
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 August 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.705883
This article is part of the Research Topic Identification and Functional Dissection of Stress-responsive Genes in Cotton View all 26 articles
 Lili Lu1,2†
Lili Lu1,2† Ghulam Qanmber2†
Ghulam Qanmber2† Jie Li1,2
Jie Li1,2 Mengli Pu3
Mengli Pu3 Guoquan Chen3
Guoquan Chen3 Shengdong Li3
Shengdong Li3 Le Liu3
Le Liu3 Wenqiang Qin2
Wenqiang Qin2 Shuya Ma2
Shuya Ma2 Ye Wang1,2
Ye Wang1,2 Quanjia Chen1*
Quanjia Chen1* Zhao Liu3*
Zhao Liu3*The APETALA2 (AP2)/ethylene response factor plays vital functions in response to environmental stimulus. The ethylene response factor (ERF) subfamily B3 group belongs to the AP2/ERF superfamily and contains a single AP2/ERF domain. Phylogenetic analysis of the ERF subfamily B3 group genes from Arabdiposis thaliana, Gossypium arboreum, Gossypium hirsutum, and Gossypium raimondii made it possible to divide them into three groups and showed that the ERF subfamily B3 group genes are conserved in cotton. Collinearity analysis identified172 orthologous/paralogous gene pairs between G. arboreum and G. hirsutum; 178 between G. hirsutum and G. raimondii; and 1,392 in G. hirsutum. The GhERF subfamily B3 group gene family experienced massive gene family expansion through either segmental or whole genome duplication events, with most genes showing signature compatible with the action of purifying selection during evolution. Most G. hirsutum ERF subfamily B3 group genes are responsive to salt stress. GhERF13.12 transgenic Arabidopsis showed enhanced salt stress tolerance and exhibited regulation of related biochemical parameters and enhanced expression of genes participating in ABA signaling, proline biosynthesis, and ROS scavenging. In addition, the silencing of the GhERF13.12 gene leads to increased sensitivity to salt stress in cotton. These results indicate that the ERF subfamily B3 group had remained conserved during evolution and that GhERF13.12 induces salt stress tolerance in Arabidopsis and cotton.
Plants are sessile organisms continuously challenged by changing environmental conditions. As a result, different signal cascades mediate plant metabolism and development to ensure adaptation to these environmental challenges (Markakis et al., 2012). Transcription factors (TFs) are key regulators that play important roles in various stress responses (Debbarma et al., 2019). In Arabidopsis, the plant specific TF APETALA2/ethylene-responsive element binding factor (AP2/ERF) constitutes one of the largest among 65 TF families (Nakano et al., 2006; Mitsuda and Ohme-Takagi, 2009). AP2 TF was first isolated from Arabidopsis in 1994 (Jofuku et al., 1994). Since then, the total number of identified plant AP2/ERF increased from 147 in Arabidopsis thaliana to 210 in Zea mays (Liu et al., 2013; Guo et al., 2016). The AP2/ERF family is a nuclear protein with 68 amino acid repeat motifs and possesses DNA-binding activity (Ohme-Takagi and Shinshi, 1995). Based on its evolutionary structure, AP2/ERF was divided into three distinct subgroups (Riechmann et al., 2000). The AP2 subfamily members (14 in Arabidopsis) contain double AP2/ERF domains, the RAV subfamily (six members) contains a single AP2/ERF domain and a B3-DNA binding domain, while another subfamily (125 members) contains only one AP2/ERF domain (Sakuma et al., 2002). Based on phylogenetic and similarity analyses of the AP2/ERF domain, these 125 members were divided into a DREB subfamily (group A; 56 members), an ERF subfamily (group B; 65 members), and others (four members). The DREB and ERF subfamilies were further subdivided into six groups, from A1–A6 and B1–B6, respectively. The ERF subfamily B3 group contains a single AP2 domain with β-1, β-2, β-3, and α-helix regions (Heyman et al., 2018; Cao et al., 2020). To date, the ERF TF family has been identified in Arabidopsis (Nakano et al., 2006), rice (Rashid et al., 2012), cotton (Jin and Liu, 2008), maize (Liu et al., 2013), soybean (Zhang et al., 2008), wheat (Zhuang et al., 2011), sorghum (Yan et al., 2013), and various other species.
Previous reports identified the roles played by ERF TFs in regulating biotic and abiotic stress responses by repressing or activating abscisic acid (ABA)-related gene expressions (Licausi et al., 2013; Mizoi et al., 2013). For example, the overexpression of AtERF4 inhibits root elongation and negatively regulates signaling of ABA and ethylene (Yang et al., 2005). AtERF7 represses ABA-responsive genes (Zhang et al., 2007), and ERF111 negatively regulates ABA responses and stress-related gene expression (Pandey et al., 2005). Similarly, AtERF13 belongs to the B3 group of the ERF subfamily, and the overexpression of AtERF13 leads to hypersensitivity to glucose, whose effect is mediated by ABA (Lee et al., 2010). AtERF15 positively regulates ABA responses in Arabidopsis (Lee et al., 2015), and BpERF13 positively responds and regulates cold-induced stress response (Lv et al., 2020). The overexpression of ERF1-V from Haynaldia villosa increases tolerance to salt stresses in wheat (Xing et al., 2017). Similarly, another wheat ERF gene, TaERF3, promotes tolerance to salt stresses. Moreover, the amount of proline and chlorophyll in TaERF3-overexpressing wheat treated with 250 mM NaCl was higher than that of control plants, while the amount of hydrogen peroxide showed an inverse pattern (Rong et al., 2014). CitERF13 is involved in the regulation of photosynthesis by transient overexpression in Nicotiana tabacum leaves (Xie et al., 2017). Further, ERF95 (or ESE1, Ethylene and Salt Inducible 1) and ERF98 regulate salt tolerance in Arabidopsis (Zhang L. et al., 2011; Zhang et al., 2012), while ERF97 (previously named AtERF14) mediates defense response in Arabidopsis (Catinot et al., 2015).
Cotton is a major source of natural fibers for the textile industry and an excellent model to study polyploidy (Malik et al., 2018; Yang et al., 2020). Cotton plants are grown in over 70 countries worldwide and play an important role in the global economy (Qiao et al., 2008). However, various abiotic stresses, such as drought and salt, affect cotton plant growth and yield (Wang et al., 2017; Chen et al., 2019; Qanmber et al., 2019b). Moreover, it has been shown that soil salinity ranging from 8–18 dS/m results in 10–55% yield loss in cotton (Satir and Berberoglu, 2016). Therefore, abiotic stresses, such as salinity, constitute a serious problem that merits further in-depth study. Transcriptomic data analysis identified 2,356 differentially expressed genes between two cotton samples (saline or water-treated), in particular among transcription factors that were classified as ERF members (17.6%). Furthermore, when compared with the control, cotton seedlings silenced with GhERF4L and GhERF54L were significantly less tolerant to salt stress (Long et al., 2019). Finally, previous studies found that GhERF1, GhERF 2, GhERF 3, GhERF6, and GhERF38 mediate salt and drought tolerance (Qiao et al., 2008; Jin et al., 2010; Ma et al., 2017).
In this study, we identified the ERF subfamily B3 group in three cotton species, namely G. arboreum, G. hirsutum, and G. raimondii. In order to study the evolution of the ERF subfamily B3 group, we studied the genes, phylogeny, sequence logos, duplication events, collinearity, selection pressure (Ka/Ks non-synonymous/synonymous), and chromosomal location. In addition, we determined the expression patterns of genes with salt stress in specific tissues. Subcellular localization analysis was performed for GhERF13.12. We overexpressed GhERF13.12 in Arabidopsis with ACC (1-aminocyclopropane-1-carboxylic acid) treatment and subsequently studied the effects of salt stress by analyzing POD activity, the content of DAB, MDA, H2O2, total chlorophyll, and soluble sugars, as well as salt-responsive gene expression. We then evaluated the role played by GhERF13.12 in salt tolerance in cotton by silencing the gene. Collectively, this study provides an understanding of the ERF subfamily B3 group and the role played by GhERF13.12 in cotton salt tolerance.
We identified the members of the ERF subfamily B3 group gene family in Arabidopsis by keyword searching in the Arabidopsis database (http://www.arabidopsis.org) and downloading the corresponding protein sequences from TAIR 10 (http://www.arabidopsis.org). These protein sequences were then used as query in Local BLASTP to identify the ERF subfamily B3 group genes in G. arboretum (Institute of Cotton Research, ICR, version 1.0) (Du et al., 2018), G. hirsutum (Nanjing Agricultural University, NAU, version 1.1) (Zhang et al., 2015), and G. raimondii (Joint Genome Institute, JGI, version 2.0) (Paterson et al., 2012). We downloaded the databases for different plant species, such as, G. raimondii, G. hirsutum, and G. arboreum, from the Cotton Genome Database (https://www.cottongen.org/) with an e-value cutoff of 1e−5 (Yu et al., 2014). Moreover, we obtained ERF subfamily B3 group gene pfam ID (PF00847) from the Pfam website (http://pfam.janelia.org/) (Finn et al., 2016) and further verified the identified ERF subfamily B3 group gene family members in G. arboreum, G. hirsutum, and G. raimondii using a hidden Markov model (HMM).
To understand the evolutionary relationship of ERF subfamily B3 group gene family members, full-length protein sequences of A. thaliana, G. arboreum, G. hirsutum, and G. raimondii were aligned in Clustal X1.81 (http://www.clustal.org/), and an evolutionary tree was constructed using the NJ method in MEGA 7.0 (Kumar et al., 2016), as described previously (Qanmber et al., 2018; Wang et al., 2018; Yu et al., 2018). To estimate the reliability of clade classification, we selected 1,000 replicates using the bootstrap method. A Poisson model with default parameters enabled us to estimate the substitution rate.
