
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 October 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.706679
This article is part of the Research TopicAdvances and Challenges of RNAi Based Technologies for Plants, Volume 2View all 13 articles
Secondary metabolic pathways in grape berries are tightly regulated by an array of molecular mechanisms, including microRNA-mediated post-transcriptional regulation. As recently discovered, before being processed into mature microRNAs (miRNAs), the primary transcripts of miRNAs (pri-miRNAs) can encode for small miRNA-encoded peptides (micropeptides – miPEPs) that ultimately lead to an accentuated downregulation of the respective miRNA-targeted genes. Although few studies about miPEPs are available, the discovery of miPEPs reveals a new layer of gene regulation at the post-transcriptional level that opens the possibility to regulate plant metabolism without resorting to gene manipulation. Here, we identified a miPEP encoded in non-mature miR164c putatively targeting grapevine transcription factor VvMYBPA1 (miPEP164c/miPEP-MYBPA1), a positive regulator of key genes in the proanthocyanidin (PA)-biosynthetic pathway, a pathway that competes directly for substrate with the anthocyanin-biosynthetic pathway. Thus, the objective of this work was to test the hypothesis that the exogenous application of miPEP164c (miPEP-MYBPA1) can modulate the secondary metabolism of grape berry cells by inhibiting PA biosynthetic pathway while simultaneously stimulating anthocyanin synthesis. The exogenous application of miPEP164c to suspension-cultured cells from grape berry (cv. Gamay) enhanced the transcription of its corresponding non-mature miR164c, with a maximum effect at 1 μM and after a period of 10 days, thus leading to a more pronounced post-transcriptional silencing of its target VvMYBPA1. This led to a significant inhibition of the PA pathway, mostly via inhibition of leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) enzymatic activities and VvLAR1 downregulation. In parallel, the anthocyanin-biosynthetic route was stimulated. Anthocyanin content was 31% higher in miPEP164c-treated cells, in agreement with the observed upregulation of VvUFGT1 transcripts and UFGT enzyme activity levels.
Although grapevines are well adapted to semi-arid climate, the increasingly more frequent combined effect of drought, high air temperature and high evaporative demand has a negative impact in grapevine yield (Chaves et al., 2010) and, if severe, also in berry quality (Teixeira et al., 2013). Therefore, berry and wine quality depend strongly on the grapevine adaptability to drought, heat and light/UV intensity. This abiotic stressors particularly impact highly regulated molecular mechanisms underlying the synthesis of several quality-related compounds, such as anthocyanins, proanthocyanidins (PAs), flavanols, and flavonols (Downey et al., 2006; Teixeira et al., 2013).
Anthocyanin and PA (condensed tannins) biosynthetic pathways compete for two common precursors, leucoanthocyanidins and anthocyanidins (Li et al., 2016). Proanthocyanidins are composed of several monomers of (+)-catechin and (−)-epicatechin, both flavan-3-ols that originate in a branch deviation of the general flavonoid pathway. Catechin synthesis is catalyzed by leucoanthocyanidin reductase (LAR) that uses leucoanthocyanidins as substrate. However, leucoanthocyanidins can also be catalyzed by leucoanthocyanidin oxygenase (LDOX), continuing the flavonoid pathway and resulting in the formation of anthocyanidins, a substrate of both UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT), in the synthesis of anthocyanins, and anthocyanidin reductase (ANR), in the synthesis of epicatechin, another building block of PAs (Gagné et al., 2009).
In grapevine, many transcription factors belonging to the R2R3-MYB family are involved in the regulation of flavonoid synthesis by inducing or silencing key biosynthetic genes along the flavonoid pathway (Deluc, 2006; Matus et al., 2009). The transcription factors VvMYB5a and VvMYB5b are already described as positive regulators of the flavonoid pathway, inducing an upregulation of late-stage berry-associated genes such as VvCHI (chalcone isomerase), VvF3′5 (flavonoid 3′,5′-hydroxylase), VvDFR (dihydroflavonol 4-reductase), VvLDOX, VvANR, and VvLAR1 leading to the synthesis of flavonols, anthocyanidins, and PAs (Cavallini et al., 2014; Pérez-Díaz et al., 2016). VvMYBPA1, expressed during flowering and early berry development, is a positive regulator of PA synthesis, by upregulating VvLDOX, VvANR, and VvLAR1 genes (Bogs et al., 2007; Cavallini et al., 2015), thus limiting the progress of the anthocyanin-biosynthetic route.
Regulation of the flavonoid pathway can also be coordinated at the post-transcriptional level by several microRNAs (miRNAs; Xie et al., 2010) that negatively regulate the expression of their target genes, either by promoting degradation of such target messenger RNAs (mRNAs) or by leading to inhibition of targeted mRNA translation (Pantaleo et al., 2010). MicroRNAs are initially transcribed as much larger primary transcripts (pri-miRNAs) and only become mature miRNA after several maturation processes (Xie et al., 2010). Like any other protein-coding gene, miRNAs are transcribed by RNA polymerase II originating the primary transcript of miRNA (pri-miRNA) that consists of a few hundred bases, a 5′cap and 3′ poly-A tail and the characteristic stem-loop structure where the miRNA sequence is inserted, and which is recognized by members of the Dicer-like1 family enzymes. This enzyme cleaves the 5′cap and 3′ poly-A tail of the primary transcript, transforming it in a precursor miRNA (pre-miRNA). DCL1 also carries out the subsequent cleavage of pre-miRNA to release the miRNA:miRNA∗ duplex which is then translocated to the nucleus by HASTY transporter where the correct miRNA strand is incorporated in a ribonuclear particle to form the RISC complex, the machinery that mediates miRNA-mediated gene silencing (Budak and Akpinar, 2015).
In a groundbreaking finding, it was discovered that, before being processed into mature miRNAs, some pri-miRNAs contain small open reading frames (ORF) that could encode for small regulatory peptides called miRNA-encoded peptides (miPEPs; Lauressergues et al., 2015). The mechanism of action of miPEPs is by enhancing the transcription and accumulation of the corresponding pri-miRNA, in a sort of positive feedback loop, that subsequently results in accentuated downregulation of the respective miRNA-targeted genes (Couzigou et al., 2015). For instance, the overexpression of miPEP171b in Medicago truncatula led to the increased accumulation of endogenous miR171b (involved in the formation of lateral roots), which resulted in significant changes in root development (Couzigou et al., 2016). Moreover, in soybean (Glycine max), it was demonstrated for the first time that the exogenous application of well-chosen, synthetic miPEP172c had a positive impact in nodule formation, by inducing the overexpression of pri-miR172c, whose correspondent miR172c accumulation results in an increase in nodule formation and consequent improvement of nitrogen fixation (Couzigou et al., 2016).
More recently, a miPEP in Arabidopsis, miPEP858, was reported by screening the 1,000 bp region upstream of pre-miR858 for small ORFs (Sharma et al., 2020) miPEP858 was able to modulate the expression of targets gene involved in plant growth and development and also on the phenylpropanoid pathway, by inducing the expression of pri-miR858.
In a very recent finding, using a fluorescein-labeled peptide, it was demonstrated that exogenously applied miPEP-156a could effectively penetrate plant seedlings through the root system and disperse systemically to the leaves of young seedlings of Brassica rapa (Erokhina et al., 2021). The application of this miPEP exerted a moderate morphological effect consisting of a growth acceleration of the main root of the seedling, that was parallel to an increase in pri-miR156a expression. These morphological and molecular level effects were apparently related with the ability of the miPEP to rapidly transfer into the cell nuclei and bind to nuclear chromatin.
Screening for small ORFs, either in the precursor sequence of miRNA or in the region upstream of such precursor, is the mainstream method for finding putative miPEPs when a miRNA is available, or a targeted gene is in mind. Yet, alternative molecular approaches for screening for miPEPs have also been successful. These include homology based computational analysis using expressed sequence tags (ESTs) of a certain species genome by blasting it against miRNA sequences already described, to find homologous of miRNAs and then repeat the same methodologies of miRNA target prediction and screening for small ORFs in the pre-miRNA sequence (Ram et al., 2019).
Recently, a miPEP in grapevine was reported, miPEP171d1, originating from a non-mature miRNA conserved within different plant species and associated with root organ development. When exogenously applied, miPEP171d1 was able to promote adventitious root formation thus enabling to overcome challenges in clonal propagation (Chen et al., 2020).
Although few studies about miPEPs are available, the discovery of miPEPs reveals a new layer of gene regulation at the post-transcriptional level that opens the possibility to regulate plant metabolism without resorting to gene manipulation.
