
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 June 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.699442
Plant-based expression platforms are currently gaining acceptance as a viable alternative for the production of recombinant proteins (RPs), but the degradation of RPs by proteases in cells hinders their superb potentials. Co-expression of a protease inhibitor (PI) shows promise as a strategy to prevent RP from proteolytic degradation in plants. However, competitive effects behind the PI-RP co-expression system may mask or obfuscate the in situ protective effects of a companion PI. Here, we explored the competitive effects by co-expressing reteplase (rPA) with three unrelated PIs, namely NbPR4, HsTIMP, and SlCYS8, in Nicotiana benthamiana leaves. Remarkably, the accumulation of rPA was significantly repressed by each of the three PIs, suggesting that the competitive effects may be common among the PIs. The repression can be attenuated by reducing the PI inoculum dose in the co-inoculation mixtures, showing a negative correlation between the PI abundance of the PI-RP system and competitive effects. Interestingly, when a replicating vector was used to modulate the relative abundance of PI and RP in vivo, rPA was still boosted even at the maximal testing dose of PI, indicating that the competitive effects reduced to an ignorable level by this in vivo approach. Furthermore, a 7- to 12-fold increase of rPA was achieved, proving that it is a useful way for stimulating the potentials of a companion PI by overcoming competitive effects. And, this approach can be applied to molecular farming for improving the RP yields of plant expression systems.
In recent years, plants are gradually regarded as a viable alternative for producing recombinant proteins (RPs; Shanmugaraj et al., 2020), due to their potential for low energy requirement, reduced animal pathogen contamination risks, and post-translational modifications (Schillberg and Finnern, 2021). The RPs can be essentially produced in plants by either stable transformation or transient expression (Gils et al., 2005). And, the latter approach is more efficient in terms of time consumption as well as batch processing (Kopertekh and Schiemann, 2019). Furthermore, the Agrobacterium-mediated methods were established to deliver the transgenes into the host cells for transiently producing RPs in plants (Zhang et al., 2020). When more than one gene should be expressed at the same time, it can be achieved conveniently by mixing the Agrobacterium cultures carrying different transgenes just before agroinfiltration in vitro. Various RPs have been expressed transiently in plants in recent years, including proteins for therapeutic, diagnostic, research, and industrial applications. Several plant-derived RPs have reached or are close to the market (Fox, 2012; Schillberg and Finnern, 2021), but the relatively low yields of RPs limited more of them to be commercialized (Zischewski et al., 2016; Schillberg and Finnern, 2021).
Proteases are pervasive in all organisms and are the essential regulators in living cells (Schaller, 2004). Hundreds of proteases are encoded by plant genomes and hold crucial functions in controlling protein turnover, regulating development, and responding to defense (Tripathi and Sowdhamini, 2006). It was reported that more than 1,000 protease genes were identified in the Arabidopsis genome (Schaller, 2004), and 975 putative proteases were annotated in agroinfiltrated Nicotiana benthamiana leaves (Grosse-Holz et al., 2018a). RPs are therefore faced with a diverse and complex proteolytic microenvironment while expressing in plants. Since proteases can degrade RPs whether in vitro (Paireder et al., 2017) or in vivo (Benchabane et al., 2008), they are the major sponsors for the low accumulation of RPs in cells.
Several approaches have been developed to escort RPs against degradation in plant cells, including subcellular targeting, stabilizing agents, gene knockdown, fusion partners, and co-expression of protease inhibitors (PIs; Mandal et al., 2016). Among them, a companion PI offers at least two significant advantages. First, it may avoid the requirements of adding PIs during protein purification processes and therefore reduce the cost of products (Robert et al., 2016). Second, transiently co-expressing PI has minimal side effects on plant tissues due to temporal and local limitations (Grosse-Holz et al., 2018b).
Some PIs were found to boost RPs accumulation in planta by either stable transformation (Komarnytsky et al., 2006; Pillay et al., 2012) or transient expression (Goulet et al., 2012; Robert et al., 2013; Jutras et al., 2016). However, in vivo protective effects of PIs on RPs are not as efficient as theoretically expected in the application. Sainsbury et al. (2013) reported that the co-expression of SlCYS8 (a tomato Cys protease inhibitor) with human alpha-1-antichymotrypsin (α1ACT) in plants resulted in a marked decrease in α1ACT accumulation instead of increasing by preventing its degradation by leaf Cys proteases as expected, suggesting an apparent competitive effect between PI and RP during pre- and/or post-translational processes (Sainsbury et al., 2013). They tried to overcome the competitive effect by fusing the SlCYS8 and α1ACT to be one chimeric protein. However, SlCYS8 acts as a stabilizing fusion partner to increase α1ACT levels instead of a protease inhibitor (Sainsbury et al., 2013). There are only a few studies available on this issue since then. Recently, it was shown that the protective effects of PIs dramatically increased when the competitive effects were eliminated by using a non-functional PI as control (Grosse-Holz et al., 2018b). Therefore, the competition may mask the potential of PIs, which causes the protective effects of a companion PI to be largely underestimated in applications.
To address this issue, three PIs, namely NbPR4, HsTIMP, and SlCYS8, that were used previously to increase RP accumulation upon co-expression (Grosse-Holz et al., 2018b), were selected for this study. And, rPA, a truncated human tissue-type plasminogen activator (tPA), was used as a model RP due to instability when expressed in plant tissues (Hidalgo et al., 2017). We explored the competitive effects behind the PI-RP system by co-expressing rPA with those unrelated PIs (belonging to different families of PIs; I25 family for SlCYS8, I43 for NbPR4, and I35 for HsTIMP) in N. benthamiana leaves. Subsequently, we demonstrated a negative correlation between the PI abundance and competitive effects by reducing PI inoculum dose in mixtures. Finally, we diminished the competitive effects to an ignorable level by co-expressing a replicating vector with PI.
