
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 July 2021
Sec. Plant Abiotic Stress
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.698443
This article is part of the Research Topic Al-induced and -activated Signals in Aluminium Resistance View all 7 articles
The C2H2-type zinc finger transcription factor SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1) plays a critical role in aluminum (Al) resistance and low phosphate (Pi) response mainly through promoting the expression of the malate transporter-encoding gene ARABIDOPSIS THALIANA ALUMINUM ACTIVATED MALATE TRANSPORTER 1 (AtALMT1). We previously showed that REGULATION OF ATALMT1 EXPRESSION 3 (RAE3/HPR1), a core component of the THO/TREX complex, is involved in the regulation of nucleocytoplasmic STOP1 mRNA export to modulate Al resistance and low Pi response. Here, we report that RAE2/TEX1, another core component of the THO complex, is also involved in the regulation of Al resistance and low Pi response. Mutation of RAE2 reduced the expression of STOP1-downstream genes, including AtALMT1. rae2 was less sensitive to Al than rae3, which was consistent with less amount of malate secreted from rae3 roots than from rae2 roots. Nevertheless, low Pi response was impaired more in rae2 than in rae3, suggesting that RAE2 also regulates AtALMT1-independent pathway to modulate low Pi response. Furthermore, unlike RAE3 that regulates STOP1 mRNA export, mutating RAE2 did not affect STOP1 mRNA accumulation in the nucleus, although STOP1 protein level was reduced in rae2. Introduction of rae1 mutation into rae2 mutant background could partially recover the deficient phenotypes of rae2. Together, our results demonstrate that RAE2 and RAE3 play overlapping but distinct roles in the modulation of Al resistance and low Pi response.
Aluminum (Al) toxicity represents a major constraint for crop production on acid soils, which comprise more than 30% of the world’s arable land (von Uexkull and Mutert, 1995). To grow on the acid soils, plants have evolved multiple mechanisms to detoxify Al. One key Al-resistance mechanism is that plants can exude organic acids, including malate, citrate, and oxalate, to chelate and detoxify Al (Ma et al., 2001; Ryan et al., 2001; Liu et al., 2014). The model plant Arabidopsis (Arabidopsis thaliana) secretes both malate and citrate for the Al detoxification, albeit malate plays a more important role in Al resistance than citrate (Hoekenga et al., 2003, 2006; Liu et al., 2009). Genes involved in the exudation of malate and citrate were first identified in crops, which encode anion transporters belonging to the Al-activated malate transporter (ALMT) family and the multidrug and toxic compound extrusion (MATE) family, respectively (Sasaki et al., 2004; Furukawa et al., 2007; Magalhaes et al., 2007). In Arabidopsis, ARABIDOPSIS THALIANA ALUMINUM ACTIVATED MALATE TRANSPORTER 1 (AtALMT1) and ARABIDOPSIS THALIANA MULTIDRUG AND TOXIC EXTRUSION (AtMATE) are mainly responsible for root malate and citrate secretion in response to Al stress, respectively (Hoekenga et al., 2006; Liu et al., 2009).
The zinc-finger transcription factor SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1) plays a critical role in Al resistance, which is achieved mainly through the regulation of AtALMT1 expression (Iuchi et al., 2007). STOP1 also induces the expression of AtMATE and ALUMINUM SENSITIVE 3 (ALS3) (Sawaki et al., 2009). ALS3 and ARABIDOPSIS THALIANA SENSITIVE TO ALUMINUM RHIZOTOXICITY (AtSTAR1) form a functional ATP-binding cassette (ABC) transporter to be localized at the tonoplast for the Al detoxification although the underlying mechanism is still unclear (Larsen et al., 2005; Huang et al., 2010; Dong et al., 2017). Recently, GLUTAMATE DEHYDROGENASE 1 AND 2 (GDH1 and GDH2), which regulate Al resistance probably through maintaining cellular pH homeostasis under Al stress conditions, are also demonstrated to be the direct targets of STOP1 (Tokizawa et al., 2021). The transcription of STOP1 is not responsive to Al stress but its encoded protein can be induced by the Al stress (Iuchi et al., 2007; Zhang et al., 2019). We recently discovered that F-box proteins REGULATION OF ATALMT1 EXPRESSION 1 (RAE1) and RAE1 HOMOLOG 1 (RAH1) can facilitate STOP1 ubiquitination and promote its degradation via the ubiquitin-26S proteasome pathway, which is important for balancing Al resistance and plant growth (Zhang et al., 2019; Fang et al., 2021b). STOP1 is also subjected to SUMO modification through the SUMO E3 ligase SIZ1-dependent and SIZI-independent pathways, and is deSUMOylated by the SUMO proteases EARLY IN SHORT DAYS 4 (ESD4) (Fang et al., 2020, 2021a; Xu et al., 2021). The altered levels of STOP1 SUMOylation would affect STOP1 activity and stability, which in turn influences the expression of STOP1-downstream genes and Al resistance (Fang et al., 2020, 2021a; Xu et al., 2021).
Besides Al resistance, STOP1-AtALMT1 pathway is also involved in the regulation of low phosphate (Pi) response (Balzergue et al., 2017; Mora-Macias et al., 2017). Under low Pi conditions, plants can remodel root architecture, such as the inhibition of primary root growth and the increase in lateral root and root hair densities, to enhance Pi uptake. In stop1 or Atalmt1, the low Pi-induced root growth inhibition is impaired (Balzergue et al., 2017; Mora-Macias et al., 2017). AtALMT1-mediated malate release is proposed to promote apoplastic iron (Fe) toxicity and consequently cause the inhibition of root growth when Pi is deficient. In addition, LOW PHOSPHATE ROOT 1 AND 2 (LPR1 and LPR2) encoding multi-copper oxidases with ferroxidase activity were demonstrated to play a critical role in low Pi-induced root growth inhibition by the promotion of Fe accumulation and callose deposition in the meristem of the primary root (Svistoonoff et al., 2007; Muller et al., 2015). Brassinosteroid (BR) signaling pathway and CLE14 peptide act upstream and downstream of LPR1/LPR2 to modulate low Pi response, respectively (Singh et al., 2014, 2018; Gutierrez-Alanis et al., 2017). The P5-type ATPase PHOSPHATE DEFICIENCY RESPONSE 2 (PDR2) interacts genetically with LPR1/LPR2 and plays a negative role in low Pi-induced inhibition of root growth (Ticconi et al., 2009).
THO is a conserved multisubunit complex in yeast (Saccharomyces cerevisiae), plants, and animals, which is functionally linked to transcription, mRNA processing, and export (Aguilera, 2005; Luna et al., 2012; Heath et al., 2016). Yeast THO complex consists of four strongly interacting subunits of Tho2, Hpr1, Mft1, and Thp2 (Chavez et al., 2000), while plant THO complex is composed of at least six subunits: HPR1/THO1/EMU1, THO2, TEX1/THO3, THO5A/B, THO6, and THO7A/B, which do not contain the orthologs of yeast Mft1 and Thp2 (Furumizu et al., 2010; Jauvion et al., 2010; Yelina et al., 2010). The core THO complex can associate with additional proteins such as the mRNA export factors UAP56 and Aly to form TREX (Heath et al., 2016). Several subunits of the plant THO complex, including HPR1, TEX1/THO3, THO6, and THO2, have been reported to regulate the accumulation of small RNAs including small interfering RNAs (siRNAs) and microRNA (miRNA), which consequently influence the plant morphology, root-associated acid phosphatase activity, female germline specification, and coumarin scopolin accumulation (Furumizu et al., 2010; Jauvion et al., 2010; Yelina et al., 2010; Francisco-Mangilet et al., 2015; Tao et al., 2016; Su et al., 2017; Doll et al., 2018). HPR1 was also documented to regulate disease resistance and senescence probably through modulating mRNA export (Pan et al., 2012). Recently, Guo et al. (2020) found that HPR1 is involved in the regulation of Al resistance partly through the modulation of nucleocytoplasmic STOP1 mRNA export. It is unclear whether other subunits of the THO complex play a similar role to HPR1 in the regulation of the Al resistance and STOP1 mRNA export.
To identify the genes involved in the regulation of AtALMT1 and/or STOP1, we previously generated a reporter line containing a fusion gene of AtALMT1 promoter with luciferase (LUC) reporter gene to screen mutants showing altered LUC signal. Through the screen, we have identified three regulation of AtALMT1 expression (RAE) genes (Zhang et al., 2019; Fang et al., 2020; Guo et al., 2020), which include RAE3 that encodes HPR1. In this study, we identified RAE2 encoding TEX1, another subunit of the THO complex, by using the same screening system. We found that RAE2 plays a similar role to RAE3 in the regulation of STOP1 protein accumulation and the expression of STOP1-downstream genes, but unlike RAE3, RAE2 does not regulate STOP1 mRNA export. Furthermore, RAE2 modulation of low Pi response involves a different mechanism from RAE3.
