- 1College of Agronomy, Northwest A&F University, Yangling, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research of CAAS, Anyang, China
Photoperiod is an important external factor that regulates flowering time, the core mechanism of which lies in the circadian clock-controlled expression of FLOWERING LOCUS T (FT) and its upstream regulators. However, the roles of the circadian clock in regulating cotton flowering time are largely unknown. In this study, we cloned two circadian clock genes in cotton, GhLUX1 and GhELF3. The physicochemical and structural properties of their putative proteins could satisfy the prerequisites for the interaction between them, which was proved by yeast two-hybrid (Y2H) and Bimolecular Fluorescent Complimentary (BiFC) assays. Phylogenetic analysis of LUXs and ELF3s indicated that the origin of LUXs was earlier than that of ELF3s, but ELF3s were more divergent and might perform more diverse functions. GhLUX1, GhELF3, GhCOL1, and GhFT exhibited rhythmic expression and were differentially expressed in the early flowering and late-flowering cotton varieties under different photoperiod conditions. Both overexpression of GhLUX1 and overexpression of GhELF3 in Arabidopsis delayed flowering probably by changing the oscillation phases and amplitudes of the key genes in the photoperiodic flowering pathway. Both silencing of GhLUX1 and silencing of GhELF3 in cotton increased the expression of GhCOL1 and GhFT and resulted in early flowering. In summary, the circadian clock genes were involved in regulating cotton flowering time and could be the candidate targets for breeding early maturing cotton varieties.
Introduction
Floral transition under favorable circumstances is necessary for the reproductive success of most plant species. Changes in day length (photoperiod) are reliable environmental signals that can be monitored by plants to ensure the proper flowering time (Song et al., 2013; Shim et al., 2017). Generally, the photoperiodic flowering pathway can be divided into three domains: light input, circadian clock, and output. CONSTANS (CO), the key activator of FLOWERING LOCUS T (FT), is regulated by both light signaling and the circadian clock. The circadian clock restricts CO transcription to late afternoon and night. In long days (LD), CO protein is stabilized by the light of late afternoon and activate the transcription of FT, In short days (SD), CO protein is degraded at night and FT transcription can’t be activated, which leads to late flowering (Kinmonth-Schultz et al., 2013).
The molecular architecture of the Arabidopsis circadian clock is comprised of multiple feedback loops. The initial model is a transcriptional feedback loop comprised of LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and TIMING OF CAB EXPRESSION 1 (TOC1). In the morning, LHY and CCA1 are expressed and repressed TOC1 transcription (Alabadi et al., 2001; Lu et al., 2009; Yakir et al., 2009). At dusk, the decreased levels of CCA1 and LHY induce TOC1 expression, which in turn represses CCA1 and LHY transcription (Gendron et al., 2012; Huang et al., 2012). An additional loop is comprised of PSEUDO-RESPONSE REGULATOR 9 (PRR9), PRR7 and PRR5, which are sequentially expressed throughout the day and redundantly repressed CCA1 and LHY expression (Nakamichi et al., 2010; Nakamichi et al., 2012; Liu et al., 2016). PRR9, PRR7 and PRR5 are reciprocally repressed by CCA1 and LHY (Adams et al., 2015). In addition, PRR9 is also repressed by the evening complex (EC) (Nagel and Kay, 2012), which is comprised of LUX ARRHYTHMO (LUX), and EARLY FLOWERING 3 (ELF3) and ELF4. The repression of the EC components by CCA1, LHY and the activation of CCA1, LHY by the EC components form another feedback loop (Nagel and Kay, 2012; Adams et al., 2015).
Since the circadian clock is comprised of multiple interconnected feedback loops, mutation and overexpression of any component of the circadian clock will change the oscillation properties (phase, period and amplitude) of other components and affect flowering time. In Arabidopsis, both cca1 mutant and lhy mutant show early flowering only under SD conditions (Mizoguchi et al., 2002), while cca1 lhy double mutant shows early flowering under both LD and SD conditions (Mizoguchi et al., 2002; Fujiwara et al., 2008). Both CCA1 overexpression and LHY overexpression delay flowering under both LD and SD conditions (Wang and Tobin, 1998; Mizoguchi et al., 2002; Lu et al., 2012). Both prr5 mutant and prr7 mutant show late flowering only under LD conditions (Yamamoto et al., 2003; Nakamichi et al., 2005, 2007), and prr5 prr7 prr9 triple mutant also shows late flowering only under LD conditions (Nakamichi et al., 2005, 2007). Both PRR5 overexpression and PRR9 overexpression promote flowering under both LD and SD conditions (Matsushika et al., 2002; Sato et al., 2002). Mutation of any EC component (ELF3, ELF4, LUX) promotes flowering more significantly under SD conditions than under LD conditions (Zagotta et al., 1996; Liu et al., 2001; Doyle et al., 2002; Hazen et al., 2005; Lu et al., 2012). Both ELF3 overexpression and ELF4 overexpression delay flowering under only LD conditions (Liu et al., 2001; McWatters et al., 2007).
The effects of the circadian clock on flowering time have also been reported in some crops. In barley, PHOTOPERIOD1 (Ppd-H1) gene, a homolog of AtPRR7, regulates photoperiodic flowering by promoting HvFT1 expression independently of HvCO1 (Turner et al., 2005; Campoli et al., 2012). Loss-of-function of HvELF3 leads to early flowering under both LD and SD conditions. HvELF3 also plays key roles in maintaining the photoperiodic sensitivity in spring barley by repressing HvFT1 (Faure et al., 2012; Boden et al., 2014). In rice, overexpression of OsCCA1 leads to late flowering (Izawa et al., 2002; Murakami et al., 2007). Loss-of-function of OsPRR37, a homolog of AtPRR7, promotes flowering. Overexpression of OsPRR37 delays flowering (Liu et al., 2018). OsELF3, promotes flowering in SDs by activating OsEhd1 and promotes flowering in LDs by repressing OsGhd7 (Zhao et al., 2012). In soybean, overexpression of GmPRR37 delays flowering and mutation of GmPRR37 promotes flowering under LD conditions (Wang et al., 2020). Overexpression of GmELF4 in Arabidopsis delays flowering (Marcolino-Gomes et al., 2017).
Upland cotton (Gossypium hirsutum) is an important cash crop for its high productivity of natural textile fiber, seed oil and protein meal (Zhang T. et al., 2015). With the increasing competition for farmland use between cotton and grain, early maturation of cotton has become a primary breeding objective to enable cotton-wheat rotation. In addition, shortened life cycle allows cotton plants to develop under suitable climatic conditions (Li et al., 2013). However, little is known about the molecular mechanisms that regulate the flowering time of cotton. Recent studies report that the two integrators of multiple flowering pathways, GhFT and its putative activator, GhCOL1 (CONSTANS-like 1), are overexpressed in Arabidopsis and the transgenic plants exhibit early flowering. Moreover, both GhCOL1 and GhFT exhibit diurnally rhythmic expression with peak in the morning (Guo et al., 2015; Cai et al., 2017). These observations imply that the circadian clock is involved in regulating cotton flowering time. In our study, two circadian clock components, GhLUX1 and GhELF3 were cloned. The physicochemical properties and tertiary structures of their protein sequences were predicted. We further analyzed the rhythmic expression patterns of GhLUX1, GhELF3, GhCOL1, and GhFT in the early flowering and late-flowering varieties under different photoperiod conditions. Finally, we characterized the roles of GhLUX1 and GhELF3 in regulating flowering time by overexpressing their coding sequences in Arabidopsis and silencing their transcripts in cotton. This work demonstrates that the circadian clock is involved in regulating cotton flowering time for the first time and lays a foundation for exploring how the interaction of multiple flowering pathways controls cotton flowering time.
Materials and Methods
Plant Materials and Growth Conditions
The early flowering cotton variety CCRI50 and the late-flowering cotton variety GX11 (Cheng et al., 2021) were grown in the constant temperature (25°C) room under the LD cycles (16 h light/8 h dark). When the fifth true leaves of cotton seedlings were fully expanded, the seedlings of CCRI50 and GX11 were divided into four portions. One portion was remained in the room under the LD cycles and the other three portions were transferred into the rooms under the SD cycles (8h light/16h dark), constant dark and constant light at 6:00, respectively. After the seedlings were entrained for 24 h under the four conditions, the first true leaves of three biological replicates of the seedlings were sampled every 4 h from 6:30 to 2:30 of the next day to extract RNA. Cotton variety GX11 were grown in the constant temperature (25°C) room under the LD cycles (16 h light/8 h dark). The seedlings at the cotyledon stage were used for VIGS experiment. Positive VIGS plants’ first and second true leaves were defoliated when the fourth true leaves were fully expanded. When the eighth true leaves were fully expanded, the fourth true leaves of three biological replicates of positive VIGS plants were sampled every 4 h from 6:30 to 2:30 of the next day to extract RNA.
To produce the plants used for genetic transformation, sterilized Arabidopsis thaliana (Columbia ecotype) seeds were sown on the 1/2 MS media with 0.8% agar, and after incubation at 4°C for 3 days, the plates were placed in the constant temperature (21°C) room under the LD cycles (16 h light/8 h dark). Ten-days-old seedlings were transplanted into pots and cultivated in the same room. The T3 lines of GhLUX1-overexpressed and GhELF3-overexpressed Arabidopsis and WT were grown under the same conditions to observe their phenotypes of flowering time, bolting time and rosette leave number. When the WT plants’ flower buds were visible, the top fourth rosette leaves of three biological replicates of WT, GhLUX1-overexpressed and GhELF3-overexpressed Arabidopsis seedlings were sampled every 3 h from 7:00 to 4:00 of the next day to extract RNA.
Tobacco (Nicotiana benthamiana) was grown in the constant temperature (21°C) room under the LD cycles (16 h light/8 h dark). Five-weeks-old tobacco plants were used for subcellular localization and BiFC experiments.
