- 1School of Agriculture Food and Wine, University of Adelaide, Glen Osmond, SA, Australia
- 2Joint International Research Laboratory of Metabolic and Developmental Sciences, Shanghai Jiao Tong University-University of Adelaide Joint Centre for Agriculture and Health, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 3School of Life Sciences and Engineering, Southwest University of Science and Technology, Mianyang, China
MADS-box genes have a wide range of functions in plant reproductive development and grain production. The ABCDE model of floral organ development shows that MADS-box genes are central players in these events in dicotyledonous plants but the applicability of this model remains largely unknown in many grass crops. Here, we show that transcript analysis of all MIKCc MADS-box genes through barley (Hordeum vulgare L.) inflorescence development reveals co-expression groups that can be linked to developmental events. Thirty-four MIKCc MADS-box genes were identified in the barley genome and single-nucleotide polymorphism (SNP) scanning of 22,626 barley varieties revealed that the natural variation in the coding regions of these genes is low and the sequences have been extremely conserved during barley domestication. More detailed transcript analysis showed that MADS-box genes are generally expressed at key inflorescence developmental phases and across various floral organs in barley, as predicted by the ABCDE model. However, expression patterns of some MADS genes, for example HvMADS58 (AGAMOUS subfamily) and HvMADS34 (SEPALLATA subfamily), clearly deviate from predicted patterns. This places them outside the scope of the classical ABCDE model of floral development and demonstrates that the central tenet of antagonism between A- and C-class gene expression in the ABC model of other plants does not occur in barley. Co-expression across three correlation sets showed that specifically grouped members of the barley MIKCc MADS-box genes are likely to be involved in developmental events driving inflorescence meristem initiation, floral meristem identity and floral organ determination. Based on these observations, we propose a potential floral ABCDE working model in barley, where the classic model is generally upheld, but that also provides new insights into the role of MIKCc MADS-box genes in the developing barley inflorescence.
Introduction
Flowers are often composed of four different floral organs organised in concentric whorls numbered from peripheral to central position. The outer whorls are sepals and petals in many dicots, including the model plant Arabidopsis, and lemma/palea and lodicules in grasses, while the inner whorls contain the male reproductive organs, the stamens, in the third whorl and the female organs, the carpels, in the fourth whorl (Ciaffi et al., 2011). Genetic studies have identified a large number of regulatory genes that control the specification of these distinct floral organs in plants (Alvarez-Buylla et al., 2010). The ABCDE model, originally proposed for Arabidopsis and Antirrhinum majus, associates the developmental determination of specific flower organs with the combinatorial activity of several classes of homeotic selector genes, most of which encode MIKCc MADS domain developmental transcription factors. Those MIKCc genes that function in the ABCDE model are divided into A-, B-, C-, D- and E-classes, which correspond with the AP1, AP3/PI, AG(C), AG(D) and SEP clades, respectively. A- and E-class genes determine the first whorl organs; A-, B- and E-class genes determine the second whorl; B-, C- and E-class genes control the third whorl; and C- and E-class genes specify the fourth whorl. D- and E-class genes are involved in ovule development within the carpel. Individual genes within a class usually act redundantly with each other in some roles, so that mutation of single members often leads to a subtle, absent or incomplete phenotype (Coen and Meyerowitz, 1991; Angenent et al., 1995; Pelaz et al., 2000). Studies demonstrate the conservation of gene homologues underlying the ABCDE model across most flowering plants, with only the AG subfamily (C-class) being present in gymnosperms (Theissen et al., 2016; Chen et al., 2017), suggesting the regulatory principles of some of these clades have been conserved during flower evolution.
The MADS-box genes have been divided into two subgroups: Type I and Type II, which are present in plants, animals and fungi (Kwantes et al., 2012; Chen et al., 2017). The encoded proteins cooperatively bind to DNA at conserved CArG boxes [CC(A/T)6GG] or [C(A/T)8G] to regulate gene expression (Theissen and Saedler, 2001). In plants, Type II MADS-box genes are called MIKC type, including MIKCc and MIKC* sub-types, an acronym of the four different domains that have been identified in all genes of this type. These transcription factors contain the conserved MADS DNA-binding domain (M), Intervening domain (I), Keratin-like domain (K) and the C-terminal domain (C), while the small c stands for classic. The closest relatives are the MIKC* genes, with the α-, β- and γ-MADS box genes completing the MADS family (Kwantes et al., 2012; Smaczniak et al., 2012). Within the MIKCc family, there are more clades than just those associated with the ABCDE model, such as the SVP-like floral repressors and the SOC1-like floral promotors (Becker and Theißen, 2003). The Type I lineage contains genes with simpler gene structure and lacking a Keratin-like domain. Their function is generally not well understood yet in plants, with some exceptions (Colombo et al., 2008; Callens et al., 2018). Additionally, MADS-box genes have also been reported to play an important role in abiotic stress, thermal regulation and plastic developmental responses in plants (Castelán-Muñoz et al., 2019; Li et al., 2021).
While the MIKCc clades and their roles in inflorescence development are generally conserved, the individual genes within a class often show no direct homology between grasses and Arabidopsis, making them co-orthologues (Ciaffi et al., 2011). More difficult to identify in grasses is the FLC-clade, which governs vernalisation and flowering time in Arabidopsis (Becker and Theißen, 2003). The FLC-clade genes in grasses are truncated and therefore could only be correctly classified using synteny and phylogenetic reconstruction (Ruelens et al., 2013). Present in grasses, but not in eudicots, is the OsMADS32 class, which is loosely related to B-class genes (Ciaffi et al., 2011).
The putative 11 MIKC-type MADS-box genes from the last common ancestor of monocots and eudicots increased to at least 24 genes in the last common ancestor of rice, wheat and maize (Bremer, 2002; Ciaffi et al., 2011). During this time of duplication and diversification in the MIKC family, the complex grass inflorescence and floral structures of the Poaceae family evolved. Changes in the copy number and expression pattern of MADS-box genes are closely associated with the morphological diversification of grass inflorescences (Ciaffi et al., 2011).
Generally, the ABCDE model of floral organogenesis can be applied to grasses as well. Functional studies in rice highlight mostly the homeotic changes defined by the model for predicted A-class genes (Wu et al., 2017), B-class genes (Nagasawa et al., 2003; Yadav et al., 2007; Yao et al., 2008), C- and D-class genes (Yamaguchi et al., 2006; Dreni et al., 2011) and E-class genes (Cui et al., 2010; Gao et al., 2010; Wu et al., 2018). The expression pattern and timing of MIKCc genes in other grasses indicate this likely extends to the whole grass family (Digel et al., 2015; Harrop et al., 2016; Feng et al., 2017; Callens et al., 2018; Zhu et al., 2018; Liu et al., 2020; Schilling et al., 2020).
A major determinant of floral organogenisis in grasses outside of the MADS-box genes is the YABBY family gene DROOPING LEAF (DL) which is involved in the regulation of carpel specification in rice (Nagasawa et al., 2003; Yamaguchi et al., 2004). Expression of DL orthologues in the carpel of maize and wheat is required to suppress the expression of B-class genes and is thus essential for floral organ specification according to the ABCDE model. Conserved expression suggests that DL function in carpel specification is a common feature in grasses (Bommert et al., 2005). While CRABS CLAW, the Arabidopsis co-orthologue of DL, has a function in carpel development, there is no homeotic change to the carpel identity in its absence (Bowman and Smyth, 1999), indicating a divergence in floral organogenesis between eudicots and grasses.
Some MIKCc genes associated with the ABCDE model have adopted additional roles in grasses, like the AP1 clade gene HvMADS14 which is a vernalisation integrator in barley (Trevaskis et al., 2007a), expression of which is important for floral transition onset. Alternatively in rice, OsMADS34 has been shown to modulate inflorescence branching (Gao et al., 2010; Kobayashi et al., 2010), which is not canonically an E-class role.
The Triticeae crop barley has a simple branchless spike-type inflorescence. During early development, the inflorescence meristem differentiates several spikelet ridges, and each ridge develops a determinate triple spikelet meristem, which in turn gives three spikelet meristems (SM; Wang et al., 2021). Each SM produces one floral meristem, always resulting in three single-flowered spikelets per rachis node (Koppolu and Schnurbusch, 2019). Inflorescence development in barley, as well as wheat, can be divided into stages by examining the development of the inflorescence meristem and noting the emergence and shape of the floral, spikelet and floret meristems followed by the sequential initiation and growth of the floral organs (Waddington et al., 1983). These Waddington stages range from the transition of the vegetative to the reproductive meristem at W1, to pollination or anthesis at W10, and include a series of developmental and cellular events. However, the transcription and regulation of ABCDE model components in barley inflorescence development and floret formation still remain unknown. Here, we performed transcript analysis of MIKCc genes through inflorescence developmental stages and in individual floral organs by quantitative reverse transcription PCR (RT-qPCR). Our findings reveal that the ABCDE model can be mostly applied to barley, while deviations point to interesting adaptations that can reveal more about inflorescence development in grasses.
Materials and Methods
Identification of MIKCc Genes
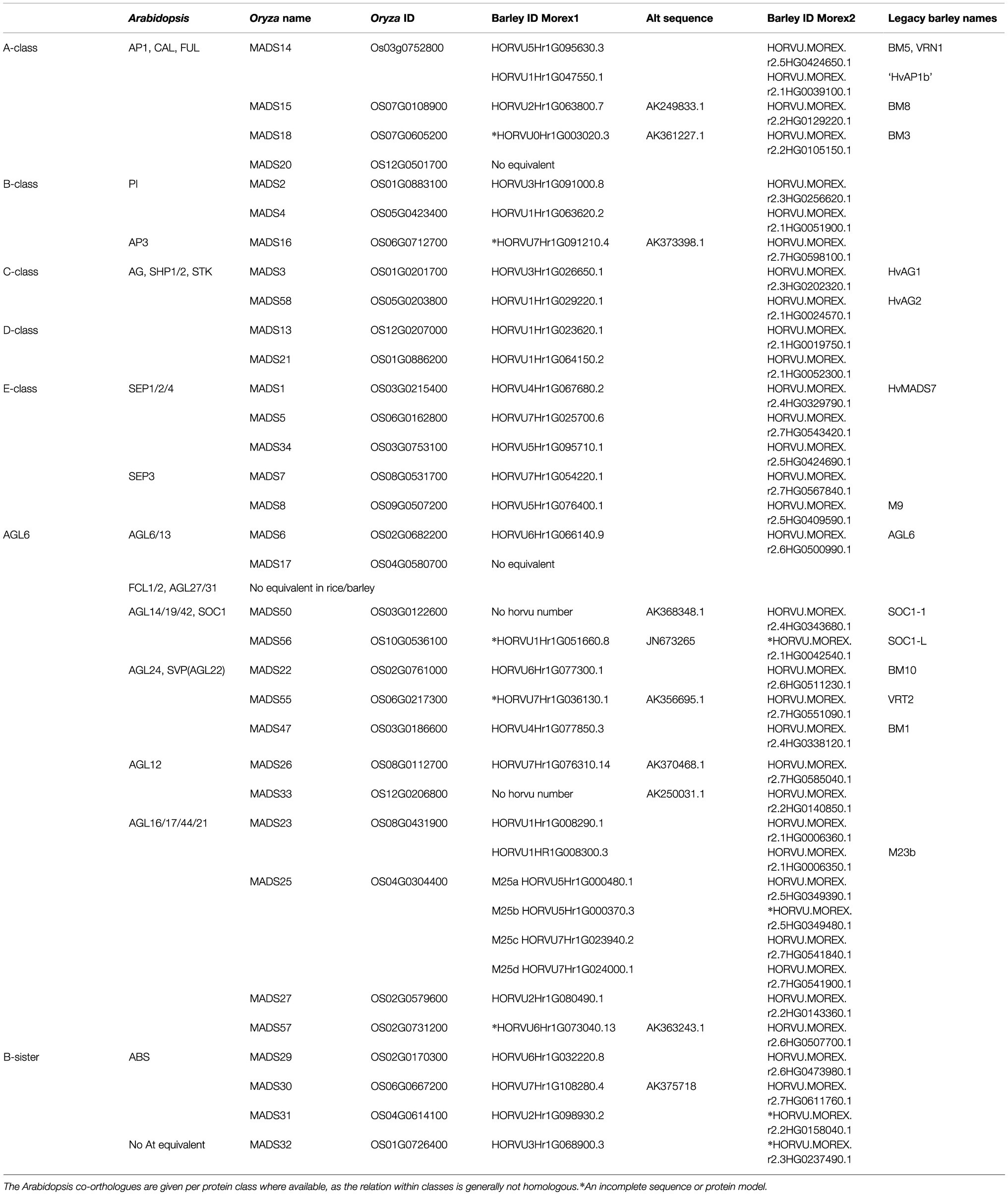
Barley MIKCc MADS-box genes were identified by name and BLAST searches, using rice homologues, in the HORVU data set1 using Geneious software version 8.1.3 (Biomatters). Additional genes, and more accurate coding sequences, were found using an online tBLASTn search of transcript data at NCBI.2 Previously identified MADS-box genes annotated as MADS-box proteins in the Uniprot database,3 IPK Gatersleben and NCBI databases were categorised using the PlantTFDB as follow-up analysis for MIKC-type MADS-box members (Jin et al., 2014; Mascher et al., 2017; Monat et al., 2019). Where no known (complete) transcript sequences were available, the FGENESH+ protein-based gene prediction tool (Solovyev, 2007) was used to identify the most likely transcripts. Genes were named after their rice homologues, rather than the previous names used in barley, to standardise naming and make functional comparisons to other grasses easier (Table 1).
Phylogenetic Analysis
MIKCc MADS-box proteins from Arabidopsis, rice, sorghum and Brachypodium were collected from published data (Arora et al., 2007; Wei et al., 2014). The sequences obtained were aligned with previously identified barley MIKCc MADS-box proteins using the MUSCLE algorithm before manual inspection and minor adjustments (Edgar, 2004). The IQ-TREE web server was used to create a maximum likelihood phylogenetic tree (Trifinopoulos et al., 2016). JTT+I+G4 was selected as the best model and bootstrap was set at 1,000 replicates.
