- 1College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Horticultural Plant Growth, Development and Quality Improvement of the Ministry of Agriculture/Department of Horticulture, Zhejiang University, Hangzhou, China
Resveratrol is a natural polyphenol compound produced in response to biotic and abiotic stresses in grape berries. However, changes in resveratrol caused by bud sport in grapes are scarcely reported. In this study, trans-resveratrol and cis-resveratrol were identified and quantified in the grape berries of ‘Summer Black’ and its bud sport ‘Nantaihutezao’ from the veraison to ripening stages using ultra performance liquid chromatography-high resolution tandem mass spectrometry (UPLC-HRMS). We found that bud sport accumulates the trans-resveratrol earlier and increases the contents of cis-resveratrol in the earlier stages but decreases its contents in the later stages. Simultaneously, we used RNA-Seq to identify 51 transcripts involved in the stilbene pathways. In particular, we further identified 124 and 19 transcripts that negatively correlated with the contents of trans-resveratrol and cis-resveratrol, respectively, and four transcripts encoding F3'5'H that positively correlated with the contents of trans-resveratrol by weighted gene co-expression network analysis (WGCNA). These transcripts may play important roles in relation to the synergistic regulation of metabolisms of resveratrol. The results of this study can provide a theoretical basis for the genetic improvement of grapes.
Introduction
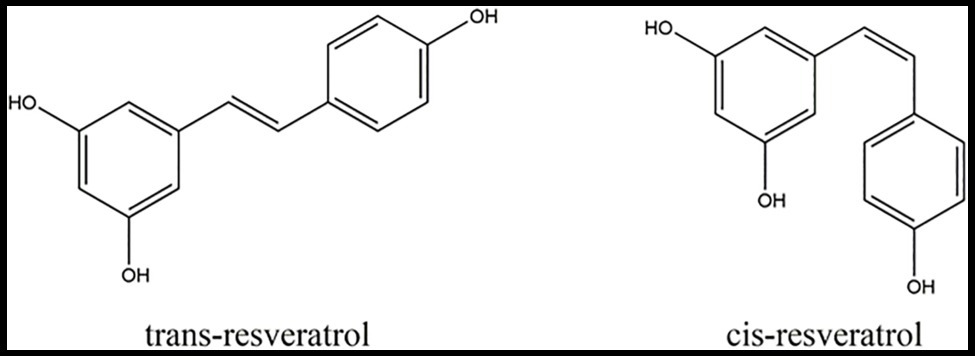
Resveratrol (C14H12O3; 3,4',5-trihydroxystilbene) is a beneficial secondary metabolite that belongs to the stilbene family, consisting of two aromatic rings joined by a methylene bridge, and exists in two isomers, trans-resveratrol and cis-resveratrol (Stervbo et al., 2007; Guerrero et al., 2010; Figure 1). The difference between the chemical structures of the cis-resveratrol and trans-resveratrol is the geometry of carbon–carbon double bond (Huang and Mazza, 2011). Resveratrol, in general, and trans-resveratrol, in particular, have many biological activities, including antifungal and antibacterial effects, as well as neuroprotective, cardioprotective, type 2 diabetes, and anticancer actions (Vitaglione et al., 2005; Yu et al., 2012; Sun et al., 2021). Apart from the above effects, resveratrol also has positive effects on the resistance of plants to biotic stress and can enhance the nutritional value of several vegetables and fruits, such as peanuts and grapes (Harikumar and Aggarwal, 2008; Giovinazzo et al., 2012).
Resveratrol is derived from the phenylpropanoid pathway that involves a series of enzymes such as phenylalanine ammonia-lyase (PAL), 4-coumarate: coenzyme A ligase (4CL), cinnamic acid 4-hydroxylase (C4H), and stilbene synthase (STS; Gatto et al., 2008; Wang et al., 2010; Duan et al., 2015). In grapes, trans-resveratrol is a leading and important stilbene compound, but cis-resveratrol has not been detected often (Liu et al., 2010). Many studies on resveratrol production have shown that its biosynthesis is induced by biotic and abiotic stresses, for example, UV-C radiation (Bais et al., 2000; Bai et al., 2019), pathogen attack (Schnee et al., 2008), salinity stress (Ismail et al., 2012), and application of chemicals, such as aluminum ions and ozone (Adrian et al., 1996; Duan et al., 2015; Bai et al., 2019). It can be induced in response to plant hormones, such as ethylene and jasmonates (Belhadj et al., 2008; D’Onofrio et al., 2009). Moreover, resveratrol can be transported to different tissues of grapevine subjected to stress and adaptation mechanisms in response to the environment, which may cause resveratrol to be converted to its isomers (Ji et al., 2014). Transcriptome analysis provides an insight into the PAL, 4CL, and STS expression profiles showing that the concentration of their transcripts increased from veraison to maturity, as well as the content of resveratrol also increases in the grape (Gatto et al., 2008). Meanwhile, the competition between STS and chalcone synthase (CHS) for the same substrates is the factor that affects the accumulation of resveratrol (Jeandet et al., 1995; Gatto et al., 2008; Yin et al., 2016; Lu et al., 2021).
Bud sport is a consequence of the genetic variation of somatic cells leading to the occurrence of qualitative and quantitative phenotypic alterations in many vegetatively propagating plants, including grapes (Zhao et al., 2015). It is an essential source for breeding new varieties when bud sports grow into branches in fruit trees. Among bud sports, most studies have focused on the molecular basis of the color changes and fruit ripening of grape (Guo et al., 2016; Bustamante et al., 2017; Xu et al., 2017). Bustamante et al. (2017) found a novel earlier ripening and sweeter taste bud mutation ‘Red Globe’, registered as ‘Pink Globe’. Compared to its parents, ‘Pink Globe’ has lower anthocyanin content and Vvufgt expression level. A similar result exists between ‘Benitaka’ and its bud mutation ‘Brazil’ (Xu et al., 2017). Guo et al. (2016) revealed many genes that were differentially expressed in the berries of ‘Kyoho’ and its early ripening mutant ‘Fengzao’ by using comparative profiling analysis. However, a comprehensive analysis of resveratrol being affected by bud sport is still lacking to date. Recently, we have identified a novel early ripening bud mutation on ‘Summer Black’ and named it ‘Nantaihutezao’ in Huzhou, Zhejiang Province, the maturity time of which is approximately 2 weeks earlier than ‘Summer Black’. There were no significant differences in the total anthocyanin contents and total soluble solid (TSS) between the two cultivars (Leng et al., 2021). We previously found that the resveratrol content and its composition in berry peel were different between the two cultivars, which might constitute a useful experimental system for the study of resveratrol metabolic mechanism and will also help us to better understand the mechanism of grape ripening underlying the bud sport variety. In this study, we performed resveratrol quantification and transcriptomic comparison between two cultivars at six time points, from veraison to ripening stages, to investigate the genetic difference. We aimed to find candidate genes related to the biosynthesis of trans-resveratrol and cis-resveratrol, which will lay a foundation for further identification of the function of genes and their improvement for fruit quality.
Materials and Methods
Sample Collection
A 3-year-old ‘Summer Black’ and its bud sport ‘Nantaihutezao’ grape berries were used as the materials in this study. The grapevines were planted in a plastic film greenhouse in Longcong village of Huzhou city in the Zhejiang province, with normal fertilizer and water management along with the control of pests and diseases. Samples were collected every 7 days from veraison (S5) to berry ripening (S10). Thirty grape berries were picked from two clusters in different positions and directions each time with no evidence of stress symptoms or disease. At the same time, the samples were immediately frozen in liquid nitrogen and then stored at −80°C for subsequent testing.
Resveratrol Profiling
Resveratrol was extracted according to a previously published method. The extracted resveratrol solution was analyzed and quantified by UPLC-HRMS and ultra-high-performance liquid chromatography-diode array detector (UPLC-DAD), respectively, following the established methods in our laboratory (Leng et al., 2020).