To perform collinearity analysis, orthologous and paralogous gene pairs were obtained as previously described (Qanmber et al., 2019b,c; Ali et al., 2020). Shortly, orthologous and paralogous gene pairs belonging to the cotton ERF subfamily B3 group genes were retrieved by an all-vs. all search in BLASTP. The results were further subjected to MCScan analysis to generate collinear blocks within and between cotton A, D, At, and Dt genomes and sub-genomes. We then used the MCScanX software to estimate collinear pairs and gene duplication event types in the cotton ERF subfamily B3 group genes, and the CIRCOS software was used to draw the final figure (Krzywinski et al., 2009; Zheng et al., 2020). Further, in order to calculate the Ka/Ks values, we aligned the protein sequences of the ERF subfamily B3 group genes using Clustal X1.81 (http://www.clustal.org/) and converted the protein sequences of the ERF subfamily B3 group genes into cDNA sequences using the PAL2NAL package (http://www.bork.embl.de/pal2nal/). Finally, we obtained synonymous (Ks) and non-synonymous (Ka) divergence level values using the “simple KaKs calculator” software in TBtools (Chen et al., 2020).
For qRT-PCR analysis, we extracted RNA using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). We assessed the concentration and quality of the RNA by NanoDrop2000 (Thermo Fisher Scientific, Waltham, MA, United States), and then synthesized 1 μg of RNA into cDNA with TransScript® RT All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (https://www.transgen.com.cn/news_show/927.html) as previously described (Xiong et al., 2019). The following parameters were used for the RT-PCR procedure: 94°C for 3 min, 28 cycles of 10 s at 98°C, 10 s at 59°C, and 10 s at 72°C. The 2 × T5 Super PCR Mix (Colony) was used. The PCR products were detected by electrophoresis in a 1.5% agarose gel. Actin2 (AT3G18780.1) and GhHis3 (AF024716) were used as internal controls, and SYBR Green was used to conduct the qRT-PCR analysis on a LightCycler 480 (Roche Diagnostics GmbH, Mannheim, Germany). Each experiment was repeated three times (biological repeats), and the relative gene expression values were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The primers used in this experiment are listed in Supplementary Table 5. Values are shown as the mean ± SE (n = 3, Student t-test, *P < 0.05, **P < 0.01, and ***P < 0.001).
For subcellular localization, full-length coding sequences of the GhERF13.12 gene were inserted in a pEG101 vector expressing YFP, and then a vector expressing the GhERF13.12-YFP fusion protein was constructed (Zhao et al., 2019). The construct in GV3101 was then transformed into 2-cm diameter leaves of 1-month-old tobacco (N. benthamiana) cells for subcellular localization analysis. The plants were placed in the dark for 24 h, and tobacco epidermal cells were observed after 72 h of inoculation using a fluorescence microscope.
For the ectopic expression of the GhERF13.12 gene in Arabidopsis, full-length cDNA sequences were amplified by PCR using gene-specific primers, cloned into a pCAMBIA-2300 vector with a constitutive Cauliflower mosaic virus 35S promoter, and then transformed into Arabidopsis via the floral dip method using the Agrobacterium tumefaciens strain GV3101 (Zhang et al., 2006). For the silencing of the GhERF13.12 gene in cotton, we cloned a 300-bp fragment of the coding sequence (CDS) with high specificity into a pTRV-RNA2 vector using XbaI and SmaI restriction sites to generate TRV::GhERF13.12. Similarly, TRV::GhCLA1 was constructed as a visual marker for monitoring silencing efficiency. TRV::00 (empty vector) was used as the negative control. We transformed these constructed vectors into the Agrobacterium tumefaciens strain GV3101.
We used the wild-type (WT) Columbia-0 (Col-0) ecotype for ectopic expression and subsequent analysis in Arabidopsis. For cotton, the G. hirsutum ZM24 variety was used to collect the samples used for tissue specific expression. Cotton plants at three distinct leaf stages were treated with 400 mM NaCl for 6, 12, and 24 h using the ZM24 variety for salt treatment and virus-induced gene silencing (VIGS) (Qanmber et al., 2019b). The cotton plant ZM9807 (salt-tolerant) was used as a positive control. Furthermore, 250 mM NaCl was used to stress Arabidopsis seedlings for 9 days. We then collected these samples, which were immediately frozen in liquid nitrogen and stored at −80°C until they were used for RNA extraction. Arabidopsis plants were grown under conditions of 22°C on a 16 h light/8 h dark photoperiod. WT and GhERF13.12 transgenic Arabidopsis seedlings were incubated in a MS medium with or without 10 μM ACC for 3 days in darkness in order to check the ethylene-evoked triple response. Cotton seeds were planted in vegetative soil and vermiculite (v/v = 1:1) in flower pots and grown at 30°C on a 16 h light/8 h dark photoperiod.
We measured various physiological indices using the Solarbio kit following the instructions of the manufacturer. A UV-1600 spectrophotometer (AOE Instruments, Shanghai, China) was used to measure absorbance (Qanmber et al., 2019b). We harvested 0.2 g fresh leaves of WT and transgenic plants. Malondialdehyde (MDA) content was measured at 450, 532, and 600 nm as previously described (Rao and Sresty, 2000). To evaluate the amount of soluble sugar, 1 ml distilled water was added into 0.2 g of leaf materials; the mixture was ground and homogenized and then boiled for 10 min in a centrifuge tube. The mixture was centrifuged at 8,000 g for 10 min at room temperature, and then the absorbance of the supernatant was measured at 620 nm. Total chlorophyll content was estimated by measuring the absorbance in the extracts at 663 and 645 nm. Peroxidase (POD) was measured as mentioned in a previous study (Wang et al., 2012). Further, 3, 3'-diaminobenzidine (DAB) was used to measure the H2O2 levels and the staining was stopped until the difference could be observed and decolorized with 75% ethanol as previously described (Zhang X. et al., 2011). Classification of salt damage levels and salt damage index was determined as previously described (Qanmber et al., 2019b). Values are the means ± SE (n = 3, Student t-test, *P < 0.05, **P < 0.01, and ***P < 0.001).
A total of 148 members of the ERF subfamily B3 group were identified in four plant species using different in silico approaches, such as the SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2015), PROSITE (http://prosite.expasy.org/), and InterProscan 63.0 (http://www.ebi.ac.uk/interpro/) (Jones et al., 2014) websites, as described in previous studies (Liu et al., 2018; Faiza et al., 2019; Li et al., 2019; Qanmber et al., 2019a). Among these, 18 ERF subfamily B3 group genes were found in A. thaliana, 18 in G. arboretum (diploid cotton), 94 in G. hirsutum (tetraploid cotton), and 18 in G. raimondii (diploid cotton) (Supplementary Table 1). The highest number of genes was, thus, found in G. hirsutum, indicating that the ERF subfamily B3 group experienced large scale expansions in this species. Notably, G. hirsutum had more ERF subfamily B3 group genes than both G. raimondii and G. arboreum, demonstrating polyploidy and massive gene duplication events in the evolutionary process. As the main object of this study was G. hirsutum, we compared the ERF subfamily B3 group genes identified in this species from NAU with JGI, ICR, and HAU genome sequence databases. As we found no differences, we proceeded with the ERF subfamily B3 group genes retrieved from NAU.
To estimate the evolutionary relationship among the ERF subfamily B3 group genes, we constructed a phylogenetic tree of this gene family in four plant species, namely, A. thaliana, G. arboreum, G. hirsutum, and G. raimondii. Phylogenetic analysis divided all the ERF subfamily B3 group genes into three groups, from A1 to A3 (Figure 1), according to Arabidopsis group classification (Nakano et al., 2006). Group A3 is the largest, containing 60 ERF subfamily B3 group genes. In contrast, group A2 is the smallest, with a total of 38 genes. Group A1 contains a total of 50 ERF subfamily B3 group genes, of which 5 are found in A. thaliana, 6 in G. arboreum, 33 in G. hirsutum, and 6 in G. raimondii. Group A2 contains a total of six, six, 21, and five ERF subfamily B3 group genes in A. thaliana, G. arboreum, G. hirsutum, and G. raimondii, respectively. Similarly, group A3 contains 7, 6, 40, and 7 ERF subfamily B3 group genes in A. thaliana, G. arboreum, G. hirsutum, and G. raimondii, respectively. Accordingly, all groups represented in the ERF subfamily B3 group are present in all species. Phylogenetic analysis demonstrated that the ERF subfamily B3 group genes found in G. hirsutum experienced maximum gene family expansion compared with other species, as the homologous/orthologous pairs clustered together across all three groups. Further, A. thaliana and G. hirsutum showed gene pairs that originated from the same node, indicating that the A. thaliana and, especially, the G. hirsutum ERF subfamily B3 group genes experienced massive gene duplication and gene family expansion. This suggests gene duplication was a major reason behind the expansion of the ERF subfamily B3 group gene family. However, it is possible that gene duplication differed between groups and plant species.