Gene Cloning and Sequence Analysis
The protein sequences of AtLUX1 (AT3G46640) and AtELF3 (AT2G25930) were, respectively, used as the queries to search against the protein databases of G. hirsutum1 using BLAST with e-value threshold set at 1e-5. The best hits were defined as GhLUX1 and GhELF3, respectively. The coding sequences of GhLUX1 and GhELF3 were amplified from the cDNA of the cotton varieties TM-1, CCRI50 and GX11, and the genomic sequences of GhLUX1 and GhELF3 were amplified from the DNA of the cotton variety TM-1 using the gene-specific primers (Supplementary Table 2). The PCR products were cloned into the pBI121 vector and sequenced. The exon-intron structures of GhLUX1 and GhELF3 were generated and visualized by submitting their genomic and coding sequences to GSDS 2.02 (Hu et al., 2015). The molecular weight, isoelectric point and grand average of hydropathicity of GhLUX1’s and GhELF3’s putative protein sequences were predicted using ExPASy3 (Artimo et al., 2012).
The protein sequences of AtLUX1 and AtELF3 were, respectively, used as the queries to search against the protein databases of 27 plant species (Supplementary Table 3) using BLAST with e-value threshold set at 1e-5. BLAST hits with scores more than 200 were considered as homologs of AtLUX1 and AtELF3. The protein sequences of all the LUXs and ELF3s were, respectively, aligned using Clustal Omega with default parameters4 (Madeira et al., 2019). The resulted alignments were used as the input files of MrBayes v3.2.5 to construct the phylogenetic trees with the evolutionary model set to the GTR substitution model and Ngen, Samplefreq set to 1,000,000, 100, respectively (Ronquist and Huelsenbeck, 2003).
The tertiary structures of GhLUX1 and GhELF3 were predicted on the I-TASSER website5 (Roy et al., 2010). The multiple sequence alignment results of all the LUXs and ELF3s were, respectively, used to calculate conservation scores of each amino acid site of GhLUX1 and GhELF3 on the Protein Residue Conservation Prediction website6 with the default parameters (Capra and Singh, 2007). The tertiary structures were visualized using PyMOL v2.3.0 and the conservation score of each amino acid site was mapped to the color of corresponding amino acid of the tertiary structures with blue corresponding to low conservation score and red corresponding to high conservation score.
DNA, RNA Extraction, and Quantitative Real-Time PCR (qRT-PCR)
Genomic DNA was extracted via the cetyl-trimethylammonium bromide (CTAB) method as described previously (Porebski et al., 1997). Total RNA was isolated using an RNAprep Pure Plant Kit (DP441) (Tiangen, Beijing, China). The RNA was used as the template for cDNA synthesis using a PrimeScriptTM RT Reagent Kit with gDNA Eraser (RR047A) (TaKaRa, Dalian, China). The qRT-PCR was performed using UltraSYBR Mixture (Low ROX) (CW2601) (CWBIO, Beijing, China) and an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The thermocycler program consisted of pre-denaturation at 95°C for 30 s followed by 40 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 32 s. The data were calculated in accordance with the 2–ΔΔCt formula, in which ΔΔCt = Ctgene – Ctreference – scale factor (the maximum of Ctgene – Ctreference of all the samples in one experiment) (Livak and Schmittgen, 2001). GhActin and AtACT2 were, respectively, used as the reference genes when analyzing samples of cotton and Arabidopsis. The gene-specific primers used for the qRT-PCR were listed in the Supplementary Table 2.
Transcription Activation and Y2H Assays
The full-length, N-terminal and C-terminal coding sequences of GhLUX1 and GhELF3 were cloned into the pGBKT7 and pGADT7 vectors with the gene-specific primers (Supplementary Table 2). Then, the combinations of pGADT7 with pGBKT7, pGBKT7-GhLUX1, pGBKT7-GhELF3, pGBKT7-GhLUX1-N, and pGBKT7-GhLUX1-C were co-transferred into the yeast strain Y2HGold which was cultured on DDO (SD/-Leu/-Trp) plates for 3 days. Three independent colonies on the DDO plates were chosen to test the transcription activations on QDO (SD/-Leu/-Trp/-His/-Ade) plates. The combinations of pGADT7, pGADT7-GhELF3, pGADT7-GhELF3-N, pGADT7-GhELF3-C with pGBKT7, pGBKT7-GhLUX1-C were co-transferred into the yeast strain Y2HGold which was cultured on DDO plates for 3 days. Three independent colonies on the DDO plates were chosen to detect the interactions on QDO plates.
Subcellular Localization and BiFC Assays
The coding sequences of GhLUX1 and GhELF3 were cloned into the pBI121-GFP vectors with the gene-specific primers (Supplementary Table 2). The recombinant vectors were transiently transformed into the leaves of 5-weeks-old tobacco plants using Agrobacterium tumefaciens strain GV3101. After the plants were placed in the dark for 2 days, the injected leaves’ fluorescence was observed using confocal laser scanning microscopy (Leica TCS SP8).
The coding sequences of GhLUX1 and GhELF3 were, respectively, cloned into the pSPYCE and pSPYNE vectors with the gene-specific primers (Supplementary Table 2). Agrobacterium solutions containing pSPYCE, pSPYNE and pSPYCE-GhLUX1 were mixed with the same volumes of Agrobacterium solutions containing pSPYNE-GhELF3, pSPYCE-GhLUX1, and pSPYNE-GhELF3, correspondingly. The following procedures were same to those used in the above subcellular localization experiment.
Arabidopsis Transformation
The recombinant pBI121 vectors (pBI121-GhLUX1 and pBI121-GhELF3) constructed in the gene cloning step were transferred into the Agrobacterium tumefaciens strain GV3101 and were transformed into Arabidopsis via the floral dip method (Clough and Bent, 1998). The positive plants were selected on 1/2MS medium containing kanamycin (50 mg/L), and further confirmed via PCR and qRT-PCR.
Virus-Induced Gene Silencing
Virus-induced gene silencing (VIGS) was performed as described previously (Gu et al., 2014). Briefly, the ∼300 bp fragments within GhLUX1’s and GhELF3’s coding sequences were cloned into the pCLCrVA vector using gene-specific primers (Supplementary Table 2). The recombinant vectors were transferred into the Agrobacterium tumefaciens strain GV3101. Solutions of Agrobacterium containing pCLCrV-GhLUX1, pCLCrV-GhELF3, pCLCrV-PDS (positive control), pCLCrVA (negative control) were, respectively, mixed with solutions of Agrobacterium containing pCLCrVB (helper vector). The mixed solutions were injected into the cotyledons of 10-d-old GX11 seedlings. When the leaves of the pCLCrVA-PDS plants became white, positive plants were detected using PCR and qRT-PCR, and then the positive plants were transplanted into large pots and used for phenotypic observation of flowering time.
Results
Characterization of Nucleotide and Putative Protein Sequences of GhLUX1 and GhELF3
The most homologous genes in G. hirsutum to AtLUX and AtELF3 were identified as GhLUX1 and GhELF3, respectively. The coding sequences of GhLUX1 and GhELF3 cloned from CCRI50 and GX11 were same to those cloned from TM-1, suggesting that the protein functions of GhLUX1 and GhELF3 might be unchanged in different cotton varieties. By comparing the coding sequences and genomic sequences, one exon and four exons were found in GhLUX1 and GhELF3, respectively (Supplementary Figure 1). The properties of putative protein sequences were listed in Supplementary Table 1. Notably, the isoelectric points (pIs) of GhLUX1 and GhELF3 were 5.28 and 8.84, respectively, suggesting they were charged oppositely in cotton cells. In addition, GhLUX1 and GhELF3 showed similar grand average of hydropathicity (GRAVY) and were both hydrophilic proteins. These properties of GhLUX1 and GhELF3 satisfied some prerequisites for the interaction between the two proteins.
Evolutionary Difference Between LUXs and ELF3s
To explore the evolutionary difference between LUXs and ELF3s, homologs of AtLUX and AtELF3 were screened in 27 plant species’ protein databases and the phylogenetic trees were constructed. There was no LUX identified in chlorophytes (C. reinhardtii) and bryophytes (P. patens). The most ancient LUX was identified in pteridophytes (S. moellendorffii). Only one LUX was found in the early species before dicots, while one to six LUXs were found in different dicots. More than one LUXs contained in some dicots (G. max, P. trichocarpa, D. carot, A. thaliana, B. rapa and four Gossypium species) had the closest phylogenetic relationships (Supplementary Figure 2A). The most ancient ELF3 was identified in the most basal lineage of angiosperms (A. trichopoda). The numbers of ELF3s increased to two or three in monocots and ELF3s in dicots diverged into two subclades (Supplementary Figure 2B). These results indicated that ELF3s might arise later than LUXs, but evolve more rapidly to perform more diverse functions in plants than LUXs.
Characterization of the Predicted Tertiary Structures of GhLUX1 and GhELF3
The tertiary structure of one protein usually implies its potential molecular functions. The tertiary structures of GhLUX1 and GhELF3 were predicted on the I-TASSER server and their conservation scores at each amino acid site were calculated on the Protein Residue Conservation Prediction website. GhLUX1 consisted of helices and coils. Two conserved regions were distributed in the N-terminus and middle part of the protein, respectively. The more conserved Myb DNA-binding domain consisted of three helices (Supplementary Figure 3A). GhELF3 was divided into an N-terminal large subunit and a C-terminal small subunit linked by a random coil. There was large open space between the large subunit and the small subunit. The helices and coils of the large subunit formed a groove, the two terminals of which were two conserved regions. The small unit consisted of helices, sheets and coils and contained two close conserved regions in its middle part (Supplementary Figure 3B).
