- Division of Biological Science and Technology, Yonsei University, Wonju, South Korea
ATBS1-INTERACTING FACTOR 2 (AIF2) is a non-DNA-binding basic-helix-loop-helix (bHLH) transcription factor. Here, we demonstrate that AIF2 negatively modulates brassinosteroid (BR)-induced, BRASSINAZOLE RESISTANT 1 (BZR1)-mediated pollen and seed formation. AIF2-overexpressing Arabidopsis plants (AIF2ox) showed defective pollen grains and seed production while two AIF2 knockout mutants, aif2-1 and aif2-1/aif4-1, displayed opposite phenotypes. Genes encoding BZR1-regulated positive factors of seed size determination (SHB1, IKU1, MINI3) were suppressed in AIF2ox and genes for negative factors (AP2 and ARF2) were enhanced. Surprisingly, BZR1-regulated pollen genes such as SPL, MS1, and TDF1 were aberrantly up-regulated in AIF2ox plants. This stage-independent abnormal expression may lead to a retarded and defective progression of microsporogenesis, producing abnormal tetrad microspores and pollen grains with less-effective pollen tube germination. Auxin plays important roles in proper development of flower and seeds: genes for auxin biosynthesis such as TCPs and YUCCAs as well as for positive auxin signalling such as ARFs were suppressed in AIF2ox flowers. Moreover, lipid biosynthesis- and sucrose transport-related genes were repressed, resulting in impaired starch accumulation. Contrarily, sucrose and BR repressed ectopic accumulation of AIF2, thereby increasing silique length and the number of seeds. Taken together, we propose that AIF2 is negatively involved in pollen development and seed formation, and that sucrose- and BR-induced repression of AIF2 positively promotes pollen production and seed formation in Arabidopsis.
Introduction
Seed development and seed size determination in plants are complicated processes controlled by diverse hormones and downstream transcription factors (Sun X. et al., 2010; Li and Li, 2016). Seeds comprise three genetically distinctive structures: the embryo giving rise to the seedling, the endosperm providing nutrients for the embryo, and the seed coat enclosing the embryo and endosperm. The endosperm arises from the central cell and constitutes the major volume of the mature seed. In Arabidopsis after fertilisation, rapid proliferation and expansion of the endosperm occurs to generate a large and multinucleated cell or syncytium until the embryo reaches the heart stage and results in a large increase in seed size or volume of the seed cavity (Sun X. et al., 2010). Several factors have been shown to control seed size by regulating endosperm growth (Li and Li, 2016). Loss-of-function mutations of HAIKU (IKU) and the WRKY transcription factor MINI-SEED 3 (MINI3) caused precocious cellularisation of the syncytial endosperm resulting in the reduction in endosperm size and embryo proliferation (Garcia et al., 2003; Luo et al., 2005; Wang et al., 2010). The recruitment of SHORT HYPOCOTYL UNDER BLUE1 (SHB1) by MINI3 to its own and IKU2 promoters upregulated their expression (Zhou et al., 2009). Arabidopsis APETALA2 (AP2) encodes a plant transcription factor having the AP domain that is negatively involved in regulation of seed size and numbers (Ohto et al., 2009). ap2 seeds underwent an early expanded growth period that was associated with delayed endosperm cellularisation and outgrowth of the endosperm central vacuole, resulting in an increase in embryo cell number and size, enlarged embryo sac, and large seeds with increased total protein and oil content (Jofuku et al., 2005; Ohto et al., 2009). Additionally, proteins involved in ubiquitin–proteasome pathways, G-protein signalling, mitogen-activated protein kinase signalling, and epigenetic regulation and paternal imprinting were substantially involved in the control of seed size and numbers (Sun X. et al., 2010; Li and Li, 2016; Li N. et al., 2019).
Plant hormones are closely involved in the regulation of reproduction, embryogenesis, and determination of seed size and yields. Auxin signalling was closely linked to endosperm development, embryo polarity, and patterning (Figueiredo and Köhler, 2018). Embryo sacs of plants selectively silenced for AUXIN RESPONSIVE FACTORs (ARFs) exhibited identity defects at the micropylar pole, and the pollen grains were morphologically aberrant and unviable (Liu et al., 2018). In addition, a loss-of-function mutant of ARF2, initially identified as mmt mutant, showed extra cell division in the integuments surrounding the ovule, leading to the formation of enlarged seed coats and seed size (Schruff et al., 2006). ABSCISIC ACID-INSENSITIVE5 (ABI5)-mediated abscisic acid (ABA) signalling pathways were negatively involved in the early stage of seed development by suppressing SHB1 expression; thus, an ABA biosynthesis-deficient mutant, aba2-1, produced seeds with increased size, mass, and embryo cell number (Cheng et al., 2014). YODA (YDA) is a mitogen-activated protein kinase, and YDA and ETHYLENE-INSENSITIVE3 (EIN3) were integral to a sugar-mediated metabolism cascade regulating seed mass by maternally controlling embryo and seed sizes (Meng et al., 2018). Transcriptome analysis of the early stage of proliferating endosperm revealed that cytokinin signalling-related genes were significantly enriched (Day et al., 2008). Indeed, triple loss-of-function mutants of cytokinin receptors, ahk2 ahk3 cre1, produced enlarged but fewer seeds per silique, and this increase in seed size was correlated with an increase in the size of the mutant embryo (Riefler et al., 2006).
Brassinosteroids (BRs) are plant steroid hormones that play crucial roles in plant growth and development via extensive signal integration through direct interactions with numerous signalling pathways (Kim and Russinova, 2020). Upon binding of BRs to BRASSINOSTEROID-INSENSITIVE 1 (BRI1), the activation of BRI1 and BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) complex and the subsequent phosphorylation of BRASSINOSTEROID SIGNALLING KINASE (BSK) initiated a signalling cascade, relaying the membrane surface signal to the nucleus to activate the positively acting transcription factors BRASSINAZOLE RESISTANT 1/BRI1 EMS SUPPRESSOR 2 (BZR1/BES2) and BZR2/BES1 (He et al., 2005; Sun Y. et al., 2010). In the absence of BRs, their growth-promoting pathways were negatively balanced through GSK3/SHAGGY-LIKE BRASSINOSTEROID-INSENSITIVE 2 (ATSK21/BIN2)-mediated BZR1/BES1 degradation (He et al., 2002) and the antagonistic BIN2-driven increase in ATBS1-INTERACTING FACTOR 2 (AIF2) stability, an atypical non-DNA-binding bHLH transcription factor acting as a negative regulator of BR-induced growth promotion (Kim et al., 2017). Other AIF2 homologues such as AIF1, AIF3, and AIF4 were also identified with their high amino acid sequence similarity, and these AIFs were functionally redundant in inhibiting plant growth (Wang et al., 2009; Ikeda et al., 2013; Kim et al., 2017).
Environmental and endogenous stimuli affecting the timing and duration of reproductive phase can significantly impact seed yields (Shirley et al., 2019). In this regard, BRs control diverse aspects of floral organ formation, embryo and seed development, and seed size determination. For example, BSK family proteins contributed to early embryonic patterning, and bsk1bsk2 double loss-of-function mutants exhibited reduced zygote cell growth, which resulted in a small basal cell followed by a small suspensor cell (Neu et al., 2019). Moreover, BR biosynthesis- (cpd) or signalling-defective (bri1-116, bin2-1) mutants had greatly reduced number of pollen grains and were defective in pollen release and exine pattern formation (Ye et al., 2010). The same study showed that BES1, a positive transcription activator for BR signalling pathways, directly bound to promoter regions of genes (SPL/NZZ, TDF1, AM5, MS1, and MS2) encoding proteins essential for anther and pollen development (Chen W. et al., 2019). BZR1 (a BES1 homologue) family transcription factors were also involved in the regulation of anther development, in a BRI1-independent manner, by upregulating SPOROCYTELESS (SPL) and its upstream activator AGAMOUS (AG) that were required for the initiation of archesporial cells (Chen L. G. et al., 2019). Furthermore, BZR1-mediated BR signalling pathways positively influenced seed numbers by regulating the expression of genes (HLL, ANT, AP2, INO) that were involved in development of ovule and ovule integument (Huang et al., 2013; Jia et al., 2020). BZR1 also directly bound to and activated positive regulators of seed development (SHB1, MINI3, and IKU2) and repressed negative regulators of seed size (AP2 and ARF2) (Jiang et al., 2013). In this study, we demonstrate that AIF2 is negatively involved in pollen development and seed formation, and that sucrose- and BR-induced repression of AIF2 bHLH transcription factor positively controls pollen production and seed size/numbers in Arabidopsis.
Materials and Methods
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana (Col-0 and WS), an AIF2 T-DNA knockout mutant (aif2-1, CS811403), aif2-1/aif4-1 double knockout mutant, AIF2 overexpressing p35S:AIF2-EGFP transgenic plants (AIF2ox, Kim et al., 2017), a BIN2 triple knockout mutant (bin2KO, bin2bil1bil2; Yan et al., 2009), a BIN2 gain-of-function mutant bin2-1 (Peng et al., 2008), arf2-7 (Okushima et al., 2005), a BZR1 gain-of-function mutant bzr1-1D (Wang et al., 2002), and a BZR1 dominant negative mutant bzr1-1DdEAR (Oh et al., 2014) were used for phenotype analysis and generation of transgenic plants. A native promoter-driven reporter plant, pAIF2:AIF2-GUS, was used to observe AIF2 expression and localisation. Seeds were surfaced-sterilised with 95% ethanol for 10 min and cold-treated in the dark at 4°C for 72 h. These sterilised seeds were then sown in pots containing Sunshine No. 5 soil (Polysciences, United States) and grown in a growth room operating under a 16 h light (100–150 μmol m–2 s–1) and 8 h dark cycle at 23–25°C.
Generation of Transgenic Plants With Different Mutant Backgrounds
For generation of transgenic plants ectopically expressing variants of AIF2-EGFP, cDNAs encoding either the full-length (AIF2ox) or C-terminus truncated forms of AIF2 (AIF2dC) were amplified using primers listed (Supplementary Table 1) and inserted upstream of the myc-tag EGFP-expressing pB7FWG2 (Karimi et al., 2002) binary vectors. Subsequently, Agrobacterium cultures carrying each construct were used to transform Col-0, aif2-1, bzr1-1D, bin2-1, and bin2bil1bil2 genetic lines, generating plants designated as p35S:AIF2FL-EGFP/Col-0 (AIF2ox), p35S::AIF2dC-EGFP/Col-0 (AIF2dC), p35S:AIF2FL-EGFP/aif2-1(AIF2ox/aif2-1), p35S:AIF2dC-EGFP/aif2-1(AIF2dC/aif2-1), p35S:AIF2FL-EGFP/bzr1-1D (AIF2ox/bzr1-1D), p35S:AIF2dC-EGFP/bzr1-1D (AIF2dC/bzr1-1D), p35S:AIF2FL-EGFP/bin2bil1bil2 (AIF2ox/bin2bil1bil2), and p35S:AIF2dC-EGFP/bin2-1 (AIF2dC/bin2-1). Pollen grains of AIF2ox or bzr1-1DdEAR were crossed to a stigma of arf2-7 or aif2-1 plants to produce the AIF2ox/arf2-7 and the bzr1-1DdEAR/aif2-1 transgenic plants, respectively.
Generation of aif2-1/aif4-1 Double Knockout Plants
The CRISPR-Cas9 system was used as described previously (Kim et al., 2016). Briefly, guide RNA sequences targetting the exon of AIF4 (At1g09250) gene were designed using the guide RNA(gRNA) design tool (Concordet and Haeussler, 2018)1 as follows: 5′-GATTGAACTCGTCTCCGGCGCGGCG-3′ and 5′-AAACCGC CGCGCCGGAGACGAGTTC-3. The complementary gRNA was then inserted into pHAtC vector, and the resulting construct was transformed into aif2-1 to generate the aif2-1/aif4-1 double knockout mutant. A deletion of guanine at base pair position 249 starting from the initiator ATG was confirmed by performing DNA sequencing for the PCR-amplified AIF4 gene.
