
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 September 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.708697
This article is part of the Research TopicMetabolism of Fruit Volatile Organic CompoundsView all 12 articles
 Zerui Yang1,2,3
Zerui Yang1,2,3 Chunzhu Xie1
Chunzhu Xie1 Ting Zhan1
Ting Zhan1 Linhuan Li1
Linhuan Li1 Shanshan Liu1
Shanshan Liu1 Yuying Huang1
Yuying Huang1 Wenli An1
Wenli An1 Xiasheng Zheng1*
Xiasheng Zheng1* Song Huang1,3*
Song Huang1,3*Trans-isopentenyl diphosphate synthases (TIDSs) genes are known to be important determinants for terpene diversity and the accumulation of terpenoids. The essential oil of Cinnamomum camphora, which is rich in monoterpenes, sesquiterpenes, and other aromatic compounds, has a wide range of pharmacological activities and has therefore attracted considerable interest. However, the TIDS gene family, and its relationship to the camphor tree (C. camphora L. Presl.), has not yet been characterized. In this study, we identified 10 TIDS genes in the genome of the C. camphora borneol chemotype that were unevenly distributed on chromosomes. Synteny analysis revealed that the TIDS gene family in this species likely expanded through segmental duplication events. Furthermore, cis-element analyses demonstrated that C. camphora TIDS (CcTIDS) genes can respond to multiple abiotic stresses. Finally, functional characterization of eight putative short-chain TIDS proteins revealed that CcTIDS3 and CcTIDS9 exhibit farnesyl diphosphate synthase (FPPS) activity, while CcTIDS1 and CcTIDS2 encode geranylgeranyl diphosphate synthases (GGPPS). Although, CcTIDS8 and CcTIDS10 were found to be catalytically inactive alone, they were able to bind to each other to form a heterodimeric functional geranyl diphosphate synthase (GPPS) in vitro, and this interaction was confirmed using a yeast two-hybrid assay. Furthermore, transcriptome analysis revealed that the CcTIDS3, CcTIDS8, CcTIDS9, and CcTIDS10 genes were found to be more active in C. camphora roots as compared to stems and leaves, which were verified by quantitative real-time PCR (qRT-PCR). These novel results provide a foundation for further exploration of the role of the TIDS gene family in camphor trees, and also provide a potential mechanism by which the production of camphor tree essential oil could be increased for pharmacological purposes through metabolic engineering.
Cinnamomum camphora is a subtropical evergreen tree species that has been widely cultivated in southern China for over 1,500years (Chen et al., 2020b). Studies have shown that essential oil extracted from the leaves of the camphor tree, which is rich in monoterpenes, sesquiterpenes, and other aromatic compounds, has a wide range of pharmacological activities, including antibacterial, antioxidant, and insecticidal properties (Yu et al., 2019).
Cinnamomum camphora subspecies can be grouped into five chemotypes according to the dominant component in their essential oils, which were extracted from their leaves: linalool, D-borneol, camphor, cineole, or nerolidol (Guo et al., 2017; Chen et al., 2018). These terpenoids have important industrial and pharmaceutical applications. For example, D-borneol is a well-established traditional Chinese medicine that is used to treat cardiovascular diseases, including stroke, coronary heart disease, and angina pectoris (Yang et al., 2020). D-Borneol has been documented in various versions of the Chinese Pharmacopoeia (Huang et al., 2016; Liang et al., 2018; Ren et al., 2018; Chai et al., 2019; Chen et al., 2019; Yang et al., 2020), and it is in high demand as a key ingredient in many traditional Chinese herbal formulas. This compound can also be used to relieve pain resulting from wounds, injuries, burns, and cuts (Yang et al., 2020). However, despite the high medicinal value of volatile terpenoids, limited plant resources, and low extraction efficiency limit the amounts of the essential oils from C. camphora that can be obtained for research or practical applications (Ma et al., 2021).
An increasing body of research has demonstrated that metabolic engineering consists of optimizing the genetic and regulatory mechanisms that govern cellular processes, and this is an effective method that can be used to increase the production of active natural products in microorganisms and plants (Choi et al., 2019; Li and Mutanda, 2019). Identifying the genes that govern production and accumulation of the essential oil in C. camphora could therefore enable the implementation of this approach to increase the essential oil yield, which would assist in meeting high demand.
Terpenoids are the largest category of plant specialized metabolites. More than 55,000 of these compounds have thus far been described, and are grouped into hemi- (C5), mono- (C10), sesqui- (C15), di- (C20), sester- (C25), tri- (C30), tetra- (C40), and poly- (C50) terpenoids according to the number of carbon atoms they contain (Dudareva et al., 2004; Pichersky et al., 2006; Ueoka et al., 2020). Despite diverse functions and structures, all terpenoids contain two universal C5 units: isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP). The mevalonic acid (MVA) pathway gives rise to IPP, and via enzymatic isomerization, to DMAPP, whereas the methylerythritol phosphate (MEP) pathway directly produces both IPP and DMAPP. IPP and DMAPP can be condensed (head to tail) by prenyltransferase (PTS) or isoprenyl diphosphate synthase (IDS), resulting in a series of prenyl diphosphates with various chain lengths. These linear precursors are then catalyzed by terpene synthase (TPS) and other modifying enzymes to form a variety of different terpenoids (Tholl and Lee, 2011; Athanasakoglou and Grypioti, 2019; Johnson et al., 2019; Johnson and Bhat, 2019; Adal and Mahmoud, 2020; Hivert et al., 2020; Miller and Bhat, 2020).
Isoprenyl diphosphate synthase is located at the branch point of the terpenoid biosynthetic pathway, and plays a vital role in the formation of diverse terpenoid structures (Jia and Chen, 2016). Differences in terpenoid synthase gene expression and the supply of precursors determine the terpenoid composition produced by plants (Dudareva et al., 2000; Johnson et al., 2019; Johnson and Bhat, 2019; Adal and Mahmoud, 2020; Miller and Bhat, 2020). Both trans- and cis- isomers of the products of IDS exist (Liang et al., 2002). Trans-IDS (TIDS) enzymes synthesize isoprenyl diphosphates (Barja and Rodríguez-Concepción, 2020) and are classified as short-chain (SC-TIDS, C10–20), medium-chain (MC-TIDS, C25–35), or long-chain (LC-TIDS, C40–50), depending on the length of the isoprenyl diphosphates that they produce (Wang et al., 2019). Cis-IDSs (CIDSs) were initially predicted to synthesize long-chain isoprenyl diphosphates (>C50) for dolichol and polyprenol production (Jia and Chen, 2016). Although, TIDSs and CIDSs have similarities in substrate preference and reaction products, they utilize different catalytic mechanisms and may be readily distinguished from one another by their primary amino acid (AA) sequences (Akhtar et al., 2013).
Short-chain-trans-isopentenyl diphosphate synthase includes homodimeric or heterodimeric geranyl diphosphate synthase (GPPS), farnesyl diphosphate synthase (FPPS), and geranylgeranyl diphosphate synthase (GGPPS; Jia and Chen, 2016), and their products provide the precursors for monoterpenes, sesquiterpenes, and diterpenes, respectively. Geranylfarnesyl diphosphate synthase (GFPPS) and polyprenyl diphosphate synthase (PPPS) are MC-TIDSs, whereas solanesyl diphosphate synthase (SPPS) is classified as an LC-TIDS (Vandermoten et al., 2009; Nagel et al., 2015; Wang et al., 2016; Kopcsayová and Vranová, 2019).
Phylogenetic analysis has revealed that plant TIDS genes can be documented into five subfamilies according to sequence identity: TIDS-a, -b, -c, and -e including the genes encoding FPPS, SPPS, PPPS, and small subunits (SSUs) of GPPS, respectively (Jia and Chen, 2016). The TIDS-d subfamily is more complex, and it includes genes encoding GGPPS, GFPPS, and PPPS that share a high sequence identity of at least 40%, with some shared identities of 55% or greater (Jia and Chen, 2016; Cui et al., 2019). Identification of entire TIDS gene families in plant species is required to determine their functions and to understand their combined effect on the specific profile of terpenoids produced.
Thus far, this level of TIDS gene family characterization has only been achieved in the model plant Arabidopsis thaliana (Kopcsayová and Vranová, 2019), in which 16 TIDS genes were identified, and in the tomato plant Solanum lycopersicum (Zhou and Pichersky, 2020), in which 10 were characterized. Studies characterizing these TIDS genes have revealed that while most of the synthase types they encode are homodimeric, GPPS may exist as either a homodimer or a heterodimer containing a small subunit (SSU) and a large subunit (LSU), or both (Rai et al., 2013; Chen et al., 2015; Adal and Mahmoud, 2020). The SSU can be further separated into two types (I and II), which are generally inactive alone. The LSU may be either inactive alone or possess GGPPS activity, while its heterodimer is an active GPPS (Burke et al., 2004; Rai et al., 2013).
Given that isoprenyl diphosphate biosynthesis is a crucial determinant for the formation of downstream terpenoid type and their yield, in-depth studies on TIDSs in the camphor tree would be instrumental in optimizing the production of these medicinally valuable products. Here, for the first time, we conducted a genome-wide analysis in the borneol chemotype of C. camphora to identify and characterize TIDS genes and the proteins they encode.
Borneol chemotype C. camphora plants were purchased from Ji’an Yu Feng Natural Species Co., Ltd. (Ji’an, China), and grown in an artificial climate box (Shanghai Yiheng Instrument Co., Ltd., Shanghai, China) at 25°C with a 12h light and then 12h dark photoperiod. We verified the origin of these plants through DNA barcodes.
We downloaded TIDS protein sequences of A. thaliana and S. lycopersicum from The Arabidopsis Information Resource (TAIR)1 and the Sol Genomics Network (Wijfjes and Smit, 2019).2 We then used the Protein Basic Logical Alignment Search Tool (BLASTP, United States National Library of Medicine)3 to compare these sequences with the C. camphora database (unpublished), using previously established thresholds for e-value (≤1e−5) and identity (Cui et al., 2019). Any genes identified were then used as queries for a second round of BLASTP searches to ensure no putative CcTIDS genes were missed. We also conducted a Hidden Markov Model (HMM) search for sequence homologs using the HMMER 3.0 program and the polyprenyl synthase domain PF00348 as a bait, and previously established e-value and identity thresholds (Wong et al., 2015; Cui et al., 2019).
The BLASTP and HMM search results were then integrated to identify candidate TIDS genes. Their sequences were then submitted to the online Pfam database and NCBI conserved domains database (CDD) in order to verify the presence of the polyprenyl synthase domain (Zhu et al., 2020). The physicochemical parameters of each CcTIDS, including molecular weight and isoelectric point, were calculated using the ExPASy online tool (Zhu et al., 2020).4 The MEME online tool5 was used to discover conserved domains in the amino acid sequence of each CcTIDS (Zhu et al., 2020). Finally, we used the chloroP (Emanuelsson et al., 1999),6 TargetP (Emanuelsson et al., 2000),7 Wolfpsort (Horton et al., 2007),8 and Plant-mPloc (Chou and Shen, 2010)9 tools to predict the subcellular localization of each CcTIDS.
