
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 17 August 2021
Sec. Plant Metabolism and Chemodiversity
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.704697
This article is part of the Research TopicPhenylpropanoid Systems Biology and BiotechnologyView all 34 articles
 Tao Yao1,2†
Tao Yao1,2† Kai Feng1,2†
Kai Feng1,2† Meng Xie3
Meng Xie3 Jaime Barros2,4
Jaime Barros2,4 Timothy J. Tschaplinski1,2
Timothy J. Tschaplinski1,2 Gerald A. Tuskan1,2
Gerald A. Tuskan1,2 Wellington Muchero1,2*
Wellington Muchero1,2* Jin-Gui Chen1,2*
Jin-Gui Chen1,2*The phenylpropanoid pathway serves as a rich source of metabolites in plants and provides precursors for lignin biosynthesis. Lignin first appeared in tracheophytes and has been hypothesized to have played pivotal roles in land plant colonization. In this review, we summarize recent progress in defining the lignin biosynthetic pathway in lycophytes, monilophytes, gymnosperms, and angiosperms. In particular, we review the key structural genes involved in p-hydroxyphenyl-, guaiacyl-, and syringyl-lignin biosynthesis across plant taxa and consider and integrate new insights on major transcription factors, such as NACs and MYBs. We also review insight regarding a new transcriptional regulator, 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, canonically identified as a key enzyme in the shikimate pathway. We use several case studies, including EPSP synthase, to illustrate the evolution processes of gene duplication and neo-functionalization in lignin biosynthesis. This review provides new insights into the genetic engineering of the lignin biosynthetic pathway to overcome biomass recalcitrance in bioenergy crops.
It is hypothesized that the first land plants possessed adaptive metabolic, physiologic, and morphologic changes as a means of coping with abiotic stresses, such as UV-B irradiation and desiccation (Niklas et al., 2017). In this scenario the phenylpropanoid pathway played a pivotal role in land colonization of early plants by yielding protective secondary metabolites including flavonoids and lignin. Many flavonoids bestowed land plants with the ability to absorb UV-B, while lignin, as the cell wall component, provided mechanical support and facilitated water transport for the vascular plants (Rensing, 2018). Recently several comparative genomics, phylogenetics, and evolutionary genetics approaches have been employed to illustrate the evolution of phenylpropanoid biosynthetic pathway (Ma and Constabel, 2019; Davies et al., 2020). In this review, we unite these current outcomes and provide a comprehensive overview of the phylogenetic occurrence of phenylpropanoid biosynthetic and lignin biosynthetic pathways and showcase the role of gene duplication and neo-functionalization contributing to land plant evolution.
To aid our understanding of the phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants, we offer a primer on lignin biosynthesis. Lignin is derived from three major hydroxycinnamyl alcohols, including p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol by radical coupling (Weng and Chapple, 2010). As such, p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monolignols are the main units for lignin polymerization. In addition, two additional non-canonical monolignols, caffeyl alcohol (C), and 5-hydroxyconiferyl (5HG) alcohol, have been found naturally in some species or can be introduced via genetic engineering (Dixon and Barros, 2019; Wang X. et al., 2020).
The lignin biosynthetic pathway has been refined and re-envisioned by several research groups over the past two decades. Based on recent studies in the model herbaceous plant Arabidopsis and the model woody plant Populus, eleven core structural enzymes of the lignin biosynthetic pathway have been identified (Boerjan et al., 2003; Vanholme et al., 2013; Zhang et al., 2020). L-phenylalanine ammonia-lyase (PAL), 4-hydroxycinnamate CoA ligase (4CL), and cinnamate 4-hydroxylase (C4H) are the three enzymes that belong to the general phenylpropanoid pathway shared by the biosynthesis of lignin and flavonoids. Generally, the initial substrate of the phenylpropanoid pathway, phenylalanine, is converted into cinnamate by PAL, C4H coverts cinnamate into p-coumarate, and p-coumarate is then activated by 4CL to form p-coumaroyl CoA.
The other eight enzymes belong to lignin-specific pathway (Figure 1), including cinnamoyl CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), coumarate 3-hydroxylase (C3H), coumaroyl shikimate 3′-hydroxylase (C3′H), ferulate/coniferaldehyde 5-hydroxylase (F5H), caffeate/5-hydroxy-coniferaldehyde 3/5-O-methyltransferase (COMT), caffeoyl CoA 3-O-methyltransferase (CCoAOMT), hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT), and caffeoyl shikimate esterase (CSE). p-coumaroyl CoA is converted into the simplest H-lignin monomer by a reductase CCR and a dehydrogenase CAD. In addition to CAD and CCR, G-lignin biosynthesis starting from p-coumarate requires C3H, COMT, and 4CL; or 4CL, HCT, C3′H, CSE, and CCoAOMT. F5H and COMT are crucial for S-lignin biosynthesis. Noticeably, aldehyde dehydrogenase (ALDH) catalyzes the opposite direction of reactions in lignin biosynthesis, which is required for ferulate and sinapate biosynthesis from coniferaldehyde and sinapaldehyde, respectively (Nair et al., 2004).
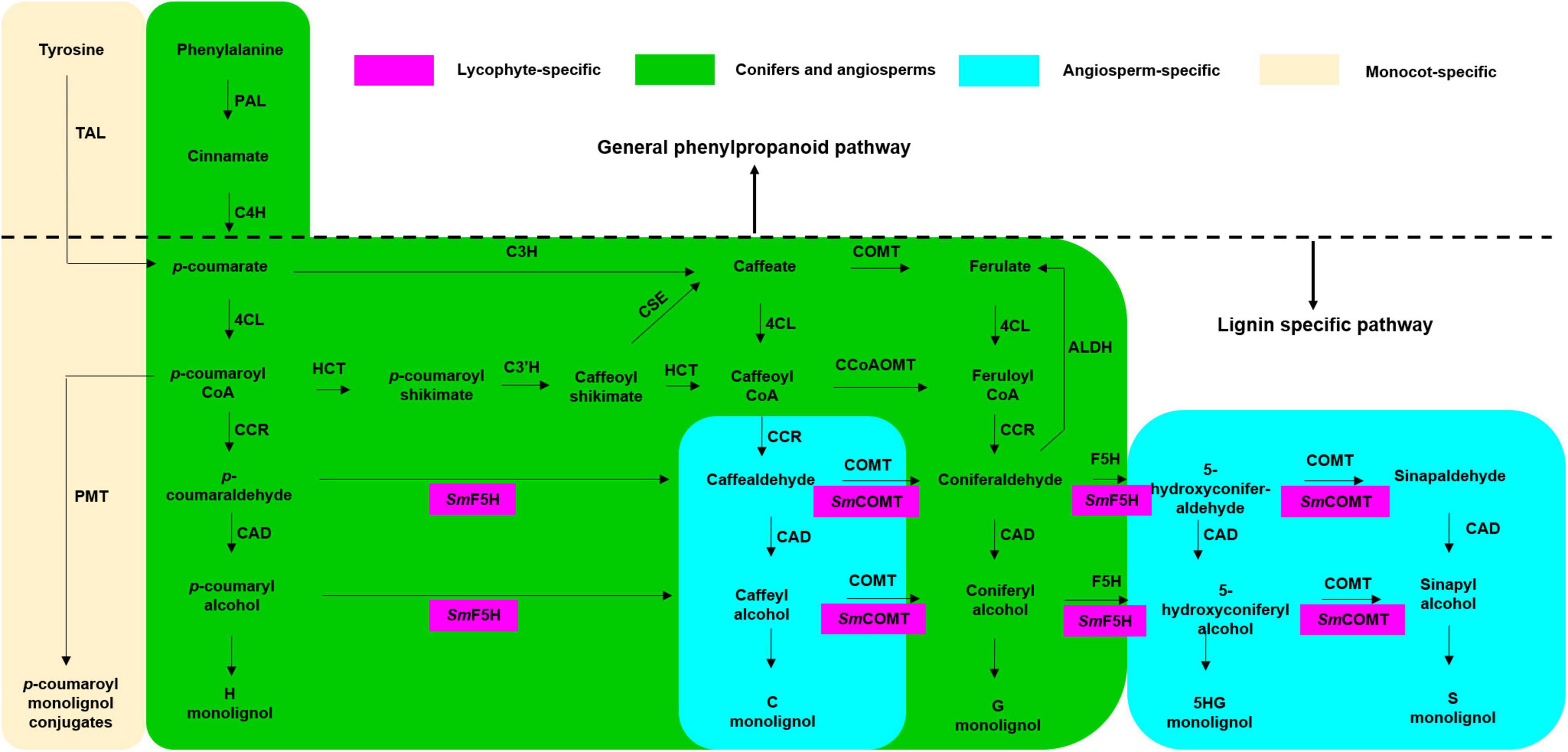
Figure 1. Schematic representation of pathways to produce H, G, and S-monolignols across different plant species. Pathways leading to monolignol biosynthesis include general phenylpropanoid pathway and lignin-specific pathway, which are separated by the dash line. S-lignin biosynthesis in lycophytes occurs in an independent pathway mediated by SmF5H and SmCOMT (highlighted in purple). Some pathways are shared by conifers and angiosperms (highlighted in green) while the others are angiosperm-specific (highlighted in blue). There is also monocot-specific pathway mediated by TAL (highlighted in yellow).
