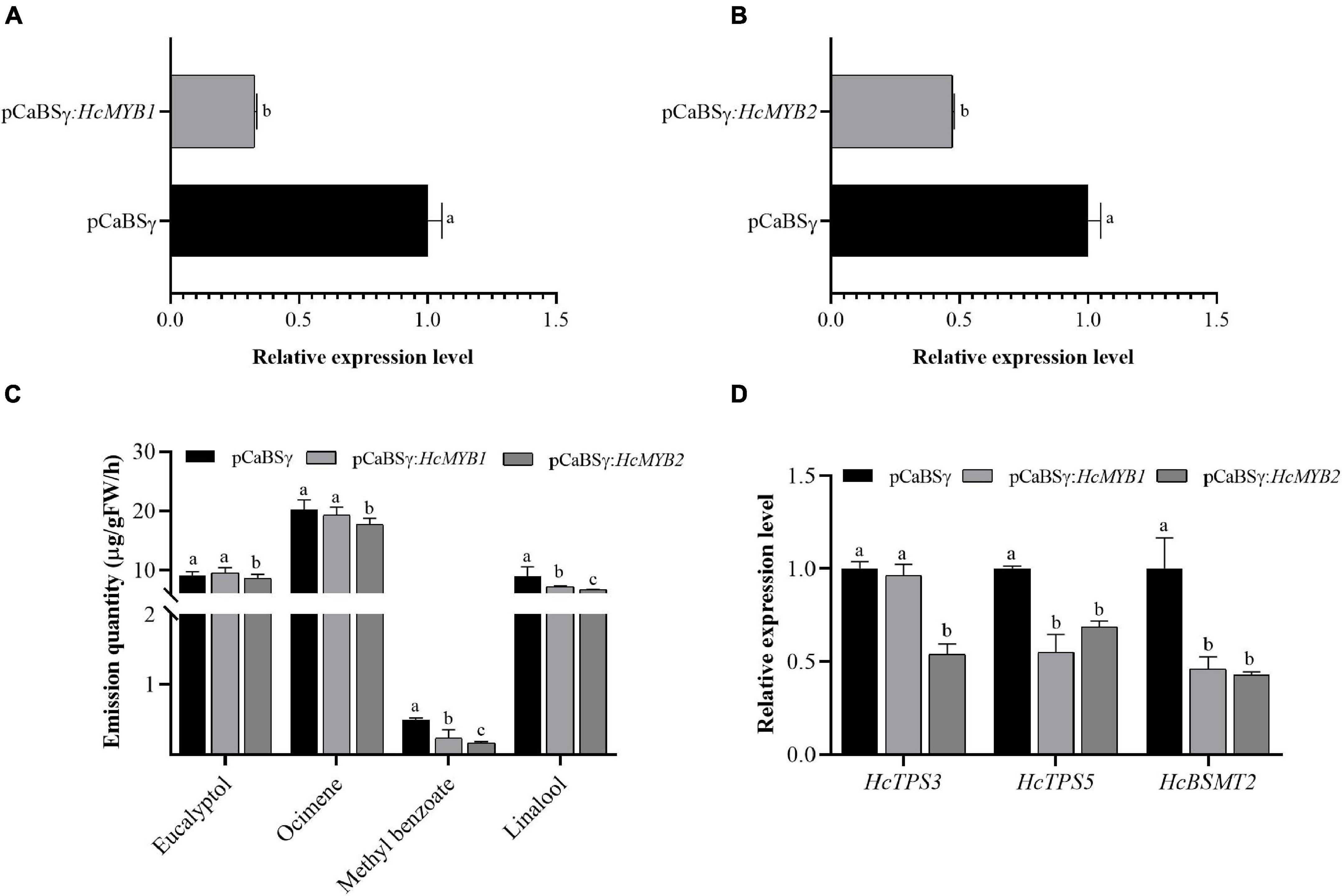
Figure 6. HcMYB1 and HcMYB2 gene silencing alter HcTPS3, HcTPS5, and HcBSMT2 expression and volatile emission in flowers. (A,B) Relative expression levels are presented as fold change values. (C) Emission levels of floral volatiles. (D) Expression levels of HcTPS3, HcTPS5, and HcBSMT2. Data represent the mean ± SD (n = 3–5). Different letters indicate significant differences (P < 0.05).
MYB TFs transcriptionally regulate several genes by binding to the MEB [(T)(T)TGAC(C/T)] sequences in their promoters (Rushton et al., 2010). In silico cis-element analysis revealed the presence of MYB-binding motifs in HcBSMTs and HcTPSs sequences. MYB-core binding motifs were present in ten out of twelve HcBSMTs. The number of MYB-binding motifs varies from one to thirteen. Interestingly, the number of MYB-bindings motifs in HcBSMT2 was highest compared to other HcBSMTs (Supplementary Table 1). Similarly, MYB-core binding motifs were found in sixty out of sixty-two HcTPSs. The promoter sequence analysis of HcBSMT2 (1131 bp) and HcTPS5 (1555 bp) revealed the presence of MYB-core binding motifs in their sequences. There were 13 and 5 copies of MYB-binding motifs in the sequences of HcBSMT2 and HcTPS5, respectively, suggesting that HcMYB1 and HcMYB2 might target these genes.
To determine whether HcMYB1 and/or HcMYB2 bind to the promoters of HcBSMT2 and HcTPS5, we performed a yeast one-hybrid (Y1H) assay. Bait strains co-expressing HcMYB1 and HcMYB2 and harboring proHcBSMT2 grew well in SD-Leu medium containing the antibiotic aureobasidin A (AbA), whereas bait strains harboring proHcTPS5 grew well only when they expressed HcMYB2 (Figures 7A,B). These results indicate that HcMYB1 and HcMYB2 bind to the HcBSMT2 promoter, while HcMYB2 binds to the HcTPS5 promoter.
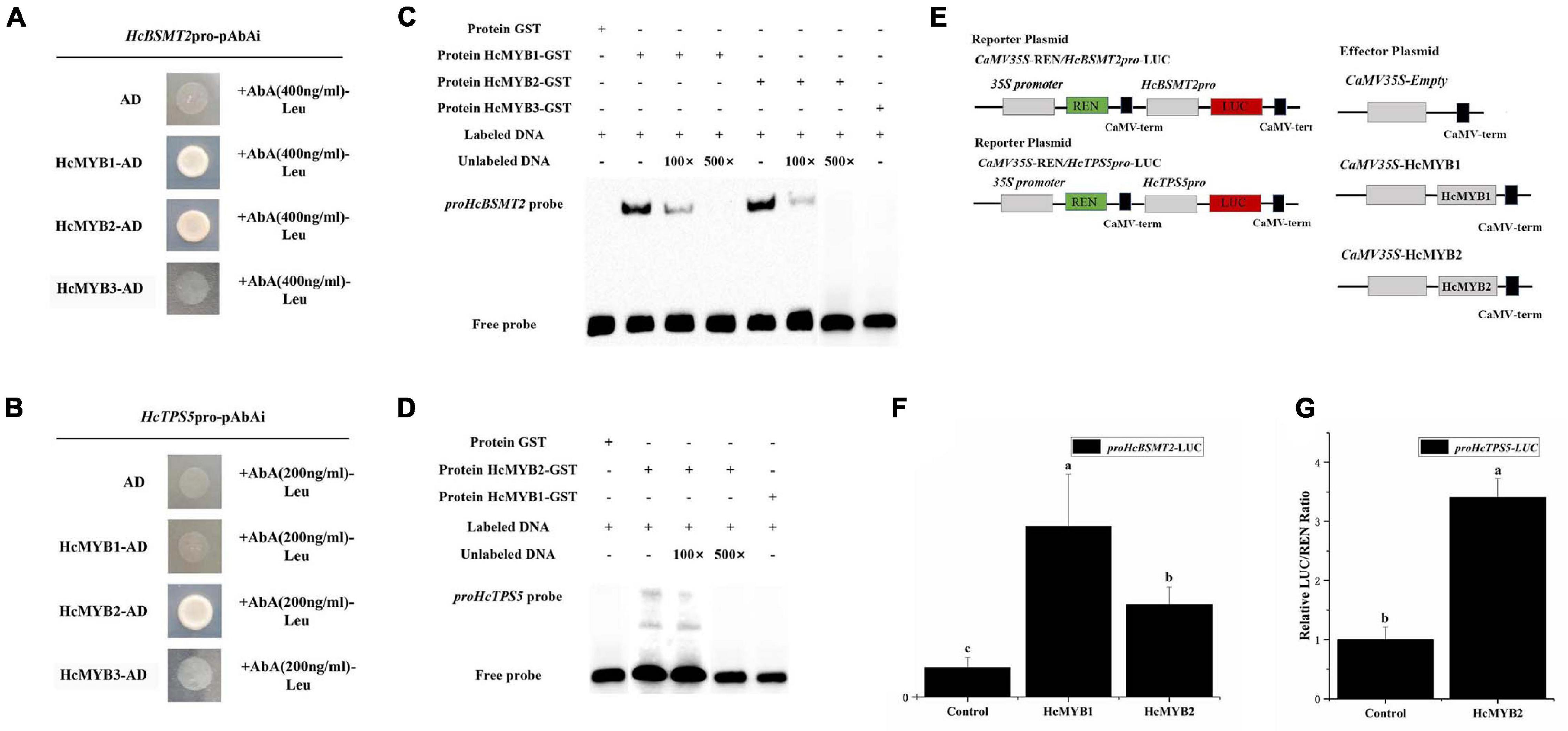
Figure 7. HcMYB1 and HcMYB2 interact with the HcBSMT2 and HcTPS5 promoters. (A) Interactions of HcMYB1 and HcMYB2 with the HcBSMT2 promoter. (B) Interactions of HcMYB1 and HcMYB2 with the HcTPS5 promoter. HcMYB3 and empty vector were used as a control. (C) Binding activity of HcMYB1 and HcMYB2 with the proHcBSMT2 probe. (D) Binding activity of HcMYB2 with the proHcTPS5 probe. (E) Diagram of the reporter and effector plasmids. (F) HcMYB1 and HcMYB2 transactivate the HcBSMT2 promoter. (G) HcMYB2 transactivates the HcTPS5 promoter. The promoter activity is presented as the ratio of LUC to REN luminescence. Different letters on bars indicate statistically significant differences (P < 0.05).