RNA Extraction and RNA-Seq
Total RNAs of pericarp were extracted using Trizol (Invitrogen) with three biological replicates according to the recommendations of the manufacturer. The RNA quantity was checked using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, US), the RNA concentration was assessed using Qubit® RNA Assay Kit in Qubit®2.0 Fluorometer (Life Technologies, US), and the RNA integrity was evaluated using an Agilent 2,100 Bioanalyzer (Agilent Technologies, US). A total of 36 cDNA libraries (three biological repetitions) were constructed using TruSeq™ RNA Sample Preparation Kit (Illumina, US) following the instructions of the manufacturer and deep sequenced by the Biomarker Biotechnology Corporation (Beijing, China), using Illumina HiSeq X-ten platform. All the raw sequence data have been deposited at NCBI Sequence Read Archive (SRA) with the accession code GSE142313.
Gene Functional Annotation and Expression Analysis
After removing adapter reads and filtering low-quality sequences, the clean reads were aligned to the reference Vitis vinifera genome (Jaillon et al., 2007) using the TopHat 2 software(Trapnell et al., 2009).1 The fragments per kilobase of exon per million mapped reads (FPKM) were used to calculate the abundance values of the transcript using the Cuffdiff program.2 Differentially expressed genes between the two cultivars at the same stage were analyzed using DESeq software (Anders and Huber, 2010), with the estimated absolute log2 fold change (FC) > 1 and false discovery rate (FDR) < 0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using KOBAS 3.0 software (Wu et al., 2006; Xie et al., 2011).
Weighted Gene Co-expression Network Analysis and Visualization
From all genes, the interesting gene modules and co-expression network analysis were identified by weighted gene co-expression network analysis (WGCNA) package in R (Langfelder and Horvath, 2008). The sample trait based on the correlation of the gene expression profile with intramodular connectivity and gene significance were used to identify critical genes in two cultivars at the different phases for further validation. A soft thresholding power of seven in the WGCNA package was prescribed as a soft threshold of the correlation matrix. Topological overlap matrix (TOM) similarity algorithm was used to convert the adjacency matrix to TOM and to hierarchically cluster genes. The Dynamic Hybrid Tree Cut algorithm was adopted to cut the hierarchal clustering tree and define modules as branches from the tree cutting (Zhan et al., 2015). Module Eigen gene was used to summarize the expression profile of each module.
Real-Time Quantitative Reverse Transcription -PCR Validation and Statistical Analysis
Real-time quantitative reverse transcription (qRT)-PCR analysis was performed to validate the RNA-Seq data according to a previous method (Leng et al., 2017). The primers are listed in Supplementary Table S1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH: NCBI accession number: CBI14856.3) gene was used as the internal control for calculating the relative expression of the mRNA. All samples were analyzed in triplicate, and the results were presented as the mean ± SE. The statistical significance of differences was determined with a t-test using SPSS16.0 statistical software package (IBM). Originpro 2016 (Microcal Software) was applied for the construction of the figures.
Results
Trans-resveratrol and cis-resveratrol in our samples were identified by using reversed-phase UPLC-HRMS, the ESI-ion trap MS and MS/MS data were compared with those found in the previous studies, and the retention times were compared with that of the standards (Figure 2). The tandem mass spectrum of the [M−H]− ion at m/z 227 for both trans-resveratrol and cis-resveratrol, and their fragmentation spectrum were dominated by the product ion at m/z 185 and 143, representing the loss of one and two ketene molecule. The retention times of trans-resveratrol and cis-resveratrol under the established conditions were 31.73 and 34.708 min, respectively.
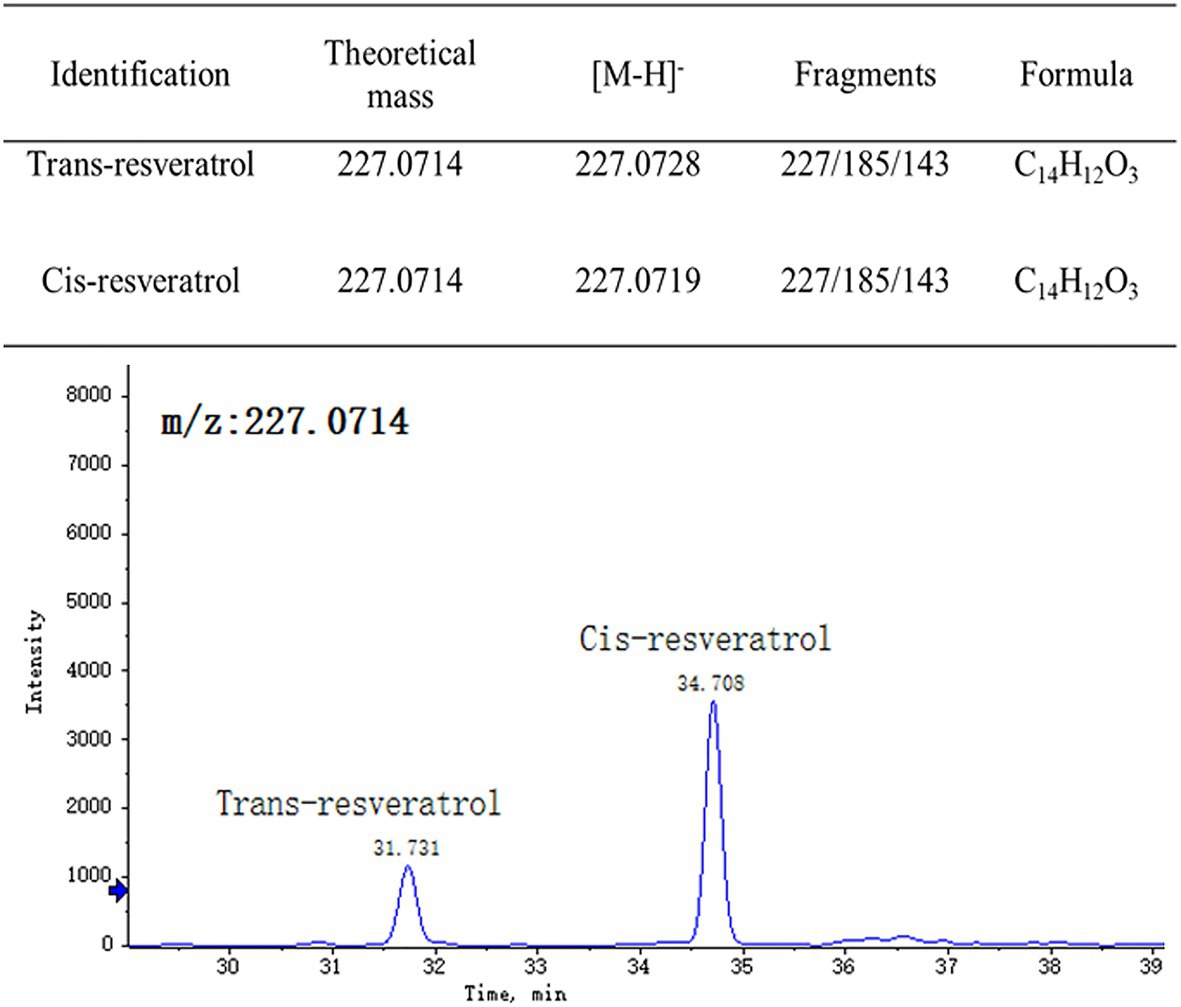
Figure 2. Chromatographic and mass spectrometric parameters of detected trans-resveratrol and cis-resveratrol in grape berry.
In this study, we compared the contents of trans-resveratrol and cis-resveratrol in the ‘Summer Black’ and ‘Nantaihutezao’ grape berry skins from veraison to ripening stages (Figure 3). The results showed that both the contents of trans-resveratrol and cis-resveratrol were accumulated with increasing time. Compared with the ‘Summer Black’, the contents of cis-resveratrol were higher in ‘Nantaihutezao’ at the S6 and S7 stages, but lower in the later stages. For trans-resveratrol, the contents in ‘Nantaihutezao’ were higher than ‘Summer Black’ during the development, but no difference in maturity. In other words, bud sport only leads to earlier accumulation of trans-resveratrol without increasing its contents.
