- 1Key Laboratory of Plant Functional Genomics of the Ministry of Education, Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, Jiangsu Key Laboratory of Crop Genetics and Physiology, College of Agriculture, Yangzhou University, Yangzhou, China
- 2Department of Biotechnology, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, China
Rice is the most important source of food worldwide, providing energy, and nutrition for more than half of the population worldwide. Rice grain quality is a complex trait that is affected by several factors, such as the genotype and environment, and is a major target for rice breeders. Cis-regulatory elements (CREs) are the regions of non-coding DNA, which play a critical role in gene expression regulation. Compared with gene knockout, CRE modifications can fine-tune the expression levels of target genes. Genome editing has provided opportunities to modify the genomes of organisms in a precise and predictable way. Recently, the promoter modifications of coding genes using genome editing technologies in plant improvement have become popular. In this study, we reviewed the results of recent studies on the identification, characterization, and application of CREs involved in rice grain quality. We proposed CREs as preferred potential targets to create allelic diversity and to improve quality traits via genome editing strategies in rice. We also discussed potential challenges and experimental considerations for the improvement in grain quality in crop plants.
Introduction
Rice provides major nutrients and energy for more than half of the population worldwide. In the last 30 years, rice yield has been continuously improved through implementing a series of breeding programs (Xu et al., 2020). Meanwhile, the demand for high quality, multiresistance, and wide adaptability of rice variants is also increasing, especially for the high-quality rice under conditions of improved living standards (Rao et al., 2014). Generally, rice grain quality is a combination of milling, appearance, eating, cooking, nutritional, and hygiene traits (Zhou et al., 2020). Therefore, to meet the needs of consumers and producers, researchers have to understand the molecular mechanisms and genetic basis that determines rice quality, and breeders and seed companies have to develop rice varieties with excellent quality and high yield. However, the direct regulation of coding genes for target traits is difficult and is often accompanied by negative effects; therefore, it cannot achieve the expected effect. For instance, Pérez et al. (2019) used CRISPR/Cas9 to introduce mutations affecting the Wx gene [encoding granule-bound starch synthase I (GBSSI)] in the rice endosperm. The amylose content (AC) declined to as low as 5% in homozygous seeds, accompanied by abnormal cellular organization in the aleurone layer and amorphous starch grain structures. Plant genetic engineering efforts to improve grain quality in crop plants using well-characterized promoter elements to modify the expression of regulatory genes and/or transcription factors (TFs) have proven to be advantageous (Yang et al., 2019). Therefore, in this perspective, we mainly discussed the identification and analysis of cis-regulatory elements (CREs) involved in rice grain quality and the modification of CREs via genome editing technologies for the improvement in rice grain quality.
Genetics and Genomics of Rice Grain Quality
In general, four main quality traits, namely, milling properties, appearance, nutritional value, and cooking quality, are widely used to assess rice grain quality (Zhou et al., 2020). With the rapid development of high-throughput technologies and functional genomics, many genes controlling important quality traits have been cloned in rice, and some molecular mechanisms have been characterized (Zhou et al., 2020). Milling quality is a complex grain trait, including the recovery of brown, milled, and head rice. Several major quantitative trait loci (QTLs) are associated with rice milling quality, such as qBRR-10, which influence brown rice recovery (Ren et al., 2016). Rice grain shape is an important index of appearance quality and yield, and several related genes and TFs have been studied (Huang et al., 2013). For example, GS3, GW5, GLW7, GW8, GS2, and GS9 are the main genes or transcription regulators affecting variations in rice grain shape and size. In addition, genes such as GL7/GW7, GL3.1, GS5, GW2, and GL3.3, which were identified in natural variants, have profound effects on grain shape and functions, and their mechanisms have been studied (Huang et al., 2013). The composition and structure of starch play crucial roles in rice grain quality, especially for eating and cooking quality. Enzymes involved in the starch synthesis pathway (including GBSS, SS, SBE, ISA, and PUL) and associated TFs (i.e., Dull, OsEBP89, OsRSR1, and OsbZIP58) have been clearly determined in rice (Zhou et al., 2020). Many major functional genes have been identified, which affect nutrients such as seed proteins, essential amino acids, vitamins, minerals, and anthocyanins (Das et al., 2020). Similarly, grain quality-related genes and their metabolic pathways have been studied in other crops, and the results have been discussed in a recent review (Birla et al., 2017). The characterization of these quality-related genes has laid the foundation for the improvement in rice quality. Although many genes related to crop grain quality have been explored, and their genetic mechanisms have been analyzed, how these genes can be better used in breeding production and commercial utilization requires further study.
Cis-Regulatory Elements and Their Mining in Plants
Fundamentally, in addition to genes, many elements present in the plant genome control the gene expression levels via interactions with DNA or regulatory proteins. Limited genetic diversity restricts the amount and effectiveness of the improvement in rice quality. Recent results showed that genetic changes in CREs of genes play important roles in shaping phenotypic diversity by altering gene expression (Swinnen et al., 2016).
