
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 August 2021
Sec. Plant Breeding
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.691576
This article is part of the Research Topic Advances in Breeding for Quantitative Disease Resistance View all 28 articles
Cucumber is a widely grown vegetable crop plant and a host to many different plant pathogens. Cucumber vein yellowing virus (CVYV) causes economic losses on cucumber crops in Mediterranean countries and in some part of India such as West Bengal and in African countries such as Sudan. CVYV is an RNA potyvirus transmitted mechanically and by whitefly (Bemisia tabaci) in a semipersistent manner. Control of this virus is heavily dependent on the management of the insect vector and breeding virus-resistant lines. DNA markers have been used widely in conventional plant breeding programs via marker-assisted selection (MAS). However, very few resistance sources against CVYV in cucumber exist, and also the lack of tightly linked molecular markers to these sources restricts the rapid generation of resistant lines. In this work, we used genomics coupled with the bulked segregant analysis method and generated the MAS-friendly Kompetitive allele specific PCR (KASP) markers suitable for CsCvy-1 selection in cucumber breeding using a segregating F2 mapping population and commercial plant lines. Variant analysis was performed to generate single-nucleotide polymorphism (SNP)-based markers for mapping the population and genotyping the commercial lines. We fine-mapped the region by generating new markers down to 101 kb with eight genes. We provided SNP data for this interval, which could be useful for breeding programs and cloning the candidate genes.
Cucumber plants, Cucumis sativus, have been cultivated as a vegetable crop across the globe for centuries (Tatlioglu, 1993). The fruit is consumed as fresh or industrialized product, and the major producing countries are China (7,033,8971 tons), Turkey (1,916,645 tons), Russia (1,626,360 tons), Ukraine (1,034,170 tons), and Iran [871,692 tons (FAO, 2019)]. As an important vegetable, cucumber is challenged by many different fungal, oomycete, bacterial, and viral pathogens (Kong et al., 2015; Słomnicka et al., 2018; Bandamaravuri et al., 2020).
One of the most devastating viral pathogens is Cucumber vein yellowing virus (CVYV), which belongs to the Potyviridae family (Lecoq et al., 2000), has an RNA genome (Janssen et al., 2005), is transmitted mechanically and by whitefly, Bemisia tabaci, in a semipersistent manner (Mansour and Al-Musa, 1993), and infects a number of cucurbit species (Gil-Salas et al., 2011). The occurrence and heavy crop losses due to CVYV infection in the open fields and under protected cucumber crops have been reported in the Mediterranean countries from Israel to Portugal (Cohen and Nitzany, 1960; Louro et al., 2004). The main symptoms of CVYV on the cucumber include vein clearing followed by vein yellowing on the youngest leaves (Cohen and Nitzany, 1960), the occasional occurrence of yellow/green mosaics on the fruit (Cuadrado et al., 2007), and eventual general necrosis of the entire infected plant (Cohen and Nitzany, 1960). Mechanical transmission of the virus allows the use of cucumber as a test and indicative plant for multiplication.
Cucumber vein yellowing virus has been classified as a quarantine viral pathogen in the EPPO A2 Action List (https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list). Control of this virus relies heavily on the application of integrated pest management (IPM) practices that incorporate the ecosystem-based strategies, including cultural practices, biological and chemical control of the vector, and the use of resistant varieties (Horowitz et al., 2011). Sanitation, use of certified virus-free seedlings, and eradicating diseased plants parts are common practices for controlling viral plant pathogens (Hilje et al., 2001; Nazarov et al., 2020). Although chemical pesticides have been used to control the whitefly insect vector, concerns to human health, occurrence of insecticide resistance, and damage to the environment led to a search for alternative measures (Sani et al., 2020). Use of microbial biological control agents (MBCA), such as entomopathogenic fungi (Faria and Wraight, 2001; Sani et al., 2020), use of barrier or trap crops (Zhang et al., 2020), and use of beneficial insects, such as predators or parasitoids (Moreno-Ripoll et al., 2014), have been considered.
The RNA-guided genome editing using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 has been also used to generate virus-resistant crops (Liu and Fan, 2014). For example, Chandrasekaran et al. (2016) used Cas9/sgRNA constructs to target the recessive eukaryotic translation initiation factor 4E (eIF4E) gene in cucumber. They reported that the homozygous T3 lines showed immunity to CVYV (Chandrasekaran et al., 2016) indicating the possibility of alternative new methods for CVYV control. Planting cultivars resistant to the whitefly and/or to the virus is one of the most important control measures in the CVYV management. In a study to identify cucumber lines resistant to whitefly, Novaes et al. (2020) screened 60 genotypes and found that accessions IAC-1214, IAC-1201, Campeiro, Japonês, IAC-1311, Kyria, and IAC-1175 displayed some low levels of attractiveness to these insects and suggested they could be included in the breeding programs to develop whitefly-resistant cucumber lines.