Total RNA Isolation and qRT-PCR Analysis
Total RNAs were extracted using a plant RNA extraction kit (Intron Biotechnology, South Korea) from flowers at different floral stages, siliques with developing seeds, or siliques isolated from in vitro-cultured flowers. To examine semi-quantitative RNA expression, the first-strand cDNA was synthesised using a ReverTra Ace qPCR RT Master Mix kit (Toyobo) according to the manufacturer’s instructions. Quantitative real-time RT-PCR (qRT-PCR) was performed by the SYBR green method using the Applied Biosystems Step One Plus System (Applied Biosystems, United States) with appropriate primers (Supplementary Table 2). The conditions for PCR amplification were as follows: 1 cycle of 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Expression of each transcript was normalised against the amount of UBC1 control in each sample. Three biological replicates were included in each experiment, and expression in each replicate was measured three times.
Protein Isolation and Western Blot Analysis
To examine expression of AIF2-EGFP in AIF2ox transgenic plants, total proteins were extracted from in vitro-cultured flowers using a homogenisation buffer (125 mM Tris-Cl, 4% sodium dodecyl sulphate, 2% β-mercaptoethanol, 1 mM phenylmethylsulfonyl, pH 7.9) and size-fractionated on 12% SDS-PAGE. Fractioned total proteins were then transferred onto a nitrocellulose membrane (Whatman, Germany) and probed against anti-GFP rabbit polyclonal antibodies (Santa Cruz Biotechnology, United States) in 5% milk/TBST (50 mM Tris-acetate, 150 mM NaCl, 0.05% Tween 20, pH 7.6). Goat anti-rabbit HRP-conjugated secondary antibody (Abcam, United Kingdom) was used to quantify the AIF2-EGFP protein. Peroxidase activity was detected using an ECL solution (Thermo Fisher Scientific Inc., United States) according to the manufacturer’s instructions.
Histochemical Staining and Microscopic Observation
For pollen grain staining, anthers were removed from newly opening flowers and stained with Alexander’s solution for 8 h at 50°C (Peterson et al., 2010), mPS-PI solution for 2 h at room temperature (Truernit et al., 2008), or pollen isolation solution containing 5μg/ml DAPI. Stained anthers or pollens were then observed using either a differential interference contrast (DIC)-equipped fluorescence microscope (Olympus BX60, Japan) or a Meta NLO-UV confocal laser scanning microscope (Zeiss LSM510, Germany).
To examine in vivo pollen tube growth, pistils were hand-pollinated with pollen grains of the same flower. The pollinated pistils were then fixed with 25% acetic acid at different times (h) after pollination, hydrated with an ethanol series, and softened with NaOH. Pollen tubes were then stained with aniline blue following a previously reported method (Mori et al., 2006) and their growth was examined using the Zeiss confocal microscope. For in vitro pollen tube growth assay, pollen grains were collected from 10–20 freshly opened flowers and grown on a solid germination medium (Boavida and McCormick, 2007) for 6 h in the dark at room temperature. Pollen tubes were photographed using a camera connected to the DIC-equipped Olympus microscope and their lengths were measured using Image J.2
To examine AIF2 expression in planta, opened flowers of pAIF2:AIF2-GUS transgenic plants were collected and fixed in 90% acetone for 20 min on ice. Staining and detection of GUS activity were performed according to the method described by Jefferson (1987). The stained floral organs were observed under the DIC-equipped Olympus microscope.
For starch staining of developing seeds, pistils were hand-pollinated with pollen grains of the same flower. Developing siliques were then collected at different days after pollination (DAP), placed in fixing solution containing 10% acetic acid and 90% ethanol (v/v), and incubated overnight in a water bath at 60°C followed by washing with 70% ethanol. Siliques were then stained in Lugol’s iodine solution for 5 min and observed under a DIC-equipped Olympus microscope.
Seed Clearing and Imaging Analysis
For determination of embryo developmental stages, siliques were fixed overnight in solution containing 10% acetic acid and 90% ethanol (v/v) and washed twice sequentially with 90% and 70% ethanol. Siliques were then cleared overnight with chloral hydrate solution (Yadegari et al., 1994). These cleared seeds from siliques were mounted in clearing solution for observation under the Olympus microscope. Afterward, the embryo area and the rest of the integument-surrounded area were measured using Image J.
In vitro Flower Culture
Flowers in an early stage of seed development (between DAP3.5 to DAP4) were cut and immediately transferred to 30% ethanol for 3 min. These sterilised flowers were placed in half-strength solid MS media containing brassinolide (BL, 10–9 M) supplemented with or without 3% sucrose (w/v). These flowers were then cultured for 9 days in a growth chamber operating under a 16 h light (100–150 μmol m–2 s–1) and 8 h dark cycle at 23–25°C. Siliques were collected from in vitro-cultured flowers to examine their phenotypes.
Measurement and Statistical Analysis
Over 100 siliques or flowers were collected from 30–40 plants and used for each experiment. All experiments were conducted in triplicate at a minimum, and the data were statistically analysed using the Student’s t-test.
Results
Overexpression of AIF2 Resulted in Defective Formation of Pollen Grains and Reduced Seed Production
Previously, we demonstrated that AIF2 were negatively involved in BR-induced growth regulation (Kim et al., 2017); nonetheless, its roles in the development of other organs are unknown. As an initial step to elucidate the roles of AIF2 in pollen and seed development, we first investigated silique phenotype of AIF2ox transgenic plants (p35S:AIF2-EGFP/Col-0). Three independent transgenic lines (AIF2ox-1 to AIF2ox-3) differentially expressed AIF2 protein, ranged from high to low levels compared with the Col-0 plants (AIF2ox-1 to AIF2ox-3, respectively) and showed retarded growth phenotypes as previously reported (Supplementary Figures 1A,B; Kim et al., 2017). Interestingly, their siliques were smaller in AIF2ox plants, and their reduction in silique length at floral stage 17 was inversely correlated with the abundance of AIF2 proteins: for instance, AIF2ox-1 showed the most severely retarded silique phenotype (Supplementary Figures 1C,D). Hereafter, we took AIF2ox-1 line for further analysis of pollen, embryo, and seed phenotypes.
Disrupting pollen/ovule development, pollination, pollen tube growth, and fertilisation results in a reduced number of seed sets and silique size. We found that AIF2-overexpressing transgenic Arabidopsis plants produced smaller and frequently unfused siliques (Figure 1A, left and middle panels). Regarding unfused siliques, INDEHISCENT (IND), SPATULA (SPT), and ALCATRAZ (ALC) are bHLH transcription factors required for proper valve margin development and later differentiation of the silique dehiscence zone, allowing seed dispersal (Girin et al., 2011; Groszmann et al., 2011; Kay et al., 2013). Accordingly, we found that IND and ALC expression was down-regulated in AIF2-overexpressing plants, whereas it was upregulated in aif2-1 and aif2-1/aif4-1 plants (Figure 1A, right panel). In addition to the short and unfused silique phenotypes, the number of ovules in a silique was also greatly reduced in AIF2ox (Figure 1B, left and middle panels), and even the fertilised eggs of AIF2ox plants produced a higher ratio of aborted seeds (Figure 1B, right panel). Consequently, the number of both siliques per plant (Figure 1C) and seeds per silique (Figure 1D) in AIF2ox plants was lower than that in wild-type Col-0, resulting in a significantly reduced total seed weight or productivity in AIF2ox plants (Figure 1E). In contrast, aif2-1/aif4-1 plants displayed opposite silique and seed phenotypes.
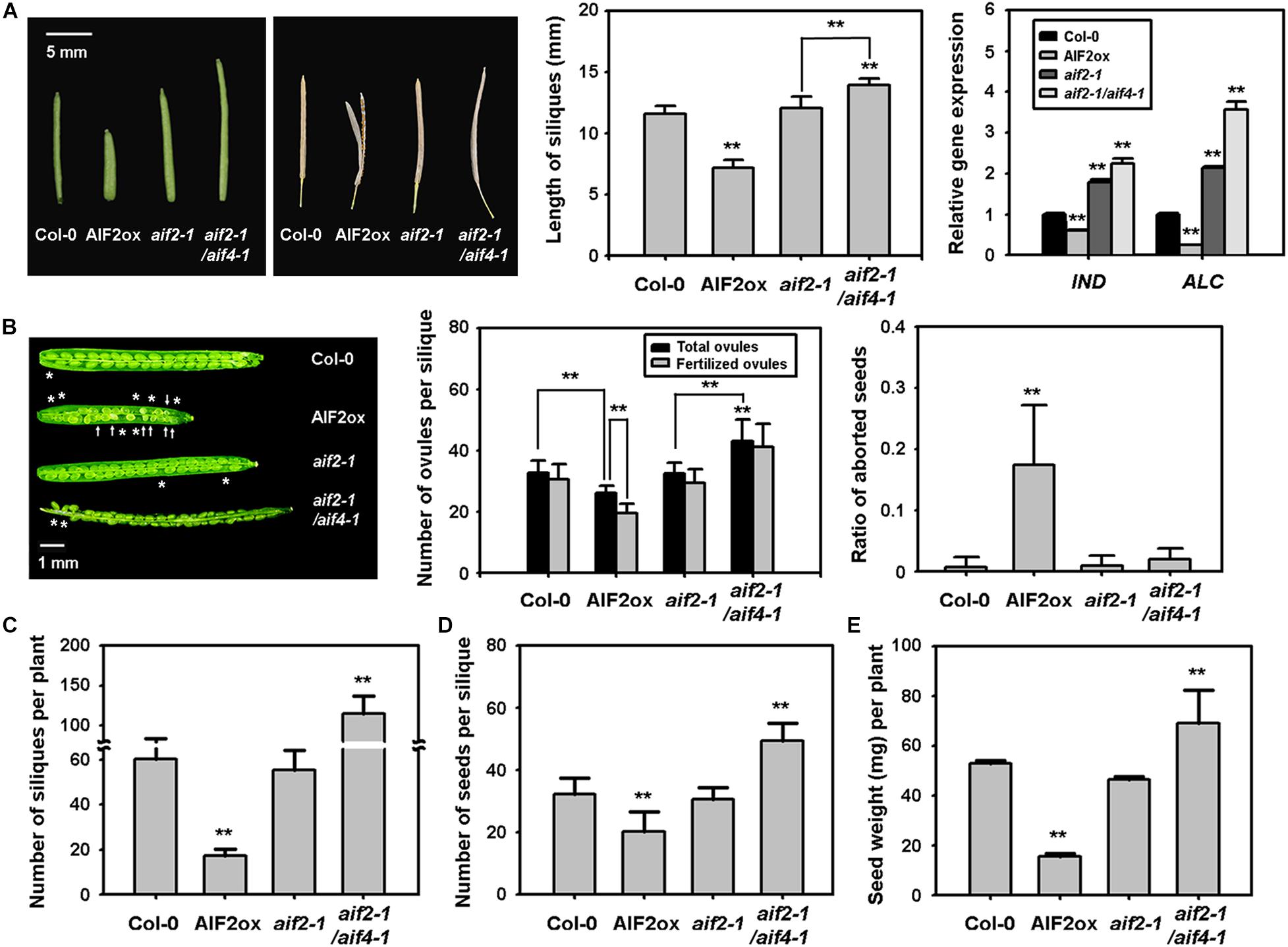
Figure 1. Defective silique phenotypes of AIF2-overexpressing p35S:AIF2-EGFP transgenic plants (AIF2ox). (A) Silique with shorter and premature dehiscence phenotypes. The picture on the left shows representative shorter and unfused gynoecia silique phenotypes. Siliques from opened flowers were measured for their length (middle) and transcript expression of genes involved in valve margin development of the carpel (right). Relative expression of each gene is normalised as a ratio of the Col-0 plants, which was set to 1. (B) Reduced ovule development and aborted seeds. The picture on the left shows regions lacking ovule development (*) and aborted fertilised eggs (arrow). Total ovules presented in the middle are calculated by the sum of visibly developed ovules and invisible empty ovule area. Fertilised ovules indicate the visible ovules. The ratio of aborted seeds on the right represents the ratio of seeds with defective phenotype (such as smaller or white) to total visible seeds. (C–E) Number of siliques (C), seeds (D), and total seed weight (E) was measured from more than 100 siliques collected from 30–40 plants. Bar graphs represent means ± SD and statistical difference from either the Col-0 control or bracketed samples is indicated by two asterisks (**) at P < 0.01.
Arabidopsis plants are propagated through self-pollination; thus, the ratio of stamen to pistil length is important for successful pollination. We found that pistils and stamens of AIF2ox plants were shorter than those of Col-0 plants (Figure 2A, 1st to 3rd panels). Two knockout plants of AIF2, aif2-1 and aif2-1/aif4-1, had longer pistils and stamen. Nonetheless, the ratio of stamen to pistil was higher in AIF2ox plants (Figure 2A, right panel) indicating that the reduced growth of the stamen or pistil in AIF2ox plants is unlikely to be the cause of the reduced seed production and retarded silique development.