SNP Analysis
A list of barley single-nucleotide polymorphisms (SNPs) was compiled using the comprehensive SNP database, recently made accessible at IPK Gatersleben4 and the barley pan-genome sets (Milner et al., 2019; Jayakodi et al., 2020).5 Gene locations in the SNP-browser were found by HORVU number where available, otherwise by position on the chromosome. Exon and amino acid changes were assessed by comparison to an alignment of cDNA sequences and chromosome fragments in Geneious 8.1.3 (Biomatters Ltd). Pan-genome predicted CDS sequences were extracted and assessed by multiple alignment in EUGENE (UniPro).6 Rice SNPs were collected using the online interface of the SNP database (Mansueto et al., 2017).7
Inflorescence Tissue Sampling
Hordeum vulgare L. variety Golden Promise was grown in a controlled environment room with 16h light at 15°C day and 10°C night temperatures, at 70% humidity, in 8cm square pots containing coco-peat standing in closed trays and watered from below every 2days. A midday light maximum of 500μmole photons m−2 s−1 was used. Inflorescence tissue samples were taken from the main stem and examined under a dissecting microscope. Immature spikes exactly matching the desired Waddington stage (Waddington et al., 1983) were immediately frozen in liquid nitrogen and stored at −80°C.
For the W1 stage, where the meristem height was less than 1mm, 30 meristems were taken per sample. To capture transcript changes through pollination, one additional stage was introduced, called W10.5, which was taken 3days after pollination. One sample represents one biological replicate, for which 25 individual meristems (IM) were collected at W1.5, 20 IM at W2, 15 IM at W2.5, 12 IM at W3.5, 10 IM at W4, 8 IM at W4.5, 6 IM at W6.5 and 5 IM at W8.5. At W9.5 and W10.5, five separate single IM were taken, and combined at a later stage, such that each final sample comes from at least five individual plants.
Additionally, floral organ samples were taken from five different plants at Waddington stage 9.5. Twelve florets were harvested for the palea/lemma, the stamens and the carpel, while 20 florets were dissected for a total of 40 lodicules.
RNA Extraction
Total RNA was extracted using the Qiagen Plant RNA Kit, Ambion Turbo RNA-free Kit and approximately 200ng of RNA used for cDNA synthesis with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions.
Real-Time RT-qPCR and Co-expression Analysis
Primers were designed across the stop codon of each gene, forward in the gene and reverse in the 3′UTR (Supplementary Table 2). This is not only to avoid problems with sequence similarity between closely related genes, but also because the RNA in this position is less likely to be degraded.
RT-qPCR was performed as described by Burton et al. (2008). The quantity of the cDNA was assessed with four standard genes (HvGAP, HvCyclophilin, HvTubulin and HvHSP70) and normalised by relative threshold cycle value over the time course and floral organ samples individually using the average expression of the best matching three standard genes (HvGAP was excluded). All RT-qPCR was performed on three independent technical repeats with similar results. Transcript correlation analysis of the normalised expression values was done using the Pearson correlation function in MeV4.9.8 Hierarchical clustering analysis was performed using pheatmap package in R.9
RNA In situ Hybridisation
Meristems were obtained as described, placed into FAA fixative solution (3.7% formaldehyde, 50% ethanol and 5% acetic acid) and vacuum infiltrated. Samples were dehydrated in an ethanol series which was subsequently swapped for D-lemonene (HistoChoice, Sigma), and finally paraffin wax (Paramat pastillated, Gurr) at 60°C. Embedded samples were cut into 6–8μm sections on a Leica RM2265 microtome and placed on lysine coated slides.
Dioxigenin labelled probes were made, in sense and antisense configuration, using the DIG labelling kit (Roche Diagnostics), following the manufacturer’s instructions. Primers used to generate the probes are listed in Supplementary Table 3.
Slides were dewaxed in D-lemonene and rehydrated in an ethanol series (2× 100%, 95% ethanol and 85 and 75% ethanol with 0.85% NaCl). The following steps were performed with an InsituPro robot (Invatis): Finalise rehydration, proteinase K digestion and re-dehydration. Re-dehydration was finalised with a reverse of the rehydration steps above, and the slides dried at 37°C. The following steps were also performed with the InsituPro robot: hybridisation, stringent washes, RNAse digestion, immunolabelling (AntiDIG-APconjugate, Roche) and washing. Substrate (NBT/BCIP, Roche) was added according to the manufacturer’s instructions and incubated overnight in the dark. Slides were fixed with ImmunoHistoMount (Sigma-Aldrich) and observed with a Nikon Ni-E optical microscope. Pictures were processed for colour, brightness and contrast in GIMP2.10.2 (www.gimp.org).
Available Public Expression Data Analysis
Transcript data for barley early inflorescence development by RNA-seq were collected from supplemental data set 3 (Digel et al., 2015), selecting only the introgression line (S42-IL017) inflorescence samples grown in long day conditions.
Transcript data for rice early inflorescence meristem types were collected from supplemental data S1 (Harrop et al., 2016), selecting the MADS-box genes by name search.
Transcript data for wheat inflorescence development were collected from Supplementary Table 4 ‘List of wheat homologues similar to rice MADS-box genes’ (Feng et al., 2017).
Results
Identification and Phylogenetic Analysis of MIKCc MADS-Box Genes in the Barley Genome
The recent barley genome assembly contains 32 MIKCc MADS-box genes annotated as expressed sequences and a pseudogene strongly resembling HvMADS14 with 97% identity, but only covering the latter 67% (Mascher et al., 2017; Monat et al., 2019). Through comparison with homologous genes from rice, and a tBLASTn search for available transcript sequences of barley, two genes labelled as HvMADS27 (HORVU1Hr1G008290.1 and HORVU1HR1G008300.3) were found to be more closely related to OsMADS23, while another two genes, HvMADS50 and HvMADS33, were identified as transcript sequences for barley, although not present in the MOREX v1 genome assembly. Additionally, a more accurate exon sequence was found for nine MIKCc genes through comparison with available transcripts for HvMADS16, HvMADS18, HvMADS55, HvMADS56 and HvMADS57, and by analysing the genomic region with FGENESH, guided by the OsMADS25 sequence, for HvMADS25a/b/c/d. This brings the total to 34 MIKCc MADS-box genes and one pseudogene in barley (Table 1).
There is a barley homologue for 31 of the 33 complete MIKCc genes in rice; missing homologues are the SQUA/AP1 gene OsMADS20 and the AGL6-like gene OsMADS17. There is only one copy of OsMADS25 in rice, but four in barley, here designated HvMADS25a, HvMADS25b, HvMADS25c and HvMADS25d. Apart from these exceptions, each barley MIKCc gene has a clear orthologue in rice. Phylogenetic analysis of encoded proteins showed that A-, B-, C-, D- and E-class proteins are conserved between rice and barley, and also with Brachypodium and sorghum, but show a divergence with eudicot Arabidopsis (Figure 1; Table 1). The FLC-like MIKCc proteins, involved in flowering time and vernalisation in Arabidopsis (Becker and Theißen, 2003), have truncated homologues in grasses. In barley, these homologues are ODDSOC1 and ODDSOC2, which are missing the C-terminal domain and part of the keratin-like domain and fail to group with the MIKCc proteins in a phylogenetic tree based on sequence similarity alone (Ruelens et al., 2013). Conversely, the MADS32 clade has no orthologous gene group in Arabidopsis, or likely in any eudicot (Figure 1). Thus the overall phylogenetic analysis showed that most barley MIKCc MADS-box proteins have a close evolutionary relationship with their orthologues in rice, Brachypodium and sorghum, but not Arabidopsis (Figure 1).
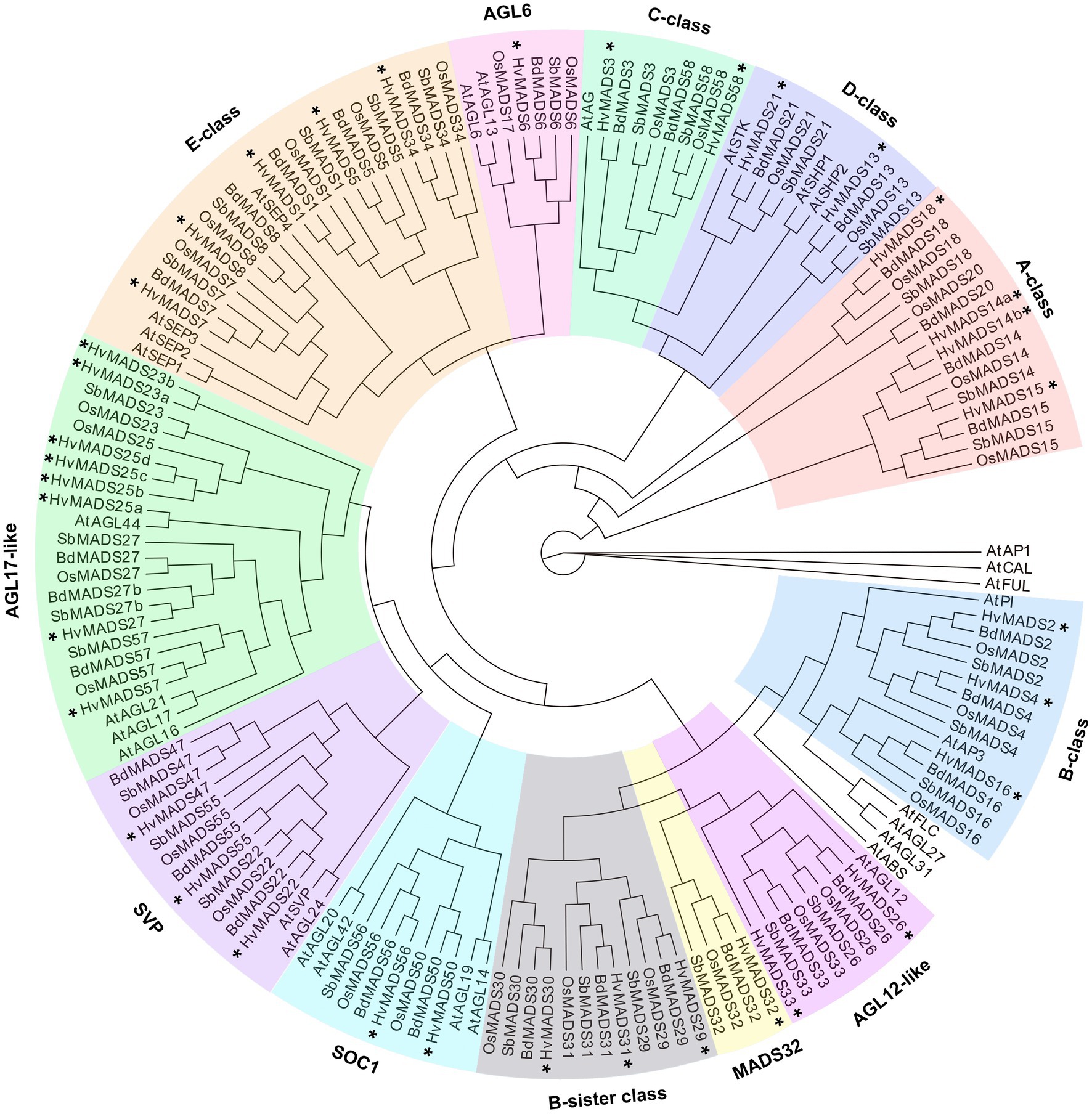
Figure 1. Phylogenetic analysis of all MIKCc-type MADS-box proteins (except for the truncated FLC-like proteins) in barley compared to the homologues in rice, Brachypodium, sorghum and Arabidopsis. The classes within the family are delimited by coloured boxes. SOC1, SUPRESSOR OF CONSTANS1-like proteins; SVP, SVP/VRT-like proteins. Stars (*) indicate the MADS-box homologues of barley.
The coding region of MIKCc MADS-box genes shows some expected conservation of intron-exon patterns within the different classes of closely related genes. Most exon patterns are long-short-short-medium-short-short-long, whereas intron length, and therefore gene length, varies more widely (Supplementary Figure 1). Some of the HvMADS25 paralogous genes have big introns (a) 0.5kbp, (b) 6kbp, (c) 10kbp and (d) 15kbp.
Low Occurrence of Single-Nucleotide Polymorphisms Shows High Conservation of MIKCc Genes’ Coding Region
To reveal the natural variation within the MIKCc MADS-box gene family during barley domestication, a comprehensive search for SNPs was performed in a database from exome sequencing of 22,626 barley cultivars, landraces and wild relatives that was recently made accessible at IPK Gatersleben, and from the barley pan-genome database that have been sequenced in 20 cultivars (Milner et al., 2019; Jayakodi et al., 2020).
The result shows that only 14 of the 34 MIKCc MADS-box genes contain any SNPs in the varieties sampled in the SNP-browser. Within these 14 genes, only half exhibit amino acid changes (Supplementary Table 1), although never in the first 110 amino acids, a region that contains the MADS domain and the Intervening domain. Remarkably, all but one of the 20 SNPs associated with HvMADS2 occur between HvMADS2 and its neighbouring gene HORVU.MOREX.r2.3HG0256630.1, suggesting active variation of the transcriptional interaction between these two genes. In the promoter region of HvMADS22, within 1kb of the start of the coding region, there are 12 SNPs of varying rarity. Three of these, 529572337T, 529572349T and 529572362A, commonly occur together and correspond to Hybernum viborg, a winter barley grown in many parts of the world (Supplementary Table 1). The results from pan-genome database show a different distribution of SNPs, but the amount is equally low in most of the clades. The more recently duplicated genes in the AGL17 clade (e.g., HvMADS25) have more abundant SNPs, as do HvMADS30 and HvMADS50 to a lesser extent. Apart from four amino acid indels among all MIKCc encoded proteins and a deletion of the last six amino acids of HvMADS16, no other variety in the coding sequence was detected across the pan-genome (Supplementary Table 1).