Cis-Regulatory Elements: Important Regulators of Gene Expression
The CREs are the noncoding DNA containing binding sites for TFs or other regulatory molecules that affect transcription, and they ultimately guide plant growth and development, cell differentiation, and responses to various stresses (Lu et al., 2018). Therefore, the whole genome identification and functional characterization of CREs involved in the DNA–protein interactions is a key aspect to understand plant transcription regulation (Lu et al., 2018). Generally, CREs in eukaryotes include promoters, enhancers, and other CREs, among which enhancers tend to be much more variable (Meng et al., 2021). Enhancers that drive transcription are independent of their distance and location from their cognate promoters, which allows a gene to be regulated by multiple remote enhancers with different spatiotemporal activities (Meng et al., 2021). The sequence structure of promoters of quality-related genes is relatively simple and conserved, and their functions have been clarified in rice. The identification and functional analysis of CREs are also in progress in rice (Swinnen et al., 2016). More comprehensive collections and analyses of CREs in rice are necessary, which will accelerate the fine-tuning of the improvement in rice quality via CRE editing.
Mining CREs Based on “Omics”
The systematic identification of CREs in plant genomes is critically important to understand the transcriptional regulation and its exploitation for the improvement in quality. Some important results have been obtained using approaches involving sequence conservation within short distances from target genes (Huang et al., 2020a). Additionally, some databases, such as PlantCARE, which store all plant transcription sites, consensus sequences, and matrices described in the literature, can also be used to predict possible CREs for one or more genes (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Several limitations are as follows: sequence conservation can be restricted, CREs that are far from their target genes will not be detected, and we have less information regarding their tissue specificity and functionality (Lu et al., 2018).
Generally, CREs are preferentially located in the accessible chromatin regions (España et al., 2017). Therefore, genomics and epigenomics are used to identify and analyze CREs. For example, the alignment of the upstream sequences of OsPLDα1 orthologs across 34 rice accessions revealed sequence variations and identified CREs involved in differential transcription of orthologs, which resulted in the low expression of OsPLDα1 and reduced free fatty acid content in the oil, facilitating good quality bran oil (Kaur et al., 2020). A genome-wide association study (GWAS) screen of 45 natural accessions and the pif4 mutant in Arabidopsis identified CREs affecting SAUR26 gene expression (Wang et al., 2021). Similar CRE mining strategies have been reported in rice (Ho and Geisler, 2019). The whole genome transcriptome profiling using microarrays was employed to discover CREs associated with drought and salinity stress tolerance in rice (Mishra et al., 2018).
In addition, CREs can be identified based on their elevated sensitivity to enzymes such as the bacterial transposase Tn5, DNase I, and micrococcal nuclease (MNase) (Zhang et al., 2016; Lu et al., 2017; Zhao et al., 2020). The coupling of chromatin accessibility assays with the high-throughput DNA sequencing, such as DNase-seq, MNase-seq, self-transcribing active regulatory region sequencing (STARR-seq), and assay for transposase-accessible chromatin using sequencing (ATAC-seq), represents an important technological development and has enabled the identification of CREs on a genome-wide scale (Lu et al., 2018). Numerous putative CREs were identified in plant species through the strategies of genetic, epigenomic, and functional molecular characterization (Lu et al., 2019). A major limitation to these assays is the lack of cell-type resolution and the paucity of information regarding which CREs function in specific tissues or cell types at the genome-wide scale, especially in endosperm tissue, which is closely related to rice quality.
Moreover, ribosome profiling (ribo-seq) provides a viable strategy to analyze active translation by determining ribosome occupancy in a transcriptome-wide manner, which can then be used to identify valid upstream open reading frames (uORFs), a type of CRE (Lulla et al., 2019). However, the large-scale identification of uORFs has not been reported in rice. Thus, ribo-seq is expected to be an effective tool to identify rice quality-related uORFs. Furthermore, combined with the multiomics data, such as RNA sequencing (RNA-seq), chromatin immunoprecipitation sequencing (ChIP-seq), and proteomics, CREs and their related TFs could be mined more accurately (Farmer et al., 2021).
The Key Roles of Cis-Regulatory Elements in the Improvement in Rice Quality
Unlike changes to protein-coding genes, which often result in easily interpretable loss-of-function alleles, the mutations of CREs offer the potential of fine-tuning gene expression without other adverse effects, leading to improved rice quality (Huang et al., 2020a; Zeng et al., 2020). Therefore, more attention should be paid to CREs and their influence on gene expression.