To confirm the binding ability of HcMYB1 and HcMYB2 to the HcBSMT2 and HcTPS5 promoters, we performed an electrophoretic mobility shift assay (EMSA) using GST-HcMYB1 and GST-HcMYB2 in Escherichia coli. The probes used for proHcTPS5 and proHcBSMT2 were 49 bp, which start from (+)1168 to (+)1216 and (+)291 to (+)339, respectively. The sequences of the probes are listed in Supplementary Table 2. Purified recombinant GST-HcMYB1 and GST-HcMYB2 fusion proteins bound to biotin-labeled probes derived from the HcBSMT2 promoter, leading to a mobility shift, whereas no mobility shift occurred in the presence of GST alone (Figure 7C). Next, we performed a competition assay, which showed that adding a 100-fold amount of unlabeled probe molecules (as compared to the labeled molecules) to the binding reaction reduced the intensity of the protein-DNA complex signal, and adding 500-fold unlabeled probes prevented any protein-DNA complex from being detected (Figure 7C). We also observed binding between HcMYB2 and a biotin-labeled probe from the HcTPS5 promoter (Figure 7D). These results endorse the aforementioned data that HcMYB1 binds to the HcBSMT2 promoter and HcMYB2 binds to the HcBSMT2 and HcTPS5 promoters.
To test the ability of HcMYB1 and HcMYB2 to activate the HcBSMT2 and HcTPS5 promoters, we performed a dual-luciferase assay. We individually cloned the promoter regions of HcBSMT2 and HcTPS5 into reporter plasmids and the ORFs of HcMYB1 and HcMYB2 into effector plasmids (Figure 7E). HcMYB1 and HcMYB2 significantly enhanced HcBSMT2 promoter activity (by 4.8-fold and 2.2-fold, respectively) compared to the control (Figure 7D). Meanwhile, HcMYB2 significantly enhanced HcTPS5 promoter activity (by 2.4-fold) compared to the control (Figure 7F). Therefore, HcMYB1 and HcMYB2 activate the HcBSMT2 promoter and HcMYB2 activates the HcTPS5 promoter, indicating that these TFs have different target genes in N. benthamiana leaves. These findings indicate that both HcMYB1 and HcMYB2 are transcriptional activators of volatile biosynthesis genes in flowers.
MYB proteins interact with many other proteins involved in hormone signal transduction, such as the jasmonic acid (JA)-responsive repressor proteins of the JASMONATE ZIM-DOMAIN (JAZ) family and the ABA signal receptor protein PYRABACTIN RESISTANCE LIKE (PYL) (Qi et al., 2014; Zhao et al., 2014). In a Y2H assay, HcMYB1 interacted with the auxin-responsive protein HcIAA4, whereas HcMYB2 did not (Figure 8A). To verify the interaction between HcMYB1 and HcIAA4, we performed a BiFC assay. Expressing the N-terminal half of YFP fused to HcMYB1 (HcMYB1-YFPN) and the C-terminal half of YFP fused to HcIAA4 (HcIAA4-YFPC) in N. benthamiana leaves resulted in fluorescence. Moreover, the reciprocal experiment with the C-terminal half of YFP fused to HcMYB1 (HcMYB1-YFPC) and the N-terminal half of YFP fused to HcIAA4 (HcIAA4-YFPN) also resulted in fluorescence and the control combinations of YFPC + HcMYB1-YFPN and HcIAA4-YFPC + YFPN did not result in fluorescence (Figure 8B). To elucidate the functional significance of the interaction between HcMYB1 and HcIAA4, we co-transformed N. benthamiana leaves with the same amounts of effectors carrying HcMYB1 and/or HcIAA4 in combination with the HcBSMT2pro-LUC reporter constructs. The effect of HcMYB1 on HcBSMT2pro expression was repressed in the presence of HcIAA4 (Figure 8C). These results demonstrate that HcMYB1 directly activates HcBSMT2 expression, which is modulated by its interacting partner HcIAA4.