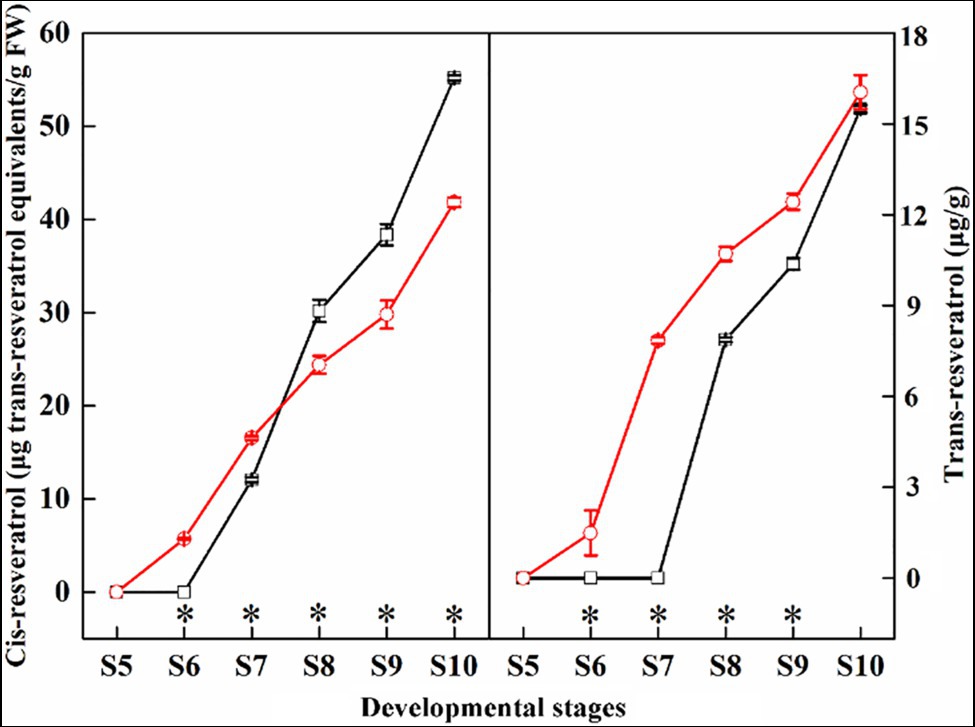
Figure 3. The changes in the contents of trans-resveratrol and cis-resveratrol during grape berry development and ripening in ‘Summer Black’ (black line) and ‘Nantaihutezao’ (red line). *Indicates the significant differences (p < 0.05, n = 3).
As the metabolism of resveratrol of the bud sport changed in this study, we performed a transcriptomic comparison between ‘Summer Black’ and ‘Nantaihutezao’ cultivars from veraison to ripening stages of grape berry to gain insights into the molecular mechanism related to the metabolism of resveratrol caused by bud sport and identify key regulatory genes. RNA-Seq was performed using the Illumina HiSeq X-ten sequencing platform. A total of 273.05 Gb clean data were obtained following quality assessment and data filtering, with more than 90% Q30 scores of clean bases, and the mapped ratios were between 66.21 and 73.65% (Leng et al., 2021). To confirm the availability of data obtained by RNA-Seq in this study, some upregulated and downregulated genes between two cultivars were selected for quantification by qRT-PCR. Those genes encoded members of the enzymes involved in the phenylpropanoid pathway, transcription factors, and some genes randomly selected for different expression levels in each stage (Figure 4). The results indicated that the qRT-PCR expression profiles were generally consistent with the RNA-Seq values, suggesting the reliability of the RNA-Seq data.
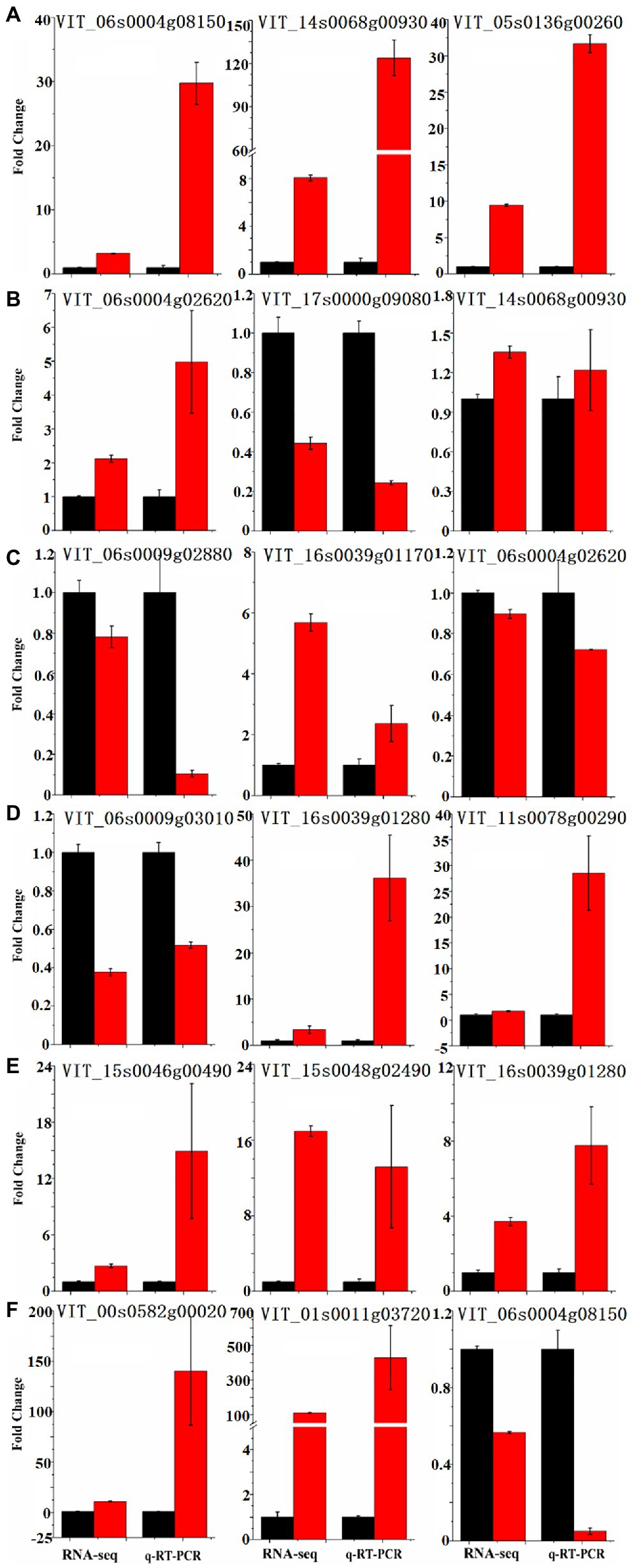
Figure 4. Real-time quantitative reverse transcription (qRT)-PCR validation of the differentially expressed genes between ‘Summer Black’ (black bar chart) and its bud sport ‘Nantaihutezao’ (red bar chart). (A) S5; (B) S6; (C) S7; (D) S8; (E) S9; and (F) S10.
According to the KEGG annotations, we found 45 differentially expressed transcripts involved in the biosynthesis of stilbene induced by bud sport during grape berry development (Figure 5). Among them, there were 27 transcripts with 26 upregulated genes and 1 downregulated gene, 39 transcripts with all upregulated genes, 35 transcripts with all upregulated genes, 12 transcripts with all upregulated genes, 34 transcripts with all upregulated genes, and 6 transcripts with 3 upregulated genes and 3 downregulated genes in the phases of S5, S6, S7, S8, S9, and S10, respectively. These genes may play key roles in the variations of trans-resveratrol and cis-resveratrol contents. In addition, three transcripts (VIT_14s0068g00930, VIT_16s0022g01070, and VIT_05s0136g00260) encoding CHS may play an important role in the synergistic regulation of metabolisms of resveratrol, which needs to be further analyzed in the future.