Genetics of resistance to CVYV have been investigated by several groups (Picó et al., 2003). A Spanish landrace of short cucumber, C. sat-10, was found to be monogenic and displaying dominant resistance to CVYV (Picó et al., 2008). Similarly, a cucumber cultivar named Kyoto-3-feet originating from Japan has been reported to be resistant to CVYV (Martín-Hernández and Picó, 2021); however, detailed information on the nature of these resistance mechanisms is not available. Cucumber hybrid lines resistant to CVYV exist in the commercial market; however, currently all the work for selecting resistant lines relies on traditional pathotesting efforts. Recently, Pujol et al. (2019) described their elegant study on the resistant accession CE0749, a CVYV-resistant long Dutch-type cucumber. They used genomics and bulked segregant analysis (BSA) (Michelmore et al., 1991) and fine-mapped a locus containing the gene CsCvy-1 locus in a 625 kb region with 24 candidate genes (Pujol et al., 2019).
Here, we described our investigations on the identification of DNA markers for fine mapping CsCvy-1 using genomics and BSA. We used both segregating F2 populations and the available commercial F1 hybrids, mapped the locus down to 101 kb with eight genes, and provided single-nucleotide polymorphism (SNP) data for the interval, which could be useful for plant breeding programs.
An F2 mapping population, generated from a cross between a susceptible (YT-189-1) and a resistant (YT-MLN-33) cucumber inbred lines (Yüksel Tohum A.S., Antalya, Turkey), was used in the phenotyping and genotyping experiments. F3 families were raised by selfing the selected lines and used to determine the genotype of the F2 lines.
The CVYV isolate used in this study was obtained from DSMZ (Braunschweig, Germany) and propagated in susceptible cucumber plants (Cucumis sativus, line YT-189-1). Virus inoculum was prepared by homogenizing 1 g infected leaves in 4 ml 0.01 M phosphate buffer (pH 7.0) containing 0.2% sodium sulfate and 0.2% diethyldithiocarbamic acid (DIECA, Sigma-Aldrich, St. Louis, MO, United States). After adding 600-mesh carborundum and active carbon, cotyledons of cucumber plants (parental lines, F1, and F3 generations), which were at the cotyledon to one-true-leaf stages, were mechanically inoculated. A second inoculation was performed 3 days after the first one to eliminate escapees. The inoculated cucumber seedlings were then kept in a growth chamber with temperature control set at 30/25°C (day/night) with a 16 h light/8 h dark photoperiod for 3 weeks and observed every 2 days. First symptoms were observed 5–7 days postinoculation (dpi), but became obvious after 12–15 dpi. After 3 weeks, no further symptom developments were observed; thus, 15 dpi was selected to be the optimal time for symptom evaluation. Plants showing clear symptoms including mosaic and vein yellowing on leaves were rated as susceptible, whereas those with no symptoms or a very light vein discoloration on only the oldest ones were accepted as resistant. A minimum of 20 plants was used per treatment.
Young leaves were collected from parental and F2 lines. Plant genomic DNA was isolated using the Wizard Magnetic Kit (Promega, Madison, WI, United States) following the instructions of manufacturer. DNA was extracted from each individual plant lines, and a gel electrophoresis was performed to determine whether high molecular weight DNAs were isolated. The resistant and susceptible bulks were generated from 20 resistant and 20 susceptible F2 individuals, respectively, as described in earlier studies (Devran et al., 2015, 2018). Genomic DNA library and sequencing have been carried out by the University of Exeter Sequencing Service after quality check of DNAs, generating 2 ×150 bp paired-end read data for each parent line and bulked (resistant and susceptible) pools with Illumina HiSeq 2500 (Illumina, Inc. San Diego, CA, USA).
As previously described (Devran et al., 2018), we took the NGS analysis approach where the raw reads were trimmed using BBDuk (filter = 27, trimk = 27; https://sourceforge.net/projects/bbmap/) to remove Illumina adapters and to quality trim both ends to Q12. Subsequently, trimmed sequences from parental lines and the bulks were mapped onto the available reference cucumber genome (V2 and V3) using BBMap (https://sourceforge.net/projects/bbmap/), and the alignment data were converted to the BAM format (Li et al., 2009). As the CsCvy-1 locus was previously mapped onto chromosome 5 (Pujol et al., 2019), the data from the interval on chromosome 5: 7,000,000–7,850,000 were extracted using SAMtools (Li et al., 2009). The variant detection has been performed using BCFtools (Li et al., 2009) and a publicly available custom script (https://github.com/davidjstudholme/SNPsFromPileups) as previously described; (Yemataw et al., 2018). Integrative Genomics Viewer (IGV) was used to visualize the alignment results (Robinson et al., 2011).