A maximum likelihood (ML) evolutionary tree was constructed using the identified C. camphora TIDS amino acid sequences and those of 10 other plant species (A. thaliana: Arabidopsis Genome Initiative, 2000; Oryza sativa: Yu et al., 2002; Physcomitrella patens: Rensing et al., 2008; Selaginella moellendorffii: Banks et al., 2011; S. lycopersicum: Tomato Genome Consortium, 2012; Amborella trichopoda: Amborella Genome Project, 2013; Picea abies: Nystedt et al., 2013; Zea mays: Jiao et al., 2017; and Cinnamomum micranthum: Chaw et al., 2019) and the Molecular Evolutionary Genetics Analysis (MEGA) tool (version 7.0; Kumar et al., 2016) with the best models of JTT+G. Bootstrap value, which indicates the reliability of each branch node, was set at 1,000 replicates. The resulting tree was visualized using EvolView v3 (Subramanian et al., 2019).
Exons and introns for each TIDS were identified using the gene transfer format (GTF) file from the C. camphora genome, which contains information regarding gene structure, and visualized using TBtools (Chen et al., 2020a). Finally, in order to identify cis-regulatory elements, TBtools was used to extract the 2,000bp upstream sequence for each TIDS gene identified in the C. camphora genome (Chen et al., 2020a). We then compared these sequences with the PlantCARE database of plant cis-acting regulatory elements (Lescot et al., 2002). Circos graphs showing the chromosomal localization and results of the synteny analysis of the CcTIDS sequences were drawn using TBtools (Chen et al., 2020a).
Total RNA was extracted from the root, stem, and leaf of C. camphora using the Plant Pure Plant RNA Kit (Aidlab, Beijing, China). cDNA was synthesized from 1μg of high-quality total RNA (OD 260/280=1.8–2.2, OD 260/230≥2.0, RIN≥6.5, and 28S:18S≥1.0) using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions (Su et al., 2019).
The primers use to amplify eight putative SC-TIDSs were designed using the Primer Premier 5 program (Supplementary Table S4). Amplification by PCR was conducted using the 2×TransStart FastPfu PCR SuperMix (TransGen Biotech, Beijing, China) and cDNA as the template. Purified PCR products were then cloned into the pEASY-blunt vector using previously described methods (Su et al., 2019).
Quantitative real-time PCR (qRT-PCR) was performed using the TransStar Tip Green qPCR SuperMix (TransGen Biotech, Beijing, China) and the CFX96 Touch Deep Well platform (Bio-Rad, United States; Yang et al., 2020). We used a previously described reaction system composition and qRT-PCR procedure (Yang et al., 2020). Primers are listed in Supplementary Table S4.
Full-length putative SC-TIDS gene sequences without stop codons were each fused with enhanced green fluorescent protein (EGFP) and ligated into the pAN580 vector (using the primers described in Supplementary Table S4). The recombinant vectors were then transformed into Arabidopsis protoplasts using polyethylene glycol (PEG; Zhou and Pichersky, 2020). Protoplast isolation and recombinant vector transformation were performed with protocols described in previous studies (Yoo et al., 2007; Wang et al., 2018). EGFP fluorescent signals were observed using a Zeiss laser scanning microscope (LSM) 800 (Zeiss, Germany) as previously described (Beck et al., 2013; Su et al., 2019).
Truncated or full length versions of the eight putative SC-TIDSs were ligated into the pET32a [polyhistidine (6x His) tag, Wego, Guanghzou, China] or pMAL-C5X (MBP tag, Wego, Guanghzou, China) expression vectors using the pEASY-Basic Seamless Cloning and Assembly Kit (TransGen Biotech) following the manufacturer’s instructions (Su et al., 2019), using the primers listed in Supplementary Table S4. Constructs were verified using Sanger sequencing. Upon sequence confirmation, recombinant plasmids were transferred into the expression strain Escherichia coli Rossetta (DE3; Huayueyang, Beijing, China). Heterologous protein expression in this strain and the purification of the fusion protein were conducted according to previously established methods (Su et al., 2019).
In order to establish the function of each SC-TIDS, a 200-μl in vitro enzymatic activity reaction containing 25mmol/L MOPSO buffer with pH of 7.0, 10mmol/L magnesium chloride, 10% glycerin, 20–50μg protein, and substrates (150μmol/L IPP+40μmol/L DMAPP or GPP or FPP, Sigma-Aldrich, United States), was conducted. Each mixture was first incubated at 30°C for 6h, and then, 200μl of 200mmol/L Tris–HCl (pH 9.5), containing 2units of bovine intestine alkaline phosphatase (18unitsmg−1; Sigma-Aldrich) and 2units of potato apyrase (25.2unitsmg−1; Sigma-Aldrich) were added (Rai et al., 2013). An overnight hydrolysis reaction was carried out at 30°C. Ethyl acetate was then used to extract the enzymatic reaction buffer in 2×400μl extractions (Su et al., 2019). The ethyl acetate extracts were concentrated to 100μl under N2, and these concentrated extracts were then used for gas chromatography–mass spectrometry (GC–MS) analysis using previously described methods (Athanasakoglou and Grypioti, 2019; Yang et al., 2020). The GPPS large and small subunits from Catharanthus roseus were also analyzed as a positive control (Rai et al., 2013).
The interactions between the CcGPPS small and large subunits were verified by the following experiments: the truncated versions of these subunits were amplified from the cDNA of C. camphora, followed by fusing to the activation domain of the pGADT7 vector or the binding domain of the pGBKT7 vector. Recombinant vectors were co-transformed into Saccharomyces cerevisiae AH109 yeast and sequentially cultivated on the synthetic dropout (SD) medium SD/-Trp/-Leu. The interaction between the two proteins was tested on SD/-Trp/-Leu/-His/-Ade medium supplemented with 150mM 3-amino-1,24-triazole (3-AT).
Based on the HMM scan and BLASTP search results, 10 full-length, protein-coding TIDS-like gene sequences were identified in C. camphora, which we numbered CcPTPS1–10 according to their locations on the chromosomes. These genes included eight putative SC-TIDS (two FPPSs, CcTIDS3 and CcTIDS9; two GPPSs, CcTIDS4 and CcTIDS5; three GGPPSs, CcTIDS1, CcTIDS2, and CcTIDS8; and one GPPS small subunit, CcTIDS10) and two putative LC-TIDS (SPPS, and CcTIDS6 and CcTIDS7), which were distributed along six of the 12 chromosomes (Figure 1). In addition, genome synteny analysis showed that three segmental duplications and no tandem duplication events likely occurred, suggesting that segmental duplication might be one of the reasons for TIDS gene family expansion (Figure 1).
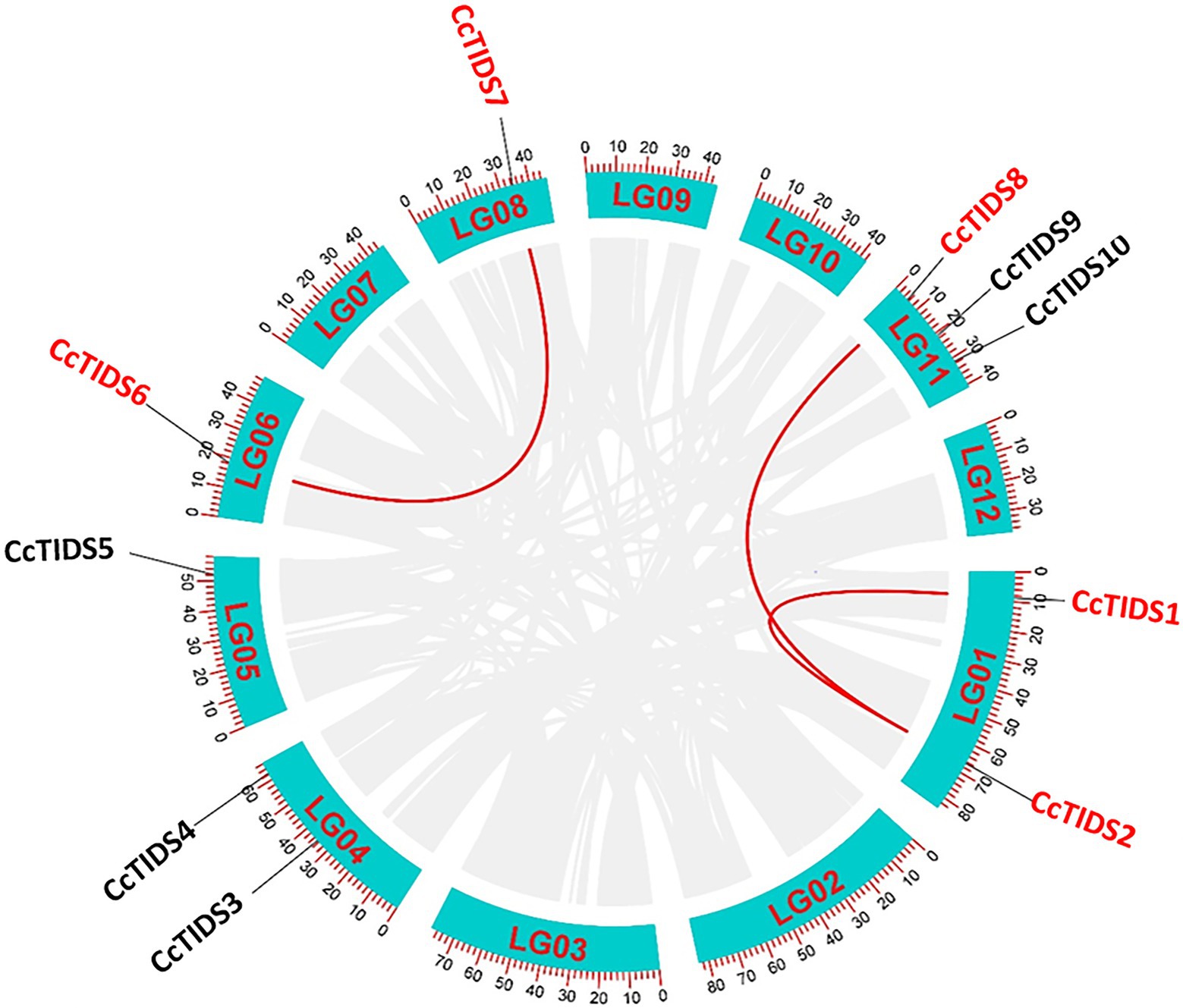
Figure 1. Circos graph showing the chromosomal locations and duplicated gene pairs of trans-isopentenyl diphosphate synthases (CcTIDS) genes in the C. camphora genome. The blue segments represent the 12 chromosomes present in this species. Duplicated genes are indicated as red lines between each gene pair.
The proteins encoded by the CcTIDS genes were found to have a minimum size of 298 AA and a maximum size of 428 AA. All of the CcTIDS proteins had an isoelectric point (pI) <7, indicating that these proteins are rich in acidic AAs. Specific information for all CcTIDS proteins is shown in Table 1.
A total of 100 TIDS-like genes were identified in 10 other plant species (Supplementary Tables S1 and S2). It can be concluded from the number of TIDS genes contained in each species that as the complexity of the species increases, the greater amount TIDS it contains (Coman et al., 2014). In addition, fewer CcTIDS genes were found in C. camphora than those in A. thaliana, Populus trichocarpa, O. sativa, and Z. mays, but equal to those in S. lycopersicum and the stout camphor tree C. micranthum.