Although lignin has not been discovered in bryophytes, nine structural gene families that are responsible for the biosynthesis of H- and G-lignin monomers occur in moss genomes (Xu et al., 2009; Table 1). Studies using the model plant Physcomitrella patens shed light on the biosynthetic pathway of phenylpropanoids and lignin. Knock-out of the CYP98 gene in P. patens, which encodes a P450 oxygenase, blocks the biosynthesis of the moss cuticle, thus affecting gametophore formation and organ fusion. C3′H is a homolog of CYP98 in higher plants. However, CYP98 in moss uses the p-coumaroyl-threonate as substrate, whereas C3′H in higher plants uses p-coumaroyl-shikimate as substrate (Schoch et al., 2001), leading to the distinct biosynthetic pathway for cuticle (Renault et al., 2017). Interestingly, no phenylpropanoid genes have been found in red algae genomes, but trace amounts of lignin have been reported in red algae, and as such, indicating that the lignin biochemical machinery preexisted the evolution of land plants (Martone et al., 2009; Brawley et al., 2017). The extant presence of lignin in red algae may also represent convergent evolution independent of lignin biosynthesis in bryophytes.
During land plant evolution, lignin appeared first in lycophytes (Renault et al., 2019) in the form of H-lignin. Interestingly, there are only low levels of H-lignin in gymnosperms and traces of H-lignin in angiosperms. In contrast, H-lignin is highly abundant in seedless vascular plants, including lycophytes and pteridophytes. Lignin is found between the cellulose matrix and forms a rigid cell wall in these plants (Espiñeira et al., 2011; Ralph et al., 2019). In gymnosperms and angiosperms, H-lignin can be enriched by down-regulation of C3′H, HCT, and CSE genes (Franke et al., 2002a; Wagner et al., 2007; Coleman et al., 2008; Li et al., 2010; Vanholme et al., 2013, Fornalé et al., 2015), though in many cases growth was negatively impacted. Interestingly, the Arabidopsis C3′H mutant ref8 showed severe growth defect that was rescued by disruption of the mediator complex units MED5a and MED5b (Bonawitz et al., 2014). These results indicate that H-lignin may represent one of the earliest forms of lignin.
G-lignin biosynthesis in pteridophytes is evolutionarily conserved. The Df4CL2 gene is a 4-coumarate:coenzyme A ligase coding gene identified from the fern species Dryopteris fragrans. Heterologous expression of this gene in tobacco increased the synthesis of lignin, demonstrating the conserved function of 4CL in D. fragrans and tobacco (Nicotiana tabacum) (Li et al., 2020). Similarly, two CCoAOMTs have been cloned from the fern species Polypodiodes amoena, and their functions in lignin biosynthesis have been confirmed via heterologous expression in Arabidopsis (Zhang X.-S. et al., 2019).
S-lignin has been identified in lycophytes such as Selaginella moellendorffii; however, its biosynthetic pathway is different from that in angiosperms (Renault et al., 2019). In angiosperm, both C3H and F5H are involved in S-lignin biosynthesis. In contrast, in lycophytes, SmF5H has dual functions that enables S-lignin to be synthesized directly from p-coumaraldehyde and p-coumaryl alcohol. Here, SmF5H and SmCOMT form a gene cluster and are responsible for S-lignin biosynthesis. Phylogenetic analyses suggest that these two genes were independently evolved from their counterparts in angiosperm (Weng et al., 2008b, 2011). Besides the well-known S-lignin biosynthesis in Selaginella, several ferns, such as Dennstaedtia bipinnata, also contain a large amount of S-lignin in the sclerotic sheaths. However, the biosynthetic pathway has not been elucidated (Logan and Thomas, 1985; Weng and Chapple, 2010). Further studies of the lignin biosynthesis-related genes in these fern species and comparation with what we have known in other species in the lineage is needed to provide insights on the evolution of the S-lignin biosynthetic pathway.
Gymnosperms diverged from angiosperms 300 million years ago (De La Torre et al., 2020). In general, gymnosperms lack the F5H gene, and therefore, gymnosperm lignin mainly contains G-monolignol and contains no or little S-monolignol (Li et al., 2001; Weng and Chapple, 2010). When Cf4CL and CfCCoAOMT were cloned from Cryptomeria fortunei, a gymnosperm, and heterologously expressed in tobacco, an angiosperm, G-lignin biosynthesis was increased, indicating that these two lignin genes can function equally well in both gymnosperms and angiosperms (Guo et al., 2019). Similarly, a CSE, LkCSE, from Larix kaempferi, can convert caffeoyl shikimate to caffeate and shikimate, supporting the conserved function of CSE between gymnosperms and angiosperms (Wang et al., 2019). Gymnosperms also produce a compression layer within xylem that enriched in H-lignin in tracheid. A recent study showed that spatial patterning of H- and G-lignin during wood formation is related to different localizations and enzyme activities of lignin polymerization enzymes, laccases (Hiraide et al., 2021). Interestingly, some gymnosperm species, such as Gentales, can also synthesize S-lignin (Renault et al., 2019). Gnetum genmon contains angiosperm-like vessels as well as tracheids and fiber tracheids (Tomlinson, 2001), and it shares the chemical compositions of lignin with angiosperms (Nawawi et al., 2016). These results suggest that the biosynthetic pathway for G-lignin is shared between gymnosperms and angiosperms. On the basis of these results we are left with two alternate hypotheses; ancient gymnosperms were able to produce S-lignin which was subsequently lost in modern gymnosperms or the occurrence of S-lignin in Gentales is a recent convergent evolutionary event. As an ancient gymnosperm, further systematic studies of lignin biosynthesis in Gentales are needed to definitively describe the evolution trajectory in gymnosperms.
Angiosperms contain the lignin composed of G-, S-, and H-lignin monomers in various ratios (Mansfield et al., 2012). The lignin biosynthetic pathways of angiosperms have been characterized using the model plants, Arabidopsis, Populus, and Brachypodium, among others. Xu et al. (2009) analyzed 10 of 11 lignin biosynthetic gene families (without CSE) across 14 plant species and 1 symbiotic fungal species using comparative genomics. The analysis revealed that the rapid expansion of these gene families occurred after the divergence between dicots and monocots 140–150 million years ago (Xu et al., 2009; Rao and Dixon, 2018).
Although the lignin biosynthetic pathways are generally conserved among angiosperms, alternative pathways have evolved in monocots. In dicots, the first enzyme in the phenylpropanoid pathway, PAL, converts phenylalanine (Phe) to cinnamate. Cinnamate is then converted to p-coumarate by the second enzyme, C4H. However, a bypass route has been discovered in monocots. PTAL was identified as a bifunctional enzyme that recognizes tyrosine (Tyr) as the substrate and converts it to p-coumarate directly in Brachypodium distachyon (Barros et al., 2016). 13C isotope feeding with BdPTAL1-RNAi transgenic plants revealed that BdPTAL1-mediated lignin biosynthesis contributed to half of the total lignin content in B. distachyon (Barros et al., 2016). Another grass-specific enzyme is p-coumaroyl-CoA:monolignol transferase (PMT) that catalyzes the incorporation of p-coumarate into the lignin polymer backbone typically found in the Poaceae family (Withers et al., 2012; Petrik et al., 2014). These findings suggest that lineage-specific lignin biosynthetic pathways have evolved independently in dicots and monocots and highlight the need to study species-specific branches in the lignin biosynthetic pathway.
The C4H gene progenitor appears to have duplicated in early seed plants, yielding two clades that are preserved in Taxaceae and most angiosperms. A second duplication event happened after the divergence of dicots and monocots. By analyzing the protein structure and function of Brachypodium C4H, it was found that each of Brachypodium C4H paralog genes can rescue the growth defect of the Arabidopsis c4h mutant, indicating that the C4Hs in monocots preserved the canonical function in lignin biosynthesis. However, the protein structures of C4Hs in B. distachyon differ from that in Arabidopsis. This newly derived C4H type in monocots has an elongated N-terminus, which alters the subcellular localization and allows the orientation of C4H to the lumen of endoplasmic reticulum (ER) through a double-spanning hairpin structure. Therefore, it is possible that an alternate C4H exists within the ER (Renault et al., 2017).
Coumarate 3-hydroxylase and C3′H catalyze the conversion of p-coumarate and p-coumaroyl shikimate into caffeate (via a bifunctional cytosolic ascorbate peroxidase, Barros et al., 2019) and caffeoyl shikimate (via a cytochrome P450 monooxygenase, Schoch et al., 2001), respectively. These enzymes play important roles in G-lignin and S-lignin biosynthesis. There is only one member of the C3′H family in Arabidopsis thaliana and two members of the cytosolic C3H family in A. thaliana and B. distachyon (Franke et al., 2002b; Barros et al., 2019). PtrC3′H3 was recognized as the homolog of Arabidopsis C3′H. However, it was proposed that PtrC3′H3 requires PtrC4H1 or PtrC4H2 to form a complex to enhance its enzymatic activity in Populus trichocarpa (Chen et al., 2011; Figure 1). Recent study showed that triple knocking-down PtrC4H1/PtrC4H2/PtrC3′H3 causes monolignol benzoate (ML-BL) conjugation and significantly reduces lignin biosynthesis while increasing H-lignin for about 70-fold (Kim et al., 2020). These findings suggest that simultaneous modification of C4H and C3H could be used for reducing biomass recalcitrance in bioenergy crops.
Phylogenetic analysis of 192 4CLs across land plants suggested that a duplication of the 4CL gene family occurred prior to the split of gymnosperms and angiosperms (Li et al., 2015). Functional divergence of the 4CL gene family, post duplication, has been broadly found in angiosperms. In fact, four members of the 4CL gene family have been reported in P. patens, but only three of them were expressed under tested conditions (Silber et al., 2008). There are four 4CL genes in Arabidopsis, five in rice, and seven in Populus. Functional analysis of these gene families revealed that only one subgroup of this gene families is involved in lignin biosynthesis, while other subgroups are involved in the biosynthesis of flavonoids or phenolics via neofunctionalization (Ehlting et al., 1999; Gui et al., 2011; Li et al., 2015; Rao et al., 2015; Table 1). Loss-of-function mutation of 4CL genes in herbaceous species causes reductions in G-lignin and increase of S/G ratios. However, knock-out 4CL1 gene in Populus led to reduction of S-lignin and decrease of S/G ratio, and the homeostasis of G-lignin was maintained by 4CL5 in 4cl1 mutant. These findings point toward a functional divergence of 4CLs between herbaceous and woody species (Xiong et al., 2019; Tsai et al., 2020).