Transcriptional Activity and Interaction of GhLUX1 and GhELF3
To examine whether GhLUX1 and GhELF3 acted as transcription factors, the transcriptional activation assay was performed in yeast cells. The yeast cells containing pGADT7 and pGBKT7-GhLUX1 plasmids could grow normally on the quadruple dropout media, whereas the yeast cells containing pGADT7 and pGBKT7-GhELF3 plasmids could not (Figure 1A), suggesting that GhLUX1 had transcriptional activity, but GhELF3 did not. Further segmentation of GhLUX1 suggested GhLUX1-N (residues 1–154) had transcriptional activity, but GhLUX1-C (residues 155–337) did not (Figure 1B). Subcellular localization assay showed that both GhLUX1-GFP and GhELF3-GFP could be transported into the nucleus of N. benthamiana cells (Figure 1C), indicating that both GhLUX1 and GhELF3 might perform their functions in the nucleus.
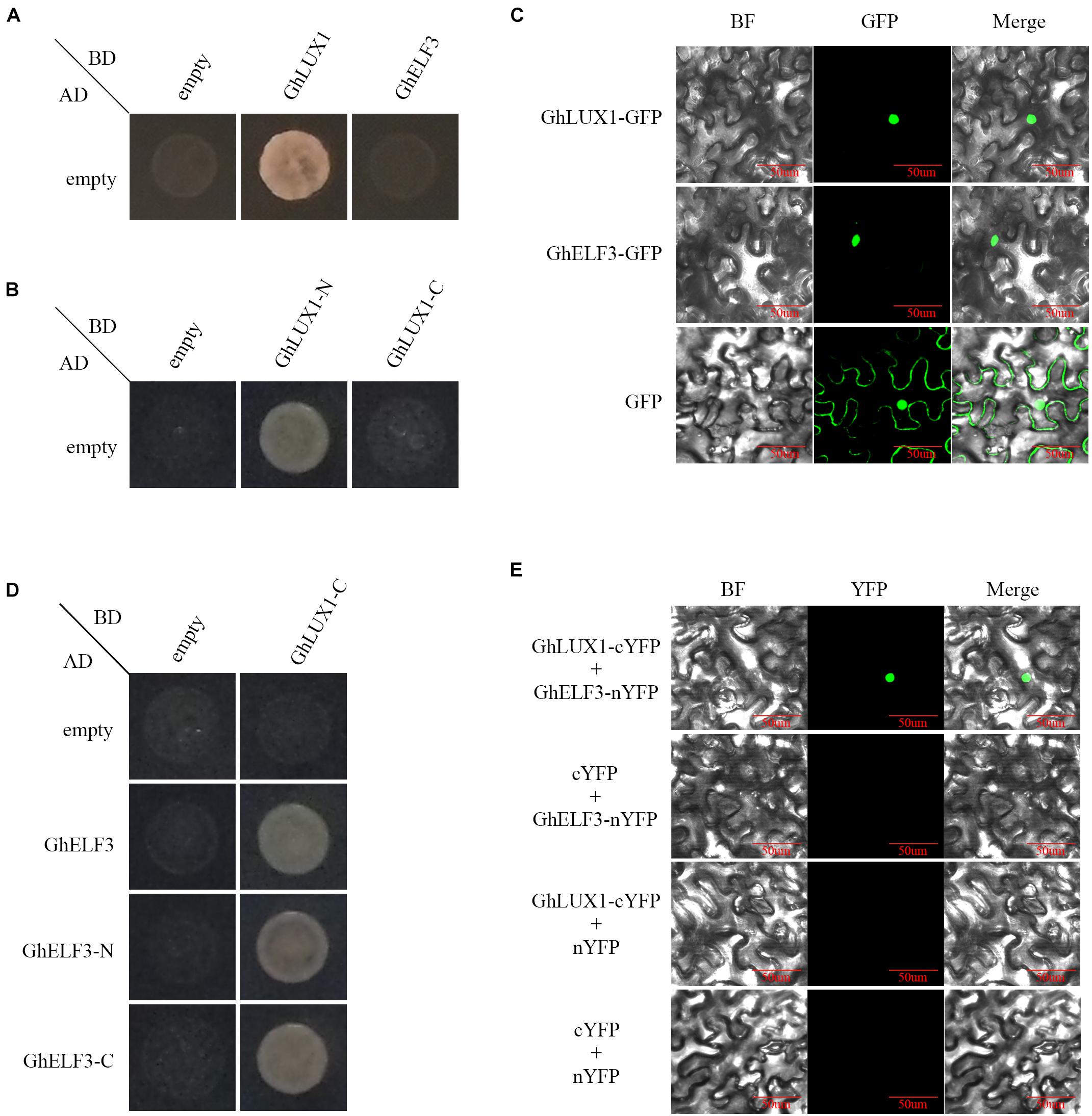
Figure 1. Transcriptional activity and interaction of GhLUX1 and GhELF3. (A) Transcriptional activity of GhLUX1 and GhELF3 in Y2HGold yeast cells. Yeast cells containing different combinations of pGADT7 and pGBKT7, pGBKT7-GhLUX1, pGBKT7-GhELF3 vectors are cultured on the SD-Trp/-Leu/-His/-Ade/medium. (B) Transcriptional activity of GhLUX1-N and GhLUX1-C in Y2HGold yeast cells. Yeast cells containing different combinations of pGADT7 and pGBKT7, pGBKT7-GhLUX1-N (residues 1–154), pGBKT7-GhLUX1-C (residues 155–337) vectors are cultured on the SD-Trp/-Leu/-His/-Ade/medium. (C) Subcellular localization of GhLUX1 and GhELF3 in N. benthamiana epidermal cells. GhLUX1-GFP, GhELF3-GFP, and GFP are transiently expressed in N. benthamiana epidermal cells and visualized under confocal microscopy. (D) GhLUX1-GhELF3 interaction in Y2HGold yeast cells. Yeast cells containing different combinations of pGADT7, pGADT7-GhELF3, pGADT7-GhELF3-N (residues 1–460), pGADT7-GhELF3-C (residues 467–705) and pGBKT7, pGBKT7-GhLUX1-C vectors are cultured on the SD-Trp/-Leu/-His/-Ade/medium. (E) GhLUX1-GhELF3 interaction in N. benthamiana epidermal cells. Different combinations of GhLUX1-cYFP, GhELF3-nYFP and empty vectors are transiently coexpressed in N. benthamiana epidermal cells and visualized under confocal microscopy.
In Arabidopsis, the evening complex (EC) was formed by the direct interactions of AtELF3 and AtLUX (residues 144–323), AtELF3 (residues 261–484) and AtELF4 (Huang and Nusinow, 2016). To examine whether GhLUX1 interacted with GhELF3, yeast two-hybrid and Bimolecular Fluorescent Complimentary (BiFC) assay were performed. Because of the auto-activations of GhLUX1 and GhLUX1-N (Figures 1A,B), GhLUX1-C was used as the bait. GhLUX1-C showed interactions with GhELF3, GhELF3-N (residues 1–460) and GhELF3-C (residues 467–705) (Figure 1D). The BiFC result showed that GhLUX1 interacted with GhELF3 in the nuclei of N. benthamiana cells (Figure 1E). The transcriptional activity of GhLUX1 and the interaction of GhLUX1 and GhELF3 in the nucleus indicated that GhLUX1 might recruit GhELF3 to the promoters of target genes to regulate their transcriptions.
Rhythmic Expression of GhLUX1, GhELF3, GhCOL1, and GhFT in LD and SD
To determine whether cotton flowering time was regulated by GhLUX1 and GhELF3, the expression patterns of GhLUX1, GhELF3, GhCOL1, and GhFT in LD (16 h light/8 h dark) and SD (8 h light/16 h dark) were compared between the early flowering variety, CCRI50 and the late-flowering variety, GX11. All the four genes exhibited rhythmic expression patterns under both photoperiod conditions and in both cotton varieties (Figure 2). Compared with GX11, CCRI50 showed lower expression levels of GhLUX1 and GhELF3 from the afternoon till the early night of LD but showed higher expression levels of GhLUX1 in the afternoon of SD and higher expression levels of GhELF3 from the night till the morning of SD (Figures 2A,B), which suggested that GhLUX1 and GhELF3 might repress flowering in LD but promote flowering in SD. This situation was similar to that LUXs and ELF3s repressed flowering in long day plant (LDP) species but promoted flowering in short day plant (SDP) species (Bu et al., 2021). In addition, CCRI50 showed higher expression levels of GhCOL1 in the morning of both LD and SD and higher expression levels of GhFT at most times of both LD and SD (Figures 2C,D), which was consistent with the roles of GhCOL1 and GhFT in promoting flowering.
