
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 July 2021
Sec. Plant Pathogen Interactions
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.701683
This article is part of the Research Topic The Role of Epigenetics in Biotic Stress in Plants View all 7 articles
N6-methyladenosine (m6A) modification is a dynamically regulated RNA modification that impacts many cellular processes and pathways. This epitranscriptomic methylation relies on the participation of RNA methyltransferases (referred to as “writers”) and demethylases (referred to as “erasers”), respectively. We previously demonstrated that the Arabidopsis thaliana protein atALKBH9B showed m6A-demethylase activity and interacted with the coat protein (CP) of alfalfa mosaic virus (AMV), causing a profound impact on the viral infection cycle. To dissect the functional activity of atALKBH9B in AMV infection, we performed a protein-mapping analysis to identify the putative domains required for regulating this process. In this context, the mutational analysis of the protein revealed that the residues between 427 and 467 positions are critical for in vitro binding to the AMV RNA. The atALKBH9B amino acid sequence showed intrinsically disordered regions (IDRs) located at the N-terminal part delimiting the internal AlkB-like domain and at the C-terminal part. We identified an RNA binding domain containing an RGxxxRGG motif that overlaps with the C-terminal IDR. Moreover, bimolecular fluorescent experiments allowed us to determine that residues located between 387 and 427 are critical for the interaction with the AMV CP, which should be critical for modulating the viral infection process. Finally, we observed that atALKBH9B deletions of either N-terminal 20 residues or the C-terminal’s last 40 amino acids impede their accumulation in siRNA bodies. The involvement of the regions responsible for RNA and viral CP binding and those required for its localization in stress granules in the viral cycle is discussed.
The addition of a methyl group to the N6 position of adenosine (m6A) is the most abundant internal modification in eukaryote mRNAs (Boccaletto et al., 2018; Covelo-Molares et al., 2018; Arribas-Hernández and Brodersen, 2020; Zhou et al., 2020). It regulates many steps of RNA metabolism, including splicing (Zhao et al., 2014), stability (Wang et al., 2014), translation (Meyer et al., 2015), nuclear-export (Zheng et al., 2013), RNA structures (Bayoumi et al., 2020), and protein/RNA interactions (Liu et al., 2015). Also, it modulates the epigenetic effects of some non-coding RNAs (ncRNA) (Meyer and Jaffrey, 2017). Since the 1970s, m6A modification has been known to tag not only cellular RNAs but also RNAs of multiple viruses (Wei and Moss, 1975; Furuichi et al., 1976; Krug et al., 1976; Dimock and Stoltzfus, 1977), although its functional relevance has remained elusive mainly due to the lack of efficient methods of m6A detection and subsequent analysis. Recent studies have demonstrated the crucial roles of m6A in the virus–host interactions; however, most of these studies have focused on animal viruses (Dang et al., 2019; Williams et al., 2019; Arribas-Hernández and Brodersen, 2020), whereas it remains very limited in plant viruses (Martínez-Pérez et al., 2017; Li et al., 2018; Arribas-Hernández and Brodersen, 2020).
In mammals, m6A methylation is catalyzed co-transcriptionally by a multicomponent m6A methyltransferase complex (MTC, also known as “writer”) (Liu et al., 2014; Ping et al., 2014). The core component of MTC is a ∼200 kDa heterodimer comprised of METTL3 and METTL14 (Huang et al., 2020). Other regulatory subunits of MTC have also been identified, including WTAP and its cofactors KIAA1429 (VIRMA), ZC3H13, and RBM15/RBM15B, which play roles in anchoring MTC to nuclear speckles and U-rich regions adjacent to m6A sites in mRNAs. Two main RNA demethylases, “erasers,” belonging to Fe(II)/2-oxoglutarate (2OG) dioxygenase superfamily, named AlkB homology 5 (ALKBH5) and FTO, have been described to remove m6A marks (Aik et al., 2014; Xu et al., 2014). The third component of the m6A modification machinery consists of “reader” proteins that recognize this modification and modulate the activity and half-life of diverse RNAs. Thus, several YTH domain family members, YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, mediate many of the phenotypic effects of this epitranscriptomic modification (Luo and Tong, 2014; Xiao et al., 2016).
By homology with mammals, functional orthologous genes of the m6A machinery have been discovered in Arabidopsis (Arribas-Hernández and Brodersen, 2020). MTA, MTB, FIP37, VIRILIZER, orthologs of METTL3, METTL14, WTAP, and KIAA1429 (human protein), respectively, have been identified as member proteins of the m6A writer complex (Zhong et al., 2008; Bodi et al., 2012; Shen et al., 2016; Růžička et al., 2017). In the Arabidopsis genome, 14 “readers” of the YTH family have been identified (ECT1-11; Evolutionarily Conserved C-Terminal Region 1–11, At4g11970 and the Cleavage and Polyadenylation Specificity Factor 30). The Arabidopsis genome encodes 14 homologs of Alk B family of “eraser” proteins (atALKBH1A-D, atALKBH2, atALKBH6, atALKBH8A-B, atALKBH9A-C, and atALKBH10A-B) (Mielecki et al., 2012; Kawai et al., 2014), of which, atALKBH9B and atALKBH10B, have been shown to present m6A demethylase activity in vitro and m6A-related functions in vivo (Duan et al., 2017; Martínez-Pérez et al., 2017). Recently the potential eraser atALKBH6 has been shown to play important roles in seed germination, seedling growth, and survival of Arabidopsis under abiotic stresses (Huong et al., 2020). Interestingly, atALKBH9B is the only m6A demethylase that is located exclusively in the cytoplasm (Mielecki et al., 2012), forming granules that colocalize with SGS3 (a component of siRNA bodies) and in some cases, associates with DCP1 (P bodies) (Ingelfinger et al., 2002; Martínez de Alba et al., 2015; Martínez-Pérez et al., 2017).
Arabidopsis genome has been described to encode more than 200 putative RNA-binding proteins (RBP) (Lorković and Barta, 2002; Abbasi et al., 2011; Ambrosone et al., 2012). RBPs are key factors in post-transcriptional gene regulation, protein synthesis, viral replication, cellular defense, and developmental regulation (Terribilini et al., 2006; Glisovic et al., 2008; Pallas and Gómez, 2013; Prall et al., 2019). RBPs are often modular and contain one or more conserved RNA-binding domains (RBD) (Lee et al., 2012; Re et al., 2014). RNA recognition motifs (RRM) and the K homology (KH) domain are the most abundant structural motifs in eukaryotes (Lorković and Barta, 2002; Chen and Varani, 2013). Other RBDs include the glycine-rich motif (GRM), the double-stranded RNA binding domain (dsRBD), DEAD- Box-, PUF, SAM-, ZnF-domains (Lunde et al., 2007; Re et al., 2014; Lee and Kang, 2016), and the RGG/RG motif (Thandapani et al., 2013). However, the majority of residues predicted to be in the protein-RNA interface are not part of a characterized RBDs (Terribilini et al., 2006). In many cases, intrinsically disordered regions (IDRs) have been identified in proteins that do not display characterized RNA-binding sites (Calabretta and Richard, 2015; Varadi et al., 2015). Among other roles, IDRs participate in protein-protein and protein-RNA interactions and are enriched in “disorder-promoting amino acids” such as G, P, or R (Gsponer and Madan Babu, 2009). In this context, IDRs can encompass diverse functional motives such as RNA binding motifs or low-complexity (LC) domains (Castello et al., 2012). These proteins often go through binding induced-folding. Thus, as a consequence of their structural felxibility, the RNA-protein interactions can experiment conformational changes in the protein structure, the RNA or both (Frankel, 1999). Additionally, proteins containing IDRs promote liquid-liquid phase separation in the assembly and degradation of RNA granules such as stress granules or P-bodies (Spector, 2006; Lin et al., 2015). Similar to what was observed in mammals, some Arabidopsis m6A readers, such as ECT2, ECT3, and ECT4 present IDRs and can form cytoplasmic granules (Scutenaire et al., 2018; Arribas-Hernández et al., 2020). Moreover, ECT2 was found to undergo a gel-like phase transition in vitro (Arribas-Hernández et al., 2018).
Alfalfa mosaic virus (AMV) belongs to the Bromoviridae family and, like the rest of the members of this family, its genome consists of three single-stranded RNAs of plus polarity (Bujarski et al., 2019). RNA1 and RNA2 encode the replicase subunits (P1 and P2), whereas RNA3 encodes the movement protein (MP) and serves as a template for the synthesis of subgenomic RNA4 (sgRNA4), which encodes the coat protein (CP) (Bol, 2005; Pallas et al., 2013). We previously demonstrated that atALKBH9B is a key factor in AMV infection since the suppression of the atALKBH9B m6A demethylation activity reduces viral accumulation. Moreover, it was shown that the CP of this virus interacts with atALKBH9B, pointing to a direct subversion of an endogenous regulatory pathway by the virus (Martínez-Pérez et al., 2017). Due to the functional relevance of atALKBH9B, the first m6A demethylase described in plants, we carried out a functional mapping of the protein to identify putative domains implicated in the diverse interactions of this m6A-demethylase with both the viral RNA and the coat protein.