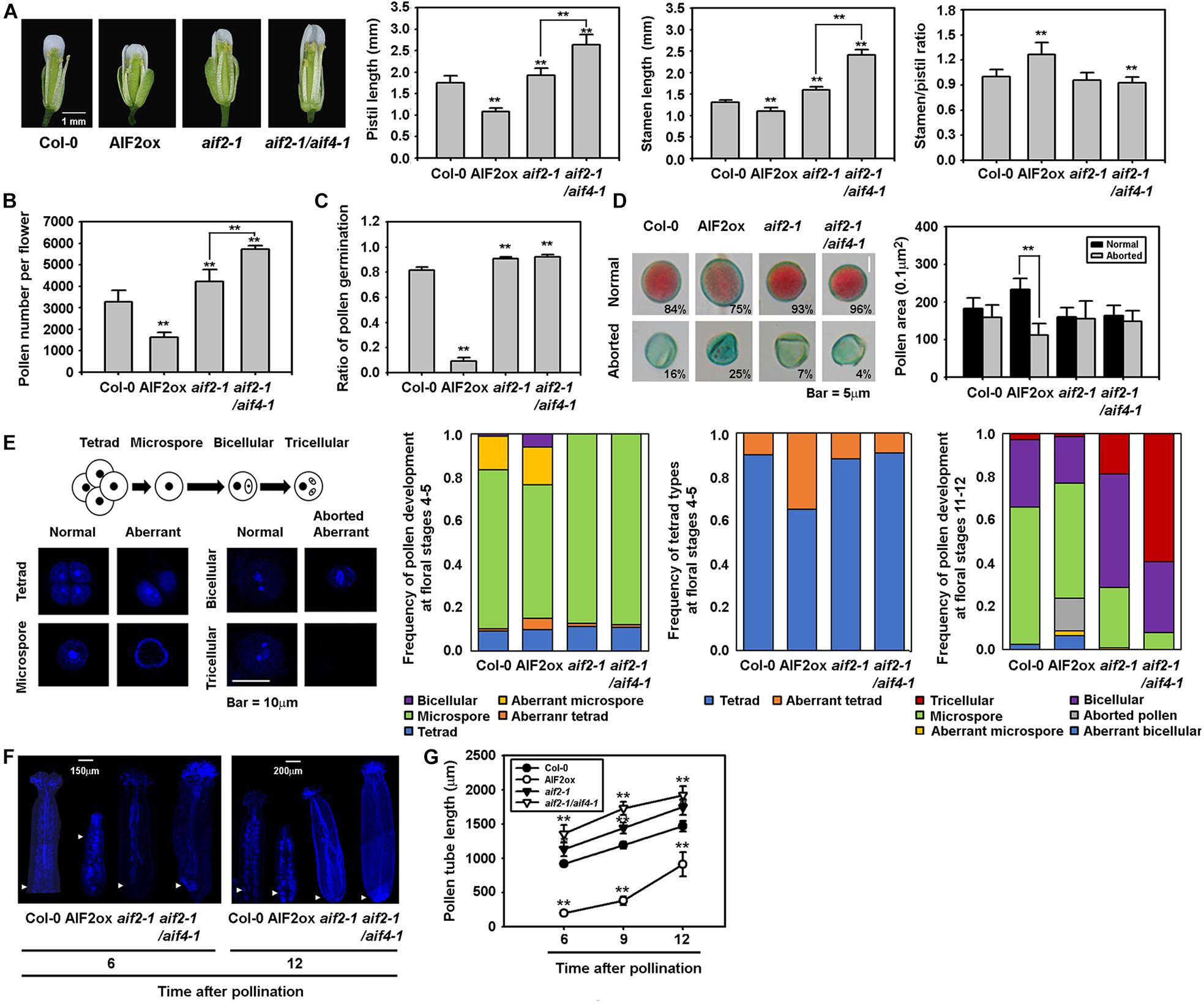
Figure 2. Retarded growth of reproductive organs and degenerated pollen production and germination found in AIF2ox plants. (A) Flowers of Col-0, AIF2ox, aif2-1 and aif2-1/aif4-1 plants and the lengths of pistils, stamens, and the ratio of stamen to pistil. Number of open flowers examined >30. (B) Measurement of pollen numbers counted under a bright microscope without pollen staining. Number of open flowers examined >20. (C) In vitro pollen germination analysis. The efficiency of germination is represented by the ratio of germinated pollens after 6 h incubation in the germination medium. Number of pollens examined >3,000 taken from 20–25 flowers. (D) Frequency and size of the Alexander solution -stained viable pollens (normal) and non-stained aborted pollens. Number of pollens examined >2,000 taken from 15–20 open flowers (E) Defective pollen development in AIF2ox flowers. Pollens in different floral stages were stained with DAPI to reveal pollen developmental stages and pollens with intact nuclei (normal), without (aborted) or abnormal nuclei having defects in mitosis and appearance (aberrant). Number of pollens examined >1,000 taken from flowers at stage 4–5 or stage 11-12. (F,G) In vivo pollen tube growth assay. Arrowhead indicates the end of aniline blue-stained pollen tubes at 6 or 12 h after hand-pollinated self-pollination in the same flowers (F) and lengths of pollen tubes at different times were measured after hand-pollination (G). n > 15 for each time point was used for analysis. Statistical difference from either the Col-0 control or bracketed samples is indicated by two asterisks (**) at P < 0.01.
Next, we examined pollen productivity and viability. AIF2 was specifically expressed in unfertilised ovules and pollen grains of pAIF2:AIF2-GUS plants but not in the petal, sepal, stigma, and style (Supplementary Figure 2A). This expression pattern of AIF2 implies that AIF2 may play a role in male- and female gametophyte development. Similarly, AIF2-EGFP proteins of AIF2ox plants were abundantly expressed in ovules and pollen grains (Supplementary Figures 2B–D). Interestingly, AIF2ox plants contained fewer pollen grains in the anthers, and this reduction in pollen numbers was inversely correlated with the expression levels of AIF2 proteins (Supplementary Figure 3). Consequently, the number of pollens harvested from all anthers from a flower was lower in AIF2ox plants, but slightly higher in aif2-1 and aif2-1/aif4-1 plants, than in Col-0 plants (Figure 2B). To test pollen viability, we performed in vitro pollen germination assay and found that the efficiency of pollen germination was dramatically reduced in AIF2ox plants (Figure 2C). More than 80% of pollen grains in Col-0 and the two AIF2 knockout plants germinated successfully and initiated pollen tube growth, whereas only 9.2% of pollen germinated in AIF2ox plants. This poor germination efficiency may due to, in part, the high frequency of aborted and smaller pollens commonly observed in AIF2ox plants (Figure 2D).
In Arabidopsis, microspore mother cell (2N) undergoes a series of meiosis I and meiosis II (microsporogenesis) to produce a tetrad of microspores (N), and each microspore develops into a bicellular pollen containing a vegetative cell and a generative cell and further to tricellular mature pollen having one vegetative cell and two sperm cells (microgametogenesis) (Figure 2E, 1st panel). We found that AIF2ox plants produced higher frequency of aberrant tetrad (36% of tetrads), a tetrad microspore having no nucleus or less microspores, in flowers of stage 4—5 (Figure 2E, 2nd and 3rd panel). These defects may lead an increased ratio of aborted/aberrant microspore and bicellular pollens (Figure 2E, 4th panel). In contrast, the ratio of normal microspore, bicellular and tricellular pollens at floral stage 11—12 was decreased. These results imply that AIF2ox plants underwent a defective microsporogenesis, thus produced less mature and viable pollens. Notably, it seemed that male gametophytogenesis progressed faster in aif2-1 and aif2-1/aif4-1 than in Col-0 plants (Figure 2E, 4th panel). Nonetheless, they had a similar ratio of normal microspore, bicellular and tricellular pollens in total. To further test pollen activity, we manually self-pollinated stigmas of test plants and measured the growth of pollen tubes. At 6 h after hand-pollination, the wild-type pollen tubes grew 917 μm on average, whereas those of aif2-1 and aif2-1/aif4-1 plants were longer and those of AIF2ox plants were shorter (Figures 2F,G). This retarded pollen growth in AIF2ox plants was also confirmed by the fact that all AIF2ox pollen tubes germinated in vitro were in the range of 0 to 150 μm (average 67.2 μm), whereas those of Col-0 and aif2-1 plants grew 240 μm and 184 μm on average, respectively (Supplementary Figure 4). Nonetheless, most pollen tubes of Col-0 (1,468 μm), AIF2ox (914 μm), aif2-1(1,746 μm), and aif2-1/aif4-1(1,920 μm) reached almost the end of the pistils at 12 h, considering the pistil lengths of Col-0 (1.74 mm), AIF2ox (1.08 mm), aif2-1 (1.92 mm), and aif2-1/aif4-1 (2.53 mm) (Figure 2A). These results suggest that pollen tube growth is unlikely the reason for reduced male sterility and seed productivity in the AIF2ox plants. Collectively, we demonstrated that the defective silique growth and seed production in AIF2-overexpressing transgenic plants were caused by the reduced amount of pollen production and less-effective pollen tube germination but not by retarded stamen/pistil growth or pollen tube elongation.
Expression Patterns of Pollen- and Auxin-Related Genes Were Significantly Modulated in AIF2-Overexpressing Plants
Timely expression of SPL/NZZ, TDF1, and MS1 is essential for early microspore mother cell formation to late pollen maturation (Yang et al., 2007; Ye et al., 2010; Chen L. G. et al., 2019; Chen W. et al., 2019). We examined transcript expression of these genes in flowers at floral stages 11/12 and 15. Two mitotic divisions of microspores and tapetum degeneration occur at floral stage 11, and desiccation of pollen grains followed by anther dehiscence occurs in flowers of floral stage 12 (Kim et al., 2001). Then, the flower opens and is self-pollinated during the stages 13 to 15. As expected, in floral stage 15 of Col-0 and the two AIF2 knockout plants, these genes were transcriptionally down-regulated compared to the transcription of these genes in stage 11 or 12 flowers (Figures 3A–C). Unexpectedly, we found that SPL and TDF1 at stage 15 maintained a higher expression both in AIF2ox and pAIF2:AIF2-GUS plants. In addition, although MS1 in floral stage 15 showed lower expression than that at stage 11/12, a relatively higher expression was maintained than that of the same floral stage in Col-0 and the two AIF2 knockout plants.