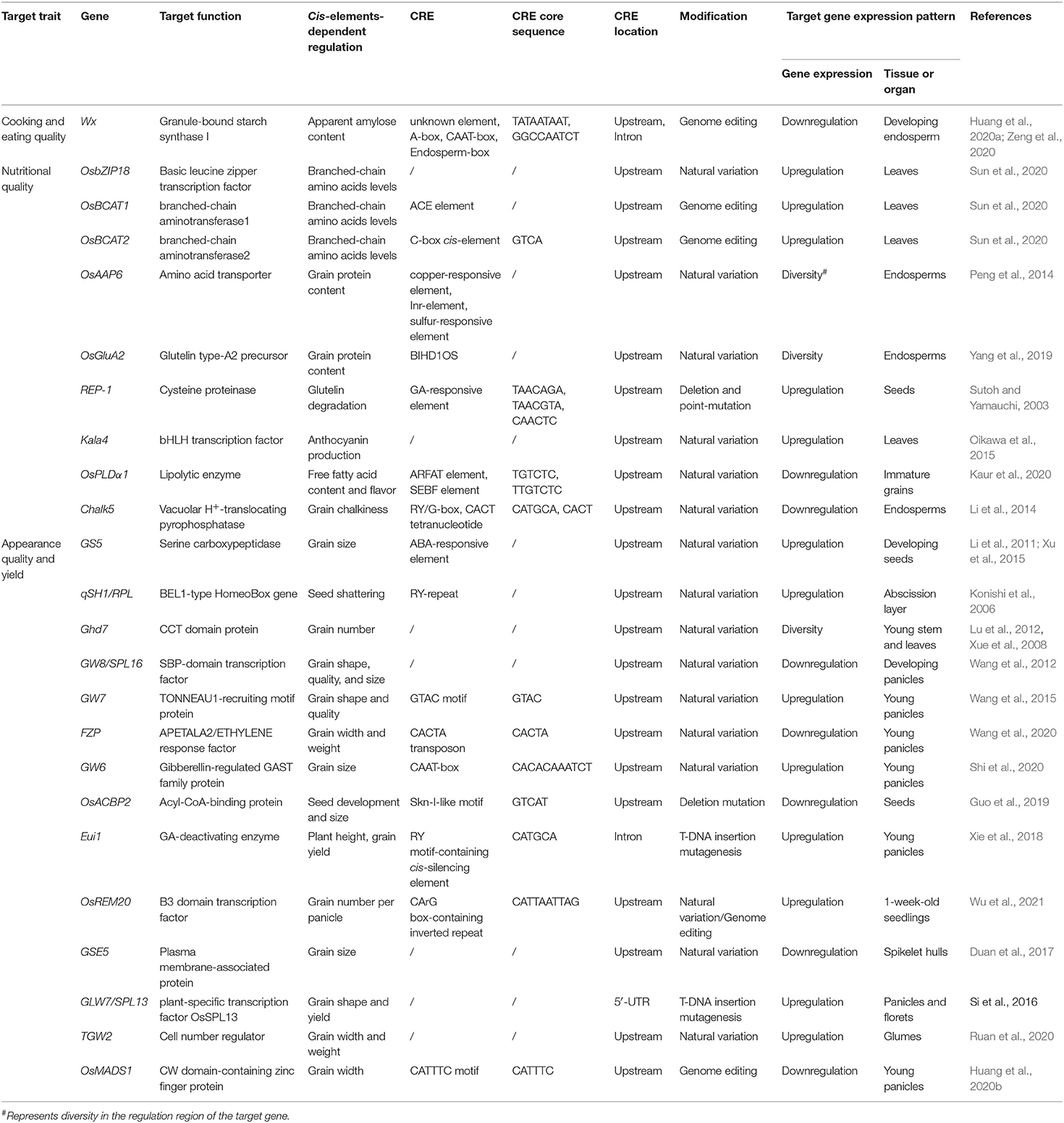
Employing both bioinformatic and experimental methods, CREs involved in gene regulation have been identified, and most of the genes affected by these CREs have been found to encode TFs that regulate plant development (España et al., 2017). Several universal CREs have been identified in the 5′ upstream region of the starch synthetase gene, lysine metabolism genes, and seed-storage protein genes in rice and/or other crops (Table 1) (Chen et al., 2012). Recent studies have shown that CREs play an important role in improving rice quality traits (Table 1).
The ARFAT and SEBF elements have been identified as CREs that might act as repressors in regulating OsPLDα1 expression, which lead to decreased free fatty acid content in oil and improve the flavor and quality of rice bran oil (Table 1) (Kaur et al., 2020). Two common variations in the potential CREs of the OsAAP6 5′-untranslated region (5′-UTR) seem to be associated with grain protein content diversity and nutritional quality, mainly in indica cultivars (Peng et al., 2014). A single nucleotide polymorphism (SNP) located in the OsGluA2 promoter region is associated with its transcript expression level and grain protein content diversity (Yang et al., 2019). For amino acids, natural variations in the OsbZIP18 promoter contribute to branched-chain amino acid levels in rice (Sun et al., 2020). Two consensus nucleotide polymorphisms in the Chalk5 promoter in rice varieties might partly account for the differences in Chalk5 mRNA levels that contribute to natural variation in grain chalkiness (Li et al., 2014). GRAIN WIDTH 7 (GW7) is an important gene that controls cell division in the spikelet hulls, and its expression is regulated by the repressive TF, i.e., GRAIN WIDTH8 (GW8). A mutation in the CRE in the promoter of GW7 led to enhanced GW7 expression and ultimately to the improved yield and grain quality (Sakamoto and Matsuoka, 2008; Wang et al., 2015). Similarly, natural variation in the promoter of GSE5 contributes to grain size diversity, and that in TGW2 determines rice grain width and weight (Duan et al., 2017; Ruan et al., 2020). A novel variation in the FRIZZLE PANICLE (FZP) gene promoter improved rice grain number and yield (Wang et al., 2020).