Taking these groundbreaking discoveries as basis, the objective of this work was to test the hypothesis that the exogenous application of a newly identified putative grapevine miPEP by our group (miPEP164c – miPEP-MYBPA1) can modulate the secondary metabolism of grape berry cells by inhibiting PA biosynthetic pathway while simultaneously stimulating anthocyanin synthesis. The micropeptide miPEP164c is putatively targeting MYBPA1, as predicted in silico, a gene encoding for a transcription factor that acts as a positive regulator of PA synthesis by activating the expression of VvLAR and VvANR, the genes encoding for the enzymes responsible for catechin and epicatechin synthesis (Bogs et al., 2007). For that, a wide array of molecular biology and classic biochemistry approaches were combined to better assess the impact of miPEP164c exogenous treatments on the transcription of key genes involved in secondary metabolic pathways, on the biochemical activity of the corresponding key enzymes, and on the final concentration of secondary metabolites such as anthocyanins and PAs.
A series of in silico analyses to identify potential miPEPs in grapevine by combining several bioinformatic tools and databases such as the bioinformatic tool psRNATarget Finder (Dai et al., 2018), a plant small regulatory RNA target predictor, with the aid of GenBank, was used to retrieve information on which, how and where (in the target RNA) miRNAs putatively regulated key genes directly or indirectly involved in the flavonoid biosynthetic pathway. We identified miR164c as a putative negative regulator of transcription factor VvMYBPA1, involved in regulation of the PA pathway and proceeded to screen it in miRbase (microRNA database; Kozomara and Griffiths-Jones, 2014) for their stem-loop sequence or pri-miRNA sequence, the non-mature sequence of the regulatory miRNA possibly harboring small ORFs corresponding to regulatory miPEPs. Finally, the obtained stem-loop sequence was then ran in a bioinformatic ORF finder tool that recognizes in the introduced sequence all possible ORFs that could translate into a small peptide, by defining several parameters based on the few miPEPs so far identified in the literature (Lauressergues et al., 2015). This several step analysis led to the identification of one ORF with 48 bp, encoding a putative miPEP of 16 amino acids, that was designated as miPEP164c.
Following in silico identification, miPEP164c and a scrambled miPEP were ordered from Smart Bioscience as 1 mg aliquot. The scrambled miPEP aminoacidic sequence is the same as miPEP164c aminoacidic composition but in a random/scrambled order: MGTISKETCSQTNQCT. Solubilization of the micropeptides was conducted as recommended by Smart-Bioscience Peptide Solubility Guidelines.1 Both miPEPs were solubilized in 200 μL of acetic acid (10%) and 800 μL of DMSO to a final concentration of 1 mg/mL. A solution of 200 μL of acetic acid (10%) and 800 μL of DMSO was used as control in the exogenous treatments.
Grape berry cell suspensions of the cultivar Gamay Freaux cv. were maintained in Gamborg B5 medium in 250 mL flasks at 25°C with constant agitation on a rotator shaker at 100 rpm and under 16 h/8 h photoperiod. The culture medium composition was as follows: 3 g/L Gamborg B5 salt mixture and vitamins; 30 g/L sucrose (3% m/v); 250 mg/L casein enzymatic hydrolysate; 0.1 mg/L α-naphthaleneacetic acid (NAA); 0.2 mg/L Kinetin, and a final pH of 5.7. The suspension-cultured cells were allowed to grow for 10-day, until the exponential phase, when they were subcultured by transferring 10 mL of cells to 30 mL of fresh medium.
Immediately after sub-cultivation, concentrations ranging from 0.1 to 2 μM of miPEP164c were added to the cell cultures, in a volume that represented no more than 0.15% (v/v) of the total volume of the cell suspension. All cell suspensions, including control cells (treated with the same volume of control solution) and cells treated with the scrambled miPEP (a control miPEP with the same aminoacidic composition as miPEP164c but with a scrambled amino acid sequence) were cultivated for 10-day with constant agitation and a 16 h/8 h photoperiod. A time-course analysis of 1 μM of miPEP164c with several time-points (1 h, 24 h, 3-, 5- and 10-day) was also conducted. Also, upon miPEP164c addition (t = 0 h), samples were taken for anthocyanin and PA base level quantifications. Cells were collected when necessary, filtered and immediately frozen with liquid nitrogen and stored at −80°C. A part of cells of each experimental condition was lyophilized.
Anthocyanins were extracted from 100 mg of grape berry cells from each experimental condition. After adding 1 mL of 90% methanol and 10% deionized H2O, the suspensions were vigorously shaken for 30 min, following centrifugation at 18000 × g for 20 min. The supernatants were collected and 200 μL of each supernatant was mixed with 1.8 mL of a solution of 25 mM KCl (pH = 1.0) and absorbance was measured at 520 and 700 nm. Total anthocyanin quantification was calculated in relation to cyanidin-3-glucoside (C-3-G) equivalents, as follows in the equation, and subsequently presented per dry weight (DW):
where MW is the molecular weight of C-3-G (449.2 g mol–1), DF is the dilution factor and ε is the molar extinction coefficient of C-3-G (26900 M–1 cm–1).
Proanthocyanidin content was determined using an adapted colorimetric vanillin-HCl assay described by Broadhurst and Jones (1978). To extract PAs, 1 mL of 100% methanol was added to 5 mg of lyophilized grape berry cells and vigorously shaken for 30 min followed by centrifugation at 18000 × g for 15 min. Supernatants were collected and diluted in a 1:1 ratio with methanol to final volume of 500 μL. The methanolic extracts were added to clean assay tubes wrapped with aluminum foil. Then, 3 mL of a solution of 4% (m/v) vanillic acid freshly prepared in methanol was added and mixed very gently. Finally, 1.5 mL of concentrated hydrochloric acid was added to each reaction tube and mixed very gently. The reactions were allowed to stand for 6 min and the absorbance of the samples was measured spectrophotometrically at 500 nm. To discard absorbance interference caused by anthocyanin presence in the samples, control reactions for each condition were prepared with 3 mL of methanol instead of vanillic acid and the absorbance measured at 500 nm was discounted from the absorbance of reaction mixtures with vanillic acid. An epigallocatechin gallate (EGCG) standard curve, with concentrations ranging from 10 to 200 μg employing the same method was always prepared for each quantification of PA content.
Protein extraction was conducted as described in Conde et al. (2016). Lyophilized grape berry cells were mixed with extraction buffer in approximately 1:1 (v/v) powder: buffer ratio. Protein extraction buffer contained 50 mM Tris–HCl, pH 8.9, 5 mM MgCl2, 1 mM EDTA, 1 mM PMSF, 5 mM DTT, and 0.1% (v/v) Triton X-100. Homogenates were thoroughly vortexed and centrifuged at 18000 × g for 15 min at 4°C. Supernatants were maintained on ice and used for all enzymatic assays. Total protein concentrations of the extracts were determined by the method of Bradford (1976) using bovine serum albumin as a standard.
The biochemical activity of UFGT was determined as described by Lister et al. (1996) adapted by Conde et al. (2016). The assay mixture contained 385 μL of 100 mM Tris–HCl reaction buffer (pH 8), 100 μL of enzyme extract, 10 μL of 50 mM UDP-glucose and the reaction was initiated with 5 μL of 100 mM quercetin as substrate for the enzyme activity (saturating concentration) to a final reaction volume of 500 μL. Each mixture was incubated for 30 min in the dark with gentle shaking. After incubation, dilutions were prepared with 100 μL of each assay mixture and 900 μL of Tris–HCl reaction buffer and absorbance was read at 350 nm immediately after (t = 0) and 30 min later (t = 30) to follow the production of quercetin 3-glucoside (ε = 21877 M–1 cm–1).
Leucoanthocyanidin reductase enzymatic activity was measured by spectrophotometrically monitoring the conversion of dihydroquercetin to (+)-catechin following the method of Gagné et al. (2009) with some adaptions. The assay mixture contained 1.7 mL of Tris–HCl buffer (0.1 M, pH 7.5), 300 μL of protein extract, 2 μL of NADPH (100 mM) and the reaction was initiated by adding 1 μL of dihydroquercetin (10 mg mL–1 in DMSO). The production of (+)-catechin (ε = 10233 M–1 cm–1) was followed at 280 nm for 30 min.
The biochemical activity of ANR was determined as described by Zhang et al. (2012) with some adaptations. The assay mixture contained 1.5 mL of PBS buffer (0.1 M, pH 6.5), 60 μL of enzyme extract, 40 μL of ascorbic acid (20 mM), 50 μL of cyanidin chloride (2 mM) and the reaction was initiated by adding 75 μL of NADPH (20 mM) followed by a 1/10 dilution with PBS reaction buffer for proper absorbance measure. The enzyme activity was monitored by measuring the rate of NADPH (ε = 6.22 mM–1 cm–1) oxidation at 340 nm for 20 min, at 45°C.