The wild-type (WT) Arabidopsis plant (Columbia background; Col-0) contains a homozygous AtALMT1 promoter-driven LUC transgene (pAtALMT1:LUC) (Zhang et al., 2019). rae2 (tex1), rae3 (hpr1-7), and rae1 (rae1-1) mutants with altered LUC signal were screened from an ethyl methanesulfonate (EMS)-mutagenized library carrying the pAtALMT1:LUC reporter gene previously (Zhang et al., 2019). The T-DNA insertion line Atalmt1 (SALK_00962) was derived from the Nottingham Arabidopsis Stock Centre (NASC).1
Arabidopsis seeds were surface-sterilized with 6% sodium hypochlorite solution and stratified at 4°C for 3 days in the dark. Seeds were then sown on 1.2% agar medium containing the full-strength Hoagland nutrient and 1% sucrose for 7 days. For luminescence signal detection, 7-day-old seedlings were used for the observation by a CCD imaging apparatus (Lumazone P1300B, Roper Scientific). Plants were grown on nutrient agar plates or one-tenth-strength Hoagland nutrient solution in a growth chamber (CU-36L4, Percival) or grown on soils in a growth room at 22°C with 14 h of light (100 μmol m–2 s–1; Philips TLD26W865 cool daylight tubes) and 10 h of darkness.
Five-day-old seedlings of WT, rae2, and rae3/hpr1 grown on 0.4% gellan gum (G1910, Sigma-Aldrich) medium containing the full-strength Hoagland nutrient and 1% sucrose were pretreated with a 2% MGRL nutrient solution containing 1% sucrose at pH 4.8 for 2 h and then exposed to the same solution containing 0 or 20 μM Al at pH 4.8 for 12 h. Root exudates were collected for malate measurement by the NAD/NADH enzymatic cycling method (Hampp et al., 1984). For the determination of Al content, three-week-old seedlings of WT and rae2 grown on the one-tenth-strength Hoagland solution were pretreated with 0.5 mM CaCl2 solution for 6 h at pH 4.8 and then treated with 0.5 mM CaCl2 solution containing 0 or 30 μM Al for 12 h at pH 4.8. Al content in roots was determined according to previous methods (Ligaba-Osena et al., 2017; Zhang et al., 2019). The Al concentration in a diluted solution was measured by inductively coupled plasma mass spectrometry (ICP-MS; PerkinElmer NexION300D).
A soaked gel medium was used to evaluate Al resistance according to a previous method (Larsen et al., 2005) with some modifications. Briefly, a nutrient gel medium consisting of 50 mL (pH 5.0) of 0.25 mM (NH4)2SO4, 1 mM KNO3, 0.2 mM KH2PO4, 2 mM MgSO4, 1 mM Ca(NO3)2, 1 mM CaSO4, 1 μM MnSO4, 5 μM H3BO3, 0.05 μM CuSO4, 0.2 μM ZnSO4, 0.02 μM NaMoO4, 0.1 μM CaCl2, 0.001 μM CoCl2, 1% sucrose, and 0.4% gellan gum (G1910, Sigma-Aldrich) was first prepared and then soaked with 30 mL of the same nutrient solution without gellan gum containing 0, 0.5, 0.75, or 1 mM AlCl3 at pH 3.6. After soaking for 2 days, the solution was removed and the gel medium was dried for seed sowing. After 7 days of growth on the gel medium, seedlings were imaged and root lengths were measured by ImageJ software. Relative root growth expressed as root length with Al treatment/root length without Al × 100 was used to assess Al resistance.
To evaluate the response to low Pi, seeds were sown on 0.4% gellan gum medium (pH 5.7) containing the full-strength Hoagland nutrient and 1% sucrose with 40 μM Fe(III)-EDTA and 0, 20, or 1,000 μM NH4H2PO4. After growth for 7 days, the seedlings were photographed and the relative root growth was used for evaluating the sensitivity to low Pi.
The rae2 mutant was crossed with WT to produce an F2 population for genetic analysis and mapping-by-sequencing. High-throughput DNA sequencing was performed by using the Illumina HisSeq4000 system that produces 150-bp paired-end reads, which were carried out by a commercial company (Shanghai Hanyu Biotech) (accession number in NCBI: SRR14277220). The MutMap method (Abe et al., 2012) was used to identify the candidate region of rae2 with slight modifications. The detailed procedure for data processing and map of candidate mutant gene had been described previously (Zhang et al., 2019). To confirm the candidate region of rae2, derived cleaved amplified polymorphic sequence (dCAPS) markers were developed and used for the linkage analysis of F2 plants with decreased LUC signal (Supplementary Table 1).
For the complementation test of rae2, a DNA fragment harboring a 2-kb promoter and gene sequence of TEX1 (At5g56130) was amplified (Supplementary Table 2) and cloned into the pCAMBIA3301 vector. The construct was transformed into rae2 mutant through Agrobacterium tumefaciens (strain GV3101)-mediated transformation method. Single-locus homozygous transgenic lines were used for the complementation test.
To compare the expression of Al-resistance genes among WT and various mutants, seedlings grown on 1.2% agar medium as described earlier for 9 days were pretreated with a 0.5 mM CaCl2 solution at pH 4.8 for 6 h and then treated with the same solution containing 0 or 30 μM AlCl3 at pH 4.8 for 12 h. Roots were excised for RNA extraction and expression analysis. To determine RAE3 expression in various tissues, roots, old and young rosette leaves, cauline leaves, stems, flowers, and siliques of Col-0 wild-type plants were harvested for RNA extraction, respectively. Total RNA were extracted by the TaKaRa MiniBEST plant RNA Extraction Kit (Cat # 9769). About 1 μg of total RNAs was used for the synthesis of the first-strand cDNAs using the HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech) after digestion with DNase I. Diluted cDNA products were subjected to the real-time RT-PCR analysis using the SYBR Green Master Mix (Vazyme Biotech) and gene-specific primers (Supplementary Table 2). The reference gene UBQ10 was used as an internal control for the expression analysis.
The RAE2/TEX1 coding sequence was amplified and fused in frame with GFP at the N- or C-terminals of RAE2 under the control of 35S promoter in pCAMBIA1300 vector. Each resultant vector was transformed into Arabidopsis protoplasts for the subcellular localization analysis. GFP fluorescence in the protoplasts was observed by a confocal laser scanning microscope (Leica SP8).
Two-week-old plants of WT, rae2, and rae3 grown on a one-tenth-strength Hoagland nutrient solution were pretreated with a 0.5 mM CaCl2 solution at pH 4.8 for 6 h and then treated with the same solution containing 0 or 30 μM AlCl3 at pH 4.8 for 12 h. Root nuclei of each line were isolated according to a previous method (Folta and Kaufman, 2006) with slight modifications. The detailed process for the root nuclei isolation has been described previously (Guo et al., 2020). The isolated nuclei were subjected to RNA extraction and expression analysis. The SAND family gene AT2G28390 was used as an internal control.
To create pRAE2:GUS transgenic lines, the 2-kb promoter of RAE2/TEX1 was amplified by a primer pair (Supplementary Table 2) and inserted into pORE-R2 vector harboring the GUS reporter gene. The construct was then introduced into Col-0 plants by Agrobacterium-mediated transformation method. A single-locus homozygous transgenic line was used for GUS activity assay. The transgenic seedlings grown on a one-tenth-strength Hoagland nutrient solution for around 3 weeks were stained with a commercialized GUS staining solution (161031; O’Biolab Co., Ltd., Beijing, China) for overnight at 37°C. To investigate whether GUS activity was affected by Al stress, seven-day-old transgenic seedlings were pretreated with a 0.5 mM CaCl2 solution at pH 4.8 for 6 h and subsequently treated with the same solution (pH 4.8) containing 0 or 30 μM Al for 12 h. The roots were then stained with the GUS staining solution for 2 h at 37°C. Stained tissues were photographed with a stereomicroscope (SZX12, Olympus) equipped with a camera (DP20, Olympus).
Twelve-old-day seedlings of Col-0 WT, rae2, and rae3 harboring pSTOP1:STOP1-HA transgene were pretreated with a 0.5mM CaCl2 solution at pH 4.8 for 6 h and then treated with the same solution containing 0 or 30 μM Al at pH 4.8 for 12 h. Total proteins were extracted from the roots or shoots of each line using a extraction buffer composed of 20 mM Tris–HCl (pH 7.4), 300 mM NaCl, 5 mM MgCl2 (pH 8.0), 5 mM DTT, 0.5% NP-40, 50 μM MG132 (A2585; APExBIO, United States), and 1× Complete Protease inhibitor tablets EDTA-free (5892791001; Roche). Immunoblot analysis was performed to compare STOP1-HA protein level using anti-HA antibody (H3663; Sigma-Aldrich).
We previously conducted a forward genetic screen on an EMS-mutagenized population, which harbors the AtALMT1 promoter-driven luciferase reporter gene (pAtALMT1:LUC), and identified a series of regulation of AtALMT1 expression (rae) mutants with altered LUC signal (Zhang et al., 2019). In this study, we characterized one of these mutants, called rae2, which displayed a decreased LUC signal similar to previously reported rae3/hpr1 mutant (Figure 1A). The expression levels of the LUC transgene and the endogenous AtALMT1 gene were then compared in the roots of the wild type (WT), rae2, and rae3 under different Al conditions. Compared with WT, rae2 showed a reduced expression of LUC and AtALMT1 at a similar level to rae3 under both 0 (–Al) or 30 μM Al (+Al) conditions (Figures 1B,C).