The lack of both natural and selected variation in these genes suggests a high rate of conservation and therefore importance for fitness and domestication. In rice, there are significantly more SNPs that change an amino acid, although many occur at a very low frequency (Supplementary Table 1; Mansueto et al., 2017).7 These findings demonstrate that MIKCc MADS-box genes are not only conserved among grass species, but also show very few SNPs and other sequence variety between the coding sequence in both wild and domesticated barley.
Global MIKCc Transcripts Are Concentrated in the Inflorescence and Caryopsis
The ABCDE-class MIKCc genes in barley, according to the transcript data accompanying the HORVU database (Mayer et al., 2012), are predominantly expressed in the developing inflorescence at 0.5 and 1.5cm (INF1 and INF2) and in developing seed (CAR1 and CAR2; Figure 2).
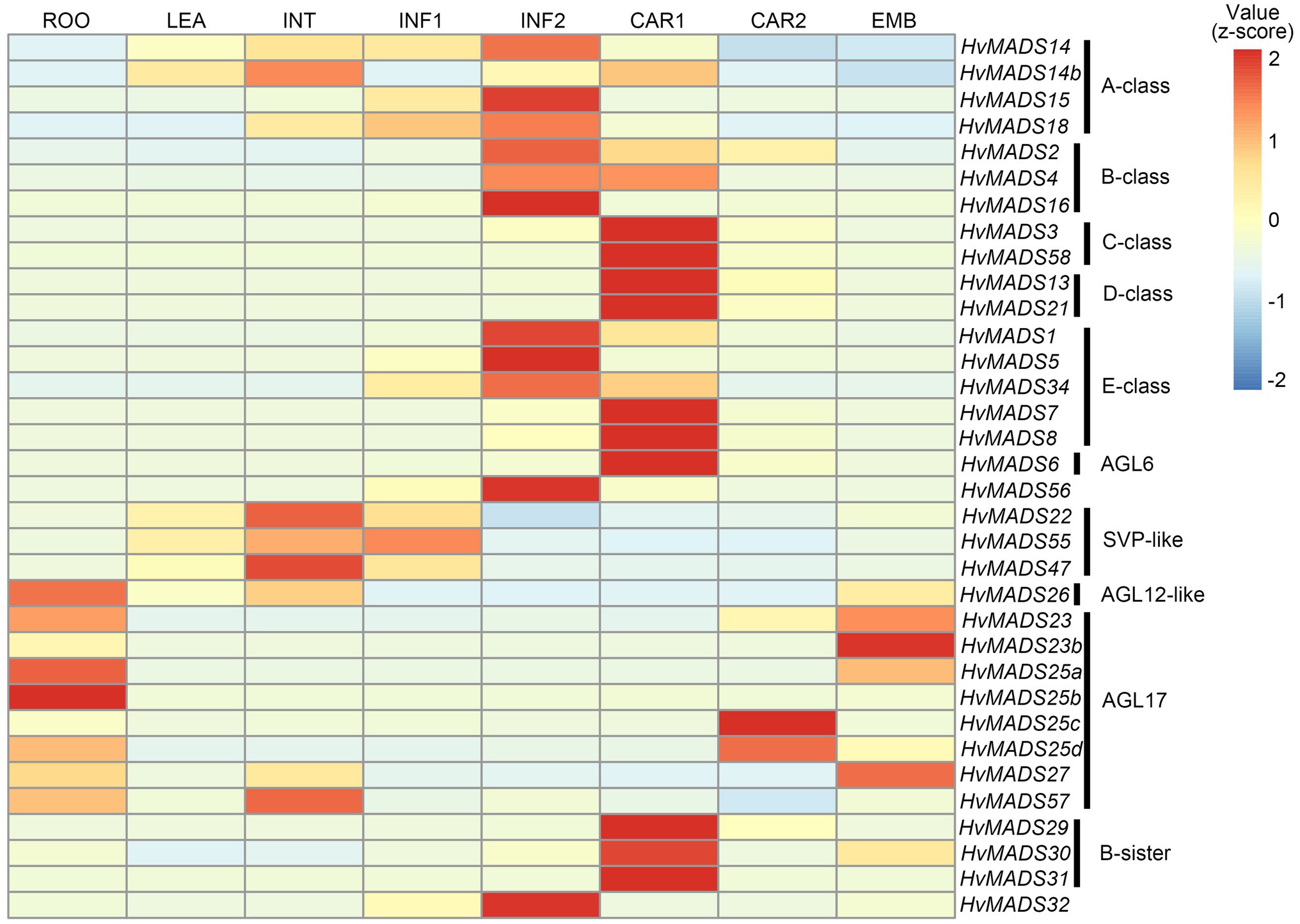
Figure 2. Hierarchical clustering analysis of MIKCc genes’ global transcript in different barley tissues. RPKM (Reads Per Kilobase Million) is normalised using a pheatmap package in R. ROO, roots 17days after planting (DAP); LEA, leaves 17 DAP; INT, third internode 42 DAP; INF1, developing inflorescence 5mm (about 30 DAP); INF2, developing inflorescence 10–15mm (about 50 DAP); CAR1, developing grain 5days after anthesis (DAA); CAR2, developing grain 15 DAA; and EMB, embryo 4days after germination. Raw data from Mayer et al. (2012).
Transcript in other barley tissues, such as leaf and root, is seen for the SVP-like genes HvMADS22/47/55. AGL17-like genes HvMADS23a/b, HvMADS25a/b/c/d and HvMADS27 are expressed mostly in the root and during late seed development; however, their overall transcript level is low (RPKM 10 or less), suggesting they may not be functional, or they are only expressed under stress conditions, like some AGL17-like genes in wheat (Schilling et al., 2020). HvMADS57 has RPKMs of 25 and 35 in root and internode, respectively, which indicates it is the most likely among the AGL17-like genes to be functional. OsMADS57 has been shown to function in cold tolerance in rice, directly targeting OsWRKY94 and OsD14 (Guo et al., 2013; Chen et al., 2018b).
Overall, among the 34 MIKCc MADS-box genes, most of them (20) are expressed in the developing inflorescence and, similar to homologues in related species, are probably involved in meristem transitions and floral organ development (Arora et al., 2007; Paollacci et al., 2007; Wei et al., 2014). However, to gain any insight into transcript similarities and differences of the MIKCc genes in barley inflorescence development, and to what extent the ABCDE model is likely conserved, a higher resolution transcript profile from floral transition to pollination and a complete set of floral organ transcript data would be required.
Transcript Profiles of MIKCc Genes in Inflorescence Development and Floral Organs
The transcript profiles of MIKCc genes through barley inflorescence development (Figures 3, 4) can be related to developmental events by Waddington stage (Figure 3A). Combined with transcript data in the floral organs at Waddington stage W9.5 (Figure 5 and Supplementary Figure 4), a comparison to established ABCDE models in other species can be made.
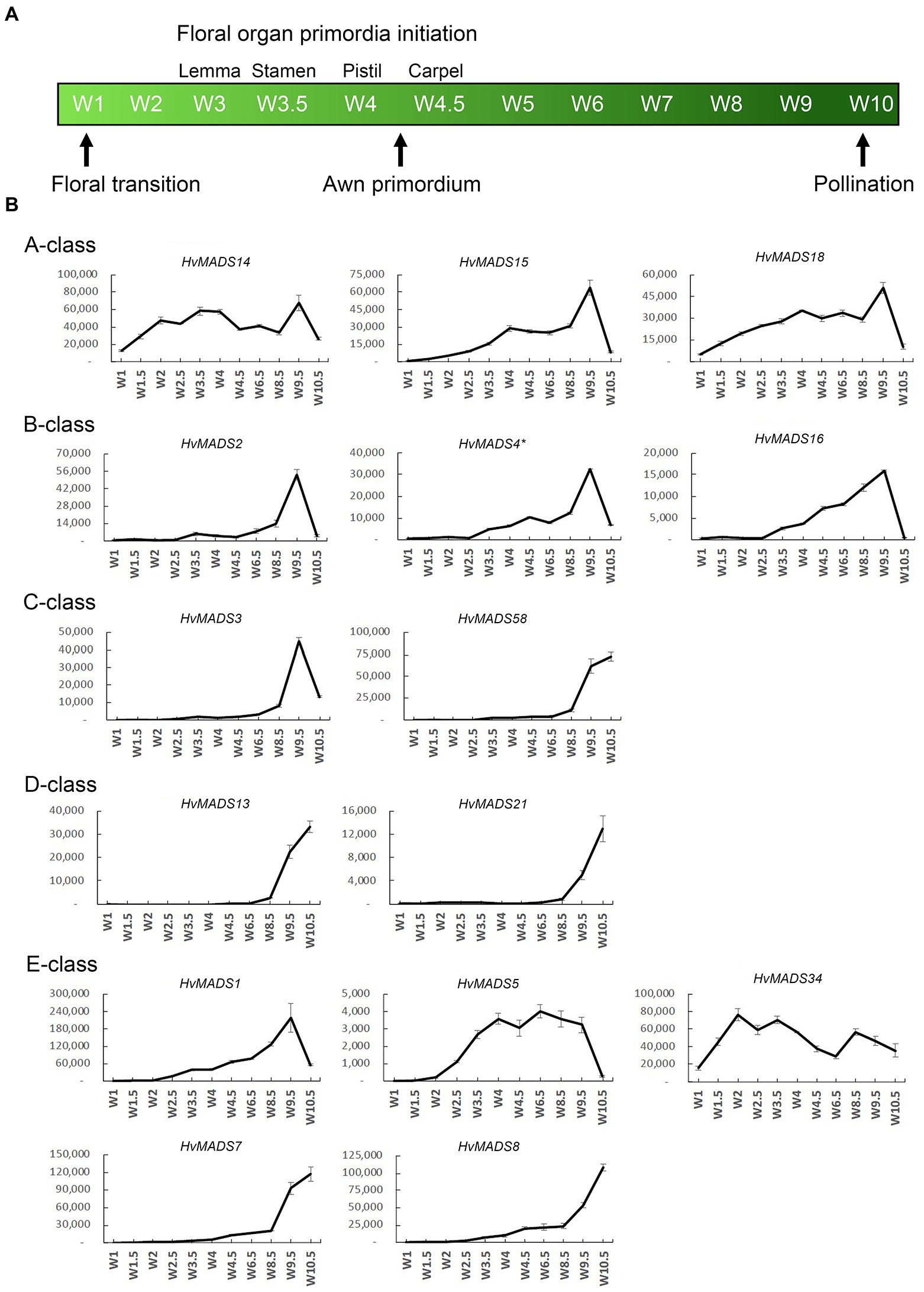
Figure 3. (A) Waddington stages for barley and their relation to major developmental steps. (B) Transcript profiles of AP1, AP3/PI, AG(C), AG(D) and SEP MIKCc MADS-box genes in the shoot apex through inflorescence development as measured by the Waddington stage in Golden Promise. Error bars represent one standard deviation, based on technical replicates. A-class: The predicted A-class function in the outer floral organs predicts that transcript starts after W3, where the lemma primordium is first formed. However, expression of all three AP1 genes increases earlier, at the floral transition W1. AP3/PI: The expression of predicted B-class genes starts to increase at W3.5, where the stamen primordia are formed, and peaks right before pollination. AG(C): HvMADS3 and HvMADS58 both start expression around W3.5 when the stamen primordia appear, however HvMADS3 peaks before pollination and declines quickly afterwards, while HvMADS58 maintains peak expression through to W10. AG(D): HvMADS13 and HvMADS21 both start significant expression only after W6.5, well after the pistil primordium is formed, which first appears at W4. Their peak expression is after pollination. E-class: There is a clear difference between the LOFSEP genes HvMADS1, HvMADS5 and HvMADS34 that express earlier and sharply drop at pollination (W10) and HvMADS7 and HvMADS8 expression, which starts later around W3.5 and continues to rise through pollination.
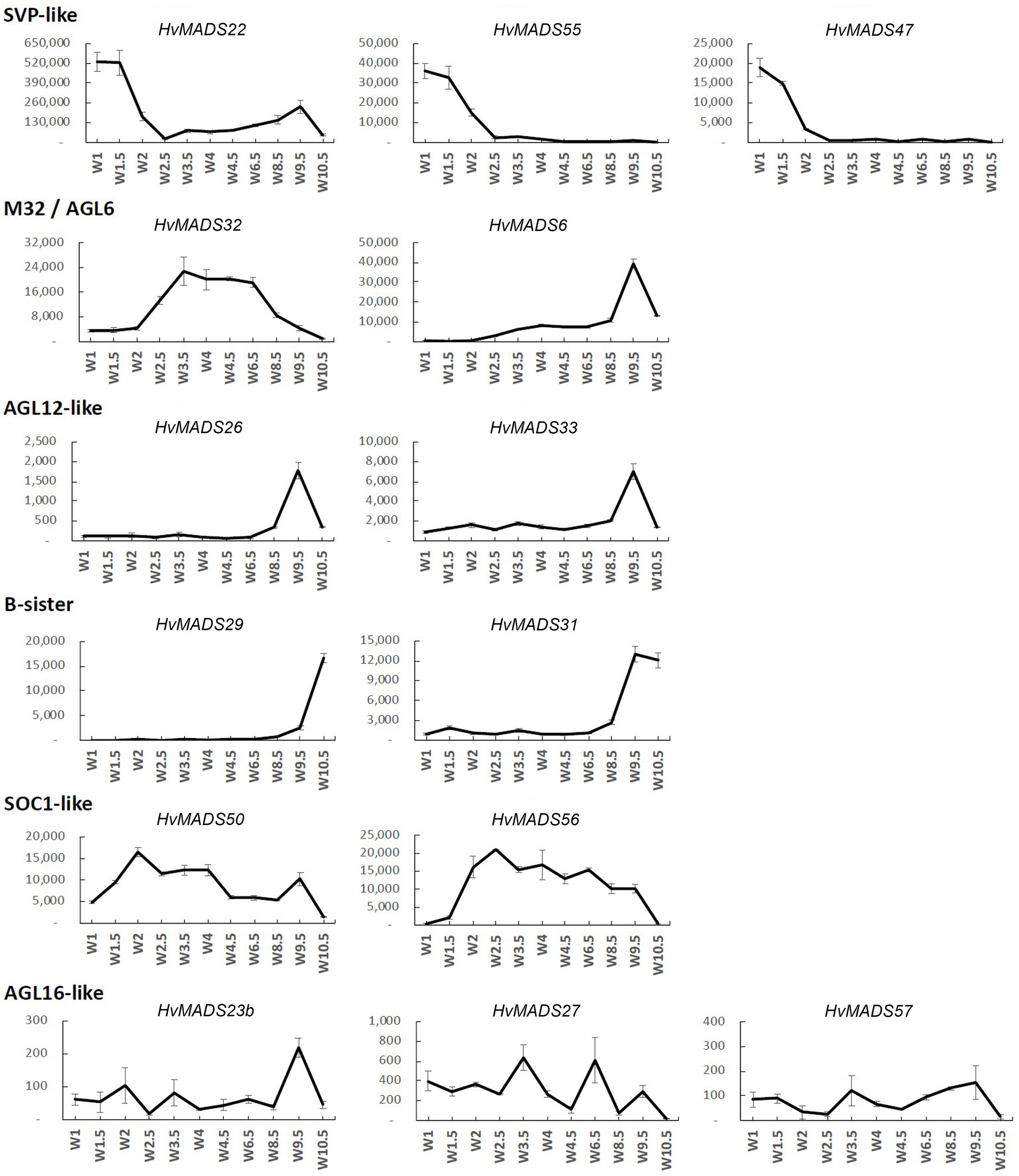
Figure 4. Transcript profile of the non-ABCDE MIKCc MADS-box genes through inflorescence development by Waddington stage in Golden Promise. Error bars represent one standard deviation, based on technical replicates.
