- 1Henan Collaborative Innovation Center of Modern Biological Breeding, Henan Institute of Science and Technology, Xinxiang, China
- 2Department of Biology, East Carolina University, Greenville, NC, United States
- 3Elizabeth City State University, Elizabeth City, NC, United States
- 4College of Life Sciences, Anhui Normal University, Wuhu, China
- 5Peanut Research Institute, Luohe Academy of Agricultural Sciences, Luohe, China
The mechanism of miRNA-mediated root growth and development in response to nutrient deficiency in peanut (Arachis hypogaea L.) is still unclear. In the present study, we found that both nitrogen (N) and potassium (K) deficiency resulted in a significant reduction in plant growth, as indicated by the significantly decreased dry weight of both shoot and root tissues under N or K deficiency. Both N and K deficiency significantly reduced the root length, root surface area, root volume, root vitality, and weakened root respiration, as indicated by the reduced O2 consuming rate. N deficiency significantly decreased primary root length and lateral root number, which might be associated with the upregulation of miR160, miR167, miR393, and miR396, and the downregulation of AFB3 and GRF. The primary and lateral root responses to K deficiency were opposite to that of the N deficiency condition. The upregulated miR156, miR390, NAC4, ARF2, and AFB3, and the downregulated miR160, miR164, miR393, and SPL10 may have contributed to the growth of primary roots and lateral roots under K deficiency. Overall, roots responded differently to the N or K deficiency stresses in peanuts, potentially due to the miRNA-mediated pathway and mechanism.
Introduction
Cultivated peanut (Arachis hypogaea L.) is one of the three major legume grain and oilseed crops in China, with a planting area of over 5.0 × 106 ha and annual pod yields of more than 1.6 × 107 t in recent years, which plays a significant role in ensuring nutritional food security in China (Zhang et al., 2020). However, since the soil has been frequently over-exploited in modern agricultural practices, nitrogen (N) and potassium (K) have been the important limiting factors. Therefore, they were often frequently supplemented in the form of fertilizers (Kuicheski et al., 2015).
There is a large amount of N fertilizer usage all over the world. Still, N fertilizer’s recovery rate or efficiency is relatively low in arable land, accounting for only 25–50% of the application amount (Sharma and Bali, 2018; Omara et al., 2019). K deficiency is very common. For example, in China, more than one-third of cultivated soil is in the state of K deficiency (soil available K content is 50–70 mg/kg) or serious K deficiency (soil available K content is less than 50 mg/kg; Wang and Wu, 2009). Therefore, it is of great significance for optimizing nutrient management, particularly the supply of N and K in balance with crop demand and for the genetic improvement of peanut to explore the response mechanism of N deficiency and K deficiency in peanut.
Since N is involved in synthesizing important components such as proteins, nucleic acids, phospholipids, chlorophyll, enzymes, vitamins, alkaloids, and certain growth hormones in plants, it is also called the element of life (Hawkesford et al., 2012; Kant, 2018). The common symptoms of N deficiency include leaf yellowing, cell division slowing down, and enzyme activity decreasing and then leading to plant growth retardation (Salvagiotti et al., 2008). Compared with N, K does not directly participate in synthesizing any chemical substances in plants, but it is the most abundant cation in plants (Salvagiotti et al., 2008). It is widely distributed in various tissues and organs of plants, accounting for about 2–10% of the dry weight of plants (Leigh and Wyn Jones, 1984). K plays a very important role in maintaining cytoplasmic charge balance, regulating enzymatic reaction, promoting protein synthesis, and maintaining cell osmotic pressure and other physiological activities (Leigh and Wyn Jones, 1984). At the same time, K also participates in a series of physiological and biochemical processes, such as stomatal movement, cell signal transduction, cell elongation, and phloem material transport (Sustr et al., 2019). Proper K application is conducive to regulating nutrient transport and distribution in peanuts, promoting nodule formation, improving nodule nitrogen fixation capacity (Wang et al., 2013), regulating carbon and N metabolism, and improving peanut yield, quality, and resistance (Helmy and Ramadan, 2014; Almeida et al., 2015; Chakraborty et al., 2016).
The root is the first plant organ to sense and absorb nutrients, and their morphological response to different growth conditions varies greatly (Gruber et al., 2013). Plant roots show a high degree of plasticity for changes in the availability of nutrient resources, and the degree of this plasticity depends on the plant species in many cases (Sun et al., 2017; Sustr et al., 2019). Apart from the role as a nutrient element, nitrate or ammonium (the main form of N that can be taken by the roots) acting as a signal regulate many physiological processes, including the development of root system architecture (RSA; Sun et al., 2017; Kant, 2018) by regulating cell division and expansion (Luo et al., 2020). The growth and development of RSA depend on K supply that affected root differentiation and elongation (Sustr et al., 2019). Sufficient cytoplasmic K level is required for protein synthesis and enzyme activity in root cells to maintain cytoplasmic pH (Walker et al., 1998) and protein anion charge (Maathuis and Sanders, 1996). In addition, the cells in the elongation region need expansion pressure, which is established by osmotically active substances (including K; Pritchard, 1994; Dolan and Davies, 2004). In the mature zone of roots, root hairs grow through K flux (Guilhem et al., 2003; Zhao et al., 2016). In addition, the RSA and root hair coverage rate in plants changed adaptively, thus improving K uptake under K limitation (Guilhem et al., 2003; Zhao et al., 2016).
Plants have evolved several physiological and molecular adaptive responses to cope with nutrient deficiency stresses (Kuicheski et al., 2015; Nguyen et al., 2015). microRNAs (miRNAs) are short (21–24 nucleotide) RNAs, which can bind to RNA-induced silencing complex (RISC), recognize its target mRNAs, and regulate the expression of the targeted genes through transcriptional cleavage and translation inhibition (Zhang et al., 2006). If miRNA and mRNA are completely complementary, miRNA cleaves the phosphodiester bond between two nucleotides of the target mRNA sequence (generally between the 10th and 11th nucleotides of miRNA), that is to guide the specific cleavage of mRNA; if not, it will cause translation inhibition (Zhang et al., 2006; Sunkar et al., 2012). Studies have shown that miRNAs can regulate the quantity and spatiotemporal accumulation of target mRNAs and thus play a key regulatory role in plant stress response (Sunkar et al., 2012; Zhang, 2015), including in nutrient deficiency stress (Kuicheski et al., 2015; Nguyen et al., 2015).
Studies have shown that nutrient availability would change the expression of miRNAs and their target mRNAs and then affect the growth and development of plants (Kuicheski et al., 2015). Furthermore, different crops respond differently to nutritional deficiencies (Kuicheski et al., 2015; Fontana et al., 2020; Thornburg et al., 2020). However, the mechanism of miRNAs related to root growth and development in response to nutrient deficiency in peanuts is still unclear. In this work, we systematically explored the response of root development and morphology to N deficiency and K deficiency and the potential miRNA-mediated mechanism during these signs of progress. The observation of the expression of the miRNAs and their targeted mRNAs in peanut plants exposed to N deficiency and K deficiency could reveal the mechanisms of the plant suffering from nutrient deficiency stress and help find candidate genes to develop important tools for plant nutrition breeding.
Materials and Methods
Plant Growth Conditions, Nutrient Stress Treatments, and Sampling
The peanut (Arachis hypogaea L. cv. Yuanza 9,102) seeds of the same size, fully mature, and free of diseases and insect pests were selected, soaked in 10% H2O2 solution for 5 min washed with deionized water five to six times. The seeds were wrapped in absorbent paper, soaked in a plastic bucket containing 2 L saturated CaSO4, and placed in the artificial climate chamber (constant temperature 28°C, blue light irradiation) for germination. When the root length of peanut seedlings was about 10 cm (4 days), the seedlings with uniform appearance were selected and planted in plastic pots containing 5 L of modified Hoagland nutrient solution (pH 6.3 ± 0.1): (1) normal N (NN) nutrient solution; (2) low N (LN) nutrient solution; (3) normal K (NK) nutrient solution; (4) low K (LK) nutrient solution. The nutrient solution group of NN consists of: 2.5 mM Ca(NO3)2, 2.5 mM KCl, 2 mM NaCl, 1 mM MgSO4, 0.5 mM KH2PO4, 0.1 mM EDTA-FeNa, 2 × 10−2 mM H3BO3, 1 × 10−3 mM ZnSO4, 1 × 10−3 mM MnSO4 2 × 10−4 mM·L−1 CuSO4, and 5 × 10−6 mM (NH4)6Mo7O24. Compared with the nutrient solution group of NN, the 2.5 mM Ca(NO3)2 was replaced by 0.05 mM Ca(NO3)2 and 2.45 mM CaCl2 in the nutrient solution group of LN. The nutrient solution group of NK consists of: 2.5 mM Ca(NO3)2, 2.5 mM KCl, 2 mM NaCl, 1 mM MgSO4, 0.5 mM NH4H2PO4, 0.1 mM EDTA-FeNa, 2 × 10−2 mM H3BO3, 1 × 10−3 mM ZnSO4, 1 × 10−3 mM MnSO4, 2 × 10−4 mM·L−1 CuSO4, and 5 × 10−6 mM (NH4)6Mo7O24. Compared with the nutrient solution group of NN, the 0.5 mM KH2PO4 was replaced by 0.5 mM NH4H2PO4 in the nutrient solution group of NK. Compared with the nutrient solution group of NK, the 2.5 mM KCl was replaced by 0.02 mM KCl and 2.48 mM NaCl in the nutrient solution group of LK. The nutrient solution was changed every 3 days to avoid nutrient depletion. The light source of the growth chamber was a biological sodium lamp. The light/dark period was 14/10 h, and the day and night temperature were 30 ± 2°C/25 ± 2°C. A certain amount of deionized water was added to the culture container every day to supplement the water consumed by transpiration. The air pump was used for ventilation for 24 h. Four and eight days after the nutrient stress treatments, the plants were collected to determine the dry and fresh weight and conduct root morphological analysis; the roots were collected to measure root respiration and root vigor. In addition, the root samples from each treatment were harvested and frozen in liquid nitrogen immediately and then stored at −80°C before RNA extraction.
Plant Growth Determination
Five peanut plants were randomly selected, and the substrate on the surface of the root and dust on the surface of leaves were washed with deionized water. After the surface water was wiped off, the peanut plants were divided into two parts: the aboveground part and the root part, and the fresh weight of each part was weighed. First, the plant height was measured with a ruler. Next, all leaves were scanned by a desktop scanner (EPSON Perfection V700 Photo), and pictures were taken. Then, WinRHIZO was used to analyze the leaf area (cm2). Finally, the samples were dried at 105°C for 30 min and then to a constant weight at 80°C. Then, the dry weights of aboveground and roots were weighed, respectively.