A ML evolutionary tree was built to determine the phylogenetic relationships between the C. camphora TIDS family and those found in the other plants (Figure 2). None of the genes apart from those in S. lycopersicum and A. thaliana has yet been functionally characterized. In order to better distinguish the functional attributes of the TIDS genes on each branch, we added several well-characterized TIDS genes when constructing the phylogenetic tree (Supplementary Table S3). The TIDS genes in the resulting tree clustered into recently described catalytically distinct subfamilies (Jia and Chen, 2016).
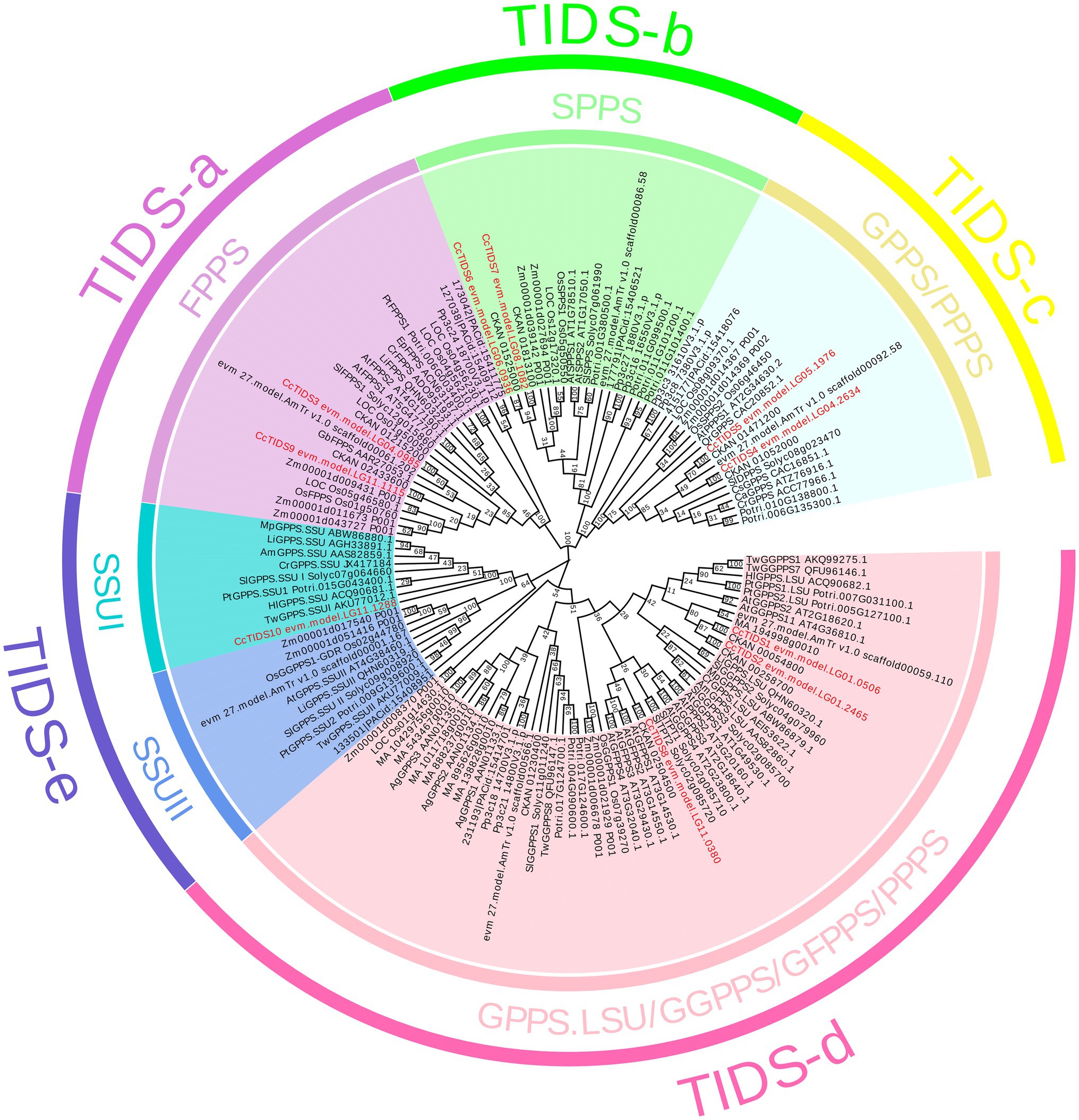
Figure 2. A phylogenetic tree constructed using putative or characterized TIDS genes from 11 sequenced land plant genomes and a neighbor joining approach. GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; LSU, large subunit; SSUI/II, small subunit I/II; GFPPS, geranylfarnesyl diphosphate synthase; PPPS, polyprenyl diphosphate synthase; and SPPS, solanesyl diphosphate synthase.
According to enzymatic activity and the relevance of genetic evolution, the subfamily encoding TIDS-e proteins was subdivided into two clades, which contained genes encoding GPPS small subunits I and II. Among them, only one single GPPS small subunit-encoding C. camphora gene, CcTIDS10, was placed (within the small subunit I clade), indicating that the genome of C. camphora only contained GPPS small subunit genes from this class. CcTIDS6 and CcTIDS7 were clustered closely with the genes encoding the TIDS-b subfamily of proteins, indicating that these two genes may function as SPPS enzymes.
CcTIDS1, CcTIDS2, and CcTIDS8 were found to belong to the TIDS-d subfamily, and therefore, may encode GGPPS, GFPPS, or PPPS enzymes. CcTIDS3 and CcTIDS9 were clustered with genes encoding TIDS-a proteins, indicating that they may encode FPPS enzymes. Although, the TIDS-c subfamily is known to contain mostly PPPS (Jia and Chen, 2016), genes encoding functionally characterized GPPS enzymes were included in this clade according to our analysis. Two CcTIDSs, CcTIDS4, and CcTTPS5, were clustered within this subfamily and were more closely related with the known GPPS-encoding genes, indicating that they may possess GPPS activity as well (Figure 2).
The evolution of gene families may be accompanied by changes in gene structure, which may provide additional information from which the diversification of gene functions can be inferred (Ye et al., 2017; Cui et al., 2019; Li et al., 2020). For this reason, we further analyzed the organization of the exons and introns of CcTIDS genes and compared them with those from A. thaliana and S. lycopersicum. Our results showed that the TIDS genes within the same subgroup mostly exhibited similar arrangements of exons and introns. For instance, CcTIDS6 and CcTIDS7, which were assigned to the TIDS-b-encoding subfamily, possessed six introns each. CcTIDS1, CcTIDS2, and CcTIDS8 were all found to contain no introns, indicating that they do not encode GFPPS, which are known to contain two introns, although, they were closely clustered with known GFPPS genes, and belong to the same subfamily (Figure 1). However, it remains unclear whether these genes encode GPPS large subunits or GGPPS proteins.
Our analysis on conserved TIDS motifs showed that TIDS belonging to the same subfamily exhibited a similar motif composition and arrangement (Figures 3A–D). Catalytically important conserved motifs were recognized in every identified TIDS sequence after multiple sequence alignment of CcTIDS, A. thaliana TIDS (AtTIDS), and S. lycopersicum TIDS (SlTIDS). Two common aspartate-rich motifs, the first and second aspartate-rich motif (FARM and SARM, respectively), are known binding sites for DMAPP (Su et al., 2019), and these were present in sequences of all the TIDS sequences except for those of CcTIDS10, AtGPPS small subunit II, and SlGPPS small subunit I.
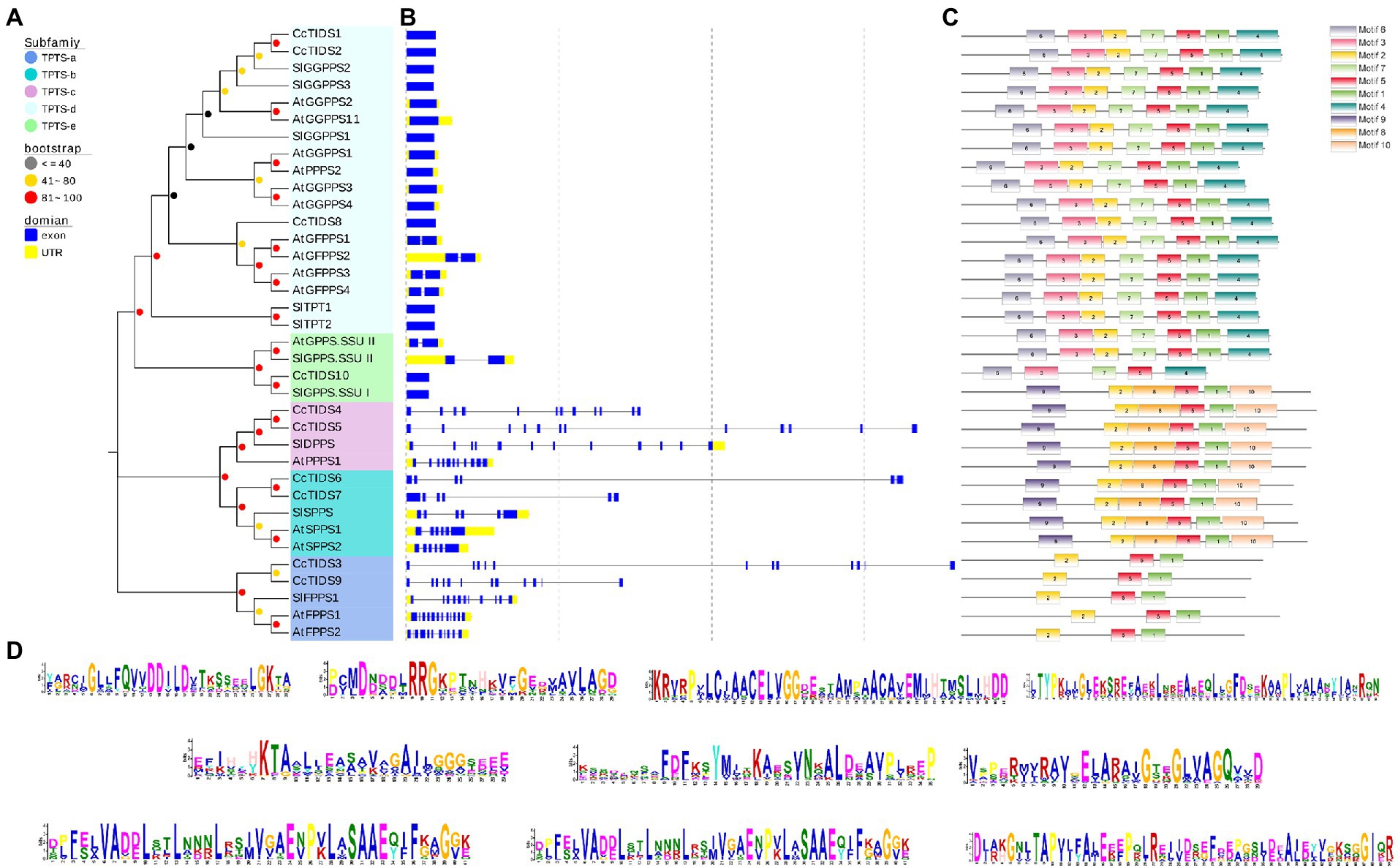
Figure 3. Tran-prenyltransferase protein subfamily analysis based on phylogenetic relationships. (A) Intron-exon structures, (B) conserved motifs, (C) analysis of C. camphora (Cc)TIDS, Arabidopsis thaliana (At)TIDS, and Solanum lycopersicum (Sl)TIDS. (D) Legend depicting the protein sequence of the corresponding motif. GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; LSU, large subunit; SSUI/II, small subunit I/II; GFPPS, geranylfarnesyl diphosphate synthase; PPPS, polyprenyl diphosphate synthase; and SPPS, solanesyl diphosphate synthase.