In a previous study, we demonstrated that atALKBH9B interacts with the viral RNA, although the kinetic parameters of this interaction, as well as the identification of the RBD, were not analyzed (Martínez-Pérez et al., 2017). To estimate the capacity of the wild-type atALKBH9B to bind the viral RNA, we first conducted Electrophoretic Mobility Shift Assays (EMSA) by incubating a constant amount (5 ng) of an RNA transcript, corresponding to the 3′ untranslated region of AMV-RNA3 (3′UTR-RNA3), with increasing concentrations of the glutathione S-transferase fusion protein (GST:atALKBH9Bwt) (Figure 1). We chose this part of the RNA3/4 molecule because it is the same as the one that specifically binds CP, allowing us to directly compare it with this specific interaction. The decrease in the chemiluminescent signal intensity corresponding to free RNA was evident at quantities exceeding 400 ng of GST:atALBKH9Bwt (lane 7, Figure 1A), suggesting the formation of a protein–RNA complex. At this point, it is important to note that the non-radioactive EMSA described here (see section “Material and Methods”) requires the transfer to a nylon membrane step to detect both free RNA and the corresponding ribonucleoprotein (RNP) complex. When a large amount of molecules binds to the RNA, as in a non-sequence specific interaction, the transfer of the RNP complex to the nylon membrane and its posterior detection is difficult or even impossible (Marcos et al., 1999; Herranz and Pallás, 2004; Salavert et al., 2020). In any case, the disappearance of the free RNA band is evidence of complex formation (Carey, 1991) and was therefore quantified by film densitometry to calculate the apparent constant dissociation (Kd) of the RNA– GST:atALBKH9Bwt interaction from the linear regression of the mean values from at least three technical replicates (Marcos et al., 1999). The Kd value of GST:atALKBH9Bwt was estimated to be 0.30 μM (Figure 1B).
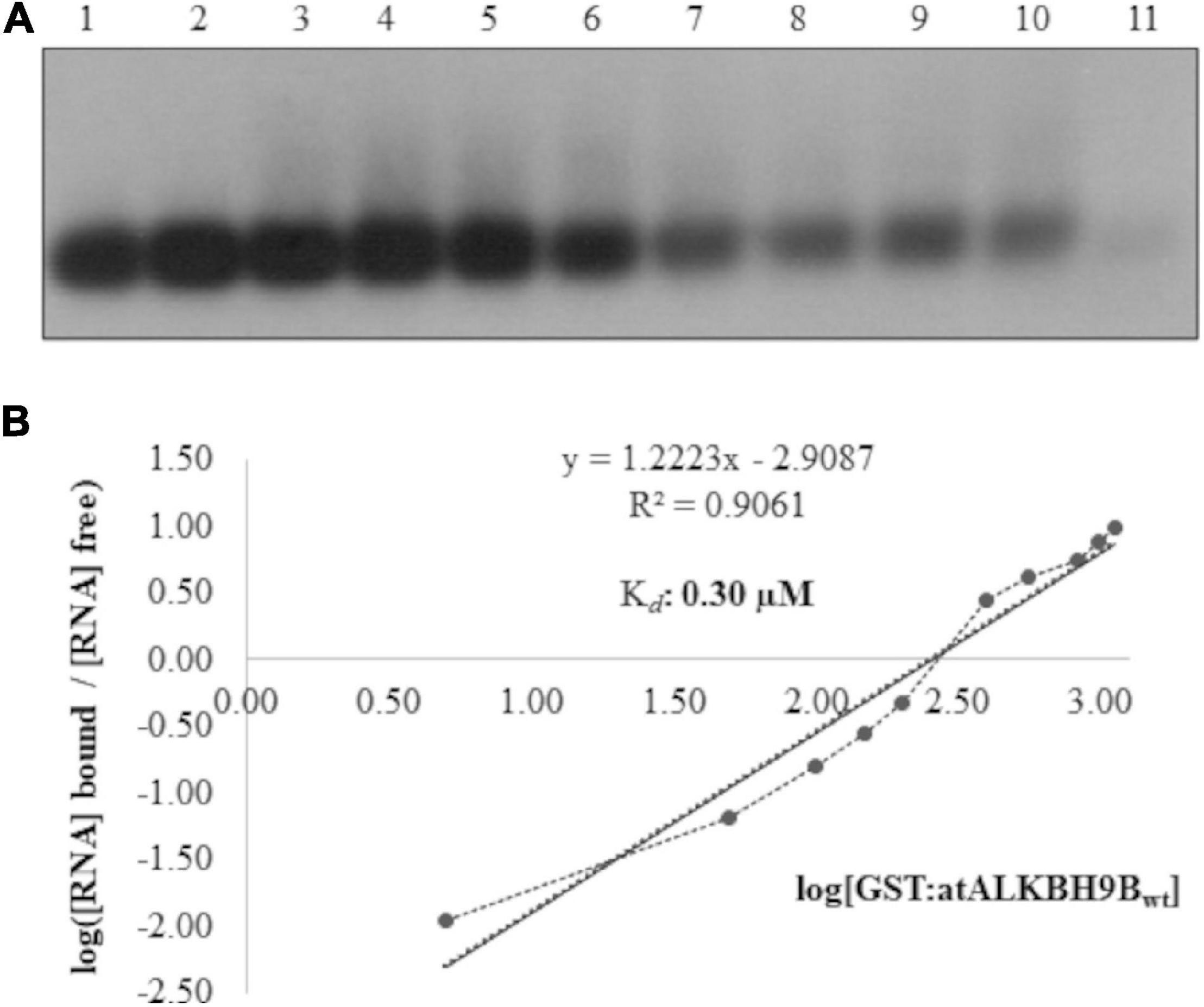
Figure 1. Analysis of RNA-protein complexes formed between purified GST:atALKBH9Bwt protein and the 3′UTR transcript of AMV-RNA3/4. (A) EMSA after incubation of 5 ng of 3′UTR transcript with no protein (lane 1) or with 5, 50, 100, 150, 200, 400, 560, 840, 980, and 1120 ng of GST:atALKBH9Bwt (lanes 2–11) corresponding to 0.01, 0.06, 0.12, 0.18, 0.24, 0.48, 0.67, 1.00, 1.17, and 1.22 μM, respectively. (B) Hill transformation. The thin line is the best fit determined by least-squares analysis, with the corresponding r coefficient and equation given in the insert. The slope of the best-fit equation determines a Hill coefficient of 1.35, indicating no or mild cooperativity, and the y-intercepted point gives the value of the Kd, which was 0.30 μM.
In order to determine the region of atALKBH9Bwt with RNA-binding activity, we first designed GST:atALKBH9B deletion mutants lacking 160 amino acids at either the N- or C-terminal part (Δ160Nt and Δ160Ct, respectively), or 187 amino acids in the internal region of the protein (Δ187 Int) (Supplementary Figure 1A). Northwestern blot assays using AMV-sgRNA4 labeled with digoxigenin showed a complete loss of RNA-binding capacity of GST:atALKBH9BΔ187Int and GST:atALKBH9BΔ160Ct proteins (Supplementary Figure 1B). Thus, we designed new GST:atALKBH9B deletion mutants affecting the internal and/or C-terminal regions: GST:Δ258Nt; GST:Δ258Nt/Δ80Ct; GST:Δ258Nt/Δ40Ct; GST:Δ258Nt/Δ20Ct and GST:Δ387Nt (Figure 2A). Northwestern blot assays showed that only mutant Δ258Nt/Δ80Ct does not bind RNA, indicating that residues between positions 427 and 467 are critical for RNA-protein interaction (Figure 2B). To confirm this observation, we evaluated the RNA-binding activity of a mutant lacking residues from 427 and 467 (GST:ΔRBD) and one comprising only the atALKBH9B 427–467 residues (GST:RBD) (Figure 2A). As shown in Figure 2B, only GST:RBD retained the RNA-binding capacity.