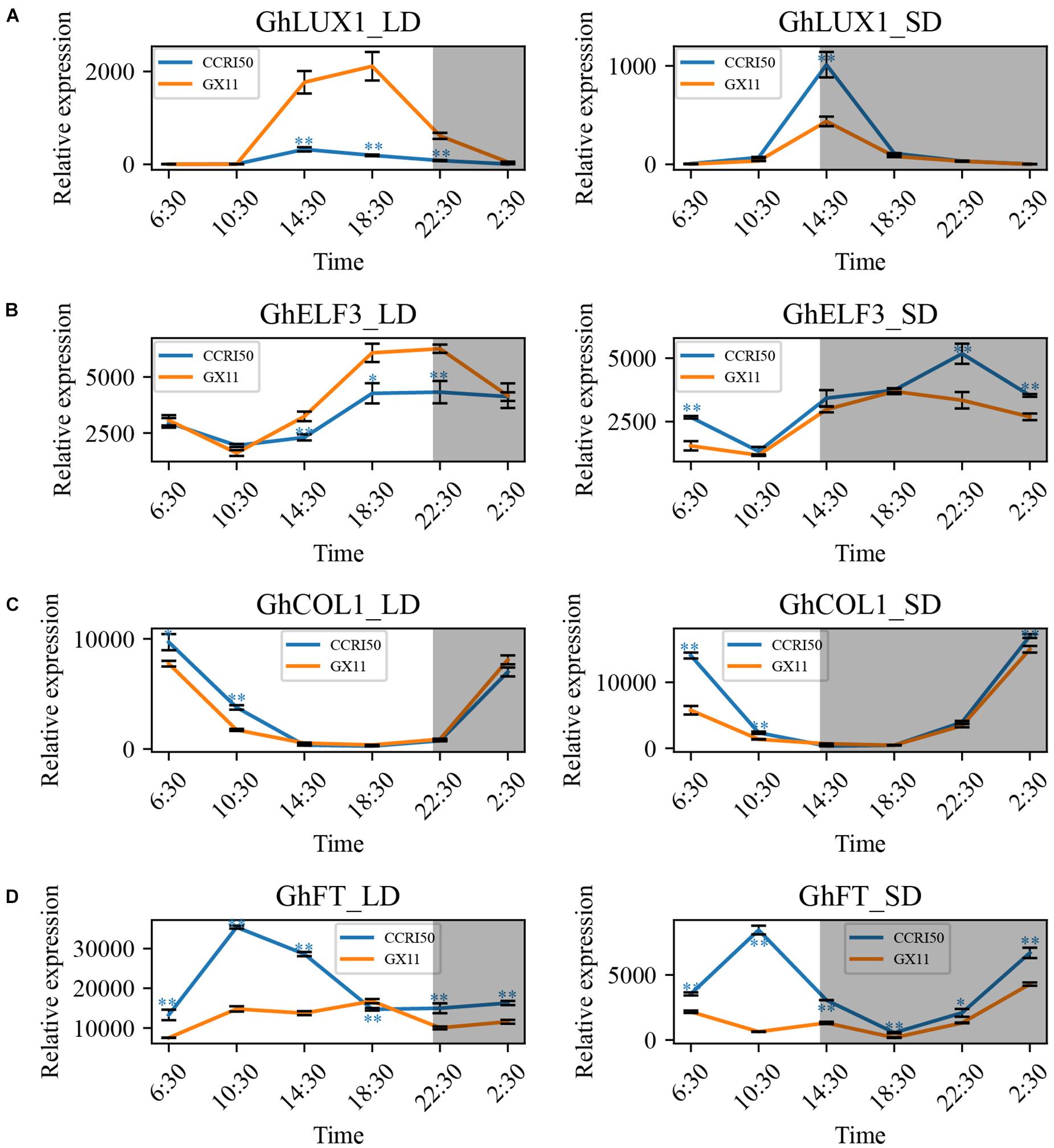
Figure 2. GhLUX1, GhELF3, GhCOL1, and GhFT are differentially expressed in late-flowering GX11 and early flowering CCRI50 under LD and SD conditions. Expression patterns of (A) GhLUX1, (B) GhELF3, (C) GhCOL1, and (D) GhFT in GX11, CCRI50 under LD and SD conditions. All the expression levels are made relative to the expression level of GhLUX1 in GX at 2:30 under SD. The data are the means ± standard errors (SEs) of three biological replicates. The asterisks indicate significant differences of comparison between GX11 and CCRI50 at each time point (∗∗P < 0.01, ∗P < 0.05, Student’s t-test). The gray shadows indicate the dark periods.
Rhythmic Expression of GhLUX1, GhELF3, GhCOL1, and GhFT in Constant Light and Dark
To exclude the effects of day-night alteration on the oscillations of GhLUX1, GhELF3, GhCOL1, and GhFT transcripts, the expression patterns of the four genes in constant light (LL) and dark (DD) were analyzed. In LL and DD, all the four genes still exhibited rhythmic expression patterns in GX11 and CCRI50 (Figure 3). Similar to the situations in LD and SD, CCRI50 showed lower expression levels of GhLUX1 and GhELF3 at the specific times of LL but showed higher expression levels of GhLUX1 and GhELF3 at the specific times of DD (Figures 3A,B). In addition, CCRI50 showed higher expression levels of GhFT at most times of LL and DD (Figure 3D). However, CCRI50 showed lower expression levels of GhCOL1 in the morning of LL but showed much higher expression levels of GhCOL1 at all the times of DD (Figure 3C). In addition, compared with the expression of GhLUX1 and GhELF3 in LD and SD, the expression of GhLUX1 in LL and DD was impaired dramatically (Figures 2A, 3A), while the expression of GhELF3 in LL and DD was just changed slightly (Figures 2B, 3B), indicating the robust oscillation of GhELF3 under different photoperiod conditions. Furthermore, the expression levels of GhFT in DD were dramatically decreased compared with those in LD, SD and LL (Figures 2D, 3D), indicating that GhFT was repressed by unknown regulators in darkness. In addition, oscillation phases of GhFT transcript were significantly different not only between two varieties but also among the four photoperiod conditions (Figures 2D, 3D). These results implied that the circadian clock could exhibit different oscillation properties in different cotton varieties and could be entrained by different photoperiods.
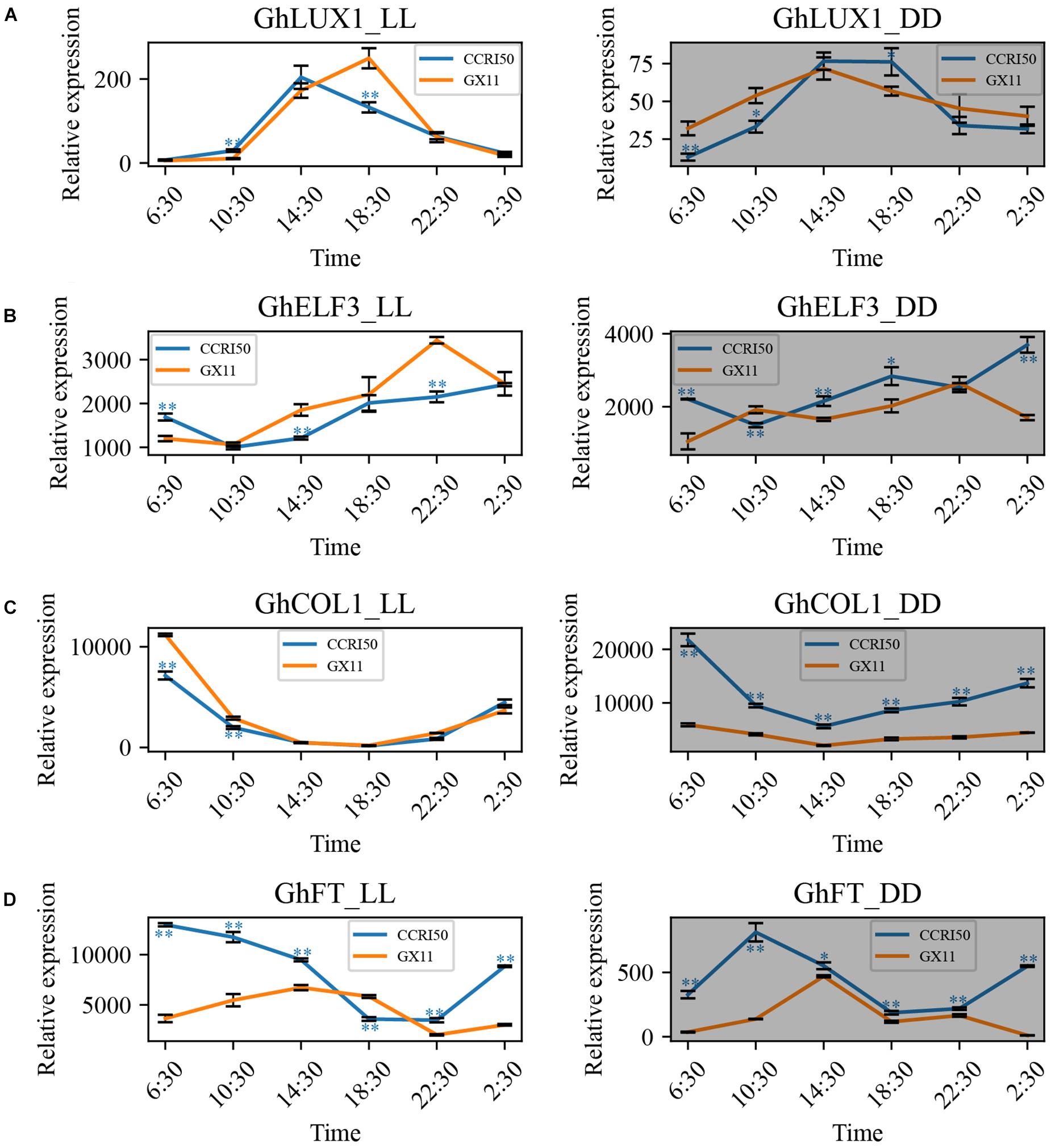
Figure 3. GhLUX1, GhELF3, GhCOL1, and GhFT persist differently rhythmic expression in late-flowering GX11 and early flowering CCRI50 under constant conditions. Expression patterns of (A) GhLUX1, (B) GhELF3, (C) GhCOL1, and (D) GhFT in GX11, CCRI50 under LL and DD conditions. All the expression levels are made relative to the expression level of GhLUX1 in GX at 2:30 under SD. The data are the means ± SEs of three biological replicates. The asterisks indicate significant differences of comparison between GX11 and CCRI50 at each time point (∗∗P < 0.01, ∗P < 0.05, Student’s t-test). The gray shadows indicate the dark periods.
Both Overexpression of GhLUX1 and Overexpression of GhELF3 in Arabidopsis Delay Flowering
To explore the functional roles of GhLUX1 and GhELF3 in regulating flowering time, coding sequences of GhLUX1 and GhELF3 driven by the 35S promoter were transformed into Arabidopsis. Three independent T3 lines with significantly higher expression levels of GhLUX1 and GhELF3 than the WT were selected to observe their flowering phenotypes (Figures 4B, 4G). All the transgenic lines exhibited later flowering than the WT did (Figures 4A,F). Compared with the WT, the GhLUX1-overexpressed lines and the GhELF3-overexpressed lines flowered 4–5.7 and 4–4.9 days later on average, respectively (Figures 4E,J). In addition, the GhLUX1-overexpressed lines and the GhELF3-overexpressed lines bolted later and had more rosette leaves compared with the WT (Figures 4C,D,H,I), which was consistent with their later flowering time. These results suggested that GhLUX1 and GhELF3 could perform similar functions to AtLUX and AtELF3, respectively, in regulating flowering time of Arabidopsis (Zagotta et al., 1996; Hazen et al., 2005).