Several of the SNPs within the interval were converted to Kompetitive Allele Specific PCR markers (KASP) by taking 100 bases either side of the SNP. KASP primers were developed using the LGC's primer picker software, Middlesex, United Kingdom. The PCRs were performed in a total volume of 15 μl that included DNA (10 ng; 5 μl), KASP Assay Mix (0.2 μl), KASP Master Mix (7.5 μl), and distilled water (2.3 μl). The KASP assay reactions were performed using the LightCycler® 480 II (Roche) using 61–55°C touchdown protocol (https://biosearch-cdn.azureedge.net/assetsv6/KASP-thermal-cycling-conditions-all-protocols.pdf). The fluorescence signal was measured for 2 min at 25°C using a FluOstar Omega Microplate Reader (BMG LABTECH, Ortenberg, Germany).
As the CsCvy-1 locus had been previously mapped (Pujol et al., 2019), we used some of the published KASP markers including CVYV-184, CVYV-187, CVYV-188, CVYV-190, and CVYV-122 in this work. Published and newly generated KASP markers were first tested on parents to confirm the identified polymorphisms and then 120 segregating F2 lines. Marker genotyping data and the viral disease phenotyping data were used to confirm the CsCvy-1 interval. As we developed new markers (Supplementary Table 1) to narrow the interval down, we also tested these markers with the commercial F1 hybrid lines, which were obtained from the relevant companies. As F2 lines are a segregating population, markers discovered using F2s may not be a reliable MAS-friendly marker. Therefore, we used F1 hybrid lines to narrow the interval further down and identify the MAS-friendly markers.
Cucumber reference genome sequences ChineseLong 9930 v2 are at http://cucurbitgenomics.org/organism/2 ChineseLong 9930 v3 at (https://ftp.ncbi.nlm.nih.gov/genomes/genbank/plant/Cucumis_sativus/latest_assembly_versions/GCA_000004075.3_Cucumber_9930_V3/). The raw sequence reads aligning to the interval have been deposited in the Sequence Read Archive (SRA) and are accessible via BioProject accession PRJNA713378.
A cross was generated between the susceptible C. sativus inbred line YT-189-1 and the resistant inbred line, YT-MLN-33. The F1 hybrid showed resistance to CVYV, indicating that resistance was dominant. The F1 was selfed to generate segregating F2 populations. A total of 120 F2 lines were taken to F3 level, and 20 F3 lines descending from each F2s were inoculated with the virus to determine accurately the phenotype of the mapping population. Disease symptoms, including mosaics and vein yellowing, were obvious on the leaves of susceptible plants at 15 dpi (Figure 1). The segregation ratio observed in this bioassay was 92:28 (resistant:susceptible, 3:1; with Chi-square = 0.05 and p ≤ 0.05), suggesting that a single locus was providing resistance to CVYV in this cross and allowing the subsequent analysis.

Figure 1. Healthy and Cucumber vein yellowing virus (CVYV)-infected cucumber leaves. Cucumber plants were mechanically inoculated at the cotyledon to one-true-leaf stages. A second inoculation was performed 3 days after the first one. The inoculated cucumber seedlings were then kept in a growth chamber with temperature control set at 30/25°C (day/night) with a 16 h light/8 h dark photoperiod for 3 weeks and observed every 2 days. The plants were evaluated for symptom development at 15 days after the first inoculation (dpi). Control plants were treated with buffer without virus in a similar manner. (A) Leaf of an uninoculated control plant, (B) leaf of a Cucumber vein yellowing virus (CVYV)-inoculated-resistant plant, and (C) leaf of a CVYV-inoculated-susceptible plant.
We used a next-generation sequencing (NGS)-based BSA approach whereby we generated bulks from DNA isolated from 20 resistant and susceptible F2 lines. We generated 150-bp paired-end Illumina HiSeq2500 sequencing data from the two parents and bulks (resistant and susceptible). A total of 390 million reads for each parent and 391 million reads for each bulk were generated. We then mapped these reads onto to the cucumber reference genome sequence (GenBank: GCA_000004075.3). However, during the course of our work, a locus designated CsCvy-1 mapped on chromosome 5 was published using BSA approach (Pujol et al., 2019). This prompted us to check whether we were mapping the same region even though we were using different breeding lines. We used published CVYV-184, CVYV-187, CVYV-188, CVYV-190, and CVYV-122 KASP markers (Pujol et al., 2019) to determine whether the resistance locus in our parental line YT-MLN-33 is linked to CsCvy-1. Our mapping data showed a clear linkage (Table 1, Supplementary Figure 1), and therefore we concentrated on chromosome 5. As we had already performed an SNP analysis using the then-available version of the reference genome sequence (GCA_000004075.2), we developed several KASP markers and mapped the CsCvy-1 locus in our segregating mapping population. To make our work comparable with the published data, we then mapped our clean NGS reads onto the updated reference genome sequence (GCA_000004075.3), concentrated on a region between SNP10218, identified in this work, and the published CVYV122 marker (Supplementary Table 2). Several of the published markers were not polymorphic for the parental lines we used, e.g., CVYV-173, CVYV-174, CVYV-175, and CVYV 176.