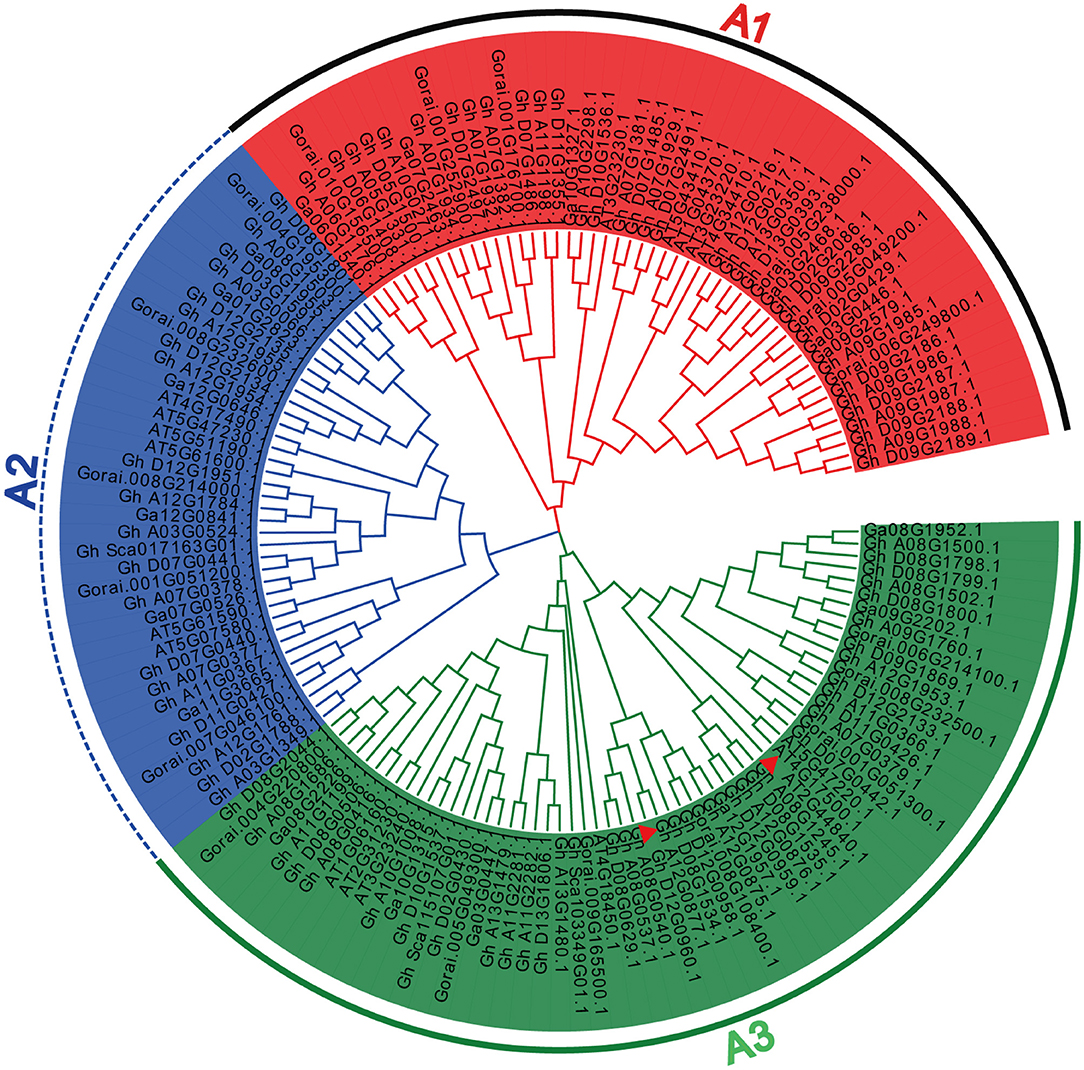
Figure 1. Phylogenetic analysis of the (ERF) subfamily B3 group gene family. The 148 genes were divided into three groups (A1–A3). These genes were found in A. thaliana, G. arboreum, G. hirsutum, and G. raimondii. The NJ method was used to build the phylogenetic tree, and topological robustness was evaluated by bootstrapping with 1,000 replications. The cotton and Arabidopsis ERF accessions are indicated. The gene marked with a red triangle corresponds to ERF13 in Arabidopsis and cotton.
To find whether the ERF subfamily B3 group genes were conserved throughout the evolutionary process, we constructed sequence logos of the ERF subfamily B3 group for G. arboreum, G. raimondii, and G. hirsutum (Supplementary Figure 1). The results predicted a very similar distribution of amino acid residues in the protein sequences of the different species, such as in the N and C terminals. These results indicate the cotton ERF subfamily B3 group genes were highly conserved in the course of evolution.
As the phylogenetic analysis evidenced the occurrence of gene duplication events during cotton evolution, we next explored which types of gene duplication took place in the ERF subfamily B3 group in each species. A total of 15 whole genome duplications (WGDs) and segmental duplications, along with two dispersed duplications, were identified in G. arboreum. We also found 13 WGDs and segmental duplications, in addition to a dispersed duplication in G. raimondii. Finally, we identified 71 WGDs and segmental duplications, 22 dispersed, and singleton duplication events in G. hirsutum (Supplementary Table 2). These results demonstrate that WGDs and segmental duplications were the major duplication type throughout the evolution of the ERF subfamily B3 group gene family in cotton.
Next, we performed collinearity analysis among three observed cotton species, namely, G. arboreum, G. hirsutum, and G. raimondii. We identified 172 orthologous and paralogous gene pairs between G. arboreum and G. hirsutum. Among these, 87 were found between the A-subgenomes of G. hirsutum and G. arbortum, while 85 orthologous/paralogous gene pairs were found between the D-subgenomes of G. hirsutum and G. arboreum. Similarly, we identified 178 orthologous/paralogous gene pairs between G. hirsutum and G. raimondii, of which 91 and 87 occurred between the A- and D-subgenomes of G. hirsutum and G. raimondii, respectively (Figure 2, Supplementary Table 3). Surprisingly, no orthologous/paralogous gene pairs were identified on chromosomes 2 and 4 of G. arboreum; chromosomes A01, A02, A04, A06, D01, and D04 of G. hirsutum; and chromosomes 2 and 12 of G. raimondii. We were also able to identify 1,392 orthologous/paralogous gene pairs in G. hirsutum within and between the A- and D-subgenomes (Supplementary Figure 2, Supplementary Table 3).

Figure 2. Multiple collinearity analysis of cotton ethylene response function (ERF) subfamily B3 group genes. Multiple collinearity analysis found collinearity blocks among G. arboretum (red), G. hirsutum (yellow), and G. raimondii (blue).
The Ka/Ks (non-synonymous/synonymous) analysis of the orthologous/paralogous gene pairs between G. arboreum, G. hirsutum, and G. raimondii, as well as within G. hirsutum, demonstrated 339 pairs between the three species with Ka/Ks < 1, including 315 pairs with Ka/Ks < 0–0.5, 24 pairs with Ka/Ks < 0.5–1, and 11 pairs with Ka/Ks > 1 (Supplementary Table 3). Similarly, all (1,392) the orthologous/paralogous gene pairs within G. hirsutum showed Ka/Ks < 1 (1,362 pairs with Ka/Ks < 0–0.5 and 30 pairs with Ka/Ks < 0.5–1) (Supplementary Table 4). We hypothesize that most orthologous/paralogous gene pairs in cotton underwent purifying selection in the course of evolution.
To understand the functional roles played by the GhERF subfamily B3 group genes, we examined the expression patterns of 15 selected GhERF subfamily B3 group genes with salt stress and measured transcript levels by qRT-PCR analysis (Figure 3). We evaluated the transcript levels at three time points (6, 12, and 24 h after salt treatment) and compared the results with control CK samples (0 h). After salt treatment, a total of five GhERF subfamily B3 group genes were upregulated, namely, Gh_A10G1008, Gh_D08G1798, Gh_A09G1985, Gh_D09G2285, and Gh_D12G0960. In contrast, the genes Gh_D05G1500, Gh_A11G2282, Gh_A12G1951, Gh_D07G0441, and Gh_D08G0629 were downregulated. Two GhERF subfamily B3 group genes (Gh_A08G1503 and Gh_A07G1934) were preferentially expressed at 12 h after the salt treatment, while all other genes showed erratic expression patterns. Interestingly, the genes Gh_A10G1008 and Gh_A11G2282, and Gh_A12G0875, Gh_D08G0629, and Gh_D12G0960 revealed different expression patterns despite being homologous, as they are present in the same sub-branches in the phylogenetic tree (Figure 1).
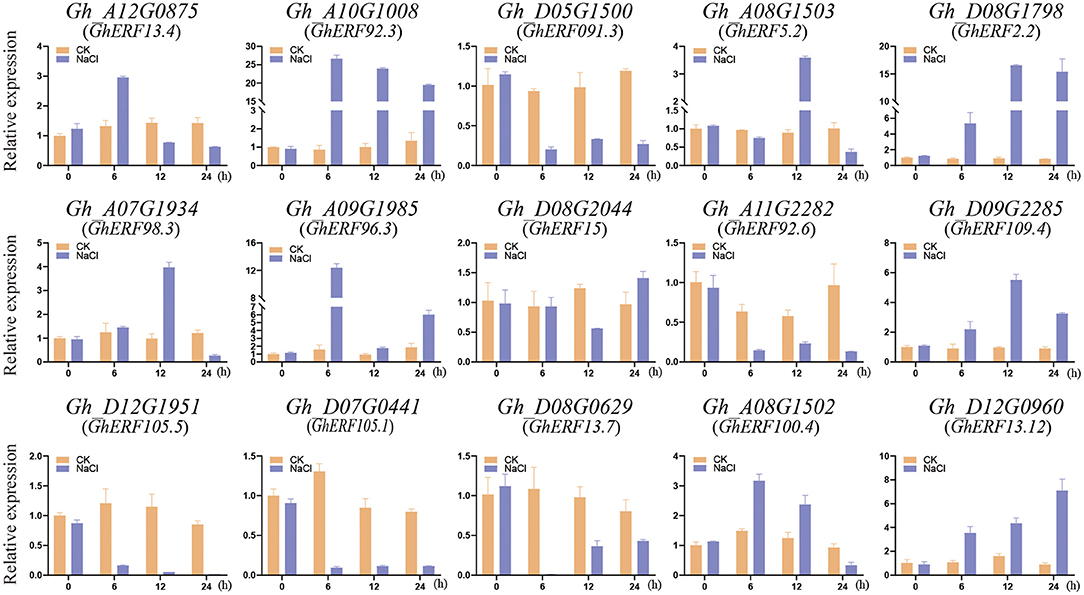
Figure 3. Transcript analysis of the GhERF subfamily B3 group genes by salt stress treatment. The response of G. hirsutum ERF subfamily B3 group genes to salt stress was examined using real time fluorescent quantitative PCR analysis. The error bars correspond to the standard deviation among three biological replicates.