The binary plasmid pCB301 (Xiang et al., 1999) was used as the backbone for all constructs. And, in vitro DNA synthesis and restriction enzyme-mediated cloning were employed to create the co-expressed constructs. For the construction of the non-replicating rPA expression vector, the CPMV expression cassette (Peyret and Lomonossoff, 2013) was synthesized (GenScript) and cloned into the pCB301 via ApaI and BamHI restrict sites to generate pCPMV-EV construct. Then the N. benthamiana codon-optimized rPA (Invitrogen) with an N-terminal PR1b signal peptide (GenBank accession no. D90197.1) was synthesized and cloned into pCPMV-EV using NruI and FspI restriction sites to generate pCPMV-rPA construct (Figure 1A). For plant-based expression of the inhibitors, synthetic sequences of the PIs gene (NbPR4, HsTIMP, or SlCYS8) were separately cloned into plasmid pCBNoX P19 (Dong et al., 2019) using NcoI and XbaI restriction sites generating pCB301-PIs constructs (Figure 1B). To obtain the replicating rPA expression vector, the rPA expression cassette was cloned into plasmid pJL TRBO-G, a TMV (Tobacco mosaic virus)-based vector (Lindbo, 2007), by PacI and NotI restriction sites to create the pTMV-rPA construct. Meanwhile, the digested DNA of pJL TRBO-G was made blunt using Klenow fragment and circularized by self-ligation yielding pTMV-EV. After construction, plasmids were transformed into Agrobacterium tumefaciens strain GV3101 via the freeze-thaw method.
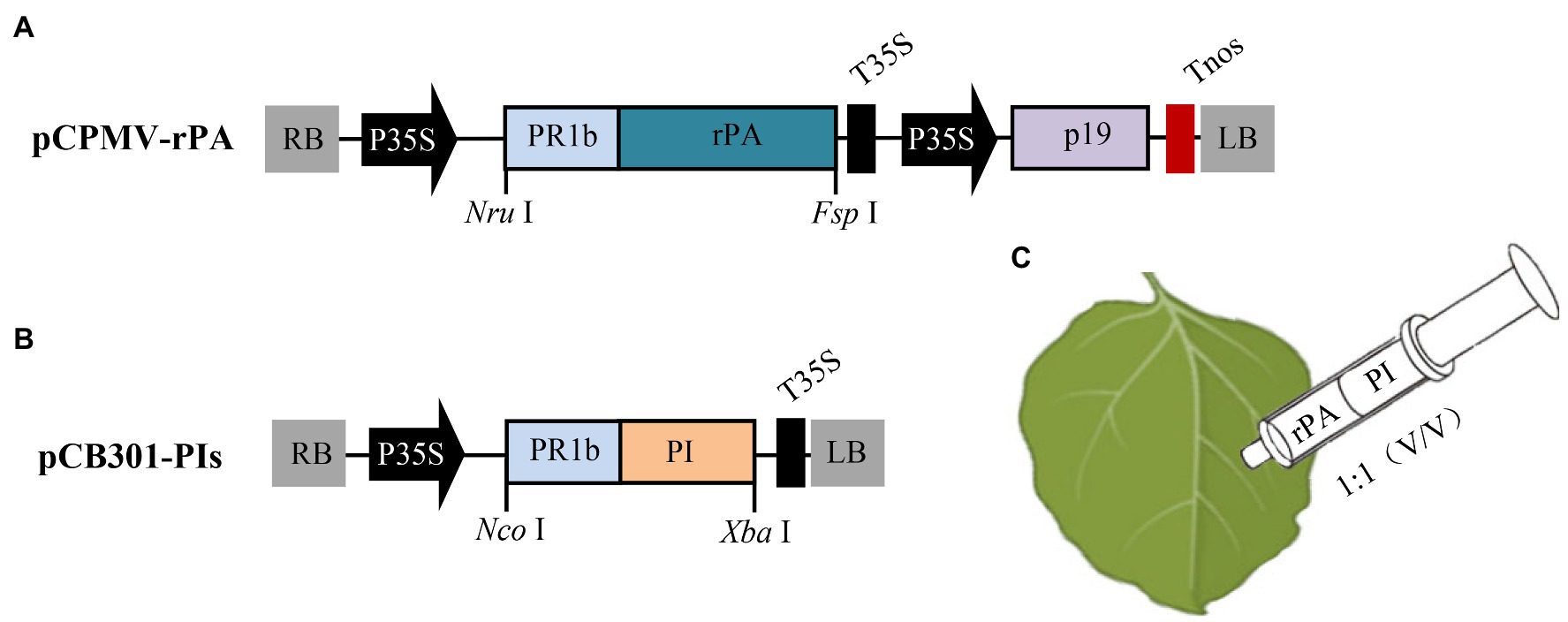
Figure 1. Schematic representation of the construction of the co-expression vectors and the co-agroinfiltration strategy. (A) The T-DNA region of pCPMV-rPA construct, a non-replicating rPA expression vector. LB and RB, the left and right T-DNA borders; P35S, CaMV 35S promoter; PR1b, the secretory signal peptide of tobacco pathogenesis-related protein 1b gene; rPA, the N. benthamiana codon-optimized rPA gene; T35S, CaMV transcription terminator; p19, the gene of silencing suppressor from TBSV (Tomato bushy stunt virus); Tnos, the terminator of nopaline synthase gene. (B) The T-DNA region of the pCB301-PI construct, for expressing protease inhibitor (PI). PI, either of NbPR4, HsTIMP, or SlCYS8; NruI, FspI, NcoI, and XbaI, restriction enzymes. (C) The agroinfiltration strategy for transiently co-expressing rPA with PI in N. benthamiana. Bacterial inoculum for the rPA and PI were mixed at an equal volume and infiltrated into leaves via a syringe.