Root Morphological Analysis
The primary root length was measured with a ruler. Then, counts were taken for the lateral root number. After that, the whole root was scanned with a desktop scanner (EPSON Perfection V700 Photo), and root pictures were taken. Finally, WinRHIZO was used to analyze the length (cm), surface area (cm2), volume (cm3), and diameter (mm) of the root samples.
Determination of Root Vitality
Root vitality was determined according to triphenyltetrazolium chloride (TTC) staining method. The roots of five seedlings from each treatment were weighed and cut into a small fragment that was ~1 cm in length. Then the root was soaked in 20 ml of 0.6% TTC solution prepared with 0.1 mol/L phosphate buffer (pH = 7.4). After 24 h, the buffer solution was poured out, and the roots were washed with deionized water three times. Then the roots were soaked in 20 ml 95% ethanol and a water bath at 85°C for 10 min to extract water-insoluble triphenylmethyl hydrazone (TTF). The absorbance of the extract was measured at 485 nm, and the root vitality was expressed as OD g−1 FW.
Root Respiration Rate Measurement
The roots of five seedlings from each treatment were weighed and put into the incubator chamber of the Clark Chlorolab2 system. Then, 2 ml saturated CaSO4 solution was added, and the oxygen (O2) consumption was measured after 10 min. The O2 consumption rate was calculated and represented by the root respiration rate, expressed as nmol min g−1 FW.
RNA Extraction and Gene Expression Analysis
About 200 mg of root samples of peanut seedlings on the 4th and 8th days of N deficiency, K deficiency, and the control treatments were used to extract total RNAs. According to our previous reports, RNA extraction and isolation were performed as outlined in the manufacturer’s protocols of the MirVana miRNA Isolation Kit (Candar-Çakir et al., 2016; Fontana et al., 2020; Yu et al., 2020). The quality of the extracted RNAs was firstly tested by Nanodrop ND-1000 (Nanodrop Technologies, Wilmington, DE, United States). One percentage agarose gel electrophoresis was used, and the quality and concentrations of RNAs were detected by micro ultraviolet spectrophotometer. Then the extracted RNA was stored at −80°C for reverse transcription.
According to the procedures of the manufacturer of TianGen miRcute Plus miRNA First Strand cDNA first Synthesis Kit (Beijing, China), the RNA samples were reverse transcribed into cDNAs with miRNA gene-specific primers and pol(T) primers of all protein-coding genes. Then the synthesized cDNA was stored at −20°C used for further analysis.
To investigate the potential mechanism of miRNA-mediated plant response to N deficiency and K deficiency, we targeted miRNAs related to plant root development and/or the dynamic balance of nutrients in other species (Nguyen et al., 2015; Li and Zhang, 2016; Zhang and Unver, 2018; Song et al., 2019). After considering all these criteria, a total of 20 miRNAs were selected for this study. They were miR156, miR160, miR162, miR164, miR165, miR166, miR167, miR169, miR171, miR172, miR319, miR390, miR393, miR395, miR399, miR396, miR778, miR827, miR847, and miR857. The expression of seven targets genes (NAC4, AFB3, ARF1, ARF2, bHLH74, SPL10, and GRF) found in the literature was also analyzed. The expression of miRNAs and their target genes were analyzed with the gene-specific forward and reverse primers according to the Tiangen miRcute Plus miRNA qPCR detection kit (Beijing, China). Each gene and miRNA expression analysis included three biological replicates and three technical replicates. The expression value was standardized with reference gene EF1A. The ΔΔCT method was used to calculate the fold change of the tested miRNAs and their target genes.
Statistical Analysis
The data of each morphological and physiological index was calculated by five biological repeats, and gene expression was obtained by three biological repeats. Statistical software SPSS version 19 was employed for ANOVA and followed by the least significant difference (LSD) test. A value of p less than 0.05 and 0.01 was considered a significant difference and extremely significant difference and marked as * and ** in the graphs.
Results
Effects of N and K Deficiency on the Growth and Development of Peanut Seedlings
As shown in Figure 1, the dry weight of shoot and root and leaf areas significantly decreased under both N and K deficiency on the 4th and 8th days. In addition, the plant height was significantly decreased under N and K deficiency on the 8th day. Compared with the seedlings cultured in the normal nutrient solutions, the dry weight of shoot, root, and total plant of the seedlings cultured in N deficiency nutrient solutions on the 8th day decreased by 29.67, 25.73, and 18.60%, respectively, and that of the seedlings cultured in K deficiency nutrient solutions on 8th day decreased by 32.97, 27.15, and 14.29%, respectively. In addition, the leaf area and the plant height of the seedlings cultured in N deficiency nutrient solutions on the 8th day decreased by 51.79 and 48.37%, respectively, and that of the seedlings cultured in K deficiency nutrient solutions on the 8th day decreased by 58.51 and 25.54%, respectively.
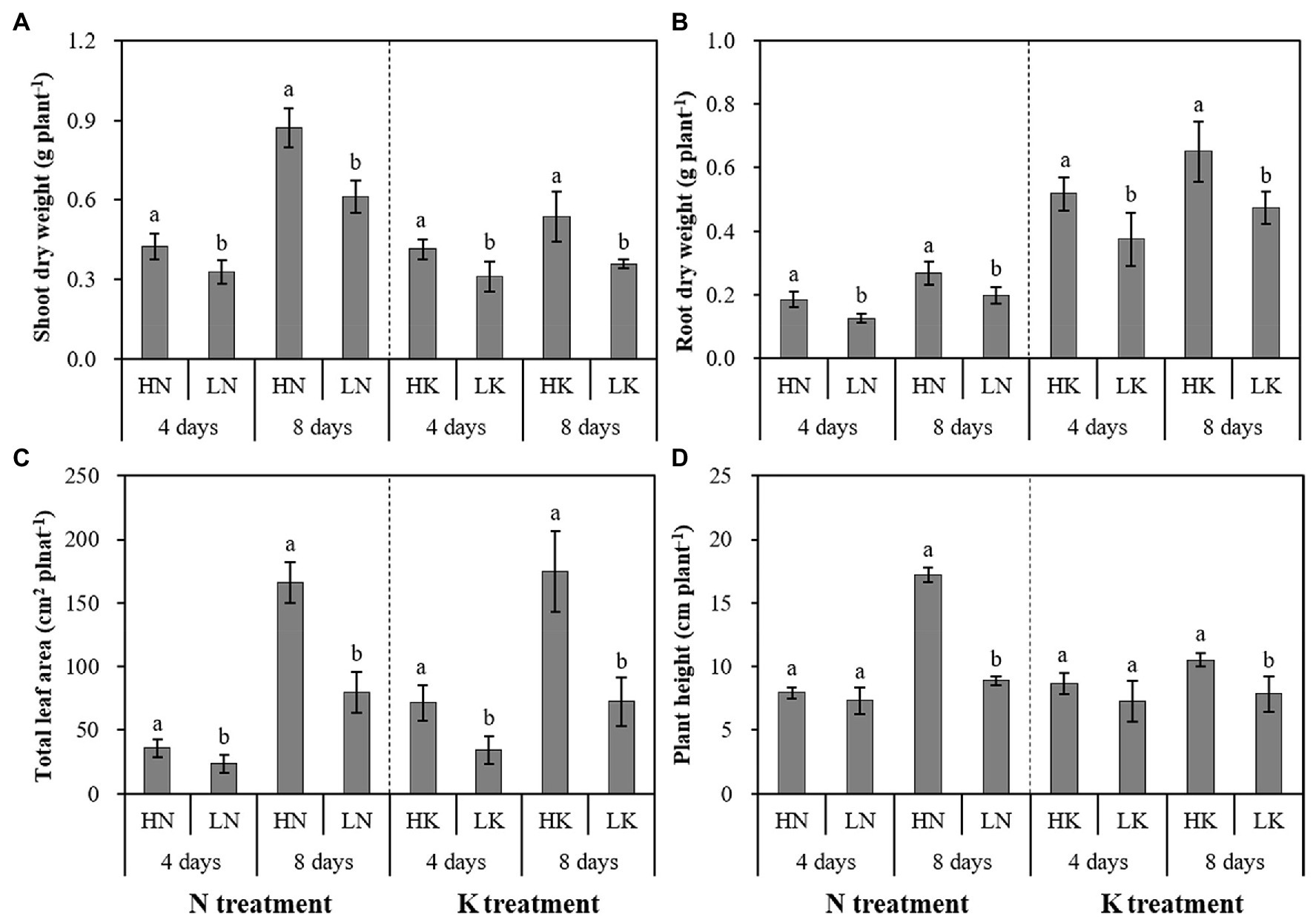
Figure 1. The effect of N deficiency and K deficiency on the shoot dry weight (A), root dry weight (B), total leaf area (C), and plant height (D) of peanut seedlings, respectively. The data are the means ± SDs (n = 5). The different letters on the bars indicate significant differences between the different N (or K) treatments and the control treatment on the same day according to the LSD test (p = 0.05). N, nitrogen; K, potassium; and LSD, least significant difference.
Effects of N and K Deficiency on Root Development of Peanut Seedlings
N and K deficiency had significant effects on root development, indicated not only by the root biomass but also the root length, size, and number of branches (Figure 2). As shown in Figure 3, compared with the seedlings cultured in the normal nutrient solutions, the total root volume and total root surface area as well as the total root length of the seedlings, cultured in N deficiency nutrient solutions on the 8th day, decreased by 50.29, 40.78, and 29.50%, respectively, and that of the seedlings cultured in K deficiency nutrient solutions on 8th day decreased by 19.06, 21.57, and 24.42%, respectively. On the other hand, compared with the seedlings cultured in the normal nutrient solutions, the average root diameter had no significant change under K deficiency conditions. At the same time, it increased significantly under 8 days of low N treatment.