In addition, two conserved cysteine-rich (CxxxC) motifs were found in CcTIDS10, AtGPPS small subunit II, and SlGPPS small subunit I, and one each in CcTIDS1, CcTIDS2, and CcTIDS8. These CxxxC motifs play important roles in the interaction between the two subunits of the plant heterodimer GPPS. Our data suggest that CcTIDS10 is most likely a GPPS small subunit that forms a heterodimer with CcTIDS1, CcTIDS2, or CcTPS8 as a GPPS large subunit (Supplementary Figure S1).
We analyzed cis-elements in the promoter of the CcTIDS genes to better understand their potential regulation and function. In total, 343 cis-acting elements were identified, and they were grouped into three categories, which were responsible for plant growth and development, phytohormone responsiveness, and stress responsiveness, according to previous research (Abdullah et al., 2018). Of these, nine belong to the plant growth and development category, of which AAGAA motifs (involved in the endosperm) and AS-1 elements (involved in shoot expression) accounted for the highest proportion (19.05%).
The greatest proportion of cis-acting regulatory elements related to phytohormone response were myelocytomatosis (MYC) elements that are associated with methyl jasmonate (MeJA), accounting for 50%. All CcTIDS genes contained at least five MYC cis-acting elements, suggesting that CcTIDS gene expression might be moderated by MeJA. Nearly half of all the cis-acting elements were associated with stress responsiveness (167/343), of which the most common three cis-acting elements were the STRE motif related to stress (19%), G-box (14%), and box 4 (9%), which are associated with responsiveness to light (Figure 4). These results suggest that CcTIDS gene expression may be induced or suppressed by MeJA, and these genes may play roles in plant responses to a variety of abiotic stressors.
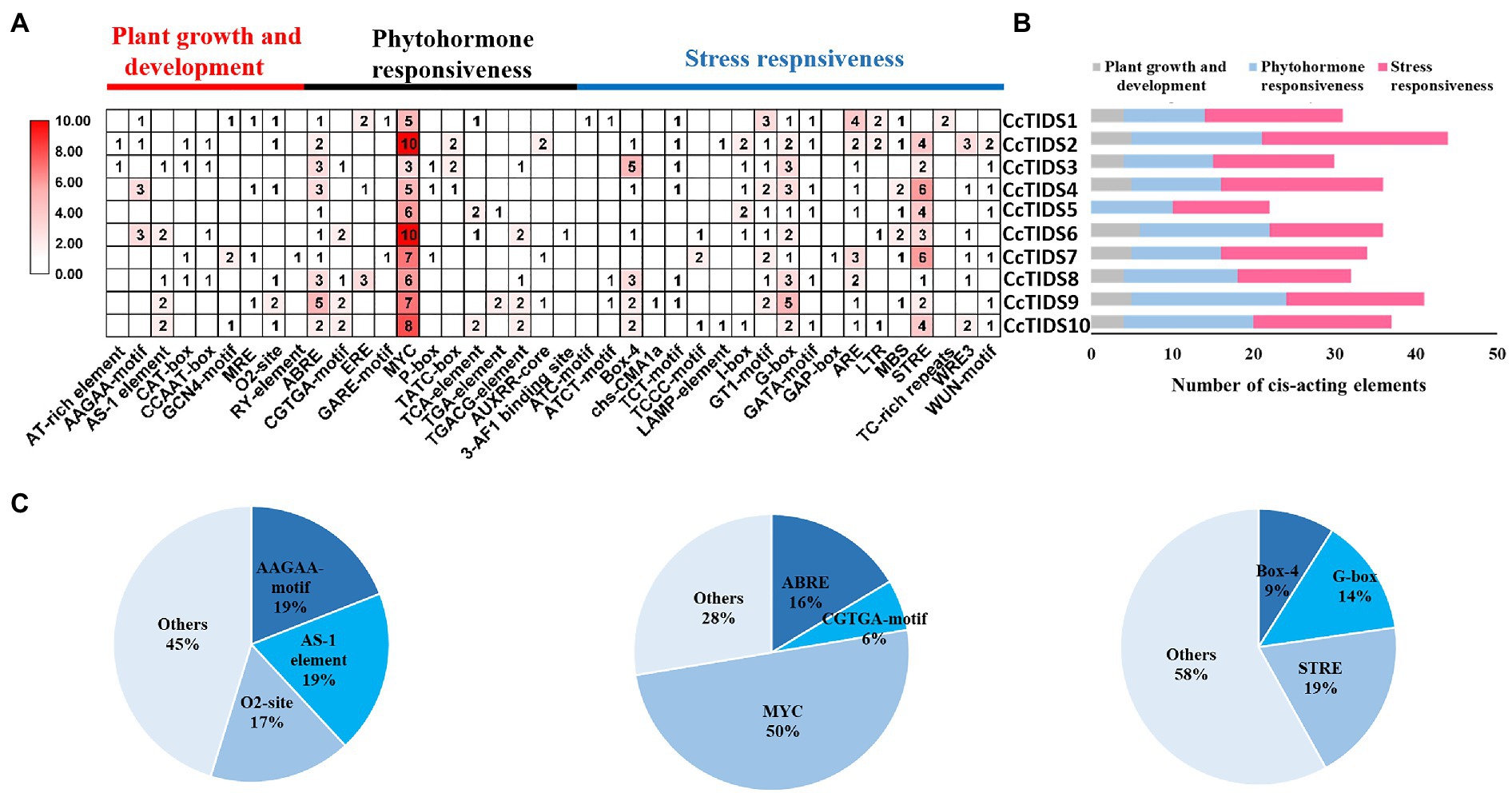
Figure 4. (A) The number of cis-acting elements in each C. camphora (Cc)TIDS gene, indicated by the intensity of the red color and numbers in the grid. (B) Histograms indicating the number of cis-acting elements in each CcTIDS gene with functions in plant growth and development, phytohormone responsiveness, or stress responsiveness. (C) Pie charts showing the ratio of different cis-acting elements in each structural category.
Our sequence alignment, evolution, and gene structure analyses all indicated that the CcTIDS6 and CcTIDS7 genes most likely encode SPPS enzymes. Hence, only the eight putative SC-TIDS-encoding genes were included in further analyses of expression patterns and subcellular localization, and an enzymatic assay.
To identify potential roles for CcTIDS genes in different tissues of C. camphora, we established a transcriptome database based on RNA sequencing of the leaves, roots, and stems (Accession number: PRJNA747104). qRT-PCR was used to verify these transcriptome data. We found that the expression profiles of CcTIDS genes were tissue-specific (Figure 5). For example, CcTIDS1 and CcTIDS2 were upregulated in the leaf tissue compared with the root and stem, whereas the expression level of CcTIDS3, 9, 8, and 10 were upregulated in the root compared with the other tissues. This may indicate that there are functional differences in CcTIDS according to the different C. camphora organs from which the tissue was derived.
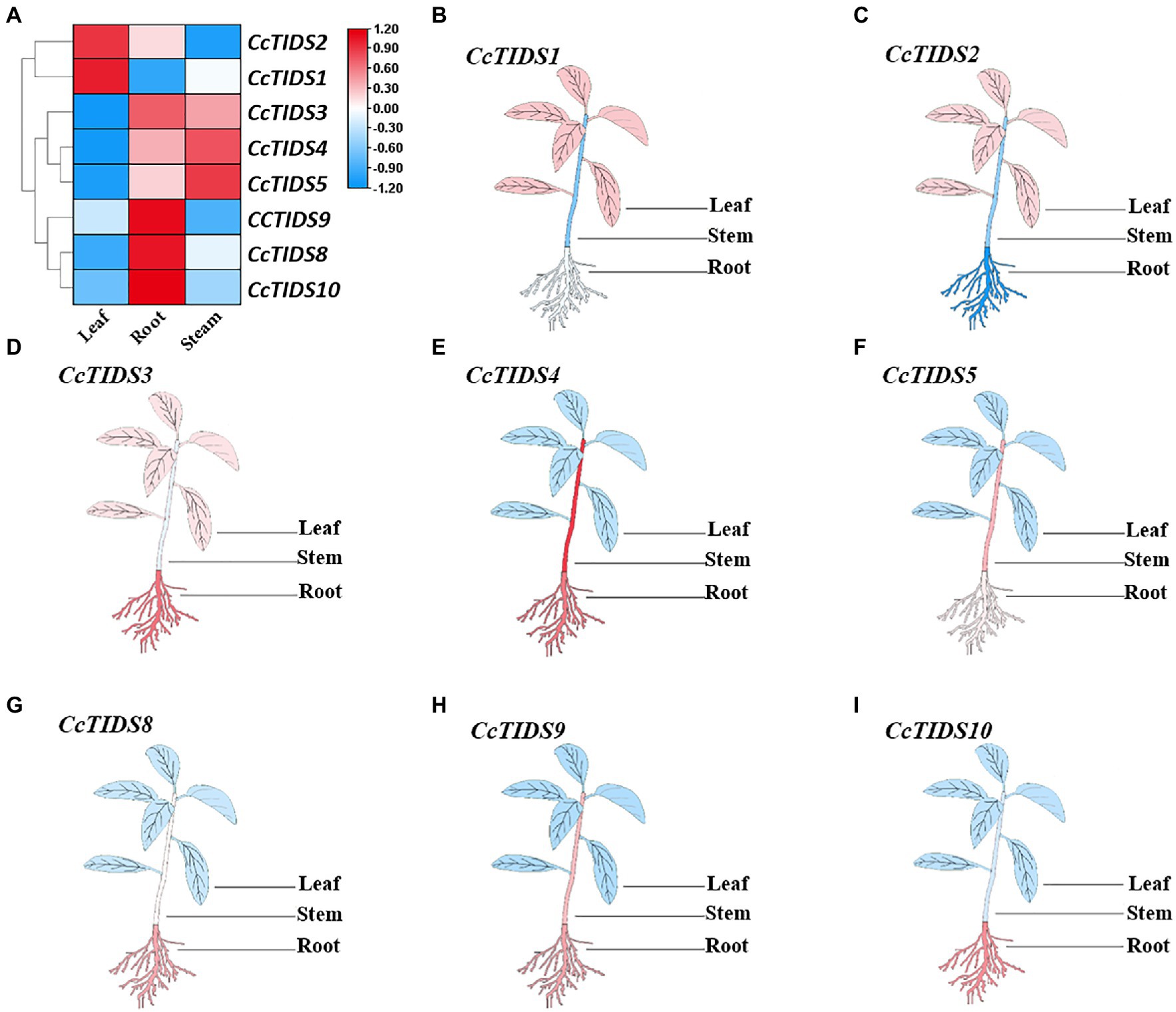
Figure 5. Differential expression of C. camphora (Cc)TIDS genes in different tissues by transcriptome and quantitative real-time PCR (qRT-PCR). (A) FPKM value of the RNA sequencing data for each gene in leaf, root, and stem tissues. (B–I) Relative expression of each gene in the different plant tissues as calculated by qRT-PCR, with red and blue representing high and low expression levels, respectively.