The recombinant A. tumefaciens were grown in liquid LB media supplying appropriate antibiotics (kanamycin, rifampicin, and gentamycin, 50 μg/ml for each) with gentle agitation for ~24 h at 28°C. Then, the cultures were centrifuged and re-suspended in an appropriate volume of 10 mM MES buffer (pH 5.6, 10 mM MgCl2, and 200 μM acetosyringone) to obtain the required density (Optical Density [OD] value at 600 nm, OD600; the OD600 value 1.2 for rPA; 1.2, 0.6, 0.4, and 0.2 for SlCYS8 separately). For co-expression, the suspensions were mixed in the ratio of 1:1 (V/V; Figure 1C), left in the dark for 2 h of incubation, and then infiltrated into N. benthamiana leaves with a syringe. For comparison, the individual mixes were infiltrated into various zones within a leaf (Figures 2A, 3A). In one experiment, 15–20 seedlings and three leaves of each seedling were infiltrated. Three independent experiments were performed.
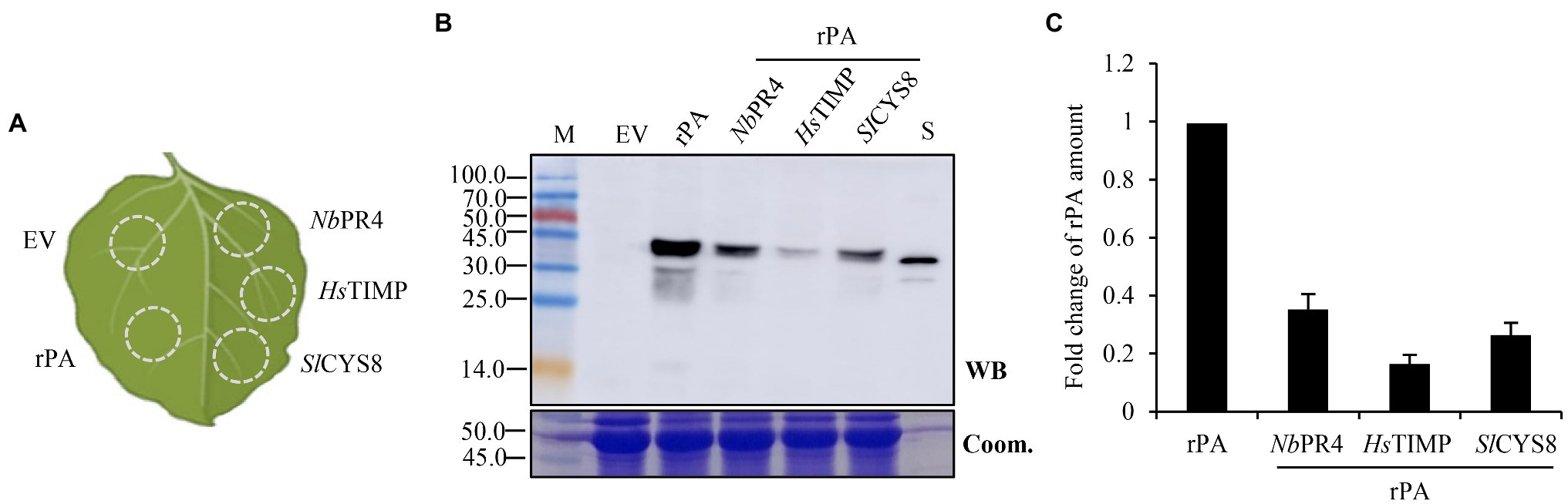
Figure 2. Co-expression of PI with a non-replicating vector (pCPMV-rPA) represses rPA accumulation in plants. (A) Schematic representation of the agroinfiltrated zones within a leaf. EV, empty vector, negative control; rPA, rPA alone; NbPR4, co-expression of rPA and NbPR4; HsTIMP, co-expression of rPA and HsTIMP; SlCYS8, co-expression of rPA and SlCYS8. (B) Western blot analysis of crude leaf protein homogenate. The Coomassie-stained large subunit of Rubisco was used as a loading control (lower panel) and the polyclonal rabbit anti-tPA antibody was used to immunodetect the recombinant rPA (upper panel). M, molecular weight marker (kDa); S, E. coli-expressed rPA, positive control. (C) ELISA assay of the relative rPA accumulation following the co-expression of PI. The data are presented as fold change of rPA amount in the absence or presence of PI and are determined from three separate batches of infiltrations. The average value for rPA alone is 69.7 μg/g +/− 0.7.