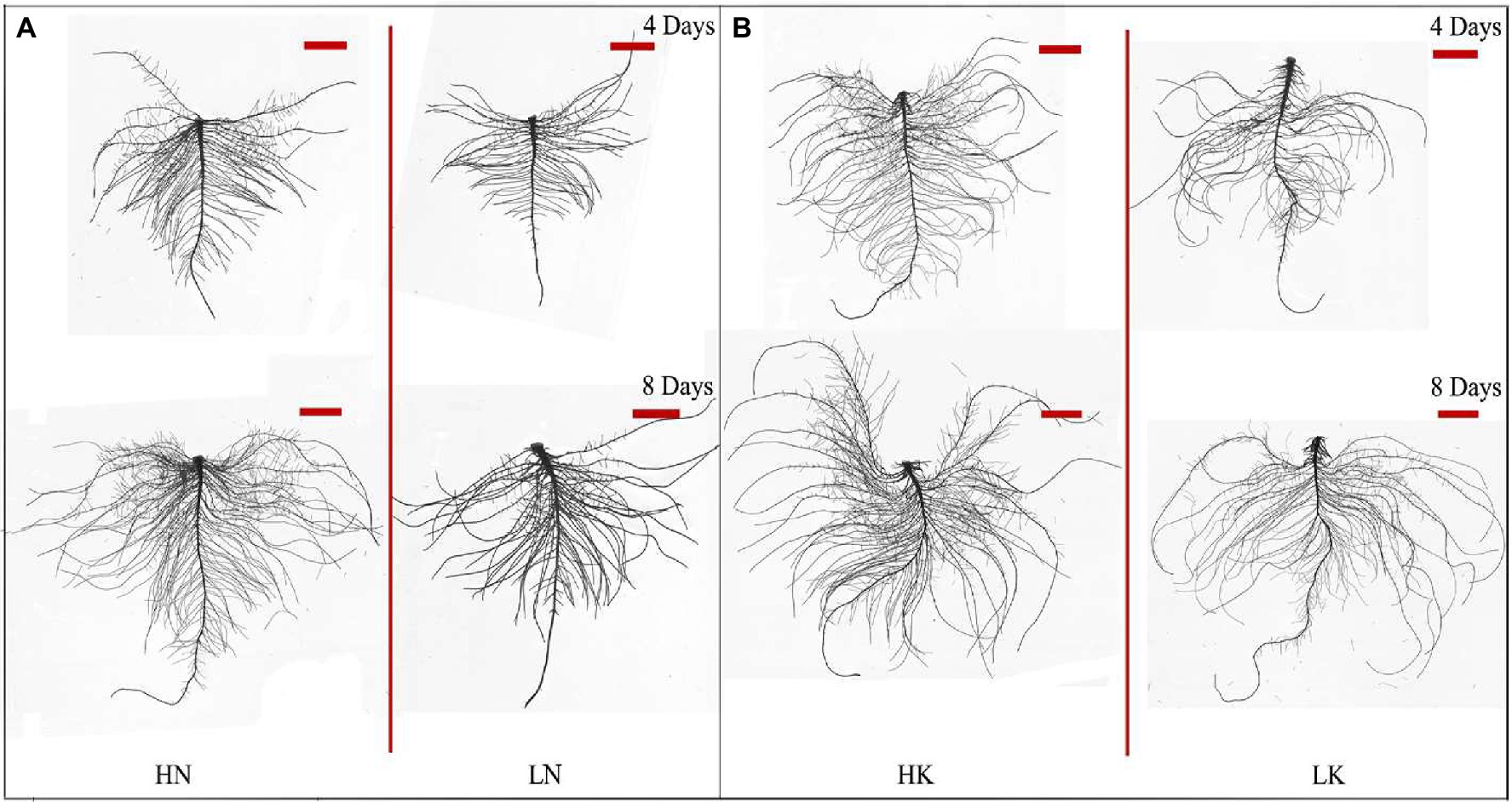
Figure 2. Root morphology difference between high N (HN) group and low N (LN) group (A) and the difference between high K (HK) group and low K (LK) group (B) after 4 and 8 days of treatments. N, nitrogen and K, potassium. The original size of the target bar is 0.2 cm high and 3.5 cm wide.
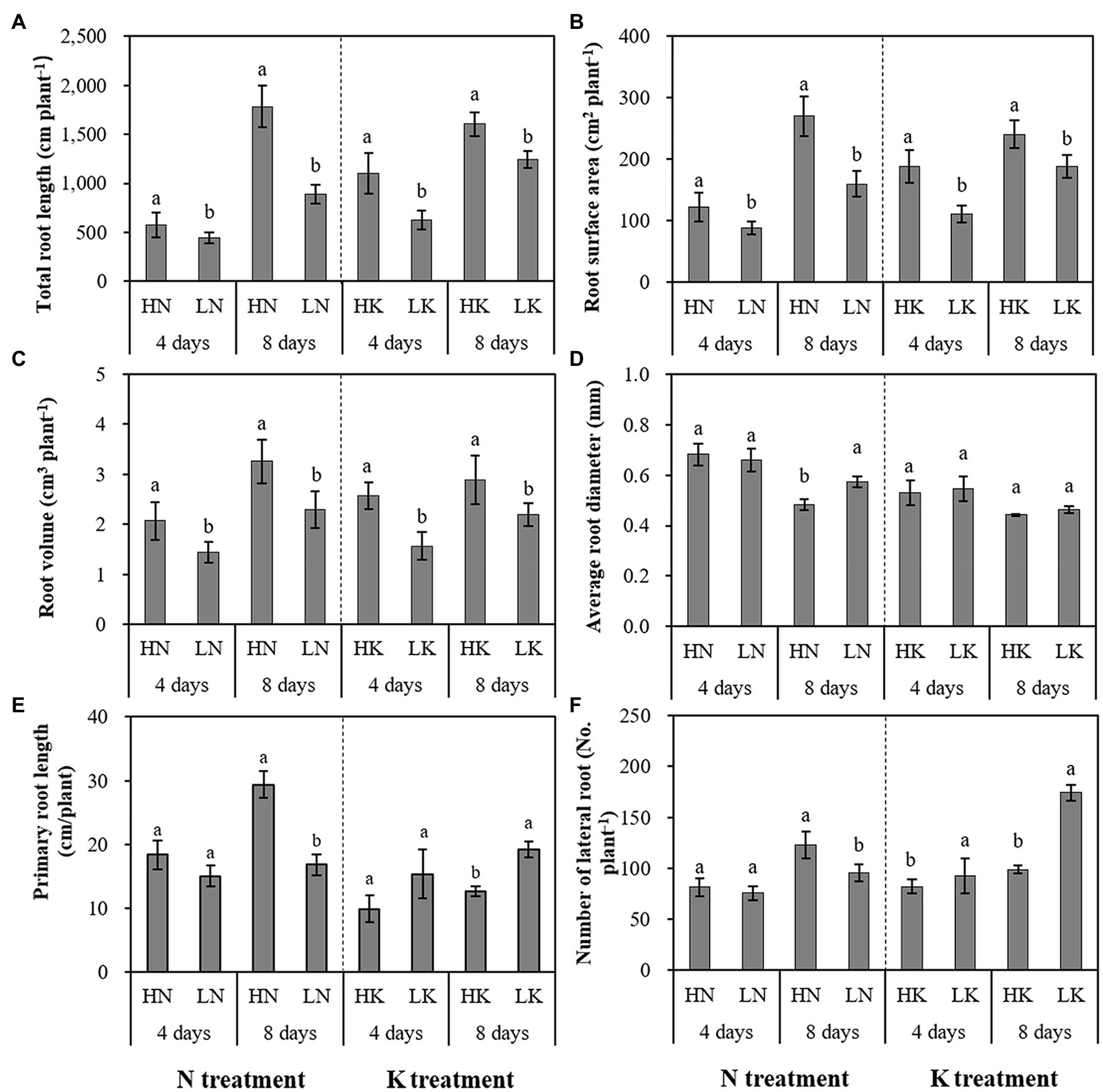
Figure 3. The effect of N deficiency and K deficiency on the total root length (A), root surface area (B), root volume (C), average root diameter (D), primary root length (E), and the number of lateral roots (F) of peanut seedlings, respectively. The data are the means ± SDs (n = 5). The different letters on the bars indicate significant differences between the different N (or K) treatments and the control treatment on the same day according to the LSD test (p = 0.05). N, nitrogen; K, potassium; and LSD, least significant difference.
N and K deficiency effects on the primary root length and lateral root number of peanut seedlings were opposite. N deficiency significantly reduced the primary root length of the seedlings on both the 4th and 8th days of N deficiency treatment. In addition, it decreased the lateral root number of the seedlings on the 8th day of N deficiency treatment. In contrast, K deficiency significantly promoted the growth of the primary root and increased the lateral root number of the seedlings on the 8th day. Compared with the seedlings cultured in the normal nutrient solutions, the primary root length and the lateral root number of the seedlings cultured in N deficiency nutrient solutions on the 8th day decreased by 44.89 and 22.49%, respectively, and that of the seedlings cultured in K deficiency nutrient solutions on 8th day increased by 59.04 and 71.46%, respectively.
N and K Deficiency Affected Root Vigor and Root Respiration in Peanut Seedlings
N and K deficiency significantly impacted the growth and development of roots and significantly affected the health of roots, which was manifested by changes in root vigor and respiration (Figure 4). On both the 4th and 8th days of treatment, the root vigor was significantly changed under both N deficiency and K deficiency treatments, and with the extension of the stress time, the root vigor showed a gradual decline trend. Compared with the seedlings cultured in the normal nutrient solutions, the root vigor of the seedlings cultured in N deficiency nutrient solutions and the seedlings cultured in K deficiency nutrient solutions on the 8th day of treatment decreased by 33.84 and 44.64%, respectively. On the 4th day of treatment, there was no significant difference between the root respiration rate of seedlings grown in N deficient nutrient solution and that of seedlings grown in a normal nutrient solution. However, with the extension of stress time, the root respiration kept decreasing. The difference of the root respiration between N deficiency treatment and the controls reached a significant level on the 8th day of treatment. The root respiration of the seedlings cultured in K deficiency nutrient solutions showed a gradual decline trend, and the difference of the root respiration between the seedlings grown in K deficient nutrient solution and the seedlings grown in normal nutrient solution reaches a significant level on both 4th and 8th days of K treatment.