Upstream Open Reading Frames: Important Cis-Regulatory Elements in the 5′ Leading Sequence
Upstream open reading frames, as translational regulatory elements, are located in the 5′-UTR of eukaryotic mRNAs, and generally inhibit the translation initiation of downstream primary ORFs (pORFs) through ribosome stalling (Kurihara, 2020). In plants, uORFs have been predicted in ~30% of the 5′-UTRs of genes, and some of these uORFs have been reported to regulate crucial growth and developmental processes (von Arnim et al., 2014). Increasing numbers of excellent crop genes have been identified and characterized (Zhou et al., 2020). Furthermore, to improve crop characteristics, many genes that regulate important traits are required to have a high translation rate, rather than their functional loss or reduction (Xu et al., 2017; Reis et al., 2020). Based on the knowledge that uORFs negatively affect the translation of the pORF, the strategy of modulating uORFs to fine-tune the translation could be used to analyze gene function and improve crop traits. For example, uORF TBF1-mediated translation enabled engineered Arabidopsis and rice broad-spectrum disease resistance without any reduction in grain yield (Xu et al., 2017).
Genome Editing Toward Better Grain QualitY via Targeting Cis-Regulatory Elements
All the earlier studies suggested that CRE modification can regulate the expression of key genes for rice quality and effectively improve grain quality. However, the identification of natural mutations in the promoter regulatory regions of the gene is time-consuming and difficult, which slows the improvement in rice quality. Recently, genome editing technology has been developed and optimized and has been applied successfully to a large number of plants, which has accelerated the identification and application of regulatory elements in gene promoter regions in rice (Huang et al., 2020a; Zeng et al., 2020). Genome editing is a versatile, relevant, and preferred technique for functional genomics, as well as crop improvement, involving introducing DNA mutations in the form of deletions and/or insertions or base substitutions in target gene sequences (Fiaz et al., 2019; Tabassum et al., 2021).
The starch synthase gene Wx is very important for rice eating and cooking quality; therefore, it has been a popular target for study. Zeng et al. (2020) disrupted the Endosperm-box, A-box, and CAAT-box of the promoter sequence and intron region of the Wx gene using genome editing, which generated new Wx alleles producing various ACs by quantitative regulation of its expression. Novel Wx alleles, in which CREs in the Wx promoter near a predicted TATA-box were edited using the CRISPR/Cas9 system, produced fine-tuned amylose levels and improved the rice grain quality (Huang et al., 2020a). The deletion mutants of the CATTTC motif exhibited the lower expression of OsMADS1 and produced narrower rice grains (Huang et al., 2020b). Wu et al. (2021) enhanced rice grain production by manipulating the CArG box-containing inverted repeat sequence of OsREM20. Although there are only a limited number of studies demonstrating successful editing of CREs for crop improvement, it is anticipated that genome editing techniques such as CRISPR/Cas9 will lead to further CRE editing to improve rice grain quality.
For uORFs, editing the uORF of LsGGP2 increased oxidative stress tolerance and the ascorbate content by ~150% in lettuce (Zhang et al., 2018). In Arabidopsis, a conserved peptide uORF (CPuORF33) was identified in the 5′-UTR of AtHB1 mRNA, which ensures a relatively low level of AtHB1 expression in aerial parts and avoids adverse phenotypes (Ribone et al., 2017). Similarly, there are several studies on the regulation of uORFs in morphogenesis, signaling pathways, and nutrient absorption stress response in Arabidopsis (Zhang et al., 2020). Reis et al. (2020) identified the PHO1 uORF in the genomes of crops such as rice, maize, barley, and wheat, which improved plant growth under inorganic phosphorus (Pi)-deficient conditions. A tandem repeat sequence in the 5′-UTR of GLW7 alters its expression by affecting transcription and translation, resulting in enhanced rice grain length and yield (Si et al., 2016). Hence, uORFs have great potential to improve rice grain quality by positively regulating target genes.
Conclusion and Future Perspectives
The aim of the plant functional genomics is to explore the key genes in plant growth and development, to determine their regulatory mechanisms, and to fine-tune the gene expression effectively, with the aim of improving traits for research and/or commercial use. Crop quality is an important trait whose further improvement requires increased research resources. Recently, several crop quality-related genes and metabolic pathways have been identified and explored (Zhou et al., 2020). In addition, the high-throughput technologies have accelerated the identification and analysis of these functional quality-related genes (Hernandez-Garcia and Finer, 2014). However, it is difficult to achieve the desired improvement in quality through the direct manipulation of these elite genes. The identification and modification of the CREs of these genes would provide an appropriate approach to modulate their expression (Huang et al., 2020c). The popularity of genome editing further confirms that CREs are good potential targets to create new alleles for the improvement in rice quality in transgene-free derivatives (Figure 1).