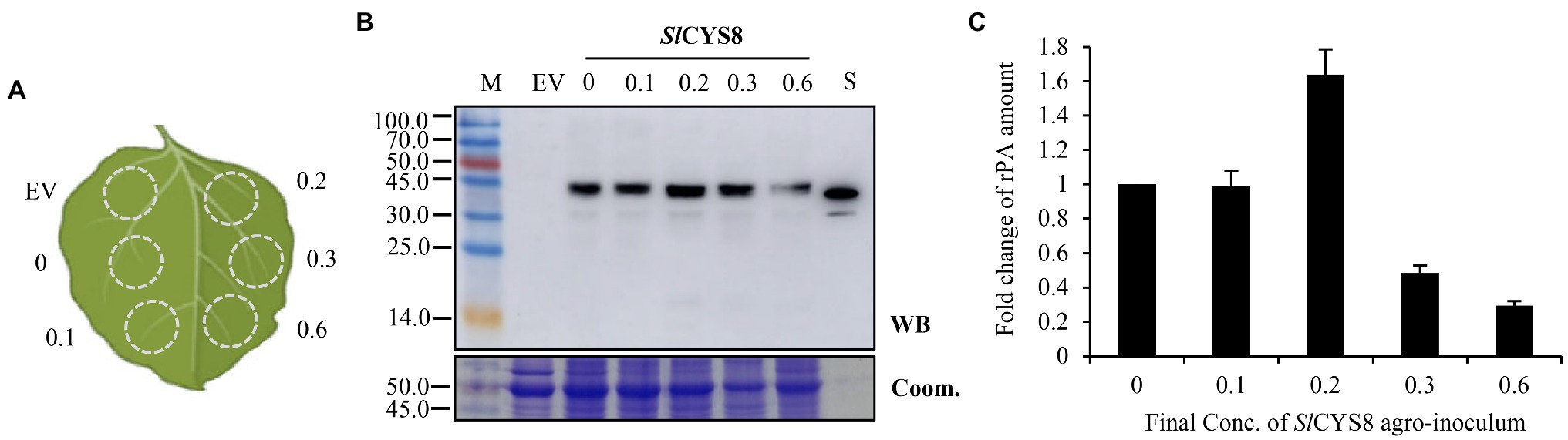
Figure 3. Modulating the dose of SlCYS8 inoculum in mixtures attenuates repression on rPA accumulation. (A) Schematic representation of the agroinfiltrated zones within a leaf. The numbers show the final OD600 value of the SlCYS8 inoculum in mixtures, while the final value of 0.6 is for the rPA construct. (B) Western blot analysis of crude leaf protein homogenate. The Coomassie-stained large subunit of Rubisco was used as a loading control (lower panel) and the polyclonal rabbit anti-tPA antibody was used to immunodetect rPA (upper panel). M, molecular weight marker (kDa); EV, empty vector, negative control; S, E. coli-expressed rPA, positive control. (C) ELISA assay of the relative rPA accumulation following SlCYS8 co-expression. The data are presented as fold change of rPA amount in the absence or presence of SlCYS8 and are determined from three separate batches of infiltrations. The average value for treatment “0” is 69.2 μg/g +/− 0.9.
The infiltrated zones of the leaf were sampled with a hole punch at 5 day post-inoculation (dpi) in all experiments, and nine discs from three individual plants were merged per sample. The leaf discs were weighed and then ground in pre-cooled buffer (50 mM Tris-HCl, 100 mM NaCl, 5 mM ethylenediaminetetraacetic acid [EDTA], 0.1% Tween 20, 0.1% protease inhibitors cocktail, pH 8.0) at a ratio of 1:4 (g/ml). The homogenates were clarified in a centrifuge at 4°C (12,000 g for 15 min), and the supernatant was collected for later assays.
A total of 10 μl of clarified leaf homogenate was mixed with loading buffer, heated for 5 min, and then separated on a 12% polyacrylamide gel. After electrophoresis, one gel was for Coomassie blue staining, and another was blotted to a 0.45 μm NC membrane (Sigma-Aldrich). The membrane was then blocked with 5% (w/v) milk in phosphate-buffered saline (PBS) for 2 h at 25°C and immersed in 1/5,000 rabbit polyclonal anti-human tissue-type plasminogen activator (tPA) antibody (Abcam) and 1/5,000 goat anti-rabbit HRP conjugated secondary antibody (Sigma-Aldrich) successively. The specific immunoreactive proteins were visualized by enhanced chemiluminescent (ECL)-solution (GE Healthcare).
The rPA accumulation was measured by using an ELISA-based assay as previously described. The ELISA plate was coated with the leaf extracts (overnight at 4°C), blocked with 5% (w/v) PBS-made milk (1 h at 37°C), and incubated in 1/10,000 rabbit polyclonal anti-tPA antibody (2 h at 37°C) and 1/5,000 goat anti-rabbit HRP conjugated secondary antibody (2 h at 37°C) successively. After each step, the plate was washed three times with PBS buffer at 5-min intervals. The plate was re-filled with 3,3',5,5'-tetramethylbenzidine (TMB) substrate solution (Solarbio), incubated at 25°C for 15–30 min, and the reaction was halted by adding 1.0 M phosphoric acid. And then, OD values at 450 nm were read by using a microplate reader (Bio-Rad). A standard curve was generated with Escherichia coli–expressed rPA for each plate and the extracts from the empty vector (pCPMV-EV or pTMV-EV) infiltrated leaves were used as a negative control. The relative accumulation level was described as a fold change of the amount of rPA in the absence and presence of PI. The observed values were obtained from the mean of three independent biological replicates and were used to perform Student’s t-test.
The rPA expression vector is a CPMV (Cowpea mosaic virus) RNA2-based construct, which allows high-level RP expression in plants without viral replication (Sainsbury and Lomonossoff, 2008). And, both expression vectors for rPA and PIs have identical T-DNA backbone and transcriptional elements. Furthermore, they all are targeted to the secretory pathway guided by PR1b signal peptides (Figures 1A,B). The vectors were mixed in equal proportion (final OD600 of 0.6 for each) and then inoculated into N. benthamiana leaves by agroinfiltration (Figure 1C).