Subcellular localization predictions using four different online software programs revealed that CcTIDS1, 2, 8, and 10 are likely to be located in plastids. However, the results for CcTIDS3, 4, 5, and 9 were inconsistent (Table 1). In order to obtain exact subcellular location information for each candidate CcTIDS, we analyzed expression patterns using fluorescence microscopy (Figure 6). Consistent with all prediction programs, the GFP signals for CcTIDS1, 2, 8, and 10 were localized mainly within chloroplasts. Fluorescently tagged CcTIDS3 and CcTIDS9 proteins were both localized in the cytoplasm, while fluorescent signals for CcTIDS4 and CcTIDS5 recombinant proteins indicated that they may be located within mitochondria.
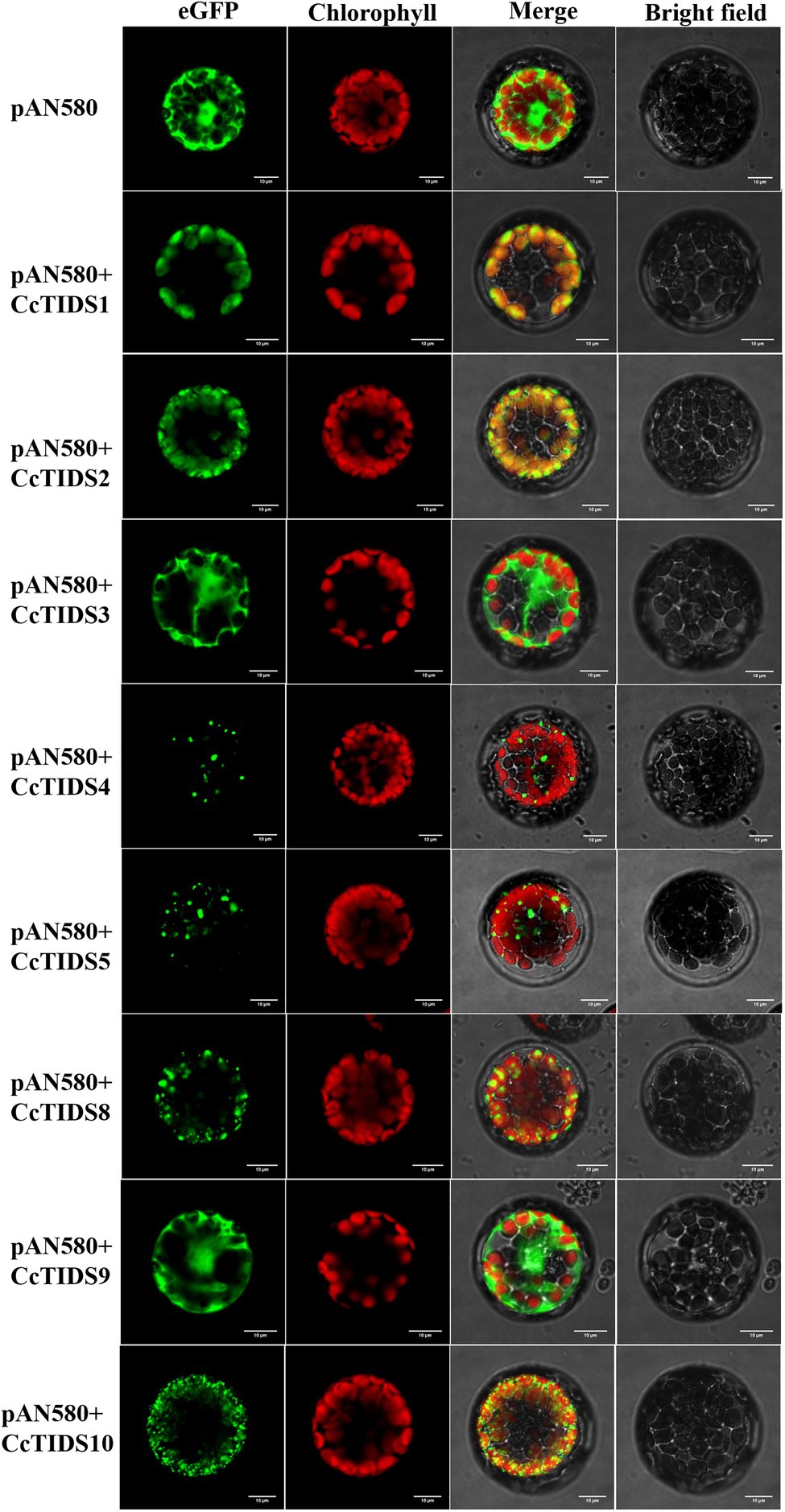
Figure 6. Subcellular localization of eight putative C. camphora trans-prenyltransferase (CcTIDS) enzymes in A. thaliana protoplasts. eGFP, green fluorescent protein fluorescence image; chlorophyll, chlorophyll autofluorescence image; bright-field, transmission image; merged, all channels (eGFP, chlorophyll, and bright-field) combined.
In vitro enzymatic assays were conducted using recombinant proteins extracted and purified from E. coli expression strains. The large subunit of CrGPPS was found to catalyze IPP and DMAPP to form geranyl diphosphate (GPP) and geranylgeranyl diphosphate (GGPP), whereas the small subunit of CrGPPS was found to be inactive alone. However, when both subunits were co-incubated, they catalyzed the formation of GPP (Supplementary Figure S2). This is accordance with the results reported in previous research, indicating that the method is valid for the verification of SC-TIDS in vitro enzymatic activity (Rai et al., 2013).
To characterize the two putative CcGPPS proteins (CcTIDS4 and 5), full-length and truncated versions of CcGPPSs were both analyzed. However, full-length recombinant CcGPPS protein was found to be completely insoluble. Therefore, soluble protein that was purified from the truncated version of each CcGPPS was used in the in vitro enzymatic activity assays. Unexpectedly, no catalytic products were detected for the truncated versions of both CcTIDS4 and CcTIDS5 under the conditions used here (Table 2).
Two full-length putative CcFPPS proteins (CcTIDS 3 and 9) lacking a transit peptide were generated. As expected, both recombinant proteins exhibited FPPS activity, producing FPP as the unique product, indicating that these two TIDS proteins are FPP synthases. Therefore, we reclassified these two TIDS proteins as CcFPPS1 and CcFPPS2 (Figure 7).
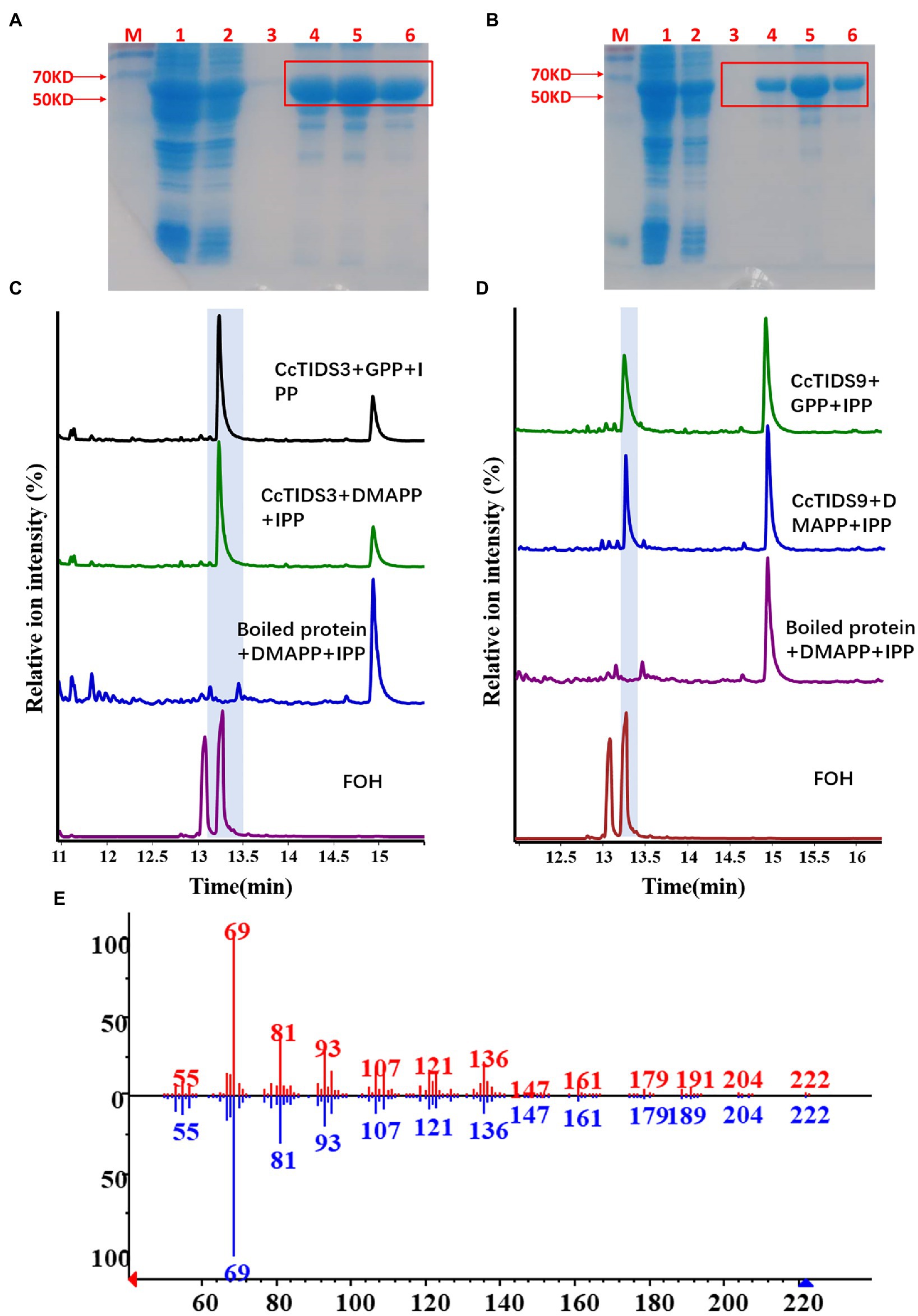
Figure 7. Gas chromatography–mass spectrometry (GC–MS) profile showing in vitro reaction products of C. camphora trans-prenyltransferases CcTIDS3 and CcTIDS9, where different prenyldiphosphates were used as substrates. Expression and purification of (A) CcTIDS3 and (B) CcTIDS9 recombinant protein from Escherichia coli Rosetta (DE3) harboring pET32a(+)–CcTIDS3/CcTIDS9, where lane 1 shows the total protein after induction, 2 shows the soluble protein, and 3–6 shows the purified CcTIDS3 or CcTIDS9 recombinant protein from the first to the fourth collected tube. The GC–MS chromatogram of the reaction products generated by (C) CcTIDS3 and (D) CcTIDS9 and the acid hydrolysis products of farnesol (FOH) standards. (E) Mass spectra of farnesol standard (shown in red) and products in the NIST14/Wiley275 library (blue). GPP, geranyl diphosphate; DMAPP, dimethylallyl diphosphate; and IPP, isopentenyl diphosphate.