Hydroxycinnamoyl transferase catalyzes the conversion of caffeoyl shikimate to caffeoyl-CoA. Down-regulation of AtHCT caused the reduction of S-lignin content in Arabidopsis (Hoffmann et al., 2004). The orthologs of HCTs are present among all the land plants, which suggests that this enzyme evolved before the occurrence of lignin. A recent study showed that P. patens HCT and Marchantia polymorpha HCT can complement the deficiency of Arabidopsis hct mutant in terms of morphology and metabolite levels, suggesting that the function of HCT is likely conserved in all embryophytes (Kriegshauser et al., 2021). It appears that gene duplication of HCT occurred in dicots that produced the HQT gene. Despite the sequence similarity between HCT and HQT, the latter is required for biosynthesizing chlorogenic acid rather than lignin in Cynara cardunculus (Sonnante et al., 2010). Knock-down of HCT led to increase of G-lignin and decrease of S-lignin and S/G ratio in Populus (Zhou et al., 2020). However, knock-down of both HCT1 and HCT2 did not drastically change lignin content or composition in B. distachyon. Meanwhile, the saccharification efficiency was greatly enhanced in the double knock-down line (Serrani-Yarce et al., 2021). These findings suggest HCT genes play different roles in some monocots compared to that of dicots.
Caffeoyl shikimate esterase is a newly discovered enzyme involved in monolignol biosynthesis. Together with 4CL, these two enzymes form a bypass pathway of monolignol biosynthesis in Arabidopsis (Vanholme et al., 2013). CSE genes cloned from Medicago truncatula and Populus deltoides have been shown to be functionally conserved with their Arabidopsis homolog (Ha et al., 2016; Saleme et al., 2017). However, the homolog of CSE gene has not been identified in most monocots, including maize and Brachypodium. Recently, the generation of cse1, cse2 single mutant and cse1/cse2 double mutant in Populus further confirmed their partial redundant roles in lignin biosynthesis. In addition to causing a 35% reduction in lignin content, the cse1/cse2 double mutant significantly improved cellulose-to-glucose transformation efficiency. As such, CSEs in Populus could be promising target genes in biorefinery although their growth penalty should be managed to avoid (de Vries et al., 2021). Noticeably, CSE has also been shown to be functional in gymnosperms, such as Larix kaempferi (Wang et al., 2019). These findings suggest that CSE may be evolved prior to the divergence of gymnosperms and angiosperms, but was lost in many monocots (Wang et al., 2019; Serrani-Yarce et al., 2021).
Caffeate/5-hydroxy-coniferaldehyde 3/5-O-methyltransferase and F5H are two key enzymes required for catalyzing the intermediates in G-lignin biosynthesis into S-lignin biosynthesis. It has been reported that simultaneously manipulating COMT and F5H resulted in a dramatic change of S-lignin biosynthesis (Wu et al., 2019). COMT and F5H in S. moellendorffii appears to have an independent origin compared to that of angiosperms. There are two F5H genes in Arabidopsis (AtF5H1/CYP84A1 and AtF5H2/CYP84A2), and only AtF5H1 has been confirmed to be involved in lignin biosynthesis (Meyer et al., 1998). Similarly, there is one functional COMT gene identified among 13 homologous genes in Arabidopsis (Raes et al., 2003). In Populus, five F5H genes have been cloned, and two of them, PtrF5H1 and PtrF5H2, were reported to be involved in lignin biosynthesis. Thirteen members of COMT gene family were identified in P. trichocarpa, but only PtrCOMT2 is highly expressed in xylem (Shi et al., 2009; Table 1). The function of F5H was shown to be conserved in monocots, such as Oryza sativa. One of three F5H genes, OsCAld5H1, was reported to greatly affect the S/G-lignin composition via over-expression or knock-out (Takeda et al., 2017, 2019). OsCAldOMT1 has been proven to be a functional COMT in rice (Lam et al., 2019). Noticeably, it not only regulates S-lignin biosynthesis, but also controls tricin-lignin biosynthesis. The dual functions of OsCAldOMT1 seems to be specific in grass species (Lam et al., 2019). CCoAOMT, another O-methyltransferase, converts feruloyl CoA to sinapoyl CoA and is required for the conversion of G-lignin into S-lignin. Genetic engineering of this enzyme led to change in G-lignin biosynthesis in Populus, alfalfa, Pinus radiata, maize, and tobacco (Zhong et al., 2000; Guo et al., 2001; Wagner et al., 2011; Li et al., 2013; Xiao et al., 2020). These studies suggest the function of CCoAOMT is likely to be conserved among all angiosperms and occurred with the advent of the angiosperms.
Cinnamoyl CoA reductase recognizes four types of cinnamoyl-CoAs, including p-coumaroyl CoA, caffeoyl CoA, feruloyl CoA, and sinapoyl CoA, and converts them into cinnamaldehydes. Phylogenetic analysis of 146 CCR genes of various land plants revealed that CCR family contains three classes: CCR, CCR-like, and DFR, and that only the CCR class contains bona fide lignin biosynthetic genes. All these three classes are distributed across land plants, including P. patens, which contains a single functional CCR gene. These results suggested that the progenitor CCR gene evolved after the advent of lycophytes (Barakat et al., 2011). Still, functional divergence within the CCR family has arisen in several species. For example, in Arabidopsis, AtCCR1 is involved in lignin biosynthesis, whereas AtCCR2 is involved in pathogen response (Lauvergeat et al., 2001; Ruel et al., 2009). Downregulation of a CCR gene, CCR2, reduces lignin biosynthesis and increases saccharification efficiency in Populus. However, it also causes severe biomass penalty (Van Acker et al., 2014). Recently, a ccr2 mutant was generated by the CRISPR/Cas9 approach that contain a null and haplo-insufficient allele in Populus. This mutant line does not have growth penalty, but still has low lignin content and improved saccharification efficiency (De Meester et al., 2020). Therefore, CCR2 gene could be a useful target that can be deployed in genetic engineering of bioenergy woody crops.
Cinnamyl alcohol dehydrogenase catalyzes the final step of monolignol biosynthesis leading to compositional differences in lignin forms. Guo et al. (2010) performed phylogenetic analysis of the CAD gene family from 52 species and classified them into three classes. Class I comprises bona fide CADs which are only present in vascular plants, suggestive of their co-occurrence with the advent of lignin. The functional characterizations of Class II and Class III CADs remain unclear (Guo et al., 2010). Within the large gene families, CADC and CADD, PtrCAD1 and OsCAD2 have been reported to be functional CAD genes involved in lignin biosynthesis in Arabidopsis, rice, and Populus. Knock-down or knock-out of these genes resulted in reduced lignin content as well as altered lignin structures (Anderson et al., 2015; Van Acker et al., 2017; Martin et al., 2019). Finally, it was reported that CAD and CCR form an enzyme complex that regulates monolignol biosynthesis in P. trichocarpa (Yan et al., 2019).
In summary, as an important branch of the phenylpropanoid pathway, structural genes of the lignin biosynthetic pathway are conserved in most embryophytes. F5H and COMT contribute to S-lignin biosynthesis and have been hypothesized to have independent origins in S. moellendorffii and angiosperms. Gene duplications and gene family expansion of lignin biosynthetic genes in angiosperms have given rise to sub-functionalization and neo-functionalization of the various members, which is consistent with their morphological and functional changes compared with lower plants.
The lignin biosynthetic pathway includes both structural genes and regulatory proteins. Transcriptional regulation, controlling the gene expression of structural genes, plays important roles in lignin biosynthesis. Such genes reflect the phylogenetic occurrence of the phenylpropanoid pathway and evolutionary trajectory of lignin biosynthesis in plants. MYBs and NACs are two major transcription factor families, comprising three layers of the hierarchical transcriptional regulatory network (Ohtani and Demura, 2019; Figure 2). Therefore, we focus on analyzing these two families of transcription factors to illustrate the evolutionary divergence of transcriptional regulation in lignin biosynthesis.