Among these 15 GhERF subfamily B3 group genes, Gh_D12G0960 was selected for subsequent analysis. The gene contains a full-length CDS region consisting of 630 bp that encodes a putative ERF protein with 209 amino acid residues. Sequence alignment analysis indicated that Gh_D12G0960 was the closest homolog of the Arabidopsis AT2G44840.1 (AtERF13), with a total of 78.33% sequence identity between both domains. We, thus, termed Gh_D12G0960 as GhERF13.12 based on this similarity. GhERF13.12 has a predicted mass of 23.4 KD, a calculated pI of 4.88, and a protein sequence that includes β-1, β-2, β-3, and α-helix regions with a typical AP2/ERF domain (highlighted with red line) in both AtERF13 and GhERF13.12, as observed in the alignment (Supplementary Figure 3).
The tissue-specific expression pattern of GhERF13.12 was observed by qRT-PCR in seven vegetative and fiber tissues, such as the root, stem, leaf, and flower and 0, 5, and 10 DPA (days post-anthesis) fiber. The transcript level of GhERF13.12 was higher in the roots and leaves, which might be related to stress responses (Figure 4A). Moreover, the subcellular localization of GhERF13.12 was determined by transient expression in tobacco leaves, and it was found in the nucleus when compared with the YFP control vector (Figure 4B).
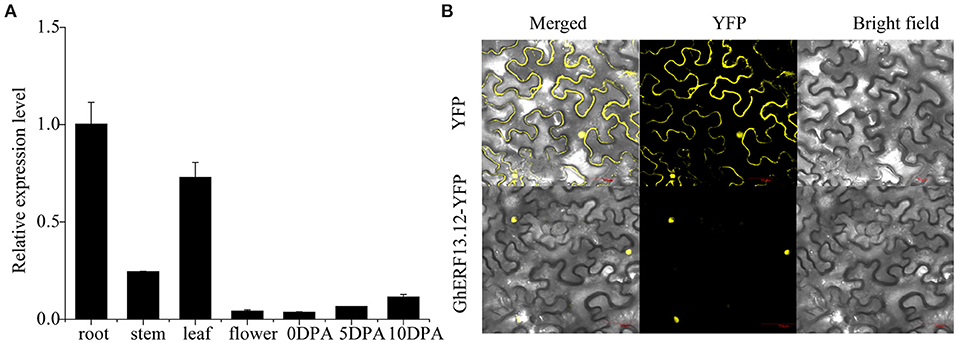
Figure 4. Transcript profile of the GhERF13.12 gene. (A) Tissue-specific expression pattern of GhERF13.12 obtained by qRT-PCR analysis in the root, stem, leaf, and flower at 0, 5, and 10 DPA (days post-anthesis). Fiber samples were taken from ZM24 cotton plants. The standard deviation among three biological replicates is represented by the error bars. (B) Subcellular localization of GhERF13.12 in tobacco leaves.
To understand the role of GhERF13.12, we overexpressed the coding sequences of the gene in Arabidopsis. The transcript level of GhERF13.12 in overexpression transgenic Arabidopsis was analyzed by RT-PCR, and we selected three transgenic lines L1–L3 for subsequent experiments (Supplementary Figure 4A). To check the ethylene-evoked triple response in homozygous lines (T3 generation), transgenic seedlings were subjected to ACC treatment (Figure 5A). In general, the results showed that the hypocotyl length in GhERF13.12 transgenic plants largely decreased by 10 μM following ACC treatment and when compared with WT plants. This indicates that ethylene administered by its precursor ACC inhibited cell elongation and the growth of GhERF13.12 transgenic seedlings (Figure 5A, Supplementary Figure 4B). These observations are in accordance with previous reports (Wu et al., 2002). To evaluate the function of GhERF13.12 in response to salt stress, the GhERF13.12 transgenic plants (L1, L2, and L3) were analyzed with salt treatment. The 20-day-old WT and GhERF13.12 transgenic plant seedlings were subjected to 250 mM salt treatment for nine consecutive days, and they showed dramatic differences: WT leaves were dry and yellow, while transgenic plant leaves were greener. As an important parameter of plant photosynthetic capacity, we measured chlorophyll content after salt treatment, and found that the GhERF13.12 overexpressing lines showed higher chlorophyll content compared with WT under stress conditions, indicating these plants are more tolerant to salt than WT plants (Figures 5B,D).
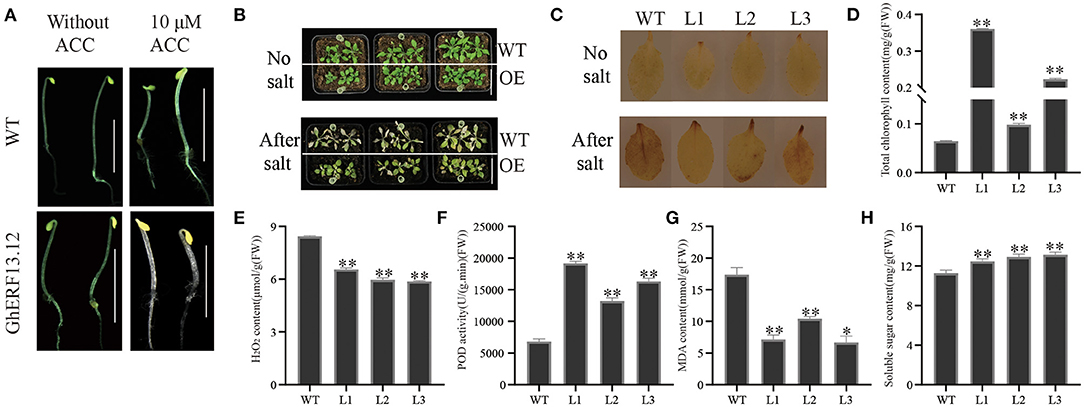
Figure 5. Ectopic expression of the GhERF13.12 gene in Arabidopsis and salt stress treatment. (A) Comparison of hypocotyl length in GhERF13.12 overexpressing lines and WT plants with and without ACC treatment. (B) Salt stress treatment of GhERF13.12 overexpressing lines and WT plants. Bar = 3 cm. (C) DAB staining of GhERF13.12 overexpressing lines and WT plants with and without salt treatment. Physiological analysis, such as (D) total chlorophyll amount, (E) H2O2 content, (F) POD activity, (G) MDA content, and (H) soluble sugar content in WT and GhERF13.12 overexpressing lines in Arabidopsis. The standard deviation among three biological replicates is shown by the error bars. *p < 0.05 and **p < 0.01 (Student's t-test). WT, wild type; L1, L2, and L3, GhERF13.12 transgenic homozygous lines.
To further elucidate the mechanism of increased salt tolerance in GhERF13.12 transgenic plants, we performed additional physiological analyses of these transgenic (with salt stress) and WT plants. We found that the amount of MDA and H2O2 of all the three lines of GhERF13.12 transgenic plants were lower than WT plants (Figures 5E,G). However, POD activity and soluble sugar content were higher in GhERF13.12 transgenic plants than WT plants (Figures 5F,H). Furthermore, the plant leaves with and without salt treatment were subjected to DAB staining, and we observed less staining with light color in transgenic plants (Figure 5C). H2O2 is one of the most important reactive oxygen species and can accumulate in plants with various environmental stresses. The results indicated that the overexpression of GhERF13.12 reduces ROS-caused damages in Arabidopsis plants under salt stress conditions by reducing H2O2 content (Li Y. et al., 2019).
In order to investigate the possible regulatory mechanism of GhERF13.12 involved in salt stress response, we selected several stress related genes and examined their expression by qRT-PCR in the GhERF13.12-overexpressing and WT Arabidopsis plants with salt stress treatment. We then examined the transcript levels of nine stress-related genes, namely, AtASA1 (ATP Sulfurylase Arabidopsis 1), AtAAO1 (abscisic aldehyde oxidase 1), AtCOR47 (Cold-Regulated 47), AtMAPKKK18 (MAPKK kinase 18), AtP5CS1 (pyrroline-5-carboxylate synthetase 1), AtP5CS2 (pyrroline-5-carboxylate synthetase 2), AtRD29B (Responsive to Desiccation 29B), AtAPX (Stromal Ascorbate Peroxidase), and AtCOR15A (Cold-Regulated 15A) (Figure 6). The qRT-PCR results demonstrated that transcript levels of all the nine genes were relatively higher in all three GhERF13.12 transgenic lines compared with WT plants under salt stress conditions. Previous studies have shown that these upregulated genes are involved in the ABA signaling pathway and proline biosynthesis pathway, and ROS scavenging (Li Y. et al., 2019). Therefore, we hypothesize that GhERF13.12 might participate in multiple processes related to salt tolerance.