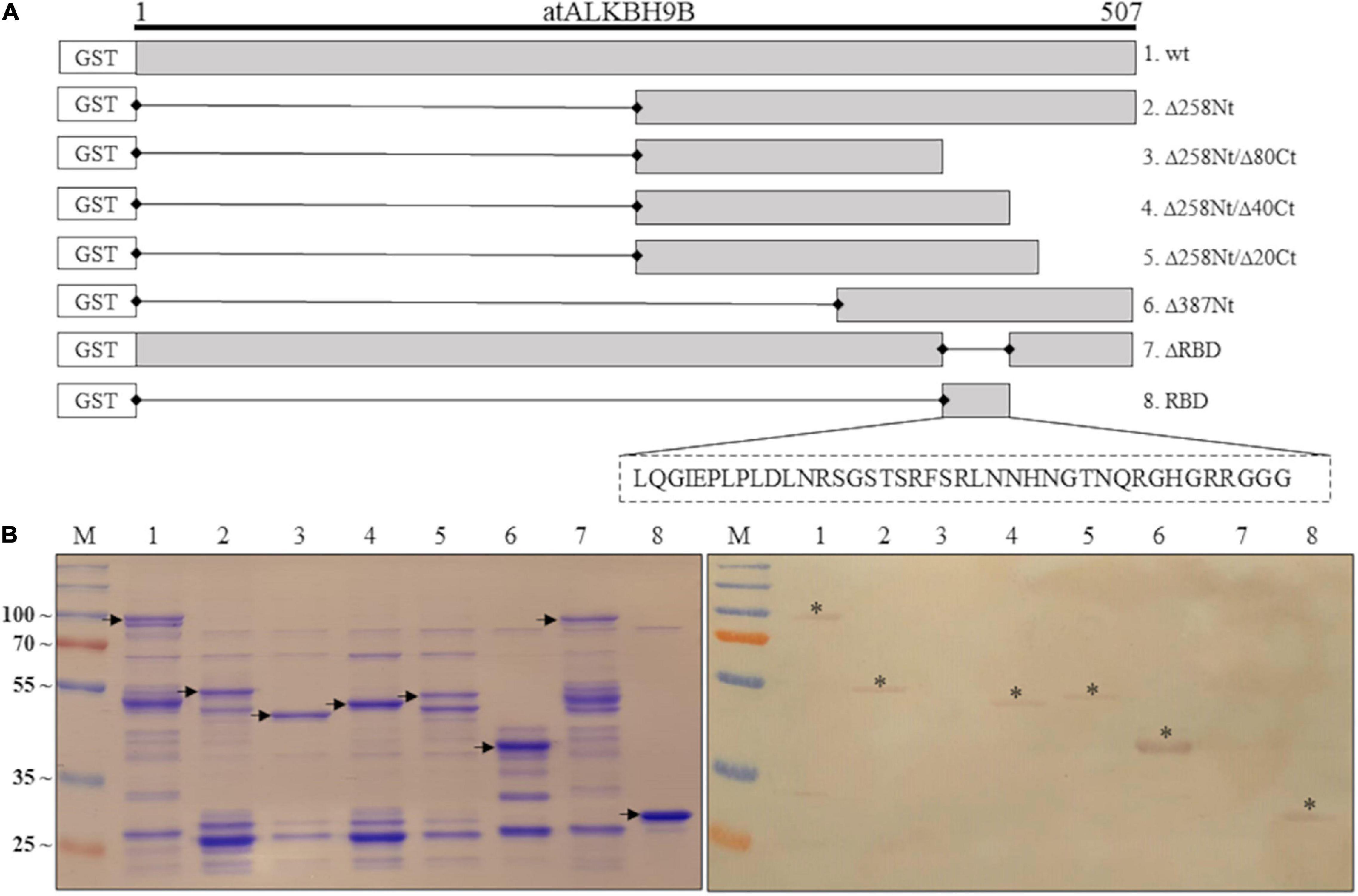
Figure 2. Analysis of the RNA-binding activity of purified GST:atALKBH9B proteins by Northwestern blot assay. (A) GST fusion proteins containing Internal or C-terminal deletions of atALKBH9B were produced in a prokaryotic system. (B) Duplicated membranes with GST:atALKBH9Bwt (lane 1) or its mutant variants (lanes 2–8), as depicted in (A), were resolved by 12% SDS–PAGE and, after transferring to nitrocellulose, incubated with 5 μg of AMV-sgRNA 4 labeled with digoxigenin. The left panel shows a Coomassie blue-stained gel, and the right panel shows results from Northwestern blot analysis. Positions of full-length GST:atALKBH9B proteins are indicated by arrows. Asterisks indicate GST:atALKBH9B fusion proteins which interact with the AMV-sgRNA4. The positions of molecular mass markers (in kDa) are shown on the left side.
To validate the northwestern blot assays, EMSAs were performed with GST:atALKBH9Bwt, GST:Δ160Nt, GST:Δ258Nt; GST:Δ258Nt/Δ80Ct; GST:Δ258Nt/Δ40Ct and GST:Δ258Nt/Δ20Ct by incubating the viral RNA with different protein concentrations. As expected, EMSA showed a decrease in chemiluminescent signal intensity corresponding to the free viral RNA in mutants containing the RBD (Supplementary Figures 2A,B), whereas those lacking the predicted RBD did not reveal RNA binding (GST:Δ258Nt/Δ80Ct). Additionally, we studied the affinity and specificity of the atALKBH9B RBD-RNA interaction by analyzing the shift in electrophoretic mobility of the 3’UTR-RNA3 (AMV) in the presence of GST:RBD fusion protein. As shown in Figure 3, increasing concentrations of GST:RBD diminished the amount of free RNA that becomes undetectable at 450 ng of the protein. A Hill transformation was used to analyze our data (Figure 3B). The Kd value of the RBD fused to GST was 0.64 μM, indicating slightly less RNA-binding than the full-length protein (GST:atALKBH9Bwt). From the linear regression adjustment, a Hill coefficient c of 2.4 was obtained; this value above 1 (c = 1 indicates no cooperativity) would be taken as an indication of positive cooperativity.
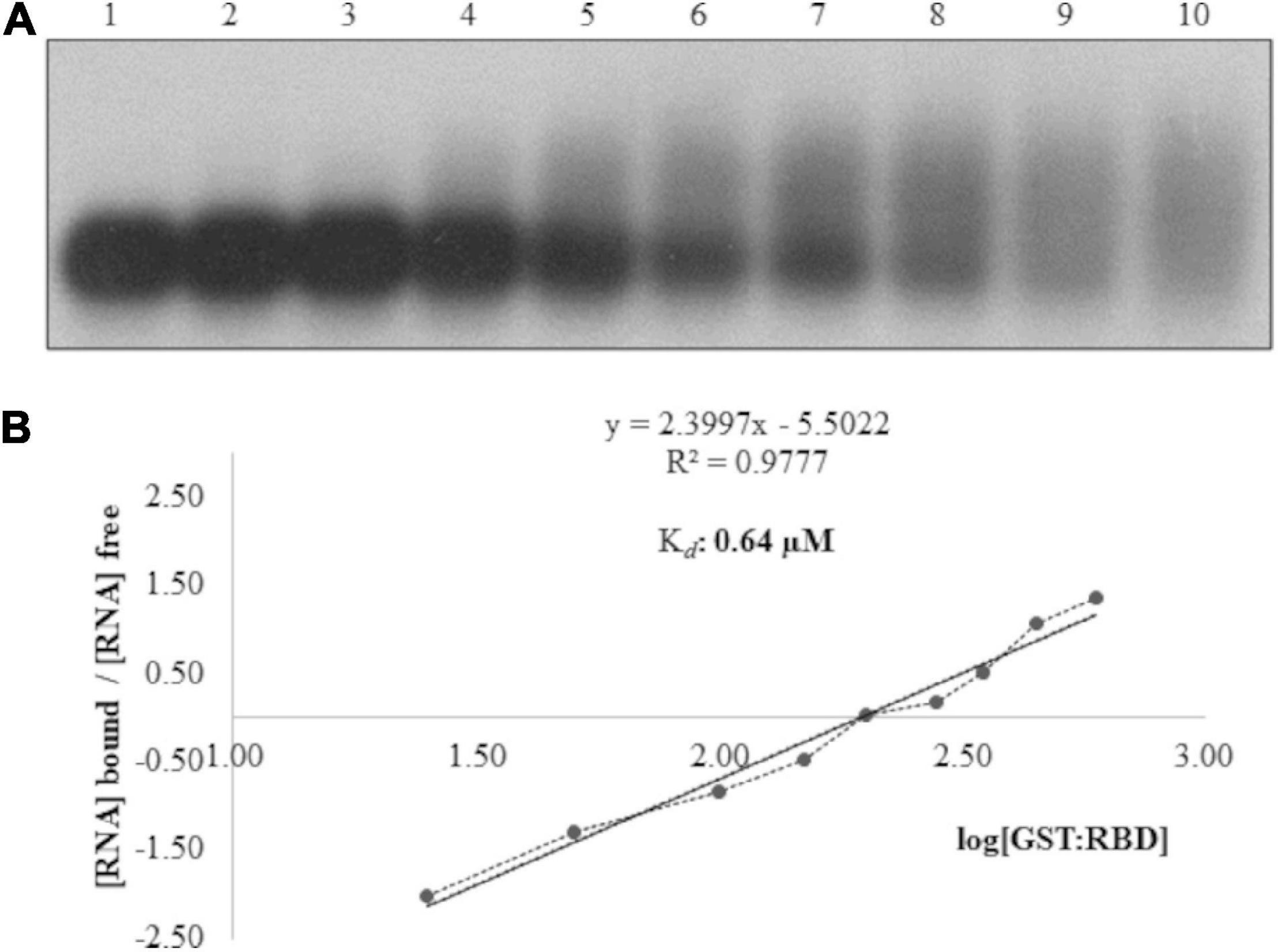
Figure 3. Analysis of RNA–protein complexes formed between purified GST:RBD of atALKBH9B protein and the 3’UTR transcript of AMV-RNA3/4. (A) EMSA after incubation of 5 ng of 3’UTR transcript with no protein (lane 1) or with 25, 50, 100, 150, 200, 280, 350, 450 and 600 ng of GST:RBD of 9B (lanes 2–10) corresponding to 0.08, 0.16, 0.33, 0.49, 0.65, 0.91, 1.14, 1.47, and 1.96 μM, respectively. (B) Hill transformation. The thin line is the best fit determined by least-squares analysis, with the corresponding r coefficient and equation given in the insert. The slope of the best-fit equation determines a Hill coefficient of 2.4, and the y intercepted point gives the value of the apparent constant dissociation (Kd), which was 0.64 μM.