It was predicted that three putative CcGGPPS proteins (CcTIDS1, 2, and 8) and one putative CcGPPS small subunit (CcTIDS10) contained a transit peptide. Therefore, truncated versions of these four proteins were fused to His or MBP tags, resulting in the formation of soluble protein. The activity of these four soluble proteins was detected in all substrate combinations. As a result, CcTIDS1 and 2 emitted a prominent chromatographic signal corresponding to geranylgeraniol (C20), demonstrating that CcTIDS1 and 2 function individually as bona fide GGPPS enzymes. Hence, these two proteins were renamed CcGGPPS1 and CcGGPPS2 (Figures 8A–F). In contrast, CcTIDS8 and 10 did not exhibit any chromatographic peak signal for farnesol (C15), geraniol (C10), or geranylgeraniol (C20), suggesting that these proteins are inactive alone (Figure 8G).
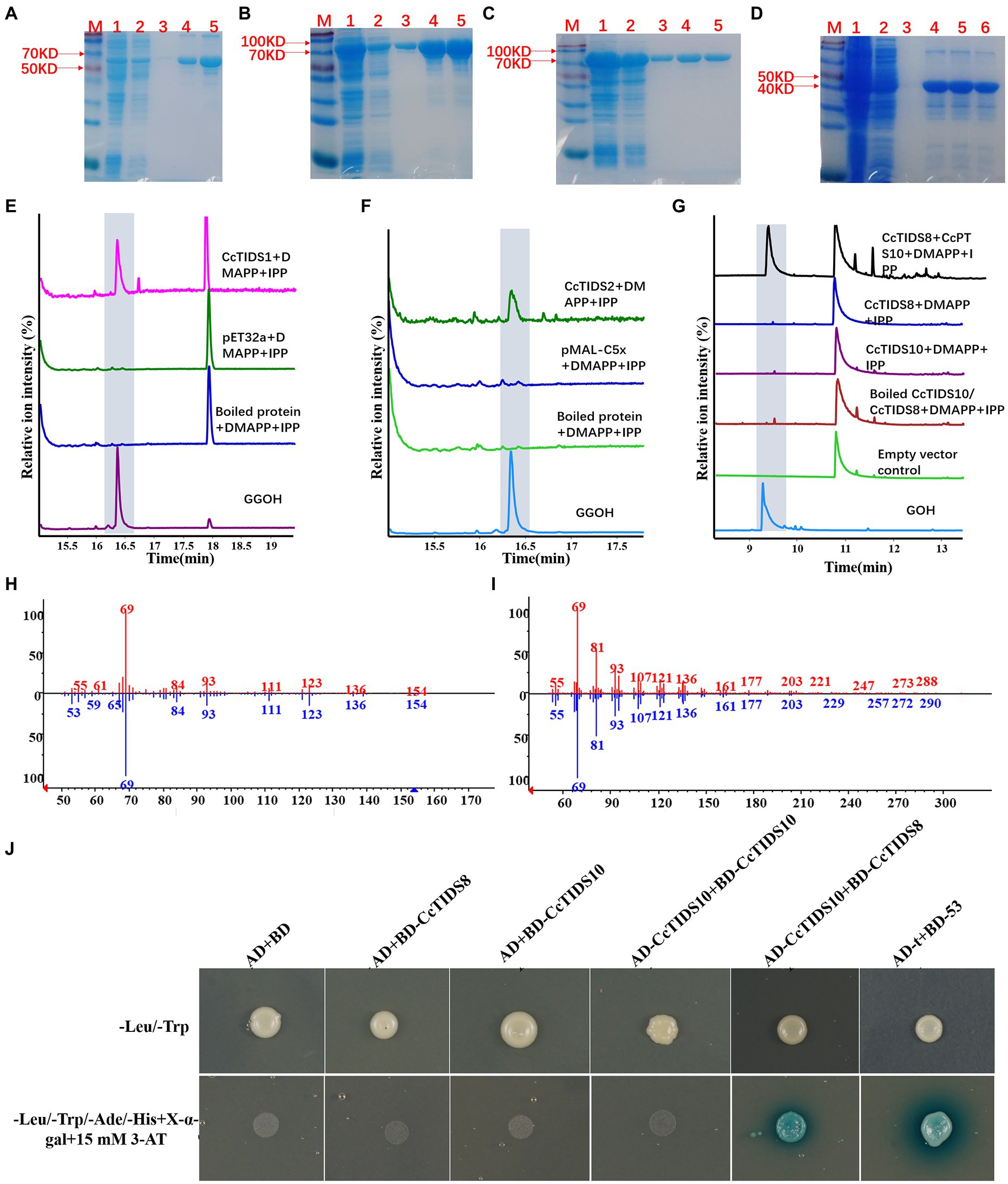
Figure 8. Gas chromatography–mass spectrometry profile showing in vitro reaction products of C. camphora trans-prenyltransferases CcTIDS1, CcTIDS2, CcTIDS8, and CcTIDS9, where different prenyldiphosphates were used as substrates. Expression and purification of (A) CcTIDS1, (B) CcTIDS2, (C) CcTIDS8, and (D) CcTIDS10 recombinant protein from E. coli Rosetta (DE3) harboring pET32a(+)–CcTIDS1/CcTIDS10 or pMAL-C5x–CcTIDS2/CcTIDS8, where lane 1 shows the total protein after induction, 2 shows the soluble protein, 3–6 show the purified CcTIDS1 or CcTIDS2, CcTIDS8, or CcTIDS10 recombinant protein from the first to the fourth collected tube. The GC–MS chromatogram of the reaction products generated by (E) CcTIDS8 and CcTIDS10, (F) CcTIDS1, and (G) CcPTS2 and the acid hydrolysis products of geraniol (GOH) and gerylgeraniol (GGOH) standards. (H) Mass spectra of GOH standard (red) and products in the NIST14/Wiley275 library (blue). (I) Mass spectra of the GGOH standard (red) and products in the NIST14/Wiley275 library (blue). (J) Confirmation of the interaction between CcTIDS8 and CcTIDS10 using a yeast two-hybrid system, where blue color indicates an interaction. Yeast cells harboring both constructs were spotted on synthetic dropout (SD) medium lacking Ade, His, Leu, and Trp (SD/−4) to test for protein interactions. AD: pGADT7; BD: pGBDT7; AD-T: pGADT7::T; BD-53: pGBKT7-53. Cells co-transformed with pGADT7::T and pGBKT7-53 were included as a positive control, and pGADT7 and pGBDT7 as a vector control.
Because CcTIDS10 was identified as the only CcGPPS small subunit, we determined whether this protein was able to interact with CcGGPPS1/2 or CcTIDS8 to form active heterodimers. Intriguingly, when CcTIDS10 was co-incubated with CcTIDS8, but not CcGGPPS1 or 2, GPP was detected as a sole product (Figures 8G-I).
A yeast two-hybrid system (Y2H) was used to confirm the interaction of CcTIDS10 with CcTIDS8. As expected, CcTIDS10 was able to interact with CcTIDS8 and itself (Figure 8J). These results, along with the co-localization of CcTIDS10 and CcTIDS8 in the plastid, confirmed that the inactive CcTIDS10 and CcTIDS8 interact to form heteromeric functional GPPS, generating GPP as the sole product. Thus, CcTIDS10 was designated as CcGPPS.small subunit (SSU), whereas CcTIDS8 was renamed as CcGPPS.large subunit (LSU).
In this study, we reported the characterization of the C. camphora TIDS gene family for the first time. Of the 10 identified TIDS genes, three (CcGGPPS1, CcGGPPS2, and CcGPPS.LSU) were clustered within the subfamily encoding TIDS-d enzymes, while one single gene, CcTIDS10, was closely related to the subfamily encoding TIDS-e enzymes. Apart from those in gymnosperms, TIDS proteins within the TIDS-d and TIDS-e subfamily have been classified as GGPPS paralogs in previous studies. As a result, many GGPPS homologs were predicted from the plant genomes (Coman et al., 2014).
The expansion of the GGPPS family in A. thaliana occurred at distinct evolutionary time points through different duplication mechanisms, including whole gene, tandem, and segmental genome duplications (Coman et al., 2014). The evolution of this A. thaliana GGPPS family likely involved neofunctionalization (with the duplicated gene developing a function that was not present in the ancestral gene), subfunctionalization (in which the duplicated and ancestral genes retain different parts of the original function of the ancestral gene), and pseudogenization (loss of function; Coman et al., 2014). Based on our fragment duplication and functional differentiation results for CcGGPPS1, CcGGPPS2, and CcGPPS.LSU, and the phylogenetic tree we constructed, these three genes may have been derived from the same ancestor, having undergone neo- or subfunctionalization.
Thus far, homodimeric GPPS has been functionally characterized in only a few plant species (Burke et al., 2004; Hsiao et al., 2008; Schmidt and Gershenzon, 2008; Gutensohn et al., 2013; Rai et al., 2013; Adal and Mahmoud, 2020). We found that the C. camphora genome contained two putative homodimeric GPPS genes (CcTIDS4 and CcTIDS5). Our evolutionary analysis suggested that these two genes were related to those encoding the functionally characterized homodimeric GPPS from A. thaliana, and subcellular localization experiments indicated that the two proteins encoded by these genes might localize to the mitochondria, consistent with earlier reports on C. roseus (Rai et al., 2013). However, there was no observable GPPS activity associated with the proteins in vitro.
Instead, CcGPPS.SSU (CcTIDS10) and CcGPPS.LSU (CcTIDS8), both identified in our study, were found to interact with one another to form a heterodimeric CcGPPS that produced GPP from IPP and DMAPP substrates in vitro. These proteins were found to be inactive alone. This is consistent with previous reports in both Mentha×piperita subunits (Burke et al., 2004), both Lavendula×intermedia subunits (Adal and Mahmoud, 2020), and A. thaliana GPPS.SSU2 (Wang and Dixon, 2009).
CcGPPS.SSU and CcGPPS.LSU were primarily expressed in roots compared with stems and leaves. Furthermore, both CcGPPS.SSU and CcGPPS.LSU were found to reside in plastids, which are the known site of monoterpene biosynthesis (Magnard et al., 2015; Yin et al., 2017). These results suggest that heteromeric CcGPPS likely participates in the biosynthesis of GPP in C. camphora.
Interestingly, all CcTIDS genes had orthologs of greater than 90% identity with the C. micranthum genome, with the sole exception of CcGPPS.SSU (Supplementary Figure S3). Cinnamomum micranthum, known as the stout camphor tree, is the sister species of C. camphora. The genome of C. micranthum with a contig N50 of 0.9Mb has been published. However, contig N50 of the genome of C. camphora is 23.89Mb (data not shown), indicating that the assembly quality of the C. camphora genome is significantly better than that of the C. micranthum genome. This lower quality may explain why the GPPS.SSU gene is missing from the C. micranthum genome.