Total RNA extraction was performed according to Reid et al. (2006) in combination with purification steps from the GRS Total Plant RNA extraction kit. After treatment with DNase I (Qiagen), cDNA was synthesized from 1 μg of total RNA using Maxima first strand cDNA synthesis kit from Thermo Fisher Scientific, following the manufacturer’s instructions. RNA concentration and purity were determined using Nanodrop and its integrity assessed in a 1% agarose gel stained with SYBR Safe (InvitrogenTM, Life Technologies).
Quantitative real-time PCR was performed with QuantiTect SYBR Green PCR Kit (Qiagen) in a CFX96 Real-Time Detection System (Bio-Rad), using 1 μL of cDNA in a final reaction volume of 10 μL per well. Specific primer pairs used for each target gene are listed in Supplementary Table 1. Melting curve analysis was performed for specific gene amplification confirmation. As reference genes, VvACT1 (actin) and VvGAPDH (glyceraldehyde-3-phosphate dehydrogenase) were selected, as these genes were proven to be very stable and ideal for qPCR normalization purposes in grapevine (Reid et al., 2006). For all experimental conditions tested, three biological replicates were used with internal triplicates. The expression values were normalized by the average of the expression of the reference genes as described by Pfaffl (2001) and analyzed using the software Bio-Rad CFX Manager (Bio-Rad).
The results were statistically analyzed by Student’s t-test using Prism vs. 6 (GraphPad Software, Inc.). For each condition, statistical differences between the mean values of miPEP-treated cells and control cells are marked with asterisks. For every experimental approach, three biological replicates (constituted by three independent treatments and respective experiments) were used.
An in silico analysis for micropeptide screening led to the selection of miPEP164c, a candidate miPEP with putative regulatory function in grape berry flavonoid biosynthesis metabolic pathway, particularly in the branch of PA synthesis. miR164c was predicted in silico to post-transcriptionally inhibit grapevine transcription factor VvMYBPA1, involved in the activation of flavonoid synthesis, specifically of PA synthesis (via LAR1, LAR2, and ANR activation). Relevant information obtained by the in silico analysis regarding the miPEP selected for this study, including its aminoacidic sequence, attributed name and respective mature miRNA name and miRbase accession number, as well as that of its precursor miRNA (pre-miRNA), is detailed in Table 1.

Table 1. Detailed information about the micropeptide identified by an in silico analysis and selected for this study and its corresponding mature miRNA and mode of action.
To confirm if miPEP164c exogenous application is indeed activating the accumulation of its in silico predicted miRNA (miR164c), gene expression analysis by real-time qPCR of the non-mature pre-miR164c was performed on cells treated with different concentrations of miPEP164c, ranging from 0.1 to 2 μM. As shown in Figure 1A, 10 days after treatment, the transcript levels of pre-miR164c were increasingly upregulated along miPEP concentration reaching a maximum effect at 1 μM (3.5-fold increase) that was approximately maintained at 2 μM of miPEP164c (3.2-fold increase). The exogenous application of a scrambled miPEP at 1 μM did not result in any changes in transcript levels of pre-miR164c after 10 days of treatment.
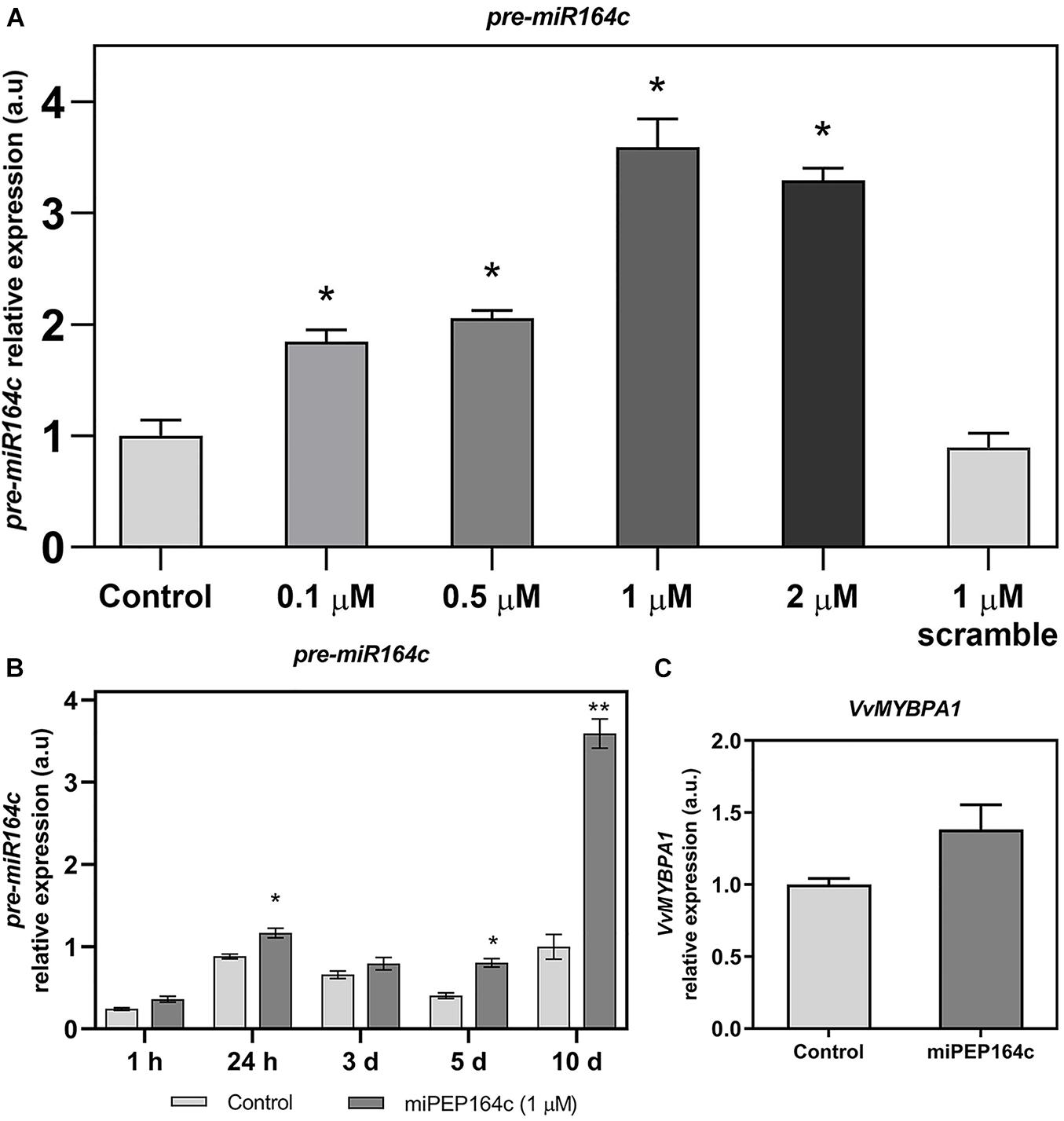
Figure 1. Steady-state transcript levels of pre-miR164c in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with various concentrations of miPEP164c (A) in suspension-cultured grape berry cells (cv. Gamay) eliciated with 1 μM of miPEP164c throughout a period of 10-day (B) and the steady-state transcript levels of VvMYBPA1 (C) in suspension-cultured grape berry cells (cv. Gamay) 10 days after elicitation with 1 μM miPEP164c. Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance in relation to the control (Student’s t-test; *P < 0.05; **P < 0.001).
A time-course analysis of the transcript levels of pre-miR164c in cells treated with 1 μM of miPEP164c through a period of 10 days (1 h, 24 h, 3-, 5- and 10-day) confirmed, as shown in Figure 2A, that the expression of pre-miR164c was slightly stimulated at 24 h and 5-day of miPEP164c treatment, but had the highest increase (3.5 fold) after 10 days of treatment (Figure 1B).