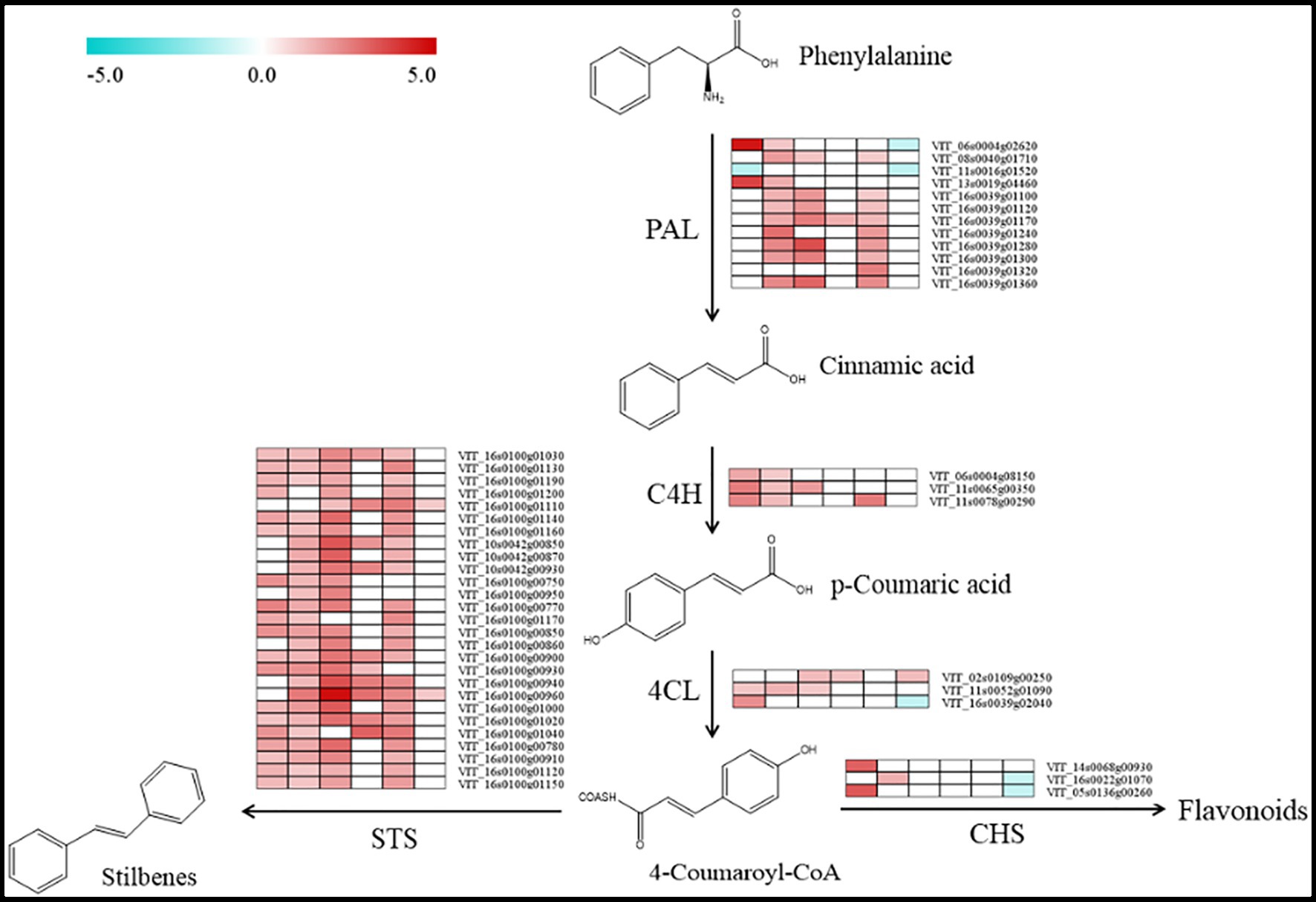
Figure 5. Biosynthetic pathway of resveratrol in grape berries. Boxes from left to right that follow the berry development were obtained by using the MultiExperiment Viewer software. The data set comprised of data regarding the changes in the relative expression of the ‘Nantaihutezao’ in relation to the ‘Summer Black’ at the same stage, which were expressed as log2 fold change.
To get a broader view on the gene expression levels involved in trans-resveratrol and cis-resveratrol metabolism pathways, respectively, modules associated with the contents of trans-resveratrol and cis-resveratrol were obtained by WGCNA (Figure 6). In this study, six main modules were identified from the RNA-Seq data. The red module contained 150 genes that were positively correlated with the contents of trans-resveratrol (cor = 0.46, p = 0.01). The modules that negatively correlated with the contents of trans-resveratrol were cyan (cor = −0.58, p = 0.001), turquoise (cor = −0.75, p = 7e-06), and gray (cor = −0.5, p = 0.008). The numbers of genes in each module are 87, 647, and 2,109 for cyan, turquoise, and gray colors, respectively. Interestingly, the turquoise module was also negatively correlated with the contents of cis-resveratrol (cor = −0.54, p = 0.004).
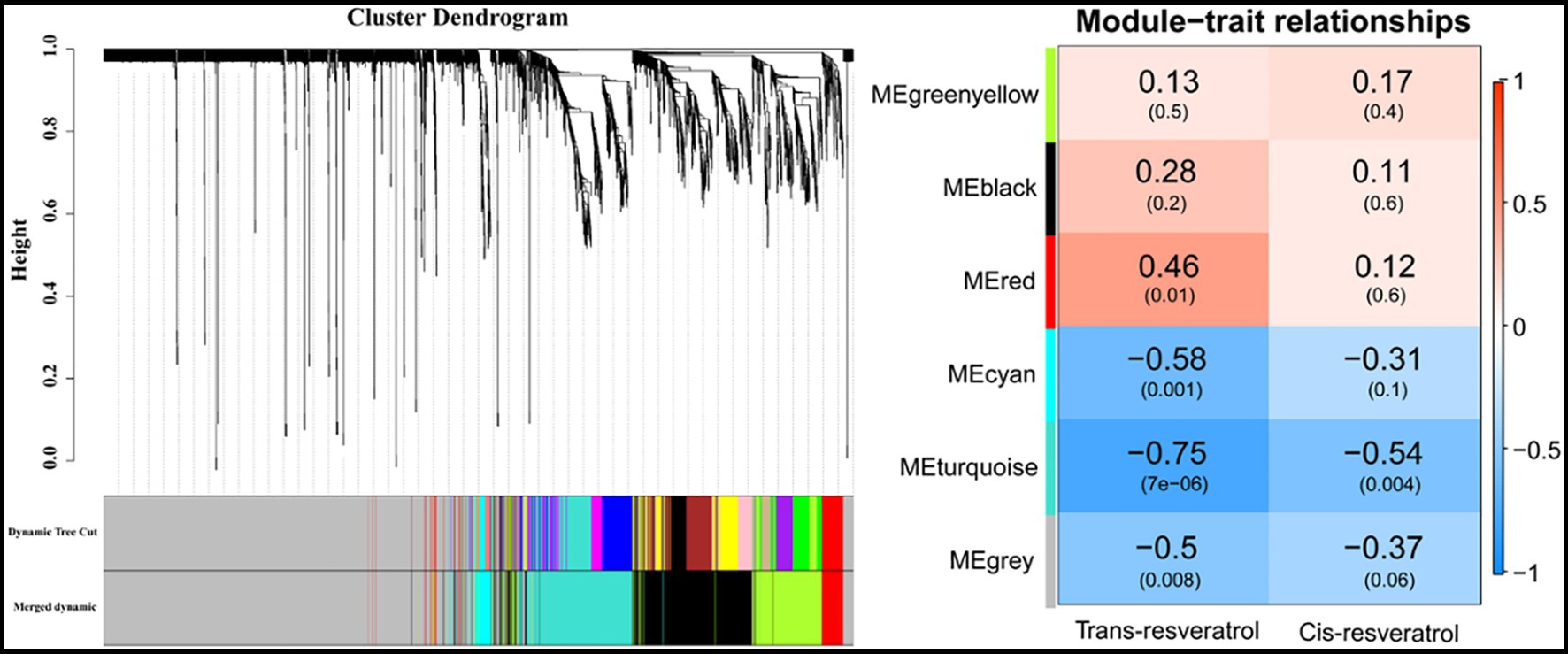
Figure 6. The relationship between the module and the contents of trans-resveratrol and cis-resveratrol was performed by weighted gene co-expression network analysis (WGCNA).