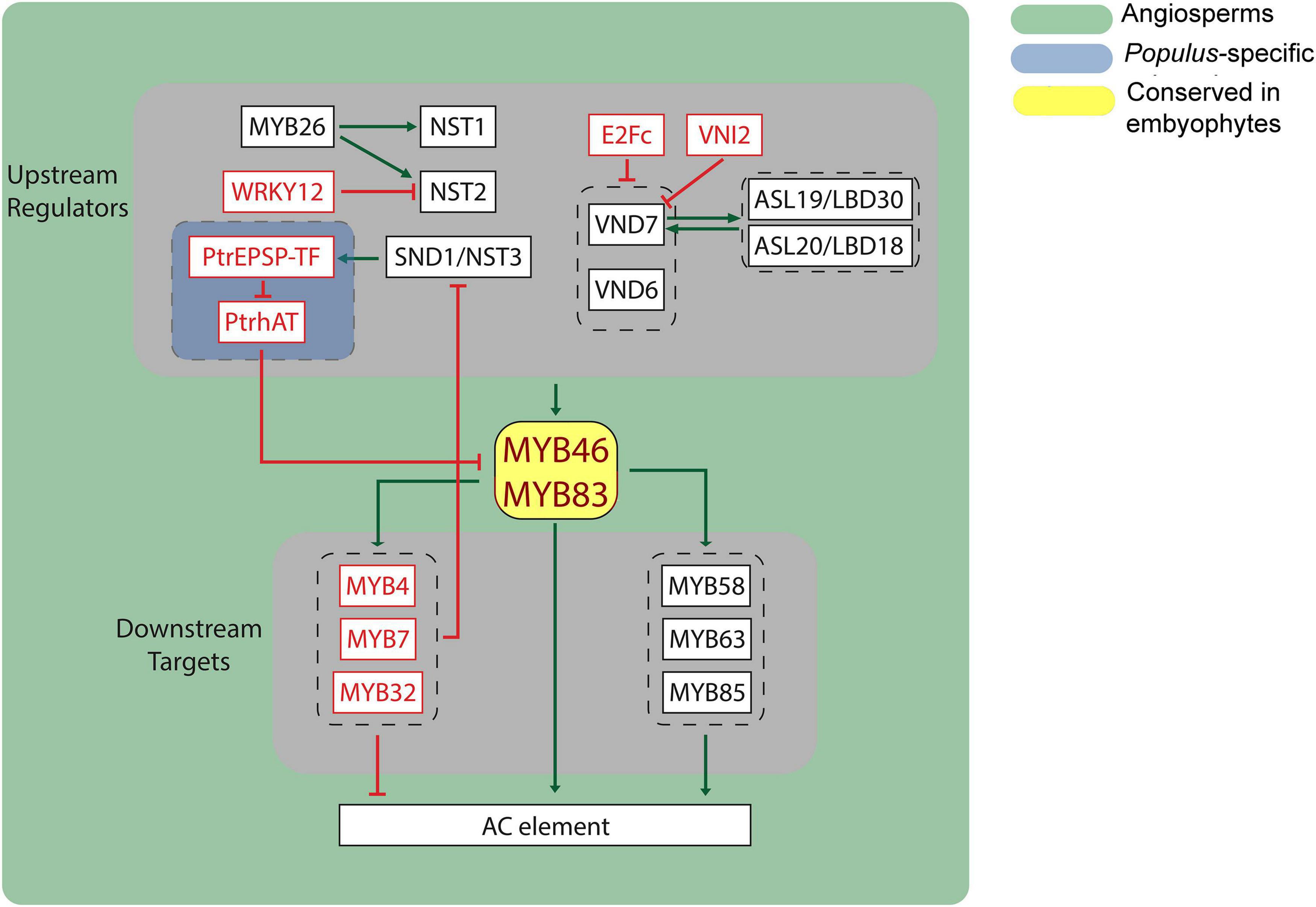
Figure 2. Transcriptional network of lignin biosynthesis in plants. Black box indicates transcriptional activator, and red box indicates transcriptional repressor. Green arrows indicate transcriptional activation, and red blunt arrows indicate transcriptional repression. AC element, recognized by MYBs, were found in most of lignin biosynthetic genes.
Transcription factor MYB46 is a central regulator in secondary cell wall formation (Zhong et al., 2007). MYB46 and MYB83 are two functionally redundant A. thaliana MYB transcription factors that act as master switches of lignin biosynthesis regulating nine out of 11 monolignol biosynthetic genes (PAL, C4H, 4CL, HCT, C3′H, CCoAOMT, F5H, CCR, and CAD) (Kim et al., 2014). Besides lignin, the biosynthesis of other secondary cell wall components, including xylan and cellulose, are also regulated by MYB46/MYB83 (McCarthy et al., 2009; Zhong and Ye, 2012; Kim et al., 2013). Several MYB46 orthologs from other plant species have also been shown to function as key regulators for secondary cell wall biosynthesis, including PtMYB4 from pine, EgMYB2 from Eucalyptus, OsMYB46 from rice, PtrMYB2, PtrMYB3, PtrMYB20, and PtrMYB21 from Populus, and ZmMYB46 from maize (Patzlaff et al., 2003; Goicoechea et al., 2005; Zhong et al., 2011, 2013). The functions of MYB46 and MYB83 in lignin biosynthesis are well-conserved in angiosperms.
The phylogenetic history of lignin related MYBs appears to coincide with the advent of the lignin biosynthetic genes, which first emerged in early land plants (Xu et al., 2014; Bowman et al., 2017). Homologs of MYB46 and MYB83 have been found in P. patens and S. moellendorffii (Zhong et al., 2010). Functional conservation of their homologs via transgenic validation has also been demonstrated in vascular plants, including gymnosperms and angiosperms (Zhao and Bartley, 2014). We hypothesis that MYB46 and MYB83 might be required for phenylpropanoid biosynthesis outside of the lignin biosynthetic pathway in non-vascular plants while playing core roles in lignin biosynthesis in all vascular plants.
Major transcription factors regulating MYB46/MYB83 are the NAC TF family proteins (Figure 2). NAC TF family proteins share a conserved NAC domain located at the N-terminal region and a highly divergent C-terminal activation domain (Olsen et al., 2005). These TFs are specific to plants and play diverse roles in plant defense, growth, and development (Olsen et al., 2005). NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2 are redundantly responsible for secondary wall thickening in anther endothecium (Mitsuda et al., 2005). A MYB family protein, MYB26, localized in the nucleus, was found to be an upstream positive regulator of NST1 and NST2. Overexpression of MYB26 was found to increase lignin deposition and the expression of NST1 and NST2 (Yang et al., 2007). Recent study shows that Xylem NAC Domain 1 (XND1) interacts with NST1 and inhibits the transcriptional activity of NST1, thus repressing secondary cell formation (Zhang Q. et al., 2019). In addition, VASCULAR-RELATED NAC-DOMAIN 6 (VND6) and VND7 directly regulate MYB46 and MYB83 expression (Zhong et al., 2008; McCarthy et al., 2009; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Overexpression of VND6 and VND7 can induce the ectopic differentiation of metaxylem-like vessels and protoxylem-like vessels, respectively (Kubo et al., 2005). Functional suppression of VND6 and VND7 caused defects in the formation of vessel elements (Kubo et al., 2005; Yamaguchi et al., 2008). In Arabidopsis, there are seven VND genes (VND1-VND7). Similar to VND6 and VND7, overexpression of VND1 to VND5 also induces ectopic secondary cell wall deposition, suggesting that all VND members contribute to lignin biosynthesis during xylem vessel development (Endo et al., 2014; Zhou et al., 2014).
A third class of TFs involved in lignin biosynthesis include the WRKY gene family. Mutation of the Arabidopsis WRKY12 gene caused secondary cell wall thickening in pith cells that is associated with ectopic deposition of lignin, xylan, and cellulose. WRKY12 mutation upregulated the transcription of downstream genes encoding the NAC domain TF NST2 and the zinc finger TF C3H14, which activate secondary wall synthesis (Wang et al., 2010). Direct binding of WRKY12 to the NST2 gene promoter led to repression of NST2 and C3H14, as defined by in vitro assays and in planta transgenic experiments (Wang et al., 2010). Interestingly, WRKY12 gene is expressed in both pith and cortex that do not have secondary wall thickening, suggesting that WRKY12 may control the parenchymatous nature of pith cells by acting as a negative regulator of secondary cell wall NACs (Wang et al., 2010). WRKY15 was reported to repress the expression of VND7 and suppress tracheary elements (TEs) differentiation through indirect regulation (Ge et al., 2020). Based on our current understanding, WRKY TFs act upstream of NACs to regulate secondary cell wall biosynthesis.
Two members of the ASYMMETRIC LEAVES2-LIKE/LATERAL ORGAN BOUNDARIES DOMAIN (ASL/LBD) family ASL19/LBD30, ASL20/LBD18 were identified to be involved in a positive feedback loop for VND7 expression that regulates TEs differentiation-related genes (Soyano et al., 2008). Overexpression of ASL19 and ASL20 induced trans-differentiation of cells from non-vascular tissues into TE-like cells, similar to those induced by VND6 or VND7 overexpression. Expression of both ASL19/LBD30 and ASL20/LBD18 are dependent on VND6 and VND7 (Soyano et al., 2008). XND1 has been reported to inactivate VND6 by physically interacting with VND6 and directing VND6 from the nucleus to the cytoplasm (Zhong et al., 2020). Another NAC transcriptional factor, VND-INTERACTING2 (VNI2), can bind to VND proteins and has been shown to function as a transcriptional repressor of VND7-mediated gene transcription (Yamaguchi et al., 2010). Recent studies show that E2Fc is a key upstream regulator of VND6 and VND7, directly targeting the genomic loci of VND6 and VND7. E2Fc is a transcriptional repressor, and transcript abundance of VND6 and VND7 were significantly increased in E2Fc knockdown Arabidopsis lines (Taylor-Teeples et al., 2015). Taken together, VND6 and VND7 represent key regulators in lignin biosynthesis whose functions are tightly regulated by various TFs (Ko et al., 2012; Schuetz et al., 2013). Phylogenetic analysis discovered close homologs of VND6 and VND7 in all vascular plants, whose functions were demonstrated to be conserved in P. trichocarpa, Zea mays, Oryza sativa, and B. distachyon (Zhong et al., 2010, 2011; Valdivia et al., 2013).
SND1/NST3 and NST1 are required for secondary wall thickening in stem fibers (Mitsuda et al., 2007). When these genes were expressed constitutively in Arabidopsis, ectopic secondary wall thickening in various tissues was induced (Mitsuda et al., 2005, 2007). Putative orthologs of NST1, NST2, and SND1/NST3 are present in the genome of Populus and are expressed in developing xylem (Mitsuda et al., 2007), implicating a role in lignin biosynthesis. The function of NST homologs in lignin biosynthesis has been confirmed in Medicago truncatula and cotton (Zhao et al., 2010; Fang et al., 2020). However, the homologs of NST proteins have not been identified in gymnosperms or earlier species, implying that these proteins may not have evolved until the appearance of angiosperms (Nakano et al., 2015).
Three MYB family proteins, MYB58, MYB63, and MYB85, whose coding genes are direct targets of MYB46, have been shown to function as direct transcriptional activators of lignin biosynthesis during secondary wall formation in Arabidopsis (Zhong et al., 2008; Ko et al., 2009; Demura and Ye, 2010; Zhou et al., 2020; Figure 2). All three MYBs cause ectopic lignin deposition when overexpressed.