Subsequently, we used PSI-BLAST (Altschul et al., 1997) to compare atALKBH9B RBD with other RNA binding proteins (RBP) from the databank, but the alignment of sequences did not reveal any significant similarities. Nonetheless, a high percentage of IDRs has been reported in viral, prokaryotic, and eukaryotic RBPs (Varadi et al., 2015). Therefore, we evaluated whether atALKBH9B presented these IDRs, using PrDOS1 (Ishida and Kinoshita, 2007). The results showed that 45.4% of all amino acids of atALKBH9B form IDRs through both N- and C-terminal regions, including most RBD (aa 427–467) (Figure 4). Remarkably, this RBD is enriched in G and R residues (20% and 15%, respectively), which are also common in the aforementioned IDRs. It is worth noting the presence of a RGxxxRGG motif and two extras RG residues that have been described in several RBPs showing characteristic disorder regions (Thandapani et al., 2013).
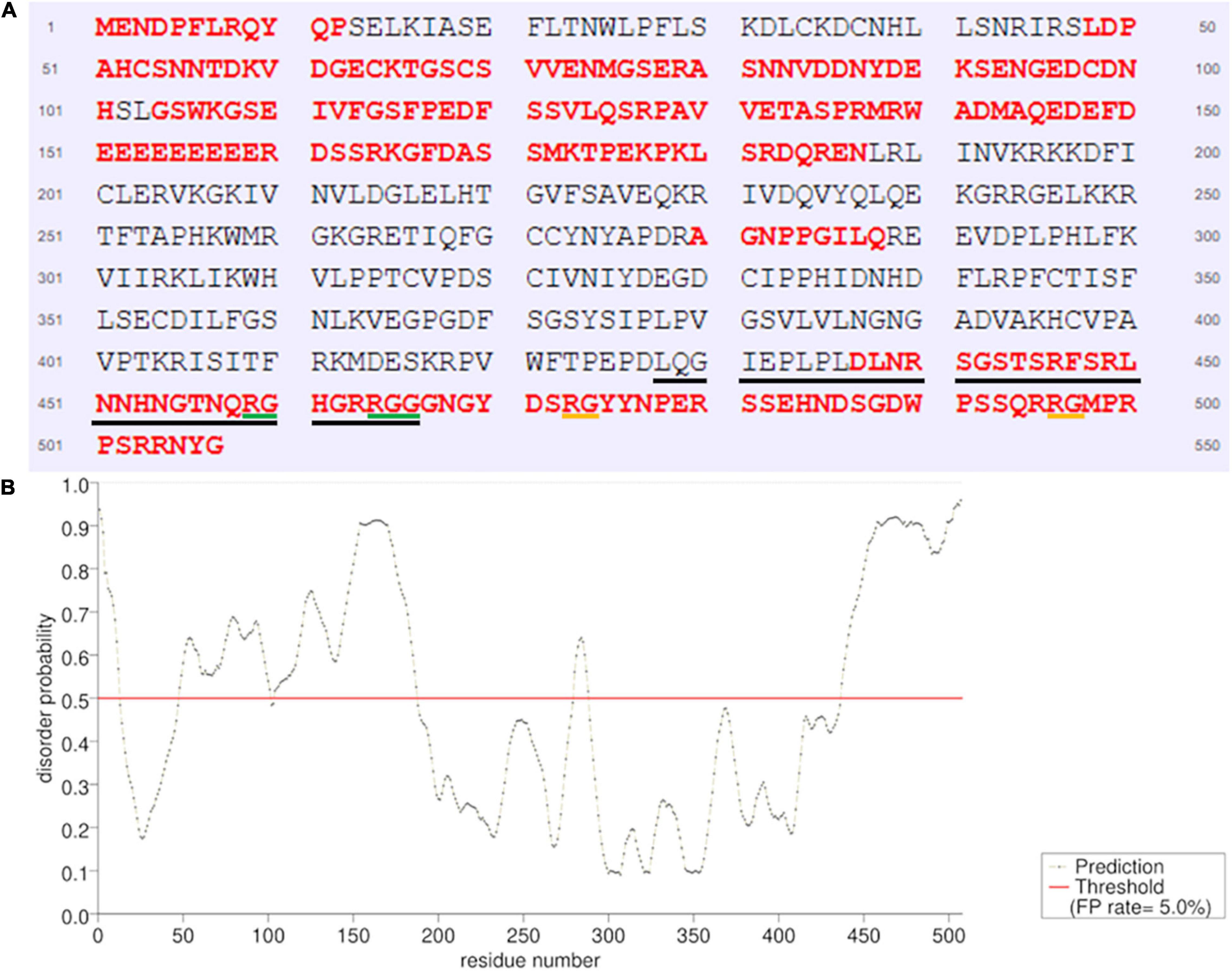
Figure 4. Prediction of natively disordered regions of atALKBH9B using PrDOS. (A) atALKBH9B amino acid sequence with disordered residues in red. The RBD located between positions 427–467 is underlined (black line). RGxxxRGG motif is underlined in green and the two extras RGs are in orange. (B) Disorder profile plot. Prediction false positive rate: 5%.
Taken together, these results indicate that atALKBH9B efficiently cooperatively binds viral RNA through an RBD that has the characteristics of the IDRs present in other viral and eukaryotic RBPs.
We previously showed that the demethylase activity of atALKBH9B affected the infectivity of AMV but not of cucumber mosaic virus (CMV), correlating with the ability of atALKBH9B to interact (or not) with their coat proteins (Martínez-Pérez et al., 2017). Since this interaction is, thus, critical for the proviral function of atALKBH9B, we decided to delineate the domain of atALKBH9B involved in the host-protein/viral-protein interaction using the Bimolecular Fluorescent Complementation (BiFC). For this purpose, we first designed three mutants corresponding to the N-terminal, central and C-terminal regions, fused to the N- and C-terminal parts of the YFP (Figure 5A, left panel [I]). These constructs were co-infiltrated with the corresponding N–CYFP:CP proteins in Nicotiana benthamiana leaves, and fluorescence was examined by confocal laser scanning microscopy (CLSM) after 48h. Leaves co-infiltrated with CYFP-9Bwt plus NYFP-CP, CYFP-9BΔ160Nt plus NYFP-CP, and CYFP:9BΔ187Int plus NYFP-CP rendered a strong YFP fluorescence signal in the cells, whereas no reconstituted YFP fluorescence was detected in leaves co-infiltrated with the pair CYFP-9BΔ160Ct plus NYFP:CP (Figure 5B and Supplementary Figure 3, upper panels).