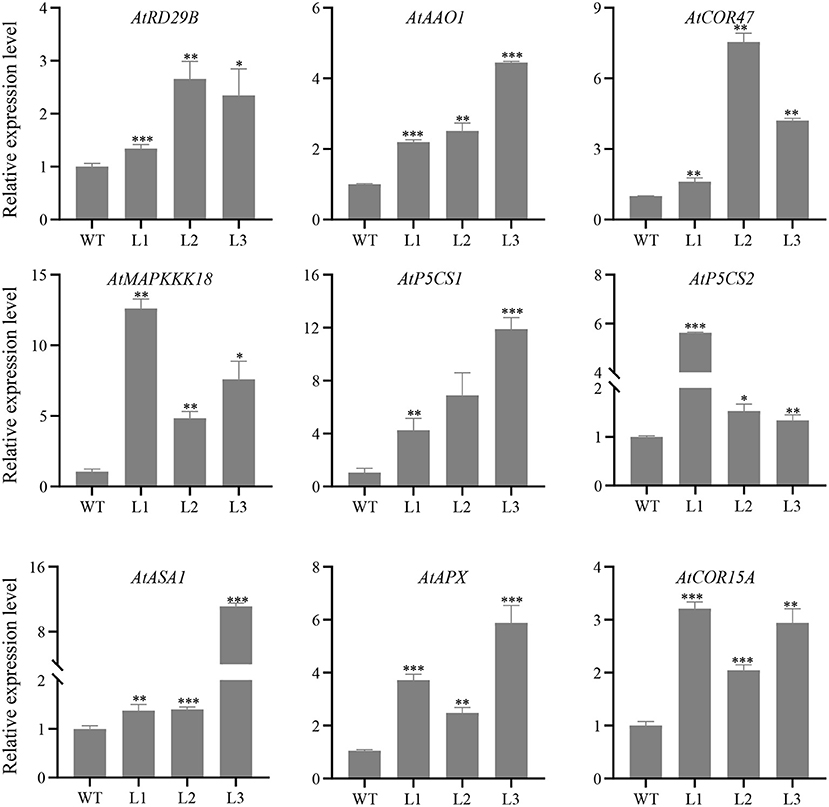
Figure 6. Transcript levels of salt stress-related marker genes. Relative expression patterns of nine genes related to salt stress were examined by qRT-PCR in WT and GhERF13.12 overexpressing lines. The standard deviation among three biological replicates is shown by the error bars. *p < 0.05, **p < 0.01, and ***p < 0.001 (Student's t-test). WT, wild type; L1, L2, and L3, GhERF13.12 transgenic homozygous lines.
We silenced the GhERF13.12 gene in cotton plants through a VIGS experiment in order to further validate the functions of this gene. We monitored the efficiency of gene silencing when the leaves of positive control seedlings showed a silencing phenotype (Supplementary Figure 5A) and found that the GhERF13.12 gene was successfully silenced (Figure 7A). Next, we applied salt stress treatment to WT, control, and GhERF13.12-silenced plants and found that the tolerance levels of the GhERF13.12-silenced plants to salt stress decreased in comparison with the WT and control plants (Figure 7B). Similarly, soluble sugar content was lower in the GhERF13.12-silenced plants than in both the WT and control plants (Figure 7F). Moreover, DAB staining, salt damage index, and MDA content were higher in the GhERF13.12 silenced plants than in the WT and control plants (Figures 7C–E). The transcript levels of salt stress response genes in the silenced plants were lower than those in the control plants (Supplementary Figure 5B). Collectively, the findings are in accordance with previous studies analyzing the ectopic expression of GhERF13.12 in Arabidopsis, which validates the authenticity of the findings.
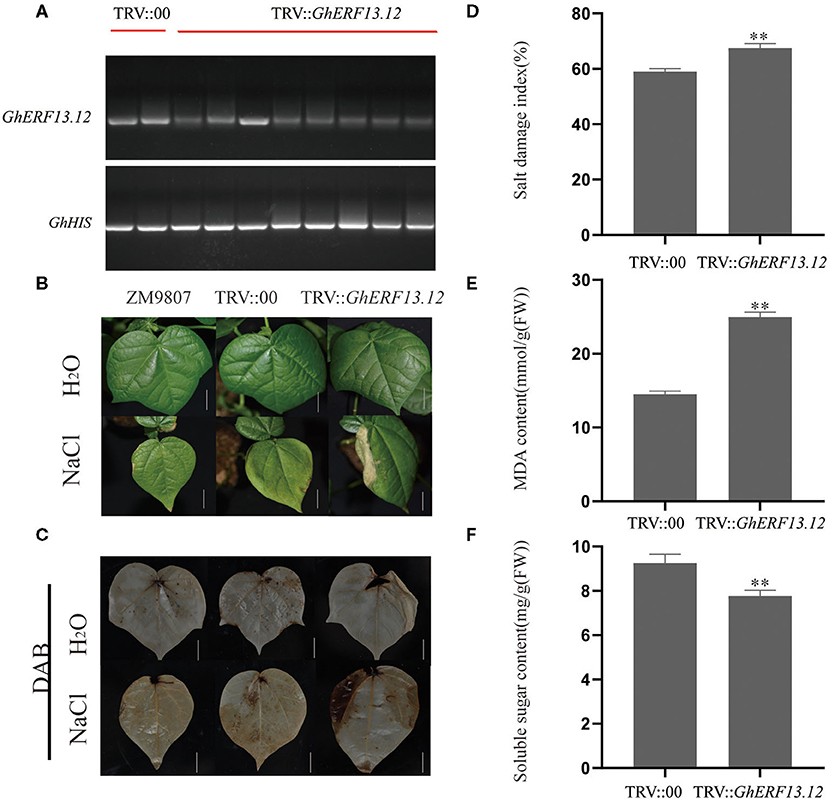
Figure 7. Gene silencing of GhERF13.12 and salt stress treatment in cotton. GhERF13.12 was silenced by virus-induced gene silencing (VIGS). (A) RT-PCR expression analysis of TRV::00 and TRV::GhERF13.12 plants to validate the silencing of the GhERF13.12 gene. GhHIS3 was used as an internal control. (B) Salt stress treatment of ZM9807, TRV::00, and TRV::GhERF13.12 plants with and without salt stress. Bar = 2.5 cm. (C) DAB staining of ZM9807, TRV::00, and TRV::GhERF13.12 plants with and without salt stress. Other parameters, such as (D) salt damage index, (E) MDA content, and (F) soluble sugar content were observed in TRV::00 and TRV::GhERF13.12 plants. The standard deviation among three biological replicates is shown by the error bars. **p < 0.01 (Student's t-test).
Researchers have conducted several studies to clarify the evolutionary history of the ERF TF family in different crop species (Nakano et al., 2006; Jin and Liu, 2008; Zhang et al., 2008; Zhuang et al., 2011; Rashid et al., 2012; Liu et al., 2013; Yan et al., 2013). Here, we report a total of 148 members of the ERF subfamily B3 group gene family, including 18 genes in A. thaliana, 18 in G. arboreum, 94 in G. hirsutum, and 18 in G. raimondii. On the basis of the Arabidopsis group classification, phylogenetic analysis divided all the cotton ERF subfamily B3 group genes into three groups ranging from A1 to A3, with A3 being the largest group and A2 being the smallest. Phylogenetic classification was similar to previous reports (Ren et al., 2017). Each group showed ERF subfamily B3 group genes from all observed species. Moreover, the GhERF subfamily B3 group genes exhibited maximum gene family expansion through massive gene duplication, as evidenced by the fact that GhERF subfamily B3 group genes homologous/orthologous gene pairs clustered together in all groups. Accordingly, phylogenetic analysis demonstrated gene duplication was a major factor behind gene family expansion in G. hirsutum. Further, sequence analysis showed similar amino acid residue distributions, illustrating that the ERF subfamily B3 group gene family was highly conserved in G. arboreum, G. hirsutum, and G. raimondii during the evolutionary process. Previously, many genome-wide analyses depicted the expansion and conservation of different gene families in cotton (Ren et al., 2017; Faiza et al., 2019; Qanmber et al., 2019b).
Cotton is an allotetraploid that provides the best model for studying the effects of polyploidy (Yang et al., 2020). The hybridization of the A genome (from G. herbaceum and G. arboreum) and the D genome (from G. raimondii) caused chromosome doubling and the subsequent emergence of nascent At/Dt (allotetraploid cotton) genomes (Li et al., 2015). Here, WGD and segmental duplication represent the main duplication type experienced by the ERF subfamily B3 group gene family in cotton. Gene duplication analysis of the HH3 and AAI gene families in cotton showed they also experienced massive WGD and segmental duplication (Qanmber et al., 2019a,c). Polyploidy contributed to gene duplication events and increased the number of ERF subfamily B3 group gene family members in G. hirsutum. Previously, WGD and segmental duplications were found to occur frequently in plants (Cannon et al., 2004), happening twice in Arabidopsis, in the Brassicaceae lineage. Cacao and cotton shared a common ancestor and experienced ancient duplication (Li et al., 2014). Gene duplication is a major force of evolution and generates new genes, some of which are redundant (Flagel and Wendel, 2009). We showed that many GhERF subfamily B3 group gene pairs were clustered, indicating ancient genome duplication. These pairs experienced rearrangements and shuffling, creating genetic diversity. Previous studies have proposed four possible fates of duplicated genes: (1) the deletion of a gene copy to remove functional redundancy during evolution; (2) sub-functionalization of both copies sharing common functions with parental copies and development of partially different functions through time; (3) one copy acquires a new function (neo-functionalization) during evolution; and (4) maintenance of essential genes for plant growth and development, which is an intermediate between neo- and sub-functionalization processes (Charon et al., 2012).
Collinearity analysis indicated 172 orthologous/paralogous gene pairs between G. arboreum and G. hirsutum, and 178 orthologous/paralogous gene pairs between G. hirsutum and G. raimondii. Overall, the GhERF subfamily B3 group gene family showed a higher number of orthologous/paralogous gene pairs with G. raimondii than with G. arboreum. Similarly, 1,392 orthologous/paralogous gene pairs were reported in G. hirsutum. These results indicate that the increase in G. hirsutum ERF subfamily B3 group genes might result from massive gene duplication due to polyploidy. Previous studies have shown that flowering plants experienced polyploidy events during evolution in order to adapt to changing environmental conditions (Ramsey and Schemske, 1998). This might explain the higher numbers of G. hirsutum ERF subfamily B3 group genes and more orthologous/paralogous gene pairs. However, post-hybridization causes gene loss in the process of improved arrangement of genomic sequences together with doubling of chromosomes (Paterson et al., 2004). In contrast, cotton experienced minor changes than Brassica (Gaeta et al., 2007) and paleopolyploid maize (Woodhouse et al., 2010).