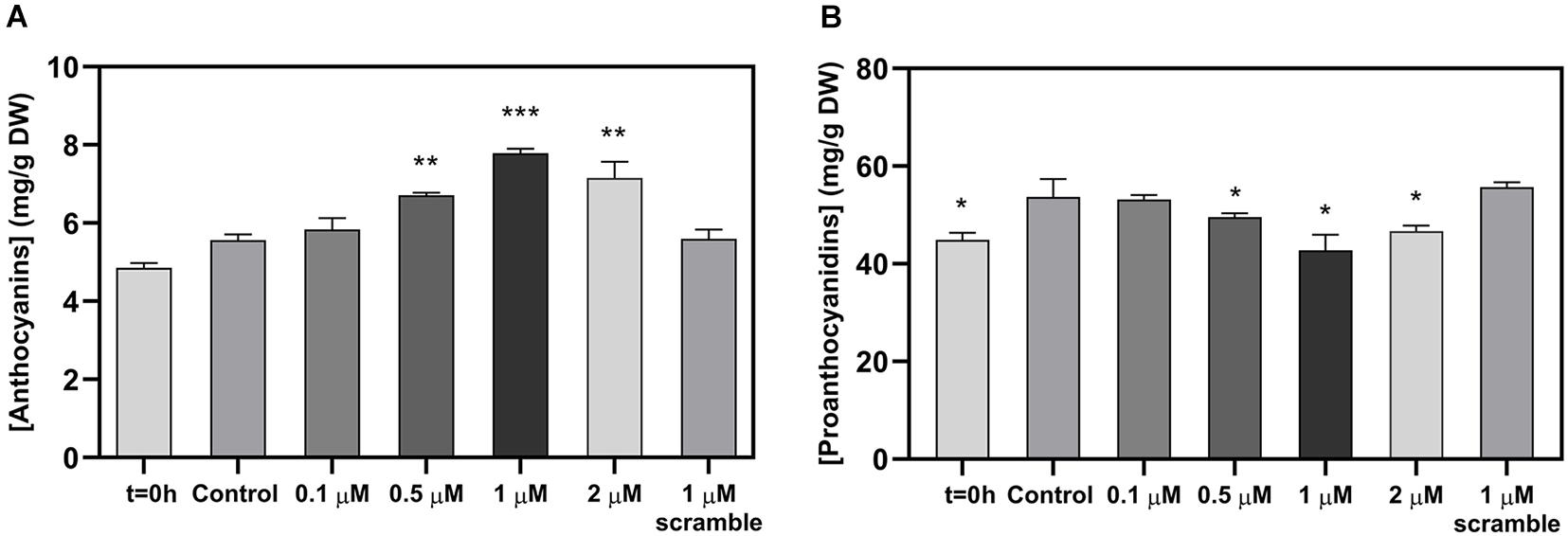
Figure 2. Effect of the exogenous application of different concentrations of miPEP164c on total anthocyanin content (A) and on total proanthocyanidin (PA) content (B) in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with miPEP164c. Anthocyanin concentration is represented as mg of cyanidin-3-glucoside (C-3-G) equivalents per g of fresh weight (FW). Asterisks indicate statistical significance in relation to the control (Student’s t-test; *P < 0.05; **P < 0.01; ***P < 0.001).
No statistically significant change in the transcript levels of VvMYBPA1 was observed after 10 days of miPEP164c treatment (Figure 1C).
Spectrophotometric quantifications of grape berry cells secondary metabolites revealed a significant increase in anthocyanin content at 0.5, 1, and 2 μM of miPEP164c after 10 days of treatment with the highest concentration of anthocyanins quantified in cells treated with 1 μM miPEP164c, with 7.4 mg of total anthocyanins per g of dry weight compared to only 5.6 mg per g of dry weight in control cells, which represented a 31% increase (Figure 2A).
In parallel, PAs concentration decreased significantly at those same concentrations of 0.5, 1, and 2 μM of miPEP164c with a more representative decrease (26%) from 54 mg of PAs per g of dry weight in control cells to 42.7 mg of PAs per g of dry weight after 10 days of treatment with 1 μM miPEP164c (Figure 2B). The exogenous application of 1 μM of the scrambled miPEP did not change the concentrations of both anthocyanins and PAs.
A time-course analysis by real-time qPCR on VvLAR1 expression in cells treated with 1 μM of miPEP164c showed a significant decrease of 20% after 10-day, while no statistically significant changes were observed on earlier time-points (Figure 3A). In agreement with the observed decrease in the VvLAR1 transcripts, LAR enzyme activity was 3-fold reduced in Gamay cells elicited with 1 μM miPEP164c, decreasing from a Vmax of 0.63 nmol min–1 mg protein–1 in control cells to a Vmax of 0.20 nmol min–1 mg protein–1 in miPEP-treated cells (Figure 3B). Regarding VvANR transcript levels, although there was a slight decrease at 24 h of miPEP exposure (20%) when compared to control cells, no changes were observed at other time-points (Figure 4A). However, a significant decrease of 27% was observed in the enzyme activity of ANR in cells treated with 1 μM miPEP164c for 10-day, from a Vmax of 14 nmol min–1 mg protein–1 in control cells to 10.31 nmol min–1 mg protein–1 in miPEP-treated cells (Figure 4B).
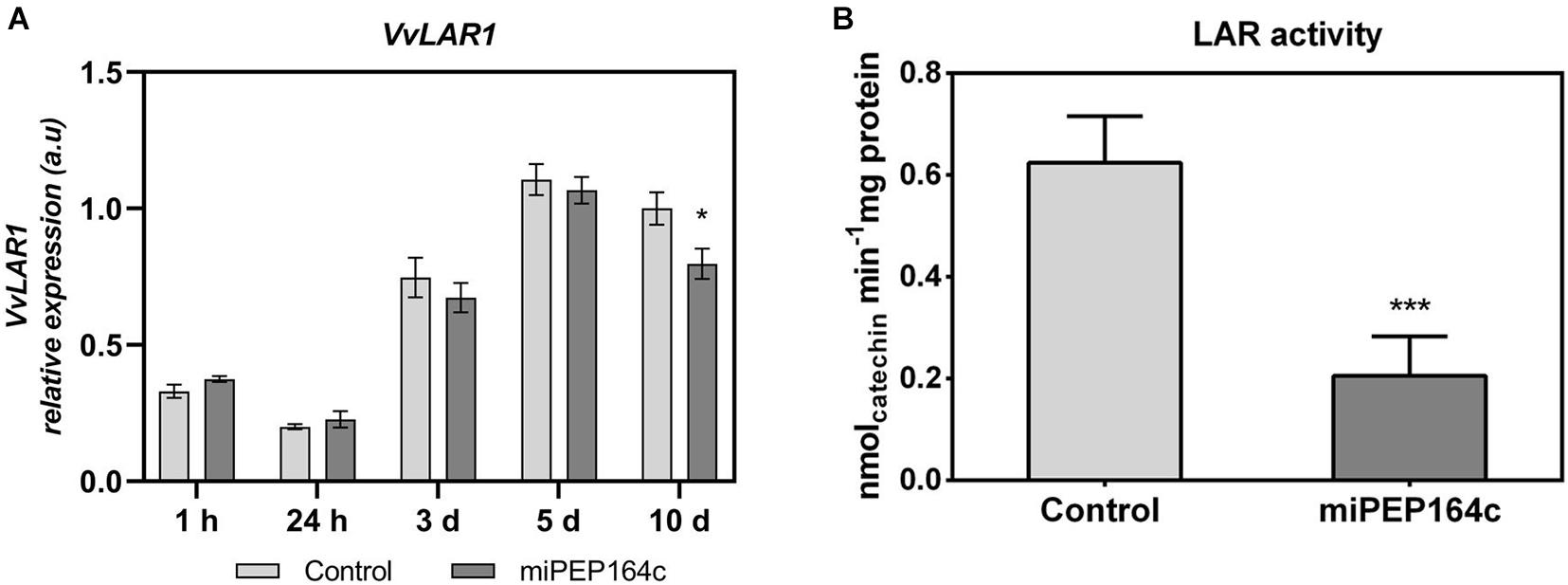
Figure 3. Steady-state transcript levels of VvLAR1 in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with various concentrations of miPEP164c (A) and the effect on the specific activity of leucoanthocyanidin reductase (LAR) (B) in suspension-cultured grape berry cells (cv. Gamay) after 10 days of elicitation with 1 μM miPEP164c. Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance in relation to the control (Student’s t-test; *P < 0.05). LAR biochemical activity represented as the Vmax in grape berry cells under miPEP164c treatment. Values are the mean ± SEM. Asterisks indicates statistical significance (Student’s t-test; ***P < 0.001).
