
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 28 July 2021
Sec. Plant Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.693272
This article is part of the Research TopicMaize Seed Development and Genetic ImprovementView all 5 articles
 Yong Wang1†
Yong Wang1† Xin-Yuan Liu1†
Xin-Yuan Liu1† Zi-Qin Huang1
Zi-Qin Huang1 Yan-Yan Li1
Yan-Yan Li1 Yan-Zhuo Yang1
Yan-Zhuo Yang1 Aqib Sayyed1
Aqib Sayyed1 Feng Sun1
Feng Sun1 Zhi-Qun Gu1
Zhi-Qun Gu1 Xiaomin Wang2
Xiaomin Wang2 Bao-Cai Tan1*
Bao-Cai Tan1*The conversion of cytidines to uridines (C-to-U) at specific sites in mitochondrial and plastid transcripts is a post-transcriptional processing event that is important to the expression of organellar genes. Pentatricopeptide repeat (PPR) proteins are involved in this process. In this study, we report the function of a previously uncharacterized PPR-DYW protein, Empty pericarp17 (EMP17), in the C-to-U editing and kernel development in maize. EMP17 is targeted to mitochondria. The loss-function of EMP17 arrests maize kernel development, abolishes the editing at ccmFC-799 and nad2-677 sites, and reduces the editing at ccmFC-906 and -966 sites. The absence of editing causes amino acid residue changes in CcmFC-267 (Ser to Pro) and Nad2-226 (Phe to Ser), respectively. As CcmFC functions in cytochrome c (Cytc) maturation, the amount of Cytc and Cytc1 protein is drastically reduced in emp17, suggesting that the CcmFC-267 (Ser to Pro) change impairs the CcmFC function. As a result, the assembly of complex III is strikingly decreased in emp17. In contrast, the assembly of complex I appears less affected, suggesting that the Nad2-226 (Phe to Ser) change may have less impact on Nad2 function. Together, these results indicate that EMP17 is required for the C-to-U editing at several sites in mitochondrial transcripts, complex III biogenesis, and seed development in maize.
The mitochondrion is a semi-autonomous organelle that provides energy and metabolites for cell activity. Plant mitochondrial genome inherits ∼5% genes from its prokaryotic ancestor, which encodes proteins of the respiratory complexes and ribosome, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs) (Clifton et al., 2004). Post-transcriptional processing of mitochondrial transcripts—including conversion of cytidines to uridines (C-to-U) editing, 5′- and 3′-terminus maturation, and intron splicing—is important for the function of the encoded proteins (Giege and Brennicke, 2001; Hammani and Giege, 2014). C-to-U editing is a major event of post-transcriptional processing, as it often restores the conserved amino acids (Kotera et al., 2005; Liu et al., 2013), generates translation start or stop codon (Kadowaki et al., 1995), regulates intron splicing (Xu et al., 2020), and enhances tRNA precursor efficient processing (Fey et al., 2002). In flowering plants, C-to-U editing occurs predominantly in mitochondrial transcripts with 300–600 editing sites (Notsu et al., 2002; Handa, 2003; Mower and Palmer, 2006; Bentolila et al., 2013; Wang et al., 2019), in comparison to the 20–40 sites in plastid transcripts (Tsudzuki et al., 2001; Tillich et al., 2005). A lack of editing often causes a deleterious impact on plant growth and development, and in some cases, even embryo lethality (Li et al., 2014; Xie et al., 2016; Wang et al., 2017; Xiao et al., 2018).
Although many factors have been identified to function in the C-to-U editing, the exact mechanism remains to be deciphered (Sun et al., 2016; Small et al., 2020). Pentatricopeptide repeat (PPR) proteins have been identified to play an essential role in editing site recognition. PPR proteins belong to one of the largest protein families in plants, with over 400 members in terrestrial plants (Lurin et al., 2004; O’Toole et al., 2008; Fujii and Small, 2011; Wei and Han, 2016). PPRs are classified into two classes: P-class and PLS-class (Small and Peeters, 2000; Lurin et al., 2004; Cheng et al., 2016). The P-class PPR proteins consist of bona fide P-motifs with 35 amino acids, while the PLS-class proteins harbor P-, L- (35–36 amino acids), and S-motifs (31 amino acids), and often carry an E, E+, and/or DYW domain at the C-terminus. As ubiquitous RNA binding factors, PPR proteins were found to take part in almost all of the post-transcriptional processing in mitochondria and plastids (Barkan and Small, 2014). The PLS-class PPR proteins mediate the C-to-U editing by specific binding to the nucleotide sequence upstream of the editing site through the PPR motifs (Tasaki et al., 2010; Okuda and Shikanai, 2012; Jiang et al., 2018). Furthermore, C-to-U editing also involves deamination of the cytidine (Blanc et al., 1995; Yu and Schuster, 1995), and the DYW domain possesses the cytidine deaminase (CDA) activity as demonstrated in PpPPR65 and PpPPR56 (Oldenkott et al., 2019; Hayes and Santibanez, 2020). The DYW domain also contains the motif of zinc-binding signature residues [HxE(x)nCxxC] commonly found in deaminases (Bhattacharya et al., 1994). Most DYW domains in higher plant PPR-DYW proteins contain the CDAs-like signature motif, which has been found to be indispensable for RNA editing (Boussardon et al., 2014; Hayes et al., 2015; Wagoner et al., 2015).
Accumulating evidence suggests that C-to-U editing is carried out by a protein complex, termed “editosome” (Sun et al., 2016; Small et al., 2020). Recent studies have shown that proteins of distinct families are involved in RNA editing in addition to the PPRs. These include multiple organelle RNA editing factors (MORFs)/RNA-editing factor interacting proteins (RIPs), organelle RRM proteins (ORRMs), organelle zinc-finger 1 (OZ1), and other proteins as reviewed by Small et al. (2020). MORFs/RIPs harboring a conserved MORF/RIP motif have been shown to be required for the editing at a large number of sites in mitochondria and nearly all sites in plastids in Arabidopsis (Bentolila et al., 2013). MORFs also selectively interact with PPR proteins and form homo- or heterodimers (Bentolila et al., 2012; Takenaka et al., 2012; Glass et al., 2015; Zehrmann et al., 2015; Haag et al., 2017). ORRM proteins, on the other hand, are required for the editing at many sites in mitochondria (ORRM2, ORRM3, ORRM4, and ORRM5) (Shi et al., 2015, 2016, 2017), and more than half of the sites in plastids in Arabidopsis (ORRM1 and ORRM6) (Sun et al., 2013; Hackett et al., 2017). ORRM proteins can interact with themselves, other ORRMs, and MORFs (Sun et al., 2013; Shi et al., 2015, 2016). ORRM1, specifically, is co-purified with OZ1, which is required for the editing at 81% plastid target Cs (Sun T. et al., 2015). A recent report showed that an active editing complex contains PPRs, RIPs, ORRM1, OZ1, and ISE2 in maize chloroplasts (Sandoval et al., 2019).
Plant mitochondria harbor two mono-hemic c-type cytochromes: Cytochrome c (Cytc) and Cytochrome c1 (Cytc1). They are the essential factors of the oxidative phosphorylation (OXPHOS) chain. Cytc1, anchored in the inner membrane, is a core subunit of complex III. On the other hand, Cytc, peripherally associated with the inner membrane, shuttles electrons from complex III to complex IV (Giege et al., 2008). After translation, apo-cytochrome c peptides are no longer functional. They have to undergo a maturation process in which the heme prosthetic groups are covalently attached to apo-Cytc1 and apo-Cytc via thioether bonds. In gram-negative bacteria, 8–9-ccm genes (ccmA to ccmI) are involved in the cytochrome c maturation (CCM), referred to as the CCM pathway or system I (Thony-Meyer et al., 1995). Plant mitochondria inherit the CCM pathway from the prokaryote ancestor (Giege et al., 2008), and in maize specifically, the CCM pathway involves seven proteins: three (CCMA, CCME, and CCMH) encoded by the nuclear genes and four (CcmB, CcmC, CcmFN, and CcmFC) by the mitochondrial genes (Clifton et al., 2004; Meyer et al., 2005; Rayapuram et al., 2008). Furthermore, loss-of-function of CCM factors often causes embryo lethality in plants. For example, a deficiency of CcmFN protein in the emp7 and ppr27 mutants results in a deficiency of Cytc and Cytc1, leading to impaired assembly of complex III and arrested seed development in maize (Sun F. et al., 2015; Liu et al., 2020).
In this study, we report the function of an uncharacterized mitochondrion-targeted PPR-DYW protein, erythropoietin-mimetic peptide 17 (EMP17), in maize. The results demonstrate that EMP17 is required for the editing at four sites of ccmFC and nad2 transcripts in mitochondria. Deficiency in the editing at ccmFC-799 and nad2-677 causes amino acid changes in the encoded protein. We provide further evidence that the Ser-to-Pro change at CcmFC-267 impairs the CcmFC activity, blocks the maturation of Cytc and Cytc1, and disrupts the assembly of complex III, which attributes to the arrest of seed development in the emp17 maize mutant.
The emp17 mutant was isolated from the UniformMu mutagenic population in nearly isogenic W22 background (McCarty et al., 2005). The plants were grown in the experimental field of Shandong University in Qingdao, Shandong province under natural conditions.
Genomic DNA (gDNA) was extracted by using a urea-phenol-chloroform-based method as described by Tan et al. (2011). The Emp17-F1 primer and Mu specific primer TIR8 (TIR8a:TIR8b:TIR8c:TIR8d = 1:2:2:1) were used to detect the Mu insertion in Emp17, and the Emp17-F1/Emp17-R1 primer pair was used to amplify the wild type Emp17.
The protein-coding region of Emp17 complementary DNA (cDNA) was placed downstream of the maize ubiquitin 1 (Ubi-1) promoter in the pUNTF vector. This pUNTF-Emp17 construct was transformed into maize inbred KN5585 via callus transformation. Positive transgenic plants of Emp17 were screened by using the Bar gene, and further verified by PCR using the Ubi-F primer anchored to the ubiquitin sequence in the vector and the Emp17 specific primer Emp17-R1.
The 420 bp, 5′-sequence of Emp17 encoding the 140 amino acids of the N-terminus region was cloned into pENTR/D-TOPO vector (Invitrogen in ThermoFisher Scientific, http://www.thermofisher.com), and then transferred to the pBI221 vector to create the Emp17N140-GFP fusion by the Gateway site-specific recombination. The resulting construct, pBI221-Emp17N140-GFP, was introduced into Arabidopsis protoplasts. MitoTracker Red was used as a marker for mitochondria. The fluorescence signals were detected under a ZEISS LSM 880 confocal microscope.