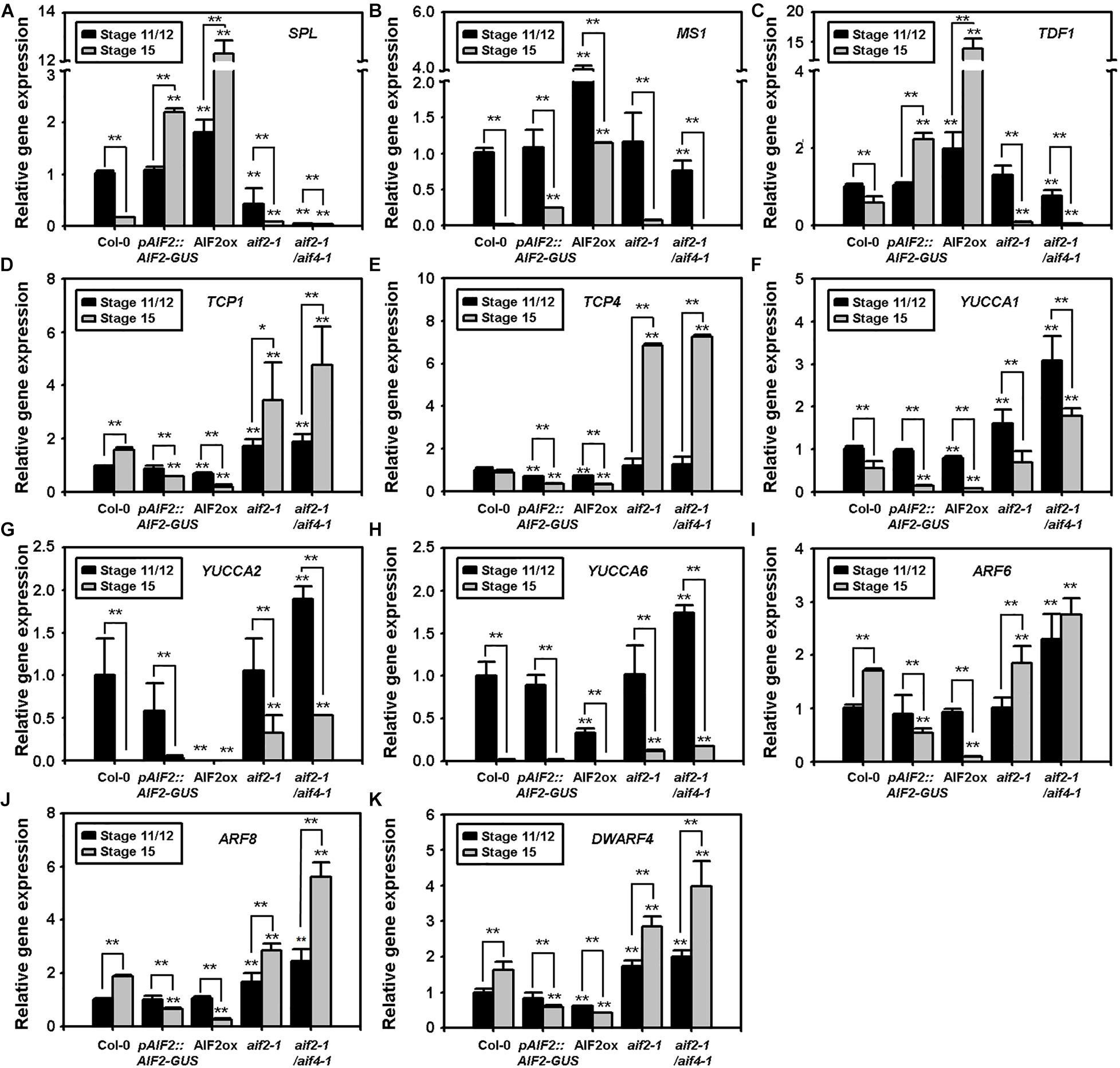
Figure 3. (A–K) Transcript expressions of pollen- and seed-regulating transcription factors (A–C), auxin biosynthesis (D–H) and signalling-related genes (I,J), and a brassinosteroid biosynthesis gene, DWF4 (K). Total RNA was isolated from flowers at floral stages 11/12 or 15, and transcript expression of genes was examined using qRT-PCR. Relative expression of each gene is normalised as a ratio of the Col-0 plants at floral stage 11/12, which was set to 1. Statistical difference from the same stage of the Col-0 flowers or bracketed samples is indicated by an asterisk (*) on bars at P < 0.05 and two asterisks (**) at P < 0.01.
Auxin plays important roles in the proper development of flower and seeds (Shirley et al., 2019); thus, the mutants defective in auxin biosynthetic genes, such as YUCCAs, show not only abnormal flowers but also defects in the embryo and endosperm of seeds (Cheng and Zhao, 2007; Figueiredo and Köhler, 2018). TEOSINTE BRANCHED 1, CYCLODEA, and PROLIFERATING CELL FACTORSs (TCPs) bind to the promoters of YUCCAs to promote their gene expression and directly upregulate auxin levels (Challa et al., 2016; Zhou et al., 2018). Moreover, TCP1 promotes BR biosynthesis by directly upregulating the expression of a BR-biosynthetic gene DWARF4 (DWF4) (Guo et al., 2010). We found that TCP1 and TCP4 genes of AIF2ox or the pAIF2:AIF2-GUS plants were relatively down-regulated at stage 11/12 compared with those of Col-0 plants at the same stage, and they were further downregulated at stage 15 (Figures 3D,E). Interestingly, these two genes were greatly upregulated at stage 15 and/or stage 11/12 of aif2-1 and aif2-1/aif4-1 plants. Similarly, three auxin biosynthetic genes (YUCCAs) and genes for two positive regulators of auxin signalling (ARFs) were down-regulated at either stage 11/12 (YUCCA1, YUCCA2, and YUCCA6) or 15 (YUCCA1, YUCCA2, ARF6, and ARF8) of AIF2ox plants while expression of these genes in the two AIF2 knockout plants at the same stage was relatively up-regulated compared with those of Col-0 plants (Figures 3F–J). Transcript expression pattern of DWF4 was also similar to that of the TCP1, so that it was relatively down-regulated when TCP1 was suppressed at stage 15 of two AIF2-expressing transgenic lines (Figure 3K). These results indicate that aberrant expressions of pollen development-, auxin-, and BR-related genes in AIF2ox plants may partially explain the observed reduction in pollen grains together with the less-effective pollen tube germination and aborted seed development.
AIF2ox Plants Differentially Regulated Transcript Expression of Seed-Forming Regulators
Previously, BZR1-mediated BR signalling pathways were shown to increase seed size by affecting the integument, endosperm, and embryo development (Jiang et al., 2013). We found that ectopic expression of AIF2 in aif2-1 plants results in smaller and lighter plant seeds. The seed length to width ratio in Col-0 or aif2-1 and aif2-1/aif4-1 plants was 1.8–2.2, and ectopic expression of AIF2 in the aif2-1 plants modified the average ratio to 1.54 (Figure 4A). This implies that seeds of AIF2-overexpressing plants were likely to be rounder rather than ellipsoidal, typical of the seeds of Col-0 and the two AIF2 knockout plants. As for a confirmation of the AIF2 functions in seed size and weight determination, we demonstrated that expression of C-terminal deleted AIF2 (AIF2dC, a dominant negative form of AIF2 function, Kim et al., 2020) obliterated this complementation effect. In addition, AIF2ox plants produced lighter seeds than those of Col-0, aif2-1, aif2-1/aif4-1, and AIF2dC-overexpressing aif2-1 plants (Figure 4B). Accordingly, we found that expression of the endosperm- and embryo-forming SHB1, IKU1, and MINI3 were greatly reduced in AIF2ox plants. In contrast, AP2 and ARF2, which negatively act in seed formation, were upregulated (Figure 4C) in the same AIF2ox plants. ARF2 is a transcriptional repressor of auxin-regulated genes, and arf2 loss-of-function mutations increased seed size and weight as well as showed late flowering phenotypes under long day conditions in Arabidopsis (Schruff et al., 2006; Choi et al., 2018). To further investigate genetically whether the increased expression of ARF2 in the AIF2ox plants was responsible for the small-seed phenotype, we crossed pollen of AIF2ox with ovules of arf2-7 plants and found that an ectopic expression of AIF2 did not modulate arf2-7 seed phenotypes (Figure 4D). These findings suggest that AIF2 acted upstream of ARF2 in negatively regulating seed shape and weight.
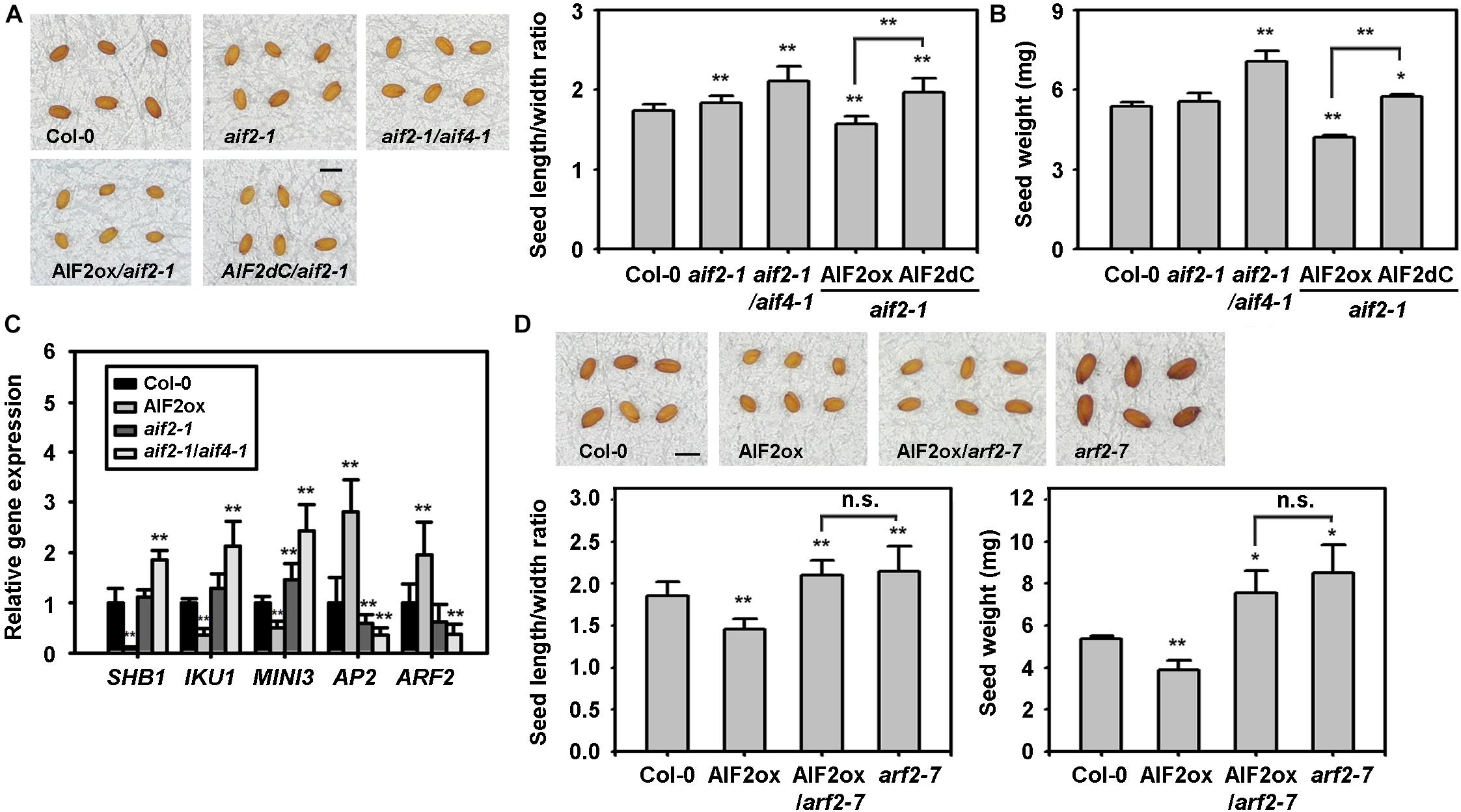
Figure 4. AIF2-mediated negative regulation of seed size and weight. (A,B) AIF2 overexpression-led smaller and rounder (less length to width ratio) (A) and lighter (B) seeds of aif2-1 loss-of-function plants and their reciprocal confirmation through complementation assay with a dominant negative form of AIF2, AIF2dC. (C) Transcript expression of seed size-related genes in siliques with developing seeds. Relative expression of each gene is normalised as a ratio of the Col-0 plants, which was set to 1. (D) ARF2-dependent AIF2 effects on seed size and weight. Scale bars in pictures represent 0.5 mm in length. Statistical difference from the Col-0 control is indicated by an asterisk (*) at P < 0.05 and two asterisks (**) at P < 0.01. n.s., non-significant. Number of seeds examined for measurement of length/width ratio and weight >800.
Reduction in seed size often results from coordinated reduction in endosperm size, embryo proliferation, and cell elongation of the maternally derived integument. AIF2 was predicted to be highly expressed in the seed coats, chalazal endosperm, and spotted areas of peripheral endosperms through the pre-globular to torpedo stages (Supplementary Figure 5).3 In contrast, its expression was low in the developing embryo. To evaluate the effects of AIF2 on the endosperm- and embryo-forming processes, we morphologically investigated the progression of seed development in AIF2ox plants. All Col-0 plants at DAP3 progressed to globular embryos, whereas none of the AIF2ox plants showed distinct globular embryos (Figure 5A). At DAP6, all Col-0 and aif2-1 plants developed into heart stage embryos. In contrast, almost half the AIF2ox plants remained as globular stage embryos. At DAP8, more than 40% of Col-0 plants had torpedo stage embryos, which further progressed in aif2-1 plants such that all embryos were at the torpedo stage. Again, most embryos of AIF2ox plants were still at the heart stage, and only 5% of the total embryos were at the torpedo stage at DAP8. These results imply that embryonic progression is severely delayed in AIF2-overexpressing plants.