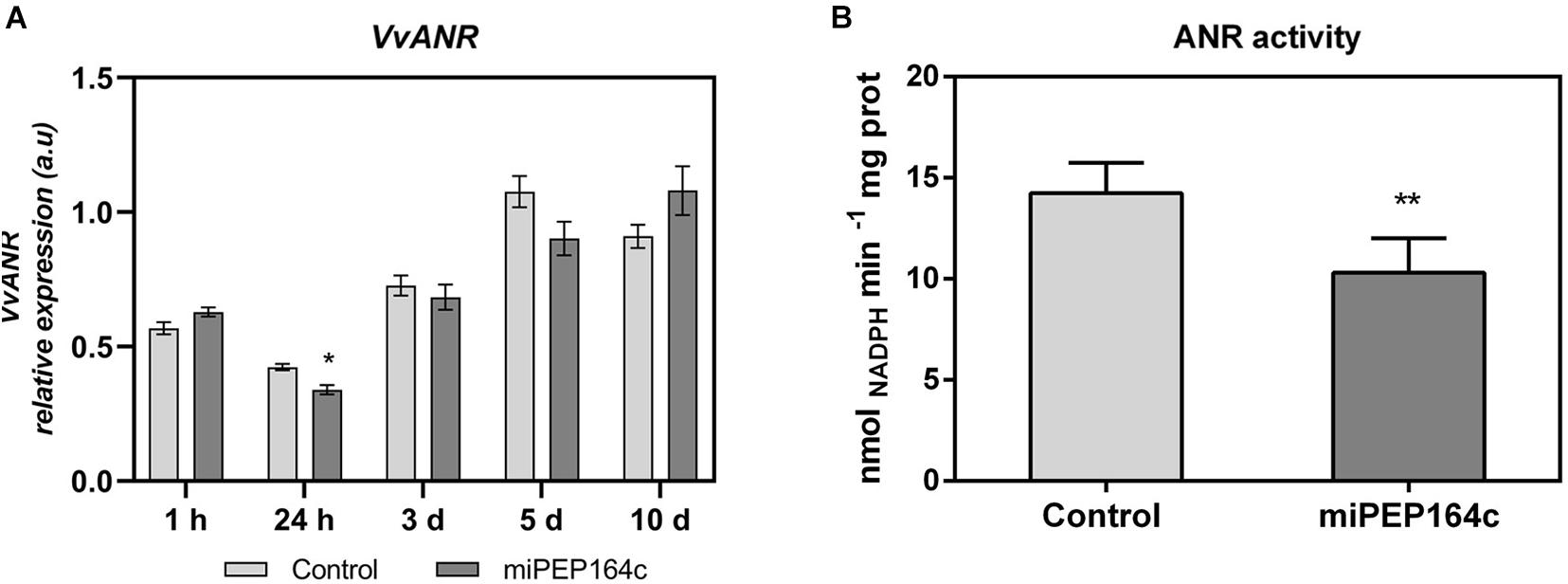
Figure 4. Steady-state transcript levels of VvANR in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with various concentrations of miPEP164c (A) and the effect on the specific activity of anthocyanidin reductase (ANR) (B) in suspension-cultured grape berry cells (cv. Gamay) after 10 days of elicitation with 1 μM miPEP164c. Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance in relation to the control (Student’s t-test; *P < 0.05). ANR biochemical activity represented as the Vmax in grape berry cells under miPEP164c treatment. Values are the mean ± SEM. Asterisks indicates statistical significance (Student’s t-test; **P < 0.01).
The expression of VvUFGT1 was strongly stimulated by miPEP164c application, reflected by a 4-fold increase in the expression levels in grape berry cells under this treatment (Figure 5A). The biochemical activity of UFGT was 3.2-fold higher in miPEP164c treated cells, in agreement with the transcriptional analysis, reaching a Vmax of 3.4 μmol h–1 mg protein–1 (Figure 5B), which corroborates with a significant increase in the total concentration of anthocyanins observed previously (Figure 2A).
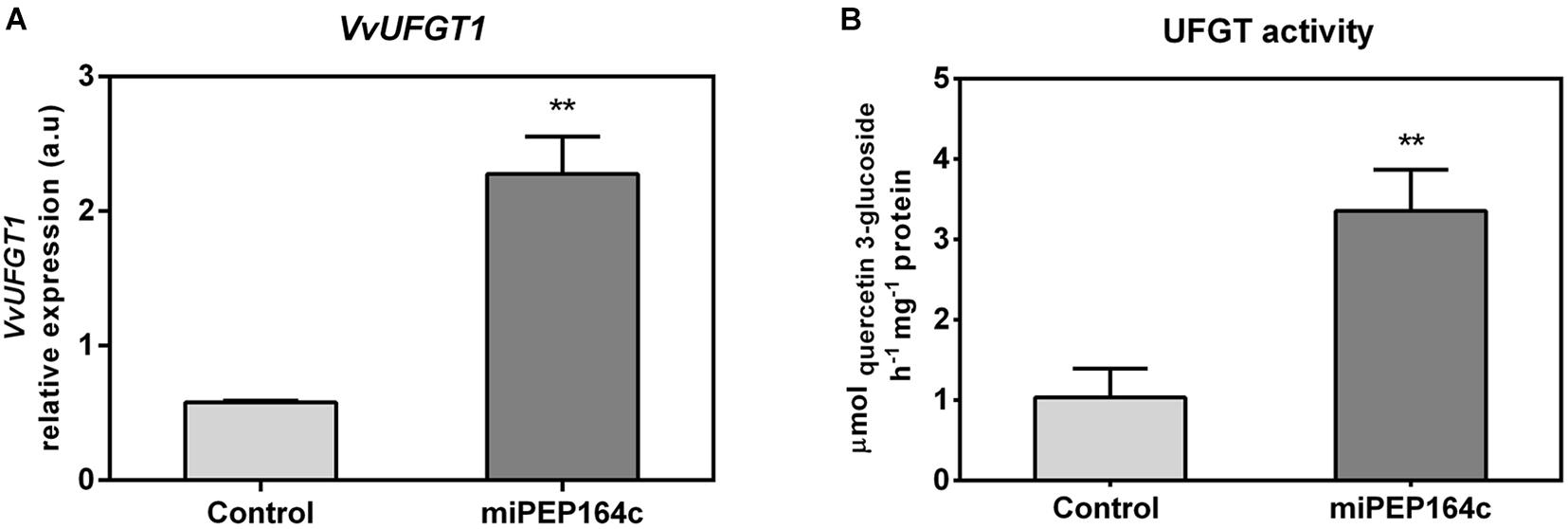
Figure 5. Steady-state transcript levels of VvUFGT1 (A) in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with 1 μM miPEP164c and the effect on the specific activity of UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT). (B) Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance (Student’s t-test; **P < 0.01). UFGT biochemical activity, represented as the Vmax in grape berry cells under miPEP164c treatment. Values are the mean ± SEM. Asterisks indicates statistical significance (Student’s t-test; **P < 0.01).
The expression levels of VvDFR were also significantly stimulated, with a 2-fold increase in grape berry cells 10 days after miPEP164c treatment (Figure 6A). VvLDOX expression was also significantly stimulated by 42% when compared to control cells (Figure 6B).
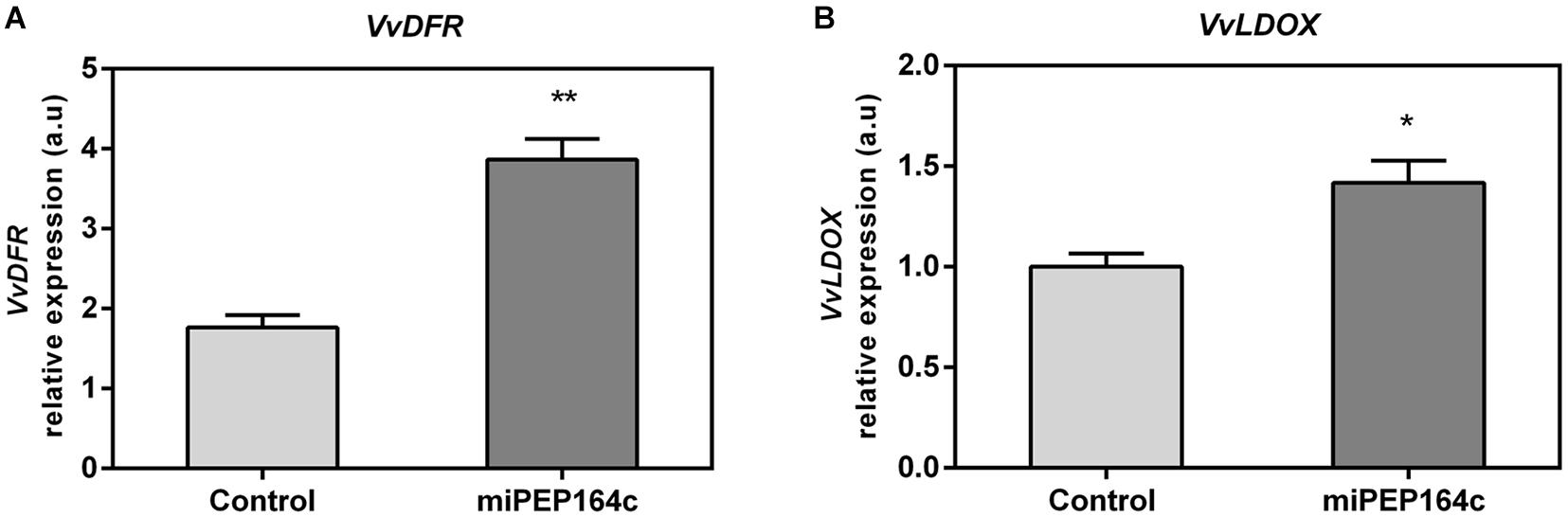
Figure 6. Steady-state transcript levels of VvDFR (A) and VvLDOX (B) in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with 1 μM miPEP164c. Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance (Student’s t-test; *P < 0.05; **P < 0.01).