Wild type and emp17 kernels were harvested at 10 and 14 days after pollination (DAP) from the selfed ears of emp17 (+/−) heterozygous plants. The kernels were fixed in 4% paraformaldehyde at 4°C for 24 h. The fixed kernels were dehydrated, cleared, infiltrated, embedded, sectioned, stained, and observed as described by Liu et al. (2013).
Total RNA was extracted from developing kernels and other tissues of maize with the RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com) according to the manufacturer’s instruction. The potential gDNA contamination was removed through Dnase I (New England Biolabs, www.neb.sg) treatment. The single-stranded cDNA was obtained by reverse transcription reaction using the Transcriptor First Strand cDNA Synthesis kit (Thermo Fisher Scientific, http://www.thermofisher.com). Quantitative real-time PCR (qRT-PCR) with SYBR green (Bio-Rad, http://www.bio-rad.com) was performed in a Roche Light Cycler 96. The relative transcript level was calculated as described in previous studies (Wang et al., 2019). ZmActin (GRMZM2G126010) was used as a control in RT-PCR and qRT-PCR. Detailed information of the primers was listed in Supplementary Table 1.
RNA sample preparation of the wild type and emp17 kernels and reverse transcription were performed as described above. The resulting cDNA was used as a template to amplify the 35 mitochondrial predicted-protein coding genes by RT-PCR. RNA editing was analyzed by directly sequencing the RT-PCR amplicons. The primers were listed in Supplementary Table 1. Three biological repetitions were analyzed.
Crude mitochondria were extracted from the wild type and emp17 kernels at 14 DAP as described previously (Li et al., 2014). Mitochondrial complexes were separated by the blue native polyacrylamide gel electrophoresis (BN-PAGE) assay. In-gel staining of complex I and supercomplex I + III2 activity was performed as described previously (Meyer et al., 2009). The assembly of complex III and complex V was detected by transferring the protein complexes to nitrocellulose membranes and hybridized with anti-Cytc1 and anti-Atp1 antibodies. In-gel staining of complex IV was detected as described by Wang et al. (2020). Crude mitochondria were extracted and protein concentration was determined by the Bradford assay kit (Bio-Rad, http://www.bio-rad.com). For Western detection of complex proteins, 8 μg protein from emp17, and 8 μg, 4 μg (1/2), and 2 μg (1/4) protein from wild type were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membranes and detected by anti-Nad9, anti-Cytc1, anti-Cytc, anti-Cox2, and anti-Atp1 as described previously (Sun F. et al., 2015).
The respiration rates were determined according to the protocol described by Wang et al. (2015). O2 consumption of the wild type and emp17 kernels was measured at dark by using a Chlorolab II liquid-phase oxygen electrode (Hansatech, http://www.hansatech-instruments.com/) in a reaction buffer containing 10 mM HEPES, 10 mM MES, and 2 mM CaCl2 (where pH was adjusted to 6.8 with KOH). Total respiration rate (Vt) was defined by the O2 consumption before adding any respiration inhibitor. The cytochrome pathway capacity (Vcyt) was determined by Vt minus the O2 consumption in the presence of 2 mM salicylhydroxamic acid (SHAM) and the alternative pathway capacity (Valt) was defined by Vt minus the O2 consumption in the presence of 2 mM potassium cyanide (KCN). All three respiration rates were expressed as nmol O2 min–1 g–1 fresh weight.
PPR proteins form one of the largest protein families in maize with over 520 members annotated in the B73 genome. Among these, 82 are classified as the PPR-DYW subgroup proteins (Wei and Han, 2016). However, only six PPR-DYW proteins in maize have been fully characterized thus far: PPR2263 (Sosso et al., 2012), EMP5 (Liu et al., 2013), EMP18 (Li et al., 2019), EMP21 (Wang et al., 2019), PPR27 (Liu et al., 2020), and DEK46 (Xu et al., 2020). GRMZM2G019689 is a PPR-DYW subgroup protein encoded by an intron-less gene, hereafter referred to as “Emp17” (Figures 1A, 2A). EMP17 consists of 645 amino acid residues, and possesses 8 PPR motifs, an E1 and E2 domain, and a DYW domain with the conserved CDAs-like signature residues (HxE(x)nCxxC) (Figure 1A and Supplementary Figure 1). qRT-PCR analysis showed that Emp17 was ubiquitously expressed in major tissues in maize, with a relatively high expression in stem and leaf (Supplementary Figure 2). Phylogenetic analysis revealed an extensive conservation in the protein sequences from Amborella trichopoda to mono- and eudicotyledonous species (Figure 1B). However, no clear orthologs were found in Oryza sativa, Arabidopsis thaliana, Brassica napus, and Gossypium hirsutum.
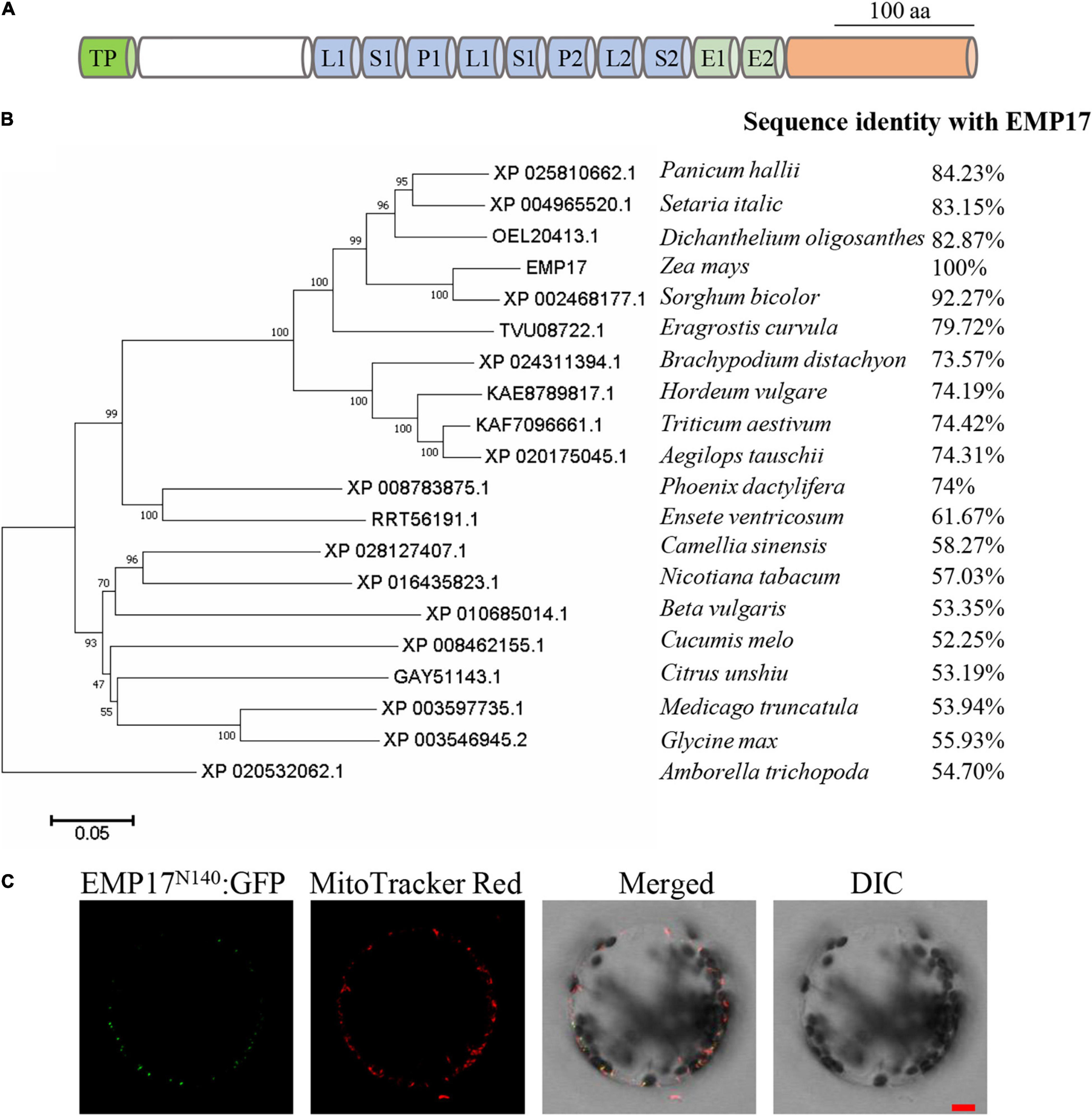
Figure 1. Emp17 encodes a canonical PPR-DYW protein localized in mitochondria. (A) The protein structure of EMP17. aa, amino acid; TP, targeted peptide; L, PPR-like long motif; S, PPR-like short motif. (B) Evolutionary relationships of putative EMP17 orthologs. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. (C) The subcellular localization of EMP17. The 140 amino acids fragment from the N-terminus of EMP17 was fused with green fluorescence protein (EMP17N140:GFP) and transiently expressed in Arabidopsis protoplasts. Fluorescence signals were observed by confocal microscope ZEISS LSM 880. The mitochondria were stained by MitoTracker Red. DIC, differential interference contrast; Bar = 10 μm.
EMP17 was predicted to localize in mitochondria by the TargetP1 and Predotar algorithms.2 To experimentally localize EMP17, the N-terminal region containing 140 aa of EMP17 was fused with the green fluorescent protein (GFP) and then transiently expressed in Arabidopsis protoplasts. GFP signals were detected in punctated dots that merged with the mitochondria stained by the MitoTracker Red (Figure 1C). No GFP signals were found in other cellular compartments (Figure 1C), indicating that EMP17 is specifically localized in mitochondria.
To study the function of EMP17, a Mutator (Mu) insertion mutant (emp17) was isolated from the UniformMu mutagenic population (McCarty et al., 2005). A Mu3 element was confirmed to be inserted at +660 bp from the translation start codon of Emp17 (Figure 2A). The selfed progenies of emp17 (+/−) heterozygous plants produced about 1/4 kernels with an empty pericarp phenotype (Figure 2B), and no wild type Emp17 transcript was detected in the embryo and endosperm of the empty pericarp kernels (Figure 2C). Linkage analysis in an F2 population showed that only plants segregating emp mutant phenotype carried Mu insertion in Emp17, indicating that the Mu insertion in Emp17 is either tightly linked to or the cause of the emp phenotype (Figure 2B and Supplementary Figure 3).