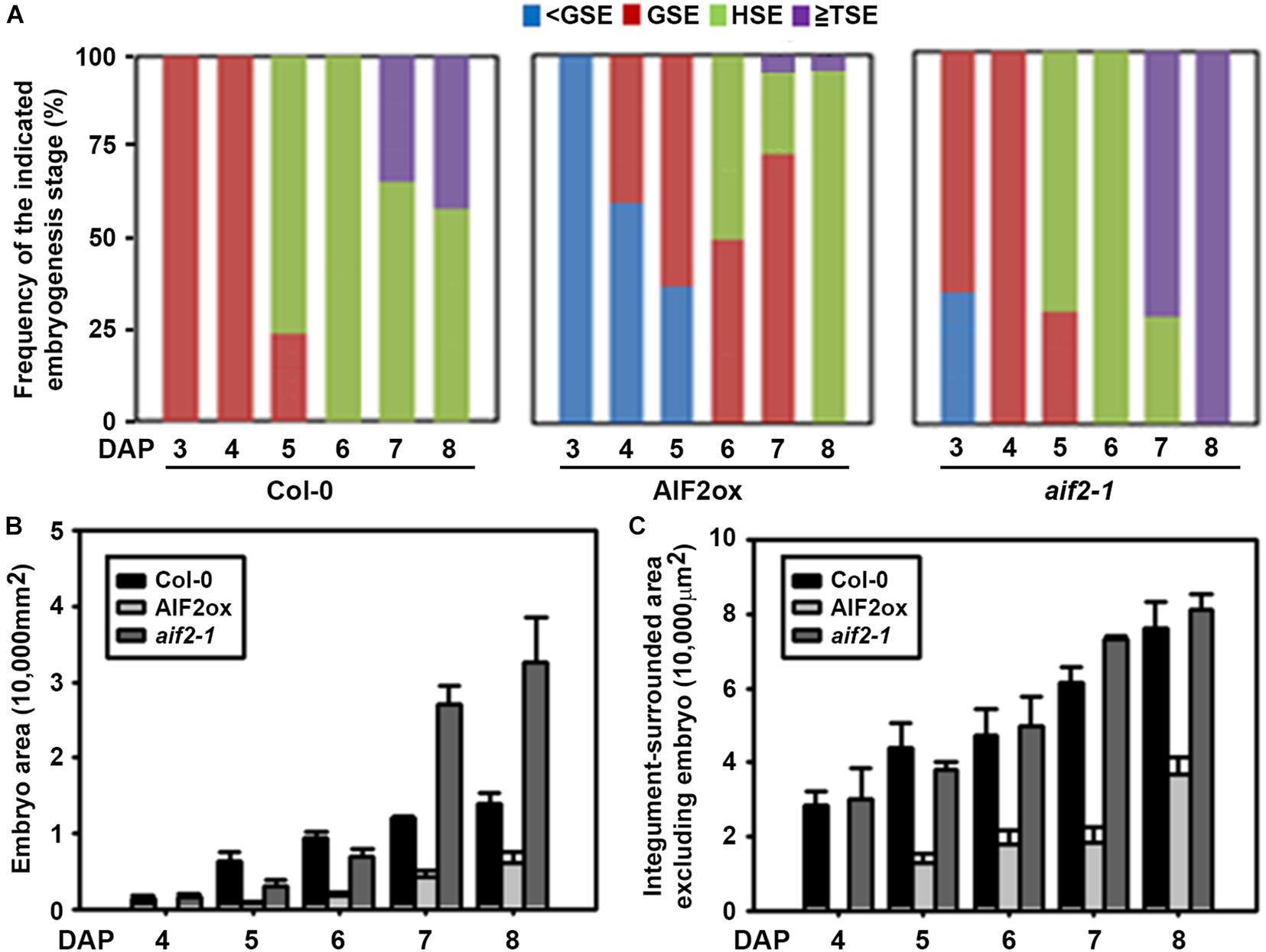
Figure 5. Delayed embryogenesis and smaller embryo development in AIF2-overexpressing p35S:AIF2-EGFP (AIF2ox) transgenic plants. (A–C) Comparison of the frequency of developmental stages of embryos (A), size of embryo area (B), and the size of remaining integument-surrounded area excluding the embryo (C) observed at different days after hand-pollinated self-pollination (DAP). <GSE, pre-globular stage; GSE, globular stage embryo; HSE, heart stage embryo; TSE, torpedo stage embryo. Number of embryos examined for each time point >150–200.
After fertilisation, the embryo grows to fill the cavity at the expense of the endosperm; thus, at maturity, the seed contains only a single layer of endosperm cells in Arabidopsis (Olsen, 2001; Sun X. et al., 2010). We found that the embryo area was increased in Col-0 plants (Figure 5B). A dramatic increase in the embryo size was especially observed at DAP5 when more than 75% of embryos progressed to the heart stage from the globular stage. The embryo area of aif2-1 at DAP7 and 8 was much larger than that of Col-0 plants, mainly because most aif2-1 embryos were in the torpedo stage. In comparison, the average embryo area in AIF2ox plants was much smaller, mainly because of their delayed embryonic progression. For example, embryos of AIF2ox seeds were at the globular or heart stage at DAP6 when all embryos of Col-0 or aif2-1 seeds were at the heart stage. Similarly, integument-surrounded seed area excluding the embryo area showed a size reduction in AIF2ox plants (Figure 5C). Collectively, our results demonstrated that AIF2-overexpressing transgenic plants suppressed genes encoding positive factors (SHB1, IKU1, MINI3) of seed size determination but promoted gene expression for negative factors (AP2 and ARF2), resulting in delayed embryogenesis and seeds with smaller size.
AIF2-Regulation of Seed Shape and Weight Is Epistatic to Those by BZR1 and BIN2
Previously, we demonstrated that AIF2 was significantly suppressed by BRI1/BZR1-mediated signalling pathways, and BIN2-mediated AIF2 phosphorylation augmented the BIN2/AIF2-mediated negative circuit of BR signalling pathways in growth-promoting cellular activities (Kim et al., 2017). In this study, BIN2 triple knockout mutant (bin2bil1bil2, bin2KO) had ellipsoidal seed shape which was almost similar to that of WS plants (insignificant increase in seed length to width ratio) (Figure 6A). However, constitutive expression of AIF2 in bin2KO background produced rounder seeds by significantly decreasing the seed length to width ratio. In contrast, BIN2 gain-of-function mutant (bin2-1) produced rounder seeds, and an ectopic expression of a C-terminal deleted AIF2 (bin2-1/AIF2dC) resulted in the ellipsoidal shape owing to an increase in seed length to width ratio. Similarly, transgenic expression of AIF2 or C-terminal-deleted AIF2 either decreased or increased seed weights in bin2KO and bin2-1, respectively (Figure 6B). These results suggest that AIF2 acted downstream of BIN2 in the regulation of seed size and weight.
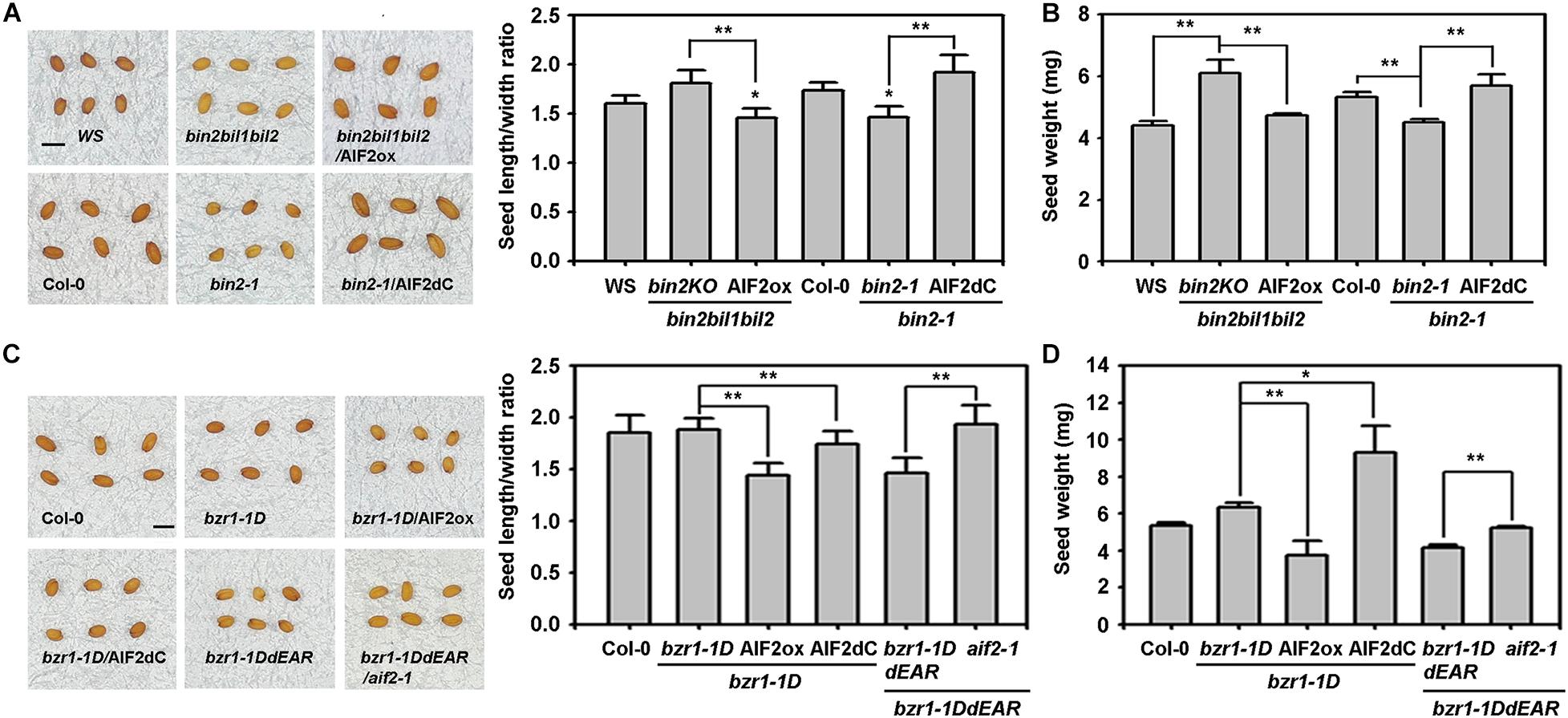
Figure 6. Genetic and functional interactions of AIF2 with BIN2 and BZR1 in the regulation of seed size and weight. (A,B) Evaluation of genetic and functional interaction of AIF2 with BIN2 in the regulation of seed size (A) and seed weight (B). Transgenic constructs ectopically expressing AIF2ox or AIF2dC were transformed into a BIN2 gain-of-function mutant, bin2-1, or a BIN2 loss-of-function mutant, bin2bil1bil2 (bin2KO), and their seed phenotypes were evaluated. (C,D) Evaluation of genetic and functional interaction of AIF2 with BZR1 in the regulation of seed size (C) and seed weight (D). Transgenic constructs ectopically expressing AIF2ox or AIF2dC were transformed into a BZR1 gain-of-function mutant, bzr1-1D, or plants constitutively expressing bzr1-1DdEAR, a loss-of-function form of BZR1, were crossed to aif2-1 to examine their seed phenotypes. Statistical difference between bracketed samples is indicated by an asterisk (*) at P < 0.05 and two asterisks (**) at P < 0.01. Number of seeds examined for measurement of length/width ratio and weight >800. Scale bars in pictures represent 0.5 mm in length.
Deletion of ERF-associated amphiphilic repression (EAR) motif at the carboxy terminus of BZR1 abolished the abilities to regulate gene expression and cell elongation (Oh et al., 2014). We found that transgenic expression of EAR-deleted bzr1-1D (bzr1-1DdEAR) produced round and light seeds, similar to the seeds of bzr1-1D that ectopically expressing AIF2ox. Again, expression of AIF2dC in bzr1-1D partially negated the AIF2 effects on seed shape determination (Figure 6C) and even greatly increased seed weights in the same plants (Figure 6D). These results imply that AIF2 acted downstream of BZR1 for seed size and weight determination. Supporting this idea, the described dominant negative effects of bzr1-1DdEAR in seed phenotypes were not functional in aif2-1 genetic background plants. Thus, AIF2-controlled seed phenotypes acted downstream of BZR1 and BIN2, and BZR1-regulated seed shape and size were contrary to that by AIF2, whereas BIN2 functioned similar to AIF2.
Transcript Suppression of Sucrose Transporter Genes and Lipid-Biosynthetic Genes in AIF2ox Plants and Subsequent Defects in Starch and Oil Accumulation
AIF2ox plants presented in this report not only delayed embryogenesis but also generated wrinkled and shrunken seeds (Supplementary Figure 6). Therefore, we examined starch accumulation in developing seeds, investigated transcript expression of proteins which promote sucrose transport and lipid biosynthesis, and scrutinised the cause of AIF2ox phenotypes.