Transcriptional analysis showed that the expression of VvGST4 under miPEP164c treatment also increased 2-fold (Figure 7A). Similarly, the transcript levels of VvMATE1 increased by 55% (Figure 7B), while the expression of VvABCC1 seemed not to be affected by treatment with this micropeptide (Figure 7C). These genes encode transporters that accumulate anthocyanins in the vacuole.
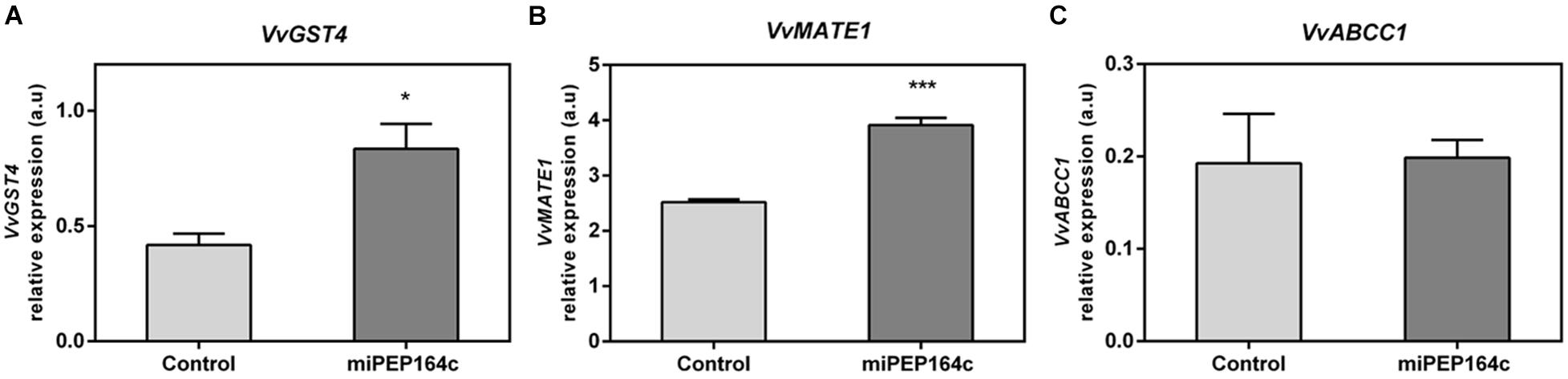
Figure 7. Steady-state transcript levels of VvGST4 (A), VvMATE1 (B) and VvABCC1 (C) in suspension-cultured grape berry cells (cv. Gamay) 10-day after elicitation with 1 μM miPEP164c. Gene expression analysis, by real-time qPCR was normalized with the expression of reference gene VvACT1 and VvGAPDH. Values are the mean ± SEM. Asterisks indicate statistical significance (Student’s t-test; *P < 0.05; ***P < 0.001).
Grape berry secondary metabolism generates, by a cascade of reactions scattered through different branches of the phenylpropanoid pathway, a wide range of bioactive compounds with key roles in plant defense responses and with several health-related benefits to humans, making them metabolites of interest for many industries (Teixeira et al., 2013). Therefore, the search for new strategies to modulate these complex pathways in the hopes of either minimizing the effects of several stress factors in the composition and quality of grape secondary metabolites or to increase the synthesis and accumulation of bioactive metabolites of interest such as antioxidant compounds like anthocyanins, is a research line of great importance, not only to the viticulture industry but also in several industries of health-promoting products (Zhang et al., 2015). In the present study, we sought to validate a new and promising strategy to modulate the secondary metabolism of Gamay grape berry cells by testing a synthetic miPEP, putatively enhancing the transcription and accumulation of miR164c and ultimately promoting a more pronounced silencing of its predicted target. Because this transcription factor is involved in the molecular activation of key genes in the PA pathway, ultimately, we wanted to evaluate if a miPEP-based treatment could regulate grape berry secondary metabolism, by activating miRNA-mediated post-transcription silencing mechanisms of specific targets. Results obtained were very promising as this treatment could represent an innovative and easy-to-apply strategy to modulate the synthesis of more quality-related compounds, resulting in crops with added-value characteristics, without the need for more drastic, time consuming and more expensive strategies, as genetic transformation of crops.
Overall, results confirmed that the exogenous application of miPEP164c is indeed enhancing the accumulation of miR164c which putatively resulted in a more pronounced post-transcriptional silencing of transcription factor VvMYBPA1 and consequently of MYBPA1-activated genes, here observed by a downregulation of VvLAR1 expression and of LAR and ANR specific activity resulting in a significant decrease of 26% of total PA content in cells exposed to exogenously applied miPEP164c for 10-day. According to our results, the regulatory effect of miPEP164c, both at anthocyanin/PA concentrations and transcriptional levels, was most significant at a concentration of 1 μM and after a period of 10-day. This suggests that the regulatory effect of miPEP164c is cumulative along a period of exposure to the peptide, at least until 10-day, but not evident in a period of hours after the peptide treatment. It is possible that this cumulative effect after a longer period of exposure is due to the several molecular steps in the mechanistic underlying miPEP regulation intracellularly and the fact that miPEP164c does not directly regulate ultimate targets as LAR and ANR within PA-synthetic pathway, which probably results in more time needed to observe its regulatory effect.
Indeed, gene expression analysis by real-time qPCR confirmed that miPEP164c increased the expression levels of the pre-miR164c in Gamay cells along its concentration until 1 μM in an overwhelmingly more evident mode after 10-day. Thus, a positive loop was established, in which a consequent increased translation into miPEP164c, ultimately results in higher levels of mature miR164c and accentuated negative regulation of the target gene VvMYBPA1. In silico analyses suggests that the mode of action of miR164c is through inhibition of translation of VvMYBPA1, not by cleavage of the target messenger RNA, due to a lack of 100% complementarity between the guide miRNA and the target mRNA (Waterhouse and Hellens, 2015). This is in agreement with our results showing that the treatment with miPEP164c did not induce any significant changes in the expression levels of VvMYBPA1.
Evidence for the involvement of post-transcriptional silencing of VvMYBPA1 mediated by miPEP164c was obtained when the MYBPA1-activated enzymes VvLAR and VvANR were clearly down-regulated. Both VvANR and VvLAR1, are key genes leading to the synthesis of PAs (Gagné et al., 2009). However, the expression of VvANR, encoding for the enzyme that synthesizes epicatechins from anthocyanidins, was not affected after 10-day of treatment, possibly to compensate the decreased activity of VvLAR, in order to ensure a certain amount of monomers for PAs biosynthesis. Also, VvANR expression may be regulated by several other regulatory proteins, such as VvMYC1, a bHLH transcription factor that physically interacts with MYB-like transcription factors like MYBPA1 and MYB5a/b to coordinate the regulation of VvANR, and therefore silencing of one transcription regulator may be overcome by another regulatory mechanism (Heppel, 2010).
The observed significant increase in anthocyanin total content in Gamay cells mediated by the application of miPEP164c corroborates our hypothesis that a miPEP164c-mediated silencing of PA synthesis would divert the carbon flow to the anthocyanin branch, due to the constant competition of both pathways for the same substrates, as reported before (Liao et al., 2015). Gene expression analysis of VvUFGT1, that glycosylates anthocyanidins into anthocyanins, revealed a strong upregulation of its expression levels in response to the elicitation with the micropeptide which goes in agreement with the observed increase of the UFGT specific activity that also increased.