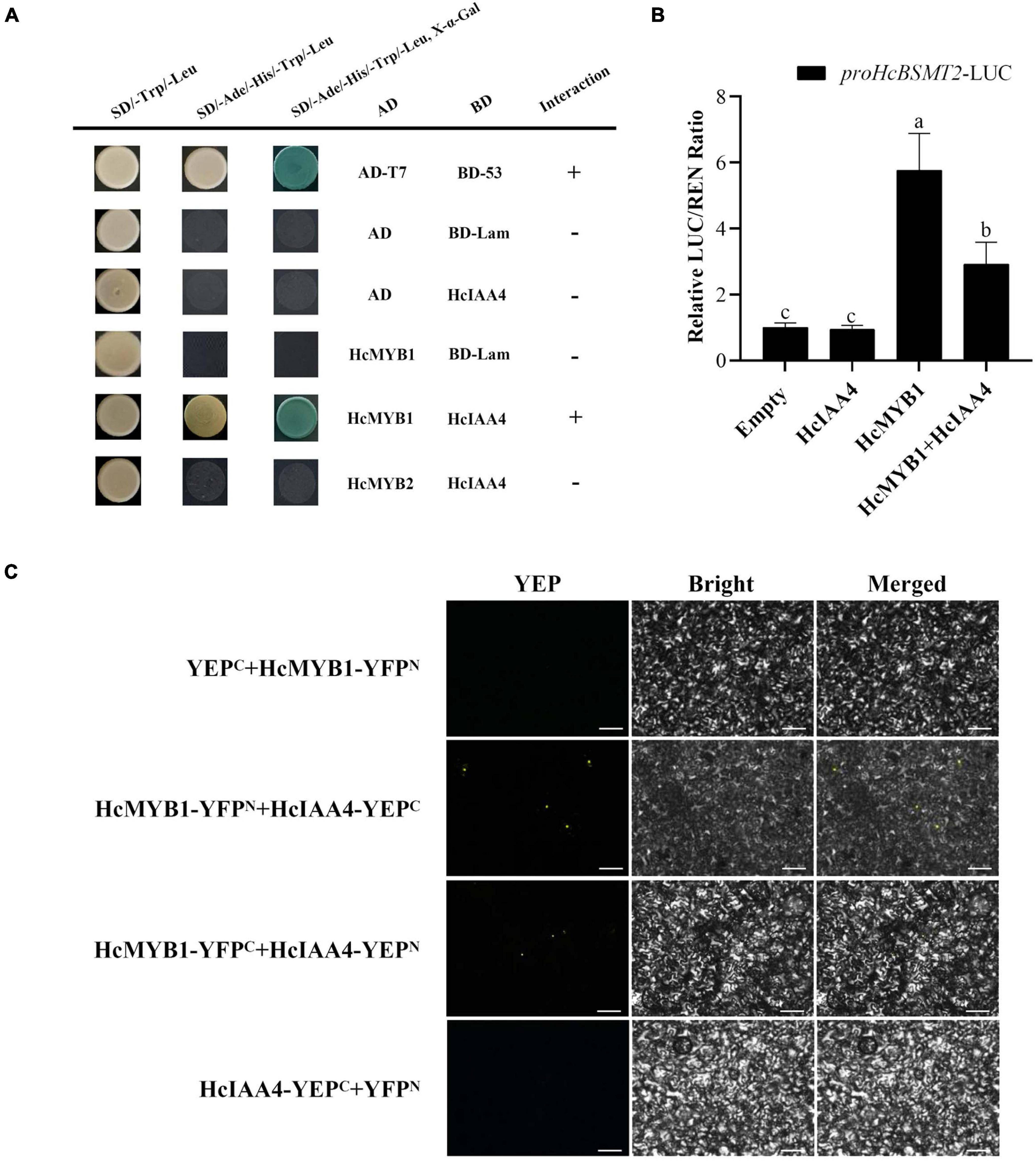
Figure 8. HcMYB1 interacts with HcIAA4. (A) Analysis of the interactions of HcIAA4, HcMYB1, and HcMYB2 via yeast two-hybrid assay. Yeast cells were transformed with DBD–HcARF4 + AD–HcMYB1, DBD–HcARF4 + AD–HcMYB2, DBD + AD–HcMYB1, DBD–HcIAA4 + pGADT7-T, pGBKT7-53 + pGADT7-T, DBD–HcIAA4 + pGADT7-T or pGBKT7-Lamin + pGADT7-T. The vectors were transformed into yeast strain Y2HGold and then transformants were screened by plating on SD/-Trp/-His/-Ade + X-α-gal medium. BD-Lam was used as a negative control. ADT7-BD53 was used as a positive control. Three independent replicates were performed. (B) BiFC assay between HcMYB1 and HcIAA4 in N. benthamiana leaves. Fusion constructs YFPC + HcMYB1-YFPN, HcMYB1-YFPN + HcIAA4-YFPC, HcMYB1-YFPC + HcIAA4-YFPN, and HcIAA4-YFPC + YFPN were co-infiltrated into N. benthamiana leaves and observed for fluorescence complementation. Bars, 5 μm. (C) The activation ability of HcMYB1 is modulated by HcIAA4. Different letters on bars indicate statistically significant differences (P < 0.05).
The transcriptional regulatory network governing floral scent emission has not been thoroughly elucidated. To date, only a few TFs that regulate the expression of scent-related genes have been identified (Katiyar et al., 2012; Abbas et al., 2021b). R2R3-MYB TFs are key regulators of the phenylpropanoid and terpenoids biosynthetic pathway in plants (Du et al., 2009; Zhu et al., 2015; Yang et al., 2020). MYB TFs in the same subgroup have similar functions (Zhu et al., 2015). Here, we used previously characterized R2R3-MYB proteins involved in secondary metabolism to construct a phylogenetic tree with HcMYB1 and HcMYB2 from H. coronarium (Figure 1B). HcMYB1 was clustered in Group III with AtMYB77, which modulates auxin signal transduction (Shin et al., 2007), and AtMYB44, a stress-responsive protein involved in senescence and ABA signaling (Jaradat et al., 2013), suggesting that HcMYB1 might play a key role in hormone signaling. Meanwhile, HcMYB2
was classified into Group I along with AmMYB305, AmMYB340, AtMYB24, AtMYB21, PhEOBII, and FaEOBII (Figure 1). These R2R3-MYB TFs regulate the metabolic pathway of the volatile compound phenylpropanoid (Uimari and Strommer, 1997; Shin et al., 2002; Li et al., 2006b; Liu et al., 2009; Spitzer-Rimon et al., 2010; Medina-Puche et al., 2015), suggesting that HcMYB2 might be involved in regulating the floral volatile metabolic pathway in H. coronarium.
The production and emission of fragrance compounds by flowers are strictly regulated during the floral lifespan and often peak when the flower is in full bloom and pollinators are active (Dudareva et al., 2013; Muhlemann et al., 2014; Abbas et al., 2017). Methyl benzoate and linalool are the main phenylpropanoids and terpenoid volatiles that contribute to flower scent in H. coronarium (Fan et al., 2003, 2007; Báez et al., 2011; Yue et al., 2015; Abbas et al., 2021a). We observed that the expression of HcMYB1 changed during flower development, with the highest expression level detected at the full bloom stage (F5) (Figure 2B. A similar expression pattern was detected in lilac (Syringa oblata) during different stages of flower development, as two R2R3-MYB TF genes were upregulated at the full-bloom stage compared to the bud stage (Zhu et al., 2015). Similarly, the expression level of FhMYB5 gradually increased during the flower developmental stages, resembling anthocyanin biosynthesis pattern and function in the flavonoid pathway in Freesia hybrida (F. hybrida) (Li et al., 2019). In Rosa hybrida, mRNA levels of a putative scent-related gene (RhMYB1) were developmentally regulated peaking at full bloom stage similar with other rose scent-related genes, such as phenylacetaldehyde synthase RhPAAS, the sesquiterpene synthase RhGDS, the alcohol acetyltransferase RhAAT and the orcinol O-methyltransferases (OOMT) (Lavid et al., 2002; Yan et al., 2011).
Interestingly, the expression patterns of HcMYB1 and HcMYB2 during development were similar to those of HcBSMT2 and HcTPS5, which are responsible for the formation of the volatiles methyl benzoate and linalool, respectively (Figure 2D). Both compounds reach their highest levels in flowers at the full bloom stage (Yue et al., 2015). Similarly, HcBSMT2 and HcTPS5 expression levels were highest in flowers during the periods when the largest amounts of volatiles were released (Supplementary Figure 5). Thus, the expression of TF genes (HcMYB1 and HcMYB2) and structural genes (HcBSMT2 and HcTPS5) was associated with flower development and the production of high levels of volatiles (Supplementary Figure 1). In F. hybrida and A. thaliana, FhMYB21L1 and FhMYB21L2, TF genes were synchronously expressed with FhTPS1 and could activate its expression significantly (Yang et al., 2020). These results suggest that HcMYB1 and HcMYB2 regulate volatile production during flower development, which is similar to the roles of R2R3-MYBs PhEOBII and FaEOBII in petunia and strawberry, respectively (Van Moerkercke et al., 2011; Medina-Puche et al., 2015).