The coding genes of three other MYB family proteins, MYB32, MYB4, and MYB7, are also directly activated by MYB46 (Ko et al., 2009). These three MYBs, sharing high sequence similarity with a conserved EAR motif, have been shown to be transcriptional repressors (Dubos et al., 2010). Trans-activation assays showed that these MYB transcription factors directly repress the expression of SND1, forming a feedback regulatory loop to maintain the abundance of SND1 (Wang et al., 2011).
KNOTTED ARABIDOPSIS THALIANA7 (KNAT7) and BEL1-LIKE HOMEODOMAIN6 (BLH6) belong to knotted-like homeobox proteins and bel1-like homeodomain proteins, respectively. KNAT7 and BLH6 were reported to be direct targets of MYB46 and MYB83 (Zhong and Ye, 2012). KNAT7 and BLH6 interact with each other and negatively regulate lignin biosynthesis while KNAT3 was reported to form heterodimer with KNAT7 to synergistically regulate lignin content and composition (Liu et al., 2014; Qin et al., 2020; Wang S. et al., 2020).
Although the first and second layers of master switches of lignin biosynthesis were shown to be conserved in vascular plants even in early land plants, the targets of MYB46/83 have not been shown to be functionally conserved in lower plants. For example, the close homologs of MYB58 and MYB63 failed to be identified in lower tracheophytes (Zhong et al., 2010). In addition, their homologs in switchgrass were found to be mainly involved in flavonoids biosynthesis rather than lignin biosynthesis. A plausible explanation is that wide expansion, promiscuous functionality, and functional diversification of the MYB family across different species have made it difficult to identify the genuine orthologs responsible for lignin biosynthesis regulation (Zhao and Bartley, 2014; Nakano et al., 2015). Furthermore, lineage-specific MYBs may contribute to lignin biosynthesis in different tracheophytes. For instance, MYB75 was found to repress secondary cell wall biosynthesis and activate anthocyanin biosynthesis in dicots but not in monocots (Zhao and Bartley, 2014).
5-enolpyruvylshikimate-3-phosphate (EPSP) synthase is a key enzyme in shikimate pathway, which is present in both plants and many prokaryotes. EPSP synthase has been a well-known herbicide target, which has been widely used in agriculture (Sammons and Gaines, 2014). Noticeably, there is only one copy of an EPSP synthase coding gene in green algae, lycophytes, and bryophytes, but duplicated genes were found in angiosperms, such as Arabidopsis and Populus (Tohge et al., 2013; Yang et al., 2017; Xie et al., 2018; Figure 3). The gene duplication in angiosperms may have given rise to neo-functionalization for the additional gene copy.
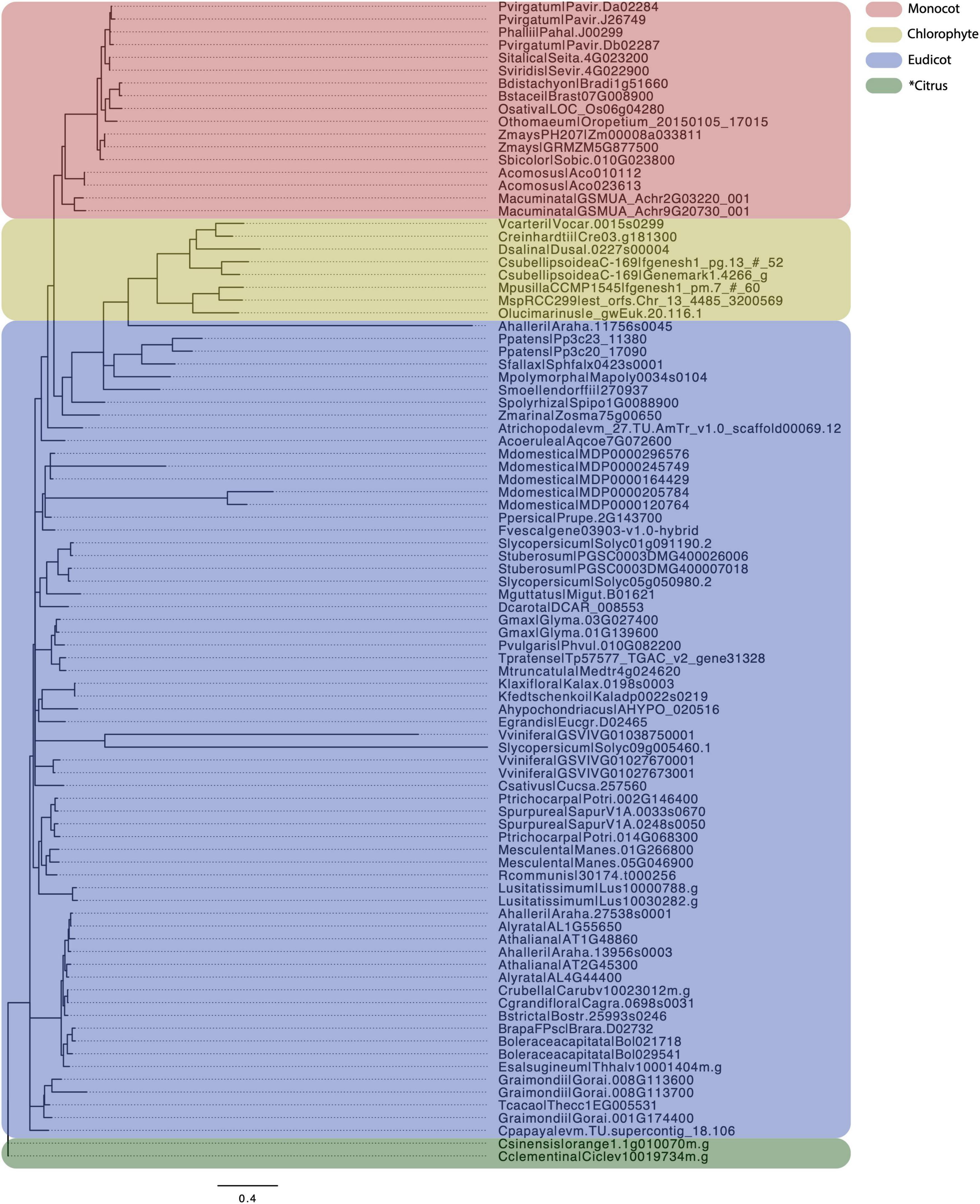
Figure 3. Molecular dating of EPSPs. A total of 91 EPSPs, identified by searching against PtrEPSP-TF in phytozome, were used for molecular dating analysis. We first used MUSCLE (Edgar, 2004) to perform multiple alignments of EPSP proteins, an in-house python script was then used to convert the amino acid alignment to nucleotide alignment, and finally TrimAL (Capella-Gutiérrez et al., 2009) was used to trim the alignment using parameters “-gt 0.8 -st 0.001,” which indicate the tolerating gaps of no more than 20% and similarity score less than 0.001. Mrbayes (Huelsenbeck and Ronquist, 2001) was used to conduct molecular dating with parameters “lset nst = 6 rates = invgamma” using the “GTR + I + Γ” model. A total of 10,000,000 mcmc generations were run after the standard deviation of split frequencies falls under 0.05. FigTree (Rambaut, 2012) was used to visualize the phylogenetic tree. Number of each node indicates the posterior probabilities. Pink, yellow, blue, and green colors separate monocot, Chlorophyte, Eudicot, and Citrus, respectively.
A recent study in P. trichocarpa discovered the transcriptional regulatory function of one EPSP synthase gene (PtrEPSP-TF) (Xie et al., 2018). Overexpression of PtrEPSP-TF led to ectopic deposition of lignin, accumulation of phenylpropanoid metabolites and differential expression of secondary cell wall biosynthetic genes. It was shown that PtrEPSP-TF accumulates in the nucleus and acts as a transcriptional repressor by directly binding to the promoter element of a hAT transposase family gene (PtrhAT). PtrhAT is also located in the nucleus and serves as a transcriptional repressor. The direct target of PtrhAT is PtrMYB021, which is a homolog of MYB46 in Arabidopsis that acts as a master switch for secondary cell wall biosynthesis, as described above. By repressing the expression of PtrhAT, PtrEPSP-TF activates the expression of PtrMYB021 and the phenylpropanoid pathway (Xie et al., 2018). In conclusion, PtrEPSP-TF/PtrhAT/PtrMYB021 form an additional regulatory loop in lignin biosynthesis in Populus.
PtrEPSP-TF distinguishes itself from ancestral EPSP synthases by carrying an additional helix-turn-helix (HTH) motif in the N-terminus (Xie et al., 2018). HTH motifs are commonly found in transcription factors as nucleic acid binding domains (Aravind et al., 2005). With the addition of the N-terminal HTH DNA binding motif, PtrEPSP-TF exhibited nuclear accumulation and functioned as a transcriptional repressor. By comparing 57 EPSP synthase isoforms from 42 plant genomes, the HTH motif was found to be almost entirely missing in EPSP synthases in non-vascular, algal, and monocots, but was found in many dicots (Xie et al., 2018). The presence of secondary cell wall is a key distinguishing feature separating dicots from algae and mosses. It is intriguing that this shikimate pathway derived-EPSP synthase isoform appears to have obtained a regulatory function modulating the expression of processes that are ubiquitous in dicots relative to other plants. With this in mind, we hypothesized that domain co-option may have occurred during the course of evolution when early dicotyledonous plants attained complex cell wall structure (Weng et al., 2008a; Tohge et al., 2013). The discovery of the additional regulatory loop of MYB46 in Populus also supports the existence of woody plant-specific regulatory mechanisms in lignin biosynthesis.
The phenylpropanoid pathway produced thousands of metabolites which are essential for plant terrestrialization and subsequent radiation. Lignins appeared as specialized metabolites with the evolution of tracheophytes. The identification of progenitors of lignin biosynthetic genes in bryophytes provides new insights into the origin of lignin biosynthesis (Kriegshauser et al., 2021). The recent progress on genome sequencing of Charophyte algae, bryophytes, lycophytes, and ferns have also provided unprecedented opportunities to study the origin of phenylpropanoid biosynthetic pathway (Szövényi et al., 2021).