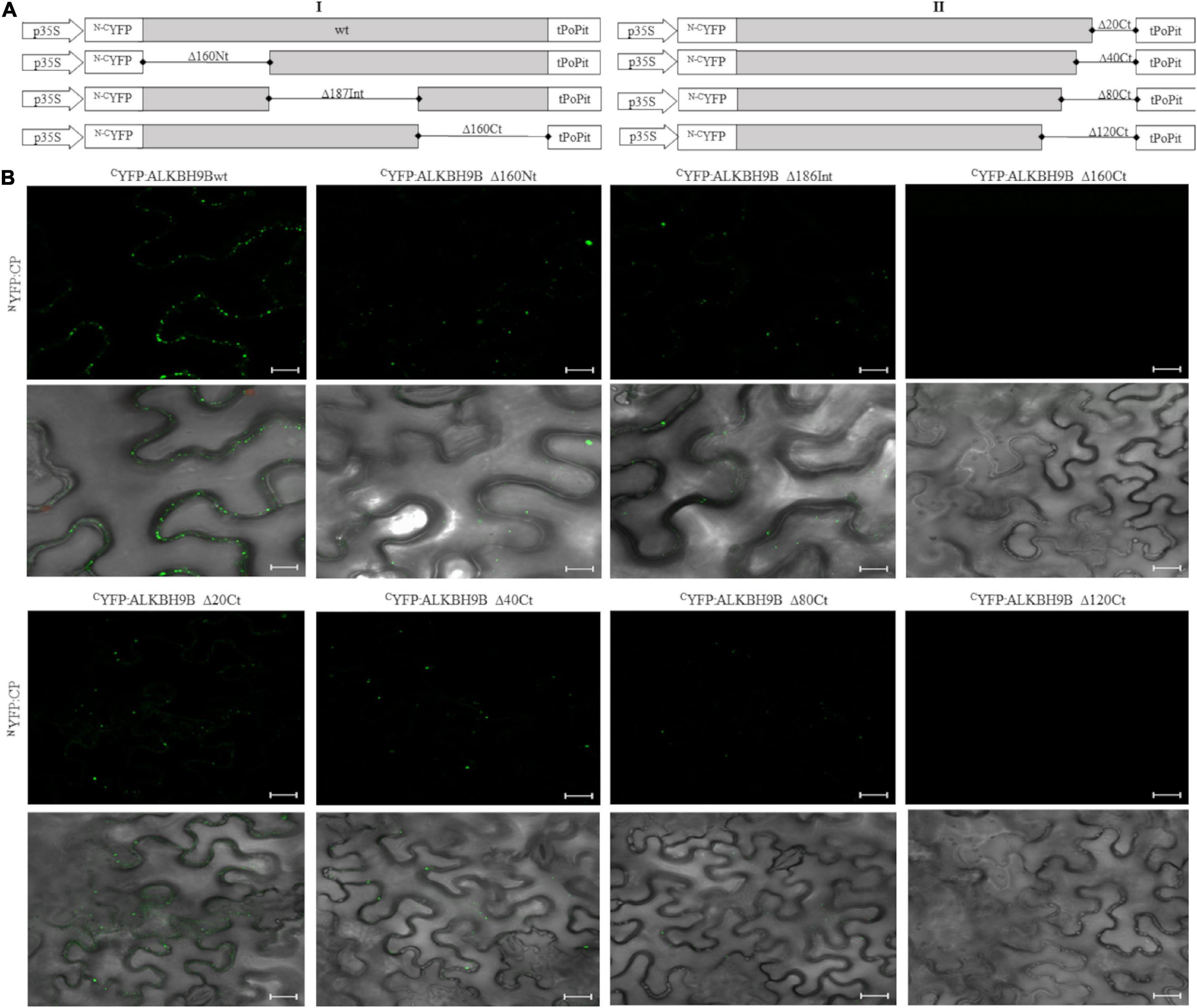
Figure 5. In vivo BiCF visualization of the interaction between N–CYFP:atALKBH9B fusion proteins and N–CYFP:CP of AMV in N. benthamiana. (A) Schematic representation of the N–CYFP:atALKBH9B fusion proteins. (B) BiFC images of epidermal cells co-infiltrated with CYFP:atALKBH9B proteins combined with the NYFP:CP, previously described in Aparicio et al. (2006). Scale bar = 20 μm.
To further delimit the domain involved in atALKBH9B/AMV-CP interaction, we designed new N–CYFP:fusion mutants by deleting residues located at the C-terminal region of atALKBH9B (Figure 5A, right panel [II]). Leaves co-infiltrated with CYFP-atALKBH9BΔ20Ct plus NYFP:CP, CYFP:atALKBH9BΔ40Ct plus NYFP:CP and CYFP:atALKBH9BΔ80Ct plus NYFP:CP produced a reconstituted YFP fluorescence signal in the cells, whereas no YFP fluorescence was detected in leaves co-infiltrated with the pair CYFP:atALKBH9BΔ120Ct plus NYFP:CP (Figure 5B and Supplementary Figure 3, lower panels). Finally, western blot assays using anti cYFP and NYFP antibodies confirmed that all atALKBH9B mutated versions fused to the cYFP and the NYFP:CP accumulated at detectable levels in the co-infiltrated tissues (Supplementary Figure 4). Overall, our results suggest that amino acids located between positions 387 and 427 of atALKBH9B are critical for the interaction with the AMV-CP.
atALKBH9B is the only protein of the 14 homologs of E. coli AlkB that is exclusively localized in the cytoplasm (Mielecki et al., 2012). Our previous studies determined that atALKBH9B can specifically colocalize with SGS3 protein (a component of siRNA bodies), forming biomolecular-condensates associated with stress granules (Martínez-Pérez et al., 2017).
To identify the region of the protein involved in this subcellular localization, we performed localization experiments in N. benthamiana leaves by expressing a series of deletion mutants of atALKBH9B fused to the GFP (Figure 6A).
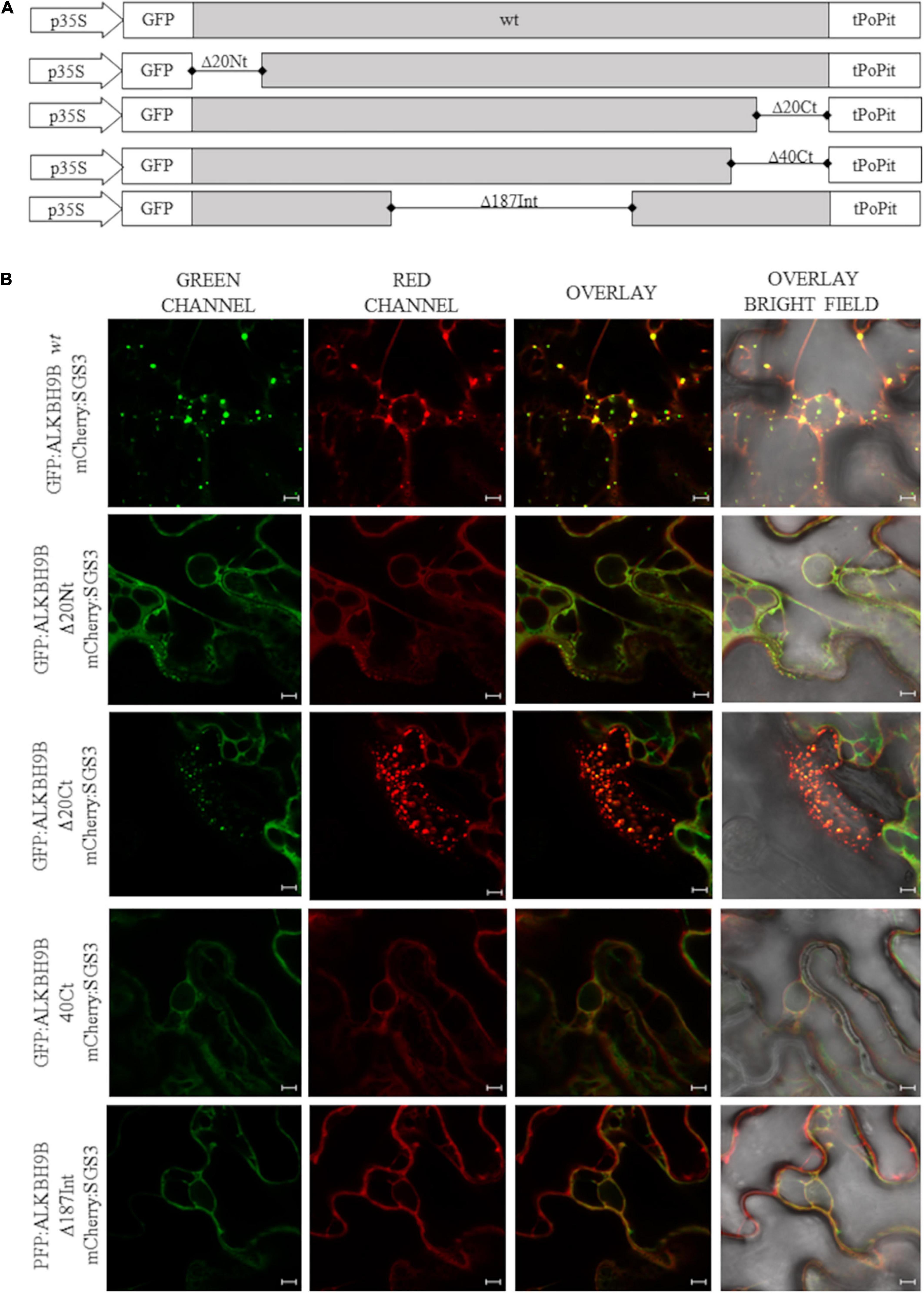
Figure 6. Subcellular localization of atALKBH9B proteins fused with GFP in infiltrated leaves (A) Schematic representation of the different GFP:atALKBH9B fusion proteins used. (B) CLSM images of N. benthamiana leaf epidermal cells co-infiltrated with agrobacterium expressing constructs indicated in (A) plus the SGS3 protein fused to the mCherry fluorescent protein (mCherry:SGS3). Scale bar = 20 μm.