The volatile biosynthesis pathway, and particularly the emission of methyl benzoate and linalool, is induced by IAA, suggesting that IAA regulates the expression of transcription factors or key enzymes involved in this pathway at the protein or transcript level. In the current study, we demonstrated that auxin induces the expression of both TF genes (HcMYB1 and HcMYB2) and key biosynthesis genes [phenylalanine ammonia lyase (PAL), BSMT, and TPS] (Figures 3B,C). IAA treatment also upregulated HcBSMT2 and HcTPS5 expression, especially at 12 h (Figure 3D). In A. thaliana, expression of AtTPS21 and AtTPS11 was induced by the phytohormones, and both inductions require AtMYC2 (Hong et al., 2012). R2R3-MYBs such as AtMYB77 and AtMYB44 are involved in the response to auxin signaling in Arabidopsis (Shin et al., 2007; Yamaguchi et al., 2013). Similarly, FaMYB10 and FaEOBII are regulated by auxin in strawberry (Perkins-Veazie, 1995; Chai et al., 2011; Jia et al., 2011). On the other hand, auxin contents decrease and FaMYB10 and FaEOBII expression increases during fruit development in strawberry (Medina-Puche et al., 2014, 2015). During flower development in H. coronarium, auxin contents increased and HcMYB1 and HcMYB2 expression increased (Figures 2C,E). The differences in auxin response patterns between the HcMYB and FaMYB genes may be due to the evolutionary distance between H. coronarium (Zingiberaceae) and F. × ananassa (Rosacea). To validate the function of auxin, the flowers were treated with PCIB which is widely used to inhibit auxin action (Oono et al., 2003). The data showed that in contrast to auxin, the emission of main floral volatiles and expression level of aforementioned key structural volatile synthesis genes significantly downregulated (Figures 4A,B). The following data endorse the abovementioned findings that auxin plays an essential role in floral scents. Furthermore, relative to the control, total IAA and JA contents significantly upregulated and downregulated under auxin and PCIB treatment, respectively (Figure 4C). Several studies showed that MYB TF respond to various phytohormones. Under JA treatment, the transcript abundance of Pinus taeda PtMYB14 and PtMYB13 rapidly increased by 14-fold and 2-fold, respectively, while Picea glauca PgMYB14 and PgMYB15 transcripts increased 4-fold and 2-fold. Furthermore, the characterization of the aforementioned TF genes reveals PtMYB14 as a putative regulator of an isoprenoid and flavonoid-oriented response in conifers (Bedon et al., 2010). In Apples, MdMYB9 and MdMYB11 were involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin (An et al., 2014). The regulatory patterns of MYBs are dependent on developmental stage, tissue type, and environmental conditions. Much remains to be learned about the mechanistic basis of the responses of MYB TFs to auxin signaling molecules during volatile formation.
The structural genes HcBSMT2 and HcTPS5 are essential for the formation of methyl benzoate and linalool, respectively, in H. coronarium flowers (Yue et al., 2015). Notably, we detected MYB-binding elements in the promoters of HcBSMT2 and HcTPS5 (Supplementary Table 1). This result is supported by the finding that HcMYB1 transactivates the HcBSMT2 promoter and that HcMYB2 transactivates the HcBSMT2 and HcTPS5 promoters (Figure 7). In certain plants, floral scent biosynthesis is dependent on transcriptional regulation, and TFs control volatile emissions (Colquhoun et al., 2011; Muhlemann et al., 2012). ODO1 was the first R2R3-type MYB transcription factor shown to regulate the benzenoid biosynthesis pathway in petunia, followed by the R2R3-MYB TFs EOBI and EOBII. ODO1 strongly influences the floral scent pathway by regulating the transcript levels of many key genes (PAL, CM, DAHPS, SAMS, and EPSPS). Meanwhile, ODO1 is directly regulated by EOBII. Moreover, EOBI directly binds to and activates the promoters of ODO1, IGS, and PAL to regulate scent production (Verdonk et al., 2005; Spitzer-Rimon et al., 2010, 2012). In F. hybrida, FhMYB5 and FhbHLH mainly contribute to the regulation of anthocyanin and proanthocyanidin via activating the expression of biosynthetic genes (FhCHS, FhCHI, FhF3H, FhF3′H, FhF3′5′H, and FhDFR) involved in the flavonoid pathway (Li et al., 2019). In spearmint, MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase (Reddy et al., 2017). Likewise, several R2R3-MYB transcription factor have been identified which are potentially involved in the regulation of flavonoid biosynthesis via controlling the expression of structural genes (Cao et al., 2021; Zhang et al., 2021). Similarly, Lilium hybrid ODO1 (LhODO1) regulates fragrance biosynthesis via regulating the expression of structural genes involved in the shikimate and benzenoid/phenylpropanoid pathway (Yoshida et al., 2018).
In H. coronarium, HcMYB1 and HcMYB2 directly activate the methyl benzoate biosynthesis gene HcBSMT2, whereas HcMYB2 activates the linalool biosynthesis gene HcTPS5. Therefore, HcMYB2 activates two different groups of volatile biosynthesis genes, suggesting it plays a dual role in controlling both the phenylpropanoid and terpenoid pathways. Several R2R3-MYB TFs regulate the biosynthesis of one or more units of phenylpropanoid-derived compounds, such as MdMYB3, AtMYB4, and AtMYB12 (Aharoni et al., 2001; Deluc et al., 2006, 2008; Luo et al., 2008; Rommens et al., 2008; Liu J. et al., 2015). Here, we examined the effects of HcMYB1 and HcMYB2 on floral scent via gene silencing (Figure 6). Linalool and methyl benzoate levels significantly decreased in flowers when HcMYB1 or HcMYB2 was silenced, confirming the direct connection between the functional activity of these two TFs and volatile biosynthesis. The silencing of HcMYB1 or HcMYB2 also led to the downregulation of key structural scent-related genes (HcTPS3, HcTPS5, and HcBSMT2) from the terpenoid and phenylpropanoid pathways (Figure 6D). Similar results were obtained in petunia, where the silencing of R2R3-MYB (ODO1) led to the downregulation of several scent-related genes (Spitzer-Rimon et al., 2012). In addition, overexpressing PAP1 from Arabidopsis modulated the accumulation of terpenoid and phenylpropanoid scent compounds in rose flowers (Zvi et al., 2012). Nevertheless, before this study, little was known about the transcriptional regulatory mechanism underlying scent compound biosynthesis in non-model fragrance plants such as H. coronarium.
MYB TFs form complexes by interacting with other proteins, such as MYB-helix-loop-helix (bHLH)-WD40 proteins involved in regulating anthocyanin biosynthesis (Zhou and Memelink, 2016). MYB TFs also interact with other proteins involved in hormone signaling pathways, such as JAZ and PYL, which are crucial components of the JA and ABA signal-transduction pathways, respectively (Qi et al., 2014; Zhao et al., 2014). However, little is known about the interactions of MYB TFs with proteins in the auxin-signaling pathway. In Arabidopsis, the auxin signaling pathway repressor Aux/IAA (AtIAA29) interacts with the TF WRKY57 to mediate leaf senescence (Jiang et al., 2014). It was also observed that both HcMYB1 and HcIAA4 showed high protein expression in flowers (Supplementary Figure 4). In the current study, we uncovered an interaction between HcMYB1 and the auxin-responsive protein HcIAA4 via Y2H and BiFC assays (Figures 8A,B). We also demonstrated that HcIAA4 represses the activity of HcMYB1 (Figure 7C). Similarly, in Arabidopsis, AtJAZ proteins interact with MYBs such as MYB75, thereby decreasing their transcriptional activity (Qi et al., 2014). In A. thaliana and F. hybrida, MYB21 interacts with MYC2 to form MYB–bHLH complex to regulate the expression of TPS genes and floral scent emission in flowers (Yang et al., 2020). In Fagopyrum tataricum, the repressive activities of FtMYBs are directly enhanced by their interactions with FtSAD2 or FtJAZ1 (Zhang et al., 2018). The identification of protein–protein interactions between MYB TFs and other proteins provides clues about the regulation of gene expression and secondary metabolism during volatile biosynthesis.
In petunia, a network comprising three R2R3 MYB TFs (EOBI, EOBII, and ODO1) regulates flower-specific genes in the phenylpropanoid volatile biosynthesis pathway (Verdonk et al., 2005; Spitzer-Rimon et al., 2010, 2012; Van Moerkercke et al., 2011). Similarly, FaMYB10 regulates FaEOBII expression in strawberry (Medina-Puche et al., 2014, 2015). In A. thaliana, MYC2 interacts with DELLA protein to regulate the expression of sesquiterpene synthase genes (TPS21 and TPS11), and the expression of TPS21 and TPS11 was modulated by phytohormones (Hong et al., 2012). Likewise, LcMYB1 interacts with LcbHLH to regulate the expression of key structural anthocyanin biosynthesis genes in Litchi chinensis (Lai et al., 2016). The roles of MYB proteins in scented ornamental plants uncovered in the current study sheds light on the evolution of this important transcription factor family, providing new insights into how they regulate the biosynthesis of secondary metabolic compounds, including terpenoids and phenylpropanoids, in plants. The key role of MYB TFs in controlling the biosynthesis of volatile compounds highlights the potential of engineering these TFs to enhance the economic value of ornamental plant species.