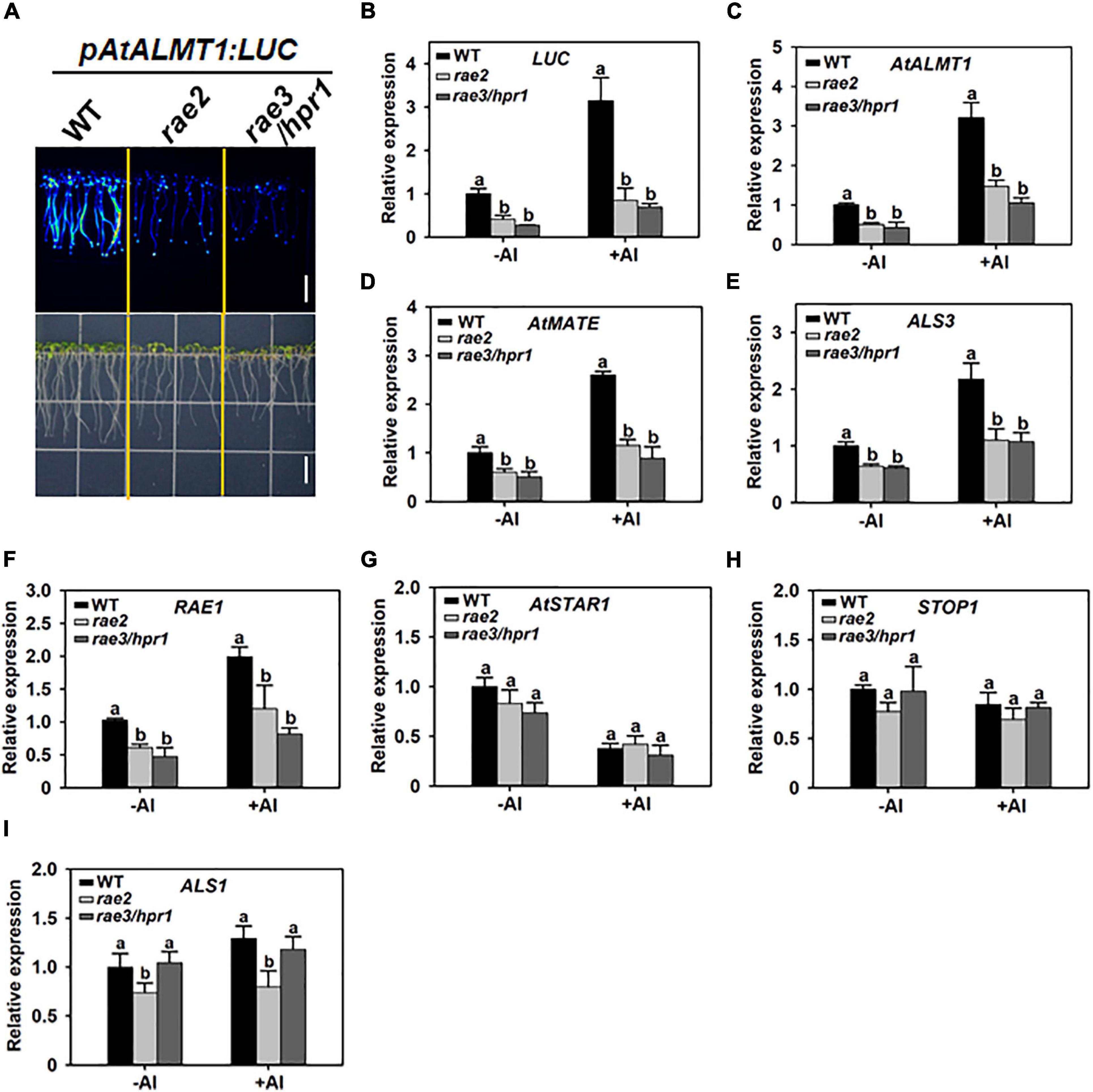
Figure 1. Mutation of RAE2 decreases the expression of STOP1-regulated genes. (A) Decreased LUC signal of pAtALMT1:LUC in rae2 mutant compared with wild type (WT). Bar = 7 mm. (B–F) Reduced mRNA expression of STOP1-regulated genes in rae2 and rae3/hpr1, which include LUC reporter gene (B), AtALMT1 (C), AtMATE (D), ALS3 (E), and RAE1 (F). (G–I) Comparison of the expression levels of non-STOP1-target genes in WT, rae2, and rae3, which include AtSTAR1 (G), STOP1 (H), and ALS1 (I). Seven-day-old seedlings were exposed to 0 (–Al) or 30 μM Al (+Al) for 12 h, and then the roots were excised for the expression analysis. Data shown are means ± SD of three biological replicates. Different letters at each treatment indicate the values significantly different (P < 0.05, ANOVA followed by Tukey’s test).
To examine whether the rae2 mutation influences the expression of other Al-resistance genes, we compared the expression levels of AtMATE, ALS3, RAE1, AtSTAR1, STOP1, and ALS1 in WT, rae2, and rae3. Like rae3, rae2 showed a decreased expression of the three STOP1-regulated genes AtMATE, ALS3, and RAE1 (Figures 1D–F), while the expression of AtSTAR1 and STOP1, which are not regulated by STOP1, was not affected in rae2 and rae3 (Figures 1G,H). The expression of ALS1 not targeted by STOP1 was slightly reduced in rae2 but not in rae3 (Figure 1I). Together, these results demonstrate that RAE2 is involved in the regulation of the expression of STOP1-downstream genes.
Because the rae2 mutation reduced AtALMT1 expression, we compared the level of malate secretion in WT, rae2, and rae3. Without Al treatment, the amount of malate secreted in rae2 decreased slightly but not significantly from that in WT (Figure 2A), although the expression of AtALMT1 was significantly decreased in rae2 compared to WT (Figure 1C), which could be attributed to the fact that Al stress is required for the trigger of AtALMT1-mediated malate secretion (Hoekenga et al., 2006; Iuchi et al., 2007). In the presence of Al, the level of Al-activated malate secretion in rae2 was lower than that in WT, but higher than that in rae3 (Figure 2A). In accordance with the decreased exudation of malate in rae2, the mutant accumulated a higher level of Al than the WT under Al treatment conditions (Figure 2B).
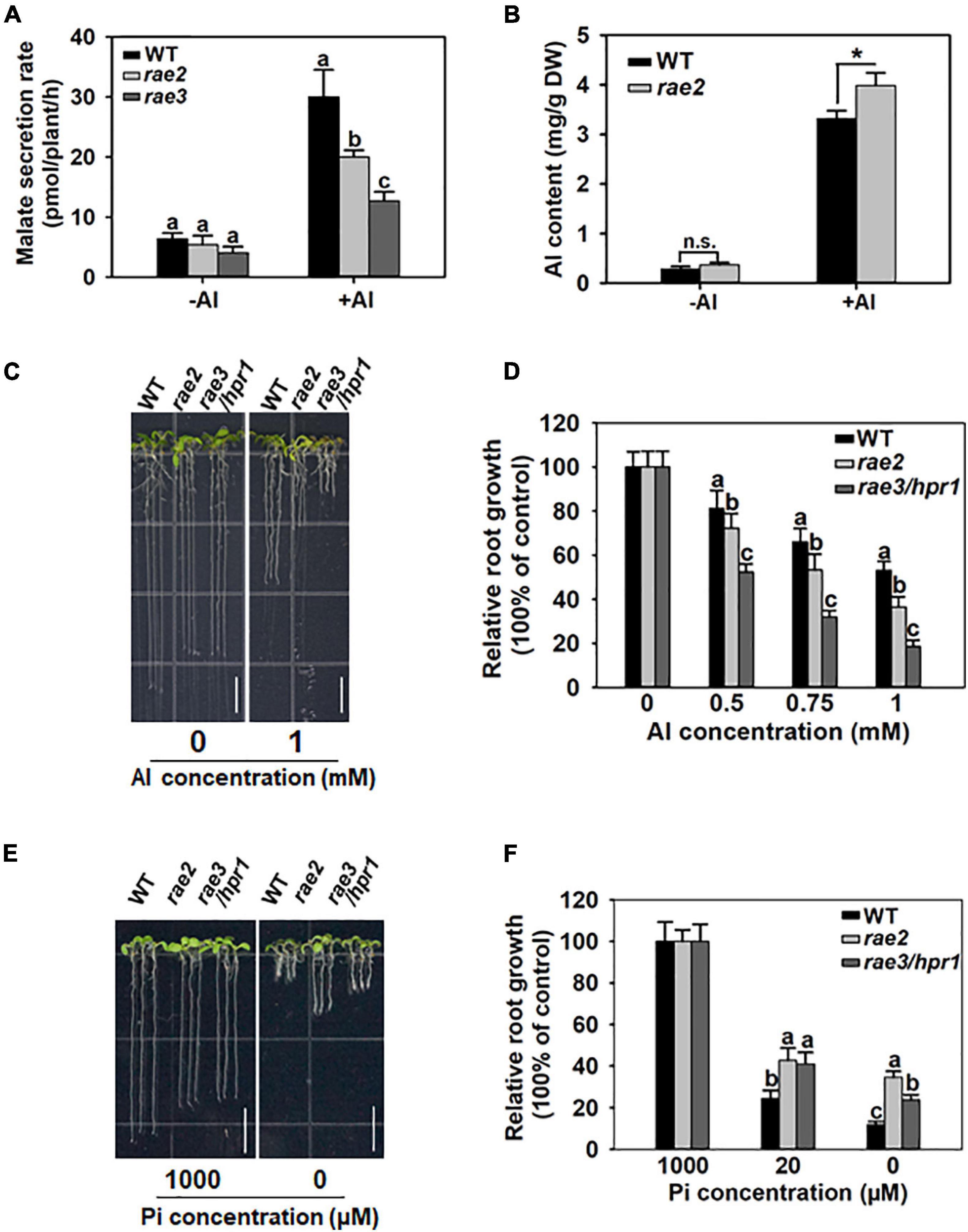
Figure 2. Mutation of RAE2 reduces Al resistance and low Pi response. (A) Malate secretion was reduced in rae2 compared to WT under +Al conditions. Five-day-old seedlings of WT, rae2, and rae3 were treated with 0 (–Al) or 20 μM Al (+Al) for 12 h, and then root exudates were collected for malate content determination. Values are means ± SD of three biological replicates. (B) rae2 mutant accumulated a higher level of Al than WT. Three-week-old plants of WT and rae2 were exposed to 0 (–Al) or 30 μM Al (+Al) at pH 4.8 for 12 h, and the roots were harvested for the measurement of Al content. Values are means ± SD of three biological replicates. Asterisks indicate that the values are statistically different (Student’s t-test, *P < 0.001). (C,D) Representative images (C) and relative root growth (D) of WT, rae2, and rae3 at different Al concentrations. Seedlings of WT, rae2, and rae3 were grown on a soaked gel medium containing 0, 0.5, 0.75, or 1 mM Al for 7 days, and then the relative root growth was calculated to determine their Al resistance. Values are means ± SD (n = 16–20). Bar = 7 mm. (E,F) Images (E) and quantitative data (F) of low Pi-response phenotype in WT, rae2, and rae3. Seedlings were grown on gellan gum medium containing 0, 20 or 1,000 μM Pi for 7 days, and the relative root growth was used to evaluate the low Pi response. Bar = 7 mm. Values are means ± SD (n = 15–20). Different letters at each treatment indicate that the values are significantly different (P < 0.05, ANOVA followed by Tukey’s test).