The genes in the red, cyan, turquoise, and gray modules were found to be significantly correlated with the contents of trans-resveratrol and cis-resveratrol. The association of separate genes within these modules with the contents of trans-resveratrol and cis-resveratrol was further investigated. For this purpose, we performed individual pathway enrichment analyses for these genes in the significantly correlated modules to hunt their related functions and obtain the significant KEGG pathways (Supplementary Figure S1). In this study, our analysis revealed 73 candidate transcripts enriched in the phenylpropanoid pathway that negatively correlated with the contents of trans-resveratrol. In addition, there are 51 transcripts enriched in the flavonoid biosynthesis and plant–pathogen interaction pathways, which may play a negative regulatory role in the metabolism of resveratrol (Figure 7A; Supplementary File S1). Furthermore, we found four transcripts VIT_06s0009g02810, VIT_06s0009g02880, VIT_06s0009g02920, and VIT_06s0009g03050 encoding flavonoid-3'5'-hydroxylase (F3'5'H) to be positively correlated with the contents of trans-resveratrol (Figure 7B; Supplementary File S2). In this study, we have found no genes that positively correlated with the contents of cis-resveratrol, but 19 candidate genes enriched in the plant hormone signal transduction pathway negatively correlated with the contents of cis-resveratrol, suggesting that these genes may participate in the biosynthesis of cis-resveratrol, but further study needs to be undertaken to determine their potential roles (Figure 7C; Supplementary File S3). Generally, these findings have revealed promising candidate genes for contributing to the variation in the contents of trans-resveratrol and cis-resveratrol.
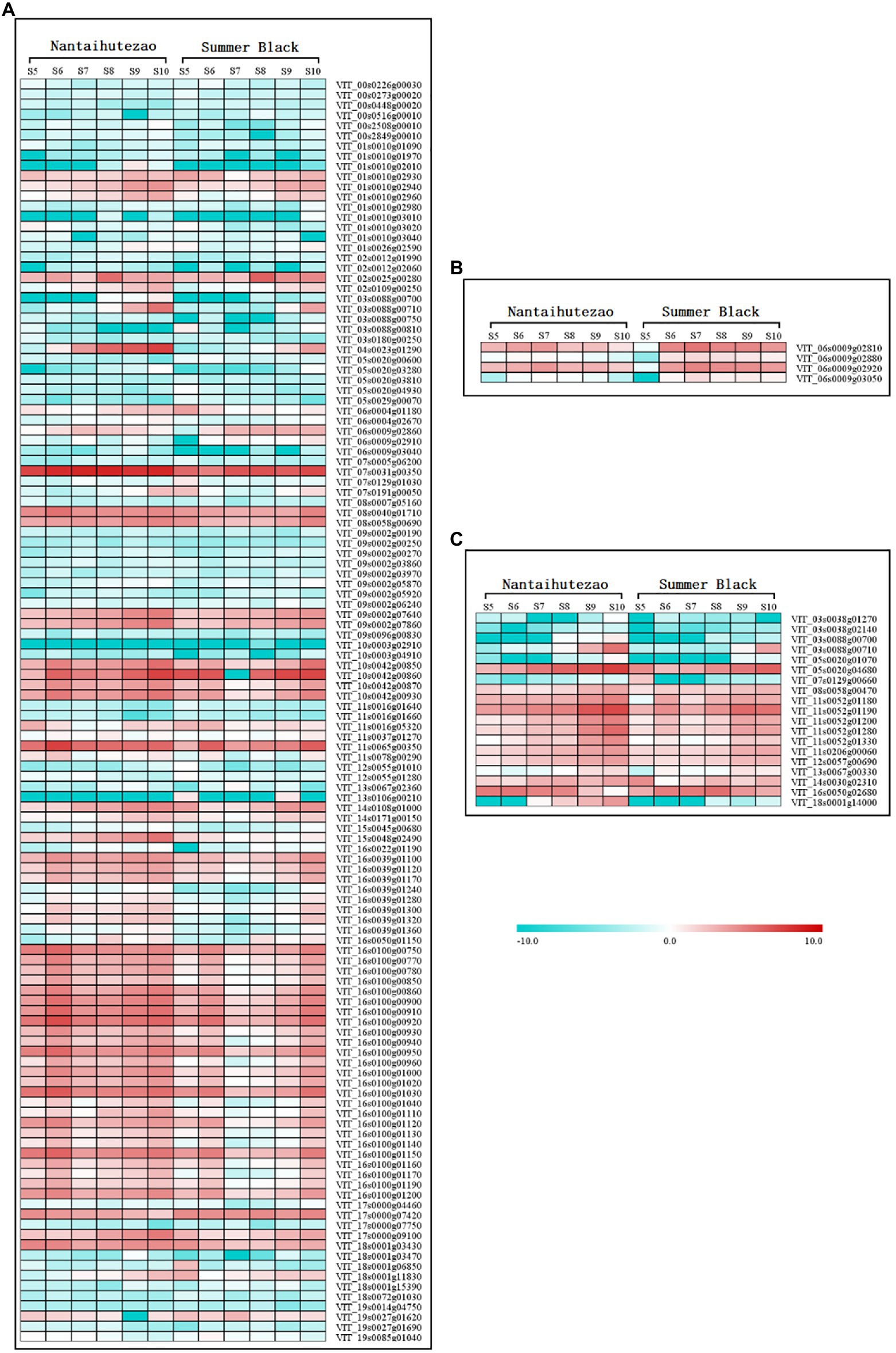
Figure 7. The expression profiles of genes enriched in the different Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the modules correlated with the contents of resveratrol. Boxes from left to right that follow the berry development were obtained by using the MultiExperiment Viewer software. The data set was normalized to the values as log2 transformed. The left set of boxes is for the bud sport ‘Nantaihutezao’, and the right set is for ‘Summer Black’. (A) Genes negatively correlated with the contents of trans-resveratrol; (B) genes positively correlated with the contents of trans-resveratrol; and (C) genes negatively correlated with the contents of cis-resveratrol.
Discussion
Many studies previously reported that the accumulation of resveratrol is well known in grape berries at veraison and continues throughout berry ripening, with significant differences among grape varieties (Gatto et al., 2008; Mei et al., 2015). In the cultivation of grapes, we discovered a bud sport on ‘Summer Black’ and named it ‘Nantaihutezao’, whose maturity time is 2 weeks earlier than that of its parents. Besides, during preliminary observation, ‘Nantaihutezao’ showed better color and fruit quality than ‘Summer Black’ (Leng et al., 2021). In terms of resveratrol, the contents of trans-resveratrol and cis-resveratrol were both accumulated with increasing time, which is consistent with previous findings. Interestingly, the contents and rates of trans-resveratrol and cis-resveratrol accumulation were significantly different due to bud sport. Considering these changes, we performed transcriptomic comparison and WGCNA to gain insights into the molecular mechanism related to the metabolism of resveratrol caused by bud sport and identify key regulatory genes.