The individual agro-inoculum was infiltrated into different sectors of the same leaf (Figure 2A). The infiltrated leaves were then sampled at 5 day post-inoculation (dpi) and used for detecting the expression of recombinant rPA. Co-infiltration with rPA and PI expression vectors resulted in detectable immunoblot bands in western blot analysis with anti-rPA antibody, except for empty vector (Figure 2B). Bands of ca. 45.0 kDa had appeared in all the samples upon co-expression. Conversely, rPA expressed by E. coli, positive control, showed a major band with a position corresponding to 39.0 kDa. Since rPA contains two potential N-glycosylation sites at positions Asp-184 and -448 (Noble and McTavish, 1996), it indicated that this plant-derived rPA was glycosylated by post-translational processing. And, the weak bands were presumably due to proteolytic trimming or cleavage of recombinant rPA (Ny et al., 1984). The accumulation of rPA was then quantified using an ELISA assay. The results revealed a 65–84% decrease of rPA accumulation upon PI co-expression (Figure 2C), suggesting that co-expression of PI significantly represses the rPA accumulation in N. benthamiana leaves. Because vectors share similar expression elements, we speculate a competition between PI and rPA during co-expression. We speculate that PI overwhelms the rPA during the competition, therefore resulting in a repression of rPA expression. And, the competitive effects may be common among PIs because all three unrelated PIs show evident repression of rPA expression.
In an attempt to overcome the competitive effects, we kept the rPA construct (pCPMV-rPA) at a final OD600 value of 0.6 and gradually reduced the dose of SlCYS8 in the co-inoculation mixtures. And, the mixes were agroinfiltrated into different sectors within the leaf, as described in Figure 3A. The results showed that the accumulation of recombinant rPA gently increased along with reducing the dose of SlCYS8 inoculum in the co-inoculation mixtures (Figures 3B,C), indicating a negative correlation between the PI abundance of the PI-RP system and competitive effects. At the final OD600 value of 0.2, a ~60% increase of rPA accumulation was achieved (Figure 3C). Interestingly, rPA accumulation was similar to the control, with the rPA alone, at OD600 of 0.1 (Figure 3C). Because the protective effects of PIs are dose dependent in vivo (Grosse-Holz et al., 2018b), we speculate that the PI molecules in cells are not enough under this PI inoculation dose and therefore the repression overwhelms the protection, even though the repression has attenuated at this moment (OD600 value of 0.1), and thus no enhancement of rPA accumulation. Likewise, the protection is over the repression at OD600 value of 0.2, resulting in a slight increment of rPA. We tested NbPR4 and HsTIMP, respectively, and got similar results (Supplementary Figure S1), suggesting that modulating PI abundance in the PI-RP system influences the outcome of the competition (repression or increment of RP accumulation).
The relative abundance of PI in the PI-RP system is critical for the result of the competition. If we use a viral replicating vector to express the RP instead of the non-replicating construct, the replication increases the RP level of the PI-RP system in inoculated cells, resulting in a relative decline of PI abundance without altering its absolute dose, thereby creating an optimal in vivo microenvironment for co-expression.
To verify this hypothesis, a replicating rPA expression vector (Figure 4A), pTMV-rPA, was constructed based on a TMV-derived vector (Lindbo, 2007). For comparison, we maintained the rPA expression cassette as same as that of the non-replicating vector, pCPMV-rPA. And, the final OD600 of the pTMV-rPA construct was kept at the same value as that of pCPMV-rPA at 0.6. The results showed that the rPA accumulation boosted along with the increment of SlCYS8 dose (Figures 4B,C), showing a positive correlation between rPA accumulation and the dose of SlCYS8 inoculum. Furthermore, the rPA accumulation was still boosted even at the maximal testing dose of the SlCYS8, suggesting that the PI-RP system had an ignorable level of competitive effects while co-expression of replicating vector with PI. Similar results were obtained when testing NbPR4 and HsTIMP (Supplementary Figure S2). In addition, a 7- to 12-fold increase of rPA accumulation was achieved at the maximal testing dose of PIs (Figure 4C; Supplementary Figure S2), proving that it is feasible to overcome the competitive effects of the PI-RP system by using this in vivo approach.