Starch is actively accumulated in the proliferating tissues, whole seed coat, ovary wall, placenta–septum region, and funiculus during early zygote and embryo development (Hedhly et al., 2016), which was also seen in the globular to torpedo stage embryos of Col-0 and aif2-1 plants (Figure 7A). In contrast, starch granules in the seed coat of AIF2ox plants were relatively weakly stained with Lugol’s iodine dye. Sucrose, the major transport form of carbohydrate in plants, is delivered via the phloem to the maternal seed coat and then secreted from the seed coat to the embryo through SWEET11, 12, and 15 efflux carriers (Chen et al., 2015). Compared to the transcript levels of SWEET11, SWEET12, and SWEET15 in Col-0, the transcript levels were greatly down-regulated in siliques of AIF2ox plants but upregulated in those of aif2-1 (except for SWEET12) (Figure 7B). We hypothesised that reduced expression of sucrose transporter genes in AIF2ox plants and the subsequent defects in starch accumulation resulted in seeds with delayed embryogenesis and wrinkled phenotype.
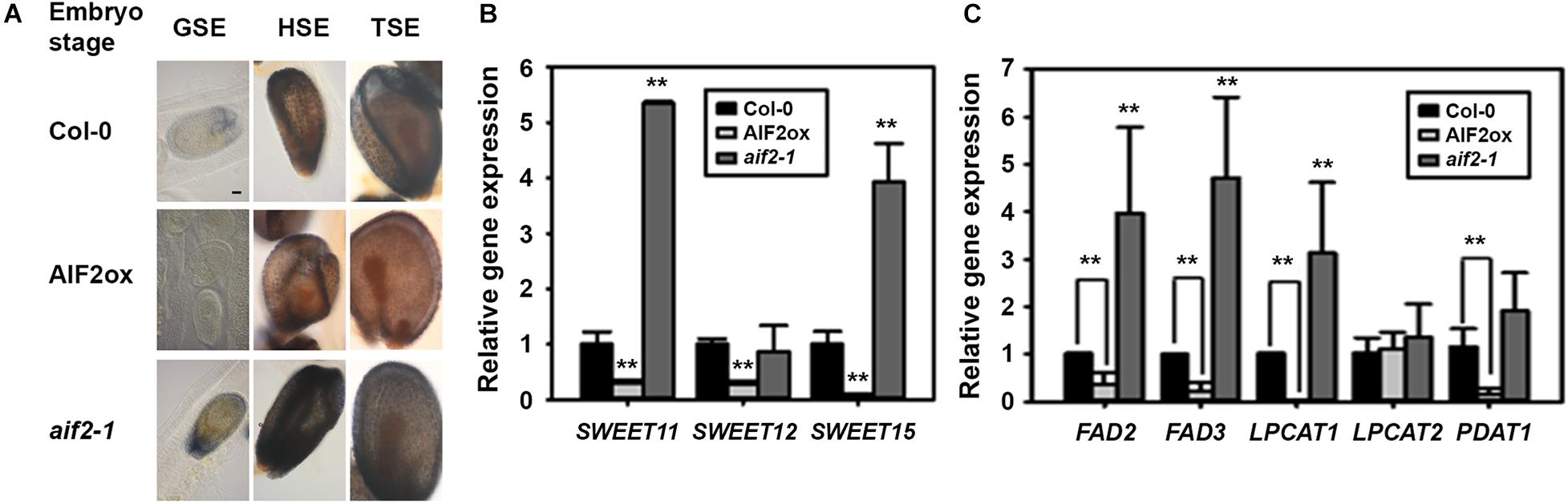
Figure 7. Reduced starch and oil accumulation in seeds of AIF2-overexpressing p35S:AIF2-EGFP (AIF2ox) transgenic plants. (A) Photographic comparison of starch accumulation in developing seeds. Siliques were stained with Lugol’s staining dye, and seeds with embryos at the same developmental stage were compared for starch accumulation. Number of seeds examined for each stage of the corresponding plants >150. Pictures show a representative image. (B,C) Total RNA was extracted from siliques with developing seeds, and the transcript expression for sucrose transport-related genes (B) and lipid biosynthesis genes (C) was measured using qRT-PCR. Relative expression of each gene is normalised as a ratio of the Col-0 plants, which was set to 1. Statistical difference from the Col-0 control is indicated by two asterisks (∗∗) at P < 0.01.
Developing embryos of Arabidopsis and oilseed rape initially accumulated mother plant-driven starch, but the starch levels were declined with increase in the rates of storage lipid and protein synthesis (Andriotis et al., 2012). Accumulation of seed oil requires the co-ordination of de novo fatty acid (FA) biosynthesis and triacylglycerol (TAG) assembly. It was known that FA desaturase 2/3 (FAD 2/3), acyl-CoA:lysophosphatidylcholine acyl transferases (LPCATs), acyl-CoA:diacylglycerol acyltransferase 1 (DGAT1), and phospholipid:diacylglycerol acetyltransferase 1 (PDAT1) were positively involved in the modification of FAs and subsequent assembly of FA-driven acyl-CoA into glycerol, producing TAGs (Zhang et al., 2009; Xu et al., 2012; Lou et al., 2014). We showed that transcript expression of FAD2, FAD3, LPCAT1, and PDAT1 was greatly suppressed in AIF2ox plants but promoted in aif2-1 plants (Figure 7C). Thus, we concluded that suppressed expression of sucrose-transporting genes (SWEET11/12/15) and lipid-biosynthesis genes (FAD2/3, LPCAT1, and PDAT1) in AIF2-overexpressing transgenic plants resulted in reduced starch and lipid accumulation in the developing seeds resulting in shrunken and small phenotypes.
Sucrose- and BR-Induced Repression of AIF2 Positively Controlled Seed and Silique Development
Sucrose is a necessary nutrient for embryo and seed development. Developing seeds form new carbon sink, generating high sugar flow from vegetative tissues to the seeds. To further confirm sucrose- and BL-mediated regulation of the AIF2ox phenotype, we examined the effects of BL and sucrose on silique phenotype and AIF2 stability in in vitro-cultured flowers. We found that the supply of BL did not result in an increase in silique length or seed numbers (Figure 8A). However, providing BL and sucrose promoted silique growth and seed production in AIF2ox, Col-0, and aif2-1 plants, but was less effective in AIF2ox plants. Non-efficient promotion of BL itself might be attributed to the lack of nutrient supply found normally in intact plants. Such rescues of silique development were accompanied with a dramatic reduction in AIF2 stability in BL- and sucrose-treated AIF2ox plants (Figure 8B). BL-induced AIF2 degradation did not seem enough to cause the substantial recovery of silique growth because of the shortage in nutrients. Accordingly, supplying sucrose together with BL to the in vitro culture medium was the most effective in increasing transcript expression of FAD3, LPCAT1, and PDAT1 (Figure 8C). Moreover, SWEET15 was upregulated by the supplementation of BL or BL with sucrose (Figure 8D). Unexpectedly, SWEET11 was highly upregulated by sucrose, and this effect was obliterated by the additional supplementation of BL. These results suggest that BR and sucrose reduced protein abundance of AIF2 transcription factor and increased starch and oil production for the successful generation of seeds.
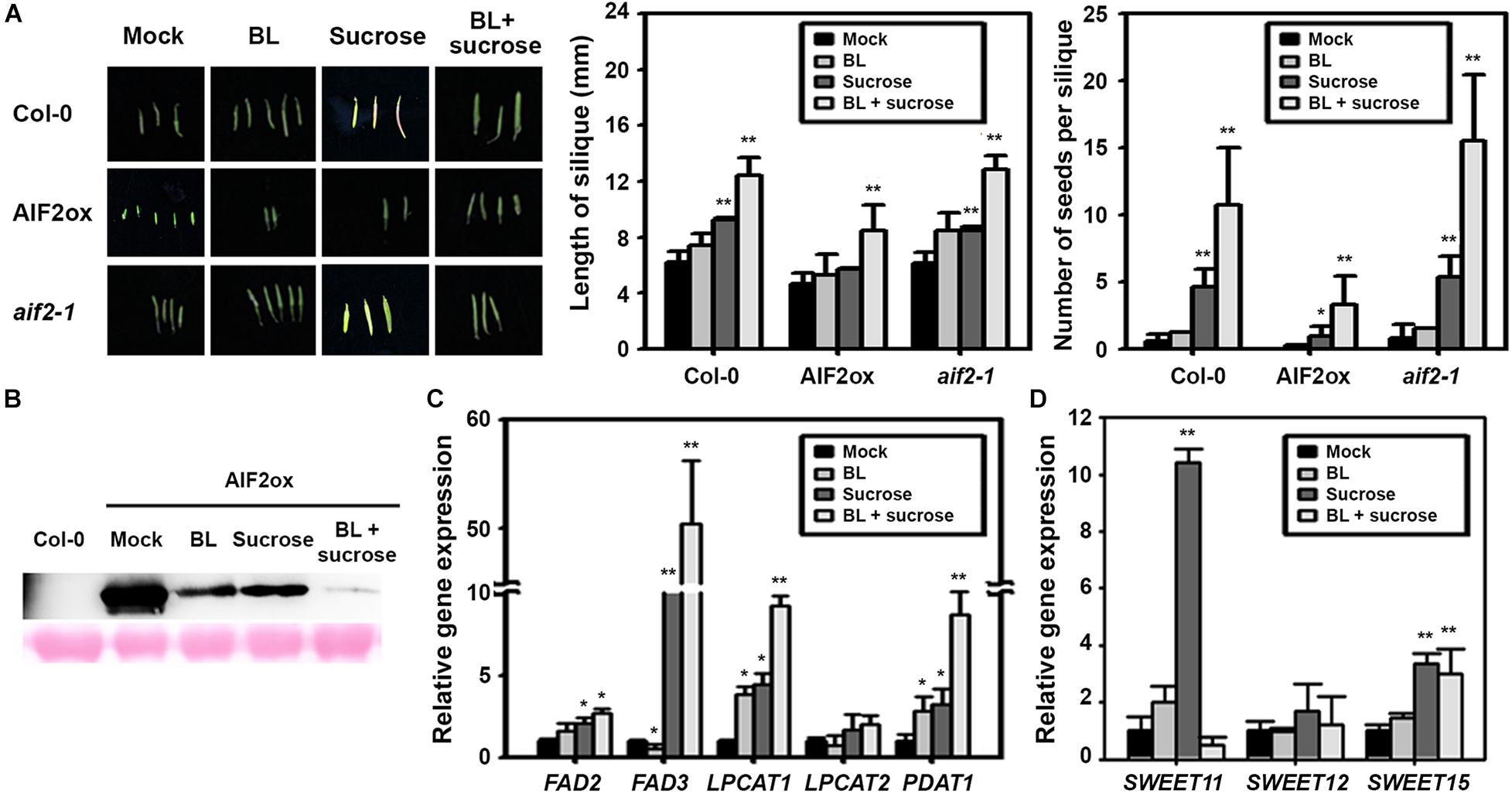
Figure 8. Sucrose- and brassinosteroid-induced suppression of AIF2 stability and silique phenotypes. Flowers in between DAP3.5 to DAP4 (n > 40 for each experiment) were collected and cultured for 9 days in solid MS media containing brassinolide (BL, 10– 9 M) supplemented with or without 3% sucrose (w/v). (A) Length and seed numbers of in vitro-cultured silique. (B) Western blot analysis of AIF2-EGFP expression in siliques of in vitro-cultured p35S:AIF2-EGFP transgenic lines (AIF2ox) (C,D) Total RNA was extracted from in vitro-cultured siliques of the AIF2ox, and the transcript expression for lipid biosynthesis genes (C) and sucrose transport-related genes (D) was measured using qRT-PCR. Relative expression of each gene is normalised as a ratio of the mock-treated plants, which was set to 1. Statistical difference from the mock-treated control is indicated by an asterisk (*) at P < 0.05 and two asterisks (**) at P < 0.01.