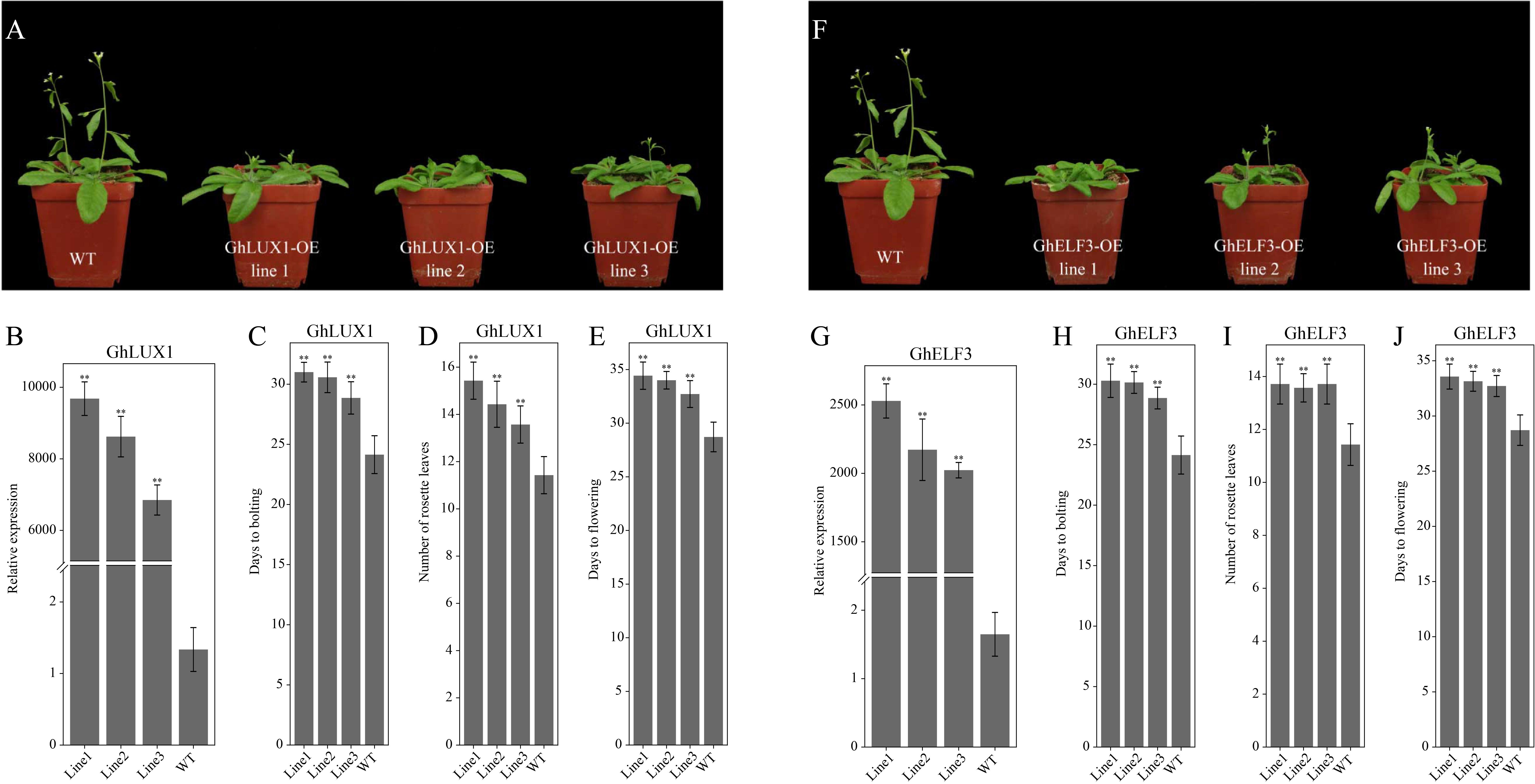
Figure 4. Both overexpression of GhLUX1 and overexpression of GhELF3 in Arabidopsis delay flowering. (A,F) Phenotypic characteristics of 4-week-old WT and transgenic lines. (B,G) Expression levels of GhLUX1 and GhELF3 in the WT and the transgenic lines. All the expression levels are made relative to the expression level of GhLUX1 in the WT. The data are the means ± SEs of three biological replicates. (C,H) Days to bolting, (D,I) rosette leaf numbers and (E,J) days to flowering of the WT and the transgenic lines (means ± SEs, n = 24 plants). The asterisks indicate significant differences compared to the WT plants at each time point (∗∗P < 0.01, Student’s t-test).
Both Overexpression of GhLUX1 and Overexpression of GhELF3 Change the Oscillations of the Circadian Clock Genes and the Key Genes in the Photoperiodic Flowering Pathway
Because the circadian clock is comprised of multiple interconnected feedback loops, we hypothesized that overexpression of GhLUX1 and overexpression of GhELF3 in Arabidopsis changed the running of the whole circadian clock. To test the hypothesis, we measured the expression levels of several core circadian clock genes (including AtLUX, AtELF3, AtELF4, AtPRR7, AtLHY, and AtCCA1) during the 24 h in the transgenic lines and the WT. All the six genes were upregulated or downregulated in the GhLUX1-overexpressed line and the GhELF3-overexpressed line compared with in the WT, although their expression trends during the 24 h were similar between the two transgenic lines and the WT (Figures 5A–F). Overexpression of GhELF3 significantly repressed the expression of four evening- or afternoon-phased clock genes (including AtLUX, AtELF3, AtELF4, and AtPRR7), while overexpression of GhLUX1 repressed the expression of the four genes to a lesser extent (Figures 5A–D). The expression of a morning-phased gene, AtLHY, was promoted in the GhLUX1-overexpressed line to a higher extent than in the GhELF3-overexpressed line (Figure 5E). The expression of AtLHY’s close homolog, AtCCA1, was promoted by overexpression of GhLUX1 but was repressed by overexpression of GhELF3 (Figure 5F). These results indicated that overexpression of GhLUX1 and overexpression of GhELF3 could change the running of the Arabidopsis circadian clock differently.
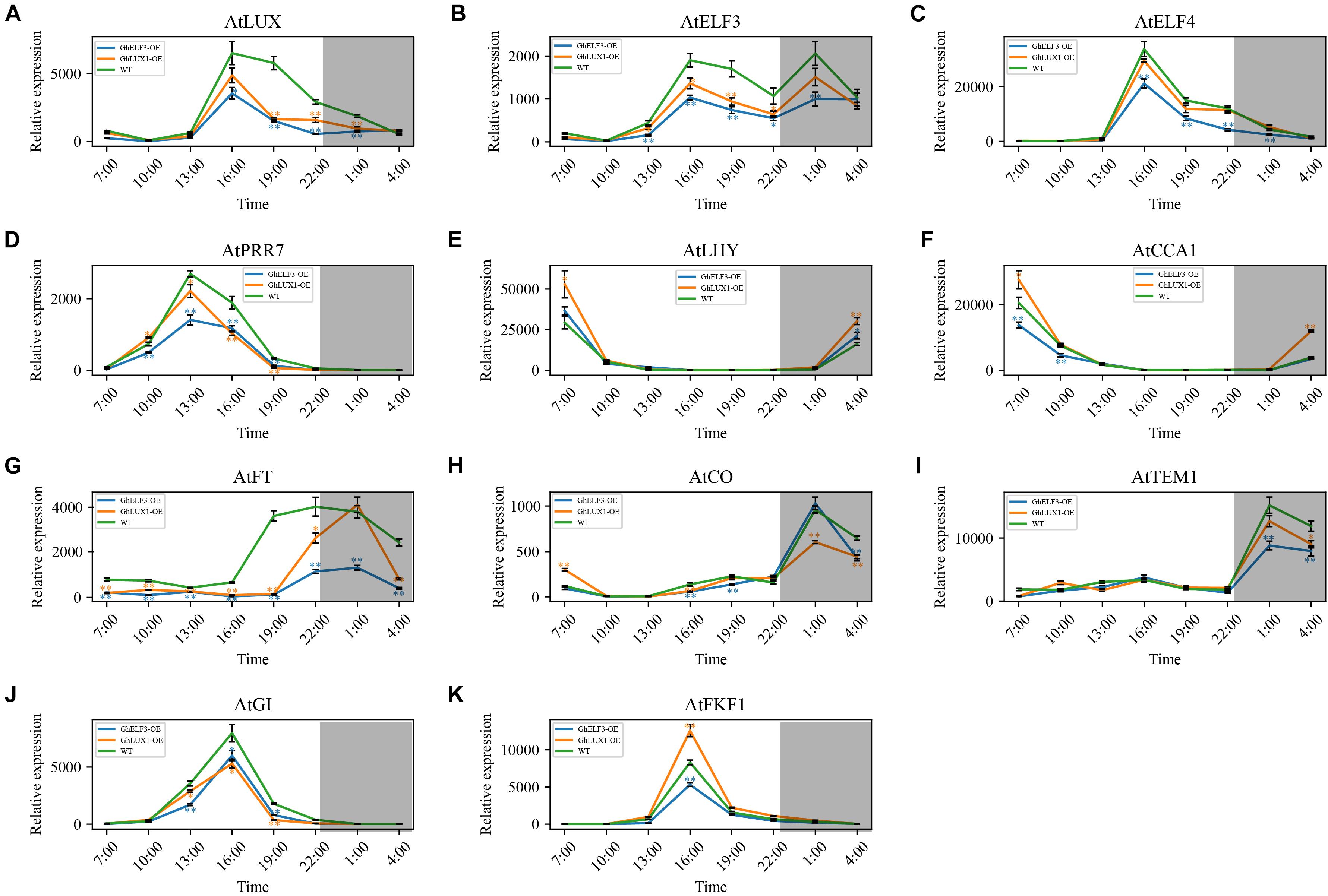
Figure 5. Both overexpression of GhLUX1 and overexpression of GhELF3 change the oscillations of the circadian clock genes and the key genes in the photoperiodic flowering pathway. Expression patterns of (A–F) the circadian clock genes and (G–K) the key genes in the photoperiodic flowering pathway in the WT and the transgenic lines. All the expression levels are made relative to the expression level of AtFKF1 in the WT at 10:00. The data are the means ± SEs of three biological replicates. The asterisks indicate significant differences compared to the WT plants at each time point (∗∗P < 0.01, ∗P < 0.05, Student’s t-test). The gray shadows indicate the dark periods.