We then compared Al resistance phenotype in WT, rae2, and rae3. In the absence of Al, the root length of rae2 and rae3 was similarly shorter than that of WT (Figure 2C). Nevertheless, under Al stress conditions, the root growth of rae2 was inhibited more than that of WT (Figures 2C,D), indicating that rae2 was more sensitive to Al toxicity than WT. Compared to rae3/hpr1, the Al-sensitive phenotype in rae2 was weaker (Figures 2C,D).
AtALMT1-mediated malate secretion has been reported to play a positive role in low phosphate (Pi)-induced root growth inhibition (Balzergue et al., 2017; Mora-Macias et al., 2017). To investigate whether the rae2 mutation affected low Pi response, we grew the seedlings of WT, rae2, and rae3 under different Pi levels. The result showed that rae2 was less sensitive to low Pi-induced inhibition of root growth than WT (Figures 2E,F), consistent with the reduced secretion of malate in rae2 compared to WT (Figure 2A). Nevertheless, although the amount of malate exudation in rae2 was more than that in rae3, rae2 was less sensitive to low Pi than rae3 (Figures 2E,F), suggesting that the rae2 mutation also influences the malate secretion-independent pathway to modulate low Pi response.
A genetic analysis of rae2 was conducted by using an F2 population from a cross between WT and rae2 mutant. Observation of LUC signal in 213 F2 plants showed that 52 plants displayed a reduced LUC signal, while the remaining 161 plants had normal LUC signal. The ratio of the number of plants with reduced LUC signal to the number with normal LUC signal fits to 1:3 (χ2 = 0.013, P = 0.91), implying that the reduced LUC expression in rae2 was controlled by a single recessive gene.
To clone the RAE2 gene, pooled DNA from the 52 F2 plants with reduced LUC signal as described earlier was subjected to the whole-genome sequencing. We used the WT DNA as a control, which was sequenced previously (Zhang et al., 2019). We mapped rae2 to a small region of chromosome 5 (Supplementary Figure 1) through MutMap method (Abe et al., 2012). We then developed two derived cleaved amplified polymorphic sequence (dCAPS) markers within the candidate region of rae2 on the basis of the mutations occurred in rae2 (Supplementary Table 1) to perform a linkage analysis in 36 F2 plants with the reduced LUC signal. The T02 marker displayed a linkage to the LUC signal phenotype with two recombinants, while the T01 marker, developed on a G-to-A substitution at +1,738 bp from the start codon of At5g56130 (TEX1), was completely linked to the mutant phenotype (Figure 3A, Supplementary Table 1). This substitution caused an amino acid change from tryptophan to stop codon in TEX1 in rae2 (Figure 3A).
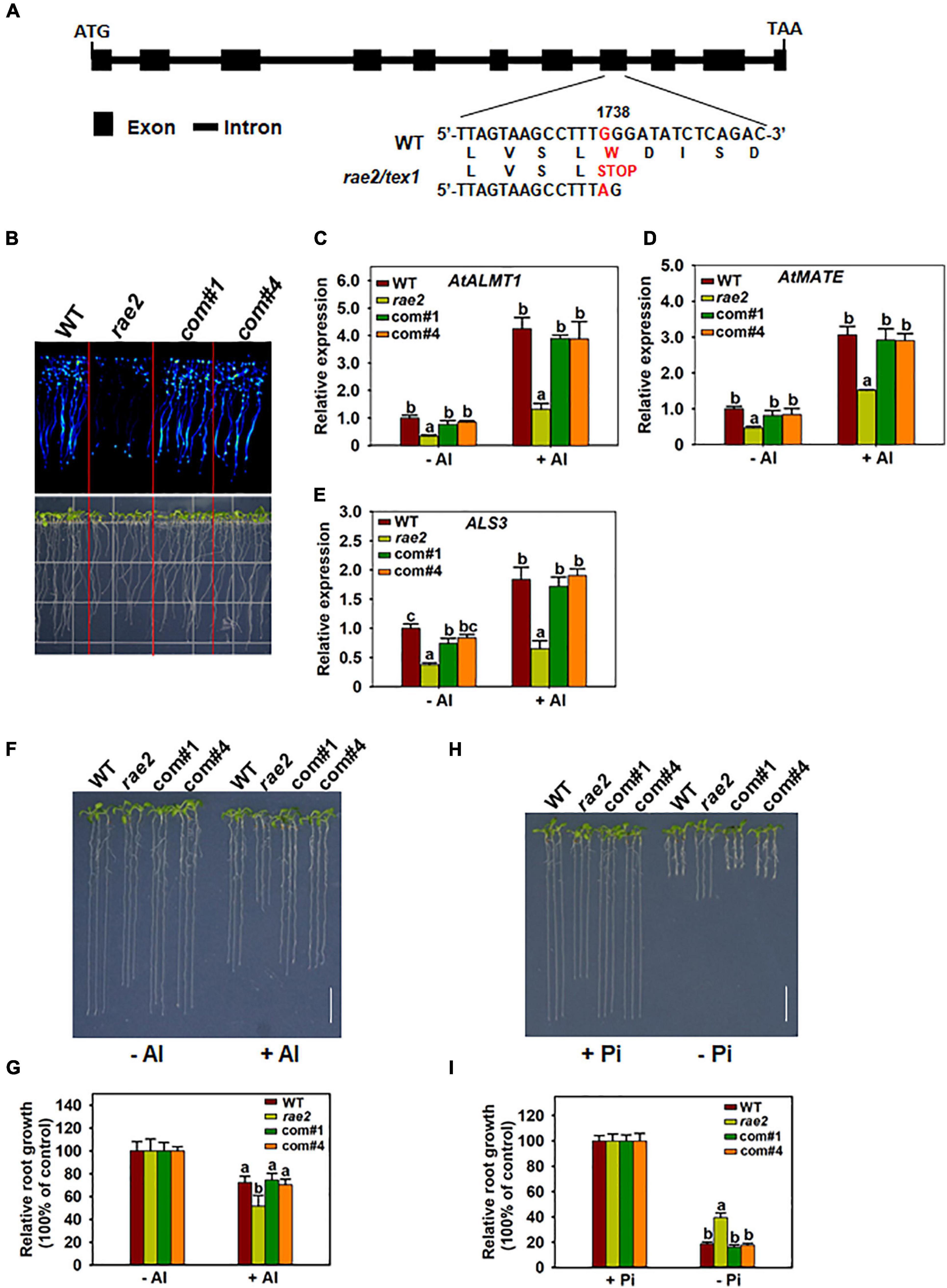
Figure 3. Complementation test of rae2 mutant. (A) Gene structure and mutation sites of RAE2. Boxes and horizontal lines between the boxes indicate the coding regions and introns, respectively. A single-nucleotide substitution from G to A was occurred at +1738 from the start codon in rae2, which causes an amino acid change from tryptophan to stop codon. (B–E) The decreased LUC signal (B) and expression of STOP-regulated genes in rae2, including AtALMT1 (C), AtMATE (D), and ALS3 (E), were rescued in two complementation lines. Roots of 7-day-old seedlings were exposed to 0 (–Al) or 30 μM Al (+Al) for 12 h for the expression analysis. Values are means ± SD (n = 3). (F,G) Complementation of Al resistance phenotype. Representative images (F) and relative root growth (G) of WT, rae2, and complementation lines at different Al concentrations. Values are means ± SD (n = 18–23) (H,I) Complementation of low Pi response phenotype. Representative images (H) and relative root growth (I) of WT, rae2, and complementation lines at different Pi concentrations. Values are means ± SD (n = 19–22). Means with different letters are significantly different (P < 0.05, ANOVA followed by Tukey’s test).
To further confirm that RAE2 is TEX1, we conducted a complementation test on rae2 by transforming a WT RAE2 with 2-kb promoter and the gene sequence into the mutant. The reduced LUC signal in rae2 was fully rescued in two independent complementation lines (Figure 3B). The decreased expression of STOP1-regulated genes, including AtALMT1, AtMATE, and ALS3, in the mutant was also recovered in the two lines (Figures 3C–E). We then compared Al resistance and low Pi response in WT, rae2, and the two complementation lines. The result showed that the reduced Al resistance and low Pi response in rae2 were fully rescued in the two complementation lines (Figures 3F–I). Together, these results indicate that mutation of TEX1 is responsible for the defective phenotypes in rae2.