Finally, it is often assumed that Ka/Ks = 1 indicates neutral evolution, Ka/Ks < 1 represents purifying selection, and Ka/Ks > 1 suggests positive selection and accelerated evolution (Conant and Wolfe, 2008). The Ka/Ks values found between orthologous/paralogous gene pairs in G. arboreum, G. hirsutum, and G. raimondii, and within G. hirsutum, indicated most experienced purifying selection during cotton evolution. Together, the findings provide comprehensive information on polyploidization, chromosomal interaction, and transfer of information via inter-genomic hereditary and different rates of evolution, and show that the observed large-scale gene family expansion of ERF subfamily B3 group gene family in G. hirsutum was due to segmental or whole genome duplication events.
Several studies elucidated the role of ERF genes in response to biotic and abiotic types of stress, especially salt stress (Pandey et al., 2005; Zhang et al., 2007, 2012; Lee et al., 2010, 2015; Zhang L. et al., 2011; Catinot et al., 2015). In this study, some genes were upregulated after salt stress treatment, suggesting the GhERF subfamily B3 group genes might play roles in salt stress response. Among these, we noted that Gh_D12G0960 (GhERF13.12) was upregulated at all measured time points. Subsequent analysis confirmed its homology with AT2G44840.1 (ERF13) and showed it contains β-1, β-2, β-3, and α-helix regions and a putative AP2/ERF domain. We further showed that the GhERF13.12 protein is located in the nucleus.
ABA is a signaling molecule that takes part in abiotic stress tolerance regulation (such as salt and drought) (Li Y. et al., 2019). AtERF15 participates in ABA response, and its transgenic lines were sensitive to high salinity and osmotic pressure at the seedling stage (Lee et al., 2015). The transcript levels of two key enzyme genes for ABA biosynthesis increased in overexpressed JERF1 transgenic rice, which was more drought-tolerant than WT plants (Zhang et al., 2010). GhERF38 participates in NaCl, mannitol, ABA responses, and reduces plant tolerance to salt stress when overexpressed in Arabidopsis (Ma et al., 2017). In this study, transcripts of AtAAO1, AtMAPKKK18, AtCOR15A, AtCOR47, and AtRD29B, all of which are involved in abscisic acid-activated signaling pathway, were upregulated in GhERF13.12 transgenic Arabidopsis under salt stress conditions. Proline is considered a compatible penetrant that can scavenge free radicals, maintain intracellular redox balance, and help maintain the integrity of cell membranes under salt and drought stress conditions, thereby improving salt and drought tolerance (Liu et al., 2014; Zhang et al., 2016; Li Y. et al., 2019; Zhao et al., 2019). P5CSs are involved in the proline glutamate biosynthesis pathway, which can increase the accumulation of proline and improve salt and drought tolerance in various plants along with a ROS scavenger that attenuates oxidative stress under high salinity conditions (Yamada et al., 2005; Liu et al., 2014). The transcript of OsP5CS increased in JERF1 transgenic rice under drought stress conditions (Zhang et al., 2010). Here, we showed that the ectopic expression of GhERF13.12 in Arabidopsis under salt stress conditions and the upregulation of AtP5CS1 and AtP5CS2 may lead to proline accumulation. Hence, GhERF13.12 may regulate gene expression in the ABA signaling and proline synthesis pathways in Arabidopsis under salt stress conditions.
Salt stress induces ionic stress, osmotic stress, and secondary stresses, especially oxidative stress in plants (Yang and Guo, 2018a). Therefore, in order to adapt to salt stress environments, plants rely on signals and pathways to re-establish cellular ion, osmotic, and ROS homeostasis (Yang and Guo, 2018b). White birch BpERF13 overexpression lines significantly increased tolerance to subfreezing treatment and reduced ROS (Lv et al., 2020). As a non-enzymatic antioxidant, ascorbic acid (AsA) contributes to ROS-scavenging and participates in salt tolerance (Wang and Huang, 2019). AsA biosynthesis enzymes are highly expressed in AtERF98 overexpressing Arabidopsis, whereby overexpressing plants exhibit salt tolerance (Zhang et al., 2012). Ascorbate peroxidase (APX) is an important enzyme that removes H2O2 in plants. The overexpression of PutAPX in Arabidopsis improves salt stress tolerance by reducing the accumulation of H2O2 (Guan et al., 2015). It was demonstrated that ATP-sulfurylase contributes to ROS scavenging and maintains reduced cellular-redox environments in plants under salt stress conditions (Anjum et al., 2015). Here, the expression levels of AtAPX and AtASA1 in GhERF13.12 transgenic plants significantly increased under salt stress conditions (Li Z. et al., 2019). In abiotic stress response, plants can undergo a series of physiological and biochemical changes, in particular in the amount of H2O2, MDA, and POD activity (Zhu, 2002). The depth of reddish-brown stain formed by the reaction between DAB and endogenous H2O2 can indicate the level of H2O2 accumulation. We compared the levels of ROS accumulation in WT and GhERF13.12 transgenic lines after salinity treatment and found more ROS accumulation in WT (Figure 5C). At the same time, this is consistent with the observed amount of H2O2 in quantitative analysis, which was lower than that of WT (Figure 5E). POD is a ROS scavenging enzyme that mediates ROS homeostasis in order to protect normal metabolism. A higher POD activity is beneficial to protect the lipid membrane from damage caused by free radicals and to avoid membrane lipid peroxidation caused by unsaturated fatty acids in the membrane (Jiang et al., 2007). We showed POD activity in transgenic plants was higher than that in WT (Figure 5F). Lipid membranes are fragile to stress-induced cellular damage, and the degree of damage is usually used to measure tolerance to the applied stress. MDA is the final product of lipid peroxidation and a reliable indicator of membrane damage caused by stress conditions. In addition, the accumulation of ROS could be insinuated by the MDA content in plant cells (Jain et al., 2001). Here, the accumulation of MDA in the WT plants was higher than in the GhERF13.12 transgenic lines after salt treatment (Figure 5G). This is in agreement with the lower amounts of MDA accumulated in WT than the GhERF13.12 silenced plants after salt treatment (Figure 7E). In addition, after salt stress treatment, the transcript levels of salt stress-related genes were lower in the GhERF13.12 silenced plants than those in the control plants. The results suggest that GhERF13.12 may act as a stress regulator that increases the scavenging enzyme activity by maintaining ROS homeostasis.
It has been reported that ~6% of land in the world is affected by salt stress, including 22% of arable land and 33% of irrigated croplands used for agricultural practices (Munns and Tester, 2008; Shrivastava and Kumar, 2015; Patel et al., 2020). Cotton is one of the most important natural fiber crops, being used as an edible oil and biofuel, and often faces salinity stress during its lifespan, which reduces fiber production and quality (Longenecker, 1974). Previous genetic engineering studies have shown that ERF genes in cotton, such as GhERF2-GhERF6 (Champion et al., 2009; Jin et al., 2010) and GhDREB1 (Gao et al., 2009), are involved in the regulation of plant response to salinity. The results provide a meaningful reference for future research on salt tolerance of upland cotton, and show that GhERF13.12 is an important candidate gene to improve cotton salt tolerance.
In this study, we identified ERF subfamily B3 group gene family members in G. arboreum, G. hirsutum, and G. raimondii. Phylogenetic analysis of ERF subfamily B3 group genes in Arabidopsis and three cotton species made it possible to divide these genes into three distinct groups. Cotton ERF subfamily B3 group genes were highly conserved during the course of evolution. The large-scale gene family expansion of the ERF subfamily B3 group gene family in G. hirsutum occurred because of segmental or whole genome duplication events, and most genes show typical signatures of purifying selection. At the same time, the GhERF13.12 sequence is related to constitutive salt stress responses, and the overexpression of GhERF13.12 transgenic plants have enhanced salt stress tolerance. Furthermore, the overexpression of GhERF13.12 upregulates transcripts of genes that participate in ABA signaling, proline biosynthesis, and ROS scavenging. These genes lead to increased ABA and proline content, and reduced ROS accumulation, improving tolerance under salt stress conditions. However, the transcript levels of these genes were downregulated in GhERF13.12 silenced plants after salt treatment. The results of this study will help to clarify the evolution of the ERF B3 subfamily genes in cotton and provide a basis for improving plant tolerance to abiotic stress. In the future, further studies are needed to understand the signal network and the exact function of the GhERF13.12 gene, along with the molecular mechanism of salt tolerance regulation.
Publicly available datasets were analyzed in this study. This data can be found at: https://www.cottongen.org/data/download/genome_diploid_A_nd_D5 and https://www.cottongen.org/data/download/genome_tetraploid/AD1#CALL.
LLu, GQ, QC, and ZL: conceptualization. GQ, GC, SL, and LLi: software. LLu and GQ: writing—original draft. YW, JL, and MP: formal analysis. MP and WQ: investigation. LLu, MP, and SM: data curation. WQ and SM: methodology. GQ and ZL: writing—review and editing. QC and ZL: supervision. All the authors have read and agreed to the published version of the manuscript.
This study was supported by the National Natural Science Foundation of China (Project 32001597) and the State Key Laboratory of Cotton Biology Open Fund (Grant No. CB2020A02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.705883/full#supplementary-material
Supplementary Figure 1. Conserved amino acid sequence in three cotton species, such as G. arboreum, G. raimondii, and G. hirsutum.