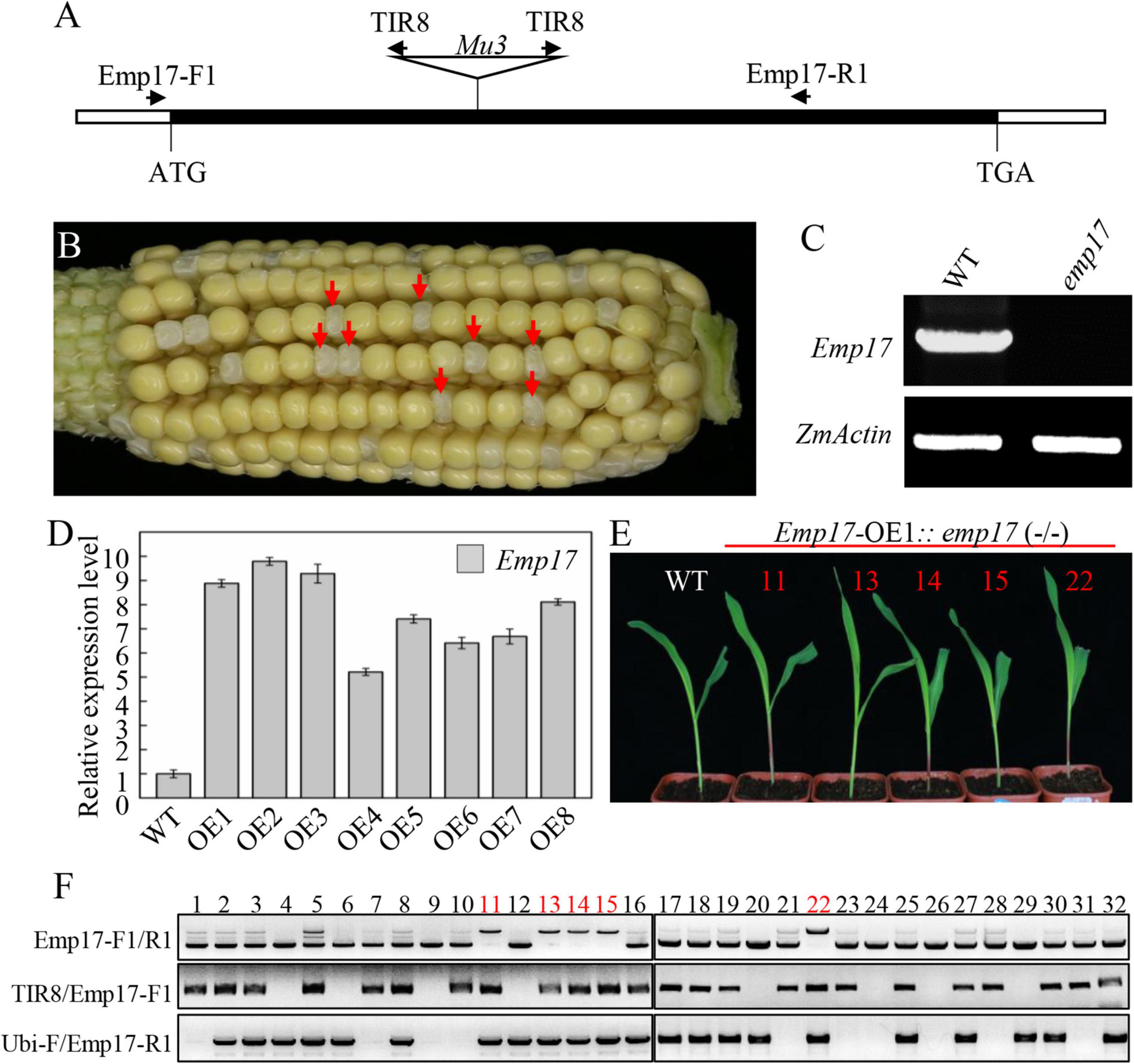
Figure 2. Mutation of Emp17 arrests maize embryogenesis and endosperm development. (A) The gene structure of Emp17. The Mu transposon insertion was marked by a triangle. (B) A selfed ear of the emp17 heterozygous plant. The empty pericarp kernels (emp17) are marked by red arrows. (C) RT-PCR analysis of Emp17 transcription profiling in wild type and emp17. (D) The expression profiles of Emp17 in wild type and Emp17 overexpression plants. (E) The phenotype of Emp17-OE1:emp17 (–/–) and wild type (WT) seedlings. (F) Genotype analysis of seedlings. The bands amplified by PCR using TIR8/Emp17-F1 primers indicate the Mu insertion in Emp17. The bands amplified by PCR using Ubi-F/Emp17-R1 primers indicate Emp17 transgenic plants. The bands amplified by PCR using Emp17-F1/R1 primers indicate Emp17 gene containing a Mu insertion (the longer bands) in the Emp17-OE1:emp17 (–/–) seedlings, and the wild type Emp17 gene (the shorter bands) in emp17 (+/−) and WT, respectively.
To confirm whether Emp17 is the causal gene for the empty pericarp phenotype, we created transgenic plants over-expressing Emp17 (Emp17-OE) in the inbred line KN5585 by placing Emp17 under the Ubi-1 promoter. Eight independent transgenic lines (Emp17-OE1 to OE8) were obtained, and the expression level of Emp17 in these lines was 5–9.5 times higher than that in the wild type as detected by qRT-PCR (Figure 2D). We crossed Emp17-OE1 with the emp17 (+/−) heterozygous plants and selfed the F1 to obtain the F2 progeny. The F2 seedlings were genotyped by PCR. To distinguish between the endogenous Emp17 and transgene Emp17, the Emp17-F1 primer was anchored to the 5’-UTR of the endogenous Emp17, ensuring that it could not anneal to the transgene that lacks this 5′-UTR sequence. The ubiquitin specific primer Ubi-F and Mu primer TIR8 were then used. Thus, the Emp17-F1/R1 primer pair amplified the endogenous Emp17, the Ubi-F/Emp17-R1 pair amplified the transgene, and the TIR8/Emp17-F1 pair detected the Mu3 insertion in Emp17. Genotyping 32 of the F2 seedlings identified 5 seedlings that were homozygous for emp17 harboring the Emp17 transgene (Figures 2E,F). The Emp17-F1/R1 primer pair amplified a larger fragment in these five emp17 (−/−) seedlings, which were proven to contain the Mu3 element. These five seedlings showed normal growth and development compared with the wild type (Figure 2E), indicating over-expression of Emp17 rescued the embryo-lethal phenotype of this mutant. These results demonstrate that Emp17 (GRMZM2G019689) is the causal gene for the empty pericarp phenotype of maize kernels in emp17.
The emp17 kernels were substantially smaller than the wild type siblings throughout kernel development. At 14 DAP, the emp17 embryo and endosperm was smaller than the wild type (Figures 3A,B). Paraffin sectioning indicated that the embryogenesis and endosperm development were severely arrested in emp17. At 10 DAP, leaf primordia (LP), shoot apical meristem (SAM), and root apical meristem (RAM) were clearly developed in the wild type embryo (Figure 3C). Conversely, the mutant embryo only proceeded to the early transition stage (Figure 3F). At 14 DAP, the wild-type embryo entered the late embryogenesis stage (Figures 3D,E), while the mutant embryo remained at the transition stage and the endosperm was arrested at the cellularization stage (Figures 3G,H).
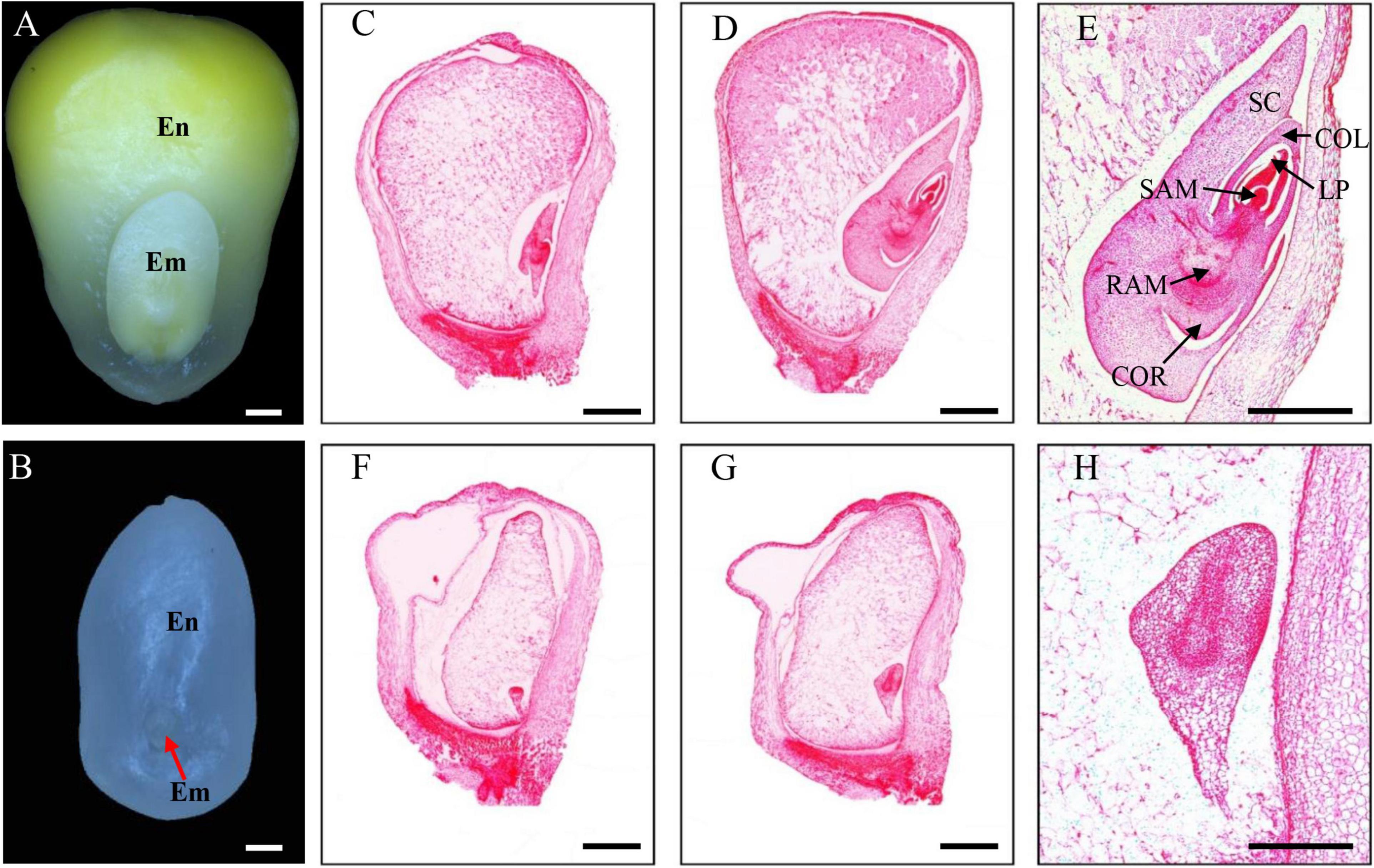
Figure 3. The emp17 mutation arrests maize embryogenesis and endosperm development. (A,B) The embryo (Em) and endosperm (En) of the wild type (A) and emp17 (B) at 14 days after pollination (DAP). (C–H) Paraffin section of WT (C–E) and emp17 (F–H) developing kernels. (C,F) 10 DAP; (D,E,G,H) 14 DAP. SC, scutellum; COL, coleoptile; LP, leaf primordia; SAM, shoot apical meristem; RAM, root apical meristem; COR, coleorhiza. Bars = 1 mm in (A–D,F,G) and 500 μm in (E,H).