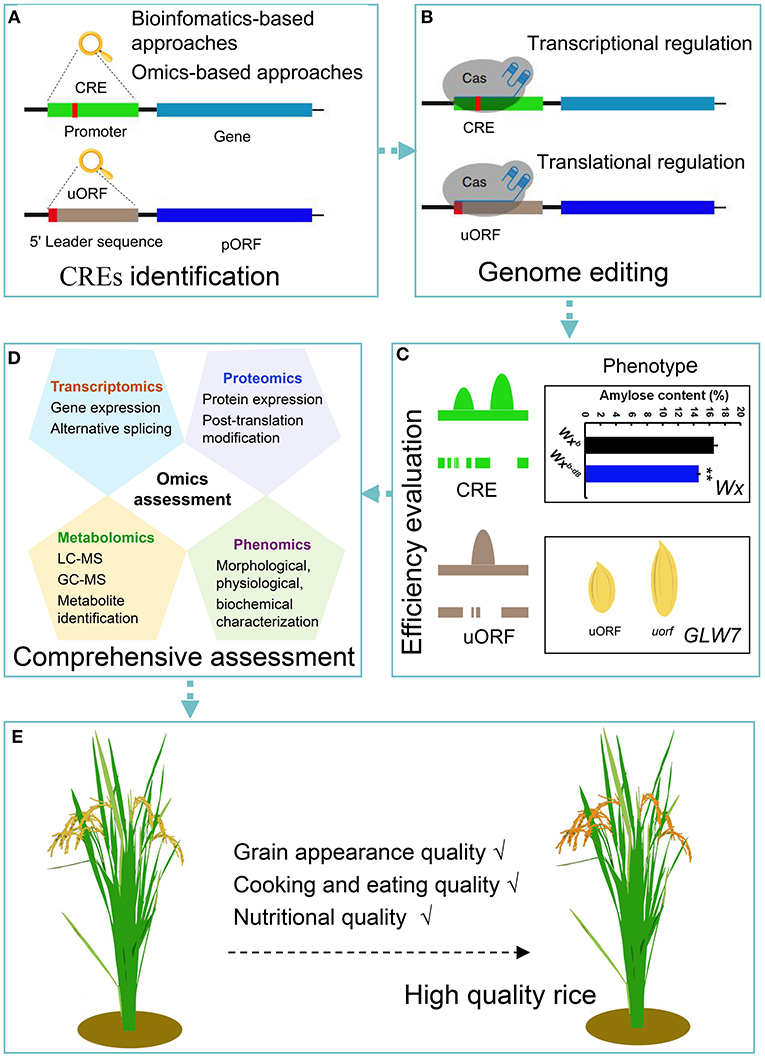
Figure 1. Schematic representation of the workflow depicting the application of genome editing approaches to obtain high grain quality rice. (A) CREs are identified by multiomics strategies and bioinformatic approaches. Cis-regulatory sequences are linear nucleotide fragments of non-coding DNA. Their localization and orientation in relation to genes and activity vary (Verter and Botha, 2010). uORFs, as translational regulatory elements, are located in the 5′ leader sequence of eukaryotic mRNAs and generally inhibit the translation initiation of downstream pORFs (Kurihara, 2020). (B) Selection of the desired target DNA sequences of CREs and uORFs, and recognition of PAM sequences for genome editing, which regulates gene expression at the transcription and translation levels, respectively. (C) Efficiency evaluation using molecular identification and phenotypic analysis. For example, a genome editing strategy was used to edit the core promoter region of the Wx gene, and the obtained Wxb−d8 mutant had a nine nucleotide deletion (Huang et al., 2020a). In the Wxb−d8 mutant, the predicted core promoter region was disrupted, which decreased Wx expression compared with the wild-type Wxb and reduced the amylose content (%) in the mature rice seeds (Huang et al., 2020a). The same method is applicable to edit the predicted uORF of GLW7 (Si et al., 2016). The deletion of predicted uORF in the GLW7 5′ leader sequence causing increased expression levels of the GLW7 protein and enhanced rice grain length. Colored peaks represent different TF-binding events within CREs or uORFs, and peak height indicates the chromatin accessibility from genomic data sets (i.e., DHase-seq and ATAC-seq). Lines with spaces beneath the TF-binding peak indicate destroyed CRE or uORF sequences. (D) Comprehensive assessment of edited rice using multiomics strategies. (E) Obtaining the desired high-quality rice. CRE, Cis-regulatory element; uORF, upstream open reading frame; pORF, primary open reading frame; PAM, protospacer adjacent motif; Wx, the Waxy gene encoding granule-bound starch synthase I, which controls amylose synthesis in rice endosperm; Wxb, Nipponbare carrying the Wxb allele; Wxb−d8, the Wxb-d8 mutant with nine nucleotide deletion of the core promoter region in Nipponbare (Wxb). GLW7, which is encoding the plant-specific TF OsSPL13, positively regulates cell size in the grain hull, resulting in enhanced rice grain length and yield (Si et al., 2016); TF, transcription factor.
Recently developed omics approaches, such as DNase-seq, ATAC-seq, and ribo-seq, have identified certain CREs and uORFs. Thus, a more comprehensive analysis of CREs in promoter regions and uORFs in 5′ leading sequences will increase opportunities for quality-related genome editing. Additionally, these techniques are complex and have limitations, especially in the study of CREs and uORFs related to endosperm traits in rice. Hence, the development of an effective and simple method or system for the CRE identification in rice seeds is essential to improve rice quality. Moreover, CREs are very short (usually only a few nucleotides), and constructing a uORF mutant requires the start codon to be modified; therefore, more precise genome editing techniques should be considered to avoid limited protospacer adjacent motif (PAM) sites, off-target mutations, and low homology-directed repair (HDR) efficiency.
In addition to the technological issues, there are significant gaps in our knowledge of gene regulation in most species. At present, studies on CREs and uORFs have mainly focused on their identification and functional analysis (Hernandez-Garcia and Finer, 2014). Given the importance of CREs for gene expression in crops, in-depth studies, such as CRE regulatory mechanisms and related metabolic connections, require further research. Thus, the analyses of the CREs related to crop quality and their transcriptional and translational regulatory modifications are essential. It is hoped that the combination of CREs and genome editing technologies will enable the simultaneous manipulation of multiple traits in rice (Figure 1).