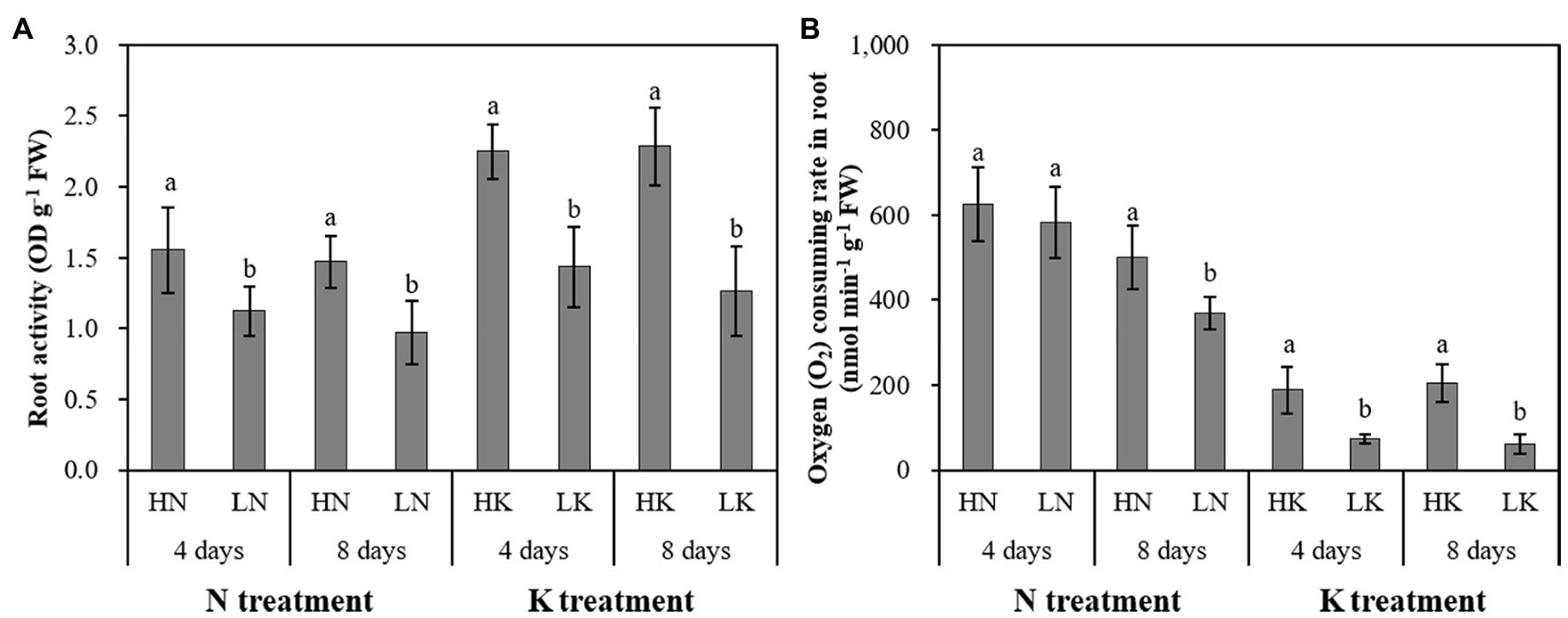
Figure 4. The effect of N deficiency and K deficiency on the root activity (A) and oxygen (O2) consuming rate in the root (B) of peanut seedlings, respectively. The data are the means ± SDs (n = 5). The different letters on the bars indicate significant differences between the different N (or K) treatments and the control treatment on the same day according to the LSD test (p = 0.05). N, nitrogen; K, potassium; and LSD, least significant difference.
N Deficiency and K Deficiency Changed the Expression of miRNAs and Their Targeted mRNAs
N and K deficiency changed the expression of miRNAs and their targeted mRNAs (Figure 5). However, the response of different miRNAs to N deficiency and K deficiency was different. On the 4th day of N deficiency treatment, except miR156, miR162, miR172, miR396, miR778, the expression of other miRNAs was inhibited, and the inhibition of the expression of miR167, miR169, miR395, miR399, and miR857 reached a significant level compared with that in the controls (Figure 5A). In contrast, on the 8th day of N deficiency treatment, except miR399, miR778, and miR857, the expression of the other tested miRNAs was induced compared with that in the controls, and the induction of the expression of miR156, miR160, miR165, miR166, miR167, miR169, miR171, miR172, miR319, miR390, miR393, miR396, miR827, and miR847 reached a significant level compared with that in the controls. On the 4th day of K deficiency treatment, all the 20 miRNAs were induced (Figure 5B). Among them, miR167 was upregulated the most by 6.32-fold, and the expression of miR156, miR164, miR165, miR166, miR169, miR393, miR396, and miR847 were also upregulated by more than threefold change. On the 8th day of treatment, although the expression of most miRNAs (except for miR160, miR164, miR171, miR393, and miR827) under K deficiency treatments were still higher than that in the control treatment, the value of fold changes was much lower than that determined on 4th day of treatments. On the 8th day of treatments, the expression of miR160, miR164, miR171, and miR393 under K deficiency treatment was significantly lower than that under the controls.
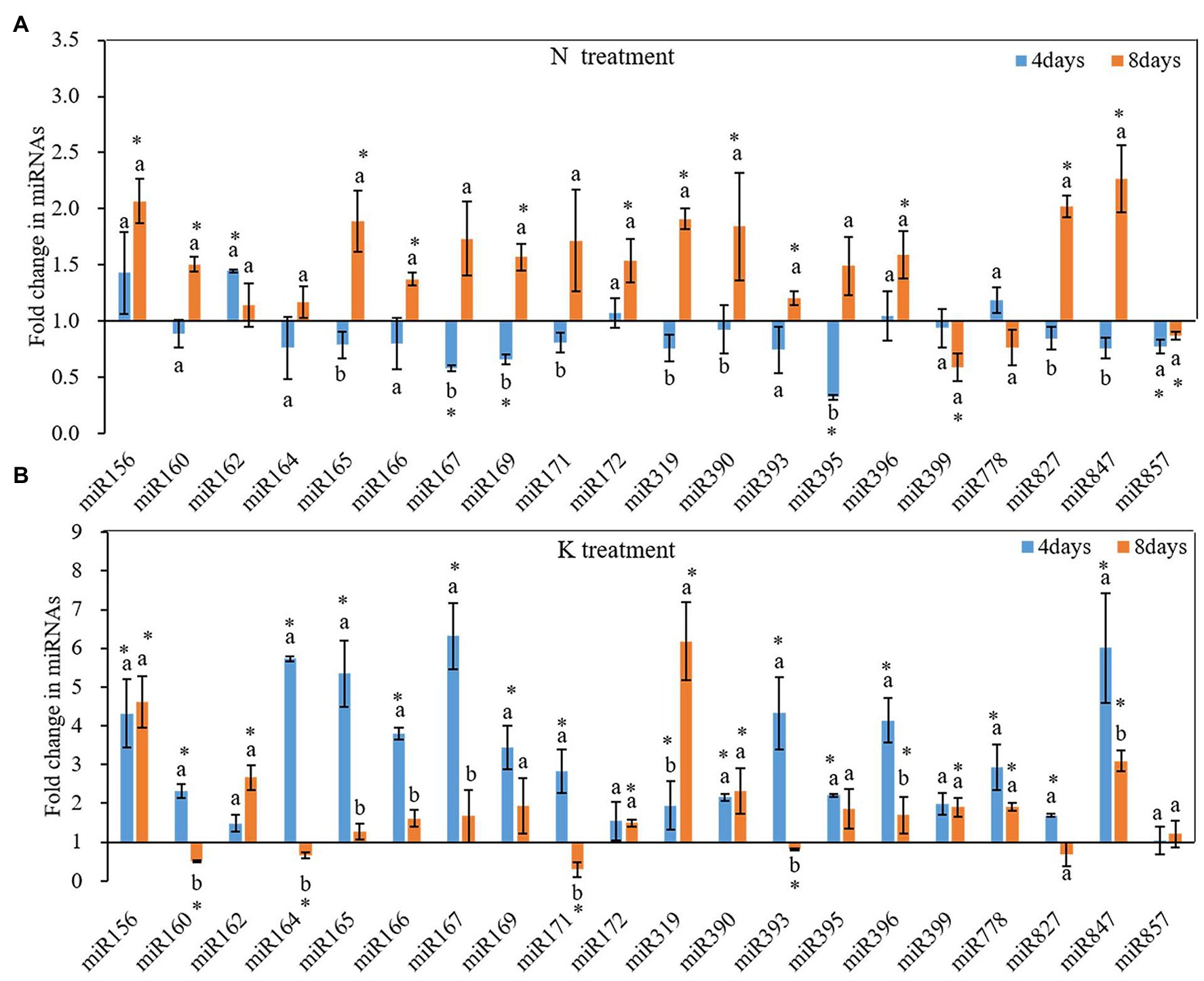
Figure 5. Fold change of microRNAs (miRNA) expression between N deficiency and the controls (A) and between K deficiency and the controls (B) in the root of peanut seedlings, respectively. Error bars represent SD (n = 3). The different letters on the bars indicate significant differences in the fold change of miRNA target expression between 4 and 8 days of treatment according to the LSD test (p = 0.05). A value of p less than 0.05 was considered as a significant difference between N deficiency and the controls (A), and between K+ deficiency and the controls (B), which marked as ∗ in the graphs. N, nitrogen; K, potassium; and LSD, least significant difference.
N and K deficiency also changed the expression of protein-coding genes (Figure 6). On the 4th day of treatments, the expression of all tested seven miRNA-targeted genes, except NAC4 and ARF2, was inhibited under N deficiency treatment, and the induction of the expression of ARF1 and GRF reached a significant level compared with that in the controls (Figure 6A). In contrast, after 8 days of treatments, the total of potential miRNA targets, except the ARF2 and GRF gene, was induced by N deficiency treatment. Compared with the control, among these genes, NAC4 had the highest expression under N deficiency treatment, and the fold change value reached 3.46. On both the 4th and 8th days of K deficiency treatment, all tested seven potential miRNA targets, except AFB3 gene on 4th day of treatments and SPL10 on 8th day of treatments, were up-expressed in the root of seedlings grown in K deficient nutrient solution than that in normal nutrient solution (Figure 6B). ARF2 was the most upregulated among the seven target genes, and the fold changes up to 6.47-fold on the 4th day of K deficiency condition. Although the expression of most potential miRNA targets in the root of seedlings grown in K deficient nutrient solution was still higher than that of the control treatment on the 8th day of treatment, the fold change was much smaller than that determined on the 4th day of treatment. ARF1, ARF2, bHLH74 and SPL10 transcription factor gene expression levels were decreased by 57.75, 75.04, 30.29, and 68.25%, respectively, on the 8th day of treatments compared with that on 4th day of treatments under K deficiency treatments.