As previously reported, most of the known PPR-DYW proteins function in the C-to-U editing of organellar RNA (Guillaumot et al., 2017; Wang et al., 2019). The maize mitochondrial genome was predicted to encode 22 electron transport chain proteins, 11 ribosomal proteins, a maturase MatR, and a membrane transporter protein MttB (Clifton et al., 2004). These 35 gene transcripts were amplified from the emp17 mutant and wild type kernels, and directly sequenced. Comparison of the sequences revealed that the C-to-U editing at ccmFC-799 and nad2-677 sites was abolished in emp17, and completely edited in the wild type (Figure 4A). In addition, the editing at ccmFC-906 and -966 sites was substantially decreased in emp17 in comparison with the wild type (Figure 4A). In the Emp17-OE1:emp17 (−/−) seedlings, the editing at nad2-677, ccmFC-799, -906, and -966 sites was restored (Figure 4A). These results indicate that the loss-of-function of Emp17 abolishes the editing at ccmFC-799 and nad2-677, and decreases the editing at ccmFC-906 and -966.
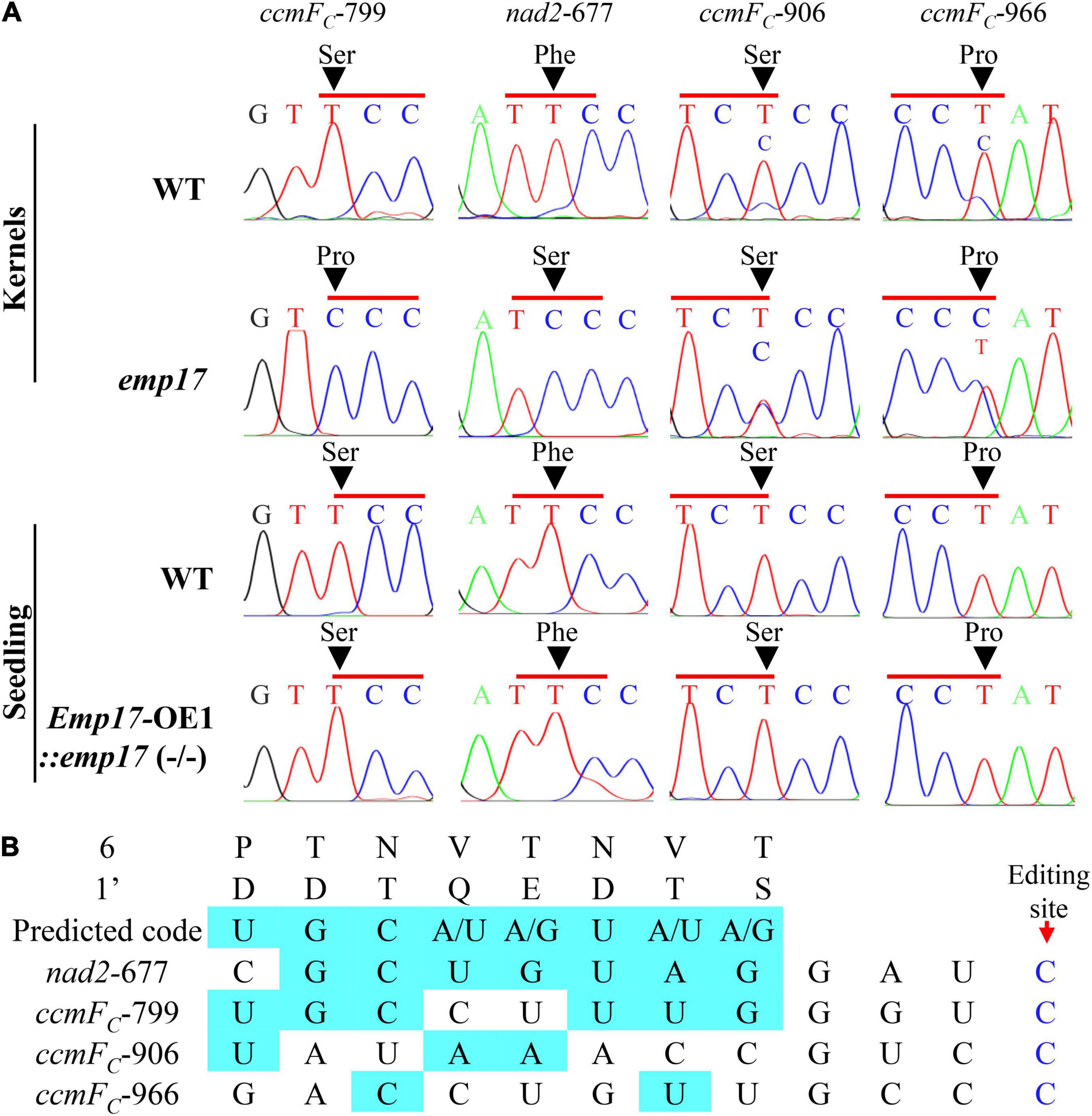
Figure 4. Loss of function of EMP17 abolishes the editing at ccmFC-799 and nad2-677 sites. (A) The defective editing sites in the emp17 mutant. Sites subject to defective editing marked by arrows. (B) Alignment of the amino acid residues at position 6 and 1′ in each PPR motif of EMP17 with -4 to -11 bp upstream sequence of these four defective editing sites.
PPR proteins recognize the target RNA sequence in a “one-PPR motif: one-nucleotide” manner, in which the sixth amino acid residue in the first PPR-motif and the 1′ amino acid residue in the next PPR-motif specifies the base of the RNA sequence (Barkan et al., 2012). Based on this code, the PPR motifs of EMP17 were aligned with the upstream sequence of nad2-677, ccmFC-799, -906, and -966 sites. The results showed that the codes aligned mostly with the ccmFC-799 and nad2-677 sites, but poorly with the ccmFC-906 and -966 sites (Figure 4B), suggesting that EMP17 probably binds strongly to the upstream sequences of the ccmFC-799 and nad2-677 sites, but weakly with the sequences of the ccmFC-906 and -966 sites.
The deficient editing at ccmFC-799 and nad2-677 in the emp17 mutant led to a Ser-to-Pro change at CcmFC-267 and a Phe-to-Ser change at Nad2-226, respectively (Figure 4A). Alignment of both gDNA and cDNA sequences of the ccmFC and nad2 orthologs indicated that these two amino acids were conserved in lower plants (Physcomitrella patens and Marchantia polymorpha), dicots (Glycine max, Nicotiana tabacum, Beta vulgaris, and Brassica napus), and monocots (Zea mays, Triticum aestivum, and Oryza sativa) (Figures 5A,B). The conservation of these two amino acid residues implies that these residues are probably important to the functional integrity of the CcmFC and Nad2 proteins.
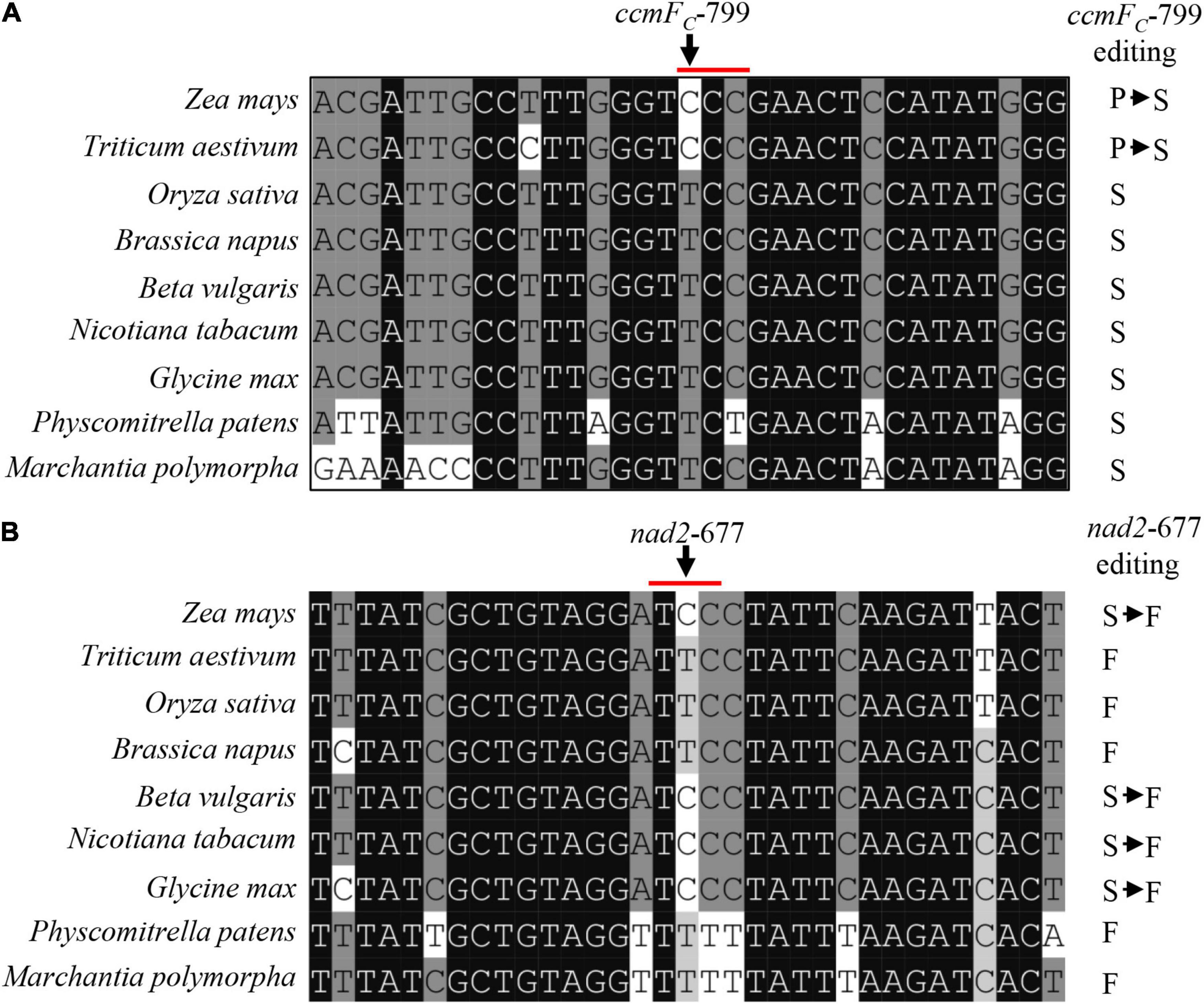
Figure 5. The amino acid residues encoded by ccmFC-799 and nad2-677 in multiple species. (A,B) Alignment of the neighboring gDNA sequences of ccmFC and nad2. The gDNA and cDNA sequences were derived from GenBank/EST and GenBank/EMBL databases. The abolished editing sites in the emp17 mutant are arrowed.