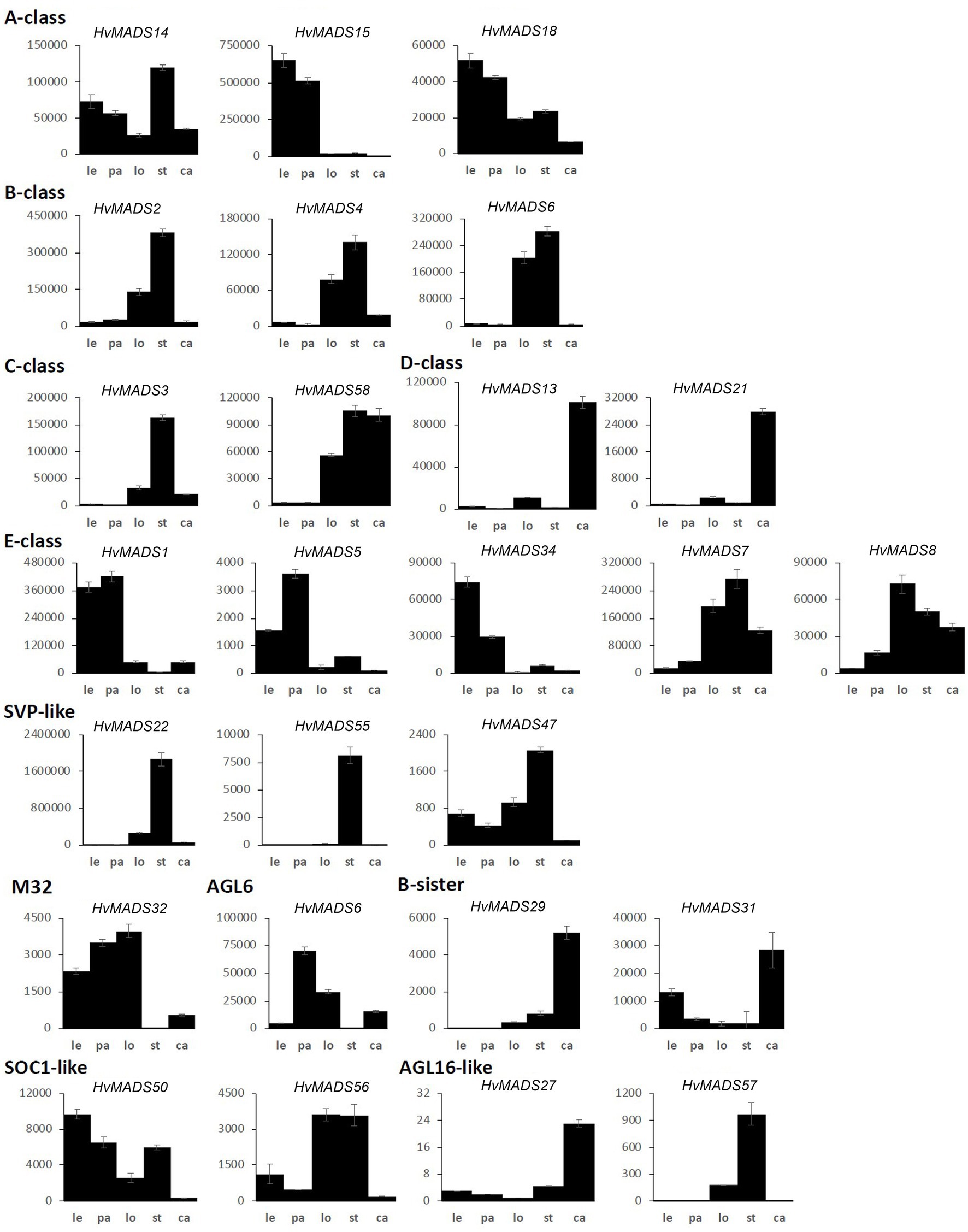
Figure 5. MIKCc MADS-box gene expression in floral organs at Waddington stage 9.5. Error bars represent one standard deviation, based on technical replicates. Le, lemma; pa, palea; lo, lodicules; st, stamens; and ca, carpel.
AP1 (A-Class)
Canonical A-class function in the outer floral organs would predict expression starting after the lemma primordium is first formed, at W3. However, expression of all three A-class genes increases before this, at the floral transition W1, indicating a role in earlier inflorescence development. HvMADS14 transcript is already present at W1, peaks at W3.5, declines into W4.5 and peaks again at W9.5, right before pollination. The early expression of HvMADS18, along with the decline at W4.5, is less pronounced than for HvMADS14, but still recognisable (Figure 3B). HvMADS15 transcript is closer to the expected profile of an A-class function gene, and transcript is indeed confined to the lemma and palea. While HvMADS14 and HvMADS18 are also expressed in the lemma and palea, their transcript is not confined there and HvMADS14 is surprisingly more strongly expressed in the stamens than in the first whorl, indicating that the barley AP1 clade genes may have an additional function diverging from the classical ABCDE model (Figure 5 and Supplementary Figure 4).
AP3/PI (B-Class)
B-class gene transcripts start to increase at W3.5, where the stamen primordia are formed, and peak right before pollination (Figure 3B). Transcript is confined to the lodicules and stamens, exactly following the ABCDE model, indicating B-class function is likely to be completely conserved in barley (Figure 5 and Supplementary Figure 4).
AG (C-Class)
HvMADS3 and HvMADS58 both start expression around W3.5 when stamen primordia appear, in accordance with the ABCDE model. However, where HvMADS3 peaks before pollination and declines quickly afterwards, HvMADS58 maintains peak expression through W10, indicating subfunctionalisation of the two genes, where HvMADS58 is responsible for the C-class function in the carpel (Figures 3, 5). Both C-class genes also show some expression in the lodicules, which does not fit with the ABCDE model (Figure 5 and Supplementary Figure 4).
AG (D-Class)
HvMADS13 and HvMADS21 both start significant expression only after W6.5, well after the pistil primordium is first formed at W4. Peak transcript is after pollination and confined to the fruit, indicating that their canonical role in ovule development and into fruit development is likely to be conserved in barley (Figure 3B).
SEP (E-Class)
There is a clear difference between the ‘LOFSEP’ genes HvMADS1, HvMADS5 and HvMADS34 that express earlier and sharply drop at pollination (W10) and HvMADS7 and HvMADS8 expression, which starts later around W3.5 and continues to rise through pollination (Figures 3A,B). Floral organ transcripts show a division along the same line, where the LOFSEP genes are mostly confined to the lemma and palea, whereas HvMADS7 and HvMADS8 are expressed in the lodicules, stamens and carpel (Figure 5 and Supplementary Figure 4). Therefore, the LOFSEP genes probably perform the E-class function in the lemma and palea, while HvMADS7 and HvMADS8 fulfil the E-class function in the other floral organs. In contrast to all other E-class genes, HvMADS34 is expressed at W1 and peaks at W2, similar to HvMADS14, hinting at a function in early inflorescence development.
SVP-Like
The three SVP-like genes, HvMADS22, HvMADS47 and HvMADS55, are highly expressed at the start of the floral transition and quickly decline to insignificant expression at W2.5, which indicates a role at this early stage. HvMADS22 has a surprising resurgence in expression to a new maximum at W9.5, exclusively in the stamens, indicating possible neofunctionalisation (Figures 4, 5 and Supplementary Figure 4).
MADS6
HvMADS6, closely related to the E-class genes, has an expression profile similar to HvMADS1 (Figure 4), but contrastingly is not expressed in the lemma, but rather in the lodicules (Figure 5 and Supplementary Figure 4). HvMADS1 and HvMADS6 may be partially redundant in E-class function, but not in the lemma and lodicules.
MADS32
HvMADS32 has no direct equivalent in Arabidopsis, and no assigned function in the original ABCDE model. The HvMADS32 transcript appears before initiation of the floral organs and uniquely declines after W6.5, unlike any other MIKCc gene (Figure 4). Floral organ expression is concentrated in the lemma, palea and lodicules (Figure 5 and Supplementary Figure 4).
B-Sister
HvMADS29 and HvMADS31 are expressed late in inflorescence development, mostly after W8.5, and are strongly expressed after pollination (Figure 4). Combined with a nearly exclusive expression in the carpel, they are likely to be involved in ovule and seed development (Figure 5 and Supplementary Figure 4).
SOC1-Like
HvMADS50 and HvMADS56 expression starts early, with a peak at W2 and W2.5, much like HvMADS14 and HvMADS34. Late expression is weaker, but only disappears after pollination (Figures 4, 5 and Supplementary Figure 4).
MIKCc Genes With Low Transcript in the Inflorescence
There was no significant transcript detected for several AGL17-like genes, including HvMADS23a and all four HvMADS25 co-orthologues during the stages of inflorescence development examined here. Global expression analyses indicated that these transcripts are more prevalent in embryo, leaf and root tissue (Figure 2). Of the AGL17-like genes that did have measurable transcript, namely, HvMADS23b, HvMADS27 and HvMADS57, low abundance and erratic profiles preclude any meaningful speculation on their function (Figure 4). The HvMADS14b pseudogene did seem to be expressed based on primer pair tests at various temperatures, but could never be sufficiently separated from the very similar and much more abundant HvMADS14 transcript to provide a clear expression profile (data not shown). The final missing profile is that of HvMADS30, a B-sister gene, for which no expression was detected.
Co-expression Profiles Reveal a Novel Regulatory Network Among MADs-Box Genes in Barley Developing Inflorescences
To quantify co-expression of MIKCc MADS-box genes, which indicates possible functional connections, a correlation analysis was performed. Correlation analysis of the transcript profiles generated by RT-qPCR of all MIKCc MADS-box genes reveals three sets, here defined as members having a correlation coefficient of over 0.9 with at least two other members, and a less cohesive pseudoset (Figures 6A,B; Supplementary Table 4). A cluster analysis of the data showed a similar result of grouping (Supplementary Figure 5). Correlation set 1 is expressed mostly at W1 and W1.5, during the floral transition, and quickly disappears after this stage (Figure 6). In contrast, the pseudoset is spread out over the whole time course, but has some members with high transcript levels between W1.5 and W3.5 where none of the other sets show strong expression. This is the window for spikelet- and floret meristem initiation and development and then follows correlation set 2, which starts at W3 and stops after W9.5, where most floral organs develop. Finally, correlation set 3 shows transcript the latest and generally has maximum expression at W10.5, after pollination. HvMADS23B, HvMADS27 and HvMADS57 did not group into any set (Figure 6).