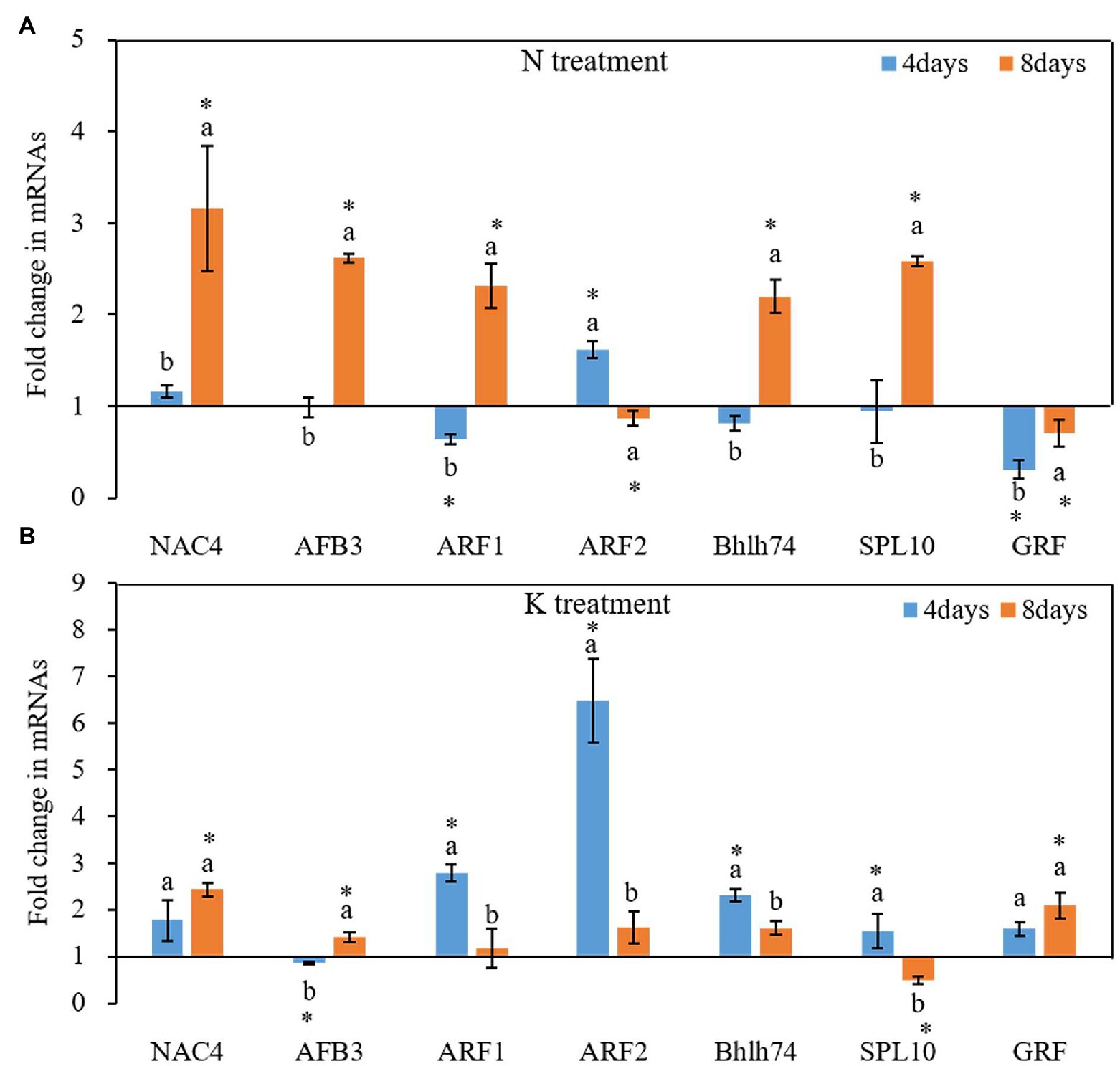
Figure 6. Fold change of miRNA target gene expression between N deficiency and the controls (A) and between K deficiency and the controls (B) in the root of peanut seedlings, respectively. The different letters on the bars indicate significant differences in the fold change of miRNA target expression between 4 and 8 days of treatment according to the LSD test (p = 0.05). A value of p less than 0.05 was considered as a significant difference between N (or K) deficiency and the controls, which marked as ∗ in the graphs. N, nitrogen; K, potassium; and LSD, least significant difference.
The Relationship Between miRNAs and Their Targeted mRNAs Under N and K Deficiency Treatments
miRNA regulates gene expression by shearing target mRNAs or inhibiting protein translation. Therefore, the expression of miRNA is negatively correlated with the expression of its targeted mRNAs. That is, if miRNA is upregulated, the expression of its target gene should be downregulated. The expression pattern of miRNAs and their target mRNA are often complex when secondary reactions and other gene regulatory factors besides miRNAs. In this case, the reverse expression relationship between the miRNAs and their target gene may not be observed. In the present study, we used linear equations to simulate the relationship between the fold change of miRNAs and their targeted mRNAs from the 4th day of treatment to the 8th day of treatment. The results showed that the expression of miRNAs had a negative linear relationship with their targets under both N deficiency and K deficiency treatment. And the reverse relationship could be described by the equation: y = −4.1416x + 8.9204 (R2 = 0.898, Figure 7A) and the equation: y = −1.2628x + 1.6193 (R2 = 0.8343, Figure 7B) under N deficiency and K deficiency treatments, respectively. It indicates that the tested miRNAs negatively regulate the expression of their targeted mRNAs.
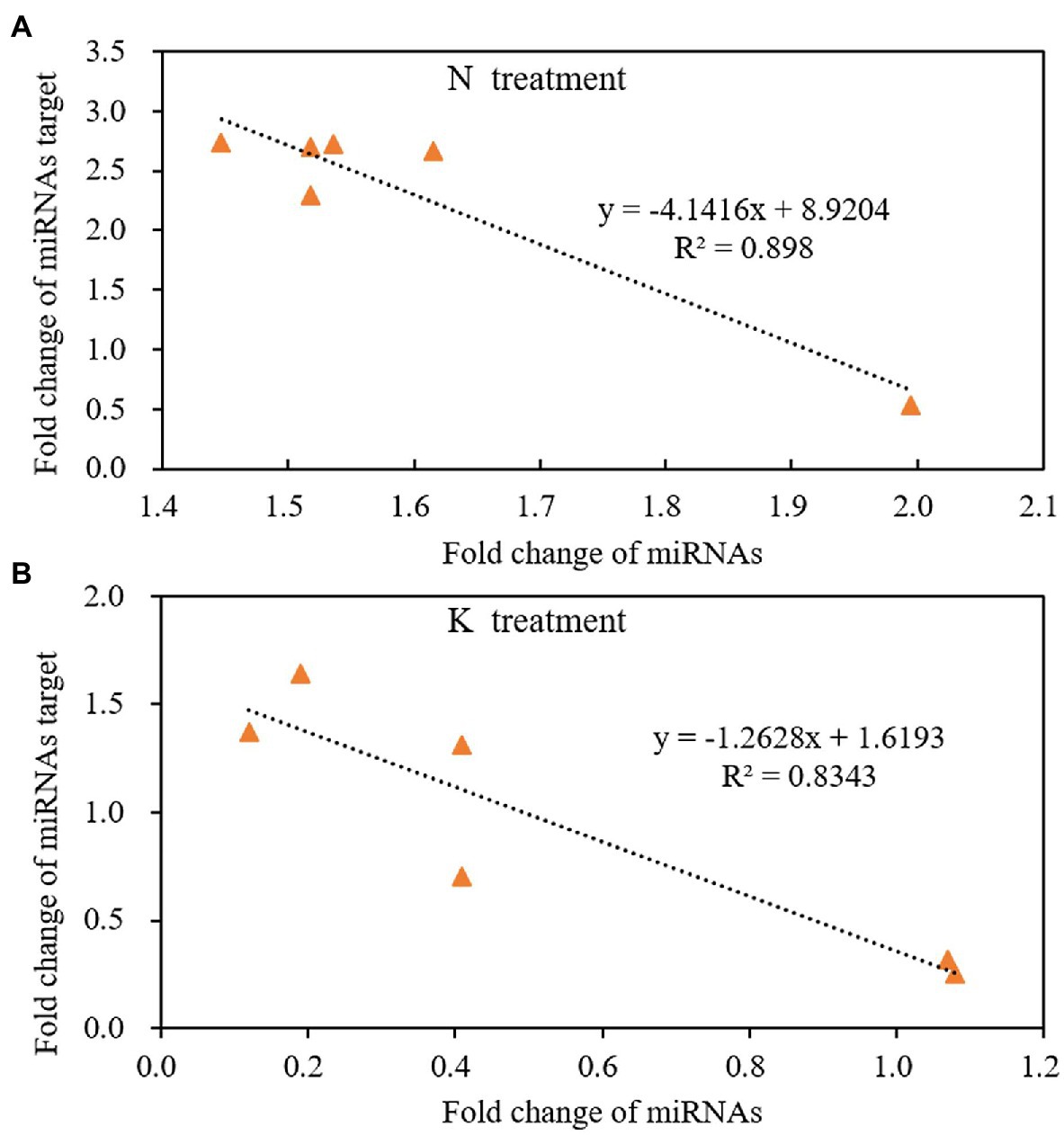
Figure 7. The reverse liner relationship of fold change between miRNAs and their targets under N treatment (A) and K treatment (B) in the root of peanut seedlings. N, nitrogen and K, potassium.
Discussion
N and K Deficiency Significantly Inhibited the Growth and Development of Peanut Seedlings
Nitrogen is the main component of many important compounds in plants. It participates in plant photosynthesis, a series of biochemical reactions, and plays an important role in biomass accumulation and yield formation in crops (Hawkesford et al., 2012; Kant, 2018). Potassium is the quality element of plants, which participates in osmotic adjustment, photosynthesis, material transport, and other processes, and it can improve the stress resistance of plants (Helmy and Ramadan, 2014; Almeida et al., 2015; Chakraborty et al., 2016). Therefore, the deficiency of these two kinds of macro-elements will lead to the inhibition of plant growth. In this study, the dry weight of shoot and root, leaf area, and plant height of peanut seedlings were significantly reduced under both 0.1 mM NO3− and 0.02 mM K+ treatment. It meant that the 0.1 mM NO3− and 0.02 mM K+ caused the low N stress and low K stress to the peanut seedlings, respectively, and inhibited the growth of peanut seedlings. These N deficiency-induced growth inhibition phenomena were also observed in other plant species, including potatoes (Xie et al., 2018). Studies have also shown that 0.02 mM K+ can induce aberrant growth and development in cotton (Fontana et al., 2020) and wheat (Thornburg et al., 2020), which probably due to the reduction of chlorophyll content and photosynthetic capacity.