Nad2 is a subunit located in the hydrophobic arm of mitochondrial complex I (Braun et al., 2014), and CcmFC is involved in the maturation of Cytc and Cytc1, which are components of mitochondrial complex III (Giege et al., 2008). To assess the impact of the amino acid residue alteration on these complexes, we analyzed the assembly and activity of mitochondrial complexes and the complex proteins in emp17. Mitochondria were isolated from the embryo and endosperm of the emp17 mutant and wild type, respectively. The mitochondrial complexes were separated with BN-PAGE. Coomassie Brilliant Blue (CBB) staining showed that the level of complex I was comparable between emp17 and wild type, but the levels of complex III and supercomplex I + III2 were remarkably decreased in emp17 (Figure 6A). Furthermore, in-gel staining of the NADH dehydrogenase activity showed consistent results (Figure 6B). Western blot analysis indicated that the level of complex III as detected by anti-Cytc1 antibody was drastically decreased in emp17 (Figure 6C). Similarly, western blotting detection of complex V using anti-Atp1 antibody and in-gel staining of complex IV activity indicated that complex V and complex IV were increased in emp17 as compared with wild type (Figures 6D,E). Western blot assays using anti-Nad9, Cytc1, Cytc, Cox2, and Atp1 antibodies showed that the levels of Cytc and Cytc1 was dramatically reduced. On the other hand, the level of Nad9 and Cox2 was substantially increased and the level of Atp1 was moderately increased in emp17 as compared with wild type (Figure 6F). These results indicate that the abolished editing at nad2-677 site causing the Phe-to-Ser change at Nad2-226 appears not to significantly affect the assembly of mitochondrial complex I. However, the Ser-to-Pro change at CcmFC-267 severely inhibits the maturation of Cytc1 and Cytc and biogenesis of complex III, implying a critical role of Ser267 to the function of CcmFC.
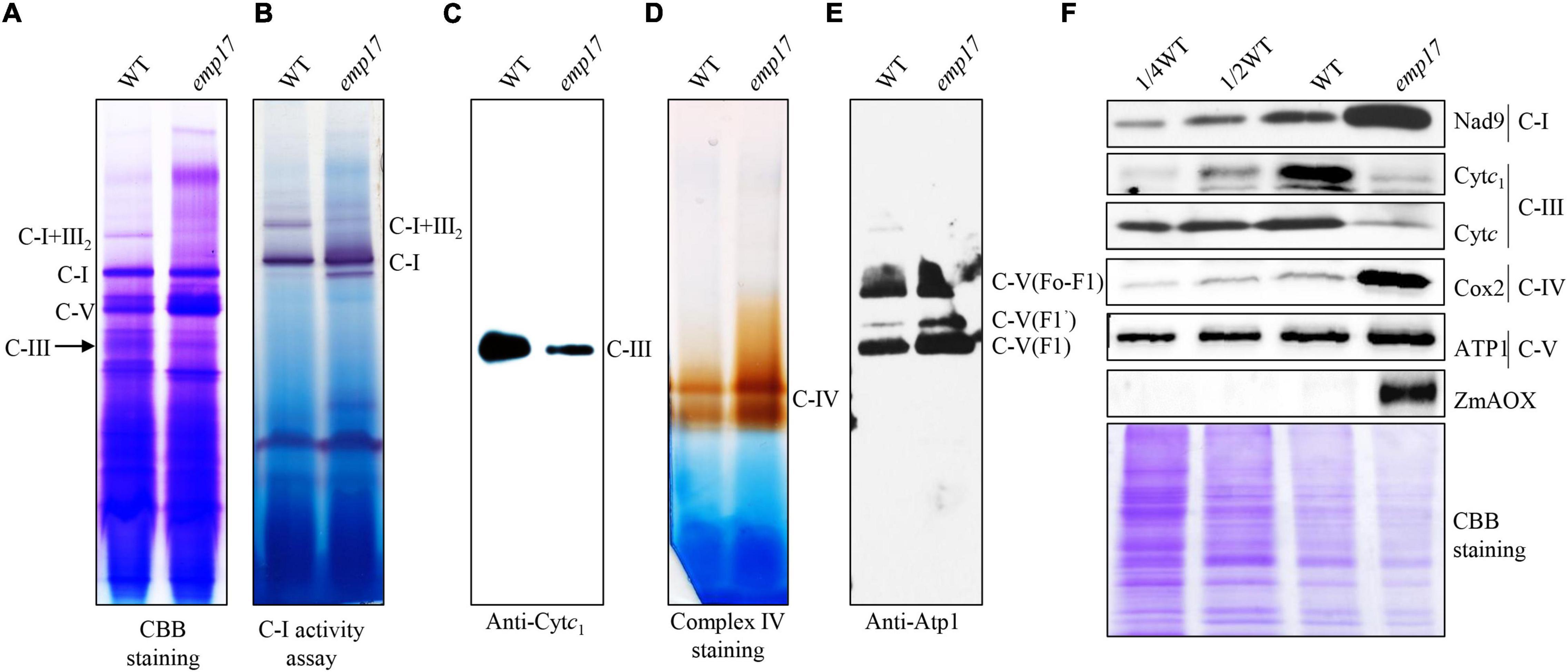
Figure 6. Mutation of Emp17 compromises the assembly of the mitochondrial complex III and supercomplex I + III2. (A) Blue native-PAGE (BN-PAGE) analysis of the assembly of complex I and supercomplex I + III2. The gel was stained with Coomassie Brilliant Blue (CBB). (B) In-gel staining of the nicotinamide adenine dinucleotide (NADH) dehydrogenase activity of complex I and supercomplex I + III2. The loading control is Dihydrolipoamide dehydrogenase (DLDH) activity. (C) Western blotting assay based on antibodies against Cytc1 (complex III). (D) In-gel activity staining of mitochondrial complex IV. (E) Western blotting assay based on antibody against Atp1 (complex V). (F) Western blotting assay with antibody against Nad9, Cytc1, Cytc, Cox2, Atp1, and ZmAOX. CBB staining was used for loading control. C-I: complex I, C-III: complex III, C-I + III2: supercomplex I + III2, C-V: complex V.
As previously reported, blocking of the cytochrome pathway often induces the alternative non-phosphorylating pathway in the respiratory chain (Yang et al., 2017; Wang et al., 2019; Xu et al., 2020). The maize genome hosts three ZmAOX genes (ZmAOX1, ZmAOX2, and ZmAOX3) (Karpova et al., 2002). As indicated by RT-PCR and qRT-PCR results, the level of ZmAOX2 and ZmAOX3 transcripts was dramatically increased in emp17, especially ZmAOX2 (Figures 7A,B). Western blotting analysis revealed that ZmAOX is expressed at low levels in the wild type kernels, but drastically enhanced in emp17 (Figure 6F).
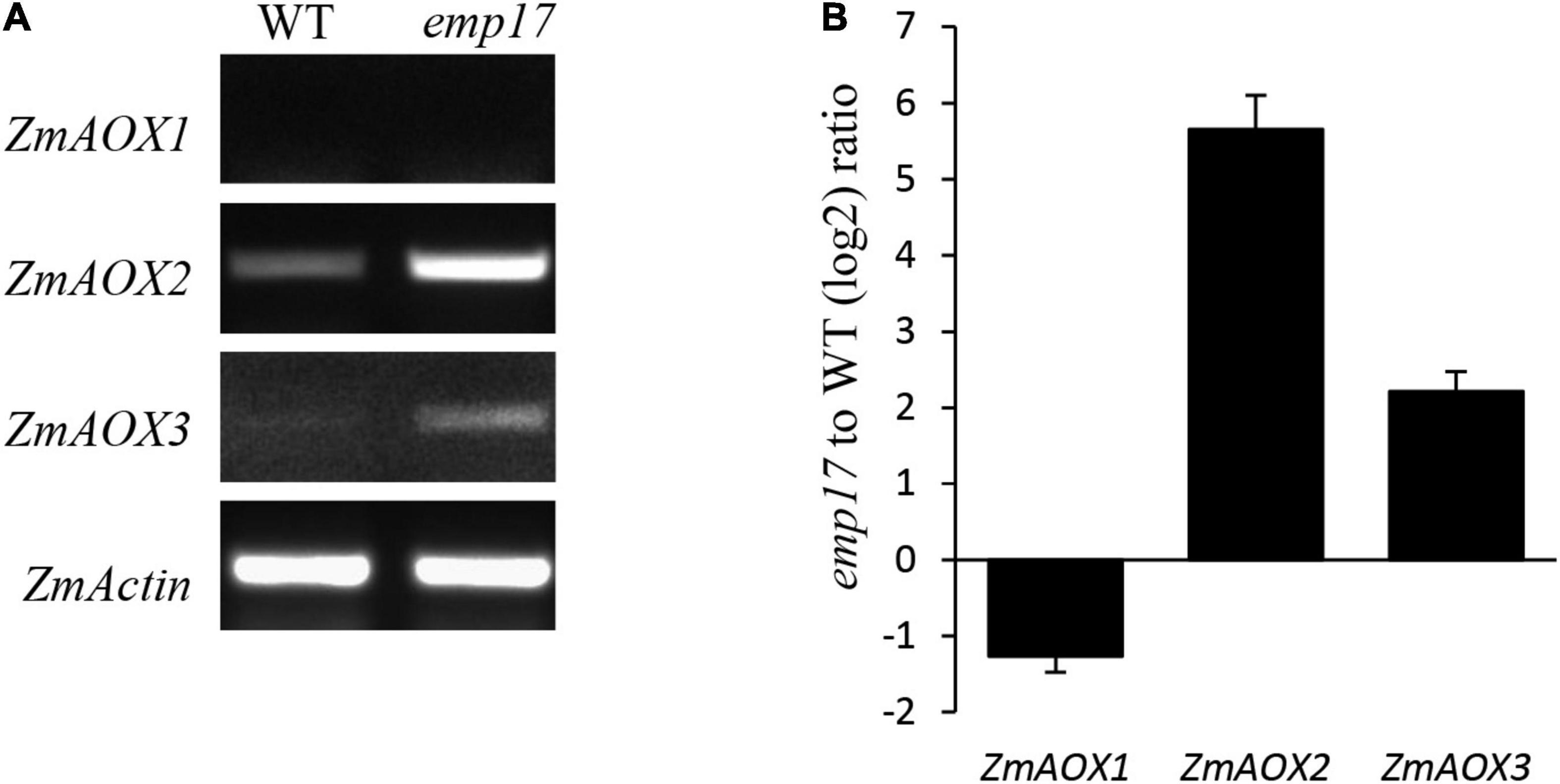
Figure 7. The expression analysis of the ZmAOX genes in wild type and emp17. (A,B) RT-PCR and qRT-PCR were used for analyzing the expression of the ZmAOX genes in wild type and emp17, respectively. RNA was extracted from 14 DAP embryos and endosperms. qRT-PCR values represent three biological replicates and are normalized against ZmActin. Error bars represent the ± SD.