In conclusion, we demonstrated that the IAA-responsive, flower-specific R2R3-MYB TFs HcMYB1 and HcMYB2 function as activators of terpenoid and phenylpropanoid biosynthesis in H. coronarium. Both HcMYB1 and HcMYB2 interact with the promoter of HcBSMT2, encoding the key enzyme for methyl benzoate biosynthesis. Furthermore, HcMYB2 regulates the expression of HcTPS5, which plays a key role in linalool biosynthesis. Finally, we showed that auxin takes part in volatile biosynthesis by regulating the expression of R2R3-HcMYB transcription factor genes via protein–protein interactions in H. coronarium. Our findings provide important insights into the roles of auxin and MYB TFs in the biosynthesis of floral scent compounds, laying the foundation for plant metabolic engineering efforts.
Hedychium coronarium plants were grown at South China Agricultural University under natural light conditions. For RNA extraction, the plant materials were harvested, immediately frozen in liquid nitrogen, and stored at −80°C. To analyze tissue-specific gene expression patterns, three tissues were used: fully open flowers, mature green leaves, and healthy rhizomes of 2-year-old H. coronarium plants. The process of flower development was divided into six stages: tight green bud stage (F1), white bud stage (F2), initial flowering stage (F3), half-open stage (F4), full-bloom stage (F5), and flower senescence (F6).
For IAA treatment, flowers at the F2 stage were cut into 35-cm pieces, placed in sterile water containing 100 μM IAA, and incubated for 12 h in a growth chamber under a 14/10 h light/dark cycle at 25°C. IAA and PCIB stock solution was prepared as per the manufacturer’s protocol. Briefly, 18.79 mg IAA powder was dissolved in 1.5 mL methanol and diluted in 100 mL sterilized water. Likewise, 321 mg PCIB powder was dissolved and diluted as aforementioned conditions. Thereafter, the detached flowers were placed in a flask containing 100 mL solution and covered with a silver sheet to protect them from degradation. The control flowers were kept in with a similar amount of volume of sterile water without IAA under the same aforementioned conditions. Volatile content was analyzed in flowers at the full-bloom stage. After analysis, all samples were frozen in liquid nitrogen and stored at −80°C for further experiments. Three to five independent experiments was performed with each experimental variant. A. thaliana and N. benthamiana plants used for subcellular localization and BiFC assays were grown in a growth chamber at 24°C under a 12/12 light/dark cycle.
The sequences of HcMYB1 and HcMYB2 were obtained from a flower RNA-seq database for H. coronarium (SRP049915). The related protein sequences were retrieved from the NCBI database2. The protein sequences were aligned, and a phylogenetic tree based on the R2R3-MYB domain was constructed using Clustal Ω (Sievers et al., 2011) and MEGA X (Kumar et al., 2018) software.
Total RNA was isolated from flowers at different stages of development and different organs using a HiPure Plant RNA Mini Kit (Magen) following the manufacturer’s protocol. Each 1 μg RNA sample was reverse transcribed using a PrimeScript RT Reagent Kit with Genomic DNA Eraser (Takara) following the manufacturer’s suggestions. To generate primers for reverse-transcription PCR (RT-PCR), the specific sequences of genes in H. coronarium were selected, and primers for the genes were designed with Primer 5.0 software (Abbas et al., 2020). The reaction mixtures (20 μl) included 10 μL pink SYBR mix, 7.2 μl distilled water, 0.4 μl each primer (10 μM), and 2.0 μl template cDNA. The expression values were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). A similar procedure was performed for all treatments using specific primers listed in Supplementary Table 2.
The full-length fragments excluding the stop codon were fused with the GFP gene in the p35S-EGFP-1 vector using SmaI at the 5′ end and SpeI at the 3′ end. The isolation and transformation of Arabidopsis protoplasts were performed as described by Yoo et al. (2007). The protoplasts were observed and photographed at 18 h after transformation under a confocal laser-scanning microscope.
Barley stripe mosaic virus (BSMV) was used for VIGS, as this system has successfully been used in monocots (Renner et al., 2009; Yuan et al., 2011). The pCaBSγ vector was linearized with ApaI before inserting the fragments. To specifically silence HcMYB1 and HcMYB2, a 280-bp fragment of each gene (from the 3′ end) was amplified by PCR from H. coronarium cDNA and inserted into pCaBSγ to produce pCaBSγ:HcMYB1 and pCaBSγ:HcMYB2, respectively. The cultures were harvested by centrifugation at 5000 rpm for 10 min and resuspended in infiltration buffer (10 mM MgCl2, 0.1 mM acetosyringone, 10 mM MES, pH 5.6) to a final OD600 of ∼1. For VIGS, flowers at the F1 stage were collected, dipped in the bacterial suspension, and vacuum infiltrated at 0.8 MPa. After the vacuum was released, the flowers were washed in deionized water, placed into liquid MS medium, and cultured under a 12/12 h light/dark cycle at 16°C for 5 days. Total floral volatile compounds were analyzed at the full-bloom stage via gas chromatography-mass spectrometer (GC-MS); the experiment was replicated three to four times.
The yeast-one-hybrid assay was performed using the Gold Yeast One Hybrid System (Clontech, Takara). To generate bait-specific reporter strains, a 1131-bp fragment (−1 to −1131 bp upstream of ATG) of the HcBSMT2 promoter and a 1555-bp fragment (−1 to −1555 bp upstream of ATG) of the HcTPS5 promoter were inserted into pAbAi to generate HcBSMT2-pAbAi and HcTPS5-pAbAi, respectively. The plasmids were integrated into the genome of yeast strain Y1H Gold (Clontech, Takara) via homologous recombination, and transformed colonies were selected on uracil-deficient synthetic dropout (SD/-Ura) medium. Different concentrations of the antibiotic AbA were used to select the bait strains. To generate the prey constructs, full-length HcMYB1 and HcMYB2 were cloned into pGAL4. The prey constructs were transformed into yeast cells harboring the bait constructs. The in vivo DNA-binding activity was determined based on the growth status of the transformed yeast cells on leucine-deficient synthetic dropout (SD/-Leu) medium supplemented with the selected concentration of AbA after 3–5 days of cultivation at 30°C.
The full-length HcIAA2 and HcIAA4 sequences were ligated into prey vector pGADT7 (AD). The coding sequences of HcIAA4, HcMYB1, and HcMYB2 were cloned into bait vector pGBKT7 (BD). The prey construct and specific bait construct were co-transformed into yeast strain Y2HGold, which harbors four reporter genes (Ade, MEL1, His, and AUR1) under the control of a GAL4-responsive promoter. The empty pGADT7 vector was used as a blank control. The positive transformants that grew on SD/-Trp medium were inoculated onto SD plates (SD/-Leu-Trp-His-Ade) and incubated for 3–5 days at 30°C; transactivation activity was confirmed by growth on these plates. Yeast colonies expressing the α-galactosidase MEL1 turned blue upon the addition of X-α-Gal substrate (Clontech, TaKaRa). Primers used to amplify the genes are listed in Supplementary Table 2.
To assay the transcriptional activities of HcMYB1 and HcMYB2, the full-length coding regions were independently fused to the GAL4 DNA-binding domain ligated to the pBD vector and used as the effectors. The reporter vector was modified from the pGreenII 0800-LUC vector, which includes the firefly luciferase gene (LUC) driven by the CaMV 35S minimal promoter with five repeats of upstream activating sequence, as well as the 35S-promoter-driven Renilla reniformis luciferase gene (REN) as an internal control. To examine the binding of HcMYB1 and HcMYB2 to the promoters of HcBSMT2 and HcTPS5, the promoters were cloned into the pGreenII 0800-LUC double-reporter vector (Hellens et al., 2005), whereas HcMYB1 and HcMYB2 were cloned into the pGreenII 62-SK vector as effectors. The effector and reporter plasmids were electroporated into Agrobacterium tumefaciens strain EHA105 and injected into N. benthamiana leaves with a needleless syringe. After 3–5 days, leaves were collected and LUC and REN activities examined via a dual-luciferase assay (Promega, United States) using a Luminoskan Ascent Microplate Luminometer (Thermo Fisher, United States) following the manufacturer’s protocol. The transactivation ability and the binding activity of HcMYB1 and HcMYB2 are indicated by the ratio of LUC to REN. Four or five measurements were carried out for each combination and three independent experiments were performed.