In the important photoperiodic flowering pathway, the key integrators, CO and FT, as well as a number of their regulators were under the control of the circadian clock. In the GhLUX1-overexpressed line and the GhELF3-overexpressed line, the expression of AtFT was repressed and delayed to the later time of the day (19:00–4:00) compared with in the WT (16:00–4:00). In addition, the expression of AtFT was repressed more strongly by GhELF3 than by GhLUX1 (Figure 5G). AtCO, the primary activator of AtFT, exhibited slightly higher expression levels in the WT than in the two overexpression lines at 16:00, which was consistent with the rapidly increasing expression of AtFT in WT and the persistent low expression of AtFT in the two overexpression lines from 16:00 to 19:00. In addition, AtCO exhibited slightly higher expression levels in the WT and the GhLUX1-overexpressed line than in the GhELF3-overexpressed line at 19:00, which was consistent with the slowly increasing expression of AtFT at high level in the WT, the dramatically increasing expression of AtFT at medium level in the GhLUX1-overexpressed line and the slowly increasing expression of AtFT at low level in the GhELF3-overexpressed line from 19:00 to 22:00. Although the expression of AtCO reached peaks in all the three lines and were repressed in the GhLUX1-overexpressed line at 1:00, the expression of AtFT began to decrease dramatically in all the three lines and was not repressed in the GhLUX1-overexpressed line at 1:00 (Figures 5G,H). This discrepancy between the expression of AtCO and AtFT at 1:00 might be explained by the degradation of AtCO protein and high expression levels of AtTEM1 (the main repressor of AtFT) at night. The expression of AtTEM1 began to increase at 22:00 and reached peaks at 1:00. Compared with in the WT, the expression of AtTEM1 was repressed to a higher extent in the GhELF3-overexpressed line than in the GhLUX1-overexpressed line (Figure 5I). We speculated that the higher AtFT was expressed in the late afternoon and early evening, the higher level of AtTEM1 was needed to repress the expression of AtFT at night. AtGI and AtFKF1 were under the control of the circadian clock, they promoted flowering not only by regulating the expression timing of AtCO but also by directly regulating the expression of AtFT (Sawa and Kay, 2011). We therefore examined whether the expression of AtGI and AtFKF1 was changed in the GhLUX1-overexpressed line and the GhELF3-overexpressed line. Compared with in the WT, the expression of AtGI was repressed in the GhLUX1-overexpressed line and the GhELF3-overexpressed line (Figure 5J). The expression of AtFKF1 was promoted in the GhLUX1-overexpressed line but repressed in the GhELF3-overexpressed line (Figure 5K). These results suggested that the circadian clock could regulate the diurnally rhythmic expression of the key genes in the photoperiodic flowering pathway to regulate flowering time.
Both Silencing of GhLUX1 and Silencing of GhELF3 in Cotton Promote Flowering
To further investigate the roles of GhLUX1 and GhELF3 in regulating flowering time of cotton, GhLUX1 and GhELF3 were silenced in cotton via virus-induced gene silencing (VIGS). The GhLUX1-silenced plants and the GhELF3-silenced plants flowered 3.6 and 5.1 days earlier on average than the control (CLCrVA) plants (Figure 6B). When the first flowers of the control plants were blooming, the second flowers of the GhLUX1-silenced plants and the GhELF3-silenced plants were blooming and had bloomed, respectively (Figure 6A). Compared with the control plants, the expression of GhLUX1 in the GhLUX1-silenced plants and the expression of GhELF3 in the GhELF3-silenced plants were significantly decreased when they were highly expressed during the 24 h (Figures 6C,D). In addition, the expression of GhLUX1 in the GhELF3-silenced plants didn’t change, while the expression of GhELF3 in the GhLUX1-silenced plants was slightly repressed at 14:30 and 18:30 (Figures 6C,D). The expression of GhFT in both the GhLUX1-silenced plants and the GhELF3-silenced plants was increased at 6:30 and 10:30, which might result from the increased expression of GhCOL1 at 2:30 and 6:30 in these plants (Figures 6E,F). These results suggested that the circadian clock might regulate cotton flowering time by regulating the expression of GhFT and GhCOL1.
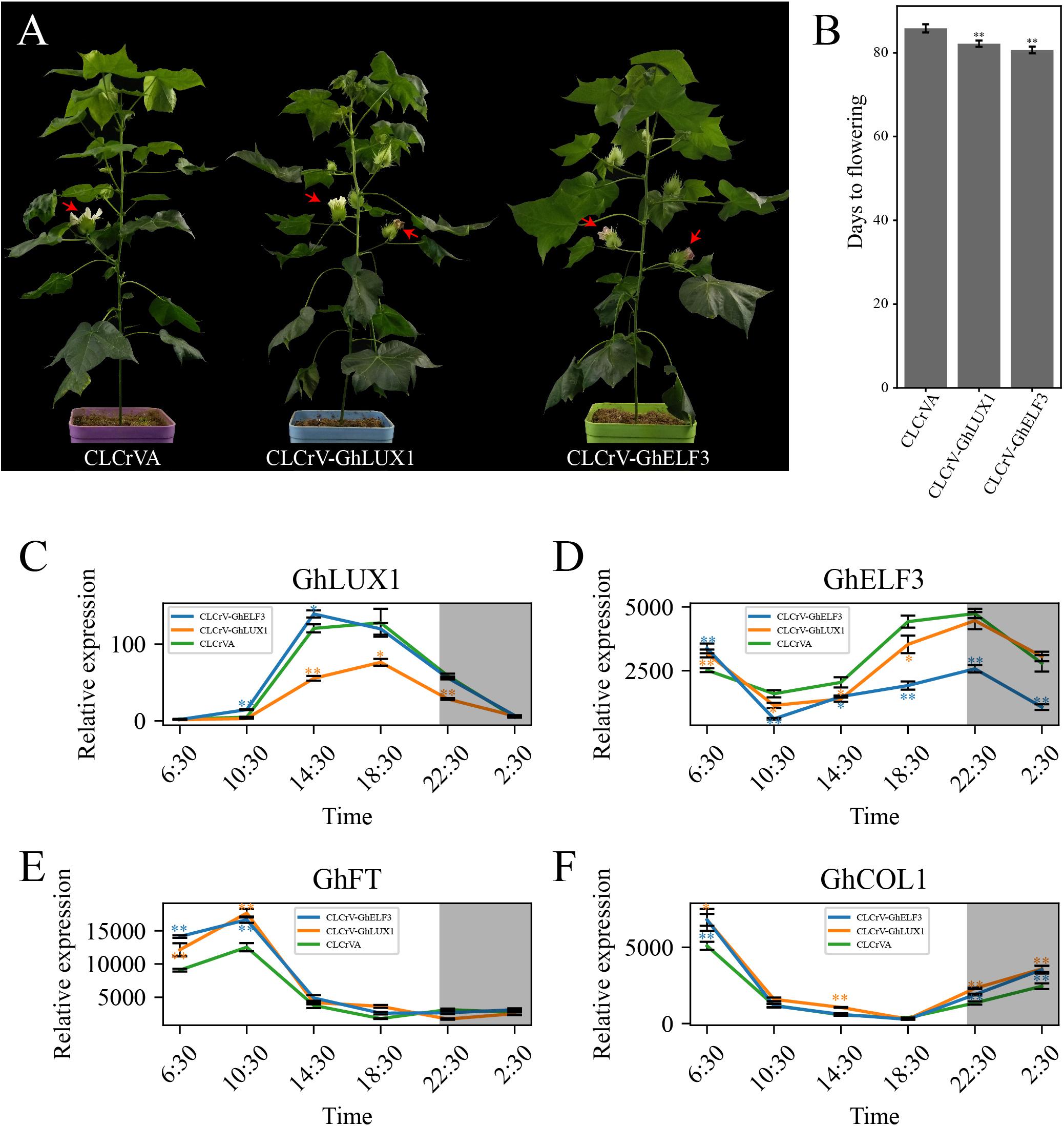
Figure 6. Both silencing of GhLUX1 and silencing of GhELF3 in cotton promote flowering. (A) Phenotypic characteristics of 86-day-old control (CLCrVA), GhLUX1-silenced (CLCrV-GhLUX1) and GhELF3-silenced (CLCrV-GhELF3) plants. (B) Days to flowering of the control plants and the gene-silenced plants (means ± SEs, n = 18 plants). (C–F) Expression patterns of GhLUX1, GhELF3, GhFT, and GhCOL1 in the control plants and the gene-silenced plants. All the expression levels are made relative to the expression level of GhLUX1 in CLCrV-GhLUX1 at 6:30. The data are the means ± SEs of three biological replicates. The asterisks indicate significant differences compared to the control plants at each time point (∗∗P < 0.01, ∗P < 0.05, Student’s t-test). The gray shadows indicate the dark periods.
Discussion
Appropriate flowering time is crucial for reproduction success and crop yield. Great efforts have been made to illuminate the complex molecular networks that control flowering time. In Arabidopsis, the important photoperiodic flowering pathway depends on the circadian clock-controlled transcription of key genes in the pathway. Here, we report that two components of the circadian clock in cotton, GhLUX1 and GhELF3 participate in flowering time regulation by affecting the transcription of GhCOL1 and GhFT (Figure 6).