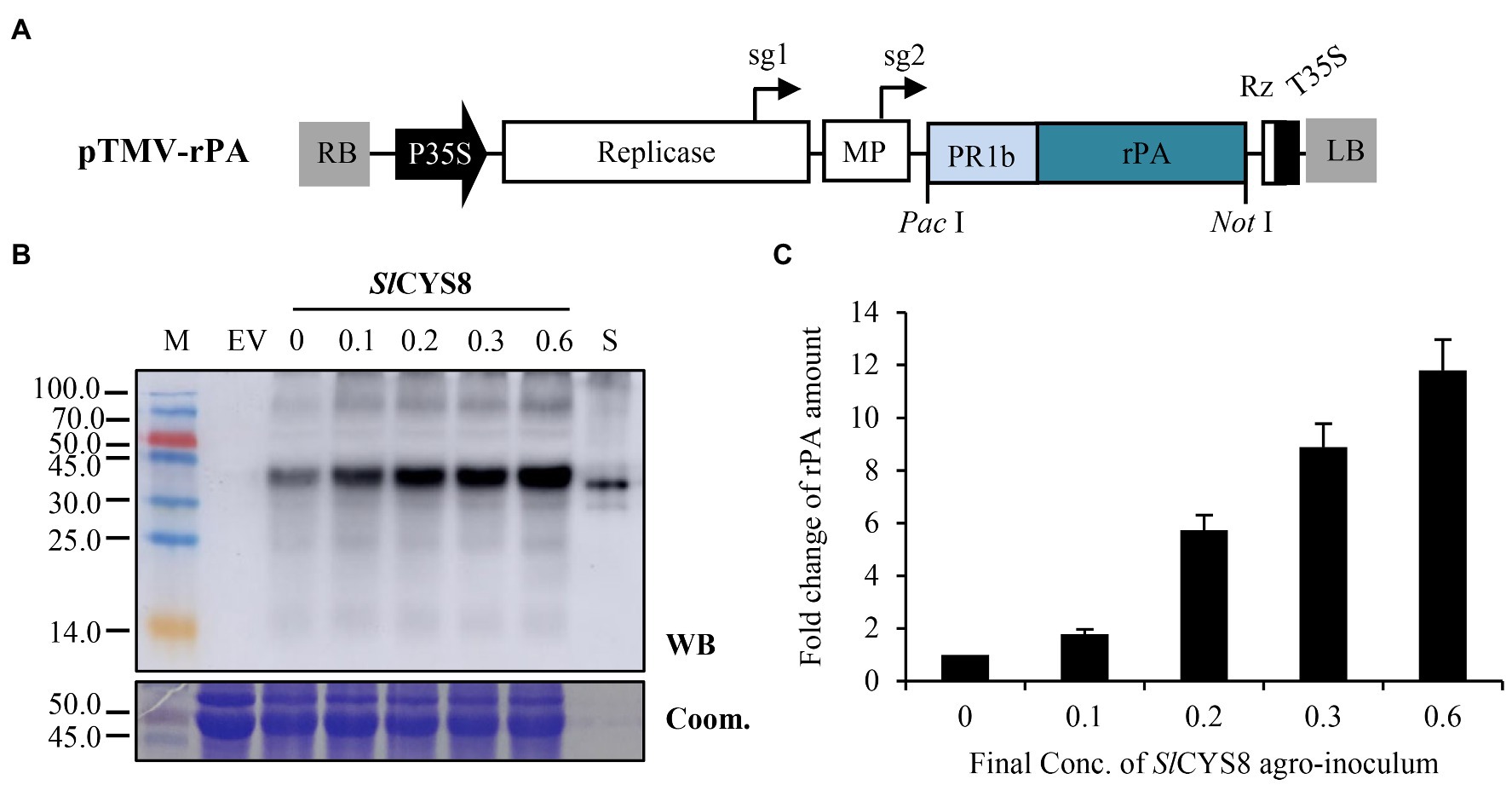
Figure 4. Co-expression of SlCYS8 with a replicating vector overcomes repression on rPA accumulation. (A) The T-DNA region of the pTMV-rPA construct, a replicating TMV-mediated rPA expression vector. Replicase, RNA-dependent RNA polymerase of TMV; sg1 or sg2, subgenomic mRNA1 or mRNA2 promotor of the TMV; MP, movement-protein of TMV; Rz, ribozyme; T35S. PacI and NotI, restriction enzymes. (B) Western blot analysis of crude leaf protein homogenate. The Coomassie-stained large subunit of Rubisco was used as a loading control (lower panel) and the polyclonal rabbit anti-tPA antibody was used to immunodetect rPA (upper panel). M, molecular weight marker (kDa); EV, empty vector, negative control; S, E. coli-expressed rPA, positive control. The numbers show the final OD600 value of the SlCYS8 inoculum in the mixtures, while the final value of 0.6 is for the rPA construct. (C) ELISA assay of the relative rPA accumulation following SlCYS8 co-expression. The data are presented as fold change of rPA amount in the absence or presence of SlCYS8 and are determined from three separate batches of infiltration. The average value for treatment “0” is 69.3 μg/g +/− 1.2.
Co-expression of PI is a promising strategy to improve the relatively low yields of RPs in plants by preventing RP from proteolytic degradation. Several PIs were demonstrated to guard RP against degradation in vivo, among which SlCYS8 is the well-characterized one. However, only ~40% of increment on monoclonal antibody (mAb) was achieved upon SlCYS8 co-expression (Robert et al., 2013). Furthermore, the co-expression of SlCYS8 was reported to markedly decrease rather than increase α1ACT accumulation (Sainsbury et al., 2013). Conversely, the accumulation of three different RPs was enhanced ~7- to ~16-fold upon co-expression with SlCYS8 when a mutant SlCYS8 (SlCYS8-Q47P), which lacks inhibitory activity (Jutras et al., 2016), was used as a negative control (Grosse-Holz et al., 2018b). Those pieces of evidence indicate that something behind the PI-RP co-expression system masks or obfuscates the practical potentials of a SlCYS8 companion or a PI companion, in other words.
For exploring the issue, three unrelated PIs, including SlCYS8, and rPA were chosen to establish the PI-RP co-expression system. To create an “authentic” PI-RP system, P19 and rPA (model RP) were expressed by a single construct (Figure 1A) instead of two separate constructs as previously described (Robert et al., 2013; Jutras et al., 2016; Grosse-Holz et al., 2018b). All the three PIs showed remarkedly repression on rPA accumulation in the leaves of N. benthamiana upon co-expression, although the strength of the repression differed among PIs (Figure 2). The results are similar to what was observed by Sainsbury et al. (2013). We speculate that there are competitive effects in the PI-RP system. This conclusion is drawn from three pieces of evidence. First, the vectors for both RP and PI share similar expression elements (T-DNA backbone, transcriptional elements, and signal peptides) but resulting in remarkable repression of RP (Figures 1A,B). Second, the repression can be attenuated by reducing the PI inoculum dose while the RP dose remains unchanged in the co-infiltration mixtures (Figure 3). Third, the repression reduces to an ignorable level when increasing RP abundance in the PI-RP system by replication (Figure 4). Moreover, the competitive effects may be common among the PIs because three testing PIs belong to different families.