In Vitis vinifera two types of anthocyanin tonoplast transporters that accumulate anthocyanins in the vacuole were identified: primary transporters from the ATP-binding cassette (ABC) family, such as the VvABCC1 who requires the presence of reduced glutathione (GSH) to properly transport anthocyanins, through the tonoplast, into the vacuole (Jiang et al., 2019); and tonoplast secondary transporters like VvMATE1 (anthoMATE) of the multidrug and toxic extrusion family that use the H+ gradient to transport mostly acylated anthocyanins (Gomez et al., 2009). Also crucial for anthocyanin stabilization and transport are the glutathione S-transferases, as the paradigmatic case of grapevine’s VvGST4, to promote anthocyanin S-conjugation with reduced glutathione for anthocyanin-stabilization purposes (Conn et al., 2008). Several studies on the role of GSTs in anthocyanin accumulation have described GSTs as escort/carrier proteins, binding anthocyanins to form a GST-anthocyanin complex, protecting them from oxidation and guiding anthocyanins from the cytosolic surface of the ER to the vacuole for proper storage mediated by tonoplast transporters such as VvMATE1 and VvABCC1 (Zhao and Dixon, 2010; Jiang et al., 2019). Our results strongly supported that anthocyanin transport capacity to the vacuole, where they are stored in grape berry cells, was also stimulated by miPEP164c application as the expression of the anthocyanin tonoplast transporter VvMATE1 and anthocyanin carrier protein VvGST4, was upregulated by this micropeptide. It is not understood how plants choose between ATP-hydrolysis-dependent or H+/Na+-gradient dependent mechanisms for transport of native metabolites or xenobiotics. However, it is believed that the conjugation ligands, such as glucose or glutathione, play a key role in the determination of which transport mechanism will be used (Zhao and Dixon, 2010). However, the expression of VvABCC1 was not affected by miPEP164c contrarily to what would be expected considering the upregulation of VvGST4 expression. This could be due to the presence of other regulatory proteins affecting the expression of VvABCC1, other phenolic substrates that also need to be transported by this mechanism, the majority of anthocyanins might not be in the glycosylated form, which is the preferred form of anthocyanins of this type of transporter, or simply because it is competing with the upregulated VvMATE1 transporter for anthocyanins (Francisco et al., 2013).
In this study, recurring to a combination of molecular and biochemical approaches, we revealed that miPEP164c exogenous application induced a strong up-regulation of genes involved in anthocyanin synthesis, transport, and accumulation in the vacuole. Additionally, miPEP164c provoked a downregulation of PA synthesis (a pathway that directly competes with the anthocyanin-biosynthetic pathway), due to a decrease in VvLAR1 expression levels with a corresponding very significant decrease in LAR total biochemical activity, accompanied also by a downregulated ANR biochemical activity.
This upregulation of the anthocyanin biosynthetic route seems to be an indirect effect of miPEP164c putatively inhibiting transcription factor MYBPA1, a known positive regulator of PA synthesis. Thus, these metabolic alterations triggered by miPEP164c clearly resulted in higher concentration of anthocyanins and lower concentration of PAs, due to miR164c-mediated negative regulation of PA-related transcription factor VvMYBPA1 and, consequently, VvLAR1 and VvANR, ultimately leading to PA synthesis inhibition and anthocyanin synthesis stimulation as these pathways directly compete for substrate, in a mechanism illustrated in Figure 8.
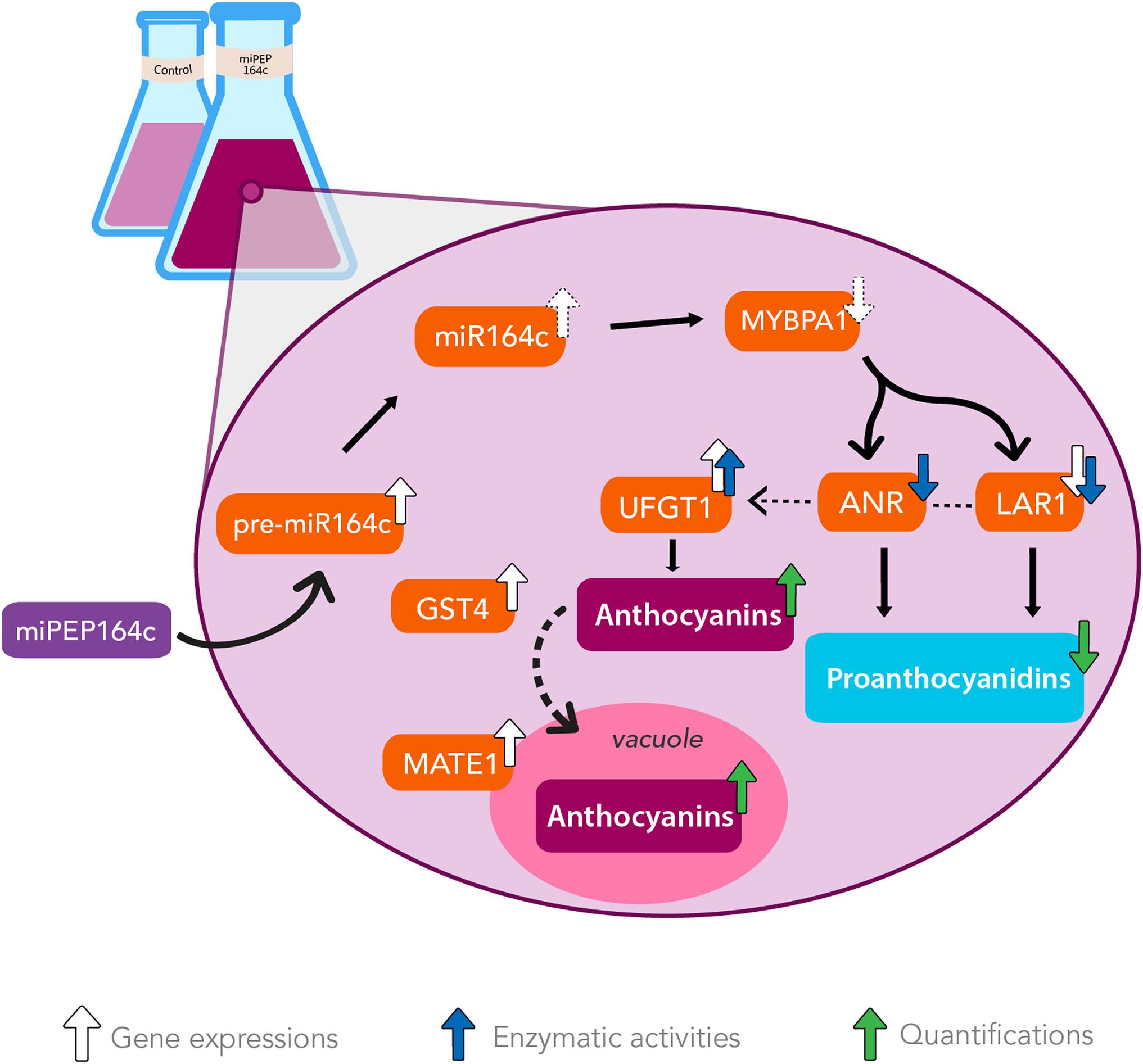
Figure 8. The exogenous application of miPEP164c increases anthocyanin synthesis and accumulation while decreasing PA synthesis by enhancing miR164c-mediated downregulation of PA-synthetic pathway in Gamay grape berry cell suspensions. Addition of miPEP164c provoked an increase in the transcription of pre-miR164c and consequently of its mature form, miR164c, which putatively led to a decrease in the translation of transcription factor MYBPA1. This inhibition in MYBPA1 translation resulted in a downregulation of VvLAR1 expression and LAR total biochemical activity, as well as ANR total biochemical activity, decreasing the intracellular concentration of PAs. This downregulation of the PA pathway indirectly led to the stimulation of the anthocyanin synthesis by increasing VvUFGT1 expression and UFGT total biochemical activity, as well as vacuolar accumulation, as shown by VvGST4 and VvMATE1 overexpression.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
MV performed the experiments and wrote the manuscript. JR and HB performed the experiments. HG advised, wrote, and reviewed the manuscript. AC conceptualized the work, performed the experiments, and wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Fundação para a Ciência e Tecnologia (FCT), under the strategic programs UID/AGR/04033/2020 and UID/BIA/04050/2020. This work was also supported by FCT and European Funds (FEDER/POCI/COMPETE2020) through the research project “MitiVineDrought—Combining “omics” with molecular, biochemical, and physiological analyses as an integrated effort to validate novel and easy-to-implement drought mitigation strategies in grapevine while reducing water use” with ref. PTDC/BIA-FBT/30341/2017 and ref. POCI-01-0145-FEDER-030341, respectively; through the research project “BerryPlastid—Biosynthesis of secondary compounds in the grape berry: unlocking the role of the plastid” with ref. POCI-01-0145-FEDER-028165 and ref. PTDC/BIA-FBT/28165/2017, respectively; and also through the FCT-funded research project “GrapeInfectomics” (PTDC/ASPHOR/28485/2017). AC was supported with a post-doctoral researcher contract/position within the project “MitiVineDrought” (PTDC/BIA-FBT/30341/2017 and POCI-01-0145-FEDER-030341). FEDER/COMPETE also supported this work through the project “AgriFoodXXI” (ref. NORTE-01-0145-FEDER-000041). MV was supported by a Ph.D. fellowship funded by FCT (SFRH/BD/144637/2019). HB was supported by a Ph.D. fellowship funded by FCT (SFRH/BD/144638/2019). This work also benefited from the networking activities within the European COST Action CA17111 “INTEGRAPE—Data Integration to maximize the power of omics for grapevine improvement,” the CoLAB VINES & WINES, and the CoLAB 4FOOD – Collaborative Laboratory for Innovation in the Food Industry.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was first available online as a pre-print version at BioRxiv with the reference: Vale et al. (2021).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.706679/full#supplementary-material
Bogs, J., Jaffe, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Broadhurst, R. B., and Jones, W. T. (1978). Analysis of condensed tannins using acidified vanillin. J. Sci. Food Agric. 29, 788–794. doi: 10.1002/jsfa.2740290908