The phenylalanine is known as the primary substrate for the biosynthesis of flavonoids and stilbenes via the phenylpropanoid pathway in most plants such as grapes (Jeandet et al., 2021; Lu et al., 2021). The biosynthesis of resveratrol was achieved through a small branch of the phenylpropanoid pathway and is considered as a competitive extension of the branch of the flavonoids (Vannozzi et al., 2018; Andi et al., 2021; Jeandet et al., 2021). Upstream of the phenylpropanoid pathway, the expression levels of PAL, C4H, 4CL, and STS genes were closely related to the contents of resveratrol. In this study, we found 12 transcripts (VIT_06s0004g02620, VIT_08s0040g01710, VIT_11s0016g01520, VIT_13s0019g04460, VIT_16s0039g01100, VIT_16s0039g01120, VIT_16s0039g01170, VIT_16s0039g01240, VIT_16s0039g01280, VIT_16s0039g01300, VIT_16s0039g01320, and VIT_16s0039g01360) encoding PAL, 3 transcripts (VIT_06s0004g08150, VIT_11s0065g00350, and VIT_11s0078g00290) encodeing C4H, and 3 transcripts (VIT_02s0109g00250, VIT_11s0052g01090 and VIT_16s0039g02040) encoding 4CL that were significantly upregulated or downregulated at some developmental stages between two cultivars. These differentially expressed transcripts might be involved in regulating the biosynthesis of resveratrol. STS is a key enzyme leading to the biosynthesis of resveratrol (Flamini et al., 2013; Lavhale et al., 2018; Vannozzi et al., 2018). We found 27 transcripts (VIT_16s0100g01030, VIT_16s0100g01130, VIT_16s0100g01190, VIT_16s0100g01200, VIT_16s0100g01110, VIT_16s0100g01140, VIT_16s0100g01160, VIT_10s0042g00850, VIT_10s0042g00870, VIT_10s0042g00930, VIT_16s0100g00750, VIT_16s0100g00950, VIT_16s0100g00770, VIT_16s0100g01170, VIT_16s0100g00850, VIT_16s0100g00860, VIT_16s0100g00900, VIT_16s0100g00930, VIT_16s0100g00940, VIT_16s0100g00960, VIT_16s0100g01000, VIT_16s0100g01020, VIT_16s0100g01040, VIT_16s0100g00780, VIT_16s0100g00910, VIT_16s0100g01120, and VIT_16s0100g01150) encoding STS that were differentially expressed at certain developmental stages, and all of them were upregulated in Nantaihutezao. These transcripts might be involved in the metabolism of resveratrol. Furthermore, CHS, a key enzyme of the flavonoid pathway, is closely related to STS and also affects the resveratrol content, because both enzymes compete for the same substrate and control the entry points into the flavonoid and stilbene pathways, respectively (Andi et al., 2021; Jeandet et al., 2021). In our data, it was found that three differentially expressed transcripts (VIT_14s0068g00930, VIT_16s0022g01070, and VIT_05s0136g00260) encoding CHS might be related to the changes in the resveratrol. For the trans-resveratrol, many previous studies have reported a relationship in the high contents with increasing concentration of some genes, such as PAL, 4CL, and STS, in the grape varieties from veraison to maturity (Degu et al., 2014; Duan et al., 2015; Suzuki et al., 2015; Wong et al., 2016; Bai et al., 2019). Conversely, cis-resveratrol was neglected because it has not been detected often, but it appears to be formed by hydrolysis of pieced or resveratrol polymers. It can also be transformed from trans-resveratrol by light-induced or enzymatic isomerization and may possess health-promoting properties (Lopez-Hernandez et al., 2007; Liu et al., 2011; Erte et al., 2020). Early studies indicated that the contents of trans-resveratrol and cis-resveratrol increase from veraison to harvesting and are induced in response to biotic and abiotic stresses. Meanwhile, PAL, 4CL, and STS expression profiles showed an increasing concentration of their transcripts and had a higher accumulation in the grape of high resveratrol contents (Gatto et al., 2008; Bai et al., 2019). Anyway, our transcriptome analysis showed that 48 differentially expressed transcripts, encoding PAL, C4H, 4CL, STS, and CHS, were important genes in the biosynthesis of stilbene during grape berry development induced by bud sport (Figure 5). Therefore, we speculated that these transcripts might play important roles in the accumulation of resveratrol.
In recent years, the accumulation of resveratrol induced by biotic and abiotic stresses, such as pathogen attack and plant hormones, has been demonstrated by many studies (Belhadj et al., 2008; Schnee et al., 2008; D’Onofrio et al., 2009). WRKY, MYB, bZIP, and ERF transcription factors have been involved in regulating STS genes to increase the resveratrol content (Riccio et al., 2020; Wang et al., 2020). In addition, PRXs genes also have been reported to participate in regulating stilbene synthesis (Park et al., 2021). Meanwhile, the phenylpropanoid pathway starts from phenylalanine, which belongs to the downstream pathway after the sucrose synthesis and degradation, glycolysis, and tricarboxylic acid cycle pathways (Dai et al., 2013). Upstream genes in those pathways also affect the resveratrol content. To further study the molecular mechanism of the biosynthesis of trans-resveratrol and cis-resveratrol, we performed WGCNA analysis to investigate functional modules that correlate with the contents of trans-resveratrol and cis-resveratrol and then enrich the genes in these functional modules into KEGG pathways. Our analysis has revealed 124 candidate transcripts enriched in the phenylpropanoid, flavonoid biosynthesis, and plant–pathogen interaction pathways (Supplementary File S1), which were proposed to play a negative regulatory role in the contents of trans-resveratrol. Among these candidate transcripts, most of them are annotated into metabolism and biosynthesis of phenylalanine, such as VIT_19s0014g04750, VIT_05s0020g03280, VIT_17s0000g09100, VIT_16s0100g01130, VIT_16s0100g01030, and VIT_00s2508g00010. VIT_19s0014g04750 is annotated as beta-glucosidase that is involved in the phenylpropanoid biosynthesis pathway. It might affect the contents of trans-resveratrol by regulating its precursor, phenylalanine. VIT_05s0020g03280 and VIT_17s0000g09100 encoded primary-amine oxidase, which performs a similar function. VIT_16s0100g01130 and VIT_16s0100g01030 encoded STS, and VIT_00s2508g00010 encoded PAL, which are directly involved in the stilbene biosynthesis pathway. Besides, numerous other transcripts are annotated into plant hormone signal transduction and plant–pathogen interaction pathways, such as VIT_12s0055g01280, VIT_03s0088g00810, and VIT_05s0020g04930. These transcripts might indirectly affect the contents of trans-resveratrol due to changes in plant hormones or pathogen attack. We found four transcripts (VIT_06s0009g02810, VIT_06s0009g02880, VIT_06s0009g02920, and VIT_06s0009g03050) encoding F3′5′H that were positively correlated with the contents of trans-resveratrol (Supplementary File S2), which belong to the biosynthesis of flavonoid. Because the competition between these genes in the biosynthesis of flavonoid and resveratrol for the same substrates was the factor to affect the resveratrol accumulation, similar to CHSs, we speculated that these enriched transcripts may have the potential role in the direct or indirect regulation of biosynthesis of trans-resveratrol. In addition, there are 19 candidate transcripts (VIT_03s0038g01270, VIT_03s0038g02140, VIT_03s0088g00700, etc.) enriched in the plant hormone signal transduction pathway (Supplementary File S3) that may participate in the biosynthesis of cis-resveratrol.
In conclusion, our findings provide hundreds of differentially expressed transcripts with obscure functions, which can provide new insights into the response of grape berries to bud sport, and also could be used in future functional and molecular biological studies of resveratrol metabolism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FL and LW designed the experiments and composed the study. FL, YY, JZ, XZ, and NS performed the experiments. FL, JS, and ZZ analyzed the data. FL and HJ contributed to reagents, materials, and analytical tools. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Natural Science Foundation of Jiangsu Province (BK20190899); the Doctor of Entrepreneurship and Innovation program of Jiangsu Province; and the Yangzhou City’s Green and Golden Phoenix Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.690095/full#supplementary-material
Supplementary Figure S1 | Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of gene enrichment analysis.