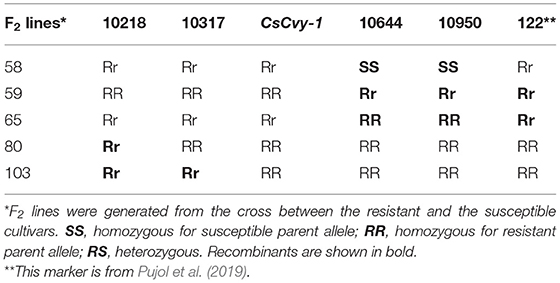
Table 1. Molecular markers used to define the interval for CsCvy-1 locus and the critical recombinant F2 lines.
As our ultimate aim was to identify a marker that is tightly linked to CsCvy-1, we wanted to narrow the interval and generate further markers to identify an MAS-friendly marker. Using NGS data from parents and bulks, we mined the data on chromosome 5: 10,218,000–11,370,000 (ChineseLong 9930 ASM407v2, Supplementary Figure 1). KASP markers were then designed and used for mapping to narrow the interval. A total of 13 new KASP markers were generated, and the locus was mapped down to a 327-kb interval between the markers 10,317 and 10,644 K using the available F2 lines (Supplementary Table 2). As the version-three reference genome sequence became available, we also used this version and mined the data on chromosome 5: 7,000,000–7,850,000 (GCA_000004075.3, Supplementary Figure 2) for SNPs. A total of 436 SNPs have been detected (Supplementary Table 3). It should be noted here that although markers developed in this work using the GCA_000004075.V2 reference map to the region, several of the newly developed ones, especially toward the marker CVYV-122, were missing when the GCA_000004075.V3 reference was used. This may have been due to misassembly of the region as there was a 394 kb was missing in the GCA_000004075.3 genome (Figure 2).
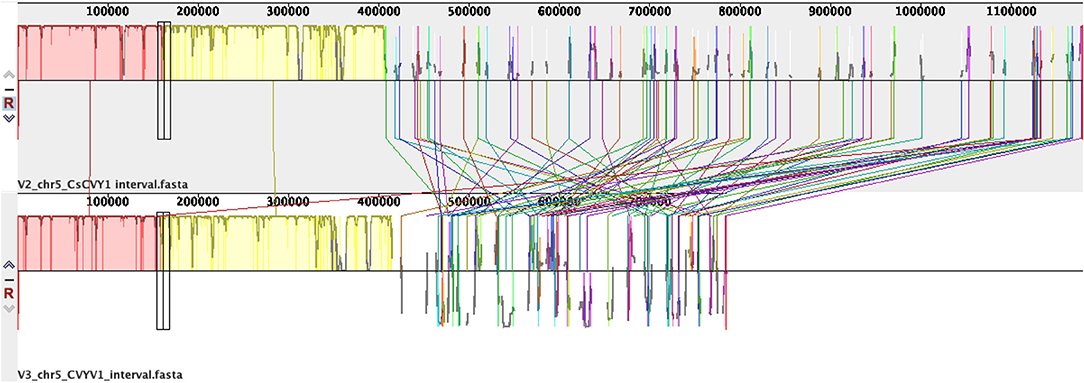
Figure 2. Pairwise sequence alignment of CsCvy-1 interval in reference genomes version two and three (GCA_000004075.2 and GCA_000004075.3). Sequences were aligned using Progressive Mauve (Darling et al., 2004).
Although we had enough number of markers to map the locus further, the number of F2 lines to bring the interval down was not sufficient to identify further recombinants. We then obtained seeds of more than 20 commercial cucumber varieties with claimed CVYV phenotype and confirmed their phenotype by testing them with the CVYV isolate. Their DNAs were screened with our newly developed markers, and we narrowed down the locus to a 101-kb interval between the markers 10,317 and 10,418 K (Supplementary Table 4, Table 2). This finding suggests that the polymorphism identified in this work has been maintained across different varieties that have been used in the commercial breeding programs. In addition, the identified polymorphisms within the interval could be used in a breeding program by checking the existence of polymorphisms in the lines used.