Since RAE2 and RAE3 encode different subunits of the THO complex and are both involved in the regulation of Al resistance and low Pi response, we attempted to generate rae2 rae3 double mutant by making a cross between the two single mutants rae2 and rae3. Genotyping 52 progenies of rae2/RAE2 rae3/rae3 plants showed that 16 progenies had RAE2/RAE2 rae3/rae3 genotype, while the remaining 36 progenies possessed rae2/RAE2 rae3/rae3 genotype. We were unable to identify rae2 rae3 double mutant plants, suggesting that mutations of both RAE2 and RAE3 would cause embryonic lethality.
RAE2 was well expressed in all the tissues examined (Figure 4A), with relatively lower expression in cauline leaves and stems compared to other tissues. The expression of RAE2 was not affected by Al stress (Figure 4B). To analyze the expression pattern of RAE2 in detail, we fused RAE2 promoter with GUS reporter gene and then introduced the construct into WT background. GUS staining in a homozygous transgenic line showed that GUS signal was preferentially detected in the tip regions of the roots (Figure 4C). In the mature root zones and leaves, GUS activity was mainly detected in the vascular tissues (Figures 4C,D). Consistent with mRNA expression data of RAE2, the level of GUS signal was not responsive to Al stress (Figure 4E).
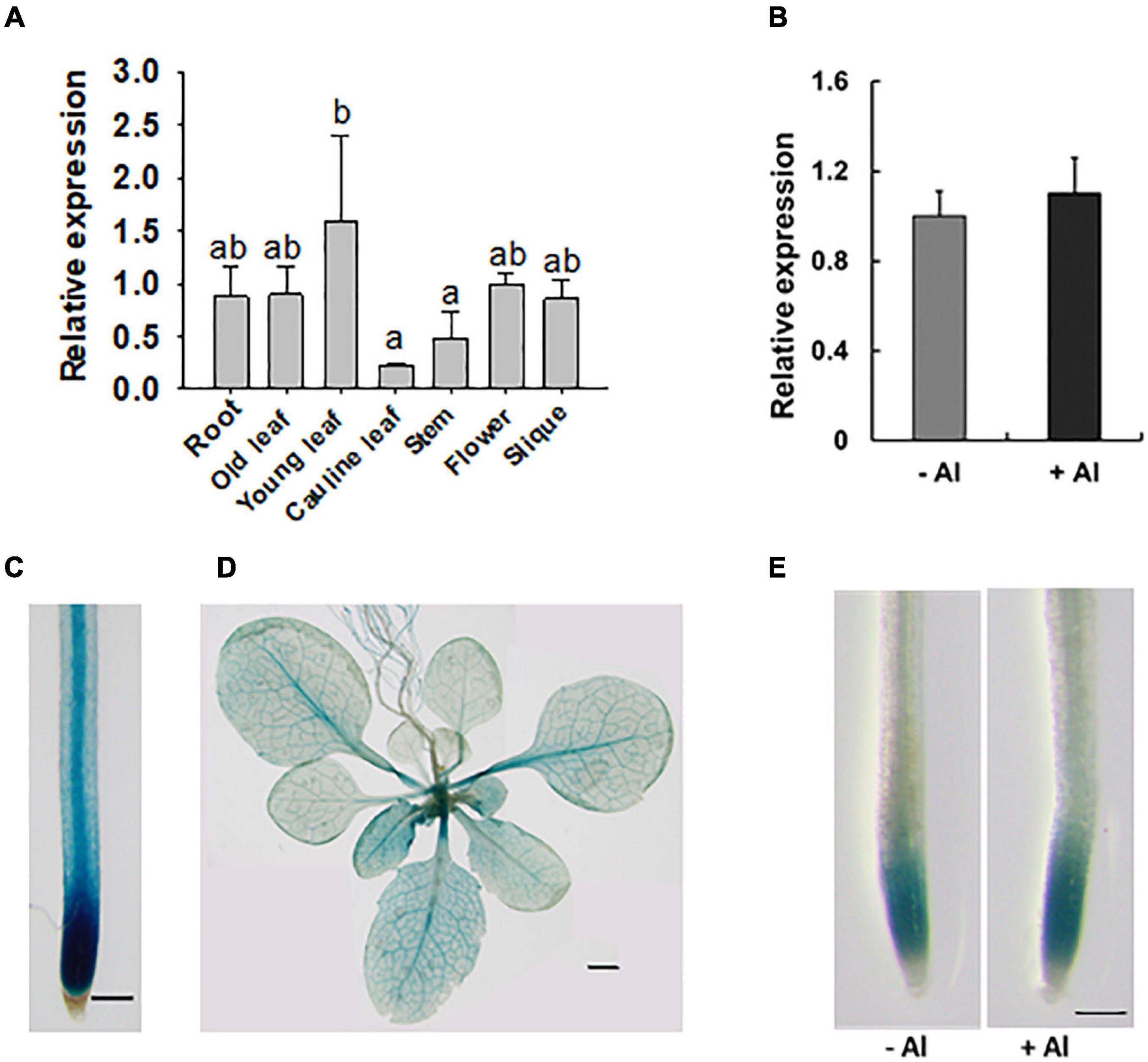
Figure 4. Expression pattern of RAE2. (A) Expression analysis of RAE2 in roots, old and young leaves, cauline leaves, stems, flowers, and siliques. Values are means ± SD (n = 3). Means with different letters are significantly different (P < 0.05, ANOVA followed by Tukey’s test). (B) Effect of Al stress on RAE2 expression. Seven-day-old seedlings were exposed to 0 (–Al) or 30 μM Al (+Al) for 12 h, and the roots were sampled for the expression analysis. Values are means ± SD (n = 3). (C–E) Detection of GUS activity in pRAE2:GUS transgenic lines. (C) GUS expression is preferentially detected in the root tips and central vascular tissues of mature root zones. Scale bar = 200 μm. (D) GUS expression in the shoots of three-week-old plants. Scale bar = 1 mm. (E) GUS expression in roots of 7-day-old seedlings in response to 0 (–Al) or 30 μM Al (+Al) for 12 h. Scale bar = 200 μm.
To determine the subcellular localization of RAE2, we made constructs of 35S:RAE2-GFP and 35S:GFP-RAE2 and then transformed each construct into Arabidopsis protoplasts. Both RAE2-GFP and GFP-RAE2 green fluorescent signals were predominantly detected in the nucleus, whereas the signal of GFP control was found in both the nucleus and the cytoplasm (Supplementary Figure 2). These results indicate that RAE2 is localized in the nucleus, which is in agreement with a previous report (Sorensen et al., 2017).
Since the expression of STOP1-regulated genes was decreased in rae2 (Figure 1), we examined whether the rae2 mutation influenced STOP1 protein accumulation. We crossed a previously generated transgenic line of pSTOP1:STOP1-HA (Zhang et al., 2019) with rae2 to introduce the transgene into the mutant background. We compared the STOP1-HA protein level in Col-0 WT, rae2, and rae3. The result showed that STOP1-HA protein level in the roots was lower in rae2 than in WT under both –Al and +Al conditions (Figure 5A). The reduced STOP1-HA protein accumulation in rae3 was more prominent than that in rae2. We also determined the STOP1-HA protein level in the shoots, and the result showed that the STOP1-HA protein accumulation was reduced in the shoots as well (Figure 5B).