Discussion
BRs control diverse aspects of floral organ formation, seed development, and seed size determination. For example, two BR signalling activators, BES1 and its homologue BZR1, positively regulated tapetum and microspore development by directly upregulating SPL/NZZ, TDF1, and MS1/2 (Ye et al., 2010; Chen L. G. et al., 2019; Chen W. et al., 2019). In contrast, the expression of SPL and MS1 was significantly reduced in BR biosynthesis- (cpd) or signalling-defective (bri1-116, bin2-1) mutants producing greatly reduced number of pollen grains (Ye et al., 2010). Surprisingly, we found that SPL and MS1 were highly upregulated in pollen- and seed-defective AIF2ox plants (Figures 3A,B). It is notable that MS1-overexpressing transgenic plants exhibited an excess deposition of wall materials and a loss of the regular structure of the pollen wall, eventually resulting in defective pollen development (Yang et al., 2007). MS1 protein was expressed in a developmentally regulated manner between late tetrad spore and microspore release and then broken down rapidly (Yang et al., 2007). Hence, it was suggested that MS1 breakdown was critical for the progression of pollen development, and the persistence of MS1 protein may serve to downregulate genes required for continued development of microspores. We showed that SPL, MS1, and TDF1 in AIF2ox plants were relatively highly expressed even in flowers of stages 11/12 and 15 (Figures 3A–C). Moreover, AIF2ox plants showed retarded and defective progression of microsporogenesis, producing aberrant tetrad microspores (Figure 2E). Thus, it is possible that the stage-independent aberrant expression of pollen-producing genes such as SPL and MS1 in AIF2ox plants may act reversibly on microspore development and viability.
The auxin biosynthetic pathway is majorly regulated by catalytic activities of multiple monooxygenases encoded by the YUCCA genes, and TCP transcription factors can directly upregulate YUCCAs to increase auxin levels (Guo et al., 2010; Challa et al., 2016). Disruption of TCPs caused phenotypes that resemble spl-D, the heterozygous gain-of-function mutants of SPL (Wei et al., 2015). In other hand, spl-D mutants showed repressed expression of YUCCA2 and YUCCA6 and produced few and small flowers and short/wrinkled siliques with shrivelled seeds that could be partially rescued by crossing with yuc6-D, a dominant mutant of YUC6 (Li et al., 2008). ARFs are a class of transcriptional modulators that regulate auxin-mediated gene expression. Likely, auxin biosynthesis-regulating genes, Arabidopsis ARF6 and ARF8, through proper microRNA167-controlled cleavage, were critically involved in regulation of both gametophyte reproduction (Wu et al., 2006) and embryonic and seed development (Yao et al., 2019). In addition, ARF2 was negatively involved in the regulation of auxin-induced flowering time and seed size (Choi et al., 2018). Notably, similar to aif2-1 plants, arf2 loss-of-function mutants produced seeds with dramatically increased size and weight (Schruff et al., 2006). Based on the above studies, we suggest that ectopic expression of SPL in AIF2ox plants together with downregulation of TCPs, YUCCAs, and ARF6/8 (Figure 3) and upregulation of ARF2 (Figure 4C) may lead to the observed defective phenotypes of pollen, embryogenesis, and seeds/siliques (Figure 9). We demonstrated that AIF2-regulation of seed size and shape was epistatic to bzr1-1D and bin2-1 genetic backgrounds (Figure 6). bin2-1 exhibited reduced fertility, aborted ovules, and short siliques similar to those of AIF2ox plants, and auxin partially rescued the infertility phenotype of bin2-1 (Li T. et al., 2019). Thus, it is probable that BIN2/AIF2 regulatory networks act via a coordinative interaction with auxin signalling pathways. In fact, rice OsSK41 (also known as OsGSK5, a BIN2 homologue) interacted with and phosphorylated OsARF4 (Hu et al., 2018). As a result, the expression of a common set of downstream genes was repressed, including some auxin-responsive genes during rice grain development; thus, the loss-of-function mutants of OsSK41 and OsARF4 showed increased grain length and weight. Further genetic analysis demonstrating in vivo functional interactions of BIN2/AIF2 and auxin signalling pathways are needed in future study.
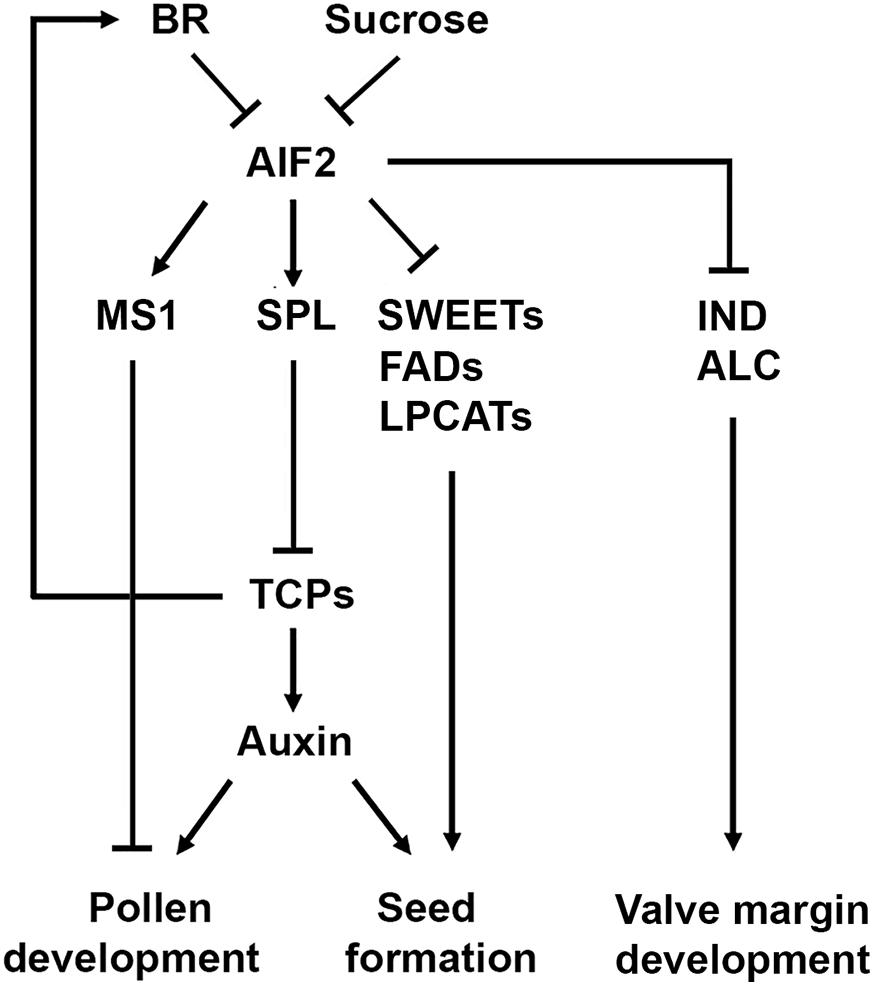
Figure 9. A diagram explaining AIF2-regulated pollen development and seed formation. Stage-independent aberrant expression of pollen-producing genes such as SPL and MS1 together with suppression of sucrose-transporting SWEETs, lipid biosynthesis-regulating FADs/LPCATs, and valve margin-forming IND/ALC in AIF2ox plants may act adversely on pollen microspore development, auxin-regulated seed formation and proper silique development. Sucrose- and BR-induced repression of AIF2 bHLH transcription factor positively controls pollen development and/or seed size and numbers.
Sucrose is delivered via the phloem to the maternal seed coat and then to the embryo through SWEET11, 12, and 15 efflux carriers (Chen et al., 2015). In addition, seeds with delayed embryogenesis and wrinkled phenotype commonly arise from defects in sucrose transport and endosperm formation (Andriotis et al., 2012; Chen et al., 2015). In this study, we further demonstrated that transcript suppression of sucrose transporter genes (SWEET11, SWEET12, and SWEET15) and lipid-biosynthesis genes (FAD2, FAD3, LPCAT1, and PDAT1) in AIF2-overexpressing plants resulted in the production of wrinkled seeds with reduced starch and oil levels (Figure 7). Similar to our AIF2-overexpressing transgenic plants, the sweet11;12;15 triple mutant (lacking the ability to provide nutrients to the embryo and endosperm) showed delayed embryo development and reduced seed weight and lipid content, and exogenously supplied sucrose promoted embryo growth of sweet11;12;15 mutants (Chen et al., 2015). Arabidopsis SWEETs such as SWEET8 and SWEET13 also played important roles in nurturing pollen grains; thus, mutation of these genes caused defective primexine deposition and pollen wall pattern formation resulting in male sterility (Sun et al., 2013).
Antisense expression of CPD, a gene involved in BR biosynthesis, in Arabidopsis impaired the ability of the plant to assimilate carbohydrates, and such transgenic plants displayed a clear reduction in starch content (Schlüter et al., 2002). Moreover, increasing BR levels in rice enhanced CO2 assimilation, favoured sucrose accumulation in the leaf, and increased assimilation of glucose to starch in the seed (Wu et al., 2008). Thus, the high expression level of SPL in AIF2ox plants and the subsequent reduction in TCP1 expression followed by the decrease in BR biosynthesis resulted from a transcriptional decrease in DWF4 may lead to attenuation of BZR1-mediated BR signalling pathways and reinforced BIN2/AIF2-mediated BR-defective signalling pathways. Supporting this idea, BR- and sucrose-regulated negative repression of AIF2 accumulation were co-related with accumulation of oil and starch and a resulting increase in seed number and silique length (Figure 8). Collectively, we propose that the impaired pollen and seed phenotypes of AIF2-overexpressing transgenic plants may be, in part, owing to the reduced capacity for sugar/starch production and defects in sugar transport during gametophyte formation, embryogenesis, and seed formation.
In this study, we demonstrated that multiple genes regulating development of pollen grains, seeds, and siliques were differentially regulated in AIF2ox plants (Figure 9). AIF2 is a non-DNA-binding bHLH transcription factor and it regulates target gene expression by binding to other DNA-binding bHLH proteins. Previously, we showed that AIF2 interacts with INDUCER OF CBF EXPRESSION 1 (ICE1), a nuclear-localised MYC-like bHLH transcription factor, via their C-termini (Kim et al., 2020). A successful formation of the AIF2–ICE1 complex, the subsequent direct upregulation of C-REPEAT BINDING FACTORS (CBFs), and the antagonistic downregulation of PIF4 were negatively involved in dark-triggered and BR-induced leaf senescence, thus, helping plants continue to grow and remain green for a long time (Kim et al., 2020). Many transcription factors with bHLH domain have been shown to regulate flower and seed development. For instance, SPT can heterodimerise with ALC, and these two proteins apparently undergo sub-functionalisation with SPT, being essential for earlier development of carpel margin tissues, and ALC, specialising in later dehiscence zone development (Groszmann et al., 2011). Therefore, future studies need to verify whether AIF2 interacts with other bHLH family proteins and whether this interaction and the resulting functions depend on age-specific binding partners of AIF2 during plant reproductive processes.
Data Availability Statement
All data associated with the paper are available in this manuscript. Novel materials used and described in the paper are available for non-commercial research purposes from the corresponding author (c29vaHdhbkB5b25zZWkuYWMua3I=).
Author Contributions
Soo-HK managed whole experimental processes and wrote this manuscript. YK performed most experiments and generated Figures 2, 3, 5–9. Sun-HK made Figures 1 and 3. D-MS generated the aif2-1/aif4-1 double knockout plants. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (grant numbers NRF-2019R1H1A2080044 and NRF-2020R1F1A1068793 to Soo-HK and NRF-2018R1A6A3A01013324 and NRF-2019R1C1C1006182 to YK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.704958/full#supplementary-material
Supplementary Figure 1 | Expression level-dependent aerial and silique phenotypes of AIF2-overexpressing independent transgenic lines (p35S:AIF2-EGFP/Col-0, AIF2ox).
Supplementary Figure 2 | Expression of the native promoter-driven and CaMV35S promoter-driven AIF2 protein in floral organs.
Supplementary Figure 3 | Reduced pollen production in AIF2ox plants.
Supplementary Figure 4 | Efficacy of in vitro-germinated pollen tube growth.
Supplementary Figure 5 | Relative expression of AIF2 (At3g06590) in developing seeds.
Supplementary Figure 6 | Morphology of seeds stained with mPS-PI, followed by confocal microscope observation.
Supplementary Table 1 | Primers used in cDNA amplification of AIF2.
Supplementary Table 2 | Primers used in quantitative real time RT-PCR analysis.
Abbreviations
AIF2, ATBS1-INTERACTING FACTOR 2; AIF2ox, AIF2 over-expressing p35S,AIF2-EGFP transgenic plants; bHLH, basic-helix-loop-helix; BL, brassinolide; BR, brassinosteroids; DAP, days after pollination.