Moreover, it is not easy to forensically detect genome editing events at the molecular level, especially as no foreign DNA exists in the line in which the regulatory element(s) are subtly edited. Given this limitation, the downstream “omics” technologies that can reveal the effects of the edits, such as proteomics and metabolomics, should be considered to fully assess the changes of proteins and/or their compositions in novel foodstuffs from the edited crops. The integration of the in-depth understanding of gene regulatory mechanisms and related networks, and genome editing to identify and modify CREs at the single nucleotide level in plant genomes, might represent a promising strategy for future crop improvement.
Author Contributions
QY, YD, and TZ organized and wrote the manuscript. JZ collected and summarized some points. QL and DZ provided critical evaluation and edited the text. All the authors have read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31801322 and 31971914), the Young Elite Scientists Sponsorship Program by the China Association for Science and Technology (2018QNRC001), the Fund of Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Jiangsu Province Government Project (BK2018003), the China Postdoctoral Science Foundation (2021M690552), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (SJCX21_1607).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ARFAT, auxin response factor; AtHB1, Arabidopsis thaliana HomeoBox 1; FZP, Frizzle panicle; GBSS, Granule-bound starch synthase; GL3.1, Grain length 3.1; GL3.3, Grain length 3.1; GLW7, Grain length and weight on chromosome 7; GS2, Grain size on chromosome 2; GS3, Grain size on chromosome 3; GS5, Grain size on chromosome 5; GS9, Grain shape on chromosome 9; GSE5, Grain size on chromosome 5; GW2, Grain width 2; GW5, QTL for grain width and weight on chromosome 5; GW7, Grain width 7; GW8, Grain width 8; ISA, Isoamylase; LsGGP2, GDP-L-galactose phosphorylase 2; OsAAP6, Amino acid permease 6; OsbZIP18, basic leucine zipper transcription factor 18; OsbZIP58, basic leucine zipper transcription factor 58; OsEBP89, Oryza sativa ethylene-responsive element binding protein, clone 89; OsGluA2, glutelin type-A2 precursor; OsMADS1, MADS-domain transcription factor; OsPLDα1, Phospholipase D alpha 1; OsREM20, Oryza sativa REPRODUCTIVE MERISTEM 20; OsRSR1, Rice starch regulator 1; OsSPL16, squamosa promoter binding protein-like 16; PHO1, PHOSPHATE1; PUL, pullulanase; qBRR-10, Brown rice rate on chromosome 10; qSH1, QTL of seed shattering in chromosome 1; SAUR26, SMALL AUXIN UP RNA gene 26; SBE, starch-branching enzyme; SEBF, silencing element binding factor; SS, soluble starch synthase; TGW2, 1000-grain weight 2.
References
Birla, D. S., Malik, K., Sainger, M., Chaudhary, D., Jaiwal, R., and Jaiwal, P. K. (2017). Progress and challenges in improving the nutritional quality of rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 57, 2455–2481. doi: 10.1080/10408398.2015.1084992
Chen, Y., Wang, M., and Ouwerkerk, P. B. F. (2012). Molecular and environmental factors determining grain quality in rice. Food Energy Secur. 1, 111–132. doi: 10.1002/fes3.11
Das, P., Adak, S., and Majumder, A. L. (2020). Genetic manipulation for improved nutritional quality in rice. Front. Genet. 11:776. doi: 10.3389/fgene.2020.00776
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
España, A. P., Santiago-Algarra, D., Pradel, L., and Spicuglia, S. (2017). High-throughput approaches to study cis-regulating elements. Biol. Aujourdhui. 211, 271–280. doi: 10.1051/jbio/2018015
Farmer, A., Thibivilliers, S., Ryu, K. H., Schiefelbein, J., and Libault, M. (2021). Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant 14, 372–383. doi: 10.1016/j.molp.2021.01.001
Fiaz, S., Ahmad, S., Noor, M. A., Wang, X., Younas, A., Riaz, A., et al. (2019). Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int. J. Mol. Sci. 20:888. doi: 10.3390/ijms20040888
Guo, Z. H., Haslam, RP, Michaelson, L. V., Yeung, E. C., Lung, S. C., Napier, J. A., et al. (2019). The overexpression of rice ACYL-CoA-BINDING PROTEIN2 increases grain size and bran oil content in transgenic rice. Plant J. 100, 1132–1147. doi: 10.1111/tpj.14503
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217–218:109–119. doi: 10.1016/j.plantsci.2013.12.007
Ho, C. L., and Geisler, M. (2019). Genome-wide computational identification of biologically significant cis-regulatory elements and associated transcription factors from rice. Plants 8:441. doi: 10.3390/plants8110441
Huang, L., Li, Q., Zhang, C., Chu, R., Gu, Z., Tan, H., et al. (2020a). Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 18, 2164–2166. doi: 10.1111/pbi.13391