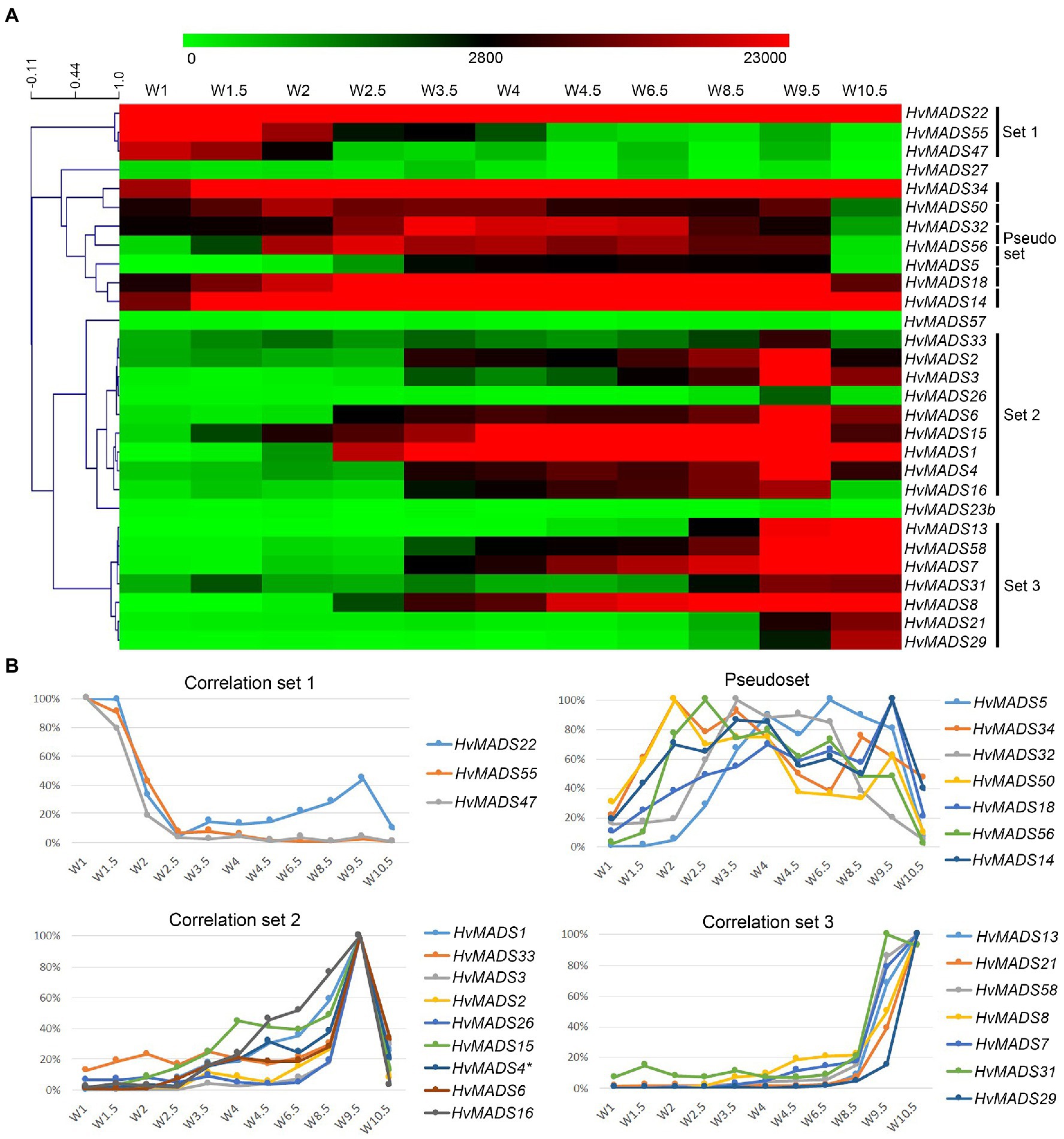
Figure 6. Transcript profiles of MIKCc MADS-box genes can be grouped into correlation sets. (A) Pearson correlation of time course expression reveals three sets and a pseudoset. Expression is represented on a logarithmic colour scale, where the maximum value is capped to provide the best visual contrast in the data set spanning orders of magnitude. Correlation tree and scale bar are presented on the left side. (B) Relative expression profiles through inflorescence development of the MADS-box genes within each correlation set.
Set 1: Floral Transition
This contains three SVP-like genes, HvMADS22, HvMADS47 and HvMADS55. Expression starts high at W1 and quickly declines to a minimum at W2.5. Remarkably, the expression of HvMADS22 is upregulated again after W2.5 and peaks at W9.5 (Figures 4, 6). As the members of this set are already a class within the MIKCc genes, it is likely that they redundantly repress further inflorescence development.
Pseudoset: Expressed During Development of the Spikelet- and Floret Meristem
Expression of genes in the pseudoset is not as closely correlated as members of the other sets, but a general pattern can still be distinguished. Transcript mostly rises between the floral transition (W1) and emergence of the floral organs (W3–W4), and for some genes, the maximum expression is also in this early time-frame (Figure 6B). The SEP clade genes in the pseudoset are LOFSEP genes HvMADS5 and HvMADS34. HvMADS34 really stands out from the other E-class genes due to the very early high level of expression peaking at W2. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1)-like genes HvMADS50 and HvMADS56 are both expressed early in barley inflorescence development with a maximum at W2 and W2.5, respectively, and show a steep decline after W4. Early expression of pseudoset members indicates a function in floral development prior to the formation of the floral organs, such as a role in the spikelet- or floret meristem. Correlation of HvMADS14, HvMADS34 and HvMADS50 early expression suggests the possibility of related functions (Figure 7A).
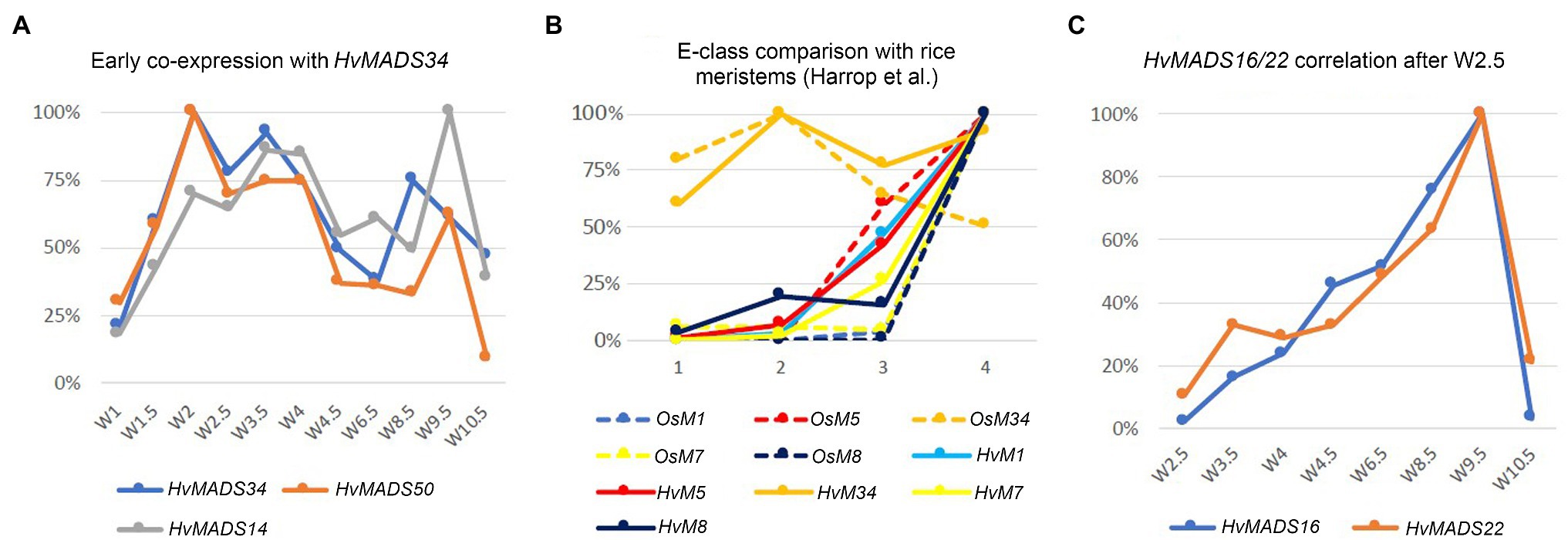
Figure 7. Co-expression within barley or with equivalent stages in rice can suggest related functions. (A) Early co-expression of HvMADS34 with HvMADS14 (A-class) and HvMADS50 (SOC1-like). (B) Comparison of early E-class gene expression in barley and rice. Analysis of the differential gene expression in the inflorescence meristem types of rice using laser microdissection followed by RNA sequencing (dashed lines) from the supplemental data of Harrop et al. (2016). While directly comparing the results of their work with expression in the early stages of the whole barley inflorescence meristem is a false equivalency, it can still provide some insights. The best matching Waddington stages W1.5–W3.5 expression data are shown (solid lines; this paper). On the x-axis, 1 is inflorescence meristem/W1.5, 2 is branch meristem/W2, 3 is elongated branch meristem/W2.5 and 4 is spikelet meristem/W3.5. (C) Correlation of the relative expression of HvMADS22 and HvMADS16 between W2.5 and W10.5.
Set 2: Lemma, Palea, Lodicule and Stamen Development
Correlation set 2 is not as uniform as sets 1 and 3. Transcripts in general appear around W3–W3.5, increase to a maximum right before anthesis at W9.5 and quickly diminish immediately after pollination at W10.5 (Figure 6B).
All three AP1 clade genes are strongly expressed in the lemma and palea, although the expression of HvMADS14 and HvMADS18 starts significantly earlier. The LOFSEP subclade, HvMADS1, HvMADS5 and HvMADS34, are strongly expressed in the lemma and palea, but hardly at all in the lodicule, stamen and carpel (Figure 5). Of these, only HvMADS1 appears in correlation set 2. The strongest difference in expression between the lemma and palea is for HvMADS6, which is expressed in the palea, but at very low levels in the lemma (Figures 5, 8). In general, this indicates A- and E-class genes are expressed in the first whorl, which is consistent with the ABCDE model in other plants.
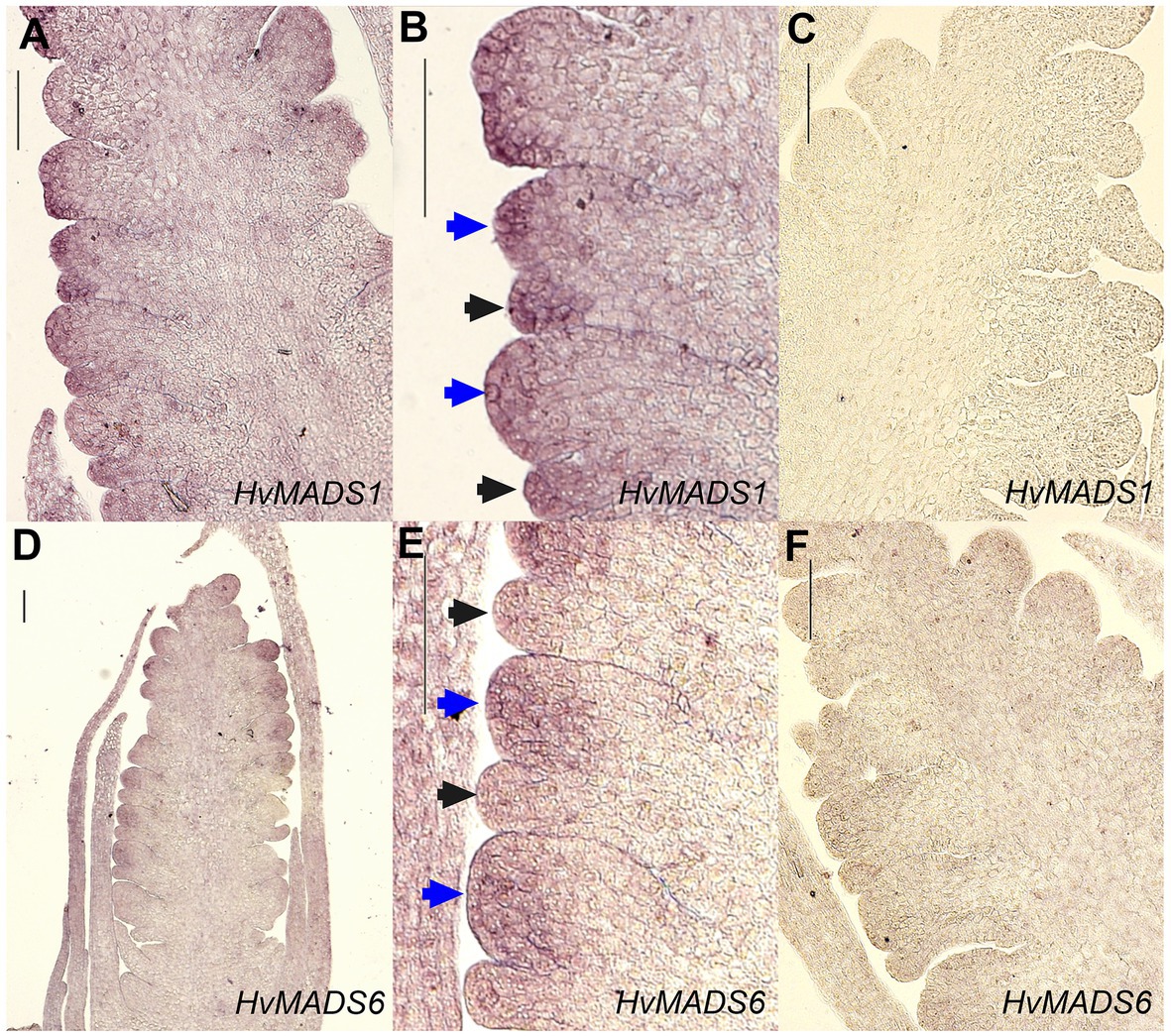
Figure 8. In situ hybridisation of HvMADS1 and HvMADS6 probes to sections of the barley inflorescence at W3. HvMADS1 is expressed in the floret meristem (blue arrowheads) and the lemma primordium (black arrowheads; A,B), but HvMADS6 is more strongly expressed in the floret meristem (D,E). (C, F) are the sense probe controls. Scale bars: 100μm.
Lodicule expression is shown for the AP1 clade genes HvMADS14 and HvMADS18, all three B-class and both C-class genes HvMADS3 and HvMADS58 plus strongly for the SEP3 subclade of the E-class genes, HvMADS7 and HvMADS8. In this tissue, the HvMADS32 transcript profile in the pseudoset is not similar to that of set 2 members, with the most notable difference being an early sharp decline after W6.5. HvMADS6 is also expressed in the lodicules. Canonically, the second whorl has A-, B- and E-class gene expression so the C-class transcript in barley lodicules is unexpected, and clashes strongly with the ABCDE model.
The AP1 clade genes HvMADS14 and HvMADS18 are both expressed in the stamens, and for HvMADS14, this is the highest expression seen in any floral organ. All three B-class genes and both C-class genes, HvMADS3 and HvMADS58 plus the E-class genes HvMADS7 and HvMADS8 are expressed here too, while the LOFSEP genes are only marginally expressed. Surprisingly, the highest expressed MIKCc gene in the stamens is HvMADS22, an SVP-like gene from correlation set 1. Ignoring the HvMADS22 expression before W2.5, the profile thereafter is very similar to that of correlation set 2, for HvMADS16 in particular (Figure 7C). The expected B-, C- and E-class expression for the third whorl is present in barley stamens, but the addition of A-class expression and HvMADS22 is unexpected.