To assess the physiological impact, we then measured the Vt, Valt, and Vcyt by using a Chlorolab II liquid-phase oxygen electrode, and specific inhibitors SHAM and KCN, respectively (Wang et al., 2015). The ratios of Valt to Vt and Vcyt to Vt in emp17 and wild type kernels were calculated. The results indicated that Vt and Vcyt in emp17 were decreased to about 18 and 10% in comparison with that in wild type, respectively (Table 1). The ratio of Valt/Vt (75%) in emp17 is higher than that by about twofold (43%) in wild type. These data confirm that mitochondrial complex III dysfunction severely reduces the cytochrome respiration and induces the alternative respiration pathway in emp17.
Putative orthologous proteins of EMP17 can be found in many sequenced plant species in the NCBI and Uniport database (Figure 1B). Phylogenetic analysis revealed a high degree of conservation of putative EMP17 orthologs in A. trichopoda and mono- and dicotyledonous species. EMP17 shared an over 50% sequence identity with most of its orthologous proteins (Figure 1B), i.e., 54.7% with that in A. trichopoda, the single living species of the sister lineage to all other extant flowering plants (Soltis et al., 2011). However, a putative EMP17 ortholog cannot be found in O. sativa, A. thaliana, B., and G. hirsutum. The most closely related protein in rice is Os12g0109300, sharing a 40.99% sequence identity with EMP17. In turn, Os12g0109300 shares an 81.94% sequence identity with maize protein GRMZM5G811022. As such, Os12g0109300 is unlikely to be an EMP17 ortholog in rice. The most closely related homologs of EMP17 are OTP82 in A. thaliana, hypothetical protein XP_013641116 in B. napus (accession number XP_013641116), and hypothetical protein (accession number KAG4215874) in G. hirsutum. All of these proteins share a less than 38% sequence identity with EMP17, much lower than that with the A. trichopoda homolog (54.7%). The results suggest that these species may have lost the Emp17 gene in the genome.
This raises the question of why this protein is conserved in some species, but lost in others. The gDNA sequence of CcmFC shows that the ccmFC-799 site is “T” in A. trichopoda and eudicots. However, in monocots, both “T” and “C” present (Supplementary Figure 5A). In all the species harboring ccmFC-799C in mitochondrial DNA, putative orthologs of EMP17 can be identified in the nuclear genomes (Phoenix dactylifera, Z. mays, Sorghum bicolor, and T. aestivum) (Supplementary Figure 5A). In the species with ccmFC-799T in the mitochondrial gene, putative ortholog of EMP17 may be lost (Supplementary Figure 5A). A consistent result can be found between the existence of EMP17 orthologs in the nuclear genome and the nad2-677 site in mitochondria as well (Supplementary Figure 5B). These results suggest that EMP17 orthologs probably exist in the early flowering plants that do not require the editing function of EMP17 at the ccmFC-799 or nad2-677 sites because both sites are “T.” But later in evolution, when the “T” was mutated to “C,” EMP17 was recruited for the editing function. For the species that maintain a “T” at this site, the Emp17 orthologs may be degenerated or lost. This notion offers a possible explanation for the disappearance of a clear EMP17 ortholog in O. sativa, A. thaliana, B. napus, G. hirsutum, and possibly in other species as well.
This study provides strong evidence that EMP17, a previously uncharacterized PPR-DYW protein, functions in the editing of mitochondrial transcripts in maize. Since only 6 out of the 82 maize PPR-DYW proteins have been fully characterized so far, the elucidation of the EMP17 function adds a new piece of information to the repertoire of this large protein family. Our results show that EMP17 is exclusively localized in mitochondria (Figure 1C), and loss-of-function in EMP17 abolishes the editing at ccmFC-799 and nad2-677 sites while reducing the editing at ccmFC-906 and ccmFC -966 sites (Figure 4A). Conversely, the expression of Emp17 restores the editing defects in the emp17 mutant (Figure 4A), demonstrating that EMP17 is required for the editing at these sites. In addition, based on the “one PPR motif: one nucleotide” recognition codes (Barkan et al., 2012), the 6,1’-amino acid residue combinations of EMP17 align well with the upstream sequences of ccmFC-799 and nad2-677 sites, and weakly with those of the ccmFC-906 and -966 sites (Figure 4B), suggesting that EMP17 may recognize its substrates specifically. Furthermore, the lack of editing is accompanied by a reduced mitochondrial complex III assembly, inhibition of the cytochrome pathway, elevated alternative pathway, and severely reduced respiration rates in the emp17 mutants. All of these results provide convincing evidence that EMP17 functions on the C-to-U editing at these sites in mitochondria and the loss of function of EMP17 impairs the cytochrome respiratory pathway.
The severely reduced assembly of complex III suggests that the impaired OXPHOS chain can likely be owed to the loss of editing at the ccmFC sites in emp17 (Figure 4A). Except for the three ccmFC sites and the nad2-677 site, no other defects were found in the transcripts that are directly associated with complex III. In the four mitochondrion-encoded CCM pathway proteins (CcmB, CcmC, CcmFN, and CcmFC) that are essential for the CCM and biogenesis of complex III (Giege et al., 2008), no defects were detected in the transcripts of ccmB, ccmC, and ccmFN. Additionally, expression levels of ccmB, ccmFC, and ccmFN in emp17 were either indistinguishable from that in wild type, or increased (ccmC) (Supplementary Figure 4). CcmFC has been shown to be important for plant growth and development, as the loss of ccmFC expression led to a deficiency of the c-type cytochromes and complex III in wtf9 (Francs-Small et al., 2012). Consistent with that, the Cytc1 and Cytc proteins were barely detectable (Figure 6F), and the assembly of complex III was severely decreased in emp17 (Figure 6C). These results suggest that the unedited ccmFC-799 disrupts the complex III assembly and leads to a dysfunction of CCM.
The editing deficiency at the ccmFC-799 site causing the Ser267-to-Pro267 change in CcmFC is probably the major cause for the inhibited kernel development in emp17. In plant mitochondria, Cytc and Cytc1 are the essential components of mitochondrial complex III in the OXPHOS chain. Maturation of Cytc and Cytc1 is crucial to mitochondrial functions, and hence, to plant growth and development. Impairment of c-type cytochrome maturation arrests seed development or plant growth. A deficiency of mature Cytc1 and Cytc in the ccmh mutant causes embryo lethality in Arabidopsis (Meyer et al., 2005). Similarly, a deficiency of Cytc1 and Cytc resulting from abolished editing at the ccmFN-1553 and -1357 sites in emp7 and ppr27, respectively, results in embryo lethality in maize (Sun F. et al., 2015; Liu et al., 2020). The lack of Cytc1 and Cytc blocks the assembly of mitochondrial complex III, decreases the cytochrome respiration rate, and elevates the alternative non-phosphorylating pathway in the emp7 and ppr27 mutants. For the emp17 mutants, we found consistent results. The assembly of complex III was severely inhibited in emp17, and Vt and Vcyt were decreased to about 18 and 10% in comparison with that in wild type, respectively (Table 1), and the expression of ZmAOX2 and ZmAOX3 was dramatically increased in emp17 compared with wild type (Figures 6F, 7A,B). Thus, the lack of Cytc1 and Cytc and severely reduced biogenesis of mitochondrial complex III blocks the cytochrome pathway and impairs the kernel development.
Our data suggest that the abolished editing at the nad2-677 site is probably not a major cause for the defective seed development in emp17. The editing deficiency at nad2-677 results in a Phe-to-Ser change at Nad2-226. As an essential component of the mitochondrial complex I, a deficiency of Nad2 impairs the assembly of complex I and arrests kernel development in maize (Xiu et al., 2016; Yang et al., 2020). However, the Phe-to-Ser change at Nad2-226 in emp17 does not significantly affect the assembly of mitochondrial complex I, or its activity as determined by in-gel NADH dehydrogenase activity (Figure 6B). Although we cannot rule out the possibility that the detected assembled complex I is in fact non-functional in the electron transfer chain, it is likely that the amino acid residue change in Nad2-226 may have a less detrimental impact on the Nad2 function.