CsCvy-1 locus mapped by Pujol et al. (2019) contained 24 genes. However, as we mapped the interval down to 101 kb, we used the annotations of the cucumber reference genome (GCA_000004075.3) to identify genes within the interval. The CsCvy-1 locus in our mapping interval contains eight predicted genes (Table 3). Although CsaV3_5G011160 encodes a cytochrome P450-like protein, CsaV3_5G011170, CsaV3_5G011190, and CsaV3_5G011230 encode unknown proteins. However, CsaV3_5G011180 encodes a serine/arginine repetitive matrix protein 2 isoform X2, CsaV3_5G011220 encodes an endo-1,4-beta-xylanase, and two genes, CsaV3_5G011200 and CsaV3_5G011210, both encode RNA-dependent RNA polymerase 1-like (RDR1-like) proteins. Interestingly, the deletion in the intragenic region of the RDR1 reported in this interval (Pujol et al., 2019) has been maintained in the resistant inbred line we used.
Here, we present genetic evidence that a single dominant locus CsCvy-1 confers resistance to CVYV infection in our inbred lines, consistent with a recent report (Pujol et al., 2019). Our genomic and molecular investigations using an F2 mapping population and also commercial resistant and susceptible varieties enabled us to map this locus down to 101-kb interval in which eight genes reside. Use of genomics allowed the identification of SNPs that could be used in breeding programs.
The plant immune system has the ability to recognize extracellular or intracellular molecules derived from plant pathogens and generate a defense response to restrict the pathogen growth or replication (Wang et al., 2008; Tör et al., 2009; Steinbrenner et al., 2015). Map-based studies usually involve the phenotyping and genotyping of a large number of individual plants in a segregating population. Using this approach, genes conferring resistance to fungal, oomycete, bacterial, and viral pathogens have been mapped, and many of them have been cloned (Tai et al., 1999; Borhan et al., 2008; Kim et al., 2017; Chen et al., 2021).
Linkage analysis plays a significant role in the cloning genes or generating markers tightly linked to the locus of interest using map-based approach. When we started this investigation, there was no published data on the chromosomal location of the gene conferring resistance to CVYV. During our SNP analysis, Pujol et al. (2019) published their work on the mapping of CsCvy-1 in cucumber using genomic approach. We used relevant markers from this published work; however, several of them were not polymorphic in our parental lines, indicating the importance of generating SNP data from the lines used in generating mapping populations. After establishing the linkage between the resistance source in our material and the CsCvy-1, it was obvious that we were mapping the same locus, and thus we zoomed into the region.
We used genomics and BSA previously to clone genes (Woods-Tör et al., 2018) and to generate MAS-friendly molecular markers (Devran et al., 2018) that are tightly linked to the gene of interest. Our experience shows that although the use of reference genomes helps the identification of variants in the region of interest, different versions of reference genome assemblies produced different results in the SNP analysis. It was the case in this study where we initially used version 2 (GCA_000004075.2) as the reference and generated markers for our mapping work. Although all the markers generated from version 2 mapped the gene, several of them were missing when version 3 (GCA_000004075.3) were used, indicating the importance of mapping for confirmation and using more than one available reference genome.
High number of individual lines in a map-based study help identify the recombinant lines, which enables narrowing the interval. It can be easy to generate large number of F2 lines from plants, such as Arabidopsis thaliana (Tör et al., 2002). However, in plants such as cucumber, it may not be possible to achieve large number of F2s. In this work, we relied on 120 F2 lines to narrow the interval down to a 327 kb. Considering the breeding efforts where many characters are collected in a “pure” line, during which many crosses are carried out and many recombination events take place, for an MAS-friendly marker, the interval needs to be very small so that the likelihood of a recombination event between the marker and the gene of interest is almost zero. Bearing this in mind, we used the commercial cucumber F1 hybrids in our phenotyping and genotyping assays and reduced the interval down to 101 kb with eight genes.
Resistance to plant pathogens could be provided by membrane-bound proteins, such as receptor-like proteins (RLPs) (Wang et al., 2008) or receptor-like kinases (RLKs) (Roux et al., 2011; Zhang et al., 2013) or by the cytoplasmic nucleotide-binding, leucine-rich repeat (NLR) immune receptors (Adachi et al., 2019). There were no classic RLP, RLK, or NLR-type genes in the 101-kb CsCvy-1 interval. Pujol et al. (2019) looked into the small variants and structural variation in the locus and argued that CsaV3_5G011180 encoding for serine/arginine repetitive matrix protein (SARMP) could be a possible candidate. In addition, Pujol et al. (2019) postulated that CsaV3_5G011200 and CsaV3_5G011210 encoding RDRs 1a and 1b had the most appealing modifications in the locus and discussed the role of RDRs in RNA silencing pathways. Leibman et al. (2018) carried out detailed investigations into the RDR1-like genes in cucumber and reported the presence of four putative RDR1-family genes. They then investigated the expression of these RDR1-like genes and their role in defense against different viruses, including Zucchini yellow mosaic virus (ZYMV), CMV, and CVYV and showed that the level of RDR1-like gene expression varied according to the virus used (Leibman et al., 2018).