The repression or increment of RP is the outcome of the competition between PI and RP. When PI overwhelms the RP during the competition, it results in a repression of RP expression (for example, α1ACT and rPA), otherwise, RP was found to increase (for example, antibody). Nevertheless, the modulating relative abundance of PI, whether in vivo or in vitro, can alter the outcome of the competition, indicating a negative correlation between the PI abundance and competitive effects. Grosse-Holz et al. (2018b) suggested that the protective effects of PI on RP are dose dependent in vivo. In the present study, RPs accumulate more with higher PI inoculum dose under an ignorable competition situation (Figure 4C; Supplementary Figure S2), also reasoning that it functions in a dose-dependent manner. Since both protective and competitive effects are related to PI abundance, modulating the relative abundance of PI seems to be a useful way to stimulate the potentials of the PI-RP system in the application. The repression can be attenuated by reducing the PI dose in the co-inoculation mixtures (Figure 2; Supplementary Figure S1). However, when repression was circumvented by this in vitro modulation, a companion PI lost most of its in situ protective effects due to inadequate dose at that moment (Figure 2; Supplementary Figure S1). Moreover, A. tumefaciens with a high density (above OD600 value of 1.0) results in leaf yellowing or wilting (Kagale et al., 2012). Therefore, the approach by modulating PI dose in vitro in co-inoculum has shortcomings in the application. Theoretically, an environment in which keeping the PI absolute abundance at a high level while the relative abundance in the PI-RP system low enough to avoid repression is useful in stimulating the protective potential of PIs. And, our studies showed that modulating PI abundance in vivo is more feasible than in vitro in terms of RP accumulation.
Sainsbury et al. (2013) speculated that competitive effects might occur during pre- and/or post-translational processes. In our study, the co-expression vectors have identical transcriptional elements (promoter and terminator), and increasing the abundance of RP mRNAs in vivo by viral replication reduces competitive effects. Those data suggest that the competitive effects may also occur during the translational processes, especially during protein synthesis. The PI mRNAs somehow are favored over the RP during translation, but this superiority is abolished when the abundance of RP mRNAs is dominant in the PI-RP system in cells.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
SW designed the experiments and wrote the manuscript. SW and ZL supervised the research. JM and XD conducted the vector construction and agroinfiltration. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Project Nos. 32060041 and 31760039).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Renier A.L. van der Hoorn (University of Oxford, United Kingdom) for kindly providing the sequence information of the PI genes (NbPR4, HsTIMP, or SlCYS8) and John A. Lindbo (Ohio State University, United States) for the Tobacco mosaic virus (TMV) RNA-based expression vector (TRBO).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.699442/full#supplementary-material
Benchabane, M., Goulet, C., Rivard, D., Faye, L., Gomord, V., and Michaud, D. (2008). Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 6, 633–648. doi: 10.1111/j.1467-7652.2008.00344.x
Dong, J., Ding, X., and Wang, S. (2019). Purification of the recombinant green fluorescent protein from tobacco plants using alcohol/salt aqueous two-phase system and hydrophobic interaction chromatography. BMC Biotechnol. 19:86. doi: 10.1186/s12896-019-0590-y
Fox, J. L. (2012). First plant-made biologic approved. Nat. Biotechnol. 30:472. doi: 10.1038/nbt0612-472
Gils, M., Kandzia, R., Marillonnet, S., Klimyuk, V., and Gleba, Y. (2005). High-yield production of authentic human growth hormone using a plant virus-based expression system. Plant Biotechnol. J. 3, 613–620. doi: 10.1111/j.1467-7652.2005.00154.x
Goulet, C., Khalf, M., Sainsbury, F., D’Aoust, M. A., and Michaud, D. (2012). A protease activity-depleted environment for heterologous proteins migrating towards the leaf cell apoplast. Plant Biotechnol. J. 10, 83–94. doi: 10.1111/j.1467-7652.2011.00643.x
Grosse-Holz, F., Kelly, S., Blaskowski, S., Kaschani, F., Kaiser, M., and van der Hoorn, R. (2018a). The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 16, 1068–1084. doi: 10.1111/pbi.12852
Grosse-Holz, F., Madeira, L., Zahid, M. A., Songer, M., Kourelis, J., Fesenko, M., et al. (2018b). Three unrelated protease inhibitors enhance accumulation of pharmaceutical recombinant proteins in Nicotiana benthamiana. Plant Biotechnol. J. 16, 1797–1810. doi: 10.1111/pbi.12916
Hidalgo, D., Abdoli-Nasab, M., Jalali-Javaran, M., Bru-Martínez, R., Cusidó, R. M., Corchete, P., et al. (2017). Biotechnological production of recombinant tissue plasminogen activator protein (reteplase) from transplastomic tobacco cell cultures. Plant Physiol. Biochem. 118, 130–137. doi: 10.1016/j.plaphy.2017.06.013