For EMSA, pGEX-4T-1 (GE Healthcare) was used to generate the GST-HcMYB1 and GST-HcMYB2 expression vectors, which were transformed into E. coli strain BM Rosetta (DE3). The expression of the recombinant fusion proteins was induced by adding 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Following incubation at 28°C for 8 h, the fusion proteins were purified using Glutathione Superflow Resin (Clontech) according to the manufacturer’s instructions. The fragments (∼50 bp) containing putative MBE-binding sequences in the HcTPS5 and HcBSMT2 promoters were labeled with biotin. EMSA was carried out using a Light Shift Chemiluminescent EMSA Kit (Thermo Scientific) as previously described (Tan et al., 2019). The purified fusion protein was incubated with biotin-labeled DNA fragments and a 100-fold molar excess of unlabeled DNA fragments with the same sequences that were used as competitors; GST protein with labeled DNA was used as a negative control. The protein-DNA complexes were separated by 5% native polyacrylamide gel electrophoresis, detected based on chemiluminescence on a ChemiDoc MP Imaging System (Bio-Rad), and transferred onto a nylon membrane.
The full-length HcMYB1 and HcIAA4 sequences were separately inserted into PUC-SPYNE and PUC-SPYCE to form HcMYB1-YFPN and HcIAA4-YFPC, respectively. The empty vector (control) and recombinant plasmids were transformed into EHA105 competent cells, and different combinations of Agrobacterium cultures were co-infiltrated into N. benthamiana leaves. The plants were cultivated in an incubator under a 16 h/8 h light/dark cycle for 3 days, and the infiltrated leaves were visualized under a Leica DM RXA2 upright fluorescence microscope as described previously (Ke et al., 2019).
The flower samples were ground to a fine powder in liquid nitrogen (N2) and transferred to 15 mL tubes containing 5 mL extraction solvent (2:1:0.002 [v/v/v] 2-propanol:H2O:HCl). The samples were sonicated for 15 min and incubated at 4°C for 30 min with shaking (100 rpm). After adding 5 mL dichloromethane, the samples were incubated under the same conditions and centrifuged at 10,000 rpm for 10 min at 4°C. The samples were concentrated in the dark via aeration of the solvent mixture using nitrogen gas, followed by the addition of 1.0 mL methanol and purification through a Sep-PakTM C18 reverse-phase extraction cartridge. The samples were dried completely, dissolved in 200 μL methanol, and filtered through a 0.22-mm PTFE filter. The IAA standards were prepared by dissolving IAA standards in methanol (Sigma, United States). Chromatographic and mass spectrometric conditions were as described previously (Niu et al., 2014). The experiment was performed in three biological and three technical replicates.
For the quantification of targeted hormones, flower samples were finely ground with liquid nitrogen following the protocol as described in Pan et al. (2010). Briefly, finely grounded flower samples and an adequate amount of internal standard (IS) were placed in 15 mL centrifuge tubes and 2-propanol/H2O/concentrated HCl (2:1:0.002, vol/vol/vol) were added to each tube followed by shaking at a speed of 5000 rpm for 10 min at 4°C. Thereafter, supernatants were transferred into a new tube and subjected under a gentle stream of highly purified nitrogen gas to a final volume of 3 mL and pH was adjusted to 8.0. Add twice the volume of petroleum ether to the solvent phase and shake at a speed of 5000 rpm for 10 min at 4°C and repeat this step. The sample solution was injected into the reverse-phase C18 Gemini HPLC column for UPLC-MS/MS analysis. The parameters of mass spectrometry for the measurement of hormones in the H. coronarium flowers are given in Supplementary Table 4. The experiment was performed in three biological and three technical replicates.
To analyze volatile compounds, a flower was placed in a 250-mL glass bottle and covered with an aluminum sheet; ethyl caprate was used as an internal standard. After 30 min of incubation, a PDMS fiber was inserted into the bottle, incubated for 30 min to adsorb volatiles, and injected into a gas chromatography-mass spectrometry system (Agilent) for volatile analysis as described previously (Yue et al., 2015). The experiment was performed in five to seven biological replicates.
All data were analyzed using LSD with Origin software. P-values < 0.05 were considered to be significant.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YK, FA, and YF conceived and designed the experiment. YK, FA, and YZ contributed to the experiment and data analysis. YK and FA wrote the manuscript. FA, YF, and RY revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Key-Areas Research and Development Program of Guangdong Province (Grant No. 2020B020220007), National Natural Science Foundation of China (Grant No. 31770738), People’s Livelihood Science and Technology Projects of Guangzhou (Grant No. 201903010054) to YF, and National Natural Science Foundation of China to RY (Grant No. 31870690).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.710826/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of six HcMYB proteins with Arabidopsis MYB protein family.
Supplementary Figure 2 | Correlation between HcMYB1 and HcMYB2 expression and the emission of volatile compounds.
Supplementary Figure 3 | A comparative analysis of transcript abundance of six HcMYB genes during flower development using RNA-seq data.
Supplementary Figure 4 | The protein expression level of HcMYB1 and HcIAA4 in different tissues.
Supplementary Figure 5 | The expression levels of key structural genes (HcTPS5 and HcBSMT2) in different tissues.
Supplementary Figure 6 | Heatmap showing expression profiles (log2TPM) of HcBSMT genes in different tissues.
Supplementary Figure 7 | Heatmap showing expression profiles (log2TPM) of HcTPS genes in different tissues.
Supplementary Table 1 | MYB-core binding motifs in the promoter regions of HcBSMTs and HcTPSs.
Supplementary Table 2 | Primers used in this study.
Supplementary Table 3 | Genes used for phylogenetic analysis and their accession numbers.
Supplementary Table 4 | The parameters index of UPLC-MS/MS used for the measurement of hormones in the H. coronarium flowers.
AbA, aureobasidin A; ABA, abscisic acid; Aux/IAA, auxin/indole-3-acetic acid; BALD, benzaldehyde dehydrogenase; BiFC, bimolecular fluorescence complementation; BSMT, salicylic acid/benzoic acid methyltransferase; cDNA, complementary DNA; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl diphosphate; FPPS, FPP synthase; GC-MS, gas chromatography-mass spectrometer; GFP, green fluorescent protein; IAA, indole-3-acetic acid; NLS, nuclear localization signal; OD, optical density; PAL, phenylalanine ammonia lyase; SD, synthetically defined medium; TPS, terpene synthase; Y2H, yeast two-hybrid; Y1H, yeast one-hybrid.