The NLR-type disease resistance genes in Arabidopsis have been reported to be clustered in the genome (Holub, 2007), and some of them function together and could be in head-to-head orientation, termed paired NLRs (Saucet et al., 2015). Further detailed studies indicated that one of them could function as a pathogen sensor, and the other member as signaling executor (Van de Weyer et al., 2019). Here, we have RDR1a and RDR1b in the interval right next to each other, functioning “like an R-gene” (Leibman et al., 2018), but it is not totally clear from expression studies whether they function together as some genetic investigations are needed. It is tempting to speculate that RDR1a and RDR1b are the most suitable candidate genes for the CVYV resistance.
Our strategy to use genomics and BSA to identify SNPs and generate molecular markers that could be employed in the selection of CsCvy-1 enabled us to screen several markers and narrowed the interval down. These SNPS and markers could be used to identify polymorphism in different backgrounds in any breeding program to select CsCvy-1. Subsequent experiments could be designed to silence both RDR1 and RDR2 genes individually and together in the same background to reveal their dependence onto each other and their contribution to CVYV defense.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA713378.
EK, ZD, and MT planned and designed the research. EK carried out pathology tests. ZD performed genotyping using markers. EÖ made crossings between plant lines and produced offspring. YH, MT, and DS analyzed and interpreted the data. EK, ZD, YH, DS, and MT wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that this study received funding from M.Y. Genetik Tarim Tek. Lab. Tic. Ltd. Sti. and Yüksel Tohum A.S. The funder had the following involvement in the study: M.Y. Genetik Tarim Tek. Lab. Tic. Ltd. Sti. provided equipments to carry out pathology and molecular tests, and Yüksel Tohum A.S. provided seeds and plant materials. The sequencing work in this project utilized equipment funded by the Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT101650MA), and BBSRC LOLA award (BB/K003240/1).
EK was employed by company M.Y. Genetik Tarim Tek. Lab. Tic. Ltd. Sti. EÖ as employed by company Yüksel Tohum Tarim San. ve Tic. A. S.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.691576/full#supplementary-material
Supplemental Figure 1. Position of SNP-based markers on chromosome 5 in ChineseLong 9930 ASM407v2.
Supplemental Figure 2. Position of SNP-based markers on chromosome 5 in ChineseLong 9930 GCA_000004075.3.
Supplemental Table 1. KASP marker sequences developed in this study and previously published work.
Supplemental Table 2. Mapping CsCvy-1 with existing and newly generated KASP markers using F2 population.
Supplemental Table 3. SNPs and their positions in CsCvy-1 locus on chromosome 5 using GCA_000004075.3. Proportions of aligned reads supporting the variant base are given.
Supplemental Table 4. Mapping CsCvy-1 with existing and newly generated KASP markers using F1 hybrids.
Adachi, H., Contreras, M. P., Harant, A., Wu, C. H., Derevnina, L., Sakai, T., et al. (2019). An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. Elife 8:121. doi: 10.7554/eLife.49956
Bandamaravuri, K. B., Nayak, A. K., Bandamaravuri, A. S., and Samad, A. (2020). Simultaneous detection of downy mildew and powdery mildew pathogens on Cucumis sativus and other cucurbits using duplex-qPCR and HRM analysis. AMB Express 10:135. doi: 10.1186/s13568-020-01071-x
Borhan, M. H., Gunn, N., Cooper, A., Gulden, S., Tör, M., Rimmer, S. R., et al. (2008). WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. MPMI 21, 757–768. doi: 10.1094/MPMI-21-6-0757
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chen, C., Jost, M., Clark, B., Martin, M., Matny, O., Steffenson, B. J., et al. (2021). BED-domain-containing NLR from wild barley confers resistance to leaf rust. Plant Biotechnol. J. 19:1206–1215. doi: 10.1111/pbi.13542
Cohen, S., and Nitzany, F. (1960). A whitefly transmitted virus of Cucurbits in Israel. Phytopathol Mediterranea 1, 44–46.
Cuadrado, I. M., Janssen, D., Velasco, L., Ruiz, L., and Segundo, E. (2007). First report of cucumber vein yellowing virus in Spain. Plant Dis. 85, 336–336. doi: 10.1094/PDIS.2001.85.3.336A
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Devran, Z., Kahveci, E., Hong, Y., Studholme, D. J., and Tör, M. (2018). Identifying molecular markers suitable for Frl selection in tomato breeding. Theor. Appl. Genet. 131:2099–2105. doi: 10.1007/s00122-018-3136-0
Devran, Z., Kahveci, E., Özkaynak, E., Studholme, D. J., and Tör, M. (2015). Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing. Mol, Breed. 35:101. doi: 10.1007/s11032-015-0294-5
FAO (2019). Available online at: http://www.fao.org/faostat/en/#data/QC (accessed June 5, 2021).