The Peanut Root System Has a Specific Response to N Deficiency and K Deficiency Stresses
Roots are the main organ for plants to absorb nutrients and water. It is also the first organ to sense the changes of rhizosphere environmental conditions and shows strong plasticity to adapt to the external environment changes (Malamy, 2005). Analysis of RSA provides good quantitative readouts that can be used to identify the genes and signaling pathways, enabling plants to sense changes in the root environment and integrate them into adaptive responses (Kellermeier et al., 2014). The traditional view is that each nutrient deficiency will lead to a typical root structure (Kellermeier et al., 2014). This study also found that the peanut root system has a different response to N deficiency and K deficiency conditions. Previous studies have shown that plants intend to develop exploratory roots with more lateral roots (Araya et al., 2014; Kant, 2018). Mohd-Radzman and coworkers demonstrated that a uniform low concentration of nitrate (e.g., ≤1 mM) enhanced the growth of both lateral and primary roots of legumes (Mohd-Radzman et al., 2013). While our result about the peanut root system has a specific response to low N stress, both the lateral and primary root growth was inhibited under N deficiency conditions. Studies in Arabidopsis showed that under mild N deficiency, the average length of the lateral root was significantly promoted, while under severe N deficiency, the total lateral root length decreased, and there was almost no lateral root formation (Mohd-Radzman et al., 2013). Forde (2014) considered that the effect of low N stress or N deficiency on root branches depends on the N nutrition status of plants and the degree of stress on plants. Based on the above results, it can be considered that different crop roots respond differently to N deficiency, and root growth is related to the degree of N deficiency stress.
Previous studies have demonstrated that K deficiency has a negative effect on root elongation and the number of first-order lateral roots (Jung et al., 2009; Kellermeier and Amtmann, 2013), but the response varies among ecotypes, cultivars, species, and even root types (Kellermeier and Amtmann, 2013; Sustr et al., 2019). In this study, low K promotes both lateral and primary root growth while reduced the total root length. Similar results were also observed in wheat (Thornburg et al., 2020), and almost the opposite result was observed in cotton (Fontana et al., 2020). Although the number of lateral roots and the primary root length increased, the root system was inhibited under K deficiency treatment. The inhibition of root growth may be related to K deficiency induced slows down cell volume growth or restricts the transport of assimilation from the phloem to underground organs (Cakmak et al., 1994a,b).
Root vitality can represent the absorption, synthesis, oxidation, and reduction capacity of the root system and is a comprehensive index reflecting the absorption function of roots. 2,3,5-triphenyltetrazolium chloride is often used as a quantitative indicator of cell activity and is a good root metabolic reactive agent (Cakmak et al., 1994a,b). TTC can receive electrons directly from the electron transport link, and its reduction is directly related to mitochondrial respiration rate (Comas et al., 2000; Ruf and Brunner, 2003). Therefore, root vitality is the use of dehydrogenase activity in the respiratory chain to reflect the physiological indicators of root life activities. It is also a reflection of the strength of respiration. In this study, N deficiency and K deficiency significantly decreased the root vitality and the O2 consuming rate, representing the root respiration rate. Fontana et al. (2020) also found that K deficiency can cause the decline of root vitality and root respiration rate in cotton seedling planted in a low K solution that contained KCl (0.02 mM) for 8 days. The study in wheat showed that low N stress weakens the key enzyme activities of the NADP dehydrogenase system in roots. The decrease of NADPH synthesis affects the respiratory and metabolic activities of roots (Zhou et al., 2018). These results showed that N deficiency and K deficiency blocked root metabolism, decreased absorption capacity, and inhibited root growth, resulting in decreased root vitality.
miRNA Response to N and K Deficiency in Plants
N and K deficiency altered the expression of certain miRNAs and miRNA-mediated genes. There were differences in the miRNA expression patterns between the plants cultivated in N deficiency nutrient solution and those cultivated in K deficiency nutrient solution. Nguyen et al. (2015) observed that under N deficiency stress, the expression of miR156, miR160, and miR171 was induced, and the expression of miR169, miR172, miR319, miR396, and miR399 were reduced in Arabidopsis roots, while the expression of these miRNAs in maize roots was opposite. Therefore, it can be seen that the miRNA expression patterns varied with crop varieties and N deficiency. In this study, all 20 tested miRNAs, except for miR160, miR164, miR171, miR393, and miR827, were upregulated after 4 and 8 days of K deficiency treatments (Figure 5B). These may be the molecular mechanism of the adaptive response of peanut roots to N and K deficiency.
Previous studies have shown that miRNAs regulate genes encoding AUXIN RESPONSE FACTORS (ARFs) after transcription (Williams et al., 2005; Yuan et al., 2019). ARFs can specifically combine with auxin response elements in the auxin response gene promoter region to activate or inhibit gene expression, thus regulating plant growth and development (Hagen and Guilfoyle, 2002). Conserved miR160 regulates root growth and gravitational properties by negatively regulating three ARFs (ARF10, 16, and 17). In plants, overexpressing mir160c, ARF10, and ARF16 mRNA expression was decreased. The roots were short and attractive, with tumor-like apex related to uncontrolled cell division of apical meristem and failure of small columnar cell differentiation (Wang et al., 2005). The conserved miR167 negatively regulates auxin signaling by targeting ARF6 and ARF8. Under the condition of high nitrate, the expression level of miR167 in the root of Arabidopsis was decreased, allowing the accumulation of ARF8 in the pericyclic cell and lateral root cap, thereby stimulating the auxin signal, promoting lateral root formation, but inhibiting root elongation (Gifford et al., 2008). Auxin responsive miR390 is specifically expressed in the basal part of lateral root primordia and the potential vascular parenchymal cells and act as a regulatory factor by triggering the biogenesis of TAS3-derived transacting short-interfering RNAs (tasiRNAs; Marin et al., 2010). These tasiRNAs can repress the expression of ARF2, ARF3, and ARF4, thus promoting the development progression of lateral root (Marin et al., 2010; Du and Scheres, 2017). In this study, miR160, miR167, and miR390 downregulated on the 4th day and upregulated on the 8th day of N deficiency treatment. While ARF upregulated on the 4th day and downregulated on the 8th day of N deficiency treatment. miR160 upregulated on the 4th day and downregulated on the 8th day of K deficiency treatment. miR167 was nearly 6.32- and 1.67-fold upregulated on the 4th and 8th days of K deficiency treatment, respectively. The expression of miR167 on the 8th day was significantly lower than that on the 4th day of K deficiency. In addition, miR390 was upregulated on both the 4th and 8th days of K deficiency, and the ARF2 expression increased on both the 4th and 8th days of K deficiency. Thus, the altered expression of miR160, miR167, and miR390/ARF2 may be related to the inhibition of lateral root under N deficiency and the enhancement of lateral root under K deficiency treatment.
In addition to ARF-mediated auxin signal transduction, miRNAs also regulated auxin perception (Yuan et al., 2019). Studies have demonstrated that conserved miR393 targets genes encoding a small number of F-box-containing auxin receptors in many plants, including TRANSPORT INHIBITOR RESPONSE1 (TIR1) and AUXIN SIGNALING F-BOX (AFBs; Yuan et al., 2019), which are important for the growth of both primary root and lateral root (Vidal et al., 2013). miR393/AFB3 has been considered a nitrate responsive regulatory module that can integrate N and auxin signaling to regulate root growth in plant response to nitrate availability (Vidal et al., 2010, 2013). In this study, miR393 was downregulated on the 4th day and upregulated on the 8th day of N deficiency treatment. The expression of AFB3 was upregulated on the 4th day and further upregulated on the 8th day of N deficiency treatment. The expression pattern of miR393 under K deficiency stress is the opposite of its expression pattern under N deficiency stress. The expression pattern of miR393 was upregulated on the 4th day and downregulated on the 8th day of K deficiency treatment. The expression pattern of AFB3 was the opposite of the expression pattern of miR393 under K deficiency stress. The different changes of miR393/AFB3 under N deficiency and K deficiency conditions may explain the different responses of lateral root number and root length under these two conditions.
miR396 is a conserved miRNA in plants, closely related to plant growth and development (Debernardi et al., 2012; Li and Zhang, 2016). It has been found that miR396 targets the GROWTH REGULATION FACTORS (GRF) gene family and bHLH74, which promote cell proliferation during leaf development (Debernardi et al., 2012). miR396 may regulate root growth by restricting cell proliferation in root apical meristems (Debernardi et al., 2012). In this study, miR396 was significantly upregulated, and its target GRF was significantly downregulated on the 8th day of N deficiency. Thus, the altered expression of miR396/GRF may be related to the inhibition of lateral root under N deficiency treatment. Under K deficiency, miR396 was significantly upregulated on both the 4th and 8th days and compared with the expression on the 4th day, the expression of miR396 on the 8th day was significantly decreased. In contrast, the expression of GRF was significantly upregulated on both the 4th and 8th days of K deficiency. Therefore, the upregulation of GRF could be the reason for the enhancement of root growth in peanut under K deficiency.
Genes of the NAC (NAMATAF-CUC) family are the auxin signal transduction genes responsible for lateral root emergence and have been identified as the target of miR164 (Guo et al., 2005). Plants with low miR164 expression have high NAC1 expression and produced more lateral roots (Guo et al., 2005). In this study, miR164 was downregulated on the 4th day and upregulated on the 8th day of N deficiency. While the expression pattern of miR164 under K deficiency stress is just the opposite of its expression pattern under N deficiency stress. The expression of NAC was upregulated on the 4th day and downregulated on the 8th day of K deficiency. Because NAC transduces auxin signal for lateral root emergence (Guo et al., 2005), the upregulation of NAC under potassium deficiency further indicated that potassium deficiency induced the emergence of lateral roots.
In plants, miR156 targets some SPL (squamosa promoter binding protein-like) genes, which are plant-specific transcription factors in many developmental processes (Gou et al., 2011; Yu et al., 2015). In Arabidopsis, 17 members of the SPL gene family, 11 of which are regulated by miR156 (Qian et al., 2017). Yu et al. (2015) found that miR156 was differentially expressed in specific cells and tissues of lateral roots and responded to auxin signal transduction pathway by regulating SPL3, SPL9, and SPL10 genes, thereby inhibiting the growth of lateral roots, in which SPL10 played a leading role. In the present study, miR156 was nearly 4.3- and 4.6-fold induced on 4th and 8th days of K deficiency treatment. The SPL10 gene was 1.54-fold upregulated on the 4th day and was 0.49-fold downregulated on the 8th day of K deficiency treatment. The upregulated miR156 and the downregulated SPL10 may be the reason for initiating lateral roots under the K deficiency condition. Additionally, the upregulated SPL10 may be the reason for the lower number of lateral roots under the N deficiency condition. Plants overexpressing miR156 produced more lateral roots, while plants with lower miR156 levels had fewer lateral roots (Yu et al., 2015). SPLs can also directly activate miR172 and induce the floral inducer FLOWERING LOCUS (FT) through miR172-mediated AP2 inhibition, promoting plant flowering (Song et al., 2019). Over-expression of miR172 confers early flowering (Wang et al., 2016) and increased nodule numbers in soybean (Wang et al., 2014). A study in Arabidopsis suggested that miR172 acts downstream of miR156 and mediated the effect of miR156 on flowering time and the change of vegetative phase (Wu et al., 2009). Further study of the functions of these miRNAs and their regulated gene network will elucidate the molecular mechanism of peanut response to nutrient deficiency, which provides a new strategy for peanut molecular improvement using advanced technology, including the CRISPR/Cas genome editing (Li et al., 2021; Zhang et al., 2021).