Set 3: Carpel and Ovule Development
Correlation set 3 contains the canonical members of a carpel and an ovule quartet: C-, D- and E-class genes. Additionally, the expression of set 3 genes peaks past pollination at W10.5. HvMADS58, an AG clade (C-class) gene that is part of correlation set 3, shows strong expression in the carpel, while the other AG (C-class) gene HvMADS3, a member of set 2, is only marginally expressed (Figure 5). The expression of D-class genes HvMADS13 and HvMADS21 starts late, even compared to other set 3 members, after W6.5, and is found almost exclusively in the carpel tissue. HvMADS7 and HvMADS8 are expressed late in floret development, and the final two genes in correlation set 3 are the B-sister genes HvMADS29 and HvMADS31.
MADS2 and MADS4 Are Covered by Neighbouring Kinase Transcripts
The HvMADS4 (HORVU1Hr1G063620) genomic sequence is completely covered by the transcript of the neighbouring gene on the opposite DNA strand, HORVU1Hr1G063610, a serine/threonine protein kinase. As a result, any primer pair that targets HvMADS4 will also amplify this protein kinase transcript. To circumvent this problem, we designed a primer pair at the end of the HvMADS4 transcript, with an alternative reverse primer just outside of the HvMADS4 transcript. Subtracting the signal of the latter pair from the former provides a more accurate representation of the expression level of HvMADS4 only, which is designated HvMADS4* here.
A similar problem occurs with HvMADS2 where the genomic span of the gene is also transcribed from the opposite direction, encoding a neighbouring kinase, HORVU3Hr1G090990. In this case however, the kinase expression was so low compared to the HvMADS2 expression that trying to subtract it did not increase accuracy significantly.
Discussion
In this study, we revealed that the MIKCc MADS-box genes in barley are highly conserved and that their expression can be grouped in correlated sets that are linked to developmental events in the spike, spikelet and floret. This suggests that floral organogenesis is regulated by the ABCDE model in barley. Phylogeny shows that the MIKCc MADS-box family in barley is highly conserved, and SNP data confirm that natural variations of MIKCc MADS-box genes do not occur frequently during barley domestication. The consistency in the number of genes in each class and the mostly one to one matching homology of MIKCc genes between barley, rice, sorghum and Brachypodium (Figure 1; Arora et al., 2007; Wei et al., 2014) suggests that the determination of floral organs, a process dominated by the MIKCc family, is probably conserved as well. In the genome of bread wheat, 195 MIKCc MADS-box genes have been identified (Schilling et al., 2020). This high number is not only due to hexaploidy and subsequent frequent retention of MIKCc genes, but also due to recent duplication events, theorised to help wheat adapt to diverse growth conditions. The redundancy and neofunctionalisation have led to much more frequent and severe changes in nucleotide sequences of MIKCc genes in wheat. Nevertheless, retention of functional homologues of each rice MIKCc gene suggests the conservation of their role in the inflorescence development of wheat (Schilling et al., 2020). This is also reflected in the similar floral organ development pathway seen among grasses. The ABCDE model has been extrapolated to rice, maize and wheat previously (Ciaffi et al., 2011), and the morphological and genetic conservation suggests that it may apply to barley as well. While the barley MIKCc genes are highly conserved, variation in their expression profiles can provide an insight into the robustness of the ABCDE model in barley.
Grouping MIKCc MADS-box genes using a temporal expression profile in the developing inflorescence and floral organs has its drawbacks. Many shoot apical meristem samples contain spikelets and florets at multiple stages, with younger meristems near the top (Supplementary Figure 3). Additionally, many of the ABCDE proteins are predicted to participate in multiple floral quartets, or even have roles before the floral organs are initiated, like some A-class and E-class genes, which also complicates deconvoluting transcript profiles. However, these expression profiles can still be divided into four distinct groups mathematically (Figures 6A,B; Supplementary Figure 5). The profile of each gene, combined with expression data from individual floral organs, gives a clear indication of whether each gene conforms to their expected role within the ABCDE model, as found for homologues in rice, wheat and other plant species. Nonconforming transcript profiles are also highlighted which may hint at subfunctionalisation, neofunctionalisation or new interactions that warrant further investigation. Our data showed that most barley MIKCc MADS-box genes are expressed at the specific developmental stage and in the predicted floral organs during barley inflorescence development. However, there are several strong deviations in the expression patterns of some genes expected to have an ABCDE-class function, indicating neofunctionalisation.
SVP-Like MIKCc Genes Likely Act as Floral Inhibitors
The high start and quick decline of HvMADS22, HvMADS47 and HvMADS55 expression is in accordance with previous RNA sequencing of barley early inflorescence meristems (Digel et al., 2015; Supplementary Figure 2A) and is similar to RT-qPCR results reported by (Trevaskis et al., 2007b). In rice, the same pattern can be observed through the progression of meristem types, where OsMADS22 and OsMADS55 expression is high in the inflorescence meristem, lower in the branch meristem and at a minimum in the spikelet meristem (Harrop et al., 2016). In the inflorescence of Setaria viridis, a member of the Panicoideae (e.g. maize and sorghum) again the same decline in early inflorescence development is observed (Zhu et al., 2018). This conserved expression pattern likely indicates a conserved function of the SVP clade in grasses. In barley, the expression of HvMADS22 peaks again at W9.5, but this re-emergence later in floret development is mirrored only in wheat (Feng et al., 2017; Supplementary Figure 2B). HvMADS22 (BM10) and HvMADS47 (BM1) act as floral inhibitors and can cause partial or full floral reversion when ectopically expressed (Trevaskis et al., 2007b). The expression profiles of HvMADS22, HvMADS47 and HvMADS55 fit the function as floral inhibitors well, except for the resurgence of HvMADS22 expression.
APETALA and LOFSEP Transcripts Dominate in the Lemma and Palea
The ABCDE model states the first floral whorl is defined by A- and E-class genes (Theissen and Saedler, 2001). However, whether the palea and lemma are true first whorl floral organs in grasses is still debated (Ciaffi et al., 2011). HvMADS14, HvMADS15, and HvMADS18 are all strongly expressed in the lemma and palea, similar to observations in Brachypodium (Wei et al., 2014) and wheat (Paollacci et al., 2007), giving each APETALA gene the potential to fill the A-class role. Since HvMADS7 and HvMADS8 are not expressed in the lemma and palea, the E-class role is likely performed by the LOFSEP genes, also seen in Brachypodium (except for the MADS7 homologue being expressed in the palea; Wei et al., 2014) and in wheat (Paollacci et al., 2007). The quadruple knockdown of OsMADS1/5/7/8 (leaving OsMADS34 as the only remaining E-class gene) transforms all floral organs in the rice floret into leaf-like structures, except for the lemma (Cui et al., 2010). As HvMADS34 is more strongly expressed in the lemma in barley, this may be the only floral organ where HvMADS34 acts in an E-class role. Accumulation of barley HvMADS1 mRNA also showed a high level in lemma (Figures 8A–C). Expression of HvMADS32, the only member of a MIKCc class unique to monocots, in the lemma and palea makes it likely to be a member of the lemma and palea floral quartet, although its unique expression profile does not match the other likely members of the quartet. Additionally, the MADS32 homologue in Brachypodium shows only weak expression in the palea and not the lemma (Wei et al., 2014). The strongest difference in expression between the lemma and palea is for HvMADS6, which is weakly expressed in the lemma (Figures 8D–F). Even though HvMADS6 does not belong to the SEPALLATA clade, it has been reported to fulfil an E-class function in plants (reviewed by Dreni and Zhang, 2016).
The lemma and palea floral quartets in barley are probably composed of APETALA and LOFSEP proteins, in accordance with predictions from the ABCDE model (Figure 9). In the palea, HvMADS6 may play an E-class role in the floral quartet, possibly resulting in the morphological differences between the lemma and palea in barley.
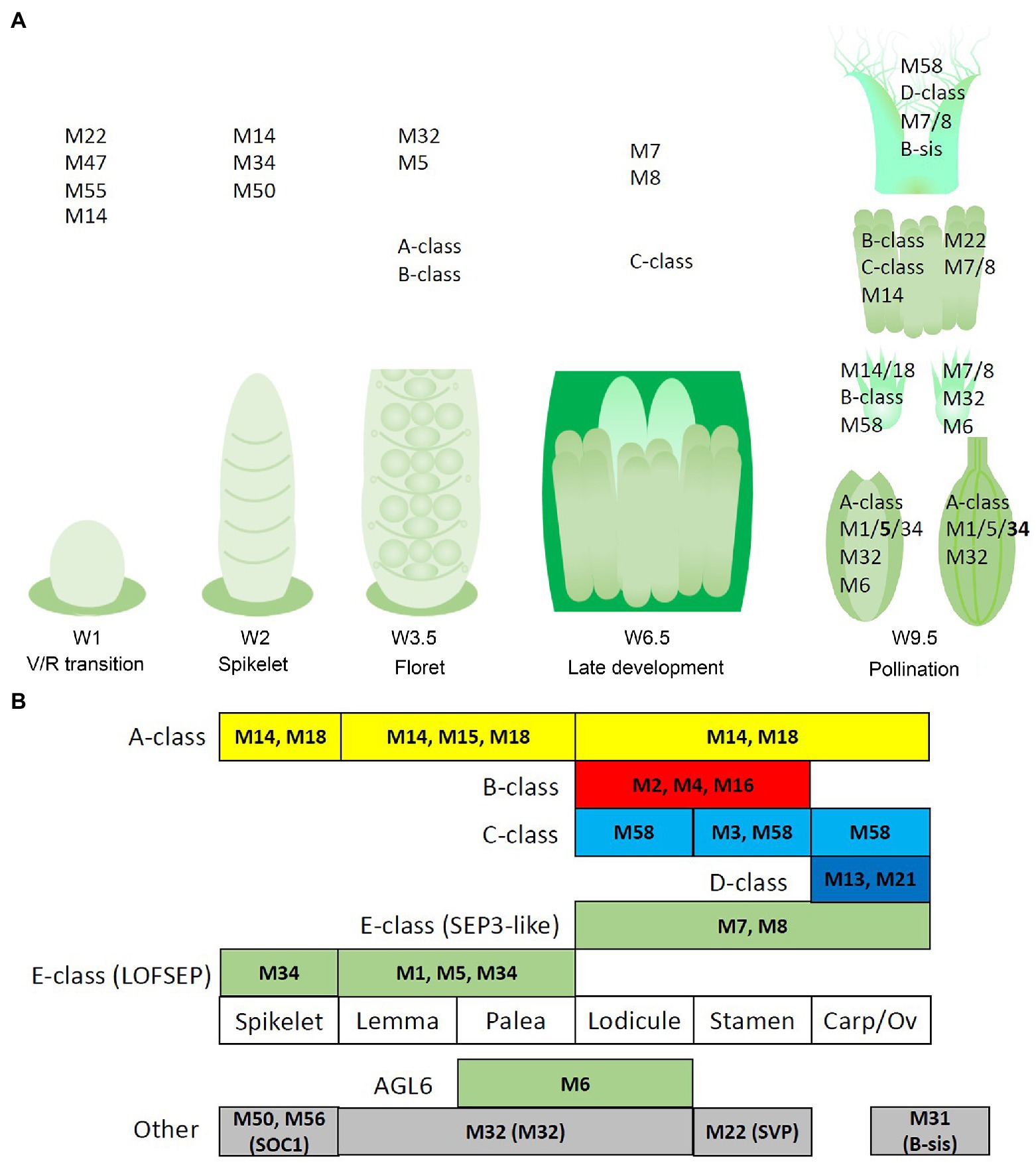
Figure 9. Potential regulatory networks of MIKCc MADS-box genes in barley inflorescence and floral development. In this figure, ‘MADS’ is abbreviated as ‘M’. (A) Prominent expression of the MIKCc MADS-box genes through developmental stages and the floral organs of barley. (B) Schematic of an adapted ABCDE model for barley floral development. Canonical ABCDE genes are depicted above the floral organs where they are expressed, while additional expressed MIKCc genes are shown below. Transcript before the start of floral organ primordia initiation is given in the leftmost column, tentatively labelled ‘spikelet’.
Lodicules Contain Predicted AP1, AP3/PI and SEP Transcript but Surprising AG (C-Class) Expression
The second whorl is canonically determined by a floral quartet consisting of one A-class, two B-class and one E-class protein. Unlike the APETALA gene MADS15 in rice and wheat (Kyozuka et al., 2000; Paollacci et al., 2007), the barley orthologue, HvMADS15, is not expressed in the lodicules. All three B-class genes are expressed in the barley lodicules, in accordance with the ABCDE model and this expression is neatly matched in Brachypodium and wheat (Paollacci et al., 2007; Wei et al., 2014). The conservation of this B-class function and mechanism in grasses is shown by the homeotic conversion of lodicules into whorl 1-like bracts in mutants of AP3/DEF subclade members OsMADS16/SUPERWOMAN1 in rice (Nagasawa et al., 2003) and SILKY in maize (Ambrose et al., 2000). The role of OsMADS2 and OsMADS4 as redundant PI/GLO seems clear from the spw1-like phenotype (homeotic conversion of stamens to carpels) of the double knockout line (Yao et al., 2008; Ciaffi et al., 2011). Moreover, the strong expression of E-class genes HvMADS7, HvMADS8 and AGL6-like gene HvMADS6 in the lodicules makes them the likely candidates for the E-class role in the lodicule-defining floral quartet in barley, which is consistent with rice homologues and maize MADS6 homologue ZAG3 (Thompson et al., 2009). Furthermore, the osmads7/8 double mutant shows aberrant lodicules (Cui et al., 2010), indicating that the LOFSEP (E-class) genes do not redundantly cover this function. Additionally, HvMADS32 is strongly expressed in the lodicules, but its transcript profile in the pseudoset is not similar to that of other potential members of the lodicule quartet, most notably its early sharp decline after W6.5. However, in rice, the osmads32 mutants do show some homeotic conversion of the lodicules (Sang et al., 2012), and a disrupted protein interaction with OsMADS2 and OsMADS4 is likely to be responsible for at least part of the OsMADS32 function (Wang et al., 2015). Therefore, HvMADS32 could have a function in lodicule determination and potentially be part of a lodicule quartet, but only in the early stages (Thompson et al., 2009).