Thus GFP:atALKBH9B mutants were transiently co-expressed by agroinfiltration with mCherry:SGS3. As expected, full length atALKBH9B co-localized with SGS3 (Figure 6B, upper panels) whereas that only atALKBH9B with a deletion of the C-terminal 20 residues (Figure 6B panel GFP:atALKBH9BΔ20Ct) accumulated in cytoplasmic granules colocalizing with SGS3. In contrast, deletion of the N-terminal 20, C-terminal 40 or internal 186 residues (Figure 6A; GFP:atALKBH9BΔ20Nt, GFP:atALKBH9BΔ40Ct and GFP:atALKBH9BΔ186Int) resulted in proteins showing a diffuse pattern throughout the cytoplasm (Figure 6B). These results indicated that different regions of the atALKBH9B sequence are involved in its subcellular localization, which could be explained assuming that the proper three-dimensional folding of the protein might be a critical requirement to form the cytoplasmic granules and consequently to colocalize with siRNA bodies.
m6A RNA methylation in plants has emerged as an important cellular process of gene regulation in development (Wan et al., 2015; Růžička et al., 2017), response to abiotic stress (Li et al., 2014), and antiviral defense (Martínez-Pérez et al., 2017). Recently, atALKBH9B, atALKBH10B, and atALKBH6 were described as m6A erasers involved in AMV infection, flowering time in Arabidopsis, and growth and abiotic stress responses, respectively (Duan et al., 2017; Martínez-Pérez et al., 2017; Huong et al., 2020).
To dissect the functional activity of atALKBH9B in plant-virus infection, a protein-mapping analysis was carried out to identify putative domains required to regulate this process. In this context, by mutagenesis and northwestern analysis, we analyzed the ALKBH9B RNA binding activity and delimited the region between residues 427 and 467 as critical for binding in vitro sgRNA4 (AMV). Moreover, EMSA analysis led us to determine that the Kd of the binding of the RBD alone (0.64 μM, Figure 3) was slightly higher than the one obtained with the full-length protein (0.30 μM, Figure 1), evidencing that other domains of the protein favor the binding to the viral RNA. Nonetheless, the RBD Kd value obtained is within the range reported for other plant viruses RBPs, such as Nla from Tobacco etch virus -TVE- (1.1–1.3 μM) (Daròs and Carrington, 1997), Turnip crinkle virus CP (0.5 μM) (Skuzeski and Morris, 1995), p7 MP of Carnation mottle virus (0.7 μM) (Marcos et al., 1999), AMV CP (0.5 μM) (Baer et al., 1994), or the MP of prunus necrotic ringspot virus (Herranz and Pallás, 2004). Additionally, Huong et al. (2020) showed the RNA-binding capabilities of atALKBH6, although their biochemical parameters were not determined.
Visual inspection and computational analysis of the atALKBH9B sequence revealed no obvious structured RBD that could justify the RNA-binding properties described above. However, recently Varadi et al. (2015) demonstrated the prevalence of IDRs in RNA binding proteins and domains. Among other functions, IDRs are implicated in protein–protein and RNA–protein interactions (Castello et al., 2012). In fact, IDRs are a type of domain that is frequently found in proteins that undergo liquid–liquid phase separation (LLPS), a process that likely contributes to the formation and stability of RNA granules (Alberti et al., 2019). This has been demonstrated in YTHDF1, YTHDF2, and YTHDF3, m6A-binding proteins, which undergo LLPS in the presence of polymethylated mRNAs. The resulting mRNA-YTHDF complexes form P-bodies and stress granules (Ries et al., 2019). Additionally, IDRs have been found to encompass diverse functional motives, e.g., well-established RNA binding activity such as RGG/RG and YGG motives, or low-complexity (LC) domains (Castello et al., 2012; Alberti et al., 2019). Both the conformational flexibility and the establishment of extended conserved electrostatic interfaces with RNAs have been proposed to provide the capability of the IDRs to specifically target different RNAs (Varadi et al., 2015). Furthermore, many proteins localized in RNA granules contain IDRs encompassing prion-like LC domains required for RNA granules assembly (Gilks et al., 2004; Reijns et al., 2008). The atALKBH9B RBD identified here is enriched with G and R (20% and 15%, respectively) and presents an RGxxxRGG motif between positions 469 and 465 (Figure 1A) or a Tri-RG motif present in proteins implicated in various cellular processes like RNA biogenesis, DNA damage signaling, and mRNA translation (Thandapani et al., 2013). Interestingly, we found that 45.4% of atALKBH9B amino acid sequence forms IDRs, located at the N-terminal part, delimiting the internal AlkB-like domain located between positions 216 and 411 and at the C-terminal part. In fact, around 77.5% of the RBD is contained in the C-terminal disordered region (Figure 4). Furthermore, the C-terminal IDR of atALKBH9B exhibits two additional RG residues at positions 473–474 and 466–497, respectively (Figure 4).
Our results can be explained considering that the RGxxxRGG motif between positions 459 and 466 plays a critical role in the protein–RNA interface. This interaction might induce the formation of a flexible structure permitting additional contacts through RG residues at positions 473–474 and 496–497 and YG residues at 506–507, enhancing the binding affinity and specificity of the interaction. For example, the splicing factor Tra2-β1 presents IDRs in the N- and C-terminal regions of the RRM. In the interface protein–RNA, this region adopts a folded structure, forming extensive contacts (Cléry et al., 2011). On the other hand, we previously reported that atALKBH9B is exclusively cytoplasmic, forming discrete granules which colocalize with siRNA bodies, and some are associated with P bodies (Martínez-Pérez et al., 2017). Here we show that deletion of the first N-terminal 20 residues or the C-terminal last 40 amino acids impedes its accumulation in siRNA bodies rendering a diffuse cytoplasmic pattern (Figure 6B). Interestingly, the deleted atALKBH9B N- and C-terminal parts are predicted to form IDRs, and the C-terminal is rich in Y and S residues, which would participate in the RNA granules formation. In this sense, mutation of LC domains in hnRNPA2 and FUS reduced the efficiency of their recruitment in hydrogel polymers in vitro (Xiang et al., 2015) and stress granules (SG) in cells (Kato et al., 2012), respectively. Moreover, phosphorylation of the SG-nucleating protein G3BP within its IDR (Ser 149) impaired its ability to induce the formation of SGs (Kedersha et al., 2016).
Finally, we found that amino acids located between positions 387 and 427 of atALKBH9B are critical for CP–AMV interaction (Figure 5 and Supplementary Figure 3). Interestingly, this region is located next to the RNA-binding site, but it is not part of the C-terminal IDR (Figure 4). Considering that atALKBH9B binds RNA to remove m6A-modification (Martínez-Pérez et al., 2017), and the CP is a multifunctional protein indispensable for the viral replication and translation (Bol, 2005; Guogas et al., 2005; Herranz et al., 2012), it may be possible that the CP binds atALKBH9B in order to modulate the vRNA binding and the m6A demethylase activity in benefit of the virus.
In summary, we have mapped the atALKBH9B regions responsible for RNA and viral CP binding and those required for its localization in stress granules. CP-binding and RNA binding are located at the protein C-terminal, the former partly overlapping the AlkB-like domain, whereas the RBD is partially embedded in the predicted IDR located at the C-terminal (Figure 7). Thus, as found in other proteins (Protter and Parker, 2016), atALKBH9B IDRs and the RBD could act cooperatively to promote the formation of RNA granules. This follows the role of both IDRs and folded domains in mediating RNA binding and oligomerization by acting together with RNAs to produce and maintain these granules (Jonas and Izaurralde, 2013; Protter and Parker, 2016). Therefore, although our results reinforce the existence of this cooperativity in RNA granules formation, the mechanisms underlying IDR-folded domain cooperativity and their potential regulation need further examination.

Figure 7. atALKBH9B multi-domain structure. Predicted IDRs, between positions 1–12, 48–101, 104–187, 280–288, and 437–507, are shown in red. AlkB-like domain, located at 216–411, is shown in gray. Black boxes in the AlkB-like domain delimit the 2-OG stabilizing motif (NxY: 324–326), motif involved in binding to iron -Fe II- (HxD/E⋅⋅⋅H: 335–340) and the substrate specificity motif (RxxxxxR: 335–411) [Alignment between the atALKBH9B and hsALKBH5 proteins, using the BLAST tool]. The AMV CP binding domain (CP-bd: 387–427) and RNA binding domain (RBD: 427–467) are shown in yellow and blue. RGxxxRGG motif between positions 469 and 465 within the RBD characterized here is shown in green.
Full-length atALKBH9B ORF and deletion mutants were subcloned into pGEX-KG (GE Healthcare Life Sciences) to generate a construct with 9B merged to the C-terminal part of the GST. GST and GST:atALKBH9B fusion proteins were expressed in BL21 (DE3) E. coli cells and purified with glutathione Sepharose 4B beads (GE Healthcare Life Sciences) according to the manufacturer’s recommendations. All protein purification procedures were performed at 4°C.