Abbas, F., Ke, Y., Yu, R., and Fan, Y. (2019). Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 249, 71–93. doi: 10.1007/s00425-018-3006-7
Abbas, F., Ke, Y., Yu, R., Yue, Y., Amanullah, S., Jahangir, M. M., et al. (2017). Volatile terpenoids: multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 246, 803–816. doi: 10.1007/s00425-017-2749-x
Abbas, F., Ke, Y., Zhou, Y., Waseem, M., Yu, Y., Ashraf, U., et al. (2020). Cloning, functional characterization and expression analysis of LoTPS5 from Lilium ‘Siberia’. Gene 756:144921. doi: 10.1016/j.gene.2020.144921
Abbas, F., Ke, Y., Zhou, Y., Yu, Y., Waseem, M., Ashraf, U., et al. (2021a). Genome-wide analysis of ARF transcription factors reveals HcARF5 expression profile associated with the biosynthesis of β-ocimene synthase in Hedychium coronarium. Plant Cell Rep. 40, 1269–1284. doi: 10.1007/s00299-021-02709-1
Abbas, F., Ke, Y., Zhou, Y., Yu, Y., Waseem, M., Ashraf, U., et al. (2021b). Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12:623742. doi: 10.3389/fpls.2021.623742
Aharoni, A., De Vos, C. R., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313x.2001.01154.x
An, X. H., Tian, Y., Chen, K. Q., Liu, X. J., Liu, D. D., Xie, X. B., et al. (2014). MdMYB9 and MdMYB11 are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 56, 650–662. doi: 10.1093/pcp/pcu205
Báez, D., Pino, J. A., and Morales, D. (2011). Floral scent composition in Hedychium coronarium J. Koenig analyzed by SPME. J. Essent. Oil Res. 23, 64–67. doi: 10.1080/10412905.2011.9700460
Bedon, F., Bomal, C., Caron, S., Levasseur, C., Boyle, B., Mansfield, S. D., et al. (2010). Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid-and flavonoid-oriented responses. J. Exp. Bot. 61, 3847–3864. doi: 10.1093/jxb/erq196
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12:691384. doi: 10.3389/fpls.2021.691384
Chai, Y. M., Jia, H. F., Li, C. L., Dong, Q. H., and Shen, Y. Y. (2011). FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 62, 5079–5089. doi: 10.1093/jxb/err207
Colquhoun, T. A., Schwieterman, M. L., Wedde, A. E., Schimmel, B. C., Marciniak, D. M., Verdonk, J. C., et al. (2011). EOBII controls flower opening by functioning as a general transcriptomic switch. Plant Physiol. 156, 974–984. doi: 10.1104/pp.111.176248
Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511. doi: 10.1104/pp.105.067231
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J. M., et al. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053. doi: 10.1104/pp.108.118919
Du, H., Zhang, L., Liu, L., Tang, X. F., Yang, W. J., Wu, Y. M., et al. (2009). Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 74, 1–11. doi: 10.1134/s0006297909010015
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Dudareva, N., Klempien, A., Muhlemann, J. K., and Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198, 16–32. doi: 10.1111/nph.12145
Dudareva, N., Negre, F., Nagegowda, D. A., and Orlova, I. (2006). Plant volatiles: recent advances and future perspectives. Crit. Rev. Plant Sci. 25, 417–440. doi: 10.1080/07352680600899973
Fan, Y. P., Yu, R., Huang, Y., and Chen, Y. (2003). Studies on the essential constituent of Hedychium flavum and H. coronarium. Acta Hortic. Sin. 30:475. doi: 10.16420/j.issn.0513-353x.2003.04.030
Fan, Y.-P., Wang, X.-R., Yu, R.-C., and Yang, P. (2007). Analysis on the aroma components in several species of Hedychium. Acta Hortic. Sin. 34, 231–234. doi: 10.16420/j.issn.0513-353x.2007.01.049
Gershenzon, J., and Dudareva, N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3, 408–414. doi: 10.1038/nchembio.2007.5
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J., and Chen, X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648. doi: 10.1105/tpc.112.098749
Jaradat, M. R., Feurtado, J. A., Huang, D., Lu, Y., and Cutler, A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13:192. doi: 10.1186/1471-2229-13-192
Jia, H. F., Chai, Y. M., Li, C. L., Lu, D., Luo, J. J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. doi: 10.1104/pp.111.177311
Jia, H., Xie, Z., Wang, C., Shangguan, L., Qian, N., Cui, M., et al. (2017). Abscisic acid, sucrose, and auxin coordinately regulate berry ripening process of the Fujiminori grape. Funct. Integr. Genomic 17, 441–457. doi: 10.1007/s10142-017-0546-z
Jiang, Y., Liang, G., Yang, S., and Yu, D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid–and auxin-mediated signaling in jasmonic acid–induced leaf senescence. Plant Cell 26, 230–245. doi: 10.1105/tpc.113.117838
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Ke, M., Gao, Z., Chen, J., Qiu, Y., Zhang, L., and Chen, X. (2018). Auxin controls circadian flower opening and closure in the waterlily. BMC Plant Biol. 18:143. doi: 10.1186/s12870-018-1357-7
Ke, Y., Abbas, F., Zhou, Y., Yu, R., Yue, Y., Li, X., et al. (2019). Genome-wide analysis and characterization of the Aux/IAA family genes related to floral scent formation in Hedychium coronarium. Int. J. Mol. Sci. 20:3235. doi: 10.3390/ijms20133235
Kim, H. J., Park, K. J., and Lim, J. H. (2011). Metabolomic analysis of phenolic compounds in buckwheat (Fagopyrum esculentum M.) sprouts treated with methyl jasmonate. J. Agric. Food Chem. 59, 5707–5713. doi: 10.1021/jf200396k
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3−MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Krizek, B. A. (2011). Auxin regulation of Arabidopsis flower development involves members of the aintegumenta-like/plethora (AIL/PLT) family. J. Exp. Bot. 62, 3311–3319. doi: 10.1093/jxb/err127
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai, B., Du, L. N., Liu, R., Hu, B., Su, W. B., Qin, Y. H., et al. (2016). Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front. Plant Sci. 7:166. doi: 10.3389/fpls.2016.00166
Lan, J. B., Yu, R. C., Yu, Y. Y., and Fan, Y. P. (2013). Molecular cloning and expression of Hedychium coronarium farnesyl pyrophosphate synthase gene and its possible involvement in the biosynthesis of floral and wounding/herbivory induced leaf volatile sesquiterpenoids. Gene 518, 360–367. doi: 10.1016/j.gene.2013.01.007
Lavid, N., Wang, J., Shalit, M., Guterman, I., Bar, E., Beuerle, T., et al. (2002). O-methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol. 129, 1899–1907. doi: 10.1104/pp.005330
Li, J., Li, X., Guo, L., Lu, F., Feng, X., He, K., et al. (2006a). A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J. Exp. Bot. 57, 1263–1273. doi: 10.1093/jxb/erj094
Li, J., Yang, X., Wang, Y., Li, X., Gao, Z., Pei, M., et al. (2006b). Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem. Biophys. Res. Commun. 341, 1155–1163. doi: 10.1016/j.bbrc.2006.01.077
Li, R., and Fan, Y. (2011). Molecular cloning and expression analysis of a terpene synthase gene, HcTPS2, in Hedychium coronarium. Plant Mol. Biol. Rep. 29, 35–42. doi: 10.1007/s11105-010-0205-1
Li, Y., Shan, X., Zhou, L., Gao, R., Yang, S., Wang, S., et al. (2019). The R2R3-MYB factor FhMYB5 from Freesia hybrida contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis. Front. Plant Sci. 9:1935. doi: 10.3389/fpls.2018.01935
Liao, W., Yang, Y., Li, Y., Wang, G., and Peng, M. (2016). Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental-stress-induced abscission. Sci. Rep. 6:32006. doi: 10.1038/srep32006
Liu, G., Ren, G., Guirgis, A., and Thornburg, R. W. (2009). The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary. Plant Cell 21, 2672–2687. doi: 10.1105/tpc.108.060079
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Liu, L., Ramsay, T., Zinkgraf, M., Sundell, D., Street, N. R., Filkov, V., et al. (2015). A resource for characterizing genome−wide binding and putative target genes of transcription factors expressed during secondary growth and wood formation in Populus. Plant J. 82, 887–898. doi: 10.1111/tpj.12850
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. doi: 10.1111/j.1365-313X.2008.03597.x
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodríguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria× ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Medina-Puche, L., Molina-Hidalgo, F. J., Boersma, M., Schuurink, R. C., López-Vidriero, I., Solano, R., et al. (2015). An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol. 168, 598–614. doi: 10.1104/pp.114.252908