Surprisingly, both barley C-class genes HvMADS3 and HvMADS58 are expressed in the lodicules, similar to homologues of wheat TaAG-1 and TaAG-2 and Brachypodium BdMADS18, but not with maize orthologues (ZAG1, ZmM2, and ZmM23; Mena et al., 1996; Paollacci et al., 2007; Wei et al., 2014). One of the central regulatory mechanisms in the ABCDE model is the antagonistic role of A- and C-class genes. In rice, the C-class genes have a role in suppressing additional lodicule formation (Yamaguchi et al., 2006). The separation of expression domains of the A-class and C-class genes, by mutual negative regulation, is one of the core tenets of the ABC model as originally devised in Arabidopsis. Here, we show that in barley, APETALA clade genes are expressed in the inner floral organs, and AGAMOUS clade transcripts show up in the second whorl floral organs, the lodicules.
Stamens Contain Transcripts of Both Predicted and Unexpected Members
The A-class genes HvMADS14 and HvMADS18 are both expressed in the stamens, where for HvMADS14, it is the highest expression in any floral organ. Similarly, the wheat homologue TaAP1-1 is expressed in all floral organs, and in Brachypodium, the AP1 gene BdMADS3 is also expressed in the stamens (Paollacci et al., 2007). In rice, OsMADS14 and OsMADS18 are expressed in the stamens, but OsMADS14 is the main actor in stamen identity (Wu et al., 2017). All three B-class genes are expressed as expected, which is similar to rice, where a knockdown of the rice B-class gene OsMADS16 or both OsMADS2 and OsMADS4 results in homeotic conversion of the stamens into carpel-like organs (Yao et al., 2008). C-class genes, HvMADS3 and HvMADS58, are strongly expressed here, similar to their counterparts in rice. Rice OsMADS3 plays a crucial role in stamen identity (Yamaguchi et al., 2006), but the relative importance of C-class genes in barley will have to be investigated further. Furthermore, transcripts of E-class genes HvMADS7 and HvMADS8 are present, where the LOFSEP genes are only marginally expressed. The homologous genes in wheat, TaSEP4 and TaSEP3, are also predominantly expressed in the inner floral organs (Paollacci et al., 2007). In rice, OsMADS7/8 double knockdown plants the stamens were affected, but not completely abolished as in the OsMADS1/5/7/8 quadruple knockdown lines, so OsMADS7 and OsMADS8 have a primary E-class function, but not an exclusive one (Cui et al., 2010). Surprisingly, the most highly expressed MIKCc gene in the stamens is HvMADS22, an SVP-like gene, which normally functions as a floral repressor and in Brachypodium the HvMADS22 homologue BdMADS30 is also strongly expressed (Wei et al., 2014). Investigating expression and phenotypic differences correlated with the SNPs variations may provide insight in the HvMADS22 role.
In summary, the canonical members of the third whorl floral quartet are expressed in barley stamens: HvMADS16 (AP3/DEF B-class), HvMADS2 or HvMADS4 (PI/GLO B-class), HvMADS3 or HvMADS58 (C-class) and HvMADS7 or HvMADS8 (E-class). However, the APETALA clade genes HvMADS14 and HvMADS18, and an SVP-like gene, HvMADS22, also show significant expression (Figure 9A). While the expression profiles for HvMADS14 and HvMADS18 do not reveal meaningful co-expression, HvMADS22 expression after W2.5 is very similar to other probable stamen quartet members (Figure 7C), suggesting the unlikely neofunctionalisation of a floral repressor in this organ.
AG, SEP, and B-Sister Genes Are Expressed in the Carpel and Ovule
Carpel fate is induced by a quartet of two C- and two E-class genes, while the ovule quartet contains one C-, two D- and one E-class gene (Theissen et al., 2016). Floral meristem determinacy (FMD) is likely to be regulated by the remnant of the floret meristem, located within the carpel samples.
Strong carpel expression of the AG (C-class) gene HvMADS58, in contrast to marginal HvMADS3 (Figure 3B) expression, is a sign of subfunctionalisation among the AG genes where the C-class role in carpel development is fulfilled primarily by HvMADS58. The wheat homologue of HvMADS3, TaAG-2, is also predominantly expressed in the stamens, compared to the carpel (Paollacci et al., 2007). An osmads58 mutant in rice develops abnormal carpels, while osmads3 carpels develop almost completely normally, showing that OsMADS58 is the primary C-class gene for carpel development (Yamaguchi et al., 2006). The late expression of D-class genes HvMADS13 and HvMADS21 is found almost exclusively in the carpel samples (collected at W9.5), which is consistent with a potential role in ovule development. HvMADS7 and HvMADS8, E-class genes of the SEP3 sub-clade, are expressed late in floret development, as in wheat (Feng et al., 2017) and potentially fulfil the E-class role for the inner floral organs. For HvMADS8 (BM9), this boundary at the lodicule has also been shown by in situ hybridisation; however, it also shows HvMADS1 (BM7) expression in the developing ovule (Schmitz et al., 2000). While HvMADS1 is expressed in the carpel at W9.5, most of it disappears after pollination. This leads to the conclusion that HvMADS7 and HvMADS8 are likely part of the carpel and ovule quartets, and HvMADS1 may have a function during ovule development or FMD. The final two genes in correlation set 3 are the B-sister genes HvMADS29 and HvMADS31. AtABS, a B-sister gene in Arabidopsis, has been linked to endothelium development and interaction with AtSEP3, and D-class genes suggest they may function in an additional floral quartet (Kaufmann et al., 2005). No expression of the B-sister gene HvMADS30 was detected. OsMADS30 is not a suitable guide for the function of its homologue in barley because the rice gene has a recent insertion and an altered expression pattern compared to related grass species (Schilling et al., 2015). However, it was recently revealed that MADS30-like gene expression is induced by biotic stress in wheat (Schilling et al., 2020), suggesting that a similar mechanism in barley may be why no HvMADS30 expression was identified. Short genes can be expressed more rapidly than long genes and can be associated with fast dividing cells, particularly in zygotic tissue (Heyn et al., 2015). B-sister genes have short introns (Supplementary Figure 1), and their expression is associated with tissues of the ovule and developing grain. Long genes take longer to express, causing a so-called ‘intron delay’ that can be of regulatory significance. Additionally, longer genes with sizable introns are often more highly expressed (Heyn et al., 2015). However, there does not seem to be any clear correlation between intron size and frequency of expression for MIKCc genes in barley.
To summarise, the canonical MIKCc members of the fourth whorl quartet are present in the barley carpel: C-class gene HvMADS58 and E-class genes HvMADS7 and HvMADS8. The ovule quartet is also represented in the carpel samples: HvMADS58 as the C-class gene, both D-class genes (HvMADS13 and HvMADS21) and two E-class genes: HvMADS7 and HvMADS8 (Figures 9A,B). The additional expression of B-sister genes in this correlation set may imply a redundant function in the carpel or the ovule-determining quartet; however, it is more likely to be related to a function in the endothelium and other ovule and early seed roles. The B-sister proteins in eudicots have been shown to interact with C-, D- and E-class proteins, and the mutant has defects in the endothelium (de Folter et al., 2006).
MIKCc Gene Expression Implies a Role in Inflorescence-, Spikelet- and Floret Meristems
MIKCc genes play an important role in grass inflorescence architecture and spikelet differentiation (Digel et al., 2015; Liu et al., 2015; Li et al., 2021; Wang et al., 2021). In our study, SVP-like genes, HvMADS22, HvMADS47 and HvMADS55, and one A-class gene HvMADS14 (VRN1), and E-class gene HvMADS34, show a high transcription level at the early inflorescence development stage before spikelet differentiation, suggesting a role for these genes in inflorescence meristem maintenance and spikelet meristem identity. APETALA1 genes in barley are likely to perform A-class functions in floral organ determination (see below); however, HvMADS14 (VRN1) has an additional role in the vernalisation response and probably in establishing and maintaining inflorescence meristem identity in barley (Trevaskis et al., 2007a). The early expression of HvMADS18, along with the decline at W4.5, is less pronounced than for HvMADS14, but still recognisable, indicating a potentially weaker redundant role in establishing and maintaining inflorescence meristem identity. HvMADS15 is part of correlation set 2 and is therefore more likely to perform an A-class function exclusively. A similar divide is present in wheat, where MADS14 and MADS18 co-homologues have reduced expression after W4, while MADS15 co-homologues do not (Feng et al., 2017).
Mutants in rice show that E-class OsMADS34 gene is involved in inflorescence branching (Gao et al., 2010; Kobayashi et al., 2010), and unsurprisingly, OsMADS34 is highly expressed in the inflorescence branch meristem of rice (Harrop et al., 2016). However, mutation of barley HvMADS34 does not show the change of inflorescence architecture (Li et al., 2021). When comparing E-class gene expression in the early inflorescence meristem between barley and rice, the early peak of HvMADS34 expression is conserved (Figures 7A,B). No other E-class gene (nor HvMADS6) could provide redundancy for a potential HvMADS34 function around stage W2, because their expression starts later in inflorescence development. We can only speculate that the early HvMADS34 expression is merely a vestigial remnant from the ancestral inflorescence, which did have a branched morphology (Remizowa et al., 2013). Recently, barley E-class MADS-box protein, HvMADS1, has been reported to be responsible for maintaining an unbranched spike architecture at high temperatures; the hvmads1 mutant shows the changed inflorescence meristem determinacy at warm temperature conditions, forming a branched inflorescence-like structure (Li et al., 2021). Thus, the Triticeae spikes merely suppress inflorescence branching is given credence by the branching phenotype of the com2 (COMPOSITUM2) and com1/bdi1 (COMPOSITUM1/BRANCHED AND INDETERMINATE SPIKELET 1) mutants in barley and the tetraploid ‘miracle wheat’ (Poursarebani et al., 2015, 2020; Shang et al., 2020) and in the loss-of-function of barley HvMADS1 mutant under high ambient temperature (Li et al., 2021) which show that most of the components for a branching inflorescence are still present in some Triticeae.
The loosely correlated genes in the pseudoset display early expression in the window between floral transition and the start of floral organ formation, when the spikelet and floret meristems are formed. The sudden downturn in transcript between W4 and W4.5 that many members of the pseudoset have in common (Figure 6B) coincides with the end of formation of new spikelet meristems at the awn primordium stage (Alqudah and Schnurbusch, 2014). This indicates that members of this pseudoset could be involved in inflorescence meristem determination, including the inflorescence-, spikelet- and floret meristems.
Adapting the ABCDE Model for Barley
Overall, the ABCDE model for grasses still follows the same basic structure as the model from Arabidopsis, the addition of DELLA notwithstanding (Ciaffi et al., 2011). The results presented here show that this generally holds true for MIKCc gene expression in barley as well, although there are some deviations. The ABCDE proteins are known to initiate floral organ fate, as shown by mutants with homeotic changes. However, their role is not limited to just the initial direction of floral organ primordia. ABCDE proteins have been shown to bind in an organ specific way to promoter regions of genes involved in growth and differentiation of floral organ tissues, up to SPOROCYTELESS, a master regulator of gametogenesis in Arabidopsis (Chen et al., 2018a). The rising expression levels of most ABCDE-class genes throughout floret development indicate this continuous affirmation of organ identity by floral quartets may be present in barley as well. These persistent roles make floral organ sample collection at W9.5 a reasonable predictor of floral organ fate determining ABCDE-class genes. However, when looking at the MIKCc genes outside the ABCDE-functions, the most strongly expressed genes are HvMADS22 in the stamens, usually classified as a floral repressor, and HvMADS32, which may be crucial for the discrete border between the outer and inner floral organs (Figure 9B). Some basal angiosperms have a more gradual transition between their floral organs, which does not fit with the ABCDE model, which results in discrete floral whorls. This is accompanied by a more gradual change in gene expression in these taxa and is captured in the ‘fading borders’ model (Buzgo et al., 2004; Soltis et al., 2007). This states that the gradually rising expression of, for example, C-class genes, and the slowly fading expression of A-class genes, results in intermediate floral organs with some characteristics from the adjacent organs. This may be the ancestral angiosperm ABC model, where only later more stringent restrictions on the expression evolved to separate the second and third whorls, resulting in the A–C antagonism in the ABCDE model for eudicots, and perhaps a different solution evolved in grasses, involving HvMADS32.
Because MIKCc proteins function in floral quartets, the next step to gain more insight into the potentially changed roles of these genes should be protein interaction studies. So far, when discussing the ABCDE model, the floral organs have often been considered indivisible units that either gain the correct identity or are homeotically converted. In the barley stamens, 10 different MIKCc genes from five classes are strongly expressed (Figure 9B), which are unlikely to form just one floral quartet. There may be variants of the stamen quartet that help define specific tissues within the stamens or even complete quartets. Alternatively, some of these MIKCc genes may have a function in stamens independent of a floral quartet structure. A somewhat similar tissue-specific expression of MIKCc genes has been shown in the ovule of rice (Kubo et al., 2013).
Altogether, these findings show that while the general setup of flowering is conserved, there are many interesting deviations in barley, and likely other grasses, that merit further research into what they mean for both the evolution of flowering in grasses and the potential adaptability of the inflorescence for crop yield breeding.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author Contributions
Study was conceived by HK and DZ. Experiments were done by HK, NS, and SK. Bioinformatics were done by HK, JSh, and JSc. Data analysis was done by HK, NS, and GL. Manuscript was written by HK, GL, and RB. Supervision and funding by RB and DZ. All authors contributed to the article and approved the submitted version.
Funding
HK was supported by a University of Adelaide Beacon postgraduate scholarship, and the work was completed as part of the Australian Research Council Discovery Project (grant no. DP170103267 and DP170103352) and the Science and Research grant of Southwest University of Science and Technology (grant no. 19zx7146).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dr. Xiuyuan Yang, Dr. Gwen Mayo and Adelaide Microscopy for help with the in situ experiment; Laura Wilkinson for the HvMADS13 primers; and Cindy Callens for the HvMADS29 primers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.705286/full#supplementary-material
Footnotes
1. ^https://webblast.ipk-gatersleben.de/barley_ibsc/downloads/
2. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^https://bridge.ipk-gatersleben.de/#snpbrowser, accessed April 2019.