Supplementary Table S1 | Primer sequences of qRT-PCR.
Supplementary File S1 | KEGG pathway enrichment analyses of genes was negatively correlated with trans-resveratrol.
Supplementary File S2 | KEGG pathway enrichment analyses of genes was positively correlated with trans-resveratrol.
Supplementary File S3 | KEGG pathway enrichment analyses of genes was negatively correlated with cis-resveratrol.
Footnotes
References
Adrian, M., Jeandet, P., Bessis, R., and Joubert, J. M. (1996). Induction of phytoalexin (resveratrol) synthesis in grapevine leaves treated with aluminium chloride (AlCl3). J. Agric. Food Chem. 44, 1979–1981. doi: 10.1021/jf950807o
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andi, S. A., Gholami, M., Ford, C. M., and Maskani, F. (2021). Impact of light irradiance on the biosynthesis of ABA-elicited phenolic compounds in suspension-cultured Vitis vinifera L. cells. Plant Cell Tissue Organ Cult. 1–14. doi: 10.1007/s11240-021-02077-4
Bai, R., Luo, Y. Y., Wang, L. X., Li, J., Wu, K. R., Zhao, G. F., et al. (2019). A specific allele of MYB14 in grapevine correlates with high stilbene inducibility triggered by Al3+ and UV-C radiation. Plant Cell Rep. 38, 37–49. doi: 10.1007/s00299-018-2347-9
Bais, A. J., Murphy, P. J., and Dry, I. B. (2000). The molecular regulation of stilbene phytoalexin biosynthesis in Vitis vinifera during grape berry development (vol 27, pg 425, 2000). Aust. J. Plant Physiol. 27, 425–433. doi: 10.1071/PP00007
Belhadj, A., Telef, N., Cluzet, S., Bouscaut, J., Corio-Costet, M. F., and Merillon, J. M. (2008). Ethephon elicits protection against Erysiphe necator in grapevine. J. Agric. Food Chem. 56, 5781–5787. doi: 10.1021/jf800578c
Bustamante, L., Saez, V., Hinrichsen, P., Castro, M. H., Vergara, C., von Baer, D., et al. (2017). Differences in Vvufgt and VvmybA1 gene expression levels and phenolic composition in table grape (Vitis vinifera L.) “Red Globe” and its somaclonal variant “Pink Globe”. J. Agric. Food Chem. 65, 2793–2804. doi: 10.1021/acs.jafc.6b04817
Dai, Z. W., Leon, C., Feil, R., Lunn, J. E., Delrot, S., and Gomes, E. (2013). Metabolic profiling reveals coordinated switches in primary carbohydrate metabolism in grape berry (Vitis vinifera L.), a non-climacteric fleshy fruit. J. Exp. Bot. 64, 1345–1355. doi: 10.1093/jxb/ers396
Degu, A., Hochberg, U., Sikron, N., Venturini, L., Buson, G., Ghan, R., et al. (2014). Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 14:188. doi: 10.1186/s12870-014-0188-4
D’Onofrio, C., Cox, A., Davies, C., and Boss, P. K. (2009). Induction of secondary metabolism in grape cell cultures by jasmonates. Funct. Plant Biol. 36, 323–338. doi: 10.1071/FP08280
Duan, D., Halter, D., Baltenweck, R., Tisch, C., Troster, V., Kortekamp, A., et al. (2015). Genetic diversity of stilbene metabolism in Vitis sylvestris. J. Exp. Bot. 66, 3243–3257. doi: 10.1093/jxb/erv137
Erte, E., Vural, N., Mehmetoglu, U., and Guvenc, A. (2020). Optimization of an abiotic elicitor (ultrasound) treatment conditions on trans-resveratrol production from Kalecik Karasi (Vitis vinifera L.) grape skin. J. Food. Sci. Technol. 58, 2121–2132. doi: 10.1007/s13197-020-04722-w
Flamini, R., Mattivi, F., De Rosso, M., Arapitsas, P., and Bavaresco, L. (2013). Advanced knowledge of three important classes of grape phenolics: anthocyanins, stilbenes and flavonols. Int. J. Mol. Sci. 14, 19651–19669. doi: 10.3390/ijms141019651
Gatto, P., Vrhovsek, U., Muth, J., Segala, C., Romualdi, C., Fontana, P., et al. (2008). Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric. Food Chem. 56, 11773–11785. doi: 10.1021/jf8017707
Giovinazzo, G., Ingrosso, I., Paradiso, A., De Gara, L., and Santino, A. (2012). Resveratrol biosynthesis: plant metabolic engineering for nutritional improvement of food. Plant Foods Hum. Nutr. 67, 191–199. doi: 10.1007/s11130-012-0299-8
Guerrero, R. F., Puertas, B., Fernandez, M. I., Palma, M., and Cantos-Villar, E. (2010). Induction of stilbenes in grapes by UV-C: comparison of different subspecies of Vitis. Innovative Food Sci. Emerg. Technol. 11, 231–238. doi: 10.1016/j.ifset.2009.10.005
Guo, D. L., Xi, F. F., Yu, Y. H., Zhang, X. Y., Zhang, G. H., and Zhong, G. Y. (2016). Comparative RNA-Seq profiling of berry development between table grape “Kyoho” and its early-ripening mutant “Fengzao”. BMC Genomics 17:795. doi: 10.1186/s12864-016-3051-1
Harikumar, K. B., and Aggarwal, B. B. (2008). Resveratrol-A multitargeted agent for age-associated chronic diseases. Cell Cycle 7, 1020–1035. doi: 10.4161/Cc.7.8.5740
Huang, X., and Mazza, G. (2011). Simultaneous analysis of serotonin, melatonin, piceid and resveratrol in fruits using liquid chromatography tandem mass spectrometry. J. Chromatogr. A 1218, 3890–3899. doi: 10.1016/j.chroma.2011.04.049
Ismail, A., Riemann, M., and Nick, P. (2012). The jasmonate pathway mediates salt tolerance in grapevines. J. Exp. Bot. 63, 2127–2139. doi: 10.1093/jxb/err426
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–U465. doi: 10.1038/nature06148
Jeandet, P., Sbaghi, M., Bessis, R., and Meunier, P. (1995). The potential relationship of phytoalexin (resveratrol) synthesis to anthocyanin content in grape skins. Vitis 34, 91–94. doi: 10.1016/0304-4238(95)00769-P
Jeandet, P., Vannozzi, A., Sobarzo-Sanchez, E., Uddin, M. S., Bru, R., Martinez-Marquez, A., et al. (2021). Phytostilbenes as agrochemicals: biosynthesis, antifungal activity, metabolic engineering and biotechnology. Nat. Prod. Rep. doi: 10.1039/D0NP00030B [Epub ahead of print]
Ji, M., Li, Q., Ji, H., and Lou, H. X. (2014). Investigation of the distribution and season regularity of resveratrol in Vitis amurensis via HPLC-DAD-MS/MS. Food Chem. 142, 61–65. doi: 10.1016/j.foodchem.2013.06.131
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lavhale, S. G., Kalunke, R. M., and Giri, A. P. (2018). Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 248, 1063–1078. doi: 10.1007/s00425-018-2965-z
Leng, F., Cao, J. P., Ge, Z. W., Wang, Y., Zhao, C. N., Wang, S. P., et al. (2020). Transcriptomic analysis of root restriction effects on phenolic metabolites during grape berry development and ripening. J. Agric. Food Chem. 68, 9090–9099. doi: 10.1021/acs.jafc.0c02488
Leng, F., Tang, D. D., Lin, Q., Cao, J. P., Wu, D., Wang, S. P., et al. (2017). Transcriptomic analyses of ascorbic acid and carotenoid metabolites influenced by root restriction during grape berry development and ripening. J. Agric. Food Chem. 65, 2008–2016. doi: 10.1021/acs.jafc.6b05322