Faria, M., and Wraight, S. P. (2001). Biological control of Bemisia tabaci with fungi. Crop Prot. 20, 767–778. doi: 10.1016/S0261-2194(01)00110-7
Gil-Salas, F. M., Peters, J., Boonham, N., Cuadrado, I. M., and Janssen, D. (2011). Yellowing disease in zucchini squash produced by mixed infections of Cucurbit yellow stunting disorder virus and Cucumber vein yellowing virus. Phytopathology 101, 1365–1372. doi: 10.1094/PHYTO-12-10-0343
Hilje, L., Costa, H. S., and Stansly, P. A. (2001). Cultural practices for managing Bemisia tabaci and associated viral diseases. Crop Prot. 20, 801–812. doi: 10.1016/S0261-2194(01)00112-0
Holub, E. B. (2007). Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10, 415–424. doi: 10.1016/j.pbi.2007.05.003
Horowitz, A. R., Antignus, Y., and Gerling, D. (2011). Management of Bemisia tabaci whiteflies, in The Whitefly, Bemisia tabaci (Homoptera: Aleyrodidae) Interaction With Geminivirus-Infected Host Plants, ed W. Thompson (Dordrecht: Springer), 293–322. doi: 10.1007/978-94-007-1524-0_11
Janssen, D., Martín, G., Velasco, L., Gómez, P., Segundo, E., Ruiz, L., et al. (2005). Absence of a coding region for the helper component-proteinase in the genome of cucumber vein yellowing virus, a whitefly-transmitted member of the Potyviridae. Arch. Virol. 150, 1439–1447. doi: 10.1007/s00705-005-0515-z
Kim, S. B., Kang, W. H., Huy, H. N., Yeom, S. I., An, J. T., Kim, S., et al. (2017). Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. 213:886–899. doi: 10.1111/nph.14177
Kong, W., Chen, N., Liu, T., Zhu, J., Wang, J., He, X., et al. (2015). Large-scale transcriptome analysis of cucumber and Botrytis cinerea during Infection. PLoS ONE 10:e0142221. doi: 10.1371/journal.pone.0142221
Lecoq, H., Desbiez, C., Delécolle, B., Cohen, S., and Mansour, A. (2000). Cytological and molecular evidence that the whitefly-transmitted Cucumber vein yellowing virus is a tentative member of the family Potyviridae. J. General Virol. 81, 2289–2293. doi: 10.1099/0022-1317-81-9-2289
Leibman, D., Kravchik, M., Wolf, D., Haviv, S., Weissberg, M., Ophir, R., et al. (2018). Differential expression of cucumber RNA-dependent RNA polymerase 1 genes during antiviral defence and resistance. Mol. Plant Pathol. 19, 300–312. doi: 10.1111/mpp.12518
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, L., and Fan, X. D. (2014). CRISPR–Cas system: a powerful tool for genome engi-neering. Plant Mol. Biol. 85, 209–218. doi: 10.1007/s11103-014-0188-7
Louro, D., Quinot, A., Neto, E., Fernandes, J. E., Marian, D., Vecchiati, M., et al. (2004). Occurrence of Cucumber vein yellowing virus in cucurbitaceous species in southern Portugal. Plant Pathol. 53, 241–241. doi: 10.1111/j.0032-0862.2004.00996.x
Mansour, A., and Al-Musa, A. (1993). Cucumber vein yellowing virus; host range and virus vector relationships. J. Phytopathol. 137, 73–78. doi: 10.1111/j.1439-0434.1993.tb01327.x
Martín-Hernández, A. M., and Picó, B. (2021). Natural resistances to viruses in cucurbits. Agronomy 11:23. doi: 10.3390/agronomy11010023
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Moreno-Ripoll, R., Gabarra, R., Symondson, W. O. C., King, R. A., and Agustí, N. (2014). Do the interactions among natural enemies compromise the biological control of the whitefly Bemisia tabaci? J. Pest Sci. 87, 133–141. doi: 10.1007/s10340-013-0522-x
Nazarov, P. A., Baleev, D. N., Ivanova, M. I., Sokolova, L. M., and Karakozova, M. V. (2020). Infectious plant diseases: etiology, current status, problems and prospects in plant protection. Acta Naturae 12, 46–59. doi: 10.32607/actanaturae.11026
Novaes, N. S., Lourenção, A. L., Bentivenha, J. P. F., Baldin, E. L. L., and Melo, A. M. T. (2020). Characterization and potential mechanisms of resistance of cucumber genotypes to Bemisia tabaci (Hemiptera: Aleyrodidae). Phytoparasitica 48, 643–657. doi: 10.1007/s12600-020-00826-3
Picó, B., Sifres, A., Martinez-Perez, E., Leiva-Brondo, M., and Nuez, F. (2008). Genetics of the resistance to CVYV in cucumber. In Modern variety breeding for present and future needs, in Proceedings of the 18th EUCARPIA General Congress, eds J. Prohens and M. L. Badenes (Valencia), 452–456.