Conclusion
In this study, both N deficiency and K deficiency caused abnormal growth and development of peanut seedlings. They changed the expression of miRNAs and their target genes related to root development. Under N deficiency condition, the lateral root initiation and emergence inhibition may be related to the upregulation of miR164, miR393, and miR396 and the downregulated AFB3 and GRF (Figure 8A). The inhibition of primary root elongation may be associated with the upregulation of miR160 under N deficiency condition. Under the K deficiency condition, the upregulated miR156, miR390, NAC4, ARF2, and AFB3, and the downregulated miR164, miR393, and SPL10 may be involved in the enhancement in the lateral root initiation and emergence (Figure 8B). Downregulated miR160 probably contributes to promoting primary root elongation. Overall, the roots of a peanut had different responses to N deficiency and K deficiency stresses, which is potentially due to the miRNAs-mediated pathway and mechanism. In conclusion, the different responses of peanut roots to nitrogen and potassium deficiency stress may be related to miRNA-mediated regulation of root growth and development.
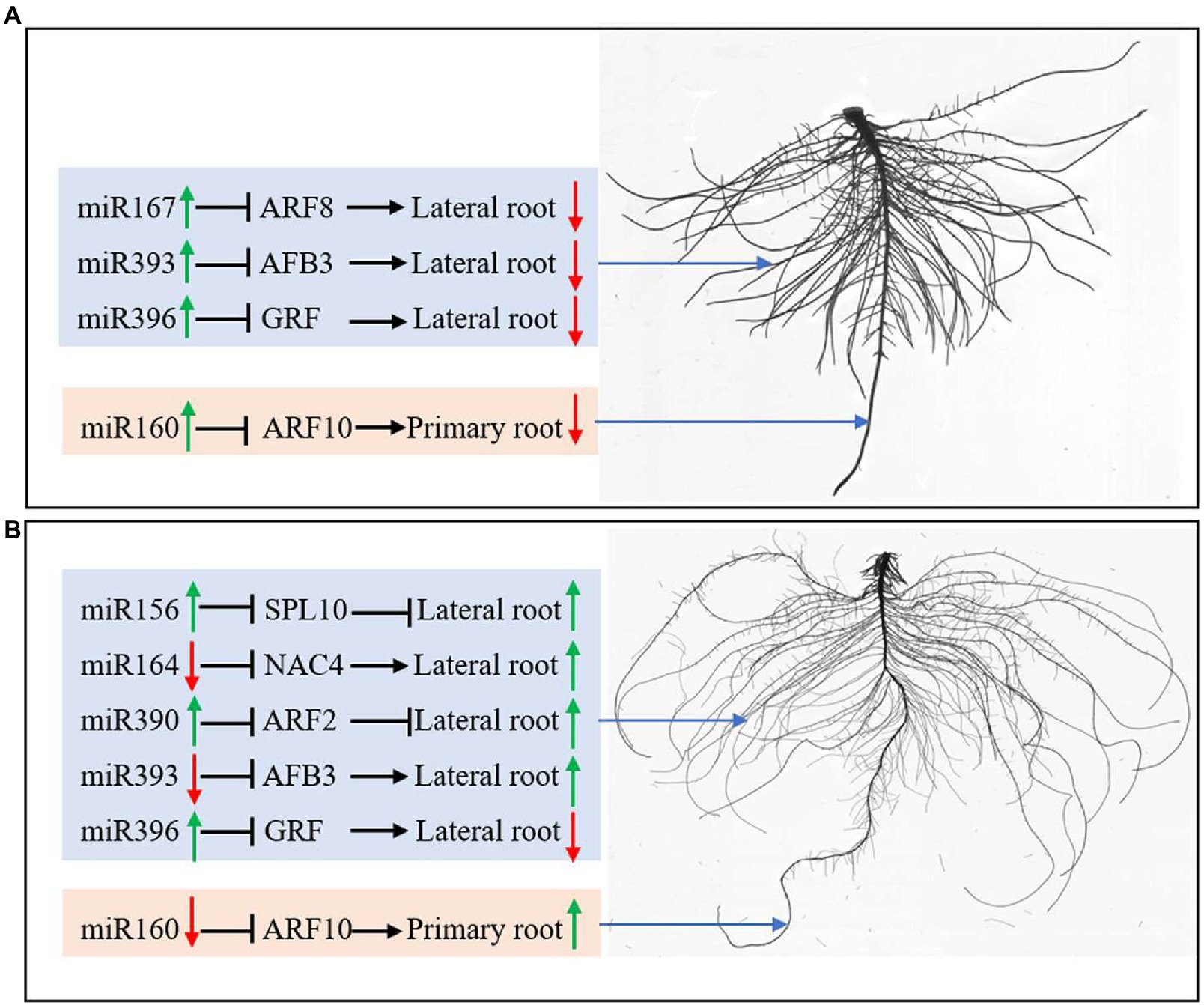
Figure 8. miRNAs regulate root development by controlling protein-coding genes’ expression under N deficiency (A) and K deficiency (B). The green arrow represents the upregulated expression of miRNAs and the enhanced growth of roots; the red arrow represents the downregulated expression of miRNAs and the inhibited growth of roots. N, nitrogen and K, potassium.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
LL: data curation, formal analysis, investigation, writing – original draft, and writing – review and editing. QL and WL: data curation and investigation. KD: methodology and investigation. CP: methodology and software. SO: investigation and validation. JL and GW: methodology, data curation, and investigation. JF and TT: investigation. IP: investigation and revision of the manuscript. FL: investigation and software. ZZ: conceptualization, methodology, resources, supervision, funding acquisition, and writing – original draft. YZ: conceptualization, methodology, resources, and supervision. XP: conceptualization, methodology, supervision, and funding acquisition. BZ: conceptualization, methodology, resources, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Science and Technology Special Project of Xinxiang City of China (ZD2020004), the Leading Talent Project in Science and Technology Innovation of Central Plain of China (214200510021), the Program for Innovative Research Team (in Science and Technology) in the University of Henan Province (21IRTSTHN023), and the Key R&D and promotion projects of Henan Province (202102110181 and 212102110070). This project was also supported by the U.S. National Science Foundation (award 1658709 to BZ and XP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Almeida, H. J., Pancelli, M. A., Prado, R. M., Cavalcante, V. S., and Cruz, F. J. R. (2015). Effect of potassium on nutritional status and productivity of peanuts in succession with sugarcane. J. Soil Sci. Plant Nutr. 15, 1–10. doi: 10.4067/S0718-95162015005000001
Araya, T., Miyamoto, M., Wibowo, J., Suzuki, A., Kojima, S., Tsuchiya, Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 111, 2029–2034. doi: 10.1073/pnas.1319953111
Cakmak, I., Hengeler, C., and Marschner, H. (1994a). Changes in phloem export of sucrose in leaves in response to phosphorus, potassium and magnesium deficiency in bean plants. J. Exp. Bot. 45, 1251–1257. doi: 10.1093/jxb/45.9.1251
Cakmak, I., Hengeler, C., and Marschner, H. (1994b). Partitioning of shoot and root dry matter and carbohydrates in bean plants susffering from phosphorus, potassium and magnesium deficiency. J. Exp. Bot. 45, 1245–1250. doi: 10.1093/jxb/45.9.1245
Candar-Çakir, B., Arican, E., and Zhang, B. H. (2016). Small RNA and degradome deep sequencing reveals drought- and tissue-specific microRNAs and their important roles in drought-sensitive and -tolerant tomato genotypes. Plant Biotechnol. J. 14, 1727–1746. doi: 10.1111/pbi.12533
Chakraborty, K., Bhaduri, D., Meena, H. N., and Kalariya, K. (2016). External potassium (K+) application improves salinity tolerance by promoting Na+-exclusion, K+-accumulation and osmotic adjustment in contrasting peanut cultivars. Plant Physiol. Biochem. 103, 143–153. doi: 10.1016/j.plaphy.2016.02.039
Comas, L. H., Eissenstat, D. M., and Lakso, A. N. (2000). Assessing root death and root system dynamics in a study of grape canopy pruning. New Phytol. 147, 171–178. doi: 10.1046/j.1469-8137.2000.00679.x
Debernardi, J. M., Rodriguez, R. E., Mecchia, M. A., and Palatnik, J. F. (2012). Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8:e1002419. doi: 10.1371/journal.pgen.1002419
Dolan, L., and Davies, J. (2004). Cell expansion in roots. Curr. Opin. Plant Biol. 7, 33–39. doi: 10.1016/j.pbi.2003.11.006
Du, Y., and Scheres, B. (2017). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. doi: 10.1093/jxb/erx223
Fontana, J. E., Wang, G., Sun, R., Xue, H., and Pan, X. (2020). Impact of potassium deficiency on cotton growth, development and potential microRNA-mediated mechanism. Plant Physiol. Biochem. 153, 72–80. doi: 10.1016/j.plaphy.2020.05.006
Forde, B. G. (2014). Nitrogen signalling pathways shaping root system architecture: an update. Curr. Opin. Plant Biol. 21, 30–36. doi: 10.1016/j.pbi.2014.06.004
Gifford, M. L., Alexis, D., Gutierrez, R. A., Coruzzi, G. M., and Birnbaum, K. D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U. S. A. 105, 803–808. doi: 10.1073/pnas.0709559105
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., and Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Gruber, B. D., Giehl, R. F., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Guilhem, D., Caroline, J., Stamatis, R., Polydefkis, H., and Liam, D. (2003). AKT1 and TRH1 are required during root hair elongation in Arabidopsis. J. Exp. Bot. 54, 781–788. doi: 10.1093/jxb/erg066
Guo, H., Xie, Q., Fei, J., and Chua, N. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hawkesford, M., Horst, W., Kichey, T., Lambers, H., Schjoerring, J., Mller, I. S., et al. (2012). “Functions of macronutrients,” in Marschners Mineral Nutrition of Higher Plants. ed. P. Marschner (Amsterdam, Netherlands: Elsevier), 135–189.