Muhlemann, J. K., Klempien, A., and Dudareva, N. (2014). Floral volatiles: from biosynthesis to function. Plant Cell Environ. 37, 1936–1949. doi: 10.1111/pce.12314
Muhlemann, J. K., Maeda, H., Chang, C. Y., San Miguel, P., Baxter, I., Cooper, B., et al. (2012). Developmental changes in the metabolic network of snapdragon flowers. PLoS One 7:e40381. doi: 10.1371/journal.pone.0040381
Niu, Q., Zong, Y., Qian, M., Yang, F., and Teng, Y. (2014). Simultaneous quantitative determination of major plant hormones in pear flowers and fruit by UPLC/ESI-MS/MS. Anal. Methods 6, 1766–1773. doi: 10.1039/c3ay41885E
Oono, Y., Ooura, C., Rahman, A., Aspuria, E. T., Hayashi, K. I., Tanaka, A., et al. (2003). p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol. 133, 1135–1147. doi: 10.1104/pp.103.027847
Pan, X., and Welti, R. and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Perkins-Veazie, P. (1995). Growth and ripening of strawberry fruit. Hortic. Rev. 17, 267–297. doi: 10.1002/9780470650585.ch8
Pichersky, E., and Dudareva, N. (2007). Scent engineering: toward the goal of controlling how flowers smell. Trends Biotechnol. 25, 105–110. doi: 10.1016/j.tibtech.2007.01.002
Qi, T., Huang, H., Wu, D., Yan, J., Qi, Y., Song, S., et al. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26, 1118–1133. doi: 10.1105/tpc.113.121731
Raguso, R. A. (2009). Floral scent in a whole−plant context: moving beyond pollinator attraction. Funct. Ecol. 23, 837–840. doi: 10.1111/j.1365-2435.2009.01643.x
Ramya, M., Kwon, O. K., An, H. R., Park, P. M., Baek, Y. S., and Park, P. H. (2017). Floral scent: regulation and role of MYB transcription factors. Phytochem. Lett. 19, 114–120. doi: 10.1016/j.phytol.2016.12.015
Ramya, M., Lee, S. Y., An, H. R., Park, P. M., Kim, N. S., and Park, P. H. (2019). MYB1 transcription factor regulation through floral scent in Cymbidium cultivar ‘Sael Bit’. Phytochem. Lett. 32, 181–187. doi: 10.1016/j.phytol.2019.06.007
Reddy, V. A., Qian, W., Dhar, N., Kumar, N., Venkatesh, P. N., Rajan, C., et al. (2017). Spearmint R2R3−MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS. LSU). Plant Biotechnol. J. 15, 1105–1119. doi: 10.1111/pbi.12701
Renner, T., Bragg, J., Driscoll, H. E., Cho, J., Jackson, A. O., and Specht, C. D. (2009). Virus-induced gene silencing in the culinary ginger (Zingiber officinale): an effective mechanism for down-regulating gene expression in tropical monocots. Mol. Plant 2, 1084–1094. doi: 10.1093/mp/ssp033
Rommens, C. M., Richael, C. M., Yan, H., Navarre, D. A., Ye, J., Krucker, M., et al. (2008). Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol. J. 6, 870–886. doi: 10.1111/j.1467-7652.2008.00362.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shin, B., Choi, G., Yi, H., Yang, S., Cho, I., Kim, J., et al. (2002). AtMYB21, a gene encoding a flower−specific transcription factor, is regulated by COP1. Plant J. 30, 23–32. doi: 10.1046/j.1365-313x.2002.01264.x
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi: 10.1105/tpc.107.050963
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high−quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Spitzer-Rimon, B., Farhi, M., Albo, B., Cna’ani, A., Ben Zvi, M. M., Masci, T., et al. (2012). The R2R3-MYB–like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24, 5089–5105. doi: 10.1105/tpc.112.105247
Spitzer-Rimon, B., Marhevka, E., Barkai, O., Marton, I., Edelbaum, O., Masci, T., et al. (2010). EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 22, 1961–1976. doi: 10.1105/tpc.109.067280
Tan, H., Man, C., Xie, Y., Yan, J., Chu, J., and Huang, J. (2019). A crucial role of GA-regulated flavonol biosynthesis in root growth of Araboidopsis. Mol. Plant 12, 521–537. doi: 10.1016/j.molp.2018.12.021
Uimari, A., and Strommer, J. (1997). Myb26: a MYB−like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 12, 1273–1284. doi: 10.1046/j.1365-313x.1997.12061273.x
Van Moerkercke, A., Haring, M. A., and Schuurink, R. C. (2011). The transcription factor emission of benzenoids II activates the MYB ODORANT1 promoter at a MYB binding site specific for fragrant petunias. Plant J. 67, 917–928. doi: 10.1111/j.1365-313X.2011.04644.x
Verdonk, J. C., Haring, M. A., van Tunen, A. J., and Schuurink, R. C. (2005). ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17, 1612–1624. doi: 10.1105/tpc.104.028837
Wu, Q., Tao, X., Ai, X., Luo, Z., Mao, L., Ying, T., et al. (2018). Contribution of abscisic acid to aromatic volatiles in cherry tomato (Solanum lycopersicum L.) fruit during postharvest ripening. Plant Physiol. Biochem. 130, 205–214. doi: 10.1016/j.plaphy.2018.06.039
Yamaguchi, N., Wu, M. F., Winter, C. M., Berns, M. C., Nole-Wilson, S., Yamaguchi, A., et al. (2013). A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24, 271–282. doi: 10.1016/j.devcel.2012.12.017
Yan, H., Zhang, H., Wang, Q., Jian, H., Qiu, X., Wang, J., et al. (2011). Isolation and identification of a putative scent-related gene RhMYB1 from rose. Mol. Biol. Rep. 38, 4475–4482. doi: 10.1007/s11033-010-0577-1
Yang, Z., Li, Y., Gao, F., Jin, W., Li, S., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71, 4140–4158. doi: 10.1093/jxb/eraa184
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2:1565. doi: 10.1038/nprot.2007.199
Yoshida, K., Oyama-Okubo, N., and Yamagishi, M. (2018). An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp.). Mol. Breed. 38, 1–14. doi: 10.1007/s11032-018-0902-2
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6:e26468. doi: 10.1371/journal.pone.0026468
Yue, Y., Wang, L., Yu, R., Chen, F., He, J., Li, X., et al. (2021). Coordinated and high-level expression of biosynthetic pathway genes is responsible for the production of a major floral scent compound methyl benzoate in Hedychium coronarium. Front. Plant Sci. 12:650582. doi: 10.3389/fpls.2021.650582
Yue, Y., Yu, R., and Fan, Y. (2015). Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genomics 16:470. doi: 10.1186/s12864-015-1653-7
Zhang, K., Logacheva, M. D., Meng, Y., Hu, J., Wan, D., Li, L., et al. (2018). Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. J. Exp. Bot. 69, 1955–1966. doi: 10.1093/jxb/ery032
Zhang, X., He, Y., Li, L., Liu, H., and Hong, G. (2021). Involvement of the R2R3-MYB transcription factors MYB21 and its homologs in regulating the stamen flavonols accumulation in Arabidopsis. J. Exp. Bot. 72, 4319–4332. doi: 10.1093/jxb/erab156
Zhao, Y., Xing, L., Wang, X., Hou, Y. J., Gao, J., Wang, P., et al. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7:ra53. doi: 10.1126/scisignal.2005051
Zhou, M., and Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.biotechadv.2016.02.004
Zhou, M., Zhang, K., Sun, Z., Yan, M., Chen, C., Zhang, X., et al. (2017). LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol. 174, 1348–1358. doi: 10.1104/pp.17.00160
Zhu, N., Cheng, S., Liu, X., Du, H., Dai, M., Zhou, D. X., et al. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236, 146–156. doi: 10.1016/j.plantsci.2015.03.023
Zvi, M. M. B., Shklarman, E., Masci, T., Kalev, H., Debener, T., Shafir, S., et al. (2012). PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol. 195, 335–345. doi: 10.1111/j.1469-8137.2012.04161.x
Keywords: Hedychium coronarium, floral scent, auxin, MYB transcription factors, biosynthesis
Citation: Ke Y, Abbas F, Zhou Y, Yu R and Fan Y (2021) Auxin-Responsive R2R3-MYB Transcription Factors HcMYB1 and HcMYB2 Activate Volatile Biosynthesis in Hedychium coronarium Flowers. Front. Plant Sci. 12:710826. doi: 10.3389/fpls.2021.710826
Received: 17 May 2021; Accepted: 13 July 2021;
Published: 03 August 2021.
Edited by:
Renata Rivera-Madrid, Scientific Research Center of Yucatán (CICY), MexicoReviewed by:
Qian Shen, Shanghai Jiao Tong University, ChinaCopyright © 2021 Ke, Abbas, Zhou, Yu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanping Fan, ZmFueWFucGluZ0BzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.