In addition, although, CcTIDS4 and CcTIDS5 did not possess GPPS activity in our study, they may perform different functions. For example, At2g34630 from A. thaliana was initially classified as a homodimeric GPPS (Bouvier et al., 2000), but further study indicated that At2g34630 silencing did not affect the production of monoterpenes, and At2g34630 was subsequently identified as a PPPS (Hsieh et al., 2011). Therefore, the functions of CcTIDS4 and 5 should be investigated in future studies.
In conclusion, this is the first comprehensive and systematic genome-wide analysis of TIDS gene families in C. camphora. In total, 10 TIDS genes in the borneol chemotype of C. camphora were identified. These genes likely expanded through segmental duplication events, and expression was found to respond to multiple abiotic stressors via cis-acting elements in the promoter region. Eight putative SC-TIDS were identified, six of which were catalytically active, including a heteromeric GPPS (composed of CcGPPS.SSU and CcGPPS.LSU), two CcFPPS (CcTIDS3 and 9), and two CcGGPPS (CcTIDS1 and 2), which catalyzed the biosynthesis of GPP, FPP, and GGPP, respectively (Figure 9).
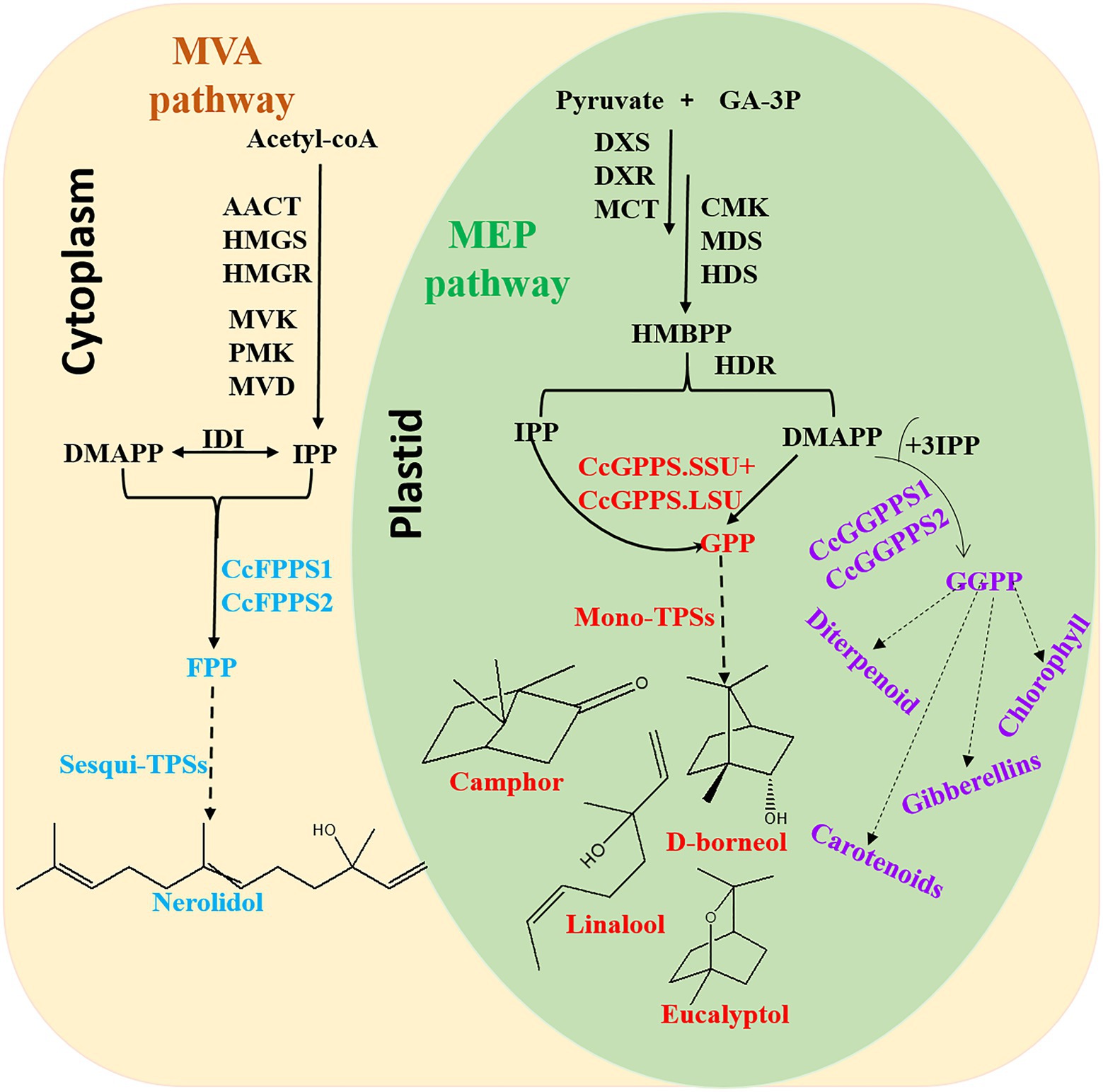
Figure 9. Overview of the terpenoid biosynthesis pathway in C. camphora. AACT, acetoacetyl-coenzyme A thiolase; HMGS, 3-hydroxy-3-methylglutaryl coenzyme A synthase; HMGR, 3-hydroxy-3-methylglutaryl coenzyme A reductase; MVK, mevalonate kinase; PMK, 5-phospho mevalonate kinase; MVD, mevalonate diphosphate decarboxylase; IDI, isopentenyl diphosphate isomerase; FPPS, farnesyl diphosphate synthase; DXS,1-deoxy-D-xylulose-5-phosphate synthase; DXR,1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol-4-(cytidyl-5-diphosphate) transferase; CMK, 4-(cytidine50-diphospho)-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS,1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase; and HDR, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase.
Finally, CcTIDS3, 8, 9, and 10 were more active in roots compared with stems and leaves, which were confirmed by both transcriptome analysis and qRT-PCR experiments. These novel insights provide the basis for further investigation of the TIDS family in C. camphora, and a good foundation from which to develop a metabolic engineering approach to increase production of pharmacologically valuable essential oils from the camphor tree.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
ZY conceived, designed, and performed the experiments, analyzed the data, prepared the figures and tables, and authored and reviewed drafts of the paper. CX and LL conceived, designed, and performed the experiments, analyzed the data, and authored and reviewed drafts of the paper. SL, YH, and WA conceived, designed, and performed the experiments, and authored and reviewed drafts of the paper. SH and XZ conceived and designed the experiments, analyzed the data, prepared the figures and tables, and authored and reviewed drafts of the paper. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (grant number: 81903741).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.708697/full#supplementary-material
1. ^https://www.arabidopsis.org/index.jsp
2. ^https://solgenomics.net/organism/Solanum_lycopersicum/genome/
3. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^https://web.expasy.org/protparam
5. ^https://meme-suite.org/meme/tools/meme
6. ^http://www.cbs.dtu.dk/services/ChloroP/
Abdullah, M., Cheng, X., Cao, Y., Su, X., Manzoor, M. A., Gao, J., et al. (2018). Zinc finger-homeodomain transcriptional factors (ZHDs) in upland cotton (Gossypium hirsutum): genome-wide identification and expression analysis in fiber development. Front. Genet. 9:357. doi: 10.3389/fgene.2018.00357
Adal, A. M., and Mahmoud, S. S. (2020). Short-chain isoprenyl diphosphate synthases of lavender (Lavandula). Plant Mol. Biol. 102, 517–535. doi: 10.1007/s11103-020-00962-8
Akhtar, T. A., Matsuba, Y., Schauvinhold, I., Yu, G., Lees, H. A., Klein, S. E., et al. (2013). The tomato cis-prenyltransferase gene family. Plant J. 73, 640–652. doi: 10.1111/tpj.12063
Amborella Genome Project (2013). The Amborella genome and the evolution of flowering plants. Science 342:1241089. doi: 10.1126/science.1241089
Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Athanasakoglou, A., and Grypioti, E. (2019). Isoprenoid biosynthesis in the diatom Haslea ostrearia. New Phytol. 222, 230–243. doi: 10.1111/nph.15586
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., dePamphilis, C., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963. doi: 10.1126/science.1203810
Barja, M. V., and Rodríguez-Concepción, M. (2020). A simple in vitro assay to measure the activity of geranylgeranyl diphosphate synthase and other short-chain prenyltransferases. Methods Mol. Biol. 2083, 27–38. doi: 10.1007/978-1-4939-9952-1_2
Beck, G., Coman, D., Herren, E., Ruiz-Sola, M. A., Rodríguez-Concepción, M., Gruissem, W., et al. (2013). Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 82, 393–416. doi: 10.1007/s11103-013-0070-z
Bouvier, F., Suire, C., d’Harlingue, A., Backhaus, R. A., and Camara, B. (2000). Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24, 241–252. doi: 10.1046/j.1365-313x.2000.00875.x
Burke, C., Klettke, K., and Croteau, R. (2004). Heteromeric geranyl diphosphate synthase from mint: construction of a functional fusion protein and inhibition by bisphosphonate substrate analogs. Arch. Biochem. Biophys. 422, 52–60. doi: 10.1016/j.abb.2003.12.003
Chai, Y., Yin, Z., Fan, Q., Zhang, Z., Ye, K., Xu, Y., et al. (2019). Protective effects of angong niuhuang pill on early atherosclerosis in apoE(−/−) mice by reducing the inflammatory response. Evid. Based Complement. Alternat. Med. 2019:9747212. doi: 10.1155/2019/9747212
Chaw, S. M., Liu, Y. C., Wu, Y. W., Wang, H. Y., Lin, C. I., Wu, C. S., et al. (2019). Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants 5, 63–73. doi: 10.1038/s41477-018-0337-0
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Fan, D., and Wang, G. (2015). Heteromeric geranyl(geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol. Plant 8, 1434–1437. doi: 10.1016/j.molp.2015.05.001
Chen, J., Tang, C., Zhang, R., Ye, S., Zhao, Z., Huang, Y., et al. (2020b). Metabolomics analysis to evaluate the antibacterial activity of the essential oil from the leaves of Cinnamomum camphora (Linn.) Presl. J. Ethnopharmacol. 253:112652. doi: 10.1016/j.jep.2020.112652
Chen, Z. X., Xu, Q. Q., Shan, C. S., Shi, Y. H., Wang, Y., Chang, R. C., et al. (2019). Borneol for regulating the permeability of the blood-brain barrier in experimental ischemic stroke: preclinical evidence and possible mechanism. Oxidative Med. Cell. Longev. 2019:2936737. doi: 10.1155/2019/2936737
Chen, C., Zheng, Y., Zhong, Y., Wu, Y., Li, Z., Xu, L. A., et al. (2018). Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genomics 19:550. doi: 10.1186/s12864-018-4941-1
Choi, K. R., Jang, W. D., Yang, D., Cho, J. S., Park, D., and Lee, S. Y. (2019). Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837. doi: 10.1016/j.tibtech.2019.01.003
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. doi: 10.1371/journal.pone.0011335
Coman, D., Altenhoff, A., Zoller, S., Gruissem, W., and Vranová, E. (2014). Distinct evolutionary strategies in the GGPPS family from plants. Front. Plant Sci. 5:230. doi: 10.3389/fpls.2014.00230
Cui, L., Yang, G., Yan, J., Pan, Y., and Nie, X. (2019). Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genomics 20:750. doi: 10.1186/s12864-019-6144-9
Dudareva, N., Murfitt, L. M., Mann, C. J., Gorenstein, N., Kolosova, N., Kish, C. M., et al. (2000). Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell 12, 949–961. doi: 10.1105/tpc.12.6.949
Dudareva, N., Pichersky, E., and Gershenzon, J. (2004). Biochemistry of plant volatiles. Plant Physiol. 135, 1893–1902. doi: 10.1104/pp.104.049981
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Guo, X., Cui, M., Deng, M., Liu, X., Huang, X., Zhang, X., et al. (2017). Molecular differentiation of five Cinnamomum camphora chemotypes using desorption atmospheric pressure chemical ionization mass spectrometry of raw leaves. Sci. Rep. 7:46579. doi: 10.1038/srep46579
Gutensohn, M., Orlova, I., Nguyen, T. T., Davidovich-Rikanati, R., Ferruzzi, M. G., Sitrit, Y., et al. (2013). Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 75, 351–363. doi: 10.1111/tpj.12212
Hivert, G., Davidovich-Rikanati, R., Bar, E., Sitrit, Y., Schaffer, A., Dudareva, N., et al. (2020). Prenyltransferases catalyzing geranyldiphosphate formation in tomato fruit. Plant Sci. 296:110504. doi: 10.1016/j.plantsci.2020.110504
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hsiao, Y. Y., Jeng, M. F., Tsai, W. C., Chuang, Y. C., Li, C. Y., Wu, T. S., et al. (2008). A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2-4D motif. Plant J. 55, 719–733. doi: 10.1111/j.1365-313X.2008.03547.x