Helmy, A. M., and Ramadan, M. F. (2014). Yield quality parameters and chemical composition of peanut as affected by potassium and gypsum applications under foliar spraying with boron. Commun. Soil Sci. Plant Anal. 45, 2397–2412. doi: 10.1080/00103624.2014.929700
Jung, J. Y., Shi, R., and Schachtman, D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21, 607–621. doi: 10.1105/tpc.108.063099
Kant, S. (2018). Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 74, 89–96. doi: 10.1016/j.semcdb.2017.08.034
Kellermeier, F., and Amtmann, C. A. (2013). Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol. 161, 1421–1432. doi: 10.1104/pp.112.211144
Kellermeier, F., Armengaud, P., Seditas, T. J., Danku, J., and Salt, D. E. (2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26, 1480–1496. doi: 10.1105/tpc.113.122101
Kuicheski, F. R., Correa, R., Gomes, I., de Lima, J., and Margis, R. (2015). NPK macronutrients and microRNA homeostasis. Front. Plant Sci. 6:451. doi: 10.3389/fpls.2015.00451
Leigh, R. A., and Wyn Jones, R. G. (1984). A hypothesis relating critical potassium concentrations for growth and distribution and functions of this ion in the plant cell. New Phytol. 97, 1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x
Li, C., Brant, E., Budak, H., and Zhang, B. H. (2021). CRISPR/Cas: a Nobel prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 22, 253–284. doi: 10.1631/jzus.B2100009
Li, C., and Zhang, B. H. (2016). MicroRNAs in control of plant development. J. Cell. Physiol. 231, 303–313. doi: 10.1002/jcp.25125
Luo, L., Zhang, Y., and Xu, G. (2020). How does nitrogen shape plant architecture? J. Exp. Bot. 15, 4415–4427. doi: 10.1093/jxb/eraa187
Maathuis, F. J. M., and Sanders, D. (1996). Mechanisms of potassium absorption by higher plant roots. Physiol. Plant. 96, 158–168. doi: 10.1111/j.1399-3054.1996.tb00197.x
Malamy, J. E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77. doi: 10.1111/j.1365-3040.2005.01306.x
Marin, E., Jouannet, V., Herz, A., Lokerse, A. S., Weijers, D., Vaucheret, H., et al. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117. doi: 10.1105/tpc.109.072553
Mohd-Radzman, N. A., Djordjevic, M. A., and Imin, N. (2013). Nitrogen modulation of legume root architecture signaling pathways involves phytohormones and small regulatory molecules. Front. Plant Sci. 4:385. doi: 10.3389/fpls.2013.00385
Nguyen, G., Rothstein, S., Spangenberg, G., and Kant, S. (2015). Role of microRNAs involved in plant response to nitrogen and phosphorous limiting conditions. Front. Plant Sci. 6:629. doi: 10.3389/fpls.2015.00629
Omara, P., Aula, L., Oyebiyi, F., and Raun, W. R. (2019). World cereal nitrogen use efficiency trends: review and current knowledge. Agrosyst. Geosci. Environ. 2, 1–8. doi: 10.2134/age2018.10.0045
Pritchard, J. (1994). The control of cell expansion in roots. New Phytol. 127, 3–26. doi: 10.1111/j.1469-8137.1994.tb04255.x
Qian, M. J., Ni, J. B., Niu, Q. F., Bai, S. L., Bao, L., Li, J. Z., et al. (2017). Response of miR156-SPL module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 8:550. doi: 10.3389/fphys.2017.00550
Ruf, M., and Brunner, I. (2003). Vitality of tree fine roots: reevaluation of the tetrazolium test. Tree Physiol. 23, 257–263. doi: 10.1093/treephys/23.4.257
Salvagiotti, F., Cassman, K. G., Specht, J. E., Walters, D. T., Weiss, A., and Dobermann, A. (2008). Nitrogen uptake, fixation and response to fertilizer N in soybeans: a review. Field Crop Res. 108, 1–13. doi: 10.1016/j.fcr.2008.03.001
Sharma, L. K., and Bali, S. K. (2018). A review of methods to improve nitrogen use efficiency in agriculture. Sustainability 1:51. doi: 10.3390/su10010051
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant—environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Sun, C., Yu, J., and Hu, D. (2017). Nitrate: a crucial signal during lateral roots development. Front. Plant Sci. 8:485. doi: 10.3389/fpls.2017.00485
Sunkar, R., Li, Y. F., and Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. doi: 10.1016/j.tplants.2012.01.010
Sustr, M., Soukup, A., and Tylova, E. (2019). Potassium in root growth and development. Plants 8:435. doi: 10.3390/plants8100435
Thornburg, T. E., Liu, J., Li, Q., Xue, H., and Pan, X. (2020). Potassium deficiency significantly affected plant growth and development as well as microRNA-mediated mechanism in wheat (Triticum aestivum L.). Front. Plant Sci. 11:1219. doi: 10.3389/fpls.2020.01219
Vidal, E. A., Araus, V., Lu, C., Parry, G., and Green, P. J. (2010). Nitrate-responsive Mir393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 7, 4477–4482. doi: 10.1073/pnas.0909571107
Vidal, E. A., Moyano, T. C., Riveras, E., Contreras-Lopez, O., and Gutierrez, R. A. (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. U. S. A. 110, 12840–12845. doi: 10.1073/pnas.1310937110
Walker, D., Black, C., and Miller, A. (1998). The role of cytosolic potassium and ph in the growth of barley roots. Plant Physiol. 118, 957–964. doi: 10.1104/pp.118.3.957
Wang, Y., Kang, Y., Wang, M., and Zhao, C. (2013). Effects of potassium application on the accumulated nitrogen source and yield of peanut. J. Nucl. Agric. Sci. 27, 126–131.
Wang, T., Sun, M. Y., Wang, X. S., Li, W. B., and Li, Y. G. (2016). Over-expression of GmGIa-regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 17:645. doi: 10.3390/ijms17050645
Wang, J., Wang, L., Mao, Y., Cai, W., Xue, H., and Chen, X. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216. doi: 10.1105/tpc.105.033076
Wang, Y., Wang, L., Zou, Y., Chen, L., Cai, Z., Zhang, S., et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26, 4782–4801. doi: 10.1105/tpc.114.131607
Wang, Y., and Wu, W. (2009). Molecular genetic mechanism of high efficient potassium uptake in plants. Chin. Bull. Bot. 44, 27–36. doi: 10.3969/j.issn.1674-3466.2009.01.003
Williams, L., Carles, C. C., Osmont, K. S., and Fletcher, J. C. (2005). A database analysis method identifies an endogenous transacting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. U. S. A. 102, 9703–9708. doi: 10.1073/pnas.0504029102
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xie, X., Li, X. Q., Zebarth, B. J., Niu, S., Tang, R., Tai, H. H., et al. (2018). Rapid screening of potato cultivars tolerant to nitrogen deficiency using a hydroponic system. Am. J. Potato Res. 95, 157–163. doi: 10.1007/s12230-017-9621-1
Yu, N., Niu, Q., Ng, K., and Chua, N. (2015). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685. doi: 10.1111/tpj.12919
Yu, J. J., Su, D., Yang, D., Dong, T., Tang, Z., Li, H., et al. (2020). Chilling and heat stress-induced physiological changes and microRNA-related mechanism in sweetpotato (Ipomoea batatas L.). Front. Plant Sci. 11:687. doi: 10.3389/fpls.2020.00687
Yuan, W., Suo, J., Shi, B., Zhou, C., Bai, B., Bian, H., et al. (2019). The barley miR393 has multiple roles in regulation of seedling growth, stomatal density, and drought stress tolerance. Plant Physiol. Biochem. 142, 303–311. doi: 10.1016/j.plaphy.2019.07.021
Zhang, B. (2015). MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. doi: 10.1093/jxb/erv013
Zhang, J. L., Geng, Y., Guo, F., Li, X. G., and Wan, S. B. (2020). Research progress on the mechanism of improving peanut yield by single-seed precision sowing. J. Integr. Agric. 19, 1919–1927. doi: 10.1016/S2095-3119(19)62763-2
Zhang, B. H., Pan, X. P., Cobb, G. P., and Anderson, T. A. (2006). Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. 289, 3–16. doi: 10.1016/j.ydbio.2005.10.036
Zhang, B. H., and Unver, T. (2018). A critical and speculative review on microRNA technology in crop improvement: current challenges and future directions. Plant Sci. 274, 193–200. doi: 10.1016/j.plantsci.2018.05.031
Zhang, D., Zhang, Z. Y., Unver, T., and Zhang, B. H. (2021). CRISPR/Cas: a powerful tool for gene function study and crop improvement. J. Adv. Res. 29, 207–221. doi: 10.1016/j.jare.2020.10.003
Zhao, S., Zhang, M. L., Ma, T. L., and Wang, Y. (2016). Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28, 3005–3019. doi: 10.1105/tpc.16.00684
Keywords: peanut, nitrogen deficiency, potassium deficiency, microRNA, gene expression
Citation: Li L, Li Q, Davis KE, Patterson C, Oo S, Liu W, Liu J, Wang G, Fontana JE, Thornburg TE, Pratt IS, Li F, Zhang Z, Zhou Y, Pan X and Zhang B (2021) Response of Root Growth and Development to Nitrogen and Potassium Deficiency as well as microRNA-Mediated Mechanism in Peanut (Arachis hypogaea L.). Front. Plant Sci. 12:695234. doi: 10.3389/fpls.2021.695234
Edited by:
Turgay Unver, FicusBio, TurkeyReviewed by:
Fangjun Li, China Agricultural University, ChinaYingpeng Han, Northeast Agricultural University, China
Copyright © 2021 Li, Li, Davis, Patterson, Oo, Liu, Liu, Wang, Fontana, Thornburg, Pratt, Li, Zhang, Zhou, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Zhang, el96eTEyM0AxNjMuY29t; Baohong Zhang, emhhbmdiQGVjdS5lZHU=; Yanzhong Zhou, enl6bGhoc0AxNjMuY29t
 Lijie Li1
Lijie Li1 Zhiyong Zhang
Zhiyong Zhang Xiaoping Pan
Xiaoping Pan Baohong Zhang
Baohong Zhang