Picó, B., Villar, C., Nuez, F., and Weber, W. E. (2003). Screening Cucumis sativus landraces for resistance to cucumber vein yellowing virus. Plant Breeding 122, 426–430. doi: 10.1046/j.1439-0523.2003.00882.x
Pujol, M., Alexiou, K. G., Fontaine, A. S., Mayor, P., Miras, M., Jahrmann, T., et al. (2019). Mapping Cucumber vein yellowing virus resistance in cucumber (Cucumis sativus L.) by using BSA-seq analysis. Front. Plant Sci. 10:1583. doi: 10.3389/fpls.2019.01583
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Sani, I., Ismail, S. I., Abdullah, S., Jalinas, J., Jamian, S., and Saad, N. (2020). A review of the biology and control of whitefly, Bemisia tabaci (hemiptera: Aleyrodidae), with special reference to biological control using entomopathogenic fungi. Insects 11:619. doi: 10.3390/insects11090619
Saucet, S. B., Ma, Y., Sarris, P. F., Furzer, O. J., Sohn, K. H., and Jones, J. D. (2015). Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 6, 6338–6312. doi: 10.1038/ncomms7338
Słomnicka, R., Olczak-Woltman, H., Korzeniewska, A., Gozdowski, D., Niemirowicz-Szczytt, K., and Bartoszewski, G. (2018). Genetic mapping of psl locus and quantitative trait loci for angular leaf spot resistance in cucumber (Cucumis sativus L.). Mol. Breeding 38, 111–119. doi: 10.1007/s11032-018-0866-2
Steinbrenner, A. D., Goritschnig, S., and Staskawicz, B. J. (2015). Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog. 11:e1004665. doi: 10.1371/journal.ppat.1004665
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U.S.A. 96, 14153–14158. doi: 10.1073/pnas.96.24.14153
Tatlioglu, T. (1993). Cucumber: Cucumis sativus L, in Genetic Improvement of Vegetable Crops, eds G. Kalloo and B. O. Bergh (Pergamon), 197–234. doi: 10.1016/B978-0-08-040826-2.50017-5
Tör, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Türk, F., et al. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. doi: 10.1105/tpc.001123
Tör, M., Lotze, M. T., and Holton, N. (2009). Receptor-mediated signalling in plants: molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654. doi: 10.1093/jxb/erp233
Van de Weyer, A. L., Monteiro, F., Furzer, O. J., Nishimura, M. T., Cevik, V., Witek, K., et al. (2019). A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178, 1260–1272.e14. doi: 10.1016/j.cell.2019.07.038
Wang, G., Ellendorff, U., Kemp, B., Mansfield, J. W., Forsyth, A., Mitchell, K., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517. doi: 10.1104/pp.108.119487
Woods-Tör, A., Studholme, D. J., Cevik, V., Telli, O., Holub, E. B., and Tör, M. (2018). A suppressor/avirulence gene combination in Hyaloperonospora arabidopsidis determines race specificity in Arabidopsis thaliana. Front. Plant Sci. 9:1957. doi: 10.3389/fpls.2018.00265
Yemataw, Z., Muzemil, S., Ambachew, D., Tripathi, L., Tesfaye, K., Chala, A., Farbos, A., et al. (2018). Genome sequence data from 17 accessions of Ensete ventricosum, a staple food crop for millions in Ethiopia. Data Brief 18, 285–293. doi: 10.1016/j.dib.2018.03.026
Zhang, W., Fraiture, M., Kolb, D., Löffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis RECEPTOR-LIKE PROTEIN30 and Receptor-Like Kinase SUPPRESSOR OF BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Keywords: CVYV, cucumber, marker assisted selection, kompetitive allele-specific PCR genotyping, plant breeding
Citation: Kahveci E, Devran Z, Özkaynak E, Hong Y, Studholme DJ and Tör M (2021) Genomic-Assisted Marker Development Suitable for CsCvy-1 Selection in Cucumber Breeding. Front. Plant Sci. 12:691576. doi: 10.3389/fpls.2021.691576
Received: 06 April 2021; Accepted: 22 June 2021;
Published: 18 August 2021.
Edited by:
Anna Maria Mastrangelo, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Shivendra Kumar, Iowa State University, United StatesCopyright © 2021 Kahveci, Devran, Özkaynak, Hong, Studholme and Tör. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmut Tör, bS50b3JAd29yYy5hYy51aw==
†ORCID: Erdem Kahveci orcid.org/0000-0002-4467-5400
Zübeyir Devran orcid.org/0000-0001-7150-284X
Ercan Özkaynak orcid.org/0000-0002-4793-7963
Yiguo Hong orcid.org/0000-0002-3352-9686
David J. Studholme orcid.org/0000-0002-3010-6637
Mahmut Tör orcid.org/0000-0002-4416-5048
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.