Dilutions of GST or GST:atALKBH9B purified proteins were electrophoresed in 12% SDS/PAGE and transferred to nitrocellulose membranes. Membranes were incubated overnight at 4°C in Renaturing Buffer (10 mM Tris⋅HCl pH 7.5, 1 mM EDTA, 0.1 M NaCl, 0.05% Triton X-100, 1X Blocking Reagent, Roche). After this, membranes were incubated with 20 mL of the same buffer containing 50 ng/μL of the AMV sgRNA 4 labeled with digoxigenin for 3 h at 25°C. For the EMSA assay, 5 ng of 3′UTR of AMV RNA 3 transcripts were heated for 5 min at 85°C and cooled at room temperature for 15 min. Different amounts of purified GST:atALKBH9B fusion proteins were added and incubated for 30 min at 4°C in a 10-μl final volume of Union Buffer (100 mM Tris-HCl pH 8.0, 1 M NaCl, 8 units of RiboLock RNase inhibitor, Thermo Fisher Scientific). Following incubations, the samples were separated through 1.2% agarose. RNAs were transferred to positively charged nylon membranes (Roche). RNAs were visualized on blots using DIG-labeled riboprobes corresponding to the 3’UTR of AMV RNA 3. Synthesis of the digoxigenin-labeled riboprobes, hybridization and digoxigenin-detection procedures were carried out as described in (Pallás et al., 1998).
N–CYFP:atALKBH9B (wild-type and deletion mutants); GFP:atALKBH9B and mCherry:SGS3 fusion proteins were cloned using the Gateway System (Invitrogen) according to the manufacturer’s recommendations. Plasmid expressing the CPAMV merged to the N, or C-terminal part of the YFP (N–CYFP:CPAMV), was previously described in Aparicio et al. (2006). All binary vectors were transformed into Agrobacterium tumefaciens C58 cells. Pairs of cultures carrying specific fusion proteins were mixed at an optical density of 0.5 each in infiltration solution (10 mM MES, pH 5.5, and 10 mM MgCl2) and co-infiltrated into N. benthamiana leaves. Laser-scanning confocal images were taken 48 h after agroinfiltration. Excitation and emission wavelengths were 488 and 508 nm for GFP, 514 and 527 nm for YFP, and 545 and 572 nm for mCherry. Expression of fusion proteins was corroborated by western blot analysis using anti-NtGFP or anti-CtGFP (Roche) antibodies were conducted following the recommendations of the manufacturer.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
VP and FA conceived the project and designed the experiments. LA-M conducted the experiments with assistance from JM-M and MM-P. LA-M, FA, and VP wrote the manuscript. All authors analyzed and discussed the results.
This work was funded by grant PID2020-115571RB-I00 from the Spanish Agencia Estatal de Investigación (AEI) and Fondo Europeo de Desarrollo Regional (FEDER). LA-M is the recipient of a Ph.D. fellowship from the Ministerio de Ciencia, Tecnología y Telecomunicaciones (MICITT) from Costa Rica.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Lorena Corachan for her excellent technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.701683/full#supplementary-material
Abbasi, N., Park, Y.-I., and Choi, S.-B. (2011). Pumilio puf domain RNA-binding proteins in Arabidopsis. Plant Signal. Behav. 6, 364–368. doi: 10.4161/psb.6.3.14380
Aik, W., Scotti, J. S., Choi, H., Gong, L., Demetriades, M., Schofield, C. J., et al. (2014). Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42, 4741–4754. doi: 10.1093/nar/gku085
Alberti, S., Gladfelter, A., and Mittag, T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434. doi: 10.1016/j.cell.2018.12.035
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ambrosone, A., Costa, A., Leone, A., and Grillo, S. (2012). Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 182, 12–18. doi: 10.1016/j.plantsci.2011.02.004
Aparicio, F., Sánchez-Navarro, J. A., and Pallás, V. (2006). In vitro and in vivo mapping of the Prunus necrotic ringspot virus coat protein C-terminal dimerization domain by bimolecular fluorescence complementation. J. Gen. Virol. 87, 1745–1750. doi: 10.1099/vir.0.81696-0
Arribas-Hernández, L., Bressendorff, S., Hansen, M. H., Poulsen, C., Erdmann, S., and Brodersen, P. (2018). An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30, 952–967. doi: 10.1105/tpc.17.00833
Arribas-Hernández, L., and Brodersen, P. (2020). Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 182, 79–96. doi: 10.1104/pp.19.01156
Arribas-Hernández, L., Simonini, S., Hansen, M. H., Paredes, E. B., Bressendorff, S., Dong, Y., et al. (2020). Recurrent requirement for the m6A-ECT2/ECT3/ECT4 axis in the control of cell proliferation during plant organogenesis. Development 147:dev189134. doi: 10.1242/dev.189134
Baer, M. L., Houser, F., Loesch-Fries, L. S., and Gehrke, L. (1994). Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 13, 727–735. doi: 10.1002/j.1460-2075.1994.tb06312.x
Bayoumi, M., Rohaim, M. A., and Munir, M. (2020). Structural and virus regulatory insights into avian N6-methyladenosine (m6A) machinery. Front. Cell Dev. Biol. 8:543. doi: 10.3389/fcell.2020.00543
Boccaletto, P., MacHnicka, M. A., Purta, E., Pitkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Bodi, Z., Zhong, S., Mehra, S., Song, J., Graham, N., Li, H., et al. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3:48. doi: 10.3389/fpls.2012.00048
Bol, J. F. (2005). Replication of Alfamo- and Ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43, 39–62. doi: 10.1146/annurev.phyto.43.101804.120505
Bujarski, J., Gallitelli, D., García-Arenal, F., Pallás, V., Palukaitis, P., Krishna Reddy, M., et al. (2019). ICTV virus taxonomy profile: Bromoviridae. J. Gen. Virol. 100, 1206–1207. doi: 10.1099/jgv.0.001282
Calabretta, S., and Richard, S. (2015). Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 40, 662–672. doi: 10.1016/j.tibs.2015.08.012
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. doi: 10.1016/j.cell.2012.04.031
Chen, Y., and Varani, G. (2013). Engineering RNA-binding proteins for biology. FEBS J. 280, 3734–3754. doi: 10.1111/febs.12375
Cléry, A., Jayne, S., Benderska, N., Dominguez, C., Stamm, S., and Allain, F. H. T. (2011). Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-β1. Nat. Struct. Mol. Biol. 18, 443–451. doi: 10.1038/nsmb.2001
Covelo-Molares, H., Bartosovic, M., and Vanacova, S. (2018). RNA methylation in nuclear pre-mRNA processing. Wiley Interdiscip. Rev. RNA 9:e1489. doi: 10.1002/wrna.1489
Dang, W., Xie, Y., Cao, P., Xin, S., Wang, J., Li, S., et al. (2019). N6-methyladenosine and viral infection. Front. Microbiol. 10:417. doi: 10.3389/fmicb.2019.00417
Daròs, J. A., and Carrington, J. C. (1997). RNA binding activity of NIa proteinase of tobacco etch potyvirus. Virology 237, 327–336. doi: 10.1006/viro.1997.8802
Dimock, K., and Stoltzfus, C. M. (1977). Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 16, 471–478. doi: 10.1021/bi00622a021
Duan, H. C., Wei, L. H., Zhang, C., Wang, Y., Chen, L., Lu, Z., et al. (2017). ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29, 2995–3011. doi: 10.1105/tpc.16.00912
Furuichi, Y., Morgan, M., Shatkin, A. J., and Darnell, J. E. (1976). The methylation of adenovirus-specific nuclear and cytoplasmic aRNA. Nucleic Acids Res. 3, 749–766. doi: 10.1093/nar/3.3.749
Gilks, N., Kedersha, N., Ayodele, M., Shen, L., Stoecklin, G., Dember, L. M., et al. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398. doi: 10.1091/mbc.E04-08-0715
Glisovic, T., Bachorik, J. L., Yong, J., and Dreyfuss, G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986. doi: 10.1016/j.febslet.2008.03.004
Gsponer, J., and Madan Babu, M. (2009). The rules of disorder or why disorder rules. Prog. Biophys. Mol. Biol. 99, 94–103. doi: 10.1016/j.pbiomolbio.2009.03.001
Guogas, L. M., Laforest, S. M., and Gehrke, L. (2005). Coat protein activation of alfalfa mosaic virus replication is concentration dependent. J. Virol. 79, 5752–5761. doi: 10.1128/jvi.79.9.5752-5761.2005
Herranz, M. C., and Pallás, V. (2004). RNA-binding properties and mapping of the RNA-binding domain from the movement protein of Prunus necrotic ringspot virus. J. Gen. Virol. 85, 761–768. doi: 10.1099/vir.0.19534-0
Herranz, M. C., Pallas, V., and Aparicio, F. (2012). Multifunctional roles for the N-terminal basic motif of Alfalfa mosaic virus coat protein: nucleolar/cytoplasmic shuttling, modulation of RNA-binding activity, and virion formation. Mol. Plant Microbe Interact. 25, 1093–1103. doi: 10.1094/MPMI-04-12-0079-R
Huang, H., Weng, H., and Chen, J. (2020). The biogenesis and precise control of RNA m6A methylation. Trends Genet. 36, 44–52. doi: 10.1016/j.tig.2019.10.011
Huong, T. T., Ngoc, L. N. T., and Kang, H. (2020). Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 21:6707. doi: 10.3390/ijms21186707
Ingelfinger, D., Arndt-Jovin, D. J., Lührmann, R., and Achsel, T. (2002). The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8, 1489–1501.