Huang, L., Sreenivasulu, N., and Liu, Q. (2020c). Waxy editing: old meets new. Trends Plant Sci. 25, 963–966. doi: 10.1016/j.tplants.2020.07.009
Huang, R., Jiang, L., Zheng, J., Wang, T., Wang, H., Huang, Y., et al. (2013). Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18, 218–226. doi: 10.1016/j.tplants.2012.11.001
Huang, Y., Bai, X., Cheng, N., Xiao, J., Li, X., and Xing, Y. (2020b). Wide Grain 7 increases grain width by enhancing H3K4me3 enrichment in the OsMADS1 promoter in rice (Oryza sativa L.). Plant J. 102, 517–528. doi: 10.1111/tpj.14646
Kaur, A., Neelam, K., Kitazumi, A., Kaur, K., Sharma, P., Mangat, G. S., et al. (2020). Novel cis-acting regulatory elements in wild Oryza species impart improved rice bran quality by lowering the expression of phospholipase D alpha1 enzyme (OsPLDα1). Mol. Biol. Rep. 47, 401–422. doi: 10.1007/s11033-019-05144-4
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Kurihara, Y. (2020). uORF shuffling fine-tunes gene expression at a deep level of the process. Plants 9:608. doi: 10.3390/plants9050608
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, Y., Fan, C., Xing, Y., Yun, P., Luo, L., Yan, B., et al. (2014). Chalk5 encodes a vacuolar H(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Lu, L., Yan, W., Xue, W., Shao, D., and Xing, Y. (2012). Evolution and association analysis of Ghd7 in rice. PLoS ONE 7:e34021. doi: 10.1371/journal.pone.0034021
Lu, Z., Hofmeister, B. T., Vollmers, C., DuBois, R. M., and Schmitz, R. J. (2017). Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res 45:e41. doi: 10.1093/nar/gkw1179
Lu, Z., Marand, A. P., Ricci, W. A., Ethridge, C. L., Zhang, X., and Schmitz, R. J. (2019). The prevalence, evolution and chromatin signatures of plant regulatory elements. Nat. Plants 5, 1250–1259. doi: 10.1038/s41477-019-0548-z
Lu, Z., Ricci, W. A., Schmitz, R. J., and Zhang, X. (2018). Identification of cis-regulatory elements by chromatin structure. Curr. Opin Plant Biol. 42, 90–94. doi: 10.1016/j.pbi.2018.04.004
Lulla, V., Dinan, A. M., Hosmillo, M., Chaudhry, Y., Sherry, L., Irigoyen, N., et al. (2019). An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat. Microbiol. 4, 280–292. doi: 10.1038/s41564-018-0297-1
Meng, F., Zhao, H., Zhu, B., Zhang, T., Yang, M., Li, Y., et al. (2021). Genomic editing of intronic enhancers Unveils their role in fine-tuning tissue-specific gene expression in Arabidopsis thaliana. Plant Cell 25:koab093. doi: 10.1093/plcell/koab093
Mishra, P., Singh, N., Jain, A., Jain, N., Mishra, V.G.P., et al. (2018). Identification of cis-regulatory elements associated with salinity and drought stress tolerance in rice from co-expressed gene interaction networks. Bioinformation 14, 123–131. doi: 10.6026/97320630014123
Oikawa, T., Maeda, H., Oguchi, T., Yamaguchi, T., Tanabe, N., Ebana, K., et al. (2015). The birth of a black rice gene and its local spread by introgression. Plant Cell 27:2401–2414. doi: 10.1105/tpc.15.00310
Peng, B., Kong, H., Li, Y., Wang, L., Zhong, M., Sun, L., et al. (2014). OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5:4847. doi: 10.1038/ncomms5847
Pérez, L., Soto, E., Farr,é, G, Juanos, J., Villorbina, G., Bassie, L., et al. (2019). CRISPR/Cas9 mutations in the rice Waxy/GBSSI gene induce allele-specific and zygosity-dependent feedback effects on endosperm starch biosynthesis. Plant Cell Rep. 38, 417–433. doi: 10.1007/s00299-019-02388-z
Rao, Y., Li, Y., and Qian, Q. (2014). Recent progress on molecular breeding of rice in China. Plant Cell Rep. 33, 551–564. doi: 10.1007/s00299-013-1551-x
Reis, R. S., Deforges, J., Sokoloff, T., and Poirier, Y. (2020). Modulation of shoot phosphate level and growth by PHOSPHATE1 upstream open reading frame. Plant Physiol. 183, 1145–1156. doi: 10.1104/pp.19.01549
Ren, D., Rao, Y., Huang, L., Leng, Y., Hu, J., Lu, M., et al. (2016). Fine mapping identifies a new QTL for brown rice rate in rice (Oryza Sativa L.). Rice 9:4. doi: 10.1186/s12284-016-0076-7
Ribone, P. A., Capella, M., Arce, A. L., and Chan, R. L. (2017). A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype. Plant Physiol. 175, 1238–1253. doi: 10.1104/pp.17.01060
Ruan, B., Shang, L., Zhang, B., Hu, J., Wang, Y., Lin, H., et al. (2020). Natural variation in the promoter of TGW2 determines grain width and weight in rice. N. Phytol. 227, 629–640. doi: 10.1111/nph.16540
Sakamoto, T., and Matsuoka, M. (2008). Identifying and exploiting grain yield genes in rice. Curr. Opin Plant Biol. 11, 209–214. doi: 10.1016/j.pbi.2008.01.009