Hsieh, F. L., Chang, T. H., Ko, T. P., and Wang, A. H. (2011). Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 155, 1079–1090. doi: 10.1104/pp.110.168799
Huang, J., Tang, X., Ye, F., He, J., and Kong, X. (2016). Clinical therapeutic effects of aspirin in combination with fufang danshen diwan, a traditional chinese medicine formula, on coronary heart disease: a systematic review and meta-analysis. Cell. Physiol. Biochem. 39, 1955–1963. doi: 10.1159/000447892
Jia, Q., and Chen, F. (2016). Catalytic functions of the isoprenyl diphosphate synthase superfamily in plants: a growing repertoire. Mol. Plant 9, 189–191. doi: 10.1016/j.molp.2015.12.020
Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M. C., Wang, B., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546, 524–527. doi: 10.1038/nature22971
Johnson, S. R., and Bhat, W. W. (2019). Promiscuous terpene synthases from Prunella vulgaris highlight the importance of substrate and compartment switching in terpene synthase evolution. New Phytol. 223, 323–335. doi: 10.1111/nph.15778
Johnson, S. R., Bhat, W. W., Bibik, J., Turmo, A., Hamberger, B., Evolutionary Mint Genomics, C., et al. (2019). A database-driven approach identifies additional diterpene synthase activities in the mint family (Lamiaceae). J. Biol. Chem. 294, 1349–1362. doi: 10.1074/jbc.RA118.006025
Kopcsayová, D., and Vranová, E. (2019). Functional gene network of prenyltransferases in Arabidopsis thaliana. Molecules 24:4556. doi: 10.3390/molecules24244556
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Gao, K., Yang, X., Khan, W. U., Guo, B., Guo, T., et al. (2020). Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus. Int. J. Biol. Macromol. 161, 999–1010. doi: 10.1016/j.ijbiomac.2020.06.056
Li, J., and Mutanda, I. (2019). Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 10:4850. doi: 10.1038/s41467-019-12879-y
Liang, P. H., Ko, T. P., and Wang, A. H. (2002). Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269, 3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x
Liang, C. J., Li, J. H., Zhang, Z., Zhang, J. Y., Liu, S. Q., and Yang, J. (2018). Suppression of MIF-induced neuronal apoptosis may underlie the therapeutic effects of effective components of Fufang Danshen in the treatment of Alzheimer's disease. Acta Pharmacol. Sin. 39, 1421–1438. doi: 10.1038/aps.2017.210
Ma, R., Su, P., Jin, B., Guo, J., Tian, M., Mao, L., et al. (2021). Molecular cloning and functional identification of a high-efficiency (+)-borneol dehydrogenase from Cinnamomum camphora (L.) Presl. Plant Physiol. Biochem. 158, 363–371. doi: 10.1016/j.plaphy.2020.11.023
Magnard, J. L., Roccia, A., Caissard, J. C., Vergne, P., Sun, P., Hecquet, R., et al. (2015). Plant volatiles. Biosynthesis of monoterpene scent compounds in roses. Science 349, 81–83. doi: 10.1126/science.aab0696
Miller, G. P., and Bhat, W. W. (2020). The biosynthesis of the anti-microbial diterpenoid leubethanol in Leucophyllum frutescens proceeds via an all-cis prenyl intermediate. Plant J. 104, 693–705. doi: 10.1111/tpj.14957
Nagel, R., Bernholz, C., Vranová, E., Košuth, J., Bergau, N., Ludwig, S., et al. (2015). Arabidopsis thaliana isoprenyl diphosphate synthases produce the C25 intermediate geranylfarnesyl diphosphate. Plant J. 84, 847–859. doi: 10.1111/tpj.13064
Nystedt, B., Street, N. R., Wetterbom, A., Zuccolo, A., Lin, Y. C., Scofield, D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. doi: 10.1038/nature12211
Pichersky, E., Noel, J. P., and Dudareva, N. (2006). Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science 311, 808–811. doi: 10.1126/science.1118510
Rai, A., Smita, S. S., Singh, A. K., Shanker, K., and Nagegowda, D. A. (2013). Heteromeric and homomeric geranyl diphosphate synthases from Catharanthus roseus and their role in monoterpene indole alkaloid biosynthesis. Mol. Plant 6, 1531–1549. doi: 10.1093/mp/sst058
Ren, L., Wang, J., Feng, L., Wang, S., and Li, J. (2018). Efficacy of suxiao jiuxin pill on coronary heart disease: a meta-analysis of randomized controlled trials. Evid. Based Complement. Alternat. Med. 2018:9745804. doi: 10.1155/2018/9745804
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Schmidt, A., and Gershenzon, J. (2008). Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69, 49–57. doi: 10.1016/j.phytochem.2007.06.022
Su, P., Gao, L., Tong, Y., Guan, H., Liu, S., Zhang, Y., et al. (2019). Analysis of the role of geranylgeranyl diphosphate synthase 8 from Tripterygium wilfordii in diterpenoids biosynthesis. Plant Sci. 285, 184–192. doi: 10.1016/j.plantsci.2019.05.013
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Tholl, D., and Lee, S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9:e0143. doi: 10.1199/tab.0143
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Ueoka, H., Sasaki, K., Miyawaki, T., Ichino, T., Tatsumi, K., Suzuki, S., et al. (2020). A cytosol-localized geranyl diphosphate synthase from Lithospermum erythrorhizon and its molecular evolution. Plant Physiol. 182, 1933–1945. doi: 10.1104/pp.19.00999
Vandermoten, S., Haubruge, E., and Cusson, M. (2009). New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cell. Mol. Life Sci. 66, 3685–3695. doi: 10.1007/s00018-009-0100-9
Wang, C., Chen, Q., Fan, D., Li, J., Wang, G., and Zhang, P. (2016). Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in brassicaceae plants. Mol. Plant 9, 195–204. doi: 10.1016/j.molp.2015.10.010
Wang, G., and Dixon, R. A. (2009). Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 106, 9914–9919. doi: 10.1073/pnas.0904069106
Wang, J., Lin, H. X., Su, P., Chen, T., Guo, J., Gao, W., et al. (2019). Molecular cloning and functional characterization of multiple geranylgeranyl pyrophosphate synthases (ApGGPPS) from Andrographis paniculata. Plant Cell Rep. 38, 117–128. doi: 10.1007/s00299-018-2353-y
Wang, H., Ma, D., Yang, J., Deng, K., Li, M., Ji, X., et al. (2018). An integrative volatile terpenoid profiling and transcriptomics analysis for gene mining and functional characterization of AvBPPS and AvPS involved in the monoterpenoid biosynthesis in amomum villosum. Front. Plant Sci. 9:846. doi: 10.3389/fpls.2018.00846
Wijfjes, R. Y., and Smit, S. (2019). Hecaton: reliably detecting copy number variation in plant genomes using short read sequencing data. BMC Genomics 20:818. doi: 10.1186/s12864-019-6153-8
Wong, W. C., Yap, C. K., Eisenhaber, B., and Eisenhaber, F. (2015). dissectHMMER: a HMMER-based score dissection framework that statistically evaluates fold-critical sequence segments for domain fold similarity. Biol. Direct 10:39. doi: 10.1186/s13062-015-0068-3
Yang, Z., An, W., Liu, S., Huang, Y., Xie, C., Huang, S., et al. (2020). Mining of candidate genes involved in the biosynthesis of dextrorotatory borneol in Cinnamomum burmannii by transcriptomic analysis on three chemotypes. PeerJ 8:e9311. doi: 10.7717/peerj.9311
Ye, J., Yang, H., Shi, H., Wei, Y., Tie, W., Ding, Z., et al. (2017). The MAPKKK gene family in cassava: genome-wide identification and expression analysis against drought stress. Sci. Rep. 7:14939. doi: 10.1038/s41598-017-13988-8
Yin, J. L., Wong, W. S., Jang, I. C., and Chua, N. H. (2017). Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 213, 1133–1144. doi: 10.1111/nph.14280
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. doi: 10.1126/science.1068037
Yu, H., Ren, X., Liu, Y., Xie, Y., Guo, Y., Cheng, Y., et al. (2019). Extraction of Cinnamomum camphora chvar. Borneol essential oil using neutral cellulase assisted-steam distillation: optimization of extraction, and analysis of chemical constituents. Ind. Crop. Prod. 141:111794. doi: 10.1016/j.indcrop.2019.111794
Zhou, F., and Pichersky, E. (2020). The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 226, 1341–1360. doi: 10.1111/nph.16431
Keywords: genome-wide identification, Cinnamomum camphora, trans-isopentenyl diphosphate synthases, functional characterization, gene family
Citation: Yang Z, Xie C, Zhan T, Li L, Liu S, Huang Y, An W, Zheng X and Huang S (2021) Genome-Wide Identification and Functional Characterization of the Trans-Isopentenyl Diphosphate Synthases Gene Family in Cinnamomum camphora. Front. Plant Sci. 12:708697. doi: 10.3389/fpls.2021.708697
Received: 12 May 2021; Accepted: 28 July 2021;
Published: 13 September 2021.
Edited by:
Jian Li, Beijing Technology and Business University, ChinaReviewed by:
Raimund Nagel, University of Leipzig, GermanyCopyright © 2021 Yang, Xie, Zhan, Li, Liu, Huang, An, Zheng and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiasheng Zheng, eHN6aGVuZ0BnenVjbS5lZHUuY24=; Song Huang, aHVhbmdubjQyMUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.