Ishida, T., and Kinoshita, K. (2007). PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464. doi: 10.1093/nar/gkm363
Jonas, S., and Izaurralde, E. (2013). The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27, 2628–2641. doi: 10.1101/gad.227843.113
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi: 10.1016/j.cell.2012.04.017
Kawai, Y., Ono, E., and Mizutani, M. (2014). Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343. doi: 10.1111/tpj.12479
Kedersha, N., Panas, M. D., Achorn, C. A., Lyons, S., Tisdale, S., Hickman, T., et al. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860. doi: 10.1083/jcb.201508028
Krug, R. M., Morgan, M. A., and Shatkin, A. J. (1976). Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 20, 45–53. doi: 10.1128/jvi.20.1.45-53.1976
Lee, D. H., Kim, D. S., and Hwang, B. K. (2012). The pepper RNA-binding protein CaRBP1 functions in hypersensitive cell death and defense signaling in the cytoplasm. Plant J. 72, 235–248. doi: 10.1111/j.1365-313X.2012.05063.x
Lee, K., and Kang, H. (2016). Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 39, 179–185. doi: 10.14348/molcells.2016.2359
Li, D., Zhang, H., Hong, Y., Huang, L., Li, X., Zhang, Y., et al. (2014). Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 32, 1169–1186. doi: 10.1007/s11105-014-0724-2
Li, Z., Shi, J., Yu, L., Zhao, X., Ran, L., Hu, D., et al. (2018). N6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 15, 87. doi: 10.1186/s12985-018-0997-4
Lin, Y., Protter, D. S. W., Rosen, M. K., and Parker, R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219. doi: 10.1016/j.molcel.2015.08.018
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Lorković, Z. J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. doi: 10.1093/nar/30.3.623
Lunde, B. M., Moore, C., and Varani, G. (2007). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490. doi: 10.1038/nrm2178
Luo, S., and Tong, L. (2014). Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. U.S.A. 111, 13834–13839. doi: 10.1073/pnas.1412742111
Marcos, J. F., Vilar, M., Pérez-Payá, E., and Pallás, V. (1999). In vivo detection, RNA-binding properties and characterization of the RNA-binding domain of the p7 putative movement protein from carnation mottle carmovirus (CarMV). Virology 255, 354–365. doi: 10.1006/viro.1998.9596
Martínez de Alba, A. E., Moreno, A. B., Gabriel, M., Mallory, A. C., Christ, A., Bounon, R., et al. (2015). In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 43, 2902–2913. doi: 10.1093/nar/gkv119
Martínez-Pérez, M., Aparicio, F., López-Gresa, M. P., Bellés, J. M., Sánchez-Navarro, J. A., and Pallás, V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. U.S.A. 114, 10755–10760. doi: 10.1073/pnas.1703139114
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi: 10.1146/annurev-cellbio-100616
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Mielecki, D., Zugaj, D.Ł, Muszewska, A., Piwowarski, J., Chojnacka, A., Mielecki, M., et al. (2012). Novel AlkB dioxygenases—alternative models for in silico and in vivo studies. PLoS One 7:e30588. doi: 10.1371/journal.pone.0030588
Pallas, V., Aparicio, F., Herranz, M. C., Sanchez-Navarro, J. A., and Scott, S. W. (2013). The molecular biology of ilarviruses. Adv. Virus Res. 87, 139–183. doi: 10.1016/b978-0-12-407698-3.00005-3
Pallas, V., and Gómez, G. (2013). Phloem RNA-binding proteins as potential components of the long-distance RNA transport system. Front. Plant Sci. 4:130. doi: 10.3389/fpls.2013.00130
Pallás, V., Más, P., and Sánchez-Navarro, J. A. (1998). Detection of plant RNA viruses by nonisotopic dot-blot hybridization. Methods Mol. Biol. 81, 461–468. doi: 10.1385/0-89603-385-6:461
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Prall, W., Sharma, B., and Gregory, B. D. (2019). Transcription is just the beginning of gene expression regulation: the functional significance of RNA-binding proteins to post-transcriptional processes in plants. Plant Cell Physiol. 60, 1939–1952. doi: 10.1093/pcp/pcz067
Protter, D. S. W., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi: 10.1016/j.tcb.2016.05.004
Re, A., Joshi, T., Kulberkyte, E., Morris, Q., and Workman, C. T. (2014). RNA–protein interactions: an overview. Methods Mol. Biol. 1097, 491–521. doi: 10.1007/978-1-62703-709-9_23
Reijns, M. A. M., Alexander, R. D., Spiller, M. P., and Beggs, J. D. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121, 2463–2472. doi: 10.1242/jcs.024976
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. doi: 10.1038/s41586-019-1374-1
Růžička, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Salavert, F., Navarro, J. A., Owen, C. A., Khechmar, S., Pallás, V., and Livieratos, I. C. (2020). Cucurbit chlorotic yellows virus p22 suppressor of RNA silencing binds single-, double-stranded long and short interfering RNA molecules in vitro. Virus Res. 279:197887. doi: 10.1016/j.virusres.2020.197887
Scutenaire, J., Deragon, J. M., Jean, V., Benhamed, M., Raynaud, C., Favory, J. J., et al. (2018). The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30, 986–1005. doi: 10.1105/tpc.17.00854
Shen, L., Liang, Z., Gu, X., Chen, Y., Teo, Z. W. N., Hou, X., et al. (2016). N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 38, 186–200. doi: 10.1016/j.devcel.2016.06.008
Skuzeski, J. M., and Morris, T. J. (1995). Quantitative analysis of the binding of turnip crinkle virus coat protein to RNA fails to demonstrate binding specificity but reveals a highly cooperative assembly interaction. Virology 210, 82–90. doi: 10.1006/viro.1995.1319
Spector, D. L. (2006). Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Nat. Rev. Mol. Cell Biol 172, 887–898. doi: 10.1016/j.cell.2006.11.026
Terribilini, M., Lee, J. H., Yan, C., Jernigan, R. L., Honavar, V., and Dobbs, D. (2006). Prediction of RNA binding sites in proteins from amino acid sequence. RNA 12, 1450–1462. doi: 10.1261/rna.2197306
Thandapani, P., O’Connor, T. R., Bailey, T. L., and Richard, S. (2013). Defining the RGG/RG Motif. Mol. Cell 50, 613–623. doi: 10.1016/j.molcel.2013.05.021
Varadi, M., Zsolyomi, F., Guharoy, M., Tompa, P., and Levy, Y. K. (2015). Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One 10:e0139731. doi: 10.1371/journal.pone.0139731
Wan, Y., Tang, K., Zhang, D., Xie, S., Zhu, X., Wang, Z., et al. (2015). Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 16:272. doi: 10.1186/s13059-015-0839-2
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wei, C. M., and Moss, B. (1975). Methylated nucleotides block 5’ terminus of vaccinia virus messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 72, 318–322. doi: 10.1073/pnas.72.1.318
Williams, G. D., Gokhale, N. S., and Horner, S. M. (2019). Regulation of viral infection by the RNA modification N6-methyladenosine. Annu. Rev. Virol. 6, 235–253. doi: 10.1146/annurev-virology-092818-015559
Xiang, S., Kato, M., Wu, L. C., Lin, Y., Ding, M., Zhang, Y., et al. (2015). The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839. doi: 10.1016/j.cell.2015.10.040
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, C., Liu, K., Tempel, W., Demetriades, M., Aik, W. S., Schofield, C. J., et al. (2014). Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J. Biol. Chem. 289, 17299–17311. doi: 10.1074/jbc.M114.550350
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. doi: 10.1038/cr.2014.151
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. doi: 10.1105/tpc.108.058883
Keywords: Ncpsdummy6-methyladenosine, RNA covalent modifications, RNA-binding proteins, plant viruses, alfamovirus, RNA demethylases, epitranscriptomics, ALKBH
Citation: Alvarado-Marchena L, Marquez-Molins J, Martinez-Perez M, Aparicio F and Pallás V (2021) Mapping of Functional Subdomains in the atALKBH9B m6A-Demethylase Required for Its Binding to the Viral RNA and to the Coat Protein of Alfalfa Mosaic Virus. Front. Plant Sci. 12:701683. doi: 10.3389/fpls.2021.701683
Received: 28 April 2021; Accepted: 09 June 2021;
Published: 05 July 2021.
Edited by:
German Martinez, Swedish University of Agricultural Sciences, SwedenReviewed by:
Susheel Sagar Bhat, Swedish University of Agricultural Sciences, SwedenCopyright © 2021 Alvarado-Marchena, Marquez-Molins, Martinez-Perez, Aparicio and Pallás. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vicente Pallás, dnBhbGxhc0BpYm1jcC51cHYuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.