Shi, C. L., Dong, N. Q., Guo, T., Ye, W. W., Shan, J. X., and Lin, H. X. (2020). A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 103, 1174–1188. doi: 10.1111/tpj.14793
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sun, Y., Shi, Y., Liu, G., Yao, F., Zhang, Y., Yang, C., et al. (2020). Natural variation in the OsbZIP18 promoter contributes to branched-chain amino acid levels in rice. N. Phytol. 228, 1548–1558. doi: 10.1111/nph.16800
Sutoh, K., and Yamauchi, D. (2003). Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J. 34, 635–645. doi: 10.1046/j.1365-313X.2003.01753.x
Swinnen, G., Goossens, A., and Pauwels, L. (2016). Lessons from domestication:targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 21, 506–515. doi: 10.1016/j.tplants.2016.01.014
Tabassum, J., Ahmad, S., Hussain, B., Mawia, A. M., Zeb, A., and Ju, L. (2021). Applications and potential of genome-editing systems in rice improvement: current and future perspectives. Agronomy 11:1359. doi: 10.3390/agronomy11071359
Verter, M., and Botha, F. C. (2010). “Synthetic promoter engineering” in Plant Developmental Biology - Biotechnological Perspectives, eds E. C. Pua, and M. R. Davey (Berlin; Heidelberg: Springer), 393–414. doi: 10.1007/978-3-642-04670-4_20
von Arnim, A. G., Jia, Q., and Vaughn, J. N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12. doi: 10.1016/j.plantsci.2013.09.006
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, S. S., Chung, C. L., Chen, K. Y., and Chen, R. K. (2020). A novel variation in the FRIZZLE PANICLE (FZP) gene promoter improves grain number and yield in rice. Genetics 215, 243–252. doi: 10.1534/genetics.119.302862
Wang, Z., Yang, L., Wu, D., Zhang, N., and Hua, J. (2021). Polymorphisms in cis-elements confer SAUR26 gene expression difference for thermo-response natural variation in Arabidopsis. N. Phytol. 229, 2751–2764. doi: 10.1111/nph.17078
Wu, X., Liang, Y., Gao, H., Wang, J., Zhao, Y., Hua, L., et al. (2021). Enhancing rice grain production by manipulating naturally evolved cis-regulatory element-containing inverted repeat sequence of OsREM20. Mol. Plant 16:S1674-2052(21)00091-5.
Xie, Y., Zhang, Y., Han, J., Luo, J., Li, G., Huang, J., et al. (2018). The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in rice. Mol. Plant 11, 720–735. doi: 10.1016/j.molp.2018.03.001
Xu, C., Liu, Y., Li, Y., Xu, X., Xu, C., Li, X., et al. (2015). Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 66, 2611–2623. doi: 10.1093/jxb/erv058
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. doi: 10.1038/nature22372
Xu, L., Yuan, S., and Man, J. (2020). Changes in rice yield and yield stability in China during the past six decades. J. Sci. Food Agric. 100, 3560–3569. doi: 10.1002/jsfa.10385
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yang, Y., Guo, M., Sun, S., Zou, Y., Yin, S., Liu, Y., et al. (2019). Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat. Commun. 10:1949. doi: 10.1038/s41467-019-09919-y
Zeng, D., Liu, T., Ma, X., Wang, B., Zheng, Z., Zhang, Y., et al. (2020). Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 18, 2385–2387. doi: 10.1111/pbi.13427
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898. doi: 10.1038/nbt.4202
Zhang, T., Marand, A. P., and Jiang, J. (2016). PlantDHS: a database for DNase I hypersensitive sites in plants. Nucleic Acids Res 44, D1148–D1153. doi: 10.1093/nar/gkv962
Zhang, T., Wu, A., Yue, Y., and Zhao, Y. (2020). uORFs: important cis-regulatory elements in plants. Int. J. Mol. Sci. 21:6238. doi: 10.3390/ijms21176238
Zhao, H., Zhang, W., Zhang, T., Lin, Y., Hu, Y., Fang, C., et al. (2020). Genome-wide MNase hypersensitivity assay unveils distinct classes of open chromatin associated with H3K27me3 and DNA methylation in Arabidopsis thaliana. Genome Biol. 21:24. doi: 10.1186/s13059-020-1927-5
Keywords: rice, cis-regulatory element, genome editing, grain quality, upstream open reading frame
Citation: Ding Y, Zhu J, Zhao D, Liu Q, Yang Q and Zhang T (2021) Targeting Cis-Regulatory Elements for Rice Grain Quality Improvement. Front. Plant Sci. 12:705834. doi: 10.3389/fpls.2021.705834
Received: 11 May 2021; Accepted: 08 July 2021;
Published: 11 August 2021.
Edited by:
Wei Wang, Henan Agricultural University, ChinaReviewed by:
Tomokazu Kawashima, University of Kentucky, United StatesShakeel Ahmad, Ayub Agriculture Research Institute, Pakistan
Copyright © 2021 Ding, Zhu, Zhao, Liu, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqing Yang, cXF5YW5nQHl6dS5lZHUuY24=; Tao Zhang, emhhbmd0YW9AeXp1LmVkdS5jbg==
 Yu Ding
Yu Ding Jiannan Zhu1
Jiannan Zhu1 Dongsheng Zhao
Dongsheng Zhao Qiaoquan Liu
Qiaoquan Liu Qingqing Yang
Qingqing Yang Tao Zhang
Tao Zhang