On the basis of current knowledge of lignin biosynthetic pathways across tracheophytes, we conclude that most lignin biosynthetic genes experienced expansions and neofunctionalization. As a result, lignin biosynthetic pathway has become increasingly complex evidenced by the existence of many alternate pathways and regulatory hierarchies. In support of this hypothesis many of the alternative pathways have been shown to be lineage specific. Lignin biosynthesis in monocots served an example of diversification. For example, PTAL-mediated by-pass route in lignin biosynthesis and PMT-mediated lignin modification are specific to monocots (Petrik et al., 2014; Barros et al., 2016). Equally, S-lignin biosynthesis in S. moellendorffii suggested that S-lignin biosynthetic pathway may be evolved multiple times or lost in gymnosperms and other pteridophytes (Weng and Chapple, 2010).
Transcriptional regulatory modules have been shown to be generally conserved for phenylpropanoid and lignin biosynthesis; however, a third layer of MYB TFs are not evolutionarily conserved and have witnessed a wide expansion of family members. Finally, newly identified TFs, such as EPSP-TF, have been shown to regulate lignin biosynthesis specifically in woody plants (Xie et al., 2018). The studies on transcriptional regulation of lignin biosynthesis represents an emerging opportunity to understand the phylogenetic occurrence of the phenylpropanoid pathway and lignin biosynthesis in plants.
In this review, we summarized the phylogenetic occurrence of lignin biosynthetic genes and related transcriptional regulation across different plant species. Comprehensively, the core enzymes in lignin biosynthesis and basal transcriptional regulatory module are conserved among embryophytes, although bryophytes do not produce lignin. With evolutionary time, lignin composition diversity has increased and has been associated with gene duplication, functional gene co-option, and neo- and sub-functionalization, which involved many structural genes and transcriptional regulators. In addition, concomitant with the increase of lignin biosynthetic complexity, is the increase in functional diversity, e.g., water conductivity and defense. As most of the current knowledge of lignin biosynthesis is based on the study of a few angiosperms, identification and functional characterization of the lignin biosynthetic pathways and their regulation in lower plants will provide a comprehensive view of their evolutionary history and lead to new insights in lignin biosynthesis.
TY and KF drafted the manuscript. MX, JB, TT, GT, WM, and J-GC revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Center for Bioenergy Innovation by the Office of Biological and Environmental Research in the U.S. Department of Energy Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anderson, N. A., Tobimatsu, Y., Ciesielski, P. N., Ximenes, E., Ralph, J., Donohoe, B. S., et al. (2015). Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27, 2195–2209. doi: 10.1105/tpc.15.00373
Aravind, L., Anantharaman, V., Balaji, S., Babu, M. M., and Iyer, L. M. (2005). The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262. doi: 10.1016/j.fmrre.2004.12.008
Barakat, A., Yassin, N. B., Park, J. S., Choi, A., Herr, J., and Carlson, J. E. (2011). Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants. Plant Sci. 181, 249–257. doi: 10.1016/j.plantsci.2011.05.012
Barros, J., Escamilla-Trevino, L., Song, L., Rao, X., Serrani-Yarce, J. C., Palacios, M. D., et al. (2019). 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 10:1994. doi: 10.1038/s41467-019-10082-7
Barros, J., Serrani-Yarce, J. C., Chen, F., Baxter, D., Venables, B. J., and Dixon, R. A. (2016). Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2:16050. doi: 10.1038/nplants.2016.50
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin Biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Wheeler, G. L., Lohr, M., Goodson, H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. U. S. A. 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, H.-C., Li, Q., Shuford, C. M., Liu, J., Muddiman, D. C., Sederoff, R. R., et al. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108, 21253–21258. doi: 10.1073/pnas.1116416109
Coleman, H. D., Park, J.-Y., Nair, R., Chapple, C., and Mansfield, S. D. (2008). RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 105, 4501–4506. doi: 10.1073/pnas.0706537105
Davies, K. M., Jibran, R., Zhou, Y., Albert, N. W., Brummell, D. A., Jordan, B. R., et al. (2020). The Evolution of Flavonoid Biosynthesis: a Bryophyte Perspective. Front. Plant Sci. 11:7. doi: 10.3389/fpls.2020.00007
De La Torre, A. R., Piot, A., Liu, B., Wilhite, B., Weiss, M., and Porth, I. (2020). Functional and morphological evolution in gymnosperms: a portrait of implicated gene families. Evol. Appl. 13, 210–227. doi: 10.1111/eva.12839
De Meester, B., Madariaga Calderón, B., de Vries, L., Pollier, J., Goeminne, G., Van Doorsselaere, J., et al. (2020). Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele. Nat. Commun. 11:5020. doi: 10.1038/s41467-020-18822-w
de Vries, L., Brouckaert, M., Chanoca, A., Kim, H., Regner, M. R., Timokhin, V. I., et al. (2021). CRISPR-Cas9 editing of CAFFEOYL SHIKIMATE ESTERASE 1 and 2 shows their importance and partial redundancy in lignification in Populus tremula x P. alba. Plant Biotechnol. J. [Epub Online ahead of print]. doi: 10.1111/pbi.13651
Demura, T., and Ye, Z.-H. (2010). Regulation of plant biomass production. Curr. Opin. Plant Biol. 13, 298–303. doi: 10.1016/j.pbi.2010.03.002
Dixon, R. A., and Barros, J. (2019). Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 9:190215. doi: 10.1098/rsob.190215
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E., and Kombrink, E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19, 9–20. doi: 10.1046/j.1365-313X.1999.00491.x
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2014). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Espiñeira, J. M., Novo Uzal, E., Gómez Ros, L. V., Carrión, J. S., Merino, F., Ros Barceló, A., et al. (2011). Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol. 13, 59–68. doi: 10.1111/j.1438-8677.2010.00345.x
Fang, S., Shang, X., Yao, Y., Li, W., and Guo, W. (2020). NST- and SND-subgroup NAC proteins coordinately act to regulate secondary cell wall formation in cotton. Plant Sci. 301:110657. doi: 10.1016/j.plantsci.2020.110657
Fornalé, S., Rencoret, J., Garcia-Calvo, L., Capellades, M., Encina, A., Santiago, R., et al. (2015). Cell wall modifications triggered by the down-regulation of Coumarate 3-hydroxylase-1 in maize. Plant Sci. 236, 272–282. doi: 10.1016/j.plantsci.2015.04.007
Franke, R., Hemm, M. R., Denault, J. W., Ruegger, M. O., Humphreys, J. M., and Chapple, C. (2002a). Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30, 47–59. doi: 10.1046/j.1365-313X.2002.01267.x
Franke, R., Humphreys, J. M., Hemm, M. R., Denault, J. W., Ruegger, M. O., Cusumano, J. C., et al. (2002b). The ArabidopsisREF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45. doi: 10.1046/j.1365-313X.2002.01266.x
Ge, S., Han, X., Xu, X., Shao, Y., Zhu, Q., Liu, Y., et al. (2020). WRKY15 suppresses tracheary element differentiation upstream of VND7 during xylem formation. Plant Cell 32, 2307–2324. doi: 10.1105/tpc.19.00689
Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., et al. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43, 553–567. doi: 10.1111/j.1365-313X.2005.02480.x
Gui, J., Shen, J., and Li, L. (2011). Functional characterization of evolutionarily divergent 4-coumarate: coenzyme A ligases in rice. Plant Physiol. 157, 574–586. doi: 10.1104/pp.111.178301
Guo, D., Chen, F., Inoue, K., Blount, J. W., and Dixon, R. A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88. doi: 10.1105/tpc.13.1.73
Guo, D.-M., Ran, J.-H., and Wang, X.-Q. (2010). Evolution of the cinnamyl/sinapyl alcohol dehydrogenase (CAD/SAD) gene family: the emergence of real lignin is associated with the origin of bona fide CAD. J. Mol. Evol. 71, 202–218. doi: 10.1007/s00239-010-9378-3
Guo, Z., Hua, H., Xu, J., Mo, J., Zhao, H., and Yang, J. (2019). Cloning and functional analysis of lignin biosynthesis genes Cf4CL and CfCCoAOMT in Cryptomeria fortunei. Genes 10:619. doi: 10.3390/genes10080619
Ha, C. M., Escamilla-Trevino, L., Yarce, J. C. S., Kim, H., Ralph, J., Chen, F., et al. (2016). An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 86, 363–375. doi: 10.1111/tpj.13177
Hiraide, H., Tobimatsu, Y., Yoshinaga, A., Lam, P. Y., Kobayashi, M., Matsushita, Y., et al. (2021). Localised laccase activity modulates distribution of lignin polymers in gymnosperm compression wood. New Phytol. 230, 2186–2199. doi: 10.1111/nph.17264.5
Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., et al. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16, 1446–1465. doi: 10.1105/tpc.020297
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Kim, H., Li, Q., Karlen, S. D., Smith, R. A., Shi, R., Liu, J., et al. (2020). Monolignol benzoates incorporate into the lignin of transgenic Populus trichocarpa depleted in C3H and C4H. ACS Sustain. Chem. Eng. 8, 3644–3654. doi: 10.1021/acssuschemeng.9b06389