Jutras, P. V., Marusic, C., Lonoce, C., Deflers, C., Goulet, M. C., Benvenuto, E., et al. (2016). An accessory protease inhibitor to increase the yield and quality of a tumour-targeting mAb in Nicotiana benthamiana leaves. PLoS One 11:e0167086. doi: 10.1371/journal.pone.0167086
Kagale, S., Uzuhashi, S., Wigness, M., Bender, T., Yang, W., Borhan, M. H., et al. (2012). TMV-gate vectors: gateway compatible tobacco mosaic virus based expression vectors for functional analysis of proteins. Sci. Rep. 2:874. doi: 10.1038/srep00874
Komarnytsky, S., Borisjuk, N., Yakoby, N., Garvey, A., and Raskin, I. (2006). Cosecretion of protease inhibitor stabilizes antibodies produced by plant roots. Plant Physiol. 141, 1185–1193. doi: 10.1104/pp.105.074419
Kopertekh, L., and Schiemann, J. (2019). Transient production of recombinant pharmaceutical proteins in plants: evolution and perspectives. Curr. Med. Chem. 26, 365–380. doi: 10.2174/0929867324666170718114724
Lindbo, J. A. (2007). TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 145, 1232–1240. doi: 10.1104/pp.107.106377
Mandal, M. K., Ahvari, H., Schillberg, S., and Schiermeyer, A. (2016). Tackling unwanted proteolysis in plant production hosts used for molecular farming. Front. Plant Sci. 7:267. doi: 10.3389/fpls.2016.00267
Noble, S., and McTavish, D. (1996). Reteplase. A review of its pharmacological properties and clinical efficacy in the management of acute myocardial infarction. Drugs 52, 589–605. doi: 10.2165/00003495-199652040-00012
Ny, T., Elgh, F., and Lund, B. (1984). The structure of the human tissue-type plasminogen activator gene: correlation of intron and exon structures to functional and structural domains. Proc. Natl. Acad. Sci. U. S. A. 81, 5355–5359. doi: 10.1073/pnas.81.17.5355
Paireder, M., Tholen, S., Porodko, A., Biniossek, M. L., Mayer, B., Novinec, M., et al. (2017). The papain-like cysteine proteinases NbCysP6 and NbCysP7 are highly processive enzymes with substrate specificities complementary to Nicotiana benthamiana cathepsin B. Biochim. Biophys. Acta Proteins Proteom. 1865, 444–452. doi: 10.1016/j.bbapap.2017.02.007
Peyret, H., and Lomonossoff, G. P. (2013). The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83, 51–58. doi: 10.1007/s11103-013-0036-1
Pillay, P., Kibido, T., du Plessis, M., van der Vyver, C., Beyene, G., Vorster, B. J., et al. (2012). Use of transgenic oryzacystatin-I-expressing plants enhances recombinant protein production. Appl. Biochem. Biotechnol. 168, 1608–1620. doi: 10.1007/s12010-012-9882-6
Robert, S., Jutras, P. V., Khalf, M., D’Aoust, M. A., Goulet, M. C., Sainsbury, F., et al. (2016). Companion protease inhibitors for the in situ protection of recombinant proteins in plants. Methods Mol. Biol. 1385, 115–126. doi: 10.1007/978-1-4939-3289-4_8
Robert, S., Khalf, M., Goulet, M. C., D’Aoust, M. A., Sainsbury, F., and Michaud, D. (2013). Protection of recombinant mammalian antibodies from development-dependent proteolysis in leaves of Nicotiana benthamiana. PLoS One 8:e70203. doi: 10.1371/journal.pone.0070203
Sainsbury, F., and Lomonossoff, G. P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218. doi: 10.1104/pp.108.126284
Sainsbury, F., Varennes-Jutras, P., Goulet, M. C., D’Aoust, M. A., and Michaud, D. (2013). Tomato cystatin SlCYS8 as a stabilizing fusion partner for human serpin expression in plants. Plant Biotechnol. J. 11, 1058–1068. doi: 10.1111/pbi.12098
Schaller, A. (2004). A cut above the rest: the regulatory function of plant proteases. Planta 220, 183–197. doi: 10.1007/s00425-004-1407-2
Schillberg, S., and Finnern, R. (2021). Plant molecular farming for the production of valuable proteins - critical evaluation of achievements and future challenges. J. Plant Physiol. 258–259:153359. doi: 10.1016/j.jplph.2020.153359
Shanmugaraj, B., I Bulaon, C. J., and Phoolcharoen, W. (2020). Plant molecular farming: a viable platform for recombinant biopharmaceutical production. Plants 9:842. doi: 10.3390/plants9070842
Tripathi, L. P., and Sowdhamini, R. (2006). Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7:200. doi: 10.1186/1471-2164-7-200
Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D. J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. doi: 10.1023/A:1006201910593
Zhang, Y., Chen, M., Siemiatkowska, B., Toleco, M. R., Jing, Y., Strotmann, V., et al. (2020). A highly efficient Agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Commun. 1:100028. doi: 10.1016/j.xplc.2020.100028
Keywords: protease inhibitor, competitive effects, transient expression, replicating vector, agroinfiltration, Nicotiana benthamiana
Citation: Ma J, Ding X, Li Z and Wang S (2021) Co-expression With Replicating Vector Overcoming Competitive Effects Derived by a Companion Protease Inhibitor in Plants. Front. Plant Sci. 12:699442. doi: 10.3389/fpls.2021.699442
Received: 23 April 2021; Accepted: 21 May 2021;
Published: 17 June 2021.
Edited by:
Joachim Hermann Schiemann, Julius Kühn-Institut, GermanyReviewed by:
Hugh S. Mason, Arizona State University, United StatesCopyright © 2021 Ma, Ding, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Wang, d2FuZ19zQG54dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.