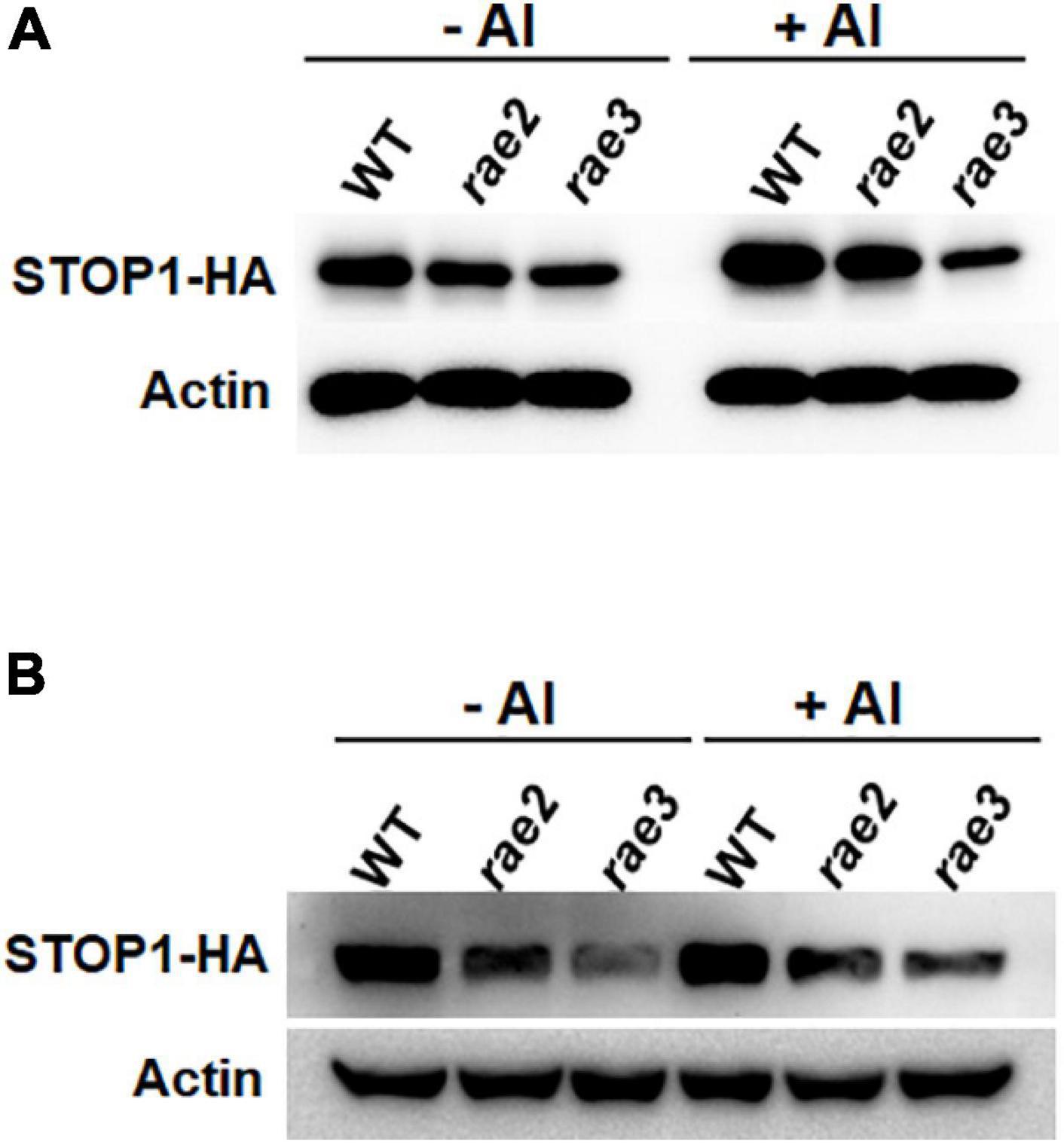
Figure 5. Mutation of RAE2 reduces STOP1 protein accumulation. (A,B) Comparison of STOP1-HA protein level in roots (A) and shoots (B) of WT, rae2, and rae3 harboring pSTOP1:STOP1-HA transgene. Seedlings with 12 days old were exposed to 0 (–Al) or 30 μM Al (+Al) for 12 h. The STOP1-HA and internal control actin was detected by anti-HA and anti-actin antibodies, respectively.
To investigate whether RAE2 modulates STOP1 protein accumulation via a similar mechanism to RAE3/HPR1, which regulates nucleocytoplasmic STOP1 mRNA export (Guo et al., 2020), we isolated the nuclei of WT, rae2, and rae3, and then quantified the expression levels of the Al-resistance genes in the nucleus. Generally, the levels of nuclear mRNA of STOP1-regulated genes were similarly decreased in rae2 and rae3 under Al stress conditions, except that the expression of AtALMT1 in the nucleus was reduced in rae2 but not in rae3 compared to WT (Figures 6A–D). The nuclear mRNA levels of AtSTAR1 and ALS1 not targeted by STOP1 were similar among the three lines (Figures 6E,F). Noticeably, although rae3 accumulated more STOP1 mRNA in the nucleus than WT as previously reported (Guo et al., 2020), the level of nuclear STOP1 mRNA in rae2 did not differ from that in WT (Figure 6G). Together, these results demonstrate that RAE2 is involved in the modulation of STOP1 protein accumulation, which might be not through regulating nucleocytoplasmic STOP1 mRNA export.
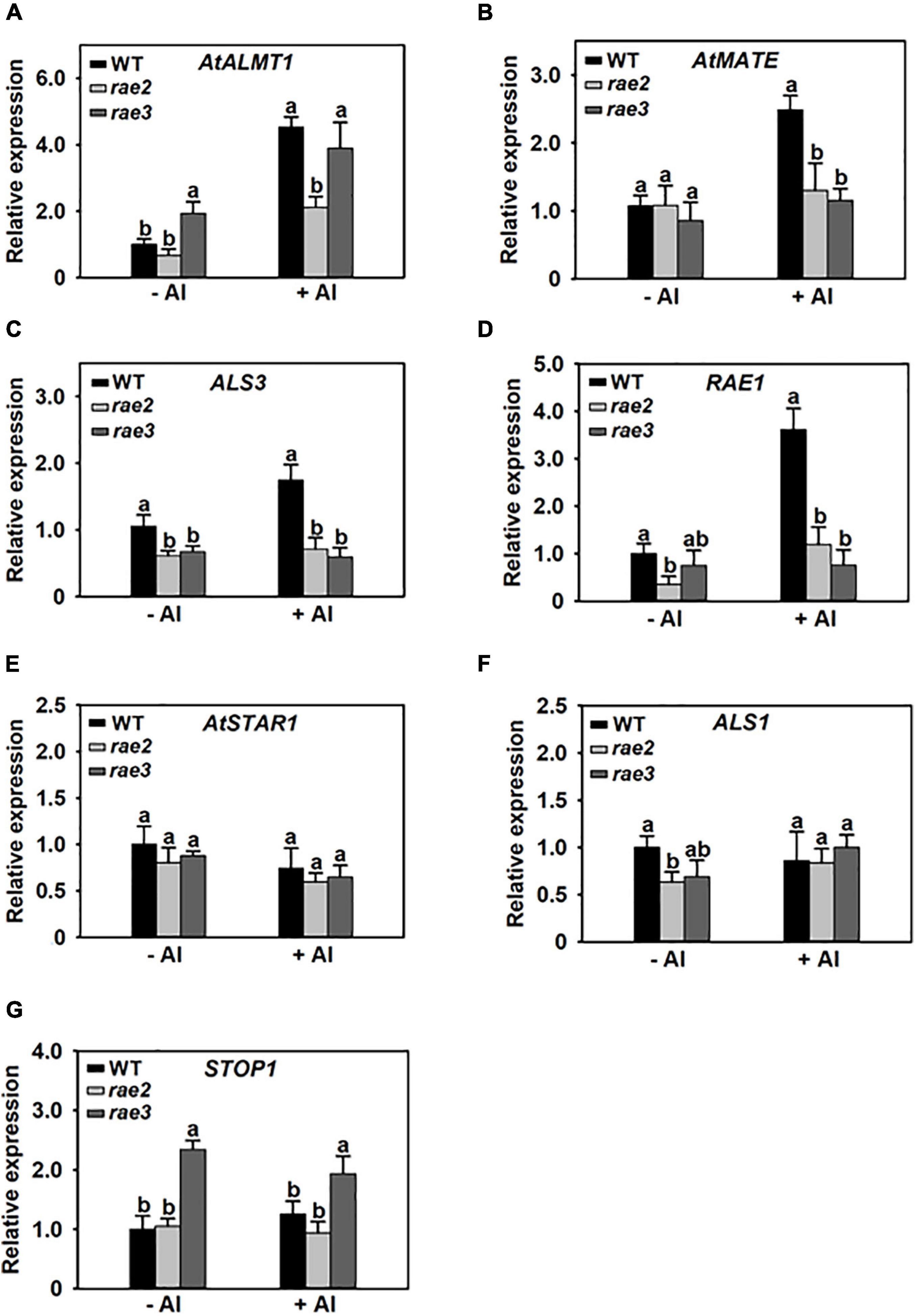
Figure 6. The rae2 mutation does not influence STOP1 mRNA export. Three-week-old plants of WT, rae2, and rae3 were treated with 0 (–Al) or 30 μM Al (+Al) for 12 h, and then the root nuclei were isolated for the real-time RT-PCR analysis of Al-resistance genes, which include AtALMT1 (A), AtMATE (B), ALS3 (C), RAE1 (D), AtSTAR1 (E), ALS1 (F), and STOP1 (G). Values are means ± SD (n = 3). Means with different letters are significantly different (P < 0.05, ANOVA followed by Tukey’s test).
To investigate whether the decreased expression of STOP1-regulated genes is responsible for the reduced Al resistance and low Pi response in rae2, we introduced rae1 mutation into the rae2 mutant background via crossing. RAE1 encodes an F-box protein that negatively regulates the expression of STOP1-downstream genes through the modulation of STOP1 stability (Zhang et al., 2019). Expression analysis showed that the decreased expression of STOP1-regulated Al resistance genes, AtALMT1 and AtMATE, in rae2 was fully recovered in rae2 rae1 double mutant compared to WT, while that of ALS3 was partially recovered (Figures 7A–C). We then compared the Al resistance and low Pi response in WT, Atalmt1, rae2, rae1, and rae2 rae1. The results showed that the rae1 mutation could partially rescue the reduced Al resistance in rae2 (Figures 7D,E). The reduced response to Pi deficiency in rae2 was also partially rescued by the introduction of rae1 mutation (Figures 7F,G).