Footnotes
References
Andriotis, V. M. E., Pike, M. J., Schwarz, S. L., Rawsthorne, S., Wang, T. L., and Smith, A. M. (2012). Altered starch turnover in the maternal plant has major effects on Arabidopsis fruit growth and seed composition. Plant Physiol. 160, 1175–1186. doi: 10.1104/pp.112.205062
Boavida, L. C., and McCormick, S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52, 570–582. doi: 10.1111/j.1365-313x.2007.03248.x
Challa, K. R., Aggarwal, P., and Nath, U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28, 2117–2130. doi: 10.1105/tpc.16.00360
Chen, L. G., Gao, Z., Zhao, Z., Liu, X., Li, Y., Zhang, Y., et al. (2019). BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant 12, 1408–1415. doi: 10.1016/j.molp.2019.06.006
Chen, L. Q., Lin, I. W., Qu, X. Q., Sosso, D., McFarlane, H. E., Londoño, A., et al. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27, 607–619. doi: 10.1105/tpc.114.134585
Chen, W., Lv, M., Wang, Y., Wang, P. A., Cui, Y., Li, M., et al. (2019). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10:4164.
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Cheng, Y., and Zhao, Y. (2007). A role for auxin in flower development. J. Integr. Plant Biol. 49, 99–104. doi: 10.1111/j.1744-7909.2006.00412.x
Choi, H. S., Seo, M., and Cho, H. T. (2018). Two TPL-binding motifs of ARF2 are involved in repression of auxin responsiveness. Front. Plant Sci. 9:372. doi: 10.3389/fpls.2018.00372
Concordet, J. P., and Haeussler, M. (2018). CRISPOR intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245.
Day, R. C., Herridge, R. P., Ambrose, B. A., and Macknight, R. C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148, 1964–1984. doi: 10.1104/pp.108.128108
Figueiredo, D. D., and Köhler, C. (2018). Auxin: a molecular trigger of seed development. Genes Dev. 32, 479–490. doi: 10.1101/gad.312546.118
Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jürgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670. doi: 10.1104/pp.102.018762
Girin, T., Paicu, T., Stephenson, P., Fuentes, S., Körner, E., O’Brien, M., et al. (2011). INDEHISCENT and SPATULA interact to specify carple and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23, 3641–3653. doi: 10.1105/tpc.111.090944
Groszmann, M., Paicu, T., Alvarez, J. P., Swain, S. M., and Smyth, D. R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68, 816–829. doi: 10.1111/j.1365-313x.2011.04732.x
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., and Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173. doi: 10.1105/tpc.109.069203
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
He, J. X., Gendron, J. M., Yang, Y., Li, J., and Wang, Z. Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190. doi: 10.1073/pnas.152342599
Hedhly, A., Vogler, H., Schmid, M. W., Galliardini, V., Santelia, D., and Grossniklaus, U. (2016). Starch turnover and metabolism during flower and early embryo development. Plant Physiol. 172, 2388–2402. doi: 10.1104/pp.16.00916
Hu, Z., Lu, S. J., Wang, M. J., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, H. Y., Jiang, W. B., Hu, Y. W., Wu, P., Zhu, J. Y., Liang, W. Q., et al. (2013). BR signal influences Arabidopsis ovule and seed numbers through regulating related genes expression by BZR1. Mol. Plant 6, 456–469. doi: 10.1093/mp/sss070
Ikeda, M., Mitsuda, N., and Takagi, O.-M. (2013). ATBS1 interacting factors negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal. Behav. 8:e23448. doi: 10.4161/psb.23448
Jia, D., Chen, L. G., Yin, G., Yang, X., Gao, Z., Guo, Y., et al. (2020). Brassinosteroids regulate outer ovule integument growth in part via the control of INNER NO OUTER by BRASSINOZOLE-RESISTANT family transcription factors. J. Integr. Plant Biol. 62, 1093–1111. doi: 10.1111/jipb.12915
Jiang, W. B., Huang, H. Y., Hu, Y. W., Zhu, S. W., Wang, Z. Y., and Lin, W. H. (2013). Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 162, 1965–1977. doi: 10.1104/pp.113.217703
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/bf02667740
Jofuku, K. D., Omidyar, P. K., Gee, Z., and Okamuro, J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3117–3122. doi: 10.1073/pnas.0409893102
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/s1360-1385(02)02251-3
Kay, P., Groszmann, M., Ross, J. J., Parish, R. W., and Swain, S. M. (2013). Modifications of a conserved regulatory network involving INDEHISCENT controls multiple aspects of reproductive tissue development in Arabidopsis. New Phytol. 197, 73–87. doi: 10.1111/j.1469-8137.2012.04373.x
Kim, H., Kim, S. T., Ryu, J., Choi, M. K., Kweon, J., Kang, B. C., et al. (2016). A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J. Integr. Plant Biol. 58, 705–712. doi: 10.1111/jipb.12474
Kim, S. Y., Hong, C. B., and Lee, I. (2001). Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J. Plant Res. 114, 301–307. doi: 10.1007/pl00013991
Kim, Y., Park, S. U., Shin, D. M., Pham, G., Jeong, Y. S., and Kim, S. H. (2020). ATBS1-INTERACTING FACTOR 2 negatively regulates dark- and brassinosteroid-induced leaf senescence through interactions with INDUCER OF CBF EXPRESSION 1. J. Exp. Bot. 71, 1475–1490. doi: 10.1093/jxb/erz533
Kim, Y., Song, J. H., Park, S. U., Jeong, Y. S., and Kim, S.-H. (2017). Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR Signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol. 58, 227–239.
Li, L. C., Qin, G. J., Tsuge, T., Hou, X. H., Ding, M. Y., Aoyama, T., et al. (2008). SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol. 179, 751–764. doi: 10.1111/j.1469-8137.2008.02514.x
Li, N., and Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, N., Xu, R., and Li, Y. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. doi: 10.1146/annurev-arplant-050718-095851
Li, T., Kang, X., Wei, L., Zhang, D., and Lin, H. (2019). A gain-of-function mutation in Brassinosteroid-insensitive 2 alters Arabidopsis floral organ development by altering auxin levels. Plant Cell Rep. 39, 259–271. doi: 10.1007/s00299-019-02489-9
Liu, Z., Miao, L., Huo, R., Song, X., Johnson, C., Kong, L., et al. (2018). ARF2-ARF4 and ARF5 are essential for female and male gametophyte development in Arabidopsis. Plant Cell Physiol. 59, 179–189. doi: 10.1093/pcp/pcx174
Lou, Y., Schwender, J., and Shanklin, J. (2014). FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J. Biochem. Chem. 289, 17996–18007. doi: 10.1074/jbc.m114.572883
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Meng, L. S., Xu, M. K., Wan, W., and Wang, J. Y. (2018). Integration of environmental and developmental (or metabolic) control of seed mass by sugar and ethylene metabolisms in Arabidopsis. J. Agric. Food Chem. 66, 3477–3488. doi: 10.1021/acs.jafc.7b05992
Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71. doi: 10.1038/ncb1345
Neu, A., Eilbert, E., Asseck, L. Y., Slane, D., Henschen, A., Wang, K., et al. (2019). Constitutive signaling activity of a receptor-associated protein links fertilization with embryonic patterning in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 116, 5795–5804. doi: 10.1073/pnas.1815866116
Oh, E., Zhu, J. Y., Ryu, H., Hwang, I., and Wang, Z. Y. (2014). TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 5:4140.
Ohto, M. A., Floyd, S. K., Fischer, R. L., Goldberg, R. B., and Harada, J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 22, 277–289. doi: 10.1007/s00497-009-0116-1
Okushima, Y., Mitina, I., Quach, H. L., and Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. doi: 10.1111/j.1365-313x.2005.02426.x
Olsen, O. A. (2001). Endosperm development: cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267. doi: 10.1146/annurev.arplant.52.1.233
Peng, P., Yan, Z., Zhu, Y., and Li, J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 2, 338–346. doi: 10.1093/mp/ssn001
Peterson, R., Slovin, J. P., and Chen, C. (2010). A simplified method for differential staining of aborted and non-aborted pollen grains. Intl. J. Plant Biol. 1, 66–69.
Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Schlüter, U., Köpke, D., Altmann, T., and Müssig, C. (2002). Analysis of carbohydrate metabolism of CPD antisense plants and brassinosteroid-deficient cbb1 mutant. Plant Cell Environ. 25, 783–791. doi: 10.1046/j.1365-3040.2002.00860.x
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signaling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Shirley, N. J., Aubert, M. K., Wilkinson, L. G., Bird, D. C., Lora, J., Yang, X., et al. (2019). Translating auxin responses into ovules, seeds and yield: insight from Arabidopsis and the cereals. J. Integr. Plant Biol. 61, 310–336. doi: 10.1111/jipb.12747
Sun, M. X., Huang, X. Y., Yang, J., Guan, Y. F., and Yang, Z. N. (2013). Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 26, 83–91. doi: 10.1007/s00497-012-0208-1
Sun, X., Shantharaj, D., Kang, X., and Ni, M. (2010). Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13, 611–620. doi: 10.1016/j.pbi.2010.08.009
Sun, Y., Fan, X.-Y., Cao, D. M., Tang, W., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Truernit, E., Bauby, H., Dubreucq, B., Grandjean, O., Runions, J., Barthelemy, J., et al. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503. doi: 10.1105/tpc.107.056069
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313x.2010.04271.x
Wang, H., Zhu, Y., Fujioka, S., Asami, T., Li, J., and Li, J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basal helix-loop-helix proteins. Plant Cell 21, 3781–3791. doi: 10.1105/tpc.109.072504
Wang, Z.-Y., Nakano, T., Gendron, J., He, H., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/s1534-5807(02)00153-3
Wei, B., Zhang, J., Pang, C., Yu, H., Guo, D., Jiang, H., et al. (2015). The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling Arabidopsis ovule development. Cell Res. 25, 121–134. doi: 10.1038/cr.2014.145
Wu, C.-Y., Trieu, A., Radhakrishnan, P., Kwok, S. F., Harris, S., Wang, J., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145. doi: 10.1105/tpc.107.055087
Wu, M.-F., Tian, Q., and Reed, J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
Xu, J., Carlsson, A. S., Francis, T., Zhang, M., Hoffman, T., Giblin, M. E., et al. (2012). Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol. 12:4. doi: 10.1186/1471-2229-12-4
Yadegari, R., de Paiva, G. R., Laux, T., Koltunow, A. M., Apuya, N., Zimmerman, J. L., et al. (1994). Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6, 1713–1729. doi: 10.2307/3869903
Yan, Z., Zhao, J., Peng, P., Chihara, R. K., and Li, J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150, 710–721. doi: 10.1104/pp.109.138099
Yang, C., Vizcay-Barrena, G., Conner, K., and Wilson, Z. A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548. doi: 10.1105/tpc.107.054981
Yao, X., Chen, J., Zhou, J., Yu, H., Ge, C., Zhang, M., et al. (2019). An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 180, 453–464. doi: 10.1104/pp.19.00127
Ye, Q., Zhu, W., Li, L., Zhang, S., Yin, Y., Ma, H., et al. (2010). Brassinosteroids control male sterility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. U.S.A. 107, 6100–6105. doi: 10.1073/pnas.0912333107
Zhang, M., Fan, J., Taylor, D. C., and Ohlrogge, J. B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21, 3885–3901. doi: 10.1105/tpc.109.071795
Zhou, Y., Zhang, D., An, J., Yin, H., Fang, S., Chu, J., et al. (2018). TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 176, 1850–1861. doi: 10.1104/pp.17.01566
Keywords: auxin, ATBS1-INTERACTING FACTOR 2, BRASSINOSTEROID-INSENSITIVE 2, brassinosteroid, BRASSINAZOLE RESISTANT 1, pollen, seed
Citation: Kim Y, Kim S-H, Shin D-M and Kim S-H (2021) ATBS1-INTERACTING FACTOR 2 Negatively Modulates Pollen Production and Seed Formation in Arabidopsis. Front. Plant Sci. 12:704958. doi: 10.3389/fpls.2021.704958
Received: 04 May 2021; Accepted: 02 July 2021;
Published: 27 July 2021.
Edited by:
Fa Cui, Ludong University, ChinaCopyright © 2021 Kim, Kim, Shin and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soo-Hwan Kim, c29vaHdhbkB5b25zZWkuYWMua3I=
 Yoon Kim
Yoon Kim Sun-Ho Kim
Sun-Ho Kim Dong-Min Shin
Dong-Min Shin Soo-Hwan Kim
Soo-Hwan Kim