Kim, W.-C., Kim, J.-Y., Ko, J.-H., Kang, H., and Han, K.-H. (2014). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 85, 589–599. doi: 10.1007/s11103-014-0205-x
Kim, W.-C., Ko, J.-H., Kim, J.-Y., Kim, J., Bae, H.-J., and Han, K.-H. (2013). MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73, 26–36. doi: 10.1111/j.1365-313x.2012.05124.x
Ko, J.-H., Kim, W.-C., and Han, K.-H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665. doi: 10.1111/j.1365-313X.2009.03989.x
Ko, J. H., Kim, W. C., Kim, J. Y., Ahn, S. J., and Han, K. H. (2012). MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 5, 961–963. doi: 10.1093/mp/sss076
Kriegshauser, L., Knosp, S., Grienenberger, E., Tatsumi, K., Gütle, D. D., Sørensen, I., et al. (2021). Function of the HYDROXYCINNAMOYL-CoA:SHIKIMATE HYDROXYCINNAMOYL TRANSFERASE is evolutionarily conserved in embryophytes. Plant Cell 33, 1472–1491. doi: 10.1093/plcell/koab044
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Lam, P. Y., Tobimatsu, Y., Matsumoto, N., Suzuki, S., Lan, W., Takeda, Y., et al. (2019). OsCAldOMT1 is a bifunctional O-methyltransferase involved in the biosynthesis of tricin-lignins in rice cell walls. Sci. Rep. 9:11597. doi: 10.1038/s41598-019-47957-0
Lauvergeat, V., Lacomme, C., Lacombe, E., Lasserre, E., Roby, D., and Grima-Pettenati, J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57, 1187–1195. doi: 10.1016/S0031-9422(01)00053-X
Li, L., Cheng, X. F., Leshkevich, J., Umezawa, T., Harding, S. A., and Chiang, V. L. (2001). The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13, 1567–1586. doi: 10.1105/tpc.010111
Li, S. S., Chang, Y., Li, B., Shao, S. L., and Zhen-Zhu, Z. (2020). Functional analysis of 4-coumarate: coA ligase from Dryopteris fragrans in transgenic tobacco enhances lignin and flavonoids. Genet. Mol. Biol. 43:e20180355. doi: 10.1590/1678-4685-gmb-2018-0355
Li, X., Bonawitz, N. D., Weng, J.-K., and Chapple, C. (2010). The Growth Reduction Associated with Repressed Lignin Biosynthesis in Arabidopsis thaliana Is Independent of Flavonoids. Plant Cell 22, 1620–1632. doi: 10.1105/tpc.110.074161
Li, X., Chen, W., Zhao, Y., Xiang, Y., Jiang, H., Zhu, S., et al. (2013). Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet. Mol. Biol. 36, 540–546. doi: 10.1590/s1415-47572013005000039
Li, Y., Kim, J. I., Pysh, L., and Chapple, C. (2015). Four isoforms of Arabidopsis 4-coumarate: coA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Liu, Y., You, S., Taylor-Teeples, M., Li, W. L., Schuetz, M., Brady, S. M., et al. (2014). BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 26, 4843–4861. doi: 10.1105/tpc.114.128322
Logan, K. J., and Thomas, B. A. (1985). Distribution of Lignin Derivatives in Plants. New Phytol. 99, 571–585. doi: 10.1111/j.1469-8137.1985.tb03685.x
Ma, D., and Constabel, C. P. (2019). MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 24, 275–289. doi: 10.1016/j.tplants.2018.12.003
Mansfield, S. D., Kim, H., Lu, F., and Ralph, J. (2012). Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7, 1579–1589. doi: 10.1038/nprot.2012.064
Martin, A. F., Tobimatsu, Y., Kusumi, R., Matsumoto, N., Miyamoto, T., Lam, P. Y., et al. (2019). Altered lignocellulose chemical structure and molecular assembly in CINNAMYL ALCOHOL DEHYDROGENASE-deficient rice. Sci. Rep. 9:17153. doi: 10.1038/s41598-019-53156-8
Martone, P. T., Estevez, J. M., Lu, F., Ruel, K., Denny, M. W., Somerville, C., et al. (2009). Discovery of Lignin in Seaweed Reveals Convergent Evolution of Cell-Wall Architecture. Curr. Biol. 19, 169–175. doi: 10.1016/j.cub.2008.12.031
McCarthy, R. L., Zhong, R., and Ye, Z.-H. (2009). MYB83 Is a Direct Target of SND1 and Acts Redundantly with MYB46 in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
Meyer, K., Shirley, A. M., Cusumano, J. C., Bell-Lelong, D. A., and Chapple, C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95, 6619–6623. doi: 10.1073/pnas.95.12.6619
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Nair, R. B., Bastress, K. L., Ruegger, M. O., Denault, J. W., and Chapple, C. (2004). The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16, 544–554. doi: 10.1105/tpc.017509
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Nawawi, D. S., Syafii, W., Akiyama, T., and Matsumoto, Y. (2016). Characteristics of guaiacyl-syringyl lignin in reaction wood in the gymnosperm Gnetum gnemon L. Holzforschung 70, 593–602. doi: 10.1515/hf-2015-0107
Niklas, K. J., Cobb, E. D., and Matas, A. J. (2017). The evolution of hydrophobic cell wall biopolymers: from algae to angiosperms. J. Exp. Bot. 68, 5261–5269. doi: 10.1093/jxb/erx215
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Ohtani, M., and Demura, T. (2019). The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 56, 82–87. doi: 10.1016/j.copbio.2018.10.002
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L. J., Smith, C., et al. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36, 743–754. doi: 10.1046/j.1365-313X.2003.01916.x
Petrik, D. L., Karlen, S. D., Cass, C. L., Padmakshan, D., Lu, F., Liu, S., et al. (2014). p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J. 77, 713–726. doi: 10.1111/tpj.12420
Qin, W., Yin, Q., Chen, J., Zhao, X., Yue, F., He, J., et al. (2020). The class II KNOX transcription factors KNAT3 and KNAT7 synergistically regulate monolignol biosynthesis in Arabidopsis. J. Exp. Bot. 71, 5469–5483. doi: 10.1093/jxb/eraa266
Raes, J., Rohde, A., Christensen, J. H., Van de Peer, Y., and Boerjan, W. (2003). Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. doi: 10.1104/pp.103.026484
Ralph, J., Lapierre, C., and Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi: 10.1016/j.copbio.2019.02.019
Rambaut, A. (2012). Figtree v1.4. http://tree.bio.ed.ac.uk/software/figtree/ (accessed January 13, 2021).
Rao, G., Pan, X., Xu, F., Zhang, Y., Cao, S., Jiang, X., et al. (2015). Divergent and overlapping function of five 4-coumarate/coenzyme A ligases from Populus tomentosa. Plant Mol. Biol. Rep. 33, 841–854. doi: 10.1007/s11105-014-0803-4
Rao, X., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Renault, H., De Marothy, M., Jonasson, G., Lara, P., Nelson, D. R., Nilsson, I., et al. (2017). Gene duplication leads to altered membrane topology of a cytochrome P450 enzyme in seed plants. Mol. Biol. Evol. 34, 2041–2056. doi: 10.1093/molbev/msx160
Renault, H., Werck-Reichhart, D., and Weng, J.-K. (2019). Harnessing lignin evolution for biotechnological applications. Curr. Opin. Biotechnol. 56, 105–111. doi: 10.1016/j.copbio.2018.10.011
Rensing, S. A. (2018). Great moments in evolution: the conquest of land by plants. Curr. Opin. Plant Biol. 42, 49–54. doi: 10.1016/j.pbi.2018.02.006
Ruel, K., Berrio-Sierra, J., Derikvand, M. M., Pollet, B., Thévenin, J., Lapierre, C., et al. (2009). Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol. 184, 99–113. doi: 10.1111/j.1469-8137.2009.02951.x
Saleme, M. D. L. S., Cesarino, I., Vargas, L., Kim, H., Vanholme, R., Goeminne, G., et al. (2017). Silencing CAFFEOYL SHIKIMATE ESTERASE Affects Lignification and Improves Saccharification in Poplar. Plant Physiol. 175, 1040–1057. doi: 10.1104/pp.17.00920
Sammons, R. D., and Gaines, T. A. (2014). Glyphosate resistance: state of knowledge. Pest Manag. Sci. 70, 1367–1377. doi: 10.1002/ps.3743
Schoch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P., et al. (2001). CYP98A3 from Arabidopsis thaliana Is a 3’-Hydroxylase of Phenolic Esters, a Missing Link in the Phenylpropanoid Pathway. J. Biol. Chem. 276, 36566–36574. doi: 10.1074/jbc.M104047200
Schuetz, M., Smith, R., and Ellis, B. (2013). Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 64, 11–31. doi: 10.1093/jxb/ers287
Serrani-Yarce, J. C., Escamilla-Trevino, L., Barros, J., Gallego-Giraldo, L., Pu, Y., Ragauskas, A., et al. (2021). Targeting hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase for lignin modification in Brachypodium distachyon. Biotechnol. Biofuels 14:50. doi: 10.1186/s13068-021-01905-1
Shi, R., Sun, Y.-H., Li, Q., Heber, S., Sederoff, R., and Chiang, V. L. (2009). Towards a Systems Approach for Lignin Biosynthesis in Populus trichocarpa: transcript Abundance and Specificity of the Monolignol Biosynthetic Genes. Plant Cell Physiol. 51, 144–163. doi: 10.1093/pcp/pcp175
Silber, M. V., Meimberg, H., and Ebel, J. (2008). Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry 69, 2449–2456. doi: 10.1016/j.phytochem.2008.06.014
Sonnante, G., D’Amore, R., Blanco, E., Pierri, C. L., De Palma, M., Luo, J., et al. (2010). Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol. 153, 1224–1238. doi: 10.1104/pp.109.150144
Soyano, T., Thitamadee, S., Machida, Y., and Chua, N. H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373. doi: 10.1105/tpc.108.061796
Szövényi, P., Gunadi, A., and Li, F.-W. (2021). Charting the genomic landscape of seed-free plants. Nat. Plants 7, 554–565. doi: 10.1038/s41477-021-00888-z
Takeda, Y., Koshiba, T., Tobimatsu, Y., Suzuki, S., Murakami, S., Yamamura, M., et al. (2017). Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 246, 337–349. doi: 10.1007/s00425-017-2692-x