5. ^https://barley-pangenome.ipk-gatersleben.de/, accessed February 2021.
7. ^http://snp-seek.irri.org/_snp.zul, accessed June 2019.
References
Alqudah, A. M., and Schnurbusch, T. (2014). Awn primordium to tipping is the most decisive developmental phase for spikelet survival in barley. Funct. Plant Biol. 41:424. doi: 10.1071/FP13248
Alvarez-Buylla, E. R., Benítez, M., Corvera-Poiré, A., Chaos Cador, A., de Folter, S., Gamboa de Buen, A., et al. (2010). Flower development. Arabidopsis Book 8:e0127. doi: 10.1199/tab.0127
Ambrose, B. A., Lerner, D. R., Ciceri, P., Padilla, C. M., Yanofsky, M. F., and Schmidt, R. J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5, 569–579. doi: 10.1016/S1097-2765(00)80450-5
Angenent, G. C., Franken, J., Busscher, M., Vandijken, A., Vanwent, J. L., Dons, H. J. M., et al. (1995). A novel class of mads box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. doi: 10.1105/tpc.7.10.1569
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Becker, A., and Theißen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/s1055-7903(03)00207-0
Bommert, P., Satoh-Nagasawa, N., Jackson, D., and Hirano, H.-Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46, 69–78. doi: 10.1093/pcp/pci504
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. doi: 10.1242/dev.126.11.2387
Bremer, K. (2002). Gondwanan evolution of the grass alliance of families (Poales). Evolution 56, 1374–1387. doi: 10.1111/j.0014-3820.2002.tb01451.x
Burton, R. A., Jobling, S. A., Harvey, A. J., Shirley, N. J., Mather, D. E., Bacic, A., et al. (2008). The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 146, 1821–1833. doi: 10.1104/pp.107.114694
Buzgo, M., Soltis, D. E., Soltis, P. S., and Ma, H. (2004). Towards a comprehensive integration of morphological and genetic studies of floral development. Trends Plant Sci. 9, 164–173. doi: 10.1016/j.tplants.2004.02.003
Callens, C., Tucker, M. R., Zhang, D., and Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459. doi: 10.1093/jxb/ery086
Castelán-Muñoz, N., Herrera, J., Cajero-Sánchez, W., Arrizubieta, M., Trejo, C., García-Ponce, B., et al. (2019). MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 10:853. doi: 10.3389/fpls.2019.00853
Chen, D., Yan, W., Fu, L. Y., and Kaufmann, K. (2018a). Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat. Commun. 9:4534. doi: 10.1038/s41467-018-07850-2
Chen, F., Zhang, X., Liu, X., and Zhang, L. (2017). Evolutionary analysis of MIKCc-type MADS-box genes in gymnosperms and angiosperms. Front. Plant Sci. 8:895. doi: 10.3389/fpls.2017.02248
Chen, L., Zhao, Y., Xu, S., Zhang, Z., Xu, Y., Zhang, J., et al. (2018b). OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 218, 219–231. doi: 10.1111/nph.14977
Ciaffi, M., Paolacci, A. R., Tanzarella, O. A., and Porceddu, E. (2011). Molecular aspects of flower development in grasses. Sex. Plant Reprod. 24, 247–282. doi: 10.1007/s00497-011-0175-y
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls – genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Colombo, M., Masiero, S., Vanzulli, S., Lardelli, P., Kater, M. M., and Colombo, L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54, 1037–1048. doi: 10.1111/j.1365-313X.2008.03485.x
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
de Folter, S., Shchennikova, A. V., Franken, J., Busscher, M., Baskar, R., Grossniklaus, U., et al. (2006). A B-sister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 47, 934–946. doi: 10.1111/j.1365-313X.2006.02846.x
Digel, B., Pankin, A., and von Korff, M. (2015). Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27, 2318–2334. doi: 10.1105/tpc.15.00203
Dreni, L., Pilatone, A., Yun, D., Erreni, S., Pajoro, A., Caporali, E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23, 2850–2863. doi: 10.1105/tpc.111.087007
Dreni, L., and Zhang, D. (2016). Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. J. Exp. Bot. 67, 1625–1638. doi: 10.1093/jxb/erw046
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, N., Song, G., Guan, J., Chen, K., Jia, M., Huang, D., et al. (2017). Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol. 174, 1779–1794. doi: 10.1104/pp.17.00310
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Guo, S., Xu, Y., Liu, H., Mao, Z., Zhang, C., Ma, Y., et al. (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4:1566. doi: 10.1038/ncomms2542
Harrop, T. W., Ud Din, I., Gregis, V., Osnato, M., Jouannic, S., Adam, H., et al. (2016). Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. Plant J. 86, 75–88. doi: 10.1111/tpj.13147
Heyn, P., Kalinka, A. T., Tomancak, P., and Neugebauer, K. M. (2015). Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 37, 148–154. doi: 10.1002/bies.201400138
Jayakodi, M., Padmarasu, S., Haberer, G., Bonthala, V. S., Gundlach, H., Monat, C., et al. (2020). The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289. doi: 10.1038/s41586-020-2947-8
Jin, J., Zhang, H., Kong, L., Gao, G., and Luo, J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–D1187. doi: 10.1093/nar/gkt1016
Kaufmann, K., Anfang, N., Saedler, H., and Theissen, G. (2005). Mutant analysis, protein-protein interactions and subcellular localization of the Arabidopsis B-sister (ABS) protein. Mol. Gen. Genomics. 274, 103–118. doi: 10.1007/s00438-005-0010-y
Kobayashi, K., Maekawa, M., Miyao, A., Hirochika, H., and Kyozuka, J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51, 47–57. doi: 10.1093/pcp/pcp166
Koppolu, R., and Schnurbusch, T. (2019). Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J. Integr. Plant Biol. 61, 278–295. doi: 10.1111/jipb.12771
Kubo, T., Fujita, M., Takahashi, H., Nakazono, M., Tsutsumi, N., and Kurata, N. (2013). Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 54, 750–765. doi: 10.1093/pcp/pct029
Kwantes, M., Liebsch, D., and Verelst, W. (2012). How MIKC* MADS-Box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302. doi: 10.1093/molbev/msr200
Kyozuka, J., Kobayashi, T., Morita, M., and Shimamoto, K. (2000). Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiol. 41, 710–718. doi: 10.1093/pcp/41.6.710
Li, G., Kuijer, H. N. J., Yang, X., Liu, H., Shen, C., Shi, J., et al. (2021). MADS1 maintains barley spike morphology at high ambient temperatures. Nat. Plants. doi: 10.1038/s41477-021-00957-3
Liu, H., Li, G., Yang, X., Kuijer, H. N. J., Liang, W., and Zhang, D. (2020). Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. Crop J. 8, 71–86. doi: 10.1016/j.cj.2019.04.005
Mansueto, L., Fuentes, R. R., Borja, F. N., Detras, J., Abriol-Santos, J. M., Chebotarov, D., et al. (2017). Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res. 45, D1075–D1081. doi: 10.1093/nar/gkw1135
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mayer, K. F. X., Waugh, R., Langridge, P., Close, T. J., Wise, R. P., Graner, A., et al. (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. doi: 10.1038/nature11543
Mena, M., Ambrose, B. A., Meeley, R. B., Briggs, S. P., Yanofsky, M. F., and Schmidt, R. J. (1996). Diversification of C-function activity in maize flower development. Science 274, 1537–1540. doi: 10.1126/science.274.5292.1537
Milner, S. G., Jost, M., Taketa, S., Mazón, E. R., Himmelbach, A., Oppermann, M., et al. (2019). Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 51, 319–326. doi: 10.1038/s41588-018-0266-x
Monat, C., Padmarasu, S., Lux, T., Wicker, T., Gundlach, H., Himmelbach, A., et al. (2019). TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 20:284. doi: 10.1186/s13059-019-1899-5
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Paollacci, A. R., Tanzarella, O. A., Porceddu, E., Varotto, S., and Ciaffi, M. (2007). Molecular and phylogenetic analysis of MADS-box genes of MIKC type and chromosome location of SEP-like genes in wheat (Triticum aestivum L.). Mol. Gen. Genomics. 278, 689–708. doi: 10.1007/s00438-007-0285-2
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Poursarebani, N., Seidensticker, T., Koppolu, R., Trautewig, C., Gawronski, P., Bini, F., et al. (2015). The genetic basis of composite spike form in barley and “miracle-wheat”. Genetics 201, 155–165. doi: 10.1534/genetics.115.176628
Poursarebani, N., Trautewig, C., Melzer, M., Nussbaumer, T., Lundqvist, U., Rutten, T., et al. (2020). COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun. 11:5138. doi: 10.1038/s41467-020-18890-y
Remizowa, M. V., Rudall, P. J., Choob, V. V., and Sokoloff, D. D. (2013). Racemose inflorescences of monocots: structural and morphogenetic interaction at the flower/inflorescence level. Ann. Bot. 112, 1553–1566. doi: 10.1093/aob/mcs246
Ruelens, P., de Maagd, R. A., Proost, S., Theißen, G., Geuten, K., and Kaufmann, K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280. doi: 10.1038/ncomms3280
Sang, X. C., Li, Y. F., Luo, Z. K., Ren, D. Y., Fang, L. K., Wang, N., et al. (2012). Chimeric floral Organs1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol. 160, 788–807. doi: 10.1104/pp.112.200980
Schilling, S., Gramzow, L., Lobbes, D., Kirbis, A., Weilandt, L., Hoffmeier, A., et al. (2015). Non-canonical structure, function and phylogeny of the B-sister MADS-box gene OsMADS30 of rice (Oryza sativa). Plant J. 84, 1059–1072. doi: 10.1111/tpj.13055
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S., and Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1111/nph.16122
Schmitz, J., Franzen, R., Ngyuen, T. H., Garcia-Maroto, F., Pozzi, C., Salamini, F., et al. (2000). Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 42, 899–913. doi: 10.1023/A:1006425619953
Shang, Y., Yuan, L., Di, Z., Jia, Y., Zhang, Z., Li, S., et al. (2020). A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 71–22, 7118–7131. doi: 10.1093/jxb/eraa416
Smaczniak, C., Immink, R. G. H., Angenent, G. C., and Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Soltis, D. E., Chanderbali, A. S., Kim, S., Buzgo, M., and Soltis, P. S. (2007). The ABC model and its applicability to basal angiosperms. Ann. Bot. 100, 155–163. doi: 10.1093/aob/mcm117
Solovyev, V. (2007). “Statistical approaches in eukaryotic gene prediction,” in Handbook of Statistical Genetics. 3rd Edn. eds. D. J. Balding, M. Bishop, and C. Cannings (Wiley-Interscience), 1616.
Theissen, G., Melzer, R., and Rumpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Thompson, B. E., Bartling, L., Whipple, C., Hall, D. H., Sakai, H., Schmidt, R., et al. (2009). Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21, 2578–2590. doi: 10.1105/tpc.109.067751
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007a). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Trevaskis, B., Tadege, M., Hemming, M. N., Peacock, W. J., Dennis, E. S., and Sheldon, C. (2007b). Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 143, 225–235. doi: 10.1104/pp.106.090860
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Waddington, S. R., Cartwright, P. M., and Wall, P. C. (1983). A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. 51, 119–130. doi: 10.1093/oxfordjournals.aob.a086434
Wang, C., Yang, X., and Li, G. (2021). Molecular insights into inflorescence meristem specification for yield potential in cereal crops. Int. J. Mol. Sci. 22:3508. doi: 10.3390/ijms22147719
Wang, H. H., Zhang, L., Cai, Q., Hu, Y., Jin, Z. M., Zhao, X. X., et al. (2015). OsMADS32 interacts with PI-like proteins and regulates rice flower development. J. Integr. Plant Biol. 57, 504–513. doi: 10.1111/jipb.12248
Wei, B., Zhang, R. Z., Guo, J. J., Liu, D. M., Li, A. L., Fan, R. C., et al. (2014). Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS One 9:e84781. doi: 10.1371/journal.pone.0116239
Wu, D., Liang, W., Zhu, W., Chen, M., Ferrandiz, C., Burton, R. A., et al. (2018). Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 176, 1646–1664. doi: 10.1104/pp.17.00704
Wu, F., Shi, X., Lin, X., Liu, Y., Chong, K., Theissen, G., et al. (2017). The ABCs of flower development: mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J. 89, 310–324. doi: 10.1111/tpj.13386
Yadav, S. R., Prasad, K., and Vijayraghavan, U. (2007). Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176, 283–294. doi: 10.1534/genetics.107.071746
Yamaguchi, T., Lee, D. Y., Miyao, A., Hirochika, H., An, G., and Hirano, H. Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18, 15–28. doi: 10.1105/tpc.105.037200
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H. Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509. doi: 10.1105/tpc.018044
Yao, S. G., Ohmori, S., Kimizu, M., and Yoshida, H. (2008). Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 49, 853–857. doi: 10.1093/pcp/pcn050
Keywords: barley, inflorescence, floral organs, meristems, MADS, transcript profiling, ABC model, floret
Citation: Kuijer HNJ, Shirley NJ, Khor SF, Shi J, Schwerdt J, Zhang D, Li G and Burton RA (2021) Transcript Profiling of MIKCc MADS-Box Genes Reveals Conserved and Novel Roles in Barley Inflorescence Development. Front. Plant Sci. 12:705286. doi: 10.3389/fpls.2021.705286
Edited by:
Fa Cui, Ludong University, ChinaReviewed by:
Luis Morales-Quintana, Autonomous University of Chile, ChileYong Jia, Murdoch University, Australia
Copyright © 2021 Kuijer, Shirley, Khor, Shi, Schwerdt, Zhang, Li and Burton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Li, Z2FuZy5saUBhZGVsYWlkZS5lZHUuYXU=; Rachel A. Burton, cmFjaGVsLmJ1cnRvbkBhZGVsYWlkZS5lZHUuYXU=
 Hendrik N. J. Kuijer
Hendrik N. J. Kuijer Neil J. Shirley
Neil J. Shirley Shi F. Khor
Shi F. Khor Jin Shi2
Jin Shi2 Gang Li
Gang Li Rachel A. Burton
Rachel A. Burton