Supplementary Figure 2. Collinearity analysis of the GhERF subfamily B3 group genes. Collinearity analysis of G. hirsutum ERF subfamily B3 group genes among and within the A- and D-subgenomes is represented by the red lines.
Supplementary Figure 3. Sequence alignment among ERF13 proteins. Sequence alignment of A. thaliana and G. hirsutum ERF13 proteins depicted β-1, β-2, β-3, and α-helix regions, and a putative AP2/ERF domain (red region).
Supplementary Figure 4. Transcript levels and hypocotyl length in GhERF13.12 transgenic Arabidopsis. (A) Transcript levels of GhERF13.12 in transgenic Arabidopsis as measured by RT-PCR. (B) Determination of hypocotyl length in GhERF13.12 transgenic Arabidopsis with or without ACC in MS.
Supplementary Figure 5. Phenotype of virus-induced gene silencing (VIGS) plants and transcript levels of salt stress related genes. (A) Silenced phenotype of positive control seedlings during VIGS experiment. (B) Transcript levels of salt stress-related genes in GhERF13.12-silenced plants with salt stress.
Supplementary Table 1. List of primers used in this study (5′-3′).
Supplementary Table 2. Gene IDs of all identified ERF subfamily B3 group genes in A. thaliana, G. arboreum, G. raimondii, and G. hirsutum.
Supplementary Table 3. Type of gene duplication in G. arboreum, G. raimondii and G. hirsutum ERF subfamily B3 group genes.
Supplementary Table 4. Cotton ethylene response factor (ERF) subfamily B3 group genes orthologous and paralogous gene pairs and their respective Ka/Ks values in G. arboreum, G. raimondii, and G. hirsutum.
Supplementary Table 5. G. hirsutum ethylene response factor (ERF) subfamily B3 group genes orthologous and paralogous gene pairs and their respective Ka/Ks values.
ERF, Ethylene Response Factor; WGD, Whole Genome Duplication; MDA, Malondialdehyde; POD, Peroxidase; ROS, Reactive Oxygen Species; NAU, Nanjing Agricultural University; JGI, Joint Genome Institute; ICR, Institute of Cotton Research.
Ali, F., Qanmber, G., Wei, Z., Yu, D., Li, Y., Gan, L., et al. (2020). Genome-wide characterization and expression analysis of geranyl geranyl diphosphate synthase genes of cotton (Gossypium spp.) in plant development and abiotic stresses. BMC Genomics. 21, 1–15. doi: 10.21203/rs.3.rs-20104/v1
Anjum, N. A., Gill, R., Kaushik, M., Hasanuzzaman, M., Pereira, E., Ahmad, I., et al. (2015). ATP-sulfurylase, sulfur-compounds, and plant stress tolerance. Front. Plant Sci. 6:210. doi: 10.3389/fpls.2015.00210
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cao, S., Wang, Y., Li, X., Gao, F., Feng, J., and Zhou, Y. (2020). Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in ammopiptanthus nanus. Plants 9:455. doi: 10.3390/plants9040455
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). Ethylene response factor 96 positively regulates A rabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate–and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Champion, A., Hebrard, E., Parra, B., Bournaud, C., Marmey, P., Tranchant, C., et al. (2009). Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and xanthomonas. Mol. Plant Pathol. 10, 471–485. doi: 10.1111/j.1364-3703.2009.00549.x
Charon, C., Bruggeman, Q., Thareau, V., and Henry, Y. (2012). Gene duplication within the green lineage: the case of TEL genes. J. Exp. Bot. 63, 5061–5077. doi: 10.1093/jxb/ers181
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, E., Zhang, X., Yang, Z., Zhang, C., Wang, X., Ge, X., et al. (2019). BR deficiency causes increased sensitivity to drought and yield penalty in cotton. BMC Plant Biol. 19:220. doi: 10.1186/s12870-019-1832-9
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Debbarma, J., Sarki, Y. N., Saikia, B., Boruah, H. P. D., Singha, D. L., and Chikkaputtaiah, C. (2019). Ethylene response factor (ERF) family proteins in abiotic stresses and crispr-Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol. Biotechnol. 61, 153–172. doi: 10.1007/s12033-018-0144-x
Du, X. M., Huang, G., He, S. P., Yang, Z. E., Sun, G. F., Ma, X. F., et al. (2018). Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
Faiza, A., QANMBER, G., Yonghui, L., Shuya, M., Lili, L., Zuoren, Y., et al. (2019). Genome-wide identification of gossypium indeterminate domain genes and their expression profiles in ovule development and abiotic stress responses. J. Cotton Res. 2, 1–16. doi: 10.1186/s42397-019-0021-6
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gaeta, R. T., Pires, J. C., Iniguez-Luy, F., Leon, E., and Osborn, T. C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417. doi: 10.1105/tpc.107.054346
Gao, S. Q., Chen, M., Xia, L. Q., Xiu, H. J., Xu, Z. S., Li, L. C., et al. (2009). A cotton (Gossypium hirsutum) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311. doi: 10.1007/s00299-008-0623-9
Guan, Q. J., Wang, Z. J., Wang, X. H., Takano, T., and Liu, S. K. (2015). A peroxisomal APX from Puccinellia tenuiflora improves the abiotic stress tolerance of transgenic Arabidopsis thaliana through decreasing of H2O2 accumulation. J. Plant Physiol. 175, 183–191. doi: 10.1016/j.jplph.2014.10.020
Guo, B., Wei, Y., Xu, R., Lin, S., Luan, H., Lv, C., et al. (2016). Genome-wide analysis of APETALA2/ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS ONE 11:e0161322. doi: 10.1371/journal.pone.0161322
Heyman, J., Canher, B., Bisht, A., Christiaens, F., and De Veylder, L. (2018). Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 131:jcs208215. doi: 10.1242/jcs.208215
Jain, M., Mathur, G., Koul, S., and Sarin, N. B. (2001). Ameliorative effects of proline on salt stress-induced lipid peroxidation in cell lines of groundnut (Arachis hypogaea L.). Plant Cell Rep. 20, 463–468. doi: 10.1007/s002990100353
Jiang, Y., Yang, B., Harris, N. S., and Deyholos, M. K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58, 3591–3607. doi: 10.1093/jxb/erm207
Jin, L.-G., and Liu, J.-Y. (2008). Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton (Gossypium hirstum). Plant Physiol. Biochem. 46, 46–53. doi: 10.1016/j.plaphy.2007.10.004
Jin, L. G., Li, H., and Liu, J. Y. (2010). Molecular characterization of three ethylene responsive element binding factor genes from cotton. J. Integr. Plant Biol. 52, 485–495. doi: 10.1111/j.1744-7909.2010.00914.x
Jofuku, K. D., Den Boer, B., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, S.-B., Lee, S.-J., and Kim, S. Y. (2015). AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 34, 71–81. doi: 10.1007/s00299-014-1688-2
Lee, S.-J., Park, J. H., Lee, M. H., Yu, J.-H., and Kim, S. Y. (2010). Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol. 10:277. doi: 10.1186/1471-2229-10-277
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, J., Yu, D., Qanmber, G., Lu, L., Wang, L., Zheng, L., et al. (2019). GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China Life Sci. 62, 63–75. doi: 10.1007/s11427-018-9307-y
Li, Y., Zhang, H., Zhang, Q., Liu, Q. C., Zhai, H., Zhao, N., et al. (2019). An AP2/ERF gene, IbRAP2-12, from sweetpotato is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Sci. 281, 19–30. doi: 10.1016/j.plantsci.2019.01.009
Li, Z., Li, J., Bing, J., and Zhang, G. (2019). The role analysis of APX gene family in the growth and developmental processes and in response to abiotic stresses in Arabidopsis thaliana. Hereditas 41, 534–547. doi: 10.16288/j.yczz.19-026
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA 2/ethylene responsive factor (AP 2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, D. G., He, S. Z., Zhai, H., Wang, L. J., Zhao, Y., Wang, B., et al. (2014). Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell Tissue Organ Cult. 117, 1–16. doi: 10.1007/s11240-013-0415-y
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9:e1003790. doi: 10.1371/journal.pgen.1003790
Liu, Z., Qanmber, G., Lu, L., Qin, W., Liu, J., Li, J., et al. (2018). Genome-wide analysis of BES1 genes in gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 61, 1566–1582. doi: 10.1007/s11427-018-9412-x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, L., Yang, W. W., Liao, P., Guo, Y. W., Kumar, A., and Gao, W. (2019). Transcriptome analysis reveals differentially expressed ERF transcription factors associated with salt response in cotton. Plant Sci. 281, 72–81. doi: 10.1016/j.plantsci.2019.01.012
Longenecker, D. E. (1974). The influence of high sodium in soils upon fruiting and shedding, boll characteristics, fiber properties, and yields of two cotton species. Soil Sci. 118, 387–396. doi: 10.1097/00010694-197412000-00007
Lv, K., Li, J., Zhao, K., Chen, S., Nie, J., Zhang, W., et al. (2020). Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Science 292:110375. doi: 10.1016/j.plantsci.2019.110375
Ma, L., Hu, L., Fan, J., Amombo, E., Khaldun, A., Zheng, Y., et al. (2017). Cotton GhERF38 gene is involved in plant response to salt/drought and ABA. Ecotoxicology 26, 841–854. doi: 10.1007/s10646-017-1815-2
Malik, W., Shah, M. S. A., Abid, M. A., Qanmber, G., Noor, E., Qayyum, A., et al. (2018). Genetic basis of variation for fiber quality and quality related biochemical traits in Bt and non-Bt colored cotton. Int. J. Agric. Biol. 20, 2117–2124. doi: 10.17957/IJAB/15.0761