The lack of editing at ccmFC-799 as a result of the Emp17 mutation constitutes a surrogate mutation of Ser267 to Pro267 in the CcmFC protein. The severe impact of this mutation on CCM and complex III assembly illustrates the importance of the Ser residue at CcmFc-267 for the CcmFC function. In bacteria and plant mitochondria, CcmF and CcmH are proposed to take part in the final step of CCM, ligating heme delivered by CcmE to apo-cytc (Giege et al., 2008; Rayapuram et al., 2008). In plant mitochondria, the ccmF gene has been split into multiple genes. For instance, ccmF is split into three genes (ccmFN1, ccmFN2, and ccmFC) in Arabidopsis (Unseld et al., 1997), and two genes (ccmFN and ccmFC) in maize (Clifton et al., 2004). Structure prediction and trypsin digestion experiments suggest that AtCcmFC has six transmembrane helices, three intermembrane space loops, and four mitochondrial matrix domains (Rayapuram et al., 2008). Transmembrane helice prediction by the TMHMM Server v. 2.03 indicates that the structure of ZmCcmFC and AtCcmFC is quite similar. The two proteins share a 78% sequence identity. Based on this structure, Ser267 is located in the third intermembrane space loop named domain 6 (D6) of CcmFC (Figure 8A). Alignment of the protein sequences shows that the Ser267 in various CcmFC proteins is highly conserved across species (Figure 5A), suggesting its importance to CcmFC function. It is known that Pro residue is a disruptor of protein α-helix and not favored in the β-sheet structures, as it has structure is limited and cannot complete the H-bonding network. For example, the Leu-to-Pro change in Nad7-279 and Atp6-213 and in the α-helix region is attributed to the destruction of the Nad7 and Atp6 function in the maize mutant emp18 and smk1 (Li et al., 2014, 2019). The Ser267 in CcmFC was predicted in a β-sheet by the Swiss-model algorithm4 (Figure 8B). Proline is not favored in β-sheet structures as it cannot complete the H-bonding network. It is possible that the Ser267-to-Pro267 mutation in CcmFC in the β-sheet may negatively impact the structural stability of D6 that leads to non-functional CcmFC. The exact function of the D6 intermembrane space loop in CcmFC is unknown, however, these results imply that the Ser267 residue in D6 plays a critical role for the function of CcmFC.
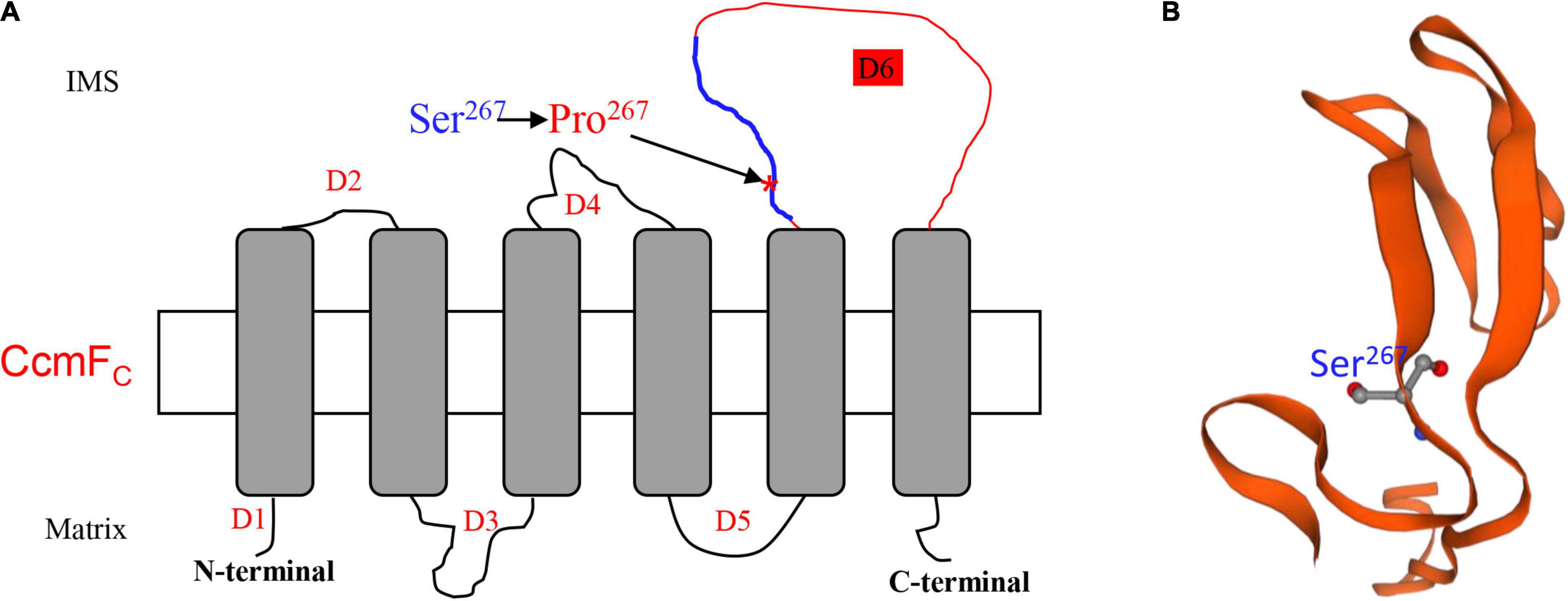
Figure 8. Ser267 is in the third intermembrane space loop named Domain 6 (D6). (A) Topology model of ZmCcmFC predicted by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The Ser267 was marked by arrow. D1-6, Domain1-6; IMS, Intermembrane space. (B) The predicted protein structure of peptide from 258th to 302th in ZmCcmFC marked by blue line in (A).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
YW and B-CT designed the research and analyzed the data. YW and X-YL conducted most of the experiments. FS performed the BN gel assay. Z-QH, Y-YL, and Z-QG participated in the linkage and genetic complementarity analysis. XW performed the respiration rate assay. YW, X-YL, Y-ZY, AS, and B-CT wrote the article. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (Project No. 31630053) and the National Key Research and Development Program of China (Grant No. 2016YFD0101003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Maize Genetic Stock Center for providing the maize stocks.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.693272/full#supplementary-material
Supplementary Figure 1 | The alignment of the DYW domains within PpPPR56, PpPPR65, and EMP17.
Supplementary Figure 2 | The transcription profiling of Emp17 in wild type.
Supplementary Figure 3 | The Linkage analysis of emp17.
Supplementary Figure 4 | Transcription profiling of the 35 mitochondrial protein-coding genes in wild type and emp17.
Supplementary Figure 5 | The co-evolutionary relationship between EMP17 and the editing sites controlled by EMP17.
Supplementary Table 1 | Primers used for the study.
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. doi: 10.1371/journal.pgen.1002910
Bentolila, S., Heller, W. P., Sun, T., Babina, A. M., Friso, G., van Wijk, K. J., et al. (2012). RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 109, E1453–E1461.
Bentolila, S., Oh, J., Hanson, M. R., and Bukowski, R. (2013). Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9:e1003584. doi: 10.1371/journal.pgen.1003584
Bhattacharya, S., Navaratnam, N., Morrison, J. R., Scott, J., and Taylor, W. R. (1994). Cytosine nucleoside/nucleotide deaminases and apolipoprotein B mRNA editing. Trends Biochem. Sci. 19, 105–106. doi: 10.1016/0968-0004(94)90200-3
Blanc, V., Litvak, S., and Araya, A. (1995). RNA editing in wheat mitochondria proceeds by a deamination mechanism. FEBS Lett. 373, 56–60. doi: 10.1016/0014-5793(95)00991-h
Boussardon, C., Avon, A., Kindgren, P., Bond, C. S., Challenor, M., Lurin, C., et al. (2014). The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 203, 1090–1095. doi: 10.1111/nph.12928
Braun, H. P., Binder, S., Brennicke, A., Eubel, H., Fernie, A. R., Finkemeier, I., et al. (2014). The life of plant mitochondrial complex I. Mitochondrion 19(Pt. B), 295–313.
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Clifton, S. W., Minx, P., Fauron, C. M., Gibson, M., Allen, J. O., Sun, H., et al. (2004). Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136, 3486–3503. doi: 10.1104/pp.104.044602
Fey, J., Weil, J., Tomita, K., Cosset, A., Dietrich, A., Small, I., et al. (2002). Role of editing in plant mitochondrial transfer RNAs. Gene 286, 21–24. doi: 10.1016/s0378-1119(01)00817-4
Francs-Small, C. C., Kroeger, T., Zmudjak, M., Ostersetzer-Biran, O., Rahimi, N., Small, I., et al. (2012). A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 69, 996–1005. doi: 10.1111/j.1365-313x.2011.04849.x
Fujii, S., and Small, I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. doi: 10.1111/j.1469-8137.2011.03746.x
Giege, P., and Brennicke, A. (2001). From gene to protein in higher plant mitochondria. C. R. Acad. Sci. Serie III 324, 209–217.
Giege, P., Grienenberger, J. M., and Bonnard, G. (2008). Cytochrome c biogenesis in mitochondria. Mitochondrion 8, 61–73. doi: 10.1016/j.mito.2007.10.001
Glass, F., Hartel, B., Zehrmann, A., Verbitskiy, D., and Takenaka, M. (2015). MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant 8, 1466–1477. doi: 10.1016/j.molp.2015.05.008
Guillaumot, D., Lopez-Obando, M., Baudry, K., Avon, A., Rigaill, G., Falcon de Longevialle, A., et al. (2017). Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. U.S.A. 114, 8877–8882. doi: 10.1073/pnas.1705780114
Haag, S., Schindler, M., Berndt, L., Brennicke, A., Takenaka, M., and Weber, G. (2017). Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9. Nucleic Acids Res. 45, 4915–4928. doi: 10.1093/nar/gkx099
Hackett, J. B., Shi, X., Kobylarz, A. T., Lucas, M. K., Wessendorf, R. L., Hines, K. M., et al. (2017). An organelle RNA recognition motif protein is required for photosystem II subunit psbF transcript editing. Plant Physiol. 173, 2278–2293.