Circadian clock genes have been found in organisms across the three domains of life: Archaea, Bacteria, and Eucarya. During evolution, reconfiguration of the circadian clock network has led to non-homologous network components utilized by different lineages. The components of transcriptional feedback loops of the clock in early plant lineages (chlorophytes and bryophytes) vs. angiosperms are apparently different (McClung, 2013). Our phylogenetic analysis of LUXs and ELF3s in 27 plant species show that both LUXs and ELF3s are not found in chlorophytes (C. reinhardtii) and bryophytes (P. patens). The most ancient LUX and ELF3 were identified in pteridophytes (S. moellendorffii) and the most basal angiosperm lineage (A. trichopoda), respectively (Supplementary Figure 2). These results indicate that LUXs and ELF3s may be sequentially added into the ancestral network of the circadian clock after the occurrence of bryophytes, which is consistent with the hypothesis that the circadian clock network is evolutionarily dynamic with new components joining and old components quitting (McClung, 2013). Given that ELF3 may act as a regulator of light input into the oscillator (McWatters et al., 2000), the occurrence of this novel regulator in the earliest angiosperm implies that light entrainment to the circadian clock may originate from or be reinforced in the higher plant species. Redundancy generated by gene duplication usually promotes regulatory neofunctionalization (redeployment of TFs into new networks) (Wohlbach et al., 2009). ELF3 numbers (two or three) in monocots are more than LUX numbers (only one) in monocots and ELF3s in dicots are divided into two subclades (Supplementary Figure 2). These results suggest that ELF3s are more redundant than LUXs and have diverged more greatly to perform more diverse functions. In Arabidopsis, ELF3 doesn’t contain any known DNA-binding domains and therefore performs its regulatory functions mainly by interacting with multiple other proteins, including phyB, COP1, BBX19, PIF4, LUX1, ELF4, NOX, SVP, TOC1, and GI (Huang and Nusinow, 2016). Although we demonstrate that GhELF3 interacts with GhLUX1 in the nucleus (Figures 1D,E), whether GhELF3 can interact with other proteins needs to be further investigated to better understand the diverse roles of GhELF3 in cotton growth and development.
The wild species of cotton are short-day plants that originated from tropical regions (Li et al., 2015). Domesticated G. hirsutum became photoperiod-insensitive during its adaptation to long-day conditions of temperate regions, while semi-domesticated races of G. hirsutum still are photoperiod-sensitive and don’t flower in LD (Zhang R. et al., 2015). Another short-day plant, G. max originated from temperate region. During adaptation to wide latitudes, the photoperiod response of G. max is changed due to artificial selection and natural variation of the circadian clock genes (Lu et al., 2017, 2020). We test whether this mechanism leads to different flowering times of cotton cultivars. Both GhLUX1 and GhELF3 are differently expressed between CCRI50 and GX11 in LD, SD, LL, and DD (Figures 2A,B, 3A,B), implying that differences in the circadian clock may contribute to different flowering times of cotton cultivars. In addition, the oscillations of GhLUX1 and GhELF3 transcripts in CCRI50 and GX11 respond to photoperiod in different manners (Figures 2A,B, 3A,B), indicating that photoperiod may regulate GhLUX1 and GhELF3 expression through different ways and photoperiod responses can be different in cotton varieties with different flowering time.
As the integrator of multiple flowering pathways, FT is transported from companion cells of leaves to shoot apical meristem and then induces the expression of floral identity genes (Guo et al., 2015). A previous study demonstrates that GhFT also functions as a flowering promoter. The diurnal oscillation of GhFT mRNA in both LD and SD implies that the transcription of GhFT is under the control of the circadian clock or/and respond to the day-night transition (Guo et al., 2015). Our expression analysis shows that GhFT mRNA oscillates diurnally not only in LD and SD, but also in LL and DD (Figures 2D, 3D), which indicates that the circadian clock persists to oscillate and controls the transcription of GhFT under constant conditions. The different oscillation properties (the timings of rise and fall during the 24 h, amplitudes, peak levels and trough levels) of GhFT mRNA under the four conditions may result from the circadian clock’s response to different photoperiods. However, the oscillation properties of GhCOL1 mRNA, especially the timing of rise and fall, are similar among the four conditions (Figures 2D, 3D), indicating that GhCOL1 may be regulated by circadian clock genes different from those regulating GhFT. Furthermore, the discrepancy between higher levels of GhCOL1 and lower levels of GhFT in DD suggests that unknown repressors of GhFT, probably homologs of AtTEM1/2 (Castillejo and Pelaz, 2008; Osnato et al., 2012; Marin-Gonzalez et al., 2015), may dominate GhFT transcription in the dark. It will be interesting to identify these repressors in cotton and explore whether they are regulated by the circadian clock. In Arabidopsis, AtFT promoter is directly bound by another circadian clock gene, AtGI, and is activated by GI in a CO-independent manner (Sawa and Kay, 2011). Further identification of other circadian clock genes in cotton will be helpful to understand the complex roles of the circadian clock in regulating cotton flowering time.
Because the core components of the plant circadian clock form multiple feedback loops and these loops interlocked with one another (Hsu and Harmer, 2014), it’s difficult to confirm the precise molecular functions of one certain component in regulating flowering time. Both overexpression of GhLUX1 and overexpression of GhELF3 in Arabidopsis alter the oscillation amplitudes of their Arabidopsis orthologs and other circadian clock components (Figures 5A–F). Furthermore, the oscillation amplitudes of the core flowering genes in the photoperiodic flowering pathway are also altered in the two transgenic lines, except that the oscillation phase of AtFT in the GhLUX1-overexpressed line is delayed rather than that the oscillation amplitude is changed (Figures 5G–K). Although GhLUX1 and GhELF3 can perform functions by forming a complex (Figures 1D,E), the different expression alterations of the circadian clock genes and the flowering genes between the GhLUX1-overexpressed line and the GhELF3-overexpressed line indicate that GhLUX1 and GhELF3 can also perform functions independently from each other. These results are helpful to understand the specific functions of different circadian clock components in orchestrating the expression of multiple flowering genes. Virus-induced silencing of GhLUX1 and silencing of GhELF3 in cotton promote flowering by upregulating GhCOL1 and GhFT (Figure 6). Untangling the complex regulation relationships between the circadian clock and flowering in cotton depends on the future identification of direct regulators of GhCOL1 and GhFT in the photoperiodic flowering pathway and other flowering pathways, and more importantly, subsequent investigation of the relationships between the circadian clock and these regulators.
In summary, GhLUX1 and GhELF3, the two components of the circadian clock, are differentially expressed in the early flowering and late-flowering cotton varieties, which also exhibit different expression oscillations of two core flowering genes, GhCOL1 and GhFT. Both overexpression of GhLUX1 and overexpression of GhELF3 in Arabidopsis delay flowering by altering the expression oscillations of multiple key genes in the photoperiodic flowering pathway. Both silencing of GhLUX1 and silencing of GhELF3 in cotton promote flowering by increasing the expression of GhCOL1 and GhFT. Our results demonstrate that the circadian clock is involved in regulating cotton flowering time and provide a theoretical basis for breeding cotton varieties with desired flowering and maturity time.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
SY and HWe designed the experiments. AW, PC, and LM performed the experiments. PH analyzed the results and wrote the manuscript. HWe and HWa revised the manuscript. All authors reviewed and approved the final manuscript.
Funding
This research was supported by the National Key R&D Program of China (2020YFD1001004) and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.691489/full#supplementary-material
Supplementary Figure 1 | Exon-intron structures of GhLUX1 and GhELF3.
Supplementary Figure 2 | Phylogenetic trees of LUXs and ELF3s in plant species.
Supplementary Figure 3 | Tertiary structures of GhLUX1 and GhELF3.
Supplementary Table 1 | Properties of the putative protein sequences of GhLUX1 and GhELF3.
Supplementary Table 2 | Primers used in this study.
Supplementary Table 3 | Information about the 27 plant species used in identifying LUXs and ELF3s.