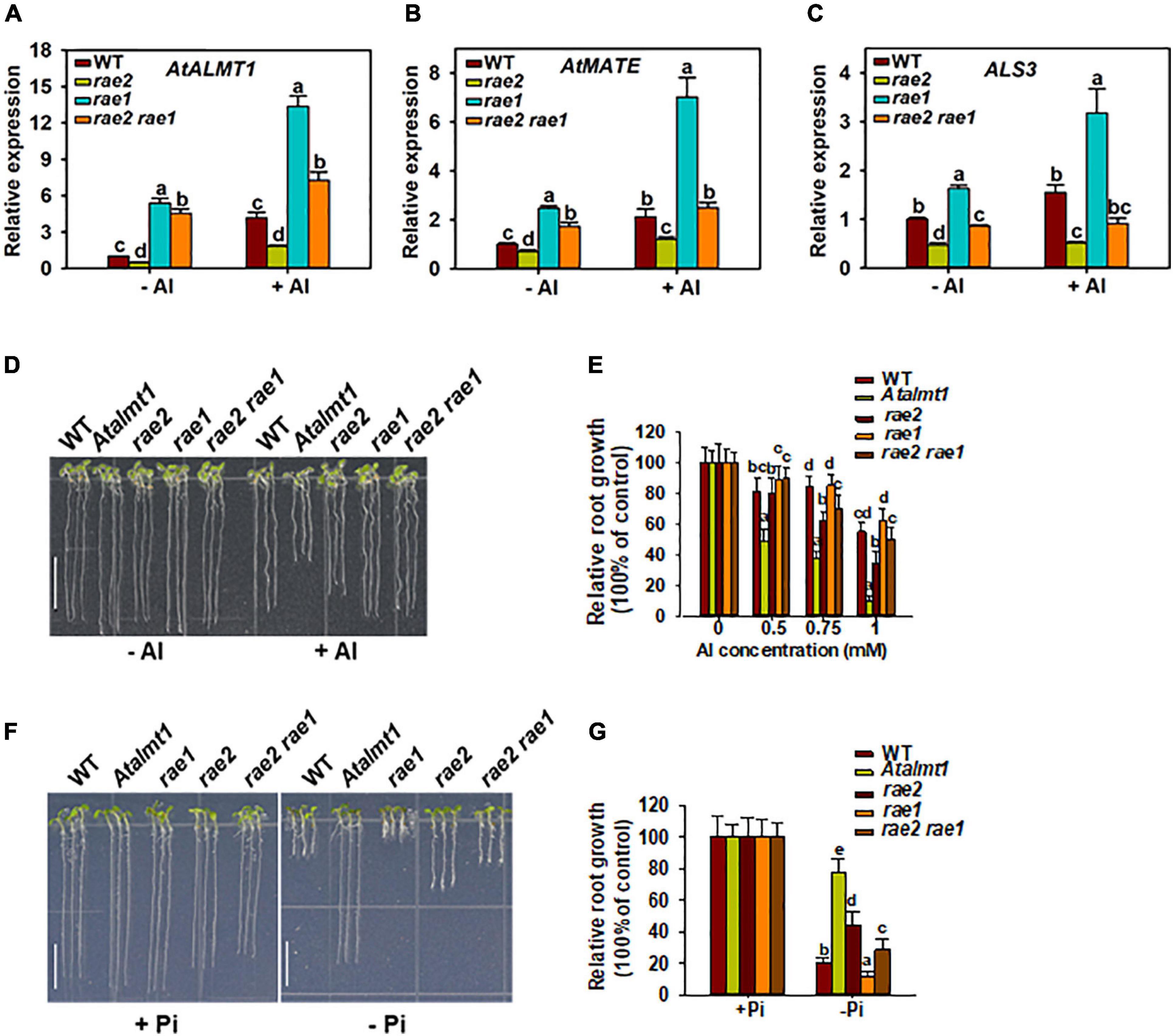
Figure 7. Introduction of rae1 mutation partially restores the defective phenotypes of rae2. (A–C) Expression analysis of STOP1-regulated genes, including AtALMT1 (A), AtMATE (B) and ALS3 (C), in WT, rae2, rae1, and rae2 rae1. Seedlings with 7 days old were exposed to 0 (–Al) or 30 μM Al (+Al) for 12 h, and then roots were excised for the expression analysis. Values are means ± SD (n = 3). (D,E) Representative images (D) and Al resistance (E) of WT, Atalmt1, rae2, rae1, and rae2 rae1 under different Al concentrations. Values are means ± SD (n = 13–18). (F,G) Representative images (F) and relative root growth (G) of WT, Atalmt1, rae2, rae1, and rae2 rae1 at 20 (–Pi) or 1,000 Pi (+Pi). Scale bar = 7 mm. Values are means ± SD (n = 39–56). Means with different letters are significantly different (P < 0.05, ANOVA followed by Tukey’s test).
The THO complex is conserved across a wide range of organisms. Components of THO have been reported to regulate mRNA export or the trafficking of small RNA precursors to be involved in diverse biological processes in plants (Furumizu et al., 2010; Jauvion et al., 2010; Yelina et al., 2010; Francisco-Mangilet et al., 2015; Tao et al., 2016; Doll et al., 2018). We previously discovered that the THO complex component RAE3/HPR1 regulates STOP1 mRNA export to be involved in the regulation of Al resistance (Guo et al., 2020). In this study, we found that another component of the THO complex, RAE2/TEX1, is also involved in regulating Al resistance. We showed that the expression of STOP1-downstream genes and STOP1 protein accumulation were reduced in rae2 and the introduction of rae1 mutation into the rae2 background could partially rescue the increased sensitivity of rae2 to Al stress, which suggests that RAE2 modulates Al resistance partially through the regulation of STOP1 protein level. Nevertheless, unlike RAE3/HPR1, RAE2 does not regulate STOP1 mRNA export (Figure 6). The underlying mechanism as to how RAE2 regulates STOP1 protein accumulation is still unknown. THO components are also reported to be able to physically associate with 5′ cap-binding complex (CBC), exon junction complex (EJC), and 3′-end processing factors, implying that THO/TREX might be involved in the regulation of 5′-capping, splicing, and 3′-end processing (Cheng et al., 2006; Johnson et al., 2009; Gromadzka et al., 2016; Heath et al., 2016). Whether RAE2 regulates STOP1 accumulation through the modulation of STOP1 mRNA processing remains to be investigated in the future.
Compared to rae3, the increased Al sensitivity in rae2 is less prominent (Figures 2C,D). Although rae2 and rae3 showed a decreased expression of AtALMT1 at a similar level (Figure 1C), the reduction in malate secretion was stronger in rae3 than in rae2 (Figure 2A), which could be attributed to the fact that RAE3 instead of RAE2 is also involved in the regulation of AtALMT1 mRNA export (Figure 6A; Guo et al., 2020). These results suggest that the different Al sensitivity between rae3 and rae2 may be due to their difference in the level of malate exudation. In contrast, although AtALMT1-mediated malate exudation plays a positive role in low Pi-triggered root growth inhibition (Balzergue et al., 2017; Mora-Macias et al., 2017), the root growth was less inhibited in rae2 than in rae3 under low Pi conditions (Figures 2E,F), suggesting that RAE2 also regulates other genes to modulate low Pi response. In addition to AtALMT1, LPR1/LPR2 and PDR2 have also been demonstrated to play critical roles in the regulation of low Pi response by modulating Fe accumulation in root tips (Svistoonoff et al., 2007; Ticconi et al., 2009; Muller et al., 2015). CLE14 peptide and BR signaling pathway components BKI1 and BES1/BZR1 were found to be involved in the low Pi response as well (Singh et al., 2014, 2018; Hirayama et al., 2018). It remains to be determined whether RAE2 is involved in the regulation of these genes required for low Pi response.
Knockout of THO2 encoding a subunit of THO complex causes embryonic or early seedling lethality (Furumizu et al., 2010; Jauvion et al., 2010; Francisco-Mangilet et al., 2015), indicating that THO2 is essential. By contrast, homozygous rae2 and rae3 mutants are viable, but the rae2 rae3 double mutant could not be obtained, suggesting that RAE2 and RAE3 play redundant roles in the regulation of plant growth and that plant THO/TREX complex is likely to be a dynamic structure in which RAE2 and RAE3 are interchangeable. Our results demonstrate that RAE2 and RAE3 play overlapping but distinct roles in the regulation of Al resistance and low Pi response. Further work is required to determine whether other subunits of the THO complex are also involved in the modulation of the response to Al and low Pi stresses.
The original contributions presented in the study are publicly available. This data can be found here: NCBI repository, accession number: SRR14277220.