Hammani, K., and Giege, P. (2014). RNA metabolism in plant mitochondria. Trends Plant Sci. 19, 380–389. doi: 10.1016/j.tplants.2013.12.008
Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31, 5907–5916. doi: 10.1093/nar/gkg795
Hayes, M. L., and Santibanez, P. I. (2020). A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 295, 3497–3505. doi: 10.1074/jbc.ra119.011790
Hayes, M. L., Dang, K. N., Diaz, M. F., and Mulligan, R. M. (2015). A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. J. Biol. Chem. 290, 10136–10142. doi: 10.1074/jbc.m114.631630
Jiang, T., Zhang, J., Rong, L., Feng, Y., Wang, Q., Song, Q., et al. (2018). ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 69, 3037–3051. doi: 10.1093/jxb/ery139
Kadowaki, K., Ozawa, K., Kazama, S., Kubo, N., and Akihama, T. (1995). Creation of an initiation codon by RNA editing in the cox1 transcript from tomato mitochondria. Curr. Genet. 28, 415–422. doi: 10.1007/bf00310809
Karpova, O. V., Kuzmin, E. V., Elthon, T. E., and Newton, K. J. (2002). Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14, 3271–3284. doi: 10.1105/tpc.005603
Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. doi: 10.1038/nature03229
Li, X. L., Huang, W. L., Yang, H. H., Jiang, R. C., Sun, F., Wang, H. C., et al. (2019). EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol. 221, 896–907. doi: 10.1111/nph.15425
Li, X.-J., Zhang, Y.-F., Hou, M., Sun, F., Shen, Y., Xiu, Z.-H., et al. (2014). Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79, 797–809. doi: 10.1111/tpj.12584
Liu, R., Cao, S. K., Sayyed, A., Yang, H. H., Zhao, J., Wang, X. M., et al. (2020). The DYW-subgroup pentatricopeptide repeat protein PPR27 interacts with ZmMORF1 to facilitate mitochondrial RNA editing and seed development in maize. J. Exp. Bot. 71, 5495–5505. doi: 10.1093/jxb/eraa273
Liu, Y. J., Xiu, Z. H., Meeley, R., and Tan, B. C. (2013). Empty Pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25, 868–883. doi: 10.1105/tpc.112.106781
Lurin, C., Andrés, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyère, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
McCarty, D. R., Settles, A. M., Suzuki, M., Tan, B. C., Latshaw, S., Porch, T., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44, 52–61. doi: 10.1111/j.1365-313x.2005.02509.x
Meyer, E. H., Giege, P., Gelhaye, E., Rayapuram, N., Ahuja, U., Thony-Meyer, L., et al. (2005). AtCCMH, an essential component of the c-type cytochrome maturation pathway in Arabidopsis mitochondria, interacts with apocytochrome c. Proc. Natl. Acad. Sci. U.S.A. 102, 16113–16118. doi: 10.1073/pnas.0503473102
Meyer, E. H., Tomaz, T., Carroll, A. J., Estavillo, G., Delannoy, E., Tanz, S. K., et al. (2009). Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151, 2187–2187.
Mower, J. P., and Palmer, J. D. (2006). Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol. Genet. Genomics 276, 285–293. doi: 10.1007/s00438-006-0139-3
Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., et al. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268, 434–445. doi: 10.1007/s00438-002-0767-1
O’Toole, N., Hattori, M., Andres, C., Iida, K., Lurin, C., Schmitz-Linneweber, C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128. doi: 10.1093/molbev/msn057
Okuda, K., and Shikanai, T. (2012). A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Res. 40, 5052–5064. doi: 10.1093/nar/gks164
Oldenkott, B., Yang, Y., Lesch, E., Knoop, V., and Schallenberg-Rudinger, M. (2019). Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2:85.
Rayapuram, N., Hagenmuller, J., Grienenberger, J. M., Bonnard, G., and Giege, P. (2008). The three mitochondrial encoded CcmF proteins form a complex that interacts with CCMH and c-type apocytochromes in Arabidopsis. J. Biol. Chem. 283, 25200–25208. doi: 10.1074/jbc.m802621200
Sandoval, R., Boyd, R. D., Kiszter, A. N., Mirzakhanyan, Y., Santibanez, P., Gershon, P. D., et al. (2019). Stable native RIP9 complexes associate with C-to-U RNA editing activity, PPRs, RIPs, OZ1, ORRM1 and ISE2. Plant J. 99, 1116–1126. doi: 10.1111/tpj.14384
Shi, X., Castandet, B., Germain, A., Hanson, M. R., and Bentolila, S. (2017). ORRM5, an RNA recognition motif-containing protein, has a unique effect on mitochondrial RNA editing. J. Exp. Bot. 68, 2833–2847. doi: 10.1093/jxb/erx139
Shi, X., Germain, A., Hanson, M. R., and Bentolila, S. (2016). RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiol. 170, 294–309. doi: 10.1104/pp.15.01280
Shi, X., Hanson, M. R., and Bentolila, S. (2015). Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Res. 43, 3814–3825. doi: 10.1093/nar/gkv245
Small, I. D., and Peeters, N. (2000). The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47.
Small, I. D., Schallenberg-Rudinger, M., Takenaka, M., Mireau, H., and Ostersetzer-Biran, O. (2020). Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 01, 1040–1056. doi: 10.1111/tpj.14578
Soltis, D. E., Smith, S. A., Cellinese, N., Wurdack, K. J., Tank, D. C., Brockington, S. F. et al. (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730. doi: 10.3732/ajb.1000404
Sosso, D., Mbelo, S., Vernoud, V., Gendrot, G., Dedieu, A., Chambrier, P., et al. (2012). PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24, 676–691. doi: 10.1105/tpc.111.091074
Sun, F., Wang, X., Bonnard, G., Shen, Y., Xiu, Z., Li, X., et al. (2015). Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 84, 283–295. doi: 10.1111/tpj.12993
Sun, T., Bentolila, S., and Hanson, M. R. (2016). The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 21, 962–973. doi: 10.1016/j.tplants.2016.07.005
Sun, T., Germain, A., Giloteaux, L., Hammani, K., Barkan, A., Hanson, M. R., et al. (2013). An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. U.S.A. 110, E1169–E1178.
Sun, T., Shi, X., Friso, G., Van Wijk, K., Bentolila, S., and Hanson, M. R. (2015). A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 11:e1005028. doi: 10.1371/journal.pgen.1005028
Takenaka, M., Zehrmann, A., Verbitskiy, D., Kugelmann, M., Hartel, B., and Brennicke, A. (2012). Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109. doi: 10.1073/pnas.1202452109
Tan, B. C., Chen, Z. L., Shen, Y., Zhang, Y. F., Lai, J. S., and Sun, S. S. M. (2011). Identification of an active new mutator transposable element in maize. G3 1, 293–302. doi: 10.1534/g3.111.000398
Tasaki, E., Hattori, M., and Sugita, M. (2010). The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFC transcript. Plant J. 62, 560–570. doi: 10.1111/j.1365-313x.2010.04175.x
Thony-Meyer, L., Fischer, F., Kunzler, P., Ritz, D., and Hennecke, H. (1995). Escherichia coli genes required for cytochrome c maturation. J. Bacteriol. 177, 4321–4326. doi: 10.1128/jb.177.15.4321-4326.1995
Tillich, M., Funk, H. T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., et al. (2005). Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43, 708–715. doi: 10.1111/j.1365-313x.2005.02484.x
Tsudzuki, T., Wakasugi, T., and Sugiura, M. (2001). Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 53, 327–332. doi: 10.1007/s002390010222
Unseld, M., Marienfeld, J. R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 15, 57–61. doi: 10.1038/ng0197-57
Wagoner, J. A., Sun, T., Lin, L., and Hanson, M. R. (2015). Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 290, 2957–2968. doi: 10.1074/jbc.m114.622084
Wang, G., Zhong, M., Shuai, B., Song, J., Zhang, J., Han, L., et al. (2017). E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 214, 1563–1578. doi: 10.1111/nph.14507
Wang, H. C., Sayyed, A., Liu, X. Y., Yang, Y. Z., Sun, F., Wang, Y., et al. (2020). SMALL KERNEL4 is required for mitochondrial cox1 transcript editing and seed development in maize. J. Integr. Plant Biol. 62, 777–792. doi: 10.1111/jipb.12856
Wang, X.-M., Chang, N., Bi, Y.-R., and Tan, B.-C. (2015). Measurement of mitochondrial respiration rate in maize (Zea mays) leaves. Bio Protoc. 5:e1483.
Wang, Y., Liu, X. Y., Yang, Y. Z., Huang, J., Sun, F., Lin, J., et al. (2019). Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 15:e1008305. doi: 10.1371/journal.pgen.1008305
Xiao, H., Zhang, Q., Qin, X., Xu, Y., Ni, C., Huang, J., et al. (2018). Rice PPS1 encodes a DYW motif-containing pentatricopeptide repeat protein required for five consecutive RNA-editing sites of nad3 in mitochondria. New Phytol. 220, 878–892. doi: 10.1111/nph.15347
Xie, T., Chen, D., Wu, J., Huang, X., Wang, Y., Tang, K., et al. (2016). Growing Slowly 1 locus encodes a PLS-type PPR protein required for RNA editing and plant development in Arabidopsis. J. Exp. Bot. 67, 5687–5698. doi: 10.1093/jxb/erw331
Xiu, Z., Sun, F., Shen, Y., Zhang, X., Jiang, R., Bonnard, G., et al. (2016). EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J. 85, 507–519. doi: 10.1111/tpj.13122
Xu, C. H., Song, S., Yang, Y. Z., Lu, F., Zhang, M. D., Sun, F., et al. (2020). DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. Plant J. 103, 1767–1782. doi: 10.1111/tpj.14862
Yang, Y. Z., Ding, S., Wang, H. C., Sun, F., Huang, W. L., Song, S., et al. (2017). The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 214, 782–795. doi: 10.1111/nph.14424
Yang, Y. Z., Ding, S., Wang, Y., Wang, H. C., Liu, X. Y., Sun, F., et al. (2020). PPR20 is required for the cis-splicing of mitochondrial nad2 intron 3 and seed development in maize. Plant Cell Physiol. 61, 370–380. doi: 10.1093/pcp/pcz204
Yu, W., and Schuster, W. (1995). Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant-mitochondria. J. Biol. Chem. 270, 18227–18233. doi: 10.1074/jbc.270.31.18227
Keywords: CcmFC, EMP17, pentatricopeptide repeat protein, mitochondrion, seed development, maize
Citation: Wang Y, Liu X-Y, Huang Z-Q, Li Y-Y, Yang Y-Z, Sayyed A, Sun F, Gu Z-Q, Wang X and Tan B-C (2021) PPR-DYW Protein EMP17 Is Required for Mitochondrial RNA Editing, Complex III Biogenesis, and Seed Development in Maize. Front. Plant Sci. 12:693272. doi: 10.3389/fpls.2021.693272
Received: 06 May 2021; Accepted: 01 July 2021;
Published: 28 July 2021.
Edited by:
Julian Eaton-Rye, University of Otago, New ZealandReviewed by:
Niaz Ahmad, National Institute for Biotechnology and Genetic Engineering, PakistanCopyright © 2021 Wang, Liu, Huang, Li, Yang, Sayyed, Sun, Gu, Wang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Cai Tan, YmN0YW5Ac2R1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.