Leng, F., Ye, Y. L., Zhu, X. H., Zhang, Y., Zhang, Z. Y., Shi, J. Y., et al. (2021). Comparative transcriptomic analysis between “Summer Black” and its bud sport “Nantaihutezao” during developmental stages. Planta 253:23. doi: 10.1007/s00425-020-03543-7
Liu, W., Liu, C. Y., Yang, C. X., Wang, L. J., and Li, S. H. (2010). Effect of grape genotype and tissue type on callus growth and production of resveratrols and their piceids after UV-C irradiation. (vol 122, pg 475, 2010). Food Chem. 124:1099. doi: 10.1016/j.foodchem.2010.07.038
Liu, W., Liu, C. Y., Yang, C. X., Wang, L. J., and Li, S. H. (2011). Effect of grape genotype and tissue type on callus growth and production of resveratrols and their piceids after UV-C irradiation. Food Chem. 122, 475–481. doi: 10.1016/j.foodchem.2010.03.055
Lopez-Hernandez, J., Paseiro-Losada, P., Sanches-Silva, A. T., and Lage-Yusty, M. A. (2007). Study of the changes of trans-resveratrol caused by ultraviolet light and determination of trans- and cis-resveratrol in Spanish white wines. Eur. Food Res. Technol. 225, 789–796. doi: 10.1007/s00217-006-0483-x
Lu, Y., Song, Y. Y., Zhu, J., Xu, X. G., Pang, B., Jin, H., et al. (2021). Potential application of CHS and 4CL genes from grape endophytic fungus in production of naringenin and resveratrol and the improvement of polyphenol profiles and flavour of wine. Food Chem. 347:128972. doi: 10.1016/j.foodchem.2020.128972
Mei, Y. Z., Liu, R. X., Wang, D. P., Wang, X., and Dai, C. C. (2015). Biocatalysis and biotransformation of resveratrol in microorganisms. Biotechnol. Lett. 37, 9–18. doi: 10.1007/s10529-014-1651-x
Park, S. C., Pyun, J. W., Jeong, Y. J., Park, S. H., Kim, S., Kim, Y. H., et al. (2021). Overexpression of VlPRX21 and VlPRX35 genes in Arabidopsis plants leads to bioconversion of trans-resveratrol to δ-viniferin. Plant Physiol. Biochem. 162, 556–563. doi: 10.1016/j.plaphy.2021.03.015
Riccio, B. V. F., Sposito, L., Carvalho, G. C., Ferrari, P. C., and Chorilli, M. (2020). Resveratrol isoforms and conjugates: a review from biosynthesis in plants to elimination from the human body. Arch. Pharm. 353:e2000146. doi: 10.1002/ardp.202000146
Schnee, S., Viret, O., and Gindro, K. (2008). Role of stilbenes in the resistance of grapevine to powdery mildew. Physiol. Mol. Plant Pathol. 72, 128–133. doi: 10.1016/j.pmpp.2008.07.002
Stervbo, U., Vang, O., and Bonnesen, C. (2007). A review of the content of the putative chemopreventive phytoalexin resveratrol in red wine. Food Chem. 101, 449–457. doi: 10.1016/j.foodchem.2006.01.047
Sun, B., Zheng, Y. L., Yang, S. K., Zhang, J. R., Cheng, X. Y., Ghiladi, R., et al. (2021). One-pot method based on deep eutectic solvent for extraction and conversion of polydatin to resveratrol from Polygonum cuspidatum. Food Chem. 343:128498. doi: 10.1016/j.foodchem.2020.128498
Suzuki, M., Nakabayashi, R., Ogata, Y., Sakurai, N., Tokimatsu, T., Goto, S., et al. (2015). Multiomics in grape berry skin revealed specific induction of the stilbene synthetic pathway by ultraviolet-C irradiation. Plant Physiol. 168, 47–59. doi: 10.1104/pp.114.254375
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Vannozzi, A., Wong, D. C. J., Holl, J., Hmmam, I., Matus, J. T., Bogs, J., et al. (2018). Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L.). Plant Cell Physiol. 59, 1043–1059. doi: 10.1093/pcp/pcy045
Vitaglione, P., Sforza, S., Galaverna, G., Ghidini, C., Caporaso, N., Vescovi, P. P., et al. (2005). Bioavailability of trans-resveratrol from red wine in humans. Mol. Nutr. Food Res. 49, 495–504. doi: 10.1002/mnfr.200500002
Wang, Y. C., Chen, H., and Yu, O. (2010). Metabolic engineering of resveratrol and other longevity boosting compounds. Biofactors 36, 394–400. doi: 10.1002/biof.126
Wang, D., Jiang, C. Y., Liu, W. D., and Wang, Y. J. (2020). The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J. Exp. Bot. 71, 3211–3226. doi: 10.1093/jxb/eraa097
Wong, D. C. J., Gutierrez, R. L., Dimopoulos, N., Gambetta, G. A., and Castellarin, S. D. (2016). Combined physiological, transcriptome, and cis-regulatory element analyses indicate that key aspects of ripening, metabolism, and transcriptional program in grapes (Vitis vinifera L.) are differentially modulated accordingly to fruit size. BMC Genomics 17:416. doi: 10.1186/s12864-016-2660-z
Wu, J. M., Mao, X. Z., Cai, T., Luo, J. C., and Wei, L. P. (2006). KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720–W724. doi: 10.1093/nar/gkl167
Xie, C., Mao, X. Z., Huang, J. J., Ding, Y., Wu, J. M., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, Y. S., Jiang, N., Zhang, Y. G., Wang, M., Ren, J. P., and Tao, J. M. (2017). A SNP in the promoter region of the VvmybA1 gene is responsible for differences in grape berry color between two related bud sports of grape. Plant Growth Regul. 82, 457–465. doi: 10.1007/s10725-017-0272-5
Yin, X. J., Singer, S. D., Qiao, H. B., Liu, Y. J., Jiao, C., Wang, H., et al. (2016). Insights into the mechanisms underlying ultraviolet-C induced resveratrol metabolism in grapevine (V-amurensis Rupr.) cv. “Tonghua-3”. Front. Plant Sci. 7:503. doi: 10.3389/fpls.2016.00503
Yu, W., Fu, Y. C., and Wang, W. (2012). Cellular and molecular effects of resveratrol in health and disease. J. Cell. Biochem. 113, 752–759. doi: 10.1002/jcb.23431
Zhan, J. P., Thakare, D., Ma, C., Lloyd, A., Nixon, N. M., Arakaki, A. M., et al. (2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27, 513–531. doi: 10.1105/tpc.114.135657
Keywords: grape, resveratrol, transcriptome, ultra performance liquid chromatography-high resolution tandem mass spectrometry, weighted gene co-expression network analysis
Citation: Leng F, Ye Y, Zhou J, Jia H, Zhu X, Shi J, Zhang Z, Shen N and Wang L (2021) Transcriptomic and Weighted Gene Co-expression Correlation Network Analysis Reveal Resveratrol Biosynthesis Mechanisms Caused by Bud Sport in Grape Berry. Front. Plant Sci. 12:690095. doi: 10.3389/fpls.2021.690095
Edited by:
Brian Farneti, Fondazione Edmund Mach, ItalyReviewed by:
Nicola Busatto, Fondazione Edmund Mach, ItalyPhilippe Jeandet, Université de Reims Champagne-Ardenne, France
Copyright © 2021 Leng, Ye, Zhou, Jia, Zhu, Shi, Zhang, Shen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Wang, bGl3YW5nQHl6dS5lZHUuY24=
 Feng Leng
Feng Leng Yunling Ye1
Yunling Ye1 Li Wang
Li Wang