Budak, H., and Akpinar, B. A. (2015). Plant miRNAs: biogenesis, organization and origins. Funct. Integr. Genomics 15, 523–531. doi: 10.1007/s10142-015-0451-2
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Cavallini, E., Zenoni, S., Finezzo, L., Guzzo, F., Zamboni, A., Avesani, L., et al. (2014). Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol. 55, 517–534. doi: 10.1093/pcp/pct190
Chaves, M. M., Zarrouk, O., Francisco, R., Costa, J. M., Santos, T., Regalado, A. P., et al. (2010). Grapevine under deficit irrigation: hints from physiological and molecular data. Ann. Bot. 105, 661–676. doi: 10.1093/aob/mcq030
Chen, Q. J., Deng, B. H., Gao, J., Zhao, Z. Y., Chen, Z. L., Song, S. R., et al. (2020). A mirna-encoded small peptide, vvi-miPEP171d1, regulates adventitious root formation. Plant Physiol. 183, 656–670. doi: 10.1104/pp.20.00197
Conde, A., Pimentel, D., Neves, A., Dinis, L.-T., Bernardo, S., Correia, C. M., et al. (2016). Kaolin foliar application has a stimulatory effect on phenylpropanoid and flavonoid pathways in grape berries. Front. Plant Sci. 7:1150. doi: 10.3389/fpls.2016.01150
Conn, S., Curtin, C., Bézier, A., Franco, C., and Zhang, W. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59, 3621–3634. doi: 10.1093/jxb/ern217
Couzigou, J., Lauressergues, D., and Combier, J. (2015). miRNA-encoded peptides (miPEPs): a new tool to analyze the roles of miRNAs in plant biology. RNA Biol. 12, 1178–1180.
Couzigou, J. M., André, O., Guillotin, B., Alexandre, M., and Combier, J. P. (2016). Use of microRNA-encoded peptide miPEP172c to stimulate nodulation in soybean. New Phytol. 211, 379–381. doi: 10.1111/nph.13991
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Deluc, L. (2006). Characterization of a Grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511. doi: 10.1104/pp.105.067231
Downey, M. O., Dokoozlian, N. K., and Krstic, M. P. (2006). Cultural practice and environmental impacts on the flavonoid composition of grapes and wine: a review of recent research. Am. J. Enol. Vitic. 57, 257–268.
Erokhina, T. N., Ryazantsev, D. Y., Samokhvalova, L. V., Mozhaev, A. A., Orsa, A. N., Zavriev, S. K., et al. (2021). Activity of chemically synthesized peptide encoded by the miR156A precursor and conserved in the Brassicaceae family plants. Biochem. Moscow 86, 551–562. doi: 10.1134/S0006297921050047
Francisco, R. M., Regalado, A., Ageorges, A., Burla, B. J., Bassin, B., Eisenach, C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. doi: 10.1105/tpc.112.102152
Gagné, S., Lacampagne, S., Claisse, O., and Gény, L. (2009). Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol. Biochem. 47, 282–290. doi: 10.1016/j.plaphy.2008.12.004
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-level, A., Souquet, J., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
Heppel, S. C. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant. 3, 509–523. doi: 10.1093/mp/ssp118
Jiang, S., Chen, M., He, N., Chen, X., Wang, N., Sun, Q., et al. (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 6:40. doi: 10.1038/s41438-019-0118-6
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, 68–73. doi: 10.1093/nar/gkt1181
Lauressergues, D., Couzigou, J.-M., Clemente, H. S., Martinez, Y., Dunand, C., Bécard, G., et al. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90–93. doi: 10.1038/nature14346
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921. doi: 10.1111/nph.13816
Liao, L., Vimolmangkang, S., Wei, G., Zhou, H., Korban, S. S., and Han, Y. (2015). Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 6:243. doi: 10.3389/fpls.2015.00243
Lister, C. E., Lancaster, J. E., and Walker, J. R. L. (1996). Developmental changes in enzymes of flavonoid biosynthesis in the skins of red and green apple cultivars. J. Sci. Food Agric. 71, 313–320. doi: 10.1002/(SICI)1097-0010(199607)71:3<313::AID-JSFA586>3.0.CO;2-N
Matus, J. T., Loyola, R., Vega, A., Peña-Neira, A., Bordeu, E., Arce-Johnson, P., et al. (2009). Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60, 853–867. doi: 10.1093/jxb/ern336
Pantaleo, V., Szittya, G., Moxon, S., Miozzi, L., Moulton, V., Dalmay, T., et al. (2010). Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 62, 960–976. doi: 10.1111/j.0960-7412.2010.04208.x
Pérez-Díaz, J. R., Pérez-Díaz, J., Madrid-Espinoza, J., González-Villanueva, E., Moreno, Y., and Ruiz-Lara, S. (2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 90, 63–76. doi: 10.1007/s11103-015-0394-y
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Ram, M. K., Mukherjee, K., and Pandey, D. M. (2019). Identification of miRNA, their targets and miPEPs in peanut (Arachis hypogaea L.). Computat. Biol. Chem. 83:107100. doi: 10.1016/j.compbiolchem.2019.107100
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., and Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. doi: 10.1186/1471-2229-6-27
Sharma, A., Badola, P. K., Bhatia, C., Sharma, D., and Trivedi, P. K. (2020). Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 6, 1262–1274. doi: 10.1038/s41477-020-00769-x
Teixeira, A., Eiras-Dias, J., Castellarin, S. D., and Gerós, H. (2013). Berry phenolics of grapevine under challenging environments. Int. J. Mol. Sci. 14, 18711–18739. doi: 10.3390/ijms140918711
Vale, M., Rodrigues, J., Badim, H., Geros, H., and Conde, A. (2021). Exogenous application of non-mature miRNA-encoded miPEP164c inhibits proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. bioRxiv [Preprint]. doi: 10.1101/2021.03.04.433997
Waterhouse, P. M., and Hellens, R. P. (2015). Plant biology: coding in non-coding RNAs. Nature 520, 41–42. doi: 10.1038/nature14378
Xie, Z., Khanna, K., and Ruan, S. (2010). Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21, 790–797. doi: 10.1016/j.semcdb.2010.03.012
Zhang, X., Liu, Y., Gao, K., Zhao, L., Liu, L., Wang, Y., et al. (2012). Characterisation of anthocyanidin reductase from Shuchazao green tea. J. Sci. Food Agric. 92, 1533–1539. doi: 10.1002/jsfa.4739
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6:8635. doi: 10.1038/ncomms9635
Keywords: non-mature miRNA-encoded peptide, miRNA regulation, grape berry secondary metabolism, post-transcriptional regulation, anthocyanins
Citation: Vale M, Rodrigues J, Badim H, Gerós H and Conde A (2021) Exogenous Application of Non-mature miRNA-Encoded miPEP164c Inhibits Proanthocyanidin Synthesis and Stimulates Anthocyanin Accumulation in Grape Berry Cells. Front. Plant Sci. 12:706679. doi: 10.3389/fpls.2021.706679
Received: 07 May 2021; Accepted: 15 September 2021;
Published: 05 October 2021.
Edited by:
Elena Baraldi, University of Bologna, ItalyReviewed by:
José-Antonio Daròs, Polytechnic University of Valencia, SpainCopyright © 2021 Vale, Rodrigues, Badim, Gerós and Conde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hernâni Gerós, Z2Vyb3NAYmlvLnVtaW5oby5wdA==; Artur Conde, YXJ0dXJjb25kZUBiaW8udW1pbmhvLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.