All authors conceived the project. C-FH drafted the manuscript. Y-FZ, JG, and YZ performed the experiments. Y-FZ and JG helped to analyze the data and wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by Shanghai Natural Science Foundation (Grant No. 20ZR1466500), National Natural Science Foundation of China (Grant No. 31870223 to C-FH), National Key Laboratory of Plant Molecular Genetics, and the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.698443/full#supplementary-material
Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178. doi: 10.1038/nbt.2095
Aguilera, A. (2005). Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17, 242–250. doi: 10.1016/j.ceb.2005.03.001
Balzergue, C., Dartevelle, T., Godon, C., Laugier, E., Meisrimler, C., Teulon, J. M., et al. (2017). Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 8:15300. doi: 10.1038/Ncomms15300
Chavez, S., Beilharz, T., Rondon, A. G., Erdjument-Bromage, H., Tempst, P., Svejstrup, J. Q., et al. (2000). A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19, 5824–5834. doi: 10.1093/emboj/19.21.5824
Cheng, H., Dufu, K., Lee, C. S., Hsu, J. L., Dias, A., and Reed, R. (2006). Human mRNA export machinery recruited to the 5’ end of mRNA. Cell 127, 1389–1400. doi: 10.1016/j.cell.2006.10.044
Doll, S., Kuhlmann, M., Rutten, T., Mette, M. F., Scharfenberg, S., Petridis, A., et al. (2018). Accumulation of the coumarin scopolin under abiotic stress conditions is mediated by the Arabidopsis thaliana THO/TREX complex. Plant J. 93, 431–444. doi: 10.1111/tpj.13797
Dong, J. S., Pineros, M. A., Li, X. X., Yang, H. B., Liu, Y., Murphy, A. S., et al. (2017). An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 10, 244–259. doi: 10.1016/j.molp.2016.11.001
Fang, Q., Zhang, J., Yang, D. L., and Huang, C. F. (2021a). The SUMO E3 ligase SIZ1 partially regulates STOP1 SUMOylation and stability in Arabidopsis thaliana. Plant Signal. Behav. 16:1899487. doi: 10.1080/15592324.2021.1899487
Fang, Q., Zhang, J., Zhang, Y., Fan, N., van den Burg, H. A., and Huang, C. F. (2020). Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32, 3921–3938. doi: 10.1105/tpc.20.00687
Fang, Q., Zhou, F., Zhang, Y., Singh, S., and Huang, C. F. (2021b). Degradation of STOP1 mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 106, 493–506. doi: 10.1111/tpj.15181
Folta, K. M., and Kaufman, L. S. (2006). Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat. Protoc. 1, 3094–3100. doi: 10.1038/nprot.2006.471
Francisco-Mangilet, A. G., Karlsson, P., Kim, M. H., Eo, H. J., Oh, S. A., Kim, J. H., et al. (2015). THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 82, 1018–1029. doi: 10.1111/tpj.12874
Furukawa, J., Yamaji, N., Wang, H., Mitani, N., Murata, Y., Sato, K., et al. (2007). An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–1091. doi: 10.1093/pcp/pcm091
Furumizu, C., Tsukaya, H., and Komeda, Y. (2010). Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16, 1809–1817. doi: 10.1261/rna.2265710
Gromadzka, A. M., Steckelberg, A. L., Singh, K. K., Hofmann, K., and Gehring, N. H. (2016). A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 44, 2348–2361. doi: 10.1093/nar/gkw009
Guo, J., Zhang, Y., Gao, H., Li, S., Wang, Z. Y., and Huang, C. F. (2020). Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 228, 179–193. doi: 10.1111/nph.16658
Gutierrez-Alanis, D., Yong-Villalobos, L., Jimenez-Sandoval, P., Alatorre-Cobos, F., Oropeza-Aburto, A., Mora-Macias, J., et al. (2017). Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Dev. Cell 41, 555–570. doi: 10.1016/j.devcel.2017.05.009
Hampp, R., Goller, M., and Fullgraf, H. (1984). Determination of compartmented metabolite pools by a combination of rapid fractionation of oat mesophyll protoplasts and enzymic cycling. Plant Physiol. 75, 1017–1021. doi: 10.1104/Pp.75.4.1017
Heath, C. G., Viphakone, N., and Wilson, S. A. (2016). The role of TREX in gene expression and disease. Biochem. J. 473, 2911–2935. doi: 10.1042/Bcj20160010
Hirayama, T., Lei, G. J., Yamaji, N., Nakagawa, N., and Ma, J. F. (2018). The putative peptide gene FEP1 regulates iron deficiency response in Arabidopsis. Plant Cell Physiol. 59, 1739–1752. doi: 10.1093/pcp/pcy145
Hoekenga, O. A., Maron, L. G., Pineros, M. A., Cancado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 9738–9743. doi: 10.1073/pnas.0602868103
Hoekenga, O. A., Vision, T. J., Shaff, J. E., Monforte, A. J., Lee, G. P., Howell, S. H., et al. (2003). Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping. A physiologically simple but genetically complex trait. Plant Physiol. 132, 936–948. doi: 10.1104/pp.103.023085
Huang, C. F., Yamaji, N., and Ma, J. F. (2010). Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 153, 1669–1677. doi: 10.1104/pp.110.155028
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Ikka, T., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 104, 9900–9905. doi: 10.1073/pnas.0700117104
Jauvion, V., Elmayan, T., and Vaucheret, H. (2010). The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22, 2697–2709. doi: 10.1105/tpc.110.076638
Johnson, S. A., Cubberley, G., and Bentley, D. L. (2009). Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3 ’ end processing factor Pcf11. Mol. Cell 33, 215–226. doi: 10.1016/j.molcel.2008.12.007
Larsen, P. B., Geisler, M. J. B., Jones, C. A., Williams, K. M., and Cancel, J. D. (2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 41, 353–363. doi: 10.1111/j.1365-313X.2004.02306.x
Ligaba-Osena, A., Fei, Z. J., Liu, J. P., Xu, Y. M., Shaff, J., Lee, S. C., et al. (2017). Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytol. 214, 830–841. doi: 10.1111/nph.14420
Liu, J. P., Magalhaes, J. V., Shaff, J., and Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Liu, J. P., Pineros, M. A., and Kochian, L. V. (2014). The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 56, 221–230. doi: 10.1111/jipb.12162
Luna, R., Rondon, A. G., and Aguilera, A. (2012). New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochim. Biophys. Acta 1819, 514–520. doi: 10.1016/j.bbagrm.2011.11.012
Ma, J. F., Ryan, P. R., and Delhaize, E. (2001). Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278. doi: 10.1016/S1360-1385(01)01961-6
Magalhaes, J. V., Liu, J., Guimaraes, C. T., Lana, U. G. P., Alves, V. M. C., Wang, Y. H., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161. doi: 10.1038/ng2074
Mora-Macias, J., Ojeda-Rivera, J. O., Gutierrez-Alanis, D., Yong-Villalobos, L., Oropeza-Aburto, A., Raya-Gonzalez, J., et al. (2017). Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc. Natl. Acad. Sci. U.S.A. 114, E3563–E3572. doi: 10.1073/pnas.1701952114
Muller, J., Toev, T., Heisters, M., Teller, J., Moore, K. L., Hause, G., et al. (2015). Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230. doi: 10.1016/j.devcel.2015.02.007
Pan, H. R., Liu, S. M., and Tang, D. Z. (2012). HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 69, 831–843. doi: 10.1111/j.1365-313X.2011.04835.x
Ryan, P., Delhaize, E., and Jones, D. (2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 527–560. doi: 10.1146/annurev.arplant.52.1.527
Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313X.2003.01991.x
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. doi: 10.1104/pp.108.134700
Singh, A. P., Fridman, Y., Friedlander-Shani, L., Tarkowska, D., Strnad, M., and Savaldi-Goldstein, S. (2014). Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol. 166, 678–688. doi: 10.1104/pp.114.245019
Singh, A. P., Fridman, Y., Holland, N., Ackerman-Lavert, M., Zananiri, R., Jaillais, Y., et al. (2018). Interdependent nutrient availability and steroid hormone signals facilitate root growth plasticity. Dev. Cell 46, 59–72. doi: 10.1016/j.devcel.2018.06.002
Sorensen, B. B., Ehrnsberger, H. F., Esposito, S., Pfab, A., Bruckmann, A., Hauptmann, J., et al. (2017). The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 93, 283–298. doi: 10.1007/s11103-016-0561-9
Su, Z. X., Zhao, L. H., Zhao, Y. Y., Li, S. F., Won, S., Cai, H. Y., et al. (2017). The THO complex non-cell-autonomously represses female germline specification through the TAS3-ARF3 module. Curr. Biol. 27, 1597–1609. doi: 10.1016/j.cub.2017.05.021
Svistoonoff, S., Creff, A., Reymond, M., Sigoillot-Claude, C., Ricaud, L., Blanchet, A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792–796. doi: 10.1038/ng2041
Tao, S. B., Zhang, Y., Wang, X. Y., Xu, L., Fang, X. F., Lu, Z. J., et al. (2016). The THO/TREX complex active in miRNA biogenesis negatively regulates root-associated acid phosphatase activity induced by phosphate starvation. Plant Physiol. 171, 2841–2853. doi: 10.1104/pp.16.00680
Ticconi, C. A., Lucero, R. D., Sakhonwasee, S., Adamson, A. W., Creff, A., Nussaume, L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U.S.A. 106, 14174–14179. doi: 10.1073/pnas.0901778106
Tokizawa, M., Enomoto, T., Ito, H., Wu, L., Kobayashi, Y., Mora-Macias, J., et al. (2021). High affinity promoter binding of STOP1 is essential for early expression of novel aluminum-induced resistance genes GDH1 and GDH2 in Arabidopsis. J. Exp. Bot. 72, 2769–2789. doi: 10.1093/jxb/erab031
von Uexkull, H. R., and Mutert, E. (1995). Global extent, development and economic-impact of acid soils. Plant Soil 171, 1–15. doi: 10.1007/BF00009558
Xu, J., Zhu, J., Liu, J., Wang, J., Ding, Z., and Tian, H. (2021). SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis. J. Integr. Plant Biol. 63, 1147–1160. doi: 10.1111/jipb.13091
Yelina, N. E., Smith, L. M., Jones, A. M. E., Patel, K., Kelly, K. A., and Baulcombe, D. C. (2010). Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13948–13953. doi: 10.1073/pnas.0911341107
Keywords: aluminum resistance, Arabidopsis thaliana, low phosphate response, STOP1, TEX1, THO/TREX complex
Citation: Zhu Y-F, Guo J, Zhang Y and Huang C-F (2021) The THO/TREX Complex Component RAE2/TEX1 Is Involved in the Regulation of Aluminum Resistance and Low Phosphate Response in Arabidopsis. Front. Plant Sci. 12:698443. doi: 10.3389/fpls.2021.698443
Received: 21 April 2021; Accepted: 18 June 2021;
Published: 12 July 2021.
Edited by:
Luis E. Hernandez, Autonomous University of Madrid, SpainReviewed by:
Paul Larsen, University of California, Riverside, United StatesCopyright © 2021 Zhu, Guo, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao-Feng Huang, aHVhbmdjZkBwc2MuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.