Markakis, M. N., De Cnodder, T., Lewandowski, M., Simon, D., Boron, A., Balcerowicz, D., et al. (2012). Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol. 12:208. doi: 10.1186/1471-2229-12-208
Mitsuda, N., and Ohme-Takagi, M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi: 10.1093/pcp/pcp075
Mizoi, J., Ohori, T., Moriwaki, T., Kidokoro, S., Todaka, D., Maruyama, K., et al. (2013). GmDREB2A; 2, a canonical dehydration-responsive element-binding protein2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161, 346–361. doi: 10.1104/pp.112.204875
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Patel, M. K., Kumar, M., Li, W. Q., Luo, Y., Burritt, D. J., Alkan, N., et al. (2020). Enhancing salt tolerance of plants: from metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cells 9:2492. doi: 10.3390/cells9112492
Paterson, A. H., Bowers, J. E., and Chapman, B. A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Nat. Acad. Sci. U.S.A. 101, 9903–9908. doi: 10.1073/pnas.0307901101
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D. C., et al. (2012). Repeated polyploidization of gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Qanmber, G., Ali, F., Lu, L., Mo, H., Ma, S., Wang, Z., et al. (2019a). Identification of histone H3 (HH3) genes in gossypium hirsutum revealed diverse expression during ovule development and stress responses. Genes 10:355. doi: 10.3390/genes10050355
Qanmber, G., Daoqian, Y., Jie, L., Lingling, W., Shuya, M., Lili, L., et al. (2018). Genome-wide identification and expression analysis of gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses. J. Cotton Research 1:1. doi: 10.1186/s42397-018-0004-z
Qanmber, G., Liu, J., Yu, D., Liu, Z., Lu, L., Mo, H., et al. (2019b). Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 20, 1750. doi: 10.3390/ijms20071750
Qanmber, G., Lu, L., Liu, Z., Yu, D., Zhou, K., Huo, P., et al. (2019c). Genome-wide identification of GhAAI genes reveals that GhAAI66 triggers a phase transition to induce early flowering. J. Exp. Bot. 70, 4721–4736. doi: 10.1093/jxb/erz239
Qiao, Z. X., Huang, B., and Liu, J. Y. (2008). Molecular cloning and functional analysis of an ERF gene from cotton (Gossypium hirsutum). Biochim. Biophys. Acta 1779, 122–127. doi: 10.1016/j.bbagrm.2007.10.003
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Rao, K. M., and Sresty, T. (2000). Antioxidative parameters in the seedlings of pigeonpea (Cajanus cajan (L.) Millspaugh) in response to Zn and Ni stresses. Plant Sci. 157, 113–128. doi: 10.1016/S0168-9452(00)00273-9
Rashid, M., Guangyuan, H., Guangxiao, Y., Hussain, J., and Xu, Y. (2012). AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evolutionary Bioinformatics 8, 321–355. doi: 10.4137/EBO.S9369
Ren, Z., Yu, D., Yang, Z., Li, C., Qanmber, G., Li, Y., et al. (2017). Genome-wide identification of the MIKC-type MADS-box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front. Plant Sci. 8:384. doi: 10.3389/fpls.2017.00384
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rong, W., Qi, L., Wang, A. Y., Ye, X. G., Du, L. P., Liang, H. X., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Satir, O., and Berberoglu, S. (2016). Crop yield prediction under soil salinity using satellite derived vegetation indices. Field Crops Res. 192, 134–143. doi: 10.1016/j.fcr.2016.04.028
Shrivastava, P., and Kumar, R. (2015). Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Wang, J., and Huang, R. F. (2019). Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 10:319. doi: 10.3389/fpls.2019.00319
Wang, L., Hu, Z., Zhu, M., Zhu, Z., Hu, J., Qanmber, G., et al. (2017). The abiotic stress-responsive NAC transcription factor SlNAC11 is involved in drought and salt response in tomato (Solanum lycopersicum L.). Plant Cell Tissue Organ Cult. 129, 161–174. doi: 10.1007/s11240-017-1167-x
Wang, L., Xu, C., Wang, C., and Wang, Y. (2012). Characterization of a eukaryotic translation initiation factor 5A homolog from Tamarix androssowii involved in plant abiotic stress tolerance. BMC Plant Biol. 12:118. doi: 10.1186/1471-2229-12-118
Wang, L., Yang, Z., Zhang, B., Yu, D., Liu, J., Gong, Q., et al. (2018). Genome-wide characterization and phylogenetic analysis of GSK gene family in three species of cotton: evidence for a role of some GSKs in fiber development and responses to stress. BMC Plant Biol. 18:330. doi: 10.1186/s12870-018-1526-8
Woodhouse, M. R., Schnable, J. C., Pedersen, B. S., Lyons, E., Lisch, D., Subramaniam, S., et al. (2010). Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homeologs. PLoS Biol. 8:e1000409. doi: 10.1371/journal.pbio.1000409
Wu, K., Tian, L., Hollingworth, J., Brown, D. C., and Miki, B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128, 30–37. doi: 10.1104/pp.010696
Xie, X. L., Xia, X. J., Kuang, S., Zhang, X. L., Yin, X. R., Yu, J. Q., et al. (2017). A novel ethylene responsive factor CitERF13 plays a role in photosynthesis regulation. Plant Sci. 256, 112–119. doi: 10.1016/j.plantsci.2016.11.001
Xing, L. P., Di, Z. C., Yang, W. W., Liu, J. Q., Li, M. N., Wang, X. J., et al. (2017). Overexpression of ERF1-V from haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8:1948. doi: 10.3389/fpls.2017.01948
Xiong, F., Zhuo, F., Reiter, R. J., Wang, L., Wei, Z., Deng, K., et al. (2019). Hypocotyl elongation inhibition of melatonin involves in repressing brassinosteroids biosynthesis in Arabidopsis. Front. Plant Sci. 10:1082. doi: 10.3389/fpls.2019.01082
Yamada, M., Morishita, H., Urano, K., Shiozaki, N., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2005). Effects of free proline accumulation in petunias under drought stress. J. Exp. Bot. 56, 1975–1981. doi: 10.1093/jxb/eri195
Yan, H., Hong, L., Zhou, Y., Jiang, H., Zhu, S., Fan, J., et al. (2013). A genome-wide analysis of the ERF gene family in sorghum. Genet. Mol. Res. 12, 2038–2055. doi: 10.4238/2013.May.13.1
Yang, Y. Q., and Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y. Q., and Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yang, Z., Qanmber, G., Wang, Z., Yang, Z., and Li, F. (2020). Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci. 25, 488–500. doi: 10.1016/j.tplants.2019.12.011
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Yu, D., Qanmber, G., Lu, L., Wang, L., Li, J., Yang, Z., et al. (2018). Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 18:350. doi: 10.1186/s12870-018-1545-5
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. doi: 10.1093/nar/gkt1064
Zhang, A. D., Liu, D. D., Hua, C. M., Yan, A., Liu, B. H., Wu, M. J., et al. (2016). The Arabidopsis gene zinc finger protein 3(ZFP3) Is involved in salt stress and osmotic stress response. PLoS ONE 11:e0168367. doi: 10.1371/journal.pone.0168367
Zhang, G., Chen, M., Chen, X., Xu, Z., Guan, S., Li, L.-C., et al. (2008). Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (glycine max L.). J. Exp. Bot. 59, 4095–4107. doi: 10.1093/jxb/ern248
Zhang, H., Li, W., Chen, J., Yang, Y., Zhang, Z., Zhang, H., et al. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. doi: 10.1007/s11103-006-9072-4
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, T. Z., Hu, Y., Jiang, W. K., Fang, L., Guan, X. Y., Chen, J. D., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, X., Henriques, R., Lin, S.-S., Niu, Q.-W., and Chua, N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641. doi: 10.1038/nprot.2006.97
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., and Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. doi: 10.1007/s11103-011-9732-x
Zhang, Z., Wang, J., Zhang, R., and Huang, R. (2012). The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71, 273–287. doi: 10.1111/j.1365-313X.2012.04996.x
Zhang, Z. J., Li, F., Li, D. J., Zhang, H. W., and Huang, R. F. (2010). Expression of ethylene response factor JERF1 in rice improves tolerance to drought. Planta 232, 765–774. doi: 10.1007/s00425-010-1208-8
Zhao, Y., Yang, Z., Ding, Y., Liu, L., Han, X., Zhan, J., et al. (2019). Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 286, 28–36. doi: 10.1016/j.plantsci.2019.05.021
Zheng, L., Wu, H., Qanmber, G., Ali, F., Wang, L., Liu, Z., et al. (2020). Genome-Wide study of the GATL gene family in Gossypium hirsutum L. Reveals that GhGATL genes act on pectin synthesis to regulate plant growth and fiber elongation. Genes 11:64. doi: 10.3390/genes11010064
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: ERF, phylogenetic analysis, gene duplication, collinearity analysis, ectopic expression, salt stress, gene silencing
Citation: Lu L, Qanmber G, Li J, Pu M, Chen G, Li S, Liu L, Qin W, Ma S, Wang Y, Chen Q and Liu Z (2021) Identification and Characterization of the ERF Subfamily B3 Group Revealed GhERF13.12 Improves Salt Tolerance in Upland Cotton. Front. Plant Sci. 12:705883. doi: 10.3389/fpls.2021.705883
Received: 06 May 2021; Accepted: 05 July 2021;
Published: 09 August 2021.
Edited by:
Hong Zhang, Texas Tech University, United StatesReviewed by:
Wenliang Xu, Central China Normal University, ChinaCopyright © 2021 Lu, Qanmber, Li, Pu, Chen, Li, Liu, Qin, Ma, Wang, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quanjia Chen, Y2hxamlhQDEyNi5jb20=; Zhao Liu, bGl1emhhb2NhYXNAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.