Footnotes
- ^ https://www.cottongen.org/cottongen_downloads/Gossypium_hirsutum/ZJU_G.hirsutum_AD1genome_v2.1/genes/TM-1_V2.1.gene.pep.fa.gz
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://web.expasy.org/protparam/
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^ https://compbio.cs.princeton.edu/conservation/score.html
References
Adams, S., Manfield, I., Stockley, P., and Carre, I. A. (2015). Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS One 10:e0143943. doi: 10.1371/journal.pone.0143943
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucl. Res. 40, W597–W603. doi: 10.1093/nar/gks400
Boden, S. A., Weiss, D., Ross, J. J., Davies, N. W., Trevaskis, B., Chandler, P. M., et al. (2014). EARLY FLOWERING3 Regulates Flowering in Spring Barley by Mediating Gibberellin Production and FLOWERING LOCUS T Expression. Plant Cell 26, 1557–1569. doi: 10.1105/tpc.114.123794
Bu, T. T., Lu, S. J., Wang, K., Dong, L. D., Li, S. L., Xie, Q. G., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. U.S.A. 8:118. doi: 10.1073/pnas.2010241118
Cai, D., Liu, H., Sang, N., and Huang, X. (2017). Identification and characterization of CONSTANS-like (COL) gene family in upland cotton (Gossypium hirsutum L.). PLoS One 12:e0179038. doi: 10.1371/journal.pone.0179038
Campoli, C., Shtaya, M., Davis, S. J., and Von Korff, M. (2012). Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12:97. doi: 10.1186/1471-2229-12-97
Capra, J. A., and Singh, M. (2007). Predicting functionally important residues from sequence conservation. Bioinformatics 23, 1875–1882. doi: 10.1093/bioinformatics/btm270
Castillejo, C., and Pelaz, S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. doi: 10.1016/j.cub.2008.07.075
Cheng, S., Chen, P., Su, Z., Ma, L., Hao, P., Zhang, J., et al. (2021). High-resolution temporal dynamic transcriptome landscape reveals a GhCAL-mediated flowering regulatory pathway in cotton (Gossypium hirsutumL.). Plant Biotechnol. J. 19, 153–166. doi: 10.1111/pbi.13449
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Doyle, M. R., Davis, S. J., Bastow, R. M., Mcwatters, H. G., Kozma-Bognar, L., Nagy, F., et al. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. doi: 10.1038/nature00954
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., Von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Fujiwara, S., Oda, A., Yoshida, R., Niinuma, K., Miyata, K., Tomozoe, Y., et al. (2008). Circadian Clock Proteins LHY and CCA1 Regulate SVP Protein Accumulation to Control Flowering in Arabidopsis. Plant Cell 20, 2960–2971. doi: 10.1105/tpc.108.061531
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Gu, Z., Huang, C., Li, F., and Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649. doi: 10.1111/pbi.12169
Guo, D., Li, C., Dong, R., Li, X., Xiao, X., and Huang, X. (2015). Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. J. Integr. Plant Biol. 57, 522–533. doi: 10.1111/jipb.12316
Hazen, S. P., Schultz, T. F., Pruneda-Paz, J. L., Borevitz, J. O., Ecker, J. R., and Kay, S. A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392. doi: 10.1073/pnas.0503029102
Hsu, P. Y., and Harmer, S. L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19, 240–249. doi: 10.1016/j.tplants.2013.11.007
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, H., and Nusinow, D. A. (2016). Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 32, 674–686. doi: 10.1016/j.tig.2016.08.002
Huang, W., Perez-Garcia, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the Core of the Arabidopsis Circadian Clock Defines the Network Structure of the Oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Devel. 16, 2006–2020. doi: 10.1101/gad.999202
Kinmonth-Schultz, H. A., Golembeski, G. S., and Imaizumi, T. (2013). Circadian clock-regulated physiological outputs: Dynamic responses in nature. Sem. Cell Devel. Biol. 24, 407–413. doi: 10.1016/j.semcdb.2013.02.006
Li, C., Zhang, Y., Zhang, K., Guo, D., Cui, B., Wang, X., et al. (2015). Promoting flowering, lateral shoot outgrowth, leaf development, and flower abscission in tobacco plants overexpressing cotton FLOWERING LOCUS T (FT)-like gene GhFT1. Front. Plant Sci. 6:454. doi: 10.3389/fpls.2015.00454
Li, J., Fan, S. L., Song, M. Z., Pang, C. Y., Wei, H. L., Li, W., et al. (2013). Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Rep. 32, 1675–1686. doi: 10.1007/s00299-013-1479-1
Liu, C., Qu, X., Zhou, Y., Song, G., Abiri, N., Xiao, Y., et al. (2018). OsPRR37 confers an expanded regulation of the diurnal rhythms of the transcriptome and photoperiodic flowering pathways in rice. Plant Cell Environ. 41, 630–645. doi: 10.1111/pce.13135
Liu, T. L., Newton, L., Liu, M. J., Shiu, S. H., and Farre, E. M. (2016). A G-Box-Like Motif Is Necessary for Transcriptional Regulation by Circadian Pseudo-Response Regulators in Arabidopsis. Plant Physiol. 170, 1168–1168. doi: 10.1104/pp.16.000890
Liu, X. L., Covington, M. F., Fankhauser, C., Chory, J., and Wanger, D. R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. doi: 10.1105/tpc.13.6.1293
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S. J., Dong, L. D., Fang, C., Liu, S. L., Kong, L. P., Cheng, Q., et al. (2020). Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52:428. doi: 10.1038/s41588-020-0604-7
Lu, S. J., Zhao, X. H., Hu, Y. L., Liu, S. L., Nan, H. Y., Li, X. M., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49:773. doi: 10.1038/ng.3819
Lu, S. X., Knowles, S. M., Andronis, C., Ong, M. S., and Tobin, E. M. (2009). CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL Function Synergistically in the Circadian Clock of Arabidopsis. Plant Physiol. 150, 834–843. doi: 10.1104/pp.108.133272
Lu, S. X., Webb, C. J., Knowles, S. M., Kim, S. H. J., Wang, Z. Y., and Tobin, E. M. (2012). CCA1 and ELF3 Interact in the Control of Hypocotyl Length and Flowering Time in Arabidopsis. Plant Physiol. 158, 1079–1088. doi: 10.1104/pp.111.189670
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucl. Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Marcolino-Gomes, J., Nakayama, T. J., Molinari, H. B. C., Basso, M. F., and Henning, L. M. M. (2017). Functional Characterization of a Putative Glycine max ELF4 in Transgenic Arabidopsis and Its Role during Flowering Control. Front. Plant Sci. 8:618. doi: 10.3389/fpls.2017.00618
Marin-Gonzalez, E., Matias-Hernandez, L., Aguilar-Jaramillo, A. E., Lee, J. H., Ahn, J. H., Suarez-Lopez, P., et al. (2015). SHORT VEGETATIVE PHASE Up-Regulates TEMPRANILLO2 Floral Repressor at Low Ambient Temperatures. Plant Physiol. 169, 1214–1224. doi: 10.1104/pp.15.00570
Matsushika, A., Imamura, A., Yamashino, T., and Mizuno, T. (2002). Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol. 43, 833–843. doi: 10.1093/pcp/pcf118
McClung, C. R. (2013). Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Devel. Biol. 24, 430–436. doi: 10.1016/j.semcdb.2013.02.007
McWatters, H. G., Bastow, R. M., Hall, A., and Millar, A. J. (2000). The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408, 716–720. doi: 10.1038/35047079
McWatters, H. G., Kolmos, E., Hall, A., Doyle, M. R., Amasino, R. M., Gyula, P., et al. (2007). ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 144, 391–401. doi: 10.1104/pp.107.096206
Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H. R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Devel. Cell 2, 629–641. doi: 10.1016/s1534-5807(02)00170-3
Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 110–121. doi: 10.1093/pcp/pcl043
Nagel, D. H., and Kay, S. A. (2012). Complexity in the Wiring and Regulation of Plant Circadian Networks. Curr. Biol. 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 Are Transcriptional Repressors in the Arabidopsis Circadian Clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kiba, T., Kamioka, M., Suzuki, T., Yamashino, T., Higashiyama, T., et al. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128. doi: 10.1073/pnas.1205156109
Nakamichi, N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698. doi: 10.1093/pcp/pci086
Nakamichi, N., Kita, M., Niinuma, K., Ito, S., Yamashino, T., Mizoguchi, T., et al. (2007). Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48, 822–832. doi: 10.1093/pcp/pcm056
Osnato, M., Castillejo, C., Matias-Hernandez, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:810. doi: 10.1038/ncomms1810
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mole. Biol. Rep. 15, 8–15. doi: 10.1007/bf02772108
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Sato, E., Nakamichi, N., Yamashino, T., and Mizuno, T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43, 1374–1385. doi: 10.1093/pcp/pcf166
Sawa, M., and Kay, S. A. (2011). GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Wang, L., Sun, S., Wu, T., Liu, L., Sun, X., Cai, Y., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation inGmPRR37affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18, 1869–1881. doi: 10.1111/pbi.13346
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217.
Wohlbach, D. J., Thompson, D. A., Gasch, A. P., and Regev, A. (2009). From elements to modules: regulatory evolution in Ascomycota fungi. Curr. Opin. Genet. Devel. 19, 571–578. doi: 10.1016/j.gde.2009.09.007
Yakir, E., Hilman, D., Kron, I., Hassidim, M., Melamed-Book, N., and Green, R. M. (2009). Posttranslational Regulation of CIRCADIAN CLOCK ASSOCIATED1 in the Circadian Oscillator of Arabidopsis. Plant Physiol. 150, 844–857. doi: 10.1104/pp.109.137414
Yamamoto, Y., Sato, E., Shimizu, T., Nakamich, N., Sato, S., Kato, T., et al. (2003). Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 44, 1119–1130. doi: 10.1093/pcp/pcg148
Zagotta, M. T., Hicks, K. A., Jacobs, C. I., Young, J. C., Hangarter, R. P., and Meekswagner, D. R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702. doi: 10.1046/j.1365-313X.1996.10040691.x
Zhang, R., Ding, J., Liu, C. X., Cai, C. P., Zhou, B. L., Zhang, T. Z., et al. (2015). Molecular Evolution and Phylogenetic Analysis of Eight COL Superfamily Genes in Group I Related to Photoperiodic Regulation of Flowering Time in Wild and Domesticated Cotton (Gossypium) Species. PLoS One 10:e0118669. doi: 10.1371/journal.pone.0118669
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Keywords: GhLUX1, GhELF3, circadian clock, cotton, flowering time
Citation: Hao P, Wu A, Chen P, Wang H, Ma L, Wei H and Yu S (2021) GhLUX1 and GhELF3 Are Two Components of the Circadian Clock That Regulate Flowering Time of Gossypium hirsutum. Front. Plant Sci. 12:691489. doi: 10.3389/fpls.2021.691489
Received: 23 April 2021; Accepted: 20 July 2021;
Published: 09 August 2021.
Edited by:
George Coupland, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Michael Gerard Muszynski, University of Hawai‘i at Mānoa, United StatesDiana Mihaela Buzas, University of Tsukuba, Japan
Copyright © 2021 Hao, Wu, Chen, Wang, Ma, Wei and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hengling Wei, aGVuZ2xpbmd3ZWlAMTYzLmNvbQ==; Shuxun Yu, eXN4MTk1MzExQDE2My5jb20=
 Pengbo Hao1
Pengbo Hao1 Aimin Wu
Aimin Wu Pengyun Chen
Pengyun Chen Hantao Wang
Hantao Wang Hengling Wei
Hengling Wei Shuxun Yu
Shuxun Yu