Takeda, Y., Suzuki, S., Tobimatsu, Y., Osakabe, K., Osakabe, Y., Ragamustari, S. K., et al. (2019). Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J. 97, 543–554. doi: 10.1111/tpj.14141
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tohge, T., Watanabe, M., Hoefgen, R., and Fernie, A. R. (2013). The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 48, 123–152. doi: 10.3109/10409238.2012.758083
Tomlinson, P. B. (2001). Reaction tissues in Gnetum gnemon—a preliminary report. IAWA J. 22, 401–413. doi: 10.1163/22941932-90000385
Tsai, C.-J., Xu, P., Xue, L.-J., Hu, H., Nyamdari, B., Naran, R., et al. (2020). Compensatory Guaiacyl Lignin Biosynthesis at the Expense of Syringyl Lignin in 4CL1-Knockout Poplar. Plant Physiol. 183, 123–136. doi: 10.1104/pp.19.01550
Valdivia, E. R., Herrera, M. T., Gianzo, C., Fidalgo, J., Revilla, G., Zarra, I., et al. (2013). Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64, 1333–1343. doi: 10.1093/jxb/ers394
Van Acker, R., Déjardin, A., Desmet, S., Hoengenaert, L., Vanholme, R., Morreel, K., et al. (2017). Different routes for conifer-and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 175, 1018–1039. doi: 10.1104/pp.17.00834
Van Acker, R., Leplé, J.-C., Aerts, D., Storme, V., Goeminne, G., Ivens, B., et al. (2014). Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. U. S. A. 111, 845–850. doi: 10.1073/pnas.1321673111
Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y., Sundin, L., Goeminne, G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602
Wagner, A., Ralph, J., Akiyama, T., Flint, H., Phillips, L., Torr, K., et al. (2007). Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc. Natl. Acad. Sci. U. S. A. 104, 11856–11861. doi: 10.1073/pnas.0701428104
Wagner, A., Tobimatsu, Y., Phillips, L., Flint, H., Torr, K., Donaldson, L., et al. (2011). CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J. 67, 119–129. doi: 10.1111/j.1365-313X.2011.04580.x
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U. S. A. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Wang, H., Zhao, Q., Chen, F., Wang, M., and Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x
Wang, S., Yamaguchi, M., Grienenberger, E., Martone, P. T., Samuels, A. L., and Mansfield, S. D. (2020). The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 101, 293–309. doi: 10.1111/tpj.14541
Wang, X., Chao, N., Zhang, M., Jiang, X., and Gai, Y. (2019). Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms. Int. J. Mol. Sci. 20:6071. doi: 10.3390/ijms20236071
Wang, X., Zhuo, C., Xiao, X., Wang, X., Docampo-Palacios, M. L., Chen, F., et al. (2020). Substrate-Specificity of LACCASE 8 Facilitates Polymerization of Caffeyl Alcohol for C-Lignin Biosynthesis in the Seed Coat of Cleome hassleriana. Plant Cell 32, 3825–3845. doi: 10.1105/tpc.20.00598
Weng, J.-K., Akiyama, T., Ralph, J., and Chapple, C. (2011). Independent recruitment of an O-methyltransferase for syringyl lignin biosynthesis in Selaginella moellendorffii. Plant Cell 23, 2708–2724. doi: 10.1105/tpc.110.081547
Weng, J.-K., and Chapple, C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285. doi: 10.1111/j.1469-8137.2010.03327.x
Weng, J. K., Li, X., Bonawitz, N. D., and Chapple, C. (2008a). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172. doi: 10.1016/j.copbio.2008.02.014
Weng, J. K., Li, X., Stout, J., and Chapple, C. (2008b). Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. U. S. A. 105, 7887–7892. doi: 10.1073/pnas.0801696105
Withers, S., Lu, F., Kim, H., Zhu, Y., Ralph, J., and Wilkerson, C. G. (2012). Identification of Grass-specific Enzyme That Acylates Monolignols with p-Coumarate. J. Biol. Chem. 287, 8347–8355. doi: 10.1074/jbc.M111.284497
Wu, Z., Wang, N., Hisano, H., Cao, Y., Wu, F., Liu, W., et al. (2019). Simultaneous regulation of F5H in COMT-RNAi transgenic switchgrass alters effects of COMT suppression on syringyl lignin biosynthesis. Plant Biotechnol. J. 17, 836–845. doi: 10.1111/pbi.13019
Xiao, Y., Li, J., Liu, H., Zhang, Y., Zhang, X., Qin, Z., et al. (2020). The Effect of Co-Transforming Eucalyptus urophylla Catechol-O-methyltransferase and Caffeoyl-CoA O-methyltransferase on the Biosynthesis of Lignin Monomers in Transgenic Tobacco. Russ. J. Plant Physiol. 67, 879–887. doi: 10.1134/S1021443720050180
Xie, M., Muchero, W., Bryan, A. C., Yee, K., Guo, H. B., Zhang, J., et al. (2018). A 5-Enolpyruvylshikimate 3-Phosphate Synthase Functions as a Transcriptional Repressor in Populus. Plant Cell 30, 1645–1660. doi: 10.1105/tpc.18.00168
Xiong, W., Wu, Z., Liu, Y., Li, Y., Su, K., Bai, Z., et al. (2019). Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 12:82. doi: 10.1186/s13068-019-1421-z
Xu, B., Ohtani, M., Yamaguchi, M., Toyooka, K., Wakazaki, M., Sato, M., et al. (2014). Contribution of NAC Transcription Factors to Plant Adaptation to Land. Science 343, 1505–1508. doi: 10.1126/science.1248417
Xu, Z., Zhang, D., Hu, J., Zhou, X., Ye, X., Reichel, K. L., et al. (2009). Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10:S3. doi: 10.1186/1471-2105-10-S11-S3
Yamaguchi, M., Kubo, M., Fukuda, H., and Demura, T. (2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. doi: 10.1111/j.1365-313X.2008.03533.x
Yamaguchi, M., Mitsuda, N., Ohtani, M., Ohme-Takagi, M., Kato, K., and Demura, T. (2011). VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66, 579–590. doi: 10.1111/j.1365-313X.2011.04514.x
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Yan, X., Liu, J., Kim, H., Liu, B., Huang, X., Yang, Z., et al. (2019). CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol. 222, 244–260. doi: 10.1111/nph.15505
Yang, C., Xu, Z., Song, J., Conner, K., Vizcay Barrena, G., and Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 Regulates Secondary Thickening in the Endothecium and Is Essential for Anther Dehiscence. Plant Cell 19, 534–548. doi: 10.1105/tpc.106.046391
Yang, X., Beres, Z. T., Jin, L., Parrish, J. T., Zhao, W., Mackey, D., et al. (2017). Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana. PLoS One 12:e0175820. doi: 10.1371/journal.pone.0175820
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., and Chen, J.-G. (2020). Transcriptional and Post-transcriptional Regulation of Lignin Biosynthesis Pathway Genes in Populus. Front. Plant Sci. 11:652. doi: 10.3389/fpls.2020.00652
Zhang, Q., Luo, F., Zhong, Y., He, J., and Li, L. (2019). Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis. J. Exp. Bot. 71, 1449–1458. doi: 10.1093/jxb/erz513
Zhang, X.-S., Ni, R., Wang, P.-Y., Zhu, T.-T., Sun, C.-J., Lou, H.-X., et al. (2019). Isolation and functional characterization of two Caffeoyl Coenzyme A 3-O-methyltransferases from the fern species Polypodiodes amoena. Plant Physiol. Biochem. 136, 169–177. doi: 10.1016/j.plaphy.2019.01.021
Zhao, K., and Bartley, L. E. (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol. 14:135. doi: 10.1186/1471-2229-14-135
Zhao, Q., Gallego-Giraldo, L., Wang, H., Zeng, Y., Ding, S.-Y., Chen, F., et al. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63, 100–114. doi: 10.1111/j.1365-313X.2010.04223.x
Zhong, R., Kandasamy, M. K., and Ye, Z.-H. (2020). XND1 regulates secondary wall deposition in xylem vessels through the inhibition of VND functions. Plant Cell Physiol. 62, 53–65. doi: 10.1093/pcp/pcaa140
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z.-H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, R., Lee, C., and Ye, Z.-H. (2010). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15, 625–632. doi: 10.1016/j.tplants.2010.08.007
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z.-H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., McCarthy, R. L., Haghighat, M., and Ye, Z.-H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8:e69219. doi: 10.1371/journal.pone.0069219
Zhong, R., Morrison, W. H. III, Himmelsbach, D. S., Poole, F. L. II, and Ye, Z. H. (2000). Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124, 563–578. doi: 10.1104/pp.124.2.563
Zhong, R., Richardson, E. A., and Ye, Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. doi: 10.1105/tpc.107.053678
Zhong, R., and Ye, Z.-H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. doi: 10.1093/pcp/pcr185
Zhou, J., Zhong, R., and Ye, Z.-H. (2014). Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9:e105726. doi: 10.1371/journal.pone.0105726
Keywords: lignin biosynthesis, tracheophytes, transcription factor, EPSP synthase, phylogenetic occurrence, lignin utilization
Citation: Yao T, Feng K, Xie M, Barros J, Tschaplinski TJ, Tuskan GA, Muchero W and Chen J-G (2021) Phylogenetic Occurrence of the Phenylpropanoid Pathway and Lignin Biosynthesis in Plants. Front. Plant Sci. 12:704697. doi: 10.3389/fpls.2021.704697
Received: 03 May 2021; Accepted: 19 July 2021;
Published: 17 August 2021.
Edited by:
Toshiaki Umezawa, Kyoto University, JapanCopyright © 2021 Yao, Feng, Xie, Barros, Tschaplinski, Tuskan, Muchero and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wellington Muchero, bXVjaGVyb3dAb3JubC5nb3Y=; Jin-Gui Chen, Y2hlbmpAb3JubC5nb3Y=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.